Abstract
Myc and Mad are basic helix-loop-helix leucine zipper (bHLH-LZ) proteins that heterodimerize with Max to bind DNA and thereby influence the transcription of Myc-responsive genes. Myc-Max dimers transactivate whereas Mad-Max-mSin3 complexes repress Myc-mediated transcriptional activation. We have previously shown that the N-terminal mSin3 binding domain and the centrally located bHLH-LZ are required for Mad1 to function during a molecular switch from proliferation to differentiation. Here we demonstrate that the carboxy terminus (CT) of Mad1 contains previously unidentified motifs necessary for the regulation of Mad1 function. We show that removal of the last 18 amino acids of Mad1 (region V) abolishes the growth-inhibitory function of the protein and the ability to reverse a Myc-imposed differentiation block. Moreover, deletion of region V results in a protein that binds DNA weakly and no longer represses Myc-dependent transcriptional activation. In contrast, deletion of the preceding 24 amino acids (region IV) together with region V restores DNA binding and transcriptional repression, suggesting a functional interplay between these two regions. Furthermore, phosphorylation within region IV appears to mediate this interplay. These findings indicate that novel regulatory elements are present in the Mad1 CT.
The elucidation of molecular switches involved in deciding basic cell fate is critical to the understanding of normal development and cancer. Although c-Myc has long been implicated as a principal component in such decisions, only recently has the significance of the role of Mad1 in the switch from proliferation to differentiation been recognized (4, 15, 23). Myc and Mad belong to a superfamily of interacting proteins that regulate cell growth and differentiation. Myc, Max, Mad1, Mxi1 (Mad2), Mad3, Mad4, and Mnt are members of this family; all contain a motif consisting of a basic DNA binding domain (BR), a helix-loop-helix (HLH), and a leucine zipper (LZ) (bHLH-LZ motif) (28). In addition to the centrally located bHLH-LZ, Mad1, Mxi1, Mad3, Mad4, and Mnt contain an N-terminal mSin3 binding domain (SID) (31, 32, 46, 54). Mad family proteins (Mad1, Mxi1, Mad3, and Mad4) also possess a moderately conserved carboxy terminus (CT) (31). For Myc and Mad to function in vivo, they must heterodimerize with Max, bind DNA in a sequence-specific manner, and influence the transcription of Myc-responsive genes (2, 3, 5, 8, 14, 25, 47, 48, 51, 58). Max is capable of forming homodimers, but heterodimerization with c-Myc, Mad, or Mxi1 is favored (4, 43, 58). Myc-Max heterodimers bind to E-box DNA elements and activate transcription (3, 36, 38), whereas Mad-Max heterodimers antagonize Myc function by a mechanism which includes the recruitment of corepressor proteins such as mSin3, N-CoR, SMRT, and histone deacetylase to these sites (1, 27, 39).
Typically, the expression of Mad1 is up-regulated during terminal differentiation, often as an immediate-early response to differentiation-induced stimuli. In parallel, the expression of c-Myc is down-regulated, and a switch from proliferation to differentiation occurs (4, 16, 18, 30, 41, 44). Murine erythroleukemia (MEL) cells induced to differentiate with N′N′-hexamethylene bisacetamide (HMBA) growth arrest in G1 and withdraw from the cell cycle. This process includes the sequestration of the transcription factor E2F and the subsequent down-regulation of downstream proteins involved in cell cycle progression, such as c-Myc, c-Myb, DNA polymerase α, cdk4, thymidylate synthase, and dihydrofolate reductase (45). Overexpression of a transfected c-myc gene in MEL cells blocks inducer-mediated differentiation by a mechanism that prevents G0/G1 arrest and exit from the cell cycle (22, 24, 34, 49). Conversely, ectopic expression of Mad1 arrests cells in G1 and provides a mechanism of exit from the cell cycle during the induction of the differentiation process (12, 52). The opposing effects exerted by c-Myc and Mad on cell cycle progression and their reciprocal regulation during cell growth and differentiation in hematopoietic cells suggest that c-Myc expression is essential for cell growth, while up-regulation of Mad1 is associated with cellular differentiation.
Previously we have shown that the SID and bHLH-LZ domains are essential for Mad1 to function during a transition from proliferation to differentiation (15). Likewise, the SID, BR, and LZ domains are required for the inhibition of Myc-Ras cotransformation (11, 37, 40). The function of the Mad1 CT, however, has not yet been clearly defined. In fact, conflicting reports regarding the function of this region exist in the literature. While one group has reported that deletion of the Mad1 CT domain is dispensable for the inhibition of Myc-Ras cotransformation (11), another group has shown that removal of the CT results in a partial reduction in transcriptional repression by Mad1 (37). Moreover, cotransfection of MEL cells with Myc and Mad1 lacking a CT domain gave rise to transfectants that lost both transgenes shortly after transfection (15), precluding us from studying the function of the CT within the context of cellular differentiation.
In this study, we sequentially deleted regions within the Mad1 CT to closely examine whether the CT of the protein plays a role in the regulation of Mad1 function. We demonstrate that deletion of the last 18 amino acids (aa) of the protein (region V) abolishes growth inhibition by Mad1, the ability to reverse a Myc-imposed differentiation block, and competence to inhibit Myc-Ras cotransformation, while deletion of region V and adjacent region IV restores Mad1 function. Furthermore, we also show that phosphorylation of the CT domain, specifically at the two putative casein kinase II (CKII) sites in region IV, impairs Mad1 DNA binding and transcriptional repression. Taken together, our results indicate that the CT domain contains novel positive and negative regulatory elements that modulate Mad1 function.
MATERIALS AND METHODS
Mutagenesis and plasmid construction.
pL1hmcneor contains an XhoI-EcoRI genomic fragment of human c-myc under the transcriptional control of the Moloney murine leukemia virus long terminal repeat (19). The full-length mad1 coding region was amplified and cloned into pMTH-β-globin neo to generate the pMTHmad1 expression vector as described previously (15). pMTHmad1 was used as a template for PCR-directed mutagenesis to generate the ΔCT, ΔII-V, ΔIII-V, ΔIV-V, ΔIV, and ΔV mutants. PCR amplification was done with primers that deleted the nucleotides encoding amino acids 156 to 221 (ΔCT), 163 to 221 (ΔII-V), 168 to 221 (ΔIII-V), 180 to 221 (ΔIV-V), 180 to 203 (ΔIV), and 204 to 221 (ΔV). The mutated mad1 cDNAs were cloned into pMTH-β-globin neo and/or pRC/CMV expression vectors. The two-step thermal cycled fusion method (35) was used to generate the human mad1 point mutations. Nucleotides encoding serines 182, 184, 205, and 214 were mutated to encode alanines (Ser-Ala). The individual expression constructs with the following combinations of Ser-Ala point mutations were cloned into the pRC/CMV vector: 182+184, 205, 214, 205+214, 182+184+205, 182+184+214, 182+184+205+214, and ΔV 182+184 (numbers refer to amino acid positions).
Transfection and cell culture.
C19 MEL cells (20) and COS-7 cells were grown in RPMI medium and Dulbecco modified Eagle medium (DMEM), respectively, containing 10% heat-inactivated fetal bovine serum (FBS) (Gibco), 100 U of penicillin, and 100 μg of streptomycin (Gibco) per ml. The C19 cell line was transfected by the Lipofectin (Gibco) method (6) with either wild-type or mutant pMTH Mad1 expression vector alone or cotransfected with pL1hmcneor. To select for individual clones, transfected cells (107) were transferred to 24-well plates at a density of 105 cells/ml. Growth medium was supplemented with Geneticin (G418; Gibco) at 600 μg/ml for selection and 200 μg/ml for maintenance. For specific experiments, expression of the transfected mad1 gene was induced by supplementing the medium with ZnCl2 (Sigma). COS-7 cells were seeded at a density of 5 × 105 cells in 60-mm dishes and cultured for 24 h prior to transfection (Lipofectin). Cells were transfected with 1 μg of pMyc3E1bLuc (a luciferase reporter plasmid containing three E-box regulatory elements) (26) together with 0.5 μg of pCH110 (a β-galactosidase expression plasmid) (Pharmacia) and 0.15 μg of pRC/CMV expression vector for wild-type Mad1 (wtMad1), ΔCT, ΔII-V, ΔIII-V, ΔIV-V, ΔIV, the Ser-Ala point mutant wtMad1(Ser182/184 to Ala182/184), or ΔV(Ser182/184 to Ala182/184) or empty vector. Harvesting of cells and determination of luciferase activity were performed with a Promega Luciferase Assay System kit according to the manufacturer's instructions. Luciferase activity was measured with a Monolight 2010 (Analytical Luminescence Laboratory) luminometer, normalized according to both protein levels and β-galactosidase activities, and expressed relative to the activity obtained with pMyc0E1bLuc (a luciferase reporter plasmid lacking E-box regulatory elements) (26).
RT-PCR analyses.
C19 MEL cells (1.5 × 107) were transfected as described above with 15 μg of plasmid DNA (pMTH) and plated in multiwell plates at a density of 5 × 105 cells/well (30 wells/transfection). Once G418-resistant cells emerged, clones were expanded for further analysis. Total cellular RNA was isolated with a PUREgene RNA isolation kit (GENTRA Systems, Inc., Minneapolis, Minn.) from 106 cells grown for 48 h in 100 μM ZnCl2 and analyzed for the expression of the transgene with a Perkin-Elmer RNA PCR kit and a modified procedure (29). Briefly, total cellular RNA was DNase treated to remove residual genomic DNA. DNase was then heat inactivated at 75°C, and 250 ng of RNA was reverse transcribed with Moloney murine leukemia virus reverse transcriptase (RT) and oligo(dT) primers (Perkin-Elmer). An aliquot of cDNA corresponding to 62.5 ng of total RNA was then PCR amplified (Amplitaq; Perkin-Elmer) with human Mad1 expression vector-specific primers and analyzed by agarose gel electrophoresis.
Differentiation assay.
The differentiation assay was performed as previously described (15). Briefly, cells were seeded at a density of 105 cells/ml and incubated for 24 h before the addition of ZnCl2 to the culture medium at a final concentration of 75 μM. HMBA (Sigma; final concentration, 3.5 mM) was added 24 h later. Cells were maintained in logarithmic phase daily by 1:1 dilution with fresh medium for up to 6 days. At specific times, cells were removed, hemoglobin was stained with acid-benzidine (Sigma), and the number of benzidine-positive cells was counted with a phase-contrast microscope.
REF transformation assay.
Rat embryo fibroblast (REF) cells were prepared from 13-day-old Fischer rat embryos by The Frederick Cancer Research and Development Center (Frederick, Md.) and grown in DMEM supplemented with 10% FBS (Gibco). After one or two passages, the cells were plated at a low density (2 × 105) and transfected by the calcium phosphate precipitation technique 24 h later. Transfection mixtures include 2 to 3 μg of pL1hmcneor (15), 2 μg of pEJras (activated c-Ha-rasVal12; American Type Culture Collection, Manassas, Va.), and 3.5 μg of pRC/CMV expression vectors for wtMad1 or the corresponding mutants or empty vector (control). The DNA concentration in each transformation mixture was adjusted to 12 μg with the pRC/CMV empty vector. Transfected cells were fed every 4 days with fresh DMEM–4% FBS, and transformed foci were scored after 15 days.
Protein isolation and immunoblotting.
Cells were sonicated in immunoprecipitation buffer (50 mM Tris [pH 8.0], 5 mM EDTA, 150 mM NaCl, 0.5% NP-40) containing 10 μg of aprotinin (Sigma) per ml, 0.1 mg of phenylmethylsulfonyl fluoride (Sigma) per ml, 50 mM sodium fluoride (Sigma), 100 mM sodium orthovanadate (Sigma), 1 mM dithiothreitol (Sigma), 0.2 mM okadaic acid (Gibco), and 1/10 volume of Complete Mini EDTA-free protease inhibitor cocktail (Boehringer Mannheim). Aliquots (100 to 150 μg) were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and proteins were electrotransferred to nitrocellulose membranes. Protein blots were immunoreacted with either c-Myc antibody (polyclonal antibody 06-303; Upstate Biotechnology, Inc.) or Mad1 antibody (polyclonal antibody sc-766; Santa Cruz), followed by horseradish peroxidase-linked secondary antibody (Amersham). Immunoreactive bands were detected by enhanced chemiluminescence (Amersham).
Coimmunoprecipitations.
The pRC/CMV expression vectors for wtMad1 and the different Mad1 CT deletion mutants or a pGEM 7 expression plasmid encoding mouse Max (p22 long form) was used in a TNT Coupled Reticulocyte Lysate System (Promega) according to the manufacturer's instructions to in vitro translate the proteins used in the coimmunoprecipitation experiments. Eight microliters of 35S-labeled in vitro-translated wtMad1 or of the different Mad1 CT deletion mutants was incubated with 1 μl of unlabeled in vitro-translated Max for 10 min at 42°C and an additional 20 min at room temperature to promote Mad-Max dimer formation. The lysates were incubated for 1 h at 4°C with protein A-agarose, and proteins binding nonspecifically to protein A were removed by centrifugation. The supernatants were incubated with 1 μg of anti-Max antiserum (polyclonal antiserum 06-525; Upstate Biotechnology) for 3 h on ice, followed by overnight incubation at 4°C with protein A-agarose. The resulting immune complexes were pelleted by brief centrifugation. The pellets were washed three times with phosphate-buffered saline–0.1% NP-40 containing 10 μg of aprotinin per ml, 0.1 mg of phenylmethylsulfonyl fluoride per ml, 50 mM sodium fluoride, 100 mM sodium orthovanadate, 1 mM dithiothreitol, 0.2 mM okadaic acid, and 1/10 volume of Complete Mini EDTA-free protease inhibitor cocktail. The immunoreacted complexes were collected by centrifugation, resuspended in SDS loading buffer, boiled for 5 min, and subjected to SDS–12.5% PAGE. The 35S-labeled Mad1 proteins binding to Max were detected by autoradiography.
EMSA.
For the electrophoretic mobility shift assay (EMSA), 8 μl of in vitro-translated wtMad1 or of the different Mad1 CT deletion mutants was preincubated with 1 μl of in vitro-translated Max for 10 min at 42°C and an additional 20 min at room temperature to promote Mad-Max dimer formation. These conditions were chosen to favor Mad-Max heterodimerization over Max homodimerization, since reticulocyte lysate-expressed Max protein is phosphorylated and therefore preferentially heterodimerizes with Mad (50). The complexes were then incubated for 30 min at room temperature in the presence of approximately 1.4 ng of a 32P-labeled CMD (specific E-box) probe (an oligonucleotide with a c-Myc consensus binding site: 5′-AGCTTCAGACCACGTGGTCGGG-3′) (10) in a buffer containing 10 mM Tris-HCl (pH 7.5), 50 mM NaCl, 0.5 mM dithiothreitol, 0.05% NP-40, 1% glycerol, and 1 μg of poly(dI-dC) · poly(dI-dC) (Pharmacia) in a total volume of 20 μl. In some experiments, 2 μl of anti-Max antiserum (polyclonal antiserum sc-197X; Santa Cruz) was added, the reaction mixture was further incubated overnight at 4°C, and the protein-DNA complexes were separated from the free probe by electrophoresis on 4% polyacrylamide in 0.5× Tris-borate-EDTA at 150 V. The free probe was run out of the gel for better separation of the complexes. In some experiments, in vitro-translated wtMad1 or Mad1 CT deletion mutants were treated with 1.5 μg of potato alkaline phosphatase (PAP) for 15 min at 37°C (7) prior to the preincubation step.
RESULTS
Regions of homology within the Mad1 CT.
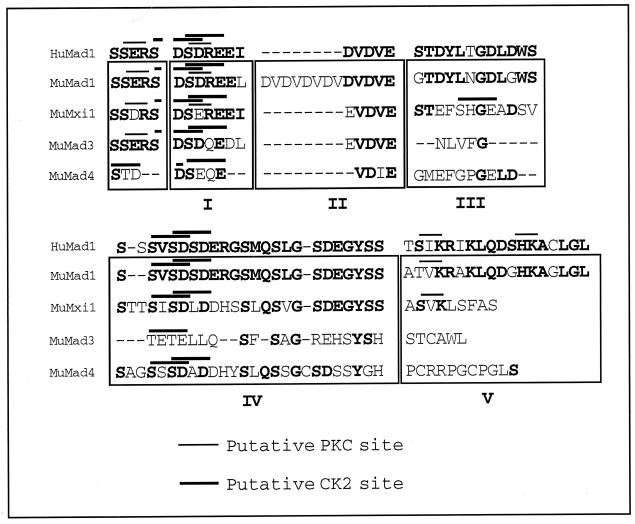
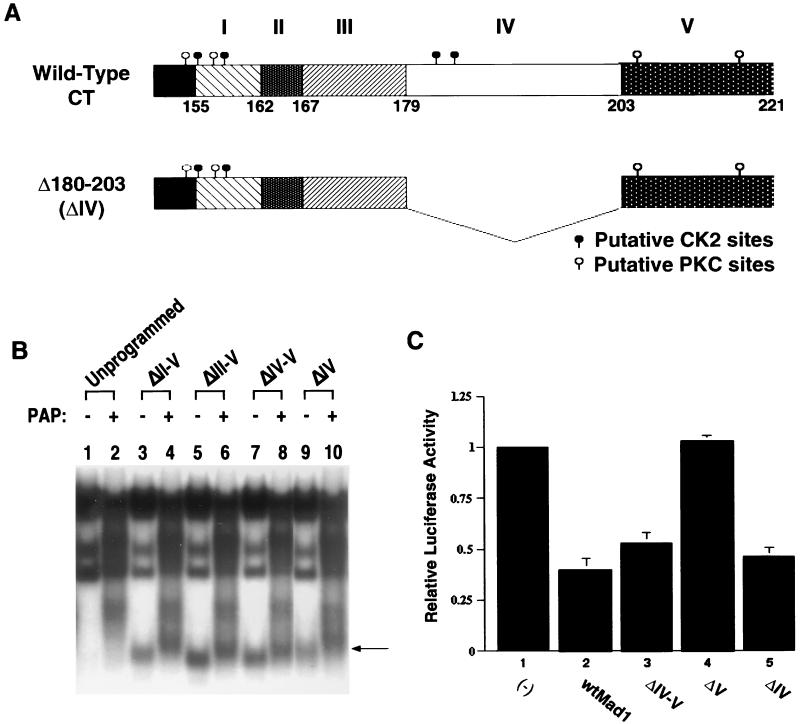
We compared the protein sequence of human Mad1 CT with that of murine Mad1, Mxi1, Mad3, and Mad4 (31) and identified five regions of homology (Fig. 1). Regions I, II, and IV share a high degree of homology across species and among murine Mad family members. Regions III and V are comparatively less conserved among the different murine family members but are fairly conserved across species. In addition, regions I to IV contain acidic residues, while region V is more basic in nature. Of interest, region II contains an acidic Asp-Val motif which is repeated twice in human Mad1 and six times in murine Mad1. Also, a number of putative CKII and protein kinase C (PKC) phosphorylation sites are clustered throughout the CT, suggesting that phosphorylation within this region may play an important role in the regulation of Mad1 function.
FIG. 1.
Sequence comparison of CT regions of human (Hu) Mad1 and murine (Mu) Mad1, Mxi1 (Mad2), Mad3, and Mad4 proteins (31). Indicated in boldface are the conserved amino acids. The roman numerals indicate the different regions of homology, and the thick and thin bars above the sequences indicate the putative CKII and PKC phosphorylation sites, respectively. Amino acids preceding region I of the CT have been included since they contain residues which are part of putative phosphorylation sites in region I.
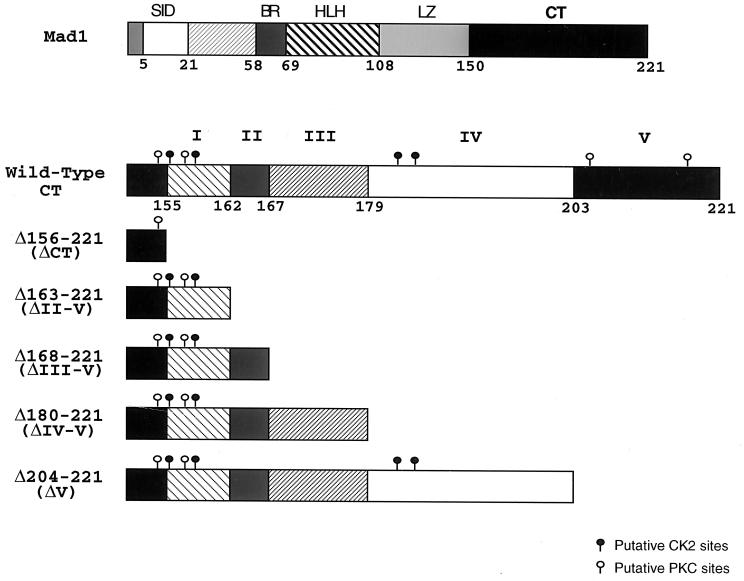
We constructed a series of human Mad1 CT deletion mutants lacking different homology boxes (Fig. 2). ΔCT was obtained by deleting the last 66 aa of the human Mad1 protein (15, 37). In addition, deletion mutants ΔII-V, ΔIII-V, ΔIV-V, and ΔV were generated by removing aa 163 to 221, 168 to 221, 180 to 221, and 204 to 221, respectively. All deletion mutants were cloned into the pMTH-β-globin-neo zinc-inducible expression vector and used in subsequent studies.
FIG. 2.
Structural and functional domains within the human Mad1 protein. (Top) Graphic representation of the structural and functional motifs present in the human Mad1 protein. The numbers represent the amino acid positions in the protein. (Bottom) Five domains (I to V) based on regions of homology within the Mad1 CT were defined by amino acid sequence comparison of human Mad1 to murine Mad1, Mxi1 (Mad2), Mad3, and Mad4 (Fig. 1). The individual CT deletion mutants were generated by PCR-directed mutagenesis of the full-length human mad1 coding region as described in Materials and Methods. The locations of putative CKII and PKC phosphorylation sites are shown.
Deletion of region V abrogates growth inhibition by Mad1.
To examine the effect of the Mad1 CT on the inhibition of cell growth, wtMad1 and the various CT mutants were individually transfected into C19 MEL cells. Since the expression of wtMad1 has been shown to be inhibitory to cell survival (13, 15, 52, 56), the ability to generate stable transfectants expressing the transgenes of the different mutants was used to identify novel functional motifs.
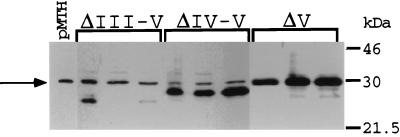
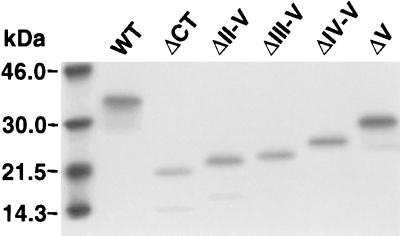
The results of two experiments are summarized in Table 1. Shown are the number of independent G418-resistant clones obtained (out of a total of 30 wells plated) and the number of clones expressing zinc-inducible levels of the transgene (determined by RT-PCR). No stable expressors were generated for either wtMad1 or ΔCT, consistent with the growth-inhibitory properties attributed to these proteins. Similarly, no clones expressing the ΔII-V transgene and very few expressing the ΔIII-V and ΔIV-V transgenes were generated. In contrast, many ΔV expressors were obtained, indicating that this truncated protein is not as potent a growth inhibitor as wtMad1, ΔCT, or the other CT mutants. To examine protein expression levels, lysates derived from the different transfectants (cultured for 3 months) were analyzed on Western blots. Three representative clones of each group are shown in Fig. 3. Protein levels in the ΔIII-V transfectants were variable and, for the most part, low, and the few ΔIV-V clones expressed fairly high levels of the mutant protein. For the ΔV transgene, many clones expressing high levels of the mutant protein were obtained.
TABLE 1.
Effect of CT deletions on Mad1 growth-inhibitory propertiesa
| Mad expression vector (residues deleted) | No. of clones that showed the indicated characteristic
|
|||
|---|---|---|---|---|
| Expt 1
|
Expt 2
|
|||
| G418 resistanceb | Expression of transgenec | G418 resistanceb | Expression of transgenec | |
| Wild type | 8 | 0 | 2 | 0 |
| ΔCT (156–221) | 3 | 0 | 2 | 0 |
| ΔII-V (163–221) | ND | ND | 8 | 0 |
| ΔIII-V (168–221) | ND | ND | 5 | 3 |
| ΔIV-V (180–221) | ND | ND | 6 | 4 |
| ΔV (204–221) | 18 | 9 | 18 | 16 |
ND, not determined. Results are from two independent experiments (1 and 2).
Numbers of G418-resistant clones out of a total of 30 wells plated.
Number of clones expressing zinc-inducible levels of the transgene (as determined by RT-PCR).
FIG. 3.
Expression of ΔIII-V, ΔIV-V, and ΔV proteins in stable transfectants. Protein lysates derived from the different MEL cell transfectants were analyzed on Western blots with antibodies directed against the full-length Mad1 protein. An extract prepared from cells transfected with the pMTH-β-globin neo empty vector was used as a negative control (pMTH). The protein extracts were prepared from cells cultured for 3 months after selection. The presence of a 30-kDa nonspecific band (only band present in the pMTH control) which comigrates with ΔV and migrates above ΔIII-V and ΔIV-V is indicated by an arrow. Representative clones for each group are shown.
Region V is required for the switch from proliferation to differentiation.
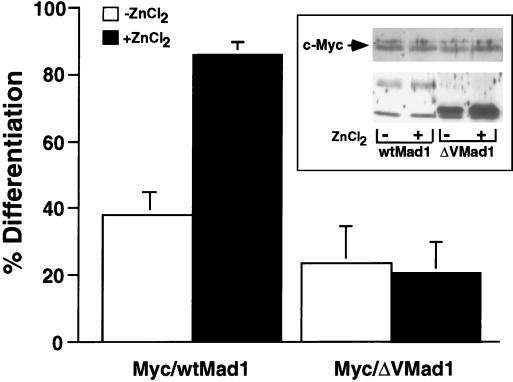
We have previously shown that Mad1 requires functional SID, BR, and LZ motifs to reverse a Myc-imposed differentiation block (15). However, due to the instability of the Myc-ΔCT Mad1 cotransfectants in culture and the inability to generate stable single transfectants expressing ΔCT Mad1, we were unable to investigate whether the CT domain is required for Mad1 to function in a differentiation switch (15). Therefore, we specifically focused on CT region V and more carefully examined its function during HMBA-induced differentiation. MEL cells were cotransfected with a constitutive c-Myc expression vector and either wtMad1 or ΔV zinc-inducible expression plasmids. In this system, c-Myc is constitutively expressed and therefore blocks the ability of the cells to differentiate upon induction with HMBA (19). Shown in Fig. 4 are the results obtained from three independent clones of c-Myc–wtMad1 and c-Myc–ΔV. In the absence of zinc, the majority of the cells (more than 60%) failed to differentiate upon induction. The addition of zinc to the culture media up-regulated the expression of wtMad1 and ΔV. While more than 80% of the cells expressing c-Myc–wtMad1 differentiated, the c-Myc–ΔV cotransfectants remained blocked. The inability to overcome the block to differentiation in these cotransfectants was not due to low levels of expression of ΔV. In fact, expression of the ΔV transgene was higher than that of the wtMad1 gene. Levels of expression of transfected c-Myc in c-Myc–wtMad1 and c-Myc–ΔV clones were similar (inset in Fig. 4). The precise mechanism by which Mad1 functions during a differentiation switch is unknown. Nonetheless, the results presented here demonstrate that CT region V is necessary for Mad1 to function in this process.
FIG. 4.
Region V mediates a molecular switch from cell proliferation to differentiation. MEL cells were stably transfected with pL1hmcneor, a constitutive c-Myc expression vector, and zinc-inducible wtMad1 or ΔV expression vector. The medium was supplemented with 75 μM ZnCl2 to induce the expression of wtMad1 and ΔV. Differentiation was chemically induced with HMBA. Results from three independent clones of both cotransfectants, each expressing comparable levels of c-Myc, are shown. The percent differentiation was determined by acid-benzidine staining of hemoglobin-positive cells, which were counted with a phase-contrast microscope. Results are presented as mean ± standard deviation. (Inset) Expression of c-Myc and Mad1 in one representative c-Myc–wtMad1 clone and one representative c-Myc–ΔV clone. Protein extracts prepared from HMBA-induced cells grown in the absence (−) or presence (+) of zinc, as indicated below the panels, were subjected to analysis by Western blotting. The blots were then immunoreacted with c-Myc- and Mad1-specific antisera.
Region V mediates Mad1 transcriptional repression.
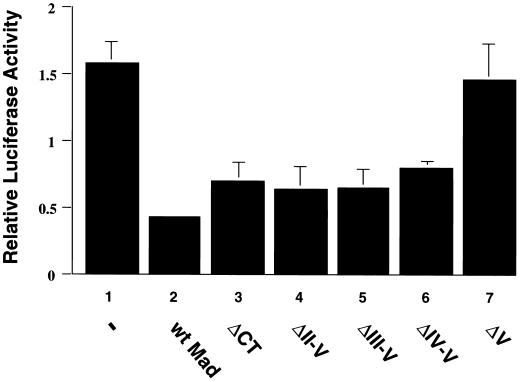
In view of the requirement of CT region V for Mad1 to inhibit growth and to function in a differentiation switch, we sought to examine the effect of this motif on the ability of Mad1 to repress c-Myc-dependent transcriptional activation. COS-7 cells were transiently cotransfected with pMyc3E1bLuc, a luciferase reporter plasmid containing three E-box regulatory elements (26), and expression vectors for either wtMad1, ΔCT, ΔII-V, ΔIII-V, ΔIV-V, or ΔV. A β-galactosidase expression plasmid was used to correct for differences in transfection efficiency. As shown in Fig. 5 and consistent with previous reports (28), wtMad1 expression resulted in the repression of transcriptional activation (compare bar 2 to bar 1). In contrast, the repressive effect of wtMad1 was lost when region V was deleted (compare bar 7 to bar 2). However, the inhibitory effect of Mad1 on transcriptional activation was restored when additional CT regions were deleted (compare bars 4, 5, and 6 to bar 2) or when the entire Mad1 CT was removed (compare bar 3 to bar 2). Similar results were obtained when the cells were cotransfected with a c-Myc expression vector and either wtMad1 or the various Mad1 mutants (data not shown).
FIG. 5.
Transcriptional repression by Mad1 CT mutants. COS-7 cells were transfected with pMycE1bLuc3 (a luciferase reporter plasmid containing three E-box regulatory elements) together with pCH110 (a β-galactosidase expression plasmid) and pRC/CMV expression vector for wtMad1, ΔCT, ΔII-V, ΔIII-V, ΔIV-V, or ΔV or empty vector (−), as indicated. Luciferase activity was normalized according to both protein levels and β-galactosidase activities and expressed relative to that obtained with pMycE1bLuc0 (a luciferase reporter plasmid lacking E-box elements). The results shown are from two independent experiments performed with triplicate samples. Results are presented as mean ± standard deviation.
These results demonstrate the importance of region V for the ability of Mad1 to antagonize c-Myc-mediated transcriptional activation. Furthermore, the data also suggest that there is an interplay among the different CT motifs.
Region V modulates Mad1 DNA binding.
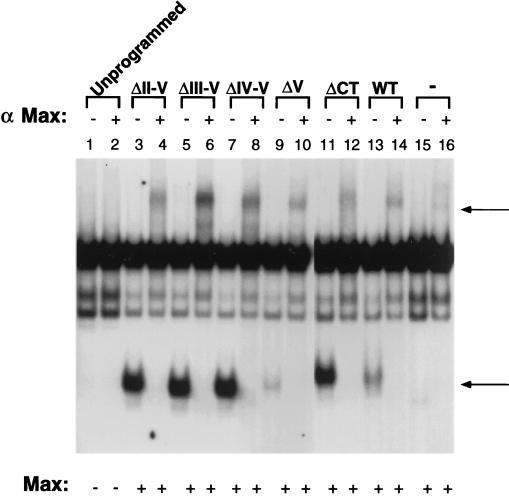
To gain further insight into the mechanism by which deletion of region V leads to a loss of Mad1 repressor function, we performed EMSAs to test for Mad1 DNA binding activity. Since Mad1 binds DNA as a heterodimeric complex with Max, in vitro-translated wtMad1, ΔCT, ΔII-V, ΔIII-V, ΔIV-V, or ΔV was preincubated with in vitro-translated Max to promote dimer formation. The extracts were then reacted with a radiolabeled E-box-containing DNA probe, and DNA binding of Mad1-Max complexes was analyzed by an EMSA (Fig. 6). wtMad1, ΔCT, ΔII-V, ΔIII-V, and ΔIV-V bound DNA efficiently. Moreover, DNA binding of ΔII-V, ΔIII-V, and ΔIV-V was more efficient than that of wtMad1. In contrast, deletion of region V resulted in a Mad1 protein with comparatively weak DNA binding activity (Fig. 6, lane 9). The specificity of the Mad1-Max heterocomplexes was confirmed by supershifting with anti-Max antiserum (Fig. 6). Similar results were obtained with protein extracts prepared from MEL cells stably transfected with various CT deletion mutants (data not shown). Of note, the weak DNA binding activity of ΔV was not due to inefficient heterodimerization with Max, since coimmunoprecipitation experiments revealed that the various Mad1-Max heterocomplexes immunoprecipitated at comparable levels with antibodies directed against Max (Fig. 7). In summary, deletion of region V impaired the ability of Mad1 to bind DNA, but removal of additional regions within the CT restored the DNA binding activity of the protein.
FIG. 6.
DNA binding activity of Mad1 CT mutants. wtMad1 and the CT deletion mutants were translated in vitro in rabbit reticulocyte lysates and preincubated with in vitro-translated Max to promote dimer formation. DNA binding of the Mad-Max complexes was analyzed by an EMSA. Supershifting with anti-Max antiserum (α Max) was performed as indicated above the lanes. The bottom arrow indicates the position of Mad-Max complexes, and the top arrow indicates the position of supershifted Mad-Max complexes. Results shown are representative of two independent experiments. WT, wild type; −, Mad1 not added.
FIG. 7.
Coimmunoprecipitation of Mad1 mutants and Max. In vitro-translated Max was incubated with 35S-labeled in vitro-translated wtMad1, ΔCT, ΔII-V, ΔIII-V, ΔIV-V, or ΔV, as indicated. The lysates were incubated as described in the legend to Fig. 6, and the Mad-Max complexes were immunoprecipitated with anti-Max antiserum. Mad1 without the LZ was used as a negative control in this assay (data not shown). The immunoprecipitates were analyzed by SDS-PAGE, and the various Mad1 proteins were detected by autoradiography. WT, wild type.
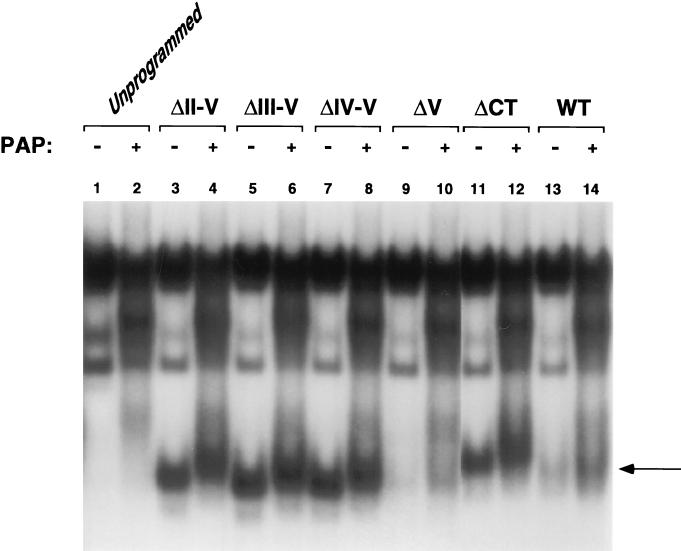
Dephosphorylation restores DNA binding activity to the ΔV protein.
The Mad1 CT contains several putative phosphorylation sites, as shown in Fig. 1 and 2. We sought to determine whether phosphorylation within this domain modulates the DNA binding activity of the protein. wtMad1, Max, and the CT mutants were expressed in vitro in rabbit reticulocyte lysate system. All extracts, with the exception of Max, were treated with PAP (7). PAP-treated and untreated protein lysates of wtMad1, ΔCT, and the various CT mutants were preincubated with Max, and binding to DNA was analyzed by an EMSA. As shown in Fig. 8, the binding of ΔII-V, ΔIII-V, ΔIV-V, and ΔCT was efficient and not significantly affected by phosphatase treatment (lanes 4, 6, 8, and 12, respectively). There was a slight shift in the migration of the above complexes, indicative of a change in the phosphorylation state of the proteins. In contrast, PAP treatment of ΔV lead to more efficient DNA binding (Fig. 8, lanes 9 and 10), comparable to that of nontreated wtMad1 (lane 13). Likewise, wtMad1 also bound DNA more efficiently after PAP treatment (Fig. 8, lanes 13 and 14).
FIG. 8.
Effect of phosphatase treatment on the DNA binding activity of the CT mutants. wtMad1 and the CT deletion mutants were translated in vitro in rabbit reticulocyte lysates and, where indicated, treated with PAP. The lysates were preincubated with in vitro-translated Max and analyzed by an EMSA. Results shown are representative of two independent experiments. The arrow indicates the position of the Mad-Max complexes. WT, wild type.
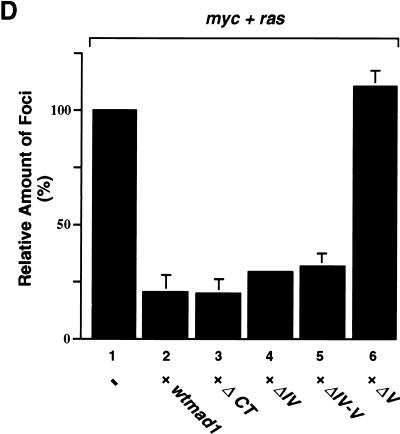
Functional interplay between regions IV and V.
Our findings suggest that in addition to the involvement of region V in the regulation of Mad1 function, there is an interplay between the different CT motifs. As we have shown earlier, deletion of region V abolished Mad1-associated functions, while removal of additional regions within the CT restored growth-inhibitory properties to the protein. More specifically, deletion of region V together with region IV rendered a Mad1 protein that bound DNA efficiently and repressed c-Myc-induced transcriptional activation (Fig. 5 and 6). Moreover, phosphorylation within region IV appeared to mediate these functions. Although phosphorylated ΔV protein (which retains region IV) did not bind DNA, phosphatase treatment restored its binding capacity to levels comparable to that of untreated wtMad1. In contrast, the DNA binding activities of ΔCT, ΔII-V, ΔIII-V, and ΔIV-V were not significantly affected by phosphatase treatment (Fig. 8). To examine more carefully the relevance of region IV in the regulation of Mad1 function, a deletion mutant lacking region IV (aa 180 to 203) was constructed (Fig. 9A). As expected, this mutant protein bound DNA efficiently regardless of its phosphorylation state (Fig. 9B, compare lane 10 to lane 9), and the binding was comparable to that of the ΔII-V, ΔIII-V, and ΔIV-V proteins (all lacking region IV). These results suggest that the regulation of phosphorylation in region IV by region V is important for DNA binding. Although dephosphorylation of ΔV (which retains region IV) restored DNA binding to levels comparable to those of wtMad1, dephosphorylation of ΔIV had no effect due to the absence of region IV in this mutant.
FIG. 9.
DNA binding, transcriptional repression, and effect on Myc-Ras cotransformation by the ΔIV mutant. (A) Graphic representation of the ΔIV mutant protein. The expression plasmid for ΔIV, which encodes a mutant protein lacking aa 180 to 203, was generated by PCR-directed mutagenesis of the full-length mad1 coding region. The mutated mad1 cDNA was subsequently cloned into the pRC/CMV expression vector. (B) DNA binding activity of ΔIV. The CT deletion mutant ΔII-V, ΔIII-V, ΔIV-V, or ΔIV was translated in vitro in the reticulocyte lysate system and treated with PAP, where indicated. DNA binding was analyzed by an EMSA. The arrow indicates the position of the Mad-Max complexes. (C) Effect of region IV on Mad1-mediated transcriptional repression. COS-7 cells were transfected with pMycE1bLuc3 and pRC/CMV expression plasmid for wtMad1, ΔIV-V, ΔV, or ΔIV or empty vector (−), as indicated. Luciferase activity was normalized as described in the legend to Fig. 5. Results are presented as mean ± standard deviation. (D) Effect of Mad1 CT mutants on Myc-Ras cotransformation. REF primary cultures were transfected with c-myc together with pEJras and either wtMad1 or one of the CT mutants (in the pRC/CMV expression vector) indicated. The number of transformed foci was scored after 15 days. Results from three independent transformation experiments are presented as mean ± standard deviation. Results represent the average number of foci relative to those obtained with myc and ras alone.
To further evaluate the effect of region IV on Mad1 function, the ability of the ΔIV mutant to repress Myc-dependent transcriptional activation was investigated. COS-7 cells were transiently cotransfected with either wtMad1, ΔIV-V, ΔV, or ΔIV expression plasmid and pMyc3E1bLuc, the luciferase reporter plasmid containing three E-box regulatory elements (Fig. 9C). As shown previously (Fig. 5), wtMad1 and ΔIV-V expression led to the repression of transcriptional activation (Fig. 9C, compare bars 2 and 3 to bar 1). Deletion of region V, however, led to a loss of the inhibitory effect of Mad1 (Fig. 9C, bar 4). In contrast, deletion of region IV rendered a protein capable of inhibiting transcriptional activation (Fig. 9C, bar 5), and this effect was comparable to that of ΔIV-V (bar 3).
We next examined the ability of the ΔIV mutant to inhibit Myc-Ras cotransformation. REF secondary cultures were cotransfected with c-myc and pEJras (c-Ha-rasVal12) in the presence or absence of either wtMad1, ΔCT, ΔIV, ΔIV-V, or ΔV. As shown in Fig. 9D and in agreement with previous results (37), the number of transformed foci obtained after 15 days was significantly reduced (∼80%) by the wtMad1 protein. Similarly, transformation was profoundly inhibited in the presence of the ΔCT, ΔIV, or ΔIV-V mutant. The number of foci in each of these cotransformations was comparable to the number obtained with wtMad1. As expected, the ΔV mutant failed to inhibit Myc-Ras cotransformation.
Together, the data indicate that region IV plays a critical role in mediating Mad1 function and that regulation through phosphorylation within this region is important for the function of the Mad1 protein.
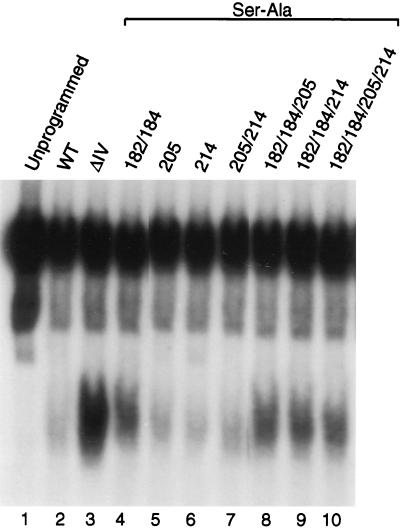
CKII phosphorylation in region IV mediates Mad1 DNA binding and transcriptional repression.
To further elucidate the effect of phosphorylation in region IV on Mad1 DNA binding, we substituted serines 182 and 184 within the putative CKII sites in this region with the nonphosphorylatable amino acid alanine (Ser182/184 to Ala182/184). wtMad1, ΔIV, and wtMad1(Ser182/184 to Ala182/184) were translated in vitro in the rabbit reticulocyte system and preincubated with in vitro-expressed Max, and the ability of the complexes to bind DNA was examined by an EMSA (Fig. 10). As shown earlier, deletion of region IV gave rise to a protein which bound DNA more efficiently than wtMad1 (Fig. 10, compare lane 3 to lane 2). This effect was partially mimicked by preventing the phosphorylation of Ser182 and Ser184. The ability of wtMad1(Ser182/184 to Ala182/184) to bind DNA increased relative to that of wtMad1 (Fig. 10, compare lane 4 to lane 2). However, this increase was less profound than that seen for the ΔIV protein.
FIG. 10.
DNA binding activities of the Mad1 phosphorylation mutants. wtMad1, ΔIV, and the Ser-Ala point mutants were translated in vitro in rabbit reticulocyte lysates, and their DNA binding activities were analyzed by an EMSA. Results shown are representative of two independent experiments. WT, wild type.
We also mutated serines 205 and 214 within the putative PKC sites in region V. However, no change in the DNA binding activity of the mutant proteins was detected (Fig. 10, lanes 5, 6, and 7) relative to that of wtMad1 (lane 2). Similarly, when the serines in region IV (serines 182 and 184) and V (serines 205 and 214) were mutated together, no appreciable changes in DNA binding were observed relative to the result obtained with mutation of serine182/184 alone. In summary, phosphorylation at the PKC sites does not appear to have an effect on Mad1 DNA binding activity. In contrast, phosphorylation at the CKII sites located in region IV appears to regulate the DNA binding activity of the protein.
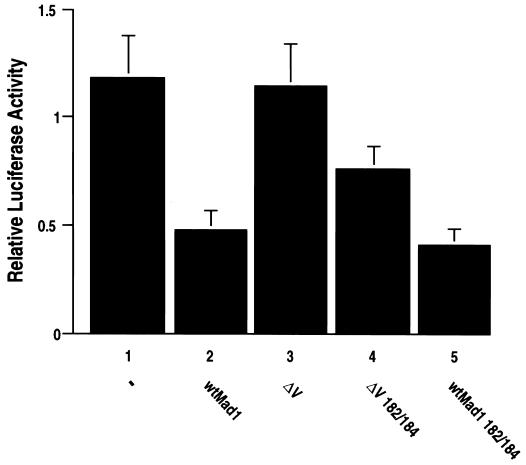
To gain further insight into the effect of phosphorylation in region IV on the function of Mad1, we investigated the ability of the phosphorylation mutants wtMad1(Ser182/184 to Ala182/184) and ΔV(Ser182/184 to Ala182/184) to suppress transactivation by Myc. COS-7 cells were transiently cotransfected with either wtMad1, ΔV, wtMad1(Ser182/184 to Ala182/184), or ΔV(Ser182/184 to Ala182/184) expression plasmid and pMyc3E1bLuc. As shown earlier (Fig. 5 and 9C), wtMad1 expression led to the repression of transcriptional activation, while the ΔV mutant had no effect. However, ΔV(Ser182/184 to Ala182/184) showed partially restored repressor activity (Fig. 11). Because wtMad1 is already a strong repressor of transcriptional activation by Myc, no noticeable increase in repression was observed with mutant wtMad1(Ser182/184 to Ala182/184). In addition, no change in repressor activity was detected with the various region V PKC phosphorylation mutants (data not shown). These findings support our hypothesis that region V masks the function of a negative regulator in region IV which appears to be modulated through phosphorylation at the CKII sites.
FIG. 11.
Transcriptional repression by Mad1 phosphorylation mutants. COS-7 cells were transfected with pMycE1bLuc3 and pRC/CMV expression vector for wtMad1, ΔV, ΔV(Ser182/184 to Ala182/184), or wtMad1(Ser182/184 to Ala182/184). An empty vector was used as a control (lane 1). The results shown are from two independent transfection experiments performed with triplicate samples. Results are presented as mean ± standard deviation.
DISCUSSION
In this report, we present evidence that the CT of human Mad1 contains novel regulatory elements important for mediating functions associated with the protein. By analyzing a series of Mad1 CT deletion mutants, we demonstrated that deletion of the last 18 aa (region V) results in a truncated protein that binds DNA inefficiently and is therefore unable to mediate a repressive effect on transcriptional activation. Furthermore, removal of region V abrogates the inhibitory effect of Mad1 on cell growth (Table 1) and results in a protein that is incapable of reversing a c-Myc-imposed block to differentiation (Fig. 4) and is unable to antagonize Myc in a Myc-Ras cotransformation assay (Fig. 9D). In contrast, deletion of region IV together with region V, as well as more extensive deletions (i.e., II-V, III-V, and IV-V), restores function to the protein. We also showed that regulation via the phosphorylation of putative CKII sites in region IV is critical for mediating Mad1 DNA binding and transcriptional repression. In support of this notion, phosphatase treatment of the ΔV protein partially restores its DNA binding activity to levels comparable to those of wtMad1. Likewise, deletion of region IV generates a protein that retains its DNA binding ability, is virtually unaffected by phosphatase treatment (Fig. 9B), and represses Myc-dependent transcriptional activation (Fig. 9C). Most importantly, amino acid substitution at positions 182 and 184 (Ser to Ala) in the ΔV mutant, which prevents phosphorylation in region IV, partially restores the ability of the protein to repress transcription (Fig. 11). Altogether, these results suggest that the two putative CKII sites located in region IV function as regulatory phosphorylation elements. In fact, deletion of region V together with region I (which also contains putative CKII sites) and region II does not restore function to the protein (unpublished observations). These results strongly support our view that of the putative phosphorylation sites located in the CT, only the two CKII sites within region IV appear to be involved in the regulation of Mad1 function.
There is evidence in the literature to indicate that CKII activity is elevated in cells induced to proliferate (9, 57). In addition, CKII phosphorylation of Max has been shown to modulate its DNA binding activity (7, 10) and, most importantly, Mxi1 repression of the Myc promoter has been reported to be dependent on consensus CKII phosphorylation sites present in the CT (42). Our results support the notion that the CT of Mad1 contains novel, discrete functional domains that are important for the regulation of Mad1 function. Furthermore, an interplay among the different motifs, specifically regions IV and V, appears to be critical in this process. Based on the data presented in this report, we believe that regions IV and V contain contrasting regulatory elements. While region IV acts as a weak negative regulator, region V appears to positively control Mad1 function. Although the precise mechanism remains to be determined, it is conceivable that an intramolecular folding mechanism similar to that described for the c-myb proto-oncogene product (c-Myb) or other eukaryotic transcription factors, such as activating transcription factor 2 and the p50 subunit of NF-κB, may be involved (17, 33, 53). Interestingly, the Mad1 CT contains a DVES motif similar to the EVES motif shown to be involved in the molecular folding described for c-Myb (Fig. 1) (17). Furthermore, the CT of Mad1 is extremely acidic, with the exception of region V, which is basic in nature. We propose that Mad1 undergoes conformational changes as a result of phosphorylation and dephosphorylation (specifically in region IV) and other posttranslational modifications. These changes might allow region V (basic) to fold back on regions I to IV (acidic) and preclude the CT from interacting with the basic DNA binding domain. As a result, Mad1 would bind DNA via the DNA binding domain and exert its repressive effect on transcriptional activation. Since amino acid substitution at positions 182 and 184 in the ΔV mutant partially restores its ability to repress transcription, it is likely that CKII phosphorylation in region IV is partly responsible for mediating this process. The fact that the amino acid substitution partially restores the function of the ΔV mutant suggests that region IV contains a repressive element that negatively regulates Mad1 function. In wtMad1, this effect is masked by region V. In contrast to the importance of the CKII phosphorylation sites in region IV, phosphorylation at the putative PKC sites in region V appears to be dispensable for Mad1 function. Although we have not excluded the possibility of an intramolecular folding mechanism in Mad1, using the yeast two-hybrid system we have been unable to show a direct interaction between the CT and the DNA-binding domain of Mad1 (unpublished observations). This finding would imply either that a direct interaction such as that described for c-Myb does not exist or perhaps that in Mad1 this mechanism requires interactions with intermediary proteins absent in yeast cells.
Previous studies have established the importance of the N-terminal SID and centrally located bHLH-LZ domains for Mad1 function. Reports from our laboratory and others have shown that the SID and bHLH-LZ are required for Mad1 to function in a molecular switch from cell proliferation to differentiation (15, 52). In addition, the importance of these domains for the repressive effect of Mad1 on transcriptional activation and Myc or Ras cotransformation is also documented (11, 37, 40). In contrast to these clearly defined functional motifs, a role for the CT of Mad1 has not yet been clearly elucidated. Previous studies have shown that removal of the entire CT does not change the growth-inhibitory function of Mad1 (11, 15). However, we found that clones coexpressing c-Myc and ΔCT are unstable (15). In light of our new findings, this instability may be due to the loss of regulatory elements within the CT, including the CKII phosphorylation sites.
Since Mad expression has been tightly associated with the transition from proliferation to differentiation, it has been proposed that a loss of Mad function may be associated with a lack of normal differentiation and tumorigenesis (12, 21, 30, 55). Therefore, a better understanding of the mechanism(s) by which Mad functions to promote a transition to differentiation may lead to novel approaches for the treatment of certain types of tumors.
ACKNOWLEDGMENTS
We thank Brian Hacker for assistance in plasmid preparation and R. Davis (University of Massachusetts Medical School, Worcester) for providing the pMycE1b0Luc and pMyc3E1bLuc plasmids. We also thank Maria Zajac-Kaye for helpful discussions.
REFERENCES
- 1.Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 2.Amati B, Brooks M W, Levy N, Littlewood T D, Evan G I, Land H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell. 1993;72:233–245. doi: 10.1016/0092-8674(93)90663-b. [DOI] [PubMed] [Google Scholar]
- 3.Amin C, Wagner A J, Hay N. Sequence-specific transcriptional activation by Myc and repression by Max. Mol Cell Biol. 1993;13:383–390. doi: 10.1128/mcb.13.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayer D E, Eisenman R N. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev. 1993;7:2110–2119. doi: 10.1101/gad.7.11.2110. [DOI] [PubMed] [Google Scholar]
- 5.Ayer D E, Kretzner L, Eisenman R N. Mad: a heterodimeric partner for Max that antagonizes Myc transcriptional activity. Cell. 1993;72:211–222. doi: 10.1016/0092-8674(93)90661-9. [DOI] [PubMed] [Google Scholar]
- 6.Bar-Ner M, Messing L T, Cultraro C M, Birrer M J, Segal S. Regions within the c-Myc protein that are necessary for transformation are also required for inhibition of differentiation of murine erythroleukemia cells. Cell Growth Differ. 1992;3:183–190. [PubMed] [Google Scholar]
- 7.Berberich S J, Cole M D. Casein kinase II inhibits the DNA-binding activity of Max homodimers but not Myc/Max heterodimers. Genes Dev. 1992;6:166–176. doi: 10.1101/gad.6.2.166. [DOI] [PubMed] [Google Scholar]
- 8.Blackwood E M, Eisenman R N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 9.Bosc D G, Lüscher B, Litchfield D W. Expression and regulation of protein kinase CK2 during the cell cycle. Mol Cell Biochem. 1999;191:213–222. [PubMed] [Google Scholar]
- 10.Bousset K, Henriksson M, Luscher-Firzlaff J M, Litchfield D W, Luscher B. Identification of casein kinase II phosphorylation sites in Max: effects on DNA-binding kinetics of Max homo- and Myc/Max heterodimers. Oncogene. 1993;8:3211–3220. [PubMed] [Google Scholar]
- 11.Cerni C, Bousset K, Seelos C, Burkhardt H, Henriksson M, Luscher B. Differential effects by Mad and Max on transformation by cellular and viral oncoproteins. Oncogene. 1995;11:587–596. [PubMed] [Google Scholar]
- 12.Chen J, Willingham T, Margraf L R, Schreiber-Agus N, DePinho R A, Nisen P D. Effects of the MYC oncogene antagonist, MAD, on proliferation, cell cycling and the malignant phenotype of human brain tumor cells. Nat Med. 1995;1:638–643. doi: 10.1038/nm0795-638. [DOI] [PubMed] [Google Scholar]
- 13.Chin L, Schreiber-Agus N, Pellicer I, Chen K, Lee H, Dudast M, Cordon-Cardo C, DePinho R A. Contrasting roles for Myc and Mad proteins in cellular growth and differentiation. Proc Natl Acad Sci USA. 1995;92:8488–8492. doi: 10.1073/pnas.92.18.8488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crouch D H, Lang C, Gillespie D A F. The leucine zipper domain of avian c-Myc is required for transformation and autoregulation. Oncogene. 1990;5:683–689. [PubMed] [Google Scholar]
- 15.Cultraro C M, Bino T, Segal S. Function of the c-Myc antagonist Mad1 during a molecular switch from proliferation to differentiation. Mol Cell Biol. 1997;17:2353–2359. doi: 10.1128/mcb.17.5.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cultraro C M, Bino T, Segal S. Regulated expression and function of the c-Myc antagonist Mad1 during a molecular switch from proliferation to differentiation. Curr Top Microbiol Immunol. 1997;224:150–158. doi: 10.1007/978-3-642-60801-8_15. [DOI] [PubMed] [Google Scholar]
- 17.Dash A B, Orrico F C, Ness S A. The EVES motif mediates both intermolecular and intramolecular regulation of c-Myb. Genes Dev. 1996;10:1858–1869. doi: 10.1101/gad.10.15.1858. [DOI] [PubMed] [Google Scholar]
- 18.Delgado M D, Lerga A, Canelles M, Gomez-Casares M T, Leon J. Differential regulation of Max and role of c-Myc during erythroid and myelomonocytic differentiation of K562 cells. Oncogene. 1995;10:1659–1665. [PubMed] [Google Scholar]
- 19.Dmitrovsky E, Kuehl W M, Hollis G F, Kirsch I R, Bender T P, Segal S. Expression of a transfected human c-myc oncogene inhibits differentiation of a mouse erythroleukaemia cell line. Nature. 1986;322:748–750. doi: 10.1038/322748a0. [DOI] [PubMed] [Google Scholar]
- 20.Dube S K, Gaedicke G, Kluge G, Weimann N, Melderis B J, Steinheider H, Crozier G, Beckmann H, Ostertag W. Hemoglobin-synthesizing mouse and human erythroleukemic cell lines as model systems for the study of differentiation and control of gene expression. In: Nakar W, Ono T, Suigimura T, Sugano H, editors. Differentiation and control of malignancy in tumor cells. Tokyo, Japan: University of Tokyo Press; 1974. pp. 99–135. [Google Scholar]
- 21.Edelhoff S, Ayer D E, Zervos A S, Steingrimsson E, Jenkins N A, Copeland N G, Eisenman R N, Brent R, Disteche C M. Mapping of two genes encoding members of a distinct subfamily of MAX interacting proteins: MAD to human chromosome 2 and mouse chromosome 6, and MXI1 to human chromosome 10 and mouse chromosome 19. Oncogene. 1994;9:665–668. [PubMed] [Google Scholar]
- 22.Einat M, Resnitzky D, Kimchi A. Close link between reduction of c-myc expression by interferon and G0/G1 arrest. Nature. 1985;313:597–600. doi: 10.1038/313597a0. [DOI] [PubMed] [Google Scholar]
- 23.Foley K P, McArthur G A, Queva C, Hurlin P J, Soriano P, Eisenman R N. Targeted disruption of the MYC antagonist MAD1 inhibits cell cycle exit during granulocyte differentiation. EMBO J. 1998;17:774–785. doi: 10.1093/emboj/17.3.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freytag S O. Enforced expression of the c-myc oncogene inhibits cell differentiation by precluding entry into a distinct predifferentiation state in G0/G1. Mol Cell Biol. 1988;8:1614–1624. doi: 10.1128/mcb.8.4.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu W, Cechova K, Tassi V, Dalla-Favera R. Opposite regulation of gene transcription and cell proliferation by c-Myc and Max. Proc Natl Acad Sci USA. 1993;90:2935–2939. doi: 10.1073/pnas.90.7.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta S, Seth A, Davis R J. Transactivation of gene expression by Myc is inhibited by mutation at the phosphorylation sites Thr-58 and Ser-62. Proc Natl Acad Sci USA. 1993;90:3216–3220. doi: 10.1073/pnas.90.8.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassig C A, Fleischer T C, Bilin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 28.Henriksson M, Luscher G. Proteins of the Myc network: essential regulators of cell growth and differentiation. Cancer Res. 1996;68:110–182. doi: 10.1016/s0065-230x(08)60353-x. [DOI] [PubMed] [Google Scholar]
- 29.Huang Z, Fasco M J, Kaminsky L S. Optimization of DNaseI removal of contaminating DNA from RNA for use in quantitative RNA-PCR. BioTechniques. 1996;20:1014–1019. doi: 10.2144/96206st02. [DOI] [PubMed] [Google Scholar]
- 30.Hurlin P J, Foley K P, Ayer D E, Eisenman R N, Hanahan D, Arbeit J M. Regulation of Myc and Mad during epidermal differentiation and HPV-associated tumorigenesis. Oncogene. 1995;11:2487–2501. [PubMed] [Google Scholar]
- 31.Hurlin P J, Queva C, Ayer D E, Copeland N G, Jenkins N A, Eisenman R N. Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. EMBO J. 1995;14:5646–5659. doi: 10.1002/j.1460-2075.1995.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurlin P J, Queva C, Eisenman R N. Mnt, a novel Max-interacting protein is coexpressed with Myc in proliferating cells and mediates repression at Myc binding sites. Genes Dev. 1997;11:44–58. doi: 10.1101/gad.11.1.44. [DOI] [PubMed] [Google Scholar]
- 33.Ji L, Arcinas M, Boxer L M. NF-κB sites function as positive regulators of expression of the translocated c-myc allele in Burkitt's lymphoma. Mol Cell Biol. 1994;14:7967–7974. doi: 10.1128/mcb.14.12.7967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaczmarek L, Hyland J K, Watt R, Rosenberg M, Baserga R. Microinjected c-myc as a competence factor. Science. 1985;228:1313–1315. doi: 10.1126/science.4001943. [DOI] [PubMed] [Google Scholar]
- 35.Kahn S M, Jiang W, Borner C, O'Driscoll K, Weinstein I B. Construction of defined deletion mutants by thermal cycled fusion: applications to protein kinase C. Tech J Methods Cell Mol Biol. 1990;2:27–30. [Google Scholar]
- 36.Kato G J, Barrett J, Villa-Garcia M, Dang C V. An amino-terminal c-Myc domain required for neoplastic transformation activates transcription. Mol Cell Biol. 1990;10:5914–5920. doi: 10.1128/mcb.10.11.5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koskinen P J, Ayer D E, Eisenman R N. Repression of Myc-Ras cotransformation by Mad is mediated by multiple protein-protein interactions. Cell Growth Differ. 1995;6:623–629. [PubMed] [Google Scholar]
- 38.Kretzner L, Blackwood E M, Eisenman R N. Myc and Max proteins possess distinct transcriptional activities. Nature. 1992;359:426–429. doi: 10.1038/359426a0. [DOI] [PubMed] [Google Scholar]
- 39.Laherty C D, Yang W, Sun J, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 40.Lahoz E G, Xu L, Schreiber-Agus N, DePinho R A. Suppression of Myc, but not E1a, transformation activity by Max-associated proteins, Mad and Mxi1. Proc Natl Acad Sci USA. 1994;91:5503–5507. doi: 10.1073/pnas.91.12.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larsson L, Pettersson M, Oberg F, Nilsson K, Luscher B. Expression of mad, mxi1, max and c-myc during induced differentiation of hematopoietic cells: opposite regulation of mad and c-myc. Oncogene. 1994;9:1247–1252. [PubMed] [Google Scholar]
- 42.Lee T C, Ziff E B. Mxi1 is a repressor of the c-myc promoter and reverses activation by USF. J Biol Chem. 1999;274:595–606. doi: 10.1074/jbc.274.2.595. [DOI] [PubMed] [Google Scholar]
- 43.Littlewood T D, Amati B, Land H, Evan G I. Max and c-Myc/Max DNA-binding activities in cell extracts. Oncogene. 1992;7:1783–1792. [PubMed] [Google Scholar]
- 44.Lymboussaki A, Kaipainen A, Hatva E, Vastrik I, Jeskanen L, Jalkanen M, Werner S, Stenback R, Alitalo R. Expression of Mad, an antagonist of Myc oncoprotein function, in differentiating keratinocytes during tumorigenesis of the skin. Br J Cancer. 1996;73:1347–1355. doi: 10.1038/bjc.1996.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marks P A, Richon V M, Kiyokawa J, Rifkind R A. Inducing differentiation of transformed cells with hybrid polar compounds: a cell cycle-dependent process. Proc Natl Acad Sci USA. 1994;91:10251–10254. doi: 10.1073/pnas.91.22.10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meroni G, Reymond A, Alcalay M, Borsani G, Tanigami A, Tonlorenzi R, Lo Nigro C, Messali S, Zollo M, Ledbetter D H, Brent R, Ballabio A, Carrozzo R. Rox, a novel bHLHZIP protein expressed in quiescent cells that heterodimerizes with Max, binds a non-canonical E box and acts as a transcriptional repressor. EMBO J. 1997;16:2892–2906. doi: 10.1093/emboj/16.10.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukherjee B, Morgenbesser S D, DePinho R A. Myc family oncoproteins function through a common pathway to transform normal cells in culture: cross-interference by Max and trans-acting dominant mutants. Genes Dev. 1992;6:1480–1492. doi: 10.1101/gad.6.8.1480. [DOI] [PubMed] [Google Scholar]
- 48.Prendergast G C, Lawe D, Ziff E B. Association of Myn, the murine homolog of Max, with c-Myc stimulates methylation-sensitive DNA binding and Ras cotransformation. Cell. 1991;65:395–407. doi: 10.1016/0092-8674(91)90457-a. [DOI] [PubMed] [Google Scholar]
- 49.Prochownik E V, Kukowska J, Rodgers C. c-myc antisense transcripts accelerate differentiation and inhibit G1 progression in murine erythroleukemia cells. Mol Cell Biol. 1988;8:3683–3695. doi: 10.1128/mcb.8.9.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prochownik E V, VanAntwerp M E. Differential patterns of DNA binding by myc and max proteins. Proc Natl Acad Sci USA. 1993;90:960–964. doi: 10.1073/pnas.90.3.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reddy C D, Dasgupta P, Saikumar P, Dudek H, Rauscher F, Reddy E P. Mutational analysis of Max: role of basic, helix-loop-helix/leucine zipper domains in DNA binding, dimerization and regulation of Myc-mediated transcriptional activation. Oncogene. 1992;7:2085–2092. [PubMed] [Google Scholar]
- 52.Roussel M F, Ashmun R A, Sherr C J, Eisenman R N, Ayer D E. Inhibition of cell proliferation by the Mad1 transcriptional repressor. Mol Cell Biol. 1996;16:2796–2801. doi: 10.1128/mcb.16.6.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sano Y, Tokitou F, Dai P, Maekawa T, Yamamoto T, Ishi S. CBP alleviates the intramolecular inhibition of ATF-2 function. J Biol Chem. 1998;273:29098–29105. doi: 10.1074/jbc.273.44.29098. [DOI] [PubMed] [Google Scholar]
- 54.Schreiber-Agus N, Chin L, Chen K, Torres R, Rao G, Guida P, Skoultchi A I, DePinho R A. An amino-terminal domain of Mxi1 mediates Anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor Sin3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 55.Shapiro D N, Valentine V, Eagle L, Yin X, Morris S W, Prochownik E V. Assignment of the human MAD and MXI1 genes to chromosomes 2p12-p13 and 10q24-25. Genomics. 1994;23:282–285. doi: 10.1006/geno.1994.1496. [DOI] [PubMed] [Google Scholar]
- 56.Vastrik I, Kaipainen A, Penttila T, Lymboussakis A, Alitalo R, Parvinen M, Alitalo K. Expression of the mad gene during cell differentiation in vivo and its inhibition of cell growth in vitro. J Cell Biol. 1995;128:1197–1208. doi: 10.1083/jcb.128.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu X, Landesman-Bollag E, Channavajhala P L, Seldin D C. Murine protein kinase CK2: gene and oncogene. Mol Cell Biochem. 1999;191:65–74. [PubMed] [Google Scholar]
- 58.Zervos A S, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]