Abstract
The transcription of ribosomal DNA, ribosomal protein (RP) genes, and 5S and tRNA genes by RNA polymerases (Pols) I, II, and III, respectively, is rapidly and coordinately repressed upon interruption of the secretory pathway in Saccharomyces cerevisiae. We find that repression of ribosome and tRNA synthesis in secretion-defective cells involves activation of the cell integrity pathway. Transcriptional repression requires the upstream components of this pathway, including the Wsc family of putative plasma membrane sensors and protein kinase C (PKC), but not the downstream Bck1–Mkk1/2–Slt2 mitogen-activated protein kinase cascade. These findings reveal a novel PKC effector pathway that controls more than 85% of nuclear transcription. It is proposed that the coordination of ribosome and tRNA synthesis with cell growth may be achieved, in part, by monitoring the turgor pressure of the cell.
The production of ribosomes and associated protein-synthetic machinery consumes substantial amounts of metabolic energy. It is estimated that yeast cells growing with a generation time of about 100 min devote at least 60% of their total nuclear transcription to the synthesis of the large rRNAs by RNA polymerase (Pol) I and as much as 50% of the initiation events by Pol II to the transcription of ribosomal protein (RP) genes (30). This level of transcription is needed to sustain the production of some 2,000 ribosomes per min and allow the doubling of the cell's ribosome content during each cell cycle. The large investment of high-energy phosphate in ribosome synthesis provides a biological rationale for the evolution of mechanisms that safeguard the cell's metabolic economy. These mechanisms couple the synthesis of ribosomes and ribosomal substrates, such as tRNAs, with the protein-synthetic needs of the cell and the availability of nutrients and/or growth factors (33, 35).
A potentially important mechanism in higher eukaryotic cells that may help to achieve metabolic economy and coordinate protein-synthetic capacity with cell growth involves transcriptional repression of Pols I and III by the tumor suppressor protein Rb (32). This activity of Rb results from its inhibition of preinitiation complex assembly and/or function through direct binding of the Pol I- and Pol III-specific transcription factors UBF and TFIIIB, respectively (2, 3, 15, 29, 34). In addition to Rb and the related pocket proteins, p107 and p130 (27), other mechanisms for controlling transcription of the protein-synthesizing machinery appear to exist. In yeast cells, for example, the production of ribosomal components and tRNAs in response to nutrient availability and growth rate is regulated predominantly at the transcriptional level in the absence of an Rb homolog (11, 24, 33, 35). These observations suggest that yeast may be a good model system in which to examine the fundamental mechanisms coordinating ribosome synthesis with cell growth.
In the past several years, studies with Saccharomyces cerevisiae have uncovered a regulatory circuit that connects the synthesis of the large rRNAs and RPs with the secretory pathway (18, 20, 21). These studies have shown that interruption of the secretory pathway at various points, from peptide insertion into the endoplasmic reticulum (ER) to vesicle fusion with the plasma membrane, causes transcriptional repression of ribosomal DNA (rDNA) and RP genes. The intracellular signaling pathway mediating this response does not involve any of the common mechanisms known to regulate ribosome biosynthesis and is distinct from the pathway that monitors protein folding in the ER (21, 22). However, continued protein synthesis and protein kinase C (PKC) are both required for repression in secretion-defective cells (21, 22). These findings, together with the fact that repression of RP genes can be induced by chlorpromazine, an agent that causes membrane stretching, provide the current working model for signal transduction in this system (22). It is proposed that a failure of the secretory pathway leads to an insufficiency in the supply of lipids and proteins which are needed for growth of the plasma membrane and cell wall synthesis. In the absence of plasma membrane growth, continued protein synthesis is thought to create a higher-than-normal intracellular pressure (turgor), effectively stretching the plasma membrane. This perturbation of the plasma membrane is suggested to activate a PKC-dependent cell integrity pathway leading to transcriptional repression of ribosome synthesis. Activation of the PKC–mitogen-activated protein (MAP) kinase cell integrity pathway following an increase in the osmotic gradient across the plasma membrane (high inside, low outside) is well documented (8). However, the role of this pathway in repressing rRNA and RP gene transcription in secretion-blocked cells has not yet been fully explored.
To date, neither the synthesis of the 5S rRNA component of the ribosome nor that of the tRNA adapter molecules of protein synthesis has been examined in secretion-blocked cells. Given the likelihood that these Pol III transcripts are coregulated with ribosome synthesis and the possibility that a common signaling pathway might control transcription by Pols I and III, as well as 50% of the activity of Pol II, we decided to investigate this issue. We show here that transcription of 5S rRNA and tRNA genes by Pol III is indeed coordinately repressed with transcription of RP genes in secretion-defective cells. In addition, we find that transcriptional repression of RP and tRNA genes under these conditions is signaled by a novel branch of the cell integrity pathway that includes plasma membrane sensors of the Wsc family and PKC but not the downstream MAP kinase cascade.
MATERIALS AND METHODS
Yeast strains and growth conditions.
Yeast strains used in this work are listed in Table 1. W303α and its congenic derivatives (312 and 169ts), strain 1783 and its congenic derivative (DL252), the KM0XX strains (where X is any number), and strain AC2c9 were all grown in yeast-peptone-dextrose (YPD). MN and ALH strains were grown in YPD containing 1 M sorbitol. Strain YK193 and its isogenic wild-type derivative (containing pFL44-SLT2-HA) were grown in synthetic complete (SC) minimal medium lacking uracil. Tunicamycin (Sigma) was dissolved at 5 mg/ml in 75% methanol and used at a final concentration of 2.5 μg/ml.
TABLE 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| W303 | matα ade2-1 leu2-3,112 ura3-1 his3-11,15 trp1-1 can1-100 ssd1-1 | 17 |
| 312 | matα ade2-1 leu2-3,112 ura3-1 trp1-1 can1-100 sly1-1 | 22 |
| 169ts | matα ade2-1 leu2-3,112 ura3-1 his3-11,15 can1-100 ssd1-1 ypt6-1 | 17 |
| 1783 | mata leu2-3,112 ura3-52 trp1-1 his4 | 13 |
| DL252 | mata leu2-3,112 ura3-52 trp1-1 his4 bck1Δ::URA3 | 13 |
| AC2c9 | MATα ura3-52 his3-11,15 leu2-3,112 trp1-1 met8-1 tfc4Δ::LEU2 pRS313-tfc4-9 | Lab stock |
| KM011 | MATa his3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 rap1::LEU2 sly1-1 pHIS3CEN6-RAP1 | 20 |
| KM013 | MATa his3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 rap1::LEU2 sly1-1 pHIS3CEN6-rap1-17 | 20 |
| KM014 | MATa his3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 rap1::LEU2 SLY1 pHIS3CEN6-RAP1 | 20 |
| KM016 | MATa his3-11,15 ade2-1 ura3-1 leu2-3,112 trp1-1 can1-100 rap1::LEU2 SLY1 pHIS3CEN6-rap1-17 | 20 |
| MN51 | MATa his4 trp1-1 ura3 SEC1 PKC1 | 22 |
| MN52 | MATa his4 trp1-1 ura3 sec1-1 PKC1 | 22 |
| MN53 | MATα his4 trp1-1 ura3 SEC1 pkc1Δ::LEU2 | 22 |
| MN54 | MATα his4 trp1-1 ura3 sec1-1 pkc1Δ::LEU2 | 22 |
| ALHWT | MATa leu2 his3 ura3 trp1 ade8 | 28 |
| ALH715 | MATa leu2 his3 ura3 trp1 ade8 wsc1::ADE8 wsc3::TRP1 | 28 |
| ALH718 | MATa leu2 his3 ura3 trp1 ade8 wsc1::ADE8 wsc2::URA3 | 28 |
| Y783WT | mata ura3-52 lys2-801 ade2-101 his3-Δ200 leu2-Δ98 slt2-Δ10::LEU2 pFL44-SLT2-HA | 19 |
| YK193 | mata ura3-52 lys2-801 ade2-101 his3-Δ200 leu2-Δ98 slt2-Δ10::LEU2 pFL44-slt2-HA K54R | 19 |
Northern analysis and quantitation of RNA levels.
Total RNA was extracted by glass-bead disruption in the presence of hot phenol (23). For Northern analysis of tRNA precursors, RNA (20 μg) was resolved on 10% polyacrylamide–8.3 M urea gels in Tris-borate-EDTA (TBE) buffer. After electrophoresis, gels were soaked for 10 min in 0.5× TBE–1 μg of ethidium bromide/ml prior to digital photography (Alpha Innotech). The RNA was then electrophoretically transferred (in 0.5× TBE at 100 V for 2 h at 4°C) to Nytran Plus membranes (Schleicher & Schuell) using a Bio-Rad Transfer apparatus. After transfer, membranes were dried for 10 min and the RNA was cross-linked by UV irradiation (0.2 J/cm). For Northern analysis of mRNAs, total RNA (10 μg) was electrophoresed on 1.5% agarose gels containing 6% formaldehyde (21). Capillary transfer of the RNA to Nytran Plus membranes was performed using 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and was followed by UV cross-linking as described above. [32P]-labeled oligonucleotide probes were prepared as described previously (26) for detecting all tRNA precursors, TCM1 and CYH2 mRNAs, U4 snRNA, and U3 small nucleolar RNA (snRNA) (probe sequences are available upon request). KAR2 and ACT1 mRNA probes were prepared by random priming and runoff transcription, respectively (22). Hybridization and washing of blots probed with [32P]-labeled oligonucleotides were performed at 37°C (except for TCM1 and CYH2 [42°C]) as described previously (26). In addition, two 30-min posthybridization washes were performed in 2× SSC–0.1% sodium dodecyl sulfate (SDS). Random-primed DNA and antisense RNA probes were used as described by Mizuta and Warner (21). Hybridization was detected using phosphor storage plates and a phosphorimager (Molecular Dynamics). Signal intensities were quantified using ImageQuant software (Molecular Dynamics). Briefly, lines one lane wide were analyzed using a peak finder to determine the area under each curve. These data were normalized for variations in loading using mature tRNA levels quantified by a peak finder from digital images of ethidium bromide-stained gels. Normalized signal intensities were expressed relative to the values obtained at the permissive temperature or prior to the addition of tunicamycin.
In vivo labeling.
Log-phase cultures (A600, 0.4 to 0.7) were grown in low-phosphate YPD at 23°C and shifted to 37°C. At various times before and after the temperature shift, 1 ml of cells was removed to tubes containing 150 μCi of carrier-free [32P]orthophosphate for a further 5-min incubation. Cells were harvested rapidly and frozen prior to the preparation of total RNA (23). RNA (5 μg) was resolved on 10% denaturing polyacrylamide gels (see above) which were fixed, dried, and exposed to phosphor storage plates.
Western analysis.
Whole-cell lysates were prepared by glass-bead breakage into radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors (26). Equal amounts of the extracts (normalized by cell number) were analyzed by Western blotting (26) using a rabbit polyclonal antibody specific for the doubly phosphorylated tripeptide, phosphothreonine-glutamic acid-phosphotyrosine, found in activated Slt2 (New England Biolabs) (28). The product specifications report no reactivity with as much as 4 μg of unphosphorylated MAP kinase. Antibody-antigen complexes were detected using the recommended horseradish peroxidase-conjugated secondary antibody and enhanced chemiluminescence (Amersham). Antibodies to TATA-binding protein (TBP) were used as a loading control (26).
RESULTS
Inhibition of Pol III gene transcription in secretion-blocked cells.
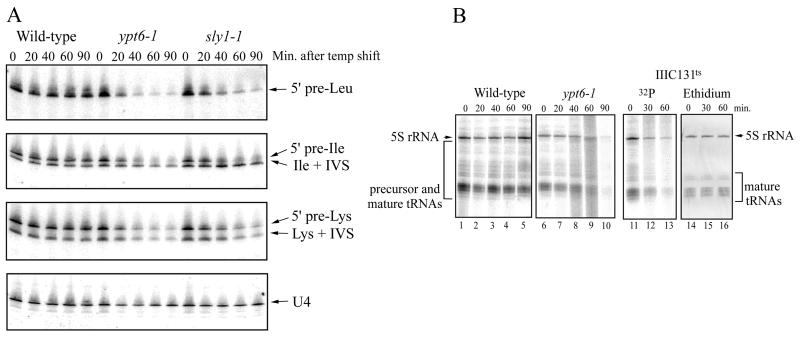
To determine whether the transcription of tRNA genes exhibit a dependence on the secretory pathway similar to that seen for rDNA and RP genes, Northern blots of total RNA from a wild-type strain and two temperature-sensitive strains with secretory mutations (ypt6-1 and sly1-1) were hybridized with 5′ flank- or intron-specific precursor tRNA probes. These probes detect very short lived tRNA precursors (1), and the resulting hybridization signals provide a reliable measure of Pol III transcription on these genes (4, 26). At the nonpermissive temperature, the ypt6-1 and sly1-1 mutations cause repression of rDNA and RP gene transcription (18, 21). YPT6 encodes a homolog of the human GTPase Rab6 and functions in yeast in ER-to-Golgi complex transport or cis-to-medial-Golgi complex transport. The SLY1 gene product is essential for protein and vesicle trafficking between the ER and the Golgi complex (12). The two mutant strains and a congenic wild-type strain were grown to mid-log phase at 23°C and then shifted to 37°C, the nonpermissive temperature. At various times before and after the temperature shift, cells were harvested and frozen rapidly to preserve the tRNA precursors.
In wild-type cells, the levels of 5′ flank- or intron-containing precursors for three tRNA genes were relatively unaffected at 37°C (Fig. 1A). However, in both secretory mutant strains, the same tRNA precursors were progressively and substantially reduced. By 90 min, most of the tRNA precursors had decreased to 5 to 10% of the level prior to the temperature shift. These changes occurred in the absence of changes in the steady-state levels of small RNAs including 5.8S rRNA, 5S rRNA, and mature tRNAs (as determined by quantitative image analysis of ethidium bromide-stained gels [data not shown]) and the Pol II-transcribed U4 snRNA (Fig. 1A). As reflected by the precursor tRNA levels, the reduction in Pol III transcription was very rapid; 50% inhibition of precursor tRNA synthesis was achieved in about 20 min (for an example, see Fig. 4B). Similar results have been obtained for other tRNA genes and are presumably representative of the entire tRNA gene family. Thus, as for the transcription of rDNA and RP genes by Pols I and II, respectively, continued transcription of tRNA genes by Pol III requires a functional secretory pathway.
FIG. 1.
Inhibition of Pol III transcription in secretion-blocked cells. (A) Northern blots of RNA (20 μg/lane) from a wild-type strain (W303α) and two temperature-sensitive secretory mutant strains (containing a ypt6-1 or sly1-1 mutation) are shown before, and at various times after, a shift from 23°C to the nonpermissive temperature of 37°C. Blots were probed sequentially with tRNA precursor-specific oligonucleotides and a U4 snRNA control oligonucleotide. The tRNALeu probe detects only 5′ flank-containing precursors, whereas the tRNAIle and tRNALys probes detect intron-containing precursors. Data were quantified using a phosphorimager and ImageQuant software as described in Materials and Methods. (B) In vivo [32P] labeling of RNA in secretory and TFIIIC mutant strains. Strains growing in low-phosphate medium were pulse-labeled for 5 min with [32P]orthophosphate at 23°C and at the indicated times after the shift to 37°C. Lanes 1 to 5 and 6 to 10 show the RNAs labeled in wild-type (W303α) and ypt6-1 strains, respectively. Loading of equal amounts of RNA (5 μg) in each lane was confirmed by staining with ethidium bromide prior to drying of the gel. The RNAs labeled in a temperature-sensitive TFIIIC mutant strain, AC2c9 (lanes 11 to 13), are compared with the negative image of the ethidium bromide-stained gel (lanes 14 to 16).
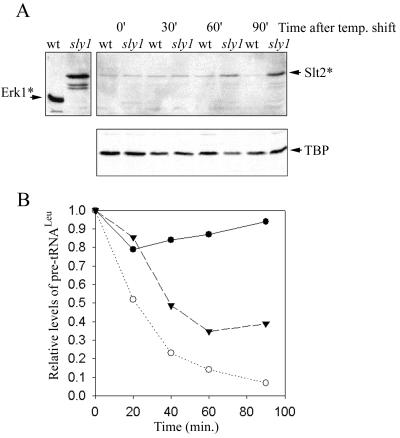
FIG. 4.
Slt2 activation and Pol III transcriptional repression in a sly1-1 strain. Wild-type (W303α) and sly1-1 (312) strains were grown at 23°C, shifted to 33°C, and harvested at various times to prepare total denaturing cell lysates for Western blotting or RNA for Northern analysis. (A) A Western blot was probed sequentially with a phosphospecific MAP kinase antibody that recognizes activated yeast Slt2 (Slt2∗) (28) and antibodies to TBP, which provide a control for protein loading. The panel at the upper left shows the signal for activated ERK1 (ERK1∗) and activated Slt2 (from an extract of 39°C heat-shocked yeast cells). (B) Northern analysis of pre-tRNALeu levels in the sly1-1 strain, shifted from 23 to 33°C (▾) or to 37°C (○) and from a wild-type strain (W303α) shifted from 23 to 37°C (●). The data at 37°C were quantified from Fig. 1A.
In contrast to tRNA precursors, the other major Pol III transcription product in yeast, 5S rRNA, has a long half-life and undergoes minimal processing. Therefore, the transcription of 5S rRNA genes was monitored by pulse-labeling with [32P]orthophosphate. Log-phase cells from a wild-type strain, a secretory mutant strain (the ypt6-1 mutation), and a strain containing a temperature sensitive mutation in the second-largest subunit of TFIIIC were incubated for 5 min with [32P]orthophosphate at the permissive temperature. This results in heavy labeling of bands corresponding to 5S rRNA and mature tRNA (Fig. 1B; compare lanes 1, 6, and 11 to lane 14). In the TFIIIC mutant strain, decreased Pol III transcription following a shift to the nonpermissive temperature reduces the labeling of these and other bands (primarily tRNA precursors [Fig. 1B, lanes 11 to 13]). In the wild-type strain, no reduction in RNA labeling is seen up to 90 min after the temperature shift (Fig. 1B, lanes 1 to 5). Conversely, in the ypt6-1 strain, labeling of 5S rRNA is severely reduced after 60 and 90 min at the nonpermissive temperature (Fig. 1B, lanes 9 and 10). Reduced labeling of tRNA precursors and mature tRNA is also seen at these times. Thus, the secretory pathway impacts the transcription of all the RNA components of the ribosome. Moreover, as demonstrated by both Northern analysis and pulse-labeling, the consequences of interrupting the secretory pathway go beyond transcriptional repression of ribosomal components to include ribosomal substrates, namely, tRNAs.
Coordinate repression of tRNA and RP gene transcription.
The use of temperature-sensitive strains to study cellular processes can sometimes lead to unanticipated synergistic interactions resulting from the simultaneous change in temperature and loss of protein function. To avoid this possibility, Pol III transcription was examined in cells treated with tunicamycin, which interferes with the secretory pathway by inhibiting the glycosylation of proteins in the ER. Addition of tunicamycin to an early-log-phase culture of a wild-type strain inhibited Pol III transcription, as demonstrated by a reduction in the level of leucine precursor tRNA (Fig. 2A). This effect of tunicamycin was delayed compared to that in secretory mutant strains. However, the gradual induction of KAR2 (a marker for the unfolded protein response pathway), which peaks at 90 min, suggests that the delay in repression reflects the slower kinetics of the drug-induced secretory block relative to secretory pathway mutations (14). Despite the delay in achieving inhibition of the secretory pathway, image analysis revealed that a 50% reduction in precursor tRNALeu synthesis occurred within one cell division (Fig. 2B). Four hours after tunicamycin addition, the level of precursor tRNALeu was 10 to 15% of the level prior to addition of the drug. A similar end point was reached with conditional secretory pathway mutations (Fig. 1A and 4B; also data not shown). Thus, the dependence of Pol III transcription on a functional secretory pathway is demonstrated by two independent methods: conditional mutations in the pathway and an inhibitor of secretory protein processing.
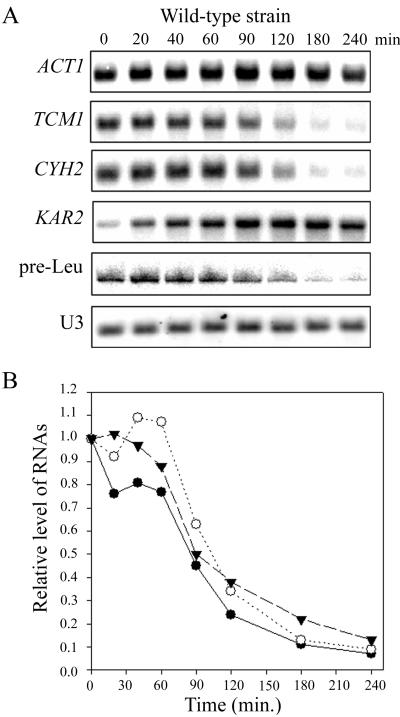
FIG. 2.
Coordinate repression of RP and tRNA gene transcription in tunicamycin-treated cells. Tunicamycin was added to a mid-log-phase culture of a wild-type strain (W303α) growing at 30°C. Cells were harvested at the indicated times for the preparation of total RNA. (A) Northern blots were hybridized to probes specific for actin (ACT1), ribosomal protein (TCM1 and CYH2), KAR2, 5′ flanked pre-tRNALeu (pre-Leu), or U3 snoRNA. (B) Quantitation of RP mRNA and pre-tRNALeu levels. Digital images of the blots from panel A were quantified using the ImageQuant line peak finder. Integrated peak areas were normalized for loading using a digital image of the ethidium bromide-stained gel taken prior to electrophoretic transfer. ●, TCM1; ○, CYH2; ▾, pre-tRNALeu.
The effect of tunicamycin was also examined for two RP mRNAs (TCM1 and CYH2), actin mRNA (ACT1), and U3 snoRNA (Fig. 2). In agreement with previous observations, RP mRNA levels decreased substantially in the presence of tunicamycin, with little or no change in the levels of actin mRNA or U3 snoRNA during the time course (20, 22). Given the naturally short half-life of RP mRNAs in wild-type cells (<10 min at 37°C) and evidence that RP mRNA stability does not change with inhibition of the secretory pathway, the decrease in RP mRNA levels can be attributed solely to repression of their transcription (17). Quantitative analysis of the Northern blots reveals virtually identical kinetics for the inhibition of the two RP mRNAs and the tRNALeu precursor (Fig. 2B), suggesting that the transcription of RP and tRNA genes is coordinately repressed in secretion-defective cells and that a common signaling pathway may mediate this response.
Components of the cell integrity pathway are required for repression of ribosome and tRNA synthesis.
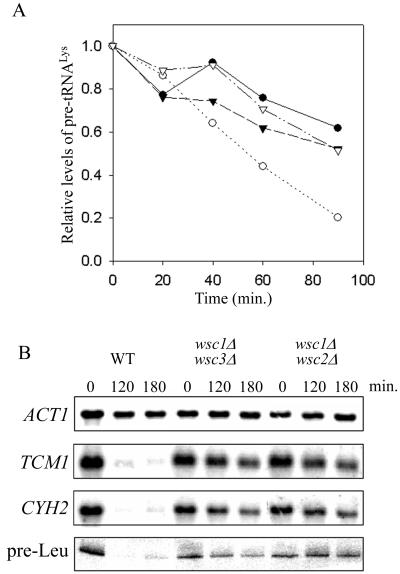
Recent studies of the regulatory circuit connecting the secretory pathway and ribosome synthesis have revealed that transcriptional repression of rDNA and RP genes is blocked in a PKC deletion (pkc1Δ) strain (22). This finding, together with the ability of a membrane-stretching agent, chlorpromazine, to repress RP genes, suggested a role for the cell integrity pathway in signaling this response (22). The cell integrity pathway mediates cell cycle-regulated cell wall synthesis and responds to a variety of environmental stimuli, including elevated temperature (37 to 39°C), hypoosmotic shock, and pheromone treatment (8). To determine whether this pathway might also signal transcriptional repression in secretion-blocked cells, we first sought to extend the results obtained with the PKC deletion to the Pol III system. Congenic strains containing all combinations of the SEC1/sec1-1 and PKC1/pkc1Δ genes were grown to mid-log phase at 23°C in YPD containing 1 M sorbitol (sorbitol suppresses the cell lysis phenotype of pkc1Δ strains but not the sec1-1 secretory defect [16, 22]). Analysis of lysine precursor tRNAs after a shift to the nonpermissive temperature shows that their levels decrease in the sec1-1 strain, reaching 20% of the level prior to the temperature shift after 90 min (Fig. 3A). However, in the sec1-1 pkc1Δ strain, the decrease was considerably less pronounced. Indeed, lysine precursor tRNA levels in the double-mutant strain were indistinguishable from those in the wild-type and pkc1Δ strains, declining only to about 60% of the level prior to the temperature shift after 90 min. Similar results were obtained using a leucine precursor tRNA probe. The results for these Pol III transcripts parallel the findings for the large rRNAs and RP mRNAs in the same strains (22). Together, these data suggest that PKC mediates transcriptional repression of the protein-synthesizing machinery in response to a defect in the secretory pathway.
FIG. 3.
Deletion of Wsc proteins or PKC1 abrogates repression of RP and tRNA genes due to a secretory defect. (A) Congenic strains (MN51 through MN54, differing at the SEC1 and PKC1 loci) were grown at 23°C in YPD plus 1 M sorbitol and shifted to 37°C. RNAs (20 μg) from cells harvested before and at various times after the temperature shift were analyzed by Northern blotting using an intron-specific lysine tRNA probe. The plotted data represent the sum of the areas determined by peak integration for both intron-containing tRNALys precursors (see, e.g., Fig. 1A). ●, PKC1 SEC1; ○, PKC1 sec1-1; ▾, pkc1Δ SEC1; ▿, pkc1Δ sec1-1. (B) Isogenic strains (ALHWT, ALH715, and ALH718) differing at the WSC loci were grown at 30°C in YPD plus 1 M sorbitol to mid-log phase before the addition of tunicamycin. RNAs from cells harvested at the indicated times were analyzed by Northern blotting using actin and RP mRNA probes and the 5′ flank pre-tRNALeu probe (as in Fig. 2A).
The farthest-upstream components of the cell integrity pathway that have been identified to date are the Wsc proteins. The four Wsc proteins and the related Mid2 gene product are putative cell surface sensors based on their localization, topology, and glycosylation (7, 25, 28). Although the functions of the Wsc proteins in cell integrity signaling are partially redundant, the phenotype of a wsc1Δ mutation is more severe than that for deletions of the other family members and results in an osmotic remedial cell lysis phenotype at 37 to 39°C. We therefore examined the effect of tunicamycin on the transcription of RP and tRNA genes in a set of isogenic strains with deletions of WSC1 and either WSC2 or WSC3. As shown by the levels of RP mRNAs and pre-tRNALeu, transcriptional repression of the corresponding genes in strain ALHWT was complete 2 h after the addition of tunicamycin. This indicates that strain ALHWT is somewhat more sensitive to tunicamycin than W303α (compare Fig. 2 and 3B). However, for both the wsc1Δ wsc2Δ and wsc1Δ wsc3Δ strains, little or no decrease in the levels of RP mRNAs or precursor tRNA was seen up to 3 h after the addition of the drug (Fig. 3B). Thus, deletion of these Wsc proteins severely impairs transcriptional repression in secretion-blocked cells. These results demonstrate that the upstream components of the cell integrity pathway, specifically the Wsc proteins and PKC, are part of a common signaling pathway mediating transcriptional repression of ribosome and tRNA synthesis.
A novel signaling pathway downstream of PKC.
The cell integrity pathway is the only PKC pathway that has been defined in yeast (8). Signaling through this pathway involves PKC activation of a specific downstream MAP kinase cascade comprising the MEK kinase Bck1, a pair of redundant MEKs (Mkk1 and Mkk2), and the MAP kinase Slt2 (Mpk1). Activation of Slt2 by the MEKs involves dual phosphorylation at a unique … TEY … sequence in the kinase domain. We therefore sought to confirm that Slt2 is activated in cells with blocks in the secretory pathway by Western blotting using a phosphospecific antibody. Wild-type and sly1-1 cells were grown at 23°C to mid-log phase and then shifted to 33°C. This temperature is nonpermissive for sly1-1 (20) and maintains a low background level of activated Slt2 in a wild-type strain (Fig. 4A). Cells collected at various times (up to 90 min after the temperature shift) were lysed under denaturing conditions, and equal amounts of the extracts were analyzed by Western blotting (26). An increase in the amount of doubly phosphorylated Slt2 was apparent in the mutant strain, but not in the wild-type strain, after 60 and 90 min at the nonpermissive temperature (Fig. 4A). Since activation of the cell integrity pathway does not affect the steady-state level of Slt2 protein (8, 13, 28), we conclude that inhibition of the secretory pathway leads to activation of the Slt2 kinase. In a parallel experiment, RNAs from cells grown under the same conditions were analyzed by Northern blotting using a leucine precursor tRNA-specific probe. This allowed a quantitative comparison of precursor tRNALeu synthesis in the sly1-1 strain at 33 and 37°C (Fig. 4B) and a qualitative comparison of Pol III transcriptional repression and Slt2 activation (compare Fig. 4A with Fig. 4B). The data show that (i) the kinetics of repression of the pre-Leu tRNA is slower, and the end point reached is higher, in the sly1-1 strain grown at 33 versus 37°C and (ii) pre-Leu tRNA synthesis is fully repressed at times when activated Slt2 is readily detected. An equivalent correlation between the activation of Slt2 and the repression of RP and tRNA genes was obtained with tunicamycin-treated wild-type cells (data not shown). Although Slt2 is clearly activated in secretion-blocked cells, these experiments do not address whether Slt2 activation is required for or is coincident with transcriptional repression.
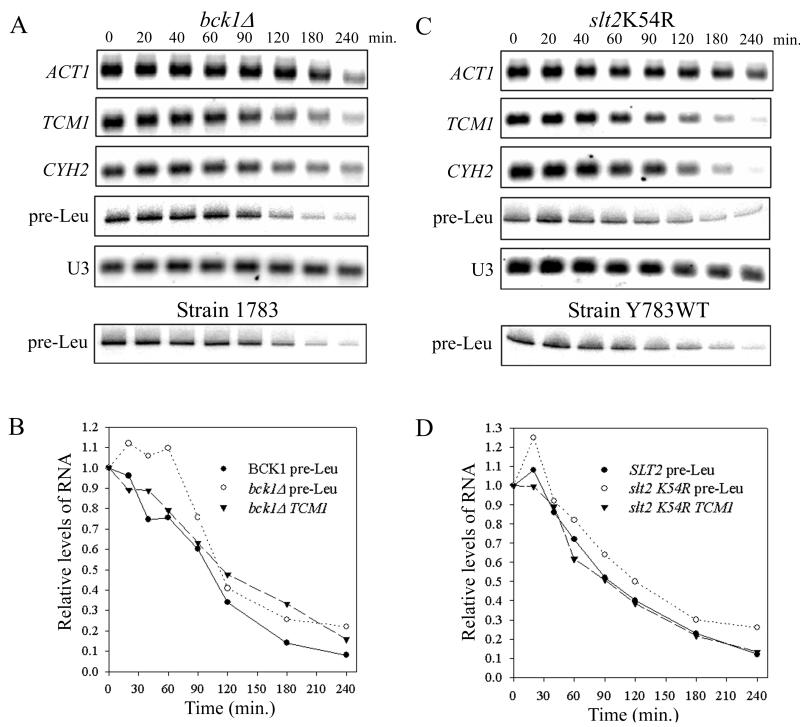
To test directly whether components of the cell integrity pathway MAP kinase cascade are necessary for transcriptional repression in response to a secretory defect, we examined the effects of tunicamycin on RP mRNA and tRNA levels in a bck1Δ strain and in a strain containing a catalytically inactive SLT2 allele, slt2 K54R (36). Neither mutation in the MAP kinase cascade affected transcriptional repression of RP or tRNA genes (Fig. 5). Quantitative analysis of the blots revealed the same kinetics of repression for pre-Leu tRNA and TCM1 mRNA in the mutant strains as for pre-Leu tRNA in isogenic wild-type strains (Fig. 5B and D) and in W303α (Fig. 2B). Moreover, repression occurs without significant effects on actin mRNA (up to 180 min) or U3 snoRNA (Fig. 5) and without changes in the steady-state levels of the abundant small RNAs (5.8S, 5S, or mature tRNA [data not shown]). Since Slt2 is dependent on PKC and downstream components of the cell integrity pathway for activation (8), and the known biochemical consequences of activating the cell integrity pathway (e.g., phosphorylation of Swi6 and Rlm1) are dependent on Slt2 (19, 31), our data suggest that the cell integrity pathway is bifurcated downstream of PKC and that a novel effector branch of the pathway mediates transcriptional repression in secretion-blocked cells.
FIG. 5.
Inactivation of the cell integrity pathway MAP kinase cascade does not affect repression of RP or tRNA genes in response to tunicamycin. (A) A strain with the MEK kinase BCK1 deleted (DL252) and an isogenic control (1783) were grown at 25°C in YPD to mid-log phase before the addition of tunicamycin. Cells were harvested at various times thereafter to prepare RNA for Northern analysis. The same probes were used as in Fig. 2A. (B) Quantitation of pre-Leu tRNA and TCM1 mRNA from panel A (for details, see Materials and Methods). (C) Strain YK193 (19), which contains a catalytically inactive SLT2 allele (slt2 K54R [36]), and its isogenic wild-type control (Y783WT) were grown and analyzed as described for panel A except that SC-uracil medium was used. (D) Quantitation of pre-Leu tRNA and TCM1 mRNA from panel C.
rap1-17 affects a Pol II-specific branch of a signaling pathway monitoring secretory function.
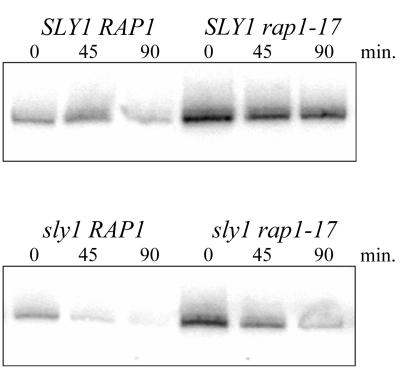
In previous work (20), the repression of RP gene transcription resulting from a defect in the secretory pathway was attenuated by a mutation in RAP1 (rap1-17) that deletes its silencing domain. This effect was observed for RP genes that contain Rap1-binding sites in their promoter (the vast majority) as well as for RP genes that do not (20). These findings suggested that Rap1 represses the transcription of RP genes without necessarily binding to their promoters. Although RAP1 is not known to play a role in Pol III transcription, the preceding observations prompted us to test whether rap1-17 could prevent the repression of Pol III transcription. Congenic strains with all combinations of the RAP1/rap1-17 and SLY1/sly1-1 genes were grown at 23°C and then shifted to 36°C to obtain cells for RNA preparation (20). Subsequent Northern analysis using a leucine precursor tRNA-specific probe showed no effect of the rap1-17 mutation on transcriptional repression by the sly1-1 secretory block (Fig. 6). Leucine precursor tRNA synthesis decreased in both the sly1-1 RAP1 and sly1-1 rap1-17 strains, such that 90 min after the temperature shift, the amount of pre-tRNALeu was only 20 to 25% of the level prior to the shift. In contrast, the amount of pre-tRNALeu in the SLY1 rap1-17 strain at this time was still greater than 72% of that in the 23°C control. Surprisingly, the rap1-17 mutation increased the level of leucine tRNA precursors about threefold (at 23°C) compared to those in the RAP1 strains, regardless of their secretory competence. This result reveals a previously unknown negative effect of RAP1 on Pol III transcription. It is clear, however, that RAP1 does not mediate signaling to the Pol III transcription machinery in response to the sly1-1 secretory defect. In light of results obtained previously for RP gene transcription in these strains (20, 22), the present findings indicate that the signaling pathway mediating the secretory defect is bifurcated downstream of PKC to produce Pol II- and Pol III-specific effector branches (Fig. 7).
FIG. 6.
RAP1 does not mediate repression of Pol III transcription in secretion-blocked cells. Congenic strains (KM011, KM013, KM014, and KM016) containing all pairwise combinations of SLY1 and sly1-1 with RAP1 and rap1-17 were grown at 23°C and shifted to 36°C. RNA (20 μg) from cells harvested before and after the temperature shift was analyzed by Northern blotting using the 5′ flank pre-tRNALeu probe. The quantified data are described in the text.
FIG. 7.
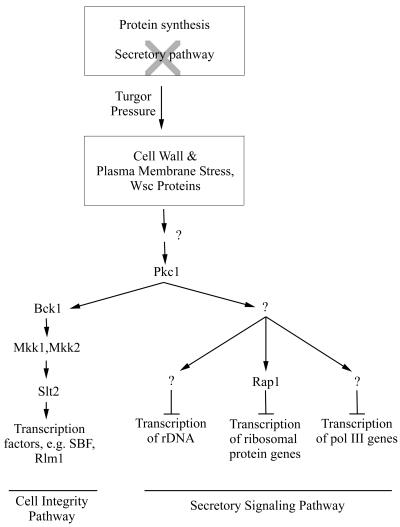
Model of the secretory signaling pathway mediating transcriptional repression of ribosome and tRNA synthesis. Turgor pressure generated by ongoing protein synthesis and a reduction or blockage in the delivery of secretory vesicles to the plasma membrane is proposed to activate the pathway. The guanine nucleotide exchange factor Rom2 and the Rho1 GTPase are possible upstream components of the secretory signaling pathway (between Wsc and Pkc1) based on studies of actin depolarization (5). The number of steps downstream of Pkc1 is unknown. Arrows indicate activation; bars indicate repression.
DISCUSSION
The energetic cost of producing the machinery for protein synthesis.
The work reported here, together with earlier studies (30), shows that interruption of the secretory pathway in yeast leads to transcriptional repression of the large rRNAs by Pol I, RP genes by Pol II, and 5S rRNA and tRNA genes by Pol III. Thus, all of the components of the ribosome as well as its tRNA substrates are subject to this repression. Repression of transcription under these conditions is not genome-wide, however, as the levels of several glycolytic mRNAs (PGK1, PYK1, and ENO1) and actin mRNA (which have half-lives of less than 20 min [10]) do not decrease in secretion-defective cells (17, 18, 20–22). Indeed, the levels of some mRNAs (e.g., KAR2) increase significantly as interruption of the secretory pathway leads to activation of the unfolded protein response pathway (Fig. 2) (14, 17, 20). In rapidly growing yeast, about 60% of the total nuclear transcription is dedicated to Pol I synthesis of the large rRNAs (30). Another 14% is carried out by Pol III in the synthesis of 5S rRNA and tRNAs (determined from the pulse-labeling experiment [Fig. 2]). Of the remaining ∼26% of nuclear transcription that is directed by Pol II, around 50% of the initiation events take place on RP genes (17, 30). Thus, we estimate that more than 85% of the total nuclear transcription can be repressed when the secretory pathway is blocked. Given the enormous amount of metabolic energy consumed with this level of transcriptional activity, it makes good biological sense for cells such as yeast to shut down the production of ribosomes and ribosomal substrates when they are unable to expand their plasma membranes. The conservation of metabolic energy achieved by this repression may help to promote cell survival under adverse conditions.
A branch of the cell integrity pathway mediates transcriptional repression.
The four Wsc proteins and the related Mid2 protein are thought to sense and signal changes in cell wall integrity resulting from heat stress, hypotonic stress, and pheromone treatment (25, 28). The Wsc proteins signal PKC either directly or indirectly (via Rom2 and Rho1 (5, 8). PKC in turn relays the signals from the Wsc proteins to the downstream MAP kinase cascade, ending in the activation of Slt2, in order to induce the expression of cell wall synthesis genes for remodeling or repair of the cell wall (8, 28). wsc1Δ and pkc1Δ mutants have an osmotic remedial cell lysis phenotype at or above 25°C that is characteristic of cell integrity pathway deletion mutants.
The results presented above demonstrate that wsc1Δ and pkc1Δ mutants are also defective in the repression of ribosome and tRNA synthesis when the secretory pathway is blocked (Fig. 3). Further evidence for a role of the Wsc proteins and PKC in signaling a secretory defect is our finding that Slt2 is activated in sly1-1 and tunicamycin-treated cells (Fig. 4A and data not shown). Slt2 activation is tightly linked to the cell integrity pathway (8): it is abolished in pkc1Δ strains (reference 13 and our unpublished data) and is largely, although not completely, defective in a wsc1 strain (residual activation in this case reflects the partially redundant function of the Wsc proteins) (28). Thus, the initial stages of the response to a secretory defect or to cell wall stress seem to be the same. However, the response to a secretory defect does not lead primarily to the activation of transcription, but to its repression, specifically toward the transcription of genes comprising the protein-synthetic machinery. Moreover, components of the cell integrity pathway MAP kinase cascade (Bck1 and Slt2) are not required for this response (Fig. 5). These observations suggest that the cell integrity pathway is bifurcated downstream of PKC and that a novel effector branch of the pathway mediates transcriptional repression (Fig. 7). Bifurcation of the signaling pathway downstream of PKC has been suggested previously, based on genetic studies (6, 9). However, the functions of the novel pathway and its components have remained obscure. We suggest that this branch of the cell integrity pathway is responsible for the repression of genes that make up the protein-synthesizing machinery. Activation of this pathway frees substantial resources needed by a cell under stress and reduces the increased stress caused by ongoing protein synthesis under conditions where the cell is unable to synthesize either plasma membrane or cell wall.
Independent evidence for a bifurcated signaling pathway below PKC has come from the observation that the depolarization of both actin and the integral plasma membrane protein β-1,3-glucan synthase upon heat shock requires Wsc1 and Rom2 (the guanine nucleotide exchange factor of Rho1) and can be induced in strains expressing hyperactivated (toxic) alleles of RHO1 and PKC1 (5). As in our experiments, BCK1 and SLT2 are not required for the depolarization response. These findings raise the possibility that a common PKC effector pathway mediates both actin depolarization and transcriptional repression of ribosome synthesis. However, there are differences in the signaling of these responses: conditions which cause actin depolarization (e.g., incubation at 37°C for 35 min) (5) do not result in PKC-dependent repression of ribosome or tRNA synthesis (Fig. 1) (22). Thus, there may be three functionally distinct branches of the PKC pathway. In any event, further branching of the PKC effector pathway mediating transcriptional repression is required to enable signaling to the Pol I, the Pol II, and the Pol III transcription machinery (Fig. 7). This is indicated by the ability of a silencing defective allele of RAP1 (rap1-17) to block repression of RP genes but not tRNA genes (Fig. 6) or rDNA (data not shown) upon inactivation of the secretory pathway.
Implications for the coordination of ribosome and tRNA synthesis with cell growth.
The finding that transcriptional repression in secretion-defective cells requires members of the Wsc family of putative plasma membrane sensors and continued protein synthesis (21), together with the ability of a membrane stretch agent, chlorpromazine, to induce repression of RP genes (22), suggests that activation of the signaling pathway in secretion-blocked cells is initiated by the elevated internal turgor pressure (Fig. 7). Presumably, this condition is extreme in most of our experiments due to the complete interruption of the secretory pathway. However, analysis of a sly1-1 mutant at 33 and 37°C shows that the kinetics and magnitude of repression of a tRNA gene are clearly different (Fig. 4B). Similarly, secretory mutants that are variably leaky lead to different levels of repression of rRNA and RP gene transcription (21). Thus, the severity of the secretion defect influences the level of transcriptional repression. It seems likely, therefore, that turgor pressure, and its effect on transcription, provides a means for balancing the production of the protein-synthesizing machinery with the delivery of secretory vesicles to the plasma membrane.
ACKNOWLEDGMENTS
We thank Roymarie Ballester, David Levin, and Mike Snyder for providing yeast strains and Pierre-Alain Delley and Mike Hall for communicating their results prior to publication.
This work was supported by NIH grants to I.M.W. (GM42728) and J.R.W. (GM25532) and by an award from the Irma T. Hirschl Trust to I.M.W.
REFERENCES
- 1.Blatt B, Feldmann H. Characterization of precursors to tRNA in yeast. FEBS Lett. 1973;37:129–133. doi: 10.1016/0014-5793(73)80441-7. [DOI] [PubMed] [Google Scholar]
- 2.Cavanaugh A H, Hempel W M, Taylor L J, Rogalsky V, Todorov G, Rothblum L I. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;374:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- 3.Chu W M, Wang Z, Roeder R G, Schmid C W. RNA polymerase III transcription repressed by Rb through its interactions with TFIIIB and TFIIIC2. J Biol Chem. 1997;272:14755–14761. doi: 10.1074/jbc.272.23.14755. [DOI] [PubMed] [Google Scholar]
- 4.Cormack B P, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 5.Delley P-A, Hall M N. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J Cell Biol. 1999;147:163–174. doi: 10.1083/jcb.147.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Errede B, Levin D E. A conserved kinase cascade for MAP kinase activation in yeast. Curr Opin Cell Biol. 1993;5:254–260. doi: 10.1016/0955-0674(93)90112-4. [DOI] [PubMed] [Google Scholar]
- 7.Gray J V, Ogas J P, Kamada Y, Stone M, Levin D E, Herskowitz I. A role for the Pkc1 MAP kinase pathway of Saccharomyces cerevisiae in bud emergence and identification of a putative upstream regulator. EMBO J. 1997;16:4924–4937. doi: 10.1093/emboj/16.16.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gustin M C, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helliwell S B, Schmidt A, Ohya Y, Hall M N. The Rho1 effector Pkc1, but not Bni1, mediates signalling from Tor2 to the actin cytoskeleton. Curr Biol. 1998;8:1211–1214. doi: 10.1016/s0960-9822(07)00511-8. [DOI] [PubMed] [Google Scholar]
- 10.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 11.Ju Q, Warner J R. Ribosome synthesis during the growth cycle of Saccharomyces cerevisiae. Yeast. 1994;10:151–157. doi: 10.1002/yea.320100203. [DOI] [PubMed] [Google Scholar]
- 12.Kaiser C A, Gimeno R E, Shaywitz D A. Protein secretion, membrane biogenesis and endocytosis. In: Broach J R, Pringle J R, Jones E W, editors. The molecular biology and cellular biology of the yeast Saccharomyces cerevisiae. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 91–228. [Google Scholar]
- 13.Kamada Y, Jung U S, Piotrowski J, Levin D E. The protein kinase C-activated MAP kinase pathway of Saccharomyces cerevisiae mediates a novel aspect of the heat shock response. Genes Dev. 1995;9:1559–1571. doi: 10.1101/gad.9.13.1559. [DOI] [PubMed] [Google Scholar]
- 14.Kohno K, Normington K, Sambrook J, Gething M J, Mori K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol Cell Biol. 1993;13:877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larminie C G, Cairns C A, Mital R, Martin K, Kouzarides T, Jackson S P, White R J. Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. EMBO J. 1997;16:2061–2071. doi: 10.1093/emboj/16.8.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levin D E, Bartlett-Heubusch E. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J Cell Biol. 1992;116:1221–1229. doi: 10.1083/jcb.116.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B, Nierras C R, Warner J R. Transcriptional elements involved in the repression of ribosomal protein synthesis. Mol Cell Biol. 1999;19:5393–5404. doi: 10.1128/mcb.19.8.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B, Warner J R. Mutation of the Rab6 homologue of Saccharomyces cerevisiae, YPT6, inhibits both early Golgi function and ribosome biosynthesis. J Biol Chem. 1996;271:16813–16819. doi: 10.1074/jbc.271.28.16813. [DOI] [PubMed] [Google Scholar]
- 19.Madden K, Sheu Y J, Baetz K, Andrews B, Snyder M. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science. 1997;275:1781–1784. doi: 10.1126/science.275.5307.1781. [DOI] [PubMed] [Google Scholar]
- 20.Mizuta K, Tsujii R, Warner J R, Nishiyama M. The C-terminal silencing domain of Rap1p is essential for the repression of ribosomal protein genes in response to a defect in the secretory pathway. Nucleic Acids Res. 1998;26:1063–1069. doi: 10.1093/nar/26.4.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizuta K, Warner J R. Continued functioning of the secretory pathway is essential for ribosome synthesis. Mol Cell Biol. 1994;14:2493–2502. doi: 10.1128/mcb.14.4.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nierras C R, Warner J R. Protein kinase C enables the regulatory circuit that connects membrane synthesis to ribosome synthesis in Saccharomyces cerevisiae. J Biol Chem. 1999;274:13235–13241. doi: 10.1074/jbc.274.19.13235. [DOI] [PubMed] [Google Scholar]
- 23.O'Connor J P, Peebles C L. In vivo pre-tRNA processing in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:425–439. doi: 10.1128/mcb.11.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Powers T, Walter P. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rajavel M, Philip B, Buehrer B M, Errede B, Levin D E. Mid2 is a putative sensor for cell integrity signaling in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:3969–3976. doi: 10.1128/mcb.19.6.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sethy-Coraci I, Moir R D, Lopez-de-Leon A, Willis I M. A differential response of wild type and mutant promoters to TFIIIB70 overexpression in vivo and in vitro. Nucleic Acids Res. 1998;26:2344–2352. doi: 10.1093/nar/26.10.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sutcliffe J E, Cairns C A, McLees A, Allison S J, Tosh K, White R J. RNA polymerase III transcription factor IIIB is a target for repression by pocket proteins p107 and p130. Mol Cell Biol. 1999;19:4255–4261. doi: 10.1128/mcb.19.6.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verna J, Lodder A, Lee K, Vagts A, Ballester R. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:13804–13809. doi: 10.1073/pnas.94.25.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voit R, Schafer K, Grummt I. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Mol Cell Biol. 1997;17:4230–4237. doi: 10.1128/mcb.17.8.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Warner J. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe Y, Takaesu G, Hagiwara M, Irie K, Matsumoto K. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol. 1997;17:2615–2623. doi: 10.1128/mcb.17.5.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White R J. Regulation of RNA polymerases I and III by the retinoblastoma protein: a mechanism for growth control? Trends Biochem Sci. 1997;22:77–80. doi: 10.1016/s0968-0004(96)10067-0. [DOI] [PubMed] [Google Scholar]
- 33.White R J. RNA polymerase III transcription. New York, N.Y: Springer-Verlag; 1998. [Google Scholar]
- 34.White R J, Trouche D, Martin K, Jackson S P, Kouzarides T. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature. 1996;382:88–90. doi: 10.1038/382088a0. [DOI] [PubMed] [Google Scholar]
- 35.Woolford J L J, Warner J. The ribosome and its synthesis. In: Broach J R, Pringle J R, Jones E W, editors. The molecular biology and cellular biology of the yeast Saccharomyces cerevisiae. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1991. pp. 587–626. [Google Scholar]
- 36.Zarzov P, Mazzoni C, Mann C. The SLT2 (MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J. 1996;15:83–91. [PMC free article] [PubMed] [Google Scholar]