Abstract
Glucocorticoids (GCs), important regulators of epidermal growth, differentiation, and homeostasis, are used extensively in the treatment of skin diseases. Using keratin gene expression as a paradigm of epidermal physiology and pathology, we have developed a model system to study the molecular mechanism of GCs action in skin. Here we describe a novel mechanism of suppression of transcription by the glucocorticoid receptor (GR) that represents an example of customizing a device for transcriptional regulation to target a specific group of genes within the target tissue, in our case, epidermis. We have shown that GCs repress the expression of the basal-cell-specific keratins K5 and K14 and disease-associated keratins K6, K16, and K17 but not the differentiation-specific keratins K3 and K10 or the simple epithelium-specific keratins K8, K18, and K19. We have identified the negative recognition elements (nGREs) in all five regulated keratin gene promoters. Detailed footprinting revealed that the function of nGREs is to instruct the GR to bind as four monomers. Furthermore, using cotransfection and antisense technology we have found that, unlike SRC-1 and GRIP-1, which are not involved in the GR complex that suppresses keratin genes, histone acetyltransferase and CBP are. In addition, we have found that GR, independently from GREs, blocks the induction of keratin gene expression by AP1. We conclude that GR suppresses keratin gene expression through two independent mechanisms: directly, through interactions of keratin nGREs with four GR monomers, as well as indirectly, by blocking the AP1 induction of keratin gene expression.
Glucocorticoids (GCs) mediate their effect through nuclear receptors, transcription factors that, depending on the presence or absence of the ligand, regulate gene expression. The glucocorticoid receptor (GR) is stored in the cytoplasm in its inactive form, bound to the heat shock protein Hsp90 (49). Ligand binding causes activation of the receptor, release from Hsp90, and its translocation to the nucleus. Activated GR binds specific DNA sequences in target genes, designated glucocorticoid response elements (GREs), and either induces or suppresses gene transcription (1, 2, 6, 57).
Recent studies have identified a group of proteins that interact with nuclear receptors called coregulators. Depending on their effect on transcription, the coregulators are designated as coactivators or corepressors (24, 56). The liganded receptor binds to the response elements and recruits coactivators (such as SRC-1, GRIP-1, NCoA, or TIF-2) that interact with cointegrators such as p-CIP and CBP/p300 (7, 15, 22, 28, 45, 67). The cointegrators bind histone acetyltransferase (HAT), which leads to the induction of transcription (39, 47, 73). In addition, CBP/p300 itself is a histone acetylase that can induce transcription without further interaction with HAT (42, 67). Although the role of the coregulators in transcriptional regulation by NRs, including GR, is a rapidly developing area of research, very little is known about the role of coregulators in active repression, i.e., in the suppression of transcription by liganded receptors.
Skin is a major target tissue for GC action. Corticosteroids, analogs of the glucocorticoid hormone, are the most commonly used therapeutic agents in dermatology (5, 58, 70). They have been used as immunosuppressive agents for T-cell or cytokine-mediated tissue damage. They suppress ICAM-1, interleukin-1 (IL-1), IL-2, IL-6, granulocyte-macrophage colony-stimulating factor, tumor necrosis factor alpha, and gamma interferon (IFN-γ), which are all components of the immune response (8, 29). In addition, GCs act as growth inhibitory agents and affect cell-cell interactions (55). However, very little is known about the molecular mechanisms of GC action in epidermis. Therefore, to begin to understand such a complex subject, we have developed a model system in which we use keratins, a family of differentially expressed epidermal genes, as reporters of GC action in epidermis. We have focused on the regulation of the keratin gene expression by GCs because this large family of epithelium-specific genes has a very precise expression pattern reflecting the physiological and pathological states of the epithelial cells (4, 17).
Initially, we focused on the following questions: what are the effects of GCs on epidermal gene expression, how are they mediated, and most importantly, how do such general and potent transcription factors specifically target and regulate gene expression in this tissue? Interestingly, we found that GCs suppress the expression of a subset of the keratin genes K5-K14, K6-K16, and K17, whose expression is altered in cutaneous diseases (64). GCs regulate these genes through two different and independent mechanisms. First, GCs directly suppress transcription through a novel mechanism that involves binding of four monomers of the GR to the keratin GREs. This mechanism substantially differs from all those previously described because it uses monomers rather than dimers of GR, it involves different coregulators, and it uses HAT to suppress transcription rather than induce it. The second, independent mechanism of GC function is by blocking the AP1-mediated induction of keratin gene expression. AP1 is commonly active in the proliferative and inflammatory processes for which GCs are usually prescribed.
The effect of GCs on keratin gene expression reflects directly their specific action in epidermal physiology because GCs target only the keratin genes expressed during the inflammatory response and wound-healing process. The mechanism through which the regulation occurs is a novel one that represents an exciting example of tissue specificity in hormone action.
MATERIALS AND METHODS
Plasmids, their growth, and their purification.
Plasmids pK14CAT, pK5CAT, pK6CAT, pK16CAT, pK3CAT, pK10CAT, pK17CAT, pK19CAT, pK14M1113, K17M1, K5M1, and pRSVZ have been described previously (27, 62). The plasmids pK8 and pK18 were gifts from R. Oshima (46), pK13 was a gift from J. Schweizer (61), plasmids containing human GR nuclear receptor and GRE-CAT were gifts from P. Chambon (18), TK-CAT plasmid was a gift from H. H. Samuels (14), plasmid containing NCoR was a gift from C. K. Glass (21), plasmid containing SRC-1 was a gift from B. O'Malley (44), CBP was from R. H. Goodman (30), and plasmid containing GRIP1 was a gift from M. Stallcup (23). Plasmids were grown in JM101 E. coli host to saturation density in Luria-Bertani medium. DNA was extracted and purified using the Mega Prep Kit from Promega.
Cloning and mutagenesis.
We have used a previously described method (63) to mutagenize the primary GRE site in the K17 promoter and create K17GREM1. The primers used for the PCRs were K17outF (5′-GGGTCTAGACAACCCATTTCCCCACCA), K17insR (5′-TTTACTAGTTTTTATTCCCCTGGGCTTTCATCACCA), K17insF (5′-TTTACTAGTGAGCAAGCCTGTTGTAATCGC), and K17outR (5′-GGGAAGCTTCATCATGGTGGCGGCGGC); K17GREM4 was previously described as K17M1 by Radoja et al. (51). K5M1 and K14M1 were also previously described (51, 65).
KRETKCAT contains the acidic signature sequence motif cloned into the TKCAT (gift from H. H. Samuels). The primers used for the PCR were KREf (5′-TTTTAAGCTTGCCCCCCAGCCACCTG) and KREr (5′-TTTTCTCGACGCTTTGCTCCTCTGT).
The insert was introduced using HindIII and SalI restriction sites. Positive clones in all cloning procedures were identified by restriction digestion and subsequently by sequencing.
Cell growth.
HeLa cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% calf serum at 37°C in a 5% CO2 atmosphere in medium containing penicillin and streptomycin as described earlier (62). The day before transfection cells were plated onto 60-mm dishes. At 4 h before transfection the medium was changed to phenol-red-free DMEM supplemented with charcoal-pretreated 10% calf serum depleted of steroids as described elsewhere (62).
Normal human foreskin epidermal keratinocytes were a generous gift from M. Simon. The cultures were initiated using 3T3 feeder layers as described earlier (52) and then frozen in liquid N2 until used. Once thawed, the keratinocytes were grown without feeder cells in defined serum-free keratinocyte medium supplemented with epidermal growth factor and bovine pituitary extract (Keratinocyte-SFM; GIBCO). Cells were expanded through two 1:4 passages before transfection and transfected at 100% confluency.
Transfection using Ca3(PO4)2.
We have generally followed the published procedure for cells that were at 80% confluence (25). At the time of transfection 3 μg of the CAT plasmid, 1 μg of the nuclear receptor expression vector plasmid, 1 μg of the pRSVZ reference plasmid, and a sufficient amount of carrier were added into each dish to bring the total to 10 μg of DNA. After transfection cells were incubated in phenol-red-free DMEM supplemented with charcoal-pretreated 10% calf serum depleted of steroids in the presence or absence of 0.1 or 1 μM dexamethasone (DEX; Sigma) in ethanol. Then, 10 nM trichostatin A (TSA) in ethanol (Wako Bioproducts) was used where indicated. The cells were harvested at 48 h after transfection by scraping them into 5 ml of phosphate-buffered saline (PBS), washed once more in PBS, and resuspended in 150 μl of 0.25 M Tris buffer at pH 7.8. All transfections were performed in duplicate plates, and each transfection experiment was repeated two to five times. The CAT and β-galactosidase assays were performed as described earlier (25, 62).
Transfection using polybrene with DMSO shock.
We used transfections with polybrene and dimethyl sulfoxide (DMSO) to transfect the DNA into the 100% confluent keratinocytes as previously described (25). On the day of transfection cells were washed and incubated in the basal medium without epidermal growth factor EGF or bovine pituitary extract. Each transfection contained 10 to 15 μg of keratin-CAT construct and 3 μg of RSVZ construct per dish. The cells were then incubated with or without 0.1 or 1 μM DEX (Sigma) dissolved in ethanol. At 36 to 48 h after transfection cells were washed twice with PBS and then harvested by scraping. The cell disruption by repeated freeze-thaw cycles, as well as the CAT and β-galactosidase assays, has also been described (25, 62).
Use of antisense oligonucleotides.
We used oligonucleotides with the sequences CATCTTGCTCGCCTCCCCCGC for human HDAC1 mRNA, ATTTCCGAGCTACGATCACCCGC for human HAT 1 (HAT1) mRNA (69) and, as a control, TGGATCATCTTCTGCCATTCT for NF-κB mRNAs. They were synthesized as phosphorothioates to prolong their half-lives in the cells. These sequences were designed to bind the initiation codon and the sequences immediately upstream, sites that commonly confer the most efficient antisense blocking. The cells were incubated in 1% delipidized medium from the beginning of transfection. The antisense DNAs were added to the transfected DNA mixture and subsequently to the medium of the cells transfected with the GR-responsive construct GRE-TK-CAT and K14CAT. Including the antisense DNA in the transfection mix has the advantage of ensuring that the cells that received the transfected DNA also received the antisense oligonucleotides. We added 5 μM concentrations of the oligonucleotides to the medium immediately after transfection and again 18 h later. Cells were harvested 36 h after transfection, and enzyme assays were performed.
Enzyme assays.
Briefly, the substrate solution contained 6 mg of o-nitrophenyl-d-galactoside (Sigma) freshly dissolved in PM buffer (66 mM Na2HPO4, 33 mM NaH2PO4, 40 mM mercaptoethanol, 2 mM MgSO4, and 0.1 mM MnCl2). The reaction mixture contained 100 μl of substrate solution, 300 μl of PM2 buffer, and 50 μl of keratinocyte cell extract or 20 to 30 μl of HeLa cell extract. It was incubated at 37°C until development of the yellow color was obvious, usually for 0.5 to 1 h. The time of the reaction was recorded, and the reaction was stopped by the addition of 0.4 ml of 1 M Na2CO3. The optical density at 420 nm was measured on a spectrophotometer (Gilford).
The CAT reaction mixture contained 69 μl of 1 M Tris HCl (pH 7.8), 1 μl of 14C-labeled chloramphenicol (Cm; 40 to 50 mCi/mmol; New England Nuclear), 20 μl of 4 mM acetyl-coenzyme A solution, 30 to 60 μl of cell extract, and enough water to bring the total reaction volume to 150 μl. After incubation at 37°C for 1 h, the mixture was extracted into 1 ml of ethyl acetate, phases were separated by brief centrifugation, the organic layer was transferred to a new tube, and the solvent was evaporated. The residue was dissolved in 30 μl of ethyl acetate and separated by thin-layer chromatography on silica gel in chloroform-methanol at 95:5. The plates were exposed to X-ray film for 12 to 24 h, and the intensity of the radioactive spots was determined using Ambis Radioanalytic System (Ambis, Inc., San Diego, Calif.). The conversion of chloramphenicol to its monoacetylated derivative was kept below 50% by varying the amount of extract or the duration of the reaction.
All CAT values were normalized for transfection efficiency by calculating the ratio of CAT activity to β-galactosidase in each transfected plate. Each transfection experiment was separately performed three or more times, with each datum point resulting from duplicate or triplicate transfections.
Northern blots.
Primary human keratinocytes were incubated in the presence or absence of 1 μM DEX for 6, 12, and 24 h. Cells were harvested by trypsinization, and the Quiagen RNeasy Kit was used to isolate total RNA according to the manufacturer's protocol. Then, 10 μg of total RNA was loaded per lane on the agarose gel. Capillary transfer to the Nylon membrane (Amersham) was performed according to a commercial protocol (Amersham). Probes K14cDNA and HPRTcDNA were labeled using a Random Primer Labeling Kit (Boehringer Mannheim). Next, 2 × 106 cpm of hybridization solution (Amersham) per ml was used to hybridize the membrane according to a commercial protocol (Amersham). The membrane was exposed to Kodak X-ray film for 18 h at −70°C.
Electrophoretic mobility shift assays (EMSAs).
Escherichia coli-expressed DNA-binding domain portions of hGR and cT3R were a gift from H. H. Samuels and have been described previously (16, 66). Oligonucleotides were synthesized on a Pharmacia Gene Assembler Plus Synthesizer. The sequence of oligonucleotides contained a 5′-GGG overhang designed for labeling. All double-stranded oligonucleotides used were gel purified before use. The oligonucleotides used in the EMSAs as probes were GRE (5′-GGGAGAACATAATGTTCT), NGRE (5′-GGGGATCCGGAAGGTCACGTCCAGGATC), K14RE (5′-GGGGCTAGCCTGTGGGTGATGAAAGCCAAGGGGAATGGAAAG), K17RE (5′-GG GTGGGAGCTGGCAGGTGGCCAGTGGTGATGAAAGCCCAAGGG), K5RE (5′-GGGTGACCGGTGAGCTCACAGCTGCCCCCCAGGCATGCCCA), K17S1 (5′-GGGGAAAC), K17S4 (5′-GGGTGGTGA), and GRE1/2 (5′-GGGAGAACA).
Double-stranded oligonucleotides corresponding to the sequences above were labeled with [α-32P]dCTP, using the Klenow fragment of E. coli DNA polymerase I. A total of 30,000 cpm of the resulting probe was mixed with 0.35 pg of purified receptor proteins and incubated first for 30 min at room temperature and then for 10 min at 4°C. In the experiments with the dose curve of GR-DBD 10, 15, 20, and 30 pg of purified protein was used. Experiments with full-size human GR were done similarly using recombinant GR from Affinity Bioreagents and following their commercial protocol. We used a monoclonal antibody raised against the DBD region of GR (Affinity Bioreagents). The incubation was done in a 30-μl volume in 25 mM Tris (pH 7.8), 500 μM EDTA, 88 mM KCl, 10 mM 2-β-mercaptoethanol, 0.1 μg of aprotinin, 0.1 μg of poly(dI-dC), 0.05% (vol/vol) Triton X-100, and 10% (vol/vol) glycerol. Samples were loaded on a 4% polyacrylamide gel and separated by electrophoresis (20 to 25 mA) at 4°C for 2 h with a buffer containing 10 mM Tris, 7.5 mM acetic acid, and 40 μM EDTA (pH 7.8). Gels were dried and exposed to X-ray film for 4 h at −70°C. The quantification of the affinity of protein binding to K17S1 and K17S4 was performed by spot densitometry using the Alpha Imager 2000 Analysis System from Alpha Innotech Corporation.
DNase I footprinting.
We have followed the general protocol described by Lakin (31). First, 1 μg of primer K17ft (5′-GCCCCCAGCCACCTGGGAGCT) was labeled by using polynucleotide kinase (Promega) and [γ-32P]dATP (Amersham). Next, 1.5 × 106 cpm of each primer was used in the primer extension reaction with K17ft (5′-GCTTGCTCCTCTGTTTCCATTCCCCTGGGCTTTCATCACCACTGGCCACCTGCCAGCTCCCAGGTGGCTGGGGGC) as a template and Klenow endonuclease (Boehringer Mannheim). The product was purified from a 2.5% agarose gel. The band corresponding to the size of the probe was cut out of the gel and eluted overnight in Tris-EDTA buffer (pH 8) at 4°C; this was followed by ethanol precipitation. Subsequently, two different reactions were performed in parallel: A/G Maxam-Gilbert sequencing (following the standard protocol) (35, 36) and DNase I footprinting. For the footprinting reaction our protocol for gel shifts was used to allow binding of the protein to the DNA: 25 μl of the binding mix (see gel shift protocol above) was combined with 50 ng of purified receptor protein and 50,000 cpm of probe. After 20 min of incubation at 4°C, 50 μl of solution containing 10 mM MgCl2 and 5 mM CaCl2 was added and incubated 1 min on ice. Next, 3 μl of the 1:25 dilution of the DNase I (5 U/ml of stock), a dilution which we have found to be optimal for our conditions, was added and incubated exactly 1 min on ice. The reaction was stopped by adding 90 μl of stop solution containing 20 mM EDTA (pH 8.0), 1% sodium dodecyl sulfate, 0.2 M NaCl, and 100 μg of yeast RNA per ml. DNA was purified by phenol extraction followed by ethanol precipitation. The pellet was resuspended in 1.4 μl of 9 M urea, 1% NP-40, and, after mixing, 4.6 μl of formamide loading buffer (USB) was added. All samples were heated at 90°C for 5 min, chilled on ice, and loaded on the 12% sequencing polyacrylamide gel as were the samples with the A/G Maxam-Gilbert sequencing reactions of the same DNA. Gels were subjected to 1,000 V of current for 1 h, dried on the gel dryer, and exposed to the X-ray film. The footprint localization was determined by the bands that were protected by the bound protein from cleavage by DNase I, which appeared on the film as “disappeared” bands when the footprinted sample lane on the gel was compared with the sample that had no protein in the mix. The protected bands were then compared with the A/G sequence lane on the same gel, revealing the nucleotides involved in binding of protein.
Immunohistology.
The forearms of healthy volunteers were treated with 0.05% clobetasole propionate (Temovate) twice a day. After 1, 2, 3, or 4 days, 4-mm punch biopsies were taken from the treated site and from an untreated adjacent site. Biopsies were embedded in Tissue Tek OCT compound (Miles Scientific), frozen in liquid nitrogen, and stored at −70°C. Frozen sections of 4 to 6 μm were cut (Fung Frigocut 2,800 E Cryostat) and then collected onto gelatin-coated slides. The sections were stained according to a standard immunofluorescent staining protocol (40). The primary antibody used was polyclonal rabbit anti-human GR antibody (Affinity Bioreagents). The secondary antibody used was anti-rabbit fluorescein isothiocyanate preabsorbed with human serum proteins (Sigma Immunochemicals). An Olympus Microscope was used to analyze the slides.
RESULTS
Physiology of the GRs in human keratinocytes.
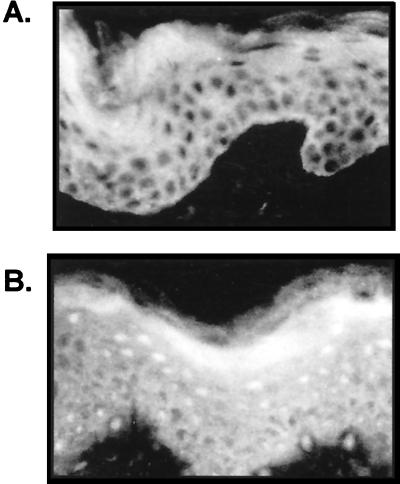
We have found that activation and nuclear translocation of GRs occur in keratinocytes, both in vivo and in vitro, in the presence of a specific ligand (Fig. 1 and data not shown). We applied clobetasole propionate, a potent synthetic steroid commonly used in dermatological therapy, topically to the skin of a volunteer. We obtained biopsies of treated and untreated skin and stained tissue sections with GR-specific antibody. GR, found in the cytoplasm of untreated skin (Fig. 1A), was activated and translocated to the nuclei in the treated skin (Fig. 1B). The activation of GR and its nuclear translocation was detected in all cell layers of the epidermis. This is a clear demonstration of activation of a transcription factor in skin caused by a topical treatment. We have found similar results in vitro using primary human keratinocytes and HeLa cells (data not shown). Within 6 h of treatment, the GR was translocated to the nuclei of the both cell types, and the GR remained nuclear during the 24-h treatment, thus demonstrating the activation and nuclear translocation of the GR both in vitro and in vivo.
FIG. 1.
Activation of the GR in human epidermis in vivo. Clobetasole propionate was applied topically to the skin of a volunteer for 4 days. Biopsies of the treated and adjacent untreated skin and were sectioned and stained with a GR-specific antibody. (A) GR is present in the cytoplasm of untreated epidermis. (B) Clobetasole propionate caused translocation of the GR from the cytoplasm to the nucleus.
Effects of GR on transcriptional regulation of keratin genes.
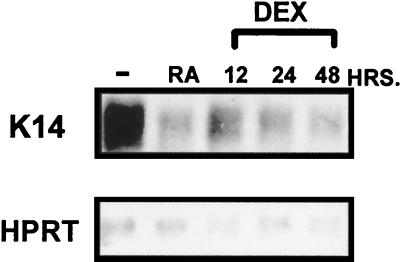
To determine the effect of GCs on gene transcription in epidermis, we have measured K14 and K16 mRNA levels during treatment of keratinocytes with DEX. Results from the Northern blot analysis are shown in Fig. 2. Interestingly, the K14 mRNA levels significantly decreased during the 12 h of treatment of keratinocytes by DEX and decrease even further after 24 and 48 h of treatment (Fig. 2). Retinoic acid (RA) also decreases K14 mRNA levels, as shown previously (Fig. 2 and reference 59). In contrast, HPRT mRNA, which was used as a control, was not significantly changed during the treatments. Similar results are obtained with K16 mRNA (data not shown). Taken together, the strong decrease of keratin mRNA levels by DEX indicates either the inhibition of keratin gene transcription or, alternatively, an increase in keratin mRNA turnover.
FIG. 2.
GCs inhibit the transcription of the endogenous K14 keratin gene. The levels of K14 mRNA were measured by using Northern blot analysis during keratinocyte treatment with DEX. The K14 mRNA levels significantly decreased during the 12-h treatment and decreased further during 24- and 48-h treatments. RA also decreases K14 mRNA levels, as shown previously (59). In contrast, HPRT mRNA, which was used as a control, was not significantly changed during the treatments.
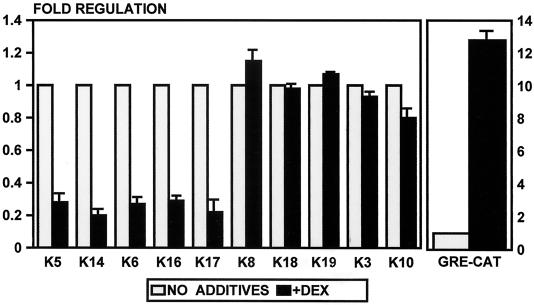
To determine if DEX inhibits keratin gene expression at the level of transcription, we tested 10 different keratin gene promoters. We cotransfected keratin gene promoter-CAT constructs into keratinocytes and HeLa cells and incubated the transfected cells in the presence or absence of DEX. We found that DEX suppressed the expression of the basal layer, as well as the disease-specific keratins K5, K14, K6, K16, and K17, three- to fivefold (Fig. 3). In contrast, the expression of the differentiation-specific keratins, K10 and K3, and the simple epithelium-specific keratins, K8, K18, and K19, was not affected by DEX (Fig. 3). The expression of all 10 of these keratins is regulated by retinoids and thyroid hormones (51, 62, 66).
FIG. 3.
GCs suppress transcription of five epidermal keratin genes. Primary human keratinocytes were cotransfected with keratin promoter-CAT constructs, and the cells were incubated in the absence or presence of DEX. DEX suppresses the expression of the basal-cell-specific keratins K5 and K14 and the disease-associated keratins K6, K16, and K17. It does not regulate the expression of the differentiation-specific keratins K3 and K10 or the simple epithelium-specific keratins K8, K18, and K19. GRE-CAT, a positive control, was induced by DEX as expected. The error bars represent the standard errors of the mean of multiple experiments, each performed with duplicate transfections.
We used GRE-CAT, the TK promoter that contains a positive GRE, as a control in our experiments and found it to be induced 12- to 15-fold by DEX (Fig. 3). We have found similar results in HeLa cells, i.e., GR suppressed K5, K6, K14, and K17, whereas it induces GP-CAT (data not shown). In addition, we have cotransfected a GR-expressing plasmid and found that the inhibition by DEX was not enhanced by the addition of exogenous GR, which means that the endogenous GR is sufficient to regulate fully the keratin gene expression (data not shown).
Identification of the GREs in keratin genes.
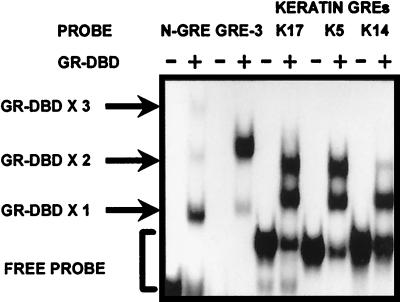
We have previously identified the RA and thyroid hormone response elements (RARE and TRE) in keratin promoters and found that they are complex sequences consisting of multiple binding sites. We hypothesized that if the RAR and T3R bind to the complex elements, the GR may recognize and bind the same sites. Therefore, we used these sequences as probes to test the binding of GR. We have used K17, K5, and K14 nGREs and recombinant GRs containing the DNA-binding domain (GR-DBD) in gel shift experiments and found that the sequences bind GRs in addition to T3R and RAR (Fig. 4). As controls in binding experiments we used the nGRE identified in the POMC gene that binds the monomer-plus-dimer of GR (10) and the consensus GRE spaced by three nucleotides (GRE-3). Both controls bound the GR as expected, i.e., the POMC nGRE bound monomer-plus-dimer of GR, whereas the GRE-3 bound a homodimer of GR.
FIG. 4.
GR binds to keratin GREs. EMSAs were performed with purified GR-DBD and K5, K17, and K14 response elements as DNA probes. The negative GRE characterized in the POMC gene (N-GRE) and the consensus GRE spaced by three nucleotides (GRE-3) were used as control DNA probes. As expected, GR binds to N-GRE in a monomer-plus-dimer formation and to GRE-3 as a homodimer. GR also binds to all three keratin GREs. GR-DBD X1, X2, and X3 refer to one, two, or three molecules of GR-DBD bound to DNA, respectively.
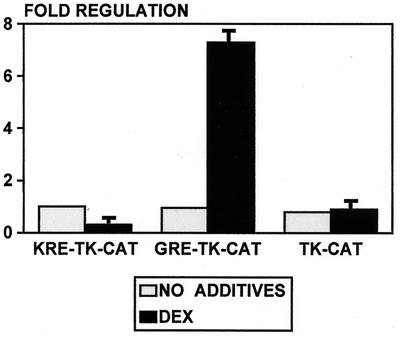
Are the keratin nGREs true negative elements or does the promoter context make keratin GREs negative? For example, the protein-protein interaction between the receptor and an adjacent protein may cause negative regulation. To test this possibility, we removed the keratin K17 nGRE from its promoter and cloned it into the TK promoter. The keratin GRE (KRE) cloned into TK promoter mediated repression by GR (Fig. 5). This means that KREs are “self-contained negative REs,” i.e., they contain all of the information necessary to instruct the receptor to repress, independently of the background and the context of the promoter. This result is very important because it proves that KREs are not promoter context dependent.
FIG. 5.
Keratin nGRE does not depend on the promoter context. nGRE from K17 promoter was cloned into the minimal TK promoter and tested for regulation by DEX in keratinocytes. The nGRE mediates the suppression of the TK promoter by DEX. The positive control plasmid GRE-TK, containing consensus GRE, similarly cloned into the TK promoter, was activated by DEX, whereas the minimal TK promoter lacking a response element was not regulated.
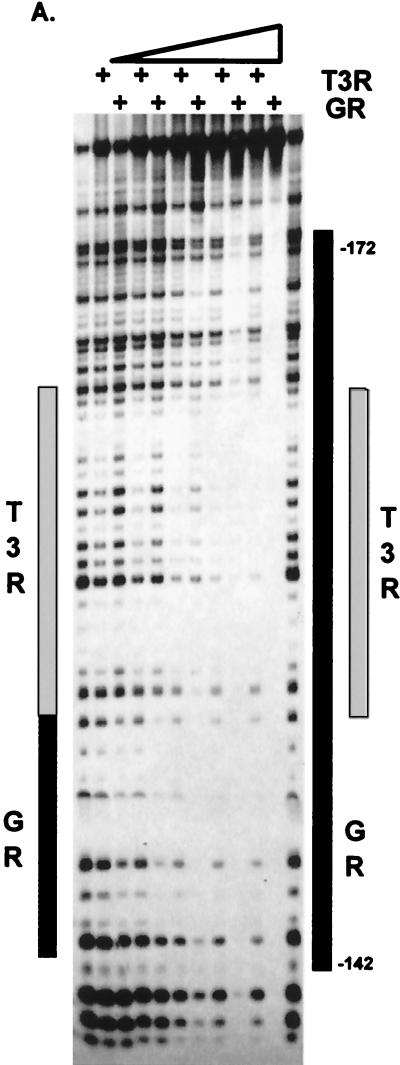
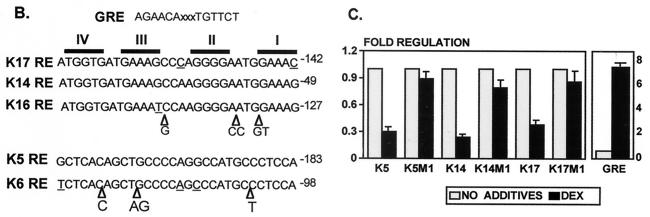
To map precisely the interactive sites between keratin GRE and GR, we have used K17GRE in DNase I footprint experiments (Fig. 6). Interestingly, GR-DBD binds initially to a single monomer binding site at positions −146 to −140. As the protein concentration increases, the GR-DBD footprint “grows” to occupy a total of four binding sites. In this respect GR is quite different from T3R. T3R binds to the same region as GR in the K17 promoter (Fig. 6A). The T3R footprint is adjacent to the initial GR binding site, and it does not grow when the protein concentration increases. The footprinting experiments identified nGREs in keratin promoters as follows: in the K5 promoter at −213 to −183, in K6 at −132 to −98, in K14 at −79 to −49, in K16 at −162 to −127, and in K17 at −172 to −142 (Fig. 6B). Importantly, we have found that the sequences responsible for GR binding in the acidic keratin promoters have >90% identity, constituting a signature sequence. In addition, the GREs in the two basic keratin genes also have a high degree of similarity. However, the acidic signature sequence is different from the basic signature sequence, although both consist of a cluster of binding sites that bind four monomers of GR.
FIG. 6.
Identification and mapping of keratin GREs. (A) Footprinting analysis. Footprints of GR and T3R on K17RE are shown. Gray rectangles represent the binding of the T3R. The black rectangle on the left represents the initial GR binding site in the K17GRE; increased amounts of GR cause an enlargement of the initial footprint marked by a black rectangle on the right. (B) Signature sequences of the keratin GREs. Keratin GREs, mapped by footprinting, have a high degree of sequence homology specific for acidic or basic keratin genes. The acidic signature sequences (top) and basic signature sequences (bottom) are shown, with differences underlined and triangles marking the insertions. The positions of the sequences in the promoter are indicated. Respective binding sites are shown on the top marked with roman numerals. The GRE consensus sequence is shown on the top for comparison. (C) Identification of the GREs in keratin promoters by site-specific mutagenesis. GREs in K5, K14, and K17 promoters were altered to create K5M1, K14M1, and K17M1, respectively, and tested for regulation by DEX in keratinocytes. Regulation by DEX in all three mutant promoters was abolished, thus confirming that the GREs identified by EMSA and footprinting mediate the regulation of keratin gene transcription by GR.
The identified nGREs in keratin promoters provide binding to the GR, but it is not clear that those binding sites function as nGREs, i.e., cause the suppression of transcription by GR. Therefore, we mutagenized the sequences of nGREs in the context of their promoters and used the mutants in cotransfection experiments. Introduced mutations altered only the sequences of the binding sites, whereas the number of the base pairs within each binding site remained unchanged (51). We have found that in all three promoters the introduced mutations, K5M1, K14M1, and K17M1, are sufficient to abolish regulation by DEX (Fig. 6C). Therefore, the identified nGREs are responsible for the regulation of keratin gene expression by GR.
Four monomers of GR bind to keratin nGREs.
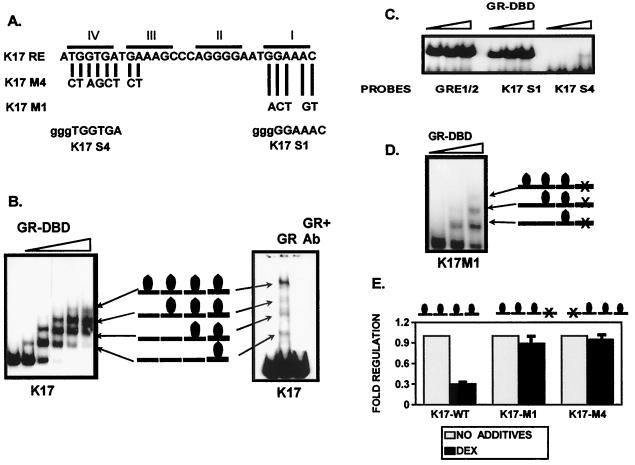
Intrigued by the growth of the footprint (Fig. 6), we examined carefully the interaction between the GR and the nGRE. The results from the gel shift assay confirmed the footprinting results (Fig. 7). The binding assay with small amounts of the purified GR-DBD initially revealed a binding pattern that is consistent with the interpretation of binding of a GR monomer (Fig. 7B; see also the Discussion). As the concentration of the receptor increases, the monomer is converted into a two-monomer unit, which further becomes three and finally four. These experiments suggested that the GR binds the keratin GREs as a monomer rather than as a homodimer. In addition, as the receptor concentration increases, the GR binds as multiple monomers (see details in Discussion). Most importantly, we have obtained the same binding pattern with the full-size GR (Fig. 7B). Just like the GR-DBD, the full-size GR also binds the keratin GRE as four monomers. The binding is specific because an antibody against the DBD of the GR blocks it. The fact that keratin GREs bind four monomers of the GR, but not homodimers, implies a new mechanism of negative regulation through which GR suppresses keratin gene expression.
FIG. 7.
Four monomers of GR bind to the keratin GREs and mediate their suppression. (A) Summary of the sequence analysis. Introduced mutations used in cotransfection experiments are shown as K17M1 and K17M4. K17S4 and K17S1 probes for the gel shift experiment are shown below with 5′ GGG overhang designed for labeling. (B) Gel shift experiment with recombinant GR-DBD (left), full-size GR (right), and a K17RE probe is shown. GR-DBD binds initially as a monomer. As the concentration of the GR-DBD increases, two monomers, then three, and finally four are bound to the GREs. A gel shift experiment with recombinant human full-size GR and a K17RE probe shows a similar binding pattern. A monoclonal GR-specific antibody raised against the DBD region blocks the binding. (C) The primary and quaternary binding sites in the K17GRE have different affinities of binding to GR. GR binds with similar affinities to the consensus GRE half-site (GRE1/2) and the primary binding site in K17GRE (K17S1), whereas the quaternary site (K17S4) binds with a significantly lower affinity in gel shift experiments. (D) Binding of the GR to four binding sites in keratin GRE is not cooperative. Mutation in the primary binding site K17M1 did not affect the binding of the GR to the remaining three binding sites in keratin K17GRE in the gel shift experiments. (E) The binding of all four monomers is required for the suppression of keratin gene transcription. Mutations introduced into the primary (K17M1) or the quaternary (K17M4) binding sites abolished regulation by DEX in cotransfection experiments.
What is the role of the four binding sites in keratin nGREs? To characterize the binding sites within the keratin nGREs, we have used primary and quaternary binding sites as separate probes in the gel shift experiments. Our results show that the primary binding site has an approximately 10-fold-higher affinity of binding to the GR than the quaternary site (Fig. 7C).
One can speculate further that the binding of GR to nGRE is cooperative, i.e., binding of the first monomer to the high-affinity primary site initiates the binding of the remaining three monomers to the further sites. To test this we have mutagenized the primary binding site and used the mutagenized GRE as a probe in the gel shift and footprinting experiments. We found that the binding of the GR to the primary site was abolished, but the binding to the remaining sites was intact (Fig. 7D). Thus, the binding is not cooperative. Finally, we asked whether all four binding sites are necessary for regulation. Therefore, we have mutagenized the primary and, separately, the quaternary binding site in the K17 promoter, creating two mutant promoters: K17GREM1 and K17GREM4 (for the position of the mutations, see Fig. 7A). Neither mutant promoter was regulated by the GR in cotransfection experiments (Fig. 7E), which means that all four sites are necessary for regulation. To test the binding of the GR to the mutants, we used them as probes in footprinting experiments, which show that in both mutants the GR binding was altered in the mutagenized regions (data not shown). The binding to unaltered sites was unaffected. Taken together, these results indicate that, although they have different affinities of binding to the GR, all four binding sites are necessary for the regulation to occur.
Role of the coregulators in the regulation of keratin genes by GR.
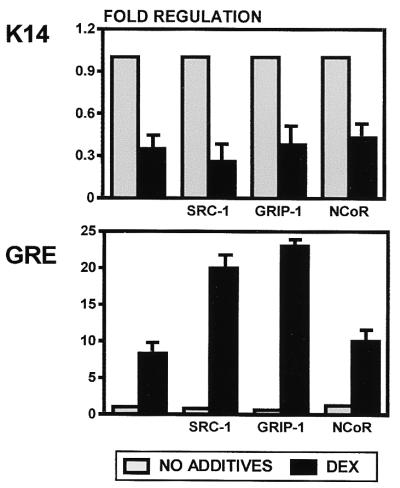
To determine whether known coregulators play a role in the suppression of keratin gene expression by GR, we have used vectors expressing coregulators SRC-1 and GRIP-1 in cotransfection experiments (Fig. 8). Interestingly, SRC-1 and GRIP-1 had no effect on DEX-mediated suppression of keratin promoters, whereas they enhanced the activation of GRE-CAT. This means that the coregulatory proteins that enhance induction of transcription on a consensus (positive) element are not involved in the suppression of keratin genes by the same receptor. Since both SRC-1 and GRIP-1 are known as coactivators, we have tested NCoR, a corepressor, in cotransfection experiments. We have found that, like SRC-1 and GRIP-1, NCoR had no effect on the keratin gene regulation by DEX (Fig. 8). As expected, NCoR did not affect GRE-CAT regulation by DEX either (Fig. 8), whereas it enhanced the repression of TRE-CAT by unliganded T3R (data not shown). We are currently searching for specific coregulators that interact with the GR in the context of keratin promoters.
FIG. 8.
Role of the coregulators in regulation of keratin gene expression by GR. We have tested coactivators SRC-1 and GRIP-1 and corepressor NCoR in cotransfection experiments with K14CAT (top) and GRE-CAT (bottom). None of the coregulators tested affected the suppression of keratin gene expression by DEX. SRC-1 and GRIP-1, but not the NCoR, enhanced the activation of the positive control, GRE-CAT, as expected.
Role of the histone acetylation in the regulation of keratin genes by GR.
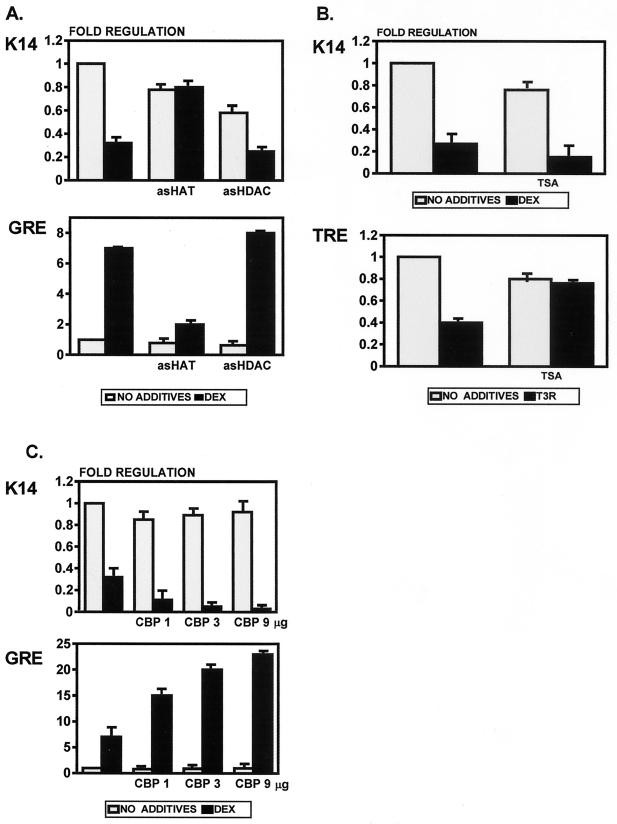
Histone acetylase, but not histone deacetylase, plays a role in the suppression of keratin gene expression by GR (Fig. 9). We have used antisense technology to test the role of histone acetylation in this regulation. Using an antisense approach one can target and block specific mRNAs, thus inhibiting the new synthesis of a particular protein. We added antisense oligonucleotides blocking HAT (AS-HAT) or HDAC (AS-HDAC) in cotransfection experiments and tested their effect on keratin gene regulation by GR. Surprisingly, we found that AS-HAT blocks the repression of keratin gene expression by DEX, whereas it blocks the induction of GRE-CAT (Fig. 9A). AS-HDAC had no effect on either keratin promoters or GRE-CAT. AS-HDAC efficiently blocked the suppression of TRE-CAT by unliganded T3R, as expected (data not shown). To confirm that HDAC does not play a role in the suppression by DEX, we have used TSA, a specific inhibitor of HDAC (41, 74). We found that TSA did not affect the suppression of K14CAT by DEX, while it did block the suppression of TRE-CAT by unliganded T3R (Fig. 9B). Due to a toxic effect on the cells, TSA decreased the basic activity of the reporter constructs. However, the fold regulation did not change in the presence of DEX, thus confirming that HDAC does not play a role in regulation of keratin gene expression by DEX. In addition, we have used a plasmid expressing CBP in cotransfection experiments to confirm our findings with HAT. CBP, as a component of a coactivator pathway on positive response elements, has acetylase activity and binds to the receptor-coregulator complex that further interacts with HAT. Therefore, if HAT is a component of the repressor complex in the regulation of keratin genes by GR (as our data indicate), CBP should be a component as well. We expected that it would enhance the suppression by DEX. Indeed, we have found that CBP does enhance suppression of keratin gene suppression by DEX three- to fivefold (Fig. 9C). The enhancement is concentration dependent. Conversely, CBP enhances the DEX-mediated activation of the positive control GRE-CAT. Taken together, this means that histone acetylation is involved in the repression of keratin gene expression by GR. It appears that both CBP and HAT are auxiliary proteins specific for the liganded receptor and not direct inducers of transcription. Furthermore, our results indicate that although HAT activity is usually associated with gene activation, it may also participate in gene repression.
FIG. 9.
HAT, but not HDAC, is involved in the suppression of keratin gene expression by DEX. (A) We added antisense oligonucleotide-blocking HAT (AS-HAT) and one blocking HDAC (AS-HDAC) into cotransfection experiments. AS-HAT blocked the repression of keratin gene expression by DEX, whereas it blocked the induction of GRE-CAT. AS-HDAC had no effect on either the keratin promoters or GRE-CAT. (B) TSA, an HDAC inhibitor, does not affect the suppression of keratin gene expression by DEX. As expected, TSA blocks the inhibition of the positive control, thyroid response element TRE-CAT, by unliganded T3R. (C) CBP enhances suppression of keratin gene expression by DEX in a dose-dependent manner, whereas it enhances activation of GRE-CAT. Three different amounts of CBP-expressing plasmid were used (1, 3, and 9 μg).
Second mechanism of inhibition of keratin gene expression by GR.
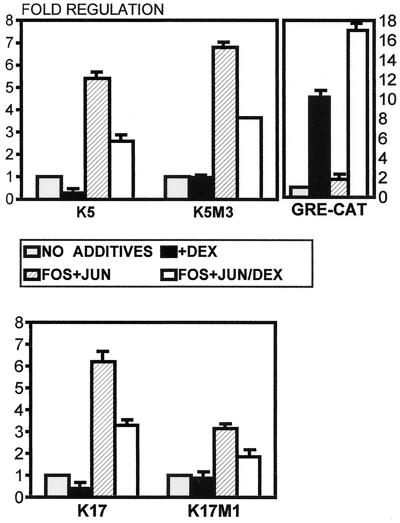
GR has been shown to interfere with AP1 in several systems (3, 20, 38, 48, 68). We have shown previously that K5, K6, K16, and K17 keratin genes contain functional AP1 sites (34, 37, 43). Therefore, we have tested the GR effect on keratin gene regulation by AP1 protein complex. We cotransfected HeLa cells with components of AP1, Fos, and Jun expressing plasmids along with K5 and K17 CAT constructs and incubated the cells in the presence or absence of DEX. We found that DEX significantly blocks the induction of K5 and K17 by AP1 (Fig. 10). Furthermore, when we used mutants of K5M1 and K17M1, which are not directly regulated by DEX (see Fig. 6C), we found that both mutant promoters were induced by AP1 and that this induction can be inhibited by DEX (Fig. 10). This means that GCs, in addition to the direct mechanism, use another, indirect mechanism of regulating keratin gene expression, namely, by inhibiting their induction by AP1.
FIG. 10.
GR blocks the induction of keratin gene expression by AP1. We have cotransfected HeLa cells with Fos and Jun expression plasmids along with K5 and K17 CAT constructs. The transfected cells were cultures in the presence or absence of DEX. DEX significantly blocked the induction of K5 and K17 by the cotransfected Fos and Jun. Furthermore, K5M1 and K17M1, which are no longer directly regulated by DEX (see Fig. 6C), were induced by AP1, and this induction was inhibited by DEX. The inhibition of the AP1 is mediated by a different mechanism that does not depend on GREs in keratin promoters.
DISCUSSION
The mechanism of suppression of keratin gene transcription by GR, described here, is an excellent example of tissue specificity in gene regulation. There are several levels of specificity evident in this particular mechanism. The first is the DNA sequence of the nGRE in keratin genes. Keratin GREs constitute the first group of native negative regulatory elements identified in a gene family. The signature sequences are only found in the five keratin genes regulated by GR, RAR, and T3R and not in the genes regulated only by RAR and T3R. This suggests that the structure of the response elements in these five keratin genes is customized to suit the purpose of binding all three receptors. The second level is the binding of four monomers of GR, arguably the most surprising result in this study. The unique property of the keratin nGRE is to allow binding of four GR monomers, and only if all four are bound does the regulation by GR occur. The third level relates to the coregulatory proteins. The simplest explanation for their failure to affect keratin gene regulation is that the GR is in a monomeric form. The coregulators tested either do not bind the monomeric GR or cannot fulfill the function if bound to the monomer. It is also possible that suppression by the liganded receptor requires its own set of coregulators that are yet to be discovered. The final level is the histone acetylation, which was another surprise. Our results indicate that histone acetylase, but not histone deacetylase, plays a role in the suppression of transcription. Taken together, every step in the path of the suppression of keratin gene expression by GR is different than those previously described. We believe that this mechanism is specifically designed for the purpose of targeting keratin genes within the target tissue, i.e., epidermis.
We identified the acidic keratin signature sequence, which was 93% identical, in the K14, K16, and K17 promoters, and the basic signature sequence, which was 91% identical, in the K5 and K6 promoters. The acidic and basic signature sequences do not have significant homology. Interestingly, the GR binds to both signature sequences in identical patterns, i.e., as four monomers, which means that, although differentially expressed, K5, K6, K14, K16, and K17 are regulated by GR through the same mechanism. Our results also emphasize the important role that the DNA sequence plays in transcriptional regulation by nuclear receptors. In this mechanism, the structure and sequence of the nGRE in keratin promoters positions the receptor in a specific configuration. We believe that the character of keratin nGRE is the initial signal for suppression.
Negative regulation, or suppression, by GR and other nuclear receptors may occur through several different mechanisms, such as direct binding of the receptor to a negative GRE, e.g., in the keratin genes and the POMC gene (10); direct interference with transcriptional machinery, e.g., in the osteocalcin gene where GRE overlaps with the TATA box (60); protein-protein interactions with positive regulators, e.g., AP1, forming an inactive complex (20, 38, 48); and induction of expression of an inhibitor of transcription, e.g., IκB, through which GR functionally inhibits those genes induced by NF-κB (9, 19). The molecular mechanism of keratin gene suppression by GR described here is one of the rare examples of active repression. Direct negative regulation proceeds through binding of the four GR monomers to the keratin GREs. There are three lines of evidence leading to this conclusion. The first arises from the gel shift and footprinting experiments. Judging by the intensity of the bands and binding patterns it is clear that binding occurs by the addition of monomers one by one, not by two dimers or a dimer and a monomer. In addition, when one site is mutagenized the remaining three sites bind three monomers (see Fig. 7D), thus confirming the independent binding of monomers. The second line of evidence arises from the mutagenesis experiments of the GR binding sites in the keratin nGRE. Regulation occurs only if all four binding sites are intact, although their affinities of binding are different. The third line of evidence arises from the structure and sequence of the binding sites. Judging by the crystal structure of the GR-DBD it is not possible to fit the receptor dimer in any two combinations of the GR binding sites in keratin nGRE (32, 33). According to the current knowledge, the spacing between the half sites has to be either three or four nucleotides, in the inverted palindrome orientation. In addition, the sequence of the consensus binding site seems to be restrictive, i.e., if it is changed from AGAACA to AGGACA, it changes the affinity of binding from a preferentially GR binding site to an ER binding site (53, 54). None of these rules, however, applies to the identified keratin binding sites. Instead, their structure, sequence, and orientation preclude the binding of GR homodimers. Consistently, no homology between nGREs in keratin genes and consensus, positive, GRE can be found. Therefore, multiple monomer binding of the GR is a novel mechanism of suppression by glucocorticoids, so far found only in the epidermal keratin genes.
The role of coregulators in this mechanism, although not defined, is evidently different than what has been described in the literature. The known general coactivators that interact with liganded GR and enhance positive regulation do not affect the function of the same receptor when it suppresses keratin gene expression. The most probable explanation is that known coregulators do not functionally interact with GR on keratin nGREs because they cannot function with monomers of GR. There is some evidence suggesting that coregulators may interact with two receptors at the same time through different regions and that their function depends on the position and alosteric conformation of the interactive sites (72). Several tissue-specific coregulators have been described recently (50, 75), raising the possibility that the coregulators involved in keratin regulatory mechanism might be epidermis specific rather than general. We are currently investigating these possibilities.
Coactivators recognize and bind to liganded receptor on one side and to CBP/p300 on another. CBP/p300, in addition to being a histone acetylase itself, can also interact with HAT, causing histone acetylation and further induction of transcription. Liganded GR suppresses keratin gene transcription, which immediately raises the question regarding the role of histone acetylation in this regulation. One can expect that the GR on keratin nGREs interacts somehow with other proteins that interact with histone deacetylase (HDAC) causing repression of transcription. Interestingly, and much to our surprise, the results show exactly the opposite: histone acetylase, rather than deacetylase, participates in the suppression of keratin gene transcription. This result is further supported by our finding that CBP as well enhances the suppression of keratin gene expression by GR. These findings are in contrast to the previously established paradigm of HAT as a member of the transcriptional activator complex (39). Our results suggest that HAT is a coregulator specific for the liganded receptor and is not directly responsible for induction of transcription. This leaves open the question of the function of the acetylation of histones per se in the regulation of transcription. It has been shown that CBP/p300 can have a repressive function as well (71). Understandably, our findings raise more questions that are subject of further studies. However, it is clear that these findings point toward multiple functions of histone acetylase in transcriptional regulation dependent on a particular mechanism rather than as a general phenomenon.
Our laboratory and others have previously shown that RA and T3 receptors regulate the expression of keratin genes (62, 66). Our finding is that GCs target for regulation only those keratin genes that are aberrantly expressed in diseased epidermis, specifically the basal-cell-specific K5-K14, the activated keratinocyte-specific K6-K16, and the “inflammation”-specific keratin K17. GCs do not affect simple or differentiation-specific keratin genes. The question one must ask is, what is the biological relevance of this regulation? Long-term topical treatment by GCs causes thinning of epidermis (58), which correlates with the suppression of K5-K14 keratin genes, markers of the basal keratinocytes. In addition, GCs are known to inhibit the wound healing process, which is attributed to their growth-inhibitory effect (70). This correlates with the suppression of K6-K16 keratin genes, markers of activated keratinocytes (27). Finally, GCs are most often used therapeutically for their anti-inflammatory effects. This correlates with the suppression of expression of the K17 keratin gene, which is present in epidermis only during IFN-γ-related inflammatory processes (26).
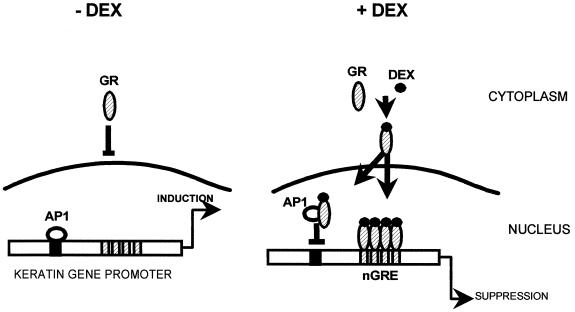
We have found that in addition to this direct regulation, the GR indirectly regulates expression of keratin genes by blocking AP1 transcription factor. This indirect regulation may be particularly important in wound healing, psoriasis, and inflammatory dermatoses, which are associated with activation of the AP1 proteins (11–13). The direct binding of the GR to GRE in the keratin promoter is not involved in this mechanism, because the indirect regulatory pathway is still functional in the K5 and K17 mutant promoters, in which the direct regulation does not occur. This means that the direct and the indirect regulatory pathways are independent of each other and may function at the same time (Fig. 11). Homodimerization of the GR is not necessary for the interaction with c-Jun, and it is tempting to speculate that monomers of GR have a dual function in keratin gene regulation: they directly inhibit the transcription by binding in multiple copies to keratin REs, and they bind to c-Jun blocking keratin induction by the AP1 complex. In addition, the dominant-negative GR used to generate transgenic mice, as described by Reichardt et al., was impaired in homodimerization, i.e., it binds to the DNA only as a monomer (53, 54). Interestingly, the inhibition of AP1 by the GR in this mouse was intact, and the animal did not have an aberrant skin phenotype, suggesting that the two mechanisms of regulation of keratin gene expression in epidermis of this mouse are intact.
FIG. 11.
Diagram of the two different regulatory pathways of suppression of keratin gene expression by monomers of GR.
Having potent and general transcription factors, such as nuclear receptors, is the nature-designed distinct way of reaching specific targets, thus differentially orchestrating gene expression in different tissues at the same time. We describe here a novel mechanism of suppression of transcription by GR that represents an example of customizing a device for the transcriptional regulation to target a specific group of genes in the target tissue, the epidermis.
ACKNOWLEDGMENTS
Our research was supported by National Institutes of Health grants AR30682, AR39176, and AR40522. M.T.-C. is a recipient of Advanced Polymer Systems Research Fellowship Award granted through the Dermatology Foundation and Rudolf Baer Foundation Research Grant.
We give special thanks to Irwin M. Freedberg for his support, enthusiasm, and dedication to this project and our work. We also give special thanks to Anita Orlin for her help, understanding, and support. We thank R. Oshima, J. Schweizer, P. Chambon, H. H. Samuels, B. W. O'Malley, M. Stullcup, R. H. Goodman, C. K. Glass, and L. Freedman for gifts of plasmids. We also thank E. Hadzic and H. H. Samuels for sharing with us their expertise in nuclear receptor purification; Laxmi Rao for her help in developing footprinting technique; and Sasa Radoja for critical reading of the manuscript.
REFERENCES
- 1.Abraham L J, Bradshaw A D, Northemann W, Fey G H. Identification of a glucocorticoid response element contributing to the constitutive expression of the rat liver alpha 1-inhibitor III gene. J Biol Chem. 1991;266:18268–18275. [PubMed] [Google Scholar]
- 2.Alroy I, Freedman L P. DNA binding analysis of glucocorticoid receptor specificity mutants. Nucleic Acids Res. 1992;20:1045–1052. doi: 10.1093/nar/20.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett T J, Vedeckis W V. Occupancy and composition of proteins bound to the AP-1 sites in the glucocorticoid receptor and c-jun promoters after glucocorticoid treatment and in different cell types. Recept Signal Transduct. 1996;6:179–193. [PubMed] [Google Scholar]
- 4.Blumenberg M. Keratinocytes: biology and differentiation. In: Arndt K A, Leboit P E, Robinson J K, Wintroub B U, editors. Cutaneous medicine and surgery. Vol. 1. Philadelphia, Pa: W. B. Saunders Company; 1996. pp. 58–74. [Google Scholar]
- 5.Budunova I V, Carbajal S, Kang H, Viaje A, Slaga T J. Altered glucocorticoid receptor expression and function during mouse skin carcinogenesis. Mol Carcinog. 1997;18:177–185. [PubMed] [Google Scholar]
- 6.Cha H H, Cram E J, Wang E C, Huang A J, Kasler H G, Firestone G L. Glucocorticoids stimulate p21 gene expression by targeting multiple transcriptional elements within a steroid responsive region of the p21waf1/cip1 promoter in rat hepatoma cells. J Biol Chem. 1998;273:1998–2007. doi: 10.1074/jbc.273.4.1998. [DOI] [PubMed] [Google Scholar]
- 7.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 8.Cronstein B N, Kimmel S C, Levin R I, Martiniuk F, Weissmann G. Corticosteroids are transcriptional regulators of acute inflammation. Trans Assoc Am Phys. 1992;105:25–35. [PubMed] [Google Scholar]
- 9.De Bosscher K, Schmitz M L, Vanden Berghe W, Plaisance S, Fiers W, Haegeman G. Glucocorticoid-mediated repression of nuclear factor-kappaB-dependent transcription involves direct interference with transactivation. Proc Natl Acad Sci USA. 1997;94:13504–13509. doi: 10.1073/pnas.94.25.13504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drouin J, Sun Y L, Chamberland M, Gauthier Y, De Lean A, Nemer M, Schmidt T J. Novel glucocorticoid receptor complex with DNA element of the hormone-repressed POMC gene. EMBO J. 1993;12:145–156. doi: 10.1002/j.1460-2075.1993.tb05640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher G J, Datta S C, Talwar H S, Wang Z Q, Varani J, Kang S, Voorhees J J. Molecular basis of sun-induced premature skin ageing and retinoid antagonism. Nature. 1996;379:335–339. doi: 10.1038/379335a0. [DOI] [PubMed] [Google Scholar]
- 12.Fisher G J, Talwar H S, Lin J, Lin P, McPhillips F, Wang Z, Li X, Wan Y, Kang S, Voorhees J J. Retinoic acid inhibits induction of c-Jun protein by ultraviolet radiation that occurs subsequent to activation of mitogen-activated protein kinase pathways in human skin in vivo. J Clin Investig. 1998;101:1432–1440. doi: 10.1172/JCI2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher G J, Voorhees J J. Molecular mechanisms of photoaging and its prevention by retinoic acid: ultraviolet irradiation induces MAP kinase signal transduction cascades that induce Ap-1-regulated matrix metalloproteinases that degrade human skin in vivo. J Investig Dermatol Symp Proc. 1998;3:61–68. [PubMed] [Google Scholar]
- 14.Forman B M, Casanova J, Raaka B M, Ghysdael J, Samuels H H. Half-site spacing and orientation determines whether thyroid hormone and retinoic acid receptors and related factors bind to DNA response elements as monomers, homodimers, or heterodimers. Mol Endocrinol. 1992;6:3429–3442. doi: 10.1210/mend.6.3.1316541. [DOI] [PubMed] [Google Scholar]
- 15.Freedman L P. Increasing the complexity of coactivation in nuclear receptor signaling. Cell. 1999;97:5–8. doi: 10.1016/s0092-8674(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 16.Freedman L P, Luisi B F, Korszun Z R, Basavappa R, Sigler P B, Yamamoto K R. The function and structure of the metal coordination sites within the glucocorticoid receptor DNA binding domain. Nature. 1988;334:543–546. doi: 10.1038/334543a0. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs E. Epidermal differentiation: the bare essentials. J Cell Biol. 1990;111:2807–2814. doi: 10.1083/jcb.111.6.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Govindan M V, Devic M, Green S, Gronemeyer H, Chambon P. Cloning of the human glucocorticoid receptor cDNA. Nucleic Acids Res. 1985;13:8293–8304. doi: 10.1093/nar/13.23.8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heck S, Bender K, Kullmann M, Gottlicher M, Herrlich P, Cato A C. I kappaB alpha-independent downregulation of NF-kappaB activity by glucocorticoid receptor. EMBO J. 1997;16:4698–4707. doi: 10.1093/emboj/16.15.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heck S, Kullmann M, Gast A, Ponta H, Rahmsdorf H J, Herrlich P, Cato A C. A distinct modulating domain in glucocorticoid receptor monomers in the repression of activity of the transcription factor AP-1. EMBO J. 1994;13:4087–4095. doi: 10.1002/j.1460-2075.1994.tb06726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinzel T, Lavinsky R M, Mullen T M, Söderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:6628–6638. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 22.Hong H, Kohli K, Garabedian M J, Stallcup M R. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horwitz K B, Jackson T A, Bain D L, Richer J K, Takimoto G S, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996;10:1167–1177. doi: 10.1210/mend.10.10.9121485. [DOI] [PubMed] [Google Scholar]
- 25.Jiang C K, Connolly D, Blumenberg M. Comparison of methods for transfection of human epidermal keratinocytes. J Investig Dermatol. 1991;97:969–973. doi: 10.1111/1523-1747.ep12491889. [DOI] [PubMed] [Google Scholar]
- 26.Jiang C K, Flanagan S, Ohtsuki M, Shuai K, Freedberg I M, Blumenberg M. Disease-activated transcription factor: allergic reactions in human skin cause nuclear translocation of STAT-91 and induce synthesis of keratin K17. Mol Cell Biol. 1994;14:4759–4769. doi: 10.1128/mcb.14.7.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang C K, Magnaldo T, Ohtsuki M, Freedberg I M, Bernerd F, Blumenberg M. Epidermal growth factor and transforming growth factor alpha specifically induce the activation- and hyperproliferation-associated keratins 6 and 16. Proc Natl Acad Sci USA. 1993;90:6786–6790. doi: 10.1073/pnas.90.14.6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 29.Kupper T. The role of cells and cytokines in immunity and inflammation. In: Oppenheim J, Shevach E, editors. Immunophysiology. New York, N.Y: Oxford University Press; 1990. pp. 285–305. [Google Scholar]
- 30.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bächinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 31.Lakin N D. Determination of DNA sequences that bind transcription factors by DNA footprinting. In: Latchman D S, editor. Transcription factors: a practical approach. New York, N.Y: IRL Press at Oxford University Press; 1993. pp. 27–46. [Google Scholar]
- 32.Luisi B F, Schwabe J W, Freedman L P. The steroid/nuclear receptors: from three-dimensional structure to complex function. Vitam Horm. 1994;49:1–47. doi: 10.1016/s0083-6729(08)61145-0. [DOI] [PubMed] [Google Scholar]
- 33.Luisi B F, Xu W X, Otwinowski Z, Freedman L P, Yamamoto K R, Sigler P B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 34.Magnaldo T, Bernerd F, Freedberg I M, Ohtsuki M, Blumenberg M. Transcriptional regulators of expression of K6 and K16, the disease-associated keratin. DNA Cell Biol. 1993;12:911–923. doi: 10.1089/dna.1993.12.911. [DOI] [PubMed] [Google Scholar]
- 35.Maxam A M, Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maxam A M, Gilbert W. A new method for sequencing DNA. Bio/Technology. 1992;24:99–103. [PubMed] [Google Scholar]
- 37.Milisavljevic V, Freedberg I M, Blumenberg M. Characterization of nuclear protein binding sites in the promoter of keratin K17 gene. DNA Cell Biol. 1996;15:65–74. doi: 10.1089/dna.1996.15.65. [DOI] [PubMed] [Google Scholar]
- 38.Miner J N, Yamamoto K R. The basic region of AP-1 specifies glucocorticoid receptor activity at a composite response element. Genes Dev. 1992;12B:2491–2501. doi: 10.1101/gad.6.12b.2491. [DOI] [PubMed] [Google Scholar]
- 39.Montminy M. Transcriptional activation. Something new to hang your HAT on. Nature. 1997;387:654–655. doi: 10.1038/42594. [DOI] [PubMed] [Google Scholar]
- 40.Mutasim D F, Vaughan A, Supapannachart N, Farooqui J. Skin explant culture: a reliable method for detecting pemphigoid antibodies in pemphigoid sera that are negative by standard immunofluorescence and immunoblotting. J Investig Dermatol. 1993;101:624–627. doi: 10.1111/1523-1747.ep12366084. [DOI] [PubMed] [Google Scholar]
- 41.Niki T, Rombouts K, De Bleser P, De Smet K, Rogiers V, Schuppan D, Yoshida M, Gabbiani G, Geerts A. A histone deacetylase inhibitor, trichostatin A, suppresses myofibroblastic differentiation of rat hepatic stellate cells in primary culture. Hepatology. 1999;29:858–867. doi: 10.1002/hep.510290328. [DOI] [PubMed] [Google Scholar]
- 42.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 43.Ohtsuki M, Flanagan S, Freedberg I M, Blumenberg M. A cluster of five nuclear proteins regulates keratin gene transcription. Gene Expr. 1993;3:201–213. [PMC free article] [PubMed] [Google Scholar]
- 44.Oñate S A, Boonyaratanakornkit V, Spencer T E, Tsai S Y, Tsai M J, Edwards D P, O'Malley B W. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J Biol Chem. 1998;273:12101–12108. doi: 10.1074/jbc.273.20.12101. [DOI] [PubMed] [Google Scholar]
- 45.Oñate S A, Tsai S Y, Tsai M J, O'Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 46.Oshima R G, Baribault H, Caulín C. Oncogenic regulation and function of keratins 8 and 18. Cancer Metastasis Rev. 1996;15:445–471. doi: 10.1007/BF00054012. [DOI] [PubMed] [Google Scholar]
- 47.Pazin M J, Kadonaga J T. What's up and down with histone deacetylation and transcription? Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 48.Pearce D, Matsui W, Miner J N, Yamamoto K R. Glucocorticoid receptor transcriptional activity determined by spacing of receptor and nonreceptor DNA sites. J Biol Chem. 1998;273:30081–30085. doi: 10.1074/jbc.273.46.30081. [DOI] [PubMed] [Google Scholar]
- 49.Pratt W B. The role of heat shock proteins in regulating the function, folding, and trafficking of the glucocorticoid receptor. J Biol Chem. 1993;268:21455–21458. [PubMed] [Google Scholar]
- 50.Puigserver P, Wu Z, Park C W, Graves R, Wright M, Spiegelman B M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 51.Radoja N, Diaz D V, Minars T J, Freedberg I M, Blumenberg M, Tomic-Canic M. Specific organization of the negative response elements for retinoic acid and thyroid hormone receptors in keratin gene family. J Investig Dermatol. 1997;109:566–572. doi: 10.1111/1523-1747.ep12337483. [DOI] [PubMed] [Google Scholar]
- 52.Randolph R K, Simon M. Characterization of retinol metabolism in cultured human epidermal keratinocytes. J Biol Chem. 1993;268:9198–9205. [PubMed] [Google Scholar]
- 53.Reichardt H M, Kaestner K H, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, Schutz G. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- 54.Reichardt H M, Kaestner K H, Wessely O, Gass P, Schmid W, Schutz G. Analysis of glucocorticoid signalling by gene targeting. J Steroid Biochem Mol Biol. 1998;65:111–115. doi: 10.1016/s0960-0760(97)00181-7. [DOI] [PubMed] [Google Scholar]
- 55.Scheinman R I, Gualberto A, Jewell C M, Cidlowski J A, Baldwin A S., Jr Characterization of mechanisms involved in transrepression of NF-kappa B by activated glucocorticoid receptors. Mol Cell Biol. 1995;15:943–953. doi: 10.1128/mcb.15.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shibata H, Spencer T E, Onate S A, Jenster G, Tsai S Y, Tsai M J, O'Malley B W. Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action. Recent Prog Horm Res. 1997;52:141–164. [PubMed] [Google Scholar]
- 57.Slater E P, Hesse H, Beato M. Regulation of transcription by steroid hormones. Ann N Y Acad Sci. 1994;733:103–112. doi: 10.1111/j.1749-6632.1994.tb17260.x. [DOI] [PubMed] [Google Scholar]
- 58.Sloan K B, Araujo O E, Flowers F P. Topical corticosteroid therapy. In: Arndt K A, Leboit P E, Robinson J K, Wintraub B U, editors. Cutaneous medicine and surgery. Vol. 1. Philadelphia, Pa: W. B. Saunders Company; 1996. pp. 160–166. [Google Scholar]
- 59.Stellmach V, Leask A, Fuchs E. Retinoid-mediated transcriptional regulation of keratin genes in human epidermal and squamous cell carcinoma cells. Proc Natl Acad Sci USA. 1991;88:4582–4586. doi: 10.1073/pnas.88.11.4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stromstedt P E, Poellinger L, Gustafsson J A, Carlstedt-Duke J. The glucocorticoid receptor binds to a sequence overlapping the TATA box of the human osteocalcin promoter: a potential mechanism for negative regulation. Mol Cell Biol. 1991;11:3379–3383. doi: 10.1128/mcb.11.6.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sutter C, Nischt R, Winter H, Schweizer J. Aberrant in vitro expression of keratin K13 induced by Ca2+ and vitamin A acid in mouse epidermal cell lines. Exp Cell Res. 1991;195:183–193. doi: 10.1016/0014-4827(91)90515-v. [DOI] [PubMed] [Google Scholar]
- 62.Tomic M, Jiang C K, Epstein H S, Freedberg I M, Samuels H H, Blumenberg M. Nuclear receptors for retinoic acid and thyroid hormone regulate transcription of keratin genes. Cell Regul. 1990;1:965–973. doi: 10.1091/mbc.1.12.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tomic-Canic M, Bernerd F, Blumenberg M. A simple method to introduce internal deletions or mutations into any position of a target DNA sequence. Methods Mol Biol. 1996;57:249–257. doi: 10.1385/0-89603-332-5:249. [DOI] [PubMed] [Google Scholar]
- 64.Tomic-Canic M, Komine M, Freedberg I M, Blumenberg M. Epidermal signal transduction and transcription factor activation in activated keratinocytes. J Dermatol Sci. 1998;17:167–181. doi: 10.1016/s0923-1811(98)00016-4. [DOI] [PubMed] [Google Scholar]
- 65.Tomic-Canic M, Sunjevaric I, Freedberg I M, Blumenberg M. Identification of the retinoic acid and thyroid hormone receptor-responsive element in the human K14 keratin gene. J Investig Dermatol. 1992;99:842–847. doi: 10.1111/1523-1747.ep12614806. [DOI] [PubMed] [Google Scholar]
- 66.Tomie-Canie M, Day D, Samuels H H, Freedberg I M, Blumenberg M. Novel regulation of keratin gene expression by thyroid hormone and retinoid receptors. J Biol Chem. 1996;271:1416–1423. doi: 10.1074/jbc.271.3.1416. [DOI] [PubMed] [Google Scholar]
- 67.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 68.Uht R M, Anderson C M, Webb P, Kushner P J. Transcriptional activities of estrogen and glucocorticoid receptors are functionally integrated at the AP-1 response element. Endocrinology. 1997;138:2900–2908. doi: 10.1210/endo.138.7.5244. [DOI] [PubMed] [Google Scholar]
- 69.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 70.Vickers C F H. Topical corticosteroids. In: Fitzpatrick T B, Eisen A Z, Wolff K, Freedberg I M, Austen K F, editors. Dermatology in general medicine. Vol. 2. New York, N.Y: McGraw-Hill, Inc.; 1987. pp. 2540–2545. [Google Scholar]
- 71.Waltzer L, Bienz M. Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature. 1998;395:521–525. doi: 10.1038/26785. [DOI] [PubMed] [Google Scholar]
- 72.Westin S, Kurokawa R, Nolte R T, Wisely G B, McInerney E M, Rose D W, Milburn M V, Rosenfeld M G, Glass C K. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- 73.Wolffe A P. Transcriptional control. Sinful repression. Nature. 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 74.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 75.Zamir I, Dawson J, Lavinsky R M, Glass C K, Rosenfeld M G, Lazar M A. Cloning and characterization of a corepressor and potential component of the nuclear hormone receptor repression complex. Proc Natl Acad Sci USA. 1997;94:14400–14405. doi: 10.1073/pnas.94.26.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]