Abstract
The coronavirus disease COVID-19 has been the cause of millions of deaths worldwide. Among the SARS-CoV-2 proteins, the non-structural protein 1 (NSP1) has great importance during the virus infection process and is present in both alpha and beta-CoVs. Therefore, monitoring of NSP1 polymorphisms is crucial in order to understand their role during infection and virus-induced pathogenicity. Herein, we analyzed how mutations detected in the circulating SARS-CoV-2 in the population of the city of Manaus, Amazonas state, Brazil could modify the tertiary structure of the NSP1 protein. Three mutations were detected in the SARS-CoV-2 NSP1 gene: deletion of the amino acids KSF from positions 141 to 143 (delKSF), SARS-CoV-2, lineage B.1.195; and two substitutions, R29H and R43C, SARS-CoV-2 lineage B.1.1.28 and B.1.1.33, respectively. The delKSF was found in 47 samples, whereas R29H and R43C were found in two samples, one for each mutation. The NSP1 structures carrying the mutations R43C and R29H on the N-terminal portion (e.g. residues 10 to 127) showed minor backbone divergence compared to the Wuhan model. However, the NSP1 C-terminal region (residues 145 to 180) was severely affected in the delKSF and R29H mutants. The intermediate variable region (residues 144 to 148) leads to changes in the C-terminal region, particularly in the delKSF structure. New investigations must be carried out to analyze how these changes affect NSP1 activity during the infection. Our results reinforce the need for continuous genomic surveillance of SARS-CoV-2 to better understand virus evolution and assess the potential impact of the viral mutations on the approved vaccines and future therapies.
Keywords: SARS-CoV-2, COVID-19, coronavirus, polymorphism, NSP1, 3D structure modeling
Impact statement
Manaus, the capital of the Amazonas state, Brazil, is suffering from a second wave of COVID-19 cases in 2021. As a result, the continuous monitoring of polymorphisms in the virus genome is essential to aid in the development of new vaccines and drugs. Here we describe three mutations of SARS-CoV-2 NSP1 detected in Manaus, Amazonas state, Brazil, and how these affect the tertiary structure of this protein. For this, we performed a modeling for this protein, which made it possible to observe that the C-terminal region of NSP1 had been severely affected by two mutations (delKSF and R29H). The intermediate variable region (residues 144 to 148) leads to changes in the C-terminal region, particularly in the delKSF structure. Therefore, future studies are necessary in order to evaluate the influence of these structural changes during infection.
Introduction
The severe respiratory syndrome caused by coronavirus 2 (SARS-CoV-2) has become a health problem that has resulted in millions of deaths worldwide. The World Health Organization reported 107,838,255 COVID-19 cases as of February, 2021, with 2,373,398 confirmed deaths. 1 In Brazil, the number of deaths has surpassed 233,520, with the Amazonas state being one of the most affected states. In December 2020, the Amazonas state suffered a second wave of infections, which caused an unexpected increase of COVID-19 hospital admissions in Manaus (3431 between 1 January and 19 January 2021 vs. 552 between 1 December and 19 December 2020). 2 This second wave coincided with the emergence of the variant of concern (VOC) P.1, which evolved from a local B.1.1.28 clade, 3 and three reinfection cases were confirmed to be caused by the new variant of concern P.1. 4 , 5 This has increased the importance of constant monitoring of SARS-CoV-2 protein polymorphisms.
Among SARS-CoV-2 proteins, the non-structural protein 1 (NSP1) has great importance during the virus infection process and is present in both alpha and beta-CoVs. In SARS-CoV-2, this protein has 180 amino acids and a molecular mass of 19,775.31 Da, and is predicted as a stable molecule (28.83 instability index) with an average life of 30 h inside the cellular environment. 6 NSP1 induces a reduction in the translation of the host cell’s RNA by binding its C-terminal region to the ribosome’s smaller subunit. This binding is interrupted when the conserved 5ʹUTR region of the viral RNA interacts with the N-terminal region of NSP1, thus allowing the translation of the viral mRNA, through a mechanism which still not yet wholly understood, 7 and inhibiting the type I interferon response. 8
These previous observations support that NSP1 plays an essential role in viral infection. Therefore, NSP1 is currently one of the targets for new antiviral drugs. 9 , 10 Accordingly, it is crucial to monitor NSP1 polymorphisms in order to understand their role during infection and virus-induced pathogenicity.
Shen et al. 11 observed that the NSP1 of Sars-CoV-2 has a greater inhibitory activity of host protein synthesis than Sars-CoV-1. They report that mutations in H165 and K164 of NSP1 interfere in this function and in the capacity of inhibiting the immune response pathways of the infected subject. Loureiro et al. 12 also reported deletions in amino acids at the positions 83 to 85 which are capable to affect NSP1 structure and folding.
Therefore, this work aimed to analyze how mutations detected in the circulating SARS-CoV-2 detected Manaus, Amazonas state, Brazil, could modify the tertiary structure of the NSP1, by performing a modeling of this protein.
Materials and methods
Samples were collected in Manaus, Amazonas state, between March and October of 2020. All samples were sequenced following the protocol described by Nascimento et al. 13 with simple modifications. The samples were amplified with Platinum SuperFi II Green PCR master mix, visualized on agarose gel electrophoresis and precipitated with PEG 8000. The amplicons were quantified by fluorimetry, and those belonging to the same sample were normalized and pooled. Next-generation sequencing libraries were constructed using the NexteraXT DNA prep kit and sequenced with the MiSeq reagent kit v2 (500 cycles). Nucleotide sequencing was accomplished using the MiSeq Illumina sequencer installed at Fiocruz Amazônia. Raw data were converted using the Illumina pipeline at BaseSpace; trimmed for quality using BBDuk and assembled with BBMap embedded in Geneious v10.2.6, using the sequence YP_009742608.1as the reference. Alignment was performed using Clustal omega (EMBL-EBI).
Before modeling, using BLAST 14 a search was made for templates structures contained in the Protein Data Bank (PDB) (http://www.pdb.org) with the similarity between the NSP1 sequence of SARS-CoV-2 isolated from Wuhan (GenBank RefSeq NC_045512.2). The selected templates were subjected to homology modeling with the Modeller 9.25. 15 After modeling the NSP1 of SARS-CoV-2 from Wuhan, the mutations found by our group were applied. Each new structure was modeled separately, generating 1000 models in Modeller 9.25. The best model selected is the one with the lowest score in the DOPE (Discrete optimized protein energy) calculation function of Modeller 9.25. The RMSD calculations were performed on the Swiss-PDBViewer v4.1 16 and the analysis of the location of the mutations and image production was performed on the UCSF Chimera. 17
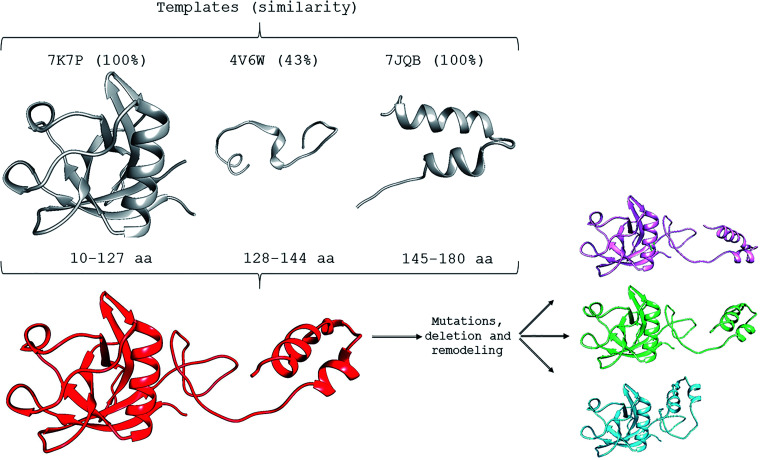
The SARS-CoV-2 NSP1 structure (PDB identification code: 7K7P), the rabbit 40S ribosome complex (7JQB), 18 , 19 and the Drosophila melanogaster 80S ribosome 20 (4V6W) were used as templates (Figure 1). After this initial modeling, fragment 3 (regions 128 to 144) was subjected to a loop refinement in the Modeller software’s advanced routine. This final approach was necessary since this region showed the least similarity with the templates. The quality of all final models was assessed using the Procheck software. 21 All structures presented structural quality with more than 90% of amino acids in favorable regions. None of the amino acids were in the prohibited region.
Figure 1.
Template structures were used to model the NSP1 structure of SARS-CoV-2 (sequence NC_045512.2). The 7K7P, 4V6W, and 7JQB templates were used in portions 10–127, 128–144, and 145–180, respectively. The percentage of local similarity in each region is indicated in parentheses. After modeling, the mutations R29H (magenta), R43C (green), and the KSF deletion (cyan) were applied and subjected to remodeling in Modeller 9.25. (A color version of this figure is available in the online journal.)
Results and discussion
Forty-nine samples were sequenced for SARS-CoV-2 NSP1 gene: 47 samples contained a deletion of the amino acids KSF from positions 141 to 143 (delKSF), SARS-CoV-2 lineage B.1.195; and one sample presented a substitution of R29H, SARS-CoV-2 lineage B.1.1.28; and one sample presented a substitution of R43C, SARS-CoV-2 lineage B.1.1.33. An alignment of the sample sequences is shown in Figure 2.
Figure 2.
Alignment of SARS-CoV-2 NSP1 (ORF1ab) from Wuhan (YP_009742608.1) and three isolates from Manaus: 1. R29H; 2. R43C; and 3. delKSF. Colors highlight amino acids. (A color version of this figure is available in the online journal.)
The NSP1 structures carrying the mutations R43C and R29H on the N-terminal portion (e.g. residues 10 to 127) showed minor backbone divergence when compared to the RefSeq model. Thus, our results are in accordance with Clark et al. 18 and Semper et al. 22 who analyzed the structure of SARS-CoV-2 NSP1 (10-126aa) and reported that the tertiary structure of NSP1 is composed of regular secondary structural elements that are arranged sequentially as β1-η1-α1-β2-η3-β3-β4-β5-β6-β7, with η1 and η2 being 310 helices.
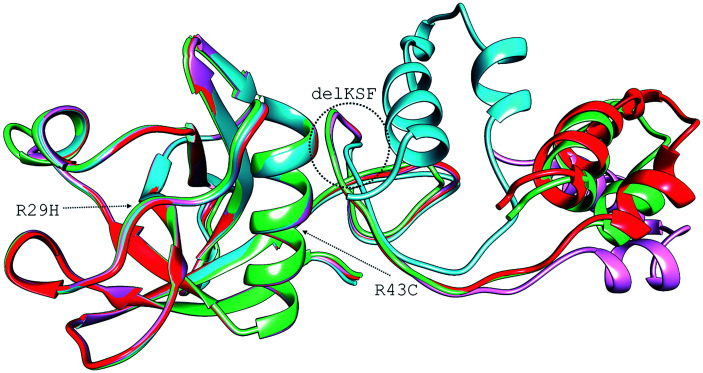
The NSP1 C-terminal region (residues 145 to 180) was severely affected in the delKSF and R29C mutants. The intermediate variable region (residues 144 to 148) led to changes in the C-terminal region, particularly in the delKSF structure (Figure 3). The RMSD (root mean square deviation) between the three complete structures and the RefSeq strain presented the following results: RefSeq:delKSF, 7.63 Å; RefSeq:R43C, 1.18 Å; and RefSeq:R29H, 4.66 Å. Since loops may play critical roles in protein-protein interactions, 23 these mutations may be interfering with NSP1-ribosome interaction by a process that has not yet been elucidated. Shi et al. 24 observed that the elongation of the linking region between the N-terminal and the C-terminal portion of NSP1 progressively reduces the ability of the 5ʹUTR of the viral RNA to escape inhibition. Therefore, the linker’s size reduction in the delKSF variant may contribute positively to the unblocking of the NSP1 C-terminal portion of the ribosome after binding the 5ʹUTR of viral RNA. Benedetti et al. 25 suggest that KSF deletion could affect the structure of the C-terminal region of the protein, which is essential for regulating viral replication and shutting down the host’s gene expression. These mutations suggest that the virus is undergoing an evolutionary process that is decreasing its pathogenicity, since they were found, in the vast majority, in asymptomatic individuals. 25
Figure 3.
Overlapping of the four NSP1 modeled structures of SARS-CoV-2. In red, the structure of the reference sequence (NC_045512.2). In magenta, the structure with an arginine mutation at position 29 by a histidine (R29H). In green, the structure with an arginine mutation at position 43 by a cysteine (R43C). In cyan, the structure with the deletion of three amino acids: lysine 141, serine 142, and phenylalanine 143 (delKSF). (A color version of this figure is available in the online journal.)
Semper et al. 22 also reported a homology-based model of the full-length NSP1 protein using PSIPRED and DISPRRED3. Their model presented a similar structure to that obtained in the present study, i.e. composed of the capped β-barrel motif, a disordered linker, and the C-terminal helix-turn-helix. They also suggest that the C-terminal domain is highly dynamic and capable of multiple configurations due to this disordered linker. Perhaps, this condition contributes to the NSP1 C-terminal binding to the ribosome mRNA entry tunnel, as observed by Thoms et al. 26
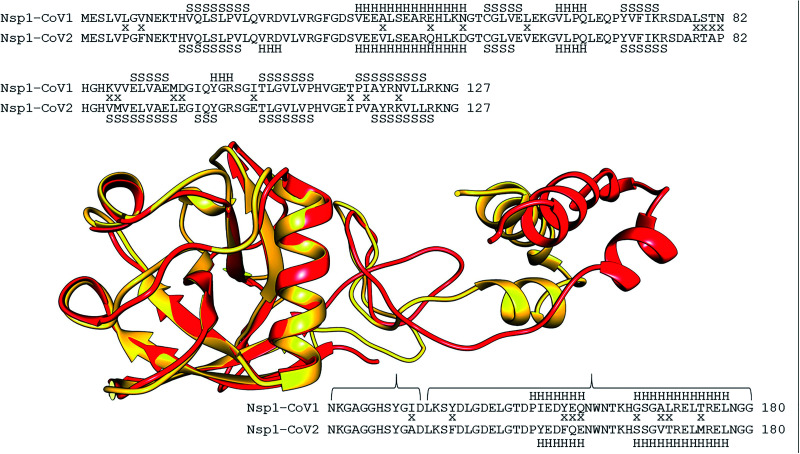
The SARS-CoV NSP1 (10-180aa) was also modeled, despite lacking a complete template. We used the following two PDB files: 2GDT, an NMR structure of the SARS-CoV-1 NSP1 27 and 7JQB, the SARS-CoV-2 NSP1, in a complex with the rabbit 40S ribosome. 19 The root mean square deviation (RMSD) between the SARS-CoV-2 RefSeq NSP1 structure and SARS-CoV-1 NSP1 modeled in this study was 6.49 Å. The alignment and overlapping between SARS-CoV-1 and SARS-CoV-2 NSP1 revealed considerable conservation in the N-terminal region (10 to 127 residues) (Figure 4). There is an α-helix (V23, R24, and D25 residues) and one β-sheet strand (I97, Q98 and Y99 residues) in SARS-CoV-2 that is not found in SARS-CoV-1. On the other hand, the Y99, G100, and R101 residues are present in SARS-CoV-1 and absent in SARS-CoV-2. Other changes in the α-helix and β-sheet strands extension were also observed. These characteristics were also observed by Kucirka et al. 28 and Semper et al. 22 who both described that the fold of SARS-CoV-2 NSP1 is generally well conserved when compared to the previously solved structure of NSP1 from SARS-CoV-1, with the following differences: the presence of an additional 310 helix (η1) in the SARS-CoV-2 structure (residues 23 to 25), extension of β-strands within the interior of the protein, and the alternative conformations of two major loops located between β3 and β4 (loop 1) and between β4 and β6 (loop 2, β4, and β5 in SARS-CoV-1), resulting in differences in the electrostatic surface potential.
Figure 4.
Alignment and overlap between the NSP1 structures of SARS-CoV (yellow) and SARS-CoV2 (red). In the alignment, divergences were indicated with an “x” between the two sequences. Above and below the two sequences, the positions of the secondary structures were shown. “H” indicates the alfa-helix and “S” the beta-sheet. The alignment of the 10–127 portion was positioned above the structures, and the 128–180 portion below. (A color version of this figure is available in the online journal.)
In conclusion, we present herein a modeling for SARS-CoV-1 and 2 NSP1 and the analysis of new variants found in Manaus, a Brazilian city that has been severely affected by COVID-19. As observed by Wathelet et al., 29 deletions or mutations in SARS-CoV-1 NSP1 could result in attenuation in infection models and restore the innate immune response in infected cells. Therefore, more studies must be done aiming to understand the correlation between the evolution of the disease in infected patients and the presence of these variants to provide evidence of how the virulence and transmissibility of these strains are affected by such polymorphisms. The genomic surveillance of SARS-CoV-2 is still necessary in order to more fully understand the virus’ evolution and evaluate the potential impact of any viral mutations, such as those addressed in this study, in regards to the approved vaccines.
ACKNOWLEDGMENTS
The authors thank the Program for Technological Development in Tools for Health—FIOCRUZ—for the use of the nucleotide sequencing facilities at ILMD—Fiocruz Amazônia.
AUTHORS’ CONTRIBUTIONS: FBZ, LAM, and FGN conceived the study; FGN and FBZ designed the study protocol; VN, VS, AC, FN, AKC, DD, GS, MM, KP, LG, MS, MJ, FON, TM, CDC, and FGN performed the molecular tests; LAM, FZ, and FGN performed the analysis and interpretation of the data; FON and MJB collected the biological samples; LAM, FBZ, JG, and FGN wrote the manuscript; LAM, FZ, JG, and FGN critically revised the manuscript for intellectual content; FGN obtained the funding for the study. All authors read and approved the final manuscript.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
ETHICAL APPROVAL: This study was approved by the Ethics Committee of the Amazonas State University, under approval number: CAAE: 25430719.6.0000.5016.
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by the Amazonas State Research Support Foundation (FAPEAM) [PCTI-EmergeSaúde/AM call N°005/2020 and Rede Genômica de Vigilância em Saúde – REGESAM]; the National Council for Scientific and Technological Development (CNPq) [Grants 440856/2016–7 and 403276/2020–9], the Coordination for the Improvement of Higher Education Personnel (CAPES) [Grants 88881.130825/2016–01 and 88887.130823/2016–00]; and Oswaldo Cruz Foundation-Inova Fiocruz (Grant VPPCB-007-FIO-18–2-30 – Geração de conhecimento).
ORCID iDs
Fernando Berton Zanchi https://orcid.org/0000-0003-3386-0069
Juliane Glória https://orcid.org/0000-0002-9122-1145
References
- 1.WHO. Weekly Operational Update on COVID-19, Emergency Situational Updates, 13 February. World Health Organization, 2021
- 2.Sabino EC, Buss LF, Carvalho MPS, Prete CA, Crispim MAE, Fraiji NA, Pereira RHM, Parag KV, Da Silva Peixoto P, Kraemer MUG, Oikawa MK, Salomon T, Cucunuba ZM, Castro MC, De Souza Santos AA, Nascimento VH, Pereira HS, Ferguson NM, Pybus OG, Kucharski A, Busch MP, Dye C, Faria NR. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet 2021; 397:452–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naveca F, Nascimento V, Souza V, Corado A, Nascimento F, Silva G, Costa Á, Duarte D, Pessoa K, Mejía M, Brandão M, Gonçalves JM, Costa L, Sampaio CD, Barros V, Silva D, Mattos M, Pontes T, Abdalla G, Santos L, Arantes J, Dezordi I, Siqueira F, Wallau M, Resende G, Delatorre P, Gräff E, Bello TG. COVID-19 epidemic in the Brazilian state of Amazonas was driven by long-term persistence of endemic SARS-CoV-2 lineages and the recent emergence of the new variant of concern P.1. Res Square 2021. DOI: 10.21203/rs.3.rs-275494/v1 [Google Scholar]
- 4.FIOCRUZ/FVS-AM. Nota Técnica Conjunta N° 09. 2021
- 5.Naveca F, da Costa C, Nascimento V, Souza V, Corado A, Nascimento F, Costa A, Duarte D, Silva G, Mejía M, Pessoa K, Gonçalves L, Brandão MJ, Jesus M, Pinto R, Silva M, Mattos T, Abdalla L, Santos JH, Costa-Filho R, Wallau GL, Siqueira MM, Delatorre E E, Gräf T, Bello G, Resende PC. SARS-CoV-2 reinfection by the new variant of concern (VOC) P.1 in Amazonas, Brazil. 2021. https://virological.org/t/sars-cov-2-reinfection-by-the-new-variant-ofconcern-voc-p-1-in-amazonas-brazil/596 (accessed Feb 20, 2021). [Google Scholar]
- 6.Min YQ, Mo Q, Wang J, Deng F, Wang H, Ning YJ. SARS-CoV-2 nsp1: bioinformatics, potential structural and functional features, and implications for drug/vaccine designs. Front Microbiol 2020; 11:587317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut M, Thiel V, Mühlemann O, Ban N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol 2020; 27:959–66 [DOI] [PubMed] [Google Scholar]
- 8.Xia H, Cao Z, Xie X, Zhang X, Chen JY, Wang H, Menachery VD, Rajsbaum R, Shi PY. Evasion of type I interferon by SARS-CoV-2. Cell Rep 2020; 33:108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lima Menezes G, da Silva RA. Identification of potential drugs against SARS-CoV-2 non-structural protein 1 (NSP1). J Biomol Struct Dyn 2020;1–11. DOI: 10.1080/07391102.2020.1792992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma A, Tiwari V, Sowdhamini R. Computational search for potential COVID-19 drugs from FDA-approved drugs and small molecules of natural origin identifies several anti-virals and plant products. J Biosci 2020; 45:100. DOI: 10.1007/s12038-020-00069-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen Z, Zhang G, Yang Y, Li M, Yang S, Peng G. Lysine 164 is critical for SARS-CoV-2 Nsp1 inhibition of host gene expression. J Gen Virol 2021; 102:001513. DOI: 10.1099/jgv.0.001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loureiro CL, Jaspe RC, D´Angelo P, Zambrano JL, Rodriguez L, Alarcon V, Delgado M, Aguilar M, Garzaro D, Rangel HR, Pujol FH. SARS-CoV-2 genetic diversity in Venezuela: predominance of D614G variants and analysis of one outbreak. PLoS One 2021; 16:e0247196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nascimento VAD, Corado ALG, Nascimento FOD, Costa Á, Kad Duarte DCG, Luz SLB, Gonçalves LMF, Jesus MS, Costa CFD, Delatorre E, Naveca FG. Genomic and phylogenetic characterisation of an imported case of SARS-CoV-2 in Amazonas state, Brazil. Mem Inst Oswaldo Cruz 2020; 115:e200310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403–10 [DOI] [PubMed] [Google Scholar]
- 15.Webb B, Sali A. Comparative protein structure modeling using modeller. Curr Protoc Bioinformatics 2016; 54:5.6.1–5.6.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 1997; 18:2714–23 [DOI] [PubMed] [Google Scholar]
- 17.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera – a visualization system for exploratory research and analysis. J Comput Chem 2004; 25:1605–12 [DOI] [PubMed] [Google Scholar]
- 18.Clark LK, Green TJ, Petit CM. Structure of nonstructural protein 1 from SARS-CoV-2. J Virol 2021; 95:e02019–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan S, Peng L, Park JJ, Hu Y, Devarkar SC, Dong MB, Shen Q, Wu S, Chen S, Lomakin IB, Xiong Y. Nonstructural protein 1 of SARS-CoV-2 is a potent pathogenicity factor redirecting host protein synthesis machinery toward viral RNA. Mol Cell 2020; 80:1055–66.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anger AM, Armache JP, Berninghausen O, Habeck M, Subklewe M, Wilson DN, Beckmann R. Structures of the human and drosophila 80S ribosome. Nature 2013; 497:80–5 [DOI] [PubMed] [Google Scholar]
- 21.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Crystallogr 1993; 26:283–91 [Google Scholar]
- 22.Semper C, Watanabe N, Savchenko A. Structural characterization of nonstructural protein 1 from SARS-CoV-2. iScience 2021; 24:101903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gavenonis J, Sheneman BA, Siegert TR, Eshelman MR, Kritzer JA. Comprehensive analysis of loops at protein-protein interfaces for macrocycle design. Nat Chem Biol 2014; 10:716–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi M, Wang L, Fontana P, Vora S, Zhang Y, Fu TM, Lieberman J, Wu H. SARS-CoV-2 Nsp1 suppresses host but not viral translation through a bipartite mechanism. SSRN Electron J 2020. DOI: 10.1101/2020.09.18.302901 [Google Scholar]
- 25.Benedetti F, Snyder GA, Giovanetti M, Angeletti S, Gallo RC, Ciccozzi M, Zella D. Emerging of a SARS-CoV-2 viral strain with a deletion in nsp1. J Transl Med 2020; 18:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, Kratzat H, Hayn M, Mackens-Kiani T, Cheng J, Straub JH, Stürzel CM, Fröhlich T, Berninghausen O, Becker T, Kirchhoff F, Sparrer KMJ, Beckmann R. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 2020; 369:1249–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Almeida MS, Johnson MA, Herrmann T, Geralt M, Wüthrich K. Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus. J Virol 2007; 81:3151–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. Variation in False-Negative rate of reverse transcriptase polymerase chain Reaction-Based SARS-CoV-2 tests by time since exposure. Ann Intern Med 2020; 173:262–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wathelet MG, Orr M, Frieman MB, Baric RS. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J Virol 2007; 81:11620. [DOI] [PMC free article] [PubMed] [Google Scholar]