ABSTRACT
Magnetotactic bacteria (MTB) are prokaryotes that form intracellular magnetite (Fe3O4) or greigite (Fe3S4) nanocrystals with tailored sizes, often in chain configurations. Such magnetic particles are each surrounded by a lipid bilayer membrane, called a magnetosome, and provide a model system for studying the formation and function of specialized internal structures in prokaryotes. Using fluorescence-coupled scanning electron microscopy, we identified a novel magnetotactic spirillum, XQGS-1, from freshwater Xingqinggong Lake, Xi’an City, Shaanxi Province, China. Phylogenetic analyses based on 16S rRNA gene sequences indicate that strain XQGS-1 represents a novel genus of the Alphaproteobacteria class in the Proteobacteria phylum. Transmission electron microscopy analyses reveal that strain XQGS-1 forms on average 17 ± 3 magnetite magnetosome particles with an ideal truncated octahedral morphology, with an average length and width of 88.3 ± 11.7 nm and 83.3 ± 11.0 nm, respectively. They are tightly organized into a single chain along the cell long axis close to the concave side of the cell. Intrachain magnetic interactions likely result in these large equidimensional magnetite crystals behaving as magnetically stable single-domain particles that enable bacterial magnetotaxis. Combined structural and chemical analyses demonstrate that XQGS-1 cells also biomineralize intracellular amorphous calcium phosphate (2 to 3 granules per cell; 90.5- ± 19.3-nm average size) and weakly crystalline calcium carbonate (2 to 3 granules per cell; 100.4- ± 21.4-nm average size) in addition to magnetite. Our results expand the taxonomic diversity of MTB and provide evidence for intracellular calcium phosphate biomineralization in MTB.
IMPORTANCE Biomineralization is a widespread process in eukaryotes that form shells, teeth, or bones. It also occurs commonly in prokaryotes, resulting in more than 60 known minerals formed by different bacteria under wide-ranging conditions. Among them, magnetotactic bacteria (MTB) are remarkable because they might represent the earliest organisms that biomineralize intracellular magnetic iron minerals (i.e., magnetite [Fe3O4] or greigite [Fe3S4]). Here, we report a novel magnetotactic spirillum (XQGS-1) that is phylogenetically affiliated with the Alphaproteobacteria class. In addition to magnetite crystals, XQGS-1 cells form intracellular submicrometer calcium carbonate and calcium phosphate granules. This finding supports the view that MTB are also an important microbial group for intracellular calcium carbonate and calcium phosphate biomineralization.
KEYWORDS: magnetotactic bacteria, biomineralization, magnetosome, calcium phosphate, calcium carbonate
INTRODUCTION
Magnetotactic bacteria (MTB) are phylogenetically and morphologically diverse prokaryotes that tend to live at or just below the oxic-anoxic transition zone (OATZ) in aquatic environments (1). They form intracellular organelles, known as magnetosomes, in which membrane-surrounded magnetite (Fe3O4) and/or greigite (Fe3S4) nanocrystals biomineralize (2, 3). Within most MTB, these magnetic particles are organized into a chain or chains that enable cells to swim along geomagnetic field lines, a unique behavior initially called magnetotaxis (2) and subsequently modified to magnetoaerotaxis (4, 5), or magnetochemotaxis (6). Taking advantage of magnetoaerotaxis or magnetochemotaxis, MTB cells efficiently shuttle up and down across the OATZ to acquire nutrients and redox power for cell growth and biomineralization so that they play roles in driving the biogeochemical cycling of elemental C, N, P, S, and Fe at the aquatic OATZ (7–9). MTB remains (i.e., magnetofossils) are also potential carriers of proxy information for paleomagnetic and paleoenvironmental reconstructions (10–12) because magnetite crystals formed by MTB generally have magnetically ideal single-domain (SD) properties and group- or species-specific morphology (13–16).
MTB also often precipitate other intracellular inclusions, such as elemental sulfur (S0) granules, nitrate-storing vacuoles, polyphosphate globules, polyhydroxybutyrates (PHBs), Ba-rich particles, and even calcium carbonate (CaCO3), which have been reported only for some strains in the Achromatium genus and the Cyanobacteria phylum (7–9, 17–26). These observations reinforce the view that MTB may be a model system for studying biomineralization and the physiological function of intracellular biominerals within prokaryotes (9, 22). However, compared to magnetosome particles, morphological and chemical features of these biominerals and their corresponding cellular purposes and molecular mechanisms within MTB are largely unknown. Also, it is unclear whether MTB precipitate calcium phosphate, even though calcium-rich polyphosphate granules are common in MTB, and calcium phosphate is widespread in Bacteria and Eukaryota, such as within cyanobacteria and animals (27, 28). Previous studies have shown that calcium phosphate and calcium-rich polyphosphate can be distinguished by their Ca/P ratios; i.e., the former have higher Ca/P ratios (1.2 to 2.2) than the latter (0.22 to 0.26) (29, 30).
Here, we report a novel magnetotactic spirillum from freshwater Xingqinggong Lake, Xi’an City, western China, identified with a fluorescence in situ hybridization-coupled scanning electron microscopy (FISH-SEM) approach (20). We present 16S rRNA gene analyses to assess its phylogenetics and discuss the significance of calcium carbonate and calcium phosphate biominerals produced intracellularly by this bacterium.
RESULTS
MTB identification and phylogenetic analysis.
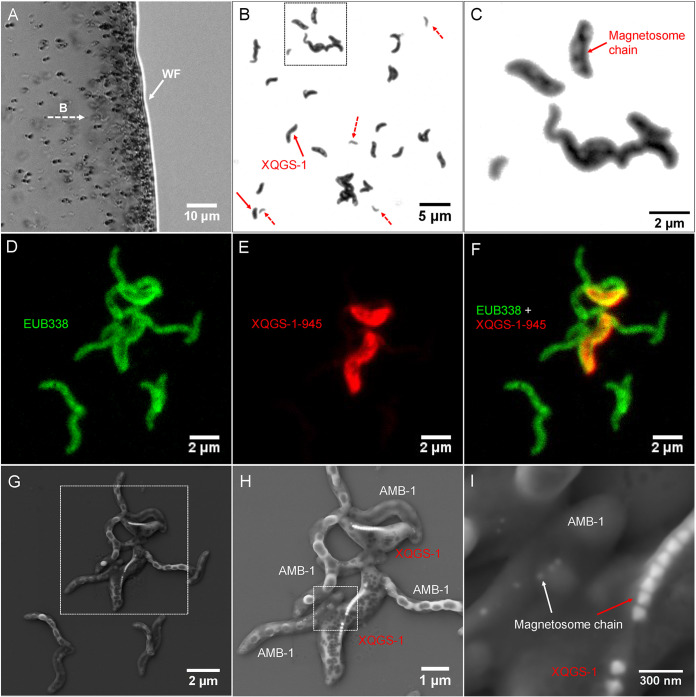
As shown in Fig. 1, living MTB cells were collected magnetically in the laboratory from a sediment microcosm (from Lake Xingqinggong) dominated by spirillum and vibrio MTB cells for molecular and microscopic analyses. Optical microscopic observations indicate that the magnetic collection is dominated by one group of spirillum bacteria (tentatively named XQGS-1) with larger sizes than those of other spirilla. Molecular analyses based on 30 16S rRNA gene sequences from this collection indicate that 22 sequences have high identities (>99%) with each other; the other 8 sequences share high identities (>97%) with known bacteria such as Methylobacillus sp. and Magnetospirillum sp. Specifically, six clones of Magnetospirillum sp. share 99.8% identity of 16S rRNA gene sequences with uncultured MTB Magnetospirillum sp. clone WYH-24 (GenBank accession number JX537774) (31). Two clones of Methylobacillus sp. share 97.1% identity of 16S rRNA gene sequences with uncultured non-MTB Methylobacillus sp. clone CH1-43 (GenBank accession number JX079398) (see Table S1 in the supplemental material).
FIG 1.
Morphological identification of strain XQGS-1 cells with a coupled FISH-SEM approach. (A) Optical micrograph of living MTB cells swimming parallel to an applied magnetic field (B) and gathering at a droplet water front (WF). (B) Optical micrograph of magnetically collected bacteria stained with crystal violet. XQGS-1 cells are indicated by solid red arrows; other small spirilla are indicated by dashed red arrows. (C) Close-up view of the bacteria indicated by the dashed-line box in panel B. A magnetosome chain structure within some XQGS-1 cells is observed by optical microscopy. (D) Fluorescence micrograph of bacteria hybridized in situ with the 5′-FAM-labeled universal bacterial probe EUB338. (E) Fluorescence micrograph of bacteria hybridized in situ with the 5′-Cy3-labeled XQGS-1-specific probe XQGS1-945. (F) Overlapped fluorescence microscopy image. (G) Coordinated SEM image of the same field of view as the one in panel D. (H) Close-up view of the bacteria indicated by the dashed-line box in panel G. (I) Close-up view of the magnetosome chains indicated by the dashed-line box in panel H.
Coupled FISH-SEM experiments (20) were then performed to identify strain XQGS-1 phylogenetically and structurally at the single-cell level (Fig. 1 and Fig. S1 and S2). Epifluorescence microscopic observations reveal that all observed bacterial cells (n = 359) are hybridized with the EUB338 probe (green), while only some cells (n = 31) hybridize with both the EUB338 and XQGS1-945 probes (red). Subsequent coordinated SEM observations reveal that all 31 cells have a similar spiral shape and form a single magnetosome chain with obviously larger cells and magnetosome particles than those for inner control cells of Magnetospirillum magneticum AMB-1 (Fig. 1F and Fig. S1G). That is, these large spirilla are cells of strain XQGS-1. SEM observations with energy-dispersive X-ray spectroscopy (EDXS) analyses further reveal that AMB-1 cells contain intracellular micrometer-sized granules with higher carbon contents than those in other cytoplasmic regions. This suggests that they might be polyhydroxybutyrate (PHB) granules (Fig. S3). Elemental Fe maps match well with magnetosome chains within XQGS-1 cells, but they could not be observed in AMB-1 cells because of spatial resolution limitations and because SEM-EDXS is a surface analytical technique that does not provide effective signals from deeper and smaller targets, e.g., small magnetosome particles within AMB-1 cells.
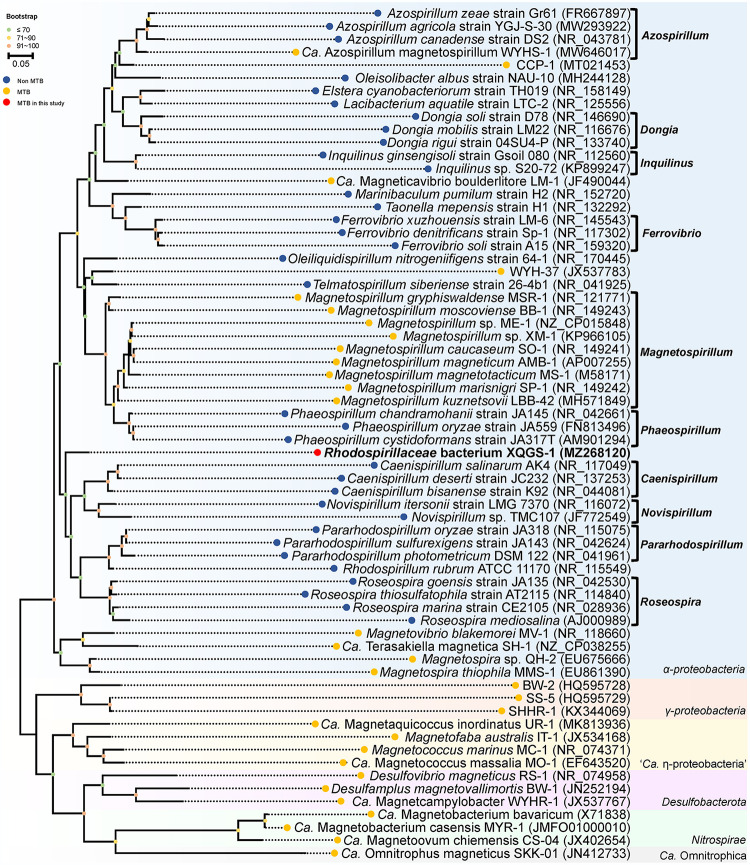
Phylogenetic analysis based on 16S rRNA gene sequences indicates that strain XQGS-1 is affiliated with the Alphaproteobacteria class of the Proteobacteria phylum (Fig. 2). Although it could be clustered on the phylogenetic tree with bacteria in the Caenispirillum and Novispirillum genera of the Rhodospirillaceae family, the bootstrap value of the node between them is <70%, which indicates that this classification is not well supported by existing data (32) (Fig. 2). Further comparative analysis indicates that strain XQGS-1 shares low 16S rRNA gene sequence identities with all reported bacteria, such as Caenispirillum salinarum (identity, 92.6%) (33), Oleiliquidispirillum nitrogeniifigens (identity, 92.5%) (34), and MTB clone WYH-37 (identity, 92.5%) (31). Such differences are less than the genus criterion level of 94.5% (35), which indicates that strain XQGS-1 represents a novel MTB species in a novel genus from the Alphaproteobacteria class.
FIG 2.
Phylogenetic tree of MTB and the Rhodospirillaceae family based on 16S rRNA gene sequences using the maximum likelihood method. Bootstrap values at nodes are given as percentages of 1,000 replicates. GenBank accession numbers are given in parentheses. The bar represents 5% sequence divergence. MTB that do not belong to the Alphaproteobacteria are selected as outgroups.
Morphological and structural features of intracellular inclusions within XQGS-1 cells.
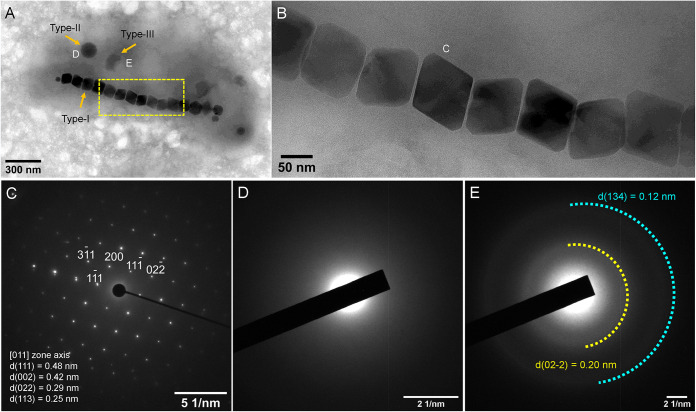
Transmission electron microscopy (TEM) observations indicate that XQGS-1 cells have spiral or vibrio shapes with an average cell length and diameter of 2.43 ± 0.48 μm and 0.84 ± 0.13 μm, respectively (n = 31). Based on morphological and structural features, three intracellular inclusion types are recognized (Fig. 3), including chain-organized magnetic nanoparticles (type I) (Fig. 3A and B), spherical granules (type II) (Fig. 3A), and irregular granules (type III) (Fig. 3A). Selected-area electron diffraction (SAED) results for individual particles indicate that XQGS-1 contains well-crystallized single magnetite crystals (Fig. 3C). Type II granules lack obvious diffraction points or rings and are amorphous (Fig. 3D). In contrast, type III inclusions produce two weak diffraction rings with calculated d-spacing values of ∼0.20 and ∼0.12 nm (Fig. 3E).
FIG 3.
Morphological and structural features of intracellular inclusions within an XQGS-1 cell. (A) TEM image of an XQGS-1 cell that contains three electron-dense particle types. (B) Close-up view of an XQGS-1 magnetosome chain indicated by the yellow dashed box in panel A. (C to E) Selected-area electron diffraction (SAED) patterns recorded from an individual magnetite particle (C) (labeled “C” in panel B), a type II granule (D) (labeled “D” in panel A), and a type III particle (E) (labeled “E” in panel A). SAED analyses indicate that the magnetic particles are well-crystallized single crystals with d-spacings of 0.48 nm and 0.42 nm, which correspond to the interplanar spacings (d) of the {111}, {002}, {022}, and {113} planes of magnetite, respectively. In contrast, the SAED pattern in panel D for type II granules has no obvious diffraction rings or spots, which is typical of amorphous phases, while the SAED pattern in panel E has two weak, smeared diffraction rings, which indicates that type III inclusions are weakly crystalline.
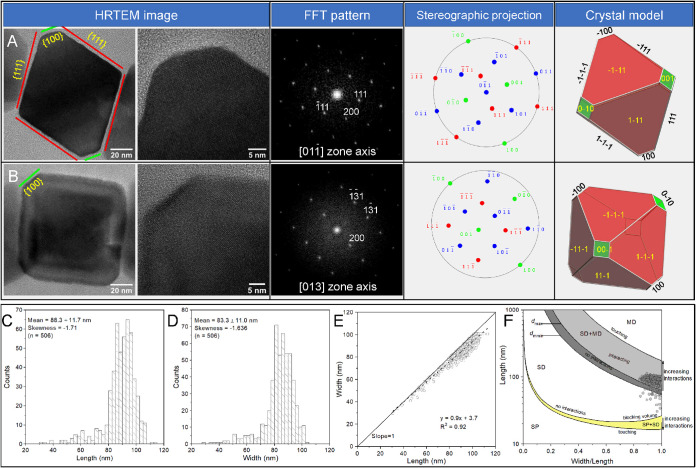
The morphology of magnetite crystals produced by XQGS-1 was further studied with high-resolution transmission electron microscopy (HRTEM) observations. Lattice imaging of particles along various zone axes, combined with crystal modeling, indicates that the magnetite particles are nearly equidimensional. An ideal crystal model of a truncated octahedron with eight large {111} faces and six small {100} faces matches almost all two-dimensional (2D) projections of individual particles (Fig. 4A and B).
FIG 4.
HRTEM images and crystal models of magnetite crystals in XQGS-1. (A and B) Representative particles recorded from the (A) and [013] (B) zone axes of magnetite. The first column shows HRTEM images of particles with labeled crystal faces, the second column shows partial enlargements of the first column, and the third through fifth columns show the corresponding fast Fourier transform (FFT) patterns, stereographic projections, and morphological models, respectively. (C to E) Histograms of crystal length (C) and width (D) for magnetite particles from strain XQGS-1 with linear crystal length-width relationships (E). The black dashed line is the linear regression result (equation shown). (F) Grain size (length) and shape (width/length) distributions of magnetite particles formed within XQGS-1 cells. Boundaries between single-domain (SD), multidomain (MD), and superparamagnetic (SP) regions for interacting and noninteracting particles are from previous studies by Butler and Banerjee (42) and Muxworthy and Williams (13), respectively.
Most magnetosome magnetite particles have sizes of between ∼80 and ∼105 nm, with an average crystal length, width, and width/length ratio of 88.3 ± 11.7 nm, 83.3 ± 11.0 nm, and 0.94 ± 0.04 (n = 506; 31 cells analyzed), respectively. Crystal length and width distributions are skewed negatively, with skewness values of −1.710 and −1.636, respectively (Fig. 4C and D). Along with a linear relationship between crystal length and width (Fig. 4E), these observations are typical of octahedral or cubo-octahedral magnetite produced by cultured and uncultured MTB (36, 37) with homothetical crystal growth (38, 39). Each XQGS-1 bacterium forms on average 17 ± 3 magnetite magnetosome particles (n = 31) that are organized tightly into a single chain along the cell long axis close to the concave side of the cell. Strong intrachain magnetic interactions constrain the magnetic moment of each particle to lie along the chain axis, so the magnetite crystals within XQGS-1 cells are likely to behave as stable SD particles (Fig. 4F) (13, 40, 41).
Chemical features of intracellular inclusions within XQGS-1 cells.
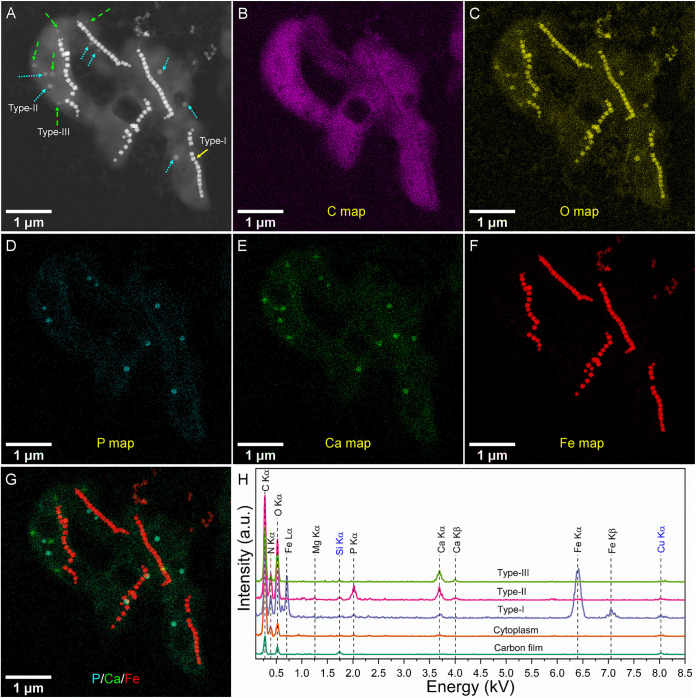
Elemental maps at subnanometer resolution were obtained using EDXS with scanning transmission electron microscopy (STEM) observations in the high-angle annular dark-field (STEM-HAADF) mode to investigate the chemical composition of intracellular inclusions within XQGS-1 cells (Fig. 5 and Fig. S4 and S5). As expected, magnetite particles are rich in Fe and O. In contrast, type II granules are rich in Ca, P, and O, with an average diameter of 90.5 ± 19.3 nm (n = 33; 14 cells analyzed), while type III inclusions are rich in Ca and O, with no P, and have an average diameter of 100.4 ± 21.4 nm (n = 36; 14 cells analyzed).
FIG 5.
EDXS elemental mapping of five XQGS-1 cells in the STEM-HAADF mode. (A) STEM-HAADF image. Solid yellow, dotted cyan, and dashed green arrows indicate type I, II, and III inclusions, respectively. (B to F) EDXS elemental maps of C (C Kα) (B), O (O Kα) (C), P (P Kα) (D), Ca (Ca Kα) (E), and Fe (Fe Kα) (F). (G) RGB (red, green, and blue) map with P (cyan), Ca (green), and Fe (red). (H) EDXS spectra for selected regions (∼50 nm) of carbon film on the TEM grid, and cytoplasm of XQGS-1 cells, with type I inclusions (magnetite particles), type II inclusions (calcium phosphate granule), and type III inclusions (calcium carbonate inclusion). a.u., arbitrary units. Kβ, X ray is emitted when vacancy in K-shell is filled by a transition from M-shell.
Semiquantitative EDXS microanalyses (30, 43, 44) indicate that the average atomic ratios of Mg to P to Ca are 2.76:35.73:61.51 for type II granules (n = 10) (Table 1) and 2.5:1.0:96.5 for type III granules (n = 10) (Table 2). Comparatively, the Mg/P/Ca atomic ratio for type II granules is significantly different from that for Ca/Mg-rich polyphosphate inclusions formed by cyanobacteria (e.g., Mg/P/Ca ratio of 16.9:66.5:16.6) (30), although they vary significantly among granules. Specifically, among the 10 granules, 6 have Ca/P ratios close to those of amorphous hydroxyapatite [Ca10(PO4)6(OH)2·nH2O], with a Ca/P ratio of 1.67, and amorphous calcium phosphates [CaxHy(PO4)z·n(H2O)] (n = 3 to 4.5), with Ca/P ratios of ∼1.2 to 2.2 (29). The other four have slightly higher (e.g., 3.33 and 3.64) or lower (e.g., 0.62 and 0.95) Ca/P ratios than those of amorphous hydroxyapatite and amorphous Ca phosphate. This indicates that some type II granules may be a mixture of calcium phosphate and other minerals, such as calcium carbonate and polyphosphate.
TABLE 1.
Mg/P/Ca atomic ratios within type II inclusions formed by strain XQGS-1 cells versus polyphosphate formed by cyanobacteriaa
| Atomic ratio |
||||
|---|---|---|---|---|
| Inclusion | Mg Kα | P Kα | Ca Kα | Ca/P ratio |
| Type II inclusion 1 | 4.92 | 31.38 | 63.7 | 2.03 |
| Type II inclusion 2 | 2.41 | 33.92 | 63.67 | 1.88 |
| Type II inclusion 3 | 0.84 | 38.37 | 60.79 | 1.58 |
| Type II inclusion 4 | 0 | 31.59 | 68.41 | 2.17 |
| Type II inclusion 5 | 5.65 | 40.11 | 54.24 | 1.35 |
| Type II inclusion 6 | 0 | 23.08 | 76.92 | 3.33 |
| Type II inclusion 7 | 2.03 | 30.56 | 67.41 | 2.21 |
| Type II inclusion 8 | 9.78 | 55.75 | 34.47 | 0.62 |
| Type II inclusion 9 | 2.01 | 21.09 | 76.9 | 3.64 |
| Type II inclusion 10 | 0 | 51.38 | 48.62 | 0.95 |
| Avg | 2.76 | 35.73 | 61.51 | 1.72 |
| Polyphosphate 1 | 14.63 | 68.73 | 16.64 | 0.24 |
| Polyphosphate 2 | 14.28 | 67.93 | 17.79 | 0.26 |
| Polyphosphate 3 | 19.31 | 64.30 | 16.39 | 0.25 |
| Polyphosphate 4 | 18.91 | 64.99 | 16.10 | 0.25 |
| Polyphosphate 5 | 17.87 | 65.20 | 16.93 | 0.26 |
| Polyphosphate 6 | 16.34 | 66.79 | 16.87 | 0.25 |
| Polyphosphate 7 | 17.49 | 67.46 | 15.05 | 0.22 |
| Polyphosphate 8 | 16.31 | 66.91 | 16.78 | 0.25 |
| Avg | 16.9 | 66.5 | 16.6 | 0.25 |
See reference 30. Positions of type II inclusions on STEM-HAADF images are shown in Fig. S5 in the supplemental material. Kα, the X ray is emitted when vacancy in K-shell is filled by a transition from L-shell.
TABLE 2.
Mg/P/Ca atomic ratios within type III inclusions formed by strain XQGS-1 cells versus calcium carbonate formed by cyanobacteriaa
| Atomic ratio |
|||
|---|---|---|---|
| Inclusion | Mg Kα | P Kα | Ca Kα |
| Type III inclusion 1 | 3.95 | 4.42 | 91.63 |
| Type III inclusion 2 | 6.64 | 0 | 93.36 |
| Type III inclusion 3 | 0 | 0 | 100 |
| Type III inclusion 4 | 3.74 | 2.75 | 93.51 |
| Type III inclusion 5 | 2.49 | 1.64 | 95.87 |
| Type III inclusion 6 | 0 | 0 | 100 |
| Type III inclusion 7 | 0 | 0 | 100 |
| Type III inclusion 8 | 3.49 | 1.51 | 94.02 |
| Type III inclusion 9 | 3.47 | 0 | 96.53 |
| Type III inclusion 10 | 0.96 | 0 | 99.04 |
| Avg | 2.5 | 1.0 | 96.5 |
| Calcium carbonate 1 | 0.68 | 0.33 | 98.99 |
| Calcium carbonate 2 | 0 | 0.81 | 99.19 |
| Calcium carbonate 3 | 0 | 0 | 100 |
| Calcium carbonate 4 | 0 | 0 | 100 |
| Calcium carbonate 5 | 0.51 | 0.71 | 98.78 |
| Calcium carbonate 6 | 1.14 | 3.13 | 95.73 |
| Calcium carbonate 7 | 0 | 0 | 100 |
| Calcium carbonate 8 | 0 | 4.59 | 95.41 |
| Calcium carbonate 9 | 1.78 | 2.14 | 96.08 |
| Calcium carbonate 10 | 0 | 4.95 | 95.05 |
| Avg | 0.4 | 1.67 | 97.9 |
See reference 30. Positions of type III inclusions on STEM-HAADF images are shown in Fig. S4 and S5 in the supplemental material. Kα, the X ray is emitted when vacancy in K-shell is filled by a transition from L-shell.
The average atomic ratio for type III inclusions is similar to that of calcium carbonate formed by cyanobacteria (e.g., Mg/P/Ca ratio of 16.9:66.5:16.6) (30) (Fig. 6 and Table 2). TEM-based SAED results and semiquantitative EDXS microanalyses demonstrate that type II granules and type III inclusions within XQGS-1 cells consist of amorphous calcium phosphate and potentially crystallized calcium carbonate, respectively.
FIG 6.
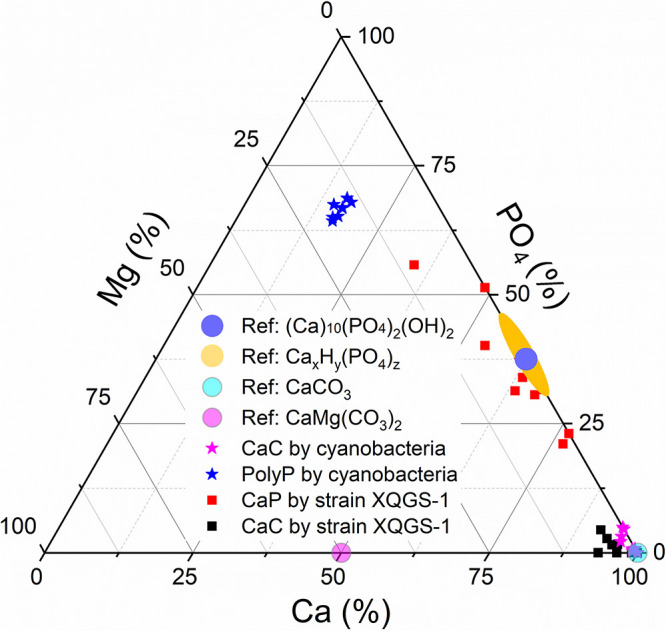
Ternary plot of Ca/Mg/PO4 ratios (atomic ratios [percent]) for calcium phosphate (CaP) (solid red squares) and calcium carbonate (CaC) (solid black squares) formed by XQGS-1 cells. Stoichiometric amorphous hydroxyapatite [Ca10(PO4)6(OH)2·nH2O] (solid blue dot), amorphous calcium phosphates [CaxHy(PO4)z·n(H2O)] (solid yellow dot), calcium carbonate (CaCO3) (solid cyan dot), and dolomite (CaMgCO3) (solid purple dot) are indicated for reference. Calcium carbonate (solid purple stars) and polyphosphate (PolyP) (solid blue stars) formed intracellularly by cyanobacteria are used for comparison (30).
DISCUSSION
Strain XQGS-1 is a novel genus that forms large magnetite magnetosomes.
MTB are phylogenetically affiliated with three classes (Alphaproteobacteria, Gammaproteobacteria, and “Candidatus Etaproteobacteria”) in the Proteobacteria phylum, the Nitrospirae phylum, the Desulfobacterota phylum (previously the Deltaproteobacteria class in the Proteobacteria phylum) (45), the “Candidatus Omnitrophica” phylum (1, 46, 47), and even other phyla across the domain Bacteria based on metagenomic and 16S rRNA approaches (48–50). With the development and application of advanced microscopic techniques, significant recent progress has been made in characterizing the chemistry and mineralogy of magnetic magnetosome particles within cultured and uncultured MTB (9, 16, 19, 20, 37, 51–53). Using the coupled FISH-SEM approach (20), we phylogenetically and structurally identified a novel MTB strain, XQGS-1, from freshwater Xingqinggong Lake, Xi’an, Shaanxi Province, northwestern China. Strain XQGS-1 has low 16S rRNA gene sequence identity to all available non-MTB bacteria and MTB strains in the Magnetospirillum genus (54–62) (Fig. 2). Bacteria closely related to strain XQGS-1 are MTB strain WYH-37 (∼92.5% sequence identity) (31) and Caenispirillum salinarum (∼92.6% sequence identity) (33), which are phylogenetically affiliated with the Rhodospirillaceae family and the Caenispirillum genus, respectively. Such differences are less than the genus criterion level of 94.5% and greater than the family criterion level of 86.5% (35). Therefore, strain XQGS-1 may represent a novel genus in the Rhodospirillaceae family of the Alphaproteobacteria class. Compared with other known MTB within the Rhodospirillaceae family (e.g., MS-1, AMB-1, and MSR-1) (54, 63, 64), XQGS-1 cells have obviously larger cell sizes with obviously larger magnetite particles.
Magnetite crystals produced by MTB generally have sizes of 30 to 120 nm, which fall well within the magnetic stable single-domain (SSD) region. However, some previous studies have shown that some uncultured MTB strains form magnetite crystals with maximum sizes or lengths of >120 nm, which might not be in the SSD state when particles are isolated. However, TEM-based electron holography and rock magnetic analyses demonstrate that such unusual magnetite crystals produced by MTB still behave as SSD particles because they are highly elongated or closely packed into a chain or chains (19, 65). Numerical calculations also indicate that magnetostatic interactions within the magnetosome chain(s) can significantly increase the threshold size or blocking volume between the SSD and multidomain (MD) states and decrease the threshold size or blocking volume between the SSD and superparamagnetic (SP) states (13). As shown in Fig. 4F, magnetite crystals produced by strain XQGS-1 still fall within the SSD region because they are tightly packed into single chains within the cell.
Previous studies of cultured MTB strains (e.g., alphaproteobacterial strains AMB-1 and MSR-1) have shown that magnetosome crystal size can be affected by environmental factors (e.g., oxygen concentrations and chemical compositions) (30, 63, 66) and that some genes play a key role in controlling magnetosome crystal size (67, 68). Compared to other alphaproteobacterial MTB strains (Table 3), which generally form cubo-octahedral or prismatic magnetite crystals with sizes of ∼30 to 60 nm, strain XQGS-1 forms truncated octahedral magnetite with obviously larger crystal sizes (e.g., 88.3- ± 11.7-nm average length and 83.3- ± 11-nm average width). It is unknown whether the larger magnetite crystals produced by strain XQGS-1 are related to specific environmental factors, size-controlling genes, or both. This point deserves future metagenomic and physiological studies.
TABLE 3.
Magnetosome particle morphologies, sizes, and width/length ratios of strain XQGS-1 and other alphaproteobacterial MTB strains
| Strain | GenBank accession no. | Magnetosome particlea |
Reference | |||
|---|---|---|---|---|---|---|
| Morphology | Mean length (nm) ± SD | Mean width (nm) ± SD | Width/length ratio | |||
| XQGS-1 | MZ268120 | Truncated octahedral | 88.3 ± 11.7 | 83.3 ± 11 | 0.94 | This study |
| Magnetospirillum magneticum AMB-1 | AP007255 | Cubo-octahedral | ∼50 | ∼45 | 0.85 | 63 |
| Magnetospirillum sp. strain XM-1 | KP966105 | 43.7 | NA | 0.85 | 59 | |
| Magnetospirillum magnetotacticum MS-1 | M58171 | 40–50 | 40–50 | 0.9 | 64 | |
| Magnetospirillum sp. strain ME-1 | CP015848 | 32.5 ± 4.5 | 28.9 ± 4.5 | 0.89 | 60 | |
| Magnetospira thiophila MMS-1 | EU861390 | 61 ± 12 | 52 ± 11 | 0.85 | 69 | |
| Magnetospirillum kuznetsovii LBB-42 | MH571849 | 38 ± 7 | NA | NA | 70 | |
| Magnetospirillum gryphiswaldense MSR-1 | NR_121771 | ∼42 | ∼42 | NA | 54 | |
| CCP-1 | MT021453 | 58 ± 4 | 40 ± 3 | NA | 22 | |
| “Ca. Azospirillum magnetospirillum” WYHS-1 | MW646017 | 34.5 ± 6.7 | 32.8 ± 6.5 | 0.95 | 26 | |
| Magnetovibrio blakemorei MV-1 | NR_118660 | Prismatic | 53 | 35 | 0.63 | 71 |
| “Ca. Terasakiella magnetica” PR-1 | CACVCF000000000 | 44 ± 13 | 34 ± 11 | 0.77 | 72 | |
| Terasakiella sp. strain SH-1 | CP038255 | 48.3 ± 8.9 | 35.7 ± 5.2 | 0.74 | 56 | |
| Magnetospira sp. strain QH-2 | EU675666 | 81 ± 23 | 58 ± 20 | 0.71 | 57 | |
NA, not applicable.
Expanded potential of MTB that produce calcium-bearing minerals.
Despite widespread bacterial calcium carbonate precipitation in soil and aquatic environments, few groups have been reported to biomineralize intracellular CaCO3, including some species from the Achromatium genus in the Gammaproteobacteria class and some strains in the Cyanobacteria phylum (24, 25). Achromatium strains are large sulfur bacteria with a worldwide distribution in the OATZ of freshwater and marine environments. Strains that produce amorphous CaCO3, such as Achromatium oxaliferum, can accumulate abundant inclusions that fill most of the cell volume (24, 73, 74). Cyanobacteria phylum strains can live in aerobic environments. Similar to Achromatium, CaCO3-producing Cyanobacteria, such as “Candidatus Gloeomargarita lithophora” and Thermosynechococcus elongatus, can absorb calcium actively and deposit it in a membrane-enveloped microcompartment (75). Two distribution patterns of CaCO3 inclusions have been discovered in Cyanobacteria strains, one with inclusions scattered throughout the cell cytoplasm and another with inclusions at cell poles (25). Recently, Monteil and coworkers reported a novel MTB strain, CCP-1, affiliated with the Alphaproteobacteria class from both the water column and sediment of the permanently stratified ferruginous Lake Pavin, France, which forms 2 to 4 amorphous CaCO3 granules with an 856- ± 95-nm average size that can occupy much of the cytoplasm (22). It has been suggested that intracellular CaCO3 inclusions could serve as ballast to adjust the vertical position of CCP-1 cells within stratified waters (22). TEM observations indicate that freshwater MTB strain XQGS-1 also forms intracellular CaCO3, although the particles are smaller and fewer than those in strain CCP-1, with a 100.4- ± 21.4-nm average size. This indicates that intracellular CaCO3 biomineralization occurs not only within MTB strain CCP-1. Genomic prediction suggests that carbonic anhydrases can transform to CO2 and release OH− to increase concentrations for CaCO3 precipitation in cyanobacteria (25), which occurs in all magnetic Alphaproteobacteria and some MTB strains in other classes or phyla (26). Together with a previous study of strain CCP-1, we confirm that some MTB strains can form intracellular CaCO3. The presence and function of such genes deserve further testing for strains CCP-1 and XQGS-1.
Our results indicate that strain XQGS-1 can also biomineralize intracellular calcium phosphate besides CaCO3 and magnetite. From the analysis of 31 XQGS-1 cells with EDXS elemental mapping in STEM-HAADF mode, we do not find polyphosphate granules, even though they are reported widely in MTB (7, 8, 18, 21, 76). One possible explanation is that entire polyphosphate granules and partial CaCO3 granules transform into calcium phosphate with the help of alkaline phosphatase (27, 30), which can explain the variable Ca/P ratios (0.62 to 3.65) in Ca-phosphate granules. Alternatively, polyphosphate and intracellular CaCO3 could have no role in Ca-phosphate biomineralization in strain XQGS-1. Further genomic analysis of strain XQGS-1 and other MTB, together with systematic chemical microanalyses of cultured and uncultured MTB, would help to test for key genes responsible for intracellular calcium phosphate and CaCO3 biomineralization within XQGS-1 and other MTB. It is worth noting that TEM-EDXS analysis enables only a semiquantitative estimation of the chemical composition of a target because it is element specific. Chemical analysis of granular mineral phases in strain XQGS-1 cells is needed in the future using synchrotron-based scanning transmission X-ray microscopy or Raman spectroscopy (9, 22, 77).
Consistent with previous observations (22, 23, 78), our results indicate that MTB have versatile biomineralization capability in forming iron oxides (e.g., magnetite, greigite, or both) and calcium-bearing minerals (e.g., CaCO3, calcium phosphate, or both). Considering their dominant occurrence in aquatic OATZ environments, MTB may also play roles in the deposition and burial of calcium-bearing minerals. Biogenic minerals such as magnetite, phosphorite, carbonate, and sulfur have been detected widely in the geological record to provide traces of life and paleoenvironmental indicators (79–82). Further work is needed to understand the expansion of calcium biomineralization among modern MTB and to identify magnetofossil and biogenic calcium-bearing mineral cooccurrences comprehensively in the geological record. Such studies would enable a better understanding of the biogeochemical roles of MTB in terms of geographical distribution and on geological time scales.
MATERIALS AND METHODS
Sampling site, MTB collection, and sample preparation.
Sediment samples were collected from freshwater Xingqinggong Lake, Xi’an City, Shaanxi Province, western China (34°15′49.8″N, 108°58′9.3″E). The sampling site had a salinity of 0.55 ppt, a pH of 7.93, and a temperature of 22°C at the time of sampling (May 2019), which were measured in situ with an HQ40d portable multiparameter meter (Hach Company, USA). Surface sediment and water were collected at a water depth of ∼1 to 2 m and stored in a 500-ml wide-mouth reagent bottle with an ∼2:1 sediment-to-water ratio. The bottles were express shipped to the laboratory and stored in darkness at ambient temperature (∼20°C) to build laboratory microcosms. In this study, 36 sediment sample bottles were collected to incubate MTB in laboratory microcosms. MTB were checked every 2 weeks using the hanging-drop technique (83), in which a small sediment drop (∼50 μl) was sampled ∼1 cm below the water-sediment interface and then dropped onto a slide. The slide was immediately turned upside down on a hollow rubber ring in a magnetic field for 10 min to allow MTB to swim to the droplet edge. In a microcosm containing dominantly spiral-shaped MTB (e.g., XQGS-1 cells), the MTB swam toward the south pole of a bar magnet (north-seeking MTB) with a concentration of ∼30 cells per μl and were selected for magnetic collection. Living MTB cells were extracted magnetically from the microcosm using a homemade magnetic separation apparatus, washed three times with MilliQ water, and then divided into three parts for TEM, molecular, and fluorescence in situ hybridization (FISH)-scanning electron microscopy (SEM) experiments according to a protocol described previously (20).
16S rRNA gene sequencing and phylogenetic analysis.
PCR amplification of 16S rRNA genes of MTB cells was performed using the universal bacterial primers 27F (5-′AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) (84), according to the method described previously by Li et al. (20). Amplified products were purified with a QIAquick gel extraction kit (Qiagen, Germany), ligated using the pMD19-T vector (TaKaRa, Japan), and then cloned into strain DH5α competent cells of Escherichia coli (Huada, Beijing, China). The 16S rRNA genes were sequenced at the Huada Genome Center, Beijing, China. After discarding short sequences (<1,300 bp), the remaining sequences were aligned with close relatives using the Gblocks 0.91b algorithm with default parameters (85) to eliminate poorly aligned positions. Along with the MTB strain studied here, MTB that have been identified phylogenetically and structurally and non-MTB strains from pure cultures that belong to the Rhodospirillaceae family were selected to build a phylogenetic tree with the NCBI taxonomy (https://www.ncbi.nlm.nih.gov/). The maximum likelihood (86) phylogenetic tree was constructed using the MEGA software package (version 7.0) (87) and a general time-reversible model with gamma-distributed plus invariant sites (GTR+G+I) to describe nucleotide evolution selected by “Find Best DNA/Protein Models” in the MEGA software package with the Bayesian information criterion (BIC). Bootstrap values were calculated with 1,000 replicates.
Coupled FISH-SEM experiments and MTB identification.
For FISH analysis, a species-specific oligonucleotide probe, XQGS1-945 (5′-CCCTTCCTCAAGCGACTTGC-3′), was designed to target the corresponding 16S rRNA gene of strain XQGS-1, with a 64°C melting temperature (Tm), measured directly during probe synthesis. Probe specificity was assessed with the probe evaluation tool ProbeMatch with no other 16S rRNA sequences in the RDP database matching (88). The universal bacterial probe EUB338 (5′-GCTGCCTCCCGTAGGAGT-3′) was used as a positive-control probe for FISH (20, 89). Probe EUB338 was synthesized and labeled fluorescently with fluorescein phosphoramidite (FAM) at the 5′ end, while probe XQGS1-945 was synthesized and labeled fluorescently with the hydrophilic sulfoindocyanine dye Cy3 at the 5′ end. Appropriate amounts of Magnetospirillum magneticum AMB-1 cells were added as inner control cells for FISH with subsequent epifluorescence microscopy and SEM observations according to the protocol described previously by Li et al. (20). Epifluorescence microscopy experiments were performed using an Olympus BX51 microscope. The same sample was then carbon coated using a Leica ACE200 low-vacuum sputter coater (Leica Microsystems, Wetzlar, Germany) and then observed with a Zeiss Ultra-55 field emission gun SEM instrument (Carl Zeiss, Germany) operating at a working voltage of 5 kV with a ∼5-mm distance.
TEM analysis.
Conventional TEM observations were performed with a JEM-F200 microscope (JEOL Ltd., Tokyo, Japan) operating at 200 kV at the Institute of Geology and Geophysics, Chinese Academy of Sciences, Beijing, China. Cell diameter, particle number, and crystal length (along the long axis) and width (perpendicular to the long axis) of magnetite particles were measured from TEM images of individual MTB cells.
High-resolution TEM (HRTEM) and scanning TEM (STEM) observations in high‐angle annular dark-field (HAADF) mode and electron diffraction structure analysis were performed with a JEM‐2100F microscope (JEOL Ltd., Tokyo, Japan) at 200 kV at JEOL Ltd., Beijing, China. This instrument is equipped with an X-Max EDXS system (Oxford Instruments, Oxford, UK), which allows microchemical analysis of submicrometer regions in TEM mode. STEM-EDXS spectrum analysis was performed using JEOL Analysis Station software; semiquantitative chemical microanalyses of individual CaCO3 and calcium phosphate inclusions relied on the use of K-factors (43).
Data availability.
The 16S rRNA gene sequence of magnetotactic spirillum strain XQGS-1 has been deposited in GenBank under accession number MZ268120.
ACKNOWLEDGMENTS
This study was supported financially by the National Natural Science Foundation of China (grants 41920104009, 41890843, 41621004, and 41425008), the Senior User Project of RVKEXUE2019GZ06 (Center for Ocean Mega-Science, Chinese Academy of Sciences), and the Australian Research Council (grants DP160100805 and DP200100765).
We thank the TEM engineers at the IGGCAS, Beijing, China (Gu Lixin and Tang Xu), for smooth running of facilities. We thank Yuan Jianzhong at JEOL (Beijing) Co. Ltd. for kindly providing access to TEM facilities.
J.L. designed the research. P.L. and X.R. did the fieldwork (sampling sediments). P.L. prepared samples and carried out microbiological experiments. J.L., Y.L., and P.L. carried out TEM experiments. J.L. and P.L. performed FISH-SEM experiments. P.L., Y.L., and J.L. carried out data and statistical analysis. J.L. and P.L. prepared the manuscript. All authors participated in the discussion of results and manuscript refinement.
Footnotes
Supplemental material is available online only.
Contributor Information
Jinhua Li, Email: lijinhua@mail.iggcas.ac.cn.
Jeremy D. Semrau, University of Michigan—Ann Arbor
REFERENCES
- 1.Lefèvre CT, Bazylinski DA. 2013. Ecology, diversity, and evolution of magnetotactic bacteria. Microbiol Mol Biol Rev 77:497–526. 10.1128/MMBR.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blakemore RP. 1975. Magnetotactic bacteria. Science 190:377–379. 10.1126/science.170679. [DOI] [PubMed] [Google Scholar]
- 3.Bazylinski DA, Frankel RB. 2004. Magnetosome formation in prokaryotes. Nat Rev Microbiol 2:217–230. 10.1038/nrmicro842. [DOI] [PubMed] [Google Scholar]
- 4.Frankel RB, Bazylinski DA, Johnson MS, Taylor BL. 1997. Magneto-aerotaxis in marine coccoid bacteria. Biophys J 73:994–1000. 10.1016/S0006-3495(97)78132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lefèvre CT, Bennet M, Landau L, Vach P, Pignol D, Bazylinski DA, Frankel RB, Klumpp S, Faivre D. 2014. Diversity of magneto-aerotactic behaviors and oxygen sensing mechanisms in cultured magnetotactic bacteria. Biophys J 107:527–538. 10.1016/j.bpj.2014.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao XG, Egli R, Petersen N, Hanzlik M, Zhao XY. 2014. Magnetotaxis and acquisition of detrital remanent magnetization by magnetotactic bacteria in natural sediment: first experimental results and theory. Geochem Geophys Geosyst 15:255–283. 10.1002/2013GC005034. [DOI] [Google Scholar]
- 7.Rivas-Lamelo S, Benzerara K, Lefèvre CT, Monteil CL, Jézéquel D, Menguy N, Viollier E, Guyot F, Férard C, Poinsot M, Skouri-Panet F, Trcera N, Miot J, Duprat E. 2017. Magnetotactic bacteria as a new model for P sequestration in the ferruginous Lake Pavin. Geochem Perspect Lett 5:35–41. 10.7185/geochemlet.1743. [DOI] [Google Scholar]
- 8.Schulz-Vogt HN, Pollehne F, Jürgens K, Arz HW, Beier S, Bahlo R, Dellwig O, Henkel JV, Herlemann DPR, Krüger S, Leipe T, Schott T. 2019. Effect of large magnetotactic bacteria with polyphosphate inclusions on the phosphate profile of the suboxic zone in the Black Sea. ISME J 13:1198–1208. 10.1038/s41396-018-0315-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J, Liu P, Wang J, Roberts AP, Pan Y. 2020. Magnetotaxis as an adaptation to enable bacterial shuttling of microbial sulfur and sulfur cycling across aquatic oxic-anoxic interfaces. J Geophys Res Biogeosci 125:e2020JG006012. 10.1029/2020JG006012. [DOI] [Google Scholar]
- 10.Kopp RE, Kirschvink JL. 2008. The identification and biogeochemical interpretation of fossil magnetotactic bacteria. Earth Sci Rev 86:42–61. 10.1016/j.earscirev.2007.08.001. [DOI] [Google Scholar]
- 11.Larrasoaña JC, Liu Q, Hu P, Roberts AP, Mata P, Civis J, Sierro FJ, Perez-Asensio JN. 2014. Paleomagnetic and paleoenvironmental implications of magnetofossil occurrences in late Miocene marine sediments from the Guadalquivir Basin, SW Spain. Front Microbiol 5:71. 10.3389/fmicb.2014.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amor M, Mathon FP, Monteil CL, Busigny V, Lefèvre CT. 2020. Iron-biomineralizing organelle in magnetotactic bacteria: function, synthesis and preservation in ancient rock samples. Environ Microbiol 22:3611–3632. 10.1111/1462-2920.15098. [DOI] [PubMed] [Google Scholar]
- 13.Muxworthy AR, Williams W. 2009. Critical superparamagnetic/single-domain grain sizes in interacting magnetite particles: implications for magnetosome crystals. J R Soc Interface 6:1207–1212. 10.1098/rsif.2008.0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pósfai M, Lefèvre C, Trubitsyn D, Bazylinski DA, Frankel RB. 2013. Phylogenetic significance of composition and crystal morphology of magnetosome minerals. Front Microbiol 4:344. 10.3389/fmicb.2013.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Menguy N, Roberts AP, Gu L, Leroy E, Bourgon J, Yang X, Zhao X, Liu P, Changela H, Pan Y. 2020. Bullet-shaped magnetite biomineralization within a magnetotactic deltaproteobacterium: implications for magnetofossil identification. J Geophys Res Biogeosci 125:e2020JG005680. 10.1029/2020JG005680. [DOI] [Google Scholar]
- 16.Li J, Menguy N, Leroy E, Roberts AP, Liu PY, Pan Y. 2020. Biomineralization and magnetism of uncultured magnetotactic coccus strain THC-1 with non-chained magnetosomal magnetite nanoparticles. J Geophys Res Solid Earth 125:e2020JB020853. 10.1029/2020JB020853. [DOI] [Google Scholar]
- 17.Bazylinski DA, Dean AJ, Williams TJ, Long LK, Middleton SL, Dubbels BL. 2004. Chemolithoautotrophy in the marine, magnetotactic bacterial strains MV-1 and MV-2. Arch Microbiol 182:373–387. 10.1007/s00203-004-0716-y. [DOI] [PubMed] [Google Scholar]
- 18.Cox BL, Popa R, Bazylinski DA, Lanoil B, Douglas S, Belz A, Engler DL, Nealson KH. 2002. Organization and elemental analysis of P-, S-, and Fe-rich inclusions in a population of freshwater magnetococci. Geomicrobiol J 19:387–406. 10.1080/01490450290098504. [DOI] [Google Scholar]
- 19.Li JH, Menguy N, Gatel C, Boureau V, Snoeck E, Patriarche G, Leroy E, Pan YX. 2015. Crystal growth of bullet-shaped magnetite in magnetotactic bacteria of the Nitrospirae phylum. J R Soc Interface 12:20141288. 10.1098/rsif.2014.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li JH, Zhang H, Menguy N, Benzerara K, Wang FX, Lin XT, Chen ZB, Pan YX. 2017. Single-cell resolution of uncultured magnetotactic bacteria via fluorescence-coupled electron microscopy. Appl Environ Microbiol 83:e00409-17. 10.1128/AEM.00409-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li JH, Zhang H, Liu PY, Menguy N, Roberts AP, Chen HT, Wang YZ, Pan YX. 2019. Phylogenetic and structural identification of a novel magnetotactic deltaproteobacterium strain WYHR-1 from a freshwater lake. Appl Environ Microbiol 85:e00731-19. 10.1128/AEM.00731-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monteil CL, Benzerara K, Menguy N, Bidaud CC, Michot-Achdjian E, Bolzoni R, Mathon FP, Coutaud M, Alonso B, Garau C, Jézéquel D, Viollier E, Ginet N, Floriani M, Swaraj S, Sachse M, Busigny V, Duprat E, Guyot F, Lefèvre CT. 2021. Intracellular amorphous Ca-carbonate and magnetite biomineralization by a magnetotactic bacterium affiliated to the Alphaproteobacteria. ISME J 15:1–18. 10.1038/s41396-020-00747-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taoka A, Kondo J, Oestreicher Z, Fukumori Y. 2014. Characterization of uncultured giant rod-shaped magnetotactic Gammaproteobacteria from a fresh water pond in Kanazawa, Japan. Microbiology (Reading) 160:2226–2234. 10.1099/mic.0.078717-0. [DOI] [PubMed] [Google Scholar]
- 24.Head IM, Gray ND, Clarke KJ, Pickup RW, Jones JG. 1996. The phylogenetic position and ultrastructure of the uncultured bacterium Achromatium oxaliferum. Microbiology (Reading) 142(Part 9):2341–2354. 10.1099/00221287-142-9-2341. [DOI] [PubMed] [Google Scholar]
- 25.Benzerara K, Skouri-Panet F, Li JH, Ferard C, Gugger M, Laurent T, Couradeau E, Ragon M, Cosmidis J, Menguy N, Margaret-Oliver I, Tavera R, Lopez-Garcia P, Moreira D. 2014. Intracellular Ca-carbonate biomineralization is widespread in cyanobacteria. Proc Natl Acad Sci USA 111:10933–10938. 10.1073/pnas.1403510111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li JH, Liu PY, Tamaxia A, Zhang H, Liu Y, Wang J, Menguy N, Zhao X, Roberts AP, Pan YX. 2021. Diverse intracellular inclusion types within magnetotactic bacteria: implications for biogeochemical cycling in aquatic environments. J Geophys Res Biogeosci 126:e2021JG006310. 10.1029/2021JG006310. [DOI] [Google Scholar]
- 27.Kathuria S, Martiny AC. 2011. Prevalence of a calcium-based alkaline phosphatase associated with the marine cyanobacterium Prochlorococcus and other ocean bacteria. Environ Microbiol 13:74–83. 10.1111/j.1462-2920.2010.02310.x. [DOI] [PubMed] [Google Scholar]
- 28.Boonrungsiman S, Gentleman E, Carzaniga R, Evans ND, McComb DW, Porter AE, Stevens MM. 2012. The role of intracellular calcium phosphate in osteoblast-mediated bone apatite formation. Proc Natl Acad Sci USA 109:14170–14175. 10.1073/pnas.1208916109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cosmidis J, Benzerara K, Nassif N, Tyliszczak T, Bourdelle F. 2015. Characterization of Ca-phosphate biological materials by scanning transmission X-ray microscopy (STXM) at the Ca L2,3-, P L2,3- and C K-edges. Acta Biomater 12:260–269. 10.1016/j.actbio.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Li JH, Oliver IM, Cam N, Boudier T, Blondeau M, Leroy E, Cosmidis J, Skouri-Panet F, Guigner J-M, Ferard C, Poinsot M, Moreira D, Lopez-Garcia P, Cassier-Chauvat C, Chauvat F, Benzerara K. 2016. Biomineralization patterns of intracellular carbonatogenesis in cyanobacteria: molecular hypotheses. Minerals 6:10. 10.3390/min6010010. [DOI] [Google Scholar]
- 31.Chen HT, Lin W, Wang YZ, Li JH, Chen GJ, Pan YX. 2013. Diversity and magnetism of magnetotactic bacteria in Lake Weiyang near Xi’an city. Quat Sci 33:88–96. [Google Scholar]
- 32.Wiesemuller B, Rothe H. 2006. Interpretation of bootstrap values in phylogenetic analysis. Anthropol Anz 64:161–165. 10.1127/anthranz/64/2006/161. [DOI] [PubMed] [Google Scholar]
- 33.Ritika C, Suresh K, Kumar PA. 2012. Caenispirillum salinarum sp. nov., a member of the family Rhodospirillaceae isolated from a solar saltern. Int J Syst Evol Microbiol 62:1698–1702. 10.1099/ijs.0.035287-0. [DOI] [PubMed] [Google Scholar]
- 34.Li F-L, Wang X-T, Shan J-J, Li S, Zhang Y-X, Li X-Z, Li D-A, Li W-J, Wang L. 2020. Oleiliquidispirillum nitrogeniifigens gen. nov., sp. nov., a new member of the family Rhodospirillaceae isolated from oil reservoir water. Int J Syst Evol Microbiol 70:3468–3474. 10.1099/ijsem.0.004200. [DOI] [PubMed] [Google Scholar]
- 35.Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer K-H, Whitman WB, Euzéby J, Amann R, Rosselló-Móra R. 2014. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645. 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 36.Devouard B, Pósfai M, Hua X, Bazylinski DA, Frankel RB, Buseck PR. 1998. Magnetite from magnetotactic bacteria: size distributions and twinning. Am Mineral 83:1387–1398. 10.2138/am-1998-11-1228. [DOI] [Google Scholar]
- 37.Zhang H, Menguy N, Wang F, Benzerara K, Leroy E, Liu P, Liu W, Wang C, Pan Y, Chen Z, Li J. 2017. Magnetotactic coccus strain SHHC-1 affiliated to Alphaproteobacteria forms octahedral magnetite magnetosomes. Front Microbiol 8:969. 10.3389/fmicb.2017.00969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li JH, Pan YX, Chen GJ, Liu QS, Tian LX, Lin W. 2009. Magnetite magnetosome and fragmental chain formation of Magnetospirillum magneticum AMB-1: transmission electron microscopy and magnetic observations. Geophys J Int 177:33–42. 10.1111/j.1365-246X.2009.04043.x. [DOI] [Google Scholar]
- 39.Jandacka P, Alexa P, Pistora J, Li JH, Vojtkova H, Hendrych A. 2013. Size distributions of nanoparticles from magnetotactic bacteria as signatures of biologically controlled mineralization. Am Mineral 98:2105–2114. 10.2138/am.2013.4429. [DOI] [Google Scholar]
- 40.Dunin-Borkowski RE, McCartney MR, Frankel RB, Bazylinski DA, Posfai M, Buseck PR. 1998. Magnetic microstructure of magnetotactic bacteria by electron holography. Science 282:1868–1870. 10.1126/science.282.5395.1868. [DOI] [PubMed] [Google Scholar]
- 41.Li JH, Ge KP, Pan YX, Williams W, Liu QS, Qin HF. 2013. A strong angular dependence of magnetic properties of magnetosome chains: implications for rock magnetism and paleomagnetism. Geochem Geophys Geosyst 14:3887–3907. 10.1002/ggge.20228. [DOI] [Google Scholar]
- 42.Butler RF, Banerjee SK. 1975. Theoretical single‐domain grain size range in magnetite and titanomagnetite. J Geophys Res 80:4049–4058. 10.1029/JB080i029p04049. [DOI] [Google Scholar]
- 43.Abad Ortega MDM, Nieto F. 2003. Quantitative EDX analysis in TEM. Practical development, limitations and standards, p 687–694. In Méndez-Vilas A (ed), Science, technology and education of microscopy: an overview. Formatex, Badajoz, Spain. [Google Scholar]
- 44.Scimeca M, Bischetti S, Lamsira HK, Bonfiglio R, Bonanno E. 2018. Energy dispersive X-ray (EDX) microanalysis: a powerful tool in biomedical research and diagnosis. Eur J Histochem 62:2841. 10.4081/ejh.2018.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waite DW, Chuvochina M, Pelikan C, Parks DH, Yilmaz P, Wagner M, Loy A, Naganuma T, Nakai R, Whitman WB, Hahn MW, Kuever J, Hugenholtz P. 2020. Proposal to reclassify the proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int J Syst Evol Microbiol 70:5972–6016. 10.1099/ijsem.0.004213. [DOI] [PubMed] [Google Scholar]
- 46.Komeili A. 2012. Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria. FEMS Microbiol Rev 36:232–255. 10.1111/j.1574-6976.2011.00315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu PY, Liu Y, Zhao X, Roberts AP, Zhang H, Zheng Y, Wang FX, Wang LS, Menguy N, Pan YX, Li JH. 2021. Diverse phylogeny and morphology of magnetite biomineralized by magnetotactic cocci. Environ Microbiol 23:1115–1129. 10.1111/1462-2920.15254. [DOI] [PubMed] [Google Scholar]
- 48.Lin W, Zhang WS, Paterson GA, Zhu QY, Zhao X, Knight R, Bazylinski DA, Roberts AP, Pan YX. 2020. Expanding magnetic organelle biogenesis in the domain Bacteria. Microbiome 8:152. 10.1186/s40168-020-00931-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uzun M, Alekseeva L, Krutkina M, Koziaeva V, Grouzdev D. 2020. Unravelling the diversity of magnetotactic bacteria through analysis of open genomic databases. Sci Data 7:252. 10.1038/s41597-020-00593-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abreu F, Leao P, Vargas G, Cypriano J, Figueiredo V, Enrich-Prast A, Bazylinski DA, Lins U. 2018. Culture-independent characterization of a novel magnetotactic member affiliated to the Beta class of the Proteobacteria phylum from an acidic lagoon. Environ Microbiol 20:2615–2624. 10.1111/1462-2920.14286. [DOI] [PubMed] [Google Scholar]
- 51.Pósfai M, Kasama T, Dunin-Borkowski RE. 2013. Biominerals at the nanoscale: transmission electron microscopy methods for studying the special properties of biominerals, p 375–433. In Nieto F, Livi KJT, Oberti R (ed), Minerals at the nanoscale. European Mineralogical Union, London, England. [Google Scholar]
- 52.Zhu XH, Hitchcock AP, Bazylinski DA, Denes P, Joseph J, Lins U, Marchesini S, Shiu H-W, Tyliszczak T, Shapiro DA. 2016. Measuring spectroscopy and magnetism of extracted and intracellular magnetosomes using soft X-ray ptychography. Proc Natl Acad Sci USA 113:E8219–E8227. 10.1073/pnas.1610260114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baumgartner J, Menguy N, Gonzalez TP, Morin G, Widdrat M, Faivre D. 2016. Elongated magnetite nanoparticle formation from a solid ferrous precursor in a magnetotactic bacterium. J R Soc Interface 13:20160665. 10.1098/rsif.2016.0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schleifer KH, Schuler D, Spring S, Weizenegger M, Amann R, Ludwig W, Kohler M. 1991. The genus Magnetospirillum gen. nov. description of Magnetospirillum gryphiswaldense sp. nov. and transfer of Aquaspirillum magnetotacticum to Magnetospirillum magnetotacticum comb. nov. Syst Appl Microbiol 14:379–385. 10.1016/S0723-2020(11)80313-9. [DOI] [Google Scholar]
- 55.Eden PA, Schmidt TM, Blakemore RP, Pace NR. 1991. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int J Syst Bacteriol 41:324–325. 10.1099/00207713-41-2-324. [DOI] [PubMed] [Google Scholar]
- 56.Du H, Zhang W, Zhang W, Zhang W, Pan H, Pan Y, Bazylinski DA, Wu L-F, Xiao T, Lin W. 2019. Magnetosome gene duplication as an important driver in the evolution of magnetotaxis in the Alphaproteobacteria. mSystems 4:e00315-19. 10.1128/mSystems.00315-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu KL, Pan HM, Li JH, Yu-Zhang K, Zhang SD, Zhang WY, Zhou K, Yue HD, Pan YX, Xiao T, Wu LF. 2010. Isolation and characterization of a marine magnetotactic spirillum axenic culture QH-2 from an intertidal zone of the China Sea. Res Microbiol 161:276–283. 10.1016/j.resmic.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 58.Popa R, Fang W, Nealson KH, Souza-Egipsy V, Berquo TS, Banerjee SK, Penn LR. 2009. Effect of oxidative stress on the growth of magnetic particles in Magnetospirillum magneticum. Int Microbiol 12:49–57. [PubMed] [Google Scholar]
- 59.Wang YZ, Lin W, Li JH, Zhang TW, Li Y, Tian JS, Gu LX, Heyden YV, Pan YX. 2015. Characterizing and optimizing magnetosome production of Magnetospirillum sp. XM-1 isolated from Xi’an City Moat, China. FEMS Microbiol Lett 362:fnv167. 10.1093/femsle/fnv167. [DOI] [PubMed] [Google Scholar]
- 60.Ke L, Chen Y, Liu P, Liu S, Wu D, Yuan Y, Wu Y, Gao M. 2018. Characteristics and optimised fermentation of a novel magnetotactic bacterium, Magnetospirillum sp. ME-1. FEMS Microbiol Lett 365:fny052. 10.1093/femsle/fny052. [DOI] [PubMed] [Google Scholar]
- 61.Dziuba M, Koziaeva V, Grouzdev D, Burganskaya E, Baslerov R, Kolganova T, Chernyadyev A, Osipov G, Andrianova E, Gorlenko V, Kuznetsov B. 2016. Magnetospirillum caucaseum sp. nov., Magnetospirillum marisnigri sp. nov and Magnetospirillum moscoviense sp. nov., freshwater magnetotactic bacteria isolated from three distinct geographical locations in European Russia. Int J Syst Evol Microbiol 66:2069–2077. 10.1099/ijsem.0.000994. [DOI] [PubMed] [Google Scholar]
- 62.Monteil CL, Grouzdev DS, Perrière G, Alonso B, Rouy Z, Cruveiller S, Ginet N, Pignol D, Lefèvre CT. 2020. Repeated horizontal gene transfers triggered parallel evolution of magnetotaxis in two evolutionary divergent lineages of magnetotactic bacteria. ISME J 14:1783–1794. 10.1038/s41396-020-0647-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J, Pan Y. 2012. Environmental factors affect magnetite magnetosome synthesis in Magnetospirillum magneticum AMB-1: implications for biologically controlled mineralization. Geomicrobiol J 29:362–373. 10.1080/01490451.2011.565401. [DOI] [Google Scholar]
- 64.Maratea D, Blakemore RP. 1981. Aquaspirillum magnetotacticum sp. nov., a magnetic spirillum. Int J Syst Bacteriol 31:452–455. 10.1099/00207713-31-4-452. [DOI] [Google Scholar]
- 65.Lins U, McCartney MR, Farina M, Frankel RB, Buseck PR. 2005. Habits of magnetosome crystals in coccoid magnetotactic bacteria. Appl Environ Microbiol 71:4902–4905. 10.1128/AEM.71.8.4902-4905.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Faivre D, Menguy N, Pósfai M, Schüler D. 2008. Environmental parameters affect the physical properties of fast-growing magnetosomes. Am Mineral 93:463–469. 10.2138/am.2008.2678. [DOI] [Google Scholar]
- 67.Yang W, Li RG, Peng T, Zhang Y, Jiang W, Li Y, Li JL. 2010. mamO and mamE genes are essential for magnetosome crystal biomineralization in Magnetospirillum gryphiswaldense MSR-1. Res Microbiol 161:701–705. 10.1016/j.resmic.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 68.Murat D, Quinlan A, Vali H, Komeili A. 2010. Comprehensive genetic dissection of the magnetosome gene island reveals the step-wise assembly of a prokaryotic organelle. Proc Natl Acad Sci USA 107:5593–5598. 10.1073/pnas.0914439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meldrum FC, Mann S, Heywood BR, Frankel RB, Bazylinski DA. 1993. Electron microscopy study of magnetosomes in two cultured vibrioid magnetotactic bacteria. Proc R Soc Lond B Biol Sci 251:237–242. [Google Scholar]
- 70.Koziaeva VV, Rusakova SA, Slobodova NV, Uzun M, Kolganova TV, Skryabin KG, Grouzdev DS. 2019. Magnetospirillum kuznetsovii sp. nov., a novel magnetotactic bacterium isolated from a lake in the Moscow region. Int J Syst Evol Microbiol 69:1953–1959. 10.1099/ijsem.0.003408. [DOI] [PubMed] [Google Scholar]
- 71.Bazylinski DA, Williams TJ, Lefèvre CT, Trubitsyn D, Fang J, Beveridge TJ, Moskowitz BM, Ward B, Schübbe S, Dubbels BL, Simpson B. 2013. Magnetovibrio blakemorei gen. nov., sp nov., a magnetotactic bacterium (Alphaproteobacteria: Rhodospirillaceae) isolated from a salt marsh. Int J Syst Evol Microbiol 63:1824–1833. 10.1099/ijs.0.044453-0. [DOI] [PubMed] [Google Scholar]
- 72.Monteil CL, Perrière G, Menguy N, Ginet N, Alonso B, Waisbord N, Cruveiller S, Pignol D, Lefèvre CT. 2018. Genomic study of a novel magnetotactic Alphaproteobacteria uncovers the multiple ancestry of magnetotaxis. Environ Microbiol 20:4415–4430. 10.1111/1462-2920.14364. [DOI] [PubMed] [Google Scholar]
- 73.Salman V, Yang T, Berben T, Klein F, Angert E, Teske A. 2015. Calcite-accumulating large sulfur bacteria of the genus Achromatium in Sippewissett Salt Marsh. ISME J 9:2503–2514. 10.1038/ismej.2015.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benzerara K, Bolzoni R, Monteil C, Beyssac O, Forni O, Alonso B, Asta MP, Lefèvre C. 2021. The gammaproteobacterium Achromatium forms intracellular amorphous calcium carbonate and not (crystalline) calcite. Geobiology 19:199–213. 10.1111/gbi.12424. [DOI] [PubMed] [Google Scholar]
- 75.Blondeau M, Sachse M, Boulogne C, Gillet C, Guigner J-M, Skouri-Panet F, Poinsot M, Ferard C, Miot J, Benzerara K. 2018. Amorphous calcium carbonate granules form within an intracellular compartment in calcifying cyanobacteria. Front Microbiol 9:1768. 10.3389/fmicb.2018.01768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu P, Tamaxia A, Liu Y, Qiu H, Pan J, Jin Z, Zhao X, Roberts AP, Pan Y, Li J. 19 April 2021. Identification and characterization of magnetotactic Gammaproteobacteria from a salt evaporation pool, Bohai Bay, China. Environ Microbiol 10.1111/1462-2920.15516. [DOI] [PubMed]
- 77.Li JH, Bernard S, Benzerara K, Beyssac O, Allard T, Cosmidis J, Moussou J. 2014. Impact of biomineralization on the preservation of microorganisms during fossilization: an experimental perspective. Earth Planet Sci Lett 400:113–122. 10.1016/j.epsl.2014.05.031. [DOI] [Google Scholar]
- 78.Isambert A, Menguy N, Larquet E, Guyot F, Valet JP. 2007. Transmission electron microscopy study of magnetites in a freshwater population of magnetotactic bacteria. Am Mineral 92:621–630. 10.2138/am.2007.2278. [DOI] [Google Scholar]
- 79.Bailey J, Corsetti F, Greene SE, Crosby C, Liu P, Orphan V. 2013. Filamentous sulfur bacteria preserved in modern and ancient phosphatic sediments: implications for the role of oxygen and bacteria in phosphogenesis. Geobiology 11:397–405. 10.1111/gbi.12046. [DOI] [PubMed] [Google Scholar]
- 80.Li JH, Benzerara K, Bernard S, Beyssac O. 2013. The link between biomineralization and fossilization of bacteria: insights from field and experimental studies. Chem Geol 359:49–69. 10.1016/j.chemgeo.2013.09.013. [DOI] [Google Scholar]
- 81.Cosmidis J, Benzerara K, Menguy N, Arning E. 2013. Microscopy evidence of bacterial microfossils in phosphorite crusts of the Peruvian shelf: implications for phosphogenesis mechanisms. Chem Geol 359:10–22. 10.1016/j.chemgeo.2013.09.009. [DOI] [Google Scholar]
- 82.Benzerara K, Menguy N. 2009. Looking for traces of life in minerals. C R Palevol 8:617–628. 10.1016/j.crpv.2009.03.006. [DOI] [Google Scholar]
- 83.Schüler D. 2002. The biomineralization of magnetosomes in Magnetospirillum gryphiswaldense. Int Microbiol 5:209–214. 10.1007/s10123-002-0086-8. [DOI] [PubMed] [Google Scholar]
- 84.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, New York, NY. [Google Scholar]
- 85.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard J-F, Guindon S, Lefort V, Lescot M, Claverie J-M, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469. 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 87.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim YJ, Teletia N, Ruotti V, Maher CA, Chinnaiyan AM, Stewart R, Thomson JA, Patel JM. 2009. ProbeMatch: rapid alignment of oligonucleotides to genome allowing both gaps and mismatches. Bioinformatics 25:1424–1425. 10.1093/bioinformatics/btp178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Amann RI, Krumholz L, Stahl DA. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol 172:762–770. 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S5, Table S1. Download aem.01556-21-s0001.pdf, PDF file, 3.1 MB (3.1MB, pdf)
Data Availability Statement
The 16S rRNA gene sequence of magnetotactic spirillum strain XQGS-1 has been deposited in GenBank under accession number MZ268120.