ABSTRACT
Furfural is a common furan inhibitor formed due to dehydration of pentose sugars, like xylose, and acts as an inhibitor of microbial metabolism. Overexpression of NADH-specific FucO and deletion of NADPH-specific YqhD had been a successful strategy in the past in conferring tolerance against furfural in Escherichia coli, which highlights the importance of oxidoreductases in conferring tolerance against furfural. In a screen consisting of various oxidoreductases, dehydrogenases, and reductases, we identified the yghA gene as an overexpression target to confer tolerance against furfural. YghA preferably used NADH as a cofactor and had an apparent Km value of 0.03 mM against furfural. In the presence of 1 g liter−1 furfural and 10% xylose (wt/vol), yghA overexpression in an ethanologenic E. coli strain SSK42 resulted in an ethanol efficiency of ∼97%, with a 5.3-fold increase in ethanol titers compared to the control. YghA also exhibited activity against the less toxic inhibitor 5-hydroxymethyl furfural, which is formed due to dehydration of hexose sugars, and thus is a formidable target for overexpression in ethanologenic strain for fermentation of sugars in biomass hydrolysate.
IMPORTANCE Lignocellulosic biomass represents an inexhaustible source of carbon for second-generation biofuels. Thermo-acidic pretreatment of biomass is performed to loosen the lignocellulosic fibers and make the carbon bioavailable for microbial metabolism. The pretreatment process also results in the formation of inhibitors that inhibit microbial metabolism and increase production costs. Furfural is a potent furan inhibitor that increases the toxicity of other inhibitors present in the hydrolysate. Thus, it is desirable to engineer furfural tolerance in E. coli for efficient fermentation of hydrolysate sugars.
KEYWORDS: Escherichia coli, ethanol, YghA, furfural, lignocellulose, tolerance
INTRODUCTION
Lignocellulosic biomass presents an inexhaustible source of biocarbon that can be channeled toward the production of compounds with industrial relevance (1, 2). This has the potential to alleviate the ever-increasing need to burn fossil fuels for energy needs. However, biomass recalcitrance is a major impediment to cost-efficient conversion of lignocellulosic biomass into biofuels. Plants have evolved complex structural and physical mechanisms which resist the breakdown of complex oligosaccharides into simpler monosaccharides (3). Among sugars, the pentose content in lignocellulose can vary from 5 to 30% (2). On a dry basis, xylan can constitute up to 24 wt% of grasses (4). In order to maximize the yield of pentose sugars and make the biomass accessible for enzymatic saccharification and microbial metabolism, lignocellulosic biomass is pretreated to remove lignin and loosen the polysaccharides. Depending upon biomass composition and/or requirements, the pretreatment can be any from physical, physicochemical, chemical, and biological processes (5–8).
Dilute acid pretreatment at higher temperatures is frequently used to maximize yield of xylose from xylan polymers (9–11). However, pretreatment conditions also result in the formation of furan aldehydes, organic acids, and aromatic compounds, which act as inhibitors of cellular metabolism (5, 12, 13) and consequently lower productivity of the biofuel compound of interest. Furfural and 5-hydroxymethyl furfural (5-HMF) are the furan aldehydes that are generated due to dehydration of xylose and glucose, respectively, under acidic conditions at high temperatures. Furfural is a key inhibitor (12) which acts synergistically with other aldehyde inhibitors (14) to inhibit microbial growth. Furfural causes DNA damage (15–17), oxidative stress (18), and redox imbalance (19) in the microbial cells. Efforts targeted toward engineering furfural tolerance in Escherichia coli have resulted in improved fermentation of lignocellulosic sugars (20). To remove furfural toxicity from growth media, both biological (21) and chemical (22, 23) treatments have been applied. The drawbacks of using biological and chemical treatments are the reduction in the nitrogen content and an increase in processing costs, respectively, of the biomass. Genome analysis of a hydrolysate-resistant E. coli strain has identified the role of genes involved in primary and secondary metabolism, RNA metabolism, sugar transport, vitamin metabolism, and antioxidant activity (24) with potential to confer tolerance against lignocellulosic inhibitors. Published literature supports the idea that a multidimensional approach to engineer furfural tolerance has been successful. Furfural tolerance has been successfully engineered in E. coli via augmenting the cellular NADPH pools (25), silencing genes using NADPH pools to detoxify furfural (26), increasing expression of NADH-specific genes (27), multidrug-resistant (MDR) efflux pumps (28), enhancing DNA repair (29, 30), membrane biogenesis (30), and stress tolerance (30, 31). Tolerance against 5-HMF, a less toxic compound than furfural, has also been achieved using MDR efflux pumps (28) and enhancing NADPH pools (32).
Microbial cells respond to furfural challenge by converting it into furfuryl alcohol. Furfuryl alcohol with a 50% inhibitory concentration (IC50) value of 4.0 g liter−1 (33) is relatively less toxic to E. coli than furfural with an IC50 value of 2.4 g liter−1 (14). Studies have used a strategy whereby deletion of aldehyde reductases (ALRs) of E. coli silenced the competing pathways with a consequent increase in titers of the biomolecule of interest (34, 35). This led us to ponder whether the activity of ALRs can be exploited to engineer tolerance in E. coli toward an aldehyde inhibitor, furfural. In the present study, we screened the collection of strains consisting of single-gene deletions against a series of reductases, oxidoreductases, and dehydrogenases to observe any significant alteration in tolerance of E. coli toward furfural. We used xylose as the primary carbon source in AM1 minimal growth medium, considering its predominance in the acid hydrolysate of biomass. Based on our screening results, we hypothesized that overexpression of yghA will confer tolerance against furfural. We report two important observations in engineered ethanologenic E. coli strain SSK42 (pTrcHisA-yghA), which has high expression levels of the yghA gene compared to the control SSK42 (pTrcHisA) strain. First, in the presence of furfural, SSK42 (pTrcHisA-yghA) has growth advantage compared to the control SSK42 (pTrcHisA) strain. Second, in contrast with SSK42 (pTrcHisA), the presence of inhibitor has no significant influence on maximum ethanol titers of SSK42 (pTrcHisA-yghA).
RESULTS
Screening of E. coli BW25113 mutants for target gene selection.
In the first screen, a relatively low concentration of sugar at 0.2% (wt/vol) was used in AM1 medium. It has been reported that any concentration of xylose above 1.8% starts to exert osmotic stress under similar microaerobic conditions (36). Thus, the nonlimiting, as well as relatively lower, concentration of sugar would help to prevent any confounding variables from influencing the tolerance of microbe toward furfural. Bacterial growth (optical density at 600 nm [OD600]) was monitored at 3 h and 6 h since the most impactful influence of gene mutation on productivity should be observed at the initial stages of growth (Fig. S2 in the supplemental material). A statistical approach was applied in order to select BW25113 mutant strains for the next screening step. Strains that displayed ≤5% variation in readings for both 3-h and 6-h time points (Table S2) were selected. The rationale was that the genetic makeup of such strain(s) is relatively better suited to withstand furfural stress and leads to a predictable growth response. Using the statistical approach, BW25113 derivatives, which had deletion in the yhhX, betB, yphC, ycjS, and yghA genes, respectively, were selected for the next screen.
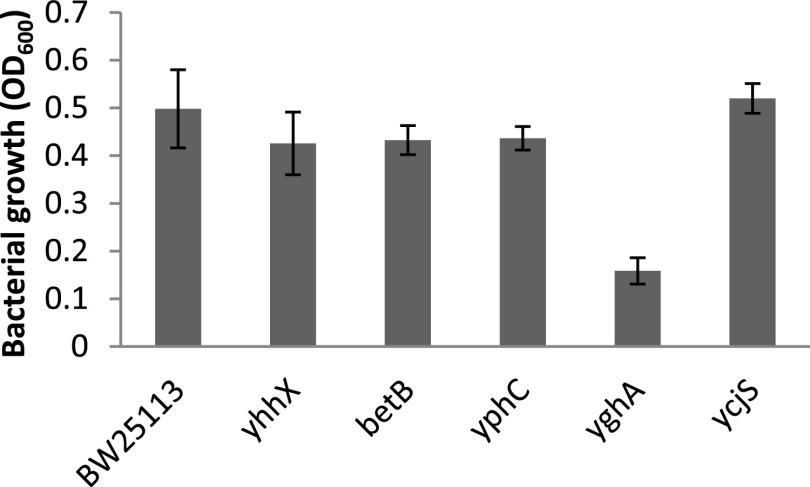
In the second screen, both osmotic and furfural stresses were increased by increasing the concentration of xylose and furfural to 5% (wt/vol) and 1.0 g liter−1, respectively. The high concentration of furfural did not result in significant growth in any of the selected strains at the end of 48 h (Fig. S3). Thus, in the third screening step, the concentration of furfural in AM1 medium was reduced to 0.5 g liter−1 while keeping the xylose concentration constant at 5% (wt/vol). At reduced furfural concentration, growth was observed in all strains at an earlier time point of 24 h (Fig. 1). Among the selected strains, the one with deletion of the yghA gene displayed a 3.15-fold decrease in biomass compared to the wild-type (WT) parent strain BW25113. Based upon this observation, it was hypothesized that overexpression of yghA shall confer tolerance against furfural in E. coli.
FIG 1.
Screening of E. coli BW25113 strain deficient in respective genes indicated in the x axis. Cultures were grown in AM1 media containing 5% xylose (wt/vol) and 0.5 g liter−1 furfural. OD600 was recorded at 24 h. Values are averages of n = 2 independent experiments. Error bars represent SD of the mean.
To validate if the deletion of yghA in BW25113ΔyghA was the only reason for reduced tolerance to furfural, pTrcHisA-yghA was transformed in strain BW25113ΔyghA and tested for its ability to tolerate furfural. The resulting strain was designated BW25113ΔyghA (pTrcHisA-yghA). As a control, an empty pTrcHisA was transformed into BW25113ΔyghA strain, and the resulting strain was labeled BW25113ΔyghA (pTrcHisA). The growth behavior of the transformed strains was monitored under similar sugar and furfural concentrations in the presence of 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) as an inducer. A 1.85-fold increase in bacterial growth (OD600) was observed in strain with cloned yghA compared to one carrying empty plasmid (Fig. S4) at 24 h. This observation suggested that the expression of yghA indeed is associated with an increase in biomass in the presence of furfural.
YghA predominantly uses NADH as a cofactor.
His-tagged YghA was overexpressed in TOP10 cells and purified to homogeneity. YghA showed activity against furfural in the presence of NADH as well as NADPH. The apparent Km values of YghA against furfural in the presence of NADH and NADPH were determined to be 0.03 and 0.05 mM, respectively, whereas the Vmax values against furfural for NADH and NADPH were calculated to be 0.003 and 0.001 mM min−1, respectively. The Kcat/Km values in the presence of NADH and NADPH were determined to be 0.42 and 0.23 mM−1 min−1, respectively. The enzyme kinetics data suggest that YghA has a preference to utilize NADH over NADPH as a cofactor in the presence of furfural as a substrate.
Furfural tolerance conferred by YghA is neutral to the tested carbon sources.
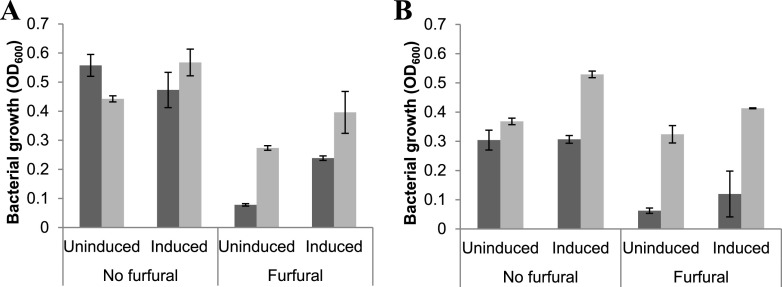
Glucose is the preferred carbon source for E. coli due to enhanced energy and reducing power yield compared to xylose. It was thus tested whether glucose as the sole carbon source results in enhanced tolerance against furfural compared to the condition where xylose is the sole carbon source (Fig. 2). In the presence of 1.0 g liter−1 furfural, biomass increase in the control strain BW25113ΔyghA (pTrcHisA) was observed only in the presence of glucose (induced condition) and not that much in xylose (neither induced nor uninduced) (Fig. 2A). The biomass increase in BW25113ΔyghA (pTrcHisA-yghA), on the other hand, was similar in both of the carbon sources in the presence of the inhibitor. With xylose as carbon source, under induced conditions, the growth reduction for BW25113ΔyghA (pTrcHisA-yghA) in the presence of furfural was 1.28-fold, while that for BW25113ΔyghA (pTrcHisA) was around 2.57-fold (Fig. 2B). BW25113ΔyghA (pTrcHisA-yghA) also showed higher growth under uninduced conditions in the presence of furfural for both xylose and glucose than BW25113ΔyghA (pTrcHisA). It suggested that leaky expression of yghA is sufficient to confer tolerance against furfural under tested conditions. Based on these results, it was concluded that yghA confers tolerance against furfural in a sugar-neutral manner.
FIG 2.
Influence of carbon source in promoting biomass formation in the E. coli BW25113ΔyghA host with either pTrcHisA (dark gray) or with pTrcHisA-yghA (light gray). Media consisted of 1 g liter−1 furfural, 0.1 mM IPTG, and either 5% (wt/vol) of glucose (A) or xylose (B). OD600 was recorded at 48 h. Values are averages of n = 2 independent experiments. Error bars represent SD of the mean.
It was observed that in the absence of furfural (Fig. 2A and B), yghA overexpression hardly resulted in any growth advantage when grown in glucose as a carbon source. However, in the presence of xylose, yghA overexpression resulted in 1.2- and 1.83-fold higher growth under uninduced and induced conditions, respectively (Fig. 2B), suggesting the supportive role of yghA during xylose metabolism beyond furfural tolerance.
The relevance of this idea was tested, under the same conditions, in the context of an E. coli strain, SSK42, whose genome has been optimized for ethanol production using xylose as the primary carbon source under microaerobic conditions (37, 38). Among other genetic changes, the promoter of PDH complex in SSK42 has been replaced by that of gapA, which allows production of an additional NADH under anaerobic conditions (37). However, this additional NADH was not able to rescue growth in the presence of furfural (Fig. S5A and B). It is difficult to ascertain a reason for cessation of growth for SSK42 in capped tube culture, as the strain has been evolutionarily adapted by passaging on AM1-xylose medium plates and then by alternating sole carbon source—either glucose or xylose—in planktonic culture (38), and the redox status of resultant SSK42 is not known. Despite the failure of YghA in improving the growth of the SSK42 strain in the presence of furfural, we observed that furfural could still be metabolized from the culture medium by SSK42 (pTrcHisA-yghA) (Fig. S5C).
In the previous studies, engineering furfural tolerance in E. coli has also been demonstrated to confer tolerance against the relatively less toxic furan aldehyde 5-HMF (28, 32). It was also found that overexpression of yghA leads to tolerance toward 5-HMF, and no 5-HMF could be detected in the media after 48 h in culture containing SSK42 (pTrcHisA-yghA) strain (Fig. S6).
Furfural clearance from medium is similar for both 0.01 and 0.1 mM IPTG concentrations.
The reduction in furfural concentration in uninduced samples (Fig. S5C) made it prudent to investigate the role of IPTG in conferring competitive advantage in clearing furfural from the medium. It is a known fact that chemical properties of IPTG lead to induction of toxicity to E. coli BL21 cells (39). Any reduction in IPTG concentration will be beneficial in reducing toxicity and ultimately contribute toward increasing ethanol productivity by strain SSK42 (pTrcHisA-yghA). Since furfural metabolism is a multigenic trait, we added chloramphenicol to the culture medium to prevent the synthesis of any new cellular proteins and reduce any confounding effect of same on the results.
It was observed that in nongrowing whole cells of SSK42 (pTrcHisA-yghA), ∼60% furfural was removed from the medium in the absence of IPTG at 2 h, representing a clearance rate of 0.60 g liter−1 h−1 (Fig. S7). While in the presence of IPTG, ∼83 to 86% furfural was removed from the medium. Interestingly, similar concentrations of furfural (0.27 to 0.34 g liter−1) at 0.01- and 0.1-mM IPTG concentrations were observed after 2 h, which represents a furfural clearance rate of 0.86 and 0.83 g liter−1 h−1, respectively. It suggests that increasing the concentration of IPTG from 0.01 to 0.1 mM does not confer any catalytic advantage via higher YghA protein levels to expedite the reduction of furfural from the media.
YghA overexpression leads to enhanced microbial and ethanol productivity in the bioreactor.
The Hungate tube is a poorly buffered environment where mixing of nutrients and maintenance of microaerobic environment is also not optimum. The results (Fig. S5C) indicate that SSK42 (pTrcHisA-yghA) is effective in clearing furfural from the media even when an increase in biomass is severely compromised. The bioreactor is a controlled environment wherein pH and mixing of nutrients can be efficiently achieved. Thus, the increase in microbial biomass in a bioreactor was tested while keeping media components similar, as in Fig. S5C. Based upon results obtained from Fig. S7, the concentration of IPTG concentration was decreased from 0.1 to 0.01 mM in the bioreactor.
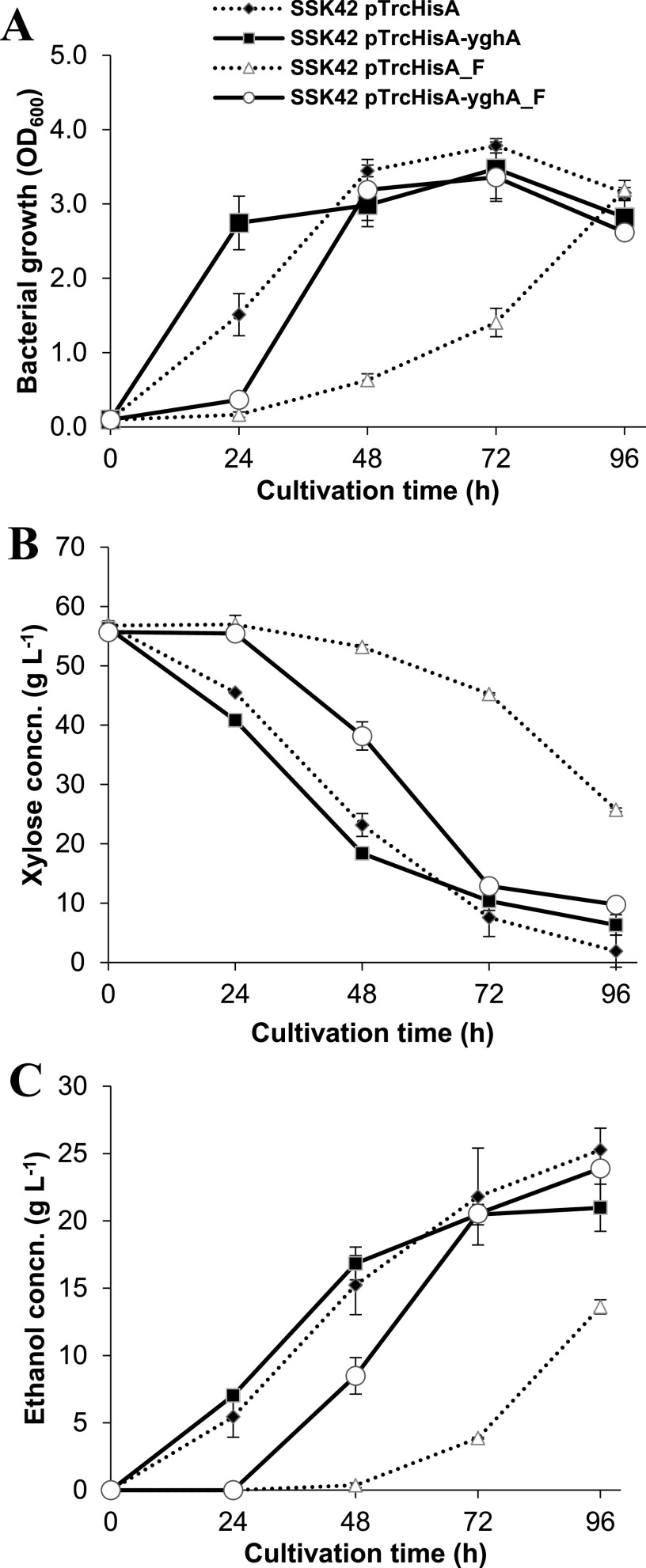
It was observed that in a controlled bioreactor environment and at 5% xylose (wt/vol) concentration, both SSK42 (pTrcHisA) and SSK42 (pTrcHisA-yghA) strains demonstrated their ability to increase the biomass in the presence of furfural (Table 1; Fig. 3A). The rate of increase in biomass for the yghA-overexpressing strain SSK42 (pTrcHisA-yghA) was 0.059 g liter−1 h−1 (1.6-fold higher than the control) and was achieved in the 24- to 48-h time period as against the value of 0.037 g liter−1 h−1 achieved in the 72- to 96-h time period for the control strain SSK42 (pTrcHisA). The corresponding difference in xylose consumption rate was also seen (Fig. 3B). The concentration of furfural at different time points is indicated in Fig. S8. When the ethanol production was evaluated in the presence of furfural and at 96 h, it was observed that the maximum ethanol titer of SSK42 (pTrcHisA-yghA) strain was 1.76-fold higher than that observed in SSK42 (pTrcHisA), and the value was comparable to the titer observed for the strains in the absence of furfural (Fig. 3C). In terms of volumetric ethanol productivity, the maximum value shifted to the 24-h earlier time point in yghA overexpressing strain and also resulted in an increase in productivity of almost 18% for SSK42 (pTrcHisA-yghA) compared to SSK42 (pTrcHisA) (Table 1). The maximum specific ethanol productivity value in the presence of 1 g liter−1 of furfural for SSK42 (pTrcHisA) was 0.35 g g−1 h−1 at 72 to 96 h, while it was 0.40 g g−1 h−1 for SSK42 (pTrcHisA-yghA) at 24 to 48 h (Table S3). The maximum ethanol yield in the case of SSK42 (pTrcHisA-yghA) was around 15% higher than that of SSK42 (pTrcHisA).
TABLE 1.
Impact of furfural on biomass and ethanol formation by SSK42 (pTrcHisA) and SSK42 (pTrcHisA-yghA) strains under low (5%) and high (10%) xylose loading in a bioreactora
| Strain | Furfural concn. (g liter−1) | Data for: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Cells |
Ethanol |
||||||||
| Final concn. (g liter−1) | Specific growth rate |
Max titer (g liter−1) | Productivity |
Yield |
|||||
| Max value (h−1) | Time period (h) | Max. value (g liter−1 h−1) | Time period (h) | g g−1 sugar | % of max. theoretical | ||||
| Strains under 5% xylose (wt/vol) loading | |||||||||
| SSK42 (pTrcHisA) | 0 | 1.89 ± 0.02 | 0.113 ± 0.018 | 0–24 | 25.27 ± 0.02 | 0.41 ± 0.03 | 24–48 | 0.46 ± 0.03 | 89.84 ± 5.54 |
| SSK42 (pTrcHisA-yghA) | 0 | 1.74 ± 0.20 | 0.138 ± 0.004 | 0–24 | 20.97 ± 1.74 | 0.41 ± 0.05 | 24–48 | 0.42 ± 0.04 | 81.79 ± 8.23 |
| SSK42 (pTrcHisA) | 1 | 1.60 ± 0.06 | 0.056 ± 0.015 | 24–48 | 13.61 ± 0.53 | 0.41 ± 0.02 | 72–96 | 0.44 ± 0.02 | 85.53 ± 4.26 |
| SSK42 (pTrcHisA-yghA) | 1 | 1.77 ± 0.04 | 0.091 ± 0.007 | 24–48 | 23.90 ± 2.99 | 0.50 ± 0.04 | 48–72 | 0.52 ± 0.08 | 101.58 ± 14.99 |
| Strains under 10% xylose (wt/vol) loading | |||||||||
| SSK42 (pTrcHisA) | 0 | 2.31 ± 0.02 | 0.127 ± 0.008 | 0–24 | 31.40 ± 4.25 | 0.50 ± 0.07 | 48–72 | 0.46 ± 0.13 | 89.63 ± 24.56 |
| SSK42 (pTrcHisA-yghA) | 0 | 2.55 ± 0.08 | 0.139 ± 0.000 | 0–24 | 39.72 ± 4.38 | 0.62 ± 0.05 | 24–48 | 0.47 ± 0.07 | 92.13 ± 14.61 |
| SSK42 (pTrcHisA) | 1 | 1.36 ± 0.20 | 0.030 ± 0.013 | 48–72 | 7.78 ± 2.69 | 0.29 ± 0.11 | 96–120 | 0.35 ± 0.07 | 68.92 ± 12.80 |
| SSK42 (pTrcHisA-yghA) | 1 | 2.75 ± 0.12 | 0.104 ± 0.003 | 24–48 | 41.58 ± 1.66 | 0.72 ± 0.13 | 48–72 | 0.50 ± 0.06 | 97.19 ± 12.64 |
Results are average of n = 2 independent experiments. Error is SD of the mean.
FIG 3.
Fermentation profile of SSK42 strain with or without the overexpression of the yghA gene in a bioreactor at 5% xylose (wt/vol) load. “F” indicates furfural treatment at 1 g liter−1. Bacterial growth (A) and xylose (B) and ethanol (C) concentrations are indicated in the respective panels. Values are averages of n = 2 independent experiments. Error bars represent SD of the mean.
An increase in sugar concentration results in osmotic stress response and is characterized by expression of soxS, sodA, and katE genes, which are also involved in combating oxidative stress (40). This oxidative stress is in addition to the reactive oxygen species (ROS) accumulation induced in microbial cell in response to the furfural challenge (18). Consequently, it is desirable from an applied microbiology perspective that the furfural tolerance of ethanologenic SSK42 (pTrcHisA-yghA) strain can also withstand the stress imposed by high sugar concentration. Thus, the growth dynamics of strains SSK42 (pTrcHisA) and SSK42 (pTrcHisA-yghA) were tested at 10% sugar (wt/vol) loading.
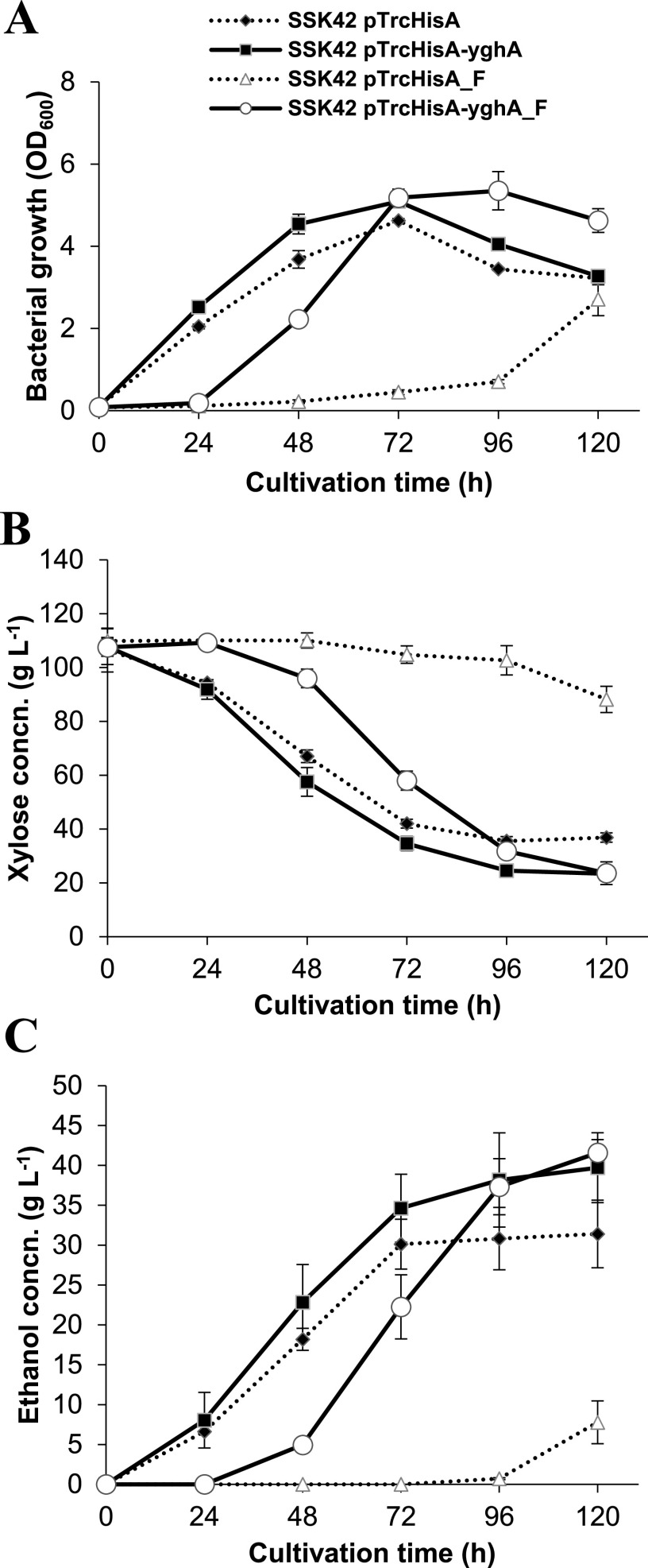
It was observed that the maximum biomass titer at 10% sugar (wt/vol) load and in the presence of furfural for strain SSK42 (pTrcHisA-yghA) was 2.75 ± 0.12 g liter−1, while for SSK42 (pTrcHisA), it was 1.36 ± 0.20 g liter−1, which represented an ∼2-fold increase in biomass concentration and 1.5-fold increase in biomass productivity for YghA-overexpressing strain (Table 1; Fig. 4A). A similar difference in xylose consumption was also observed (Fig. 4B). The difference in maximum titers of ethanol was much more remarkable. The maximum ethanol titers for SSK42 (pTrcHisA-yghA) and SSK42 (pTrcHisA) were 41.58 ± 1.66 and 7.78 ± 2.69 g liter−1, respectively, which represented a 5.3-fold higher ethanol titer. For SSK42 (pTrcHisA-yghA), a 1.4-fold increase in ethanol yield per gram of xylose consumed was also observed compared to SSK42 (pTrcHisA). The maximum theoretical yields for ethanol in the cases of SSK42 (pTrcHisA-yghA) and SSK42 (pTrcHisA) were 97.19 ± 12.64 and 68.92 ± 12.80%, respectively. It represents an ∼30% increase in the efficiency of ethanol production. The increase in ethanol productivity value was also, remarkably, 2.5-fold higher for SSK42 (pTrcHisA-yghA) than SSK42 (pTrcHisA). At the end of 96 h, the average ethanol productivity was 0.39 g liter−1 h−1 (Fig. 4C), which was comparable with the 0.42 g liter−1 h−1 value obtained by overexpression of SDR pump mdtJI (28). The maximum specific ethanol productivity value in the presence of 1 g liter−1 of furfural for SSK42 (pTrcHisA) was 0.34 g g−1 h−1 at 96 to 120 h, while it was 0.39 g g−1 h−1 for SSK42 (pTrcHisA-yghA) at 48 to 72 h (Table S3). Our fermentation results prove that overexpression of YghA with relatively higher affinity toward NADH compared to NADPH is a useful strategy to confer tolerance against furfural at high sugar loadings.
FIG 4.
Fermentation profile of SSK42 strain with or without the overexpression of the yghA gene in a bioreactor at increased osmotic stress exerted by 10% xylose (wt/vol). Bacterial growth (A) and xylose (B) and ethanol (C) concentrations are indicated in the respective panels. “F” indicates furfural treatment at 1 g liter−1. Values are averages of n = 2 independent experiments. Error bars represent SD of the mean.
DISCUSSION
In this study, we used a three-step screening strategy to investigate a set of E. coli oxidoreductases, dehydrogenases, and reductases that can confer tolerance against an aldehyde inhibitor, furfural, which is commonly generated during pretreatment of lignocellulose. Since, at the first screening step, either deletion or overexpression of the respective gene would be advantageous to the microbe to withstand furfural stress, our statistical approach was a nonbiased one to identify potential gene targets. Additionally, the stepwise increase in stresses (both osmotic and inhibitor) was helpful in keeping confounding factors at bay in the first screening step and select strains for the next step of screening (Fig. S2 in the supplemental material). We found that the previously poorly characterized oxidoreductase YghA is able to confer tolerance against 1 g liter−1 furfural and ∼10% xylose (wt/vol) as sugar source in an ethanologenic E. coli strain SSK42. Other screening studies have identified overexpression of thyA (29), mdtJI (28), and groESL and lpcA (30) to be effective in conferring tolerance against a concentration range of 0.75 to 1.25 g liter−1of furfural.
Compared to glucose, xylose is a poor source of energy and reducing equivalents that are essential for microbial growth. The problem is further compounded under oxygen limitation where the cells are mainly dependent upon substrate-level phosphorylation and transhydrogenases for generation of energy and reducing equivalents, respectively. Under anaerobic conditions, the switch from glucose to xylose results in the reduction of cellular generation of NADH/FADH2 and NADPH by 1.4- and 2.8-fold, respectively (41). This makes NADPH a scarce source for anabolic reactions under oxygen limitation, and its diversion for furfural detoxification further depletes cellular NADPH pools and compromises an increase in biomass. Compared to NADPH, YghA has relatively higher affinity for using NADH as a cofactor for furfural detoxification, which should be beneficial in increasing cellular NADPH pools. The low apparent Km values of YghA for furfural (0.03 mM) in the presence of NADH also reflect in our data where expression at 0.01 mM IPTG concentration was sufficient to confer tolerance against furfural, and a further increase in YghA expression conferred no additional growth advantage in the presence of furfural. On the other hand, with an apparent Km of 0.4 mM against furfural, overexpression of NADH-specific oxidoreductase FucO has also been reported to confer tolerance against furfural (27). Less is known about the function of YghA in the native E. coli host, and no structural studies have been carried out on it. In an earlier study, YghA has been defined to be an NADP+-dependent aldehyde reductase with activity toward butyraldehyde and decanal (34). YghA has been reported to harbor a NAD(P) binding Rossman fold domain (42). Based on our results, we would further broaden the use of cofactor and aldehyde substrate by YghA.
Interestingly, the tolerance conferred by an oxidoreductase (YghA) against furfural and the maximum ethanol titers obtained are comparable to that offered by polyamines (43) and a small multidrug resistance (SDR) pump (28). This observation further lends credence to the fact that microbial metabolism harbors a multidimensional capability to efficiently counter the metabolic challenge of furfural, which exerts its toxicity by inhibiting different metabolic targets of cellular machinery.
MATERIALS AND METHODS
Strains and media.
All strains used in this study are listed in Table S1 in the supplemental material. A set of mutants of strain BW25113 with a deletion of one additional gene was used in this study (44). The said strains were obtained from Coli Genetics Stock Center (CGSC), Yale University, New Haven, CT. The workhorse microbial strain of this study is SSK42 (genotype E. coli B PgapA PDH Δldh ΔfrdA ΔpflB), which has undergone extensive evolutionary adaptation in AM1 minimal media with either glucose or xylose as carbon sources (35, 44). The kanamycin resistance cassette was removed from SSK42 using temperature-sensitive pCP20, which harbors the FLP recombinase gene (45). The genome of E. coli B served as a template for PCR amplification of yghA gene using primer set YghA_pTrcHis_NheI, CCCGCTAGCATGTCTCATTTAAAAGACCCGACC, and YghA_pTrcHisA_BamHI, CCCGGATCCTTAACCTAAATGCTCGCCG. The open reading frame (ORF) of yghA was cloned into pTrcHisA at Nhe1 and BamH1 restriction sites to obtain the pTrcHisA-yghA construct. For physiological analysis, strains were grown in AM1 medium having composition (NH4)2HPO4 (19.92 mM), NH4H2PO4 (7.56 mM), KCl (2.00 mM), MgSO4·7H2O (1.50 mM), betaine HCl (1.00 mM), FeCl3·6H2O (8.88 μM), CoCl2·6H2O (1.26 μM), CuCl2·2H2O (0.88 μM), ZnCl2 (2.20 μM), Na2MoO4·2H2O (1.24 μM), H3BO3 (1.21 μM), MnCl2·4H2O (2.50 μM), and CaCl2·2H2O (1.36 μM) (46). Concentrations of xylose used have been mentioned at the respective experiments, and ampicillin was used at 12.5 mg liter−1.
Culture conditions.
Glycerol stocks of microbial strains were revived on petri plates containing AM1 media with 2% xylose (wt/vol) as carbon source. Two additional restreaks, each from a well-isolated colony, were performed, and a third streak was used to start primary overnight cultures, which were grown in Hungate tubes with capacity of 18 ml in a shaker incubator at 37°C and 150 rpm. In these tubes, 10 ml AM1 medium containing xylose, at the concentration mentioned against respective experiment, was used. All secondary cultures were seeded at OD600 of 0.1. Expression of yghA was induced using either 0.01 or 0.1 mM IPTG as indicated against the respective experiment.
Screening of E. coli BW25113 strains.
Wild-type E. coli BW25113 was screened for sensitivity to the following concentrations of furfural: 0, 0.25, 0.50, 0.75, 1.00, 1.25, 1.50, 1.75, 2.00, 2.25, and 2.50 g liter−1. One set consisted of 0.2% xylose (wt/vol) as the sole carbon source, while another set consisted of 0.2% (wt/vol) glucose. For both sugar sets, the 50% lethal dose (LD50) concentration of furfural was determined to be 0.25 g liter−1 at 3 h of cultivation (Fig. S1 in the supplemental material). Furfural tolerance in the presence of xylose was of our primary interest; thus, it was included for further screening. The first screen of 54 E. coli BW25113 mutant and BW25113 parent strains was performed at 0.25 g liter−1 furfural and 0.2% xylose (wt/vol). Selected strains from the first screens were subjected to a second screen consisting of 1 g liter−1 furfural and 5% xylose (wt/vol). In the second screen, none of the selected strains displayed a significant increase in biomass at the end of 48 h. Thus, the same selected set of strains was subjected to the third screen consisting of 0.5 g liter−1 furfural and 5% xylose (wt/vol) in AM1 media. A significant change in biomass was observed in strains at 24 h at the reduced furfural concentration. All observations were recorded at least in duplicate.
Influence of IPTG concentration on furfural removal.
Cultures of SSK42 (pTrcHisA) and SSK42 (pTrcHisA-yghA) strains were grown overnight in capped tubes in the presence of either 0, 0.01, or 0.1 mM IPTG concentration. After around 16 h, cultures were treated with 50 mg liter−1 chloramphenicol for 2 h. Furfural removal was then monitored in nongrowing whole-cell cultures at an OD600 of 2.0 in the medium consisting of 5% xylose (wt/vol), 12.5 mg liter−1 ampicillin, 50 mg liter−1 chloramphenicol, 2.0 g liter−1 furfural, and either 0, 0.01, or 0.1 mM IPTG. Tubes were incubated in 37°C at 150 rpm and sampled at indicated time points.
Purification of YghA.
E. coli TOP10 harboring 6×His tagged pTrcHisA-yghA was cultured overnight in LB medium in the presence of ampicillin. The secondary culture was started using 1% of the overnight culture and induced using 0.1 mM IPTG at OD600 of ∼0.4. Cells were harvested after 4 h and stored at −80°C overnight. Pellets were resuspended further in lysis buffer (20 mM Tris-HCl [pH 7.5], 500 mM NaCl, and 5 mM imidazole). The cells were lysed using a microtip sonicator. After centrifugation of lysate at ∼9,000 × g (1 h and 4°C), the supernatant was filtered (0.45 μm) and purified via Ni-nitrilotriacetic acid (Ni-NTA) metal affinity chromatography. The purified protein was dialyzed in 100 mM sodium phosphate buffer, and the purity was checked on SDS-PAGE gel. The concentration of YghA was determined by bicinchoninic acid (BCA) protein assay kit (G-Biosciences, MO, USA).
Fermentation.
Primary cultures were started in 250-ml shake flasks with 100 ml AM1 medium with 5% xylose (wt/vol), 12.5 mg liter−1 ampicillin, and 0.01 mM IPTG. After around 16 h growth, cells were harvested. Fermenters were seeded at OD600 of 0.1 in 300 ml AM1 media consisting of 1 g liter−1 furfural, 12.5 mg liter−1 ampicillin, 0.01 mM IPTG, and either 5% or 10% xylose (wt/vol) as sugar source. pH was maintained at 7.0 using 2N KOH. Air in headspace was pumped in at 0.03 liter per minute (LPM) for the first 24 h and then increased to 0.04 LPM for the remaining fermentation period.
Analysis.
Xylose and ethanol concentration was determined using Shimadzu HPLC with Aminex HPX-87H (300 by 7.8 mm) column and refractive index (RI) detector. The column temperature was maintained at 60°C, and that of the detector was maintained at 50°C. We used 4 mM H2SO4 as a mobile phase at a flow rate of 0.6 ml min−1. A reference standard of 1 g liter−1 for each metabolite was obtained from Absolute Standards, USA. Biomass was estimated by recording optical density at 600 nm using a UV-visible (UV-Vis) spectrophotometer (Ultrospec 3100; Amersham Biosciences). The concentration of furfural was estimated using the UV-Vis method as reported before (47).
ACKNOWLEDGMENTS
We would like to thank Juhi Sharma and the late Mohammad Asad for their kind help in enzyme assays.
A senior research fellowship was provided to S.B.J. by the Indian Council of Medical Research. This study was funded by Department of Biotechnology, Government of India, via Bioenergy Centre grant no. BT/PR/Centre/03/2011-Phase II.
Footnotes
Supplemental material is available online only.
Contributor Information
Syed Shams Yazdani, Email: shams@icgeb.res.in.
Pablo Ivan Nikel, Novo Nordisk Foundation Center for Biosustainability.
REFERENCES
- 1.Alonso DM, Bond JQ, Dumesic JA. 2010. Catalytic conversion of biomass to biofuels. Green Chem 12:1493–1513. doi: 10.1039/c004654j. [DOI] [Google Scholar]
- 2.Hahn-Hägerdal B, Galbe M, Gorwa-Grauslund MF, Lidén G, Zacchi G. 2006. Bio-ethanol - the fuel of tomorrow from the residues of today. Trends Biotechnol 24:549–556. doi: 10.1016/j.tibtech.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 4.Hinman ND, Schell DJ, Riley J, Bergeron PW, Walter PJ. 1992. Preliminary estimate of the cost of ethanol production for ssf technology. Appl Biochem Biotechnol 34–35:639–649. doi: 10.1007/BF02920584. [DOI] [Google Scholar]
- 5.Sun Y, Cheng J. 2002. Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour Technol 83:1–11. doi: 10.1016/s0960-8524(01)00212-7. [DOI] [PubMed] [Google Scholar]
- 6.Mosier N, Wyman C, Dale B, Elander R, Lee YY, Holtzapple M, Ladisch M. 2005. Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour Technol 96:673–686. doi: 10.1016/j.biortech.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 7.Saha BC, Cotta MA. 2010. Comparison of pretreatment strategies for enzymatic saccharification and fermentation of barley straw to ethanol. N Biotechnol 27:10–16. doi: 10.1016/j.nbt.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 8.Saha BC. 2003. Hemicellulose bioconversion. J Ind Microbiol Biotechnol 30:279–291. doi: 10.1007/s10295-003-0049-x. [DOI] [PubMed] [Google Scholar]
- 9.Hendriks ATWM, Zeeman G. 2009. Pretreatments to enhance the digestibility of lignocellulosic biomass. Bioresour Technol 100:10–18. doi: 10.1016/j.biortech.2008.05.027. [DOI] [PubMed] [Google Scholar]
- 10.Saha BC, Iten LB, Cotta MA, Wu YV. 2005. Dilute acid pretreatment, enzymatic saccharification and fermentation of wheat straw to ethanol. Process Biochem 40:3693–3700. doi: 10.1016/j.procbio.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Taherzadeh MJ, Karimi K. 2007. Bioethanol review. BioResources 2:472–499. [Google Scholar]
- 12.Mills TY, Sandoval NR, Gill RT. 2009. Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli. Biotechnol Biofuels 2:26. doi: 10.1186/1754-6834-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zha Y, Westerhuis JA, Muilwijk B, Overkamp KM, Nijmeijer BM, Coulier L, Smilde AK, Punt PJ. 2014. Identifying inhibitory compounds in lignocellulosic biomass hydrolysates using an exometabolomics approach. BMC Biotechnol 14:22. doi: 10.1186/1472-6750-14-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaldivar J, Martinez A, Ingram LO. 1999. Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol Bioeng 65:24–33. doi:. [DOI] [PubMed] [Google Scholar]
- 15.Khan QA, Shamsi FA, Hadi SM. 1995. Mutagenicity of furfural in plasmid DNA. Cancer Lett 89:95–99. doi: 10.1016/0304-3835(95)90163-9. [DOI] [PubMed] [Google Scholar]
- 16.Hadi SM, Shahabuddin, Rehman A. 1989. Specificity of the interaction of furfural with DNA. Mutat Res Lett 225:101–106. doi: 10.1016/0165-7992(89)90125-5. [DOI] [PubMed] [Google Scholar]
- 17.Rahman SA, Hadi SM. 1991. Reaction of furfural and methylfurfural with DNA: use of single-strand-specific nucleases. Food Chem Toxicol 29:719–721. doi: 10.1016/0278-6915(91)90131-P. [DOI] [PubMed] [Google Scholar]
- 18.Allen SA, Clark W, McCaffery JM, Cai Z, Lanctot A, Slininger PJ, Liu ZL, Gorsich SW. 2010. Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol Biofuels 3:2–10. doi: 10.1186/1754-6834-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ask M, Bettiga M, Mapelli V, Olsson L. 2013. The influence of HMF and furfural on redox-balance and energy-state of xylose-utilizing Saccharomyces cerevisiae. Biotechnol Biofuels 6:22. doi: 10.1186/1754-6834-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Yomano LP, Lee JY, York SW, Zheng H, Mullinnix MT, Shanmugam KT, Ingram LO. 2013. Engineering furfural tolerance in Escherichia coli improves the fermentation of lignocellulosic sugars into renewable chemicals. Proc Natl Acad Sci USA 110:4021–4026. doi: 10.1073/pnas.1217958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X, Wang Y, Liu W, Bao J. 2011. Biological removal of inhibitors leads to the improved lipid production in the lipid fermentation of corn stover hydrolysate by Trichosporon cutaneum. Bioresour Technol 102:9705–9709. doi: 10.1016/j.biortech.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 22.Martinez A, Rodriguez ME, Wells ML, York SW, Preston JF, Ingram LO. 2001. Detoxification of dilute acid hydrolysates of lignocellulose with lime. Biotechnol Prog 17:287–293. doi: 10.1021/bp0001720. [DOI] [PubMed] [Google Scholar]
- 23.Martinez A, Rodriguez ME, York SW, Preston JF, Ingram LO. 2000. Effects of Ca(OH) 2 treatments (“overliming”) on the composition and toxicity of bagasse hemicellulose hydrolysates. Biotechnol Bioeng 69:526–536. doi:. [DOI] [PubMed] [Google Scholar]
- 24.Warner JR, Reeder PJ, Karimpour-Fard A, Woodruff LBA, Gill RT. 2010. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat Biotechnol 28:856–862. doi: 10.1038/nbt.1653. [DOI] [PubMed] [Google Scholar]
- 25.Jilani SB, Dev C, Eqbal D, Jawed K, Prasad R, Yazdani SS. 2020. Deletion of pgi gene in E. coli increases tolerance to furfural and 5-hydroxymethyl furfural in media containing glucose-xylose mixture. Microb Cell Fact 19:153. doi: 10.1186/s12934-020-01414-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller EN, Jarboe LR, Yomano LP, York SW, Shanmugam KT, Ingram LO. 2009. Silencing of NADPH-dependent oxidoreductase genes (yqhD and dkgA) in furfural-resistant ethanologenic Escherichia coli. Appl Environ Microbiol 75:4315–4323. doi: 10.1128/AEM.00567-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Miller EN, Yomano LP, Zhang X, Shanmugam KT, Ingram LO. 2011. Increased furfural tolerance due to overexpression of NADH-dependent oxidoreductase FucO in Escherichia coli strains engineered for the production of ethanol and lactate. Appl Environ Microbiol 77:5132–5140. doi: 10.1128/AEM.05008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurgan G, Panyon LA, Rodriguez-Sanchez Y, Pacheco E, Nieves LM, Mann R, Nielsen DR, Wanga X. 2019. Bioprospecting of native efflux pumps to enhance furfural tolerance in ethanologenic Escherichia coli. Appl Environ Microbiol 85:e02985-18. doi: 10.1128/AEM.02985-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng H, Wang X, Yomano LP, Shanmugam KT, Ingram LO. 2012. Increase in furfural tolerance in ethanologenic Escherichia coli LY180 by plasmid-based expression of ThyA. Appl Environ Microbiol 78:4346–4352. doi: 10.1128/AEM.00356-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glebes TY, Sandoval NR, Reeder PJ, Schilling KD, Zhang M, Gill RT. 2014. Genome-wide mapping of furfural tolerance genes in Escherichia coli. PLoS One 9:e87540. doi: 10.1371/journal.pone.0087540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glebes TY, Sandoval NR, Gillis JH, Gill RT. 2015. Comparison of genome-wide selection strategies to identify furfural tolerance genes in Escherichia coli. Biotechnol Bioeng 112:129–140. doi: 10.1002/bit.25325. [DOI] [PubMed] [Google Scholar]
- 32.Miller EN, Turner PC, Jarboe LR, Ingram LO. 2010. Genetic changes that increase 5-hydroxymethyl furfural resistance in ethanol-producing Escherichia coli LY180. Biotechnol Lett 32:661–667. doi: 10.1007/s10529-010-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaldivar J, Martinez A, Ingram LO. 2000. Effect of alcohol compounds found in hemicellulose hydrolysate on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol Bioeng 68:524–530. doi:. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez GM, Atsumi S. 2014. Toward aldehyde and alkane production by removing aldehyde reductase activity in Escherichia coli. Metab Eng 25:227–237. doi: 10.1016/j.ymben.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fatma Z, Jawed K, Mattam AJ, Yazdani SS. 2016. Identification of long chain specific aldehyde reductase and its use in enhanced fatty alcohol production in E. coli. Metab Eng 37:35–45. doi: 10.1016/j.ymben.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 36.Purvis JE, Yomano LP, Ingram LO. 2005. Enhanced trehalose production improves growth of Escherichia coli under osmotic stress. Appl Environ Microbiol 71:3761–3769. doi: 10.1128/AEM.71.7.3761-3769.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munjal N, Mattam AJ, Pramanik D, Srivastava PS, Yazdani SS. 2012. Modulation of endogenous pathways enhances bioethanol yield and productivity in Escherichia coli. Microb Cell Fact 11:145. doi: 10.1186/1475-2859-11-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jilani SB, Venigalla SSK, Mattam AJ, Dev C, Yazdani SS. 2017. Improvement in ethanol productivity of engineered E. coli strain SSY13 in defined medium via adaptive evolution. J Ind Microbiol Biotechnol 44:1375–1384. doi: 10.1007/s10295-017-1966-4. [DOI] [PubMed] [Google Scholar]
- 39.Dvorak P, Chrast L, Nikel PI, Fedr R, Soucek K, Sedlackova M, Chaloupkova R, Lorenzo V, Prokop Z, Damborsky J. 2015. Exacerbation of substrate toxicity by IPTG in Escherichia coli BL21(DE3) carrying a synthetic metabolic pathway. Microb Cell Fact 14:201–215. doi: 10.1186/s12934-015-0393-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smirnova GV, Zakirova ON, Oktyabr'skii ON. 2001. The role of antioxidant systems in the response of Escherichia coli to heat shock. Microbiology 70:512–518. doi: 10.1023/A:1012391601933. [DOI] [PubMed] [Google Scholar]
- 41.Gonzalez JE, Long CP, Antoniewicz MR. 2017. Comprehensive analysis of glucose and xylose metabolism in Escherichia coli under aerobic and anaerobic conditions by 13C metabolic flux analysis. Metab Eng 39:9–18. doi: 10.1016/j.ymben.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Kothari A, Krummenacker M, Latendresse M, Mun L, Shearer AG, Ong Q, Paley S, Schro I, Subhraveti P, Travers M, Weerasinghe D, Weiss V, Collado-Vides J, Gunsalus RP, Paulsen I, Karp PD. 2013. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res 41:D605–D612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geddes RD, Wang X, Yomano LP, Miller EN, Zheng H, Shanmugam KT, Ingram LO. 2014. Polyamine transporters and polyamines increase furfural tolerance during xylose fermentation with ethanologenic Escherichia coli strain LY180. Appl Environ Microbiol 80:5955–5964. doi: 10.1128/AEM.01913-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez A, Grabar TB, Shanmugam KT, Yomano LP, York SW, Ingram LO. 2007. Low salt medium for lactate and ethanol production by recombinant Escherichia coli B. Biotechnol Lett 29:397–404. doi: 10.1007/s10529-006-9252-y. [DOI] [PubMed] [Google Scholar]
- 47.Martinez A, Rodriguez ME, York SW, Preston JF, Ingram LO. 2000. Use of UV absorbance to monitor furans in dilute acid hydrolysates of biomass. Biotechnol Prog 16:637–641. doi: 10.1021/bp0000508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3, Fig. S1 to S8. Download aem.01855-21-s0001.pdf, PDF file, 0.9 MB (926.3KB, pdf)