Abstract
Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) directly interacts with host's epithelial and immune cells, leading to inflammatory response induction, which is considered the hallmark of infection. The host immune system is programmed to facilitate the clearance of viral infection by establishing a modulated response. However, SARS-CoV-2 takes the initiative and its various structural and non-structural proteins directly or indirectly stimulate the uncontrolled activation of injurious inflammatory pathways through interaction with innate immune system mediators. Upregulation of cell-signaling pathways such as mitogen-activate protein kinase (MAPK) in response to recognition of SARS-CoV-2 antigens by innate immune system receptors mediates unbridled production of proinflammatory cytokines and cells causing cytokine storm, tissue damage, increased pulmonary edema, acute respiratory distress syndrome (ARDS), and mortality. Moreover, this acute inflammatory state hinders the immunomodulatory effect of T helper cells and timely response of CD4+ and CD8+ T cells against infection. Furthermore, inflammation-induced overproduction of Th17 cells can downregulate the antiviral response of Th1 and Th2 cells. In fact, the improperly severe response of the innate immune system is the key to conversion from a non-severe to severe disease state and needs to be investigated more deeply. The virus can also modulate the protective immune responses by developing immune evasion mechanisms, and thereby provide a more stable niche. Overall, combination of detrimental immunostimulatory and immunomodulatory properties of both the SARS-CoV-2 and immune cells does complicate the immune interplay. Thorough understanding of immunopathogenic basis of immune responses against SARS-CoV-2 has led to developing several advanced vaccines and immune-based therapeutics and should be expanded more rapidly. In this review, we tried to delineate the immunopathogenesis of SARS-CoV-2 in humans and to provide insight into more effective therapeutic and prophylactic strategies.
Keywords: Virus, SARS-CoV-2, Inflammatory, Immune evasion, Immunotherapy, Vaccine
Nomenclature
- ADE
antibody-dependent enhancement
- ACE2
angiotensin-converting enzyme 2
- DMV
double membrane vesicle
- GRP78
glucose Regulated Protein 78
- HE
hemagglutinin-esterase
- HAT
human airway trypsin-like protease genome
- IKKε
inhibitor of κB kinase ε
- MEV
multi-epitope vaccine
- nAb
neutral antibody
- RBD
receptor-binding domain
- PAMP
pathogen associated molecular pattern
- PID
predicted intrinsic disorder
- PRR
pattern recognition receptor
- ORF
open reading frame
- ssRNA
sense single-stranded RNA
- TBK1
TANK-binding kinase 1
- TLR
Toll like receptor
- TMPRSS2
transmembrane protease serine 2
- VIC
Viral-immunity cycle
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) or human coronavirus 2019 (HCoV-19) is the causative agent of coronavirus disease 2019 (COVID-19) (Tian et al., 2020). SARS-CoV-2 causes mild to severe infections in the respiratory tract and may lead to mortality in some patients, especially senescent individuals or those with underlying diseases such as diabetes. The current global pandemic caused by SARS-CoV-2 shows that in contrast to middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV-2 is more contagious, which may be due to the capacity of SARS-CoV-2 to pass from one host to the next by easily crossing the respiratory mucosa (Keshavarz et al., 2021; Omrani et al., 2020).
The combination of mucous membrane and immune system constitutes a natural barrier against the virus. The initial innate immune responses induced by type I interferon (IFN), complement proteins, and cytokines/chemokines limit the replication and spread of SARS-CoV-2, and also mediate activation of the downstream adaptive immune responses (Kikkert, 2020). However, viruses can evade these responses via the expression of specific structural and functional components for creating ‘stealth’ or ‘camouflage’ effects. For example, SARS-CoV directly infects epithelium to pass respiratory mucosa (Richt et al., 2012) and exploits some immune cells as shelters and vehicles (Liu et al., 2016), both of which help the virus to be less exposed to the immune system. In severe cases of SARS, macrophages and T cells are infected (Dandekar and Perlman, 2005), production of proinflammatory cytokines is induced, and accumulation of monocyte-macrophages and neutrophils in the lung is increased, which all are associated with extensive lung damage (Kindler and Thiel, 2016).
Similar to SARS-CoV, SARS-CoV-2 replication leads to the production of proinflammatory mediators, which possibly induces aggressive inflammation (Huang et al., 2020). Hence, understanding the molecular interaction between the immune system and SARS-CoV-2 provides a framework for treatment of viral infection, and demonstrates how the virus-specific immune responses lead to viral immune escape. Here, we describe the immune responses and immunopathogenesis of COVID-19, suggesting a hint for developing novel procedures to manage SARS-CoV-2 infection and to decrease related mortalities.
2. Overview of SARS-CoV-2
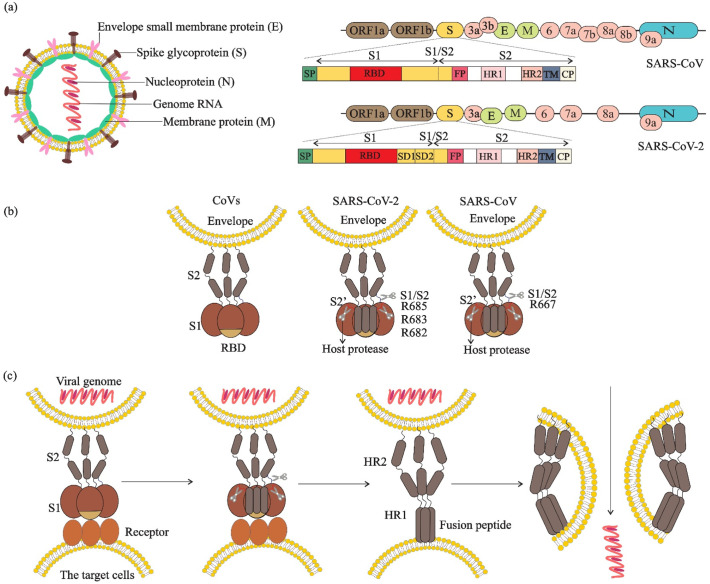
SARS-CoV-2 virions with solar corona microscopic morphology possess positive sense single-stranded RNA genome (+ssRNA) and nucleoprotein (N) to form a ribonucleoprotein complex (RNP). The RNP is embedded with the envelope composed of a lipid membrane containing membrane protein (M), envelope protein (E), and spike glycoprotein (S) (Artika et al., 2020). The S protein comprises S1 and S2 subunits for binding to receptors on human cells. In constant to other coronaviruses, the hemagglutinin-esterase (HE) protein, which facilitates the release of viruses from cells, does not express in SARS-CoV-2 (Chan et al., 2020) (Fig. 1 a). The successful infection by SARS-CoV-2 occurs through recognition of receptor on the surface of target cell, binding of S protein to the receptor, anchoring the viral envelope to the target cell, and ultimately the viral genome internalization (Plemper, 2011) (Fig. 1 b, c). During fusion reaction, the host proteases including trypsin, tryptase Clara, human airway trypsin-like protease (HAT), and transmembrane protease serine 2 (TMPRSS2) catalyze the binding of S glycoproteins to the host cell by proteolytic priming (Wang et al., 2020a).
Fig. 1.
Virus particles, complete genome sequences, spike structure, cleavage sites and fusion reaction of SARS-CoV and SARS-CoV-2. a) Virions contain a +ssRNA genome of 26–32 kb in size. The genome ORF1a/b encodes polyproteins, which form the viral replicase transcriptase complex. The other ORFs on the genome encode four main structural proteins: S, E, N and M proteins, as well as several accessory proteins. b) The spike protein structure is composed of an extracellular (EC) domain, a transmembrane anchor domain and a short intracellular tail. EC domain contains two functional subunits, a receptor-binding subunit (S1) and a membrane-fusion subunit (S2), S1 contains two independent domains, an N-terminal domain (S1-NTD) and receptor binding domain (RBD), the S1/S2 cleavage site is shown in its uncleaved, native state and resides in an unstructured region between S1 and S2, the S2’ cleavage site is exposed only after receptor binding. c) Fusion reaction; S1 attaches to a receptor on the target cells, induces a conformational change in the S, exposing cleavage sites between S1 and S2. In SARS-CoV-2, the trimeric S protein then cleaves into S1 and S2 subunits by cellular proteases (scissors), the fusion peptide (FP) latches onto the target membrane, anchoring the virus and cell together. The heptad repeat 2 (HR2) then folds to interact with the heptad repeat 1 (HR1), bringing the membranes together. The successful refolding of enough adjacent S2s leads to fusion of viral and cell membranes and release of the viral genome into the target cell cytoplasm.
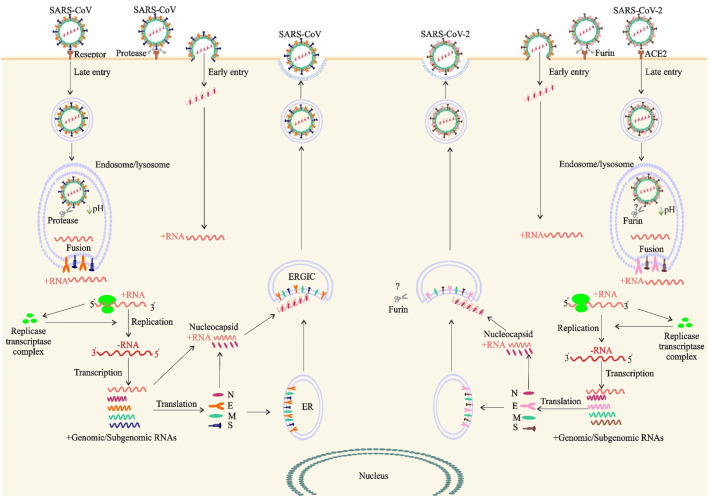
The fusion and gene delivery is dependent on the location and the type of proteolytic enzyme, and can be occurred at the cell surface (non-endosomal priming) and on subcellular membranes (endosomal priming) (Lukassen et al., 2020). In non-endosomal priming, after viral membrane fusion to the host cell surface, viral RNA directly releases into the cytoplasm. Nevertheless, in endosomal priming, the virus first enters into the host cell by means of endosomes, and then releases its genetic material into the cytoplasm (Fig. 2 ). Akin to SARS-CoV, SARS-CoV-2 exploits the non-endosomal priming but with 10- to 20-fold higher affinity (Wrapp et al., 2020). The S glycoprotein of SARS-CoV-2 attaches to angiotensin-converting enzyme 2 (ACE2) receptor on target cells via receptor-binding domain (RBD) in S1 subunit (Letko et al., 2020), priming by TMPRSS2 (Lukassen et al., 2020). However, in vitro blocking of TMPRSS2 showed that SARS-CoV-2 has not been completely inhibited (Hoffmann et al., 2020; Kawase et al., 2012). Moreover, entry of SARS-CoV-2 to 293/hACE2 cells mainly occurs through endocytosis, which is mediated by PIKfyve, TPC2, and cathepsin L, representing the higher flexibility of S protein of SARS-CoV-2 than SARS-CoV (Coutard et al., 2020).
Fig. 2.
The life cycle of SARS-CoV and SARS-CoV-2 in host cells. Both viruses enter target cells through fusion at the cell surface (early entry) or in the endocytic compartment (late entry). Entry route depends on which proteases activate the spike proteins. In early entry, the virus is cleaved at S protein by cell-surface transmembrane serine proteases (TTSPs). If an S protein is unable to be cleaved by transmembrane proteases, due to S protein sequence or lack of protease expression on target cell, the virus must undergo endocytosis and be activated by Cathepsin in the endosome/lysosome. The S proteins of SARS-CoV and SARS-CoV-2 bind to cellular receptor angiotensin-converting enzyme 2 (ACE2). Early and late entry lead to release of the viral +ssRNA genome to the cytoplasm. ORF1a and ORF1ab are translated and processed to form the RNA replicase–transcriptase complex for driving the production of negative-sense Full-length RNAs [(−) RNA]. Full-length (−) RNA is transcribed into four sub-genomic mRNAs which during translation encode viral structural proteins including S, E, M and N. Viral nucleocapsids are assembled from genomic RNA and N protein in the cytoplasm, followed by budding into the lumen of the endoplasmic reticulum (ER)–Golgi intermediate compartment (ERGIC). Virions are then released from the infected cell through exocytosis and unlike nonenveloped viruses, do not lyse cells.
Furin-mediated pre-cleavage at the S1/S2 site of the S protein (see Fig. 1) plays an important role in viral fusion (Coutard et al., 2020). Furin is expressed by secretory pathway of infected cells of organs and tissues in brain, lung, gastrointestinal tract, liver, pancreas, reproductive tissues, and thus makes these tissues sensitive to SARS-CoV-2 infection (Wang et al., 2020a). The stronger transmissibility and higher infectivity of SARS-CoV-2 is due to the furin-mediated pre-cleavage mutation, which increases binding affinity of SARS-CoV-2 to its receptor (Lukassen et al., 2020; Wrapp et al., 2020). Moreover, S protein binds to integrins as an alternative receptor through a conserved RGD (403–405: Arg-Gly-Asp) motif present in its receptor-binding domain which can promote virus entry and infection of the host cell through activating transducing pathways including phosphatidylinositol-3 kinase (PI–3 K) or mitogen-activate protein kinase (MAPK) (Sigrist et al., 2020). Another cell surface receptor, Neuropilin-1 (NRP1) can bind to furin-cleaved proteins such as SARS-CoV-2 S protein and facilitate virus attachment and cell entry which significantly enhances SARS-CoV-2 infectivity and pathogenesis (Cantuti-Castelvetri et al., 2020).
Both of SARS-CoV and SARS-CoV-2 attach to ACE2 receptor on host cells surfaces in respiratory and gastrointestinal tract (Hoffmann et al., 2020) (Fig. 2). The highest level of ACE2 expression in nasal epithelial cells (specifically goblet/secretory cells) and ciliated cells, as early viral targets, constitutes standing by reservoirs helping more efficient SARS-CoV-2 transmission (Sungnak et al., 2020). Furthermore, ACE2 expression has been reported in high, medium and low levels in different body tissues, which all can be considered as potent alternative targets outside of lung (Di Sotto et al., 2018). However, one study has reported a negative correlation between the high expression level of ACE2 in Asian females and young people with COVID-19 severity and fatality (Wang et al., 2020b). Interestingly, SARS-CoV-2 S protein binds to CD147-spike protein, a receptor that expresses on the surface of host cells (Wang et al., 2020b). Studies on pseudovirus and live virus infection showed that T cells, with very low levels of ACE2, were infected through S protein-mediated membrane fusion of SARS-CoV-2, which indicates the involvement of another receptor, rather than ACE2, in infection of such immune cells (Wang et al., 2020c).
3. Immune response to SARS-CoV-2 infection
The immune system is able to direct its antiviral responses, which are a sequential set of steps leading to elimination of viruses and infected cells by non-specific or specific responses.
3.1. Innate immune responses
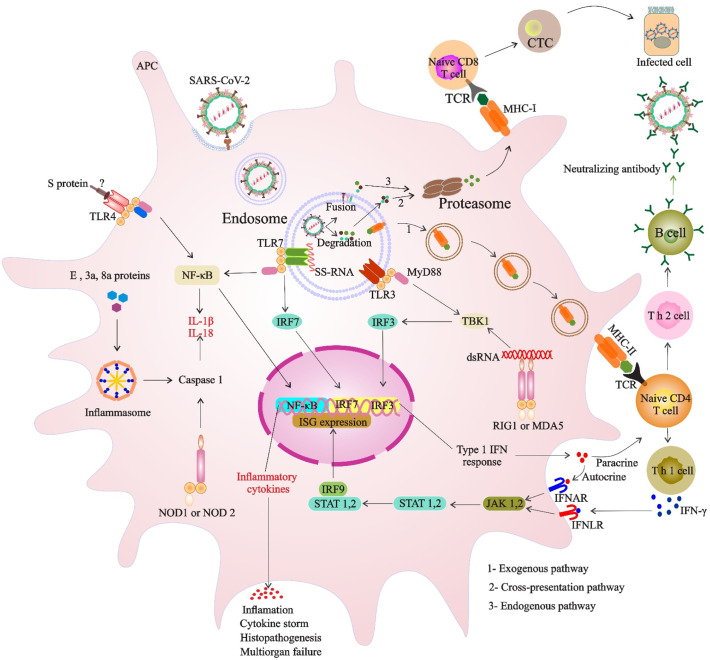
Antiviral immune response is initiated through recognition of the molecular signatures of viral particles, known as pathogen-associated molecular patterns (PAMPs), by pattern recognition receptors (PRRs) of innate immune cells. Toll-like receptors (TLRs) as the PRR sensors are residents of the cell surface (TLR1, 2, 4, 5, and 6), of the endosome (TLRs 3, 7, 8, 9, and 10), and of the cytosol (NOD-like receptors (NLRs) and retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs)) (Chan and Gack, 2016). During the infection process of MERS-CoV, SARS-CoV, and SARS-CoV-2, the endosomal or the cytosolic PRR sensors can recognize distinct forms of viral RNA including ssRNA, 5′ triphosphate RNA, and double-stranded RNA (dsRNA) as PAMPs (Alosaimi et al., 2020). The link between the PRRs and these PAMPs leads to activation of downstream signaling cascades, production of types I and III interferons (IFNs), and secretion of proinflammatory cytokines by multiple myeloid lineages and plasmacytoid dendritic cells (DCs), and therefore grants them a vital antiviral role (Perlman and Netland, 2009). Then, autocrine IFN-I mediates the expression of IFN-stimulated genes (ISGs) and establishes a so-called antiviral state to inhibit virus infection (Prompetchara et al., 2020). Proinflammatory responses locally recruit a variety of innate macrophages, neutrophils, Natural killer (NK) cells and DCs (Kindler and Thiel, 2016) (Fig. 3 ). Consequently, these responses and their balance give rise to clearance of viral infection. However, proinflammatory responses could also be injurious, especially when hypercytokinemia has formerly been triggered. Impairment of innate immune response in COVID-19-induced pneumonia has been shown to lead to poor outcome. In comparison of COVID-19 and non-COVID-19 patients with community-acquired pneumonia, mechanical ventilation was significantly longer in COVID-19 patients, which was associated with increased plasma concentrations of CXCL10, granulocyte-macrophage colony-stimulating factor (GM-CSF) and C—C chemokine ligand 5 (CCL5) in these patients (Blot et al., 2020).
Fig. 3.
Coronavirus immunity cycle. Following SARS-CoV-2 uptake in the endosome and virion degradation the viral antigens are presented in (1) the exogenous pathway, (2) cross-presentation pathway and (3) endogenous pathway; partial genome transcription may provide a source of antigen for priming T-cells by MHC class-1 antigen processing following endogenous pathway. Viral pathogen-associated molecular patterns (PAMPs), using endosomal or cytosolic PRRs, as well as the production of cytokines such as IFN-1, can promote potent cellular mediated immune responses. Structural or nonstructural proteins might be recognized by TLR-4 or inflammasome, leading to the activation of proinflammatory cytokines. Abbreviations: APC, antigen-presenting cells; IFN-1, interferon 1; ssRNA, single-stranded RNA; TBK1, TANK-binding kinase 1.
3.2. Adaptive immune responses
Severe Acute respiratory syndrome coronavirus induce the adaptive immune responses to recruit T cells for killing the virus-infected cells and B cells for producing the pathogen-specific antibodies, which proceeds until reaching an appropriate level of response (Newton et al., 2016) (Fig. 3). During the virus incubation and non-severe stages, the adaptive immunity in most people is primed to induce a significant immune response against SARS-CoV-2 infection (Qin et al., 2020; Tan et al., 2020a). Infiltration of CD4+ and CD8+ T cells in the lungs and bronchoalveolar lavage fluid (BALF) of BALB/C mice sensitized to SARS-COV-2 infection has been shown, which type I IFN pathway is essential for efficient response of these T cells. Indeed, increasing resistance to SARS-COV-2 infection, rapid clearance of this infection, and reducing COVID-19 immunopathogenesis and severity depend on the response of these specific T cells (Zhuang et al., 2021).
The increase in white blood cell (WBC) counts, neutrophil-lymphopenia ratio (NLR) and T lymphopenia, and decline in the number of monocytes, eosinophils, basophils, B cells, T cells, NK cells, memory helper T cells, and regulatory T cells (Tregs) have been reported in the patients with CoVs infection (Qin et al., 2020). Lymphopenia, as the representative of immunological dysregulation, is a common feature of SARS-CoV-2 infection and can be used as a biomarker for evaluation of hospitalization period, treatment effect, and outcomes of COVID-19 patients (Bermejo-Martin et al., 2020; Tan et al., 2020a). Moreover, decrease in the counts of T cells, especially CD8+ T cells, as well as increase in the levels of interleukin (IL)-6, IL-10, IL-2 and IFN-γ in the peripheral blood in the severe cases, and the neutrophil-to-CD8+ T cell ratio (N8R) were identified as important determinants of prognosis for severe SARS-CoV-2 disease (Tan et al., 2020a; Xu et al., 2020). Finally, SARS-CoV-2 neutral antibodies (nAbs) can act against receptor binding domains or subdomains engaged in membrane fusion or virus entry, which needs to be investigated more deeply (Jiang et al., 2020a).
The clinical signs of severe COVID-19 patients have shown the elevated plasma proinflammatory factors such as IL-1β, IL-8, and tumor necrosis factor-alpha (TNF-α) (Huang et al., 2020). However, an increased level of serum C-reactive protein (CRP), serum amyloid A (SAA), procalcitonin (PCT), D-dimer, and creatine kinase, indicates sustained inflammatory response and disturbed coagulation mechanism after infection with SARS-CoV-2. In addition, a high number of TH17 cells in peripheral blood and cytokines involved in TH17-type responses by SARS-CoV-2 suggests the presence of a TH17-type cytokine storm (Wu and Yang, 2020). Furthermore, these cytokines induce the production of IL-21 and IL-22, supporting the TH17 cell maintenance and mediating antimicrobial peptides production in the mucosal organs, respectively. The high expression of inflammatory mediators can generate a feedback cycle between cytokines and immune cells, which may lead to cytokine storms, more severe respiratory complications and higher case fatality rate (Alosaimi et al., 2020). It has proven that the extreme decrease of lymphocytes and the augmented levels of cytokines, in particular IL-2 and IL-6, are reliable indicators of severe COVID-19 (Tan et al., 2020b). Also according to another survey, COVID-19 patients admitted to the intensive care unit (ICU) have higher serum levels of IL-6, CRP and procalcitonin, which IL-6 and CRP have been identified as the strongest predictors of disease severity in patients admitted with COVID-19 (Broman et al., 2021).
Adaptive immunity to the S protein of SARS-CoV-2, T cell responses and specific cytokine patterns produced by T helper cells (TH1, TH2, and TH17) are keys to understanding the immunopathology of COVID-19 and vaccine design (Weiskopf et al., 2020). Functional assays have also indicated that peptides derived from the M protein of SARS-CoV-2 are involved in T-cell responses in most of the COVID-19 patients (Mateus et al., 2020). Circulating SARS-CoV-2-specific T cells (CD8+ and CD4+) were identified in COVID-19 convalescent patients (Mateus et al., 2020). Recently, pre-existing SARS-CoV-2 cross-reactive memory T cells are also detected in 28 to 50% of non-exposed individuals (Grifoni et al., 2020; Mateus et al., 2020). Accumulating evidence has indicated the presence of pre-existing memory CD4+ T cells in blood samples than cross-reactive CD8+ memory T cells (Lipsitch et al., 2020), suggesting their exposure to some of common coronaviruses (Grifoni et al., 2020).Studies have demonstrated a significant decline in the number of CD3+ T cells, CD8+ T cells and natural killer cells, as well as a slight decrease in CD4+ T cells count in patients with COVID-19, introducing the counts of CD8+ and CD4+ T cells as a diagnosis and prognosis marker for COVID-19 disease (Jiang et al., 2020b). Also function of some special subtypes of T cells can be affected during SARS-CoV-2 infection. For example, activation of mucosa associated invariant T (MAIT) cells is associated with the severity of COVID-19 disease. MAIT cells are involved in the immune response against SARS-CoV-2 through an innate-like response independent of HLA-presented peptide antigens and may be involved in COVID-19 immunopathogenesis. Evidences suggest that in COVID-19 patients these cells become highly activated and their number in the bloodstream decreases. On the other hand, a significant increase in MAIT cells and IL-17A proinflammatory cells in the respiratory tract of these patients is observed that the presence of MAIT cell CD69high and CXCR3low immune types are associated with poor outcome (Parrot et al., 2020). Severe malfunction of MAIT cells in COVID-19 patients also has been reported in another study, where these innate-like T cells could express high levels of proinflammatory cytokines, including IL-17A and TNFα. Drastic changes in the expression profile of cytokines leading to a defective antiviral function in MAIT cells of COVID-19 patients can be associated with the immunopathogenesis of this disease (Deschler et al., 2021).
4. SARS-CoV-2-immunoescape
In order to disrupt innate immune responses and establish the viral infection, CoVs employ various mechanisms such as hiding of PRR agonists or suppressing the activation of PRRs and their downstream signaling cascades (Chan and Gack, 2016; Kikkert, 2020). The production of three types of IFNs (type I: IFN-α and -β, type II: IFN-γ, and type III: IFN lambdas) is an important early host immune defense mechanism; however, their expression is downregulated by SARS-CoV and SARS-CoV-2 (Frieman et al., 2008). IFN III, as the primary local defending agent, protects epithelial surfaces of lung cells from low doses of pathogenicity (Wells and Coyne, 2018; Zhou et al., 2018). In higher doses of the virus, the second line of defense, type I IFN machinery, covers broader areas of the tissue (Kikkert, 2020). The unique aspect of the type III IFN is not inducing inflammation as much as type I IFN and is probably responsible for the protection of epithelial tissue from immunopathology (Galani et al., 2017).
Regarding the tricks employed by SARS-CoV-2 to hamper IFN-I responses, it has been demonstrated that upon infection with SARS-CoV-2, nonstructural protein 6 (nsp6) and nsp13 bind to TANK binding kinase 1 (TBK1) and inhibit the phosphorylation of interferon regulatory factor 3 (IRF3) or TBK1, respectively. Moreover, open reading frame 6 (ORF6) was also indicated to target importin Karyopherin α 2 (KPNA2), resulting in IRF3 nuclear translocation. Of note, suppression of IFN-I signaling through SARS-CoV-2 nsp1 and nsp6 is highly more potent than those observed in SARS-CoV and MERS-CoV (Xia et al., 2020).
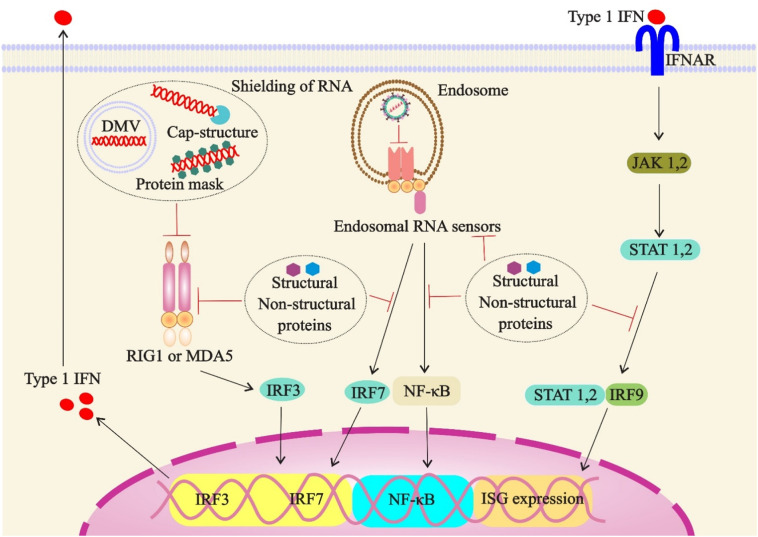
The SARS-CoV, MERS-CoV and SARS-CoV-2 share the same structural (e.g., M, N, E) and non-structural proteins such as open reading frame (ORF) proteins that can inhibit type I IFNs signaling at several levels (Prompetchara et al., 2020). These proteins are capable of shielding viral RNAs from innate immune sensors by creating double membrane vesicles (DMVs) (Oudshoorn et al., 2017), and adding a cap or a cap-like structure to the 5′-end of RNAs (Chen et al., 2011) (Fig. 4 ). They also mask the viral dsRNA by protein 4a (p4a) (Batool et al., 2017), and mediate alternative replication inside the nucleus, owing to the absence of RNA sensors (i.e., RIG-I-like sensors or TLRs) (Kikkert, 2020).
Fig. 4.
Likely mechanisms of suppression of the type 1 interferon response during SARS-CoV-2 infection. Structural and nonstructural proteins of virus or double membrane vesicles (DMVs) can shield PAMPs from immune sensors. Structural and nonstructural proteins also inactivate immune sensors or components of downstream type I IFN signaling. Abbreviations: DMV, double membrane vesicle; ISG, IFN-stimulated gene; MDA5, melanoma differentiation-associated protein 5.
Structural proteins also can directly interact with the inhibitor of κB kinase ε (IKKε)and TANK-binding kinase 1 (TBK1) in the cytoplasm and decrease the STAT1 phosphorylation, which thereby suppresses the IFN-1 anti-viral responses (De Wit et al., 2016; Siu et al., 2014; Siu et al., 2009; Yang et al., 2015).The CoVs'EndoU (endoribonuclease U) can inactivate the host immune sensors by cleaving a 5′-polyuridines sequence of negative-sense viral RNA, which acts as MDA5 (melanoma differentiation-associated protein 5)-dependent PAMP (Hackbart et al., 2020). In addition, CoVs proteins can interfere with host mRNA translations and cleave the cellular (innate immune) factors by SARS-CoV nsp1protein (Kikkert, 2020).
The absence of ORF 3b together with significant changes in ORF6 of SARS-CoV-2 viral protein have made this virus much higher sensitive to type I IFN compared to SARS-CoV (Lokugamage et al., 2020). ORF6 and ORF8 of SARS-CoV-2 can inhibit type I interferon (IFN-β) activation, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) pathway, the interferon-stimulated response element (ISRE) and interferon-stimulated genes (ISGs) such as ISG54 and ISG56 (Zhang et al., 2020). Furthermore, downregulation of MHC-I surface expression on various cell types is driven through direct interaction of ORF8 of SARS-CoV-2 with MHC-I molecules, which thus leads to the disturbance of antigen presentation and of the function of CTLs in recognition and elimination of infected cells (Zhang et al., 2020).As recently shown, the papain-like protease (PLpro) of SARS-CoV-2, the important factor in generating functional replicase complex, preferentially can reduce type I interferon responses via cleavage of the ubiquitin-like interferon-stimulated gene 15 protein (ISG15) from interferon responsive factor 3 (IRF3), and also post-translational modifications on host proteins (Devaraj et al., 2007).Also viral NSP3 protease with cleavage of IRF3exacerbates the reduction of type I interferon response and immunodepression during SARS-CoV-2 infection. Furthermore, cleavage of vital modulators of inflammatory pathways including NLRP12 and TAB1 by viral NSP5 protease leads to increased cytokine production and severe inflammatory response observed in COVID-19 patients. Interestingly, the absence of a cleavage motif in IRF3 and NLRP12 has been shown to inhibit the cleavage of these proteins by SARS-CoV-2 proteases and reduce the disease severity (Moustaqil et al., 2020). Although delayed IFN-I responses and pathogenesis of SARS-CoV-2and SARS-CoV infections share to a certain extent, the viral load of virus and the levels of IFN-I play important roles in immunopathogenesis of them (Lingeswaran et al., 2020). Recently, it has been shown that some of the SARS-CoV-2 proteins including NSP1, NSP3, NSP12, NSP13, NSP14, ORF3, ORF6 and M protein can restrain the Sendai virus-induced activation of IFN-β promoter (Lei et al., 2020; Zhang et al., 2020). Generally, passive and active properties of structural and nonstructural proteins shared by SARS-CoV and SARS-CoV-2can interfere with multiple steps during the initial innate immune responses, including detection of RNA sensors, signaling pathway of type I IFN production, and paracrine activation downstream of type I IFN (Fig. 4).
Table 1.
Summary of antiretroviral, viral entry, cell/plasma and alternative strategies against severe COVID-19 (Ky and Mann, 2020; Lythgoe and Middleton, 2020).
| Treatment strategies | Drug name | Mechanism of action |
|---|---|---|
| Receptor agonists | Losartan | Anti-RAS |
| Antimalarial | Camostat | Targeting viral entry |
| Hydroxychloroquine | Targeting viral entry | |
| Chloroquine phosphate | Targeting viral entry | |
| Antiviral | Umifenovir (Arbidol) | Targeting viral replication |
| Remdesivir | Targeting viral replication | |
| Lopinavir-ritonavir | Targeting viral replication | |
| Arbidol or lopinavir-ritonavir | Targeting viral replication | |
| Darunavir-cobicistat | Targeting viral replication | |
| Cell and plasma | Mesenchymal stem cells (MSCs) | Tissue repair |
| Plasma | Neutralizing antibodies | |
| Alternative | Bevacizumab | Anti-VEGF |
5. Inflammatory signaling pathways of SARS-CoV-2
Mechanisms initiating inflammatory effects in response to SARS-CoV-2 infections are majorly associated with the activation of innate immune signaling (Chan et al., 2020). It has been demonstrated that the expression of genes involved in interferon signaling, neutrophil degranulation and the innate immune response in the lungs of rhesus macaque monkeys increase following SARS-CoV-2 infection, while collagen production-inducing pathways are downregulated. In COVID-19 patients, aging is an important risk factor for poor prognosis and increased mortality, so that IFN type I and notch signaling increase significantly in the lungs of juvenile infected macaques compared to old infected macaques. While in older monkeys with COVID-19 disease an increase in neutrophil counts and neutrophil/lymphocyte ratio is observed (Rosa et al., 2021). SARS-CoV-2 can be recognized through NOD-like receptor protein 3 (NLRP3) on the cell membrane followed by formation of NLRP3 inflammasome, mediated by the adaptor protein apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC) and caspase-1, to induce the pro-inflammatory responses (He et al., 2016) (Fig. 5 a). SARS-CoV-2 and SARS-CoV may promote an inflammatory response by inflammasome NLRP3 activation via cell pyroptosis and structural proteins (Paniri and Akhavan-Niaki, 2020; Zheng and Kanneganti, 2020). The rise of IL-1β in patients infected with SARS-CoV-2 can be the result of NLRP3 inflammasome activation, suggesting the cell pyroptotic activity of SARS-CoV-2 (Huang et al., 2020).
Fig. 5.
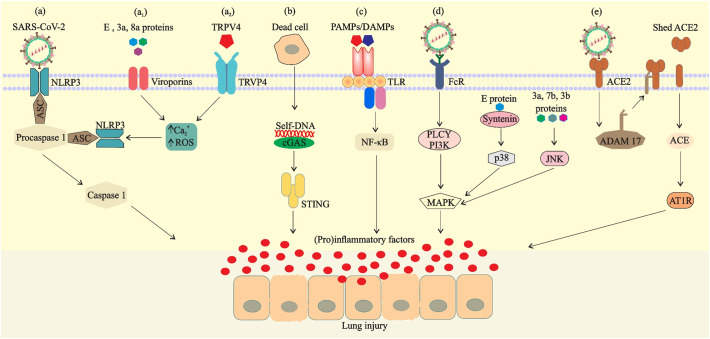
Possible mechanisms of SARS-CoV-2-mediated inflammatory responses. a) NOD-like receptor protein 3 (NLRP3) is activated by virus, Ca2+ influx or ROS (induced via viroporins (a1) or TRPV4 (a2)) and binds to the precursor of caspase-1 (procaspase-1) through the adaptor protein ASC in the cell to form a multiprotein complex, thereby activating caspase-1. b) Entry of danger-associated molecular pattern (self- DNA) into the cytoplasm from the nucleus of mitochondria or dead cells activates the cGAS–STING pathway. c) Binding of viral PAMPs/DAMPs to the TLRs and activation of transcription factors for inducing proinflammatory factors. d) Viral structural proteins or binding of virus-Ab complex to FcR can also activate proinflammatory responses via MAPK signaling. e) Binding of the spike protein to ACE2 induces ADAM 17 activity, thereby reducing the number of ACE2 expressed on the cell surface.
The activity of 3a, E, and 8a proteins as ion-channel (IC) or viroporins in SARS-CoV has been previously identified (Castaño-Rodriguez et al., 2018). The IC activity in the membrane of the endoplasmic reticulum (ER)-Golgi intermediate compartment (ERGIC) leads to the release of Ca2 + into the cytosol, activation of the inflammasome complex, and subsequently the release of pro-inflammatory cytokines such as TNF-α, IL-1β, and IL-6 (Farag et al., 2020) (Fig. 5a1). The transient receptor potential channel subfamily V member 4 (TRPV4) cation is a mechanosensitive Ca2 + channel that is widely expressed in all tissues as well as in immune cells (Michalick and Kuebler, 2020). In macrophages, TRPV4 mediates pro-inflammatory functions and anti-inflammatory effects (Hamanaka et al., 2010; Liedtke et al., 2011). In lung alveolar epithelium, TRPV4 has been shown to act as a key regulator of epithelial barrier function and the alveolar-capillary barrier (Kuebler et al., 2020). During infection, TRPV4 activation through pathogen-associated signals can lead to pro-inflammatory phenotypes and thereby boosts the acute inflammatory responses (Scheraga et al., 2020) (Fig. 5a2). Thus, loss of the TRPV4 barrier function can lead to the formation of alveolar edema (Alvarez et al., 2006). It is believed that TRPV4 hyperactivation plays a key role in inflammatory diseases by damaging the alveolar-capillary barrier, and so has currently been considered as a target for developing therapeutic strategies against COVID-19 (Kuebler et al., 2020).
The DNA sensors, cyclic GMP-AMP synthase (cGAS) and STING (stimulator of interferon genes), regulate the expression of inflammatory IFN-I/III mediators (Channappanavar et al., 2016; Sun et al., 2009). Dysfunction of these pathways in response to lesions of cells and tissues, autoimmune diseases, and STING-associated vasculopathy (SAV) gives rise to human inflammatory disease (Liu et al., 2014). Self-DNA as DAMP (danger associated molecular patterns) is released from damaged tissues and apoptotic cells associated with the viral infection, and can induce cGAS-STING-IFN pathway, which subsequently leads to cytokine storm (Benmerzoug et al., 2019) (Fig. 5 b). In addition, SARS-CoV-2 S protein and other virulence factors can modulate the IL-1β secretion through activation of TLR4 and thus exacerbate the lung inflammation (Conti et al., 2020) (Fig. 5 c).
Infectious virus-antibody complexes interact with the Fc receptor (FcR) or other cells receptors and promote viral cellular uptake, and then activate the MAPK pathway as a mediator of pro-inflammatory responses (Chan et al., 2020). Furthermore, the MAPK pathway can be activated through antibody-dependent enhancement (ADE) mechanism by employing anti-spike IgG (Anti-S-IgG), PDZ-binding motif (PBM) of E protein, and structural proteins to induce inflammatory responses (Alam et al., 2020) (Fig. 5 d). For instance, PBM enables SARS-CoV and SARS-CoV-2 to bind to over 400 cellular proteins for the control of cell function and virulence (Alam et al., 2020; Castaño-Rodriguez et al., 2018). Indeed, the interaction of the viral PBM with syntenin PDZ motifs activates the p38-MAPK pathway and promotes the acute pro-inflammatory response (Jimenez-Guardeño et al., 2014). Additionally, structural 3a and 7a proteins, by promoting the JNK activation, induce the inflammation and aggravate SARS-CoV infection (Kanzawa et al., 2006; Varshney and Lal, 2011). Altogether, these features trigger the cytokine storm, leading to increased edema in the lungs and thereby causing acute respiratory distress syndrome (ARDS) (Alam et al., 2020).
During acute lung injury, ACE2 can play a protective role (Hamming et al., 2004; Kuba et al., 2005). However, some studies have shown that binding of SARS-CoV-2 to ACE2 using SARS-CoV S protein in the mouse model downregulates the ACE2 expression and neutralizes its protective effect (Glowacka et al., 2010). It has been demonstrated that the cell entry of the virus induces the ADAM-17-dependent shedding of ACE2 N-terminal domain (Oudit et al., 2009), and this depletion leads to dysfunction of the renin-angiotensin system (RAS) and enhancement in inflammation and vascular permeability (Chan et al., 2020) (Fig. 5 e).
6. Immunopathogenesis of COVID-19
The SARS-CoV-2 life cycle is not cytolytic, and viral replication and assembly are temporally linked with epithelial cells; nevertheless, the damage to the lungs of COVID-19 patients has been observed in severe cases (Abdulamir and Hafidh, 2020; Lu et al., 2020; Zhou et al., 2020a). So, what is the cause of this damage? It can be the outcome of intense inflammatory responses associated with immune dysregulation, direct infection of immune cells, or autoimmune reactions (Dandekar and Perlman, 2005). Although the exact immune mechanism that brings about mortality among COVID-19 patients is still unknown, however, immune responses in COVID-19 infection are induced in both mild and severe phases (Shi et al., 2020).
Upon SARS-CoV-2 infection, the respiratory epithelial cells and alveolar macrophages initiate an innate immune response through secretion of inflammatory factors (Alosaimi et al., 2020), leading to the recruitment of neutrophils and inflammatory monocyte-macrophages (IMMs) to the infection site (Channappanavar and Perlman, 2020; Kindler and Thiel, 2016). In our previous review we have discussed about mechanisms of influenza infection-induced metabolic perturbations leading to aberrant immune responses (Keshavarz et al., 2020). In fact, by interfering the host's immunometabolic balance viruses can tailor the immune response for their propagation and survival. Changes in the bioenergetics and mitochondrial dysfunction of monocytes are evident in patients with COVID-19 pneumonia. During these changes monocytes develop extensive disorders in the glycolysis pathway, cellular respiration and the structure and function of mitochondria. These metabolic changes are accompanied by a significant increase in different subsets of monocytes with higher expression levels of inhibitory checkpoints PD-1 or PDL-1 and increased plasma levels of inflammatory cytokines and chemokines including GM-CSF, IL-18, CCL2, CXCL10 and osteopontin. This can reveal the important role of monocytes in exacerbating inflammation, pathogenesis and mortality due to COVID-19 disease and worse prognosis in these patients (Gibellini et al., 2020). The process of these changes is also similar to the immunometabolic paralysis seen during sepsis (Rubio et al., 2019). High amounts of IL-1B, IFN-γ, chemokine CXCL10, and monocyte chemoattractant protein-1 (MCP-1) in the peripheral blood induce T-helper 1 (Th1)-mediated immune responses, in order to modulate the uncontrolled inflammatory responses (Huang et al., 2020). Findings indicate an increase in the expression of HIF1α and its transcriptionally regulated genes in blood myeloid cells of patients with severe COVID-19 disease. In these patients a shift toward an increase in circulating immature myeloid cells occurs, that has been shown to be caused by a physiological response called emergency myelopoiesis. These myeloid cells together with bronchoalveolar cells express HIF1α and its regulated genes, which are associated with inflammation, virus identification, and metabolism. Increased expression of HIF1α in COVID-19 patients and its role in inflammatory processes, immunometabolism and expression of TLRs, can make it a suitable molecular marker to assess the severity of COVID-19 and a preferential option for targeted treatment of this disease (Taniguchi-Ponciano et al., 2021). SARS-CoVs also stimulate antigen-presenting cells (APCs), which contribute to pro-inflammatory cytokine milieu and activate naïve CD4+ T cells. The regulated immune responses and regenerative processes can eliminate the virus (in non-severe stages of the disease) and preclude the disease progression to severe stages (Shi et al., 2020) (Fig. 6 a). However, reduced/delayed IFN-I signaling and the high level of viral and immune responses, all expand the number of recruited immune cells, which is associated with local inflammation and lung damage (Channappanavar and Perlman, 2020; Roberts et al., 2007) (Fig. 6 b). Significant increase in inflammatory factors and inducers of vasculit is and vascular remodeling have been observed in COVID-19 patients, some of which are significantly associated with severe disease and ICU admission. Factors associated with the Th2 response including IL-4 and IL-13 may increase, with IL-4 increasing in men more than women. Overall, an intertwined network of cytokines and growth factors leads to vascular and stromal remodeling, activation of innate immunity and activation of type 2 immune responses, which are involved in COVID-19 immunopathogenesis and increased severity of the disease (Petrey et al., 2021).
Fig. 6.
Non-severe and severe COVID19. a) During non-severe stage, innate and specific adaptive immune responses can eliminate the virus and prevent disease progression to severe stages. b) Nevertheless, high viral load and propagation, followed by dysregulated immune response and massive destruction of the affected tissues can induce innate inflammation in the lungs that is largely mediated by inflammatory monocyte-macrophages (IMMs). Massive accumulation of pathogenic inflammatory IMMs and exuberant inflammation increase the severity of disease and lead to lung damage in the severe stages of the disease.
Severe inflammation associated with cytokine storm can change the differentiation and activity of lymphocytes. Analysis of some parameters related to lymphocytes has shown the increased expression of T cells exhaustion markers such as PD-1, Tim-3 and NKG2A and the decreased number of total T cells and NK cells (Bell et al., 2019; Zheng et al., 2020). These conditions induce apoptosis in T cells that have sensitized to IFN-I and reduce the number of virus-specific CD8+ and CD4+ T cells (Channappanavar and Perlman, 2020). Virus-induced inflammatory factor storms can generate serious pathological changes including fibrosis, alveolar-capillary barrier damage, hyaluronic acid (HA) membrane formation, and HA production that are collectively linked to ARDS (Bell et al., 2019; Hallgren et al., 1989). Therefore, optimization and modulation of immune responses may be a promising treatment for severe COVID-19 patients.
SARS-CoV-2 has shown the heterogeneity of clinical signs and symptoms such as those observed to be akin to Kawasaki disease and toxic shock syndrome, namely multisystem inflammatory syndrome (MIS) that first recognized in children (MIS-C) (Godfred-Cato et al., 2020; Riollano-Cruz et al., 2020) and then was reported in adults (MIS-A) (Shaigany et al., 2020). Regardless of the role of acute viral infection in this occurrence, some highlight the mediating role of IgG antibody in enhancement of the disease post-infectiously. The latter postulation is strengthen by the facts that (1) in a number of countries and after a peak of SARS-CoV-2, a delay has been observed before there port of MIS-C cases; and (2) frequently, unlike RT-PCR assay, SARS-CoV-2 antibody testing of children with MIS-C has been resulted positive (Rowley, 2020). Comparison of severe clinical signs of COVID-19 in children has identified a wide range of symptoms from pneumonia to MIS-C. MIS-C is the most common clinical form in children with severe COVID-19 disease. Instead of the classic COVID-19 disease with severe respiratory involvement, these children are more likely to have gastrointestinal symptoms, shock and immune response dysregulation and to need vasoactive drugs, and are less likely to need mechanical ventilation (García-Salido et al., 2020). Many possibilities including injury from systemic inflammation, acute viral myocarditis, hypoxia, stress cardiomyopathy, and with less probability coronary artery (CA)-mediated ischemia have been proposed to describe this situation (Sperotto et al., 2020). Moreover, the observed cardiac dysfunction in some patients may be due to the combinatorial adverse effect of these mechanisms. It is also likely that the variability in clinical presentation be due to the involvement of distinct mechanisms in different patients (Godfred-Cato et al., 2020).
More recent studies have suggested certain immunological conditions as leading causes of MIS-C. SARS-CoV-2-associated acute MIS-C has been characterized by higher B-cell plasmablast and double-negative B-cell frequencies accompanied by severe activation of some other immune cells including neutrophils, monocytes and memory CD8+ T-cells. Furthermore, systemic activation of complement factors and increased plasma C5b-9 level were related to acute MIS-C progression (Syrimi et al., 2021). Such possible role for hyper-activated complement system in pathogenesis of MIS-C also has been proposed elsewhere (Pombo et al., 2021). A study on several children with SARS-CoV-2 infection and mild to severe MIS-C has revealed that CDR3-independent interaction of unprocessed SARS-CoV-2 spike protein with T cell receptor (TCR) causes extensive proliferation and activation of TCRβ variable gene 11–2 (TRBV11–2) T cells which is associated with cytokine storm and MIS-C severity. Moreover, all patients had HLA class I alleles A02, B35, and C04 simultaneously which was associated with TRBV11–2 T cells expansion and increased susceptibility to severe MIS-C (Porritt et al., 2021). Activation of NF-κB by SARS-CoV-2 S and N proteins induces production of proinflammatory cytokines like IL-6 which shows positive correlation with disease severity. In fact, SARS-CoV-2-associated MIS-C can be a result of cytokine release syndrome (CRS) and these are tightly related due to their inflammatory basis (Que et al., 2021).
7. Immune-based therapies in COVID-19
Therapy strategies such as inhibition of SARS-CoV-2 fusion/entry, disruption of replication, suppression of excessive inflammatory response, and combination strategies are racing to develop a treatment against COVID-19 (Horby et al., 2020; Roth et al., 2020). Recently, thousands of clinical trials have been conducted to evaluate the effectiveness of those strategies for the treatment of COVID-19 all around the world. Among those clinical trials, inhaled nitric oxide has shown a beneficial effect in eliciting antiviral responses against SARS-CoV-2 (Alvarez et al., 2020). In another trial, danoprevir (Ganovo), which is a potent protease inhibitor and has been approved and marketed to treat chronic hepatitis C patients, has been shown to be effective against SARS-CoV-2, which may be due to the structure similarity between chymotrypsin-like protease of SARS-CoV-2 with HCV proteases (Chen et al., 2020). Hydroxychloroquine, an antimalarial drug, has also been reported to be efficient in reduction of viral load among COVID-19 patients and its efficiency was further enhanced by azithromycin (Gautret et al., 2020).
Table 1, Table 2, Table 3 have summarized the ongoing clinical trials for treatment of COVID-19. However, the balance of innate and adaptive immune responses is a decisive factor in the treatment of COVID-19, which can be achieved by immunomodulatory agents with or without combination with antivirals (Table 3). The imbalanced IFN-mediated immune response is considered as a key factor in the COVID-19 severity (Xu et al., 2020), especially in the elderly than in children and adults, which may be due to the earlier induction of IFNs in children and their less developed immune system (Vabret et al., 2020; Zhou et al., 2018). Furthermore, inhibition of cytokine storm by the regulation of immune responses using immunomodulatory agents such as agonists for suppression of IL-6, IL-1β, and TH17 type responses is possible. Moreover, IL-10-mediated hampering of IFN-γ, IL-12, and TNF-α, by polarizing immune responses toward Th2 and away from Th1 pathways (Fadel et al., 2020), might minimize the lung injuries and increase the survival chance. The efficacy of antagonism of cytokine/chemokine signaling pathways such astocilizumab, sarilumab, siltuximab for IL-6 signaling pathway, and gimsilumab for targeting GM-CSF are now being tested for managing COVID-19 outbreak (Vabret et al., 2020). In a study on a 50 years old patient with cytokine storm and ARDS induced by COVID-19, administration of anakinra, an IL-1 receptor antagonist commonly used to treat auto-inflammatory disorders in adult patients, reduced oxygen demand and improved inflammatory and ferritin markers (Nemchand et al., 2020). Recently, corticosteroids such as dexamethasone, which can reduce inflammation through inhibition of pro-inflammatory transcription factors, have demonstrated to reduce the hospitalization period of COVID-19 patients (Gralinski and Menachery, 2020; Horby et al., 2020). Neutrophil extracellular traps (NETs) are web-like structures containing antimicrobial granules that prevent the spread of pathogens in the bloodstream (Papayannopoulos, 2018), but excessive increase of NETs in the serum of COVID-19 patients leads to endothelial dysfunction and microvascular immunothrombosis and is associated with multi-organ damage and high mortality (Middleton et al., 2020). Fostamatinib, a spleen tyrosine kinase inhibitor, is a potential treatment for COVID-19 disease that can inhibit the release of NETs induced by the plasma of COVID-19 patients (Strich et al., 2021).
Table 2.
Summary of combination strategies against severe COVID-19 (Ky and Mann, 2020; Lythgoe and Middleton, 2020).
| Treatment strategies | Drug name | Mechanism of action |
|---|---|---|
| Antiviral/Antimalarial | Lopinavir, ritonavir, hydroxychloroquine | Viral replication and viral entry |
| Remdesivir, hydroxychloroquine remdesivir; hydroxychloroquine | Viral replication and viral entry | |
| oseltamivir, chloroquine, | ||
| Darunavir, ritonavir, lopinavir, oseltamivir, favipiravir | Viral replication and viral entry | |
| Antiviral/immunomodulators | Lopinavir þ ritonavir; ribavirin; IFN-b1b | Antiretroviral and immunomodulatory |
| Favipiravir þ tocilizumab | Antiretroviral and immunomodulatory | |
| Antiviral/Antimalarial/immunomodulators | Remdesivir; lopinavir þ ritonavir; IFN-b1a; hydroxychloroquine | Antiretroviral, viral entry, and immunomodulatory |
Table 3.
Summary of immunomodulatory agents against severe COVID-19.
| Therapeutic agents | Summary | References |
|---|---|---|
| Poly ICLC, Interferon alpha | Interferon-inducing agents and innate anti-viral response | Saxena et al., 2019; Wang et al., 2020a, Wang et al., 2020b, Wang et al., 2020c |
| Suramin | Inhibition of cGAS-STING-IFN production pathway | (Wang et al., 2018, Wang et al., 2020e) |
| Tocilizumab (TCZ) | IL6R antagonists, inhibition of IL-6 that is correlated with cytokine storm | (Emery et al., 2008; Norelli et al., 2018) |
| Fedratinib | Inhibition of the TH17 type response that contributes to the cytokine storm | Wu and Yang (2020) |
| Amantadine, hexamethylene amiloride 9, 10, SB2035805 | Inhibition of proinflammatory response arisen from E protein functions | Alam et al. (2020) |
| Lymphocyte Antigen 6 Family Member E (LY6E) | LY6E impairs coronavirus fusion and confers immune control of viral disease | (Pfaender et al., 2020) |
| Melatonin | The efficacy in the inhibition of NLRP3 inflammasome, reducing the infiltration of macrophages and neutrophils into the lung | (Acuña-Castroviejo et al., 2020) |
| Hydroxychloroquine (HCQ) | Inhibiting the cytokine storm by reducing CD154 expression in T cells | (Zhou et al., 2020b) |
| Anti-NKG2A mAb, Monalizumab | Targeting NKG2A may prevent the functional exhaustion of cytotoxic lymphocytes | (Yaqinuddin and Kashir, 2020) |
| Thalidomide | Regulating immunity, inhibiting the inflammatory cytokine surge | Chen et al. (2020) |
| Jakotinib, hydrochloride, Baricitinib | a JAK inhibitor as well as an AAK1 inhibitor | Zhang et al. (2020) |
| Chloroquine (QC), Tocilizumab, Baricitinib | To quench the cytokine storm | (Askanase et al., 2020) |
| Anakinra | Interleukin 1 receptor antagonist | (Shetty et al., 2020) |
| Intravenous immunoglobulin (IVIg) | Blocking Fc gamma receptor mediated response | (Shetty et al., 2020) |
| Mesenchymal stem cells (MSCs) | Powerful immunomodulatory for innate and adaptive immune system | (Leng et al., 2020) |
| IL-10 | Polarizes immune system toward Th2 | Abdulamir and Hafidh (2020) |
| IL-37, IL-38 | Inhibit IL-1β and other proinflammatory IL-family members | Conti et al. (2020) |
| Etanercept, adalimumab | TNF-α inhibitor | (Russell et al., 2020) |
The gold standard strategy for improving COVID-19 infection can be followed through inhibition of the hyperinflammation and subsequent repairing of damaged cells, which can be achieved by means of cell therapy and regenerative medicine (Parhizkar Roudsari et al., 2020; Roudsari et al., 2020). Applying mesenchymal stem cells (MSCs)-based approach is a hopeful strategy to reduce inflammation via releasing prostaglandin E2 (PGE2), transforming growth factor-beta (TGF-β), nitric oxide (NO), and indoleamine 2,3-dioxygenase (IDO), and repairing damaged tissue by keratinocyte growth factor (KGF), vascular endothelial growth factor (VEGF) and hepatocyte growth factor (HGF) (Golchin et al., 2020; Parhizkar Roudsari et al., 2020). Moreover, cell therapy based on NK-cells using NCT04280224, CYNK-001, and NKG2D-ACE2 CAR-NK cells is currently recruiting COVID-19 patients (Bonam et al., 2020; Market et al., 2020). Immune-based therapy with IFN (ChiCTR: ChiCTR 2,000,029,600) with specific timing and dose has also been adopted for treatment of SARS-CoV-2 infection (Zhou et al., 2020c).
Besides, the oral administration of Olumiant (baricitinib), a JAK1/JAK2 suppressor under evaluation in phase 3 in adult hospitalized COVID-19 patients, has shown potential in decreasing the serum levels of pro-inflammatory cytokines, rapid recovery of circulating T and B cell frequencies, and increasing the antibody production against the SARS-CoV-2 spike protein, and thereby reduction in the need for oxygen therapy (Bronte et al., 2020). Leronlimab, the C—C chemokine receptor type 5 inhibitor developed against HIV-1 infection, is another drug that is in early stages of clinical trials for treatment of COVID-19 disease (Yang et al., 2020).
Using immunoglobulins and plasma therapy have improved clinical outcomes (Zhou et al., 2020c) and is recommended for healthcare systems of COVID-19 (Chen and Xia, 2020; Verma et al., 2020). Generally, until the production of an efficient immunotherapy agent such as vaccine, which produces a durable response through inducing immunological memory, the reduction in severity of the disease is in demand for administration of passive immunotherapy agents. For that purpose, produced neutralizing antibodies against structural proteins of SARS-CoV-2 such as monoclonal antibodies, polyclonal antibodies, and convalescent plasma from recovered COVID-19 patients are currently under investigation (Vabret et al., 2020).
8. Talented SARS-CoV-2Epitopes for vaccination
SARS-CoV-2 dramatically differs in transmissibility compared to other coronaviruses (Goh et al., 2020), which may be due to the differences in predicted intrinsic disorder (PID), an index for the rigidity of viral shells (Dandekar and Perlman, 2005). PID analysis has demonstrated that the outer shell of SARS-CoV-2is the hardest among the coronavirus family members and so is more resilient in body fluids and the environment (Dandekar and Perlman, 2005; Kindler and Thiel, 2016). This property and fecal-oral-respiratory way of the virus transmission (Huang et al., 2020), together with durability of the virus outside the body for longer period justify the upper rate of SARS-CoV-2 spread than of MERS-CoV and SARS-CoV. As it is an emerging virus, many target antigen selection and vaccine platform for SARS-CoV-2 are inherited from previous antibody-mediated protection studies on SARS-CoV and MERS-CoV (Prompetchara et al., 2020). Accordingly, the SARS-CoV RBD recombinant protein represents a potential candidate for the production of SARS-CoV-2 heterologous vaccine (Gralinski and Menachery, 2020). The non-self-sequences with minimal risk of cross-reactivity are considered ideal immunogenic epitopes; thus, most investigations have focused on the surface S glycoprotein of HCoVs, due to its high immunogenicity and ability to exert an efficient neutralizing effect (Gralinski and Menachery, 2020; Lucchese, 2020). Recently, 66 epitopes out of 107 human-foreign spike protein pentapeptides have been offered for this purpose, providing experimental proof for the immunogenic potential of non-self-peptides (Lucchese, 2020). B-cell and MHC class I and class II allele epitopes have been identified as best immunogenic peptides based on the number of alleles and antigenicity scores. The antigenic differences or invalidation of the most SARS-CoV spike protein antibodies for SARS-CoV-2 can be attributed to then on-conserved regions of SARS-CoV-2 spike protein sequence in comparison to SARS-CoV (Lv et al., 2020; Tian et al., 2020; Zheng and Song, 2020). However, the S2 domain of SARS-CoV and SARS-CoV-2 is a highly conserved and immunogenic target; thus, mAbs targeting the S2 domain of SARS-CoV can cross-react with SARS-CoV-2 and serve as efficient antigens for inducing both humoral and cellular immunity against SARS-CoV-2.
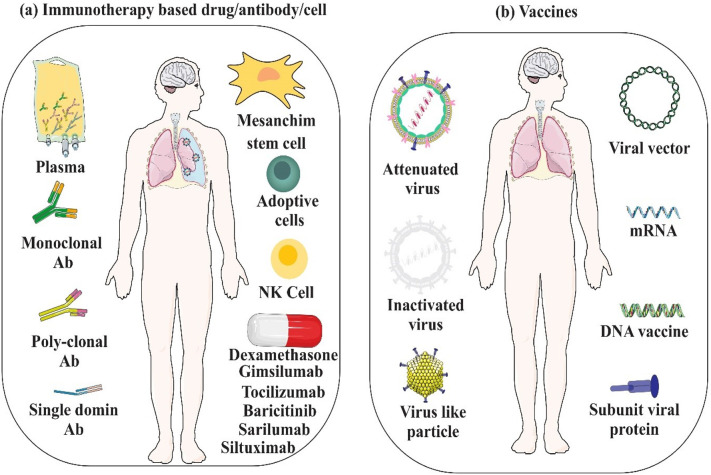
On the other hand, due to the scarcity of cross-reactive antibodies binding both SARS-CoV-2 and SARS-CoV S proteins and providing cross-neutralizing responses, development of new antibodies and vaccines specific of SARS-CoV-2 seems necessary (Lv et al., 2020). The antiviral properties of viral vaccines are often based on antibody-mediated protection; although they can be efficient against virus infection, owing to their poor induction of T cell responses, the possibility of rapid mutation in the surface proteins of the virus (Rosendahl Huber et al., 2014), and the ability of the virus in entering quiescently (Shin et al., 2020), those vaccines may not be able to provide the thorough immunity. The current vaccine methodologies are based on the development of personalized vaccines like those developed against some cancers or populationalized vaccinomics, in which the design of peptides are based on viral genome and the immune characteristics of the individual or the target populations such as epitopes with the capacity to bind to multiple HLA alleles, or CD8+ and CD4+ T cell epitopes against S and M proteins (De Groot et al., 2020). Moreover, due to their high potential in eliciting cellular and humoral immunity, design of multi-epitope vaccines (MEV) such as a 55,426.35 kDa vaccine protein, which has been designed against SARS-CoV-2 using immunoinformatics approaches, is of importance (ul Qamar et al., 2020). Using novel synthetic biology approaches that exploit reverse genetics (Gibson et al., 2010) and transformation-associated recombination (TAR) cloning (Kouprina and Larionov, 2008) to reconstruct the virus of target, biomedical scientists are now able to traceSARS-CoV-2 functional and structural evolution forthwith, helping them in immediate responding to appearing alterations and also in efficient vaccine development (Nhu Thao et al., 2020). Many technologies for manufacturing of COVID-19 vaccine, which initially look promising, are summarized in Fig. 7 b.
Fig. 7.
Immunotherapy and immunization against COVID-19. a) Immunotherapy based drugs/antibodies/cells can directly/indirectly suppress virus particles, and modulate immune system in severe COVID-19. b) Strategies for developing COVID-19 vaccines.
So far, a variety of these technologies have been employed successfully to develop effective vaccines against SARS-CoV-2 infection. The BBIBP-CorV vaccine is one of the recently developed vaccines manufactured based on the inactivated virus platform. In an early study in several mammalian species, the vaccine induced the production of high levels of neutralizing antibodies and provided effective protection against SARS-CoV-2 challenge in rhesus macaques (Wang et al., 2020d). This vaccine was then approved in subsequent randomised, double-blind, placebo-controlled clinical trials in terms of high safety and immunogenicity (Xia et al., 2021). Another successful vaccine developed after the SARS-CoV-2 pandemic to combat the COVID-19 severity and mortality was the ChAdOx1 nCoV-19 (AZD1222) vaccine which is a replication-deficient recombinant chimpanzee adenovirus vector expressing the SARS-CoV-2 spike protein gene (Folegatti et al., 2020). In animal studies, this vaccine significantly reduced viral load and also showed no adverse side effects (Marsh et al., 2021). The ChAdOx1 nCoV-19 vaccine has also shown excellent results in several blinded, randomised, controlled clinical trials and its safety and efficacy against symptomatic and severe COVID-19 has been proven in different age groups (Falsey et al., 2021; Voysey et al., 2021). However, there are some recent reports about post-vaccination venous sinus thrombosis and thrombocytopenia syndrome that have raised little doubt about continued general vaccination with ChAdOx1 nCoV-19 and indicate that this vaccine may need to be used with more cautions (Douxfils et al., 2021; Mehta et al., 2021; Wolf et al., 2021). Despite of these rare post-vaccination lethal adverse events, comprehensive studies have shown considerable benefit of vaccination with this vaccine in preventing COVID-19 mortality especially in people over 55 years of age (Kiem et al., 2021). The recombinant adenovirus vector technology also has been employed for development of another SARS-CoV-2 vaccine named Ad26.CoV2·S which can induce potent neutralizing antibody response and Th1 IFN-γ type cellular immunity in mice (Bos et al., 2020). The results of clinical trials of this vaccine are also very satisfactory with acceptable safety and induction of potent immune response that can protect against severe COVID-19 disease, hospitalization and mortality (Sadoff et al., 2021a; Sadoff et al., 2021b). Continuous efforts since the COVID-19 pandemic have also led to the development of two highly effective vaccines produced as nanoparticle–encapsulated, nucleoside-modified, mRNA encoding SARS-CoV-2 spike protein, including BNT162b2 (Walsh et al., 2020) and mRNA-1273 (Corbett et al., 2020a). BNT162b2 vaccine has successfully passed all clinical trials with 95% protection against Covid-19 and a reassuring safety in ≥16 years old participants (Polack et al., 2020; Walsh et al., 2020). The other mRNA vaccine, mRNA-1273 is able to elicit highly protective immune response in nonhuman primates (Corbett et al., 2020b), and also subsequent clinical trials have shown a favorable safety profile and 94.1% efficacy in severe COVID-19 prevention for this vaccine (Baden et al., 2021; Jackson et al., 2020). Although very rare cases of allergic reactions as anaphylaxis has been reported after administration of the first dose of these two mRNA vaccines which prevented the injection of the second dose (COVID, C, Team, R, 2021a, COVID, C, Team, R, 2021b).
9. Conclusion
COVID-19 is a respiratory disease that results from an undesirable interaction between SARS-CoV-2 and the host's immune system, which together promote the duration, and severity of the disease. Indeed, it is quite obvious now that the extensive tissue and organ damage and high mortality following SARS-CoV-2 infection cannot be attributed to the limited pathogenic effect of viral propagation alone, and instead, the outcome of the disease caused by this virus is forcefully dependent to dilemma of protective or pathogenic host immune response. This emphasizes on the central role of the improperly severe inflammatory aspects of immune response in the pathogenesis of the COVID-19 disease which take the initiate and disrupt the regulative and protective role of CD4+ and CD8+ T cells. This phenomenon is strongly supported by the beneficial effects of immunomodulatory therapy on controlling disease severity. Over the past 2 years, substantial progress has been made in deciphering the mechanisms of COVID-19 immunopathogenesis and new advanced vaccines and immunotherapeutic agents have been developed and evaluated in clinical trials which should be extended more seriously. Also continues monitoring of SARS-CoV-2 genomic evolution and antigenic changes can facilitate updating the existing vaccines as soon as emergence of new highly pathogenic mutant strains. However, despite the relative success of these methods in infection control, therapeutic approaches based on the host's genetic susceptibility and personalized medicine seem to be the missing link of universal efforts for COVID-19 prevention and control. Future studies also can focus on a comprehensive understanding of factors complicating the COVID-19 control process especially the immune evasion mechanisms of the virus, to develop more updated specific vaccines and even theranostic agents.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and materials
Not applicable.
Funding
Not applicable.
Declaration of competing interest
The authors declare that they have no competing interests.
Acknowledgements
Not applicable.
References
- Abdulamir A.S., Hafidh R.R. The possible immunological pathways for the variable Immunopathogenesis of COVID--19 infections among healthy adults, elderly and children. Electronic Journal of General Medicine. 2020;17 [Google Scholar]
- Acuña-Castroviejo D., Escames G., Figueira J.C., de la Oliva P., Borobia A.M., Acuña-Fernández C. Clinical trial to test the efficacy of melatonin in COVID-19. J. Pineal Res. 2020;69:12683. doi: 10.1111/jpi.12683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam I., Kamau A., Kulmanov M., Arold S.T., Pain A., Gojobori T., Duarte C.M. bioRxiv; 2020. Functional Pangenome Analysis Suggests Inhibition of the Protein E as a Readily Available Therapy for COVID-2019. (2020.2002.2017.952895) [Google Scholar]
- Alosaimi B., Hamed M.E., Naeem A., Alsharef A.A., AlQahtani S.Y., AlDosari K.M., Alamri A.A., Al-Eisa K., Khojah T., Assiri A.M., et al. MERS-CoV infection is associated with downregulation of genes encoding Th1 and Th2 cytokines/chemokines and elevated inflammatory innate immune response in the lower respiratory tract. Cytokine. 2020;126 doi: 10.1016/j.cyto.2019.154895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez D.F., King J.A., Weber D., Addison E., Liedtke W., Townsley M.I. Transient receptor potential vanilloid 4–mediated disruption of the alveolar septal barrier: a novel mechanism of acute lung injury. Circ. Res. 2006;99:988–995. doi: 10.1161/01.RES.0000247065.11756.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez R.A., Berra L., Gladwin M.T. American Thoracic Society; 2020. Home Nitric Oxide Therapy for COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artika I.M., Dewantari A.K., Wiyatno A. Molecular biology of coronaviruses: current knowledge. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e04743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askanase A.D., Khalili L., Buyon J.P. Thoughts on COVID-19 and autoimmune diseases. Lupus Sci. Med. 2020;7 doi: 10.1136/lupus-2020-000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batool M., Shah M., Patra M.C., Yesudhas D., Choi S. Structural insights into the Middle East respiratory syndrome coronavirus 4a protein and its dsRNA binding mechanism. Sci. Rep. 2017;7:11362. doi: 10.1038/s41598-017-11736-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell T.J., Brand O.J., Morgan D.J., Salek-Ardakani S., Jagger C., Fujimori T., Cholewa L., Tilakaratna V., Östling J., Thomas M. Defective lung function following influenza virus is due to prolonged, reversible hyaluronan synthesis. Matrix Biol. 2019;80:14–28. doi: 10.1016/j.matbio.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmerzoug S., Ryffel B., Togbe D., Quesniaux V.F. Self-DNA sensing in lung inflammatory diseases. Trends Immunol. 2019;40:719–734. doi: 10.1016/j.it.2019.06.001. [DOI] [PubMed] [Google Scholar]
- Bermejo-Martin J.F., Almansa R., Menéndez R., Mendez R., Kelvin D.J., Torres A. Lymphopenic community acquired pneumonia as signature of severe COVID-19 infection. J. Inf. Secur. 2020;80:e23–e24. doi: 10.1016/j.jinf.2020.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blot M., Bour J.-B., Quenot J.P., Bourredjem A., Nguyen M., Guy J., Monier S., Georges M., Large A., Dargent A. The dysregulated innate immune response in severe COVID-19 pneumonia that could drive poorer outcome. J. Transl. Med. 2020;18:1–14. doi: 10.1186/s12967-020-02646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonam S.R., Kaveri S.V., Sakuntabhai A., Gilardin L., Bayry J. Adjunct immunotherapies for the management of severely ill COVID-19 patients. Cell Reports Medicine. 2020;1 doi: 10.1016/j.xcrm.2020.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos R., Rutten L., van der Lubbe J.E., Bakkers M.J., Hardenberg G., Wegmann F., Zuijdgeest D., de Wilde A.H., Koornneef A., Verwilligen A. Ad26 vector-based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 spike immunogen induces potent humoral and cellular immune responses. npj Vaccines. 2020;5:1–11. doi: 10.1038/s41541-020-00243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman N., Rantasärkkä K., Feuth T., Valtonen M., Waris M., Hohenthal U., Rintala E., Karlsson A., Marttila H., Peltola V. IL-6 and other biomarkers as predictors of severity in COVID-19. Ann. Med. 2021;53:410–412. doi: 10.1080/07853890.2020.1840621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronte V., Ugel S., Tinazzi E., Vella A., De Sanctis F., Canè S., Batani V., Trovato R., Fiore A., Petrova V. Baricitinib restrains the immune dysregulation in patients with severe COVID-19. J. Clin. Invest. 2020;130 doi: 10.1172/JCI141772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;eabd2985 doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño-Rodriguez C., Honrubia J.M., Gutiérrez-Álvarez J., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Fernandez-Delgado R., Verdia-Báguena C., Queralt-Martín M., et al. Role of severe acute respiratory syndrome coronavirus Viroporins E, 3a, and 8a in replication and pathogenesis. mBio. 2018;9 doi: 10.1128/mBio.02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S., Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes & Infections. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Y.K., Gack M.U. Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 2016;14:360–373. doi: 10.1038/nrmicro.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Perlman S. In: MERS Coronavirus: Methods and Protocols. Vijay R., editor. Springer US; New York, NY: 2020. Evaluation of activation and inflammatory activity of myeloid cells during pathogenic human coronavirus infection; pp. 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R., Fehr A.R., Vijay R., Mack M., Zhao J., Meyerholz D.K., Perlman S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Xia R. Early experience with convalescent plasma as immunotherapy for COVID-19 in China: Knowns and unknowns. Vox Sang. 2020;115:507–514. doi: 10.1111/vox.12968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zhang Z., Wang L., Huang Z., Gong F., Li X., Chen Y. MedRxiv; 2020. First clinical Study Using HCV Protease Inhibitor Danoprevir to Treat Naive And Experienced COVID-19 Patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Su C., Ke M., Jin X., Xu L., Zhang Z., Wu A., Sun Y., Yang Z., Tien P., et al. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 2′-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti P., Ronconi G., Caraffa A., Gallenga C.E., Ross R., Frydas I., Kritas S.K. Induction of pro-inflammatory cytokines (IL-1 and IL-6) and lung inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): anti-inflammatory strategies. J. Biol. Regul. Homeost. Agents. 2020;34:327–331. doi: 10.23812/CONTI-E. [DOI] [PubMed] [Google Scholar]
- Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O’Connell S., Bock K.W., Minai M. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID, C, Team, R Allergic reactions including anaphylaxis after receipt of the first dose of Moderna COVID-19 vaccine—United States, December 21, 2020–January 10, 2021. Morb. Mortal. Wkly Rep. 2021;70:125. doi: 10.15585/mmwr.mm7004e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COVID, C, Team, R Allergic reactions including anaphylaxis after receipt of the first dose of Pfizer-BioNTech COVID-19 vaccine—United States, December 14–23, 2020. Morb. Mortal. Wkly Rep. 2021;70:46. doi: 10.15585/mmwr.mm7002e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar A.A., Perlman S. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 2005;5:917–927. doi: 10.1038/nri1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot A.S., Moise L., Terry F., Gutierrez A.H., Hindocha P., Richard G., Hoft D.F., Ross T.M., Noe A.R., Takahashi Y., et al. Better epitope discovery, precision immune engineering, and accelerated vaccine design using Immunoinformatics tools. Front. Immunol. 2020:11. doi: 10.3389/fimmu.2020.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit E., Van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschler S., Kager J., Erber J., Fricke L., Koyumdzhieva P., Georgieva A., Lahmer T., Wiessner J.R., Voit F., Schneider J. Mucosal-associated invariant T (MAIT) cells are highly activated and functionally impaired in COVID-19 patients. Viruses. 2021;13:241. doi: 10.3390/v13020241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj S.G., Wang N., Chen Z., Chen Z., Tseng M., Barretto N., Lin R., Peters C.J., Tseng C.-T.K., Baker S.C. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2007;282:32208–32221. doi: 10.1074/jbc.M704870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Sotto A., Checconi P., Celestino I., Locatelli M., Carissimi S., De Angelis M., Rossi V., Limongi D., Toniolo C., Martinoli L., et al. Antiviral and antioxidant activity of a hydroalcoholic extract from Humulus lupulus L. Oxidative Med. Cell. Longev. 2018;2018:5919237. doi: 10.1155/2018/5919237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douxfils J., Favresse J., Dogné J.-M., Lecompte T., Susen S., Cordonnier C., Lebreton A., Gosselin R., Sié P., Pernod G. Hypotheses behind the very rare cases of thrombosis with thrombocytopenia syndrome after SARS-CoV-2 vaccination. Thromb. Res. 2021;203:163–171. doi: 10.1016/j.thromres.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P., Keystone E., Tony H., Cantagrel A., Van Vollenhoven R., Sanchez A., Alecock E., Lee J., Kremer J. IL-6 receptor inhibition with tocilizumab improves treatment outcomes in patients with rheumatoid arthritis refractory to anti-tumour necrosis factor biologicals: results from a 24-week multicentre randomised placebo-controlled trial. Ann. Rheum. Dis. 2008;67:1516–1523. doi: 10.1136/ard.2008.092932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadel R., Morrison A., Vahia A., Smith Z.R., Chaudhry Z., Bhargava P., Miller J., Kenney R., Alangaden G., Ramesh M.S. medRxiv; 2020. Early Short Course Corticosteroids in Hospitalized Patients with COVID-19. (2020.2005.2004.20074609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey A.R., Sobieszczyk M.E., Hirsch I., Sproule S., Robb M.L., Corey L., Neuzil K.M., Hahn W., Hunt J., Mulligan M.J. Phase 3 safety and efficacy of AZD1222 (ChAdOx1 nCoV-19) Covid-19 vaccine. N. Engl. J. Med. 2021 doi: 10.1056/NEJMoa2105290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag N.S., Breitinger U., Breitinger H.G., El Azizi M.A. Viroporins and inflammasomes: a key to understand virus-induced inflammation. Int. J. Biochem. Cell Biol. 2020;122 doi: 10.1016/j.biocel.2020.105738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Heise M., Baric R. SARS coronavirus and innate immunity. Virus Res. 2008;133:101–112. doi: 10.1016/j.virusres.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani I.E., Triantafyllia V., Eleminiadou E.-E., Koltsida O., Stavropoulos A., Manioudaki M., Thanos D., Doyle S.E., Kotenko S.V., Thanopoulou K. Interferon-λ mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity. 2017;46:875–890.e876. doi: 10.1016/j.immuni.2017.04.025. [DOI] [PubMed] [Google Scholar]
- García-Salido A., de Carlos Vicente J.C., Hofheinz S.B., Ramírez J.B., Barrio M.S., Gordillo I.L., Yuste A.H., Pardellans C.G., Tejedor M.C.-M., Labarga B.H. Severe manifestations of SARS-CoV-2 in children and adolescents: from COVID-19 pneumonia to multisystem inflammatory syndrome: a multicentre study in pediatric intensive care units in Spain. Crit. Care. 2020;24:1–13. doi: 10.1186/s13054-020-03332-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautret P., Lagier J.-C., Parola P., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gibellini L., De Biasi S., Paolini A., Borella R., Boraldi F., Mattioli M., Lo Tartaro D., Fidanza L., Caro-Maldonado A., Meschiari M. Altered bioenergetics and mitochondrial dysfunction of monocytes in patients with COVID-19 pneumonia. EMBO Molecular Medicine. 2020;12 doi: 10.15252/emmm.202013001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Glass J.I., Lartigue C., Noskov V.N., Chuang R.-Y., Algire M.A., Benders G.A., Montague M.G., Ma L., Moodie M.M. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Herzog P., Pfefferle S., Steffen I., Muench M.O., Simmons G., Hofmann H., Kuri T., Weber F. Differential downregulation of ACE2 by the spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus NL63. J. Virol. 2010;84:1198–1205. doi: 10.1128/JVI.01248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfred-Cato S., Bryant B., Leung J., Oster M.E., Conklin L., Abrams J., Roguski K., Wallace B., Prezzato E., Koumans E.H., et al. COVID-19-associated multisystem inflammatory syndrome in children - United States, March-July 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:1074–1080. doi: 10.15585/mmwr.mm6932e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh G.K., Dunker A.K., Foster J.A., Uversky V.N. Rigidity of the outer Shell predicted by a protein intrinsic disorder model sheds light on the COVID-19 (Wuhan-2019-nCoV) infectivity. Biomolecules. 2020;10 doi: 10.3390/biom10020331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golchin A., Seyedjafari E., Ardeshirylajimi A. Mesenchymal stem cell therapy for COVID-19: present or future. Stem Cell Rev. Rep. 2020;16:427–433. doi: 10.1007/s12015-020-09973-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12 doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e1415. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbart M., Deng X., Baker S.C. Coronavirus endoribonuclease targets viral polyuridine sequences to evade activating host sensors. Proc. Natl. Acad. Sci. 2020;117:8094–8103. doi: 10.1073/pnas.1921485117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgren R., Samuelsson T., Laurent T.C., Modig J. Accumulation of hyaluronan (hyaluronic acid) in the lung in adult respiratory distress syndrome. Am. Rev. Respir. Dis. 1989;139:682–687. doi: 10.1164/ajrccm/139.3.682. [DOI] [PubMed] [Google Scholar]
- Hamanaka K., Jian M.-Y., Townsley M.I., King J.A., Liedtke W., Weber D.S., Eyal F.G., Clapp M.M., Parker J.C. TRPV4 channels augment macrophage activation and ventilator-induced lung injury. Am. J. Phys. Lung Cell. Mol. Phys. 2010;299:L353–L362. doi: 10.1152/ajplung.00315.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M., Lely A., Navis G.V., Van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Hara H., Núñez G. Mechanism and regulation of NLRP3 Inflammasome activation. Trends Biochem. Sci. 2016;41:1012–1021. doi: 10.1016/j.tibs.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and Is blocked by a clinically proven protease inhibitor. Cell. 2020;vol. 181:271–280.e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P., Lim W.S., Emberson J., Mafham M., Bell J., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E., et al. medRxiv; 2020. Effect of Dexamethasone in Hospitalized Patients with COVID-19: Preliminary Report. (2020.2006.2022.20137273) [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J. An mRNA vaccine against SARS-CoV-2—preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Guo Y., Luo Q., Huang Z., Zhao R., Liu S., Le A., Li J., Wan L. T-cell subset counts in peripheral blood can be used as discriminatory biomarkers for diagnosis and severity prediction of coronavirus disease 2019. J. Infect. Dis. 2020;222:198–202. doi: 10.1093/infdis/jiaa252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Guardeño J.M., Nieto-Torres J.L., DeDiego M.L., Regla-Nava J.A., Fernandez-Delgado R., Castaño-Rodriguez C., Enjuanes L. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzawa N., Nishigaki K., Hayashi T., Ishii Y., Furukawa S., Niiro A., Yasui F., Kohara M., Morita K., Matsushima K. Augmentation of chemokine production by severe acute respiratory syndrome coronavirus 3a/X1 and 7a/X4 proteins through NF-κB activation. FEBS Lett. 2006;580:6807–6812. doi: 10.1016/j.febslet.2006.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase M., Shirato K., van der Hoek L., Taguchi F., Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 2012;86:6537–6545. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavarz M., Solaymani-Mohammadi F., Namdari H., Arjeini Y., Mousavi M.J., Rezaei F. Metabolic host response and therapeutic approaches to influenza infection. Cell. Mol. Biol. Lett. 2020;25:1–19. doi: 10.1186/s11658-020-00211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavarz M., Tavakoli A., Zanganeh S., Mousavi M.J., Vahdat K., Mahmudpour M., Nabipour I., Darabi A., Keshmiri S. Future Virology; 2021. Clinical Characteristics of Outpatients and Inpatients with COVID-19 in Bushehr: A Report from the South of Iran. [Google Scholar]
- Kiem C.T., Andronico A., Bosetti P., Paireau J., Alter L., Boëlle P.-Y., Fontanet A., Lévy-Bruhl D., Cauchemez S. Benefits and risks associated with different uses of the COVID-19 vaccine Vaxzevria: a modelling study, France, may to September 2021. Eurosurveillance. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.26.2100533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkert M. Innate immune evasion by human respiratory RNA viruses. Journal of Innate Immunity. 2020;12:4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E., Thiel V. SARS-CoV and IFN: too little, too late. Cell Host Microbe. 2016;19:139–141. doi: 10.1016/j.chom.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouprina N., Larionov V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat. Protoc. 2008;3:371–377. doi: 10.1038/nprot.2008.5. [DOI] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuebler W.M., Jordt S.-E., Liedtke W.B. Urgent reconsideration of lung edema as a preventable outcome in COVID-19: inhibition of TRPV4 represents a promising and feasible approach. Am. J. Phys. Lung Cell. Mol. Phys. 2020;318:L1239–L1243. doi: 10.1152/ajplung.00161.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ky B., Mann D.L. COVID-19 clinical trials: a primer for the cardiovascular and cardio-oncology communities. JACC: Basic Trans. Science. 2020;5:501–517. doi: 10.1016/j.jacbts.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L., et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Z., Zhu R., Hou W., Feng Y., Yang Y., Han Q., Shan G., Meng F., Du D., Wang S., Fan J. Transplantation of ACE2-mesenchymal stem cells improves the outcome of patients with COVID-19 pneumonia. Aging Dis. 2020;11:216–228. doi: 10.14336/AD.2020.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke W., Weber D.S., Eyal F.G., Clapp M.M., Hamanaka J.C.K., Jian M.-Y., Townsley M.I., King J.A. TRPV4 channels augment macrophage activation and. Qual. Res. 2011;11:443–446. doi: 10.1152/ajplung.00315.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingeswaran M., Goyal T., Ghosh R., Suri S., Mitra P., Misra S., Sharma P. Inflammation, immunity and Immunogenetics in COVID-19: a narrative review. Indian J. Clin. Biochem. 2020;35:260–273. doi: 10.1007/s12291-020-00897-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M., Grad Y.H., Sette A., Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat. Rev. Immunol. 2020;20:709–713. doi: 10.1038/s41577-020-00460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wei Q., Nishiura K., Peng J., Wang H., Midkiff C., Alvarez X., Qin C., Lackner A., Chen Z. Spatiotemporal interplay of severe acute respiratory syndrome coronavirus and respiratory mucosal cells drives viral dissemination in rhesus macaques. Mucosal Immunol. 2016;9:1089–1101. doi: 10.1038/mi.2015.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Jesus A.A., Marrero B., Yang D., Ramsey S.E., Montealegre Sanchez G.A., Tenbrock K., Wittkowski H., Jones O.Y., Kuehn H.S. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 2014;371:507–518. doi: 10.1056/NEJMoa1312625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage K.G., Hage A., Schindewolf C., Rajsbaum R., Menachery V.D. bioRxiv; 2020. SARS-CoV-2 Is Sensitive to Type I Interferon Pretreatment. (2020.2003.2007.982264) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchese G. Epitopes for a 2019-nCoV vaccine. Cell. Mol. Immunol. 2020;17:539–540. doi: 10.1038/s41423-020-0377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W., et al. bioRxiv; 2020. SARS-CoV-2 Receptor ACE2 and TMPRSS2 Are Predominantly Expressed in a Transient Secretory Cell Type in Subsegmental Bronchial Branches. 2020.2003.2013.991455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv H., Wu N.C., Tsang O.T.-Y., Yuan M., Perera R.A.P.M., Leung W.S., So R.T.Y., Chan J.M.C., Yip G.K., Chik T.S.H., et al. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lythgoe M.P., Middleton P. Ongoing clinical trials for the management of the COVID-19 pandemic. Trends Pharmacol. Sci. 2020;41:363–382. doi: 10.1016/j.tips.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Market M., Angka L., Martel A.B., Bastin D., Olanubi O., Tennakoon G., Boucher D.M., Ng J., Ardolino M., Auer R.C. Flattening the COVID-19 curve with natural killer cell based immunotherapies. Front. Immunol. 2020;11:1512. doi: 10.3389/fimmu.2020.01512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh G., McAuley A., Au G., Riddell S., Layton D., Singanallur N., Layton R., Payne J., Durr P., Bender H. ChAdOx1 NCoV-19 (AZD1222) vaccine candidate significantly reduces SARS-CoV-2 shedding in ferrets. npj Vaccines. 2021;6:67. doi: 10.1038/s41541-021-00315-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P.R., Mangion S.A., Benger M., Stanton B.R., Czuprynska J., Arya R., Sztriha L.K. Brain, Behavior, and Immunity; 2021. Cerebral Venous Sinus Thrombosis and Thrombocytopenia after COVID-19 Vaccination–a Report of Two UK Cases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalick L., Kuebler W.M. TRPV4-a missing link between mechanosensation and immunity. Front. Immunol. 2020;11:413. doi: 10.3389/fimmu.2020.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton E.A., He X.-Y., Denorme F., Campbell R.A., Ng D., Salvatore S.P., Mostyka M., Baxter-Stoltzfus A., Borczuk A.C., Loda M. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood, The Journal of the American Society of Hematology. 2020;136:1169–1179. doi: 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustaqil M., Ollivier E., Chiu H.-P., Van Tol S., Rudolffi-Soto P., Stevens C., Bhumkar A., Hunter D.J., Freiberg A.N., Jacques D. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerging Microbes & Infections. 2020:1–27. doi: 10.1080/22221751.2020.1870414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemchand P., Tahir H., Mediwake R., Lee J. Cytokine storm and use of anakinra in a patient with COVID-19. BMJ Case Reports CP. 2020;13 doi: 10.1136/bcr-2020-237525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A.H., Cardani A., Braciale T.J. The host immune response in respiratory virus infection: balancing virus clearance and immunopathology. Semin. Immunopathol. 2016;38:471–482. doi: 10.1007/s00281-016-0558-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nhu Thao T.T., Labroussaa F., Ebert N., V’kovski P., Stalder H., Portmann J., Kelly J., Steiner S., Holwerda M., Kratzel A., et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature. 2020;582:561–565. doi: 10.1038/s41586-020-2294-9. [DOI] [PubMed] [Google Scholar]
- Norelli M., Camisa B., Barbiera G., Falcone L., Purevdorj A., Genua M., Sanvito F., Ponzoni M., Doglioni C., Cristofori P., Traversari C. Monocyte-derived IL-1 and IL-6 are differentially required for cytokine-release syndrome and neurotoxicity due to CAR T cells. Nat. Med. 2018;24:739–748. doi: 10.1038/s41591-018-0036-4. [DOI] [PubMed] [Google Scholar]
- Omrani M., Keshavarz M., Nejad Ebrahimi S., Mehrabi M., McGaw L.J., Ali Abdalla M., Mehrbod P. Potential natural products against respiratory viruses: a perspective to develop anti-COVID-19 medicines. Front. Pharmacol. 2020;11:2115. doi: 10.3389/fphar.2020.586993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudit G., Kassiri Z., Jiang C., Liu P., Poutanen S., Penninger J., Butany J. SARS-coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur. J. Clin. Investig. 2009;39:618–625. doi: 10.1111/j.1365-2362.2009.02153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudshoorn D., Rijs K., Limpens R.W.A.L., Groen K., Koster A.J., Snijder E.J., Kikkert M., Bárcena M. Expression and cleavage of Middle East respiratory syndrome coronavirus nsp3-4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with coronaviral RNA replication. mBio. 2017;8 doi: 10.1128/mBio.01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniri A., Akhavan-Niaki H. Emerging role of IL-6 and NLRP3 inflammasome as potential therapeutic targets to combat COVID-19: role of lncRNAs in cytokine storm modulation. Life Sci. 2020;257:118114. doi: 10.1016/j.lfs.2020.118114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat. Rev. Immunol. 2018;18:134. doi: 10.1038/nri.2017.105. [DOI] [PubMed] [Google Scholar]
- Parhizkar Roudsari P., Alavi-Moghadam S., Payab M., Sayahpour F.A., Aghayan H.R., Goodarzi P., Mohamadi-jahani F., Larijani B., Arjmand B. Auxiliary role of mesenchymal stem cells as regenerative medicine soldiers to attenuate inflammatory processes of severe acute respiratory infections caused by COVID-19. Cell Tissue Bank. 2020;21:405–425. doi: 10.1007/s10561-020-09842-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrot T., Gorin J.-B., Ponzetta A., Maleki K.T., Kammann T., Emgård J., Perez-Potti A., Sekine T., Rivera-Ballesteros O., Gredmark-Russ S. MAIT cell activation and dynamics associated with COVID-19 disease severity. Science Immunology. 2020;5 doi: 10.1126/sciimmunol.abe1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrey A.C., Qeadan F., Middleton E.A., Pinchuk I.V., Campbell R.A., Beswick E.J. Cytokine release syndrome in COVID-19: innate immune, vascular, and platelet pathogenic factors differ in severity of disease and sex. J. Leukoc. Biol. 2021;109:55–66. doi: 10.1002/JLB.3COVA0820-410RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaender S., Mar K.B., Michailidis E., Kratzel A., Boys I.N., V’kovski P., Fan W., Kelly J.N., Hirt D., Ebert N., Stalder H., Kleine-Weber H., Hoffmann M., Hoffmann H.-H., Saeed M., Dijkman R., Steinmann E., Wight-Carter M., McDougal M.B., Hanners N.W., Pöhlmann S., Gallagher T., Todt D., Zimmer G., Rice C.M., Schoggins J.W., Thiel V. LY6E impairs coronavirus fusion and confers immune control of viral disease. Nat. Microbiol. 2020;5:1330–1339. doi: 10.1038/s41564-020-0769-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper R.K. Cell entry of enveloped viruses. Current Opinion in Virology. 2011;1:92–100. doi: 10.1016/j.coviro.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Marc G.P., Moreira E.D., Zerbini C. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;384:1576–1577. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo F., Seabra C., Soares V., Sá A.J., Ferreira I., Mendes M. COVID-19-related multisystem inflammatory syndrome in a young adult. European Journal of Case Reports in Internal Medicine. 2021;8 doi: 10.12890/2021_002520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porritt R.A., Paschold L., Rivas M.N., Cheng M.H., Yonker L.M., Chandnani H., Lopez M., Simnica D., Schultheiß C., Santiskulvong C. HLA class I–associated expansion of TRBV11-2 T cells in multisystem inflammatory syndrome in children. J. Clin. Invest. 2021;131 doi: 10.1172/JCI146614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- ul Qamar M.T., Rehman A., Ashfaq U.A., Awan M.Q., Fatima I., Shahid F., Chen L.-L. 2020.2002.2028.970343. bioRxiv; 2020. Designing of a next generation multiepitope based vaccine (MEV) against SARS-COV-2: Immunoinformatics and <em>in silico</em> approaches. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que Y., Hu C., Wan K., Hu P., Wang R., Luo J., Li T., Ping R., Hu Q., Sun Y. Cytokine release syndrome in COVID-19: a major mechanism of morbidity and mortality. Int. Rev. Immunol. 2021:1–14. doi: 10.1080/08830185.2021.1884248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richt J.A., Rockx B., Ma W., Feldmann F., Safronetz D., Marzi A., Kobasa D., Strong J.E., Kercher L., Long D., et al. Recently emerged swine influenza a virus (H2N3) causes severe pneumonia in Cynomolgus macaques. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riollano-Cruz M., Akkoyun E., Briceno-Brito E., Kowalsky S., Reed J., Posada R., Sordillo E.M., Tosi M., Trachtman R., Paniz-Mondolfi A. Multisystem inflammatory syndrome in children related to COVID-19: a New York City experience. J. Med. Virol. 2020;93:424–433. doi: 10.1002/jmv.26224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Deming D., Paddock C.D., Cheng A., Yount B., Vogel L., Herman B.D., Sheahan T., Heise M., Genrich G.L., et al. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog. 2007;3 doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa B.A., Ahmed M., Singh D.K., Choreño-Parra J.A., Cole J., Jiménez-Álvarez L.A., Rodríguez-Reyna T.S., Singh B., Gonzalez O., Carrion R. IFN signaling and neutrophil degranulation transcriptional signatures are induced during SARS-CoV-2 infection. Communications Biology. 2021;4:1–14. doi: 10.1038/s42003-021-01829-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendahl Huber S., van Beek J., de Jonge J., Luytjes W., Van Baarle D. T cell responses to viral infections – opportunities for peptide vaccination. Front. Immunol. 2014;5 doi: 10.3389/fimmu.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A., Luetke S., Meinberger D., Hermes G., Sengle G., Koch M., Streichert T., Klatt A.R. bioRxiv; 2020. LL-37 fights SARS-CoV-2: the vitamin D-inducible peptide LL-37 inhibits binding of SARS-CoV-2 spike protein to its cellular receptor angiotensin converting enzyme 2 in vitro. [Google Scholar]
- Roudsari P.P., Alavi-Moghadam S., Payab M., Sayahpour F.A., Aghayan H.R., Goodarzi P., Mohamadi-Jahani F., Larijani B., Arjmand B. Auxiliary role of mesenchymal stem cells as regenerative medicine soldiers to attenuate inflammatory processes of severe acute respiratory infections caused by COVID-19. Cell Tissue Bank. 2020;21:1. doi: 10.1007/s10561-020-09842-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley A.H. Understanding SARS-CoV-2-related multisystem inflammatory syndrome in children. Nat. Rev. Immunol. 2020;20:453–454. doi: 10.1038/s41577-020-0367-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio I., Osuchowski M.F., Shankar-Hari M., Skirecki T., Winkler M.S., Lachmann G., La Rosée P., Monneret G., Venet F., Bauer M. Current gaps in sepsis immunology: new opportunities for translational research. Lancet Infect. Dis. 2019;19:e422–e436. doi: 10.1016/S1473-3099(19)30567-5. [DOI] [PubMed] [Google Scholar]
- Russell B., Moss C., George G., Santaolalla A., Cope A., Papa S., Van Hemelrijck M. Associations between immune-suppressive and stimulating drugs and novel COVID-19—a systematic review of current evidence. Ecancermedicalscience. 2020;14:1022. doi: 10.3332/ecancer.2020.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B. Safety and efficacy of single-dose Ad26. COV2. S vaccine against Covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff J., Le Gars M., Shukarev G., Heerwegh D., Truyers C., de Groot A.M., Stoop J., Tete S., Van Damme W., Leroux-Roels I. Interim results of a phase 1–2a trial of Ad26. COV2. S Covid-19 vaccine. N. Engl. J. Med. 2021;384:1824–1835. doi: 10.1056/NEJMoa2034201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena M., Sabado R.L., La Mar M., Mohri H., Salazar A.M., Dong H., Correa Da Rosa J., Markowitz M., Bhardwaj N., Miller E. Poly-ICLC, a TLR3 agonist, induces transient innate immune responses in patients with treated HIV-infection: a randomized double-blinded placebo controlled trial. Front. Immunol. 2019;10:725. doi: 10.3389/fimmu.2019.00725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheraga R.G., Southern B.D., Grove L.M., Olman M.A. The role of TRPV4 in regulating innate immune cell function in lung inflammation. Front. Immunol. 2020;11:1211. doi: 10.3389/fimmu.2020.01211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaigany S., Gnirke M., Guttmann A., Chong H., Meehan S., Raabe V., Louie E., Solitar B., Femia A. An adult with Kawasaki-like multisystem inflammatory syndrome associated with COVID-19. Lancet. 2020;396:e8–e10. doi: 10.1016/S0140-6736(20)31526-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty R., Ghosh A., Honavar S.G., Khamar P., Sethu S. Therapeutic opportunities to manage COVID-19/SARS-CoV-2 infection: present and future. Indian J. Ophthalmol. 2020;68:693–702. doi: 10.4103/ijo.IJO_639_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., Bucci E., Piacentini M., Ippolito G., Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A., Schulz L., Widera M., Mehdipour A.R., Tascher G., et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020;587:657–662. doi: 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist C.J., Bridge A., Le Mercier P. A potential role for integrins in host cell entry by SARS-CoV-2. Antivir. Res. 2020;177:104759. doi: 10.1016/j.antiviral.2020.104759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.-L., Kok K.-H., Ng M.-H.J., Poon V.K., Yuen K.-Y., Zheng B.-J., Jin D.-Y. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3· TANK· TBK1/IKKϵ complex. J. Biol. Chem. 2009;284:16202–16209. doi: 10.1074/jbc.M109.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.-L., Chan C.-P., Kok K.-H., Woo P.C.-Y., Jin D.-Y. Suppression of innate antiviral response by severe acute respiratory syndrome coronavirus M protein is mediated through the first transmembrane domain. Cell. Mol. Immunol. 2014;11:141–149. doi: 10.1038/cmi.2013.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperotto F., Friedman K.G., Son M.B.F., VanderPluym C.J., Newburger J.W., Dionne A. Cardiac manifestations in SARS-CoV-2-associated multisystem inflammatory syndrome in children: a comprehensive review and proposed clinical approach. Eur. J. Pediatr. 2020:1–16. doi: 10.1007/s00431-020-03766-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strich J.R., Ramos-Benitez M.J., Randazzo D., Stein S.R., Babyak A., Davey R.T., Suffredini A.F., Childs R.W., Chertow D.S. Fostamatinib inhibits neutrophils extracellular traps induced by COVID-19 patient plasma: a potential therapeutic. J. Infect. Dis. 2021;223:981–984. doi: 10.1093/infdis/jiaa789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Li Y., Chen L., Chen H., You F., Zhou X., Zhou Y., Zhai Z., Chen D., Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrimi E., Fennell E., Richter A., Vrljicak P., Stark R., Ott S., Murray P.G., Al-Abadi E., Chikermane A., Dawson P. The immune landscape of SARS-CoV-2-associated Multisystem Inflammatory Syndrome in Children (MIS-C) from acute disease to recovery. iScience. 2021;24 doi: 10.1016/j.isci.2021.103215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Wang Q., Zhang D., Ding J., Huang Q., Tang Y.-Q., Wang Q., Miao H. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduction and Targeted Therapy. 2020;5:33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M., Liu Y., Zhou R., Deng X., Li F., Liang K., Shi Y. Immunopathological characteristics of coronavirus disease 2019 cases in Guangzhou, China. Immunology. 2020;160:261–268. doi: 10.1111/imm.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi-Ponciano K., Vadillo E., Mayani H., Gonzalez-Bonilla C.R., Torres J., Majluf A., Flores-Padilla G., Wacher-Rodarte N., Galan J.C., Ferat-Osorio E. Increased expression of hypoxia-induced factor 1α mRNA and its related genes in myeloid blood cells from critically ill COVID-19 patients. Ann. Med. 2021;53:197–207. doi: 10.1080/07853890.2020.1858234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Li C., Huang A., Xia S., Lu S., Shi Z., Lu L., Jiang S., Yang Z., Wu Y. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg Microbes Infect. 2020;9:382–385. doi: 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney B., Lal S.K. SARS-CoV accessory protein 3b induces AP-1 transcriptional activity through activation of JNK and ERK pathways. Biochemistry. 2011;50:5419–5425. doi: 10.1021/bi200303r. [DOI] [PubMed] [Google Scholar]
- Verma H.K., Farran B., Bhaskar L.V. Convalescent plasma transfusion a promising therapy for coronavirus diseases 2019 (COVID-19): current updates. Antibody Therapeutics. 2020;3:115–125. doi: 10.1093/abt/tbaa010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Sooreshjani M.A., Mikek C., Opoku-Temeng C., Sintim H.O. Suramin potently inhibits cGAMP synthase, cGAS, in THP1 cells to modulate IFN-β levels. Future Med. Chem. 2018;10:1301–1317. doi: 10.4155/fmc-2017-0322. [DOI] [PubMed] [Google Scholar]
- Wang H., Zhang Y., Huang B., Deng W., Quan Y., Wang W., Xu W., Zhao Y., Li N., Zhang J. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell. 2020;182:713–721.e719. doi: 10.1016/j.cell.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Chen W., Zhou Y.-S., Lian J.-Q., Zhang Z., Du P., Gong L., Zhang Y., Cui H.-Y., Geng J.-J., et al. bioRxiv; 2020. SARS-CoV-2 Invades Host Cells Via a Novel Route: CD147-Spike Protein. (2020.2003.2014.988345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Qiu Y., Li J.-Y., Zhou Z.-J., Liao C.-H., Ge X.-Y. A unique protease cleavage site predicted in the spike protein of the novel pneumonia coronavirus (2019-nCoV) potentially related to viral transmissibility. Virol. Sin. 2020;35:337–339. doi: 10.1007/s12250-020-00212-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xu W., Hu G., Xia S., Sun Z., Liu Z., Xie Y., Zhang R., Jiang S., Lu L. SARS-CoV-2 infects T lymphocytes through its spike protein-mediated membrane fusion. Cell. Mol. Immunol. 2020;1 doi: 10.1038/s41423-020-0498-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Li D., Liu T., Wang H., Luo F., Liu Y. Subcutaneous injection of IFN alpha-2b for COVID-19: an observational study. BMC Infect. Dis. 2020;20:723. doi: 10.1186/s12879-020-05425-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., et al. medRxiv; 2020. Phenotype of SARS-CoV-2-specific T-cells in COVID-19 patients with acute respiratory distress syndrome. (2020.2004.2011.20062349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells A.I., Coyne C.B. Type III interferons in antiviral defenses at barrier surfaces. Trends Immunol. 2018;39:848–858. doi: 10.1016/j.it.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M.E., Luz B., Niehaus L., Bhogal P., Bäzner H., Henkes H. Thrombocytopenia and intracranial venous sinus thrombosis after “COVID-19 vaccine AstraZeneca” exposure. J. Clin. Med. 2021;10:1599. doi: 10.3390/jcm10081599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Yang X.O. TH17 responses in cytokine storm of COVID-19: an emerging target of JAK2 inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020;53:368–370. doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H., Cao Z., Xie X., Zhang X., Chen J.Y.-C., Wang H., Menachery V.D., Rajsbaum R., Shi P.-Y. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhang Y., Wang Y., Wang H., Yang Y., Gao G.F., Tan W., Wu G., Xu M., Lou Z. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2021;21:39–51. doi: 10.1016/S1473-3099(20)30831-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., Liu S., Zhao P., Liu H., Zhu L. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Fulcher J.A., Ahn J., Berro M., Goodman-Meza D., Dhody K., Sacha J.B., Naeim A., Yang O.O. Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America. 2020. Clinical characteristics and outcomes of COVID-19 patients receiving compassionate use Leronlimab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Ye F., Zhu N., Wang W., Deng Y., Zhao Z., Tan W. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci. Rep. 2015;5:17554. doi: 10.1038/srep17554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaqinuddin A., Kashir J. Innate immunity in COVID-19 patients mediated by NKG2A receptors, and potential treatment using Monalizumab, Cholroquine, and antiviral agents. Med. Hypotheses. 2020;140:109777. doi: 10.1016/j.mehy.2020.109777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang J., Chen Y., Luo B., Yuan Y., Huang F., Yang T., Yu F., Liu J., Liu B., et al. 2020. The ORF8 Protein of SARS-CoV-2 Mediates Immune Evasion through Potently Downregulating MHC-I. bioRxiv. (2020.2005.2024.111823) [Google Scholar]
- Zheng H.-Y., Zhang M., Yang C.-X., Zhang N., Wang X.-C., Yang X.-P., Dong X.-Q., Zheng Y.-T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Kanneganti T.-D. The regulation of the ZBP1-NLRP3 inflammasome and its implications in pyroptosis, apoptosis, and necroptosis (PANoptosis) Immunol. Rev. 2020;297:26–38. doi: 10.1111/imr.12909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Song L. Novel antibody epitopes dominate the antigenicity of spike glycoprotein in SARS-CoV-2 compared to SARS-CoV. Cell. Mol. Immunol. 2020;17:536–538. doi: 10.1038/s41423-020-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.-h, Wang Y.-n, Chang Q.-y, Ma P., Hu Y., Cao X. Type III interferons in viral infection and antiviral immunity. Cell. Physiol. Biochem. 2018;51:173–185. doi: 10.1159/000495172. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dai S.M., Tong Q. COVID-19: a recommendation to examine the effect of hydroxychloroquine in preventing infection and progression. J. Antimicrob. Chemother. 2020;75:1667–1670. doi: 10.1093/jac/dkaa114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–890.e882. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z., Lai X., Sun J., Chen Z., Zhang Z., Dai J., Liu D., Li Y., Li F., Wang Y. Mapping and role of T cell response in SARS-CoV-2–infected mice. J. Exp. Med. 2021;218 doi: 10.1084/jem.20202187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.