Abstract
Calpains are a family of Ca2+-dependent intracellular cysteine proteases, including the ubiquitously expressed μ- and m-calpains. Both μ- and m-calpains are heterodimers, consisting of a distinct large 80-kDa catalytic subunit, encoded by the genes Capn1 and Capn2, and a common small 28-kDa regulatory subunit (Capn4). The physiological roles and possible functional distinctions of μ- and m-calpains remain unclear, but suggested functions include participation in cell division and migration, integrin-mediated signal transduction, apoptosis, and regulation of cellular control proteins such as cyclin D1 and p53. Homozygous disruption of murine Capn4 eliminated both μ- and m-calpain activities, but this did not affect survival and proliferation of cultured embryonic stem cells or embryonic fibroblasts, or the early stages of organogenesis. However, mutant embryos died at midgestation and displayed defects in the cardiovascular system, hemorrhaging, and accumulation of erythroid progenitors.
The two forms of calpain found as stable proteins in mammalian tissues are known as μ-calpain and m-calpain (7, 39, 43, 53, 58). The enzymes are cytosolic thiol proteases, absolutely dependent on Ca2+ for activity. They both consist of an 80-kDa large subunit (from the genes Capn1 and Capn2, respectively), each of which forms a heterodimer with a common 28-kDa small subunit (Capn4). The structure of these enzymes was first predicted from their amino acid sequence (42) and has been established very recently by X-ray crystallography (25, 56). The large subunits contain a thiol protease region (domains I and II) related to the papain and cathepsin families, a C2-like domain III, and a C-terminal, calmodulin-like domain (domain IV), which binds Ca2+ at several EF hands. The small subunit consists of an N-terminal domain V, containing ∼30% glycine residues, and a C-terminal Ca2+-binding domain VI very similar to domain IV (6, 31). The two subunits are bound to each other mainly through contacts between domains IV and VI (25).
Historically it has been assumed that the calpain small subunit is essential for μ- and m-calpain activity. The two enzymes are invariably found as heterodimers in tissue extracts, and expression of the m-calpain large subunit alone in Escherichia coli gave a low yield of soluble protein with no activity (21). Expression of the μ-calpain large subunit alone has, however, been reported to yield some calpain activity, or to give an insoluble product which was active following renaturation (37, 62), suggesting that at least one function of the small subunit is to act as a chaperone in assisting folding of the large subunit (58, 69, 70). It has also been reported that Ca2+ activation of calpain involves subunit dissociation and that the large subunits formed in this way can be active as monomers (58). There are some reports which do not agree with this hypothesis (12, 71), but the question should be considered open.
In recent years, several other calpain-related cDNA sequences have been described, but there is little information on the occurrence or function of the corresponding proteins (5, 9, 20, 33). Calpain 3 or p94 (Capn3) has attracted great interest since deficiency of calpain 3 causes limb girdle muscular dystrophy 2A (3, 18, 23, 28, 45). The natural substrates of calpain 3 are not known, and the protein has an extremely short half-life in vivo, making it very difficult to assay. In the context of this study on disruption of the Capn4 gene, it may be noted that calpain 3 does not appear to require a calpain small subunit or to be Ca2+ dependent. Calpain 3 mRNA is expressed briefly in human fetal heart but later only in skeletal muscle, while in the mouse embryo, it could not be detected before day 11.5 of gestation (embryonic day 11.5 [E11.5]) (3, 18, 23). The conventional calpains have not been extensively studied in the embryo, but m-calpain was detected in chick embryonic fibroblasts (30), and expression in the mouse embryo of mRNAs for Capn5, -6, and -11 has recently been described (10).
The physiological roles of the calpains remain unclear (7, 43). The presence and conservation of μ- and m-calpains in almost all mammalian cells suggest that these enzymes are essential, but the absence of fully specific calpain inhibitors has so far prevented unambiguous proof of a particular physiological role. Many of the calpain inhibitors commonly used in earlier work have been shown also to inhibit the proteasome (36) or cathepsins or to inhibit entirely different enzymes, for example, a protein tyrosine phosphatase (49). The calpain genes have no major regulatory features in their promoter regions and are usually considered to be housekeeping enzymes. The relative levels of μ- and m-calpains and of their inhibitor, calpastatin, vary from tissue to tissue, again suggesting some degree of regulation and importance to the cell (59). Their Ca2+ dependence suggests a link to signal transduction (19), and the common assumption that the calpains become membrane bound on exposure to Ca2+ (35, 39) suggests that cytoskeletal proteins may be among the favored substrates. The list of proposed calpain substrates is very long (19, 63), and many reports have suggested that calpain may be involved in cell adhesion, spreading, and migration (26, 29, 49), myoblast fusion (4, 11), cell cycle control (36), and mitosis (50).
Apoptosis is another cell function for which there are conflicting reports about the involvement of calpain. The existence of numerous caspases (41), the problems of inhibitor specificity, and the multiplicity of apoptosis pathways and experimental cell systems have led to many conflicting reports about the role of calpain (34, 46, 66, 68). Several members of the Bcl-2 and Bax families, which are important in apoptosis, can be cleaved both by caspases and by calpain (14). Calpain was reported to be involved in radiation-induced apoptosis, upstream of caspase 3 (64), but caspase activation was found to be upstream of calpain activation in drug-induced apoptosis (67). Calpain activity was found to cause apoptosis of neutrophils, but Fas antigen-induced apoptosis was independent of calpain (54, 55).
With the intention of resolving some of these questions, we have therefore generated both heterozygous Capn4+/− and homozygous Capn4−/− embryonic stem (ES) cells and have attempted to generate the corresponding mice. The work was based on the assumption that loss of a functional Capn4 gene would abolish both μ- and m-calpain activity. The physiological consequences of this mutation were impossible to predict, and it was not known whether any other forms of calpain, or other protease systems entirely, might compensate for the absence of the classical μ- and m-calpains.
The results have shown, as predicted, that homozygous Capn4−/− mouse cells lack μ- and m-calpain activity as measured by casein zymography, but even in the absence of detectable calpain activity, homozygous Capn4−/− ES cells and embryonic fibroblasts grow and divide apparently normally. However, the large subunit genes have not been altered and are still transcribed, although the resultant large subunit proteins are unstable in the absence of the small subunit. It is not impossible that these isolated large subunits may still exert some calpain activity, even if transient and unregulated, but no assay to detect this activity unambiguously is available.
Capn4+/− mice were viable, fertile, and phenotypically normal, but the Capn4−/− embryos died at midgestation, with indications of defects in both vasculogenesis and erythropoiesis. Embryonic development is thus absolutely dependent on the presence of a functional Capn4 gene, and we assume therefore on the presence of normal levels of at least one of μ- and m-calpains in their normal heterodimeric form.
MATERIALS AND METHODS
Disruption of the calpain small subunit gene.
Genomic and cDNA clones of the mouse calpain small subunit (Capn4) gene have been described elsewhere (1) (accession no. AF058297, AF058298, and AF139373). The gene contains 11 exons. The left (upstream) arm of the targeting vector consisted of a 2-kb genomic PvuII fragment, extending from intron 6 to the middle of exon 9. The right (downstream) arm was a 4-kb BamHI-HindIII genomic DNA fragment extending from intron 9 and including exons 10 and 11, to a point ∼1.5 kb downstream of the polyadenylation signal. These DNA segments were inserted on either side of the PGK-neo cassette in pPNT (Fig. 1) (61). This construct replaces approximately 450 bp, including the 3′-terminal half of Capn4 exon 9, with the PGK-neo cassette. The thymidine kinase cassette was included in the vector to permit negative selection of nonhomologous recombination events with ganciclovir (GANC). This plasmid was linearized by NotI digestion for electroporation into R1 ES cells, and transformed cells were selected for G418 and GANC resistance by growth in the presence of 0.2 mg of G418 (Gibco/BRL, Burlington, Ontario, Canada) per ml and 2 μM GANC (Syntex, Inc.) for 8 to 9 days (24). Homozygous mutant ES cells (Capn4−/−) were obtained by means of further growth and selection of heterozygous (Capn4+/−) ES cells in the presence of 1 mg of G418 per ml.
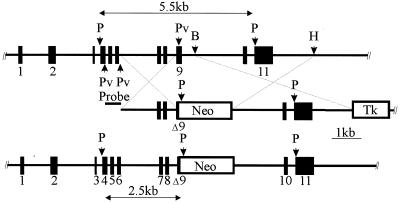
FIG. 1.
Targeted disruption of the calpain small subunit gene. Schematic representations show the murine Capn4 allele (upper), the targeting vector (middle), and the targeted allele (lower). The Capn4 gene was cloned from the 129SvJ mouse strain (1). Positions of PstI (P), PvuII (Pv), BamHI (B), and HindIII (H) restriction endonuclease sites are indicated. Homologous recombination replaces a 441-bp PvuII-BamHI fragment containing the 3′ end of Capn4 exon 9 with the neo cassette. Tk, thymidine kinase.
Genotyping.
Initial screening for Capn4+/− ES cell clones was carried out by PCR, using a sense-strand primer located just upstream of the 2-kb left arm of homology and an antisense-strand primer located within the PGK-neo cassette. Tentatively identified Capn4+/− ES cell clones were characterized by Southern blot hybridization of PstI-digested genomic DNA with the 487-bp PvuII fragment located upstream of the 2-kb left arm of homology. This probe recognized a 5.5-kb PstI fragment from the wild-type Capn4 locus and a 2.5-kb PstI fragment from the targeted Capn4 locus (Fig. 1).
Germ line transmission.
Germ line transmission of the Capn4 mutant allele was achieved by the “darning needle” aggregation chimera method (40). Briefly, eight-cell embryos were recovered from CD1 matings at a point prior to blastomere compaction. Acid Tyrode's buffer was used to remove the zona pellucida, and the embryos were placed in smooth depressions made with a darning needle in 35-mm-diameter culture dishes. Clumps of 8 to 12 ES cells were placed adjacent to each embryo within the depressions. During overnight culture, the ES cells were absorbed into the compacting morula, and those embryos which developed further to the blastocyst stage were transferred into the uteri of surrogate mothers. At term, chimeric animals were identified first by black eye pigmentation and subsequently by patches of agouti coat color. Chimeric males were initially bred with CD1 females to identify males capable of germ line transmission of the mutant allele. These males were subsequently bred with 129SvJ females to establish the mutant allele in an inbred state.
Northern blotting.
Total RNA was isolated from ES cells using TRIzol reagent (Gibco/BRL) according to the manufacturer's instructions, and poly(A)+ RNA was fractionated using the polyATtract system (Promega, Madison, Wis.). Samples of poly(A)+ RNA were resolved on 1.2% formaldehyde agarose gels and transferred to nylon membranes (65). The membranes were probed with [α-32P]dATP-labeled cDNA fragments (15) for calpain subunits as follows: for the mouse m-calpain large subunit, EST ms22d05.ri (mouse calpain 2; accession no. AA168283), obtained from IMAGE Consortium, Lawrence Livermore National Laboratory; for the mouse μ-calpain large subunit, a rat μ-calpain large subunit cDNA cloned in this laboratory with kind assistance from H. Sorimachi (52); and for the mouse calpain small subunit, a cDNA coding for the C-terminal domain VI of the mouse calpain small subunit and the 3′ untranslated region (1). Equal gel loading was confirmed both by ethidium bromide staining and by stripping the blots and reprobing with an actin cDNA.
Separation and analysis of μ- and m-calpain activities.
Calpain activities in cell and tissue extracts were detected by casein zymography (2, 44). Cultured cells were scraped into lysis buffer (50 mM HEPES [pH 7.6], 150 mM NaCl, 1% Triton X-100, 5 mM EDTA, 10 mM 2-mercaptoethanol, 0.1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin per ml) without trypsin treatment. E9.5-E10.5 embryos were homogenized by pipetting up and down in this buffer. After centrifugation to remove particulate debris, aliquots representing 30 μg of protein from cell extracts, or 20 to 40% of each soluble embryo extract, were mixed with nondenaturing gel loading buffer and electrophoresed in nondenaturing polyacrylamide gels containing casein. Calpain activity was detected by incubating the gels overnight with Ca2+, followed by fixation and staining with Coomassie brilliant blue. Calpain activities were observed as colorless regions cleared of casein in a blue background.
Gel electrophoresis, immunoblotting, and antibodies.
Protein samples denatured in sodium dodecyl sulfate (SDS) sample buffer were run on SDS-polyacrylamide gels (9%) in Tris-Tricine buffer (48) and blotted onto Immobilon membranes as previously described (60). A polyclonal antibody which binds the large (rat Capn2) subunit of rat and mouse m-calpain has been described elsewhere (47). Antibodies to the mitogen-activated protein kinase ERK1 (K-23) were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif.).
ES cell culture.
Fetal bovine serum was from HyClone Laboratories Inc. (Logan, Utah) and was tested for its ability to support ES cell growth. Gelatin was from Sigma-Aldrich Canada (Oakville, Ontario, Canada). All other reagents for tissue culture were from Gibco/BRL. Murine ES cells were maintained on gelatinized plates at 37°C under 5% CO2 in ES medium (Dulbecco modified Eagle medium [high glucose] supplemented with 15% fetal bovine serum, 0.1 mM nonessential amino acids, antibiotics [penicillin, 100 U/ml; streptomycin, 100 μg/ml; amphotericin, 25 μg/ml], 2 mM l-glutamine, 1 mM sodium pyruvate, 0.1 mM 2-mercaptoethanol, and 1,000 U of recombinant leukemia inhibitory factor per ml).
Histological analysis.
Time of fertilization was determined by observation of copulation plugs, and noon of that day was defined as E0.5. Embryos were dissected from pregnant Capn4+/− females that had been bred with Capn4+/− males, and the yolk sacs were separated and used for genotype analysis. Embryos were fixed overnight in 4% paraformaldehyde in phosphate-buffered saline (pH 7.2), dehydrated through graded ethanol, and embedded in paraffin wax. Serial cross or sagittal 7-μm sections were dewaxed in xylene, rehydrated through graded ethanol, and stained with hematoxylin and eosin. TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) assays were performed on dewaxed sections using the DeadEnd colorimetric apoptosis detection system from Promega according to the manufacturer's instructions. Photographs were taken with a Kodak DC120 digital camera mounted on a Nikon inverted microscope using an MDS 120 Universal C-mount adapter.
RESULTS
The murine Capn4 gene (1) was disrupted by replacement of the 3′-terminal half of exon 9 with a PGK-neo cassette (61) (Fig. 1). This construct may give rise by alternative splicing of the RNA transcript to several possible forms of the small subunit, all having deletions or disruptions of the 30 to 40 C-terminal amino acids. These altered subunits are unlikely to be functional, since the C-terminal residues are known to be essential for activity and for heterodimer formation with the large subunit (13, 25).
ES cells.
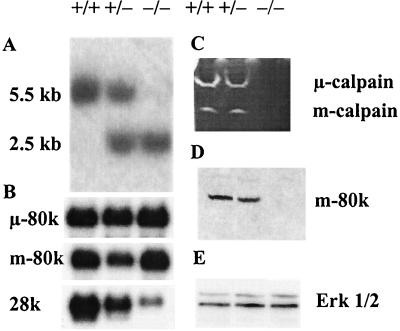
Two independent targeted Capn4+/− ES cell clones were generated, and one of these was converted to the homozygous mutant state (Capn4−/−) by further selection of cells in elevated G418 concentrations. The ES cell genotypes were characterized by Southern blotting, showing the predicted wild-type 5.5-kb and targeted 2.5-kb PstI bands (Fig. 2A). Northern blot analysis (Fig. 2B) showed that the levels of μ-calpain 80-kDa subunit (Capn1) mRNA were equal in all three lines. The levels of m-calpain 80-kDa subunit (Capn2) mRNA were equal in Capn4+/+ and Capn4−/− ES cells but were slightly reduced in some Capn4+/− ES cell lines. The level of the calpain small subunit (Capn4) mRNA was significantly lower in Capn4+/− ES cells than in Capn4+/+ ES cells, consistent with the presence of only one correct allele. In Capn4−/− ES cells, a trace amount of an mRNA of approximately the same size was detected, which is assumed to represent an unstable or low abundance mRNA arising from the disrupted Capn4 gene.
FIG. 2.
Analysis of ES cells. (A) Southern blotting. A 487-bp exon 5/6-containing PvuII fragment (Fig. 1) was used as a probe in Southern blot hybridization analysis of DNA isolated from Capn4+/+, Capn4+/−, and Capn4−/− ES cells. PstI fragments of 5.5 or 2.5 kb are diagnostic of the wild-type or targeted Capn4 allele, respectively. (B) Northern blotting. Equal amounts of poly(A)+ RNA were electrophoresed on 1.2% formaldehyde agarose gels and transferred to nylon membranes. The membranes were probed with [α-32P]dATP-labeled cDNA fragments specific for the mouse μ- and m-calpain large subunits and the mouse calpain small subunit. The final washes were in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS at 52°C. Equal gel loading was confirmed both by ethidium bromide staining and by stripping the blots and reprobing with an actin cDNA. (C) Casein zymogram. ES cells were lysed in lysis buffer (see Materials and Methods). The lysates were centrifuged to remove cell debris, and portions of the supernatants containing 30 μg of protein were run on casein zymograms. After electrophoresis, the gels were rinsed and incubated overnight in the presence of 5 mM Ca2+ and 10 mM 2-mercaptoethanol. (D and E) Immunoblot analysis. Equal amounts of protein denatured in SDS sample buffer were run on SDS-polyacrylamide gels (9%) in Tris-Tricine buffer and then blotted onto Immobilon. The calpain m-calpain large subunit (80 kDa) (D) and the control Erk1/2 kinase proteins (42 and 44 kDa) (E) were detected with specific antibodies, followed by horseradish peroxidase-labeled second antibodies and enhanced chemiluminescence detection. The Capn4 genotype of the cells represented in each lane is shown as +/+, +/−, or −/−.
Calpain activity was assayed in ES cell lysates by casein zymography. Both μ- and m-calpain activities were observed in wild-type Capn4+/+ ES cells and in heterozygous Capn4+/− cells, but neither activity was detectable in Capn4−/− cells (Fig. 2C).
Immunoblotting analysis showed that the m-calpain large subunit protein was present in equal amounts in Capn4+/+ and Capn4+/− ES cells but was either absent or detectable only in trace amounts in Capn4−/− ES cells (Fig. 2D). Reduced levels of m-calpain large subunit protein were also seen in Capn4−/− embryos (Fig. 4C). No satisfactory immunoblots have been obtained for the μ-calpain large subunit or for the small subunit, owing to the lack of appropriate antibodies. Commercial antibodies claiming specificity for these mouse proteins were purchased but were found to bind only to irrelevant proteins in wild-type mouse tissue extracts. As a control, the expression of Erk1/2 kinase was also assessed by immunoblotting (Fig. 2E). Approximately equal protein loading in the immunoblot analysis was also demonstrated by Coomassie blue staining (data not shown).
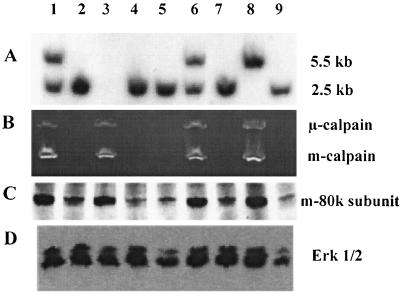
FIG. 4.
Analysis of E10.5 embryos. (A) Genotypes of individual embryos were determined by Southern blotting analysis of DNA from yolk sacs as described in the legend to Fig. 2A. No result was obtained for embryo 3, which is, however, seen in panels B and C to be either +/+ or +/−. (B) Calpain activity determined in individual embryo lysates as described in the legend to Fig. 2C. (C and D) Immunoblotting analysis of mouse m-calpain large subunit (C) and Erk1/2 kinase (D), performed as described in the legend to Fig. 2.
Growth rate.
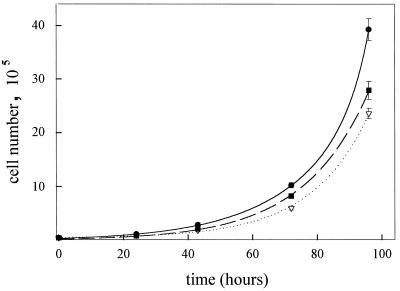
The growth rates of the different ES cell lines in ES medium were measured over 4 days, and the results are shown in Fig. 3. A simple exponential fit of the data to an equation of the form y = y0.eat gave population doubling times as follows: Capn4+/+ ES cells, 13.1 ± 0.7 h; Capn4+/− ES cells, 14.0 ± 0.2 h; and Capn4−/− ES cells, 12.7 ± 0.4 h.
FIG. 3.
ES cell growth curves. Several six-well plates were prepared with ES cells grown in ES medium. The cells were harvested with trypsin at 20- to 24-h intervals up to 96 h and counted by means of a Coulter counter. The data points and error bars (not all of which are visible) indicate the average cell number and range of three observations. The lines are exponential curves fitted to the data for Capn4+/+ (●), Capn4+/− (▿), and Capn4−/− (■) ES cells.
Embryonic fibroblasts.
Primary fibroblast cultures were established from E9.5 embryos, and the growth characteristics of Capn4−/− embryonic fibroblasts were indistinguishable from those of Capn4+/+ or Capn4+/− genotypes. Casein zymography of fibroblast extracts to detect calpain activity, and immunoblotting to detect the m-calpain large subunit, all gave results entirely consistent with those for the ES cells and embryos of each genotype (data not shown).
Transgenic mice.
Capn4+/− ES cell clones were used to produce chimeric animals by the aggregation method (40), and germ line transmission of the targeted Capn4 allele was achieved with two independent ES cell clones. The Capn4+/− F1 animals showed no apparent defects either in gross anatomy or by histological analysis. The F2 weanlings (n = 225) from heterozygous intercrosses consisted of 67% Capn4+/−, 33% Capn4+/+, and no Capn4−/− animals; in contrast, embryos (n = 252) isolated from heterozygous intercrosses at E9.5 and E10.5 showed the expected Mendelian ratio of Capn4−/− (24%), Capn4+/− (48%), and Capn4+/+ (28%) (Fig. 4A). This suggested that the Capn4−/− animals might have died during embryonic development or perinatally. Furthermore, the average litter size at weaning age was 8.3, compared to an average of 9.6 embryos isolated from pregnant mice. Indeed, all Capn4−/− embryos isolated after E11.5 were dead or moribund. Analysis of calpain activity in E10.5 embryos by means of casein zymography revealed μ- and m-calpain activities in Capn4+/+ and Capn4+/− embryos but not in Capn4−/− embryos (Fig. 4B).
Immunoblotting showed that the mouse m-calpain large subunit protein was present at normal levels in wild-type and heterozygous embryos and was present in trace amounts in homozygous embryos (Fig. 4C). The control immunoblot of Erk1/2 kinase in these embryo extracts is shown in Fig. 4D.
Histological analysis of embryos.
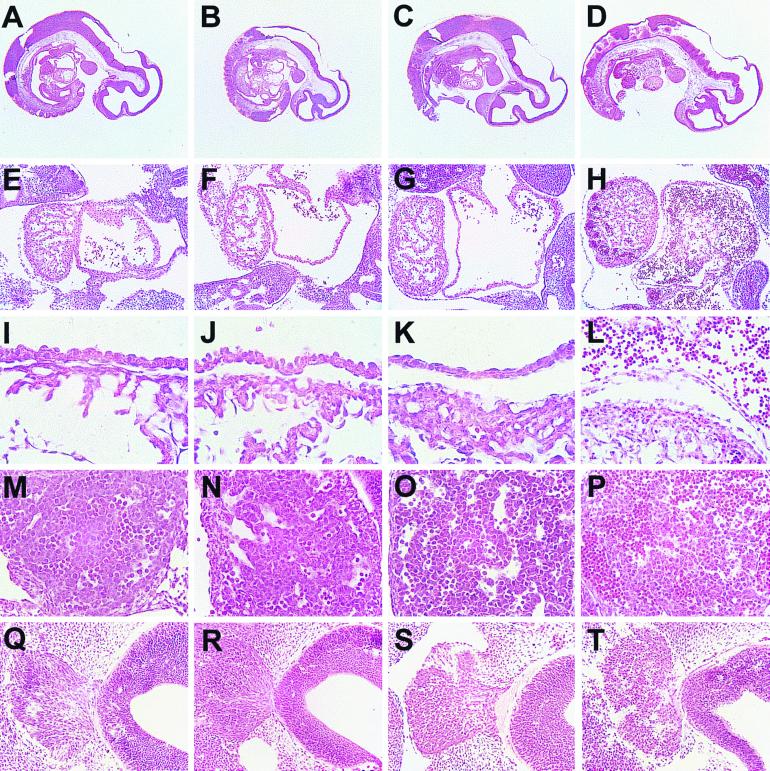
Examination of embryos isolated at different stages of development revealed that all Capn4−/− embryos older than E11.5 were dead or dying. However at earlier times, live Capn4−/− embryos were found in the expected Mendelian ratios, and E9.5 Capn4−/− embryos were indistinguishable from their Capn4+/− and Capn4+/+ littermates (data not shown). At E10.5, although Capn4−/− embryos were still alive, the degree of yolk sac vasculature was reduced, and there was hemorrhaging into the space between the embryo and the amnion. Histological analysis of serial sections prepared from E10.5 Capn4−/− embryos revealed that the cardiovascular system appeared to have developed normally up to this stage, but that the extensive morphogenic events in the heart chambers and vessels leading to and from the heart did not appear to be proceeding normally (Fig. 5). In particular, the common atrial chamber had begun to collapse, and the asymmetric restructuring of the sinus venosus did not appear to be occurring. Endothelial cells in the atrial walls appeared to be rounding up at E10.5, and by E11.5 they had delaminated (Fig. 5I to L). Another striking observation was the large accumulation of nucleated erythroid cells within the heart chambers, blood vessels, and developing liver (Fig. 5D, H, L, and P). This accumulation of erythroid cells correlated with the death of Capn4−/− embryos at or around E11.5. We also observed other, more subtle defects which invite further study. For example, histological analysis suggested the presence of more apoptotic cells among migrating neural crest population from the trigeminal ganglion and within the adjacent fourth ventricle (Fig. 5Q to T). By E11.5 the neural crest no longer displayed a morphology typical of migrating cells, and there appeared to be a higher proportion of apoptotic cells. However, in situ TUNEL analysis at earlier embryonic stages (E9.5 to E10.5) failed to detect significant differences in the number of apoptotic cells (data not shown).
FIG. 5.
Histological analysis of E10.5 and E11.5 embryos. (A to D) Midsagittal sections showing the heart, branchial arches, dorsal aorta, sinus venosus, and liver primordium. (E to H) Sagittal sections through the heart showing ventricle (left) and atrium (right). (I to L) Higher-power magnification of heart sagittal sections showing the atrial wall (upper) and a portion of the ventricle (lower). (M to P) Sagittal sections through the liver primordium. (Q to T) Sagittal sections through the fourth ventricle (right) and adjacent trigeminal neural crest (left). (A, E, I, M, and Q) Capn4+/+ E10.5 embryos; (B, F, J, N, and R) Capn4−/− E10.5 embryos; (C, G, K, O, and S) Capn4+/+ E11.5 embryos; (D, H, L, P, and T) Capn4−/− E11.5 embryos.
DISCUSSION
The Capn4 gene was disrupted on the basis of the assumption that absence of a functional calpain small subunit would abolish the activity of the classical μ- and m-calpains. This assumption was found to be essentially correct. Casein zymography showed that Capn4+/+ and Capn4+/− cells and embryos contained normal levels of calpain activity, but no calpain activity could be detected in extracts of Capn4−/− ES cells, embryonic fibroblasts, or embryos. The casein zymogram method is not quantitative, but it represents the only feasible means to separate and detect the activities of both μ- and m-calpains in very small samples, such as ES cell or fibroblast cultures, or single E9.5-E10.5 mouse embryos. Methods for small-scale separation and quantitative assay of μ- and m-calpains have been described, but they require two column steps (hydrophobic and ion exchange) for each sample, to separate the enzymes from each other and from calpastatin, and require at least 0.5 g of tissue or cultured cells (27, 30).
The embryonic lethality at E11.5 also indicates clearly that at least one of the two μ- and m-calpain activities is absolutely required for the later stages of fetal development. However, Capn4−/− embryos developed normally until midgestation, and cells lacking calpain activity still proliferated at normal rates and remained attached to the tissue culture substrate. This suggests either that calpain is not required for cell growth and division and for some forms of cell adhesion, a conclusion which does not agree with some previous work, or that these cells still contained some residual calpain activity.
Calpain activity could persist in Capn4−/− cells by two mechanisms: by an unexpected activity of the disrupted small subunit, which is highly improbable; or by a residual activity of the isolated large subunits, which cannot presently be excluded. With respect to the small subunit, Northern blotting showed that a trace amount of a Capn4-derived transcript was still present in Capn4−/− ES cells. The sequence of this putative residual Capn4 transcript has not been determined, but the three most likely possibilities are: (i) transcription termination in the PGK-neo cassette; (ii) splicing from exon 8 to cryptic splice sites in the beginning of the neo coding sequence (38) followed by termination in the PGK-neo cassette; or (iii) splicing around the PGK-neo cassette from exon 8 to exon 10. In all three cases, the resulting transcripts would encode small subunits that lacked critical portions of the C terminus. The disrupting PGK-neo cassette is located within exon 9 in the codon for residue 230 of the natural 268-amino-acid small subunit. Translational readthrough from exon 9 into the PGK-neo cassette would result in replacement of the natural 38 C-terminal residues with 98 heterologous amino acid residues encoded by sequences in the PGK promoter. Translational readthrough from the end of exon 8 into the neo coding region would result in replacement of the natural 68 C-terminal residues with an undetermined heterologous sequence derived from the neo coding region. Splicing of exon 8 to exon 10 would generate a protein lacking residues 204 to 242. These hypothetical small subunit peptides are most unlikely to combine with the calpain large subunit to form active enzyme, since the natural C-terminal portion of the small subunit is essential for heterodimer formation and activity (13). The instability of the large subunits, discussed below, is also a proof that no functional small subunit was present.
With respect to the large subunits, their mRNAs were present at normal levels in cells of all genotypes, but only trace levels of the m-calpain large subunit could be detected by immunoblotting. The same is almost certainly true for the μ-calpain large subunit, although only poor immunoblot evidence could be obtained. It is clear that the calpain large subunits in Capn4−/− cells are unstable in the absence of functional small units and are rapidly turned over. There is no unambiguous assay available to measure the possible in vivo activity of these large subunits (particularly at trace levels); thus, it remains unresolved whether the isolated large subunits simply fail to fold correctly in the absence of the small subunit and are rapidly degraded, or whether they exert some transient and unregulated activity prior to (auto)degradation. The latter case would be similar to that of calpain 3, which has a very short half-life and can be assayed only by observation of characteristic autolysis products (23). If it is assumed that the isolated calpain large subunits in Capn4−/− cells possess some short-lived activity, it is also necessary to assume that this activity is sufficient to support normal growth and adhesion of Capn4−/− ES cells and fibroblasts but not sufficient for full development of the fetus. The idea of a dose effect is rather unsatisfactory, and it is attractive to speculate that the difference between cell survival and tissue survival reflects some more specific calpain-dependent cell-cell interaction.
It was also not clear at the outset whether the activities of other calpain-related genes, or of other protease systems such as caspases and the proteasome, might compensate for the absence of μ- and m-calpains, but compensation was not observed. The death at midgestation of all Capn4−/− embryos shows clearly that the presence of the calpain small subunit is essential for normal fetal development. We assume that the absence of the small subunit exerts its effect by causing the loss of normal μ- and m-calpain activities.
Embryonic lethality at or around E10 has become a common observation in mouse knockout studies, and defects in the cardiovascular system are frequently observed in these cases (8, 16, 17, 22, 32, 51, 57). In the Capn4−/− mice, embryonic death coincided with a critical stage in heart development when septation of both heart chambers is occurring as well as formation of the atrioventricular canal. Histological analysis of Capn4−/− embryos showed that the cell linings of both atrium and ventricle were losing their integrity by E10.5 and were delaminated by E11.5 (Fig. 5J and L). As the dual-chambered atrium was not observed in E10.5 Capn4−/− embryos, we have tentatively concluded that embryonic death may have resulted from a failure in heart morphogenesis. We cannot rule out other possibilities, including hemorrhaging and the apparent accumulation of erythroid cells that is observed at the time of embryonic death. These erythroid cells appear to be arising in the primitive liver because we observed large islands of erythropoiesis in the E11.5 liver (Fig. 5P). However, the erythroid cells accumulating in the vasculature and throughout the embryo retain their nuclei. This raises the interesting possibility of a blockage in erythroid development as a result of loss of calpain activity, which will be investigated further. By E11.5 there were more dead cells in the brain ventricles and neural crest of Capn4−/− embryos than in the controls, and these latter cells did not appear to be migrating normally (Fig. 5T). However, this may not be directly related to a role for calpain activity in programmed cell death because in situ TUNEL analysis of embryos failed to reveal differences in the proportion of apoptotic cells at slightly earlier stages (E9.5 to E10.5).
While the essential functions that are disrupted in the absence of normal calpain activity remain unknown, it is important to note that the presence of the calpain small subunit is not required for the growth of ES cells or of embryonic fibroblasts. This appears to rule out a strict requirement for μ- or m-calpain in general cell functions including cell cycle regulation, survival signaling pathways, adhesion, and migration. Such a conclusion would disagree with previous work from several laboratories, and further study of these cell lines is required to establish the importance of calpain in these phenomena and in processing many of the reported calpain substrates. The activity of at least one of the two μ- or m-calpain activities is, however, clearly essential to survival and development of the embryo beyond midgestation. This observation increases the importance of experiments designed to disrupt each of the Capn1 and Capn2 genes alone. Our results do not provide the hoped-for demonstration of a physiological function for calpain, but they have provided homozygous cells for further work in tissue culture and point the way to alternative mouse models of calpain function. It has been established very clearly that calpain is essential to life.
ACKNOWLEDGMENTS
We thank Andras Nagy for providing the R1 ES cell line, Janet Rossant for the 129SvJ mouse genomic DNA library, Ralph Zirngibl for modification of the pPNT vector and for helpful discussions, and Marion Arnold for histology.
This work was supported by grants from the Medical Research Council of Canada, the Heart and Stroke Foundation of Canada, and the National Cancer Institute of Canada. P.A.G. is an MRC scholar.
REFERENCES
- 1.Arthur J S, Greer P A, Elce J S. Structure of the mouse calpain small subunit gene. Biochim Biophys Acta. 1998;1388:247–252. doi: 10.1016/s0167-4838(98)00166-6. [DOI] [PubMed] [Google Scholar]
- 2.Arthur J S C, Mykles D L. Calpain zymography with casein or FITC-casein. In: Elce J C, editor. Methods in molecular biology. 144. Calpain protocols and methods. Totowa, N.J: Humana Press Inc.; 2000. pp. 109–116. [DOI] [PubMed] [Google Scholar]
- 3.Baghdiguian S, Martin M, Richard I, Pons F, Astier C, Bourg N, Hay R T, Chemaly R, Halaby G, Loiselet J, Anderson L V, Lopez de Munain A, Fardeau M, Mangeat P, Beckmann J S, Lefranc G. Calpain 3 deficiency is associated with myonuclear apoptosis and profound perturbation of the IκBα/NF-κB pathway in limb-girdle muscular dystrophy type 2A. Nat Med. 1999;5:503–511. doi: 10.1038/8385. [DOI] [PubMed] [Google Scholar]
- 4.Balcerzak D, Poussard S, Brustis J J, Elamrani N, Soriano M, Cottin P, Ducastaing A. An antisense oligodeoxyribonucleotide to m-calpain mRNA inhibits myoblast fusion. J Cell Sci. 1995;108:2077–2082. doi: 10.1242/jcs.108.5.2077. [DOI] [PubMed] [Google Scholar]
- 5.Barnes T M, Hodgkin J. The tra-3 sex determination gene of Caenorhabditis elegans encodes a member of the calpain regulatory protease family. EMBO J. 1996;15:4477–4484. [PMC free article] [PubMed] [Google Scholar]
- 6.Blanchard H, Grochulski P, Li Y, Arthur J S, Davies P L, Elce J S, Cygler M. Structure of a calpain Ca2+-binding domain reveals a novel EF-hand and Ca2+-induced conformational changes. Nat Struct Biol. 1997;4:532–538. doi: 10.1038/nsb0797-532. [DOI] [PubMed] [Google Scholar]
- 7.Carafoll E, Molinari M. Calpain: a protease in search of a function? Biochem Biophys Res Commun. 1998;247:193–203. doi: 10.1006/bbrc.1998.8378. [DOI] [PubMed] [Google Scholar]
- 8.Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, Declercq C, Pawling J, Moons L, Collen D, Risau W, Nagy A. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- 9.Dear T N, Moller A, Boehm T. CAPN11: a calpain with high mRNA levels in testis and located on chromosome 6. Genomics. 1999;59:243–247. doi: 10.1006/geno.1999.5859. [DOI] [PubMed] [Google Scholar]
- 10.Dear T N, Boehm T. Diverse mRNA expression patterns of the mouse calpain genes Capn5, Capn6 and Capn11 during development. Mech Dev. 1999;89:201–209. doi: 10.1016/s0925-4773(99)00214-2. [DOI] [PubMed] [Google Scholar]
- 11.Dourdin N, Balcerzak D, Brustis J J, Poussard S, Cottin P, Ducastaing A. Potential m-calpain substrates during myoblast fusion. Exp Cell Res. 1999;246:433–442. doi: 10.1006/excr.1998.4325. [DOI] [PubMed] [Google Scholar]
- 12.Dutt P, Arthur J S, Croall D E, Elce J S. m-Calpain subunits remain associated in the presence of calcium. FEBS Lett. 1998;436:367–371. doi: 10.1016/s0014-5793(98)01167-3. [DOI] [PubMed] [Google Scholar]
- 13.Elce J S, Davies P L, Hegadorn C, Maurice D H, Arthur J S C. The effects of truncations of the small subunit on m-calpain activity and heterodimer formation. Biochem J. 1997;326:31–38. doi: 10.1042/bj3260031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fadeel B, Zhivotovsky B, Orrenius S. All along the watchtower: on the regulation of apoptosis regulators. FASEB J. 1999;13:1647–1657. doi: 10.1096/fasebj.13.13.1647. [DOI] [PubMed] [Google Scholar]
- 15.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Addendum. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- 16.Ferrara N, Carver-Moore K, Chen H, Dowd M, Lu L, O'Shea K S, Powell-Braxton L, Hillan K J, Moore M W. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- 17.Fong G H, Rossant J, Gertsenstein M, Breitman M L. Role of the Flt-1 receptor tyrosine kinase in regulating the assembly of vascular endothelium. Nature. 1995;376:66–70. doi: 10.1038/376066a0. [DOI] [PubMed] [Google Scholar]
- 18.Fougerousse F, Durand M, Suel L, Pourquie O, Delezoide A L, Romero N B, Abitbol M, Beckmann J S. Expression of genes (CAPN3, SGCA, SGCB, and TTN) involved in progressive muscular dystrophies during early human development. Genomics. 1998;48:145–156. doi: 10.1006/geno.1997.5160. [DOI] [PubMed] [Google Scholar]
- 19.Fox J E B, Saido T C. Calpain in signal transduction. In: Wang K K W, Yuen P W, editors. Calpain: pharmacology and toxicology of calcium-dependent protease. Philadelphia, Pa: Taylor & Francis; 1999. pp. 103–126. [Google Scholar]
- 20.Franz T, Vingron M, Boehm T, Dear T N. Capn7: a highly divergent vertebrate calpain with a novel C-terminal domain. Mamm Genome. 1999;10:318–321. doi: 10.1007/s003359900995. [DOI] [PubMed] [Google Scholar]
- 21.Graham-Siegenthaler K, Gauthier S, Davies P L, Elce J S. Active recombinant rat calpain II. Bacterially produced large and small subunits associate both in vivo and in vitro. J Biol Chem. 1994;269:30457–30460. [PubMed] [Google Scholar]
- 22.Henkemeyer M, Rossi D J, Holmyard D P, Puri M C, Mbamalu G, Harpal K, Shih T S, Jacks T, Pawson T. Vascular system defects and neuronal apoptosis in mice lacking ras GTPase-activating protein. Nature. 1995;377:695–701. doi: 10.1038/377695a0. [DOI] [PubMed] [Google Scholar]
- 23.Herasse M, Ono Y, Fougerousse F, Kimura E, Stockholm D, Beley C, Montarras D, Pinset C, Sorimachi H, Suzuki K, Beckmann J S, Richard I. Expression and functional characteristics of calpain 3 isoforms generated through tissue-specific transcriptional and posttranscriptional events. Mol Cell Biol. 1999;19:4047–4055. doi: 10.1128/mcb.19.6.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill D P, Wurst W. Screening for novel pattern formation genes using gene trap approaches. Methods Enzymol. 1993;225:664–681. doi: 10.1016/0076-6879(93)25043-2. [DOI] [PubMed] [Google Scholar]
- 25.Hosfield C M, Elce J S, Davies P L, Jia Z. Crystal structure of calpain reveals the structural basis for Ca2+-dependent protease activity and a novel mode of enzyme activation. EMBO J. 1999;18:6880–6889. doi: 10.1093/emboj/18.24.6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huttenlocher A, Palecek S P, Lu Q, Zhang W, Mellgren R L, Lauffenburger D A, Ginsberg M H, Horwitz A F. Regulation of cell migration by the calcium-dependent protease calpain. J Biol Chem. 1997;272:32719–32722. doi: 10.1074/jbc.272.52.32719. [DOI] [PubMed] [Google Scholar]
- 27.Karlsson J O, Gustavsson S, Hall C, Nilsson E. A simple one-step procedure for the separation of calpain I, calpain II and calpastatin. Biochem J. 1985;231:201–204. doi: 10.1042/bj2310201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinbara K, Sorimachi H, Ishiura S, Suzuki K. Skeletal muscle-specific calpain, p49: structure and physiological function. Biochem Pharmacol. 1998;56:415–420. doi: 10.1016/s0006-2952(98)00095-1. [DOI] [PubMed] [Google Scholar]
- 29.Kulkarni S, Saido T C, Suzuki K, Fox J E. Calpain mediates integrin-induced signaling at a point upstream of Rho family members. J Biol Chem. 1999;274:21265–21275. doi: 10.1074/jbc.274.30.21265. [DOI] [PubMed] [Google Scholar]
- 30.Kwak K B, Chung S S, Kim O M, Kang M S, Ha D B, Chung C H. Increase in the level of m-calpain correlates with the elevated cleavage of filamin during myogenic differentiation of embryonic muscle cells. Biochim Biophys Acta. 1993;1175:243–249. doi: 10.1016/0167-4889(93)90212-8. [DOI] [PubMed] [Google Scholar]
- 31.Lin G-D, Chattopadhyay D, Maki M, Wang K K W, Carson M, Jin L, Yuen P-W, Takano E, Hatanaka M, DeLucas L J, Narayana S V L. Crystal structure of calcium bound domain VI of calpain at 1.9Å resolution and its role in enzyme assembly, regulation, and inhibitor binding. Nat Struct Biol. 1997;4:539–547. doi: 10.1038/nsb0797-539. [DOI] [PubMed] [Google Scholar]
- 32.Lyons I, Parsons L M, Hartley L, Li R, Andrews J E, Robb L, Harvey R P. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 33.Matena K, Boehm T, Dear N. Genomic organization of mouse Capn5 and Capn6 genes confirms that they are a distinct calpain subfamily. Genomics. 1998;48:117–120. doi: 10.1006/geno.1997.5133. [DOI] [PubMed] [Google Scholar]
- 34.McGinnis K M, Gnegy M E, Park Y H, Mukerjee N, Wang K K. Procaspase-3 and poly(ADP)ribose polymerase (PARP) are calpain substrates. Biochem Biophys Res Commun. 1999;263:94–99. doi: 10.1006/bbrc.1999.1315. [DOI] [PubMed] [Google Scholar]
- 35.Mellgren R L. Calcium-dependent proteases: an enzyme system active at cellular membranes? FASEB J. 1987;1:110–115. doi: 10.1096/fasebj.1.2.2886390. [DOI] [PubMed] [Google Scholar]
- 36.Mellgren R L, Zhang W, Lu Q, Lane R D. Involvement of calpains in cell cycle G1- to S-phase. In: Wang K K W, Yuen P W, editors. Calpain: pharmacology and toxicology of calcium-dependent protease. Philadelphia, Pa: Taylor & Francis; 1999. pp. 161–178. [Google Scholar]
- 37.Meyer S L, Bozyczko-Coyne D, Mallya S K, Spais C M, Bihovsky R, Kawooya J K, Lang D M, Scott R W, Siman R. Biologically active monomeric and heterodimeric recombinant human calpain I produced using the baculovirus expression system. Biochem J. 1996;314:511–519. doi: 10.1042/bj3140511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyers E N, Lewandoski M, Martin G R. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 39.Molinari M, Carafoli E. Calpain: a cytosolic proteinase active at the membranes. J Membr Biol. 1997;156:1–8. doi: 10.1007/s002329900181. [DOI] [PubMed] [Google Scholar]
- 40.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder J C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakagawa T, Zhu H, Morishima N, Li E, Xu J, Yankner B A, Yuan J. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 42.Ohno S, Emori Y, Imajoh S, Kawasaki H, Kisaragi M, Suzuki K. Evolutionary origin of a calcium-dependent protease by fusion of genes for a thiol protease and a calcium-binding protein? Nature. 1984;312:566–570. doi: 10.1038/312566a0. [DOI] [PubMed] [Google Scholar]
- 43.Ono Y, Sorimachi H, Suzuki K. Structure and physiology of calpain, an enigmatic protease. Biochem Biophys Res Commun. 1998;245:289–294. doi: 10.1006/bbrc.1998.8085. [DOI] [PubMed] [Google Scholar]
- 44.Raser K J, Posner A, Wang K K W. Casein zymography: a method to study mu-calpain, m-calpain, and their inhibitory agents. Arch Biochem Biophys. 1995;319:211–216. doi: 10.1006/abbi.1995.1284. [DOI] [PubMed] [Google Scholar]
- 45.Richard I, Broux O, Allamand V, Fougerousse F, Chiannilkulchai N, Bourg N, Brenguier L, Devaud C, Pasturaud P, Roudaut C, et al. Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2a. Cell. 1995;81:27–40. doi: 10.1016/0092-8674(95)90368-2. [DOI] [PubMed] [Google Scholar]
- 46.Ruiz-Vela A, Gonzalez de Buitrago G, Martinez-A C. Implication of calpain in caspase activation during B cell clonal deletion. EMBO J. 1999;18:4988–4998. doi: 10.1093/emboj/18.18.4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samis J A, Back D W, Graham E J, DeLuca C I, Elce J S. Constitutive expression of calpain II in the rat uterus during pregnancy and involution. Biochem J. 1991;276:293–299. doi: 10.1042/bj2760293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 49.Schoenwaelder S M, Burridge K. Evidence for a calpeptin-sensitive protein-tyrosine phosphatase upstream of the small GTPase Rho. A novel role for the calpain inhibitor calpeptin in the inhibition of protein-tyrosine phosphatases. J Biol Chem. 1999;274:14359–14367. doi: 10.1074/jbc.274.20.14359. [DOI] [PubMed] [Google Scholar]
- 50.Schollmeyer J E. Calpain II involvement in mitosis. Science. 1988;240:911–913. doi: 10.1126/science.2834825. [DOI] [PubMed] [Google Scholar]
- 51.Shalaby F, Rossant J, Yamaguchi T P, Gertsenstein M, Wu X F, Breitman M L, Schuh A C. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature. 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- 52.Sorimachi H, Amano S, Ishiura S, Suzuki K. Primary sequences of rat mu-calpain large and small subunits are, respectively, moderately and highly similar to those of human. Biochim Biophys Acta. 1996;1309:37–41. doi: 10.1016/s0167-4781(96)00135-2. [DOI] [PubMed] [Google Scholar]
- 53.Sorimachi H, Ishiura S, Suzuki K. Structure and physiological function of calpains. Biochem J. 1997;328:721–732. doi: 10.1042/bj3280721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Squier M K, Cohen J J. Calpain, an upstream regulator of thymocyte apoptosis. J Immunol. 1997;158:3690–3697. [PubMed] [Google Scholar]
- 55.Squier M K, Sehnert A J, Sellins K S, Malkinson A M, Takano E, Cohen J J. Calpain and calpastatin regulate neutrophil apoptosis. J Cell Physiol. 1999;178:311–319. doi: 10.1002/(SICI)1097-4652(199903)178:3<311::AID-JCP5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 56.Strobl S, Fernandez-Catalan C, Braun M, Huber R, Masumoto H, Nakagawa K, Irie A, Sorimachi H, Bourenkow G, Bartunik H, Suzuki K, Bode W. The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium. Proc Natl Acad Sci USA. 2000;97:588–592. doi: 10.1073/pnas.97.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suri C, Jones P F, Patan S, Bartunkova S, Maisonpierre P C, Davis S, Sato T N, Yancopoulos G D. Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell. 1996;87:1171–1180. doi: 10.1016/s0092-8674(00)81813-9. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki K, Sorimachi H. A novel aspect of calpain activation. FEBS Lett. 1998;433:1–4. doi: 10.1016/s0014-5793(98)00856-4. [DOI] [PubMed] [Google Scholar]
- 59.Thompson V F, Goll D E. Purification of μ-calpain, m-calpain, and calpastatin from animal tissues. In: Eke J S, editor. Methods in molecular biology. 144. Calpain protocols and methods. Totowa, N.J: Humana Press Inc.; 2000. pp. 3–16. [PubMed] [Google Scholar]
- 60.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tybulewicz V L, Crawford C E, Jackson P K, Bronson R T, Mulligan R C. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell. 1991;65:1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- 62.Vilei E M, Calderara S, Anagli J, Berardi S, Hitomi K, Maki M, Carafoli E. Functional properties of recombinant calpain I and of mutants lacking domains III and IV of the catalytic subunit. J Biol Chem. 1997;272:25802–25808. doi: 10.1074/jbc.272.41.25802. [DOI] [PubMed] [Google Scholar]
- 63.Wang K K W, Yuen P W. Calpain substrates, assay methods, regulation, and its inhibitor agents. In: Wang K K W, Yuen P W, editors. Calpain: pharmacology and toxicology of calcium-dependent protease. Philadelphia, Pa: Taylor & Francis; 1999. pp. 77–101. [Google Scholar]
- 64.Waterhouse N J, Finucane D M, Green D R, Elce J S, Kumar S, Alnemri E S, Litwack G, Khanna K, Lavin M F, Watters D J. Calpain activation is upstream of caspases in radiation-induced apoptosis. Cell Death Differ. 1998;5:1051–1061. doi: 10.1038/sj.cdd.4400425. [DOI] [PubMed] [Google Scholar]
- 65.Wilson G M, Roberts E A, Deeley R G. Modulation of LDL receptor mRNA stability by phorbol esters in human liver cell culture models. J Lipid Res. 1997;38:437–446. [PubMed] [Google Scholar]
- 66.Wolf B B, Goldstein J C, Stennicke H R, Beere H, Amarante-Mendes G P, Salvesen G S, Green D R. Calpain functions in a caspase-independent manner to promote apoptosis-like events during platelet activation. Blood. 1999;94:1683–1692. [PubMed] [Google Scholar]
- 67.Wood D E, Newcomb E W. Caspase-dependent activation of calpain during drug-induced apoptosis. J Biol Chem. 1999;274:8309–8315. doi: 10.1074/jbc.274.12.8309. [DOI] [PubMed] [Google Scholar]
- 68.Yadav S S, Sindram D, Perry D K, Clavien P A. Ischemic preconditioning protects the mouse liver by inhibition of apoptosis through a caspase-dependent pathway. Hepatology. 1999;30:1223–1231. doi: 10.1002/hep.510300513. [DOI] [PubMed] [Google Scholar]
- 69.Yoshizawa T, Sorimachi H, Tomioka S, Ishiura S, Suzuki K. A catalytic subunit of calpain possesses full proteolytic activity. FEBS Lett. 1995;358:101–103. doi: 10.1016/0014-5793(94)01401-l. [DOI] [PubMed] [Google Scholar]
- 70.Yoshizawa T, Sorimachi H, Tomioka S, Ishiura S, Suzuki K. Calpain dissociates into subunits in the presence of calcium ions. Biochem Biophys Res Commun. 1995;208:376–383. doi: 10.1006/bbrc.1995.1348. [DOI] [PubMed] [Google Scholar]
- 71.Zhang W, Mellgren R L. Calpain subunits remain associated during catalysis. Biochem Biophys Res Commun. 1996;227:890–896. [PubMed] [Google Scholar]