Abstract
The concepts, methodologies and applications of some of the major molecular or DNA markers commonly used in plant science have been presented. The general principles of molecular marker techniques have been elucidated with detailed explanation of some notable basic concepts associated with marker applications: marker polymorphism, dominant or co-dominant mode of inheritance, agronomic trait-marker linkage, genetic mutations and variation. The molecular marker methods that have been extensively reviewed are RFLP, RAPD, SCAR, AFLP, SSR, CpSSR, ISSR, RAMP, SAMPL, SRAP, SSCP, CAPS, SNP, DArT, EST, and STS. In addition, the practicality of the retrotransposon-based marker methods, IRAP, REMAP, RBIP, and IPBS, have been discussed. Moreover, some salient characteristics of DNA markers have been compared and the various marker systems classified as PCR- or non-PCR-based, dominantly or co-dominantly inherited, locus specific or non-specific as well as at the levels of marker polymorphism and efficiency of marker reproducibility. Furthermore, the principles and methods of the following DNA markers have been highlighted: Penta-primer amplification refractory mutation system (PARMS), Conserved DNA-Derived Polymorphism (CDDP), P450-based analogue (PBA) markers, Tubulin-Based Polymorphism (TBP), Inter-SINE amplified polymorphism (ISAP), Sequence specific amplified polymorphism (S-SAP), Intron length polymorphisms (ILPs), Inter small RNA polymorphism (iSNAP), Direct amplification of length polymorphisms (DALP), Promoter anchored amplified polymorphism (PAAP), Target region amplification polymorphism (TRAP), Conserved region amplification polymorphism (CoRAP), Start Codon Targeted (SCoT) Polymorphism, and Directed Amplification of Minisatellite DNA (DAMD). Some molecular marker applications that have been recently employed to achieve various objectives in plant research have also been outlined. This review will serve as a useful reference resource for plant breeders and other scientists, as well as technicians and students who require basic know-how in the use of molecular or DNA marker technologies.
Keywords: Molecular marker, Plant genetic diversity, Polymorphism, Basic concept, Genetic mutation
Molecular marker; Plant genetic diversity; Polymorphism; Basic concept; Genetic mutation
1. Introduction
The era of molecular marker development and applications begun in the 1980s. This landmark in plant genomic research was followed by the achievement of PCR-based DNA markers a decade on. Since then, the applications of many molecular markers have been reported in various aspects of plant molecular breeding and genomics (Nadeem et al., 2018). The PCR technique enables specific DNA sequences to be practically amplified from genomic DNA sections using specific or arbitrary designed oligonucleotide primers. Molecular markers constitute very useful tools currently available for research in plant improvement. These markers are mainly nucleic acids that are polymorphic among individuals or populations (Collard et al., 2005). Genetically, genotypes exhibit contrasting pool of fragments as a result of point mutations in oligonucleotide priming sites. In some cases, the distance between the termini sequences altered by insertion or deletion mutation events could lead to polymorphism.
DNA marker protocols mediated by PCR applications have become commonly used in plant genomic analysis. Moreover, advances in different aspects of plant genetics have enhanced deeper insight into molecular markers, plant species genetic diversity and tremendously aided the success of plant molecular breeding. Current rapid advances in molecular marker technology have produced novel techniques that are greatly facilitating research in almost every sphere of crop development and improvement. DNA markers have been very valuable in revealing the extent and distribution of variation in a diversity of crop species (Hailu and Asfere, 2020) and aiding tremendously in different plant germplasm conservation and management. Molecular marker systems have also facilitated the assembly of genetic maps and speed plant breeding by marker assisted-selection (Jiang, 2013; Collard et al., 2005). Molecular or DNA markers also aid in the introduction of desirable traits from wild relatives to cultivated species by providing vital phylogenetic information. Furthermore, the application of molecular marker technology enables differences in gene sequence to be directly observed and described. Ultimately, revealed gene sequence information facilitates the cloning and manipulation of genes of interest in crop research and improvement. The advent of molecular marker technologies introduced such a degree of precision that hitherto was impossible to achieve in plant breeding (Reddy et al., 2021; Nadeem et al., 2018; Collard et al., 2005).
This review encompasses the basic concepts, methodologies and applications of some of the molecular marker techniques currently widely in use in many laboratories. The molecular marker methods extensively discussed are RFLP, RAPD, SCAR, AFLP, SSR, CpSSR, ISSR, RAMP, SAMPL, SRAP, SSCP, CAPS, SNP, DArT, EST, STS, IRAP, REMAP, RBIP, and IPBS. Practically, there is no ideal molecular marker method that meets all expectations and does not pose one form of challenge or another with its application. It is, therefore, always important to take into consideration some essential factors in the selection of the appropriate DNA marker techniques that will enable the achievement of particular set of research objectives (Nadeem et al., 2018). The choice of which marker technique to apply depends very much on the understanding of the set objective, level of anticipated genetic variation and data to be generated from the study samples, sample size to be worked with, accessibility of probes or primer sets, availability of the technical know-how and appropriate facilities, time and financial considerations (Anne, 2006).
Well established molecular marker techniques such as the Arbitrarily Amplified DNA (AAD) markers for example Amplified Fragment Length Polymorphism (AFLP), Inter Simple Sequence Repeat (ISSR), and Random Amplified Polymorphic DNA (RAPD) are more popular and extensively used in plant genomic research. Compared to the popular traditional AAD markers, newly developed molecular marker techniques are less incorporated into plant breeding programs. Nonetheless, an appreciable number of plant molecular breeding works are recently using these new advanced molecular marker methods to achieve varied research objectives. Over the years, molecular marker research has focused largely on the development of molecular markers that are more efficient for the genomic analysis of crops of economic interest. On the contrary, little research resources have been devoted to producing molecular markers for the genomic investigation of under-utilized crops that do not attract significant economic importance. Consequently, in most under-utilized crops there is still woeful lack of sequence information or data to aid primer design. In such crops, therefore, some DNA marker techniques are still not applicable. It is, however, envisaged that in the near future, these crops will also be covered when DNA sequencing cost reduces significantly and the development of molecular markers becomes cheaper. This overview of molecular marker methods will further broaden the understanding and enable more effective and efficient use of DNA marker approaches in plant breeding to drive sustainable agricultural production and use.
2. The concept of molecular or DNA markers
A molecular or DNA marker is the difference in DNA nucleotide sequence—between individual organisms or species—that is in proximity or tightly linked to a target gene that expresses a trait. Typically, the target gene, expressed trait or biological function and the associated tightly linked molecular marker are inherited together (Collard et al., 2005). The specific genomic location of the molecular marker within chromosomes referred to as locus or loci may be known or unknown. It is noteworthy that molecular or DNA markers do not influence traits associated with the expression or function of the linked gene or genes. The tight association of molecular markers to a trait or gene of particular biological function, makes the markers serve as practical signs or flags that signal a particular gene locus and aid the detection or identification of the associated traits whether the genes involved are known or unknown and whether the gene(s) can be detected or not (Reddy et al., 2021). DNA markers can be useful for telling the individual genotypic differences in same or different species, if differences referred to as polymorphisms, exist in the marker nucleotide sequences between or among individuals or species. Molecular marker polymorphisms are due to varied type of mutations of DNA that create nucleotide sequence differences between or among organisms (Lincoln et al., 2018).
Generally, marker polymorphisms in organisms are caused by point mutations arising from single nucleotide substitutions, rearrangements involving insertions or deletions, DNA section duplication, translocations and inversions as well as mistakes in replication of DNA that are tandemly repeated (Selvakumari et al., 2017). Molecular marker signals that are used to reveal genotypic differences between individuals due to marker sequence differences are called polymorphic markers. On the other hand, DNA markers that cannot be used to differentiate between or among genotypes are referred to as monomorphic markers. The characteristics of a good and very useful DNA marker are that the marker is ubiquitous and evenly distributed throughout the genome, easy to assay, cost effective, multiplexed and can be automated. An ideal molecular marker must also be highly polymorphic, co-dominant in expression to enable effective discrimination between homozygotes and heterozygotes, should be highly reproducible and possible to share data generated among laboratories. Additional characteristics of a very good molecular DNA marker are that the marker creates no detrimental effect on phenotype, is genome-specific in nature, and multi-functional (Kordrostami and Rahimi, 2015).
Practically, a molecular marker is not just the associated polymorphism but the totality of the detailed protocols or procedures for its detection or identification. More often than not, a molecular marker has been considered from just the narrow view of differences in DNA sequences between individuals or polymorphism. It is, however, insightful to note that in some cases a molecular marker may practically simply be a primer or a set of primers, restriction enzyme(s) or a combination of primers and enzymes or other relevant components, coupled with the procedures for running the marker. The implication is that for a DNA section to be considered a molecular marker, a complete package of the set of primers, restriction enzymes or other relevant components as well as the established detailed procedure for the detection of that particular molecular marker must be known or available. Without such complete collection of marker specific information, a sequence polymorphism cannot be relevant as a molecular marker. Indeed, it is this complete collection of information that practically defines a molecular marker completely.
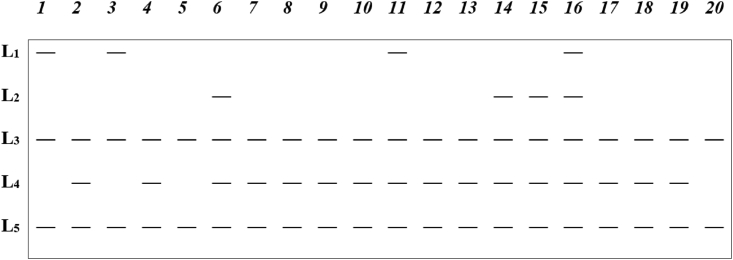
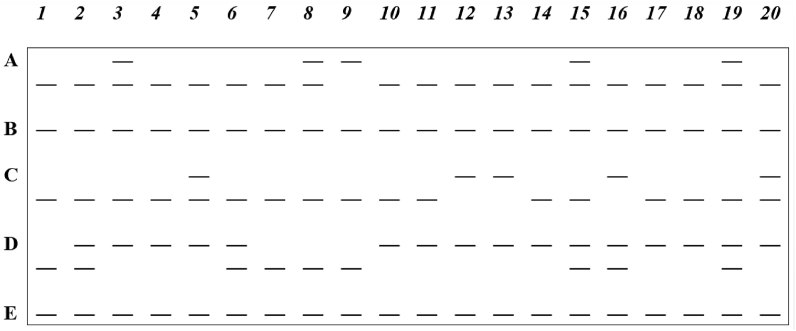
DNA markers are the most ubiquitous across the genome of organisms and are usually found in regions of the genome that do not code for proteins. These molecular markers are, therefore, considered neutral but are the most informative and widely used for many plant breeding studies (Collard et al., 2005). The major practical advantage associated with DNA markers that has enhanced their widespread use stems from the high abundance of these markers across genomes. Besides, environmental impact or influence on the developmental cycle of an organism on DNA markers is usually minimal or nil (Nadeem et al., 2018). DNA markers that are polymorphic can be classified as dominant (Figure 1) or co-dominant molecular markers based on mode of gene action and inheritance. As depicted in Figure 1, three loci namely L1, L2 and L4 were polymorphic because they revealed differences among the accessions. On the other hand, two loci, L3 and L5 were monomorphic since at those loci the accessions could not be differentiated. Those polymorphic molecular markers that can be used to differentiate between homozygote and heterozygote individuals for a trait are referred to as co-dominant markers (Figure 2). A hypothetical electrophoresis gel with twenty accessions (labelled 1 to 20) of a plant species analyzed using a co-dominant marker that detected five loci: A, B, C, D, and E, is shown in Figure 2. Three loci, A, C and D were polymorphic because they revealed differences among the accessions. On the other hand, two loci, B and E were monomorphic because at those loci the accessions could not be differentiated. The following polymorphic accessions showed one heterozygous locus: Accession 2-locus D; Accession 3-locus A; Accession 5-locus C; Accession 8-locus A; Accession 6-locus D; Accession 16-locus D; Accession 20-locus C. In addition, polymorphic accessions that possessed two heterozygous loci are Accession 15-loci A and D; Accession 19-loci A and D. It should also be noted that accessions 1, 4, 7, 9, 10, 11, 12, 13, 14, 17, 18 were monomorphic with only one band in each of the five loci. Unlike co-dominant markers, dominant polymorphic markers are unable to discriminate between homozygotes and heterozygotes (Figure 1). In the detection of molecular markers by gel electrophoresis, co-dominant markers are observed on the gel as DNA bands of many different alleles whereas a dominant marker only has two alleles represented as present or absent of bands. Practically, the variations in band sizes visualized on the electrophoretic gels define marker alleles. Co-dominant polymorphic markers may generate many different alleles that represent the homozygote dominant, the recessive and heterozygote individuals (Figure 2), whereas dominant markers produce only two alleles depicting homozygote dominants combined with heterozygotes as one composite present band and recessives as absent of band or vice versa (Figure 1).
Figure 1.
A hypothetical electrophoresis gel with twenty accessions (1–20) of a plant species analyzed using a dominant marker which detected five loci L1, L2, L3, L4 and L5. (—) depict DNA bands or marker alleles.
Figure 2.
A hypothetical electrophoresis gel with twenty accessions (1–20) of a plant species analyzed using a co-dominant marker which detected five loci: A, B, C, D and E. (—) depict DNA bands or marker alleles.
DNA markers are categorized into various classes depending on the detection method: hybridization, polymerase chain reaction (PCR) and DNA sequence dependent molecular markers. Some salient characteristics of major molecular markers have been compared (Table 1). The PCR technique for amplification of a section of DNA in a large quantity was developed by Cary Mullis in 1983. The subsequent automation of PCR (Mullis et al., 1986) was a major technological breakthrough in genome and molecular biology related research. PCR has since been a very useful technique to plant molecular breeders for DNA marker development and analysis. Important considerations for achieving successful product amplification in any PCR-based marker system are the quality and type of Taq DNA polymerase that is used. This is because these attributes of the enzyme determine its efficiency since low-quality DNA polymerase is only capable of producing short PCR fragments, whereas high-fidelity DNA polymerase will generate longer PCR products (Moazen et al., 2012). In this regard, specialized Taq polymerases have been designed for various PCR applications that are more efficient in driving PCR than standard Taq polymerase. For instance, to minimize challenges such as primer dimers and non-specific generation of PCR products, Hot Start Taq has been designed with an inhibitor to make the enzyme inactive at low temperatures. Hot Start Taq is only active and efficient after heating to 95 °C. Such specialized Taq polymerases with high-fidelity are industrially available and enable easier generation of PCR fragments even from for example high-GC templates. Specialized Taq polymerases can also catalyze the attainment of PCR products that are longer than 1 Kb and efficiently yield PCR fragments from samples with some amount of inhibitors (Moazen et al., 2012).
Table 1.
Comparing some salient characteristics of major molecular marker techniques.
| Marker | PCR-Based | Mode of inheritance | Locus specificity | Level of polymorphism | Reproducibility |
|---|---|---|---|---|---|
| RFLP | No | Co-dominant | Yes | Low-Medium | High |
| RAPD | Yes | Dominant | No | Medium-High | Low |
| SCAR | Yes | Co-dominant | Yes | High | High |
| AFLP | Yes | Dominant | No | High | Medium |
| SSR | Yes | Co-dominant | Yes | High | High |
| ISSR | Yes | Dominant | Yes | High | Medium |
| CpSSR | Yes | Co-dominant | Yes | Low | High |
| SAMPL | Yes | Co-dominant | Yes | High | High |
| SRAP | Yes | Dominant | No | Medium-High | Low |
| SSCP | Yes | Co-dominant | Yes | High | High |
| CAPS | Yes | Co-dominant | Yes | High | High |
| SNP | Yes | Co-dominant | Yes | Extremely High | High |
| DArT | No | Dominant | Yes | High | High |
| EST | Yes | Dominant | Yes | High | High |
| STS | Yes | Co-dominant | Yes | High | High |
| RAMP | Yes | Co-dominant | Yes | High | High |
Many different types of DNA marker systems are available and used efficiently in crop breeding studies in various crops of food, medicinal and industrial value. DNA or molecular markers are broadly classified by Poczai et al. (2013): Arbitrarily amplified DNA markers (AAD) including AFLP, ISSR, and RAPD; Conserved DNA based markers (CDM) for instance, Conserved DNA-Derived Polymorphism (CDDP), P450-based analogue (PBA) markers, and Tubulin-Based Polymorphism (TBP); Transposable element based markers (TEM) comprising IRAP, REMAP, Inter-SINE amplified polymorphism (ISAP), IPBS and S-SAP. Resistance-gene based markers (RGM) for example RGAP; Targeted fingerprinting markers (TFM) including Direct amplification of length polymorphisms (DALP), Promoter anchored amplified polymorphism (PAAP), SRAP, Target region amplification polymorphism (TRAP), Conserved region amplification polymorphism (CoRAP) and Start Codon Targeted (SCoT) Polymorphism.
The following section presents a review of the basic concepts, methodologies and applications of a number of the most common and widely used DNA marker techniques in crop improvement (Table 2). A wide diversity of molecular marker methods are currently available for genotyping a variety of plant genomes. Molecular or DNA markers are being increasingly used in basic genomic studies and applied plant breeding. The considerations for deciding on the particular molecular marker to use are based on the plant species to be studied, the goal of the research work and availability of the requisite resources.
Table 2.
DNA marker applications for improvement in various plant species.
| Application of molecular markers | Molecular marker used | Plant species | References |
|---|---|---|---|
| Genetic diversity, DNA fingerprint and germplasm conservation | DArT; ISSR and RAPD; CDDP; RAPD and ISSR | Maize (Zea mays); Castor Bean (Ricinus communis); Musa L. (Musaceae); Gloriosa superba; rice (Oryza sativa L.) | Badu-Apraku et al. (2021); Kim et al. (2021); Igwe et al. (2021); Tilwari and Sharma (2021); Mazumdar et al. (2020), |
| Assessment of heterosis in breeding | SSR and SNP; AFLP, RAPD and SSR; SNP; EST and SSR | Cotton (Gossypium hirsutum); Maize (Zea mays); Maize (Zea mays); wheat (Triticum aestivum); Rice (Oryza sativa) | Geng et al. (2021); Esan et al. (2021); Tomkowiak et al. (2020); Nie et al. (2019); Pavani et al. (2018) |
| Identification of haploid/diploid plants | SSR | Maize (Zea mays) | Rádi et al. (2020) |
| Bulk segregant analysis | SNP; SNP; SSR | Castor bean; Rice (Oryza sativa); Soybean | Wang et al. (2021); Yang et al. (2021); Sreenivasa et al. (2020) |
| Marker-assisted selection | SSR; SRAP; | Cassava (Manihot esculenta); Camellia oleifera; | Olasanmi et al. (2021); Feng et al. (2020) |
| Genetic and physical mapping | SNP; SSR; SNP; SSR | Cotton (Gossypium hirsutum); Safflower (Carthamus tinctorius); Pineapple guava (Acca sellowiana); Cassava (Manihot esculenta) | Feng et al. (2021); Jegadeeswaran et al. (2021); Quezada et al. (2021),Adjebeng-Danquah et al. (2020). |
| QTL mapping and characterization | SSR and AFLP; SSR; ISSR; SNP | Rubber; Cotton (Gossypium hirsutum); Capsicum annuum and C. frutescens; Peach (Prunus persica); wheat (Triticum aestivum) | Hou et al. (2021); Li et al. (2021); Olatunji and Afalayan (2019); Rawandoozi et al. (2021),Asif et al. (2020) |
| Association mapping | SSR; SSR; SNP | Maize (Zea mays); Chickpea; Maize (Zea mays) | Jha et al. (2021); Kim et al. (2021a); López-Malvar et al. (2021) |
| Marker assisted back cross breeding | SNP; EST and SNP | Rice (Oryza sativa); Cowpea (Vigna unguiculata) | Cheng et al. (2017); Batieno et al. (2016) |
| Hybrid identification | SSR; EST and SSR | Citrus (Citrus aurantifolia and Citrus limon); Elymus sibiricus | Guzmán et al. (2017); Zhao et al. (2017) |
2.1. Random fragment length polymorphism (RFLP)
RFLP is polymorphism that is dependent on DNA sequence length variation generated by restriction enzyme cleavage of genomic DNA at random but specific recognition sites of restriction enzymes coupled with DNA probe hybridization in Southern blotting. RFLP reveals variation in DNA sequences by the presence or absence of fragments of varying base pair sizes or lengths that are produced from the restriction of DNA samples using specific endonucleases (Konzen et al., 2017). Restriction endonucleases are enzymes isolated from bacteria. These enzymes cut DNA at specific recognition sites into shorter fragment sizes. Industrially isolated restriction enzymes are usually used by scientists to digest double stranded DNA at particular sites with specific enzyme recognition DNA sequences. Each restriction endonuclease recognizes different four to six base-pair restriction sites and, therefore, cuts DNA at diverse but specific recognition sequences. By implication, variations in the number of restriction site repeats and randomness of the distribution of particular recognition site of a restriction enzyme among individuals, generate varying number and fragment lengths after restriction digestion. The randomness of the distribution of restriction enzyme recognition cut sites creates differences in length between the restriction recognition sequences among individuals. The length of the random DNA restriction fragments produced after enzyme digestion, therefore, differ among organisms. Usually, RFLP bands correspond to DNA fragments of size range 2–10 kb.
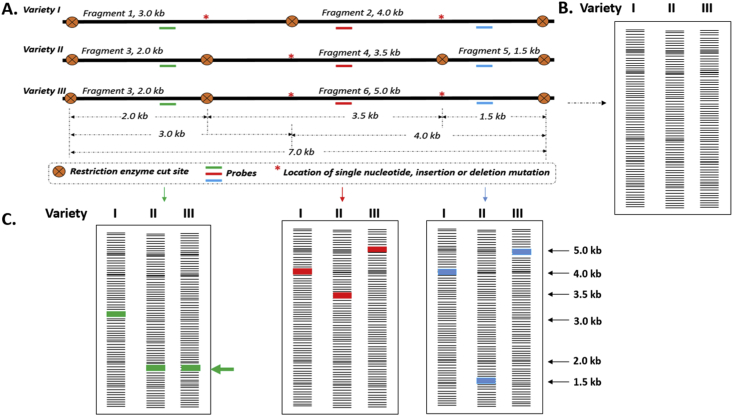
The observed differences in nucleic acid sequences and restriction enzyme cut sites imply that genetic differences or RFLP polymorphisms exist in the DNA sequence of the particular individuals. Variation in RFLP DNA band patterns in individuals arise from mutations that are mainly due to single nucleotide base-pair deletions and or insertions. These mutations create new restriction enzyme recognition sequences or delete existing enzyme recognition sequences, and thus, produce fragments of varied lengths and polymorphism. The recognition site of a restriction enzyme is lost when even just a single nucleotide base alteration is introduced in an existing enzyme recognition restriction site in the DNA. Similarly, such mutations could create new restriction enzyme recognition cleavage sites in the genome. A single base difference between two individuals or among many individuals could, therefore, result in either the presence of a new restriction site or absence of a pre-existing restriction site. In principle, the creation or deletion of restriction enzyme recognition sequences caused by insertions or deletion mutations within restriction recognition sites, serve as the main basis for RFLP (Figure 3A). Restriction enzyme digestion of DNA into fragments, gel separation of restriction products and detection of variation in Varieties I, II and III are depicted in Figure 3. In Variety I, restriction enzyme digestion produced Fragment 1, 3.0 kb and Fragment 2, 4.0 kb. In Variety II, Fragment 3, 200 bps; Fragment 4, 3.5 kb and Fragment 5, 1.5 kb were obtained. In Variety III, Fragment 3, 2.0 kb and Fragment 6, 5.0 kb were produced. The variation in length of restriction enzyme digestion products among the varieties is the result of the presence or absence of mutations among the different varieties leading to the loss of restriction enzyme sites or introduction of new restriction sites in the DNA sequences (Figure 3A).
Figure 3.
Restriction enzyme digestion of DNA into fragments, gel separation of restriction products and detection of variation in Varieties I, II and III. (A). Analysis of a section of DNA (long thick black line) of three Varieties I, II and III using a restriction enzyme and three designed probes (green, red and blue short lines). Location of mutations is shown as red asterisk. (B). Gel electrophoresis of the restriction enzyme digested DNA samples of the three Varieties I, II and III. The restriction digestion procedure generates a huge number of fragments with varying lengths in the reaction solution. Upon electrophoresis, the huge number of DNA fragments show virtually as lane of DNA smear on the gel for each variety. (C). Southern hybridization using three designed probes to detect variation among the three varieties I, II and III.
A generalized procedure of RFLP analysis is described briefly (Konzen et al., 2017). Firstly, pure DNA is isolated from usually the leaf tissues of the individuals to be tested. RFLP analysis require the extraction of sufficient amount of DNA. Achieving this can be quite laborious. For this reason, in some cases, PCR is used to amplify a DNA section of interest, over a duration of 2–3 h, to obtain good quantities of DNA required for efficient RFLP analysis. Where practicable, the PCR strategy cuts significantly the time involved in sample analysis. The isolated DNA or PCR product is digested to produce restriction fragments using selected restriction enzymes. The restriction digestion procedure produces a huge number of fragments with varying lengths in the reaction solution. Next, the obtained restriction DNA fragments are analyzed using agarose or polyacrylamide gel electrophoresis (PAGE) to reveal DNA fragment size variations. DNA fragments are negatively charged and separate based on their size and charge during electrophoresis.
Electrophoresis is carried out by loading the enzyme digested DNA samples in wells created in a gel placed in an electrophoretic tank fixed with positive and negative electrodes. An electric current is applied to the electrophoretic tank filled with running buffer solution to cause the DNA fragments to drift towards the positive electrode. Shorter fragments migrate at a higher speed through the matrix of a gel than the longer fragments and thus, create distinct DNA band profiles. In a further step, the DNA bands are made visible by gel staining using luminescent dyes such as ethidium bromide. The fragments are visualized under UV light illumination as bands of different lengths in the gel. Due to the huge number of DNA fragments in the digestion reaction solution, the fragments show virtually as lane of DNA smear in the gel for each individual and reveals no information about the individuals being studied (Figure 3B). Hence, the essence of the use of probes to detect existing variation in samples by targeting specific fragments with same complementary sequences as the used probe(s). To detect and identify genetic variation in the DNA captured in the electrophoretic gel, Southern blot hybridization with labelled DNA probe(s) is carried out (Southern, 1975).
A short DNA sequence in the form of an RFLP probe is designed to bind to fragments in the digested DNA sample in the electrophoresis gel. Typically, probes comprise short single- or low-copy genomic DNA or clones of cDNA. RFLP probes are locus specific and made up of sequences homologous to unique regions of the genome. The probe and target DNA Southern blotting reveals patterns unique to a genotype at a specific locus (Figure 3C). Southern hybridization using three designed probes to detect variation among the three Varieties I, II and III is shown in Figure 3C. The green probe bound to 3.0 kb fragment 1 in Variety I. In Variety II and III, the green probe bound to similar Fragment 3 of size 2.0 kb (Arrowed green) in both varieties. This probe is, therefore, unable to discriminate between Varieties II and III. The implication is that the green probe is not well designed and cannot be considered as a good probe. On the other hand, the red probe bound different fragment lengths: 4.0 kb, 3.5 kb and 5.0 kb in Variety I, II and III respectively and revealed variation among all the varieties. Similarly, probe blue hybridized to different fragment sizes: 4.0 kb. 1.5 kb and 5.0 kb in Variety I, II and III respectively and could clearly distinguish the three varieties. Unlike the green probe, the red and blue probes could, therefore, be considered as good probes since they were able to detect variation and separate all the varieties. In related species, heterologous labelled probes may be used. Usually, probes are labelled with a radioactive isotope. Alternatively, non-radioactive stains could be used. Generally, genomic or complementary DNA (cDNA) libraries are used as very useful resource for probe design. Designed probes of RFLP markers are typically conserved as clones ligated in suitable plasmid vectors. Subsequently, the DNA probes can be conveniently isolated from the plasmid vector constructs in which the probes are contained. RFLP probe development involves the restriction of genomic DNA with restriction enzymes that are methylation-sensitive such as PstI (Wolff et al., 1994). The basis for the choice of PstI is that expressed genes are usually not methylated and, therefore, PstI clones have the advantage of enriching the created library for single-copy or low-copy expressed gene sequences (Wolff et al., 1994). The digested DNA is separated in agarose gel based on size variation. The obtained fragments ranging 500 to 2000 bps are excised, isolated from the gel and cloned into a suitable plasmid vector. Southern blots of the fragment inserts are hybridized to restriction endonuclease digested total genomic DNA of different genotypes. Probe clones are screened and those probes that form hybrid complex with single-copy sequences are selected. Notably, in species that are known to exhibit moderate to high polymorphism, the application of up to four restriction endonucleases enhances the efficiency of polymorphism detection and identification.
Most RFLP markers are co-dominant and thus, able to detect both alleles in a heterozygous sample. RFLP markers are moderately polymorphic, highly locus-specific and highly reproducible. RFLP markers are also highly abundant in the genome and randomly distributed. However, RFLP analysis is practically a laborious, time-consuming and technically demanding procedure. Large quantity of purified, high molecular weight DNA is essential for each DNA digestion. Moreover, RFLPs marker analysis not amenable to automation. Besides, in plant species where suitable probes are lacking, the financial implication for probe design could be a drawback. RFLP is now obsolete due to the development of technically less demanding and cheaper polymerase chain reaction (PCR) based DNA marker profiling technologies. Extensively, RFLPs have been used to reveal genetic variation and phylogenetic associations in individuals as well as gene mapping studies.
2.2. Randomly amplified polymorphic DNA (RAPD)
The RAPD marker application protocol involves arbitrary oligonucleotide short primers usually 8 to 15 nucleotides in length that by PCR, randomly amplify DNA sections of large genomic DNA (Babu et al., 2021). In RAPD analysis, prior DNA sequence information is not an important requirement because of the use of arbitrary primers. As much as possible, a primer sequence selected for RAPD must have at least 40% GC (Guanine and Cytosine) content but 50%–80% of GC is usually preferred (Premkrishnan and Arunachalam, 2012). At this GC content, the primer will be able to function efficiently at the annealing temperature that facilitates the operation of DNA polymerase to effect DNA elongation (Cao et al., 2015). In addition, the primer must not have present in it a palindromic sequence (Premkrishnan and Arunachalam, 2012). Primer sequences are selected based on their highly polymorphic characteristics by reviewing earlier works described by various researchers. The selected RAPD primers can then be purchased from a number of different companies (e.g. Operon Biotechnologies— http://www.operon.com/or Integrated DNA Technologies, Inc., Coralville, IA, U.S.), either as primer sets or as individual primers, using the selected primer sequences obtained from literature review of earlier studies. However, because the polymorphism of RAPD primers can vary even within same cultivars, usually a large set of about 50–100 primers are preliminarily tested with the cultivar of interest. Out of these primers, a fewer number comprising about 10–20 that constitute the most informative or highly polymorphic are selected for the main analysis.
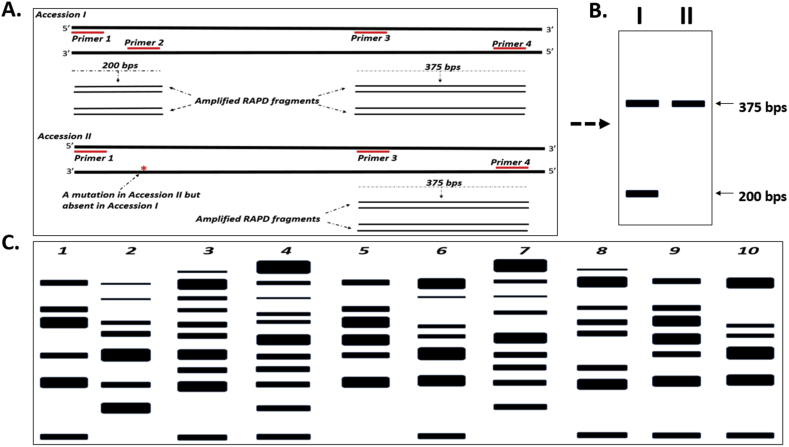
In many other marker systems, two primers comprising forward and reverse short DNA sequences, are required in the same PCR reaction. In RAPD PCR, however, one oligonucleotide primer is used per reaction and one copy occurs in forward primer orientation and another copy of the same primer assumes a reverse primer orientation. Commonly, RAPD primers are capable of generating PCR products from between one and ten genomic DNA sites concurrently. RAPD PCR fragment lengths usually range between 0.5 and 5 kb. It is worthy to note that primers may successfully amplify a portion of DNA or fail to amplify PCR fragments. The success or otherwise of the RAPD PCR depends on the presence, distribution and location of primer complementary sequences on the template genomic DNA. In instances where the two primer copies anneal at a distance too far apart or the 3′ ends of the two primer copies do not occur in the proper orientation to each other on the template, no PCR fragment is amplified (Zakiyah et al., 2019). Moreover, in cases where mutation alters a section that was previously complementary to a primer, the annealing of that primer at the altered site will be disrupted and hence, no PCR product will be produced. Individuals with such mutation effects will exhibit different DNA banding patterns of amplified DNA sections in an electrophoretic gel (Figures 4A and B). In Accession I (Figure 4A), the binding of primers 1 and 2 to the depicted DNA section produced fragment of size 200 bps. Similarly, the binding of primers 3 and 4 to the section of DNA of Accession I, amplified another fragment of size 375 bps. Two different RAPD fragment sizes were, thus produced in Accession I. In Accession II, a mutation indicated as red asterisk is present at the binding site of primer 2. The primer 2 binding site is, therefore, lacking in Accession II resulting in the amplification of only one RAPD fragment (350 bps size) in that accession. The 375 bp fragment is monomorphic because it could not differentiate between the accessions. The 200 bps fragment, however, is polymorphic since it discriminated between the accessions.
Figure 4.
RAPD variation between two plant Accessions I and II. (A). A section of the double strand DNA of Accessions I and II are shown as two long parallel thick black lines. (B). Gel electrophoresis RAPD pattern of Accessions I and II showing two bands (200 and 375 bps fragments) in Accession I but one band in Accession II (375 bps fragment). (C). Hypothetical banding patterns resulting from gel electrophoresis of RAPD PCR products of ten accessions (1–10) of a plant species.
PCR amplification occurs when two annealing template DNA locations have similar sequences, appropriately at a close distance of separation from each other and anneal to the template facing each other in orientation. Successful fragment PCR amplification is, therefore, very much linked to the nucleotide constitution of the template DNA sequence, genome size or complexity of the species as well as the sequence and length of the primer(s) used (Zakiyah et al., 2019). RAPD fragments are easily separated in agarose gels by electrophoresis. The gel is stained with ethidium bromide or any other appropriate stain and visualized by ultraviolet (UV) illumination (Baghel and Bansal, 2017). Complex patterns of PCR products are generated by the random annealing of the arbitrary RAPD primers to various locations in the genome of the target organism (Figure 4C). The variation in the banding patterns or the polymorphism shown for each accession (Figure 4C), arises from the presence and/or absence of different types of mutations that determine the binding or non-binding of the respective primers to different sections of the DNA of each of the accessions. RAPD polymorphisms are detected as present or absent of DNA bands mainly due to sequence size variations between primer binding sites with the target DNA of individuals (Baghel and Bansal, 2017). In principle, RAPD polymorphisms arise primarily from variation in the primer hybridization or annealing positions in the target genome. It is, however, useful to note that not all primer annealing sites within the target genome will necessarily produce amplified PCR fragments. Therefore, practically, RAPD polymorphisms are fragment length variations in actually generated PCR products from in between primer annealing sites in the target genome (Figure 4B).
The RAPD technique has been quite convenient and widely used because it is easy and quick to assay. This marker also has the advantage of neither requiring DNA probes nor prior sequence information for primer design. Such sequence data information is not important because random primers are commercially available. Besides, because PCR is used in RAPD analysis, only little amount of purified, high molecular weight DNA is needed. Essentially, contamination of sample DNA templates must be avoided. This precautionary measure is important because the short random primers used in RAPDs, can easily amplify DNA fragments in variety of organisms. The RAPD procedure is automatable and quick. However, the reproducibility of RAPD profiles is very problematic. Therefore, in order to reproduce RAPD profiles, it is absolutely critical to maintain very strict and consistent PCR reaction conditions. Basically, a strict adherence to RAPD protocols is vital due to the extreme sensitivity of RAPD profile generation to PCR reaction conditions (Vekariya et al., 2017). Invariably, the low reproducibility of marker profiles makes RAPDs inefficient to compare or use between or among laboratories working on similar research objectives.
Besides, being a dominant marker, it is difficult to tell if an amplified DNA locus is heterozygous or homozygous during RAPDs profile interpretation (Rajesh et al., 2014). Another drawback associated with RAPD analysis is that the technique is locus non-specific and the interpretation of electrophoretic gel patterns is quite confusing. Also, in some instances, the bands consist of co-migration of different amplified products thus, making band identification difficult to assign and problematic to analyze. Furthermore, the complex patterns of RAPD markers also pose challenges in the consistent scoring of electrophoretic images and mixture interpretation. Moreover, RAPD PCR fragments with similar lengths may be non-homologous or constitute the same DNA sequences. Another important shortcoming is that data quality is limited because RAPD is a dominant marker. Recent modifications have, however, improved the RAPD technique into more efficient marker methods like SCAR, SRAP and CAPS (Yang et al., 2014; Babu et al., 2021). These improved marker variants of RAPDs overcome the associated disadvantages of RAPDs and complement the efficiency in the applications of the marker.
2.3. Sequence characterized amplified regions (SCAR)
SCAR marker was first developed and initially applied to studies of downy mildew resistance genes in lettuce by Paran and Michelmore (1993). SCAR is an improved variant of RAPDs. The modification and conversion of RAPDs into a co-dominant, more locus specific and reproducible SCAR marker, enhances marker reliability (Yang et al., 2014). A SCAR is basically a PCR mediated technique that identifies genomic DNA fragment at a single locus using a pair of specific 15–30 bp oligonucleotide primers designed from nucleotide sequences derived from cloned polymorphic RAPDs fragments. SCAR marker procedures are technically simple and easy to carry out. The main limitation, however, is that sequence data from RAPDs polymorphic fragment is needed in order to design SCAR PCR primers. Obviously, this requirement for the prior knowledge of sequence information presents a hindrance to the use of SCAR. Compared to RAPDs primers, SCAR primers are longer. The constraints of low reproducibility that is associated with RAPDs analysis is surmounted in SCARs with the use of longer PCR primers. SCARs are, therefore, more locus specific and more reproducible compared to RAPDs (Yang et al., 2014). The reason for enhanced reproducibility is that SCAR PCR is less sensitive to reaction conditions (Cheng et al., 2016). In addition, being a PCR-based molecular marker, only little amount of target DNA is needed for SCAR analysis. SCARs are co-dominant inherited markers and detect mono-locus. Comparatively, SCARs are more informative than RAPDs which are dominant markers (Yang et al., 2014). Nonetheless, the likelihood is there for SCARs to exhibit dominance in some instances where at a section of sequence variation, one or both PCR primers partially overlap.
Cho et al. (2015) described briefly the procedure for converting RAPDs into SCARs. Firstly, RAPD PCR is performed and polymorphisms associated with length variation are detected by electrophoresis using stained agarose gel, followed by visualization of DNA bands under UV light illumination. Polymorphic DNA bands are cut from the agarose gel and purified. The purified DNA fragments are cloned into appropriate plasmid vector and then sequenced to determine the nucleotide sequences of the fragments. The obtained sequence data of the polymorphic DNA fragments is analyzed by comparing the sequences with known DNA sequences that are available at the NCBI (National Center for Biotechnology Information) database for sequence uniqueness. Next, the obtained nucleotide sequences of the polymorphic DNAs are used to guide the design and synthesis of specific pairs of internal SCAR primers (Cho et al., 2015). Apart from RAPDs, the usefulness of AFLP and ISSR markers in genetic applications has been greatly expanded by similarly converting polymorphic fragments of these markers to SCARs. SCARs are mostly applied in gene mapping research objectives (Boyd et al., 2019) and marker assisted selection.
2.4. Amplified fragment length polymorphism (AFLP)
AFLPs are DNA fragments of size range 80–500 bps, derived from restriction enzyme digestion reaction, followed by attachment of oligonucleotide adapters to restriction fragments and amplification of a subset of the fragments by selective PCR. Thus, AFLP marker analysis protocol in part combines the RFLP and PCR technologies to carry out digestion of DNA and PCR amplification (Sorkheh et al., 2007). Restriction enzymes act with high degree of specificity and hence, enhance the generation of a reproducible set of DNA fragments. The AFLP electrophoretic patterns are genetically due to diversity arising from restriction enzyme recognition sites or genetic variation in genomic regions that occur in between. Foremost procedure in AFLP analysis is the isolation of high quality DNA from tissues of the research organism. The isolated genomic DNA is then restricted with two enzymes.
The next step involves the ligation of adapters to the restriction fragments. The adapter attached fragments are amplified by PCR under stringent annealing conditions. The primers used are designed with sequences complementary to the adapter restriction site sequence and additional selective nucleotides at their 3′-ends. The resulting PCR products include only those fragments that have complementary nucleotides extending beyond the restriction recognition site. The PCR products are analyzed by denaturing polyacrylamide gel electrophoresis to reveal the existing genetic polymorphisms. AFLP are dominant markers and polymorphisms are detected as present or absent of electrophoretic DNA bands in polyacrylamide gels. The bands are recorded as present or absent based on particular sizes generated for each sample. The four main steps of the AFLP technique including fragmentation of DNA by restriction digestion, attachment of adapters and ligation, restriction fragment selective PCR and gel electrophoresis analysis, have been described in more detail (Blears et al., 1998).
2.4.1. Generation of restriction fragments by enzyme digestion of genomic DNA
In AFLP, a frequent cutter restriction enzyme and a rare cutter restriction enzyme are used for fragmentation of isolated genomic DNA into shorter pieces. Restriction fragments are generated by combining a 6- to 8-base recognition rare-cutting restriction enzyme (Example ApaI, AseI, EcoRI, HindIII, and PstI), and a frequent-cutting restriction enzyme of 4-base recognition (Example MseI and TaqI). MseI with the recognition sequence TTAA is the preferred frequent cutter restriction enzyme. The reason is that most eukaryotic genomes are adenine and thymine rich (Rajewska et al., 2012). The essence of using two enzymes is to enable the generation of restriction fragments for PCR amplification and to subsequently produce a number of band profiles that are practically of manageable complexity. The frequent-cutter enables the required fragment sizes between 100 and 1000 bps to be obtained for efficient PCR amplification. The restriction digestion produces three main groups of fragments. These include rare-cutter enzyme generated fragments with both ends cut, frequent-cutter enzyme generated fragments with both ends cut and the third group constitute those fragments with one end cut by either the rare-cutter or the frequent-cutter. Usually, the bulk of about 90% of the restriction fragments generated are those with frequent-cutter sites on both ends.
2.4.2. Joining of adapters to restriction fragments and ligation
Adapters are enzyme specific and composed of two oligonucleotides (10–30 base pairs) that are in part complementary with each other. Under appropriate conditions in a reaction solution, the two oligonucleotides form a double-stranded conformation with ends that anneal to the sticky ends of the respective restriction enzyme sites. Invariably, adapters are short DNA sequences that are enzyme specifically designed to aid the detection and identification of an unknown DNA sequence. Adapters and the restriction fragments are combined and ligated together in a reaction catalyzed by T4 DNA ligase. The adapter sequences and the restriction recognition sequences serve as primer annealing sites in subsequent selective PCR. The ligation procedure usually causes loss of the original restriction enzyme recognition site due to base changes incorporated into the adapter sequence. The practical significance of the loss of enzyme restriction sites is that it enables restriction and ligation reactions to be performed in the same tube simultaneously. The advantage is that any fragment-to-fragment ligation is removed by enzyme restriction. Moreover, non-phosphorylated adapters are commonly used in order to prevent adapter-to-adapter joining and ligation. These two aspects of the protocol ensure that virtually all restriction fragments have adapters ligated to their ends.
2.4.3. Selective PCR of restriction fragments
In the AFLP protocol, following the restriction-ligation reaction, selective generation of a concise subset of restriction fragments is carried out by PCR. The objective of this selective amplification is to decrease the complexity of the mixture of fragments in the PCR reaction solution and subsequently in the gel following electrophoresis (Vos et al., 1995). A less complex band profiles in the gel facilitates more accurate scoring and data analysis. Two AFLP primers are used for the selective PCR amplification. One primer comprises a 5′ section complementary to the adapter and the adjacent rare-cutter restriction site sequence with 3′ selective (1–3 bps) nucleotides extension. The second primer also has a 5′ end that is complementary to the adapter and the frequent-cutter recognition site sequence with an additional 3′ selective nucleotides (1–3 bps) extension. The primer 3′-end extension with selective nucleotide addition enable a subset of the total number of restriction fragments to be amplified. The selective nucleotide extensions also enable additional polymorphisms to be detected beyond those polymorphisms that are possible with only restriction site analysis. With each selective nucleotide addition, the density of the DNA profiles generated decreases. The profile patterns obtained with each selective nucleotide addition when compared is just a sub profile of the preceding pattern. This observation, nonetheless, holds for nucleotide additions of maximum three selective bases. Comparing profiles obtained with three selective base primers extensions and profiles generated with four selective base primer extensions, Vos et al. (1995) demonstrated the amplification of additional DNA bands in the profiles generated with four selective base primer extension. This implies a loss of selectivity and an indication of mismatches tolerance during annealing by primers with 4-base extensions but not primers with 3-base extensions.
Selective amplification is carried out under stringent annealing conditions and only template fragments with nucleotides complementary to the primer selective nucleotide extension beyond the restriction site are amplified. Generally, the rare- and frequent-cutter enzyme sticky end restriction fragments are preferentially amplified instead of the more predominant frequent-cutter fragments. This differential amplification arises because the primer that targets the rare-cutting restriction site and adapter is designed to have a higher annealing temperature compared to that of the frequent-cutter associated primer. Furthermore, two primers are engaged in the amplification of the fragments cut by both rare- and frequent-cutter enzymes and thus, avoid the creation of an inverted repeat at the ends (Vos et al., 1995). It is, therefore, very important to note that PCR conditions are a very critical aspect of the AFLP technique. Besides, an enhanced effectiveness of the primer matching the rare-cutter site enables only a practically manageable fraction of the fragments to be competently amplified to allow efficient detection of polymorphism. The complexity of the mixture of amplified fragments depends on the genome complexity of the species being studied, the types of restriction enzymes used for the DNA fragmentation reaction, and the number and nature of the selective nucleotides incorporated into the primers.
2.4.4. Gel electrophoresis, visualization and analysis
Following the step of denaturation by heating at 90–95 °C for 3–5 min, is the procedure of AFLP gel electrophoresis (Blears et al., 1998). The denatured samples are either labeled radioactively or non-radioactively with a fluorescent dye. The prepared samples are loaded into a 4.5% or 5% polyacrylamide gels for electrophoretic separation of the samples. The developed AFLP DNA band profiles are visualized by silver staining. Compared to other markers such as RAPDs and RFLPs or other currently used PCR mediated molecular markers, AFLP is superior in revealing higher number of polymorphic loci DNA bands per reaction (Sorkheh et al., 2007). The markers produced are reliable and reproducible due to the stringent annealing conditions used. An impediment that hinder the success of AFLP is the necessity for good quality DNA of high molecular weight. The marker reveals mainly dominant alleles, and it is also known that co-migrating fragments belonging to different loci do not have similar sequence homology. A high number of bands with varying intensity are observed in AFLP profiling. It is, therefore, essential in AFLP analysis to employ a certain set of strict but subjective criteria as guidelines for scoring of bands (Blears et al., 1998). AFLPs have been applied frequently to investigate genetic identity and phylogenetic relationships in many species because the marker generates highly informative profiles (Akash et al., 2013). AFLP is highly abundant and densely dispersed randomly across the genome and, therefore, widely valued for gene mapping research objectives as well as construction of high-resolution genetic maps.
2.5. Simple sequence repeats (SSRs)
SSRs are simple sequences that arrange in tandem repeats of mononucleotide, dinucleotide, trinucleotide, tetranucleotide, pentanucleotide and hexanucleotide motifs (Koelling et al., 2012) that are highly abundant, polymorphic and randomly dispersed throughout the nuclear, chloroplast and mitochondrial genomes of many species (Kalia et al., 2011). The most frequently occurring motifs are the mono-, di-, tri- and tetra nucleotides. Evolutionarily, it has been hypothesized that SSRs resulted from either single-strand DNA slippage, double-strand DNA recombination or mobility of transposable genetic DNA molecules and nucleotide disparities (Koelling et al., 2012). SSRs have also been confirmed to be present in protein-coding genes and ESTs (Yan et al., 2017). SSRs constitute one of the classes of microsatellites. Other classes of microsatellites include simple sequences that arrange in short tandem repeats and polymorphic microsatellite length variations. Of these classes of microsatellites, SSRs show lesser number of repetitions per locus but exhibit higher polymorphism (Padmakar et al., 2015). The reason is that the application of PCR in the analysis of the SSR technique enhances the capacity to detect higher level of polymorphism in SSRs in microsatellite regions (Padmakar et al., 2015).
Polymorphic SSR profiles in plants arise from variation in the number of repeats of the SSR motifs between or among individuals in a population. The presently increased availability and accessibility of large EST data, developed full genome sequences and libraries, serve as good guide for the easy design of SSR primers and as well in some species, aid the quick identification of repeat motifs. SSRs are searched and identified from genomic libraries or within genes using EST databases (Yan et al., 2017). Compared to RAPDs or ISSRs analysis where single primers are used per PCR reaction, SSR marker loci are generated with two-primer PCR reactions. One primer anneals to the DNA template in a forward direction and the other primer hybridizes to the template in a reverse orientation. Both primers anneal complementary to specific DNA sequences flanking the SSR sequence (Mei et al., 2015, Figure 5).
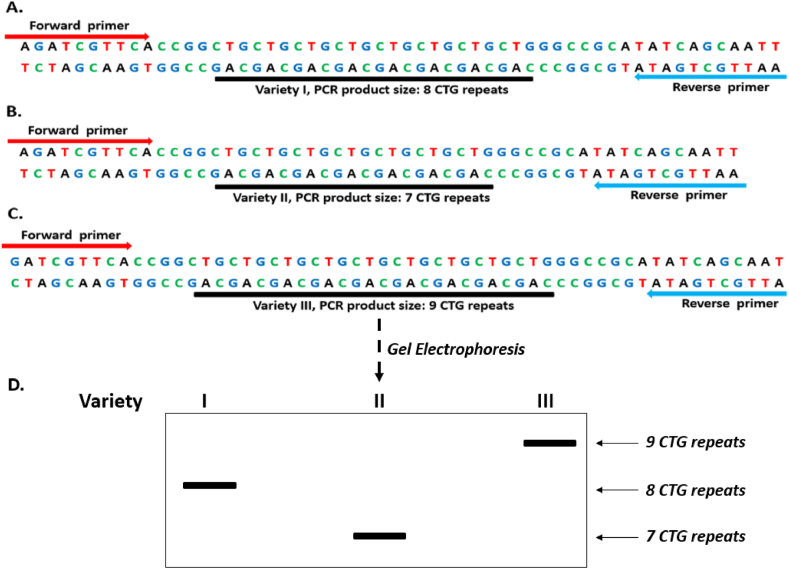
Figure 5.
SSR variation among three plant Varieties I, II, and III with 8, 7 and 9 CTG microsatellite repeats respectively in (A), (B) and (C). (D). Gel electrophoresis SSR profile of the three Varieties I, II and III showing polymorphic bands due to differences in the number of CTG repeats among the Varieties I (8 CTG repeats), II (7 CTG repeats) and III (9 CTG repeats).
In brief, SSR marker development involves first the creation of a library of SSR motifs and subsequently, the discovery of particular SSR motifs (Singh et al., 2020). Afterwards, the identification of favourable genomic sections is carried out and primer design is done to enable PCR to be undertaken. SSR PCR fragments separation is accomplished with agarose or polyacrylamide gel electrophoresis. Next, the assessment and evaluation of gel banding patterns are done. Finally, SSR profile analysis and interpretation are performed for investigation of polymorphism. SSRs are locus-specific markers that are inherited in a co-dominant pattern, an attribute of the marker that presents a major advantage for efficient plant mapping and population genetic studies (Kumar et al., 2021; Padmakar et al., 2015). SSRs are very informative but the marker discovery or development stage, which includes DNA sequencing, can be quite expensive. Locus-specific SSR markers are more informative and so their development serves as a very useful genetic resource. However, the development of such SSR specific-loci involves isolation of individual loci and their characterization. To achieve that, the steps to follow include construction of DNA library of microsatellite probes, library screening and sequencing of positive clones. Lastly, PCR primer synthesis and optimization are accomplished. Another impediment is that non-specific PCR products and generation of monomorphic loci cause generally quite low recovery rate of useful SSRs. In addition, as a prerequisite for efficient genotyping, there is the need for a high resolution gel equipment system. Unfortunately, large scale SSR marker analysis is restricted by its dependence on gel or capillary electrophoresis for profiling and recording.
2.6. Chloroplast simple sequence repeat (cpSSRs) microsatellites
Microsatellites in the form of SSRs are randomly distributed in all plant nuclear and chloroplast genomes. Comparatively, chloroplast genomes exhibit lower rate of genetic mutation than do nuclear genomes. It is, therefore, more daunting to detect appreciable amount of sequence variations in chloroplast genomes. However, cpSSRs provide higher polymorphism levels and thus make the marker attractive for easy genotyping and population genetic studies (Moore et al., 2010; Drew et al., 2014). Notably, cpSSRs show varied polymorphism levels across species and loci. The application of cpSSRs is popular because these markers are easier to isolate than nuclear microsatellites. CpSSRs are predominantly mononucleotide tandem repeat motifs, typically repeated 8–15 times. It has been established that cpSSRs in chloroplast non-coding genomic regions show intra-species variation in microsatellite repeat number and hence, provide polymorphism (Moore et al., 2010; Drew et al., 2014). Non-coding regions of cpDNA genomes comprising mainly introns and intergenic spacers are more targeted for genetic diversity characterization (Shaw et al., 2007). The focus on non-coding regions stems from the fact that cpDNA genomes have been found to evolve slowly and thus, not very informative for evolutionary and population genetic evaluation. Moreover, non-coding regions are predominantly repetitive genomic sections that occur in tandem in the form microsatellites (Dong et al., 2012). Compared to single nucleotide variations, non-coding genomic regions have been found to reveal higher allelic variation (Haasl and Payseur, 2011). Genetically, chloroplast chromosomes are non-recombinants and are inherited uniparental. The implication of the non-recombining chromosome inheritance is that all cpSSR loci are linked. Though, the mode of inheritance is largely uniparental, cases of biparental inheritance exist and lead to heteroplasmy (Bai et al., 2014).
To aid in expanding the application and to make cpSSRs more informative, the use of genus-specific cpSSR primers to achieve cross-species amplification is helpful. Comparing studies that used species-specific primers and those that used universal primers, it was found that the later primers generated significantly lesser number of polymorphic loci. Universal cpSSR primers, therefore, though potentially useful, are limited to producing only a small number of polymorphic profiles. The attraction of using cpDNA markers is enhanced by the capability of universal primers to efficiently amplify homologous genomic sections in many different species (Shaw et al., 2007). Meanwhile, in wild plant species, de novo sequencing of non-coding cpDNA has been found to be the best means for developing cpSSRs. Moreover, the complexity of the genetic diversity is linked to hypervariable sections of chloroplast DNA (cpDNA). Practically, it is essential that the alleles of cpSSR are sequenced. It is also necessary that the reliability of cpSSR assays are validated.
Chloroplast genomes (cpDNA) have been very informative and enabled the development of appreciable data for the establishment of phylogenetic relationships within and among species (Moore et al., 2010; Drew et al., 2014). CpSSR markers are ascribed several advantages such as easy identification and analysis, occurrence and abundance in gene-rich regions, efficient transferability among closely or distantly related plant species and extensive use for the estimation of genetic diversity. These markers are useful tools for identifying maternal parent putative hybrid progeny. Nonetheless, cpSSR markers may be deficient in identifying hybrids as these chloroplast associated markers provide only maternal data (Li et al., 2020). Other main drawbacks that hinder the effective use of cpSSRs include lack of variation in some species or occurrence of very narrow genetic diversity, unavailability of general primers, the effects of size homoplasy, heteroplasmy phenomenon, and interspecific hybridization mediated cytoplasmic introgression. Nonetheless, cpDNA have been very informative and enabled the development of appreciable data for the establishment of phylogenetic relationships within and among species (Moore et al., 2010; Drew et al., 2014). The marker has also been used to aim at tracking uniparental gene flow patterns in populations (Bai et al., 2014).
2.7. Inter simple sequence repeat (ISSR)
ISSRs are genomic sections flanked by simple sequence repeat or microsatellite sequences. Microsatellite sequences are genomic regions that consist of simple sequence motifs of short DNA sequences—usually di-, tri- and tetra- or penta-nucleotides in size—repeated multiple times in tandem. ISSRs are ubiquitous and randomly distributed throughout the genome. The ISSR marker system is based on the generation of multilocus markers through the PCR amplification of genomic regions flanked at each end by identical microsatellites repeat sections that are oppositely oriented. The size of the intervening DNA nucleotide sequence between the microsatellites must be within a size range that practically allows for successful PCR amplification (Sarwat et al., 2016). A foremost consideration for successful ISSR analysis is the need to obtain high quality DNA. In addition, it is important to standardize the quantity of template DNA used in each PCR reaction. The application of consistent amount of DNA in each PCR is essential in order to obtain consistent concentrations of amplification products, uniform and reproducible band intensities across samples. Generally, ISSR analysis starts with PCR amplification using an ISSR primer with isolated genomic DNA as template. ISSR-PCR is followed by agarose or polyacrylamide gel electrophoresis of PCR products and visualization. The next step is scoring of the ISSR bands and finally, data analysis.
Unlike many other marker systems, ISSR-PCR reaction is a single-primer PCR amplification (Sharafi et al., 2017). Some molecules of the primer serve as forward primers and copies of the same primer orient and anneal to the template DNA in the opposite direction as reverse primers. ISSR primers are usually long ranging between 15 and 30 bases in size and designed to comprise repetitive simple DNA sequences that target genomic microsatellite sections. In ISSR analysis, the primers used may be unanchored primers or primers anchored at either the 3′ or 5′ end. The primer anchor is normally made up of 1–4 degenerate nucleotides that overlap the flanking microsatellite sequences (Sharafi et al., 2017). Usually, anchored primers are preferred for ISSR-PCR. Unanchored ISSR primers may be unstable along the microsatellite region, produce inconsistent amplification during PCR, and thus hamper the reproducibility of ISSR profiles (Figure 6). ISSRs can have reproducibility problems. ISSR-PCR is usually done with high annealing temperature of between 45 and 60 °C, determined based on the ISSR primer melting temperature. A touch-down PCR reaction profile covering this range of annealing temperatures (45–60 °C) helps to avoid having to try several different temperatures with ISSR primers that are difficult to optimize using the standard PCR method. ISSRs are highly reliable because of high annealing temperature of the primers and longer sequence products (Yuan et al., 2019).
Figure 6.
Schematic illustration of the design and annealing of ISSR PCR primers. (A). Unanchored GAGAGAGAGA primer. Unanchored primer can anneal anywhere in the CT dinucleotide repeat region on the template DNA leading to slippage and eventually ultimately smear formation in electrophoretic gels. (B). 3′ anchored GAGAGAGAGANN primer. The primer is anchored at the 3′ end with two nucleotides (NN). The anchor nucleotides guide the primer to anneal at specific regions and not anneal arbitrarily on the template DNA. The anchorage enables the generation of clear bands. (C). 5′ NNGAGAGAGAGA anchored primers. The primer is anchored at the 5′ end with two nucleotides (NN). The anchor enables the primer to anneal at specific regions and not anneal arbitrarily on the template DNA. The primer amplifies part of the repeat region in addition, leading to larger band sizes.
The ISSR multilocus markers comprise averagely 10–60 fragments that are generated simultaneously from multiple loci during PCR. Moreover, the ISSR PCR products occur between 100 and 3000 bps in range, representing the distance variation between adjacent and oppositely oriented microsatellite regions (Mohanty et al., 2012, Figure 7). The ISSR-PCR DNA products are profiled by electrophoresis in agarose or polyacrylamide gel and visualized under UV light illumination. The bands are recorded as present or absent based on particular sizes. ISSR markers are highly polymorphic and characterized as dominant markers (Yadav et al., 2019). However, to expand their usefulness, ISSRs can also be transformed into co-dominant markers such as Sequence Characterized Amplified Regions. ISSRs are simple, technically easier, less demanding to perform and low-cost compared to other dominant marker systems.
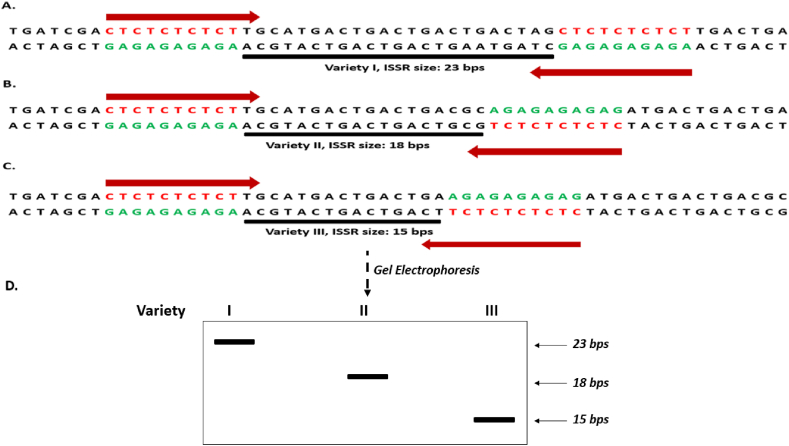
Figure 7.
A hypothetical scheme of the concept of an ISSR marker defined by the intervening DNA sequence between the SSR, CTCTCTCTCT. (A), (B) and (C) show length polymorphism in three Varieties I, II and III respectively using a single primer depicted as thick orange arrows. (D). An electrophoretic gel depicting the band profile arising from variation in inter SSR nucleotides differences (hypothetically 23, 18 and 15 bps respectively) among the three Varieties I, II and III.
The main advantage of ISSRs is that no prior knowledge of DNA data is needed to determine primer sequences. In addition, the application of PCR makes it possible for only small amount of DNA to be used in ISSR analytical procedures. Typically, 10–50 ng of good quality DNA is sufficient for each reaction. ISSR marker applications circumvent most of the practical limitations of SSR, AFLP and RAPD analysis (Padmakar et al., 2015; Ali et al., 2015). Some technique variations of ISSR analysis include SPAR and DAMD. SPAR-PCR is driven by one primer designed to contain only the core motif of the target repetitive simple sequence region. DAMD PCR reaction on the other hand, also involves one primer that is created using only an SSR core motif. ISSR markers create highly polymorphic data and useful in many research objectives. ISSR has been applied to generate valuable information in closely related or distant species in studies of genetic diversity and phylogeny, genome mapping, and evolutionary biology (Thakur et al., 2016; Guo, 2014).
2.8. Microsatellites and RAPD/AFLP marker modifications
2.8.1. Randomly amplified microsatellite polymorphisms (RAMP)
RAMP is a PCR-assisted technique which combines the advantages of the SSR and RAPD marker methods. Comparatively, SSR markers reveal higher level of polymorphism than many other molecular markers. However, SSR markers are quite labour intensive to carry out. RAPD markers are more cost effective, however, they detect low level of polymorphism as compared to that of SSR markers. RAMP markers were, therefore, developed taking cognizance of the merits and demerits of the two marker systems. RAMP markers resolve to a large extent, the major challenges posed by microsatellite and RAPD markers (Liu et al., 2020). The RAMP technique uses a microsatellite matching primer which amplifies a section of isolated DNA with or without the activity of RAPD primers. The SSR or microsatellite targeted primers are designed to have radiolabeled 5′ anchor and 3′ repeats. The melting temperature of the 5′ anchored primers is kept at 10–15 °C higher than the melting temperature of the RAPD primers. The temperature manipulation enables a more effective hybridization of the anchored primer to the complementary sections of the template DNA (Liu et al., 2020). The PCR products obtained are separated with agarose gel electrophoresis. RAMP markers are widely dispersed in the genome of many plant species, present higher polymorphism and are technically cost effective. RAMPs have been proven to be valuable PCR-based DNA marker and have successfully been applied for molecular characterization and genetic relationship studies in various plants species (Salazar et al., 2014).
2.8.2. Selectively amplified microsatellite polymorphic loci (SAMPL)
SAMPL was developed taking into consideration the high multiplex ratio for AFLPs (Vos et al., 1995) and the high level of genetic polymorphism associated with SSR. The SAMPL marker protocol involves PCR driven by one AFLP primer—specific to adapter nucleotide sequences linked to DNA restriction fragments—matched with another primer complementary to SSR sequences in order to reveal genetic polymorphisms (Morgante and Vogel, 1994). SAMPL analysis incorporates restriction fragments selection to simplify and minimize the complexity of the multilocus SSR profiles generated. This strategy enhances the convenience and effectiveness of the marker for the analysis of large genomes.
The SAMPL technique involves a few main steps as described by Gupta et al. (2005). Adapters are first ligated with the aid of DNA ligase to restriction DNA fragments generated by a two-enzyme digestion of DNA into smaller fragment sizes usually a rare-cutter and another a frequent-cutter restriction enzymes. Following restriction digestion, a subset of the restriction fragments with their ends linked to adapter sequences are selectively amplified using primers complementary to the adapter sequences. Furthermore, a selective amplification is carried out to obtain a subset of the amplified DNA fragments containing SSR sequences. The selective amplification procedure is enhanced by the inclusion of additional selective nucleotides at the 3′-ends of the adapter primers that extend into DNA fragments (Gupta et al., 2005). The PCR amplified products are profiled in polyacrylamide gel to enable higher band resolution and observed under UV light illumination. It is noteworthy that the complexity of SAMPL profiles is cut down considerably by the incorporation of an affinity capture step aided by magnetic beads that take away a fraction of the restriction fragments.
2.9. Sequence related amplified polymorphism (SRAP)
SRAP involves the production of PCR copies of the coding regions of genomes using short arbitrary PCR primers complementary to open reading frames (ORFs). Li and Quiros (2001) first created and reported the application of SRAP markers. SRAP specifically amplifies coding regions based on a unique combination of two different arbitrarily designed primers. The forward primer is complementary to Guanine and Cytosine rich exon sections whereas the reverse primer is complementary to Adenine and Thymine rich promoter, intron, and spacer regions. The primers are usually of size 17–18 nucleotides for SRAP markers. The primers are designed such that 13–14 bps serve as core sequences and the initial 10–11 bases from the 5′ end of the primer function as filler sequences (Alzahib et al., 2021). These filler sequences do not occur in any specific composition. In the forward primer, a GC-rich core sequence CCGG is placed next after the filler sequences. Similarly, for the reverse primer, an AT-rich core sequence AATT is introduced next after the filler sequences. In both primers, following these AT- and GC-rich sequences, at the 3′ ends are placed three random selective nucleotides (Li and Quiros, 2001; Budak et al., 2004). Technically, fluorescent labeled forward primer can be used and PCR products profiled by capillary electrophoresis and scored.
SRAP employs specific annealing temperature PCR conditions in a two-step procedure (Li and Quiros, 2001). A step of one cycle of initial 4-minute denaturation at 94 °C, is followed by a template generation phase which covers five cycles of 1-minute denaturation at 94 °C, low-temperature annealing of 35 °C, and elongation step at 72 °C. This template generation phase then gives way to template amplification phase for 35 cycles with a higher annealing temperature of 50 °C and finally ends with an extended elongation step of 2-minute, 72 °C (Budak et al., 2004; Li and Quiros, 2001). The PCR amplified products are profiled by gel electrophoresis, visualized through autoradiography, and scored by the presence of DNA band or absence of it in the gel profile (Alwala et al., 2008; Alghamdi et al., 2012).
The SRAP marker technology is efficient, robust and highly variable, simple and less technically demanding process. SRAP is also inexpensive and effective with high reproducibility and versatility (Alzahib et al., 2021). SRAPs are dominant markers in nature. Generation of SRAP data and its analysis reveal valuable information for assessing genetic diversity of large germplasm collections and identification of quantitative trait loci in different taxa (Wanga et al., 2015). As an ORF-based marker that targets functional genes, SRAP is very valuable in marker assisted crop breeding and improvement (Bhatt et al., 2017; Ge and Daizhen 2015).
2.10. Single strand conformational polymorphism (SSCP)
SSCP is a mutation detection and screening approach that enables the identification of mutant variants in many different species. SSCP is a technically simple and easy method that is based on electrophoretic mobility differences of single stranded DNAs for detecting unknown mutations and accurate analysis of allelic and mutational sequence variations. SSCP has proven to be an efficient technique that is commonly applied to detect single nucleotide substitutions, small deletions and insertions as well as micro-inversions. Single nucleotide changes alter and cause differential mobility of single stranded DNA in non-denaturing gels. This characteristic of single stranded DNA is relied on in the application of SSCP for the detection of mutations. The principle of detection of sequence variations or mutations is based on the differences in movement of single stranded wild-type and mutant PCR fragments during gel electrophoresis. The method depends on differential gel electrophoretic mobility based on unique folding of denatured single stranded DNA.
Denatured single strands of DNA with different nucleotide sequences fold into different secondary structures or conformations. DNA fragments with even just a single nucleotide change exhibit measurable gel electrophoretic mobility differences DNA when subjected to non-denaturing SSCP analysis. A slight difference in mobility of the experimental sample relative to a normal control or wild-type fragment indicates a mutation. The variation in strand fold shape or conformation gives rise to different rates of migration speed within gel during electrophoresis. The folding of DNA strands is so nucleotide specific that even one base difference affect the form of folding and results in different conformations. The resulting secondary structures of DNA produces unique conformations determined by the nature of the primary nucleotide sequence. The type of secondary structure formed dictate the mobility of the fragment on non-denaturing acrylamide gel electrophoresis.
DNA isolation from target organisms is an important step in the protocol of SSCP PCR and analysis. In this technique, PCR is used to produce DNA amplicons referred to as PCR-SSCP fragments. The SSCP procedure is efficient for the detection of single nucleotide polymorphic mutations for PCR product size range between 450 and 500 bps. The DNA fragments are denatured by 90–95 °C heat treatment for 3–5 min or by chemical denaturants to separate the initially double-stranded fragments into single strands. This step is immediately followed by rapid cooling by putting the denatured samples on ice to cause renaturation but prevent the single DNA strands from reassuming double-stranded DNA states. The formed single DNA strands undergo folding into a variation of conformational structures. Some of the single DNA strands fail to re-anneal with their complementary strands but rather form intra-strand secondary structures by themselves. Finally, scanning for mutations and analysis of polymorphic bands are performed. To boost the efficiency for scanning, radiolabeled PCR fragments are assessed in mutation detection enhancement gels. Alternatively, non-isotopic SSCP assessment is undertaken using precast gels. It is estimated that SSCP exhibits approximately as high as between 80 and 90% detection of potential point mutations. SSCP is a low-cost, technically quite simple to carry out and is potentially high-throughput. The technique has proven useful in the detection and analysis of many diseases and associated causal organisms. SSCP is also amenable to appropriate modifications for mutation studies in genes of any organisms.
2.11. Cleaved amplified polymorphic sequences (CAPS)
The methodology of CAPS marker application combines PCR amplification and restriction enzyme digestion of PCR products to detect variation in the form of sequence polymorphism among organisms (Babu et al., 2017; Reyes et al., 2015; Barth et al., 2002). CAPS is, therefore, based on polymorphism arising from differences in DNA restriction enzyme recognition sequences in individuals. Differences in genomes among organisms is created by single nucleotide substitution mutations arising from insertions and deletions that cause the introduction or removal of restriction enzyme recognition sites. Based on the similarity between RFLPs and the CAPS techniques in combining PCR, restriction enzyme cleavage and fragment length polymorphism, CAPS marker is also referred to as PCR-RFLP. However, compared to the complex Southern blot hybridization and the arduous radioactive probe detection procedures involved in RFLPs, CAPS analysis is simpler and less cumbersome.
PCR is applied to amplify DNA fragments from target genotypes using primers of size usually 20–25 bps. The PCR primers used are designed from DNA sequences available in genebanks for instance NCBI, cDNA sequences or sequences obtained from isolated and cloned polymorphic RAPD bands. It is also noteworthy that to enhance the likelihood of detecting polymorphisms, the primers used for CAPS-PCR are designed targeting introns or 3′ un-translated regions. Typically, the PCR amplified products of the gene or genomic region of interest, range between 300 and 2000 bps. PCR amplification is followed by restriction of the obtained PCR fragments separately with suitable and appropriate number of restriction enzymes. Some of the restriction enzymes may be more polymorphic than others. It is, therefore, useful to screen polymorphism with a number of restriction enzymes. The digested PCR products are then screened for polymorphism by gel electrophoresis separation of the restriction fragments (Figure 8).
Figure 8.
CAPS marker assisted selection of Genotypes I, II and III based on (T/A) single nucleotide substitution polymorphism. The nucleotide substitution T/A in the EcoRV cut site are shown in red. (A). PCR produces only the A allele (in blue) in Genotype I, only the T allele (in green) in Genotype III, but produces both the T and A alleles in Genotype II. The T allele has EcoRV cut site (GATATC) but not the A allele. (B). The T allele in Genotype II or Genotype III gives rise to two fragments of size 330 bps and 350 bps after EcoRV restriction digestion. (C). Gel electrophoresis of EcoRV restriction digested products, therefore, produces one band in Genotype I (PCR fragment not cut, 680 bps), two bands in Genotype III (PCR fragment cut into 330 and 350 bps) and three bands in Genotype II (one PCR fragment not cut 680 bps, other fragment cut into two, 330 and 350 bps fragments).
Alternatively, the PCR product can be cloned with suitable plasmid vectors and sequenced. The sequence information enables the choice of the set of restriction enzymes appropriate for the efficient restriction digestion of the PCR product. The prepared PCR product is separated either by agarose gel electrophoresis or polyacrylamide gel electrophoresis. Using a dye such as ethidium bromide, the electrophoretic gel is appropriately stained and the CAPS profile generated is visualized. The DNA band profiles generated represent polymorphisms in the form of the presence or absence—induced by single point mutations—of restriction endonuclease recognition sites. Differences in fragment lengths generated as a result of variation in restriction enzyme sites location, reveal polymorphisms as different band size profiles on gels.
CAPS polymorphisms are more daunting to unravel compared to other restriction fragment associated techniques such as RFLPs because of size limitations of amplified fragments which ranges 300 to 2000 bps. In addition, knowledge of DNA sequence of the genomic region of interest is necessary for the development of CAPS PCR primers. However, CAPS markers are versatile so some of the mentioned difficulties associated with the marker can be overcome by combining CAPS with SSCP, SCAR, AFLP or RAPD (Arif et al., 2019; Barth et al., 2002) and thus, enhance the detection of DNA polymorphisms. CAPS are polymorphic co-dominant and informative markers. Comparatively, CAPS genotypes are easily identified, scored and analyzed. The CAPS molecular method is one of the very widely used procedures for the detection of single nucleotide substitutions or polymorphisms. The objectives of mutation detection and genomic variation studies in many plant species have been successfully achieved using CAPS (Arif et al., 2019). Another advantage is that little amounts of DNA are sufficient to attain efficient PCR amplification. Besides, the marker has high reproducibility of DNA bands profile data (Babu et al., 2017; Reyes et al., 2015). However, the main disadvantage in the application of the CAPS method is that, it is restricted to single point mutations that introduce restriction endonuclease recognition sites or interrupt existing sites leading to loss of restriction recognition sites.
2.12. Single nucleotide polymorphisms (SNPs)
SNPs are genetic polymorphisms that arise from particular single base pair positional differences in DNA sequences among individuals of a species or different species. Compared to other genetic markers, SNPs are the highest frequently occurring form of DNA sequence polymorphisms in organisms (Manivannan et al., 2021). SNPs are commonly located in coding and non-coding as well as intergenic regions of genomes at varying abundance across each of these genomic regions with a frequency of one SNP per every 100–300 bps of DNA (Fusari et al., 2008). Invariably, a single-nucleotide base is the fundamental and smallest genetic unit of inheritance in organisms. SNPs variation is created when a recognition site of a restriction enzyme is present on one allele and that particular recognition sequence is absent on the other allele. The presence or absence of SNPs results via the occurrence of mutation events such as transitions or transversions as well as insertions or deletions based on nucleotides substitutions. Furthermore, the restriction due to presence or non-restriction as a result of the absence of restriction enzyme recognition sites in the DNA region, gives rise to differences in DNA fragments of various length polymorphisms upon enzyme digestion and electrophoretic separation in gels.
There are many methods of SNP genotyping that uses allelic distinction techniques and detection platforms. Different kinds of SNP genotyping assays are currently available for genetic diversity detection and identification. Most of these assays involve the application of molecular mechanisms notably, extension of primers in PCR, invasive cleavage involving restriction digestion, oligonucleotide joining by ligation and allele-specific hybridization (Bruse et al., 2008). Until recently, SNP detection was mainly achieved by the Sanger dideoxy-sequencing method. Sanger sequencing, however, poses considerable constraints such as producing far more data volumes than necessary, inefficient in detecting SNPs in heterozygous DNA templates, time-consuming and very expensive. These practical challenges triggered the rapid development and evolution of many alternative gel-based SNP variation detection approaches that are technically easier to handle. Some of such molecular marker techniques include CAPS, Allele Specific PCR (AS-PCR) and SSCP. In all these techniques, SNP detection is based on the electrophoresis of PCR products in agarose or polyarcylamide gel. The use of gels, however, presents serious limitations to the achievement of high throughput.
The alternatives to surmounting the difficulties associated with gel-based marker techniques are the now high-throughput SNP genotyping platforms, which employ mainly the applications of mini-sequencing, heteroduplex analysis and allele specific hybridization. Methods like next generation sequencing (NGS), Genotyping-by-Sequencing (GBS), chip-based NGS, and allele-specific PCR comprise some of the most recently developed high-throughput genotyping technologies that have been applied to reveal very informative SNPs. These markers have, therefore, become the most attractive in relation to the many other available molecular markers for genotyping (Long et al., 2017). SNPs discovery is accomplished by creating sequence alignments and analysis using sequence data stored in databases. Currently, many SNPs techniques being developed aim at achieving at a go, large scale analysis. Most of such SNP methods, like the traditional techniques, still require a marker specific amplification reaction, marker specific oligonucleotide primers, or the use of probes like Taqman or Molecular Beacon (Ewart et al., 2019) and sequencing. SNP markers are characteristically biallelic and stable over many generations. In addition, SNP markers are amenable to automation, high throughput and, therefore, a popular and convenient technique for molecular plant breeding applications.
2.13. Diversity array technology (DArT)
DArT is a generic microarray-based hybridization, high-throughput and reproducible approach for detecting the presence and absence of individual fragments for DNA fingerprinting analysis. The technique has proven so efficient and very effective that even a single DArT assay is capable of genotyping simultaneously thousands of polymorphic loci distributed over the genome (van Deventer et al., 2020). A good quality genomic DNA of 50–100 ng amount is enough for purposes of DArT analysis. Unlike other marker systems, previous sequence data is not required for the application of the DArT technique in the evaluation of traits of interest. Another advantage of DArT is that it is very cost effective. DArT overcomes many of the limitations of currently available marker technologies (Onley et al., 2021).
DArT involves the screening of a randomized library of fragments created purposely to improve the chances of detecting SNPs mainly across the genome in the form of insertion or deletions. Essentially, DArT involves the construction of a genomic library which constitutes the genomic representation (van Deventer et al., 2020). DArT libraries are constructed with the best suitable individuals appropriate for the intended research purpose, using either single or pooled samples. For achievement of the objective of mapping studies, individuals used are usually the parents of a segregating population. On the other hand, in genetic variation evaluation, the DNA is obtained from either domesticated crop varieties or their known wild relatives. The next important step is the printing of the genomic library on microarray chips. Following the preparation of the microarray chips, is the labelling of the genomic representations. The next procedure is hybridization of the labelled genomic representation on the microarray chips. Finally, the chip is scanned and then data analysis is carried out. Therefore, the general concept upon which DArT works basically depends on genome complexity simplification by restriction digestion reactions, followed by hybridization of the restriction fragments to microarray chips to produce simultaneously an assay of thousands of markers across a genome (Onley et al., 2021).
Generally, in DArT marker analysis, initial discovery array experiments are undertaken in order to identify markers in the genomic library. The identified markers are then used to create genotyping arrays by re-arraying on new slides to enable high-throughput detection of thousands of markers in large populations. The simplification in genome complexity to enable efficient genotyping has now been markedly improved to involve cutting edge technologies such as next generation sequencing (NGS). The application of this approach has enabled effective achievement of quick SNP detection in diverse organisms (Sansaloni et al., 2011). Similarly, based on enhanced approaches, NGS has been proposed as a means to genotype coupled with RAD (Restriction-associated DNA) sequencing and recently, by a similar method generally termed GBS (Genotyping-by-Sequencing) (Zhu et al., 2019).
For the discovery and to facilitate scoring of marker data and analysis, an identical platform is usually created. DArT markers are scored with high accuracy and the DArT microarray platform itself allows flexibility of applications. In principle, for the objective of assembling polymorphic markers to create genotyping arrays of a single genotype, the discovered polymorphic markers are normally used for genotyping (Onley et al., 2021). However, in the absence of such objective, discovered markers need not be genotyped. The DArT method presents many merits compared to other molecular marker technologies. Most marker technologies are gel based and dependent on electrophoresis separation of fragments, resulting in low throughput. Array-based methods for instance DArT, are capable of separating very high DNA fragment densities as high as over many tens of thousands (>10,000), whereas polyacrylamide gels, could only accommodate and separate between 50-150 DNA fragments (van Deventer et al., 2020). DArT is, therefore, conveniently a higher throughput method with the advantage of enabling parallel marker data analysis instead of the limited capacity encountered with serial data analysis.
2.14. Expressed sequence tag (EST)
EST is complementary DNA (cDNA) generated by using reverse transcriptase to drive the conversion of messenger RNA (mRNA) into DNA. The development of ESTs is necessary for enhancing the stability of mRNAs. This is because the chemistry of nucleic acids is such that nucleic acids in the form of DNA or cDNA molecules are more stable than nucleic acids as RNA molecules. Complementary DNA (cDNA) consists of only exon—protein coding section of an RNA transcript—DNA sequences from mRNA with non-protein coding intron sequences excised or spliced. Introns are non-protein coding DNA sequences that interrupt the continuity of sequences that code for proteins. ESTs are created by cDNA sequencing. Sequencing of few hundreds of nucleotides is carried out from either the 5′ or 3′ end of the cDNA to generate 5′ ESTs or 3′ ESTs, respectively (Huang et al., 2017). The 5′ ESTs arise from the exons. Exons are known to be conserved usually across species and maintain same sequence constitution within a gene family. The 3′ ESTs on the other hand, are more associated with non-protein coding introns or un-translated regions (UTRs). Comparatively, introns or UTRs are less conserved across species than exhibited in coding sequences. The major impediments in identifying genes from genomic sequences depend on the species of organisms involved, size and complexity of the genome and the frequency of occurrence of introns (Huang et al., 2017). ESTs are produced from cDNA libraries. Currently, there are several millions of ESTs from a diversity of organisms available in many databases.
ESTs have been very informative in gene discovery by providing valuable information on gene function. ESTs have aided in the design of DNA microarrays and associated probes in gene function research. Besides, many highly valuable EST-based molecular markers, such as Restriction Fragment Length Polymorphism, Single Nucleotide Polymorphism, Inter Simple Sequence Repeats and Cleaved Amplified Polymorphic Sequences have been achieved (Xiao et al., 2020). EST derived single or low-copy RFLPs applications have proven very useful in the creation of high-density genetic and physical maps (Xia et al., 2007). ESTs also enable in silico production of SNP and SSR markers (Bhardwaj et al., 2016). SNPs are developed from ESTs comprising mainly 3′-untranslated regions (UTRs) of cDNA clones, in order to optimize the likelihood of discovery of nucleotide variations linked to existing genetic diversity in species of organisms. In this approach, primer pairs complementary to the EST sequences are designed. PCR amplification of corresponding regions from several genotypes is undertaken and sequenced. Sequence comparison across the genotypes is then performed to reveal SNPs. Alternatively, potential SNPs can be identified computationally by using sets of ESTs obtained from different cultivars and comparing the sequences in an alignment (Bhardwaj et al., 2016).
EST based SSRs are identified using pattern-finding computer software applications. Primer pairs are designed from available sequence data to determine variation in length polymorphism among cultivars of interest. ESTs have been found to be very useful resource for SSR marker development because typically, in various plant species, SSRs of size at least 20 bps suitable for marker development can be screened from ESTs. Comparatively, EST derived SSRs are normally associated more with regions conserved and transcribed than un-transcribed regions (Bushakra et al., 2015), and hence speculated to be easier transferred among closely related genera. EST derived SSRs are more functionally linked to differential gene expression compared to SSRs of non-coding genomic origin (Bushakra et al., 2015).
2.15. Sequence tagged site (STS)
STS is a DNA section that is characteristically short and exhibits sequence uniqueness. The exact STS sequence is usually found only at a location and nowhere else in the genome. The exact STS is any genomic locus defined by its primer sequences. Olson et al. (1989) developed and first reported STS markers as DNA landmarks during the human genome physical mapping project. STS was later found useful and adopted in plant genomic studies. An STS can be developed for any genomic site, provided that locus can be cloned and sequenced. STS markers are co-dominant, technically simple to carry out and highly reproducible. The marker is also suitable for high throughput and automation (Olson et al., 1989). STS markers are, therefore, very useful when integrated with a stable robust and reliable DNA marker linked to genes of a trait of agronomic interest.
Zhang et al. (2013) successfully generated STS in the form of SCARs from individual RAPD markers. RAPD polymorphic bands were cloned and the nucleotide constitution of the terminal ends were determined by sequencing. The obtained sequences were used to design primers for specific amplification of desired fragments. Similarly, RAPD-like adapted Random Amplified Microsatellite Site (RAMS) employs primers designed to target repetitive sequences of microsatellites in amplification reactions. In their genetic diversity studies using RAMS, Cardona et al. (2018) established that RAMS marker generates a high incidence of polymorphism and more reproducible and transferable products. In addition, SPAR polymorphic banding profiles are developed with core motifs of microsatellites. STS is also analyzed with PCR primers that are developed based on either the nucleotide sequences of RFLP probes or generated AFLP fragments. However, the transformation of AFLP markers for STS marker applications is technically quite difficult, particularly in polyploids such as hexaploid wheat (Prins et al., 2001). Nonetheless, the technique has been applied to obtain successful results in species of several crops (Cardona et al., 2018).
2.16. Retrotransposon-based markers
Retrotransposons have at their ends long terminal repeats (LTRs) that are exceedingly conserved sequences (Wu, 2017). Retrotransposons are the most predominant mobile genetic molecules in eukaryotes (Kalendar et al., 2010, 2018). These mobile genetic molecules or elements are frequently located in genomic regions next to known genes. The sequences of several retrotransposons have been produced. Analysis of many retrotransposons within and between plant species show that retrotransposons exhibit high level of heterogeneity and insertion polymorphism (Kalendar et al., 2017). Moreover, high copy numbers of retrotransposons are known to be dispersed in plant genomes. The abundance and widespread random distribution of retrotransposon molecules throughout diverse plant genomes have been explored for DNA marker studies (Kalendar et al., 2010). The LTRs sequences are used to guide primer design for retrotransposon-based marker analysis. Characteristically, retrotransposon insertions are irreversible, and thus, appropriate particularly for phylogenetic studies. Several modifications and variations of retrotransposon-based markers are available (Kalendar et al., 2017; Kalendar and Schulman, 2014; Schulman et al., 2012). The following are the main retrotransposon-based markers described in this review: IRAP, REMAP, RBIP, and IPBS. The comparisons of some important attributes of these markers have been outlined in Table 3. Other useful molecular markers that have also been recently applied in various plant species have been presented (Table 4).
Table 3.
Comparison of some important attributes of retrotransposon-based molecular markers.
| Marker | PCR-based | Detection | Specificity | Polymorphism | Reproducibility |
|---|---|---|---|---|---|
| IRAP | Yes | Multi-locus | High | Medium | Medium |
| REMAP | Yes | Multi-locus | High | Medium | Medium |
| RBIP | Yes | Single-locus | High | High | High |
| IPBS | Yes | Single-locus | High | High | High |
Table 4.
Other DNA marker systems recently applied in various plant species.
| Molecular marker system | Working concept of marker | Marker applications | References |
|---|---|---|---|
| Penta-primer amplification refractory mutation system (PARMS) | SNP fluorescence scanning genotyping technology. Universal fluorescent primer pairs, allele-specific primer pairs, and reverse shared primer pairs are used to amplify SNP loci. | Development of PARMS marker of the TAC1 gene in rice. | Gao et al. (2021); Lu et al., (2020); Zhang et al. (2019); Ye et al., (2001) |
| Conserved DNA-Derived Polymorphism (CDDP) | Conserved genes representing protein sequences of preserved amino acids are amplified with short universal or degenerate primers to reveal length polymorphism. | Genetic stability assessment in long-term micropropagated rose Rosa damascene trigintipetala; Bananas and plantains, Musa L. (Musaceae Juss.) genetic diversity and population assessment. | Igwe et al. (2021); El Dessoky et al. (2020); Poczai et al. (2013) |
| P450-based analogue (PBA) markers | Polymorphism is determined by differences in random distribution of cytochrome P450 regions amplified with universal primers that target the CYP or heme-binding sites in plants. | Genetic variability assessment in drumstick (Moringa oleifera Lam.) accessions. | Ravi et al. (2020); Poczai et al. (2013); Rana et al. (2005) |
| Tubulin-Based Polymorphism (TBP) | Single degenerate primer pairs complementary to conserved sections of the tubulin exons, anneal and amplify intervening introns. | Genotyping different genetic resources in the legume family. | Braglia et al. (2020); Poczai et al. (2013) |
| Inter-SINE amplified polymorphism (ISAP) | ISAP primers randomly target various locations within SINE elements and amplify intervening genomic regions | Differentiation of common poplar genotypes; Genotyping of Cucumis melo accessions; Tomato breeding programs | Reiche et al. (2021); Sormin et al. (2021),Pantchev et al. (2019); Poczai et al. (2013) |
| Sequence specific amplified polymorphism (S-SAP) | Adapters are ligated to restriction enzyme digested fragments. LTR and adapter specific target primers with selective nucleotide extensions are used for PCR amplification. | Genetic diversity and population structure of Eryngium maritimum; Genetic variability and marker assisted selection in Mexican guava | Leviòa et al. (2019); Campos-Rivero et al. (2017); Poczai et al. (2013) |
| Intron length polymorphisms (ILPs) | Detected by PCR using primers designed to anchor exons flanking target introns. | Salt tolerance genes identification, association mapping and virus-induced gene silencing in cotton; High-throughput genetic diversity analysis in rice. |
Cai et al. (2017); Badoni et al. (2016) |
| Inter small RNA polymorphism (iSNAP) | Small RNAs complementary primer and a primer complementary to flanking regions are applied to produce polymorphic profiles. | Construct a rice genetic map and fingerprint eight tobacco varieties | Gui et al. (2011); Poczai et al. (2013) |
| Direct amplification of length polymorphisms (DALP) | A primer with the −40 USP core and 3′ selective nucleotides is paired as forward primer with the common M13 sequencing primer to create polymorphic profiles. | Differentiation of Panax ginseng from P. quinquefolius | Poczai et al. (2013); Ha et al. (2001) |
| Promoter anchored amplified polymorphism (PAAP) | Short PCR primers are incorporated with degenerate regions that anneal to plant promoter regions. These are then used to detect polymorphism. | Divergence assessment in soybean | Mokate et al. (2017); Poczai et al. (2013) |
| Target region amplification polymorphism (TRAP) | An arbitrary SRAP primer and a fixed primer complementary to ESTs are combined to generate polymorphic profiles. | Genetic Structure Analysis in Sugarcane (Saccharum spp.); Discern genetic variability in sweet sorghum | Junior et al. (2020); Khidr et al. (2020); Poczai et al. (2013) |
| Conserved region amplification polymorphism (CoRAP) | Arbitrary primers are designed from ESTs and combined with fixed primer incorporated with the plant intron associated sequence core CACGC. | Genetic variability and relationships among Salvia ecotypes | Fabriki-Ourang and Yousefi-Azarkhanian (2018); Poczai et al. (2013) |
| Start Codon Targeted (SCoT) Polymorphism | Random primers with ATG start codons incorporated are used either alone or in combination to amplify polymorphic fragments. | Genetic diversity assessment of Iranian Aegilops triuncialis accessions; Assessing genetic diversity in Crepedium acuminatum. | Khodaee et al. (2021); Thakur et al. (2021); Poczai et al. (2013) |
| Directed Amplification of Minisatellite DNA (DAMD) | A PCR mediated marker method that uses a single primer complementary only to the core motif of a minisatellite. | Genetic variability in selected date palm (Phoenix dactylifera L.) cultivars | Purayil et al. (2018) |
2.16.1. Inter-retrotransposon amplified polymorphism (IRAP)
IRAP is a retrotransposon-based marker that is generated based on the proximity of two retrotransposons using outward facing PCR primers that are complementary and anneal to their LTRs sequences. Kalendar et al. (2010) developed and first reported the application of IRAP. In carrying out IRAP analysis, DNA fragments intervening two adjacent LTR of retrotransposons are PCR generated with primers that target the 3′ ends of the LTR sequences. El zayat et al. (2021) identified that LTR sequences usually occur in three orientations: tail-to-tail, head-to-head and head–to-tail orientations. In the head-to-head orientation of LTR sequences, identical sequences separated by short intervening distances in different strands are amplified with primers oriented away from each other. PCR is applied to amplify the LTR intervening sequences using single primer as forward and reverse per reaction. Similarly, in the tail-to-tail, the location and orientation of the LTRs is reverse that of the head-to-head orientation. The LTRs are amplified using a single primer oriented in opposite direction towards each other. Finally, for the head-to-tail oriented LTRs, two primers are used, a 5′- and 3′-primer (El zayat et al., 2021). Agarose gel electrophoresis is used to reveal the IRAP profile. A single IRAP amplification reaction can generate several PCR products of varied sizes ranging 300 to 3000 bps (Kalendar et al., 2010). IRAP is a dominant and multiplex approach that is applied to produce retrotransposon insertion site variations in various plant species.
2.16.2. Retrotransposon microsatellite amplification polymorphisms (REMAP)
REMAP is one of the retrotransposon-based DNA marker methods that involves the detection of variation in the intervening genomic sequences between a retrotransposon long terminal repeat (LTR) and a closely located SSR or microsatellite in genomes (Kahl, 2001). REMAP profile development is a PCR-assisted technique. In this marker method, microsatellite complementary primers are used in conjunction with specific primers complementary to the retrotransposon LTR (El zayat et al., 2021). Therefore, REMAP fragments or sequences are generated by amplifying the intervening sequences between retrotransposons and microsatellites using a primer complementary to a sequence of an LTR and another primer complementary to a simple sequence repeat motif. The primers for REMAP PCR are designed such that those primers that target the microsatellite loci contain sequences complementary to the respective repeat motif with a 2–4 nucleotides 3′ end anchor in order to prevent primer slippage arising from arbitrary annealing between microsatellite motifs (Nadeem et al., 2018). REMAP PCR products can be profiled on high-resolution agarose gel by electrophoresis. REMAPs are dominant and multiplex marker systems that are convenient for assessment of variation in retrotransposon insertion sites. REMAP is commonly applied in the analysis of the genetic diversity in populations of diverse plant species.
2.16.3. Retrotransposon-based insertion polymorphism (RBIP)
In RBIP analysis, the insertion of retrotransposon anywhere in a genome is investigated mediated by PCR. The development of RBIP for detection of points of retrotransposon insertion is realized via the design of primers complementary to sections of LTRs and also primers complementary to the retrotransposon and sequence of flanking DNA (Metcalfe et al., 2015). The procedure of RBIP for revealing of retrotransposon insertion polymorphisms is performed by carrying out two different types of PCR. One PCR uses a primer that is complementary to the retrotransposon and another primer complementary to the flanking DNA sequence. The other PCR involves designed primers with 3′ and 5′ ends flanking both sections of the insertion site of the retrotransposons. RBIP marker represents a single locus and co-dominant (Nadeem et al., 2018). Polymorphism in RBIP is usually detected by gel electrophoresis or hybridization, using agarose or dot assays respectively. In high-throughput RBIP analysis, data is obtained usually by fluorescent scoring from generated tagged microarray marker profiles (Ghonaim et al., 2020). A major challenge with RBIP characterization is the need for information on sequence of sections adjoining the retrotransposon insertion sites. The advantages of RBIP are that it can easily be automated and sample throughput can be increased through the use of technologies that do not involve gel analysis such as TaqMan™ or DNA chip technology.
2.16.4. Inter-primer binding site (IPBS)
IPBS marker is a useful tool for analyzing variations in primer binding site (PBS) intervals in LTR retrotransposons between or among individuals using PCR. PBS occurs universally in all LTR-based retrotransposon sequences (Barut et al., 2020; Kalendar et al., 2010). PBS are conserved regions that are found in the LTRs of retrotransposons and are usually located next to the 5′ LTR sequences. LTR sequences, on the other hand, show wide variations across LTR retrotransposon families (Kalendar et al., 2017). LTR retrotransposons are reverse transcribed with the binding of the PBS region by the 3′ terminal sequences of tRNA. A tRNA complement exhibits universal genomic presence and are generally dispersed in all LTR retrotransposons and retroviruses. In LTR retrotransposons, tRNA complements serve as the reverse transcriptase primer binding site. Inter-Primer Binding Site (IPBS) basically relies on the primer binding site. The function of the PBS primers is to bind LTRs and effect the amplification of diverse sequences (Barut et al., 2020).
Across genomes, retrotransposons occur as mixed, inverted, nested or truncated in the chromosome (Flavia et al., 2017). The amplification of retrotransposons is achieved with conservative PBS primers. The IPBS technology is a marked improvement on other retrotransposon analysis methods because IPBS circumvents the necessity for prior sequence knowledge (Kantar et al., 2021; Arystanbekkyzy et al., 2018; Yaldiz et al., 2018). This advantage has made the use of this marker quite attractive because sequence data or information require additional investment in cloning and sequencing of LTR. Recently, the applications of IPBS marker technique have become popular for the inspection of genetic diversity and relationships in various plant species (Kantar et al., 2021; Ali et al., 2019; Yaldiz et al., 2018).
3. Conclusion
The principles and methodology of the most popular and extensively used molecular markers for crop breeding and improvement have been elucidated. In all, thirty-four markers have been presented and probably constitutes one of the largest and most elaborate coverage of molecular marker overview in a single presentation. The molecular marker techniques discussed comprise primarily the well-established Arbitrarily Amplified DNA (AAD) marker methods, the microsatellite-based marker techniques and the retrotransposon-based molecular marker approaches. Indeed, molecular or DNA marker techniques provide tremendous opportunities for molecular genomic research. However, these markers should not be viewed as a substitute for the other agro-morphological or biochemical markers but rather, molecular markers should be applied as complementing techniques in genomics and plant breeding to provide a more complete understanding of the diversity in available germplasm and ways in which such diversity can best be exploited to enhance agricultural production, sustainable food and nutrition supply.
Declarations
Author contribution statement
The author conceptualized, reviewed the relevant literature and prepared this article.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
No data was used for the research described in the article.
Declaration of interests statement
The author declares no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The author expresses his profound gratitude to the Biotechnology and Nuclear Agriculture Research Institute (BNARI) of the Ghana Atomic Energy Commission (GAEC) as well as the Graduate School of Nuclear and Allied Sciences (SNAS) of the University of Ghana, for making available their facilities that significantly aided in producing this review material. The author also extends appreciation to the many references he used to support this review work.
References
- Adjebeng-Danquah J., Manu-Aduening J., Asante I.K., Agyare R.Y., Gracen V. Genetic diversity and population structure analysis of Ghanaian and exotic cassava accessions using simple sequence repeat (SSR) markers. Heliyon. 2020;6(1) doi: 10.1016/j.heliyon.2019.e03154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akash M.W., Shiyab S.M., Saleh M.I. Yield and AFLP analyses of inter-landrace variability in okra (Abelmoschus esculentus L.) Life Sci. J. 2013;10(2):2771–2779. [Google Scholar]
- Alghamdi S., Al-Faifi S., Migdadi H., Khan M.E., El-Harty M.A. Molecular diversity assessment using sequence related amplified polymorphism (SRAP) markers in Vicia faba L. Int. J. Mol. Sci. 2012;13:16457–16471. doi: 10.3390/ijms131216457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali E.M., Tohidfar M., Karimi M. Determination of genetic uniformity in transgenic cotton plants using DNA markers (RAPD and ISSR) and SDS-PAGE. J. Plant Mol. Breed. 2015;3(2):36–43. [Google Scholar]
- Ali F., Yılmaz A., Nadeem M.A., Habyarimana E., Subaşı I., Nawaz M.A., Chaudhary H.J., Shahid M.Q., Ercişli S., Zia M., Chung G., Baloch F.S. Mobile genomic element diversity in world collection of safflower (Carthamus tinctorius L.) panel using İPBS retrotransposon markers. PloS One. 2019;14(2) doi: 10.1371/journal.pone.0211985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwala S., Kimbeng C.A., Williams W., Kang M.S. Molecular markers associated with resistance to Aspergillus flavus in maize grain: QTL and discriminant analyses. J. New Seeds. 2008;9:1–18. [Google Scholar]
- Alzahib R.H., Migdadi H.M., Ghamdi A.A.A., Alwahibi M.S., Afzal M., Elharty E.H., Alghamdi S.S. Exploring genetic variability among and within hail tomato landraces based on sequence-related amplified polymorphism markers. Diversity. 2021;13:135. [Google Scholar]
- Anne C. Choosing the right molecular genetic markers for studying biodiversity: from molecular evolution to practical aspects. Genetica. 2006;127:101–120. doi: 10.1007/s10709-005-2485-1. [DOI] [PubMed] [Google Scholar]
- Arif M., Aristya G., Kasiamdari R. Genetic diversity of strawberry cultivars in banyuroto, Magelang, Indonesia based on cleaved amplified polymorphic sequence. Biodiversitas. 2019;20(6):1721–1728. [Google Scholar]
- Arystanbekkyzy M., Nadeem M.A., Akta S.H., Yeken M.Z., Zencirci N. Phylogenetic and taxonomic relationship of Turkish wild and cultivated Emmer (Triticum turgidum ssp. dicoccoides) revealed by iPBS-Retrotransposons markers. Int. J. Agric. Biol. 2018:1814–9596. [Google Scholar]
- Asif A.A., Garcia M., Tilbrook J., Brien C., Dowling K. Identification of salt tolerance QTL in wheat RIL mapping population using destructive and non- destructive phenotyping. Funct. Plant Biol. 2020;48(2):131–140. doi: 10.1071/FP20167. [DOI] [PubMed] [Google Scholar]
- Babu B.K., Mathur R.K., Kumar P.N., Ramajayam D., Ravichandran G., Venu M.V.B. Development, identification and validation of CAPS marker for SHELL trait which governs dura, pisifera and tenera fruit forms in oil palm (Elaeis guineensis Jacq.) PloS One. 2017;12(2) doi: 10.1371/journal.pone.0171933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu K.N., Sheeja T.E., Minoo D., Rajesh M.K., Samsudeen K., Suraby E.J., Kumar I.P.V. Random amplified polymorphic DNA (RAPD) and derived techniques. Methods Mol. Biol. 2021;2222:219–247. doi: 10.1007/978-1-0716-0997-2_13. [DOI] [PubMed] [Google Scholar]
- Badoni S., Das S., Sayal Y.K., Gopalakrishnan S., Singh A.K., Rao A.R.…Tyagi A.K. Genome-wide generation and use of informative intron-spanning and intron-length polymorphism markers for high-throughput genetic analysis in rice. Sci. Rep. 2016;6 doi: 10.1038/srep23765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badu-Apraku B., Garcia-Oliveira A.L., Petroli C.D. Genetic diversity and population structure of early and extra-early maturing maize germplasm adapted to sub- Saharan Africa. BMC Plant Biol. 2021;21:96. doi: 10.1186/s12870-021-02829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baghel S., Bansal Y.K. In vitro regeneration of Guizotia abyssinica Cass. And evaluation of genetic fidelity through RAPD markers. South Afr. J. Bot. 2017;109:294–307. [Google Scholar]
- Bai W.N., Wang W.T., Zhang D.Y. Contrasts between the phylogeographic patterns of chloroplast and nuclear DNA highlight a role for pollen-mediated gene flow in preventing population divergence in an East Asian temperate tree. Mol. Phylogenet. Evol. 2014;81:37–48. doi: 10.1016/j.ympev.2014.08.024. [DOI] [PubMed] [Google Scholar]
- Barth S., Melchinger A.E., Lubberstedt T.H. Genetic diversity in Arabidopsis thaliana L. Heynh. Investigated by cleaved amplified polymorphic sequence and inter-simple sequence repeat ISSR) Mol. Ecol. 2002;11(3):495–505. doi: 10.1046/j.0962-1083.2002.01466.x. [DOI] [PubMed] [Google Scholar]
- Barut M., Nadeem M.A., Karaköy T., Baloch F.S. DNA fingerprinting and genetic diversity analysis of world quinoa germplasm using İPBS-retrotransposon marker system. Turk. J. Agric. For. 2020;44(5):479–491. [Google Scholar]
- Batieno B.J., Danquah E., Tignegre J., Huynh B., Drabo I., Close T.J., Ofori K., Roberts P., Ouedraogo T.,J. Application of marker-assisted backcrossing to improve cowpea (Vigna unguiculata L. Walp) for drought tolerance. J. Plant Breed Crop Sci. 2016;8(12):273–286. [Google Scholar]
- Bhardwaj A., Dhar Y., Asif M. In Silico identification of SNP diversity in cultivated and wild tomato species: insight from molecular simulations. Sci. Rep. 2016;6:38715. doi: 10.1038/srep38715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt J., Kumar S., Patel S. Sequence-related amplified polymorphism (SRAP) markers based genetic diversity analysis of cumin genotypes. Ann. Agrar. Sci. 2017;15:434–438. [Google Scholar]
- Blears, De Grandis S.A., Lee H., Trevors J.T. Amplified fragment length polymorphism (AFLP): a review of the procedure and its applications. MJ J. Indust. Microbiol. Biotechnol. 1998;21:99–114. [Google Scholar]
- Boyd M., Panoyan M.A., Michael P. Development and characterization of species- diagnostic ISSR and SCAR DNA markers for differentiating red maple (Acer rubrum) and silver maple (A. saccharinum) Genome. 2019;62:527–535. doi: 10.1139/gen-2019-0037. [DOI] [PubMed] [Google Scholar]
- Braglia L., Gavazzi F., Morello L. On the applicability of the Tubulin-Based Polymorphism (TBP) genotyping method: a comprehensive guide illustrated through the application on different genetic resources in the legume family. Plant Methods. 2020;16:86. doi: 10.1186/s13007-020-00627-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruse S.E., Moreau M.P., Azaro M.A., Zimmerman R., Hoffman A., Linda M., Brzustowicz L.M. Improvements to bead based oligonucleotide ligation SNP genotyping assays. Biotechniques. 2008;45(5):559–571. doi: 10.2144/000112960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budak H., Shearman B., Gaussoin R., Dweikat I. Application of sequence-related amplified polymorphism markers for characterization of turfgrass species. J. Hortic. Sci. 2004;39 doi: 10.1007/s00122-003-1428-4. [DOI] [PubMed] [Google Scholar]
- Bushakra J.M., Lewers K.S., Staton M.E. Developing expressed sequence tag libraries and the discovery of simple sequence repeat markers for two species of raspberry (Rubus L.) BMC Plant Biol. 2015;15:258. doi: 10.1186/s12870-015-0629-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C., Wu S., Niu E., Cheng C., Guo W. Identification of genes related to salt stress tolerance using intron-length polymorphic markers, association mapping and virus- induced gene silencing in cotton. Sci. Rep. 2017;7(1):528. doi: 10.1038/s41598-017-00617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Rivero G., Caz&ares-S&anchez E., Tamayo-Ord&oñez M.C., Tamayo-Ord&oñez Y.J., Padilla-Ram&irez J.S., Quiroz-Moreno A., S&anchez-Teyer L.F. Application of sequence specific amplified polymorphism (SSAP) and simple sequence repeat (SSR) markers for variability and molecular assisted selection (MAS) studies of the Mexican guava. Afr. J. Agric. Res. 2017;12(29):2372–2387. [Google Scholar]
- Cao X., Wu Z., Zhou R. A novel random amplified polymorphic DNA-based strategy for genetic diversity analysis and identification of tomatoes. Genet. Mol. Res. 2015;14(1):1650–1661. doi: 10.4238/2015.March.6.11. [DOI] [PubMed] [Google Scholar]
- Cardona C.C.C., Morillo Y., Conronado A.C.M., Ochoa I. Genetic diversity in oil palm (Elaeis guineensis Jacq) using RAM (random amplified microsatellites) Bragantia. 2018;77(4):546–556. Epub. 18, 2018. [Google Scholar]
- Cheng J.L., Li J., Qi Y.M. Development of novel SCAR markers for genetic characterization of Lonicera japonica from high GC-RAMP-PCR and DNA cloning. Genet. Mol. Res. 2016;15(10):4238. doi: 10.4238/gmr.15027737. [DOI] [PubMed] [Google Scholar]
- Cheng A., Ismail I., Osman M., Hashim H., Zainual N.S.M. Rapid and targeted introgression of fgr gene through marker-assisted backcrossing in rice (Oryza sativa L.) Genome. 2017;60(12) doi: 10.1139/gen-2017-0100. [DOI] [PubMed] [Google Scholar]
- Cho K.H., Noh J.H., Park S.J. Development of sequence characterized amplified region markers for the identification of grapevine cultivars. Hortic. Sci. 2015;50(12):1744–1750. [Google Scholar]
- Collard B.C.Y., Jahufer M.Z.Z., Brouwer J.B. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica. 2005;142:169–196. [Google Scholar]
- Dong Y.H., Yao H.H., Lin Z.H., Zhang L.L. Characterization of 62 polymorphic EST-SSR markers in the blood clam (Tegillarca granosa) and their cross-amplification in Scapharca subcrenata. Conserv. Genet. Resour. 2012;4:991–997. [Google Scholar]
- Drew B.T., Ruhfel B.R., Smith S.A., Moore M.J., Briggs B.G. Another look at the root of the angiosperms reveals a familiar tale. Syst. Biol. 2014;63:368–382. doi: 10.1093/sysbio/syt108. [DOI] [PubMed] [Google Scholar]
- El Dessoky D.S., Attia A.O., Ismail I.A., Alotaibi S.S., Aljuaid B.S. Molecular assessment of genetic stability using CDDP and DNA-barcoding assays in long-term micropropagated rose plant. Pakistan J. Biol. Sci. 2020;23:1176–1183. doi: 10.3923/pjbs.2020.1176.1183. [DOI] [PubMed] [Google Scholar]
- El zayat M.A.S., Hassan A.H., Nishawy E. Patterns of genetic structure and evidence of Egyptian Citrus rootstock based on informative SSR, LTR-IRAP and LTR-REMAP molecular markers. J. Genet. Eng. Biotechnol. 2021;19:29. doi: 10.1186/s43141-021-00128-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esan V.I., Omilani O.O., Olajide A.A., Adebayo R., Sangoyomi T.E. Genetic diversity, advance, heritability and heterosis estimation in F1 hybrids and parental lines of maize. Int. J. Plant Breed. Genet. 2021;15:1–6. [Google Scholar]
- Ewart K.M., Johnson R.N., Ogden R., Joseph L., Frankham G.J., Lo N. Museum specimens provide reliable SNP data for population genomic analysis of a widely distributed but threatened cockatoo species. Mol. Ecol. Resour. 2019;19(6):1578–1592. doi: 10.1111/1755-0998.13082. [DOI] [PubMed] [Google Scholar]
- Fabriki-Ourang S., Yousefi-Azarkhanian M. Genetic variability and relationships among Salvia ecotypes/species revealed by TRAP-CoRAP markers. Biotechnol. Biotechnol. Equip. 2018;32(6):1486–1495. [Google Scholar]
- Feng J., Jiang Y., Yang Z., Chen S., El-Kassaby Y.A., Chen H. Marker-assisted selection in hybrid population. Silvae Genet. 2020;69(1):63–72. [Google Scholar]
- Feng J., Zhang X., Zhang M. Physical mapping and InDel marker development for the restorer gene Rf2 in cytoplasmic male sterile CMS-D8 cotton. BMC Genom. 2021;22:24. doi: 10.1186/s12864-020-07342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavia M., Tommaso G., Marilena C., Andrea C., Lucia N. Genome-wide analysis of LTR- retrotransposon diversity and its impact on the evolution of the genus Helianthus (L.) BMC Genom. 2017;18:634. doi: 10.1186/s12864-017-4050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusari C.M., Lia V.V., Hopp H.E. Identification of Single Nucleotide Polymorphisms and analysis of Linkage Disequilibrium in sunflower elite inbred lines using the candidate gene approach. BMC Plant Biol. 2008;8:7. doi: 10.1186/1471-2229-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Liang H., Huang J. Development of the PARMS marker of the TAC1 gene and its utilization in rice plant architecture breeding. Euphytica. 2021;217:49. [Google Scholar]
- Ge D., Daizhen Z. Application of sequence-related amplified polymorphism to genetic diversity analysis in Limonium sinense. J. Genet. 2015;94:35–38. doi: 10.1007/s12041-015-0524-y. [DOI] [PubMed] [Google Scholar]
- Geng X., Qu Y., Jia Y. Assessment of heterosis based on parental genetic distance estimated with SSR and SNP markers in upland cotton (Gossypium hirsutum L.) BMC Genom. 2021;22:123. doi: 10.1186/s12864-021-07431-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghonaim M., Kalendar R., Barakat H. High-throughput retrotransposon-based genetic diversity of maize germplasm assessment and analysis. Mol. Biol. Rep. 2020;47:1589–1603. doi: 10.1007/s11033-020-05246-4. [DOI] [PubMed] [Google Scholar]
- Gui Y., Yan G., Bo S., Tong Z., Wang Y. iSNAP: a small RNA-based molecular marker technique. Plant Breed. 2011 [Google Scholar]
- Guo X.H. Genetic diversity analysis of okra (Abelmoschus esculentus L.) by inter-simple sequence repeat (ISSR) markers. Genet. Mol. Res. 2014;13(2):3165–3175. doi: 10.4238/2014.April.25.1. [DOI] [PubMed] [Google Scholar]
- Gupta A.K., Kang B., Roy J., Rajora Om. Large scale development of selectively amplified microsatellite polymorphic loci (SAMPL) markers in spruce (Picea) Mol. Ecol. Notes. 2005;5(3):481–483. [Google Scholar]
- Guzmán M.J.L.F., Rodríguez L.F.G.P.G., Mariscal, Suárez P.A.P., Santos M.O. Identifying hybrids of Citrus aurantifolia × Citrus limon using simple sequence repeats (SSR) markers. Rev. Mex. Cienc. Agríc. 2017;8(6) [Google Scholar]
- Ha W.-Y., Yau F.C.-F., But P.P.-H., Wang J., Shaw P.-C. Direct amplification of length polymorphism analysis differentiates Panax ginseng from P. quinquefolius. Planta Med. 2001;67(6):587–589. doi: 10.1055/s-2001-16483. [DOI] [PubMed] [Google Scholar]
- Haasl R., Payseur B. Multi-locus inference of population structure: a comparison between single nucleotide polymorphisms and microsatellites. Heredity. 2011;106:158–171. doi: 10.1038/hdy.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailu G., Asfere Y. The role of molecular markers in crop improvement and plant breeding programs: a review. Agric. J. 2020;15(6):171–175. [Google Scholar]
- Hou B., Li W., Gao X., Zhang X., Feng S., Wu Y. Construction and integration of genetic map and quantitative trait locus (QTL) mapping of yield-related traits, Acta Agriculturae Scandinavica. Sect. B Soil Plant Sci. 2021 [Google Scholar]
- Huang X., Yang L., Jin Y., Lin J. Generation, Annotation, and Analysis of a Large-Scale Expressed Sequence Tag Library from Arabidopsis pumila to Explore Salt-Responsive Genes. Front. Plant Sci. 2017;8(955) doi: 10.3389/fpls.2017.00955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IAEA (International Atomic Energy Agency), Vienna . A Manual; 2002. Mutant Germplasm Characterization Using Molecular Markers. IAEA-TCS-19 ISSN 1018–5518. [Google Scholar]
- Igwe D.O., Ihearahu O.C., Osano A.A. Genetic diversity and population assessment of Musa L. (Musaceae) employing CDDP markers. Plant Mol. Biol. Rep. 2021 [Google Scholar]
- Jegadeeswaran M., Kadirvel P., Srinivas P.S., Senthilvel S., Selvaraj V.M. Genetic mapping reveals a major QTL associated with tolerance to the aphid, Uroleucon compositae (Theobald) in safflower (Carthamus tinctorius) Plant Breed. 2021 [Google Scholar]
- Jha U., Jha R., Bohra A., Manjunatha L., Saabale P., Parida S.…Singh N. Plant Genetic Resources: Characterization and Utilization. 2021. Association mapping of genomic loci linked with Fusarium wilt resistance (Foc2) in chickpea; pp. 1–8. [Google Scholar]
- Jiang G.L. In: Plant Breeding from Laboratories to Fields. Andersen S.B., editor. IntechOpen; Rijeka, Croatia: 2013. Molecular markers and marker-assisted breeding in plants; pp. 45–83. [Google Scholar]
- Junior C.A.D.K., Manechini J.R.V., Corrêa R.X. Genetic structure analysis in sugarcane (saccharum spp.) using target region amplification polymorphism (TRAP) markers based on sugar- and lignin-related genes and potential application in core collection development. Sugar Tech. 2020;22:641–654. [Google Scholar]
- Kahl G. Wiley VCH; Weinheim: 2001. The Dictionary of Gene Technology. [Google Scholar]
- Kalendar R., Schulman A.H. In: Molecular Plant Taxonomy: Methods and Protocols, Methods in Molecular Biology. Pascale Besse., editor. Vol. 1115. Humana Press; 2014. Transposon based tagging: IRAP, REMAP, and iPBS; pp. 233–255. [DOI] [PubMed] [Google Scholar]
- Kalendar R., Antonius K., Smýkal P., Schulman A.H. İPBS: a universal method for DNA fingerprinting and retrotransposon isolation. Theor. Appl. Genet. 2010;121(8):1419–1430. doi: 10.1007/s00122-010-1398-2. [DOI] [PubMed] [Google Scholar]
- Kalendar R., Aizharkyn K.S., Khapilina O.N., Amenov A.A., Tagimanova D.S. Plant diversity and transcriptional variability assessed by retrotransposon-based molecular markers. Russ. J. Genet.: Appl. Res. 2017;21(1):128–134. [Google Scholar]
- Kalendar R., Amenov A., Daniyarov A. Use of retrotransposon-derived genetic markers to analyse genomic variability in plants. Funct. Plant Biol. 2018;46(1):15–29. doi: 10.1071/FP18098. [DOI] [PubMed] [Google Scholar]
- Kalia R.K., Rai M.K., Kalia S., Singh R., Dhawan A.K. Microsatellite markers: an overview of the recent progress in plants. Euphytica. 2011;177:309–334. [Google Scholar]
- Kantar F., Yemşen S.N., Bülbül C., Yılmaz N., Mutlu N. Phenotypic and iPBS- retrotransposon marker diversity in okra (Abelmoschus esculentus (L.) Moench) germplasm. Biotech Stud. 2021;30(1):7–15. [Google Scholar]
- Khidr Y.A., Mekuriaw S.A., Hegazy A.E. Suitability of target region amplified polymorphism (TRAP) markers to discern genetic variability in sweet sorghum. J. Genet. Eng. Biotechnol. 2020;18:59. doi: 10.1186/s43141-020-00071-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodaee L., Azizinezhad R., Etminan A.R., Khosroshahi M. Assessment of genetic diversity among Iranian Aegilops triuncialis accessions using ISSR, SCoT, and CBDP markers. J. Genet. Eng. Biotechnol. 2021;19:5. doi: 10.1186/s43141-020-00107-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Lei P., Wang A., Liu S., Zhao Y., Huang F., Yu Z., Zhu G., He Z., Tan D. Vol. 11. 2021. Genetic diversity of Castor bean (Ricinus communis L.) revealed by ISSR and RAPD markers; p. 457. [Google Scholar]
- Kim H.R., Sa K.J., Nam-Gung M., Park K.J., Ryu S.-H. Genetic characterization and association mapping in near-isogenic lines of waxy maize using seed characteristics and SSR markers. Genes Genom. 2021;43(1):79–90. doi: 10.1007/s13258-020-01030-7. [DOI] [PubMed] [Google Scholar]
- Koelling J., Coles M.C., Matthews P.D., Schwekendiek A. Development of new microsatellite markers (SSRs) for Humulus lupulus. Mol. Breed. 2012;30:479–484. [Google Scholar]
- Konzen E.R., Peron R., Ito M.A. Molecular identification of bamboo genera and species based on RAPD-RFLP markers. Silva Fenn. 2017;51(4):1691. [Google Scholar]
- Kordrostami M., Rahimi M. Paper Presented at Conference on Genetics in the Third Millennium. Vol. 13. 2015. Molecular markers in plants: concepts and applications; pp. 4024–4031. [Google Scholar]
- Kumar S.P.J., Susmita C., Agarwal D.K. Assessment of genetic purity in rice using polymorphic SSR markers and its economic analysis with grow-out-test. Food Anal. Meth. 2021;14:856–864. [Google Scholar]
- Leviòa B., Rostoks N., Syed N.H., Flavell A.J., Levins G. Genetic diversity and structure of northern populations of the declining coastal Planteryngium maritimum. Proc. Latv. Acad. Sci. Sect. A B. 2019;73(5):446–454. (722) [Google Scholar]
- Li G., Quiros C. Sequence-related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: its application to mapping and gene tagging in Brassica. Theor. Appl. Genet. 2001;103:455–461. [Google Scholar]
- Li C., Zheng Y., Huang P. Molecular markers from the chloroplast genome of rose provide a complementary tool for variety discrimination and profiling. Sci. Rep. 2020;10:12188. doi: 10.1038/s41598-020-68092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Pan Z., He S. QTL mapping of agronomic and economic traits for four F2 populations of upland cotton. J. Cotton Res. 2021;4:3. [Google Scholar]
- Lincoln M., Sunil K.V., Saugato S., Jawaharlal K. Potential applications of molecular markers in plant. Curr. Trends Biomed. Eng. Biosci. 2018;12(4):555844. [Google Scholar]
- Liu X., Du J., Asaduzzaman Khan A., Cheng J., Wei C. Analysis of genetic diversity and similarities between different Lycium varieties based on ISSR analysis and RAMP-PCR markers. World Acad. Sci. J. 2020;2:83–90. [Google Scholar]
- Long Y.M., Chao W.S., Ma G.J. An innovative SNP genotyping method adapting to multiple platforms and throughputs. Theor. Appl. Genet. 2017;130:597–607. doi: 10.1007/s00122-016-2838-4. [DOI] [PubMed] [Google Scholar]
- López-Malvar A., Butron A., Malvar R.A. Association mapping for maize stover yield and saccharification efficiency using a multiparent advanced generation intercross (MAGIC) population. Sci. Rep. 2021;11:3425. doi: 10.1038/s41598-021-83107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Hou J., Ouyang Y., Luo H., Zhao J. A direct PCR–based SNP marker– assisted selection system (D-MAS) for different crops. Mol. Breed. 2020;40(1):9. [Google Scholar]
- Manivannan A., Choi S., Jun T.-H., Yang E.-Y., Kim J.-H. Genotyping by sequencing-based discovery of SNP markers and construction of linkage map from F5 population of pepper with contrasting powdery mildew resistance trait. BioMed Res. Int. 2021;2021:8. doi: 10.1155/2021/6673010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar S.R., Hoque H., Sinha B., Chowdhury W.R., Hasan M.N. Genetic variability analysis of partially salt tolerant local and inbred rice (Oryza sativa L.) through molecular markers. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e04333. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Z., Zhang C., Khan A.M. Angelica Sinensis (Oliv.) Diels Varieties of Metcalfe. 2015. Efficiency of improved RAPD and ISSR markers in assessing genetic diversity and relationships. [Google Scholar]
- Metcalfe C.J., Oliveira S.G., Gaiarsa J.W., Aitken K.S., Monalisa S., Carneiro M.S. Using quantitative PCR with retrotransposon-based insertion polymorphisms as markers in sugarcane. J. Exp. Bot. 2015;66(14):4239–4250. doi: 10.1093/jxb/erv283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazen F., Rastegari A., Hoseini S.M., Panjehpour M., Miroliaei M. Optimization of Taq DNA polymerase enzyme expression in Escherichia coli. Adv. Biomed. Res. 2012;1:82. doi: 10.4103/2277-9175.103004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty S., Joshi R.S., Subudhi E. Genetic stability assessment of micro propagated mango ginger (Curcuma amada Roxb.) through RAPD and ISSR markers. Res J Med Plants. 6, 529–536. Electron. J. Biotechnol. 2012;18(2):96–102. [Google Scholar]
- Mokate Y.S., Chimote V.P., Deshmukh M.P., Gawai K., Kale A.A., Jadhav A.S. Use of promoter-anchored RAPD analysis for divergence assessment in soybean as against conventionally used ISSR assay. Appl. Biol. Res. 2017;19:1–9. [Google Scholar]
- Moore M.J., Soltis P.S., Bell C.D., Burleigh J.G., Soltis D.E. Proceedings of the National Academy of Sciences, USA. Vol. 107. 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots; pp. 4623–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M., Vogel J. Compound microsatellite primers for the detection of genetic polymorphisms. US Patent Application. 1994;08/326456 [Google Scholar]
- Mullis K.F., Faloona F., Scharf S., Saiki R., Horn G., Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harbor Symp. Quant. Biol. 1986;51:263–273. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- Nadeem M.A., Nawaz M.A., Shahid M.Q. DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2018;32(2):261–285. [Google Scholar]
- Nie Y., Ji W., Ma S. Assessment of heterosis based on genetic distance estimated using SNP in common wheat. Agronomy. 2019;9(2):66. [Google Scholar]
- Olasanmi B., Kyallo M., Yao N. Marker-assisted selection complements phenotypic screening at seedling stage to identify cassava mosaic disease-resistant genotypes in African cassava populations. Sci. Rep. 2021;11:2850. doi: 10.1038/s41598-021-82360-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olatunji T.L., Afalayan A.J. Evaluation of genetic relationship among varieties of Capsicum annuum L. and Capsicum frutescens L. in West Africa using ISSR markers. Heliyon. 2019;5 doi: 10.1016/j.heliyon.2019.e01700. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M., Hood L., Cantor C.H., Botstein D. A common language for physical mapping of the human genome. Science. 1989;24:1434–1435. doi: 10.1126/science.2781285. [DOI] [PubMed] [Google Scholar]
- Onley I.R., Austin J.J., Mitchell K.J. Sex assignment in a non-model organism in the absence of field records using Diversity Arrays Technology (DArT) data. Conserv. Genet. Res. 2021 [Google Scholar]
- Padmakar B., Sailaja D., Aswath C. Molecular exploration of guava (Psidium guajava L.) genome using SSR and RAPD markers: a step towards establishing linkage map. J. Hortic. Sci. 2015;10(2):130–135. [Google Scholar]
- Pantchev I., Sibel A., Sarsu F., Tomlekova N. Applicability of ISAP, ISSR and SSR markers in tomato breeding programs. Veg. Crops Russia. 2019:24–26. [Google Scholar]
- Paran I., Michelmore R.W. Development of reliable PCR-based markers linked to downy mildew resistance genes in lettuce. Theor. Appl. Genet. 1993;85:985–993. doi: 10.1007/BF00215038. [DOI] [PubMed] [Google Scholar]
- Pavani M., Sundaram R.M., Ramesha M.S. Prediction of heterosis in rice based on divergence of morphological and molecular markers. J. Genet. 2018;97:1263–1279. [PubMed] [Google Scholar]
- Poczai P., Varga I., Laos M. Advances in plant gene-targeted and functional markers: a review. Plant Methods. 2013;9:6. doi: 10.1186/1746-4811-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkrishnan B.V., Arunachalam V. Silico RAPD priming sites in expressed sequences and iSCAR markers for oil palm. Int. J. Genom. 2012;2012:5. doi: 10.1155/2012/913709. Article ID 913709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins R., Groenewald J.Z., Marais G.F., Snape J., Koebner R.M.D. Vol. 103. 2001. AFLP and STS tagging of Lr19, a gene conferring resistance to leaf rust in wheat Theoretical and Applied Genetics; pp. 618–624. (4) [Google Scholar]
- Purayil F.T., Robert G.A., Gothandam K.M. Genetic variability in selected date palm (Phoenix dactylifera L.) cultivars of United Arab Emirates using ISSR and DAMD markers. 3. Biotech. 2018;8:109. doi: 10.1007/s13205-018-1108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quezada M., Amadeu R.R., Vignale B., Cabrera D., Pritsch C., Garcia A.A.F. Construction of a high-density genetic map of acca sellowiana (berg.) burret, an outcrossing species, based on two connected mapping populations. Front. Plant Sci. 2021 doi: 10.3389/fpls.2021.626811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rádi F., Török K., Nagymihály M. Improved reliability in production of maize inbred lines by the combination of the R1-Navajo marker with flow cytometry or microsatellite genotyping. Cereal Res. Commun. 2020;48:423–430. [Google Scholar]
- Rajesh M.K., Jerard B.A., Preethi P. Application of RAPD markers in hybrid verification in coconut. Crop Breed. Appl. Biotechnol. 2014;14(1):36–41. [Google Scholar]
- Rajewska M., Wegrzyn K., Konieczny I. AT-rich region and repeated sequences – the essential elements of replication origins of bacterial replicons. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2012;2(36):408–434. doi: 10.1111/j.1574-6976.2011.00300.x. [DOI] [PubMed] [Google Scholar]
- Rana M.K., Munjal V.D., Sonika S., Bhat K.V. Utility of P450-based-analogue (PBAs) markers for diversity analysis of linseed (Linum usitatissimum L.) cultivars. Indian J. Plant Genet. Res. 2005;18(2):194–199. [Google Scholar]
- Ravi R.S.D., Siril E.A., Nair B.R. The efficiency of Cytochrome P450 gene-based markers in accessing genetic variability of drumstick (Moringa oleifera Lam.) accessions. Mol. Biol. Rep. 2020;47(4):2929–2939. doi: 10.1007/s11033-020-05391-w. [DOI] [PubMed] [Google Scholar]
- Rawandoozi Z.J., Hartmann T.P., Carpenedo S. Mapping and characterization QTLs for phenological traits in seven pedigree-connected peach families. BMC Genom. 2021;22:187. doi: 10.1186/s12864-021-07483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy B.V., Reddy C.V.C.M., Sekhar A.C., Reddy P.C.O., Rajasekhar P., Srinivasulu K. Role of molecular based markers methods and their applications in crop improvement. Plant Cell Biotechnol. Mol. Biol. 2021;22(23-24):38–54. [Google Scholar]
- Reiche B., Koegler A., Morgenstern K., Brueckner M., Weber B. Application of retrotransposon-based Inter-SINE Amplified Polymorphism (ISAP) markers for the differentiation of common poplar genotypes. Can. J. For. Res. 2021 [Google Scholar]
- Reyes P.A., Ochoa J.C., Montoya C., Daza E., Ayala I.M., Romero H.M. Development and validation of a bi-directional allele-specific PCR tool for differentiation in nurseries of dura, tenera and pisifera oil palms. Agron. Colomb. 2015;33(1):5–10. [Google Scholar]
- Salazar J.A., Rasouli M., Martínez-Gómez P. Random amplified microsatellite polymorphism (RAMP) application in prunus characterization and mapping. Acta Hortic. 2014;1028:61–64. [Google Scholar]
- Sansaloni C., Petroli C., Jaccoud D., Carling J., Detering F., Grattapaglia D., Kilian A. Diversity arrays technology (DArT) and next-generation sequencing combined: genome- wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus. BMC Proc. 2011;5(7):1–2. [Google Scholar]
- Sarwat M., Srivastava S., Khan T.H. RAPD and ISSR polymorphism in the medicinal plants: Ocimum sanctum, O basilicum and O gratissimum. IJPPR. 2016;8(8):1417–1424. [Google Scholar]
- Schulman A., Flavell A., Paux E., Noel Ellis T.H. The application of LTR retrotransposons as molecular markers in plants. Methods Mol. Biol. 2012;859:115–153. doi: 10.1007/978-1-61779-603-6_7. [DOI] [PubMed] [Google Scholar]
- Selvakumari E., Jenifer J., Priyadharshini S. Application of DNA fingerprinting for plant identification. J. Acad. Ind. Res. 2017;5:10. [Google Scholar]
- Sharafi A., Abkenar A., Sharafi A. Molecular genetic diversity assessment of citrus species grown in Iran revealed by SSR, ISSR and CAPS molecular markers. J. Sci. Res. 2017;2(22):22–27. [Google Scholar]
- Shaw J., Lickey E.B., Schilling E.E., Small R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am. J. Bot. 2007;94(3):275–288. doi: 10.3732/ajb.94.3.275. [DOI] [PubMed] [Google Scholar]
- Singh R.B., Mahenderakar M.D., Jugran A.K., Singh R.K., Srivastava R.K. Assessing genetic diversity and population structure of sugarcane cultivars, progenitor species and genera using microsatellite (SSR) markers. Gene. 2020;753:144800. doi: 10.1016/j.gene.2020.144800. [DOI] [PubMed] [Google Scholar]
- Sorkheh K., Shiran B., Aranzana M.J., Mohammadi S.A., Martínez-Gomez P. Application of amplified fragment length polymorphism (AFLPs) analysis to plant breeding and genetics: procedures, applications and prospects. J. Food Agric. Environ. 2007;5(1):197–204. [Google Scholar]
- Sormin S.Y.M., Purwantoro A., Setiawan A.G., Teo C.H. Vol. 22. 2021. Application of inter-SINE amplified polymorphism (ISAP) markers for genotyping of Cucumis melo accessions and its transferability in Coleus spp. p. 5. [Google Scholar]
- Southern E.M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sreenivasa V., Lal S., Kiran Babu P., Mahadeva Swamy H., Yadav R., Talukdar A., Rathod D. Inheritance and mapping of drought tolerance in soybean at seedling stage using bulked segregant analysis. Plant Genet. Resour. Charact. Util. 2020;18(2):63–70. [Google Scholar]
- Thakur J., Dwivedi M.D., Sourabh P. Genetic homogeneity revealed using SCoT, ISSR and RAPD markers in micro propagated Pittosporum eriocarpum Royle- an endemic and endangered medicinal plant. PloS One. 2016;11(7) doi: 10.1371/journal.pone.0159050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J., Dwivedi M.D., Singh N., Uniyal P.L., Goel S., Pandey A.K. Applicability of start Codon targeted (SCoT) and inter simple sequence repeat (ISSR) markers in assessing genetic diversity in Crepidium acuminatum (D. Don) szlach. J. Appl. Res. Med. Arom. Plant. 2021;23:100310. [Google Scholar]
- Tilwari A., Sharma R. Random amplified polymorphic DNA and inter simple sequence repeat markers reveals genetic diversity between micro propagated, wild and field cultivated genotypes of Gloriosa superba: an endangered medicinal plant. Mol. Biol. Rep. 2021;48:2437–2452. doi: 10.1007/s11033-021-06278-0. [DOI] [PubMed] [Google Scholar]
- Tomkowiak A., Bocianowski J., Kwiatek M., Kowalczewski P.L. De Gruyter; 2020. Dependence of the Heterosis Effect on Genetic Distance, Determined Using Various Molecular Markers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deventer R., Rhode C., Marx M., Roodt-Wilding R. The development of genome- wide single nucleotide polymorphisms in blue wildebeest using the DArTseq platform. Genomics. 2020;112(5):3455–3464. doi: 10.1016/j.ygeno.2020.04.032. [DOI] [PubMed] [Google Scholar]
- Vekariya S., Taviad K., Acharya R.N. Development of random amplified polymorphic DNA markers for authentication of Croton tiglium Linn. J. Phytopharmacol. 2017;6(3):164–166. [Google Scholar]
- Vos P., Hogers R., Bleeker M., Reijans M., Tvandev Lee T. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yu A., Li F. Bulked segregant analysis reveals candidate genes responsible for dwarf formation in woody oilseed crop castor bean. Sci. Rep. 2021;11:6277. doi: 10.1038/s41598-021-85644-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanga G., Guoa Y., Zhaoa Y. Construction of a molecular genetic map for hawthorn based on SRAP markers. Biotechnol. Biotechnol. Equip. 2015;29(3):441–447. [Google Scholar]
- Wolff K., Rijn J.P.-V., Hofstra H. RFLP analysis in chrysanthemum. I. Probe and primer development. Theor. Appl. Genet. 1994;88(3-4):472–478. doi: 10.1007/BF00223663. [DOI] [PubMed] [Google Scholar]
- Wu J. Vol. 56. Anhui Agric. Univ.; 2017. Construction and Application of Identification and Analysis Process of Full-Length LTR-Retrotransposons (D) [Google Scholar]
- Xia Z., Tsubokura Y., Hoshi M., Hanawa M., Yano C. An integrated high-density linkage map of soybean with RFLP, SSR, STS, and AFLP markers using a single F2 population. DNA Res. 2007;14(6):257–269. doi: 10.1093/dnares/dsm027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao N., Wang H., Yao W. Development and evaluation of SSR markers based on large scale full-length transcriptome sequencing in sugarcane. Trop. Plant Biol. 2020;13:343–352. [Google Scholar]
- Yadav S., Shah V., Mod B. Genetic diversity analysis between different varieties of chickpea (Cicer arietinum L.) using SSR markers. Int. J. Appl. Sci. Biotechnol. 2019;7(2):236–242. [Google Scholar]
- Yaldiz G., Camlica M., Nadeem M.A., Nawaz M.A., Baloch F.S. Genetic diversity assessment in Nicotiana tabacum L. with iPBS-retrotransposons. Turk. J. Agric. For. 2018;42(3):154–164. [Google Scholar]
- Yan Z.Z., Yan R., Fan W., Kai L., Zhang D.Y., Qi Y., Zhang Y.F., Zhao Y.F., Zhang J.Y. The development and screening of EST-SSR markers in Melilotus albus. Pratacult. Sci. 2017 [Google Scholar]
- Yang L., Khan M.A., Mei Z. Development of RAPD-SCAR markers for Lonicera japonica (Caprifoliaceae) variety authentication by improved RAPD and DNA cloning. Rev. Biol. Trop. 2014;62(4):1649–1657. doi: 10.15517/rbt.v62i4.13493. [DOI] [PubMed] [Google Scholar]
- Yang L., Lei L., Li P., Wang J., Wang C. Identification of candidate genes conferring cold tolerance to rice (Oryza sativa L.) at the bud-bursting stage using bulk segregant analysis sequencing and linkage mapping. Front. Plant Sci. 2021 doi: 10.3389/fpls.2021.647239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S., Dhillon S., Ke X., Collins A.R., Day I.N. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001;29(17):E88. doi: 10.1093/nar/29.17.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C.Y., Zhang C., Wang P., Hu S., Chang H.P. In vitro propagation of chia (Salvia hispanica L.) and assessment of fidelity using random amplified polymorphic DNA and inter simple sequence repeat molecular markers. J. Appl. Biol. Biotechnol. 2019;7(1):42–47. [Google Scholar]
- Zakiyah N., Handoyo T., Kim K.M. Genetic diversity analysis of Indonesian aromatic rice varieties (Oryza sativa L.) using RAPD. J. Crop Sci. Biotechnol. 2019;22:55–63. [Google Scholar]
- Zhang W., Yin L., Shasha Wei S., Deng Z., Yi J.R., Wu, Qin C.H.E.N. RAPD marker conversion into a SCAR marker for rapid identification of Johnsongrass [Sorghum halepense (L.) pers.] Not. Bot. Horti Agrobot. Cluj-Napoca. 2013;41:306–312. [Google Scholar]
- Zhang W., Tan L., Sun H., Zhao X., Liu F. Natural variations at TIG1 encoding a TCP transcription factor contribute to plant architecture domestication in rice. Mol. Plant. 2019;12(8):1075–1089. doi: 10.1016/j.molp.2019.04.005. [DOI] [PubMed] [Google Scholar]
- Zhao X., Zhang J., Zhang Z. Hybrid identification and genetic variation of Elymus sibiricus hybrid populations using EST-SSR markers. Hereditas. 2017;154:15. doi: 10.1186/s41065-017-0053-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Niu E., Shi A., Mou B. Genetic diversity analysis of olive germplasm (Olea europaea L.) with genotyping-by-sequencing technology. Front. Genet. 2019;10:755. doi: 10.3389/fgene.2019.00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.