Abstract
Irritable bowel syndrome (IBS) results from disordered brain–gut interactions. Identifying susceptibility genes could highlight the underlying pathophysiological mechanisms. We designed a digestive health questionnaire for UK Biobank and combined identified cases with IBS with independent cohorts. We conducted a genome-wide association study with 53,400 cases and 433,201 controls and replicated significant associations in a 23andMe panel (205,252 cases and 1,384,055 controls). Our study identified and confirmed six genetic susceptibility loci for IBS. Implicated genes included NCAM1, CADM2, PHF2/FAM120A, DOCK9, CKAP2/TPTE2P3 and BAG6. The first four are associated with mood and anxiety disorders, expressed in the nervous system, or both. Mirroring this, we also found strong genome-wide correlation between the risk of IBS and anxiety, neuroticism and depression (rg > 0.5). Additional analyses suggested this arises due to shared pathogenic pathways rather than, for example, anxiety causing abdominal symptoms. Implicated mechanisms require further exploration to help understand the altered brain–gut interactions underlying IBS.
Subject terms: Genome-wide association studies, Irritable bowel syndrome, Psychiatric disorders
Genome-wide association analysis of irritable bowel syndrome identifies genetic susceptibility loci and highlights shared pathways with mood and anxiety disorders.
Main
IBS is common worldwide and typically presents in early adulthood with symptoms including abdominal pain, bloating and bowel dysfunction1. Symptom intensity varies over time and between individuals but IBS has been reported, in severe cases, to affect quality of life as much as renal impairment or diabetes2.
IBS accounts for approximately half of all referrals to gastroenterology clinics3. Although of doubtful clinical value, many patients undergo multiple investigations, including colonoscopies, to exclude conditions such as Crohn’s disease or cancer. To the frustration of patients and clinicians alike, all tests are characteristically normal. The healthcare costs, combined with indirect employment costs, of IBS amount to at least €13 billion (£11.7 billion) annually in Europe4.
Attempts have been made to identify positive clinical diagnostic features and reduce investigations. The widely used and recently revised Rome III criteria define IBS as unexplained abdominal pain or discomfort eased by defecation, with altered stool form or frequency, for more than six months5. Three main subtypes are recognized: constipation-predominant (IBS-C), diarrhea-predominant (IBS-D) or ‘mixed’/alternating constipation and diarrhea (IBS-M). Individuals with functional constipation and functional diarrhea share the disordered bowel pattern with IBS but do not suffer from pain.
The commonly used IBS treatments, ranging from dietary exclusion to psychoactive medications, are relatively ineffective and their variety reflects the uncertain etiology6. Behavioral therapies, while more effective (number needed to treat = 4), are not widely available7. Given the high frequency, impact and cost of IBS, there is a pressing need for improved pathophysiological understanding to enable better therapeutic approaches.
The relative importance of peripheral and gut versus central and psychological factors to IBS etiology is uncertain. The consensus view is that IBS results from abnormal brain–gut interactions. Recent epidemiological data suggested that, in individuals developing both IBS and psychological features, the former preceded the latter in two thirds of cases and the latter preceded the former in one third8. IBS is associated with abnormalities of central pain processing but also increased gut permeability, mast cell activation, disordered motility and dysbiosis9. Up to one in ten cases are triggered after infection, so-called postinfectious IBS (PI-IBS)10.
IBS aggregates in families, with individuals being two to three times more likely to develop IBS if they have an affected relative. Estimates of heritability from twin studies range widely from 0 up to 57%11. Twin studies have indirectly investigated whether IBS and mental health conditions share a genetic basis but have proved inconclusive12. Although genetic association studies have provided pathogenic and therapeutic insights for many conditions13, only one variant (rs10512344) has previously been identified at genome-wide significance in IBS, with only modest replication14. Larger datasets are clearly required to characterize the recognized heritable component of IBS; the use of large population-based cohorts had been proposed11,15. Broad meta-analysis of cases from different cohorts and case definitions is also a proven method of increasing power16. Between 2006 and 2010, UK Biobank (UKB) recruited half a million people aged 40–69 years. Participants underwent baseline assessment and consented to long-term follow-up, including questionnaires, and linkage to routinely collected health data. All participants underwent genome-wide SNP genotyping. Therefore, UKB provides a powerful epidemiological resource for exploring risk factors for health outcomes17,18.
The main aim of the current study was to identify genetic risk factors for IBS through an analysis involving over 250,000 affected individuals. We report robustly validated genetic susceptibility loci for IBS and provide evidence of its shared genetic etiology with mood and anxiety disorders.
Results
Epidemiology
We designed a digestive health questionnaire (DHQ) for the UKB website, with a link e-mailed to 332,793 UKB participants with valid e-mail addresses. A total of 171,061 (51.4%) responses were received and analyzed (Supplementary Fig. 1). The DHQ included validated instruments for IBS diagnosis (Rome III symptom criteria), the IBS symptom severity score (IBS-SSS, measured using the IBS Severity Scoring System)19 and the Patient Health Questionnaire 12 Somatic Symptom score (PHQ-12)20. It also asked about previous IBS diagnosis, environmental exposures and associated conditions (including anxiety or depression, based on treatment sought or offered).
After sample exclusions and quality control, we identified a total of 40,548 UKB participants of European ancestry (Fig. 1a) who met the diagnostic criteria for IBS (based on DHQ Rome III symptom data, self-report of previous medical IBS diagnosis or electronic medical records; see Methods and Supplementary Tables 1 and 2). The demographics of the DHQ respondents are presented in Table 1. Females were affected by IBS more commonly than males (72.1%). IBS-M, with hard and loose stools present at least ‘sometimes’ (alternating), was the most common subtype in patients defined according to the Rome III criteria (55.0%).
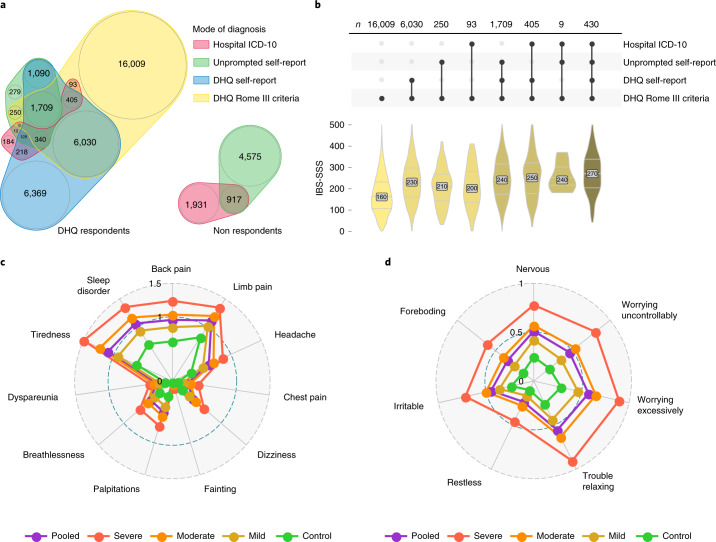
Fig. 1. Diagnostic modalities and comorbidities of IBS.
a, Venn diagram of overlap between UKB IBS cases by different diagnostic modality, split by DHQ respondents and nonrespondents. The areas and numbers indicate the sample size. Most participants with current symptoms (DHQ Rome III, yellow) did not report being diagnosed with IBS either when listing medical conditions unprompted at UKB enrollment (unprompted self-report, green) or when asked specifically about a previous IBS diagnosis when completing the DHQ (DHQ self-report, blue). Conversely, many participants previously diagnosed with IBS, even those formally recorded during a hospital admission (hospital ICD-10, red), did not have symptoms sufficient for Rome III criteria IBS diagnosis at the time of their DHQ response. b, Among individuals experiencing IBS symptoms (DHQ Rome III positive), those previously diagnosed by a clinician had greater symptom severity, with an increase in the number of IBS diagnostic modalities (connected dots, middle; top: sample size is shown) being associated with an increase in symptom severity score (IBS-SSS, bottom). Distributions are colored by the number of diagnoses and the groups shown are mutually exclusive. For post-hoc statistics, see Supplementary Table 3. c, Severity of different somatic symptoms in the past three months among digestively healthy controls and IBS cases (classified as mild, moderate and severe based on IBS-SSS). Mean scores for PHQ-12 items ranked from 0 (not bothered at all) to 2 (bothered a lot) are shown. Pooled refers to all UKB cases in the discovery cohort. d, As above, for symptoms of anxiety in the last two weeks, measured using average scores for GAD-7 items ranked from 0 (never bothered) to 3 (bothered nearly every day).
Table 1.
Demographics, symptom severity, family history and associated conditions for IBS patients diagnosed via different modalities and with different IBS subtypes
| Scores among DHQ participants completing the relevant sections | Among DHQ participants (%) | Atopy (%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group | n | Male (%) | Female (%) | Age (years) | Age (s.d.) | Mean IBS-SSS | Mean PHQ-12 | Mean GAD-7 | Family history of IBS | Childhood antibiotic exposure | Born by Cesarean section | Treatment for anxiety offered or sought | Treatment for depression offered or sought | Pooled asthma/hay fever/eczema |
| Controls (DHQ respondents) | 72,356 | 50.3 | 49.7 | 65.3 | 7.5 | 33 | 4.0 | 1.5 | 9.5 | 9.6 | 2.5 | 16.1 | 18.0 | 18.1 |
| Hospital ICD-10 | 4,237 | 23.4 | 76.6 | 65.9 | 7.9 | 202* | 7.4* | 3.6* | 31.8* | 24.0* | 2.5 | 43.0* | 43.6* | 29.3* |
| Unprompted self-report | 9,309 | 25.7 | 74.3 | 65.2 | 8.0 | 196* | 7.1* | 3.3* | 30.7* | 19.8* | 2.3 | 41.1* | 40.0* | 29.6* |
| DHQ, Rome III criteria | 24,845 | 27.9 | 72.1 | 63.9 | 7.7 | 194* | 7.1* | 3.3* | 24.3* | 20.4* | 2.6 | 33.7* | 35.1* | 24.0* |
| DHQ, self-report | 16,289 | 26.2 | 73.8 | 64.0 | 7.7 | 177* | 6.9* | 3.1* | 29.0* | 21.6* | 2.7 | 39.7* | 39.3* | 25.9* |
| Pooled (any of the four definitions above) | 40,548 | 27.9 | 72.1 | 64.3 | 7.8 | 173* | 6.8* | 3.1* | 24.0* | 20.0* | 2.6 | 34.3* | 35.2* | 24.8* |
| DHQ, Rome III criteria, type C | 3,989 | 16.9 | 83.1 | 64.5 | 7.8 | 190* | 6.8* | 3.2* | 22.2* | 19.2* | 2.2 | 32.3* | 34.1* | 21.4* |
| DHQ, Rome III criteria, type D | 6,506 | 33.2 | 66.8 | 63.8 | 7.7 | 185* | 6.5* | 3.0* | 23.8* | 19.1* | 2.8 | 32.1* | 33.0* | 24.0* |
| DHQ, Rome III criteria, type M | 13,666 | 28.5 | 71.5 | 63.8 | 7.8 | 204* | 7.5* | 3.6* | 25.6* | 21.5* | 2.6 | 35.2* | 36.9* | 24.8* |
| DHQ, Rome III criteria, type U | 672 | 31.0 | 69.0 | 65.6 | 7.5 | 124* | 5.6* | 2.4* | 16.7* | 17.7* | 2.8 | 27.5* | 24.9* | 23.8* |
| DHQ, postinfectious | 860 | 24.2 | 75.8 | 62.2 | 7.8 | 196* | 7.4* | 3.6* | 30.3* | 27.7* | 3.5 | 42.0* | 42.4* | 26.6* |
| DHQ, functional constipation | 3,502 | 33.9 | 66.1 | 67.1 | 7.4 | 64* | 4.7* | 1.8* | 12.5* | 11.0 | 2.5 | 23.1* | 25.4* | 19.0 |
| DHQ, functional diarrhea | 5,386 | 61.0 | 39.0 | 65.1 | 7.5 | 49* | 4.3* | 1.8* | 11.0* | 10.7* | 2.6 | 20.3* | 22.0* | 20.5* |
Digestively healthy controls and functional constipation as well as diarrhea groups are shown for reference. Gastrointestinal symptoms are captured by the IBS-SSS (range 0–500), while somatic symptoms are captured by the (modified) PHQ-12 (range 0–22). The GAD-7 score captures symptoms of anxiety (range 0–21). The single asterisk marks significant differences from the control group after adjusting for age, sex, DHQ participation and (Bonferroni) multiple testing at P < 0.05/108 (two-sided logistic regression test). Age and sex differences were not tested.
A total of 24,845 respondents reported current abdominal symptoms meeting standard diagnostic criteria (DHQ Rome III, Fig. 1) at the time of the survey, providing a point prevalence of IBS of 14.5%. Of these, only 8,836 (35.6%) had a hospital-documented IBS diagnosis, via an IBS ICD-10 code (International Statistical Classification of Diseases and Related Health Problems, 10th revision), or reported having been diagnosed with IBS by a doctor (DHQ self-report and/or unprompted self-report). They reported greater gastrointestinal symptom severity (quantified via IBS-SSS) than those not medically diagnosed (odds ratio (OR) and 95% confidence interval (CI) = 1.07 (1.07–1.08) per IBS-SSS unit); Fig. 1b and Supplementary Table 3).
Risk factors and associated conditions
As reported previously, a family history of IBS was more common in cases than controls (24.0 versus 9.5%, OR and 95% CI = 3.73 (3.60–3.88)). However, birth by Cesarean section was not (2.6 versus 2.5%, OR and 95% CI = 1.02 (0.94–1.11); Table 1). A significantly higher proportion of cases with IBS recalled receiving long-term or recurrent antibiotics during childhood compared with controls (20.0 versus 9.6%, OR and 95% CI = 2.22 (2.13–2.30)). The severity of current IBS symptoms correlated positively with recalled childhood antibiotic exposure (OR and 95% CI = 1.04 (1.04–1.04) per IBS-SSS unit) and family history (OR and 95% CI = 1.05 (1.05–1.06)). Participants with anxiety also reported increased antibiotics use in childhood (18.4%, OR and 95% CI = 1.64 (1.59–1.70); Supplementary Table 4).
Regarding comorbidities, as documented previously, the rates of appendicectomy, cholecystectomy and hysterectomy were all increased in IBS (Supplementary Table 5), as were the rates of atopic disease (Table 1). Anxiety and depression were each approximately twice as common (Table 1 and Supplementary Table 4); 34.3% of cases reported treatment for anxiety compared with 16.1% of controls. This effect was more prominent in individuals medically diagnosed with IBS.
The median PHQ-12 score was 4 (interquartile range (IQR) = 2–6) in controls and 6 (IQR = 4–9) in pooled cases (7 in all four constituent subgroups, Supplementary Table 6; see also ‘Median scores among pooled and individual diagnoses’ in the Supplementary Note). The PHQ-12 score correlated with IBS symptom severity (Pearson’s correlation = 0.40 (95% CI = 0.39–0.41) among 31,402 IBS cases completing all PHQ-12 and IBS-SSS questions), with back pain, limb pain and tiredness driving this association (Fig. 1c and Supplementary Figs. 2 and 3). Among UKB participants with previous generalized anxiety disorder-7 (GAD-7) scores (n = 79,430; Supplementary Table 7) or Patient Health Questionnaire-9 (PHQ-9) depression scores (n = 79,087, Supplementary Table 8; see Supplementary Note for definitions), these scores were consistently higher in cases with IBS than controls (OR and 95% CI = 1.14 (1.14–1.15) per GAD-7 unit and 1.15 (1.15–1.16) per PHQ-9 unit) and correlated with IBS-SSS (Pearson’s correlations and 95% CIs among cases = 0.24 (0.22–0.25) and 0.27 (0.25–0.28), respectively; Fig. 1d and Supplementary Fig. 4).
The respective prevalence for functional constipation and functional diarrhea (that is, bowel disturbance without abdominal pain or discomfort; see ‘Definitions of IBS cases’ in the Supplementary Note) were 6.4% and 11.7% (Table 1). Somatic symptoms (PHQ-12) and treatment for anxiety or depression were less strongly associated with functional diarrhea than with IBS-D (excess OR and 95% CI = 1.24 (1.22–1.26) per PHQ-12 unit and 1.58 (1.46–1.72), respectively), with similar effects for functional constipation and IBS-C (Table 1).
Genetics
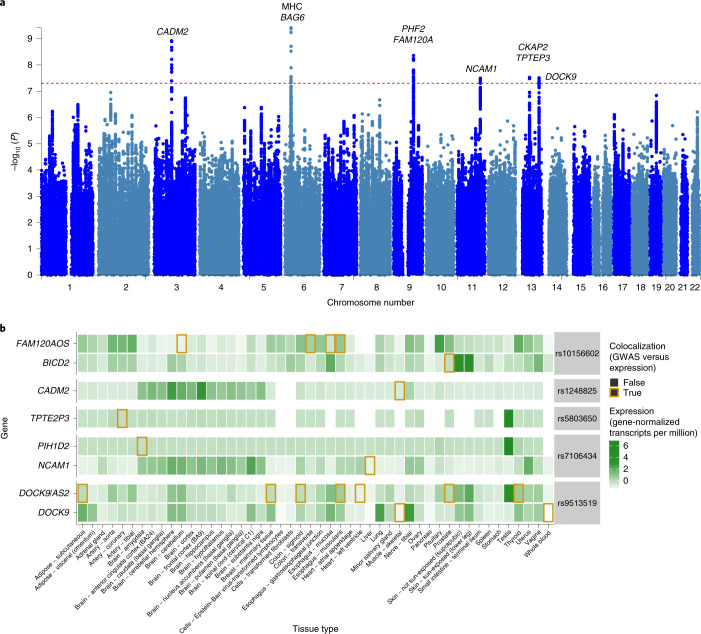
We identified six independent IBS susceptibility loci at genome-wide significance (P < 5 × 10−8) in a discovery cohort totaling 53,400 cases and 433,201 controls (Fig. 2a and Supplementary Fig. 5). This resulted from pooling IBS cases across all case definitions to maximize power, in a meta-analysis of data from UKB (40,548 cases and 293,220 controls; Supplementary Tables 1 and 2) and the international collaborative Bellygenes initiative (12,852 cases and 139,981 controls; Methods and Supplementary Table 9). Using data from an independent panel from 23andMe (Supplementary Note), all six loci were replicated at Bonferroni significance (P < 0.0083) with the same direction of effect (Table 2). All were found on autosomal chromosomes (none on the X chromosome) and conferred modest ORs < 1.05. Three out of six loci also had reported associations with mood and anxiety disorders and related phenotypes21–25.
Fig. 2. Genome-wide association results for IBS.
a, Manhattan plot showing the distribution of IBS-associated SNPs across the genome. The dashed line indicates the genome-wide significance threshold at P = 5 × 10−8. P values are from a two-sided test, after inverse variance-weighted fixed-effects meta-analysis. See Supplementary Fig. 5 for zoomed-in visualizations with linkage disequilibrium data, posterior probabilities of causality and transcript annotations. b, Expression of genes implicated in IBS etiology through causal effects of associated variants on gene expression across a range of tissues. Darker green indicates higher gene expression and the golden outlines indicate tissue–gene combinations where the IBS-associated variant was known to influence gene expression in this tissue (colocalization posterior > 0.5). Genes with at least one colocalization event across tissues with expression quantitative trait locus data from the GTEx dataset are shown.
Table 2.
Variants associated with IBS, their effect measured in the discovery cohort and P values for association in the discovery cohort, the replication cohort and the meta-analysis of these two
| Variant | Effect (discovery) | P values | Annotation | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chromosome number | Position | Alleles | Frequency | OR | 95% CI | Discovery | Replication | Meta-analysis | Mapped gene | Previously implicated in |
| rs1248825 | 3 | 84,993,411 | C/A | 0.33 | 1.05 | 1.03–1.07 | 1.20 × 10−9 | 4.90 × 10−8 | 7.48 × 10−15 | CADM2 | Personality traits (risk-taking, neuroticism, anxiety)21, cannabis use22 |
| rs2736155 | 6 | 31,605,199 | G/C | 0.48 | 1.05 | 1.02–1.07 | 3.88 × 10−10 | 8.28 × 10−6 | 3.19 × 10−12 | BAG6 | |
| rs10156602 | 9 | 96,345,328 | G/A | 0.63 | 1.04 | 1.02–1.06 | 4.36 × 10−9 | 1.18 × 10−8 | 3.04 × 10−15 | PHF2, FAM120AOS | Neuroticism23, depression23, autism24 |
| rs7106434 | 11 | 112,860,579 | C/T | 0.41 | 1.04 | 1.02–1.06 | 3.19 × 10−8 | 2.27 × 10−5 | 9.17 × 10−11 | NCAM1 | Neuroticism23, depression25, cannabis use22, anorexia nervosa40 |
| rs5803650 | 13 | 53,939,598 | CT/C | 0.48 | 1.05 | 1.03–1.07 | 2.97 × 10−8 | 2.25 × 10−8 | 6.31 × 10−14 | CKAP2, TPTE2P3 | |
| rs9513519 | 13 | 99,610,146 | G/A | 0.62 | 1.04 | 1.02–1.06 | 3.09 × 10−8 | 4.20 × 10−5 | 2.31 × 10−10 | DOCK9 | |
The reported frequencies and effects are those of the second allele. The second allele is defined such that it increases IBS risk. Allele frequencies are taken from UKB. Previous associations were obtained from the literature and GWAS Catalog (Supplementary Note).
We undertook genetic fine-mapping to establish plausible causal variants (Supplementary Fig. 5) and used several techniques to identify candidate causal genes within IBS risk loci (Supplementary Table 10; see ‘Gene mapping’ in the Supplementary Note). Among the genes implicated (Table 2) were two encoding neural adhesion molecules: neural cell adhesion molecule 1 (NCAM1) and cell adhesion molecule 2 (CADM2). Ranking tissues according to enrichment for risk gene expression (Supplementary Fig. 6), the brain came top of the list (LDSC applied to specifically expressed genes26 coefficient = 8.32 × 10−10, s.e.m. = 4.5 × 10−10, P = 0.03). However, this result was not statistically significant after correcting for multiple testing, which may in part reflect lack of power due to low SNP heritability. Using expression colocalization analysis as a separate method to implicate specific gene–tissue combinations, we found evidence that the six IBS-associated variants regulate gene expression across a number of tissues, with many genes particularly expressed in the brain (Fig. 2b).
One association mapped to the major histocompatibility complex (MHC) class 3 region close to BAG cochaperone 6 (BAG6). The signal is not driven by human leukocyte antigen (HLA) alleles and is independent of known MHC associations with ulcerative colitis, celiac disease or microscopic colitis (Supplementary Fig. 7 and Supplementary Tables 11 and 12) (refs. 27–30). It is also independent of lead variants for neuroticism at this locus (highest r2 = 0.51)23.
Eight additional loci showed genome-wide significant association with various IBS definitions (Methods) but not the whole discovery cohort, of which five were replicated in the 23andMe data (Supplementary Fig. 8 and Supplementary Tables 13 and 14). These require further study. The female-specific signal identified previously14 for unprompted self-reported IBS in the UKB was also observed in our female-specific analysis of unprompted self-reported data but was not detected in female-specific analyses of any other case definitions from UKB or Bellygenes initiative, nor replicated in the 23andMe unstratified analyses of both sexes (Supplementary Table 15), possibly suggesting survey-specific factors playing a role. Specific candidate gene associations previously reviewed in the literature31,32 also did not show significant evidence of association after multiple testing correction (all P > 0.015).
LDSC estimated a modest but significant genome-wide SNP heritability for IBS of 5.77% (s.e.m. = 0.35%) in the discovery cohort, with no evidence of population stratification (LDSC intercept = 0.9951, s.e.m.= 0.007). This was consistent across case definitions within UKB (h2 range of 5.42–7.71%), with similar values seen in the Bellygenes (h2 = 3.14%, s.e.m. = 0.74%) and 23andMe cohorts (h2 = 5.39%, s.e.m.= 0.02%).
IBS-C showed weak genetic correlation with functional constipation, as did IBS-D with functional diarrhea (Supplementary Fig. 9). IBS-C and IBS-D correlated with each other but there were no cross-correlations, that is, IBS-C did not correlate with functional diarrhea. Heritability for the IBS subtypes was comparable with IBS overall; IBS subtypes showed similar genetic correlation with mental health and personality traits (Supplementary Table 16).
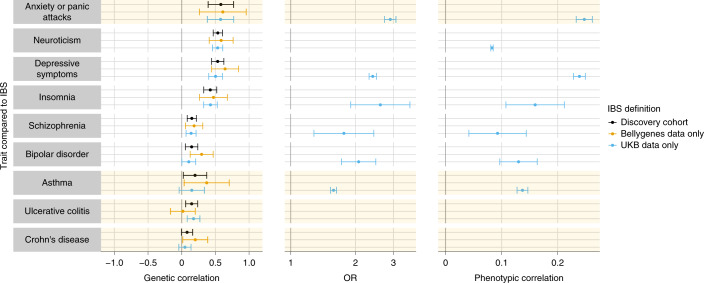
We compared the overlap between susceptibility with IBS and 751 other traits and diseases listed in the LD Hub33. The strongest correlations in genome-wide risk were with mood and anxiety disorders and related phenotypes, including anxiety (rg = 0.58, s.e.m. = 0.10), neuroticism (rg = 0.54, s.e.m. = 0.04), depression (rg = 0.53, s.e.m. = 0.05) and insomnia (rg = 0.42, s.e.m. = 0.05)33. Across the genome, the same alleles that predisposed to IBS also predisposed to mood and anxiety disorders. The correlations were consistent regardless of the mode of diagnosis of anxiety or depression (Supplementary Fig. 10) (refs. 34,35). We calculated phenotypic correlations for these traits on a comparable liability scale (Fig. 3 and Supplementary Table 16). Mostly, the phenotypic and genotypic correlations mirrored each other, although genetic correlations were often larger. Notably, other digestive diseases presenting with similar symptoms, including celiac disease (rg = 0.03, s.e.m. = 0.08, P = 0.69) and Crohn’s disease (rg = 0.08, s.e.m. = 0.04, P = 0.06), were not genetically correlated with IBS.
Fig. 3. Genetic and phenotypic correlations between IBS and other traits.
Correlations of genetic risk (coheritability estimates, left) and phenotype (ORs, middle, and liability-scale correlation, right) between IBS and other traits. The subset of all LD Hub traits (excluding rapid GWAS results) with significant genetic correlation (two-tailed coheritability test as implemented in the LDSC with unadjusted P < 0.05) in both UKB data (light blue, 40,548 cases and 293,220 controls) and the independent Bellygenes cohort (orange, 12,852 cases and 139,981 controls) is shown, as well as the meta-analysis used for discovery (black, 53,400 cases and 433,201 controls). IBS had a similar genetic risk profile to traits such as neuroticism, depression and insomnia (for which P values were significant in all datasets after multiple testing correction for the number of traits tested). Traits highlighted in yellow were added manually given their clinical relevance. Anxiety was not initially included since it is only available in the LD Hub as a rapid GWAS result. Among UKB participants, we present ORs (middle) and phenotypic correlations values (right) for these traits. Sample sizes (restricted to UKB participants who were either cases or controls in the discovery cohort; n = 333,768) were as follows: anxiety or panic attacks, 8,714; neuroticism, 271,423 (scores); depressive symptoms, 24,311; insomnia, 326; schizophrenia, 574; bipolar disorder, 1,207; asthma, 41,178.
We also ran higher-specificity (IBS cases meeting at least 2 of the 4 UKB case definitions, 11,201 cases and 293,220 controls) and high-severity (IBS-SSS > 300, 4,296 cases and 72,356 controls) analyses in UKB. The former produced no new associations. The latter, while being more heritable (liability scale h2 = 0.42, s.e.m. = 0.05, Cochran’s Q = 51.7, P = 6.31 × 10−13 compared with the discovery cohort IBS), produced one association (rs9947289, P = 2.80 × 10−8) that did not replicate (P = 0.57 in the 23andMe data; Supplementary Table 13). Both of these phenotypes recapitulated the same genetic correlation with mood and anxiety disorders as found in the discovery cohort (Supplementary Fig. 11).
To explore the role of shared genetic risk versus direct phenotypic overlap, we compared the genome-wide association study (GWAS) results for IBS having removed participants with anxiety to the GWAS results for anxiety having removed participants with IBS (for anxiety definitions, see Supplementary Tables 17 and 18). The genetic correlation between IBS and anxiety attenuated but remained strong (rg = 0.31, s.e.m. = 0.06; Supplementary Fig. 12). We next used bidirectional Mendelian randomization36 with an independent anxiety GWAS37, as well as genome-wide latent variable Mendelian randomization38, to explore directionality. Multiple models could explain our data (Supplementary Table 19) but they were best explained by shared genetic risk pathways rather than causal effects between the two traits. Similar complex causal relationships were evident between IBS and mental health and personality traits other than anxiety (Supplementary Table 19).
Discussion
The importance of this study lies in its scale and therefore the robustness of its genetic results. We have identified replicable genetic associations for IBS, providing new biological insights, while demonstrating that overall its heritability is modest. Two observations are particularly striking: the genetic overlap between IBS and mood and anxiety disorders and the lack of signals implicating genes expressed specifically in the gut or overlapping other intestinal disorders. Our findings suggest that, with respect to the genetically determined risk for IBS, neuronal pathways play a dominant role.
Increasing abdominal symptom severity correlated with increasing PHQ-12 somatic symptom scores, particularly for the domains of tiredness, back pain, limb pain and headache (Fig. 1). Multifocal pain suggests either poor coping skills, perhaps relating to psychological comorbidity, or visceral hypersensitivity from aberrant antinociceptive mechanisms39. By contrast, the painless bowel disorders, functional constipation and functional diarrhea, were less strongly associated with raised PHQ-12 scores or psychological comorbidity.
IBS showed the strongest genome-wide overlap with psychological traits: anxiety, neuroticism, depression and schizophrenia (Fig. 3). GAD-7 anxiety scores correlated with IBS severity (Fig. 1) and 34.3% of cases with IBS had sought or had been treated for anxiety versus 16.1% of controls (Table 1). Although the phenotypic correlation was strong, the genetic correlation appeared quantitatively even greater (Fig. 3). Furthermore, this genetic correlation between IBS and anxiety persisted even after eliminating data from individuals with phenotypic overlap (that is, between GWAS for ‘IBS excluding anxiety’ and ‘anxiety excluding IBS’; Supplementary Fig. 12). Thus, their co-occurrence probably reflects shared etiologic pathways between IBS and anxiety rather than one condition simply causing the other. This conclusion was supported by the Mendelian randomization analysis.
Four out of six of the confirmed IBS loci implicated genes influencing mood or anxiety disorders, genes expressed in the nervous system or both. These include NCAM1 (also associated with neuroticism, anxiety, mood disorders and anorexia nervosa)23,25,40, CADM2 (also associated with neuroticism, anxiety and cannabis use)21,22, PHD finger protein 2 (PHF2)/family with sequence similarity 120A (FAM120A) (also associated with neuroticism, depression and autism)23,24 and dedicator of cytokinesis 9 (DOCK9). Brain expression of NCAM1 and CADM2 was implicated in our colocalization analysis (Fig. 2b and Supplementary Table 10): both regulate neural circuit formation and influence changes in white matter microstructure found in both mood disorders and IBS25,41,42. PHF2 and DOCK9 also play key roles in brain development43,44. Of note, NCAM1, PHF2 and DOCK9 are also expressed in the rich network of nerve fibers and ganglia of the gut, while CADM2 is not45. Predominant brain expression, combined with the coassociation of IBS with several psychological traits, perhaps most strongly implicates the central nervous system as the site where these gene variants exert their action. However, the genetic variants may also be acting peripherally for the subset expressed in the enteric nervous system, which shares many neurotransmitters, signaling pathways and anatomical properties as well as rich communication with the brain.
The MHC signal is independent of known HLA associations with ulcerative colitis and celiac disease; in fact, it localizes to BAG6 (Supplementary Fig. 7 and Supplementary Tables 10–12). BAG6 is known to chaperone misfolded proteins, regulate membrane protein dynamics and affect diverse processes from apoptosis to antigen presentation46,47. Functional exploration of BAG6 may yield new IBS pathophysiological insights unconnected to the nervous system.
IBS genome-wide SNP heritability was just 5.8% (s.e.m. < 0.01) in the European ancestry population in this study and the effect sizes of our susceptibility loci were modest (OR < 1.05). Earlier genetic studies of IBS were underpowered to detect such small effects. By comparison, SNP heritability estimates for Crohn’s disease, ulcerative colitis and anxiety are 41%, 23% and 26%, respectively48,49. Previous IBS heritability estimates, from family and twin studies, varied widely at 0–57% (ref. 11). Our results indicate that the genetic contribution to IBS heritability is modest and imply that additional environmental factors, including dysbiosis, diet, stress and learned behaviors, all potentially shared within families, play a more prominent role.
Regarding dysbiosis, we noted increased childhood exposure to antibiotics among IBS cases (20.0%) versus controls (9.6%). While there are clearly biases inherent to recall of events from childhood, this result is corroborated by previous studies specifically set up to address this question50. Interestingly, we saw the same association with anxiety (18.4%). Among possible explanations, childhood antibiotics might increase the risk of IBS (and perhaps anxiety) by embedding a dysbiotic gut flora and disturbing the balance of short-chain fatty acid metabolites known to influence microglial development and mood50,51. Equally, anxiety in late adulthood might influence recall of childhood antibiotic exposure, and familial anxiety might lead parents to take their offspring to the doctor repeatedly for minor ailments, resulting in recurrent antibiotic exposure. While enteric infection can alter the baseline gut microbiota and trigger PI-IBS, in the UKB PI-IBS closely mirrored ‘conventional’ IBS in terms of symptom severity, frequency of family history and association with psychological traits, suggesting that the infectious ‘seed’ falls on fertile ground to trigger IBS in predisposed individuals.
One question is whether the neuronal emphasis of our results derives from our strategy of combining multiple IBS definitions to increase statistical power, including pooling ‘opposite’ subtypes (for example, IBS-C and IBS-D), that is, whether gut-specific effects might be lost in the pooling such that the brain remains the common link between these. However, the heritability of IBS subtypes is comparable with IBS overall; IBS-C and IBS-D share approximately 50% of their genetic susceptibility and each of the subgroups also individually genetically correlates with mental health and personality traits (Supplementary Table 16). Furthermore, subtype GWAS identified only one significant signal in IBS-C and none in IBS-D, suggesting an absence of strong subtype-specific, possibly gut-focused genetic effects.
Aside from the pooling strategy, justified by our LDSC analysis (Methods), other potential weaknesses include the use of Rome III criteria instead of the more restricted Rome IV criteria, since the former were the standard at the time of study design52, the fact the IBS diagnosis was made based on Rome III symptoms reported via the DHQ rather than by medical review for nearly half of cases in the UKB cohort and the limited age range and ancestry of UKB. However, we believe that the fact that all of the loci identified at genome-wide significance thresholds in the discovery panel replicated in the independent 23andMe panel validates both the findings and the approach taken.
Our GWAS and the results of our polygenic analyses provide important new insights. Individual loci identified by the GWAS implicate new target genes within previously under-researched pathways (for example, neuronal adhesion). Mendelian randomization and genome-wide correlation analyses demonstrate shared genetic risk pathways between anxiety and IBS that are independent of the comorbidity between these two traits. This may point toward a mechanistic rationale for the efficacy of psychoactive medications and behavioral therapies and suggest that more attention should be paid to identifying new therapeutics that target neuronal function. We anticipate that future research will build on our discoveries, both by investigating the target genes identified and exploring the shared genetic risk across traits to improve our understanding of the disordered brain–gut interactions that characterize IBS.
Methods
For details of the cohorts, descriptive statistics, association analysis methodologies and functional interpretation of associations, see the Supplementary Note.
Ethics oversight
The UKB DHQ was approved as a substantial amendment to the UKB protocol by the North West – Haydock Research Ethics Committee, reference no. 11/NW/038e. The Bellygenes initiative study received ethical approval from the Stockholm Ethics Examination Authority (EPN ID 2016/1620-31/2) and Monash University Human Research Ethics Committee (MUHREC ID 20326). Written informed consent was obtained from all participants. This study did not award compensation to any participant.
Data and study participants
Our discovery cohort combined cases of IBS identified in UKB with cases from the Bellygenes initiative. Replication was sought in an independent panel from 23andMe. Cases ascertained in UKB met at least one of the following four conditions (the DHQ is viewable online – UKB resource 595): (1) DHQ Rome III: met Rome III symptom criteria for IBS diagnosis without other diagnostic explanations for these symptoms; (2) DHQ ‘self-report’: answered ‘yes’ to the question ‘Have you ever been diagnosed with IBS?’; (3) Unprompted ‘self-report’: self-reported IBS diagnosis in response to question ‘Has a doctor ever told you that you have any … serious medical conditions?’; (4) hospital ICD-10: linked hospital episode statistics indicating inpatient or day-case admission with clinician diagnosis of IBS entered as main or secondary ICD-10 diagnosis.
Participants with conditions such as celiac disease, inflammatory bowel disease or previous intestinal resectional surgery that could result in IBS-like symptoms were excluded from both cases and digestively healthy controls to avoid signal contamination. For detailed case and control inclusion and exclusion criteria, see Supplementary Tables 1 and 2, respectively. To maximize sample size, cases from the 4 UKB groups were pooled (n = 40,548). This approach was supported by demonstrating high genetic correlations between them using LDSC48 following a separate GWAS on each (minimum pairwise rg = 0.70, s.e.m. = 0.06; Supplementary Fig. 13) and by previous literature on the consistency of genetic results obtained from different diagnostic definitions in UKB16.
We then meta-analyzed IBS GWAS data from UKB (40,548 cases) and Bellygenes initiative (12,852 cases and 139,981 controls; Supplementary Table 9), an international collaboration studying IBS genetics based on electronic medical records, specialist diagnoses form tertiary clinics and questionnaire data (including Rome III criteria) across multiple cohorts, having again demonstrated high genetic correlation between them (rg = 0.998, s.e.m. = 0.129). This produced a total discovery cohort of 53,400 cases and 433,201 controls. Evidence of replication was sought in a large 23andMe dataset (Supplementary Note). 23andMe cases (n = 205,252) self-reported being diagnosed or treated for IBS while controls (n = 1,384,055) did not.
Analyses of IBS subtypes were conducted solely using UKB DHQ data based on standard definitions of IBS-C, IBS-D, IBS-M and IBS-U according to the frequency of hard or lumpy stools versus loose, mushy or watery stools. Functional constipation and functional diarrhea cases were identified similarly, and with the same exclusions per IBS cases, but (in contrast to the Rome III definition of IBS) needed to have responded ‘never’ when asked about the frequency of abdominal pain in the last three months. Likewise, analyses of IBS severity (using the IBS-SSS) and associated somatic symptoms (using the PHQ-12) were restricted to DHQ respondents. Anxiety and depression were identified among UKB participants based on previously surveyed responses to GAD-7 anxiety and PHQ-9 depression questionnaires, self-report of diagnosis with depression or anxiety/panic attack, diagnostic codes for major depression and phobic or generalized anxiety disorder in electronic healthcare records or reporting of treatment being sought or offered for these conditions in our DHQ (Supplementary Note).
Statistical analysis
Association between IBS and nongenetic risk factors, including risk factors assayed by recall from the DHQ, was tested using logistic regression conditioning on age and sex (Supplementary Note, ‘Nongenetic associations’).
Standard genetic quality control was carried out to remove samples with poor genotype quality and variants with poor genotyping or imputation performance. Only participants of European ancestry were included in the discovery dataset due to the limited number of non-European ancestry participants. GWAS were conducted using a linear mixed model (BOLT-LMM v.2.3.2)53 to control for population stratification and relatedness. Meta-analysis of GWAS summary statistics was carried out using METAL (March 2011 release)54. The UKB GWAS was stratified into DHQ respondents and nonrespondents, with results meta-analyzed to avoid genetic confounding with questionnaire response (Supplementary Fig. 14).
We assigned loci to candidate genes using annotations from FUMA v.1.3.455, as well as from a colocalization analysis using Coloc v.3.2-156 on multi-tissue expression data from the Genotype-Tissue Expression (GTEx) consortium56,57. We calculated SNP heritability and coheritability (rg, genetic correlation) using univariate and bivariate LDSC48 against a range of traits via the LD Hub website33. Other statistical analyses were carried out using R v.3.6.1; any P values were obtained from two-sided tests unless otherwise specified.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41588-021-00950-8.
Supplementary information
Supplementary Note and Figs. 1–14.
Supplementary Tables 1–22.
Acknowledgements
This research was conducted using the UK Biobank resource under application no. 17670. We acknowledge the contribution made by all participants in the cohorts studied. We also thank all 23andMe employees and participants involved in its research. The research leading to these results received funding from the European Union Seventh Framework Programme under grant no. 313010 (Biobanking and Biomolecular Research Infrastructure—Large Prospective Cohorts). We received funding support from the National Institute for Health Research Biomedical Research Centres based at Cambridge University Hospital NHS Trust and the University of Cambridge (no. BRC-1215-20014); the University of Oxford (no. BRC-1215-20008); Nottingham University Hospitals NHS Trust and the University of Nottingham (no. BRC-1215-20003); and the University Hospital of South Manchester and University of Manchester (no. BRC-1215-20007). This research was funded in whole, or in part, by Wellcome Trust grant nos. 215097/Z/18/Z, 203141/Z/16/Z, 093885/Z/10/Z, 098051, 100956/Z/13/Z and 280750/Z/17/Z. For the purpose of open access, we have applied a CC BY public copyright licence to any author accepted manuscript version arising from this submission. Additional support was received from the Li Ka Shing Foundation and the Kennedy Trust for Rheumatology Research. M.C. received past and current support from the National Institutes of Health (NIH) for genetic studies in IBS (nos. R01 DK 92179 and 115950). A.-I.V.’s contribution was supported by a Kennedy Trust Prize Studentship. This work received support from European Research Council Starting Grant no. 715772, a Nederlandse Organisatie voor Wetenschappelijk (NOW)-VIDI grant, Netherlands Heart Foundation CVON grant no. 2018-27 and NWO Gravitation grant Exposome-NL. The study was supported by grants from the Swedish Research Council (VR project no. 2017-02403), the Health Department of the Basque Government (grant no. 2015111133) and the Spanish Ministry of Economy and Competitiveness (Instituto Salud Carlos III grant no. FIS PI17/00308) to M.D.A. This work received infrastructure support from the Deutsche Forschungsgemeinschaft Cluster of Excellence ‘Precision Medicine in Chronic Inflammation’ (no. EXC2167). The GTEx Project was supported by the Common Fund of the Office of the Director of the NIH and by the National Cancer Institute, National Human Genome Research Institute, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health and National Institute of Neurological Disorders and Stroke. The data used for the colocalization analyses were obtained from the GTEx Portal on 16 June 2019. The Lifelines Biobank initiative has been made possible by funds from Fonds Economische Structuurversterking, Samenwerkingsverband Noord Nederland and Ruimtelijk Economisch Programma. The UK Household Longitudinal Study, a source of UK controls in this work, is led by the Institute for Social and Economic Research at the University of Essex and funded by the Economic and Social Research Council. The survey was conducted by NatCen and the genome-wide scan data were analyzed and deposited by the Wellcome Trust Sanger Institute. Information on how to access the data can be found on the Understanding Society website (https://www.understandingsociety.ac.uk/). The Nord-Trøndelag Health Study (HUNT study) is a collaboration between the HUNT Research Centre (Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology), Trøndelag County Council, the Central Norway Regional Health Authority and the Norwegian Institute of Public Health. We are grateful to D. Levy and all Million Veteran Program (MVP) staff, researchers and volunteers, who provided anxiety association summary statistics for this work, and especially participants who previously served their country in the military and now generously agreed to enroll in the MVP study. We thank S. Berens, F. Kraus, E. Stroe-Kunold and W. Herzog of the Heidelberg outpatient clinic and the supporting staff of each site of the IBS Net Germany. The data and analyses presented in the current publication are based on the use of study data downloaded from the database of Genotypes and Phenotypes (dbGaP) website (https://www.ncbi.nlm.nih.gov/gap/) under phs000674.v2.p2 (now superseded by phs000674.v3.p3) and under phs000428.v2.p2.
Author contributions
N.A.K., C.A.A., L.M., A.Z., P.J.W., R.S., M.D., L.J. and M.P. designed the study. J.H., G.F., A.F., G.A., E.N.-J., K.H., T.E., M.C., G.B., the 23andMe Research Team and the Bellygenes Initiative collected the data. C.E., T.Z., F.B., L.M., A.-I.V., M.H., M.Z., A.H.S., M.T.-L. and L.J. conducted the analyses. A.F., G.A., K.H., T.E., A.Z., G.M., M.D. and M.P. guided the analysis. N.A.K., C.A.A., M.C., P.J.W., R.S., G.M., M.D. and M.P. interpreted the data. J.S. and S.S. replicated the results. C.E., A.-I.V. and L.J. produced the figures and tables. C.E., N.A.K., P.J.W., R.S., M.D., L.J. and M.P. wrote the manuscript.
Data availability
Genome-wide summary statistics have been deposited to the European Bioinformatics Institute GWAS Catalog (https://www.ebi.ac.uk/gwas/) under accession no. GCST90016564. Individual-level data on the DHQ responses, along with matching genotype, electronic health record and survey data, are available via an application to the UK Biobank Access Management System (https://bbams.ndph.ox.ac.uk/ams/). Individual-level data for 23andMe were not shared as part of this project to protect the privacy of 23andMe participants. Data used for the analyses pertinent to the Bellygenes initiative include both individual-level and aggregate data. Individual-level data from the following sources can be obtained via applications to the respective biobanks and cohorts: TWINGENE (https://ki.se/en/research/swedish-twin-registry-for-researchers); HUNT (https://www.ntnu.edu/hunt/data); Michigan Genomics Initiative (https://precisionhealth.umich.edu/our-research/michigangenomics/); Estonian Genome Center of the University of Tartu (https://genomics.ut.ee/en/biobank.ee/data-access); Lifelines (https://www.lifelines.nl/researcher/how-to-apply); Gene-Environment and Gene-Gene Interaction Research Application (dbGaP Study accession no. phs000674.v2.p2, now superseded by phs000674.v3.p3). Data from IBS patients from tertiary centers can be requested from Mauro D’Amato at mdamato@cicbiogune.es and may be made available depending on specific material and data transfer agreements with principal investigators at respective collaborating institutions.
Code availability
All software used is publicly available at the URLs or references cited. The R code for additional analyses is available at 10.5281/zenodo.5048820.
Competing interests
C.A.A. and L.J. are paid consultants for Genomics. C.A.A. is a paid consultant for Celgene. J.S. and S.S. are employed by and hold stock or stock options in 23andMe. Members of the 23andMe research team are employed by and hold stock or stock options in 23andMe. P.J.W. has acted as a consultant or received research funding from Danone, Allergan, Ironwood Pharmaceuticals and Salix Pharmaceuticals, all outside of the submitted work. R.S. has accepted research grants from Sanofi and Zespri International and speaker fees from Alfawasserman. G.M. is a director of and shareholder in Genomics and partner in Peptide Groove LLP. The other authors declare no competing interests.
Footnotes
Peer review information Nature Genetics thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Tenghao Zheng, Nicholas A. Kennedy.
A list of authors and their affiliations appears at the end of the paper.
These authors jointly supervised this work: Mauro D’Amato, Luke Jostins, Miles Parkes.
Contributor Information
Mauro D’Amato, Email: mdamato@cicbiogune.es.
Luke Jostins, Email: luke.jostins@kennedy.ox.ac.uk.
Miles Parkes, Email: miles.parkes@addenbrookes.nhs.uk.
23andMe Research Team:
Michelle Agee, Stella Aslibekyan, Adam Auton, Robert K. Bell, Katarzyna Bryc, Sarah K. Clark, Sarah L. Elson, Kipper Fletez-Brant, Pierre Fontanillas, Nicholas A. Furlotte, Pooja M. Gandhi, Karl Heilbron, Barry Hicks, David A. Hinds, Karen E. Huber, Ethan M. Jewett, Yunxuan Jiang, Aaron Kleinman, Keng-Han Lin, Nadia K. Litterman, Marie K. Luff, Jey C. McCreight, Matthew H. McIntyre, Kimberly F. McManus, Joanna L. Mountain, Sahar V. Mozaffari, Priyanka Nandakumar, Elizabeth S. Noblin, Carrie A. M. Northover, Jared O’Connell, Aaron A. Petrakovitz, Steven J. Pitts, G. David Poznik, J. Fah Sathirapongsasuti, Anjali J. Shastri, Janie F. Shelton, Chao Tian, Joyce Y. Tung, Robert J. Tunney, Vladimir Vacic, Xin Wang, and Amir S. Zare
The Bellygenes Initiative:
Purna Kashyap, Lin Chang, Emeran Mayer, Margaret Heitkemper, Gregory S. Sayuk, Tamar Ringel-Kulka, Yehuda Ringel, William D. Chey, Shanti Eswaran, Juanita L. Merchant, Robert J. Shulman, Luis Bujanda, Koldo Garcia-Etxebarria, Aldona Dlugosz, Greger Lindberg, Peter T. Schmidt, Pontus Karling, Bodil Ohlsson, Susanna Walter, Åshild O. Faresjö, Magnus Simren, Jonas Halfvarson, Piero Portincasa, Giovanni Barbara, Paolo Usai-Satta, Matteo Neri, Gerardo Nardone, Rosario Cuomo, Francesca Galeazzi, Massimo Bellini, Anna Latiano, Lesley Houghton, Daisy Jonkers, Alexander Kurilshikov, Rinse K. Weersma, Mihai Netea, Jonas Tesarz, Annika Gauss, Miriam Goebel-Stengel, Viola Andresen, Thomas Frieling, Christian Pehl, Rainer Schaefert, Beate Niesler, Wolfgang Lieb, Kurt Hanevik, Nina Langeland, Knut-Arne Wensaas, Sverre Litleskare, Maiken E. Gabrielsen, Laurent Thomas, Vincent Thijs, Robin Lemmens, Lukas Van Oudenhove, and Mira Wouters
Supplementary information
The online version contains supplementary material available at 10.1038/s41588-021-00950-8.
References
- 1.Ford AC. Commentary: estimating the prevalence of IBS globally––past, present and future. Aliment. Pharmacol. Ther. 2020;51:198–199. doi: 10.1111/apt.15508. [DOI] [PubMed] [Google Scholar]
- 2.Gralnek IM, Hays RD, Kilbourne A, Naliboff B, Mayer EA. The impact of irritable bowel syndrome on health-related quality of life. Gastroenterology. 2000;119:654–660. doi: 10.1053/gast.2000.16484. [DOI] [PubMed] [Google Scholar]
- 3.Soubieres A, Wilson P, Poullis A, Wilkins J, Rance M. Burden of irritable bowel syndrome in an increasingly cost-aware National Health Service. Frontline Gastroenterol. 2015;6:246–251. doi: 10.1136/flgastro-2014-100542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hillilä MT, Färkkilä NJ, Färkkilä MA. Societal costs for irritable bowel syndrome—a population based study. Scand. J. Gastroenterol. 2010;45:582–591. doi: 10.3109/00365521003637211. [DOI] [PubMed] [Google Scholar]
- 5.Longstreth GF, et al. Functional bowel disorders. Gastroenterology. 2006;130:1480–1491. doi: 10.1053/j.gastro.2005.11.061. [DOI] [PubMed] [Google Scholar]
- 6.Pacheco RL, et al. What do Cochrane systematic reviews say about the management of irritable bowel syndrome? Sao Paulo Med. J. 2019;137:82–91. doi: 10.1590/1516-3180.2018.053740119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Windgassen S, et al. The journey between brain and gut: a systematic review of psychological mechanisms of treatment effect in irritable bowel syndrome. Br. J. Health Psychol. 2017;22:701–736. doi: 10.1111/bjhp.12250. [DOI] [PubMed] [Google Scholar]
- 8.Koloski NA, Jones M, Talley NJ. Evidence that independent gut-to-brain and brain-to-gut pathways operate in the irritable bowel syndrome and functional dyspepsia: a 1-year population-based prospective study. Aliment. Pharmacol. Ther. 2016;44:592–600. doi: 10.1111/apt.13738. [DOI] [PubMed] [Google Scholar]
- 9.Enck P, et al. Irritable bowel syndrome. Nat. Rev. Dis. Primers. 2016;2:16014. doi: 10.1038/nrdp.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jalanka J, Salonen A, Fuentes S, de Vos WM. Microbial signatures in post-infectious irritable bowel syndrome––toward patient stratification for improved diagnostics and treatment. Gut Microbes. 2015;6:364–369. doi: 10.1080/19490976.2015.1096486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saito YA. The role of genetics in IBS. Gastroenterol. Clin. North Am. 2011;40:45–67. doi: 10.1016/j.gtc.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bengtson M-B, Aamodt G, Vatn MH, Harris JR. Co-occurrence of IBS and symptoms of anxiety or depression, among Norwegian twins, is influenced by both heredity and intrauterine growth. BMC Gastroenterol. 2015;15:9. doi: 10.1186/s12876-015-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson MR, et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 14.Bonfiglio F, et al. Female-specific association between variants on chromosome 9 and self-reported diagnosis of irritable bowel syndrome. Gastroenterology. 2018;155:168–179. doi: 10.1053/j.gastro.2018.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Amato M. Genes and functional GI disorders: from casual to causal relationship. Neurogastroenterol. Motil. 2013;25:638–649. doi: 10.1111/nmo.12173. [DOI] [PubMed] [Google Scholar]
- 16.DeBoever C, et al. Assessing digital phenotyping to enhance genetic studies of human diseases. Am. J. Hum. Genet. 2020;106:611–622. doi: 10.1016/j.ajhg.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sudlow C, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manolio TA, Bailey-Wilson JE, Collins FS. Genes, environment and the value of prospective cohort studies. Nat. Rev. Genet. 2006;7:812–820. doi: 10.1038/nrg1919. [DOI] [PubMed] [Google Scholar]
- 19.Francis CY, Morris J, Whorwell PJ. The irritable bowel severity scoring system: a simple method of monitoring irritable bowel syndrome and its progress. Aliment. Pharmacol. Ther. 1997;11:395–402. doi: 10.1046/j.1365-2036.1997.142318000.x. [DOI] [PubMed] [Google Scholar]
- 20.Spiller RC, et al. The Patient Health Questionnaire 12 Somatic Symptom scale as a predictor of symptom severity and consulting behaviour in patients with irritable bowel syndrome and symptomatic diverticular disease. Aliment. Pharmacol. Ther. 2010;32:811–820. doi: 10.1111/j.1365-2036.2010.04402.x. [DOI] [PubMed] [Google Scholar]
- 21.Boutwell B, et al. Replication and characterization of CADM2 and MSRA genes on human behavior. Heliyon. 2017;3:e00349. doi: 10.1016/j.heliyon.2017.e00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasman JA, et al. GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat. Neurosci. 2018;21:1161–1170. doi: 10.1038/s41593-018-0206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagel M, Watanabe K, Stringer S, Posthuma D, van der Sluis S. Item-level analyses reveal genetic heterogeneity in neuroticism. Nat. Commun. 2018;9:905. doi: 10.1038/s41467-018-03242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrovska J, et al. The NCAM1 gene set is linked to depressive symptoms and their brain structural correlates in healthy individuals. J. Psychiatr. Res. 2017;91:116–123. doi: 10.1016/j.jpsychires.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 26.Finucane HK, et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 2018;50:621–629. doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goyette P, et al. High-density mapping of the MHC identifies a shared role for HLA-DRB1*01:03 in inflammatory bowel diseases and heterozygous advantage in ulcerative colitis. Nat. Genet. 2015;47:172–179. doi: 10.1038/ng.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monsuur AJ, et al. Effective detection of human leukocyte antigen risk alleles in celiac disease using tag single nucleotide polymorphisms. PLoS ONE. 2008;3:e2270. doi: 10.1371/journal.pone.0002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green HD, et al. Genome-wide association study of microscopic colitis in the UK Biobank confirms immune-related pathogenesis. J. Crohns Colitis. 2019;13:1578–1582. doi: 10.1093/ecco-jcc/jjz104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Westerlind H, et al. Dense genotyping of immune-related loci identifies HLA variants associated with increased risk of collagenous colitis. Gut. 2017;66:421–428. doi: 10.1136/gutjnl-2015-309934. [DOI] [PubMed] [Google Scholar]
- 31.Camilleri M, Katzka DA. Irritable bowel syndrome: methods, mechanisms, and pathophysiology. Genetic epidemiology and pharmacogenetics in irritable bowel syndrome. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;302:G1075–G1084. doi: 10.1152/ajpgi.00537.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camilleri M, et al. Neuropeptide S receptor induces neuropeptide expression and associates with intermediate phenotypes of functional gastrointestinal disorders. Gastroenterology. 2010;138:98–107.e4. doi: 10.1053/j.gastro.2009.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng J, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ripke S, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol. Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wray NR, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13:e1007081. doi: 10.1371/journal.pgen.1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levey DF, et al. Reproducible genetic risk loci for anxiety: results from ~200,000 participants in the Million Veteran Program. Am. J. Psychiatry. 2020;177:223–232. doi: 10.1176/appi.ajp.2019.19030256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darrous, L., Mounier, N. & Kutalik, Z. Simultaneous estimation of bi-directional causal effects and heritable confounding from GWAS summary statistics. Preprint at medRxivhttps://www.medrxiv.org/content/10.1101/2020.01.27.20018929v3 (2020). [DOI] [PMC free article] [PubMed]
- 39.Whitehead WE, Palsson O, Jones KR. Systematic review of the comorbidity of irritable bowel syndrome with other disorders: what are the causes and implications? Gastroenterology. 2002;122:1140–1156. doi: 10.1053/gast.2002.32392. [DOI] [PubMed] [Google Scholar]
- 40.Watson HJ, et al. Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nat. Genet. 2019;51:1207–1214. doi: 10.1038/s41588-019-0439-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kolkova K, Novitskaya V, Pedersen N, Berezin V, Bock E. Neural cell adhesion molecule-stimulated neurite outgrowth depends on activation of protein kinase C and the Ras-mitogen-activated protein kinase pathway. J. Neurosci. 2000;20:2238–2246. doi: 10.1523/JNEUROSCI.20-06-02238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frei JA, Andermatt I, Gesemann M, Stoeckli ET. The SynCAM synaptic cell adhesion molecules are involved in sensory axon pathfinding by regulating axon–axon contacts. J. Cell Sci. 2014;127:5288–5302. doi: 10.1242/jcs.157032. [DOI] [PubMed] [Google Scholar]
- 43.Pappa S, et al. PHF2 histone demethylase prevents DNA damage and genome instability by controlling cell cycle progression of neural progenitors. Proc. Natl Acad. Sci. USA. 2019;116:19464–19473. doi: 10.1073/pnas.1903188116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shi L. Dock protein family in brain development and neurological disease. Commun. Integr. Biol. 2013;6:e26839. doi: 10.4161/cib.26839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 46.Kawahara H, Minami R, Yokota N. BAG6/BAT3: emerging roles in quality control for nascent polypeptides. J. Biochem. 2013;153:147–160. doi: 10.1093/jb/mvs149. [DOI] [PubMed] [Google Scholar]
- 47.Mock J-Y, et al. Bag6 complex contains a minimal tail-anchor-targeting module and a mock BAG domain. Proc. Natl Acad. Sci. USA. 2015;112:106–111. doi: 10.1073/pnas.1402745112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Purves KL, et al. A major role for common genetic variation in anxiety disorders. Mol. Psychiatry. 2020;25:3292–3303. doi: 10.1038/s41380-019-0559-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krogsgaard LR, Engsbro AL, Bytzer P. Antibiotics: a risk factor for irritable bowel syndrome in a population-based cohort. Scand. J. Gastroenterol. 2018;53:1027–1030. doi: 10.1080/00365521.2018.1500638. [DOI] [PubMed] [Google Scholar]
- 51.Silva YP, Bernardi A, Frozza RL. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front. Endocrinol. (Lausanne) 2020;11:25. doi: 10.3389/fendo.2020.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmulson MJ, Drossman DA. What is new in Rome IV. J. Neurogastroenterol. Motil. 2017;23:151–163. doi: 10.5056/jnm16214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loh P-R, Kichaev G, Gazal S, Schoech AP, Price AL. Mixed-model association for biobank-scale datasets. Nat. Genet. 2018;50:906–908. doi: 10.1038/s41588-018-0144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lonsdale J, et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Note and Figs. 1–14.
Supplementary Tables 1–22.
Data Availability Statement
Genome-wide summary statistics have been deposited to the European Bioinformatics Institute GWAS Catalog (https://www.ebi.ac.uk/gwas/) under accession no. GCST90016564. Individual-level data on the DHQ responses, along with matching genotype, electronic health record and survey data, are available via an application to the UK Biobank Access Management System (https://bbams.ndph.ox.ac.uk/ams/). Individual-level data for 23andMe were not shared as part of this project to protect the privacy of 23andMe participants. Data used for the analyses pertinent to the Bellygenes initiative include both individual-level and aggregate data. Individual-level data from the following sources can be obtained via applications to the respective biobanks and cohorts: TWINGENE (https://ki.se/en/research/swedish-twin-registry-for-researchers); HUNT (https://www.ntnu.edu/hunt/data); Michigan Genomics Initiative (https://precisionhealth.umich.edu/our-research/michigangenomics/); Estonian Genome Center of the University of Tartu (https://genomics.ut.ee/en/biobank.ee/data-access); Lifelines (https://www.lifelines.nl/researcher/how-to-apply); Gene-Environment and Gene-Gene Interaction Research Application (dbGaP Study accession no. phs000674.v2.p2, now superseded by phs000674.v3.p3). Data from IBS patients from tertiary centers can be requested from Mauro D’Amato at mdamato@cicbiogune.es and may be made available depending on specific material and data transfer agreements with principal investigators at respective collaborating institutions.
All software used is publicly available at the URLs or references cited. The R code for additional analyses is available at 10.5281/zenodo.5048820.