Summary
Few validated protocols are available for large-scale collection, storage, and analysis of microbiome samples from the vagina, skin, and mouth. To prepare for a large-scale study on the female microbiome by remote self-sampling, we investigated the impact of sample collection, storage, and host DNA depletion on microbiome profiling. Vaginal, skin, and saliva samples were analyzed using 16S rRNA gene amplicon and metagenomic shotgun sequencing, and qPCR. Of the two tested storage buffers, the eNAT buffer could keep the microbial composition stable during various conditions. All three tested host DNA-depletion approaches showed a bias against Gram-negative taxa. However, using the HostZERO Microbial DNA and QIAamp DNA Microbiome kits, samples still clustered according to body site and not by depletion approach. Therefore, our study showed the effectiveness of these methods in depleting host DNA. Yet, a suitable approach is recommended for each habitat studied based on microbial composition.
Subject areas: Body substance sample, Microbiology, Omics
Graphical abstract

Highlights
-
•
Lysis buffer keeps the microbial composition stable during various storage conditions
-
•
Host DNA depletion introduces a larger bias toward Gram-negative taxa
-
•
The HostZERO Microbial DNA kit performed best in human DNA depletion for metagenomics
-
•
Body site-specific approach based on microbial composition is needed to minimize bias
Body substance sample; Microbiology; Omics
Introduction
The importance of microbial communities outside the gut is increasingly recognized, resulting in an increased number of studies on previously underexplored body sites such as the vagina, upper airways, and skin (HMP, 2019). To aim for high-resolution microbiome analysis, including taxonomic identification up to species or strain level and an accurate functional identification, metagenomic sequencing is preferred over 16S rRNA gene amplicon sequencing (Quince et al., 2017). The vagina, upper airways, and skin require tailored protocols and can be sampled via different approaches, of which the most commonly used and least invasive method is a swab. However, previous research has shown that various types of swabs can have an underestimated effect on, for example, the collection of specific host epithelial cells (Daley et al., 2006; Zasada et al., 2020). This represents challenges for metagenomic shotgun sequencing owing to the high proportion of human genome reads in relation to microbial reads (Marotz et al., 2018). These high numbers of human DNA can lead to a decreased sensitivity for microbial detection in samples and consequently increased costs for metagenomic shotgun sequencing. Host DNA depletion prior to DNA extraction is therefore preferable, especially for low bacterial biomass samples such as the skin with a high number of human cells. However, host DNA depletion should be evaluated carefully and separately per body site, owing to phenotypic and physiological differences of these mucocutaneous body sites that shape their microbial communities (Anderson et al., 2014; Byrd et al., 2018). For example, in the vagina, epithelial cells contain large cytoplasmic stores of glycogen as a substrate for Lactobacillus spp. that produce lactic acid and maintain a low vaginal pH of between 3.8 and 4.5 (Anderson et al., 2014). In the mouth and saliva, the importance of food and digestive/(post)prandial enzymes is also an important factor to consider (Tenovuo, 1998). The skin varies from dry to moist to oily and is influenced by sweat glands, hair follicles, and sebaceous glands with different microbes residing at different depths (Byrd et al., 2018).
Although several practical guidelines have been formulated for feasible high-quality cost-efficient gut microbiome research such as reviewed by Vandeputte et al. (2017), information on the influence of crucial factors such as storage conditions and microbial extraction methods for large-scale studies on vaginal, skin, and saliva microbiome samples is limited. An additional challenge is the application of metagenomic shotgun sequencing that is emerging as a highly promising approach because of the possibility of assessing bacterial, fungal, and viral taxonomy and function. However, it requires additional optimization for cost efficiency and reproducibility in larger trials. Based on previous studies (Costea et al., 2017; Hallmaier-Wacker et al., 2018; Quince et al., 2017), we identified two aspects that currently represent bottlenecks in microbiome research through metagenomic shotgun sequencing: the storage conditions and DNA extraction method. The goal was to optimize these methods for our large-scale Citizen Science project, Isala (www.isala.be/en/), using the approach depicted in Figure 1. First, investigating storage conditions is crucial since immediate processing of samples is not possible in a large-scale Citizen Science project for logistic reasons. We therefore tested two different swabs for longer storage, the eNAT swab and the MSwab. These swabs were selected because of their use in previous studies, easy accessibility, and low cost (Kaan et al., 2020; Susic et al., 2020). In addition, we tested three different methods that include DNA extraction and host DNA depletion in comparison with the QIAamp PowerFecal DNA kit, which contains no host DNA depletion step, as reference standard used in many microbiome projects with similar respiratory and vaginal samples (Bartolomaeus et al., 2021; Boeck et al., 2017; Ferrarese et al., 2020; Lebeer et al., 2018; Lim et al., 2020). First, we tested host DNA depletion by osmotic lysis of mammalian cells and treatment with propidium monoazide (PMA), prior to lysis of the microbial cells and DNA extraction using the QIAamp PowerFecal DNA kit, as described by Marotz et al. (2018). In addition, we evaluated the efficacy of host depletion of two commercial kits with host DNA depletion (the QIAamp DNA Microbiome kit and the HostZERO Microbial DNA kit), which were already used in previous work (Figure 1) (Heravi et al., 2020; Lim et al., 2017). The obtained microbiome yield and profiles were analyzed via 16S rRNA gene amplicon sequencing and shallow metagenomic shotgun sequencing, qPCR, and DNA quantification methods. Our aim was to derive a cost-effective, hands-on, robust pipeline using shallow metagenomic shotgun sequencing that is useful for small, medium, and large-scale microbiome research flows and to obtain critical insights in the importance of biological differences between sampling sites.
Figure 1.
Overview of the study workflow
(A) Three types of samples (vaginal, skin, and saliva swabs) were self-collected by study participants (n = 23) for microbiome analysis. Different factors influencing the microbiome analyses were studied, starting with (B) swab methods for sample collection (eNAT or MSwab) and storage conditions with analysis at four time points (T1, T2, T3, T4). Next, (C) four DNA extraction and host DNA depletion methods were compared. These included the QIAamp PowerFecal DNA kit (without host DNA depletion) or QIAamp PowerFecal DNA kit with host DNA depletion by cell lysis and treatment with propidium monoazide (PMA). In addition, the QIAamp DNA Microbiome kit and the HostZERO Microbial DNA kit with host DNA depletion were tested. Finally, (D) microbiome sequencing was performed with either 16S rRNA gene amplicon sequencing or shallow metagenomic shotgun sequencing with (E) subsequent corresponding bioinformatics analysis.
Results
Human DNA concentrations significantly lower after host DNA depletion
To investigate the impact of microbial enrichment in samples from different body sites, we asked eight healthy female volunteers to self-sample their vagina, skin, and saliva with two different swabs, namely, the eNAT swab and the MSwab (Figure 1B), resulting in 48 swabs. Initially, DNA was extracted within 3 h after sampling to evaluate microbial enrichment with four DNA extraction and host DNA depletion methods (Figure 1C), resulting in 192 samples for DNA quantification, qPCR, and sequencing.
Total DNA concentrations for all samples were first measured spectrophotometrically (Qubit 3.0 Fluorometer) (Figures 2A–2C). The total DNA concentrations ranged from 0.05 to 39.90 ng/μL for vaginal swabs, 0.05 to 28.00 ng/μL for saliva swabs, and 0.05 to 0.26 ng/μL for skin swabs. For all body sites, the concentrations were considerably lower after host DNA depletion with all three methods compared with no host DNA depletion (QIAamp PowerFecal DNA kit) (Figures 2A–2C). There was no significant effect of the swab buffer on the total DNA concentration for any of the body sites.
Figure 2.
Influence of microbial DNA enrichment methods on total, human, and bacterial DNA in vaginal, saliva, and skin samples measured with Qubit Fluorometer and qPCR
Three microbial DNA enrichment methods were compared with a standard microbial DNA extraction using the QIAamp PowerFecal DNA kit: Propidium monoazide (PMA) + QIAamp PowerFecal DNA kit, QIAamp Microbiome kit and HostZERO Microbial DNA Kit. Two different swabs with a lysis storage buffer were used for sampling: FLOQSwab with 1 mL eNAT medium and FLOQSwab with 1 mL MSwab medium.
(A–C) Total double-stranded DNA quantification with Qubit Fluorometer for vaginal (A), saliva (B), and skin swabs (C). Concentrations too low to measure have the theoretical value of 0.05 ng/μL.
(D–I) qPCR-based DNA quantification based on specific primers for human (D–F) and bacterial DNA (G–I). Values below the detection limit for qPCR (Ct-values <38, red dotted line) were replaced by a hypothetical value below the limit. Statistics were performed with Kruskal-Wallis test with Dunn's multiple comparisons test against the QIAamp PowerFecal DNA kit, ∗ = p < 0.05, ∗∗ = p < 0.001 (p values in Table S1).
Subsequent quantitative polymerase chain reaction (qPCR) analysis with universal human, bacterial, and fungal primers was performed on a subset of the samples (Figures 2D–2F). Without host DNA depletion, vaginal samples contained the highest concentration of human DNA for both tested swabs (2.440 ng/μL, eNAT; 2.364 ng/μL, MSwab), followed by saliva (0.651 ng/μL, eNAT; 0.814 ng/μL, MSwab) and skin (0.002 ng/μL, eNAT; 0.001 ng/μL, MSwab) samples (Figures 2A–2C). Of interest, without host DNA depletion, saliva samples contained approximately 2-fold (eNAT) and 4-fold (MSwab) more bacterial DNA than vaginal samples. Skin samples contained the lowest concentration of bacterial DNA (0.0002 ng/μL, eNAT; 0.0002 ng/μL; MSwab), which was 345-fold (eNAT) and 230-fold (MSwab) less than vaginal samples. It is noteworthy that none of the methods extracted high concentrations of fungal DNA from these healthy donors (Figure S1).
We compared the levels of obtained human and bacterial DNA between the two different swabs and four different DNA extraction and host DNA depletion methods (Figure 2). No significant differences were detected in the amount of human DNA or bacterial DNA between the two swabs after immediate processing for all investigated body sites. However, the four microbial enrichment methods performed differently in terms of human DNA depletion. The HostZERO Microbial DNA kit appeared the most effective host DNA depletion method for vaginal and saliva samples based on qPCR data. Compared with the QIAamp PowerFecal DNA kit (no host DNA depletion), no significant decrease in bacterial DNA concentrations was found, whereas human DNA concentrations were significantly decreased. For the already very low human DNA concentrations in skin samples (0.05–0.26 ng/μL), no significant decrease in human DNA was observed for any of the methods. The skin bacterial DNA load did not significantly decrease for the HostZERO Microbial DNA kit with the eNAT swab, but a significant decrease in bacterial DNA was observed for the MSwab.
For a successful shotgun sequencing run, samples must contain a minimum starting concentration of DNA (1–100 ng in maximum 30 μL recommended by the manufacturer for the Nextera DNA Flex Library Prep kit, Illumina). Since many samples, especially the skin samples, contained very low DNA concentrations after host DNA depletion, we examined whether we could use a lower starting concentration for shotgun sequencing than recommended. A commercial bacterial mock community (BEI Resources), composed with the DNA of 20 different species, was added to the library in three different starting concentrations (0.5, 1, 500 ng in 30 μL) to examine the library sizes and taxonomic profiles (Figure S2). Here, we observed no effect of a lower starting DNA concentration, suggesting that it is possible to use less than the requested 1 ng DNA to start the library preparation protocol. An additional bottleneck in working with low biomass samples is accurate equimolar pooling of shotgun libraries, with the aim of achieving approximately equal read counts across the samples. The total DNA concentration in the library was measured spectrophotometrically with the Qubit Fluorometer and with the Fragment Analyzer, and compared with the final read concentration (i.e., reads per pooled volume) per sample (Figure S3). Both methods predicted the final read concentration relatively poorly, with the Qubit Fluorometer making a slightly better prediction. This shows that equimolar pooling is still an important challenge for metagenomic shotgun sequencing of low biomass samples.
Lysis buffer conserved microbial profiles over longer storage
When performing a large-scale Citizen Science project, several logistic factors such as storage conditions and transport should be considered. We therefore aimed to select a suitable transport buffer for longer sample storage at a participant's home (4°C) and transport with postal services (room temperature). Hereto, additional vaginal eNAT and MSwabs from five healthy women were collected and placed at four storage conditions with different storage durations and temperatures (total of 40 samples processed with the HostZERO Microbial DNA kit), as schematically visualized in Figure 1B. For the first set of samples (T1), both host DNA depletion and microbial isolation were finished on the day of sampling (approximately 3 h after sampling). The second sample set (T2) was stored for 3 weeks at 4°C, followed by 3 days at room temperature (mimicking package transport by the postal service), after which both host DNA depletion and microbial isolation were executed. The third sample set (T3) started with the same storage conditions as T2 but only the host DNA depletion step was finished right after storage conditions. The host DNA depleted samples were then stored for 3 days at −80°C, after which the microbial DNA extraction step was completed. The fourth sample set (T4) also started with the same storage conditions as T2, followed by storing the aliquot at −20°C for 3 days, after which both host DNA depletion and microbial isolation were finished. We evaluated the impact of these different storage conditions on the variability in DNA concentration and detected taxonomic microbial community composition.
All vaginal samples stored under different conditions were analyzed using 16S rRNA gene amplicon sequencing (Figure 3). The microbial community of the eNAT samples showed an almost identical taxonomic composition across all storage conditions for each participant (n = 5). For the MSwab samples, a notable bias was clearly prominent in the three longer storage conditions, indicating that the taxonomic composition is influenced by storage. For example, the detected relative abundance of an important genus in the vaginal niche (Gardnerella 1) was clearly increased in participant 0002 and 0004 after longer storage. The beta-diversity by Bray-Curtis dissimilarities on relative abundances and the alpha diversity with inverse Simpson index also showed a swab-dependent taxonomic bias, with higher dissimilarities for the MSwab, when evaluating how longer storage could affect the detected microbiome composition (Figures S4 and S5).
Figure 3.
Relative taxonomic abundances based on 16S rRNA gene amplicon sequencing of vaginal samples to evaluate the influence of different swabs (eNAT and MSwab) and storage conditions (T1–T4)
Bar charts of taxonomic composition of the samples on ASV-level at the four time points grouped per participant. Top 11 ASVs are visualized, the rest is grouped together in residual. A first set of swabs were stored at 4°C and processed within 3 h after sampling (T1). All other samples were stored for 3 weeks at 4°C, followed by 3 days at room temperature. A second set of swabs was processed immediately at T2, whereas the last two sets of swabs were frozen for 3 days after T2. The third set was frozen at −20°C and entirely processed (T3). The last set of samples first underwent host DNA depletion at T2 and after 3 days storage at −80°C a DNA extraction (T4).
Enrichment methods differ in discriminatory power of vagina, skin, and saliva samples
Since the eNAT swab provided the most stable microbial profiles in terms of taxonomic composition, independent of the storage conditions, this swab was selected as the most ideal in the study protocol. Next, we aimed to select a microbial enrichment method with host DNA depletion suitable for vaginal, skin, and saliva samples based on 16S rRNA gene amplicon sequencing. 16S rRNA gene amplicon sequencing data of vaginal, saliva, and skin eNAT swabs from eight healthy female volunteers was extracted within 3 h after sampling with four DNA extraction and host DNA depletion methods (Figure 1C), resulting in a total of 96 samples. A hierarchical clustering dendrogram was used to distinctly visualize which samples contain a similar taxonomic composition. Most samples clustered according to their mucocutaneous niche and not by the used microbial enrichment method (Figure 4A), suggesting that the biological microbial composition of body sites is still prevailing. For the QIAamp PowerFecal DNA kit, QIAamp DNA Microbiome kit and HostZERO Microbial DNA kit, the taxonomic profiling was mostly driven by the body site and not by extraction method or participant since samples processed using these methods cluster according to the body site (Figure 4A). Only for the propidium monoazide (PMA) + QIAamp PowerFecal DNA kit, the vaginal, skin, and saliva samples clustered more together, suggesting that the mechanical and technical effects of the PMA + QIAamp PowerFecal DNA kit overrode the biological differences among the mucocutaneous niches.
Figure 4.
Impact of enrichment methods on clustering of 96 vagina, skin, and saliva samples based on 16S rRNA gene amplicon sequencing data
(A) Dendrogram with strip colored per microbial enrichment method (red, QIAamp PowerFecal DNA kit; blue, PMA + QIAamp PowerFecal DNA kit; green, QIAamp DNA Microbiome Kit; and yellow, HostZERO Microbial DNA kit) and rectangles colored per body site (pink, vagina; light blue, skin; light green, saliva). The dendrogram visualizes the result of average-linkage hierarchical clustering on pairwise Bray-Curtis distances between the samples.
(B) Bray-Curtis dissimilarities between each taxonomic profile and the profile from the same participant resulting from the QIAamp PowerFecal DNA extraction (golden standard), grouped by body site to evaluate how different microbial enrichment methods could affect the detected microbiome composition. Statistics were performed with the Wilcoxon test (p values are visualised on the figure). Data are represented as mean ± SEM.
The summed differences in read counts for all taxa across the different participants were analyzed with pairwise Bray-Curtis (BC) dissimilarities, comparing the three microbial enrichment methods with the standard QIAamp PowerFecal DNA kit for each participant (Figure 4B). A beta-diversity of 0 indicates that the bacterial composition between the used methods is the same, whereas a beta-diversity of 1 means that the composition differs completely. Compared with the skin and saliva samples, the vaginal samples showed the lowest mean BC dissimilarity over all the tested methods, respectively, 58.9% for PMA + QIAamp PowerFecal DNA kit, 21.0% for QIAamp DNA Microbiome kit, and 30.9% for HostZERO Microbial DNA kit. This indicates that the vaginal samples within a participant were more similar over the different microbial enrichment methods compared with the skin and saliva samples. For all body sites, the PMA + QIAamp PowerFecal DNA kit showed the largest BC dissimilarity, respectively, 43.2% for vagina, 65.5% for skin, and 78.0% for saliva samples. These differences in dissimilarity based on beta-diversity analysis indicate that the strategies to eliminate host DNA might unequally influence the presence of bacterial taxa. Regarding the host DNA depletion efficiency and BC dissimilarities, both QIAamp DNA Microbiome kit and HostZERO Microbial DNA kit showed similar performance for vaginal and skin samples. Therefore, we continued using this host DNA depletion kit based on our experience of time-efficiency in the laboratory and cost per sample. The HostZERO Microbial DNA kit performed considerably better on both criteria by cutting work time and cost in half.
Host DNA was effectively depleted for shotgun metagenomic sequencing
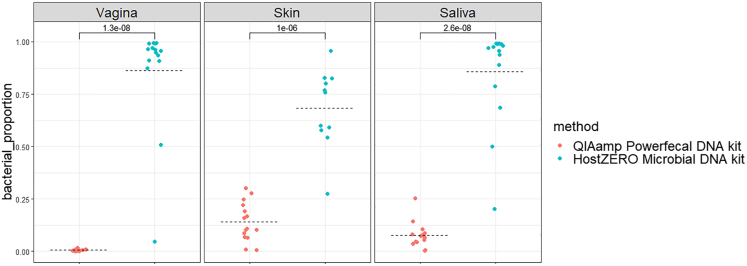
After our detailed qPCR and 16S rRNA gene amplicon sequencing data analysis, we aimed to evaluate the HostZERO Microbial DNA kit for its effectivity in reducing host reads in metagenomic sequencing and compared this with the QIAamp PowerFecal DNA kit (without host DNA depletion). Therefore, a new pool of vaginal, skin, and saliva eNAT swabs from fifteen healthy women were collected and extracted with both HostZERO Microbial DNA kit and QIAamp PowerFecal DNA kit within 3 h after sampling, resulting in 90 samples for sequencing. Without host DNA depletion, vaginal samples contained 95.9% of human reads, skin samples 92.4%, and saliva samples 92.3%. The HostZERO Microbial DNA kit could deplete human reads from vaginal, skin, and saliva samples considerably, resulting in 13.6%, 31.6%, and 14.4% human reads in vaginal, skin, and saliva samples, respectively (Figure 5). Subsequently, shallow shotgun sequencing confirmed that host DNA contamination was significantly lower for the HostZERO Microbial DNA kit compared with the QIAamp PowerFecal DNA kit for vaginal, skin, and saliva samples, concluding that the host DNA depletion was successful.
Figure 5.
Bacterial proportion of shallow shotgun sequencing data per body habitat (vagina, skin, saliva) and microbial enrichment method without (pink, QIAamp PowerFecal DNA kit) and with (blue, HostZERO Microbial DNA kit) host DNA depletion.
Statistics were performed with the Wilcoxon test (p values are visualised on the figure).
In addition, the decrease in total DNA concentration after host DNA depletion steps caused only skin samples (4 of 15 samples) to fail quality control during library preparations for shotgun sequencing, compared with saliva and vaginal samples. Therefore, we evaluated the extent to which bacterial read concentrations were reduced along with the host DNA. Here, we noticed an increase in bacterial read concentration after host DNA depletion for saliva and mainly vaginal samples, which was not the case for skin samples (Figure S6).
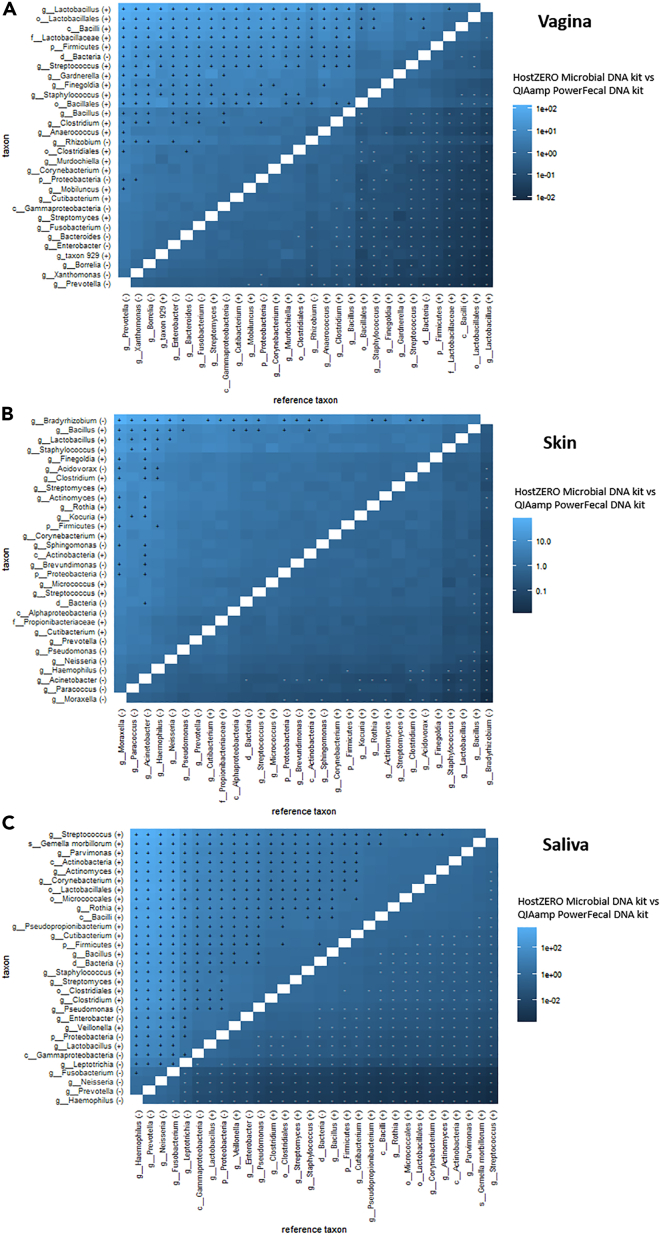
Taxonomic bias of enrichment methods toward Gram-negative taxa
After we evaluated the effectiveness of host DNA depletion for vaginal, skin, and saliva samples, we also evaluated the taxonomic impact of host DNA depletion. Here, we compared the detected taxonomic profiles and looked at the differential abundance per taxon while comparing both methods (Figure 6). Overall, there was significant differential abundance of most taxa between both kits, with the HostZERO Microbial DNA kit significantly reducing the relative abundance of mostly Gram-negative taxa. When looking for overlapping taxa in the microbial community of the different body sites, some taxa were found to be similarly reduced, such as Prevotella spp., Fusobacterium spp., and Gammaproteobacteria in both vaginal and saliva samples, and Haemophilus spp. and Neisseria spp. in both skin and saliva samples. When focusing on body site-specific key taxa, several other Gram-negative taxa showed to be significantly different abundant. For the vaginal samples, the Gram-negative taxa such as Bacteriodes spp., Xanthomonas spp., and Enterobacter spp. were significantly reduced after host DNA depletion (Figure 6A). The effects on saliva samples were also highly dependent on the kit used, with the HostZERO Microbial DNA kit significantly reducing the abundance of Gram-negative taxa such as Staphylococcus spp. and Pseudomonas spp. (Figure 6C). Finally, Acinetobacter spp. and Moraxella spp. were significantly less represented after host DNA depletion in the skin samples (Figure 6B). This was also confirmed by the pairwise BC dissimilarities, comparing the HostZERO Microbial DNA kit with the standard QIAamp PowerFecal DNA kit for each participant (Figure S7). Here, the vaginal samples showed the lowest mean BC dissimilarity, whereas the saliva samples within a participant were the least similar over the two enrichment methods compared with the skin and vaginal samples.
Figure 6.
Each cell of the heatmap represents the differential abundance of a taxon when comparing two methods (HostZERO Microbial DNA kit versus QIAamp PowerFecal DNA kit), relative to another taxon, on which a Wilcoxon test (+ in heatmap relates to p < 0.05 and - relates to p ≥ 0.05) was performed.
The entire plot was also corrected for multiple testing (QIAamp PowerFecal DNA kit and the HostZERO Microbial DNA kit). The color represents the median of all pairwise differences of log ratios between two groups of samples (two-sample Hodges-Lehmann estimator). (A) Vagina; (B) Skin; (C) Saliva. (−), Gram-negative taxon; (+), Gram-positive taxon; ∗taxon 929, Peptostreptococcaceae bacterium oral taxon 929.
Discussion
Citizen Science-based studies that strongly rely on active collaboration of citizens to generate scientific insights are increasingly gaining interest. However, several logistic and technical challenges need to be evaluated in order to conduct a large-scale project of the human microbiome, such as the Isala project that our group launched on March 24, 2020 (www.isala.be/en/). In this study, we therefore evaluated sample collection and storage conditions for vaginal, skin, and saliva swabs. In addition, the effectiveness of host DNA depletion and microbial enrichment was evaluated for four different approaches. Particular attention was given to methods that could significantly decrease human DNA, since this is important for metagenomic shotgun sequencing, while not substantially decreasing bacterial DNA or introducing a significant taxonomic bias.
Under ideal laboratory conditions, samples are processed immediately after collection, which is not feasible in a large-scale Citizen Science study. We therefore investigated the effects of storage time and temperature on the microbial profiles of vaginal samples, using two different commercial swabs. First, we evaluated the eNAT swab, which contains a lysis buffer specifically made for longer stabilization and preservation of microbial DNA and RNA (Young et al., 2020). We also evaluated the MSwab, which contains a weaker lysis buffer and is focused on collection and transport of clinical samples to preserve microbial viability (Lei et al., 2020). We observed a clear swab buffer-dependent taxonomic shift in microbial composition of vaginal samples over time when using the MSwab and thus would recommend to use the eNAT swab when longer storage of vaginal swabs at various temperatures is preferred. This finding confirms the applicability of the eNAT swab for microbiome analysis as described by Young et al. (2020).
In addition to sample collection and storage, several factors related to sample processing need to be considered. Here, we specifically aimed to optimize the workflow for metagenomic shotgun sequencing, since this technique allows a high-resolution microbiome analysis (Hillmann et al., 2018). An important challenge for this technique is the proportion of human genome reads in relation to microbial reads. As expected by the physiological characteristics of the mucocutaneous niches and observations of other studies (Bjerre et al., 2019; Marotz et al., 2018), we confirmed via qPCR that skin swabs do not contain as many human cells as vaginal and saliva swabs. Although the vaginal mucosa and skin epidermis resemble each other structurally, an important difference is the higher permeability to water and proteins of the vaginal stratum corneum and the absence of keratin bundles in vaginal epithelium (Boskey et al., 2001; Wylie and Henderson, 1969). Also, the specific mucous polysaccharide structure and mucin composition at the wet epithelium of the mouth and vagina differ considerably from that of the skin. Mucus is composed of water, mucins, globular proteins, salts, DNA, lipids, cells, and cellular debris and forms a dense, viscoelastic layer over epithelial cells, which can result in a higher concentration of human DNA and cells on swabs (Leal et al., 2017; Lethem et al., 1990). In addition to containing relatively low human DNA concentrations, skin samples generally also contain a low bacterial biomass (Bjerre et al., 2019; Grogan et al., 2019). In our study, DNA concentrations from 0.05 to 0.26 ng/μL were obtained for skin swabs. This indicates that skin samples might be more difficult to be accurately analyzed by metagenomic shotgun sequencing in combination with host DNA depletion methods.
Because the impact of host DNA is so crucial in shotgun sequencing, an in-depth evaluation of host DNA depletion mechanisms on microbial profiling of different body sites was performed through 16S rRNA gene amplicon and shallow metagenomic shotgun sequencing. Previous studies have demonstrated that vaginal, skin, and saliva samples have distinct microbial patterns, making them ideal for use as microbial fingerprinting tool for diagnostic and forensic purposes (Costello et al., 2009; Tackmann et al., 2018). Most samples in our study clustered according to the mucocutaneous niche (i.e., vagina, skin, and saliva) and not by microbial enrichment method, indicating that the distinct microbial patterns could be equally profiled with most of the used methods, with the exception of PMA in combination with the QIAamp PowerFecal DNA kit. PMA is a cell membrane-impermeable DNA intercalator that can fragment and eliminate the exposed human DNA from downstream analysis and was introduced previously as an adequate host DNA depletion method (Marotz et al., 2018). Our results, however, suggest that this approach is not suitable for forensic and diagnostic fingerprinting of mucocutaneous samples since the method introduces a significant taxonomic bias. In addition, we hypothesize that swabs that are stored in a lysis buffer as opposed to a non-lysing buffer might have more free-floating bacterial DNA that could be depleted faster than anticipated.
Beta-diversity analysis also suggested that the microbial enrichment methods unequally influenced the detected abundance of specific bacterial taxa in our samples. Furthermore, this taxonomic bias depended on the biological differences between the studied body sites. As such, the HostZERO Microbial DNA kit significantly reduced the relative abundance of Gram-negative taxa within skin and saliva samples (e.g., Haemophilus spp., Neisseria spp. and Prevotella spp.) when compared with the QIAamp PowerFecal DNA kit. The taxonomic bias represents a disadvantage, as previous research has suggested that both Gram-negative bacteria (e.g., Prevotella, Veillonella, Neisseria, Haemophilus, Campylobacter, Fusobacterium, Mycoplasma) and Gram-positive bacteria (e.g., Streptococcus, Rothia, Actinomyces) represent an important taxonomic and functional part of the human salivary microbiome (Dzidic et al., 2018; Hasan et al., 2014; Marotz et al., 2018). Of note, Salonen et al. (2010) demonstrated that recovery of the Gram-negative bacteria is often impaired by the lysis used in the DNA extraction method. The identification of Porphyromonas gingivalis, for instance, in caries and periodontal samples was significantly variable depending on the DNA extraction kit used, whereas Streptococcus mutans detection was not significantly affected. The authors proposed that bacteria with a thin or absent cell wall might be lysed together with human cells, resulting in a skewed microbiome profile. Indeed, in our study this effect can also be observed for the HostZERO Microbial DNA kit. This was much more pronounced within salivary microbial communities and less within the vaginal microbial communities, which mostly contained Gram-positive bacteria. To avoid significant skewing of the detected taxonomic abundances toward bacteria more resistant to lysis, it is crucial to choose an appropriate kit and tailor the protocol to the expected microbiome composition (Hardwick et al., 2018). We advise against using any of the three tested host DNA depletion methods for skin and saliva samples, as more research focused on minimizing the taxonomic bias of Gram-negative bacteria is necessary. We do recommend the QIAamp DNA Microbiome kit and HostZERO Microbial DNA kit for vaginal samples that mostly contain Gram-positive bacteria. However, it is important to mention that a taxonomic bias by microbial DNA isolation cannot entirely be avoided. We therefore recommend researchers to apply a method that minimizes this bias as much as possible. In addition, we highlight that it is important to apply the same protocol throughout the study, to minimize potential batch effect associated with the experimental flow of microbiome analysis.
Depending on the study population, women have a vaginal microbiome that is generally dominated by Gram-positive Lactobacillus spp. (i.e., L. crispatus, L. iners, L. gasseri, and L. jensenii) or by non-lactobacilli (e.g., Bifidobacterium, Gardnerella, Atopobium, Prevotella, and Streptococcus) (France et al., 2020; Ravel et al., 2011). Even though the sample size was relatively small, the vaginal microbiome profiles in our study still resembled those reported in the literature, namely, mainly dominated by L. crispatus, L. gasseri, and L. jensenii. For the saliva samples, Willis et al. (2018) found that the salivary microbiome mostly clusters into two community types, dominated by the Gram-negative Neisseria or Prevotella, but it also contains large proportions of Gram-positive Streptococcus taxa (e.g., Streptococcus salivarius and Streptococcus oralis). Although in our study the Gram-negative taxa were strongly depleted with the HostZERO Microbial DNA kit, using the QIAamp PowerFecal DNA kit we found both Gram-negative (e.g., Neisseria spp., Prevotella spp., and Veilonella spp.) and Gram-positive (e.g., Streptococcus spp.) taxa that presented a more expected salivary microbiome profile. At last, for the skin samples, Costello et al. (2009) and Grice et al. (2009) studied the microbiome of various skin body sites and reported that the four most detected phyla were Actinobacteria (51.8%), Firmicutes (24.4%), Proteobacteria (16.5%), and Bacteroidetes (6.3%). Moist body sites, such as the inner elbow that was sampled in this study, were dominated by Corynebacteria, Propionibacterium, and Staphylococcus species (Grice et al., 2009). In our 16S rRNA gene amplicon sequencing data, Propionibacterium were less detected than Corynebacterium and Staphylococcus species, which is a known bias when sequencing the V4 region of 16S rRNA gene. In the metagenomic shotgun data, Cutibacterium acnes was clearly present. Although Gram-negative Acinetobacteria were not the most abundant on moist skin sites, it was clear that these were strongly depleted in the host DNA depleted samples with the HostZERO Microbial DNA kit.
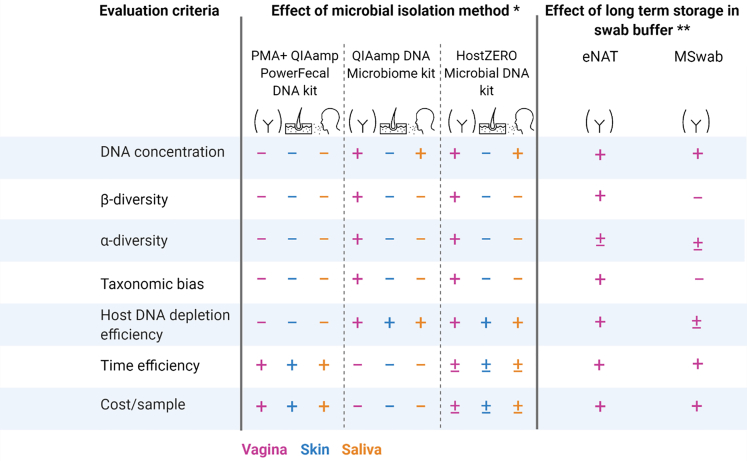
Our results show that the HostZERO Microbial DNA kit was most ideal for vaginal samples, since we obtained an effective host DNA depletion and no remarkable taxonomic bias, as summarized in Figure 7. For skin and saliva samples, we recommend the QIAamp PowerFecal DNA kit since less taxonomic bias was observed compared with processing with the HostZERO Microbial DNA kit. In addition, the HostZERO Microbial DNA kit showed a higher risk for low-biomass samples (i.e., skin samples) failing quality control during downstream processing. Respectively, the HostZERO Microbial DNA kit was not an added value to the low biomass skin samples and even increased their risk to fail in metagenomic shotgun sequencing. For the saliva samples, a selection was made with the aim to microbial fingerprint as accurately as possible, and therefore recovering as much taxa as possible. Finally, the HostZERO Microbial DNA kit method scored best from all host DNA depletion methods regarding cost, time, and labor efficiency per sample.
Figure 7.
Evaluation of different microbial isolation methods and swab buffers for storage for microbiome sequencing of vaginal, skin, and saliva samples
Legend: +, good; +/−, medium; -, poor kit performance (based on result section) when compared with the QIAamp PowerFecal DNA kit as standard method for the effect of microbial isolation methods, and compared with no storage (processing on day of sampling) for the effect of longer storage in swab buffer. ∗Short-term storage, ∗∗all samples performed with the HostZERO Microbial DNA kit.
Conclusion
In conclusion, we evaluated a sample collection method for efficient transport and longer storage of microbiome samples on various temperatures. The eNAT buffer keeps the microbial composition stable during various storage conditions, in contrast to the MSwab buffer. In addition, our findings emphasize that the microbial community composition of mucocutaneous body sites such as the vagina, skin, and mouth (saliva) should, next to the aim of the study, always be taken into account when implementing a microbiome analysis protocol, and especially a host DNA depletion method for metagenomic shotgun sequencing. For vaginal and saliva samples, two of three tested host DNA depletion methods successfully reduce human DNA while not or barely reducing bacterial DNA. But all three host depletion methods drastically changed the detected microbial compositions of the mucocutaneous samples, except the HostZERO Microbial DNA kit and QIAamp DNA Microbiome kit for the vaginal samples. Overall, the HostZERO Microbial DNA kit was the best-performing host DNA depletion method, as it even enriches bacterial reads for vaginal and saliva samples. However, it is important to recognize that microbial DNA isolation methods introduce a taxonomic bias that cannot entirely be avoided. The aim should therefore always be to minimize this bias as much as possible and be aware of the taxa toward which this bias extends. Finally, the molecular and biological impact of storage over a longer period of time (>6 months up to years) in combination with host DNA depletion approaches should be further investigated.
Limitations of the study
More research is needed not only on the taxonomic bias toward the over- and/or underrepresentation of Gram-positive and Gram-negative bacteria but also on the sensitivity and specificity of the existing methods toward viruses, archaea, and fungi. We therefore acknowledge that our study had some limitations:
-
−
Limited sample size of 322 samples and 23 participants
-
−
Limited storage time conditions tested (up to 4 weeks) and this only for vaginal swabs with mainly L. crispatus and L. iners profiles. To evaluate long-term storage (>6 months to years), more research is needed including different time points and sampling sites.
-
−
Two of three host DNA depletion methods were only evaluated with qPCR and 16S rRNA gene amplicon sequencing (no shotgun metagenomic sequencing).
-
−
Our focus was mainly on bacteria and not on viruses, archaea, and fungi
-
−
Equimolar pooling for shotgun sequencing by both Qubit and the Fragment analyzer was not optimal. Since this is still a bottleneck in the entire field of microbiome sequencing, more research and optimization are needed.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Genomic DNA from Microbial Mock Community B (Even, High Concentration), V5.1H | BEI Resources | HM-276D |
| Lacticaseibacillus rhamnosus GG | American Type Culture Collection (ATCC) | ATCC53103 |
| Candida albicans | Centre of Microbial and Plan Genetics (CMPG), KULeuven | SC5314 |
| Biological samples | ||
| Human vaginal swabs | University of Antwerp | N/A |
| Human saliva swabs | University of Antwerp | N/A |
| Human skin swabs | University of Antwerp | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| eNATTM | COPAN Diagnostics | 6U073S01 |
| MSwabTM | COPAN Diagnostics | 404C.R |
| de Man, Rogosa and Sharpe (MRS) | Difco | BD288210 |
| SeaKemR LE Agarose | Lonza | 50004 |
| 1.0N NaOH | Illumina | NA |
| PhiX sequencing control v3 | Illumina | FC-110-3001 |
| Trizma base | Sigma-Aldrich | 77-86-1 |
| Ethylenediaminetetraaceticacid (EDTA) | Sigma-Aldrich | 60-00-4 |
| Tween-20 | Biorad | 170-6531 |
| Propidium monoazide (PMAxxTM) | Biotium | 40069 |
| PowerSYBR® Green PCR Master Mix | Applied Biosystems | 13266519 |
| Critical commercial assays | ||
| QIAamp Powerfecal DNA kit | Qiagen | 12830-50 |
| Agencourt AMPure XP | Beckman Coulter | A63881 |
| HostZERO Microbial DNA Kit | Zymo Research | D4310 |
| QIAamp DNA Microbiome Kit | Qiagen | 51704 |
| Nextera™ DNA Flex Library Prep | Illumina | 20015829 |
| High Sensitivity NGS Fragment Kit | Agilent | DNF-474 |
| NucleoSpin 96 Tissue kit | Machery-Nagel | MN 740609.50 |
| Deposited data | ||
| DNA-Seq data | This paper | PRJEB45093 |
| R code | This paper | github.com/Sarahtopia/isala_pilot |
| Experimental models: Cell lines | ||
| Human Caco-2 epithelial cells | ATCC | HTB-37 |
| Oligonucleotides | ||
| Human housekeeping gene PPIA (PPIA_F) GCT TGC TGG CAG TTA GAT GTC |
Jacobsen et al. (2014) | N/A |
| Human housekeeping gene PPIA (PPIA_R) AGA GGT CTG TTA AGG TGG GC |
Jacobsen et al. (2014) | N/A |
| Bacterial 16S rRNA gene V4 region (515F) GTG CCA GCM GCC GCG GTA A |
Kozich et al. (2013) | N/A |
| Bacterial 16S rRNA gene V4 region (806R) GGA CTA CHV GGG TWT CTA AT |
Kozich et al. (2013) | N/A |
| Bacterial 16S rRNA gene (338F) ACT CCT ACG GGA GGC AGC AG |
Ovreås et al. (1997) | N/A |
| Bacterial 16S rRNA gene (518R) ATT ACC GCG GCT GCT GG |
Ovreås et al. (1997) | N/A |
| Fungal ITS gene (ITS86F) GTGAATCATCGAATCTTTGAA |
Op De Beeck et al. (2014) | N/A |
| Fungal ITS gene (ITS4R) TCCTCCGCTTATTGATATGC-3 |
Op De Beeck et al. (2014) | N/A |
| Software and algorithms | ||
| GraphPad Prism | GraphPad Software | https://www.graphpad.com/ |
| DADA2, version 1.6.0 | https://doi.org/10.1038/nmeth.3869 | https://benjjneb.github.io/dada2/index.html |
| R version 3.4.4 | https://www.r-project.org/ | |
| Tidyamplicons | github.com/Swittouck/tidyamplicons | |
| Other | ||
| MiSeq Desktop sequencer | Illumina | M00984 |
| StepOne Plus Real-Time PCR System (v.2.0) | Applied Biosystems | N/A |
| Qubit 3.0 Fluorometer | Life Technologies | Q33216 |
| EVETM Automatic cell counter | NanoEntek | EVE-MC |
| 5200 Fragment Analyzer System | Agilent | M5310AA |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sarah Lebeer (sarah.lebeer@uantwerpen.com).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Human subjects
For the microbiome analysis, human subjects were recruited and self-sampled in a standardized way. Vaginal, saliva and skin swabs were obtained from 23 healthy premenopausal adult participants (100% female, average age 27). This study was approved by the Ethics Committee of the Antwerp University Hospital/University of Antwerp (registration number B300201942076, registered 18 November 2019, ClinicalTrials.gov Identifier: NCT04319536). The written informed consent was obtained from all participants prior to sampling.
Sample collection
Vaginal, skin (elbow bend) and saliva samples were collected with the eNATTM and the MSwabTM (Copan, Brescia, Italy). Vaginal swabs were turned around 2-3 times to acquire enough biomass. For the skin samples, participants were required to not wash the elbow and knee region for 8 to 12 hours before sampling. Skin samples (elbow bend) were collected by turning around the swab while brushing an indicated area of 20 cm2, for 30 seconds. Prior to sampling, the swab was soaked in a vial of sterile pre-moisture buffer (50 mM Tris buffer [pH 7.6], 1 mM EDTA [pH 8.0], and 0.5% Tween-20). Lastly, for the saliva samples participants were required to not brush their teeth, eat, or drink 1 hour before sampling. They spit 2-3 times in a sterile 50 mL falcon and turned the swab around in the saliva to acquire enough biomass. All swabs were transferred immediately after sampling to the vial which contained the commercial transport buffer. At last, all samples were stored at 4°C until further processing approximately 3 hours later.
Method details
Host DNA depletion and microbial DNA extraction
Before further processing, all samples were vortexed for 15-30 seconds and divided into four aliquots. DNA from the first aliquot was extracted with the PowerFecal® DNA Isolation Kit (Qiagen, Hilden, Germany) according to the instructions of the manufacturer. The second aliquot was also processed with the PowerFecal® DNA Isolation Kit (Qiagen), however prior to DNA extraction samples were treated with propidium monoazide (lyPMA) according to Marotz et al. (2018), with the adjustment that we started from 200 μL swab buffer instead of fresh saliva. The third aliquot was processed with the QIAamp DNA Microbiome Kit (Qiagen) according to the instructions of the manufacturer. The fourth aliquot was processed with the HostZERO Microbial DNA Kit (Zymo Research, California, United States). DNA concentration of all samples was measured using the Qubit 3.0 Fluorometer (Life Technologies, Ledeberg, Belgium) according to the instructions of the manufacturer. The mock community DNA was obtained through BEI Resources, NIAID, NIH as part of the Human Microbiome Project: Genomic DNA from Microbial Mock Community B (Even, High Concentration), v5.1H, for Whole Genome Shotgun Sequencing, HM-276D. Microbial composition of the mock community DNA can be found in Table S2.
Sample storage conditions
To evaluate the optimal storage conditions for the samples two vaginal swabs of each participant were collected (eNATTM and the MSwabTM), aliquoted and later processed with the HostZERO Microbial DNA kit (Zymo Research). For the first aliquot (T1), both host DNA depletion and microbial isolation were finished on the day of sampling (approximately 3 hours after sampling). The second aliquot (T2) was stored for 3 weeks at 4°C, followed by 3 days at room temperature (mimicking package transport by the postal service), after which the both host DNA depletion and microbial isolation were executed. The third aliquot (T3), started with the same storage conditions as T2 but only the host DNA depletion step was finished right after storage conditions. The host DNA depleted sample was then stored for 3 days at -80°C, after which the microbial DNA extraction step was completed. The fourth aliquot (T4) also started with the same storage conditions as T2, followed by storing the aliquot at -20°C for 3 days, after which both host DNA depletion and microbial isolation were finished.
Quantitative PCR (qPCR)
qPCR was used for estimation of absolute bacterial, fungal and human DNA concentrations in samples after DNA extraction. qPCR was performed in duplicate on a 20-fold dilution (to avoid interference of PCR inhibitors) of total DNA isolated from the samples, using the StepOnePlus Real-Time PCR System (v.2.0; Applied Biosystems®, Foster City, California, USA), SYBR® Green chemistry (PowerUp™ SYBR® Green Master Mix, Applied Biosystems®, Foster City, California, USA) and primers and PCR conditions as indicated in Resource table and Tables S3, S4 and S5. All primers were designed on the basis of published sequences and chemically synthesized by Integrated DNA Technologies (IDT, Leuven, Belgium). Primers for the human housekeeping gene PPIA were used to estimate human DNA concentration (PPIA_F and PPIA_R, (Jacobsen et al., 2014)). Universal bacterial 16S rRNA primers were used to estimate bacterial DNA concentration (338F and 518R, (Ovreås et al., 1997)). Universal fungal ITS primers were used to estimate fungal DNA concentration (ITS86F en ITS4R, (Op De Beeck et al., 2014)). Standard curves were used to estimate bacterial, fungal or human DNA concentrations in the samples and derived from serially diluted DNA from an overnight culture of Lacticaseibacillus rhamnosus GG (primers 338F and 518R), Candida albicans (primers ITS86F and ITS4R) and human Caco-2 epithelial cells (primers PPIA_F and PPIA_R). Bacterial concentration was determined by plating out on MRS growth medium and human cell concentration was determined by cell counter.
16S rRNA gene amplicon sequencing
Illumina MiSeq 16S rRNA gene amplicon sequencing was performed on the extracted DNA using all different host DNA depletion and DNA extraction methods. No less than 5 μl of each bacterial DNA sample was used to amplify the V4 region of the 16S rRNA gene. All DNA samples and negative controls of both PCR (PCR grade water) and the DNA extraction kit (Figure S8) were included. Standard barcoded forward (515F) and reverse primer (806R) were used. These primers were altered for dual index paired-end sequencing, as described in Kozich et al. (2013). The resulting PCR products were checked on a 1% agarose gel. The PCR products were purified using the Agencourt AMPure XP Magnetic BeadCapture Kit (Beckman Coulter, Suarlee, Belgium) and the concentration of all samples was measured using the Qubit 3.0 Fluorometer. Next, a library was prepared by pooling all PCR samples in equimolar concentrations. This library was loaded onto a 0.8% agarose gel and purified using the NucleoSpin Gel and PCR clean-up (Macherey-Nagel). The final concentration of the library (2 nM) was measured with the Qubit 3.0 Fluorometer. Afterwards the 5 μl of the library was denatured with 0.2N NaOH (Illumina, San Diego California United States), diluted to 6 pM and spiked with 10% PhiX control DNA (Illumina). Dual-index paired-end sequencing was performed on the V4 region of the 16S rRNA gene using a MiSeq Desktop sequencer (M00984, Illumina).
Shallow metagenomic shotgun sequencing
Library preparation for metagenomic shotgun sequencing was performed using the Nextera™ DNA Flex Library Prep (Illumina), according to the instructions of the manufacturer. For the Nextera™ DNA Flex Library Prep, 2 – 30 μL DNA sample was used to obtain input DNA with a start amount between 1 and 100 ng. For the Nextera™ XT DNA Library Preparation kit, 1 ng DNA samples in 5 μL was used as input DNA. For both protocols, when the 1 ng input DNA could not be obtained for a certain DNA sample, the library preparation was continued with the highest available amount of input DNA. Pooling of the libraries was done individually using the Qubit 3.0 Fluorometer. During library preparation, library quality was checked using the 5200 Fragment Analyzer System with Agilent High Sensitivity NGS Fragment Kit (DNF-474). 22μL NGS Diluent Marker solution was mixed with 2μL library and ran on the Fragment Analyzer, according the instruction of the manufacturer. The NGS DNA Ladder was used as standard.
Bioinformatic analysis
Quality control and processing of 16S rRNA amplicon reads was performed using the R package DADA2, version 1.6.0. Reads with more than two expected errors were removed; no trimming was performed. Forward and reverse reads were denoised per sample using the DADA2 algorithm. Reads were then merged; during this process, read pairs with sequence conflicts were removed. Chimeras were detected and removed with the removeBimeraDenovo function. The merged and denoised reads (amplicon sequence variants or ASVs) were taxonomically annotated from the phylum to the genus level with the assignTaxonomy function using the EzBioCloud reference 16S rRNA database (Yoon et al., 2017). Non-bacterial ASVs (e.g. mitochondria and chloroplasts) were removed. ASVs with a length greater than 299 bases were also removed. Samples contained on average 22,287 high-quality reads per sample. All data handling and visualization was performed in R version 3.4.4 (R Core Team, 2020) using the tidyverse set of packages and the in-house package tidyamplicons (github.com/Swittouck/tidyamplicons) (De Boeck et al., 2020).
Metagenomic shotgun reads were processed with Kraken2 (Wood and Salzberg, 2014). Forward and reverse read pairs were classified from the phylum to the species level using the MiniKraken2 v2 reference database. This database was constructed from the bacterial, archaeal and viral genomes in NCBI RefSeq, in addition to the human genome version GRCh38 (to detect human contamination). Reads classified to non-bacterial taxa were removed. Based on these classifications, a read count table was constructed where the columns represent taxa and the rows represent samples. Taxa were either species or higher-level taxa for reads that were unclassified at one or more ranks. Samples contained on average 39,164 high-quality bacterial reads per sample Parsing of Kraken2 output was implemented in R v3.6.1 (R Core Team, 2020).
Quantification and statistical analysis
Statistical details can be found in the figure legends. Statistical analysis for the Qubit and qPCR data (Figure 2) was performed in Graphpad Prism 9, using a nonparametric Kruskal-Wallis with Dunn’s multiple comparisons. Statistical analysis of pairwise Bray-Curtis dissimilarities (Figure 4A)and bacterial proportions (Figure 5) was performed using Wilcoxon tests in R-studio. Differential abundances (Figure 6) were tested using our in-house codifab method: for each pairwise combination of taxa, the logratio of their relative abundances was calculated for each sample. For each combination of taxa, the difference of their logratio between two groups of samples was expressed as the median of all pairwise differences between the two sample groups (Hodge-Lehmann estimator). Next, for each combination of taxa, a Wilcoxon rank-sum test was performed to compare their logratio between two groups of samples. This resulted in nˆ2 p-values for n taxa; these were corrected for multiple testing using the method of Benjamini and Yekutieli (Benjamini and Yekutieli, 2001).
Additional resources
This study was approved by the Ethics Committee of the Antwerp University Hospital/University of Antwerp: registration number B300201942076, registered 18 November 2019, ClinicalTrials.gov Identifier: https://clinicaltrials.gov/ct2/show/NCT04319536.
This study was part of a large-scale Citizen Science project investigating the female microbiome in Belgium. More information about the goals and preliminary results of this project: https://isala.be/en/
Acknowledgments
First, we would like to thank all participants for donating samples. In addition, we would also like to thank the following colleagues of our research group for their contributions: Ines Tuyaerts, Nele Van de Vliet, and Eline Cauwenberghs for their help with sample processing and Vincent Greffe for his help with data visualization. S.A. was supported by the University Research Fund (BOF-DOCPRO 37054) of the University of Antwerp. L.D. was supported by Baekeland of VLAIO (HBC.2020.2873). I.S. was supported by the IOF-POC University of Antwerp funding ReLACT (FFI190115). S.W. was supported by FWO aspirant fundamental research (file number 72798). W.V.B. and S.L. were supported by the European Research Council grant (Lacto-be 852600).
Author contributions
All authors worked on the conceptualization of the research project. The experiments were performed by S.A. and L.D. Data-analyses and visualization were performed by S.A., L.D., S.W., and W.V.B. Data interpretation was done by S.A., L.D., I.S., S.W., W.V.B., I.D.B., and S.L. The original draft was written by S.A., L.D., I.S., and S.L. All authors contributed to reviewing and editing of the paper. All authors proofread and approved the manuscript.
Declaration of interests
S.L. is a member of the scientific advisory board of YUN NV. L.D. was funded by VLAIO through a Baekeland mandate in collaboration with YUN NV. The remaining authors have no conflicts of interest to declare.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103306.
Supplemental information
Data and code availability
-
•
All sequencing data were deposited in ENA: PRJEB45093 (https://www.ebi.ac.uk/ena/browser/view/PRJEB45093).
-
•
The R code generated during this study can be found on GitHub at github.com/Sarahtopia/isala_pilot.
References
- Anderson D.J., Marathe J., Pudney J. The structure of the human vaginal stratum corneum and its role in immune defense. Am. J. Reprod. Immunol. 2014;71:618–623. doi: 10.1111/aji.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomaeus T.U.P., Birkner T., Bartolomaeus H., Löber U., Avery E.G., Mähler A., Weber D., Kochlik B., Balogh A., Wilck N. Quantifying technical confounders in microbiome studies. Cardiovasc. Res. 2021;117:863–875. doi: 10.1093/cvr/cvaa128. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
- Bjerre R.D., Hugerth L.W., Boulund F., Seifert M., Johansen J.D., Engstrand L. Effects of sampling strategy and DNA extraction on human skin microbiome investigations. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-019-53599-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck I. De, Wittouck S., Wuyts S., Oerlemans E.F.M., van den Broek M.F., Vandenheuvel D., Vanderveken O., Lebeer S. Comparing the healthy nose and nasopharynx microbiota reveals continuity as well as. Front. Microbiol. 2017;8:1–11. doi: 10.3389/fmicb.2017.02372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boskey E.R., Cone R.A., Whaley K.J., Moench T.R. Origins of vaginal acidity: high D/L lactate ratio is consistent with bacteria being the primary source. Hum. Reprod. 2001;16:1809–1813. doi: 10.1093/humrep/16.9.1809. [DOI] [PubMed] [Google Scholar]
- Byrd A.L., Belkaid Y., Segre J.A. The human skin microbiome. Nat. Publish. Group. 2018;16:143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- Costea P.I., Zeller G., Sunagawa S., Pelletier E., Alberti A., Levenez F., Tramontano M., Driessen M., Hercog R., Jung F.E. Towards standards for human fecal sample processing in metagenomic studies. Nat. Biotechnol. 2017;35:1069–1076. doi: 10.1038/nbt.3960. [DOI] [PubMed] [Google Scholar]
- Costello E.K., Lauber C.L., Hamady M., Fierer N., Gordon J.I., Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley P., Castriciano S., Chernesky M., Smieja M. Comparison of flocked and rayon swabs for collection of respiratory epithelial cells from uninfected volunteers and symptomatic patients. J. Clin. Microbiol. 2006;44:2265–2267. doi: 10.1128/JCM.02055-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boeck I., van den Broek M.F.L., Allonsius C.N., Spacova I., Wittouck S., Martens K., Wuyts S., Cauwenberghs E., Jokicevic K., Vandenheuvel D. Lactobacilli have a niche in the human nose. Cell Rep. 2020;31:107674. doi: 10.1016/j.celrep.2020.107674. [DOI] [PubMed] [Google Scholar]
- Dzidic M., Boix-Amorós A., Selma-Royo M., Mira A., Collado M. Gut microbiota and mucosal immunity in the neonate. Med. Sci. 2018;6:56. doi: 10.3390/medsci6030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrarese R., Zuppardo R.A., Puzzono M., Mannucci A., Amato V., Ditonno I., Patricelli M.G., Raucci A.R., Clementi M., Elmore U. Oral and fecal microbiota in lynch syndrome. J. Clin. Med. 2020;9:2735. doi: 10.3390/jcm9092735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- France M., Ma B., Gajer P., Brown S., Humphrys M., Holm J., Waetjen L.E., Brotman R., Ravel J. VALENCIA: a nearest centroid classification method for vaginal microbial communities based on composition. Microbiome. 2020:1–15. doi: 10.21203/rs.2.24139/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice E.A., Kong H.H., Conlan S., Deming C.B., Davis J., Young A.C., NISC Comparative Sequencing Program. Bouffard G.G., Blakesley R.W., Murray P.R., Green E.D. Topographical and temporal diversity of the human skin microbiome. Science (New York, N.Y.) 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan M.D., Bartow-McKenney C., Flowers L., Knight S.A.B., Uberoi A., Grice E.A. Research techniques made simple: profiling the skin microbiota. J. Invest. Dermatol. 2019;139:747–752.e1. doi: 10.1016/j.jid.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmaier-Wacker L.K., Lueert S., Roos C., Knauf S. The impact of storage buffer, DNA extraction method, and polymerase on microbial analysis. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-24573-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick S.A., Chen W.Y., Wong T., Kanakamedala B.S., Deveson I.W., Ongley S.E., Santini N.S., Marcellin E., Smith M.A., Nielsen L.K. Synthetic microbe communities provide internal reference standards for metagenome sequencing and analysis. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-018-05555-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan N.A., Young B.A., Minard-Smith A.T., Saeed K., Li H., Heizer E.M., McMillan N.J., Isom R., Abdullah A.S., Bornman D.M. Microbial community profiling of human saliva using shotgun metagenomic sequencing. PLoS One. 2014;9:97699. doi: 10.1371/journal.pone.0097699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heravi F.S., Zakrzewski M., Vickery K., Hu H. Host DNA depletion efficiency of microbiome DNA enrichment methods in infected tissue samples. J. Microbiol. Methods. 2020;170:105856. doi: 10.1016/j.mimet.2020.105856. [DOI] [PubMed] [Google Scholar]
- Hillmann B., Al-ghalith G.A., Shields-cutler R.R., Zhu Q., Gohl D.M., Beckman K.B., Knight R., Knights D. Evaluating the information content of shallow shotgun metagenomics. mSystems. 2018;3:1–12. doi: 10.1128/mSystems.00069-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HMP A review of 10 years of human microbiome research activities at the US National Institutes of Health, Fiscal Years 2007-2016. Microbiome. 2019;7:1–19. doi: 10.1186/s40168-019-0620-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen A.V., Yemaneab B.T., Jass J., Scherbak N. Reference gene selection for qPCR is dependent on cell type rather than treatment in colonic and vaginal human epithelial cell lines. PLoS One. 2014;9:e115592. doi: 10.1371/journal.pone.0115592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaan A.M., Buijs M.J., Brandt B.W., Keijser B.J.F., de Ruyter J.C. Home sampling is a feasible method for oral microbiota analysis for infants and mothers. J. Dent. 2020;100:100023. doi: 10.1016/j.jdent.2020.103428. [DOI] [PubMed] [Google Scholar]
- Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal J., Smyth H.D.C., Ghosh D. International Journal of Pharmaceutics. Vol. 532. Elsevier B.V.; 2017. Physicochemical properties of mucus and their impact on transmucosal drug delivery; pp. 555–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeer Sarah, Oerlemans Eline, Claes Ingmar, Wuyts Sander, Henkens Tim, Spacova Irina, van den Broek Marianne, Tuyaerts Ines, Wittouck Stijn, De Boeck Ilke. Topical cream with live lactobacilli modulates the skin microbiome and reduce acne symptoms. BioRXiv. 2018:1–28. doi: 10.1360/zd-2013-43-6-1064. [DOI] [Google Scholar]
- Lei T., Yang J., Becker A., Ji Y. Methods in Molecular Biology. Vol. 2069. 2020. Identification of target genes mediated by two-component regulators of staphylococcus aureus using RNA-seq technology. [DOI] [PubMed] [Google Scholar]
- Lim Y., Totsika M., Morrison M., Punyadeera C. The saliva microbiome profiles are minimally affected by collection method or DNA extraction protocols. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-017-07885-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lethem M., James S., Marriott C., Burke J. The origin of DNA associated with mucus glycoproteins in cystic fibrosis sputum. Eur. Respir. J. 1990;3:19–23. [PubMed] [Google Scholar]
- Lim M.Y., Park Y.S., Kim J.H., Nam Y. Do. Evaluation of fecal DNA extraction protocols for human gut microbiome studies. BMC Microbiol. 2020;20:1–7. doi: 10.1186/s12866-020-01894-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marotz C.A., Sanders J.G., Zuniga C., Zaramela L.S., Knight R., Zengler K. Improving saliva shotgun metagenomics by chemical host DNA depletion. Microbiome. 2018;6:42. doi: 10.1186/s40168-018-0426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Op De Beeck M., Lievens B., Busschaert P., Declerck S., Vangronsveld J., Colpaert J.V. Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS One. 2014;9:e97629. doi: 10.1371/journal.pone.0097629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovreås L., Forney L., Daae F.L., Torsvik V. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments. Appl. Environ. Microbiol. 1997;63:3367–3373. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C., Walker A.W., Simpson J.T., Loman N.J., Segata N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017;35:833–844. doi: 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- R Core Team . 2020. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Ravel J., Gajer P., Abdo Z., Schneider G.M., Koenig S.S.K., McCulle S.L., Karlebach S., Gorle R., Russell J., Tacket C.O. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. U S A. 2011;108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A., Nikkilä J., Jalanka-Tuovinen J., Immonen O., Rajilić-Stojanović M., Kekkonen R.A., Palva A., de Vos W.M. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J. Microbiol. Methods. 2010;81:127–134. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Susic D., Davis G., O’ Sullivan A.J., McGovern E., Harris K., Roberts L.M., Craig M.E., Mangos G., Hold G.L., El-Omar E.M., Henry A. Microbiome Understanding in Maternity Study (MUMS), an Australian prospective longitudinal cohort study of maternal and infant microbiota: study protocol. BMJ Open. 2020;10:e040189. doi: 10.1136/bmjopen-2020-040189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tackmann J., Arora N., Sebastian T., Schmidt B., Frederico J., Rodrigues M. Ecologically informed microbial biomarkers and accurate classification of mixed and unmixed samples in an extensive cross- study of human body sites. Microbiome. 2018:1–16. doi: 10.1186/s40168-018-0565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenovuo J. Antimicrobial function of human saliva - how important is it for oral health? Acta Odontol. Scand. 1998;56:250–256. doi: 10.1080/000163598428400. [DOI] [PubMed] [Google Scholar]
- Vandeputte D., Tito R.Y., Vanleeuwen R., Falony G., Raes J. Practical considerations for large-scale gut microbiome studies. FEMS Microbiol. Rev. 2017;41:S154–S167. doi: 10.1093/femsre/fux027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis J.R., González-Torres P., Pittis A.A., Bejarano L.A., Cozzuto L., Andreu-Somavilla N., Alloza-Trabado M., Valentín A., Ksiezopolska E., Company C. Citizen science charts two major “stomatotypes” in the oral microbiome of adolescents and reveals links with habits and drinking water composition. Microbiome. 2018;6:218. doi: 10.1186/s40168-018-0592-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D.E., Salzberg S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie J.G., Henderson A. Identity and glycogen-fermenting ability of lactobacilli isolated from the vagina of pregnant women. J. Med. Microbiol. 1969;2:363–366. doi: 10.1099/00222615-2-3-363. [DOI] [PubMed] [Google Scholar]
- Yoon S.H., Ha S.M., Kwon S., Lim J., Kim Y., Seo H., Chun J. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.R., Jenkins K., Araujo-Perez F., Seed P.C., Kelly M.S. Long-term stability of microbiome diversity and composition in fecal samples stored in eNAT medium. MicrobiologyOpen. 2020;9:1–7. doi: 10.1002/mbo3.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasada A.A., Zacharczuk K., Woźnica K., Główka M., Ziółkowski R., Malinowska E. The influence of a swab type on the results of point-of-care tests. AMB Express. 2020;10 doi: 10.1186/s13568-020-00978-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All sequencing data were deposited in ENA: PRJEB45093 (https://www.ebi.ac.uk/ena/browser/view/PRJEB45093).
-
•
The R code generated during this study can be found on GitHub at github.com/Sarahtopia/isala_pilot.