Abstract
In all eukaryotes, the initiation of DNA synthesis requires the formation of prereplicative complexes (pre-RCs) on replication origins, followed by their activation by two S-T protein kinases, an S-phase cyclin-dependent kinase (S-CDK) and a homologue of yeast Dbf4-Cdc7 kinase (Dbf4p-dependent kinase [DDK]). Here, we show that yeast DDK activity is cell cycle regulated, though less tightly than that of the S-CDK Clb5-Cdk1, and peaks during S phase in correlation with Dbf4p levels. Dbf4p is short-lived throughout the cell cycle, but its instability is accentuated during G1 by the anaphase-promoting complex. Downregulating DDK activity is physiologically important, as joint Cdc7p and Dbf4p overexpression is lethal. Because pre-RC formation is a highly ordered process, we asked whether S-CDK and DDK need also to function in a specific order for the firing of origins. We found that both kinases are activated independently, but we show that DDK can perform its function for DNA replication only after S-CDKs have been activated. Cdc45p, a protein needed for initiation, binds tightly to chromatin only after S-CDK activation (L. Zou and B. Stillman, Science 280:593–596, 1998). We show that Cdc45p is phosphorylated by DDK in vitro, suggesting that it might be one of DDK's critical substrates after S-CDK activation. Linking the origin-bound DDK to the tightly regulated S-CDK in a dependent sequence of events may ensure that DNA replication initiates only at the right time and place.
Evolution has selected organisms that replicate their genomes both rapidly and accurately. To complete S phase swiftly without loss of accuracy, eukaryotes have subdivided their large genomes into many replication units. The yeast Saccharomyces cerevisiae utilizes 250 to 400 origins distributed along its 16 chromosomes, whereas tens of thousands of origins are probably needed to replicate the human genome. The firing of that many origins must be tightly controlled so that each piece of DNA is duplicated only once per cell cycle (12). To achieve this feat, the initiation of DNA replication follows strict rules dictated by cell cycle progression and cyclin-dependent kinase (CDK) activity. For instance, replication-competent origins are formed only during the G1 phase when the CDK activity level is low, but they require a high level of CDK activity for firing. As firing destroys origin competence, reinitiation is prevented until the next oscillation (drop plus rise) of CDK activity, usually at the G1-S transition. However, DNA replication is also flexible. Not all origins are activated at the same time during S phase (20), and many origins do not fire every cell cycle (23, 74). In metazoans, there is also considerable variation in the number of origins a cell utilizes and thus in S phase length: in Drosophila, DNA synthesis lasts for about 3 min during the rapid embryonic cell cycles, but in somatic tissues synthesis takes much longer (about 10 h), mainly because there are fewer initiation events (4). Thus, origin firing is modulated in the realms of time, space, and efficiency, the basis of which modulation is largely unknown. Nonetheless, origins do not fire stochastically but in a predictable manner from one cell cycle to the next, according to a specific replication program (33). Deviation from this program may lead to incomplete or delayed replication of certain loci and genetic instability (26). Therefore, there is considerable interest in understanding the molecular mechanisms that control the initiation and fine-tuning of DNA replication.
The initiation of DNA replication is portrayed as a two-step process which begins with the formation of replication-competent origins in G1 (12). This involves the sequential binding of the origin-recognition complex (ORC), Cdc6p, and the minichromosome maintenance complex (MCM) onto autonomously replicating sequences, i.e., the forming of prereplicative complexes (pre-RCs) (2, 14, 72). In the second step, these origins are activated by two sets of S-T protein kinases, an S-phase CDK (S-CDK) and the Dbf4-Cdc7 kinase (Dbf4p-dependent kinase [DDK]). How these two kinases trigger the initiation of DNA replication is currently not clear. One role of S-CDKs is to load Cdc45p, another protein needed for initiation, onto chromatin (48, 77). One or more additional steps catalyzed by S-CDKs and DDK then lead to recruitment of the single-stranded DNA binding protein RPA and the DNA polymerase–α-primase complex (48, 73).
Cyclin-CDK complexes are required for the initiation of DNA replication in all eukaryotes. In human, frog, and fly, cyclin E (cycE)-Cdk2, cycA-Cdk2, and cycA-Cdc2 kinases exhibit S-phase-promoting activity (40, 42, 68). In budding and fission yeast, this activity is conveyed by the B-type cyclin-CDK complexes Clb5p-Cdc28p, Clb6p-Cdc28p, and cig2-cdc2 (19, 22, 63). Activation of S-CDKs in late G1 phase is tightly regulated both transcriptionally and posttranslationally, in particular by specific CDK inhibitors like Sic1p and rum1 (46, 62). The destruction of these inhibitors in late G1, mediated by G1 CDKs and ubiquitin-dependent proteolysis, leads to rapid activation of S-CDKs and S-phase entry (3, 21, 62). Deletion of SIC1 or CLB5,6 advances or delays S-phase entry, respectively, indicating that CDKs are the prime temporal regulators of S phase within the cell cycle (61–63). Cells lacking CLB5, which rely on Clb6-Cdk1 for DNA replication, fail to activate late origins (17), suggesting that S-CDKs also act at the level of origins to regulate firing throughout S phase. This might occur via delayed deposition of Cdc45p onto late origins (1).
CDKs are required for but not sufficient to trigger DNA synthesis, which also involves the S-T kinase composed of the Cdc7p catalytic and Dbf4p regulatory moieties (DDK; reviewed in references 36 and 64). Structural and functional homologues of Cdc7p and Dbf4p have now been found in various eukaryotes, suggesting that DDK function is universally required for S-phase entry (6, 7, 34, 35, 43, 47, 60). Dbf4p binds to ARS1 by one-hybrid assay (18) and shows a punctate nuclear staining resembling replication foci (43, 52). Consistent with this localization, Cdc7p was shown to be required throughout the S phase for the firing of late origins (5, 16). Confirming earlier proposals (32, 75), recent reports show that Dbf4p is an unstable protein which peaks during S phase and confers kinase activity on Cdc7p (7, 10, 50).
Thus both S-CDKs and DDK seem to trigger the initiation of DNA replication at the level of origins. CDKs phosphorylate many initiation factors, but thus far a direct demonstration that these events are causal for initiation is still lacking. Proposed DDK substrates are less numerous, and Mcm2p is the leading candidate (6, 43, 44, 50). Surprisingly, a single recessive mutation in MCM5 (bob1-1) completely bypasses the requirement of Cdc7p and Dbf4p for S phase (27, 32). mcm5/bob1 cells lacking genes for Cdc7p or Dbf4p grow quite normally, suggesting that DDK might control initiation rather than be intrinsically required for it. Accordingly, Cdc7p is not essential for premeiotic S phase (30). It is not currently known why two kinases are needed for the initiation of DNA replication in the mitotic cycle, in which order they function, or what their critical molecular targets are. Here, we show that DDK activity is cell cycle regulated, although less tightly than that of the S-CDK Clb5-Cdk1. In spite of its moderate fluctuation, DDK activity is kept in check by both limited Cdc7p synthesis and high Dbf4p turnover. Increasing DDK activity by overexpression of both Cdc7p and Dbf4p is lethal. DDK is already active before S-CDKs are turned on, but, crucially, it cannot trigger DNA replication until after S-CDKs have been activated. Thus, DDK acts downstream of S-CDKs for the initiation of DNA replication. Cdc45p becomes tightly associated with origins after S-CDK activation (1, 77). Consistently, we find that DDK phosphorylates Cdc45p in vitro. Thus, our data support a double-trigger model for the initiation of DNA replication. S-CDKs which are abundant and tightly regulated would act globally to prime DNA replication on many origins. Then the limiting and origin-bound DDK would act locally, downstream of S-CDKs, to remove a block of firing at the level of individual origins. By acting sequentially, S-CDK and DDK would ensure that the initiation of DNA replication occurs both at the right time and place.
MATERIALS AND METHODS
Strains and media.
Yeast strains used in this study are listed in Table 1. Except for E1004, they are either congenic or backcrossed at least four times to W303 and they have been constructed using classical genetic techniques (37). Strains were grown either in YEP medium supplemented with 50 mg of adenine per liter and 2% dextrose (YEPD), raffinose (YEPRaf), or galactose (YEPGal) or in supplemented minimal medium, as described previously (37). Plates contained 2% agar (Difco). Yeast transformation was performed by the lithium acetate method.
TABLE 1.
Strain list
| Strain | Genotype | Source or reference |
|---|---|---|
| E001 | MATa ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3-1 GAL | W303-1a |
| E171 | MATα cdc4-1 HA6-CLB5 | 62 |
| E533 | MATa DBF4-myc18/LEU2 | M. Shirayama |
| E539 | MATa cdc4-1 DBF4-myc18/LEU2 | E171 × E533 |
| E540 | MATα cdc15-2 DBF4-myc18/LEU2 | This study |
| E542 | MATa cdc28-4 HA6-CLB5 DBF4-myc18/LEU2 | This study |
| E550 | MATα TRP1 cdc7-1 DBF4-myc18/LEU2 | This study |
| E552 | MATα cdc23-1 DBF4-myc18/LEU2 | This study |
| E633 | MATa dbf4::HIS3 URA3::GAL-DBF4 | This study |
| E639 | MATa cdc7-1 URA3::GAL-SIC1(7x) TRP1 | This study |
| E645 | MATa URA3::GAL-DBF4-myc6 YCplac22/Tet-CDC7 | This study |
| E784 | MATa cdc4-1 dbf4::HIS3 URA3::GAL-DBF4 | This study |
| E793 | MATa YCplac22/Tet-CDC7 YCplac33 | This study |
| E794 | MATa YEplac195/Tet-DBF4-myc6 YCplac22 | This study |
| E795 | MATa YCplac22 YCplac33 | This study |
| E796 | MATa YEplac195/Tet-DBF4-myc6 YCplac22/Tet-CDC7 | This study |
| E809 | MATa bar1::hisG YEplac195/Tet-DBF4-myc6 | This study |
| E810 | MATa apc1-1 bar1::hisG YEplac195/Tet-DBF4-myc6 | This study |
| E935 | MATa TRP1 cdc7-1 HA6-CLB5 | E171 |
| E992 | MATa bar1::hisG URA3::GAL-CLB2 DBF4-myc18/LEU2 | This study |
| E994 | MATa cdc23-1 URA3::GAL-CLB2 DBF4-myc18/LEU2 | This study |
| E1004a | MATa bar1 bob1-1 cdc7::HIS3 | 27 |
Not congenic with W303.
Plasmid and strain constructions.
Standard procedures were used for DNA manipulations (58). PCR cloning was performed with Pwo (Boehringer) or Vent (New England Biolabs) DNA polymerases. The Tet-CDC7 plasmid (D577) was constructed by cloning CDC7 into pCM189 (24) and transferring tTA-tetO7-CDC7 into YCplac22 (25). Tet-DBF4-myc6 (D485) was constructed by cloning DBF4 into pCM190/YEplac195 (24), followed by insertion of a myc6 cassette before the stop codon. The same PCR product was cloned behind the GAL1-10 promoter in YIplac211 to produce GAL-DBF4 (D537). A myc6-NotI cassette was then inserted at the C terminus to produce GAL-DBF4-myc6 (D479). To produce Mcm2p in Escherichia coli, MCM2 was cloned in a pET11d-derived vector (70) containing six histidine codons behind the AUG, yielding plasmid D468. The plasmid expressing His6-Cdc45p (D692) was constructed similarly. DBF4 was disrupted in a diploid strain (E622) by replacing a 1.8-kb BglII-BspE1 fragment with HIS3. This strain was then transformed with D537 and sporulated, and His+ Ura+ spores were obtained by tetrad dissection to yield strain E633.
Cell synchronizations.
Centrifugal elutriations were performed as described previously (63). For synchronization in late G1 by pheromone, MATa cells (4 × 106/ml) were arrested for 2 to 3 h at 25°C, with 1 μg of α-factor per ml (0.1 μg/ml for bar1 strains) added twice to the medium at 1.5-h intervals, and released either by filtration and washing or by the addition of pronase (50 μg/ml; Calbiochem). Nocodazole arrest was performed for 2.5 h, with nocodazole at a concentration of 30 μg/ml in 1% dimethyl sulfoxide, unless stated otherwise. Cell cycle progression was monitored by flow cytometry (FACScan) according to a method described previously (19).
RNA analysis.
Total RNA isolation and Northern analysis were performed as described previously (11), with 10 μg of RNA per lane. Probes were prepared by random priming of PCR products of DBF4 (positions −298 to +2112), CLB5 (positions +1 to +1328), or CMD1 (positions +1 to +442).
Bacterial protein expression and antibody production.
Cdc7p was produced in Escherichia coli BL21-DE3 containing pET3a-CDC7 (D433) and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 37°C (70). Inclusion bodies were purified, resuspended in Laemmli buffer, boiled, and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The band containing Cdc7p was used to immunize rabbits. Polyclonal antibodies were affinity-purified on polyvinylidene difluoride (PVDF) strips containing Cdc7p as described previously (58). His6-Mcm2p and His6-Cdc45p were produced in E. coli BL21-DE3 transformed with D468 and D692, respectively. Cells (volume, 500 ml; A600 = 0.8) were induced with 1 mM IPTG for 60 min at 25°C, washed with cold phosphate-buffered saline (PBS), and resuspended in 20 ml of RJD buffer (10 mM HEPES [pH 7.6], 50 mM NaCl, 5 mM MgCl2, 15% glycerol, 0.05% NP-40) supplemented with Complete protease inhibitor mix (Boehringer). This suspension was frozen in liquid N2, thawed at 37°C, and sonicated five times on ice for 30 s each time, at 2-min intervals. The lysate was clarified at 10,000 × g for 10 min at 4°C. A 50% slurry of Talon metal affinity resin (0.8 ml; Clontech) was added to the supernatant for 90 min at 4°C. Bound proteins were washed with RJD buffer plus 4 mM imidazole for Mcm2p or 1 mM imidazole for Cdc45p, and were eluted with 1 ml of RJD buffer plus 300 mM imidazole.
SDS-PAGE and Western blotting.
Protein extracts for Western blotting were prepared from mid-log-phase cultures (108 cells) that were fixed by adding trichloroacetic acid (TCA) (final concentration, 10%) directly to the medium, harvested by centrifugation, and washed once in 10% TCA. Cell pellets were frozen in liquid N2 and stored at −70°C. Yeast pellets were thawed on ice and resuspended in 200 μl of 10% TCA. Zirconium beads (1 volume; diameter, 0.5 mm; Biospec Products Inc.) were added, and the cells were broken by vigorous vortexing for 10 min on a Vibrax (VXR; Ika Laboratories) multimixer. Total cell lysates were transferred to new tubes, and the beads were washed twice with 200-μl volumes of 10% TCA. Extracts were pooled, and proteins were precipitated by centrifugation (10 min at 1,000 × g). Pellets were resuspended in Laemmli loading dye buffered with 0.17 M Tris base, boiled for 10 min, and spun for 10 min at 1,000 × g to remove insoluble material. Protein concentrations were determined using the Bradford protein assay (Sigma). Routinely, 20 to 100 μg of proteins were loaded onto an SDS–8% PAGE gel and blotted semidry (3 h at 1 mA/cm2 in 39 mM glycine, 48 mM Tris base, 0.1% SDS, 20% methanol) onto a PVDF membrane (Millipore). After transfer, proteins were revealed with 0.2% amido-black. Immunological detection was performed essentially as described previously (58). Membranes blocked in PBS, 0.1% Tween, and 3% nonfat milk were incubated with antibodies in the same solution for 1 h at room temperature. Antibodies were used at the following dilutions: mouse anti-Myc (9E10), 1/2,000; anti-polyhistidide (Sigma), 1/2,500; rabbit polyclonal anti-Cdc7p (affinity-purified), 1/100; anti-Sic1p, 1/5,000; anti-Swi6p, 1/100,000; anti-Clb2p, 1/2,000; secondary antibodies coupled to horseradish peroxidase (Sigma), 1/5,000 (anti-mouse) or 1/10,000 (anti-rabbit).
Protein extraction and immunoprecipitations.
Mid-log-phase cells (108 cells) were harvested by centrifugation, washed once in ice-cold Stop mix buffer (0.9% NaCl, 1 mM NaN3, 10 mM EDTA, 50 mM NaF), frozen in liquid N2, and stored at −70°C. Cell pellets were thawed and resuspended in 2 volumes of ice-cold breaking buffer (50 mM Tris-Cl [pH 7.5], 15 mM MgCl2, 200 mM NaCl, 1% NP-40, 1 mM EDTA, 1 mM dithiothreitol) supplemented with an inhibitor cocktail (60 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, 20 μg of leupeptin per ml, 40 μg of aprotinin per ml, 0.1 mM Na-orthovanadate, 15 mM paranitrophenylphosphate, 5 mM EGTA). After addition of zirconium beads (1 volume), cells were lysed by vortexing at 4°C for 8 min on a multimixer. Extracts were clarified twice by centrifugation at 15,000 × g for 10 min at 4°C. Unless stated otherwise, immunoprecipitations (IPs) were performed on 0.1 mg of proteins for 1 h at 4°C on a rotating wheel with antibodies (anti-Myc or anti-Cdc7p) diluted to 1/10, in a total volume of 30 μl. Immune complexes were then precipitated by incubation for 1 h at 4°C with 16 μl of protein A-Sepharose beads (50% slurry; Pharmacia) preblocked with 10 mg of bovine serum albumin per ml. Beads were recovered by spinning for 1 min at 500 × g and washed five times with 0.5 ml of breaking buffer.
Cdc7p protein kinase assay.
Anti-Myc or anti-Cdc7p immune complexes were washed twice with 0.5 ml of 25 mM morpholinepropanesulfonic acid (MOPS) (pH 7.2) to remove detergents. An 8-μl volume of kinase buffer(25 mM MOPS [pH 7.2], 15 mM MgCl2, 5 mM EGTA, 5 μCi of [γ-32P]ATP at a specific activity of 5,000 Ci/mmol, with or without 10 μM ATP) and 0.2 μg of the substrate were added to the beads. The reaction mixture (total volume, 30 μl) was then incubated at room temperature with slight agitation for 30 min, and the reaction was terminated by the addition of 14 μl of 3× Laemmli loading buffer. After boiling, phosphoproteins were resolved by SDS-PAGE, either fixed and dried or transferred to PVDF membranes, and revealed by autoradiography. Equal amounts of Cdc7p were present in the immune complexes, and the assay was linear with respect to enzyme level (data not shown).
RESULTS
Assay to measure endogenous Dbf4-Cdc7 kinase activity.
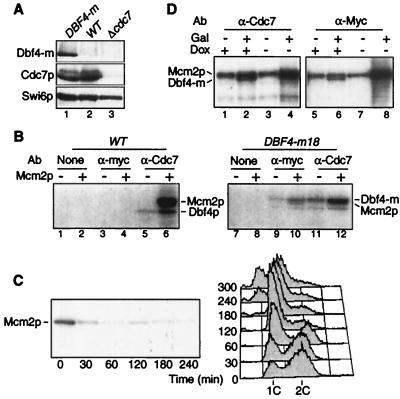
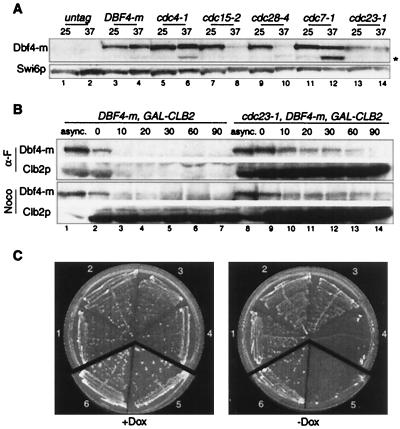
To study the regulation of DDK during the cell cycle, we devised an IP kinase assay able to detect endogenous Dbf4-Cdc7 kinase activity. The assay uses either an anti-Myc monoclonal antibody (MAb) (9E10) against Dbf4p tagged at its C-terminus with 6 or 18 myc epitopes (Dbf4-m), or an affinity-purified polyclonal antibody directed against Cdc7p. Each antibody recognizes a single band at the expected size in Western blots of whole-cell extracts (Fig. 1A, lane 1). These bands correspond to Dbf4-m and Cdc7p, as they are absent in extracts from untagged and cdc7Δ bob1 strains, respectively, and stronger in overproducing strains (Fig. 1A, lanes 2 and 3, and data not shown). Tagged strains which contain a single copy of DBF4-m integrated at the DBF4 locus grow as wild-type strains do by the criteria of morphology, doubling time, and fluorescent-activated cell scanner (FACS) profile (data not shown), suggesting that the Dbf4-m protein is fully functional.
FIG. 1.
Dbf4-Cdc7 kinase assay. (A) Western blot of whole-cell extracts from myc-tagged DBF4-m (E533) (lane 1), wild-type (W303) (lane 2) or cdc7Δ bob1-1 (E1004) (lane 3) strains probed with anti-Myc (9E10), anti-Cdc7p and anti-Swi6p (as a loading control) antibodies. (B) Kinase assay on immunoprecipitates from wild-type (W303) (lanes 1 to 6) or DBF4-m (E533) (lanes 7 to 12) strains using either anti-Myc or anti-Cdc7p antibodies. Phosphorylation of endogenous Dbf4p and recombinant Mcm2p (0.2 μg, even lanes) was monitored. Lanes 1, 2, 7, and 8 are controls with no antibody. (C) Dbf4-Cdc7 kinase activity depends on Dbf4p. dbf4Δ GAL-DBF4 cells (E633) grown on YEPGal medium were transferred at time zero to YEPD medium to turn off DBF4 synthesis. Dbf4-Cdc7 associated kinase activity (blot) and cell cycle distribution by FACS (graph) were analyzed at various times thereafter. (D) Overexpression of Dbf4p and Cdc7p in vivo increases kinase activity. pGAL-DBF4-m6 pTet-CDC7 cells (E645) grown on YEPRaf plus doxycycline (Dox) (10 μg/ml) medium were transferred for 2 h to medium containing 2% galactose and/or lacking doxycycline to induce synthesis of Dbf4p and/or Cdc7p, respectively. Kinase activity was measured using anti-Cdc7 (lanes 1 to 4) or anti-Myc (lanes 5 to 8) antibodies. WT, wild type; Ab, antibody.
To test for kinase activity, we incubated extracts from untagged or DBF4-m cells with the anti-Myc or anti-Cdc7p antibodies, precipitated the immune complexes with protein A-Sepharose and monitored the phosphorylation of recombinant Mcm2p, a known substrate of the Dbf4-Cdc7 kinase (44). Anti-Cdc7p immunoprecipitates showed kinase activity towards endogenous Dbf4p and added Mcm2p (Fig. 1B, lanes 5, 6, 11, and 12). The strongest band corresponded to Mcm2p, as the band is absent when Mcm2p is omitted (compare Fig. 1B, lanes 5 and 6). The weaker band corresponded to cellular Dbf4p, as it demonstrated lower mobility in the DBF4-m strain (Fig. 1B, lanes 5 and 11). While anti-Cdc7p immunoprecipitates showed kinase activity in extracts from both strains, the anti-Myc immunoprecipitates showed kinase activity only on the DBF4-m strain (Fig. 1B, lanes 3, 4, 9, and 10). This kinase also phosphorylated Mcm2p and Dbf4-m, but in this case the two substrates almost comigrated due to the 18 myc epitopes on Dbf4-m. No signal was detected when antibodies were omitted (Fig. 1B, lanes 1 and 2 and 7 and 8) indicating that the kinase does not bind nonspecifically to protein A beads. Thus our assay is specific for endogenous Dbf4-Cdc7 kinase.
To strengthen this point even further, we inactivated Dbf4-Cdc7 kinase in vivo using temperature-sensitive (ts) dbf4-1 and cdc7-1 strains. No phosphorylation of Dbf4p or Mcm2p was detected in extracts from either ts strain when the reaction was performed at 37°C (see Fig. 4B and 7), indicating that kinase activity requires wild-type DBF4 and CDC7.
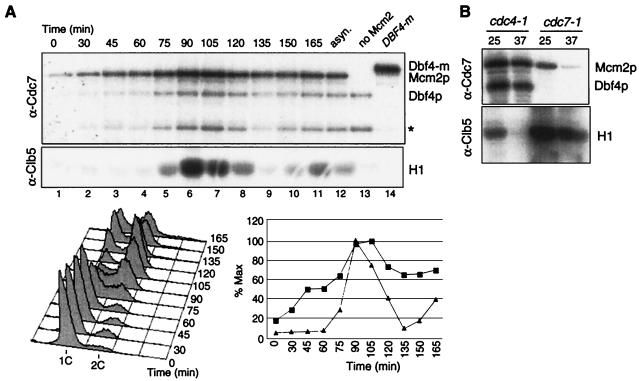
FIG. 4.
Clb5-Cdk1 and Dbf4-Cdc7 kinases are activated independently. (A) Cdc7p- and Clb5p-associated kinase activities were measured during a synchronous culture of wild-type cells (W303) synchronized by centrifugal elutriation. Mcm2p was used as a substrate in the Dbf4-Cdc7 kinase assay (except for the last two lanes, where it was omitted) and histone H1 was used in the Clb5-Cdk1 assay. Asterisk, unidentified phosphoprotein. Cell cycle progression and quantitation of kinase activities (squares, Dbf4-Cdc7; triangles, Clb5-Cdk1) are indicated in the graph. (B) Dbf4-Cdc7 and Clb5-Cdk1 kinases are active in cdc4- and cdc7-arrested cells, respectively. cdc4-1 (E171) and cdc7-1 (E935) cells containing HA6-CLB5 were grown in YEPD medium at 25°C and arrested for 3 h at 37°C. Cdc7p- (top) or Clb5p-associated proteins (bottom) were immunoprecipitated using anti-Cdc7p or anti-hemagglutinin (12CA5) antibodies, respectively, and used for kinase assay with Mcm2p or histone H1 as substrates.
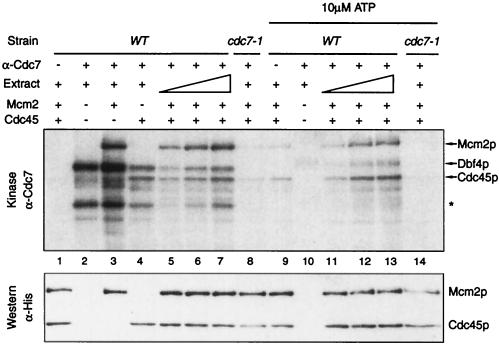
FIG. 7.
Cdc45p is phosphorylated by Dbf4-Cdc7 in vitro. Extracts from wild type (E001) or cdc7-1 (E935) cells grown at the permissive temperature were incubated with anti-Cdc7p antibodies, immunoprecipitated, and used in kinase assays (at 37°C) in the absence (lanes 1 to 8) or presence (lanes 9 to 14) of excess cold ATP. IPs were performed from 100 μg or 50 to 200 μg (lanes 5 to 7 and 11 to 13) of total extract. Recombinant His6-Mcm2p and/or His6-Cdc45p substrates were added as indicated. Lanes 1 and 9 are control IPs with no antibody added. All reactions were performed at 37°C. Asterisk, an unidentified phosphoprotein that might correspond to Cdc7p. After autoradiography, the membrane was probed with anti-polyhistidine antibodies (bottom panel) to reveal the presence of Mcm2p and Cdc45p, which comigrate with radioactive bands.
Dbf4-Cdc7 kinase is regulated by Dbf4p levels.
Akin to cyclins for CDKs, Dbf4p is required to activate the Cdc7p catalytic subunit (7, 32, 50). If Dbf4p were an unstable protein, another means to inactivate the Dbf4-Cdc7 kinase would be to deplete Dbf4p by shutting off its synthesis. Therefore, we constructed a strain in which DBF4 was deleted but kept alive by a pGAL-DBF4 construct integrated at the URA3 locus. This strain proliferates in galactose-containing medium (GAL promoter on) but dies on glucose (GAL promoter off). When cells growing asynchronously in YEPGal medium were transferred to glucose, Dbf4-Cdc7-associated kinase activity dropped to background levels within 30 min (Fig. 1C). This demonstrates that Dbf4p is an unstable activator of Cdc7p with a half-life of less than 30 min. The decrease of kinase activity in vitro was followed by an accumulation of cells with 1C DNA (Fig. 1C), indicating that Dbf4-Cdc7 activity also dropped in vivo and that initiation of DNA replication was impaired. Strikingly, at 4 h after glucose addition many cells showed less than 1C DNA, suggesting that they underwent a reductional anaphase followed by cell division, without replicating their DNA. This resembles Cdc6p depletion (54) and is indicative of a complete failure both to initiate DNA replication and to refrain from the segregation of unreplicated chromosomes (34). In the case of Cdc6p, this is due to the lack of pre-RC formation, but in the case of Dbf4p it is probably because pre-RCs do not fire. This suggests that specific replication structures, and not pre-RCs, refrain from mitosis when DNA replication is incomplete (45).
Thus, depleting Dbf4p in vivo decreases Mcm2p phosphorylation in vitro. To see if the converse is true, i.e., if Cdc7p and/or Dbf4p overexpression leads to an increase in kinase activity, we constructed a strain containing an integrated copy of GAL-DBF4m as well as a centromeric plasmid with CDC7 under the control of a tetracycline-regulatable promoter (24). Because these constructs can be induced independently of each other by galactose addition or doxycycline removal, respectively, we could test the effects of Dbf4p and/or Cdc7p overexpression in the same strain. Figure 1D shows that Dbf4p or Cdc7p induction alone did not increase Mcm2p phosphorylation. In contrast, when both subunits were coexpressed, Dbf4-Cdc7 kinase activity on Mcm2p increased 3.5-fold (Fig. 1D, lanes 4 and 8 and data not shown). This differs from Cdk1 and cyclins (where overexpression of the latter alone causes hyperactivation) and suggests that neither Dbf4p nor Cdc7p is present in large excess over its partner in cycling cells.
Dbf4-Cdc7 kinase activity is cell cycle regulated. (i) Northern analysis.
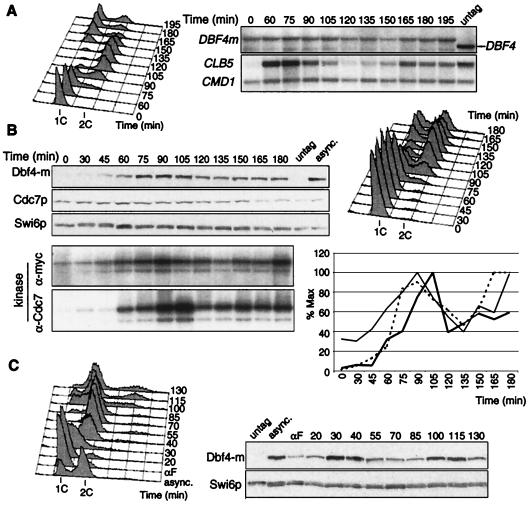
Having designed a reliable assay, we set out to analyze Dbf4-Cdc7 kinase regulation during the cell cycle. As transcriptional regulation of cyclins closely mirrors their associated CDK activity, we first analyzed DBF4 mRNA levels in a synchronous culture of DBF4m cells. Early G1 cells obtained by centrifugal elutriation were inoculated in fresh medium, and aliquots were taken at regular intervals for Northern and FACS analysis. DNA replication began at 60 min and was finished by 105 min; cell division was completed by 165 min (Fig. 2A). The strong fluctuation of CLB5 mRNA levels, which peaked at the initiation of DNA replication and plummeted in G2, denoted the good synchrony of the culture. In contrast, DBF4 mRNA was present throughout the cell cycle, with only a very slight increase at G1-S relative to the CMD1 mRNA used as loading control. The myc-coding sequence did not alter DBF4 mRNA synthesis or stability, as similar results were obtained with an untagged strain (data not shown). These data indicate that DBF4 is not a G1-S-specific transcript, in contrast to previous suggestions (9). CDC7 transcripts do not fluctuate (65; data not shown).
FIG. 2.
Cell cycle regulation of Dbf4-Cdc7 kinase activity. (A) Northern blot analysis of synchronous DBF4-m cells (E533) obtained by centrifugal elutriation. The filter was probed with DBF4 and reprobed with CLB5 and CMD1 as synchrony and loading controls, respectively. The rightmost lane (untag) is RNA from exponentially growing wild-type cells, showing probe specificity. Cell cycle progression is indicated by FACS analysis (left). (B) Western blot (top) and Dbf4-Cdc7 kinase activity (bottom) of DBF4-m cells (E533) synchronized by centrifugal elutriation. Whole-cell extracts from TCA-fixed cells were analyzed by Western blot using anti-Myc, anti-Cdc7p or anti-Swi6p antibodies. Dbf4-m and Dbf4-Cdc7-associated kinase activities were measured on Dbf4p. The graph shows quantitation of total Dbf4p normalized against Swi6p (dotted line) and of kinase activities using anti-Myc (thin line) or anti-Cdc7p (bold line) antibodies. (C) Dbf4p fluctuation after pheromone release. DBF4-m cells (E533) were arrested by α-factor for 2 h at 25°C, washed, and released into YEPD medium at 30°C. Swi6p, loading control. untag, untagged strain; asynch., asynchronous culture.
(ii) Western and kinase analysis.
To see whether Dbf4-Cdc7 kinase is regulated posttranscriptionally, we performed a similar experiment but monitored Dbf4p and Cdc7p levels as well as their associated kinase activities. Figure 2B shows that Cdc7p was present at constant levels throughout the cell cycle. In contrast, Dbf4p was rare in early G1 cells, and its levels rose in late G1 (60 min), peaked during S phase (90 min) and decreased thereafter (135 min). The low levels of Dbf4p in early G1 cells, despite the presence of its mRNA, indicate that cell cycle-regulated mechanisms, either translational or proteolytic, operate to limit Dbf4p levels in G1. Dbf4p- and Cdc7p-associated kinase activities closely mirrored Dbf4p levels. Elutriated early G1 cells showed little Dbf4-Cdc7 kinase activity, which increases in late G1, reaches a maximum during S phase and decreases moderately during mitosis. We conclude that Dbf4-Cdc7 kinase activity is cell cycle regulated, though less tightly than that of Clb5- or Clb2-associated kinase (see Fig. 4A), most likely via its association with Dbf4p.
To confirm Dbf4p's cell cycle fluctuation using a different synchronization protocol, we released DBF4m cells from α-factor arrest. Dbf4p levels were low but detectable in the pheromone-arrested cells, peaked during S phase (30 to 40 min after release) and decreased in G2-M (Fig. 2C). Taken together with the promoter shutoff and Northern experiments (Fig. 1C and 2A), these data are consistent with the notion that Dbf4p is synthesized throughout the cell cycle but is more stable during S phase.
Posttranscriptional regulation of Dbf4p.
To test the idea that Dbf4p is more unstable at certain cell cycle stages, we analyzed Dbf4-m levels in cells arrested by various cdc mutations (Fig. 3A). The Dbf4-m protein was detected in every tagged strain at a permissive temperature (25°C), but at 37°C only in the wild-type, cdc4, cdc7, and cdc23 strains. Little or no Dbf4p was seen in cdc28-4- and cdc15-2-arrested cells. This suggests that Dbf4p is less stable in pre-Start G1 and in postanaphase cells. We also noticed that Dbf4p tends to break down rapidly, particularly when cells are incubated at 37°C. This problem was partly circumvented by rapid TCA fixation of the cells in the culture medium.
FIG. 3.
APC-dependent and -independent proteolysis of Dbf4p. (A) Dbf4p steady-state levels in wild-type or various cdc mutants growing at 25°C or arrested for 2.5 h at 37°C. Lanes: 1 and 2, wild type (untag) (W303); 3 and 4, DBF4-m (E533); 5 and 6, cdc4-1 (E539); 7 and 8, cdc15-2 (E540), 9 and 10, cdc28-4 (E542); 11 and 12, cdc7-1 (E550); 13 and 14, cdc23-1 (E552). The asterisk indicates cleavage of Dbf4p. (B) Half-life of endogenous Dbf4p and GAL-induced Clb2p in wild-type (E992) and cdc23-1 (E994) cells arrested in G1 or in G2-M. Cells growing in YEPRaf medium at 25°C (async.) were arrested by α-factor (α-F) or nocodazole (Noco). After 2 h, galactose (1%) was added to induce Clb2p synthesis, first at 25°C (30 min) and then at 37°C (30 min), during which APC was inactivated (in the cdc23-1 strain). Glucose (1%) and cycloheximide (10 μg/ml) were added at time zero to all cultures (which remained arrested and at 37°C) to repress CLB2 transcription and general protein synthesis, respectively. The fate of Dbf4-m and Clb2p in the same cells following cycloheximide addition was monitored by Western blot using anti-Myc and anti-Clb2p antibodies. (C) Joint overexpression of Dbf4p and Cdc7p is lethal in wild-type cells, while Dbf4p overexpression is lethal in an apc1-1 strain. Wild type cells containing Tet-CDC7 (streak 1, E793), Tet-DBF4-m (2, E794), both plasmids together (4, E796) or empty vectors (3, E795) were streaked on selective plates containing or lacking doxycycline (Dox) and incubated for 4 days at 25°C. Similarly, wild type (streak 6, E809) or apc1-1 (streak 5, E810) cells containing the Tet-DBF4-m plasmid were grown on plates with or without doxycycline (2 μg/ml). Additional material and quantitation of the data can be found at our web site (www.igm.cnrs-mop.fr/schwob.html).
Most cell cycle-regulated proteolysis in yeast is accounted for by two ubiquitin ligases, SCF and anaphase-promoting complex-cyclosome (APC/C) (53). SCFCdc4 targets several G1/S regulators for destruction, including the CDK inhibitor Sic1p and the initiation factor Cdc6p. The accumulation of Dbf4p in cdc4-arrested cells could indicate that it is a target of SCFCdc4, or that Dbf4p is stable at this particular cell cycle stage. We favor the latter hypothesis because Dbf4p also accumulates in cells blocked in late G1 by expression of a nondegradable Sic1p (data not shown). The accumulation of Dbf4p in cdc7- or hydroxyurea-arrested cells, in which APC is inactive, suggests instead that it might be an APC substrate. To test APC's contribution directly, we compared the decay of endogenous Dbf4-m after protein synthesis shutoff in wild-type cells and cdc23 mutants arrested either in G1 (α-factor) or G2 (nocodazole). If Dbf4p proteolysis depends on APC, then its levels should decrease less rapidly in cdc23-1 mutant cells. Moreover, Dbf4p should be stable in nocodazole-arrested cells in which APC is inactive. As an internal control for APC's inactivation, we used a strain that also expresses Clb2p, a well-known APC substrate, from the GAL promoter. Wild-type and mutant cells were arrested for 2 h in α-factor or nocodazole at 25°C, after which galactose was added and the cultures were shifted to 37°C to induce Clb2p and inactivate APC. After 30 min at 37°C, glucose and cycloheximide were added to repress CLB2 transcription and global protein synthesis, respectively. The amount of Dbf4p in whole-cell extracts at different times after the addition of cycloheximide was determined by Western blot analysis, and half-lives were measured by densitometry. Dbf4p is highly unstable in G1 (half-life of 5 min) and stabilized by inactivation of Cdc23p (Fig. 3B and data not shown). Thus, APC contributes to Dbf4p instability. However, while Clb2p levels do not decrease for the entire time course, Dbf4p still decays in the cdc23 mutant (15-min half-life), suggesting that Dbf4p is also degraded independently of APC. The latter was confirmed with nocodazole, where, in stark contrast to Clb2p, which is stable, Dbf4p levels still decreased, for both wild-type and mutant strains. These results, seen on physiological levels, clearly establish that while APC contributes to Dbf4p turnover, it cannot solely account for it. SCFCdc4 is not responsible for this residual instability (data not shown), and further investigation will be needed to define its nature.
Combined Dbf4p and Cdc7p overexpression is lethal.
One way to address the biological significance of Dbf4p's instability during the cell cycle is to see whether its stabilization or overexpression might have any effect on cell division. To this end, cells were transformed with plasmids expressing either DBF4 or CDC7 alone, or both simultaneously from a tetracycline-regulatable promoter. Figure 3C shows that cells overexpressing each protein individually formed colonies like the wild type, but that cells overproducing both Cdc7p and Dbf4p were unable to form colonies on selective plates. In liquid cultures, Dbf4-Cdc7 kinase activity on Mcm2p increased threefold during the first 4 to 6 h of induction and slowly decreased afterwards (data not shown). The inhibition of cell proliferation was maximal after 8 to 12 h of derepression and due, at least in part, to loss of the Tet-DBF4 and Tet-CDC7 plasmids. Currently, we have no explanation for this lethality, which needs to be confirmed with integrated constructs. Toxicity was observed in wild-type cells only when both subunits were coexpressed, suggesting that it might be due to an excess of bona fide Dbf4-Cdc7 kinase activity. Consistent with Dbf4p being an APC substrate, Dbf4p overexpression alone was lethal at 25°C in an apc1-1 mutant (Fig. 3C). This suggests that high Dbf4p levels can also have adverse effects independently of Cdc7p overexpression.
S-CDK and DDK are activated independently.
Both Dbf4-Cdc7 and Clb-Cdk1 kinases are required for the initiation of DNA replication, but it is not clear whether one kinase is needed to activate the other. In a first attempt to address this question, we compared the timing of activation of both kinases in a synchronous culture of elutriated wild-type cells. Clb5p and Cdc7p-associated proteins were precipitated using specific antibodies, and their kinase activities were measured using histone H1 and Mcm2p, respectively, as the substrates (Fig. 4A). Clb5-Cdk1 kinase activity was tightly cell cycle regulated, with a sharp increase observable at the time of S phase (75 to 105 min) and strong downregulation in M-G1 (135 to 150 min). Like that of Clb5-Cdk1, Dbf4-Cdc7 kinase activity peaked during S phase, but its regulation was less stringent. Phosphorylation of Mcm2p by Dbf4-Cdc7 increased gradually during G1 (30 to 60 min) and decreased somewhat in G2-M (120 to 135 min). Thus, regulation of Dbf4-Cdc7 kinase activity cannot account for its precise, late-G1 execution point. Importantly, Dbf4-Cdc7 kinase was already active before Clb5-Cdk1 activity was detectable, suggesting that the latter is not needed to activate the former.
To confirm this point, we measured Clb5- and Cdc7-associated kinase activities in cdc4- and cdc7-arrested cells. The former arrest at the G1-S boundary due to persistence of the Clb-Cdk1 inhibitor Sic1p (62). As expected, Clb5-Cdk1 and Dbf4-Cdc7 activities were lost in the cdc4 and cdc7 arrests, respectively. However the reciprocal was not true. Clb5-Cdk1 is active in the cdc7 block, and Dbf4-Cdc7 kinase is active in cdc4-arrested cells (Fig. 4B). Thus, neither kinase requires the other for its activation. The fact that Clb5-Cdk1 kinase is active in cdc7-arrested cells was expected because such cells form round buds and a mitotic spindle that necessitate Clb-Cdk1 activity (8, 63, 71). Dbf4-Cdc7 kinase activity in cdc4-arrested cells was more controversial (32, 75). Our observation that Dbf4-Cdc7 kinase is active in cells lacking Clb-Cdk1 activity was confirmed using anti-Myc antibodies in a DBF4-m cdc4 strain, as well as in cells arrested at the G1-S boundary with a dominant GAL-SIC1 construct (data not shown). In all cases, we found high levels of Dbf4p and associated kinase activity. We conclude that S-CDKs and DDK are activated in late G1 independently of each other.
Clb5-Cdk1 function is independent of Dbf4-Cdc7 activity.
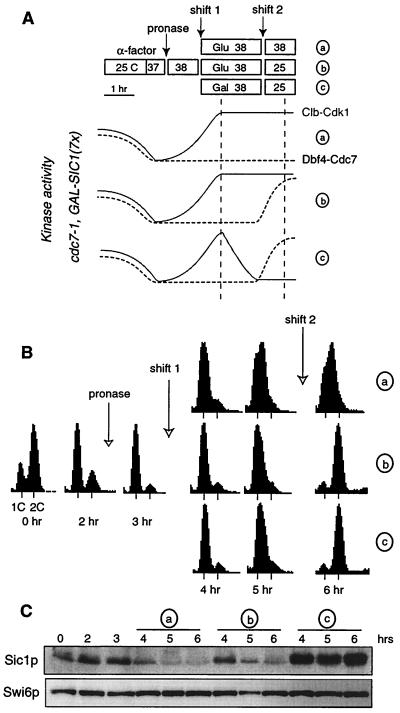
Despite their independent activation, these two kinases may still function sequentially for the initiation of DNA replication. The order between two steps in a pathway can be addressed in vivo by the method of reciprocal shifts (28). Briefly, if two activities A (e.g., Clb5-Cdk1) and B (e.g., Dbf4-Cdc7) are connected to each other in a dependent sequence A→B, then it should be possible to complete step A without prior activation of B, but not step B without prior activation of A. In a first experiment, we tested if Clb5-Cdk1 was able to perform its function while Dbf4-Cdc7 was kept inactive (Fig. 5A). This was done by scoring DNA replication after reactivation of Dbf4-Cdc7 in conditions restrictive for Clb-Cdk1 (a shift from cdc7− CLB+ to CDC7+ clb−). One kinase was inactivated using a ts allele (cdc7-1), while the other was turned off by expression of the Clb-Cdk1 inhibitor Sic1p under the control of the GAL promoter (seven integrated copies). To allow for Sic1p accumulation, its induction preceded Dbf4-Cdc7 reactivation. Specifically, cdc7-1 GAL-SIC1(7×) cells were presynchronized for 2 h by α-factor in YEPRaf medium (GAL promoter off) and then released at 38°C by the addition of pronase, which degrades α-factor. After 1 h, budded cells began to appear, indicating that cells progressed towards the cdc7 arrest point. At this time the culture was split in three parts and maintained at 38°C for a further 2 h to ensure that most, if not all, cells reached a cdc7− CLB+ status. Glucose was added to the first two cultures (designated a and b) (Fig. 5A), while galactose was added in the third (designated c) to induce Sic1p. By the end of these 2 h, Sic1p levels in the third culture were equivalent to those in cdc4-arrested cells and therefore sufficient to inactivate Clb-Cdk1 kinases completely (Fig. 5C). Cultures b and c were then shifted to 25°C to reactivate Dbf4-Cdc7, while the first was kept at 38°C to show that cells did not replicate without Dbf4-Cdc7 reactivation (Fig. 5B). In contrast, the shift to 25°C led to full replication within 1 h, indicating reversibility of the cdc7 arrest (culture b). Notably, the cells in galactose (culture c) also replicated their DNA despite inhibitory levels of Sic1p at the time of the shift. This increase in DNA content is not seen if only one kinase is activated (data not shown), and therefore represents bona fide DNA replication. Thus, the requirement of Clb5-Cdk1 for DNA replication had been fulfilled while Dbf4-Cdc7 was inactive and was no longer necessary after Dbf4-Cdc7 reactivation.
FIG. 5.
Clb5-Cdk1 performs its function for DNA replication independently of Dbf4-Cdc7. (A) Schematic outlay of the cdc7− Clb+→clb− Cdc7+ reciprocal shift experiment. cdc7-1 GAL-SIC1(7x) cells (E639) were presynchronized with α-factor as indicated, released at 38°C to inactivate Cdc7p, and then either maintained arrested (a), shifted to 25°C to reactivate Dbf4-Cdc7 (b), or shifted to 25°C after Clb5-Cdk1 was inactivated by Sic1p induction (c). Cells were preincubated for 30 min at 37°C before α-factor release to anticipate the heat-shock response that would otherwise delay Sic1p degradation (57). Kinase activities are depicted as solid (Clb-Cdk1) and dotted (Dbf4-Cdc7) curves. Vertical lines materialize the shift in activities. (B) FACS profiles indicate that cultures b and c replicate their DNA with similar kinetics. The slight rightward shift of the 1C peak in culture a is likely due to cell enlargement or cdc7-1 leakiness but not to bona fide replication. (C) Western blot analysis of Sic1p and Swi6p (as loading control). Additional controls cited as data not shown can be found at our web site (www.igm.cnrs-mop.fr/schwob.html).
Despite the technical difficulties of taking cells rapidly through a cln−→CLB+→clb− regimen, two lines of evidence suggest that the appropriate conditions were met. First, cells eventually replicated their DNA, indicating that Clb-Cdk1 kinases were active at some point of the time course and, moreover, that the event they triggered was not reversed during their subsequent inactivation. Second, the vast majority of cells show elongated, cdc4-like buds after 2 h in the galactose- but not in the glucose-containing medium (data not shown). This strongly suggests that Clb-Cdk1 kinases were inactive at the time of the shift at 25°C. It is still possible that low levels of Clb-Cdk1 kinases triggered DNA replication, but we find this unlikely because S phase was equally efficient after the downshift regardless of Sic1p induction. To circumvent the problems linked to slow Sic1p degradation and poor induction from the GAL promoter at 38°C, we have since repeated the experiment using a dbf4Δ Tet-DBF4 GAL-SIC1(4×) strain. Cells were first arrested in a cdc7− state by depleting Dbf4p with doxycycline for 3 h at 25°C; then galactose was added, and the doxycycline was removed to inhibit Clb-Cdk1 and reactivate Dbf4-Cdc7, respectively. In this case, Sic1p levels increased rapidly, but cells still replicated their DNA once Dbf4-Cdc7 was reactivated (data not shown). These data lead us to conclude that S-CDKs perform their function for DNA replication independently of Dbf4-Cdc7.
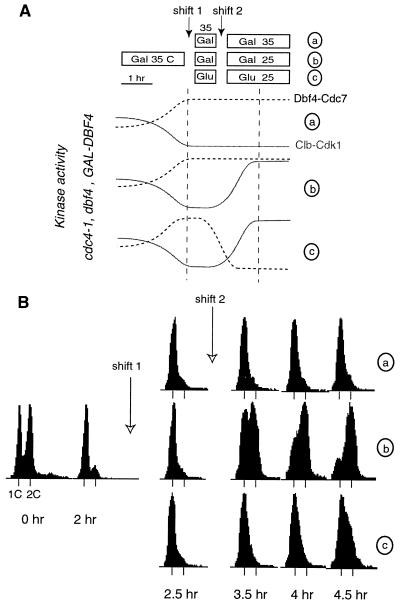
Dbf4-Cdc7 function requires prior Clb-Cdk1 activity.
The reciprocal experiment (clb− CDC7+→CLB+ cdc7−) was performed to test if Dbf4-Cdc7 function could be completed before Clb5-Cdk1 activation. In this case, we used a strain (cdc4-1 dbf4Δ GAL-DBF4) in which Clb5-Cdk1 kinases can be inactivated at 35°C by the cdc4-1 mutation, while the Dbf4-Cdc7 kinase is controlled by galactose-dependent DBF4 expression (Fig. 6A). Cells were grown at 25°C in galactose-containing medium and arrested for 2 h at 35°C in a cdc4 (clb−) state, in which cells have elongated buds and unreplicated DNA but active Dbf4-Cdc7 (Fig. 4B). At this time, the culture was again split in three parts, which all remained at 35°C for a further 30 min, while the third (culture c) was transferred to glucose-containing medium. Previous experiments indicated that turning off DBF4 transcription for 30 min was sufficient to deplete cellular Dbf4p (Fig. 1C). At this point, cultures b and c were shifted at 25°C to degrade Sic1p and reactivate Clb5-Cdk1 kinases. As expected, the cells left at 35°C (culture a) did not replicate their DNA, whereas those in galactose at 25°C (culture b; DBF4 on) did replicate within 60 to 90 min (Fig. 6B). This confirms that cdc4 arrest is both tight and reversible in our conditions. Remarkably culture c, in which Dbf4p was depleted before Clb-Cdk1 kinases were reactivated, did not replicate at all for 90 min and replicated only slightly thereafter. This small increase in DNA content might be due to incomplete Dbf4p depletion or to some Cdc7p-independent origin firing (55). Thus Dbf4-Cdc7 kinase cannot perform its function unless Clb-Cdk1 kinases are also active or have been previously active.
FIG. 6.
Dbf4-Cdc7 requires prior Clb-Cdk1 activation to promote DNA replication. (A) Schematic outlay of the clb− Cdc7+→cdc7− Clb+ reciprocal shift experiment. A culture of cdc4-1 dbf4Δ GAL-DBF4 cells (E784) growing in YEPGal was shifted to 35°C to inactivate Clb-Cdk1 kinases as indicated, and then either maintained arrested (a), shifted to 25°C to reactivate Clb5-Cdk1 (b), or shifted after Dbf4-Cdc7 kinase was inactivated by depletion of Dbf4p (2% glucose) (c). (B) FACS profiles indicate that culture b replicated its DNA whereas culture c did not replicate or replicated very inefficiently.
As Dbf4-Cdc7 is required throughout S phase, an alternative interpretation for the incomplete DNA replication in culture c could be that initiation took place but that Dbf4p depletion prevented the firing of late origins (5, 16). This is unlikely for two reasons: first, the failure to activate late origins delays S-phase completion only slightly (17), not for more than 60 min as shown in Fig. 6; second, DNA replication occurred when Clb-Cdk1 reactivation was anticipated by 30 min without changing the timing of Dbf4p depletion (data not shown), indicating that DDK activation was not too short to allow replication in culture c. Anyhow, the Dbf4-Cdc7 kinase was fully active in the cdc4 arrest and this was clearly insufficient to promote rapid DNA replication after Clb-Cdk1 reactivation. Thus, DNA replication does not take place normally when Clb-Cdk1 kinases are activated only after Dbf4-Cdc7 activity has been turned off. The simplest explanation for this data is that Dbf4-Cdc7 function for DNA replication requires prior Clb-Cdk1 activation.
Dbf4-Cdc7 kinase phosphorylates Cdc45p in vitro.
One explanation for the requirement of Clb-Cdk1 activity prior to the Dbf4-Cdc7-mediated step would be that the former provides a substrate for the latter. Dbf4p and Cdc7p are nonabundant proteins which are recruited to origins via interaction with ORC (18, 52). Therefore, only proteins bound to origins are likely to be phosphorylated efficiently by Dbf4-Cdc7. Cdc45p is a good candidate, because it is loaded onto origins in a Clb5-Cdk1 dependent manner (77). We therefore tested if Cdc45p could be phosphorylated by Dbf4-Cdc7. Recombinant Cdc45p (His6-Cdc45p) was purified from bacteria and used as a substrate in our Dbf4-Cdc7 kinase assay. We find that Cdc45p is phosphorylated by Dbf4-Cdc7 to the same extent as Mcm2p (Fig. 7, lanes 4 to 7 and 11 to 13). Interestingly, Dbf4p autophosphorylation decreases when Cdc45p, not Mcm2p, is added to the reaction (Fig. 7, lanes 2 to 4) and when excess ATP is present (Fig. 7, lanes 10 to 13). Phosphorylation depends on the amount of Dbf4p in the extract (Fig. 2 and Fig. 7, lanes 5 to 7 and 11 to 13) and is lost at 37°C in the cdc7-1 ts strain (Fig. 7, lanes 8 and 14). However, arguing that phosphorylation of Cdc45p by Dbf4-Cdc7 is biologically relevant will require confirmation in vivo and by mutational analysis. Nonetheless, the observation that Cdc45p is phosphorylated by Dbf4-Cdc7 in vitro, along with the dependencies of Cdc45p loading and Dbf4-Cdc7 function upon Clb-Cdk1 activity (reference 77 and this paper) and the interdependence of CDC7 and CDC45 (51), suggests that Cdc45p could be Dbf4-Cdc7's critical substrate for the initiation of DNA replication.
DISCUSSION
In all eukaryotes, the initiation of DNA replication requires the activation of two protein kinases, an S-phase CDK (S-CDK) (19, 22, 62, 68) and a homologue of the yeast Dbf4-Cdc7 kinase (DDK) (39, 43, 47, 56). While CDKs have received considerable attention, little was known on the regulation of DDK probably because metazoan homologues and physiological substrates were discovered only recently (44, 60). We devised a sensitive and specific assay first to analyze the regulation of Dbf4-Cdc7 kinase during the cell cycle and second to ask whether S-CDK and DDK need to function in a precise order for the initiation of DNA replication. As reported recently (50), we find that Dbf4-Cdc7 kinase activity is cell cycle regulated, low in G1, maximal during S phase, and decreasing thereafter, exactly mirroring Dbf4p levels. However, Dbf4-Cdc7 activity never completely disappeared during vegetative growth, unlike that of Clb5-Cdk1 (the S-CDK), which oscillates with a large amplitude. This comparatively modest fluctuation reflects the weak cell cycle-dependent transcriptional and posttranslational regulation acting on Dbf4p. In contrast to the commonly held view, DBF4 transcripts are not G1-S specific and they fluctuate only slightly, in stark contrast with CLB5 mRNA (Fig. 2A) (50, 67). Strikingly, DBF4 mRNA is abundant in elutriated early G1 cells, indicating either that the MluI cell cycle box (MCB) element in its promoter is not functional or that DBF4 transcripts are stable. The latter is unlikely, because transcriptional shutoff leads to rapid depletion of Dbf4p (Fig. 1C). Thus transcriptional regulation cannot account for the lack of Dbf4p in these early G1 cells. Instead, we find that Dbf4p is very unstable throughout the cell cycle (half-life, 15 min), but even more so during G1 (half-life, 5 min). This increased instability is due to the APC/C (Fig. 3B) (10, 50), which might endow Dbf4p with periodicity.
Is there a biological significance to Dbf4p instability? Unlike critical regulators such as Sic1p, Pds1p, or Clb2p, whose destruction is necessary for cell cycle progression, Dbf4p apparently does not need to be fully degraded each cell cycle. Indeed, DDK activity is detectable throughout the cell cycle and ectopic DBF4 expression has no effect (Fig. 2 and 4 and data not shown). Consistent with this idea, it has been shown that at the time of cytokinesis, cells contain enough Dbf4p for at least two additional divisions (39). But then, why do cells bother to turn over Dbf4p rapidly? It has to be noted that incomplete degradation does not mean that turnover is useless. Overlapping mechanisms might exist to prevent excess Dbf4-Cdc7 kinase from causing adverse effects. For example, Dbf4p binds to replication origins in a regulated fashion which might limit spatially the juxtaposition of Dbf4-Cdc7 with its molecular targets (18, 52). Alternatively, Dbf4p instability might be needed to keep DDK activity at low levels, which could be important for cell homeostasis without immediate effect on cell cycle progression. Interestingly, high Cdc7p levels influence the rate of induced mutagenesis (29, 65). We find that while overexpression of either Cdc7p or Dbf4p alone has little effect on cell proliferation, overexpression of both subunits together markedly increases kinase activity in vitro and is detrimental to cells (Fig. 1D and 3C). This effect requires overexpression of both subunits and is therefore likely to be the consequence of Dbf4-Cdc7 kinase hyperactivation. Moreover, it suggests that at the time of the toxic effect (perhaps S phase) (38), neither Cdc7p nor Dbf4p is in large excess over its partner. We estimated, by comparing signals of purified and endogenous Cdc7p on Western blots, that cells contain roughly 300 to 600 Cdc7p molecules, i.e., not more than twice the number of active origins (R. Nougarède, unpublished data). This limited number is surprising in light of earlier experiments which suggested that cells contain 200 times more Cdc7p than is needed for a single division (65). If both estimates are not grossly incorrect, it would imply that cells could replicate with less than one Cdc7p molecule per origin. As Dbf4-Cdc7 kinase acts throughout S phase at the level of individual origins, this might be possible, for example, by recycling Cdc7p from early to late origins or by regrouping several origins around a single Dbf4-Cdc7 complex. Accordingly, Dbf4p adopts a punctate staining in the nucleus resembling replication foci, unlike Clb5p, which is diffuse (52).
In our hands Dbf4-Cdc7 kinase activity correlates strictly with Dbf4p levels over a 10-fold range, strengthening the notion that Dbf4p controls Dbf4-Cdc7 activation (7, 32, 50). Endogenous Dbf4p (which coimmunoprecipates with Cdc7p) as well as exogenously added Mcm2p is phosphorylated proportionally to the amount of Dbf4p present in the extract. Numerous genetic and physical interactions place the MCM complex as the leading candidate for Cdc7 kinase targets. Of its six subunits, Mcm2p appears to be the preferred substrate (6, 50), although some reports have indicated otherwise (44, 60). We have not tested other MCM subunits but find that Cdc45p purified from bacteria is phosphorylated by Dbf4-Cdc7 in vitro as efficiently as Mcm2p is. This is the first biochemical evidence that Cdc45p might be a target of Dbf4-Cdc7. Support for this notion comes from the observation that Cdc45p is required for the initiation of DNA replication in a step interdependent with that of Dbf4-Cdc7 (51, 76). Cdc45p interacts with Mcm2p and Mcm5p, but their association with chromatin is regulated differently (31, 77). Loading is independent of Dbf4-Cdc7 (77), but dissociation from chromatin occurs with kinetics similar to those of Dbf4p (52). A mutation in Mcm5 (bob1) bypasses the requirement of Dbf4-Cdc7 kinase for DNA replication (27, 32). If Dbf4-Cdc7 and Cdc45 were to mediate a single event (the same event, e.g., a conformational change in the MCM complex), then we would expect that the bob1 mutation also bypasses cdc45 mutants. We find this is not the case (data not shown), indicating that Cdc45p has a role for DNA replication independent of Dbf4-Cdc7, even though their functions were shown to be interdependent (51).
Understanding how eukaryotes initiate DNA replication will require the identification of S-CDK and DDK targets. In conceptual terms it is also crucially important to establish whether these two kinases act independently of each other or in a concerted manner to fire replication origins. Cell cycle progression results from successive events that occur in a precise order. Order can be imposed by timing or dependency mechanisms, but the latter seem to predominate in somatic cells (49). The formation of replication-competent origins also obeys such dependency rules, as follows: MCMs bind to origins only after ORC and Cdc6p, whereas stable Cdc45p association requires MCMs as well as S-CDK activity (13). The result is the following order of dependent events: ORC→Cdc6p→MCM→Cdc45p. We wanted to find out if S-CDK and DDK also have to follow such a strict order to fire pre-RCs. In their seminal paper, Hereford and Hartwell (28) proposed that Cdc7p acts downstream of Cdc4. This was not the result of reciprocal shifts but was inferred from the double mutant phenotype and from the dependency of the Cdc4 step, but not the Cdc7p step, on protein synthesis. Since we know that Cdc4 is required to activate S-CDKs, these results indicate that S-CDKs and DDK differ in their requirements for activation, but do not address their order of function. To untangle this issue, we first analyzed when precisely the two kinases are activated during the cell cycle. We found that Clb5-Cdk1 activity is more tightly regulated than that of Dbf4-Cdc7 (Fig. 4A). As a consequence, Dbf4-Cdc7 is already active before Clb5-Cdk1 activity is turned on and is still active after Clb5-Cdk1 activity is turned off. This was expected because precocious or retarded activation of Clb-Cdk1 kinases is sufficient to advance or delay initiation, respectively (61–63). Moreover, we show that Clb5-Cdk1 kinase is fully active in a cdc7 block and vice versa, which demonstrates that the two kinases are activated independently. However, a kinase can be active long before it actually performs its function. We used conditional alleles and regulatable promoters to control Clb5-Cdk1 (S-CDK) and Dbf4-Cdc7 (DDK) activities in classical reciprocal shifts experiments in vivo (28). These show that Dbf4-Cdc7 kinase, in spite of being activated earlier, cannot perform its function for DNA replication before Clb5-Cdk1 is turned on. Hence, S phase does not take place when the S-CDK is activated after DDK has been turned off. In other words, fulfillment of the Dbf4-Cdc7 kinase requirement for S phase necessitates prior Clb5-Cdk1 activation. The converse is not true: S-CDK activity is not needed any longer for S when it is turned off before DDK is turned on. Thus, Clb5-Cdk1 can perform its essential function independently of Dbf4-Cdc7. Taken together, the results establish a dependent pathway (Clb5-Cdk1→Dbf4-Cdc7→S phase) in which S-CDK must act prior to DDK for the initiation of DNA replication. What could such a dependency mean in concrete terms? S-CDKs might be required, for example, to bring a substrate in proximity to DDK. Cdc45p would fulfill these criteria, as it is loaded onto origins by S-CDK, phosphorylated (in vitro) by DDK, and displaced from chromatin at the same time as Dbf4p (52, 77). Alternatively, S-CDKs might alter the conformation of the pre-RC in such a way as to allow its activation by DDK.
Do our data imply that the sole function of S-CDKs is to prepare the ground for DDK? We believe this is not the case. Indeed, the bob1/mcm5 mutation which bypasses DDK function does not bypass cdc4 mutants for viability (32) or for DNA replication (F. Della Seta, unpublished data). Because Cdc4p's sole function for DNA replication is to degrade Sic1p (62), the bob1 mutation clearly cannot bypass S-CDK function. Thus S-CDKs are also needed for DNA replication independently of Dbf4-Cdc7. This is why we favor a sequential model for the roles of S-CDK and DDK, whereby DDK impinges on the initiation mechanism only after S-CDKs have performed their function, i.e., concomitantly with or after Cdc45p loading (Fig. 8). In contrast to that of S-CDKs (15, 69), DDK activity is dispensable for S phase under certain circumstances, such as in the mcm5/bob1 mutant or during meiosis (30, 32). Therefore, we see the role of DDK more as a regulatory appendage to the core mechanism of initiation driven by S-CDKs. As DDK is limiting and bound to origins, such a regulatory step just before the initiation of DNA replication would allow the controlled firing of individual origins throughout S phase. It has been shown that DNA damage and stalled replication forks block the firing of late origins in a Rad53- and Mec1-dependent manner (59, 66) which involves phosphorylation and displacement of Dbf4p from chromatin (10, 52). Therefore, we would like to propose a double-trigger mechanism for the initiation of DNA replication, whereby S-CDKs that are abundant but tightly regulated act globally to give the go-ahead signal for S phase, and DDK which is less cell cycle regulated but limited in amount and concentrated at origins would act locally to give the pace at which individual origins fire. These two modes of temporal and spatial regulation would join to ensure that DNA replication initiates at the right time and place and is properly modulated under changing physiological conditions.
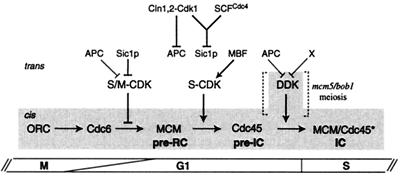
FIG. 8.
Model for the contributions of S-CDK and DDK in the initiation of DNA replication. CDKs are tightly regulated, negatively in G1 by APC/C and Sic1p and positively in late G1 by MBF-dependent cyclin transcription (41). The former allows pre-RC formation and the latter allows pre-RC activation. Negative S-CDK control is broken when G1 CDKs (Cln1,2-Cdk1) trigger both the inactivation of APC and the proteolysis of Sic1p (with help of the E3 ubiquitin ligase SCFCdc4). S-CDKs, which are abundant, act globally to convert the pre-RC in a preinitiation complex (pre-IC) by loading Cdc45p onto origins. Soon after or throughout S phase, the inactive pre-IC is changed in an active initiation complex (IC) by the action of DDK, possibly by phosphorylating MCMs and Cdc45p. DDK is scant but localized at origins and its activity is maintained at low levels by APC-dependent and -independent (X) Dbf4p proteolysis. The DDK regulation module, which acts downstream of S-CDKs, is dispensable in the mcm5-bob1 mutant or during meiosis. Shading indicates cis factors that are bound to origins.
ACKNOWLEDGMENTS
We thank C. Hardy, M. Shirayama, B. Stillman, and W. Zachariae for yeast strains, M. Aldea and C. Mann for plasmids, and K. Nasmyth and M. Tyers for antibodies. Suggestions and critical reading of the manuscript by A. Devault, R. Hipskind, and P. Pasero are gratefully acknowledged.
Research was funded by grants from the Centre National de la Recherche Scientifique (CNRS), Fondation pour la Recherche Médicale (FRM), and Association pour la Recherche sur le Cancer (ARC). F.D.S. benefited from an exchange program between CNRS and Université de Nancy. R.N. was supported by a Ph.D. fellowship from MENRT.
REFERENCES
- 1.Aparicio O M, Stout A M, Bell S P. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc Natl Acad Sci USA. 1999;96:9130–9135. doi: 10.1073/pnas.96.16.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell S P, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 3.Benito J, Martin-Castellanos C, Moreno S. Regulation of the G1 phase of the cell cycle by periodic stabilization and degradation of the p25(rum1) CDK inhibitor. EMBO J. 1998;17:482–497. doi: 10.1093/emboj/17.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumenthal A B, Kriegstein H J, Hogness D S. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38:205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- 5.Bousset K, Diffley J F X. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown G W, Kelly T J. Purification of Hsk1, a minichromosome maintenance protein kinase from fission yeast. J Biol Chem. 1998;273:22083–22090. doi: 10.1074/jbc.273.34.22083. [DOI] [PubMed] [Google Scholar]
- 7.Brown G W, Kelly T J. Cell cycle regulation of Dfp1, an activator of the Hsk1 protein kinase. Proc Natl Acad Sci USA. 1999;96:8443–8448. doi: 10.1073/pnas.96.15.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byers B, Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harbor Symp Quant Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- 9.Chapman J W, Johnston L H. The yeast gene DBF4, essential for entry into S phase, is cell cycle regulated. Exp Cell Res. 1989;180:419–428. doi: 10.1016/0014-4827(89)90068-2. [DOI] [PubMed] [Google Scholar]
- 10.Cheng L, Collyer T, Hardy C F. Cell cycle regulation of DNA replication initiator factor Dbf4p. Mol Cell Biol. 1999;19:4270–4278. doi: 10.1128/mcb.19.6.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cross F R, Tinkelenberg A H. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- 12.Diffley J F. Once and only once upon a time: specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- 13.Diffley J F. Replication control: choreographing replication origins. Curr Biol. 1998;8:R771–R773. doi: 10.1016/s0960-9822(07)00483-6. [DOI] [PubMed] [Google Scholar]
- 14.Diffley J F, Cocker J H, Dowell S J, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- 15.Dirick L, Goetsch L, Ammerer G, Byers B. Regulation of meiotic S phase by Ime2 and a Clb5,6-associated kinase in Saccharomyces cerevisiae. Science. 1998;281:1854–1857. doi: 10.1126/science.281.5384.1854. [DOI] [PubMed] [Google Scholar]
- 16.Donaldson A D, Fangman W L, Brewer B J. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev. 1998;12:491–501. doi: 10.1101/gad.12.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donaldson A D, Raghuraman M K, Friedman K L, Cross F R, Brewer B J, Fangman W L. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell. 1998;2:173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- 18.Dowell S J, Romanowski P, Diffley J F. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science. 1994;265:1243–1246. doi: 10.1126/science.8066465. [DOI] [PubMed] [Google Scholar]
- 19.Epstein C B, Cross F R. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 1992;6:1695–1706. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- 20.Fangman W L, Brewer B J. A question of time: replication origins of eukaryotic chromosomes. Cell. 1992;71:363–366. doi: 10.1016/0092-8674(92)90505-7. [DOI] [PubMed] [Google Scholar]
- 21.Feldman R M, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 22.Fisher D L, Nurse P. A single fission yeast mitotic cyclin B p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1 cyclins. EMBO J. 1996;15:850–860. [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman K L, Brewer B J, Fangman W L. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells. 1997;2:667–678. doi: 10.1046/j.1365-2443.1997.1520350.x. [DOI] [PubMed] [Google Scholar]
- 24.Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 25.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 26.Hansen R S, Canfield T K, Fjeld A D, Mumm S, Laird C D, Gartler S M. A variable domain of delayed replication in FRAXA fragile X chromosomes: X inactivation-like spread of late replication. Proc Natl Acad Sci USA. 1997;94:4587–4592. doi: 10.1073/pnas.94.9.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardy C F, Dryga O, Seematter S, Pahl P M, Sclafani R A. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci USA. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hereford L M, Hartwell L H. Sequential gene function in the initiation of Saccharomyces cerevisiae DNA synthesis. J Mol Biol. 1974;84:445–461. doi: 10.1016/0022-2836(74)90451-3. [DOI] [PubMed] [Google Scholar]
- 29.Hollingsworth R E, Jr, Ostroff R M, Klein M B, Niswander L A, Sclafani R A. Molecular genetic studies of the Cdc7 protein kinase and induced mutagenesis in yeast. Genetics. 1992;132:53–62. doi: 10.1093/genetics/132.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hollingsworth R E, Jr, Sclafani R A. Yeast pre-meiotic DNA replication utilizes mitotic origin ARS1 independently of CDC7 function. Chromosoma. 1993;102:415–420. doi: 10.1007/BF00360406. [DOI] [PubMed] [Google Scholar]
- 31.Hopwood B, Dalton S. Cdc45p assembles into a complex with Cdc46p/Mcm5p, is required for minichromosome maintenance, and is essential for chromosomal DNA replication. Proc Natl Acad Sci USA. 1996;93:12309–12314. doi: 10.1073/pnas.93.22.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson A L, Pahl P M, Harrison K, Rosamond J, Sclafani R A. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol Cell Biol. 1993;13:2899–2908. doi: 10.1128/mcb.13.5.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson D A, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140:1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.James S W, Bullock K A, Gygax S E, Kraynack B A, Matura R A, MacLeod J A, McNeal K K, Prasauckas K A, Scacheri P C, Shenefiel H L, Tobin H M, Wade S D. nimO, an Aspergillus gene related to budding yeast Dbf4, is required for DNA synthesis and mitotic checkpoint control. J Cell Sci. 1999;112:1313–1324. doi: 10.1242/jcs.112.9.1313. [DOI] [PubMed] [Google Scholar]
- 35.Jiang W, Hunter T. Identification and characterization of a human protein kinase related to budding yeast Cdc7p. Proc Natl Acad Sci USA. 1997;94:14320–14325. doi: 10.1073/pnas.94.26.14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnston L H, Masai H, Sugino A. First the CDKs, now the DDKs. Trends Cell Biol. 1999;9:249–252. doi: 10.1016/s0962-8924(99)01586-x. [DOI] [PubMed] [Google Scholar]
- 37.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 38.Kilbey B J. Mutagenesis in yeast-misreplication or misrepair? Mol Gen Genet. 1984;197:519–521. doi: 10.1007/BF00329955. [DOI] [PubMed] [Google Scholar]
- 39.Kitada K, Johnston L H, Sugino T, Sugino A. Temperature-sensitive cdc7 mutations of Saccharomyces cerevisiae are suppressed by the DBF4 gene, which is required for the G1/S cell cycle transition. Genetics. 1992;131:21–29. doi: 10.1093/genetics/131.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knoblich J A, Sauer K, Jones L, Richardson H, Saint R, Lehner C F. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 1994;77:107–120. doi: 10.1016/0092-8674(94)90239-9. [DOI] [PubMed] [Google Scholar]
- 41.Koch C, Nasmyth K. Cell cycle regulated transcription in yeast. Curr Opin Cell Biol. 1994;6:451–459. doi: 10.1016/0955-0674(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 42.Krude T, Jackman M, Pines J, Laskey R A. Cyclin/Cdk-dependent initiation of DNA replication in a human cell-free system. Cell. 1997;88:109–119. doi: 10.1016/s0092-8674(00)81863-2. [DOI] [PubMed] [Google Scholar]
- 43.Kumagai H, Sato N, Yamada M, Mahony D, Seghezzi W, Lees E, Arai K I, Masai H. A novel growth- and cell cycle-regulated protein, ASK, activates human Cdc7-related kinase and is essential for G1/S transition in mammalian cells. Mol Cell Biol. 1999;19:5083–5095. doi: 10.1128/mcb.19.7.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lei M, Kawasaki Y, Young M R, Kihara M, Sugino A, Tye B K. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J J, Deshaies R J. Exercising self-restraint: discouraging illicit acts of S and M in eukaryotes. Cell. 1993;74:223–226. doi: 10.1016/0092-8674(93)90413-k. [DOI] [PubMed] [Google Scholar]
- 46.Martin-Castellanos C, Labib K, Moreno S. B-type cyclins regulate G1 progression in fission yeast in opposition to the p25rum1 cdk inhibitor. EMBO J. 1996;15:839–849. [PMC free article] [PubMed] [Google Scholar]
- 47.Masai H, Miyake T, Arai K. hsk1+, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J. 1995;14:3094–3104. doi: 10.1002/j.1460-2075.1995.tb07312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. EMBO J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murray A W, Kirschner M W. Dominoes and clocks: the union of two views of the cell cycle. Science. 1989;246:614–621. doi: 10.1126/science.2683077. [DOI] [PubMed] [Google Scholar]
- 50.Oshiro G, Owens J C, Shellman Y, Sclafani R A, Li J J. Cell cycle control of Cdc7p kinase activity through regulation of Dbf4p stability. Mol Cell Biol. 1999;19:4888–4896. doi: 10.1128/mcb.19.7.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Owens J C, Detweiler C S, Li J J. CDC45 is required in conjunction with CDC7/DBF4 to trigger the initiation of DNA replication. Proc Natl Acad Sci USA. 1997;94:12521–12526. doi: 10.1073/pnas.94.23.12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pasero P, Duncker B P, Schwob E, Gasser S M. A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation competent origins of replication. Genes Dev. 1999;13:2159–2176. doi: 10.1101/gad.13.16.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peters J M. SCF and APC: the Yin and Yang of cell cycle regulated proteolysis. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- 54.Piatti S, Lengauer C, Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reynolds A E, McCarroll R M, Newlon C S, Fangman W L. Time of replication of ARS elements along yeast chromosome III. Mol Cell Biol. 1989;9:4488–4494. doi: 10.1128/mcb.9.10.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roberts B T, Ying C Y, Gautier J, Maller J L. DNA replication in vertebrates requires a homolog of the Cdc7 protein kinase. Proc Natl Acad Sci USA. 1999;96:2800–2804. doi: 10.1073/pnas.96.6.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rowley A, Johnston G C, Butler B, Werner-Washburne M, Singer R A. Heat shock-mediated cell cycle blockage and G1 cyclin expression in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:1034–1041. doi: 10.1128/mcb.13.2.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 59.Santocanale C, Diffley J F. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 60.Sato N, Arai K, Masai H. Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: in vitro phosphorylation of MCM subunits by a putative human homologue of Cdc7. EMBO J. 1997;16:4340–4351. doi: 10.1093/emboj/16.14.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneider B L, Yang Q H, Futcher A B. Linkage of replication to start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- 62.Schwob E, Böhm T, Mendenhall M D, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233–244. doi: 10.1016/0092-8674(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 63.Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- 64.Sclafani R A, Jackson A L. Cdc7 protein kinase for DNA metabolism comes of age. Mol Microbiol. 1994;11:805–810. doi: 10.1111/j.1365-2958.1994.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 65.Sclafani R A, Patterson M, Rosamond J, Fangman W L. Differential regulation of the yeast CDC7 gene during mitosis and meiosis. Mol Cell Biol. 1988;8:293–300. doi: 10.1128/mcb.8.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 67.Spellman P T, Sherlock G, Zhang M Q, Iyer V R, Anders K, Eisen M B, Brown P O, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strausfeld U P, Howell M, Descombes P, Chevalier S, Rempel R E, Adamczewski J, Maller J L, Hunt T, Blow J J. Both cyclin A and cyclin E have S-phase promoting (SPF) activity in Xenopus egg extracts. J Cell Sci. 1996;109:1555–1563. doi: 10.1242/jcs.109.6.1555. [DOI] [PubMed] [Google Scholar]
- 69.Stuart D, Wittenberg C. CLB5 and CLB6 are required for premeiotic DNA replication and activation of the meiotic S/M checkpoint. Genes Dev. 1998;12:2698–2710. doi: 10.1101/gad.12.17.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 71.Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher A B, Nasmyth K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- 72.Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- 73.Tanaka T, Nasmyth K. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J. 1998;17:5182–5191. doi: 10.1093/emboj/17.17.5182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yamashita M, Hori Y, Shinomiya T, Obuse C, Tsurimoto T, Yoshikawa H, Shirahige K. The efficiency and timing of initiation of replication of multiple replicons of Saccharomyces cerevisiae chromosome VI. Genes Cells. 1997;2:655–665. doi: 10.1046/j.1365-2443.1997.1530351.x. [DOI] [PubMed] [Google Scholar]
- 75.Yoon H J, Loo S, Campbell J L. Regulation of Saccharomyces cerevisiae CDC7 function during the cell cycle. Mol Biol Cell. 1993;4:195–208. doi: 10.1091/mbc.4.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zou L, Mitchell J, Stillman B. CDC45, a novel yeast gene that functions with the origin recognition complex and Mcm proteins in initiation of DNA replication. Mol Cell Biol. 1997;17:553–563. doi: 10.1128/mcb.17.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zou L, Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]