Summary
The performance of deep learning in disease detection from high-quality clinical images is identical to and even greater than that of human doctors. However, in low-quality images, deep learning performs poorly. Whether human doctors also have poor performance in low-quality images is unknown. Here, we compared the performance of deep learning systems with that of cornea specialists in detecting corneal diseases from low-quality slit lamp images. The results showed that the cornea specialists performed better than our previously established deep learning system (PEDLS) trained on only high-quality images. The performance of the system trained on both high- and low-quality images was superior to that of the PEDLS while inferior to that of a senior corneal specialist. This study highlights that cornea specialists perform better in low-quality images than the system trained on high-quality images. Adding low-quality images with sufficient diagnostic certainty to the training set can reduce this performance gap.
Subject areas: Ocular surface, Ophthalmology, Artificial intelligence
Graphical abstract
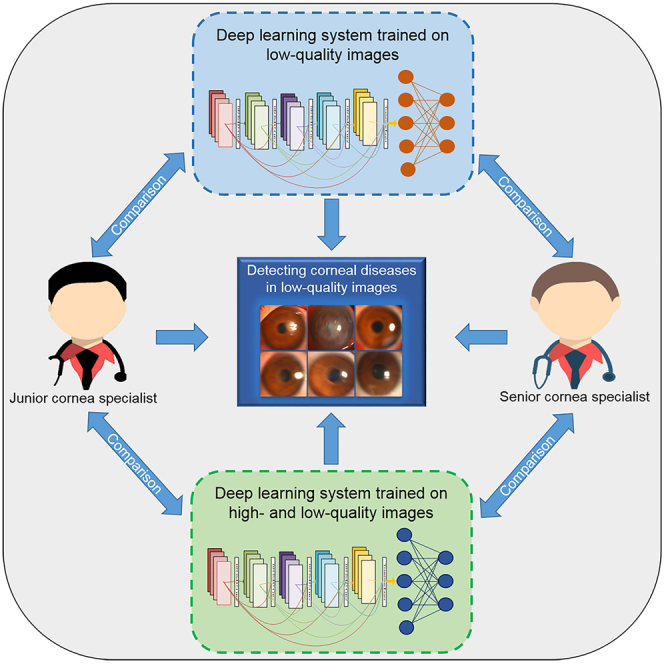
Highlights
-
•
Deep learning performs poorly in low-quality images for detecting corneal diseases
-
•
Corneal specialists perform better than the PEDLS in low-quality images
-
•
The performance of the NDLS is better than that of the PEDLS in low-quality images
-
•
Adding low-quality images to the training set can improve the system's performance
Ocular surface; Ophthalmology; Artificial intelligence
Introduction
Recently deep learning has attained remarkable performance in disease screening and diagnosis (Cheung et al., 2021; Hosny and Aerts, 2019; Li et al., 2020a, Li et al., 2020b, Li et al., 2020c, Li et al., 2020d; Matheny et al., 2019; Zhou et al., 2021). The performance of deep learning is comparable with and even superior to that of human doctors in many clinical image analyses (Li et al., 2021a, Li et al., 2021b, Li et al., 2021c, Li et al., 2021d Li et al., 2020a, Li et al., 2020b, Li et al., 2020c, Li et al., 2020d; Li et al., 2019; Ting et al., 2017; Xie et al., 2020; Zhang et al., 2020). For example, the accuracy of a deep learning system in distinguishing coronavirus pneumonia from computed tomography images reached the level of senior radiologists (87.5% versus 84.5%; p > 0.05) and exceeded the level of junior radiologists (87.5% versus 65.6%; p < .05) (Zhang et al., 2020). In discerning corneas with contraindications for refractive surgery based on corneal tomographic images, comparable accuracy was observed between a deep learning system and refractive surgeons (95% versus 92.8; p = 0.72) (Xie et al., 2020). Our previous study also demonstrated that a senior cornea specialist and a deep learning system had similar performance (accuracy: 96.7% versus 97.3%; p = 0.50) in screening for keratitis from slit lamp images (Li et al., 2021a, Li et al., 2021b, Li et al., 2021c, Liu et al., 2020).
For facilitating feature extraction, most studies only utilize high-quality images to establish deep learning systems (Cheung et al., 2021; Esteva et al., 2017; Li et al., 2020a, Li et al., 2020b, Li et al., 2020c, Li et al., 2020d; Li et al., 2021a, Li et al., 2021b, Li et al., 2021c, Liu et al., 2020; Luo et al., 2019; Xie et al., 2020; Zhang et al., 2020). Although deep learning acquires good performance in high-quality images, its performance was poor in low-quality images, which were inevitable in real clinical scenarios due to many factors such as patient noncompliance, hardware imperfections, and operator errors (Li et al., 2020a, Li et al., 2020b, Li et al., 2020c, Li et al., 2020d; Li et al., 2021a, Li et al., 2021b, Li et al., 2021c, Liu et al., 2020; Trucco et al., 2013). For instance, in screening for lattice degeneration/retinal breaks, glaucomatous optic neuropathy, and retinal exudation/drusen, deep learning systems achieved area under the receiver operating characteristic curves (AUCs) of 0.990, 0.995, and 0.982 in high-quality fundus images, respectively, whereas achieved AUCs of 0.635, 0.853, and 0.779 in low-quality fundus images, respectively (Li et al., 2020a, Li et al., 2020b, Li et al., 2020c, Li et al., 2020d).
To date, whether human doctors also perform poorly in low-quality images is not well investigated. If the performance of human doctors in low-quality images is better than deep learning, this result exposes a vulnerability of deep learning systems, and further studies are needed to build more robust deep learning systems. If the performance of the human doctors was similar to that of deep learning systems in low-quality images, it indicates that the detection of diseases from low-quality images may be inherently difficult.
To explore this issue, our study aimed to compare the performance of a previously established deep learning system (PEDLS) (Li et al., 2021a, Li et al., 2021b, Li et al., 2021c, Liu et al., 2020) with that of cornea specialists in low-quality slit lamp images for classifying keratitis, other corneal abnormalities, and normal cornea. In addition, this study investigated whether the performance of the deep learning system in low-quality images would be improved by training a deep learning network with both high- and low-quality images.
Results
Data characteristics
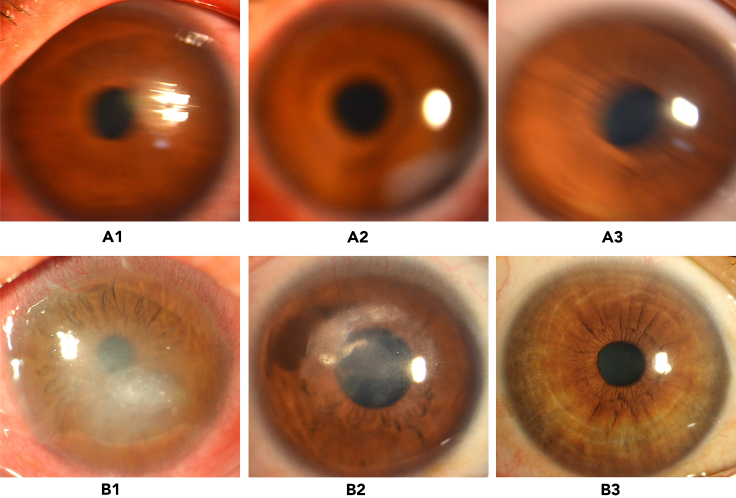
In total, 12,411 high-quality images (keratitis = 5,586; other corneal abnormalities = 2,293; and normal cornea = 4,532) and 1,705 low-quality images (keratitis = 628; other corneal abnormalities = 516; and normal cornea = 561) were used in this study. Representative examples of low- and high-quality images are shown in Figure 1. The detailed information on the development dataset and external test dataset is described in Table 1.
Figure 1.
Representative examples of low- and high-quality slit lamp images
(A) Low-quality images: (A1) keratitis; (A2) other corneal abnormalities; (A3) normal cornea.
(B) High-quality images: (B1) keratitis; (B2) other corneal abnormalities; (B3) normal cornea.
Table 1.
Summary of datasets
| Item | Development dataset |
External test dataset | ||
|---|---|---|---|---|
| Training dataset | Validation dataset | Internal test dataset | ||
| Keratitis | ||||
| No. of high-quality imagesa | 2,185/5,505 (39.7) | 511/1,257 (40.7) | 483/1,208 (40.0) | 2,407/6,146 (39.2) |
| No. of low-quality imagesa | 367/5,505 (6.7) | 75/1,257 (6.0) | 81/1,208 (6.7) | 105/6,146 (1.7) |
| Other corneal abnormalities | ||||
| No. of high-quality imagesa | 585/5,505 (10.6) | 136/1,257 (10.8) | 130/1,208 (10.8) | 1,442/6,146 (23.5) |
| No. of low-quality imagesa | 299/5,505 (5.4) | 61/1,257 (4.9) | 69/1,208 (5.7) | 87/6,146 (1.4) |
| Normal cornea | ||||
| No. of high-quality imagesa | 1756/5,505 (31.9) | 408/1,257 (32.5) | 373/1,208 (30.9) | 1995/6,146 (32.5) |
| No. of low-quality imagesa | 313/5,505 (5.9) | 66/1,257 (5.3) | 72/1,208 (6.0) | 110/6,146 (1.8) |
Data are no. of images/total no. (%) unless otherwise indicated.
Comparison of the PEDLS against corneal specialists in low-quality images
For the classification of keratitis, other corneal abnormalities, and normal cornea, the cornea specialist with 3 years of experience achieved accuracies of 82.8% (78.5–87.0), 69.9% (64.7–75.0), and 81.8% (77.4–86.1), respectively, and the senior cornea specialist with 7 years of experience achieved accuracies of 93.7% (91.0–96.4), 93.0% (90.2–95.9), and 94.7% (92.2–97.2), respectively, whereas the PEDLS achieved accuracies of 69.9% (64.7–75.0), 60.6% (55.1–66.1), and 70.9% (65.7–76.0), respectively, in low-quality images from the external test dataset. The overall performance of the PEDLS is lower than that of the cornea specialists (p < .05) (Table 2).
Table 2.
Comparison of deep learning systems with cornea specialists in low-quality images
| One-versus-rest classification | PEDLS | NDLS | Cornea specialist A | Cornea specialist B | P1 | P2 | P3 | P4 | P5 |
|---|---|---|---|---|---|---|---|---|---|
| Keratitis versus others + normal | |||||||||
| Sensitivity (95% CI) | 70.5% (61.8–79.2) | 77.1% (69.1–85.2) | 72.4% (63.8–80.9) | 92.4% (87.3–97.5) | 0.815 | <0.001 | 0.383 | 0.001 | 0.016 |
| Specificity (95% CI) | 69.5% (63.1–76.0) | 90.9% (86.8–94.9) | 88.3% (83.8–92.8) | 94.4% (91.2–97.6) | <0.001 | <0.001 | 0.487 | 0.248 | <0.001 |
| Accuracy (95% CI) | 69.9% (64.7–75.0) | 86.1% (82.2–90.0) | 82.8% (78.5–87.0) | 93.7% (91.0–96.4) | <0.001 | <0.001 | 0.220 | 0.001 | <0.001 |
| Others versus keratitis + normal | |||||||||
| Sensitivity (95% CI) | 46.0% (35.5–56.4) | 80.5% (72.1–88.8) | 59.8% (49.5–70.1) | 90.8% (84.7–96.9) | 0.065 | <0.001 | 0.011 | 0.093 | <0.001 |
| Specificity (95% CI) | 66.5% (60.2–72.8) | 87.4% (83.0–91.9) | 74.0% (68.1–79.8) | 94.0% (90.8–97.1) | 0.073 | <0.001 | <0.001 | 0.034 | <0.001 |
| Accuracy (95% CI) | 60.6% (55.1–66.1) | 85.4% (81.5–89.4) | 69.9% (64.7–75.0) | 93.0% (90.2–95.9) | 0.009 | <0.001 | <0.001 | 0.004 | <0.001 |
| Normal versus keratitis + others | |||||||||
| Sensitivity (95% CI) | 35.5% (36.5–44.4) | 74.5% (66.4–82.7) | 68.2% (59.5–76.9) | 89.1% (83.3–94.9) | <0.001 | <0.001 | 0.360 | 0.009 | <0.001 |
| Specificity (95% CI) | 91.1% (87.1–95.2) | 87.5% (82.8–92.2) | 89.6% (85.3–93.9) | 97.9% (95.9–99.9) | 0.728 | 0.001 | 0.636 | <0.001 | 0.092 |
| Accuracy (95% CI) | 70.9% (65.7–76.0) | 82.8% (78.5–87.0) | 81.8% (77.4–86.1) | 94.7% (92.2–97.2) | 0.001 | <0.001 | 0.826 | <0.001 | <0.001 |
PEDLS, previously established deep learning system; NDLS, new deep learning system; CI, confidence interval. “Others” denotes other corneal abnormalities. “Normal” denotes normal cornea. P1 indicates the p value calculated between the PEDLS and cornea specialist A using the McNemar test. P2 indicates the p value calculated between the PEDLS and cornea specialist B using the McNemar test. P3 indicates the p value calculated between the NDLS and cornea specialist A using the McNemar test. P4 indicates the p value calculated between the NDLS and cornea specialist B using the McNemar test. P5 indicates the p value calculated between the PEDLS and NDLS using the McNemar test. Cornea specialist A has 3 years of clinical experience. Cornea specialist B has 7 years of clinical experience.
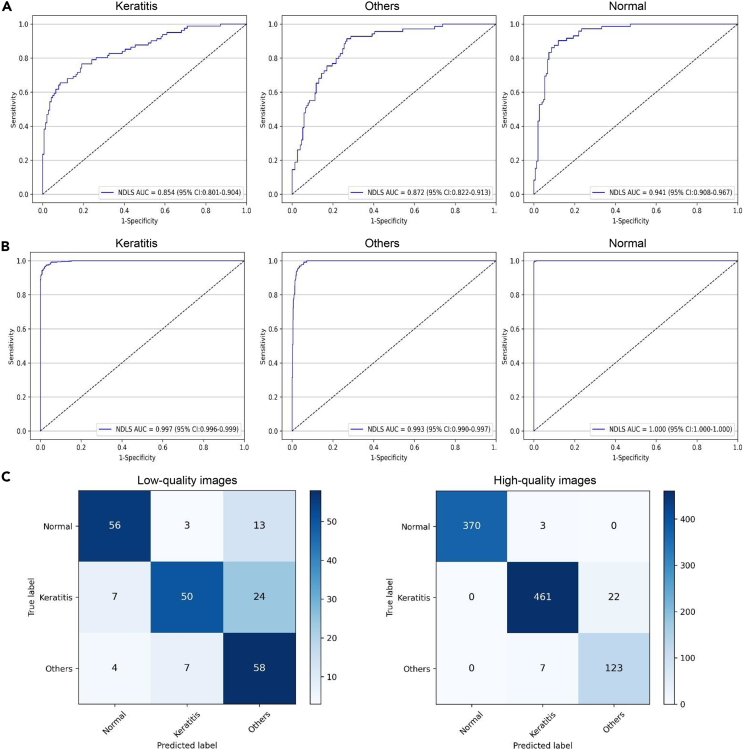
Performance of the NDLS in the internal and external test datasets
In the internal text dataset, the NDLS achieved AUCs of 0.854 (95% confidence interval [CI], 0.801 to 0.904), 0.872 (95% CI, 0.822 to 0.913), and 0.941 (95% CI, 0.908 to 0.967), respectively, for classifying keratitis, other corneal abnormalities, and normal cornea from low-quality images and achieved AUCs of 0.997 (95% CI, 0.996 to 0.999), 0.993 (95% CI, 0.990 to 0.997), and 1.000 (95% CI, 1.000 to 1.000), respectively, for classifying keratitis, other corneal abnormalities, and normal cornea from high-quality images (Figure 2).
Figure 2.
Performance of the NDLS in the internal test dataset
(A) Receiver operating characteristic curves (ROCs) of the NDLS for discerning keratitis, other corneal abnormalities, and normal cornea in low-quality images.
(B) ROCs of the NDLS for discerning keratitis, other corneal abnormalities, and normal cornea in high-quality images.
(C) Confusion matrixes of the NDLS in low- and high-quality images. NDLS, new deep learning system. “Others” denotes other corneal abnormalities. “Normal” denotes normal cornea.
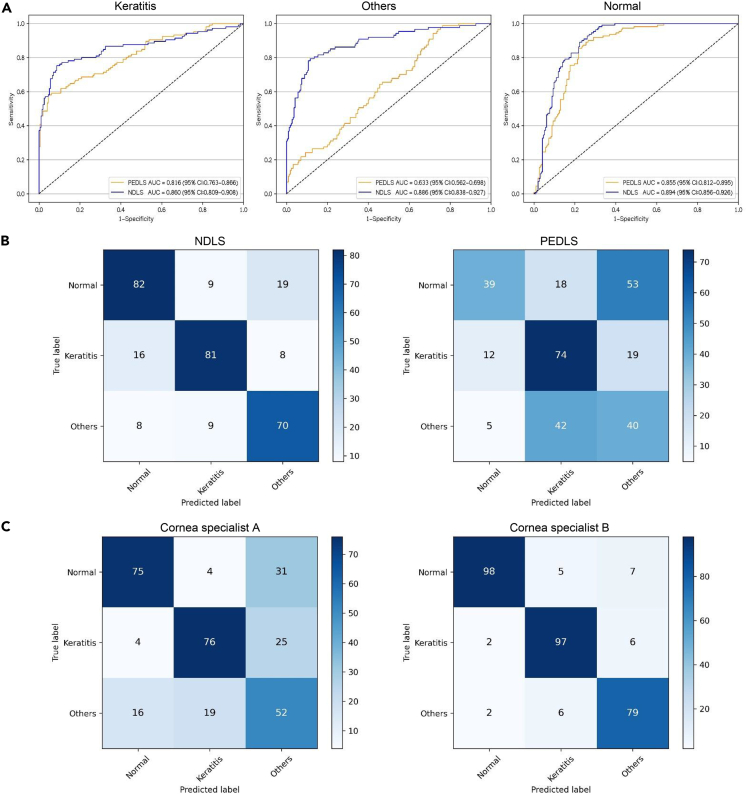
In the external text dataset, the AUCs of NDLS were 0.860 (95% CI, 0.809 to 0.908), 0.886 (95% CI, 0.838 to 0.927), and 0.894 (95% CI, 0.856 to 0.926), respectively, for classifying keratitis, other corneal abnormalities, and normal cornea from low-quality images (Figure 3) and were 0.988 (95% CI, 0.986 to 0.991), 0.976 (95% CI, 0.972 to 0.979), and 0.990 (95% CI, 0.988 to 0.992), respectively, for classifying keratitis, other corneal abnormalities, and normal cornea from high-quality images (Figure S1).
Figure 3.
Performance of the NDLS, PEDLS, and cornea specialists in low-quality images from the external test dataset
(A) Receiver operating characteristic curves of the NDLS and PEDLS for discerning keratitis, other corneal abnormalities, and normal cornea in low-quality images.
(B) Confusion matrixes of the NDLS and PEDLS in low-quality images.
(C) Confusion matrixes of the cornea specialists in low-quality images. NDLS, new deep learning system. PEDLS, previously established deep learning system.
Further information including accuracies, sensitivities, and specificities of the NDLS in the internal and external test datasets is displayed in Table 3.
Table 3.
Performance of the NDLS in the internal and external test datasets
| One-versus-rest classification | Internal test dataset |
External test dataset |
||
|---|---|---|---|---|
| High-quality images | Low-quality images | High-quality images | Low-quality images | |
| Keratitis versus others + normal | ||||
| Sensitivity (95% CI) | 95.4% (93.6–97.3) | 74.1% (64.5–83.6) | 90.7% (89.6–91.9) | 77.1% (69.1–85.2) |
| Specificity (95% CI) | 98.0% (96.8–99.2) | 92.9% (88.7–97.1) | 97.8% (97.3–98.3) | 90.9% (86.8–94.9) |
| Accuracy (95% CI) | 96.8% (95.6–97.9) | 86.0% (81.5–90.6) | 94.9% (94.3–95.4) | 86.1% (82.2–90.0) |
| Others versus keratitis + normal | ||||
| Sensitivity (95% CI) | 94.6% (90.7–98.5) | 84.1% (75.4–92.7) | 90.7% (89.2–92.2) | 80.5% (72.1–88.8) |
| Specificity (95% CI) | 97.4% (96.4–98.5) | 83.0% (77.1–89.0) | 93.6% (92.9–94.4) | 87.4% (83.0–91.9) |
| Accuracy (95% CI) | 97.1% (96.0–98.1) | 83.3% (78.4–88.2) | 92.9% (92.3–93.6) | 85.4% (81.5–89.4) |
| Normal versus keratitis + others | ||||
| Sensitivity (95% CI) | 99.2% (98.3–100) | 77.8% (68.2–87.4) | 94.0% (92.9–95.0) | 74.5% (66.4–82.7) |
| Specificity (95% CI) | 100% (100-100) | 92.0% (87.7–96.3) | 96.9% (96.3–97.4) | 87.5% (82.8–92.2) |
| Accuracy (95% CI) | 99.7% (99.4–100) | 87.4% (83.0–91.8) | 95.9% (95.4–96.4) | 82.8% (78.5–87.0) |
“Others” denotes other corneal abnormalities. “Normal” denotes normal cornea. NDLS, new deep learning system; CI, confidence interval.
Comparison of NDLS with PEDLS and cornea specialists in low-quality images
In low-quality images from the external test dataset, the overall performance of the NDLS was greater than that of the PEDLS for detecting keratitis, other corneal abnormalities, and normal cornea (p < .05) (Table 2). The corresponding ROCs and confusion matrices of the NDLS and PEDLS are described in Figures 3A and 3B. The t-SNE technique showed that the features of each category learned by the NDLS were more separable than those of the PEDLS (Figure S2A).
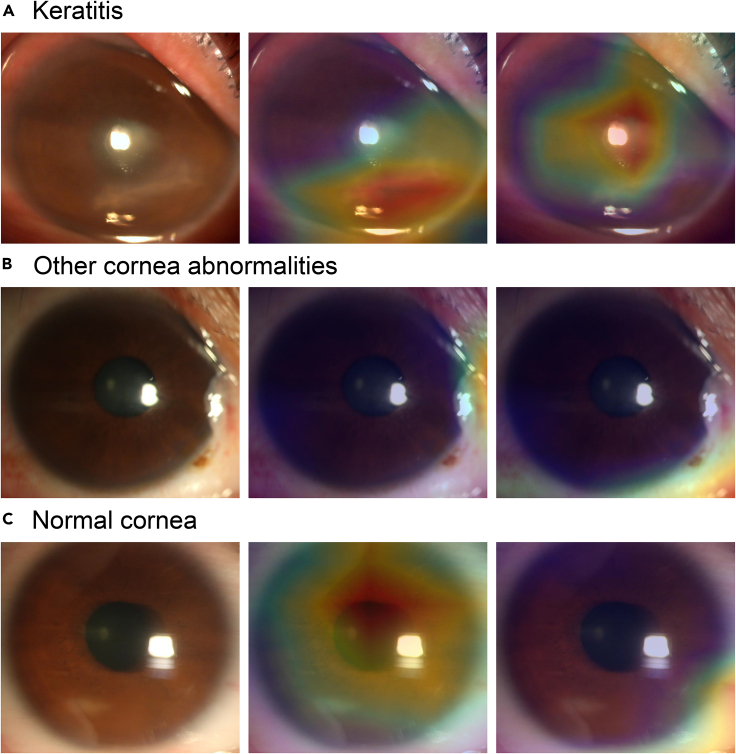
Representative examples of the heatmaps of the NDLS and PEDLS in low-quality images are shown in Figure 4. For abnormal cornea findings (keratitis and other corneal abnormalities), the heatmaps of the NDLS effectively displayed the highlighted visualization on the lesion regions, whereas the heatmaps of the PEDLS highlighted the other regions. For normal cornea, the heatmaps of the NDLS highlighted the region of the cornea, whereas the heatmaps of the PEDLS showed the highlighted visualization on the region of the conjunctiva.
Figure 4.
Representative examples of the heatmaps of the NDLS and PEDLS in low-quality images
(A) Keratitis.
(B) Other corneal abnormalities.
(C) Normal cornea. The images are shown in the order of original images (left), corresponding heatmaps of the NDLS (middle), and corresponding heatmaps of the PEDLS (right) for each category. NDLS, new deep learning system. PEDLS, previously established deep learning system.
The accuracies of the NDLS in detecting keratitis and normal cornea were comparable to that of the cornea specialist with 3 years of experience (p > 0.05), whereas the accuracy of the NDLS in detecting other corneal abnormalities is greater than that of the cornea specialist with 3 years of experience (p < .05) in low-quality images from the external test dataset (Table 2). The overall performance of the NDLS was lower than that of the cornea specialist with 7 years of experience (p < .05) for classifying keratitis, other corneal abnormalities, and normal cornea in low-quality images from the external test dataset (Table 2).
Comparison between NDLS and PEDLS in high-quality images
In high-quality images from the external test dataset, the overall performance of the NDLS was lower than that of the PEDLS for detecting keratitis, other corneal abnormalities, and normal cornea (p < .05) (Table S1). The corresponding ROCs and confusion matrices of the NDLS and PEDLS are presented in Figure S1. The t-SNE technique showed that the features of each category learned by the NDLS in high-quality images were less separable than those of the PEDLS (Figure S2B).
Discussion
In this study, we conducted performance comparisons between a deep learning system and cornea specialists for classifying keratitis, other corneal abnormalities, and normal cornea in low-quality images. We found that the performance of cornea specialists in low-quality images greatly exceeded that of the PEDLS trained on high-quality images. This result indicates that the PEDLS is not so robust as the cornea specialists in detecting abnormal cornea findings from low-quality slit lamp images. The reason is that the deep learning system might perceive the noise in low-quality images as part of the object and its texture while human experts often treat the noise as a layer in front of the image (Geirhos et al., 2017). Besides, human experts, through experience and evolution, were exposed to some low-quality images and thus have an advantage over the PEDLS (Geirhos et al., 2017).
To improve the resiliency of a deep learning system in real-world settings, we trained the NDLS using both high- and low-quality images. The NDLS achieved higher accuracies than those of the PEDLS in identifying keratitis (86.1% versus 69.9%), other corneal abnormalities (85.4% versus 60.6%), and normal cornea (82.8% versus 70.9%) from low-quality images. It demonstrates that the performance of the deep learning network on low-quality images could be improved if low-quality images with sufficient diagnostic certainty are added to the training set. In addition, heatmaps were generated to interpret the decision-making rationales of the PEDLS and NDLS in low-quality images. As shown in Figure 4, the heatmaps of the NDLS are more interpretable than those of the PEDLS, and this further substantiates the effectiveness of the NDLS.
Performance comparison between the NDLS and cornea specialists in identifying keratitis, other corneal abnormalities, and normal cornea from low-quality images was conducted in the present study. The results showed that the performance of the NDLS in low-quality images was comparable with that of the cornea specialist with 3-year clinical experience. However, the performance of the cornea specialist with 7-year clinical experience still exceeded that of the NDLS in low-quality images. A possible explanation is that the senior cornea specialist may have more prior experience in analyzing low-quality images and therefore has better performance. Increasing the sample size of low-quality images in a training set could potentially reduce a performance gap between the deep learning system and the senior cornea specialist in low-quality images.
Although the NDLS has higher performance than the PEDLS in low-quality images, the NDLS performance in high-quality images was slightly lower than that of the PEDLS. This illustrates that adding low-quality images to the training set brings the deep learning system some noise, which has negative influence on the system in screening for abnormal cornea findings from high-quality images. Further research is required to find an approach that would not decrease the performance of a deep learning system in high-quality images while increasing its performance in low-quality images.
In summary, this study shows that cornea specialists achieve higher accuracies than those of the PEDLS in detecting keratitis, other corneal abnormalities, and normal cornea from low-quality slit lamp images. The performance of the system (NDLS) in low-quality images is improved when adding low-quality images with sufficient diagnostic certainty to the training set. However, its performance is still below that of the senior cornea specialist. Further studies are needed to further reduce and close this performance gap and develop a robust deep learning system that can perform well in both high- and low-quality images.
Limitations of the study
A potential limitation of this study is that we only confirmed the cornea specialists had greater performance in low-quality slit lamp images than that of the deep learning system. Whether this phenomenon also appears in other types of clinical images is not investigated and left for future work.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and algorithms | ||
| DenseNet | Huang et al. (2017) | https://github.com/liuzhuang13/DenseNet |
| Previously established deep learning system (PEDLS) | Li et al., 2021a, Li et al., 2021b, Li et al., 2021c, Li et al., 2021d | https://github.com/jiangjiewei/Keratitis-Source |
| New deep learning system (NDLS) | This study | https://github.com/jiangjiewei/CD-system-Source |
| Gradient-weighted Class Activation Mapping (Grad-CAM) | Selvaraju et al. (2017) | https://github.com/ramprs/grad-cam/ |
| PyTorch | Version 1.6.0 | https://pytorch.org/docs/stable/ |
| Matplotlib | Version 3.3.1 | https://matplotlib.org/3.3.1/ |
| Scikit-learn | Version 0.23.2 | https://scikit-learn.org/stable/whats_new/v0.23 |
| Python | Version 3.7.8 | https://www.python.org/downloads/release/python-378/ |
Resource availability
Lead contact
Further information and requests should be directed to the lead contact, Dr. Wei Chen (chenwei@eye.ac.cn).
Materials availability
This study did not generate new unique reagents.
Method details
Study approval
This study was approved by Ningbo Eye Hospital (NEH) Ethics Review Committee (protocol number 2020-qtky-017) and conducted following the Declaration of Helsinki. Because deidentified images were used, the review committee denoted that patient consent was not required in this study.
Information of the PEDLS
The information including the technical, clinical details, and performance of the PEDLS for classifying keratitis, other corneal abnormalities, and normal cornea has been described previously (Li et al., 2021a, Li et al., 2021b, Li et al., 2021c, Liu et al., 2020). Notably, this PEDLS was developed and evaluated only based on high-quality slit lamp images and a total of 302 low-quality slit lamp images from external test datasets were excluded (Li et al., 2021a, Li et al., 2021b, Li et al., 2021c, Liu et al., 2020). The image quality was considered “poor” if the cornea was blurred and/or distorted.
PEDLS vs. corneal specialists in low-quality slit lamp images
Two cornea specialists who had 4 and 7 years of clinical experience were recruited to investigate their performance in low-quality slit lamp images for classifying keratitis, other corneal abnormalities, and normal cornea. The cornea specialists were not informed of any clinical information related to these images. The low-quality images from external test datasets were used to compare the performance of PEDLS to that of the corneal specialists.
Developing a deep learning system with images of both high and low quality
Both high- and low-quality images with clear diagnoses from NEH were used to develop a new deep learning system (NDLS) for the classification of keratitis, other corneal abnormalities, and normal cornea. The images were randomly divided into training (70%), validation (15%), and internal test (15%) datasets at the subject level. No overlap between these datasets was allowed.
In the image preprocessing phase, pixel values of slit lamp images were normalized to a range of 0–1, and the size of the slit lamp images was resampled to a resolution of 224 × 224 pixels. Data augmentation was applied to increase the heterogeneity of the training dataset, avoiding overfitting and bias during the training process (Bloice et al., 2019). The training dataset was increased to 6-fold of the original size (from 5,505 to 33,030) using horizontal and vertical flips, random cropping, and random rotations around the image center.
The NDLS was trained using the DenseNet121 algorithm which exhibited the optimal performance in detecting keratitis, other corneal abnormalities, and normal cornea in our previous study (Li et al., 2021a, Li et al., 2021b, Li et al., 2021c, Liu et al., 2020). The DenseNet121 has 8.1 × 106 trainable parameters and contains 121 layers densely connected through jointing all preceding layers into subsequent layers to achieve strengthened feature propagation and alleviate the vanishing-gradient problem (Huang et al., 2017). Weights pre-trained on the ImageNet database of 1.4 million images were used to initialize the DenseNet121 architecture (Russakovsky et al., 2015). Transfer learning was performed because it could improve the accuracy of image-based deep learning (Kermany et al., 2018). In this study, we set the learning rate of the parameters of the Softmax classification layer to 10 times that of other layers' parameters. This transfer learning technology guaranteed that the parameters of the Softmax classification layer were fully trained while the parameters of other layers were only fine-tuned using slit lamp images.
The NDLS was built in Python programming language leveraging PyTorch (version 1.6.0, https://pytorch.org/docs/stable/) as a backend. The adaptive moment estimation (ADAM) optimizer was utilized for training and the hyper-parameters were set as follows: learning rate = 0.001, β1 = 0.9, β2 = 0.999, weight decay = 1 × 10−4. During the training process, the cross-entropy loss and accuracy were calculated on the validation dataset after each epoch for monitoring the performance. After 80 epochs, the training was stopped due to the absence of further improvement in both cross-entropy loss and accuracy. The model with the lowest loss on the validation dataset was saved as the optimal model. The images of the external test dataset used to evaluate the performance of the deep learning model in this study were assembled from Zhejiang Eye Hospital (ZEH), Jiangdong Eye Hospital (JEH), and Ningbo Ophthalmic Center (NOC). The running time of the DenseNet121 in the whole training process is 2.30 hours and the average time that the model needs in testing every image is 0.18 seconds with NVIDIA RTX 2080Ti GPU. The process of the development and assessment of the NDLS is displayed in Figure S3.
Visualization heatmap
Gradient-weighted Class Activation Mapping (Grad-CAM) was utilized to create "visual explanations" for the decisions from the deep learning system by superimposing a visualization layer at the end of the CNN model (Selvaraju et al., 2017). This technique leverages the gradients of any target concept, flowing into the last convolutional layer to create a localization map highlighting crucial regions in the image for predicting the concept. Redder regions denote more significant features on the system’s classification.
NDLS vs. PEDLS and NDLS vs. cornea specialists in low-quality images
The low-quality images from the external test dataset were used to compare the performance of the NDLS to that of PEDLS to investigate whether training a deep learning network with both high- and low-quality images could improve its performance in low-quality images for classifying keratitis, other corneal abnormalities, and normal cornea. The t-distributed stochastic neighbor embedding (t-SNE) technique was employed to show the embedding features of each category learned by the deep learning system in a two-dimensional space (van der Maaten and Hinton, 2008). Also, a performance comparison was conducted between the NDLS and corneal specialists in low-quality images.
NDLS vs. PEDLS in high-quality images
The high-quality images from the external test dataset were used to compare the performance of the NDLS to that of PEDLS to investigate whether training a deep learning network with both high- and low-quality images could improve/decline its performance in high-quality images for the classification of keratitis, other corneal abnormalities, and normal cornea.
Statistical analysis
The one-versus-rest strategy was employed to evaluate the performance of the deep learning systems and cornea specialists. The receiver operator characteristic (ROC) curves were plotted using the packages of matplotlib (version 3.3.1, https://matplotlib.org/3.3.1/) and Scikit-learn (version 0.23.2, https://scikit-learn.org/stable/whats_new/v0.23). The 2-sided 95% confidence intervals (CIs) were Wilson score intervals for sensitivity, specificity, and accuracy, and were Delong intervals for AUC. The proportion comparisons were conducted using the McNemar test. Statistical analyses were performed using Python 3.7.8 (https://www.python.org/downloads/release/python-378/, Wilmington, Delaware, USA). All statistical tests were 2-sided, and findings were considered statistically significant at p < .05.
Additional resources
This study did not generate additional data.
Acknowledgments
This study received funding from the National Key R&D Program of China (grant no. 2019YFC0840708), Medical Health Science and Technology Project of Zhejiang Provincial Health (grant no. 2022RC069), Ningbo Science & technology program (grant no. 2021S118), Ningbo Medical Technology Program (grant no. 2020Y54), and the National Natural Science Foundation of China (grant no. 81970770). The funding organizations played no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author contributions
Conception and design: Z.L., J.J., Q.Z., and W.C. Funding obtainment: Z.L. and W.C. Provision of study data: H.W., S.W., and W.C. Collection and assembly of data: Z.L., Q.Z., W.Q., X.L., H.W., and S.W. Data analysis and interpretation: Z.L., J.J., L.G., W.Q, and W.C. Manuscript writing: all authors. Final approval of the manuscript: all authors.
Declaration of interests
The authors report no declarations of interest.
Published: November 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103317.
Contributor Information
Qinxiang Zheng, Email: zhengqinxiang@aliyun.com.
Wei Chen, Email: chenwei@eye.ac.cn.
Supplemental information
Data and code availability
Restrictions apply to the availability of the development and external test datasets, which were used with the permission of the participants for the present study. Anonymized data may be available for research purposes from the corresponding authors on reasonable request. The data are not publicly available due to hospital regulation restrictions and patient privacy concerns. The code used in this study can be accessed at GitHub (https://github.com/jiangjiewei/CD-system-Source).
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.
References
- Bloice M.D., Roth P.M., Holzinger A. Biomedical image augmentation using Augmentor. Bioinformatics. 2019;35:4522–4524. doi: 10.1093/bioinformatics/btz259. [DOI] [PubMed] [Google Scholar]
- Cheung C.Y., Xu D., Cheng C., Sabanayagam C., Tham Y., Yu M., Rim T.H., Chai C.Y., Gopinath B., Mitchell P. A deep-learning system for the assessment of cardiovascular disease risk via the measurement of retinal-vessel calibre. Nat. Biomed. Eng. 2021;5:498–508. doi: 10.1038/s41551-020-00626-4. [DOI] [PubMed] [Google Scholar]
- Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M., Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geirhos R., Janssen D.H.J., Schütt H.H., Rauber J., Bethge M., Wichmann F.A. 2017. Comparing deep neural networks against humans: object recognition when the signal gets weaker.https://arxiv.org/abs/1706.06969 [Google Scholar]
- Hosny A., Aerts H. Artificial intelligence for global health. Science. 2019;366:955–956. doi: 10.1126/science.aay5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. 2017 IEEE Conference on Computer Vision and Pattern Recognition. CVPR; 2017. Densely connected convolutional networks; pp. 2261–2269. [Google Scholar]
- Kermany D.S., Goldbaum M., Cai W., Valentim C., Liang H., Baxter S.L., McKeown A., Yang G., Wu X., Yan F. Identifying medical diagnoses and treatable diseases by Image-Based deep learning. Cell. 2018;172:1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- Li Z., Guo C., Lin D., Nie D., Zhu Y., Chen C., Zhao L., Wang J., Zhang X., Dongye M. Deep learning for automated glaucomatous optic neuropathy detection from ultra-widefield fundus images. Br. J. Ophthalmol. 2020;105:1548–1554. doi: 10.1136/bjophthalmol-2020-317327. [DOI] [PubMed] [Google Scholar]
- Li Z., Guo C., Nie D., Lin D., Zhu Y., Chen C., Wu X., Xu F., Jin C., Zhang X. Deep learning for detecting retinal detachment and discerning macular status using ultra-widefield fundus images. Commun. Biol. 2020;3:15. doi: 10.1038/s42003-019-0730-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Guo C., Nie D., Lin D., Zhu Y., Chen C., Xiang Y., Xu F., Jin C., Zhang X. Development and evaluation of a deep learning system for screening retinal hemorrhage based on Ultra-Widefield fundus images. Transl. Vis. Sci. Technol. 2020;9:3. doi: 10.1167/tvst.9.2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Guo C., Nie D., Lin D., Zhu Y., Chen C., Zhang L., Xu F., Jin C., Zhang X. A deep learning system for identifying lattice degeneration and retinal breaks using ultra-widefield fundus images. Ann. Transl. Med. 2019;7:618. doi: 10.21037/atm.2019.11.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Guo C., Nie D., Lin D., Zhu Y., Chen C., Zhao L., Wu X., Dongye M., Xu F. Deep learning from "passive feeding" to "selective eating" of real-world data. NPJ Digit. Med. 2020;3:143. doi: 10.1038/s41746-020-00350-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Guo C., Nie D., Lin D., Cui T., Zhu Y., Chen C., Zhao L., Zhang X., Dongye M. Automated detection of retinal exudates and drusen in ultra-widefield fundus images based on deep learning. Eye (Lond) 2021 doi: 10.1038/s41433-021-01715-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Jiang J., Chen K., Chen Q., Zheng Q., Liu X., Weng H., Wu S., Chen W. Preventing corneal blindness caused by keratitis using artificial intelligence. Nat. Commun. 2021;12:3738. doi: 10.1038/s41467-021-24116-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Jiang J., Chen K., Zheng Q., Liu X., Weng H., Wu S., Chen W. Development of a deep learning-based image quality control system to detect and filter out ineligible slit-lamp images: a multicenter study. Comput. Methods Programs Biomed. 2021;203:106048. doi: 10.1016/j.cmpb.2021.106048. [DOI] [PubMed] [Google Scholar]
- Li Z., Jiang J., Zhou H., Zheng Q., Liu X., Chen K., Weng H., Chen W. Development of a deep learning-based image eligibility verification system for detecting and filtering out ineligible fundus images: a multicentre study. Int. J. Med. Inform. 2021;147:104363. doi: 10.1016/j.ijmedinf.2020.104363. [DOI] [PubMed] [Google Scholar]
- Liu Y., Jain A., Eng C., Way D.H., Lee K., Bui P., Kanada K., de Oliveira M.G., Gallegos J., Gabriele S. A deep learning system for differential diagnosis of skin diseases. Nat. Med. 2020;26:900–908. doi: 10.1038/s41591-020-0842-3. [DOI] [PubMed] [Google Scholar]
- Luo H., Xu G., Li C., He L., Luo L., Wang Z., Jing B., Deng Y., Jin Y., Li Y. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case-control, diagnostic study. Lancet Oncol. 2019;20:1645–1654. doi: 10.1016/S1470-2045(19)30637-0. [DOI] [PubMed] [Google Scholar]
- Matheny M.E., Whicher D., Thadaney I.S. Artificial intelligence in health care: a report from the national academy of medicine. JAMA. 2019;323:509–510. doi: 10.1001/jama.2019.21579. [DOI] [PubMed] [Google Scholar]
- Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M. ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 2015;115:211–252. [Google Scholar]
- Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. IEEE International Conference on Computer Vision. ICCV; 2017. Grad-CAM: visual explanations from deep networks via gradient-based localization. 2017; pp. 618–626. [Google Scholar]
- Ting D., Cheung C.Y., Lim G., Tan G., Quang N.D., Gan A., Hamzah H., Garcia-Franco R., San Y.I., Lee S.Y. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318:2211–2223. doi: 10.1001/jama.2017.18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trucco E., Ruggeri A., Karnowski T., Giancardo L., Chaum E., Hubschman J.P., Al-Diri B., Cheung C.Y., Wong D., Abramoff M. Validating retinal fundus image analysis algorithms: issues and a proposal. Invest. Ophthalmol. Vis. Sci. 2013;54:3546–3559. doi: 10.1167/iovs.12-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L., Hinton G. Visualizing Data using t-SNE. J. Mach Learn. Res. 2008:2579–2605. [Google Scholar]
- Xie Y., Zhao L., Yang X., Wu X., Yang Y., Huang X., Liu F., Xu J., Lin L., Lin H. Screening candidates for refractive surgery with corneal Tomographic-Based deep learning. JAMA Ophthalmol. 2020;138:519–526. doi: 10.1001/jamaophthalmol.2020.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Liu X., Shen J., Li Z., Sang Y., Wu X., Zha Y., Liang W., Wang C., Wang K. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;182:1360. doi: 10.1016/j.cell.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Yang Y., Yu C., Liu J., Duan X., Weng Z., Chen D., Liang Q., Fang Q., Zhou J. Ensembled deep learning model outperforms human experts in diagnosing biliary atresia from sonographic gallbladder images. Nat. Commun. 2021;12:1259. doi: 10.1038/s41467-021-21466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Restrictions apply to the availability of the development and external test datasets, which were used with the permission of the participants for the present study. Anonymized data may be available for research purposes from the corresponding authors on reasonable request. The data are not publicly available due to hospital regulation restrictions and patient privacy concerns. The code used in this study can be accessed at GitHub (https://github.com/jiangjiewei/CD-system-Source).
Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.