Abstract
Graph machine learning (GML) is receiving growing interest within the pharmaceutical and biotechnology industries for its ability to model biomolecular structures, the functional relationships between them, and integrate multi-omic datasets — amongst other data types. Herein, we present a multidisciplinary academic-industrial review of the topic within the context of drug discovery and development. After introducing key terms and modelling approaches, we move chronologically through the drug development pipeline to identify and summarize work incorporating: target identification, design of small molecules and biologics, and drug repurposing. Whilst the field is still emerging, key milestones including repurposed drugs entering in vivo studies, suggest GML will become a modelling framework of choice within biomedical machine learning.
Keywords: graph machine learning, drug discovery, drug development
Introduction
The process from drug discovery to market costs, on average, well over $1 billion and can span 12 years or more [1–3]; due to high attrition rates, rarely can one progress to market in less than ten years [4, 5]. The high levels of attrition throughout the process not only make investments uncertain but require market approved drugs to pay for the earlier failures. Despite an industry-wide focus on efficiency for over a decade, spurred on by publications and annual reports highlighting revenue cliffs from ending exclusivity and falling productivity, significant improvements have proved elusive against the backdrop of scientific, technological and regulatory change [2]. For the aforementioned reasons, there is now a greater interest in applying computational methodologies to expedite various parts of the drug discovery and development pipeline [6], see Figure 1.
Figure 1 .
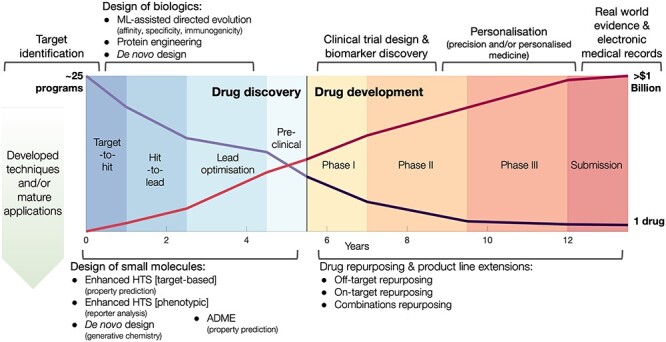
Timeline of drug development linked to potential areas of application by GML methodologies. Preclinical drug discovery applications are shown on the left side of the figure (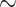 5.5 years), and clinical drug development applications are shown on the right hand side of the figure (
5.5 years), and clinical drug development applications are shown on the right hand side of the figure (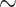 8 years). Over this period, for every
8 years). Over this period, for every 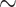 25 drug discovery programmes, a single successful drug reaches market approval. Applications listed in the top of half of the figure are less developed in the context of GML with limited experimental validation. Financial, timeline and success probability data are taken from Paul et al. [5].
25 drug discovery programmes, a single successful drug reaches market approval. Applications listed in the top of half of the figure are less developed in the context of GML with limited experimental validation. Financial, timeline and success probability data are taken from Paul et al. [5].
Digital technologies have transformed the drug development process generating enormous volumes of data. Changes range from moving to electronic lab notebooks [7], electronic regulatory submissions, through increasing volumes of laboratory, experimental and clinical trial data collection [8] including the use of devices [9, 10] to precision medicine and the use of ‘big data’ [11]. The data collected about therapies extend well beyond research and development to include hospital, specialist and primary care medical professionals’ patient records — including observations taken from social media, e.g. for pharmacovigilance [12, 13]. There are innumerable online databases and other sources of information including scientific literature, clinical trials information, through to databases of repurposable drugs [14, 15]. Technological advances now allow for greater -omic profiling beyond genotyping and whole genome sequencing (WGS); standardization of microfluidics and antibody tagging has made single-cell technologies widely available to study both the transcriptome, e.g. using RNA-seq [16], the proteome (targeted), e.g. via mass cytometry [17], or even multiple modalities together [18].
One of the key characteristics of biomedical data that is produced and used in the drug discovery process is its inter-connected nature. Such data structure can be represented as a graph, a mathematical abstraction ubiquitously used across disciplines and fields in biology to model the diverse interactions between biological entities that intervene at the different scales. At the molecular scale, proteins and other biomolecules can be represented as graphs capturing spatial and structural relationships between their amino acid residues [19, 20] and small molecule drugs as graphs relating their constituent atoms and chemical bonding structure [21, 22]. At an intermediary scale, interactomes are graphs that capture specific types of interactions between biomolecular species (e.g. metabolites, mRNA, proteins) [23], with protein–protein interaction (PPI) graphs being perhaps most commonplace. Finally, at a higher level of abstraction, knowledge graphs can represent the complex relationships between drugs, side effects, diagnosis, associated treatments and test results [24, 25] as found in electronic medical records (EMR).
Within the last decade, two emerging trends have reshaped the data modelling community: network analysis and deep learning. The ‘network medicine’ paradigm has long been recognized in the biomedical field [26], with multiple approaches borrowed from graph theory and complex network science applied to biological graphs such as PPIs and gene regulatory networks (GRNs). Most approaches in this field were limited to handcrafted graph features such as centrality measures and clustering. In contrast, deep neural networks, a particular type of machine learning algorithms, are used to learn optimal tasks-specific features. The impact of deep learning was ground-breaking in computer vision [27] and natural language processing [28] but was limited to specific domains by the requirements on the regularity of data structures. At the convergence of these two fields is graph machine learning (GML) a new class of ML methods exploiting the structure of graphs and other irregular datasets (point clouds, meshes, manifolds, etc).
The essential idea of GML methods is to learn effective feature representations of nodes [29, 30] (e.g. users in social networks), edges (e.g. predicting future interactions in recommender systems) or entire graphs [31] (e.g. predicting properties of molecular graphs). In particular, graph neural networks (GNNs) [32–34], which are deep neural network architectures specifically designed for graph-structure data, are attracting growing interest. GNNs iteratively update the features of the nodes of a graph by propagating information from their neighbours. These methods have already been successfully applied to a variety of tasks and domains such as recommendation in social media and E-commerce [35–38], traffic estimations in Google Maps [39], misinformation detection in social media [40], and various domains of natural sciences including modelling fluids, rigid solids, and deformable materials interacting with one another [41] and event classification in particle physics [42, 43].
In the biomedical domain, GML has now set the state of the art for mining graph-structured data including drug–target–indication interaction and relationship prediction through knowledge graph embedding [30, 44, 45]; molecular property prediction [21, 22], including the prediction of absorption, distribution, metabolism and excretion (ADME) profiles [46]; early work in target identification [47] to de novo molecule design [48, 49]. Most notably, Stokes et al. [50] used directed message passing GNNs operating on molecular structures to propose repurposing candidates for antibiotic development, validating their predictions in vivo to propose suitable repurposing candidates remarkably structurally distinct from known antibiotics. Therefore, GML methods appear to be extremely promising in applications across the drug development pipeline.
Compared to previous review papers on GML [51–55] for the machine learning community or general reviews on ML within drug development more broadly [56–58], the focus of our paper is both for biomedical researchers without extensive ML backgrounds and ML experts interested in biomedical applications — with a thematic focus on GML. We provide an introduction to the key terms and building blocks of graph learning architectures (Definitions & Machine Learning on Graphs) and contextualize these methodologies within the drug discovery and development pipeline from an industrial perspective for method developers without extensive biological expertize (Drug Development Applications) before providing a closing discussion (Discussion).
Definitions
Notations and preliminaries of graph theory
We denote a graph 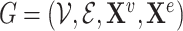 where
where 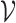 is a set of
is a set of  nodes, or vertices, and
nodes, or vertices, and 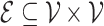 is a set of
is a set of 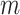 edges. Let
edges. Let 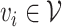 denote a node and
denote a node and 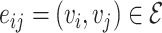 denote an edge from node
denote an edge from node 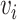 to node
to node 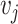 . When multiple edges can connect the same pair of nodes, the graph is called a multigraph. Node features are represented by
. When multiple edges can connect the same pair of nodes, the graph is called a multigraph. Node features are represented by 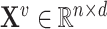 and
and 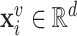 are the
are the 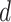 features of node
features of node 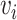 . Edge features, or attributes, are similarly represented by
. Edge features, or attributes, are similarly represented by 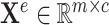 where
where 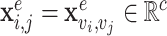 . We may also denote different nodes as
. We may also denote different nodes as  and
and  such that
such that 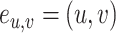 is the edge from
is the edge from 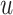 to
to 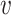 with attributes
with attributes 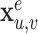 . Note that under this definition, undirected graphs are defined as directed graphs with each undirected edge represented by two directed edges.
. Note that under this definition, undirected graphs are defined as directed graphs with each undirected edge represented by two directed edges.
The neighbourhood 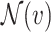 of node
of node 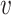 , sometimes referred to as one-hop neighbourhood, is the set of nodes that are connected to it by an edge,
, sometimes referred to as one-hop neighbourhood, is the set of nodes that are connected to it by an edge, 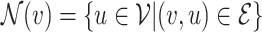 , with shorthand
, with shorthand 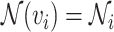 used for compactness. The cardinality of a node’s neighbourhood is called its degree and the diagonal degree matrix,
used for compactness. The cardinality of a node’s neighbourhood is called its degree and the diagonal degree matrix, 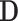 , has elements
, has elements  .
.
Two nodes 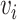 and
and 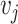 in a graph
in a graph 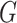 are connected if there exists a path in
are connected if there exists a path in 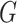 starting at one and ending at the other, i.e. there exists a sequence of consecutive edges of
starting at one and ending at the other, i.e. there exists a sequence of consecutive edges of 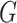 connecting the two nodes. A graph is connected if there exists a path between every pair of nodes in the graph. The shortest path distance between
connecting the two nodes. A graph is connected if there exists a path between every pair of nodes in the graph. The shortest path distance between 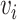 and
and 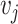 is defined as the number of edges in the shortest path between the two nodes and denoted by
is defined as the number of edges in the shortest path between the two nodes and denoted by 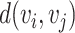 .
.
A graph 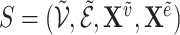 is a subgraph of
is a subgraph of  if and only if
if and only if  and
and 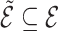 . If it also holds that
. If it also holds that 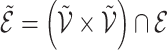 , then
, then 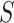 is called an induced subgraph of
is called an induced subgraph of 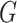 .
.
The adjacency matrix 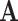 typically represents the relations between nodes such that the entry on the
typically represents the relations between nodes such that the entry on the 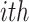 row and
row and 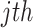 column indicates whether there is an edge from node
column indicates whether there is an edge from node 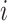 to node
to node 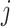 , with
, with 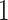 representing that there is an edge, and
representing that there is an edge, and 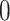 that there is not (i.e.
that there is not (i.e. 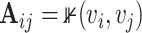 ). Most commonly, the adjacency matrix is a square (from a set of nodes to itself), but the concept extends to bipartite graphs where an
). Most commonly, the adjacency matrix is a square (from a set of nodes to itself), but the concept extends to bipartite graphs where an 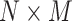 matrix can represent the edges from one set of
matrix can represent the edges from one set of 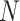 nodes to another set of size
nodes to another set of size 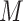 , and is sometimes used to store scalar edge weights. The Laplacian matrix of a simple (unweighted) graph is
, and is sometimes used to store scalar edge weights. The Laplacian matrix of a simple (unweighted) graph is 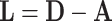 . The normalized Laplacian
. The normalized Laplacian  is often preferred, with a variant defined as
is often preferred, with a variant defined as 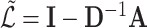 .
.
Knowledge graph
The term knowledge graph is used to qualify a graph that captures 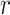 types of relationships between a set of entities. In this case,
types of relationships between a set of entities. In this case, 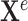 includes relationship types as edge features. Knowledge graphs are commonly introduced as sets of triplets
includes relationship types as edge features. Knowledge graphs are commonly introduced as sets of triplets 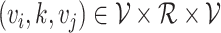 , where
, where 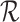 represents the set of relationships. Note that multiple edges of different types can connect two given nodes. As such, the standard adjacency matrix is ill-suited to capture the complexity of a knowledge graph. Instead, a knowledge graph is often represented as a collection of adjacency matrices
represents the set of relationships. Note that multiple edges of different types can connect two given nodes. As such, the standard adjacency matrix is ill-suited to capture the complexity of a knowledge graph. Instead, a knowledge graph is often represented as a collection of adjacency matrices 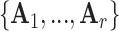 , forming an adjacency tensor, in which each adjacency matrix
, forming an adjacency tensor, in which each adjacency matrix 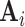 captures one type of relationship.
captures one type of relationship.
Random walks
A random walk is a sequence of nodes selected at random during an iterative process. A random walk is constructed by considering a random walker that moves through the graph starting from a node 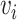 . At each step, the walker can either move to a neighbouring node with probability
. At each step, the walker can either move to a neighbouring node with probability 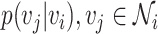 or stay on node
or stay on node 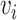 with probability
with probability 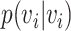 . The sequence of nodes visited after a fixed number of steps
. The sequence of nodes visited after a fixed number of steps 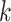 gives a random walk of length
gives a random walk of length 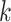 . Graph diffusion is a related notion that models the propagation of a signal on a graph. A classic example is heat diffusion [59], which studies the propagation of heat in a graph starting from some initial distribution.
. Graph diffusion is a related notion that models the propagation of a signal on a graph. A classic example is heat diffusion [59], which studies the propagation of heat in a graph starting from some initial distribution.
Graph isomorphism
Two graphs 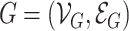 and
and 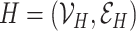 are said to be isomorphic if there exists a bijective function
are said to be isomorphic if there exists a bijective function 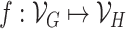 such that
such that 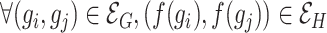 . Finding if two graphs are isomorphic is a recurrent problem in graph analysis that has deep ramifications for machine learning on graphs. For instance, in graph classification tasks, it is assumed that a model needs to capture the similarities between pairs of graphs to classify them accurately.
. Finding if two graphs are isomorphic is a recurrent problem in graph analysis that has deep ramifications for machine learning on graphs. For instance, in graph classification tasks, it is assumed that a model needs to capture the similarities between pairs of graphs to classify them accurately.
The Weisfeiler-Lehman (WL) graph isomorphism test [60] is a classical polynomial-time algorithm in graph theory. It is based on iterative graph recolouring, starting with all nodes of identical ‘colour’ (label). At each step, the algorithm aggregates the colours of nodes and their neighbourhoods and hashes the aggregated colour into unique new colours. The algorithm stops upon reaching a stable colouring. If at that point, the colourings of the two graphs differ, the graphs are deemed non-isomorphic. However, if the colourings are the same, the graphs are possibly (but not necessarily) isomorphic. In other words, the WL test is a necessary but insufficient condition for graph isomorphism. There exist non-isomorphic graphs for which the WL test produces identical colouring and thus considers them possibly isomorphic; the test is said to fail in this case [61].
Machine Learning on Graphs
Most machine learning methods that operate on graphs can be decomposed into two parts: a general-purpose encoder and a task-specific decoder [62]. The encoder embeds a graph’s nodes, or the graph itself, in a low-dimensional feature space. To embed entire graphs, it is common first to embed nodes and then apply a permutation invariant pooling function to produce a graph level representation (e.g. sum, max or mean over node embeddings). The decoder computes an output for the associated task. The components can either be combined in two-step frameworks, with the encoder pre-trained in an unsupervised setting, or in an end-to-end fashion. The end tasks can be classified following multiple dichotomies: supervised/unsupervised, inductive/transductive and node-level/graph-level.
Supervised/unsupervised task. This is the classic dichotomy found in machine learning [63]. Supervised tasks aim to learn a mapping function from labelled data such that the function maps each data point to its label and generalizes to unseen data points. In contrast, unsupervised tasks highlight unknown patterns and uncover structures in unlabelled datasets.
Inductive/transductive task. Inductive tasks correspond to supervised learning discussed above. Transductive tasks expect that all data points are available when learning a mapping function, including unlabelled data points [32]. Hence, in the transductive setting, the model learns both from unlabelled and labelled data. In this respect, inductive learning is more general than transductive learning, as it extends to unseen data points.
Node-level/graph-level task. This dichotomy is based on the object of interest. A task can either focus on the nodes within a graph, e.g. classifying nodes within the context set by the graph, or focus on whole graphs, i.e. each data point corresponds to an entire graph [31]. Note that node-level tasks can be further decomposed into node attribute prediction tasks [32] and link inference tasks [30]. The former focuses on predicting properties of nodes while the latter infers missing links in the graph.
As an illustration, consider the task of predicting the chemical properties of small molecules based on their chemical structures. This is a graph-level task in a supervised (inductive) setting whereby labelled data is used to learn a mapping from chemical structure to chemical properties. Alternatively, the task of identifying groups of proteins that are tightly associated in a PPI graph is an unsupervised node-level task. However, predicting proteins’ biological functions using their interactions in a PPI graph corresponds to a node-level transductive task.
Further, types of tasks can be identified, e.g. based on whether we have static or varying graphs. Biological graphs can vary and evolve along a temporal dimension resulting in changes to composition, structure and attributes [64, 65]. However, the classifications detailed above are the most commonly found in the literature. We review below the existing classes of GML methods.
Traditional approaches
Graph statistics
In the past decades, a flourish of heuristics and statistics have been developed to characterize graphs and their nodes. For instance, the diverse centrality measures capture different aspects of graphs connectivity. The closeness centrality quantifies how closely a node is connected to all other nodes, and the betweenness centrality measures how many shortest paths between pairs of other nodes a given node is part of. Furthermore, graph sub-structures can be used to derive topological descriptors of the wiring patterns around each node in a graph. For instance, motifs [66] and graphlets [67] correspond to sets of small graphs used to characterize local wiring patterns of nodes. Specifically, we can derive a feature vector with length corresponding to the number of considered motifs (or graphlets) where the  element indicates the frequency of the
element indicates the frequency of the  motif.
motif.
These handcrafted features can provide node, or graph, representations that can be used as input to machine learning algorithms. A popular approach has been the definition of kernels based on graph statistics that can be used as input to support vector machines (SVM). For instance, the graphlet kernel [68] captures node wiring patterns similarity, and the WL kernel [69] captures graph similarity based on the WL algorithm discussed in Section 2.
Random walks
Random-walk based methods have been a popular, and successful, approach to embed a graph’s nodes in a low-dimensional space such that node proximities are preserved. The underlying idea is that the distance between node representations in the embedding space should correspond to a measure of distance on the graph, measured here by how often a given node is visited in random walks starting from another node. Deepwalk [29] and node2vec [70] are arguably the most famous methods in this category.
In practice, Deepwalk simulates multiple random walks for each node in the graph. Then, given the embedding 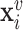 of a node
of a node 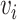 , the objective is to maximize the log probability
, the objective is to maximize the log probability  for all nodes
for all nodes 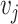 that appear in a random walk within a fixed window of
that appear in a random walk within a fixed window of 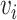 . The method draws its inspiration from the SkipGram model developed for natural language processing [71].
. The method draws its inspiration from the SkipGram model developed for natural language processing [71].
DeepWalk uses uniformly random walks, but several follow-up works analyze how to bias these walks to improve the learned representations. For example, node2vec biases the walks to behave more or less like certain search algorithms over the graph. The authors report a higher quality of embeddings with respect to information content when compared to Deepwalk.
Geometric approaches
Geometric models for knowledge graph embedding posit each relation type as a geometric transformation from source to target in the embedding space. Consider a triplet 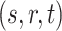 ,
, 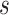 denoting the source node and
denoting the source node and 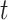 denoting the target node. A geometric model learns a transformation
denoting the target node. A geometric model learns a transformation 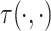 such that
such that 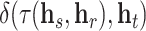 is small, with
is small, with 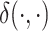 being some notion of distance (e.g. Euclidean distance) and
being some notion of distance (e.g. Euclidean distance) and 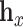 denoting the embedding of entity
denoting the embedding of entity 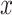 . The key differentiating choice between these approaches is the form of the geometric transformation
. The key differentiating choice between these approaches is the form of the geometric transformation 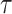 .
.
TransE [72] is a purely translational approach, where 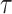 corresponds to the sum of the source node and relation embeddings. In essence, the model enforces that the motion from the embedding
corresponds to the sum of the source node and relation embeddings. In essence, the model enforces that the motion from the embedding 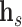 of the source node in the direction given by the relation embedding
of the source node in the direction given by the relation embedding 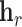 terminates close to the target node’s embedding
terminates close to the target node’s embedding 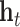 as quantified by the chosen distance metric. Due to its formulation, TransE is not able to account effectively for symmetric relationships or one-to-many interactions.
as quantified by the chosen distance metric. Due to its formulation, TransE is not able to account effectively for symmetric relationships or one-to-many interactions.
Alternatively, RotatE [30] represents relations as rotations in a complex latent space. Thus, 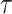 applies a rotation matrix
applies a rotation matrix 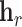 , corresponding to the relation, to the embedding vector of the source node
, corresponding to the relation, to the embedding vector of the source node 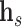 such that the rotated vector
such that the rotated vector 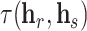 lies close to the embedding vector
lies close to the embedding vector 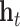 of the target node in terms of Manhattan distance. The authors demonstrate that rotations can correctly capture diverse relation classes, including symmetry/anti-symmetry.
of the target node in terms of Manhattan distance. The authors demonstrate that rotations can correctly capture diverse relation classes, including symmetry/anti-symmetry.
Matrix/tensor factorization
Matrix factorization is a common problem in mathematics that aims to approximate a matrix 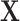 by the product of
by the product of 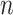 low-dimensional latent factors,
low-dimensional latent factors, 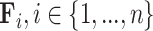 . The general problem can be written as
. The general problem can be written as
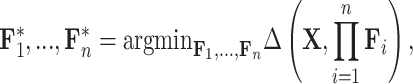 |
where 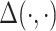 represents a measure of the distance between two inputs, such as Euclidean distance or Kullback-Leibler divergence. In machine learning, matrix factorization has been extensively used for unsupervised applications such as dimensionality reduction, missing data imputation and clustering. These approaches are especially relevant to the knowledge graph embedding problem and have set state-of-the-art (SOTA) results on standard benchmarks [73].
represents a measure of the distance between two inputs, such as Euclidean distance or Kullback-Leibler divergence. In machine learning, matrix factorization has been extensively used for unsupervised applications such as dimensionality reduction, missing data imputation and clustering. These approaches are especially relevant to the knowledge graph embedding problem and have set state-of-the-art (SOTA) results on standard benchmarks [73].
For graphs, the objective is to factorize the adjacency matrix 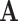 , or a derivative of the adjacency matrix (e.g. Laplacian matrix). It can effectively be seen as finding embeddings for all entities in the graph on a low-dimensional, latent manifold under user-defined constraints (e.g. latent space dimension) such that the adjacency relationships are preserved under dot products.
, or a derivative of the adjacency matrix (e.g. Laplacian matrix). It can effectively be seen as finding embeddings for all entities in the graph on a low-dimensional, latent manifold under user-defined constraints (e.g. latent space dimension) such that the adjacency relationships are preserved under dot products.
Laplacian eigenmaps, introduced by Belkin et al. [74], is a fundamental approach designed to embed entities based on a similarity derived graph. Laplacian eigenmaps uses the eigendecomposition of the Laplacian matrix of a graph to embed each of the 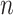 nodes of a graph
nodes of a graph 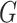 in a low-dimensional latent manifold. The spectral decomposition of the Laplacian is given by equation
in a low-dimensional latent manifold. The spectral decomposition of the Laplacian is given by equation 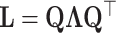 , where
, where 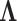 is a diagonal matrix with entries corresponding to the eigenvalues of L and column
is a diagonal matrix with entries corresponding to the eigenvalues of L and column 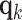 of
of 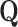 gives the eigenvector associated to the
gives the eigenvector associated to the 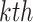 eigenvalue
eigenvalue 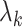 (i.e.
(i.e.  ). Given a user defined dimension
). Given a user defined dimension 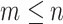 , the embedding of node
, the embedding of node 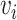 is given by the vector
is given by the vector 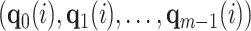 , where
, where 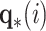 indicates the
indicates the 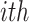 entry of vector
entry of vector  .
.
Nickel et al. [75] introduced RESCAL to address the knowledge graph embedding problem. RESCAL’s objective function is defined as
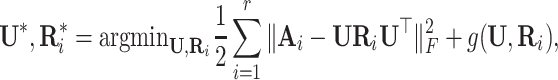 |
where 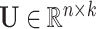 and
and 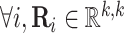 , with
, with 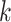 denoting the latent space dimension. The function
denoting the latent space dimension. The function 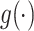 denotes a regularizer, i.e. a function applying constraints on the free parameters of the model, on the factors
denotes a regularizer, i.e. a function applying constraints on the free parameters of the model, on the factors  and
and  . Intuitively, factor
. Intuitively, factor 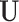 learns the embedding of each entity in the graph and factor
learns the embedding of each entity in the graph and factor 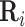 specifies the interactions between entities under relation
specifies the interactions between entities under relation 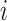 . Yang et al. [76] proposed DistMult, a variation of RESCAL that considers each factor
. Yang et al. [76] proposed DistMult, a variation of RESCAL that considers each factor 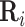 as a diagonal matrix. Trouillon et al. [77] proposed ComplEx, a method extending DistMult to the complex space taking advantage of the Hermitian product to represent asymmetric relationships.
as a diagonal matrix. Trouillon et al. [77] proposed ComplEx, a method extending DistMult to the complex space taking advantage of the Hermitian product to represent asymmetric relationships.
Alternatively, some existing frameworks leverage both a graph’s structural information and the node’s semantic information to embed each entity in a way that preserves both sources of information. One such approach is to use a graph’s structural information to regularize embeddings derived from the factorization of the feature matrix 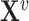 [78, 79]. The idea is to penalize adjacent entities in the graph to have closer embeddings in the latent space, according to some notion of distance. Another approach is to jointly factorize both data sources, for instance, introducing a kernel defined on the feature matrix [80, 81].
[78, 79]. The idea is to penalize adjacent entities in the graph to have closer embeddings in the latent space, according to some notion of distance. Another approach is to jointly factorize both data sources, for instance, introducing a kernel defined on the feature matrix [80, 81].
Graph neural networks
GNNs were first introduced in the late 1990s [82–84] but have attracted considerable attention in recent years, with the number of variants rising steadily [32–34, 44, 85–89]. From a high-level perspective, GNNs are a realization of the notion of group invariance, a general blueprint underpinning the design of a broad class of deep learning architectures. The key structural property of graphs is that the nodes are usually not assumed to be provided in any particular order, and any functions acting on graphs should be permutation invariant (order-independent); therefore, for any two isomorphic graphs, the output of said functions are identical. A typical GNN consists of one or more layers implementing a node-wise aggregation from the neighbour nodes; since the ordering of the neighbours is arbitrary, the aggregation must be permutation invariant. When applied locally to every node of the graph, the overall function is permutation equivariant, i.e. its output is changed in the same way as the input under node permutations.
GNNs are amongst the most general class of deep learning architectures currently in existence. Popular architectures such as DeepSets [90], transformers [91] and convolutional neural networks [92] can be derived as particular cases of GNNs operating on graphs with an empty edge set, a complete graph, and a ring graph, respectively. In the latter case, the graph is fixed and the neighbourhood structure is shared across all nodes; the permutation group can therefore be replaced by the translation group, and the local aggregation expressed as a convolution. While a broad variety of GNN architecture exists, their vast majority can be classified into convolutional, attentional and message-passing ‘flavours’ — with message-passing being the most general formulation.
Message passing networks
A message passing-type GNN layer is comprised of three functions: (1) a message passing function Msg that permits information exchange between nodes over edges; (2) a permutation-invariant aggregation function Agg that combines the collection of received messages into a single, fixed-length representation (3) and an update function Update that produces node-level representations given the previous representation and the aggregated messages. Common choices are a simple linear transformation for Msg, summation, simple- or weighted-averages for Agg and multilayer perceptrons (MLP) with activation functions for the Update function, although it is not uncommon for the Msg or Update function to be absent or reduced to an activation function only. Where the node representations after layer 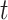 are
are 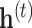 , we have
, we have
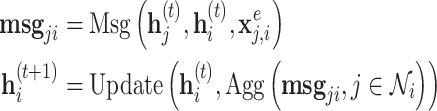 |
or, more compactly,
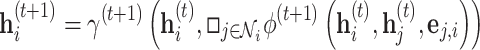 |
where 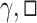 and
and 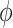 are the update, aggregation and message passing functions, respectively, and
are the update, aggregation and message passing functions, respectively, and  indicates the layer index [93]. The design of the aggregation
indicates the layer index [93]. The design of the aggregation  is important: when chosen to be an injective function, the message passing mechanism can be shown to be equivalent to the colour refinement procedure in the WL algorithm [86]. The initial node representations,
is important: when chosen to be an injective function, the message passing mechanism can be shown to be equivalent to the colour refinement procedure in the WL algorithm [86]. The initial node representations, 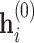 , are typically set to node features,
, are typically set to node features,  . Figure 2 gives a schematic representation of this operation.
. Figure 2 gives a schematic representation of this operation.
Figure 2 .
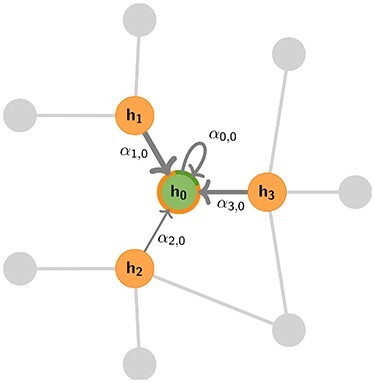
Illustration of a general aggregation step performed by a GNN for the central node (green) based on its direct neighbours (orange). Messages may be weighted depending on their content, the source or target node features or the attributes of the edge they are passed along, as indicated by the thickness of incoming arrows.
Graph convolutional network
The graph convolutional network (GCN) [32] can be decomposed in this framework as
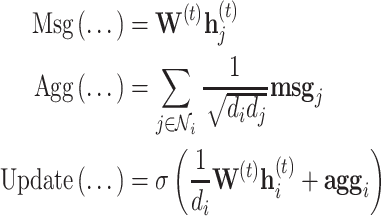 |
where 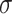 is some activation function, usually a rectified linear unit (ReLU). The scheme is simplified further if we consider the addition of self-loops, that is, an edge from a node to itself, commonly expressed as the modified adjacency
is some activation function, usually a rectified linear unit (ReLU). The scheme is simplified further if we consider the addition of self-loops, that is, an edge from a node to itself, commonly expressed as the modified adjacency 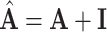 , where the aggregation includes the self-message and the update reduces to
, where the aggregation includes the self-message and the update reduces to
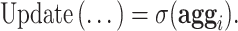 |
With respect to the notations in Figure 2, for GCN we have 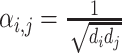 .
.
As the update depends only on a node’s local neighbourhood, these schemes are also commonly referred to as neighbourhood aggregation. Indeed, taking a broader perspective, a single-layer GNN updates a node’s features based on its immediate or one-hop neighbourhood. Adding a second GNN layer allows information from the two-hop neighbourhood to propagate via intermediary neighbours. By further stacking GNN layers, node features can come to depend on the initial values of more distant nodes, analogous to the broadening the receptive field in later layers of convolutional neural networks—the deeper the network, the broader the receptive field (see Figure 3). However, this process is diffusive and leads to features washing out as the graph thermalizes. This problem is solved in convolutional networks with pooling layers, but an equivalent canonical coarsening does not exist for irregular graphs.
Figure 3 .
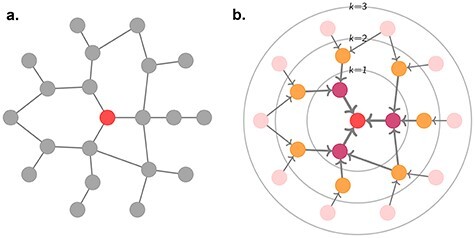
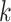 -hop neighbourhoods of the central node (red). Typically, a GNN layer operates on the 1-hop neighbourhood, i.e. nodes with which the central node shares an edge, within the
-hop neighbourhoods of the central node (red). Typically, a GNN layer operates on the 1-hop neighbourhood, i.e. nodes with which the central node shares an edge, within the 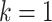 circle. Stacking layers allows information from more distant nodes to propagate through intermediate nodes.
circle. Stacking layers allows information from more distant nodes to propagate through intermediate nodes.
Graph attention network
Graph attention networks (GAT) [33] weight incoming messages with an attention mechanism and multi-headed attention for train stability, including self-loops, the message, aggregation and update functions
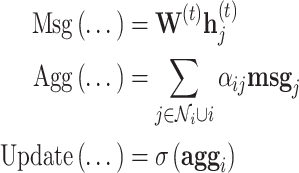 |
are otherwise unchanged. Although the authors suggest the attention mechanism is decoupled from the architecture and should be task specific, in practice, their original formulation is most widely used. The attention weights, 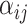 are softmax normalized, that is
are softmax normalized, that is
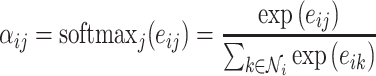 |
where 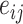 is the output of a single layer feed-forward neural network without a bias (a projection) with LeakyReLU activations, that takes the concatenation of transformed source- and target-node features as input,
is the output of a single layer feed-forward neural network without a bias (a projection) with LeakyReLU activations, that takes the concatenation of transformed source- and target-node features as input,
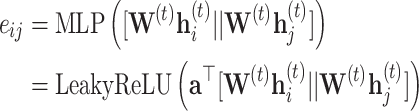 |
where 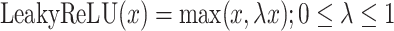 .
.
Relational graph convolutional networks
At many scales of systems biology, the relationships between entities have a type, a direction, or both. For instance, the type of bonds between atoms, binding of two proteins, and gene regulatory interactions are essential to understanding the systems in which they exist. This idea is expressed in the message passing framework with messages that depend on edge attributes. Relational graph convolutional networks (R-GCNs) [44] learn separate linear transforms for each edge type, which can be viewed as casting the graph as a multiplex graph and operating GCN-like models independently on each layer, as shown in Figure 4.
Figure 4 .
A multi-relational graph (A) can be parsed into layers of a multiplex graph (B). The R-GCN learns a separate transform for each layer, and the self-loop and messages are passed according to the connectivity of the layers. For example, node (A) passes a message to node (B) in the top layer, receives a message from (B) in the middle layer, and does not communicate with (B) in the bottom layer.
The R-GCN model decomposes to
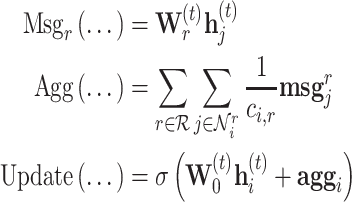 |
for edge types 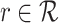 , with separate transforms
, with separate transforms 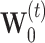 for self-loops, and problem-specific normalization constant
for self-loops, and problem-specific normalization constant 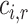 .
.
The different types of GNNs above illustrate some approaches to define message passing on graphs. Note that there is no established best scheme for all scenarios and that each specific application might require a different scheme.
Graph pooling
Geometric deep learning approaches machine learning with graph-structured data as the generalization of methods designed for learning with grid and sequence data (images, time-series; Euclidean data) to non-Euclidean domains, i.e. graphs and manifolds [94]. This is also reflected in the derivation and naming conventions of popular GNN layers as generalized convolutions [32, 44]. Modern convolutional neural networks have settled on the combination of layers of 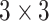 kernels interspersed with
kernels interspersed with 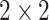 max-pooling. Developing a corresponding pooling workhorse for GNNs is an active area of research. The difficulty is that, unlike Euclidean data structures, there are no canonical up- and down-sampling operations for graphs. As a result, there are many proposed methods that centre around learning to pool or prune based on features [95–99], and learned or non-parametric structural pooling [100–103]. However, the distinction between featural and structural methods is blurred when topological information is included in the node features.
max-pooling. Developing a corresponding pooling workhorse for GNNs is an active area of research. The difficulty is that, unlike Euclidean data structures, there are no canonical up- and down-sampling operations for graphs. As a result, there are many proposed methods that centre around learning to pool or prune based on features [95–99], and learned or non-parametric structural pooling [100–103]. However, the distinction between featural and structural methods is blurred when topological information is included in the node features.
The most successful feature-based methods extract representations from graphs directly either for cluster assignments [95, 99] or for top-k pruning [96–98]. DiffPool uses GNNs both to produce a hierarchy of representations for overall graph classification and to learn intermediate representations for soft cluster assignments to a fixed number of pools [95]. Figure 5 presents an example of this kind of pooling. top-k pooling takes a similar approach, but instead of using an auxiliary learning process to pool nodes, it is used to prune nodes [96, 97]. In many settings, this simpler method is competitive with DiffPool at a fraction of the memory cost.
Figure 5 .
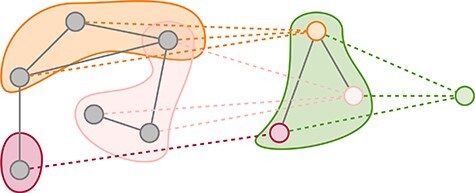
A possible graph pooling schematic. Nodes in the original graph (left, grey) are pooled into nodes in the intermediate graph (centre) as shown by the dotted edges. The final pooling layer aggregates all the intermediate nodes into a single representation (right, green). DiffPool could produce the pooling shown [95].
Structure-based pooling methods aggregate nodes based on the graph topology and are often inspired by the processes developed by chemists and biochemists for understanding molecules through their parts. For example, describing a protein in terms of its secondary structure ( -helix,
-helix,  -sheets) and the connectivity between these elements can be seen as a pooling operation over the protein’s molecular graph. Figure 6 shows how a small molecule can be converted to a junction tree representation, with the carbon ring (in pink) being aggregated into a single node. Work on decomposing molecules into motifs bridges the gap between handcrafted secondary structures and unconstrained learning methods [103]. Motifs are extracted based on a combined statistical and chemical analysis, where motif templates (i.e. graph substructures) are selected based on how frequently they occur in the training corpus and molecules are then decomposed into motifs according to some chemical rules. More general methods look to concepts from graph theory such as minimum cuts [100, 102] and maximal cliques [101] on which to base pooling. Minimum cuts are graph partitions that minimize some objective and have obvious connections to graph clustering, whilst cliques (subsets of nodes that are fully connected) are in some sense at the limit of node community density.
-sheets) and the connectivity between these elements can be seen as a pooling operation over the protein’s molecular graph. Figure 6 shows how a small molecule can be converted to a junction tree representation, with the carbon ring (in pink) being aggregated into a single node. Work on decomposing molecules into motifs bridges the gap between handcrafted secondary structures and unconstrained learning methods [103]. Motifs are extracted based on a combined statistical and chemical analysis, where motif templates (i.e. graph substructures) are selected based on how frequently they occur in the training corpus and molecules are then decomposed into motifs according to some chemical rules. More general methods look to concepts from graph theory such as minimum cuts [100, 102] and maximal cliques [101] on which to base pooling. Minimum cuts are graph partitions that minimize some objective and have obvious connections to graph clustering, whilst cliques (subsets of nodes that are fully connected) are in some sense at the limit of node community density.
Figure 6 .
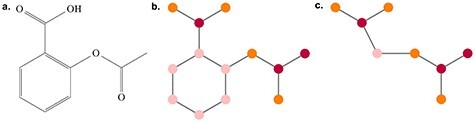
Illustration of (A) the molecule aspirin, (B) its basic graph representation and (C) the associated junction tree representation. Colours on the node correspond to atom types.
Drug Development Applications
The process of discovering a drug and making it available to patients can take up to 10 years and is characterized by failures, or attrition, see Figure 1. The early discovery stage involves target identification and validation, hit discovery, lead molecule identification and then optimization to achieve the desired characteristics of a drug candidate [114]. Pre-clinical research typically comprises both in vitro and in vivo assessments of toxicity, pharmacokinetics (PK), pharmacodynamics (PD) and efficacy of the drug. Providing good pre-clinical evidence is presented, the drug then progresses for human clinical trials normally through three different phases of clinical trials. In the following subsections, we explore how GML can be applied to distinct stages within the drug discovery and development process.
In Table 1, we provide a summary of how some of the key work reviewed in this section ties to the methods discussed in Section 3. Table 2 outlines the underlying key data types and databases therein. Biomedical databases are typically presented as general repositories with minimal reference to downstream applications. Therefore, additional processing is often required for a specific task; to this end, efforts have been directed towards processed data repositories with specific endpoints in mind [115, 116].
Table 1.
Exemplar references linking applications from Section 4 to methods described in Section 3. The entries in the data types column refers to acronyms defined in Table 2. The last column indicates the presence of follow up experimental validation. *The authors consider a mesh graph over protein surfaces.
| Relevant application | Ref. | Method type | Task level | ML approach | Data types | Exp. val? |
|---|---|---|---|---|---|---|
| 4.1 Target identification | ||||||
| − | [47] | Geometric (§3.2) | Node-level | Unsupervised | Di, Dr, GA | |
| 4.2 Design of small molecules therapies | ||||||
| Molecular property prediction | [21] | GNN (§3.4) | Graph-level | Supervised | Dr | |
| [104] | GNN (§3.4) | Graph-level | Supervised | Dr | ||
| [22] | GNN (§3.4) | Graph-level | Supervised | Dr | ||
| Enhanced high throughput screens | [50] | GNN (§3.4) | Graph-level | Supervised | Dr | ✓ |
| De novo design | [105] | GNN (§3.4) | Graph-level | Unsupervised | Dr | |
| [48] | Factorisation (§3.3) | Graph-level | Semi-supervised | Dr | ✓ | |
| 4.3 Design of new biological entities | ||||||
| ML-assisted directed evolution | − | − | − | − | − | |
| Protein engineering | [49] | GNN (§3.4) | Subgraph-level | *Supervised | PS | |
| De novo design | [106] | GNN (§3.4) | Graph-level | Supervised | PS | ✓ |
| 4.4 Drug repurposing | ||||||
| Off-target repurposing | [107] | Factorisation (§3.3) | Node-level | Unsupervised | Dr, PI | |
| [108] | GNN (§3.4) | Graph-level | Supervised | Dr, PS | ||
| [109] | Factorisation (§3.3) | Node-level | Unsupervised | Dr, Di | ||
| On-target repurposing | [110] | GNN (§3.4) | Node-level | Supervised | Dr, Di | |
| [111] | Geometric (§3.2) | Node-level | Unsupervised | Dr, Di, PI, GA | ||
| Combination repurposing | [112] | GNN (§3.4) | Node-level | Supervised | Dr, PI, DC | |
| [113] | GNN (§3.4) | Graph-level | Supervised | Dr, DC | ✓ | |
Table 2.
Different types of data relevant to drug discovery and development applications with associated databases.
Target identification
Target identification is the search for a molecular target with a significant functional role(s) in the pathophysiology of a disease such that a hypothetical drug could modulate said target culminating with beneficial effect [132, 133]. Early targets included G-protein coupled receptors (GPCRs), kinases, and proteases and formed the major target protein families amongst first-in-class drugs [134] — targeting other classes of biomolecules is also possible, e.g. nucleic acids. For an organ-specific therapy, an ideal target should be strongly and preferably expressed in the tissue of interest, and preferably a three-dimensional structure should be obtainable for biophysical simulations.
There is a range of complementary lines of experimental evidence that could support target identification. For example, a phenomenological approach to target identification could consider the imaging, histological or -omic presentation of diseased tissue when compared to matched healthy samples. Typical differential presentation includes chromosomal aberrations (e.g. from WGS), differential expression (e.g. via RNA-seq) and protein translocation (e.g. from histopathological analysis) [135]. As the availability of -omic technologies increases, computational and statistical advances must be made to integrate and interpret large quantities of high dimensional, high-resolution data on a comparatively small number of samples, occasionally referred to as panomics [136, 137].
In contrast to a static picture, in vitro and in vivo models are built to examine the dynamics of disease phenotype to study mechanism. In particular, genes are manipulated in disease models to understand key drivers of a disease phenotype. For example, random mutagenesis could be induced by chemicals or transposons in cell clones or mice to observe the phenotypic effect of perturbing certain cellular pathways at the putative target protein [133]. As a targeted approach, bioengineering techniques have been developed to either silence mRNA or remove the gene entirely through genetic editing. In modern times, CRISPR is being used to knockout genes in a cleaner manner to prior technologies, e.g. siRNA, shRNA, TALEN [138–140]. Furthermore, innovations have led to CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa) that allow for suppression or overexpression of target genes [141].
To complete the picture, biochemical experiments observe chemical and protein interactions to inform on possible drug mechanisms of action [132], examples include: affinity chromatography, a range of mass spectrometry techniques for proteomics, and drug affinity responsive target stability assays [142–144]. X-ray crystallography and cryogenic electron microscopy (cryo-EM) can be used to detail structures of proteins to identify druggable pockets [145]; computational approaches can be used to assess the impacts of mutations in cancer resulting in perturbed crystal structures [146]. Yeast two-hybrid or three-hybrid systems can be employed to detail genomic PPI or RNA–protein interaction [147, 148].
Systems biology aims to unify phenomenological observations on disease biology (the ‘-omics view’), genetic drivers of phenotypes (driven by bioengineering) through a network view of interacting biomolecules [149]. The ultimate goal is to pin down a ‘druggable’ point of intervention that could hopefully reverse the disease condition. One of the outcomes of this endeavour is the construction of signalling pathways; for example, the characterization of the TGF- , PI3K/AKT and Wnt-dependent signalling pathways have had profound impacts on oncology drug discovery [150–152].
, PI3K/AKT and Wnt-dependent signalling pathways have had profound impacts on oncology drug discovery [150–152].
In contrast to complex diseases, target identification for infectious disease requires a different philosophy. After eliminating pathogenic targets structurally similar to those within the human proteome, one aims to assess the druggability of the remaining targets. This may be achieved using knowledge of the genome to model the constituent proteins when 3D structures are not already available experimentally. The Blundell group has shown that 70-80% of the proteins from Mycobacterium tuberculosis and a related organism, Mycobacterium abscessus (infecting cystic fibrosis patients), can be modelled via homology [153, 154]. By examining the potential binding sites, such as the catalytic site of an enzyme or an allosteric regulatory site, the binding hotspot can be identified and the potential value as a target estimated [155]. Of course, target identification is also dependent on the accessibility of the target to a drug, as well as the presence of efflux pumps — and metabolism of any potential drug by the infectious agent.
From systems biology to machine learning on graphs
Organisms, or biological systems, consist of complex and dynamic interactions between entities at multiple scales. At the submolecular level, proteins are chains of amino acid residues which fold to adopt highly specific conformational structures. At the molecular scale, proteins and other biomolecules physically interact through transient and long-timescale binding events to carry out regulatory processes and perform signal amplification through cascades of chemical reactions. By starting with a low-resolution understanding of these biomolecular interactions, canonical sequences of interactions associated with specific processes become labelled as pathways that ultimately control cellular functions and phenotypes. Within multicellular organisms, cells interact with each other forming diverse tissues and organs. A reductionist perspective of disease is to view it as being the result of perturbations of the cellular machinery at the molecular scale that manifest through aberrant phenotypes at the cellular and organismal scales. Within target identification, one is aiming to find nodes that upon manipulation lead to a causal sequence of events resulting in the reversion from a diseased to a healthy state.
It seems plausible that target identification will be the greatest area of opportunity for machine learning on graphs. From a genetics perspective, examining Mendelian traits and genome-wide association studies linked to coding variants of drug targets have a greater chance of success in the clinic [156, 157]. However, when examining PPI networks, Fang et al. [158] found that various protein targets were not themselves ‘genetically associated’, but interacted with other proteins with genetic associations to the disease in question. For example in the case of rheumatoid arthritis (RA), tumour necrosis factor (TNF) inhibition is a popular drug mechanism of action with no genetic association — but the interacting proteins of TNF including CD40, NFKBIA, REL, BIRC3 and CD8A have variants that are known to exhibit a genetic predisposition to RA.
Oftentimes, systems biology has focused on networks with static nodes and edges, ignoring faithful characterization of underlying biomolecules that the nodes represent. With GML, we can account for much richer representations of biology accounting for multiple relevant scales, for example, graphical representation of molecular structures (discussed in Sections 4.2 and 4.3), functional relationships within a knowledge graph (discussed in Section 4.4), and expression of biomolecules. Furthermore, GML can learn graphs from data as opposed to relying on pre-existing incomplete knowledge [159, 160]. Early work utilizing GML for target identification includes Pittala et al. [47], whereby a knowledge graph link prediction approach was used to beat the in house algorithms of open targets [161] to rediscover drug targets within clinical trials for Parkinson’s disease.
The utilization of multi-omic expression data capturing instantaneous multimodal snapshots of cellular states will play a significant role in target identification as costs decrease [162, 163] — particularly in a precision medicine framework [136]. Currently, however, only a few panomic datasets are publicly available. A small number of early adopters have spotted the clear utility in employing GML [164], occasionally in a multimodal learning [165, 166], or causal inference setting [167]. These approaches have helped us move away from the classical Mendelian ‘one gene – one disease’ philosophy and appreciate the true complexity of biological systems.
Design of small molecule therapies
Drug design broadly falls into two categories: phenotypic drug discovery and target-based drug discovery. Phenotypic drug discovery (PDD) begins with a disease’s phenotype without having to know the drug target. Without the bias from having a known target, PDD has yielded many first-in-class drugs with novel mechanisms of action [168]. It has been suggested that PDD could provide the new paradigm of drug design, reducing costs substantially and increasing productivity [169]. However, drugs found by PDD are often pleiotropic and impose greater safety risks when compared to target-oriented drugs. In contrast, best-in-class drugs are usually discovered by a target-based approach.
For target-based drug discovery, after target identification and validation, ‘hit’ molecules would be identified via high-throughput screening of compound libraries against the target [114], typically resulting in a large number of possible hits. Grouping these into ‘hit series’ and they become further refined in functional in vitro assays. Ultimately, only those selected via secondary in vitro assays and in vivo models would be the drug ‘leads’. With each layer of screening and assays, the remaining compounds should be more potent and selective against the therapeutic target. Finally, lead compounds are optimized by structural modifications, to improve properties such as PKPD, typically using heuristics, e.g. Lipinski’s rule of five [170]. In addition to such structure-based approach, fragment-based (FBDD) [171, 172] and ligand-based drug discovery (LBDD) have also been popular [173, 174]. FBDD enhances the ligand efficiency and binding affinity with fragment-like leads of  150 Da, whilst LBDD does not require 3D structures of the therapeutic targets.
150 Da, whilst LBDD does not require 3D structures of the therapeutic targets.
Both phenotypic- and target-based drug discovery comes with their own risks and merits. While the operational costs of target ID may be optional, developing suitable phenotypic screening assays for the disease could be more time-consuming and costly [169, 175]. Hence, the overall timeline and capital costs are roughly the same [175].
In this review, we make no distinction between new chemical entities (NCE), new molecular entities (NME) or new active substances (NAS) [176].
Modelling philosophy
For a drug, the base graph representation is obtained from the molecule’s SMILES signature and captures bonds between atoms, i.e. each node of the graph corresponds to an atom and each edge stands for a bond between two atoms [21, 22]. The features associated with atoms typically include its element, valence and degree. Edge features include the associated bond’s type (single, double, triple), its aromaticity, and whether it is part of a ring or not. Additionally, Klicpera et al. [22] consider the geometric length of a bond and geometric angles between bonds as additional features. This representation is used in most applications, sometimes complemented or augmented with heuristic approaches.
To model a graph structure, Jin et al. [105] used the base graph representation in combination with a junction tree derived from it. To construct the junction tree, the authors first define a set of molecule substructures, such as rings. The graph is then decomposed into overlapping components, each corresponding to a specific substructure. Finally, the junction tree is defined with each node corresponding to an identified component and each edge associates overlapping components.
Jin et al. [103] then extended their previous work by using a hierarchical representation with various coarseness of the small molecule. The proposed representation has three levels: (1) an atom layer, (2) an attachment layer and (3) a motif layer. The first level is simply the basic graph representation. The following levels provide the coarse and fine-grain connection patterns between a molecule’s motifs. Specifically, the attachment layer describes at which atoms two motifs connect, while the motif layer only captures if two motifs are linked. Considering a molecule base graph representation 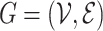 , a motif is defined as a subgraph of
, a motif is defined as a subgraph of 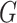 induced on atoms in
induced on atoms in 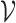 and bonds in
and bonds in 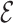 . Motifs are extracted from a molecule’s graph by breaking bridge bonds.
. Motifs are extracted from a molecule’s graph by breaking bridge bonds.
Kajino [177] opted for a hypergraph representation of small molecules. A hypergraph is a generalization of graphs in which an edge, called a hyperedge, can connect any number of nodes. In this setting, a node of the hypergraph corresponds to a bond between two atoms of the small molecule. In contrast, a hyperedge then represents an atom and connects all its bonds (i.e. nodes).
Molecular property prediction
Pharmaceutical companies may screen millions of small molecules against a specific target, e.g. see GlaxoSmithKline’s DNA-encoded small molecule library of 800 million entries [178]. However, as the end result will be optimized via a skilled medicinal chemist, one should aim to substantially cut down the search space by screening only a representative selection of molecules for optimization later. One route towards this is to select molecules with heterogeneous chemical properties using GML approaches. This is a popular task with well-established benchmarks such as QM9 [179] and MD17 [180]. Top-performing methods are based on GNNs.
For instance, using a graph representation of drugs, Duvenaud et al. [21] have shown substantial improvements over non-graph-based approaches for molecule property prediction tasks. Specifically, the authors used GNNs to embed each drug and tested the predictive capabilities of the model on diverse benchmark datasets. They demonstrated improved interpretability of the model and predictive superiority over previous approaches which relied on circular fingerprints [181]. The authors use a simple GNN layer with a read-out function on the output of each GNN layer that updates the global drug embedding.
Alternatively, Schutt et al. [182] introduced SchNet, a model that characterizes molecules based on their representation as a list of atoms with interatomic distances, that can be viewed as a fully connected graph. SchNet uses learned embeddings for each atom using two modules: (1) an atom-wise module and (2) an interaction module. The former applies a simple MLP transformation to each atom representation input, while the latter updates the atom representation based on the representations of the other atoms of the molecule and using relative distances to modulate contributions. The final molecule representation is obtained with a global sum pooling layer over all atoms’ embeddings.
With the same objective in mind, Klicpera et al. [22] recently introduced DimeNet, a novel GNN architecture that diverges from the standard message passing framework presented in Section 3. DimeNet defines a message coefficient between atoms based on their relative positioning in 3D space. Specifically, the message from node 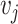 to node
to node 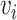 is iteratively updated based on
is iteratively updated based on 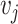 ’s incoming messages as well as the distances between atoms and the angles between atomic bonds. DimeNet relies on more geometric features, considering both the angles between different bonds and the distance between atoms. The authors report substantial improvements over SOTA models for the prediction of molecule properties on two benchmark datasets.
’s incoming messages as well as the distances between atoms and the angles between atomic bonds. DimeNet relies on more geometric features, considering both the angles between different bonds and the distance between atoms. The authors report substantial improvements over SOTA models for the prediction of molecule properties on two benchmark datasets.
Most relevant to the later stages of preclinical work, Feinberg et al. extended previous work on molecular property prediction [104] to include ADME properties [46]. In this scenario, by only using structures of drugs predictions were made across a diverse range of experimental observables, including half-lives across in vivo models (rat, dog), human Ether-à-go-go-Related Gene protein interactions and IC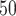 values for common liver enzymes predictive of drug toxicity.
values for common liver enzymes predictive of drug toxicity.
Enhanced high-throughput screens
Within the previous section, chemical properties were a priori defined. In contrast, Stokes et al. [50] leveraged results from a small phenotypic growth inhibition assay of 2335 molecules against Escherichia coli to infer antibiotic properties of the ZINC15 collection of >107 million molecules. After ranking and curating hits, only 23 compounds were experimentally tested — leading to halicin being identified. Of particular note was that the Tanimoto similarity of halicin when compared its nearest neighbour antibiotic, metronidazole, was only 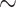 0.21 — demonstrating the ability of the underlying ML to generalize to diverse structures.
0.21 — demonstrating the ability of the underlying ML to generalize to diverse structures.
Testing halicin against a range of bacterial infections, including Mycobacterium tuberculosis, demonstrated broad-spectrum activity through selective dissipation of the 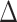 pH component of the proton motive force. In a world first, Stokes et al. showed efficacy of an AI-identified molecule in vivo (Acinetobacter baumannii infected neutropenic BALB/c mice) to beat the standard of care treatment (metronidazole) [50].
pH component of the proton motive force. In a world first, Stokes et al. showed efficacy of an AI-identified molecule in vivo (Acinetobacter baumannii infected neutropenic BALB/c mice) to beat the standard of care treatment (metronidazole) [50].
De novo design
A more challenging task than those previously discussed is de novo design of small molecules from scratch; that is, for a fixed target (typically represented via 3D structure) can one design a suitable and selective drug-like entity?
In the landmark paper, Zhavoronkov et al. [48] created a novel chemical matter against discoidin domain receptor 1 (DDR1) using a variational autoencoder style architecture. Notably, they penalize the model to select structures similar to disclosed drugs from the patent literature. As the approach was designed to find small molecules for a well-known target, crystal structures were available and subsequently utiliszed. Additionally, the ZINC dataset containing hundreds of millions of structures was used (unlabelled data) along with confirmed positive and negative hits for DDR1.
In total, six compounds were synthesized with four attaining <1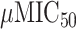 values. Whilst selectivity was shown for two molecules of DDR1 when compared to DDR2, selectivity against a larger panel of off-targets was not shown. Whilst further development (e.g. PK or toxicology testing) was not shown, Zhavoronkov et al. demonstrated de novo small molecule design in an experimental setting [48]. Arguably, the recommendation of an existing molecule is a simpler task than designing one from scratch.
values. Whilst selectivity was shown for two molecules of DDR1 when compared to DDR2, selectivity against a larger panel of off-targets was not shown. Whilst further development (e.g. PK or toxicology testing) was not shown, Zhavoronkov et al. demonstrated de novo small molecule design in an experimental setting [48]. Arguably, the recommendation of an existing molecule is a simpler task than designing one from scratch.
Design of new biological entities
New biological entities (NBE) refer to biological products or biologics, that are produced in living systems [183]. The types of biologics are very diversified, from proteins (>40 amino acids), peptides, antibodies, to cell and gene therapies. Therapeutic proteins tend to be large, complex structured and are unstable in contrast to small molecules [184]. Biologic therapies typically use cell-based production systems that are prone to post-translational modification and are thus sensitive to environmental conditions requiring mass spectrometry to characterize the resulting heterogeneous collection of molecules [185].
In general, the target-to-hit-to-lead pathway also applies to NBE discovery, with similar procedures like high-throughput screening assays. Typically, an affinity-based high-throughput screening method is used to select from a large library of candidates using one target. One must then separately study off-target binding from similar proteins, peptides and immune surveillance [186].
Modelling philosophy
Focusing on proteins, the consensus to derive the protein graph representation is to use pairwise spatial distances between amino acid residues, i.e. the protein’s contact map, and to apply an arbitrary cut-off or Gaussian filter to derive adjacency matrices [19, 20, 187], see Figure 7.
Figure 7 .
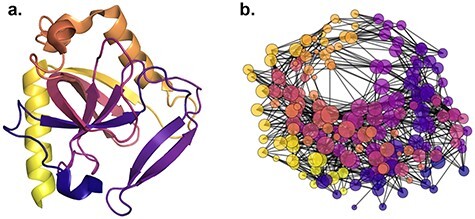
Illustration of (A) a protein (PDB accession: 3EIY) and (B) its graph representation derived based on intramolecular distance with cut-off threshold set at 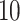 Å.
Å.
However, protein structures are substantially more complex than small molecules and, as such, there are several resulting graph construction schemes. For instance, residue-level graphs can be constructed by representing the intramolecular interactions, such as hydrogen bonds, that compose the structure as edges joining their respective residues. This representation has the advantage of explicitly encoding the internal chemistry of the biomolecule, which determines structural aspects such as dynamics and conformational rearrangements. Other edge constructions can be distance-based, such as K-NN (where a node is joined to its 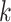 most proximal neighbours) [19, 20] or based on Delaunay triangulation. Node features can include structural descriptors, such as solvent accessibility metrics, encoding the secondary structure, distance from the centre or surface of the structure and low-dimensional embeddings of physicochemical properties of amino acids. It is also possible to represent proteins as large molecular graphs at the atomic level in a similar manner to small molecules. Due to the plethora of graph construction and featurization schemes available, tools are being made available to facilitate the pre-processing of said protein structure [188].
most proximal neighbours) [19, 20] or based on Delaunay triangulation. Node features can include structural descriptors, such as solvent accessibility metrics, encoding the secondary structure, distance from the centre or surface of the structure and low-dimensional embeddings of physicochemical properties of amino acids. It is also possible to represent proteins as large molecular graphs at the atomic level in a similar manner to small molecules. Due to the plethora of graph construction and featurization schemes available, tools are being made available to facilitate the pre-processing of said protein structure [188].
One should note that sequences can be considered as special cases of graphs and are compatible with graph-based methods. However, in practice, language models are preferred to derive protein embeddings from amino acids sequences [189, 190]. Recent works suggest that combining the two can increase the information content of the learnt representations [187, 191]. Several recurrent challenges in the scientific community aim to push the limit of current methods. For instance, the CAFA [192] and CAPRI [193] challenges aim to improve protein functional classification and PPI prediction.
ML-assisted directed evolution
Display technologies have driven the modern development of NBEs; in particular, phage display and yeast display are widely used for the generation of therapeutic antibodies. In general, a peptide or protein library with diverse sequence variety is generated by PCR, or other recombination techniques [194]. The library is ‘displayed’ for genotype–phenotype linkage such that the protein is expressed and fused to surface proteins while the encoding gene is still encapsulated within the phage or cell. Therefore, the library could be screened and selected, in a process coined ‘biopanning’, against the target (e.g. antigen) according to binding affinity. Thereafter, the selected peptides are further optimized by repeating the process with a refined library. In phage display, selection works by physical capture and elution [195]; for cell-based display technologies (like yeast display), fluorescence-activated cell sorting is utilized for selection [196].
Due to the repeated iteration of experimental screens and the high number of outputs, such display technologies are now being coupled to ML systems for greater speed, affinity and further in silico selection [197, 198]. As of yet, it does not appear that advanced GML architectures have been applied in this domain, but promising routes forward have been recently developed. For example, Hawkins-Hooker et al. [199] trained multiple variational autoencoders on the amino acid sequences of 70 000 luciferase-like oxidoreductases to generate new functional variants of the luxA bacterial luciferase. Testing these experimentally led to variants with increased enzyme solubility without disrupting function. Using this philosophy, one has a grounded approach to refine libraries for a directed evolution screen.
Protein engineering
Some proteins have reliable scaffolds that one can build upon, for example, antibodies whereby one could modify the variable region but leave the constant region intact. For example, Deac et al. [200] used dilated (á trous) convolutions and self-attention on antibody sequences to predict the paratope (the residues on the antibody that interact with the antigen) as well as a cross-modal attention mechanism between the antibody and antigen sequences. Crucially, the attention mechanisms also provide a degree of interpretability to the model.
In a protein-agnostic context, Fout et al. [19] used Siamese architectures [201] based on GNNs to predict at which amino acid residues are involved in the interface of a protein–protein complex. Each protein is represented as a graph where nodes correspond to amino acid residues and edges connect each residue to its 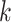 closest residues. The authors propose multiple aggregation functions with varying complexities and following the general principles of a diffusion convolutional neural network [202]. The output embeddings of each residue of both proteins are concatenated all-to-all and the objective is to predict if two residues are in contact in the protein complex based on their concatenated embeddings. The authors report a significant improvement over the method without the GNN layers, i.e. directly using the amino acid residue sequence and structural properties (e.g. solvent accessibility, distance from the surface).
closest residues. The authors propose multiple aggregation functions with varying complexities and following the general principles of a diffusion convolutional neural network [202]. The output embeddings of each residue of both proteins are concatenated all-to-all and the objective is to predict if two residues are in contact in the protein complex based on their concatenated embeddings. The authors report a significant improvement over the method without the GNN layers, i.e. directly using the amino acid residue sequence and structural properties (e.g. solvent accessibility, distance from the surface).
Gainza et al. [49] recently introduced molecular surface interaction fingerprinting (MaSIF) for tasks such as, binding pocket classification or protein interface site prediction. The approach is based on GML applied on mesh representations of the solvent-excluded protein surface, abstracting the underlying sequence and internal chemistry of the biomolecule. In practice, MaSIF first discretizes a protein surface with a mesh where each point (vertex) is considered as a node in the graph representation. Then, the protein surface is decomposed into overlapping small patches based on the geodesic radius, i.e. clusters of the graph. For each patch, geometric and chemical features are handcrafted for all nodes within the patch. The patches serve as bases for learnable Gaussian kernels [203] that locally average node-wise patch features and produce an embedding for each patch. The resulting embeddings are fed to task-dependent decoders that, for instance, give patch-wise scores indicating if a patch overlaps with an actual protein-binding site.
De novo design
One of the great ambitions of bioengineering is to design proteins from scratch. In this case, one may have an approximate structure in mind, e.g. to inhibit the function of another endogenous biomolecule. This motivates the inverse protein-folding problem, identifying a sequence that can produce a pre-determined protein structure. For instance, Ingraham et al. [191] leveraged an autoregressive self-attention model using graph-based representations of structures to predict corresponding sequences.
Strokach et al. [106] leveraged a deep GNN to tackle protein design as a constraint satisfaction problem. Predicted structures resulting from novel sequences were initially assessed in silico using molecular dynamics and energy-based scoring. Subsequent in vitro synthesis of sequences led to structures that matched the secondary structure composition of serum albumin evaluated using circular dichroism.
With a novel amino acid sequence that could generate the desired shape of an arbitrary protein, one would then want to identify potential wanted and unwanted effects via functional annotations. These include enzyme commission (EC) numbers, a hierarchy of labels to characterize the reactions that an enzyme catalyzes — previously studied by the ML community [204, 205].
Zamora et al. [20] developed a pipeline based on graph representations of a protein’s amino acid residues for structure classification. The model consists of the sequential application of graph convolutional blocks. A block takes as input two matrices corresponding to residue features and coordinates, respectively. The block first uses a layer to learn Gaussian filters applied on the proteins’ spatial distance kernel, hence deriving multiple graph adjacency matrices. These are then used as input graphs in a GNN layer which operates on the residue features. The block then performs a 1D average pooling operation on both the feature matrix and the input coordinate matrix, yielding the outputs of the block. After the last block, the final feature matrix is fed to a global attention pooling layer which computes the final embedding of the protein used as input to an MLP for classification. The model performs on par with existing 3D-CNN-based models. However, the authors observe that it is more interpretable, enabling the identification of substructures that are characteristic of structural classification.
Recently, Gligorijevic et al. [187] proposed DeepFRI, a model that predicts a protein’s functional annotations based on structure and sequence information. The authors define the graph representation of a protein-based on the contact map between its residues. They first use a pre-trained language module that derives 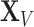 , each protein’s graph is then fed to multiple GCN layers [32]. The outputs of each GCN layer are concatenated to give the final embedding of a protein that is fed to an MLP layer giving the final functional predictions. The authors report substantial improvements over SOTA methods. Furthermore, they highlight the interpretability of their approach, demonstrating the ability to associate specific annotations with particular structures.
, each protein’s graph is then fed to multiple GCN layers [32]. The outputs of each GCN layer are concatenated to give the final embedding of a protein that is fed to an MLP layer giving the final functional predictions. The authors report substantial improvements over SOTA methods. Furthermore, they highlight the interpretability of their approach, demonstrating the ability to associate specific annotations with particular structures.
Drug repurposing
The term drug repurposing is used to describe the use of an existing drug, whether approved or in development as a therapy, for an indication other than the originally intended indication. Considering that only 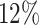 of drugs that reach clinical trials receive FDA approval, repurposed drugs offer an attractive alternative to new therapies as they are likely to have shorter development times and higher success rates with early stages of drug discovery already completed. It has been estimated that repurposed treatments account for 30% of newly FDA approved drugs and their associated revenues [206], and that up to 75% of entities could be repurposed [207]. Note that we incorporate product line extensions within drug repurposing whereby one wishes to identify secondary indications, different formulations for an entity, or partner drugs for combination therapies.
of drugs that reach clinical trials receive FDA approval, repurposed drugs offer an attractive alternative to new therapies as they are likely to have shorter development times and higher success rates with early stages of drug discovery already completed. It has been estimated that repurposed treatments account for 30% of newly FDA approved drugs and their associated revenues [206], and that up to 75% of entities could be repurposed [207]. Note that we incorporate product line extensions within drug repurposing whereby one wishes to identify secondary indications, different formulations for an entity, or partner drugs for combination therapies.
As such, there is a major interest in using in silico methods to screen and infer new treatment hypotheses [208]. Drug repurposing relies on finding new indications for existing molecules, either by identifying actionable pleiotropic activity (off-target repurposing), similarities between diseases (on-target repurposing), or by identifying synergistic combinations of therapies (combination repurposing). Well-known examples include: ketoconazole (Nizoral) used to treat fungal infections via enzyme CYP51A1 and now used to treat Cushing syndrome via off-target interactions with CYP17A1 (steroid synthesis/degradation), NR3C4 (androgen receptor) and NR3C1 (glucocorticoid receptor); sildenafil (Viagra) originally intended for pulmonary arterial hypertension on-target repurposed to treat erectile dysfunction; and Genvoya, a combination of emtricitabine, tenofovir alafenamide (both reverse transcriptase inhibitors), elvitegravir (an integrase inhibitor) and cobicistat (a CYP3A inhibitor to improve PK) to treat human immunodeficiency virus.
Off-target repurposing
Estimates suggest that each small molecule may interact with tens, if not hundreds of proteins [209]. Due to small molecule pleiotropy — particularly from first-in-class drugs [169] — off-targets of existing drugs can be a segue to finding new indications.
A variety of traditional techniques are used to identify missing drug–target interactions. For instance, when the structure of the protein is known, these stem from biophysical simulations, i.e. molecular docking or molecular dynamics. Depending on how one thresholds sequence similarity between the  21 000 human proteins, structural coverage whereby a 3D structure of the protein exists ranges from 30 (
21 000 human proteins, structural coverage whereby a 3D structure of the protein exists ranges from 30 (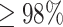 seq. sim.) to 70% (
seq. sim.) to 70% (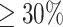 seq. sim.) [210]. However,
seq. sim.) [210]. However, 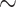 34% of proteins are classified as intrinsically disordered proteins with no 3D structure [211, 212]. Besides, drugs seldom have a fixed shape due to the presence of rotatable bonds. There is now a growing body of GML literature to infer missing drug–target interactions both with and without relying on the availability of a 3D protein structure.
34% of proteins are classified as intrinsically disordered proteins with no 3D structure [211, 212]. Besides, drugs seldom have a fixed shape due to the presence of rotatable bonds. There is now a growing body of GML literature to infer missing drug–target interactions both with and without relying on the availability of a 3D protein structure.
Requiring protein structures, Torng et al. [108] focused on the task of associating drugs with protein pockets they can bind to. Drugs are represented based on their atomic structures and protein pockets are characterized with a set of key amino acid residues connected based on Euclidean distance. Drug embeddings are obtained with the GNN operator from Duvenaud et al. [21]. To derive embeddings of protein pockets, the authors first use two successive graph autoencoders with the purposes of (1) deriving a compact feature vector for each residue, and (2) deriving a graph-level representation of the protein pocket itself. These autoencoders are pre-trained, with the encoder of the first serving as input to the second. Both encoders are then used as input layers of the final model. The association prediction between a drug and a protein pocket is then obtained by feeding the concatenation of the drug and pocket representations to an MLP layer. The authors report improved performance against the previous SOTA model based on a 3D-CNN operating on a grid-structure representation of the protein pocket [213].
A range of GML methods for drug–target interaction do not require protein structure. For instance, Gao et al. [214] use two encoders to derive embeddings for proteins and drugs, respectively. For the first encoder, recurrent neural networks are used to derive an embedding matrix of the protein-based on its sequence and functional annotations. For the second encoder, each drug is represented by its underlying graph of atoms and the authors use GNNs to extract an embedding matrix of the graph. They use three layers of a graph isomorphism network [86] to build their subsequent architecture. Finally, a global attention pooling mechanism is used to extract vector embeddings for both drugs and proteins based on their matrix embeddings. The two resulting vectors are fed into a Siamese neural network [201] to predict their association score. The proposed approach is especially successful compared to baseline for cold-start problems where the protein and/or drug are not present in the training set.
Alternatively, Nascimento et al. [215] introduce KronRLS-MKL, a method that casts drug–target interaction prediction as a link prediction task on a bi-partite graph capturing drug–protein binding. The authors define multiple kernels capturing either drug similarities or protein similarities based on multiple sources of data. The optimization problem is posed as a multiple kernel learning problem. Specifically, the authors use the Kronecker operator to obtain a kernel between drug–protein pairs. The kernel is then used to predict a drug–protein association based on their similarity to existing drug–target link. Crichton et al. [216] cast the task in the same setting. However, the authors use existing embedding methods, including node2vec [70], deepwalk [29] and LINE [217], to embed nodes in a low-dimensional space such that the embeddings capture the local graph topology. The authors feed these embeddings to a machine learning model trained to predict interactions. The underlying assumption is that a drug will be embedded closer to its protein targets.
Similarly, Olayan et al. [107] propose DDR to predict drug–target interactions. The authors first build a graph where each node represents either a drug or a protein. In addition to drug–protein edges, an edge between two drugs (or two proteins) represents their similarity according to a predefined heuristic from multiple data sources. DDR embeds each drug–protein pair based on the number of paths of predefined types that connect them within the graph. The resulting embeddings are fed to a random forest algorithm for drug–target prediction.
Recently, Mohamed et al. [218] proposed an end-to-end knowledge graph embedding model to identify off-target interactions. The authors construct a large knowledge graph encompassing diverse data pertaining to drugs and proteins, such as associated pathways and diseases. Using an approach derived from DistMult and ComplEx, the authors report state-of-the-art results for off-target prediction.
On-target repurposing
On-target repurposing takes a holistic perspective and uses known targets of a drug to infer new putative indications based on diverse data. For instance, one can identify functional relationships between a drug’s targets and genes associated with a disease. Also, one may look for similarities between diseases — especially those occurring in different tissues. Hypothetically, one could prospectively find repurposing candidates in the manner of Fang et al. [158] by finding a missing protein-protein interactions between a genetically validated target and a drug’s primary target. Knowledge graph completion approaches have been particularly effective in addressing these tasks.
For instance, Yang et al. [109] introduced bounded nuclear norm regularization (BNNR). The authors build a block matrix with a drug similarity matrix, a disease similarity matrix, and a disease–drug indication matrix. The method is based on the matrix completion property of singular value thresholding algorithm applied to the block matrix. BNNR incorporates regularization terms to balance approximation error and matrix rank properties to handle noisy drug–drug and disease–disease similarities. It also adds a constraint that clips the association scores to the interval 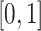 . The authors report performance improvements when compared to competing approaches.
. The authors report performance improvements when compared to competing approaches.
Alternatively, Wang et al. [110] recently proposed an approach to predict new drug indications based on two bipartite graphs, capturing drug–target interactions and disease–gene associations, and a PPI graph. Their algorithm is composed of an encoder module, relying on GAT [33], and an MLP decoder module. The encoder derives drug and disease embeddings through the distillation of information along the edges of the graphs. The input features for drugs and diseases are based on similarity measures. On the one hand, drug features correspond to the Tanimoto similarities between its SMILES representation and that of the other drugs. On the other hand, a disease’s features are defined by its similarity to other diseases computed based on MeSH-associated terms.
Combination repurposing
Combination drugs have been particularly effective in diseases with complex aetiology or an evolutionary component where resistance to treatment is common, such as infectious diseases. If synergistic drugs are found, one can reduce dose whilst improving efficacy [219, 220]. Strategically, combination therapies provide an additional way to extend the indications and efficacy of available entities. They can be used for a range of purposes, for example, convenience and compliance as a fixed-dose formulation (e.g. valsartan and hydrochlorothiazide for hypertension [221]), to achieve synergies (e.g. co-trimoxazole: trimethoprim and sulfamethoxazole for bacterial infections), to broaden spectrum (e.g. for treatment of infections by an unknown pathogen), or to combat disease resistance (e.g. multi-drug regimens for drug-sensitive and drug-resistant tuberculosis). The number of potential pairwise combinations of just two drugs makes a brute force empirical laboratory testing approach a lengthy and daunting prospect. To give a rough number, there exist around 4000 approved drugs which would require 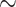 8 million experiments to test all possible combinations of two drugs at a single dose. Besides, there are limitless ways to change the dosage and the timing of treatments, as well as the delivery method.
8 million experiments to test all possible combinations of two drugs at a single dose. Besides, there are limitless ways to change the dosage and the timing of treatments, as well as the delivery method.
Arguably some of the first work using GML to model combination therapy was DECAGON by Zitnik et al. [112] used to model polypharmacy side-effects via a multi-modal graph capturing drug–side effect–drug triplets in addition to PPI interactions. In contrast, Deac et al. [222] forwent incorporation of a knowledge graph instead modelling drug structures directly and using a coattention mechanism to achieve a similar level of accuracy. Typically architectures predicting drug–drug antagonism can be minimally adapted for prediction of synergy. However, more nuanced architectures are emerging combining partial knowledge of drug–target interactions with target–disease machine learning modules [113].
Outlook
In the last year to address the unprecedented COVID-19 global health crisis, multiple research groups have explored graph-based approaches to identify drugs that could be repurposed to treat SARS-CoV-2 [111, 223–225]. For instance, Morselli et al. [224] proposed an ensemble approach combining three different graph-based association strategies. The first two are similar in principle. First, each drug and each disease is represented by the set of proteins that it targets. Second, the association between a drug and a disease is quantified based on the distance between the two sets on a PPI graph. The two approaches differ on whether the distance is quantified with shortest paths or random walks. The last strategies rely on GNNs for multimodal graphs (knowledge graph). The graph contains PPIs, drug–target interactions, disease–protein associations and drug indications. The formulation of the GNN layer is taken from the DECAGON model [112], an architecture similar to the R-GCN model [44].
Alternatively, Zeng et al. [111] use RotatE to identify repurposing hypotheses to treat SARS-CoV-2 from a large knowledge graph constructed from multiple data sources and capturing diverse relationships between entities such as drugs, diseases, genes and anatomies. Additionally, Ioannidis et al. [225] proposed a modification of the RGCN architecture to handle few-shot learning settings in which some relations only connect a handful of nodes.
In the rare disease arena, Sosa et al. [226] introduced a knowledge graph embedding approach to identify repurposing hypotheses for rare diseases. The problem is cast as a link prediction problem in a knowledge graph. The authors use the Global Network of Biological Relationships [227], a knowledge graph built through literature mining and that contains diverse relationships between diseases, drugs and genes. Due to the uncertainty associated with associations obtained from literature mining, the authors use a knowledge graph embedding approach design to account for uncertainty [228]. Finally, the highest ranking associations between drugs and rare diseases are investigated, highlighting literature and biological support.
Drug repurposing is now demonstrating itself as a first use case of GML methods likely to lead to new therapies within the coming years. Outside of the pharmaceutical industry, GML methods recommending nutraceuticals [229] may also offer fast routes to market through generally recognized as safe regulatory designations.
Discussion
We have discussed how GML has produced the state-of-the-art results both on graph-level problems for the description of drugs and other biomolecules, and node-level problems for the navigation of knowledge graphs and representation of disease biology. With the design, synthesis and testing of de novo small molecules [48], the in vitro and in vivo testing of drug repurposing hypotheses [50], and target identification frameworks being conceptualized [47], we are potentially entering into a golden age of validation for GML within drug discovery and development.
A few key hurdles limit lossless representation of biology. At the molecular level, bonds can be either rotatable single bonds or fixed bonds; accurately representing the degrees of freedom of a molecule is a topic of active research [230]. At the cellular level, expression of mRNA and proteins exhibit stochastic dynamics [231, 232]. A pathway is not expressed in a binary fashion: some proteins may only have the potential to be expressed, e.g. via unspliced pre-mRNA, and meanwhile, proteases are actively recycling unwanted proteins. Historically, most -omic platforms have recorded an average ‘bulk’ signal; however, with the recent single-cell revolution, GML offers a principled approach to the characterization of signalling cascades.
GML is still in its infancy, and underlying theoretical guarantees and limitations are under active research. For instance, deeper GNNs suffer from oversmoothing of features and oversquashing of information. Oversmoothing is the phenomenon of features washing out through repeated rounds of message passing and aggregation [233]. The inverse, having too few layers to exchange information globally, is referred to as under-reaching [234]. These issues have limited the expressivity of traditional GNNs [235, 236]. To alleviate this, a promising direction is to incorporate global information in the model, for instance, by using contrastive approaches [31, 237], by augmenting the framework with relative positioning to anchor nodes [238, 239], or by implementing long-range information flow [230].
Due to the problem of missing data within biomedical knowledge graphs, we envisage opportunities for active learning to be deployed to label critical missing data points to explore experimentally and therefore reduce model uncertainty [240]. Due to the significant expense associated with drug discovery and development, integrating in silico modelling and experimental research is of great strategic importance. While active learning has previously led to biased datasets in other settings, modern techniques are addressing these drawbacks [241, 242].
Finally, because GML allows for the representation of unstructured multimodal datasets, one can expect to see tremendous advances made within data integration. Most notably, highly multiplexed single-cell omic technologies are now being expanded in spatial settings [243, 244]. In addition, CRISPR screening data with associated RNA sequencing readouts are emerging as promising tools to identify key genes controlling a cellular phenotype [245].
Key Points
Historically, analysis of biomolecular interaction and gene regulatory networks has been of huge academic interest, but with limited translatable results within drug discovery and development.
Network medicine has offered promising results using handcrafted graph features but lacked any principled solution to the problem of integrating disparate biological data sources: structural data (drugs and biomolecules), functional relationships (inhibition, activation, etc) and expression (from RNA-seq, proteomics, etc).
Deep learning has now been applied to a number of areas within biomedical research, in particular, achieving superior-to-physician results in the interpretation of biomedical images, e.g. histopathological specimens.
Graph ML blends the techniques of network topology analysis with deep learning to learn effective feature representations of nodes.
Graph ML has been applied to problems within drug discovery and development to huge success with emerging experimental results: design of small molecules, prediction of drugtarget interactions, prediction of drugdrug interactions and drug repurposing have all been tasks showing considerable success and improvement over simpler non-graph ML methods.
Supplementary Material
Acknowledgments
We gratefully acknowledge support from William L. Hamilton, Benjamin Swerner, Lyuba V. Bozhilova and Andrew Anighoro.
Contributor Information
Thomas Gaudelet, Relation Therapeutics, London, UK.
Ben Day, Relation Therapeutics, London, UK; The Computer Laboratory, University of Cambridge, UK.
Arian R Jamasb, Relation Therapeutics, London, UK; The Computer Laboratory, University of Cambridge, UK; Department of Biochemistry, University of Cambridge, UK.
Jyothish Soman, Relation Therapeutics, London, UK.
Cristian Regep, Relation Therapeutics, London, UK.
Gertrude Liu, Relation Therapeutics, London, UK.
Jeremy B R Hayter, Relation Therapeutics, London, UK.
Richard Vickers, Relation Therapeutics, London, UK.
Charles Roberts, Relation Therapeutics, London, UK; Juvenescence, London, UK.
Jian Tang, Mila, the Quebec AI Institute, Canada; HEC Montreal, Canada.
David Roblin, Relation Therapeutics, London, UK; Juvenescence, London, UK; The Francis Crick Institute, London, UK.
Tom L Blundell, Department of Biochemistry, University of Cambridge, UK.
Michael M Bronstein, Relation Therapeutics, London, UK; Department of Computing, Imperial College London, UK; Twitter, UK.
Jake P Taylor-King, Relation Therapeutics, London, UK; Juvenescence, London, UK.
References
- 1. DiMasi JA, Grabowski HG, Hansen RW. Innovation in the pharmaceutical industry: new estimates of R&D costs. J Health Econ 2016;47:20–33. [DOI] [PubMed] [Google Scholar]
- 2. Steedman M, Taylor K. Ten years on - measuring return from pharmaceutical innovation 2019 Technical report. Deloitte, 2019. https://www2.deloitte.com/uk/en/pages/life-sciences-and-healthcare/articles/measuring-return-from-pharmaceutical-innovation.html. [Google Scholar]
- 3. Wouters OJ, McKee M, Luyten J. Estimated research and development investment needed to bring a new medicine to market, 2009-2018. JAMA 2020;323(9):844–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martin L, Hutchens M, Hawkins C. Clinical trial cycle times continue to increase despite industry efforts. Nat Rev Drug Discov 2017;16:157. [DOI] [PubMed] [Google Scholar]
- 5. Paul SM, Mytelka DS, Dunwiddie CT, et al. . How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nat Rev Drug Discov 2010;9(3):203–14. [DOI] [PubMed] [Google Scholar]
- 6. Réda C, Kaufmann E, Delahaye-Duriez A. Machine learning applications in drug development. Comput Struct Biotechnol J 2020;18:241–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nishida E, Ishita E, Watanabe Y, et al. . Description of research data in laboratory notebooks: challenges and opportunities. Proc Assoc Inf Sci Technol 2020;57(1):e388. [Google Scholar]
- 8. Surae S. Data-driven transformation in drug discovery. Drug Discovery World 2018. https://www.ddw-online.com/the-data-driven-transformation-in-drug-discovery-784-201808/. [Google Scholar]
- 9. Coran P, Goldsack JC, Grandinetti CA, et al. . Advancing the use of mobile technologies in clinical trials: recommendations from the clinical trials transformation initiative. Digital Biomarkers 2019;3(3):145–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marquis-Gravel G, Roe MT, Turakhia MP, et al. . Technology-enabled clinical trials: transforming medical evidence generation. Circulation 2019;140(17):1426–36. [DOI] [PubMed] [Google Scholar]
- 11. Hulsen T, Jamuar SS, Moody AR, et al. . From big data to precision medicine. Front Med 2019;6:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sloane R, Osanlou O, Lewis D, et al. . Social media and pharmacovigilance: a review of the opportunities and challenges. Br J Clin Pharmacol 2015;80(4):910–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sarker A, Ginn R, Nikfarjam A, et al. . Utilizing social media data for pharmacovigilance: a review. J Biomed Inform 2015;54:202–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Corsello SM, Bittker JA, Liu Z, et al. . The drug repurposing hub: a next-generation drug library and information resource. Nat Med April 2017;23(4):405–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pan P, Verbaanderd C, Sukhatme V, et al. . Redo_db: the repurposing drugs in oncology database. ecancermedicalscience 2018;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heath JR, Ribas A, Mischel PS. Single-cell analysis tools for drug discovery and development. Nat Rev Drug Discov 2016;15(3):204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spitzer MH, Nolan GP. Mass cytometry: single cells, many features. Cell 2016;165(4):780–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McGinnis CS, Patterson DM, Winkler J, et al. . Multi-seq: sample multiplexing for single-cell rna sequencing using lipid-tagged indices. Nat Methods 2019;16(7):619–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fout A, Byrd J, Shariat B, et al. . Protein interface prediction using graph convolutional networks. In: Advances in Neural Information Processing Systems. Red Hook, NY, USA: Curran Associates, Inc., 2017;6530–9.
- 20. Zamora-Resendiz R, Crivelli S. Structural learning of proteins using graph convolutional neural networks. bioRxiv 2019;610444. [Google Scholar]
- 21. Duvenaud DK, Maclaurin D, Iparraguirre J, et al. . Convolutional networks on graphs for learning molecular fingerprints. In: Advances in Neural Information Processing Systems. Neural Information Processing Systems (NeurIPS) Proceedings, Curran Associates, Inc., 2015;2224–32. https://papers.nips.cc/paper/2015/hash/f9be311e65d81a9ad8150a60844bb94c-Abstract.html. [Google Scholar]
- 22. Klicpera J, Groß J, Günnemann S. Directional message passing for molecular graphs. arXiv 2020. https://arxiv.org/abs/2003.03123. [Google Scholar]
- 23. Han J-DJ. Understanding biological functions through molecular networks. Cell Res 2008;18(2):224–37. [DOI] [PubMed] [Google Scholar]
- 24. Zhu W, Razavian N. Variationally regularized graph-based representation learning for electronic health records. In: Proceedings of the Conference on Health, Inference, and Learning, ACM, 2021. doi: 10.1145/3450439.3451855. [DOI] [Google Scholar]
- 25. Choi E, Xu Z, Li Y, et al. . Learning the graphical structure of electronic health records with graph convolutional transformer. Proceedings of the AAAI Conference on Artificial Intelligence 2020;34:606–13. [Google Scholar]
- 26. Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet 2011;12(1):56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Voulodimos A, Doulamis N, Doulamis A, et al. . Deep learning for computer vision: a brief review. Comput Intell Neurosci 2018;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Young T, Hazarika D, Poria S, et al. . Recent trends in deep learning based natural language processing. IEEE Comput Intell Mag 2018;13(3):55–75. [Google Scholar]
- 29. Perozzi B, Al-Rfou R, Deepwalk SS. Online learning of social representations. In: Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. New York, USA: Association for Computing Machinery, 2014;701–10. [Google Scholar]
- 30. Sun Z, Deng Z-H, Nie J-Y, et al. . Rotate: Knowledge graph embedding by relational rotation in complex space. In: International Conference on Learning Representations (ICLR), 2018. https://openreview.net/forum?id=HkgEQnRqYQ.
- 31. Sun F-Y, Hoffmann J, Verma V, et al. . Infograph: unsupervised and semi-supervised graph-level representation learning via mutual information maximization. ICLR 2020. https://arxiv.org/abs/1908.01000.
- 32. Kipf TN, Welling M. Semi-supervised classification with graph convolutional networks. ICLR 2017. https://arxiv.org/abs/1609.02907.
- 33. Veličković P, Cucurull G, Casanova A, et al. . Graph attention networks. In: 6th International Conference on Learning Representations, ICLR 2018 – Conference Track Proceedings, 2018. https://openreview.net/forum?id=rJXMpikCZ.
- 34. Gilmer J, Schoenholz SS, Riley PF, et al. . Neural message passing for quantum chemistry. In: Proceedings of Machine Learning Research, 2017;1263–72. http://proceedings.mlr.press/v70/gilmer17a.html.
- 35. Pal A, Eksombatchai C, Zhou Y, et al. . PinnerSage: multi-modal user embedding framework for recommendations at pinterest. In: Proceedings of the 26th ACM SIGKDD Conference on Knowledge Discovery & Data Mining (KDD `20). Virtual Event, CA, USA. ACM, New York, NY, USA, 2020;10. https://doi/10.1145/3394486.3403280. [Google Scholar]
- 36. Yang Hongxia. Aligraph: a comprehensive graph neural network platform. In: In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining (KDD `19). New York, NY, USA: Association for Computing Machinery, 2019;3165–66. 10.1145/3292500.3340404. [DOI] [Google Scholar]
- 37. Rossi E, Frasca F, Chamberlain B, et al. . Sign: scalable inception graph neural networks. Graph Representation Learning and Beyond (GRL+) Workshop at the 37th International Conference on Machine Learning, ICML. 2020. https://arxiv.org/abs/2004.11198.
- 38. Rossi E, Chamberlain B, Frasca F, et al. . Temporal graph networks for deep learning on dynamic graphs 2020. https://arxiv.org/abs/2006.10637.
- 39. Lange O, Perez L. Traffic prediction with advanced graph neural networks, 2020. https://deepmind.com/blog/article/traffic-prediction-with-advanced-graph-neural-networks.
- 40. Monti F, Frasca F, Eynard D, et al. . Fake news detection on social media using geometric deep learning. 2019. https://arxiv.org/abs/1902.06673.
- 41. Sanchez-Gonzalez A, Godwin J, Pfaff T, et al. . Learning to simulate complex physics with graph networks. International Conference on Machine Learning. Advances in Neural Information Processing Systems (NeurIPS 2020), 2020;33:8459–68. [Google Scholar]
- 42. Shlomi J, Battaglia P, et al. . Graph neural networks in particle physics. Mach Learn: Sci Technol 2020;2(2).https://iopscience.iop.org/article/10.1088/2632-2153/abbf9a. [Google Scholar]
- 43. Choma Nicholas, Monti Federico, Gerhardt Lisa, et al. . Graph neural networks for icecube signal classification. 2018 17th IEEE International Conference on Machine Learning and Applications (ICMLA). Institute of Electrical and Electronics Engineers Inc, 2018, pp. 386–91. doi: 10.1109/ICMLA.2018.00064. [DOI] [Google Scholar]
- 44. Schlichtkrull Michael, Kipf Thomas N, Bloem Peter, Van Den Berg Rianne, Titov Ivan, and Welling Max. Modeling relational data with graph convolutional networks. European Semantic Web Conference, 593–607. Springer, 2018. [Google Scholar]
- 45. Balazevic Ivana, Allen Carl, and Hospedales Timothy. Tucker: Tensor factorization for knowledge graph completion. Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP). Hong Kong, China: Association for Computational Linguistics, 5188–5197, 2019. [Google Scholar]
- 46. Feinberg EN, Joshi E, Pande VS, et al. . Improvement in admet prediction with multitask deep featurization. J Med Chem 2020;63(16):8835. [DOI] [PubMed] [Google Scholar]
- 47. Pittala S, Koehler W, Deans J, et al. . Relation-weighted link prediction for disease gene identification. In: 4th Knowledge Representation and Reasoning Meets Machine Learning Workshop (KR2ML), NeurIPS 2020. https://arxiv.org/abs/2011.05138.
- 48. Zhavoronkov A, Ivanenkov YA, Aliper A, et al. . Deep learning enables rapid identification of potent ddr1 kinase inhibitors. Nat Biotechnol 2019;37(9):1038–40. [DOI] [PubMed] [Google Scholar]
- 49. Gainza P, Sverrisson F, Monti F, et al. . Deciphering interaction fingerprints from protein molecular surfaces using geometric deep learning. Nat Methods 2020;17(2):184–92. [DOI] [PubMed] [Google Scholar]
- 50. Stokes JM, Yang K, Swanson K, et al. . A deep learning approach to antibiotic discovery. Cell 2020;180(4):688–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nickel M, Murphy K, Tresp V, et al. . A review of relational machine learning for knowledge graphs. Proc IEEE 2015;104(1):11–33. [Google Scholar]
- 52. Zhou J, Cui G, Zhang Z, et al. . Graph neural networks: a review of methods and applications. AI Open 2018;1:57. [Google Scholar]
- 53. Wu Z, Pan S, Chen F, et al. . A comprehensive survey on graph neural networks. IEEE Trans Neural Netw Learn Systems 2020;32(1):4. [DOI] [PubMed] [Google Scholar]
- 54. Hamilton WL, Ying R, Leskovec J. Representation learning on graphs: methods and applications. IEEE Data Engineering Bulletin 2017.https://arxiv.org/abs/1709.05584.
- 55. Zhang Z, Cui P, Zhu W. Deep learning on graphs: a survey. IEEE Trans Knowl Data Eng 2020. https://arxiv.org/abs/1812.04202. [Google Scholar]
- 56. Talevi A, Morales JF, Hather G, et al. . Machine learning in drug discovery and development. Part 1: a primer. CPT Pharmacometrics Syst Pharmacol 2020;9(3):129–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vamathevan J, Clark D, Czodrowski P, et al. . Applications of machine learning in drug discovery and development. Nat Rev Drug Discov 2019;18(6):463–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rifaioglu AS, Atas H, Martin MJ, et al. . Recent applications of deep learning and machine intelligence on in silico drug discovery: methods, tools and databases. Brief Bioinform 2019;20(5):1878–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kondor Risi Imre and Lafferty John. Diffusion kernels on graphs and other discrete structures. Proceedings of the 19th International Conference on Machine Learning (ICML), 315–22, 2002. http://people.cs.uchicago.edu/∼risi/papers/diffusion-kernels.pdf.
- 60. Weisfeiler B, Lehman AA. A reduction of a graph to a canonical form and an algebra arising during this reduction. Nauchno-Technicheskaya Informatsia 1968;2(9). [Google Scholar]
- 61. Berkholz Christoph and Grohe Martin. Limitations of algebraic approaches to graph isomorphism testing. International Colloquium on Automata, Languages, and Programming, 155–166. Springer, 2015. [Google Scholar]
- 62. Chami I, Abu-El-Haija S, Perozzi B, et al. . Machine learning on graphs: a model and comprehensive taxonomy. 2020. https://arxiv.org/abs/2005.03675.
- 63. Murphy KP. Machine Learning: A Probabilistic Perspective. MIT Press, 2012. [Google Scholar]
- 64. Othmer HG, Scriven LE. Instability and dynamic pattern in cellular networks. J Theor Biol 1971;32(3):507–37. [DOI] [PubMed] [Google Scholar]
- 65. Praktiknjo SD, Obermayer B, Zhu Q, et al. . Tracing tumorigenesis in a solid tumor model at single-cell resolution. Nat Commun 2020;11(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Milo R, Shen-Orr S, Itzkovitz S, et al. . Network motifs: simple building blocks of complex networks. Science 2002;298(5594):824–7. [DOI] [PubMed] [Google Scholar]
- 67. Pržulj N, Corneil DG, Jurisica I. Modeling interactome: scale-free or geometric? Bioinformatics 2004;20(18):3508–15. [DOI] [PubMed] [Google Scholar]
- 68. Shervashidze N, S. Vishwanathan, T Petri, K Mehlhorn, and K Borgwardt. Efficient graphlet kernels for large graph comparison. Proceedings of the Twelth International Conference on Artificial Intelligence and Statistics, in PMLR, 2009;5:488–95. [Google Scholar]
- 69. Shervashidze N, Schweitzer P, Van Leeuwen EJ, et al. . Weisfeiler-lehman graph kernels. J Mach Learn Res 2011;12(9):2539. [Google Scholar]
- 70. Grover Aditya and Leskovec Jure. Node2vec: scalable feature learning for networks. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD `16). Association for Computing Machinery, New York, NY, USA, 2016:855–64. doi: 10.1145/2939672.2939754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mikolov T, Chen K, Corrado G, et al. . Efficient estimation of word representations in vector space. In: Proceedings of the International Conference on Learning Representations (ICLR). 2013. https://arxiv.org/abs/1301.3781.
- 72. Bordes Antoine, Usunier Nicolas, Garcia-Duran Alberto, Weston Jason, and Yakhnenko Oksana. Translating embeddings for modeling multi-relational data. Advances in NeurIPS Proceedings, Curran Associates, Inc., 2013;2787–2795.
- 73. Rossi A, Firmani D, Matinata A, et al. . Knowledge graph embedding for link prediction: a comparative analysis. ACM Trans. Knowl. Discov. Data 2020;15. https://dl.acm.org/doi/abs/10.1145/3424672. [Google Scholar]
- 74. Belkin Mikhail and Niyogi Partha. Laplacian eigenmaps and spectral techniques for embedding and clustering. In: Thomas G. Dietterich, Suzanna Becker, Zoubin Ghahramani (eds). Advances in Neural Information Processing Systems 14: Proceedings of the 2001 Conference. Cambridge, MA, US: MIT Press, 2002;585–591. 10.7551/mitpress/1120.003.0080. [DOI]
- 75. Nickel Maximilian, Tresp Volker, and Kriegel Hans-Peter. A three-way model for collective learning on multi-relational data. International Conference on Learning Representations. Proceedings of the 28 th International Conference on Machine Learning, Bellevue, WA, USA, 2011;Vol.11:809–16. [Google Scholar]
- 76. Yang B, Yih W-t, He X, et al. . Embedding entities and relations for learning and inference in knowledge bases. In: Proceedings of the International Conference on Learning Representations (ICLR) 2015.
- 77. Trouillon T, Welbl J, Riedel S, et al. . Complex embeddings for simple link prediction. Int Conf Mach Learn (ICML) 2016;48:2071. [Google Scholar]
- 78. Cai D, He X, Han J, et al. . Graph regularized nonnegative matrix factorization for data representation. IEEE Trans Pattern Anal Mach Intell 2011;33(8):1548–60. [DOI] [PubMed] [Google Scholar]
- 79. Chang Shiyu, Han Wei, Tang Jiliang, Qi Guo-Jun, Aggarwal Charu C, and Huang Thomas S. Heterogeneous network embedding via deep architectures. In Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD `15). Association for Computing Machinery, New York, NY, USA, 2015;119–28. doi: 10.1145/2783258.2783296. [DOI]
- 80. Huang X, Li J, Hu X. Label informed attributed network embedding. In Proceedings of the Tenth ACM International Conference on Web Search and Data Mining (WSDM `17). New York, NY, USA: Association for Computing Machinery, 2017;731–39. doi: 10.1145/3018661.3018667. [DOI]
- 81. Huang Xiao, Li Jundong, and Xia Hu. Accelerated attributed network embedding. Proceedings of the 2017 SIAM international conference on data mining, 633–641. SIAM, 2017. [Google Scholar]
- 82. Sperduti A, Starita A. Supervised neural networks for the classification of structures. IEEE Trans Neural Netw 1997;8(3):714–35. [DOI] [PubMed] [Google Scholar]
- 83. Gori Marco, Monfardini Gabriele, and Scarselli Franco. A new model for learning in graph domains. Proceedings of IEEE International Joint Conference on Neural Networks, 2005, Vol. 2, 729–734. IEEE, 2005. [Google Scholar]
- 84. Merkwirth C, Lengauer T. Automatic generation of complementary descriptors with molecular graph networks. J Chem Inf Model 2005;45(5):1159–68. [DOI] [PubMed] [Google Scholar]
- 85. Hamilton Will, Ying Zhitao, and Leskovec Jure. Inductive representation learning on large graphs. Advances in Neural Information Processing Systems. Long Beach, CA, USA: NeurIPS, Curran Associates, Inc., 2017;1024–1034. [Google Scholar]
- 86. Xu K, Hu W, Leskovec J, et al. . How powerful are graph neural networks? International Conference on Learning Representations 2018. https://arxiv.org/abs/1810.00826.
- 87. Xu K, Li C, Tian Y, et al. . Representation learning on graphs with jumping knowledge networks. International Conference on Machine Learning 2018;80:5453–62. [Google Scholar]
- 88. Maron H, Ben-Hamu H, Shamir N, et al. . Invariant and equivariant graph networks. International Conference on Learning Representations 2018. https://arxiv.org/abs/1812.09902.
- 89. Chami I, Ying Z, Ré C, et al. . Hyperbolic graph convolutional neural networks. Advances in Neural Information Processing Systems 2019;32:4868–79. [PMC free article] [PubMed] [Google Scholar]
- 90. Zaheer M, Kottur S, Ravanbakhsh S, et al. . Deep sets. In: NIPS. Red Hook, NY, USA: Curran Associates, Inc., 2017.
- 91. Vaswani A, Shazeer N, Parmar N, et al. . Attention is all you need. In: NIPS. Red Hook, NY, USA: Curran Associates, Inc., 2017.
- 92. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521(7553):436–44. [DOI] [PubMed] [Google Scholar]
- 93. Fey Matthias and Lenssen Jan E.. Fast graph representation learning with PyTorch Geometric. ICLR Workshop on Representation Learning on Graphs and Manifolds. International Conference on Learning Representations (ICLR), 2019. https://rlgm.github.io/papers/2.pdf. [Google Scholar]
- 94. Bronstein MM, Bruna J, LeCun Y, et al. . Geometric deep learning: going beyond euclidean data. IEEE Sig Process Mag 2017;34(4):18–42. [Google Scholar]
- 95. Ying Z, You J, Morris C, et al. . Hierarchical graph representation learning with differentiable pooling. Advances in Neural Information Processing Systems 2018;31:4800–10. [Google Scholar]
- 96. Cangea C, Veličković P, Jovanović N, et al. . Towards sparse hierarchical graph classifiers. Workshop on Relational Representation Learning (R2L), NIPS 2018. https://arxiv.org/abs/1811.01287.
- 97. H Gao and S Ji. Graph U-Nets. Proceedings of the 36th International Conference on Machine Learning, in Proceedings of Machine Learning Research, 2019;97:2083–92. Available from http://proceedings.mlr.press/v97/gao19a.html. [Google Scholar]
- 98. Lee J, Lee I, Kang J. Self-attention graph pooling. Proceedings of the 36th International Conference on Machine Learning, PMLR 2019;97:3734. [Google Scholar]
- 99. Bodnar C, Cangea C, Liò P. Deep graph mapper: seeing graphs through the neural lens. 2020. https://arxiv.org/abs/2002.03864. [DOI] [PMC free article] [PubMed]
- 100. Boykov Y, Veksler O. Graph cuts in vision and graphics: theories and applications. In: Handbook of Mathematical Models in Computer Vision. Springer, 2006;79–96. [Google Scholar]
- 101. Luzhnica E, Day B, Lio P. Clique pooling for graph classification. 2019. https://arxiv.org/abs/1904.00374.
- 102. Bianchi Filippo Maria, Grattarola Daniele, and Alippi Cesare. Spectral clustering with graph neural networks for graph pooling. In: Proceedings of the 37th International Conference on Machine Learning, PMLR. 2729–2738. ACM, 2020119:874. [Google Scholar]
- 103. Jin W, Barzilay R, Jaakkola T. Hierarchical generation of molecular graphs using structural motifs. PMLR, 2020. arXiv preprint arXiv:2002.03230. [Google Scholar]
- 104. Feinberg EN, Sur D, Wu Z, et al. . Potentialnet for molecular property prediction. ACS Central Sci 2018;4(11):1520–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Jin W, Barzilay R, Jaakkola T. Junction tree variational autoencoder for molecular graph generation. International Conference on Machine Learning 2018;2323–32.
- 106. Strokach A, Becerra D, Corbi-Verge C, et al. . Fast and flexible protein design using deep graph neural networks. Cell Syst 2020;11(4):402–11. [DOI] [PubMed] [Google Scholar]
- 107. Olayan RS, Ashoor H, Bajic VB. Ddr: efficient computational method to predict drug–target interactions using graph mining and machine learning approaches. Bioinformatics 2018;34(7):1164–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Torng W, Altman RB. Graph convolutional neural networks for predicting drug-target interactions. J Chem Inf Model 2019;59(10):4131–49. [DOI] [PubMed] [Google Scholar]
- 109. Yang M, Luo H, Li Y, et al. . Drug repositioning based on bounded nuclear norm regularization. Bioinformatics 2019;35(14):i455–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wang Z, Zhou M, Arnold C. Toward heterogeneous information fusion: bipartite graph convolutional networks for in silico drug repurposing. Bioinformatics 2020;36(Supplement_1):i525–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Zeng X, Song X, Ma T, et al. . Repurpose open data to discover therapeutics for covid-19 using deep learning. J Proteome Res 2020. [DOI] [PubMed] [Google Scholar]
- 112. Zitnik M, Agrawal M, Leskovec J. Modeling polypharmacy side effects with graph convolutional networks. Bioinformatics 2018;34(13):i457–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Jin W, Barzilay R, Jaakkola T. Modeling drug combinations based on molecular structures and biological targets arXiv preprint arXiv:2011.04651. 2020.
- 114. Hughes JP, Rees S, Barrett Kalindjian S, et al. . Principles of early drug discovery. Br J Pharmacol 2011;162(6):1239–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Huang Kexin, Fu Tianfan, Gao Wenhao, Zhao Yue, Roohani Yusuf, Leskovec Jure, Coley Connor, Xiao Cao, Sun Jimeng, and Zitnik Marinka. Therapeutics data commons: machine learning datasets for therapeutics. https://zitniklab.hms.harvard.edu/TDC/, November 2020.
- 116. Walsh Brian, Mohamed Sameh K, and Nováček Vít: A knowledge graph for relational learning on biological data. In: Proceedings of the 29th ACM International Conference on Information & Knowledge Management (CIKM `20). Association for Computing Machinery, New York, NY, USA, 2020;3173–80. doi: 10.1145/3340531.3412776. [DOI] [Google Scholar]
- 117. Mendez D, Gaulton A, Bento AP, et al. . ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res 2019;47(D1):D930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wishart David S, Feunang Yannick D, Guo An C, Lo Elvis J, Marcu Ana, Grant Jason R, Sajed Tanvir, Johnson Daniel, Li Carin, Sayeeda Zinat, Assempour Nazanin, Iynkkaran Ithayavani, Liu Yifeng, Maciejewski Adam, Gale Nicola, Wilson Alex, Chin Lucy, Cummings Ryan, Le Diana, Pon Allison, Knox Craig, and Wilson Michael. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res, 46(D1):D1074–D1082, November 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Corsello SM, Bittker JA, Liu Z, et al. . The drug repurposing hub: a next-generation drug library and information resource. Nat Med April 2017;23(4):405–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kim S, Chen J, Cheng T, et al. . Pubchem in 2021: new data content and improved web interfaces. Nucleic Acids Res 2021;49(D1):D1388–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Sterling T, Irwin JJ. Zinc 15–ligand discovery for everyone. J Chem Inf Model 2015;55(11):2324–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zagidullin B, Aldahdooh J, Zheng S, et al. . Drugcomb: an integrative cancer drug combination data portal. Nucleic Acids Res 2019;47(W1):W43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Tatonetti NP, Patrick PY, Daneshjou R, et al. . Data-driven prediction of drug effects and interactions. Sci Transl Med 2012;4(125):125ra31–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Berman HM, Westbrook J, Feng Z, et al. . The protein data bank. Nucleic Acids Res 2000;28(1):235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Stark C. BioGRID: a general repository for interaction datasets. Nucleic Acids Res January 2006;34(90001):D535–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Szklarczyk D, Gable AL, Lyon D, et al. . String v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47(D1):D607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kanehisa M, Furumichi M, Sato Y, et al. . Kegg: integrating viruses and cellular organisms. Nucleic Acids Res 2021;49(D1):D545–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Fabregat A, Jupe S, Matthews L, et al. . The reactome pathway knowledgebase. Nucleic Acids Res 2018;46(D1):D649–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. The gene ontology resource: enriching a gold mine. Nucleic Acids Res 2021;49(D1):D325–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Schriml LM, Mitraka E, Munro J, et al. . Human disease ontology 2018 update: classification, content and workflow expansion. Nucleic Acids Res 2019;47(D1):D955–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Piñero J, Bravo À, Queralt-Rosinach N, et al. . Disgenet: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 2016;gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Schenone M, Dančík V, Wagner BK, et al. . Target identification and mechanism of action in chemical biology and drug discovery. Nat Chem Biol 2013;9(4):232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Titov DV, Liu JO. Identification and validation of protein targets of bioactive small molecules. Bioorg Med Chem 2012;20(6):1902–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Eder J, Sedrani R, Wiesmann C. The discovery of first-in-class drugs: origins and evolution. Nat Rev Drug Discov 2014;13(8):577–87. [DOI] [PubMed] [Google Scholar]
- 135. Paananen J, Fortino V. An omics perspective on drug target discovery platforms. Brief Bioinform . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Sandhu C, Qureshi A, Emili A. Panomics for precision medicine. Trends Mol Med 2018;24(1):85–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Matthews H, Hanison J, Nirmalan N. “omics” -informed drug and biomarker discovery: opportunities, challenges and future perspectives. Proteomes 2016;4(3):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Boettcher M, McManus MT. Choosing the right tool for the job: Rnai, talen, or crispr. Mol Cell 2015;58(4):575–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Smith I, Greenside PG, Natoli T, et al. . Evaluation of rnai and crispr technologies by large-scale gene expression profiling in the connectivity map. PLoS Biol 2017;15(11):e2003213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Peretz L, Besser E, Hajbi R, et al. . Combined shRNA over CRISPR/cas9 as a methodology to detect off-target effects and a potential compensatory mechanism. Sci Rep 2018;8:93. 10.1038/s41598-017-18551-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Sage C, Lawo S, Panicker P, et al. . Dual direction crispr transcriptional regulation screening uncovers gene networks driving drug resistance. Sci Rep 2017;7(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Cuatrecasas P, Wilchek M, Anfinsen CB. Selective enzyme purification by affinity chromatography. Proc Natl Acad Sci U S A 1968;61(2):636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Lomenick B, Jung G, Wohlschlegel JA, et al. . Target identification using drug affinity responsive target stability (darts). Curr Prot Chem Biol 2011;3(4):163–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Ong S-E, Mann M. Mass spectrometry–based proteomics turns quantitative. Nat Chem Biol 2005;1(5):252–62. [DOI] [PubMed] [Google Scholar]
- 145. Shoemaker SC, Ando N. X-rays in the cryo-electron microscopy era: structural biology’s dynamic future. Biochemistry 2018;57(3):277–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Malhotra S, Alsulami AF, Heiyun Y, et al. . Understanding the impacts of missense mutations on structures and functions of human cancer-related genes: a preliminary computational analysis of the cosmic cancer gene census. PloS One 2019;14(7):e0219935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Hamdi A, Colas P. Yeast two-hybrid methods and their applications in drug discovery. Trends Pharmacol Sci 2012;33(2):109–18. [DOI] [PubMed] [Google Scholar]
- 148. Licitra EJ, Liu JO. A three-hybrid system for detecting small ligand–protein receptor interactions. Proc Natl Acad Sci 1996;93(23):12817–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Butcher EC, Berg EL, Kunkel EJ. Systems biology in drug discovery. Nat Biotechnol 2004;22(10):1253–9. [DOI] [PubMed] [Google Scholar]
- 150. Akhurst RJ, Hata A. Targeting the tgfβ signalling pathway in disease. Nat Rev Drug Discov 2012;11(10):790–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Hennessy BT, Smith DL, Ram PT, et al. . Exploiting the pi3k/akt pathway for cancer drug discovery. Nat Rev Drug Discov 2005;4(12):988–1004. [DOI] [PubMed] [Google Scholar]
- 152. Janssens N, Janicot M, Perera T. The wnt-dependent signaling pathways as target in oncology drug discovery. Invest New Drugs 2006;24(4):263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Ochoa-Montaño B, Mohan N, Blundell TL. Chopin: a web resource for the structural and functional proteome of mycobacterium tuberculosis. Database 2015;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Skwark MJ, Torres PHM, Copoiu L, et al. . Mabellini: a genome-wide database for understanding the structural proteome and evaluating prospective antimicrobial targets of the emerging pathogen mycobacterium abscessus. Database 2019;2019:baz113. 10.1093/database/baz113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Blundell TL. A personal history of using crystals and crystallography to understand biology and advanced drug discovery. Crystals 2020;10(8):676. [Google Scholar]
- 156. King EA, Davis JW, Degner JF. Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. PLoS Genet 2019;15(12):e1008489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Nelson MR, Tipney H, Painter JL, et al. . The support of human genetic evidence for approved drug indications. Nat Genet 2015;47(8):856–60. [DOI] [PubMed] [Google Scholar]
- 158. Fang H, De Wolf H, Knezevic B, et al. . A genetics-led approach defines the drug target landscape of 30 immune-related traits. Nat Genet 2019;51(7):1082–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Wang Y, Sun Y, Liu Z, et al. . Dynamic graph cnn for learning on point clouds. ACM Trans Graphics 2019;38(5):1–12. [Google Scholar]
- 160. Kazi A, Cosmo L, Navab N, et al. . Differentiable graph module (dgm) graph convolutional networks arXiv preprint arXiv:2002.04999. 2020. [DOI] [PubMed]
- 161. Carvalho-Silva D, Pierleoni A, Pignatelli M, et al. . Open targets platform: new developments and updates two years on. Nucleic Acids Res 2019;47(D1):D1056–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Nicora G, Vitali F, Dagliati A, et al. . Integrated multi-omics analyses in oncology: a review of machine learning methods and tools. Front Oncol 2020;10:1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Sánchez-Valle J, Tejero H, Fernández JM, et al. . Interpreting molecular similarity between patients as a determinant of disease comorbidity relationships. Nat Commun 2020;11(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Wang T, Shao W, Huang Z, et al. . Moronet: multi-omics integration via graph convolutional networks for biomedical data classification. bioRxiv 2020. [Google Scholar]
- 165. Nguyen ND, Wang D. Multiview learning for understanding functional multiomics. PLoS Comput Biol 2020;16(4):e1007677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Ma T, Zhang A. Integrate multi-omics data with biological interaction networks using Multi-view Factorization AutoEncoder (MAE). BMC Genomics 2019;20:944. 10.1186/s12864-019-6285-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Pfister N, Williams EG, Peters J, et al. . Stabilizing variable selection and regression arXiv preprint arXiv:1911.01850. 2019.
- 168. Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov 2011;10(7):507–19. [DOI] [PubMed] [Google Scholar]
- 169. Moffat JG, Vincent F, Lee JA, et al. . Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat Rev Drug Discov 2017;16(8):531–43. [DOI] [PubMed] [Google Scholar]
- 170. Lipinski CA, Lombardo F, Dominy BW, et al. . Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 1997;23(1-3):3–25. [DOI] [PubMed] [Google Scholar]
- 171. Blundell TL, Jhoti H, Abell C. High-throughput crystallography for lead discovery in drug design. Nat Rev Drug Discov 2002;1(1):45–54. [DOI] [PubMed] [Google Scholar]
- 172. Murray CW, Blundell TL. Structural biology in fragment-based drug design. Curr Opin Struct Biol 2010;20(4):497–507. [DOI] [PubMed] [Google Scholar]
- 173. Erlanson DA, McDowell RS, O’Brien T. Fragment-based drug discovery. J Med Chem 2004;47(14):3463–82. [DOI] [PubMed] [Google Scholar]
- 174. Acharya C, Coop A, Polli JE, et al. . Recent advances in ligand-based drug design: relevance and utility of the conformationally sampled pharmacophore approach. Curr Comput Aided Drug Des 2011;7(1):10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Zheng W, Thorne N, McKew JC. Phenotypic screens as a renewed approach for drug discovery. Drug Discov Today 2013;18(21-22):1067–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Branch SK, Agranat I. “new drug” designations for new therapeutic entities: new active substance, new chemical entity, new biological entity, new molecular entity. J Med Chem 2014;57(21):8729–65. [DOI] [PubMed] [Google Scholar]
- 177. Kajino H. Molecular hypergraph grammar with its application to molecular optimization. International Conference on Machine Learning 2019;3183–91.
- 178. Clark MA, Acharya RA, Arico-Muendel CC, et al. . Design, synthesis and selection of dna-encoded small-molecule libraries. Nat Chem Biol 2009;5(9):647–54. [DOI] [PubMed] [Google Scholar]
- 179. Ramakrishnan R, Dral PO, Rupp M, et al. . Quantum chemistry structures and properties of 134 kilo molecules. Scientific Data 2014;1(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Chmiela S, Tkatchenko A, Sauceda HE, et al. . Machine learning of accurate energy-conserving molecular force fields. Sci Adv 2017;3(5):e1603015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Glen RC, Bender A, Arnby CH, et al. . Circular fingerprints: flexible molecular descriptors with applications from physical chemistry to adme. IDrugs 2006;9(3):199. [PubMed] [Google Scholar]
- 182. Schütt KT, Sauceda HE, Kindermans P-J, et al. . Schnet–a deep learning architecture for molecules and materials. J Chem Phys 2018;148(24):241722. [DOI] [PubMed] [Google Scholar]
- 183. Shire SJ. Formulation and manufacturability of biologics. Curr Opin Biotechnol 2009;20(6):708–14. [DOI] [PubMed] [Google Scholar]
- 184. Patel PK, King CR, Feldman SR. Biologics and biosimilars. J Dermatol Treat 2015;26(4):299–302. [DOI] [PubMed] [Google Scholar]
- 185. Mo J, Tymiak AA, Chen G. Structural mass spectrometry in biologics discovery: advances and future trends. Drug Discov Today 2012;17(23-24):1323–30. [DOI] [PubMed] [Google Scholar]
- 186. Kumar A, Kiran.. Characterization of protein-protein and protein-peptide interactions: implication for biologics design (February 2, 2020). Proceedings of International Conference on Drug Discovery (ICDD) 2020. Available at SSRN: https://ssrn.com/abstract=3530208. [Google Scholar]
- 187. Gligorijevic V, Renfrew PD, Kosciolek T, et al. . Structure-based function prediction using graph convolutional networks. bioRxiv 2020;5(9):786236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Jamasb AR, Lio P, Blundell T. Graphein - a python library for geometric deep learning and network analysis on protein structures. In: bioRxiv, 2020.
- 189. Rao Roshan, Bhattacharya Nicholas, Thomas Neil, Duan Yan, Chen Peter, Canny John, Abbeel Pieter, and Song Yun. Evaluating protein transfer learning with tape. Advances in Neural Information Processing Systems, Curran Associates Inc., 2019;9689–9701. [PMC free article] [PubMed]
- 190. Rives A, Goyal S, Meier J, et al. . Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. bioRxiv USA: National Academy of Sciences, 2019;622803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Ingraham John, Garg Vikas, Barzilay Regina, and Jaakkola Tommi. Generative models for graph-based protein design. Advances in Neural Information Processing Systems, 2019;32:15820–15831. [Google Scholar]
- 192. Radivojac P, Clark W, Oron T, et al. . A large-scale evaluation of computational protein function prediction. Nat Methods 2013;10:221–27. 10.1038/nmeth.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Lensink MF, Nadzirin N, Velankar S, et al. . Modeling protein-protein, protein-peptide, and protein-oligosaccharide complexes: Capri 7th edition. Proteins: Struct Funct Bioinform 2020;88(8):916–38. [DOI] [PubMed] [Google Scholar]
- 194. Galán A, Comor L, Horvatić A, et al. . Library-based display technologies: where do we stand? Mol Biosyst 2016;12(8):2342–58. [DOI] [PubMed] [Google Scholar]
- 195. Nixon AE, Sexton DJ, Ladner RC. Drugs derived from phage display: from candidate identification to clinical practice. In: MAbs, Vol. 6. Taylor & Francis, 2014;73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Bradbury ARM, Sidhu S, Dübel S, et al. . Beyond natural antibodies: the power of in vitro display technologies. Nat Biotechnol 2011;29(3):245–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Rickerby HF, Putintseva K, Cozens C. Machine learning-driven protein engineering: a case study in computational drug discovery. Eng Biol 2020;4(1):7–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Yang KK, Wu Z, Arnold FH. Machine-learning-guided directed evolution for protein engineering. Nat Methods 2019;16(8):687–94. [DOI] [PubMed] [Google Scholar]
- 199. Hawkins-Hooker A, Depardieu F, Baur S, et al. . Generating functional protein variants with variational autoencoders. BioRxiv 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Deac A, VeliČković P, Sormanni P. Attentive cross-modal paratope prediction. J Comput Biol 2019;26(6):536–45. [DOI] [PubMed] [Google Scholar]
- 201. Bromley Jane, Guyon Isabelle, LeCun Yann, Säckinger Eduard, and Shah Roopak. Signature verification using a “siamese” time delay neural network. Advances in Neural Information Processing Systems, 1994;737–744.
- 202. Atwood J, Towsley D. Diffusion-convolutional neural networks. Advances in Neural Information Processing Systems. Curran Associates, Inc., 2016;1993–2001. [Google Scholar]
- 203. Monti Federico, Boscaini Davide, Masci Jonathan, Rodola Emanuele, Svoboda Jan, and Bronstein Michael M. Geometric deep learning on graphs and manifolds using mixture model cnns. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 5115–5124, 2017.
- 204. Ryu JY, Kim HU, Lee SY. Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers. Proc Natl Acad Sci USA: National Academy of Sciences, 2019;116(28):13996–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Dalkiran A, Rifaioglu AS, Martin MJ, et al. . Ecpred: a tool for the prediction of the enzymatic functions of protein sequences based on the ec nomenclature. BMC Bioinformatics 2018;19(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Pillaiyar T, Meenakshisundaram S, Manickam M, et al. . A medicinal chemistry perspective of drug repositioning: recent advances and challenges in drug discovery. Eur J Med Chem 2020;112275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Nosengo N. New tricks for old drugs. Nature 2016;534(7607):314–6. [DOI] [PubMed] [Google Scholar]
- 208. Hodos RA, Kidd BA, Shameer K, et al. . In silico methods for drug repurposing and pharmacology. Wiley Interdiscip Rev Syst Biol Med 2016;8(3):186–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Zhou H, Gao M, Skolnick J. Comprehensive prediction of drug-protein interactions and side effects for the human proteome. Sci Rep 2015;5:11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Somody JC, MacKinnon SS, Windemuth A. Structural coverage of the proteome for pharmaceutical applications. Drug Discov Today 2017;22(12):1792–9. [DOI] [PubMed] [Google Scholar]
- 211. Deiana A, Forcelloni S, Porrello A, et al. . Intrinsically disordered proteins and structured proteins with intrinsically disordered regions have different functional roles in the cell. PloS One 2019;14(8):e0217889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Uversky VN. Intrinsically disordered proteins and their “mysterious”(meta) physics. Front Phys 2019;7:10. [Google Scholar]
- 213. Ragoza M, Hochuli J, Idrobo E, et al. . Protein–ligand scoring with convolutional neural networks. J Chem Inf Model 2017;57(4):942–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Gao Kyle Yingkai, Fokoue Achille, Luo Heng, Iyengar Arun, Dey Sanjoy, and Zhang Ping. Interpretable drug target prediction using deep neural representation. Proceedings of the 27th International Joint Conference on Artificial Intelligence, Vol. 2018, 3371–3377, 2018. [Google Scholar]
- 215. Nascimento AC, Prudêncio RB, Costa IG. A multiple kernel learning algorithm for drug-target interaction prediction. BMC Bioinformatics 2016;17:46. 10.1186/s12859-016-0890-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Crichton G, Guo Y, Pyysalo S, et al. . Neural networks for link prediction in realistic biomedical graphs: a multi-dimensional evaluation of graph embedding-based approaches. BMC Bioinformatics 2018;19(1):176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Tang Jian, Qu Meng, Wang Mingzhe, Zhang Ming, Yan Jun, and Mei Qiaozhu. Line: large-scale information network embedding. Proceedings of the 24th International Conference on World Wide Web, 1067–1077, 2015.
- 218. Mohamed SK, Nováček V, Nounu A. Discovering protein drug targets using knowledge graph embeddings. Bioinformatics Oxford, England: Oxford University Press, 2020;36(2):603–10. [DOI] [PubMed] [Google Scholar]
- 219. Keith CT, Borisy AA, Stockwell BR. Multicomponent therapeutics for networked systems. Nat Rev Drug Discov 2005;4(1):71–8. [DOI] [PubMed] [Google Scholar]
- 220. He L, Kulesskiy E, Saarela J, et al. . Methods for high-throughput drug combination screening and synergy scoring. In: Cancer Systems Biology. Springer, 2018;351–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. DiPette DJ, Skeete J, Ridley E, et al. . Fixed-dose combination pharmacologic therapy to improve hypertension control worldwide: clinical perspective and policy implications. J Clin Hypertens US: Wiley Periodicals Inc., 2019;21:4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Deac A, Huang Y-H, Veličković P, et al. . Drug-drug adverse effect prediction with graph co-attention arXiv preprint arXiv:1905.00534. 2019.
- 223. Zhou Y, Hou Y, Shen J, et al. . Network-based drug repurposing for novel coronavirus 2019-ncov/sars-cov-2. Nat Cell Discov 2020;6(1):1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Gysi DM, Do Valle Í, Zitnik M, et al. . Network medicine framework for identifying drug repurposing opportunities for Covid-19 arXiv, pages arXiv–2004. 2020. [DOI] [PMC free article] [PubMed]
- 225. Ioannidis VN, Zheng D, Karypis G. Few-shot link prediction via graph neural networks for covid-19 drug-repurposing. ICML 2020 Workshop on Graph Representation Learning and Beyond 2020. https://arxiv.org/abs/2007.10261.
- 226. Sosa DN, Derry A, Guo M, et al. . A literature-based knowledge graph embedding method for identifying drug repurposing opportunities in rare diseases. Pacific Symposium on Biocomputing 2019;25:463–25. [PMC free article] [PubMed] [Google Scholar]
- 227. Percha B, Altman RB. A global network of biomedical relationships derived from text. Bioinformatics 2018;34(15):2614–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Chen X, Chen M, Shi W, et al. . Embedding uncertain knowledge graphs. Proceedings of the AAAI Conference on Artificial Intelligence 2019;33:3363–70. [Google Scholar]
- 229. Veselkov K, Gonzalez G, Aljifri S, et al. . Hyperfoods: machine intelligent mapping of cancer-beating molecules in foods. Sci Rep 2019;9(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Flam-Shepherd D, Tony W, Friederich P, et al. . Neural message passing on high order paths arXiv preprint arXiv:2002.10413. 2020.
- 231. Kholodenko BN. Cell-signalling dynamics in time and space. Nat Rev Mol Cell Biol 2006;7(3):165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Raj A, Oudenaarden AV. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 2008;135(2):216–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Oono Kenta and Suzuki Taiji. Graph neural networks exponentially lose expressive power for node classification. International Conference on Learning Representations, 2019.
- 234. Barceló Pablo, Kostylev Egor V, Monet Mikael, Pérez Jorge, Reutter Juan, and Silva Juan Pablo. The logical expressiveness of graph neural networks. International Conference on Learning Representations, ICLR, 2019. [Google Scholar]
- 235. Dehmamy Nima, Barabási Albert-László, and Yu Rose. Understanding the representation power of graph neural networks in learning graph topology. Advances in NeurIPS Proceedings, Curran Associates, Inc., 2019;15413–15423. [Google Scholar]
- 236. Chen Z, Chen L, Villar S, et al. . Can graph neural networks count substructures? NeurIPS Proceedings Vancouver, Canada: Curran Associates, Inc., 2020. arXiv preprint arXiv:2002.04025. [Google Scholar]
- 237. Velickovic P, Fedus W, Hamilton WL, et al. . Deep graph infomax. In: ICLR, 2019.https://arxiv.org/abs/1809.10341.
- 238. You J, Ying R, Leskovec J. Position-aware graph neural networks. In: Chaudhuri K, Salakhutdinov R (eds). International Conference on Machine Learning PMLR, ML Research Press, 2019;7134–43. [Google Scholar]
- 239. Li P, Wang Y, Wang H, et al. . Distance encoding–design provably more powerful gnns for structural representation learning. 2020;33. arXiv preprint arXiv:2009.00142. [Google Scholar]
- 240. Sverchkov Y, Craven M. A review of active learning approaches to experimental design for uncovering biological networks. PLoS Comput Biol 2017;13(6):e1005466. 10.1371/journal.pcbi.1005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Gudovskiy D, Hodgkinson A, Yamaguchi T, et al. . Deep active learning for biased datasets via fisher kernel self-supervision. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition 2020;9041–9:9041.
- 242. Aggarwal Umang, Popescu Adrian, and Hudelot Céline. Active learning for imbalanced datasets. The IEEE Winter Conference on Applications of Computer Vision, 1428–1437, 2020.
- 243. Burgess DJ. Spatial transcriptomics coming of age. Nat Rev Genet 2019;20:317. 10.1038/s41576-019-0129-z. [DOI] [PubMed] [Google Scholar]
- 244. Baharlou H, Canete NP, Cunningham AL, et al. . Mass cytometry imaging for the study of human diseases-applications and data analysis strategies. Front Immunol 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Daniloski Z, Jordan TX, Wessels H-H, et al. . Identification of required host factors for sars-cov-2 infection in human cells. Cell 2020;184(1):92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


