Abstract
Staphylococcus aureus is the leading cause of secondary infections in hospitals and a challenging pathogen in food industries. Decades after it was first reported, β-lactam-resistant S. aureus remains a subject of intense research owing to the ever-increasing issue of drug resistance. S. aureus bacteriophages (phages) or their encoded products are considered an alternative to antibiotics as they have been shown to be effective in treating some S. aureus-associated infections. In this review, we present a concise collection of the literature on the pathogenic potential of S. aureus and examine the prospects of using S. aureus phages and their encoded products as antimicrobials.
Keywords: Methicillin-resistant Staphylococcus aureus (MRSA), Staphylococcus bacteriophage, Antimicrobial resistance, Bacteriophage therapy, Phage-encoded antimicrobial
Introduction
Staphylococcus aureus was first identified and described in 1880 by Alexander Ogston, a surgeon who isolated the bacterium from a surgical abscess [1]. S. aureus is a Gram-positive bacterium and facultative anaerobe with low genomic G + C content. It is a common commensal of humans and animals and may be found colonizing nares, axilla, perineum, skin, and multiple other body sites; however, nasal carriage is of greater concern because it is the most common source of MRSA for disease spread [2, 3]. It is estimated that approximately 15% of the population persistently carry S. aureus in the anterior nares; some populations have higher rates of S. aureus colonization including health care workers and hospitalized patients [4]. People harboring this bacterium are at greater risk of developing S. aureus infections [1, 5]. In humans, S. aureus can infect most organs and is a leading cause of death by infection [6]. It is one of the major causes of morbidity and mortality in hospitals in part because it can produce biofilms on indwelling devices such as catheters and medical implants, from which bacterial cells may detach resulting in secondary and disseminated infections [5]. S. aureus is one of the most successful pathogens due to the vast arsenal of virulence factors, including anchor proteins, secreted toxins and enzymes, polysaccharides, and immune system modulators. These are often located on mobile genetic elements (MGEs), which help diversify the virulence and resistance genes via horizontal gene transfer. Methicillin, a semi-synthetic β-lactam antibiotic, was first applied to treat S. aureus infections, but the bacterium soon developed resistance and became known as methicillin-resistant S. aureus (MRSA). Several reports have described vancomycin-resistant S. aureus (VRSA) and vancomycin-intermediate S. aureus (VISA); however, vancomycin remains in the list of first choice antibiotics for treating S. aureus severe infections [7]. The World Health Organization (WHO) has identified several bacterial pathogens, including MRSA, as leading causes of nosocomial infections; these infections are mainly caused by inappropriate and irrational use of antimicrobials/antibiotics in the healthcare facilities and food industries. In 2015, the WHO introduced an action plan against antimicrobial resistance, wherein one of the important focal points was the development of novel antimicrobial products to combat multidrug-resistant strains [8]. A study has demonstrated that fluoroquinolone antibiotics, such as ciprofloxacin, can induce an SOS response, which causes mobilization of several pathogenic islands (PIs), such as SaPIbov1, a prophage-encoded Shiga toxin, which induces production of phages and toxins [9]. Antibiotics not only cause selective pressure and propagation of resistance genes, but they may also be harmful to the health because most antibiotics are broad spectrum, and thus disrupt the normal microbiota and may cause gut dysbiosis [10]. Antibiotics that target protein synthesis can severely disrupt mitochondria and therefore affect normal physiologic functions [11].
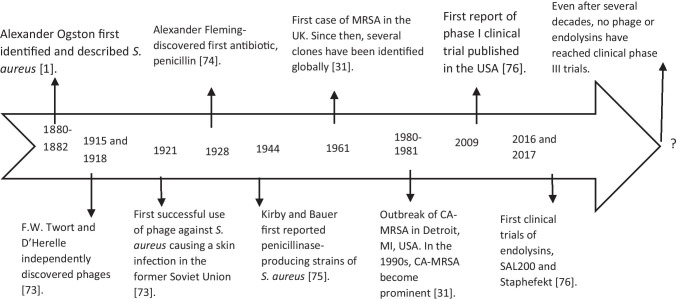
S. aureus has the ability to rapidly develop resistance to nearly any antibiotic drug coming into clinical use [12]. Studies on S. aureus have intrigued researchers worldwide, especially with regard to its evolution, as some types and subtypes of MRSA emerged and spread globally. MRSA is a major threat both in healthcare and outside healthcare settings (i.e., in communities). According to the reports of Centers for Disease Control and Prevention (CDC), USA, and SENTRY Antimicrobial Surveillance Program, there has been a decrease in MRSA infections [13, 14]. Still infections due to MRSA and methicillin-susceptible S. aureus (MSSA) are of major concern because people with MRSA infections are 64% more likely to die than people with MSSA infections [15]; also, CDC reported a slight increase in the MSSA infections and suggested that any kind of S. aureus infection is a serious concern. Bacteria can be resistant to current antibiotics, and with the slow progression in discovering new antimicrobials, among important candidates for treating MRSA infections include bacteriophages (i.e., phage viruses that kill bacteria). Phages were discovered earlier than antibiotics, and although their antibacterial effects were known, they were not used as antibacterial treatments because of the popularity gained by penicillin as a broad-spectrum antibiotic during World War II. However, in some parts of the world, such as in the former Soviet Union, the use of phages as therapeutics has continued as a common practice integrated into their healthcare system [16]. Western countries and other parts of the world only recognized the potential of phages as an alternative to antibacterial therapy several decades after their discovery with the description of more accurate studies involving phage efficacy, pharmacodynamics, and genomics [17]. Figure 1 presents a timeline of the major events in the history of S. aureus and the applications of its phages as antimicrobials.
Fig. 1.
Timeline of important events in the history of S. aureus, bacteriophages, and antibiotics. For several decades, bacteriophages received no attention from scientific communities in many countries, and their applications were confined to Eastern Europe. However, they have slowly gained momentum, especially regarding S. aureus. However, much work remains to be done as no phase III trials have been reported thus far
In this review, we highlight the health problems associated with MRSA, and describe the effectiveness of phages and phage proteins against this important nosocomial pathogen.
What makes S. aureus a successful pathogen?
The presence of a plethora of virulence factors that help the bacteria to invade host tissues, and of several MGEs carrying resistance determinants rendered the S. aureus versatile as a hospital pathogen. All S. aureus isolates have at least one prophage, but most have four [18]. The increase in drug-resistant S. aureus results from the inevitable genetic response to selective pressure by antimicrobial therapy [19]. Horizontal gene transfer is responsible for the resistance to most antibiotics, including methicillin and vancomycin [12]. Other resistance mechanisms, such as random mutations, confer resistance to fluoroquinolones, daptomycin, and linezolid [12].
S. aureus is not naturally competent for transformation (a genetic exchange mechanism), but the recently discovered cryptic sigma factor H makes it competent for transformation under certain conditions [20]. Interestingly, the activation of this factor was linked to the transfer of MGEs and may play important role in the evolution of antimicrobial resistance in S. aureus [20].
S. aureus is a clonal species, but it exhibits high genetic recombination within its core genome, especially in regions involving MGEs including a conjugative transposon, ICE6013; the staphylococcal cassette chromosome (SCC); and the genomic island, vSAα. This core genome transfer enables the bacterium to adapt through homologous recombination at the species level [21].
Virulence factors
Virulence factors can be grouped according their functions. The host tissue colonization proteins (A) comprise surface-associated proteins such as fibrinogen-binding proteins, fibronectin-binding proteins and collagen-binding proteins [5]. The host immune system evasion factors (B) include capsular polysaccharides, protein A, and leukocidins [5]. The toxins that damage host tissues (C) include α-toxins, β-toxins, δ-toxins, γ-toxins and Panton-Valentine leukocidin (PVL) [5] and, the superantigens (D) comprise enterotoxins (staphylococcal enterotoxins; SEs), toxic shock syndrome toxin (TSST-1), and staphylococcus like-superantigen proteins (SSL). Superantigens nonspecifically activate T-cells without normal antigen presentation and trigger a downstream immune response [5]. Superantigen enterotoxins are important to the food industry because they cause poisoning and severe emetic responses. The two types of enterotoxins are the classical and newly discovered enterotoxins [22]. The classical enterotoxins include SEA–SEE, which are the causative agents of 95% of food poisoning cases [22]. The newly identified enterotoxins (e.g., SEG) account for the other 5% of these cases [22]. SEs are usually found in MGEs, which potentially facilitate their disseminated [20]. Additional virulence factors include coagulase, proteases, lipases, DNases, fatty acid-metabolizing enzymes, among others [5].
Regulation of the virulence and toxin genes in S. aureus determines the course of disease development. The accessory gene regulator (agr) is a peptide quorum-sensing regulator, which is considered the main S. aureus global virulence regulator [23]. Four agr gene-polymorphisms were identified thus far [24]. A recent study on the hospital-associated (HA)-MRSA strain, USA100, implicated the agr-II gene in the regulation of a number of virulence-associated genes, and the expression of toxins involved in skin disease [23]. Additionally, the Agr-II system of USA 100 MRSA was also associated with the optimal bacterium surviving in sublethal antibiotic doses [23]. Besides Agr, other regulators can globally affect the bacterial virulence and biofilm development, including the two-component system SaeRS, the transcriptional regulator SarA, and a number of SarA homologs (e.g., MgrA, SarT, SarU and SarZ) [25]. More recently, a number of non-coding RNAs has been recognized as important virulence regulators, and many of them named Spr (small pathogenicity island RNA rNAs) are expressed from genes located in pathogenicity or resistance island [26]. The virulence regulatory systems of S. aureus form a complex network of regulators, commonly acting in synchrony with Agr and other regulatory systems [27].
Resistance genes
S. aureus strains that are resistant to methicillin or oxacillin are resilient to all beta-lactam antibiotics. Frequently, these strains are also resistant to other antibiotic classes such as macrolides, aminoglycosides, and fluoroquinolones [19]. The core genome of S. aureus is highly clonal, thereby few clonal lineages of MRSA predominate in different regions. However, approximately 15–20% of the S. aureus genome is composed of MGEs, which include plasmids, bacteriophages, transposons, PIs, and SCCs [28]. Haaber et al. (2017) presented a detailed account of the role of these MGEs in spreading resistance in S. aureus. These MGEs can house several toxins, superantigen and resistance determinants. The transfer usually occurs via conjugation and transduction during colonization of the host tissue. S. aureus also forms biofilms that promote the transfer of resistance genes [28]. Extensive literature is available on the mechanisms of transfer of resistance genes in laboratory settings, but little is known about the process associated with the actual conditions that may promote the increase of these transfers.
Molecular basis of MRSA
Methicillin-susceptible S. aureus (MSSA) contains penicillin-binding proteins (PBPs) which are membrane-bound proteins characterized by their affinity to penicillin. When beta-lactam antibiotics bind to PBPs, the peptidoglycan is disrupted, killing the S. aureus cells. However, in MRSA, an alternative form of PBP called PBP2a or PBP2′, encoded by the mecA gene, is present in the SCCmec element. The alternative protein has low affinity for β-lactam antibiotics such as methicillin or oxacillin. Therefore, the peptidoglycan remains undisturbed, and the MRSA survives [12, 19]. The earliest report of MRSA appeared in the year 1961 and since then it has spread across the globe [4, 19]. Regarding the evolution of healthcare-associated MRSA (HA-MRSA), it is now clear from the genomic studies that the SCCmec have entered in many independent events in the great majority of MRSA lineages [29].
The International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) is dedicated to classifying MRSA. There are several types and allotypes of SCCmec. Currently, the IWG-SCC recognizes 13 SCCmec types [30]. Essentially, all SSCmec elements carry either mecA or mecC genes and have two complexes: the mec gene complex and the cassette chromosome recombinase (ccr) gene complex [30]. MRSA can be classified as hospital-associated (HA), community-associated (CA), or livestock-associated (LA) according to the strain’s origin and the molecular typing (multilocus sequence typing-MLST and SCCmec typing). HA-MRSA is most frequently associated with patients previously admitted to hospitals, who are older and have one or more comorbidity [1]. HA-MRSA may cause severe infections including pneumonia, invasive infections and bacteremia, being often resistant to non-beta-lactam antibiotics [31]. Community-acquired MRSA (CA-MRSA) can affect healthy individuals (i.e., populations not previously admitted to hospitals), is more susceptible to non-beta-lactam antibiotics, commonly produces Panton-Valentine leucocidin (PVL), and causes distinct clinical manifestations [31, 32]. Strains of CA-MRSA are predominantly associated with skin and soft tissue infections, but may be also involved in necrotizing pneumonia and severe sepsis [31, 32].
MRSA has a complex epidemiology, and the demarcation between HA-MRSA and CA-MRSA sometimes is not an easy task [1, 31]. Some reports have confirmed that CA-MRSA has shifted from communities to hospitals and has acquired more resistance determinants [31, 32]. For example, MRSA clones carrying SCCmec type IV, especially USA300, have caused nosocomial MRSA outbreaks in the USA [31, 32]. In addition, community onset infections caused by HA-MRSA had been reported [31]. Regarding the origin and spread of CA-MRSA, it is postulated that CA-MRSA clones have arisen in the community by the horizontal transfer of SCCmec elements and PVL genes to the genomes of MSSA strains prevalent in a population [31].
MRSA strains associated with livestock animals (i.e., LA-MRSA) are additional threats. The sequence type (ST) 398 is the most studied MRSA strain in primarily domesticated animals. Humans in contact with these animals may become colonized, and severe LA-MRSA infections in humans have been reported [33, 34]. House companion animals with infections caused by MRSA lineages similar to those of human origin have also been reported. These observations suggest that domestic animals may serve as reservoirs for MRSA infections in humans and vice-versa [33].
Most common MRSA types worldwide
A MRSA lineage is generally defined as a group of strains with an identical sequence type and SCCmec type [19]. The most common lineages of HA-MRSA are ST239(CC8)-SCCmecIII (Brazilian clone), ST22-SCCmecIV (EMRSA15) and ST5(CC5)-SCCmecII (USA100) [35]. Among CA-MRSA the main linages are ST8(CC8)-SCCmecIV (USA300), ST30(CC30)-SCCmecIV, ST59(CC59)-SCCmecIV and ST1(CC1)-SCCmecIV (USA400) [35]. Finally, the most common LA-MRSA belongs to the lineage ST398(ST398)-SCCmecV [35]. These clonal types differ in their origins, genotypes, virulence and resistance genes, and clinical manifestations. For example, the most frequent CA-MRSA strains carry the lukSF-genes for PVL, and USA100 has more resistance profiles but is less toxic and less virulent than is USA300 [31, 32]. Similarly, USA300 was more virulent than USA400 in a rat model of necrotizing pneumonia. This increased virulence was associated with the upregulation of major virulence factors including PVL and α-toxins [36].
The most common molecular techniques used to identify MRSA strains and understand their spread and molecular evolution are PCR-based tests (SCCmec typing), pulsed-field gel electrophoresis (PFGE)-banding patterns and DNA-sequencing (MLST, spa typing and whole genome sequencing approaches [29, 35].
S. aureus phages
Similar to all bacterial viruses, S. aureus phages are either lysogenic or lytic based on their life cycles. A temperate phage integrates its genome into its host (known as a prophage) and can propagate along with its host until unsuitable conditions occur, such as application of mitomycin-C or ultraviolet rays. Conversely, a lytic phage never integrates its genome into the host genome; it replicates and synthesizes its components, assembles all the components, and exits by disrupting its host. With the advent of transmission electron microscopy and whole-genome sequencing, the classification of bacterial and archaeal viruses has evolved, and the International Committee on Taxonomy of Viruses has developed a better classification system for phages.
Staphylococcal phages were previously used only to type S. aureus strains (phage typing), and most literature related to these phages is based on the phage’s properties that render S. aureus pathogenic (that is, evolution as a pathogen). S. aureus phages were previously classified according to their reaction with a polyclonal antibody, and 11 serological groups were identified [18, 37]. Kwan et al. (2005) studied the genomes of 27 S. aureus phages and identified three classes. Most class I phages contain small genomes of ~ 20 kbp, with short noncontractile tails. Class II phages contain genomes of ~ 40 kbp with long noncontractile tails, and class III phages contain genomes of ~ 125 kbp with long contractile tails [38]. All S. aureus phages belong to the order Caudovirales [18, 37, 38]. S. aureus phages have linear double-stranded DNA, and, based on their tail morphology, can be grouped into three families: Myoviridae, with long contractile double sheath tails, Siphoviridae, with long noncontractile tails, and Podoviridae, with small contractile tails [18]. Most temperate phages of S. aureus belong to the Siphoviridae family and are considered less important for application-based purposes. S. aureus phages in the Myoviridae and Podoviridae families are mostly lytic. These families are important for downstream applications because a highly potent lytic phage is desirable as it subverts the essential metabolic pathways of the host [38]. An updated and comprehensive study on the diversity and evolution of 205 staphylococcal phages has been done by Oliveira et al. 2019; they grouped the staphylococcal phages into four major clusters (A-D clusters) and several sub-clusters and one singleton [39].
Myoviridae phages of S. aureus
Myoviridae phages are thought to be the most promising candidates for phage therapy because the phages of this group have broad host ranges (i.e., they are infective against several pathogenic S. aureus strains). Further, these phages are considered safe because most of them lack virulence- or resistance-associated genes [18, 38]. The Myoviridae family includes the historical phage, Twort, which was first discovered by Twort in 1915 [38]. Phage K is the best representative of Myoviridae. Phages K, G1, Twort Au2 and 812 are highly similar [38]. Phage K can infect both coagulase-negative and coagulase-positive staphylococci [40]. It has a large genome of 127,395 bp and contains introns, a notable feature in bacteriophages with large genomes [40]. Because they are safe and have a broad host range, these phages are present in many commercial phage preparations. Several commercial phages belong to Myoviridae, including Fersis, BFC-1 and Pyophage, which are made by the Eliava Institute, Tbilisi, Georgia, which is indicated for otolaryngological diseases and skin infections caused by staphylococci and streptococci. These commercial phage preparations are often cocktail of phages that act against several pathogens.
Siphoviridae phages of S. aureus
Staphylococcus phages in Siphoviridae display a temperate lifestyle and exist in the S. aureus genome as prophages. They are involved in the evolution of pathogenic S. aureus [41]. Goerke et al. (2009) designed a classification scheme for S. aureus Siphoviridae phages according to the integrase gene sequences. Most prophages are clustered under seven major groups: Sa1int to Sa7int. The most prevalent prophage belongs to Sa3int [41]. Sa3int prophages insert into the hlb gene; hence, they are called HIL-converting phages. Sa3int genome carries the immune evasion cluster (IEC) encoding the immunomodulatory proteins Sea, Sak, Scin and Chips, thereby helping S. aureus evade innate immunity and colonize the host tissue [41].
A few lytic Siphoviridae phages also exist; for example, phage SA97 is a Siphoviridae phage that carries a lysogeny module but lacks the important repressor gene, cI, which is involved in lysogenization. Thus, a Siphoviridae phage may also be considered in clinical applications [42].
Podoviridae phages of S. aureus
A few phages are known from this family, including 44AHJD, P-68, SAP-2, S24-1, S-13′, and phiAGO1.3. A recent study by Glowacka-Rutkowska et al. (2019) on phage phiAGO1.3 from the Podoviridae family demonstrated that phiAGO1.3 was not only active against most clinical S. aureus isolates, but it was related to the attenuation of bacterial virulence in the phiAGO1.3-infect cells that became resistant to this phage. The effect was associated with a truncated and inactive ArlS protein, which is part of the ArlRS two-component system [43]. The resistant bacteria could also become infected by another broad-range phage of the genus Kayvirus [43]. Moreover, phiAGO1.3, after being in equilibrium with its host (i.e., emergence of phage resistance), could still infect S. aureus in an in vivo model [43].
Phage receptors binding on S. aureus
Initially, phages interact with specific receptors located in the bacterial cell wall. Upon successful phage-receptor recognition device interaction, the phage can enter the bacterium for subsequent infection events. S. aureus has a thick peptidoglycan cell wall composed of repeating units of disaccharide N-acetyl-muramic acid (MurNAc) and N-acetyl-glucosamine (GlcNAc); the layers of these polymers are linked by tetrapeptides. The Gram-positive bacterial cell wall is interspersed with anionic polymers: wall teichoic acids (WTA) and lipoteichoic acid [44]. These polymers are also water-soluble, comprising glycerol or ribitol moieties [44]. The S. aureus cell wall is involved in cell division, biofilm formation, nasal colonization, and invasive infections [45]. It is also the primary site for phage attachment [44, 45]. Mutational studies on cell wall biosynthetic enzymes have revealed the actual site of phage attachment. WTA is the attachment site for staphylococcal phages [46]. The level of glycosylation also plays a crucial role in phage attachment; for example, TarM and TarS gene products play roles in WTA glycosylation. S. aureus lacking either α-o-GlcNAc or β-o-GlcNAc remains susceptible to the phages of serogroup B (i.e., staphylococcal phages of Siphoviridae), but because both types of WTA glycosylation are lacking, the phages are not adsorbed [46]. Staphylococcal phages of the family Podoviridae bind to β-o-GlcNAc of the WTA [47]. Myoviridae S. aureus phages utilizes backbone of WTA, but ɸSA039 (a member of the group) requires the β-GlcNAc residue [46].
Regarding phage receptor-binding proteins (RBPs), a detailed study was conducted on the Siphoviridae phage ϕ11 in which the ϕ11 phage baseplate protein Gp45, codified by the gp45 gene, was identified as the RBPs. The ϕ11 phage binds to GlcNAc on the cell wall [47]. Gp45 then forms a trimer; hence, six trimers assemble along the baseplate core and attach to the glycan moiety [47]. This assembly is conserved among most glycan-recognizing Siphoviridae phages [47]. Another notable example, the Twort-like ϕSA012 phage, has two RBPs that enable its wide host range [48], which are encoded by the open-reading frame (ORF)105 and ORF103. The ORFs products are bound to the WTA backbone and α-GlcNAc, respectively [48].
Disadvantages of whole bacteriophages as antimicrobial agents
The application of whole phages may have some disadvantages including a possible transfer of virulence- or resistance-associated genes via transduction. Most phage genomes are not fully annotated; for example, Cui et al. (2017) assessed the safety of the Myoviridae family, and ~ 70% of the ORFs had unknown functions that may be involved in disseminating resistance or virulence genes [49]. Thus, applications of whole phages should be avoided. Further, phages and bacteria are constantly evolving, and several historically important phages may become ineffective as the pathogenic bacteria evolve. For example, phage K is effective against many coagulase-positive and coagulase-negative staphylococci but ineffective against several MRSA strains [50]. Moreover, purification of the bacteriophages from lysate is a crucial step; if improperly purified, the chances of contamination with bacterial toxins can worsen an infection [51]. Another point of concern is that phages could be cleared from the bloodstream rapidly, since phage-neutralizing antibodies would hinder phage therapy [52]. However, some studies suggest that phages remain in circulation for a longer period of time [52]. Phage basically is a nucleoprotein particle and can induce immune responses. But in fact, phage induces anti-inflammatory effects that increase the capacity of the phage to suppress bacterial numbers and thus infection [53].
Phage-encoded products as antimicrobials
Phages with double-stranded DNA genomes encode special bacterial cell wall-degrading enzymes, which are endolysins and phage tail-associated murein lytic enzymes (TAMEs) or virion-associated peptidoglycan hydrolases (VAPGHs) [54, 55]. TAMEs or VAPGHs are part of the structural components of the phage tail; they hydrolyze the cell wall from the outside after being adsorbed by the host and slightly degrade the bacterial cell wall to access the inside of the cell [55]. Endolysins are phage-encoded peptidoglycan hydrolases that degrade the host at the end of the host’s life cycle. Holins are proteins that help endolysins access the bacterial cytoplasmic membrane [54, 56]. Endolysins and VAPGHs/TAMEs are useful in exogenous therapeutic applications.
Recombinant endolysins are more useful for exogenous application against Gram-positive bacteria than against Gram-negative bacteria because Gram-positive bacteria lack an outer protective layer, whereas Gram-negative bacteria have a thick extra outer membrane layer [56, 57]. Gram-negative bacterial phages have spanins that are essential to degrade both internal (IM) and outer (OT) membranes [54]. Phage endolysins of Gram-positive bacteria are reported to have modular organization, whereas those of Gram-negative bacteria have globular organization. Most bioengineering strategies exploit the modular organization [57]. Therefore, the prospects of creating various recombinant endolysins by shuffling these modules may lead to better antimicrobials. Endolysins have two important domains: The N-terminal domain (i.e., the catalytic domain) and the C-terminal domain (i.e., the cell wall-binding domain [CBD] with a linker region) [54, 58]. VAPGHs lack a CBD [55]. Recombinant catalytic domains are determined based on the cleavage site of the peptidoglycan [54, 56]. The most common catalytic domains of S. aureus phage endolysins are cysteine, histidine-dependent amidohydrolase peptidase (CHAP), and N-acetylmuramoyl-L-alanine amidase. SH3b domains is the common CBD [54, 56]. LysK, Twort, ϕH5, and ϕ11 are the best characterized S. aureus phage endolysins and all have CHAP-amidase-SH3b domains [59]. Recombinant endolysins have shown strong staphylolytic activity both in vitro and in vivo, and have biofilm-disrupting capabilities [56]. A report showed that a truncated version of endolysin was sufficient for staphylolytic activity; for example, Twort endolysin required the CHAP domain, although the presence of the SH3b domain was unnecessary but could enhance the overall activity [59]. Importantly, phage lysins are better than antibiotics in disrupting biofilms because some antibiotics can have limited penetration through biofilm, which can lead to bacterial adaptation resulting in increased bacterial survival and biofilm accumulation [60].
Phage proteins can become more effective when applied with other antimicrobial substances. For example, a study revealed that the combination of LysSA97 with carvacrol, a chemical component in the essential oils of oregano (Origanum vulgaris) and thyme (Thymus vulgaris), acted synergistically to inactivate S. aureus in food products for food preservation [61]. Refer to Table 1 for more recently studied phage encoded lytic proteins.
Table 1.
Examples of recombinant phage-encoded cell wall lytic enzymes (lysins) against S. aureus
| S. no | Recombinant lysin | Domains/modules | Remarks | Reference |
|---|---|---|---|---|
| 1 | LysKΔamidase | N-terminal 220 a.a., C-terminal 105 a.a | Kills several MRSA and MSSA and several coagulase-negative staphylococci | [62] |
| 2 | φH5 | CHAP, amidase and SH3b domains | Effective against S. aureus in pasteurized milk, important as food preservative | [63] |
| 3 | Twort (PlyTW) | CHAP (12–141 a.a.), amidase-2 (179–318 a.a.) and SH3b-5 (382–449 a.a.) domains | Effective against mastitis-causing S. aureus | [59] |
| 4 | ϕ11 | CHAP, amidase and SH3b domains | Acts in several S. aureus strains and coagulase-negative staphylococci. Effective in eradicating biofilm and useful in surface decontamination | [60] |
| 5 | LysSA97 | N-terminal CHAP, amidase-3 and C-terminal putative CBD | Effective for several S. aureus food strains. Acts against biofilm | [61] |
| 6 | CHAPk | Truncated version of LysK, having only a CHAP domain | Kills MRSA strains especially in mastitis, some coagulase-negative staphylococci. Acts as biofilm disrupters | [64] |
| 7 | Chimeric phage lysins | CBD LysK and lysostaphin CBD fused to catalytic λSA from streptococcal endolysin, that is λSA2-E-LysK-SH3b and λSA2-E-Lyso-SH3b, respectively | Effective against strain causing mastitis | [65] |
| 8 | Triple-acting lytic enzymes | Different constructs by shuffling follow domains from two enzymes, glycine-glycyl M3 endopeptidase from lysostaphin and a CHAP and amidase from LysK. SH3b CBD domains are present in both | Different constructs showed varying potentials as lytic enzymes and were effective even against intracellular S. aureus infections and biofilm | [58] |
| 9 | Protein 17 | Virion-associated component of P68 phage. N-terminus has catalytic activity; C-terminus has substrate recognition | First virion-associated component from Podoviridae, shows strong lytic ability against several clinical S. aureus strains | [66] |
| 10 | P128 (chimeric protein) | Phage K tail-associated region combined with SH3b domain of lysostaphin | Effective against several MRSA strains | [67] |
Apart from the cell wall lytic enzymes, phage genome encodes a plethora of proteins that can inhibit bacterial growth. For example, in a genomic study involving 26 S. aureus phages, 31 proteins were identified with the ability of inhibiting bacterial growth. The interaction of these proteins with their host targets may open prospects for new antibacterials inspired in these phage products [68].
Advantages of phage-encoded proteins over whole phages
The disadvantages related to applications of whole phages lead to exploring phage-encoded enzymes. These enzymes have several advantages; for example, lytic enzymes eliminate any chances of transferring resistance genes. Improperly purified phages may be toxic, but phage lytic proteins that are synthesized through recombinant DNA technologies are highly purified. Resistance to recombinant endolysins has not widely been reported [54, 56–58]. Moreover, recombinant multi-domain enzymes with different targets on the peptidoglycan reduce the chances of resistance development [58]. For example, triple-acting lytic enzymes are a better option because, first, it is effective against intracellular S. aureus infections, and second, the bacteria will have difficulty to develop resistance against the lytic domains simultaneously [58]. Further, these enzymes cause lowly immunogenic reactions [56, 58], and commonly display species-level specificity, which helps prevent selective pressure on the commensal bacteria [69]. Thus, these enzymes seem to be safer than are antibiotics for therapeutic practices.
Limitations of recombinant phage lysins
First, although there is no evidence of severe immunogenic response to phage lysins, it may evoke a mild immune response. Second, numerous labs report different lytic activity for the same protein depending on the enzyme assay used [59], but there should not be inconsistency.
Phages and their products currently in use and regulatory issues
Several phages are in clinical use but restricted to Eastern Europe. However, they are now slowly gaining recognition in the West. In the USA, the Food and Drug Administration has approved some phages. In food industries, these commercialized phage preparations usually combine two or more phages. For example, the combination of P68 (Podoviridae family) and K*710 (Myoviridae family), developed by Novolytics Ltd., UK, in its gel form for human applications [34]. It is effective against several MRSA strains as well as LA-MRSA [34]. Another important commercial phage is Stafal, manufactured by Bohemia Pharmaceuticals, Czech Republic. Stafal comprises a broad range of phages from the Myoviridae family, genus Kayvirus [70]. It is effective against several biofilm-producing MRSA and MSSA strains. However, despite its ability to eliminate sessile cells, tenfold greater concentration of the phage preparation was required to kill cells embedded in the biofilm matrix in comparison with the killing rate observed for platonic cells [70]. Multiple pathogenic bacteria may cause disease symptoms; therefore, phage preparations with phages targeting several pathogenic bacteria in a single preparation are commercially available through Brimrose Technology Corporation, Sparks Glencoe, MD, USA and Eliava Pharmacy, Tbilisi, GA, USA. Regarding phage-encoded products, a lysin of note is CF-301, which is directed against MRSA and has completed phase 1 human trials [71]. It is the first lysin to enter phase 2 clinical trials for treating patients with bacteremia or endocarditis [71]. Phages have been recognized as antimicrobials for over a century, but their applications in clinics are less accepted as legitimate treatments, primarily because of regulatory issues. These issues include whether the phages are regulated like antibiotic drugs, which is difficult because phages can replicate inside bacterial cells, although using phage-encoded products prevents such issues. Additionally, antimicrobial-producing companies have little interest in phages because of patent issues. However, in some areas, they are now being considered alternatives for treating untreatable infections. For example, in the USA, a provision called the Compassionate Use Program allows unauthorized medicine when all other treatment options fail, and the use of phage-based therapies have been recommended for those patients facing antimicrobial failures [72]. Moreover, in most compassionate cases, the synergistic application of phages and antibiotics is encouraged, and the sequence of their administration also matters. For example, to combat S. aureus infections, the best results are obtained when phage therapy precedes antibiotic therapy [72]. Some diseases, such as osteoarticular infections, are deep-seated and difficult to treat with antibiotics. In these cases, phage therapy may be the best option [72].
Concluding remarks
Antibiotics are still considered effective against some MRSA-related infections, and vancomycin and daptomycin are among the first chosen antibiotics. However, with the emergence of VISA and VRSA, alternative antimicrobials are urgently needed. The irrational use of antibiotics has affected not only humans but also the environment as we constantly interact with our surroundings. Phages serve as natural tools to lessen the deleterious impact of antibiotics and make up for the paucity of new effective antibiotics. Unlike antibiotics, phages are narrow and specific in action, are self-replicative, and are abundant in nature; hence, they can be easily isolated as and when required. However, care must be taken when using phages; for example, the preparation must be ultrapure, and the phages must contain no resistance/virulence genes or integrase genes because these genes could be transferred to commensal bacteria through transduction. Additionally, phage-encoded products are attractive tools for fighting and treating pathogenic S. aureus. A famous English proverb states that “An enemy’s enemy is a friend”. This is apt for bacterial viruses that can serve as weapons against pathogenic bacteria. The prospects for phages and phage-encoded products are very encouraging in using phages as therapeutics with respect to S. aureus-related infections.
Acknowledgements
We are thankful to Environmental Virology Cell, CSIR-NEERI, Nagpur and Director CSIR-NEERI for providing the research facility and infrastructure. Financial assistance to Akanksha Rai in form of fellowship from the University Grants Commission (UGC), India is gratefully acknowledged. We thank Traci Raley, MS, ELS, for editing a draft of this manuscript.
Author contribution
Conceptualization: Krishna Khairnar and Akanksha Rai; Writing and original draft preparation: Akanksha Rai; Writing, reviewing, and editing: Akanksha Rai and Krishna Khairnar.
Funding
Financial assistance as fellowship from the University Grants Commission (UGC), India.
Declarations
Ethics approval
Not applicable.
Conflict of interest
The authors declare no competing interests.
Footnotes
Responsible Editor: Agnes M.S. Figueiredo
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Williams REO. Healthy carriage of Staphylococcus aureus: its prevalence and importance. Bacteriol Rev. 1963;27(1):56. doi: 10.1128/br.27.1.56-71.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wertheim HF, Melles DC, Vos MC, Van Leeuwen W, Van Belkum A, Verbrugh HA, Nouwen JL. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5(12):751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 4.Taylor TA, Unakal CG (2020) Staphylococcus aureus. [Updated 2020 Aug 23]. In: StatPearls. StatPearls Publishing, Treasure Island (FL). Available from: https://www.ncbi.nlm.nih.gov/books/NBK441868/
- 5.Foster T. Staphylococcus. In: Baron S, editor. Medical Microbiology. 4. Galveston: The University of Texas Medical Branch; 1996. pp. 187–197. [Google Scholar]
- 6.Wilde AD, Snyder DJ, Putnam NE, Valentino MD, Hammer ND, Lonergan ZR, et al. Bacterial hypoxic responses revealed as critical determinants of the host-pathogen outcome by TnSeq analysis of Staphylococcus aureus invasive infection. PLoS Pathog. 2015;11(12):e1005341. doi: 10.1371/journal.ppat.1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGuinness WA, Malachowa N, DeLeo FR. Focus: infectious diseases: vancomycin-resistance in Staphylococcus aureus. Yale J Biol Med. 2017;90(2):269–281. [PMC free article] [PubMed] [Google Scholar]
- 8.WHO (2020) Global action plan on AMR https://www.who.int/antimicrobial-resistance/global-action-plan/en/ accessed 29 January 2020
- 9.Úbeda C, Maiques E, Knecht E, Lasa Í, Novick RP, Penadés JR. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol Microbiol. 2005;56(3):836–844. doi: 10.1111/j.1365-2958.2005.04584.x. [DOI] [PubMed] [Google Scholar]
- 10.Singh R, Sripada L, Singh R. Side effects of antibiotics during bacterial infection: mitochondria, the main target in host cell. Mitochondrion. 2014;16:50–54. doi: 10.1016/j.mito.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Keeney KM, Yurist-Doutsch S, Arrieta MC, Finlay BB. Effects of antibiotics on human microbiota and subsequent disease. Annu Rev Microbiol. 2014;68:217–235. doi: 10.1146/annurev-micro-091313-103456. [DOI] [PubMed] [Google Scholar]
- 12.Pantosti A, Sanchini A, Monaco M. Mechanisms of antibiotic resistance in Staphylococcus aureus. Future Microbiol. 2007;2:323–334. doi: 10.2217/17460913.2.3.323. [DOI] [PubMed] [Google Scholar]
- 13.CDC (2019) Vital signs www.cdc.gov/vitalsigns/staph/index.html accessed on 05 April 2021
- 14.Diekema DJ, Pfaller MA, Shortridge D, Zervos M, Jones RN. Twenty-year trends in antimicrobial susceptibilities among Staphylococcus aureus from the SENTRY Antimicrobial Surveillance Program. Open Forum Infect Dis. 2019;6(Suppl 1):S47–S53. doi: 10.1093/ofid/ofy270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance accessed on 05 April 2021
- 16.Furfaro LL, Payne MS, Chang BJ. Bacteriophage therapy: clinical trials and regulatory hurdles. Front Cell Infect Microbiol. 2018;8:376. doi: 10.3389/fcimb.2018.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merril CR, Scholl D, Adhya SL. The prospect for bacteriophage therapy in Western medicine. Nat Rev Drug Discov. 2003;2(6):489–497. doi: 10.1038/nrd1111. [DOI] [PubMed] [Google Scholar]
- 18.Deghorain M, Van Melderen L. The Staphylococci phages family: an overview. Viruses. 2012;4(12):3316–3335. doi: 10.3390/v4123316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyon BR, Skurray RON. Antimicrobial resistance of Staphylococcus aureus: genetic basis. Microbiol Rev. 1987;51(1):88–134. doi: 10.1128/mr.51.1.88-134.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morikawa K, Takemura AJ, Inose Y, Tsai M, Ohta T, Msadek T. Expression of a cryptic secondary sigma factor gene unveils natural competence for DNA transformation in Staphylococcus aureus. PLoS Pathog. 2012;8(11):e1003003. doi: 10.1371/journal.ppat.1003003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everitt RG, Didelot X, Batty EM, Miller RR, Knox K, Young BC, Bowden R, Auton A, Votintseva A, Larner-Svensson H, Charlesworth J. Mobile elements drive recombination hotspots in the core genome of Staphylococcus aureus. Nat Commun. 2014;5(1):1–9. doi: 10.1038/ncomms4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alibayov B, Zdenkova K, Sykorova H, Demnerova K. Molecular analysis of Staphylococcus aureus pathogenicity islands (SaPI) and their superantigens combination of food samples. J Microbiol Methods. 2014;107:197–204. doi: 10.1016/j.mimet.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Grundstad ML, Parlet CP, Kwiecinski JM, Kavanaugh JS, Crosby HA, Cho YS, Heilmann K, Diekema DJ, Horswill AR. Quorum sensing, virulence, and antibiotic resistance of USA100 methicillin-resistant Staphylococcus aureus isolates. mSphere. 2019;4(4):e00553–19. doi: 10.1128/mSphere.00553-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarraud S, Lyon GJ, Figueiredo AMS, Lina G, Vandenesch F, Etienne J, Muir TW, Novick RP. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcusaureus. J Bacteriol. 2011;193(24):7027. doi: 10.1128/JB.06355-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung AL, Nishina KA, Trotonda MP, Tamber S. The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Biol. 2008;40(3):355–361. doi: 10.1016/j.biocel.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pichon C, Felden B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. PNAS. 2005;102(40):14249–14254. doi: 10.1073/pnas.0503838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desgranges E, Marzi K, Moreau K, Romby P, Caldelari I. Chapter 35 Noncoding RNA. In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Braunstein M, Rood JI, editors. Gram-positive Pathogens. 3. New York: Wiley; 2019. [Google Scholar]
- 28.Haaber J, Penadés JR, Ingmer H. Transfer of antibiotic resistance in Staphylococcus aureus. Trends Microbiol. 2017;25(11):893–905. doi: 10.1016/j.tim.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 29.Deurenberg RH, Stobberingh EE. The evolution of Staphylococcus aureus. Infect Genet Evol. 2008;8(6):747–763. doi: 10.1016/j.meegid.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 30.Baig S, Johannesen TB, Overballe-Petersen S, Larsen J, Larsen AR, Stegger M. Novel SCCmec type XIII (9A) identified in an ST152 methicillin-resistant Staphylococcus aureus. Infect Genet Evol. 2018;61:74–76. doi: 10.1016/j.meegid.2018.03.013. [DOI] [PubMed] [Google Scholar]
- 31.David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23(3):616–687. doi: 10.1128/cmr.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boucher HW, Corey GR. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2008;46(Supplement 5):S344–S349. doi: 10.1086/533590. [DOI] [PubMed] [Google Scholar]
- 33.Pantosti A. Methicillin-resistant Staphylococcus aureus associated with animals and its relevance to human health. Front Microbiol. 2012;3:127. doi: 10.3389/fmicb.2012.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verstappen KM, Tulinski P, Duim B, Fluit AC, Carney J, Van Nes A, Wagenaar JA. The effectiveness of bacteriophages against methicillin-resistant Staphylococcus aureus ST398 nasal colonization in pigs. PLoS ONE. 2016;11(8):e0160242. doi: 10.1371/journal.pone.0160242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Figueiredo AMS, Ferreira FA. The multifaceted resources and microevolution of the successful human and animal pathogen methicillin-resistant Staphylococcus aureus. Mem Inst Oswaldo Cruz. 2014;109(3):265–278. doi: 10.1590/0074-0276140016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montgomery CP, Boyle-Vavra S, Adem PV, Lee JC, Husain AN, Clasen J, Daum RS. Comparison of virulence in community-associated methicillin-resistant Staphylococcus aureus pulsotypes USA300 and USA400 in a rat model of pneumonia. J Infect Dis. 2008;198(4):561–570. doi: 10.1086/590157. [DOI] [PubMed] [Google Scholar]
- 37.Xia G, Wolz C. Phages of Staphylococcus aureus and their impact on host evolution. Infect Genet Evol. 2014;21:593–601. doi: 10.1016/j.meegid.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 38.Kwan T, Liu J, DuBow M, Gros P, Pelletier J. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci USA. 2005;102(14):5174–5179. doi: 10.1073/pnas.0501140102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliveira H, Sampaio M, Melo LD, Dias O, Pope WH, Hatfull GF, Azeredo J. Staphylococci phages display vast genomic diversity and evolutionary relationships. BMC Genomics. 2019;20(1):357. doi: 10.1186/s12864-019-5647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Flaherty S, Coffey A, Edwards R, Meaney W, Fitzgerald GF, Ross RP. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting Gram-positive bacteria with a low G+ C content. J Bacteriol. 2004;186(9):2862–2871. doi: 10.1128/JB.186.9.2862-2871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goerke C, Pantucek R, Holtfreter S, Schulte B, Zink M, Grumann D, Bröker BM, Doskar J, Wolz C. Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J Bacteriol. 2009;191(11):3462–3468. doi: 10.1128/jb.01804-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang Y, Shin H, Lee JH, Park CJ, Paik SY, Ryu S. Isolation and genome characterization of the virulent Staphylococcus aureus bacteriophage SA97. Viruses. 2015;7(10):5225–5242. doi: 10.3390/v7102870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Głowacka-Rutkowska A, Gozdek A, Empel J, Gawor J, Żuchniewicz K, Kozińska A, Dębski J, Gromadka R, Łobocka M. The ability of lytic staphylococcal podovirus vB_SauP_phiAGO1. 3 to coexist in equilibrium with its host facilitates the selection of host mutants of attenuated virulence but does not preclude the phage antistaphylococcal activity in a nematode infection model. Front Microbiol. 2019;9:3227. doi: 10.3389/fmicb.2018.03227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rakhuba DV, Kolomiets EI, Dey ES, Novik GI. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol J Microbiol. 2010;59(3):145–155. doi: 10.33073/pjm-2010-023. [DOI] [PubMed] [Google Scholar]
- 45.Winstel V, Xia G, Peschel A. Pathways and roles of wall teichoic acid glycosylation in Staphylococcus aureus. Int J Med Microbiol. 2014;304(3–4):215–221. doi: 10.1016/j.ijmm.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 46.Azam AH, Tanji Y. Peculiarities of Staphylococcus aureus phages and their possible application in phage therapy. Appl Microbiol Biotechnol. 2019;103:4279–4289. doi: 10.1007/s00253-019-09810-2. [DOI] [PubMed] [Google Scholar]
- 47.Koç C, Xia G, Kühner P, Spinelli S, Roussel A, Cambillau C, Stehle T. Structure of the host-recognition device of Staphylococcus aureus phage ϕ11. Sci Rep. 2016;6(1):1–11. doi: 10.1038/srep27581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takeuchi I, Osada K, Azam AH, Asakawa H, Miyanaga K, Tanji Y. The presence of two receptor-binding proteins contributes to the wide host range of staphylococcal Twort-like phages. Appl Environ Microbiol. 2016;82(19):5763–5774. doi: 10.1128/AEM.01385-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cui Z, Guo X, Dong K, Zhang Y, Li Q, Zhu Y, Zeng L, Tang R, Li L. Safety assessment of Staphylococcus phages of the family Myoviridae based on complete genome sequences. Sci Rep. 2017;7(1):1–8. doi: 10.1038/srep41259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ajuebor J, Buttimer C, Arroyo-Moreno S, Chanishvili N, Gabriel EM, O’Mahony J, McAuliffe O, Neve H, Franz C, Coffey A. Comparison of Staphylococcus phage K with close phage relatives commonly employed in phage therapeutics. Antibiotics. 2018;7(2):37. doi: 10.3390/antibiotics7020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merabishvili M, Pirnay JP, Verbeken G, Chanishvili N, Tediashvili M, Lashkhi N, Glonti T, Krylov V, Mast J, Van Parys L, Lavigne R. Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS ONE. 2009;4(3):e4944. doi: 10.1371/journal.pone.0004944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sulakvelidze A, Kutter E (2005) Bacteriophage therapy. In: Kutter E, Sulakvelidze A (eds) Bacteriophage biology and applications, 1st edn. CRC Press, Boca Raton, p 381–436
- 53.Van Belleghem JD, Clement F, Merabishvili M, Lavigne R, Vaneechoutte M. Pro- and anti-inflammatory responses of peripheral blood mononuclear cells induced by Staphylococcus aureus and Pseudomonas aeruginosa phages. Sci Rep. 2017;7(1):8004. doi: 10.1038/s41598-017-08336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gutiérrez D, Fernández L, Rodríguez A, García P. Are phage lytic proteins the secret weapon to kill Staphylococcus aureus? MBio. 2018;9(1):e01923–e2017. doi: 10.1128/mBio.01923-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adhya S, Merril CR, Biswas B. Therapeutic and prophylactic applications of bacteriophage components in modern medicine. Cold Spring Harb Perspect Biol. 2014;4(1):a012518. doi: 10.1101/cshperspect.a012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmelcher M, Shen Y, Nelson DC, Eugster MR, Eichenseher F, Hanke DC, Loessner MJ, Dong S, Pritchard DG, Lee JC, Becker SC. Evolutionarily distinct bacteriophage endolysins featuring conserved peptidoglycan cleavage sites protect mice from MRSA infection. J Antimicrob Chemother. 2015;70(5):1453–1465. doi: 10.1093/jac/dku552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Love MJ, Bhandari D, Dobson RC, Billington C. Potential for bacteriophage endolysins to supplement or replace antibiotics in food production and clinical care. Antibiotics. 2018;7(1):17. doi: 10.3390/antibiotics7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Becker SC, Roach DR, Chauhan VS, Shen Y, Foster-Frey J, Powell AM, Bauchan G, Lease RA, Mohammadi H, Harty WJ, Simmons C. Triple-acting lytic enzyme treatment of drug-resistant and intracellular Staphylococcus aureus. Sci Rep. 2016;6(1):1–10. doi: 10.1038/srep25063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Becker SC, Swift S, Korobova O, Schischkova N, Kopylov P, Donovan DM, Abaev I. Lytic activity of the staphylolytic Twort phage endolysin CHAP domain is enhanced by the SH3b cell wall binding domain. FEMS Microbiol Lett. 2015;362(1):1–8. doi: 10.1093/femsle/fnu019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sass P, Bierbaum G. Lytic activity of recombinant bacteriophage φ11 and φ12 endolysins on whole cells and biofilms of Staphylococcus aureus. Appl Environ Microbiol. 2007;73(1):347–352. doi: 10.1128/AEM.01616-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang Y, Yoon H, Kang DH, Chang PS, Ryu S. Endolysin LysSA97 is synergistic with carvacrol in controlling Staphylococcus aureus in foods. Int J Food Microbiol. 2017;244:19–26. doi: 10.1016/j.ijfoodmicro.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 62.Zhou Y, Zhang H, Bao H, Wang X, Wang R. The lytic activity of recombinant phage lysin LysKΔamidase against staphylococcal strains associated with bovine and human infections in the Jiangsu province of China. Res Vet Sci. 2017;111:113–119. doi: 10.1016/j.rvsc.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 63.Obeso JM, Martínez B, Rodríguez A, García P. Lytic activity of the recombinant staphylococcal bacteriophage ΦH5 endolysin active against Staphylococcus aureus in milk. Int J Food Microbiol. 2008;128(2):212–218. doi: 10.1016/j.ijfoodmicro.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 64.Fenton M, Keary R, McAuliffe O, Ross RP, O’Mahony J, Coffey A. Bacteriophage-derived peptidase CHAP (K) eliminates and prevents Staphylococcal biofilms. Int J Microbiol. 2013;2013:625341. doi: 10.1155/2013/625341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmelcher M, Powell AM, Becker SC, Camp MJ, Donovan DM. Chimeric phage lysins act synergistically with lysostaphin to kill mastitis-causing Staphylococcus aureus in murine mammary glands. Appl Environ Microbiol. 2012;78(7):2297–2305. doi: 10.1128/aem.07050-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takáč M, Bläsi U. Phage P68 virion-associated protein 17 displays activity against clinical isolates of Staphylococcus aureus. Antimicrob Agents Chemother. 2005;49(7):2934–2940. doi: 10.1128/AAC.49.7.2934-2940.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paul VD, Rajagopalan SS, Sundarrajan S, George SE, Asrani JY, Pillai R, Chikkamadaiah R, Durgaiah M, Sriram B, Padmanabhan S. A novel bacteriophage Tail-Associated Muralytic Enzyme (TAME) from Phage K and its development into a potent antistaphylococcal protein. BMC Microbiol. 2011;11(1):1–11. doi: 10.1186/1471-2180-11-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu J, Dehbi M, Moeck G, Arhin F, Bauda P, Bergeron D, Callejo M, Ferretti V, Ha N, Kwan T, et al. Antimicrobial drug discovery through bacteriophage genomics. Nat Biotechnol. 2004;22:185–191. doi: 10.1038/nbt932. [DOI] [PubMed] [Google Scholar]
- 69.Kashani HH, Schmelcher M, Sabzalipoor H, Hosseini ES, Moniri R. Recombinant endolysins as potential therapeutics against antibiotic-resistant Staphylococcus aureus: current status of research and novel delivery strategies. Clin Microbiol Rev. 2018;31(1):e00071–17. doi: 10.1128/CMR.00071-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dvořáčková M, Růžička F, Benešík M, Pantůček R, Dvořáková-Heroldová M. Antimicrobial effect of commercial phage preparation Stafal® on biofilm and planktonic forms of methicillin-resistant Staphylococcus aureus. Folia Microbiol. 2019;64(1):121–126. doi: 10.1007/s12223-018-0622-3. [DOI] [PubMed] [Google Scholar]
- 71.Fischetti VA. Development of phage lysins as novel therapeutics: a historical perspective. Viruses. 2018;10(6):310. doi: 10.3390/v10060310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patey O, McCallin S, Mazure H, Liddle M, Smithyman A, Dublanchet A. Clinical indications and compassionate use of phage therapy: personal experience and literature review with a focus on osteoarticular infections. Viruses. 2019;11(1):18. doi: 10.3390/v11010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wittebole X, De Roock S, Opal SM. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence. 2014;5(1):226–235. doi: 10.4161/viru.25991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tan SY, Tatsumura Y. Alexander Fleming (1881–1955): discoverer of penicillin. Singapore Med J. 2015;56(7):366–367. doi: 10.11622/smedj.2015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Investig. 2003;111(9):1265–1273. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abdelkader K, Gerstmans H, Saafan A, Dishisha T, Briers Y. The preclinical and clinical progress of bacteriophages and their lytic enzymes: the parts are easier than the whole. Viruses. 2019;11(2):96. doi: 10.3390/v11020096. [DOI] [PMC free article] [PubMed] [Google Scholar]