Abstract
Introduction
Deep learning techniques are gaining momentum in medical research. Evidence shows that deep learning has advantages over humans in image identification and classification, such as facial image analysis in detecting people’s medical conditions. While positive findings are available, little is known about the state-of-the-art of deep learning-based facial image analysis in the medical context. For the consideration of patients’ welfare and the development of the practice, a timely understanding of the challenges and opportunities faced by research on deep-learning-based facial image analysis is needed. To address this gap, we aim to conduct a systematic review to identify the characteristics and effects of deep learning-based facial image analysis in medical research. Insights gained from this systematic review will provide a much-needed understanding of the characteristics, challenges, as well as opportunities in deep learning-based facial image analysis applied in the contexts of disease detection, diagnosis and prognosis.
Methods
Databases including PubMed, PsycINFO, CINAHL, IEEEXplore and Scopus will be searched for relevant studies published in English in September, 2021. Titles, abstracts and full-text articles will be screened to identify eligible articles. A manual search of the reference lists of the included articles will also be conducted. The Preferred Reporting Items for Systematic Reviews and Meta-Analyses framework was adopted to guide the systematic review process. Two reviewers will independently examine the citations and select studies for inclusion. Discrepancies will be resolved by group discussions till a consensus is reached. Data will be extracted based on the research objective and selection criteria adopted in this study.
Ethics and dissemination
As the study is a protocol for a systematic review, ethical approval is not required. The study findings will be disseminated via peer-reviewed publications and conference presentations.
PROSPERO registration number
CRD42020196473.
Keywords: public health, information technology, health informatics, telemedicine, biotechnology & bioinformatics
Strengths and limitations of this study.
This systematic review protocol follows the Preferred Reporting Items for Systematic Review and Meta-Analysis Protocols guidelines.
By examining the characteristics and effects of deep learning-based facial image analysis in medical research, this systematic review bridges the gap in the literature.
This review is limited to evidence on the use and application of deep learning technologies in patients’ facial image identification and classification.
Non-English databases will not be searched, which might limit the representativeness of the results.
Background
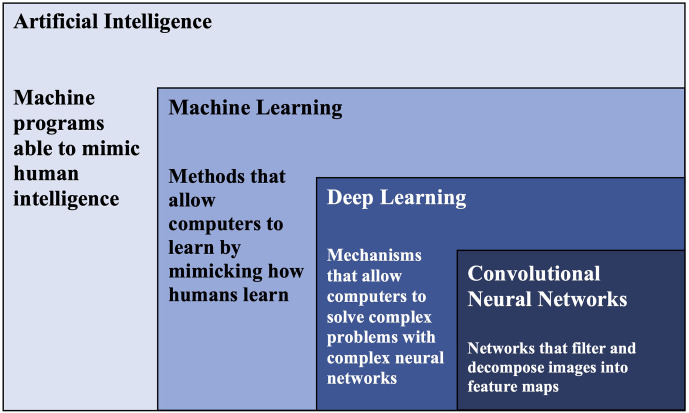
As disease manifestations often show in various places in the human body, such as Down syndromes can change patients’ facial features, researchers have been investigating whether analysing appearance features can facilitate early disease detection and identification.1–5 One promising field is deep learning-based facial analysis.6–8 Deep learning represents a powerful range of artificial intelligence (AI) algorithm that allows computers to tackle complex problems via capitalising on neural networks, such as convolutional neural networks (CNNs), that are rich in neurons, layers and interconnectivity (see figure 1).9 Simply put, deep learning is a mechanism that allows computers to solve complex problems by neural network architecture. This ability to develop complex network structures gives deep learning a distinctive advantage: it can automatically transform raw data input into meaningful features that enable pattern identification.10 Deep learning technique has revolutionary potential in practical and research fields.11 In practice, as deep learning effectively identifies objects, traffic signs and faces, its adaptations have been widely applied in designing robots and self-driving cars.12–15 Deep learning has also been widely adopted in biomedical and clinical research, particularly in the field of medical imaging.16–19
Figure 1.
Relationship between artificial intelligence, machine learning, deep learning and convolutional neural networks.
Medical conditions are often diagnosed by means of tests, such as biopsy and diagnostic imaging. An example list of diseases that have been analysed by deep learning technologies could be found in table 1. As diagnostic imaging is non-invasive and can facilitate personalised medicine, it is a preferred test option for patients and healthcare practitioners.20 21 This, in turn, has contributed to the exponential growth of medical imaging data and the increasing need for boosting medical image processing power to formulate diagnosis swiftly.21 22 Compared with traditional computer aided diagnosis for analysing medical imaging, such as hand-crafted radiomics for tumour detection, deep learning methods are superior in their ability to process large quantities of medical images accurately and cost-effectively, without exerting a heavy workload on radiologists.23–27 Evidence shows that deep learning-based medical image analysis was able to increase accuracy rates in various disease contexts, such as the identification of spinal disorder1 and lung cancer histology,28 classification of skin lesion29 and chronic gastritis,30 and the prediction of tumour-related genes31 and vascular diseases.32
Table 1.
An example list of diseases that have been analysed by deep learning techniques
| Disease context | Deep learning technique |
| Acromegaly | Convolutional neural network (along with Generalized Linear Models; K-nearest neighbors; Support Vector Machines; forests of randomized trees)63 |
| Cancer | Convolutional neutral network64 |
| Cornelia de Lange syndrome | DeepGestalt technology7 |
| Coronary artery disease | Convolutional neural network65 |
| Down syndrome | Independent component analysis66 |
| Facial dermatological disorders | Convolutional neural network67 |
| Keratinocytic skin cancer | Convolutional neutral network68 |
| Inherited retinal degenerations | Convolutional neural network69 |
| Noonan syndrome | DeepGestalt technology33 |
| Pain intensity | Convolutional neutral network70 |
| Neurological disorders | Convolutional neutral network71 |
Applying the deep learning technique to perform facial recognition and analysis tasks, researchers found that the technique yielded superior results in identifying and classifying faces of people with cancer from those without.6 Similarly, examining facial phenotypes of people with genetic disorders, findings indicate that the technique was effective and was able to yield an optimal 91% top-10 accuracy.33 Evidence further indicates that, for some tasks involving identifying and classifying facial images, deep learning techniques have often performed on par or better than human beings.5 7 10 34–36 Comparing clinical and deep learning evaluations of microdeletion syndrome facial phenotypes, researchers found that deep learning outperformed clinical evaluations in terms of sensitivity and specificity by 96%.35 These findings combined suggest that deep learning-based facial analysis technology has great potential to address complex medical challenges prevalent in healthcare. However, there has not been any systematic review on the state-of-the-art applications of deep learning-based facial analysis in non-invasively evaluating medical conditions. Therefore, to bridge this gap, we aim to systematically review the literature and identify the characteristics and effects of deep learning-based facial analysis techniques applied in medical research.
Methods and analysis
This systematic review was registered with the International Prospective Register of Systematic Reviews database or PROSPERO a priori to improve research rigour.37 38 The principles of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses protocol was adopted to guide this systematic review.39 Our search strategy incorporated medical subject heading (MeSH) and keyword terms for the concept of deep learning and facial analysis. The search strategy was developed in consultation with an academic librarian, and subsequently will be deployed to target databases, including PubMed, PsycINFO, CINAHL, IEEEXplore and Scopus (table 2). The search will be initiated in September 2021. Studies will be limited to journal articles published in English. We will adopt two additional search mechanisms to locate eligible articles: (1) a manual search of the reference list of the included articles will be performed and (2) a reverse search of papers that cited articles included in the final review via Google Scholar. An academic librarian will facilitate the search process, helping administer the search and download the citation records to Rayyan (http://rayyan.qcri.org).
Table 2.
Example PubMed search strategy
| Concept | Search string |
| Deep learning | “deep learning”[MeSH] OR “deep learning”[TIAB] OR “artificial intelligence” [MeSH] OR “artificial intelligence” [TIAB] OR “machine learning”[MeSH] OR “machine learning”[TIAB] OR “convolutional neural network”[MeSH] OR “convolutional neural network”[TIAB] OR “convolutional neural networks”[TIAB] |
| Facial image analysis | “face detect*” OR “facial detect*” OR “face recogn*” OR “facial recogn*” OR “face extract*” or “facial extract*” OR “face analys*” OR “facial analys*” OR “face dysmorphology” OR “facial dysmorphology” OR “face phenotype*” OR “facial phenotype*” OR “face feature*” OR “facial feature*” OR “face2gene” OR “gestalt theory” OR “face photograph*” OR “facial photograph*” OR “facial expression” |
Inclusion and exclusion criteria
The inclusion criteria were developed a priori and listed in table 3. Studies will be excluded articles if they (1) did not report findings on human beings (eg, studies on mice), (2) did not focus on full facial features (eg, research on retina or lip-cleft), (3) did not conduct research in a medical context (eg, in the context of criminology) and (4) did not report empirical findings (eg, editorial or comment papers).
Table 3.
Study inclusion criteria
| Data type | Inclusion criteria |
| Participants | Individuals younger or older than 18 years old |
| Research context | Medical research or healthcare |
| Analytical technique | Deep learning algorithms-based facial image analysis |
| Language | English |
| Study type | Quantitative empirical study |
| Outcome | Report empirical and original findings on the application of deep learning-based facial image analysis in medical context (eg, accuracy of facial image analysis in detecting Down syndrome) |
Risk of bias assessment
To ensure the quality of included studies, a risk of bias assessment will be conducted independently by two reviewers, using the Cochrane Collaboration evaluation framework.40 The framework has seven domains: (1) random sequence generation, (2) allocation concealment, (3) blinding of participants and personnel, (4) blinding of outcome assessment, (5) incomplete outcome data, (6) selective reporting and (7) any other source of bias. The risk of bias will be evaluated independently by two reviewers. Potential discrepancies regarding the risk of bias will be resolved via group discussions till a consensus is reached.
Data extraction
Two reviewers will independently examine the citations and select studies for inclusion. Discrepancies will be resolved by group discussions till a consensus is reached. Data will be extracted based on the research objective and selection criteria adopted in this study. For articles that meet the inclusion criteria, the reviewers will extract the following information from the included papers: research objective/questions, disease context, sample characteristics (eg, characteristics of facial records), AI characteristics (eg, algorithm adopted), and empirical findings.
Data synthesis and analysis
If eligible studies share enough similarities to be pooled, a meta-analysis will be conducted to gain further insights into the data. Main clinical, methodological, as well as statistical differences will be carefully considered to determine the heterogeneity of the eligible studies. If eligible studies are found heterogeneous, a narrative synthesis will be conducted to summarise the data. A summary of the data extracted will be organised to synthesise key results. Both tables and graphs will be used to represent the key characteristics of eligible articles. Descriptive analysis will be performed on categorical variables. In this review, we will undertake a narrative approach to synthesise data. In other words, in addition to shedding light on key information like the sensitivity, specificity, overall accuracy of the deep learning technologies in analysing facial images (as opposed to clinicians’ analyses), we will also provide detailed analysis of the disease contexts and the techniques applied to chart the state-of-the-art of deep learning technologies in facial image analyses.
Ethics and dissemination
As the study is a protocol for a systematic review, ethical approval is not required. The study findings will be disseminated via peer-reviewed publications and conference presentations.
Patient and public involvement
The nature of the study, which is a review and analysis of previously published data, dictates that there is limited to no meaningful need for patient and public involvement in the design, delivery or dissemination of the research findings.
Discussion
Though a growing body of research has applied deep learning-based facial image analysis in the medical context for disease detection, diagnosis and prognosis, to date, no systematic review has investigated the state-of-the-art application of deep learning-based facial image analysis recognition in addressing medical diagnoses and clinical states. Therefore, to bridge this gap, we aim to systematically review the literature and present the characteristics, challenges, as well as opportunities in deep learning-based facial analysis techniques applied in medical research. To better organise the research findings, we developed a framework that illustrates the main causes for abnormal facial expressions in patients. It is important to note that we are identifying medical states and conditions and not individuals.
After reviewing the literature,5 41–55 we identified the following four preliminary categories of causes for short-term or long-term abnormal facial expressions in people: (1) gene-related factors, (2) neurological factors, (3) psychiatric conditions and (4) medication-induced triggers. Genetic-related factors, such as the presence or mutation of a certain gene, are the most studied cause for abnormal facial changes in individuals.5 7 35 Down syndrome, which is affected by the presence of a third copy of chromosome 21, is an example of genetic-related factors that can cause individuals’ abnormal facial changes.5 Neurological factors can also cause individuals’ facial phenotypes. Stroke or transient ischaemic attack is an example of neurological factors, which can occur either prior to or after the onset of the disease.56 57 The third cause for abnormal facial changes centres on individuals’ psychiatric conditions or mental illnesses, especially psychotic disorders such as the Tourette syndrome (facial tics). Last but not the least, medication-induced triggers, such as the neuroleptic malignant syndrome (caused by antipsychotic medications), can also cause abnormal facial changes in people. Details of this framework can be found in table 4. This framework will be used in the planned systematic review study to guide the data extraction process.
Table 4.
Main causes for abnormal facial expressions
| Cause | Definition and example |
| Gene-related factors | Gene-related factors are causes for individuals’ abnormal facial changes that root in the presence or mutation of one or a set of genes. Examples: Down syndrome (genetic root: presence of a third copy of chromosome 21) or Cornelia de Lange syndrome (genetic root: NIPBL or SMC1A, SMC3, RAD21 or HDAC8, BRD4 and ANKRD11 genes).5 41 42 55 |
| Neurological factors | Neurological factors are defined as reasons that are associated with individuals’ congenital or acquired disorders of nerves and the nervous system. Neurological factors can either be related to genetic or non-genetic factors, caused by irregularity in nerves associated with the brain or the face. Examples: Neurological factors with genetic causes (eg, Rett syndrome, MECP2 gene; Cervical or Cranial dystonia, GNAL gene) and without (eg, embouchure dystonia, Oromandibular dystonia)46 47; due to nerves associated with the brain (eg, stroke) or the face (Bell’s palsy or facial paralysis, Hemifacial Spasm).43–45 |
| Psychiatric conditions | Psychiatric conditions, especially psychotic disorders, have the potential to cause abnormal facial expressions among individuals. Psychiatric conditions could be broadly defined as mental illnesses, whereas psychotic disorder factors are causes to abnormal facial expressions that root in individuals’ impaired sense of reality. Examples: Non-drug-related Tourette syndrome (facial tics) or autism (facial expression limitation).48–50 |
| Medication-induced triggers | Medication-induced triggers could be understood as causes to individuals’ short-term or long-term abnormal facial changes due to their adverse reactions to a certain medication of a type of medications. Examples: Neuroleptic malignant syndrome (antipsychotic drugs), tardive dyskinesia (antipsychotic medications) or drug-related Tourette syndrome.51–54 |
Overall, insights gained from this study will be able to provide a much-needed understanding of the characteristics, challenges, as well as opportunities in the context of deep learning-based facial image analysis technologies applied in disease detection, diagnosis and prognosis. In addition to gaining a connected and comprehensive understanding of the current application of facial image analysis, results of the study will also be able to shed light on whether, similar to facial recognition used in non-medical58 59 and medical contexts,60 61 whether or to what degree is systematic bias is present in the application of deep learning technologies for facial image analysis. A biased and inaccurate facial image analysis system will not only exert unwarranted, though avoidable, disparities on patients (eg, gender inequality),60 it will also alienate the patients from the much-needed deep-learning-assisted medical opportunities their health and well-being can benefit from.62 Therefore, for the consideration of patients’ welfare and the development of the clinical practice, a timely understanding of the scope of the research literature as well as the challenges and opportunities faced by research on deep-learning-based facial image analysis is much needed.
Supplementary Material
Acknowledgments
The authors wish to thank Emme Lopez, for her assistance in the search strategy development. Furthermore, the authors are very grateful for the constructive input offered by the editors and reviewers.
Footnotes
Correction notice: This article has been corrected since it first published. Affiliation for 'Peng Jia' has been updated.
Contributors: ZS developed the research idea and drafted the manuscript. BL, FS, JG, SS, JW, PJ and XH reviewed and revised the manuscript.
Funding: This work was supported by the United Nations Development Program (UNDP) South-South Cooperation: Learning from China's Experience to improve the Ability of Response to COVID-19 in Asia and the Pacific Region; The Joint Pilot Project between the Ministry of Industry and Information Technology and the National Health Commission of the People’s Republic of China: The Development, Standardization, and Application of 5G-Powered and Cloud-Based Virtual Critical Care and Management.
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Ethics statements
Patient consent for publication
Not applicable.
References
- 1.Yang J, Zhang K, Fan H, et al. Development and validation of deep learning algorithms for scoliosis screening using back images. Commun Biol 2019;2): :390. 10.1038/s42003-019-0635-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang X, Liu J, Wu C, et al. Artificial intelligence in tongue diagnosis: using deep convolutional neural network for recognizing unhealthy tongue with tooth-mark. Comput Struct Biotechnol J 2020;18:973–80. 10.1016/j.csbj.2020.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lam C, Yi D, Guo M, et al. Automated detection of diabetic retinopathy using deep learning. AMIA Jt Summits Transl Sci Proc 2018;2017:147–55. [PMC free article] [PubMed] [Google Scholar]
- 4.Martin D, Croft J, Pitt A, et al. Systematic review and meta-analysis of the relationship between genetic risk for schizophrenia and facial emotion recognition. Schizophr Res 2020;218:7–13. 10.1016/j.schres.2019.12.031 [DOI] [PubMed] [Google Scholar]
- 5.Kruszka P, Porras AR, Sobering AK, et al. Down syndrome in diverse populations. Am J Med Genet A 2017;173:42–53. 10.1002/ajmg.a.38043 [DOI] [PubMed] [Google Scholar]
- 6.Liang B, Yang N, He G, et al. Identification of the facial features of patients with cancer: a deep learning–based pilot study. J Med Internet Res 2020;22:e17234. 10.2196/17234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Latorre-Pellicer A, Ascaso Ángela, Trujillano L, et al. Evaluating face2gene as a tool to identify cornelia de lange syndrome by facial phenotypes. Int J Mol Sci 2020;21. 10.3390/ijms21031042. [Epub ahead of print: 04 Feb 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myers L, Anderlid B-M, Nordgren A, et al. Clinical versus automated assessments of morphological variants in twins with and without neurodevelopmental disorders. Am J Med Genet A 2020;182:1177–89. 10.1002/ajmg.a.61545 [DOI] [PubMed] [Google Scholar]
- 9.Deng L, Yu D. Deep learning: methods and applications. FNT in Signal Processing 2013;7:197–387. 10.1561/2000000039 [DOI] [Google Scholar]
- 10.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436–44. 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 11.Gilani SZ, Mian A. Learning from millions of 3D scans for large-scale 3D face recognition. IEEE/CVF Conference on Computer Vision and Pattern Recognition, 2018. [Google Scholar]
- 12.Algabri R, Choi M-T. Deep-learning-based indoor human following of mobile robot using color feature. Sensors 2020;20. 10.3390/s20092699. [Epub ahead of print: 09 May 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin J, Apuroop KGS, Tamilselvam YK, et al. Table cleaning task by human support robot using deep learning technique. Sensors 2020;20:1698. 10.3390/s20061698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balado J, Martínez-Sánchez J, Arias P, et al. Road environment semantic segmentation with deep learning from MLS point cloud data. Sensors 2019;19:3466. 10.3390/s19163466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naqvi RA, Arsalan M, Batchuluun G, et al. Deep learning-based gaze detection system for automobile drivers using a NIR camera sensor. Sensors 2018;18:456. 10.3390/s18020456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu G, Hua J, Wu Z, et al. Automatic classification of esophageal lesions in endoscopic images using a convolutional neural network. Ann Transl Med 2020;8:486. 10.21037/atm.2020.03.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chartrand G, Cheng PM, Vorontsov E, et al. Deep learning: a primer for radiologists. Radiographics 2017;37:2113–31. 10.1148/rg.2017170077 [DOI] [PubMed] [Google Scholar]
- 18.Jeong Y, Kim JH, Chae H-D, et al. Deep learning-based decision support system for the diagnosis of neoplastic gallbladder polyps on ultrasonography: preliminary results. Sci Rep 2020;10:7700. 10.1038/s41598-020-64205-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sánchez Fernández I, Yang E, Calvachi P, et al. Deep learning in rare disease. detection of tubers in tuberous sclerosis complex. PLoS One 2020;15:e0232376. 10.1371/journal.pone.0232376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marro A, Bandukwala T, Mak W. Three-dimensional printing and medical imaging: a review of the methods and applications. Curr Probl Diagn Radiol 2016;45:2–9. 10.1067/j.cpradiol.2015.07.009 [DOI] [PubMed] [Google Scholar]
- 21.Atutornu J, Hayre CM. Personalised medicine and medical imaging: opportunities and challenges for contemporary health care. J Med Imaging Radiat Sci 2018;49:352–9. 10.1016/j.jmir.2018.07.002 [DOI] [PubMed] [Google Scholar]
- 22.Afshar P, Mohammadi A, Plataniotis KN, et al. From handcrafted to deep-learning-based cancer radiomics: challenges and opportunities. IEEE Signal Process Mag 2019;36:132–60. 10.1109/MSP.2019.2900993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antropova N, Huynh BQ, Giger ML. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med Phys 2017;44:5162–71. 10.1002/mp.12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giger ML. Machine learning in medical imaging. J Am Coll Radiol 2018;15:512–20. 10.1016/j.jacr.2017.12.028 [DOI] [PubMed] [Google Scholar]
- 25.Sahiner B, Pezeshk A, Hadjiiski LM, et al. Deep learning in medical imaging and radiation therapy. Med Phys 2019;46:e1–36. 10.1002/mp.13264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang S, Yang J, Fong S, et al. Artificial intelligence in cancer diagnosis and prognosis: opportunities and challenges. Cancer Lett 2020;471:61–71. 10.1016/j.canlet.2019.12.007 [DOI] [PubMed] [Google Scholar]
- 27.Litjens G, Sánchez CI, Timofeeva N, et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep 2016;6:26286. 10.1038/srep26286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moitra D, Mandal RK. Prediction of non-small cell lung cancer histology by a deep ensemble of convolutional and bidirectional recurrent neural network. J Digit Imaging 2020;33:895–902. 10.1007/s10278-020-00337-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahbod A, Schaefer G, Wang C, et al. Transfer learning using a multi-scale and multi-network ensemble for skin lesion classification. Comput Methods Programs Biomed 2020;193:105475. 10.1016/j.cmpb.2020.105475 [DOI] [PubMed] [Google Scholar]
- 30.Li Z, Togo R, Ogawa T, et al. Chronic gastritis classification using gastric X-ray images with a semi-supervised learning method based on tri-training. Med Biol Eng Comput 2020;58:1239–50. 10.1007/s11517-020-02159-z [DOI] [PubMed] [Google Scholar]
- 31.Liu S, Shah Z, Sav A, et al. Isocitrate dehydrogenase (IDH) status prediction in histopathology images of gliomas using deep learning. Sci Rep 2020;10:7733. 10.1038/s41598-020-64588-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim YD, Noh KJ, Byun SJ, et al. Effects of hypertension, diabetes, and smoking on age and sex prediction from retinal fundus images. Sci Rep 2020;10:4623. 10.1038/s41598-020-61519-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurovich Y, Hanani Y, Bar O, et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat Med 2019;25:60–4. 10.1038/s41591-018-0279-0 [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Kosinski M. Deep neural networks are more accurate than humans at detecting sexual orientation from facial images. J Pers Soc Psychol 2018;114:246–57. 10.1037/pspa0000098 [DOI] [PubMed] [Google Scholar]
- 35.Kruszka P, Addissie YA, McGinn DE, et al. 22Q11.2 deletion syndrome in diverse populations. Am J Med Genet A 2017;173:879–88. 10.1002/ajmg.a.38199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elmas M, Gogus B. Success of face analysis technology in rare genetic diseases diagnosed by whole-exome sequencing: a single-center experience. Mol Syndromol 2020;11:4–14. 10.1159/000505800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu C, Cheng L-L, Liu Y, et al. Protocol registration or development may benefit the design, conduct and reporting of dose-response meta-analysis: empirical evidence from a literature survey. BMC Med Res Methodol 2019;19:78. 10.1186/s12874-019-0715-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dos Santos MBF, Agostini BA, Bassani R, et al. Protocol registration improves reporting quality of systematic reviews in dentistry. BMC Med Res Methodol 2020;20:57. 10.1186/s12874-020-00939-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moher D, Shamseer L, Clarke M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Rev 2015;4:1. 10.1186/2046-4053-4-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higgins JP, et al. Cochrane handbook for systematic reviews of interventions. John Wiley & Sons, 2019. [Google Scholar]
- 41.Basel-Vanagaite L, Wolf L, Orin M, et al. Recognition of the cornelia de Lange syndrome phenotype with facial dysmorphology novel analysis. Clin Genet 2016;89:557–63. 10.1111/cge.12716 [DOI] [PubMed] [Google Scholar]
- 42.Cha S, Lim JE, Park AY, et al. Identification of five novel genetic loci related to facial morphology by genome-wide association studies. BMC Genomics 2018;19:481. 10.1186/s12864-018-4865-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richmond S, Howe LJ, Lewis S, et al. Facial genetics: a brief overview. Front Genet 2018;9:462. 10.3389/fgene.2018.00462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spencer CR, Irving RM. Causes and management of facial nerve palsy. Br J Hosp Med 2016;77:686–91. 10.12968/hmed.2016.77.12.686 [DOI] [PubMed] [Google Scholar]
- 45.Fasano A, Tinazzi M. Functional facial and tongue movement disorders. In: Handbook of clinical neurology. Elsevier, 2016: 353–65. [DOI] [PubMed] [Google Scholar]
- 46.Hagberg B. Clinical manifestations and stages of Rett syndrome. Ment Retard Dev Disabil Res Rev 2002;8:61–5. 10.1002/mrdd.10020 [DOI] [PubMed] [Google Scholar]
- 47.Tan N-C, Chan L-L, Tan E-K. Hemifacial spasm and involuntary facial movements. QJM 2002;95:493–500. 10.1093/qjmed/95.8.493 [DOI] [PubMed] [Google Scholar]
- 48.Liu T-L, Wang P-W, Yang Y-HC, et al. Association between facial emotion recognition and bullying involvement among adolescents with high-functioning autism spectrum disorder. Int J Environ Res Public Health 2019;16:5125. 10.3390/ijerph16245125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boutrus M, Gilani SZ, Alvares GA, et al. Increased facial asymmetry in autism spectrum conditions is associated with symptom presentation. Autism Res 2019;12:1774–83. 10.1002/aur.2161 [DOI] [PubMed] [Google Scholar]
- 50.Gill CE, Kompoliti K. Clinical features of tourette syndrome. J Child Neurol 2020;35:166–74. 10.1177/0883073819877335 [DOI] [PubMed] [Google Scholar]
- 51.Kim DD, Barr AM, Chung Y, et al. Antipsychotic-associated symptoms of tourette syndrome: a systematic review. CNS Drugs 2018;32:917–38. 10.1007/s40263-018-0559-8 [DOI] [PubMed] [Google Scholar]
- 52.Thomas CS. A study of facial dysmorphophobia. Psychiatr Bull 1995;19:736–9. 10.1192/pb.19.12.736 [DOI] [Google Scholar]
- 53.Phillips KA, Cash TF, Smolak L. Body image and body dysmorphic disorder eating disorders and obesity: a comprehensive Handbook. 2, 2002: 115. [Google Scholar]
- 54.Reese HE, McNally RJ, Wilhelm S. Facial asymmetry detection in patients with body dysmorphic disorder. Behav Res Ther 2010;48:936–40. 10.1016/j.brat.2010.05.021 [DOI] [PubMed] [Google Scholar]
- 55.White JJ, Mazzeu JF, Hoischen A, et al. DVL3 alleles resulting in a -1 frameshift of the last exon mediate autosomal-dominant robinow syndrome. Am J Hum Genet 2016;98:553–61. 10.1016/j.ajhg.2016.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Onder H, Albayrak L, Polat H. Frontal lobe ischemic stroke presenting with peripheral type facial palsy: a crucial diagnostic challenge in emergency practice. Turk J Emerg Med 2017;17:112–4. 10.1016/j.tjem.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han Y-Y, Qi D, Chen X-D, et al. Limb-shaking transient ischemic attack with facial muscles involuntary twitch successfully treated with internal carotid artery stenting. Brain Behav 2020;10:e01679. 10.1002/brb3.1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buolamwini J, Gebru T. Gender shades: Intersectional accuracy disparities in commercial gender classification. In: Proceedings of the 1st conference on Fairness, accountability and transparency, A.F. PMLR: Proceedings of Machine Learning Research, 2018: 77–91. [Google Scholar]
- 59.Drozdowski P, Rathgeb C, Dantcheva A, et al. Demographic bias in biometrics: a survey on an emerging challenge. IEEE Transactions on Technology and Society 2020;1:89–103. 10.1109/TTS.2020.2992344 [DOI] [Google Scholar]
- 60.Parikh RB, Teeple S, Navathe AS. Addressing bias in artificial intelligence in health care. JAMA 2019;322:2377–8. 10.1001/jama.2019.18058 [DOI] [PubMed] [Google Scholar]
- 61.Obermeyer Z, Powers B, Vogeli C, et al. Dissecting racial bias in an algorithm used to manage the health of populations. Science 2019;366:447–53. 10.1126/science.aax2342 [DOI] [PubMed] [Google Scholar]
- 62.Garcia RV. The harms of demographic bias in feep face recognition research. International Conference on Biometrics (ICB), 2019. [Google Scholar]
- 63.Kong X, Gong S, Su L, et al. Automatic detection of acromegaly from facial photographs using machine learning methods. EBioMedicine 2018;27:94–102. 10.1016/j.ebiom.2017.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liang B, Yang N, He G, et al. Identification of the facial features of patients with cancer: a deep learning-based pilot study. J Med Internet Res 2020;22:e17234. 10.2196/17234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin S, Li Z, Fu B, et al. Feasibility of using deep learning to detect coronary artery disease based on facial photo. Eur Heart J 2020;41:4400–11. 10.1093/eurheartj/ehaa640 [DOI] [PubMed] [Google Scholar]
- 66.Zhao Q, Okada K, Rosenbaum K, et al. Digital facial dysmorphology for genetic screening: hierarchical constrained local model using ICA. Med Image Anal 2014;18:699–710. 10.1016/j.media.2014.04.002 [DOI] [PubMed] [Google Scholar]
- 67.Goceri E. Deep learning based classification of facial dermatological disorders. Comput Biol Med 2021;128:104118. 10.1016/j.compbiomed.2020.104118 [DOI] [PubMed] [Google Scholar]
- 68.Han SS, Moon IJ, Lim W, et al. Keratinocytic skin cancer detection on the face using region-based convolutional neural network. JAMA Dermatol 2020;156:29–37. 10.1001/jamadermatol.2019.3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Camino A, Wang Z, Wang J, et al. Deep learning for the segmentation of preserved photoreceptors on en face optical coherence tomography in two inherited retinal diseases. Biomed Opt Express 2018;9:3092–105. 10.1364/BOE.9.003092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bargshady G, Zhou X, Deo RC, et al. Enhanced deep learning algorithm development to detect pain intensity from facial expression images. Expert Syst Appl 2020;149:113305. 10.1016/j.eswa.2020.113305 [DOI] [Google Scholar]
- 71.Yolcu G. Deep learning-based facial expression recognition for monitoring neurological disorders. IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.