Abstract
PIWIs are regulatory proteins that belong to the Argonaute family. Piwis are primarily expressed in gonads and protect the germline against the mobilization and propagation of transposable elements (TEs) through transcriptional gene silencing. Vertebrate genomes encode up to four Piwi genes: Piwil1, Piwil2, Piwil3 and Piwil4, but their duplication history is unresolved. We leveraged phylogenetics, synteny and expression analyses to address this void. Our phylogenetic analysis suggests Piwil1 and Piwil2 were retained in all vertebrate members. Piwil4 was the result of Piwil1 duplication in the ancestor of gnathostomes, but was independently lost in ray-finned fishes and birds. Further, Piwil3 was derived from a tandem Piwil1 duplication in the common ancestor of marsupial and placental mammals, but was secondarily lost in Atlantogenata (Xenarthra and Afrotheria) and some rodents. The evolutionary rate of Piwil3 is considerably faster than any Piwi among all lineages, but an explanation is lacking. Our expression analyses suggest Piwi expression has mostly been constrained to gonads throughout vertebrate evolution. Vertebrate evolution is marked by two early rounds of whole genome duplication and many multigene families are linked to these events. However, our analyses suggest Piwi expansion was independent of whole genome duplications.
Keywords: RNAi, Gene duplication, Argonaute gene family, Selection, Transcriptomics, Phylogenetics, Synteny
Introduction
The Piwi gene family has become one of the more charismatic gene families in recent decades due to their role in defending the genome against transposable elements (TEs) and viruses (Siomi et al., 2011; Sun et al., 2017; Kolliopoulou et al., 2019a; Kolliopoulou et al., 2019b). PIWI proteins belong to a larger family of Argonaute proteins that include two major clades, the AGOs and PIWIs (Carmell et al., 2002). All members of the Argonaute family contain a PAZ and PIWI domain and participate in RNA-induced silencing complexes (RISCs) with small RNAs. AGOs are ubiquitously expressed and bind microRNAs (Tang, 2005) to target and silence mRNAs. In vertebrates, PIWI proteins are generally restricted to the germline, bind their own class of small RNAs, PIWI interacting RNAs (piRNAs), and regulate TEs and viruses through transcriptional silencing (Girard et al., 2006; Lau et al., 2006; Brennecke et al., 2007; Sun et al., 2017). The details of PIWI function vary among animals, but most seem to be involved in a model described as the ‘ping-pong cycle’ where PIWIs cleave RNA into piRNAs, the cleaved piRNAs form riboprotein complexes with the PIWIs and direct these complexes to bind complementary transcripts. These complexes can then cleave newly bound transcripts and repeat the cycle. Secondary piRNAs can also be used to guide PIWI proteins to TE loci in the genome and initiate methylation (Aravin et al., 2008; Kuramochi-Miyagawa et al., 2008; Kuramochi-Miyagawa et al., 2008; Rozhkov, Hammell & Hannon, 2013). In addition to the ping-pong cycle, the diversity of piRNAs is increased by 5′-to-3′ phasing from the site of initial piRNA formation (Han et al., 2015; Mohn, Handler & Brennecke, 2015; Ozata et al., 2019).
The bulk of vertebrate PIWI protein functional analyses are restricted to experiments in mice. From these experiments, we understand that the expression of Piwis varies temporally during germ cell development and spermatogenesis. Piwil2 (Mili) is the first Piwi to become expressed at embryonic day 12.5 (E12.5) in developing testes and is linked to the post-transcriptional silencing of TEs (Aravin et al., 2007). Piwil4 (Miwi2) is expressed between E14.5 and post-natal day 3 (P3). piRNAs from PIWIL2 are loaded onto PIWIL4 and PIWIL4 initiates the de novo establishment of methylation marks among TE loci in gonocytes during this time (Carmell et al., 2007; Aravin et al., 2008; Molaro et al., 2014; Zoch et al., 2020). PIWIL4 is also expressed in undifferentiated spermatogonia in adults, although the link to TEs in these cell types is still under investigation (Carrieri et al., 2017; Vasiliauskaite et al., 2018). Piwil1 (Miwi) is the last Piwi gene expressed at approximately P14, during the pachytene stage of prophase I (Girard et al., 2006). However, most piRNAs associated with PIWIL1 are derived from non-coding regions and PIWIL1 plays a relevant role cleaving/removing mRNAs during later stages of spermatogenesis (Reuter et al., 2011; Li et al., 2013; Gou et al., 2015; Wu et al., 2020). Some placental mammals, including Primates and Laurasiatherians have a 4th paralog, Piwil3. Piwil3 is the least studied of the four paralogs, likely because this paralog is absent from the mouse genome. However, experiments in cattle suggest Piwil3 is largely expressed in oocytes and early embryos and PIWIL3 generates TE derived ping-pong piRNAs (Roovers et al., 2015; Tan et al., 2020).
The copy number of the Piwi gene family varies among animals, for example C.elegans encodes two Piwis from a lineage specific duplication (Wynant, Santos & Vanden Broeck, 2017), Drosophila melanogaster encodes three Piwis (Lewis et al. 2018), but some turbellaria flatworms could have up to eight lineage specific paralogs (Fontenla, Rinaldi & Tort, 2021). The varying rates of sequence evolution and gene turnover has made the animal Piwi phylogeny difficult to resolve and multiple contradicting Piwi gene trees that do not mirror species relationships have been presented (Kerner et al., 2011; Wynant, Santos & Vanden Broeck, 2017; Fontenla, Rinaldi & Tort, 2021). However, a general theme of Piwi phylogenetics suggests there are two major subfamilies, Piwil1 and Piwil2. Some trees suggested vertebrate piwis are not directly orthologous to insect Piwis (Wynant, Santos & Vanden Broeck, 2017), others found that Drosophila Ago3 is orthologous to vertebrate Piwil2 but the Piwi and Aub genes of Drosophila do not have a vertebrate ortholog (Kerner et al., 2011; Jehn et al., 2018; Fontenla, Rinaldi & Tort, 2021).
Consistent with the two subfamily generalization, vertebrate genomes encode at least two copies of the Piwi family, a copy from the Piwil1 (-iwi) group and another from the Piwil2 (-ili) group, however vertebrates have a maximum of four Piwi paralogs (See above) and questions remain about the timing and mechanism of vertebrate Piwi duplication. Here, we are interested in leveraging phylogenetic and synteny analyses to clarify the duplication history of the vertebrate Piwi family as well as understanding the patterns of selection and expression among major lineages.
Methods
Sequence acquisition
We used Ensembl v.101 (Yates et al., 2020) and NCBI release 239 (Sharma et al., 2018) to collect known Piwi and Piwi-like coding sequences (CDS) from representative species of all major groups of vertebrates. Specifically, our sampling included cyclostomes (jawless fishes; lamprey) and gnathostomes (jawed vertebrates) from chondrichthyes (cartilaginous fishes), ray-finned fishes, lobe-finned fishes and tetrapods (amphibians, reptiles, birds and mammals). To help understand the ancestral state and focus on changes that occurred within vertebrates, we included Piwis from closely related deuterostomes as outgroups (Table S1, Fig. S1). The longest CDS was selected if there were multiple transcripts for a Piwi gene. Unidentified CDSs that shared similarity to Piwis were queried against the NCBI non redundant protein database via BLAST (Altschul et al., 1990) to confirm homology. If a Piwi paralog was absent or poorly annotated in Ensembl, we queried the NCBI for the paralog using human Piwi sequences as BLAST queries.
Phylogenetic reconstruction
All Piwi sequences were translated to amino acids and aligned using the LINSI strategy in MAFFT v7 (Katoh & Standley, 2013). We constructed phylogenetic trees from the amino acid alignment with two methods. First, we used IQ-Tree2 (Minh et al., 2020) and let IQ-Tree2 find the best fitting substitution model (Kalyaanamoorthy et al., 2017). Ten independent phylogenetic analyses were run simultaneously to explore tree space and we selected the tree with the highest likelihood score. Node support for the best scoring tree was evaluated with the ultrafast bootstrap method (Hoang et al., 2018) and the SH-like approximate likelihood ratio test (SH-aLRT) (Guindon et al., 2010). In both cases, we used 1,000 pseudoreplicates. Second, we used ExaBayes v1.5.1 (Aberer, Kobert & Stamatakis, 2014) to conduct Bayesian analyses. We ran four simultaneous chains for 1,000,000 generations sampling every 500 generations, using the same substitution model chosen by IQ-Tree2 but without additional model parameters. Chains were considered as having converged once the average standard deviation of split frequency among runs was less than 0.01. We summarized results with a majority-rule consensus tree from the set of sampled trees after the first 25% were discarded. We repeated these tree construction analyses on a reduced alignment generated after removing poorly aligned regions using Gblocks and allowed up to five contiguous non-conserved positions and columns to contain gaps; -b3 = 5 -b5 = a (Castresana, 2000) to test the impact of alignment quality on our results.
Synteny analyses
To understand the conservation of synteny as well as gene gain and loss, we examined the position of protein coding genes up and downstream of Piwi genes in representative vertebrates. We used gene coordinate information from Ensembl v.101 as well as estimates of orthology and paralogy from the Compara database (Herrero et al. 2016). If a Piwi gene was presumed missing, we used BLAST in Ensembl with human Piwi queries to locate any Piwi derived fragments near expected positions. If no fragments were found, we concluded the gene was missing, but if fragments were found and a full annotation was lacking, we presumed the Piwi to be pseudogenized.
Estimating dN/dS rates and selection tests
For each Piwi paralog in each major lineage with multiple samples (birds, mammals, and ray-finned fishes), we made independent codon alignments by translating DNA sequences to amino acid sequences, generating LINSI amino acid alignments and ‘reverse translating’ alignments into codon alignments using a custom Python script (available at github.com/mike2vandy/PiwiGeneFamily along with other scripts used). We specifically conducted site-tests for diversifying selection (Goldman & Yang, 1994) among alignments using the codeml module of PAML v4.9. We calculated the likelihood of models that allow dN/dS to vary among codon positions (M1a, M2a, M8a and M8). We used the likelihood ratio test (LRT) to test for significant differences between nested models that do not allow selection to those that do (M1a vs. M2a, M8a vs. M8). We also estimated a single dN/dS rate for each alignment using codeml’s M0 one-ratio model and pairwise dN/dS distances using the modified Nei-Gojobori (proportional) method in MEGA-X (Kumar et al., 2018). We noticed an intronic sequence that included stop codons was retained in the manatee Piwil4 sequence. We suspect this was an annotation artifact and because of it, this sequence was not included in selection tests and dN/dS estimates.
To view changes in dN/dS along the mammalian Piwil1 and Piwil3 branches we constructed a phylogenetic tree of mammalian Piwil1 and Piwil3 and used a free-ratio model to estimate dN/dS rates on all branches (Yang, 1998). We also used the adaptive branch-site random effects likelihood (aBSREL; (Smith et al., 2015)) approach implemented in Datamonkey v2.2 (Weaver et al., 2018) which detects episodes of positive selection on all designated foreground branches. We performed a LRT between the null model (dN/dS = 1) against the alternative where a branch was undergoing some form of selection (dN/dS ≠ 1). All Piwil3 branches were labeled as foreground and tested.
Expression analyses
To assess historical changes in Piwi expression through vertebrate evolution, we collected RNASeq data from representative tissues (brain, heart, kidney, liver, ovary and testes) and species (elephant shark, spotted gar, clawed frog, spiny toad, chicken, opossum, cow, and human) from NCBI’s short read archive (SRA; Leinonen, Sugawara & Shumway, 2011). Accession numbers for species and tissues can be found in Table S2. For all species, we used appropriate Ensembl’s CDSs downloaded from BioMart as a mapping reference. We only used the longest CDS per gene. Prior to mapping, RNASeq reads were cleaned with Trimmomatic v0.38 (Bolger, Lohse & Usadel, 2014) using the trimming and adapter removal parameters: HEADCROP:5, SLIDINGWINDOW:5:30, MINLEN:50, and ILLUMINACLIP:2:30:10. RNASeq reads were mapped to reference CDS libraries using default parameters of RSEM v1.3.1 (Li & Dewey, 2011) and Bowtie v1.2.2 (Langmead et al., 2009) to estimate gene expression in units of transcripts per million (TPM).
Results
Phylogenetic analysis
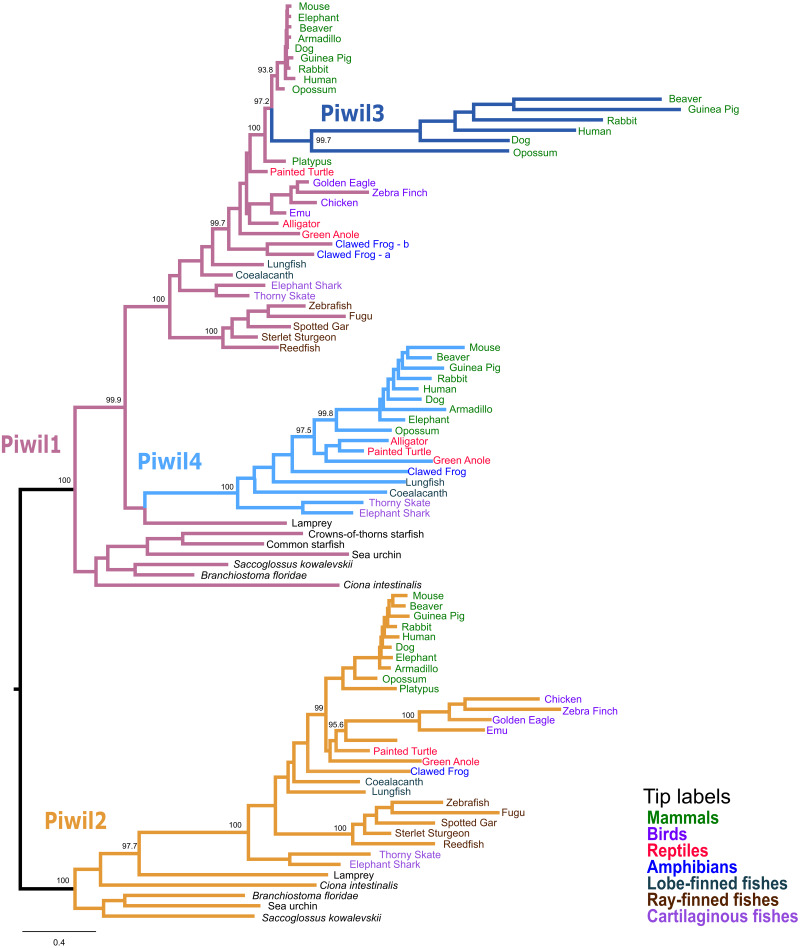
Our first goal was to reconstruct the Piwi phylogeny from vertebrate representatives and deuterostome outgroups using both maximum likelihood (ML) and Bayesian strategies. We collected 168 sequences from 63 species (Table S1). We generated trees in three different strategies: a tree of select representative species (Fig. 1), a tree from all collected sequences (Fig. S2), and because the N-terminus was difficult to align, we also estimated a tree from the most alignable regions of the amino acid alignment which included the PAZ and PIWI domains (Fig. S3). All trees were relatively similar and sequences fell into the two major clades, the Piwil1 group and Piwil2 group (Fig. 1). There have been no additional duplications of Piwil2 among deuterostomes and the subfamily is monophyletic. By contrast, Piwil4 and the mammalian specific Piwil3 were derived from independent duplications of a vertebrate Piwil1 (Fig. 1). These additional duplications within the Piwil1 clade indicate that the gnathostome Piwil1 is not a 1:1 ortholog of the ancestral Piwil1 gene. However, we have retained the traditional nomenclature for simplicity.
Figure 1. Phylogenetic reconstruction of the vertebrate Piwi family.
A phylogenetic tree constructed from a reduced dataset to summarize relationships among Piwi paralogs. The IQ-Tree2 model chosen was LG+F+I+G4 (LG model using empirical base Frequencies, a proportion of Invariable sites, and a discrete Gamma model with four rate categories). A tree constructed from all species used is presented in Fig. S2. Piwi paralogs are color coded on the tree and tip labels are color coded for major gnathostome groups. The displayed tree was the maximum likelihood tree constructed with IQ-Tree2. Numbers by nodes correspond to support values from the ultrafast bootstrap routine in IQ-Tree2.
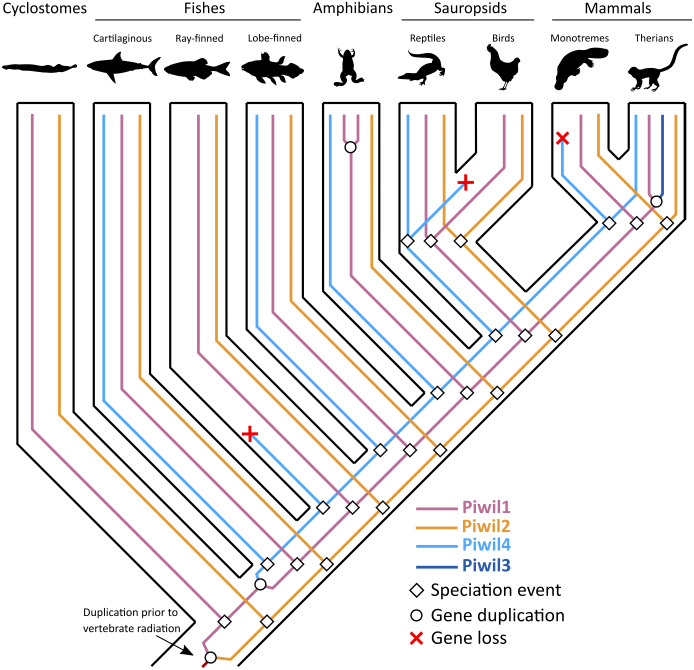
The Piwil4 gene of vertebrates was likely derived from an ancient duplication of Piwil1 in the ancestor of gnathostomes, however a lamprey Piwil1-like sequence was found sister to gnathostome Piwil4 in both ML and Bayesian trees. Therefore, we can interpret the Piwil4 duplication in two different ways. The Piwil4 duplication occurred in the ancestor of vertebrates, but Piwil1 was lost in lamprey and the lamprey sequence is an ortholog of Piwil4. Alternatively, the Piwil4 duplication occurred in gnathostomes, but the lamprey Piwil1-like gene is artificially sister to Piwil4. Piwil4 was present in cartilaginous and lobe-finned fishes, mammals and reptiles, but was absent in ray-finned fishes and birds (Fig. 1; Fig. S1).
Our analyses suggest Piwil3 is the result of a duplication of Piwil1 in the common ancestor of therian mammals given its presence in marsupial and placental mammals and its absence from Monotreme genomes (Fig. 1). However, not all placental mammals have retained Piwil3. Piwil3 was present in marsupials (opossum and koala), Laurasiatherians (bat, dog, and cow) and most Euarchontoglires (primates, rodents and relatives), but we were unable to find it in Afrotherians (elephant and manatee), Xenarthrans (armadillo) and mouse-like rodents which included the kangaroo rat, deer mouse and house mouse (Fig. S1).
An additional copy of Piwil1 (Piwil1a and Piwil1b) had been previously described in the clawed frog (Wilczynska et al., 2009), but our analyses also revealed this additional paralog was present in the spiny toad (Fig. S2). Interestingly, a third intact Piwil1 copy was identified in the spiny toad (Fig. S2). This copy is most likely a retrogene, diagnosed by a lack of introns in the coding sequence (Fig. S4A).
Synteny analyses
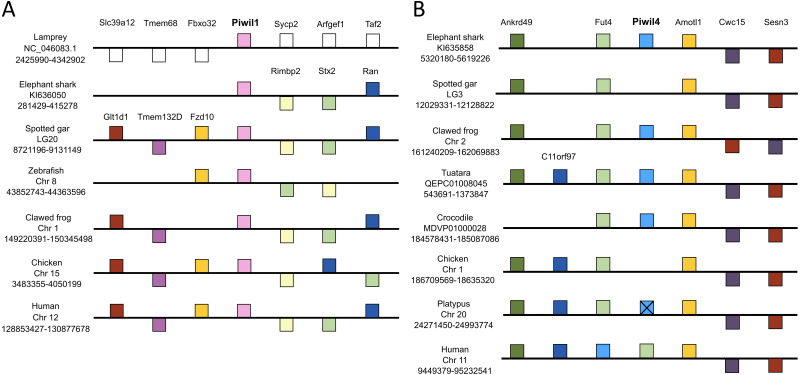
We used genomic coordinates to identify genes up- and down-stream of Piwi genes to resolve or confirm homology and infer the duplication mechanism behind the vertebrate Piwi expansion. Synteny analyses included representative species with contiguous regions for at least 10 genes up and downstream of each Piwi. In most cases, this included the elephant shark, spotted gar, zebra fish, clawed frog and chicken and human, but varied among Piwis. The lamprey assembly on Ensembl lacked contiguity around the Piwi genes, therefore we queried sea lamprey coordinate data from a GFF file at https://genomes.stowers.org/sealamprey (Smith et al. 2018). Unfortunately, there was a lack of synteny among Piwi genes between cyclostomes and gnathostomes, but synteny around Piwil1, Piwil2 and Piwil4 has generally been conserved among gnathostomes (Fig. 2; Fig. S5). Synteny around Piwil2 was less conserved than Piwil1 and observed three different sets of upstream genes, however Slc39a14 and Ppp3cc were consistently downstream of Piwil2 in all lineages (Fig. S5).
Figure 2. Synteny comparisons of vertebrate Piwil1 and Piwil4.
Organization of genes up and downstream of (A) Piwil1 and (B) Piwil4. Distances are not drawn to scale. White boxes represent genes that are not homologous to any other genes in the synteny block. Boxes on top of the black line reflect genes in forward orientation relative to Piwi genes and boxes below the line are in the opposite orientation. An “X” indicates pseudogenization and empty space between Fut4 and Amotl1 in 2B indicates an absent Piwil4.
Piwil1 was flanked by Rimbp2, Stx2, and Ran on the 3′end in all gnathostomes, but synteny was only conserved upstream of Piwil1 among bony and jawed vertebrates where Piwil1 is generally flanked by Fzd10, Tmem132d and Glt1d1 (Fig. 2A). By contrast, synteny around Piwil4 has been conserved throughout all gnathostome evolution. Although Piwil4 was lost in ray-finned fish and birds, the overall locus has remained intact (Fig. 3B). In the spotted gar, the distance between Fut4 and Amotl1 was 18,372 bases and there were no BLAST hits to Piwi genes in that region. The same was true for the chicken, where the distance between Fut4 and Amotl1 was 45,906 and lacked sequences similar to Piwil4. By contrast, the same region in the crocodile covers 157,135 bases and includes a Piwil4 gene (Fig. 3B). Interestingly, Piwil4 is intact in all mammals except the platypus. The Piwil4 locus in platypus does contain a lncRNA annotation (ENSOANT00000069592) between Fut4 and Amotl1 with similarity to Piwil4. However, a complete gene with an open reading frame could not be recovered. Therefore we suspect that Piwil4 has been pseudogenized in the platypus (Fig. 2B) but has not been fully purged from the genome.
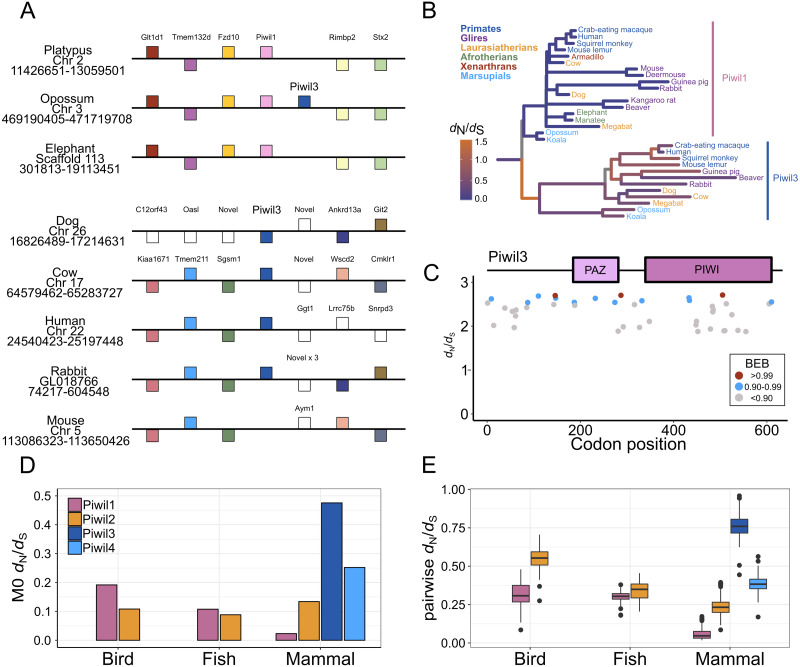
Figure 3. Evolutionary properties of Piwil3.
(A) Synteny around the Piwil3 gene. White boxes represent genes that are not homologous to any other genes in the synteny block, and empty space between Sgsm1 and Aym1 in the mouse locus indicates a missing Piwil3. (B) Evolutionary rate measured as d N/d S mapped to branches of a mammalian Piwil1 and Piwil3 phylogenetic tree calculated from a free-ratio model in Codeml. (C) Codon sites under selection estimated by Bayes Empirical Bayes (BEB) from Codeml model M2a. M2a sites were chosen over M8 because M2a is more conservative and sites under selection in M8 are also identified in M2a. (D) One-ratio d N/d S estimates among Piwis in major lineages calculated using model M0 in Codeml. (E) Pairwise d N/d S distances among Piwis and major lineages.
Piwil1 and Piwil4 were not flanked by any co-duplicating gene families (Fig. 2), an indicative signal of whole genome or segmental duplications (Campanini et al., 2015; Opazo et al., 2015). Furthermore, both teleosts and salmonid fishes, which have experienced additional whole genome duplication events (Jaillon et al., 2004; Macqueen & Johnston, 2014; Lien et al., 2016), lacked any additional Piwi paralogs (Fig. S2).
Piwil3 is a direct neighbor of Piwil1 in marsupials which would suggest that the Piwil1 and Piwil3 genes of therian mammals are co-orthologs of the ancestral mammalian Piwil1 gene derived from a tandem duplication of Piwil1 in their common ancestor (Fig. 3A). Interestingly, the position of Piwil3 is conserved among marsupials, but Piwil3 is in a novel location in placental mammals where flanking genes vary among lineages (Fig. 3A). Taking these results together, the expansion of the Piwi gene family in vertebrates can best be explained by tandem duplication followed by a translocation due to lineage-specific genomic rearrangements.
Piwil3 and selection tests
We observed very long branches among Piwil3 paralogs (Fig. 1) and decided to test the role diversifying selection has played on the evolution of Piwil3. Using PAML’s free ratio model, we found that dN/dS rates among Piwil3 sequences were considerably faster than rates among therian Piwil1 sequences (Fig. 3B). Since the free-ratio model does not offer any additional information besides changes in dN/dS and is not statistically robust, we used site-tests among Piwil3 sequences and found that models allowing selection (M2a and M8) were significantly better fits to the data than models that disallow positive selection (M1a and M8a) (Table S3). Codons throughout the entire gene, including the conserved PAZ and PIWI domains (Fig. S6), exhibited signatures of positive selection (Fig. 3C; Fig. S7) according to Bayes Empirical Bayes (BEB) estimation of site posterior probabilities. A separate branch-site test, aBSREL in HyPhy, suggested 17 out of 22 tested Piwil3 branches have undergone episodic positive selection (Fig. S8).
We then compared Piwil3 patterns of evolution to the other paralogs within tetrapod groups (birds, mammals and ray-finned fishes). Site-tests of diversifying selection suggested no other Piwi paralog is evolving in a diversifying pattern. In all cases except Piwil3, models allowing positive selection were not improved over models restricting selection (Table S3). Further, one-ratio (Fig. 3D) and pairwise (Fig. 3E) dN/dS, although likely inflated due to saturation at synonymous sites, indicated Piwil3 is the fastest evolving Piwi among all lineages while Piwil1 in mammals is highly conserved.
Historical Piwi expression
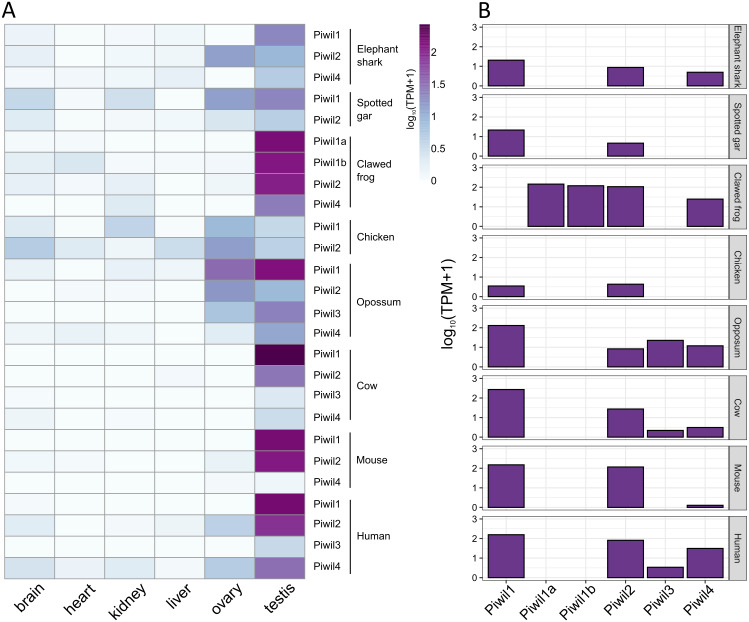
Our last goal was to identify any differences in Piwi expression among lineages, and consistent with previous results, Piwi expression was largely localized to the gonads, specifically testis, although Piwis were sporadically expressed at low levels among somatic tissues (Fig. 4A). Aravin et al. (2008) reported that Piwil4 is only expressed for a short period of time during testis development in mice, but Piwil4 has since been detected in adult undifferentiated spermatogonia (Carrieri et al., 2017; Vasiliauskait et al., 2018). Consistently, Piwil4 was detected in the adult testes of all species examined, but lowly detected in the mouse (Fig. 4B). In addition, we detected weak expression of Piwil3 in the testis of the opossum, human, and cattle. Both Piwil1 paralogs were expressed in the testis of the clawed frog (Fig. 4B). We were also able to measure Piwi expression from the brain, ovary, and testes, in the spiny toad which had an additional Piwil1 retrogene. The spiny toad expression pattern was unique as Piwil4 and Piwil1a-retro were the only Piwis expressed in the testis, but Piwil1a and Piwil1a-retro were expressed in the ovary (Fig. S4B). Piwil1b nor Piwil2 were expressed in any library (Fig. S4B).
Figure 4. Piwi expression characteristics.
(A) Expression of Piwi genes from both somatic and gonadal tissue from representatives of vertebrate lineages. Expression values were normalized by taking the log10 of transcripts per million (TPM) +1. (B) Bar plot of Piwi expression in just testes.
Discussion
Understanding the relationships among gene family members is an active area of research in evolutionary biology and the availability of whole-genome sequences allows the opportunity to address unresolved questions. The Piwi gene family is a charismatic family of proteins that function as a genome defense mechanism against invasive elements that include TEs and viruses and function in a similar fashion as CRiSPR/Cas9. However, the paralog relationships and duplication timing in vertebrates has not been thoroughly resolved.
Consistent with previous work, Piwil2 was found to be monophyletic among deuterostomes (Kerner et al., 2011), but the Piwil1 group has experienced additional duplications among vertebrates giving rise to the gnathostome Piwil4 and the therian Piwil3 paralogs (Fig. 1). Based on our phylogenetic and synteny analyses, we propose a model where Piwil4 originated from the duplication of Piwil1 early in vertebrate evolutionary history, at least as early as the ancestor of gnathostomes (Fig. 5). Unfortunately, the exact timing of Piwil4 origination is unresolved given that orthology could not be accurately inferred between cyclostomes and gnathostomes. Cyclostome genomes possess nucleotide biases and are the product of an additional cyclostome specific whole-genome triplication (Nakatani et al., 2021) and resolving orthology between gnathostome and cyclostome genes is often difficult (Qiu et al., 2011; Kuraku, 2013; Smith et al., 2013; Campanini et al., 2015; Opazo et al., 2015). However, given that 1- Piwil4 is secondarily lost, 2- all vertebrates have a Piwil1, and 3- there are only two Piwi paralogs in the lamprey genome, we propose that Piwil4 probably emerged in the gnathostome lineage, after the divergence between gnathostomes and cyclostomes. Piwil4 was then secondarily lost in the common ancestor of ray-finned fishes, retained in Sarcopterygii and its descendants, lobe-finned fishes, Sarcopterygii, and its descendants, but lost again in birds and monotremes (Fig. 5). Piwil3 originated in the common ancestor marsupial and placental mammals as a tandem duplication, but the retention and position of Piwil3 is variable among placental mammals (Fig. 3A).
Figure 5. An evolutionary hypothesis describing the history of gene gain and loss among vertebrates.
The common ancestor of vertebrates had two Piwis (an ancestral Piwil1 and Piwil2). Piwil2 has not undergone any gain or loss during vertebrate evolution. Most likely, Piwil4 was derived from a tandem duplication of Piwil1 in the common ancestor of gnathostomes, but was independently lost in ray-finned fishes, birds, and monotreme mammals. Piwil3 was derived from a tandem duplication of Piwil1 in the common ancestor of therian mammals. Source credits: Lamprey/cyclostome, Gareth Monger, http://phylopic.org/image/f9313512-3ce4-4349-86bc-060d4faa013e/ (CC 3.0, http://creativecommons.org/licenses/by/3.0/); Lobe-finned fish, Maija Karala, http://phylopic.org/image/202c2ad3-48a7-471d-87ef-c6d8406640e8/ (CC NC 3.0, http://creativecommons.org/licenses/by-nc-sa/3.0/); Amphibian, Sarah Werning, http://phylopic.org/image/cd0f49a1-4adf-448e-859c-b703a73b9481/ (CC 3.0, http://creativecommons.org/licenses/by/3.0/) Monotreme, Sarah Werning, http://phylopic.org/image/cd0f49a1-4adf-448e-859c-b703a73b9481/ (CC 3.0, http://phylopic.org/image/b406c409-2735-4a3d-a7aa-8afe0b6e72dc/); Therian, Bogdan Bocianowski, http://phylopic.org/image/2d078b25-e6a0-4beb-a5d3-5d6f16be8ebf/) (CC 3.0, http://creativecommons.org/licenses/by/3.0/); Cartilaginous fish, Public Domain: http://phylopic.org/image/545d45f0-0dd1-4cfd-aad6-2b835223ea0d/; ray-finned fish, Public Domain: http://phylopic.org/image/6f4c653a-1da1-4e02-85ef-6344b2d8e02a/; Reptile, Public Domain: http://phylopic.org/image/dffda000-77cb-4251-b837-0cd2ab21ed5b/; Chicken, Public Domain: http://phylopic.org/image/aff847b0-ecbd-4d41-98ce-665921a6d96e/.
Piwil3 was not identified in the elephant or armadillo genome. Under the most parsimonious scenario, Xenarthra and Afrotheria belong to the monophyletic group Atlantogenata (Foley, Springer & Teeling, 2016), and Piwil3 was lost in the common ancestor (Fig. S1). In addition, Piwil3 was also secondarily lost in some mouse-like rodents. Among Glires, rabbits, guinea pigs and beavers have retained Piwil3 (Fig. 3B), while the deer mouse, kangaroo rat, and house mouse lost Piwil3. The beaver is nested within the mouse-like rodent clade that includes the deer mouse, house mouse, and kangaroo rat (Blanga-Kanfi et al., 2009; Fabre et al., 2012), and encodes a Piwil3 while the remaining three species lack Piwil3 (Fig. 3B). This points to a complex scenario of gene retention (See Fig. S1) that would require further investigation to unveil.
We performed synteny analyses in an attempt to identify the duplication mechanism, be it whole genome, segmental, or tandem duplication. Piwil1 and Piwil2 were already present in the ancestor of deuterostomes so these paralogs are the product of a duplication early in animal evolution (Kerner et al., 2011), and we could not detect any co-duplicating gene families neighboring Piwil1 or Piwil4 (Fig. 2) that would point to segmental or whole genome duplication (Catchen, Conery & Postlethwait, 2009). In addition, Piwil3 is a direct neighbor of Piwil1 in the opossum genome, which likely reflects the ancestral state. Piwil3 likely migrated to a novel locus through non-homologous recombination in the ancestor of placental mammals, and synteny has not been conserved (Fig. 3A). From this evidence we conclude tandem duplication of Piwil1 followed by non-homologous recombination drove the expansion of the Piwi gene family.
The additional duplication events within the Piwil1 group likely led to novel functions among paralogs. PIWI proteins in mice bind piRNAs of different sizes and knockouts among paralogs yield discrete phenotypes (Aravin et al., 2007; Aravin et al., 2008, Kuramochi-Miyagawa et al. 2008). Unfortunately the exact function of all paralogs in all major lineages is unknown, but expression initiation varies among lineages which points to slightly different functions. For example, Piwil1 expression begins in primordial germ cells at E7 in zebrafish (Hsu et al., 2018) and E6 in chicken (Kim et al., 2012), but Piwil1 expression begins during meiosis at P14 in mice (Girard et al., 2006). Results from our dN/dS analyses suggest differences in selective pressure that could be related to these functional differences. Given the expression and dN/dS differences, we can hypothesize that the function of PIWIL1 removing mRNAs in later stages of meiosis could be unique to mammals. We also hypothesize there may be some functional redundancy between PIWIL2 and PIWIL4. Because PIWIL4 localizes to the nucleus and PIWIL2 localizes to the cytoplasm, it is thought that PIWIL2 is responsible for loading PIWIL4 with secondary piRNAs and PIWIL4 “marks” TE loci for subsequent methylation (Carmell et al., 2007; Aravin et al., 2008). Interestingly, there is some evidence that both PIWIL2 and PIWIL4 function to silence TE expression through methylation pathways (Manakov et al., 2015), although this claim is contested in Zoch et al. (2020). However, PIWIL2 does localize to the nucleus in zebrafish (Houwing, Berezikov & Ketting, 2008) and although untested, could potentially play a role in TE methylation (Goll & Halpern, 2011). Since both ray-finned fishes and birds lack Piwil4, it would be of interest to determine if the lack of Piwil4 has resulted in any measurable effect on TE methylation or test a methylation initiation function for PIWIL2 in these lineages.
Selection tests revealed evidence that Piwil3 is evolving in a fast and diversifying manner in all mammalian lineages (Fig. 3B; Fig. S8) and no Piwi in any other tested lineage is evolving in a diversifying manner (Table S3). The fast rate of Piwil3 was previously revealed in simian primates (Wynant, Santos & Vanden Broeck, 2017), but we demonstrated this phenomenon is not exclusive to primates. It is unclear why Piwil3 would exhibit an increased rate of evolution. Genes that tend to evolve in this fashion are often associated with immunity/defense against pathogens and reproduction/sexual selection (Kosiol et al., 2008; Park et al., 2011; Vandewege et al. 2013; Grueber, Wallis & Jamieson, 2014; Vandewege, Sotero-Caio & Phillips, 2020). Further, RNAi/antiviral genes have higher rates of evolution in insects relative to other genes (Obbard et al., 2009; Wynant, Santos & Vanden Broeck, 2017). Piwil1 and Piwil2, among other piRNA pathway genes, have high dN/dS rates and appear to be evolving under positive selection in teleost fishes that have wide TE diversity (Yi et al., 2014). Since Piwil3 is possibly part of a genome immune system, it could be plausible Piwil3 adapts to lineage specific genome stressors. However, Piwil3 is not present in all mammalian lineages and mutant knockout experiments have not been conducted to understand its precise function, so there is currently no good explanation as to why Piwil3 is evolving so fast.
In general, our expression results were consistent with the literature. Piwis exhibited the highest expression in germline tissues (Fig. 4A) and Piwil1 and Piwil2 were the most expressed in adult testis (Fig. 4B). We also detected the expression of Piwil4 in adult testis among all gnathostomes that encode a Piwil4, albeit expression was lowest in the mouse (Fig. 4B). In mice, Piwil4 expression coincides with gonocyte methylation erasure and re-establishment (Aravin et al., 2008; Carmell, 2008; Molaro et al., 2014) where the function is linked to homeostatic and regenerative spermatogenesis (Carrieri et al., 2017; Vasiliauskait et al., 2018). There were some inconsistencies regarding ovarian Piwi expression. Piwi expression has been previously detected in the clawed frog (Wilczynska et al., 2009) zebrafish (Houwing et al., 2007; Houwing, Berezikov & Ketting, 2008), humans and cattle (Roovers et al. 2015). Piwil3 has only been described in maturing oocytes and early embryos (Tan et al., 2020) but we found it expressed in testis (Fig. 4A) although it has not been described in testis. Nonetheless, all four Piwis were expressed in both opossum gonads suggesting Piwil3 localization to gonads was already established in the common ancestor of therian mammals.
Conclusions
We present an evolutionary study of the Piwi gene family in vertebrates that is generally composed of two to four paralogs with emphasis on resolving the phylogenetic relationships and duplication history. Our study included representative species from all of the major groups of vertebrates and deuterostome outgroups. Piwil2 has not undergone any additional duplications through vertebrate evolution, but Piwil4 and Piwil3 are derived from independent duplications of the vertebrate Piwil1 at different times. Piwil4 likely first appeared in the ancestor of gnathostomes, but was secondarily lost in ray-finned fishes and birds, and Piwil3 is therian specific. The most likely mechanism yielding these novel paralogs was tandem duplication followed by non-homologous recombination. Piwi genes are largely evolving at different rates among lineages and expression has largely been constrained to the gonads through all of vertebrate history.
Supplemental Information
The phylogenetic tree was generated using TimeTree (Kumar et al. 2017) from a list of species used in the study. Branches are color coded by major gnathostome lineages. The presence of a Piwi paralog is indicated by a red circle and absent paralog is reflected by a white circle. An x2 or x3 by a Piwi indicates 2 or 3 copies of a paralog. Although Asterias rubens was used in the study, it is absent from the tree as As. rubens relationships are lacking from TimeTree, but the Piwi presence/absence mirrors Acanthaster planci and As. rubens is sister to Ac. planci (See Table S1).
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35:15471549 DOI 10.1093/molbev/msy096.
The IQ-Tree model chosen was JTT+F+R6 (JTT model using empirical base frequencies and a FreeRate model with 6 rate categories). Piwi paralogs are color coded on the tree and tip labels are color coded to by major gnathostome groups. The displayed tree was constructed with IQ-Tree2. Numbers next to nodes reflect nodal support derived from the ultrafast bootstrap routine/ SH-aLRT/and posterior probability from the Bayesian phylogeny that mirrored the displayed tree.
A phylogenetic tree reconstructed from the most alignable residues see Fig. S2 for additional description. The IQ-Tree model chosen was also JTT+F+R6.
(A) Location and intron/exon boundaries of Piwil1 paralogs in the spiny toad. (B) Piwi expression among paralogs measured from available samples.
Organization of genes up and downstream of Piwil2. Distances are not drawn to scale. White boxes represent genes that are not homologous to any other genes in the synteny block. Boxes on top of the black line reflect genes in forward orientation relative to Piwi genes and boxes below the line are in the opposite orientation.
A sequence LOGO constructed from an amino acid alignment of all Piwi paralogs found in human, dog, chicken, spotted gar, and elephant shark. PAZ and PIWI domains are labeled. Highly gaped regions in the 5′section were removed prior to making the LOGO.
All sites tested for selection in codeml under model M2a presented in Fig. 3C. Sites labeled by an asterisk were found under selection with BEB >0.9. PAZ and PIWI domains are highlighted.
Piwil3 branches that had evidence of episodic diversifying selection are presented thicker. Significance was assessed using the LRT at a threshold of p ≤ 0.05, after correcting for multiple testing.
The list of species, common names, major lineage, and transcript accession numbers for sequences used in this study.
Likelihood ratio tests suggest models allowing selection in codon evolution were improved over models restricting selection are bolded. Codon positions where BEB >0.9 are listed, bolded positions in M8 were also found under selection in M2a.
Acknowledgments
The color palette was inspired by Prince and the Revolution’s Purple Rain (1984).
Funding Statement
Support for this work came from a US Dept. of Education HSI-STEM grant P031C110114-15. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
David Ray is an Academic Editor for PeerJ.
Author Contributions
Javier Gutierrez performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Roy Platt, Juan C. Opazo, David A. Ray and Federico Hoffmann conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Michael Vandewege conceived and designed the experiments, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
The SRA libraries used in expression analyses (Table S2) are available at Genbank.
The code used in this study is available at GitHub: github.com/mike2vandy/PiwiGeneFamily.
References
- Aberer, Kobert & Stamatakis (2014).Aberer AJ, Kobert K, Stamatakis A. ExaBayes: massively parallel bayesian tree inference for the whole-genome era. Molecular Biology and Evolution. 2014;31:2553–2556. doi: 10.1093/molbev/msu236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul et al. (1990).Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Aravin et al. (2008).Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Molecular Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin et al. (2007).Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Blanga-Kanfi et al. (2009).Blanga-Kanfi S, Miranda H, Penn O, Pupko T, De Bry RW, Huchon D. Rodent phylogeny revised: analysis of six nuclear genes from all major rodent clades. BMC Evolutionary Biology. 2009;9:71. doi: 10.1186/1471-2148-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, Lohse & Usadel (2014).Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke et al. (2007).Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Campanini et al. (2015).Campanini EB, Vandewege MW, Pillai NE, Tay B-H, Jones JL, Venkatesh B, Hoffmann FG. Early evolution of vertebrate mybs: an integrative perspective combining synteny, phylogenetic, and gene expression analyses. Genome Biology and Evolution. 2015;7:3009–3021. doi: 10.1093/gbe/evv197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell (2008).Carmell M. PIWI proteins are essential for spermatogenesis and repression of transposons in the mouse male germline. Biology of Reproduction. 2008;78:160–160. doi: 10.1093/biolreprod/78.s1.160a. [DOI] [Google Scholar]
- Carmell et al. (2007).Carmell MA, Girard A, van de Kant HJG, Bourc’his D, Bestor TH, De Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Developmental Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Carmell et al. (2002).Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes and Development. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Carrieri et al. (2017).Carrieri C, Comazzetto S, Grover A, Morgan M, Buness A, Nerlov C, O’Carroll D. A transit-amplifying population underpins the efficient regenerative capacity of the testis. Journal of Experimetnal Medicine. 2017;214:1631–1641. doi: 10.1084/jem.20161371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana (2000).Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Catchen, Conery & Postlethwait (2009).Catchen JM, Conery JS, Postlethwait JH. Automated identification of conserved synteny after whole-genome duplication. Genome Research. 2009;19:1497–1505. doi: 10.1101/gr.090480.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre et al. (2012).Fabre P-H, Hautier L, Dimitrov D, Douzery EJP. A glimpse on the pattern of rodent diversification: a phylogenetic approach. BMC Evolutionary Biology. 2012;12:88. doi: 10.1186/1471-2148-12-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley, Springer & Teeling (2016).Foley NM, Springer MS, Teeling EC. Mammal madness: is the mammal tree of life not yet resolved? Philosophical Transactions of the Royal Society B. 2016;371:20150140. doi: 10.1098/rstb.2015.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenla, Rinaldi & Tort (2021).Fontenla S, Rinaldi G, Tort JF. Lost and found: piwi and argonaute pathways in flatworms. Front. Cell. Infect. Microbiol. 2021;11:653695. doi: 10.3389/fcimb.2021.653695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard et al. (2006).Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Goldman & Yang (1994).Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Molecular Biology and Evolution. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Goll & Halpern (2011).Goll MG, Halpern ME. DNA methylation in zebrafish. Progress in Molecular Biology and Translational Science. 2011;101:193–218. doi: 10.1016/b978-0-12-387685-0.00005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou et al. (2015).Gou L-T, Dai P, Yang J-H, Xue Y, Hu Y-P, Zhou Y, Kang J-Y, Wang X, Li H, Hua M-M, Zhao S, Hu S-D, Wu L-G, Shi H-J, Li Y, Fu X-D, Qu L-H, Wang E-D, Liu M-F. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Research. 2015;25:266–266. doi: 10.1038/cr.2015.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueber, Wallis & Jamieson (2014).Grueber CE, Wallis GP, Jamieson IG. Episodic positive selection in the evolution of avian toll-like receptor innate immunity genes. PLOS ONE. 2014;9:e89632. doi: 10.1371/journal.pone.0089632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon et al. (2010).Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Han et al. (2015).Han BW, Wang W, Li C, Weng Z, Zamore PD. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science. 2015;348:817–821. doi: 10.1126/science.aaa1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero et al. (2016).Herrero J, Muffato M, Beal K, Fitzgerald S, Gordon L, Pignatelli M, Vilella AJ, Searle SMJ, Amode R, Brent S, Spooner W, Kulesha E, Yates A, Flicek P. Ensembl comparative genomics resources. Database: The Journal of Biological Databases and Curation. 2016;2016:baw053. doi: 10.1093/database/baw053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang et al. (2018).Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing, Berezikov & Ketting (2008).Houwing S, Berezikov E, Ketting RF. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO Journal. 2008;27:2702–2711. doi: 10.1038/emboj.2008.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing et al. (2007).Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RHA, Hannon GJ, Draper BW, Ketting RF. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Hsu et al. (2018).Hsu C-W, Pan Y-J, Wang Y-W, Tong S-K, Chung B-C. Changes in the morphology and gene expression of developing zebrafish gonads. General and Comparative Endocrinology. 2018;265:154–159. doi: 10.1016/j.ygcen.2018.01.026. [DOI] [PubMed] [Google Scholar]
- Jaillon et al. (2004).Jaillon O, Aury J-M, Brunet F, Petit J-L, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, Nicaud S, Jaffe D, Fisher S, Lutfalla G, Dossat C, Segurens B, Dasilva C, Salanoubat M, Levy M, Boudet N, Castellano S, Anthouard V, Jubin C, Castelli V, Katinka M, Vacherie B, Biémont C, Skalli Z, Cattolico L, Poulain J, De Berardinis V, Cruaud C, Duprat S, Brottier P, Coutanceau J-P, Gouzy J, Parra G, Lardier G, Chapple C, McKernan KJ, McEwan P, Bosak S, Kellis M, Volff J-N, Guigó R, Zody MC, Mesirov J, Lindblad-Toh K, Birren B, Nusbaum C, Kahn D, Robinson-Rechavi M, Laudet V, Schachter V, Quétier F, Saurin W, Scarpelli C, Wincker P, Lander ES, Weissenbach J, Roest Crollius H. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Jehn et al. (2018).Jehn J, Gebert D, Pipilescu F, Stern S, Kiefer JST, Hewel C, Rosenkranz D. genes and piRNAs are ubiquitously expressed in mollusks and show patterns of lineage-specific adaptation. Communications Biology. 2018;1:137. doi: 10.1038/s42003-018-0141-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy et al. (2017).Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh & Standley (2013).Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner et al. (2011).Kerner P, Degnan SM, Marchand L, Degnan BM, Vervoort M. Evolution of RNA-binding proteins in animals: insights from genome-wide analysis in the sponge Amphimedon queenslandica. Molecular Biology and Evolution. 2011;28:2289–2303. doi: 10.1093/molbev/msr046. [DOI] [PubMed] [Google Scholar]
- Kim et al. (2012).Kim TH, Yun TW, Rengaraj D, Lee SI, Lim SM, Seo HW, Park TS, Han JY. Conserved functional characteristics of the PIWI family members in chicken germ cell lineage. Theriogenology. 2012;78:1948–1959. doi: 10.1016/j.theriogenology.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Kolliopoulou et al. (2019a).Kolliopoulou A, Santos D, Taning CNT, Wynant N, Van den Broeck J, Smagghe G, Swevers L. PIWI pathway against viruses in insects. Wiley Interdisciplinary Reviews: RNA. 2019a;2019:e1555. doi: 10.1002/wrna.1555. [DOI] [PubMed] [Google Scholar]
- Kolliopoulou et al. (2019b).Kolliopoulou A, Santos D, Taning CNT, Wynant N, Van den Broeck J, Smagghe G, Swevers L. PIWI pathway against viruses in insects. Wiley Interdisciplinary Reviews: RNA. 2019b;10:e1555. doi: 10.1002/wrna.1555. [DOI] [PubMed] [Google Scholar]
- Kosiol et al. (2008).Kosiol C, Vinar T, Da Fonseca RR, Hubisz MJ, Bustamante CD, Nielsen R, Siepel A. Patterns of positive selection in six Mammalian genomes. PLOS Genetics. 2008;4:e1000144. doi: 10.1371/journal.pgen.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar et al. (2018).Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku (2013).Kuraku S. Impact of asymmetric gene repertoire between cyclostomes and gnathostomes. Seminars in Cell and Developmental Biology. 2013;24:119–127. doi: 10.1016/j.semcdb.2012.12.009. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa et al. (2008).Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, Matsuda Y, Kimura T, Okabe M, Sakaki Y, Sasaki H, Nakano T. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes & Development. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead et al. (2009).Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau et al. (2006).Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- Leinonen, Sugawara & Shumway (2011).Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database Collaboration The sequence read archive. Nucleic Acids Research. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis et al. (2018).Lewis SH, Quarles KA, Yang Y, Tanguy M, Frézal L, Smith SA, Sharma PP, Cordaux R, Gilbert C, Giraud I, Collins DH, Zamore PD, Miska EA, Sarkies P, Jiggins FM. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nature Ecology & Evolution. 2018a;2:174–181. doi: 10.1038/s41559-017-0403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li & Dewey (2011).Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2013).Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, Han BW, Xu J, Moore MJ, Schimenti JC, Weng Z, Zamore PD. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Molecular Cell. 2013;50:67–81. doi: 10.1016/j.molcel.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien et al. (2016).Lien S, Koop BF, Sandve SR, Miller JR, Kent MP, Nome T, Hvidsten TR, Leong JS, Minkley DR, Zimin A, Grammes F, Grove H, Gjuvsland A, Walenz B, Hermansen RA, Schalburg Kvon, Rondeau EB, Di Genova A, Samy JKA, Olav Vik J, Vigeland MD, Caler L, Grimholt U, Jentoft S, Våge DI, De Jong P, Moen T, Baranski M, Palti Y, Smith DR, Yorke JA, Nederbragt AJ, Tooming-Klunderud A, Jakobsen KS, Jiang X, Fan D, Hu Y, Liberles DA, Vidal R, Iturra P, Jones SJM, Jonassen I, Maass A, Omholt SW, Davidson WS. The Atlantic salmon genome provides insights into rediploidization. Nature. 2016;533:200–205. doi: 10.1038/nature17164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macqueen & Johnston (2014).Macqueen DJ, Johnston IA. A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proceedings of the Royal Society B: Biological Sciences. 2014;281:20132881. doi: 10.1098/rspb.2013.2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manakov et al. (2015).Manakov SA, Pezic D, Marinov GK, Pastor WA, Sachidanandam R, Aravin AA. MIWI2 and MILI have differential effects on piRNA biogenesis and DNA methylation. Cell Reports. 2015;12:1234–1243. doi: 10.1016/j.celrep.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh et al. (2020).Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Haeseler Avon, Lanfear R. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic Era. Molecular Biology and Evolution. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn, Handler & Brennecke (2015).Mohn F, Handler D, Brennecke J. Noncoding RNA, piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science. 2015;348:812–817. doi: 10.1126/science.aaa1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaro et al. (2014).Molaro A, Falciatori I, Hodges E, Aravin AA, Marran K, Rafii S, McCombie WR, Smith AD, Hannon GJ. Two waves of de novo methylation during mouse germ cell development. Genes and Development. 2014;28:1544–1549. doi: 10.1101/gad.244350.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani et al. (2021).Nakatani Y, Shingate P, Ravi V, Pillai NE, Prasad A, McLysaght A, Venkatesh B. Reconstruction of proto-vertebrate, proto-cyclostome and proto-gnathostome genomes provides new insights into early vertebrate evolution. Nature Communications. 2021;12:4489. doi: 10.1038/s41467-021-24573-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard et al. (2009).Obbard DJ, Gordon KHJ, Buck AH, Jiggins FM. The evolution of RNAi as a defence against viruses and transposable elements. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2009;364:99–115. doi: 10.1098/rstb.2008.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opazo et al. (2015).Opazo JC, Lee AP, Hoffmann FG, Toloza-Villalobos J, Burmester T, Venkatesh B, Storz JF. Ancient Duplications and Expression Divergence in the Globin Gene Superfamily of Vertebrates: insights from the Elephant Shark Genome and Transcriptome. Molecular Biology and Evolution. 2015;32:1684–1694. doi: 10.1093/molbev/msv054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozata et al. (2019).Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, Zamore PD. PIWI-interacting RNAs: small RNAs with big functions. Nature Reviews Genetics. 2019;20:89–108. doi: 10.1038/s41576-018-0073-3. [DOI] [PubMed] [Google Scholar]
- Park et al. (2011).Park SH, Podlaha O, Grus WE, Zhang J. The microevolution of V1r vomeronasal receptor genes in mice. Genome Biology and Evolution. 2011;3:401–412. doi: 10.1093/gbe/evr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu et al. (2011).Qiu H, Hildebrand F, Kuraku S, Meyer A. Unresolved orthology and peculiar coding sequence properties of lamprey genes: the KCNA gene family as test case. BMC Genomics. 2011;12:325. doi: 10.1186/1471-2164-12-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter et al. (2011).Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480:264–267. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- Roovers et al. (2015).Roovers EF, Rosenkranz D, Mahdipour M, Han C-T, He N, Chuva De Sousa Lopes SM, van der Westerlaken LAJ, Zischler H, Butter F, Roelen BAJ, Ketting RF. Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell Reports. 2015;10:2069–2082. doi: 10.1016/j.celrep.2015.02.062. [DOI] [PubMed] [Google Scholar]
- Rozhkov, Hammell & Hannon (2013).Rozhkov NV, Hammell M, Hannon GJ. Multiple roles for Piwi in silencing Drosophila transposons. Genes and Development. 2013;27:400–412. doi: 10.1101/gad.209767.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma et al. (2018).Sharma S, Ciufo S, Starchenko E, Darji D, Chlumsky L, Karsch-Mizrachi I, Schoch CL. The NCBI biocollections database. Database. 2018;2018:bay006. doi: 10.1093/database/bay006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi et al. (2011).Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nature Reviews Molecular Cell Biology. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Smith et al. (2013).Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, Yandell MD, Manousaki T, Meyer A, Bloom OE, Morgan JR, Buxbaum JD, Sachidanandam R, Sims C, Garruss AS, Cook M, Krumlauf R, Wiedemann LM, Sower SA, Decatur WA, Hall JA, Amemiya CT, Saha NR, Buckley KM, Rast JP, Das S, Hirano M, McCurley N, Guo P, Rohner N, Tabin CJ, Piccinelli P, Elgar G, Ruffier M, Aken BL, Searle SMJ, Muffato M, Pignatelli M, Herrero J, Jones M, Brown CT, Chung-Davidson Y-W, Nanlohy KG, Libants SV, Yeh C-Y, McCauley DW, Langeland JA, Pancer Z, Fritzsch B, De Jong PJ, Zhu B, Fulton LL, Theising B, Flicek P, Bronner ME, Warren WC, Clifton SW, Wilson RK, Li W. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nature Genetics. 2013;45:415–421. doi: 10.1038/ng.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith et al. (2018).Smith JJ, Timoshevskaya N, Ye C, Holt C, Keinath MC, Parker HJ, Cook ME, Hess JE, Narum SR, Lamanna F, Kaessmann H, Timoshevskiy VA, Waterbury CKM, Saraceno C, Wiedemann LM, Robb SMC, Baker C, Eichler EE, Hockman D, Sauka-Spengler T, Yandell M, Krumlauf R, Elgar G, Amemiya CT. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nature Genetics. 2018;50:270–277. doi: 10.1038/s41588-017-0036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith et al. (2015).Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Pond SLKosakovsky. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Molecular Biology and Evolution. 2015;32:1342–1353. doi: 10.1093/molbev/msv022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun et al. (2017).Sun YH, Xie LH, Zhuo X, Chen Q, Ghoneim D, Zhang B, Jagne J, Yang C, Li XZ. Domestic chickens activate a piRNA defense against avian leukosis virus. Elife. 2017;6:e24695. doi: 10.7554/eLife.24695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan et al. (2020).Tan M, Van Tol HTA, Rosenkranz D, Roovers EF, Damen MJ, Stout TAE, Wu W, Roelen BAJ. PIWIL3 forms a complex with TDRKH in mammalian oocytes. Cells. 2020;9:1356. doi: 10.3390/cells9061356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang (2005).Tang G. siRNA and miRNA: an insight into RISCs. Trends in Biochemical Sciences. 2005;30:106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Vandewege et al. (2013).Vandewege MW, Phillips CJ, Wickliffe JK, Hoffmann FG. Evolution of the ABPA subunit of androgen-binding protein expressed in the submaxillary glands in New and Old World rodent taxa. Journal of Molecular Evolution. 2013;76:324–331. doi: 10.1007/s00239-013-9561-4. [DOI] [PubMed] [Google Scholar]
- Vandewege, Sotero-Caio & Phillips (2020).Vandewege MW, Sotero-Caio CG, Phillips CD. Positive selection and gene expression analyses from salivary glands reveal discrete adaptations within the ecologically diverse bat family phyllostomidae. Genome Biology and Evolution. 2020;12:1419–1428. doi: 10.1093/gbe/evaa151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiliauskaite et al. (2018).Vasiliauskaite L, Berrens RV, Ivanova I, Carrieri C, Reik W, Enright AJ, O’Carroll D. Defective germline reprogramming rewires the spermatogonial transcriptome. Nature Structural & Molecular Biology. 2018;25:394–404. doi: 10.1038/s41594-018-0058-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver et al. (2018).Weaver S, Shank SD, Spielman SJ, Li M, Muse SV, Kosakovsky Pond SL. Datamonkey 2.0: a modern web application for characterizing selective and other evolutionary processes. Molecular Biology and Evolution. 2018;35:773–777. doi: 10.1093/molbev/msx335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynska et al. (2009).Wilczynska A, Minshall N, Armisen J, Miska EA, Standart N. Two Piwi proteins, Xiwi and Xili, are expressed in the Xenopus female germline. RNA. 2009;15:337–345. doi: 10.1261/rna.1422509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu et al. (2020).Wu P-H, Fu Y, Cecchini K, Özata DM, Arif A, Yu T, Colpan C, Gainetdinov I, Weng Z, Zamore PD. The evolutionarily conserved piRNA-producing locus pi6 is required for male mouse fertility. Nature Genetics. 2020;52:728–739. doi: 10.1038/s41588-020-0657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynant, Santos & Vanden Broeck (2017).Wynant N, Santos D, Vanden Broeck J. The evolution of animal Argonautes: evidence for the absence of antiviral AGO Argonautes in vertebrates. Scientific Reports. 2017;7:9230. doi: 10.1038/s41598-017-08043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang (1998).Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Molecular Biology and Evolution. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yates et al. (2020).Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, Bhai J, Billis K, Boddu S, Marugán JC, Cummins C, Davidson C, Dodiya K, Fatima R, Gall A, Giron CG, Gil L, Grego T, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, Kay M, Lavidas I, Le T, Lemos D, Martinez JG, Maurel T, McDowall M, McMahon A, Mohanan S, Moore B, Nuhn M, Oheh DN, Parker A, Parton A, Patricio M, Sakthivel MP, Abdul Salam AI, Schmitt BM, Schuilenburg H, Sheppard D, Sycheva M, Szuba M, Taylor K, Thormann A, Threadgold G, Vullo A, Walts B, Winterbottom A, Zadissa A, Chakiachvili M, Flint B, Frankish A, Hunt SE, Iisley G, Kostadima M, Langridge N, Loveland JE, Martin FJ, Morales J, Mudge JM, Muffato M, Perry E, Ruffier M, Trevanion SJ, Cunningham F, Howe KL, Zerbino DR, Flicek P. Ensembl 2020. Nucleic Acids Research. 2020;48:D682–D688. doi: 10.1093/nar/gkz1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi et al. (2014).Yi M, Chen F, Luo M, Cheng Y, Zhao H, Cheng H, Zhou R. Rapid evolution of piRNA pathway in the teleost fish: implication for an adaptation to transposon diversity. Genome Biology and Evolution. 2014;6:1393–1407. doi: 10.1093/gbe/evu105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoch et al. (2020).Zoch A, Auchynnikava T, Berrens RV, Kabayama Y, Schöpp T, Heep M, Vasiliauskaite L, Pérez-Rico YA, Cook AG, Shkumatava A, Rappsilber J, Allshire RC, O’Carroll D. SPOCD1 is an essential executor of piRNA-directed de novo DNA methylation. Nature. 2020;584:635–639. doi: 10.1038/s41586-020-2557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The phylogenetic tree was generated using TimeTree (Kumar et al. 2017) from a list of species used in the study. Branches are color coded by major gnathostome lineages. The presence of a Piwi paralog is indicated by a red circle and absent paralog is reflected by a white circle. An x2 or x3 by a Piwi indicates 2 or 3 copies of a paralog. Although Asterias rubens was used in the study, it is absent from the tree as As. rubens relationships are lacking from TimeTree, but the Piwi presence/absence mirrors Acanthaster planci and As. rubens is sister to Ac. planci (See Table S1).
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35:15471549 DOI 10.1093/molbev/msy096.
The IQ-Tree model chosen was JTT+F+R6 (JTT model using empirical base frequencies and a FreeRate model with 6 rate categories). Piwi paralogs are color coded on the tree and tip labels are color coded to by major gnathostome groups. The displayed tree was constructed with IQ-Tree2. Numbers next to nodes reflect nodal support derived from the ultrafast bootstrap routine/ SH-aLRT/and posterior probability from the Bayesian phylogeny that mirrored the displayed tree.
A phylogenetic tree reconstructed from the most alignable residues see Fig. S2 for additional description. The IQ-Tree model chosen was also JTT+F+R6.
(A) Location and intron/exon boundaries of Piwil1 paralogs in the spiny toad. (B) Piwi expression among paralogs measured from available samples.
Organization of genes up and downstream of Piwil2. Distances are not drawn to scale. White boxes represent genes that are not homologous to any other genes in the synteny block. Boxes on top of the black line reflect genes in forward orientation relative to Piwi genes and boxes below the line are in the opposite orientation.
A sequence LOGO constructed from an amino acid alignment of all Piwi paralogs found in human, dog, chicken, spotted gar, and elephant shark. PAZ and PIWI domains are labeled. Highly gaped regions in the 5′section were removed prior to making the LOGO.
All sites tested for selection in codeml under model M2a presented in Fig. 3C. Sites labeled by an asterisk were found under selection with BEB >0.9. PAZ and PIWI domains are highlighted.
Piwil3 branches that had evidence of episodic diversifying selection are presented thicker. Significance was assessed using the LRT at a threshold of p ≤ 0.05, after correcting for multiple testing.
The list of species, common names, major lineage, and transcript accession numbers for sequences used in this study.
Likelihood ratio tests suggest models allowing selection in codon evolution were improved over models restricting selection are bolded. Codon positions where BEB >0.9 are listed, bolded positions in M8 were also found under selection in M2a.
Data Availability Statement
The following information was supplied regarding data availability:
The SRA libraries used in expression analyses (Table S2) are available at Genbank.
The code used in this study is available at GitHub: github.com/mike2vandy/PiwiGeneFamily.