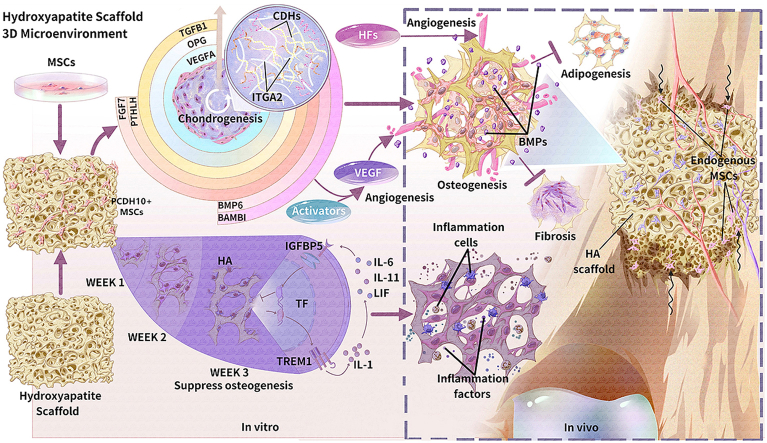
Abstract
The osteogenic microenvironment of bone-repairing materials plays a key role in accelerating bone regeneration but remains incompletely defined, which significantly limits the application of such bioactive materials. Here, the transcriptional landscapes of different osteogenic microenvironments, including three-dimensional (3D) hydroxyapatite (HA) scaffolds and osteogenic medium (OM), for mesenchymal stromal cells (MSCs) in vitro were mapped at single-cell resolution. Our findings suggested that an osteogenic process reminiscent of endochondral ossification occurred in HA scaffolds through sequential activation of osteogenic-related signaling pathways, along with inflammation and angiogenesis, but inhibition of adipogenesis and fibrosis. Moreover, we revealed the mechanism during OM-mediated osteogenesis involves the ZBTB16 and WNT signaling pathways. Heterogeneity of MSCs was also demonstrated. In vitro ossification of LRRC75A+ MSCs was shown to have better utilization of WNT-related ossification process, and PCDH10+ MSCs with superiority in hydroxyapatite-related osteogenic process. These findings provided further understanding of the cellular activity modulated by OM conditions and HA scaffolds, providing new insights for the improvement of osteogenic biomaterials. This atlas provides a blueprint for research on MSC heterogeneity and the osteogenic microenvironment of HA scaffolds and a database reference for the application of bioactive materials for bone regeneration.
Keywords: Single-cell RNA sequencing, Bone regeneration microenvironment, MSC heterogeneity, PCDH10, Bone tissue engineering
Graphical abstract
Highlights
-
•
A single-cell transcriptome of stem cells is mapped for bone regeneration.
-
•
PCDH10+ cells accelerate hydroxyapatite-related osteogenesis.
-
•
Osteogenic medium shows adverse effects on hydroxyapatite-related osteogenesis.
-
•
A time window for cell modification of hydroxyapatite in vitro is critical.
-
•
The hypoxic and inflammatory microenvironment of biomaterials deserves attention.
1. Introduction
Bone grafting is an increasing requirement in surgical procedure, correlated with the rapid increase in musculoskeletal disease occurrence because of an aging population with longer life span. The application of bone autografts and allografts is limited for several reasons, including donor site morbidity, source availability, reduced bioactivity, and risk of pathogen transmission [1,2]. While autologous bone grafting is considered as the “gold standard” for bone defect repair, advances have been made in the generation of bone substitutes, with the potential to overcome many drawbacks. Thus, there is an intense need for high quality tissue engineering of bone grafts.
Various strategies have been explored to generate bone grafts via tissue engineering approaches. Structure of biomaterials with a microarchitecture design with atomic composition similar to native bone tissue is crucial for successful bone regeneration. For this reason, many studies focus on the modification of intrinsic scaffold properties to improve both osteoinduction and osteoconduction capability [3,4]. However, a lack of clear understanding of the biological processes involved has limited the development the use of bioactive biomaterials. Although hydroxyapatite scaffolds are widely applied in research studies and clinical practice [5,6], there is an increasing need for improvement as the standard hydroxyapatite materials show low bioactivity [7].
Biomaterials can be modified to provide vital biological cues within the material, growth factors and cells, with the aim of improving osteointegration in bone regeneration, which is a prerequisite in the development of the engineered materials [2,8]. Restoration of an osteogenic microenvironment with an integration of stem cells with the biomaterials is an exciting approach and a key factor driving the development to improve the bioactivity of biomaterials. Mesenchymal stromal cells (MSCs), a widely used cell type with outstanding advantages, show a broad range of potential applications in osteogenesis and organoid construction [[9], [10], [11]]. Aiming to fabricate bioactive hydroxyapatite resembling allogeneic freeze-dried bone, we have biofabricated hydroxyapatite scaffold together with extracellular matrix secreted by cells showing better osteogenesis in vivo [12].
Although we have gained a good understanding of osteogenic biomaterials with respect to surface and spatial structures, chemical composition and biological cues, the cellular processes involved in the bioactivity of the materials remain poorly understood, limiting clinical applications [2]. The bioactivity of biomaterials guides cell behaviors and responses, in building and refining the complex microenvironments. Several studies have revealed specific osteogenic microenvironments [13]. However, detailed landscapes of osteogenic microenvironments at a high resolution are limited but needed to accelerate bone tissue engineering development. In addition, there is a need to understand the heterogeneity of MSCs and other cells used, and how subtypes can influence the responses to the biomaterials [14,15]. Single-cell RNA (scRNA) sequencing provides a strategy to decode cell phenotypes and differentiation lineage at single-cell resolution [15,16], providing a comprehensive and high-resolution interrogation of the MSC microenvironments for bone tissue engineering.
In this study, scRNA sequencing was used to profile 61,718 single cells in different osteogenic microenvironments commonly applied in fundamental research and clinical studies. We characterized and further revealed the heterogeneity of MSCs derived from Wharton's jelly of the human umbilical cord (WJMSCs) and decoded the osteogenic microenvironments of three-dimensional (3D) hydroxyapatite (HA) scaffolds and osteogenic medium (OM) conditions, providing new insights supporting the application of MSCs and bioactive materials for bone regeneration. Our dataset also provides a valuable tool for further investigations in bone research.
2. Materials and methods
Detailed information of key resources involved in this study were listed in Supplementary Table 1.
2.1. Preparation of calcined bovine bone scaffolds
Bovine vertebral cancellous bone was calcined and sintered according to the protocol of Juang HY et al. [12,17]. Briefly, the bovine bone was heated at a rate of 5 °C/min to 900 °C for 6 h and was then sintered at a temperature of 1100 °C for 1 h. After cooling to room temperature at a rate of 5 °C/min, the bone was modified to cylindrical shape at dimensions of 5 (diameter) × 3 (height) mm. The calcined bone scaffolds were then sterilized before use.
2.2. Primary culture of WJMSCs and expansion in vitro
Human umbilical cords used for this study were obtained with written informed consent from the parents (the seventh affiliated hospital of SYSU, Shenzhen, China) with the approval of the ethics committee (Permit number: 2019SYSUSH-031). WJMSCs were isolated from Wharton's jelly of human umbilical cords (n = 3, two males and one female) by adherent culture method and cultured in Prim® human MSC SF-M supplemented with 1% primocin as primary passage (P0). After cell density reached about 80% confluence, cells were dissociated with 0.25% Trypsin-EDTA incubated at 37 °C for 4 min. During the following entire process, the cells were then inoculated at a cell density of 5000 cells/cm2 and cultivated in 175 cm2 T-flasks with 30 mL of Dulbecco's Modified Eagle Media-Nutrient Mixture F-12 (DMEM/F12) containing 2 mM glutamine and supplemented with 10% FBS and 1% of penicillin-streptomycin, and cultured in a 5% CO2, 37 °C humidified incubator for cell expansion. WJMSCs at passage 3–5 were used for further experiments.
2.3. Flow cytometry analysis and tri-lineage differentiation for WJMSCs identification
For flow cytometry analysis of cell surface markers, WJMSCs at passage 4 were digested by 0.25% Trypsin-EDTA for 4 min in 37 °C humidified incubator to form a single-cell suspension. Antibodies used for flow cytometry are listed in the Key Resources table. Cells were resuspended in 100 μL cell staining buffer and nonspecific antibody binding was blocked by human Fc blocker antibody. Then 3 μL of each antibody was added to cells in each sample. Cells were stained for 20 min on ice, washed with cell staining buffer and resuspended at a final concentration of 1 × 106 cells/mL. Flow cytometry was performed on a Beckman CytoFlex instrument and data were analyzed using CytExpert. Cells were gated to exclude debris and doublets, and then analyzed for identification of cell surface markers.
For osteogenic and adipogenic differentiation, WJMSCs at passage 4 were inoculated at a cell density of 5000 cells/cm2 and cultivated in 6-well plates with 2 mL DMEM/F12 supplemented with 10% fetal bovine serum, 1% glutamine and 1% penicillin-streptomycin, and cultured in a 5% CO2, 37 °C humidified incubator. Medium was replaced with osteogenic/adipogenic induction medium at 80% confluence of WJMSCs and changed every 3 days. Staining of Alizarin Red S and Oil Red was performed after 3 weeks of differentiation. For chondrogenic differentiation, WJMSCs at passage 4 were centrifuged into pellets at a cell density of 50,000 cells/pellet and cultivated in tubes with 2 mL chondrogenic induction medium or the above-mentioned DMEM/F12 medium as control in a 5% CO2, 5% O2, 37 °C humidified incubator. Medium was changed every 3 days. Staining of Alcian Blue was proceeded for frozen sections of pellets after differentiation induction for 3 weeks.
2.4. Cell seeding and culturing on scaffolds
Bovine bone scaffolds were soaked in 2 mL cell suspension at a concentration of 1 × 107 cells/mL, and then were treated with a low atmospheric pressure condition for uniform cell distribution inside the scaffolds for 2 min as previously reported [12,18]. After cell seeding for 4 h, the cell-scaffold composites were transferred to 24-well plates with DMEM/F12 medium supplemented with 10% FBS. Two days later, the medium was changed with corresponding medium (OM and DMEM/F12 with 10% FBS) in 5% CO2 at 37 °C, which was labelled as week 0. The medium was changed every 3 days. Cell distribution in the scaffold was evaluated at week 0, 1, 2 and 3 after the seeding. Cell adhesion and morphology on the porous structure was observed by fluorescence staining of Calcein AM and propidium iodide under a fluorescence microscope.
2.5. Isolation of cells and magnetic cell sorting for dead cell removal
The cell-scaffold composites were gently crushed and cut into small fragments. MSCs from scaffolds were isolated by digestion with 0.25% Trypsin-EDTA for 5 min at 37 °C with agitation (120 rpm). After digestions, all cells were filtered through a 40 μm filter into a collection tube. For cells planted in 6-well plates, the same digestion method was adopted. Dead cells were removed by magnetic cell sorting with dead cell removal kit according to the manufacturer's protocol. Cell viability evaluation by Calcein AM and propidium iodide was applied after magnetic cell sorting and the cell debris removal procedure. Generally, samples proceeded to single-cell RNA sequencing when cell viability was higher than 80% without debris and aggregates.
2.6. Live cell imaging
The cells cultured on HA scaffolds at week 3 were isolated, cultivated in 24-well plates with 1 mL DMEM/F12 supplemented with 10% fetal bovine serum, 1% glutamine and 1% penicillin-streptomycin, and cultured in a 5% CO2, 37 °C automated live cell imager (Lionheart FX). Cells were then imaged every 10min for 2 days according to the manufacturer's instructions.
2.7. scRNA sequencing
Single cells were encapsulated into emulsion droplets using Chromium Controller. scRNA sequencing libraries were constructed using Chromium Single Cell 3′ v3 Reagent Kit according to the manufacturer's protocol. Briefly, cells were examined under a microscope and counted with a hemocytometer. Cells were then loaded in each channel with a target output of 6000 cells. Reverse transcription and library preparation were performed on C1000 Touch Thermal Cycler with 96-Deep Well Reaction Module. Amplified cDNA and final libraries were evaluated on an Agilent Bio-Analyzer using a High Sensitivity DNA Kit. Individual libraries were sequenced on Illumina Noveseq 6000 platform in 150 bp pair-ended manner at a sequencing depth of 50,000 raw read pairs per cell.
2.7.1. Pre-processing of scRNA sequencing data
Sequencing results were demultiplexed and converted to FASTQ format using Illumina bcl2fastq software. The Cell Ranger Single Cell Software Suite was used to perform sample demultiplexing, barcode processing and single-cell 3′ gene counting. An internal collection of numerical multidimensional datasets by velocyto from cellranger data was extracted as loom files. The cDNA insert was aligned to the GRCh38 reference genome. Only confidently mapped, non-PCR duplicates with valid barcodes and unique molecular identifiers were used to generate the gene-barcode matrix that contained 61,718 cells. Further analysis including quality filtering, the identification of highly variable genes, dimensionality reduction, standard unsupervised clustering algorithms and the discovery of differentially expressed genes was performed using the Scanpy R package. To exclude low quality cells and genes, we removed 1764 cells that had fewer than 300 detected genes and excluded 15,835 genes that were expressed in fewer than 50 cells. To exclude cells that were extreme outliers in terms of library complexity and that might possibly include multiple cells or doublets, we calculated the distribution of genes detected per cell and removed any cells that had more than 7000 genes. We also removed cells with more than 20% of the transcripts coming from mitochondrial genes. After quality filtering, 54,407 cells with 17,703 genes were left for further analysis. After removing unwanted cells from the dataset, we normalized the data by the total expression, multiplied by a scale factor of 10,000 and log-transformed the result. We used a linear regression through regress_out method to correct the observed batch effects among independent experiments.
2.7.2. Dimensionality reduction
We performed dimensionality reduction using gene expression data for a subset of variable genes. The variable genes were selected based on dispersion of binned variance to mean expression ratios setting as 0.25 using Find Variable Genes function of Scanpy package. Next, we performed principal component analysis (PCA) and reduced the data to the top 30 PCA components (number of components was chosen based on standard deviations of the principal components in a plateau region of an “elbow plot” and changed with different datasets).
2.7.3. Clustering and sub-clustering
We used graph-based clustering of the PCA reduced data with the Louvain Method after computing a shared nearest neighbor graph [19]. We visualized the clusters on a 2D view produced with uniform manifold approximation and projection (UMAP). For sub-clustering, we applied the same procedure of finding variable genes, dimensionality reduction, and clustering to the restricted set of data.
2.7.4. Differential expression of gene signatures
For each cluster, we used the T-Test to find rank genes that had significantly different expression when compared to the remaining clusters (paired tests when indicated) after multiple hypothesis testing correction.
2.7.5. Reconstructing pseudotime analysis and cell development trajectories
To infer the developmental progression of cells across multiple times, we used the Monocle algorithms implemented in R package for pseudotime analysis. We limited the dataset to only interested cells for the analysis. We selected the high dispersion genes across cells with mean expression ≥0.1 and empirical dispersion ≥0.1 as the ordering genes for the trajectory reconstruction, using the nonlinear reconstruction algorithm DDRTree. At the same time, we used the velocity algorithms implemented in Python package for cell development trajectories.
2.7.6. Dynamically expressed genes across pseudotime
Genes that varied across pseudotime were calculated using the Differential Gene Test function in Monocle and a cut-off of adjusted P value < 0.1 was applied. This was applied on the entire pseudotime range and also on the pseudotime intervals specific to limited cell clusters in order to avoid limitation to the genes characterized by monotonic changes across the inferred trajectory. Further branch analysis was applied using BEAM algorithms.
2.7.7. Functional enrichment analysis
GO enrichment analyses were performed using ClusterProfiler in R package and a cut-off of adjusted P value < 0.05 was applied, applying the Benjamini–Hochberg false discovery rate correction for multiple testing.
2.7.8. Lineage differentiation potency evaluation
A single numeric value representative of the aggregation of normalized expression of multiple marker genes in a specific category was defined as ‘score’ [20]. We calculated osteo/chondrogenic, adipogenic, antiadipogenic, proangiogenic and antiangiogenic scores by using the marker genes listed in Supplementary Table 2.
3. Results
3.1. Single-cell transcriptomic overview of MSCs in different osteogenic microenvironments
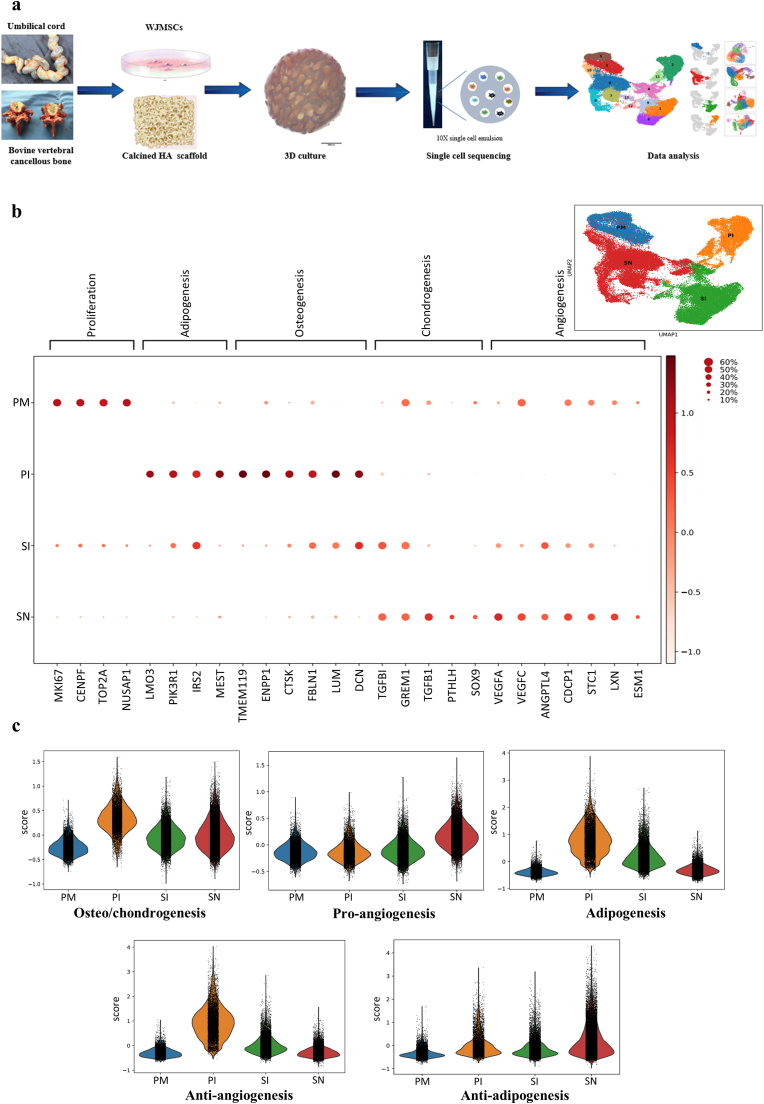
To understand the molecular features of MSC responses to different osteogenic microenvironments, we generated HA scaffolds from cancellous bone of bovine vertebrae, identified MSCs from Wharton's jelly of the human umbilical cord, embedded MSCs on HA scaffolds, and subjected the cells to 3D culture through typical procedures [17,18] (Supplementary Figs. 1a–d). scRNA sequencing of MSCs in different microenvironments was performed based on the 10X Genomics platform (Fig. 1a). To study development differences in distinct osteogenic microenvironments, we compared the transcriptomes of MSCs among a group of 2D monolayer-cultured cells with OM induction, marked as the PI (plate culture-OM induction) group; a group of cells 3D-cultured on calcined HA scaffolds with OM induction, marked as the SI (scaffold culture-OM induction) group; a group of cells 3D-cultured in HA scaffolds with basal medium, marked as the SN (scaffold culture-no induction) group; and a group of primary noninduced MSCs, marked as the PM (primary MSCs) group. Integrated and independent approaches were applied at the same time for joint analysis of differentially expressed genes (DEGs) (Supplementary Fig. 1e). A clear separation of clusters of input cells with distinct functions was observed (Fig. 1b).
Fig. 1.
Overview of the scRNA sequencing results for MSCs in different microenvironments. a, Schematic of 3D coculture and cell isolation for scRNA sequencing based on 10X Genomics. b, UMAP visualization of MSCs from 10X Genomics sequencing using 3′ chemistry showing the distinct distributions of clusters of cells in different microenvironments (right top. the colors indicate the different microenvironments; PM, primary noninduced MSCs cultured in plates; PI, plate-cultured MSCs with OM induction; SN, 3D-cultured MSCs on HA scaffolds without OM induction; SI, 3D-cultured MSCs on HA scaffolds with OM induction) and normalized expression of representative signature genes for typical functions (the gene expression frequency is indicated by the spot size, and the expression level is indicated by the color intensity). c, Lineage differentiation potential evaluation on the basis of related genes among the DEGs (Supplementary Table 2) showing the distinct functions of the different groups.
In general, MSCs cultured on the HA scaffolds (the SN group) obtained higher proangiogenic and antiadipogenic differentiation potential and exhibited lower antiangiogenic activity than the cells in the PI and SI groups (Fig. 1c). The results of GO analysis (Supplementary Figs. 2a–d) revealed that MSCs in the PM group presented prominent cell division and proliferation functions. The ossification and bone mineralization functions were highlighted in the cells of the PI group in line with the osteo/chondrogenic differentiation potential. The cells in the SN group exhibited significant immunoregulatory functions and proangiogenic effects coupled with osteogenic functions, suggesting the existence of different osteogenic mechanisms in the SN group. Unexpectedly, the hypoxic response of the cells in the SI group implied a potential conflict related to the combination of the HA scaffolds and OM.
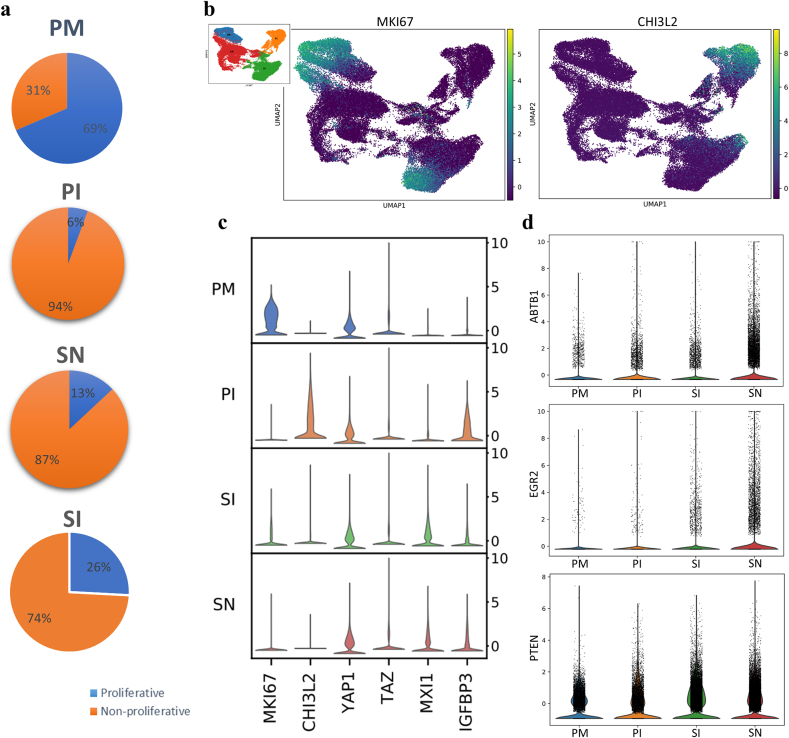
3.2. HA inhibits MSC proliferation through the MXI1/IGFBP31 and ABTB1-EGR2/PTEN pathways
To search for proliferating cells under the different conditions, we assessed the expression of MKI67 as a representative cell cycle gene (Fig. 2a–c). The cells in the PM group demonstrated significant proliferative activity with continuous high expression of cell cycle genes, which was in line with the GO analysis (Fig. 1b, Supplementary Fig. 2a). The proliferation of most cells in the PI group was significantly repressed (Fig. 2a), probably through CHI3L2, a proliferation suppressor gene [21] upregulated by OM (Fig. 2b–c). Cell contact inhibition [22] appears to play a more important role than differentiation [23] in the proliferation regulation by OM since a large proportion of cells (26%) continued to proliferate on the scaffold in the presence of OM (Fig. 2a), while no upregulation of YAP/TAZ expression, key molecules of Hippo signaling, was observed (Fig. 2c). The regulation of proliferation by OM had little connection with the Hippo signaling pathway, which has been reported to be critical in cell contact-mediated inhibition of proliferation [24]. In addition, cell proliferation on the HA scaffold was also reduced (Fig. 2a) through different pathways, including MXI1/IGFBP3 [25] (Fig. 2c) and ABTB1-EGR2/PTEN [26] (Fig. 2d). Interestingly, the combination of the HA scaffold and OM contained more proliferative cells (26%) than the HA scaffold or OM alone, and sustained CHI3L2 inhibition was observed in non-proliferative cells (Fig. 2a–b). This suggests a potential conflict of this combination with regard to proliferation regulation.
Fig. 2.
Cell proliferation regulation of MSCs in different microenvironments. a, Proportions of proliferative cells in different groups calculated in independent analysis of scRNA sequencing data. b, Expression of MKI67 and CHI3L2 represented by UMAP visualization (CHI3L2 was upregulated specifically in non-proliferative cells in the SI group). c, Violin visualization showing the expression of the indicated proliferation-regulating genes involved in the Hippo (YAP/TAZ) and MIX1/IGFBP3 signaling pathways. d, Simultaneous upregulation of ABTB1-EGR2/PTEN expression in the SN group.
3.3. Comprehensive analysis of the microenvironment of 3D HA-cultured MSCs in vitro
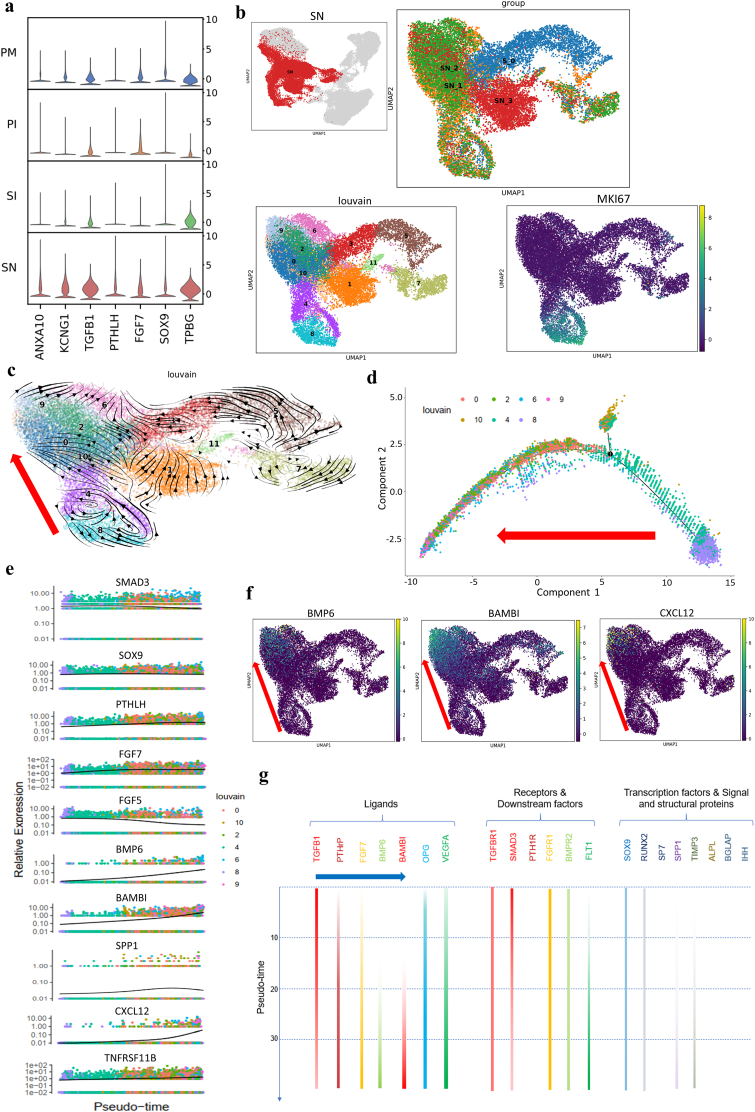
3.3.1. HA-related osteogenesis through sequential activation of the TGF-β, PTHrP and BMP signaling pathways is probably initiated by ANXA10
To explore the molecular mechanism of HA scaffold-mediated osteogenesis, we screened related molecules, signaling pathways and downstream factors (Fig. 3a). ANXA10, a calcium ion-binding molecule and atypical calcium channel [27], and the potassium channel KCNG1 were upregulated distinctively in the cells in the SN group, which could be a result of a high local calcium ion concentration within the HA scaffold [28]. In addition, the expression of parathyroid hormone-like hormone (PTHLH), a crucial autocrine calcium ion regulatory molecule and PTHrP signaling factor, correlated with ANXA10 expression. However, the expression of a typical PTHLH receptor (PTH1R) and upstream factor (IHH) [29] were not dysregulated in the SN group (data not included in DEGs), indicating that PTHLH probably binds to other receptors [30] to activate the PTHrP signaling pathway via an IHH-independent pathway.
Fig. 3.
Osteogenic process of MSCs cultured on HA scaffolds. a, Stacked violin visualization of the indicated genes highly expressed during HA scaffold-mediated osteogenesis. b, UMAP visualization of subclusters at different time points and MKI67 expression in the SN group. Clusters 8 and 4 represent proliferative cells in a relatively primitive state (S_0, SN at week 0; SN_1, SN at week 1; SN_2, SN at week 2; SN_3, SN at week 3). c, Velocity visualization of subclusters showing the cell development trajectory in the SN group and the direction of osteogenic development (red arrow, clusters 8, 4, 10, 0, 2, 9, 6). d, Monocle pseudotime trajectory showing the progression of osteogenic-lineage clusters (clusters 0, 2, 4, 6, 8, 9, and 10 according to velocity analysis and monocle pseudotime analysis; in Supplementary Fig. 3a, the red arrow shows the cell evolution direction). e, Pseudotime kinetics of the indicated genes during the development of osteogenic-lineage clusters. SOX9 and TNFRSF11B (OPG) showed steady expression levels; FGF5 and SMAD3 showed descending expression tendencies; FGF7 and PTHLH showed increasing expression levels and reached plateaus at the early stage; BMP6, BAMBI, SPP1 and CXCL12 showed elevated expression at the late stage. f, Expression distribution in UMAP visualization of the indicated genes (the red arrow shows the osteogenic direction). g, Summary of key signaling molecules involved in osteogenesis in the SN group showing the chronological activation of osteogenic signaling pathways (blue arrow). The expression level is indicated by the color intensity. The expression levels of genes not included in the pseudotime kinetics are supplied in Supplementary Fig. 3b because of low expression differences, and expression-deficiency strips show a lack of expression of PTH1R, SP7, ALPL, BGLAP and IHH.
We further evaluated the evolution of osteogenesis (Fig. 3b–c) and screened out osteogenic-lineage clusters (Fig. 3d, Supplementary Fig. 3a). The continuous high expression of TGFB1 with declining expression of the downstream molecule SMAD3 (Fig. 3a and e, Supplementary Fig. 3b) showed a tendency opposite those of other signaling pathway molecules (PTHLH, FGF7, BMP6), indicating that TGF-β signaling probably acted as an initial factor for HA scaffold-mediated osteogenesis. The following increases in FGF7 and PTHLH expression correlated with the expression of TGF-β and SOX9, suggesting the occurrence of a TGF-β- and SOX9-driven response during this osteogenic process defined as endochondral ossification [31,32] (Fig. 3a and e). On the other hand, the upregulated expression of PTHLH and TPBG (Fig. 3a) inhibited WNT-mediated ossification [33,34] and sustained preliminary chondrogenesis. Meanwhile, BMP6 signaling in a small group of cells was activated in parallel with the inhibition of TGF-β signaling by the TGF-β pseudoreceptor BAMBI and activation of the key osteogenic factor SPP1 in the cells at the later stage of differentiation, suggesting that BMP6 signaling played a critical role in the initiation of ossification (Fig. 3e–f). During this process, FGF5 expression showed typical downregulation, while there was a concurrent increase in the expression of BMP6 (Fig. 3e). This sequential activation of FGF and BMP signaling is very important since it has been shown that these molecules act in an antagonistic way when regulating chondrocyte proliferation and the process of hypertrophic differentiation [35]. Furthermore, CXCL12 was specifically upregulated at the late stage in osteogenic-lineage clusters (Fig. 3e–f). CXCL12, also named SDF-1, acts as a key regulator of MSC chemotaxis [36] and endothelial cell activation and migration [37], suggesting that the HA scaffold may have a proangiogenic function if applied in vivo.
According to these results, we speculated that the microenvironment of the HA scaffold activated the TGF-β signaling pathway to synergistically activate the ANXA10-dependent PTHrP signaling pathway and FGF signaling pathway, leading to the promotion of endochondral ossification. Following the activation of BMP signaling and inhibition of TGF-β signaling, terminal ossification was initiated (Fig. 3g). Moreover, upregulated CXCL12 expression attracted endogenous MSCs and endothelial cells to the regeneration site in vivo and promoted the coupling of osteogenesis and angiogenesis. This indicates that a strategy involving different cytokines for tissue engineering at different periods may be helpful.
3.3.2. Components and remodeling of extracellular matrix
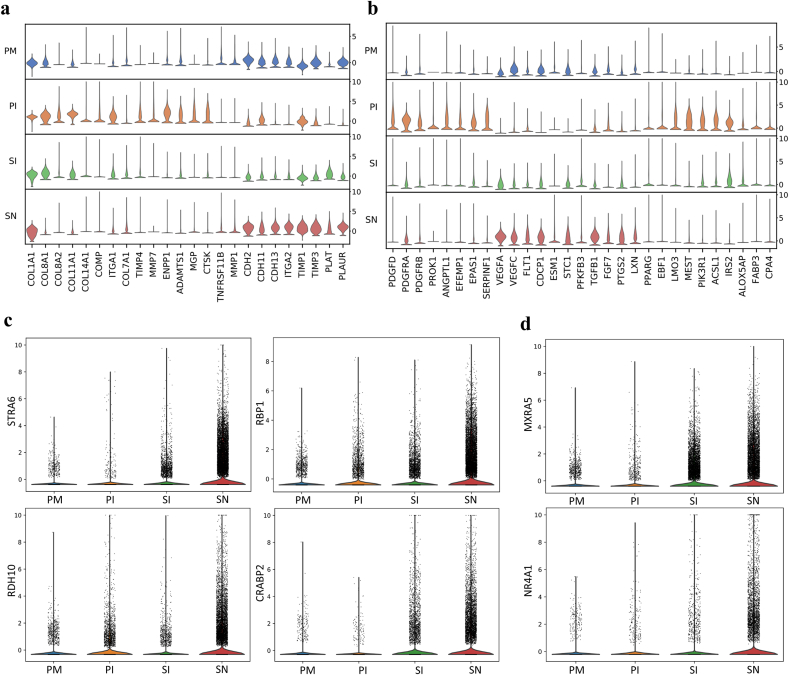
The extracellular matrix plays predominant roles in biomineralization and bone remodeling [38]. Extracellular matrix composition was evaluated at the gene level (Fig. 4a). Generally high expression of COL1A1 was observed, which indicated the induction of an osteogenic process. Under OM conditions, the expression of COL8A1, COL8A2, COL11A1, COL14A1, COMP and the corresponding ITGA1 was upregulated, while COL7A1 expression was downregulated. The downregulation of COL7A1 expression could be a response to the inhibition of TGF-β signaling [39] by OM. In addition, COL11A1 and COMP, which are believed to interact with COL2 at the protein level and are involved in chondrogenesis, were also upregulated by OM. This implies that in addition to chondrogenesis, COL11 and COMP might be involved in mineralization and bone matrix remodeling during osteogenic differentiation. On the other hand, the expression of CDHs and ITGA2 was increased in the HA microenvironment. CDHs are likely key molecules that take part in HA-induced osteogenesis, which suggests that calcium generated by HA scaffolds plays an important role in HA-related bioactivity. In particular, CDH 11, the so-called OB-cadherin that is highly expressed in osteoblasts [40], was significantly upregulated, indicating that CDH 11 might be crucial during HA-induced endochondral ossification.
Fig. 4.
Functional genetic phenotypes of MSCs in different osteogenic microenvironments. a, Stacked violin visualization of the indicated genes involved in extracellular matrix formation and matrix remodeling. b, Stacked violin visualization showing the key antiangiogenic factors (highly expressed in the PI group, including ANGPTL1, EFEMP1, EPAS1 and SERPINF1), proangiogenic factors (highly expressed in the SN group, including VEGFA, VEGFC, FLT1, CDCP1, ESM1, STC1, PFKFB3, TGFB1, FGF7, PTGS2 and LXN) and adipogenic factors (highly expressed in the PI group, including PPARG, EBF1, LMO3, MEST, PIK3R1, ACSL1, IRS2, ALOX5AP, FABP3 and CPA4) related to OM and the HA scaffold. c, Violin presentation of the expression of key molecules in the STRA6/RBP1 signaling pathway. d, Violin visualization of antifibrogenic regulators.
In contrast to those under OM induction, the cells in the HA scaffold highly expressed TIMP1 and TIMP3, resulting in inhibition of MMPs for improvement of mineralization and matrix aggregation. In addition, PLAT and PLAUR were upregulated for extracellular matrix remodeling. OPG (TNFRSF11B) expression in cells on the HA scaffold was continually observed (Figs. 3e and 4a). These results suggested that OPG/RANKL could be part of a potential regulatory pathway for osteogenesis by HA scaffolds. Specific extracellular matrix components and remodeling modes were revealed, which will guide the reconstruction of extracellular matrix during tissue engineering.
3.3.3. Molecular network of the proangiogenic microenvironment
Generally, biomaterials-based bone regeneration depends equally on osteogenesis and coupled neovascular ingrowth mediated by endothelial cells [41]. Several angiogenic activators and antagonists intersected at VEGF signaling (Fig. 4b). The expression of several VEGF-related angiogenic genes by cells in the PI group was upregulated, including PDGFD, PDGFRB, PDGFRA and PROK1 (EG-VEGF). However, VEGFA and VEGFC expression was significantly suppressed, while the expression of the antiangiogenic genes ANGPTL1, EFEMP1, EPAS1 and SERPINF1, all of which are VEGF regulators, was upregulated, indicating that OM inhibits angiogenesis through the VEGF signaling pathway by upregulating VEGF antagonists. Our results are in line with a previous study that dexamethasone, a main composition in OM, was thought to be the key reason for the inhibition of angiogenesis induced by OM through VEGF signaling pathway [42].
In contrast, the cells in the SN group highly expressed VEGFA and VEGFC along with FLT1 (a soluble VEGF receptor), a regulator of VEGF-induced angiogenesis, showing a more balanced proangiogenic microenvironment than the cells in the PI group. This likely also maintains osteogenesis at an early stage since the VEGF/FLT1 balance determines the fate of MSC differentiation [43,44]. In addition, VEGF signaling activators (CDCP1, ESM1, PFKFB3, STC1), growth factors (TGF-β, FGF7), inflammation chemokines (CXCL12, PTGS2) and hematopoietic factors (LXN) were activated and formed an intricate proangiogenic network. These results demonstrate a complicated interaction of various angiogenic activators that builds a proangiogenic microenvironment, which could promote functional cell migration and neovascular ingrowth in vivo. Then, VEGF expression is enhanced along with neovascular ingrowth, which further accelerates the coupling of angiogenesis and osteogenesis due to the combined effect of BMP and dominantly expressed VEGF [43].
3.3.4. Regulation of adipogenesis and fibrogenesis
In addition to osteogenesis and chondrogenesis, adipogenic activity was assessed under different conditions. The expression of critical adipogenic genes was elevated during OM stimulation, and CPA4, a negative feedback regulator of adipogenesis, was upregulated (Fig. 4b). This could be explained by the inclusion of a steroid as one of the OM medium components, as this steroid is also known to initiate adipogenesis [45]. The expression of STRA6/RBP1 signaling-related genes, which are strong inhibitors of cellular responses to insulin and adipogenesis when activated at the early stage of differentiation [46,47], was elevated by the HA scaffold. These results imply that the HA scaffold can activate the STRA6-RBP1-RDH10-CRABP2 signaling pathway and inhibit adipogenesis (Fig. 4c). It is reported that the differentiation toward an osteoblast phenotype occurs at the expense of an adipocyte phenotype [48]. Thus, bone regeneration would benefit from the inhibited adipogenic activity of HA scaffold.
Fibrosis is a major barrier for tissue regeneration, especially for the integration of the biomaterial with the surrounding tissues at the interface. TGF-β is a master regulator of fibrosis [49], which suggests that endochondral ossification in the HA scaffold through TGF-β signaling might be accompanied by fibrosis. However, the expression of MXRA5 and NR4A1, which are fibrosis inhibitors and TGF-β signaling regulators [50,51], was specifically upregulated in the SN group, indicating that HA leads to an antifibrotic response (Fig. 4d). These results reveal the potential key molecules of fibrosis inhibition by HA. Therefore, HA could improve osteogenic integration potential for healing of the tissue between bone and biomaterials by reducing fibrosis.
3.3.5. Hypoxic response in the HA scaffold microenvironment
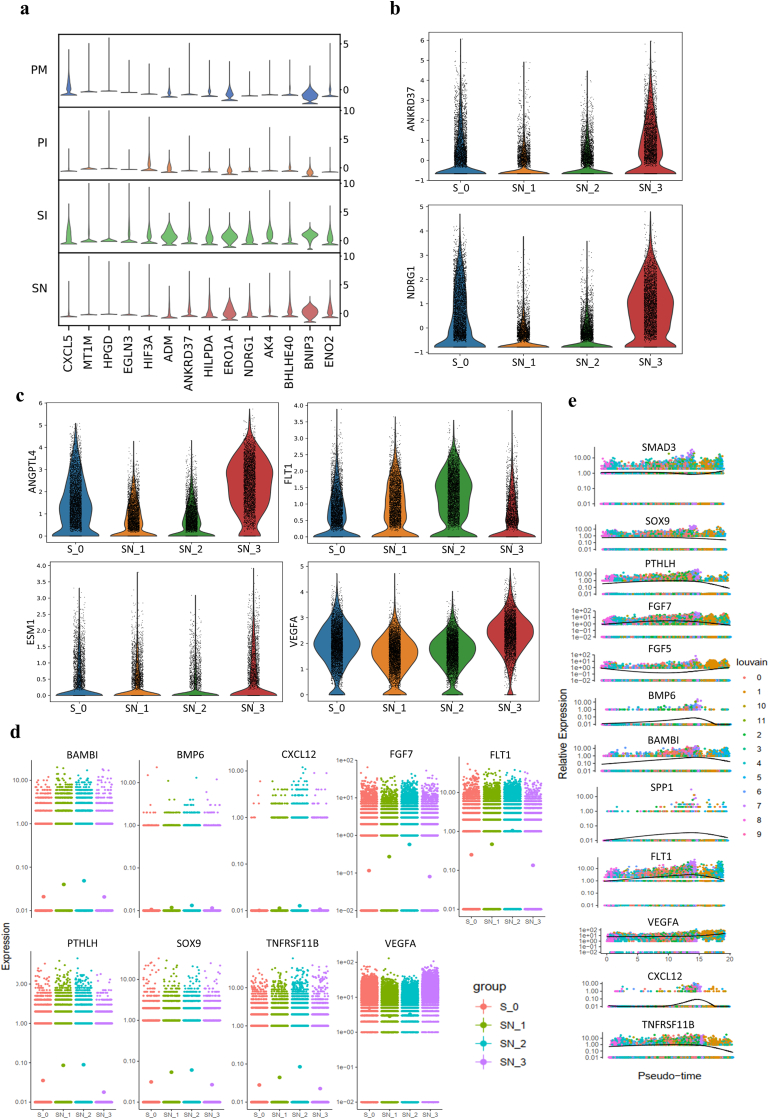
Hypoxia-related genes expression was significantly upregulated when the cells were cultured on the HA scaffold (Fig. 5a). Especially, the expression of key hypoxic genes (ANKRD37, NDRG1) dramatically enhanced in the SN group at week 3 (Fig. 5b). In addition, the cells in the SN group at week 3 showed upregulation of specific angiogenesis genes (ANGPTL4, ESM1, VEGFA) while angiogenic inhibitor FLT1 was downregulated and the same tendency of VEGFA/FLT1 was observed by pseudotime analysis (Fig. 5c–e). This gene expression pattern is expected to accelerate neovascular ingrowth as a response to the enhanced hypoxia. Furthermore, the aforementioned key signaling pathways for osteogenesis was interrupted along with significant downregulation of the aforementioned key osteogenic factors (Fig. 5d–e). This means the enhanced hypoxic response after in vitro culture for 3 weeks will hinder the osteogenic process, which indicates that a proper time window for the culture of cells on HA scaffolds in vitro is critical for the maintenance of an osteogenic microenvironment. This is in line with what we previously found in vivo [12].
Fig. 5.
Hypoxic response of MSCs on HA scaffolds. a, Stacked violin visualization of the indicated genes involved in the hypoxic response. Violin visualization of key hypoxic (b) and angiogenic (c) genes showing distinct upregulation at week 3 (FLT1, an angiogenic inhibitor, was downregulated). d, Expression tendencies of the indicated signaling molecules at different time points visualized with a gene jitter plot in Monocle showing significant downregulation of osteogenic signaling molecules with upregulation of VEGFA expression at week 3. e, Pseudotime kinetics of the indicated genes during the development of all clusters in the SN group.
3.3.6. Inflammation microenvironment
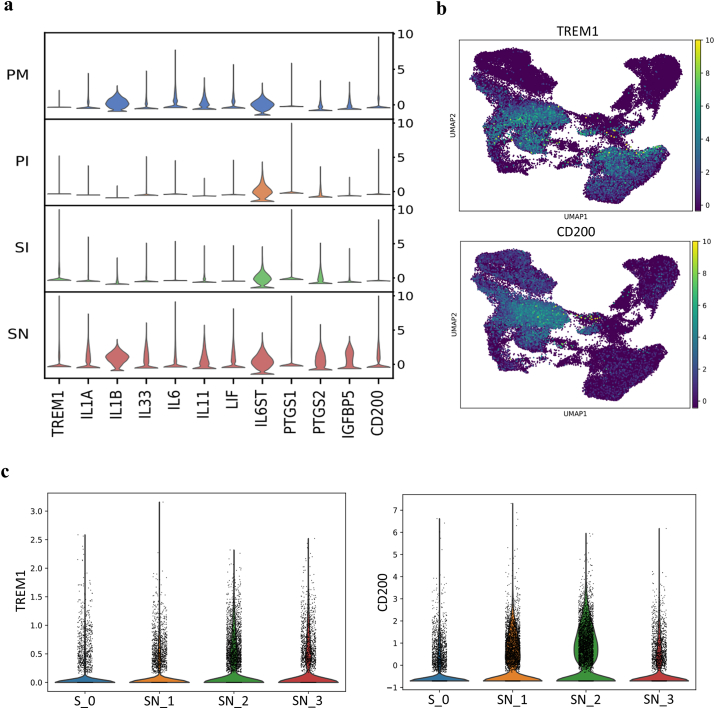
To further investigate the inflammatory microenvironment, key inflammatory factors were assessed (Fig. 6a). TREM1 (triggering receptor expressed on myeloid cells 1), a critical triggering receptor of the inflammatory cascade normally expressed on neutrophils and monocytes, was significantly and specifically upregulated in the cells on the HA scaffold (Fig. 6a–b). This indicated that TREM1 may be a key inflammatory regulator of MSCs in the HA-mediated inflammatory microenvironment, as it has been reported to function in epithelial cells [52]. TREM1 is thought to play a critical role in amplifying an uncontrolled inflammatory response and is involved in the promotion of TNF, IL and PTGS secretion [52,53]. The IL1/IL6 axis [54] of the cells in the SN group might have been activated by TREM1 with upregulation of IL1-type cytokines (IL1, IL33), IL6-type cytokines (IL6, IL11, LIF) and COXs (PTGS1, PTGS2) (Fig. 6a). Moreover, the increased expression of IGFBP5 accompanied by inflammatory factors (Fig. 6a) would inhibit osteoblast differentiation via interaction of IGFBP5 with upregulated IL6 [55]. These proinflammatory microenvironment would be sustained at a steady level with high expression of the critical immunosuppressor CD200, which is known to be upregulated in WJMSCs [56] (Fig. 6a–b). CD200 was downregulated significantly in the SN group at week 3 (Fig. 6c). The reduced expression of this immunosuppressor could disrupt the inflammatory balance, corresponding to the enhanced hypoxic reaction at week 3.
Fig. 6.
Inflammatory regulation of MSCs on HA scaffolds. a, Stacked violin visualization of the indicated genes involved in the inflammatory reaction. b, UMAP presentation of key proinflammatory factor and immunosuppressor involved in HA-mediated inflammation. c, Violin presentation of key proinflammatory factor and immunosuppressor over time (CD200 was downregulated significantly in the SN group at week 3).
In brief, these results demonstrated that the cells cultured on the HA scaffold exhibited an inflammatory microenvironment in vitro, which was enhanced over time and further inhibited subsequent osteogenesis.
3.3.7. PCDH10+ MSCs show superiority in HA-related osteogenic processes
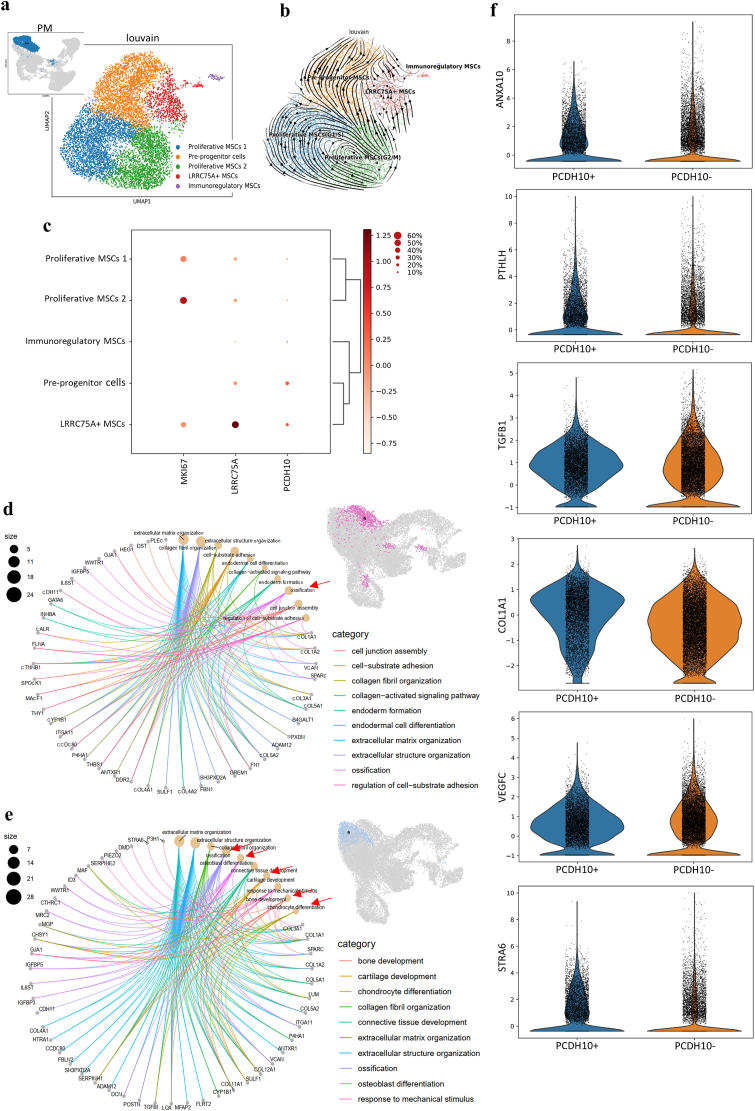
WJMSCs were classified into five functional clusters defined as the proliferative MSCs 1 (G1/S), the proliferative MSCs 2 (G2/M), the LRRC75A+ MSCs, the pre-progenitor cell (PCDH10+ MSCs) cluster and the immunoregulatory MSCs according to the marker genes and GO analysis (Fig. 7a–c, Supplementary Fig. 4, 5a-b). The results of pseudotime and diffusion trajectory analysis showed an independent evolution of LRRC75A+ MSCs, which was different from that of pre-progenitor cells (Fig. 7b, Supplementary Fig. 5b). GO enrichment analysis results revealed that LRRC75A+ MSCs showed typical ossification markers, while pre-progenitor cells (PCDH10+ MSCs) showed a combination of osteogenesis and chondrogenesis (Supplementary Fig. 5a). The different origins and functions of the MSCs suggested that the different clusters of MSCs were involved in different osteogenic differentiation processes. The results also implied that a specialized MSC subcluster for immunoregulation exists that performs functions in addition to the secretion of immunoregulatory factors, which is performed by most stem cells. These findings offering new insight into this emerging aspect of MSC function.
Fig. 7.
Heterogeneity of MSCs and PCDH10+ MSCs in HA microenvironments. a, UMAP visualization showing the functional clusters of MSCs (identified in Supplementary Figs. 4 and 5). b, Velocity visualization of MSCs showing the different cell development trajectories in the PM group (blue, proliferative MSCs 1; yellow, progenitor MSCs; green, proliferative MSCs 2; red, LRRC75A + MSCs; purple, immunoregulatory MSCs). c, Dot plot presenting the normalized expression of representative signature genes of different clusters and pre-progenitor cells identified as PCDH10+/LRRC75A- MSCs (the gene expression frequency is indicated by the spot size, and the expression level is indicated by the color intensity). GO analysis showing the different osteogenic fates in clusters 6 (d) and 9 (e) in the SN group (the red arrow shows the main osteogenic function). f, PCDH10+ cells in the SN group showing upregulated expression of key HA-related osteogenic molecules.
Interestingly, the cells in the SN group showed significant heterogeneity in cell behaviors (Supplementary Video). Two clusters of cells with distinct osteogenic fates (clusters 6 and 9) in the SN group were observed (Fig. 3c). Cluster 6, which highly expressed LRRC75A, was thought to be derived from LRRC75A+ MSCs and did not increase in proportion over time in the HA microenvironment (Supplementary Figs. 5c–d). Cluster 6 presented an ossification function according to GO analysis (Fig. 7d). The cells of cluster 9 showed both osteogenic and chondrogenic functions as pre-progenitor cells (Fig. 7e), indicating a strong connection with endochondral ossification. In addition, the aforementioned HA-related osteogenic factors (ANXA10, PTHLH, TGFB1), extracellular matrix molecule (COL1A1), proangiogenic factors (VEGFC) and antiadipogenic factor (STRA6) were upregulated in PCDH10+ cells (Fig. 7f) in the SN group, indicating that PCDH10+ MSCs showed superiority in HA-related osteogenic processes and might originate from chondro-osteogenic pre-progenitor cells (PCDH10+ MSCs).
3.4. OM does not accelerate HA-related osteogenesis
3.4.1. OM induced osteogenesis
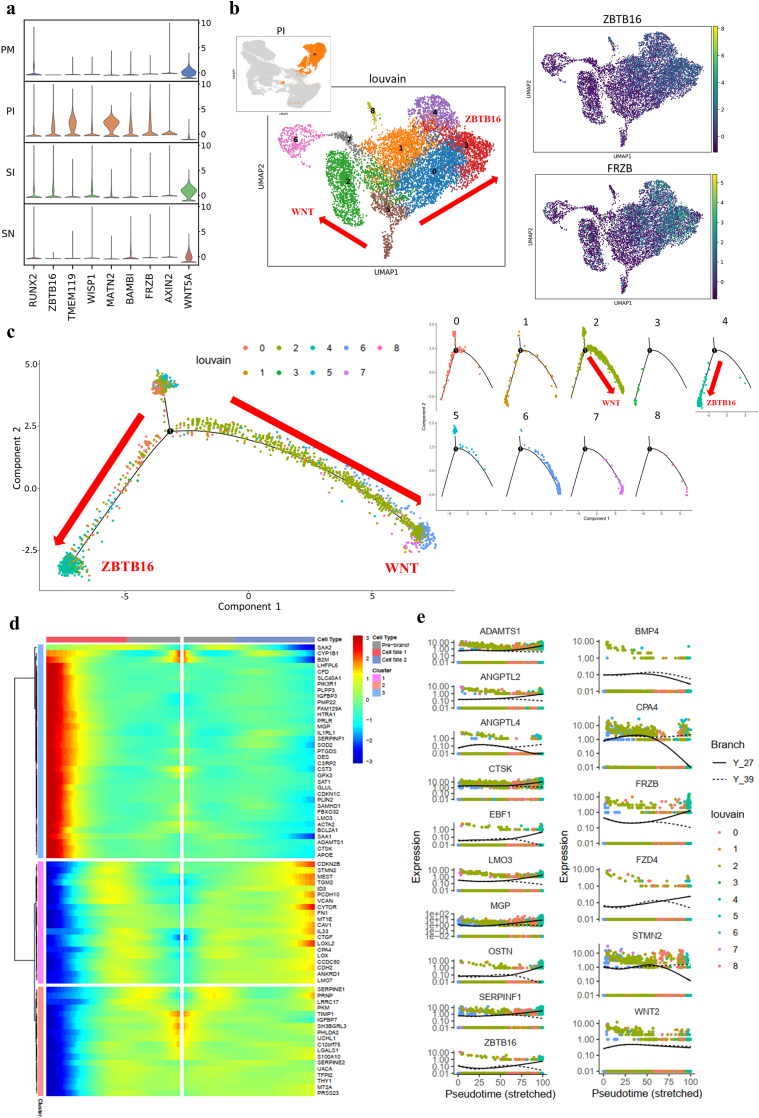
The expression of RUNX2 and SP7 (data not included in DEGs), key signaling pathway molecules induced by OM components during osteogenesis [57], was not enhanced during OM induction (Fig. 8a). In contrast, the expression of transcriptional suppressor zinc finger and BTB domain containing 16 (ZBTB16), an important marker independent of BMP and WNT signaling at the later stage of osteoblastic differentiation of stem cells [58], was significantly elevated along with the upregulated expression of TMEM119 and WISP1, leading to OM-mediated osteoblastogenesis [59,60] (Fig. 8a). ZBTB16 seemed to function via a RUNX2-and SP7-independent pathway. In addition, the upregulated expression of TGF-β inhibitors (MATN2 and BAMBI) [61,62] and WNT inhibitors (FRZB and AXIN2) indicated that inhibition of the TGF-β and WNT signaling pathways occurred in parallel with the activation of ZBTB16 (Fig. 8a).
Fig. 8.
Osteogenic process of MSCs during OM induction. a, Stacked violin visualization of key genes involved in OM-mediated osteogenesis. b, Distribution of target gene expression in the cells in the PI group at different time points and distinct cell fates matched in UMAP and cell trajectory visualization (the red arrow shows the trajectory direction; the direction with high expression of ZBTB16 is marked as ZBTB16, and the direction with low expression of the WNT inhibitor FRZB is marked as WNT). c, Cell development trajectory of MSCs induced for 3 weeks in the PI group (PI_3) as determined by Monocle analysis (the red arrow shows the direction of trajectory). d, Gene expression heatmap of the cells in the PI_3 group in a branch-dependent manner showing distinct genetic phenotypes of the ZBTB16 and WNT branches. e, Pseudotime kinetics of the indicated genes from the root of fate 1 (ZBTB16, solid line) and fate 2 (WNT, dashed line).
Meanwhile, WNT5A, a reciprocal inhibitor of the canonical WNT signaling pathway, was repressed in the cells of the PI group (Fig. 8a). This implies that canonical WNT signaling played a role in the OM-mediated osteogenesis, even though WNT inhibitors (FRZB and AXIN2) were activated. According to the cell development trajectory determined by independent Monocle analysis of the PI group, cells with distinct fates marked as ZBTB16 (clusters 0, 3, and 4) and WNT (clusters 1 and 2) were present during OM induction (Fig. 8b). The expression of ZBTB16 was restricted to limited clusters, and the results of GO analysis showed a continual function of ZBTB16-mediated osteogenesis from chondrogenesis to osteogenesis (Supplementary Fig. 6). We further analyzed the gene expression differences of branching trajectories and screened the signature genes for different trajectories (Fig. 8c–d). OSTN plays a role in bone formation and negatively regulates osteoblast differentiation [63]. Our results showed that OSTN acted as a late marker of ZBTB16-mediated osteogenesis, which was accompanied by matrix remodeling (CTSK, MGP, ADAMTS1) and proangiogenic (ANGPTL2, ANGPTL4)/adipogenic (LMO3, EBF1) tendencies (Fig. 8, Fig. 9a). However, SERPINF1, which encodes PEDF in humans, one of the strongest inhibitors of angiogenesis [64], was significantly upregulated, and such upregulation may block endochondral ossification. Therefore, ZBTB16-mediated osteogenesis induced by OM presented endochondral ossification-like differentiation and was completely at odds with the traditional understanding that OM promotes direct intramembranous ossification.
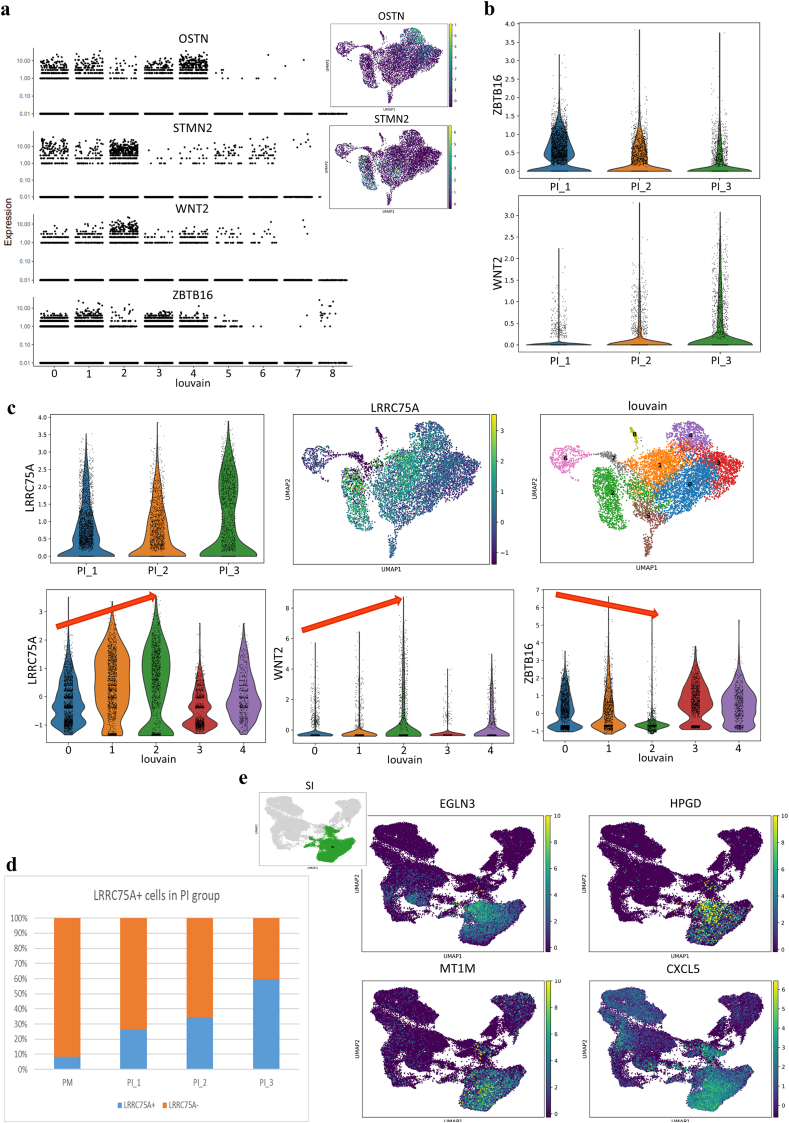
Fig. 9.
The ZBTB16/WNT2 transition, LRRC75A + MSC function during OM induction, and enhanced hypoxic and inflammatory reactions induced by HA scaffolds combined with OM. a, Marker genes of different cell fates presenting in UMAP and gene jitter plots according to Monocle analysis.b, Expression tendencies of the indicated genes during distinct osteogenic processes (PI_1, PI at week 1; PI_2, PI at week 2; PI_3, PI at week 3).c, Violin and UMAP visualization showing that LRRC75A was highly expressed in limited clusters (clusters 1 and 2) in the PI group (the red arrow shows that the expression tendencies of LRRC75A and WNT2 were the same and were totally opposite those of ZBTB16).d, Proportions of LRRC75A + MSCs in the PI group at different time points showing a steadily ascending tendency.e, UMAP visualization of the indicated hypoxia and inflammation genes specifically highly expressed in the SI group.
In addition, the expression of ZBTB16 and WNT inhibitors (FRZB and FZD4) decreased over time, while WNT signaling pathway was obviously activated by upregulation of WNT2 and downregulation of WNT inhibitors (Fig. 8, Fig. 9b). Moreover, STMN2, which is a novel WNT signaling target and modulates osteogenesis and the osteogenic/adipogenic balance [65], was analyzed as a marker gene of WNT-mediated osteogenesis (Fig. 9a). The results revealed a correlation of STMN2, WNT2 and BMP4 expression along with significant adipogenesis repression caused by increased expression of CPA4, which was totally different from the change observed for ZBTB16 (Fig. 8e).
In summary, there existed two distinct OM-induced osteogenic phases. OM initiated osteogenic differentiation via a ZBTB16-mediated endochondral-ossification-like pathway at the early stage, but ZBTB16-mediated osteogenesis was reduced over time, leading to a predominantly WNT-regulated osteogenic process at the late stage of osteogenesis. The sequential activation of ZBTB16 and WNT signaling implied a potential mechanism of cooperative osteogenesis involving ZBTB16 activation accompanied by high expression of WNT inhibitors.
The expression of matrix remodeling-related genes showed large differences between OM induction and HA scaffold culture (Fig. 4a). During OM induction, the expression of MMP7 was enhanced, while the expression of MMP1 was repressed; these findings indicate that MMP7, instead of MMP1, is involved in OM-induced matrix remodeling. In addition, the expression of ENPP1, which produces an inhibitor of hydroxyapatite crystal formation (pyrophosphate, PPi), is a regulator of the phosphate/PPi ratio [66], was specifically upregulated by OM. Other bone resorption promotors, including ADAMTS1, MGP and CTSK, were also upregulated to accelerate mineralization remodeling. Therefore, remarkable differences in extracellular matrix components and matrix remodeling existed between the 3D osteogenic scaffold and 2D osteogenic medium microenvironments. On the other hand, LRRC75A+ MSCs seemed to be significantly functional during OM induction. The number of LRRC75A+ cells increased over time under OM conditions, and the LRRC75A+ cells exhibited the same expression areas and tendencies for WNT signaling but the opposite results for ZBTB16 signaling in the PI group (Fig. 9c–d).
3.4.2. Enhanced hypoxia and inflammation responses for HA + OM culture
Moreover, the cells in the SI group presented upregulation of HIF3A and HA scaffold-related hypoxic genes (Fig. 5a) and showed no advantages for osteogenesis and angiogenesis (Fig. 1c). The HIF regulator EGLN3 and the inflammation-activating factors HPGD, MT1M, CXCL5 were upregulated (Fig. 9e), in line with the GO analysis results (Supplementary Fig. 2d). These results indicated the presence of an accelerated inflammatory microenvironment supporting inflammation activation and inflammatory cell migration in the SI group. The combination of HA scaffold culture and OM induction may lead to an uncontrollable hypoxia response and inflammation. The non-physiological combination of HA scaffolds with OM induction for bone regeneration needs to be assessed in vivo.
4. Discussion
The potential of MSCs in different in vitro osteogenic microenvironments and the complexity of MSC heterogeneity remained poorly defined for differentiation along the osteogenic lineage. Using single-cell transcriptomic sequencing, we have decoded the cellular heterogeneity and responses of MSCs on hydroxyapatite as a supporting material. Our findings provided a comprehensive understanding of MSC responses to hydroxyapatite-related osteogenic microenvironments correlating cellular outcomes to the transcriptional landscape, including cell proliferation and differentiation, extracellular matrix remodeling, hypoxia and inflammation responses, in relationship to MSC heterogeneity.
Our high-resolution osteogenic transcriptional landscape provided new insights for HA-mediated osteogenesis that can inform future studies. Owing to its natural presence in bone, hydroxyapatite scaffold has been widely used in tissue engineering for bone that possesses high mechanical stress property and excellent biocompatibility [67]. However, the low bioactivity and brittle nature of standard hydroxyapatite scaffolds have significantly limited its clinical application. Through biofabrication together with MSCs, application potentials of hydroxyapatite scaffolds can be significantly improved [12]; but how the presence of MSCs can enhance function of the scaffolds is not understood.
In this study, large-scale scRNA sequencing during osteogenic differentiation of MSC revealed HA scaffold invokes a proliferation inhibition mechanism involving MXI1/IGFBP3 and ABTB1-EGR2/PTEN pathways. This is consistent with the concept that progenitor cells exit the cell cycle prior to differentiation [23,68]. Furthermore, in the osteogenic lineage, a gene expression profile reminiscent of endochondral ossification was observed, with chronological activation of the TGF-β, SOX9, PTHrP, FGF and BMP signaling pathways. Interestingly, this process was accompanied with the establishment of a proangiogenic microenvironment, consistent with the in vivo endochondral ossification process. The inhibition of the WNT signaling pathway and activation of OPG/RANKL further support a favoring of the endochondral ossification process over intramembranous ossification. It is possible that in the presence of HA scaffold, the differentiation program directs a transition similar to the recent discovery that hypertrophic chondrocytes can transit to become osteoblasts [69].
In vivo, various endogenous cells may be attracted to the regeneration area at injury sites by the bioactive microenvironment enabling both angiogenic and osteogenic processes. A modified matrix assembly of CDHs through ITGA2 with matrix remodeling by PLAT/PLAUR may contribute to HA scaffold-driven bone regeneration.
Together, our finding highlights the importance of intervention timing and the choice of bioactive molecules, the combination of which will provide insights in the design of precise modification strategies for tissue-engineered bone. Furthermore, considering the intervening fibrous layer between biomaterials and surrounding tissues as reported previously [70], enhancement of fibrotic inhibition could improve the integration of biomaterials to accelerate bone regeneration.
The inflammatory response is a double-edged sword for tissue regeneration, known to be up regulated in the presence of the HA scaffold as an explanation for the reduced osteogenesis after 3 weeks of 3D culture base on our previous study [12]. The finding of this study suggests there could be a productive window that can be captured, balancing the osteogenic and inflammatory microenvironments for the construction of bioactive material for clinical application. Further, additional strategies such as mechanical loading for musculoskeletal system regeneration, can be taken into consideration to regulate the immunoinflammatory response of cells on biomaterials [71].
Even though OM is already widely applied in osteogenic biomaterials and acts as a classic method for in vivo osteogenesis [[71], [72], [73]], there is a general lack of attention on the potential risks. Unexpectedly, OM resulted in an excessive hypoxic and inflammatory responses of MSCs on HA scaffold with no enhanced osteogenesis in this study. The combination of HA scaffold and OM should be evaluated further with the potential adverse effects being a prime focus.
In addition, there is a need to understand the heterogeneity of MSCs, and how subtypes can influence the responses to the biomaterials. Here, MSCs were classified into several functional clusters single-cell transcriptomic profiling, including cycling-related, LRRC75A+, immunoregulatory and pre-progenitor clusters. The poorly defined LRRC75A+ MSCs, which were probably involved in WNT signaling, may play an important role in OM-mediated osteogenesis and showed a reduced tendency towards endochondral ossification, suggesting that LRRC75A might be a potential novel osteogenic marker. PCDH10, a calcium-dependent adhesion transmembrane protein, has been reported to be involved in tumor suppression [74,75]. Knowledge about PCDH10, which belongs to the nonclustered protocadherin family, is still in its infancy [76]. In addition, the function of PCDH10 in MSCs remains unclear. In this study, PCDH10+ MSCs exhibited superiority in hydroxyapatite-related osteogenic processes. These results indicate that PCDH10 might be a specific marker for MSCs and PCDH10+ MSCs could potentially be applied for bone regeneration and precise biomodification of bone repair materials, especially with regard to hydroxyapatite-related osteogenesis.
Given the concerns and potential difficulties related to the certification of medical devices containing biologics, there is increasing attention on the modification of intrinsic properties of biomaterials to achieve superior osteogenic bioactivity, such as atomic composition, nanostructure, porosity and so on [[3], [4], [5], [6], [7]]. The combination of precise biomodification and excellent intrinsic properties of bone biomaterials can be of great potential in the near future.
However, it should be noted that there are several different sources of MSCs and biomaterials. This study only addressed the condition of specific HA biomaterials and WJMSCs. The difference of subpopulations may exist among different MSC types. The properties of HA scaffolds used in this study may be different from those used in clinical practice. Cell behavior will potentially be different in biomaterials with different properties. Moreover, this study is limited to analysis at the gene level. Further experiments for verification of selected markers at the tissue, cell, and protein levels are warranted.
5. Conclusions
The heterogeneity of WJMSCs and the comprehensive microenvironments of a specific bioactive hydroxyapatite material for bone regeneration were addressed. PCDH10+ MSCs showed superiority in hydroxyapatite-related osteogenic processes involved in endochondral osteogenesis, which provides new insights for bone regeneration and precise biomodification of biomaterials with regard to the heterogeneity of MSCs. A proper time window for coculture of MSCs and hydroxyapatite scaffold in vitro was critical for the optimal bioactivity of the biomaterials. The combination of this hydroxyapatite material and osteogenic medium under a specific 3D culture condition in vitro resulted in obvious hypoxic and inflammatory responses of MSCs with no advantage of osteogenesis. The established single-cell transcriptional atlas provides foundational knowledge for a better understanding of MSC heterogeneity and responses to different osteogenic microenvironments. These results will be invaluable for future studies on bioactive materials and tissue regeneration as a reference dataset.
Data availability
The raw sequencing data are deposited in NCBI Sequence Read Archive (SRA), accession number PRJNA623696.
CRediT authorship contribution statement
Peng Guo: Methodology, Formal analysis, Investigation, Writing – original draft. Xizhe Liu: Formal analysis, Investigation, Writing – review & editing. Penghui Zhang: Methodology, Investigation, Writing – original draft. Zhongyuan He: Methodology, Investigation. Zhen Li: Conceptualization, Writing – review & editing. Mauro Alini: Conceptualization, Writing – review & editing. R. Geoff Richards: Conceptualization, Writing – review & editing. Sibylle Grad: Conceptualization, Writing – review & editing. Martin J. Stoddart: Conceptualization, Writing – review & editing. Guangqian Zhou: Data curation, Writing – review & editing. Xuenong Zou: Conceptualization, Visualization, Funding acquisition. Danny Chan: Conceptualization, Data curation, Writing – review & editing. Wei Tian: Visualization, Funding acquisition. Dafu Chen: Conceptualization, Project administration, Funding acquisition. Manman Gao: Conceptualization, Data curation, Writing – review & editing, Project administration. Zhiyu Zhou: Conceptualization, Data curation, Writing – review & editing, Supervision, Project administration, Funding acquisition. Shaoyu Liu: Conceptualization, Resources, Supervision, Project administration, Funding acquisition.
Acknowledgements
This study was supported by the National Key R&D Program of China (Grant no. 2017YFC1105000), the National Natural Science Foundation of China (Grant no. 81772400, 31900583, 31430030), the Fundamental Research Funds for the Central Universities (Grant no. 19ykzd05), the Natural Science Foundation of Guangzhou City (Grant no. 201704030082, 201807010031), the Foundation of Shenzhen Committee for Science and Technology Innovation (Grant no. JCYJ20190809142211354, GJHZ20180929160004704), the Sanming Project of Medicine in Shenzhen (Grant no. SZSM201911002), and the Beijing Municipal Health Commission (Grant no. BMHC-2021-X, BMHC-2019-9, BMHC-2018-4, PXM2020_026275_000002). Special thanks are extended to Dr. Cheng Ruijuan for technical support.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.08.009.
Contributor Information
Peng Guo, Email: guopeng@mail2.sysu.edu.cn.
Xizhe Liu, Email: gzxzhliu@qq.com.
Penghui Zhang, Email: zhangph7@mail2.sysu.edu.cn.
Zhongyuan He, Email: hezhy37@mail2.sysu.edu.cn.
Zhen Li, Email: zhen.li@aofoundation.org.
Mauro Alini, Email: mauro.alini@aofoundation.org.
R. Geoff Richards, Email: Geoff.richards@aofoundation.org.
Sibylle Grad, Email: sibylle.grad@aofoundation.org.
Martin J. Stoddart, Email: martin.stoddart@aofoundation.org.
Guangqian Zhou, Email: gqzhou@szu.edu.cn.
Xuenong Zou, Email: zxnong@hotmail.com.
Danny Chan, Email: chand@hku.hk.
Wei Tian, Email: tianweijst@vip.163.com.
Dafu Chen, Email: chendafujst@126.com.
Manman Gao, Email: gaomanm@mail2.sysu.edu.cn.
Zhiyu Zhou, Email: zhouzhy23@mail.sysu.edu.cn.
Shaoyu Liu, Email: gzsyliu@tom.com.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Agarwal R., Garcia A.J. Biomaterial strategies for engineering implants for enhanced osseointegration and bone repair. Adv. Drug Deliv. Rev. 2015;94:53–62. doi: 10.1016/j.addr.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ho-Shui-Ling A., Bolander J., Rustom L.E., et al. Bone regeneration strategies: engineered scaffolds, bioactive molecules and stem cells current stage and future perspectives. Biomaterials. 2018;180:143–162. doi: 10.1016/j.biomaterials.2018.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim H.D., Amirthalingam S., Kim S.L., et al. Biomimetic materials and fabrication approaches for bone tissue engineering. Adv Healthc Mater. 2017;6(23) doi: 10.1002/adhm.201700612. [DOI] [PubMed] [Google Scholar]
- 4.Li M., Xiong P., Yan F., et al. An overview of graphene-based hydroxyapatite composites for orthopedic applications. Bioact Mater. 2018;3(1):1–18. doi: 10.1016/j.bioactmat.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oliveira H.L., Da Rosa W.L.O., Cuevas-Suarez C.E., et al. Histological evaluation of bone repair with hydroxyapatite: a systematic review. Calcif. Tissue Int. 2017;101(4):341–354. doi: 10.1007/s00223-017-0294-z. [DOI] [PubMed] [Google Scholar]
- 6.Campana V., Milano G., Pagano E., et al. Bone substitutes in orthopaedic surgery: from basic science to clinical practice. J. Mater. Sci. Mater. Med. 2014;25(10):2445–2461. doi: 10.1007/s10856-014-5240-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chai F., Raoul G., Wiss A., et al. [Bone substitutes: classification and concerns] Rev Stomatol Chir Maxillofac. 2011;112(4):212–221. doi: 10.1016/j.stomax.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Wang W., Yeung K.W.K. Bone grafts and biomaterials substitutes for bone defect repair: a review. Bioact Mater. 2017;2(4):224–247. doi: 10.1016/j.bioactmat.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bunpetch V., Zhang Z.Y., Zhang X., et al. Strategies for MSC expansion and MSC-based microtissue for bone regeneration. Biomaterials. 2019;196:67–79. doi: 10.1016/j.biomaterials.2017.11.023. [DOI] [PubMed] [Google Scholar]
- 10.Andrzejewska A., Lukomska B., Janowski M. Concise review: mesenchymal stem cells: from roots to boost. Stem Cell. 2019;37(7):855–864. doi: 10.1002/stem.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shang F., Yu Y., Liu S., et al. Advancing application of mesenchymal stem cell-based bone tissue regeneration. Bioact Mater. 2021;6(3):666–683. doi: 10.1016/j.bioactmat.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma J., Guo W., Gao M., et al. Biomimetic matrix fabricated by LMP-1 gene-transduced MC3T3-E1 cells for bone regeneration. Biofabrication. 2017;9(4) doi: 10.1088/1758-5090/aa8dd1. [DOI] [PubMed] [Google Scholar]
- 13.Yin S., Zhang W., Zhang Z., et al. Recent advances in scaffold design and material for vascularized tissue-engineered bone regeneration. Adv Healthc Mater. 2019;8(10) doi: 10.1002/adhm.201801433. [DOI] [PubMed] [Google Scholar]
- 14.Zhou W., Lin J., Zhao K., et al. Single-cell profiles and clinically useful properties of human mesenchymal stem cells of adipose and bone marrow origin. Am. J. Sports Med. 2019;47(7):1722–1733. doi: 10.1177/0363546519848678. [DOI] [PubMed] [Google Scholar]
- 15.Baryawno N., Przybylski D., Kowalczyk M.S., et al. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell. 2019;177(7):1915–1932. doi: 10.1016/j.cell.2019.04.040. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tikhonova A.N., Dolgalev I., Hu H., et al. The bone marrow microenvironment at single-cell resolution. Nature. 2019;569(7755):222–228. doi: 10.1038/s41586-019-1104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juang H.Y., Hon M.H. Effect of calcination on sintering of hydroxyapatite. Biomaterials. 1996;17(21):2059–2064. doi: 10.1016/0142-9612(96)88882-x. [DOI] [PubMed] [Google Scholar]
- 18.Uemura T., Dong J., Wang Y., et al. Transplantation of cultured bone cells using combinations of scaffolds and culture techniques. Biomaterials. 2003;24(13):2277–2286. doi: 10.1016/s0142-9612(03)00039-5. [DOI] [PubMed] [Google Scholar]
- 19.Satija R., Farrell J.A., Gennert D., et al. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33(5):495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwalie P.C., Dong H., Zachara M., et al. A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature. 2018;559(7712):103–108. doi: 10.1038/s41586-018-0226-8. [DOI] [PubMed] [Google Scholar]
- 21.Areshkov P.O., Avdieiev S.S., Balynska O.V., et al. Two closely related human members of chitinase-like family, CHI3L1 and CHI3L2, activate ERK1/2 in 293 and U373 cells but have the different influence on cell proliferation. Int. J. Biol. Sci. 2012;8(1):39–48. doi: 10.7150/ijbs.8.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eagle H., Levine E.M. Growth regulatory effects of cellular interaction. Nature. 1967;213(5081):1102–1106. doi: 10.1038/2131102a0. [DOI] [PubMed] [Google Scholar]
- 23.Soufi A., Dalton S. Cycling through developmental decisions: how cell cycle dynamics control pluripotency, differentiation and reprogramming. Development. 2016;143(23):4301–4311. doi: 10.1242/dev.142075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pavel M., Renna M., Park S.J., et al. Contact inhibition controls cell survival and proliferation via YAP/TAZ-autophagy axis. Nat. Commun. 2018;9(1):2961. doi: 10.1038/s41467-018-05388-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ko J.Y., Yoo K.H., Lee H.W., et al. Mxi1 regulates cell proliferation through insulin-like growth factor binding protein-3. Biochem. Biophys. Res. Commun. 2011;415(1):36–41. doi: 10.1016/j.bbrc.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Unoki M., Nakamura Y. Growth-suppressive effects of BPOZ and EGR2, two genes involved in the PTEN signaling pathway. Oncogene. 2001;20(33):4457–4465. doi: 10.1038/sj.onc.1204608. [DOI] [PubMed] [Google Scholar]
- 27.Gerke V., Moss S.E. Annexins: from structure to function. Physiol. Rev. 2002;82(2):331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- 28.Rogina A., Antunovic M., Milovac D. Biomimetic design of bone substitutes based on cuttlefish bone-derived hydroxyapatite and biodegradable polymers. J. Biomed. Mater. Res. B Appl. Biomater. 2019;107(1):197–204. doi: 10.1002/jbm.b.34111. [DOI] [PubMed] [Google Scholar]
- 29.Tiet T.D., Alman B.A. Developmental pathways in musculoskeletal neoplasia: involvement of the Indian Hedgehog-parathyroid hormone-related protein pathway. Pediatr. Res. 2003;53(4):539–543. doi: 10.1203/01.PDR.0000054688.93486.18. [DOI] [PubMed] [Google Scholar]
- 30.Clemens T.L., Cormier S., Eichinger A., et al. Parathyroid hormone-related protein and its receptors: nuclear functions and roles in the renal and cardiovascular systems, the placental trophoblasts and the pancreatic islets. Br. J. Pharmacol. 2001;134(6):1113–1136. doi: 10.1038/sj.bjp.0704378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu M., Chen G., Li Y.P. TGF-beta and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016;4:16009. doi: 10.1038/boneres.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serra R., Karaplis A., Sohn P. Parathyroid hormone-related peptide (PTHrP)-dependent and -independent effects of transforming growth factor beta (TGF-beta) on endochondral bone formation. J. Cell Biol. 1999;145(4):783–794. doi: 10.1083/jcb.145.4.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kagermeier-Schenk B., Wehner D., Ozhan-Kizil G., et al. Waif1/5T4 inhibits Wnt/beta-catenin signaling and activates noncanonical Wnt pathways by modifying LRP6 subcellular localization. Dev. Cell. 2011;21(6):1129–1143. doi: 10.1016/j.devcel.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 34.Huang J., Wang L., Jiang M., et al. PTHLH coupling upstream negative regulation of fatty acid biosynthesis and Wnt receptor signal to downstream peptidase activity-induced apoptosis network in human hepatocellular carcinoma by systems-theoretical analysis. J. Recept. Signal Transduct. Res. 2012;32(5):250–256. doi: 10.3109/10799893.2012.700717. [DOI] [PubMed] [Google Scholar]
- 35.Minina E., Kreschel C., Naski M.C., et al. Interaction of FGF, Ihh/Pthlh, and BMP signaling integrates chondrocyte proliferation and hypertrophic differentiation. Dev. Cell. 2002;3(3):439–449. doi: 10.1016/s1534-5807(02)00261-7. [DOI] [PubMed] [Google Scholar]
- 36.Zheng J., Li H., He L., et al. Preconditioning of umbilical cord-derived mesenchymal stem cells by rapamycin increases cell migration and ameliorates liver ischaemia/reperfusion injury in mice via the CXCR4/CXCL12 axis. Cell Prolif. 2019;52(2) doi: 10.1111/cpr.12546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agarwal P., Isringhausen S., Li H., et al. Mesenchymal niche-specific expression of Cxcl12 controls quiescence of treatment-resistant leukemia stem cells. Cell Stem Cell. 2019;24(5):769–784. doi: 10.1016/j.stem.2019.02.018. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nudelman F., Pieterse K., George A., et al. The role of collagen in bone apatite formation in the presence of hydroxyapatite nucleation inhibitors. Nat. Mater. 2010;9(12):1004–1009. doi: 10.1038/nmat2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vindevoghel L., Lechleider R.J., Kon A., et al. SMAD3/4-dependent transcriptional activation of the human type VII collagen gene (COL7A1) promoter by transforming growth factor beta. Proc. Natl. Acad. Sci. U. S. A. 1998;95(25):14769–14774. doi: 10.1073/pnas.95.25.14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alimperti S., Andreadis S.T. CDH2 and CDH11 act as regulators of stem cell fate decisions. Stem Cell Res. 2015;14(3):270–282. doi: 10.1016/j.scr.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang M., Lin R., Wang X., et al. 3D printing of Haversian bone-mimicking scaffolds for multicellular delivery in bone regeneration. Sci Adv. 2020;6(12) doi: 10.1126/sciadv.aaz6725. eaaz6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chai M., Gu C., Shen Q., et al. Hypoxia alleviates dexamethasone-induced inhibition of angiogenesis in cocultures of HUVECs and rBMSCs via HIF-1alpha. Stem Cell Res. Ther. 2020;11(1):343. doi: 10.1186/s13287-020-01853-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan C.K., Seo E.Y., Chen J.Y., et al. Identification and specification of the mouse skeletal stem cell. Cell. 2015;160(1–2):285–298. doi: 10.1016/j.cell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy M.P., Koepke L.S., Lopez M.T., et al. Articular cartilage regeneration by activated skeletal stem cells. Nat. Med. 2020 doi: 10.1038/s41591-020-1013-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arutyunyan I.V., Rzhaninova A.A., Volkov A.V., et al. Effect of dexamethasone on differentiation of multipotent stromal cells from human adipose tissue. Bull. Exp. Biol. Med. 2009;147(4):503–508. doi: 10.1007/s10517-009-0548-5. [DOI] [PubMed] [Google Scholar]
- 46.Muenzner M., Tuvia N., Deutschmann C., et al. Retinol-binding protein 4 and its membrane receptor STRA6 control adipogenesis by regulating cellular retinoid homeostasis and retinoic acid receptor alpha activity. Mol. Cell Biol. 2013;33(20):4068–4082. doi: 10.1128/MCB.00221-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Q., Graham T.E., Mody N., et al. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005;436(7049):356–362. doi: 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
- 48.Li Y., Jin D., Xie W., et al. PPAR-gamma and wnt regulate the differentiation of MSCs into adipocytes and osteoblasts respectively. Curr. Stem Cell Res. Ther. 2018;13(3):185–192. doi: 10.2174/1574888X12666171012141908. [DOI] [PubMed] [Google Scholar]
- 49.Morikawa M., Derynck R., Miyazono K. TGF-beta and the TGF-beta family: context-dependent roles in cell and tissue physiology. Cold Spring Harb Perspect Biol. 2016;8(5) doi: 10.1101/cshperspect.a021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poveda J., Sanz A.B., Fernandez-Fernandez B., et al. MXRA5 is a TGF-beta1-regulated human protein with anti-inflammatory and anti-fibrotic properties. J. Cell Mol. Med. 2017;21(1):154–164. doi: 10.1111/jcmm.12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palumbo-Zerr K., Zerr P., Distler A., et al. Orphan nuclear receptor NR4A1 regulates transforming growth factor-beta signaling and fibrosis. Nat. Med. 2015;21(2):150–158. doi: 10.1038/nm.3777. [DOI] [PubMed] [Google Scholar]
- 52.Liu F., Zhang X., Zhang B., et al. TREM1: a positive regulator for inflammatory response via NF-kappaB pathway in A549 cells infected with Mycoplasma pneumoniae. Biomed. Pharmacother. 2018;107:1466–1472. doi: 10.1016/j.biopha.2018.07.176. [DOI] [PubMed] [Google Scholar]
- 53.Bouchon A., Facchetti F., Weigand M.A., et al. TREM-1 amplifies inflammation and is a crucial mediator of septic shock. Nature. 2001;410(6832):1103–1107. doi: 10.1038/35074114. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y., Wang K., Han G.C., et al. Neutrophil infiltration favors colitis-associated tumorigenesis by activating the interleukin-1 (IL-1)/IL-6 axis. Mucosal Immunol. 2014;7(5):1106–1115. doi: 10.1038/mi.2013.126. [DOI] [PubMed] [Google Scholar]
- 55.Peruzzi B., Cappariello A., Del Fattore A., et al. c-Src and IL-6 inhibit osteoblast differentiation and integrate IGFBP5 signalling. Nat. Commun. 2012;3:630. doi: 10.1038/ncomms1651. [DOI] [PubMed] [Google Scholar]
- 56.Najar M., Raicevic G., Jebbawi F., et al. Characterization and functionality of the CD200-CD200R system during mesenchymal stromal cell interactions with T-lymphocytes. Immunol. Lett. 2012;146(1–2):50–56. doi: 10.1016/j.imlet.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 57.Langenbach F., Handschel J. Effects of dexamethasone, ascorbic acid and beta-glycerophosphate on the osteogenic differentiation of stem cells in vitro. Stem Cell Res. Ther. 2013;4(5):117. doi: 10.1186/scrt328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marofi F., Vahedi G., Solali S., et al. Gene expression of TWIST1 and ZBTB16 is regulated by methylation modifications during the osteoblastic differentiation of mesenchymal stem cells. J. Cell. Physiol. 2019;234(5):6230–6243. doi: 10.1002/jcp.27352. [DOI] [PubMed] [Google Scholar]
- 59.Mizuhashi K., Kanamoto T., Ito M., et al. OBIF, an osteoblast induction factor, plays an essential role in bone formation in association with osteoblastogenesis. Dev. Growth Differ. 2012;54(4):474–480. doi: 10.1111/j.1440-169X.2012.01333.x. [DOI] [PubMed] [Google Scholar]
- 60.French D.M., Kaul R.J., D'Souza A.L., et al. WISP-1 is an osteoblastic regulator expressed during skeletal development and fracture repair. Am. J. Pathol. 2004;165(3):855–867. doi: 10.1016/S0002-9440(10)63348-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Korpos E., Deak F., Kiss I. Matrilin-2, an extracellular adaptor protein, is needed for the regeneration of muscle, nerve and other tissues. Neural Regen Res. 2015;10(6):866–869. doi: 10.4103/1673-5374.158332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Onichtchouk D., Chen Y.G., Dosch R., et al. Silencing of TGF-beta signalling by the pseudoreceptor BAMBI. Nature. 1999;401(6752):480–485. doi: 10.1038/46794. [DOI] [PubMed] [Google Scholar]
- 63.Thomas G., Moffatt P., Salois P., et al. Osteocrin, a novel bone-specific secreted protein that modulates the osteoblast phenotype. J. Biol. Chem. 2003;278(50):50563–50571. doi: 10.1074/jbc.M307310200. [DOI] [PubMed] [Google Scholar]
- 64.Becker J., Semler O., Gilissen C., et al. Exome sequencing identifies truncating mutations in human SERPINF1 in autosomal-recessive osteogenesis imperfecta. Am. J. Hum. Genet. 2011;88(3):362–371. doi: 10.1016/j.ajhg.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chiellini C., Grenningloh G., Cochet O., et al. Stathmin-like 2, a developmentally-associated neuronal marker, is expressed and modulated during osteogenesis of human mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2008;374(1):64–68. doi: 10.1016/j.bbrc.2008.06.121. [DOI] [PubMed] [Google Scholar]
- 66.Gao M.M., Su Q.N., Liang T.Z., et al. Transcriptional activation of ENPP1 by osterix in osteoblasts and osteocytes. Eur. Cell. Mater. 2018;36:1–14. doi: 10.22203/eCM.v036a01. [DOI] [PubMed] [Google Scholar]
- 67.Farokhi M., Mottaghitalab F., Samani S., et al. Silk fibroin/hydroxyapatite composites for bone tissue engineering. Biotechnol. Adv. 2018;36(1):68–91. doi: 10.1016/j.biotechadv.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 68.Buttitta L.A., Edgar B.A. Mechanisms controlling cell cycle exit upon terminal differentiation. Curr. Opin. Cell Biol. 2007;19(6):697–704. doi: 10.1016/j.ceb.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsushita Y., Ono W., Ono N. Growth plate skeletal stem cells and their transition from cartilage to bone. Bone. 2020;136:115359. doi: 10.1016/j.bone.2020.115359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jang C.H., Cho Y.B., Choi C.H., et al. Comparision of osteoconductivity of biologic and artificial synthetic hydroxyapatite in experimental mastoid obliteration. Acta Otolaryngol. 2014;134(3):255–259. doi: 10.3109/00016489.2013.859397. [DOI] [PubMed] [Google Scholar]
- 71.Zhang P., Liu X., Guo P., et al. Effect of cyclic mechanical loading on immunoinflammatory microenvironment in biofabricating hydroxyapatite scaffold for bone regeneration. Bioact Mater. 2021;6(10):3097–3108. doi: 10.1016/j.bioactmat.2021.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li L., Yu M., Li Y., et al. Synergistic anti-inflammatory and osteogenic n-HA/resveratrol/chitosan composite microspheres for osteoporotic bone regeneration. Bioact Mater. 2021;6(5):1255–1266. doi: 10.1016/j.bioactmat.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ribeiro V.P., Silva-Correia J., Nascimento A.I., et al. Silk-based anisotropical 3D biotextiles for bone regeneration. Biomaterials. 2017;123:92–106. doi: 10.1016/j.biomaterials.2017.01.027. [DOI] [PubMed] [Google Scholar]
- 74.Wang K.H., Liu H.W., Lin S.R., et al. Field methylation silencing of the protocadherin 10 gene in cervical carcinogenesis as a potential specific diagnostic test from cervical scrapings. Canc. Sci. 2009;100(11):2175–2180. doi: 10.1111/j.1349-7006.2009.01285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ying J., Li H., Seng T.J., et al. Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal, esophageal and multiple other carcinomas with frequent methylation. Oncogene. 2006;25(7):1070–1080. doi: 10.1038/sj.onc.1209154. [DOI] [PubMed] [Google Scholar]
- 76.Kim S.Y., Yasuda S., Tanaka H., et al. Non-clustered protocadherin. Cell Adhes. Migrat. 2011;5(2):97–105. doi: 10.4161/cam.5.2.14374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data are deposited in NCBI Sequence Read Archive (SRA), accession number PRJNA623696.