Abstract
Plant nucleotide-binding leucine-rich repeat (NLR) receptors mediate immune responses by directly or indirectly sensing pathogen-derived effectors. Despite significant advances in the understanding of NLR-mediated immunity, the mechanisms by which pathogens evolve to suppress NLR activation triggered by cognate effectors and gain virulence remain largely unknown. The agronomically important immune receptor RB recognizes the ubiquitous and highly conserved IPI-O RXLR family members (e.g., IPI-O1) from Phytophthora infestans, and this process is suppressed by the rarely present and homologous effector IPI-O4. Here, we report that self-association of RB via the coiled-coil (CC) domain is required for RB activation and is differentially affected by avirulence and virulence effectors. IPI-O1 moderately reduces the self-association of RB CC, potentially leading to changes in the conformation and equilibrium of RB, whereas IPI-O4 dramatically impairs CC self-association to prevent RB activation. We also found that IPI-O1 associates with itself, whereas IPI-O4 does not. Notably, IPI-O4 interacts with IPI-O1 and disrupts its self-association, therefore probably blocking its avirulence function. Furthermore, IPI-O4 enhances the interaction between RB CC and IPI-O1, possibly sequestering RB and IPI-O1 and subsequently blocking their interactions with signaling components. Taken together, these findings considerably extend our understanding of the underlying mechanisms by which emerging virulent pathogens suppress the NLR-mediated recognition of cognate effectors.
Key words: RB, nucleotide-binding leucine-rich repeat receptor, self-association, coiled-coil domain, Phytophthora infestans, effector
The virulence effector IPI-O4 blocks NLR RB-mediated recognition of its cognate effector IPI-O1 by suppressing the self-association of both the RB coiled-coil domain and IPI-O1. IPI-O1 and IPI-O4 exhibit striking differences in subcellular localization, self-association status, and effects on RB self-association. IPI-O4 probably sequesters the RB-IPI-O1 complex and subsequently blocks the interactions of these proteins with signaling components to prevent RB activation.
Introduction
Plants are constantly exposed to a wide range of microbial pathogens and have developed effective and sophisticated defense mechanisms to cope with pathogen attacks. The first layer of plant defense is mediated by cell-surface-localized pattern-recognition receptors (PRRs) that recognize highly conserved pathogen-associated molecular patterns (PAMPs) to induce PAMP-triggered immunity (PTI) (Jones and Dangl, 2006). Microbial pathogens have evolved virulence proteins known as effectors that interfere with host immune responses. Correspondingly, plants have developed intracellular nucleotide-binding leucine-rich repeat (NLR) receptors that detect their cognate avirulence effectors and induce the second layer of plant defense, termed effector-triggered immunity (ETI). This rapid and robust activation of defense signaling pathways shows significant overlap with PTI. ETI is often associated with localized programmed cell death, known as the hypersensitive response (HR) that restricts pathogen growth (Jones and Dangl, 2006; Jones et al., 2016). Two recent studies suggest that PTI and ETI mutually potentiate each other to mount stronger immune responses against invading pathogens (Ngou et al., 2021; Yuan et al., 2021).
The presence of Toll/interleukin-1 receptor (TIR), coiled-coil (CC), or RPW8-like CC-type motifs in the N-terminal domains of plant NLR proteins defines three major subfamilies, the TNLs, CNLs, and RNLs (Shao et al., 2016). The N-terminal CC domains have been implicated in the recognition of cofactors and cognate effectors (Rairdan et al., 2008; Chen et al., 2012; Lewis et al., 2013) or the mediation of downstream signaling (Rairdan et al., 2008; Maekawa et al., 2011; Wang et al., 2015), leading to the activation of immune responses. The CC domains alone of several NLRs are sufficient to induce cell death; these include NRG1 from Nicotiana benthamiana (Qi et al., 2018), ADR1 from Arabidopsis (Collier et al., 2011), Rx from tomato (Rairdan et al., 2008), Rp1-D21 from maize (Wang et al., 2015), Sr33 and Sr35 from wheat (Casey et al., 2016), and MLA10 from barley (Maekawa et al., 2011). Plant NLRs contain a central, highly conserved nucleotide-binding adaptor shared by APAF-1, certain R gene products, and the CED-4 (NB-ARC) domain, which acts as a molecular switch that defines the activation state of NLRs (Tameling et al., 2002; Leipe et al., 2004; Williams et al., 2014). The NB-ARC domain has been proposed to participate in the initiation of downstream signaling to activate immunity (Ade et al., 2007; Rairdan et al., 2008; De Oliveira et al., 2016; Hu et al., 2017). The C-terminal LRR domain determines effector recognition specificity for several NLRs (Dodds et al., 2001; Ravensdale et al., 2012), and in some cases, it mediates effector binding (Jia et al., 2000; Krasileva et al., 2010). Dimerization or higher-order oligomerization through N-terminal CC or TIR domains has been reported in a number of NLRs and was proposed to be a common signaling event that activates downstream defense responses (Mestre and Baulcombe, 2006; Ade et al., 2007; Gutierrez et al., 2010; Maekawa et al., 2011; Chen et al., 2012; Magnan and Baldi, 2014). The recently solved structures of ZAR1, RPP1, and ROQ1 reveal that plant NLRs form tetrameric or pentameric resistosome complexes to trigger NLR activation. These findings have substantially advanced our understanding of the biochemical mechanism that underlies NLR activation and signaling (Wang et al., 2019a, 2019b; Ma et al., 2020; Martin et al., 2020).
Phytophthora infestans, the causal agent of the devastating late blight disease, secretes a diverse array of effector proteins to manipulate host defense responses and facilitate colonization. All oomycete avirulence effectors identified to date belong to the RXLR family with the N-terminal Arg-X-Leu-Arg motif, which is required for translocation into plant cells (Whisson et al., 2007; Win et al., 2007; Jiang et al., 2008). The late blight resistance gene RB (also known as Rpi-blb1), cloned from the wild potato species Solanum bulbocastanum, encodes a CNL and confers broad-spectrum resistance to a wide range of P. infestans strains (Song et al., 2003; Van Der Vossen et al., 2003; Bhaskar et al., 2008). The majority of the in planta-induced O (IPI-O) type of RXLR effectors are recognized by RB and its orthologs in wild potato species, such as S. stoloniferum and S. papita, thereby triggering immune responses (Vleeshouwers et al., 2008; Wang et al., 2008; Champouret et al., 2009; Xie et al., 2015). The IPI-O family effectors are highly diverse and are grouped into three classes based on their amino acid sequences (Pieterse et al., 1994; Champouret et al., 2009; Halterman et al., 2010). Two closely related class I effectors, IPI-O1 and IPI-O2, and the class II member IPI-O3 all induce HR when co-expressed with RB in potato and N. benthamiana plants, whereas the class III effector IPI-O4 is not recognized by RB and instead suppresses the RB-mediated HR induced by IPI-O1 (Champouret et al., 2009; Halterman et al., 2010; Chen and Halterman, 2011; Chen et al., 2012). A survey of a large number of P. infestans isolates collected from Central America, Southeast Asia, and the United States found that IPI-O1 and IPI-O2 were present in all isolates, with the exception of US1, which lacks IPI-O2, whereas IPI-O4 was much rarer and was detected in only 6 of 41 isolates (Champouret et al., 2009; Halterman et al., 2010).
Although the exact function of IPI-O1 is unknown, its ubiquitous presence suggests that it is essential for pathogen fitness and plays a major role in pathogenesis (van West et al., 1998; Chen and Halterman, 2017). This reinforces the possibility that RB may remain effective against P. infestans during its widespread deployment in cultivated varieties. Furthermore, transgenic plants carrying RB exhibit a limited level of susceptibility to P. infestans, which can sporulate to a small degree. A field test of these stable RB potato lines showed markedly increased resistance against late blight in Toluca Valley, Mexico, the center of origin and diversity of P. infestans (Lozoya-Saldana et al., 2005; Bhaskar et al., 2008). The so-called partial or rate-limiting resistance conferred by RB exerts less selection pressure that may not elicit rapid pathogen adaptation and may thus contribute to more extended durability (Joseph et al., 2007; Halterman et al., 2008; Chen and Halterman, 2011; Halterman and Middleton, 2012). RB and its homologs from wild potato species constitute a wealth of resources for late blight resistance breeding (Tiwari et al., 2015) and have been widely used for marker-assisted selection of late blight resistance in potato (Colton et al., 2006; Pankin et al., 2011; Sokolova et al., 2011; Tiwari et al., 2013). A transgenic approach combined with conventional breeding has been shown to effectively enhance and broaden resistance against P. infestans with little or no yield cost (Shandil et al., 2017; Mambetova et al., 2018). Several recent studies show that cisgenic transformation of RB or stacking of RB with multiple late blight resistance genes, such as Rpi-blb2, Rpi-blb3, and Rpi-vnt1.1, provided significantly improved or even complete resistance against late blight (Jo et al., 2014; Haesaert et al., 2015; Ghislain et al., 2019; Rakosy-Tican et al., 2020).
Although significant progress has been made in understanding NLR-mediated recognition of cognate effectors and activation of immune responses, the underlying mechanisms by which these processes are overcome by pathogens are largely unknown. The present RB/IPI-O1/IPI-O4 system provides an ideal and unique model with which to investigate how ETI is inhibited by a virulence effector. It was previously shown that the RB CC domain associates with itself and with the two effectors IPI-O1 and IPI-O4. Even in the presence of IPI-O1, RB-mediated resistance continues to be suppressed by IPI-O4 (Chen et al., 2012). However, it remains unclear how IPI-O4 blocks RB-mediated recognition of IPI-O1. In this study, we reveal a previously unrecognized mechanism in which the virulence effector IPI-O4 suppresses NLR RB-mediated recognition of its cognate effector IPI-O1 by impairing the self-association of both RB CC and IPI-O1, which probably prevents the formation of interaction interfaces required for the assembly of the resistosome complex. We found that, despite their high homology, IPI-O1 and IPI-O4 exhibit striking differences in subcellular localization, self-association status, and effects on RB self-association. Furthermore, RB, IPI-O1, and IPI-O4 associate with each other, and IPI-O4 promotes the interaction between RB CC and IPI-O1. Our results highlight how an evolved virulence effector overcomes NLR-mediated recognition of its cognate effector.
Results
RB, IPI-O1, and IPI-O4 function in distinct subcellular compartments
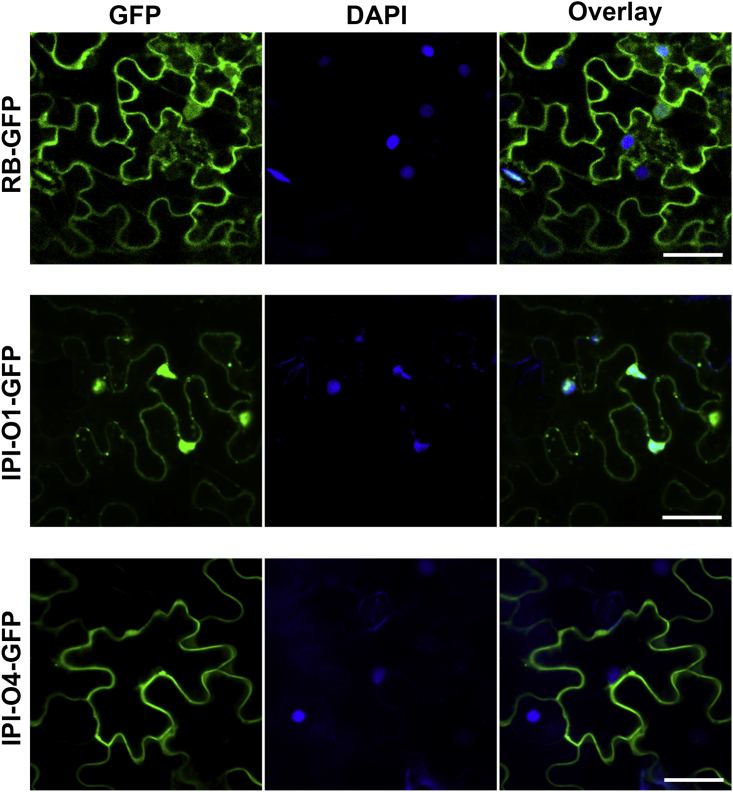
To determine the subcellular localization of RB, its cognate avirulence effector IPI-O1, and the virulence effector IPI-O4, we constructed C-terminal GFP-tagged RB, IPI-O1, and IPI-O4 and expressed them in N. benthamiana. Fluorescence microscopy showed that RB was primarily localized in the cytoplasm and the nucleus. Similarly, its cognate effector IPI-O1 was also detected in both the cytoplasm and the nucleus. By contrast, unlike RB and IPI-O1, the virulence effector IPI-O4 appears to be exclusively localized in the cytoplasm (Figure 1), suggesting that IPI-O1 and IPI-O4 may function in different subcellular compartments.
Figure 1.
Subcellular localizations of RB, IPI-O1, and IPI-O4.
Microscopy analysis shows that both RB and IPI-O1 are localized in the cytoplasm and the nucleus, whereas IPI-O4 is primarily localized in the cytoplasm. RB, IPI-O1, or IPI-O4 fused to GFP at the C terminus was expressed in N. benthamiana. Confocal images were taken at 48 h post infiltration (hpi) (left). DAPI staining depicts nuclei in blue (middle). An overlay of GFP and DAPI fluorescence signals is shown on the right. Scale bars correspond to 50 μm. The experiments were repeated three times with similar results.
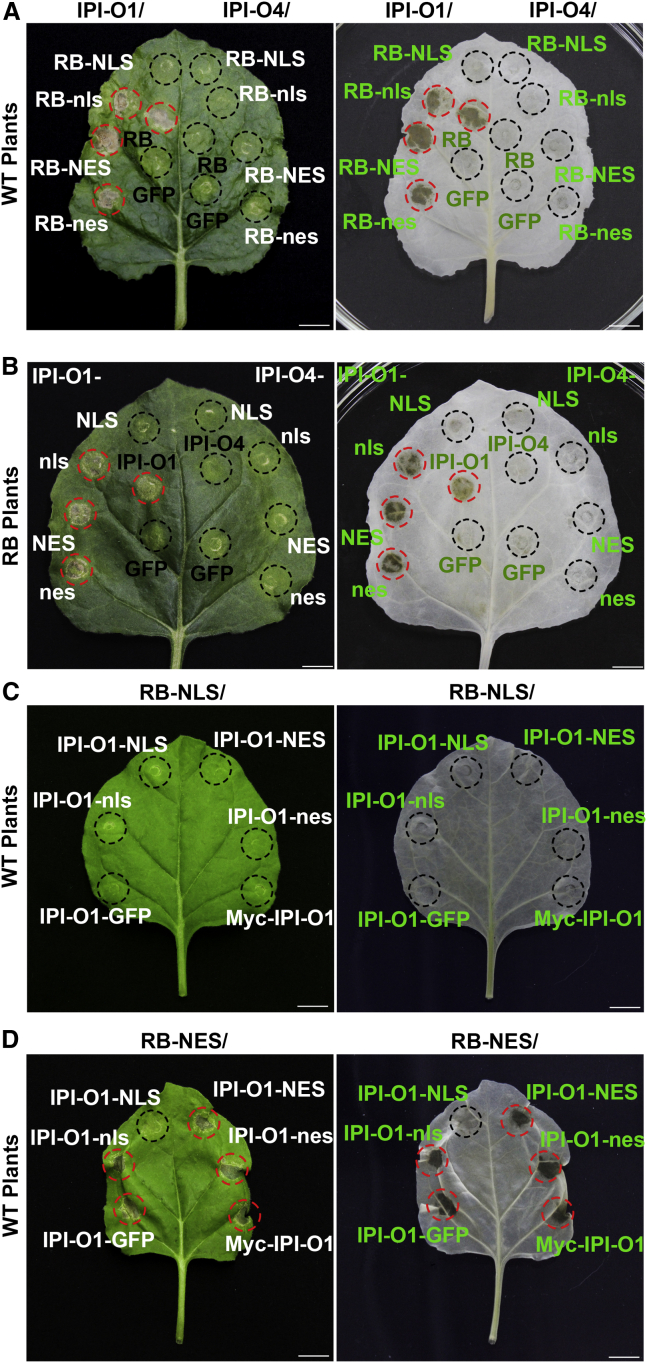
To investigate the impacts of subcellular localization of RB and IPI-O1 on their functions, RB was fused to GFP, followed by a nuclear localization sequence (NLS) from the large T antigen of the SV40 virus or a mutated nonfunctional NLS (nls) that served as a negative control. Expression of RB-GFP-NLS or RB-GFP-nls in N. benthamiana revealed that the fluorescence signal of RB-GFP-NLS was exclusively localized in the nucleus, whereas that of RB-GFP-nls was localized in both the cytoplasm and the nucleus (Supplemental Figure 1). When co-expressed with IPI-O1, RB-GFP-NLS was unable to trigger HR, but RB-GFP-nls retained HR-inducing activity (Figure 2A). Next, we fused RB to a nuclear export signal sequence (NES) derived from a human immunodeficiency virus-1 Rev protein or to a mutated NES (nes) in which critical leucine residues were substituted with alanines. As expected, RB-GFP-NES was excluded from the nucleus and detected only in the cytoplasm, whereas RB-GFP-nes was localized in both the nucleus and the cytoplasm (Supplemental Figure 1). In the presence of IPI-O1, RB-GFP was able to elicit HR when fused to either the intact NES or the nonfunctional nes (Figure 2A). These data indicate that the cytoplasmic pool of RB alone is sufficient for HR elicitation.
Figure 2.
Cytoplasmic localization of RB and IPI-O1 is required for their functions.
(A) Cytoplasmic localization of RB is required for IPI-O1-triggered HR. RB fused at the C terminus to GFP together with WT or mutated nuclear localization or export signals (NLS/nls or NES/nes) was co-expressed with either Myc-IPI-O1 or Myc-IPI-O4 in N. benthamiana.
(B) Cytoplasmic localization of IPI-O1 is required for RB-mediated HR. IPI-O1 or IPI-O4 fused at the C terminus to GFP together with WT or mutated NLS/nls or NES/nes was expressed in RB transgenic N. benthamiana plants.
(C and D) Cytoplasmic localization of both RB and IPI-O1 is required for their HR activities. RB and IPI-O1 fusion constructs from (A and B), respectively, were co-expressed in the indicated pairwise combinations in N. benthamiana plants. Myc-IPI-O1 was included as a control for the HR assays. Cell death induced at 48 hpi was visualized before (left) and after ethanol destaining (right). The infiltrated area is shown with a black circle and HR with a red circle. Scale bars correspond to 1 cm. All the experiments were repeated at least six times with similar results.
To determine the role of the subcellular location of IPI-O effectors in RB-mediated HR, IPI-O1 and IPI-O4 were fused to GFP together with functional or nonfunctional NLS/NES signal peptides and transiently expressed in RB transgenic N. benthamiana plants. Similar to the RB fusions, exclusively nuclear-localized IPI-O1-GFP-NLS failed to induce HR, but IPI-O1-nls retained HR-inducing activity (Figure 2B and Supplemental Figure 2). When IPI-O1-GFP was fused to NES, it localized predominantly to the cytoplasm and still triggered HR. Consistently, IPI-O1 tagged with the mutated nes showed the same localization as RB-GFP and induced HR (Figure 2B and Supplemental Figure 2). By contrast, IPI-O4 was unable to induce RB-mediated HR, regardless of whether it was fused to functional or nonfunctional NLS/NES (Figure 2B). These results suggest that cytoplasmic localization is essential for the elicitation activity of IPI-O1.
To further demonstrate that cytoplasmic localization of both RB and IPI-O1 is required to trigger HR, different combinations of RB and IPI-O1 fusion proteins with NLS or NES were co-expressed in plants. Consistently, the HR occurred only when RB and IPI-O1 were both targeted to the cytoplasm (Figure 2C and 2D). In addition, we fused RB and IPI-O1 to the hormone-binding domain of the mammalian glucocorticoid receptor (GR) that can sequester fusion proteins predominantly in the cytoplasm (Dittmar et al., 1997; García et al., 2010; Bai et al., 2012; Xu et al., 2014). The RB-GFP-GR fusion mediated a clear HR phenotype in the presence of IPI-O1 (Supplemental Figure 4A). Similar to the RB-GFP-GR fusion, IPI-O1 fused to GR was capable of mounting RB-mediated HR in the RB transgenic N. benthamiana plants (Supplemental Figure 4B). Immunoblotting showed that all RB and IPI-O1 fusion proteins accumulated to detectable levels (Supplemental Figure 4C and 4D). These results again confirmed that the cytoplasmic localization of RB and IPI-O1 is sufficient to induce RB-mediated HR.
Targeting RB or IPI-O1 to the plasma membrane compromises their HR-inducing activities
IPI-O effectors have been proposed to disrupt the cell wall–plasma membrane (PM) association in plants via the RGD motif to promote infection (Bouwmeester et al., 2011; Gouget et al., 2006; Liu et al., 2015; Senchou et al., 2004). A co-localization analysis using the mCherry-tagged PM-localized marker NbHir3.1 (Li et al., 2019) indicated that IPI-O1 and IPI-O4, but not RB, are partially localized to the PM (Supplemental Figure 5). These findings, together with evidence that RB associates with IPI-O1 and IPI-O4 (Chen et al., 2012), prompted us to investigate whether RB, IPI-O1, and IPI-O4 have any PM-associated functions. To examine the impact of exclusive PM localization on RB function, we fused RB to the PM association domain of Rop10 to restrict RB to the PM. The C-terminal domain of Rop10, a member of the ROP small GTPases in Arabidopsis, is sufficient for association with the PM via S-acylation. A substitution of the non-polar residues LSNIL with the charged residues REDER blocks an S-acylation-dependent membrane association (Lavy and Yalovsky, 2006; Chen et al., 2017). When co-expressed with IPI-O1, RB fused to the Rop10 tag did not induce significant HR cell death compared with the unfused RB-GFP control, whereas RB fused to the mutated Rop10 tag (mRop10) triggered normal HR cell death (Supplemental Figure 6A).
Next, we targeted IPI-O1 to the PM by fusing it to the Rop10 tag and expressed the fusion in RB transgenic N. benthamiana plants. The PM-targeted IPI-O1-GFP-Rop10 failed to induce HR. By contrast, IPI-O1 fused to an mRop10 tag retained HR-inducing activity (Supplemental Figure 6B). Immunoblotting showed that all RB and IPI-O1 fusion proteins accumulated to detectable levels (Supplemental Figure 6C and 6D). These results indicate that the forced PM association of RB or IPI-O1 significantly disrupts their HR activities.
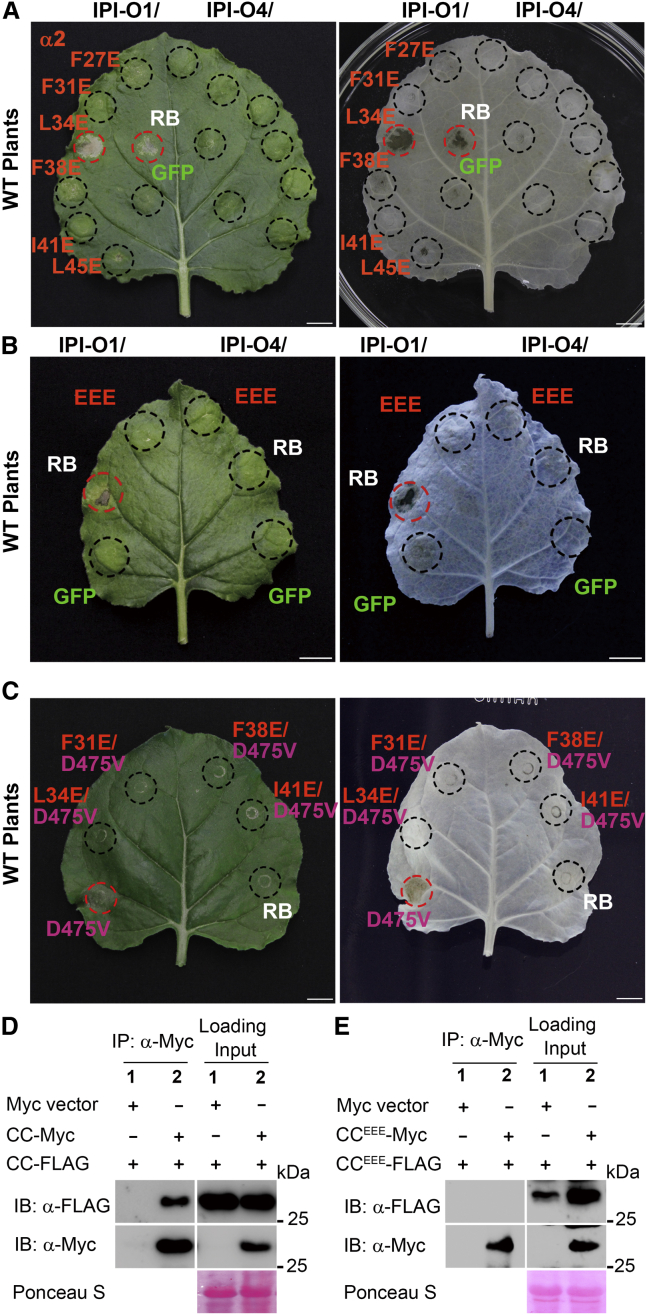
The heptad repeats of the RB CC domain are required for self-association and HR induction
The RB CC domain associates with itself but not with other domains of RB (Chen et al., 2012). The CC domains are characterized by heptad repeats of hydrophobic residues that form binding surfaces for α helices and secondary structures involved in helix–helix associations (Chambers et al., 1990; Maekawa et al., 2011; Mason and Arndt, 2004; Wróblewski et al., 2018). Secondary and 3D structure modeling of the CC domains from RB and other NLRs containing EDVID motifs, including MLA10, Sr33, Rx, and ZAR1, with well-defined patterns obtained by crystallization, NMR, or cryoelectron microscopy structural analysis, predicted the presence of four α helices consisting of heptad repeats of hydrophobic amino acids (Supplemental Figures 7 and 8) (Bentham et al., 2018; Casey et al., 2016; Hao et al., 2013; Maekawa et al., 2011; Wang et al., 2019a, 2019b). The structure and pattern of the heptad repeats are essential for the α-helical arrangement and CC self-association, and substitution of their amino acids with hydrophilic amino acids may thermodynamically destabilize the potential for self-association (Bai et al., 2012; Bentham et al., 2018; Maekawa et al., 2011; Wróblewski et al., 2018). To examine whether the α-helical conformation and self-association are essential for RB function, we replaced the hydrophobic residues with hydrophilic glutamic acids in the first (L10E, L13E, L17E, L21E, and L24E), second (F27E, F31E, L34E, F38E, I41E, and L45E), third (L62E, L65E, and V72E), or fourth α helix (V98E, F101E, L116E, L119E, and F126E) of the RB CC domain (Figure 3A and Supplemental Figure 9). Most RB variants, except for L34E and F101E, diminished or completely abrogated HR induction when co-expressed with IPI-O1, whereas none of the RB variants induced HR in the presence of IPI-O4 (Figure 3A and Supplemental Figure 9). In the second α helix, F31, L34, and I41 are conserved hydrophobic residues of the CC domain and are reported to be essential for the function and self-association of RPM1 (El Kasmi et al., 2017). We therefore tested the HR-inducing activity of the RB variant that carried the F31E/L34E/I41E (EEE) triple mutation in the α2 helix. As expected, the triple mutant failed to activate HR in the presence of IPI-O1 or IPI-O4 (Figure 3B). These data suggest that the heptad repeats are required for RB-mediated HR.
Figure 3.
Mutations in the heptad repeats of RB abolish its function and disrupt self-association of the RB CC domain
(A) RB and its variants carrying mutations in the heptad repeats from the second α helix were each co-expressed with Myc-IPI-O1 or Myc-IPI-O4 in N. benthamiana.
(B) RB and the F31E/L34E/I41E triple mutant (EEE) were each co-expressed with Myc-IPI-O1 or Myc-IPI-O4 in N. benthamiana.
(C) Cell death-inducing activity of the RB variants carrying the autoactive D475V mutation in the MHD motif in combination with mutations in the heptad repeats of RB CC. Cell death induced at 48 hpi was visualized before (left) and after ethanol destaining (right). The infiltrated area is shown with a black circle and HR with a red circle. Scale bars correspond to 1 cm. The experiments in (A, B, and C) were repeated at least six times with similar results.
(D) The RB CC domain associates with itself in vivo. Total proteins were extracted from N. benthamiana plants expressing RB CC-3×FLAG and RB CC-4×Myc. Immunoprecipitation was performed with an anti-Myc antibody, and immunoblots were probed with anti-Myc or anti-FLAG antibodies.
(E) The CC domain carrying the triple F31E/L34E/I41E mutation (CCEEE) is unable to self-associate in vivo. Total proteins were extracted from N. benthamiana plants expressing CCEEE-3×FLAG and CCEEE-4×Myc. Immunoprecipitation was performed with an anti-Myc antibody, and immunoblots were probed with anti-Myc or anti-FLAG antibodies. Ponceau S staining of immunoblots served as a loading control. The experiments in (D and E) were repeated three times with similar results.
To further examine the requirement for the heptad repeats in the RB-mediated immune response, we used an RB autoactivation variant produced by MHD D475V mutation. Transient expression assays revealed that the D475V variant autoactivated in the absence of IPI-O1 (Supplemental Figure 10A) and even in the presence of IPI-O4 (Supplemental Figure 10B). We next introduced the D475V mutation into the context of the heptad repeat variants, including F31E, L34E, F38E, and I41E, and we monitored their cell death-inducing activities. All four heptad repeat variants suppressed effector-independent autoactivation mediated by the RB variant D475V (Figure 3C). Immunoblotting showed that the D475V variant accumulated to a detectable level (Supplemental Figure 11). These findings indicate that the autoimmunity induced by RB D475V is dependent on the formation of a proper structure of α helices maintained by heptad repeats within the CC domain.
Dimerization or higher-order oligomerization through N-terminal CC domains has been reported for a number of NLRs and is thought to be a signaling event that activates downstream defense responses (Mestre and Baulcombe, 2006; Ade et al., 2007; Gutierrez et al., 2010; Maekawa et al., 2011; Chen et al., 2012; Wang et al., 2020). To investigate whether substitutions of hydrophobic residues in the heptad repeats perturb RB CC self-association, co-immunoprecipitation (co-IP) assays were performed using RB CC and its F31E/L34E/I41E (CCEEE) variant in N. benthamiana. As shown in Figure 3D, RB CC-FLAG was only immunoprecipitated by RB CC-Myc but not by the vector alone. By contrast, the CCEEE variant was unable to self-associate, indicating that the triple mutation in the heptad repeats abolished the self-association ability of the RB CC domain (Figure 3E). Collectively, these data suggest that substitutions of hydrophobic residues in the heptad repeats of the CC domain can disrupt CC self-association, probably by destabilizing the α-helical structure, thereby leading to a compromised immune response.
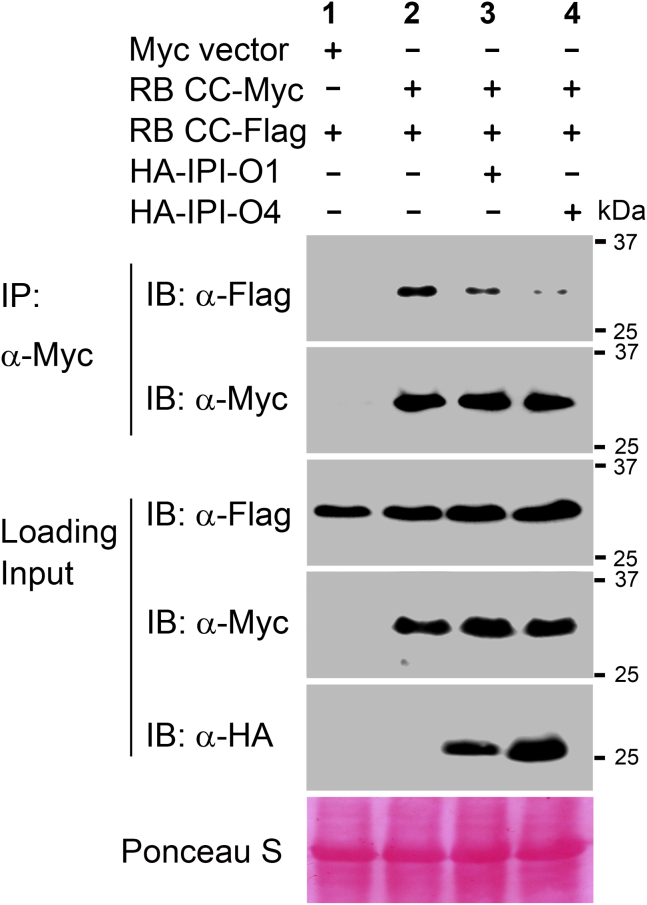
IPI-O1 and IPI-O4 differentially affect RB self-association
Although the RB CC domain was shown to interact with IPI-O1 and IPI-O4 (Chen et al., 2012), how the two functionally distinct effectors affect RB function remains to be elucidated. Because RB CC self-association is essential for RB activation, we examined the effects of the two IPI-O effectors on the self-association status of RB using co-IP assays. As shown in Figure 4, CC self-association was moderately reduced by IPI-O1 but was nearly completely blocked by IPI-O4. These results suggest that the avirulence effector IPI-O1 may largely maintain the self-association status of the RB CC domain while probably opening the closed and inactive conformation. By contrast, the virulence effector IPI-O4 greatly compromises CC self-association, thus leading to the suppression of RB activation. To further examine the effects of IPI-O4 on CC self-association, we monitored CC self-association status with increasing amounts of IPI-O4. Our results showed that the disruptive effect of IPI-O4 on CC self-association was positively correlated with IPI-O4 expression level (Supplemental Figure 12).
Figure 4.
IPI-O1 and IPI-O4 differentially affect self-association of the RB CC domain.
Total proteins were extracted from N. benthamiana plants expressing RB CC-3×FLAG and RB CC-4×Myc together with 3×HA-IPI-O1 or 3×HA-IPI-O4. Immunoprecipitation was performed with an anti-Myc antibody, and immunoblots were probed with anti-FLAG, anti-Myc, or anti-HA antibodies. Ponceau S staining of immunoblots served as a loading control. The experiments were repeated three times with similar results.
IPI-O1 interacts with itself and IPI-O4
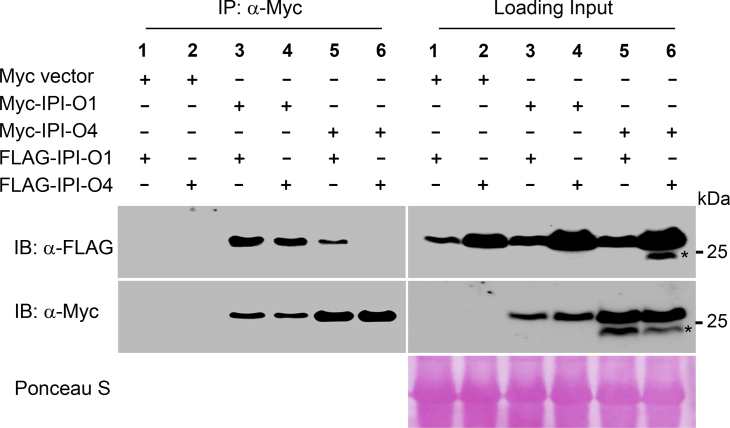
Some Phytophthora RXLR effectors, such as PexRD2 and Avr3a from P. infestans and PsAvh240 from P. sojae, have been shown to form dimers or oligomers (Boutemy et al., 2011; Guo et al., 2019; Wawra et al., 2012). To investigate whether the two IPI-O effectors self-associate, we performed co-IP assays in N. benthamiana. As shown in Figure 5, Myc-tagged IPI-O1 immunoprecipitated FLAG-tagged IPI-O1, but no detectable self-association was observed for IPI-O4, suggesting that IPI-O1 and IPI-O4 differ not only in subcellular localization but also in self-association status. The class III IPI-O effector IPI-O4 is relatively rare in the P. infestans population and often coexists with prevalent class I IPI-O effectors, including IPI-O1 (Champouret et al., 2009; Halterman et al., 2010). We therefore also tested whether the two coexisting IPI-O effectors interact with each other. Indeed, our co-IP assay showed that IPI-O1 associated with IPI-O4. The reciprocal co-IP assay also confirmed that IPI-O1 was found in the IPI-O4 precipitate (Figure 5). These data indicate that IPI-O1, but not IPI-O4, associates with itself and that IPI-O1 also interacts with IPI-O4.
Figure 5.
IPI-O1 interacts with itself and IPI-O4 in planta.
Total proteins were extracted from N. benthamiana plants expressing 4×Myc-tagged IPI-O1 or IPI-O4 together with 3×FLAG-tagged IPI-O1 or IPI-O4. Immunoprecipitation was performed with an anti-Myc antibody, and immunoblots were probed with anti-FLAG or anti-Myc antibodies. Ponceau S staining of immunoblots served as a loading control. Asterisks indicate nonspecific bands. The experiments were repeated twice with similar results.
IPI-O4 disrupts IPI-O1 self-association but enhances the RB CC-IPI-O1 interaction
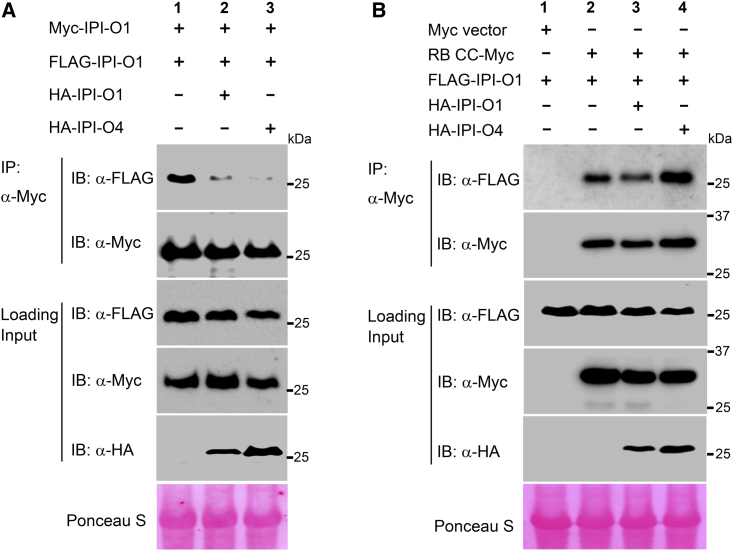
Multiple lines of evidence suggest that dimerization is required for the virulence function of effectors from Phytophthora pathogens (Boutemy et al., 2011; Wawra et al., 2012; King et al., 2014; Guo et al., 2019). Given that IPI-O1 but not IPI-O4 associates with itself and that IPI-O1 also interacts with IPI-O4, we propose that IPI-O4 may interfere with the avirulence function of IPI-O1 by perturbing IPI-O1 self-association. Therefore, we monitored the IPI-O1 self-association status in the presence or absence of IPI-O4. In a control competition assay, Myc-tagged IPI-O1 was shown to associate with FLAG-tagged IPI-O1, and this association could be reduced to some extent by the addition of HA-tagged IPI-O1. Notably, IPI-O4 significantly impaired IPI-O1 self-association (Figure 6A). Reciprocal co-IP further confirmed that IPI-O4 severely abolished IPI-O1 self-association in planta (Supplemental Figure 13). These results suggest that the virulence effector IPI-O4 perturbs the self-association of not only NLR RB but also its cognate effector IPI-O1.
Figure 6.
IPI-O4 perturbs the self-association of IPI-O1 but enhances the association of RB CC with IPI-O1.
(A) IPI-O4 perturbs the self-association of IPI-O1 in planta. Total proteins were extracted from N. benthamiana plants expressing 4×Myc-IPI-O1 and 3×FLAG-IPI-O1 together with 3×HA-tagged IPI-O1 or IPI-O4. Immunoprecipitation was performed with an anti-Myc antibody, and immunoblots were probed with anti-FLAG, anti-Myc, or anti-HA antibodies.
(B) IPI-O4 enhances the association of RB CC with IPI-O1 in vivo. Total proteins were extracted from N. benthamiana plants expressing RB CC-4×Myc and 3×FLAG-IPI-O1 together with 3×HA-tagged IPI-O1 or IPI-O4. Immunoprecipitation was performed with an anti-Myc antibody, and immunoblots were probed with anti-FLAG, anti-Myc, or anti-HA antibodies. Ponceau S staining of immunoblots served as a loading control. The experiments were repeated three times with similar results.
The virulence effector IPI-O4 not only eludes recognition by RB but also suppresses the RB-mediated resistance elicited by IPI-O1 (Halterman et al., 2010; Chen and Halterman, 2011; Chen et al., 2012). Having discovered that IPI-O4 can perturb the self-association of both RB and its cognate effector IPI-O1, we next sought to explore whether IPI-O4 influences RB CC-IPI-O1 recognition to suppress RB-mediated immunity. We examined the association between RB CC and IPI-O1 in the presence or absence of IPI-O4. As shown in Figure 6B, the interaction between Myc-tagged RB CC and FLAG-tagged IPI-O1 was reduced by competition from IPI-O1 fused to a different epitope tag (HA-IPI-O1). To our surprise, we observed increased interaction between RB CC and IPI-O1 in the presence of IPI-O4, indicating that IPI-O4 enhances rather than suppresses the CC-IPI-O1 interaction (Figure 6B).
Discussion
Plants and their pathogens are perpetually engaged in a continuous coevolutionary race for survival. Host plants must adapt to combat newly emerging pathogens, while pathogens constantly evolve to overcome plant innate immunity. An overall understanding of the mechanism that underlies the suppression of NLR-mediated avirulence effector recognition by rapidly evolving pathogens is essential to developing new recognition specificities and implementing durable resistance. Here, we revealed a mechanism by which pathogens evolve to gain new virulence and circumvent NLR-mediated recognition of an avirulence effector. We show that the avirulence effector IPI-O1 and the virulence effector IPI-O4 differ in subcellular localization, self-association status, and, in particular, effects on RB self-association mediated by the CC domain. We demonstrate that IPI-O4 disturbs the self-association of both RB CC and IPI-O1 to block RB activation. In addition, IPI-O4 enhances the interaction between RB CC and IPI-O1 and may thus sequester RB and IPI-O1 to prevent their interaction with downstream components. Ultimately, these events probably prevent the formation of interaction interfaces that are required for the assembly of a resistosome complex.
Cytoplasmic localization of both RB and IPI-O1 is required for cell death induction
Nucleocytoplasmic distribution is of particular importance for the recognition and downstream signaling pathways of a number of plant NLRs (Slootweg et al., 2010). Several NLRs can be activated in only the nuclear or cytoplasmic compartment (Elmore et al., 2011; Du et al., 2015). Others show dual localization in both the nucleus and the cytoplasm, and their localization can change upon recognition of their cognate effectors (Shen et al., 2007; Wirthmueller et al., 2007).
Our subcellular localization studies revealed that both RB and its cognate effector IPI-O1 are localized in the nucleus and cytoplasm, exhibiting similar nucleocytoplasmic distribution patterns. By contrast, the virulence effector IPI-O4 was localized exclusively in the cytoplasm (Figure 1). Furthermore, we found that RB fused to an NES was predominantly localized in the cytoplasm and able to trigger HR (Figure 2A), suggesting that RB possesses HR-inducing ability in the cytoplasm. The nucleocytoplasmic distribution and requirement for cytoplasmic localization of RB and IPI-O1 to induce cell death suggest that RB may recognize IPI-O1 primarily in the cytoplasm (Figure 2C and 2D). This differs from the finding that the race-specific NLR protein R1 recognizes its cognate effector AVR1 in the nucleus (Du et al., 2015). Additional analysis of RB and IPI-O1 proteins fused to GR or Rop10 tags provided further evidence that proper cytoplasmic distribution is essential for RB-mediated recognition of IPI-O1 to trigger immune responses (Supplemental Figures 4 and 6).
Self-association of RB is essential for the activation of immune responses
Similar to their vertebrate analogs, plant NLRs can oligomerize via homomeric or heteromeric interactions upon activation (Maekawa et al., 2011; Césari et al., 2014; Wróblewski et al., 2018). Self-association of the CC domain is required for CNL proteins to form homo- and heterodimers and activate defense responses (Maekawa et al., 2011; Casey et al., 2016; El Kasmi et al., 2017). The CC domains of several CNLs with solved structures form helical bundle structures and are maintained by heptad repeats of hydrophobic residues that mediate self-association (Maekawa et al., 2011; Hao et al., 2013; Casey et al., 2016; Wang et al., 2019a, 2019b).
Homology-based structural modeling of the CC domains of RB and other NLRs identified four α helices consisting of conserved heptad repeats of hydrophobic amino acids (Supplemental Figures 7 and 8), which are essential for the formation of self-associated complexes (Chambers et al., 1990; Mason and Arndt, 2004; Maekawa et al., 2011; Wróblewski et al., 2018). Structure-guided mutagenesis of the heptad repeats in the RB CC domain with thermodynamically unfavorable alanine destabilized the α-helical structure. HR assays using RB variants with single amino acid substitutions revealed that the majority of mutations in the heptad repeats abolished HR-inducing activity. The exceptions were the L34E and F101E derivatives, which still retained a wild-type (WT) level of HR-inducing activity (Figure 3A and Supplemental Figure 9). Overall, these results suggested that the heptad repeats have an essential role in maintaining self-association of the RB CC domain.
The D475V substitution in the MHD motif autoactivates immunity even in the absence of the IPI-O1 effector. We introduced the autoactive D475V mutation into several heptad repeat variants, including F31E, L34E, F38E, and I41E, to test the combined effects of double mutations on HR elicitation (Figure 3B). All the amino acid substitutions in the heptad repeats suppressed the autoimmune response caused by the D475V mutation. Although the L34E mutation did not alter IPI-O1-induced HR, it still blocked the autoactivated HR conferred by the D475V mutation (Figure 3A and 3C and Supplemental Figure 10). Furthermore, we found that the CC domain of the triple loss-of-function RB F31E/L34E/I41E mutant was no longer able to interact with itself (Figure 3E), confirming the essential role of heptad repeats in the maintenance of CC self-association (El Kasmi et al., 2017). These data suggest that the structure and integrity of the heptad repeats are essential for RB activation.
IPI-O1 and IPI-O4 differentially affect self-association of the RB CC domain
An increasing number of studies have shown that self-association is important for the activation of plant NLRs. For example, several CNLs, such as MLA10, Sr33, Sr35, RPS5, Rx, and RPP7, can self-associate to form dimers or higher-order oligomers (Moffett et al., 2002; Ade et al., 2007; Maekawa et al., 2011; Casey et al., 2016; Cesari et al., 2016; Li et al., 2020). Similarly, the TNLs N, SNC1, RPP7, L6, and RPS4 dimerize or form higher-order complexes either before or after activation (Mestre and Baulcombe, 2006; Bernoux et al., 2011; Xu et al., 2014; Schreiber et al., 2016; Zhang et al., 2017). Structural and functional studies of the CNL ZAR1 showed that, in the resting state, ZAR1 interacts with the adaptor protein RKS1, and the ZAR1-RKS1 complex recognizes the uridylated decoy protein PBL2 (PBL2UMP) to form a tertiary complex at an intermediate state. Upon activation, the ZAR1-RKS1-PBL2UMP complex forms a wheel-like pentameric resistosome. The ZAR1 CC domain undergoes fold switching to form an α-helical barrel that interacts with the ZAR1 LRR and winged-helix domains and contributes to resistosome pentamerization (Wang et al., 2019a, 2019b). The ZAR1 resistosome has recently been shown to form calcium-permeable channels, enabling the calcium influx that activates downstream immune responses and cell death (Bi et al., 2021). Our 3D structural modeling of the RB CC domain based on its structural alignment with ZAR1 indicates that the RB CC domain may also form a pentameric complex (Supplemental Figure 8). It remains to be determined whether the effector IPI-O1 or other components contribute to the formation of the higher-order RB structure and whether the RB resistosome exhibits calcium channel activities to trigger immune signaling and cell death.
Despite significant progress in our understanding of NLR function, the underlying mechanisms by which effectors, particularly virulence effectors, modulate NLR self-association to trigger or suppress NLR-mediated resistance are largely unknown. The present RB/IPI-O1/IPI-O4 system is unique in that the virulence effector IPI-O4 not only blocks RB-mediated recognition of its cognate effector IPI-O1 but also interacts with the RB CC domain (Chen et al., 2012). The autonomous cell death induced by the D475V mutation in the MHD domain of RB could not be suppressed by IPI-O4 (Supplemental Figure 10B). This result indicates that suppression may occur during the recognition process rather than in the downstream signaling pathway. With the availability of both avirulence and virulence effectors that are closely related and differ only in 20 amino acids (Champouret et al., 2009; Chen et al., 2012; Win et al., 2007), we were able to determine their differential effects on the status of RB self-association mediated by the CC domain. It appears that the avirulence effector IPI-O1 can change the conformational equilibrium of CC self-association, which may allow the recruitment of signaling proteins to trigger immunity. By contrast, the virulence effector IPI-O4 largely impairs CC self-association and consequently suppresses RB activation (Figure 4). Rearrangement of the NLR CC domains has been shown to be a key event during oligomerization that is required for NLR-mediated resistance (Maekawa et al., 2011; Wang et al., 2019a, 2019b). Disruption of RB CC self-association may result in a dimer or oligomer adopting an unfavorable conformation, thereby preventing it from interacting with signaling components for assembly of the active resistosome.
Virulence effector IPI-O4 suppresses RB-mediated recognition of IPI-O1 probably by disrupting IPI-O1 self-association
Multiple lines of evidence suggest that self-association of Phytophthora RXLR effectors is important for their virulence functions. The P. infestans effector Avr3a dimerizes, and the N-terminal RXLR leader region facilitates its dimerization. Mutations that prevent dimer formation fail to promote pathogen growth (Wawra et al., 2012, 2017). Recently, a crystallographic study of the RXLR effector PsAvh240 from the soybean pathogen P. sojae revealed that PsAvh240 forms a WY domain-mediated handshake-like dimer that is required for its virulence function (Guo et al., 2019). The WY domain of the P. infestans effector PexRD2 displays a core α-helical fold that supports its self-association. Bioinformatics analysis suggests that the WY domain is highly conserved and has been identified in 44% of the annotated Phytophthora RXLR effectors, either as a single copy or as tandem repeats (Boutemy et al., 2011). Notably, the avirulent effector IPI-O1 but not the virulent effector IPI-O4 exhibits self-association (Figure 5). Given that the Y motif was not predicted in IPI-O1, we propose that IPI-O1 self-association is likely to be mediated by the W motif. Sequence alignment indicated that 7 out of 20 polymorphic amino acids between IPI-O1 and IPI-O4 reside in the W motif, which is consistent with the finding that the C-terminal domain of the RXLR effector family is under positive selection (Win et al., 2007; Champouret et al., 2009). Furthermore, the two residues at positions 129 and 135, which determine the recognition specificity of IPI-O1 and IPI-O4, are either within or right next to the W motif (Chen et al., 2012).
A structural analysis of the P. infestans PexRD2 effector revealed that this effector exists as a homodimer in vitro and oligomerizes in planta (Boutemy et al., 2011). It remains to be determined whether IPI-O1 adopts an oligomeric state inside plant cells and whether IPI-O1 associates with RB as a dimer or a higher-order oligomer. Having demonstrated that IPI-O1 and IPI-O4 differ in subcellular localization (Figure 1) and self-association status (Figure 5), we provide biochemical evidence that IPI-O4 interacts with IPI-O1 and abolishes IPI-O1 self-association in planta (Figure 6A and Supplemental Figure 13). Our findings suggest that perturbation of the self-association ability of avirulent effectors by newly evolved effectors is likely to be an evolutionary strategy by which pathogens adapt to suppress the recognition of avirulent effectors mediated by their corresponding NLRs. Furthermore, in the presence of IPI-O4, the interaction between RB CC and IPI-O1 was surprisingly enhanced (Figure 6B). A possible explanation is that IPI-O4 may suppress the function of IPI-O1 and RB by tightly sequestering or inactivating IPI-O1 and RB to prevent their interaction with signaling components, leading to inhibition of RB-mediated immunity.
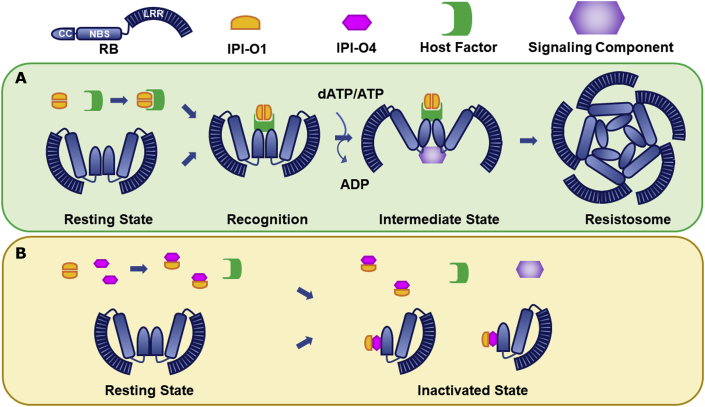
Collectively, our results indicate that the suppression of RB-mediated recognition of IPI-O1 and downstream immune signaling by the virulence effector IPI-O4 is likely to be achieved by perturbing the self-association of RB CC and IPI-O1, thereby potentially preventing the formation of interaction surfaces required for recruiting signaling components to assemble an active resistosome complex. We propose that, in the resting state, RB associates with itself via the CC domain, stabilizing RB in a closed and inactive conformation. Upon P. infestans infection, the effector IPI-O1 is delivered into the cytoplasm and recognized by RB via the CC domain. Interaction with IPI-O1 changes the self-association status of RB, probably leading to an open and active conformation that enables the recruitment of yet-to-be-identified signaling components and triggers immune responses (Figure 7A). In the presence of IPI-O4, self-association of both RB CC and IPI-O1 is largely prevented, thereby blocking the formation of interaction interfaces that are required for recruiting signaling components for the assembly of the RB resistosome complex (Figure 7B). Because IPI-O4 enhances the interaction of RB CC with IPI-O1 (Figure 6B), it is also plausible to speculate that IPI-O4 may sequester RB and IPI-O1 to prevent their association with signaling molecules, ultimately leading to suppression of RB-mediated immunity triggered by IPI-O1. However, these results do not favor the model that IPI-O4 blocks RB-mediated recognition of IPI-O1 by out-competing IPI-O1 for interaction with the RB CC domain. Because the cytoplasmic localization of both RB and IPI-O1 is required for RB activation in response to IPI-O1, and IPI-O4 is localized exclusively in the cytoplasm, it appears that RB-mediated recognition of IPI-O1 and suppression of this process by IPI-O4 occur mainly in the cytoplasm.
Figure 7.
A working model of NLR receptor RB-mediated recognition of its cognate effector IPI-O1 and its suppression by the virulence effector IPI-O4.
(A) In the resting state, RB self-associates via the CC domain, adopting a closed and inactive conformation. When IPI-O1 is present, it is recognized by RB via the CC domain, probably through an unidentified host factor, and RB adopts an open conformation, potentially leading to the recruitment of signaling components to assemble an active resistosome complex.
(B) When IPI-O4 is present, it perturbs the self-association of both RB and IPI-O1 and enhances the RB/IPI-O1 interaction, thereby probably preventing the formation of interaction interfaces that are required for recruitment of signaling components and assembly of the resistosome.
Future work will be focused on determining the nature and dynamic changes of self-association of IPI-O1 and RB upon activation, identifying host targets of the IPI-Os, and elucidating their roles in recognition of IPI-O1, activation of RB-mediated resistance, and downstream signaling. Results from these studies will provide fundamental guidance for the rational engineering of durable and broad-spectrum late blight resistance in potato.
Methods
Plant materials
WT N. benthamiana plants and transgenic N. benthamiana plants expressing RB (Halterman et al., 2010; Chen et al., 2012) were grown in growth rooms at 22°C under a 16 h light/8 h dark cycle and a light intensity of 120 μmol m−2 s−1.
Site-directed mutagenesis
Mutants of RB, IPI-O1, or IPI-O4 were generated by circular PCR using pDONR207 constructs harboring RB, IPI-O1, or IPI-O4 as templates. Primers containing the desired nucleotide substitutions were designed using the QuikChange Primer Design program (Agilent Technologies), and primer pairs complementary to the opposite strands of the target sequences were extended during temperature cycling using KAPA HiFi DNA Polymerase (KAPA Biosystems). After thermal cycling, the PCR products were treated with DpnI at 37°C for 1 h to digest the methylated plasmid DNA template, followed by transformation into E. coli. All the constructs were verified by sequencing. Primers used for site-directed mutagenesis are listed in Supplemental Table 1.
Plasmid construction
To generate C-terminal GFP fusion constructs, RB, IPI-O1, and IPI-O4 were amplified by PCR and cloned into pDONR207. After sequencing confirmation, the resulting constructs were cloned into the binary destination vector pJG186 by LR reactions. pJG186 was constructed by inserting a Gateway cloning cassette and a C-terminal GFP tag into the binary vector pCAMBIA1300 containing a rubisco small subunit gene (RbCS) terminator.
To generate C-terminal Myc-tagged and FLAG-tagged fusion constructs, the RB CC domain harboring the first 165 amino acid sequence of RB was amplified by PCR and cloned into pDONR207. After sequencing confirmation, the resulting constructs were cloned into the binary destination vectors pJG100 and pJP603 by LR reactions. pJG100 was constructed by inserting a Gateway cloning cassette and a C-terminal 4×Myc tag into the binary vector pCAMBIA1300 containing an RbCS terminator. pJP603 was constructed by inserting a Gateway cloning cassette and a C-terminal 3×FLAG tag into the binary vector pCAMBIA1300 containing an RbCS terminator.
To generate N-terminal HA-tagged, Myc-tagged, and FLAG-tagged fusion constructs, the RXLR effectors IPI-O1 and IPI-O4 were amplified by PCR and cloned into pDONR207. After sequencing confirmation, the resulting constructs were cloned into the binary destination vectors pJG99, pJP638, and pJP601 by LR reactions. pJG99 was constructed by inserting a Gateway cloning cassette and an N-terminal 3×HA tag into the binary vector pCAMBIA1300 containing an RbCS terminator. pJP638 was constructed by inserting a Gateway cloning cassette and an N-terminal 4×Myc tag into the binary vector pCAMBIA1300 containing an RbCS terminator. pJP601 was constructed by inserting a Gateway cloning cassette and an N-terminal 3×FLAG tag into the binary vector pCAMBIA1300 containing an RbCS terminator.
For the nuclear and cytoplasmic localization analysis, the SV40 T-Ag NLS (QPKKKRKVGG) motif and the PK1 NES (NELALKLAGLDINK) motif, as well as the mutated nls (QPKKTRKVGG) and nes (NELALKAAGADANK) motifs, were fused to the GFP tag at the C terminus to construct the pCAMBIA1300-based Gateway-compatible binary destination vectors pJP757 (GFP-NLS), pJP758 (GFP-nls), pJP759 (GFP-NES), and pJP760 (GFP-nes), respectively. The RB, IPI-O1, and IPI-O4 genes in pDONR207 were cloned into these binary destination vectors by LR reactions for subcellular localization analysis.
To make GR fusion constructs, the hormone-binding domain of the rat GR was cloned from pMPGWB114 (Ishizaki et al., 2015) and fused together with a Gateway cassette to the C terminus of GFP in pCAMBIA1300 to generate the Gateway-compatible binary destination vector pJP771 (GFP-GR). The RB and IPI-O1 genes were cloned into pJP771 by LR reactions to obtain RB-GFP-GR and IPI-O1-GFP-GR.
For membrane association analysis, the Rop10 tag and a mutant form of the Rop10 tag (mRop10) in which the five non-polar residues LSNIL in the GC-CCG box were replaced by REDER were amplified by PCR and fused together with a Gateway cassette to the C terminus of GFP in pCAMBIA1300 to generate the Gateway-compatible binary destination vectors pJP763c (GFP-Rop10) and pJP766b (GFP-mRop10). The RB and IPI-O1 coding sequences in pDONR207 were cloned into pJP763c and pJP766b by LR reactions to obtain RB-GFP-Rop10, RB-GFP-mRop10, IPI-O1-GFP-Rop10, and IPI-O1-GFP-mRop10.
To generate the Hir3.1-mCherry fusion construct, the NbHir3.1 gene was amplified by PCR and cloned into pDONR207. After sequencing confirmation, the resulting construct was cloned into the binary destination vector pJG185 by an LR reaction. pJG185 was constructed by inserting a Gateway cloning cassette and a C-terminal mCherry tag into the binary vector pCAMBIA1300 containing an RbCS terminator. Primers used for plasmid construction are listed in Supplemental Table 1.
Confocal microscopy
Tissues from agroinfiltrated N. benthamiana leaves were collected, mounted with water on glass slides, and analyzed with a 20× air objective (UPLSAPO 20×) using a laser-scanning Olympus SD-OSR Spinning Disk Confocal Microscope with super resolution (Olympus). PM co-localization was analyzed with a silicone oil objective (UPLSAPO 100×S) using the Olympus SD-OSR Spinning Disk Confocal Microscope. mCherry-tagged NbHir3.1 was used as a PM-localized marker for co-localization analysis in N. benthamiana. For nuclear staining, tissues were treated with 10 μg/ml of DAPI solution containing 0.1% Triton X-100. The laser excitation wavelengths were 561 nm for mCherry, 488 nm for GFP, and 405 nm for DAPI. Fluorescence intensity profiles were analyzed with Fiji software (Schindelin et al., 2012) and plotted with Microsoft Excel.
HR induction assays in N. benthamiana plants
For HR cell death assays, 4×Myc-IPI-O1, 4×Myc-IPI-O4, or its variants were transiently expressed in 4- to 6-week-old RB transgenic N. benthamiana plants (Halterman et al., 2010; Chen et al., 2012) or co-expressed with RB or its derivatives in WT N. benthamiana plants. Cell death was monitored at 48 hpi.
Protein extraction, Western blotting, and co-IP
For protein expression analysis, total proteins were extracted from N. benthamiana tissues with buffer containing 150 mM Tris–HCl, 50 mM NaCl, 1 mM EDTA, 2 mM CaCl2, 5 mM MgCl2, 0.15% NP-40, 0.1% Triton X-100 (pH 7.5), 10 mM dithiothreitol, 1 mM phenylmethanesulfonyl fluoride (PMSF), plant protease inhibitor cocktail (VWR), and 20 μM MG132. Protein samples were separated on 10% Bis-Tris PAGE gels, transferred electrophoretically to 0.45-μm Amersham Protran Premium nitrocellulose membranes (GE Healthcare), and detected with anti-GFP (Takara), anti-HA, anti-FLAG, or anti-Myc (BioLegend) antibodies.
For co-IP assays, protein extracts were incubated with an anti-Myc antibody (1:2000, Sigma-Aldrich) at 4°C for 16 h followed by incubation at 4°C for 4 h with protein A/G magnetic beads (VWR) equilibrated with the extraction buffer. The beads were washed six times with 1 ml of extraction buffer, and immunoprecipitated samples were analyzed by western blotting with anti-HA, anti-FLAG, or anti-Myc (BioLegend) antibodies.
Bioinformatic analysis and homology-based structural modeling
Protein sequences of the CC domains of RB and other CNLs, including MLA10, Sr33, Rx, and ZAR1, were aligned with ClustalW2 (Larkin et al., 2007) and refined using the predicted secondary structure of the RB CC domain and the 3D structures of the above-mentioned reference proteins. To predict the secondary structure of the RB CC domain, the first 185 amino acid sequence of RB (Chen et al., 2012) was subjected to several methods of structural analysis, including CCHMM_PROF (Bartoli et al., 2009), SCRATCH (Magnan and Baldi, 2014), RaptorX (Wang et al., 2016), JPred4 (Drozdetskiy et al., 2015), and PSIPRED (Buchan et al., 2013), to generate the secondary structure, and the consensus was used to increase the prediction reliability as described previously (Wróblewski et al., 2018). Homology-based 3D models of the monomeric, dimeric, and pentameric RB CC structures were built using Modeller version 9.25 (Fiser et al., 2000; Webb and Sali, 2016) with optimized alignments of the first 120 amino acids of RB to the reference PDB structures of Sr33 (PDB: 2NCG), MLA10 (PDB: 3QFL), and ZAR1 (PDB: 6J5T). Insertion loops were generated for the missing amino acids in the template, and non-missing residues were kept stable. The global models were evaluated with the DOPE module and then visualized using PyMOL version 2.3 (PyMOL Molecular Graphics System, Schrödinger).
Author contributions
J.S. and J.Z. designed the experiments. J.Z. performed the experiments and acquired the data. J.Z. and J.S. analyzed the data and wrote the paper.
Funding
This work was supported by a start-up fund from Texas A&M AgriLife Research and a Hatch Project from the USDA National Institute of Food and Agriculture to J.S. (TEX0-1-9675).
Acknowledgments
We thank Dennis Halterman for providing the RB transgenic N. benthamiana line and Yule Liu for providing the pJG185 and pJG186 vectors. No conflict of interest declared.
Published: August 25, 2021
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
References
- Ade J., DeYoung B.J., Golstein C., Innes R.W. Indirect activation of a plant nucleotide binding site–leucine-rich repeat protein by a bacterial protease. Proc. Natl. Acad. Sci. 2007;104:2531–2536. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S., Liu J., Chang C., Zhang L., Maekawa T., Wang Q., Xiao W., Liu Y., Chai J., Takken F.L.W. Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog. 2012;8:e1002752. doi: 10.1371/journal.ppat.1002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli L., Fariselli P., Krogh A., Casadio R. CCHMM_PROF: a HMM-based coiled-coil predictor with evolutionary information. Bioinformatics. 2009;25:2757–2763. doi: 10.1093/bioinformatics/btp539. [DOI] [PubMed] [Google Scholar]
- Bentham A.R., Zdrzałek R., De la Concepcion J.C., Banfield M.J. Uncoiling CNLs: structure/function approaches to understanding CC domain function in plant NLRs. Plant Cell Physiol. 2018;59:2398–2408. doi: 10.1093/pcp/pcy185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernoux M., Ve T., Williams S., Warren C., Hatters D., Valkov E., Zhang X., Ellis J.G., Kobe B., Dodds P.N. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe. 2011;9:200–211. doi: 10.1016/j.chom.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar P.B., Raasch J.A., Kramer L.C., Neumann P., Wielgus S.M., Austin-Phillips S., Jiang J. Sgt1, but not Rar1, is essential for the RB-mediated broad-spectrum resistance to potato late blight. BMC Plant Biol. 2008;8:8. doi: 10.1186/1471-2229-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi G., Su M., Li N., Liang Y., Dang S., Xu J., Hu M., Wang J., Zou M., Deng Y. The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell. 2021;184:3528–3541.e3512. doi: 10.1016/j.cell.2021.05.003. [DOI] [PubMed] [Google Scholar]
- Boutemy L.S., King S.R., Win J., Hughes R.K., Clarke T.A., Blumenschein T.M., Kamoun S., Banfield M.J. Structures of Phytophthora RXLR effector proteins: a conserved but adaptable fold underpins functional diversity. J. Biol. Chem. 2011;286:35834–35842. doi: 10.1074/jbc.M111.262303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester K., de Sain M., Weide R., Gouget A., Klamer S., Canut H., Govers F. The lectin receptor kinase LecRK-I.9 is a novel Phytophthora resistance component and a potential host target for a RXLR effector. PLoS Pathog. 2011;7:e1001327. doi: 10.1371/journal.ppat.1001327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan D.W.A., Minneci F., Nugent T.C.O., Bryson K., Jones D.T. Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 2013;41:W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey L.W., Lavrencic P., Bentham A.R., Cesari S., Ericsson D.J., Croll T., Turk D., Anderson P.A., Mark A.E., Dodds P.N. The CC domain structure from the wheat stem rust resistance protein Sr33 challenges paradigms for dimerization in plant NLR proteins. Proc. Natl. Acad. Sci. 2016;113:12856–12861. doi: 10.1073/pnas.1609922113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S., Moore J., Chen C., Webb D., Periyannan S., Mago R., Bernoux M., Lagudah E.S., Dodds P.N. Cytosolic activation of cell death and stem rust resistance by cereal MLA-family CC–NLR proteins. Proc. Natl. Acad. Sci. 2016;113:10204–10209. doi: 10.1073/pnas.1605483113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Césari S., Kanzaki H., Fujiwara T., Bernoux M., Chalvon V., Kawano Y., Shimamoto K., Dodds P., Terauchi R., Kroj T. The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 2014;33:1941–1959. doi: 10.15252/embj.201487923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers P., Pringle C.R., Easton A.J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J. Gen. Virol. 1990;71:3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- Champouret N., Bouwmeester K., Rietman H., van der Lee T., Maliepaard C., Heupink A., van de Vondervoort P.J.I., Jacobsen E., Visser R.G.F., van der Vossen E.A.G. Phytophthora infestans isolates lacking class I ipiO variants are virulent on Rpi-blb1 potato. Mol. Plant Microbe Interact. 2009;22:1535–1545. doi: 10.1094/mpmi-22-12-1535. [DOI] [PubMed] [Google Scholar]
- Chen T., Liu D., Niu X., Wang J., Qian L., Han L., Liu N., Zhao J., Hong Y., Liu Y. Antiviral resistance protein Tm-22 functions on the plasma membrane. Plant Physiol. 2017;173:2399–2410. doi: 10.1104/pp.16.01512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Halterman D.A. Phenotypic characterization of potato late blight resistance mediated by the broad-spectrum resistance gene RB. Phytopathology. 2011;101:263–270. doi: 10.1094/phyto-04-10-0119. [DOI] [PubMed] [Google Scholar]
- Chen Y., Halterman D.A. Phytophthora infestans effectors IPI-O1 and IPI-O4 each contribute to pathogen virulence. Phytopathology. 2017;107:600–606. doi: 10.1094/phyto-06-16-0240-r. [DOI] [PubMed] [Google Scholar]
- Chen Y., Liu Z., Halterman D.A. Molecular determinants of resistance activation and suppression by Phytophthora infestans effector IPI-O. PLoS Pathog. 2012;8:e1002595. doi: 10.1371/journal.ppat.1002595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S.M., Hamel L.-P., Moffett P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol. Plant Microbe Interact. 2011;24:918–931. doi: 10.1094/mpmi-03-11-0050. [DOI] [PubMed] [Google Scholar]
- Colton L.M., Groza H.I., Wielgus S.M., Jiang J. Marker-assisted selection for the broad-spectrum potato late blight resistance conferred by gene RB derived from a wild potato species. Crop Sci. 2006;46:589–594. doi: 10.2135/cropsci2005.0112. [DOI] [Google Scholar]
- De Oliveira A.S., Koolhaas I., Boiteux L.S., Caldararu O.F., Petrescu A.J., Oliveira Resende R., Kormelink R. Cell death triggering and effector recognition by Sw-5 SD-CNL proteins from resistant and susceptible tomato isolines to Tomato spotted wilt virus. Mol. Plant Pathol. 2016;17:1442–1454. doi: 10.1111/mpp.12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar K.D., Demady D.R., Stancato L.F., Krishna P., Pratt W.B. Folding of the glucocorticoid receptor by the heat shock protein (hsp) 90-based chaperone machinery: THE ROLE OF p23 IS TO STABILIZE RECEPTOR·hsp90 HETEROCOMPLEXES FORMED BY hsp90·p60·hsp70. J. Biol. Chem. 1997;272:21213–21220. doi: 10.1074/jbc.272.34.21213. [DOI] [PubMed] [Google Scholar]
- Dodds P.N., Lawrence G.J., Ellis J.G. Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell. 2001;13:163–178. doi: 10.1105/tpc.13.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy A., Cole C., Procter J., Barton G.J. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Berg J., Govers F., Bouwmeester K. Immune activation mediated by the late blight resistance protein R1 requires nuclear localization of R1 and the effector AVR1. New Phytol. 2015;207:735–747. doi: 10.1111/nph.13355. [DOI] [PubMed] [Google Scholar]
- El Kasmi F., Chung E.-H., Anderson R.G., Li J., Wan L., Eitas T.K., Gao Z., Dangl J.L. Signaling from the plasma-membrane localized plant immune receptor RPM1 requires self-association of the full-length protein. Proc. Natl. Acad. Sci. 2017;114:E7385–E7394. doi: 10.1073/pnas.1708288114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore J.M., Lin Z.-J.D., Coaker G. Plant NB-LRR signaling: upstreams and downstreams. Curr. Opin. Plant Biol. 2011;14:365–371. doi: 10.1016/j.pbi.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A., Do R.K.G., Šali A. Modeling of loops in protein structures. Protein Sci. 2000;9:1753–1773. doi: 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García A.V., Blanvillain-Baufumé S., Huibers R.P., Wiermer M., Li G., Gobbato E., Rietz S., Parker J.E. Balanced nuclear and cytoplasmic activities of EDS1 are required for a complete plant innate immune response. PLoS Pathog. 2010;6:e1000970. doi: 10.1371/journal.ppat.1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M., Byarugaba A.A., Magembe E., Njoroge A., Rivera C., Román M.L., Tovar J.C., Gamboa S., Forbes G.A., Kreuze J.F. Stacking three late blight resistance genes from wild species directly into African highland potato varieties confers complete field resistance to local blight races. Plant Biotechnol. J. 2019;17:1119–1129. doi: 10.1111/pbi.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouget A., Senchou V., Govers F., Sanson A., Barre A., Rougé P., Pont-Lezica R., Canut H. Lectin receptor kinases participate in protein-protein interactions to mediate plasma membrane-cell wall adhesions in Arabidopsis. Plant Physiol. 2006;140:81–90. doi: 10.1104/pp.105.066464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Wang H., Yang B., Jiang W., Jing M., Li H., Xia Y., Xu Y., Hu Q., Wang F. Phytophthora sojae effector PsAvh240 inhibits host aspartic protease secretion to promote infection. Mol. Plant. 2019;12:552–564. doi: 10.1016/j.molp.2019.01.017. [DOI] [PubMed] [Google Scholar]
- Gutierrez J.R., Balmuth A.L., Ntoukakis V., Mucyn T.S., Gimenez-Ibanez S., Jones A.M.E., Rathjen J.P. Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J. 2010;61:507–518. doi: 10.1111/j.1365-313X.2009.04078.x. [DOI] [PubMed] [Google Scholar]
- Haesaert G., Vossen J.H., Custers R., De Loose M., Haverkort A., Heremans B., Hutten R., Kessel G., Landschoot S., Van Droogenbroeck B. Transformation of the potato variety Desiree with single or multiple resistance genes increases resistance to late blight under field conditions. Crop Prot. 2015;77:163–175. doi: 10.1016/j.cropro.2015.07.018. [DOI] [Google Scholar]
- Halterman D.A., Middleton G. Presence of the potato late blight resistance gene RB does not promote adaptive parasitism of Phytophthora infestans. Am. J. Plant Sci. 2012;3:360–367. doi: 10.4236/ajps.2012.33043. [DOI] [Google Scholar]
- Halterman D.A., Kramer L.C., Wielgus S., Jiang J. Performance of transgenic potato containing the late blight resistance gene RB. Plant Dis. 2008;92:339–343. doi: 10.1094/pdis-92-3-0339. [DOI] [PubMed] [Google Scholar]
- Halterman D.A., Chen Y., Sopee J., Berduo-Sandoval J., Sánchez-Pérez A. Competition between Phytophthora infestans effectors leads to increased aggressiveness on plants containing broad-spectrum late blight resistance. PLoS One. 2010;5:e10536. doi: 10.1371/journal.pone.0010536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W., Collier S.M., Moffett P., Chai J. Structural basis for the interaction between the potato virus X resistance protein (Rx) and its cofactor ran GTPase-activating protein 2 (RanGAP2) J. Biol. Chem. 2013;288:35868–35876. doi: 10.1074/jbc.M113.517417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L., Wu Y., Wu D., Rao W., Guo J., Ma Y., Wang Z., Shangguan X., Wang H., Xu C. The coiled-coil and nucleotide binding domains of BROWN PLANTHOPPER RESISTANCE14 function in signaling and resistance against planthopper in rice. Plant Cell. 2017;29:3157–3185. doi: 10.1105/tpc.17.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki K., Nishihama R., Ueda M., Inoue K., Ishida S., Nishimura Y., Shikanai T., Kohchi T. Development of Gateway binary vector series with four different selection markers for the liverwort Marchantia polymorpha. PLoS One. 2015;10:e0138876. doi: 10.1371/journal.pone.0138876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., McAdams S.A., Bryan G.T., Hershey H.P., Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R.H.Y., Tripathy S., Govers F., Tyler B.M. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. 2008;105:4874–4879. doi: 10.1073/pnas.0709303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo K.-R., Kim C.-J., Kim S.-J., Kim T.-Y., Bergervoet M., Jongsma M.A., Visser R.G.F., Jacobsen E., Vossen J.H. Development of late blight resistant potatoes by cisgene stacking. BMC Biotechnol. 2014;14:50. doi: 10.1186/1472-6750-14-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Jones J.D.G., Vance R.E., Dangl J.L. Intracellular innate immune surveillance devices in plants and animals. Science. 2016;354:aaf6395. doi: 10.1126/science.aaf6395. [DOI] [PubMed] [Google Scholar]
- Joseph C.K., Kelly Z., Joseph C., William W.K., David S.D. Late blight resistance of RB transgenic potato lines. J. Am. Soc. Hortic. Sci. 2007;132:783–789. doi: 10.21273/jashs.132.6.783. [DOI] [Google Scholar]
- King S.R.F., McLellan H., Boevink P.C., Armstrong M.R., Bukharova T., Sukarta O., Win J., Kamoun S., Birch P.R.J., Banfield M.J. Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKKε to suppress plant immune signaling. Plant Cell. 2014;26:1345–1359. doi: 10.1105/tpc.113.120055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva K.V., Dahlbeck D., Staskawicz B.J. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell. 2010;22:2444–2458. doi: 10.1105/tpc.110.075358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lavy M., Yalovsky S. Association of Arabidopsis type-II ROPs with the plasma membrane requires a conserved C-terminal sequence motif and a proximal polybasic domain. Plant J. 2006;46:934–947. doi: 10.1111/j.1365-313X.2006.02749.x. [DOI] [PubMed] [Google Scholar]
- Leipe D.D., Koonin E.V., Aravind L. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 2004;343:1–28. doi: 10.1016/j.jmb.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Lewis J.D., Lee A.H.-Y., Hassan J.A., Wan J., Hurley B., Jhingree J.R., Wang P.W., Lo T., Youn J.-Y., Guttman D.S. The Arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proc. Natl. Acad. Sci. 2013;110:18722–18727. doi: 10.1073/pnas.1315520110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Habring A., Wang K., Weigel D. Atypical resistance protein RPW8/HR triggers oligomerization of the NLR immune receptor RPP7 and autoimmunity. Cell Host Microbe. 2020;27:405–417.e406. doi: 10.1016/j.chom.2020.01.012. [DOI] [PubMed] [Google Scholar]
- Li S., Zhao J., Zhai Y., Yuan Q., Zhang H., Wu X., Lu Y., Peng J., Sun Z., Lin L. The hypersensitive induced reaction 3 (HIR3) gene contributes to plant basal resistance via an EDS1 and salicylic acid-dependent pathway. Plant J. 2019;98:783–797. doi: 10.1111/tpj.14271. [DOI] [PubMed] [Google Scholar]
- Liu Z., Persson S., Sánchez-Rodríguez C. At the border: the plasma membrane–cell wall continuum. J. Exp. Bot. 2015;66:1553–1563. doi: 10.1093/jxb/erv019. [DOI] [PubMed] [Google Scholar]
- Lozoya-Saldana H., Belmar-Diaz C., Bradeen J., Helgeson J. Characterization of Phytophthora infestans isolates infecting transgenic and somatic hybrid potatoes resistant to the pathogen in the Toluca Valley, Mexico. Am. J. Potato Res. 2005;82:79. [Google Scholar]
- Ma S., Lapin D., Liu L., Sun Y., Song W., Zhang X., Logemann E., Yu D., Wang J., Jirschitzka J. Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science. 2020;370:eabe3069. doi: 10.1126/science.abe3069. [DOI] [PubMed] [Google Scholar]
- Maekawa T., Cheng W., Spiridon L.N., Töller A., Lukasik E., Saijo Y., Liu P., Shen Q.-H., Micluta M.A., Somssich I.E. Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe. 2011;9:187–199. doi: 10.1016/j.chom.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Magnan C.N., Baldi P. SSpro/ACCpro 5: almost perfect prediction of protein secondary structure and relative solvent accessibility using profiles, machine learning and structural similarity. Bioinformatics. 2014;30:2592–2597. doi: 10.1093/bioinformatics/btu352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mambetova S., Kirk W.W., Rosenzweig N., Douches D.S. Characterization of late blight resistance potato breeding lines with the RB gene from Solanum bulbocastanum. Am. J. Potato Res. 2018;95:564–574. doi: 10.1007/s12230-018-9664-y. [DOI] [Google Scholar]
- Martin R., Qi T., Zhang H., Liu F., King M., Toth C., Nogales E., Staskawicz B.J. Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Science. 2020;370:eabd9993. doi: 10.1126/science.abd9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason J.M., Arndt K.M. Coiled coil domains: stability, specificity, and biological implications. ChemBioChem. 2004;5:170–176. doi: 10.1002/cbic.200300781. [DOI] [PubMed] [Google Scholar]
- Mestre P., Baulcombe D.C. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell. 2006;18:491–501. doi: 10.1105/tpc.105.037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett P., Farnham G., Peart J., Baulcombe D.C. Interaction between domains of a plant NBS–LRR protein in disease resistance-related cell death. EMBO J. 2002;21:4511–4519. doi: 10.1093/emboj/cdf453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngou B.P.M., Ahn H.-K., Ding P., Jones J.D.G. Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature. 2021;592:110–115. doi: 10.1038/s41586-021-03315-7. [DOI] [PubMed] [Google Scholar]
- Pankin A., Sokolova E., Rogozina E., Kuznetsova M., Deahl K., Jones R., Khavkin E. Allele mining in the gene pool of wild Solanum species for homologues of late blight resistance gene RB/Rpi-blb1. Plant Genet. Resour. 2011;9:305–308. doi: 10.1017/s1479262111000414. [DOI] [Google Scholar]
- Pieterse C.M.J., Derksen A.-M.C.E., Folders J., Govers F. Expression of the Phytophthora infestans ipiB and ipi0 genes in planta and in vitro. Mol. Gen. Genet. 1994;244:269–277. doi: 10.1007/bf00285454. [DOI] [PubMed] [Google Scholar]
- Qi T., Seong K., Thomazella D.P.T., Kim J.R., Pham J., Seo E., Cho M.-J., Schultink A., Staskawicz B.J. NRG1 functions downstream of EDS1 to regulate TIR-NLR-mediated plant immunity in Nicotiana benthamiana. Proc. Natl. Acad. Sci. 2018;115:E10979–E10987. doi: 10.1073/pnas.1814856115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan G.J., Collier S.M., Sacco M.A., Baldwin T.T., Boettrich T., Moffett P. The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell. 2008;20:739–751. doi: 10.1105/tpc.107.056036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakosy-Tican E., Thieme R., König J., Nachtigall M., Hammann T., Denes T.-E., Kruppa K., Molnár-Láng M. Introgression of two broad-spectrum late blight resistance genes, Rpi-blb1 and Rpi-blb3, from Solanum bulbocastanum dun plus race-specific R genes into potato pre-breeding lines. Front. Plant Sci. 2020;11:699. doi: 10.3389//fpls.2020.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravensdale M., Bernoux M., Ve T., Kobe B., Thrall P.H., Ellis J.G., Dodds P.N. Intramolecular interaction influences binding of the flax L5 and L6 resistance proteins to their AvrL567 ligands. PLoS Pathog. 2012;8:e1003004. doi: 10.1371/journal.ppat.1003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber K.J., Bentham A., Williams S.J., Kobe B., Staskawicz B.J. Multiple domain associations within the Arabidopsis immune receptor RPP1 regulate the activation of programmed cell death. PLoS Pathog. 2016;12:e1005769. doi: 10.1371/journal.ppat.1005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senchou V., Weide R., Carrasco A., Bouyssou H., Pont-Lezica R., Govers F., Canut H. High affinity recognition of a Phytophthora protein by Arabidopsis via an RGD motif. Cell. Mol. Life Sci. 2004;61:502–509. doi: 10.1007/s00018-003-3394-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shandil R.K., Chakrabarti S.K., Singh B.P., Sharma S., Sundaresha S., Kaushik S.K., Bhatt A.K., Sharma N.N. Genotypic background of the recipient plant is crucial for conferring RB gene mediated late blight resistance in potato. BMC Genet. 2017;18:22. doi: 10.1186/s12863-017-0490-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z.-Q., Xue J.-Y., Wu P., Zhang Y.-M., Wu Y., Hang Y.-Y., Wang B., Chen J.-Q. Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol. 2016;170:2095. doi: 10.1104/pp.15.01487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q.-H., Saijo Y., Mauch S., Biskup C., Bieri S., Keller B., Seki H., Ülker B., Somssich I.E., Schulze-Lefert P. Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science. 2007;315:1098–1103. doi: 10.1126/science.1136372. [DOI] [PubMed] [Google Scholar]
- Slootweg E., Roosien J., Spiridon L.N., Petrescu A.-J., Tameling W., Joosten M., Pomp R., van Schaik C., Dees R., Borst J.W. Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell. 2010;22:4195–4215. doi: 10.1105/tpc.110.077537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova E., Pankin A., Beketova M., Kuznetsova M., Spiglazova S., Rogozina E., Yashina I.d., Khavkin E. SCAR markers of the R-genes and germplasm of wild Solanum species for breeding late blight-resistant potato cultivars. Plant Genet. Resour. 2011;9:309–312. doi: 10.1017/s1479262111000347. [DOI] [Google Scholar]
- Song J., Bradeen J.M., Naess S.K., Raasch J.A., Wielgus S.M., Haberlach G.T., Liu J., Kuang H., Austin-Phillips S., Buell C.R. Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc. Natl. Acad. Sci. 2003;100:9128–9133. doi: 10.1073/pnas.1533501100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling W.I.L., Elzinga S.D.J., Darmin P.S., Vossen J.H., Takken F.L.W., Haring M.A., Cornelissen B.J.C. The tomato R gene products I-2 and mi-1 are functional ATP binding proteins with ATPase activity. Plant Cell. 2002;14:2929–2939. doi: 10.1105/tpc.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari J.K., Devi S., Sharma S., Chandel P., Rawat S., Singh B.P. Allele mining in Solanum germplasm: cloning and characterization of RB-homologous gene fragments from late blight resistant wild potato species. Plant Mol. Biol. Reporter. 2015;33:1584–1598. doi: 10.1007/s11105-015-0859-9. [DOI] [Google Scholar]
- Tiwari J.K., Siddappa S., Singh B.P., Kaushik S.K., Chakrabarti S.K., Bhardwaj V., Chandel P. Molecular markers for late blight resistance breeding of potato: an update. Plant Breed. 2013;132:237–245. doi: 10.1111/pbr.12053. [DOI] [Google Scholar]
- Van Der Vossen E., Sikkema A., Hekkert B.t.L., Gros J., Stevens P., Muskens M., Wouters D., Pereira A., Stiekema W., Allefs S. An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 2003;36:867–882. doi: 10.1046/j.1365-313X.2003.01934.x. [DOI] [PubMed] [Google Scholar]
- van West P., de Jong A.J., Judelson H.S., Emons A.M.C., Govers F. The ipiO gene of Phytophthora infestans is highly expressed in invading hyphae during infection. Fungal Genet. Biol. 1998;23:126–138. doi: 10.1006/fgbi.1998.1036. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers V.G.A.A., Rietman H., Krenek P., Champouret N., Young C., Oh S.-K., Wang M., Bouwmeester K., Vosman B., Visser R.G.F. Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS One. 2008;3:e2875. doi: 10.1371/journal.pone.0002875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.-F., Ji J., Ei-Kasmi F., Dangl J.L., Johal G., Balint-Kurti P.J. Molecular and functional analyses of a maize autoactive NB-LRR protein identify precise structural requirements for activity. PLoS Pathog. 2015;11:e1004674. doi: 10.1371/journal.ppat.1004674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Hu M., Wang J., Qi J., Han Z., Wang G., Qi Y., Wang H.-W., Zhou J.-M., Chai J. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science. 2019;364:eaav5870. doi: 10.1126/science.aav5870. [DOI] [PubMed] [Google Scholar]
- Wang J., Wang J., Hu M., Wu S., Qi J., Wang G., Han Z., Qi Y., Gao N., Wang H.-W. Ligand-triggered allosteric ADP release primes a plant NLR complex. Science. 2019;364:eaav5868. doi: 10.1126/science.aav5868. [DOI] [PubMed] [Google Scholar]
- Wang J., Chen T., Han M., Qian L., Li J., Wu M., Han T., Cao J., Nagalakshmi U., Rathjen J.P. Plant NLR immune receptor Tm-22 activation requires NB-ARC domain-mediated self-association of CC domain. PLoS Pathog. 2020;16:e1008475. doi: 10.1371/journal.ppat.1008475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Allefs S., van den Berg R.G., Vleeshouwers V.G.A.A., van der Vossen E.A.G., Vosman B. Allele mining in Solanum: conserved homologues of Rpi-blb1 are identified in Solanum stoloniferum. Theor. Appl. Genet. 2008;116:933–943. doi: 10.1007/s00122-008-0725-3. [DOI] [PubMed] [Google Scholar]
- Wang S., Li W., Liu S., Xu J. RaptorX-Property: a web server for protein structure property prediction. Nucleic Acids Res. 2016;44:W430–W435. doi: 10.1093/nar/gkw306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawra S., Agacan M., Boddey J.A., Davidson I., Gachon C.M., Zanda M., Grouffaud S., Whisson S.C., Birch P.R., Porter A.J. Avirulence protein 3a (AVR3a) from the potato pathogen Phytophthora infestans forms homodimers through its predicted translocation region and does not specifically bind phospholipids. J. Biol. Chem. 2012;287:38101–38109. doi: 10.1074/jbc.M112.395129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawra S., Trusch F., Matena A., Apostolakis K., Linne U., Zhukov I., Stanek J., Kozminski W., Davidson I., Secombes C.J. The RxLR motif of the host targeting effector AVR3a of Phytophthora infestans is cleaved before secretion. Plant Cell. 2017;29:1184–1195. doi: 10.1105/tpc.16.00552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics. 2016;54:5.6.1–5.6.37. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson S.C., Boevink P.C., Moleleki L., Avrova A.O., Morales J.G., Gilroy E.M., Armstrong M.R., Grouffaud S., van West P., Chapman S. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- Williams S.J., Sohn K.H., Wan L., Bernoux M., Sarris P.F., Segonzac C., Ve T., Ma Y., Saucet S.B., Ericsson D.J. Structural basis for assembly and function of a heterodimeric plant immune receptor. Science. 2014;344:299–303. doi: 10.1126/science.1247357. [DOI] [PubMed] [Google Scholar]
- Win J., Morgan W., Bos J., Krasileva K.V., Cano L.M., Chaparro-Garcia A., Ammar R., Staskawicz B.J., Kamoun S. Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell. 2007;19:2349–2369. doi: 10.1105/tpc.107.051037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirthmueller L., Zhang Y., Jones J.D.G., Parker J.E. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr. Biol. 2007;17:2023–2029. doi: 10.1016/j.cub.2007.10.042. [DOI] [PubMed] [Google Scholar]
- Wróblewski T., Spiridon L., Martin E.C., Petrescu A.-J., Cavanaugh K., Truco M.J., Xu H., Gozdowski D., Pawłowski K., Michelmore R.W. Genome-wide functional analyses of plant coiled–coil NLR-type pathogen receptors reveal essential roles of their N-terminal domain in oligomerization, networking, and immunity. PLoS Biol. 2018;16:e2005821. doi: 10.1371/journal.pbio.2005821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Si W., Gao R., Zhang X., Yang S. Evolutionary analysis of RB/Rpi-blb1 locus in the Solanaceae family. Mol. Genet. Genomics. 2015;290:2173–2186. doi: 10.1007/s00438-015-1068-9. [DOI] [PubMed] [Google Scholar]
- Xu F., Cheng Y.T., Kapos P., Huang Y., Li X. P-Loop-dependent NLR SNC1 can oligomerize and activate immunity in the nucleus. Mol. Plant. 2014;7:1801–1804. doi: 10.1093/mp/ssu097. [DOI] [PubMed] [Google Scholar]
- Yuan M., Jiang Z., Bi G., Nomura K., Liu M., Wang Y., Cai B., Zhou J.-M., He S.Y., Xin X.-F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature. 2021;592:105–109. doi: 10.1038/s41586-021-03316-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Bernoux M., Bentham A.R., Newman T.E., Ve T., Casey L.W., Raaymakers T.M., Hu J., Croll T.I., Schreiber K.J. Multiple functional self-association interfaces in plant TIR domains. Proc. Natl. Acad. Sci. 2017;114:E2046–E2052. doi: 10.1073/pnas.1621248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.