ABSTRACT
Plasmablasts represent a specialized class of antibody-secreting effector B cells that transiently appear in blood circulation following infection or vaccination. The expansion of these cells generally tends to be massive in patients with systemic infections such as dengue or Ebola that cause hemorrhagic fever. To gain a detailed understanding of human plasmablast responses beyond antibody expression, here, we performed immunophenotyping and RNA sequencing (RNA-seq) analysis of the plasmablasts from dengue febrile children in India. We found that plasmablasts expressed several adhesion molecules and chemokines or chemokine receptors that are involved in endothelial interactions or homing to inflamed tissues, including skin, mucosa, and intestine, and upregulated the expression of several cytokine genes that are involved in leukocyte extravasation and angiogenesis. These plasmablasts also upregulated the expression of receptors for several B-cell prosurvival cytokines that are known to be induced robustly in systemic viral infections such as dengue, some of which generally tend to be relatively higher in patients manifesting hemorrhage and/or shock than in patients with mild febrile infection. These findings improve our understanding of human plasmablast responses during the acute febrile phase of systemic dengue infection.
IMPORTANCE Dengue is globally spreading, with over 100 million clinical cases annually, with symptoms ranging from mild self-limiting febrile illness to more severe and sometimes life-threatening dengue hemorrhagic fever or shock, especially among children. The pathophysiology of dengue is complex and remains poorly understood despite many advances indicating a key role for antibody-dependent enhancement of infection. While serum antibodies have been extensively studied, the characteristics of the early cellular factories responsible for antibody production, i.e., plasmablasts, are only beginning to emerge. This study provides a comprehensive understanding of the transcriptional profiles of human plasmablasts from dengue patients.
KEYWORDS: plasmablasts, dengue, human, RNA-seq, immunophenotyping, India
INTRODUCTION
Dengue is emerging as one of the most dangerous mosquito-borne human hemorrhagic fever viral diseases. Worldwide, over 100 million clinically significant cases are estimated to occur each year (1). This acute systemic infection results in clinical disease ranging from mild self-limiting febrile illness to more severe and sometimes life-threatening dengue hemorrhagic fever (DHF) or dengue shock syndrome (DSS), especially among children (2, 3). While the precise mechanisms of this varied disease spectrum are still being elucidated, studies conducted so far suggest that these diverse pathological manifestations have a multifactorial basis. DHF/DSS is typically shown to be associated with secondary heterologous dengue infections (4–6), massive T- and B-cell responses (7–9), and exaggerated induction of vascular permeability factors such as vascular endothelial growth factor (VEGF) and interleukin-6 (IL-6) that are transiently induced at high levels during the acute febrile phase (10–13). Antibodies appear to have both protective and pathological roles in dengue. While neutralizing antibodies protect, sub-neutralizing or non-neutralizing antibodies are believed to contribute to increased viral uptake via Fc receptors facilitating replication in cells through a process called antibody-dependent enhancement of infection (ADE) (14, 15).
Although antibody responses to dengue have been extensively studied (16–21), a detailed understanding of the cells responsible for this first wave of the antibody response during the acute febrile phase, i.e., plasmablasts or antibody-secreting cells (ASCs), is only beginning to emerge (8, 22). Accumulating evidence suggests that systemic infections such as dengue and Ebola induce a massive and transient plasmablast response during the acute febrile phase compared to localized infections such as influenza or even vaccinations (8, 22–26). In dengue, the plasmablast response usually peaks around the time of subsidence of fever and serious symptoms (8, 22, 23, 27). Interestingly, it has been shown that the expansion of these plasmablasts is relatively higher in patients with complicated dengue than in those with milder symptoms (8). What contributes to this massive plasmablast response in these viral hemorrhagic fevers remains unclear.
Originally, it was thought that plasmablasts primarily circulate in blood while contributing to humoral immunity (28, 29). However, historical studies in fish models suggested that these cells can also home to tissues (30). More recent studies in patients with multiple sclerosis indicated that blood-brain barrier crossing of the plasmablasts and their intrathecal immunoglobulin G (IgG) production may have physiological relevance and serve as an early indication of prolonged disease (31). Interestingly, it was shown that the trafficking programs imprinted in plasmablasts could differ depending upon the route of immunization (32). What spectrum of adhesion/homing molecules are expressed on human plasmablasts generated during systemic viral infections such as dengue thus requires detailed investigation (33).
While historically plasmablasts are best known for their antibody-secreting effector functions, studies in murine models suggest that plasmablasts can also express cytokines such as IL-10 or IL-35 that have far-reaching consequences during bacterial/parasitic infection or autoimmunity (34–36). However, what cytokines are expressed by human plasmablasts remain to be defined. Considering these factors, in this study, we performed detailed immunophenotyping and transcriptional profiling of the plasmablasts from dengue-confirmed febrile children.
RESULTS
Transcriptional profiling of plasmablasts from dengue patients.
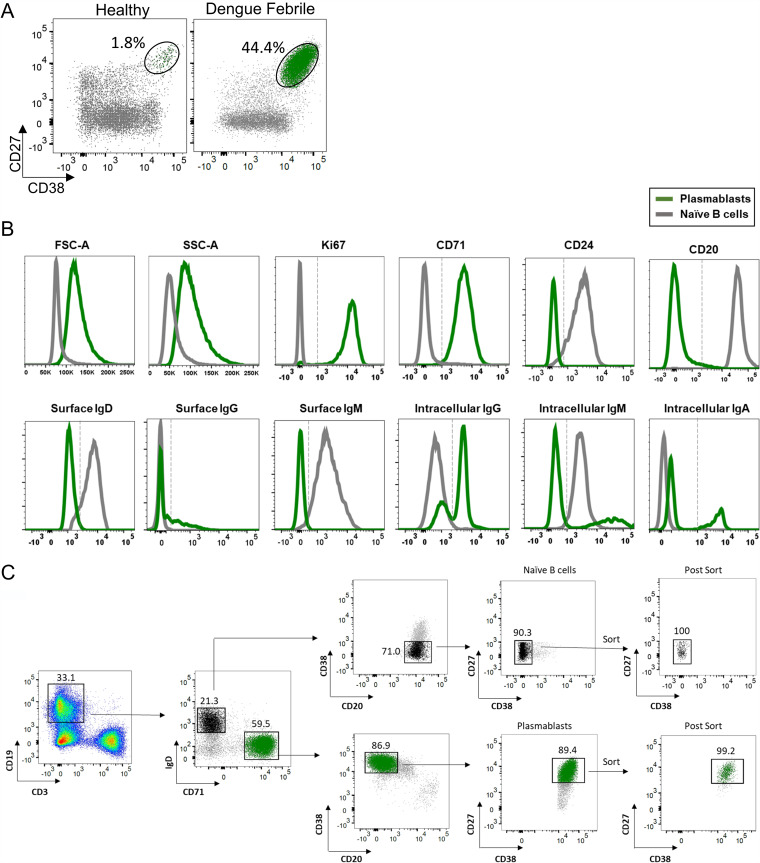
Although many studies have conducted transcriptional profiling of human plasma cells (37–45), transcriptional profiling of human plasmablasts is surprisingly limited (24, 40, 43, 45, 46). Two recent studies that have performed single-cell RNA-seq analysis of plasmablasts from dengue patients primarily focused on the expression patterns of antibody genes (47, 48). To gain a detailed understanding of the features of the plasmablasts beyond antibody expression, we sorted plasmablasts from 8 dengue patients and performed a global transcriptomic analysis using bulk RNA sequencing (Gene Expression Omnibus [GEO] accession number GSE171487). The characteristics of these patients are shown in Table 1. All the patients were children aged 8 to 12 years with confirmed dengue febrile illness 5 to 7 days after the onset of illness. Four patients had severe dengue (SD), and four had dengue with warning signs (DW). All showed a massive expansion of plasmablasts with frequencies ranging from 33% to 61% of total B cells (Table 1 and Fig. 1A). Consistent with previous studies (8, 22, 23, 27, 49–51), functional enzyme-linked immunosorbent spot (ELISpot) analysis showed that the majority of plasmablasts were IgG antibody-secreting cells (ASCs) that were primarily dengue specific (Table 1). These cells were blasting, as evidenced by high forward scatter (FSC) and side scatter (SSC), and were highly proliferating, as indicated by expression of the proliferating cell nuclear antigen Ki67 and transferrin receptor CD71, which is a key molecule for iron uptake in proliferating cells (52), and downregulated expression of the heat-stable antigen CD24, which is known to regulate B-cell receptor-mediated signals and to be heavily downregulated when B cells differentiate into antibody-secreting cells (53–55) (Fig. 1B). As expected, they showed little or no expression of CD20, a key molecule involved in efficient signaling and calcium mobilization following ligation of the B-cell receptor (BCR) or other surface proteins of B cells (56, 57). Consistent with the appreciation that plasmablasts actively secrete antibodies, we observed little or no surface IgD, IgM, or IgG, but these cells were strongly positive for the intracellular expression of IgG, IgM, or IgA (Fig. 1B, bottom row). We used a select combination of these markers to stringently identify and sort the plasmablasts. The sorting strategy is shown in Fig. 1C. The post-sort purity was >99%. For comparison of transcriptional profiles, we simultaneously sorted plasmablasts and naive B cells from the same patients.
TABLE 1.
Characteristics of the dengue patients from whom plasmablasts were sorted for RNA-seq analysis
| Patient | Age (yrs) | No. of days after onset of clinical symptoms | Disease severitya | Plasmablast frequency (%)b | No. of IgG ASCsc/106 PBMCs (% dengue specific) | No. of IgM ASCsc/106 PBMCs (% dengue specific) |
|---|---|---|---|---|---|---|
| 1 | 8.4 | 6 | SD | 58.7 | 0.55 × 106 (41.2) | 0.032 × 106 (2.5) |
| 2 | 11 | 6 | SD | 61.0 | 0.09 × 106 (70.4) | 0.002 × 106 (26.7) |
| 3 | 8.8 | 4 | SD | 49.3 | 0.066 × 106 (98) | 0.014 × 106 (2.8) |
| 4 | 11.6 | 4 | SD | 56.5 | 0.08 × 106 (60.1) | 0.012 × 106 (7.4) |
| 5 | 11.7 | 7 | DW | 33.4 | 0.076 × 106 (71.2) | 0.017 × 106 (5.6) |
| 6 | 11.2 | 7 | DW | 60.4 | 0.043 × 106 (58.3) | 0.0024 × 106 (0) |
| 7 | 9.5 | 7 | DW | 41.8 | 0.032 × 106 (48.1) | 0.0052 × 106 (6.9) |
| 8 | 12 | 5 | DW | 50.9 | 0.032 × 106 (66.7) | 0.012 × 106 (1.7) |
Disease grade classification was done according to 2009 WHO criteria (28). SD, severe dengue; DW, dengue with warning signs.
Percentage of B cells.
ASCs, antibody-secreting cells.
FIG 1.
Characterization of plasmablast expansion in dengue patients. (A, right) Example of flow cytometric staining of plasmablasts (green) gated on total B cells (gray) in an acute dengue febrile patient. (Left) For comparison, a healthy subject is shown. (B) Phenotypes of plasmablasts compared to naive B cells shown as overlay histograms. (C) Flow cytometry plots showing the gating strategy employed for bulk sorting of plasmablasts and naive B cells from 8 dengue patients analyzed by RNA-seq. CD19+ B cells that were alive and blasting were then analyzed for IgD and CD71 expression. Cells that were IgD+ CD71− (black) and IgD− CD71+ B cells (green) were then analyzed for their surface expression of CD20, CD27, and CD38. Naive B cells (black) (top) were sorted as CD20+, followed by CD27+ CD38− cells. Plasmablasts (green) (bottom) were sorted as CD20− and CD27+ CD38+ cells. Example dot plots of sort purity are shown as postsort analysis.
Because plasmablasts are primarily antibody-secreting cells, we first asked what portion of the total transcripts accounted for immunoglobulin (Ig) genes. Of the 98.4 million reads of plasmablasts, 27.9% of the total reads (27.5 million) were represented by 407 Ig genes, confirming that Ig gene transcription was highly active in plasmablasts. In contrast, the Ig gene reads contributed only 2.8% (3.7 million) of the total reads of naive B cells (129.6 million). A total of 130 Ig genes that were differentially expressed in the plasmablasts compared to naive B cells are shown in Fig. S1 in the supplemental material. Heavy chains of the IgD, IgE, IgM, IgA, and IgG subtypes are marked by dotted boxes. Corroborating our immunophenotyping analysis, plasmablasts substantially downregulated IgD and upregulated IgG, IgA, and IgM.
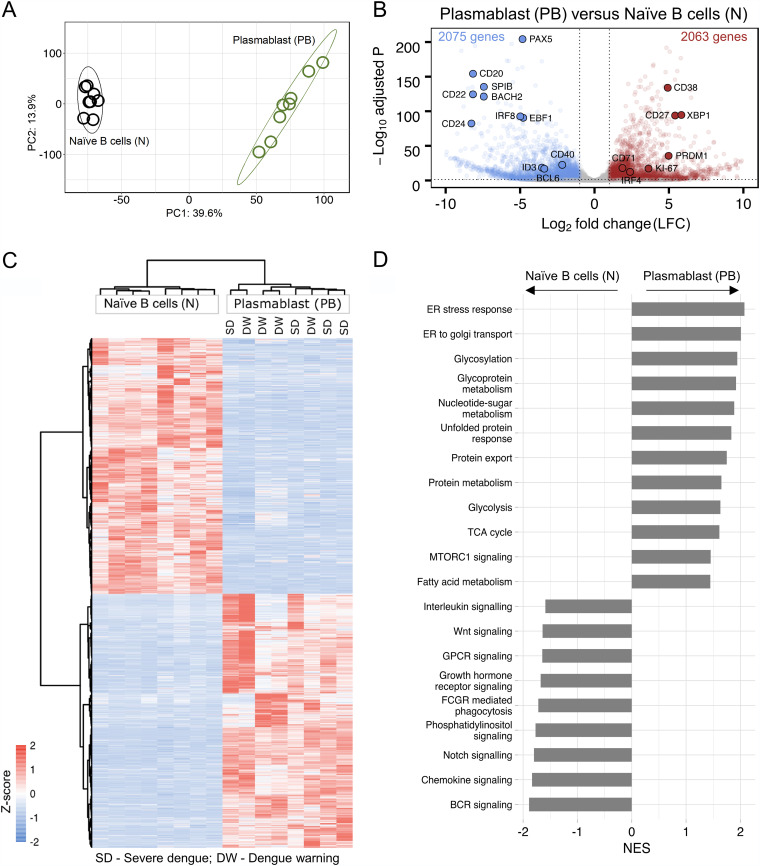
Even after excluding the Ig reads, we found that the plasmablasts continued to show a highly distinct transcriptional profile compared to naive B cells (Fig. 2A). A total of 4,138 non-Ig genes were found differentially expressed in the plasmablasts compared to naive B cells, of which 2,063 genes were upregulated and 2,075 genes were downregulated. Analysis of select genes that are typically expected to be upregulated in plasmablasts (XBP1, CD27, PRDM1, CD38, IRF4, MKI67/Ki-67, and CD71) and select genes that are typically expected to be downregulated in plasmablasts (CD20, CD40, BCL6, ID3, EBF1, PAX5, IRF8, SPIB, BACH2, CD22, and CD24) showed the expected patterns, thereby validating our analysis (Fig. 2B and Table S1). Interestingly, the differentially expressed genes in the plasmablasts did not cluster depending on the disease severity (Fig. 2C), suggesting that these Ig-independent qualitative features of the plasmablasts have little or no impact on disease severity.
FIG 2.
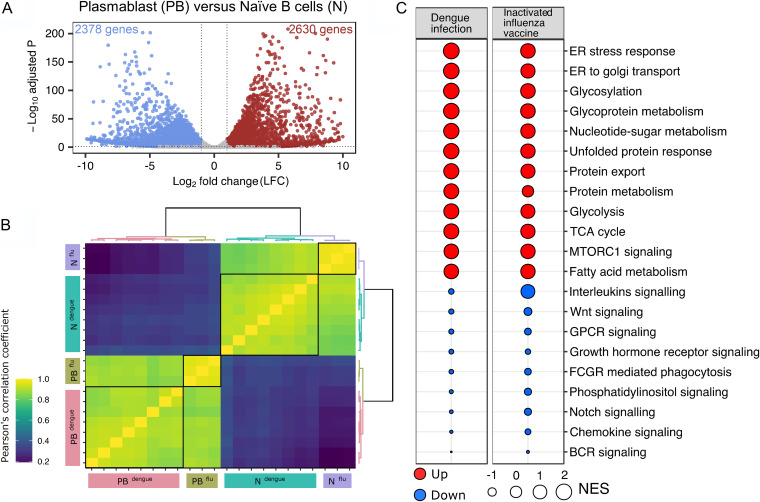
Transcriptional profiling of the plasmablasts from dengue patients. (A) Principal-component analysis (PCA) score plot of normalized counts of 14,773 genes showing clustering of plasmablasts (green) and naive B cells (black) from eight different acute dengue febrile subjects. (B) Volcano plot with −log-adjusted P values on the y axis and log2 fold changes on the x axis showing 4,138 differentially expressed genes that are significantly upregulated (red dots) (right), and downregulated (blue dots) (left) in plasmablasts versus naive B cells. Gray dots are either nonsignificant, nondifferential, or both (for significant, adjusted P value of <0.05 and P value of <0.01; for differential, log2 fold change of greater than or equal to1 or less than or equal to −1). Selected genes with known expression in plasmablasts and naive B cells are labeled in the plot with larger dots and their symbol. (C) Heat map showing hierarchical clustering of 4,138 differentially expressed genes in plasmablasts compared to naive B cells from eight acute dengue febrile patients. z-scores of normalized counts were taken for the heat map. The Ward.D2 method was used for clustering (for significant, adjusted P value of <0.05 and P value of <0.01; for differential, log2 fold change greater than or equal to 1 or less than or equal to −1). The disease severity of the individual patients is indicated at the top. (D) Gene set enrichment analysis (GSEA) performed using gene sets derived from GO biological process, Hallmark, KEGG, and Reactome gene sets from MSigDB. Significant pathways with an FDR q value of <25% are shown. Enriched term names are manually collapsed. Positive and negative normalized enrichment scores (NES) correlate with upregulated and downregulated pathways, respectively, in plasmablasts compared to naive B cells. ER, endoplasmic reticulum; TCA, tricarboxylic acid; GPCR, G-protein-coupled receptor; FCGR, Fc gamma receptor; BCR, B-cell receptor.
Gene set enrichment analysis (GSEA) of the top 20 enriched terms of the differentially expressed genes showed that the upregulated genes in plasmablasts were significantly associated with processes related to cellular metabolism, protein synthesis, glycosylation, transport, and secretion (false discovery rate [FDR] <25%) (Fig. 2D and Table S2). In contrast, the downregulated genes in plasmablasts were significantly associated with processes related to signaling through BCR or through other external ligands such as notch, phosphatidylinositol, growth hormone receptor, G-protein-coupled receptors, WNT, and interleukins. This pattern is consistent with the concept that plasmablasts are highly differentiated short-lived antibody-producing effector cells that are largely refractory to BCR signaling or other external stimuli.
The differentially expressed genes in each of the biological processes are provided in Table S3. Additionally, heat maps of the differentially expressed genes associated with proliferation, metabolism, antibody synthesis, glycosylation, growth factors, transcription factors, and interferon-stimulating genes (ISGs); their log2 fold changes (LFCs); and average counts of naive B cells and plasmablasts are shown in Fig. S2. Taken together, these data confirm that plasmablasts are highly proliferating metabolically active cellular factories equipped with the machinery needed for antibody synthesis, glycosylation, and secretion while downregulating the machinery involved in signaling through B-cell receptors and many other external stimuli.
Plasmablasts expressed several cytokines involved in angiogenesis.
It is known that B cells produce different cytokines depending on differentiation status (58–61). In contrast to naive B cells that typically express cytokines such as lymphotoxin B (LTB), tumor necrosis factor alpha (TNF-α), IL-6, or IL-10 (58–61), emerging studies in murine models show that plasma cells/plasmablasts can express cytokines such as IL-17, IL-10, or IL-35 (34, 36). The expression of these cytokines appears to have far-reaching consequences because mice in which IL-35 was knocked out exclusively in B cells could not recover from experimental autoimmune encephalomyelitis (EAE) but had higher resistance to Salmonella enterica serovar Typhimurium infection (34). However, so far, virtually nothing is known about cytokine expression by human plasmablasts during systemic infections such as dengue.
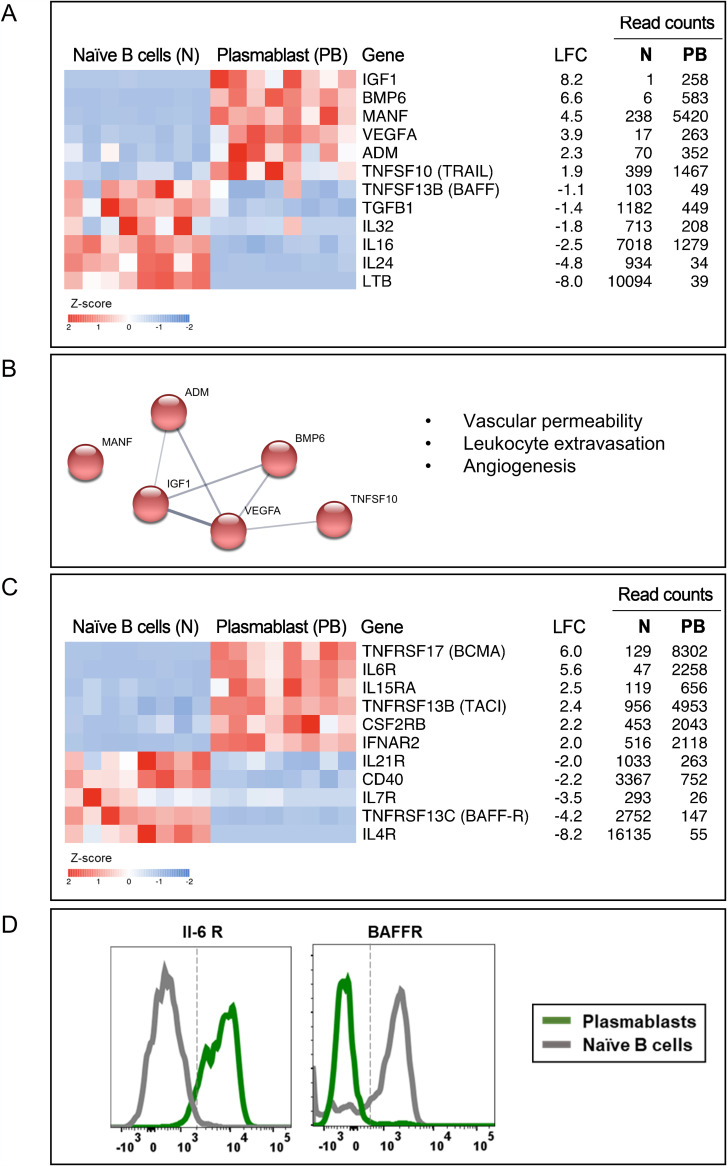
Considering this, we asked what cytokine genes are expressed in human plasmablasts responding to dengue infection. We found that 33 cytokine genes were differentially expressed between plasmablasts and naive B cells (Table S4). Select genes of interest are shown in Fig. 3A. Notably, we found that many cytokines that are constitutive to naive B cells were downregulated in plasmablasts (LTB [−8.0], IL-16 [−2.4], TGFB1 [−1.4], IL-32 [−1.8], IL-24 [−4.8], and TNFSF13B [BAFF] [−1.1]). On the other hand, plasmablasts robustly expressed several other cytokines. Notable among these include VEGFA (+3.9), ADM (+2.3), ADM2 (+3.3), BMP6 (+6.6), IGF1 (+8.2), MANF (+4.5), and TNFSF10 (+1.9) (Fig. 3A and Table S5). Interestingly, we found that many of these upregulated cytokines share functionally overlapping processes related to vascular permeability, leukocyte extravasation, and angiogenesis (Fig. 3B). Of interest is VEGF, a well-known cytokine that is involved in both physiological angiogenesis and wound repair in addition to playing a key role in vascular permeability under pathological conditions (62–66). It is interesting to note that VEGF is classically thought to play a pivotal role in mediating plasma leakage in dengue because its levels were found to be relatively higher in patients with DHF and/or DSS (67–69). Interestingly, ADM and ADM2, which are also upregulated, are known to synergize VEGFA actions on endothelial cells (70, 71). Other upregulated genes are also implicated in angiogenesis. BMP-6 is known to increase vascular hyperpermeability (72); IGF1 promotes endothelial cell migration, tube formation, and the production of the vasodilator nitric oxide (73, 74); and MANF activates VEGF signaling pathways (75). TNFSF10 (TRAIL) is also reported to have functional properties similar to those of VEGF (76). By viewing our data in juxtaposition with the known functions of these genes, it appears that plasmablasts from dengue patients upregulate several cytokine genes involved in angiogenesis.
FIG 3.
Plasmablasts from dengue patients express several cytokines involved in angiogenesis. (A) Heat maps showing select genes related to cytokines. (B) Network analysis performed in STRING describing associations between the upregulated cytokines in plasmablasts based on text mining, annotated database, experimental evidence, and homology. Protein-protein associations are represented by the edges, and the thickness of the edges corresponds to confidence in the evidence used to deduce the associations (thick, high confidence; thin, low confidence). Red nodes represent the cytokines involved in vascular permeability, leukocyte extravasation, and angiogenesis. (C) Heat map of select cytokine receptors. For both heat maps in panels A and C, a gradient of high to low gene expression levels based on z-score-normalized counts is indicated from red to blue. On the right of the heat maps are gene names with common names indicated in parentheses, log2 fold changes (LFC), average normalized counts of naive B cells, and average normalized counts of plasmablasts. Each column in the heat map represents an individual patient. (D) Flow cytometric staining for select cytokine receptors.
Plasmablasts expressed receptors for several B-cell prosurvival cytokines that are known to be induced at high levels during the acute febrile phase of systemic viral infections.
We observed that plasmablasts robustly upregulated genes encoding receptors for several cytokines that are produced at high levels during systemic viral infection, many of which are known to provide prosurvival signals to B cells. Notable among these are the receptors for the cytokines IL-6 (IL-6R) (+5.6), IL-15 (IL-15RA) (+2.5), type I interferon (IFNAR2) (+2.0), and IL-5/IL-3/colony-stimulating factor (CSF2RB) (+2.2) (Fig. 3C). Flow cytometry confirmation of select cytokine receptors is shown in Fig. 3D. It is interesting to note that some of these cytokines, for example, IL-6, are known to be induced at a very high level specifically in patients manifesting dengue hemorrhage and/or shock compared to patients with mild febrile dengue infection (10–13, 77) and that plasmablast expansion also tends to be massive in patients manifesting dengue hemorrhage and/or shock compared to patients with mild febrile dengue infection (8), implying that, perhaps, the massive expansion of the plasmablasts seen in patients with hemorrhage or shock may be mediated by these cytokines, at least in part.
Interestingly, we found that the plasmablasts downregulated TNFRSF13C/BAFF-R (−4.2) that binds monomeric and multimeric forms of BAFF to mediate the survival of transitional and naive B cells (78, 79) while upregulating TNFRSF13B/TACI (+2.4) and TNFRSF17/BCMA (+6.0) that bind multimeric forms of BAFF or APRIL to mediate proliferation, survival, as well as the maturation and differentiation of activated B cells into antibody-secreting cells (Fig. 3C).
The plasmablasts also downregulated receptors for the major homeostatic survival cytokines for B cells, IL-7 (IL-7R) (−3.5); the Tfh-associated cytokine IL-21 (IL-21R) (−2.0); and the Th2-associated cytokine IL-4 (IL-4R) (−8.2), suggesting that these cells, for the most part, are likely to have become refractory to B-cell-mediated survival signals provided under normal homeostatic conditions or during germinal center follicular reactions or Th2-associated immune responses. Additionally, these cells also downregulated CD40 (−2.2), indicating that, perhaps, these cells are likely to have become refractory to T-cell help (Fig. 3C), although it is unclear whether these cells originate in a T-cell-dependent or -independent manner.
Taken together, these findings suggest that plasmablasts, for the most part, become inept to receive signals from prosurvival cytokines available under normal homeostatic conditions while acquiring receptors for prosurvival cytokines that are induced robustly under the conditions of the acute febrile phase of infection.
Plasmablasts expressed several adhesion molecules, chemokines, and chemokine receptors involved in endothelial interactions and homing to skin or mucosal tissues, including intestine.
Examination of chemokines and chemokine receptors showed that the plasmablasts expressed several adhesion molecules, chemokines, and chemokine receptors that are involved in vascular endothelial cell interactions, adhesion, rolling, diapedesis, extravasation, and cell migration to inflammatory tissues, skin, mucosa, and intestine. An extensive list of these genes is provided in Table S6. Heat maps of select adhesion molecules are shown in Fig. 4A, heat maps of select chemokines/chemokine receptors are shown in Fig. 4B, flow cytometric staining confirmation of select chemokine receptors is shown in Fig. 4C, and string analysis of functional protein network associations of the select molecules is shown in Fig. 4D.
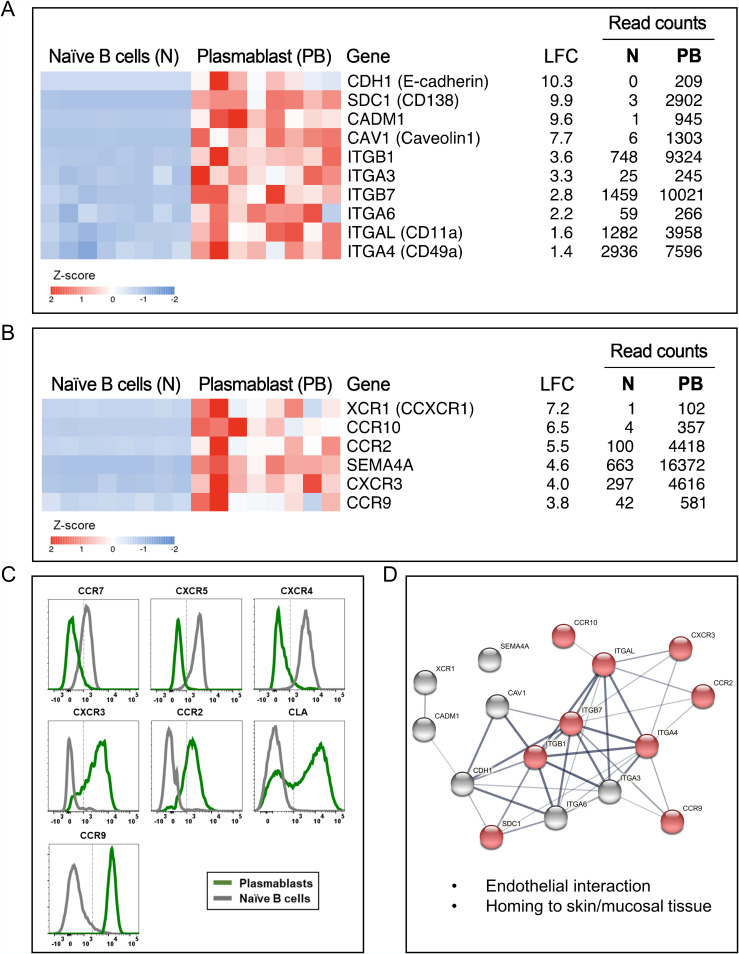
FIG 4.
Plasmablasts from dengue patients express several adhesion molecules, chemokines, and chemokine receptors involved in endothelial interactions and homing to skin or mucosal tissues, including intestine. (A to C) Heat maps showing select genes related to cell adhesion molecules (CAMs) (A) and chemokine and chemokine receptors (B) and flow cytometric confirmation of select chemokine receptors (C). In the heat maps in panels A and B, a gradient from high to low gene expression levels based on z-score-normalized counts is indicated from red to blue. At the right sides of the heat maps are gene names with common names in parentheses, log2 fold changes (LFC), average normalized counts of naive B cells, and average normalized counts of plasmablasts. The genes are arranged according to descending LFC values. Each column in the heat map represents an individual patient. (D) Network analysis performed in STRING describing associations between the select upregulated CAMs, chemokines, and chemokine receptors in plasmablasts based on text mining, annotated database, experimental evidence, and homology. Protein-protein associations are represented by the edges, and the thickness of the edges corresponds to confidence in the evidence used to deduce the associations (thick, high confidence; thin, low confidence). Each node represents the select CAMs, chemokines, and chemokine receptors, whereas the red nodes highlight the genes involved in vascular endothelial cell interactions and cell migration to skin, mucosa, intestine, and inflamed tissues.
Notable among the adhesion molecules (Fig. 4A) were CDH1 (E-cadherin) (+10.3), which is a well-characterized molecule involved in cell-cell adhesion (80, 81); SDC1 (CD138) (+9.9), which is known to have pleiotropic functions in cell adhesion (82, 83), homing (40), wound healing (84, 85), and angiogenesis (86, 87); and the cell adhesion molecule CADM1 (+9.6), and CAV1 (Caveolin 1) (+7.7), which are implicated in pleiotropic functions including cell invasion, signaling, intracellular organelle communication, and mediating endothelial dysfunction by attenuating nitric oxide under conditions of hyperlipidemia (88–90). Additionally, the plasmablasts also showed upregulation of a wide spectrum of integrins such as ITGB1 (+3.6), ITGA3 (+3.3), ITGB7 (+2.8), ITGA6 (+2.2), ITGAL/CD11a (+1.6), and ITGA4/CD49D (+1.4) that are known to be involved in cell-to-cell or cell-to-extracellular matrix (ECM) adhesion (91).
Notable among the chemokines and chemokine receptors (Fig. 4B and C) are CCR9 (+3.8), and CCR10 (+6.5), which are chemotactic receptors for mucosal chemokines such as CCL25 (generally expressed on the endothelium of the small intestine)/CCL27 (skin keratinocytes)/CCL28 (expressed on a wide variety of mucosal epithelial cells) (32, 92–94). Plasmablasts also upregulated CCR2 (+5.5) and CXCR3 (+4.0), the two major chemokine receptors for a variety of chemotactic factors such as CCL2, CCL7, CCL9, CCL10, CCL11, and CCL12 that are produced in inflamed tissues.
In addition to being equipped with the ability to home to skin, mucosal, and intestinal tissues, it is interesting to note that the plasmablasts also expressed XCR1/CCXCR1/lymphotactin receptor (+7.2) (Fig. 4B). XCR1 is known to bind to lymphotactin/XCL1, which are emerging as cytokines produced by activated NK cells, NK-T cells, and T cells (95, 96). This suggested that, perhaps, these cells are likely to be equipped with the ability to migrate toward other activated lymphocytes. Additionally, these cells also expressed a semaphorin, SEMA4A (+4.6), that functions in the regulation of the immune system, remodeling of the ECM, and angiogenesis (97–100).
Taken together, these findings show that plasmablasts are equipped with the ability to not only interact with endothelial cells of inflamed tissue but also produce several mediators involved in angiogenesis and homing to skin or mucosa as well as intestine.
Transcriptional profiles of plasmablasts responding to systemic dengue viral infection were qualitatively like those of plasmablasts responding to an inactivated influenza vaccine.
We next wondered whether the transcriptional signature that we observed in plasmablasts responding to systemic viral infection could differ from that of plasmablasts responding to an inactivated vaccine. To address this, we focused on plasmablasts derived from subjects who received an inactivated influenza vaccine. In previous studies, we showed transcriptional profiles of plasmablasts derived from flu vaccinees, but these studies were based on microarray analysis (24). Hence, we retrieved our influenza study samples and performed RNA-seq analysis of the plasmablasts versus naive B cells. Similar to what we observed for dengue, plasmablasts from flu vaccinees showed a highly distinct transcriptional profile beyond Ig genes compared to naive B cells. A total of 5,008 non-Ig genes were differentially expressed in flu vaccinee plasmablasts compared to naive B cells, as shown in a volcano plot (Fig. 5A) and Table S7. A sample-to-sample correlation analysis of naive B cells or plasmablasts from dengue patients and naive B cells or plasmablasts from flu vaccinees showed that plasmablasts from dengue and flu were qualitatively similar (Fig. 5B). Comparison of median differences in z-scores (between plasmablasts and naive B cells) of differentially expressed genes showed a positive, significant, and very large correlation in dengue and flu (rho = 0.95; P < 0.001). Additionally, a majority of GSEA terms for flu plasmablasts versus naive B cells showed normalized enrichment scores (NESs) similar to those of dengue-specific terms (Fig. 5C). Like dengue, many of the cytokine and adhesion molecules involved in angiogenesis are also upregulated in plasmablasts of the flu vaccinees. Together, these findings suggest that while there could be differences in the extent of the expansion of the human plasmablasts in different scenarios (8, 22–26), their transcriptional profiles are more likely to be qualitatively similar.
FIG 5.
Transcriptional profile of plasmablasts derived from individuals that received an inactivated influenza vaccine. (A) Volcano plot with −log-adjusted P values on the y axis and log2 fold changes on the x axis. Scattered dots represent genes. Red dots are 2,630 genes that are significantly upregulated and blue dots are 2,378 genes that are significantly downregulated in plasmablasts versus naive B cells (total of 5,008 genes). Gray dots are either nonsignificant, nondifferential, or both (for significant, adjusted P value of <0.05; for differential, log2 fold change greater than or equal to 1 or less than or equal to −1). (B) Sample-to-sample correlation heat map depicting Pearson’s correlation coefficients among samples of dengue and flu. Grouping patterns are represented by agglomerative hierarchical clustering by using the Ward.D2 method. The color key depicts a strong correlation for values of >0.8 (green) or a weak correlation for values of <0.4 (blue). (C) Comparison of GSEA terms enriched for plasmablasts versus naive B cells in flu with the terms enriched for plasmablasts versus naive B cells in dengue. The size of the bubble is proportional to the normalized enrichment scores (NES), and red and blue represent terms with positive and negative NES, respectively.
DISCUSSION
Our study provides a detailed understanding of the plasmablast gene expression profiles in dengue patients. One of the intriguing observations that have emerged from numerous human studies over the past decade is that the expansion of the plasmablasts tends to be massive in systemic viral infections that cause hemorrhagic fevers such as dengue or Ebola compared to many other localized infections such as influenza or inactivated vaccines (24–26). More intriguingly, it is shown that the expansion of the plasmablasts was much more massive in patients with complicated dengue disease such as hemorrhagic fever and/or shock than in patients with mild dengue febrile illness (8). Our findings that plasmablasts robustly upregulate cytokine receptors to several B-cell prosurvival cytokines that are induced at very high levels during systemic viral infections and that some of these cytokines tend to be induced at much higher levels in DHF/DSS cases than in mild febrile dengue cases provide clues on the potential mechanisms contributing to this massive expansion of plasmablasts typically seen in dengue. Alternatively, one can argue that the higher expansion of the plasmablasts seen in DHF/DSS cases could be due to sustained stimulation by abundant levels of antigen that may persist for a longer time in these cases. Although stronger stimulation at the beginning of infection cannot be excluded, it should be noted that the plasmablasts, once generated, downregulate their B-cell receptor (BCR) and its associated signaling machinery and hence are unlikely to receive continued antigen-mediated signals. This viewpoint is further strengthened considering a previous study by Kwissa et al., who showed that B-cell signatures were not associated with high viral loads (101). Since the levels of cytokines could differ depending on the type of infection or immunization, further studies are needed to understand how well the expansion of plasmablasts correlates with the kinetics and levels of these pro-B-cell survival cytokines induced during the acute febrile phase of systemic infections.
It is interesting to note that while maximal expansion of the plasmablasts can differ depending on the context (as low as approximately 7% of the B cells in flu vaccinees and as high as 80% to 90% in dengue and Ebola infections) (24–26), we find that the transcriptional features of the plasmablasts were qualitatively similar in systemic dengue viral infection compared to inactivated influenza vaccinees. These findings combined with our observation that the plasmablasts upregulate key genes involved in inflammatory homing, vascular permeability, leukocyte extravasation, and angiogenesis raise several important questions regarding the role of plasmablasts in the pathophysiology of viral hemorrhagic fevers. Do these massively expanding cells contribute to vascular permeability, a hallmark feature of hemorrhagic fevers? Or is the expression of these genes simply indicative of the potential of these cells to home to inflamed tissues to confer localized immunity? Further studies are needed to address these critical questions.
It is important to note that many of these vascular permeability factors can also be produced by other innate cell types (102, 103). However, it remains to be determined whether the level of expression in plasmablasts is comparable to that produced by other innate cells. Also, studies are needed to determine whether the plasmablasts constitutively secrete these cytokines or secrete them only in a localized manner when interacting with endothelial cells. Hence, further studies are needed to understand what the potential physiological effects of the plasmablast-endothelial interactions on plasma leakage could be. It is noteworthy that many of the cytokines that we found expressed in the plasmablasts can have a role in both pathological as well as physiological aspects of angiogenesis. For example, VEGFA is a classic vascular permeability factor that also has positive effects on endothelial cells, including the promotion of blood vessel formation under steady state (64–66). Detailed studies are thus needed to address the impact of plasmablasts on vascular permeability.
Most studies so far, including this study, have focused on characterizing plasmablast responses in blood circulation. It is currently unclear and intriguing as to why a cell population that primarily secretes soluble antibodies needs to home to tissue. Historical studies that looked at plasmablast responses in trout immunized with keyhole limpet hemocyanin (KLH) showed that these cells can indeed migrate to lymphoid organs as well as tissues (30). More recent studies suggest that plasmablasts can express homing markers and that the expression of these could differ depending on the route of immunization (32, 33). Additionally, studies in multiple sclerosis patients suggest that intrathecal homing of the plasmablasts and their IgG secretion may be pathogenic as these can serve as specific markers of long-term persistence of disease (31). In dengue, so far, we are unaware of studies that have examined plasmablasts in tissues. These studies combined with our findings highlight the need to carefully examine the precise role of plasmablast responses in dengue.
MATERIALS AND METHODS
Dengue patient recruitment.
The clinical site for this study is the Department of Pediatrics of the All India Institute of Medical Sciences (AIIMS), New Delhi, India. Patients included in this study were selected from ongoing studies performed between 2012 and 2019. The study was approved by the institutional ethical committee, and informed consent/assent was obtained from the study participants prior to recruitment to the study. All patients included in the study were negative for malaria antigen and chikungunya IgM. The demographics of the patients from whom the plasmablasts were sorted are provided in Table 1.
Dengue confirmation.
Dengue virus infection was confirmed by a combination of methods, including reverse transcription (RT)-PCR, as described previously (12); an NS1 ELISA (enzyme-linked immunosorbent assay) (catalog number IR031096; J Mitra); and/or a dengue IgM ELISA (catalog number 01PE20; PanBio).
Disease classification.
Based on extensive clinical laboratory tests and evaluation, the attending physicians at each clinical site classified the dengue disease grade based on 2009 WHO guidelines as dengue infection without warning signs (DI), dengue with warning signs (DW), and severe dengue (SD) (104) at the time of recruitment.
PBMC and plasma isolation.
Blood samples were collected in Vacutainer CPT tubes (catalog number 362761; Becton, Dickinson). Peripheral blood mononuclear cells (PBMCs) and plasma were separated as described previously (22). The PBMCs were washed extensively in 1% complete RPMI (RPMI 1640 containing 1% fetal calf serum [FCS], 1× penicillin-streptomycin [Pen/Strep], and 1× glutamine) and used for analytical flow cytometry and ex vivo dengue-specific ELISpot assays.
Flow cytometry staining and analysis.
Extensively washed PBMCs were stained with fluorochrome-labeled antibodies and the fixable viable dye eFluor 780 (catalog number 65-0865-14; eBioscience) for live/dead exclusion. For plasmablast analysis, cells were stained with CD19 (catalog number 302252; BioLegend), CD20 (catalog number 302312; BioLegend), CD38 (catalog number 555460; BD), and CD27 (catalog number 563092; BD) and acquired on a BD FACSCanto II instrument. Plasmablast staining was analyzed using FlowJo software (TreeStar Inc.) starting with a lymphocyte gate that included the blasting cells, and plasmablasts were described as cells that are CD3−, CD20−, CD19+/int, CD27+, and CD38+. The plasmablasts were further stained with Ki67 (catalog number 561283; BD), CD71 (catalog number 555537; BD), IgM (catalog number 314512; BioLegend), IgG (catalog number 561298; BD), IgD (catalog number 555778; BD), CXCR5 (catalog number FAB190P; R&D), CXCR4 (catalog number 555974; BD), CXCR3 (catalog number 560831; BD), CD24 (catalog number 555427; BD), CCR7 (catalog number 560765; BD), CCR2 (catalog number FAV151P; R&D), CLA (catalog number 321312; BioLegend), BAFFR (catalog number 316916; BioLegend), IL-6R (catalog number 551850; BD), and CCR9 (catalog number 358907; BioLegend) for their phenotype. For intracellular protein staining, cells were permeabilized with Cytofix/Cytoperm buffer (BD), followed by staining for 60 min with antibodies that were diluted in Perm/Wash buffer (catalog number 554723; BD).
Dengue-specific ELISpot assay.
A dengue-specific ELISpot assay was performed on PBMCs isolated as previously described (22). Briefly, 96-well ELISpot plates (catalog number MSIP45; Millipore) were coated overnight with either dengue 2 fixed virus antigen (catalog number EL-22-02; Microbix) or polyvalent total immunoglobulin (catalog number H17000; Invitrogen). Before adding PBMCs, the plates were washed thoroughly and blocked for 2 h at 37°C with RPMI 1640 containing 10% fetal bovine serum (FBS). A total of 0.1 × 106 extensively washed PBMCs were then added to the plate, diluted 3-fold until the last well had a cell number of 45 cells, and incubated overnight at 37°C undisturbed. The cells were removed, and plates were washed with 1× phosphate-buffered saline (PBS) plus 0.05% Tween 20 and developed with goat anti-human IgG biotin (catalog number 13-4998-83; eBioscience), IgM biotin (catalog number B1265; Sigma), and avidin horseradish peroxidase (HRP) (catalog number 18-4100-51; eBioscience). The spots were developed with 3-amino-9-ethylcarbazole (AEC) substrate (catalog number 551951; BD) and scanned using an automated ELISpot counter (CTL [Cellular Technologies Ltd.]).
Fluorescence-activated cell sorter (FACS) analysis of naive B cells and plasmablasts.
PBMCs isolated from peripheral whole blood of dengue patients were stained with the relevant antibodies at 4°C for 30 min, washed thoroughly, suspended in PBS containing 2% FCS, and sorted on a BD FACSAria III instrument (BD) with high-forward-scatter gates to account for the larger blasting effector lymphocytes. CD19+ CD20+ IgD+ CD71− CD27− CD38− naive B cells and CD19+/int CD20− IgD− CD71+ CD27+ CD38+ plasmablasts were isolated to a purity of >90%.
RNA isolation and library preparation.
The sorted cells were washed thoroughly, and the cell pellet was suspended in RLT buffer (Qiagen) and stored at −80°C until RNA extraction. All the samples were processed simultaneously for RNA extraction. RNA was isolated from sorted B cells for each patient using the RNeasy microkit (Qiagen) with on-column DNase digestion. RNA quality was assessed using an Agilent bioanalyzer, and 500 pg of total RNA was used as the input for cDNA synthesis using the Clontech SMART-Seq v4 ultralow-input RNA kit (TaKaRa Bio) according to the manufacturer’s instructions. Amplified cDNA was fragmented and appended with dual-indexed barcodes using the Nextera XT DNA library preparation kit (Illumina). Libraries were validated by capillary electrophoresis on an Agilent 4200 TapeStation, pooled at equimolar concentrations, and sequenced on an Illumina HiSeq3000 instrument at 100SR, yielding approximately 20 million reads per sample.
RNA-seq data analysis.
Sequenced reads were mapped to human genome GRCh38. FastQC was used to monitor the quality of sequences. Adaptor sequences were trimmed. Quantification of transcripts was done by using the featurecount option of the Subread package. Raw counts from 60,448 genes were produced as an output from which 407 immunoglobulin genes were removed for further analysis. Differential expression analysis was performed with a total of 60,041 genes using the DESeq2 package (105). Low-count genes (45,268) with raw counts of <10 in more than five samples were filtered out. Among the 14,773 genes, those with an adjusted P value (Benjamini-Hochberg method) of <0.05 and a P value of <0.01 were declared significant. A log2 fold change greater than or equal to +1 was used as a threshold to declare genes upregulated, whereas a log2 fold change of less than or equal to −1 was used for assigning downregulated genes. Furthermore, genes with low average normalized counts of 100 or lower in both cell types (plasmablasts and naive B cells) were excluded from the differentially expressed gene list. Principal-component analysis (PCA) plots were generated from the 14,773 genes. For heat maps, normalized counts transformed to their z-scores were used. Hierarchical clustering was created using Euclidean distance with the Ward.D2 method. All analyses were performed in R studio software using the DESeq2, pheatmap, dendextend, dplyr, reshape2, ggplot2, biomaRt, prcomp, and rgl packages. For cytokine genes, a total of 263 genes were analyzed from ImmPort.
Gene set enrichment analysis.
GSEA software v4.0.3 from the Broad Institute (106, 107) was used for the enrichment analysis. Our expression data set was run against Hallmark, KEGG pathway, Reactome pathway, and Gene Ontology (GO) biological process gene sets from the Broad Institute’s Molecular Signature Database (MSigDB). One thousand gene set permutations were used to estimate the nominal P value of the enrichment score (ES). To account for gene set size, the ES was normalized to obtain a normalized enrichment score (NES) for each term. The false discovery rate (FDR) was used to account for multiple testing, and terms with an FDR of <25% were considered.
Protein network analysis.
Protein-protein associations among the upregulated genes in cytokines, cell adhesion molecules, chemokines, and chemokine receptors were calculated using STRING software v11.0. (108). Each direct and indirect association was scored based on experimental evidence, curated database, text mining, coexpression, and protein homology. Functional overlap of the genes was analyzed based on the functional enrichment of the biological process.
Correlation analysis.
For sample-to-sample correlation analysis of dengue and flu subjects, the log-normalized counts of a total of 4,126 differentially expressed genes in dengue plasmablasts versus naive B cells were used for calculating Pearson’s correlation coefficient. A heat map was made using the heatmaply package in R studio. The Ward.D2 method was used for clustering. Furthermore, to calculate the overall correlation between dengue and flu, differentially expressed genes in dengue were used and compared with their expression profiles in flu. Pearson’s correlation coefficient was calculated using the median z-score difference between plasmablasts and naive B cells of dengue and flu. Those with positive and negative median z-score differences were assigned as upregulated and downregulated genes, respectively.
Statistical analysis.
All data were tabulated using MS Excel and analyzed using GraphPad Prism software. For analysis of groups, an unpaired two-tailed t test was used to determine statistical significance, and P values were interpreted as indicated in the figures (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
Data availability.
The dengue RNA-seq data set has been deposited in the Gene Expression Omnibus (GEO) with the accession number GSE171487.
ACKNOWLEDGMENTS
This work was supported by U.S. National Institutes of Health grant ICIDR 1UO1A/115654; NIH-DBT, Human Immunology Project Consortium (HIPC), grant BT/PR30260/MED/15/194/2018; and Government of India Department of Biotechnology grant DBT BT/PR5132/MED/15/85/2012.
We thank Aditya Rathi (ICGEB-TACF) for FACS sorting of the PBMCs. We are very thankful to Steven Bosinger and Kathryn Pellegrini, Yerkes Genomics Core, Emory University, Atlanta, GA, for help with RNA sequencing studies. We thank Satendra Singh and Ajay Singh, ICGEB, New Delhi, for technical support.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Kaja Murali-Krishna, Email: murali.kaja@emory.edu.
Anmol Chandele, Email: chandeleanmol@gmail.com.
Bryan R. G. Williams, Hudson Institute of Medical Research
REFERENCES
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srikiatkhachorn A, Rothman AL, Gibbons RV, Sittisombut N, Malasit P, Ennis FA, Nimmannitya S, Kalayanarooj S. 2011. Dengue—how best to classify it. Clin Infect Dis 53:563–567. 10.1093/cid/cir451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malavige GN, Ogg GS. 2017. Pathogenesis of vascular leak in dengue virus infection. Immunology 151:261–269. 10.1111/imm.12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halstead S. 2019. Recent advances in understanding dengue. F1000Res 8:F1000 Faculty Rev-1279. 10.12688/f1000research.19197.1. [DOI] [Google Scholar]
- 5.Grange L, Simon-Loriere E, Sakuntabhai A, Gresh L, Paul R, Harris E. 2014. Epidemiological risk factors associated with high global frequency of inapparent dengue virus infections. Front Immunol 5:280. 10.3389/fimmu.2014.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.OhAinle M, Balmaseda A, Macalalad AR, Tellez Y, Zody MC, Saborio S, Nunez A, Lennon NJ, Birren BW, Gordon A, Henn MR, Harris E. 2011. Dynamics of dengue disease severity determined by the interplay between viral genetics and serotype-specific immunity. Sci Transl Med 3:114ra128. 10.1126/scitranslmed.3003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duangchinda T, Dejnirattisai W, Vasanawathana S, Limpitikul W, Tangthawornchaikul N, Malasit P, Mongkolsapaya J, Screaton G. 2010. Immunodominant T-cell responses to dengue virus NS3 are associated with DHF. Proc Natl Acad Sci USA 107:16922–16927. 10.1073/pnas.1010867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Bates TM, Cordeiro MT, Nascimento EJ, Smith AP, Soares de Melo KM, McBurney SP, Evans JD, Marques ET, Jr, Barratt-Boyes SM. 2013. Association between magnitude of the virus-specific plasmablast response and disease severity in dengue patients. J Immunol 190:80–87. 10.4049/jimmunol.1103350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green S, Pichyangkul S, Vaughn DW, Kalayanarooj S, Nimmannitya S, Nisalak A, Kurane I, Rothman AL, Ennis FA. 1999. Early CD69 expression on peripheral blood lymphocytes from children with dengue hemorrhagic fever. J Infect Dis 180:1429–1435. 10.1086/315072. [DOI] [PubMed] [Google Scholar]
- 10.Malavige GN, Gomes L, Alles L, Chang T, Salimi M, Fernando S, Nanayakkara KD, Jayaratne S, Ogg GS. 2013. Serum IL-10 as a marker of severe dengue infection. BMC Infect Dis 13:341. 10.1186/1471-2334-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bozza FA, Cruz OG, Zagne SM, Azeredo EL, Nogueira RM, Assis EF, Bozza PT, Kubelka CF. 2008. Multiplex cytokine profile from dengue patients: MIP-1beta and IFN-gamma as predictive factors for severity. BMC Infect Dis 8:86. 10.1186/1471-2334-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singla M, Kar M, Sethi T, Kabra SK, Lodha R, Chandele A, Medigeshi GR. 2016. Immune response to dengue virus infection in pediatric patients in New Delhi, India—association of viremia, inflammatory mediators and monocytes with disease severity. PLoS Negl Trop Dis 10:e0004497. 10.1371/journal.pntd.0004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.John DV, Lin YS, Perng GC. 2015. Biomarkers of severe dengue disease—a review. J Biomed Sci 22:83. 10.1186/s12929-015-0191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halstead SB. 2014. Dengue antibody-dependent enhancement: knowns and unknowns. Microbiol Spectr 2:AID-0022-2014. 10.1128/microbiolspec.AID-0022-2014. [DOI] [PubMed] [Google Scholar]
- 15.Katzelnick LC, Gresh L, Halloran ME, Mercado JC, Kuan G, Gordon A, Balmaseda A, Harris E. 2017. Antibody-dependent enhancement of severe dengue disease in humans. Science 358:929–932. 10.1126/science.aan6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wahala WM, Silva AM. 2011. The human antibody response to dengue virus infection. Viruses 3:2374–2395. 10.3390/v3122374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Alwis R, Beltramello M, Messer WB, Sukupolvi-Petty S, Wahala WMPB, Kraus A, Olivarez NP, Pham Q, Brien JD, Brian J, Tsai W-Y, Wang W-K, Halstead S, Kliks S, Diamond MS, Baric R, Lanzavecchia A, Sallusto F, de Silva AM. 2011. In-depth analysis of the antibody response of individuals exposed to primary dengue virus infection. PLoS Negl Trop Dis 5:e1188. 10.1371/journal.pntd.0001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halstead SB, Rojanasuphot S, Sangkawibha N. 1983. Original antigenic sin in dengue. Am J Trop Med Hyg 32:154–156. 10.4269/ajtmh.1983.32.154. [DOI] [PubMed] [Google Scholar]
- 19.Koraka P, Suharti C, Setiati TE, Mairuhu AT, Van Gorp E, Hack CE, Juffrie M, Sutaryo J, Van Der Meer GM, Groen J, Osterhaus AD. 2001. Kinetics of dengue virus-specific serum immunoglobulin classes and subclasses correlate with clinical outcome of infection. J Clin Microbiol 39:4332–4338. 10.1128/JCM.39.12.4332-4338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valdes K, Alvarez M, Pupo M, Vazquez S, Rodriguez R, Guzman MG. 2000. Human dengue antibodies against structural and nonstructural proteins. Clin Diagn Lab Immunol 7:856–857. 10.1128/CDLI.7.5.856-857.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Churdboonchart V, Bhamarapravati N, Peampramprecha S, Sirinavin S. 1991. Antibodies against dengue viral proteins in primary and secondary dengue hemorrhagic fever. Am J Trop Med Hyg 44:481–493. 10.4269/ajtmh.1991.44.481. [DOI] [PubMed] [Google Scholar]
- 22.Wrammert J, Onlamoon N, Akondy RS, Perng GC, Polsrila K, Chandele A, Kwissa M, Pulendran B, Wilson PC, Wittawatmongkol O, Yoksan S, Angkasekwinai N, Pattanapanyasat K, Chokephaibulkit K, Ahmed R. 2012. Rapid and massive virus-specific plasmablast responses during acute dengue virus infection in humans. J Virol 86:2911–2918. 10.1128/JVI.06075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zompi S, Montoya M, Pohl MO, Balmaseda A, Harris E. 2012. Dominant cross-reactive B cell response during secondary acute dengue virus infection in humans. PLoS Negl Trop Dis 6:e1568. 10.1371/journal.pntd.0001568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellebedy AH, Jackson KJ, Kissick HT, Nakaya HI, Davis CW, Roskin KM, McElroy AK, Oshansky CM, Elbein R, Thomas S, Lyon GM, Spiropoulou CF, Mehta AK, Thomas PG, Boyd SD, Ahmed R. 2016. Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat Immunol 17:1226–1234. 10.1038/ni.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McElroy AK, Akondy RS, Davis CW, Ellebedy AH, Mehta AK, Kraft CS, Lyon GM, Ribner BS, Varkey J, Sidney J, Sette A, Campbell S, Stroher U, Damon I, Nichol ST, Spiropoulou CF, Ahmed R. 2015. Human Ebola virus infection results in substantial immune activation. Proc Natl Acad Sci USA 112:4719–4724. 10.1073/pnas.1502619112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, James J, Air GM, Capra JD, Ahmed R, Wilson PC. 2008. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453:667–671. 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balakrishnan T, Bela-Ong DB, Toh YX, Flamand M, Devi S, Koh MB, Hibberd ML, Ooi EE, Low JG, Leo YS, Gu F, Fink K. 2011. Dengue virus activates polyreactive, natural IgG B cells after primary and secondary infection. PLoS One 6:e29430. 10.1371/journal.pone.0029430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fink K. 2012. Origin and function of circulating plasmablasts during acute viral infections. Front Immunol 3:78. 10.3389/fimmu.2012.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter MJ, Mitchell RM, Meyer Sauteur PM, Kelly DF, Truck J. 2017. The antibody-secreting cell response to infection: kinetics and clinical applications. Front Immunol 8:630. 10.3389/fimmu.2017.00630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bromage ES, Kaattari IM, Zwollo P, Kaattari SL. 2004. Plasmablast and plasma cell production and distribution in trout immune tissues. J Immunol 173:7317–7323. 10.4049/jimmunol.173.12.7317. [DOI] [PubMed] [Google Scholar]
- 31.Schluter M, Oswald E, Winklmeier S, Meinl I, Havla J, Eichhorn P, Meinl E, Kumpfel T. 2021. Effects of natalizumab therapy on intrathecal immunoglobulin G production indicate targeting of plasmablasts. Neurol Neuroimmunol Neuroinflamm 8:e01030. 10.1212/NXI.0000000000001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seong Y, Lazarus NH, Sutherland L, Habtezion A, Abramson T, He XS, Greenberg HB, Butcher EC. 2017. Trafficking receptor signatures define blood plasmablasts responding to tissue-specific immune challenge. JCI Insight 2:e90233. 10.1172/jci.insight.90233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pattanapanyasat K, Khowawisetsut L, Chuansumrit A, Chokephaibulkit K, Tangnararatchakit K, Apiwattanakul N, Techasaensiri C, Thitilertdecha P, Sae-Ung T, Onlamoon N. 2018. B cell subset alteration and the expression of tissue homing molecules in dengue infected patients. J Biomed Sci 25:64. 10.1186/s12929-018-0467-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen P, Roch T, Lampropoulou V, O’Connor RA, Stervbo U, Hilgenberg E, Ries S, Dang VD, Jaimes Y, Daridon C, Li R, Jouneau L, Boudinot P, Wilantri S, Sakwa I, Miyazaki Y, Leech MD, McPherson RC, Wirtz S, Neurath M, Hoehlig K, Meinl E, Grutzkau A, Grun JR, Horn K, Kuhl AA, Dorner T, Bar-Or A, Kaufmann SHE, Anderton SM, Fillatreau S. 2014. IL-35-producing B cells are critical regulators of immunity during autoimmune and infectious diseases. Nature 507:366–370. 10.1038/nature12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsumoto M, Baba A, Yokota T, Nishikawa H, Ohkawa Y, Kayama H, Kallies A, Nutt SL, Sakaguchi S, Takeda K, Kurosaki T, Baba Y. 2014. Interleukin-10-producing plasmablasts exert regulatory function in autoimmune inflammation. Immunity 41:1040–1051. 10.1016/j.immuni.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 36.Bermejo DA, Jackson SW, Gorosito-Serran M, Acosta-Rodriguez EV, Amezcua-Vesely MC, Sather BD, Singh AK, Khim S, Mucci J, Liggitt D, Campetella O, Oukka M, Gruppi A, Rawlings DJ. 2013. Trypanosoma cruzi trans-sialidase initiates a program independent of the transcription factors RORgammat and Ahr that leads to IL-17 production by activated B cells. Nat Immunol 14:514–522. 10.1038/ni.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarte K, Zhan F, De Vos J, Klein B, Shaughnessy J, Jr.. 2003. Gene expression profiling of plasma cells and plasmablasts: toward a better understanding of the late stages of B-cell differentiation. Blood 102:592–600. 10.1182/blood-2002-10-3161. [DOI] [PubMed] [Google Scholar]
- 38.Valor LM, Rodriguez-Bayona B, Ramos-Amaya AB, Brieva JA, Campos-Caro A. 2017. The transcriptional profiling of human in vivo-generated plasma cells identifies selective imbalances in monoclonal gammopathies. PLoS One 12:e0183264. 10.1371/journal.pone.0183264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhan F, Tian E, Bumm K, Smith R, Barlogie B, Shaughnessy J, Jr.. 2003. Gene expression profiling of human plasma cell differentiation and classification of multiple myeloma based on similarities to distinct stages of late-stage B-cell development. Blood 101:1128–1140. 10.1182/blood-2002-06-1737. [DOI] [PubMed] [Google Scholar]
- 40.Lugar PL, Love C, Grammer AC, Dave SS, Lipsky PE. 2012. Molecular characterization of circulating plasma cells in patients with active systemic lupus erythematosus. PLoS One 7:e44362. 10.1371/journal.pone.0044362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mei HE, Wirries I, Frolich D, Brisslert M, Giesecke C, Grun JR, Alexander T, Schmidt S, Luda K, Kuhl AA, Engelmann R, Durr M, Scheel T, Bokarewa M, Perka C, Radbruch A, Dorner T. 2015. A unique population of IgG-expressing plasma cells lacking CD19 is enriched in human bone marrow. Blood 125:1739–1748. 10.1182/blood-2014-02-555169. [DOI] [PubMed] [Google Scholar]
- 42.Ramos-Amaya A, Rodriguez-Bayona B, Lopez-Blanco R, Andujar E, Perez-Alegre M, Campos-Caro A, Brieva JA. 2015. Survival of human circulating antigen-induced plasma cells is supported by plasma cell-niche cytokines and T follicular helper lymphocytes. J Immunol 194:1031–1038. 10.4049/jimmunol.1402231. [DOI] [PubMed] [Google Scholar]
- 43.Lin W, Zhang P, Chen H, Chen Y, Yang H, Zheng W, Zhang X, Zhang F, Zhang W, Lipsky PE. 2017. Circulating plasmablasts/plasma cells: a potential biomarker for IgG4-related disease. Arthritis Res Ther 19:25. 10.1186/s13075-017-1231-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cocco M, Stephenson S, Care MA, Newton D, Barnes NA, Davison A, Rawstron A, Westhead DR, Doody GM, Tooze RM. 2012. In vitro generation of long-lived human plasma cells. J Immunol 189:5773–5785. 10.4049/jimmunol.1103720. [DOI] [PubMed] [Google Scholar]
- 45.Jourdan M, Caraux A, De Vos J, Fiol G, Larroque M, Cognot C, Bret C, Duperray C, Hose D, Klein B. 2009. An in vitro model of differentiation of memory B cells into plasmablasts and plasma cells including detailed phenotypic and molecular characterization. Blood 114:5173–5181. 10.1182/blood-2009-07-235960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waickman AT, Gromowski GD, Rutvisuttinunt W, Li T, Siegfried H, Victor K, Kuklis C, Gomootsukavadee M, McCracken MK, Gabriel B, Mathew A, Grinyo IEA, Fouch ME, Liang J, Fernandez S, Davidson E, Doranz BJ, Srikiatkhachorn A, Endy T, Thomas SJ, Ellison D, Rothman AL, Jarman RG, Currier JR, Friberg H. 2020. Transcriptional and clonal characterization of B cell plasmablast diversity following primary and secondary natural DENV infection. EBioMedicine 54:102733. 10.1016/j.ebiom.2020.102733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rouers A, Appanna R, Chevrier M, Lum J, Lau MC, Tan L, Loy T, Tay A, Sethi R, Sathiakumar D, Kaur K, Bohme J, Leo YS, Renia L, Howland SW, Singhal A, Chen J, Fink K. 2021. CD27(hi)CD38(hi) plasmablasts are activated B cells of mixed origin with distinct function. iScience 24:102482. 10.1016/j.isci.2021.102482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rouers A, Chng MHY, Lee B, Rajapakse MP, Kaur K, Toh YX, Sathiakumar D, Loy T, Thein TL, Lim VWX, Singhal A, Yeo TW, Leo YS, Vora KA, Casimiro D, Lim B, Tucker-Kellogg L, Rivino L, Newell EW, Fink K. 2021. Immune cell phenotypes associated with disease severity and long-term neutralizing antibody titers after natural dengue virus infection. Cell Rep Med 2:100278. 10.1016/j.xcrm.2021.100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Appanna R, Kg S, Xu MH, Toh YX, Velumani S, Carbajo D, Lee CY, Zuest R, Balakrishnan T, Xu W, Lee B, Poidinger M, Zolezzi F, Leo YS, Thein TL, Wang CI, Fink K. 2016. Plasmablasts during acute dengue infection represent a small subset of a broader virus-specific memory B cell pool. EBioMedicine 12:178–188. 10.1016/j.ebiom.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Priyamvada L, Cho A, Onlamoon N, Zheng NY, Huang M, Kovalenkov Y, Chokephaibulkit K, Angkasekwinai N, Pattanapanyasat K, Ahmed R, Wilson PC, Wrammert J. 2016. B cell responses during secondary dengue virus infection are dominated by highly cross-reactive, memory-derived plasmablasts. J Virol 90:5574–5585. 10.1128/JVI.03203-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu M, Hadinoto V, Appanna R, Joensson K, Toh YX, Balakrishnan T, Ong SH, Warter L, Leo YS, Wang CI, Fink K. 2012. Plasmablasts generated during repeated dengue infection are virus glycoprotein-specific and bind to multiple virus serotypes. J Immunol 189:5877–5885. 10.4049/jimmunol.1201688. [DOI] [PubMed] [Google Scholar]
- 52.Jabara HH, Boyden SE, Chou J, Ramesh N, Massaad MJ, Benson H, Bainter W, Fraulino D, Rahimov F, Sieff C, Liu ZJ, Alshemmari SH, Al-Ramadi BK, Al-Dhekri H, Arnaout R, Abu-Shukair M, Vatsayan A, Silver E, Ahuja S, Davies EG, Sola-Visner M, Ohsumi TK, Andrews NC, Notarangelo LD, Fleming MD, Al-Herz W, Kunkel LM, Geha RS. 2016. A missense mutation in TFRC, encoding transferrin receptor 1, causes combined immunodeficiency. Nat Genet 48:74–78. 10.1038/ng.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mensah FFK, Armstrong CW, Reddy V, Bansal AS, Berkovitz S, Leandro MJ, Cambridge G. 2018. CD24 expression and B cell maturation shows a novel link with energy metabolism: potential implications for patients with myalgic encephalomyelitis/chronic fatigue syndrome. Front Immunol 9:2421. 10.3389/fimmu.2018.02421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vale AM, Schroeder HW, Jr.. 2010. Clinical consequences of defects in B-cell development. J Allergy Clin Immunol 125:778–787. 10.1016/j.jaci.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vlkova M, Fronkova E, Kanderova V, Janda A, Ruzickova S, Litzman J, Sediva A, Kalina T. 2010. Characterization of lymphocyte subsets in patients with common variable immunodeficiency reveals subsets of naive human B cells marked by CD24 expression. J Immunol 185:6431–6438. 10.4049/jimmunol.0903876. [DOI] [PubMed] [Google Scholar]
- 56.Pavlasova G, Mraz M. 2020. The regulation and function of CD20: an “enigma” of B-cell biology and targeted therapy. Haematologica 105:1494–1506. 10.3324/haematol.2019.243543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van de Ven AA, Compeer EB, Bloem AC, van de Corput L, van Gijn M, van Montfrans JM, Boes M. 2012. Defective calcium signaling and disrupted CD20-B-cell receptor dissociation in patients with common variable immunodeficiency disorders. J Allergy Clin Immunol 129:755–761.e7. 10.1016/j.jaci.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 58.Duddy M, Niino M, Adatia F, Hebert S, Freedman M, Atkins H, Kim HJ, Bar-Or A. 2007. Distinct effector cytokine profiles of memory and naive human B cell subsets and implication in multiple sclerosis. J Immunol 178:6092–6099. 10.4049/jimmunol.178.10.6092. [DOI] [PubMed] [Google Scholar]
- 59.Fillatreau S. 2018. B cells and their cytokine activities implications in human diseases. Clin Immunol 186:26–31. 10.1016/j.clim.2017.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lund FE. 2008. Cytokine-producing B lymphocytes—key regulators of immunity. Curr Opin Immunol 20:332–338. 10.1016/j.coi.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vazquez MI, Catalan-Dibene J, Zlotnik A. 2015. B cells responses and cytokine production are regulated by their immune microenvironment. Cytokine 74:318–326. 10.1016/j.cyto.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson KE, Wilgus TA. 2014. Vascular endothelial growth factor and angiogenesis in the regulation of cutaneous wound repair. Adv Wound Care (New Rochelle) 3:647–661. 10.1089/wound.2013.0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takahashi H, Shibuya M. 2005. The vascular endothelial growth factor (VEGF)/VEGF receptor system and its role under physiological and pathological conditions. Clin Sci (Lond) 109:227–241. 10.1042/CS20040370. [DOI] [PubMed] [Google Scholar]
- 64.Bates DO. 2010. Vascular endothelial growth factors and vascular permeability. Cardiovasc Res 87:262–271. 10.1093/cvr/cvq105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weis SM, Cheresh DA. 2005. Pathophysiological consequences of VEGF-induced vascular permeability. Nature 437:497–504. 10.1038/nature03987. [DOI] [PubMed] [Google Scholar]
- 66.Funyu J, Mochida S, Inao M, Matsui A, Fujiwara K. 2001. VEGF can act as vascular permeability factor in the hepatic sinusoids through upregulation of porosity of endothelial cells. Biochem Biophys Res Commun 280:481–485. 10.1006/bbrc.2000.4148. [DOI] [PubMed] [Google Scholar]
- 67.Thakur P, Chakravarti A, Aggarwal S, Uppal B, Bhalla P. 2016. Elevated levels of vascular endothelial growth factor in adults with severe dengue infection. Virusdisease 27:48–54. 10.1007/s13337-015-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tseng CS, Lo HW, Teng HC, Lo WC, Ker CG. 2005. Elevated levels of plasma VEGF in patients with dengue hemorrhagic fever. FEMS Immunol Med Microbiol 43:99–102. 10.1016/j.femsim.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 69.Srikiatkhachorn A, Ajariyakhajorn C, Endy TP, Kalayanarooj S, Libraty DH, Green S, Ennis FA, Rothman AL. 2007. Virus-induced decline in soluble vascular endothelial growth receptor 2 is associated with plasma leakage in dengue hemorrhagic fever. J Virol 81:1592–1600. 10.1128/JVI.01642-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, Xu Y, Ma J, Pang X, Dong M. 2017. Adrenomedullin promotes angiogenesis in epithelial ovarian cancer through upregulating hypoxia-inducible factor-1alpha and vascular endothelial growth factor. Sci Rep 7:40524. 10.1038/srep40524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hollander LL, Guo X, Salem RR, Cha CH. 2015. The novel tumor angiogenic factor, adrenomedullin-2, predicts survival in pancreatic adenocarcinoma. J Surg Res 197:219–224. 10.1016/j.jss.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 72.Benn A, Bredow C, Casanova I, Vukicevic S, Knaus P. 2016. VE-cadherin facilitates BMP-induced endothelial cell permeability and signaling. J Cell Sci 129:206–218. 10.1242/jcs.179960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bach LA. 2015. Endothelial cells and the IGF system. J Mol Endocrinol 54:R1–R13. 10.1530/JME-14-0215. [DOI] [PubMed] [Google Scholar]
- 74.Lin S, Zhang Q, Shao X, Zhang T, Xue C, Shi S, Zhao D, Lin Y. 2017. IGF-1 promotes angiogenesis in endothelial cells/adipose-derived stem cells co-culture system with activation of PI3K/Akt signal pathway. Cell Prolif 50:e12390. 10.1111/cpr.12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gao B, Deng J, Zhang X, Sun H, Jia G, Li J, Zhang K, Wan C, Wang L, Yan LJ, Cai Z, Ma J. 2020. Effects of mesencephalic astrocyte-derived neurotrophic factor on cerebral angiogenesis in a rat model of cerebral ischemia. Neurosci Lett 715:134657. 10.1016/j.neulet.2019.134657. [DOI] [PubMed] [Google Scholar]
- 76.Secchiero P, Gonelli A, Carnevale E, Corallini F, Rizzardi C, Zacchigna S, Melato M, Zauli G. 2004. Evidence for a proangiogenic activity of TNF-related apoptosis-inducing ligand. Neoplasia 6:364–373. 10.1593/neo.03421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Soo KM, Khalid B, Ching SM, Tham CL, Basir R, Chee HY. 2017. Meta-analysis of biomarkers for severe dengue infections. PeerJ 5:e3589. 10.7717/peerj.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Batten M, Groom J, Cachero TG, Qian F, Schneider P, Tschopp J, Browning JL, Mackay F. 2000. BAFF mediates survival of peripheral immature B lymphocytes. J Exp Med 192:1453–1466. 10.1084/jem.192.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mackay F, Browning JL. 2002. BAFF: a fundamental survival factor for B cells. Nat Rev Immunol 2:465–475. 10.1038/nri844. [DOI] [PubMed] [Google Scholar]
- 80.van Roy F, Berx G. 2008. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci 65:3756–3788. 10.1007/s00018-008-8281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen A, Beetham H, Black MA, Priya R, Telford BJ, Guest J, Wiggins GA, Godwin TD, Yap AS, Guilford PJ. 2014. E-cadherin loss alters cytoskeletal organization and adhesion in non-malignant breast cells but is insufficient to induce an epithelial-mesenchymal transition. BMC Cancer 14:552. 10.1186/1471-2407-14-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ishikawa T, Kramer RH. 2010. Sdc1 negatively modulates carcinoma cell motility and invasion. Exp Cell Res 316:951–965. 10.1016/j.yexcr.2009.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beauvais DM, Ell BJ, McWhorter AR, Rapraeger AC. 2009. Syndecan-1 regulates alphavbeta3 and alphavbeta5 integrin activation during angiogenesis and is blocked by synstatin, a novel peptide inhibitor. J Exp Med 206:691–705. 10.1084/jem.20081278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fears CY, Woods A. 2006. The role of syndecans in disease and wound healing. Matrix Biol 25:443–456. 10.1016/j.matbio.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 85.Stepp MA, Pal-Ghosh S, Tadvalkar G, Pajoohesh-Ganji A. 2015. Syndecan-1 and its expanding list of contacts. Adv Wound Care (New Rochelle) 4:235–249. 10.1089/wound.2014.0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Asuthkar S, Velpula KK, Nalla AK, Gogineni VR, Gondi CS, Rao JS. 2014. Irradiation-induced angiogenesis is associated with an MMP-9-miR-494-syndecan-1 regulatory loop in medulloblastoma cells. Oncogene 33:1922–1933. 10.1038/onc.2013.151. [DOI] [PubMed] [Google Scholar]
- 87.Lamorte S, Ferrero S, Aschero S, Monitillo L, Bussolati B, Omede P, Ladetto M, Camussi G. 2012. Syndecan-1 promotes the angiogenic phenotype of multiple myeloma endothelial cells. Leukemia 26:1081–1090. 10.1038/leu.2011.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cortez VS, Cervantes-Barragan L, Song C, Gilfillan S, McDonald KG, Tussiwand R, Edelson BT, Murakami Y, Murphy KM, Newberry RD, Sibley LD, Colonna M. 2014. CRTAM controls residency of gut CD4+CD8+ T cells in the steady state and maintenance of gut CD4+ Th17 during parasitic infection. J Exp Med 211:623–633. 10.1084/jem.20130904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Campos A, Burgos-Ravanal R, Gonzalez MF, Huilcaman R, Lobos Gonzalez L, Quest AFG. 2019. Cell intrinsic and extrinsic mechanisms of caveolin-1-enhanced metastasis. Biomolecules 9:314. 10.3390/biom9080314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Navarro A, Anand-Apte B, Parat MO. 2004. A role for caveolae in cell migration. FASEB J 18:1801–1811. 10.1096/fj.04-2516rev. [DOI] [PubMed] [Google Scholar]
- 91.Harjunpaa H, Llort Asens M, Guenther C, Fagerholm SC. 2019. Cell adhesion molecules and their roles and regulation in the immune and tumor microenvironment. Front Immunol 10:1078. 10.3389/fimmu.2019.01078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Trivedi PJ, Bruns T, Ward S, Mai M, Schmidt C, Hirschfield GM, Weston CJ, Adams DH. 2016. Intestinal CCL25 expression is increased in colitis and correlates with inflammatory activity. J Autoimmun 68:98–104. 10.1016/j.jaut.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morales J, Homey B, Vicari AP, Hudak S, Oldham E, Hedrick J, Orozco R, Copeland NG, Jenkins NA, McEvoy LM, Zlotnik A. 1999. CTACK, a skin-associated chemokine that preferentially attracts skin-homing memory T cells. Proc Natl Acad Sci USA 96:14470–14475. 10.1073/pnas.96.25.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lazarus NH, Kunkel EJ, Johnston B, Wilson E, Youngman KR, Butcher EC. 2003. A common mucosal chemokine (mucosae-associated epithelial chemokine/CCL28) selectively attracts IgA plasmablasts. J Immunol 170:3799–3805. 10.4049/jimmunol.170.7.3799. [DOI] [PubMed] [Google Scholar]
- 95.Kroczek RA, Henn V. 2012. The role of XCR1 and its ligand XCL1 in antigen cross-presentation by murine and human dendritic cells. Front Immunol 3:14. 10.3389/fimmu.2012.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Matsuo K, Kitahata K, Kawabata F, Kamei M, Hara Y, Takamura S, Oiso N, Kawada A, Yoshie O, Nakayama T. 2018. A highly active form of XCL1/lymphotactin functions as an effective adjuvant to recruit cross-presenting dendritic cells for induction of effector and memory CD8(+) T cells. Front Immunol 9:2775. 10.3389/fimmu.2018.02775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Iragavarapu-Charyulu V, Wojcikiewicz E, Urdaneta A. 2020. Semaphorins in angiogenesis and autoimmune diseases: therapeutic targets? Front Immunol 11:346. 10.3389/fimmu.2020.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Toyofuku T, Zhang H, Kumanogoh A, Takegahara N, Suto F, Kamei J, Aoki K, Yabuki M, Hori M, Fujisawa H, Kikutani H. 2004. Dual roles of Sema6D in cardiac morphogenesis through region-specific association of its receptor, Plexin-A1, with off-track and vascular endothelial growth factor receptor type 2. Genes Dev 18:435–447. 10.1101/gad.1167304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bielenberg DR, Pettaway CA, Takashima S, Klagsbrun M. 2006. Neuropilins in neoplasms: expression, regulation, and function. Exp Cell Res 312:584–593. 10.1016/j.yexcr.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 100.Suzuki K, Kumanogoh A, Kikutani H. 2008. Semaphorins and their receptors in immune cell interactions. Nat Immunol 9:17–23. 10.1038/ni1553. [DOI] [PubMed] [Google Scholar]
- 101.Kwissa M, Nakaya HI, Onlamoon N, Wrammert J, Villinger F, Perng GC, Yoksan S, Pattanapanyasat K, Chokephaibulkit K, Ahmed R, Pulendran B. 2014. Dengue virus infection induces expansion of a CD14(+)CD16(+) monocyte population that stimulates plasmablast differentiation. Cell Host Microbe 16:115–127. 10.1016/j.chom.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jaipersad AS, Lip GY, Silverman S, Shantsila E. 2014. The role of monocytes in angiogenesis and atherosclerosis. J Am Coll Cardiol 63:1–11. 10.1016/j.jacc.2013.09.019. [DOI] [PubMed] [Google Scholar]
- 103.Sidibe A, Ropraz P, Jemelin S, Emre Y, Poittevin M, Pocard M, Bradfield PF, Imhof BA. 2018. Angiogenic factor-driven inflammation promotes extravasation of human proangiogenic monocytes to tumours. Nat Commun 9:355. 10.1038/s41467-017-02610-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.WHO. 2009. Dengue guidelines for diagnosis, treatment, prevention and control. WHO, Geneva, Switzerland. [PubMed] [Google Scholar]
- 105.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. 2003. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34:267–273. 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 108.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. 2019. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2<br>. Download jvi.00610-21-s0001.pdf, PDF file, 5.8 MB (5.8MB, pdf)
Tables S1 to S7<br>. Download jvi.00610-21-s0002.xlsx, XLSX file, 2.2 MB (2.2MB, xlsx)
Data Availability Statement
The dengue RNA-seq data set has been deposited in the Gene Expression Omnibus (GEO) with the accession number GSE171487.