Abstract
Activated tumor necrosis factor alpha (TNF-α) receptor 1 (TNFR1) recruits TNFR1-associated death domain protein (TRADD), which in turn triggers two opposite signaling pathways leading to caspase activation for apoptosis induction and NF-κB activation for antiapoptosis gene upregulation. Here we show that Stat1 is involved in the TNFR1-TRADD signaling complex, as determined by employing a novel antibody array screening method. In HeLa cells, Stat1 was associated with TNFR1 and this association was increased with TNF-α treatment. TNFR1 signaling factors TRADD and Fas-associated death domain protein (FADD) were also found to interact with Stat1 in a TNF-α-dependent process. Our in vitro recombinant protein-protein interaction studies demonstrated that Stat1 could directly interact with TNFR1 and TRADD but not with FADD. Interaction between Stat1 and receptor-interacting protein (RIP) or TNFR-associated factor 2 (TRAF2) was not detected. Examination of Stat1-deficient cells showed an apparent increase in TNF-α-induced TRADD-RIP and TRADD-TRAF2 complex formation, while interaction between TRADD and FADD was unaffected. As a consequence, TNF-α-mediated I-κB degradation and NF-κB activation were markedly enhanced in Stat1-deficient cells, whereas overexpression of Stat1 in 293T cells blocked NF-κB activation by TNF-α. Thus, Stat1 acts as a TNFR1-signaling molecule to suppress NF-κB activation.
Tumor necrosis factor alpha (TNF-α) is a pleiotropic cytokine that can elicit dual but opposing reactions from many different cell types: to live or to die (1, 20). Two types of TNF receptors (TNFR1 and TNFR2) have been characterized; TNFR1 has been found to be responsible for most of the biological properties of TNF-α. Studies of TNF-α signaling events have revealed that activated TNFR1 forms signaling complexes with a number of proteins, one of which is TNFR1-associated death domain protein (TRADD) (11). On the one hand, the TNFR1-TRADD complex can form the death-initiated signaling complex (DISC) by recruiting Fas-associated death domain protein (FADD), which leads to caspase activation and apoptosis (12). On the other hand, the TNFR1-TRADD complex can recruit receptor-interacting protein (RIP) and/or TNFR-associated factor 2 (TRAF2), leading to NF-κB activation (12, 13). NF-κB activation will turn on antiapoptotic genes and inhibit TNF-α-induced cell death (25, 27). Therefore, blocking NF-κB activation increases TNF-α-induced cell death, whereas enhanced NF-κB activity protects cells from TNF-α-induced death (2, 24, 26). It is conceivable that in order to induce apoptosis, TNF-α needs not only to form the DISC and trigger the caspase activation cascade but also to minimize the activation of NF-κB as much as possible. Little is known about the mechanism(s) by which TNFRs suppress the NF-κB activation pathway while forming the DISC to trigger apoptosis.
Ample evidence suggests that tyrosine phosphorylation events are involved in TNF-α signal transduction. Tyrosine phosphorylation of phosphatidylinositol (PI) 3-kinase was recently found to be involved in NF-κB activation by TNF-α (20). Although the tyrosine kinases responsible for TNF-α-induced protein phosphorylation remain unidentified, the association of the tyrosine kinase JAK with TNFR1 was observed (8). In 3T3-L1 adipocytes, Stat1 was tyrosine phosphorylated upon TNF-α stimulation, but its DNA binding activity was undetectable (8). Previous studies have indicated that TNF-α triggers less apoptosis in Stat1-deficient cells (16). Like TNF-α, some growth factors or cytokines can quickly induce Stat tyrosine phosphorylation with poor or no DNA binding activity detected (4, 9, 19). Many SH2-containing enzymes (e.g., SHP-2) have signaling effects in addition to their catalytic activities. Similarly, Stat proteins are SH2-containing transcription factors and may function as both signal transducer and transcription factor, as the name indicates. Through its association with TNF-α signaling factors, Stat1 may act as a signal transducer rather than a transcription activator.
In this study, we examined the role of Stat1 in TNF-α signal transduction. We screened Stat1-interacting proteins using a newly designed antibody array with which different antibodies against TNF-α signaling factors were immobilized. We have obtained strong evidence that Stat1 is a component of the TNFR1-TRADD signaling complex. By binding to TRADD, Stat1 attenuates the interactions of TRADD with RIP and TRAF2 without disturbing TRADD-FADD interaction. In cells lacking Stat1, TRADD-RIP or TRADD-TRAF2 interaction was enhanced and led to markedly enhanced NF-κB transcriptional activation in response to TNF-α. Consistently in 293T cells, transient overexpression of Stat1 blocked NF-κB activation by TNF-α. Therefore, by binding to the TNFR1-TRADD signaling complex, Stat1 favors DISC formation for apoptosis induction and prevents the signaling-complex formation required for NF-κB activation.
MATERIALS AND METHODS
Cell culture and whole-cell-extract preparation.
Cultures of the epithelial cell lines HeLa, A431, 293T, 2fTGH, U3A, and U3A-S1 were all grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. In U3A-S1 cells, the expression of Stat1 (wild type or Y701F mutant form) was restored by stable transfection with an expression vector as described previously (5). Confluent cells were treated with different cytokines as indicated below for 30 min or left untreated. Whole-cell extracts were then prepared with radioimmunoprecipitation assay buffer containing freshly added proteinase inhibitors by following the manufacturer's protocol (Santa Cruz Biotechnology).
Antibodies and antibody array screening.
Antibodies against TNFR signaling molecules used for making the antibody arrays and for the immunoprecipitations were obtained from Santa Cruz Biotechnology. They were anti-TNFR1 (goat polyclonal immunoglobulin G [IgG], against the C terminus), anti-TNFR2 (goat polyclonal IgG, against the C terminus), anti-FADD (goat polyclonal IgG, against the N terminus), anti-TRADD (goat polyclonal IgG, against the C terminus), anti-RIP (goat polyclonal IgG, against the C terminus), and anti-TRAF2 (rabbit polyclonal IgG, against the C terminus). One hundred polyclonal or monoclonal antibodies, including these against TNFR signaling proteins (0.5 μg each), were immobilized on polyvinylidene difluoride (PVDF) membranes (5 by 5 cm) at predetermined positions. The antibody array membranes were then incubated with 5% milk at room temperature for 2 h, followed by incubation with whole-cell lysates from HeLa cells (2 × 107) treated or not with recombinant human TNF-α (10 ng/ml) for 30 min. After incubation for 2 h, the membranes were washed three times with phosphate-buffered saline and blotted with horseradish peroxidase (HRP)-conjugated anti-Stat1 monoclonal antibody (against the Stat1 N terminus; Transduction Laboratory) for an additional 2 h, followed by three washes and enhanced chemiluminescence (ECL) detection.
cDNA constructs.
Full-length Stat1, N-terminal Stat1 fragments (representing N-terminal amino acids 1 to 488), and C-terminal Stat1 fragments (amino acids 489 to 750) were subcloned into the pET-28a vector (Novagen) for expression of recombinant His6-Stat1 proteins. All His6-expressing proteins were confirmed by Western blotting with anti-His6 antibody. The TNFR1 cytoplasmic domain (TNFR1C), TRADD, and FADD were subcloned into the pGEX-2T vector (Amersham Pharmacia) for expression of recombinant glutathione transferase (GST) proteins. GST-expressing proteins were confirmed by Western blotting with antibodies against TNFR1, TRADD, and FADD, respectively.
Recombinant GST protein interaction, immunoprecipitation, and immunoblotting.
GST fusion proteins were purified from bacteria using glutathione-agarose beads according to the manufacturer's instructions (Sigma). Whole-cell extracts prepared as described above were incubated with either GST fusion proteins (2 μg) or different antibodies (1 μg) conjugated with protein G agarose at 4°C from 6 h to overnight. The precipitates were washed three times with lysis buffer and resolved by sodium dodecyl sulfate–10 to 12% polyacrylamide gel electrophoresis, transferred to PVDF membranes, and immunoblotted with different antibodies. Immunoblot analyses were carried out using ECL as instructed (Amersham Pharmacia). Different GST fusion proteins were incubated with bacterially expressed recombinant His6-Stat1 proteins at 4°C overnight (see Fig. 2C). The GST precipitates were immunoanalyzed with anti-His6 monoclonal antibody (Amersham Pharmacia).
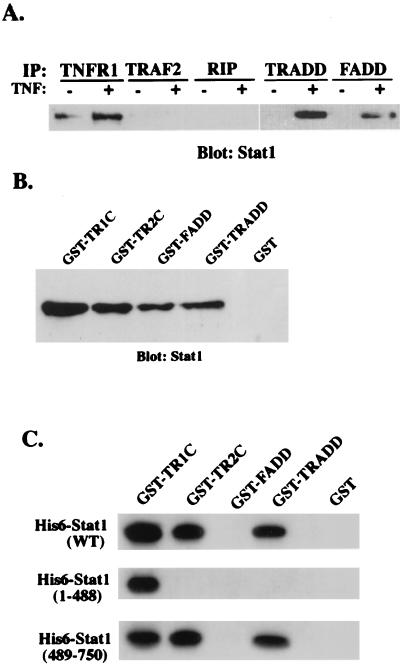
FIG. 2.
Stat1 is a component of the TNFR1-TRADD complex. (A) Whole-cell lysates of HeLa cells (2 × 107) treated or not with TNF-α (10 ng/ml) for 30 min were incubated with protein G-agarose beads conjugated with anti-TNFR1, anti-TRADD, anti-FADD, anti-RIP, or anti-TRAF2 antibody. After a washing, the bead-associated proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Western blotting with anti-Stat1 antibody. IP, immunoprecipitation. (B) Purified forms of GST-TNFR1C, GST-TNFR2C, GST-FADD, GST-TRADD, and GST alone on glutathione-agarose beads were incubated with TNF-α-treated HeLa cell lysates for 2 h at room temperature. After a washing, proteins bound to the beads were subjected to immunoblot analysis with anti-Stat1 antibody. (C) Bacterially expressed His6 epitope-tagged full-length Stat1 (WT) or N-terminally truncated (amino acids 1 to 488) and C-terminally truncated (amino acids 489 to 750) Stat1 proteins were incubated with GST-TNFR1C, GST-TNFR2C, GST-FADD, GST-TRADD, and GST alone, respectively. Precipitated proteins were detected by anti-His6 monoclonal antibody (Amersham Pharmacia).
NF-κB activity measurements.
For electrophoretic mobility shift assays (EMSAs), we used a radiolabeled double-stranded oligonucleotide corresponding to the NF-κB binding site in the mouse immunoglobulin enhancer. For Northern blotting assays, blots containing 20 μg of total RNA per lane at various time points were hybridized with specific probes that were 32P labeled with a randomly primed DNA labeling kit (Boehringer Mannheim). All probes were generated by cutting from their cDNA expression vectors. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as an equal RNA loading control. For NF-κB reporter gene assays, cells (2fTGH, U3A, and 293T) were transiently transfected with luciferase reporter construct 2×κB alone or together with TRADD, Stat1, and TRADD+Stat1 by the Lipofectamine method (Gibco/BRL). Twenty-four hours after transfection, cells were left untreated or treated with TNF-α (10 ng/ml) for 6 h. Luciferase activities were determined three times and normalized to the basal (untreated) levels.
RESULTS
Stat1 is a component of the TNFR1-TRADD signaling complex.
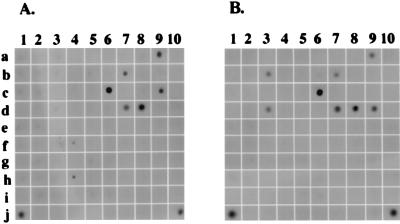
To identify Stat1-interacting molecules, we designed an antibody array that permits simultaneous detection of multiple protein-protein interactions under physiological conditions. The antibody array designed for this study contained 100 antibodies against different proteins, including TNFRs and their downstream signaling molecules. Individual antibodies were immobilized on PVDF membranes. By using these uniquely designed antibody array membranes, we screened for possible interactions between Stat1 and molecules involved in TNFR signaling pathways. Whole-cell extracts from HeLa cells that had been treated or not with recombinant human TNF-α for 30 min were incubated with antibody array membranes for 2 h. The arrays were then probed with HRP-conjugated anti-Stat1 antibody in order to identify Stat1-associated proteins. As can be seen in Fig. 1, Stat1 proteins were detected at several spots on the membrane, indicating the presence of Stat1-interacting proteins. While some interactions were unaffected by TNF-α treatment, others were either enhanced or reduced by TNF-α treatment (Fig. 1). For instance, Stat1 protein was detected by immobilized antibodies against TNFR1 and TNFR2 in both TNF-α-treated and untreated samples (Fig. 1, spots 7d and 8d). Moreover, Stat1 was detected by antibodies against TNFR1 adapter protein TRADD and TRADD binding protein FADD in TNF-α-treated cell lysates (Fig. 1B, spots 9d and 3b). As a positive control, Stat1 was detected at the position immobilized with antibody against CREB binding protein (CBP) (also called p300) (Fig. 1, spot 1j), indicating that the antibody array designed here was able to detect the interaction between Stat1 and CBP which was shown previously (3, 26). The protein-protein interactions observed here were also detected with other cell lines (data not shown).
FIG. 1.
Screening of Stat1-interacting proteins using a novel antibody array. PVDF membranes immobilized with different polyclonal or monoclonal antibodies were incubated with whole-cell lysates from HeLa cells (2 × 107) that were left untreated (A) or treated recombinant human TNF-α (10 ng/ml) for 30 min (B). After incubation for 2 h, the membranes were washed with phosphate-buffered saline and blotted with HRP-conjugated anti-Stat1 antibody followed by ECL detection. In addition to the antibodies against TNFR1 (7d), TNFR2 (8d), FADD (3b), and TRADD (9d), antibodies that also showed up positively in the array were P-CAM polyclonal antibody (Pharmingen) (9c), EGF receptor polyclonal antibody (Gibco/BRL) (6c), β-catenin monoclonal antibody (Transduction Laboratory) (10j), PI 3-kinase P85α polyclonal antibody (Santa Cruz Biotechnology) (7b), and CBP polyclonal antibody (Santa Cruz Biotechnology) (1j).
Interactions between Stat1 and individual TNF-α signaling molecules identified by the antibody array were confirmed by coimmunoprecipitation and GST fusion protein pulldown assays. In coimmunoprecipitation assays, we precipitated proteins from lysates of control and TNF-α-treated HeLa cells with TNFR1-specific antibody and analyzed them by Western blotting for Stat1. As shown in Fig. 2A, Stat1 was associated with TNFR1 and this association was greatly enhanced by treating the cells with TNF-α. Interestingly, although HeLa cells express a low but detectable level of TNFR2 (10), Stat1 was recovered from the TNFR2 immunoprecipitates under the same conditions (not shown). We then examined the interactions between Stat1 and TRADD. Although the same amounts of TRADD were immunoprecipitated from treated and untreated cells, only precipitates from TNF-α-treated cells contained Stat1 (Fig. 2A). Similarly, the association of Stat1 with FADD was detected only in the lysates of TNF-α-treated cells (Fig. 2A). Although a weak Stat1 signal was detected in the spot immobilized with antibody against TRAF2 in antibody array screening (Fig. 1A, spot 4f), Stat1 did not coprecipitate with TRAF2 or RIP in HeLa cells treated or not with TNF-α (Fig. 2A). Coprecipitation of other Stat family members (e.g., Stat3 and Stat5) with TRADD and FADD was not detected (data not shown), although they have previously been shown to be activated by TNF-α in adipocytes (8). To further confirm the interactions between Stat1 and TNFR signaling proteins, we constructed GST fusion proteins containing the intracellular domains of TNFR1 and TNFR2 (GST-TNFR1C and GST-TNFR2C) as well as GST fused to full-length TRADD and FADD. Recombinant GST fusion proteins were purified with glutathione-agarose beads and used to pull down Stat1 from cell lysates. Stat1 of TNF-α-treated HeLa cells was consistently found to interact with GST-TNFR1C, GST-TNFR2C, GST-FADD, and GST-TRADD, but not with GST alone (Fig. 2B). Thus, Stat1 appears to bind the TNFR1-TRADD-FADD complex as well as the TNFR2 cytoplasmic domain in cells.
In order to determine whether Stat1 directly interacts with these individual TNFR signaling molecules, we assessed the ability of purified GST-TNFR1C, GST-TNFR2C, GST-TRADD, and GST-FADD to bind recombinant histidine epitope (His6)-tagged full-length Stat1 and N-terminally (1 to 488 residues) and C-terminally (489 to 750 residues) truncated Stat1 in vitro. GST-TNFR1C, GST-TNFR2C, and GST-TRADD but not GST-FADD could precipitate His6-Stat1 (Fig. 2C). Interestingly, although only the C-terminal region of Stat1 could interact with TRADD and TNFR2C, both C-terminally and N-terminally truncated Stat1 proteins bound TNFR1C, suggesting that these two regions of Stat1 may cooperate for maximal binding of TNFR1. Thus, Stat1 interacts directly with TRADD but indirectly with FADD.
TNF-α-activated Stat1 does not translocate into nuclei and bind DNA.
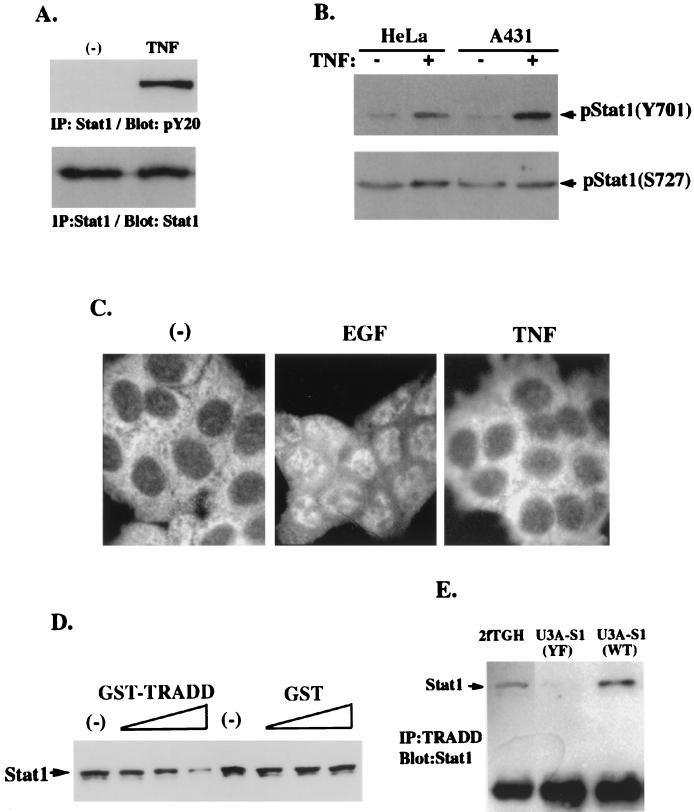
Stats recruited to ligand-bound receptors can be tyrosine phosphorylated and serve as activators of transcription after translocation into nuclei. The above results prompted us to investigate whether Stat1 was tyrosine phosphorylated and transcriptionally active upon TNF-α stimulation. Stat1 was immunoprecipitated with its specific antibody and subsequently blotted with antiphosphotyrosine antibody. As shown in Fig. 3A, TNF-α indeed induced tyrosine phosphorylation of Stat1 in HeLa cells. By using antibodies that can specifically recognize phospho-Stat1, we confirmed that the Y701 residue of Stat1 was phosphorylated upon TNF-α treatment in the HeLa and A431 cell lines (Fig. 3B), whereas Stat1 was constitutively phosphorylated at the S727 residue in these cells. However, in EMSA, DNA binding activity was undetectable with either nuclear extracts or whole-cell lysates prepared from TNF-α-treated HeLa and A431 cells incubated with the probe for Stat1 binding site M67SIE (data not shown). Taken together with previous data (8), these results suggested that Stat1 recruited by TNFR1 might function as a signal transducer rather than as an activator of transcription. We then examined Stat1 nuclear translocation in response to TNF-α treatment. In Fig. 3C, as a control, Stat1 in A431 cells treated with epidermal growth factor (EGF) exclusively translocated into nuclei. With TNF-α treatment, however, Stat1 nuclear translocation was not observed. It was recently reported that by binding to another protein factor, Stat protein might lose its DNA binding activity (6). Preincubation of GST-TRADD, but not GST alone, with whole-cell lysates of EGF-treated A431 cells reduced the amount of Stat1 bound to DNA (Fig. 3D). To confirm the role of residue Y701 of Stat1 in this TRADD-Stat1 interaction, we performed immunoprecipitation by using U3A cell lines restored with comparable levels of wild-type Stat1 and Y701F-Stat1. As expected, Stat1 was coimmunoprecipitated with TRADD in the parental 2fTGH cell line and the U3A cell line, which was restored with wild-type Stat1 (Fig. 3E), whereas Stat1-TRADD interaction was greatly reduced in the U3A cell line expressing Y701F-Stat1 (Fig. 3E). Therefore, tyrosine phosphorylation at the Y701 residue in response to TNF-α stimulation may be critical for Stat1 to interact with TRADD, and Stat1-TRADD complex formation may prevent Stat1 from nuclear translocation and binding to DNA.
FIG. 3.
TNF-α-phosphorylated Stat1 does not translocate into nuclei due to its interaction with TRADD. (A) Stat1 proteins were immunoprecipitated from whole-cell lysates of HeLa cells that received TNF-α treatment for 30 min or were left untreated. The immunoprecipitated Stat1 proteins were subjected to Western blotting with anti-pY20 or anti-Stat1 antibody. (B) Whole-cell lysates prepared from HeLa and A431 cells treated or not with TNF-α. Phospho-Stat1 was detected by immunoblotting with antibodies against Stat1-pY701 and Stat1-pS727. (C) A431 cells received no treatment (−) or were treated with EGF or TNF-α for 1 h and fixed with acetone (Santa Cruz Biotechnology protocol). Acetone-fixed cells were stained with anti-Stat1 antibody and examined with a fluorescent microscope. (D) Whole-cell lysates of EGF-treated A431 cells were preincubated with increasing amounts of GST-TRADD or GST alone and monitored by precipitation with SIE oligomer-agarose beads. After being washed, SIE bead precipitates were analyzed with Stat1 immunoblotting. (E) Parental 2fTGH cells, U3A-S1 WT cells expressing wild-type Stat1, and U3A-S1 YF cells expressing Y701F-Stat1 were incubated with TNF-α for 30 min. Whole-cell lysates prepared from these cells were incubated with agarose beads conjugated with anti-TRADD antibody. The TRADD immunoprecipitates (IP) were then analyzed with Stat1 immunoblotting.
Stat1-TNFR1-TRADD complex formation inhibits NF-κB activity.
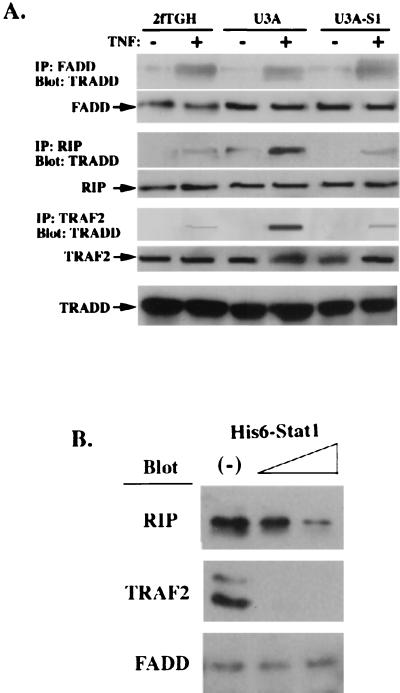
By binding to TNFR1, TRADD bifurcates for both FADD-mediated caspase activation and RIP- and/or TRAF2-mediated NF-κB activation (11, 12). The observation that Stat1 binds to TRADD and FADD but not to RIP and TRAF2 raised the possibility that Stat1 plays a role in either stabilizing the death signaling complex (TRADD-FADD) or preventing TRADD-RIP complex formation. In cells lacking Stat1 (U3A), although TRADD-FADD complex formation was unaffected (Fig. 4A), TRADD-RIP and TRADD-TRAF2 interactions in response to TNF-α were enhanced compared with those in either parental cells (2fTGH) or cells with Stat1 reconstituted (U3A-S1) (Fig. 4A). Competitive protein binding assays confirmed these results. In the presence of increasing amounts of His6-Stat1, the amounts of RIP and TRAF2 precipitated with GST-TRADD were markedly reduced (Fig. 4B). Under the same conditions, the level of FADD recovered from GST-TRADD precipitates was not apparently affected (Fig. 4B). Thus, by binding to the TNFR1-TRADD complex, Stat1 may inhibit TRADD-RIP and TRADD-TRAF2 interactions without detriment to TRADD-FADD formation.
FIG. 4.
Stat1 inhibits TRADD interaction with RIP and TRAF2 but not with FADD. (A) FADD, RIP, and TRAF2 were immunoprecipitated (IP) from whole-cell lysates prepared from 2fTGH, U3A, and U3A-S1 cells treated or not with TNF-α. FADD, RIP, and TRAF2 precipitates were then subjected to TRADD immunoblot analysis. The levels of FADD, RIP, TRAF2, and TRADD in these cell lysates were examined by blotting with their specific antibodies as indicated. (B) Whole-cell lysates prepared from TNF-α-treated HeLa cells were incubated with purified GST-TRADD and different amounts of recombinant His6-Stat1 protein (0.1 or 0.3 μg). GST-TRADD precipitates were subjected to anti-RIP, anti-TRAF2, and anti-FADD antibody immunoblot analysis.
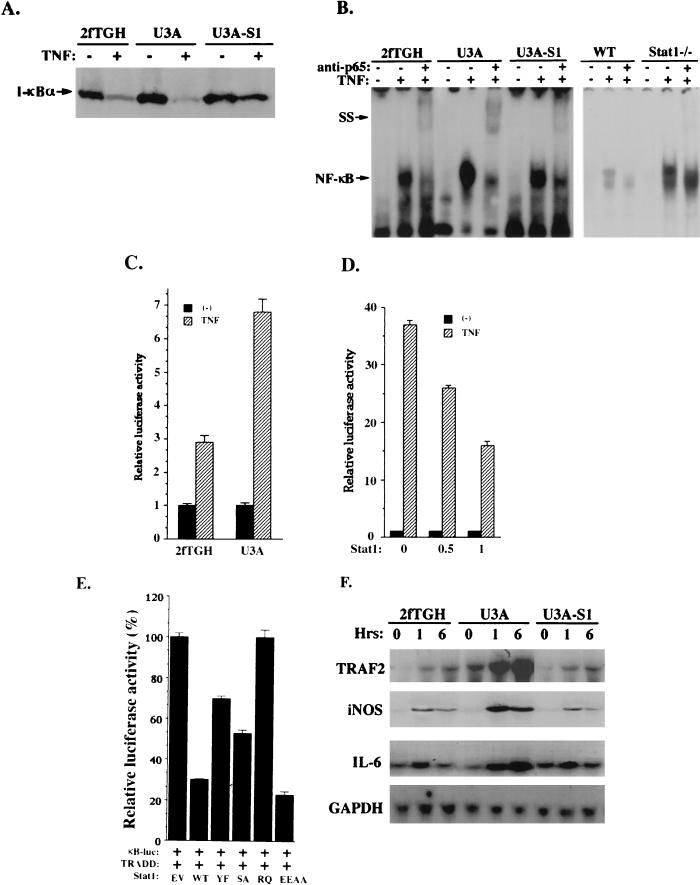
Both RIP and TRAF2 were considered to play roles in NF-κB activation through their interactions with TRADD. We hypothesized that by blocking TRADD-RIP and/or TRADD-TRAF2 interactions, Stat1 may inhibit NF-κB activation. To test this hypothesis, I-κB degradation and NF-κB activation in response to TNF-α were compared in 2fTGH, U3A, and U3A-S1 cells. Stat1-deficient U3A cells became more sensitive than 2fTGH cells and U3A-S1 cells to TNF-α in I-κB degradation (Fig. 5A). We then examined the DNA binding activity of NF-κB in these cells. Nuclear extracts were prepared from 2fTGH, U3A, and U3A-S1 cells treated or not with TNF-α and analyzed in EMSA with the NF-κB binding site as the probe. As expected, TNF-α-mediated NF-κB DNA binding activity was enhanced by a factor of 3 to 3.5 in U3A cells compared to 2fTGH and U3A-S1 cells (Fig. 5B, left panel). A similar but more striking result was obtained with Stat1 knockout mouse fibroblasts (Fig. 5B, right panel) even though comparable levels of NF-κB (p50 and p65) were present in these cells (data not shown). We suspected that the enhanced NF-κB DNA binding activity observed in Stat1-deficient cells might lead to enhanced NF-κB transcriptional activity. In order to confirm this, we examined the ability of TNF-α to activate the κB-dependent reporter construct upon transfection into U3A and 2fTGH cells. A luciferase reporter construct (pBIIx Luc) containing two copies of the NF-κB binding sequence was transfected into U3A and 2fTGH cells. TNF-α treatment resulted in an approximately sevenfold induction of luciferase activity in U3A transfectants but only a threefold induction in 2fTGH transfectants under the same conditions (Fig. 5C). Overexpression of Stat1 in 293T cells markedly reduced NF-κB activity in response to TNF-α, as indicated by the analysis of NF-κB-dependent luciferase activity (Fig. 5D).
FIG. 5.
Stat1 inhibits NF-κB activation by TNF-α. (A) Protein immunoblot analysis of I-κB-α in whole-cell extracts from 2fTGH, U3A, and U3A-S1 cells treated or not with TNF-α. (B) EMSA were performed as described previously (5) with nuclear extracts prepared from 2fTGH, U3A, and U3A-S1 cells with or without TNF-α treatment. The probe was derived from the NF-κB binding site in the promoter of the I-κB gene. The NF-κB gel shift complex can be supershifted (SS) by the antibody against NF-κB (p65) as indicated. WT, wild type. (C) U3A and 2fTGH cells (3 × 105) were transiently transfected with the 2×κB luciferase reporter construct. Twenty-four hours after transfection, cells were left untreated or treated with TNF-α and luciferase activities were determined. (D) In 293T cells (3 × 105), the 2×κB luciferase reporter was cotransfected with the empty vector (−) or Stat1 expression construct at different concentrations (0.5 and 1 μg). Luciferase activities were analyzed after TNF-α treatment. (E) In 293T cells, the 2×κB luciferase reporter was cotransfected with TRADD and Stat1. Luciferase activities were analyzed after TNF-α treatment. Stat1 proteins tested here include the wild type (WT) and Y701→F mutant (YF), S727→A mutant (SA), R602→Q mutant (RQ), and E495E496→AA mutant (EEAA) forms. (F) Cells were treated with TNF-α for different periods as indicated. Total RNA was isolated and subjected (20 μg) to analysis with probes for the TRAF2, inducible nitric oxide synthase, and interleukin-6 genes. To standardize for the amount of RNA loaded, filters were also probed with the GAPDH gene.
The effect of Stat1 on TRADD-mediated NF-κB activation was also tested. Cotransfection of wild-type Stat1 with TRADD markedly reduced NF-κB activation by TRADD (Fig. 5E). However, SH2 domain mutant Stat1 (R602→Q) completely lost its inhibitory effect on TRADD-dependent NF-κB activation, suggesting that the SH2 domain of Stat1 may play an important role in its association with TNFR1 and/or TRADD. Y701F-Stat1 was less effective than S727A-Stat1 in blocking NF-κB activation by TRADD, further supporting the notion that Y701 of Stat1 plays an important role in Stat1-TRADD interaction, as indicated by the immunoprecipitation data shown in Fig. 3E. In contrast, DNA binding domain mutant Stat1 (E495E496→AA) could still effectively block TRADD in NF-κB activation.
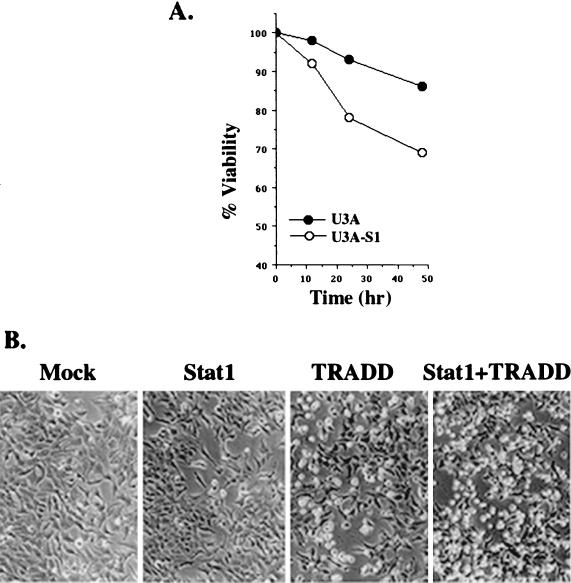
NF-κB activation by TNF-α can upregulate a variety of genes, including those involved in antiapoptosis. The gene expression patterns of TRAF2 as well as those of inducible nitric oxide synthase and interleukin-6 in response to TNF-α were evaluated in the 2fTGH, U3A, and U3A-S1 cell lines. As shown in Fig. 5F, U3A cells became more sensitive to TNF-α stimulation in the expression of all these genes. NF-κB-dependent TRAF2 expression has been recently shown to play an important role in antiapoptosis (23). TRAF2 induction was most striking in U3A cells but was modest in both 2fTGH and U3A-S1 cells. Consequently, less cell death was induced by TNF-α in Stat1-deficient U3A cells than in U3A-S1 cells with Stat1 reintroduced (Fig. 6A), consistent with a previous report (16). Cell death induced by transfection of TRADD alone was increased by cotransfection of TRADD with Stat1 in 293T cells (Fig. 6B) and COS cells (data not shown). Therefore, by binding to TRADD, Stat1 promotes apoptosis induction, presumably due to its inhibitory effect on NF-κB pathway activation.
FIG. 6.
Stat1 promotes cell death induction. (A) U3A and U3A-S1 cells were cultured in six-well dishes (106/well) and treated with TNF-α (50 ng/ml) for 12, 24, and 48 h. Cell viability was determined by trypan blue exclusion. Data reflect the average of three experiments. (B) 293T cells in six-well plates (3 × 105/well) received transient transfection with an empty vector (Mock), Stat1, TRADD, or Stat1 plus TRADD. Photographs were taken 30 h after transfection.
DISCUSSION
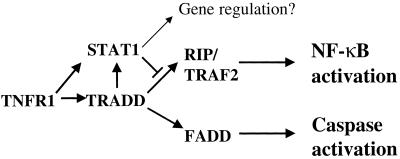
The data presented here demonstrate that Stat1 recruitment by TNFR1 can block NF-κB activation, serving as an important mechanism for facilitating TNF-α-mediated apoptosis induction. Indeed, in some cells, the NF-κB pathway is inactive in response to TNF-α (26). TRADD can interact with a number of TNF-α signaling proteins, and the balance between the TRADD-FADD and TRADD-RIP or TRADD-TRAF2 signaling complexes decides whether cells live or die. Although TRADD independently interacted with Stat1, RIP, and TRAF2, coprecipitation of Stat1 with RIP or TRAF2 was not detected. This indicates that Stat1 competes with RIP and TRAF2 to bind TRADD. An enhanced TRAF2 expression level in Stat1-deficient cells can be responsible for enhanced NF-κB activation. Nevertheless, in the presence of Stat1, interaction between TRADD and TRAF2 or between TRADD and RIP was dramatically blocked. Stat1 may therefore affect NF-κB activation at two levels: (i) by blocking interaction between TRAF2 and TRADD and (ii) by blocking TRAF2 gene expression in cells. However, NF-κB activation by TNF-α was not impaired in TRAF2 knockout animals even though TRAF2 has been considered as one possible link to NF-κB activation (17, 28). Therefore, NF-κB-dependent TRAF2 upregulation by TNF-α may be mainly involved in an NF-κB-independent antiapoptosis process. RIP is a serine/threonine kinase and has been considered as an important mediator of NF-κB activation by TRADD. In RIP-deficient cells, a failure of TNF-α to activate the transcription factor NF-κB was reported, suggesting that RIP-TRADD interaction plays a more important role in NF-κB activation by TNF-α (15). However, Stat1 facilitates TNF-α-mediated apoptosis induction at the TNFR1-TRADD complex formation-proximal step, either by displacing RIP and/or TRAF2 from TRADD or by disrupting other components required to execute the NF-κB activation signals (Fig. 7). Therefore, Stat1 should be a useful reagent for dissecting NF-κB activation initiated by other events, without excluding direct or indirect interaction between Stat1 and NF-κB (22). Although Stat1 and NF-κB can interact in nuclei with some nuclear factors, such as CBP for gene regulation, Stat1 does not translocate into nuclei for transcriptional activation in cells that have received TNF-α stimulation. Moreover, Stat1 suppresses NF-κB activity at a stage above I-κB degradation (Fig. 5A). Therefore, it is unlikely that enhanced NF-κB activity in Stat1-deficient cells is due to the lack of competition between Stat1 and NF-κB for a common nuclear factor.
FIG. 7.
Model of Stat1 in TNFR1 signal transduction. TNFR1-recruited Stat1 can form a complex with TRADD. By binding to TRADD, Stat1 prevents TRADD from further interacting with RIP and TRAF2 without affecting the association between TRADD and FADD. Consequently, the TRADD-mediated NF-κB activation process is blocked. Thus, Stat1 promotes TNF-α-induced apoptosis, presumably due to its inhibitory effect on NF-κB activation.
Stat1 deficiency on the one hand downregulates proapoptosis genes, such as caspase genes (5, 16), and on the other hand enhances NF-κB activity leading to the upregulation of antiapoptosis genes, such as the TRAF2 gene. It therefore adds an additional mechanism underlying TNF-α-mediated protection. Although Stat1 undergoes tyrosine phosphorylation upon TNF-α stimulation, its DNA binding activity is extremely weak or undetectable. These results are in agreement with a previous study (8). Some growth factor or cytokine receptors, such as hepatocyte growth factor receptor, CD40, and angiotensin II receptor, have been reported to recruit and induce tyrosine phosphorylation of Stat proteins that exhibit poor or undetectable DNA binding activity (4, 9, 19). Thus, Stat proteins may primarily serve as signal transducers when recruited by these receptors. It has been recently reported that tyrosine-phosphorylated Stat proteins lose their DNA binding activity by binding to other protein factors, such as PIAS (6). Similarly, Stat1 did not translocate into nuclei upon TNF-α stimulation, probably due to its formation of a complex with TRADD in the cytoplasm. Stat proteins can be recruited by a variety of growth factor or cytokine receptors in a constitutive or stimulation-dependent manner (7). Although there is no apparent homology between TNFR1 and TNFR2 in their cytoplasmic domains, Stat1 coprecipitated with them both. It is currently unclear which motifs of these receptors are responsible for recruiting Stat.
The reduced interaction between Y701F-Stat1 and TRADD (Fig. 3E) is consistent with the reduced effect of Y701F-Stat1 on TRADD-dependent NF-κB activation observed in Fig. 5E. These results suggest that tyrosine phosphorylation at the Y701 residue of Stat1 may be necessary for Stat1 to interact with TRADD. Currently, it is unclear whether tyrosine phosphorylation at residues other than Y701 is involved in mediating protein-protein interaction. The role of Stat proteins as adapters mediating signal transduction has been previously observed. In alpha interferon receptor, for instance, Stat2 serves as the adapter for Stat1 activation and Stat3 functions to couple another signaling pathway, i.e., the PI 3-kinase, to the receptor (18, 21). Our results provide evidence that tyrosine-phosphorylated Stat1 is involved in TNFR1 signal transduction. Rather than functioning as a nuclear transcription factor, Stat1 forms complexes with TRADD in the cytoplasm and regulates TRADD-mediated NF-κB activation.
The antibody array technology applied here allows for massive parallel protein analysis, a rapid and efficient method for large-scale protein-protein interaction examination. Antibody arrays can greatly increase the speed of experimental progress and will be valuable in the study of functional proteomics.
ACKNOWLEDGMENTS
We thank D. V. Goeddel for the TNFR1, TNFR2, and TRADD expression vectors, V. Dixit for the FADD expression vector, X.-Y. Fu for the Stat1 expression vector, S. Ghosh for the NF-κB site luciferase reporter construct (pBIIx Luc), G. Stark for the Stat1-deficient human cell line U3A and its parental cell line 2fTGH, and David Levy for the Stat1 knockout mouse fibroblasts. We also thank Alicia Chung and Michelle Embree for their critical reading of the manuscript.
Support was provided by Brown University School of Medicine (Y.E.C.).
Y.W. and T.R.W. contributed equally to this work.
REFERENCES
- 1.Baker S J, Reddy E P. Transducers of life and death: TNF receptor superfamily and associated proteins. Oncogene. 1996;12:1–9. [PubMed] [Google Scholar]
- 2.Beg A A, Baltimore D. An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D'Andrea A, Livingston D M. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 4.Boccaccio C, Ando M, Tamagnone L, Bardelli A, Michieli P, Battistini C, Comoglio P M. Induction of epithelial tubules by growth factor HGF depends on the STAT pathway. Nature. 1998;391:285–288. doi: 10.1038/34657. [DOI] [PubMed] [Google Scholar]
- 5.Chin Y E, Kitagawa M, Kuida K, Flavell R A, Fu X-Y. Activation of the STAT signaling pathway can cause expression of caspase 1 and apoptosis. Mol Cell Biol. 1997;17:5328–5337. doi: 10.1128/mcb.17.9.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung C D, Liao J, Liu B, Rao X, Jay P, Bert P, Shuai K. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- 7.Darnell J E., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 8.Guo D, Dunbar J D, Yang C H, Pfeffer L M, Donner D B. Induction of Jak/STAT signaling by activation of the type 1 TNF receptor. J Immunol. 1998;160:2742–2750. [PubMed] [Google Scholar]
- 9.Hanissian S H, Geha R S. Jak3 is associated with CD40 and is critical for CD40 induction of gene expression in B cells. Immunity. 1997;6:379–387. doi: 10.1016/s1074-7613(00)80281-2. [DOI] [PubMed] [Google Scholar]
- 10.Heller R A, Song K, Fan N, Chang R A. The p70 tumor necrosis factor receptor mediates cytotoxicity. Cell. 1992;70:47–56. doi: 10.1016/0092-8674(92)90532-h. [DOI] [PubMed] [Google Scholar]
- 11.Hsu H, Xiong J, Goeddel D V. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 12.Hsu H, Shu H-B, Pan M-G, Goeddel D V. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 13.Hsu H, Huang J, Shu H-B, Baichwal V, Goeddel D V. TNF-dependent recruitment of the protein kinase RIP to the TNF receptor-1 signaling complex. Immunity. 1996;4:387–396. doi: 10.1016/s1074-7613(00)80252-6. [DOI] [PubMed] [Google Scholar]
- 14.Karras J G, Wang Z, Huo L, Frank D A, Rothstein T L. Induction of STAT protein signaling through the CD40 receptor in B lymphocytes: distinct STAT activation following surface Ig and CD40 receptor engagement. J Immunol. 1997;159:4350–4355. [PubMed] [Google Scholar]
- 15.Kelliher M A, Grimm S, Ishida Y, Kuo F, Stanger B Z, Leder P. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity. 1998;8:297–303. doi: 10.1016/s1074-7613(00)80535-x. [DOI] [PubMed] [Google Scholar]
- 16.Kumar A, Commane M, Flickinger T W, Horvath C M, Stark G R. Defective TNF-alpha-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science. 1997;278:1630–1632. doi: 10.1126/science.278.5343.1630. [DOI] [PubMed] [Google Scholar]
- 17.Lee S Y, Reichlin A, Santana A, Sokol K A, Nussenzweig M C, Choi Y. TRAF2 is essential for JNK but not NF-kappaB activation and regulates lymphocyte proliferation and survival. Immunity. 1997;7:703–713. doi: 10.1016/s1074-7613(00)80390-8. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Leung S, Kerr I M, Stark G R. Functional subdomains of STAT2 required for preassociation with the alpha interferon receptor and for signaling. Mol Cell Biol. 1997;17:2048–2056. doi: 10.1128/mcb.17.4.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marrero M B, Schieffer B, Paxton W G, Heerdt L, Berk B C, Delafontaine P, Bernstein K E. Direct stimulation of Jak/STAT pathway by the angiotensin II AT1 receptor. Nature. 1995;375:247–250. doi: 10.1038/375247a0. [DOI] [PubMed] [Google Scholar]
- 20.Ozes O N, Mayo L D, Gustin J A, Pfeffer S R, Pfeffer L M, Donner D B. NF-kappaB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature. 1999;401:82–85. doi: 10.1038/43466. [DOI] [PubMed] [Google Scholar]
- 21.Pfeffer L M, Mullersman J E, Pfeffer S R, Murti A, Shi W, Yang C H. STAT3 as an adaptor to couple phosphatidylinositol 3-kinase to the IFNAR1 chain of the type I interferon receptor. Science. 1997;276:1418–1420. doi: 10.1126/science.276.5317.1418. [DOI] [PubMed] [Google Scholar]
- 22.Shen C-H, Stavnezer J. Interaction of Stat6 and NF-κB: direct association and synergistic activation of interleukin-4-induced transcription. Mol Cell Biol. 1998;18:3395–3404. doi: 10.1128/mcb.18.6.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith C A, Farrah T, Goodwin R G. The TNF receptor superfamily of cellular and viral proteins: activation, costimulation, and death. Cell. 1994;76:959–962. doi: 10.1016/0092-8674(94)90372-7. [DOI] [PubMed] [Google Scholar]
- 24.Van Antwerp D J, Martin S J, Kafri T, Green D R, Verma I M. Suppression of TNF-alpha-induced apoptosis by NF-kappaB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 25.Wang C Y, Mayo M W, Korneluk R G, Goeddel D V, Baldwin A S., Jr NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281:1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 26.Wang C Y, Mayo M W, Baldwin A S., Jr TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-kappaB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 27.Wu M X, Ao Z, Prasad K V, Wu R, Schlossman S F. IEX-1L, an apoptosis inhibitor involved in NF-kappaB-mediated cell survival. Science. 1998;281:998–1001. doi: 10.1126/science.281.5379.998. [DOI] [PubMed] [Google Scholar]
- 28.Yeh W C, Shahinian A, Speiser D, Kraunus J, Billia F, Wakeham A, de la Pompa J L, Ferrick D, Hum B, Iscove N, Ohashi P, Rothe M, Goeddel D V, Mak T W. Early lethality, functional NF-kappaB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity. 1997;7:715–725. doi: 10.1016/s1074-7613(00)80391-x. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath C M, Darnell J E., Jr Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]