Abstract
Arsenic is a metalloid that is toxic to plants. Arsenate (As(V)), the prevalent chemical form of arsenic, is a phosphate (Pi) analog and is incorporated into plant cells via Pi transporters. Here, we found that the MYB40 transcription factor played important roles in the control of Arabidopsis As(V) resistance. The expression of MYB40 was induced by As(V) stress. MYB40-overexpressing lines had an obvious As(V)-resistant phenotype and a reduced As(V)/Pi uptake rate, whereas myb40 mutants were sensitive to As(V) stress. Upon exposure to As(V), MYB40 directly repressed the expression of PHT1;1, which encodes a main Pi transporter. The As(V)-resistant phenotypes of MYB40-overexpressing lines were impaired by overexpression of PHT1;1, demonstrating an epistatic genetic relationship between MYB40 and PHT1;1. Moreover, overexpression of MYB40 enhanced, and disruption of MYB40 reduced, thiol-peptide contents. Upon exposure to As(V), MYB40 positively regulated the expression of PCS1, which encodes a phytochelatin synthase, and ABCC1 and ABCC2, which encode the major vacuolar phytochelatin transporters. Together, our data demonstrate that AtMYB40 acts as a central regulator of As(V) responses, providing a genetic strategy for enhancing plant As(V) tolerance and reducing As(V) uptake to improve food safety.
Key words: AtMYB40, arsenate, uptake, detoxification, transcription
The transcription factor MYB40 plays an important role in Arabidopsis resistance to arsenate stress. During arsenate stress, MYB40 directly represses PHT1;1 to reduce As(V) uptake into plant cells and directly upregulates PCS1 expression to enhance the content of phytochelatins (PCs), which form complexes with As(III). MYB40 also modulates As(III)-PC2 uptake into the vacuole by indirectly upregulating the expression of ABCC1 and ABCC2.
Introduction
Arsenic (As) is ubiquitous in the environment and is one of the most toxic metalloids found in soils (Zhao et al., 2010; Kumar et al., 2015). People experience As poisoning by drinking As-contaminated water and ingesting crops cultivated in As-polluted soils (Zhao et al., 2010). Arsenic is nonessential and toxic to plants, but it is impossible to entirely block its entry into plants because it shares transporters with essential or beneficial elements, such as phosphate (Pi) and silicon (Catarecha et al., 2007; Ma et al., 2008).
Arsenic is found primarily in inorganic forms, and arsenate (As(V)) and arsenite (As(III)) are the most common oxidation forms (Chao et al., 2014). In aerobic soils, As(V) is the predominant species and is taken up mainly via Pi transporters (Shin et al., 2004; Catarecha et al., 2007; Wu et al., 2011; Remy et al., 2012; Castrillo et al., 2013; DiTusa et al., 2016). In Arabidopsis thaliana, PHT1;1 and PHT1;4 are the main Pi transporters, and the pht1;1Δ4Δ double mutant shows a 75% reduction in Pi uptake capacity relative to wild-type plants (Shin et al., 2004). Upon exposure to As(V), both the pht1;1 mutant and the pht1;1Δ4Δ double mutant displayed significant As(V)-resistant phenotypes (Shin et al., 2004). Later, based on As(V) toxicity screening, the pht1;1-3 mutant was identified, which harbors a missense mutation in PHT1;1 (Catarecha et al., 2007). The pht1;1-3 mutant showed an As(V)-tolerant phenotype and enhanced As accumulation (Catarecha et al., 2007). As(V) represses the expression of PHT1;1 and influences the relocalization of PHT1;1 (Castrillo et al., 2013). In fact, some proteins that modulate the expression or localization of PHT1;1 are also involved in responses to As(V) stress (González et al., 2005; Castrillo et al., 2013; Wang et al., 2014; Su et al., 2015).
After being taken up into the roots, As(V) is rapidly reduced to As(III) (Pickering et al., 2000). Several arsenate reductases have been identified in plants, such as ATQ1 (also known as HAC1) from Arabidopsis (Chao et al., 2014; Sánchez-Bermejo et al., 2014), PvACR2 from Pteris vittata (Ellis et al., 2006), and OsACR2.1 and OsACR2.3 from Oryza sativa (Duan et al., 2007). Loss of ATQ1 leads to a lack of As(III) efflux ability in roots and a significant increase in As accumulation in shoots (Chao et al., 2014). As(III) is either extruded from the roots or detoxified through complexation and vacuolar sequestration (Zhao et al., 2010). The complexation of As(III) by phytochelatins (PCs) is an important mechanism of As detoxification. Upon exposure to As stress, synthesis and accumulation of PCs are induced (Grill et al., 1987; Sneller et al., 1999; Schmöger et al., 2000). The Arabidopsis phytochelatin synthase PCS1-defective mutant cad1-3, which lacks the function of PC synthase, produces few PCs and shows As(V)-hypersensitive phenotypes (Howden et al., 1995; Ha et al., 1999). Overexpression of AtPCS1 leads to enhanced As(V) tolerance and increased PC accumulation (Lee et al., 2003; Li et al., 2004). Plant vacuoles are the final detoxification stores for As, and PC–As(III) complexes are transported into the vacuoles by ABCC-type transporters (Song et al., 2010, 2014). The Arabidopsis abcc1 abcc2 double mutant is sensitive to As(V) stress, and AtPCS1 AtABCC1 co-overexpression lines show As(V)-tolerant phenotypes (Lee et al., 2003). As is translocated through the xylem to the shoots and is then loaded from the xylem into the phloem and seeds. A recent report showed that the inositol transporters AtINT2 and AtINT4 contribute to As loading into the phloem (Duan et al., 2015).
Upon exposure to As stress, the transcript levels of numerous genes, including key genes involved in As uptake, PC biosynthesis, and As translocation, undergo significant changes that help plants to survive sub-optimal growth conditions (Li et al., 2004; Abercrombie et al., 2008; Sung et al., 2009; Castrillo et al., 2013; Chao et al., 2014; Sánchez-Bermejo et al., 2014; Kumar et al., 2015). Several articles have reported the relevant regulatory mechanisms. Disruption of ACR1 in yeast results in As(III) and As(V) hypersensitivity, and ACR1 encodes a transcription factor that regulates transcription of ACR3, which encodes an arsenite transporter (Bobrowicz et al., 1997; Ghosh et al., 1999). Arabidopsis WRKY6 is an arsenate-responsive transcription factor that mediates arsenate/phosphate transporter gene expression and arsenate-induced transposon activation (Castrillo et al., 2013).
In this study, we found that the transcription factor AtMYB40 played an important role in As resistance. MYB40 was induced by As(V) stress, and MYB40-overexpressing lines were As(V) resistant. Upon exposure to As(V), MYB40 repressed the expression of the As(V)/Pi transporter gene PHT1;1, thereby decreasing As(V)/Pi uptake, and positively regulated the expression of PCS1, ABCC1, and ABCC2 to enhance As detoxification ability.
Results
MYB40 is an As(V)-responsive transcription factor
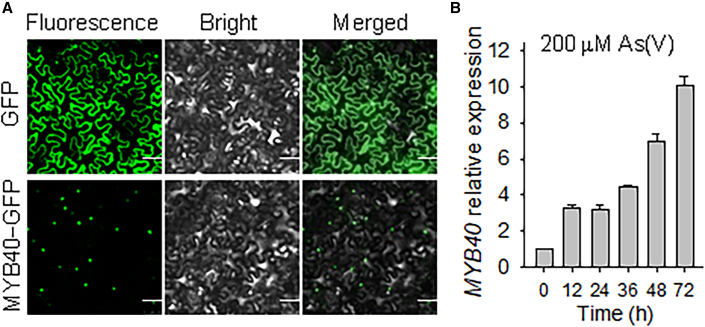
Arabidopsis MYB40 belongs to the R2R3 MYB sub-family (Stracke et al., 2001). To test the subcellular location of MYB40, the coding region of MYB40 was fused with the 3′ end of the GFP reporter gene and expressed under the Super promoter. The GFP gene alone under the Super promoter was used as a control. MYB40−GFP and GFP alone were transiently expressed in Nicotiana benthamiana leaves. The MYB40−GFP fusion protein was localized in the nucleus, and GFP alone was localized in the cytoplasm and nucleus (Figure 1A). When exposed to As(V) stress, the transcript level of MYB40 was clearly elevated (Figure 1B). These data indicate that MYB40 is an As(V)-responsive MYB transcription factor.
Figure 1.
MYB40 is an As(V) response transcription factor.
(A) Subcellular localization of MYB40–GFP fusion protein in Nicotiana benthamiana leaves. GFP alone was used as the control. Scale bar corresponds to 50 μm.
(B) qRT–PCR analysis of MYB40 in wild-type plants under As(V) stress. Seven-d-old wild-type seedlings were transferred to ½ MS medium containing 200 μM As(V), then harvested at the indicated time for qRT–PCR analysis. The qRT–PCR was performed with three technical replicates, and the experiment was repeated at least three times with similar results.
Overexpression of MYB40 enhances Arabidopsis As(V) resistance
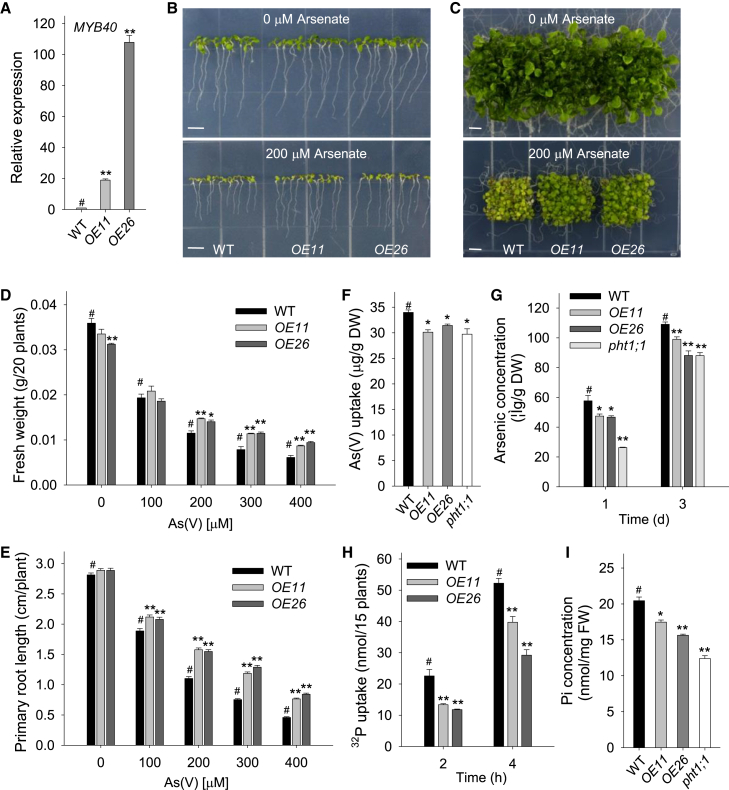
To test the function of MYB40 on As(V) tolerance, MYB40-overexpression lines were generated. The coding sequence of MYB40 was expressed under the cauliflower mosaic virus 35S promoter, and two homozygous single-copy MYB40-overexpressing lines (OE11 and OE26) were obtained (Figure 2A). There were no obvious differences among the tested genotypes when grown on ½ Murashige and Skoog (MS) medium with 0 μM As(V) (Figure 2B and 2C, top panel). When germinated and grown on ½ MS medium with 200 μM As(V) for 7 d, the MYB40-overexpressing lines (OE11 and OE26) showed As(V)-tolerant phenotypes with longer primary roots than wild-type seedlings (Figure 2B, bottom panel). When grown for 15 d, wild-type plants displayed obvious As(V) toxicosis symptoms, with stunted growth and yellow leaves, whereas the OE11 and OE26 lines remained healthy, with larger, green leaves (Figure 2C, bottom panel). Fresh weight and root length were also measured under As(V) stress. Upon exposure to different concentrations of As(V), the OE11 and OE26 lines had much higher fresh weights and longer primary roots than wild-type plants (Figure 2D and 2E), indicating that overexpression of MYB40 enhances Arabidopsis As(V) tolerance.
Figure 2.
Overexpression of MYB40 increases Arabidopsis As(V) resistance and represses As(V)/Pi uptake.
(A) qRT–PCR analysis of MYB40 expression in the MYB40-overexpressing lines (OE11 and OE26) and wild-type plants (WT). The qRT–PCR was performed with three technical replicates, and the experiment was repeated at least three times with similar results.
(B and C) Phenotypic comparison. MYB40-overexpressing lines and WT plants were germinated and grown on ½ MS medium containing 0 or 200 μM As(V) for 7 d (B) and 15 d (C), and then representative photographs were taken. Scale bar corresponds to 0.5 cm.
(D and E) Fresh weight and primary root length analysis. The MYB40-overexpressing lines and WT plants were germinated and grown on ½ MS medium containing 0, 100, 200, 300, and 400 μM As(V) for 7 d, and then the fresh weight (D) and primary root length (E) were measured. The values are means ± SE; n = 3 in (D) and (n ≥ 40) in (E).
(F) As(V) uptake analysis. Seven-d-old plants were transferred to ½ MS medium with 200 μM As(V) for 6 h, then harvested for As(V) uptake analysis. Data are shown as mean ± SE; n = 3.
(G) As content measurement. Seven-d-old plants were transferred to ½ MS medium with 200 μM As(V) for 1 or 3 d, then harvested for As content measurement. Data are shown as mean ± SE; n ≥ 5.
(H) Pi uptake was monitored over a 4-h period in 7-d-old MYB40-overexpressing lines and WT plants. Data are shown as mean ± SE; n = 3.
(I) Pi concentration of 7-d-old MYB40-overexpressing lines and WT plants grown on MS medium. Data are shown as mean ± SE; n = 3.
Asterisks in (A, D, E, F, G, H, and I) indicate significant differences compared with WT plants (#) by Student's t-test: ∗P < 0.05, ∗∗ P < 0.01.
As MYB40-overexpressing lines displayed As(V)-resistant phenotypes (Figure 2B and 2C), and As(V) is taken up via Pi transporters from the environment (Shin et al., 2004; Catarecha et al., 2007; Remy et al., 2012), we hypothesized that MYB40 may repress As(V) uptake. First, the As(V) uptake rate was measured. After exposure to 200 μM As(V) for 6 h, the OE11 and OE26 lines had lower As(V) uptake rates than the wild-type plants, similar to the pht1;1 mutant, which also showed an As(V)-tolerant phenotype (Shin et al., 2004) (Figure 2F). We further hypothesized that MYB40 represses Pi uptake and, as a result, the As(V) uptake is reduced. We therefore measured Pi uptake over a 4-h period. The Pi uptake ability of OE11 and OE26 was significantly lower than that of wild-type plants (Figure 2G). These data indicate that MYB40 enhances Arabidopsis As(V) tolerance at least in part by repressing As(V)/Pi uptake.
Disruption of MYB40 reduces Arabidopsis As(V) resistance
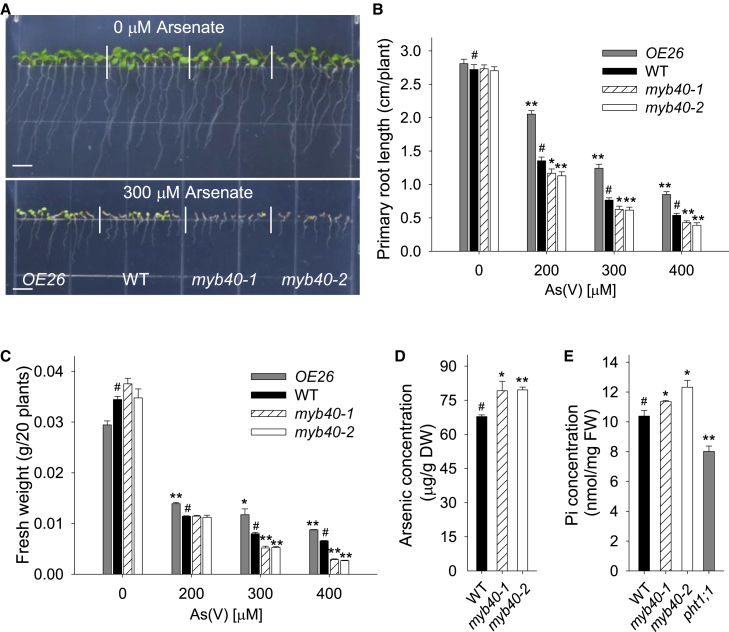
In an attempt to determine the roles of MYB40 in Arabidopsis As(V) resistance, we used CRISPR/Cas9 technology to generate the myb40 mutant, as there was no T-DNA insertion mutant at the ABRC. A pair of sgRNA targets in the MYB40 gene was selected (Supplemental Figure 1A and 1B). The CRISPR construct was transformed into wild-type Arabidopsis, and two homozygous myb40 mutants, named myb40-1 and myb40-2, were obtained. The myb40-1 mutant contained a 323-bp deletion in the MYB40 gene, resulting in the deletion of most of the first two exons and the first intron (Supplemental Figure 1C). The myb40-2 mutant had a nucleotide deletion in the C1 site and a nucleotide insertion in the C2 site (Supplemental Figure 1D), which led to a frameshift mutation. Both myb40-1 and myb40-2 were null mutants.
When germinated and grown on ½ MS medium without As(V), the myb40-1 and myb40-2 mutants had no obvious differences compared with wild-type plants (Figure 3A, top panel). When germinated and grown on ½ MS medium containing 300 μM As(V) for 7 d, the growth of myb40-1 and myb40-2 mutants was dramatically impaired relative to wild-type plants (Figure 3A, bottom panel). In the presence of As(V), the myb40-1 and myb40-2 mutants had significantly shorter primary roots and reduced fresh weights relative to wild-type plants (Figure 3B and 3C). In contrast to MYB40-overexpressing lines, the myb40-1 and myb40-2 mutants had higher As accumulation and Pi contents than wild-type plants when exposed to As(V) (Figure 3D and 3E). These data demonstrate that disruption of MYB40 results in an increased sensitivity to As(V).
Figure 3.
Disruption of MYB40 results in Arabidopsis sensitivity to As(V).
(A) Phenotypic comparison. The myb40 mutants, OE26, and WT plants were germinated and grown on ½ MS medium with or without 300 μM As(V) for 7 d, and then representative photographs were taken. Scale bar corresponds to 0.5 cm.
(B and C) Primary root length and fresh weight analysis. The myb40 mutants, OE26, and WT plants were germinated and grown on ½ MS medium containing 0, 200, 300, and 400 μM As(V) for 7 d, and then the primary root length (B) and fresh weight (C) were measured. The values are means ± SE; n ≥ 20 in (B) and n = 3 in (C).
(D and E) As and Pi content measurement. Seven-d-old seedlings were transferred to ½ MS medium with 300 μM As(V) for 3 d, then harvested for measurement of As content (D) and Pi content (E). Data are shown as mean ± SE; n = 3.
Asterisks in (B, C, D, and E) indicate significant differences compared with WT plants (#) by Student's t-test: ∗P < 0.05, ∗∗P < 0.01.
MYB40 negatively regulates PHT1;1 expression
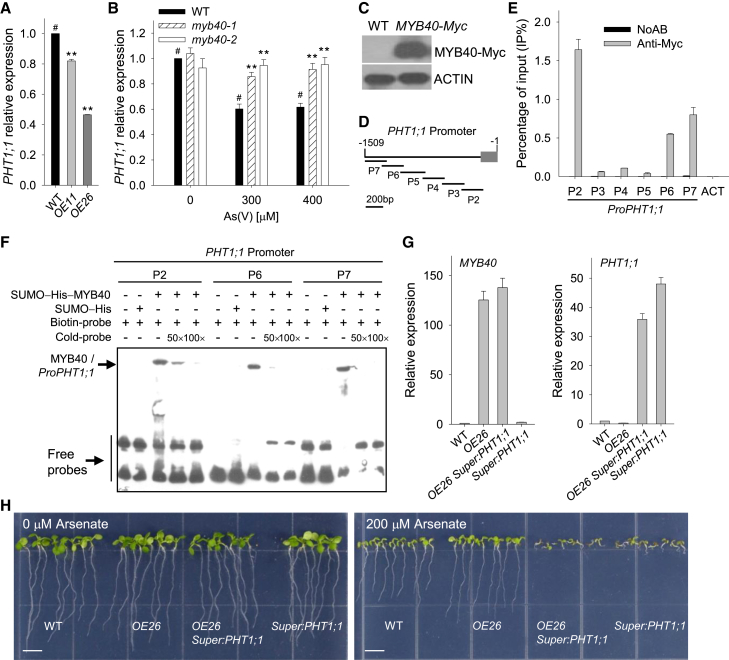
Arsenate is a structural homolog of Pi and is transported into plant cells mainly through the Pi transporters PHT1;1 and PHT1;4 in Arabidopsis (Shin et al., 2004; Catarecha et al., 2007). The pht1;1 mutant is insensitive to As(V) (Shin et al., 2004), and the PHT1;1-overexpressing line shows an As(V)-hypersensitive phenotype (Wang et al., 2014). The MYB40-overexpressing lines displayed As(V)-resistant phenotypes and had lower Pi uptake rates (Figure 2), similar to the pht1;1 mutant (Shin et al., 2004; Catarecha et al., 2007). We then hypothesized that MYB40, as an MYB transcription factor, downregulates PHT1;1 expression. The qRT–PCR results showed that the transcript levels of PHT1;1 were much lower in MYB40-overexpressing lines than in wild-type plants (Figure 4A). PHT1;1 expression was also tested in the myb40 mutants. After treatment with 300 or 400 μM As(V) for 2 d, PHT1;1 expression was repressed in the wild-type plants. By contrast, transcript levels of PHT1;1 in the myb40 mutants were similar to those in wild-type plants without As(V) treatment (Figure 4B). These data indicate that MYB40 downregulates PHT1;1 expression.
Figure 4.
MYB40 represses PHT1;1 expression in the Arabidopsis response to As stress.
(A) qRT–PCR analysis of PHT1;1 expression in MYB40-overexpressing lines and WT plants. The qRT–PCR was performed with three technical replicates, and the experiment was repeated at least three times with similar results.
(B) qRT–PCR analysis of PHT1;1 in the myb40 mutants under As(V) stress. Seven-d-old seedlings were transferred to ½ MS medium with 0, 300, and 400 μM As(V) for 2 d, then harvested for RNA extraction. The qRT–PCR was performed with three technical replicates, and the experiment was repeated at least three times with similar results. Asterisks in (A and B) indicate significant differences compared with WT plants (#) by Student's t-test: ∗∗P < 0.01.
(C) Immunoblot analysis of MYB40 protein in the MYB40-Myc transgenic line. Protein extract was analyzed by immunoblotting using anti-Myc antibody. Actin was used as the loading control.
(D) Diagram of the PHT1;1 promoter showing relative positions and sizes of different fragments. The adenine residue of the translational start codon ATG was assigned position +1, and the numbers flanking the sequences of the PHT1;1 promoter fragments are based on this number. The gray box indicates the untranslated region (UTR).
(E) ChIP–qPCR assay of MYB40 binding to the PHT1;1 promoter in vivo. Seven-d-old MYB40-Myc seedlings were transferred to ½ MS medium containing 200 μM As(V) for 2 d, then harvested for ChIP–qPCR using anti-Myc antibody. Data are shown as mean ± SE; n = 3.
(F) EMSA of MYB40 binding to the PHT1;1 promoter in vitro. Each biotin-labeled DNA probe was incubated with SUMO–His–MYB40 protein. An excess of unlabeled probe with the same sequence was added to compete with the labeled probe.
(G) The expression of MYB40 and PHT1;1 was tested by qRT–PCR in OE26, OE26 Super:PHT1;1 co-overexpression, Super:PHT1;1, and WT plants. The qRT–PCR was performed with three technical replicates, and the experiment was repeated at least three times with similar results.
(H) Phenotypic comparison. Plants were germinated and grown on ½ MS medium containing 0 or 200 μM arsenate for 7 d, and then representative photographs were taken. Scale bar corresponds to 0.5 cm.
Because MYB40 is a transcription factor (Figure 1), we investigated whether MYB40 bound to the PHT1;1 promoter. To test this binding, an MYB40-Myc transgenic line (Figure 4C) was generated for chromatin immunoprecipitation (ChIP). The PHT1;1 promoter was separated into six fragments (named P2–P7) (Figure 4D) and amplified by PCR using the primers listed in Supplemental Table 1. The 7-d-old MYB40-Myc seedlings were transferred to 200 μM As(V) for 2 d, then harvested for the ChIP assay. Chromatin immunoprecipitated with anti-Myc antibody was enriched in P2, P6, and P7 of the PHT1;1 promoter (Figure 4E), indicating that MYB40 could bind to the PHT1;1 promoter in vivo. An electrophoretic mobility shift assay (EMSA) was also conducted. The recombinant SUMO–His–MYB40 protein bound to the P2, P6, and P7 fragments of the PHT1;1 promoter, and this binding was effectively reduced by the addition of unlabeled competitors with the same sequence; the SUMO–His protein did not bind to the PHT1;1 promoter (Figure 4F), indicating that MYB40 bound to the PHT1;1 promoter in vitro. Together, these data demonstrate that MYB40 directly represses PHT1;1 expression by binding to the PHT1;1 promoter.
The MYB40-overexpressing line (OE26) was crossed with the PHT1;1-overexpressing line Super:PHT1;1 (Wang et al., 2014) to obtain the OE26 Super:PHT1;1 co-overexpression line (Figure 4G). When germinated and grown on ½ MS medium without As(V) (0 μM arsenate), there were no obvious phenotypic differences among the genotypes (Figure 4H, left panel). In the presence of 200 μM As(V), the OE26 Super:PHT1;1 co-overexpression line showed As(V)-hypersensitive phenotypes, similar to Super:PHT1;1 (Figure 4H, right panel), demonstrating that overexpression of PHT1;1 abolished the As(V)-tolerant phenotypes of the MYB40-overexpressing line.
Overexpression of MYB40 enhances, and disruption of MYB40 reduces, thiol-peptide contents
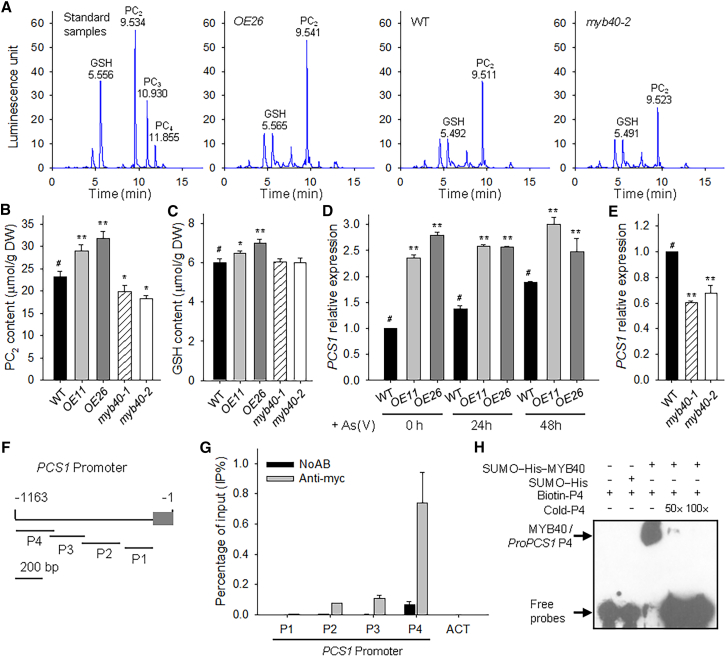
After being transported into plant cells, As(V) is reduced to As(III) by a reductase and is detoxified through complex formation with PCs and glutathione (GSH) (Zhao et al., 2010; Kumar et al., 2015). The PCS1 gene (also named CAD1) encodes a PC synthase in Arabidopsis (Ha et al., 1999; Vatamaniuk et al., 1999). The cad1 mutant is hypersensitive to As(V) stress (Ha et al., 1999), and overexpression of PCS1 increases thiol-peptide accumulation and As tolerance (Lee et al., 2003; Li et al., 2004). This led us to investigate whether MYB40 was involved in As detoxification. Seven-d-old MYB40-overexpressing lines, myb40 mutants, and wild-type seedlings were transferred to ½ MS medium containing 200 μM As(V) for 7 d, then harvested for measurement of PCs and GSH. Using GSH, PC2, PC3, and PC4 standards, fluorescent HPLC analysis clearly identified the mBBr-labeled peptides in different genotypes (Figure 5A). The level of PC2 was significantly increased in OE11 and OE26, and decreased in the myb40-1 and myb40-2 mutants, relative to wild-type seedlings (Figure 5A and 5B). GSH is a substrate for PC synthesis and plays roles in As detoxification (Zhao et al., 2010; Kumar et al., 2015). The GSH contents were higher in the MYB40-overexpressing lines (OE11 and OE26) than in wild-type plants when exposed to As(V) (Figure 5C).
Figure 5.
MYB40 modulates thiol-peptide accumulation and PCS1 expression in the Arabidopsis response to As stress.
(A) HPLC–MS chromatograms of OE26, myb40-2 mutant, and WT plants exposed to 200 μM As(V) for 7 d.
(B and C) Thiol-peptide measurements. Seven-d-old seedlings of different genotypes were transferred to ½ MS medium containing 200 μM As(V) for 7 d, then harvested for thiol-peptide measurement. The levels of PC2(B) and GSH (C) in whole seedlings were measured by fluorescent HPLC. Data are shown as mean ± SE; n = 3.
(D) qRT–PCR analysis of PCS1 expression in MYB40-overexpressing lines. Seven-d-old plants were transferred to ½ MS medium containing 200 μM As(V) and harvested at the indicated time. The qRT–PCR was performed with three technical replicates, and the experiment was repeated at least three times with similar results.
(E) qRT–PCR analysis of PCS1 in the myb40 mutants under As(V) stress. The 7-d-old seedlings were transferred to ½ MS medium with 200 μM As(V) for 2 d, then harvested for RNA extraction. The qRT–PCR was performed with three technical replicates, and the experiment was repeated at least three times with similar results.
(F) Diagram of the PCS1 promoter showing relative positions and sizes of different fragments. The adenine residue of the translational start codon ATG was assigned position +1, and the numbers flanking the sequences of the PCS1 promoter fragments are based on this number. The gray box indicates the UTR.
(G) ChIP–qPCR assay of MYB40 binding to the PCS1 promoter in vivo. Seven-d-old MYB40-Myc seedlings were transferred to ½ MS medium containing 200 μM As(V) for 2 d, then harvested for ChIP–qPCR assay using anti-Myc antibody. Data are shown as mean ± SE; n = 3.
(H) EMSA of MYB40 binding to the P4 fragment of the PCS1 promoter in vitro. Each biotin-labeled DNA probe was incubated with SUMO–His–MYB40 protein. An excess of unlabeled probe with the same sequence was added to compete with the labeled probe. Asterisks in (B, C, D, and E) indicate significant differences compared with WT plants (#) by Student's t-test: ∗P < 0.05, ∗∗P < 0.01.
MYB40 positively regulates PCS1 expression
PCS1 expression is induced under As(V) stress (Sung et al., 2009), and MYB40-overexpressing lines had higher thiol-peptide contents (Figure 5A–5C). We further hypothesized that MYB40 modulates PCS1 expression. Upon exposure to As(V), the expression of PCS1 was induced in wild-type seedlings, but it did not change in OE11 and OE26 (Figure 5D). The transcript levels of PCS1 in OE11 and OE26 were much higher than those in wild-type plants, and they were also higher than those of wild-type seedlings treated with As(V) for 48 h (Figure 5D). In contrast to MYB40-overexpressing lines, the transcript levels of PCS1 in the myb40-1 and myb40-2 mutants were obviously lower than those in wild-type plants under As(V) stress (Figure 5E). These data indicate that MYB40 positively modulates PCS1 expression under As(V) stress.
The PCS1 promoter was separated into four fragments (named P1–P4) (Figure 5F), and a ChIP assay was performed with MYB40-Myc seedlings treated with 200 μM As(V) for 2 d. Chromatin immunoprecipitated with anti-Myc antibody was enriched in the P4 fragment of the PCS1 promoter (Figure 5G), indicating that MYB40 bound to the PCS1 promoter in vivo. EMSA was also conducted to detect the binding of MYB40 to the P4 fragment of the PCS1 promoter. The SUMO–His–MYB40 protein bound to the P4 fragment of the PCS1 promoter, and this binding was effectively reduced by an unlabeled competitor with the same sequence (Figure 5H), indicating that MYB40 bound to the P4 fragment of the PCS1 promoter in vitro. Together, these data demonstrate that MYB40 is a major transcription factor that positively modulates PCS1 expression under As(V) stress.
MYB40 modulates ABCC1 and ABCC2 expression
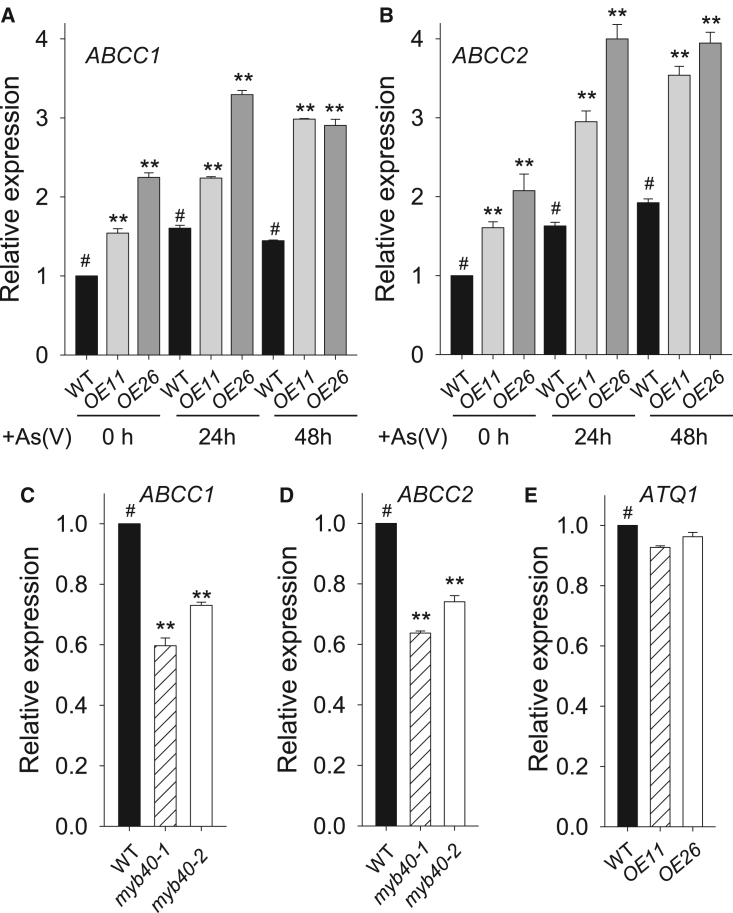
The PC–As(III) complexes are transported into vacuoles, which act as final detoxification stores for As (Zhao et al., 2010; Kumar et al., 2015). Arabidopsis ABCC1 and ABCC2 are two major vacuolar PC transporters (Song et al., 2010). The abcc1 abcc2 double mutant is hypersensitive to As(V) stress, and the ABCC1 PCS1 co-overexpression line shows As(V)-tolerant phenotypes (Song et al., 2010). We therefore tested the expression of ABCC1 and ABCC2. Upon exposure to As(V), expression of ABCC1 and ABCC2 was induced, and their transcript levels were significantly elevated in OE11 and OE26 (Figure 6A and 6B) and reduced in the myb40-1 and myb40-2 mutants (Figure 6C and 6D) relative to wild-type plants, indicating that MYB40 positively modulates the expression of ABCC1 and ABCC2.
Figure 6.
MYB40 positively modulates ABCC1 and ABCC2 expression.
(A and B) qRT–PCR analysis of ABCC1 and ABCC2 expression in MYB40-overexpressing lines and WT plants. Seven-d-old seedlings were transferred to ½ MS medium containing 200 μM As(V) and harvested at the indicated time.
(C and D) qRT–PCR analysis of ABCC1 and ABCC2 in the myb40 mutants under As(V) stress. Seven-d-old seedlings were transferred to ½ MS medium containing 200 μM As(V) for 2 d, then harvested for RNA extraction.
(E) qRT–PCR analysis of ATQ1 expression in the MYB40-overexpressing lines and WT plants.
The qRT–PCR was performed with three technical replicates, and the experiment was repeated at least three times with similar results. Asterisks in (A, B, C, and D) indicate significant differences compared with WT plants (#) by Student's t-test: ∗∗P < 0.01.
Intracellular As(III) can be effluxed from plant cells (Zhao et al., 2010; Kumar et al., 2015). Previous reports show that ATQ1 (also named HAC1) encodes an As(V) reductase that plays important roles in As(III) efflux from roots and limitation of As loading into the xylem (Chao et al., 2014; Sánchez-Bermejo et al., 2014). ATQ1 is induced by As(V) stress (Chao et al., 2014), and ATQ1 expression was therefore tested in MYB40-overexpressing lines. The qRT–PCR results showed that the transcript level of ATQ1 in MYB40-overexpressing lines was similar to that in wild-type plants (Figure 6E), indicating that MYB40 does not modulate ATQ1 expression.
Discussion
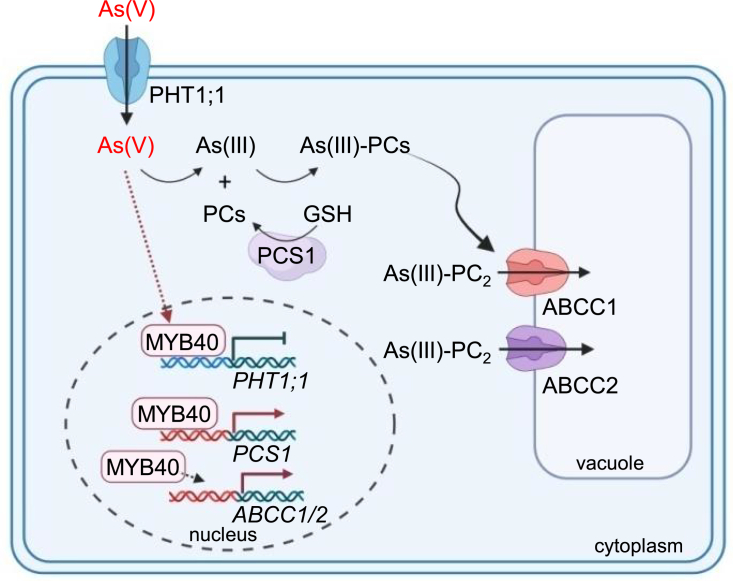
Arsenate is a Pi mimic and toxic to plants, and it is impossible to entirely block its entry into plants. Previous reports showed that the expression of thousands of genes was modulated under As(V) stress (Abercrombie et al., 2008; Castrillo et al., 2013), but the mechanism of this transcriptional regulation is not clear. This study provides evidence that the transcription factor MYB40 plays an important role in Arabidopsis response to As(V) stress (Figure 7). The expression of MYB40 was induced during As(V) stress, and MYB40 then directly repressed PHT1;1 expression to reduce As(V) uptake into plant cells and directly upregulated PCS1 expression to enhance the content of PCs, which formed complexes with As(III). MYB40 also indirectly upregulated the expression of ABCC1 and ABCC2, which encode two major vacuolar PC transporters that carry As(III)-PC2 from the cytoplasm into the vacuole. Previous reports showed that, under As(V) stress, PHT1;1 expression was repressed (Castrillo et al., 2013), and PCS1 expression was induced (Sung et al., 2009). Repressing PHT1;1 expression or increasing PCS1 expression can enhance Arabidopsis As(V) resistance (Li et al., 2004; Su et al., 2015). These data suggest that MYB40 functions as a positive regulator in Arabidopsis resistance to As(V) stress by reducing As(V) uptake and enhancing PC synthesis through direct repression of PHT1;1 expression and upregulation of PCS1 expression.
Figure 7.
A proposed working model for MYB40 in the Arabidopsis response to As stress.
During As(V) stress, the expression of MYB40 is induced, and MYB40 then directly represses PHT1;1 expression to reduce As(V) uptake into plant cells. It also directly upregulates PCS1 expression to enhance the contents of PCs, which form complexes with As(III). MYB40 indirectly upregulates the expression of ABCC1 and ABCC2, which encode two major vacuolar PC transporters that carry As(III)-PC2 from the cytoplasm into the vacuole. Created with BioRender.com.
MYB40 was localized in the nucleus and had transcriptional activity (Figure 1A and 1B), confirming that it was a transcription factor. The expression of MYB40 was significantly induced by As(V) stress, and the induction level of MYB40 was related to the duration of As(V) stress exposure (Figure 1B). Overexpression of MYB40 enhanced Arabidopsis As(V) resistance (Figure 2), and myb40 mutants showed As(V)-sensitive phenotypes (Figure 3), demonstrating that MYB40 was an As(V)-responsive transcription factor. The myb40 mutants did not show serious As(V)-sensitive phenotypes, and the expression levels of its target genes were altered by 42%–57% for PHT1;1 and by 32%–39% for PCS1 (Figures 4B and 5E). MYB40 has two transcripts, AT5G14340.1 and AT5G14340.2. Both myb40 mutants were disrupted at AT5G14340.1 but not at AT5G14340.2. We hypothesized that, because AT5G14340.2 also plays a role in Arabidopsis response to As(V) stress, knocking out both AT5G14340.1 and AT5G14340.2 might cause stronger phenotypes. The role of AT5G14340.2 under As(V) stress requires further experimental verification.
As a structural homolog of Pi, As(V) is taken up into plant cells through Pi transporters (Shin et al., 2004; Catarecha et al., 2007; Wu et al., 2011; Remy et al., 2012; Castrillo et al., 2013; DiTusa et al., 2016). The MYB40-overexpressing lines showed As(V)-tolerant phenotypes and had reduced As(V)/Pi uptake rates (Figure 2), suggesting that MYB40 enhanced Arabidopsis As(V) tolerance in part by repressing As(V)/Pi uptake. There are at least nine Pi transporters in Arabidopsis, and PHT1;1 is most highly expressed in roots of plants grown on Pi-sufficient medium (Mudge et al., 2002). During As(V) stress, the expression of PHT1;1 is clearly repressed (Castrillo et al., 2013). The pht1;1 mutant shows As(V)-tolerant phenotypes (Shin et al., 2004; Catarecha et al., 2007), and overexpression of PHT1;1 results in As(V) hypersensitivity (Wang et al., 2014), indicating that uptake of As(V) occurs mainly through the Pi transporter PHT1;1 in Arabidopsis. The expression of PHT1;1 was clearly repressed in the MYB40-overexpressing lines, and the repression level of PHT1;1 was closely related to MYB40 expression levels (Figure 4A). Upon exposure to As(V), PHT1;1 expression was repressed in wild-type plants, and this repression was abolished in the myb40 mutants (Figure 4B). Further EMSA and ChIP results showed that MYB40 could bind to the PHT1;1 promoter in vitro and in vivo (Figure 4E and 4F). All these data demonstrate that MYB40 can enhance Arabidopsis As(V) resistance at least in part through regulation of PHT1;1-dependent As(V)/Pi uptake.
A previous report demonstrated that the WRKY6 transcription factor restricts As(V) uptake in Arabidopsis (Castrillo et al., 2013). When grown on 15 μM Pi supplemented with 15 μM As(V) for 7 d, a WRKY6-overexpressing line displayed As(V)-resistant phenotypes, and no visible differences were observed in the wrky6 mutant relative to wild-type plants (Castrillo et al., 2013). WRKY6 can bind to the PHT1;1 promoter to repress PHT1;1 expression (Castrillo et al., 2013). When germinated and grown on ½ MS medium with 200 μM As(V), the WRKY6-overexpressing line and the wrky6 mutant showed no obvious differences compared with wild-type plants (data not shown), whereas the MYB40-overexpressing lines were markedly tolerant, and the myb40 mutants sensitive, to As(V) stress (Figures 2 and 3). This suggested that MYB40 and WRKY6 have different functions in Arabidopsis responses to As(V) stress. WRKY6 expression was clearly induced by As(V), and WRKY6 transcripts accumulated to their highest level after exposure to As(V) for 3 h (Castrillo et al., 2013). MYB40 was also induced by As(V) and, unlike WRKY6, its transcript level was still increased, even after exposure to As(V) for 72 h (Figure 1B). These data indicate that MYB40 and WRKY6 modulate Arabidopsis arsenate tolerance at different times.
After uptake into plant cells, As(V) is readily reduced to As(III), which is then detoxified by complexation with thiol-rich peptides and sequestered in the vacuoles (Zhao et al., 2010). PCS1 (also named CAD1) is the main PC synthase in Arabidopsis (Ha et al., 1999; Vatamaniuk et al., 1999), and the expression of PCS1 was clearly induced by As(V) (Sung et al., 2009). The cad1 mutant shows As(V)-hypersensitive phenotypes (Ha et al., 1999), and overexpression of PCS1 increases thiol-peptide accumulation and As tolerance (Lee et al., 2003; Li et al., 2004), indicating that transcriptional regulation of PCS1 is important for plant As(V) resistance. However, until now the mechanism has been unclear. Our results demonstrate that the MYB40 transcription factor positively regulates PCS1 by binding to the PCS1 promoter under As(V) stress. When plants were grown on ½ MS medium with added As(V), thiol-peptide contents were enhanced in the MYB40-overexpressing lines and reduced in the myb40 mutant relative to wild-type plants (Figure 5A–5C). The transcript level of PCS1 was clearly enhanced in the MYB40-overexpressing lines and reduced in the myb40 mutants when exposed to As(V) (Figure 5D and 5E). Furthermore, EMSA and ChIP results showed that MYB40 could bind to the PCS1 promoter in vitro and in vivo (Figure 5G and 5H). These data demonstrate that MYB40 elevates thiol-peptide contents by positively regulating PCS1 expression under As(V) stress.
The plant vacuoles are the final detoxification stores for As. ABCC1 and ABCC2 are the major vacuolar PC transporters in Arabidopsis, and the abcc1 abcc2 double mutant shows As(V)-hypersensitive phenotypes (Song et al., 2010). The ABCC1-overexpressing lines did not exhibit increased As(V) tolerance, whereas the PCS1 ABCC1 co-overexpression lines had increased As tolerance (Song et al., 2010). The transcript levels of ABCC1 and ABCC2 were significantly enhanced in the MYB40-overexpressing lines and reduced in the myb40 mutants under As(V) stress (Figure 6), indicating that MYB40 modulated the expression of ABCC1 and ABCC2. These data demonstrate that MYB40 plays important roles in As detoxification by positively regulating the expression of PCS1 and ABCC1/2, which function in a concerted way in the As detoxification pathway.
A previous report showed that the transcription factor WRKY6 downregulated PHT1;1 expression and restricted arsenate-induced transposon activation under As(V) stress (Castrillo et al., 2013). A WRKY6-GFP-overexpressing line showed markedly enhanced root length, whereas the wrky6-TNDA insertion line showed root length similar to that of wild-type plants under As(V) stress (Castrillo et al., 2013). Both MYB40 and WRKY6 directly downregulated PHT1;1 expression under As(V) stress. Both MYB40-overexpressing lines and WRKY6-GFP-overexpressing lines showed obvious As(V)-resistant tolerant phenotypes (Castrillo et al., 2013; Figures 2 and 4), and their mutants showed mild or no As(V)-sensitive phenotypes, probably owing to functional redundancy between MYB40 and WRKY6.
Transcription factor binding of cis-elements is often associated with accessible chromatin regions. Lu et al. (2017) reported that more than 90% of putative accessible chromatin regions were found within 3 kb upstream of a transcription start site (TSS), more than 75% of accessible chromatin regions were found within 1 kb upstream of the TSS, and accessible chromatin regions were mostly enriched around the TSS. In addition, the accessible chromatin region of PCS1 was within 500 bp upstream of the TSS of PCS1 (Lu et al., 2017). These results were obtained from whole seedlings or roots of Arabidopsis grown under normal conditions on ½ LS medium with 1% sugar under long day light conditions (16 h light/8 h dark) (Lu et al., 2017). In this work, under As(V) stress, MYB40 bound mainly to the 718–993 bp upstream of the TSS of PCS1 in vivo (Figure 5G). These data suggest that Arabidopsis PCS1 can be regulated by the binding of different transcription factors to specific DNA regions in response to environmental stimuli.
Methods
Plant materials and growth conditions
The wild-type plant used in this study was Arabidopsis thaliana Col-0. The PHT1;1 T-DNA insertion line Salk_088586c, named pht1;1, was ordered from the ABRC. The Super:PHT1;1 line used in the study was described previously (Wang et al., 2014). The MYB40-overexpressing lines were generated by cloning the coding sequence of MYB40 into the pCXSN vector under the 35S promoter (Chen et al., 2009a). The myb40 mutants were generated by CRISPR/Cas9 technology. A pair of sgRNA targets (C1: AACCGTGCTGTGACAAAATTGG; and C2: TCACTCTCAACTTGGCAACCG) in the MYB40 gene was selected and cloned into the pHEE2A-TRI vector (Wang et al., 2015). The MYB40-Myc transgenic line was generated by cloning the coding sequence of MYB40 into the pCAMBIA1300-Myc vector. The constructs were introduced into Arabidopsis by Agrobacterium-mediated transformation (Agrobacterium strain GV3101) using the floral-dip method (Clough and Bent, 1998), and homozygous transgenic lines were obtained.
Arabidopsis seeds were surface sterilized and kept at 4°C for 72 h in darkness before germination. Then, the seeds were plated on ½ MS medium containing 1.5% (w/v) sucrose and 0.8% (w/v) agar and grown at 22°C with 100 μmol m−2 s−1 of illumination for a 16-h daily light period, unless otherwise indicated.
For the As(V) treatment, sodium arsenate was added to the ½ MS medium at the described concentrations.
As content and As(V) uptake assay
Arabidopsis plants were germinated and grown on ½ MS medium with different concentrations of As(V) as described. Plants were dried, then mineralized with HNO3 in a pressure digester, and the As contents were determined by atomic fluorescence spectrometry.
For the As(V) uptake assay, 7-d-old seedlings grown on ½ MS medium were transferred to ½ MS with 200 μM As(V) for 6 h, and their As contents were measured.
Quantification of thiol-peptide content
Seven-d-old seedlings were transferred to ½ MS medium containing 200 μM As(V) for 7 d, then harvested for thiol-peptide content measurements. Thiol-peptide compounds, including GSH, PC2, PC3, and PC4, were analyzed using fluorescence-detection HPLC, essentially as described previously (Li et al., 2004).
Pi concentration and Pi uptake assay
Arabidopsis plants were germinated and grown on MS medium for 7 d, then harvested for Pi concentration measurement as described previously (Chen et al., 2009b).
For the Pi uptake assay, 7-d-old Arabidopsis seedlings grown on MS medium were transferred to Pi uptake solution containing 500 μM Pi supplemented with 0.2 μCi 32P orthophosphate. A group of 15 seedlings was used as one biological sample.
Subcellular localization
MYB40 fused to GFP was cloned into the pCAMBIA1300:GFP vector, named GFP, to create a MYB40–GFP construct. The plasmids (MYB40–GFP and GFP) were transformed into Agrobacterium GV3101, and transient expression assays were conducted as described previously (Chen et al., 2009b). GFP fluorescence in the transformed leaves was imaged using a confocal laser scanning microscope (Leica TCS SP5II).
qRT–PCR assay
qRT–PCR was performed using SYBR Green PCR Master Mix (Life Technologies) on a 7500 Real-Time PCR System (Applied Biosystems) following the manufacturer's protocol. Actin2/8 expression was used as an internal control. The qPCR analysis was performed with three technical replicates, and each experiment was repeated at least three times with similar results. The primers used are listed in Supplemental Table 1.
ChIP–qPCR assay
Seven-d-old MYB40-Myc seedlings were transferred to ½ MS medium containing 200 μM As(V) for 2 d, then harvested for the ChIP–qPCR assay. The ChIP–qPCR assay was conducted as described previously (Chen et al., 2009b), and the primers used are listed in Supplemental Table 1. Three independent experiments were performed with similar results. Data are mean values of three replicates ± SE from one experiment.
EMSA assay
The coding sequence of MYB40 was amplified and cloned into the pET-28a-SUMO vector (Novagen). The recombinant plasmid was introduced into E. coli strain BL21. The SUMO–His–MYB40 protein was purified using Ni-Sepharose 6 Fast Flow (GE Healthcare), and the protein concentration was determined by the Bio-Rad protein assay. The pET-28a-SUMO vector was also introduced into E. coli strain BL21, and protein with a SUMO–His tag was purified. This purified protein was named SUMO–His and was used as a control in EMSA.
The EMSA assay was conducted using a LightShift Chemiluminescent EMSA Kit (Pierce) following the manufacturer's protocol. The recombinant SUMO–His–MYB40 protein and SUMO–His protein were purified from E. coli. Fragments of the PHT1;1 and PCS1 promoters were obtained by PCR using biotin-labeled or unlabeled primers (Supplemental Table 1). Biotin-unlabeled fragments with the same sequences were used as competitors, and the SUMO–His protein alone was used as the negative control.
Funding
This work was supported by grants from the National Natural Science Foundation of China (nos. 31970273 and 31670245) and the Beijing Outstanding University Discipline Program.
Author contributions
Y.C. and H.-Y.W. conducted the experiments. Y.-F.C. designed the experiments. Y.C., H.-Y.W., and Y.-F.C. analyzed the data and wrote the article.
Acknowledgments
We thank Dr. Zhen Li for assistance with thiol-peptide content measurement. We thank all laboratory members for their valuable comments. The authors declare no conflicts of interest.
Published: August 19, 2021
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Accession numbers
Sequence data from this article can be found in the EMBL and GenBank data libraries under the following accession numbers: MYB40 (TAIR: AT5G14340), PHT1;1 (TAIR: AT5G43350), PCS1 (TAIR: AT5G44070), ABCC1 (TAIR: AT1G30400), ABCC2 (TAIR: AT2G34660), and ATQ1 (TAIR: AT1G492252G21045).
Supplemental information
References
- Abercrombie J.M., Halfhill M.D., Ranjan P., Rao M.R., Saxton A.M., Yuan J.S., Stewart C.N., Jr. Transcriptional responses of Arabidopsis thaliana plants to As(V) stress. BMC Plant Biol. 2008;8:87. doi: 10.1186/1471-2229-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrowicz P., Wysocki R., Owsianik G., Goffeau A., Ułaszewski S. Isolation of three contiguous genes, ACR1, ACR2 and ACR3, involved in resistance to arsenic compounds in the yeast Saccharomyces cerevisiae. Yeast. 1997;13:819–828. doi: 10.1002/(SICI)1097-0061(199707)13:9<819::AID-YEA142>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Castrillo G., Sanchez-Bermejo E., de Lorenzo L., Crevillen P., Fraile-Escanciano A., Tc M., Mouriz A., Catarecha P., Sobrino-Plata J., Olsson S. WRKY6 transcription factor restricts arsenate uptake and transposon activation in Arabidopsis. Plant Cell. 2013;25:2944–2957. doi: 10.1105/tpc.113.114009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catarecha P., Segura M.D., Franco-Zorrilla J.M., Garcia-Ponce B., Lanza M., Solano R., Paz-Ares J., Leyva A. A mutant of the Arabidopsis phosphate transporter PHT1;1 displays enhanced arsenic accumulation. Plant Cell. 2007;19:1123–1133. doi: 10.1105/tpc.106.041871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao D.Y., Chen Y., Chen J., Shi S., Chen Z., Wang C., Danku J.M., Zhao F.J., Salt D.E. Genome-wide association mapping identifies a new arsenate reductase enzyme critical for limiting arsenic accumulation in plants. PLoS Biol. 2014;12:e1002009. doi: 10.1371/journal.pbio.1002009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Songkumarn P., Liu J., Wang G.L. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 2009;150:1111–1121. doi: 10.1104/pp.109.137125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.F., Li L.Q., Xu Q., Kong Y.H., Wang H., Wu W.H. The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell. 2009;21:3554–3566. doi: 10.1105/tpc.108.064980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- DiTusa S.F., Fontenot E.B., Wallace R.W., Silvers M.A., Steele T.N., Elnagar A.H., Dearman K.M., Smith A.P. A member of the phosphate transporter 1 (Pht1) family from the arsenic-hyperaccumulating fern Pteris vittata is a high-affinity arsenate transporter. New Phytol. 2016;209:762–772. doi: 10.1111/nph.13472. [DOI] [PubMed] [Google Scholar]
- Duan G.L., Hu Y., Schneider S., McDermott J., Chen J., Sauer N., Rosen B.P., Daus B., Liu Z., Zhu Y.G. Inositol transporters AtINT2 and AtINT4 regulate arsenic accumulation in Arabidopsis seeds. Nat. Plants. 2015;2:15202. doi: 10.1038/nplants.2015.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan G.L., Zhou Y., Tong Y.P., Mukhopadhyay R., Rosen B.P., Zhu Y.G. A CDC25 homologue from rice functions as an arsenate reductase. New Phytol. 2007;174:311–321. doi: 10.1111/j.1469-8137.2007.02009.x. [DOI] [PubMed] [Google Scholar]
- Ellis D.R., Gumaelius L., Indriolo E., Pickering I.J., Banks J.A., Salt D.E. A novel arsenate reductase from the arsenic hyperaccumulating fern Pteris vittata. Plant Physiol. 2006;141:1544–1554. doi: 10.1104/pp.106.084079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh M., Shen J., Rosen B.P. Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U S A. 1999;96:5001–5006. doi: 10.1073/pnas.96.9.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González E., Solano R., Rubio V., Leyva A., Paz-Ares J. PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell. 2005;17:3500–3512. doi: 10.1105/tpc.105.036640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill E., Winnacker E.L., Zenk M.H. Phytochelatins, a class of heavy-metal-binding peptides from plants, are functionally analogous to metallothioneins. Proc. Natl. Acad. Sci. U S A. 1987;84:439–443. doi: 10.1073/pnas.84.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S.B., Smith A.P., Howden R., Dietrich W.M., Bugg S., O'Connell M.J., Goldsbrough P.B., Cobbett C.S. Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe. Plant Cell. 1999;11:1153–1164. doi: 10.1105/tpc.11.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden R., Goldsbrough P.B., Andersen C.R., Cobbett C.S. Cadmium-sensitive, cad1 mutants of Arabidopsis thaliana are phytochelatin deficient. Plant Physiol. 1995;107:1059–1066. doi: 10.1104/pp.107.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Dubey R.S., Tripathi R.D., Chakrabarty D., Trivedi P.K. Omics and biotechnology of arsenic stress and detoxification in plants: current updates and prospective. Environ. Int. 2015;74:221–230. doi: 10.1016/j.envint.2014.10.019. [DOI] [PubMed] [Google Scholar]
- Lee S., Moon J.S., Ko T.S., Petros D., Goldsbrough P.B., Korban S.S. Overexpression of Arabidopsis phytochelatin synthase paradoxically leads to hypersensitivity to cadmium stress. Plant Physiol. 2003;131:656–663. doi: 10.1104/pp.014118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Dhankher O.P., Carreira L., Lee D., Chen A., Schroeder J.I., Balish R.S., Meagher R.B. Overexpression of phytochelatin synthase in Arabidopsis leads to enhanced arsenic tolerance and cadmium hypersensitivity. Plant Cell Physiol. 2004;45:1787–1797. doi: 10.1093/pcp/pch202. [DOI] [PubMed] [Google Scholar]
- Lu Z., Hofmeister B.T., Christopher V., Dubois R.M., Schmitz R.J. Combining ATAC-seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Res. 2017;45:e41. doi: 10.1093/nar/gkw1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.F., Yamaji N., Mitani N., Xu X.Y., Su Y.H., McGrath S.P., Zhao F.J. Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc. Natl. Acad. Sci. U S A. 2008;105:9931–9935. doi: 10.1073/pnas.0802361105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge S.R., Rae A.L., Diatloff E., Smith F.W. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 2002;31:341–353. doi: 10.1046/j.1365-313x.2002.01356.x. [DOI] [PubMed] [Google Scholar]
- Pickering I.J., Prince R.C., George M.J., Smith R.D., George G.N., Salt D.E. Reduction and coordination of arsenic in Indian mustard. Plant Physiol. 2000;122:1171–1177. doi: 10.1104/pp.122.4.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy E., Cabrito T.R., Batista R.A., Teixeira M.C., Sa-Correia I., Duque P. The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 2012;195:356–371. doi: 10.1111/j.1469-8137.2012.04167.x. [DOI] [PubMed] [Google Scholar]
- Sánchez-Bermejo E., Castrillo G., del Llano B., Navarro C., Zarco-Fernández S., Martinez-Herrera D.J., Leo-del Puerto Y., Muñoz R., Cámara C., Paz-Ares J. Natural variation in arsenate tolerance identifies an arsenate reductase in Arabidopsis thaliana. Nat. Commun. 2014;5:4617. doi: 10.1038/ncomms5617. [DOI] [PubMed] [Google Scholar]
- Schmöger M.E.V., Oven M., Grill E. Detoxification of arsenic by phytochelatins in plants. Plant Physiol. 2000;122:793–801. doi: 10.1104/pp.122.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Shin H.S., Dewbre G.R., Harrison M.J. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 2004;39:629–642. doi: 10.1111/j.1365-313X.2004.02161.x. [DOI] [PubMed] [Google Scholar]
- Sneller F.E.C., Heerwaarden L.M.V., Kraaijeveld-smit F.J.L., Bookum W.M.T., Koevoets P.L.M., Schat H., Verkleij J.A.C. Toxicity of arsenate in Silene vulgaris, accumulation and degradation of arsenate-induced phytochelatins. New Phytol. 1999;144:223–232. [Google Scholar]
- Song W.Y., Park J., Mendoza-Cozatl D.G., Suter-Grotemeyer M., Shim D., Hortensteiner S., Geisler M., Weder B., Rea P.A., Rentsch D. Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc. Natl. Acad. Sci. U S A. 2010;107:21187–21192. doi: 10.1073/pnas.1013964107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.Y., Yamaki T., Yamaji N., Ko D., Jung K.H., Fujii-Kashino M., An G., Martinoia E., Lee Y., Ma J.F. A rice ABC transporter, OsABCC1, reduces arsenic accumulation in the grain. Proc. Natl. Acad. Sci. U S A. 2014;111:15699–15704. doi: 10.1073/pnas.1414968111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Sung D.Y., Kim T.H., Komives E.A., Mendoza-Cózatl D.G., Schroeder J.I. ARS5 is a component of the 26S proteasome complex, and negatively regulates thiol biosynthesis and arsenic tolerance in Arabidopsis. Plant J. 2009;59:802–813. doi: 10.1111/j.1365-313X.2009.03914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T., Xu Q., Zhang F.C., Chen Y., Li L.Q., Wu W.H., Chen Y.F. WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acquisition in Arabidopsis. Plant Physiol. 2015;167:1579–1591. doi: 10.1104/pp.114.253799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatamaniuk O.K., Mari S., Lu Y.P., Rea P.A. AtPCS1, a phytochelatin synthase from Arabidopsis: isolation and in vitro reconstitution. Proc. Natl. Acad. Sci. U S A. 1999;96:7110–7115. doi: 10.1073/pnas.96.12.7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Xu Q., Kong Y.H., Chen Y., Duan J.Y., Wu W.H., Chen Y.F. Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiol. 2014;164:2020–2029. doi: 10.1104/pp.113.235077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.P., Xing H.L., Dong L., Zhang H.Y., Han C.Y., Wang X.C., Chen Q.J. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015;16:144. doi: 10.1186/s13059-015-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Ren H., McGrath S.P., Wu P., Zhao F.J. Investigating the contribution of the phosphate transport pathway to arsenic accumulation in rice. Plant Physiol. 2011;157:498–508. doi: 10.1104/pp.111.178921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F.J., McGrath S.P., Meharg A.A. Arsenic as a food chain contaminant: mechanisms of plant uptake and metabolism and mitigation strategies. Annu. Rev. Plant Biol. 2010;61:535–559. doi: 10.1146/annurev-arplant-042809-112152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.