Abstract
Circular RNAs (circRNAs) are a novel class of widespread noncoding RNAs that regulate gene expression in mammals. Recent studies demonstrate that functional peptides can be encoded by short open reading frames in noncoding RNAs, including circRNAs. However, the role of circRNAs in various physiological and pathological states, such as cancer, is not well understood. In this study, through deep RNA sequencing on human endometrial cancer (EC) samples and their paired adjacent normal tissues, we uncovered that the circRNA hsa-circ-0000437 is significantly reduced in EC compared with matched paracancerous tissue. The hsa-circ-0000437 contains a short open reading frame encoding a functional peptide termed CORO1C-47aa. Overexpression of CORO1C-47aa is capable of inhibiting angiogenesis at the initiation stage by suppressing endothelial cell proliferation, migration, and differentiation through competition with transcription factor TACC3 to bind to ARNT and suppress VEGF. CORO1C-47aa directly bound to ARNT through the PAS-B domain, and blocking the association between ARNT and TACC3, which led to reduced expression of VEGF, ultimately lead to reduced angiogenesis. The antitumor effects of CORO1C-47aa on EC progression suggest that CORO1C-47aa has potential value in anticarcinoma therapies and warrants further investigation.
Keywords: hsa-circ-0000437, endometrial cancer, functional peptide, angiogenesis, VEGF
Abbreviations: ChIP, chromatin immunoprecipitation; circRNA, circular RNA; EC, endometrial cancer; IRES, internal ribosomal entrance site; mRNP, mRNA-protein particle; ncRNA, noncoding RNA; OE, overexpression; sORF, short open reading frame; TACC3, transforming acidic coiled-coil 3
Endometrial cancer (EC) is the fifth most common malignancy in women worldwide accounting for 1% to 2% of all deaths from cancer (1, 2). The incidence of EC has steadily increased and is noted to be much higher in developed countries than in developing countries in the past 10 years, and nearly 11.7% of the cases occur in China (3). The risk factors for developing EC include age, high socioeconomic status, and factors related to an excess of estrogen exposure, diabetes, or hypertension, but relatively little is known about inherited risk factors for this disease (4). Thus, there is a need for a deeper understanding of the molecular pathways involved in the occurrence and development of EC.
Noncoding RNAs (ncRNAs) represent the majority of transcripts in a cell and are known to be involved in the development of EC (5, 6). Circular RNAs (circRNAs) are a novel class of widespread ncRNAs that regulate gene expression in mammals. They are closed RNA transcripts generated by back-splicing of a single pre-mRNA. About 85% of circRNAs are aligned in sense orientation to known protein-coding genes and span 1 to 5 exons of the housed protein-coding genes. The expression of circRNAs is often highly conserved across species (7). They are naturally resistant to degradation by exonucleases, thus representing a class of stable RNAs (8, 9). Some circRNAs are known to harbor microRNAs (miRNAs) and function as a sponge to neutralize miRNA (8, 10). For instance, the circRNA CiRS-7 can function as a sponge of miR-7, and the circRNA of testes-specific Sry gene acts as a sponge for miR-138 in mice (11). Besides this, some circRNAs have also been reported to bind to and sequester RNA-binding proteins. For example, circ-Foxo3 interacted with both p21 and CDK2 to form a ternary complex, resulting in the inhibition of cell cycle progression (12). However, the biological function of most circRNAs is still undetermined.
With the development of ribosome nascent-chain complex-bound mRNAs sequencing (RNC-seq) and Ribosome footprints profiling (Ribo-seq), recent studies have demonstrated that functional peptides can be encoded by short open reading frames (sORFs) in ncRNAs (13, 14). Consequently, owing to its cytoplasmic location, the sORFs of some circRNAs can be translated through the internal ribosome entry sites by N6-methyladenosine (15). For example, both circZNF609 and circMbl were reported to be translatable (16). Circ-FBXW7, driven by an internal ribosome entry site, encodes a novel 21-kDa protein (17). circPINTexon2, generated from the long ncRNA LINC-PINT, contains a sORF encoding a tumor-suppressive peptide in human cells (18).
In this work, we generated deep RNA sequencing data from four EC samples and their paired adjacent normal tissues and identified approximately 3169 circRNA candidates. We found that the human circRNA hsa-circ-0000437 is differentially expressed in cancer and paracancerous tissues. Hsa-circ-0000437 contains a sORF encoding a peptide that is termed CORO1C-47aa. We further analyzed the functional role of this peptide in EC development and determined that CORO1C-47aa competes with TACC3 to interact with the PAS-B domain of ARNT, leading to transcriptional suppression of VEGF and inhibition of angiogenesis in EC.
Results
circRNAs expression in endometrial cancer and matched noncancerous tissues
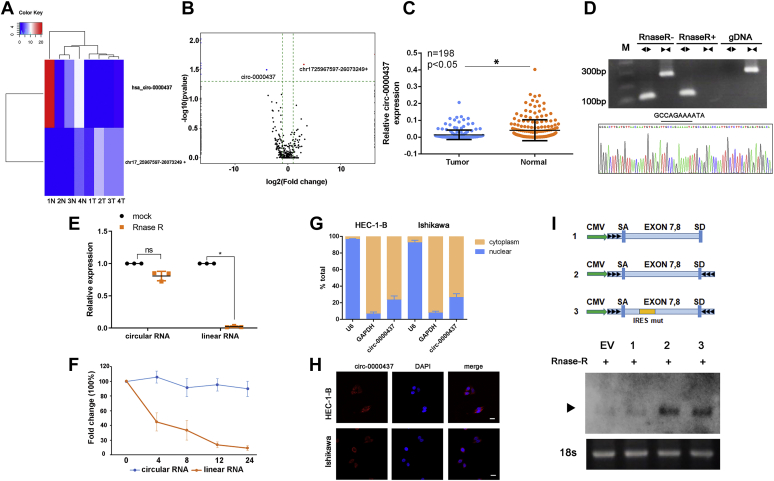
To identify the circRNAs that are potentially involved in the pathogenesis of EC, we performed RNA-Seq analysis on the ribosomal RNA-depleted total RNA from four human EC samples and their paired adjacent noncancerous tissues. The number of candidate circRNAs identified from all samples varied from 89 to 675 with the lower amount in three of four tumors compared with their matched adjacent normal tissues (Fig. S1A). About 65.9% (2087/3169) of the identified circRNAs is known circRNAs in the publicly available circRNA database, circBase (http://www.circbase.org) (19) (Fig. S1B). Most of all identified circRNAs originated from protein-coding exons and others aligned with introns, intergenic region (Fig. S1C). The median length of circRNAs is close to 600 nt, and there was no significant difference between the tumor group and the normal group (Fig. S1D). Next, we compared the expression of circRNAs in EC and their matched adjacent normal tissues. Unsupervised clustering of all identified circRNAs results in clustering of tumors versus noncancerous controls (Fig. 1A). The supervised analysis further identified hsa-circ-0000437 as the top differentially expressed circRNA that showed significant lower expression in tumors than matched normal control (log2FC = −4, p < 0.05, Fig. 1B), and circRNA chr17-25967597-26073249+ as the most significantly elevated circRNA in tumors versus normal controls (log2FC = 3, p < 0.05) (Fig. 1B).
Figure 1.
The characterization of circ-0000437.A, heat map of all differentially expressed circRNAs between normal and tumor tissues. B, the different expressions of circRNAs are shown in the volcano plot. The blue, red, and black points represent downregulated, upregulated, and no statistically significant difference circRNAs in the endometrial cancer tissues compared with the adjacent normal tissues, respectively. The blue indicated circ-0000437. The red indicated chr17-25967597-26073249+. C, the expression level of circ-0000437 was analyzed by qRT-PCR and normalized to GAPDH. The circ-0000437 was expressed at a low level in the endometrial cancer tissues compared with matched adjacent normal tissues. Data are represented as mean ± SD from three independent experiments. D, divergent primers amplify circular RNAs in cDNA but not genomic DNA (gDNA). Convergent primers amplified the PCR products from linear mRNA, which disappeared after RNase R digestion. Next, Sanger sequencing was used to confirm the circ-0000437 junction. E, the circ-0000437 was resistant to RNase R treatment compared with the linear mRNA. Data are represented as mean ± SD from three independent experiments. F, qRT-PCR for the abundance of circRNA and mRNA in cells treated with actinomycin D at indicated time point. Data are represented as mean ± SD from three independent experiments. G, circ-0000437 was detected in different cell fractions. Nuclear and cytoplasmic RNA was extracted, and junction primers were used for circ-0000437 detection. U6 was used as internal control of nuclear RNA, and GAPDH was used as internal control for cytoplasmic RNA. Data are represented as mean ± SD from three independent experiments. H, fluorescence in situ hybridization with junction-specific probes indicates the cellular localization of circ-0000437 (the scale bars represent 100 μm). I, 1, exons of the circ-0000437 gene were cloned between splicing acceptor (SA) and splicing donor (SD) sequences with upstream flanking repeat sequences. 2, exon 4 and 5 sequences of the circ-0000437 gene were cloned between SA and SD with both sides of flanking repeat sequences. 3, compared with vector 2, the putative internal ribosomal entrance site sequences were mutated. Total RNA was treated with RNase R and subjected to Northern blotting using circ-0000437 junction probes.
Expression of circ-0000437 is reduced in EC tissues
To validate whether circ-0000437 is differentially expressed in EC compared with their matched adjacent noncancerous tissues, we performed RT-qPCR assay using two divergent primer sets that amplify only the circular product of hsa-circ-0000437 to determine the levels of circ-0000437 in a total of 198 paired EC samples and matched noncancerous tissues. Circ-0000437 was significantly underexpressed in EC compared with their matched adjacent noncancerous tissues (Fig. 1C, n = 198, p < 0.05). However, the other circRNA chr17-25967597-26073249+ cannot be detected by qRCR (data not shown). Circ-0000437 originates from the circularization of the seventh and eighth exons of its host gene CORO1C. To determine whether circ-0000437 is expressed in EC, we designed two sets of primers for hsa-circ-0000437 locus: a divergent set that amplifies only the circular transcript and a convergent set that amplifies the linear transcript. Our results showed that the circular form was amplified using the divergent primers. cDNA and genomic DNA of EC were used as templates, and as expected, no amplification was observed with the divergent primers on genomic DNA (Fig. 1D). Furthermore, in contrast to the linear control RNAs, the circRNA was resistant to the treatment of RNase R (Fig. 1E), a highly processive 3′ to 5′ exoribonuclease that does not act on circRNAs (20, 21), thus validating the circular nature of circ-0000437. Sanger sequencing of the circular PCR product of hsa-circ-0000437 confirmed the sequence of circ-0000437 (Fig. 1D, lower panel) that matched the reported sequence in circbase database.
Biological characterization of circ-0000437
Next, we investigated the stability of transcripts from the hsa-circ-0000437 locus. Cells were treated with actinomycin D, a transcription inhibitor, and harvested at indicated time points for total RNA isolation. There was little change in circ-0000437 levels following incubation with actinomycin D for 24 h, while the quantity of the linear transcript from the same locus decreased dramatically with only 10% to 20% remaining 24 h after treatment (Fig. 1F).
To determine the cellular localization of circ-0000437, we isolated the cytoplasmic and nuclear fraction of EC cells. The levels of the nuclear control transcript (U6) and cytoplasmic control transcript (GAPDH mRNA) were also quantitated by RT-qPCR, which indicated a successful fractionation of the nuclear and cytoplasmic fractions (Fig. 1G). The qRT-PCR analysis of circ-0000437 by real-time RT-qPCR showed that the circ-0000437 was mostly present in the cytosol (Fig. 1G). Next, RNA fluorescence in situ hybridization (FISH) assays were performed to verify the cellular localization of circ-0000437. In agreement with the cellular fractionation data, circ-0000437 was mainly detected in the cytoplasm of EC cells via FISH assays (Fig. 1H). Endogenous circ-0000437 was further detected using a junction-specific biotin-labeled probe in Northern blotting (Fig. 1I, EV). We also overexpressed circ-0000437 using a synthetic circRNA expression vector, which was induced by side flanking repeat sequences with splicing acceptor and donor. However, the control vector that lacked the downstream flanking sequences was not overexpressed (Fig. 1I).
It is known that circRNAs can act as miRNA sponges to regulate gene expression in different cell types (22). To determine whether circ-0000437 regulates EC development through the same mechanism, we performed RNA immunoprecipitation assays using an anti-AGO2 antibody. CDR1as, a molecular sponge of miR-7 (16), was enriched in AGO2 immunoprecipitate (Fig. S2A), validating a successful pulldown of AGO2–RNA interaction complexes. However, circ-0000437 was not detected in AGO2 immunoprecipitate (Fig. S2A), suggesting that circ-0000437 may regulate the occurrence and development of EC tumors through other mechanisms.
Circ-0000437 encodes a peptide
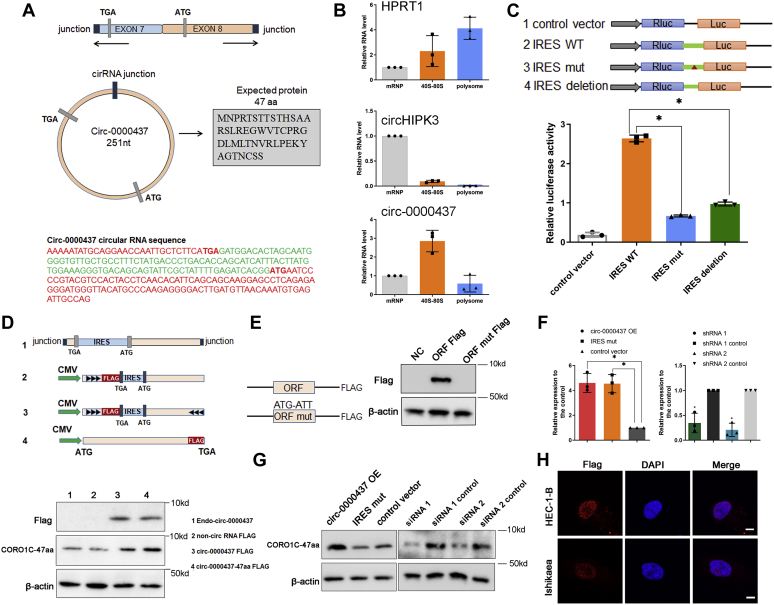
As some circRNAs have been reported to encode functional peptides, we searched the genomic sequence of cir-0000437 and found a putative 144-nt sORF with the potential to encode a 47-aa peptide in circ-0000437, spanning from the AUG codon in the exon 8 of the host gene over the splice junction to a STOP codon in the exon 7 of the host gene, which we termed as CORO1C-47aa (Fig. 2A and Fig. S3, A and B). Moreover, no homologous nucleotide sequences with the human 144-nt ORF sequences that encode CORO1C-47aa peptide were found in any other species. To determine whether this putative sORF (circORF) is functional, we first used sucrose gradient fractionation to test circ-0000437 loading onto polysomes. The mRNA-protein particles (mRNPs) were separated into three fractions: nonribosome (mRNPs without any ribosome), 40S-80S (mRNPs associated with ribosome but not being translated), and polysome (mRNPs being actively translated). Quantitation of HRPT1 and a circRNA with no ORF, circ-HIPK3, in these three fractions by qRT-PCR showed that HRPT1, but not circ-HIPK3, was abundantly detected in the polysome fraction (Fig. 2B), indicating a successful fractionation of mRNPs. Accordingly, a small but significant portion of circ-0000437 was sedimented in the polysome fraction (Fig. 2B), suggesting that cicr-0000437 may be actively translated.
Figure 2.
The coding ability of circ-0000437.A, the circular RNA sequence of circ-0000437 and the putative open reading frame (ORF) in circ-0000437. The green sequence is the internal ribosomal entrance site (IRES) and the red one is the sequencer of ORF. B, fraction of circ-0000437 or HPRT mRNA (positive control) and circHIPK3-1 (negative control) associated with mRNP, 40S-80S and polysome as measured by qRT-PCR after sucrose fractionation of C2C12 lysates. The values are represented as mean and standard error of three independent fractionations. C, the putative IRES activity in circ-0000437 was tested. The relative luciferase activity of Luc/Rluc in the above vectors was tested. Values are the average ± SD of three independent experiments. D, the vector set for detecting circ-0000437 encoded protein. 1, endo-circ-0000437, illustration showing how endogenous circ-0000437 formed. 2, non-circRNA FLAG: exons of circ-0000437 were cloned without splicing acceptor, splicing donor, FLAG tag was moved to the stop codon of the ORF. 3, circ-0000437-FLAG: exons of circ-0000437 were cloned between splicing acceptor, splicing donor, and side flanking repeat sequences. FLAG tag was moved to the stop codon of the ORF. 4, circ-000043747aa FLAG: 47-aa ORF was cloned downstream of a linear CMV promoter. E, left, diagram of the FLAG fusion constructs used for transfection. The start codon ATG of the circ-0000437 ORF is mutated to ATT. Right, ORF-FLAG fusion protein levels were determined by Western blotting with anti-FLAG antibody. F, relative expression levels of circ-0000437 as determined by qPCR in HEC-1-B cells transfected with circ-0000437 or circ-0000437-targeting siRNAs. Results are shown as means ± SD relative to vector control. G, the protein level of CORO1C-47aa were detected by Western blot in HEC-1-B cells transfected with circ-0000437 or circ-0000437-targeting siRNAs. H, immunofluorescence microscopy images of the cellular localization of the CORO1C-47aa protein in HEC-1-B and Ishikawa cells (the scale bars represent 50 μm).
Recent studies have demonstrated that the translation of circRNAs depends on the presence of an internal ribosomal entrance site (IRES), a translation regulatory sequence that is required for 5′-cap independent translation (23). circ-000437 was matched in circRNADb, one database for human circRNAs. circRNADb provided the IRES elements of circ-000437. Accordingly, we identified a putative IRES upstream of the translation start site of circ-0000437 (Fig. 2A, bottom panel, sequence in green color; Fig. S3C). To verify whether this candidate IRES is functional, we cloned the full-length wildtype or mutated putative circ-0000437 IRES sequences between the Renilla luciferase (Rluc) and Firefly luciferase (Luc) reporter genes of a dual-luciferase vector system (Fig. 2C and Fig. S3B). Therefore, the ratio of Luc/Rluc activity will indicate the efficiency of the translation of firefly luciferase. Consequently, the luciferase assay results showed that cells transfected with the full-length wildtype circ-0000437 IRES induced the Luc/Rluc activity dramatically compared with the empty control (Fig. 2C). In contrast, mutation or partial deletion of the IRES significantly reduced the activity of firefly luciferase induced by wildtype IRES (Fig. 2C), suggesting that the IRES of circ-0000437 is bioactive.
To test the protein-coding ability of circ-0000437, we cloned the full-length circ-0000437 into a pLCDH-ciR vector (plasmid 3) that was able to produce circular transcripts. We also constructed two more FLAG-tagged plasmids: one has disrupted splicing site (plasmid 2) and the other has linearized circ-0000437-aa sequencing (plasmid 4) (Fig. 2D, upper panel). As the FLAG-coding sequence is put immediately upstream of the STOP codon, a flagged protein can be produced only upon the formation of a circular transcript in plasmids 2 and 3. Transfection of the wildtype circ-0000437 overexpression (OE, plasmid 3) increased the expression of circ-0000437 by 4.6-fold in HEC-1B and Ishikawa EC cells (Fig. 2F, left panel; Fig. S2C), which was not impacted by introducing a mutation into the IRES. Consequently, the CORO1C-47aa can be detected in either circ-0000437 OE vector (plasmid 3) or its linearized counterpart (plasmid 4) transfected cells by anti-FLAG antibody. Moreover, we generated an antibody against the 47-aa peptide, which detected the expression of both endogenous and FLAG-tagged proteins (Fig. 2D, down panel). Next, we determined whether an in-frame mutation of the ATG codon of CORO1C-47aa can impair the initiation of translation. We constructed an ORF mutation expression vector, in which the sequences encoding start codons ATGGTG were mutated to ATTGTT, and a FLAG-tag to the C terminus of CORO1C-47aa. Expression of CORO1C-47aa was detected in cells transfected with the wildtype CORO1C-47aa-FLAG plasmid, but not in cells transfected with the plasmid carrying ORF mutation, by anti-FLAG Western blotting (Fig. 2E). Because CORO1C-47aa is downregulated in EC tissues compared with normal EC tissues (Fig. S2E), we generated cell lines with stable overexpression of either wildtype circ-0000437 OE vector or a circ-000043 OE plasmid with IRES mutation. Although the IRES mutation did not influence the expression of circ-0000437 (Fig. 1I), only overexpression of wildtype circ-0000437 OE plasmid led to upregulation of CORO1C-47aa (Fig. 2G). Through FLAG-labeling immunofluorescence assay, we found that CORO1C-47aa was predominantly localized inside the nucleus of both HEC-1-B and Ishikawa EC cells (Fig. 2H).
Furthermore, we used two siRNAs that specifically target the circular junction of circ-0000437. These two siRNAs successfully reduced both circ-0000437 expression (Fig. 2F, right panel) and the CORO1C-47aa peptide level with no significant impact on the expression of the host gene CORO1C (Fig. S2, F and G). The expression of neither circ-0000437 nor the 47-aa peptide was altered by two siRNAs specifically designed to target and silence CORO1C gene. Furthermore, stable overexpression of CORO1C did not elevate CORO1C-47aa expression (Fig. S2, H and I). Taken together, these results demonstrated that the circ-0000437 encodes the peptide CORO1C-47aa independently of its host gene.
Effects of ectopic CORO1C-47aa expression on EC cell proliferation
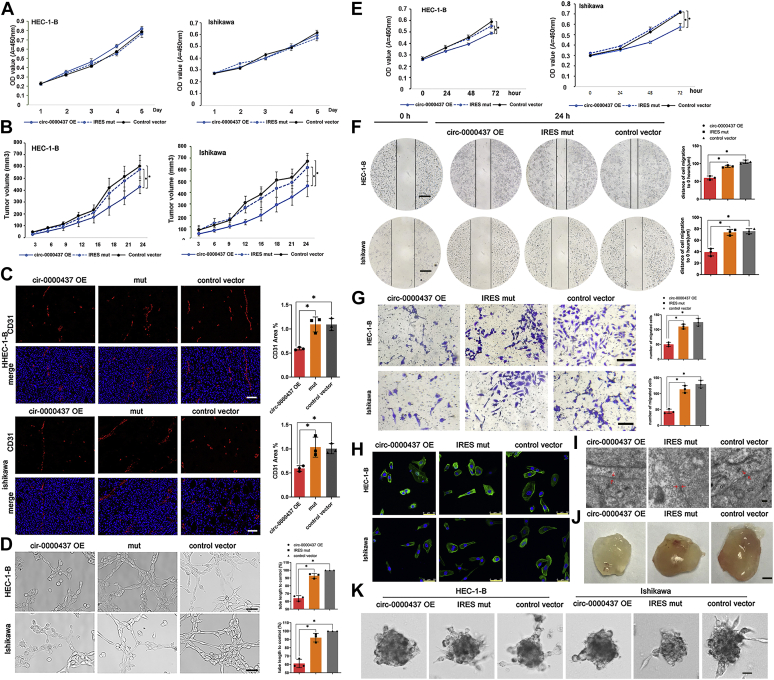
To determine the impact of CORO1C-47aa on cell proliferation, we first performed CCK-8 assays on the HEC-1B and Ishikawa cells that stably expressed CORO1C-47aa. The results from the CCK-8 assays revealed no significant difference in the cell proliferation of cells overexpressing either wildtype or IRES mutant circ-0000437 compared with the control cells (Fig. 3A). Next, we examined the biological effect of CORO1C-47aa overexpression on tumor growth in mice. The subcutaneous xenografts of EC cells overexpressing the wildtype circ-0000437 plasmid significantly grew slower than those of their counterparts expressing empty vector or IRES mutant circ-0000437 plasmid (Fig. 3B, n = 5), suggesting that CORO1C-47aa overexpression, but not the circRNA transcript itself, suppressed the tumor growth. The tumor volume doubling time (200 mm3 → 400 mm3) increased from 5 days in control HEC-1B cells to 8 days in CORO1C-47aa overexpression cells (p < 0.05). The tumor volume doubling time (200 mm3 → 400 mm3) increased from 6 days in control Ishikawa cells to 9 days in CORO1C-47aa overexpression cells (p < 0.05).
Figure 3.
Biological functions of CORO1C-47aa.A, cell proliferation analysis of circ-0000437 OE, circ-0000437 IRES mutation, and control endometrial cancer cells at different time points (mean ± SD, n = 3). B, tumor growth in xenograft mice subcutaneously implanted endometrial cancer cells that were transfected with indicated constructs (mean ± SD, n = 5, ∗p < 0.05). C, CD31 staining was performed on xenograft mice tumor tissue sections from circ-0000437 OE, circ-0000437 IRES mutation, and control groups. CD31 staining was quantified using ImageJ. Each value is the mean ± SD of three independent experiments (the scale bars represent 100 μm). D, an endothelial tube formation assay was performed using HUVEC and conditioned media from HEC-1-B and Ishikawa cells that stably express circ-0000437 OE, circ-0000437 IRES mutation (mean ± SD, n = 3, ∗p < 0.05) (the scale bars represent 100 μm). E, cell proliferation analysis of circ-0000437 OE, circ-0000437 IRES mutation, and control HUVEC cells at different time points (mean ± SD, n = 3). F, a wound healing assay was performed using HUVEC and conditioned media from HEC-1-B and Ishikawa cells that stably express circ-0000437 OE, circ-0000437 IRES mutation (mean ± SD, n = 3, ∗p < 0.05) (the scale bars represent 200 μm). G, an endothelial migration assay was performed using HUVEC and conditioned media from HEC-1-B and Ishikawa cells that stably express circ-0000437 OE, circ-0000437 IRES mutation (mean ± SD, n = 3, ∗p < 0.05) (the scale bars represent 100 μm). H, phalloidine-FITC staining of microfilament and immunofluorescence staining for HUVEC cells and conditioned media from HEC-1-B and Ishikawa cells that stably express circ-0000437 OE, circ-0000437 IRES mutation. The nucleus was counterstained with DAPI (the scale bars represent 50 μm). I, representative photographs of electron microscopy in the indicated experimental conditions (the scale bars represent 200 nm). J, representative photographs of Matrigel plugs in the indicated experimental conditions (the scale bars represent 5 mm). K, representative microphotographs of spheroids incubated with CORO1C-47aa (the scale bars represent 100 μm). IRES, internal ribosomal entrance site; OE, overexpression.
Given that CORO1C-47a decreased the growth of EC cells in vivo but not in vitro, we speculated that CORO1C-47aa may negatively regulate tumor development via controlling the formation of blood vessels running through endometrium tumors. To address whether the growth inhibition by CORO1C-47aa was attributed to the disruption of angiogenesis, we analyzed the expression of CD31, a marker of microvessel density (24), by immunofluorescence. Compared with the IRES mutated and control groups, the expression of CD31 was significantly reduced in the CORO1C-47aa overexpression group (Fig. 3C). In addition, an endothelial tube formation assay was performed to determine whether CORO1C-47aa overexpression influences endothelial network formation on matrix matrigel. The results showed that CORO1C-47aa overexpression significantly impaired the ability of HUVEC cells to form capillary tubes compared with the control (Fig. 3D).
CORO1C-47aa represses tumor angiogenesis
Angiogenesis was inseparable from endothelial cell proliferation, migration, and differentiation. Therefore, we cultured endothelial cells in condition medium from HEC-1-B and Ishikawa cells transfected with CORO1C-47aa overexpression vectors. First, we performed HUVEC cells proliferation assay. The endothelial cell proliferation was significantly decreased in the condition medium collected from either cell line overexpressing CORO1C-47aa (Fig. 3E). Next, to investigate the influence of CORO1C-47aa expression on HUVEC cell migration, we conducted wound healing assays and transwell assays using HUVEC cells cultured in conditioned medium. As shown in Figure 3, F and G, both the wound healing assays and transwell assays demonstrated that the migration capabilities of HUVEC cells were significantly suppressed in the conditioned medium collected from cells with CORO1C-47aa overexpression compared with those transfected with an empty vector or an IRES mutation plasmid.
Furthermore, we next determined the effect of CORO1C-47aa on the endothelial cell cytoskeleton. In HUVEC cells cultured in condition medium from empty vector–expressing cells, F-actin fluorescent staining showed that the skeleton surrounding the cells formed into a nest column (Fig. 4H), which is not conducive to cell migration (25). In contrast, the skeleton of HUVEC cells treatment with CORO1C-47aa-overexpressing medium failed to form apparent rearrangement of F-actin (Fig. 3H). We next determined the intercellular junction of HUVEC cells by electron microscopy. Tight intercellular connections were detected in both the control and IRES mutation groups, whereas the intercellular connections of HUVEC cells were significantly induced with decreased intercellular spacing when cultured in medium from CORO1C-47aa-overexpressing cells (Fig. 3I).
Figure 4.
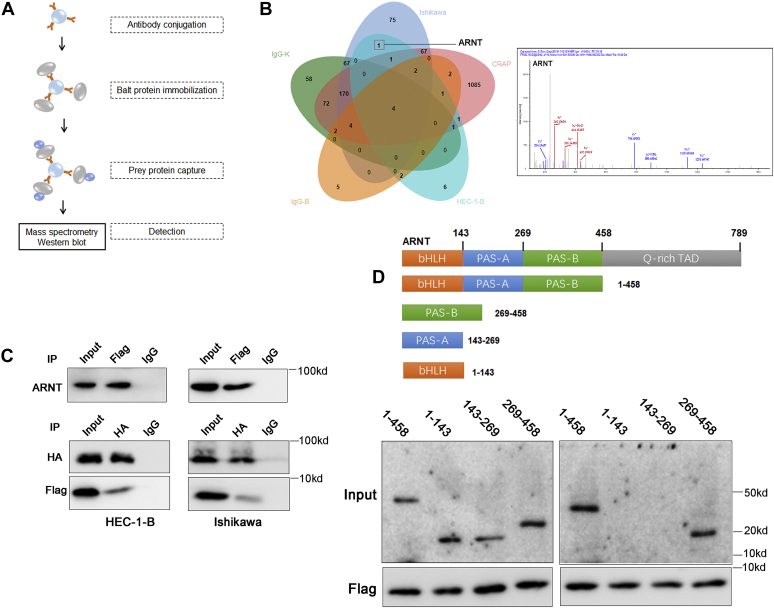
CORO1C-47aa competitively binds to ARNT and inhibits the binding of TACC3 to its PAS-B ligand.A, diagram of the coimmunoprecipitation experiments. B, mass spectrometric analysis of the coimmunoprecipitation experiments. C, top, CORO1C-47aa-FLAG plasmid was transfected into cells, and the CORO1C-47aa-FLAG complexes were coimmunoprecipitated by anti-FLAG antibody, and only ARNT was detected. Bottom, CORO1C-47aa-FLAG and ARNT-HA plasmids were transfected into cells, and the ARNT-HA complexes were coimmunoprecipitated by anti-HA antibody, and CORO1C-47aa-FLAG was detected. D, coimmunoprecipitation assays revealed the interaction of CORO1C-47aa with ARNT via the PAS-B region of ARNT.
To further support the notion that CORO1C-47aa can suppress angiogenesis, Matrigel plug assays were performed to measure neovascularization in mice. As shown in Figure 3J, the neovascularization in mice injected subcutaneously with Matrigel is significantly inhibited in the CORO1C-47aa overexpression group (Fig. S4, A and B). Finally, the effect of CORO1C-47aa was evaluated in an in vitro tridimensional model of angiogenesis in which HUVEC spheroids invade a three-dimensional fibrin matrix, generating endothelial sprouts as a result of the localized breakdown of the extracellular matrix that occurs together with HUVEC migration and growth (26). As shown in Figure 3K and Fig. S4C, CORO1C-47aa caused a reduction of HUVEC sprouting. Taken together, these results indicated that CORO1C-47aa is capable of inhibiting angiogenesis at the initiation stage by suppressing endothelial cell proliferation, migration, and differentiation.
Mass spectrometry identified the interaction between CORO1C-47aa and ARNT
To explore the potential molecular mechanisms underlying the antiangiogenesis functions of CORO1C-47aa, we performed coimmunoprecipitation using an anti-FLAG antibody in HEC-1-B and Ishikawa cells overexpressing CORO1C47aa-FLAG to identify the interacting molecules of CORO1C-47aa by mass spectrometry (Fig. 4A). We identified 76 and 7 specific CORO1C-47aa-interacting proteins in Ishikawa and HEC-1B cells, respectively (Fig. 4B and Table S3). Only one protein, Aryl hydrocarbon receptor nuclear translocator (ARNT), interacted with CORO1C-47aa-FLAG in both cell lines. Subsequent IP using the anti-FLAG antibody confirmed the interaction between CORO1C-47aa-FLAG and endogenous ARNT in both cell lines (Fig. 4C, upper panel). The reciprocal IP using an anti-HA antibody in cells cotransfected with ARNT and CORO1C-47aa also identified FLAG-tagged CORO1C-47aa (Fig. 4C, bottom panel), further confirming the interaction between CORO1C-47aa and ARNT.
ARNT is the obligate heterodimeric partner for the basic helix–loop–helix (bHLH)-PAS (PER-ARNT-SIM) proteins aryl hydrocarbon receptor (AhR) and hypoxia-inducible factor-α (HIF-α), which serve as environmental sensors for xenobiotics and hypoxia, respectively (27). To identify the key functional domain that mediates the interaction, we constructed plasmids expressing truncated ARNT proteins with deletion of bHLH or PAS domains (Fig. 4D, left panel). We divide the functional domains of ARNT into three parts: 1 to 458 contains bHLH, PAS-A, PAS-B, three functional domains; 269 to 458 contains the PAS-B domain; 143 to 269 contains the PAS-A domain; 1 to 143 contains the bHLH domain. These truncated proteins were expressed at comparable levels as the wildtype ARNT protein, but deletion of the PAS-A and PAS-B domains abrogated the interaction (Fig. 4D, right panel). Moreover, only in PAS-B, but not PAS-A, the domain itself is sufficient for maintaining the interaction with FLAG-CORO1C-47aa, suggesting that CORO1C-47aa bound to ARNT through its PAS-B domain (269–458 aa) (Fig. 4D).
CORO1C-47aa competes with TACC3 to bind to ARNT and suppress VEGF via its PAS-B domain
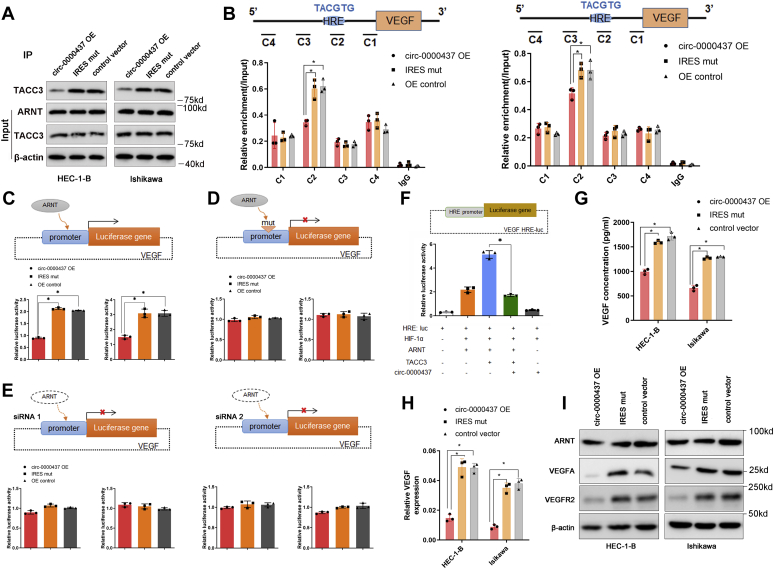
Previous research has shown that the PAS-B domain of ARNT interacts directly with the coactivator transforming acidic coiled-coil 3 (TACC3) and promotes tumor angiogenesis through upregulating the expression of the HIF target gene VEGF (28, 29). To determine whether CORO1C-47aa competes with TACC3 to bind to ARNT and suppress VEGF expression, we performed IP assays using anti-ARNT antibody in EC cells overexpressing either an empty vector or the circ-0000437 vector with or without the capability to translate CORO1C-47aa. In both HEC-1B and Ishikawa cells, overexpression of wildtype circ-0000437, but not empty vector or the circ-0000437 with mutant IRES, significantly reduced the interaction between ARNT and TACC3 (Fig. 5A). To further investigate the interactions between cis-regulatory elements in the HRE promoter and the ARNT and TACC3 proteins, we performed chromatin immunoprecipitation (ChIP) in EC cells. Both ARNT and TACC3 bound to the promoter of VEGF with significant enrichment around the regions of the VEGF HRE in HEC-1-B cells, which was downregulated by overexpression of CORO1C-47aa (Fig. 5B).
Figure 5.
CORO1C-47aa suppressing VEGF expression.A, coimmunoprecipitation assays revealed that CORO1C-47aa competitively bound to ARNT inhibits the binding of TACC3 to its PAS-B ligand. B, chromatin immunoprecipitation (ChIP) assays showed enrichment of ARNT and TACC3 at VEGF in circ-0000437 OE, circ-0000437 IRES mutation, and control EC cells. Coprecipitated DNA was analyzed by qPCR using amplicons C1 to C4 (mean ± SD, n = 3, ∗p < 0.05). C, luciferase reporter assay for VEGF promoter in circ-0000437 OE, circ-0000437 IRES mutation, and control EC cells. The reporter constructs expressing the luciferase gene under full-length VEGF gene promoter (mean ± SD, n = 3, ∗p < 0.05). D, luciferase reporter assay for VEGF promoter in circ-0000437 OE, circ-0000437 IRES mutation, and control EC cells. The reporter constructs expressing the luciferase gene under VEGF promoter deleted (mean ± SD, n = 3, ∗p < 0.05). E, luciferase reporter assay for VEGF promoter in circ-0000437 OE, circ-0000437 IRES mutation, and control EC cells. The reporter constructs expressing the luciferase gene under downregulated ARNT cells (mean ± SD, n = 3, ∗p < 0.05). F, luciferase reporter assay for VEGF HRE promoter in the indicated vectors. G, the inhibitory effect of CORO1C-47aa on VEGF secretion in cancer cells. H, the mRNA level of VEGF in circ-0000437 OE, circ-0000437 IRES mutation, and control EC cells. I, the protein level of ARNT, VEGFA, VEGFR2 in circ-0000437 OE, circ-0000437 IRES mutation, and control EC cells. EC, endometrial cancer; IRES, internal ribosomal entrance site; OE, overexpression.
To evaluate the effect of CORO1C-47aa on VEGF transcription, we then cloned the proximal promoter region of VEGF (HRE) into the pGL3-basic luciferase vector to construct a reporter plasmid (pGL3-VEGF HRE). We also constructed another plasmid, pGL3-VEGF ΔHRE, with the HRE sequences being mutated. The luciferase activity of EC cells cotransfected with pGL3-VEGF HRE and an empty vector was 2-fold higher than that of EC cells transfected with pGL3-VEGF ΔHRE and an empty vector (Fig. 5, C and D, gray bar), indicating that the VEGF HRE is functional. Accordingly, re-expression of CORO1C-47aa by transfecting the wildtype circ-0000437 vector significantly reduced the luciferase activity driven by VEGF HRE without any effect on that of pGL3-VEGF ΔHRE (Fig. 5C). Furthermore, the same experiment was performed in ARNT downregulated cells, and no significant difference was found in the luciferase activity (Fig. 5E). Because TACC3 depletion significantly decreased the activation of a VEGF hypoxia response element luciferase reporter (29), we cotransfected the pGL3-VEGF HRE reporter vector together with or without TACC3, ARNT, and CORO1C-47aa. The result showed that coexpression of HIF1a and ARNT activated the transcription of luciferase and simultaneous overexpression of TACC3 dramatically increased the luciferase activity, which was completely attenuated by CORO1C-47aa overexpression (Fig. 5F), suggesting that CORO1C-47aa competes with TACC3 in the activation of VEGF promoter.
To explore if CORO1C-47aa can decrease the secretion of VEGFA in the cell microenvironment, the level of VEGFA was determined by ELISA. The results showed that CORO1C-47aa upregulation resulted in decreased VEGFA secretion in HEC-1-B and Ishikawa cells, whereas CORO1C-47aa downregulation led to higher VEGFA production (Fig. 5G). The mRNA level of VEGF also decreased in the CORO1C-47aa-overexpressing cells (Fig. 5H). We finally performed Western blot assays to examine the protein levels of VEGFA and VEGFR2 in EC cells. As shown in Figure 5I, the expression levels of VEGFA and VEGFR2 proteins were reduced in the CORO1C-47aa-overexpressing cells, whereas the expression levels of ARNT protein were not altered. These observations support that CORO1C-47aa competitively binds to ARNT and inhibits the coactivator effect of TACC3 on VEGF promoter, resulting in the suppression of VEGF expression.
Discussion
Accumulating evidence indicates that large numbers of circRNAs exist in the human transcriptome, which may have important roles in regulating cellular function (8, 30, 31). However, their involvement in the development of tumors, especially endometrial cancer, remains mostly undescribed. Emerging studies have indicated the coding potential of some circRNAs, and their translated proteins may have a role in the pathogenesis of cancer. Most of the circRNAs reported so far are exon-containing circRNAs. Our profiling of four EC and their adjacent normal tissues identified that circ-0000437 was underexpressed in EC and could encode one 47aa peptide.
Some circRNAs possess miRNA-binding sites and function as sponges to arrest miRNA functions. CDR1as serves as a molecular sponge of miR-7 and conducts important functions in different biological or pathological progress (32). circNT5E functions as an oncogenic circRNA and a sponge of miR-422a in glioblastoma (33). When we overexpressed circ-0000437, the mRNA expression level of its host gene CORO1C was not changed. Meanwhile, our AGO2 RNA immunoprecipitation assays showed that circ-0000437 could not bind to AGO2 and form the RISC complex, suggesting that it does not function as a miRNA sponge.
Recently, several lines of evidence suggest that ncRNAs may encode functional peptides. It has been discovered that the lncRNA HOXB-AS3 encodes a conserved 53-aa peptide that suppresses colon cancer growth (34). Circ-ZNF609 has also been reported to contain an open reading frame spanning from the start codon and terminating at an in-frame stop codon, created upon circularization (16). It has been further suggested that circRNAs can be translated in vitro and in vivo in a cap-independent manner (35). In our study, we identified and validated a 144-nt small ORF that encodes a highly conserved 47-aa peptide termed CORO1C-47aa in circ-0000437, spanning the splice junction from the putative AUG of the host gene to a STOP codon. In agreement with the notion that an IRES is required for translation initiation in 5′-cap-independent coding RNAs (17, 18), we further verified the existence of a functional IRES inside the circ-0000437, spanning from the termination to the initiation codons, that drives IRES-dependent translation of CORO1C-47aa. Thus, it remains possible that additional peptides are encoded in the many RNAs that are currently annotated as ncRNAs (36).
In our study, based on a number of experiments we found that expression of CORO1C-47aa repressed HUVEC cell proliferation and tumor angiogenesis. Angiogenesis plays a major role in EC tumor growth, development and progression, and metastasis (37, 38). It has been reported that antiangiogenic therapy holds promise for treating malignancies, including endometrial cancer (39). As an angiogenic factor, VEGFA plays a crucial role in promoting the proliferation and migration of endothelial cells and increasing the permeability of tumor-related blood vessels through binding the tyrosine kinase receptors flt-1 (VEGFR-1) and KDR (VEGFR-2) (40). Our Western blotting results verified that the expression levels of VEGFA and VEGFR2 proteins were decreased significantly by overexpressing CORO1C-47aa. Microvascular density has been commonly used to measure tumor-related angiogenesis (41). In comparison with control nude mice, those in the CORO1C-47aa upregulated group had decreased CD31–microvascular density, suggesting that CORO1C-47aa can inhibit angiogenesis. Future studies are necessary to determine whether high levels of circ-0000437 correlate with a better prognosis in endometrial cancer as well as other cancer types. Given that CORO1C-47aa is very small in size, it may also hold the potential for further therapeutic development.
HIF1 is the key transcriptional effector of the hypoxia response in eukaryotes, coordinating the expression of genes involved in angiogenesis (42). The regulation of HIF1 target genes including VEGF depends on the participation of several transcription cofactors (29). Although the C-terminal transactivation domain of HIF-α subunit plays a major role in this process by directly interacting with the global transcription coactivator CBP/p300, the PAS-B domain of ARNT (HIF1-β) provides an additional recruitment site by binding to the coactivator transforming TACC3 (43). Our IP-MS study in CORO1C47aa-FLAG overexpressing cells identified ARNT as the only common interactor of CORO1C-47aa. Immune coprecipitation assays by using anti-ARNT antibody showed that the enrichment of TACC3 was remarkably reduced in CORO1C-47aa-overexpressing cells. The luciferase reporter result showed that TACC3 facilitates the recruitment of ARNT protein to HRE sites and promotes VEGF gene transcription, which was attenuated by overexpression of CORO1C-47aa, suggesting that CORO1C-47aa competes with TACC3 to bind to the PAS-B domain of ARNT and suppress VEGF transcription and secretion. Thus, our study identified an additional mechanism of how VEGF expression is regulated during hypoxia response, which warrants further testing in other cell types to ascertain whether this is a common regulation pathway for hypoxia–VEGF biology.
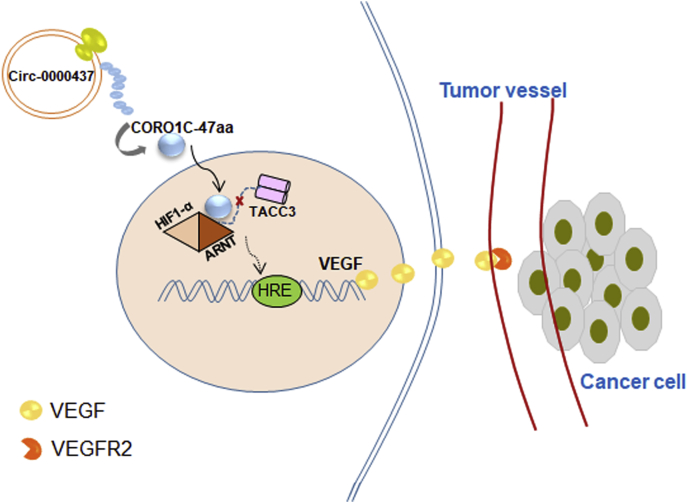
In summary, we have demonstrated that CORO1C-47aa may function as a negative regulator in tumor angiogenesis via blocking the association between ARNT and TACC3 and then reduces the expression of VEGFA. This process leads to a decrease in both expression and secretion of VEGFA, which ultimately leads to reduced angiogenesis (Fig. 6). These findings show a new correlation between CORO1C-47aa and tumor angiogenesis and unveil the mechanisms of CORO1C-47aa in protecting against carcinogenesis. As revealed by this research, it is emphasized that CORO1C-47aa is an important target in conceiving effective antiangiogenesis treatment of EC.
Figure 6.
Graphical representation of the function of CORO1C-47aa in EC. CORO1C-47aa translated from circ-0000437 functions as a negative regulator in tumor angiogenesis via blocking the association between ARNT and TACC3 and then reduces the expression of VEGFA. This process leads to a decrease in both expression and secretion of VEGFA, which ultimately leads to reduced angiogenesis.
Experimental procedures
Study subjects
The subjects participating in this study are homogenous groups of ethnic Han Chinese. To validate the RNA expression data, we analyzed a total of 198 paired EC tissues and matched noncancerous tissues obtained during surgical resection from the Peking University Third Hospital (Beijing, China) and affiliated hospitals of Soochow University (Suzhou, China). Four pairs of human EC and matched noncancerous tissues for RNA-Seq were obtained from the affiliated hospitals of Soochow University. None of the patients received chemotherapy or radiotherapy treatment before the surgery. The Medical Ethics Committee of Peking University Third Hospital and Soochow University approved this study. These studies abide by the Declaration of Helsinki principles. The clinical characteristics of all the patients are listed in Table S1.
RNA extraction and real-time quantitative polymerase chain reaction
RNase R treatment was performed as follows: 1 to 10 mg of total RNA was diluted in 20 ml of water with 4u RNase R/mg unless differently stated and 2 ml of enzyme buffer (Epicenter), then incubated 15 min at 37 °C and purified by phenol–extraction. Quantitative RT-PCR was performed to examine the expression level of mRNA as described (6). The primer sequences are shown in Table S2.
RNA-Seq and chimeric transcripts detection
RNA-Seq was performed on RNAs extracted from four EC samples and their peripheral normal tissues as described (8). Briefly, total RNAs were treated with RNase R to degrade the linear RNAs as described above and purified using RNeasy MinElute Cleanup Kit (Qiagen). Next, a strand-specific library was constructed by using a VAHTS Total RNA-seq (H/M/R) Library Prep Kit from Illumina (Vazyme) and sequenced on the Illumina HiSeq4000 platform according to the manufacturer’s instructions. The RNA-Seq data were analyzed using the find-circ-enhance authentication tool. The 20-mers from both ends of the unmapped reads were extracted and aligned to the reference genome to find unique anchor positions within the splice site. In order to ensure the reliability of project analysis results, all circRNA identified should be screened by basic conditions. The final circRNA (candidate circRNA) identifications were output using the screening conditions of numReads greater than 2 and numUniq greater than 2 (HRA000058).
Animals and cell culture
Female BALB/c nude mice that were 4 to 5 weeks of age were purchased from the Shanghai Laboratory Animal Center at the Chinese Academy of Sciences. All the mice experiments were conducted in accordance with the guidelines approved by the Laboratory Animal Center of Soochow University.
Human EC cell lines (Ishikawa and HEC-1-B) were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences and Shanghai Institute of Cell Biology. The cell lines were passaged for less than 6 months after resuscitation. Ishikawa and HEC-1-B cells were cultured in RPMI-1640 medium (Hyclone), which was supplemented with 10% fetal bovine serum (Gibco-BRL) and 1× antibiotics/antimycotics (Solarbio), at 37 °C in the presence of 5% CO2.
Northern blot assay
Northern blot was performed using Invitrogen Northern Max according to the manufacturer’s instructions. In brief, total RNA was extracted from the corresponding group of cells using TRIzol. RNA with RNase R treatment was separated by 1% formaldehyde denatured agarose gel and then transferred from gel to nylon membrane (GE Healthcare). RNA was immobilized on the membrane by baking it at 80 °C for 2 h. Biotin-labeled probes were prepared by Ribo RNAmax-T7 kit. The membrane was hybridized by a denatured biotin-labeled probe at 68 °C overnight. The membrane was then washed with 2 × SSC 0.1% SDS twice 30 min, then once 30 min, and once 1 h with 0.2 × SSC 0.1% SDS at the hybridization temperature. Finally, the biotin-labeled probe was detected by Chemiluminescent Biotin-labeled Nucleic Acid Detection Kit (Beyotime) and analyzed using Quantity One or Image Lab software (Bio-Rad).
RNA fluorescence in situ hybridization
To detect circ-0000437 RNA in EC cells, RNA FISH experiments were performed with FISH Detection Kit (Ribo) in EC cell lines according to the protocol. Cells cultured on coverslips were fixed with 4% paraformaldehyde at 4 °C for 15 min and washed three times with PBS. The samples were subsequently incubated with Pre-Hybridization Buffer at 37 °C for 30 min and Hybridization Buffer with FISH probes at 37 °C overnight in the dark using a RiboTM Fluorescent In Situ Hybridization Kit (RiboBio). On the next day, the coverslips were washed three times with wash buffer I (4 × SSC with 0.1% Tween-20), once each with wash buffer II (2 × SSC) and wash buffer III (1 × SSC) at 42 °C in the dark for 5 min and once with PBS at room temperature. Then, the cells were stained with DAPI in the dark for 10 min. The has-circ-000437-Cy3 FISH probes were designed and synthesized by RiboBio Co, Ltd. Human U6 FISH probes (RiboBio) and Human 18S FISH probes (RiboBio) were used as nuclear and cytoplasmic controls, respectively.
Coimmunoprecipitation
We performed coimmunoprecipitation experiments using a Thermo Kit (26149, Thermo) according to the manufacturer’s instructions. The lysates were applied to columns containing 10 μg of immobilized antibodies covalently linked to an amine-active resin and incubated overnight at 4 °C. Then the coimmunoprecipitate was eluted and analyzed by SDS-PAGE or mass spectrometry along with the controls.
Mass spectrometry
The peptide samples were analyzed on Thermo Fisher LTQ Obitrap ETD mass spectrometry, Briefly, samples were loaded onto an HPLC chromatography system named Thermo Fisher Easy-nLC 1000 equipped with a C18 column (1.8 mm, 0.15 × 1.00 mm). Solvent A contained 0.1% formic acid and solvent B contained 100% acetonitrile. The elution gradient was from 4% to 18% in 182 min, 18% to 90% in 13 min solvent B at a flow rate of 300 nl/min. Mass spectrometry analyses were carried out at AIMSMASS Co, Ltd in the positive-ion mode with an automated data-dependent MS/MS analysis with full scans (350–1600 m/z) acquired using FTMS at a mass resolution of 30,000 and the ten most intense precursor ions were selected for MS/MS. The MS/MS was acquired using higher-energy collision dissociation at 35% collision energy at a mass resolution of 15,000.
Raw MS files were analyzed by Proteome Discoverer 1.4 (Thermo); the parameter used for data analysis included trypsin as the protease with a maximum of two missed cleavages allowed. The mass tolerance for precursor ions and fragment ions was set to 20 ppm and 0.05 D, respectively. The search included variable modifications of methionine oxidation and deamidation and fixed modification of carbamidomethyl cysteine. Sequence database search was based on Uniprot-Homo sapiens. The minimal peptide length was set to six amino acids, and a maximum of two miscleavages was allowed. The false discovery rate was set to 0.01 for peptide and protein identifications.
Chromatin immunoprecipitation
We performed ChIP assay using the EZ ChIPTM Chromatin Immunoprecipitation Kit (Millipore) according to its manual. The cells were fixed with 1.0% formaldehyde, and the nuclei were released using lysis buffer. At 10 min to terminate the cross-linking, the chromatin was sheared into small uniform fragments by ultrasonic cell disruption system, and the protein–DNA complexes were then immunoprecipitated overnight at 4 °C using anti-TACC3, anti-ARNT, and control IgG (normal rabbit IgG) antibodies, respectively. DNA–protein cross-links were reversed, and chromatin DNA was purified and subjected to PCR analysis. The primer sequences are shown in Table S2.
Western blotting
Western blotting was performed as per standard protocols. Proteins were separated on SDS-polyacrylamide gel and transferred to nitrocellulose membrane. Membranes were incubated overnight at 4 °C with primary antibodies (FLAG tag and anti-CORO1C47aa) and visualized with a Phototope Horseradish Peroxidase Western Blot Detection kit (Thermo Fisher). A polyclonal antibody against the CORO1C-47aa peptide produced by circ-0000437 was made by ChinaPeptides Co, Ltd. The other primary antibodies: ARNT (A-3: sc-17811, Santa Cruz), TACC3 (ab138262, Abcam), VEGFA (ab1316, Abcam), VEGFR2 (2479S, CST), FLAG tag (F1804, Sigma) and HA tag (clone 12CA5, Santa Cruz). Normal mouse IgG (sc-2025) was from Santa Cruz. Antibody against beta-ACTIN (66009-1-Ig) was from Proteintech. Anti-mouse IgG (CST 7056) and Anti-rabbit IgG (CST 7054) were used, respectively.
Luciferase reporter assay
The VEGF promoter sequence (−2000 to ∼−1) was cloned into the pGL3-basic vector (Promega). The mutant of this sequence (TACGTG→CTAGCA) was also cloned into the pGL3-basic vector. Cells were seeded at 1 × 105 cells per well in 24-multiwell plates (BD Biosciences). Sixteen hours later, cells were cotransfected using Lipofectamin 3000 (Life Technologies) with 800 ng reporter plasmid and 160 ng pRL-SV40 (Luciferase Assay System; Promega). Twenty-four hours later, cells were collected to detect Renilla luciferase activity with the Dual-Luciferase Reporter Assay System (Promega) in TD-20/20 luminometer (Turner Biosystems). The results were expressed as a ratio of luciferase activity to pRL-SV40.
Tube formation assay
Matrigel (10 μl, Corning) was thawed on ice at 4 °C overnight and loaded into each well of a precooled μ-Slide Angiogenesis(Ibidi)and the plate was incubated at 37 °C for 30 min for hardening. Then, 15,000 HUVECs in 100 μl conditioned cell culture medium were plated onto the precoated Matrigel for 6 h. The resulting capillary-like structures were then photographed with a microscope and counted with ImageJ software.
Immunohistochemistry
Immunofluorescence analysis of CD31 (ab28364, Abcam) was performed using an immunofluorescence staining kit (Beyotime Institute of Biotechnology) according to the manufacturer’s instructions.
Enzyme-linked immunosorbent assay
The supernatants of culture medium samples in each group were pooled, then tested according to the protocol described by human VEGF ELISA kit (ab222510, Abcam).
In vitro proliferation, wound healing, and migration assays
Cells were seeded in 96-well plates (2000 cells per well) and cultured for a certain period of time as indicated in Results section. Cell viability was measured using CCK-8 assays (Dojindo). Each experiment with six replicates was repeated three times.
Wound healing assays were conducted by creating identical wound areas for anchorage-dependent HUVEC cells using 10-μl sterile pipette tips. Cells were seeded onto 6-well plates and coincubated with mitomycin C for 24 h and grown to confluence as a monolayer. Then cells were washed with PBS and scraped a vertical cross with constant width in the center of each well. After 24 h growth, cells in the scraped wound were observed and photographed.
Migration assay was done in a 24-well Millicell chamber in triplicate. The 0.8-μm pore inserts were coated with 30 μg of Matrigel (BD Biosciences). Cells (2 × 105) were added to the coated filters in serum-free medium. After 24 h at 37 °C in an incubator at 5% CO2, cells that migrated through the filters were fixed with methanol and stained with crystal violet. Cell numbers in three random fields were counted.
HUVEC sprouting assay
Spheroids were generated as described (44). Briefly, spheroids were embedded into a collagen gel and then rapidly transferred into a 24-well plate and allowed to polymerize at 37 °C incubators for 30 min, and various conditioned media were applied on top of the gel. The spheroids were cultured for 24 h, then angiogenesis was quantified by counting the number of sprouts for each spheroid.
Matrigel plug assay
Seven-week-old BALB/c nude mice were injected subcutaneously with 500 μl of liquid Matrigel containing HUVEC cells cultured in different condition mediums. Two weeks after injection, mice were sacrificed and plugs were harvested and photographed.
Statistical analysis
All statistical analyses were performed using GraphPad Prism Software. The differences among the groups were assessed with a paired, two-tailed Student’s t test. p < 0.05 was considered significant. All experimental assays were performed in triplicate.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (45) partner repository with the dataset identifier PXD026046. The URL is http://www.ebi.ac.uk/pride. Additional data that support the findings of this study are contained within the article and supplementary material. Any additional information or data are available upon request.
Supporting information
This article contains supporting information (6, 8, 44).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank all individuals who took part in this research. This work was supported by the grants from the National Natural Science Foundation of China (81872120).
Author contributions
F. L., Y. C., and H. L. conceptualization; S. D., L. Y., N. L., X. C., L. J., Y. Z., F. L., Y. C., and H. L. methodology; S. D., L. Y., and N. L. software; Y. C., N. L., and X. C. validation; L. J., H. L., and Y. Z. formal analysis; Y. C., S. D., N. L., X. C., H. L., F. L., Y. Z., and L. J. investigation; F. L., Y. C., N. L., X. C., and L. J. data curation; F. L., Y. C., and H. L. writing – original draft; F. L. and H. L. supervision; F. L., Y. C., L. J., Y. Z., and H. L. project administration; F. L. and H. L. funding acquisition.
Edited by Ronald Wek
Supporting information
References
- 1.Lee T.Y., Martinez-Outschoorn U.E., Schilder R.J., Kim C.H., Richard S.D., Rosenblum N.G., Johnson J.M. Metformin as a therapeutic target in endometrial cancers. Front. Oncol. 2018;8:341. doi: 10.3389/fonc.2018.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.He Y., Tao X., Huang F., Jia N., Du Y., Yu J., Feng W. Clinicopathologic features of endometrial cancer in Chinese patients younger than 50 years with a family history of cancer. Medicine. 2018;97 doi: 10.1097/MD.0000000000012968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potischman N., Hoover R.N., Brinton L.A., Siiteri P., Dorgan J.F., Swanson C.A., Berman M.L., Mortel R., Twiggs L.B., Barrett R.J., Wilbanks G.D., Persky V., Lurain J.R. Case-control study of endogenous steroid hormones and endometrial cancer. J. Natl. Cancer Inst. 1996;88:1127–1135. doi: 10.1093/jnci/88.16.1127. [DOI] [PubMed] [Google Scholar]
- 5.Li N., Zheng J., Li H., Deng J., Hu M., Wu H., Li W., Li F., Lan X., Lu J., Zhou Y. Identification of chimeric TSNAX-DISC1 resulting from intergenic splicing in endometrial carcinoma through high-throughput RNA sequencing. Carcinogenesis. 2014;35:2687–2697. doi: 10.1093/carcin/bgu201. [DOI] [PubMed] [Google Scholar]
- 6.Li W., Li H., Zhang L., Hu M., Li F., Deng J., An M., Wu S., Ma R., Lu J., Zhou Y. Long non-coding RNA LINC00672 contributes to p53 protein-mediated gene suppression and promotes endometrial cancer chemosensitivity. J. Biol. Chem. 2017;292:5801–5813. doi: 10.1074/jbc.M116.758508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Servick K. Circular RNAs hint at new realm of genetics. Science. 2017;355:1363. doi: 10.1126/science.355.6332.1363. [DOI] [PubMed] [Google Scholar]
- 8.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., Loewer A., Ziebold U., Landthaler M., Kocks C., Le Noble F. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 9.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 11.Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., Goodfellow P., Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 12.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aspden J.L., Eyre-Walker Y.C., Phillips R.J., Amin U., Mumtaz M.A., Brocard M., Couso J.P. Extensive translation of small open reading frames revealed by poly-ribo-seq. Elife. 2014;3 doi: 10.7554/eLife.03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrews S.J., Rothnagel J.A. Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 2014;15:193–204. doi: 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y., Wong C.C., Xiao X., Wang Z. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M., Laneve P., Rajewsky N., Bozzoni I. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e29. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Zhou H., Huang S., Xie B., Zhang N. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang M., Zhao K., Xu X., Yang Y., Yan S., Wei P., Liu H., Xu J., Xiao F., Zhou H., Yang X., Huang N., Liu J., He K., Xie K. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018;9:4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glazar P., Papavasileiou P., Rajewsky N. circBase: A database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vincent H.A., Deutscher M.P. Substrate recognition and catalysis by the exoribonuclease RNase R. J. Biol. Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- 21.Vincent H.A., Deutscher M.P. Insights into how RNase R degrades structured RNA: Analysis of the nuclease domain. J. Mol. Biol. 2009;387:570–583. doi: 10.1016/j.jmb.2009.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu H., Guo S., Li W., Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert W.V., Zhou K., Butler T.K., Doudna J.A. Cap-independent translation is required for starvation-induced differentiation in yeast. Science. 2007;317:1224–1227. doi: 10.1126/science.1144467. [DOI] [PubMed] [Google Scholar]
- 24.Chen H., Cong Q., Du Z., Liao W., Zhang L., Yao Y., Ding K. Sulfated fucoidan FP08S2 inhibits lung cancer cell growth in vivo by disrupting angiogenesis via targeting VEGFR2/VEGF and blocking VEGFR2/Erk/VEGF signaling. Cancer Lett. 2016;382:44–52. doi: 10.1016/j.canlet.2016.08.020. [DOI] [PubMed] [Google Scholar]
- 25.Dorland Y.L., Huveneers S. Cell-cell junctional mechanotransduction in endothelial remodeling. Cell. Mol. Life Sci. 2017;74:279–292. doi: 10.1007/s00018-016-2325-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiodelli P., Rezzola S., Urbinati C., Federici Signori F., Monti E., Ronca R., Presta M., Rusnati M. Contribution of vascular endothelial growth factor receptor-2 sialylation to the process of angiogenesis. Oncogene. 2017;36:6531–6541. doi: 10.1038/onc.2017.243. [DOI] [PubMed] [Google Scholar]
- 27.Kewley R.J., Whitelaw M.L., Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int. J. Biochem. Cell Biol. 2004;36:189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- 28.Partch C.L., Gardner K.H. Coactivator recruitment: A new role for PAS domains in transcriptional regulation by the bHLH-PAS family. J. Cell Physiol. 2010;223:553–557. doi: 10.1002/jcp.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Partch C.L., Gardner K.H. Coactivators necessary for transcriptional output of the hypoxia inducible factor, HIF, are directly recruited by ARNT PAS-B. Proc. Natl. Acad. Sci. U. S. A. 2011;108:7739–7744. doi: 10.1073/pnas.1101357108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu C.Y., Li T.C., Wu Y.Y., Yeh C.H., Chiang W., Chuang C.Y., Kuo H.C. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 2017;8:1149. doi: 10.1038/s41467-017-01216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G., Liang L., Gu J., He X., Huang S. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu L., Gong X., Sun L., Zhou Q., Lu B., Zhu L. The circular RNA Cdr1as act as an oncogene in hepatocellular carcinoma through targeting miR-7 expression. PLoS One. 2016;11 doi: 10.1371/journal.pone.0158347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Zhang J., Liu H., Hou L., Wang G., Zhang R., Huang Y., Chen X., Zhu J. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol. Cancer. 2017;16:151. doi: 10.1186/s12943-017-0719-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang J.Z., Chen M., Chen D., Gao X.C., Zhu S., Huang H., Hu M., Zhu H., Yan G.R. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol. Cell. 2017;68:171–184.e176. doi: 10.1016/j.molcel.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 35.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E., Shenzis S., Samson M., Dittmar G., Landthaler M., Chekulaeva M. Translation of CircRNAs. Mol. Cell. 2017;66:9–21.e27. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson D.M., Anderson K.M., Chang C.L., Makarewich C.A., Nelson B.R., McAnally J.R., Kasaragod P., Shelton J.M., Liou J., Bassel-Duby R., Olson E.N. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen H.X., Xu X.X., Tan B.Z., Zhang Z., Zhou X.D. MicroRNA-29b inhibits angiogenesis by targeting VEGFA through the MAPK/ERK and PI3K/Akt signaling pathways in endometrial carcinoma. Cell. Physiol. Biochem. 2017;41:933–946. doi: 10.1159/000460510. [DOI] [PubMed] [Google Scholar]
- 38.Stefansson I.M., Salvesen H.B., Akslen L.A. Vascular proliferation is important for clinical progress of endometrial cancer. Cancer Res. 2006;66:3303–3309. doi: 10.1158/0008-5472.CAN-05-1163. [DOI] [PubMed] [Google Scholar]
- 39.Lee I.I., Maniar K., Lydon J.P., Kim J.J. Akt regulates progesterone receptor B-dependent transcription and angiogenesis in endometrial cancer cells. Oncogene. 2016;35:5191–5201. doi: 10.1038/onc.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aghajanian C., Sill M.W., Darcy K.M., Greer B., McMeekin D.S., Rose P.G., Rotmensch J., Barnes M.N., Hanjani P., Leslie K.K. Phase II trial of bevacizumab in recurrent or persistent endometrial cancer: A Gynecologic Oncology Group study. J. Clin. Oncol. 2011;29:2259–2265. doi: 10.1200/JCO.2010.32.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan Z., Chen K., Wu W., Zhou Y., Zhu J., Wu G., Cao L., Zhang X., Guan H., Yang Y., Zhang W., Li J. Overexpression of HOXC10 promotes angiogenesis in human glioma via interaction with PRMT5 and upregulation of VEGFA expression. Theranostics. 2018;8:5143–5158. doi: 10.7150/thno.27310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park J.H., Yoon J., Park B. Pomolic acid suppresses HIF1alpha/VEGF-mediated angiogenesis by targeting p38-MAPK and mTOR signaling cascades. Phytomedicine. 2016;23:1716–1726. doi: 10.1016/j.phymed.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 43.Guo Y., Scheuermann T.H., Partch C.L., Tomchick D.R., Gardner K.H. Coiled-coil coactivators play a structural role mediating interactions in hypoxia-inducible factor heterodimerization. J. Biol. Chem. 2015;290:7707–7721. doi: 10.1074/jbc.M114.632786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stabile H., Mitola S., Moroni E., Belleri M., Nicoli S., Coltrini D., Peri F., Pessi A., Orsatti L., Talamo F., Castronovo V., Waltregny D., Cotelli F., Ribatti D., Presta M. Bone morphogenic protein antagonist Drm/gremlin is a novel proangiogenic factor. Blood. 2007;109:1834–1840. doi: 10.1182/blood-2006-06-032276. [DOI] [PubMed] [Google Scholar]
- 45.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (45) partner repository with the dataset identifier PXD026046. The URL is http://www.ebi.ac.uk/pride. Additional data that support the findings of this study are contained within the article and supplementary material. Any additional information or data are available upon request.