ABSTRACT
The TREX-2 complex is essential for the general nuclear mRNA export in eukaryotes. TREX-2 interacts with the nuclear pore and transcriptional apparatus and links transcription to the mRNA export. However, it remains poorly understood how the TREX-2-dependent nuclear export is connected to the subsequent stages of mRNA trafficking. Here, we show that the PCID2 subunit of Drosophila TREX-2 is present in the cytoplasm of the cell. The cytoplasmic PCID2 directly interacts with the NudC protein and this interaction maintains its stability in the cytoplasm. Moreover, PCID2 is associated with the cytoplasmic mRNA and microtubules. The PCID2 knockdown blocks nuclear export of mRNA and also affects the general mRNA transport into the cytoplasm. These data suggest that PCID2 could be the link between the nuclear TREX-2-dependent export and the subsequent cytoplasmic trafficking of mRNA.
KEYWORDS: Cytoplasmic mRNA transport, mRNA trafficking, PCID2, TREX-2
Introduction
Post-transcriptional mRNA biogenesis is a multistep process playing an important role in the implementation of cellular functions. The mRNA processing and maturation partially occur co-transcriptionally and some of the RNA binding proteins responsible for its final fate bind to the mRNA during transcription [1]. The mRNP particle containing mature mRNA with the associated proteins, is transported through the nuclear pore by protein complexes responsible for the mRNA nuclear export [2,3]. Following mRNA nuclear export, mRNP undergoes remodelling on the cytoplasmic side of the nuclear envelope (for review, see [4]). For further transport, mRNP particles bind to the microtubule-associated motor proteins and are transported along the microtubules to their destination sites. While different stages and components of mRNA trafficking have been characterized [5], many of them still remain unknown.
The THSC/TREX-2 mRNA nuclear export complex is highly conserved in eukaryotes [6–10]. It was first found in yeast [6] and later described in Drosophila (designated as AMEX), humans and plants [11,12,13,14,15]. TREX-2 is essential for the general nuclear mRNA export, and depletion of the TREX-2 subunits causes an accumulation of mRNA in the nucleus [6, 9,11,15,16,17,18,19].
The Yeast TREX-2 includes Sac3, Thp1, Sus1 (two molecules), Cdc31 and Sem1 subunits [17,20] (Table 1). Sac3 (human GANP) is a scaffold which organizes the other subunits into a complex [14]. TREX-2 is localized in the nucleoplasm and interacts with the nuclear pore complex [11,15,17,21]. It also interacts with the transcriptional apparatus, thus linking transcription to the mRNA export [10,11,14,22]. However, despite of the important role TREX-2 played in the nuclear mRNA export, the exact mechanism through which TREX-2 is linked to the cytoplasmic mRNA transport, remains unknown.
Table 1.
The TREX-2 subunits in different species
| Orthologous TREX-2/THSC/AMEX complex | |||
|---|---|---|---|
| H. sapiens | D. melanogaster | S. cerevisiae | A. thaliana |
| GANP | Xmas-2 | Sac3 | SAC3 |
| ENY2 | ENY2 | Sus1 | SUS1 |
| PCID2 | PCID2 | Thp1 | THP1 |
| DSS1 | Sem1p | Sem1 | DSS1 |
| CETN2/CETN3 | Cdc31 | CEN1/CEN2 | |
The TREX-2 complex purified from Drosophila [23] contained the Xmas-2, PCID2 and ENY2 subunits (homologs of the yeast Sac3, Thp1, and Sus1, respectively). Xmas-2 and ENY2 are localized in the nucleoplasm and interact with nuclear pores; their knockdowns lead to a general block of nuclear mRNA export [11]. The Drosophila PCID2 was first identified in a genome-wide screen for proteins involved in mRNA nuclear export. It was shown to shuttle between the nucleus and the cytoplasm, and is associated with the polysome fraction [24]. PCID2 also contains the PCI domain, which functions in protein–protein interactions [25]. The crystallization of the yeast TREX-2 demonstrates that the α-helix containing the region of Sac3 (dXmas-2) and the C-terminal region of Thp1 (dPCID2) form a platform that is predicted to interact with mRNA [13,26].
Here, we show that PCID2, together with mRNA, is transferred from the nucleus to the cytoplasm of the cell. While transitioning through the nuclear pore, PCID2 leaves behind the TREX-2 complex and interacts with the NudC protein, and this interaction stabilizes PCID2 in the cytoplasm. Cytoplasmic PCID2, together with the NudC, is associated with mRNA and microtubules, suggesting its participation in the mRNA trafficking. A knockdown of PCID2 caused a general block in the nuclear mRNA export and affected the normal mRNA distribution pattern in the cytoplasm.
Materials and methods
Antibodies
Polyclonal antibodies against PCID2 (Ab1) (amino acid fragment 278–395), NudC (amino acid fragment 166–496), Ubiquitin (2 repeats of 76 aa) were raised in our laboratory by immunizing rabbits with the corresponding His6-tagged protein fragments. Polyclonal anti-Dynactin antibodies (p150 subunit) (amino acid fragment 835–1265) were raised in our laboratory by immunizing rats with the corresponding His6-tagged protein fragment. All rabbit antibodies were affinity purified. We also used an already described polyclonal anti-Xmas-2 antibodies raised in our laboratory [11]. An anti-β-tubulin antibody obtained by M. Klymkowsky and the antibody against lamin Dmo (obtained by Fisher P.A.) were obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD, and maintained at the Department of Biological Sciences, University of Iowa. Anti-α-tubulin antibodies were also obtained from the Developmental Studies Hybridoma Bank and were deposited to the DSHB by J. Frankel/E.M. Nelsen. Cy3-conjugated goat anti-rabbit IgG antibody (Amersham) and Alexa Fluor 488-conjugated goat anti-mouse IgG antibody (Molecular Probes) were used as secondary antibodies.
Purification of the PCID2-containing complex from the cytoplasm
The cytoplasmic material was extracted from the 0 to 12 hour Drosophila embryos, as described earlier [27].
The cytoplasmic extract was separated by chromatography on a MonoQ HR 16/10 column, two times on a MonoQ 5/50 GL, and finally, on a Superdex 200 10/300 GL column. The purification procedure is shown in Fig. 3B. The columns were equilibrated with HEMG buffer (25 mM HEPES-KOH at pH 7.6, 12.5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 1 mM DTT) containing 150 mM NaCl (HEMG-150). Immunoaffinity purification was performed on a column with immobilized anti-PCID2 antibodies; the unbound proteins were washed out with HEMG-500 buffer containing 0.1% NP-40. For details of the purification procedure and MALDI-TOF MS experiments, see [28,29]. The Superdex 200 10/300 GL column was calibrated with an HMW Calibration Kit (GE Healthcare). The void volume of the column was 5.0 mL, and the volume of each fraction was 0.5 mL.
Figure 3.
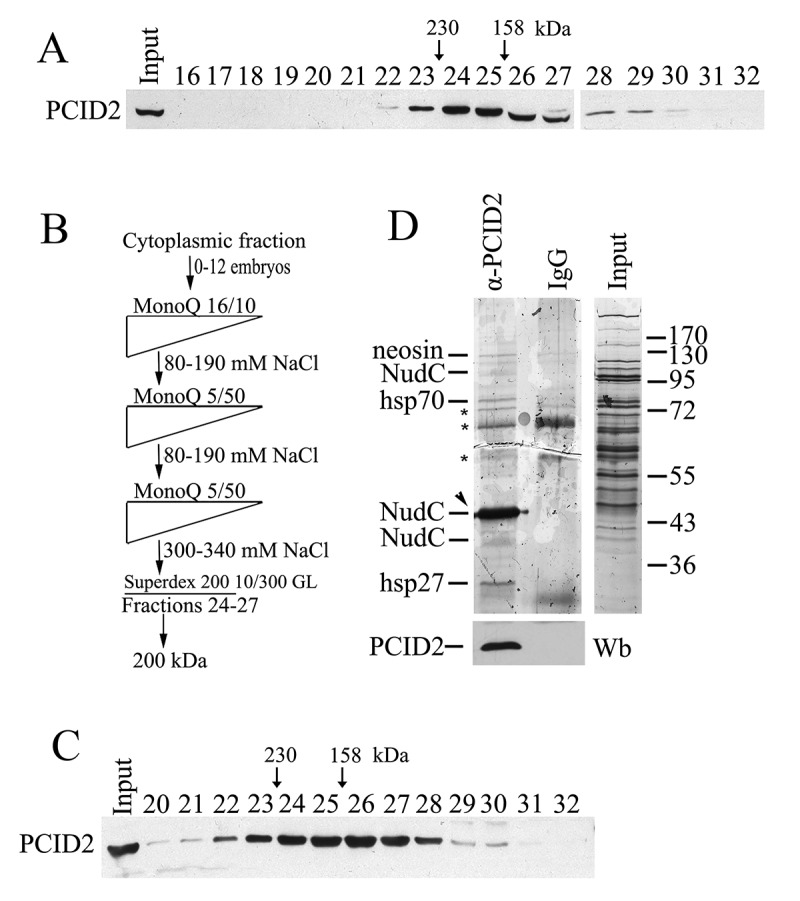
PCID2 interacts with NudC in the cytoplasm
(a) The cytoplasmic extract from Drosophila embryos (0–12 h) treated with DNase I and RNase, was fractionated on a Superdex 200 10/300 GL gel filtration column, and the fractions were analysed for the presence of PCID2 by Western blot analysis. Fraction numbers are indicated. The void volume corresponds to fraction 10. (b) The schematic of purification of PCID2-interacting proteins from the cytoplasmic extract. At each step, the proteins were eluted with a NaCl gradient, and the fractions were analysed for the presence of PCID2 by Western blotting. The peak PCID2-containing fractions were collected and loaded into the next column. After fractionation on a Superdex 200 10/300 GL column, the material was loaded onto an immunosorbent with anti-PCID2 antibodies, washed, and eluted with acid glycine. (c) The migration profile of PCID2 on the Superdex 200 10/300 GL column at the final step of purification. (d) Preparation of PCID2-associated proteins purified from the cytoplasmic extract of embryos (silver staining). Fractions 23–28 (Fig. 3c) from the last step of fractionation were incubated with affinity-purified anti-PCID2 antibodies covalently bound to protein A Sepharose. Proteins were eluted, resolved by 9% SDS-PAGE, and analysed by mass spectrometry. The control immunoprecipitation of the same material with IgG and the protein extract (Input) are shown. The non-specific bands which were also found in IgG are indicated by asterisks. The preparation also contains the trace amounts of neosin, Hsp70, and Hsp26 which may specifically co-precipitate with PCID2. The amount of PCID2 in preparation was not high as it mostly remained bound to antibodies. The PCID2 band coincides with the NudC band (indicated by arrowhead) as both proteins have the same electrophoretic mobility in the gel. To confirm the presence of PCID2 the same preparations were stained with anti-PCID2 antibodies (bottom panel). The preparation also contains the additional weak bands of NudC which are likely NudC dimer (upper band) and the result of partial degradation or represent a different NudC isoform (lower band).
Drosophila cell culture extracts
Drosophila Schneider line 2 (S2) cells were maintained at 25°C in Schneider’s insect medium (Sigma) containing 10% fetal bovine serum (HyClone, United States). To extract proteins, S2 cells were lysed in 10 mM HEPES, pH 7.9, containing 5 mM MgCl2, 0.5% NP-40, 0.45 M NaCl, 1 mM DTT, and complete protease inhibitor cocktail (Roche). Immunoprecipitation was performed as described in [11], with preliminary treatment of the extract with DNase I (USB, 0.6 U/mL) and RNase (Stratagene, 10 U/mL). Proteins from S2 cells were separated into the nuclear and cytoplasmic fractions (for RIP immunoprecipitation) using lysis buffer [40 mM HEPES, pH 7.8, containing 2.5 mM MgCl2, 100 mM NaCl, 1 mM DTT, and complete protease inhibitor cocktail (Roche)]. For nuclear lysis, we used the WLB solution (10 mM HEPES, pH 7.0, with 100 mM KCl, 5 mM MgCl2, 25 mM EDTA, 0,5% NP-40, 1% Triton X-100, 0.1% SDS, and 10% glycerol) [30]. Prior to immunoprecipitation, the extract was treated with DNase I (USB, 0.6 U/mL).
RNA immunoprecipitation
Co-IP of Ras2, and tubulin56D mRNA with antibodies was performed as described previously [30] from the lysate of S2 cells. No cross-linking reagent was used. For immunoprecipitation, the antibody–protein A sepharose beads were incubated with the lysate for 2 h at 4°C in NT2 buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.05% NP-40) containing 4 U/mL RiboLock (Thermo Scientific), 1% BSA, and 1 mg/mL ssDNA. After washing, TRI Reagent (Molecular Research Center, Inc) was added to extract RNA. An oligo-dT primer was used for reverse transcription. The levels of Ras2, tubulin56D mRNA were measured by qPCR using primers: 5ʹ-ATGCAAACGTACAAACTGGTGG-3ʹ and 5ʹ-GTCGCACTTGTTACCCACCATC-3ʹ; 5ʹ-CGAGAACACGGACGAGACCTACTG-3ʹ and 5ʹ-GGAATCGGAGGCAGGTGGTTACG-3ʹ. Each experiment was performed in at least three repeats, and the mean value was calculated.
FISH
Staining of total polyA RNA was carried out as reported previously [31]. The Cy3-labelled oligo-dT probe of 50 bp was used. In brief, S2 cells were allowed to adhere to coverslips, washed once in 1× PBS, and fixed with 3.7% paraformaldehyde in PBS for 10 min. After fixation, cells were washed in 1× PBS, permeabilized, and washed in 1× PBS again. For hybridization, the coverslips were incubated in hybridization buffer (2× SSC, 20% formamide, 0.2% BSA, 10% dextran sulphate, 1 mg/ml of total yeast tRNA) supplemented with 0.1 pmol/ml oligo(dT) labelled Cy3 probe.
Drosophila cell culture and RNAi knockdown experiments
Drosophila S2 cells were cultured at 25°C in Schneider’s insect medium (Sigma) containing 10% fetal bovine serum (HyClone). The RNAi experiments followed a published protocol [32]. We used 5–7 µg of dsRNA per 1–3 × 106 cells; dsRNA was synthesized with an Ambion MEGAscript T7 kit. The expression of the target genes was measured by qPCR and Western blot analysis. dsRNA corresponding to a fragment of green fluorescent protein (GFP) was used as a negative control. The following primers were used for the synthesis: GFP GPF 5ʹ-CGACTCACTATAGGGAGACGTAAACGGCCACAAGTTCAGC-3ʹ and 5ʹ-CGACTCACTATAGGGAGAGATGCCGTTCTTCTGCTTGTCG-3ʹ; Xmas2 5ʹ-GAATTAATACGACTCACTATAGGGAGAATGACCTGCACCGTAAG-3ʹ and 5ʹ-GAATTAATACGACTCACTATAGGGAGACCGGTTGTAGTTCATAG-3ʹ;
PCID2 5ʹ- CGACTCACTATAGGGAGAGTAGGTAGACGGGCTATGTTCG-3ʹ and 5ʹ- CGACTCACTATAGGGAGACAGTTTGTTGTGAGCATGTGAG-3ʹ;
NudC-N 5ʹ-CGACTCACTATAGGGAGACATTATTTCTACATGTGCCGGG-3ʹ and 5ʹ- CGACTCACTATAGGGAGAATTCGGCATCAGCTTACCCA-3ʹ; NudC-C 5ʹ- CGACTCACTATAGGGAGAAATGGCTGCACCTTGGAAAA-3ʹ and 5ʹ- CGACTCACTATAGGGAGAGCATTAGTTTATTGTGTGGT-3ʹ;
Ubi 5ʹ- CGACTCACTATAGGGAGAACCATCACCCTCGAGGTAGAGCCC-3ʹ and 5ʹ- CGACTCACTATAGGGAGATCCTCCACGGAGACGGAGCA-3ʹ.
Immunostaining
Immunostaining of S2 cells was performed as described earlier [11] using rabbit anti-PCID2, rabbit anti-NudC, mouse β-tubulin and corresponding secondary antibodies (Molecular Probes). The results were examined using an LSM 510 META (Zeiss) confocal microscope or a Nikon Eclipse Ti fluorescence microscope (Nikon) equipped with a CFI60 × 63 objective, and recorded using a DS-Qi2 camera.
Quantitation of fluorescence
Quantitation of fluorescence was performed using the ImageJ program according to the recommendations of the ImageJ User Guide [33].
Statistical analysis
Error bars represent SD for at least three independent experiments. All data are presented as means ± SD, and Student’s t-test was used to compare the control and treatment groups. An asterisk (*) indicates statistical significance with p-value < 0.05; (**) indicates statistical significance with p-value < 0.01.
GST Pull-down assay
The coding sequences of PCID2 and NudC were cloned into the constructs in frame with the His tag, which was cloned in pET22b (Novagene, United States) and into the constructs in frame with the GST epitope, which was cloned in pGEX-5X-1 vector (GE Healthcare Life Sciences). Expression of the recombinant His- or GST-tagged proteins was performed in E.coli BL21 cells at 20°C for 24 h. The cells were collected by centrifugation and stored at −70°C. Recombinant proteins with the His-tag were purified from cell lysates by binding to Ni-NTA (Invitrogen) and elution at rising imidazole concentrations. Recombinant proteins with the GST epitope tag were purified from the cell lysates by binding to the Glutathione SepharoseTM high performance medium and subsequently eluted by displacement with glutathione according to the manufacturer’s recommendations (GE Healthcare).
GST-fused protein immobilized on the Glutathione SepharoseTM high performance medium (GE Healthcare) was washed extensively with WLBPD-100 buffer (0.1% Triton X-100, 100 mM NaCl, 1× PBS, PIC). His-PCID2 protein was added to the resin, and the interaction assay was carried out in a final volume of 600 mL. After incubation, beads were washed five times with WLBPD-1000 (1% Triton X-100, 1 M NaCl, 1× PBS, PIC) [34]. Western blotting and immunoprecipitation were carried out using the standard protocols.
Results
PCID2 is essential for general mRNA transport, both in the nucleus and in the cytoplasm
The Drosophila nuclear TREX-2 complex purified in our previous study, contained Xmas-2, ENY2, and PCID2 (42 kDa). It was shown that the Xmas-2 and ENY2 subunits of the Drosophila TREX-2 are localized in the nucleoplasm, are associated with the nuclear pore, and participate in the nuclear export of mRNA [11]. But the role of Drosophila PCID2 in the mRNA export was not investigated.
To investigate the role of PCID2 in the nuclear mRNA export, we performed a knockdown of the Xmas-2 and PCID2 factors in S2 cells. The distribution of the total mRNA in knockdown cells was studied using a Cy3-labelled oligo-dT probe. The images of the cells were taken on a fluorescent microscope (Fig. 1a). We have quantified the distribution of the resulting fluorescent signal along the path from the nucleus to the cytoplasm. The distribution of the signal for individual knockdown cells is demonstrated in Fig. 1b [33]. A representative sample of ten randomly chosen cells from each knockdown experiment are also shown (Supplementary Fig. 1b).
Figure 1.
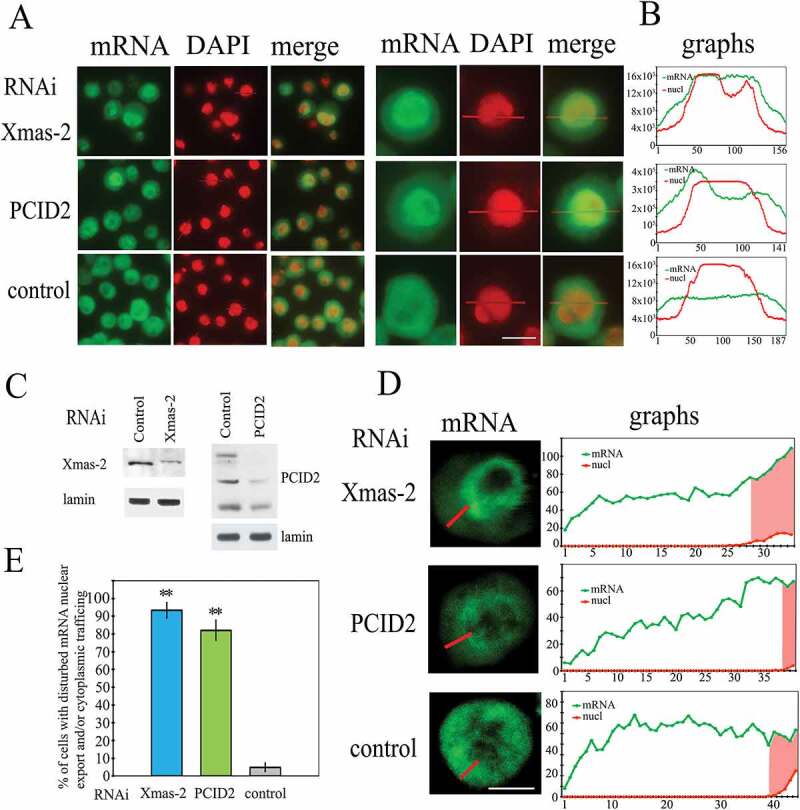
The effect of the RNAi-mediated knockdown of PCID2 and Xmas-2 on the poly(A) RNA distribution in S2 cells
(a) The distribution of mRNA in control, Xmas-2, and PCID2 RNAi knockdown in S2 cells. The poly(A) RNA was stained using a fluorescent-labelled oligo dT probe. Cells were co-stained with DAPI to visualize nuclei. Note that here and in the figures below, for better visualization of merged images, DAPI was pseudo-coloured in red. The fluorescent microscope images of the fields with several cells and the image of one representative cell are shown. Scale bar here and on other images, 10 µm. (b) Quantitation of the level of fluorescent signal in the corresponding cell was performed using ImageJ program. The intensity of the signal (Y-axis) in each point (x-axis) is demonstrated. Red graphs show the distribution of DAPI signal, while green graphs indicate the distribution of the oligo dT signal. The larger fields with signal quantification are shown in Supplementary Fig. 1. (c) The levels of Xmas-2 and PCID2 proteins in the RNAi knockdown cells. (d) The confocal microscopy images of single representative cells and quantification of the level of fluorescent signal in their cytoplasm performed using the ImageJ program. Green graphs indicate the distribution of oligo dT signal; the nuclei on the graphs are shown in red. (e) The percentages of cells with affected mRNA nuclear export (Xmas-2), or nuclear export and mRNA trafficking (PCID2). In control, knockdown cells, with both nuclear export and trafficking defects were calculated. Each RNAi knockdown experiment was performed in four replicates. About 300 cells were examined in blind count in each of the replicates, and the mean value was calculated. Error bars represent SD for at least three independent experiments. All data are presented as means ± SD, and Student’s t-test was used to compare the control and treatment groups. An asterisk (*) indicates the statistical significance with p-value < 0.05; (**) indicates the statistical significance with p-value < 0.01.
As demonstrated earlier, the RNAi knockdown of Xmas-2 and ENY2 subunits of the TREX-2 cause a general block of the nuclear mRNA export in S2 cells [11]. Indeed, in the Xmas-2 RNAi knockdown cells, most of the mRNA is accumulated in the nuclei, while mRNA distribution was rather uniform in the control wild-type Drosophila cells. Similar to the Xmas-2 RNAi, the RNAi knockdown of PCID2, caused an accumulation of mRNA in the nucleus (Fig. 1a,b, Supplementary Fig. 1a,b).
In Xmas-2 or PCID2 RNAi knockdown cells, some amount of mRNA was also observed in the cytoplasm that could be explained by an incomplete elimination of these factors by the knockdowns (Fig. 1c). To find out whether the Xmas-2 or PCID2 RNAi affected cytoplasmic mRNA trafficking, an analysis of the mRNA distribution in the cytoplasm of the knockdown cells was performed by fluorescence quantitation of the confocal microscope images (Fig. 1d). The graphs demonstrate a uniform distribution of mRNA in the cytoplasm of the control cells. The distribution of mRNA in the cytoplasm of the Xmas-2 knockdown cell, was uniform and similar to that in the control cells, indicating that Xmas-2 does not participate in the cytoplasmic transport of mRNA. Importantly, the distribution profile of mRNA in the cytoplasm of PCID2-depleted cells was in the form of a gradient with maximal accumulation of mRNA close to the nuclear envelope, indicating that PCID2 also could be involved in the maintenance of cytoplasmic mRNA transport.
Numerical analysis of the normal cells and the affected cells after knockdown, demonstrated that a high percentage of cells had impaired nuclear export or the cytoplasmic mRNA trafficking (Fig. 1e).
PCID2 is present in the cytoplasm and is associated with the nuclear membrane of S2 cells
We have raised rabbit polyclonal antibodies against the C-terminal PCID2 fragment (Ab). The affinity-purified antibodies recognized three bands (about 42, 47, and 52 kDa) on Western blot in the S2 cell extract (Supplementary Fig. 2a). The size of the lower band (about 42 kDa) corresponded to a calculated size of PCID2 (about 45 kDa), while the other bands were about 5 and 10 kDa higher (Fig. 2A, Supplementary Fig. 2a). A blocking with the immunizing peptide was performed to further confirm that all three detected bands indeed represented PCID2 (Supplementary Fig. 2b).
Figure 2.
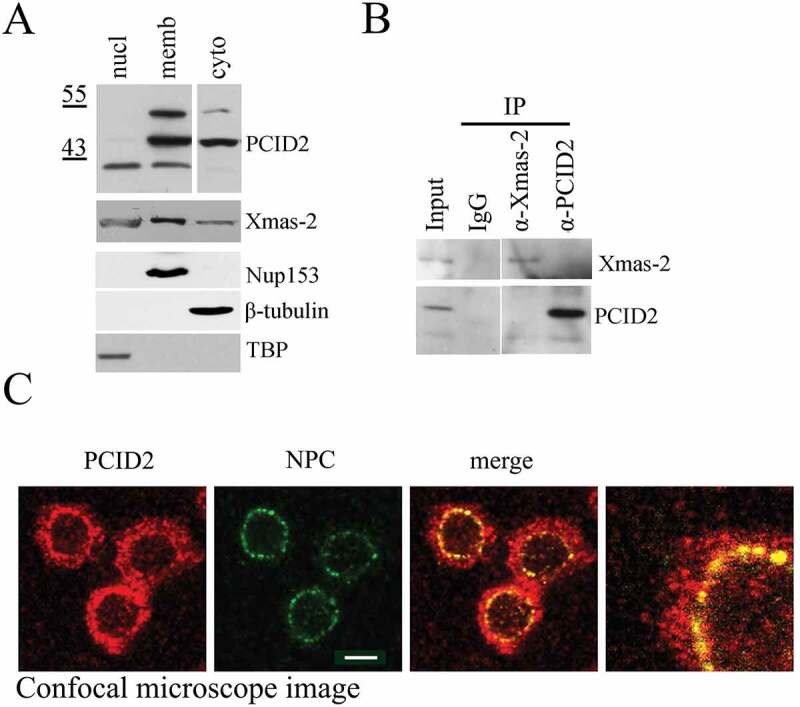
Characterization of Drosophila PCID2 protein
(a) Western-blot demonstrating the distribution of PCID2 and Xmas-2 in the cell. The same S2 cells were fractionated into nuclear (nucl), cytoplasmic (cyto), and membrane (memb) fractions and 1/10 of each extract was loaded on SDS PAGE. Western-blot was developed with anti-PCID2 or anti-Xmas-2 antibodies. The same extracts were stained with antibodies against Nup153, tubulin, and TBP to confirm the specificity of fractionation (the lower panels). (b) The results of immunoprecipitation experiments with antibodies against PCID2 and Xmas-2 or with IgG (control) from the cytoplasmic extract of S2 sells are shown on Fig. 2a. Blots were stained with antibodies against Xmas-2 and PCID2. (c) Confocal microscope image of the Drosophila S2 cells immunostained with antibodies against PCID2 and NPC and the merged and scaled up images. Scale bar, 10 μm.
Next, the origin of the PCID2 forms was investigated. The PCID2 gene contains no introns and no PCID2 isoforms were predicted in the Drosophila Genome Database (http://flybase.org/reports/FBtr0076102), thus suggesting that the observed PCID2 forms likely could be a result of different posttranslational modifications. The differences in the molecular weight of PCID2 bands indicated that it could be ubiquitinated. To test this hypothesis, we performed immunoprecipitation from the S2 cell extract with anti-PCID2 antibodies. Equal aliquots of the precipitate were resolved on SDS PAGE, and separate parts of the membrane were stained with either anti-PCID or anti-Ubiquitin (Ub) antibodies (Supplementary Fig. 2c). While anti-PCID2 antibodies recognized all three PCID2 bands in the precipitate, the anti-Ub antibodies recognized only two higher PCID2 bands, suggesting that they indeed corresponded to the ubiquitinated PCID2 forms. To confirm this result, the effect of the ubiquitin gene RNAi knockdown on the size and/or intensity of the PCID2 bands in the S2 cells was studied (Supplementary Fig. 2d). The knockdown did not affect the 42 kDa band; however, it eliminated two upper PCID2 bands. The differences in the molecular weights of the two upper PCID2 bands led to a suggestion that they could represent mono- and di-ubiquitinated forms.
To investigate the distribution of PCID2 forms in the cell, different cellular fractions were isolated as described previously [23]. The nuclear extract contained only the lower (42 kDa) PCID2 band, while the cytoplasmic fraction contained the 47 kDa band (Fig. 2A). All three PCID2 bands were associated with the membrane fraction, the higher band being membrane fraction-specific. We compared the distribution of PCID2 with that of the Xmas-2 subunit of TREX-2 (Fig. 2a). Xmas-2 was also present in all three fractions, its higher amount was detected in the nuclear membrane fraction that correlated with the association of Xmas-2 with NPC. Since some amount of Xmas-2 was also detected in the cytoplasm, we tested whether PCID2 interacts with Xmas-2 in the cytoplasmic extract in co-immunoprecipitation experiment (Fig. 2b). However, no interaction was observed, indicating that PCID2 is present in the cytoplasm independently of the TREX-2 complex.
Next, the confocal images of the S2 cells stained with the antibodies against PCID2 and NPC, confirmed that PCID2 was present in both nuclei and cytoplasm, and colocalized with the nuclear pore complex on the nuclear envelope (Fig. 2c). The same distribution was observed for the FLAG-tagged PCID2 (Supplementary Fig. 2e).
The cytoplasmic PCID2 interacts with the NudC chaperone protein
Next, we were interested in identifying proteins that interact with PCID2 in the cytoplasm. The cytoplasmic extract was prepared from the Drosophila 0–12 h embryos according to a published protocol [27]. Fractionation of the extract on a Superdex 200 10/300 GL gel filtration column indicated that cytoplasmic PCID2 migrated in several fractions. Its concentration was the highest in fractions 24–27, which corresponded to molecular mass of ~200 kDa (Fig. 3a). Several steps of chromatographic purification of the PCID2-containing complex were performed (Fig. 3b). At the last step, the PCID2-containing fractions were collected and loaded onto the Superdex 200 10/300 GL column (Fig. 3c). The PCID2 migrated in the same Superdex 200 10/300 GL fractions as during fractionation of the crude extract, suggesting that the PCID2-containing complex remained stable during purification. The PCID2-positive Superdex 200 10/300 GL fractions were collected, and the PCID2-containing complex was precipitated with anti-PCID2 antibodies covalently bound to Protein A. The precipitated proteins were eluted from the resin, resolved on SDS-PAGE, and identified by MALDI-TOF MS analysis. The preparation contained trace amounts of neosin, Hsp70, and Hsp26 which may specifically co-precipitate with PCID2. However, the prevailing was a single protein band that specifically co-precipitated with PCID2 and corresponded to the NudC protein (Fig. 3d). The amount of PCID2 in preparation (Fig. 3d) is low, as it mostly remained bound to the antibody-conjugated resin. In addition, PCID2 and NudC have the same electrophoretic mobility. Thus, to confirm the presence of PCID2, the same preparations were stained with the anti-PCID2 antibodies (Fig. 3d, lower panel).
NudC, the nuclear distribution protein C, is a cytoplasmic protein that is found in different organisms, from S. pombe to humans. It is associated with the motor dynein complex and microtubules and is involved in multiple processes connected with the cytoskeleton (for review, see [35]. In addition, NudC was shown to possess chaperone activity [36] and contains a highly conserved p23 domain responsible for chaperone function [35]. It was shown that this protein is essential for stability several proteins (Lis1, cofilin1) in the cytoplasm.
To continue our investigation, we have raised, and affinity purified anti-NudC antibodies in rabbits. They recognized a band of molecular weight similar to NudC, in the protein extract from S2 cells (Fig. 4a, Supplementary Fig. 3a). A knockdown of NudC resulted in a decrease in the intensity of this band, thus confirming the specificity of antibodies (see Fig. 5a,b below). On the Superdex 200 10/300 GL gel-filtration column, NudC migrated in the same fractions as PCID2, confirming their presence in the same complex (Supplementary Fig. 3b,c). The immunoprecipitation experiments from the whole S2 extract demonstrated a strong interaction of NudC with the cytoplasmic PCID2, but not with other PCID2 forms (Fig. 4a). Both proteins depleted each other from the extract, suggesting that most of the cytoplasmic PCID2 is associated with NudC. We generated bacterially expressed GST-tagged NudC and His-tagged PCID2, which also co-precipitated, thus strongly indicating their direct interaction (Fig. 4b, Supplementary Fig. 3d). The interaction of NudC and PCID2 expressed in bacteria also indicates that ubiquitylation of PCID2 is not required for interaction but may have distinct functions.
Figure 4.
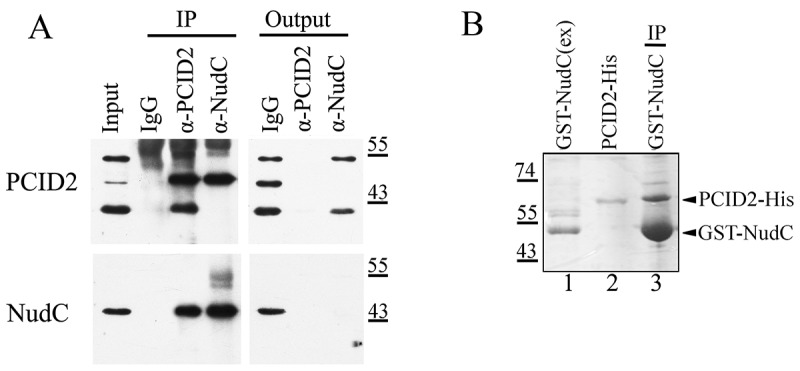
NudC specifically interacts with the cytoplasmic form of PCID2
(a) Co-immunoprecipitation from the S2 cell lysate was performed with antibodies against PCID2, NudC, or with IgG. Equal amounts of the extract (Inp) and precipitated proteins (IP) were analysed. The membrane was developed with antibodies against PCID2 or NudC.(b) The His-tagged PCID2 was purified from bacteria (lane 2) and incubated with bacterial extract containing GST-tagged NudC (lane 1). Please, note that for a better identification of the GST-tagged NudC, the extract used for incubation, was diluted (1:20). The extract was further incubated with Glutathione Sepharose, and the precipitated proteins (lane 3) were resolved on the SDS-PAGE and visualized by Coomassie staining.
Figure 5.
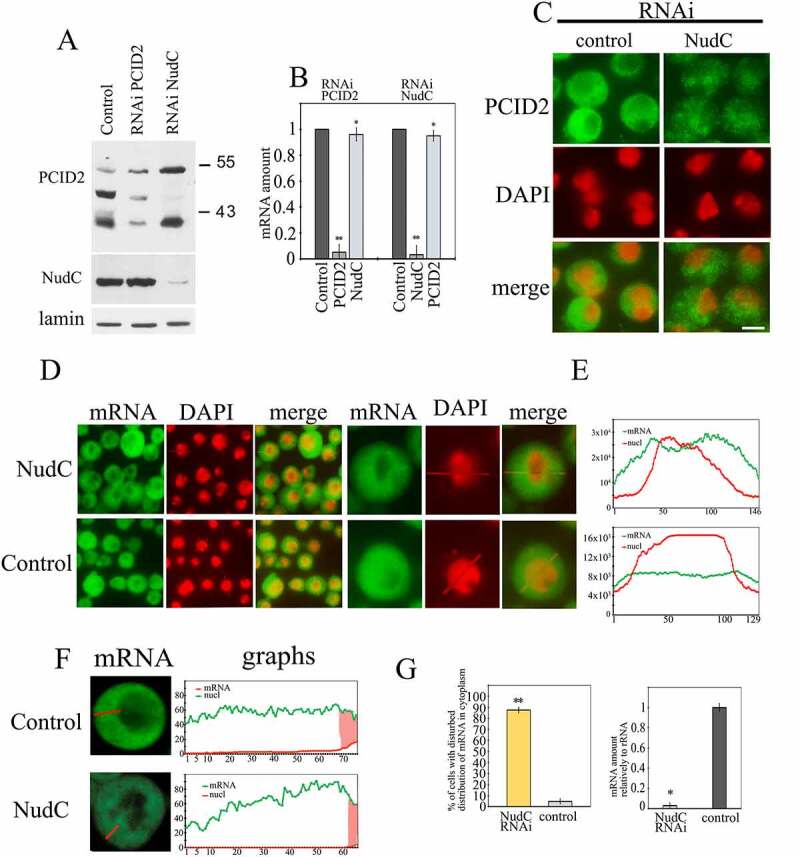
The effect of NudC knockdown on PCID2 stability and mRNA transport
The RNAi knockdown of NudC or control RNAi with GFP fragment was performed in S2 cells. (a) Western-blot demonstrates the level of PCID2 and NudC in the control, PCID2, or NudC RNAi knockdown S2 cells. The PCID2 and NudC were detected in the protein extracts using the corresponding antibodies. Lamin was used as a loading control. (b) The level of PCID2 and NudC transcripts in control RNAi, PCID2 RNAi knockdown, or NudC RNAi knockdown. Expression level was normalized to that in the control knockdown. Error bars represent SD for at least three independent experiments. All data are presented as means ± SD, and Student’s t-test was used to compare the control and treatment groups. An asterisk (*) indicates the statistical significance with p-value < 0.05; (**) indicates the statistical significance with p-value < 0.01. (c) Control and NudC RNAi S2 cells were immunostained with anti-PCID2 antibodies and fluorescent secondary antibodies, and co-stained with DAPI (pseudo-collored in red). Scale bar, 10 μm. The larger fields are shown in Supplementary Fig. 3e. (d–g) The effect of NudC knockdown on poly(A) mRNA distribution in the cell. The poly(A) RNA was stained using a fluorescent-labelled oligo dT probe. (d) The fluorescent microscope images of the fields with several cells and the image of one representative cell are shown. (e) Quantitation of the level of fluorescent signal performed using the ImageJ program. The intensity of the signal (Y-axis) in each point (x-axis) is shown. Red graphs show the distribution of DAPI signal, while green graphs indicate the distribution of oligo dT signal. The larger fields are shown in Supplementary Fig. 4. (f) The confocal microscopy images of single representative cell and quantitation of the level of fluorescent signal in cytoplasm performed using the ImageJ program. Green graphs indicate the distribution of oligo dT signal; the nuclei are shown in red. Scale bar, 10 µm. (g) The percentages of cells with damaged mRNA trafficking (left panel). Each RNAi knockdown experiment was performed in four replicates. About 300 cells were examined in blind count in each of the replicates, and the mean value was calculated. The efficiency of NudC knockdown (the level of corresponding transcripts) is demonstrated on the right panel. The level of each transcript in the knockdown cells relative to the control cells is shown. The level of transcription was normalized against the ribosomal RNA. Error bars represent SD for at least three independent experiments. All data are presented as means ± SD, and Student’s t-test was used to compare the control and treatment groups. An asterisk (*) indicates the statistical significance with p-value < 0.05; (**) indicates the statistical significance with p-value < 0.01.
NudC stabilizes PCID2 in the cytoplasm
Next, the role of NudC and PCID2 interaction was studied. The RNAi knockdown of PCID2 affected neither the protein nor the transcription level of NudC (Fig. 5a,b). On the contrary, NudC knockdown led to a specific elimination of the cytoplasmic PCID2 form (Fig. 5a). This effect occurred at the protein level, since RNAi knockdown of NudC did not affect the PCID2 transcription (Fig. 5b). As NudC was suggested to be a chaperone [36], it could be possible that NudC plays a role of a stabilization factor for PCID2 in the cytoplasm. Immunostaining of S2 cells with anti-PCID2 antibodies also demonstrated that RNAi knockdown of NudC strongly decreased the PCID2 level in the cytoplasm (Fig. 5c, Supplementary Fig. 3e).
We have further investigated the participation of PCID2 in the mRNA transport in the cytoplasm and the involvement of NudC in this process. The NudC RNAi knockdown was performed in the S2 cells and mRNA distribution was examined under a fluorescent microscope using oligo(dT) probe (Fig. 5d,e, Supplementary Fig. 4). In the NudC knockdown cells, mRNA was not uniformly distributed in the cytoplasm, as it was in the wild type cells. Instead, its distribution was in a gradient with a maximal accumulation of mRNA around the nuclear envelope and resembled that in the PCID2 knockdown cells. A more precise studying of the distribution of mRNA in the cytoplasm by the confocal microscope imaging (Fig. 5f), confirmed the similarity between the cytoplasmic mRNA distribution in PCID2 (Fig. 1d) and NudC knockdown cells (Fig. 5f). Our quantification of cells having cytoplasmic mRNA trafficking defects indicated that NudC is involved in the maintenance of cytoplasmic mRNA transport (Fig. 5g).
PCID2 is associated with mRNA and microtubules in the cytoplasm
To confirm PCID2 participation in the mRNA transport into the cytoplasm, we verified interactions of PCID2 with Ras2 and tubulin56D mRNAs in the cytoplasmic extracts, making use of the RNA immunoprecipitation experiments (RIP). Previously, Ras2 and tubulin56D mRNAs were shown to be associated with TREX-2 complex in the nuclear extract of S2 cells [23,37]. RIP experiments demonstrated that, as it was expected for the subunit of the TREX-2 complex, PCID2 was associated with the Ras2 and tubulin56D mRNAs in the nucleus (Fig. 6a).
Figure 6.
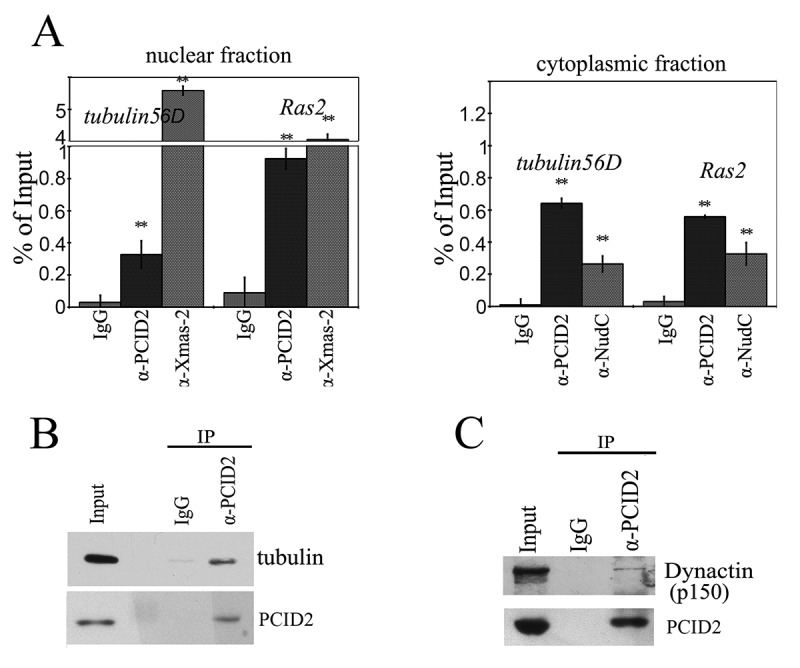
PCID2 interacts with cytoplasmic mRNA and the cytoskeleton
(a) The results of immunoprecipitation of tubulin 56D and Ras2 mRNA from the nuclear and cytoplasmic fractions of S2 cells performed with anti-PCID2 and anti-NudC antibodies (IgG was used as a negative control). The results of RNA immunoprecipitation are shown as a percentage of input. Error bars represent SD for at least three independent experiments. All data are presented as means ± SD, and Student’s t-test was used to compare the control and treatment groups. An asterisk (*) indicates the statistical significance with p-value < 0.05; (**) indicates the statistical significance with p-value < 0.01. (b) Anti-PCID2 antibodies co-precipitate tubulin from the cytoplasmic extract of S2 cells. Anti-β-tubulin antibodies were used in Western blot. Equal amounts of the extract (Inp) and precipitated proteins (IP) were analysed. (c) Anti-PCID2 antibodies co-precipitate the p150 subunit of Dynactin complex from the cytoplasmic extract of S2 cells. Equal amounts of the extract (Inp) and precipitated proteins (IP) were analysed.
The interactions of PCID2 with Ras2 and tubulin56D mRNAs were also found in the cytoplasm (Fig. 6a). The anti-NudC antibody also precipitated a noticeable amount of cytoplasmic Ras2 and tubulin56D mRNAs.
The close interaction between PCID2 and NudC led us to a hypothesis that, similar to NudC, PCID2 could be associated with tubulin and motor proteins. Indeed, we have found that PCID2 interacted with tubulin in the co-immunoprecipitation experiments (Fig. 6b). We have also observed an interaction between PCID2 and the dynactin subunit (p150) of the dynein–dynactin complex (Fig. 6c). The dynactin complex is a multiprotein assembly required for dynein motor to provide the cargo movement along the microtubules. It includes a number of proteins that have domains capable of binding dynein, microtubules, and cargos [38].
Discussion
The evolutionarily conserved TREX-2 complex of eukaryotes is essential for the general mRNA nuclear export [11–13,15,20]. It interacts with the transcription apparatus and the nuclear pore complex, and links transcription to mRNA export [11,14,15,17,21]. Here, we demonstrated that one of the core TREX-2 subunits, PCID2, is also present in the cytoplasm where it is associated with the NudC protein (Fig. 7). We further demonstrated that cytoplasmic PCID2 is associated with mRNA, tubulin, and motor proteins, and together with NudC, is essential for the cytoplasmic mRNA trafficking.
Figure 7.
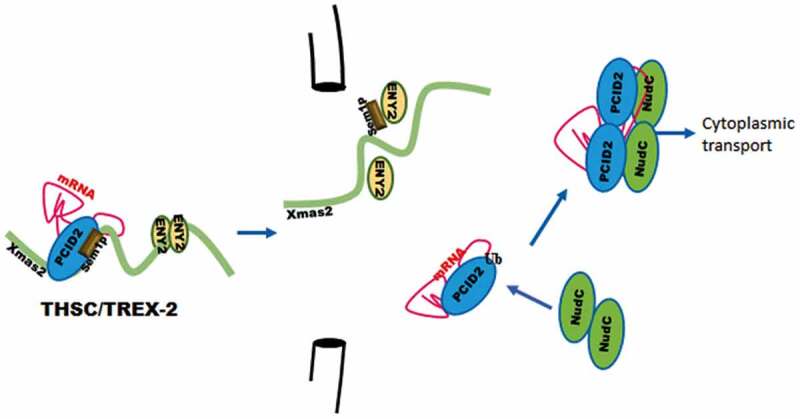
PCID2 subunit of the TREX-2 mRNA complex participates in nuclear export and cytoplasmic trafficking of mRNA
PCID2 subunit of TREX-2 complex is essential for general mRNA nuclear export and interacts with nuclear pore. In cytoplasm, PCID2 interacts with NudC protein. Together with NudC, PCID2 is associated with cytoplasmic mRNA, and is essential for general mRNA trafficking in cytoplasm.
The interaction between PCID2 and NudC is essential for the PCID2 stability in the cytoplasm, which correlates with the chaperone function of NudC shown previously [36,39].
Several studies demonstrated that similar to PCID2, NudC is associated with the motor proteins. The Drosophila NudC interacts with Lis1 [40–42] which facilitates association of dynein and dynactin with mRNA [43]. Mammalian NudC mediates interactions of the dynein and dynactin complex with kinesin-1 and supports their transport by kinesin-1 [44], for review see [35]. This indicates that NudC, similar to PCID2, is involved in the tubulin-associated mRNA trafficking.
Our study demonstrated that PCID2 is associated with mRNA, both in the nuclei and in the cytoplasm. This stable association could be explained by a direct interaction of PCID2 with mRNA. Indeed, the Drosophila PCID2, as well as its homologs in other organisms, contains the C-terminal RNA-interacting domain [13,25]. In addition, PCID2 contains the PCI domain, which is a universal domain supporting the protein-protein interactions [25]. It is possible, that PCI domain mediates PCID2 interactions with different targets, and that PCID2 acts as an adaptor for other factors that determine mRNA nuclear export, and the localization of transcripts in the cytoplasm.
We have found that non-ubiquitinated, mono-, or di-ubiquitinated forms of PCID2 are present in the cell. The unmodified PCID2 is present in the nucleus as a subunit of the TREX-2 complex, while mono-ubiquitinated form is specific for the cytoplasm. All three PCID2 forms were found in the nuclear membrane fraction, suggesting that ubiquitination is associated with the nuclear envelope, in particular, with NPC. This colocalization of Drosophila PCID2 with NPC was shown in this study. The association of PCID2 homologs, as well as of other TREX-2 subunits with NPC was also demonstrated in yeasts, Drosophila, and humans [11,15,17,21]. It could be suggested that ubiquitination of PCID2 regulates its dissociation from the TREX-2 complex, or PCID2 shuttling between the nucleus and the cytoplasm, which was demonstrated previously [24].0
The function of PCID2 in the cytoplasmic mRNA transport may be conserved in evolution, as some data on the cytoplasmic PCID2 localization were obtained for human cells [45,46]. According to the Human Protein Atlas, a moderate cytoplasmic localization of PCID2 was detected in most normal and cancer human tissues (www.proteinatlas.org). The role of PCID2 in the cytoplasmic transport in mammalian and yeast models is a question for future studies.
The PCID2 protein was shown to have important functions in mammalian cells. It is essential for the B-cell differentiation and survival, as it regulates the MAD2 (mitotic checkpoint protein 2) mRNA export [46]. PCID2 also interacts with BRCA2 protein and stabilizes it [45]. An increased expression level of PCID2 correlates with an unfavourable prognosis for renal as well as head and neck cancers. Here we presented data showing that PCID2 participates not only in the mRNA export from the nucleus to the cytoplasm but also in the subsequent cytoplasmic trafficking of mRNA. Whether the latter function of PCID2 is evolutionarily conserved in mammalian cells remains an open question.
Supplementary Material
Acknowledgments
We are grateful to José González Castaño and Vladic Mogila for critical reading the manuscript, to A. Brechalov for his help in purification of the complex and I. Toropygin for his assistance with MALDI experiments. We thank the Center for Precision Genome Editing and Genetic Technologies for Biomedicine, IGB RAS (Acta Pure protein purification system (GE Healthcare), Nanodrop, LightCycler96 (Roche)).
Funding Statement
This study was supported by program ‘Postgenome technologies and prospects for biomedical solutions’, Russian Academy of Sciences. Part of this work (sequencing) was supported by grant [075-15-2019-1660] from the Ministry of Science and Higher Education of the Russian Federation for the Center for Precision Genome Editing and Genetic Technologies for Biomedicine, Engelhardt Institute of Molecular Biology, Russian Academy of Sciences.
Disclosure statement
The authors declare no conflict of interest.
Author contributions
S.G.G. and D.V.K. conceived the project and edited the manuscript. D.V.K. supervised and designed the project, wrote the original draft, carried out statistical and fluorescence microscopy data analysis, performed FISH, RNA immunoprecipitation and coimmunoprecipitations. M.M.K. produced the antibodies to PCID2 and cultivated S2 cell culture, E.N.N. maintained Drosophila stocks. A.A.G. generated the antibodies to NudC and Dynactin, performed western blot analysis, purified the PCID2 containing protein complex, prepared the S2 and embryonic extracts, performed immunocytochemical stainings, molecular cloning, extraction and purification of the recombinant proteins, performed the RNAi knockdown, GST Pull-Down and coimmunoprecipitation assays.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Schlautmann LP, Gehring NHA.. Day in the life of the exon junction complex. Biomolecules. 2020;10:866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Xie Y, Ren Y.. Mechanisms of nuclear mRNA export: a structural perspective. Traffic. 2019;20:829–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Borden KLB. The nuclear pore complex and mRNA export in cancer. Cancers (Basel). 2020;13:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Di Liegro CM, Schiera G, Di Liegro I. Regulation of mRNA transport, localization and translation in the nervous system of mammals (Review). Int J Mol Med. 2014;33:747–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gagnon JA, Mowry KL. Molecular motors: directing traffic during RNA localization. Crit Rev Biochem Mol Biol. 2011;46:229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Fischer T, Strasser K, Racz A, et al. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. Embo J. 2002;21:5843–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kohler A, Exporting HE. RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8:761–773. [DOI] [PubMed] [Google Scholar]
- [8].Luna R, Gonzalez-Aguilera C, Aguilera A. Transcription at the proximity of the nuclear pore: a role for the THP1-SAC3-SUS1-CDC31 (THSC) complex. RNA Biol. 2009;6:145–148. [DOI] [PubMed] [Google Scholar]
- [9].Fischer T, Rodriguez-Navarro S, Pereira G, et al. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nat Cell Biol. 2004;6:840–848. [DOI] [PubMed] [Google Scholar]
- [10].Gonzalez-Aguilera C, Tous C, Gomez-Gonzalez B, et al. THP1-SAC3-SUS1-CDC31 complex works in transcription elongation-mRNA export preventing RNA-mediated genome instability. Mol Biol Cell. 2008;19:4310–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kurshakova MM, Krasnov AN, D V K, et al. SAG A and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. Embo J. 2007;26:4956–4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lu Q, Tang X, Tian G, et al. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. Plant J. 2010;61:259–270. [DOI] [PubMed] [Google Scholar]
- [13].Ellisdon AM, Dimitrova L, Hurt E, et al. Structural basis for the assembly and nucleic acid binding of the TREX-2 transcription-export complex. Nat Struct Mol Biol. 2012;19:328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jani D, Lutz S, Hurt E, et al. Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export. Nucleic Acids Res. 2012;40:4562–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wickramasinghe VO, Stewart M, Laskey RA. GANP enhances the efficiency of mRNA nuclear export in mammalian cells. Nucleus. 2010;1:393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kopytova DV, Orlova AV, Krasnov AN, et al. Multifunctional factor ENY2 is associated with the THO complex and promotes its recruitment onto nascent mRNA. Genes Dev. 2010;24:86-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rodriguez-Navarro S, Fischer T, Luo M-J, et al. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116:75–86. [DOI] [PubMed] [Google Scholar]
- [18].Faza MB, Kemmler S, Jimeno S, et al. Sem1 is a functional component of the nuclear pore complex-associated messenger RNA export machinery. J Cell Biol. 2009;184:833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wilmes GM, Bergkessel M, Bandyopadhyay S, et al. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell. 2008;32:735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jani D, Lutz S, Marshall NJ, et al. Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export. Mol Cell. 2009;33:727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Umlauf D, Bonnet J, Waharte F, et al. The human TREX-2 complex is stably associated with the nuclear pore basket. J Cell Sci. 2013;126:2656–2667. [DOI] [PubMed] [Google Scholar]
- [22].Pascual-Garcia P, Govind CK, Queralt E, et al. Sus1 is recruited to coding regions and functions during transcription elongation in association with SAGA and TREX2. Genes Dev. 2008;22:2811–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kopytova D, Popova V, Kurshakova M, et al. ORC interacts with THSC/TREX-2 and its subunits promote Nxf1 association with mRNP and mRNA export in Drosophila. Nucleic Acids Res. 2016;44:4920–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Farny NG, Hurt JA, Silver PA. Definition of global and transcript-specific mRNA export pathways in metazoans. Genes Dev. 2008;22:66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ellisdon AM, Stewart M. Structural biology of the PCI-protein fold. Bioarchitecture. 2012;2:118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Stewart M. Structure and Function of the TREX-2 Complex. Subcell Biochem. 2019;93:461–470. [DOI] [PubMed] [Google Scholar]
- [27].Moritz M. Preparing cytoplasmic extracts from Drosophila embryos. CSH Protoc. 2007;2007:4712. [DOI] [PubMed] [Google Scholar]
- [28].Boehm AK, Saunders A, Werner J, et al. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;23:7628–7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Vorobyeva NE, N V S, J V N, et al. Transcription coactivator SAYP combines chromatin remodeler Brahma and transcription initiation factor TFIID into a single supercomplex. Proc Natl Acad Sci U S A. 2009;106:11049–11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Baroni TE, S V C, George AD, et al. Advances in RIP-chip analysis : RNA-binding protein immunoprecipitation-microarray profiling. Methods Mol Biol. 2008;419:93–108. [DOI] [PubMed] [Google Scholar]
- [31].Herold A, Klymenko T, Izaurralde E. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA. 2001;7:1768–1780. [PMC free article] [PubMed] [Google Scholar]
- [32].Clemens JC, Worby CA, Simonson-Leff N, et al. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci U S A. 2000;97:6499–6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kurshakova M, Maksimenko O, Golovnin A, et al. Evolutionarily conserved E(y)2/Sus1 protein is essential for the barrier activity of Su(Hw)-dependent insulators in Drosophila. Mol Cell. 2007;27:332–338. [DOI] [PubMed] [Google Scholar]
- [35].Fu Q, Wang W, Zhou T, et al. Emerging roles of NudC family: from molecular regulation to clinical implications. Sci China Life Sci. 2016;59:455–462. [DOI] [PubMed] [Google Scholar]
- [36].Zhu X-J, Liu X, Jin Q, et al. The L279P mutation of nuclear distribution gene C (NudC) influences its chaperone activity and lissencephaly protein 1 (LIS1) stability. J Biol Chem. 2010;285:29903–29910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Popova VV, Glukhova AA, Georgieva SG, et al. Interactions of the TREX-2 complex with mRNP particle of β-tubulin 56D gene. Mol Biol (Mosk). 2016;50:1030–1038. [DOI] [PubMed] [Google Scholar]
- [38].Roberts AJ, Kon T, Knight PJ, et al. Functions and mechanics of dynein motor proteins. Nat Rev Mol Cell Biol. 2013;14:713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhang C, Zhang W, Lu Y, et al. NudC regulates actin dynamics and ciliogenesis by stabilizing cofilin 1. Cell Res. 2016;26:239–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Riera J, Rodríguez R, Carcedo MT, et al. Isolation and characterization of nudC from mouse macrophages, a gene implicated in the inflammatory response through the regulation of PAF-AH(I) activity. FEBS Lett. 2007;581:3057–3062. [DOI] [PubMed] [Google Scholar]
- [41].Aumais JP, Tunstead JR, McNeil RS, et al. NudC associates with Lis1 and the dynein motor at the leading pole of neurons. J Neurosci. 2001;21:RC187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Morris SM, Albrecht U, Reiner O, et al. The lissencephaly gene product Lis1, a protein involved in neuronal migration, interacts with a nuclear movement protein, NudC. Curr Biol. 1998;8:603–606. [DOI] [PubMed] [Google Scholar]
- [43].Dix CI, Soundararajan HC, Dzhindzhev NS, et al. Lissencephaly-1 promotes the recruitment of dynein and dynactin to transported mRNAs. J Cell Biol. 2013;202:479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yamada M, Toba S, Takitoh T, et al. mNUDC is required for plus-end-directed transport of cytoplasmic dynein and dynactins by kinesin-1. Embo J. 2010;29:517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Bhatia V, Barroso SI, Garcia-Rubio ML, et al. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature. 2014;511:362–365. [DOI] [PubMed] [Google Scholar]
- [46].Nakaya T, Kuwahara K, Ohta K, et al. Critical role of Pcid2 in B cell survival through the regulation of MAD2 expression. J Immunol. 2010;185:5180–5187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


