Positive interactions between bacteria, often described to be rare, occur commonly and primarily as parasitisms.
Abstract
Interspecies interactions shape the structure and function of microbial communities. In particular, positive, growth-promoting interactions can substantially affect the diversity and productivity of natural and engineered communities. However, the prevalence of positive interactions and the conditions in which they occur are not well understood. To address this knowledge gap, we used kChip, an ultrahigh-throughput coculture platform, to measure 180,408 interactions among 20 soil bacteria across 40 carbon environments. We find that positive interactions, often described to be rare, occur commonly and primarily as parasitisms between strains that differ in their carbon consumption profiles. Notably, nongrowing strains are almost always promoted by strongly growing strains (85%), suggesting a simple positive interaction–mediated approach for cultivation, microbiome engineering, and microbial consortium design.
INTRODUCTION
Microbial communities are composed of multiple species that interact with one another in a variety of ways as part of the “struggle for existence” (1). Interactions between species can be negative, where a species inhibits another species’ growth through nutrient exploitation and chemical warfare (2), or positive, where a species promotes another species’ growth by increasing nutrient availability and creating new niches (3). The bidirectional interaction between two species is determined by the two one-way interactions between the species. For example, a mutualism occurs when two species positively affect each other. The overall distribution of positive and negative interactions within a microbial community profoundly affects the community’s structure, stability, and productivity (4–6). These properties, in turn, shape a community’s ability to perform vital functions for the environment (7–10) and for host organisms (11–14). Despite the importance of the distribution of microbial interactions, the relative prevalence of positive and negative interactions in nature remains largely unknown.
Positive interactions are generally thought to be rare. Experimental evidence from coculture studies points to a dominance of negative interactions (2, 15). For example, evidence of positive interactions was found in <10% of pairs of bacteria isolated from tree holes (15). However, these results are subject to strong experimental biases, such as the use of a single environment, although microbial interactions can differ markedly across environmental conditions (16–18), and the use of strains that each grow individually in the environmental conditions being tested. Metabolic modeling, which can simulate millions of interactions across myriad environments, as well as limited experimental evidence, suggests that positive interactions emerge via environment-dependent secretions and can facilitate otherwise nongrowing species (18–21). In addition, evolutionary theories such as the Black Queen hypothesis argue that such secretion-mediated positive interactions are selected for (22). Together, these findings suggest that positive interactions among microbes may be common and play an important role in shaping microbial communities, but these theories have not yet been thoroughly tested experimentally.
Quantifying the prevalence of positive interactions and determining the conditions in which they occur could improve our ability to predict and control the ecology of microbial communities (23, 24). Positive interactions are predicted to enhance a community’s diversity and productivity but decrease its stability (6, 25, 26). Therefore, a better understanding of these interactions would enhance our ability to manipulate and manage communities, with widespread applicability in environmental conservation (27), crop health (28), and human health (29). Nevertheless, the data required for quantifying the distribution of interactions across environments are still lacking because of methodological limitations that frustrate comprehensive sampling of interactions under many conditions (30). Inferring interactions from metagenomic sequencing remains an outstanding challenge (31, 32), and directly measuring interactions at scale is difficult to perform using existing experimental paradigms.
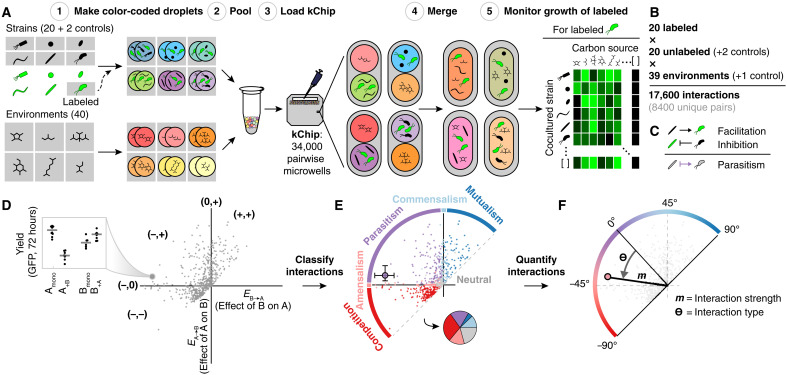
To gain a broad understanding of how species interact across a wide range of environments, we used a combinatorial screening platform called kChip (33–35) to measure >150,000 bidirectional bacterial cocultures among 20 different soil bacterial strains across 40 environments with differing carbon source identity or concentration. The kChip generates cocultures at an ultrahigh-throughput scale by rapidly and randomly combining droplets containing microbial cultures and/or medium components within microwells (Fig. 1, A to D, and fig. S1) (33). Here, we paired unlabeled (wild-type) and green fluorescent protein (GFP)–labeled versions of 20 Gammaproteobacteria isolated from soil (6 Enterobacterales and 14 Pseudomonadales; table S1 and figs. S2 and S3). We selected these bacterial strains from our larger pool of fluorescently labeled soil isolates, maximizing for phylogenetic diversity (table S1 and Materials and Methods). We cocultured each strain pair in each of the 33 single-carbon sources [0.5% (w/v)], a mix of these, 5 of the 33 at reduced concentration [0.05% (w/v)], and a no-carbon control. Carbon sources were drawn from several biochemical classes including carbohydrates, amino acids, and carboxylates (table S2). We measured the effect of each unlabeled strain on the growth of each labeled strain in each carbon source, giving a total of 17,600 combinations [20 labeled strains, (20 + 2 control) unlabeled strains, and (39 + 1 control) carbon sources]. Each combination appeared >10 times on average, and only 3% appeared <3 times and were excluded from further analysis (fig. S4). Measured one-way interactions were used to classify each bidirectional interaction qualitatively (Fig. 1E) and quantitatively (Fig. 1F).
Fig. 1. High-throughput interaction assay and analysis.
(A) Steps to assay the effect of multiple unlabeled species on a single-label species across carbon sources on each kChip. Color-coded droplets, each containing either a labeled + unlabeled coculture or a single carbon source, were generated (step 1), pooled together (step 2), and loaded onto a kChip (step 3). Each kChip contained an array of microwells that randomly paired coculture droplets with carbon source droplets. After imaging the color codes to identify the inputs per microwell, droplet pairs were merged via exposure to an electric field (step 4), and the growth of the labeled strain was measured at 0, 24, and 72 hours (step 5). (B) Overall size of the kChip screen. (C) Using data across kChips, bidirectional interactions were deduced by combining data where each strain within a given pair was the labeled strain. (D to F) Framework for kChip data analysis. Each bidirectional interaction was described qualitatively (interaction classification) and quantitatively (interaction strength, m; interaction type, ϴ).
Our data provide direct experimental evidence that positive interactions are indeed common and occur primarily as parasitisms, where one species’ growth is improved at the expense of the other’s. More broadly, we found that interactions strongly depend on the environment via differences in the carbon consumption preferences of the interacting strains. Notably, we found that strongly growing partners consistently enabled the growth of strains that were unable to grow in monoculture (85%), suggesting a simple strategy for cultivation, microbiome engineering, and design of microbial consortia.
RESULTS
Positive interactions occur frequently
The effect of each unlabeled strain on each fluorescently labeled strain was classified as positive (yield increase compared to monoculture), negative (yield decrease compared to monoculture), or 0 if there was no evidence for an effect (Fig. 1, C and D, and fig. S5) (see Materials and Methods). The size of an effect was calculated as log2 of the ratio of growth (fluorescence) in coculture to monoculture. Bidirectional interactions were classified as mutualism (+,+), commensalism (0,+), parasitism (−,+), amensalism (−,0), competition (−,−), or neutralism (0,0) (Fig. 1E and fig. S6). We focused our analysis on the 72-hour time point by which cultures typically reached saturation (33). We interpret the fluorescence at saturation as “yield,” the overall biomass of a fluorescently labeled strain grown on a given carbon source. Thus, the measured interactions reflect changes in strains’ overall growth yield rather than growth rates.
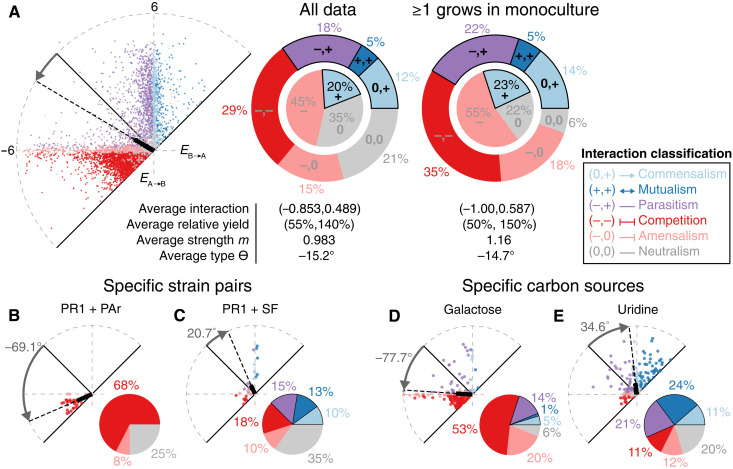
Positive interactions were common overall (Fig. 2A). Excluding cases when neither strain within a pair grew detectably in monoculture, >40% of cocultures that contained at least one strain facilitated the other’s growth, and more than half of these cases occurred within a parasitism (22% parasitisms, 14% commensalisms, and 5% mutualisms). There were also many cocultures that contained only negative interactions (35% competitions and 18% amensalisms). Relatively few interactions were neutralisms (6%), although neither strain grew in monoculture for 21% of pairs.
Fig. 2. Positive interactions occurred commonly and depended on strain pair and carbon source.
(A) Left: All bidirectional interactions on 33 distinct carbon sources. Middle: Interaction classification of all data. The inner circle represents one-way interaction classifications; the outer ring represents two-way interaction classifications. Right: Interaction classification excluding cases in which both strains comprising a coculture showed no detectable growth as monocultures on a given carbon source. (B and C) Interactions of two example cocultures on all carbon sources. PAr, Pseudomonas arsenicoxydans; PR1, Pseudomonas rhodesiae #1; SF, Serratia fonticola. (D and E) Interactions of all cocultures on two example carbon sources. Colors indicate interaction classification (legend). All data are at 72 hours.
The extent to which interacting pairs influenced each other’s growth also varied quantitatively across strain and carbon environments. For example, in a parasitism, the facilitated strain may have increased in yield more than the inhibited strain decreased or vice versa. To capture these quantitative differences, we measured the magnitude m (strength) and angle Ө (type) of each bidirectional interaction in polar coordinates (Fig. 1F). Ө represented the relative effect of cocultured strains on each other (with Ө = −90° indicating that both strains inhibited each other equally, Ө = 0° indicating a balanced parasitism with equal and opposite effects, and Ө = 90° indicating that both strains facilitated each other equally; no Ө is assigned to neutralisms).
The average interaction was a parasitism (Ө = −14.7° and m = 1.16) in which one strain was facilitated at the expense of the other strain whose growth was inhibited (Fig. 2A). On average, the inhibited strain was hurt (~50% growth compared to monoculture) more than the facilitated strain was helped (150% growth compared to monoculture). The results at 24 hours were similar and included even more positive interactions, with 54% of cocultures containing ≥1 one-way positive interaction (20% parasitisms, 25% commensalisms, and 9% mutualisms), although the average interaction, also a parasitism, favored the facilitated strain (~30% growth for one strain and 170% for the other as compared to monocultures) (fig. S7).
We have validated the kChip interaction assay by measuring a subset of the interactions in microtiter plates [(7 strains + 1 control) and four carbon sources, generating 256 combinations, or <2% of the total combination constructed on kChip]. Monoculture fold growth based on GFP and OD600 (optical density at 600 nm) measurements in microtiter plates were strongly correlated (fig. S8A). In addition, there was strong agreement between monoculture growth based on GFP measurements in microtiter plates and kChip (fig. S8B), as well as between one-way effects and interaction types calculated from these measurements (fig. S8, C and D). Last, as expected, intrastrain interactions measured on kChip (i.e., those between labeled strains and their unlabeled counterparts) were typically competitions (90% when excluding nongrowing strains) that reduced the yield of the labeled strain by 50% on average (fig. S9).
The occurrence of positive interactions differs among strain pairs and among carbon sources
The prevalence of positive interactions differed significantly among strain pairs: Positive interactions never occurred for some pairs, while, for others, they occurred in a large fraction of the tested environments (Fig. 2, B and C). Many pairs were capable of producing all six interaction types across the carbon source library (Fig. 2C), indicating that it can be challenging to predict the interaction of a particular pair in a given environment based on the pair’s interaction in a different environment.
Positive interactions were also more common on certain carbon sources than others. In galactose, for example, a majority of bidirectional interactions were competitions (53%), with only 20% containing ≥1 positive interaction (Fig. 2D). In uridine, however, competitions were relatively rare (11%), with 56% of bidirectional interactions containing ≥1 positive interaction (Fig. 2E). Several carbon sources were capable of producing all interaction classes across our strain set (fig. S10).
The strong dependence of the interaction type on strain identity and carbon source indicated a need to examine our dataset broadly to detect patterns governing the occurrence of positive interactions. We therefore tested whether characteristics of the environmental conditions or the strain pairs could explain the interactions that we observed.
Positive interactions occurred independently of environmental characteristics
Broad environmental characteristics, including carbon source biochemical class and number of carbon atoms, did not appear to dictate the occurrence of positive interactions. We found that the variation in interaction classification between carbon sources within a class was not significantly different than variation between classes [pairwise permutational multivariate analysis of variance (PERMANOVA), P > 0.05; Fig. 3A]. We hierarchically clustered all interactions for all strain pairs and carbon sources and found that carbon sources did not group by biochemical class [although tricarboxylic acid (TCA) cycle carboxylate ions grouped more adjacently] (fig. S11). We further found that the occurrence of positive interactions was only weakly correlated to the number of carbon atoms that each carbon source had (Spearman correlation, 0.32; P = 0.07) (Fig. 3B). For example, serine, a compound with only three carbon atoms, enabled more positive interactions than all other carbon sources except uridine.
Fig. 3. Positive interactions depended strongly on strain properties.
(A) Interaction classification by carbon source organized into biochemical categories. Bar colors indicate interaction classification (legends are provided in Fig. 2A). (B) Average interaction type by number of carbon atoms. The dot color indicates the biochemical class. (C) Interaction classification by phylogenetic relatedness of the taxonomic pairs. En, Enterobacterales. Ps, Pseudomonadales. (D) Interaction type by phylogenetic relatedness of the taxonomic pairs, averaged across all carbon sources. (E) Interaction classification by pairwise Euclidean metabolic distance (binned). Bin 0 represents with self-interactions. Bins 1 to 8 each contain roughly equal numbers of bidirectional interactions. (F) Interaction type by metabolic distance. All data are at 72 hours.
Positive interactions increase with strain dissimilarity
We found that positive interactions were more common among taxonomically dissimilar strains. We compared interactions within and between the two taxonomic orders represented in our set, Enterobacterales (En) and Pseudomonadales (Ps), since pairwise phylogenetic distances, calculated from full-length 16S ribosomal RNA (rRNA) gene sequences (fig. S3), distributed bimodally (fig. S12). More positive interactions occurred among interorder pairs (En + Ps) than intra-order pairs (En + En and Ps + Ps) (Fig. 3C): Twenty-six percent of En + En pairs and 20% of Ps + Ps pairs contained ≥1 positive interaction, compared to 47% for En + Ps pairs. The average interaction type was negative for intra-order cocultures (−32° for En + En and −45° for Ps + Ps) but positive for interorder cocultures (4.5° for En + Ps) (Fig. 3D) without significant differences in interaction strength (fig. S13).
The fraction of positive interactions, especially parasitisms, increased with the metabolic distance between the interacting species (Fig. 3E). Metabolic differences between strains were calculated as the Euclidean distances between strains’ carbon source utilization profiles, given by each strain’s ability to grow on each carbon source in monoculture (fig. S2). The distribution of metabolic distances between all strains was a continuous bell-shaped distribution (fig. S12), which captured more finely graded functional differences among strains than bimodal phylogenetic distances. Intrastrain mutualisms (i.e., those between labeled strains and their unlabeled counterparts) occurred in a small fraction of strains (3.7%), which we hypothesize was caused by higher starting density [a phenomenon known as an Allee effect (36)]. Interactions between metabolically similar strains were also typically competitions (68% for the most metabolically similar but nonidentical strains) (Fig. 3E). As metabolic distance increased, the fraction of pairs that exhibited ≥1 positive interaction increased monotonically from 0.2 to 61%, with a growing fraction of these positive interactions occurring within parasitisms. This result indicated a strong and directional dependence of positive interaction frequency on the functional dissimilarity of strain pairs.
As metabolic distance increased, the average interaction type Ө also became increasingly positive (Spearman correlation, 0.75; P < < 0.01) (Fig. 3F). This trend reflects both shifts in the frequency of interaction classes and quantitative changes in the relative strength of interactions’ components. Notably, among parasitisms, the average Ө within each metabolic distance bin increased, indicating an increasingly large facilitative effect relative to the reciprocal inhibitory effect (fig. S13). Since the prevalence of interaction types was correlated with strains’ metabolic capabilities, we next tested whether interactions in a given environment were dependent on the interacting strains’ abilities to grow in monoculture in that environment.
Monoculture yields shape occurrence of positive interactions in coculture
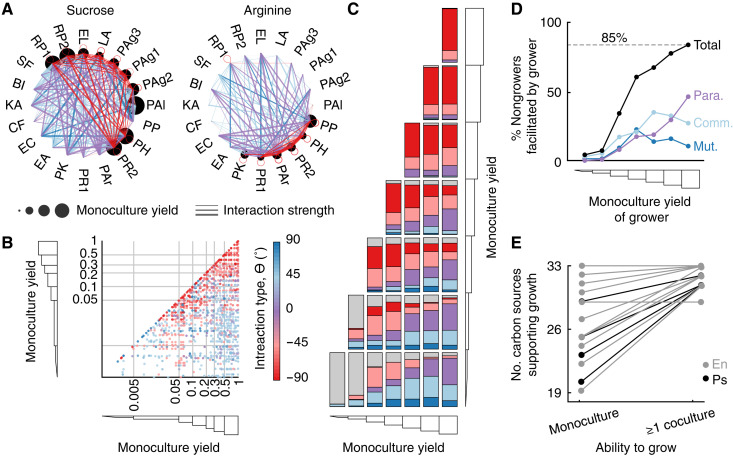
Interaction networks for each carbon source suggested that monoculture yield indeed related to interactions (examples in Fig. 4A and all networks in fig. S14). Regardless of the carbon sources or strain pair, positive interactions (often in the form of parasitisms) commonly occurred between strong and weak growers and sometimes occurred between weak growers; competition commonly occurred between strong growers (Fig. 4B).
Fig. 4. Carbon source utilization capabilities shape interactions.
(A) Example interaction networks for two carbon sources, each with different subsets of strains that could use it. The size of the circular nodes represents each strain’s growth in monoculture (normalized to each strain’s maximum monoculture yield across all carbon sources). Edge color represents interaction classification. Edge thickness represents interaction strength (m) (B) Interaction types (color) by the monoculture yields of both strains in each coculture. Gray lines indicate monoculture growth bins. (C) Interaction classification for each pair of monoculture growth bins. (D) Fractions of nongrowers that are obligately facilitated by strains with different monoculture yields. Line colors represent the interaction classifications in which the facilitation occurs (Mut., mutualism; Comm., commensalism; Para., parasitism; Total, total fraction of nongrowers facilitated). (E) The total number of carbon sources on which each strain can grow in monoculture and in at least one coculture. Each line represents one strain. All data are at 72 hours.
These patterns were found to be robust in a systematic analysis of interactions as a function of monoculture yield (Fig. 4, B and C). The frequencies of mutualisms, commensalisms, and parasitisms all increased with the difference between the monoculture yields of interacting strains (individually normalized to the maximum observed growth of each strain across all carbon sources; off-diagonal in Fig. 4C and fig. S15B). By contrast, for interactions of strain pairs able to grow on a carbon source equally well (diagonal in Fig. 4C and fig. S15A), competitions were consistently prevalent and increased in frequency with monoculture yield. Among interactions between the strongest growers, 77% were competitions where both species were inhibited compared to their monoculture growth. Moreover, in 99% of pairs of the strongest growers, at least one species was inhibited (77% competitions, 18% amensalisms, and 4% parasitisms). Interaction at 24 hours followed the same trend (fig. S16), with the overall increased prevalence of positive interactions (fig. S7) likely reflecting the lack of high monoculture yield values. Overall, interactions appeared to depend heavily on the two interacting strains’ individual abilities to grow in a specific environment, and this dependence explained the interaction variability exhibited by certain strain pairs and certain carbon sources.
These results were also consistent with the fact that there were far fewer positive interactions on low-concentration carbon sources (11% with ≥1 positive interaction, as opposed to 35% for their high-concentration counterparts) (fig. S17). The low-concentration carbon sources supported low-to-midrange monoculture yields that were similar among the strains (fig. S2). There were few instances of strong growers paired with weak growers, the regime where positive interactions otherwise emerged (fig. S17). Consequently, the interaction classifications on low-concentration carbon sources were nearly as different from their high-concentration counterparts as the high-concentration carbon sources were to each other (fig. S18). Unlike these low-concentration carbon sources, a mix of 33 carbon sources produced consistently high monoculture yields (most similar to glucose) (fig. S19). As a result, interactions occurred only between strong growers and were consequently highly negative (85% competitions, with 99% containing ≥1 inhibition) (fig. S19). For each strain pair, the interaction type Ө on the mixed carbon source condition was, with a single exception [the species pair Pseudomonas rhodesiae #1 and Pseudomonas arsenicoxydans (PR1 + PAr)], always lower than its average interaction across all carbon sources (fig. S19). Together, differences in monoculture yields provided a common statistical explanation for differences in the interaction distributions across time points, carbon source types, and carbon source concentrations.
Nongrowing strains are typically facilitated by strongly growing strains
To quantify the prevalence of obligate facilitation, where a nongrowing strain required a facilitating partner to grow on a given carbon source, we examined interactions between strains that could grow strongly on a carbon source and those that could not (bottom row in Fig. 4C and fig. S15C). As the yield of the growing strain increased, the frequency of facilitation of the nongrower increased markedly. When paired with the strongest growers, nongrowers were facilitated 85% of the time. Moreover, the relative fraction of these positive interactions that occurred as parasitisms also increased (Fig. 4D), making up 55% of obligate facilitations between the strongest growers and nongrowers. Among these parasitisms, the average interaction type also became increasingly positive (fig. S16): The magnitude of the positive interaction increased monotonically without the inhibited strain experiencing the same degree of yield loss (fig. S20).
At least one coculture partner could usually be found to support the growth of otherwise nongrowing strains on almost any carbon source. We observed a total of 125 cases where a given carbon source did not support detectable growth of a given strain in monoculture (Fig. 4E and fig. S21). By contrast, we found only 21 instances of a given carbon source never supporting the growth of a given strain despite its coculture with any partner strain. This common obligate facilitation provides a potential explanation for how biodiversity can be supported when few carbon sources are available or when only a subset of strains can use available carbon sources.
DISCUSSION
In this study, we performed comprehensive pairwise coculturing of 20 culturable soil microbes across 40 different carbon source environments. By measuring interactions across many environments, our study produced many instances in which at least one of the two cocultured strains grew poorly or not at all in monoculture, a regime in which we found that positive interactions were far more likely to occur than previously measured (2, 15). The study also produced instances where both strains grew well and antagonism was common, a result consistent with previous large-scale studies of bacterial interactions (37, 38).
Our study unearthed the wealth of positive interactions that occur in our system. While mutualisms were relatively rare (5%), commensalisms (12%) and parasitisms (18%) were common and accounted for the majority of cases where total coculture yield was greater than the sum of monocultures (24%) (fig. S22), a criterion previously used to classify cooperative interactions (15). The prevalence of these positive interactions corroborates predictions from large-scale metabolic models (19, 20, 39, 40). Our results are also consistent with the predictions of theories such as the Black Queen hypothesis, which asserts that interspecies cross-feeding of “leaky” public goods is evolutionarily selected for (22, 41). Last, our results generalize smaller-scale demonstrations that cocultured strains (42) and spent media (43) can induce growth of fastidious bacteria. Together, positive interactions increasingly appear to play a dominant role in driving community properties, such as resistance to invasion and productivity (3, 22), and in supporting microbial biodiversity (44). Positive interactions can also shape the phylogenetic composition of communities. In particular, common facilitation among phylogenetically distinct strains, as shown in our data, may promote phylogenetic diversity within communities, potentially leading to phylogenetic overdispersion (45, 46).
Interactions in our system varied significantly across environments and time points (fig. S20), suggesting that properties of natural communities can display considerable spatial and temporal variability. While interactions did not appear to significantly depend on properties intrinsic to the environment itself, they nonetheless strongly depended on the environment via the ability of each strain to individually grow in it: Negative interactions were frequent between strong growers, while positive interactions occurred commonly between strong and weak growers across all time points and environments. Given the widespread differences in growth that occur among bacteria, these results suggest that positive interactions may occur commonly in nature.
A variety of mechanisms could explain the prevalence of positive interactions in our system. First, facilitated strains might have grown on components of accumulating dead cells, although this is unlikely given the time scale of the coculture experiment (47). Second, the facilitator might have secreted carbon source–degrading enzymes that increased overall carbon availability. This mechanism is consistent with the general prevalence of positive interactions in dimeric and trimeric sugars (Fig. 3A) but may not explain positive interactions in simple carbon sources such as monomeric sugars and TCA cycle intermediates (Fig. 3, A and B). Third, the facilitator may have excreted incompletely oxidized metabolites that were used by the facilitated strain (20, 41, 48). Such “overflow metabolism” would allow strains to indirectly benefit from the biochemical transformation capabilities of their facilitators. Exploitation of newly created niches could explain the positive interactions that we observed on simple carbon sources (e.g., the excretion of short-chain fatty acids as a by-product of incomplete monosaccharide oxidation). It may also explain the rarity of positive interactions on lower carbon source concentrations since respiration is known to be favored over fermentation under such conditions and overflow is less likely to occur (48).
Despite the high throughput of our experiment, our system did not capture real-world bacterial diversity or environmental complexity. Our strain library was limited to two taxonomic orders isolated from topsoil. We only chose strains that grew on a minimal medium as part of our culturing protocol, possibly biasing our dataset in a variety of ways, e.g., against obligate facilitations for interactions involving amino acid or vitamin auxotrophies, which are known to be common (49). Strains were also pregrown on glucose as the sole carbon source before construction of coculture/carbon source combinations, imposing glucose consumption as a requisite for inclusion in our strain library and potentially affecting bacterial physiology (e.g., lag phase) and interactions. Moreover, while our carbon source library represented a variety of carbon source types, it was limited to soluble compounds, excluding many polymers on which metabolically driven positive interactions may be more common. Whether our results extend across additional phylogenetic groups (i.e., those occurring within soil and in other microbial ecosystems) and nutrient environments (i.e., across different and/or multiple carbon sources, concentrations thereof, and noncarbon nutrient requirements) should be investigated in follow-up studies to generalize trends observed in our system.
Our results indicate that knowledge of how strains grow individually in an environment can be strongly predictive of how they interact in that environment. In contrast, knowing how the same strains interact in a different environment or how different strains interact in the same environment does not appear to be very informative. Last, our results suggest that a potential strategy for inducing the growth of a nongrowing or weakly growing strain, independent of growth medium, is to coculture it with a strongly growing strain. Here, we uncovered several general, statistical rules governing microbial community structure and function. These rules deepen our understanding of microbial community ecology and are crucial to enable the efficient design and control of beneficial microbial communities.
MATERIALS AND METHODS
Strain isolation from soil samples
Soil samples (two ~12-cm columns of topsoil, ~4 cm in diameter) were collected from multiple locations in Greater Boston on 12 November 2017 (5.6°C) (specific locations are listed in table S1). Each sample was diluted in phosphate-buffered saline (PBS) within a few hours of collection (5 g of soil vortexed in 40 ml of PBS). Single strains were first isolated by streaking 70 μl of dilutions of this mixture (10−1, 10−2, 10−3, and 10−4) on 20 different solid (agar) media [tryptic soy broth (TSB; Bacto), 1% (v/v) TSB, lysogeny broth (LB; Bacto), 1% (v/v) LB, nutrient broth (NB; Bacto), 1% (v/v) NB, M9 salts (Sigma-Aldrich) + 0.5% (w/v) glucose, M9 salts + 0.005% (w/v) glucose, M9 salts + 0.005% (w/v) glucose + 0.2% (w/v) casamino acids, M9 salts + 0.005% (w/v) glucose + 0.002% (w/v) casamino acids, M9 salts + 0.5% (w/v) glucose + 0.2% (w/v) casamino acids (pH 4 and 5), M9 salts + 0.005% (w/v) glucose + 0.002% (w/v) casamino acids (pH 4 and 5), actinomycete isolation agar (Teknova), Brucella agar (Teknova), Streptomyces medium (Teknova), Campylobacter medium (Teknova), Bordetella medium (Teknova), and ATCC medium 1111 (Teknova)].
Strains were selected on the basis of the following criteria: growth in LB liquid medium of transferred colony (25°C), frozen glycerol stock revival in LB (OD600 > 0.1) (30°C), and subsequent growth on M9 + 0.5% (w/v) glucose (OD600 > 0.1) (30°C). We kept 96-well plates of isolates [LB, 25% (v/v) glycerol] at −80°C. Isolates included in the coculture experiment (table S1) were selected on the basis of robust revival from glycerol stocks and in subsequent culturing steps and the ability to label the strains via constitutive expression of GFP.
Strain labeling
The soil bacterial isolates were fluorescently labeled with the commercially available plasmid pMRE132 containing GFP2 (50). pMRE132 is a broad-host range plasmid used to constitutively express fluorescent protein genes in bacteria. First, the soil isolates and the Escherichia coli carrying pMRE132 (Ec-pMRE) were grown to saturation [5 ml of LB media (Bacto) in 50-ml Falcon tubes, loose caps, at 30°C for 24 hours, and chloramphenicol (15 mg/liter; Sigma-Aldrich) for Ec-pMRE]. Second, the saturated culture (SC) of each soil isolate was mixed with the SC of Ec-pMRE (500 μl of each). This and subsequent steps were done in 2-ml 96 deep-well plates (Eppendorf) with a VIAFLO 96 liquid handler (Integra) to increase throughput. The mixed SCs were concentrated to 10× by centrifuging (1 min, 7000 relative centrifugal force), discarding 900 μl of the supernatant, and resuspending. Immediately after, 10 μl of each SC was spotted, in three replicates, onto nutrient agar [5 g of peptone, 3 g of yeast extract, and 15 g of agar (Bacto) in 1 liter of water]. The agar plates were incubated (24 hours, 30°C) to allow for conjugation and transfer of the plasmid from Ec-pMRE to the soil isolates. The bacterial lawns were picked with sterile wooden applicators (Puritan), suspended in PBS (Corning), and mixed by pipetting 30 times with VIAFLO. The suspensions were then serially diluted (100, 10−1, and 10−2) and spotted onto minimal media agar to select for the transconjugants [1× M9 minimal salts (Teknova), glucose (5 g/liter), chloramphenicol (15 mg/liter; Sigma-Aldrich), and 15 g of agar (Bacto)]. Fluorescent colonies were picked and streaked onto minimal media agar to counterselect Ec-pMRE. Last, SC from single colonies were frozen with 50% glycerol and stored at −80°C.
Strain identification and calculation of phylogenetic distance
Each bacterium isolated from soil was classified phylogenetically with its 16S rRNA gene sequence. The full 16S gene sequences (~1500 base pairs) were obtained via Sanger sequencing (Quintara Biosciences), quality threshold-trimmed, and classified with SeqMatch (51). Sulfolobus solfataricus, a thermophilic archaeon, was used in the phylogenetic reconstruction as an outgroup species to root the tree. MUSCLE (52) with default parameters was used to align the sequences. PhyML-SMS (53, 54) with default parameters was used to select GTR + G + I as the best model and to infer the tree. The inferred branching order and branch lengths in the phylogenetic tree are the maximum likelihood estimates given the sequence alignment. The pairwise phylogenetic distances between strains were calculated directly from the phylogenetic tree with R (Phytools).
Medium construction
The medium used in the coculture experiment was an M9 minimal medium consisting of 1× M9 salts (Teknova), 1× trace metals (Teknova), 0.1 mM calcium chloride, and 2 mM magnesium sulfate. We additionally added 0.05% (w/v) bovine serum albumin (BSA) to the medium to improve the retention of fluorescent dyes used in the droplet color codes (33) (see the “Input color coding” section) and, presumably, other small molecules (55). We previously showed that addition of BSA does not affect bacterial growth (33).
A bank of kChip-deployable carbon source growth substrates was developed from which libraries could be readily created. Carbon compounds in this bank met the following criteria: The compounds were soluble at 2% (w/v) in water, the solutions were emulsifiable using Bio-Rad QX200 cartridges, and the integrity of the color code dye signals were maintained despite the presence of the carbon compound (see the Input color coding section). A total of 33 carbon compounds were chosen that passed these criteria, representing a diversity of growth substrates including monosaccharides, oligosaccharides, polysaccharides, carboxylate ions, amino acids, sugar alcohols, and a nucleic acid (fig. S2).
In the coculture experiment, a total of 40 environmental conditions were used. These included the 33 chosen compounds at 0.5% (w/v), 5 of these compounds (glucose, glycerol, pyruvate, proline, and sucrose) at 0.05% (w/v), an even mix of all 33 compounds [totaling 0.5% (w/v)], and a no-carbon control.
Carbon source plates consisting of the 40 carbon source conditions in water (4× experiment concentration) were frozen at −20°C and thawed at the onset of each round of the experiment (see the “kChip coculture construction” section in the Supplementary Materials). We previously demonstrated preservation of the frozen carbon substrate plates by showing tight correspondence in the growth of E. coli K-12 MG1655 on the freshly prepared carbon substrate plate and plates stored in −20°C for 3 and 15 days (33).
Microbial culturing
All labeled and unlabeled monocultures initially underwent two pre-experiment regrowth cycles (“starter phase” in a rich medium and “preculture phase” in minimal medium) and, at the onset of the experiment (“experiment phase”), were normalized to a starting density of OD600 = 0.02 in carbonless minimal medium. In the starter phase, glycerol stocks of the unlabeled and labeled strains were inoculated into 525 μl (0.8-ml-deep 96-well plate) of LB medium (25°C, 220 rpm, 16 hours). Inoculations from glycerol stocks were conducted via pin replicator [sterilized via 70% (v/v) ethanol bath and flame treatment between inoculations]. In the preculture phase, all cultures were washed in carbonless M9 medium two times and then diluted (1:50) into 1-ml M9 medium with 0.5% (w/v) glucose (25°C, 220 rpm, 24 hours). Last, the experimental phase began by washing cells three times in a carbonless M9 medium to remove residual glucose and normalizing to a starting OD600 of 0.02 (or ~20 cells per droplet depending on the strain).
Input color coding
Every unique input to the coculture screen (e.g., a bacterial culture or environmental condition) received a “color code,” or unique ratio of three fluorescent dyes (standardized to a total final dye concentration of 2.5 μM), before generating droplets (fig. S1B). Each set of three dyes collectively labeled each specific input. The dyes included Alexa Fluor 555, Alexa Fluor 594, and Alexa Fluor 647, all of which have distinct excitation and emission spectra, and dyes did not interfere with GFP. We previously showed that the color-coding dyes do not affect bacterial growth (33).
Droplet preparation and pooling
Each 1-nl droplet contained either a labeled/unlabeled pair or a single carbon source (Fig. 1A). The labeled strain was projected to each unlabeled strain (1:1 ratio) just before making droplets. Droplets were produced on a Bio-Rad QX200 Droplet Generator (which generated roughly 20,000 ~1-nl emulsifications prepared per 20-μl input for eight inputs at a time; 3 min per eight-input cartridge). The continuous phase was a fluorocarbon oil (3M Novec 7500). For droplet-making, 2% (w/w) fluorosurfactant (RAN Biotech 008 FluoroSurfactant) was added to stabilize droplets.
For each kChip loading (see the kChip coculture construction section in the Supplementary Materials), about 5000 droplets for each of the labeled/unlabeled inputs (22 + 2 empty controls or a total of 24 × 5000 = 120,000 droplets) and about 3000 droplets per carbon source (39 + 1 empty control or a total of 40 × 3000 = 120,000 droplets) were pooled, generating a total of about 240,000 droplets, where half contained cultures and the other half contained carbon sources. As a result, about a half of randomly generated pairwise droplet combinations on the kChip contained one labeled/unlabeled pair (premerge OD600 = 0.04 for each strain; postmerge OD600 = 0.02 for each strain or ~20 cells per droplet) and one carbon source droplet [final concentration, 0.5% (w/v) or 0.05% (w/v)]. Please refer to Supplementary Materials and Methods for a detailed description of kChip loading and fluorescence imaging.
Bootstrap resampling and interaction classification
To measure EB→A, the effect of unlabeled strain B on labeled strain A for a given carbon source environment, the log2 of the ratio of its yield of A in coculture (median of A+B replicates) to monoculture (median of Amono replicates) was calculated (Fig. 1D and fig. S5A). In instances where either of these values fell below the detection limit (DL), they were replaced with the DL, which was calculated as the 90% percentile of the distribution of A+B when no carbon source was present. Several examples of this calculation are provided in fig. S5B. Positive values indicated facilitation; negative values indicated inhibition, and 0 indicated no detected effect.
The coordinate (EB→A, EA→B) represented both sides of an interaction. To qualitatively classify this interaction, this point was plotted on a Cartesian plane (and, as necessary, reflected to the left of the identity line y = x). Uncertainty was calculated via bootstrapping: 1000 calculations were performed for EB→A and EA→B via resampling A+B, Amono, B+A, and Bmono. The 25th and 75th percentiles of the resulting distributions were plotted (Fig. 1E and fig. S6A). An interaction was classified as mutualism (+,+) if both sets of uncertainty bars fell within the first quadrant, as a parasitism if they both fell within the second quadrant (+,–), and as a competition if they both fell within the third quadrant (−,–). Error bars passing over quadrants indicated that a classification for at least one of the two effects did not adequately separate from no effect. If an uncertainty bar passed over the y axis, then an interaction was classified as a commensalism (+,0); if an uncertainty bar passed over the x axis, then the interaction was classified as an amensalism (−,0), and if both uncertainty bars passed over the x and y axes, then the interaction was classified as a neutralism (0,0). Examples of these pairwise classifications are shown in fig. S6A.
The point (EB→A, EA→B) occupied a radial continuum of possible pairwise interactions, with the magnitude m and the angle Ө of this point in polar coordinates providing a quantitative description of the interaction strength and type, respectively (Fig. 1F). The value Ө specifically represented the relative size of the effects of two strains on each other. To determine Ө, the line y = −x was assigned to 0° (representing a balanced parasitism of equal and opposite effects). Values −90° < Ө < −45° quantified competition, with −90° indicating that two strains inhibited each other equally; −45° indicated amensalism; −45° < Ө < 0° quantified parasitism, where the inhibitory effect outweighed the facilitative effect; 0° < Ө < 45° quantified parasitism, where the facilitative effect outweighed the inhibitory effect; 45° indicated commensalism; and 45° < Ө < 90° quantified mutualism, with 90° indicating that two strains facilitated each other equally. The distribution of m and Ө is given in fig. S10A (and separated by carbon source in fig. S10D). Ө for all pairwise interactions is hierarchically clustered in fig. S11.
For a given set of interactions (e.g., all pairs among a phylogenetic group), the mean interaction [μ(EB→A), μ(EA→B)] was sometimes calculated (fig. 6SB). The average interaction magnitude m and average interaction type Ө of this average interaction were also calculated. The variance in a set of interactions can also be calculated as an interaction diversity metric. This analytical framework was applied to the entire dataset (Fig. 2 and fig. S7) and to subsets of the data organized by properties of the environments, such as biochemical classification (Fig. 3B), or properties of strains, such as phylogenetic distances (Fig. 3D). The distribution of the average m and average Ө for interactions grouped by coculture pair and by carbon source are given in fig. S10 (B and C, respectively).
Binning interactions by metabolic distance or monoculture growth
Using measured resource utilization profiles (monoculture growth values across all carbon sources normalized to the maximum growth per strain, per time point) (fig. S2), the Euclidean distance between each resource utilization profile was computed for each strain pair (fig. S3C) as a measure of metabolic similarity. Whereas the phylogenetic distances among pairs produced a bimodal distribution that reflected the two taxonomic orders, the metabolic distance values produced a continuous bell-shaped distribution (fig. S12).
We binned the 190 possible cocultures into eight discrete metabolic distance bins of increasing dissimilarity and counted interaction types in each bin across all carbon sources (Fig. 3E and fig. S13). (Cocultures of labeled strains cocultured with their unlabeled counterparts, for which the metabolic distance was 0, were grouped as bin #0.) In the center of the metabolic distance distribution (bins #3, #4, #5, and #6), each bin spanned 0.25 Euclidean distance units. Because there were fewer data points nearer the tails of the distribution, bins #2 and #7 each spanned 0.5 Euclidean distance units, and bins #1 and #8 spanned 1 Euclidean distance unit. The resulting bins each had roughly equal numbers of data points.
Unlike binning by metabolic distance, binning by monoculture growth disregarded any larger interaction patterns within a given strain pair, i.e., each interaction generated among a pair across each carbon source was independently binned only by the degree to which each strain grew on the given carbon source. Normalized monoculture yields (fig. S2) were placed into one of seven bins (cutoffs: 0, 0.005, 0.05, 0.1, 0.2, 0.3, 0.5, and 1). The first bin, [0 0.005), represented undetectable growth (within background noise) as qualified as “no growth” in analyses of obligate facilitation (Fig. 4D). With the exception of the second bin, [0.005 0.05), which spanned a decade, bin cutoffs were roughly based on exponential doublings.
Interaction networks and binning
Interaction networks were constructed for all pairwise interactions occurring per carbon source (examples in Fig. 4A and all networks in fig. S14). The nodes, each representing a strain, were arranged concentrically by carbon utilization similarity (same order as in fig. S2). The size of the node corresponded linearly to the normalized monoculture growth (fig. S2). Edges between nodes, each representing a pairwise interaction, were colored by interaction classification (with neutralisms not shown). The thickness of the edge corresponded to interaction strength m.
Acknowledgments
We thank S. Kuehn and N. Kashtan for critical comments on the manuscript; A. Perez Escudero, O. Cordero, E. Alm, P. Sabeti, and C. Ackerman for useful discussions; M. Hoffman for assistance; and the Jupyter, NumPy, SciPy, scikit-image, scikit-learn, and pandas open-source development teams.
Funding: This work was supported by the NSF Graduate Research Fellowship Program [to J.K. (fellow ID 2016220942) and A.K. (fellow ID 2013164251)], a Siebel Scholars Foundation grant (to J.K.), the MIT Institute for Medical Engineering and Science Broshy Fellowship (to A.K.), a Career Award at the Scientific Interface from the Burroughs Wellcome Fund (to P.C.B.) (grant no. 1010240), an MIT Deshpande Center Innovation Grant (to P.C.B.), a Bridge Project grant from the Dana Farber/Harvard Cancer Center and the Koch Institute for Integrative Cancer Research at MIT (to P.C.B.), a technology development seed grant from the Merkin Institute for Transformative Technologies in Healthcare at the Broad Institute (to P.C.B.), and a grant from the United States–Israel Binational Science Foundation (to J.F. and P.C.B.) (grant no. 2017179).
Author contributions: This work was conceptualized by all authors. The kChip experimental platform was designed by J.K., A.K., and P.C.B. Microbial isolation was performed by J.K. and A.O. Microbial strain labeling was performed by A.O. Software was written by J.K. and A.K. Formal analysis was conducted by J.K. with feedback from A.O., P.C.B., and J.F. The experimental investigation was carried out by J.K. and A.O. Experimental data were curated by J.K., and phylogenetic data were curated by A.O. The original manuscript draft and data visualization were prepared by J.K. The manuscript was reviewed and edited by all the authors. The study was supervised by J.G., P.C.B., and J.F.
Competing interests: J.K. and P.C.B. are equity holders in a microbiome company, Concerto Biosciences (Allston, MA). P.C.B. is a consultant to and an equity holder in two companies in the microfluidics industry, 10X Genomics (Pleasanton, CA) and GALT (San Carlos, CA). The Broad Institute and MIT may seek to commercialize aspects of this work, and related applications for intellectual property have been filed. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Materials and Methods
Figs. S1 to S22
Tables S1 and S2
Legend for data S1
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.G. F. Gauze, The Struggle for Existence (The Williams & Wilkins Company, 1934). [Google Scholar]
- 2.Ghoul M., Mitri S., The ecology and evolution of microbial competition. Trends Microbiol. 24, 833–845 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Bruno J. F., Stachowicz J. J., Bertness M. D., Inclusion of facilitation into ecological theory. Trends Ecol. Evol. 18, 119–125 (2003). [Google Scholar]
- 4.Konopka A., Lindemann S., Fredrickson J., Dynamics in microbial communities: Unraveling mechanisms to identify principles. ISME J. 9, 1488–1495 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newell P. D., Douglas A. E., Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster. Appl. Environ. Microbiol. 80, 788–796 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allesina S., Tang S., Stability criteria for complex ecosystems. Nature 483, 205–208 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Schimel J. P., Schaeffer S. M., Microbial control over carbon cycling in soil. Front. Microbiol. 3, 348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arrigo K. R., Marine microorganisms and global nutrient cycles. Nature 437, 349–355 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Bardgett R. D., Freeman C., Ostle N. J., Microbial contributions to climate change through carbon cycle feedbacks. ISME J. 2, 805–814 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Zhou J., Xue K., Xie J., Deng Y., Wu L., Cheng X., Fei S., Deng S., He Z., Van Nostrand J. D., Luo Y., Microbial mediation of carbon-cycle feedbacks to climate warming. Nat. Clim. Chang. 2, 106–110 (2012). [Google Scholar]
- 11.Weller D. M., Raaijmakers J. M., Gardener B. B. M., Thomashow L. S., Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40, 309–348 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Mendes R., Kruijt M., de Bruijn I., Dekkers E., van der Voort M., Schneider J. H. M., Piceno Y. M., DeSantis T. Z., Andersen G. L., Bakker P. A. H., Raaijmakers J. M., Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Hayat R., Ali S., Amara U., Khalid R., Ahmed I., Soil beneficial bacteria and their role in plant growth promotion: A review. Ann. Microbiol. 60, 579–598 (2010). [Google Scholar]
- 14.Lynch S. V., Pedersen O., The human intestinal microbiome in health and disease. N. Engl. J. Med. 375, 2369–2379 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Foster K. R., Bell T., Competition, not cooperation, dominates interactions among culturable microbial species. Curr. Biol. 22, 1845–1850 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Klitgord N., Segrè D., Environments that induce synthetic microbial ecosystems. PLOS Comput. Biol. 6, e1001002 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoek T. A., Axelrod K., Biancalani T., Yurtsev E. A., Liu J., Gore J., Resource availability modulates the cooperative and competitive nature of a microbial cross-feeding mutualism. PLOS Biol. 14, e1002540 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuñiga C., Li C.-T., Yu G., Al-Bassam M. M., Li T., Jiang L., Zaramela L. S., Guarnieri M., Betenbaugh M. J., Zengler K., Environmental stimuli drive a transition from cooperation to competition in synthetic phototrophic communities. Nat. Microbiol. 4, 2184–2191 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Magnúsdóttir S., Heinken A., Kutt L., Ravcheev D. A., Bauer E., Noronha A., Greenhalgh K., Jäger C., Baginska J., Wilmes P., Fleming R. M. T., Thiele I., Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol. 35, 81–89 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Pacheco A. R., Moel M., Segrè D., Costless metabolic secretions as drivers of interspecies interactions in microbial ecosystems. Nat. Commun. 10, 103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldford J. E., Lu N., Bajić D., Estrela S., Tikhonov M., Sanchez-Gorostiaga A., Segrè D., Mehta P., Sanchez A., Emergent simplicity in microbial community assembly. Science 361, 469–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris J. J., Lenski R. E., Zinser E. R., The Black Queen hypothesis: Evolution of dependencies through adaptive gene loss. MBio 3, e0036-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marsland R., Cui W., Mehta P., A minimal model for microbial biodiversity can reproduce experimentally observed ecological patterns. Sci. Rep. 10, 3308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qian J. J., Akçay E., The balance of interaction types determines the assembly and stability of ecological communities. Nat. Ecol. Evol. 4, 356–365 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Coyte K. Z., Schluter J., Foster K. R., The ecology of the microbiome: Networks, competition, and stability. Science 350, 663–666 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Bunin G., Ecological communities with Lotka-Volterra dynamics. Phys. Rev. E 95, 042414 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Jariyal M., Jindal V., Mandal K., Gupta V. K., Singh B., Bioremediation of organophosphorus pesticide phorate in soil by microbial consortia. Ecotoxicol. Environ. Saf. 159, 310–316 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Kavino M., Manoranjitham S. K., In vitro bacterization of banana (Musa spp.) with native endophytic and rhizospheric bacterial isolates: Novel ways to combat Fusarium wilt. Eur. J. Plant Pathol. 151, 371–387 (2018). [Google Scholar]
- 29.Wallace B. D., Redinbo M. R., The human microbiome is a source of therapeutic drug targets. Curr. Opin. Chem. Biol. 17, 379–384 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nai C., Meyer V., From axenic to mixed cultures: Technological advances accelerating a paradigm shift in microbiology. Trends Microbiol. 26, 538–554 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Weiss S., Van Treuren W., Lozupone C., Faust K., Friedman J., Deng Y., Xia L. C., Xu Z. Z., Ursell L., Alm E. J., Birmingham A., Cram J. A., Fuhrman J. A., Raes J., Sun F., Zhou J., Knight R., Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J. 10, 1669–1681 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao H.-T., Gibson T. E., Bashan A., Liu Y.-Y., Inferring human microbial dynamics from temporal metagenomics data: Pitfalls and lessons. Bioessays 39, 1600188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kehe J., Kulesa A., Ortiz A., Ackerman C. M., Thakku S. G., Sellers D., Kuehn S., Gore J., Friedman J., Blainey P. C., Massively parallel screening of synthetic microbial communities. Proc. Natl. Acad. Sci. U.S.A. 116, 12804–12809 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulesa A., Kehe J., Hurtado J. E., Tawde P., Blainey P. C., Combinatorial drug discovery in nanoliter droplets. Proc. Natl. Acad. Sci. U.S.A. 115, 6685–6690 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ackerman C. M., Myhrvold C., Thakku S. G., Freije C. A., Metsky H. C., Yang D. K., Ye S. H., Boehm C. K., Kosoko-Thoroddsen T.-S. F., Kehe J., Nguyen T. G., Carter A., Kulesa A., Barnes J. R., Dugan V. G., Hung D. T., Blainey P. C., Sabeti P. C., Massively multiplexed nucleic acid detection with Cas13. Nature 582, 277–282 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaul R. B., Kramer A. M., Dobbs F. C., Drake J. M., Experimental demonstration of an Allee effect in microbial populations. Biol. Lett. 12, 20160070 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cordero O. X., Wildschutte H., Kirkup B., Proehl S., Ngo L., Hussain F., Le Roux F., Mincer T., Polz M. F., Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science 337, 1228–1231 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Russel J., Røder H. L., Madsen J. S., Burmølle M., Sørensen S. J., Antagonism correlates with metabolic similarity in diverse bacteria. Proc. Natl. Acad. Sci. U.S.A. 114, 10684–10688 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zelezniak A., Andrejev S., Ponomarova O., Mende D. R., Bork P., Patil K. R., Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl. Acad. Sci. U.S.A. 112, 6449–6454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Libby E., Hébert-Dufresne L., Hosseini S.-R., Wagner A., Syntrophy emerges spontaneously in complex metabolic systems. PLOS Comput. Biol. 15, e1007169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D’Souza G., Shitut S., Preussger D., Yousif G., Waschina S., Kost C., Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat. Prod. Rep. 35, 455–488 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Jeffrey Morris J., Kirkegaard R., Szul M. J., Johnson Z. I., Zinser E. R., Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “Helper” heterotrophic bacteria. Appl. Environ. Microbiol. 74, 4530–4534 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka Y., Hanada S., Manome A., Tsuchida T., Kurane R., Nakamura K., Kamagata Y., Catellibacterium nectariphilum gen. nov., sp. nov., which requires a diffusible compound from a strain related to the genus Sphingomonas for vigorous growth. Int. J. Syst. Evol. Microbiol. 54, 955–959 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Gross K., Positive interactions among competitors can produce species-rich communities. Ecol. Lett. 11, 929–936 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Cavender-Bares J., Kozak K. H., Fine P. V. A., Kembel S. W., The merging of community ecology and phylogenetic biology. Ecol. Lett. 12, 693–715 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Nemergut D. R., Schmidt S. K., Fukami T., O’Neill S. P., Bilinski T. M., Stanish L. F., Knelman J. E., Darcy J. L., Lynch R. C., Wickey P., Ferrenberg S., Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kolter R., Siegele D. A., Tormo A., The stationary phase of the bacterial life cycle. Annu. Rev. Microbiol. 47, 855–874 (1993). [DOI] [PubMed] [Google Scholar]
- 48.Basan M., Hui S., Okano H., Zhang Z., Shen Y., Williamson J. R., Hwa T., Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 528, 99–104 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zengler K., Zaramela L. S., The social network of microorganisms—How auxotrophies shape complex communities. Nat. Rev. Microbiol. 16, 383–390 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schlechter R. O., Jun H., Bernach M., Oso S., Boyd E., Muñoz-Lintz D. A., Dobson R. C. J., Remus D. M., Remus-Emsermann M. N. P., Chromatic bacteria—A broad host-range plasmid and chromosomal insertion toolbox for fluorescent protein expression in bacteria. Front. Microbiol. 9, 3052 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cole J. R., Wang Q., Fish J. A., Chai B., McGarrell D. M., Sun Y., Brown C. T., Porras-Alfaro A., Kuske C. R., Tiedje J. M., Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O., New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Lefort V., Longueville J.-E., Gascuel O., SMS: Smart model selection in PhyML. Mol. Biol. Evol. 34, 2422–2424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Skhiri Y., Gruner P., Semin B., Brosseau Q., Pekin D., Mazutis L., Goust V., Kleinschmidt F., El Harrak A., Brian Hutchison J., Mayot E., Bartolo J.-F., Griffiths A. D., Taly V., Baret J.-C., Dynamics of molecular transport by surfactants in emulsions. Soft Matter 8, 10618 (2012). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Figs. S1 to S22
Tables S1 and S2
Legend for data S1
Data S1