Abstract
As organelles of the innate immune system, Inflammasomes activate caspase-1 and other inflammatory caspases that cleave gasdermin D (GSDMD). Caspase-1 also cleaves inactive precursors of the interleukin (IL)-1 family to generate mature cytokines such as IL-1β and IL-18. Cleaved GSDMD forms transmembrane pores to enable the release of IL-1 and to drive lytic cell death through pyroptosis1–9. Here we report cryo-electron microscopy structures of the pore and the prepore of GSDMD. These structures reveal the different conformations of the two states, as well as extensive membrane-binding elements including a hydrophobic anchor and three positively charged patches. The GSDMD pore conduit is predominantly negatively charged. By contrast, IL-1 precursors possess an acidic domain that is proteolytically removed by caspase-110. When permeabilized by GSDMD pores, unlysed liposomes release positively charged and neutral cargoes faster than negatively charged ones of similar sizes, and the pores favour the passage of IL-1β and IL-18 over that of their precursors. Consistent with these findings, living - but not pyroptotic - macrophages preferentially release mature IL-1β upon perforation by GSDMD. Mutation of the acidic residues of GSDMD compromises this preference, hindering intracellular retention of the precursor and secretion of the mature cytokine. Therefore, the GSDMD pore mediates IL-1 release by electrostatic filtering, which suggests the importance of charge in addition to size in the transport of cargoes across this large channel.
Proteins of the gasdermin (GSDM) family are implicated in many inflammatory diseases and cancer, and represent promising therapeutic targets11. GSDMs have a functional N-terminal fragment (NT) and an auto-inhibitory C-terminal fragment (CT). Processing at the NT-CT linker by inflammatory caspases and other enzymes12–17 liberates GSDM-NT for pore formation3–6. GSDMD pores mediate the release of interleukin (IL)-1 family cytokines7,8 and programmed cell death including pyroptosis3–6,17–19. However, despite the functional importance of GSDMD, the mechanisms of GSDMD pore formation and the role of the pore as an IL-1 secretion conduit are yet to be elucidated.
Structures of the GSDMD pore and prepore
We optimized human GSDMD samples by 1) replacing a 17-residue segment between GSDMD-NT and -CT with the 3C protease site, 2) using phosphatidic acid (PA)-containing liposomes, 3) solubilizing the pores in C12E8 detergent, and 4) further purifying by size exclusion chromatography (Extended Data Fig. 1a–h). As the 3C site does not affect pore formation3, we denoted our construct as wildtype (WT) for simplicity. Further screening of mutants3,20 identified GSDMD L192E as less aggregated and having less orientation preference on cryo-electron microscopy (cryo-EM) grids (Extended Data Fig. 1c–e, 2a, b, 3a, b). Both WT and L192E datasets yielded similar densities representative of β-barrel pores and rings without the β-barrel or prepores (Extended Data Fig. 3c). Models were built into the L192E densities of the pore and the prepore, which had 3.9 Å and 6.9 Å resolutions, respectively, measured by Fourier shell correlation (FSC) at 0.143 (Extended Data Fig. 4, Extended Data Table 1).
GSDMD assemblies are 31- to 34-fold symmetric (Extended Data Fig. 2b, 3b), in comparison with the 26- to 28-fold symmetry of GSDMA320, suggesting oligomerization variability between GSDMs and modest size plasticity of a given GSDM. The 33-subunit GSDMD pore has inner and outer diameters of around 215 Å and 310 Å, respectively, 10–20% larger than the 180 Å and 280 Å diameters of the 27-subunit GSDMA3 pore (Fig. 1a). This difference is independent of the acidic lipid in the liposomes and the detergent for pore extraction (Extended Data Fig. 1i). Like GSDMA3, each pore-form GSDMD subunit comprises two inserted β-hairpins (HPs, “fingers”) and a globular domain (“palm”) (Fig. 1b). There is a 16° angle difference between the globular domains of pore-form GSDMD and GSDMA3, suggesting a flexible junction between the β-barrel and the globular domain (Fig. 1c).
Fig. 1 |. GSDMD architecture and conformational changes.

a, Ribbon diagram and dimensions of the 33-subunit human GSDMD pore structure fitted into its cryo-EM density. The pore features a large transmembrane β-barrel and a globular domain on the cytosolic side. b, Structure of the pore-form GSDMD subunit fitted into the cryo-EM density. The structure resembles a human left hand, with the globular domain as the “palm”, the α1 helix as the “thumb”, the membrane-inserted β-hairpins as the “fingers”, and the β1-β2 loop as the “wrist”. c, Flexible junction between the globular domain and the β-barrel, revealed by alignment of GSDMD and GSDMA3 pores at the β-barrel. The central axes are misaligned due to different pore sizes. The globular domain of the GSDMD pore is more membrane-distal than that of GSDMA3 by an approximate angle of 16°. d, Ribbon diagram and dimensions of the 33-subunit GSDMD prepore structure superimposed with its cryo-EM density. e, Prepore-to-pore transition of a GSDMD subunit. The two structures are aligned by their central axes and overlaid at the α1 helices. As the β-strands insert into the membrane, the globular domain rotates away from the membrane by approximately 38°.
The GSDMD prepore is 40 Å shorter in height than the pore (Fig. 1a, d). Prepore subunits resemble auto-inhibited GSDMD-NT (globular), and the globular domain is preserved during pore formation (Extended Data Fig. 5a). However, the globular domain exhibits a 38° rigid-body movement away from the membrane during pore formation, as visualized by aligning the pore and prepore by their central axes and the α1 helix or “thumb” (Fig. 1e, Supplementary Video 1, 2). The inward rotation makes the pore slightly smaller in diameter than the prepore (Fig. 1a, d). Meanwhile, residues that form the β-sheet (Extension domains 1 and 2 – ED1 and ED2) form HPs through refolding (Extended Data Fig. 5b, c). GSDMA3 undergoes a similar rigid-body rotation (Extended Data Fig. 5d), suggesting structural conservation of GSDM prepore-to-pore transition. Besides the prepore, other structural intermediates are possible such as arc- and slit-shaped oligomers observed under atomic force microscopy (AFM)6,21. The exact mechanism of GSDMD assembly in cells remains elusive.
Membrane contact sites
The GSDMA3 study identified the positively charged α1 helix (basic patch 1 – BP1) as critical for interaction with acidic lipids20. However, the GSDMD prepore structure suggested a more membrane-proximal motif, the β1-β2 loop, which features a hydrophobic tip flanked by basic residues (“wrist”, basic patch 2 – BP2) (Fig. 1b, 2a). The loop remains ordered in the pore (Fig. 2b). We posit that the hydrophobic tip partially inserts into the lipid bilayer as an anchor, while the surrounding basic residues interact with acidic lipids. Liposome-based terbium (Tb3+) leakage assays showed compromised membrane permeabilization ability of the W48E and W50E human GSDMD (Extended Data Fig. 6a), and double mutant on equivalent residues (F49G/W50G) in mouse GSDMD was defective in pore formation22, confirming the importance of the anchor. The hydrophobic tip is conserved both in sequence and structurally among GSDMs (Extended Data Fig. 6b–d). Consistently, L47E and W49E mutations in GSDMA3 reduced, and the triple mutation L47E/F48E/W49E abolished, pore formation (Extended Data Fig. 6e).
Fig. 2 |. Membrane interaction by multiple contact sites.
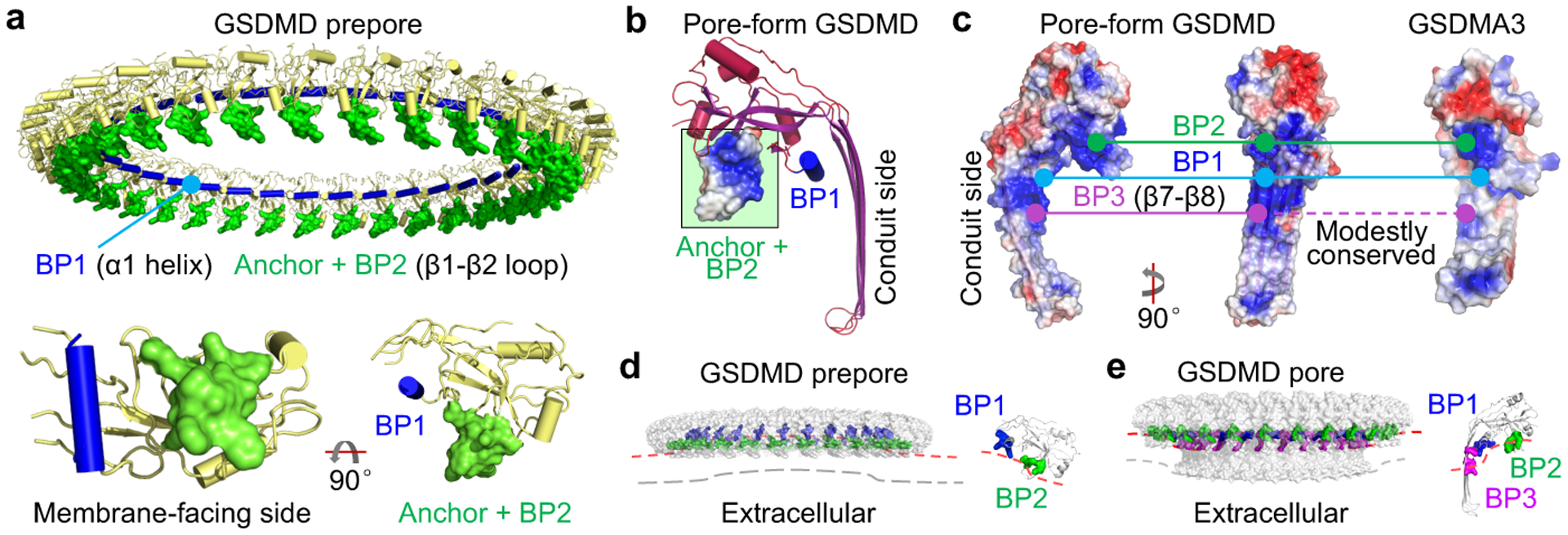
a, Membrane docking by the β1-β2 loop, the most membrane-proximal feature of the prepore. The loop contains a hydrophobic anchor flanked by basic residues of BP2. The relative positions of BP1 and the anchor-BP2 region are displayed using an individual prepore subunit in two orientations. b, Pore-form GSDMD with the hydrophobic anchor and BP2 boxed in green and electrostatic surface shown locally. c, Locations and conservation of the three BPs in GSDMD, at α1, β1-β2 (excluding the hydrophobic tip), and β7-β8 regions, respectively. Continuous line: Conserved. Dashed line: Modestly conserved. d, e, Potential membrane distortion around GSDMD prepore (d) and pore (e). A subunit of each is enlarged to show the inferred local curvature. The contrasting curvatures indicate convexity-to-concavity membrane remodelling.
While BP1 and BP2 are highly conserved (Extended Data Fig. 6b) and both located in the globular domain, there is a less conserved basic patch (BP3) on the β7-β8 hairpin of the GSDMD pore subunit (Fig. 2c). Potential bound lipid density adjacent to BP3 supports its involvement in membrane interaction (Extended Data Fig. 6f). To validate BP2 and BP3 in GSDMD, we generated the quadruple mutant R42E/K43E/K51E/R53E in BP2, and single mutants R174E and K204E in BP3. All mutations compromised and the BP2 mutation eliminated pore formation (Extended Data Fig. 6g). Likewise, BP2 quadruple mutation in GSDMA3 (R41E/K42E/R43E/K44E) resulted in loss of function (Extended Data Fig. 6h).
Full-length GSDMs do not bind lipids3,22, and therefore the membrane contact sites should be protected in auto-inhibited GSDMs. After cleavage of the inter-domain linker, GSDM-NT and -CT stay associated as a non-covalent complex in the absence of lipids3. Based on the auto-inhibited GSDMD structure, residues 242–283 of the inter-domain linker are disordered22. Because residues Q241 and T284 are located near the β1-β2 loop (anchor and BP2), the linker possibly hovers over and masks the loop. Cleavage of the linker may therefore unmask β1-β2 for lipid interaction (Extended Data Fig. 6i). Thus, the β1-β2 loop is likely the initial membrane engagement site. Furthermore, if the BPs co-interact with acidic lipids, the membrane would be convex next to the GSDMD prepore (curved towards the cytosol) and concave (curved towards extracellular) next to the pore (Fig. 2d, e). Interestingly, GSDMD-perforated membranes can be repaired by the ESCRT-III machinery23, which assembles favourably on concave membranes24. It is therefore likely that by pore formation GSDMD primes its removal from the membrane through negative feedback.
Predominantly acidic conduit
Electrostatics calculations revealed negative potentials originating from the globular domain and β-barrel and decaying towards the GSDMD pore centre (Fig. 3a, b). There are four solvent-exposed acidic patches (APs) near the conduit (Fig. 3a, Extended Data Fig. 7a), with AP1 (β2-β3, 5 D/E) and AP4 (β11, 4 D/E) in the globular domain and AP2 (β3, 2 D/E) and AP3 (β7, 3 D/E) in the β-barrel (Fig. 3a). Like GSDMD, the GSDMA3 pore conduit is also predominantly acidic (Fig. 3a). Alanine mutants AP1 and AP2 were equally functional and formed same sized pores compared to WT GSDMD or GSDMA3 (Extended Data Fig. 7b–e). Electrostatically, AP1 and AP2 mutations locally neutralize the cargo release path (Fig. 3a, b).
Fig. 3 |. Pore conduit and cargo transport.
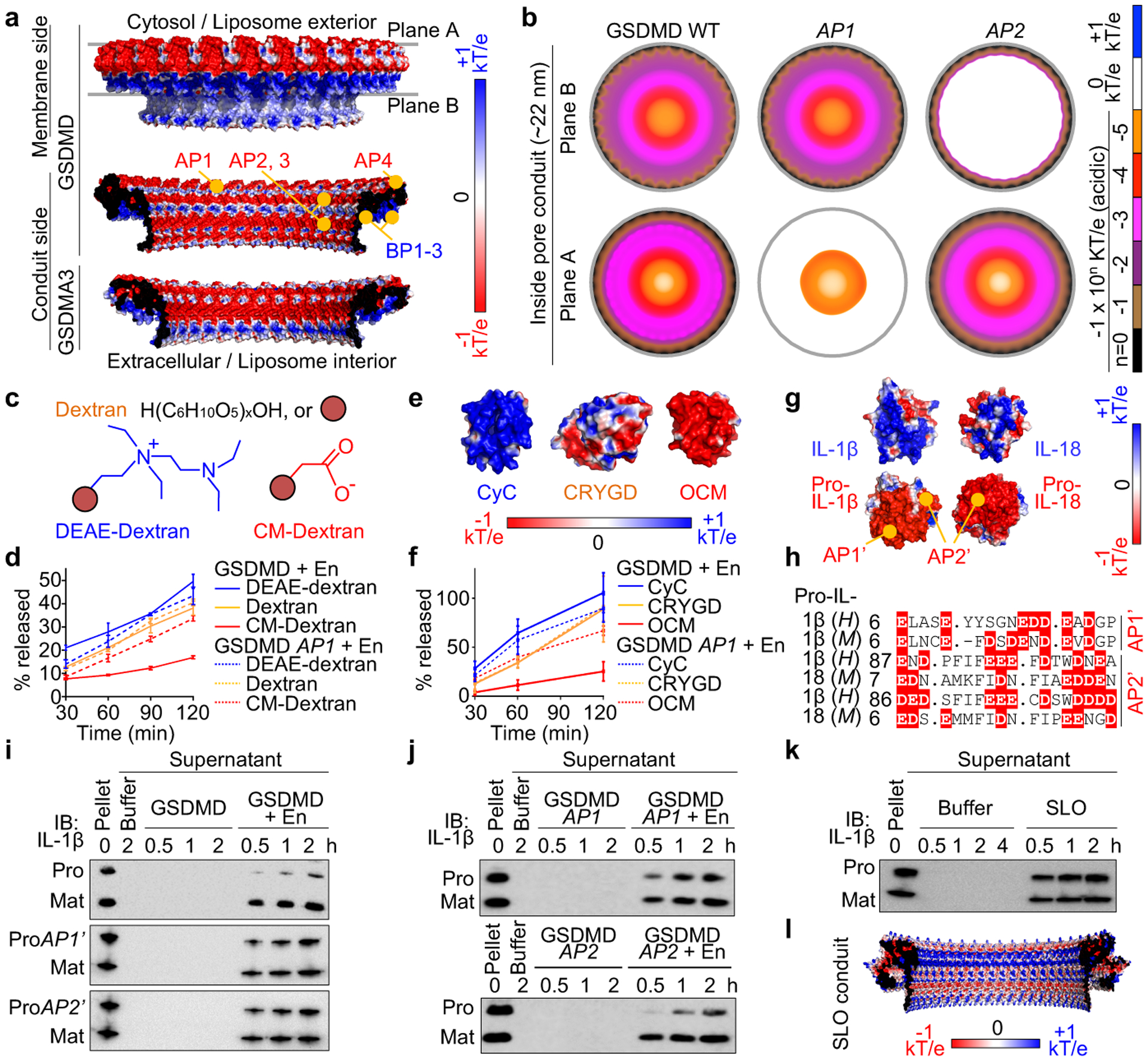
a, Electrostatics surface (−1 to +1 kT/e) of the GSDMD pore, with the membrane-bound side formed by BPs and the solvent-exposed side formed by APs. The GSDMA3 pore conduit is similarly acidic. b, Negative electrostatic coverage of the GSDMD pore conduit. Modelled AP1 (5 D/E to A) and AP2 (2 D/E to A) pores have locally neutral conduits. c, d, Cartoons (c) and release of dextran cargoes from liposomes perforated by GSDMD, quantified by FITC fluorescence (n = 3 biological replicates) (d). e, f, Electrostatics surfaces of three protein cargoes (PDB: 2B4Z, 1H4A, 1TTX) (e) and their release from liposomes permeabilized by GSDMD, evaluated by immunoblot quantification (n = 3 biological replicates) (f). g, h, Basification of IL-1β and IL-18 through caspase-1-induced maturation (g) and AP’s of the precursors shown by aligned sequences (h). H: Human. M: Mouse. Dashes: Gaps. Dots: Strings of omitted residues. i, j, Release of pro- (WT and AP’-mutant) and mature and IL-1β from liposomes permeabilized by WT GSDMD (i) and the reciprocal experiments (j). ProAP1’: 8 D/E to K. ProAP2’: 11 D/E to K. k, Release of pro- and mature IL-1β from liposomes perforated by SLO. l, Electrostatics surface of the modelled SLO pore conduit. Data shown in d and f are mean ± s.d.. Data shown in i-k are representative of two independent experiments.
Previously, a size exclusion mechanism demonstrated that charge-neutral small dextrans but not large ones can exit from GSDMD-permeabilized liposomes7. As soluble cargoes may possess a charged surface, we tested whether the acidic conduit of the GSDMD pore influences their transport. Of note, although pores formed by GSDMD subunits outside liposomes have a reverse geometry compared to those formed inside cells, cargo release in either case would experience equivalent total electrostatic interaction along the trajectory (Fig. 3a, b). Using a sub-lytic concentration of GSDMD that caused liposome permeabilization but not lysis (minimal LDH release) (Extended Data Fig. 8a), we found that small dextrans (40 kDa) carrying different charges (neutral unmodified; basic diethylaminoethyl (DEAE)-modified; and acidic carboxymethyl (CM)-modified) showed various release rates, with CM-dextran least secreted (Fig. 3c, d). The GSDMD AP1 mutant markedly enhanced CM-dextran release and minimally affected the neutral and basic counterparts (Fig. 3d). We then examined three small proteins – cytochrome C (CyC, basic), γ-crystallin D (CRYGD, neutral), and oncomodulin (OCM, acidic) – of similar hydrated diameters but contrasting surface electrostatics (Fig. 3e). Consistently, OCM was most slowly released from unlysed liposomes with GSDMD pores, and the AP1 mutant abolished this retardation effect (Fig. 3f, Extended Data Fig. 8b). Release rates of the neutral and basic dextrans, and of CyC and CRYGD were not obviously different. Therefore, the GSDMD pore repels acidic cargoes but does not appear to robustly favour basic over neutral ones. These observations may explain GSDMD-dependent rapid release of Rac-1, HMGB1, and CyC, all of which are non-acidic8,25,26.
A prominent biological role of the GSDMD pore is to mediate unconventional protein secretion of IL-1β and IL-187,8, IL-1 family cytokines that lack the signal sequence required for ER/Golgi-dependent secretion. Precursor (pro) and mature IL-1β and IL-18 are similar in size and both much smaller than the GSDMD pore; interestingly, these cytokines basify during proteolytic maturation (Fig. 3g, h). Basic, mature IL-1β was reported to accumulate at cell membrane PIP2 ruffles for secretion10. To eliminate the potential effect of PIP2 binding, we encapsulated pro- and mature IL-1β and IL-18 into PIP2-free liposomes to study their GSDMD-dependent release (Extended Data Fig. 8c). Mature IL-1β and IL-18 were secreted substantially faster than their precursors (Fig. 3i, Extended Data Fig. 8d, e), as if the pore acts as a “filter” to repel the precursors. Indeed, GSDMD and GSDMA3 AP1 and AP2 mutants enhanced pro-IL-1β release (Fig. 3i, j, Extended Data Fig. 8f, g), an effect that may be rationalized as a result of decreased repulsion along part of the cargo trajectory (Fig. 3b). Reciprocally, the AP1’ (8 D/E to K) and AP2’ (11 D/E to K) mutants of pro-IL-1β were released at higher rates than WT pro-IL-1β (Fig. 3i). Using a simplified mathematical relationship, we estimated that the electrostatic potential of the GSDMD pore conduit needs to be around −0.15 kT/e on average to account for the observed charge effect (Fig. 3k, Extended Data Fig. 8h–k), a value within the range deduced structurally (Fig. 3a, b). To further confirm that the rate differences resulted from GSDM pores, we tested β-barrel-forming toxins streptolysin O (SLO) and perfringolysin O (PFO). Under a sub-lytic condition (lack of bulky 2 MDa dextran release from liposomes), SLO and PFO did not induce preferential release of mature IL-1β (Fig. 3k, Extended Data Fig. 8l, m). Consistently, SLO and PFO pore conduits are neither predominantly acidic nor basic according to homology models based on the structure of pneumolysin27 (Fig. 3l, Extended Data Fig. 8n).
Electrostatically filtered IL-1 release
GSDMD-dependent IL-1 release occurs independently of pyroptosis, as treatment with glycine to inhibit cell membrane rupture allowed the release of IL-1β, but not LDH (a hallmark of pyroptosis)7,8. In light of minimal pro-IL-1β release in the liposome experiments, we wondered whether this phenomenon is observable cellularly. Indeed, while mature IL-1β was readily secreted from immortalized mouse bone-marrow derived macrophages (iBMDMs) with (20 μM nigericin-induced for 30 min) or without (glycine-protected) pyroptosis, pro-IL-1β was largely retained in living but not pyroptotic cells (Fig. 4a, b). To further assess whether the GSDMD pore directly influences IL-1β release, we expressed comparable levels of WT, AP1-mutant, or AP2-mutant GSDMD in GSDMD-knockout (KO) iBMDMs, and WT, AP1’-mutant, or AP2’-mutant pro-IL-1β in IL-1β-KO iBMDMs (Extended Data Fig. 9a). All cells except the KO and empty vector (EV) controls were prone to nigericin-induced pyroptosis and protected by glycine (Extended Data Fig. 9b). In comparison with WT iBMDMs, cells expressing AP1- or AP2-mutant GSDMD, or AP1’- or AP2’-mutant pro-IL-1β showed enhanced release of pro-IL-1β under glycine protection (Fig. 4c).
Fig. 4 |. Preferential IL-1β release from macrophages.
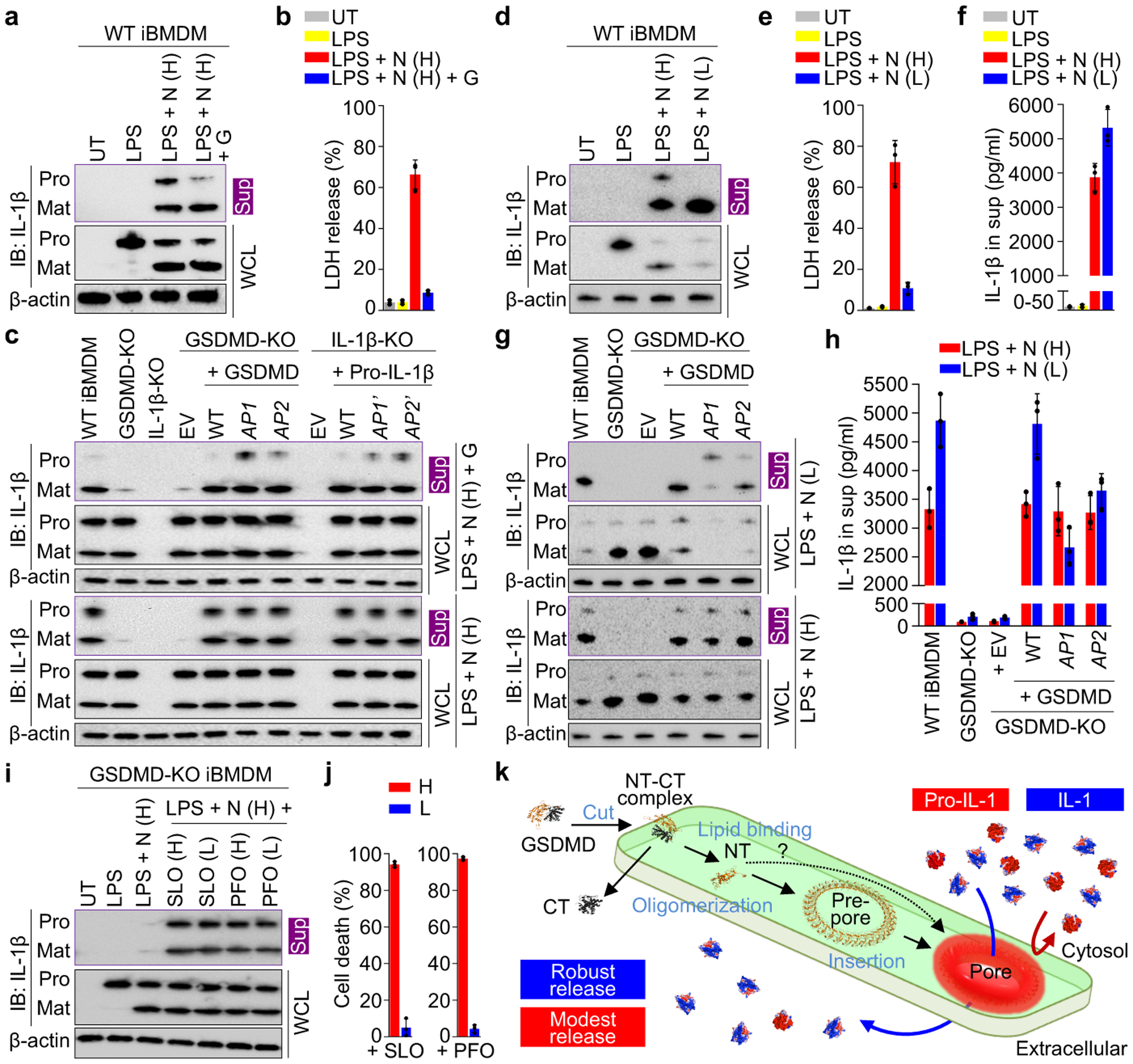
a, b, Preferential release of mature IL-1β from glycine-protected living iBMDMs permeabilized by GSDMD, shown by immunoblotting (a) and LDH release (n = 3 biological replicates) (b). UT: Untreated. N: Nigericin. G: Glycine. Uppercase H: High dose at 20 μM. Sup: Supernatant. WCL: Whole cell lysate. Pro: Precursor. Mat: Mature. c, Comparison of IL-1β release across GSDMD-KO iBMDMs expressing with WT or AP-mutant GSDMD, and across pro-IL-1β-KO iBMDMs expressing WT or AP’-mutant pro-IL-1β. EV: Empty vector, a mock transduction control. d-f, Release of mature IL-1β from living iBMDMs without glycine protection, characterized by immunoblotting (d), LDH release (n = 3 biological replicates) (e), and ELISA (n = 3 biological replicates) (f). Uppercase L: Low dose at 0.5 μM. g, h, Comparison of IL-1β release across GSDMD-KO iBMDMs expressing WT or AP-mutant GSDMD without glycine protection, evaluated by immunoblotting (g) and ELISA (n = 3 biological replicates) (h). i, j, Comparable leakage of pro-IL-1β and mature IL-1β from GSDMD-KO iBMDMs perforated by SLO and PFO (i) at cytotoxic and non-toxic concentrations shown by ATP-based cell death (n = 3 biological replicates) (j). Uppercase H: High dose at 625 nM. Uppercase L: Low dose at 0.16 nM. k, Schematic diagram for GSDMD pore formation and IL-1 release. The question mark indicates other possible assembly mechanisms. Data shown in b, e, f, h, and j are mean ± s.d.. Data shown in a and d are representative of three, and data shown in c, g, and i two, independent experiments.
Given that pro-IL-1β is inactive and unable to bind to the IL-1 receptor, we postulated that the GSDMD pore retains pro-IL-1β for processing to achieve sustained secretion of mature IL-1β. To test this hypothesis, we weakly stimulated the iBMDMs with low-dose nigericin (0.5 μM) to allow sustained IL-1β secretion with little cell death (12 h), in the absence of the osmoprotectant glycine (Fig. 4d, e). The amount of secreted mature IL-1β was higher with low-dose nigericin treatment compared to high-dose, likely due to minimal pyroptosis and continuous IL-1β release (Fig. 4f). Under the weaker stimulation, cells expressing GSDMD AP1 and AP2 mutants not only released pro-IL-1β like pyroptotic cells, but also secreted markedly less mature IL-1β than WT cells (Fig. 4g, h, Extended Data Fig. 9c). Therefore, by electrostatically impeding pro-IL-1β release, the GSDMD pore plays an important role in sustained IL-1β secretion from living cells. Furthermore, we expressed in GSDMD-KO iBMDMs a caspase-1-cleavable GSDMA3 chimera (A3chim) and its equivalent AP mutants (Extended Data Fig. 9d, e) and found that GSDMA3 also exercises this charge-based mechanism (Extended Data Fig. 9f–o). In contrast, SLO and PFO induced similar secretion of pro-IL-1β and IL-1β from GSDMD-KO iBMDMs with or without cell death (Fig. 4i, j), like in the liposome experiments (Fig. 3k, Extended Data Fig. 8l, m).
In summary, the structures here provide mechanistic insights into GSDMD pore formation and IL-1 release. It remains debated whether IL-1α utilizes GSDMD as a major secretion pathway. As both pro- and mature IL-1α are acidic, the pore may deter, if not abolish, their passage. Supporting this prediction, previous studies demonstrated a distinct secretory route of IL-1α28,29 and lack of its secretion under glycine protection30. Additionally, it is likely that charge endows GSDM pores with differential electrostatics to fine-tune the exit of damage-associated molecular patterns (DAMPs). Lastly, electrostatics modulates cargo transport but may not provide absolute gating given the large sizes of GSDM pores. Other mechanisms that contribute to DAMP release from GSDM-perforated living cells await discovery.
Methods
Generation of full-length GSDMs.
The coding sequence of full-length human GSDMD was cloned into the pDB.His.MBP vector after the N-terminal His6-MBP tag and a linker cleavable by tobacco etch virus protease (TEV). Residues 259–275 between GSDMD-NT (residues 1–241) and -CT (residues 284–484) were then replaced by the human rhinovirus 3C protease (3C) site (LEVLFQ/GP). As this construct does not contain mutation in GSDMD-NT, we call it “WT” GSDMD to be concise, and all mutants were generated on this background. All mutations in this study were introduced using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) or Gibson Assembly Master Mix (New England BioLabs), and all plasmids were verified by sequencing. Transformed E. coli BL21 (DE3) cells were grown in LB medium supplemented with 50 μg/mL kanamycin at 37 °C, induced at OD600 0.6 by 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG), and incubated for 18 h at 18 °C before harvesting. Cells were pelleted by centrifugation at 3,500 rpm for 30 min and resuspended in Buffer A (40 mM HEPES at pH 7.0, 150 mM NaCl) supplemented with 5 mM imidazole for lysis by sonication. His6-MBP-tagged GSDMD was enriched on Ni-NTA beads (Qiagen) and eluted by Buffer A supplemented with 500 mM imidazole. For GSDMD pore and prepore reconstitution, the His6-MBP tag was cleaved off by His6-TEV at 4 °C overnight. His6-MBP and His6-TEV were removed using a Ni-NTA column and the flow-through containing GSDMD was further purified using a Superdex 200 (10/300) size exclusion column (GE Healthcare Life Sciences) equilibrated with Buffer A. Engineered mouse GSDMA3 containing the 3C site, GSDMA3 chimeras, and their mutants were cloned and purified following a previous protocol20. Suggestions on the design of caspase-1-cleavable GSDMA3 were kindly provided by Dr. Feng Shao. Three GSDMA3 chimeras were designed as follow: 1) A3chim: GSDMA3-NT (residues 1–240), followed by the linker of pro-IL-1β (residues 101–122), followed by GSDMA3-CT (residues 263–464); 2) A3chim2: GSDMA3-NT, followed by the linker of GSDMD (residues 266–287), followed by GSDMA3-CT; 3) A3chim3: GSDMA3-NT, followed by the linker of GSDMD, followed by GSDMD-CT (residues 288–487). The AP mutations were then introduced to A3chim. A3chim3 was poorly expressed and not further evaluated. All purified full-length GSDMs were concentrated to approximately 10 mg/ml before use.
Reconstitution and purification of GSDMD and GSDMA3 assemblies.
1,2-dioleoyl-sn-glycero-3-phosphate (PA, in the case of GSDMD) or 1’,3’-bis[1,2-dioleoyl-sn-glycerol-3-phospho]-glycerol (CL, in the case of GSDMA3) was mixed with 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC) (Avanti Polar Lipids) at a molar ratio of 1:4. Besides PA and CL, other acidic lipids such as 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (PS) (Avanti Polar Lipids) were tested in combination with PC at various ratios including 1:4 but these recipes yielded suboptimal samples. The solvent chloroform was evaporated under nitrogen gas and resuspended in Buffer A to approximately 5 mM lipid concentration by vigorous vortexing. The liposomes formed were extruded through a 100 nm filter (Whatman Nuclepore) for 30 passes to generate unilamellar vesicles. Purified GSDMD was added at a final concentration of around 50 μM, followed by addition of ample 3C to cleave GSDMD for pore formation. The reaction proceeded on ice for 3 h before the liposomes loaded with GSDMD-NT pores were pelleted at 200,000 g for 1 h in a cold ultracentrifuge. The supernatant containing mostly full-length GSDMD, GSDMD-CT, and 3C was discarded, and the pellet was solubilized by Buffer A supplemented with 1% C12E8 (Anatrace) to extract GSDMD assemblies. To remove aggregates, solubilized GSDMD samples were subject to fractionation using a Superose 6 (10/300) size exclusion column (GE Healthcare Life Sciences) equilibrated with Buffer A supplemented with 0.2% C12E8. GSDMA3 assemblies were reconstituted and purified following established procedures20.
IL-1 purification.
All IL-1-related liposomal experiments were conducted using pure, monomeric, tag-free, freshly purified precursor or mature IL-1β and IL-18. To obtain the IL-1 cargoes, coding sequences for full-length murine pro-IL-1β and pro-IL-18, WT or charge-mutant, were inserted into a pET28a vector immediately following the N-terminal His6-SUMO tag. Sequences coding the pro-domains of pro-IL-1β and pro-IL-18 (residues 1–117 and 1–35, respectively) were then truncated out to generate vectors expressing His6-SUMO-tagged mature IL-1β and IL-18. Plasmids were transformed into E. coli BL21 (DE3) RipL cells for expression in LB medium supplemented with 50 μg/mL kanamycin. Protein expression was induced with 0.2 mM IPTG at OD600 0.8. The culture was then incubated at 18 °C for 12 h under vigorous shaking. Pelleted cells were resuspended in Buffer A for sonication, and lysates were incubated with Ni-NTA beads to enrich His6-SUMO-tagged proteins. The His6-SUMO tag was cleaved off on column using His6-tagged ULP1 protease overnight at 4 °C. Flowthrough containing tag-free IL-1s was further purified by gel filtration using the Superdex 75 (10/300) column (GE Healthcare Life Sciences) equilibrated with Buffer A. Only fractions under the monomeric chromatograph peaks were collected and used directly without further concentration.
SLO and PFO purification.
Sequences encoding SLO (residues 78–571) and PFO (residues 30–500) were cloned into a pET-28 vector to append an N-terminal His6-tag. The plasmids were transformed into E. coli BL21 (DE3). Expression was induced by 1 mM IPTG at 37 °C at OD600 1.0. The bacteria were incubated at 37 °C for 4 h under vigorous shaking before they were harvested, resuspended in Buffer A, and lysed by sonication. Proteins eluted from Ni-NTA beads by Buffer A supplemented with 500 mM imidazole were further purified using a Superdex 200 (10/300) column (GE Healthcare Life Sciences) equilibrated with Buffer A supplemented with 5 mM dithiothreitol (DTT). Fractions containing pure SLO or PFO were pooled and concentrated to approximately 10 mg/ml before use.
Caspase-1 purification.
Human caspase-1 p20 (residues 120–297) and p10 (residues 317–404) subunits were individually cloned into the pET-21a vector and expressed in E. coli BL21 (DE3). Expression was induced with 1 mM IPTG for 4 h at 37 °C. Bacterial pellets were washed twice with buffer containing 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 0.1% Triton-X-100 and 1 M guanidinium hydrochloride (GdnCl), and insoluble contents were pelleted by centrifugation at 17,000 g for 30 min. Inclusion bodies were collected and washed twice with buffer containing 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, and 1 M GdnCl, and then solubilized in buffer containing 6 M GdnCl, 25 mM Tris-HCl (pH 7.5), 5 mM EDTA, and 100 mM tris(2-carboxyethyl)phosphine (TCEP). Approximately equal molar amounts of each subunit were mixed to a total volume of 6 mL and diluted into 250 mL of buffer containing 100 mM HEPES (pH 8.0), 100 mM NaCl, 100 mM sodium malonate, 20 % sucrose, 0.5 M NDSB-201, and 10 mM TCEP. The refolded proteins were concentrated and dialyzed overnight over buffer containing 30 mM sodium acetate (pH 5.9), 5 mM TCEP, 5 % (v/v) glycerol, and then purified by HiTrap SP cation exchange chromatography (GE Healthcare Life Sciences) with buffer containing 30 mM sodium acetate (pH 5.9), 1 M NaCl, 5 mM TCEP and 10% (v/v) glycerol. To test cleavage, approximately equimolar amounts of purified caspase-1 and full-length GSDMs were mixed and incubated on ice for 4 h before SDS-PAGE analysis.
EM imaging and data processing.
GSDM samples were checked for morphology and homogeneity using negative-staining EM. 5-μl samples were applied to glow-discharged formvar-coated copper grids (Electron Microscopy Sciences), washed with 30 μL Buffer A, stained with 1% uranyl formate for 30 s, and blotted dry. Imaging was performed on a Tecnai G2 Spirit BioTWIN electron microscope (FEI) equipped with a 2k CCD camera (Advanced Microscopy Techniques) at the Electron Microscopy Facility at Harvard Medical School. For cryo-EM grid preparation, GSDMD samples (3 μL, 1–3 mg/mL protein concentration) were applied to plasma glow-discharged 300-mesh gold lacey carbon grids coated with ultrathin carbon film (Ted Pella), using a Vitrobot (FEI) set at blotting force 2, blotting time 7 s, 100% humidity, and 4 °C. Blotted grids were immediately plunged into liquid ethane and transferred to liquid nitrogen for storage. Two cryo-EM datasets were collected using Titan Krios electron microscopes (FEI) equipped with K3 cameras (Gatan) and the SerialEM software, one dataset on WT and the other on L192E GSDMD samples (Extended Data Table 1). Both datasets were processed in RELION 3.031. Raw movies were corrected for beam-induced motion using MotionCor232, and per-micrograph defocus was determined using CTFFIND433. Motion- and CTF-corrected micrographs were examined manually to discard images of unsatisfactory quality. The GSDMD pores and prepores stacked double-ring particles, likely an artefact of detergent solubilization, and therefore all 3D reconstructions were conducted on density-subtracted single rings. Afterwards, CTF refinement was performed to determine per-particle defocus level. Local resolution distribution was determined using ResMap34.
Model building and structure analysis.
The WT and the L192E GSDMD datasets yielded similar cryo-EM densities for both the pore and the prepore. The L192E densities at 3.9 Å (the 33-fold symmetric pore) and 6.9 Å (the 33-fold symmetric prepore) resolutions by gold-standard FSC (0.143) were used for model building. In building the pore model, the structure of GSDMD-NT in the auto-inhibited state (PDB: 6N9O) was docked into the globular domain part of the pore map in UCSF Chimera35. Then, extensive manual remodelling of the β-barrel domain was performed in Coot36. The model was refined against the map in real space using PHENIX37. For the prepore model, 33 copies of the auto-inhibited GSDMD-NT structure were fitted into the prepore map as rigid bodies. No manual remodelling or refinement was performed due to limited prepore map resolution. Structural models of pro-IL-1β and pro-IL-18 were generated based on crystal structures of IL-1β (PDB: 9ILB) and IL-18 (PDB: 3WO2), using the I-TASSER server38. Structural models of GSDMD AP1- and AP2-mutant pores were generated using in silico mutagenesis PyMOL Molecular Graphics System. Structural models of SLO and PFO pores were generated by the SWISS-MODEL server based on homology to pneumolysin (PDB: 5LY6). Values of electrostatic potentials at a physiological salt concentration of 150 mM and an aqueous dielectric constant of 78 were obtained using the Adaptive Poisson-Boltzmann Solver (APBS)39 plugin in PyMOL. All structural representations were generated using UCSF Chimera and PyMOL.
Terbium release assay.
Terbium (Tb3+) liposome leakage assays were performed following an established protocol40. Briefly, CL, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (PE), and PC (Avanti Polar Lipids) were mixed at a mass ratio of 5:8:4. The solvent chloroform was evaporated under nitrogen gas and the lipids were resuspended in Buffer B (20 mM HEPES at pH 7.0, 150 mM NaCl, 50 mM sodium citrate, and 15 mM TbCl3). Tb3+-loaded liposomes were then extruded through a 100-nm filter (Whatman Nuclepore). To remove unencapsulated Tb3+ and exchange exterior buffer, the extruded liposomes were subject to gel filtration using a Superose 6 (10/300) column equilibrated with Buffer C (20 mM HEPES at pH 7.0, 150 mM NaCl, 50 μM DPA). Liposomes were incubated with GSDMs with or without activating enzyme (En) treatment. GSDMD contained an N-terminal His6-MBP tag, and a mixture of TEV and 3C at a molar ratio of 1:1 was applied, with TEV to remove His6-MBP and 3C to cleave GSDMD. GSDMA3 contained a SUMO tag, and a mixture of ULP1 and 3C at a molar ratio of 1:1 was applied, with ULP1 to remove SUMO and 3C to cleave GSDMA3. In both cases, the molar ratio of lipid:GSDM:En was set to 400:5:2, with GSDM at 0.5 μM. For simplicity, the MBP and SUMO tags are not displayed in figures. Reactions on 384-well plates (Corning) were monitored by fluorescence (545 nm) using a SpectraMax M5 plate reader (Molecular Devices), with excitation at 274 nm, for 20 min at 2-min intervals.
Pore-dependent cargo release from liposomes.
Freshly purified precursor and mature IL-1s were loaded into PS-containing liposomes (25–75% PS, the rest being PC, in the case of GSDMD and GSDMA3 pore formation) or cholesterol-containing liposomes (25% cholesterol + 25% PS + 50% PC, in the case of SLO and PFO pore formation) in a similar fashion to the Tb3+ encapsulation procedure, except that Buffer A was used throughout and no extrusion was performed. The content of PS or cholesterol is 25% unless otherwise denoted in the text or figure legends. Unloaded proteins were removed by repeatedly pelleting the liposomes at 20,000 g for 15 min in a cold centrifuge and replacing the supernatant with Buffer A three times. Differently charged fluorescein isothiocyanate (FITC)-conjugated dextrans (MW 40 kDa, radii around 40 Å, Sigma Aldrich) – neutral (FD40), CM-modified (53379), and DEAE-modified (01649) – and three other small proteins (MW 12–23 kDa, diameters 30–40 Å) – cytochrome C (CyC, C2037, Sigma-Aldrich), γ-crystallin D (CRYGD, AR39126PU-N, OriGene Technologies), and oncomodulin (OCM, RPU43138, Biomatik) – were loaded using the same method. The concentrations of the loaded cargoes were approximately 1 μM for proteins and 0.01 mg/ml (or 0.25 μM) for dextrans. To ensure that cargoes were released through the pores rather than pore-induced membrane lysis, we used a low sub-lytic concentration (1x) for each pore-forming protein - GSDMD (1x = 0.5 μM), GSDMA3 (1x = 0.5 μM), SLO (1x = 0.1 μM), and PFO (1x = 0.1 μM) - evident in the minimal release of encapsulated LDH (CyQUANT LDH Cytotoxicity Kit, Invitrogen) or bulky dextrans (MW 2 MDa, Sigma-Aldrich, FD2000) during a 2-h monitoring period. The molar ratios of MBP-GSDMD:TEV:3C and of SUMO-GSDMA3:ULP1:3C were kept at 5:1:1. The MBP and SUMO tags are not shown in figures for simplicity. No activating enzymes (En) were required for pore formation by SLO and PFO. One aliquot of each reaction was pelleted at time zero, and the pellet was washed three times and dissolved to the original volume with Buffer A containing 1% Triton X-100 (Sigma-Aldrich), as a measure of total cargoes loaded into the liposomes. At indicated time points, the liposomes were pelleted for supernatant collection. For experiments involving IL-1s and the other protein cargoes, the harvested supernatants were subject to SDS-PAGE and standard immunoblotting. The primary antibodies were goat polyclonal anti-IL-1β (R&D Systems, AF401NA, 1:500), rabbit polyclonal anti-IL-18 (Genetex, GTX32675, 1:1000), rabbit polyclonal anti-CyC (PA19586, Thermo Fisher Scientific, 1:500), rabbit polyclonal anti-CRYGD (OriGene Technologies, TA332876, 1:500), and rabbit polyclonal anti-OCM (Biomatik, CAU21955, 1:1000). The incubation conditions for the primary antibodies were 4 °C overnight or room temperature for 1 h. The secondary antibodies were HRP-conjugated anti-goat IgG (Santa Cruz Biotechnology, sc-2354, 1:5000), anti-rabbit IgG (Cell Signaling Technology, 7074S, 1:5000), and anti-mouse IgG (Abcam, ab97040, 1:5000) all used at room temperature for 2 h. The Pierce ECL substrate (Thermo Fisher Scientific) was used for detection. Immunoblots were quantified using ImageJ41, and IL-1 release rate curves were obtained by fitting hyperbolic functions based on the data points in GraphPad Prism 6. The initial IL-1 release rates were extrapolated as the derivatives of the fitted functions at time zero. For experiments involving dextrans, the amounts of released dextrans in collected supernatants were measured by FITC fluorescence with excitation at 493 nm and emission at 518 nm using a SpectraMax M5 plate reader (Molecular Devices). The maximum fluorescence (100%) was set to the lysed liposomes collected at time zero. Each type of dextran was compared to its own maxima to account for potential variability in FITC labelling.
IL-1 secretion from cells.
iBMDMs from WT C57BL/6 mice and GSDMD-KO iBMDMs were kindly supplied by Drs. Jonathan Kagan and Judy Lieberman (Boston Children’s Hospital, Boston, MA, USA) and have been authenticated and tested for mycoplasma contamination in previous studies4,7. To generate IL-1β-KO iBMDMs, il-1β gRNA (5’-CAATGAGTGATACTGCCTGC-3’) was cloned into LentiCRISPR-v2 hygro as previously described42. The plasmid was then transfected into HEK293T cells with pSPAX2 and pCMV-VSV-G at 2:2:1 ratio using calcium phosphate method. Supernatant collected 2 d later was used to transduce mouse iBMDM cells. After 2 d, hygromycin (300 μg/ml) was added to select for positive cells for 10 d. Cells were then subcloned in 96-well plates and screened for IL-1β expression by immunoblotting using anti-IL-1β (R&D Systems, AF401NA, 1:500). To express WT or charge-mutant GSDMD, A3chim (C-terminally Flag-tagged), and pro-IL-1β in GSDMD-KO and IL-1β-KO cells, respectively, pLVX-puro plasmids containing the respective cDNAs, or without as a control, were transfected into HEK293T cells (Lenti-X Packaging System, Takara Bio). Supernatants after 2 d were collected to transduce KO iBMDMs. The cells were then subject to selection by puromycin (Sigma, 4 μg/ml). Expression of GSDMD, A3chim, and pro-IL-1β were verified by immunoblotting using anti-GSDMD (Abcam, ab209845, 1:500), anti-Flag (Cell Signaling Technology, 14793S, 1:1000), and anti-IL-1β (R&D Systems, AF401NA,1:500), respectively. All iBMDMs were maintained in Dulbecco’s Modified Eagle’s medium (DMEM) with L-glutamine (Thermo Fisher Scientific, 10569-004), supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, 16000-044), at 37 °C with 5% CO2. For NLRP3 inflammasome activation, iBMDMs were primed with LPS (1 μg/mL, InvivoGen, tlrl-b5lps) for 4 h. Post priming, glycine (5 mM) treatment was carried out for 1 h prior to NLRP3 activation using 20 μM nigericin (Sigma-Aldrich, N7143) for 30 min. Alternatively, LPS-primed iBMDMs were treated with 0.5 μM nigericin to allow sustained survival and IL-1β release (12 h) without glycine protection. For SLO- or PFO-mediated IL-1β release, purified SLO or PFO was added to LPS-primed nigericin (20 μM)-stimulated GSDMD-KO iBMDMs at 0.16 nM or 625 nM final concentrations for 3 h. In all cases, the supernatant was carefully collected without disturbing the cells. Cell debris was further removed by centrifugation at 15,000 rpm, and the clarified supernatant was concentrated using 10,000 MWCO spin columns. Standard immunoblotting following SDS-PAGE was performed to detect precursor or mature IL-1β using anti-IL-1β (AF401NA, R&D Systems, 1:500), with β-actin (Santa Cruz Biotechnology, sc-47778, 1:1000) as a loading control. Quantification of IL-1β in the supernatant using ELISA (ThermoFisher Scientific, 88-7013-88). Remaining supernatant and lysate were used to analyse LDH release (CyQUANT LDH Cytotoxicity Assay kit, Invitrogen), with the maximum (100%) set to the LDH level in lysed whole cells.
Mathematical modelling.
The electrostatic interaction between the pore and IL-1 was simplified as E⋅q, where E is the average electrostatic potential of the pore (kT/e) and q is the net charge of an IL-1 cargo (e). The maximum or initial rate of IL-1 release (r) was approximated as proportional to exp[-Eq/(kT)], where k is the Boltzmann constant and T is the temperature. Assumptions of this approximation include 1) the cargo size is small relative to the pore, 2) the cargo travels through the pore centre where most flux occurs, and 3) the electrostatic effect on the cargo is averaged over all directions. According to this relationship, the ratio of the rates (R) between differently charged IL-1 cargoes is R = exp[−EΔq/(kT)], or ln(R) = −EΔq/(kT). A set of measured rate ratios were then plotted against Δq (difference in net charge) to obtain the fitted electrostatic potential E that accounts for the rate differences.
Data availability.
Atomic coordinates of the 33-fold symmetric human GSDMD pore structure have been deposited in the Protein Data Bank (PDB) under accession number 6VFE. The cryo-EM density maps of the 33-fold symmetric pore and the pre-pore have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers 21160 and 21161, respectively. All other data are available from the corresponding authors upon reasonable request. Several structural coordinates in the PDB database were used in this study, which can be located by accession numbers 6CB8, 5B5R, 6N9O, 9ILB, 3WO2, 5LY6, 2B4Z, 1H4A, and 1TTX.
Extended Data
Extended Data Fig. 1 |. Reconstitution and purification of GSDMD assemblies.
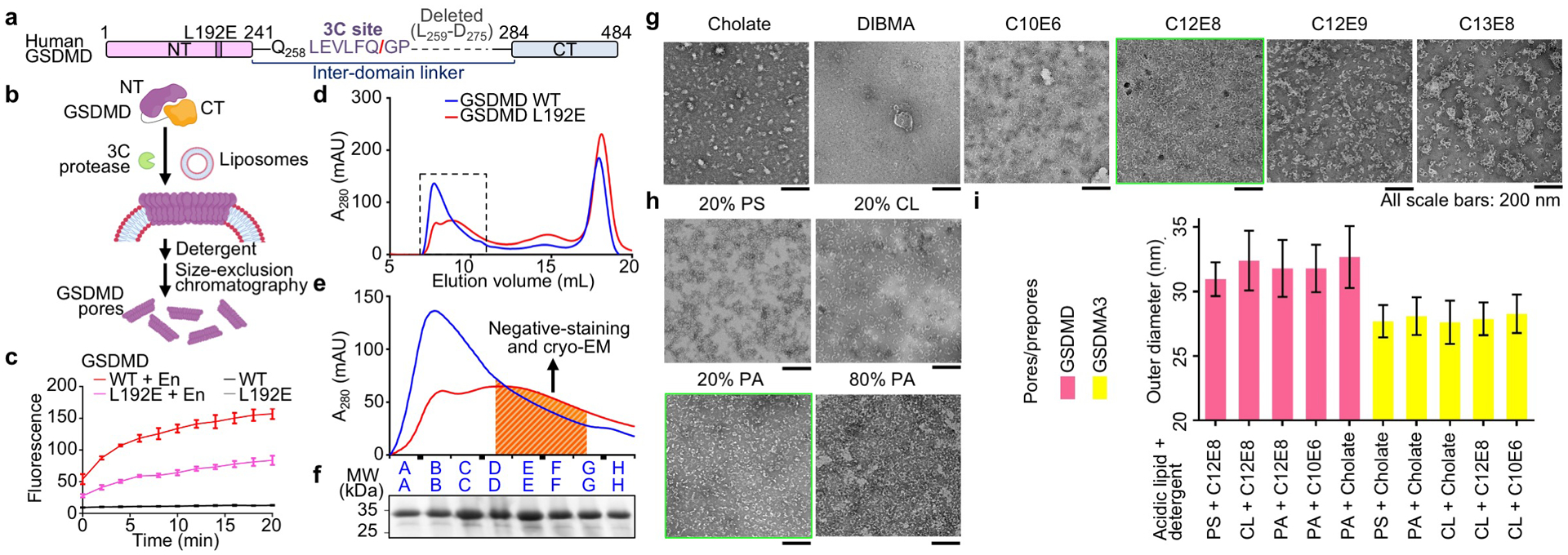
a, Optimized construct for human GSDMD referred to as “WT” GSDMD for convenience. The N-terminal MBP tag and the TEV-cleavable linker between MBP and GSDMD-NT are not shown. b, Schematic of GSDMD pore and prepore reconstitution. c, Reduced, but not abolished, activity of the GSDMD L192E mutant shown by Tb3+ leakage (n = 3 biological replicates). En: Activating enzyme. d, e, Size-exclusion chromatography profiles (d), and their locally enlarged views (e). The dashed box encloses the fractions containing the majority of GSDMD WT or L192E assemblies. The shaded fractions containing the least aggregated particles (e) were used for EM data collection. f, SDS-PAGE showing WT GSDMD-NT at around 30 kDa from the corresponding fractions in (e). g, Detergent screen. Non-ionic detergents known as ethylene glycol monoethers, with formula CxEy, yielded stable GSDMD pores. C12E8 was chosen as the final solubilizing agent. All scale bars: 200 nm. h, GSDMD pores extracted by 1% C12E8 from liposomes containing different types and amounts (%) of acidic lipids. Liposomes containing 20% PA were chosen. All scale bars: 200 nm. i, Sizes of GSDMD and GSDMA3 assemblies reconstituted on liposomes containing different types of acidic lipids (20%) and extracted by different types of detergents (1% CxEy, or 50 mM cholate), shown by outer diameters measured under negative-stain EM (from left to right, n = 64, 42, 77, 73, 14, 34, 45, 23, 21, and 18 particles). Data shown in c and i are mean ± s.d.. Data shown in f–h are representative of three independent experiments.
Extended Data Fig. 2 |. Cryo-EM data processing for the WT GSDMD dataset.
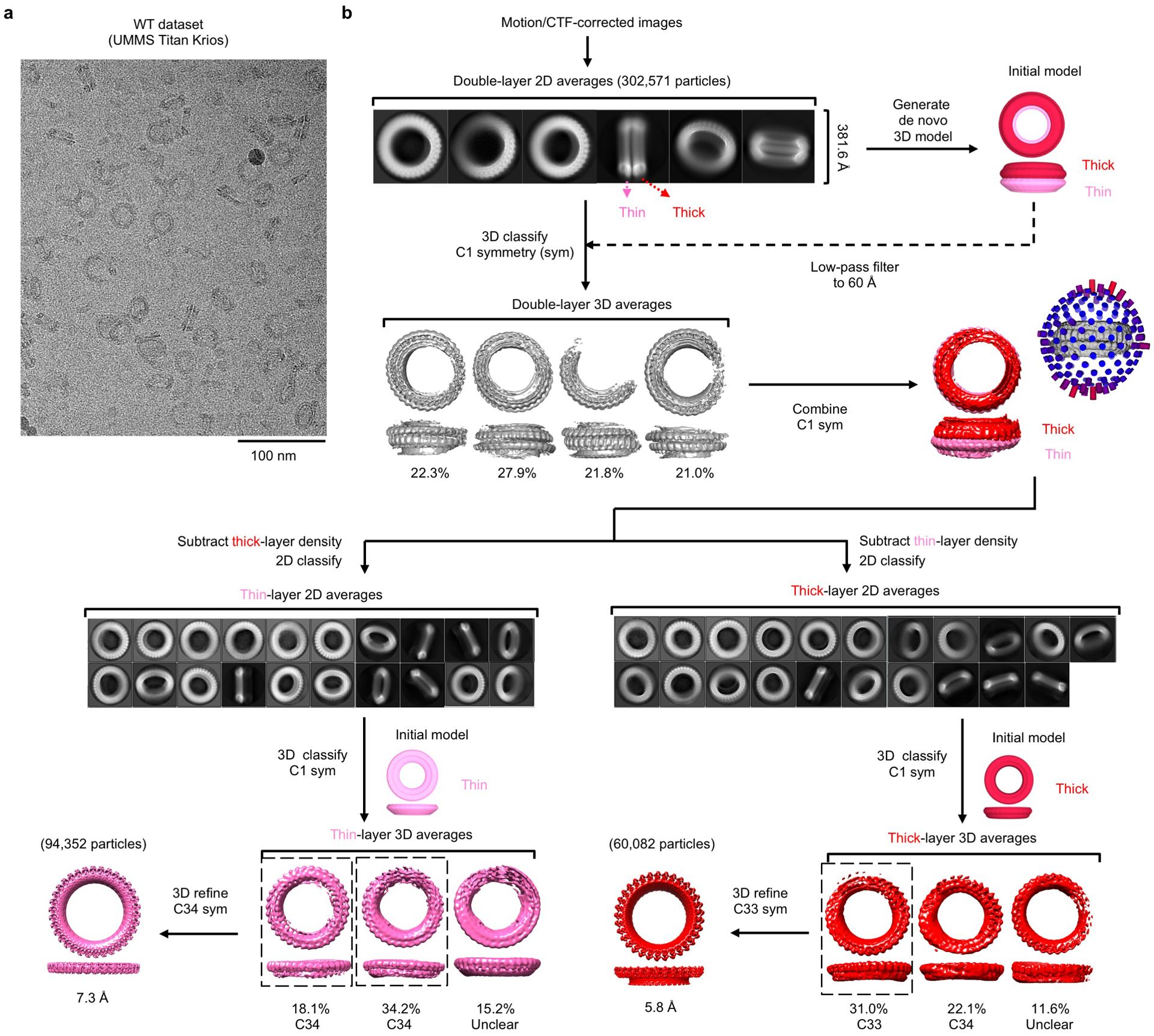
a, A cryo-EM image of the WT GSDMD sample. Scale bar: 100 nm. b, Processing of the WT GSDMD cryo-EM dataset. Initial 2D classes showed a ring-stacking phenomenon, which added to structural heterogeneity and posed challenges to symmetry determination. Density subtraction was therefore performed, followed by 3D reconstruction of each ring without assumption of symmetry, after which particle symmetry could be determined for certain 3D classes. These classes were then refined with their respective symmetry imposed to yield final cryo-EM maps. Data shown in a are representative of three independent experiments.
Extended Data Fig. 3 |. Cryo-EM data processing for the L192E GSDMD dataset.
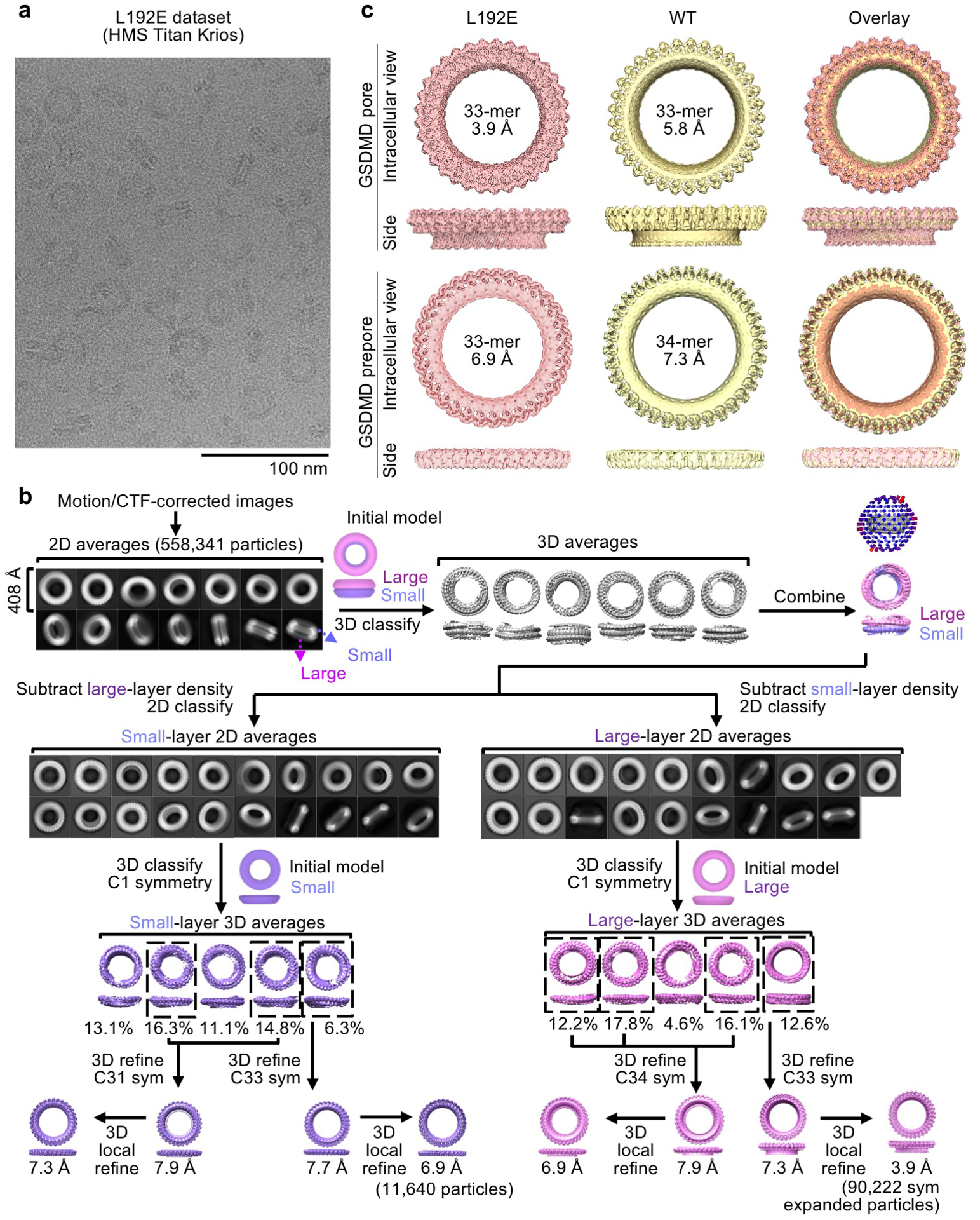
a, A cryo-EM image of the L192E GSDMD sample. Scale bar: 100 nm. b, Processing of the L192E GSDMD cryo-EM dataset. The L192E dataset was first processed following the procedures for the WT dataset. Cryo-EM maps obtained from 3D refinement with symmetry imposed were further classified without alignment to remove heterogeneous particles. Then, the best 3D classes were refined again to improve resolutions. A 3D reconstruction at 7.3 Å was further improved by symmetry expansion, 3D classification without alignment, 3D local refinement, and per-particle CTF refinement to reach the final map at 3.9 Å resolution. c, Similarity of cryo-EM maps generated from the WT and L192E datasets. Due to the higher resolutions, maps from the L192E dataset were chosen for model building. Data shown in a are representative of three independent experiments.
Extended Data Fig. 4 |. Analysis of cryo-EM densities and models.

a, Half-map-to-half-map and model-to-map Fourier shell correlation (FSC) for the 33-fold symmetric GSDMD pore and prepore from the L192E dataset. Horizontal dashed lines represent FSC cut-offs at 0.5 and 0.143. b, c, Pore-form (b) and prepore-form (c) GSDMD subunits fitted into their respective cryo-EM density. Arrows indicate secondary structural elements specified by residue numbers. d, Close-up views of the pore-form GSDMD structure fitted into its cryo-EM density at six representative locations denoted by residue numbers.
Extended Data Fig. 5 |. β-hairpin extension and prepore-to-pore transition.
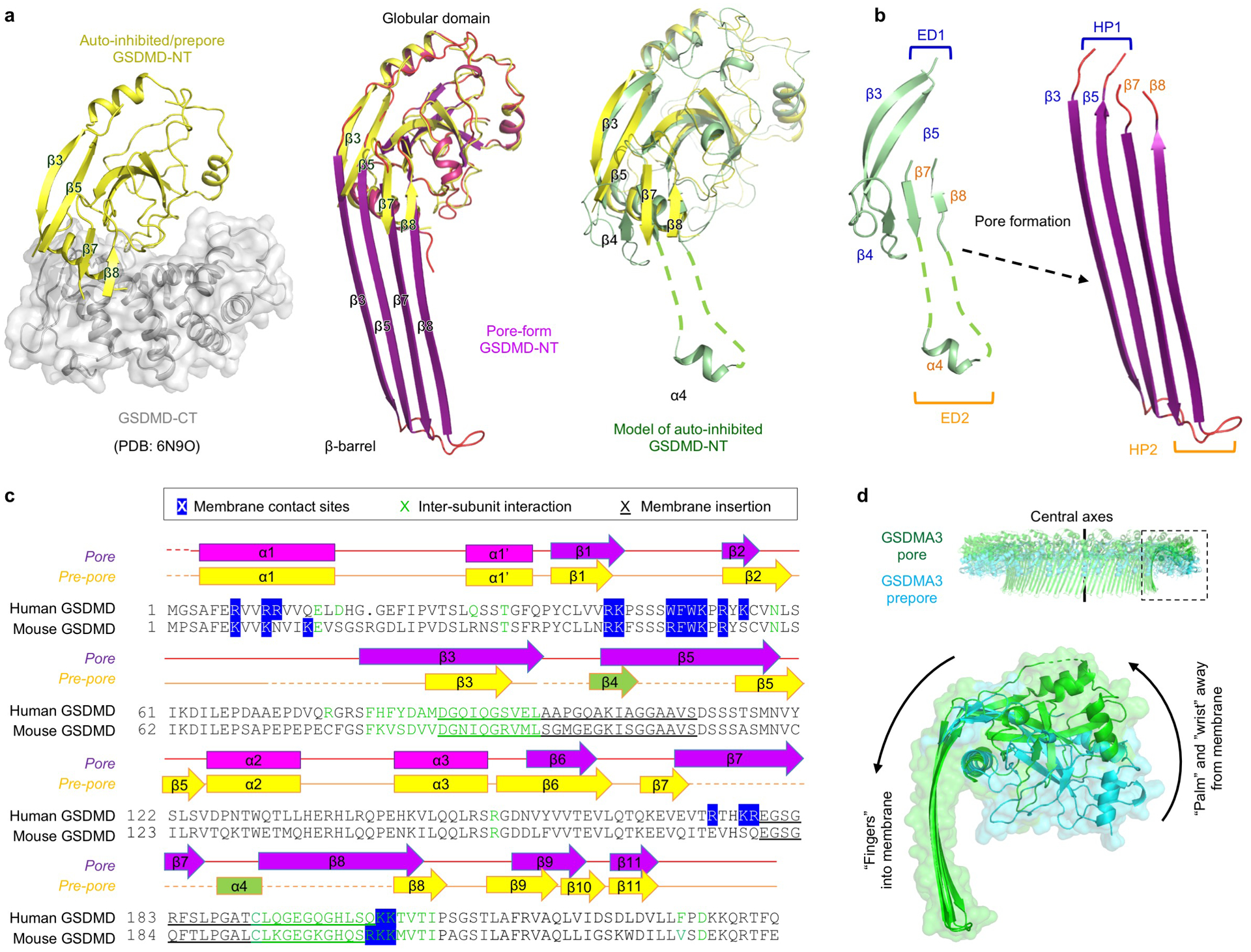
a, Comparison between auto-inhibited, prepore-form, and pore-form GSDMD. The auto-inhibited GSDMD-NT was obtained from the crystal structure of full-length GSDMD (PDB: 6N9O). The β4 strand and α4 helix are invisible in the crystal structure and were modelled based on the crystal structure of full-length GSDMA3 (PDB: 5B5R). b, Formation of β-hairpins (HPs). The β3-β4-β5 region constitute the first extension domain (ED1), which transforms into HP1 by refolding. The β7-α4-β8 region represents ED2 and becomes HP2. c, Sequence alignment of human and mouse GSDMD with secondary structures and key residues denoted. Blue highlight: Responsible for lipid binding, through either hydrophobic or charged interactions. Green: At inter-subunit interfaces. Underscore: Important for membrane insertion. d, Conserved rigid-body movement of the globular domain (“palm”) towards the membrane-distal direction during GSDM pore formation, shown by alignment of the GSDMA3 pore structure (PDB: 6CB8) and prepore model at their central axes.
Extended Data Fig. 6 |. Hydrophobic anchor and basic patches of GSDMs.
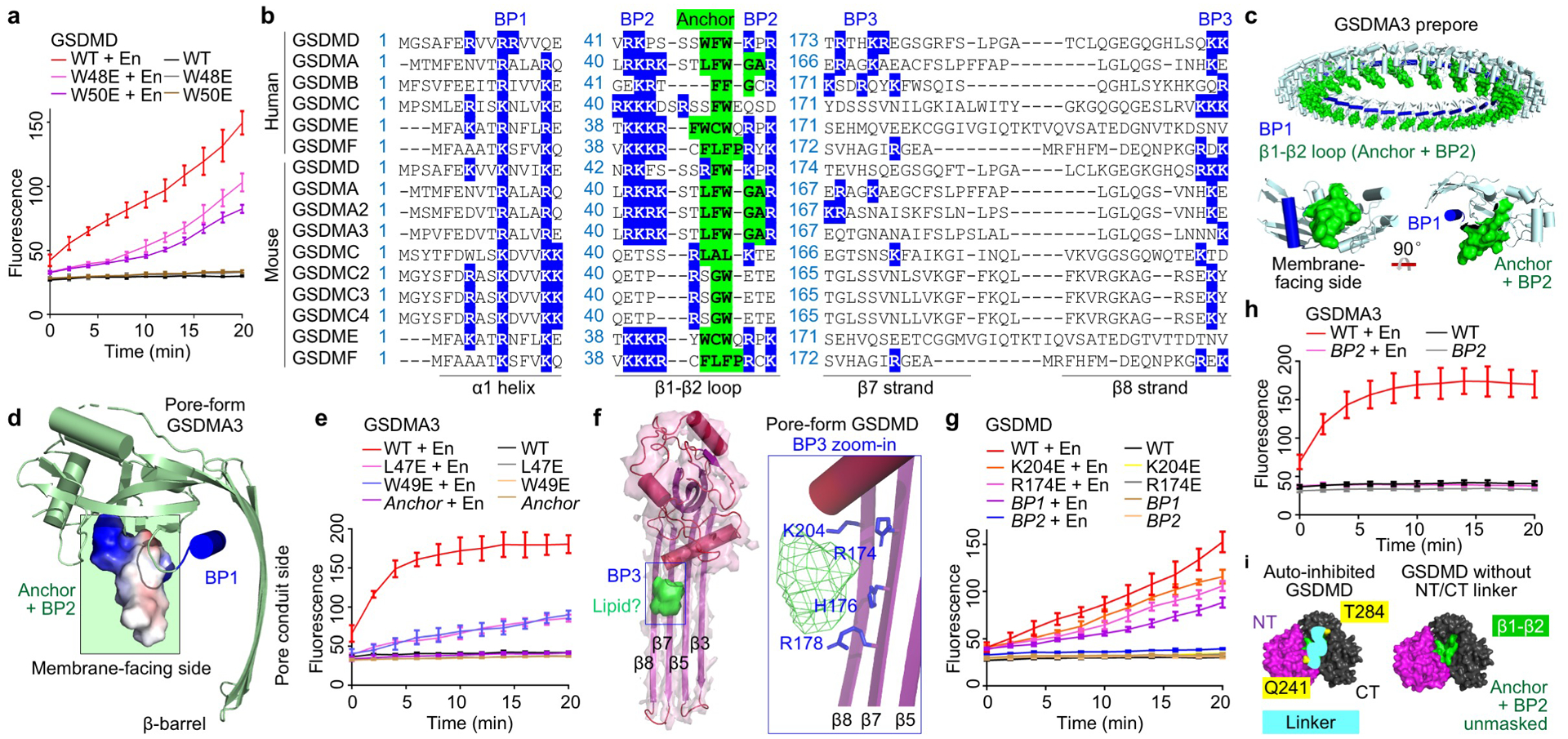
a, Effects of mutations in the hydrophobic anchor on GSDMD pore formation assessed by Tb3+ leakage from liposomes (n = 3 biological replicates). En: Activating enzyme. b, GSDM sequences aligned at the hydrophobic anchor and BPs. Blue highlight: Basic residues at BPs. Green highlight: Hydrophobic residues of the anchor. Dashes: Gaps. c, The GSDMA3 prepore model with the β1-β2 loop highlighted in green and BP1 shown in blue. A GSDMA3 prepore subunit is also shown in two orientations. d, A side view of pore-form GSDMA3 (PDB: 6CB8), with electrostatic surface shown around the β1-β2 loop. The anchor and BP2 are boxed in green. e, Impairment of the pore-forming ability of GSDMA3 by mutations in the hydrophobic anchor, shown by Tb3+ leakage (n = 3 biological replicates). Anchor: L47F48W49 mutated to E. f, A cryo-EM density blob that likely represents heads of phospholipids near BP3. Basic residues in BP3 point towards the blob. g, Effects of mutations in BP1 (R7R10R11 to E), BP2 (R42K43K51R53 to E), and BP3 (K204E or R174E) on GSDMD activity evaluated by Tb3+ leakage (n = 3 biological replicates). h, Importance of BP2 for GSDMA3 pore formation, shown by Tb3+ leakage (n = 3 biological replicates). BP2: R41K42R43K44 mutated to E. i, Exposure of the hydrophobic anchor and BP2 upon removal of the GSDMD-NT/CT inter-domain linker. Surface representations are shown for auto-inhibited GSDMD (PDB: 6N9O) with and without the inferred linker (cyan curve connecting Q241 and T284). Purple: GSDMD-NT. Black: GSDMD-CT. Green: Anchor-BP2 region. Yellow: Two ends of the linker. Data shown in a, e, g, and h are mean ± s.d..
Extended Data Fig. 7 |. GSDM acidic patches and their mutations.
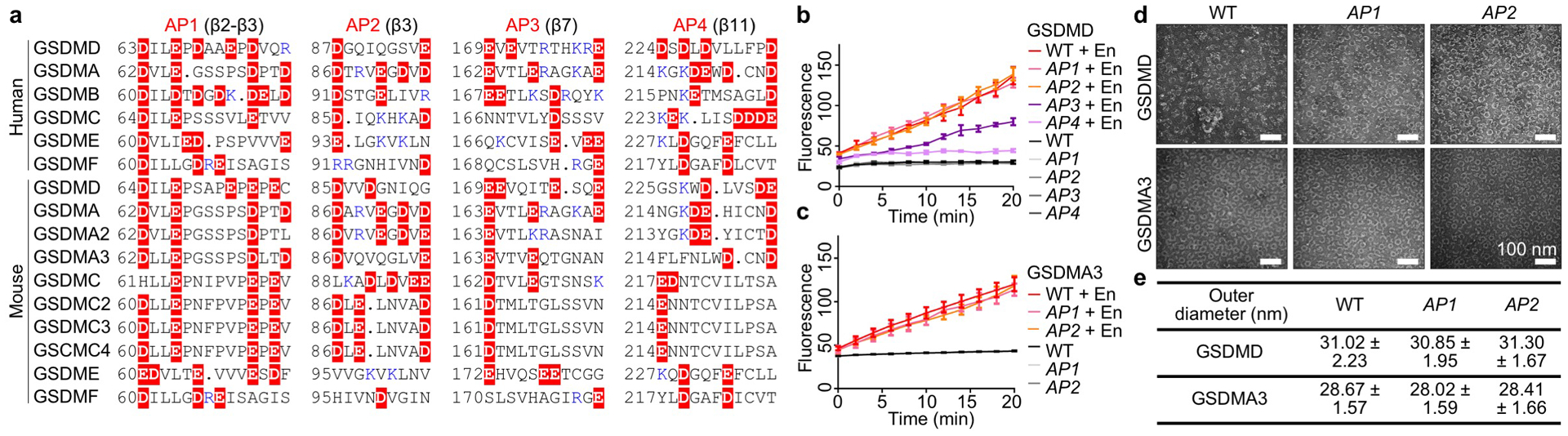
a, Locations of APs shown by aligned GSDM sequences. Dots: Strings of omitted uncharged residues. Red highlight: Acidic residues. Blue: Basic residues. Of note, the basic residues near the APs may face the membrane (such as those in BP3) and therefore do not necessarily weaken the acidity of the pore conduit. b, Assessment of alanine mutations of GSDMD APs 1 through 4 by Tb3+ leakage (n = 3 biological replicates). c, Assessment of alanine mutations of GSDMA3 AP1 and AP2 by Tb3+ liposome leakage (n = 3 biological replicates). d, Negative-staining EM images of WT and AP-mutant GSDMD and GSDMA3 assemblies, solubilized from liposomes using C12E8 and cholate, respectively. All scale bars: 100 nm. e, Outer diameters of WT or AP-mutant GSDMD and GSDMA3 assemblies, measured under negative-staining EM (n = 50 particles per group). Data shown in b, c, and e are mean ± s.d.. Data shown in d are representative of three independent experiments.
Extended Data Fig. 8 |. Liposome experiments and electrostatics analysis.
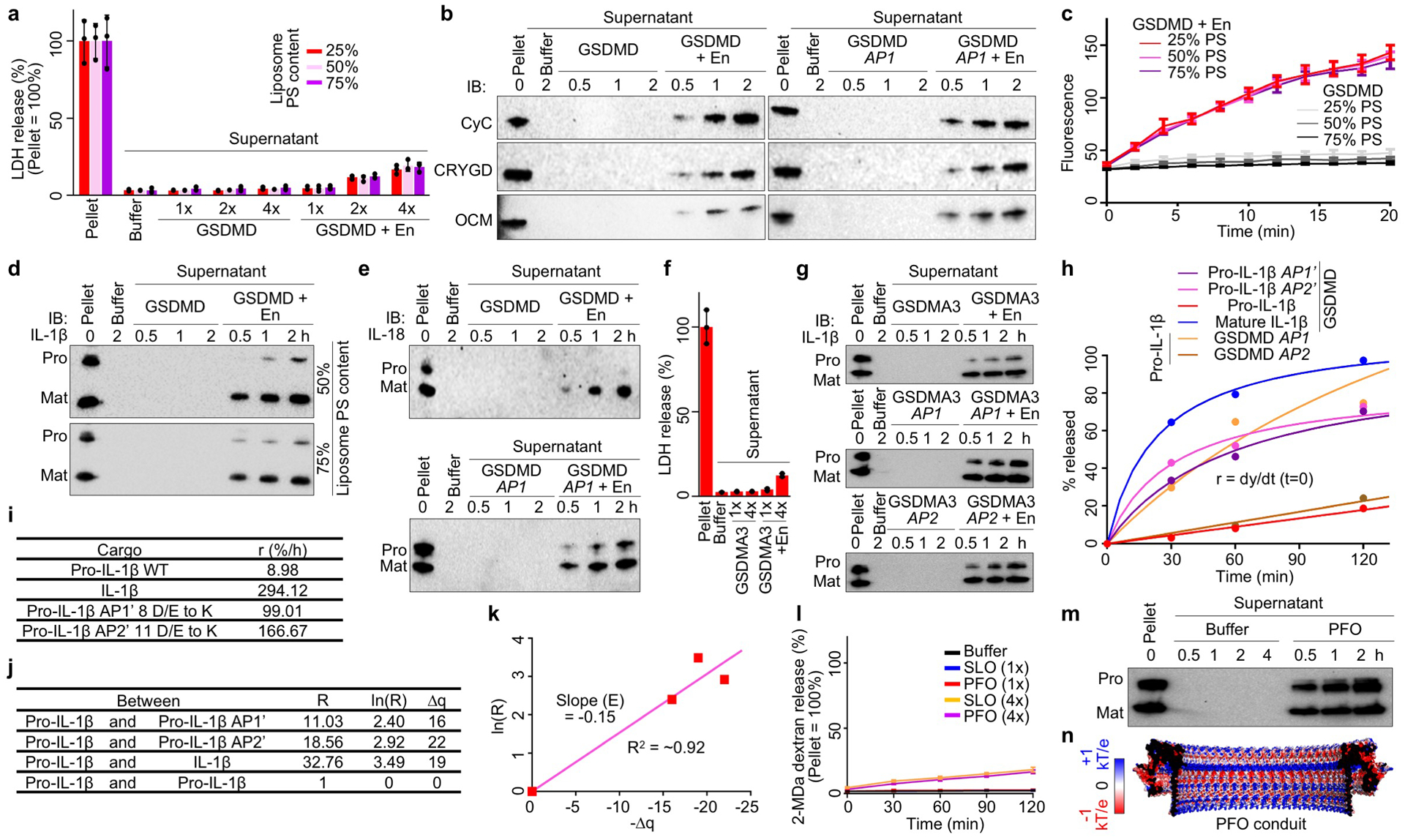
a, Unlysed liposomes (25–75% PS) evident in minimal LDH release when GSDMD was added at a sub-lytic concentration (1x = 0.5 μM) (n = 3 biological replicates). En: Activating enzyme. b, Release of CyC, CRYGD, and OCM from liposomes permeabilized by GSDMD shown by immunoblotting. c, Similar rates of GSDMD pore formation on liposomes of various acidic lipid contents (25–75% PS), according to Tb3+ release (n = 3 biological replicates). d, Preferential IL-1β release from liposomes (50% and 75% PS) perforated by GSDMD shown by immunoblotting. e, Release of pro-IL-18 and IL-18 from liposomes permeabilized by GSDMD shown by immunoblotting. f, Unlysed liposomes evident in minimal LDH release when GSDMA3 was activated at a sub-lytic concentration (1x = 0.5 μM) (n = 3 biological replicates). g, Release of pro- and mature IL-1β from liposomes perforated by GSDMA3 shown by immunoblotting. h, Release rates of IL-1β cargoes through GSDMD pores shown by fitted hyperbolic functions. i, Initial release rates (lowercase r) extrapolated from h. j, Charge differences among the cargoes (Δq) and rate ratios (uppercase R). k, Plot of ln(R) versus Δq for estimating the electrostatic potential (E) of the GSDMD pore conduit. l, Unlysed liposomes shown by lack of release of encapsulated bulky FITC-labelled dextrans (2 MDa) when SLO or PFO was added at a sub-lytic concentration (1x = 0.1 μM) (n = 3 biological replicates). m, Similar release of pro- and mature IL-1β from liposomes permeabilized by PFO. n, Electrostatics surfaces of the modelled PFO pore conduit. Data shown in a, c, f, and l are mean ± s.d.. Data shown in b are representative of three, and data shown in d, e, g, and m two, independent experiments.
Extended Data Fig. 9 |. Macrophages experiments.
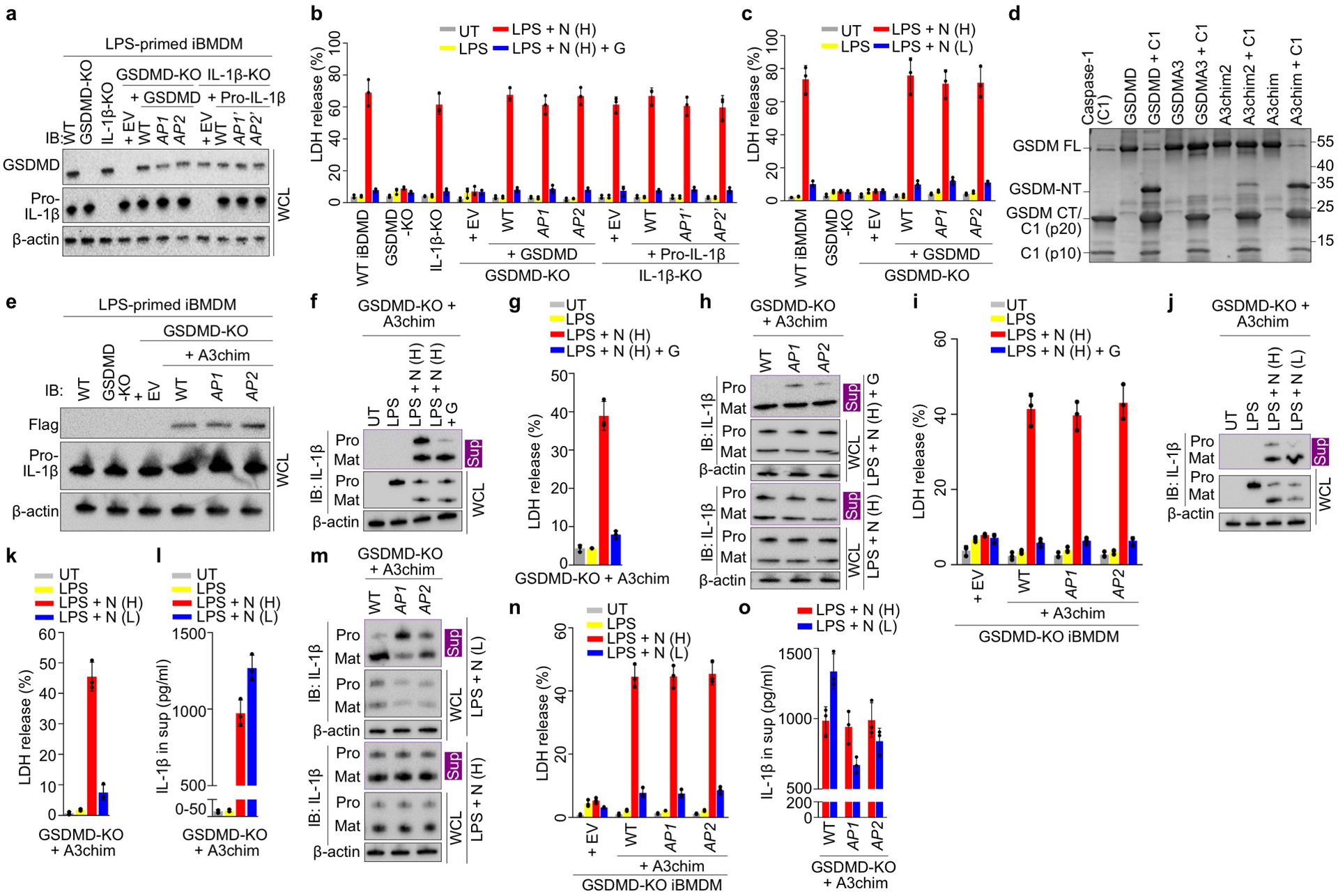
a, Comparable protein expression shown by immunoblotting. b, c, Similar sensitivity to pyroptosis and death evasion by glycine protection (b) or low-dose nigericin treatment (c), shown by LDH release (n = 3 biological replicates). d, Cleavage of engineered GSDMA3 chimera (A3chim) by caspase-1 (C1), shown by SDS-PAGE of proteolysis reactions using purified proteins. e, Comparable expression of A3chim (Flag-tagged) and its AP mutants in GSDMD-KO cells. AP1: 4 D/E to A. AP2: 2 D/E to A. f, g, Preferential release of mature IL-1β from glycine-protected living iBMDMs permeabilized by A3chim, shown by immunoblotting (f) and LDH release (n = 3 biological replicates) (g). h, i, IL-1β release from GSDMD-KO iBMDMs expressing with WT or AP-mutant A3chim under glycine protection, shown by immunoblotting (h) and LDH release (n = 3 biological replicates) (i). j-l, IL-1β release from living GSDMD-KO iBMDMs expressing A3chim stimulated by low-dose nigericin, characterized by immunoblotting (j), LDH release (n = 3 biological replicates) (k), and ELISA (n = 3 biological replicates) (l). m, n, IL-1β release from low-dose nigericin-stimulated GSDMD-KO iBMDMs expressing WT or AP-mutant A3chim, evaluated by immunoblotting (m), LDH release (n = 3 biological replicates) (n), and ELISA (n = 3 biological replicates) (o). Data shown in b, c, g, i, k, l, n, and o are mean ± s.d.. Data shown in a, d, e, f, h, j, and m are representative of two independent experiments.
Extended Data Table 1 |.
Cryo-EM data collection, refinement, and validation statistics
| WT pore | WT prepore | L192E pore (EMDB-21160) (PDB 6VFE) | L192E prepore (EMDB-21161) | |
|---|---|---|---|---|
| Data collection and processing | ||||
| Magnification | 81,000 | 81,000 | 105,000 | 105,000 |
| Voltage (kV) | 300 | 300 | 300 | 300 |
| Electron exposure (e–/Å2) | 48.58 | 48.58 | 63.25 | 63.25 |
| Defocus range (μm) | −1.0 to −2.5 | −1.0 to −2.5 | −0.8 to −2.5 | −0.8 to −2.5 |
| Pixel size (Å) | 0.53 | 0.53 | 0.85 | 0.85 |
| Symmetry imposed | C33 | C34 | C33 | C33 |
| Initial particle images (no.) | 60,082 | 94,352 | 41,413 | 21,200 |
| Final particle images (no.) | 60,082 | 94,352 | 2,734 | 11,640 |
| Map resolution (Å) | 5.8 | 7.3 | 3.9 | 6.9 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 60.0–5.8 | 60.0–7.3 | 60.0–3.9 | 60.0–6.9 |
| Refinement | ||||
| Initial model used (PDB code) | 6N90 | 6N90 | ||
| Model resolution (Å) | 3.7/4.2 | 7.3/10.6 | ||
| FSC threshold | 0.143/0.5 | 0.143/0.5 | ||
| Model resolution range (Å) | 59.0–3.4 | 45.9–3.4 | ||
| Map sharpening B factor (Å2) | −168.81 | −652.00 | ||
| Model composition | ||||
| Non-hydrogen atoms | 62,238 | 46,299 | ||
| Protein residues | 7,953 | 5,742 | ||
| Ligands | 0 | 0 | ||
| B factors (Å2) | ||||
| Protein | 73.57 | 215.68 | ||
| Ligand | N/A | N/A | ||
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.007 | 0.005 | ||
| Bond angles (°) | 0.943 | 1.153 | ||
| Validation | ||||
| MolProbity score | 2.81 | 3.00 | ||
| Clashscore | 6.96 | 26.07 | ||
| Poor rotamers (%) | 6.76 | 4.32 | ||
| Ramachandran plot | ||||
| Favored (%) | 89.82 | 87.35 | ||
| Allowed (%) | 10.18 | 12.65 | ||
| Disallowed (%) | 0.00 | 0.00 |
Supplementary Material
Acknowledgements
We thank Drs. Barry Honig, Tom Rapoport, Feng Shao, Jonathan Kagan, Douglas Golenbock, Stephen Blacklow, Andrew Kruse, and Maofu Liao for discussions. For structural data collection, we thank Drs. Richard Walsh, Sarah Sterling, Shaun Rawson, and Zongli Li at the Harvard Cryo-EM Center for Structural Biology and Drs. Kangkang Song, Kyounghwan Lee, and Chen Xu at the Cryo-EM Core Facility at University of Massachusetts Medical School. This work was supported by US National Institutes of Health grants R01AI139914 (H.W. and J.L.), DP1HD087988 (H.W.), R01AI124491 (H.W.), R01CA240955 (J.L.), R01DK095045 (A.G.), R01DK099465 (A.G.), and 5T32HL066987-18 (T.M.-F.). S.X. received the Albert J. Ryan fellowship. J.R. and Z.Z. received postdoctoral fellowships from the Charles A. King Trust.
Footnotes
Competing interests
H.W. and J.L. are co-founders of Ventus Therapeutics. The other authors declare no competing interests.
References
- 1.Kayagaki N et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671, doi: 10.1038/nature15541 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Shi J et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665, doi: 10.1038/nature15514 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Ding J et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116, doi: 10.1038/nature18590 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Liu X et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158, doi: 10.1038/nature18629 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aglietti RA et al. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc Natl Acad Sci U S A 113, 7858–7863, doi: 10.1073/pnas.1607769113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sborgi L et al. GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J 35, 1766–1778, doi: 10.15252/embj.201694696 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evavold CL et al. The Pore-Forming Protein Gasdermin D Regulates Interleukin-1 Secretion from Living Macrophages. Immunity 48, 35–44 e36, doi: 10.1016/j.immuni.2017.11.013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heilig R et al. The Gasdermin-D pore acts as a conduit for IL-1beta secretion in mice. Eur J Immunol 48, 584–592, doi: 10.1002/eji.201747404 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Kayagaki N et al. NINJ1 mediates plasma membrane rupture during lytic cell death. Nature, doi: 10.1038/s41586-021-03218-7 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Monteleone M et al. Interleukin-1beta Maturation Triggers Its Relocation to the Plasma Membrane for Gasdermin-D-Dependent and -Independent Secretion. Cell Rep 24, 1425–1433, doi: 10.1016/j.celrep.2018.07.027 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Xia S, Zhang Z, Wu H & Lieberman J Channeling inflammation: Gasdermins in physiology and disease. Nat Rev Drug Discov, doi: 10.1038/s41573-021-00154-z (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orning P et al. Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science 362, 1064–1069, doi: 10.1126/science.aau2818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarhan J et al. Caspase-8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection. Proc Natl Acad Sci U S A 115, E10888–E10897, doi: 10.1073/pnas.1809548115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burgener SS et al. Cathepsin G Inhibition by Serpinb1 and Serpinb6 Prevents Programmed Necrosis in Neutrophils and Monocytes and Reduces GSDMD-Driven Inflammation. Cell Rep 27, 3646–3656 e3645, doi: 10.1016/j.celrep.2019.05.065 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Z et al. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature 579, 415–420, doi: 10.1038/s41586-020-2071-9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Z et al. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science 368, doi: 10.1126/science.aaz7548 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Sollberger G et al. Gasdermin D plays a vital role in the generation of neutrophil extracellular traps. Sci Immunol 3, doi: 10.1126/sciimmunol.aar6689 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Wang Q et al. A bioorthogonal system reveals antitumour immune function of pyroptosis. Nature 579, 421–426, doi: 10.1038/s41586-020-2079-1 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Chen KW et al. Noncanonical inflammasome signaling elicits gasdermin D-dependent neutrophil extracellular traps. Sci Immunol 3, doi: 10.1126/sciimmunol.aar6676 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Ruan J, Xia S, Liu X, Lieberman J & Wu H Cryo-EM structure of the gasdermin A3 membrane pore. Nature 557, 62–67, doi: 10.1038/s41586-018-0058-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulvihill E et al. Mechanism of membrane pore formation by human gasdermin-D. EMBO J 37, doi: 10.15252/embj.201798321 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Z et al. Crystal Structures of the Full-Length Murine and Human Gasdermin D Reveal Mechanisms of Autoinhibition, Lipid Binding, and Oligomerization. Immunity 51, 43–49 e44, doi: 10.1016/j.immuni.2019.04.017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruhl S et al. ESCRT-dependent membrane repair negatively regulates pyroptosis downstream of GSDMD activation. Science 362, 956–960, doi: 10.1126/science.aar7607 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Lee IH, Kai H, Carlson LA, Groves JT & Hurley JH Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proc Natl Acad Sci U S A 112, 15892–15897, doi: 10.1073/pnas.1518765113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davis MA et al. Calpain drives pyroptotic vimentin cleavage, intermediate filament loss, and cell rupture that mediates immunostimulation. Proc Natl Acad Sci U S A 116, 5061–5070, doi: 10.1073/pnas.1818598116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers C et al. Gasdermin pores permeabilize mitochondria to augment caspase-3 activation during apoptosis and inflammasome activation. Nat Commun 10, 1689, doi: 10.1038/s41467-019-09397-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Pee K et al. CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin. Elife 6, doi: 10.7554/eLife.23644 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tapia VS et al. The three cytokines IL-1beta, IL-18, and IL-1alpha share related but distinct secretory routes. J Biol Chem 294, 8325–8335, doi: 10.1074/jbc.RA119.008009 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin-Sanchez F et al. Inflammasome-dependent IL-1beta release depends upon membrane permeabilisation. Cell Death Differ 23, 1219–1231, doi: 10.1038/cdd.2015.176 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Batista SJ et al. Gasdermin-D-dependent IL-1alpha release from microglia promotes protective immunity during chronic Toxoplasma gondii infection. Nat Commun 11, 3687, doi: 10.1038/s41467-020-17491-z (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheres SH RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530, doi: 10.1016/j.jsb.2012.09.006 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332, doi: 10.1038/nmeth.4193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol 192, 216–221, doi: 10.1016/j.jsb.2015.08.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kucukelbir A, Sigworth FJ & Tagare HD Quantifying the local resolution of cryo-EM density maps. Nat Methods 11, 63–65, doi: 10.1038/nmeth.2727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen EF et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612, doi: 10.1002/jcc.20084 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132, doi: 10.1107/S0907444904019158 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221, doi: 10.1107/S0907444909052925 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy A, Kucukural A & Zhang Y I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5, 725–738, doi: 10.1038/nprot.2010.5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker NA, Sept D, Joseph S, Holst MJ & McCammon JA Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98, 10037–10041, doi: 10.1073/pnas.181342398 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia S, Ruan J & Wu H Monitoring gasdermin pore formation in vitro. Methods Enzymol 625, 95–107, doi: 10.1016/bs.mie.2019.04.024 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider CA, Rasband WS & Eliceiri KW NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675, doi: 10.1038/nmeth.2089 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanjana NE, Shalem O & Zhang F Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784, doi: 10.1038/nmeth.3047 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates of the 33-fold symmetric human GSDMD pore structure have been deposited in the Protein Data Bank (PDB) under accession number 6VFE. The cryo-EM density maps of the 33-fold symmetric pore and the pre-pore have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers 21160 and 21161, respectively. All other data are available from the corresponding authors upon reasonable request. Several structural coordinates in the PDB database were used in this study, which can be located by accession numbers 6CB8, 5B5R, 6N9O, 9ILB, 3WO2, 5LY6, 2B4Z, 1H4A, and 1TTX.


