Abstract
All biological processes arise through the coordinated actions of biochemical pathways. How such functional diversity is achieved by a finite cast of molecular players remains a central mystery in biology. Spatial compartmentation – the idea that biochemical activities are organized around discrete spatial domains within cells – was first proposed nearly 40 years ago and has become firmly rooted in our understanding of how biochemical pathways are regulated to ensure specificity. However, directly interrogating spatial compartmentation and its mechanistic origins has only really become possible in the last 20 or so years, following technological advances such as the development of genetically encoded fluorescent biosensors. These powerful molecular tools permit direct, real-time visualization of dynamic biochemical processes in native biological contexts, and are essential for probing the spatial regulation of biochemical activities. In this Account, we review our lab’s efforts in developing and using biosensors to map the spatial compartmentation of intracellular signaling pathways and illuminate key mechanisms that establish the boundaries of an intricate biochemical activity architecture. We first discuss the role of regulatory fences, wherein the dynamic activation and deactivation of diffusible messengers produces diverse signaling compartments. For example, we used biosensors for the Ca2+ effector calmodulin and its downstream target calcineurin to reveal a spatial gradient of calmodulin that controls the temporal dynamics of calcineurin signaling. Our studies using cAMP biosensors have similarly elucidated fenced cAMP domains generated by competing production and degradation pathways, ranging in size from cell-spanning gradients to nanoscale hotspots. Second, we describe the role played by intracellular membranes in creating unique signaling platforms with distinctive pathway regulation, as revealed through studies using subcellularly targeted fluorescent biosensors. Using biosensors to visualize subcellular ERK pathway activity, for example, led us to discover a local signaling circuit that mediates distinct plasma membrane ERK dynamics versus global ERK signaling. Similarly, our work developing biosensors to monitor subcellular mTOR complex 1 (mTORC1) signaling allowed us to not only clarify the presence of mTORC1 activity in the nucleus but also identify a novel mechanism governing mTORC1 activation in this location. Finally, we detail how molecular assemblies enable precise spatial tuning of biochemical activity, through investigations enabled by cutting-edge advances in biosensor design. We recently identified liquid-liquid phase separation as a major factor in cAMP compartmentation aided by a new strategy for targeting biosensors to endogenously expressed proteins via genome editing, for instance, and have also been able to directly visualize nanometer-scale PKA signalosomes using an entirely new class of biosensors specifically developed for dynamic superresolution imaging of live-cell biochemical activities. Our work provides key insights into the molecular logic of spatially regulated signaling and lays the foundation for broader exploration of biochemical activity architectures across multiple spatial scales.
Graphical Abstract

INTRODUCTION
Networks of biochemical reactions underlie all critical life functions. These diverse pathways are responsible for regulating cellular metabolism, growth, survival, and decision-making, among other processes. However, the molecular machinery that comprises these networks is not floating aimlessly about the cell, engaging in random, haphazard interactions. Much like cells are divided into physical compartments defined by macromolecular structures, so too have we learned that the biochemical machinery is spatially compartmentalized to ensure the specificity and fidelity of cellular responses to dynamic environmental inputs. First proposed nearly 40 years ago,5 this idea of compartmentalized biochemical activities has blossomed into a model whereby the actions of biomolecules are spatially organized into a complex and intricate biochemical activity architecture6 coordinated across multiple spatial scales, ranging from individual molecules to entire cells and even tissues.
Our knowledge of how spatial compartmentation regulates biochemical activities has paralleled the expanding molecular toolkit for direct, real-time visualization of dynamic biochemical processes in native biological contexts, such as in living cells. While various optical probes have been developed over the years, few have been more influential in shaping the concept of biochemical activity architecture than genetically encoded fluorescent biosensors.6,7 These modular and versatile tools couple the intrinsic, genetically encoded fluorescence of green fluorescent protein (GFP) and its relatives8 with naturally occurring or engineered molecular switches to convert specific molecular events into a measurable optical readout. The molecular switch functions as a sensing unit, whereby a biochemical input triggers a conformational change that alters the fluorescence of a reporting unit. While classic designs modulate fluorescence resonance energy transfer (FRET) between a pair of fluorescent proteins (FPs) flanking the sensing unit, biosensors have come to span multiple distinct classes featuring diverse readouts.7 The ongoing evolution of biosensor design has been essential for probing activity architectures with ever-increasing detail.
Here, we discuss several examples of our recent work developing and using fluorescent biosensors, as well as other genetically encoded tools, to probe the biochemical activity architecture of intracellular signaling networks. Our efforts have succeeded in illuminating a number of molecular processes that provide boundary conditions for spatially compartmentalized signaling: regulatory fences produced through activation/deactivation, physical barriers defined by cellular membranes, and discrete molecular assemblies.
DYNAMIC REGULATORY FENCING
Early, dye-based fluorescent indicators were among the first chemical biology tools to permit spatially resolved visualization of intracellular signaling dynamics in living cells, revealing the formation of striking gradients by the ubiquitous messenger calcium (Ca2+).9 These pioneering approaches ultimately inspired some of the first genetically encoded fluorescent biosensors, which have in turn fueled deep explorations into the formation of biochemical activity gradients by numerous signaling molecules, revealing a heterogeneous biochemical landscape in which cells dynamically compartmentalize diffusible signals across different spatial scales through the action of pathway regulators (i.e., dynamic regulatory fences).
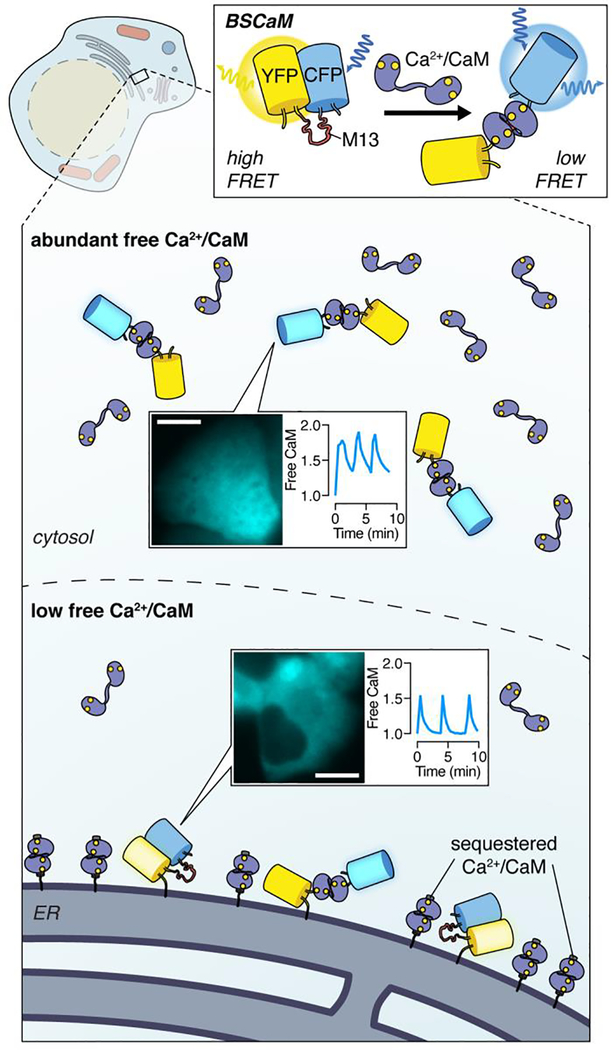
Initial designs for genetically encoded Ca2+ indicators (GECIs) universally relied on calmodulin (CaM), a major Ca2+ effector protein that undergoes a dramatic conformational change upon Ca2+ binding, as the Ca2+-sensing domain. Tethering CaM to a short peptide that selectively interacts with Ca2+-bound CaM (Ca2+/CaM) yields a robust molecular switch for controlling an optical readout such as FRET between a pair of FPs.10 However, CaM itself is a key regulator of diverse protein targets,11 and though most cellular CaM is constitutively bound to targets,12 in principle, a small pool of free Ca2+/CaM is freely exchangable. While countless studies have used GECIs to investigate spatial Ca2+ signaling,13 comparatively little has been revealed about the spatial regulation of Ca2+/CaM. Notably, sandwiching only a Ca2+/CaM-binding peptide between a FRET FP pair enables monitoring of Ca2+/CaM directly (Figure 1). When we used this FRET-based CaM biosensor14 to visualize Ca2+/CaM dynamics in pancreatic β-cells undergoing Ca2+ oscillations, we detected much higher free Ca2+/CaM levels in the cytosol than near the endoplasmic reticulum (ER) surface.15 Given the identical Ca2+ oscillation dynamics we observed at these locations, our data suggested that free CaM is unevenly distributed throughout the cell, presumably due to the differential abundance of CaM-sequestering proteins (Figure 1). This apparent CaM gradient had profound consequences on downstream Ca2+/CaM-dependent phosphatase calcineurin (CaN) activity. Using a FRET-based sensor in which the intrinsic, dephosphorylation-triggered conformational change in a CaN substrate modulates the relative distance between a pair of FPs,15,16 we detected contrasting step-like and oscillatory CaN activity dynamics from the cytosol and ER, which we were able to invert by inhibiting and overexpressing CaM, respectively. Our study, alongside the parallel discovery of Ca2+/CaM nuclear shuttling,17 shows how CaM actively encodes context-dependent spatial information during dynamic signaling events, modifying the prevalent view of CaM as passive adaptor. This example also demonstrated a more universal role for spatial gradients of regulatory biomolecules in patterning downstream signaling.
Figure 1. Visualizing calmodulin dynamics reveals a signaling gradient.
Using BSCaM, a FRET-based biosensor for measuring free Ca2+-bound calmodulin (Ca2+/CaM), we revealed the nonuniform distribution of this key regulatory molecule between the cytosol and ER surface in pancreatic β-cells. CFP fluorescence images illustrate BSCaM localization in MIN6 pancreatic β-cells. Scale bars, 10 μm.
Spatial patterning plays a central role in 3’,5’-cyclic adenosine monophosphate (cAMP) signaling, where cAMP gradients were first visualized using fluorophore-conjugated subunits of cAMP-dependent protein kinase (PKA) in neurons.18 Polarized cAMP/PKA signaling is now strongly implicated in neuronal symmetry breaking, specifically to promote axon formation,19 and we recently became interested in how cAMP/PKA compartmentalization evolves across neuronal development. We set out to study this process in hippocampal neurons using our FRET-based indicator of cAMP using Epac (ICUE).20 Like two other unimolecular cAMP biosensors developed around the same time,21 ICUE contains a fragment of the cAMP-dependent guanine-nucleotide exchange factor Epac, which undergoes a hinge-like conformational opening upon cAMP binding. In embryonic rat hippocampal neurons cultured for 5 days in vitro (DIV5), ICUE revealed an axon-directed gradient of cAMP accumulation, indicated by progressively higher responses towards distal regions of the axon, while no such gradients were observed in younger, DIV3 hippocampal neurons.22 cAMP is synthesized by adenylate cyclases (ACs) and degraded by phosphodiesterases (PDEs), and we found that PDE activity was critical for mediating cAMP responses at both developmental stages: Inhibiting PDE activity suppressed the cAMP gradient found in DIV5 neurons and actually produced a cAMP gradient in DIV3 neurons.22
PDEs play a central role in defining the spatial boundaries of cAMP signaling,23 often creating sharp barriers that block cAMP diffusion between cellular regions. For example, we and others previously identified a pool of PKA that resides in the nucleus,24,25 which we also discovered forms a complex with PDE4D5 via the A-kinase anchoring protein (AKAP) AKAP95.26 To probe this nuclear PKA pool, we took advantage of soluble AC (sAC),27 a non-transmembrane AC isoform that is selectively activated by bicarbonate, which we targeted to different subcellular regions to induce local cAMP production.24 By also localizing ICUE20 either throughout the nucleus or directly to AKAP95, we found that nuclear-ICUE, but not AKAP95-ICUE, detected cAMP originating from the plasma membrane, whereas both sensors responded to cAMP generated within the nucleus.26 Our results suggested that PDE4D5 protects AKAP95-anchored PKA from spurious activation, and indeed, disrupting PDE4 activity allowed AKAP95-tethered ICUE to sense membrane-generated cAMP.26 Acute PDE4 inhibition in fact produced an immediate response from AKAP95-ICUE in the absence of any other treatment, suggesting that PDE4 basally excludes cAMP from this microdomain. Inhibiting PDE3, another major PDE isoform, similarly allowed AKAP95-ICUE to detect membrane-generated cAMP but had no effect on basal cAMP levels,26 suggesting that PDE3 activity functions to prevent cAMP from exiting the plasma membrane compartment, consistent with our previous work.28 Thus, our study further demonstrated how different PDE isoforms create multi-tier regulatory fences23 to enable precise spatial control of cAMP/PKA signaling.
Whereas PDEs create sinks that consume cAMP,29 localized cAMP production by ACs can conversely generate cAMP hotspots.30 Recently, we observed how the juxtaposition of these opposing activities can yield discrete signaling domains. In pancreatic β-cells, glucose triggers intracellular Ca2+ and cAMP oscillations to drive insulin secretion,31 and we previously showed that β-cell cAMP/PKA and Ca2+ form a tightly coupled oscillatory signaling circuit.32 Given that AKAP79 and its rodent ortholog AKAP150 (AKAP79/150) anchor PKA to L-type voltage-gated Ca2+ channels (LTCCs),33,34 which control β-cell Ca2+ oscillations,35 we sought to investigate the role of AKAP79/150 in this oscillatory circuit. We utilized a similar approach to that described above by expressing an enhanced version of the FRET-based cAMP sensor Epac2-camps (Figure 2),36 which contains a minimal fragment of Epac2 as the cAMP-sensing unit,21 tethered directly to AKAP79 in a mouse β-cell line (MIN6). Using this probe together with a red-fluorescent GECI,37 we observed cAMP levels to oscillate in-phase with cytosolic Ca2+.1 Conversely, globally plasma membrane-tethered Epac2-camps revealed out-of-phase cAMP and Ca2+ oscillations, consistent with our previous observations in the cytosol (Figure 2).32 Importantly, given that overexpression of AKAP79, or any scaffold, may perturb signaling complexes by titrating out binding partners, we had to carefully control for biosensor expression levels during these experiments.1 More advanced strategies are expected to limit this concern (see below).
Figure 2. Discrete patterning of oscillatory phase by cAMP production and degradation.
Studying subcellular cAMP dynamics in pancreatic β-cells using the cAMP sensor citrine/cerulean (Ci/Ce) Epac2-camps reveals that, whereas plasma membrane (e.g., global) cAMP signaling is dominated by Ca2+-activated PDE1C, causing cAMP to oscillate out-of-phase with Ca2+, AKAP79/150 assembles a signaling complex in which Ca2+-stimulated AC8 activity locally overcomes PDE activity, allowing cAMP to oscillate in-phase with Ca2+. Confocal images show YFP fluorescence from AKAP (left)- and plasma membrane (right)-localized Epac2-camps in MIN6 pancreatic β-cells. Scale bars, 10 μm.
Ca2+ and cAMP are coupled through several Ca2+-regulated PDEs and ACs, with Ca2+−activatable PDE1C and AC8 being highly expressed in β-cells and implicated in insulin secretion.38 Constructing a mathematical model of this pathway, we found that the balance between these two enzymes determined the cAMP/PKA and Ca2+ phase relationship. Specifically, PDE1C was predicted to drive global Ca2+/cAMP coupling and produce out-ofphase oscillations, as Ca2+ elevations promote cAMP degradation, whereas relative AC8 activity was predicted to tune the phase relationship by increasing cAMP production during Ca2+ elevations (Figure 2). As predicted, we found that treating oscillating MIN6 cells with a selective PDE1C inhibitor totally abolished cAMP oscillations despite continued Ca2+ oscillations. Conversely, overexpressing AC8 rendered plasma membrane Epac2-camps in-phase with Ca2+, while AKAP79-Epac2-camps became out-of-phase upon AC8 knockdown.1 AC8 directly interacts with AKA79/150 in β-cells,39 and given previous evidence of AKAP79/150 plasma membrane nanoclustering,4,40 we hypothesized that AC8 clustering may be important for spatial compartmentation of oscillatory phase. Using stochastic optical reconstruction microscopy (STORM)41 superresolution imaging in MIN6 cells, we observed endogenous AKAP150 and AC8 to form discrete, highly colocalized, plasma membrane nanoclusters.1 Additional modeling predicted that reducing the density of AC8 molecules per cluster would produce out-of-phase cAMP signaling, and indeed, we found that overexpressing an AC8 N-terminal fragment to disrupt the AKAP79/150 interaction39 not only disrupted AC8 membrane clustering but also yielded out-of-phase cAMP oscillations from AKAP79-Epac2-camps.1 Our results suggested the formation of a spatially distinct signaling microdomain, likely positioned directly at sites of Ca2+ influx through LTCCs (Figure 2),40 in which AKAP-mediated AC8 nanoclustering allows precise control of local cAMP dynamics and thereby global Ca2+ oscillations.1 Thus, we revealed how dynamic fencing via closely juxtaposed positive and negative regulators can establish minute signaling compartments with highly divergent behaviors.
INTRINSIC MEMBRANE COMPARTMENTS
Lipid membranes are a keystone of cellular organization. The plasma membrane defines the boundary between the cell and its environment, while in eukaryotes, a network of endomembranes creates functionally specialized organelles. Signals originating at the plasma membrane must be transduced throughout the cell to regulate specific processes at these distinct subcellular sites. Targeting genetically encoded biosensors to different membrane structures using various localization tags has proven invaluable for elucidating how this inherent compartmentation shapes biochemical activity architectures. Our own work has revealed not only the diverse spatial activity profiles exhibited by signaling molecules associated with different membrane compartments but also the distinct molecular mechanisms governing this spatial signaling diversity.
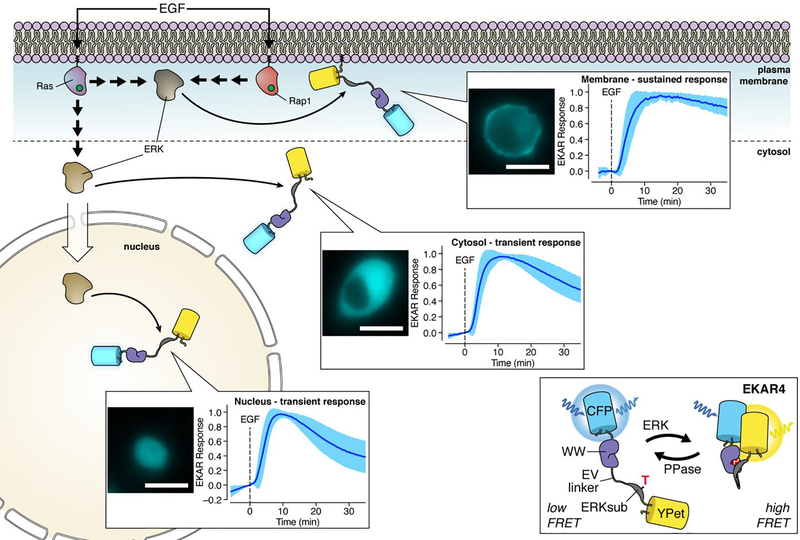
Global extracellular response kinase (ERK) signaling has long been known to exhibit different temporal dynamics in response to distinct upstream inputs, with epidermal growth factor (EGF) and nerve growth factor (NGF) stimulation notably inducing transient and sustained ERK signaling, respectively.28,42 Recently, we also discovered that local ERK signaling can display similarly divergent temporal dynamics in response to a single input.43 In this study, we utilized our newly developed 4th-generation, FRET-based ERK kinase activity reporter (EKAR4), which contains the same sensing unit found in previous EKARs,44,45 wherein an ERK-specific substrate/docking sequence is tethered to the phosphoamino acid-binding WW domain via an extended linker, but reverses the order of the FRET FP pair to help enhance the response.43 As with all FRET-based kinase activity reporters (KARs) that follow this design,7 phosphorylation of the substrate peptide induces binding by the phosphoamino acid-binding domain (PAABD), resulting in a conformational change that alters FRET (Figure 3). By targeting EKAR4 to the plasma membrane, cytosol, or nucleus in PC12 cells, we found that EGF stimulation induced markedly sustained ERK activity at the plasma membrane, versus the more transient activity pattern detected in the cytosol and nucleus.43 Although subcellular targeting may affect the dynamic range of the biosensor, the ratiometric FRET readout cancels some experimental variations and allows for quantification and comparison between different experiments.
Figure 3. Local control of plasma membrane ERK signaling.
Subcellularly targeted variants of our improved FRET-based ERK kinase activity reporter (EKAR4) revealed that epidermal growth factor (EGF) stimulation triggers sustained, local plasma membrane ERK activation through both Ras and Rap1 GTPase in PC12 cells. A separate pool of ERK regulated only by Ras is responsible for transient, global ERK signaling. CFP fluorescence images illustrate EKAR4 localization in PC12 cells. Scale bars, 10 μm.
Although typically mediated by Ras GTPases (e.g., HRas, KRas, or NRas), ERK signaling can also be triggered by direct binding and activation of B-Raf by Rap1 GTPase,46 though conflicting reports have made this non-canonical Rap1/B-Raf/ERK axis somewhat controversial. To probe deeper into the molecular mechanism underlying sustained plasma membrane ERK activity, we utilized a suite of tools to monitor and perturb Rap1 activation. We first imaged a bimolecular FRET-based Rap1 activation biosensor 47 in which Rap1 is fused to mCerulean3 (donor) and the Rap1-binding domain (RBD) of RalGDS is fused to YPet (acceptor) such that binding of activated Rap1 to the RBD will yield a FRET increase, to directly observe EGF-induced Rap1 activation in PC12 cells.43 Monitoring the membrane translocation of GFP-tagged RalGDS48 further allowed us to confirm the activation of endogenous plasma membrane Rap1. We also overexpressed a Rap1 GTPase-activating protein (Rap1GAP) to disrupt Rap1 activation, thus abolishing both Rap1 FRET and RalGDS responses, and observed the specific attenuation of the plasma membrane EKAR4 response with no effect in the cytosol.43 Similarly disrupting Ras activation completely abolished the EGF-induced cytosolic EKAR4 response but only delayed the plasma membrane EKAR4 response, which ultimately reached the same magnitude observed in control cells.43 These results suggest that Ras controls initial plasma membrane ERK activation, whereas Rap1 is required for the sustained response, and further call to mind our previous observation that growth factor-induced cAMP/PKA signaling is restricted to the plasma membrane and required for proper ERK signaling in PC12 cells,28 hinting at a possible local regulatory circuit, given the ability of PKA to phosphorylate and activate Rap1.46
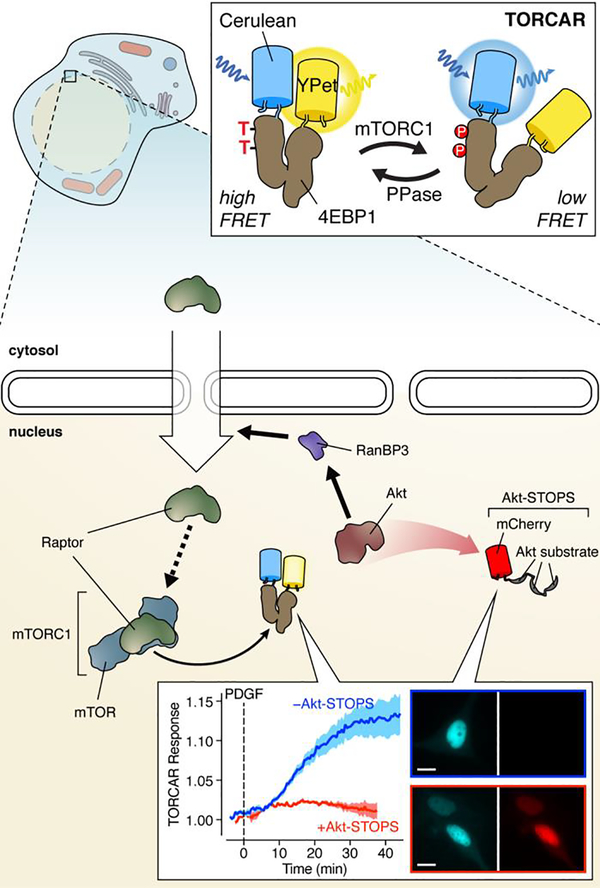
The sensitivity and selectivity afforded by subcellular biosensor targeting often illuminate previously obscured spatial dimensions of key signaling pathways. A telling example comes from our recent efforts to begin unravelling the spatiotemporal regulation of the mechanistic target of rapamycin (mTOR) complex 1 (mTORC1). mTORC1 is a critical signaling node that integrates diverse inputs, including growth factor and nutrient signals, as well as cellular stress and energy status, to regulate cellular metabolism.49 Both growth factor and nutrient stimuli trigger mTORC1 activation on the lysosome surface,49 the most well-studied site of mTORC1 signaling. Yet mTORC1 has also been linked to other subcellular compartments.50 One of the most disputed sites of mTORC1 activity is the nucleus, with conflicting reports painting a unclear picture of nuclear mTORC1 signaling.50 We hoped to address the question of compartmentalized mTORC1 signaling by developing a FRET-based mTORC1 activity reporter (TORCAR).51 After initially pursuing the substrate/PAABD switch design found in other KARs, we ultimately succeeded in utilizing the full-length eukaryotic translation initiation factor 4E binding protein 1 (4EBP1), an mTORC1 substrate which undergoes an intrinsic conformational change upon phosphorylation, sandwiched between a Cerulean donor and YPet acceptor (Figure 4). When targeted to the nucleus, TORCAR exhibited robust FRET responses in cells treated with either growth factor or nutrient stimulation.2,51
Figure 4. Regulation of nuclear mTORC1 activation by nuclear Akt activity.
By combining real-time visualization of mTORC1 activity using the FRET sensor TORCAR with local perturbation of Akt activity using Akt-STOPS, we discovered that nuclear Akt signaling controls nuclear mTORC1 activation via nuclear recruitment of the mTORC1 subunit Raptor. Inset CFP (left) and RFP (right) images show the nuclear localization of TORCAR and Akt-STOPS, respectively, in NIH3T3 cells. Images from cells both without (top) and with (bottom) Akt-STOPS expression are shown. Scale bars, 10 μm.
Our results provided the first clear evidence of endogenous mTORC1 signaling within the nucleus and immediately raised questions as to how nuclear mTORC1 activity is regulated. Specifically, given the central role played by Akt in promoting mTORC1 signaling,52 we wondered if Akt also controls nuclear mTORC1 activity. To investigate this, we developed a genetically encoded tool containing 3 tandem copies of a high-affinity Akt substrate peptide, designed to buffer and thus disrupt Akt signaling in a location-specific manner, which we termed Akt Substrate-based Tandem Occupancy Peptide Sponge (Akt-STOPS) (Figure 4).2 When we examined the FRET-response from an Akt activity reporter (AktAR), in which an Akt-specific substrate peptide and FHA1 domain are sandwiched between a FRET pair,51,53 we found that Akt-STOPS targeted to the nucleus specifically blocked the FRET-response from nuclear AktAR without affecting cytosolic AktAR. Using Akt-STOPS, we then observed that selectively inhibiting nuclear Akt activity completely abolished the growth factor-induced nuclear TORCAR response (Figure 4). We also found that growth factor stimulation induced the nuclear trafficking of Raptor, a principal mTORC1 subunit, with nuclear Raptor being both necessary and sufficient to promote nuclear mTORC1 signaling. Interestingly, Akt has been shown to regulate nuclear trafficking by phosphorylating nuclear Ran binding protein 3 (RanBP3) to facilitate nuclear import,54 and we in fact observed that expressing Akt-STOPS in the nucleus potently disrupted RanBP3 phosphorylation, highlighting an unconventional mechanism for spatiotemporal mTORC1 regulation via recruitment of a scaffolding protein to a specific subcellular location (Figure 4).2 We expect future studies using organelle-targeted sensors to reveal similarly novel spatial dimensions to key signaling pathways.
MOLECULAR ASSEMBLY
The cell interior is teeming with signaling molecules that can each engage with multiple targets. Ensuring that these diverse components participate in productive interactions, while avoiding unwanted interactions, is crucial for efficient information transfer during intracellular signaling. In addition to compartmentalizing signaling activities through boundaries defined by dynamic regulatory fences or organelle membranes, cells can precisely tune the organization of their biochemical activity architectures by collecting the signaling machinery into micro- and nanoscale molecular assemblies.
Perhaps the most striking examples of molecular assembly in cells are so-called membraneless organelles.55 Like their membrane-bound counterparts, these structures regulate discrete cellular processes by establishing biochemical environments distinct from the bulk intracellular milieu, but are formed via liquid-liquid phase separation (LLPS), a process whereby an initially homogeneous mixture spontaneously partitions into separate phases with distinct molecular compositions,56 similar to oil droplets in water. Biomolecular LLPS is typically governed by intrinsic disorder and multivalent binding,55 common features of signaling proteins.57 Multiple signaling proteins have been shown to undergo LLPS, indicating a potentially important role in the spatial regulation of intracellular signaling, though the precise impact is often unclear. Recently, we observed that the type I PKA regulatory (R) subunit, RIα, undergoes LLPS to form biomolecular condensates enriched in cAMP and PKA activity, which play a key role in cAMP compartmentation (Figure 5).3 Our efforts were greatly aided by a suite of molecular approaches designed to visualize and track the molecular behavior and signaling dynamics of endogenously expressed proteins. First, by utilizing an endogenous tagging approach based on split superfolder GFP,58 in which the 11th β-strand of GFP (GFP11) is expressed from a specific genomic locus via CRISPR/Cas9 editing and then allowed to spontaneously reconstitute with overexpressed strands 1–10 (GFP1–10), we were able to observe the formation of dynamic, liquid-like droplets by endogenously expressed RIα in living cells (Figure 5), findings that were corroborated by in vitro studies using purified RIα.3
Figure 5. A key role for phase separation in cAMP compartmentation.
The PKA regulatory subunit RIα forms phase-separated droplets that sequester cAMP, which is essential for PDEs to effectively compartmentalize cAMP signaling. We directly visualized cAMP levels within RIα droplets using FluoSTEP-ICUE, in which a FRET-based cAMP sensor is reconstituted at an endogenously expressed protein (e.g., RIα) via split FP complementation. Confocal image of GFP fluorescence shows phase-separated droplets formed by endogenously labeled RIα in HEK293T cells. Scale bar, 10 μm.
Notably, RIα droplet formation mirrored the dynamics of cAMP elevation, and we also observed the PKA catalytic (C) subunit to co-phase separate with RIα, prompting us to investigate cAMP/PKA signaling within these endogenous droplets. We thus devised fluorescent sensors targeted to endogenous proteins (FluoSTEPs), a novel strategy for targeting FRET-based biosensors to proteins of interest based on the aforementioned split FP approach. Specifically, using GFP1–10 as the FRET donor and RFP as the FRET acceptor allows us to recruit a sensor to an endogenous GFP11-tagged protein (e.g., RIα) such that the biosensor is only functional when correctly localized. Applying this strategy to our cAMP sensor ICUE (Figure 5), we detected elevated cAMP levels within RIα liquid droplets and also found that cAMP accumulation in newly forming droplets outpaced than seen in diffuse regions.3 We were similarly able to detect PKA activity enrichment within RIα droplets using a FluoSTEP version of our A-kinase activity reporter (AKAR),59 in which a PKA substrate peptide linked to an FHA1 domain serves as the molecular switch. FluoSTEPs are ideal for overcoming the limitations associated with large biosensor fusions (see above), although success strongly depends on the endogenous expression level of the target protein. Nevertheless, future improvements, such as brighter split-FPs, will undoubtedly yield FluoSTEPs with enhanced signals, further increasing their utility to probe signaling microdomains without perturbing their composition.
Further supporting these observations, we found that 99% of a fluorescently labeled cAMP analogue localized to RIα liquid droplets in vitro, indicating substantial enrichment. This dynamic sequestration of cAMP enabled RIα condensates to form traps that strongly buffer cAMP and promote compartmentalized signaling, which we revealed by taking advantage of a previously described cAMP compartmentation assay in which a FRET-based cAMP sensor is fused to an active PDE catalytic domain.29 Specifically, when we fused ICUE to the PDE4D2 catalytic domain (PDE4D2cat), we observed cAMP production to only induce a FRET response when PDE4 activity was inhibited, indicating that all cAMP in the vicinity of PDE4D2cat-ICUE was degraded. In contrast, disrupting RIα LLPS revealed clear cAMP elevations even in the absence of PDE inhibition, an effect that was lost when RIα was knocked out and enhanced when RIα was overexpressed.3 The prominent role of PDEs in cAMP compartmentation23 has long proven difficult to reconcile with the previously reported rates of cAMP diffusion (fast) and PDE catalysis (slow).3,29 Our study thus filled in crucial details regarding spatial cAMP signaling by revealing that RIα works in tandem with PDEs to establish a cAMP compartmentation system (Figure 5).
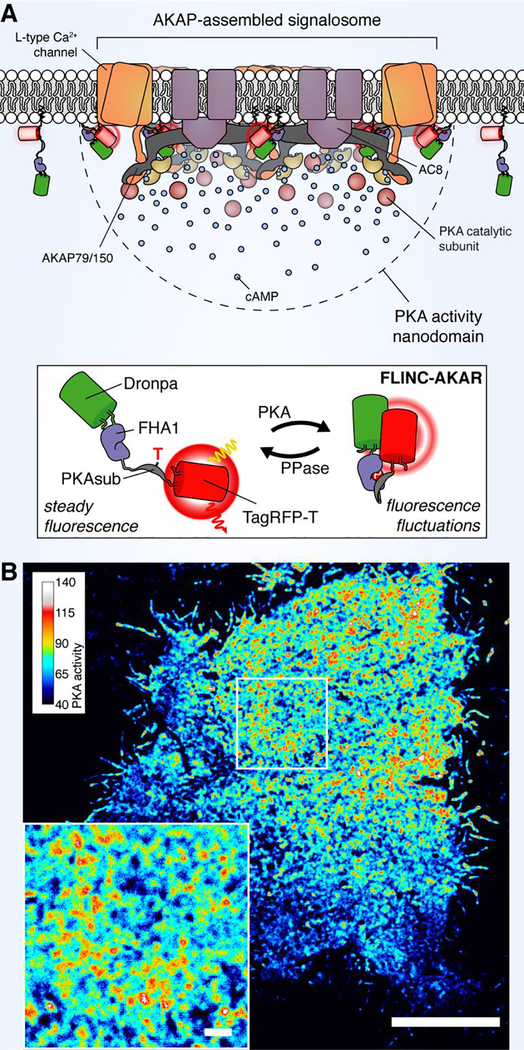
While LLPS often produces micron-scale biomolecular complexes, molecular assembly at the nanometer-scale is typically driven by multivalent scaffolds, proteins that bind and recruit various signaling enzymes to specific targets and subcellular locations.60 Scaffold proteins are involved in multiple pathways, including MAPK,61 GTPase,62 PKC,63 and PKA64 signaling. Arguably the most well-studied scaffolds are AKAPs, a diverse but functionally related class of proteins that bind and anchor PKA throughout the cell.64 AKAPs are thought to regulate local PKA signaling through the formation of multi-protein signalosomes that integrate PKA with upstream regulators (e.g., PDEs, ACs), downstream targets (e.g., ion channels, cell-surface receptors), and even orthogonal signaling enzymes (e.g., PKC, CaN, ERK).64 While previous studies have utilized biosensors to probe the regulation of PKA activity dynamics within AKAP signalosomes, our recent work using a first-in-class superresolution reporter of PKA activity was able to demonstrate in remarkable detail the precision with which AKAPs shape local PKA signaling.4
This study arose from our serendipitous observation that the red-emitting FP TagRFP-T65 exhibits stochastic fluorescence fluctuations (e.g., blinking) when brought into close proximity with the green-emitting FP Dronpa,66 a phenomenon we called Fluorescence fLuctuation INcrease by Contact (FLINC).4 Fluorophores that display stochastic fluctuations between high-and low-intensity states (i.e., photochromism) are the basis for many superresolution imaging techniques;67 therefore, by sandwiching a PKA-dependent molecular switch composed of a PKA substrate peptide and FHA1 domain tethered by an extended linker44 between Dronpa and TagRFP-T, we were able to generate a superresolution biosensor, FLINC-AKAR, that shows increased blinking behavior in response to PKA activity elevations (Figure 6).4 The response from FLINC-AKAR is imaged via photochromic stochastic optical fluctuation imaging (pcSOFI),68 an accessible live-cell superresolution imaging technique that we developed previously. As with similar techniques, each superresolution map is constructed from a long image series, necessitating trade-offs between spatial and temporal resolution in order to minimize photobleaching. Nevertheless, FLINC offers unprecedented opportunities to visualize biochemical activity dynamics at molecular length-scales. Indeed, this approach enabled us to observe that PKA activity along the plasma membrane was confined to discrete hotspots, measuring ~350 nm in diameter, upon pathway stimulation.4 Notably, we found that blocking AKAP-PKA interactions using a peptide disruptor69 greatly reduced the sensor response following stimulation and largely abolished the appearance of highly active PKA hotspots, suggesting that AKAP anchoring plays a critical role in creating these submicron-scale PKA activity domains.4
Figure 6. Nano-scale organization of PKA activity by AKAP clustering.
(A) We have used a new, superresolution PKA activity reporter FLINC-AKAR to reveal that PKA activity is organized into discrete clusters along the plasma membrane, which likely correspond to clusters of AKAP-assembled molecular signalosomes. (B) A superresolution image of a representative HeLa cell expressing FLINC-AKAR, illustrating clear activity hotspots following PKA stimulation (magnified in inset). Scale bar, 10 μm (inset, 1 μm).
The canonical model of PKA activation holds that C subunits dissociate from the holoenzyme following cAMP binding to R subunits.70 Given that AKAPs exclusively anchor PKA via R subunits,64 our results highlighted longstanding questions about how, specifically, AKAPs are able to confine diffusible PKA C. While the canonical model of PKA activation was recently called into question by studies suggesting that C and R don’t fully dissociate under physiological conditions,71 this point has proven somewhat controversial.72 Furthermore, our FLINC-AKAR imaging revealed that PKA activity microdomains form even under maximal, nonphysiological signaling conditions during which C subunits are still thought to dissociate.4,71 In pursuing this question, we performed additional superresolution imaging using STORM, which revealed that AKAP79, a well-characterized membrane AKAP,73 forms discrete plasma membrane clusters that colocalize with PKA activity microdomains, visualized simultaneously via either 2-color STORM imaging of phosphorylated PKA substrates or FLINC-AKAR imaging.4 Based on these findings, we proposed a new model of PKA spatial regulation in which AKAP clustering promotes local enrichment of R subunits, leading to enhanced recapture of C subunits to locally confine PKA activity.
CONCLUSIONS AND OUTLOOK
Here, we have explored our lab’s use of molecular tools such as genetically encoded fluorescent biosensors to decipher the molecular logic governing the spatial regulation of intracellular signaling pathways. Our work emphasizes the contribution of multiple processes operating across multiple spatial scales in defining a global biochemical activity architecture to regulate cellular function. Critically, the evolving toolkit of genetically encoded biosensors has been indispensable in our efforts to traverse these different spatial scales. On their own, sensors based on early, now classic designs have helped reveal the formation of biochemical activity gradients spanning entire cells. Through increasingly sophisticated methods of biosensor targeting, we have been able to uncover the importance of organelle membranes in shaping the biochemical landscape within cells and have even gained a new window into the role of liquid-like, membraneless organelles. Finally, an entirely new class of biosensors based on fluorescence fluctuations is providing molecular-scale information through superresolution imaging of biochemical activity dynamics. Spatial regulation is a universal feature of biochemical pathways, and these techniques are readily generalized to investigate pathways and compartments not mentioned here. Indeed, we hope to see a continued expansion in the application of these approaches to capture the full scope of biochemical activity architectures.
Following on advances in elucidating the cellular and subcellular patterning of biochemical networks, we now find ourselves in the midst of an accelerating shift toward illuminating the tissue- and organism-scale dimensions of these activity architectures. Tracking biochemical activity dynamics in vivo presents a unique set of challenges, and biosensor engineering efforts are already shifting to meet these needs. Given the optical complexity of biological tissue and the subtle biochemical changes associated with physiological processes, a chief concern is maximizing biosensor dynamic range and signal-to-noise ratio (SNR), which determine how sensitively a biosensor can respond to a biochemical change and how robustly that response can be measured. To this end, biosensor engineering efforts increasingly favor incorporating a molecular switch directly into the barrel of a single FP. Combined with directed evolution, this approach can yield sensors with orders-of-magnitude higher dynamic range and sensitivity than FRET-based designs, enabling fairly robust in vivo imaging.7 We were recently able to apply this design to engineer a suite of high-SNR, excitation-ratiometric kinase activity reporters (ExRai-KARs) that vastly outperform their FRET-based counterparts,74 using high-throughput screening to ultimately obtain an ultrasensitive PKA sensor (ExRai-AKAR2) capable of rapid, in vivo kinase activity imaging in the brains of awake mice.75 Future efforts to continue broadening the range of activities detected by this growing sensor class are likely to prove essential for mapping the biochemical activity architectures of living organisms (Figure 7).
Figure 7. Illuminating activity architecture across scales.
Continuing advances in biosensor engineering should illuminate biochemical activity architectures across molecular, cellular, and organism scales.
ACKNOWLEDGEMENTS
This work was supported by R35 CA197622, R01 DK073368, and R01 GM111665 to J.Z.
Biographies
Jin Zhang attended Tsinghua University for her undergraduate studies, and pursued her graduate studies in Chemistry at the University of Chicago. After completing her postdoctoral work at the University of California, San Diego, she joined the faculty of Johns Hopkins University School of Medicine in 2003. She was promoted to Professor of Pharmacology, Neuroscience and Oncology in 2013. In 2015 she moved back to UC San Diego and is currently a member of the Moores Cancer Center and a Professor in Departments of Pharmacology, Bioengineering and Chemistry & Biochemistry at UC San Diego. Research in her lab focuses on developing enabling technologies to probe active molecules in their native environment and characterizing how these active molecules change in diseases including cancer. Professor Zhang has received many awards including the National Institutes of Health (NIH) Director’s Pioneer Award (2009), the John J. Abel Award in Pharmacology from American Society for Pharmacology and Experimental Therapeutics (ASPET) (2012), the Pfizer Award in Enzyme Chemistry from American Chemical Society (2012), and National Institute of Cancer Outstanding Investigator Award (2015). She was elected as a Fellow of the American Association for the Advancement of Science (AAAS) in 2014 and as a Fellow of the American Institute for Medical and Biological Engineering (AIMBE) in 2019. She serves on the ASPET council as past-Secretary/Treasurer and as co-director of Cell Signaling San Diego.
Sohum Mehta received a B.S. in Biology, with a minor in Fine Arts, from the George Washington University before pursuing graduate studies at the Johns Hopkins University, where he received a Ph.D. in Biology for studies of calcineurin signaling in the yeast Saccharomyces cerevisiae. Sohum originally joined Jin Zhang’s lab in 2009 as a postdoctoral fellow at the Johns Hopkins University School of Medicine and later moved with the lab in 2015 to UC San Diego, where he continues to serve as a senior researcher. His research focuses on developing novel genetically encoded tools for the sensitive and multiplexed visualization of intracellular signaling.
Footnotes
The authors declare no competing financial interests.
KEY REFERENCES
Tenner, B., Getz, M., Ross, B., Ohadi, D., Bohrer, C.H., Greenwald, E., Mehta, S., Xiao, J., Rangamani, P., and Zhang, J. (2020). Spatially compartmentalized phase regulation of a Ca2+-cAMP-PKA oscillatory circuit. Elife 9, e55013.1 Using a FRET-based cAMP biosensor, we reveal how pancreatic beta cells generate contrasting local and global cAMP signaling domains with distinct temporal dynamics by juxtaposing the competing activities of cAMP production and degradation at the plasma membrane.
Zhou, X., Zhong, Y., Molinar-Inglis, O., Kunkel, M.T., Chen, M., Sun, T., Zhang, J., Shyy, J.Y.-J., Trejo, J., Newton, A.C., and Zhang, J. (2020). Location-specific inhibition of Akt reveals regulation of mTORC1 activity in the nucleus. Nat Commun 11, 6088.2 Combining a subcellularly targeted FRET-based mTOR complex 1 (mTORC1) activity reporter with location-specific disruption of Akt activity, we identify a novel mechanism governing local activation of mTORC1 signaling within the nuclear compartment.
Zhang, J.Z., Lu, T.-W., Stolerman, L.M., Tenner, B., Yang, J.R., Zhang, J.-F., Falcke, M., Rangamani, P., Taylor, S.S., Mehta, S., and Zhang, J. (2020). Phase Separation of a PKA Regulatory Subunit Controls cAMP Compartmentation and Oncogenic Signaling. Cell 182, 1531–1544.e15.3 We discover the formation of liquid-like biomolecular condensates by a cAMP-dependent protein kinase (PKA) regulatory subunit and reveal, using fluorescent sensors targeted to endogenous proteins, how these molecular assemblies dynamically sequester cAMP and control spatial signaling.
Mo, G.C.H., Ross, B., Hertel, F., Manna, P., Yang, X., Greenwald, E., Booth, C., Plummer, A.M., Tenner, B., Chen, Z., Wang, Y., Kennedy, E.J., Cole, P.A., Flemming, K.G., Palmer, A., Jimenez, R., Xiao, J., Dedecker, P., and Zhang, J. (2017). Genetically encoded biosensors for visualizing live-cell biochemical activity at super-resolution. Nat Methods 14, 427–434.4 Our serendipitous discovery of a novel photophysical behavior, fluorescence fluctuation increase by contact, yields a new class of biosensors for dynamic, superresolution activity imaging and allows us to directly visualize the nanoscale organization of PKA signaling by molecular assembly.
REFERENCES
- (1).Tenner B; Getz M; Ross B; Ohadi D; Bohrer CH; Greenwald E; Mehta S; Xiao J; Rangamani P; Zhang J Spatially Compartmentalized Phase Regulation of a Ca2+-CAMP-PKA Oscillatory Circuit. Elife 2020, 9 (In press.), e55013. 10.7554/elife.55013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Zhou X; Zhong Y; Molinar-Inglis O; Kunkel MT; Chen M; Sun T; Zhang J; Shyy JY-J; Trejo J; Newton AC; Zhang J Location-Specific Inhibition of Akt Reveals Regulation of MTORC1 Activity in the Nucleus. Nat Commun 2020, 11 (1), 6088. 10.1038/s41467-020-19937-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Zhang JZ; Lu T-W; Stolerman LM; Tenner B; Yang JR; Zhang J-F; Falcke M; Rangamani P; Taylor SS; Mehta S; Zhang J Phase Separation of a PKA Regulatory Subunit Controls CAMP Compartmentation and Oncogenic Signaling. Cell 2020, 182 (6), 1531–1544.e15. 10.1016/j.cell.2020.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Mo GCH; Ross B; Hertel F; Manna P; Yang X; Greenwald E; Booth C; Plummer AM; Tenner B; Chen Z; Wang Y; Kennedy EJ; Cole PA; Fleming KG; Palmer A; Jimenez R; Xiao J; Dedecker P; Zhang J Genetically Encoded Biosensors for Visualizing Live-Cell Biochemical Activity at Super-Resolution. Nat Methods 2017, 14 (4), 427–434. 10.1038/nmeth.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Steinberg SF; Brunton LL Compartmentation of G Protein-Coupled Signaling Pathways in Cardiac Myocytes. Pharmacol Toxicol 2001, 41 (1), 751–773. 10.1146/annurev.pharmtox.41.1.751. [DOI] [PubMed] [Google Scholar]
- (6).Mehta S; Zhang J Illuminating the Cell’s Biochemical Activity Architecture. Biochemistryus-us 2017, 56 (39), 5210–5213. 10.1021/acs.biochem.7b00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Greenwald EC; Mehta S; Zhang J Genetically Encoded Fluorescent Biosensors Illuminate the Spatiotemporal Regulation of Signaling Networks. Chem Rev 2018, 118 (24), 11707–11794. 10.1021/acs.chemrev.8b00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Rodriguez EA; Campbell RE; Lin JY; Lin MZ; Miyawaki A; Palmer AE; Shu X; Zhang J; Tsien RY The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins. Trends Biochem Sci 2017, 42 (2), 111–129. 10.1016/j.tibs.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Mehta S; Zhang J Dynamic Visualization of Calcium-Dependent Signaling in Cellular Microdomains. Cell Calcium 2015, 58 (4), 333–341. 10.1016/j.ceca.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Miyawaki A; Griesbeck O; Heim R; Tsien RY Dynamic and Quantitative Ca2+ Measurements Using Improved Cameleons. Proc National Acad Sci 1999, 96 (5), 2135–2140. 10.1073/pnas.96.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Klee CB; Crouch TH; Richman PG Calmodulin. Annu Rev Biochem 1980, 49 (1), 489–515. 10.1146/annurev.bi.49.070180.002421. [DOI] [PubMed] [Google Scholar]
- (12).Persechini A; Stemmer PM Calmodulin Is a Limiting Factor in the Cell. Trends Cardiovas Med 2002, 12 (1), 32–37. 10.1016/s1050-1738(01)00144-x. [DOI] [PubMed] [Google Scholar]
- (13).Pal A; Tian L Imaging Voltage and Brain Chemistry with Genetically Encoded Sensors and Modulators. Curr Opin Chem Biol 2020, 57, 166–176. 10.1016/j.cbpa.2020.07.006. [DOI] [PubMed] [Google Scholar]
- (14).Persechini A; Cronk B The Relationship between the Free Concentrations of Ca2+ and Ca2+-Calmodulin in Intact Cells. J Biol Chem 1999, 274 (11), 6827–6830. 10.1074/jbc.274.11.6827. [DOI] [PubMed] [Google Scholar]
- (15).Mehta S; Aye-Han N-N; Ganesan A; Oldach L; Gorshkov K; Zhang J Calmodulin-Controlled Spatial Decoding of Oscillatory Ca2+ Signals by Calcineurin. Elife 2014, 3, e03765. 10.7554/elife.03765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Newman RH; Zhang J Visualization of Phosphatase Activity in Living Cells with a FRET-Based Calcineurin Activity Sensor. Mol Biosyst 2008, 4 (6), 496–501. 10.1039/b720034j. [DOI] [PubMed] [Google Scholar]
- (17).Ma H; Groth RD; Cohen SM; Emery JF; Li B; Hoedt E; Zhang G; Neubert TA; Tsien RW ΓCaMKII Shuttles Ca2+/CaM to the Nucleus to Trigger CREB Phosphorylation and Gene Expression. Cell 2014, 159 (2), 281–294. 10.1016/j.cell.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Bacskai B; Hochner B; Mahaut-Smith M; Adams; Kaang B; Kandel E; Tsien R Spatially Resolved Dynamics of CAMP and Protein Kinase A Subunits in Aplysia Sensory Neurons. Science 1993, 260 (5105), 222–226. 10.1126/science.7682336. [DOI] [PubMed] [Google Scholar]
- (19).Shelly M; Lim BK; Cancedda L; Heilshorn SC; Gao H; Poo M Local and Long-Range Reciprocal Regulation of CAMP and CGMP in Axon/Dendrite Formation. Science 2010, 327 (5965), 547–552. 10.1126/science.1179735. [DOI] [PubMed] [Google Scholar]
- (20).DiPilato LM; Zhang J The Role of Membrane Microdomains in Shaping Β2-Adrenergic Receptor -Mediated CAMP Dynamics. Mol Biosyst 2009, 5 (8), 832–837. 10.1039/b823243a. [DOI] [PubMed] [Google Scholar]
- (21).Nikolaev VO; Bünemann M; Hein L; Hannawacker A; Lohse MJ Novel Single Chain CAMP Sensors for Receptor-Induced Signal Propagation. J Biol Chem 2004, 279 (36), 37215–37218. 10.1074/jbc.c400302200. [DOI] [PubMed] [Google Scholar]
- (22).Gorshkov K; Mehta S; Ramamurthy S; Ronnett GV; Zhou F-Q; Zhang J AKAP-Mediated Feedback Control of CAMP Gradients in Developing Hippocampal Neurons. Nat Chem Biol 2017, 13 (4), 425–431. 10.1038/nchembio.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Stangherlin A; Zaccolo M Phosphodiesterases and Subcellular Compartmentalized CAMP Signaling in the Cardiovascular System. Am J Physiol-heart C 2012, 302 (2), H379–H390. 10.1152/ajpheart.00766.2011. [DOI] [PubMed] [Google Scholar]
- (24).Sample V; DiPilato LM; Yang JH; Ni Q; Saucerman JJ; Zhang J Regulation of Nuclear PKA Revealed by Spatiotemporal Manipulation of Cyclic AMP. Nat Chem Biol 2012, 8 (4), 375–382. 10.1038/nchembio.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Jean-Alphonse FG; Wehbi VL; Chen J; Noda M; Taboas JM; Xiao K; Vilardaga J-P Β2-Adrenergic Receptor Control of Endosomal PTH Receptor Signaling via Gβγ. Nat Chem Biol 2017, 13 (3), 259–261. 10.1038/nchembio.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Clister T; Greenwald EC; Baillie GS; Zhang J AKAP95 Organizes a Nuclear Microdomain to Control Local CAMP for Regulating Nuclear PKA. Cell Chem Biol 2019, 26 (6), 885–891.e4. 10.1016/j.chembiol.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Buck J; Sinclair ML; Schapal L; Cann MJ; Levin LR Cytosolic Adenylyl Cyclase Defines a Unique Signaling Molecule in Mammals. Proc National Acad Sci 1999, 96 (1), 79–84. 10.1073/pnas.96.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Herbst KJ; Allen MD; Zhang J Spatiotemporally Regulated Protein Kinase A Activity Is a Critical Regulator of Growth Factor-Stimulated Extracellular Signal-Regulated Kinase Signaling in PC12 Cells. Mol Cell Biol 2011, 31 (19), 4063–4075. 10.1128/mcb.05459-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Bock A; Annibale P; Konrad C; Hannawacker A; Anton SE; Maiellaro I; Zabel U; Sivaramakrishnan S; Falcke M; Lohse MJ Optical Mapping of CAMP Signaling at the Nanometer Scale. Cell 2020, 182 (6), 1519–1530.e17. 10.1016/j.cell.2020.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Cooper DMF; Tabbasum VG Adenylate Cyclase-Centred Microdomains. Biochem J 2014, 462 (2), 199–213. 10.1042/bj20140560. [DOI] [PubMed] [Google Scholar]
- (31).Tengholm A; Gylfe E Oscillatory Control of Insulin Secretion. Mol Cell Endocrinol 2009, 297 (1–2), 58–72. 10.1016/j.mce.2008.07.009. [DOI] [PubMed] [Google Scholar]
- (32).Ni Q; Ganesan A; Aye-Han N-N; Gao X; Allen MD; Levchenko A; Zhang J Signaling Diversity of PKA Achieved via a Ca2+-CAMP-PKA Oscillatory Circuit. Nat Chem Biol 2011, 7 (1), 34–40. 10.1038/nchembio.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Oliveria SF; Dell’Acqua ML; Sather WA AKAP79/150 Anchoring of Calcineurin Controls Neuronal L-Type Ca2+ Channel Activity and Nuclear Signaling. Neuron 2007, 55 (2), 261–275. 10.1016/j.neuron.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Nichols CB; Rossow CF; Navedo MF; Westenbroek RE; Catterall WA; Santana LF; McKnight GS Sympathetic Stimulation of Adult Cardiomyocytes Requires Association of AKAP5 With a Subpopulation of L-Type Calcium Channels. Circ Res 2010, 107 (6), 747–756. 10.1161/circresaha.109.216127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Yang S-N; Berggren P-O β-Cell CaV Channel Regulation in Physiology and Pathophysiology. Am J Physiol-endoc M 2005, 288 (1), E16–E28. 10.1152/ajpendo.00042.2004. [DOI] [PubMed] [Google Scholar]
- (36).Everett KL; Cooper DMF An Improved Targeted CAMP Sensor to Study the Regulation of Adenylyl Cyclase 8 by Ca2+ Entry through Voltage-Gated Channels. Plos One 2013, 8 (9), e75942. 10.1371/journal.pone.0075942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Akerboom J; Calderón NC; Tian L; Wabnig S; Prigge M; Tolö J; Gordus A; Orger MB; Severi KE; Macklin JJ; Patel R; Pulver SR; Wardill TJ; Fischer E; Schüler C; Chen T-W; Sarkisyan KS; Marvin JS; Bargmann CI; Kim DS; Kügler S; Lagnado L; Hegemann P; Gottschalk A; Schreiter ER; Looger LL Genetically Encoded Calcium Indicators for Multi-Color Neural Activity Imaging and Combination with Optogenetics. Front Mol Neurosci 2013, 6, 2. 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Dou H; Wang C; Wu X; Yao L; Zhang X; Teng S; Xu H; Liu B; Wu Q; Zhang Q; Hu M; Wang Y; Wang L; Wu Y; Shang S; Kang X; Zheng L; Zhang J; Raoux M; Lang J; Li Q; Su J; Yu X; Chen L; Zhou Z Calcium Influx Activates Adenylyl Cyclase 8 for Sustained Insulin Secretion in Rat Pancreatic Beta Cells. Diabetologia 2015, 58 (2), 324–333. 10.1007/s00125-014-3437-z. [DOI] [PubMed] [Google Scholar]
- (39).Willoughby D; Masada N; Wachten S; Pagano M; Halls ML; Everett KL; Ciruela A; Cooper DMF AKAP79/150 Interacts with AC8 and Regulates Ca2+-Dependent CAMP Synthesis in Pancreatic and Neuronal Systems*. J Biological Chem 2010, 285 (26), 20328–20342. 10.1074/jbc.m110.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Zhang J; Carver CM; Choveau FS; Shapiro MS Clustering and Functional Coupling of Diverse Ion Channels and Signaling Proteins Revealed by Super-Resolution STORM Microscopy in Neurons. Neuron 2016, 92 (2), 461–478. 10.1016/j.neuron.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Rust MJ; Bates M; Zhuang X Sub-Diffraction-Limit Imaging by Stochastic Optical Reconstruction Microscopy (STORM). Nat Methods 2006, 3 (10), 793–796. 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Marshall CJ Specificity of Receptor Tyrosine Kinase Signaling: Transient versus Sustained Extracellular Signal-Regulated Kinase Activation. Cell 1995, 80 (2), 179–185. 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- (43).Keyes J; Ganesan A; Molinar-Inglis O; Hamidzadeh A; Zhang J; Ling M; Trejo J; Levchenko A; Zhang J Signaling Diversity Enabled by Rap1-Regulated Plasma Membrane ERK with Distinct Temporal Dynamics. Elife 2020, 9, e57410. 10.7554/elife.57410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Komatsu N; Aoki K; Yamada M; Yukinaga H; Fujita Y; Kamioka Y; Matsuda M Development of an Optimized Backbone of FRET Biosensors for Kinases and GTPases. Mol Biol Cell 2011, 22 (23), 4647–4656. 10.1091/mbc.e11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Sparta B; Pargett M; Minguet M; Distor K; Bell G; Albeck JG Receptor Level Mechanisms Are Required for Epidermal Growth Factor (EGF)-Stimulated Extracellular Signal-Regulated Kinase (ERK) Activity Pulses. J Biol Chem 2015, 290 (41), 24784–24792. 10.1074/jbc.m115.662247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Takahashi M; Li Y; Dillon TJ; Stork PJS Phosphorylation of Rap1 by CAMP-Dependent Protein Kinase (PKA) Creates a Binding Site for KSR to Sustain ERK Activation by CAMP. J Biol Chem 2017, 292 (4), 1449–1461. 10.1074/jbc.m116.768986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).O’Shaughnessy EC; Stone OJ; LaFosse PK; Azoitei ML; Tsygankov D; Heddleston JM; Legant WR; Wittchen ES; Burridge K; Elston TC; Betzig E; Chew T-L; Adalsteinsson D; Hahn KM Software for Lattice Light-Sheet Imaging of FRET Biosensors, Illustrated with a New Rap1 BiosensorSoftware for Lattice Light-Sheet FRET Imaging. J Cell Biology 2019, 218 (9), 3153–3160. 10.1083/jcb.201903019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Wang Z; Dillon TJ; Pokala V; Mishra S; Labudda K; Hunter B; Stork PJS Rap1-Mediated Activation of Extracellular Signal-Regulated Kinases by Cyclic AMP Is Dependent on the Mode of Rap1 Activation. Mol Cell Biol 2006, 26 (6), 2130–2145. 10.1128/mcb.26.6.2130-2145.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Dibble CC; Manning BD Signal Integration by MTORC1 Coordinates Nutrient Input with Biosynthetic Output. Nat Cell Biol 2013, 15 (6), 555–564. 10.1038/ncb2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Betz C; Hall MN Where Is MTOR and What Is It Doing There?Where Is MTOR and What Is It Doing There? J Cell Biology 2013, 203 (4), 563–574. 10.1083/jcb.201306041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Zhou X; Clister TL; Lowry PR; Seldin MM; Wong GW; Zhang J Dynamic Visualization of MTORC1 Activity in Living Cells. Cell Reports 2015, 10 (10), 1767–1777. 10.1016/j.celrep.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Jewell JL; Guan K-L Nutrient Signaling to MTOR and Cell Growth. Trends Biochem Sci 2013, 38 (5), 233–242. 10.1016/j.tibs.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Gao X; Zhang J Spatiotemporal Analysis of Differential Akt Regulation in Plasma Membrane Microdomains. Mol Biol Cell 2008, 19 (10), 4366–4373. 10.1091/mbc.e08-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Yoon S-O; Shin S; Liu Y; Ballif BA; Woo MS; Gygi SP; Blenis J Ran-Binding Protein 3 Phosphorylation Links the Ras and PI3-Kinase Pathways to Nucleocytoplasmic Transport. Mol Cell 2008, 29 (3), 362–375. 10.1016/j.molcel.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Banani SF; Lee HO; Hyman AA; Rosen MK Biomolecular Condensates: Organizers of Cellular Biochemistry. Nat Rev Mol Cell Bio 2017, 18 (5), 285–298. 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Hyman AA; Weber CA; Jülicher F Liquid-Liquid Phase Separation in Biology. Annu Rev Cell Dev Bi 2014, 30 (1), 39–58. 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- (57).Wright PE; Dyson HJ Intrinsically Disordered Proteins in Cellular Signalling and Regulation. Nat Rev Mol Cell Bio 2015, 16 (1), 18–29. 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Kamiyama D; Sekine S; Barsi-Rhyne B; Hu J; Chen B; Gilbert LA; Ishikawa H; Leonetti MD; Marshall WF; Weissman JS; Huang B Versatile Protein Tagging in Cells with Split Fluorescent Protein. Nat Commun 2016, 7 (1), 11046. 10.1038/ncomms11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Zhang J; Hupfeld CJ; Taylor SS; Olefsky JM; Tsien RY Insulin Disrupts β-Adrenergic Signalling to Protein Kinase A in Adipocytes. Nature 2005, 437 (7058), 569–573. 10.1038/nature04140. [DOI] [PubMed] [Google Scholar]
- (60).Langeberg LK; Scott JD Signalling Scaffolds and Local Organization of Cellular Behaviour. Nat Rev Mol Cell Bio 2015, 16 (4), 232–244. 10.1038/nrm3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Kolch W Coordinating ERK/MAPK Signalling through Scaffolds and Inhibitors. Nat Rev Mol Cell Bio 2005, 6 (11), 827–837. 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]
- (62).Lamas I; Merlini L; Vještica A; Vincenzetti V; Martin SG Optogenetics Reveals Cdc42 Local Activation by Scaffold-Mediated Positive Feedback and Ras GTPase. Plos Biol 2020, 18 (1), e3000600. 10.1371/journal.pbio.3000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Tobias IS; Newton AC Protein Scaffolds Control Localized Protein Kinase Cζ Activity. J Biol Chem 2016, 291 (26), 13809–13822. 10.1074/jbc.m116.729483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Wong W; Scott JD AKAP Signalling Complexes: Focal Points in Space and Time. Nat Rev Mol Cell Bio 2004, 5 (12), 959–970. 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- (65).Shaner NC; Lin MZ; McKeown MR; Steinbach PA; Hazelwood KL; Davidson MW; Tsien RY Improving the Photostability of Bright Monomeric Orange and Red Fluorescent Proteins. Nat Methods 2008, 5 (6), 545–551. 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Ando R; Mizuno H; Miyawaki A Regulated Fast Nucleocytoplasmic Shuttling Observed by Reversible Protein Highlighting. Science 2004, 306 (5700), 1370–1373. 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- (67).Dedecker P; Schryver FCD; Hofkens J Fluorescent Proteins: Shine on, You Crazy Diamond. J Am Chem Soc 2013, 135 (7), 2387–2402. 10.1021/ja309768d. [DOI] [PubMed] [Google Scholar]
- (68).Dedecker P; Mo GCH; Dertinger T; Zhang J Widely Accessible Method for Superresolution Fluorescence Imaging of Living Systems. Proc National Acad Sci 2012, 109 (27), 10909–10914. 10.1073/pnas.1204917109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Wang Y; Ho TG; Bertinetti D; Neddermann M; Franz E; Mo GCH; Schendowich LP; Sukhu A; Spelts RC; Zhang J; Herberg FW; Kennedy EJ Isoform-Selective Disruption of AKAP-Localized PKA Using Hydrocarbon Stapled Peptides. Acs Chem Biol 2014, 9 (3), 635–642. 10.1021/cb400900r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Martin BR; Deerinck TJ; Ellisman MH; Taylor SS; Tsien RY Isoform-Specific PKA Dynamics Revealed by Dye-Triggered Aggregation and DAKAP1α-Mediated Localization in Living Cells. Chem Biol 2007, 14 (9), 1031–1042. 10.1016/j.chembiol.2007.07.017. [DOI] [PubMed] [Google Scholar]
- (71).Smith FD; Esseltine JL; Nygren PJ; Veesler D; Byrne DP; Vonderach M; Strashnov I; Eyers CE; Eyers PA; Langeberg LK; Scott JD Local Protein Kinase A Action Proceeds through Intact Holoenzymes. Science 2017, 356 (6344), 1288–1293. 10.1126/science.aaj1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Walker-Gray R; Stengel F; Gold MG Mechanisms for Restraining CAMP-Dependent Protein Kinase Revealed by Subunit Quantitation and Cross-Linking Approaches. Proc National Acad Sci 2017, 114 (39), 10414–10419. 10.1073/pnas.1701782114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Dodge K; Scott JD AKAP79 and the Evolution of the AKAP Model. Febs Lett 2000, 476 (1–2), 58–61. 10.1016/s0014-5793(00)01671-9. [DOI] [PubMed] [Google Scholar]
- (74).Mehta S; Zhang Y; Roth RH; Zhang J; Mo A; Tenner B; Huganir RL; Zhang J Single-Fluorophore Biosensors for Sensitive and Multiplexed Detection of Signalling Activities. Nat Cell Biol 2018, 20 (10), 1215–1225. 10.1038/s41556-018-0200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Zhang J-F; Liu B; Hong I; Mo A; Roth RH; Tenner B; Lin W; Zhang JZ; Molina RS; Drobizhev M; Hughes TE; Tian L; Huganir RL; Mehta S; Zhang J An Ultrasensitive Biosensor for High-Resolution Kinase Activity Imaging in Awake Mice. Nat Chem Biol 2021, 17 (1), 39–46. 10.1038/s41589-020-00660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]