Abstract
There have been numerous human studies reporting associations between the intestinal microbiome and functional gastrointestinal disorders (FGIDs), and independently animal studies have explored microbiome-driven mechanisms underlying FGIDs. However, there is often a disconnect between human and animal studies, which hampers translation of microbiome findings to the clinic. Changes in the microbiota composition of patients with FGIDs are generally subtle, while changes in microbial function, reflected in the fecal metabolome, appear to be more precise indicators of disease subtype-specific mechanisms. While we have made significant progress in characterizing the microbiome, to effectively translate microbiome science in a timely manner, we need concurrent and iterative longitudinal studies in humans and animals to determine the precise microbial functions that can be targeted to address specific pathophysiological processes in FGIDs. A systems approach integrating multiple data layers rather than evaluating individual data layers of symptoms, physiological changes, or –omics data in isolation will allow for validation of mechanistic insights from animal studies while also allowing new discovery. Patient stratification for clinical trials based on functional microbiome alterations and/or pathophysiological measurements may allow for more accurate determination of efficacy of individual microbiome-targeted interventions designed to correct an underlying abnormality. In this review, we outline current approaches and knowledge, and identify gaps, to provide a potential roadmap for accelerating translation of microbiome science toward microbiome-targeted personalized treatments for FGIDs.
Keywords: IBS, multi-omics, microbial therapeutics, synthetic biology, metabolomics
Introduction
The precise causes of functional gastrointestinal disorders (FGIDs) remain largely unknown and are multifactorial and varied among patients. The most common FGID, irritable bowel syndrome (IBS), alone affects 12% of the population and costs the US an estimated $30 billion annually.1 FGIDs are defined based on patient symptoms and encompass all regions of the gastrointestinal (GI) tract. The pathophysiology of FGIDs is complex, but mounting evidence in recent years suggests that the microbiome may play an important role in the development, persistence, and modulation of these symptoms.
The role of gut microbiome in determining FGIDs is not entirely novel as in about 10% of IBS patients the symptoms can be traced back to an episode of infectious gastroenteritis followed by persistent alterations in the microbiota structure that are associated with a change in bowel pattern (generally diarrhea in this case) and abdominal pain or discomfort.2 The advent of next-generation sequencing has allowed us to investigate changes in gut microbiota in a culture-independent manner, significantly improving the resolution and depth of measurements. This has led to several studies examining changes in gut microbiota composition and function in FGID patients compared to healthy controls, but the findings between studies have been inconsistent. Perhaps the most convincing evidence for a role of the microbiome comes from the observation of changes in GI transit, visceral hypersensitivity and changes in intestinal permeability seen in germ free (GF) mice following transplantation of stool from patients with either constipation-predominant IBS (IBS-C) or diarrhea-predominant IBS (IBS-D) patients.3-6
We appreciate that the origin of FGIDs is highly multifactorial and is best approached from a systems view with nonlinear and self-reinforcing contributions from patient genetics, epigenetics, brain networks,7 the enteric nervous system,8 environmental and lifestyle factors and their interactions with the gut microbiome.9 This review focuses on the potential contributions of the microbiome. However, to translate this knowledge, we need to consider the microbiome in the context of systems biology involving host genetics, physiological responses, and the environment. Hence in this review we will briefly summarize the current state,9-15 highlighting the gaps and areas for improvement as we design the next phase of investigations to identify microbial drivers and potential therapeutic strategies. We focus on IBS in the review as it is the best studied FGID but the same principles will also apply to other FGIDs where we are still in early stages of investigation.
What is our current understanding of the gut microbiome as a factor underlying IBS?
Changes in gut microbiota composition associated with IBS
The advent of next-generation sequencing has fueled an increase in efforts to identify changes in the gut microbiome related to IBS. We reviewed the human literature on IBS primarily focusing on studies using next-generation microbiome sequencing (Table 1) and found a lack of consistent compositional differences in colonic mucosal or luminal microbiota that reliably distinguish IBS patients from healthy controls. We found Streptococcus levels were higher in the stool,16-18 while Proteobacteria levels were higher in the mucosa of IBS patients compared to healthy controls18, 19 and lower alpha diversity was associated with IBS symptom severity.16, 18, 20, 21 A recent study in IBS patients within a large population based colonoscopy cohort of Swedish patients did not find any significant differences in the luminal or mucosa associated microbiome in IBS patients compared to healthy controls.22 A more comprehensive view was provided by a systematic review which compiled data from 24 studies done prior to 2018 and found that while there was some overlap, none of the studies reported the same differences in gut microbiota. They found the overall diversity was decreased or not changed and relative abundance of members of the Enterobacteriaceae, Lactobacillaceae, and Bacteroides were increased, while uncultured Clostridiales, Faecalibacterium, and Bifidobacterium were decreased in patients with IBS compared with controls.10 This study was a commendable effort to identify fecal and colonic mucosal microbial signatures underlying IBS, but brought to light some of the major challenges with the current approaches.
Table 1:
Summary of main findings from gut microbiota composition studies in IBS
| Microbiota variable |
IBS subtype |
Summary of finding | References |
|---|---|---|---|
| Alpha diversity | Both | Reduced in cross-sectional samples or samples with high symptom severity | 16, 18, 20, 21 |
| Microbiota stability | IBS-C | Larger temporal variability of fecal community than healthy control samples | 18 |
| Proteobacteria | Both | Elevated in mucosal community from biopsy samples | 18, 19 |
| Streptococci | Both | Elevated in fecal community, positively associated with high symptom severity | 16-18 |
| Ruminococci | Both | Elevated in phylogenetic microarray studies, but not reported in sequencing studies | 19, 89 |
| Lachnospiraceae | Both | Elevated in fecal community, implicated in reduced fecal hypoxanthine levels | 16, 18, 90, 91 |
| Methanobrevibacter smitthii | Both | Functionally implicated in constipation but differentially abundant in very few sequencing studies | 89 |
| Alistipes | Both | Bile tolerant bacteria, have been associated to pain | 19 |
There was significant heterogeneity among the studies, which is not entirely surprising given several shared prevailing weaknesses. These weaknesses included a lack of consistent methods for processing and analyzing microbiome samples, lack of rigorous statistical testing such as failure to correct for multiple hypotheses and inappropriate statistical tests, lack of multicenter data, and a lack of information on diet and other relevant covariates. Several of the studies were likely underpowered, as there was a median of 20 patients per group, which also precluded appropriate stratification by IBS subtype and appropriate matching among cases and controls. In addition, compositional heterogeneity among studies could also be a result of differences in geography 23, 24 and the cross-sectional nature of such studies,25 which fails to capture dynamic alterations in the microbiome.
The vast majority of studies have focused on the colonic microbiome as represented by a fecal sample, while the small intestinal microbiome has largely been overlooked even though small intestinal bacteria have been implicated in functional GI symptoms. A study that included duodenal and sigmoid biopsies found biopsy samples from the two sites were more similar in their microbial composition in IBS-D patients compared to healthy controls, although this could potentially be attributed to faster GI transit in IBS-D patients.26 Indeed, streptococci and Proteobacteria groups that are elevated in IBS patients in multiple studies are both facultative anaerobes that are associated with the upper GI tract rather than the colon. A recent study in patients presenting with symptoms of diarrhea, abdominal pain, or bloating undergoing esophagogastroduodenoscopy for suspected small intestinal bacterial overgrowth found that changes in small intestinal microbial composition may underlie functional GI symptoms and that diet can drive changes in small intestinal microbial diversity, intestinal permeability and appearance of GI symptoms.27 However in this study a subset (~22%) carried an organic diagnosis and/or had history of GI surgery (~23%), while another study found no differences in the small intestinal microbiome specifically in IBS patients.28 The role of small intestinal microbiome as a driver of physiologic changes and symptoms needs to be further explored in the context of IBS and other FGIDs.
There is an increasing realization that other microbial members such as fungi may also play a role in IBS pathogenesis.29 A recent study in healthy subjects and IBS patients found that the fungal species Saccharomyces cerevisiae and Candida albicans, which dominate the human mycobiome, were increased in IBS patients.30 The study also explored potential fungi-related mechanisms using an animal model of IBS and found a role for the Dectin-1/Syk pathway in driving visceral hypersensitivity and an increased histamine release by mast cells upon stimulation with fungi.30 In animal models, treatment with antifungals (fluconazole and nystatin) 30 or a combination of peppermint and caraway oils, which has both antifungal and antibacterial properties, reversed visceral hypersensitivity.31 These studies highlight the importance of considering the composite microbiome including bacteria, fungi, bacteriophages and parasites and their interactions.
Changes in gut microbiota-related metabolites associated with IBS
An increasing awareness of the functional redundancy that exists among microbes has called into question the role of studying microbial composition alone. It could be that IBS is taxonomically diverse but functionally more coherent; meaning it does not matter as much who is there but rather what are the biological functions being perform by the community. This realization has fueled an increased focus on changes in microbial function that underlie IBS, most often using untargeted metabolomics.
A recent review summarized findings from all observational and interventional studies that employ metabolomics to identify functional differences in gut microbiota between healthy subjects and IBS patients. The review found, analogously to the compositional differences, that while each study identified differences in metabolites, some of which overlapped among studies, a common metabolomic profile could not be identified in IBS patients.32 The reasons for this heterogeneity were the same as mentioned for studies reporting changes in microbial composition, suggesting inherent limitations in clinical study design. Another recent meta-analysis assessed differences in short chain fatty acids (SCFAs) among IBS patients in 15 studies and found propionate and butyrate were reduced in IBS-C patients, while butyrate was increased in IBS-D patients in comparison to healthy controls.18, 33-35
SCFAs, particularly acetate, butyrate, and propionate, are versatile signaling molecules by which bacteria can exert their effects on GI function. SCFAs have effects on varied aspects of GI physiology such as contractility, visceral pain, and barrier function,36, 37 making it important to consider the dynamic alterations in their levels based on microbial activity. For example, acetate, butyrate, and propionate have distinct and concentration-dependent effects on intestinal contractility and the serotonergic system in animal models and colonoids.35, 38-40 In addition to SCFAs, bacteria produce proteases and neurotransmitters such as gamma-aminobutyric acid (GABA), dopamine, and norepinephrine, which may contribute to visceral pain by directly stimulating host receptors within the intestine.41, 42 Changes in the microbial capacity for conversion of primary to secondary bile acids has also been implicated in the pathogenesis of bile acid diarrhea in a subset of patients with IBS-D.18, 43-47 The lack of consistent subtype-specific metabolomic signatures in IBS patients – in spite of the known effects of bacterial metabolites on physiological processes involved in symptoms – may reflect a phenomenon in which patients of the same symptom-based subtype have different etiopathogenesis. An alternative approach that might aid in predicting therapeutic responses would be to stratify patients using targeted metabolomics of specific pathways relevant to IBS symptoms.
Gut microbiota-related mechanisms underlying IBS
The majority of mechanistic studies have been done in animal models including germ free and gnotobiotic rodent models and involve manipulations of gut microbes and microbial products with a focus on specific physiologic processes relevant to IBS. These include intestinal secretion and absorption, intestinal permeability and barrier function, motility, visceral sensation, immune activation, and central nervous system responses.9 The effects on host physiology of microbial processes such as production of SCFAs and deconjugation of bile acids described above 9 depend on the cell types involved (e.g. epithelial, immune, neuronal) and the specific region of the intestine. Physiological changes related to the gut microbiota or an intervention directed at the gut microbiota have also been reported in human studies 9 and are summarized below in Table 2 (reprint from 9).
Table 2:
Summary of pathophysiologic mechanisms related to gut microbiota in patients with FGIDs (reprint from9)
| Study | Study population |
Intervention | Sample | Mechanism studied |
Role of microbiota |
|---|---|---|---|---|---|
| Shin et al, 201892 | 60 IBS-D | L gasseri BNR 17 vs pcbo | Fecal | Transit | Transit significantly ↑ during 8 wk with L gasseri BNR17 |
| Tap et al, 201720 | 110 IBS, 39 HV | NA | Fecal, mucosal | Transit, GBA | ↑Transit with Clostridiales vs Prevotella and Bacteroides enterotypes No association between HADS and enterotype |
| Acosta et al, 201693 | 24 nonconstipat ed IBS | Rifaximin vs pcbo | Fecal | Transit, permeability, SCFA and bile acid production | No significant effects of rifaximin on permeability, bile acids, SCFAs Rifaximin associated with ↑ascending colon emptying, and colonic transit at 48 h |
| Dior et al, 201694 | 15 HV, 15 IBS-C, 16 IBS-D |
NA | Fecal | Fecal bile acids | ↓Bacterial deconjugation of bile acids in IBS-D and IBS-C feces vs HV |
| Le Neve et al, 201695 | 100 IBS | NA | Fecal | Sensation, transit | Response to lactulose challenge associated with rectal sensitivity but not with fecal microbiota or transit |
| Chumpitazi et al, 201496 | 12 IBS children | LFSFD | Fecal | Transit, metabolite composition | LFSD response associated with ↑abundance of Sporobacter and Subdoligranulum and ↓ Bacteroides, but not with transit Stool metabolites (L-urobilin, cholate) associated with response and microbiome composition |
| Jeffery et al, 201297 | 37 IBS, 20 HV | NA | Fecal | Sensation, transit, GBA | Proteobacteria associated with ↑mental component and pain threshold Actinomycetales associated inversely with depression Desulfohalobiaceae and Methanobacteriaceae associated with transit |
| Labus et al, 201798 | 29 IBS, 23 HV | NA | Fecal | GBA | No correlations between anxiety or depression symptom scores and microbial parameters; Clostridia and Bacteroidia correlated with sensory integration regions |
| Liu et al, 201699 | 40 IBS, 15 depression, 25 IBS and depression, 20 HV | NA | Fecal | GBA, immune | ↑Bacteroidetes and ↓Firmicutes in IBS-D, depression, and IBS-D with depression; Colonic mucosa inflammation associated with ↑ Bacteroides or Prevotella |
| Azpiroz et al, 2017100 | 79 IBS | scFOS vs pcbo | Fecal | GBA, sensation | scFOS reduced anxiety scores and increased fecal Bifidobacteria No significant difference in rectal sensory threshold for scFOS vs pcbo |
| Le Gall et al, 2011101 | 10 IBS, 13 UC, 22 HV | NA | Fecal | Fecal metabolites | Correlation between gut microbiota profile and metabolite composition |
| Heitkemper et al, 2018102 | 93 IBS | NA | Fecal | Permeability | Higher stool TFF3 associated with lower permeability and microbial diversity Christensenellaceae related inversely to stool TFF3 |
| Bednarska et al, 2017103 | 32 IBS, 15 HV | NA | Mucosal | Immune, permeability | Increased permeability to E coli strain HS and S typhimurium in IBS biopsy specimens vs controls ↑Plasma VIP in IBS vs HV ↑Tryptase and mast cells in IBS biopsy specimens vs HV |
| Valentin et al, 2017104 | 15 IBS-D | SBI | Duoden al brushing, fecal | Immune, permeability, metabolism | Bile acid synthesis, tryptophan metabolism, permeability, and stool microbiome not significantly different with SBI Changes in β diversity analysis, increased ↑ Proteobacteria Burkholderiales, Firmicutes Catonella, and unclassified genus organisms with SBI in duodenal microbiome |
| Ko et al, 2013105 | 53 IBS-D | Herbal (GJS), probiotic (Duolac7S; Cell Biotech Co, Ltd, Gimpo, Korea), pcbo | Fecal | Permeability | GJS with DuoLac7 ↑ B lactis, L rhamnosus, L plantarum No significant difference observed in permeability |
| Crouzet et al, 20136 | 3 IBS-C, 2 HV | NA | Fecal | Rectal sensitivity | IBS with rectal hypersensitivity have ↓bifidobacteria, ↑Enterobacteriaceae, and ↑H2-using sulfide-producing bacteria vs HV |
| Shulman et al, 2017106 | 103 IBS children | Fiber vs placebo | Fecal | GBA, permeability | No differences in psychological symptoms, permeability, or microbiome between groups |
| Compare et al, 2017107 | 10 IBS-D, 10 HV (ex vivo) | LC-DG, postbiotic | Mucosal | Immune | ↑IL1α, IL6, and IL8 messenger RNA, TLR-4 protein expression with ↓IL10 messenger RNA levels in PI-IBS-D vs HV LC-DG and PB ↓messenger RNA levels of proinflammatory cytokines and TLR-4 but ↑IL10 after LPS stimulation |
| Hustoft et al, 2017108 | 20 IBS-D or IBS-M | Low FODMAP diet, FOS vs pcbo | Fecal | Immune, SCFA | ↓IL6 and IL8, fecal bacteria (Actinobacteria, Bifidobacterium, Faecalibact erium prausnitzii), total SCFAs, and n-butyric acid on LFD FOS supplement then ↑levels of these bacteria, but cytokines and SCFAs unchanged |
| McIntosh et al, 2017109 | 37 IBS | Low vs high FODMAP | Fecal | Urinary metabolites | Significant correlations between relative bacterial abundance and symptoms and urinary metabolites (histamine, p-hydroxybenzoic acid) |
| Sundin et al, 2015110 | 11 PI-IBS, 10 HV (ex vivo) | NA | Mucosal | Immune | IL1β ↑ in PI-IBS vs HV after stimulation with Subdoligranulum variabile; IL10 ↓ in HV vs PI-IBS after stimulation with Eubacterium limosum |
| Sundin et al, 2015111 | 13 PI-IBS, 19 IBS, 16 HV | NA | Fecal, mucosal | Immune, GBA | Naive CD8+ CD45RA+ intraepithelial lymphocytes and lamina propria lymphocytes correlated negatively with mucosal microbial diversity Fecal microbial diversity correlated negatively with HADS |
| Pinto-Sanchez et al, 2017112 | 44 IBS | BL vs pcbo | Fecal | GBA, immune, urinary metabolites, neurotransmitter s, and neurotrophins | BL ↓depression and was associated with ↓limbic reactivity No difference in fecal microbiota, serum markers of inflammation, neurotrophins, and neurotransmitters Reduced urine methylamines and aromatic amino acid metabolites with BL |
| Parthasarathy et al,217 2017 | 25 CC, 25 HV | NA | Fecal | Transit | Reproducibility of fecal microbiota lower in normal transit vs slow-transit constipation |
| Parthasarathy et al, 2016113 | 25 CC, 25 HV | NA | Fecal, mucosal | Transit | Fecal microbiota profile associated with colonic transit; genera from Firmicutes correlated with faster colonic transit |
| Tian et al, 2017114 | 60 STC | FMT | NA | Transit | FMT associated with faster transit vs control treatment |
BL, Bifidobacterium longum NCC3001; cc, chronic constipation; FODMAP, fermentable oligosaccharides, disaccharides, monosaccharides, and polyols; GBA, gut–brain axis; HADS, Hospital Anxiety and Depression Scale; HV, healthy volunteer; IBS-M, irritable bowel syndrome mixed subtype; LC-DG, Lactobacillus casei DG; LFSD, low fermentable substrate diet; LPS, lipopolysaccharide; pcbo, placebo; PI-IBS, postinfectious irritable bowel syndrome; SBI, serum-derived bovine immunoglobulin/protein isolate; scFOS, short-chain fructooligosaccharide; TFF3, urine trefoil factor 3.
Current microbiota-directed therapeutic interventions in IBS
There has been a considerable focus on investigations to better understand the mechanisms by which gut microbiota modulate GI physiology, but in parallel a large number of therapeutic interventions directed at gut microbiota have also been explored. The positive aspect of such a concurrent approach is the opportunity to learn from each other as we launch the next set of investigations but unfortunately for now the two lines of investigation have been largely disconnected. Therefore, it is not surprising that the majority of the current microbiota-directed therapeutic strategies which include probiotics and synbiotics, prebiotics and dietary interventions, antibiotics, and more recently fecal microbiota transplant (FMT) have failed to show consistent results. Antibiotics such as rifaximin have shown improvement in global IBS symptoms in non-constipated IBS patients but the mechanism and specific microbial populations that are being targeted by these antibiotics remain unclear.48
Probiotics are defined as live organisms that confer health benefits to the host when administered in adequate amounts. A recent technical review focused on efficacy of probiotics in GI diseases, commissioned by American Gastroenterology Association (AGA), reviewed the literature from inception till December 2018 and after screening 1617 titles and abstracts and assessing 216 full-text articles, identified 55 interventional studies using probiotics in adults and children with IBS that met the rigor of high quality placebo controlled randomized controlled trials.49 This included a collective 5301 subjects and 44 different probiotic formulations. While individual formulations showed symptom improvement in single clinical trials, the overall certainty of evidence for use of probiotics to treat IBS was low. The AGA guidelines based on this technical review acknowledge the significant knowledge gaps and in contrast to other guidelines which suggest probiotics may be beneficial in IBS, recommend use of probiotics only in the context of clinical trials. This difference in recommendations was primarily driven by the fact that the technical review considered the evidence at the level of individual species/strain, and not the composite group of probiotics because biological effects of individual strains or formulations can vary significantly.
The most recent monograph from the American College of Gastroenterology (ACG) reviewed the evidence in support of dietary interventions, prebiotics, and synbiotics in the management of IBS.11 Based on the review of available literature a weak recommendation was made for the use of low FODMAP (fermentable oligo- di- monosaccharides and polyols) diet, and against the use of gluten-free or exclusion diets. Both recommendations were based on very low quality of evidence given the relatively small number of patients in each trial, heterogeneity, issues with blinding and a high risk of bias.11 One important concern raised in the review regarding low FODMAP, which is the most prevalent dietary intervention, was the potential harmful effect on the gut microbiome and a lack of assessment of long term efficacy which invites speculation about the potential for long-term harm resulting from gut microbiota alterations in order to achieve short-term symptomatic benefit.
Prebiotics (food or dietary supplements that alter the microbial community composition/function with the goal of improving health) and synbiotics (live microorganisms and substrate(s) selectively utilized by host microorganisms, which confer a health benefit on the host) 50 were also evaluated but the number of studies were limited (1 for prebiotic and 2 for synbiotics). Based on very low quality of evidence, a recommendation was made against the use of these products. Interestingly, poorly fermentable psyllium fiber but not wheat bran was found to be effective based on moderate quality of evidence and was strongly recommended for symptom improvement in IBS.11 This is particularly interesting given that recent studies show a beneficial effect of fiber, especially psyllium, on intestinal barrier function.51
Finally, there is accumulating evidence on the efficacy of FMT and 5 randomized controlled trials have already been conducted, of which three have shown short term symptom improvement with administration of FMT. However, as with other interventions, these studies vary in the dose, duration of treatment, route of administration, outcomes measured, donor characteristics, and study population which makes it difficult to generalize the results or make clinical recommendations regarding the use of FMT in IBS. The lack of efficacy of the current microbiome therapies highlights the large gap in our understanding of where the microbiome fits within the pathophysiology of IBS as these interventions generally do not address an underlying disease mechanism.
How do we advance the field to make meaningful progress towards mechanism-based therapeutics?
Beyond cause and effect: reinforcement as a way of perpetuating disease phenotypes
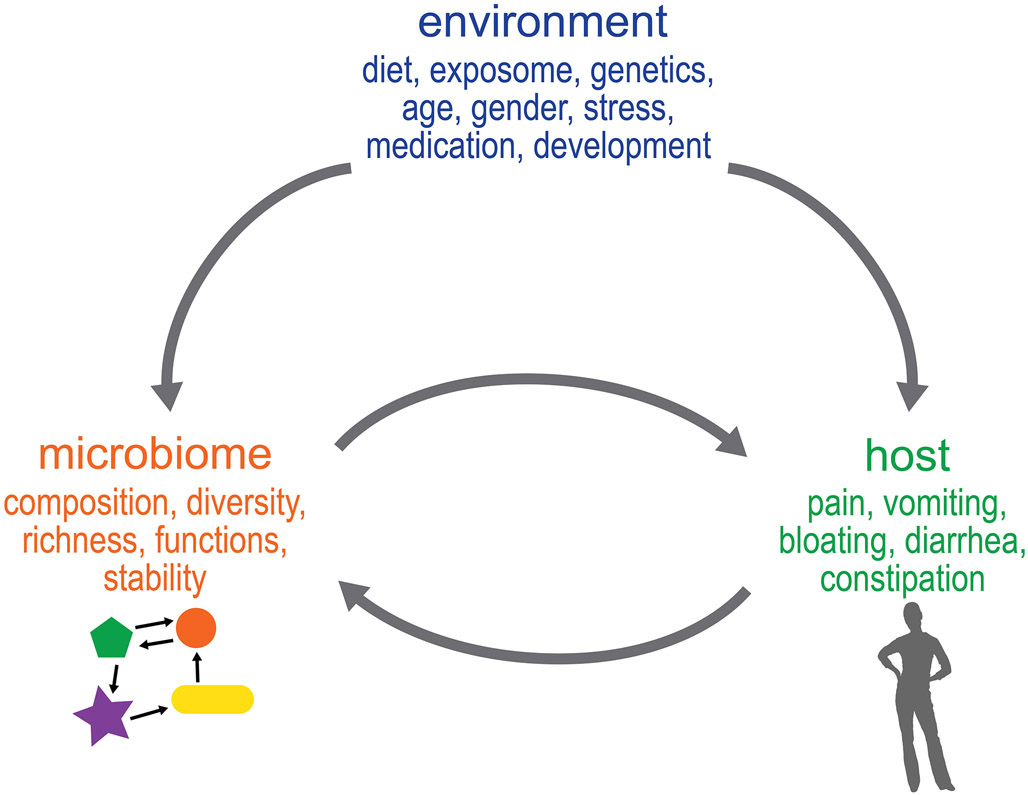
One of the challenges in understanding the role of the gut microbiome in chronic diseases has been the assumption that differences in the gut microbiome among disease and healthy individuals represent either a cause or effect, rather than a complex mutually reinforcing series of interactions between the environment, host, and microbiome (Figure 1). A myriad of risk factors for IBS such as host genetics, gender, early life trauma, diet, and stress can affect host functions as well as the microbiome but it is not easy to distinguish if these effects are independent of each other, or rather, inter-dependent. Hence it may be better to think of at least some of these interactions as a continuum where changes are being mutually reinforced to drive a phenotype.
Figure 1: Beyond cause and effect: Mutual reinforcement of changes in host function and gut microbiota may underlie FGID symptoms.
Environmental factors have been associated with changes in the microbiome and the host but it is difficult to discern if these effects are independent or inter-dependent due to constant mutual reinforcement of these changes by host and gut microbiota.
A good example to illustrate this phenomenon is diet. Dietary intolerances are common in IBS and patients often report that certain foods, or simply eating, exacerbate or initiate symptoms including bloating, pain, and altered bowel pattern.52 While there is evidence suggesting that a subset of IBS patients may have food allergies that contribute to symptoms, non-allergic food intolerances and hypersensitivity are much more likely, such as non-celiac gluten sensitivity.53 Diet affects the gut microbiome by determining resource availability,54 while the gut microbiome can affect dietary intake and preferences, either through influencing signaling mechanisms affecting satiety 55 or driving avoidance of foods that lead to increased pain, diarrhea, or flatulence.56-59 Eliminating specific foods believed to be driving symptoms, could in turn affect the microbial populations that rely on those foods,60-64 and possibly reinforce the aversive reaction.13, 65 As an example: bloating, which is common in FGIDs especially with constipation, is often attributed to increased gas production as a result of bacterial fermentation of dietary carbohydrates, even though the prevailing evidence suggests it is likely due to visceral hypersensitivity rather than increased total gas in the intestine.66 However, this often leads to avoidance of fermentable carbohydrates, resulting in decreased SCFA production, which in turn can decrease GI motility and exacerbate constipation and bloating, as SCFAs are known to promote GI motility and intestinal barrier function.35, 38, 67, 68 The slower transit would favor the growth of slower growing microorganisms (e.g. methanogens), resulting in a change in the metabolic end products such as SCFAs and gases including H2, CO2 and CH4 produced by gut microbiota, which in turn can perpetuate constipation and bloating.69 This concept is supported by a recent study in an animal model where pharmacologically induced constipation (PIC) in mice altered the microbial metabolic profile and decreased fecal butyrate production as also seen in IBS-C patients. Germ-free mice colonized with either microbiota from PIC mice or IBS-C patients exhibited longer GI transit time and lower butyrate levels compared to control mice.4 Thus one can view this as a self-reinforcing positive feedback loop of constipation leading to lower SCFA production and lower SCFA production leading to decreased GI motility and fluid secretion. Conceivably, the increased methane production associated with constipation results from similar mutual reinforcement. Diet can also accelerate GI transit (e.g., via fiber, which adds bulk and osmotically draws water into the intestine)70-72 and the resulting alterations in GI transit, in turn, alter the gut microbiome by favoring rapidly growing microbes, consistent with ecological principles of r/K selection in response to environmental disturbance.73, 74 This is in line with findings from several human studies which have found an association between transit time or stool consistency and the microbiome.75 Hence, rather than considering elements such as the microbiome as being relevant only when they are the cause of a diseases (e.g. FGIDs), one needs to focus on understanding how it fits within the broader pathophysiology. This is especially relevant as we start to develop more mechanism-based interventions targeting the microbiome.
Roadmap for improving future discovery pipeline based on lessons learnt from current strategies
As highlighted above, interpretation of data in FGID studies is complicated by the fact that both IBS and the microbiome exhibit dynamic alterations over time. Hence a snapshot of observations from cross-sectional studies lacks the temporal resolution needed to understand the role of the gut microbiome in driving physiological changes and symptoms in IBS. In addition, the majority of studies are focused on a single factor, for example microbial composition, without integrating other -omics data or additional clinical/environmental data. As such, the impact of an individual factor cannot be contextualized in the absence of other variables that can affect the disease phenotype. Hence there is a need for improved study design and data integration and we propose some strategies for this below.
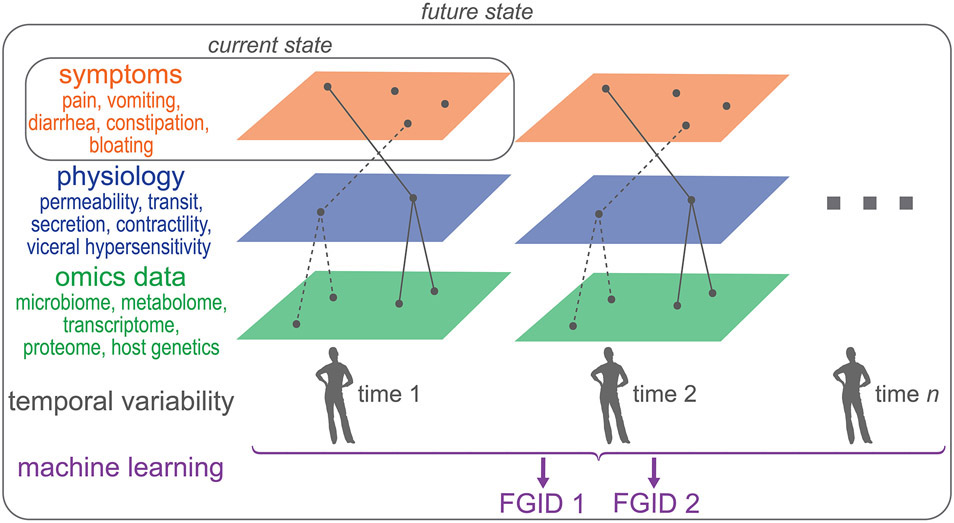
To identify novel microbiota-driven mechanisms and therapeutic targets with a high degree of confidence, we need to combine the strategies employed by individual studies as part of more comprehensive studies that include multiple data layers (Figure 2). The data first needs to be integrated within each layer (Figure 2; e.g. –omics, symptoms, physiology) and then across different layers. As an example, the concurrent use of microbial composition and metabolomics provides better separation between IBS patients and healthy controls, highlighting the benefit of integrating different types of data within the – omics data layer.76 To build on this further, metagenomics and metabolomics can be combined with host gene expression, genetics, and epigenetic changes in a biologically plausible way. For instance, if a microbe which is differentially abundant in a subset of patients is known to produce compound A, this can be verified directly using the metabolomics dataset. If compound A is known to have an effect on host gene expression, this can be directly inspected in the paired dataset as well. Such a targeted integration approach complements non-targeted discovery strategies to identify and validate novel pathways using datasets collected from the same study cohort. The integration of different types of –omics data is complex, but provides greater likelihood of identifying a biologically plausible mechanism. To validate findings from the integrated -omics data layer, we need to measure host physiological parameters that are predicted to be affected based on the omics data. We then need to determine if those physiological changes in turn are the primary driver of patient symptoms. Together all these data layers represent a cross-section of a patient’s disease state, which are appropriate for acute and stable disease states. However in chronic diseases with waxing and waning symptoms, we need longitudinal measurements of these data layers to assess dynamic changes over time (Figure 2).
Figure 2: From symptom-based to a systems approach for classification of FGIDs.
FGIDs are currently classified based on symptoms, which provides a framework for clinical studies but as we move towards more comprehensive profiling of patients using multi-omics and physiologic changes in addition to symptoms, we can envision a future state which will allow for stratification of patients by integrating multiple data layers collected longitudinally. Such a stratification strategy will not only allow for development of mechanism-based targeted therapeutics but will also allow identification of appropriate patient cohorts which are most likely to benefit from such a therapy.
In a recent study, we used such an approach by integrating longitudinally collected multi-omics measurements (including the metagenome, metabolome, host transcriptome, genetic and epigenome), with changes in host physiology and extensive clinical metadata including diet and symptoms.18 IBS subtypes and symptom severity were associated with specific changes in the gut microbiome and metabolome, and similar changes were seen at the time of self-identified flares in a subset of patients. We found that longitudinal sampling was important to overcome heterogeneity seen with cross-sectional microbiome studies given the fluctuating nature of symptoms in IBS. We initially used a targeted approach to determine the relevance of previously known bacterial metabolites that affect host physiology. SCFAs, previously shown to alter the host serotonergic pathway,35, 40 were significantly decreased in subjects with IBS-C with a corresponding decrease in secretory response to serotonin in colonic biopsies from IBS-C subjects and these changes were independent of fiber intake. We also identified two gene regions in Blautia obeum which were strongly associated with butyrate and significantly decreased in IBS-C subjects. While the therapeutic effect of Blautia obeum in IBS-C patients has not been investigated in clinical trials, a parallel, double-blinded, randomized, placebo-controlled study reported that 12 weeks of microencapsulated butyrate supplementation improved abdominal pain, stool consistency, and constipation relative to placebo in IBS patients (based on Rome III criteria, all subtypes) based on per protocol analysis.77
In IBS-D patients, basal secretion was increased in colonic biopsies with a corresponding increase in secretagogues, chenodeoxycholic acid (usually converted to lithocholic acid by gut microbiota) and the bacterial metabolite tryptamine which was recently found to be a 5HT4R agonist.78 Finally, by integrating multiple host and microbiome data layers we also identified a novel pathway involving purine metabolism which may be important in the pathophysiology of IBS. Stool hypoxanthine levels were consistently decreased over time in both IBS subtypes along with an increased functional capacity for hypoxanthine breakdown by the microbiome and the colonic epithelium, and upregulation of the purine salvage pathway in the colonic epithelium. We identified a specific microbial gene region responsible for potential utilization of hypoxanthine and confirmed the ability of bacteria to deplete luminal hypoxanthine in gnotobiotic mice. Potential benefits of restoring hypoxanthine levels in IBS patients have not been explored, although hypoxanthine is known to be an energy source for colonocytes and plays an important role in maintaining the integrity of the intestinal barrier;79, 80 its role in IBS will be need to be investigated in future work. In summary, this study is an example of how a systems approach can be leveraged to confidently identify microbial drivers of a disease state.
Moving towards therapeutic strategies based on individualized mechanisms
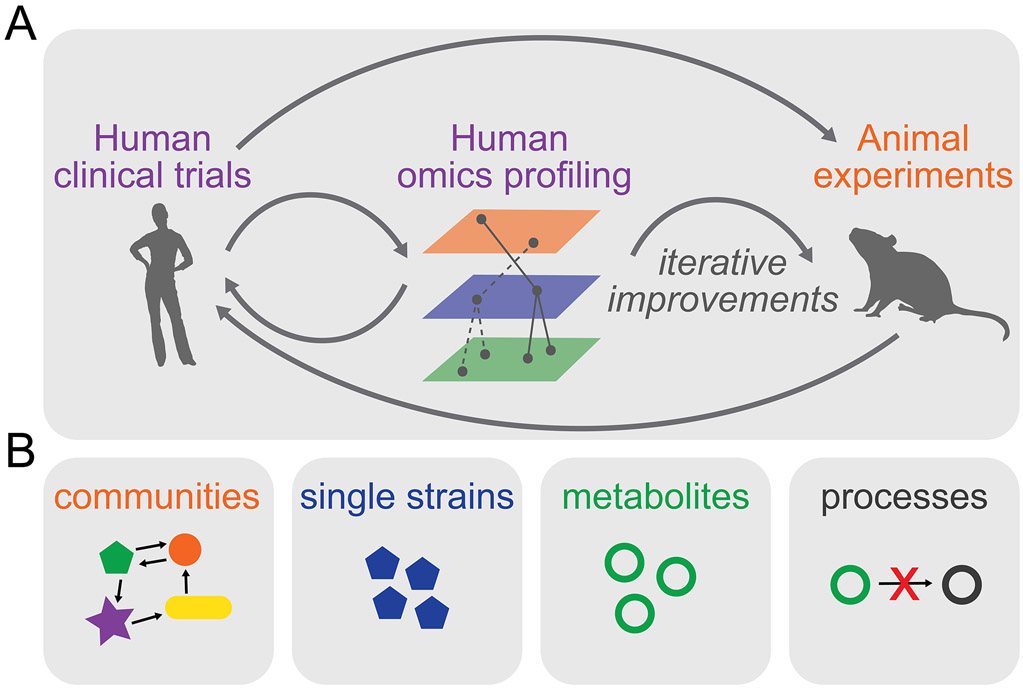
FGIDs are diagnosed based on symptom-based criteria which can result in a heterogeneous disease population with similar symptoms but different etiopathogenesis. This heterogeneity might be acceptable to help identify patients for inclusion in clinical studies focused on relieving symptoms but can prove challenging when testing therapeutics that address a specific mechanism. As an example, only a subset of IBS patients’ exhibit differences in microbial diversity/abundance but this is not taken into consideration when selecting patients for inclusion in clinical trials of therapeutics such as FMT that aim to restore microbial diversity. The current microbiota-directed therapeutics in IBS are not always informed by experimental studies and even when they do aim to restore a specific mechanism, the test population is selected based on symptoms and not mechanism. This consideration and the lack of knowledge as to which patient is likely to benefit from a treatment may explain why clinical trials of microbiome therapeutics have failed to show an effect. However, the vast number of clinical studies done thus far serve as a rich source of data and have allowed us to identify the challenges and limitations. Building on this existing knowledge, we outlined a systems approach above to improve discovery of novel microbial drivers of disease that can be verified experimentally in animal or in vitro model systems, and then tested in humans; this is an iterative process that would allow development of more robust therapeutics (Figure 3A). As the field moves towards large well designed longitudinal human studies, AI-based approaches can be used to integrate –omics data with physiology and symptoms to identify unique features of responders and non-responders for individual therapies. Such insight would allow for better stratification of therapeutics, resulting in targeted restoration of disrupted processes in specific patient populations (Figure 3A).
Figure 3: Iterative cycles of improvement to develop the next generation of individualized mechanism-based microbiome therapeutics.
A) A systems approach for discovery allows development of novel hypotheses that can be mechanistically explored in animal models or used to refine therapeutics for human studies. The data from experimental models can aid in development of mechanism based therapeutics targeting specific microbial processes. A similar approach applied to de novo clinical trials can help identify unique characteristics of responders and non-responders that can be iteratively tested to develop novel therapeutics for subsets of patients. B) Main approaches for therapeutic manipulation of the microbiome.
As discussed previously, microbial metabolites could be key drivers of the pathophysiology of IBS. The processes involved in production and consumption of these metabolites could thus be targets for novel therapeutics. In correcting microbiome alterations, the challenge is to modulate taxa or metabolite levels in a specific and tailored way. The main approaches to therapeutic manipulation of the microbiome include introducing optimized communities, introducing single strains (which can be native or synthetic), and removing or inhibiting specific microbes or microbial processes. The choice of approach will rely on knowledge on specific disease mechanisms to be targeted.
Ecological community-based approaches
Ecological approaches involve either replacing an entire community with a novel community associated with health or introducing a consortium of bacteria that perform specific functions (e.g., production of metabolites) (Figure 3B). Designing such communities, while also ensuring desired output over time, requires in depth knowledge of ecologic and metabolic interactions among microbial strains in the presence of varied substrates. As an example, a community of microbes designed to produce SCFAs may perform differently based on the types of fiber that are included in the diet as shown recently in a human study utilizing dietary fibers to boost SCFA production.81
Some of the challenges in designing such communities are anticipating ecological ripple effects (i.e. hard to predict distant changes), 82, 83 historical contingency (such as the positive reinforcement seen between GI transit and microbiota alterations), and the relative stability of the gut microbiome. In addition, the gut microbiome seems to experience phylogenetic under-dispersion, where species recruitment is more likely if close phylogenetic relatives are present.84 Interspecies competition and niche occupation can limit changes to the microbial ecosystem once it has been stably established.
Single-strain approaches
Introducing specific microbes into the host (either from natural sources or genetically engineered) may restore host functions by producing or competitively consuming critical metabolites or microbiota-derived signaling molecules (Figure 3B). Genetic engineering of bacteria can be used to optimize production or consumption of metabolites. In a recent study Bacteroides thetaiotaomicron was engineered to constitutively overproduce tryptamine under control of a phage promoter, resulting in increased intestinal secretion by tryptamine-driven activation of 5HT4R.78 A similar engineering approach to introduce a unique nutrient utilization pathway can aid in stable colonization of a non-native strain by varying levels of the specific nutrient in diet.85 Synthetic biology tools can also be utilized to develop more elaborate bacterial systems that sense changes in the environment and respond by producing or consuming specific metabolites, or by delivering therapeutic effectors to specific sites.86
Alternatively, conjugate vaccines, phage therapies, and narrow-spectrum antibiotics can be used to target single strains or species, while keeping in mind the potential ecological ripple effects.
Microbial metabolites
Microbial metabolites (Figure 3B) may also be used by themselves as therapeutics instead of using microbial communities or strains to change metabolite levels.87 The advantage of dosing metabolites is extensive prior experience with manufacturing small molecules and modeling their pharmacokinetics and pharmacodynamics, while the drawbacks include delivery at the proper site, short half-lives, and pleiotropic effects that the metabolites may have on other microbiome members or the host.
Removing or inhibiting microbial processes
Microbial metabolite levels can also be manipulated by inhibiting the processes used for their production or consumption (Figure 3B). Inhibitors of enzymatic pathways that are microbe-specific can be targeted to prevent accumulation of specific molecules without removing taxa from a community and with little expected cross-talk with human pathways.88 In cases where the host and microbes share similar pathways, existing enzyme inhibitors could be re-formulated to prevent their absorption, ensuring their effect is limited to the intestinal lumen.
Conclusions
We have made significant progress over the last decade in identifying gut microbiome changes associated with IBS, investigating microbiota-driven mechanisms relevant to IBS, and evaluating microbiome therapeutics in interventional trials. While this has provided a rich framework of data, it has also allowed us to step back and look at the shortcomings that need to be addressed in future studies. In this review we have tried to provide a summary of our current understanding and outlined steps that can help inform future research. We need to consider more robust study design for human studies which includes extensive metadata collection and relevant controls (e.g. household controls), standardize the collection and processing of microbiome samples, and improve data and metadata sharing practices. The integration of multiple data layers collected longitudinally can provide a more robust discovery pipeline and provide more meaningful stratification of patients for therapeutic trials of mechanism-based microbiome therapeutics.
Acknowledgements:
We would like to acknowledge Lyndsay Busby for secretarial assistance. This work was made possible by funding from NIH DK114007 and the Center for Individualized Medicine, Mayo Clinic, Rochester, MN (PCK) and Medical Scientist Training Program T32GM007281 (MF).
Abbreviations:
- FGID
functional gastrointestinal disorder
- IBS
irritable bowel syndrome
- GABA
gamma-aminobutyric acid
- SCFA
short chain fatty acid
- FMT
fecal microbiota transplant
- GI
gastrointestinal
- GABA
gamma-aminobutyric acid
- FODMAP
fermentable oligo- di- monosaccharides and polyols
- PIC
pharmacologically induced constipation
- AI
artificial intelligence
Footnotes
Disclosures: Purna C. Kashyap serves on the advisory board of Novome Biotechnologies and is an ad hoc consultant for Otsuka Pharmaceuticals, Pendulum Therapeutics and IP group Inc.
References:
- 1.Chey WD, Kurlander J, Eswaran S. Irritable bowel syndrome: a clinical review. JAMA 2015;313:949–58. [DOI] [PubMed] [Google Scholar]
- 2.Jalanka-Tuovinen J, Salojarvi J, Salonen A, et al. Faecal microbiota composition and host-microbe cross-talk following gastroenteritis and in postinfectious irritable bowel syndrome. Gut 2014;63:1737–45. [DOI] [PubMed] [Google Scholar]
- 3.De Palma G, Lynch MD, Lu J, et al. Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Sci Transl Med 2017;9. [DOI] [PubMed] [Google Scholar]
- 4.Touw K, Ringus DL, Hubert N, et al. Mutual reinforcement of pathophysiological host-microbe interactions in intestinal stasis models. Physiol Rep 2017;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edogawa S, Edwinson AL, Peters SA, et al. Serine proteases as luminal mediators of intestinal barrier dysfunction and symptom severity in IBS. Gut 2020;69:62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crouzet L, Gaultier E, Del'Homme C, et al. The hypersensitivity to colonic distension of IBS patients can be transferred to rats through their fecal microbiota. Neurogastroenterol Motil 2013;25:e272–82. [DOI] [PubMed] [Google Scholar]
- 7.Mayer EA, Labus JS, Tillisch K, et al. Towards a systems view of IBS. Nat Rev Gastroenterol Hepatol 2015;12:592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furness JB. The enteric nervous system and neurogastroenterology. Nat Rev Gastroenterol Hepatol 2012;9:286–94. [DOI] [PubMed] [Google Scholar]
- 9.Shin A, Preidis GA, Shulman R, et al. The Gut Microbiome in Adult and Pediatric Functional Gastrointestinal Disorders. Clin Gastroenterol Hepatol 2019;17:256–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pittayanon R, Lau JT, Yuan Y, et al. Gut Microbiota in Patients With Irritable Bowel Syndrome-A Systematic Review. Gastroenterology 2019;157:97–108. [DOI] [PubMed] [Google Scholar]
- 11.Ford AC, Moayyedi P, Chey WD, et al. American College of Gastroenterology Monograph on Management of Irritable Bowel Syndrome. Am J Gastroenterol 2018;113:1–18. [DOI] [PubMed] [Google Scholar]
- 12.Klem F, Wadhwa A, Prokop LJ, et al. Prevalence, Risk Factors, and Outcomes of Irritable Bowel Syndrome After Infectious Enteritis: A Systematic Review and Meta-analysis. Gastroenterology 2017;152:1042–1054 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whelan K, Martin LD, Staudacher HM, et al. The low FODMAP diet in the management of irritable bowel syndrome: an evidence-based review of FODMAP restriction, reintroduction and personalisation in clinical practice. J Hum Nutr Diet 2018;31:239–255. [DOI] [PubMed] [Google Scholar]
- 14.Xu D, Chen VL, Steiner CA, et al. Efficacy of Fecal Microbiota Transplantation in Irritable Bowel Syndrome: A Systematic Review and Meta-Analysis. Am J Gastroenterol 2019;114:1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohkusa T, Koido S, Nishikawa Y, et al. Gut Microbiota and Chronic Constipation: A Review and Update. Front Med (Lausanne) 2019;6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carroll IM, Ringel-Kulka T, Siddle JP, et al. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil 2012;24:521–30, e248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durban A, Abellan JJ, Jimenez-Hernandez N, et al. Instability of the faecal microbiota in diarrhoea-predominant irritable bowel syndrome. FEMS Microbiol Ecol 2013;86:581–9. [DOI] [PubMed] [Google Scholar]
- 18.Mars RAT, Yang Y, Ward T, et al. Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome. Cell 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saulnier DM, Riehle K, Mistretta TA, et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 2011;141:1782–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tap J, Derrien M, Tornblom H, et al. Identification of an Intestinal Microbiota Signature Associated With Severity of Irritable Bowel Syndrome. Gastroenterology 2017;152:111–123 e8. [DOI] [PubMed] [Google Scholar]
- 21.Hollister EB, Cain KC, Shulman RJ, et al. Relationships of Microbiome Markers With Extraintestinal, Psychological Distress and Gastrointestinal Symptoms, and Quality of Life in Women With Irritable Bowel Syndrome. J Clin Gastroenterol 2020;54:175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hugerth LW, Andreasson A, Talley NJ, et al. No distinct microbiome signature of irritable bowel syndrome found in a Swedish random population. Gut 2020;69:1076–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deschasaux M, Bouter KE, Prodan A, et al. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat Med 2018;24:1526–1531. [DOI] [PubMed] [Google Scholar]
- 24.He Y, Wu W, Zheng HM, et al. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat Med 2018;24:1532–1535. [DOI] [PubMed] [Google Scholar]
- 25.Poyet M, Groussin M, Gibbons SM, et al. A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research. Nat Med 2019;25:1442–1452. [DOI] [PubMed] [Google Scholar]
- 26.Li G, Yang M, Jin Y, et al. Involvement of shared mucosal-associated microbiota in the duodenum and rectum in diarrhea-predominant irritable bowel syndrome. J Gastroenterol Hepatol 2018;33:1220–1226. [DOI] [PubMed] [Google Scholar]
- 27.Saffouri GB, Shields-Cutler RR, Chen J, et al. Small intestinal microbial dysbiosis underlies symptoms associated with functional gastrointestinal disorders. Nat Commun 2019;10:2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sundin J, Aziz I, Nordlander S, et al. Evidence of altered mucosa-associated and fecal microbiota composition in patients with Irritable Bowel Syndrome. Sci Rep 2020;10:593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu Y, Zhou G, Qin X, et al. The Potential Role of Gut Mycobiome in Irritable Bowel Syndrome. Front Microbiol 2019;10:1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Botschuijver S, Roeselers G, Levin E, et al. Intestinal Fungal Dysbiosis Is Associated With Visceral Hypersensitivity in Patients With Irritable Bowel Syndrome and Rats. Gastroenterology 2017;153:1026–1039. [DOI] [PubMed] [Google Scholar]
- 31.Botschuijver S, Welting O, Levin E, et al. Reversal of visceral hypersensitivity in rat by Menthacarin((R)) , a proprietary combination of essential oils from peppermint and caraway, coincides with mycobiome modulation. Neurogastroenterol Motil 2018;30:e13299. [DOI] [PubMed] [Google Scholar]
- 32.Bennet SM, Keshteli AH, Bercik P, et al. Application of metabolomics to the study of irritable bowel syndrome. Neurogastroenterol Motil 2020;32:e13884. [DOI] [PubMed] [Google Scholar]
- 33.Sun Q, Jia Q, Song L, et al. Alterations in fecal short-chain fatty acids in patients with irritable bowel syndrome: A systematic review and meta-analysis. Medicine (Baltimore) 2019;98:e14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gargari G, Taverniti V, Gardana C, et al. Fecal Clostridiales distribution and short-chain fatty acids reflect bowel habits in irritable bowel syndrome. Environ Microbiol 2018;20:3201–3213. [DOI] [PubMed] [Google Scholar]
- 35.Bhattarai Y, Schmidt BA, Linden DR, et al. Human-derived gut microbiota modulates colonic secretion in mice by regulating 5-HT3 receptor expression via acetate production. Am J Physiol Gastrointest Liver Physiol 2017;313:G80–G87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Litvak Y, Byndloss MX, Baumler AJ. Colonocyte metabolism shapes the gut microbiota. Science 2018;362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Thiel IAM, Botschuijver S, de Jonge WJ, et al. Painful interactions: Microbial compounds and visceral pain. Biochim Biophys Acta Mol Basis Dis 2020;1866:165534. [DOI] [PubMed] [Google Scholar]
- 38.Hurst NR, Kendig DM, Murthy KS, et al. The short chain fatty acids, butyrate and propionate, have differential effects on the motility of the guinea pig colon. Neurogastroenterol Motil 2014;26:1586–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suply E, de Vries P, Soret R, et al. Butyrate enemas enhance both cholinergic and nitrergic phenotype of myenteric neurons and neuromuscular transmission in newborn rat colon. Am J Physiol Gastrointest Liver Physiol 2012;302:G1373–80. [DOI] [PubMed] [Google Scholar]
- 40.Reigstad CS, Salmonson CE, Rainey JF 3rd, et al. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J 2015;29:1395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Strandwitz P. Neurotransmitter modulation by the gut microbiota. Brain Res 2018;1693:128–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lomax AE, Pradhananga S, Sessenwein JL, et al. Bacterial modulation of visceral sensation: mediators and mechanisms. Am J Physiol Gastrointest Liver Physiol 2019;317:G363–G372. [DOI] [PubMed] [Google Scholar]
- 43.Shin A, Camilleri M, Vijayvargiya P, et al. Bowel functions, fecal unconjugated primary and secondary bile acids, and colonic transit in patients with irritable bowel syndrome. Clin Gastroenterol Hepatol 2013;11:1270–1275 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Camilleri M. Peripheral mechanisms in irritable bowel syndrome. N Engl J Med 2012;367:1626–35. [DOI] [PubMed] [Google Scholar]
- 45.Duboc H, Rainteau D, Rajca S, et al. Increase in fecal primary bile acids and dysbiosis in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil 2012;24:513–20, e246-7. [DOI] [PubMed] [Google Scholar]
- 46.Jeffery IB, Das A, O'Herlihy E, et al. Differences in Fecal Microbiomes and Metabolomes of People With vs Without Irritable Bowel Syndrome and Bile Acid Malabsorption. Gastroenterology 2020;158:1016–1028 e8. [DOI] [PubMed] [Google Scholar]
- 47.Zhao L, Yang W, Chen Y, et al. A Clostridia-rich microbiota enhances bile acid excretion in diarrhea-predominant irritable bowel syndrome. J Clin Invest 2020;130:438–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pimentel M, Lembo A, Chey WD, et al. Rifaximin therapy for patients with irritable bowel syndrome without constipation. N Engl J Med 2011;364:22–32. [DOI] [PubMed] [Google Scholar]
- 49.Preidis GA, Weizman AV, Kashyap PC, et al. AGA Technical Review on the Role of Probiotics in the Management of Gastrointestinal Disorders. Gastroenterology 2020;159:708–738 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanson KS, Gibson GR, Hutkins R, et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat Rev Gastroenterol Hepatol 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Llewellyn SR, Britton GJ, Contijoch EJ, et al. Interactions Between Diet and the Intestinal Microbiota Alter Intestinal Permeability and Colitis Severity in Mice. Gastroenterology 2018;154:1037–1046 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gibson PR, Barrett JS, Muir JG. Functional bowel symptoms and diet. Intern Med J 2013;43:1067–74. [DOI] [PubMed] [Google Scholar]
- 53.Ebina T, Takahashi K, Homma M, et al. Alkaline phosphatase activity of HeLa-S3 cells persistently infected with hemadsorption type 2 virus. Virology 1969;39:597–9. [DOI] [PubMed] [Google Scholar]
- 54.Walter J, Maldonado-Gomez MX, Martinez I. To engraft or not to engraft: an ecological framework for gut microbiome modulation with live microbes. Curr Opin Biotechnol 2018;49:129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fetissov SO. Role of the gut microbiota in host appetite control: bacterial growth to animal feeding behaviour. Nat Rev Endocrinol 2017;13:11–25. [DOI] [PubMed] [Google Scholar]
- 56.Bunyavanich S, Shen N, Grishin A, et al. Early-life gut microbiome composition and milk allergy resolution. J Allergy Clin Immunol 2016;138:1122–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Savage JH, Lee-Sarwar KA, Sordillo J, et al. A prospective microbiome-wide association study of food sensitization and food allergy in early childhood. Allergy 2018;73:145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zar S, Kumar D, Benson MJ. Food hypersensitivity and irritable bowel syndrome. Aliment Pharmacol Ther 2001;15:439–49. [DOI] [PubMed] [Google Scholar]
- 59.Monsbakken KW, Vandvik PO, Farup PG. Perceived food intolerance in subjects with irritable bowel syndrome-- etiology, prevalence and consequences. Eur J Clin Nutr 2006;60:667–72. [DOI] [PubMed] [Google Scholar]
- 60.Staudacher HM, Lomer MC, Anderson JL, et al. Fermentable carbohydrate restriction reduces luminal bifidobacteria and gastrointestinal symptoms in patients with irritable bowel syndrome. J Nutr 2012;142:1510–8. [DOI] [PubMed] [Google Scholar]
- 61.Ward RE, Ninonuevo M, Mills DA, et al. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol Nutr Food Res 2007;51:1398–405. [DOI] [PubMed] [Google Scholar]
- 62.Pastell H, Westermann P, Meyer AS, et al. In vitro fermentation of arabinoxylan-derived carbohydrates by bifidobacteria and mixed fecal microbiota. J Agric Food Chem 2009;57:8598–606. [DOI] [PubMed] [Google Scholar]
- 63.Makelainen H, Forssten S, Saarinen M, et al. Xylo-oligosaccharides enhance the growth of bifidobacteria and Bifidobacterium lactis in a simulated colon model. Benef Microbes 2010;1:81–91. [DOI] [PubMed] [Google Scholar]
- 64.Silk DB, Davis A, Vulevic J, et al. Clinical trial: the effects of a trans-galactooligosaccharide prebiotic on faecal microbiota and symptoms in irritable bowel syndrome. Aliment Pharmacol Ther 2009;29:508–18. [DOI] [PubMed] [Google Scholar]
- 65.Eigenmann PA, Caubet JC, Zamora SA. Continuing food-avoidance diets after negative food challenges. Pediatr Allergy Immunol 2006;17:601–5. [DOI] [PubMed] [Google Scholar]
- 66.Agrawal A, Whorwell PJ. Review article: abdominal bloating and distension in functional gastrointestinal disorders--epidemiology and exploration of possible mechanisms. Aliment Pharmacol Ther 2008;27:2–10. [DOI] [PubMed] [Google Scholar]
- 67.Ploger S, Stumpff F, Penner GB, et al. Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann N Y Acad Sci 2012;1258:52–9. [DOI] [PubMed] [Google Scholar]
- 68.Fukumoto S, Tatewaki M, Yamada T, et al. Short-chain fatty acids stimulate colonic transit via intraluminal 5-HT release in rats. Am J Physiol Regul Integr Comp Physiol 2003;284:R1269–76. [DOI] [PubMed] [Google Scholar]
- 69.Triantafyllou K, Chang C, Pimentel M. Methanogens, methane and gastrointestinal motility. J Neurogastroenterol Motil 2014;20:31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hillemeier C. An overview of the effects of dietary fiber on gastrointestinal transit. Pediatrics 1995;96:997–9. [PubMed] [Google Scholar]
- 71.Eswaran S, Muir J, Chey WD. Fiber and functional gastrointestinal disorders. Am J Gastroenterol 2013;108:718–27. [DOI] [PubMed] [Google Scholar]
- 72.Muller M, Canfora EE, Blaak EE. Gastrointestinal Transit Time, Glucose Homeostasis and Metabolic Health: Modulation by Dietary Fibers. Nutrients 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kashyap PC, Marcobal A, Ursell LK, et al. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology 2013;144:967–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pianka ER. On r- and K-Selection. The American Naturalist 1970;104:592–597. [Google Scholar]
- 75.Roager HM, Hansen LB, Bahl MI, et al. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat Microbiol 2016;1:16093. [DOI] [PubMed] [Google Scholar]
- 76.Shankar V, Reo NV, Paliy O. Simultaneous fecal microbial and metabolite profiling enables accurate classification of pediatric irritable bowel syndrome. Microbiome 2015;3:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Banasiewicz T, Krokowicz L, Stojcev Z, et al. Microencapsulated sodium butyrate reduces the frequency of abdominal pain in patients with irritable bowel syndrome. Colorectal Dis 2013;15:204–9. [DOI] [PubMed] [Google Scholar]
- 78.Bhattarai Y, Williams BB, Battaglioli EJ, et al. Gut Microbiota-Produced Tryptamine Activates an Epithelial G-Protein-Coupled Receptor to Increase Colonic Secretion. Cell Host Microbe 2018;23:775–785 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee JS, Wang RX, Goldberg MS, et al. Microbiota-Sourced Purines Support Wound Healing and Mucous Barrier Function. iScience 2020;23:101226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee JS, Wang RX, Alexeev EE, et al. Hypoxanthine is a checkpoint stress metabolite in colonic epithelial energy modulation and barrier function. J Biol Chem 2018;293:6039–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao L, Zhang F, Ding X, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018;359:1151–1156. [DOI] [PubMed] [Google Scholar]
- 82.Costello EK, Stagaman K, Dethlefsen L, et al. The application of ecological theory toward an understanding of the human microbiome. Science 2012;336:1255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lemon KP, Armitage GC, Relman DA, et al. Microbiota-targeted therapies: an ecological perspective. Sci Transl Med 2012;4:137rv5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Darcy JL, Washburne AD, Robeson MS, et al. A phylogenetic model for the recruitment of species into microbial communities and application to studies of the human microbiome. ISME J 2020;14:1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shepherd ES, DeLoache WC, Pruss KM, et al. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature 2018;557:434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Charbonneau MR, Isabella VM, Li N, et al. Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat Commun 2020;11:1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wong AC, Levy M. New Approaches to Microbiome-Based Therapies. mSystems 2019;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roberts AB, Gu X, Buffa JA, et al. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nat Med 2018;24:1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pozuelo M, Panda S, Santiago A, et al. Reduction of butyrate- and methane-producing microorganisms in patients with Irritable Bowel Syndrome. Sci Rep 2015;5:12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hollister EB, Oezguen N, Chumpitazi BP, et al. Leveraging Human Microbiome Features to Diagnose and Stratify Children with Irritable Bowel Syndrome. J Mol Diagn 2019;21:449–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maharshak N, Ringel Y, Katibian D, et al. Fecal and Mucosa-Associated Intestinal Microbiota in Patients with Diarrhea-Predominant Irritable Bowel Syndrome. Dig Dis Sci 2018;63:1890–1899. [DOI] [PubMed] [Google Scholar]
- 92.Shin SP, Choi YM, Kim WH, et al. A double blind, placebo-controlled, randomized clinical trial that breast milk derived-Lactobacillus gasseri BNR17 mitigated diarrhea-dominant irritable bowel syndrome. J Clin Biochem Nutr 2018;62:179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Acosta A, Camilleri M, Shin A, et al. Effects of Rifaximin on Transit, Permeability, Fecal Microbiome, and Organic Acid Excretion in Irritable Bowel Syndrome. Clin Transl Gastroenterol 2016;7:e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dior M, Delagreverie H, Duboc H, et al. Interplay between bile acid metabolism and microbiota in irritable bowel syndrome. Neurogastroenterol Motil 2016;28:1330–40. [DOI] [PubMed] [Google Scholar]
- 95.Le Neve B, Brazeilles R, Derrien M, et al. Lactulose Challenge Determines Visceral Sensitivity and Severity of Symptoms in Patients With Irritable Bowel Syndrome. Clin Gastroenterol Hepatol 2016;14:226–33 e1-3. [DOI] [PubMed] [Google Scholar]
- 96.Chumpitazi BP, Hollister EB, Oezguen N, et al. Gut microbiota influences low fermentable substrate diet efficacy in children with irritable bowel syndrome. Gut Microbes 2014;5:165–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jeffery IB, O'Toole PW, Ohman L, et al. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut 2012;61:997–1006. [DOI] [PubMed] [Google Scholar]
- 98.Labus JS, Hollister EB, Jacobs J, et al. Differences in gut microbial composition correlate with regional brain volumes in irritable bowel syndrome. Microbiome 2017;5:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu Y, Zhang L, Wang X, et al. Similar Fecal Microbiota Signatures in Patients With Diarrhea-Predominant Irritable Bowel Syndrome and Patients With Depression. Clin Gastroenterol Hepatol 2016;14:1602–1611 e5. [DOI] [PubMed] [Google Scholar]
- 100.Azpiroz F, Dubray C, Bernalier-Donadille A, et al. Effects of scFOS on the composition of fecal microbiota and anxiety in patients with irritable bowel syndrome: a randomized, double blind, placebo controlled study. Neurogastroenterol Motil 2017;29. [DOI] [PubMed] [Google Scholar]
- 101.Le Gall G, Noor SO, Ridgway K, et al. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J Proteome Res 2011;10:4208–18. [DOI] [PubMed] [Google Scholar]
- 102.Heitkemper MM, Cain KC, Shulman RJ, et al. Stool and urine trefoil factor 3 levels: associations with symptoms, intestinal permeability, and microbial diversity in irritable bowel syndrome. Benef Microbes 2018;9:345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bednarska O, Walter SA, Casado-Bedmar M, et al. Vasoactive Intestinal Polypeptide and Mast Cells Regulate Increased Passage of Colonic Bacteria in Patients With Irritable Bowel Syndrome. Gastroenterology 2017;153:948–960 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Valentin N, Camilleri M, Carlson P, et al. Potential mechanisms of effects of serum-derived bovine immunoglobulin/protein isolate therapy in patients with diarrhea-predominant irritable bowel syndrome. Physiol Rep 2017;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ko SJ, Han G, Kim SK, et al. Effect of korean herbal medicine combined with a probiotic mixture on diarrhea-dominant irritable bowel syndrome: a double-blind, randomized, placebo-controlled trial. Evid Based Complement Alternat Med 2013;2013:824605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shulman RJ, Hollister EB, Cain K, et al. Psyllium Fiber Reduces Abdominal Pain in Children With Irritable Bowel Syndrome in a Randomized, Double-Blind Trial. Clin Gastroenterol Hepatol 2017;15:712–719 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Compare D, Rocco A, Coccoli P, et al. Lactobacillus casei DG and its postbiotic reduce the inflammatory mucosal response: an ex-vivo organ culture model of post-infectious irritable bowel syndrome. BMC Gastroenterol 2017;17:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hustoft TN, Hausken T, Ystad SO, et al. Effects of varying dietary content of fermentable short-chain carbohydrates on symptoms, fecal microenvironment, and cytokine profiles in patients with irritable bowel syndrome. Neurogastroenterol Motil 2017;29. [DOI] [PubMed] [Google Scholar]
- 109.McIntosh K, Reed DE, Schneider T, et al. FODMAPs alter symptoms and the metabolome of patients with IBS: a randomised controlled trial. Gut 2017;66:1241–1251. [DOI] [PubMed] [Google Scholar]
- 110.Sundin J, Rangel I, Repsilber D, et al. Cytokine Response after Stimulation with Key Commensal Bacteria Differ in Post-Infectious Irritable Bowel Syndrome (PI-IBS) Patients Compared to Healthy Controls. PLoS One 2015;10:e0134836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sundin J, Rangel I, Fuentes S, et al. Altered faecal and mucosal microbial composition in post-infectious irritable bowel syndrome patients correlates with mucosal lymphocyte phenotypes and psychological distress. Aliment Pharmacol Ther 2015;41:342–51. [DOI] [PubMed] [Google Scholar]
- 112.Pinto-Sanchez MI, Hall GB, Ghajar K, et al. Probiotic Bifidobacterium longum NCC3001 Reduces Depression Scores and Alters Brain Activity: A Pilot Study in Patients With Irritable Bowel Syndrome. Gastroenterology 2017;153:448–459 e8. [DOI] [PubMed] [Google Scholar]
- 113.Parthasarathy G, Chen J, Chia N, et al. Reproducibility of assessing fecal microbiota in chronic constipation. Neurogastroenterol Motil 2017;29:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tian H, Ge X, Nie Y, et al. Fecal microbiota transplantation in patients with slow-transit constipation: A randomized, clinical trial. PLoS One 2017;12:e0171308. [DOI] [PMC free article] [PubMed] [Google Scholar]