Abstract
Background
Non-alcoholic fatty liver disease (NAFLD) is affecting more people globally. Indeed, NAFLD is a spectrum of metabolic dysfunctions that can progress to hepatocellular carcinoma (NAFLD-HCC). This development can occur in a non-cirrhotic liver and thus, often lack clinical surveillance. The aim of this study was to develop non-invasive surveillance method for NAFLD-HCC.
Methods
Using comprehensive ultra-high-performance liquid chromatography mass-spectrometry, we investigated 1,295 metabolites in serum from 249 patients. Area under the receiver operating characteristic curve was calculated for all detected metabolites and used to establish their diagnostic potential. Logistic regression analysis was used to establish the diagnostic score.
Findings
We show that NAFLD-HCC is characterised by a complete rearrangement of the serum lipidome, which distinguishes NAFLD-HCC from non-cancerous individuals and other HCC patients. We used machine learning to build a diagnostic model for NAFLD-HCC. We quantified predictive metabolites and developed the NAFLD-HCC Diagnostic Score (NHDS), presenting superior diagnostic potential compared to alpha-fetoprotein (AFP). Patients’ metabolic landscapes show a progressive depletion in unsaturated fatty acids and acylcarnitines during transformation. Upregulation of fatty acid transporters in NAFLD-HCC tumours contribute to fatty acid depletion in the serum.
Interpretation
NAFLD-HCC patients can be efficiently distinguished by serum metabolic alterations from the healthy population and from HCC patients related to other aetiologies (alcohol and viral hepatitis). Our model can be used for non-invasive surveillance of individuals with metabolic syndrome(s), allowing for early detection of NAFLD-HCC. Therefore, serum metabolomics may provide valuable insight to monitor patients at risk, including morbidly obese, diabetics, and NAFLD patients.
Funding
The funding sources for this study had no role in study design, data collection, data analyses, interpretation or writing of the report as it is presented herein.
Keywords: NAFLD, HCC, metabolomics, lipidomics, biomarker discovery
Abbreviations: AC, acylcarnitines; AV-HCC, alcohol- and viral-associated HCC; AFP, alpha-fetoprotein; AA, amino acids; ArAA, aromatic amino acids; BA, bile acids; BMI, body mass index; BCAAs, branch-chain amino acids; Cer, ceramides; ChoE, cholesteryl esters; CTRL, control; DEM, differentially expressed metabolites; DG, diglycerides; LPC, lysophosphatidylcholines; LPE, lysophosphatidylethanolamines; LPI, lysophosphatidylinositols; CMH, monohexylceramides; MUFA, monounsaturated fatty acids; OB-NAFLD, morbidly obese bariatric surgery NAFLD; NAFLD, non-alcoholic fatty liver disease; NAFL, non-alcoholic fatty liver; NAS, non-alcoholic steatohepatitis; NAE, N-acyl ethanolamines; oxFA, oxidized fatty acids; PC, phosphatidylcholines; PE, phosphatidylethanolamines; PI, phosphatidylinositols; PUFA, polyunsaturated fatty acids; SFA, saturated fatty acids; SM, sphingomyelins; S1P, sphingosine-1-phosphate; TG, triglycerides
Research in context.
Evidence before this study
Non-alcoholic fatty liver disease (NAFLD) is a group of metabolic disorders that can progress to hepatocellular carcinoma (HCC) often without a background of cirrhosis. Non-cirrhotic NAFLD patients are not under surveillance for HCC development. Some metabolomic studies distinguish stages of NAFLD (steatosis, steatohepatitis and cirrhosis). However, NAFLD versus early hepatocarcinogenesis (NAFLD-HCC) in a metabolic background has not been investigated.
Added value of this study
This is the first metabolomic study with comprehensive serum lipidomics, investigating a broad-range of lipid classes in NAFLD and NAFLD-HCC. This study provides evidence of lipidomic utility in NAFLD-HCC detection, presents the NAFLD-HCC Diagnostic Score (NHDS) as a putative screening tool for HCC development in a metabolic background and provides a comprehensive lipidomic landscape of NAFLD-HCC patients serum. We show that fatty acid (FA) uptake is increased in NAFLD-HCC tumours, leading to FA depletion in the circulation.
Implications of all the available evidence
Nearly 30% of newly diagnosed HCC patients have underlying metabolic liver disease. Unlike in the case of viral hepatitis or alcoholic-related HCC, NAFLD-HCC often develops without cirrhosis and thus, patients are not under surveillance. This demonstration of effectiveness using a non-invasive, blood-based assay in detection of resectable (early) NAFLD-HCC should encourage consideration for surveillance of this rapidly increasing, at-risk population.
Alt-text: Unlabelled box
1. Introduction
Non-alcoholic fatty liver disease (NAFLD) involves a spectrum of metabolic diseases affecting 24% of the population globally [1]. NAFLD is associated with obesity and type 2 diabetes [2], and represents a spectrum of non-malignant conditions that range from hepatic steatosis to non-alcoholic steatohepatitis (NASH) with fibrosis and ultimately cirrhosis [3]. NAFLD is emerging as a leading risk factor of hepatocellular carcinoma (HCC) in both men and women (hazard ratio ∼17) [4], but tools to detect the progression of NAFLD to HCC remain inadequate.
HCC is the fourth most common cause of cancer-related death worldwide [5] with rapidly increasing incidence and mortality rates [6,7]. The incidence of NAFLD-related HCC (NAFLD-HCC) varies between 0·5% and 19·5%, depending on the disease state (presence or absence of cirrhosis) and geography [7], [8], [9]. Indeed, the prevalence of NAFLD-HCC is increasing compared to alcohol- and viral hepatitis-related HCC (AV-HCC) [8,10]. Importantly, accumulating evidence suggests that NAFLD-HCC may develop in a non-cirrhotic background [9,11]. This presents a major clinical challenge as non-cirrhotic NAFLD patients currently are not under surveillance for developing HCC [7,11]. Furthermore, NAFLD-HCC is characterised by unique mutational signatures and an immune environment, which plays an important role in the response to checkpoint inhibitors and may lead to therapeutic failure [12], [13], [14]. Therefore, non-invasive serum biomarkers are urgently needed to monitor NAFLD, its progression to HCC, and to distinguish NAFLD-HCC from other aetiologies.
Serum metabolomics provides insight into holistic metabolic changes and can potentially predict hepatic pathological lipid abundance [15] and is reflective of the liver pathology [16]. However, metabolomic profiles are highly dependent on ethnicity and risk factors such as obesity and diabetes [17], [18], [19], [20]. The understanding of the biology underlying NAFLD-to-HCC progression is still lacking. Therefore, analysis of the metabolic shifts in the liver and in circulation at the stage of NAFLD and during malignant progression (NAFLD-HCC) is crucial for the development of new diagnostics and therapeutic modalities [21].
The aim of this study was to develop and validate a non-invasive biomarker for surveillance and early detection of NAFLD-HCC. Here, we used a comprehensive serum metabolomics-based approach to identify unique NAFLD-HCC biomarkers, allowing us to distinguish early NAFLD-HCC from patients with NAFLD or HCC on a background of alcohol or viral hepatitis (AV-HCC).
2. Patients and Methods
2.1. Study population
A total of 249 patient samples were collected in this international and multicentre study. All samples were collected and stored according to REMARK for biomarker analysis. Our study cohort was divided into discovery and validation sets. The proportion of NAFLD-HCC patients in this study (17·3%) is reflecting the reported prevalence of NAFLD-HCC [7], [8], [9]. The discovery set included serum samples from 196 patients from Spain and France: 27 patients with NAFLD-HCC, 32 patients with AV-HCC, 102 morbidly obese NAFLD patients undergoing bariatric surgery (OB-NAFLD), and 35 healthy subjects described previously [22]. Based on unsupervised clustering (Supplementary Materials) and clinical information, we classified 9 obese bariatric surgery patients with a NAS<3 and liver fibrosis score <2 as controls (CTRL). The validation set included serum samples from 37 NAFLD patients from Chile and Spain and plasma sampled from 16 NAFLD-HCC patients in Brazil and Denmark. NAFLD patients from the validation set were overweight (body mass index (BMI>25)) or obese (BMI) >30), however, significantly leaner compared to patients in the OB-NAFLD group (p<0·0001). All patients had biopsy-proven NAFLD. The NAFLD-HCC group consisted of patients who were diagnosed non-alcoholic steatohepatitis (NASH) based on liver biopsy, self-reported alcohol consumption (<20 g/day) and hepatitis B or C serology (hepatitis B surface antigen, hepatitis B surface antibody, hepatitis B core antibody, and hepatitis C antibody). All patients were qualified for curative liver resection. Serum was obtained from all patients prior to surgery, including patients undergoing bariatric surgery or liver resection. The clinical and biochemical representation of the study population is presented in Supplementary Table 1. The NAFLD-HCC patients were diagnosed before surgery with imaging (33%), biopsy (26%), both (15%), or medical concilium (11%). For remaining, 15% of patients’ diagnostic method was not reported. After removal tumours were classified by a skilled pathologist. In all patients’ blood was collected before surgical intervention.
2.2. Ethics
The study was performed following individual patient consent, local institutional review board (IRB) approval, and assessed by the Committee on Health and Research Ethics for the Capital Region of Denmark for use of archival material no. 17029679. All patient data sets were anonymised.
2.3. Metabolomic analyses
Comprehensive metabolomics including 1295 metabolites was performed on serum samples with three platforms as described previously [23]. Briefly, metabolite extraction was accomplished by fractionating the samples into pools of species with similar physicochemical properties, using appropriate combinations of organic solvents. Two UHPLC-time of flight-MS based platforms analysing methanol and chloroform/methanol serum extracts were combined with the amino acid measurement using an UHPLC-single quadrupole-MS based analysis. Platform used for the analysis methanol extraction was optimized for the profiling of fatty acids, oxidized fatty acids, acyl carnitines, lysoglycerophospholipids (monoacylglycerophospholipids and monoetherglycerophospholipids), free sphingoid bases, bile acids, and steroid sulfates. The chloroform/methanol extract platform provided coverage over glycerolipids (di- and triglycerides), cholesterol esters, sphingolipids (ceramides and sphingomyelins), and glycerophospholipids (diacylglycerophospholipids and 1-ether, 2-acylglycerophospholipids).
Metabolite extraction procedures, chromatographic separation conditions, and mass spectrometric detection conditions have been previously described [23]
Data pre-processing generated a list of chromatographic peak areas for the metabolites detected in each sample injection. An approximated linear detection range was defined for each identified metabolite, assuming similar detector response levels for all metabolites belonging to a given chemical class represented by a single standard compound. Metabolites for which more than 30% of data points were found outside their corresponding linear detection range were not used for statistical analyses. Intra and inter batch data normalisation was performed following the procedure described by Martinez-Arranz et al [24].
For fatty acid profiling we used tissue samples matched to serum samples from 9 CTRL, 32 NAFLD, 20 NASH, 27 surrounding liver tissues, and 27 tumour tissues. Methanol with internal standards was added to liver tissue (30:1, v/w) and the homogenization of the resulting mixture was then performed using a Precellys 24 homogenizer at 6500 rpm for 26 seconds. After brief vortex mixing, the samples were incubated for 1h at -20 °C. Samples were centrifuged at 18000 x g for 15 minutes at 4 °C. 120 μL of supernatant were collected from every sample and transferred to vials for UHPLC-MS analysis.
Absolute quantitation of Linoleyl carnitine, Linoleic acid, Osbond acid, 9 (Z),12 (Z)-Hexadecadienoic acid, 9 (E)-Tetradecanoic acid, Tetradecadienoic acid, Hexadecatrienoic acid, Hydroxyoctadecadienoic acid, 1-Hydroxy-2-Linoleoyl-sn-Glycero-3- Phosphatidylcholine 1-Linoleoyl-2-Hydroxy-sn-Glycero-3-Phosphatidylcholine, and 1-Hydroxy-2-Docosapentaenoyl-sn-Glycero-3-Phosphatidylcholine was also performed. Stock standard solution of Linoleyl carnitine, Linoleic acid, osbond acid, 9 (Z),12 (Z)-Hexadecadienoic acid, 9 (E)-Tetradecanoic acid, (2E, 4E)-2,4-Tetradecadienoic acid, 7 (Z),10 (Z),13 (Z)-Hexadecatrienoic acid, 9- (S)-HODE and 18:0 Lyso PC were prepared individually in methanol at a concentration level of approximately 100, 1000 or 10000 μg/mL, depending on the compound. The working standard was prepared by mixing the appropriate amount of each standard solution in methanol to reach a final concentration of approximately 100 μg/mL. Calibration standards were prepared by consecutive dilutions in methanol, ranged between 0·0005-5 μg/mL for FFAox13 and 0·005-50 μg/mL for the rest of the standards in the calibration curve. Likewise, internal standards (IS) stock solution of Octadecanoyl (18,18,18-D3)-L-Carnitine, Linoleic-9,10,12,13-D4 acid, Hexadecatrienoic 7 (Z),10 (Z), [13]14-D6 acid, and 18:1-d7 Lyso PC were prepared at a concentration of 1000 or 5000 μg/mL. The IS working solution was prepared by mixing the appropriate amount of each stock solution in methanol to reach the desire concentration. Quality control (QC) samples (reference sera samples commercially available) were treated according to the same protocol that serum and plasma samples and were analysed against the calibration curve. The intra-day precision was determined by analysing five replicates.
To estimate the concentration of validated metabolites in the discovery set, the measured absolute values of metabolites (normalised to IS and standard curves) were plotted against the normalised chromatographic peak areas for 58 samples. Simple linear regression was applied to generate the equation for each metabolite and estimate R-square (Goodness of fit). The equations used for concentration estimation are presented in Supp. Table 2.
2.4. Gene expression analysis
To estimate expression of fatty acid transport and metabolism genes we used 48 NAFLD-HCC tumour tissues and 47 matched, surrounding tissues. RNA was isolated from 20 mg of frozen tissue with AllPrep DNA/RNA/Protein Mini Kit (Qiagen) according to manufactures recommendation and sequenced Illumina PE150 (Novogene). Fastq files were trimmed with Trimmomatic-0-2·36 (TruSeq3-PE illumina adapters cut). Reads were annotated with STAR-2.5.1a to hg38 canonical genome assembly 1 pass mode, with default parameters. Gene reference (Gencode.v30) was used for gene abundancy estimation is and normalised by gene length (transcript per million, TPM). Differential expression between tumour and surrounding tissue was established with t-test (p<0·05). The Level 3 TCGA gene expression data (LIHC.rnaseqv2__illuminahiseq_rnaseqv2) from TCGA [25] were downloaded from Firebrowse (http://firebrowse.org/). Clinical data were obtained from (https://www.cbioportal.org/). Total of 12 tumour samples had identified NAFLD as one of aetiologies. For surrounding liver tissue only two matched NAFLD samples were available in the dataset, thus we included ‘Other’ (n=4) and ‘No History of Primary Risk Factors’ (n=19) as controls.
2.5. Statistics
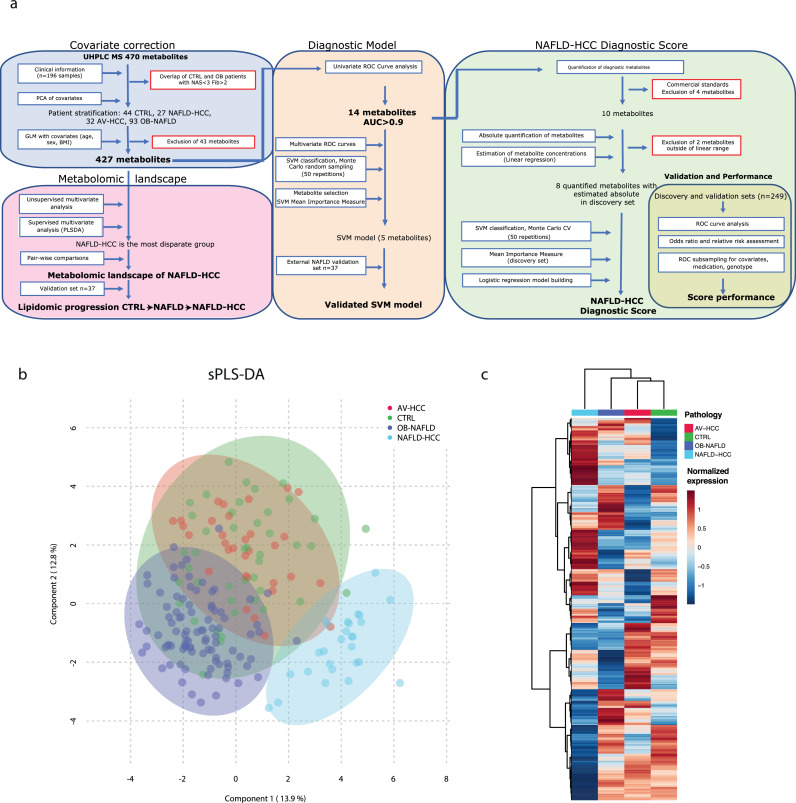
We identified 0·3% of missing values in the matrix with the maximum of missing values in one feature equal to 15%. The missing values were estimated by the k-nearest neighbour method with Metaboanalyst 4.0 [26]. Data were quantile-normalised, log-transformed before analysis (Metaboanalyst 4.0). The covariate testing and correction was performed with linear regression priori to analysis, and age, sex and BMI were included as covariates in differential expression analysis (R 4.0.4) The outline of statistical approaches is presented in Figure 1a.
Figure 1.
Workflow of statistical approaches and multivariate analysis between the patient groups. a. Schematic outline of the analytical steps applied post-detection and normalisation of metabolites b. The 3 components that based on spares partial least squares discriminant analysis (sPLS-DA model) of all detected metabolites show samples separation. c. Unsupervised hierarchical clustering of patient subgroups (each subgroup is averaged) across all metabolites (Ward's method clustering).
Differences in biochemical parameters were established with one-way ANOVA, Kruskal-Wallis test with Tukey's multiple comparisons, or Fisher exact test for categorical data (Prism 8.2.0).
The covariate testing and correction was performed with linear regression priori to analysis, and age, sex and BMI were included as covariates in differential expression analysis (R 4.0.4).
Data sets per metabolic class were calculated as the sum of the normalised areas of all the metabolites with the same chemical characteristics. The outlier analysis was performed before one-way ANOVA or Kruskal-Wallis test with Tukey's or Dunn's multiple testing (Prism 8.2.0) depending on data normality.
Multivariate analyses were performed with package mixOmics [27] or Metaboanalyst 4.0. The sparse partial least squares discriminant analysis (sPLS-DA) was used to investigate the best separation between the groups for multiple groups and orthogonal partial least squares discriminant analysis (oPLS-DA) for pair-wise compresence.
Hierarchical cluster analysis was performed with Ward's algorithm using Euclidean distances.
The receiver operating characteristic (ROC) curves were generated for pair-wise comparison and areas under the curves (AUC) were estimated with Metaboanalyst 4.0 or Prism 8.2.0
The diagnostic model was constructed using ROC curves generated by Monte-Carlo cross-validation (MCCV) using balanced sub-sampling. In each MCCV, two-thirds (2/3) of the samples were used to evaluate the feature importance and the classification model was validated on 1/3 of the samples that were left out. The procedure was repeated 50 times to calculate the performance and confidence interval of each model detecting the optimal number of features for best accuracy. The linear support vector machine (SVM) method was used for sample classification. Feature selection was performed with SVM Mean Importance Measure. The average accuracy of the model was based on 100 cross validations. Model performance was measured with 1000 permutations and both area under the ROC curve and predictive accuracy p<0·001 (Metaboanalyst 4.0).
NAFLD-HCC diagnostic score (NHDS). In the discovery set of 196 samples, 8 metabolites [1] had commercial standards allowing absolute quantification, [2] highly correlated between measured concentration and peak areas. These were used to build an SVM model to identify the optimal number of metabolites and select features (Mean Importance Measure) distinguishing NAFLD-HCC. The concentrations of selected metabolites were used to build a logistic regression model (with an additional 10-fold Cross Validation) retaining metabolites in the equation with p<0·05. Model performance was tested with 1000x permutations and both AUC and predictive accuracy reaching p<0·001 (Metaboanalyst 4.0). A logistic model was generated based on metabolite concentrations in the discovery set data establishing the NAFLD-HCC diagnostic score (NHDS) and the cut-off value. The NHDS was calculated for the validation set. ROC curves between the groups were computed and compared (Prism 8.2.0).
The odds ratio and relative risk for NHDS, clinical and biochemical features were established using Prism 8.2.0. We generated confusion matrixes for the features in discovery and validation sets as well as pulled patient sets using a cut-off value of age (50 years), BMI=30, and reference ranges for biochemical features. Statistical significance was measured with Fisher's exact test (p<0·05), the relative risk was calculated with Koopman asymptotic score and odds ratio with the Baptista-Pike method.
Role of covariates on the performance of NHDS. Patients were stratified in groups according to the underlying condition (diabetes, dyslipidaemia, cirrhosis (NAFLD-HCC)), reported medication (statin and paracetamol) use or NAFLD risk-genotype of Patatin-like phospholipase domain-containing protein 3 (PNPLA3), Transmembrane 6 Superfamily Member 2 (TM6SF2), Membrane-Bound O-acyltransferase Domain Containing 7 (MBOAT7).
Pearson correlation was applied for pattern hunter analysis to detect metabolites significantly correlating with progressive increase or decline. Metabolites were considered as correlated with pattern when (q<0·05) and abs (r)> 0·3.
Pathway overrepresentation analysis was performed with the Integrated Molecular Pathway Level Analysis (IMPaLA) [28]. As the majority of our metabolites belong to lipids, which to a high extend are not included in metabolic pathways, we used all detected serum metabolites with HMDB identifiers as background in the enrichment analysis. Lipid pathway enrichment analysis was performed with BioPAN software and default settings.
2.6. Role of funding sources
The funding sources for this study had no role in study design, data collection, data analyses or interpretation nor writing of the report as it is presented herein.
3. Results
3.1. Clinical and biochemical features of studied populations
We analysed the metabolic composition of serum obtained from 249 patients divided into a discovery set comprising patients with NAFLD-HCC (n=27), non-cancerous individuals (n=137) and 32 patients with alcohol- or viral-related HCC (AV-HCC). The control (CTRL) group included 44 individuals (35 healthy subjects [22] and 9 bariatric surgery patients with a NAFLD-activity score (NAS)<3 and fibrosis score <2) and 93 morbidly obese NAFLD patients awaiting bariatric surgery (OB-NAFLD). Furthermore, we independently validated our findings in serum from 37 patients with NAFLD and in plasma from 16 patients with NAFLD-HCC. Importantly, all NAFLD-HCC patients had no prior history of viral hepatitis or excessive alcohol consumption. The clinical, pathological, and biochemical features of all patients are summarized in Supp. Table 1 and Supp. Fig. 1.
We observed a significant difference in mean age (NAFLD-HCC patients were significantly older compared to CTRL, OB-NAFLD and AV-HCC), BMI (significantly higher in OB-NAFLD compared to CTRL, AV-HCC and NAFLD-HCC), as well as gender ratio (significantly higher prevalence of HCC among males, but equal between NAFLD-HCC and AV-HCC). Nevertheless, unsupervised principal component analysis (PCA) showed that the metabolic profiles grouped independently of these covariates (Supp. Fig. 2). Additionally, the presence of underlying diabetes, cirrhosis, and level of fibrosis did not affect patients’ grouping in unsupervised clustering. Further, the analysis of alpha-fetoprotein (AFP) or other liver biochemical features (alanine aminotransferase (ALT), gamma-glutamyl transferase (GGT), alkaline phosphatase (AP), bilirubin, albumin, and prothrombin activity) to follow the liver function showed extensive variability already in non-cancerous patients, but generally remained within the range of their references (Supp. Fig. 1). As such, these markers presented limited potential for diagnosing NAFLD-HCC (Table 1). To investigate whether metabolomic changes in NAFLD-HCC are aetiology-specific, we compared the profiles of NAFLD-HCC patients and 32 patients with AV-HCC. As such, the metabolic profiles of patients with AV-HCC showed significant overlap, but a clear separation from the profiles of patients with NAFLD-HCC (Supp. Fig. 2h). Compared to patients with AV-HCC, NAFLD-HCC patients were older (p<0·05) and less likely to develop HCC on a cirrhotic background (respectively 90% compared to 30%, Fisher's exact test p<0·0001). However, no significant differences were observed between NAFLD-HCC and AV-HCC in measurements of the liver function (AFP, AP, ALT, GGT, and bilirubin) or diabetes. Interestingly, tumours obtained from NAFLD-HCC patients were larger in size compared to tumours from patients with AV-HCC (Mann Whitney test, p=0·0006) and more frequently displayed microvascular invasion (41% NAFLD-HCC compared to 20% AV-HCC, Fisher's exact test p=0·016).
Table 1.
The area under the curve of the receiver operating characteristic curves for the model, biochemical parameters, and individual metabolites
| Model | AUC |
||
|---|---|---|---|
| CTRL | OB-NAFLD | AV-HCC | |
| 0·989 | 0·997 | 0·999 | |
| Biochemical features | |||
| AFP | 0·786 | NA | 0·613 |
| ALT | 0·776 | 0·733 | 0·570 |
| GGT | 0·927 | 0·865 | 0·562 |
| AP | 0·781 | 0·606 | 0·515 |
| Bilirubin | 0·619 | 0·859 | 0·539 |
| Individual metabolites | |||
| AC (18:2n-6) | 0·957 | 0·950 | 0·961 |
| PC (16:0/17:0) | 0·992 | 0·991 | 0·942 |
| Linoleic acid | 0·910 | 0·990 | 0·968 |
| Osbond acid | 0·944 | 0·964 | 0·965 |
| Palmitolinoleic acid | 0·915 | 0·977 | 0·948 |
| MUFA (14:1n-5trans) | 0·934 | 0·962 | 0·979 |
| PUFA (14:2n-x) | 0·978 | 0·971 | 0·998 |
| PUFA (16:3n-x) | 0·963 | 0·964 | 1·000 |
| Hydroxy-octadecadienoic acid | 0·931 | 0·982 | 0·966 |
| PC (0:0/18:2) | 0·924 | 0·909 | 0·925 |
| PC (18:2/0:0) | 0·900 | 0·925 | 0·973 |
| PC (0:0/22:5) | 0·914 | 0·930 | 0·987 |
| TG (47:0) | 0·978 | 0·968 | 0·933 |
| TG (45:1) | 0·955 | 0·933 | 0·903 |
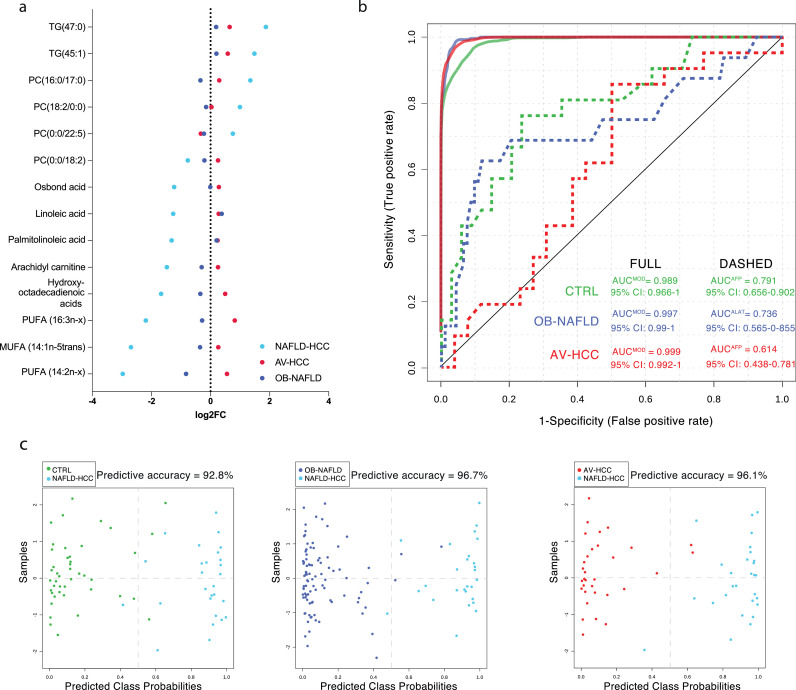
Area under the curve (AUC) for biochemical features and individual metabolites calculated as AUC of the receiver operating characteristic (ROC) curves and AUC from support vector machine (SVM)-driven modelling· Abbreviations: alpha-fetoprotein (AFP), alanine aminotransferase (ALT), gamma-glutamyl transferase (GGT), alkaline phosphatase (AP), acylcarnitine (AC), phosphatidylcholines (PC), monounsaturated fatty acids (MUFA), polyunsaturated fatty acids (PUFA) and triglycerides (TG).
3.2. NAFLD-HCC patients present a disparate serum metabolome
To establish a serum-based metabolomic landscape of NAFLD-HCC, we performed detailed metabolomics using a comprehensive library of 1295 metabolites covering amino acids (AA), glycerophospholipids, fatty acyls, sterols, sphingolipids, and glycerolipids. In total, we detected 470 metabolites. Due to the differences in age, sex and BMI between the groups, we performed covariate correction, including age, sex and BMI, and as a result 43 metabolites were excluded from further analysis (Fig. 1a). Next, sparse partial least squares discriminant analysis (sPLS-DA) revealed that NAFLD-HCC patients metabolically are the most distinct group compared to CTRL, OB-NAFLD, and patients with AV-HCC (Fig. 1b). Besides, NAFLD-HCC patients were the most dissimilar as seen by unsupervised hierarchical clustering (Fig. 1c). Interestingly, AV-HCC and NAFLD-HCC subgroups showed low similarity and clustered far apart (Fig. 1b, Supp. Fig. 2h), suggesting that unique metabolic programs may be driven by differences in the disease aetiology (NAFLD versus alcohol or viral hepatitis background).
3.3. Diagnostic potential of serum metabolomics
Serum metabolomics has been successfully used as a diagnostic tool to discriminate liver diseases [21,22]. Here, we investigated the potential of distinguishing NAFLD-HCC not only from healthy individuals and NAFLD patients, but also from AV-HCC. Thus, to generate a predictive metabolite signature, we first used receiver operating characteristic (ROC) curves and calculated area under the curve (AUC) for each metabolite as a contrast test between NAFLD-HCC and the respective comparative groups (CTRL, OB-NAFLD, and AV-HCC). As such, 89 metabolites presented an AUC>0·75, distinguishing NAFLD-HCC patients from each of the other control groups (healthy or disease). Among the metabolites in the DEM signature, 14 metabolites presented a superior AUC>0·9 in all contrast tests (Table 1). These metabolites and their fold change compared to CTRL are presented in Fig. 2a. Importantly, these 14 metabolites individually present AUCs markedly superior to alpha-fetoprotein (AFP: AUCNAFLD-HCC vs CTRL=0·791 and AUCNAFLD-HCC vs AV-HCC=0·614) and all biochemical measurements for the liver function (ALT: AUCNAFLD-HCC vs CTRL=0·776, AUCNAFLD-HCC vs NAFLD=0·733, and AUCNAFLD-HCC vs AV-HCC=0·570 or GGT: AUCNAFLD-HCC vs CTRL= 0·927, AUCNAFLD-HCC vs NAFLD=0·865, and AUCNAFLD-HCC vs AV-HCC=0·562), discriminating NAFLD-HCCs from all controls and AV-HCC (HCCs of different aetiology other than NAFLD).
Figure 2.
The diagnostic potential of lipidomics. a. The waterfall plot presenting the 14 metabolites signature distinguishing NAFLD-HCC patients. Metabolites are presented as the fold change to CTRL. b. ROC curves of 5-metabolite based model distinguishing NAFLD-HCC patients from other groups in compresence to biochemical tests. c. The predicted class probabilities (average of the cross-validation) for each sample using the best classifier (based on AUC).
Next, we assessed if a combination of the 14 metabolites would increase the diagnostic potential. As such, we used support vector machine (SVM) modelling and determined that a panel of 5 metabolites yielded the optimal predictive accuracy (Supp. Fig. 3a). Indeed, the model based on the 5 metabolites reached an AUC>0·98 (Fig. 2b) for any of the contrasts (compared to all controls and AV-HCC) with a predictive accuracy greater than 90% (Fig. 2c). Importantly, the accuracy of the diagnostic panel was confirmed in the validation set and a model performance with an AUC=0·91, including matching NAFLD-HCC patients according to BMI (Supp. Fig. 3b).
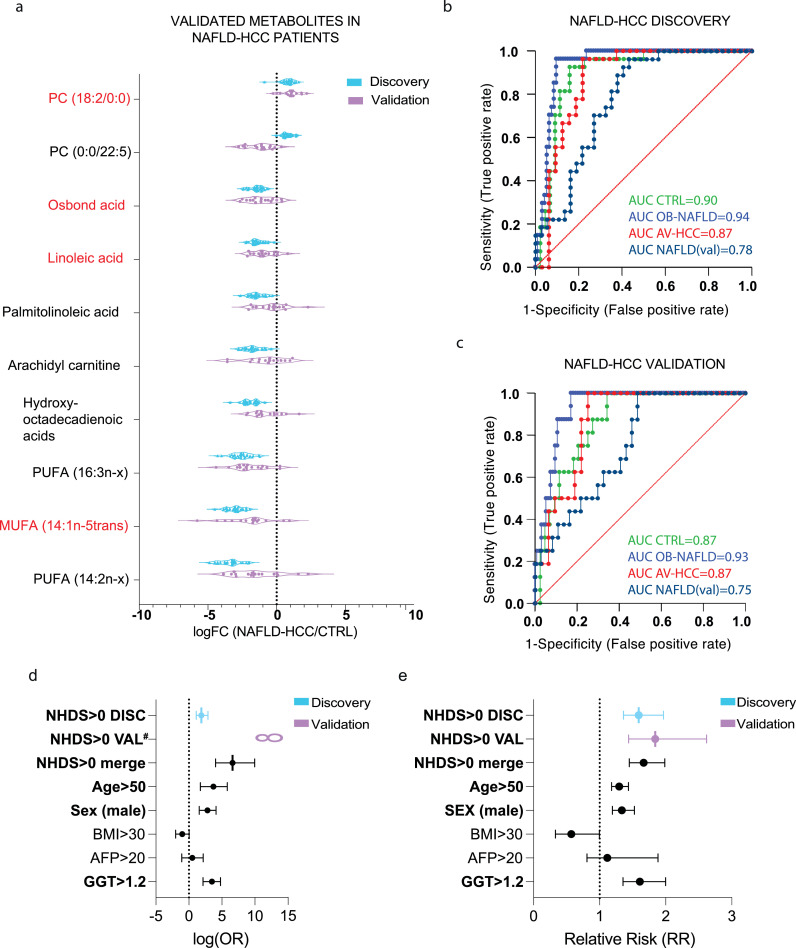
3.4. Validation and quantification of diagnostic metabolites
To reinforce the clinical relevance of the metabolite panel in the diagnosis of NAFLD-HCC patients, we next validated and quantified the abundance of each metabolite. Also, we established their reference concentration range. Among the 14 metabolites with the greatest diagnostic value, only 10 of them have commercial standards available. Also, each metabolite needs to be detected within the linear range, which allows for absolute quantification. As such, we established the abundance of metabolites in the validation set (Fig. 3a). All metabolites except PC (0:0/22:5) showed a similar trend as in the discovery set. Overall, the validation set showed a great variability, which can be due to difference in geographical sampling or availability of plasma instead of serum. Moreover, we utilised the quantification of each metabolite to generate linear regression models to assess their concentration levels in the discovery set. The linear dependence and estimated concentrations are presented in Supp. Table 2. Lastly, among the validated metabolites in the panel, which are based in SVM modelling and the mean importance measure, we determined the optimal set of metabolites (linoleic acid, osbond acid, monounsaturated fatty acid MUFA (14:1n-5trans), and PC (18:2/0:0)) to build the NAFLD-HCC Diagnostic Score (NHDS). The NHDS is dependent on the absolute concentration level of each metabolite in the score. The NHDS performed with an AUC>0·75 for any given contrast (Fig. 3b-c) as well as presented suitable power according to sample size (Supp. Table 3). Furthermore, we established the odds ratio (OR) and relative risk (RR) for the NHDS (at cut-off value 0) in the discovery, validation, and combined cohort. The NHDS>0 was predictive of NAFLD-HCC in the discovery set (Fisher's exact test, p=0·01) with relative risk RR=1·18, odds ratio OR=8·87 and in validation set (Fisher's exact test, p<0.0001) with RR=2·23 and infinite OR. Taken together, the NHDS>0 was predictive of NAFLD-HCC with the highest odds ratio OR=97·55 (95% CI=16·40 to 999·7) (Fig. 3d) and relative risk RR=1·67 (95% CI=1·45 to 1·99) (Fig. 3e). Moreover, we next tested if underlying conditions, such as diabetes, dyslipidaemias, cirrhosis effect the performance of NHDS (Supp. Table 4). Interestingly, no DEMs were detected between cirrhotic and non-cirrhotic NAFLD-HCC patients in direct pair-wise comparison. Next, we considered if the use of medication (statin and paracetamol) and the presence of known NAFLD risk-genotypes (Patatin-like phospholipase domain-containing protein 3 (PNPLA3), Transmembrane 6 Superfamily Member 2 (TM6SF2), Membrane Bound O-Acyltransferase Domain Containing 7 (MBOAT7)) effected the performance of NHDS. Indeed, the model showed significant performance (AUC: 0·888-1, p<0·0001) regardless of the underlying condition, medication used and genotypes (Supp. Table 4). Also, the NHDS predicted NAFLD-HCC independent of tumour stage (BCLC and TNM), degree of NAFLD and sex (Supp. Table 4).
Figure 3.
Validation of metabolites and NAFLD-HCC Diagnostic Score (NHDS). a. The relative abundances of 10 validated metabolites in discovery (blue) and validation (purple) sets are expressed as a fold change against CTRL. Metabolites included in NHDS are marked in red. b. ROC curves of NHDS in the discovery set. c. ROC curves of NHDS in the NAFLD-HCC validation set. d. The forest plot presenting odds ratios (Fisher's exact test) of NHDS at the established cut-off value as well as clinical and biochemical characteristics for discovery, validation, and pulled (discovery and validation) sets. AFP and GGT were available only in the discovery set. Error bars present 95% CI, bold font was used for statistically significant (Fisher's exact test, p<0•05) factors #The Odds ratio for NHDS in validation set was infinite e. The forest plot representing the relative risk (Fisher's exact test) of NHDS at the established cut-off value as well as clinical and biochemical characteristics for discovery, validation and pulled (discovery and validation) sets. AFP and GGT were available only in the discovery set. Error bars present 95% CI, bold font was used for statistically significant (Fisher's exact test, p<0•05) factors
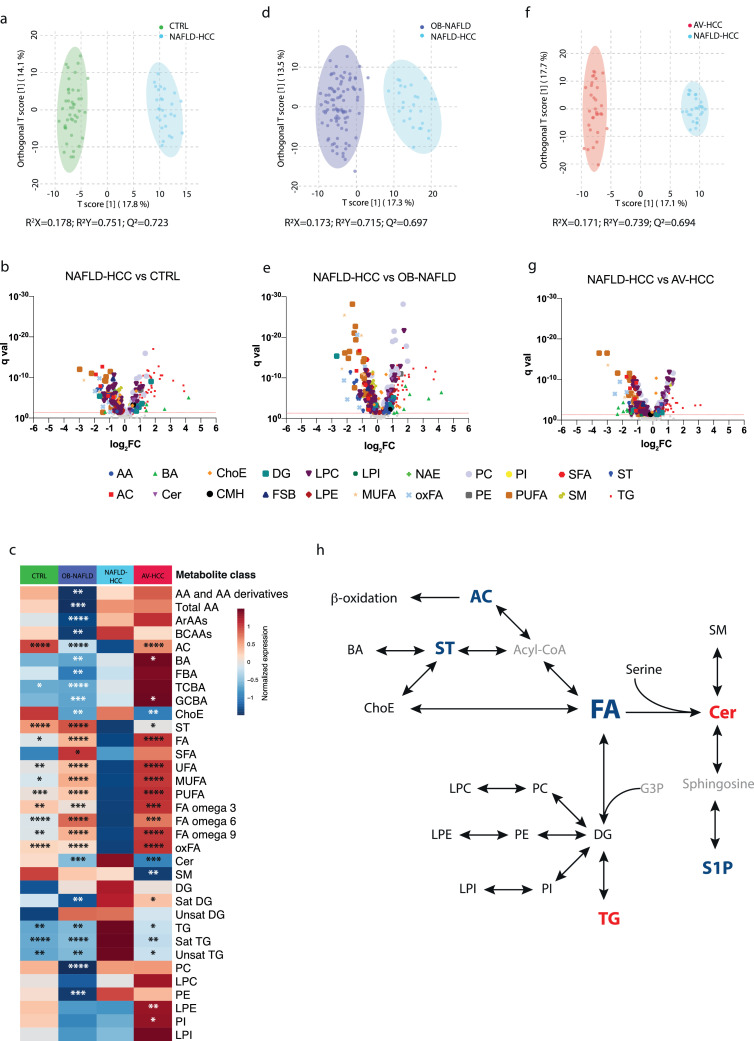
3.5. Unique metabolomic profile of NAFLD-HCC patients
To investigate differences in the metabolic profiles between NAFLD-HCC patients and the other groups, we performed a series of pair-wise tests. As such, after the covariate correction (age, sex, BMI) orthogonal partial least squares discriminant analysis (oPLS-DA) showed substantial separation of NAFLD-HCC patients from CTRL (R2X=0·178, R2Y=0·751, Q2=0·723) (Fig. 4a). The differential expression analysis identified 274 significantly different (t-test, false-discover rate (FDR) corrected p<0·05) metabolites (DEMs), among which 152 metabolites were depleted and 122 metabolites were increased (Fig. 4b, Supp. Table 5). Next, we performed pathway overrepresentation analysis of the depleted metabolites using integrated molecular pathway level analysis (IMPaLA) [28] and identified linoleic acid metabolism and G protein-coupled receptor (GPCR) signalling as the most impaired networks (Supp. Table 6). Contrary, cholesterol synthesis, membrane fluidity and trafficking, and glycerophospholipid metabolism were among the most upregulated pathways (Supp. Table 6). In addition to relate individual metabolites to their processes, we compared unique classes of metabolites with the same chemical characteristics. As such, we defined a unique depletion of acylcarnitines (AC), sterol lipids (ST), and fatty acids (FA; especially oxidized fatty acids (oxFA) and omega-6 FA) in NAFLD-HCC, while saturated triglycerides (TG) were upregulated (Fig. 4c, Supp. Fig. 4). Furthermore, we utilised Bioinformatics Methodology For Pathway Analysis (BioPAN) [29] for lipid pathway enrichment analysis. We observed a significant activation of reactions converting sphingomyelins (SM) to ceramides (Cer), a process catalysed by sphingomyelin phosphodiesterases (SMPD2 and SMPD3), as well as phosphatidylcholines (PC) to diglycerides (DG), which is catalysed by the sphingomyelin synthases (SGMS1 and SGMS2) (Supp. Fig. 5a). Similarly, significant alterations were revealed in the activity of FA desaturases and elongases with specific activation of fatty acid desaturase 1 (FADS1) and impairment in FADS2, stearoyl-CoA desaturase 1 (SCD1) and elongation of very long chain fatty acid (ELOVL) elongases (ELOVL2, and ELOVL5) (Supp. Fig. 6a).
Figure 4.
Pair-wise comparison between NAFLD-HCC and other groups. a. The orthogonal partial least squares discriminant analysis (oPLS-DA) showing separation between NAFLD-HCC and CTRL. b. Volcano plot representing DEMs between NAFLD-HCC and CTRL samples. The FDR p=0•05 is marked with a red dashed line. c. Differences between NAFLD-HCC and other groups across major metabolic classes. Statistical significance between NAFLD-HCC and other groups was established with one-way ANOVA or Kruskal-Wallis tests *p<0•05, **p<0•01, ***p<0•001, ****p<0•0001 d. The oPLA-DA model showing separation between NAFLD-HCC and OB-NAFLD. e. Volcano plot representing DEMs between NAFLD-HCC and OB-NAFLD. f. The oPLS-DA model showing separation between NAFLD-HCC and AV-HCC. g. Volcano plot representing DEMs between NAFLD-HCC and AV-HCC. h. Simplified relationship between major metabolic classes. Classes marked in blue are downregulated and red upregulated in NAFLD-HCC patients. DEMs were identified by t-test followed by FDR correction). Abbreviations: acylcarnitines (AC), amino acids (AA), aromatic amino acids (ArAAs), bile acids (BA), branched-chain amino acids (BCAAs), ceramides (Cer), cholesteryl esters (ChoE), diacylglycerols (DG), diacylglycerophosphocholine (DAPC), diacylglycerophosphoethanolamine (DAPE), fatty acids (FA), fatty acids omega 3 (FA omega 3), fatty acids omega 6 (FA omega 6), fatty acids omega 9 (FA omega 9), free bile acids (FBA), free sphingoid bases (FSB), glycine-conjugated bile acids (GCBA), lysophosphatidylcholines (LPC), lysophosphatidylethanolamines (LPE), lysophosphatidylinositols (LPI), monohexosylceramides (CMH), monounsaturated fatty acids (MUFA), N-acyl ethanolamines (NAE), oxidized fatty acids (oxFA), phosphatidylcholines (PC), phosphatidylethanolamines (PE), phosphatidylinositols (PI), polyunsaturated fatty acids (PUFA), saturated diacylglycerols (Sat.DG), saturated fatty acids (SFA), saturated triacylglycerols (Sat.TG), sphingomyelins (SM), steroids (ST), taurine-conjugated bile acids (TCBA), triacylglycerols (TG), unsaturated fatty acids (UFA).
Next, we compared OB-NAFLD and NAFLD-HCC patients. Indeed, oPLS-DA analysis showed moderate separation (R2X=0·173, R2Y=0·715, Q2=0·697) (Fig. 4d). The differential expression analysis identified 316 DEMs with 154 metabolites upregulated and 162 downregulated (Fig. 4e, Supp. Table 7). The overrepresentation analysis identified cholesterol and bile acid (BA) metabolism, aminoacyl-tRNA biosynthesis, as well as protein and glucose metabolism as significantly upregulated in NAFLD-HCC compared to OB-NAFLD. Contrary, FA biosynthesis and GPCR signalling were both downregulated (Supp. Table 8). Furthermore, OB-NAFLD significantly differed from NAFLD-HCC in specific classes of metabolites. As such, AA, BA, Cer, TG, PC, and phosphatidylethanolamines (PE) were all significantly upregulated in NAFLD-HCC compared to OB-NAFLD. Contrary, FA and AC were reduced in NAFLD-HCC (Fig. 4c, Supp. Fig. 4). Moreover, the pathway enrichment analysis identified two reactions (SM→Cer and DG→PE) as the most dynamic, whereas the opposite direction (Cer→SM and PE→PC→DG) was repressed (Supp. Fig. 5b). Moreover, ELOVL3 and ELOVL6 presented an increased activity, whereas ELOVL2, SCD1, and SCD3 were suppressed (Supp. Fig. 6b). Contrary, reactions catalysed by FADS1, FADS2, and ELOVL5 displayed a mixed activity.
Lastly, we compared the metabolomes of NAFLD-HCC to HCCs with alcohol and/or viral aetiology (AV-HCC). The oPLS-DA model showed moderate separation (R2X=0·171, R2Y=0·739, Q2=0·694) (Fig. 4f). As expected, the AV-HCC group showed greater heterogeneity compared to NAFLD-HCC, which partly can be due to the mixed etiology. Differential expression analysis identified 257 DEMs (125 downregulated and 132 upregulated) in NAFLD-HCC compared to AV-HCC (Fig. 4g, Supp. Table 9), showing an overall impairment in the free FA (FFA) biosynthesis. Contrary, insulin resistance, glycerophospholipid, and choline metabolism were upregulated in NAFLD-HCC (Supp. Table 10). Next, we compared metabolic classes between NAFLD-HCC and AV-HCC patients and observed significant depletion of AC, BA, FA, ST, lysophosphatidylethanolamines (LPE), and phosphatidylinositols (PI) in NAFLD-HCC. Contrary, cholesteryl esters (ChoE), SM, Cer, and TG were all increased in NAFLD-HCC compared to AV-HCC (Fig. 2c, Supp. Fig. 4). Moreover, lipid pathway enrichment analysis showed an increased reaction activity in the path SM→Cer (Supp. Fig. 5c) with the enzyme activities of FADS2, ELOVL5, and ELOVL2 significantly reduced (Supp. Fig. 6c).
Taken together, the serum of NAFLD-HCC patients is characterized by a significant depletion of FA reflective of a significantly lower FA biosynthesis with decreased FA desaturase and elongase activities. A depletion in both AC and ST with concurrent higher TG and Cer abundance is suggestive of a unique metabolic reprogramming in NAFLD-HCC patients. The altered SM:Cer ratio could be the result of an increased activity in the enzymes SMPD2 and SMPD3 or reduced activity of SGMS1, SGMS2, CERT1 in NAFLD-HCC patients. A simplified association between the lipid classes and their deregulation in NAFLD-HCC is presented in Fig. 4h.
3.6. NAFLD-HCC tumours are responsible for fatty acid deterioration in circulation
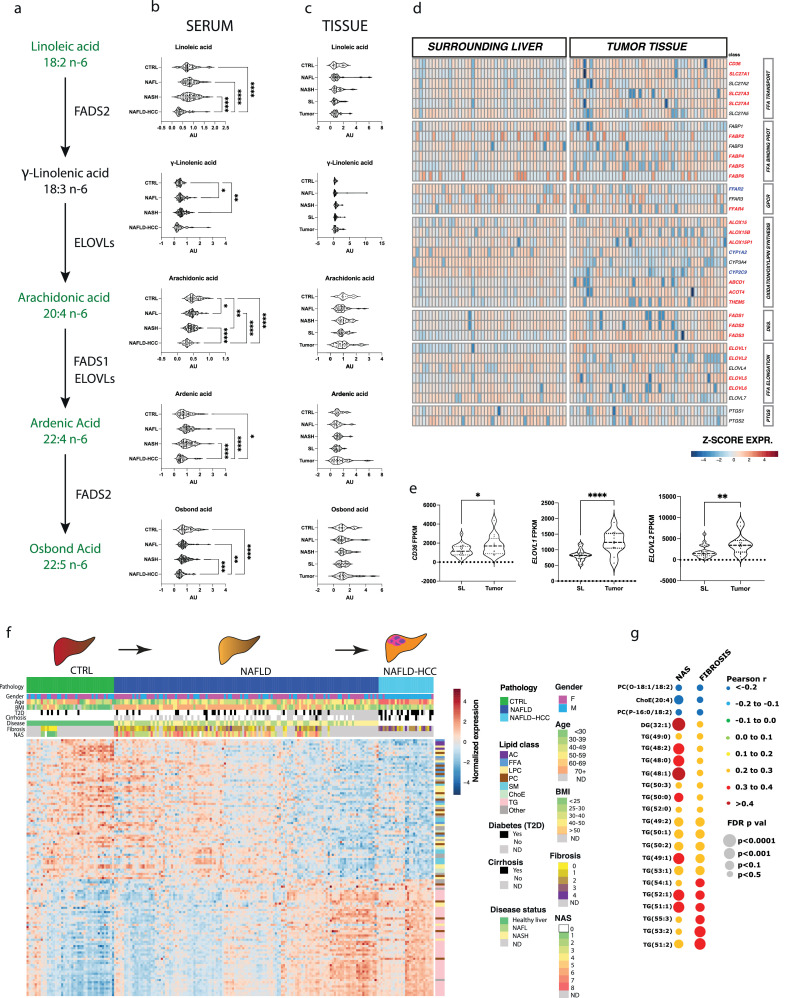
We observed significant depletion of omega-6 unsaturated fatty acids in the serum of NAFLD-HCC patients. Interestingly, essential linoleic acid (LA) as well as its metabolite osbond acid showed significant difference and diagnostic potential. As such, we investigated the concentration of LA and its elongation and desaturation pathway (Fig. 5a). We first pooled the NAFLD discovery and validation patient sets and based on the liver histology, we stratified them into NAFL (NAS<6, n=58) and NASH subsets (n=72). Indeed, we observed significant depletion of LA, arachidonic acid, ardenic acid and osbond acid in the serum from NAFLD-HCC patients (Fig. 5b). Interestingly, these FAs remained unchanged in serum-matched tumours compared to the normal tumour-adjacent tissues, healthy CTRLs as well as NAFL and NASH patients (Fig. 5c). As such, we hypothesized that tumour tissue might be responsible for the metabolic sink of FFA. To that end, we measured the expression of FA transporters, FA binding proteins, G-protein coupled receptor for FA, FA elongases, FA desaturases and genes involved in oxylipin and prostaglandin synthesis (Fig. 5d). NAFLD-HCC tumours showed a significant increase in the expression of FA transporters (CD36, SLC27A1, SLC27A3, SLC27A4), FA binding proteins (FABP2, FABP4, FABP5, FABP6), FA elongases (ELOVL1, ELOVL2, ELOVL5, ELOVL6), FA desaturases (FADS1-3). Similarly, LA lipoxygenases (ALOX15 family) were upregulated in tumours while cytochrome P450 enzymes (CYPs) with LA affinity were downregulated. Furthermore, we validated overexpression of CD36 and FA elongases in NAFLD-HCC samples in The Cancer Genome Atlas (Fig. 5e). Collectively, these data suggest that NAFLD-HCC tumours significantly depend on exogenous FAs, increasing their uptake and metabolism.
Figure. 5.
The metabolism of linoleic acid in serum and tissue and the progressive metabolic perturbations in CTRL to NAFLD to NAFLD-HCC trajectory. a. The schematic representation of LA metabolism (Fatty Acid Desaturase 1 (FADS1), Fatty Acid Desaturase 2 (FADS2), Elongation of Very Long chain fatty acids (ELOVLs)). b. Violin plots representing the abundance of LA metabolites in serum. Expression was tested with one-way ANOVA or Kruskal-Wallice test *p<0•05, **p<0•01, ***p<0•001, ****p<0•0001 c. Violin plots representing the abundance of LA metabolites in tissue (NAFLD-HCC surrounding liver (SL, n=27); NAFLD-HCC tumour tissue (T, n=27); CTRL n=9; NAFL n=32; NASH n=20). d. The heatmap representing gene expression of fatty acid transport and metabolism genes in NAFLD-HCC surrounding liver tissue and NAFLD-HCC tumours. Genes marked in red are significantly (t-test, p<0•05) upregulated and blue downregulated in tumour compared to matched surrounding liver tissue. e. The mRNA expression of CD36, ELOVL1 and ELOVL2 in NAFLD-HCC tumours in TCGA (n=12) compared to SL (n=25). f. The heatmap presenting the expression of metabolites statistically significantly correlated with disease progression. g. The bubble plot presenting Pearson's correlation of metabolites with NAS and fibrosis level in NAFLD patients.
3.7. The serum lipidome landscape reflects the progression to NAFLD-HCC
We investigated whether the serum lipidome reflects the progressive nature of NAFLD-HCC development. It has been previously reported, that major lipidomic alterations are observed in the development of steatosis, but not progression to NASH [16]. We performed pair-wise comparisons between NAFL and NASH adjusting for age, sex and BMI and found no DEMs, which is in accordance with recent findings [16]. As such, we investigated the CTRL→NAFLD→NAFLD-HCC axis employing a pattern hunter approach with Pearson correlation to designate the metabolic rearrangements in the subgroups as part of the developing disease (Fig. 5f). A total of 412 lipids were detected in the pooled discovery and validation NAFLD samples.
As such, we found a total of 121 lipids progressively altered (Pearson's correlation FDR p<0·05, r>0·3; 66 negatively, and 55 positively) following this axis (Supp. Table 11). The negatively correlated metabolites include AC, ChoE, PUFA, LPC, and ST subclasses. Among the metabolites positively correlated, we found an overrepresentation of TG (42 out of 55 metabolites). Importantly, 18 TGs significantly correlate with increasing NAS and fibrosis scores in OB-NAFLD patients, suggesting their importance in the progressive deterioration of the liver (Fig. 5g). Lastly, we tested whether any of these metabolites correlated with tumour size, recurrence, microvascular invasion, or liver cirrhosis in NAFLD-HCC patients, however, none of the metabolites reached statistical significance. This suggests that these metabolites are associated with NAFLD-HCC, but not directly involved the cancer progression.
4. Discussion
A sedentary lifestyle and overnutrition have led to an epidemic of obesity, diabetes, and NAFLD that soon may become the leading causes of HCC development. Metabolic reprogramming is at the core of NAFLD progression to HCC [21]. However, due to the need of invasive techniques for NAFLD diagnosis, our understanding of NAFLD-HCC is limited and the underlying metabolomic landscape remains elusive. The present study provides the first comprehensive analysis covering 22 classes of lipids and amino acids and identifying complex relationships in the gradual progression of NAFLD versus NAFLD-HCC patient subsets. Importantly, this study was performed in patients with biopsy-proven liver histology before therapy. To our knowledge, it is the first study aiming to non-invasively differentially diagnose NAFLD-HCC from HCC of other aetiologies. The current study is focused on early-stage, resected NAFLD-HCC patients, which is important for detection of metabolic changes at early-stage of the disease and for its application in screening of patients at-risk. Our findings warrant investigation in patients with advanced disease.
The diagnosis of NAFLD-HCC is challenging as patients with metabolic syndrome are not screened for HCC [7,11]. Indeed, often only patients with chronic liver disease of viral aetiology are offered HCC screening. Increasing levels of liver enzymes are suggestive of liver damage but are not specific to hepatocarcinogenesis. Additionally, the diagnostic capacity of AFP [30] for HCC is limited and was observed to be particularly inadequate for NAFLD-HCC patients (Fig. 3d, e, Supp. Fig. 1). As such, we have applied machine learning approaches to pin-point a combination of metabolites that offers the best diagnostic potential for NAFLD-HCC patients. We found that a combination of 5 metabolites accurately distinguishes NAFLD-HCC patients from healthy individuals (AUC=0·989), OB-NAFLD patients (AUC=0·997), and patients with AV-HCC (AUC=0·999). This model performed well against a validation set of NAFLD patients (AUC=0·905). Furthermore, to bring metabolomics to clinical practice, it is crucial to establish methods for absolute quantification of metabolites as well as their reference concentration ranges. Here, we were able to validate 10 metabolites using commercially available internal standards and measure their concentrations. As such, we built a score (NHDS) based on the absolute concentrations of 4 metabolites (3 unsaturated FA and 1 phosphatidylcholine) that can be applied on both serum and plasma measurements and performs superior to AFP and GGT (liver function) (Fig. 3D, E). The NHDS performed superior to recently developed GALAD test (AUCnon-viral/alcoholic =0·88) [31] and similarly to APAC Score AUCNAFLD/NASH=0·95 [32], both scores were developed in predominantly viral and alcohol-related cohorts.
The unsaturated fatty acids (mono- (MUFA) and polyunsaturated (PUFA)) significantly differed between NAFLD-HCC and other groups (Fig. 4) with lower levels in NAFLD-HCC. Indeed, cancer cells show higher dependency on unsaturated fatty acids to proliferate [33], and lipid-dependence was previously reported in HCC [34]. Thus, we can speculate that NAFLD-HCC tumours act as sinks for unsaturated fatty acids from the blood. Indeed, the increased transport of FA by CD36 has been observed in widely investigated in the context of NAFLD and NASH [35]. As such, its further augmentation in NAFLD-HCC tumours might be responsible for reduced serum FA making it potential therapeutic target. The essential omega-6 FA (linoleic acid/LA) is a precursor to long-chain metabolites, including arachidonic acid, which is a substrate in the prostaglandin synthesis and thus, depletion of LA may cause an altered signalling and inflammatory response [36]. The role of LA in carcinogenesis remains controversial. On the one hand, LA and its derivatives have shown a tumour-suppressive role in colorectal cancer [37]. Conversely, LA accumulation (in a murine model) increases oxidative stress and causes a selective loss of intrahepatic CD4+ T cells, leading to NAFLD-mediated hepatocarcinogenesis [38]. Interestingly, serum LA is significantly depleted in NAFLD-HCC patients (Fig. 2a, Fig. 5b), suggesting that the murine model not fully mimic the human disease, and possibly further emphasizing the potential limitation of murine studies in this field. Interestingly, the LA abundance in tissues is restored (Fig. 5c) likely due to increased uptake by the tumour (Fig. 5d). Indeed, increased metabolism of LA and arachidonic acid have been implicated in viral-associated HCC in the Korean population [39]. The liver is the key organ for carnitine synthesis and metabolism [40]. As such, the significantly decreased levels of serum acylcarnitines suggests an increased β-oxidation of FFA [41], which could be caused by increased energy demands of NAFLD-HCC tumours. Furthermore, we found elevated levels of bile acids in NAFLD-HCC patients compared to OB-NAFLD patients, but lower than AV-HCC (Fig. 4c). That can be result of altered BA metabolism in cirrhotic patients owning to altered gut microbiome in cirrhosis [42].
HCC is a progressive disease developing over decades. We observed a gradual attenuation of acylcarnitines, lysophosphatidylcholines, and unsaturated FA and an increase in phosphatidylcholines and triglycerides during the progression of liver disease. Interestingly, some of the triglycerides that were found to be augmented in NAFLD versus NAFLD-HCC were previously associated with NAS and fibrosis scores in NASH [43,44] (Fig. 5g), but were not associated with tumour size. Although, we have presented a successful metric, demonstrating the ability of the metabolite panel in diagnosing NAFLD-HCC patients and in distinguishing these patients from AV-HCC, results must be interpreted with caution given the sample size. The NHDS needs to be tested in a large cohort, including advanced nonresectable and ethnically diverse HCC patients in the prospective longitudinal cohorts. However, stratification of HCC based on their metabolome may in the future be an approach to evaluate cryptogenic HCC patients. It is notable that NAFLD-HCC patients were significantly older compared to other groups which limited the power to detect metabolic differences.
In conclusion, the depletion of unsaturated FA, and the increase of triglycerides are at the core of deregulated metabolic networks in NAFLD-HCC, leading to altered signalling and likely different nutrient utilisation by cancer cells. These changes can be exploited for non-invasive surveillance of the ‘at risk’ population for early HCC detection in the background of metabolic syndrome.
Declaration of Competing Interest
Drs. Alonso and Arretxe are employed by OWL Metabolomics (One Way Liver, S.L). Prof. Banales is a member of the scientific advisory board of OWL Metabolomics. Remaining authors declare no conflict of interest.
Acknowledgments
Contributors
Conceptualization: ML, JBA. Data curation: ML, ASL, EZ, RJA, EE, MJP, MPS, FKK, MRG, MAJ, SKO, GEV, TD, CPO, BS, RIR, JMB, JBA. Formal Analysis: ML, EA, EZ, CA. Writing – original draft: ML, JBA. All authors read and approved the manuscript. ML, JBA have verified the underlying data.
Acknowledgment
We thank the participating patients and their families as well as clinical staff at all contributing centres. We thank the French Liver Biobanks network – INCa (BB-0033-00085) for the contribution of NAFLD-HCC samples (participating hospitals: Beaujon, Bordeaux, Grenoble, Henri-Mondor, Paris-SUD, and Rennes). We thank the section of Biostatistics at Department of Public Health, University of Copenhagen for statistical advice. The results are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. We thank all funding sources: The laboratory of JBA is supported by the Novo Nordisk Foundation (14040, 0058419), Danish Cancer Society (R98-A6446, R167-A10784, R278-A16638), and the Danish Medical Research Council (4183-00118A, 1030-00070B). Data used for validation in this study provided by JMB was funded by the Spanish Ministry of Economy and Competitiveness and ’Instituto de Salud Carlos III’ grants (PI18/01075, Miguel Servet Programme CON14/00129 and CPII19/00008) co-financed by ’Fondo Europeo de Desarrollo Regional’ (FEDER); CIBERehd, Spain; IKERBASQUE, Basque foundation for Science, Spain; BIOEF (Basque Foundation for Innovation and Health Research: EiTB Maratoia BIO15/CA/016/BD); Department of Health of the Basque Country (2017111010), Euskadi RIS3 (2019222054, 2020333010); Department of Industry of the Basque Country (Elkartek: KK-2020/00008), AECC Scientific Foundation and European Commission Horizon 2020 program (ESCALON project no.: 825510). Similarly, MAJ was funded by grants from the Fondo Nacional De Ciencia y Tecnología de Chile (FONDECYT #1191145 to M.A.) and the Comisión Nacional de Investigación, Ciencia y Tecnología (CONICYT, AFB170005, CARE Chile UC).
Data sharing
The data used in this study will according to IRBs be made available upon request if the requestor has an approved protocol.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103661.
Appendix. Supplementary materials
References
- 1.Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15(1):11–20. doi: 10.1038/nrgastro.2017.109. [DOI] [PubMed] [Google Scholar]
- 2.Younossi ZM, Blissett D, Blissett R, Henry L, Stepanova M, Younossi Y. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology. 2016;64(5):1577–1586. doi: 10.1002/hep.28785. [DOI] [PubMed] [Google Scholar]
- 3.Hardy T, Oakley F, Anstee QM, Day CP. Nonalcoholic Fatty Liver Disease: Pathogenesis and Disease Spectrum. Annu Rev Pathol. 2016;11:451–496. doi: 10.1146/annurev-pathol-012615-044224. [DOI] [PubMed] [Google Scholar]
- 4.Kim GA, Lee HC, Choe J, Kim MJ, Lee MJ, Chang HS. Association between non-alcoholic fatty liver disease and cancer incidence rate. J Hepatol. 2017 doi: 10.1016/j.jhep.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 5.Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sayiner M, Golabi P, Younossi ZM. Disease Burden of Hepatocellular Carcinoma: A Global Perspective. Dig Dis Sci. 2019;64(4):910–917. doi: 10.1007/s10620-019-05537-2. [DOI] [PubMed] [Google Scholar]
- 7.Noureddin M, Rinella ME. Nonalcoholic Fatty liver disease, diabetes, obesity, and hepatocellular carcinoma. Clin Liver Dis. 2015;19(2):361–379. doi: 10.1016/j.cld.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pais R, Fartoux L, Goumard C, Scatton O, Wendum D, Rosmorduc O. Temporal trends, clinical patterns and outcomes of NAFLD-related HCC in patients undergoing liver resection over a 20-year period. Aliment Pharmacol Ther. 2017;46(9):856–863. doi: 10.1111/apt.14261. [DOI] [PubMed] [Google Scholar]
- 9.Anstee QM, Reeves HL, Kotsiliti E, Govaere O, Heikenwalder M. From NASH to HCC: current concepts and future challenges. Nat Rev Gastroenterol Hepatol. 2019 doi: 10.1038/s41575-019-0145-7. [DOI] [PubMed] [Google Scholar]
- 10.Bertot LC, Adams LA. Trends in hepatocellular carcinoma due to non-alcoholic fatty liver disease. Expert Rev Gastroenterol Hepatol. 2019;13(2):179–187. doi: 10.1080/17474124.2019.1549989. [DOI] [PubMed] [Google Scholar]
- 11.Orci LA, Sanduzzi-Zamparelli M, Caballol B, Sapena V, Colucci N, Torres F. Incidence of Hepatocellular Carcinoma in Patients with Nonalcoholic Fatty Liver Disease: A Systematic Review, Meta-analysis, and Meta-regression. Clin Gastroenterol Hepatol. 2021 doi: 10.1016/j.cgh.2021.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Pfister D, Núñez NG, Pinyol R, Govaere O, Pinter M, Szydlowska M. NASH limits anti-tumour surveillance in immunotherapy-treated HCC. Nature. 2021 doi: 10.1038/s41586-021-03362-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudek M, Pfister D, Donakonda S, Filpe P, Schneider A, Laschinger M. Auto-aggressive CXCR6+ CD8 T cells cause liver immune pathology in NASH. Nature. 2021 doi: 10.1038/s41586-021-03233-8. [DOI] [PubMed] [Google Scholar]
- 14.Pinyol R, Torrecilla S, Wang H, Montironi C, Pique-Gili M, Torres-Martin M. Molecular characterization of hepatocellular carcinoma in patients with non-alcoholic steatohepatitis. J Hepatol. 2021 doi: 10.1016/j.jhep.2021.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parker BL, Calkin AC, Seldin MM, Keating MF, Tarling EJ, Yang P. An integrative systems genetic analysis of mammalian lipid metabolism. Nature. 2019;567(7747):187–193. doi: 10.1038/s41586-019-0984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ooi GJ, Meikle PJ, Huynh K, Earnest A, Roberts SK, Kemp W. Hepatic lipidomic remodeling in severe obesity manifests with steatosis and does not evolve with non-alcoholic steatohepatitis. J Hepatol. 2021 doi: 10.1016/j.jhep.2021.04.013. [DOI] [PubMed] [Google Scholar]
- 17.Vos T, Flaxman AD, Naghavi M, Lozano R, Michaud C, Ezzati M. Years lived with disability (YLDs) for 1160 sequelae of 289 diseases and injuries 1990-2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2163–2196. doi: 10.1016/S0140-6736(12)61729-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cirulli ET, Guo L, Leon Swisher C, Shah N, Huang L, Napier LA. Profound Perturbation of the Metabolome in Obesity Is Associated with Health Risk. Cell Metab. 2019;29(2):488–500. doi: 10.1016/j.cmet.2018.09.022. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Valkengoed IGM, Argmann C, Ghauharali-van der Vlugt K, Aerts J, Brewster LM, Peters RJG. Ethnic differences in metabolite signatures and type 2 diabetes: a nested case-control analysis among people of South Asian, African and European origin. Nutr Diabetes. 2017;7(12):300. doi: 10.1038/s41387-017-0003-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Floegel A, Stefan N, Yu Z, Muhlenbruch K, Drogan D, Joost HG. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62(2):639–648. doi: 10.2337/db12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satriano L, Lewinska M, Rodrigues PM, Banales JM, Andersen JB. Metabolic rearrangements in primary liver cancers: cause and consequences. Nat Rev Gastroenterol Hepatol. 2019;16(12):748–766. doi: 10.1038/s41575-019-0217-8. [DOI] [PubMed] [Google Scholar]
- 22.Banales JM, Inarrairaegui M, Arbelaiz A, Milkiewicz P, Muntane J, Munoz-Bellvis L. Serum metabolites as diagnostic biomarkers for cholangiocarcinoma, hepatocellular carcinoma and primary sclerosing cholangitis. Hepatology. 2018 doi: 10.1002/hep.30319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barr J, Caballeria J, Martinez-Arranz I, Dominguez-Diez A, Alonso C, Muntane J. Obesity-dependent metabolic signatures associated with nonalcoholic fatty liver disease progression. J Proteome Res. 2012;11(4):2521–2532. doi: 10.1021/pr201223p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Arranz I, Mayo R, Perez-Cormenzana M, Minchole I, Salazar L, Alonso C. Enhancing metabolomics research through data mining. J Proteomics. 2015;127(Pt B):275–288. doi: 10.1016/j.jprot.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network. Electronic address wbe, Cancer Genome Atlas Research N. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169(7):1327–1341. doi: 10.1016/j.cell.2017.05.046. e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G. Metaboanalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46(W1):W486–WW94. doi: 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohart F, Gautier B, Singh A, Le Cao KA. mixOmics: An R package for 'omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13(11) doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cavill R, Kamburov A, Ellis JK, Athersuch TJ, Blagrove MS, Herwig R. Consensus-phenotype integration of transcriptomic and metabolomic data implies a role for metabolism in the chemosensitivity of tumour cells. PLoS Comput Biol. 2011;7(3) doi: 10.1371/journal.pcbi.1001113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen A, Rudge SA, Zhang Q, Wakelam MJ. Using lipidomics analysis to determine signalling and metabolic changes in cells. Curr Opin Biotechnol. 2017;43:96–103. doi: 10.1016/j.copbio.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Muehlemann M, Miller KD, Dauphinee M, Mizejewski GJ. Review of Growth Inhibitory Peptide as a biotherapeutic agent for tumor growth, adhesion, and metastasis. Cancer Metastasis Rev. 2005;24(3):441–467. doi: 10.1007/s10555-005-5135-2. [DOI] [PubMed] [Google Scholar]
- 31.Yang JD, Addissie BD, Mara KC, Harmsen WS, Dai J, Zhang N. GALAD Score for Hepatocellular Carcinoma Detection in Comparison with Liver Ultrasound and Proposal of GALADUS Score. Cancer Epidemiol Biomarkers Prev. 2019;28(3):531–538. doi: 10.1158/1055-9965.EPI-18-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambrecht J, Porsch-Ozcurumez M, Best J, Jost-Brinkmann F, Roderburg C, Demir M. The APAC Score: A Novel and Highly Performant Serological Tool for Early Diagnosis of Hepatocellular Carcinoma in Patients with Liver Cirrhosis. J Clin Med. 2021;10(15) doi: 10.3390/jcm10153392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roongta UV, Pabalan JG, Wang X, Ryseck RP, Fargnoli J, Henley BJ. Cancer cell dependence on unsaturated fatty acids implicates stearoyl-CoA desaturase as a target for cancer therapy. Mol Cancer Res. 2011;9(11):1551–1561. doi: 10.1158/1541-7786.MCR-11-0126. [DOI] [PubMed] [Google Scholar]
- 34.Senni N, Savall M, Cabrerizo Granados D, Alves-Guerra MC, Sartor C, Lagoutte I. beta-catenin-activated hepatocellular carcinomas are addicted to fatty acids. Gut. 2019;68(2):322–334. doi: 10.1136/gutjnl-2017-315448. [DOI] [PubMed] [Google Scholar]
- 35.Rada P, Gonzalez-Rodriguez A, Garcia-Monzon C, Valverde AM. Understanding lipotoxicity in NAFLD pathogenesis: is CD36 a key driver? Cell Death Dis. 2020;11(9):802. doi: 10.1038/s41419-020-03003-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gomes RN, Felipe da Costa S, Colquhoun A. Eicosanoids and cancer. Clinics (Sao Paulo) 2018;73(suppl 1):e530s. doi: 10.6061/clinics/2018/e530s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu X, Yu H, Ma Q, Shen S, Das UN. Linoleic acid suppresses colorectal cancer cell growth by inducing oxidant stress and mitochondrial dysfunction. Lipids Health Dis. 2010;9:106. doi: 10.1186/1476-511X-9-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma C, Kesarwala AH, Eggert T, Medina-Echeverz J, Kleiner DE, Jin P. NAFLD causes selective CD4 (+) T lymphocyte loss and promotes hepatocarcinogenesis. Nature. 2016;531(7593):253–257. doi: 10.1038/nature16969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jee SH, Kim M, Kim M, Yoo HJ, Kim H, Jung KJ. Metabolomics Profiles of Hepatocellular Carcinoma in a Korean Prospective Cohort: The Korean Cancer Prevention Study-II. Cancer Prev Res (Phila) 2018;11(5):303–312. doi: 10.1158/1940-6207.CAPR-17-0249. [DOI] [PubMed] [Google Scholar]
- 40.Simcox J, Geoghegan G, Maschek JA, Bensard CL, Pasquali M, Miao R. Global Analysis of Plasma Lipids Identifies Liver-Derived Acylcarnitines as a Fuel Source for Brown Fat Thermogenesis. Cell Metab. 2017;26(3):509–522. doi: 10.1016/j.cmet.2017.08.006. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams SH, Hoppel CL, Lok KH, Zhao L, Wong SW, Minkler PE. Plasma acylcarnitine profiles suggest incomplete long-chain fatty acid beta-oxidation and altered tricarboxylic acid cycle activity in type 2 diabetic African-American women. J Nutr. 2009;139(6):1073–1081. doi: 10.3945/jn.108.103754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sydor S, Best J, Messerschmidt I, Manka P, Vilchez-Vargas R, Brodesser S. Altered Microbiota Diversity and Bile Acid Signaling in Cirrhotic and Noncirrhotic NASH-HCC. Clin Transl Gastroenterol. 2020;11(3):e00131. doi: 10.14309/ctg.0000000000000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayo R, Crespo J, Martinez-Arranz I, Banales JM, Arias M, Minchole I. Metabolomic-based noninvasive serum test to diagnose nonalcoholic steatohepatitis: Results from discovery and validation cohorts. Hepatol Commun. 2018;2(7):807–820. doi: 10.1002/hep4.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Y, Oresic M, Leivonen M, Gopalacharyulu P, Hyysalo J, Arola J. Noninvasive Detection of Nonalcoholic Steatohepatitis Using Clinical Markers and Circulating Levels of Lipids and Metabolites. Clin Gastroenterol Hepatol. 2016;14(10):1463–1472. doi: 10.1016/j.cgh.2016.05.046. e6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.