Abstract
A large number of physiological processes in the adult liver are regulated by nuclear receptors that require heterodimerization with retinoid X receptors (RXRs). In this study, we have used cre-mediated recombination to disrupt the mouse RXRα gene specifically in hepatocytes. Although such mice are viable, molecular and biochemical parameters indicate that every one of the examined metabolic pathways in the liver (mediated by RXR heterodimerization with PPARα, CARβ, PXR, LXR, and FXR) is compromised in the absence of RXRα. These data demonstrate the presence of a complex circuitry in which RXRα is integrated into a number of diverse physiological pathways as a common regulatory component of cholesterol, fatty acid, bile acid, steroid, and xenobiotic metabolism and homeostasis.
Members of the nuclear receptor family regulate a broad range of developmental and physiological processes by binding to DNA response elements and regulating transcriptional activation (7). The retinoid X receptors (RXRs) are unique among the nuclear receptors in that they bind DNA as a homodimer and are required as a heterodimeric partner for a number of additional nuclear receptors to bind DNA (19). The latter receptors, termed the class II nuclear receptor subfamily, include many which are established or implicated as important regulators of gene expression in the liver (reviewed below). There are three RXR genes (18), coding for RXRα, -β, and -γ, all of which are able to heterodimerize with any of the class II receptors, although there appear to be preferences for distinct RXR subtypes by partner receptors in vivo (6).
The physiological processes that are regulated by the class II receptors in the liver are diverse. LXRα is activated by oxycholesterol and promotes cholesterol metabolism (22). FXR (also known as RIP14) is part of an interrelated process, in that FXR is activated by bile acids (the end product of cholesterol metabolism) (17, 21, 34), which serve to inhibit cholesterol catabolism. CARβ is involved in phenobarbital induction of the cytochrome P450 2B10 (Cyp2B10) gene (9), which encodes a drug- and xenobiotic-metabolic enzyme. Interestingly, CARβ has constitutively activity, but becomes inactive in response to the steroid androstane (8). PXR is activated by a wide spectrum of steroids, and induces the Cyp3A gene (Cyp3A1 in mice; Cyp3A4 in humans) which encodes a broad-spectrum oxidase that is responsible for steroid degradation and for the catabolism of numerous pharmaceutical compounds (2, 11). PPARα is activated by leukotriene B4 and by synthetic peroxisome proliferators, such as fibrates, and controls the expression of several genes which are related to fatty acid metabolism and processing (28). Although not discussed further in the context of this study, other class II nuclear receptors, including the retinoic acid and thyroid hormone receptors, are also likely to be important regulators of liver metabolism.
In the adult liver, RXRα is the most abundant of the three RXRs (18), suggesting that it might have a prominent role in hepatic functions that involve regulation by class II nuclear receptors. Conventional gene targeting approaches have only partially addressed this issue. While all three RXR genes have been individually mutated in the mouse germ line, mice lacking both RXRβ and RXRγ together (14) are viable and are not known to suffer from any impairment of liver function. In contrast, lack of RXRα results in midgestation embryonic lethality caused by insufficient heart development (10, 30), thereby preventing assessment of the role of RXRα in postnatal liver physiology. To address this role, in the present study, we have used cre/lox-mediated recombination to selectively mutate the RXRα gene in adult hepatocytes, and thereby explore the involvement of RXRα in the diverse range of physiological pathways that are regulated by class II nuclear receptors in the liver.
MATERIALS AND METHODS
Mice.
The derivations of the conventional loss-of-function RXRα allele (identified as ko), the conditional RXRα allele (identified as flox), the albumin-cre (Alb-cre) transgenic line, and the conditional lacZ reporter line ROSA26R have all been described previously (4, 25, 29, 30). Liver-specific mutation of the RXRα gene was achieved by crossing the Alb-cre transgene against the conditional allele. Experimental animals used in this study carried one allele of the Alb-cre transgene and at the RXRα locus were either RXRαflox/flox or RXRαflox/ko. Wild-type animals used in this study were of either completely wild-type genotype (RXRαwt/wt) or carried one RXRα conditional allele (RXRαflox/wt) and did not carry the cre transgene. In the absence of cre-mediated recombination, the conditional flox allele is functionally identical to the wild-type allele. Mice were housed in standard cages under a 6 a.m.–6 p.m. light-dark cycle and under standard conditions were fed either Purina PicoLab Rodent Diet 20 or Harlan Teklad 7001 diet. The latter diet was provided to control animals in high-cholesterol-diet studies, and Harlan Teklad TD86295, containing 2% cholesterol by weight in a 7001 base, was provided to experimental mice. For analysis of Cyp7A mRNA induction by dietary cholesterol, age- and littermate-matched male mice were housed individually for at least 2 weeks before beginning the high-cholesterol diet. Two hours before the onset of the light cycle at the end of the seventh night of treatment, mice were sacrificed, and liver tissue was isolated and frozen and then processed as described below.
Specificity of cre-mediated recombination.
For Southern blotting, liver DNA was isolated from dissected embryonic or postnatal stage animals by standard procedures. Genomic DNA was restriction digested with BamHI and BglII and then separated on an agarose gel and transferred to nylon membranes. The probe used in hybridizations was a HindIII-SmaI fragment of the mouse genomic RXRα locus previously described as probe C (4), which hybridizes to a region outside the two loxP sites and therefore recognizes the wild-type RXRα allele, the unrecombined conditional RXRα allele, and the conditional RXRα allele after recombination. For PCR analysis of specificity, genomic DNA was isolated from various tissues and assayed by PCR amplification with primers P1 and P3 as described previously (4). The sensitivity of this assay was determined by amplifying samples with known ratios of recombined liver genomic DNA and nonrecombined tail genomic DNA. For histochemical staining in animals carrying the conditional ROSA26R lacZ reporter gene, adult tissue was isolated and in some cases bisected manually to improve stain penetration and then fixed and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) by standard procedures. For RXRα RNA analysis, total liver RNA was converted into cDNA by random priming and then amplified for 32 cycles with primers corresponding to nucleotides 552 to 574 and 952 to 931 of X66223.1 in GenBank.
Northern blotting analysis.
All mice used as a source of RNA were male mice 2 to 4 months of age. Total liver RNA was isolated by guanidinium-phenol extraction. Twenty micrograms of total RNA per lane was resolved by electrophoresis on 1.2% agarose gels containing 2.2 M formaldehyde and then transferred to nylon membranes by capillary blotting. Probe cDNA fragments were labeled by random priming and hybridized to membranes in 7% (wt/vol) sodium dodecyl sulfate, 0.5 M sodium phosphate (pH 6.5), 1 mM EDTA, and 1 mg of bovine serum albumin per ml at 68°C overnight. The membranes were washed twice in 1% sodium dodecyl sulfate, 50 mM NaCl, and 1 mM EDTA at 68°C for 15 min each and autoradiographed with intensifying screens. The amount of mRNA expressed in individual samples was quantitated by densitometry and then normalized with the level of 18S rRNA; the mean and standard deviation for eight samples were calculated to validate the statistical significance of observed changes. Gene probes used were ApoAI and CIII (provided by J. Auwerx), Cyp4A1 (provided by F. Gonzalez), liver fatty acid-binding protein (LFABP; provided by J. Gordon), acylcoenzyme A (acyl-CoA) oxidase (provided by T. Osumi), catalase (purchased from American Type Culture Collection) Cyp2B10 (provided by M. Negishi), Cyp3A1 (provided by F. Gonzalez), and Cyp7A (provided by L. Chan). The RXR heterodimeric partner genes studied were mouse retinoic acid receptors (RARs) and RXRs (provided by R. Evans), PPARα (provided by S. Green), LXRα (provided by D. Mangelsdorf), CARβ (provided by B. Forman), PXR (provided by B. Blumberg), FXR/RIP14 (provided by D. Moore), and HNF4 (provided by F. Sladek).
Liver function, composition, and morphology.
Liver function, including blood triglyceride and cholesterol levels, was evaluated by automated analysis of major serum components. Blood samples were obtained by retroorbital extraction. Hepatic cholesterol and triglyceride were measured as previously described (22). For hematoxylin-eosin histological analysis, pieces of tissue from equivalent regions of the liver were dissected, formalin fixed, paraffin embedded, sectioned at a 5-μm thickness, and stained by standard procedures. For neutral lipid staining, oil red O was dissolved in isopropanol and mixed with water immediately prior to use. Tissue was frozen in O.C.T. (Tissue-Tek) and cryostat sectioned at a 5-μm thickness, rinsed in water, stained, rinsed in isopropanol and then water, and then stained with hematoxylin and mounted with Fisher aqueous mountant. A polarizing filter was used to visualize cholesterol crystals.
RESULTS
Generation of mice lacking RXRα in hepatocytes.
To selectively disrupt RXRα gene function in postnatal hepatocytes, we have utilized a previously described (4) conditional (“floxed”) allele of RXRα. This allele was constructed such that small loxP sequences were inserted into introns flanking the fourth exon of the gene; this exon encodes the majority of the DNA binding domain of the RXRα protein, which is essential for RXRα function. This allele is fully functional and effectively wild type in the absence of cre-mediated recombination, although upon recombination, the fourth exon is specifically deleted. To mutate the RXRα gene in hepatocytes, we crossed the floxed RXRα allele against a transgenic line in which cre recombinase is expressed under the control of the albumin promoter (25). The albumin gene is expressed exclusively in hepatocytes, beginning in midgestation and continuing for the life of the animal, and the specificity of the albumin promoter is very well defined in transgenic studies (24). Because the albumin promoter is continually expressed in hepatocytes at all stages, cre-mediated mutation of the RXRα gene is expected to be a unidirectional and cumulative process.
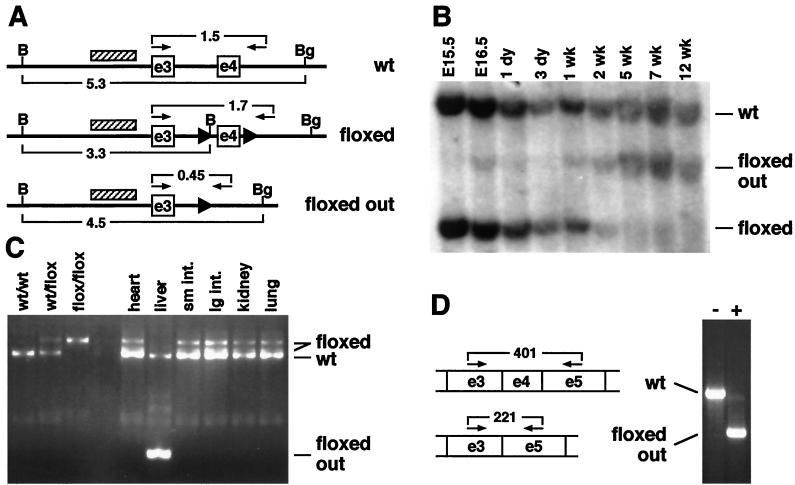
To document the timing and extent of recombination of the conditional RXRα gene under the direction of the Alb-cre transgene, genomic DNA was isolated from livers of transgenic embryos and pups, and analyzed by Southern blotting. The probe used in these hybridizations recognizes all three RXRα alleles (wild-type, nonrecombined floxed, and the recombined floxed-out alleles). In the assay shown in Fig. 1B, transgenic heterozygous animals (i.e., bearing one wild-type RXRα allele and one conditional allele) were examined, such that the wild-type RXRα allele serves as an internal control, and the extent of recombination of the conditional RXRα allele can be monitored by comparison between the nonrecombined and recombined bands on the Southern blot. Recombination of the conditional RXRα allele was first evident in this assay at embryonic day 16.5 and increased to a maximum level by 5 weeks postnatal. By quantitation of band intensities, at 5 weeks and beyond, greater than 80% of the conditional RXRα alleles are recombined. This number represents close to if not complete recombination in hepatocytes and is similar to that observed with a different target gene (25); the residual amount of nonrecombined signal presumably represents nonhepatocyte cell types which are also present in the tissue samples. Using a more sensitive PCR assay (data not shown), recombination was first detected at low levels at embryonic day 14.5, but not at embryonic day 12.5, and no recombination was ever observed at any stage in animals lacking the cre transgene.
FIG. 1.
Efficiency of recombination of the conditional RXRα allele by Alb-cre. (A) Diagrammatic representation of the chromosomal organization of the RXRα alleles described in this study (adapted from reference 4). The upper line represents the wild-type (wt) allele, the middle line represents the conditional (floxed) allele, and the bottom line represents the conditional allele after cre-mediated recombination (floxed out). Exons 3 and 4 are indicated as boxes (e3 and e4, respectively), and loxP sites are indicated by solid triangles. Brackets underneath each line indicate the sizes of restriction fragments after BamHI (B) and BglII (Bg) double digestion, as illustrated in Southern blots (panel B). The cross-hatched block indicates the location of the probe used in Southern blots. Small arrows and brackets above each line indicate PCR primers and sizes of amplification products, as illustrated in panel C. Sizes are in kilobases. The diagram approximates but is not drawn to scale. (B) Time course of recombination. Genomic DNA from liver tissue from Alb-cre transgenic, RXRαflox/wt animals at the indicated embryonic (E; measured from day of conception) or postnatal (measured from birth) time points was isolated and analyzed by Southern blotting. The wild-type (wt) band, which is not affected by recombination, serves as an internal standard; the positions of the nonrecombined (floxed) and recombined (floxed out) conditional RXRα fragments are indicated. (C) Specificity of recombination. Genomic DNA from the indicated tissues of a 7-wk-old Alb-cre transgenic, RXRαflox/wt animal was analyzed by PCR amplification. The primers used amplify to different product lengths the wild-type, nonrecombined, and recombined RXRα alleles. In samples from RXRαflox/wt animals in which the conditional allele is not recombined, there is an additional band intermediate in size between the wild-type and floxed products, which represents incomplete extension in one cycle from the conditional allele and subsequent extension on the wild-type template in a later amplification cycle. To the left are size standards from genomic tail DNA from unrelated animals. sm. int., small intestine; lg. int., large intestine. (D) RNA analysis of the conditional allele. To the left is a diagrammatic representation of a portion of the transcripts derived from the conditional allele (above; note this transcript is identical to a wild-type transcript) and from the conditional allele lacking exon 4 after recombination (below). The arrows and brackets indicate primer locations and sizes (in base pairs) of amplification products. To the right is the result of reverse transcription-PCR analysis of RXRα mRNA using total liver RNA derived from 3-month-old mice which were RXRαflox/flox and either did not (−) or did (+) carry the Alb-cre transgene. In cre-transgenic liver tissue, the predominant product is deleted for exon 4 sequences, but a trace amount of normal-size product is also seen, presumably representing expression of wild-type RXRα mRNA in nonhepatocyte cell types.
To confirm the tissue specificity of recombination, a variety of tissues were isolated from 7-week-old mice and assayed by PCR amplification for the presence of the recombined RXRα allele. As shown in Fig. 1C, no tissue other than liver displayed any sign of recombination. The sensitivity of this assay is such that a positive signal would be detected if recombination occurred in 1% of the cells of each tissue (data not shown). To further confirm the specificity of recombination, the Alb-cre transgene was crossed against the conditional ROSA26R reporter gene in which β-galactosidase is expressed, potentially in any tissue, but dependent upon cre-mediated recombination. By this method, even a very low level of recombination would be detectable at single-cell resolution. With the exception of the liver, no other fetal or adult tissue showed any evidence of recombination (data not shown). Thus, recombination driven by the Alb-cre transgene is specific for hepatocytes and is sufficiently efficient to cause maximal recombination of the conditional RXRα allele by 5 weeks after birth. RNA analysis (Fig. 1D) indicated that the RXRα allele was still expressed in the livers of adult animals in which cre-mediated recombination had occurred, except that the resultant transcript was deleted for exon 4 sequences, as expected.
Matings were then made to create mice lacking RXRα function in hepatocytes. Such mice were born and reared normally, were viable at least for over 1 year, and under standard housing conditions, appeared externally to be normal.
Absence of RXRα results in altered hepatic gene expression and physiology.
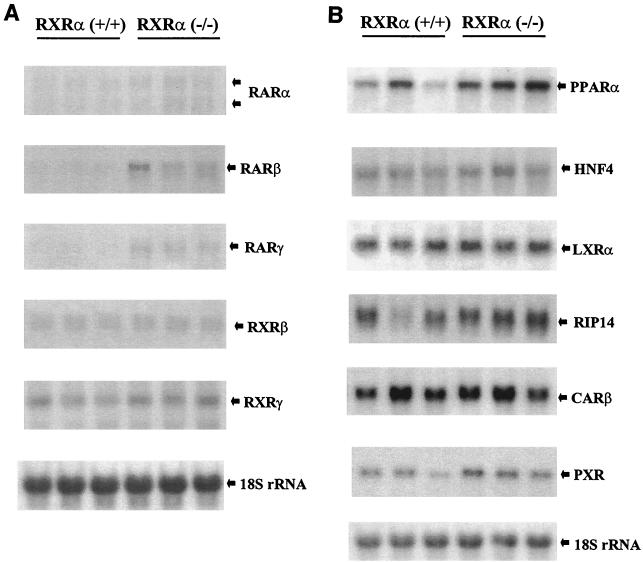
We first addressed with mice fed a standard rodent diet and not subjected to physiological stress whether absence of RXRα altered the basal expression pattern of any of the class II nuclear receptor genes that are expressed in the liver (Fig. 2). The expression of RXRβ and RXRγ was unchanged in mutant tissue relative to the wild type, indicating that a compensatory up-regulation of these RXR genes does not occur in the absence of RXRα. The expression of RXRα, LXRα, FXR/RIP14, CARβ, and HNF4 was also unchanged. The hepatic level of PXR ranged up to twofold higher in mutant livers relative to levels in wild-type animals, but this increase was not statistically significant. However, twofold increases in the levels of RARβ, RARγ, and PPARα were observed. It is currently uncertain to what extent these increases are physiologically relevant.
FIG. 2.
Expression of nuclear receptor genes in RXRα-deficient liver tissue. (A) Retinoid receptor genes. (B) Additional class II heterodimeric partners of RXR. Blots were prepared with RNA isolated from wild-type (+/+) and deficient (−/−) liver tissue and sequentially hybridized with the probes indicated. RNA was isolated from eight wild-type and eight mutant livers to control for variations in diet, hormonal status, and/or other physiological parameters that might influence gene expression, although these and other panels in this study present the results from three samples per genotype. RIP14 is also known as FXR.
We hypothesized that the consequences of the absence of RXRα in the liver, although not lethal or morphologically apparent, would be manifested in an alteration in molecular and biochemical parameters of hepatic physiological function. To address this issue, we asked whether pathways that are regulated by class II nuclear receptors as RXR heterodimeric partners are altered in the absence of RXRα.
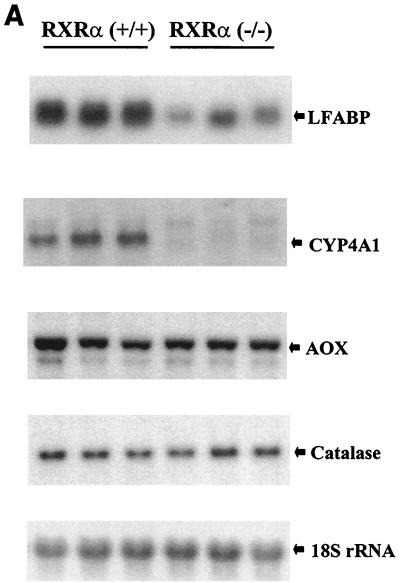
LFABP, Cyp4A1, acyl-CoA oxidase, and catalase are involved in fatty acid metabolic pathways, and their gene promoters are regulated by PPARα (28). In hepatocyte-specific RXRα mutant mice, the levels of LFABP and Cyp4A1 mRNA were markedly reduced (four- and fivefold, respectively) (Fig. 3A). In contrast, the expression of acyl-CoA oxidase and catalase was unchanged in the mutant mice.
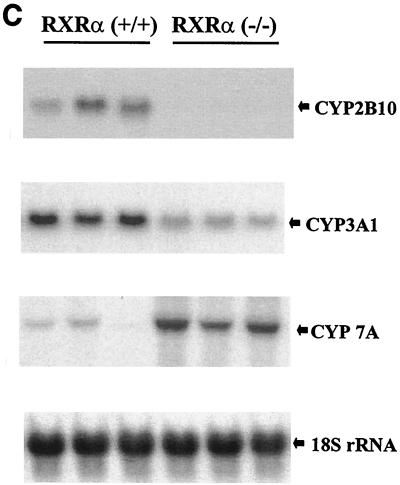
FIG. 3.
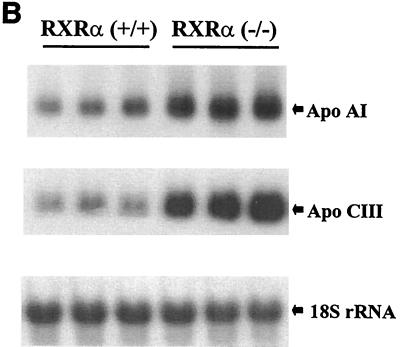
Target gene expression in RXRα-deficient liver tissue. (A) PPARα target genes. (B) Apolipoprotein genes. (C) Target genes for LXRα, FXR, CARβ, and PXR. Experimental details are as described for Fig. 2.
Apolipoproteins are involved in transport of lipids in serum. In transfection assays, the ApoCIII promoter is regulated by multiple nuclear receptors, including RXR homodimers, RXR-RAR, RXR-PPAR, HNF4, ARP-1, EAR-2, and others (15). Interestingly, the ApoCIII and ApoAI promoters are positively regulated by retinoids through RXR-RAR heterodimers and RXR homodimers and negatively regulated by fibrates through RXR-PPARα heterodimers. In liver tissue from mice lacking RXRα, expression of the ApoAI and CIII genes was increased more than three- and sixfold, respectively (Fig. 3B). ApoAI expression is also elevated in PPARα-deficient mice (23), consistent with RXRα being the principal PPARα heterodimeric partner in hepatocytes.
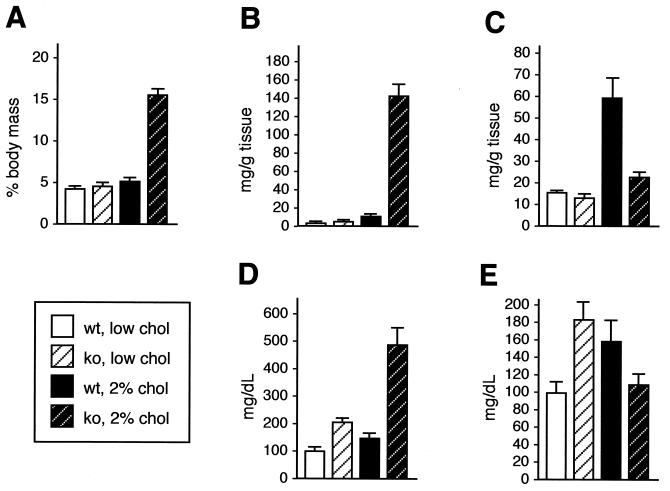
Plasma ApoCIII concentrations are positively correlated with plasma triglyceride levels (16). In mice lacking RXRα in the liver, serum triglyceride levels were substantially elevated compared to those of wild-type controls, increasing from 1.6- to 2.0-fold in four separate determinations (weighted average of 1.8-fold higher; n = 17 wild type and 21 knockout mice [Fig. 4E]). These data confirm that the increased expression of ApoCIII was manifested in a corresponding physiological increase in serum triglyceride levels and that RXRα is an essential regulatory component of fatty acid homeostasis. Hepatic triglyceride levels (Fig. 4C) were slightly lower in normally fed RXRα-deficient mice (12.7 ± 1.3 mg/g) compared to the wild type (15.5 ± 0.3 mg/g), possibly reflecting the increased mobilization of hepatic triglyceride into peripheral circulation.
FIG. 4.
Lipid parameters in normal and RXRα-deficient mice fed a standard or high-cholesterol diet. (A) Liver/body mass ratio. (B) Hepatic cholesterol. (C) Hepatic triglyceride. (D) Serum cholesterol. (E) Serum triglyceride. The data shown were obtained from age-matched female mice of the indicated genotypes maintained for 3 months either with a low-cholesterol or high-cholesterol diet; where investigated, results for male mice were generally very similar to those for females. The results shown are means ± standard errors.
Cyp2B10 is the mouse cytochrome P450 enzyme that is most effectively induced by phenobarbital. Transfection and DNA binding assays have indicated that phenobarbital induction is mediated through binding of RXR-CARβ heterodimers to DR-4 sites in the Cyp2B10 promoter (9). The expression of hepatic Cyp2B10 mRNA was reduced at least 10-fold in the absence of RXRα as compared to that of wild-type mice (Fig. 3C), suggesting that in normal mice under basal conditions, the RXRα-CARβ heterodimer is an activator of the Cyp2B10 gene.
Expression of the Cyp2B and -3A subfamilies is coordinately regulated in humans, nonhuman primates, and rodents. The tissue samples with the highest relative levels of Cyp2B also tend to display the greatest amounts of immunoreactive Cyp3A (3), and CARβ transactivates the human Cyp3A4 gene in HepG2 cells (32). Cyp3A is also a target gene for recognition by the RXR-PXR heterodimer (2, 11). We therefore examined the expression of the mouse Cyp3A1 gene, the homolog of human Cyp3A4. In the absence of RXRα, there was a fourfold reduction in the level of hepatic Cyp3A1 message compared with that of the wild type (Fig. 3C). These data suggest that the RXR-CARβ and the RXR-PXR pathways are both compromised in the RXRα-deficient mouse liver.
Bile acid synthesis is a pivotal pathway in maintaining the balance between cholesterol supply and disposal. The rate-limiting reaction of the classical bile acid synthesis pathway is the 7α-hydroxylation of cholesterol (27), which is catalyzed by cholesterol 7α-hydroxylase, a product of the liver-specific Cyp7A gene. Transcription of the Cyp7A gene is positively controlled by a cholesterol-inducible LXRα pathway and negatively regulated by bile acids through an FXR pathway (26). In RXRα knockout mouse livers, expression of the Cyp7A gene was increased more than eightfold compared with that in wild-type animals (Fig. 3C). Thus, the primary regulation of the Cyp7A gene in normally fed (low-cholesterol diet) mice appears to be inhibitory control mediated by RXRα-FXR that is alleviated in the absence of RXRα. Hepatic cholesterol was marginally elevated in RXRα mutant mice (4.4 ± 0.3 mg/g) relative to the level in the wild type (3.5 ± 0.4 mg/g) (Fig. 4B), while serum cholesterol was doubled in the mutant mice (Fig. 4D). These results indicate that cholesterol homeostasis is significantly altered in the absence of RXRα.
In summary, the results presented above demonstrate that the basal activity of every one of the examined pathways in the adult liver which involves class II receptors is compromised in the absence of RXRα.
Failure in cholesterol homeostasis in RXRα-deficient mice.
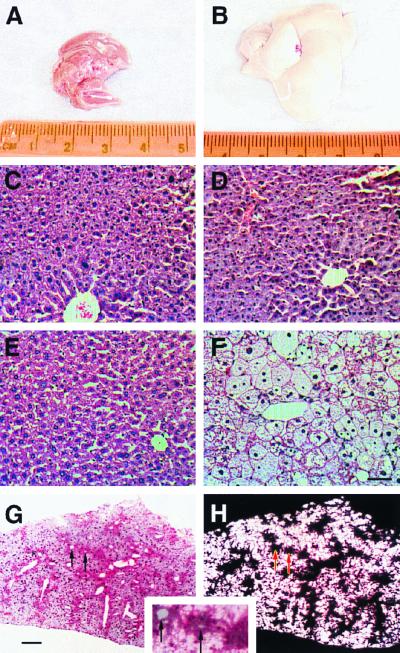
In order to define the extent to which the absence of RXRα compromises not only the basal status of hepatic class II nuclear receptor pathways, but also the response to physiological stress, we challenged our mice with a high-cholesterol (2% by weight) diet. Excess dietary cholesterol is accommodated in normal mice by increasing bile acid production and consequent fecal elimination, a process which requires LXRα (22). Our approach was similar to the previous evaluation of the LXRα−/− mouse, and in a manner which is very similar to the LXRα knockout phenotype, the livers of mice lacking RXRα specifically in hepatocytes and fed the high-cholesterol diet became progressively fatty and enlarged. After 1 week of treatment, the livers of mutant mice were already obviously more pale than those of wild-type mice or mutant mice provided with a low-cholesterol diet (data not shown), and by 1 month of treatment, the livers of these mice were almost completely white (Fig. 5A and B). The liver/body weight ratio in liver-specific RXRα mutant mice maintained on the high-cholesterol diet for 1 month was twice that of normal controls fed the same diet (data not shown), and after 3 months (the longest period measured in this study), it was triple that of control mice (Fig. 4A). Even with such pronounced hepatomegaly, mutant mice on the 2% cholesterol diet were normal in externally apparent parameters and were not moribund, lethargic, or sick.
FIG. 5.
Morphology and histology of mouse livers. Mice were fed either a low-cholesterol diet (A, C, and E) or a 2% cholesterol diet (B, D, F, G, and H) for 1 (A and B) or 2 (C to H) months. (A and B) Morphology of whole livers from hepatocyte-specific RXRα mutant mice. The normal appearance of livers from mutant mice on a standard diet (A) is identical to that of wild-type mice fed either diet (data not shown). (C and D) Histology of wild-type mouse livers. Challenge with a high-cholesterol diet does not alter histology. (E and F) Histology of mutant mouse livers. Although the untreated mutant tissue is of normal appearance (E), there is a substantial increase in hepatocyte cell size and vacuole material in high-cholesterol-fed mice (F). Panels C to F are hematoxylin-and-eosin stained and are at the same magnification; the scale bar in panel F is 50 μm. (G and H) Oil red O staining of liver tissue. When viewed under normal light (G), hepatic triglyceride (darker red staining) is preferentially located close to blood vessels, whereas hepatic cholesterol (lighter staining) is accumulated more distally. Panel H is the same section as panel G, but viewed with polarized light to illuminate cholesterol crystals (which appear white). The inset is a close-up view of a superimposition of panels G and H. Arrows in panels G and H and the inset are provided for register and point to the same two small blood vessels. The scale bar in panel G is 100 μm.
To more specifically define the composition of hepatic lipid in these mice, liver samples were assayed for cholesterol and triglyceride levels (Fig. 4B and C). Hepatic cholesterol increased three- to fourfold in wild-type mice after 3 months on the high-cholesterol diet. In contrast, there was an over 30-fold increase in hepatic cholesterol in experimental mice, demonstrating the inability of mutant mice to process and eliminate dietary cholesterol. Hepatic triglyceride levels increased fourfold in normal mice on the high-cholesterol diet, whereas the increase in mutant mice was less than twofold. The physiological significance of the latter observation is uncertain, although LXRα−/− mice (22) also show an attenuated hepatic triglyceride response to the high-cholesterol diet.
Histological sections revealed a large increase in hepatocyte cell size and vacuole material in high-cholesterol-fed mutant liver tissue (Fig. 5E and F). Tissue sections from wild-type animals showed minimal accumulation of lipid when fed the high-cholesterol diet (Fig. 5C and D). The accumulated hepatic cholesterol in mutant mice crystallized during histology processing for oil red O staining, becoming visible upon illumination with polarized light (Fig. 5G and H). Tissue from wild-type mice on either diet and from mutant mice on a low-cholesterol diet did not contain any crystalline cholesterol (data not shown). Interestingly, because oil red O preferentially stains triglyceride, we noted in high-cholesterol-fed mutant mice that triglyceride and cholesterol were stored in separate domains of the liver, with triglyceride mostly accumulated in the centrilobular region closest to major blood vessels and cholesterol mostly stored more peripherally.
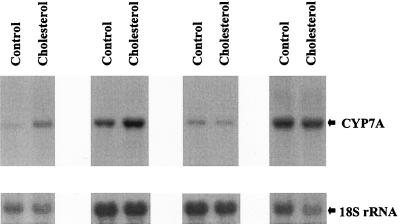
We then addressed the extent to which expression of Cyp7A is modulated by dietary cholesterol in the absence of RXRα. We assayed Cyp7A message levels in eight liver-specific RXRα homozygous mice fed either a standard diet or a 2% cholesterol diet for 1 week. As noted above, the livers of mutant mice fed the high-cholesterol diet were noticeably more pale after 1 week than the normal-appearing livers of mutant mice provided with a low-cholesterol diet, indicative of the onset of cholesterol accumulation. Cyp7A message level was increased by cholesterol exposure in the RXRα homozygous mutant mice, albeit by less than twofold on average (Fig. 6). This is an attenuated response compared to the four- to fivefold increase in Cyp7A expression in wild-type mice and the absence of increase in LXRα−/− mice subjected to the same challenge. The residual increase in Cyp7A might be a consequence of a residual level of activity of the feed-forward LXRα pathway mediated by the low hepatic levels of RXRβ and/or RXRγ still present in these mice.
FIG. 6.
Attenuated induction of Cyp7A by dietary cholesterol in RXRα-deficient mice. Hepatic Cyp7A message levels in RXRα-deficient mice provided with a low (control)- or high-cholesterol diet for 7 days are shown. There is a two- to threefold induction of Cyp7A between the first two pairs of animals and no induction in the second two pairs. The mice used in these assays are derived from a mixed-strain background, which might account for some of the variability in the level and inducibility of Cyp7A expression.
Hepatic cholesterol is packaged with apolipoproteins and exported through the circulatory system for uptake by peripheral tissues. As noted above, mutant mice on a standard low-cholesterol diet exhibited a twofold increase in basal serum cholesterol level relative to wild-type mice on the same diet. When challenged with the high-cholesterol diet, serum cholesterol levels in wild-type mice increased by 40%, whereas the increase in RXRα-deficient mice was 2.4-fold (Fig. 4D), or almost 5-fold higher than the level in wild-type mice on a normal diet. Serum cholesterol levels in mutant mice on the high-cholesterol diet for 1 month and 3 months were identical (data not shown). LXRα−/− mice fed a high-cholesterol diet also exhibited a twofold increase in serum cholesterol commensurate with their increased hepatic cholesterol load, although the baseline level of serum cholesterol in normally fed LXRα−/− mice was equivalent to that in wild-type mice.
DISCUSSION
The RXRs were initially identified on the basis of their ability to promote DNA binding of certain nuclear receptors in cell-free assays (33). Numerous biochemical and transfection assays performed since these initial observations have supported the model in which RXRs are obligate heterodimeric partners of the class II receptors and in fact have served to define the class II subfamily as requiring heterodimerization with RXR. Nonetheless, the in vivo evidence for the role of heterodimerization has so far only been established for RXR-RAR, based primarily on the analysis of RXR and RAR compound mutant mice (10, 20). In this study, we extend this principle beyond the more closely related retinoid receptor subfamily (i.e., RXRs and RARs) to include LXRα, PPARα, CARβ, PXR, and FXR, with the demonstration that physiological pathways regulated by each of these receptors are compromised in the absence of RXRα.
In addition to confirming the previously suspected role of RXR heterodimeric partners in regulating hepatic physiology, our results also provide insight into which of the nuclear receptor pathways are most critical in the regulation of certain pathways in vivo. For example, Cyp7A expression is upregulated by LXR and downregulated by FXR; the substantial increase in expression we observed after RXRα mutation suggests that in normal mice on a standard diet, the dominant regulation of the Cyp7A gene is negative regulation via the RXRα-FXR heterodimer. Similarly, of the multiple nuclear receptor complexes which have been defined by transfection assays to regulate the ApoAI and ApoCIII genes (PPARα, HNF4, ARP, EAR, etc.), our results demonstrate the importance of the RXRα-PPARα complex, although they do not necessarily discount the involvement of these additional receptors in apolipoprotein expression under basal or stressed conditions. In some cases, some ambiguity in defining the involvement of potentially competing receptor pathways remains. For example, the fourfold decrease in Cyp3A1 expression we observed as a consequence of RXRα mutation could result from reduction of an RXRα-CARβ or RXRα-PXR complex, because both heterodimers have been implicated in Cyp3A1 regulation. It is also possible, perhaps even likely, that both receptors simultaneously regulate the basal level of this target gene.
Some of the target genes we explored gave surprising results. The gene coding for acyl-CoA oxidase is a prototype PPARα target gene, being one of the first of the fatty acid oxidation pathway genes to be defined as fibrate inducible and with an element in its promoter that was among the first to define the PPAR response element (12). In vivo, however, acyl-CoA oxidase expression is not altered by RXRα mutation. Similarly, the ApoCIII promoter is inhibited by fibrate (similar to the response of ApoAI) and contains a well-defined PPARα response element. The increase in ApoCIII expression we observed in the absence of RXRα is consistent with removal of an inhibitory regulatory agent, which might be the RXRα-PPARα heterodimer. However, in PPARα−/− mice, the ApoCIII gene is normally expressed (23). While we cannot account for these results at this time, one possibility is that these genes are controlled by other regulatory pathways in vivo, either independent of or in addition to the known pathways. There may also be compensatory pathways that become active in the absence of RXRα and influence gene expression in a secondary manner. It is also possible, at least for acyl-CoA oxidase, that the presence of RXRβ or RXRγ allows normal expression of the gene coding for this product to occur, despite the alteration in expression of other PPARα target genes that occurs in the absence of RXRα.
The RARβ gene is regulated through a response element that is activated by RXR heterodimers with RAR (31). In the absence of RXRα, expression of the RARβ gene is increased (Fig. 2A), which is consistent with the removal of an inhibitory agent. Possibly, this inhibitor is RXRα-RAR, which like certain other nuclear receptor heterodimers has been demonstrated to have silencing activity in the absence of ligand via recruitment of transcriptional corepressors (5). In this context, it will be particularly interesting to explore thyroid hormone-regulated gene expression in our mice, because the thyroid receptor has a very strong transcriptional repression domain in the absence of ligand (13).
We chose to examine the cholesterol pathway in detail because the LXRα knockout phenotype has been characterized and is particularly striking. Thus, challenge with a high-cholesterol diet offers the most direct way to define RXRα as the in vivo partner of one of its class II partners, not only under standard conditions, but also under physiological stress. In fact, the liver phenotypes of both types of animals in response to a high-cholesterol diet are quite similar, being manifested in an accumulation of hepatic cholesterol and an elevated serum cholesterol level. It should be noted that there are differences between the two animal models as well (in basal hepatic and serum lipid content, in the extent of hepatic cholesterol accumulation after the high-cholesterol diet, and in Cyp7A expression). We cannot at the moment explain these differences, although we suggest that the absence of RXRα alters multiple physiological pathways in addition to LXRα that collectively account for the differences seen. Nonetheless, our results indicate that RXRα is required for at least a large component of the normal cholesterol responsiveness through LXRα. There may be pharmaceutical relevance to this observation, because the RXR-LXR heterodimer is activated independently by retinoids and by oxycholesterols (35). It may be possible, for example, to stimulate the elimination of cholesterol with RXRα-specific agonists.
The results of this study also indicate that the primary site of LXRα action in the process of cholesterol metabolism is in hepatocytes. Bile acid homeostasis is a highly dynamic process involving synthesis in hepatocytes, storage and secretion in the gall bladder, and reabsorption in the intestine. In principle, cholesterol homeostasis might primarily require LXRα action in the gall bladder or intestine (where LXRα is abundantly expressed), with hepatic events being a secondary or minor aspect of cholesterol physiology. The cholesterol phenotype seen in LXRα mutant mice does not address this issue, because the LXRα gene is mutated in all tissues of the animal. However, by specifically disrupting RXRα, the LXRα partner receptor, in hepatocytes, the critical role of LXRα in the liver is established.
Many of the class II nuclear receptor genes have been mutated in the mouse germ line, providing the opportunity to compare the hepatic phenotypes of these knockouts with the RXRα liver-specific mutant phenotype, as we have done here for LXRα. We have found that certain components of the fibrate-inducible PPARα pathway are also compromised in the absence of RXRα (Y. Wan et al., unpublished observations), consistent with the alterations we observed in the expression profile of PPARα target genes in mice maintained on a standard diet and without physiological challenge. Perhaps more valuable will be the opportunity to address with our mice the hepatic role of class II receptors for which germ line mutations have not yet been established, including the FXR, CARβ, and PXR receptors and their cognate ligands.
The central role of RXR as a heterodimeric partner for at least 10 different vertebrate class II nuclear receptor gene subfamilies (1) regulating very diverse developmental and physiological processes has been somewhat enigmatic. This relationship is evolutionarily broad, in that the Drosophila ultraspiracle locus encodes an RXR homolog which is required as the heterodimeric partner of the ecdysone receptor (36). One of the most intriguing aspects of our study is the realization that multiple metabolic pathways in the mouse liver are coordinately dependent upon RXRα. Fatty acid and cholesterol metabolism in particular are ancient processes required in all metazoan organisms that must regulate lipid absorption, storage, and utilization. The heterodimeric dependence of class II nuclear receptors on RXR may have initially evolved specifically to coordinately regulate these fundamental metabolic pathways. Once in place, the class II subfamily may have expanded and evolved to accommodate new functions and ligands (such as retinoic acid, which is unique to vertebrates, and ecdysteroids, which are found in insects), yet preserved the requirement for RXR heterodimerization.
ACKNOWLEDGMENTS
We thank M. Stallcup for helpful comments during the preparation of the manuscript.
This work was supported in its early phase by a pilot/feasibility project grant (to H.M.S.) as a component of the USC Research Center for Liver Diseases (Public Health Service grant DK48522) and by Public Health Service grant CA53596 (to Y.W.). D.J.M. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Blumberg B, Evans R M. Orphan nuclear receptors—new ligands and new possibilities. Genes Dev. 1998;12:3149–3155. doi: 10.1101/gad.12.20.3149. [DOI] [PubMed] [Google Scholar]
- 2.Blumberg B, Sabbagh W, Juguilon H, Bolado J, van Meter C M, Ong E S, Evans R M. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 1998;12:3195–3205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang T K, Weber G F, Crespi C L, Waxman D J. Differential activation of cyclophosphamide and ifosphamide by cytochromes P-450 2B and 3A in human liver microsomes. Cancer Res. 1993;53:5629–5637. [PubMed] [Google Scholar]
- 4.Chen J, Kubalak S W, Chien K R. Ventricular muscle-restricted targeting of the RXRalpha gene reveals a non-cell-autonomous requirement in cardiac chamber morphogenesis. Development. 1998;125:1943–1949. doi: 10.1242/dev.125.10.1943. [DOI] [PubMed] [Google Scholar]
- 5.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 6.Chiba H, Clifford J, Metzger D, Chambon P. Distinct retinoid X receptor-retinoic acid receptor heterodimers are differentially involved in the control of expression of retinoid target genes in F9 embryonal carcinoma cells. Mol Cell Biol. 1997;17:3013–3020. doi: 10.1128/mcb.17.6.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans R M. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forman B M, Tzameli I, Choi H S, Chen J, Simha D, Seol W, Evans R M, Moore D D. Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature. 1998;395:612–615. doi: 10.1038/26996. [DOI] [PubMed] [Google Scholar]
- 9.Honkakoski P, Zelko I, Sueyoshi T, Negishi M. The nuclear orphan receptor CAR-retinoid X receptor heterodimer activates the phenobarbital-responsive enhancer module of the CYP2B gene. Mol Cell Biol. 1998;18:5652–5658. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kastner P, Grondona J M, Mark M, Gansmuller A, LeMeur M, Decimo D, Vonesch J L, Dolle P, Chambon P. Genetic analysis of RXR alpha developmental function: convergence of RXR and RAR signaling pathways in heart and eye morphogenesis. Cell. 1994;78:987–1003. doi: 10.1016/0092-8674(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 11.Kliewer S A, Moore J T, Wade L, Staudinger J L, Watson M A, Jones S A, McKee D D, Oliver B B, Willson T M, Zetterstrom R H, Perlmann T, Lehmann J M. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 12.Kliewer S A, Umesono K, Noonan D J, Heyman R A, Evans R M. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koenig R J. Thyroid hormone receptor coactivators and corepressors. Thyroid. 1998;8:703–713. doi: 10.1089/thy.1998.8.703. [DOI] [PubMed] [Google Scholar]
- 14.Krezel W, Dupe V, Mark M, Dierich A, Kastner P, Chambon P. RXR gamma null mice are apparently normal and compound RXR alpha +/−/RXR beta −/−/RXR gamma −/− mutant mice are viable. Proc Natl Acad Sci USA. 1996;93:9010–9014. doi: 10.1073/pnas.93.17.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavrentiadou S N, Hadzopoulou C M, Kardassis D, Zannis V I. Binding specificity and modulation of the human ApoCIII promoter activity by heterodimers of ligand-dependent nuclear receptors. Biochemistry. 1999;38:964–975. doi: 10.1021/bi981068i. [DOI] [PubMed] [Google Scholar]
- 16.Le N-A, Gibson J C, Ginsberg H N. Independent regulation of plasma apolipoprotein C-II and C-III concentrations in very low density and high density lipoproteins: implications for the regulation of the catabolism of these lipoproteins. J Lipid Res. 1988;29:669–677. [PubMed] [Google Scholar]
- 17.Makishima M, Okamoto A Y, Repa J J, Tu H, Learned R M, Luk A, Hull M V, Lustig K D, Mangelsdorf D J, Shan B. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 18.Mangelsdorf D J, Borgmeyer U, Heyman R A, Zhou J Y, Ong E S, Oro A E, Kakizuka A, Evans R M. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev. 1992;6:329–344. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- 19.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 20.Mendelsohn C, Lohnes D, Decimo D, Lufkin T, LeMur M, Chambon P, Mark M. Function of the retinoic acid receptors (RARs) during development. II. Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development. 1994;120:2749–2771. doi: 10.1242/dev.120.10.2749. [DOI] [PubMed] [Google Scholar]
- 21.Parks D J, Blanchard S G, Bledsoe R K, Chandra G, Consler T G, Kliewer S A, Stimmel J B, Willson T M, Zavacki A M, Moore D D, Lehmann J M. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 22.Peet D J, Turley S D, Ma W, Janowski B A, Lobaccaro J M, Hammer R E, Mangelsdorf D J. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 23.Peters J M, Hennuyer N, Staels B, Fruchart J C, Fievet C, Gonzalez F J, Auwerx J. Alterations in lipoprotein metabolism in peroxisome proliferator-activated receptor alpha-deficient mice. J Biol Chem. 1997;272:27307–27312. doi: 10.1074/jbc.272.43.27307. [DOI] [PubMed] [Google Scholar]
- 24.Pinkert C A, Ornitz D M, Brinster R L, Palmiter R D. An albumin enhancer located 10 kb upstream functions along with its promoter to direct efficient, liver-specific expression in transgenic mice. Genes Dev. 1987;1:268–276. doi: 10.1101/gad.1.3.268. [DOI] [PubMed] [Google Scholar]
- 25.Postic C, Shiota M, Niswender K D, Jetton T L, Chen Y, Moates J M, Shelton K D, Lindner J, Cherrington A D, Magnuson M A. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 26.Russell D W. Nuclear orphan receptors control cholesterol catabolism. Cell. 1999;97:539–542. doi: 10.1016/s0092-8674(00)80763-1. [DOI] [PubMed] [Google Scholar]
- 27.Russell D W, Setchell K D R. Bile acid biosynthesis. Biochemistry. 1992;31:4737–4749. doi: 10.1021/bi00135a001. [DOI] [PubMed] [Google Scholar]
- 28.Schoonjans K, Staels B, Auwerx J. Role of the peroxisome proliferator-activated receptor (PPAR) in mediating the effects of fibrates and fatty acids on gene expression. J Lipid Res. 1996;37:907–925. [PubMed] [Google Scholar]
- 29.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 30.Sucov H M, Dyson E, Gumeringer C L, Price J, Chien K R, Evans R M. RXR alpha mutant mice establish a genetic basis for vitamin A signaling in heart morphogenesis. Genes Dev. 1994;8:1007–1018. doi: 10.1101/gad.8.9.1007. [DOI] [PubMed] [Google Scholar]
- 31.Sucov H M, Murakami K K, Evans R M. Characterization of an autoregulated response element in the mouse retinoic acid receptor type beta gene. Proc Natl Acad Sci USA. 1990;87:5392–5396. doi: 10.1073/pnas.87.14.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The repressed nuclear receptor CAR responds to phenobarbital in activating the human CYP2B6 gene. J Biol Chem. 1999;274:6043–6046. doi: 10.1074/jbc.274.10.6043. [DOI] [PubMed] [Google Scholar]
- 33.Umesono K, Giguere V, Glass C K, Rosenfeld M G, Evans R M. Retinoic acid and thyroid hormone induce gene expression through a common responsive element. Nature. 1988;336:262–265. doi: 10.1038/336262a0. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, Chen J, Hollister K, Sowers L C, Forman B M. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 35.Willy P J, Umesono K, Ong E S, Evans R M, Heyman R A, Mangelsdorf D J. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- 36.Yao T P, Segraves W A, Oro A E, McKeown M, Evans R M. Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell. 1992;71:63–72. doi: 10.1016/0092-8674(92)90266-f. [DOI] [PubMed] [Google Scholar]