Abstract
Female breast cancer has become the most commonly occurring cancer worldwide. Although it has a good prognosis under early diagnosis and appropriate treatment, breast cancer metastasis drastically causes mortality. The process of metastasis, which includes cell epithelial–mesenchymal transition, invasion, migration, and colonization, is a multistep cascade of molecular events directed by gene mutations and altered protein expressions. Ubiquitin modification of proteins plays a common role in most of the biological processes. E3 ubiquitin ligase, the key regulator of protein ubiquitination, determines the fate of ubiquitinated proteins. E3 ubiquitin ligases target a broad spectrum of substrates. The aberrant functions of many E3 ubiquitin ligases can affect the biological behavior of cancer cells, including breast cancer metastasis. In this review, we provide an overview of these ligases, summarize the metastatic processes in which E3s are involved, and comprehensively describe the roles of E3 ubiquitin ligases. Furthermore, we classified E3 ubiquitin ligases based on their structure and analyzed them with the survival of breast cancer patients. Finally, we consider how our knowledge can be used for E3s’ potency in the therapeutic intervention or prognostic assessment of metastatic breast cancer.
Keywords: ubiquitination, E3 ligase, breast cancer, metastasis, systematic review
1 Introduction
According to the latest global cancer statistics by the International Agency for Research on Cancer, female breast cancer has overtaken lung cancer and has become the most commonly occurring cancer worldwide, which accounts for about 11.7% of all new cancer cases (1). Breast cancers are highly heterogeneous, and they are classified into subtypes as Luminal A, Luminal B, HER2 (human epithelial growth factor receptor 2) positive, and basal-like (triple negative breast cancer, TNBC), with respect to the presence or absence of hormone receptors such as estrogen receptor (ER) and progesterone receptor (PR), and one oncogenic biomarker, HER2. These molecular subtypes help determine which patients are likely to respond to targeted therapies (2). Despite having good prognosis with early diagnosis and appropriate treatment, breast cancer is still the leading cause of mortality among women (3). Breast cancer–leading deaths are mostly attributed to metastasis. As mammary epithelial cells which acquire deregulated proliferation, if these malignant cells remain contained within the ducts or lobules of breast, the patients’ survival has been reported to be nearly ~98% within 5 years. In contrast, the patients’ 5-year survival rate with distant metastases at the time of diagnosis decreased to only 23% (2). Furthermore, approximately one-third of female breast cancer patients with no lymph node involvement at the time of diagnosis will develop distal metastasis (4).
1.1 The Multistep Cascade of Breast Cancer Cell Alternations in Metastasis
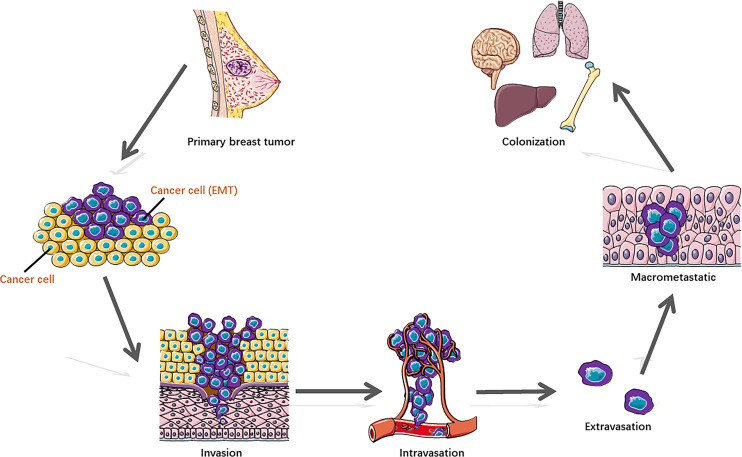
Some breast cancer cells acquire metastatic potential in a very early stage, which gains the capability to spread from the breast tissue, enter the blood or lymphatic vessels, and disseminate to distal organs, preferentially to the lung, liver, brain, and bone. Metastasis represents the multistep cascade of cancer cell alterations accompanied by structural and functional changes. It is well recognized that distant metastasis colonization consists of sequential steps ( Figure 1 ) (2, 5), including cell detachment from the primary tumor site involving epithelial–mesenchymal transition (EMT) (6); migration and invasion into surrounding tissue; penetration of the basal membrane (trans-endothelial intravasation) into the vasculature of blood and/or lymphatic vessels to be circulating tumor cells (CTCs); extravasation of CTCs to secondary sites as disseminated tumor cells (DTCs) (7); dissemination to distant organs; and formation of a micro-metastatic niche and construction of macrometastases. In addition, to survive and initiate the secondary cancer foci, cells need the capability of evading immune defenses, delivering to distant sites, adapting to supportive niches such as angiogenesis (8), and inducting retro-differentiation to gain stemness (9).
Figure 1.
Distant metastasis colonization consists of sequential steps.
1.2 The E3 Ubiquitin Ligases
Ubiquitination is a post-translational modification of proteins, which is essential for nearly all biological processes, including cell growth, autophagy, apoptosis, and differentiation. It is a three-step enzymatic cascade: i) A ubiquitin-activating enzyme (E1) mediates the activation of the carboxyl-terminal glycine residue of ubiquitin in an ATP-dependent manner. ii) The activated ubiquitin is then transferred to E1 followed by the transfer of ubiquitin to a thiolester of a ubiquitin-conjugating enzyme (E2) to format the thiolester linkage. iii) A ubiquitin protein ligase (E3) confers substrate specificity by recognizing the target proteins and mediating the conjugation of ubiquitin molecules to a lysine residue on the targeted protein via an isopeptide bond (10). E3’s ability to specifically recognize and target substrates makes it a key regulator in the ubiquitin process.
The fate of ubiquitinated proteins is dependent on the different types of ubiquitin linkage. It has been characterized as mono-ubiquitination or poly-ubiquitinated chains. Mono-ubiquitination involves the transfer of a single ubiquitin to a substrate. E3 ligases can also connect several ubiquitin molecules together using the C-terminus of one subunit and one of the seven lysine (K) residues (K6, K11, K27, K29, K33, K48, K63) or the N-terminal methionine (M1) on the other, to form a homotypic or branched ubiquitin chain (11). The recruitment of polyubiquitinated proteins to the proteasome is a classic protein turnover pathway ( Figure 2A ). Ubiquitin-dependent proteasomal degradation involves the polyubiquitination of substrate catalyzed by E1, E2, and especially E3 (12, 13). Subsequently, polymers linked through K11 or K48 (14, 15) trigger substrate protein degradation by the ATP-dependent 26S proteasome (16).
Figure 2.
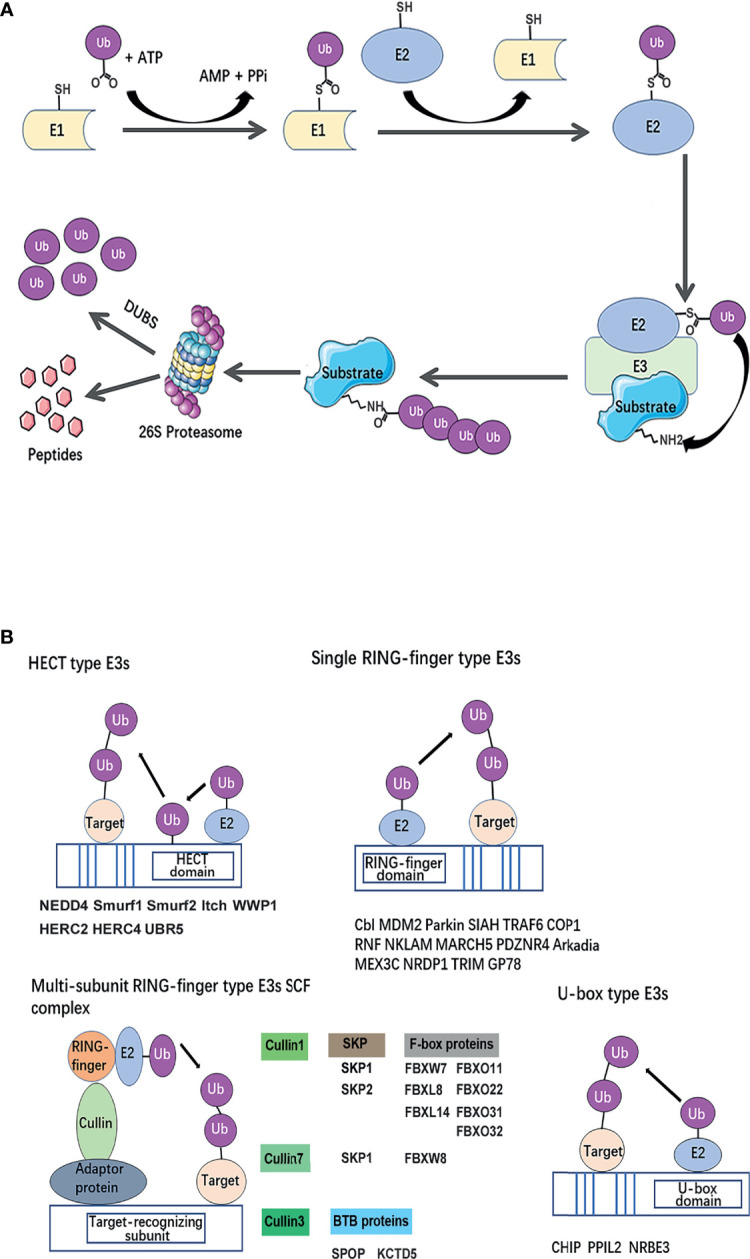
Classical mechanism and structural basis of E3 ubiquitin ligases. (A) The ubiquitin-proteasome system. A ubiquitin-activating enzyme (E1) mediates the activation of the carboxyl-terminal glycine residue of ubiquitin in an ATP-dependent manner. To format the thiolester linkage, the activated ubiquitin is then transferred to E1 followed by the transfer of ubiquitin to a thiolester of a ubiquitin-conjugating enzyme (E2). Ubiquitin protein ligase (E3) confers substrate specificity by recognizing the target proteins and mediating the conjugation of ubiquitin molecules to a lysine residue on the targeted protein via an iso-peptide bond. Polyubiquitinated substrate recognized by 26S proteasome for degradation. (B) Classification of breast cancer metastasis–related ubiquitin E3 ubiquitin ligases based on a structural basis. The canonical E3s are classified into two canonical types: HECT and RING. HECT E3s contains HECT domains which consist of a E2-interacting N-lobe and a catalytic Cys residue containing C-lobe involved in ubiquitin transfer. RING-type E3s mediate the direct transfer of ubiquitin from E2 to substrate, including single RING-finger- type E3s, RING-like (Ubox)-type E3s, and multi-subunit RING-finger-type E3s. The Cullin-RING ubiquitin ligase (CRL) family is composed of a multi-unit. Each type of E3s or members of E3 complexes has been summarized and listed below the schematic diagram of the relevant category.
Besides, different ubiquitin topologies adopt distinct structural conformations. Signals can be transduced by different types of ubiquitin modification, including mono-ubiquitination, multi-monoubiquitination, homotypic ubiquitin chains, and heterotypic ubiquitin chains, which send various ‘codes’ to precisely exert degradative and non-degradative functions including modification of protein trafficking, interaction, and signal transduction (17, 18). For example, linkages of homotypic K11 and K48 can drive proteasomal degradation; K27 linkages have been implicated in regulating DNA repair (19) and autoimmunity (20); K33 linkages were proposed to regulate trafficking; and mono-ubiquitination can prevent protein interactions (21). SMAD4 (SMAD family member 4) engages its signal partner SMAD2 (SMAD family member 2) after removing its own mono-ubiquitination (18). In addition, the mono-ubiquitination of histone H2A promotes transcriptional silencing (22), while the mono-ubiquitination of histone H2B mediates transcriptional elongation (23).
The canonical E3s are classified into two types: Homologous to the E6AP Carboxyl Terminus (HECT) family (24) and Really Interesting New Gene (RING) finger family (25). HECT E3 ligases were named accordingly to identify protein E6AP (E6-associated protein), and they are characterized by a conserved C-terminal ~350 aa HECT domain and various N-terminal substrate-binding domains (26). HECT E3 ligases mediate a two-step ubiquitin transfer process, in which ubiquitin is firstly transferred from the E2-Ub intermediate to the E3 active cysteine residue before transferring to the substrate lysine residue ( Figure 2B ) (27). In contrast, The RING core is a small domain of 40–70 residues. A cross-braced pattern of conserved cysteine and histidine residues coordinating two zinc ions maintains the native fold. The RING domain brings the E2∼Ub conjugate into the proximity of the substrate bound via substrate-recognition domains ( Figure 2B ). RING E3s are further divided into two subtypes: single RING and multi-unit RING family. The Cullin-RING ubiquitin ligase (CRL) family is composed of a multi-unit. CRLs utilize Cullin proteins as a central scaffold which binds to a RING-box protein (Rbx) and an adaptor protein–substrate receptor complex through its C- and N-termini. Most recognized CRLs are known as the SCF (Skp-Cullin-F-box) complex, in which Cullin interacts with Skp (S-phase kinase associated protein) proteins and utilizes various F-box proteins, to recruit substrates and initiate ubiquitin ligation (28). Besides, U-box E3s are also categorized as RING-type E3s, but their molecular structure subtly differs in that zinc-bound sites are replaced by a hydrophobic core (29) ( Figure 2B ).
E3s target a broad spectrum of substrates. In cancer, the aberrant functions of E3s are linked to deregulated oncoproteins or tumor suppressors and affect the biological behavior of cells. Many E3s have been reported to be associated with breast cancer metastasis over the past few decades. In this review, we provided an overview of E3 ubiquitin ligases that have been found to be deregulated in breast cancer metastasis and summarized the multistep cascade of breast cancer cell alterations in metastasis, in which these E3 ubiquitin ligases are involved. Furthermore, we classified E3s based on their structures and analyzed the correlation of E3s with the survival of breast cancer patients. Finally, we considered their potency to the therapeutic intervention or prognostic assessment of metastatic breast cancer.
2 Methods
2.1 Search Strategy and Selection Criteria
The literatures involved in this study were searched from the databases PubMed and Medline (last search updated on January 1st, 2021). The key words used in the searching were “E3 ubiquitin”, “breast cancer” and “Metastasis” or “dissemination”/“EMT”/“invasion”/“migration”/“intravasation”/“CTC”. All the searching results were imported in the Endnote software to eliminate duplicates.
By scanning the titles and abstracts and further reading the full text, the irrelevant and retracted papers were excluded. The reference list of all selected articles was scanned to identify potentially relevant reports. The search results followed these including and excluding criteria:
Including criteria: 1) research limits to the E3 ubiquitin ligases; 2) research related to human breast cancer; 3) research focused on metastasis or steps in the metastasis mechanisms of breast cancer; 4) studies provided sufficient experimental evidence or clinical data to support thesis; 5) peer-reviewed and formally published original literatures
Excluding criteria: 1) research of the regulation of E3 ubiquitin ligases; 2) research of ubiquitin-like modifiers including SUMOylation; 3) retracted articles; 4) reviews or letter to editors.
2.2 Data Extraction
The above searches were performed and reviewed by two authors independently. The following items were recorded from each study: E3 name, gene ID, substrate, role in breast cancer metastasis, cellular function, signal pathway, verification by cell biological experiments, breast cancer cell lines, and clinical significance (expression difference and survival analysis). These records were cross-checked and double- checked by another author.
2.3 Survival Analysis
The association between the specific E3s’ ubiquitin ligase expression and survival in breast cancer was first analyzed using the PrognoScan database (30) as in previously mentioned methods (31). Then, we used a Kaplan–Meier plotter (32) to validate and illustrate as a Kaplan–Meier plot, in which the distant metastasis–free survival (DMFS) curves for high (red) and low (black) expression groups dichotomized at the optimal cut-point were plotted. The logrank P-value and the hazard ratio with 95% confidence intervals were calculated. The threshold was adjusted to logrank P‐values at <0.05.
3 Results
3.1 Description of Collected Studies
As shown in the brief flow chart ( Figure 3 ), the records on E3 ubiquitin ligases involved in breast cancer metastasis were screened from both the PubMed (N = 199) and Medline (N = 103) databases. After duplicate elimination, 202 records remained. By reading the titles and abstracts of these records, we excluded 91 literatures for the following reasons: studies in other cancer types (N = 23); irrelevant to metastasis (N = 28) or irrelevant to E3 ubiquitin ligase (N = 12); focused on the regulation of E3 ubiquitin ligase (N = 17) or de-ubiquitination (N = 3); and reviews (N = 4) and retracted articles (N = 4). Other studies had been validated by reading the full texts. Finally, a total of 111 literatures were included in our analysis.
Figure 3.
Brief flow chart for literatures searching and selection. “N” refers to “number of studies”.
3.2 Roles of E3 Ubiquitin Ligases Played in the Multiple Steps of Breast Cancer Metastasis
We further extracted key information from these literatures, including E3 name, gene ID, substrate, roles in breast cancer metastasis, cellular function (experimentally verified), signal pathway, breast cancer cell lines, and clinical significance. This detailed information of 54 E3 ubiquitin ligases/ligase complexes in total have been summarized in Table 1 . By their molecular roles’ participation in the multiple steps of breast cancer metastasis, we briefly presented them as follows.
Table 1.
E3 ubiquitin ligases that played important roles in the multiple steps of breast cancer metastasis .
| E3 | Substrate | Inhibit/Promote Metastasis | Cellular Function | Molecular Pathway | References |
|---|---|---|---|---|---|
| Arkadia | Ski | Inhibit | EMT | TGF-beta | (33) |
| ASB13 | SNAI2 | Inhibit | Migration | Hippo–YAP | (34) |
| BCA2 | Autoubiquitination | Promote | Migration and invasion | EGFR | (35, 36) |
| Cbl | FAK and EGFR | Promote/inhibit | Cell detachment/EMT and migration | FAK, RANKL/RANK EGFR-ERK/Akt | (37–40) |
| CHIP | Pfn1 | Promote | Migration | ROCK1/Pfn1 | (41, 42) |
| COP1 | c-Jun | Inhibit | Migration | ETV1, GSK3β/c-Jun | (43, 44) |
| Cullin1 | N/A | Promote | EMT, migration, invasion, and tube formation | PI3K/AKT, NF-κB | (45) |
| Cullin3/SPOP | PR, Erα | Inhibit | Migration | PR, ERα | (46, 47) |
| Cullin3/KCTD5 | N/A | Promote | Migration and invasion | TRPM4 | (48) |
| Cullin7 | N/A | Promote | Invasion | N/A | (49) |
| FBXW7 | NICD1/Notch1 | Promote/inhibit (indirect evidence) | Migration and invasion | NOTCH/p62 | (50, 51) |
| FBXL8 | N/A | Promote | Migration and invasion | CCND2/IRF5 | (52) |
| FBXL14 | CDCP1 | Inhibit | EMT, migration, and invasion | PI3K/AKT | (53) |
| FBXO11 | SNAI1 | Inhibit | Emt | SNAI1 and p53/p21/BCL2 | (54, 55) |
| FBXO22 | HDM2 | Inhibit | Migration and invasion | p53/p21, SNAIL | (56) |
| FBXO31 | Slug | Inhibit | Invasion | N/A | (57) |
| FBXO32 | KLF4 | Promote | EMT, migration, and invasion | N/A | (58) |
| GP78 | HSPA5 | Inhibit | Migration and invasion | N/A | (59, 60) |
| HACE1 | Rac1 | Inhibit | Migration and invasion | N/A | (61) |
| HectD1 | ACF7 | Inhibit | EMT, migration, and invasion | N/A | (62) |
| HERC2 | BRCA1 | Promote (indirect evidence) | Invasion (indirect evidence) | BRCA1 | (63) |
| HERC4 | LATS1 | Promote | Migration and invasion | LATS1 | (64) |
| HRD1 | IGF-1R | Inhibit | EMT, migration, and invasion | IL6/NF-κB | (65) |
| ITCH | c-Jun, p73, p63, and ErbB4; RASSF1A; histone H1.2 | Promote | EMT and invasion | Hippo | (66–68) |
| MARCH5 | N/A | Promote | Migration and invasion | Mitochondrial | (69) |
| MDM2 | P53; RB; Foxo3a | Promote | EMT, migration, and invasion | p53/p21 | (70–73) |
| MEX3C | PTEN | Promote | EMT | TWIST1, SNAI1, and YAP1 | (74) |
| NEDD4 | Robo1 | Promote | Migration and invasion (indirect evidence) | FAK and Src | (75–77) |
| NKLAM | N/A | Inhibit | Tumor immunity | NK killing activity | (78) |
| NRBE3 | RB | Promote | Migration and invasion | E-cadherin | (79) |
| NRDP1 | ErbB3 and ErbB4 | Inhibit (indirect evidence) | Migration and invasion (indirect evidence) | ER | (80) |
| Parkin | HIF-1α | Inhibit | Migration and invasion | HRE/VHL | (81) |
| PDZRN4 | Kidins220 (predict) | Inhibit | Migration and invasion | N/A | (82) |
| PPIL2 | SNAI1 | Inhibit | EMT, migration, and invasion | SNAI1 | (83) |
| RNF8 | TWIST | Promote | EMT, migration, and invasion | TWIST; GSK3β | (84, 85) |
| RNF20 | Histone H2B | Promote (luminal)/inhibit (in basal-like) | Migration | ER (in luminal) and NF-kB (in basal-like) | (86, 87) |
| RNF144A | HSPA2 | Inhibit | Migration and invasion | HSPA2 | (88) |
| RNF208 | Vimentin | Inhibit | Migration and invasion | Vimentin | (89) |
| SIAH1/2 | p27 | Promote | Migration and invasion | Rb | (90) |
| SKP2 | AKT | Promote | Indirect evidence | PI3K/AKT | (91) |
| SCF-JFK | ING4 | Promote | EMT, invasion, and angiogenesis | NF-κB | (92) |
| β-Trcp | PRLr/βCatenin | Promote | EMT, migration, and invasion | PRL,Wnt | (93, 94) |
| Smurf1 | RhoA p120-catenin and TRAF4 | Promote | EMT, migration, and invasion | TGFβ | (95–99) |
| Smurf2 | Smurf1, CNKSR2 | Promote/inhibit | Migration and invasion | TGFβ, PI3K/AKT | (100–104) |
| TRAF6 | H2AX | Promote | Migration and invasion | HIF1α | (105, 106) |
| TRIM8 | Erα | Inhibit | Migration and invasion | Erα | (107) |
| TRIM11 | Erα | Promote | Migration | Erα | (108) |
| TRIM44 | N/A | Promote | Migration | NF-κB | (109) |
| TRIM47 | N/A | Promote | EMT, migration, and invasion | PI3K/Akt | (110) |
| UBR5 | N/A | Promote | EMT, migration, and invasion | STAT3, TGFα, P38MAPK | (111, 112) |
| UBR7 | H2B | Inhibit | EMT, migration, and invasion | Wnt/β-catenin | (113) |
| WWP1 | CXCR4 (indirect evidence) | Inhibit | Migration | TGFβ/Smad | (114) |
| xIAP | TAK1 | Promote | Invasion | TGFβ/Smad | (115) |
| UBE2O | AMPkα2 | Promote | EMT, migration, invasion, and stemness | AMPK/mTOR | (116) |
ASB13, ankyrin repeat and SOCS box containing 13; BCA2, breast cancer associated gene 2; Cbl‐b, Cbl proto-oncogene B; CHIP, STIP1 homology and U-box containing protein 1; CXCR4, C-X-C motif chemokine receptor 4; SPOP, speckle type BTB/POZ protein; KCTD5, K+ channel tetramerization domain 5; FBXW7, F-box and WD repeat domain containing 7; FBXL8, F-box and leucine rich repeat protein 8; FBXL14, F-box and leucine rich repeat protein 14; FBXO11, F-box protein 11; FBXO22, F-box protein 22; FBXO31, F-box protein 31; FBXO32, F-box protein 32; GP78, autocrine motility factor receptor; HACE1, HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1; HECTD1, HECT domain E3 ubiquitin protein ligase 1; HERC2, HECT and RLD domain containing E3 ubiquitin protein ligase 2; HERC4, HECT and RLD domain containing E3 ubiquitin protein ligase 4; MARCHF5, membrane associated ring-CH-type finger 5; MEX3C, mex-3 RNA binding family member C; NKLAM, Natural killer lytic-associated molecule; NRBE3, New RB E3 ubiquitin ligase; NRDP1, also known as RNF41, ring finger protein 41; Parkin, parkin RBR E3 ubiquitin protein ligase; PDZRN4, PDZ domain containing ring finger 4; PPIL2, peptidylprolyl isomerase like 2; RNF, RING Finger Protein; RASSF1, Ras association domain family member 1; SIAH1/2, siah E3 ubiquitin protein ligase 1 and 2; SKP2, S-phase kinase associated protein 2; SCF, Skp-Cullin-F-box) complex; β-Trcp, beta-transducin repeat containing E3 ubiquitin protein ligase; SMURF1 and SMURF2, SMAD specific E3 ubiquitin protein ligase 1 and 2; TRAF6, TNF receptor associated factor 6; TRIM8, TRIM11, TRIM44, TRIM47: tripartite motif containing 8, 11, 44 and 47; UBR5 and UBR7, ubiquitin protein ligase E3 component n-recognin 5 and 7; TWIST, twist family bHLH transcription factor 1; WWP1, WW domain containing E3 ubiquitin protein ligase 1; xIAP, X-linked inhibitor of apoptosis; UBE2O, ubiquitin conjugating enzyme E2 O; N/A, Not Available in the reference literature.
3.2.1 Breast Cancer Cell Epithelial–Mesenchymal Transition
EMT is thought to be a hallmark in tumor metastasis (117, 118). By the loss of cell–cell conjunction, the epithelial cells acquire migratory, invasive properties and trans-differentiate to mesenchymal phenotypic cells (9). Carcinoma cells undergoing EMT can escape from primary tumor sites, enter the circulation, and then move out to invade distant sites where secondary tumors or metastases begin to form (117, 119).
Not all breast cancer cells are typical EMT cells. In a study of breast cancer aggressiveness, by profiling EMT-like subclones from MCF-7 breast cancer cells based on EMT properties, comparing their gene-expressing differences with other non-EMT-like subclones, FBXO11 (F-box protein 11, a member of the E3 ubiquitin ligase complex) was screened out. It is concluded that FBXO11 is a candidate molecular alternative to the canonical EMT-dependent aggressiveness in highly differentiated luminal tumors (54). The specific roles suggest that E3 ligase FBXO11 targets transcription factor SNAI1 (snail family transcriptional repressor 1) protein which induces EMT, for ubiquitination-dependent degradation (55), and regulates the p53/p21/BCL2 pathway (54).
In cancer cells, the loss of E-cadherin results in EMT and contributes to increased metastasis and chemoresistance (120, 121). The absence of the ubiquitin protein ligase E3 component n-recognin 5 (UBR5) has been characterized to trigger aberrant EMT in TNBC, principally via an abrogated expression of E-cadherin, which resulted in severe lung metastasis in UBR5 knockout mice. The E-cadherin transcription repressors slug, twist, and zinc finger E-box-binding homeobox 1/2 (ZEB1/2) are overexpressed in multiple drug-resistant (MDR) breast cancer cells, making them more metastatic (122). The E3 ubiquitin ligase Casitas B lymphoma-b (Cbl-b) was reported to prevent tumor metastasis by maintaining the epithelial phenotype in MDR breast cancer cells. Cbl-b inhibits MDR breast cancer cell migration by specifically targeting epidermal growth factor receptor (EGFR) ubiquitination-dependent degradation, which prevents metastatic breast cancer cell EMT by inhibition of the EGFR-ERK/Akt-miR-200c-ZEB1 axis (37).
Not only restricted in the degradation of substrates, an RNA-binding E3 ligase, mex-3 RNA binding family member C (MEX3C), catalyzed protein PTEN (phosphatase and tensin homolog) K27-linked polyubiquitination, leading to its enzymatic functions’ switch to serine/threonine phosphatase activity (74). Consequently, switched PTEN promotes EMT.
The roles of E3 ubiquitin ligases played in breast cancer cell EMT include regulating transcription factors in EMT-inducing gene expression, abrogated expressing of key proteins, and affecting the activities of substrate proteins. The specific modification is not only as an E3 ubiquitin ligase to influence protein stability but also to induce the change of protein catalytic activities.
3.2.2 Breast Cancer Cell Invasion and Migration
Escaping from the surrounding tissues of the primary tumor, invading the blood or lymphatic vessels (intravasation), and migrating are essential steps in breast cancer metastasis. In this analysis, most of the E3 ubiquitin ligases (50 out of 54) have been detected to promote or inhibit the invasion and migration of breast cancer cells. Ubiquitin E3 ligases for tumor suppressors play important roles in tumorigenesis. The E3 ligase MDM2 has been characterized to target both tumor suppressor p53 and RB for proteasomal degradation, which promotes breast cancer cell invasion and migration via the p53/p21 pathway (70, 71). Nedd4 and carboxyl terminus of Hsc-70-interacting protein (CHIP) are two E3 ubiquitin ligases which function as regulators of tumor suppressor PTEN (123, 124). CHIP has been confirmed to promote breast cancer cell migration (41); the role of Nedd4 in breast cancer cell migration is not clear.
In the migration process, β-catenin and E-cadherin are two key proteins that function in cell–cell adhesion. SCFβTrcp has been characterized to be the E3 ligase complex responsible for β-catenin degradation (93). As for E-cadherin, MDM2 has also been identified as responsible for its ubiquitin-dependent degradation (71). The E3 ligase new RB-E3 ligase protein (NRBE3) promotes breast cancer cell migration via E-cadherin but is not dependent on its E3 ligase property (79). Although it has been speculated that UBR5 might regulate E-cadherin through targeting substrates for proteasome-dependent degradation, both in vivo and in vitro ubiquitin assays were required for validation (111).
3.2.3 Breast Cancer Cell Stemness
Breast cancers are heterogenous. Although breast cancer stem cells (BCSCs) account for a very small percentage, they have the greatest ability of self-renewal and potential of unlimited differentiation capacity into heterogeneous tumor cell populations, all of which contribute to regenerate tumor at original or distant sites in vivo. Cancer cell stemness properties can be identified by several stem cell markers such as CD34, CD44, CD123, CD133, Oct4, Sox2, Nanog, ABCG2, and MYC and the conduction of cell stemness sphere assays (116, 125).
Not only is MYC a stem cell marker; it is also a well-characterized oncoprotein that is upregulated in 30–50% of breast cancer patients. MYC is regulated by AMPKα (AMP-activated protein kinase alpha subunit)/mTORC1 (mechanistic target of rapamycin kinase) axis. Ubiquitin-conjugating enzyme E2O (UBE2O), a large E2 ubiquitin-conjugating enzyme that represents both E2 and E3 ligase activities, is found to promote AMPKα ubiquitination and degradation and then to activate the mTORC1-MYC signal pathway in breast cancer cells. By detection of stem cell markers and observation of cell stemness sphere formation, it is suggested that UBE2O endowed breast cells with cancer stemness properties (116).
In a study of the characteristics of BCSCs by selectively sorting cells with stem cell markers, another E3 ligase, Cbl, has been found to be involved in maintaining cancer cell stemness. EGFR is important for cancer stem cell maintenance and metastasis. Its turnover relies on the ubiquitin pathway. Cbl-c, a member of Cbl family, can target EGFR for k-63 linked ubiquitination and lysosomal degradation. By interfering with the binding of EGFR and its E3 ubiquitin ligase Cbl-c, a membrane protein sarcoglycan epsilon (SGCE) inhibits Cbl-c ubiquitin ability and stabilizes EGFR and then promotes breast cancer cell stemness (38).
Whether it is in regulating stem property–maintaining key proteins, or in the analysis of BCSCs’ characteristics, E3 ubiquitin ligases have been found to play roles in regulating the pluripotency of BCSCs.
3.2.4 Angiogenesis
In the process of metastasis, to survive and initiate the secondary cancer foci, cells need the capability of adapting to supportive niches such as angiogenesis (8). Two members of E3 ligases complex were found to be functional in angiogenesis.
The Skp–CUL1–F-box ubiquitin ligase complex is one of the best-characterized multi-subunit RING finger complexes composed of four subunits. F-box protein 42 (Fbxo42, also known as JFK) is one of the F-box family proteins. It is demonstrated that JFK targets ING4 for ubiquitination and degradation through the assembly of an SCFJFK ubiquitin ligase. ING4 is a member of the inhibitor of growth (ING) protein family, defined as tumor suppressors by directly interacting with p53 and promotes the transactivation of p53 and negatively regulates NF-κB-responsive gene transcription. The NF-κB pathway regulates the expression of several prominent pro-angiogenic factors, including IL-6, IL-8, CCL5, and COX-2. Degradation of ING4 by E3 ligase SCFJFk results in the destabilization of NF-κB signaling and promotes angiogenesis. In breast cancer animal models, it was also observed that JFK-mediated metastasis takes the roots of lungs (92).
Another member of the SCF complex is Cullin1. In the tube formation assay, the knockdown of Cullin1 significantly decreased the number of complete tubule structures formed by human umbilical vein endothelial cells (HUVECs) in vitro. In the tail vein metastasis animal model, MDA-MB-231 cells with Cullin1 stable knockdown showed reduced vascularization and micro-vessels in matrigel plug (45). Cullin1 regulates the zeste 2 polycomb repressive complex 2 subunit (EZH2), which enhances cytokine expression through the NF-κB pathway. The cytokine expression further results in aggravating the breast cancer cell metastasis through the PI3K–AKT–mTOR signaling pathway. Due to the lack of ubiquitin assays, it is unclear whether EZH2 is the substrate protein of Cullin1. Besides, in the SCF E3 ubiquitin complexes, Cullin1 often acts as a ‘scaffold’ protein; what the target-recognizing subunit in this complex is also needs to be clarified further.
Both JFK and Cullin1 regulated angiogenesis in breast cancer metastasis, through the NF-κB pathway. It is noticed that three other E3 ubiquitin ligases, hydroxymethylglutaryl-coenzyme A reductase degradation protein 1 (HRD1) (65), ring finger protein 20 (RNF20) (86), and tripartite motif containing 44 (TRIM44) (109), were also found regulating the NF-κB pathway. We need to pay attention to whether they are responsible for angiogenesis in breast cancer metastasis in addition to their existing roles.
3.2.5 Immunity Response/Rescue
The multiple steps of metastasis rely on reciprocal interactions between breast cancer cells and the microenvironment. Immune cells and their mediators are known to facilitate metastasis within the microenvironment (126). Natural killer (NK) cells play an essential role in the defense against viruses or microbial pathogens and malignancies produced by the body itself. In the process of immune response, NK cells execute anti-pathogen or anti-tumor activities that rely on the direct cytolytic activity of these cells and produce various cytokines (127). An E3 ubiquitin ligase, the natural killer lytic-associated molecule (NKLAM), has been characterized to play a major role in the cytolytic activity of NK cells and to control tumor development, dissemination, and distant metastasis in vivo. The target substrate of NKLAM in NK cells has not been directly determined yet (78).
Recently, ubiquitin protein ligase E3 component N-recognin 5 (UBR5) has been found to induce a CD8+ T cell–mediated immune response. The loss of UBR5 in breast cancer cells causes the appearance of certain putative immunogens’ strong CD8+ T cell–mediated response in a paracrine manner (112). UBR5 has been previously found to be involved in TNBC metastasis (111). According to these two studies, the loss of UBR5 caused reduced angiogenesis and triggered aberrant EMT depending on the EMT regulators’ inhibitor of DNA binding 1 and 3 (ID1 and ID3), which limited the metastasis of breast cancer. Although it was indicated that UBR5 executed its biological function principally via abrogated expression of E-cadherin, its specific ubiquitin substrate still needs validation through both in vivo and in vitro ubiquitin assays.
More and more evidences suggest that E3 ubiquitin ligases participate in the regulation of immunosuppression. Whether E3 can be used as combined targets of tumor immunotherapy in the future needs further research (126, 128).
Thus, the identifying regulators of each step in the process above should provide insights into the mechanisms that control breast cancer metastasis and hence patient survival.
3.3 Classification of Breast Cancer Metastasis–Related E3s Based on the Structure
Ubiquitination is a ubiquitous form of post-translational modification of proteins. In this process, E3s specifically bind to the substrate proteins, mediate the ligation of ubiquitin molecules and affect the specific proteins' turnover and functions. Thus, it is important to clarify the mechanism in target drug research and development (129). Do the mechanisms of these identified breast cancer metastasis E3 ligases have commonalities?
We classified the previously summarized E3 ligases of classical families based on their structures. The results suggested that the E3s involved in breast cancer metastasis belong to diversified classes, such as the HECT family, RING family, and U-box family ( Figure 2B ). There are eight E3 ligases that belong to the HECT family and are further classified into three subfamilies, including SMAD-specific E3 ubiquitin protein ligase 1 (Smurf1), SMAD-specific E3 ubiquitin protein ligase 2 (Smurf2), itchy E3 ubiquitin protein ligase (Itch) and WW domain–containing E3 ubiquitin protein ligase 1 (WWP1), NEDD4, belonging to the NEDD4 family, which contains a WW domain, C2 domain, and HECT domain; HECT and RLD domain which contains E3 ubiquitin protein ligases 2 and 4 (HERC2 and HERC4), as the HERC family, which contains the common RCC1-like domain (RLD) and HECT domain; and UBR5, as the “other” family, which contains a UBA domain, zinc finger domain, and HECT domain.
RING E3 ligases are further divided into two subtypes: single RING and multi-unit RING family. Several E3 ligases belong to single RING type (listed in Figure 2B , single RING family), performing a single-step ubiquitin transfer from the E2-Ub to the substrate, which work as allosteric activators. As for multi-subunit RING type, The Cullin-RING ubiquitin ligase (CRL) family is composed of a multi-unit. Most recognized CRLs are known as the SCF (SKP-Cullin-F-box) complex, in which Cullin1 interacts with Skp1 or Skp2 and utilizes various F-box proteins (shown in Figure 2B , F-box proteins) to recruit substrates and initiate ubiquitin ligation. Cullin7 also forms a SCF complex with SKP1 and F-box and WD repeat domain containing 8 (FBXW8). Cullin3-RBX1 (Ring-box1) E3 ubiquitin ligase complex requires BTB (Bric-a-brac-Tramtrack-Broad complex) domain protein as an adaptor. Two BTB proteins, speckle-type BTB/POZ protein (SPOP) and K+ channel tetramerization domain 5 (KCTD5), forms a complex with Cullin3, respectively.
Three E3 ubiquitin ligases, CHIP, NRBE3, and peptidylprolyl isomerase like 2 (PPIL2) belong to the U-box family, which contains a U-box domain. U-box E3s are also categorized as RING-type E3s, but their molecular structure subtly differs in that zinc-bound sites are replaced by a hydrophobic core (29).
There are still many E3 ligases that cannot be characterized into classical types. Ankyrin repeat and SOCS box containing 13 (ASB13) belongs to the ankyrin repeat and suppressor of cytokine signaling (SOCS) box (Asb), which contains six-ankyrin repeat domain (34, 130). The UBR-box is a 70-residue zinc finger domain present in the UBR family of E3 ubiquitin ligases. Unlike UBR5, which also contains an HECT domain, the structures responsible for UBR7 executing its E3 role need to be verified by in vitro ubiquitin assays (113, 131, 132). Intriguingly, UBE2O, an E2/E3 hybrid ubiquitin-protein ligase, displays both E2 ubiquitin conjugating enzyme and E3 ubiquitin ligase activities (116, 133). Compared to classical types, the specific catalytic mechanism of several non-classical E3s will need more efforts to be figured out in the future.
3.4 Analysis and Summary of E3 Ubiquitin Ligases Significantly Correlated With Breast Cancer Patients’ Survival
Since the E3s are involved in breast cancer metastasis, are they associated with patients’ survival? Through the analysis of literatures, we found that out of the 54 E3 ligases, 31 have been reported to be related to the survival of patients ( Table 2 , from “ASB13” to “UBE2O”). The other 23 E3 ubiquitin ligases lack clinical data to determine whether they have clinical significance. Although some of them have verified the specific roles involved in metastasis by cellular experiments and animal models, these E3 ligases’ relationships with breast cancer patients’ survival need to be elucidated. Therefore, we used publicly available clinical data to conduct survival analysis (only in which the literature were claimed to have been verified by molecular and animal experiments).
Table 2.
E3s are significantly correlated with patients’ survival.
| E3s | Clinical Significance | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Population | DetectionType | Method of Detection | Expressing Difference | Categorization | Survival Analysis | HR | 95%CI | Logrank P | |
| ASB13 | NA | mRNA | RNA-seq | Expressing difference | High vs. Low | OS | 0.67 | NA | 5.7e–09 |
| BCA2 | 3,554 | mRNA | RNA-seq | Expressing difference (in subtype cell lines) | High vs. Low | OS (in subtypes) | LuminalA: 1.21 | 1–1.46 | 0.046 |
| LuminalB: 1.41 | 1.14–1.75 | 0.0018 | |||||||
| Basal: 1.25 | 0.96–1.64 | 0.1 | |||||||
| HER2+: 1.54 | 1.01–2.34 | 0.044 | |||||||
| Cbl‐b | 292 | Protein | IHC | Expressing difference | Positive vs. Negative |
OS, DFS |
OS: 0.550 | 0.341–0.888 | 0.013 |
| DFS: 0.616 | 0.414–0.917 | 0.016 | |||||||
| COP1 | 105 | Protein | IHC | Expressing difference | Positive vs. Negative |
OS, RFS | RR: 0.65 | 0.149–6.732 | P < 0.001 |
| Cullin7 | 103 | Protein | IHC | Expressing difference | Positive vs. Negative |
OS | NA | NA | P < 0.05 |
| FBXL14 | 1,764 | mRNA | RNA-seq | Expressing difference | High vs. Low | RFS | NA | NA | P < 0.0001 |
| FBXO11 | OS: 1,402 RFS: 3,951 MFS: NA |
mRNA | RNA-seq | Expressing difference | High vs. Low | OS, | OS: 1.37 | 1.08–1.74 | 0.01 |
| RFS | RFS: 1.46 | 1.31–1.63 | P < 0.0001 | ||||||
| MFS | MFS: 0.64 | NA | 0.04 | ||||||
| FBXO22 | 410 | Protein | IHC | Expressing difference | Positive vs. Negative |
OS, DFS |
OS: 0.604 | 0.398–0.918 | 0.018 |
| DFS: 0.536 | 0.315–0.912 | 0.021 | |||||||
| GP78 | 108 | Protein | IHC | Expressing difference | Positive vs. Negative |
OS, DFS | NA | NA | <0.001 |
| HACE1 | 1,764 | mRNA | RNA-seq | Expressing difference | High vs. Low | RFS | 1.40 | 1.20–1.64 | <0.0001 |
| HectD1 | 1,864 | mRNA | RNA-seq | Expressing difference | High vs. Low | OS | NA | NA | 1.00e–16 |
| HERC4 | 161 | mRNA | RNA-seq | Expressing difference | High vs. Low | OS | NA | NA | 0.029 |
| HRD1 | 170 | Protein | IHC | Expressing difference | Positive vs. Negative |
OS | NA | NA | <0.01 |
| ITCH | OS: 1,115 | mRNA | RNA-seq | Expressing difference | Low vs. High | OS, RFS, DMFS, PPS | OS: 1.47 | 1.15–1.87 | 0.0016 |
| RFS: 3,455; | RFS: 1.39 | 1.24–1.56 | 2.2e–08 | ||||||
| DMFS: 1,609 | DMFS: 1.39 | 1.14–1.71 | 0.0013 | ||||||
| PPS: 351 | PPS: 1.3 | 1–1.68 | 0.047 | ||||||
| MARCH5 | RNA: 1,081; IHC: 65 | mRNA Protein |
RNA-seq IHC |
Expressing difference | High vs. Low Positive vs. Negative |
OS | NA | NA | 0.048 0.029 |
| Parkin | RNA: 3,951 IHC: 168 |
mRNA Protein |
RNA-seq IHC |
Expressing difference | High vs. Low Tumor vs. non-tumor |
RFS, DMFS | NA | NA | 2.4e–13 0.0090 |
| PDZRN4 | 81 | mRNA Protein |
RNA-seq IHC, WB |
Expressing difference | Low vs. High | OS, DFS | OS: 1.663 | 1.013–2.731 | 0.044 |
| DFS: 1.840 | 1.126–3.007 | 0.015 | |||||||
| RNF8 | IHC: 202 RNA: 3,315 |
Protein mRNA |
IHC RNA-seq |
Expressing difference | Low vs. High | OS, RFS, DMFS, PPS | OS: 1.31 | 1.02–1.68 | 0.035 |
| RFS: 1.15 | 1.03–1.29 | 0.013 | |||||||
| DMFS: 1.43 | 1.17–1.76 | 0.00056 | |||||||
| PPS: 1.22 | 0.93–1.6 | 0.16 | |||||||
| RNF144A | 166 | Protein | IHC | Expressing difference | Low vs. High | OS, DMFS | NA | NA | <0.05 |
| RNF208 | 3,951 | mRNA | qRT-PCR RNA-seq |
Expressing difference in subtype | Low vs. High | RFS | 0.76 | 0.69–0.85 | <0.001 |
| SIAH2 | 235 | Protein | IHC | Expressing difference | Low vs. High | OS (ER+) | 0.68 | 0.52–0.89 | <0.005 |
| SKP2 | 80 | Protein | IHC | Expressing difference in subtype | Low vs. High | OS (Her2+) | NA | NA | 0.0002 |
| SCF-JFK | NA | mRNA | RNA-seq | Expressing difference | Low vs. High | OS | LuminalA: 0.94 | NA | 0.035 |
| Basal: 7.24 | 0.035 | ||||||||
| TRAF6 | 212 | Protein | IHC | Expressing difference | Low vs. High | DMFS | NA | NA | <0.001 |
| TRIM8 | IHC: 91 RNA: NA |
Protein mRNA |
IHC RNA-seq |
Expressing difference | Low vs. High | OS | All type: 0.69 | 0.58–0.81 | 3.8e–06 |
| ER+: 0.71 | 0.53–0.96 | 0.025 | |||||||
| TRIM11 | NA | mRNA | RNA-seq | Expressing difference | Low vs. High | OS | OS: 1.63 | 1.2–2.22 | 0.005 |
| RFS | RFS: 1.57 | 1.01–2.42 | 0.027 | ||||||
| TRIM44 | 129 | Protein | IHC | Expressing difference | Low vs. High | OS | NA | NA | 0.031 |
| DMFS | NA | NA | 0.027 | ||||||
| UBR5 | IHC: 54 RNA: NA |
Protein mRNA |
IHC RNA-seq |
Expressing difference | Tumor vs. non-tumor Low vs. High |
mRNA OS | NA | NA | 0.011 |
| UBR7 | 47 | mRNA | RNA-seq | Expressing difference | Low vs. High | DMFS | 0.31 | 0.1–0.98 | 0.036 |
| WWP1 | 33 and 179 | Protein | IHC | Expressing difference | Positive vs. Negative |
DMFS | NA | NA | <0.05 |
| UBE2O | RNA: 3,951 IHC: 50 |
mRNA Protein |
RNA-seq IHC, WB |
Expressing difference | Low vs. High | OS, DMFS | IHC OS: NA | NA | P < 0.05 |
| mRNA OS: 1.63 | 1.3–2.04 | 1.5e–05 | |||||||
| DMFS: 1.54 | 1.25–1.89 | 4e–05 | |||||||
| Arkadia | 2,765 | mRNA | RNA-seq | NA | Low vs. High | DMFS | 0.73 | 0.62–0.87 | 0.00046 |
| MDM2 | 2,765 | mRNA | RNA-seq | Expressing difference | Low vs. High | DMFS | All type: 0.81 | 0.69–0.96 | 0.015 |
| LuminalA: 1.5 | 1.14–1.99 | 0.0041 | |||||||
| Basal: 0.69 | 0.5–0.94 | 0.019 | |||||||
| NKLAM | 2,765 | mRNA | RNA-seq | NA | Low vs. High | DMFS | 0.77 | 0.66–0.9 | 0.00086 |
| PPIL2 | 2,765 | mRNA | RNA-seq | Expressing difference | Low vs. High | DMFS | 0.78 | 0.65–0.94 | 0.0085 |
| Smurf1 | 2,765 | mRNA | RNA-seq | NA | Low vs. High | DMFS | 1.34 | 1.11–1.62 | 0.0022 |
| Smurf2 | 2,765 | mRNA | RNA-seq | NA | Low vs. High | DMFS | 1.52 | 1.3–1.78 | 1.9e–07 |
IHC, immunohistochemical (IHC) staining; WB, western blot; OS, overall survival; DFS, disease-free survival; RFS, relapse-free survival; DMFS, distant metastasis-free survival; PPS, post-progression survival; BCSS, breast cancer–specific survival; HR, hazard ratio; 95%CI, 95% confidence intervals; RR, relative risk; NA, not available in the reference literature.
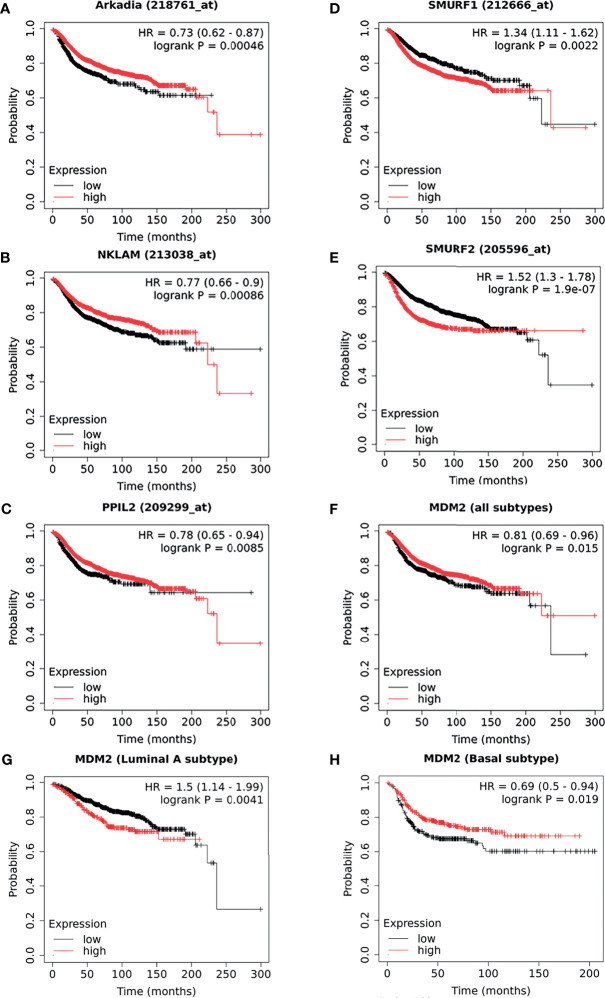
By comparing with molecular mechanism, five E3 ligases have been found to be significantly correlated with the DMFS of breast cancer patients. As illustrated in Kaplan–Meier plots ( Figure 4 ), we conducted the survival analysis of Arkadia ( Figure 4A ), NKLAM ( Figure 4B ), PPIL2 ( Figure 4C ), Smurf1 ( Figure 4D ), and Smurf2 ( Figure 4E ), which is consistent with the laboratory mechanism previously found. Patients with high expression of Arkadia (Hazard Ratio: 0.73; Logrank P: 0.00046), NKLAM (Hazard Ratio: 0.77; Logrank P: 0.00086), and PPIL2 (Hazard Ratio: 0.78; Logrank P: 0.0085), which have an inhibitory effect on metastasis, have higher survival probability. Patients with high expression of Smurf1 (Hazard Ratio: 1.34; Logrank P: 0.0022) and Smurf2 (Hazard Ratio: 1.52; Logrank P: 1.9e-07) have lower survival probability. Combining this analysis results with summarized literature reports, E3 ubiquitin ligases which are significantly correlated with patients’ survival were presented in Table 2 .
Figure 4.
The survival curves comparing patients with high and low expressions of E3s in breast cancer patients. As a supplement to the literatures, the survival analysis of Arkadia (A), NKLAM (B), PPIL2 (C), Smurf1 (D), and Smurf2 (E), which were illustrated as a Kaplan–Meier plot. Distant metastasis–free survival curves for high (red) and low (black) expression groups dichotomized at the optimal cut-point. The survival analysis of MDM2 in breast cancer was shown as all types (F), luminal A (G) subtype, and basal subtype (H), respectively. Logrank P-value and the hazard ratio with 95% confidence intervals was calculated. The threshold was adjusted to logrank P‐values at < 0.05.
Interestingly, we found that some results of survival analysis were contrary to the molecular mechanism. For example, if all subtypes of breast patients were considered together, a higher expression of MDM2 showed better survival (Hazard Ratio: 0.81; Logrank P: 0.015) ( Figure 4F ), which was obviously contrary to known molecular mechanisms. MDM2 has been well proven to be an E3 ubiquitin ligase that simultaneously targets tumor suppressor protein RB and P53 to degradation. We further analyzed different subtypes and found that it was opposite in patients with luminal A (Hazard Ratio: 1.5; Logrank P: 0.0041) ( Figure 4G ) and basal subtype (Hazard Ratio: 0.69; Logrank P: 0.019) ( Figure 4H ). This suggests that the heterogeneity of breast cancer cannot be ignored. In addition, it might be affected by the expression level or mutation of ubiquitin substrates, such as the status of p53 mutation or loss of RB. The context-dependent role of E3s and breast cancer subtypes need to be considered more in future survival analyses.
4 Discussion
In the past decades, dozens of studies have demonstrated that many E3 ubiquitin ligases play very important roles in breast cancer metastasis. E3 ligases were involved in the multiple steps of breast cancer metastasis, including EMT, invasion and migration, cell stemness, angiogenesis, and immunity response in the tumor microenvironment.
Typical E3s’ functions comprise of recognition and recruiting a specific protein to be modified and then catalyzing ubiquitin molecule discharge from an active-site cysteine onto the recruited substrate or a substrate-linked ubiquitin. Through this posttranslational modification, E3 ligases can alter the fate of their protein substrates, transducing different signals, which is critical for breast cancer metastasis. For example, the E3 ligase Cbl-b mediates the ubiquitination and degradation of EGFR, which inhibits metastatic breast cancer cells’ EMT. And another E3 ligase, MEX3C, catalyzes the tumor suppressor PTEN with K27-linked polyubiquitination and alters its enzymatic function, which leads to the accumulation of the master regulators of EMT, including twist family bHLH transcription factor 1 (TWIST1), Yes1-associated transcriptional regulator (YAP1), and SNAI1.
In light of the vital roles of E3 ubiquitin ligases in breast cancer metastasis, targeting them for cancer therapy has gained increasing attention. Notably, bortezomib, a proteasome inhibitor, was approved by the Food and Drug Administration (FDA) of United States to treat multiple myeloma and certain lymphomas, which encourages more and more researchers to screen the small molecular inhibitors of particular E3 ligases for anti-metastatic breast cancer. We queried with the key words of each E3 ubiquitin ligase names and their aliases in the AACT (Public Access to Aggregate Content of ClinicalTrials.gov, https://aact.ctti-clinicaltrials.org/) database, which is a publicly available relational database that contains information (protocol and result data elements) about every study registered in ClinicalTrials.gov. However, only the inhibitor for MDM2 has been registered for breast cancer treatment in undergoing clinical trials. It offers great opportunity for future pharmacological exploitation.
Though it is believed that inhibiting and redirecting ubiquitination in vivo are new therapeutic strategies, especially specific inhibitors of E3 ubiquitin ligase will be discovered and developed as a novel class of anticancer drugs in the foreseeable future, we are still facing significant challenges so far. Firstly, the specific substrate of E3 ligase had been elucidated in few studies and needs to be identified, especially in the process of breast cancer metastasis. Recent advances in high-throughput screening chemical methods have revolutionized our ability to match E3 ubiquitin ligases with their cellular targets (134), like the UBAIT (ubiquitin-activated interaction traps strategy), which relies on a ubiquitin molecule covalently fused to the E3 ligase of interest being charged onto E2 enzymes. Using the affinity enrichment of tagged UBAITs with following mass spectrometry can identify substrates of several E3s (135). Both in vivo and in vitro ubiquitin assays are also suggested to be used for the validation of E3 ligase substrates. Secondly, the substrate recognition specificity of E3 ligases needs to be understand more deeply in breast cancer metastasis, which is critical for the efficient small-molecule inhibition of substrate degradation. Comparing proteins modified in cell lysates versus when an E3 ligase is depleted will allow the identification of substrates (134, 136). Thirdly, structural bases and ubiquitin mechanisms facilitate with further exploit pharmacological strategy. For example, typical HECT family E3s harbor catalytic cysteines that first receive ubiquitin molecule from a bound E2~Ub intermediate and then directly deliver the ubiquitin to the substrate protein, which can pharmacologically target the catalytic cysteines of E3s (134). Instead, there are still several non-classical E3s whose structure and specific ubiquitin transfer mechanisms remain unknown. Finally, E3 ligases exhibit distinct or even opposite functions in different breast cancer subtypes, suggesting that a subtype-specific approach to E3 ligase substrates and inhibitor screening is required. A good example is K+ channel tetramerization domain 10 (KCTD10), a BTB protein, as an adaptor protein that forms E3 ubiquitin ligase complex with Cullin3. CUL3/KCTD10 ubiquitinated RhoB for K63-linked ubiquitin degradation and promote HER2-positive breast cancer cell proliferation (137, 138). Two downstream proteins of RhoB, RAC1 (Rho GTPase) and CNKSR1 (connector enhancer of kinase suppressor of Ras1), were found to be significantly correlated with the prognosis of HER2-positive breast cancer patients (138, 139). Beyond small-molecule inhibitors, proteolysis-targeting chimeras (PROTACs), which can induce the recruitment of E3 to target protein, have recently emerged as significant future therapeutic opportunities (140, 141).
In addition, breast cancer cell–secreted exosomes have been found to play roles in the microenvironment and enhance the invasiveness of recipient cells, which contribute to breast cancer invasion through the EGFR signaling (142, 143). An exosome-mediated delivery of the intrinsic PTEN-stabilizing factor PTEN-CT (Carbon Terminus) has been found to protect PTEN from E3 ligase–mediated proteasomal degradation and then inhibit breast cancer cell proliferation and migration (144). It suggests that not only intracellular ubiquitination but also intercellular ubiquitination (like exosome- mediated migratory delivery) should be followed with interest in the future.
With >700 E3 ligases in the human genome, including but not limited to the 54 E3s that have been identified to be involved in breast cancer metastasis, it has become clear that some of them are promising therapeutic targets or prognostic markers for breast cancer.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Author Contributions
YW, JG and JL designed this study. YW contributed to the conception, acquisition, analysis, and interpretation of data and drafted the manuscript. JD and YZ contributed to the acquisition of data. Schematic diagrams were drawn by JD. JG and JL contributed to the interpretation of data and critically revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (82003001, 82003227); China Postdoctoral Science Foundation (2019M663516,2021T140487); Post-Doctor Research Project, Sichuan University (20826041D4022); Post-Doctor Research Project, West China Hospital, Sichuan University (2018HXBH067); Sichuan Provincial Research Foundation for Basic Research (2020YFS0272); and Sichuan Science and Technology Program (2020YJ0046).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We sincerely appreciate all the contributors to references listed. We also thank Prof. Haiquan Lu (Advanced Medical Research Institute, Cheeloo College of Medicine, Shandong University) and Prof. Yongkang Yan (Institute for Cell Engineering, Johns Hopkins University School of Medicine) for their valuable comments and suggestions. Schematic figures were produced using Servier Medical Art (https://smart.servier.com/).
References
- 1. Ferlay JEM, Lam F, Colombet M, Mery L, Piñeros M, Znaor A, et al. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer; (2020) Available at: https://gco.iarc.fr/today. [Google Scholar]
- 2. Fahad Ullah M. Breast Cancer: Current Perspectives on the Disease Status. Adv Exp Med Biol (2019) 1152:51–64. doi: 10.1007/978-3-030-20301-6_4 [DOI] [PubMed] [Google Scholar]
- 3. Ferlay J EM, Lam F, Colombet M, Mery L, Piñeros M, Znaor A, et al. World Cancer Day:Breast Cancer Overtakes Lung Cancer in Terms of Number of New Cancer Cases Worldwide. Lyon, France: International Agency for Research on Cancer; (2020). Available at: https://iarc.who.int/news-events/world-cancer-day-2021/. [Google Scholar]
- 4. Weigelt B, Peterse JL, van 't Veer LJ. Breast Cancer Metastasis: Markers and Models. Nat Rev Cancer (2005) 5(8):591–602. doi: 10.1038/nrc1670 [DOI] [PubMed] [Google Scholar]
- 5. Mansour M, Haupt S, Chan AL, Godde N, Rizzitelli A, Loi S, et al. The E3-Ligase E6AP Represses Breast Cancer Metastasis via Regulation of ECT2-Rho Signaling. Cancer Res (2016) 76(14):4236–48. doi: 10.1158/0008-5472.Can-15-1553 [DOI] [PubMed] [Google Scholar]
- 6. Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Ng Eaton E, et al. Distinct EMT Programs Control Normal Mammary Stem Cells and Tumour-Initiating Cells. Nature (2015) 525(7568):256–60. doi: 10.1038/nature14897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hayes DF, Smerage J. Is There a Role for Circulating Tumor Cells in the Management of Breast Cancer? Clin Cancer Res (2008) 14(12):3646–50. doi: 10.1158/1078-0432.CCR-07-4481 [DOI] [PubMed] [Google Scholar]
- 8. Carmeliet P, Jain RK. Principles and Mechanisms of Vessel Normalization for Cancer and Other Angiogenic Diseases. Nat Rev Drug Discov (2011) 10(6):417–27. doi: 10.1038/nrd3455 [DOI] [PubMed] [Google Scholar]
- 9. Melzer C, von der Ohe J, Hass R. Breast Carcinoma: From Initial Tumor Cell Detachment to Settlement at Secondary Sites. BioMed Res Int (2017) 2017:8534371. doi: 10.1155/2017/8534371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Komander D, Rape M. The Ubiquitin Code. Annu Rev Biochem (2012) 81:203–29. doi: 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- 11. Mark KG, Rape M. Ubiquitin-Dependent Regulation of Transcription in Development and Disease. EMBO Rep (2021) 22(4):e51078. doi: 10.15252/embr.202051078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pickart CM. Mechanisms Underlying Ubiquitination. Annu Rev Biochem (2001) 70:503–33. doi: 10.1146/annurev.biochem.70.1.503 [DOI] [PubMed] [Google Scholar]
- 13. Pickart CM. Back to the Future With Ubiquitin. Cell (2004) 116(2):181–90. doi: 10.1016/S0092-8674(03)01074-2 [DOI] [PubMed] [Google Scholar]
- 14. Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, et al. A Multiubiquitin Chain Is Confined to Specific Lysine in a Targeted Short-Lived Protein. Science (1989) 243(4898):1576–83. doi: 10.1126/science.2538923 [DOI] [PubMed] [Google Scholar]
- 15. Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of Ubiquitin-Chain Formation by the Human Anaphase-Promoting Complex. Cell (2008) 133(4):653–65. doi: 10.1016/j.cell.2008.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Berndsen CE, Wolberger C. New Insights Into Ubiquitin E3 Ligase Mechanism. Nat Struct Mol Biol (2014) 21(4):301–7. doi: 10.1038/nsmb.2780 [DOI] [PubMed] [Google Scholar]
- 17. Dittmar G, Selbach M. Deciphering the Ubiquitin Code. Mol Cell (2017) 65(5):779–80. doi: 10.1016/j.molcel.2017.02.011 [DOI] [PubMed] [Google Scholar]
- 18. Yau R, Rape M. The Increasing Complexity of the Ubiquitin Code. Nat Cell Biol (2016) 18(6):579–86. doi: 10.1038/ncb3358 [DOI] [PubMed] [Google Scholar]
- 19. Gatti M, Pinato S, Maiolica A, Rocchio F, Prato MG, Aebersold R, et al. RNF168 Promotes Noncanonical K27 Ubiquitination to Signal DNA Damage. Cell Rep (2015) 10(2):226–38. doi: 10.1016/j.celrep.2014.12.021 [DOI] [PubMed] [Google Scholar]
- 20. Liu J, Han C, Xie B, Wu Y, Liu S, Chen K, et al. Rhbdd3 Controls Autoimmunity by Suppressing the Production of IL-6 by Dendritic Cells via K27-Linked Ubiquitination of the Regulator NEMO. Nat Immunol (2014) 15(7):612–22. doi: 10.1038/ni.2898 [DOI] [PubMed] [Google Scholar]
- 21. Yuan WC, Lee YR, Lin SY, Chang LY, Tan YP, Hung CC, et al. K33-Linked Polyubiquitination of Coronin 7 by Cul3-KLHL20 Ubiquitin E3 Ligase Regulates Protein Trafficking. Mol Cell (2014) 54(4):586–600. doi: 10.1016/j.molcel.2014.03.035 [DOI] [PubMed] [Google Scholar]
- 22. Uckelmann M, Sixma TK. Histone Ubiquitination in the DNA Damage Response. DNA Repair (Amst) (2017) 56:92–101. doi: 10.1016/j.dnarep.2017.06.011 [DOI] [PubMed] [Google Scholar]
- 23. Weake VM, Workman JL. Histone Ubiquitination: Triggering Gene Activity. Mol Cell (2008) 29(6):653–63. doi: 10.1016/j.molcel.2008.02.014 [DOI] [PubMed] [Google Scholar]
- 24. Rotin D, Kumar S. Physiological Functions of the HECT Family of Ubiquitin Ligases. Nat Rev Mol Cell Biol (2009) 10(6):398–409. doi: 10.1038/nrm2690 [DOI] [PubMed] [Google Scholar]
- 25. Deshaies RJ, Joazeiro CA. RING Domain E3 Ubiquitin Ligases. Annu Rev Biochem (2009) 78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- 26. Singh S, Ng J, Sivaraman J. Exploring the “Other” Subfamily of HECT E3-Ligases for Therapeutic Intervention. Pharmacol Ther (2021) 224:107809. doi: 10.1016/j.pharmthera.2021.107809 [DOI] [PubMed] [Google Scholar]
- 27. Wang M, Pickart CM. Different HECT Domain Ubiquitin Ligases Employ Distinct Mechanisms of Polyubiquitin Chain Synthesis. EMBO J (2005) 24(24):4324–33. doi: 10.1038/sj.emboj.7600895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Petroski MD, Deshaies RJ. Function and Regulation of Cullin-RING Ubiquitin Ligases. Nat Rev Mol Cell Biol (2005) 6(1):9–20. doi: 10.1038/nrm1547 [DOI] [PubMed] [Google Scholar]
- 29. Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL. Structural Insights Into the U-Box, a Domain Associated With Multi-Ubiquitination. Nat Struct Biol (2003) 10(4):250–5. doi: 10.1038/nsb906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mizuno H, Kitada K, Nakai K, Sarai A. PrognoScan: A New Database for Meta-Analysis of the Prognostic Value of Genes. BMC Med Genomics (2009) 2:18. doi: 10.1186/1755-8794-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Y, Song Z, Leng P, Liu Y. A Systematic Analysis Reveals Gene Expression Alteration of Serum Deprivation Response (SDPR) Gene Is Significantly Associated With the Survival of Patients With Cancer. Oncol Rep (2019) 42(3):1161–72. doi: 10.3892/or.2019.7212 [DOI] [PubMed] [Google Scholar]
- 32. Nagy A, Munkacsy G, Gyorffy B. Pancancer Survival Analysis of Cancer Hallmark Genes. Sci Rep (2021) 11(1):6047. doi: 10.1038/s41598-021-84787-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Le Scolan E, Zhu Q, Wang L, Bandyopadhyay A, Javelaud D, Mauviel A, et al. Transforming Growth Factor-Beta Suppresses the Ability of Ski to Inhibit Tumor Metastasis by Inducing Its Degradation. Cancer Res (2008) 68(9):3277–85. doi: 10.1158/0008-5472.Can-07-6793 [DOI] [PubMed] [Google Scholar]
- 34. Fan H, Wang X, Li W, Shen M, Wei Y, Zheng H, et al. ASB13 Inhibits Breast Cancer Metastasis Through Promoting SNAI2 Degradation and Relieving Its Transcriptional Repression of YAP. Genes Dev (2020) 34(19-20):1359–72. doi: 10.1101/gad.339796.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang W, Qu M, Wang J, Zhang X, Zhang H, Wu J, et al. Autoubiquitination of Feline E3 Ubiquitin Ligase BCA2. Gene (2018) 638:1–6. doi: 10.1016/j.gene.2017.09.059 [DOI] [PubMed] [Google Scholar]
- 36. Amemiya Y, Azmi P, Seth A. Autoubiquitination of BCA2 RING E3 Ligase Regulates Its Own Stability and Affects Cell Migration. Mol Cancer Res (2008) 6(9):1385–96. doi: 10.1158/1541-7786.MCR-08-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu L, Zhang Y, Qu X, Che X, Guo T, Cai Y, et al. E3 Ubiquitin Ligase Cbl-B Prevents Tumor Metastasis by Maintaining the Epithelial Phenotype in Multiple Drug-Resistant Gastric and Breast Cancer Cells. Neoplasia (2017) 19(4):374–82. doi: 10.1016/j.neo.2017.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao L, Qiu T, Jiang D, Xu H, Zou L, Yang Q, et al. SGCE Promotes Breast Cancer Stem Cells by Stabilizing EGFR. Adv Sci (Weinh) (2020) 7(14):1903700. doi: 10.1002/advs.201903700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fan Y, Qu X, Ma Y, Liu Y, Hu X. Cbl-B Promotes Cell Detachment via Ubiquitination of Focal Adhesion Kinase. Oncol (2016) 12(2):1113–8. doi: 10.3892/ol.2016.4730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang L, Teng Y, Fan Y, Wang Y, Li W, Shi J, et al. The E3 Ubiquitin Ligase Cbl-B Improves the Prognosis of RANK Positive Breast Cancer Patients by Inhibiting RANKL-Induced Cell Migration and Metastasis. Oncotarget (2015) 6(26):22918–33. doi: 10.18632/oncotarget.4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu J, Wang H, Li W, Liu K, Zhang T, He Z, et al. E3 Ubiquitin Ligase CHIP Attenuates Cellular Proliferation and Invasion Abilities in Triple-Negative Breast Cancer Cells. Clin Exp Med (2020) 20(1):109–19. doi: 10.1007/s10238-019-00594-3 [DOI] [PubMed] [Google Scholar]
- 42. Choi YN, Lee SK, Seo TW, Lee JS, Yoo SJ. C-Terminus of Hsc70-Interacting Protein Regulates Profilin1 and Breast Cancer Cell Migration. Biochem Biophys Res Commun (2014) 446(4):1060–6. doi: 10.1016/j.bbrc.2014.03.061 [DOI] [PubMed] [Google Scholar]
- 43. Ouyang M, Wang H, Ma J, Lü W, Li J, Yao C, et al. COP1, the Negative Regulator of ETV1, Influences Prognosis in Triple-Negative Breast Cancer. BMC Cancer (2015) 15:132. doi: 10.1186/s12885-015-1151-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shao J, Teng Y, Padia R, Hong S, Noh H, Xie X, et al. COP1 and GSK3β Cooperate to Promote C-Jun Degradation and Inhibit Breast Cancer Cell Tumorigenesis. Neoplasia (2013) 15(9):1075–85. doi: 10.1593/neo.13966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huang YF, Zhang Z, Zhang M, Chen YS, Song J, Hou PF, et al. CUL1 Promotes Breast Cancer Metastasis Through Regulating EZH2-Induced the Autocrine Expression of the Cytokines CXCL8 and IL11. Cell Death Dis (2018) 10(1):2. doi: 10.1038/s41419-018-1258-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang N, Sun P, Xu Y, Li H, Liu H, Wang L, et al. The GPER1/SPOP Axis Mediates Ubiquitination-Dependent Degradation of Erα to Inhibit the Growth of Breast Cancer Induced by Oestrogen. Cancer Lett (2020) 498:54–69. doi: 10.1016/j.canlet.2020.10.019 [DOI] [PubMed] [Google Scholar]
- 47. Gao K, Jin X, Tang Y, Ma J, Peng J, Yu L, et al. Tumor Suppressor SPOP Mediates the Proteasomal Degradation of Progesterone Receptors (PRs) in Breast Cancer Cells. Am J Cancer Res (2015) 5(10):3210–20. [PMC free article] [PubMed] [Google Scholar]
- 48. Rivas J, Díaz N, Silva I, Morales D, Lavanderos B, Álvarez A, et al. KCTD5, a Novel TRPM4-Regulatory Protein Required for Cell Migration as a New Predictor for Breast Cancer Prognosis. FASEB J (2020) 34(6):7847–65. doi: 10.1096/fj.201901195RRR [DOI] [PubMed] [Google Scholar]
- 49. Qiu N, He Y, Zhang S, Hu X, Chen M, Li H. Cullin 7 Is a Predictor of Poor Prognosis in Breast Cancer Patients and Is Involved in the Proliferation and Invasion of Breast Cancer Cells by Regulating the Cell Cycle and Microtubule Stability. Oncol Rep (2018) 39(2):603–10. doi: 10.3892/or.2017.6106 [DOI] [PubMed] [Google Scholar]
- 50. Ahn JS, Ann EJ, Kim MY, Yoon JH, Lee HJ, Jo EH, et al. Autophagy Negatively Regulates Tumor Cell Proliferation Through Phosphorylation Dependent Degradation of the Notch1 Intracellular Domain. Oncotarget (2016) 7(48):79047–63. doi: 10.18632/oncotarget.12986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chiang CH, Chu PY, Hou MF, Hung WC. MiR-182 Promotes Proliferation and Invasion and Elevates the HIF-1α-VEGF-A Axis in Breast Cancer Cells by Targeting FBXW7. Am J Cancer Res (2016) 6(8):1785–98. [PMC free article] [PubMed] [Google Scholar]
- 52. Chang SC, Hsu W, Su EC, Hung CS, Ding JL. Human FBXL8 Is a Novel E3 Ligase Which Promotes BRCA Metastasis by Stimulating Pro-Tumorigenic Cytokines and Inhibiting Tumor Suppressors. Cancers (2020) 12(8):07. doi: 10.3390/cancers12082210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cui YH, Kim H, Lee M, Yi JM, Kim RK, Uddin N, et al. FBXL14 Abolishes Breast Cancer Progression by Targeting CDCP1 for Proteasomal Degradation. Oncogene (2018) 37(43):5794–809. doi: 10.1038/s41388-018-0372-3 [DOI] [PubMed] [Google Scholar]
- 54. Bagger SO, Hopkinson BM, Pandey DP, Bak M, Brydholm AV, Villadsen R, et al. Aggressiveness of Non-EMT Breast Cancer Cells Relies on FBXO11 Activity. Mol Cancer (2018) 17(1):171. doi: 10.1186/s12943-018-0918-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zheng H, Shen M, Zha YL, Li W, Wei Y, Blanco MA, et al. PKD1 Phosphorylation-Dependent Degradation of SNAIL by SCF-FBXO11 Regulates Epithelial-Mesenchymal Transition and Metastasis. Cancer Cell (2014) 26(3):358–73. doi: 10.1016/j.ccr.2014.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bai J, Wu K, Cao MH, Yang Y, Pan Y, Liu H, et al. SCF(FBXO22) Targets HDM2 for Degradation and Modulates Breast Cancer Cell Invasion and Metastasis. Proc Natl Acad Sci USA (2019) 116(24):11754–63. doi: 10.1073/pnas.1820990116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Manne RK, Agrawal Y, Bargale A, Patel A, Paul D, Gupta NA, et al. A MicroRNA/Ubiquitin Ligase Feedback Loop Regulates Slug-Mediated Invasion in Breast Cancer. Neoplasia (2017) 19(6):483–95. doi: 10.1016/j.neo.2017.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen D, Yuan M, Ye Q, Wang X, Xu J, Shi G, et al. Cyanidin-3-O-Glucoside Inhibits Epithelial-to-Mesenchymal Transition, and Migration and Invasion of Breast Cancer Cells by Upregulating KLF4. Food Nutr Res (2020) 64:4240–50. doi: 10.29219/fnr.v64.4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chang YW, Chen HA, Tseng CF, Hong CC, Ma JT, Hung MC, et al. De-Acetylation and Degradation of HSPA5 Is Critical for E1A Metastasis Suppression in Breast Cancer Cells. Oncotarget (2014) 5(21):10558–70. doi: 10.18632/oncotarget.2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chang YW, Tseng CF, Wang MY, Chang WC, Lee CC, Chen LT, et al. Deacetylation of HSPA5 by HDAC6 Leads to GP78-Mediated HSPA5 Ubiquitination at K447 and Suppresses Metastasis of Breast Cancer. Oncogene (2016) 35(12):1517–28. doi: 10.1038/onc.2015.214 [DOI] [PubMed] [Google Scholar]
- 61. Kim I, Shin SH, Lee JE, Park JW. Oxygen Sensor FIH Inhibits HACE1-Dependent Ubiquitination of Rac1 to Enhance Metastatic Potential in Breast Cancer Cells. Oncogene (2019) 38(19):3651–66. doi: 10.1038/s41388-019-0676-y [DOI] [PubMed] [Google Scholar]
- 62. Duhamel S, Goyette MA, Thibault MP, Filion D, Gaboury L, Cote JF. The E3 Ubiquitin Ligase HectD1 Suppresses EMT and Metastasis by Targeting the +TIP ACF7 for Degradation. Cell Rep (2018) 22(4):1016–30. doi: 10.1016/j.celrep.2017.12.096 [DOI] [PubMed] [Google Scholar]
- 63. Peng Y, Dai H, Wang E, Lin CC, Mo W, Peng G, et al. TUSC4 Functions as a Tumor Suppressor by Regulating BRCA1 Stability. Cancer Res (2015) 75(2):378–86. doi: 10.1158/0008-5472.Can-14-2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xu Y, Ji K, Wu M, Hao B, Yao KT, Xu Y. A miRNA-HERC4 Pathway Promotes Breast Tumorigenesis by Inactivating Tumor Suppressor LATS1. Protein Cell (2019) 10(8):595–605. doi: 10.1007/s13238-019-0607-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xu YM, Wang HJ, Chen F, Guo WH, Wang YY, Li HY, et al. HRD1 Suppresses the Growth and Metastasis of Breast Cancer Cells by Promoting IGF-1R Degradation. Oncotarget (2015) 6(40):42854–67. doi: 10.18632/oncotarget.5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Salah Z, Itzhaki E, Aqeilan RI. The Ubiquitin E3 Ligase ITCH Enhances Breast Tumor Progression by Inhibiting the Hippo Tumor Suppressor Pathway. Oncotarget (2014) 5(21):10886–900. doi: 10.18632/oncotarget.2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pefani DE, Pankova D, Abraham AG, Grawenda AM, Vlahov N, Scrace S, et al. TGF-β Targets the Hippo Pathway Scaffold RASSF1A to Facilitate YAP/SMAD2 Nuclear Translocation. Mol Cell (2016) 63(1):156–66. doi: 10.1016/j.molcel.2016.05.012 [DOI] [PubMed] [Google Scholar]
- 68. Chang L, Shen L, Zhou H, Gao J, Pan H, Zheng L, et al. ITCH Nuclear Translocation and H1.2 Polyubiquitination Negatively Regulate the DNA Damage Response. Nucleic Acids Res (2019) 47(2):824–42. doi: 10.1093/nar/gky1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tang H, Peng S, Dong Y, Yang X, Yang P, Yang L, et al. MARCH5 Overexpression Contributes to Tumor Growth and Metastasis and Associates With Poor Survival in Breast Cancer. Cancer Manag Res (2019) 11:201–15. doi: 10.2147/cmar.S190694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gao C, Xiao G, Piersigilli A, Gou J, Ogunwobi O, Bargonetti J. Context-Dependent Roles of MDMX (MDM4) and MDM2 in Breast Cancer Proliferation and Circulating Tumor Cells. Breast Cancer Res (2019) 21(1):5. doi: 10.1186/s13058-018-1094-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yang JY, Zong CS, Xia W, Wei Y, Ali-Seyed M, Li Z, et al. MDM2 Promotes Cell Motility and Invasiveness by Regulating E-Cadherin Degradation. Mol Cell Biol (2006) 26(19):7269–82. doi: 10.1128/mcb.00172-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chou CC, Lee KH, Lai IL, Wang D, Mo X, Kulp SK, et al. AMPK Reverses the Mesenchymal Phenotype of Cancer Cells by Targeting the Akt-MDM2-Foxo3a Signaling Axis. Cancer Res (2014) 74(17):4783–95. doi: 10.1158/0008-5472.Can-14-0135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Xian J, Cheng Y, Qin X, Cao Y, Luo Y, Cao Y. Progress in the Research of P53 Tumour Suppressor Activity Controlled by Numb in Triple-Negative Breast Cancer. J Cell Mol Med (2020) 24(13):7451–9. doi: 10.1111/jcmm.15366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hu Q, Li C, Wang S, Li Y, Wen B, Zhang Y, et al. LncRNAs-Directed PTEN Enzymatic Switch Governs Epithelial-Mesenchymal Transition. Cell Res (2019) 29(4):286–304. doi: 10.1038/s41422-018-0134-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yu JM, Sun W, Wang ZH, Liang X, Hua F, Li K, et al. TRIB3 Supports Breast Cancer Stemness by Suppressing FOXO1 Degradation and Enhancing SOX2 Transcription. Nat Commun (2019) 10(1):5720. doi: 10.1038/s41467-019-13700-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang L, Qin Y, Wu G, Wang J, Cao J, Wang Y, et al. PRRG4 Promotes Breast Cancer Metastasis Through the Recruitment of NEDD4 and Downregulation of Robo1. Oncogene (2020) 39(49):7196–208. doi: 10.1038/s41388-020-01494-7 [DOI] [PubMed] [Google Scholar]
- 77. Tsai CF, Cheng YK, Lu DY, Wang SL, Chang CN, Chang PC, et al. Inhibition of Estrogen Receptor Reduces Connexin 43 Expression in Breast Cancers. Toxicol Appl Pharmacol (2018) 338:182–90. doi: 10.1016/j.taap.2017.11.020 [DOI] [PubMed] [Google Scholar]
- 78. Hoover RG, Gullickson G, Kornbluth J. Natural Killer Lytic-Associated Molecule Plays a Role in Controlling Tumor Dissemination and Metastasis. Front Immunol (2012) 3:393. doi: 10.3389/fimmu.2012.00393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zheng T, Lu M, Wang T, Zhang C, Du X. NRBE3 Promotes Metastasis of Breast Cancer by Down-Regulating E-Cadherin Expression. Biochim Biophys Acta Mol Cell Res (2018) 1865(12):1869–77. doi: 10.1016/j.bbamcr.2018.09.003 [DOI] [PubMed] [Google Scholar]
- 80. Hatakeyama J, Wald JH, Rafidi H, Cuevas A, Sweeney C, Carraway KL, 3rd. The ER Structural Protein Rtn4A Stabilizes and Enhances Signaling Through the Receptor Tyrosine Kinase ErbB3. Sci Signal (2016) 9(434):ra65. doi: 10.1126/scisignal.aaf1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Liu J, Zhang C, Zhao Y, Yue X, Wu H, Huang S, et al. Parkin Targets HIF-1alpha for Ubiquitination and Degradation to Inhibit Breast Tumor Progression. Nat Commun (2017) 8(1):1823. doi: 10.1038/s41467-017-01947-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lu YL, Yang X, Liu YK. Reduced PDZRN4 Promotes Breast Cancer Progression and Predicts Poor Prognosis. Int J Clin Exp Pathol (2019) 12(1):142–53. [PMC free article] [PubMed] [Google Scholar]
- 83. Jia Z, Wang M, Li S, Li X, Bai XY, Xu Z, et al. U-Box Ubiquitin Ligase PPIL2 Suppresses Breast Cancer Invasion and Metastasis by Altering Cell Morphology and Promoting SNAI1 Ubiquitination and Degradation. Cell Death Dis (2018) 9(2):63. doi: 10.1038/s41419-017-0094-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lee HJ, Li CF, Ruan D, Powers S, Thompson PA, Frohman MA, et al. The DNA Damage Transducer RNF8 Facilitates Cancer Chemoresistance and Progression Through Twist Activation. Mol Cell (2016) 63(6):1021–33. doi: 10.1016/j.molcel.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kuang J, Li L, Guo L, Su Y, Wang Y, Xu Y, et al. RNF8 Promotes Epithelial-Mesenchymal Transition of Breast Cancer Cells. J Exp Clin Cancer Res (2016) 35(1):88. doi: 10.1186/s13046-016-0363-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Tarcic O, Granit RZ, Pateras IS, Masury H, Maly B, Zwang Y, et al. RNF20 and Histone H2B Ubiquitylation Exert Opposing Effects in Basal-Like Versus Luminal Breast Cancer. Cell Death Differ (2017) 24(4):694–704. doi: 10.1038/cdd.2016.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, et al. The Histone H2B-Specific Ubiquitin Ligase RNF20/hBRE1 Acts as a Putative Tumor Suppressor Through Selective Regulation of Gene Expression. Genes Dev (2008) 22(19):2664–76. doi: 10.1101/gad.1703008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Yang YL, Zhang Y, Li DD, Zhang FL, Liu HY, Liao XH, et al. RNF144A Functions as a Tumor Suppressor in Breast Cancer Through Ubiquitin Ligase Activity-Dependent Regulation of Stability and Oncogenic Functions of HSPA2. Cell Death Differ (2020) 27(3):1105–18. doi: 10.1038/s41418-019-0400-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Pang K, Park J, Ahn SG, Lee J, Park Y, Ooshima A, et al. RNF208, an Estrogen-Inducible E3 Ligase, Targets Soluble Vimentin to Suppress Metastasis in Triple-Negative Breast Cancers. Nat Commun (2019) 10(1):5805. doi: 10.1038/s41467-019-13852-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Adam MG, Matt S, Christian S, Hess-Stumpp H, Haegebarth A, Hofmann TG, et al. SIAH Ubiquitin Ligases Regulate Breast Cancer Cell Migration and Invasion Independent of the Oxygen Status. Cell Cycle (2015) 14(23):3734–47. doi: 10.1080/15384101.2015.1104441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, et al. The Skp2-SCF E3 Ligase Regulates Akt Ubiquitination, Glycolysis, Herceptin Sensitivity, and Tumorigenesis. Cell (2012) 149(5):1098–111. doi: 10.1016/j.cell.2012.02.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yan R, He L, Li Z, Han X, Liang J, Si W, et al. SCF(JFK) Is a Bona Fide E3 Ligase for ING4 and a Potent Promoter of the Angiogenesis and Metastasis of Breast Cancer. Genes Dev (2015) 29(6):672–85. doi: 10.1101/gad.254292.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhang L, Zhou F, Li Y, Drabsch Y, Zhang J, van Dam H, et al. Fas-Associated Factor 1 Is a Scaffold Protein That Promotes β-Transducin Repeat-Containing Protein (β-TrCP)-Mediated β-Catenin Ubiquitination and Degradation. J Biol Chem (2012) 287(36):30701–10. doi: 10.1074/jbc.M112.353524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Plotnikov A, Varghese B, Tran TH, Liu C, Rui H, Fuchs SY. Impaired Turnover of Prolactin Receptor Contributes to Transformation of Human Breast Cells. Cancer Res (2009) 69(7):3165–72. doi: 10.1158/0008-5472.Can-08-4033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wu Q, Li G, Wen C, Zeng T, Fan Y, Liu C, et al. Monoubiquitination of P120-Catenin Is Essential for Tgfβ-Induced Epithelial-Mesenchymal Transition and Tumor Metastasis. Sci Adv (2020) 6(4):eaay9819. doi: 10.1126/sciadv.aay9819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yu L, Liu X, Cui K, Di Y, Xin L, Sun X, et al. SND1 Acts Downstream of TGFbeta1 and Upstream of Smurf1 to Promote Breast Cancer Metastasis. Cancer Res (2015) 75(7):1275–86. doi: 10.1158/0008-5472.CAN-14-2387 [DOI] [PubMed] [Google Scholar]
- 97. Kwon A, Lee HL, Woo KM, Ryoo HM, Baek JH. SMURF1 Plays a Role in EGF-Induced Breast Cancer Cell Migration and Invasion. Mol Cells (2013) 36(6):548–55. doi: 10.1007/s10059-013-0233-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang X, Jin C, Tang Y, Tang LY, Zhang YE. Ubiquitination of Tumor Necrosis Factor Receptor-Associated Factor 4 (TRAF4) by Smad Ubiquitination Regulatory Factor 1 (Smurf1) Regulates Motility of Breast Epithelial and Cancer Cells. J Biol Chem (2013) 288(30):21784–92. doi: 10.1074/jbc.M113.472704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Xie Y, Avello M, Schirle M, McWhinnie E, Feng Y, Bric-Furlong E, et al. Deubiquitinase FAM/USP9X Interacts With the E3 Ubiquitin Ligase SMURF1 Protein and Protects It From Ligase Activity-Dependent Self-Degradation. J Biol Chem (2013) 288(5):2976–85. doi: 10.1074/jbc.M112.430066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. David D, Surendran A, Thulaseedharan JV, Nair AS. Regulation of CNKSR2 Protein Stability by the HECT E3 Ubiquitin Ligase Smurf2, and Its Role in Breast Cancer Progression. BMC Cancer (2018) 18(1):284. doi: 10.1186/s12885-018-4188-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. David D, Jagadeeshan S, Hariharan R, Nair AS, Pillai RM. Smurf2 E3 Ubiquitin Ligase Modulates Proliferation and Invasiveness of Breast Cancer Cells in a CNKSR2 Dependent Manner. Cell Div (2014) 9:2. doi: 10.1186/1747-1028-9-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhang L, Zhou F, García de Vinuesa A, de Kruijf EM, Mesker WE, Hui L, et al. TRAF4 Promotes TGF-β Receptor Signaling and Drives Breast Cancer Metastasis. Mol Cell (2013) 51(5):559–72. doi: 10.1016/j.molcel.2013.07.014 [DOI] [PubMed] [Google Scholar]
- 103. Jin C, Yang YA, Anver MR, Morris N, Wang X, Zhang YE. Smad Ubiquitination Regulatory Factor 2 Promotes Metastasis of Breast Cancer Cells by Enhancing Migration and Invasiveness. Cancer Res (2009) 69(3):735–40. doi: 10.1158/0008-5472.CAN-08-1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Fukunaga E, Inoue Y, Komiya S, Horiguchi K, Goto K, Saitoh M, et al. Smurf2 Induces Ubiquitin-Dependent Degradation of Smurf1 to Prevent Migration of Breast Cancer Cells. J Biol Chem (2008) 283(51):35660–7. doi: 10.1074/jbc.M710496200 [DOI] [PubMed] [Google Scholar]
- 105. Li Z, Wang D, Lu J, Huang B, Wang Y, Dong M, et al. Methylation of EZH2 by PRMT1 Regulates Its Stability and Promotes Breast Cancer Metastasis. Cell Death Differ (2020) 27:3226–42. doi: 10.1038/s41418-020-00615-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Rezaeian AH, Li CF, Wu CY, Zhang X, Delacerda J, You MJ, et al. A Hypoxia-Responsive TRAF6-ATM-H2AX Signalling Axis Promotes HIF1α Activation, Tumorigenesis and Metastasis. Nat Cell Biol (2017) 19(1):38–51. doi: 10.1038/ncb3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Tian Z, Tang J, Liao X, Gong Y, Yang Q, Wu Y, et al. TRIM8 Inhibits Breast Cancer Proliferation by Regulating Estrogen Signaling. Am J Cancer Res (2020) 10(10):3440–57. doi: 10.21203/rs.3.rs-42521/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Tang J, Luo Y, Tian Z, Liao X, Cui Q, Yang Q, et al. TRIM11 Promotes Breast Cancer Cell Proliferation by Stabilizing Estrogen Receptor α. Neoplasia (2020) 22(9):343–51. doi: 10.1016/j.neo.2020.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Kawabata H, Azuma K, Ikeda K, Sugitani I, Kinowaki K, Fujii T, et al. TRIM44 Is a Poor Prognostic Factor for Breast Cancer Patients as a Modulator of NF-κb Signaling. Int J Mol Sci (2017) 18(9):1931–46. doi: 10.3390/ijms18091931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wang Y, Liu C, Xie Z, Lu H. Knockdown of TRIM47 Inhibits Breast Cancer Tumorigenesis and Progression Through the Inactivation of PI3K/Akt Pathway. Chem Biol Interact (2020) 317:108960. doi: 10.1016/j.cbi.2020.108960 [DOI] [PubMed] [Google Scholar]
- 111. Liao L, Song M, Li X, Tang L, Zhang T, Zhang L, et al. E3 Ubiquitin Ligase UBR5 Drives the Growth and Metastasis of Triple-Negative Breast Cancer. Cancer Res (2017) 77(8):2090–101. doi: 10.1158/0008-5472.Can-16-2409 [DOI] [PubMed] [Google Scholar]
- 112. Song M, Wang C, Wang H, Zhang T, Li J, Benezra R, et al. Targeting Ubiquitin Protein Ligase E3 Component N-Recognin 5 in Cancer Cells Induces a CD8+ T Cell Mediated Immune Response. Oncoimmunology (2020) 9(1):1746148. doi: 10.1080/2162402x.2020.1746148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Adhikary S, Chakravarti D, Terranova C, Sengupta I, Maitituoheti M, Dasgupta A, et al. Atypical Plant Homeodomain of UBR7 Functions as an H2BK120Ub Ligase and Breast Tumor Suppressor. Nat Commun (2019) 10(1):1398. doi: 10.1038/s41467-019-08986-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Subik K, Shu L, Wu C, Liang Q, Hicks D, Boyce B, et al. The Ubiquitin E3 Ligase WWP1 Decreases CXCL12-Mediated MDA231 Breast Cancer Cell Migration and Bone Metastasis. Bone (2012) 50(4):813–23. doi: 10.1016/j.bone.2011.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Neil JR, Tian M, Schiemann WP. X-Linked Inhibitor of Apoptosis Protein and Its E3 Ligase Activity Promote Transforming Growth Factor-{Beta}-Mediated Nuclear Factor-{Kappa}B Activation During Breast Cancer Progression. J Biol Chem (2009) 284(32):21209–17. doi: 10.1074/jbc.M109.018374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Liu X, Ma F, Liu C, Zhu K, Li W, Xu Y, et al. UBE2O Promotes the Proliferation, EMT and Stemness Properties of Breast Cancer Cells Through the UBE2O/Ampkα2/Mtorc1-MYC Positive Feedback Loop. Cell Death Dis (2020) 11(1):10. doi: 10.1038/s41419-019-2194-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kang Y, Pantel K. Tumor Cell Dissemination: Emerging Biological Insights From Animal Models and Cancer Patients. Cancer Cell (2013) 23(5):573–81. doi: 10.1016/j.ccr.2013.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Kalluri R, Weinberg RA. The Basics of Epithelial-Mesenchymal Transition. J Clin Invest (2009) 119(6):1420–8. doi: 10.1172/JCI39104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wang Y, Zhou BP. Epithelial-Mesenchymal Transition in Breast Cancer Progression and Metastasis. Chin J Cancer (2011) 30(9):603–11. doi: 10.5732/cjc.011.10226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Angelucci C, Maulucci G, Lama G, Proietti G, Colabianchi A, Papi M, et al. Epithelial-Stromal Interactions in Human Breast Cancer: Effects on Adhesion, Plasma Membrane Fluidity and Migration Speed and Directness. PloS One (2012) 7(12):e50804. doi: 10.1371/journal.pone.0050804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Lu L, Zhou D, Jiang X, Song K, Li K, Ding W. Loss of E-Cadherin in Multidrug Resistant Breast Cancer Cell Line MCF-7/Adr: Possible Implication in the Enhanced Invasive Ability. Eur Rev Med Pharmacol Sci (2012) 16(9):1271–9. [PubMed] [Google Scholar]
- 122. Galvan JA, Zlobec I, Wartenberg M, Lugli A, Gloor B, Perren A, et al. Expression of E-Cadherin Repressors SNAIL, ZEB1 and ZEB2 by Tumour and Stromal Cells Influences Tumour-Budding Phenotype and Suggests Heterogeneity of Stromal Cells in Pancreatic Cancer. Br J Cancer (2015) 112(12):1944–50. doi: 10.1038/bjc.2015.177 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 123. Ahmed SF, Deb S, Paul I, Chatterjee A, Mandal T, Chatterjee U, et al. The Chaperone-Assisted E3 Ligase C Terminus of Hsc70-Interacting Protein (CHIP) Targets PTEN for Proteasomal Degradation. J Biol Chem (2012) 287(19):15996–6006. doi: 10.1074/jbc.M111.321083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, et al. NEDD4-1 Is a Proto-Oncogenic Ubiquitin Ligase for PTEN. Cell (2007) 128(1):129–39. doi: 10.1016/j.cell.2006.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Bao B, Prasad AS. Targeting CSC in a Most Aggressive Subtype of Breast Cancer TNBC. Adv Exp Med Biol (2019) 1152:311–34. doi: 10.1007/978-3-030-20301-6_17 [DOI] [PubMed] [Google Scholar]
- 126. Zhou X, Sun SC. Targeting Ubiquitin Signaling for Cancer Immunotherapy. Signal Transduct Target Ther (2021) 6(1):16. doi: 10.1038/s41392-020-00421-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Huntington ND, Cursons J, Rautela J. The Cancer-Natural Killer Cell Immunity Cycle. Nat Rev Cancer (2020) 20(8):437–54. doi: 10.1038/s41568-020-0272-z [DOI] [PubMed] [Google Scholar]
- 128. Sambi M, Qorri B, Harless W, Szewczuk MR. Therapeutic Options for Metastatic Breast Cancer. Adv Exp Med Biol (2019) 1152:131–72. doi: 10.1007/978-3-030-20301-6_8 [DOI] [PubMed] [Google Scholar]
- 129. Zheng N, Shabek N. Ubiquitin Ligases: Structure, Function, and Regulation. Annu Rev Biochem (2017) 86:129–57. doi: 10.1146/annurev-biochem-060815-014922 [DOI] [PubMed] [Google Scholar]
- 130. Liu P, Verhaar AP, Peppelenbosch MP. Signaling Size: Ankyrin and SOCS Box-Containing ASB E3 Ligases in Action. Trends Biochem Sci (2019) 44(1):64–74. doi: 10.1016/j.tibs.2018.10.003 [DOI] [PubMed] [Google Scholar]
- 131. Li C, Beauregard-Lacroix E, Kondratev C, Rousseau J, Heo AJ, Neas K, et al. UBR7 Functions With UBR5 in the Notch Signaling Pathway and Is Involved in a Neurodevelopmental Syndrome With Epilepsy, Ptosis, and Hypothyroidism. Am J Hum Genet (2021) 108(1):134–47. doi: 10.1016/j.ajhg.2020.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Munoz-Escobar J, Kozlov G, Gehring K. Crystal Structure of the UBR-Box From UBR6/FBXO11 Reveals Domain Swapping Mediated by Zinc Binding. Protein Sci (2017) 26(10):2092–7. doi: 10.1002/pro.3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Hormaechea-Agulla D, Kim Y, Song MS, Song SJ. New Insights Into the Role of E2s in the Pathogenesis of Diseases: Lessons Learned From UBE2O. Mol Cells (2018) 41(3):168–78. doi: 10.14348/molcells.2018.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Henneberg LT, Schulman BA. Decoding the Messaging of the Ubiquitin System Using Chemical and Protein Probes. Cell Chem Biol (2021) 28(7):889–902. doi: 10.1016/j.chembiol.2021.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. O'Connor HF, Swaim CD, Canadeo LA, Huibregtse JM. Ubiquitin-Activated Interaction Traps (UBAITs): Tools for Capturing Protein-Protein Interactions. Methods Mol Biol (2018) 1844:85–100. doi: 10.1007/978-1-4939-8706-1_7 [DOI] [PubMed] [Google Scholar]
- 136. Bakos G, Yu L, Gak IA, Roumeliotis TI, Liakopoulos D, Choudhary JS, et al. An E2-Ubiquitin Thioester-Driven Approach to Identify Substrates Modified With Ubiquitin and Ubiquitin-Like Molecules. Nat Commun (2018) 9(1):4776. doi: 10.1038/s41467-018-07251-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Murakami A, Maekawa M, Kawai K, Nakayama J, Araki N, Semba K, et al. Cullin-3/KCTD10 E3 Complex Is Essential for Rac1 Activation Through RhoB Degradation in Human Epidermal Growth Factor Receptor 2-Positive Breast Cancer Cells. Cancer Sci (2019) 110(2):650–61. doi: 10.1111/cas.13899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Maekawa M, Higashiyama S. KCTD10 Biology: An Adaptor for the Ubiquitin E3 Complex Meets Multiple Substrates: Emerging Divergent Roles of the Cullin-3/KCTD10 E3 Ubiquitin Ligase Complex in Various Cell Lines. Bioessays (2020) 42(8):e1900256. doi: 10.1002/bies.201900256 [DOI] [PubMed] [Google Scholar]
- 139. Nishiyama K, Maekawa M, Nakagita T, Nakayama J, Kiyoi T, Chosei M, et al. CNKSR1 Serves as a Scaffold to Activate an EGFR Phosphatase via Exclusive Interaction With RhoB-GTP. Life Sci Alliance (2021) 4(9):1–22. doi: 10.26508/lsa.202101095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Qi SM, Dong J, Xu ZY, Cheng XD, Zhang WD, Qin JJ. PROTAC: An Effective Targeted Protein Degradation Strategy for Cancer Therapy. Front Pharmacol (2021) 12:692574. doi: 10.3389/fphar.2021.692574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Barghout SH. Targeted Protein Degradation: An Emerging Therapeutic Strategy in Cancer. Anticancer Agents Med Chem (2021) 21(2):214–30. doi: 10.2174/1871520620666200410082652 [DOI] [PubMed] [Google Scholar]
- 142. Yu DD, Wu Y, Shen HY, Lv MM, Chen WX, Zhang XH, et al. Exosomes in Development, Metastasis and Drug Resistance of Breast Cancer. Cancer Sci (2015) 106(8):959–64. doi: 10.1111/cas.12715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Kosaka N, Yoshioka Y, Tominaga N, Hagiwara K, Katsuda T, Ochiya T. Dark Side of the Exosome: The Role of the Exosome in Cancer Metastasis and Targeting the Exosome as a Strategy for Cancer Therapy. Future Oncol (2014) 10(4):671–81. doi: 10.2217/fon.13.222 [DOI] [PubMed] [Google Scholar]
- 144. Ahmed SF, Das N, Sarkar M, Chatterjee U, Chatterjee S, Ghosh MK. Exosome-Mediated Delivery of the Intrinsic C-Terminus Domain of PTEN Protects It From Proteasomal Degradation and Ablates Tumorigenesis. Mol Ther (2015) 23(2):255–69. doi: 10.1038/mt.2014.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.