Abstract
Skin injury is repaired through a multi-phase wound healing process of tissue granulation and re-epithelialization. Any failure in the healing process may lead to chronic non-healing wounds or abnormal scar formation. Although significant progress has been made in developing novel scaffolds and/or cell-based therapeutic strategies to promote wound healing, effective management of large chronic skin wounds remains a clinical challenge. Keratinocytes are critical to re-epithelialization and wound healing. Here, we investigated whether exogenous keratinocytes, in combination with a citrate-based scaffold, enhanced skin wound healing. We first established reversibly immortalized mouse keratinocytes (iKera), and confirmed that the iKera cells expressed keratinocyte markers, and were responsive to UVB treatment, and were non-tumorigenic. In a proof-of-principle experiment, we demonstrated that iKera cells embedded in citrate-based scaffold PPCN provided more effective re-epithelialization and cutaneous wound healing than that of either PPCN or iKera cells alone, in a mouse skin wound model. Thus, these results demonstrate that iKera cells may serve as a valuable skin epithelial source when, combining with appropriate biocompatible scaffolds, to investigate cutaneous wound healing and skin regeneration.
Keywords: Keratinocytes, Skin tissue engineering, Reversible immortalization, SV40 large T antigen, PPCN, Skin wound healing
Graphical abstract
Highlights
-
•
A reversibly immortalized mouse keratinocyte (i.e., iKera) line was established.
-
•
The iKera cells maintain the characteristics of keratinocytes without tumorigenecity.
-
•
A combination of thermoresponsive scaffold PPCN and iKera cells facilitates re-epithelization and cutaneous wound healing.
-
•
The iKera cells are a valuable cell source for skin tissue engineering studies.
1. Introduction
Skin is the largest organ of the body, accounting for about 15 % of the human body by mass. The primary functions of skin are to serve as a protective barrier against biological and chemical agents, to moderate temperature, and to retain body fluids [[1], [2], [3]]. To fulfill these functions, skin is composed of several cell types, such as keratinocytes, fibroblasts, and melanocytes, which are organized into three layers: epidermis, dermis, and hypodermis [1]. Epidermis is the outermost layer of skin with 15–20 layers of keratinocytes [2,3]. Epidermis plays a critical role in forming an intact barrier to protect the body from dehydration and external insults including pathogenic bacteria, viruses, allergens, and UVB irradiation, and is pathologically associated with skin injury, skin sensitivity, psoriasis, scleroderma and pigmentation, and other skin disorders [[2], [3], [4]].
Skin wound healing is a dynamic and complex multi-phase process that requires coordinated interactions between growth factors, cytokines, chemokines and various cells, leading to and the activation of keratinocytes through the intact basement membrane to act on dermal fibroblasts and melanocytes together [1,2,5]. Epithelialization is an important part of wound healing, and is the decisive parameter for successful wound closure [6], although any failure in the multi-phase process may cause the wounds to become chronic ones with abnormal scar formation, which may significantly impact a patient's quality of life due to repetitive treatments and considerable medical costs [1]. Thus, effective management of large chronic non-healing wounds remains a major clinical challenge in reconstruction and rehabilitation medicine.
Nonetheless, significant progress has been made in developing novel therapeutic approaches for wound treatment, as well as advanced and regenerative dressings to promote wound closure within a clinically relevant timeframe, through the use of novel scaffolds and/or cell-based therapeutic strategies [1]. Numerous scaffold materials, especially hydrogels and extracellular matrix (ECM), have been investigated for skin wound healing and/or skin regeneration [[7], [8], [9]]. It has been recently reported that extracellular citrate can elevate cell energy and metabologenic status, which should be beneficial for the survival and proliferation of the cells embedded in the scaffolds [[10], [11], [12]]. One interesting citrate-based biodegradable polymer gel is poly (polyethyleneglycol citrate-co-N-isopropylacrylamide) (PPCN), which is thermoresponsive and exerts intrinsic antioxidant and anti-inflammation activities [13].
Significant efforts have been devoted to the development of stem cell-based therapeutics, such as the possible use of embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), and various sources of mesenchymal stem cells (MSCs), to treat wounds, although the use of these progenitor cells may be limited due to the concerns about their availability and/or tumorigenicity [1,8,9]. Thus, more practical sources of skin cells need to be explored for effective wound treatment. Here we focused on keratinocytes because they make up the first barrier of the skin and play a critical role in the re-epithelialization process [2]. However, adult keratinocytes are difficult to isolate and maintain for long-term culture [14].
To overcome the limited life span of culturing primary keratinocytes, we established the reversibly immortalized mouse keratinocytes (iKera) cells by stably expressing SV40 large T antigen (SV40 T), which can be reversed by the FLP recombinase or silencing SV40 T. The iKera cells expressed keratinocyte markers and were responsive to UVB-induced inhibition of cell proliferation. While retaining long-term proliferative activity in vitro, the iKera cells were non-tumorigenic. In a proof-of-concept experiment, we demonstrated that the citrate-based scaffold PPCN embedded with iKera cells provided more effective cutaneous wound healing and re-epithelization than that of either PPCN or iKera cells alone in a mouse skin wound model. Therefore, our results demonstrate that the iKera cells may serve as a valuable epithelial cell source, in combination with the appropriate biocompatible scaffolds, for cutaneous wound healing and skin tissue engineering.
2. Materials and methods
2.1. Synthesis and characterization of the citrate-based polymer PPCN
The citrate-based polymer poly (polyethylene glycol citrate-co-N-isopropylacrylamide) (PPCN) was synthesized by employing a simple two-step method as previously reported [13]. First, poly (polyethylene glycol citrate) acrylate prepolymer (PPCac) was prepared by the polycondensation of citric acid, poly (ethylene glycol) (PEG) and glycerol 1,3-diglycerolate diacrylate (GDD) with a feed ratio of 5:9:1. After melting, the mixture was stirred at 140 °C for 45 min with nitrogen purge. Subsequently, PPCac was dissolved in 1,4-dioxane and equal amount of N-isopropylacrylamide (NIPAM) was added to the solution. The free radical polymerization of PPCac and NIPAM was conducted at 65 °C for 8 h using azobisisobutyronitrile (AIBN) as an initiator. The chemical and thermoresponsive characteristics were analyzed for all newly synthesized batches as reported [13]. Specifically, the 1H NMR spectrum was used to confirm the chemical structure of PPCN by employing a X500 Bruker Avance III NMR spectrometer, using DMSO‑d6 as solvent and tetramethylsilane (TMS) as reference. Fourier transform infrared spectra (FT-IR) were recorded by using Nicolet iS50 spectrometer with attenuated total reflection (ATR). The wavenumber ranged from 400 to 4000 cm−1 with the resolution of 4 cm−1 and scans of 32. The rheological properties of PPCN were studied by TA Discovery HR 20 hybrid rheometer by using temperature ramp from 20 to 40 °C with heating rate at 10 °C/min. Studies were conducted at an angular frequency of 10 rad/s and 2 % strain. PPCN has a lower critical solution temperature (LCST) of 29.7 °C and starts to solidify at above 30 °C [13,15,16].
Prior to use, PPCN powder was dissolved in Milli-Q water at 100 mg/mL. The pH of the solution was adjusted to 7.4 with 1 M sodium hydroxide solution. The PPCN solution was dialyzed for 2 days against deionized (DI) water at 4 °C with a molecular weight cut-off (MWCO) of 2 kDa and then lyophilized for 2 days. Neutralized PPCN powder was dissolved in phosphate-buffered saline (PBS) (stock solution at 100 mg/mL), sterilized by syringe filtration, and kept at 4 °C. In order to enhance cell adhesion ability of the PPCN, cells were resuspended in sterile PBS containing 0.1 % gelatin prior to mixing with PPCN stock solution (usually at 1:1 vol/vol ratio) as previously described [[15], [16], [17], [18], [19]].
2.2. Cell culture and chemicals
HEK-293 cells were obtained from ATCC (American Type Culture Collection, Manassas, VA, USA). HEK-293 derivatives 293pTP and RAPA lines were previously described [20,21]. The iMCs are SV40 T-immortalized mouse melanocytes as previously characterized [22]. All cells were maintained in high glucose complete Dulbecco's modified Eagle's medium (DMEM) supplemented with 10 % fetal bovine serum (Lonsa Science SRL), 100 units of penicillin and 100 μg of streptomycin at 37 °C in 5 % CO2 as described [[23], [24], [25]]. Unless indicated otherwise, all chemicals were purchased from Sigma-Aldrich (St Louis, MO, USA), Thermo Fisher Scientific (Waltham, MA, USA), or Solarbio (Beijing, China).
2.3. Isolation of mouse primary keratinocytes (kera) and dermal fibroblasts (Fib)
The use and care of experimental animals was approved by the Research Ethics and Regulations Committee of Chongqing Medical University, Chongqing, China. The experiment animals were obtained from and housed in the Experimental Animal Research Center of Chongqing Medical University. The mouse primary keratinocytes (Kera) and dermal fibroblasts (Fib) were isolated from the skin tissues of newborn CD1 mice as described [14]. Briefly, newborn CD1 mice were euthanized, washed with sterile PBS, and sterilized with 70 % ethanol. The back skin was cut along the dorsal midline, loosened, and pulled to the ventral side.
For the primary keratinocyte isolation, the flattened skin was kept dermis-side-down to float in cell culture dishes containing 0.25 % trypsin-EDTA overnight to separate the dermis layer and epidermis layer. The epidermis layer was collected for primary keratinocyte (Kera) isolation. The attached cells were passaged once reaching subconfluence (designated as P0, 1, 2, and 3 to reflect the number of passages) for establishing immortalized stable line (see below) or in vitro assays.
For the recovery of primary dermal fibroblasts, the full thickness skin was minced into small pieces, directly seeded onto cell culture dishes, and cultured at 37 °C in 5 % CO2 in high glucose complete DMEM supplied with 10 % FBS. The medium was changed every 24 h to remove non-adherent cells. The adherent cells were passaged once reaching subconfluence (designated as P0, 1, 2, and 3 to reflect the number of passages) for in vitro assays.
2.4. Establishment of the reversibly immortalized keratinocyte cell line (iKera)
The isolated mouse primary keratinocytes (less than 3 passages) were used to generate the immortalized keratinocytes (iKera). Briefly, the retroviral vector SSR#41 that expresses SV40 large T antigen (SV40 T) flanked with FRT sites was co-transfected with pCLAmpho packaging vector into HEK-293 cells to produce packaged retrovirus as described [[26], [27], [28], [29], [30], [31], [32], [33], [34], [35]]. Subconfluent mouse primary keratinocytes were infected with the packaged SSR#41 retrovirus. The infected keratinocytes were selected with hygromycin B (0.3 mg/mL, Invitrogen) for 7–10 days, and replated every 3 days. The resultant immortalized keratinocyte cell line was designated as iKera, which has been passaged for more than 30 generations.
2.5. RNA isolation and touchdown-quantitative real-time PCR (TqPCR)
Total RNA was isolated by using the TRIZOL Reagent (Invitrogen, China), and subjected to reverse transcription using hexamer and M-MuLV reverse transcriptase (New England Biolabs, Ipswich, MA). The cDNA products were used as PCR templates. Gene-specific PCR primers were designed by using Primer3 program (Table S1). TqPCR was carried out by using 2x SYBR Green qPCR Master Mix (Bimake, Shanghai, China) on the CFX-Connect unit (Bio-Rad Laboratories, Hercules, CA) as described [[36], [37], [38], [39]]. All TqPCR reactions were done in triplicate. Gapdh was used as a reference gene. Quantification of gene expression was carried out by using the 2−ΔΔCq method as described [38,[40], [41], [42]].
2.6. WST-1 cell proliferation assay
Exponentially growing cells were seeded in 96-well plates (2000 cells/well). At the indicated time points, the Premixed WST-1 Reagent (Clontech, Mountain View, CA) was added, followed by incubating at 37 °C for 120 min and reading absorbance at 450 nm using a microplate reader (Biotek, EON, USA) as described [[43], [44], [45], [46]].
2.7. Crystal violet cell viability assay
Exponentially growing cells were seeded in 24-well plates (6000 cells/well). At the indicated time points, the cells were gently washed with PBS and stained with 0.5 % crystal violet/formalin solution for 10 min. The stained cells were washed with tape water and air-dried for scanning. For quantitative analysis, the stained cells were dissolved in 10 % acetic acid and measured for absorbance at 592 nm as described [[47], [48], [49], [50], [51]].
2.8. Differentiation of iKera cells induced by all-trans retinoic acid (ATRA)
Exponentially growing iKera cells were seeded in 60 mm dishes (1 × 106 cells) and treated with 1 μM ATRA or DMSO. Total RNA was isolated and TqPCR was used to test the expression of genes related to keratinocyte differentiation at days 3 and 5.
2.9. Tumorigenicity analysis of the iKera cells in athymic nude mice
The use and care of the athymic nude mice were approved by the Research Ethics and Regulations Committee of Chongqing Medical University, Chongqing, China. The animals were obtained from and housed in the Experimental Animal Research Center of Chongqing Medical University. The iKera-FLuc cells were stably transduced with the firefly luciferase-expressing retroviral vector pSEB-FLuc as described [31,52,53]. Exponentially growing iKera-FLuc cells were resuspended and injected subcutaneously into the flanks of athymic nude mice (6-week old, male, 2 × 106 cells per injection, and 4 sites per mouse). Potential subcutaneous mass growth was assessed at 3, 7 and 14 days after implantation by whole body bioluminescence imaging using the IVIS Lumina Series III In Vivo Imaging System (PerkinElmer, Waltham, MA). The acquired data were quantitatively analyzed by the Living Image Software (PerkinElmer).
2.10. Construction and generation of recombinant adenoviruses
All recombinant adenoviruses used in this study were generated by using the AdEasy technology [[54], [55], [56]]. The adenoviral vector Ad-FLP was constructed and characterized in our previous studies [49,50,57,58]. To construct the adenoviral vector expressing siRNAs targeting SV40 T, we employed our previously developed pSOS-related FAMSi system, in which siRNA expression is driven by the converging human U6 and H1 promoters as described [17,19,59,60]. Three siRNA sites targeting SV40 T were designed by using Invitrogen's BLOCK-iT RNAi Designer and/or Dharmacon Horizon Discovery's siDESIGN programs (Table S2). The oligo cassettes for the three siRNAs were subcloned into a single adenoviral shuttle vector to generate recombinant adenoviral vector and subsequently the adenovirus Adsi-LTA in 293pTP or RAPA cells. Adsi-LTA also co-expresses the RFP marker gene. Adenoviral vector expresses RFP (Ad-RFP) or GFP (Ad-GFP) alone was used as a mock adenovirus control [40,61]. For all adenoviral infections, polybrene (8 μg/mL) was added to enhance infection efficiency as reported [62,63].
2.11. Transmission electron microscope (TEM) analysis
Approximately 2 × 106 cells were collected by centrifugation at 1,200 rpm for 10 min. The cell pellets were fixed with 2.5 % glutaraldehyde and stored at 4 °C. The cell pellets were washed with 0.1 M sodium cacodylate and further fixed with 4 % osmium tetroxide for 1 h, followed by serial dehydration in ascending concentrations of acetone (from 35 % to 100 %). The samples were infiltrated with a mixture of acetone and resin at the ratio of 1:1 for 1 h, 1:2 for 2 h and 100 % resin overnight as reported [64]. Finally, the samples were polymerized at 60 °C for 36 h prior sectioning to prepare the 90 nm thick sections using the ultra-microtome Leica EM UC7 (Leica, Germany). The ultrathin sections were stained with lead citrate/uranyl acetate, and analyzed under a transmission electron microscope (Hitachi 7500, Japan).
2.12. Immunofluorescence (IF) staining
The IF staining was carried out as previously reported [24,25,27,65,66]. Briefly, cells seeded and treated in chamber slides were fixed with 4 % paraformaldehyde for 15 min at RT, treated with 0.5 % Triton X-100 for 20 min, and blocked with 5 % goat serum (1:10 dilution) for 20 min at RT, followed by incubating with primary antibodies against SV40 T (1:50 dilution; Santa Cruz; Cat# sc-147), Krt10 (1:50 dilution; Bimake; Cat# A5266), Krt14 (1:50 dilution; Bimake; Cat# A5434), and Krt15 (1:50 dilution; Bimake; Cat# A5627) overnight. After being washed, the cells were incubated with DyLight 594, goat anti-mouse IgG (1:200 dilution; Abbkine; Cat# A23410) or goat anti-rabbit IgG/APC (1:200 dilution; Bioss; Cat# bs-0295G-APC). The nuclei were counterstained with DAPI (10 μg/mL). Minus primary antibody or control IgG was used as a negative control. IF results were recorded by using a laser confocal microscope (Leica TCS SP8).
2.13. Preparation of conditioned medium (CM) and treatment of immortalized keratinocytes (iKera), primary dermal fibroblasts (Fib) or immortalized melanocytes (iMCs)
Conditioned medium (CM) was prepared as previously described [67,68]. Briefly, 2 × 106 iKera cells were plated in 10 cm2 cell culture dishes. 293 cells were set up in the same fashion as the control conditioned medium. The supernatants were collected from the cultured iKera cells (iKera-CM) or 293 cells (293-CM) at 48 h, and filtered for culturing the primary dermal fibroblasts (Fib) or melanocytes (iMCs). Subconfluent Fib cells and iMCs were treated with iKera-CM or 293-CM for the indicated time points for further analysis. 2 × 106 Fib cells were plated in 10 cm2 cell culture dishes. 293 cells were set up in the same fashion as the control conditioned medium. The supernatants were collected from the cultured Fib cells (Fib-CM) or 293 cells (293-CM) at 48 h, and filtered for culturing the iKera cells. Subconfluent iKera cells were treated with Fib-CM or 293-CM for the indicated time points for further analysis.
2.14. Transwell co-culture of iKera and iMCs and melanin secretion assay
Melanin production was assessed qualitatively and quantitatively. Briefly, 5 × 105 of iMCs per well were seeded in 6-well plates, while 5 × 105 iKera or 293 cells/well were seeded in 0.4 μm transwell inserts for co-culturing experiments. Melanin secretion was analyzed under bright field microscope at indicated time points as described [22]. For quantitative analysis, the iMCs from iKera or 293 co-culture were collected for visual examination and lysed with 1 M NaOH for 5 min, followed by absorbance determination at 470–490 nm as described [69].
2.15. Mouse cutaneous wound healing model and PPCN-iKera based therapy
The experimental mouse skin wound healing model was established as described [70]. The use and care of animals, and the experimental procedures were approved by the Research Ethics and Regulations Committee of Chongqing Medical University. Briefly, 12 male 6-week-old athymic nude mice were divided into four groups. After the animals were anaesthetized, the dorsal skin of each mouse was disinfected with 70 % alcohol; and 1-cm diameter full-thickness cutaneous wound was created at the dorsum part of each mouse. In the Blank (sham) group, the wounds were treated with 100 μL PBS/mouse as a control. In the iKera group, the wounds were treated with 2 × 106 iKera cells in 100 μL PBS/mouse. In the PPCN group, the wounds were treated with 100 μL PPCN/mouse. In the PPCN + iKera group, the wounds were treated with 2 × 106 iKera cells in 100 μL PPCN/mouse. All wounds were dressed with sterile gauze. The mice were given antibiotics orally in drinking water. Wound sizes were measured at days 0, 3, 6 and 9, respectively. The average areas of wound openness in each group at the indicated time points were calculated by using the Image-Pro Plus software (Media Cybernetic, Rockville, USA).
2.16. H & E staining, Masson's trichrome staining and sirius red staining
The skin wound healing samples (i.e., 1-cm diameter from the center of the cutaneous wound) were retrieved from athymic nude mice at the endpoint day 9. The samples were fixed, paraffin-embedded, and sectioned along maximum diameter. Serial sections were deparaffinized, rehydrated, and subjected to H & E staining, Masson's trichrome staining (Masson's Trichrome Stain Kit, G1340, Solarbio, China), and Sirius red staining (Picro Sirius Red solution, G1471, Solarbio, China) as described [71,72].
2.17. Immunohistochemical (IHC) staining
IHC staining protocol was carried out by using the IHC staining (SP Kit, SP-9001, ZSGB-Bio, China) as described [73,74]. Briefly, the sections were deparaffinized and subjected to immunostaining with the primary antibodies against Krt10 (1:50 dilution; Bimake; Cat# A5266), Krt15 (1:50 dilution; Bimake; Cat# A5627), Involucrin (1:50 dilution; Bimake; Cat# A5788), SV40 T (1:50 dilution; Santa Cruz; Cat# sc-147). The Biotin labeled goat anti-mouse IgG (SP Kit, SP-9000, ZSGB-Bio, China) or goat anti-rabbit IgG (SP Kit, SP-9001, ZSGB-Bio, China) were used to visualize the presence of the proteins of interest. Hematoxylin was used to stain the nuclei. Sections incubated without primary antibodies were used negative controls. The results were recorded under a bright field microscope (Leica DM4B/Nikon 80i).
2.18. Statistical analysis
All experiments were performed at least three times and/or repeated three batches of independent experiments. Data were analyzed using GraphPad Prism 7 and presented as the mean ± standard deviations (SD). Statistical significance was determined by one-way analysis of variance and the student's t-test for the comparisons between groups. A value of p < 0.05 was considered statistically significant.
3. Results
3.1. Reversibly immortalized mouse keratinocytes (iKera cells) can be effectively established via the stable expression of SV40 T (SV40 T)
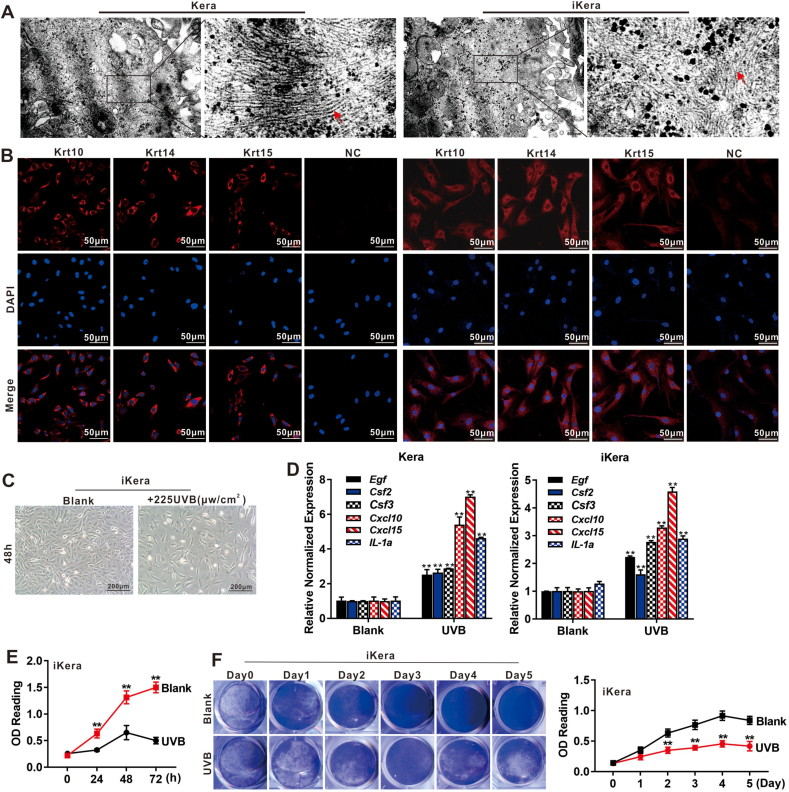
Skin consists of the epidermis, dermis, hair follicle and sebaceous gland; and the epidermis is the outermost layer of skin and consists of 15–20 layers of keratinocytes [2,3]. To establish a constant source of keratinocytes for scaffold-based skin wound healing studies, we sought to establish a reversibly immortalized keratinocyte line. We first isolated the mouse primary keratinocytes (designated as Kera cells) from newborn mice as described in methods (Fig. 1A, a to f). As SV40 large T antigen (SV40 T) is one of the most commonly used immortalization genes [27,50,75], the retroviral vector SSR#41, which expresses a hygromycin B-resistance gene and SV40 T flanked with FRT sites, was used to generate reversibly immortalized keratinocyte line, designated as iKera cells (Fig. 1B). The Kera (0 P0) and iKera cells were morphologically similar in vitro (Fig. 1C). The expression of SV40 T in iKera cells was verified by TqPCR (Fig. S1A) and IF staining (Fig. 1D, and the negative control was shown in Fig. S1B) in comparison with that in Kera cells. Unlike Kera cells, the iKera cells could be passaged and maintained high proliferative activity for at least 30 generations at the time when this work was reported. We further demonstrated that iKera cells exhibited higher proliferation rate and cell viability, compared with Kera cells (P1) in WST-1 assay (Fig. 1E) and crystal violet cell viability assay (Fig. 1F). These results indicate that the SV40 T-mediated immortalization strategy was effective to establish the iKera cells as a long-term keratinocyte line.
Fig. 1.
Establishment of the SV40 T-mediated reversibly immortalized mouse keratinocytes or iKera cells. (A) Steps of harvesting primary mouse keratinocytes: (a) removal of lower forelimbs, hind limbs and tail close to the body; (b) the freshly stripped skin tissues; (c) overnight digestion of the skin tissues with trypsin-EDTA; (d) the peeled keratin cambium tissues; (e) the cultured keratin cambium tissues after being minced into small pieces; and (f) the recovered and proliferating primary keratinocytes (Kera). (B) The schematic representation of SV40 T-mediated retroviral vector SSR#41. The SV40 T gene is flanked with the FRT sites, allowing the removal of SV40 T upon the expression of FLP recombinase. (C) The morphologic comparison of Kera (P0) and iKera cells under phase contrast microscope. (D) Expression of SV40 T. Immunofluorescence (IF) staining was used to assess the expression and localization of SV40 T in Kera (P0) and iKera cells. The nuclei were stained with DAPI. Representative results are shown. (E) Cell proliferation assay. WST-1 was used to assess the cell proliferation of Kera (P1) and iKera cells at 0, 24, 48, 72 and 96 h, respectively. “**” p < 0.01, “*” p < 0.05, Kera cells vs iKera cells. (F) Cell viability assay. Crystal violet staining assay and quantitative analysis were used to assess the viability and proliferation of Kera (P1) and iKera cells at days 0, 1, 2, 3, 4, 5 and 6. “**” p < 0.01, Kera cells vs iKera cells. (G) Subconfluent iKera cells were infected with Ad-FLP, Ad-GFP, Adsi-LTA and Ad-RFP, respectively. The fluorescence signals were recorded at 36 h after infection. (H) Removal and knockdown of SV40 T expression in iKera cells. The iKera cells were infected with Ad-FLP, Ad-GFP, Adsi-LTA or Ad-RFP for 48 h. Total RNA was isolated for TqPCR analysis of the expression of SV40 T in iKera cells. “*” p < 0.05, Ad-FLP group vs Ad-GFP group, “**” p < 0.01, Adsi-LTA group vs Ad-RFP group. (I) Effect of SV40 T removal and silencing on cell proliferation of iKera cells. Subconfluent iKera cells were infected with Ad-FLP, Ad-GFP, Adsi-LTA, or Ad-RFP. WST-1 was used to assess the cells proliferation at 0 h, 24 h, 48 h, 72 h and 96 h. “**” p < 0.01, Ad-FLP group vs Ad-GFP group, Adsi-LTA treated group vs Ad-RFP treated group. (J) Cell viability upon the removal or silencing of SV40 T in iKera cells. Subconfluent iKera cells were infected with Ad-FLP, Ad-GFP, Adsi-LTA or Ad-RFP. Crystal violet cell viability assay and quantitative analysis were used to assess cell viability and proliferation of the iKera cells. “**” p < 0.01, “*” p < 0.05, Ad-FLP group vs Ad-GFP group, or Adsi-LTA group vs Ad-RFP group. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
As shown in Fig. S1C, keratinocytes can be divided into five layers from inside to outside: basal layer, spinous layer, granular layer, hyaline layer and stratum corneum layer [2]. To test the mature and differentiation potential of the iKera cells, we treated the iKera cells with 1 μM ATRA for 3 and 5 days respectively, and found that the characteristic genes related to basal layer (Krt5, Krt14 and Krt15), spinous layer (Krt1 Krt10 and IVL), granular layer (IVL and LOR) and stratum corneum layer (LOR) were highly expressed, compared with that of the control group by qPCR (Fig. S1D), suggesting that the iKera cells may retain the mature and differentiation potential while maintaining long-term proliferative activity.
To test whether the immortalization phenotype of the iKera cells was reversible, we employed two strategies: 1) using a recombinant adenovirus overexpressing FLP recombinase (namely Ad-FLP) and 2) using a recombinant adenovirus expressing siRNAs to silence SV40 T (namely Adsi-LTA). The iKera cells could be effectively transduced by these adenoviruses, along with control Ad-GFP or Ad-RFP virus (Fig. 1G). The TqPCR analysis indicates that SV40 T expression was significantly reduced in the iKera cells transduced with Ad-FLP or Adsi-LTA, compared with that treated with the control viruses (Fig. 1H). The removal of SV40 T by Ad-FLP or silencing SV40 T by Adsi-LTA was shown to significantly decrease the proliferation rate of the iKera cells after 48 h as assessed by WST-1 assay (Fig. 1I). Both qualitative and quantitative crystal violet cell viability assays further confirmed that the removal of SV40 T antigen or silencing SV40 T expression in the iKera cells led to a significant decrease in cell viability and proliferation (Fig. 1J). Even though SV40 T was likely partially removed or silenced in the above assays, these results strongly suggest that the SV40 T-mediated immortalization phenotype of the iKera cells may be reversible.
3.2. The iKera cells retain the ultrastructure of keratin filaments, express characteristic markers of keratinocytes and are responsive to UVB irradiation-induced inhibition of cell proliferation
We first compared the ultrastructure features between primary Kera and the iKera cells. As profilaggrin is synthesized at the granular layer and stored in keratohyalin granules, and proteolyzed and broken down into many filaggrin monomers when granular cells differentiate to cornified cells, leading to the disappearance of the nucleus and cell organelles, but retention of keratin filaments, a prominent characteristic of keratinocytes [76]. TEM ultrastructure analysis revealed that abundant keratin filaments were presented in both Kera and iKera cells (Fig. 2A).
Fig. 2.
The ultrastructural and functional characteristics of iKera cells. (A) The ultrastructure of the iKera and primary Kera cells. TEM was used to assess the ultrastructure of the Kera and iKera cells. The keratin filaments are indicated by red arrows. Representative results are shown. (B) Expression of keratinocyte markers in the iKera cells. IF was used to assess the expression and localization of Krt10, Krt14 and Krt15 in Kera and iKera cells. The nuclei were counter-stained with DAPI. Representative results are shown. NC, negative control (minus primary antibody or control IgG). (C) The iKera cells are sensitive to UVB. The iKera cells were treated with 225 μw/cm2 UVB for 48 h and observed under light microscope at 48 h. Representative results are shown. (D) UVB-induced gene expression in the Kera and iKera cells. Subconfluent Kera (P1) and iKera cells were treated with 225 μw/cm2 UVB for 48 h. Total RNA was isolated for TqPCR analysis to assess the expression of the irradiation responsive genes. “**” p < 0.01, UVB treated group vs Blank group. (E) Effect of UVB radiation on cell proliferation of the iKera cells. Subconfluent iKera cells were seeded in 96-well plates and treated with 225 μw/cm2 UVB. WST-1 was used to assess the cells proliferation at 0 h, 24 h, 48 h and 72 h. “**” p < 0.01, UVB treated group vs Blank group. (F) Effect of UVB radiation on cell viability of the iKera cells. Subconfluent iKera cells were seeded in 24-well plates and treated with 225 μw/cm2 UVB. Crystal violet cell viability assay and quantitative analysis were used to assess cell viability and proliferation of the iKera cells at days 0, 1, 2, 3, 4, and 5. “**” p < 0.01, UVB treated group vs Blank group. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We next conducted qPCR analysis and determined the expression of the well-established keratinocyte markers [77,78]. We found that the expression of most of these marker genes, especially Tnf-α, Tgf-α, Krt1, Krt5, Krt14 and Krt15, was readily detected in both Kera and iKera cells (Fig. S1E). Immunofluorescence staining assay further confirmed the expression of Krt10, Krt14 and Krt15 in the Kera and iKera cells (Fig. 2B). Collectively, these results indicate the iKera cells possess structural and marker expression characteristics of keratinocytes.
We analyzed the effect of UVB irradiation on iKera cell proliferation. When subconfluent iKera cells were treated with different doses of UVB irradiation, we found that cell proliferation was inhibited by UVB in a dose-dependent and time-dependent fashion (Fig. S1F).We found that treatment with 225 μw/cm2 UVB began to suppress the proliferation of iKera cells at 48 h (Fig. 2C). It has been reported that in response to DAMPs (damage-associated molecular patterns) released by host cells during UVB irradiation or wounding, keratinocytes produce a variety of pro-inflammatory cytokines or chemokines [4,77]. We found that the expression of most pro-inflammatory cytokine or chemokine genes related to radiation, such as Egf, Csf2, Csf3, Cxcl10, Cxcl15 and IL-1a, was significantly induced upon 225 μw/cm2 UVB treatment in both Kera and iKera cells at 48 h (Fig. 2D). Meanwhile, the WST-1 assay confirmed that UVB treatment significantly inhibited the cell proliferation and viability of the iKera cells from 24 h to 72 h (Fig. 2E), and the crystal violet staining confirmed that UVB treatment significantly inhibited the cell viability of the iKera cells from Day 2–5 (Fig. 2F) [4,77]. Collectively, these results demonstrated that the iKera cells, were highly similar to primary keratinocytes at ultrastructure level and in marker expression, and were sensitive and responsive to UVB irradiation treatment.
3.3. The iKera cells promote melanogenesis and the proliferation of melanocytes and dermal fibroblasts
As it has been reported that keratinocytes can stimulate the proliferation and melanogenesis of melanocytes [79], we found that, compared with that of the control conditioned medium, the conditioned medium prepared from iKera cells significantly enhanced cell proliferation and viability of the melanocyte iMCs as assessed by WST-1 assay (Fig. 3A), and crystal violet assay (Fig. 3B). Co-culture of the iMCs and iKera cells was shown to promote the secretion and production of melanin of iMCs cells, compared with the control cells, both qualitatively (Fig. 3C) and quantitatively (Fig. 3D). Furthermore, co-culture with the iKera cells significantly impacted the expression of the genes related to synthesis and secretion of melanin, such as Mmp1, 3, 14, 16 and 20 (Fig. 3E).
Fig. 3.
The effect of iKera cells on melanocytes and dermal fibroblasts. (A) The effect of the iKera cells on melanocyte proliferation. Subconfluent iMCs were plated in 96-well plates and cultured with the conditioned medium (CM) of iKera cells. WST-1 assay was used to assess iMCs proliferation 0 h, 24 h, 48 h and 72 h. “**” p < 0.01, the iKera cells CM cultured group vs the control medium group. (B) The effect of the iKera cells on melanocyte viability and proliferation. Subconfluent iMCs were seeded in 24-well plates and cultured with the CM of iKera cells. Crystal violet cell viability assay and quantitative analysis were carried out to assess the cell viability and proliferation of iMCs at days 0, 1, 2, 3, 4, and 5. “**” p < 0.01, the iKera cell CM cultured group vs the control medium group. (C) & (D) The effect of the iKera cells on melanocyte maturation and differentiation. Subconfluent iMCs were co-seeded with iKera cells in 6-well plates. The melanin production was monitored under phase contrast microscope at 0, 24, 48, and 72 h (C). Alternatively, the cells were collected at 72 h and lysed for the quantitative analysis of melanin (D). Representative results are shown. “**” p < 0.01, iKera co-cultured group vs control group. (E) The effect of the iKera cells on the expression of melanin secretion-associated genes in melanocytes. Subconfluent iMCs cells were seeded in 6-well plates and co-cultured with the iKera cells. Total RNA was isolated at 36 h and 72 h for TqPCR analysis of the genes related to melanin secretion. “**” p < 0.01, “*” p < 0.05, iKera co-cultured group vs the iMCs only group. (F & G) Subconfluent iKera or fibroblast (Fib) cells were seeded in 96-well plates (F) or 24-well plates (G), and cultured with the CM of Fib cells or iKera cells. WST-1 assay was used to assess cells proliferation of iKera at 0, 24, 48 and 72 h after cultured with Fib CM, “**” p < 0.01, Fib CM group vs the control medium group (F panel a), WST-1 assay was used to assess cells proliferation of Fib cells at 0, 24, 48 and 72 h after cultured with iKera CM, “**” p < 0.01, iKera CM group vs the control medium group (F panel b). Alternatively, the crystal violet cell viability assay and quantitative analysis were used to assess cells proliferation of iKera cells (a) or Fib cells (b) at days 0, 1, 2, 3, 4, and 5. “**” p < 0.01, “*” p < 0.05, Fib or iKera CM group vs the control medium group (G panel a and b). (H) The effect of iKera cells on the expression of dermal fibroblast markers. Subconfluent Fib cells were cultured with iKera CM for 36 h and 72 h. Total RNA was isolated for TqPCR analysis to assess the expression of fibroblast marker genes. “**” p < 0.01, the iKera CM group vs the control medium group. (I) The iKera cells are non-tumorigenic. Subconfluent iKera cells stably tagged with FLuc (iKera-FLuc) were subcutaneously injected into the flanks of athymic nude mice as described in the Methods. The animals were subjected to Xenogen IVIS 200 whole body bioluminescence imaging at days 3, 7 and 14. Representative results are shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
During cutaneous wound healing process, the activated keratinocytes produce numerous signaling molecules such as IFN-γ, TGF-β, while dermal fibroblasts (Fib) synthesize collagen, and fibronectin, and act on both dermal melanocytes and dermal fibroblasts to repair wounds [2,5], allowing the migration of dermal fibroblasts along the fibrin network and the wound edges and the initiation of re-epithelialization from the wound edges [80]. We found that conditioned medium prepared from iKera cells promoted the cell proliferation of primary dermal fibroblasts (Fib), or vice versa, as assessed by WST-1 assay (Fig. 3F), or crystal violet cell viability assay (Fig. 3G). Since it has been reported that keratinocytes can promote dermal fibroblasts to synthesize and secrete growth factors and cytokines [81], we found that iKera conditioned medium impacted the expression of growth factor and cytokine genes, such as Egfr, IL1r1, Mmp1, P38, Kgf, and Csf2 in the dermal fibroblasts (Fib) as assessed by TqPCR analysis (Fig. 3H). Collectively, these results demonstrate that the iKera cells possess the similar biological characteristics of primary keratinocytes.
As there is a likelihood that immortalized cells may acquire tumorigenic potential [82], we sought to test if the iKera cells had a tendency to form tumors in vivo. The iKera cells were first stably labeled with firefly luciferase (namely iKera-FLuc) with retroviral vector, and injected subcutaneously into the flanks of athymic nude mice. Through whole body bioluminescence imaging using Xenogen IVIS 200, the bioluminescence signal was readily detected at day 3 after injection, but significantly decreased at day 7, and completely disappeared at day 14 after injection (Fig. 3I). The animals were monitored for an extended period, and no detectable subcutaneous masses were observed for up to six weeks, indicating that the iKera cells were not tumorigenic in vivo. Collectively, the above in vitro and in vivo results demonstrate that the iKera cells maintain long-term proliferative activity while retaining the biological characteristics of primary keratinocytes, suggesting that the iKera cells may be used as a valuable source of keratinocytes for optimizing scaffold-based skin wound healing strategy or skin tissue engineering studies.
3.4. The characteristics of the citrate-based scaffold PPCN
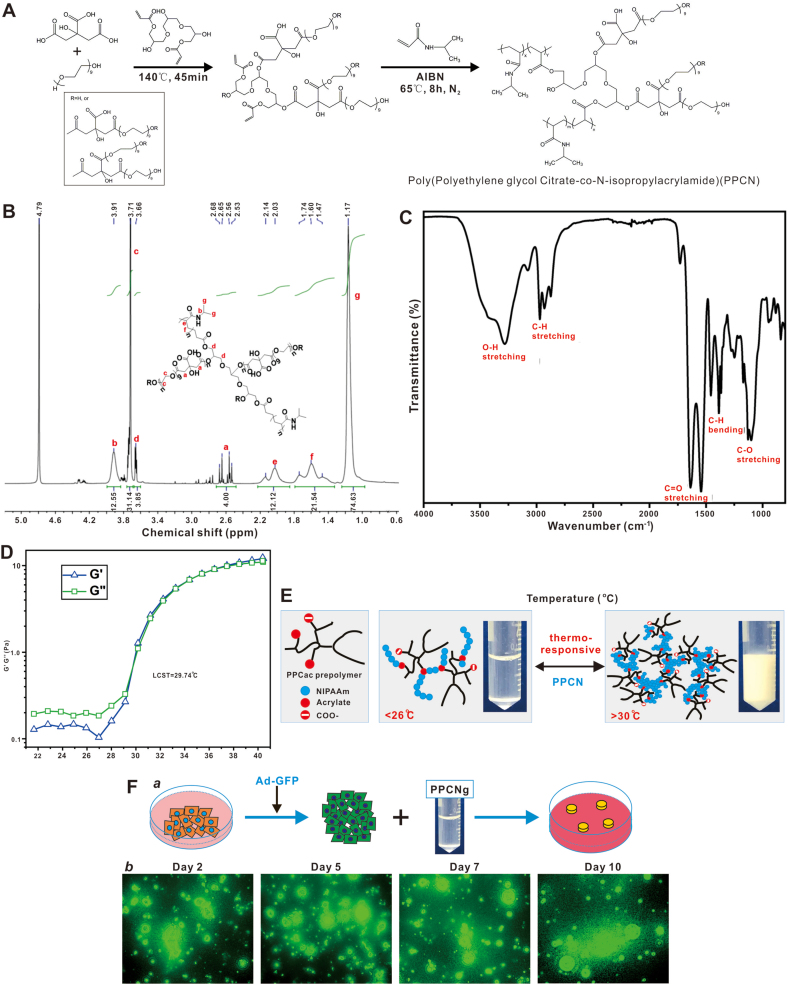
The citrate-based polymer gel PPCN [(polyethylene glycol citrate-co-N-isopropylacrylamide)] has a hierarchical architecture of micropores and can accommodate live cells in vivo [13]. The synthesis of PPCN was previously described [13] and is shown in Fig. 4A. The newly synthesized PPCN was further characterized. Specifically, the proton nuclear magnetic resonance (1H NMR) was used to confirm the chemical structure of PPCN. The composition of PPCN is verified by 1H NMR (600 MHz, DMSO‑d6, ppm): 3.91 (s, 1H), 3.81 (t, 2H), 3.73 (s, 4H), 3.66 (t, 2H), 2.53–2.68 (q, 4H), 2.03–2.14 (d, 1H), 1.47–1.74 (t, 2H), 1.17 (s, 6H) (Fig. 4B). PPCN were further characterized by FT-IR to verify its structure. The absorption of O–H stretching was observed at 3280 cm−1. The peaks displayed from 2873 to 2971 cm−1 were corresponding to symmetric and asymmetric stretching vibration of C–H bond. The C O stretching of both ester and carboxylate groups are located at 1639 and 1544 cm−1. The peaks located at 1455 and 1387 cm−1 were C–O–H vibration and C–H bending. The strong absorption bands of C–O stretching vibration were shown at 1102 cm−1. The wavenumber ranged from 400 to 4000 cm−1 with the resolution of 4 cm−1 and scans of 32 (Fig. 4C). These results validated the chemical structure of PPCN used in the study.
Fig. 4.
Characterization of the citric acid-based scaffold material PPCN. (A) The chemical synthesis scheme of PPCN. The synthesis scheme of PPCN was modified from Yang J et al. [13]. (B) Proton NMR spectrum of PPCN. The composition of PPCN is verified by 1H NMR (600 MHz, DMSO‑d6, ppm): 3.91 (s, 1H), 3.81 (t, 2H), 3.73 (s, 4H), 3.66 (t, 2H), 2.53–2.68 (q, 4H), 2.03–2.14 (d, 1H), 1.47–1.74 (t, 2H), 1.17 (s, 6H). (C). FT-IR spectrum of PPCN. PPCN was characterized by FT-IR to verify its structure. The absorption of O–H stretching was observed at 3280 cm−1. The peaks displayed from 2873 to 2971 cm−1 were corresponding to symmetric and asymmetric stretching vibration of C–H bond. The C O stretching of both ester and carboxylate groups are located at 1639 and 1544 cm−1. The peaks located at 1455 and 1387 cm−1 were C–O–H vibration and C–H bending. The strong absorption bands of C–O stretching vibration were shown at 1102 cm−1. (D) Temperature ramp rheological features of PPCN. The temperature ramp rheological curves of 100 mg/mL PPCN aqueous solution are shown. At low temperature (<29 °C), the loss modulus (G″) is higher than storage modulus (G′), indicating a liquid-like material. Above 29 °C, both G′ and G″ dramatically increase. After a crossover of G′ and G″, the storage modulus is higher than loss modulus, indicating the hydrogel formation. The crossover of moduli is corresponding to lower critical solution temperature (LCST) of PPCN, which is 29.74 °C. (E) The thermoresponsive feature of the citrate-based polymer scaffold PPCN. The PPCN gel has a critical solution temperature of 26 °C, and transforms to solid form above 30 °C. The polymeric structure of PPCN was modified from Yang J et al. [13]. (F) Biocompatibility of PPCN. The iKera cells were infected with Ad-GFP for 24 h, collected and mixed with PPCN/gelatin, followed by seeding in a cell culture dish (a). GFP signal from the infected viable iKera cells was recorded at the indicated time points (b). Representative images are shown.
The rheological properties of PPCN were analyzed by TA Discovery HR 20 hybrid rheometer. The temperature ramp rheological curves revealed that at low temperature (<29 °C), the loss modulus (G″) was higher than storage modulus (G′), indicating a liquid-like material. Above 29 °C, both G′ and G″ dramatically increase. After a crossover of G’ and G”, the storage modulus was higher than loss modulus, indicating the hydrogel formation. The crossover of moduli is corresponding to lower critical solution temperature (LCST) of PPCN, which is 29.74 °C (Fig. 4D). The thermorepsonsiveness of PPCN was further shown in Fig. 4E, i.e., in liquid form under 26 °C and transient to solid form above 30 °C. When iKera cells were transduced with Ad-GFP adenovirus and mixed with gelatin-containing PPCN to form 3-D culture in vitro (Fig. 4F, panel a), GFP signal from the entrapped iKera cells lasted at least up to 10 days (Fig. 4F, panel b), supporting the excellent biocompatibility of PPCN.
3.5. The iKera cells facilitate citrate-based scaffold PPCN-promoted cutaneous wound healing in vivo
We previously used PPCN gel as a vehicle for the in vivo delivery of various types of mesenchymal stem cells for bone regeneration studies [15,19,23,29,49,72,83,84]. Here, we tested whether PPCN could serve both as a scaffold and vehicle for delivering iKera cells to promote skin wound healing in a mouse model. The experimental mouse skin wound healing model was established as described [70]. The animals were divided into four groups: Blank, PPCN, iKera cells, and iKera + PPCN (Fig. 5A, panels a-d). While the skin wounds failed to close completely at day 9, the wounds were almost completely closed in other three groups at day 9, and the combination of iKera and PPCN provided the fastest closure of the cutaneous wounds in the mouse model (Fig. 5A panels bc vs. d & Fig. 5B). These results indicate that while the iKera cell alone or PPCN scaffold alone may promote cutaneous wound healing to certain extents, PPCN loaded with the iKera cells can effectively facilitate skin wound healing in the mouse model.
Fig. 5.
The iKera cells entrapped in the citrate-based scaffold PPCN promote skin wound healing. (A) The effects of the iKera cells and/or PPCN gel on skin wound healing. The 1-cm diameter full-thickness skin wound was created at the dorsum part of each mouse, and the mice were divided into four groups: Blank (a; 100 μL sterile PBS), PPCN (b; 100 μL PPCN), iKera cells (c; 2 × 106 of iKera cells in 100 μL PBS), and iKera + PPCN (d; 2 × 106 iKera cells in 100 μL PPCN, mixed at 20 °C). (B) Quantitative measurement of skin wound healing rate. The opening areas of skin wounds were measured and calculated at days 0, 3, 6 and 9. “**” p < 0.01, “*” p < 0.05, the PPCN group, the iKera group, or the iKera + PPCN group compared with that of the Blank group at respective time points; “##” p < 0.01, the iKera + PPCN group compared with that of the PPCN group at day 9; “^^” p < 0.01, the iKera + PPCN group compared with that of the iKera group at day 9.
3.6. The combination of iKera cells and PPCN scaffold yields optimal histologic restoration and re-epithelialization during skin wound healing
Severe acute skin loss injury usually leads to serious host response involved in tissue granulation and re-epithelialization, which is characterized by a rapid proliferation of fibroblasts to deposit randomly oriented collagen fibers to fill the tissue defect, and the migration of keratinocytes and contraction of myofibroblasts to restore the barrier, resulting in a fibrotic scar [7,8,85]. The fibrotic scar is a disorganized and flawed tissue with limited or no native skin functions such as sensation and elasticity. Thus, the histologic features of skin wound healing are important criteria to assess the restoration of native skin functions.
In order to accurately and quantitatively assess the wound healing efficiency and functional restoration, the retrieved samples were carefully processed, embedded and sectioned as illustrated in Fig. 6A. H & E stain analysis revealed that, at both lower and higher magnifications, the wounding area was partially covered with a thin layer of disorganized fibrotic tissue in the blank control group (Fig. 6B-a), whereas the wounding area was almost completely covered with a thick layer of disorganized fibrotic tissue in the PPCN group (Fig. 6B-b). While the addition of the iKera cells partially improved the re-epithelization of the wounding area in the iKera group (Fig. 6B-c), the combination of iKera cells and PPCN scaffold yielded rather well-organized full thickness skin features and re-epithelization of the wounding area, compared with that of the normal skin tissue (Fig. 6B- d vs. e). It is noteworthy that a significant restoration of skin accessory structures in the dermis region of the wound areas was observed in both the iKera cell group and the iKera + PPCN group (Fig. 6B).
Fig. 6.
Histologic and epithelization evaluation of iKera and/or PPCN-mediated skin wound healing. (A) Schematic representation of the region of interest for histologic evaluation. Skin tissue encompassing the whole wound healing area (dotted yellow circle) was harvested for tissue processing, while the center region of wound healing (dotted red cycle and highlighted blue area) was the sampling center for histologic comparison of different groups. (B) H & E histologic analysis. The skin samples (a to d) were prepared as described in (A) and subjected to paraffin-embedding, sectioning, and H & E staining. Normal mouse skin control (e) was prepared from the healthy full-thickness skin of the hip area. The keratinocyte layer was indicated with blue arrows, while representative skin appendages were indicated with blue circles. (C) & (D) Masson staining and Sirius red staining. The sections prepared from (A) were subjected to Masson staining (C) and Sirius red staining (D). The epidermal fibers in the epidermis layer (red staining) were indicated with green arrows (C), while the collagen fibers in dermis layer were indicated with yellow asterisk signs “*” (C & D). (E) Expression of epithelization markers. The retrieved and embedded skin samples were subjected to immunohistochemical (IHC) staining with antibodies against Krt15, Krt10 and Involucrin. Different magnifications were recorded under bright field microscope (see extended data in Fig. S2). Control IgG was used as a negative control (Fig. S2B). Representative results are shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Masson's trichrome is a commonly-used specialty staining method to distinguish cells from surrounding connective tissue. Masson staining showed that a lot of collagen fibers (stained in blue) generated at the wound healing in the dermis layer in other four groups, compared with that of the normal group. Meanwhile, the stratum corneum in the blank group and the PPCN group were thicker than that in both the iKera cell group and the iKera + PPCN group (Fig. 6C).
Similarly, Sirius red staining revealed that significant amounts of collagen deposition in the reticular dermis region were observed in all groups, compared with that in the normal control skin dermis (Fig. 6D), indicating the existence of active wound repair process. Collectively, the histologic analysis results demonstrate that, even though the PPCN alone or the iKera cells alone was shown to promote wound closure by forming disorganized multi-layered structures, only did the combination of the iKera and PPCN group show the effective restoration of the well-organized full thickness skin-like structures.
We also examined the expression of keratinocyte differentiation markers. IHC staining analysis indicated that the expression of keratinocyte markers Krt15, Krt10 and Involucrin was readily detected in all groups (Fig. 6E), compared with that of the negative controls (Fig. S2), although high levels of expression were constantly observed in the iKera + PPCN group (Fig. 6E) [13].
To track down the fate or status of the implanted iKera cells, we performed the IHC staining of the retrieved samples with anti-SV40 T antibody. Our IHC results showed that, while there was no positive staining observed in the blank, PPCN and normal groups, there was weak to modest positive staining in the stratum corneum of the wound healing center in the iKera + PPCN group. A strong positive staining was observed in the dermis tissue surrounding the center of wound that was not completely healed in the iKera group (Fig. 7A & B). These results indicate that the implanted iKera cells concentrated in the stratum corneum above the dermis of unhealed skin. While the exact mechanism underlying iKera cells in skin regeneration is not clear, we speculate that iKera cells may promote re-epithelialization of skin wound by directly participating in the wound repair process, and/or interact with dermal fibroblasts to accelerate wound contraction and hence healing process.
Fig. 7.
Location of the iKera cells at the skin injury repair site. The retrieved and embedded skin samples from the Blank group (a), the PPCN group (b), the iKera group, the iKera + PPCN group (d) and the Normal group (e) were subjected to immunohistochemical (IHC) staining with the anti-SV40 T antibody. Two areas of interest (A & B) are shown in the diagrams. Different magnifications were documented under a bright field microscope. Control IgG was used as a negative control (Fig. S2B). Representative results were shown.
4. Discussion
4.1. The iKera cells retain the characteristics of keratinocytes and lend themselves as a valuable resource to cell-based skin tissue engineering research
In this study, we established the iKera cells and demonstrated these cells were non-tumorigenic in athymic nude mice. More interestingly, the iKera cells were only weakly detected at the repaired skin wound in the iKera + PPCN group as assessed by anti-SV40 T IHC staining, even though the SV40 T was not removed in the implanted iKera cells. These results strongly suggest that the iKera cells may actively participate in the repair process and become more differentiated or less proliferative keratinocytes. Thus, the iKera cells should have an acceptable biosafety profile for in vivo use.
As a proof-of-concept experiment, we demonstrated that the citrate-based scaffold PPCN embedded with iKera cells provided more effective cutaneous wound healing and re-epithelization than that of either PPCN or iKera cells alone in a mouse skin wound model. While the skin injury model does not represent a full cycle of skin replacement in mice (~28 days), our model does reflect the natural healing process without any external force. Since the in vivo studies were only carried out in athymic nude mice, in which the potential impacts of T lymphocyte immunity, melanocytes and hair follicle cells in epidermis were ignored. Thus, other animal models are needed to further explore the role of keratinocytes in skin wound healing process. It is also noteworthy that, since we demonstrated iKera cells were sensitive to UVB irradiation and actively communicated with melanocytes and dermal fibroblasts, the iKera cells may have applications in skin burn injury, skin radiation injury, pigmentation, scar hyperplasia and other skin related disorders through skin tissue engineering.
4.2. Keratinocyte availability may be essential to the quality of skin wound healing
Normal wound healing is a dynamic multi-phase process that requires coordinated interactions between growth factors and various cells [1,8]. As the major cellular component of the epidermis, keratinocytes are epithelial cells of ectodermal origin and responsible for the production of the major extracellular protein keratin of the epidermis, thus playing a critical role in re-epithelialization [1,8]. Thus, keratinocytes play an essential role in cutaneous wound healing. Nonetheless, numerous efforts have been devoted to explore the potential use of stem cells to improve the rate and quality of wound healing and/or skin regeneration [1,8,9].
The use of embryonic stem cells (ESCs) is limited due to the concerns about immunogenicity and tumorigenicity, as well as possible ethical controversy and legal restrictions [86]. Induced pluripotent stem cells (iPSCs) are reprogrammed adult somatic cells and have potential use for skin wound healing, although it remains a challenge to effectively direct the iPSCs to differentiate into various skin cells including keratinocytes [87]. Furthermore, possible tumorigenic potential of iPSCs remains a concern. Mesenchymal stem cells (MSCs) are somatic progenitor cells that can self-renew and differentiate into multiple cell types including skin cells [88]. It has been reported that MSCs induced wound healing with little immunoreactivity in the host after local transplantation or systemic administration [1,7,8]. While MSCs are most commonly derived from adult bone marrow, they can also be isolated from many other tissues such as adipose tissue, umbilical cord blood, or peripheral blood [1,7,8]. Since undifferentiated MSCs can produce cytokines and exert immunomodulatory functions, MSCs have long been studied for their beneficial effects on wound healing [1,7]. However, the overall efficacy of MSC-based therapies is often difficult to assess due to the phenotypic variation of MSCs used. The isolation process of MSCs is time-consuming, and yet limited numbers of autologous MSCs can be harvested for large wounds. Furthermore, it remains a challenge to drive MSCs to differentiate into skin cells with high efficiency, and thus these progenitor cells have limited use in point-of-care settings.
In this study, we demonstrated that the reversibly immortalized keratinocytes iKera can serve as a valuable epithelial cell source for cutaneous wound healing and skin regeneration. Interestingly, a commercially available point-of-care product known as ReCell Autologous Cell Harvesting Device is an autologous skin cell suspension spray system, which uses non-cultured autologous skin cells harvested from a patient [7]. The harvested skin cells were suspended in solution and sprayed on the wound, allowing the cells to adhere to the target tissue surface [89]. Such cell suspension was shown to predominantly contain keratinocytes (~64 %) and dermal fibroblasts (~30 %), with a small population of melanocytes (~3.5 %) [89]. A comparative clinical study of ReCell and autologous split-thickness skin grafting (STSG) in the treatment of acute burns revealed that ReCell was as effective as STSG, while requiring almost 40 times less donor tissue, as well as less pain at the ReCell donor sites and improved appearance relative to STSG [7,90]. It is conceivable that reversibly immortalized or transiently immortalized keratinocytes, along with dermal fibroblasts and melanocytes, may serve a valuable allogeneic or xenogeneic cell source for wound repair if potential recipient immune response can be mitigated.
4.3. Citrate-based scaffold may provide metabonegenic advantage for cell-based therapy to treat chronic skin wounds
Numerous scaffold materials, especially hydrogels, have been investigated for skin wound healing and/or skin regeneration [7]. It is conceivable that biomaterial scaffolds such as the extracellular matrix (ECM) or individual components of the ECM including collagen, laminin or hyaluronan, are more appealing for skin wound healing since they are more responsive to and optimized for the physiologic and biomechanical requirements of wound tissue [8,9].
In this study, we focused on the citrate-based, biodegradable, and thermoresponsive polymer gel PPCN [13]. Citrate is normally an integral component in native bone and stored in bone matrix, which can be released during bone resorption. Citrate is also a key intermediate metabolite in the tricarboxylic acid (TCA) cycle, and thus plays crucial regulatory roles in maintaining cell energy homeostasis [[10], [11], [12]]. In fact, it was reported that extracellular citrate uptake through citrate transporter SLC13a5 supported osteogenic differentiation via regulation of energy-producing metabolic pathways, leading to elevated cell energy status that fuels the high metabolic demands of hMSCs osteodifferentiation [12]. We have recently demonstrated that PPCN-based scaffolds effectively supported bone formation from BMP9-stimulated MSCs [18,51,72,91]. Interestingly, the incorporation of copper metal organic framework nanoparticles (HKUST-1 NPs) within the antioxidant thermoresponsive citrate-based PPCN hydrogel induced angiogenesis, collagen deposition, and re-epithelialization during wound healing in diabetic mice [92]. Thus, it is conceivable that citrate-based scaffold materials may provide metabologenic advantage of cell-based therapies or tissue engineering to treat chronic wound healing.
5. Conclusion
In this study, we established the reversibly immortalized keratinocytes (iKera) from mouse epidermis tissues by stably expressing SV40 large T antigen (SV40 T), which is flanked with FRT sites. The immortalization phenotype of the iKera cells could be effectively reversed by the FLP recombinase or silencing SV40 T. The iKera cells exhibited typical characteristics of keratinocytes by expressing keratinocyte markers, and were highly responsive to UVB-induced inhibition of cell proliferation, as well as ATRA-induced differentiation. While retaining long-term proliferative activity in culture, the iKera cells were non-tumorigenic in xenograft tumor formation assay. Using the iKera cells, we demonstrated that the citrate-based scaffold PPCN embedded with iKera cells provided more effective cutaneous wound healing and re-epithelization than that of either PPCN or iKera cells alone, in a mouse skin wound model. Therefore, our results demonstrate that the reversibly immortalized keratinocyte iKera cells can serve as a valuable epithelial cell source, in combination with the biocompatible scaffolds, for skin tissue engineering to treat skin burn injury, skin radiation injury, pigmentation, scar hyperplasia and other skin related disorders.
Authors’ contributions
JF, TCH, JZ, Hao-W and KY conceived the project and oversaw the study. JZ, Hao-W, KY, LA, YL PZ and YG performed most of the experiments. Huifeng-W, CD, and GAA synthesized and characterized the PPCN. SS, NN, DS, FH, QL, CC, WW, BHS, and GAA provided technical supports and/or research resources/reagents. JZ, Hao-W, JF, TCH, LS, and YG collected and analyzed the collected data. JF, TCH, SHH, RRR, RCH, and HHL secured funding supports. JF, JZ, TCH, RCH, HHL, RRR, SHH, and LS drafted the manuscript. All authors reviewed and revised the manuscript, and approved the final version for submission.
Declaration of competing interest
All the authors declare that they have no potential conflicts of interest.
Acknowledgments
The reported study was supported in part by research grants from the 2019 Chongqing Support Program for Entrepreneurship and Innovation (No. cx2019113) (JF), the 2019 Science and Technology Research Plan Project of Chongqing Education Commission (KJQN201900410) (JF), the 2019 Youth Innovative Talent Training Program of Chongqing Education Commission (No. CY200409) (JF), the 2019 Funding for Postdoctoral Research (Chongqing Human Resources and Social Security Bureau No.298) (JF) and the National Key Research and Development Program of China (2016YFC1000803). RRR, TCH and GAA were partially funded by the National Institutes of Health (DE030480). WW was supported by the Medical Scientist Training Program of the National Institutes of Health (T32 GM007281). This project was also supported in part by The University of Chicago Cancer Center Support Grant (P30CA014599) and the National Center for Advancing Translational Sciences of the National Institutes of Health through Grant Number UL1 TR000430. TCH was also supported by the Mabel Green Myers Research Endowment Fund, The University of Chicago Orthopaedics Alumni Fund, and The University of Chicago SHOCK Fund. Funding sources were not involved in the study design; in the collection, analysis and/or interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.07.022.
Abbreviations
- iKera
mouse immortalized keratinocytes
- Kera
mouse primary keratinocytes
- iKera-FLuc
iKera cells stably expressing firefly luciferase
- P0
Passage 0
- SV40 T
SV40 large T antigen
- TqPCR
touchdown quantitative real-time PCR
- LSCM
laser scanning confocal microscopy
- IF
immunofluorescence
- ATRA
all-trans retinoic acid
- Ad-FLP
Adenoviral vector expressing FLP recombinase
- Ad-GFP
Adenoviral vector expressing green fluorescent protein (GFP)
- Ad-siLTA
Adenoviral vector expressing siRNAs that silence SV40 T
- Ad-RFP
Adenoviral vector expressing red fluorescent protein (RFP)
- TEM
Transmission electron microscope
- iMCs
immortalized mouse melanocytes
- Fib
mouse primary dermal fibroblasts
- PPCN
poly (polyethyleneglycol citrate-co-N-isopropylacrylamide)
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
Fig. S1. (A) Expression of SV40 T in the iKera cells. Total RNA was isolated from subconfluent Kera and iKera cells. TqPCR analysis was carried out to detect the expression of SV40 T in Kera and iKera cells. “**” p < 0.01, Kera cells vs iKera cells. (B) Negative control (NC) for Figure 1D. IF of SV40 T in the Kera and iKera cells. (C) The cuticle structure of skin. From inside to outside, keratinocytes can be divided into five layers: basal layer, spinous layer, granular layer, hyaline layer and stratum corneum layer. (D) ATRA-induced differentiation of iKera cells. The iKera cells were treated with ATRA (1μM) or DMSO for 3 and 5 days, respectively, and qPCR was used to test the expression of skin marker genes for basal layer (Krt5, Krt14 and Krt15), spinous layer (Krt1 Krt10 and IVL), granular layer (IVL and LOR) and stratum corneum layer (LOR). “**” p < 0.01, ATRA group vs DMSO group. (E) Expression of keratinocyte markers in Kera and iKera cells. Total RNA was isolated from subconfluent Kera and iKera cells and subjected to TqPCR analysis. All samples were normalized with Gapdh.(F) Dose-dependent response of the iKera cells to UVB-induced inhibition of cell survival. The iKera cells were treated with UVB at energy levels of 0, 37.5, 75, 225 and 450μw/cm2, and observed under light microscope at 0, 3, 24, 36, 48 and 72h. The optimal UVB treatment conditions are highlighted in a red box. Representative results are shown
figs2.
Fig. S2. (A) Expression of epithelialized marker genes in skin wounds in vivo. The retrieved skin samples were subjected to IHC staining of Krt15, Krt10 and Involucrin, the morphological and staining changes were observed under light microscope. (B) The stains without primary antibody and/or with control IgG were used as negative controls. Representative results are shown
References
- 1.Coalson E., Bishop E., Liu W., Feng Y., Spezia M., Liu B., Shen Y., Wu D., Du S., Li A.J., Ye Z., Zhao L., Cao D., Li A., Hagag O., Deng A., Liu W., Li M., Haydon R.C., Shi L., Athiviraham A., Lee M.J., Wolf J.M., Ameer G.A., He T.C., Reid R.R. Stem cell therapy for chronic skin wounds in the era of personalized medicine: from bench to bedside. Genes Dis. 2019;6:342–358. doi: 10.1016/j.gendis.2019.09.008. https://10.1016/j.gendis.2019.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gantwerker E.A., Hom D.B. Skin: histology and physiology of wound healing. Facial Plast Surg Clin North Am. 2011;19:441–453. doi: 10.1016/j.fsc.2011.06.009. https://10.1016/j.fsc.2011.06.009 [DOI] [PubMed] [Google Scholar]
- 3.Boer M., Duchnik E., Maleszka R., Marchlewicz M. Structural and biophysical characteristics of human skin in maintaining proper epidermal barrier function. Postepy Dermatol Alergol. 2016;33:1–5. doi: 10.5114/pdia.2015.48037. https://10.5114/pdia.2015.48037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohania D., Chandel S., Kumar P., Verma V., Digvijay K., Tripathi D., Choudhury K., Mitten S.K., Shah D. Ultraviolet radiations: skin defense-damage mechanism. Adv. Exp. Med. Biol. 2017;996:71–87. doi: 10.1007/978-3-319-56017-5_7. https://10.1007/978-3-319-56017-5_7 [DOI] [PubMed] [Google Scholar]
- 5.Idrus R.B., Rameli M.A., Low K.C., Law J.X., Chua K.H., Latiff M.B., Saim A.B. Full-thickness skin wound healing using autologous keratinocytes and dermal fibroblasts with fibrin: bilayered versus single-layered substitute. Adv. Skin Wound Care. 2014;27:171–180. doi: 10.1097/01.ASW.0000445199.26874.9d. https://10.1097/01.ASW.0000445199.26874.9d [DOI] [PubMed] [Google Scholar]
- 6.Rodrigues M., Kosaric N., Bonham C.A., Gurtner G.C. Wound healing: a cellular perspective. Physiol. Rev. 2019;99:665–706. doi: 10.1152/physrev.00067.2017. https://10.1152/physrev.00067.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J.R., Navarro J., Coburn J.C., Mahadik B., Molnar J., Holmes JHt, Nam A.J., Fisher J.P. Current and future perspectives on skin tissue engineering: key features of biomedical research, translational assessment, and clinical application. Adv Healthc Mater. 2019;8 doi: 10.1002/adhm.201801471. https://10.1002/adhm.201801471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nourian Dehkordi A., Mirahmadi Babaheydari F., Chehelgerdi M., Raeisi Dehkordi S. Skin tissue engineering: wound healing based on stem-cell-based therapeutic strategies. Stem Cell Res. Ther. 2019;10:111. doi: 10.1186/s13287-019-1212-2. https://10.1186/s13287-019-1212-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dash B.C., Xu Z., Lin L., Koo A., Ndon S., Berthiaume F., Dardik A., Hsia H. Stem cells and engineered scaffolds for regenerative wound healing. Bioengineering (Basel). 2018;5 doi: 10.3390/bioengineering5010023. https://10.3390/bioengineering5010023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma C., Gerhard E., Lu D., Yang J. Citrate chemistry and biology for biomaterials design. Biomaterials. 2018;178:383–400. doi: 10.1016/j.biomaterials.2018.05.003. https://10.1016/j.biomaterials.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma C., Kuzma M.L., Bai X., Yang J. Biomaterial-based metabolic regulation in regenerative engineering. Adv. Sci. 2019;6:1900819. doi: 10.1002/advs.201900819. https://10.1002/advs.201900819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma C., Tian X., Kim J.P., Xie D., Ao X., Shan D., Lin Q., Hudock M.R., Bai X., Yang J. Citrate-based materials fuel human stem cells by metabonegenic regulation. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E11741–E11750. doi: 10.1073/pnas.1813000115. https://10.1073/pnas.1813000115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J., van Lith R., Baler K., Hoshi R.A., Ameer G.A. A thermoresponsive biodegradable polymer with intrinsic antioxidant properties. Biomacromolecules. 2014;15:3942–3952. doi: 10.1021/bm5010004. https://10.1021/bm5010004 [DOI] [PubMed] [Google Scholar]
- 14.Lichti U., Anders J., Yuspa S.H. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat. Protoc. 2008;3:799–810. doi: 10.1038/nprot.2008.50. https://10.1038/nprot.2008.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye J., Wang J., Zhu Y., Wei Q., Wang X., Yang J., Tang S., Liu H., Fan J., Zhang F., Farina E.M., Mohammed M.K., Zou Y., Song D., Liao J., Huang J., Guo D., Lu M., Liu F., Liu J., Li L., Ma C., Hu X., Haydon R.C., Lee M.J., Reid R.R., Ameer G.A., Yang L., He T.C. A thermoresponsive polydiolcitrate-gelatin scaffold and delivery system mediates effective bone formation from BMP9-transduced mesenchymal stem cells. Biomed. Mater. 2016;11 doi: 10.1088/1748-6041/11/2/025021. https://10.1088/1748-6041/11/2/025021 [DOI] [PubMed] [Google Scholar]
- 16.Zhao C., Zeng Z., Qazvini N.T., Yu X., Zhang R., Yan S., Shu Y., Zhu Y., Duan C., Bishop E., Lei J., Zhang W., Yang C., Wu K., Wu Y., An L., Huang S., Ji X., Gong C., Yuan C., Zhang L., Liu W., Huang B., Feng Y., Zhang B., Dai Z., Shen Y., Wang X., Luo W., Oliveira L., Athiviraham A., Lee M.J., Wolf J.M., Ameer G.A., Reid R.R., He T.C., Huang W. Thermoresponsive citrate-based graphene oxide scaffold enhances bone regeneration from BMP9-stimulated adipose-derived mesenchymal stem cells. ACS Biomater. Sci. Eng. 2018;4:2943–2955. doi: 10.1021/acsbiomaterials.8b00179. https://10.1021/acsbiomaterials.8b00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X., Yuan C., Huang B., Fan J., Feng Y., Li A.J., Zhang B., Lei Y., Ye Z., Zhao L., Cao D., Yang L., Wu D., Chen X., Liu B., Wagstaff W., He F., Wu X., Luo H., Zhang J., Zhang M., Haydon R.C., Luu H.H., Lee M.J., Moriatis Wolf J., Huang A., He T.C., Zeng Z. Developing a versatile shotgun cloning strategy for single-vector-based multiplex expression of short interfering RNAs (siRNAs) in mammalian cells. ACS Synth. Biol. 2019;8:2092–2105. doi: 10.1021/acssynbio.9b00203. https://10.1021/acssynbio.9b00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B., Yang L., Zeng Z., Feng Y., Wang X., Wu X., Luo H., Zhang J., Zhang M., Pakvasa M., Wagstaff W., He F., Mao Y., Qin K., Ding H., Zhang Y., Niu C., Wu M., Zhao X., Wang H., Huang L., Shi D., Liu Q., Ni N., Fu K., Athiviraham A., Moriatis Wolf J., Lee M.J., Hynes K., Strelzow J., El Dafrawy M., Xia Y., He T.C. Leptin potentiates BMP9-induced osteogenic differentiation of mesenchymal stem cells through the activation of JAK/STAT signaling. Stem Cell. Dev. 2020;29:498–510. doi: 10.1089/scd.2019.0292. https://10.1089/scd.2019.0292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He F., Ni N., Zeng Z., Wu D., Feng Y., Li A.J., Luu B., Li A.F., Qin K., Wang E., Wang X., Wu X., Luo H., Zhang J., Zhang M., Mao Y., Pakvasa M., Wagstaff W., Zhang Y., Niu C., Wang H., Huang L., Shi D., Liu Q., Zhao X., Fu K., Reid R.R., Wolf J.M., Lee M.J., Hynes K., Strelzow J., El Dafrawy M., Gan H., He T.C., Fan J. FAMSi: a synthetic biology approach to the fast assembly of multiplex siRNAs for silencing gene expression in mammalian cells. Mol. Ther. Nucleic Acids. 2020;22:885–899. doi: 10.1016/j.omtn.2020.10.007. https://10.1016/j.omtn.2020.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu N., Zhang H., Deng F., Li R., Zhang W., Chen X., Wen S., Wang N., Zhang J., Yin L., Liao Z., Zhang Z., Zhang Q., Yan Z., Liu W., Wu D., Ye J., Deng Y., Yang K., Luu H.H., Haydon R.C., He T.C. Overexpression of Ad5 precursor terminal protein accelerates recombinant adenovirus packaging and amplification in HEK-293 packaging cells. Gene Ther. 2014;21:629–637. doi: 10.1038/gt.2014.40. https://10.1038/gt.2014.40 [DOI] [PubMed] [Google Scholar]
- 21.Wei Q., Fan J., Liao J., Zou Y., Song D., Liu J., Cui J., Liu F., Ma C., Hu X., Li L., Yu Y., Qu X., Chen L., Yu X., Zhang Z., Zhao C., Zeng Z., Zhang R., Yan S., Wu X., Shu Y., Reid R.R., Lee M.J., Wolf J.M., He T.C. Engineering the rapid adenovirus production and amplification (RAPA) cell line to expedite the generation of recombinant adenoviruses. Cell. Physiol. Biochem. 2017;41:2383–2398. doi: 10.1159/000475909. https://10.1159/000475909 [DOI] [PubMed] [Google Scholar]
- 22.Yang K., Chen J., Jiang W., Huang E., Cui J., Kim S.H., Hu N., Liu H., Zhang W., Li R., Chen X., Kong Y., Zhang J., Wang J., Wang L., Shen J., Luu H.H., Haydon R.C., Lian X., Yang T., He T.C. Conditional immortalization establishes a repertoire of mouse melanocyte progenitors with distinct melanogenic differentiation potential. J. Invest. Dermatol. 2012;132:2479–2483. doi: 10.1038/jid.2012.145. https://10.1038/jid.2012.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui J., Zhang W., Huang E., Wang J., Liao J., Li R., Yu X., Zhao C., Zeng Z., Shu Y., Zhang R., Yan S., Lei J., Yang C., Wu K., Wu Y., Huang S., Ji X., Li A., Gong C., Yuan C., Zhang L., Liu W., Huang B., Feng Y., An L., Zhang B., Dai Z., Shen Y., Luo W., Wang X., Huang A., Luu H.H., Reid R.R., Wolf J.M., Thinakaran G., Lee M.J., He T.C. BMP9-induced osteoblastic differentiation requires functional Notch signaling in mesenchymal stem cells. Lab. Invest. 2019;99:58–71. doi: 10.1038/s41374-018-0087-7. https://10.1038/s41374-018-0087-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H., Cao Y., Shu L., Zhu Y., Peng Q., Ran L., Wu J., Luo Y., Zuo G., Luo J., Zhou L., Shi Q., Weng Y., Huang A., He T.C., Fan J. Long non-coding RNA (lncRNA) H19 induces hepatic steatosis through activating MLXIPL and mTORC1 networks in hepatocytes. J. Cell Mol. Med. 2020;24:1399–1412. doi: 10.1111/jcmm.14818. https://10.1111/jcmm.14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang B., Huang L.F., Zhao L., Zeng Z., Wang X., Cao D., Yang L., Ye Z., Chen X., Liu B., He T.C., Wang X. Microvesicles (MIVs) secreted from adipose-derived stem cells (ADSCs) contain multiple microRNAs and promote the migration and invasion of endothelial cells. Genes Dis. 2020;7:225–234. doi: 10.1016/j.gendis.2019.04.005. https://10.1016/j.gendis.2019.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Westerman K.A., Leboulch P. Reversible immortalization of mammalian cells mediated by retroviral transfer and site-specific recombination. Proc. Natl. Acad. Sci. U. S. A. 1996;93:8971–8976. doi: 10.1073/pnas.93.17.8971. https://10.1073/pnas.93.17.8971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang E., Bi Y., Jiang W., Luo X., Yang K., Gao J.L., Gao Y., Luo Q., Shi Q., Kim S.H., Liu X., Li M., Hu N., Liu H., Cui J., Zhang W., Li R., Chen X., Shen J., Kong Y., Zhang J., Wang J., Luo J., He B.C., Wang H., Reid R.R., Luu H.H., Haydon R.C., Yang L., He T.C. Conditionally immortalized mouse embryonic fibroblasts retain proliferative activity without compromising multipotent differentiation potential. PloS One. 2012;7 doi: 10.1371/journal.pone.0032428. https://10.1371/journal.pone.0032428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamplot J.D., Liu B., Yin L., Zhang W., Wang Z., Luther G., Wagner E., Li R., Nan G., Shui W., Yan Z., Rames R., Deng F., Zhang H., Liao Z., Liu W., Zhang J., Zhang Z., Zhang Q., Ye J., Deng Y., Qiao M., Haydon R.C., Luu H.H., Angeles J., Shi L.L., He T.C., Ho S.H. Reversibly immortalized mouse articular chondrocytes acquire long-term proliferative capability while retaining chondrogenic phenotype. Cell Transplant. 2015;24:1053–1066. doi: 10.3727/096368914X681054. https://10.3727/096368914X681054 [DOI] [PubMed] [Google Scholar]
- 29.Shu Y., Yang C., Ji X., Zhang L., Bi Y., Yang K., Gong M., Liu X., Guo Q., Su Y., Qu X., Nan G., Zhao C., Zeng Z., Yu X., Zhang R., Yan S., Lei J., Wu K., Wu Y., An L., Huang S., Gong C., Yuan C., Liu W., Huang B., Feng Y., Zhang B., Dai Z., Shen Y., Luo W., Wang X., Haydon R.C., Luu H.H., Reid R.R., Wolf J.M., Lee M.J., He T.C., Li Y. Reversibly immortalized human umbilical cord-derived mesenchymal stem cells (UC-MSCs) are responsive to BMP9-induced osteogenic and adipogenic differentiation. J. Cell. Biochem. 2018;119:8872–8886. doi: 10.1002/jcb.27140. https://10.1002/jcb.27140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M., Chen Y., Bi Y., Jiang W., Luo Q., He Y., Su Y., Liu X., Cui J., Zhang W., Li R., Kong Y., Zhang J., Wang J., Zhang H., Shui W., Wu N., Zhu J., Tian J., Yi Q.J., Luu H.H., Haydon R.C., He T.C., Zhu G.H. Establishment and characterization of the reversibly immortalized mouse fetal heart progenitors. Int. J. Med. Sci. 2013;10:1035–1046. doi: 10.7150/ijms.6639. https://10.7150/ijms.6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bi Y., He Y., Huang J., Su Y., Zhu G.H., Wang Y., Qiao M., Zhang B.Q., Zhang H., Wang Z., Liu W., Cui J., Kang Q., Zhang Z., Deng Y., Li R., Zhang Q., Yang K., Luu H.H., Haydon R.C., He T.C., Tang N. Functional characteristics of reversibly immortalized hepatic progenitor cells derived from mouse embryonic liver. Cell. Physiol. Biochem. 2014;34:1318–1338. doi: 10.1159/000366340. https://10.1159/000366340 [DOI] [PubMed] [Google Scholar]
- 32.Bi Y., Huang J., He Y., Zhu G.H., Su Y., He B.C., Luo J., Wang Y., Kang Q., Luo Q., Chen L., Zuo G.W., Jiang W., Liu B., Shi Q., Tang M., Zhang B.Q., Weng Y., Huang A., Zhou L., Feng T., Luu H.H., Haydon R.C., He T.C., Tang N. Wnt antagonist SFRP3 inhibits the differentiation of mouse hepatic progenitor cells. J. Cell. Biochem. 2009;108:295–303. doi: 10.1002/jcb.22254. https://10.1002/jcb.22254 [DOI] [PubMed] [Google Scholar]
- 33.Wang J., Zhang H., Zhang W., Huang E., Wang N., Wu N., Wen S., Chen X., Liao Z., Deng F., Yin L., Zhang J., Zhang Q., Yan Z., Liu W., Zhang Z., Ye J., Deng Y., Luu H.H., Haydon R.C., He T.C., Deng F. Bone morphogenetic protein-9 effectively induces osteo/odontoblastic differentiation of the reversibly immortalized stem cells of dental apical papilla. Stem Cell. Dev. 2014;23:1405–1416. doi: 10.1089/scd.2013.0580. https://10.1089/scd.2013.0580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu S., Wang J., Ye J., Zou Y., Zhu Y., Wei Q., Wang X., Tang S., Liu H., Fan J., Zhang F., Farina E.M., Mohammed M.M., Song D., Liao J., Huang J., Guo D., Lu M., Liu F., Liu J., Li L., Ma C., Hu X., Lee M.J., Reid R.R., Ameer G.A., Zhou D., He T. Bone morphogenetic protein 9 (BMP9) induces effective bone formation from reversibly immortalized multipotent adipose-derived (iMAD) mesenchymal stem cells. Am J Transl Res. 2016;8:3710–3730. [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X., Chen L., Wu K., Yan S., Zhang R., Zhao C., Zeng Z., Shu Y., Huang S., Lei J., Ji X., Yuan C., Zhang L., Feng Y., Liu W., Huang B., Zhang B., Luo W., Wang X., Liu B., Haydon R.C., Luu H.H., He T.C., Gan H. Establishment and functional characterization of the reversibly immortalized mouse glomerular podocytes (imPODs) Genes Dis. 2018;5:137–149. doi: 10.1016/j.gendis.2018.04.003. https://10.1016/j.gendis.2018.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q., Wang J., Deng F., Yan Z., Xia Y., Wang Z., Ye J., Deng Y., Zhang Z., Qiao M., Li R., Denduluri S.K., Wei Q., Zhao L., Lu S., Wang X., Tang S., Liu H., Luu H.H., Haydon R.C., He T.C., Jiang L. TqPCR: a touchdown qPCR assay with significantly improved detection sensitivity and amplification efficiency of SYBR green qPCR. PloS One. 2015;10 doi: 10.1371/journal.pone.0132666. https://10.1371/journal.pone.0132666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan J., Wei Q., Liao J., Zou Y., Song D., Xiong D., Ma C., Hu X., Qu X., Chen L., Li L., Yu Y., Yu X., Zhang Z., Zhao C., Zeng Z., Zhang R., Yan S., Wu T., Wu X., Shu Y., Lei J., Li Y., Zhang W., Haydon R.C., Luu H.H., Huang A., He T.C., Tang H. Noncanonical Wnt signaling plays an important role in modulating canonical Wnt-regulated stemness, proliferation and terminal differentiation of hepatic progenitors. Oncotarget. 2017;8:27105–27119. doi: 10.18632/oncotarget.15637. https://10.18632/oncotarget.15637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu W., Deng Z., Zeng Z., Fan J., Feng Y., Wang X., Cao D., Zhang B., Yang L., Liu B., Pakvasa M., Wagstaff W., Wu X., Luo H., Zhang J., Zhang M., He F., Mao Y., Ding H., Zhang Y., Niu C., Haydon R.C., Luu H.H., Wolf J.M., Lee M.J., Huang W., He T.C., Zou Y. Highly expressed BMP9/GDF2 in postnatal mouse liver and lungs may account for its pleiotropic effects on stem cell differentiation, angiogenesis, tumor growth and metabolism. Genes Dis. 2020;7:235–244. doi: 10.1016/j.gendis.2019.08.003. https://10.1016/j.gendis.2019.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X., Zhao L., Wu X., Luo H., Wu D., Zhang M., Zhang J., Pakvasa M., Wagstaff W., He F., Mao Y., Zhang Y., Niu C., Wu M., Zhao X., Wang H., Huang L., Shi D., Liu Q., Ni N., Fu K., Hynes K., Strelzow J., El Dafrawy M., He T.C., Qi H., Zeng Z. Development of a simplified and inexpensive RNA depletion method for plasmid DNA purification using size selection magnetic beads (SSMBs) Genes Dis. 2021;8:298–306. doi: 10.1016/j.gendis.2020.04.013. https://10.1016/j.gendis.2020.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L., Luo Q., Shu Y., Zeng Z., Huang B., Feng Y., Zhang B., Wang X., Lei Y., Ye Z., Zhao L., Cao D., Yang L., Chen X., Liu B., Wagstaff W., Reid R.R., Luu H.H., Haydon R.C., Lee M.J., Wolf J.M., Fu Z., He T.C., Kang Q. Transcriptomic landscape regulated by the 14 types of bone morphogenetic proteins (BMPs) in lineage commitment and differentiation of mesenchymal stem cells (MSCs) Genes Dis. 2019;6:258–275. doi: 10.1016/j.gendis.2019.03.008. https://10.1016/j.gendis.2019.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao J., Wei Q., Fan J., Zou Y., Song D., Liu J., Liu F., Ma C., Hu X., Li L., Yu Y., Qu X., Chen L., Yu X., Zhang Z., Zhao C., Zeng Z., Zhang R., Yan S., Wu T., Wu X., Shu Y., Lei J., Li Y., Zhang W., Wang J., Reid R.R., Lee M.J., Huang W., Wolf J.M., He T.C., Wang J. Characterization of retroviral infectivity and superinfection resistance during retrovirus-mediated transduction of mammalian cells. Gene Ther. 2017;24:333–341. doi: 10.1038/gt.2017.24. https://10.1038/gt.2017.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shu Y., Wu K., Zeng Z., Huang S., Ji X., Yuan C., Zhang L., Liu W., Huang B., Feng Y., Zhang B., Dai Z., Shen Y., Luo W., Wang X., Liu B., Lei Y., Ye Z., Zhao L., Cao D., Yang L., Chen X., Luu H.H., Reid R.R., Wolf J.M., Lee M.J., He T.C. A simplified system to express circularized inhibitors of miRNA for stable and potent suppression of miRNA functions. Mol. Ther. Nucleic Acids. 2018;13:556–567. doi: 10.1016/j.omtn.2018.09.025. https://10.1016/j.omtn.2018.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang X., Wu X., Zhang Z., Ma C., Wu T., Tang S., Zeng Z., Huang S., Gong C., Yuan C., Zhang L., Feng Y., Huang B., Liu W., Zhang B., Shen Y., Luo W., Wang X., Liu B., Lei Y., Ye Z., Zhao L., Cao D., Yang L., Chen X., Haydon R.C., Luu H.H., Peng B., Liu X., He T.C. Monensin inhibits cell proliferation and tumor growth of chemo-resistant pancreatic cancer cells by targeting the EGFR signaling pathway. Sci. Rep. 2018;8:17914. doi: 10.1038/s41598-018-36214-5. https://10.1038/s41598-018-36214-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng Y., Zhang J., Wang Z., Yan Z., Qiao M., Ye J., Wei Q., Wang J., Wang X., Zhao L., Lu S., Tang S., Mohammed M.K., Liu H., Fan J., Zhang F., Zou Y., Liao J., Qi H., Haydon R.C., Luu H.H., He T.C., Tang L. Antibiotic monensin synergizes with EGFR inhibitors and oxaliplatin to suppress the proliferation of human ovarian cancer cells. Sci. Rep. 2015;5:17523. doi: 10.1038/srep17523. https://10.1038/srep17523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu X., Liu F., Zeng L., He F., Zhang R., Yan S., Zeng Z., Shu Y., Zhao C., Wu X., Lei J., Zhang W., Yang C., Wu K., Wu Y., An L., Huang S., Ji X., Gong C., Yuan C., Zhang L., Feng Y., Huang B., Liu W., Zhang B., Dai Z., Wang X., Liu B., Haydon R.C., Luu H.H., Gan H., He T.C., Chen L. Niclosamide exhibits potent anticancer activity and synergizes with sorafenib in human renal cell cancer cells. Cell. Physiol. Biochem. 2018;47:957–971. doi: 10.1159/000490140. https://10.1159/000490140 [DOI] [PubMed] [Google Scholar]
- 46.Zhao L., Huang L., Zhang J., Fan J., He F., Zhao X., Wang H., Liu Q., Shi D., Ni N., Wagstaff W., Pakvasa M., Fu K., Tucker A.B., Chen C., Reid R.R., Haydon R.C., Luu H.H., Shen L., Qi H., He T.C. The inhibition of BRAF activity sensitizes chemoresistant human ovarian cancer cells to paclitaxel-induced cytotoxicity and tumor growth inhibition. Am J Transl Res. 2020;12:8084–8098. [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang F., Li Y., Zhang H., Huang E., Gao L., Luo W., Wei Q., Fan J., Song D., Liao J., Zou Y., Liu F., Liu J., Huang J., Guo D., Ma C., Hu X., Li L., Qu X., Chen L., Yu X., Zhang Z., Wu T., Luu H.H., Haydon R.C., Song J., He T.C., Ji P. Anthelmintic mebendazole enhances cisplatin's effect on suppressing cell proliferation and promotes differentiation of head and neck squamous cell carcinoma (HNSCC) Oncotarget. 2017;8:12968–12982. doi: 10.18632/oncotarget.14673. https://10.18632/oncotarget.14673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao D., Lei Y., Ye Z., Zhao L., Wang H., Zhang J., He F., Huang L., Shi D., Liu Q., Ni N., Pakvasa M., Wagstaff W., Zhao X., Fu K., Tucker A.B., Chen C., Reid R.R., Haydon R.C., Luu H.H., He T.C., Liao Z. Blockade of IGF/IGF-1R signaling axis with soluble IGF-1R mutants suppresses the cell proliferation and tumor growth of human osteosarcoma. Am J Cancer Res. 2020;10:3248–3266. [PMC free article] [PubMed] [Google Scholar]
- 49.Hu X., Li L., Yu X., Zhang R., Yan S., Zeng Z., Shu Y., Zhao C., Wu X., Lei J., Li Y., Zhang W., Yang C., Wu K., Wu Y., An L., Huang S., Ji X., Gong C., Yuan C., Zhang L., Liu W., Huang B., Feng Y., Zhang B., Haydon R.C., Luu H.H., Reid R.R., Lee M.J., Wolf J.M., Yu Z., He T.C. CRISPR/Cas9-mediated reversibly immortalized mouse bone marrow stromal stem cells (BMSCs) retain multipotent features of mesenchymal stem cells (MSCs) Oncotarget. 2017;8:111847–111865. doi: 10.18632/oncotarget.22915. https://10.18632/oncotarget.22915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang N., Zhang W., Cui J., Zhang H., Chen X., Li R., Wu N., Chen X., Wen S., Zhang J., Yin L., Deng F., Liao Z., Zhang Z., Zhang Q., Yan Z., Liu W., Ye J., Deng Y., Wang Z., Qiao M., Luu H.H., Haydon R.C., Shi L.L., Liang H., He T.C. The piggyBac transposon-mediated expression of SV40 T antigen efficiently immortalizes mouse embryonic fibroblasts (MEFs) PloS One. 2014;9 doi: 10.1371/journal.pone.0097316. https://10.1371/journal.pone.0097316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li R., Zhang W., Yan Z., Liu W., Fan J., Feng Y., Zeng Z., Cao D., Haydon R.C., Luu H.H., Deng Z.L., He T.C., Zou Y. Long non-coding RNA (LncRNA) HOTAIR regulates BMP9-induced osteogenic differentiation by targeting the proliferation of mesenchymal stem cells (MSCs) Aging (N Y) 2021;13:4199–4214. doi: 10.18632/aging.202384. https://10.18632/aging.202384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luo W., Zhang L., Huang B., Zhang H., Zhang Y., Zhang F., Liang P., Chen Q., Cheng Q., Tan D., Tan Y., Song J., Zhao T., Haydon R.C., Reid R.R., Luu H.H., Lee M.J., El Dafrawy M., Ji P., He T.C., Gou L. BMP9-initiated osteogenic/odontogenic differentiation of mouse tooth germ mesenchymal cells (TGMCS) requires Wnt/beta-catenin signalling activity. J. Cell Mol. Med. 2021;25:2666–2678. doi: 10.1111/jcmm.16293. https://10.1111/jcmm.16293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao J., Wei Q., Zou Y., Fan J., Song D., Cui J., Zhang W., Zhu Y., Ma C., Hu X., Qu X., Chen L., Yu X., Zhang Z., Wang C., Zhao C., Zeng Z., Zhang R., Yan S., Wu T., Wu X., Shu Y., Lei J., Li Y., Luu H.H., Lee M.J., Reid R.R., Ameer G.A., Wolf J.M., He T.C., Huang W. Notch signaling augments BMP9-induced bone formation by promoting the osteogenesis-angiogenesis coupling process in mesenchymal stem cells (MSCs) Cell. Physiol. Biochem. 2017;41:1905–1923. doi: 10.1159/000471945. https://10.1159/000471945 [DOI] [PubMed] [Google Scholar]
- 54.Luo J., Deng Z.L., Luo X., Tang N., Song W.X., Chen J., Sharff K.A., Luu H.H., Haydon R.C., Kinzler K.W., Vogelstein B., He T.C. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat. Protoc. 2007;2:1236–1247. doi: 10.1038/nprot.2007.135. https://10.1038/nprot.2007.135 [DOI] [PubMed] [Google Scholar]
- 55.Breyer B., Jiang W., Cheng H., Zhou L., Paul R., Feng T., He T.C. Adenoviral vector-mediated gene transfer for human gene therapy. Curr. Gene Ther. 2001;1:149–162. doi: 10.2174/1566523013348689. https://10.2174/1566523013348689 [DOI] [PubMed] [Google Scholar]
- 56.Lee C.S., Bishop E.S., Zhang R., Yu X., Farina E.M., Yan S., Zhao C., Zheng Z., Shu Y., Wu X., Lei J., Li Y., Zhang W., Yang C., Wu K., Wu Y., Ho S., Athiviraham A., Lee M.J., Wolf J.M., Reid R.R., He T.C. Adenovirus-mediated gene delivery: potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis. 2017;4:43–63. doi: 10.1016/j.gendis.2017.04.001. https://10.1016/j.gendis.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng Q., Chen B., Wang H., Zhu Y., Wu J., Luo Y., Zuo G., Luo J., Zhou L., Shi Q., Weng Y., Huang A., He T.C., Fan J. Bone morphogenetic protein 4 (BMP4) alleviates hepatic steatosis by increasing hepatic lipid turnover and inhibiting the mTORC1 signaling axis in hepatocytes. Aging (N Y) 2019;11:11520–11540. doi: 10.18632/aging.102552. https://10.18632/aging.102552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Denduluri S.K., Scott B., Lamplot J.D., Yin L., Yan Z., Wang Z., Ye J., Wang J., Wei Q., Mohammed M.K., Haydon R.C., Kang R.W., He T.C., Athiviraham A., Ho S.H., Shi L.L. Immortalized mouse achilles tenocytes demonstrate long-term proliferative capacity while retaining tenogenic properties. Tissue Eng. C Methods. 2016;22:280–289. doi: 10.1089/ten.tec.2015.0244. https://10.1089/ten.tec.2015.0244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo Q., Kang Q., Song W.X., Luu H.H., Luo X., An N., Luo J., Deng Z.L., Jiang W., Yin H., Chen J., Sharff K.A., Tang N., Bennett E., Haydon R.C., He T.C. Selection and validation of optimal siRNA target sites for RNAi-mediated gene silencing. Gene. 2007;395:160–169. doi: 10.1016/j.gene.2007.02.030. https://10.1016/j.gene.2007.02.030 [DOI] [PubMed] [Google Scholar]
- 60.Deng F., Chen X., Liao Z., Yan Z., Wang Z., Deng Y., Zhang Q., Zhang Z., Ye J., Qiao M., Li R., Denduluri S., Wang J., Wei Q., Li M., Geng N., Zhao L., Zhou G., Zhang P., Luu H.H., Haydon R.C., Reid R.R., Yang T., He T.C. A simplified and versatile system for the simultaneous expression of multiple siRNAs in mammalian cells using Gibson DNA Assembly. PloS One. 2014;9 doi: 10.1371/journal.pone.0113064. https://10.1371/journal.pone.0113064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mao Y., Ni N., Huang L., Fan J., Wang H., He F., Liu Q., Shi D., Fu K., Pakvasa M., Wagstaff W., Tucker A.B., Chen C., Reid R.R., Haydon R.C., Ho S.H., Lee M.J., He T.-C., Yang J., Shen L., Cai L., Luu H.H. Argonaute (AGO) proteins play an essential role in mediating BMP9-induced osteogenic signaling in mesenchymal stem cells (MSCs) Genes & Diseases. 2021 doi: 10.1016/j.gendis.2021.04.004. https://10.1016/j.gendis.2021.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao C., Wu N., Deng F., Zhang H., Wang N., Zhang W., Chen X., Wen S., Zhang J., Yin L., Liao Z., Zhang Z., Zhang Q., Yan Z., Liu W., Wu D., Ye J., Deng Y., Zhou G., Luu H.H., Haydon R.C., Si W., He T.C. Adenovirus-mediated gene transfer in mesenchymal stem cells can be significantly enhanced by the cationic polymer polybrene. PloS One. 2014;9 doi: 10.1371/journal.pone.0092908. https://10.1371/journal.pone.0092908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang N., Zhang H., Zhang B.Q., Liu W., Zhang Z., Qiao M., Zhang H., Deng F., Wu N., Chen X., Wen S., Zhang J., Liao Z., Zhang Q., Yan Z., Yin L., Ye J., Deng Y., Luu H.H., Haydon R.C., Liang H., He T.C. Adenovirus-mediated efficient gene transfer into cultured three-dimensional organoids. PloS One. 2014;9 doi: 10.1371/journal.pone.0093608. https://10.1371/journal.pone.0093608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Law J.X., Chowdhury S.R., Saim A.B., Idrus R.B.H. Platelet-rich plasma with keratinocytes and fibroblasts enhance healing of full-thickness wounds. J. Tissue Viability. 2017;26:208–215. doi: 10.1016/j.jtv.2017.05.003. https://10.1016/j.jtv.2017.05.003 [DOI] [PubMed] [Google Scholar]
- 65.Zhu Y., Shi Q., Peng Q., Gao Y., Yang T., Cheng Y., Wang H., Luo Y., Huang A., He T.C., Fan J. A simplified 3D liver microsphere tissue culture model for hepatic cell signaling and drug-induced hepatotoxicity studies. Int. J. Mol. Med. 2019;44:1653–1666. doi: 10.3892/ijmm.2019.4321. https://10.3892/ijmm.2019.4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu X., Li Z., Zhang H., He F., Qiao M., Luo H., Zhang J., Zhang M., Mao Y., Wagstaff W., Zhang Y., Niu C., Zhao X., Wang H., Huang L., Shi D., Liu Q., Ni N., Fu K., Haydon R.C., Reid R.R., Luu H.H., He T.-C., Wang Z., Liang H., Zhang B.-Q., Wang N. Modeling colorectal tumorigenesis using the organoids derived from conditionally immortalized mouse intestinal crypt cells (ciMICs) Genes & Diseases. 2021 doi: 10.1016/j.gendis.2021.01.004. https://10.1016/j.gendis.2021.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou L., An N., Jiang W., Haydon R., Cheng H., Zhou Q., Breyer B., Feng T., He T.C. Fluorescence-based functional assay for Wnt/beta-catenin signaling activity. Biotechniques. 2002;33:1126–1128. doi: 10.2144/02335dd07. https://10.2144/02335dd07 30, 32 passim. [DOI] [PubMed] [Google Scholar]
- 68.Li R., Yan Z., Ye J., Huang H., Wang Z., Wei Q., Wang J., Zhao L., Lu S., Wang X., Tang S., Fan J., Zhang F., Zou Y., Song D., Liao J., Lu M., Liu F., Shi L.L., Athiviraham A., Lee M.J., He T.C., Zhang Z. The prodomain-containing BMP9 produced from a stable line effectively regulates the differentiation of mesenchymal stem cells. Int. J. Med. Sci. 2016;13:8–18. doi: 10.7150/ijms.13333. https://10.7150/ijms.13333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu D.N. Methodology for evaluation of melanin content and production of pigment cells in vitro. Photochem. Photobiol. 2008;84:645–649. doi: 10.1111/j.1751-1097.2007.00228.x. https://10.1111/j.1751-1097.2007.00228.x [DOI] [PubMed] [Google Scholar]
- 70.Masson-Meyers D.S., Andrade T.A.M., Caetano G.F., Guimaraes F.R., Leite M.N., Leite S.N., Frade M.A.C. Experimental models and methods for cutaneous wound healing assessment. Int. J. Exp. Pathol. 2020;101:21–37. doi: 10.1111/iep.12346. https://10.1111/iep.12346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao C., Qazvini N.T., Sadati M., Zeng Z., Huang S., De La Lastra A.L., Zhang L., Feng Y., Liu W., Huang B., Zhang B., Dai Z., Shen Y., Wang X., Luo W., Liu B., Lei Y., Ye Z., Zhao L., Cao D., Yang L., Chen X., Athiviraham A., Lee M.J., Wolf J.M., Reid R.R., Tirrell M., Huang W., de Pablo J.J., He T.C. A pH-triggered, self-assembled, and bioprintable hybrid hydrogel scaffold for mesenchymal stem cell based bone tissue engineering. ACS Appl. Mater. Interfaces. 2019;11:8749–8762. doi: 10.1021/acsami.8b19094. https://10.1021/acsami.8b19094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Z., Liu J., Zeng Z., Fan J., Huang S., Zhang L., Zhang B., Wang X., Feng Y., Ye Z., Zhao L., Cao D., Yang L., Pakvasa M., Liu B., Wagstaff W., Wu X., Luo H., Zhang J., Zhang M., He F., Mao Y., Ding H., Zhang Y., Niu C., Haydon R.C., Luu H.H., Lee M.J., Wolf J.M., Shao Z., He T.C. lncRNA Rmst acts as an important mediator of BMP9-induced osteogenic differentiation of mesenchymal stem cells (MSCs) by antagonizing Notch-targeting microRNAs. Aging (N Y) 2019;11:12476–12496. doi: 10.18632/aging.102583. https://10.18632/aging.102583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu N., Jiang D., Huang E., Liu X., Li R., Liang X., Kim S.H., Chen X., Gao J.L., Zhang H., Zhang W., Kong Y.H., Zhang J., Wang J., Shui W., Luo X., Liu B., Cui J., Rogers M.R., Shen J., Zhao C., Wang N., Wu N., Luu H.H., Haydon R.C., He T.C., Huang W. BMP9-regulated angiogenic signaling plays an important role in the osteogenic differentiation of mesenchymal progenitor cells. J. Cell Sci. 2013;126:532–541. doi: 10.1242/jcs.114231. https://10.1242/jcs.114231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang X., Wang F., Zhao C., Yang S., Cheng Q., Tang Y., Zhang F., Zhang Y., Luo W., Wang C., Zhou P., Kim S., Zuo G., Hu N., Li R., He T.C., Zhang H. Dentinogenesis and tooth-alveolar bone complex defects in BMP9/GDF2 knockout mice. Stem Cell. Dev. 2019;28:683–694. doi: 10.1089/scd.2018.0230. https://10.1089/scd.2018.0230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jha K.K., Banga S., Palejwala V., Ozer H.L. SV40-Mediated immortalization. Exp. Cell Res. 1998;245:1–7. doi: 10.1006/excr.1998.4272. https://10.1006/excr.1998.4272 [DOI] [PubMed] [Google Scholar]
- 76.Ishida-Yamamoto A., Igawa S., Kishibe M. Molecular basis of the skin barrier structures revealed by electron microscopy. Exp. Dermatol. 2018;27:841–846. doi: 10.1111/exd.13674. https://10.1111/exd.13674 [DOI] [PubMed] [Google Scholar]
- 77.Barrientos S., Stojadinovic O., Golinko M.S., Brem H., Tomic-Canic M. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008;16:585–601. doi: 10.1111/j.1524-475X.2008.00410.x. https://10.1111/j.1524-475X.2008.00410.x [DOI] [PubMed] [Google Scholar]
- 78.Green H., Easley K., Iuchi S. Marker succession during the development of keratinocytes from cultured human embryonic stem cells. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15625–15630. doi: 10.1073/pnas.0307226100. https://10.1073/pnas.0307226100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hirobe T., Hasegawa K., Furuya R., Fujiwara R., Sato K. Effects of fibroblast-derived factors on the proliferation and differentiation of human melanocytes in culture. J. Dermatol. Sci. 2013;71:45–57. doi: 10.1016/j.jdermsci.2013.03.012. https://10.1016/j.jdermsci.2013.03.012 [DOI] [PubMed] [Google Scholar]
- 80.Zomer H.D., Trentin A.G. Skin wound healing in humans and mice: challenges in translational research. J. Dermatol. Sci. 2018;90:3–12. doi: 10.1016/j.jdermsci.2017.12.009. https://10.1016/j.jdermsci.2017.12.009 [DOI] [PubMed] [Google Scholar]
- 81.Werner S., Krieg T., Smola H. Keratinocyte-fibroblast interactions in wound healing. J. Invest. Dermatol. 2007;127:998–1008. doi: 10.1038/sj.jid.5700786. https://10.1038/sj.jid.5700786 [DOI] [PubMed] [Google Scholar]
- 82.Maqsood M.I., Matin M.M., Bahrami A.R., Ghasroldasht M.M. Immortality of cell lines: challenges and advantages of establishment. Cell Biol. Int. 2013;37:1038–1045. doi: 10.1002/cbin.10137. https://10.1002/cbin.10137 [DOI] [PubMed] [Google Scholar]
- 83.Song D., Zhang F., Reid R.R., Ye J., Wei Q., Liao J., Zou Y., Fan J., Ma C., Hu X., Qu X., Chen L., Li L., Yu Y., Yu X., Zhang Z., Zhao C., Zeng Z., Zhang R., Yan S., Wu T., Wu X., Shu Y., Lei J., Li Y., Zhang W., Wang J., Lee M.J., Wolf J.M., Huang D., He T.C. BMP9 induces osteogenesis and adipogenesis in the immortalized human cranial suture progenitors from the patent sutures of craniosynostosis patients. J. Cell Mol. Med. 2017;21:2782–2795. doi: 10.1111/jcmm.13193. https://10.1111/jcmm.13193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang J., Liao J., Zhang F., Song D., Lu M., Liu J., Wei Q., Tang S., Liu H., Fan J., Zou Y., Guo D., Huang J., Liu F., Ma C., Hu X., Li L., Qu X., Chen L., Weng Y., Lee M.J., He T.C., Reid R.R., Zhang J. NEL-like molecule-1 (Nell1) is regulated by bone morphogenetic protein 9 (BMP9) and potentiates BMP9-induced osteogenic differentiation at the expense of adipogenesis in mesenchymal stem cells. Cell. Physiol. Biochem. 2017;41:484–500. doi: 10.1159/000456885. https://10.1159/000456885 [DOI] [PubMed] [Google Scholar]
- 85.Wong V.W., Gurtner G.C., Longaker M.T. Wound healing: a paradigm for regeneration. Mayo Clin. Proc. 2013;88:1022–1031. doi: 10.1016/j.mayocp.2013.04.012. https://10.1016/j.mayocp.2013.04.012 [DOI] [PubMed] [Google Scholar]
- 86.Coraux C., Hilmi C., Rouleau M., Spadafora A., Hinnrasky J., Ortonne J.P., Dani C., Aberdam D. Reconstituted skin from murine embryonic stem cells. Curr. Biol. 2003;13:849–853. doi: 10.1016/s0960-9822(03)00296-3. https://10.1016/s0960-9822(03)00296-3 [DOI] [PubMed] [Google Scholar]
- 87.Ohyama M., Okano H. Promise of human induced pluripotent stem cells in skin regeneration and investigation. J. Invest. Dermatol. 2014;134:605–609. doi: 10.1038/jid.2013.376. https://10.1038/jid.2013.376 [DOI] [PubMed] [Google Scholar]
- 88.Krause D.S., Theise N.D., Collector M.I., Henegariu O., Hwang S., Gardner R., Neutzel S., Sharkis S.J. Multi-organ, multi-lineage engraftment by a single bone marrow-derived stem cell. Cell. 2001;105:369–377. doi: 10.1016/s0092-8674(01)00328-2. https://10.1016/s0092-8674(01)00328-2 [DOI] [PubMed] [Google Scholar]
- 89.Wood F.M., Giles N., Stevenson A., Rea S., Fear M. Characterisation of the cell suspension harvested from the dermal epidermal junction using a ReCell(R) kit. Burns. 2012;38:44–51. doi: 10.1016/j.burns.2011.03.001. https://10.1016/j.burns.2011.03.001 [DOI] [PubMed] [Google Scholar]
- 90.Holmes Iv J.H., Molnar J.A., Carter J.E., Hwang J., Cairns B.A., King B.T., Smith D.J., Cruse C.W., Foster K.N., Peck M.D., Sood R., Feldman M.J., Jordan M.H., Mozingo D.W., Greenhalgh D.G., Palmieri T.L., Griswold J.A., Dissanaike S., Hickerson W.L. A comparative study of the ReCell(R) Device and autologous spit-thickness meshed skin graft in the treatment of acute burn injuries. J. Burn Care Res. 2018;39:694–702. doi: 10.1093/jbcr/iry029. https://10.1093/jbcr/iry029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee C.S., Bishop E.S., Dumanian Z., Zhao C., Song D., Zhang F., Zhu Y., Ameer G.A., He T.C., Reid R.R. Bone morphogenetic protein-9-stimulated adipocyte-derived mesenchymal progenitors entrapped in a thermoresponsive nanocomposite scaffold facilitate cranial defect repair. J. Craniofac. Surg. 2019;30:1915–1919. doi: 10.1097/SCS.0000000000005465. https://10.1097/SCS.0000000000005465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiao J., Chen S., Yi J., Zhang H., Ameer G.A. A cooperative copper metal-organic framework-hydrogel system improves wound healing in diabetes. Adv. Funct. Mater. 2017;27 doi: 10.1002/adfm.201604872. https://10.1002/adfm.201604872 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.