Supplemental Digital Content is available in the text.
Keywords: aortic valve stenosis, endothelial cells, nucleocytoplasmic transport, palmdelphin
Background:
PALMD (palmdelphin) belongs to the family of paralemmin proteins implicated in cytoskeletal regulation. Single nucleotide polymorphisms in the PALMD locus that result in reduced expression are strong risk factors for development of calcific aortic valve stenosis and predict severity of the disease.
Methods:
Immunodetection and public database screening showed dominant expression of PALMD in endothelial cells (ECs) in brain and cardiovascular tissues including aortic valves. Mass spectrometry, coimmunoprecipitation, and immunofluorescent staining allowed identification of PALMD partners. The consequence of loss of PALMD expression was assessed in small interferring RNA-treated EC cultures, knockout mice, and human valve samples. RNA sequencing of ECs and transcript arrays on valve samples from an aortic valve study cohort including patients with the single nucleotide polymorphism rs7543130 informed about gene regulatory changes.
Results:
ECs express the cytosolic PALMD-KKVI splice variant, which associated with RANGAP1 (RAN GTP hydrolyase activating protein 1). RANGAP1 regulates the activity of the GTPase RAN and thereby nucleocytoplasmic shuttling via XPO1 (Exportin1). Reduced PALMD expression resulted in subcellular relocalization of RANGAP1 and XPO1, and nuclear arrest of the XPO1 cargoes p53 and p21. This indicates an important role for PALMD in nucleocytoplasmic transport and consequently in gene regulation because of the effect on localization of transcriptional regulators. Changes in EC responsiveness on loss of PALMD expression included failure to form a perinuclear actin cap when exposed to flow, indicating lack of protection against mechanical stress. Loss of the actin cap correlated with misalignment of the nuclear long axis relative to the cell body, observed in PALMD-deficient ECs, Palmd−/− mouse aorta, and human aortic valve samples derived from patients with calcific aortic valve stenosis. In agreement with these changes in EC behavior, gene ontology analysis showed enrichment of nuclear- and cytoskeleton-related terms in PALMD-silenced ECs.
Conclusions:
We identify RANGAP1 as a PALMD partner in ECs. Disrupting the PALMD/RANGAP1 complex alters the subcellular localization of RANGAP1 and XPO1, and leads to nuclear arrest of the XPO1 cargoes p53 and p21, accompanied by gene regulatory changes and loss of actin-dependent nuclear resilience. Combined, these consequences of reduced PALMD expression provide a mechanistic underpinning for PALMD’s contribution to calcific aortic valve stenosis pathology.
Clinical Perspective.
What Is New?
Endothelial PALMD (palmdelphin) forms a complex with RANGAP1 (RAN GTPase activating protein 1), an essential actor in RAN-dependent release of cargo carried by XPO1 (exportin1), and thereby regulates nucleocytoplasmic transport.
Reduced PALMD expression interferes with RANGAP1 cytoplasmic localization and leads to nuclear arrest of XPO1 cargo, especially p53 and p21, correlating with broad changes in gene regulation.
Gene regulatory changes elicited by PALMD deficiency affect actin-dependent biological processes such as the formation of the nucleus-protecting actin cap, required for nuclear resilience under mechanical stress.
What Are the Clinical Implications?
Patients with calcific aortic valve stenosis with the single nucleotide polymorphism rs7543130 express reduced PALMD mRNA levels in valve endothelial cells, which show hallmarks of PALMD deficiency such as loss of cytoplasmic RANGAP1, altered nuclear morphology, and nuclear arrest of p53 and p21.
Gene regulatory changes affecting actin reorganization are detected in seemingly healthy regions of calcifying valves, in agreement with disturbed actin-dependent processes being an early event instigating the calcific process.
PALMD is prominently expressed in endothelial cells, and the presence of the PALMD single nucleotide polymorphism rs7543130 correlates with both aberrant endothelium and calcific aortic valve stenosis, suggesting that endothelial cell dysfunction is essential in development of calcific aortic valve stenosis.
PALMD (palmdelphin) belongs to the paralemmin family, including also PALM (paralemmin)-1, -2 and -3, of which PALM1 is implicated in regulation of cell shape, plasma membrane dynamics, and cell motility.1 PALMD exists as 2 splice variants with alternative C termini, PALMD-CaaX (membrane anchored) and PALMD-KKVI (cytosolic).2 PALMD is expressed in neurons as well as in other cell types, and proliferation of basal neuronal progenitors involves PALMD-CaaX binding to adducin-α, a component of the plasma membrane-associated cytoskeleton.3 PALMD is also implicated in other processes such as myoblast differentiation4 and protection against p53-dependent DNA damage.5 Single nucleotide polymorphisms (SNPs) in the PALMD locus, associated with reduced PALMD expression, are strongly linked to the development of life-threatening calcific aortic valve stenosis (CAVS).6–8 How PALMD deficiency predisposes for the development of CAVS has remained unknown.
Endothelial cells (ECs) are continuously exposed to mechanical forces exerted by blood flow, blood cells, and stretching of cardiovascular tissues. These forces are detected by the endothelium through complex mechanosensory mechanisms.9 Adaptation to changes involves altered gene regulation induced via communication between the cytoskeleton and the nuclear envelope.10 The perinuclear actin cap, frequently disrupted in cancer and laminopathies, has been assigned an important role in nuclear mechanosensation and mechanotransduction.11
Here, we show that PALMD deficiency interferes with formation of the perinuclear actin cap, accompanied by impaired nuclear resilience in ECs exposed to flow or deforming forces. The nuclear fragility is accompanied by broad gene regulatory changes in PALMD-deficient cells. The underlying mechanism involves disturbed XPO1 (Exportin1, also CRM1 [denoted chromosomal region maintenance 1])–dependent nucleocytoplasmic shuttling of cargo such as p53 and p21 in ECs silenced for PALMD or isolated from aortic valves of patients with CAVS carrying the PALMD SNP rs7543130. Nucleocytoplasmic shuttling is regulated by the RANGAP1 (RAN GTPase activating protein 1)–mediated GTP/GDP cycling of the small GTPase RAN, through which cargo is released from XPO1.12 We show that RANGAP1 associates with PALMD in healthy ECs. In PALMD deficiency, RANGAP1 and XPO1 become confined to the perinuclear region, and the XPO1 cargoes p53 and p21 are arrested in the nucleus. In conclusion, loss of PALMD disturbs normal nucleocytoplasmic shuttling, which results in gene regulatory changes including perturbed actin arrangements, thereby predisposing to pathologies such as CAVS.
Methods
Data Availability
The data that support the findings reported in this study are available from the corresponding author on reasonable request.
The mass spectrometry analysis data set has been deposited to the ProteomeXchange Consortium via the PRIDE13 partner repository with the dataset identifier PXD024887.
The RNA sequence metadata on human ECs have been deposited with the Swedish Science for Life Laboratories and can be accessed through https://doi.org/10.17044/scilifelab.14270159.
Requests for the human CAVS transcript array data set should be directed to Magnus Bäck (magnus.back@ki.se).
Requests for Palmd gene-modified mice and Palmd antimouse antibodies should be directed to Manfred W. Kilimann (kilimann@em.mpg.de).
Cell Culture and Transfections
Human umbilical vein ECs (HUVECs; ScienCell) were maintained on 1% gelatin (Sigma)–coated tissue culture plastic in EC medium MV2 (PromoCell) with Supplement Pack Endothelial Cell GM MV2 (PromoCell). When indicated, cells were treated for 1 hour with 100 nmol/L KPT-185 (Cat. 26074, Cayman) to inhibit XPO1.14
Human valve ECs (hVECs) and human valve interstitial cells (hVICs) were isolated from human aortic valves as described before15,16 with modifications (see the Supplemental Material). Human coronary vascular smooth muscle cells from 3 individuals were purchased from Lonza (Lonza Group AG).
Palmd–/– Mice
A constitutive Palmd knockout model (Palmd−/−; official Mouse Genome Informatics allele designation: Palmd<tm1.2Kili>, MGI:6477926) was generated by TaconicArtemis. The Palmd−/− strain was maintained on the C57Bl/6 background through heterozygous crossing and was fertile, with normal life span when unchallenged; the design and phenotypes of this strain will be given elsewhere (M.W. Kilimann, unpublished data, 2021). Animal husbandry and procedures were in accordance with institutional guidelines and approved by the Institutional Review Board for animal experimentation at Uppsala University (permit 5.8.18–06789/2018). Mice were anaesthetized with ketamine (Intervet)/xylazine (Elanco Denmark ApS; 100 mg/kg:20 mg/kg body weight) or isoflurane (Isoba [Zoetis], 3% induction and 1.5% maintenance). Mice used for this study were 14-week-old sex-matched males and females, showing no differences between sexes and on the basis of comparison of Palmd+/+ versus Palmd−/− littermates. Researchers were blinded to the genotype until analyses were completed.
Determination of Major Axis and Nuclear Misalignment
Nuclear misalignment was assessed by defining the major axis (MA) of the cell either by VE-cadherin immunostaining (static in vitro cultures) or by flow direction (laminar flow in vitro or blood flow in vivo). Nuclear MA was defined by Hoechst 33342 staining or by scanning electron microscopy imaging. Lines representing the MA of the nucleus and the cell body, respectively, were manually drawn in ImageJ, and the angle of inclination (nucleus relative the cell body) was calculated. The cosine (COS) of the nuclear/cell body MA angle attained a value of 1 when MAs were perfectly aligned or values moving toward 0 as angles became increasingly misaligned. For determining the percentage misaligned nuclei, thresholds were set at COS(±41o)=0.75 and, for scanning electron microscopy images, COS(±32o)=0.85, defining the most misaligned populations in the experimental conditions. Angle analyses were done on blinded samples performed by 2 investigators independently.
Microchannel Migration
HUVECs were grown in 6-well dishes to 60% to 80% confluence. Cells were transfected with siControl or siPALMD as described. Two days after transfection, cells were stained with the nuclear dye NucRed Live 647 (ThermoFisher) for 10 minutes at room temperature in cell culture medium at a final concentration of 2 drops/mL. Cells were detached, suspended at 106 cells/mL, and seeded in polydimethylsiloxane (PDMS) microchannel devices 5 h before starting image acquisition. PDMS channels were coated with fibronectin overnight and incubated with cell culture medium before cells were seeded. PDMS channels (designed with 12 µm width × 7 µm height channels and 4 µm width × 7 µm height constrictions; see Kidiyoor et al17 for details on testing channel dimensions) were mounted on 2-well Laboratory-TeK II 155379 chamber glasses for live cell imaging to allow simultaneous acquisition of siControl and siPALMD cells migrating inside the channels.
Live cell imaging inside microchannels was performed on an UltraVIEW VoX spinning-disk confocal system (PerkinElmer) equipped with an EclipseTi inverted microscope (Nikon) with a Nikon Perfect Focus System and a Hamamatsu CCD camera (C9100-50) and driven by Volocity software (Improvision; Perkin Elmer). All images were acquired through a 40× oil objective while cells were maintained at 37°C in a humidified atmosphere of 5% CO2. Images were collected every 15 minutes for 18 hours. Three optical sections were acquired every 2 µm for the 647 channel (1.5% laser power, 50 ms exposure time, binning 2, sensitivity 73). One reference bright field image of the microchannels was captured as a reference, and the Z-stack maximum intensity projections, merged with the bright field image, were used to analyze cells during migration.
Human Samples
Valves from 58 patients undergoing surgical aortic valve replacement because of CAVS were included if the valve was assessed tricuspid by the surgeon. All patients gave informed consent, and the study was approved by the Institutional Review Board for human studies (ethical permit 2012/1633). Clinical data were obtained from electronic health care records, and echocardiographic parameters were obtained from the latest available preoperative transthoracic echocardiography (used in the decision to perform surgical aortic valve replacement).
Valves were dissected into healthy, intermediate, and calcified tissue as previously described.18 Healthy tissue was defined as macroscopically noncalcified, transparent, pliable tissue. Intermediate tissue was defined as nontransparent albeit pliable tissue. Calcified tissue was defined as solid, nonpliable, thickened, nontransparent tissue. Healthy, intermediate, and calcified sections from cusps were collected for each patient, followed by storage at –80°C.
Total RNA from the dissected patient tissues was extracted using TissueLyzer and the RNeasy Tissue Mini Kit (Qiagen). RNA concentration and quality were assessed using a 2100 Bioanalyzer (Agilent) and a NanoDrop (Thermo Scientific).
Statistical Analysis
GraphPad Prism 9.0.0 was used for statistical analyses. For parametric comparison, the unpaired Student t test was used to compare means between 2 experimental groups. For nonparametric comparisons, assuming that values were not normally distributed, the Mann-Whitney U test was used. One-way ANOVA was used to compare >2 treatment groups, and 2-way ANOVA was used to compare several treatment groups examined with regard to 2 or more independent variables. The threshold for statistical significance was set at α=0.05. The statistical method used is mentioned in the corresponding figure legend. For image analyses, processing and quantifications, Image Laboratory 4.1 (BioRad), ImageJ/FIJI, Cell Profiler, and Excel software were used.
Results
Endothelial Expression of PALMD
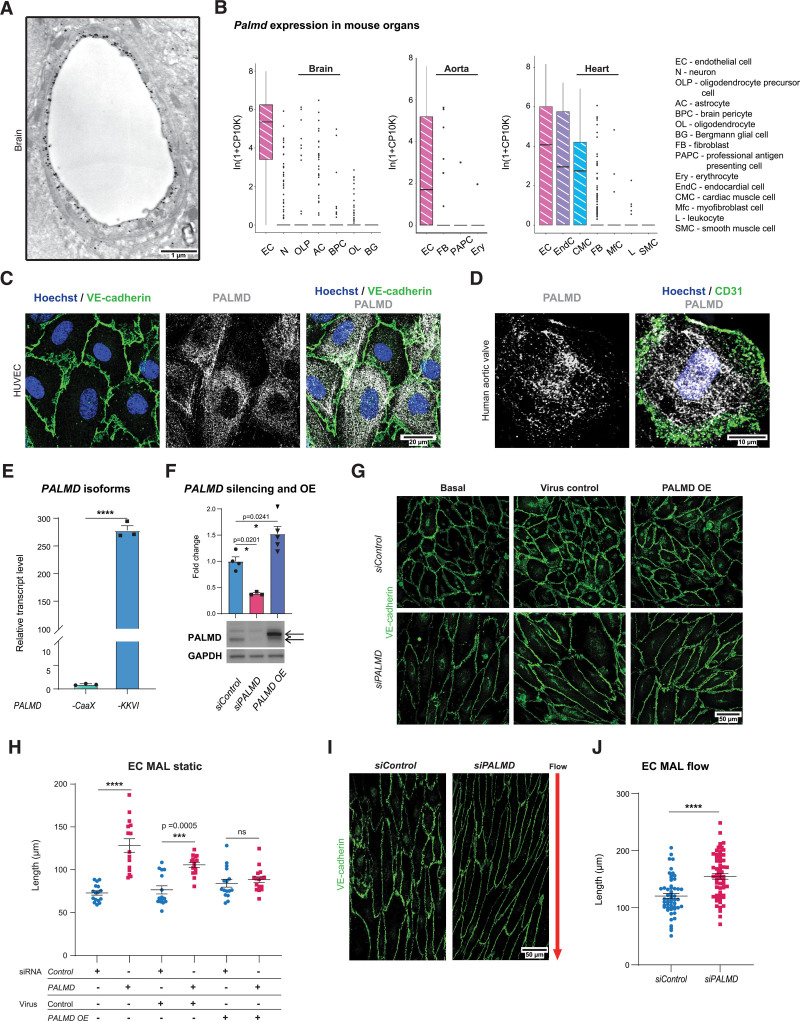
In an initial anti-Palmd immunogold transmission electron microscopic survey of the mouse brain, Palmd reactivity was detected in the cytoplasm of capillary ECs, concentrated at the lumenal plasma membrane (Figure 1A, Figure S1A). Building on this finding, mining of the single cell mouse RNA sequence database Tabula Muris19 confirmed predominant endothelial Palmd expression in the brain, the aorta, and the heart (Figure 1B). In the heart, Palmd is also expressed in endocardial cells and cardiomyocytes (Figure 1B).
Figure 1.
PALMD is expressed in endothelial cells and affects cell shape. A, Palmd immuno-electron microscopy of a mouse cerebellum capillary. B, Palmd mRNA expression in mouse brain, aorta, and heart from Tabula Muris; values normalized and log-transformed (In[1+CP10K]) as described.19 Box plot showing 25th to 75th percentiles of expression as boxes and dots representing outliers. Cell type abbreviations and full designations are given. C, HUVECs, immunostaining for VE-cadherin (green) and PALMD (gray). Hoechst 33342 shows nuclei. D, Human aortic valve en face immunostaining for CD31 (green) and PALMD (gray). Hoechst 33342 shows nuclei. E, HUVECs, qPCR of PALMD mRNA splice variants. n=3. ****P<0.0001, unpaired t test. F, PALMD immunoblot shows PALMD silencing and PALMD overexpression (OE) in HUVECs. Quantification of PALMD levels. n=3. *P=0.0201; *P=0.0241, 1-way ANOVA with Tukey correction. Black arrows in the blot indicate Myc-DDK–tagged PALMD protein (upper) and endogenous PALMD (lower), respectively. GAPDH was used for protein normalization. G, HUVECs siControl and siPALMD transduced with control or PALMD lentiviral vectors as indicated. VE-cadherin (green). OE, PALMD overexpression. H, Quantification of major axis length (MAL) in G. n=3, 15 to 16 fields of view (FoV)/condition. ***P=0.0005; ****P<0.0001. Two-way ANOVA with Tukey correction. I, HUVECs exposed to 10 dyn laminar flow, 48 hours. Arrow, flow direction. VE-cadherin (green). J, Quantification of MAL in I. n=3, 55 to 60 FoVs/condition. ****P<0.0001, Mann-Whitney. Mean±SEM shown in all graphs.
PALMD immunostaining revealed broad cytoplasmic distribution, occasionally assuming a fibrillar pattern, in primary HUVECs (Figure 1C) and in human aortic valve ECs of the intact valve (Figure 1D). Prompted by the lumen-proximal distribution of PALMD in the transmission electron microscopic analyses (Figure 1A), expression of the 2 PALMD splice variants was examined. As shown in Figure 1E, the cytosolic PALMD-KKVI form was 300-fold more abundant than the membrane-anchored PALMD-CaaX form in HUVECs.
A marked effect of PALMD silencing in HUVECs (denoted siPALMD; Figure 1F shows efficiency of PALMD silencing) was the increase in the MA length (MAL), both in static and laminar flow conditions (Figure 1G–1J). The increased MAL was not an off-target effect, as lentiviral overexpression of PALMD (Figure 1F shows level of PALMD overexpression) restored a normal MAL (Figure 1G and 1H). Under laminar flow, the MAL increased in siControl HUVECs, from a mean of 72.87 µm in static condition (Figure 1H) to 120.6 µm in flow (Figure 1J), but flow-exposed siPALMD cells increased their MAL even further (Figure 1I and 1J). The EC MAL was also increased in vivo, in the descending aorta of Palmd−/− mice. See Figure S1B for validation of loss of Palmd expression in Palmd−/− tissue; Figure S1C and S1D show the increase in EC MAL in the Palmd−/− aorta.
We further explored the in vitro behavior of siPALMD ECs and observed reduced adhesion and migration compared with siControl ECs (Figure S1E and S1F), whereas proliferation was unaffected during a 36-hour observation period (Figure S1G). In agreement with impaired adhesion, siPALMD ECs displayed lower levels of active integrin β1 and phosphorylated FAK than siControl ECs (Figure S1H–S1K). Moreover, atomic force microscopy measurements showed that although nuclear stiffness was unaffected by PALMD-deficiency, the nonnuclear stiffness measured on the cell surface away from the nucleus was lowered in siPALMD ECs (Figure S1L and S1M). Combined, these analyses showed that PALMD regulates EC adhesion, cell stiffness, and cell shape.
The PALMD-Dependent Transcriptome
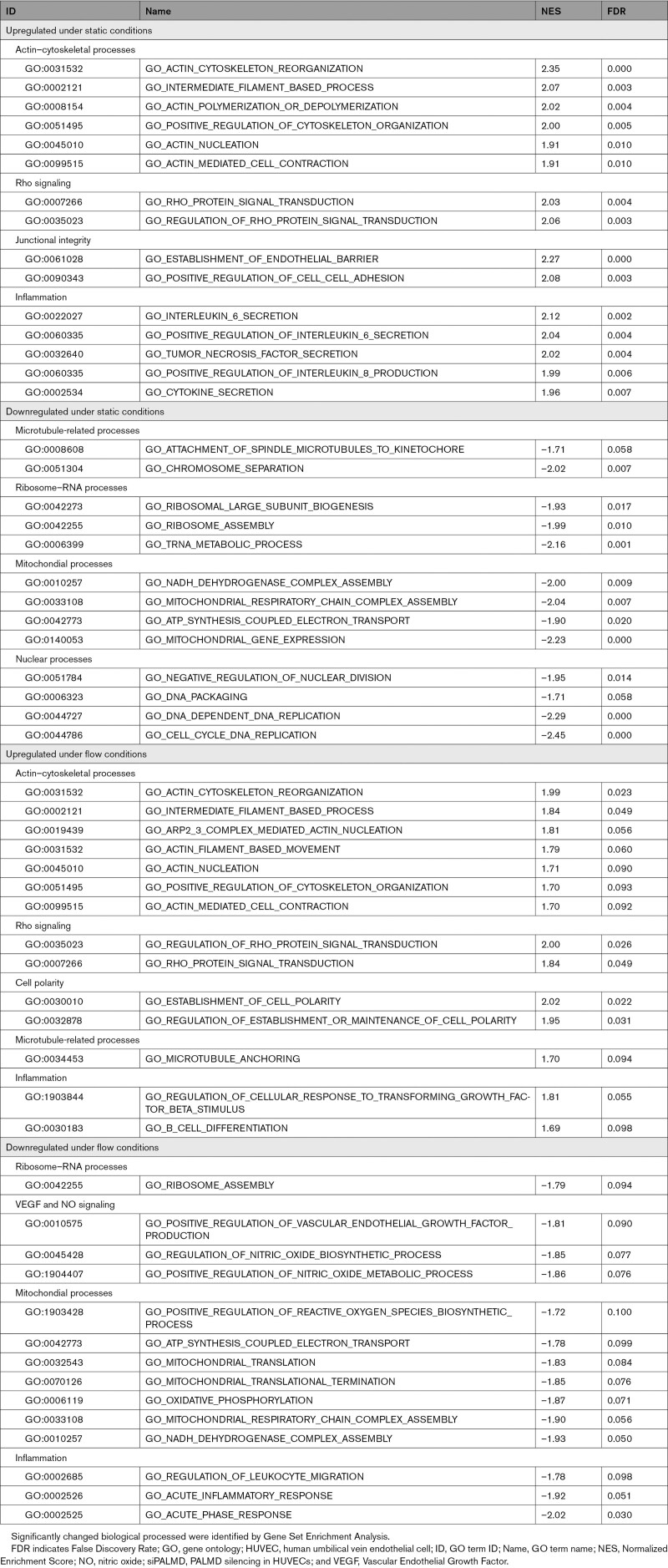
To guide further analyses of PALMD-dependent endothelial biology, RNA sequencing was performed on siPALMD and siControl ECs, kept under static or laminar flow conditions. Differentially expressed genes (DEGs) were identified as unique to each condition or shared between static and flow-treated siPALMD cells. There were 124/149 upregulated DEGs and 79/112 downregulated DEGs in static/flow conditions in the PALMD-silenced ECs compared with control (Figure S2A and S2B; see Data Set S1 for complete lists of DEGs). Gene ontology (GO) term enrichment analysis showed upregulation of several actin-related processes (eg, GO:0031532; Actin cytoskeleton reorganization) and cytoskeletal processes (eg, GO:0051495; Positive regulation of cytoskeleton organization) on PALMD silencing (Table 1, Data Set S2). Rho signaling (eg, GO:0007266; Rho protein signal transduction) was also significantly enhanced, whereas ribosomal and mitochondrial function were significantly downregulated in the absence of PALMD (Table 1). Combined, the affected GO terms implicate PALMD as highly involved in cytoskeletal control and function.
Table 1.
Top Up- or Downregulated GO Processes in siPALMD HUVECs in Static or Flow Conditions
PALMD Is Required for Formation of the Actin Cap and Nuclear Resilience
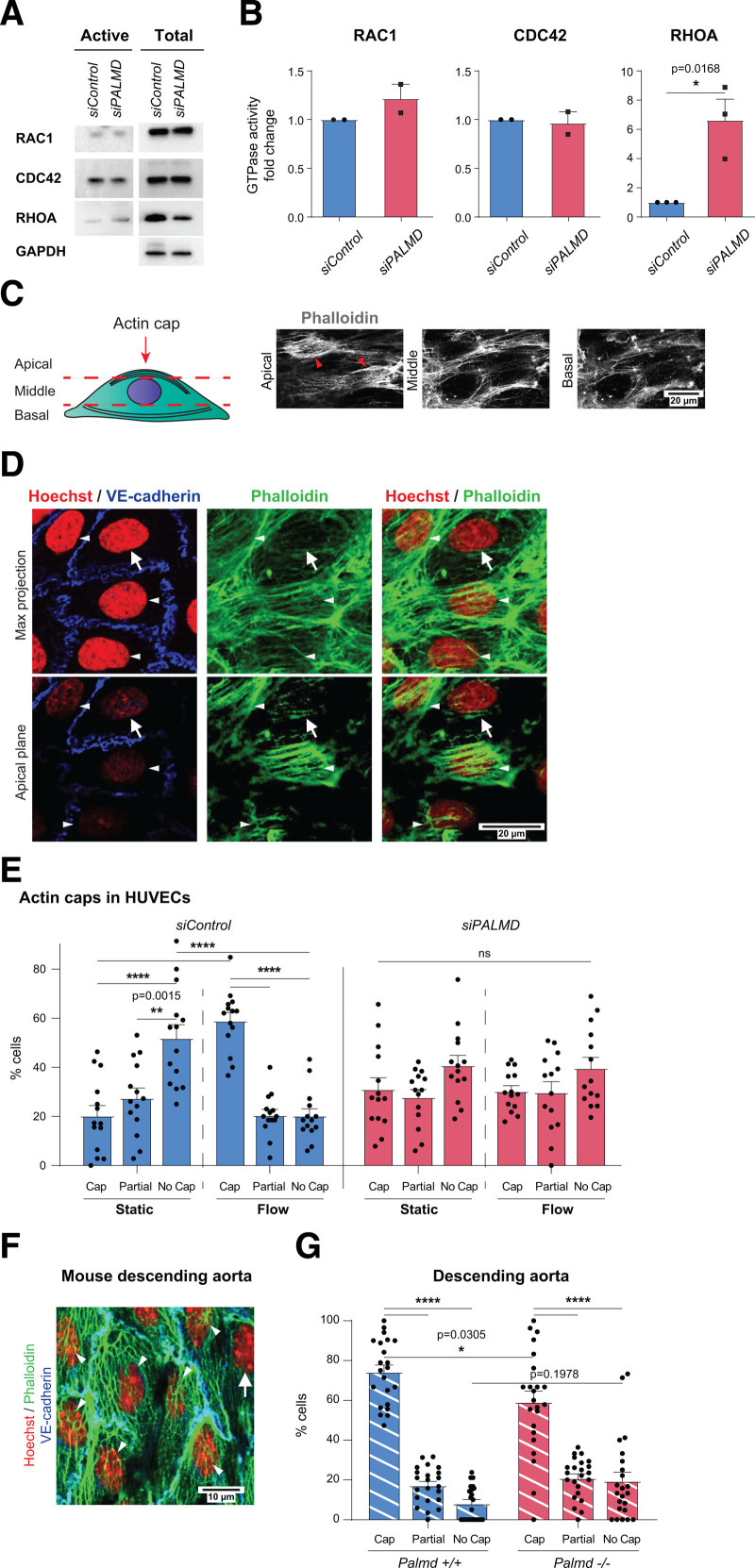
The RHO GTPase members RHOA, RAC1, and CDC42 are major regulators of the actin cytoskeleton.20 RAC1 and CDC42 expression and activity remained unchanged in siPALMD ECs. RHOA expression levels decreased slightly whereas its relative activity increased 6-fold in the PALMD-silenced static HUVECs relative to control (Figure 2A and 2B). The marked EC elongation and reduced adhesion with PALMD silencing (Figure 1G–1J, Figure S1E) made any unequivocal estimation of changes in stress fibers and cortical actin arrangement in siPALMD cells challenging. Instead, we focused on the actin cap, which is composed of perinuclear actin fibers whose assembly requires RHOA activity.21 HUVECs in static and laminar flow conditions were examined at apical, middle, and basal focal planes (Figure 2C). The actin cap was visualized in the apical plane in a fraction of the phalloidin-stained ECs as actin fibers above the nucleus, oriented parallel to the nuclear major axis (Figure 2D). In static cultures, the majority, 52%, of cells lacked an actin cap (“no cap,” Figure 2E). Remaining siControl cells displayed a complete actin cap (20%) or a partial cap with a few actin strands (28%). Strikingly, on exposure to laminar flow, 59% of siControl cells acquired an actin cap (ie, a 3-fold increase in ECs with actin cap in response to flow). In contrast, siPALMD cells showed a relatively equal distribution of caps, partial caps, and no caps in both static and flow conditions (Figure 2E). Also in the Palmd−/− descending aorta, fewer ECs displayed a perinuclear actin cap than in littermate Palmd+/+ mice (Figure 2F and 2G).
Figure 2.
PALMD regulates perinuclear actin cap formation. A and B, RAC1, CDC42, and RHOA activation assays in siControl, siPALMD HUVECs (left, A). Total lysates with GAPDH for equal loading (right, A). GTPase activities (B). n=3 (RHOA) and 2 (RAC1, CDC42). *P=0.0168, unpaired t test. C, Schematic showing focal points (apical, middle, and basal) defining the actin cap (arrowheads) at the apical point in phalloidin-stained HUVECs. D, Detection of VE-cadherin (blue), phalloidin (green), and Hoechst 33342 (red) in siControl, siPALMD HUVECs with actin caps indicated (white arrowheads) and the lack of actin cap (arrow) by maximum (max) projection (upper) and at the apical plane (lower). E, siControl, siPALMD HUVECs in static and laminar flow. Actin cap status: cap, partial, or no cap. n=3 experiment, 14 fields of view (FoV)/condition. **P=0.0015, ****P<0.0001. Two-way ANOVA with Tukey correction. F, Mouse Palmd+/+ aorta en face, VE-cadherin (blue), Hoechst 33342 (red), and phalloidin (green). Actin cap (arrowheads), without cap (arrow). G, Quantification of actin cap status in Palmd+/+ and Palmd−/− aorta. n=6 aortas and 22 to 23 FoV/genotype. *P=0.0305; ****P<0.0001, 2-way ANOVA with Tukey correction. Mean±SEM shown in all graphs.
PALMD-RANGAP1 Complex Formation Steers Subcellular Localization of XPO1
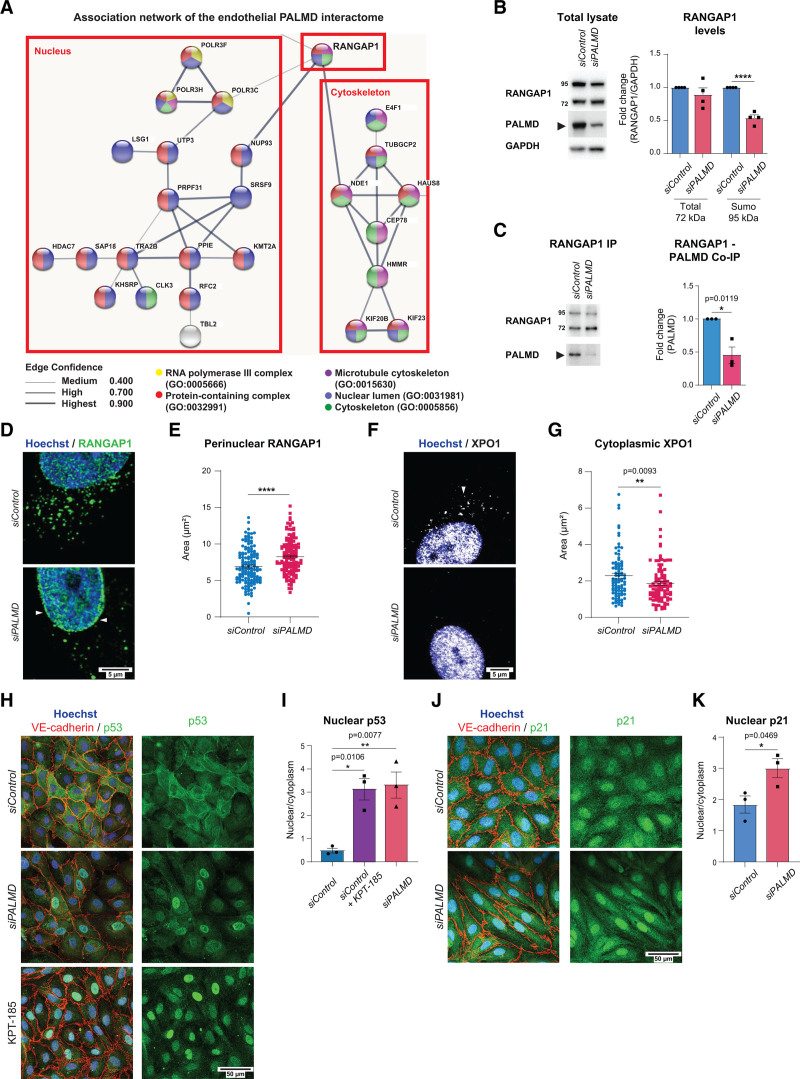
To understand the mechanism of action of PALMD, we explored its binding partners by mass spectrometry. HUVECs were transduced with a lentiviral construct encoding Myc-tagged PALMD (PALMD overexpression; see Figure 1F), and protein complexes were captured using Myc-trap, not recognizing endogenous Myc. The 136 proteins in the endothelial PALMD interactome were identified by liquid chromatography/mass spectrometry (see Data Set S3 for full list of identified proteins). Forty-seven proteins in the PALMD interactome formed an association network with 2 main clusters enriched for GO terms associated with either the nucleus or the cytoskeleton (Figure 3A). Of note, the 2 subclusters are centered around RANGAP1 (Figure 3A; Data Set S3).
Figure 3.
PALMD interacts with RANGAP1 and affects XPO1-dependent nucleocytoplasmic transport. A, Association network built on the endothelial PALMD interactome. Colors indicate GO terms that include the indicated proteins. Line thickness indicates the confidence score of the known associations. B, RANGAP1 and PALMD immunoblotting on siControl and siPALMD total lysates. Quantification (right) of RANGAP1 (total, 72 kDa and sumoylated, 95 kDa) levels normalized to GAPDH. n=4. ****P<0.0001; unpaired t test. C, RANGAP1 IP and immunoblotting for RANGAP1 and PALMD on from siControl and siPALMD HUVECs shows RANGAP1 (PALMD coimmunoprecipitation; Co-IP). Right, Level of PALMD Co-IP set to 1 in siControl. n=3. *P=0.0119, unpaired t test. D and E, Immunostaining showing perinuclear RANGAP1 (arrowheads), in siControl and siPALMD HUVECs. Quantification in E. n=3, 146 nuclei/condition. ****P<0.0001, Mann-Whitney. F and G, Immunostaining showing cytoplasmic XPO1 (arrowhead) in siControl and siPALMD HUVECs. Quantification in G. n=3 expt, 90 to 100 nuclei/condition. **P=0.0093, Mann-Whitney. H and I, Immunostaining for p53 (green), VE-cadherin (red) in siControl and siPALMD HUVECs treated or not with XPO1 inhibitor KPT-185; Hoechst33342 shows nuclei (H). Quantification of nuclear to cytoplasmic ratio in the presence and absence of PALMD (I). n=3, *P=0.0106, **P=0.0077, 1-way ANOVA with Tukey correction. J and K, Immunostaining for p21 (green) and VE-cadherin (red) in siControl and siPALMD HUVECs; Hoechst33342 shows nuclei (J). Quantification of nuclear to cytoplasmic ratio in the presence and absence of PALMD (K). n=3, *P=0.0469, unpaired t test. Mean±SEM shown in all graphs.
RANGAP1 is essential in the nucleocytoplasmic transport of proteins and RNA through regulation of the small GTPase RAN.22 RANGAP1 exists in a 72-kDa cytoplasmic pool and a 95-kDa sumoylated nuclear pore pool.23 Sumo-RANGAP1 is required for release of XPO1-cargo through RAN GTPase activity.22 On siPALMD, the 95-kDa sumoylated form of RANGAP1 decreased in total lysates, whereas levels of the 72-kDa form remained unaffected (Figure 3B). Immunoprecipitation of RANGAP1 allowed coimmunoprecipitation of PALMD, which decreased in siPALMD ECs (Figure 3C), verifying complex formation. There was a marked effect on RANGAP1 subcellular localization by silencing of PALMD, with a shift from the cytoplasm to the perinuclear region (Figure 3D and 3E). The subcellular localization of XPO1 was also affected in siPALMD cells, with a significant reduction in the cytoplasmic pool (Figure 3F and 3G).
To explore whether silencing of PALMD would affect the release of XPO1 cargo, the transcriptional regulators and known XPO1 cargoes p53 and p2124 were examined. p53 and p21 were selected for these analyses because they are distributed between the nucleus and cytoplasm in healthy ECs. Indeed, in siControl HUVECs, p53 was distributed broadly in the nucleus and the cytoplasm, whereas in siPALMD ECs, p53 was largely confined to the nucleus. A similar nuclear arrest of p53 was seen when cells were treated with the XPO1 inhibitor KPT-185 (Figure 3H and 3I).14 Also, p21 showed preferential localization to the nucleus in PALMD-silenced cells (Figure 3J and 3K). Combined, these results indicate that reduced PALMD expression disturbs nucleocytoplasmic shuttling.
In further agreement with a biological connection between PALMD and RANGAP1, silencing RANGAP1 (denoted siRANGAP1; see Figure S3A for efficiency of silencing) in static cultures resulted in a significantly increased MAL (Figure S3B and S3C). Moreover, in siRANGAP1 ECs, the subcellular localization of PALMD shifted toward the plasma membrane (Figure S3D), reminiscent of the membrane-proximal localization of Palmd in brain capillaries (Figure 1A). Still, the expression pattern of PALMD splice variants was unaffected and remained cytosolic (Figure S3E and S3F).
PALMD Is Required for Nuclear Resilience
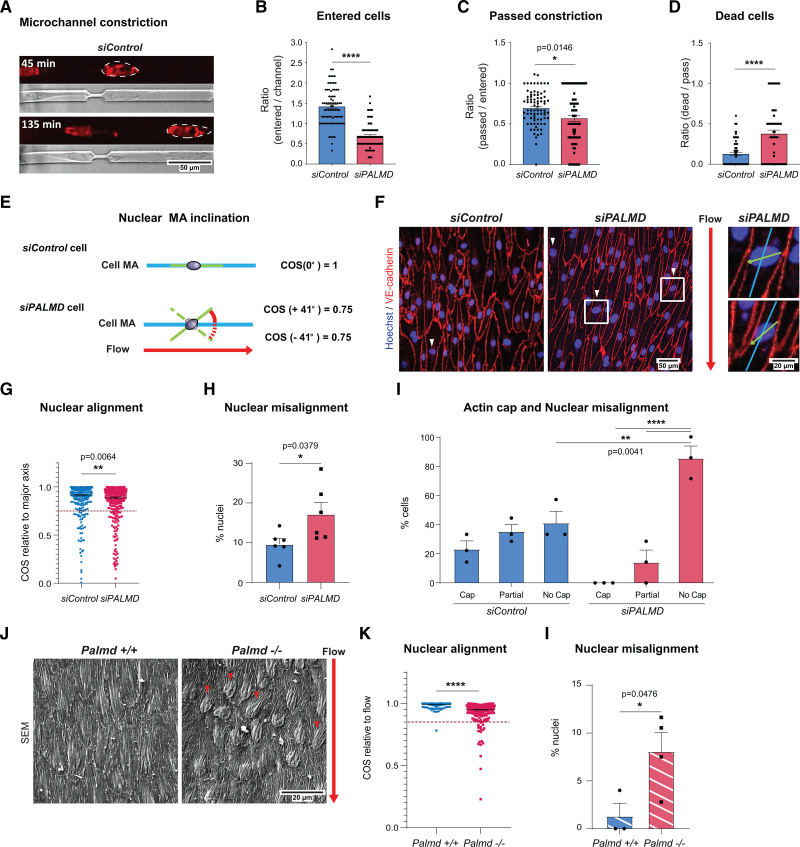
Several observations including (1) the enrichment of cytoskeletal and nuclear associated GO terms within the siPALMD transcriptome and the PALMD interactome, (2) PALMD’s complex formation with RANGAP1 and its role in nucleocytoplasmic transport, and (3) the lack of actin cap formation in flow-exposed PALMD-deficient ECs all converged on a role for PALMD in the coordination of nuclear-cytoplasmic dynamics.11 Therefore, nuclear stress in the absence of flow was assessed by tracking the ability of cells to migrate through microchannels with 4-µm constrictions (Figure 4A).17 The ability to enter and migrate through such constrictions correlates with nuclear deformability and the presence of an actin cap.25,26 On PALMD silencing, fewer cells entered the channels or passed the constriction point (Figure 4B and 4C). While attempting to pass the constriction, about 40% of the siPALMD cells died, a 4-fold increase relative to siControl cells (Figure 4D). Damage to the nuclear envelope in conjunction with mechanical stress may be the underlying reason for increased cell death.27,28 These results support a role for PALMD in nuclear resilience to mechanical stress.
Figure 4.
PALMD confers resilience to mechanical stress. A, Microchannel restriction point migration. Nuclei, NucRedTM live 647 (outlined) at time points after initiation. B and C, siControl, siPALMD HUVECs entering channel (B), or passing constriction (C). n=4, 74 to 80 channels/100 to 200 cells/expt. ****P<0.0001; *P=0.0146; unpaired t test. D, Dead/passing cells. n=4, 66 to 79 cells/expt. ****P<0.0001; unpaired t test. E, Schematic showing cell body and nuclear major axis (MA) with proper alignment (top). Angle indicating nuclear MA misalignment (below) in cosine (COS). Laminar flow direction indicated by red arrow. F, siControl, siPALMD HUVECs in laminar flow; red arrow indicates flow direction. Arrowheads indicate markedly misaligned nuclei. Boxed regions magnified to the right, showing cell body major axis (blue) and nuclear major axis (green) relative to flow. G, Nuclear alignment shown with COS as outlined in E. Dotted line defines cutoff for nuclear misalignment; COS=0.75. n=3, 416 to 420 nuclei analyzed in 2 fields of view (FoV)/expt. **P=0.0064, Mann-Whitney. H, siControl, siPALMD HUVECs, percent misaligned nuclei with COS≤0.75 as shown in G. n=3, 2 FoV/expt. *P=0.0379; unpaired t test. I, siControl, siPALMD HUVECs with actin cap, partial or no cap in cells with misaligned nuclei; COS≤0.85. n=3. **P=0.0041; ****P<0.0001, 2-way ANOVA with Tukey correction. J, Mouse Palmd+/+ and −/− aorta en face scanning electron microscopy. Red arrow indicates blood flow direction, and arrowheads indicate markedly misaligned nuclei relative to flow. K, Nuclear alignment shown with COS as outlined in E. Dotted line, COS=0.85. n=3 to 4 mice, 240 to 279 nuclei. ****P<0.0001, Mann-Whitney. L, Aorta, percent misaligned nuclei with COS≤0.85 as shown in K. n=3 to 4 mice/genotype. *P=0.0476, unpaired t test. Mean±SEM shown in all graphs.
Because the perinuclear actin cap is implicated in the control of nuclear shape and rotation,21 we next asked whether flow-induced alignment of EC nuclei in relation to the cell body was affected by loss of PALMD (Figure 4E). Indeed, in PALMD-deficient cells, nuclear alignment (expressed as the COS of the nucleus inclination angle relative to the cell body MA) was significantly increased compared with siControl ECs (Figure 4F–4H). The percentage of cells with marked misalignment between the nuclear and cell body MA (cut-off indicated in Figure 4G with a dashed line), nearly doubled in siPALMD cells (Figure 4H). A correlation between the presence of the actin cap and nuclear misalignment was observed in a subset of flow-treated siControl and siPALMD cells (COS≤0.75), scored for their actin cap status. The siControl cells showed similar distributions of cap, partial cap, and no cap among cells with misaligned nuclei. In contrast, in the siPALMD cells, most cells with misaligned nuclei lacked the actin cap, indicating a link between the suppressed cap formation and nuclear misalignment under PALMD deficiency (Figure 4I). The effect of Palmd deficiency on nuclear alignment was further explored in vivo in the mouse aorta by scanning electron microscopy analysis. Quantification of nuclear angles in relation to the flow direction showed an increased misalignment in Palmd−/− compared with wildtype aortic ECs (Figure 4J–4L). These results demonstrate that Palmd is also involved in nuclear alignment in vivo.
CAVS Is Accompanied by Nuclear Misalignment and RANGAP1 Relocalization
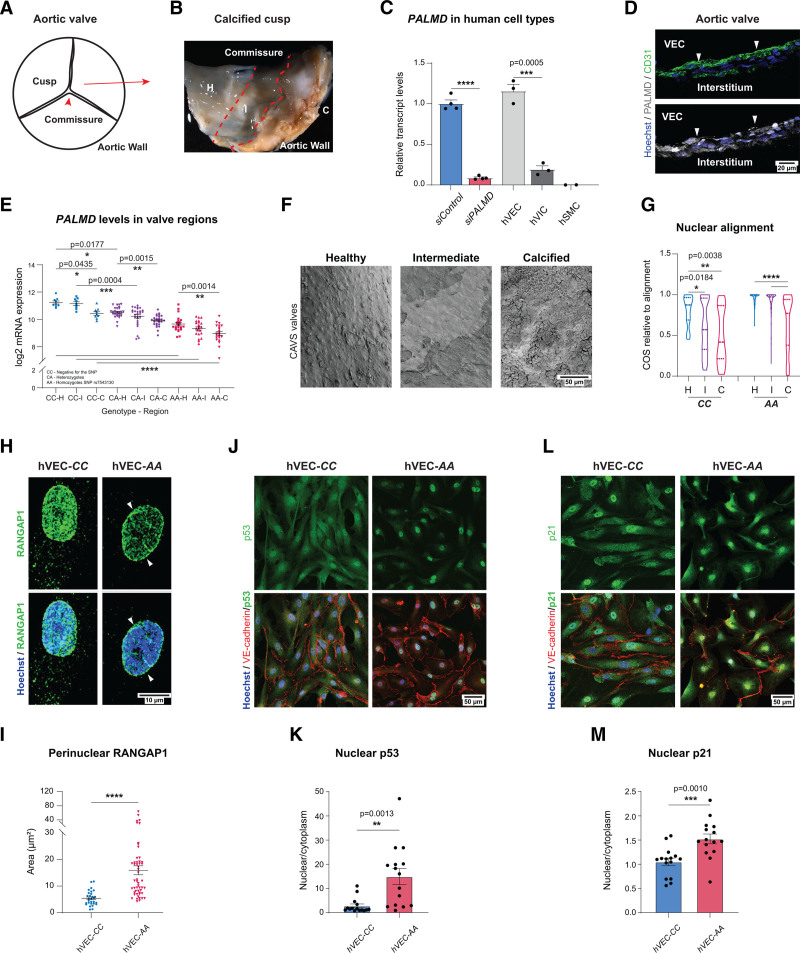
The human CAVS condition involves progressive fibro-calcific remodeling of the aortic valve cusps.29 The calcific depositions in CAVS are readily apparent macroscopically, allowing the identification of healthy, intermediate, and calcified parts of the afflicted cusps in surgically removed stenotic aortic valves (Figure 5A and 5B).
Figure 5.
PALMD-deficiency effects on EC in CAVS. A, Schematic of the aortic tricuspid CAVS valve regions: H, healthy; I, intermediate; C, calcified. B, Image of CAVS valve showing macroscopically H, I, and C regions. C, PALMD expression in siControl and siPALMD HUVECs (static condition), human valve endothelial cells (hVECs), human valve interstitial cells (hVICs), and human smooth muscle cells (hSMCs). n=2 (hSMCs), 3 (hVECs, HVICs), 4 (HUVECs). ***P=0.0005; ****P<0.0001; unpaired t test. D, CAVS valve transversal section, CD31 (green), PALMD (gray), Hoechst 33342 (blue), nuclei. PALMD in VECs (arrowheads). E, PALMD expression in 58 patients with CAVS grouped according to genotype; negative for the allele (CC), heterozygotes (CA), and homozygotes for the PALMD SNP rs7543130 (AA), analyzed in H, I, and C regions. n = 9 (CC), 27 (CA), and 22 (AA). *P=0.0435; *P=0.0177; **P=0.0015; **P=0.0014; ***P=0.0004; ****P<0.0001, 2-way ANOVA with Tukey correction. F, CAVS samples of CC and AA (SNP rs7543130), en face scanning electron microscopy. G, Nuclear MA alignment relative to flow direction, with cosine (COS) in CC and AA genotypes. n=50 (H)/170 (I)/30 (C) nuclei. *P=0.0184; **P=0.0038; ****P<0.0001, 1-way ANOVA with Tukey correction. H and I, hVECs of CC and AA genotype showing perinuclear RANGAP1. n=CC (29 nuclei), AA (52 nuclei). ****P<0.0001, Mann-Whitney. J and K, Immunostaining for p53 (green), VE-cadherin (red) in hVECs with CC and AA genotypes. Hoechst33342 (blue) shows nuclei (J). Quantification (K) shows p53 nuclear to cytoplasmic ratio. n=3, 15 fields of view (FoV).**P=0.0013, unpaired t test. L and M, Immunostaining for p21 (green), VE-cadherin (red) in patient-derived VECs with CC and AA genotypes. Hoechst33342 (blue) shows nuclei (L). Quantification (M) shows p21 nuclear to cytoplasmic ratio. n=3, 15 FoV. ***P=0.0010, unpaired t test. Mean±SEM shown in all graphs.
Analysis of different isolated valvular cell types showed preferential PALMD expression in isolated hVECs at levels similar to those seen in HUVECs (Figure 5C). In human VICs, PALMD expression was an order of magnitude lower, and expression was undetectable in human smooth muscle cells. Transversal sectioning and immunostaining confirmed colocalization of PALMD with endothelial CD31 in the human aortic valve (Figure 5D).
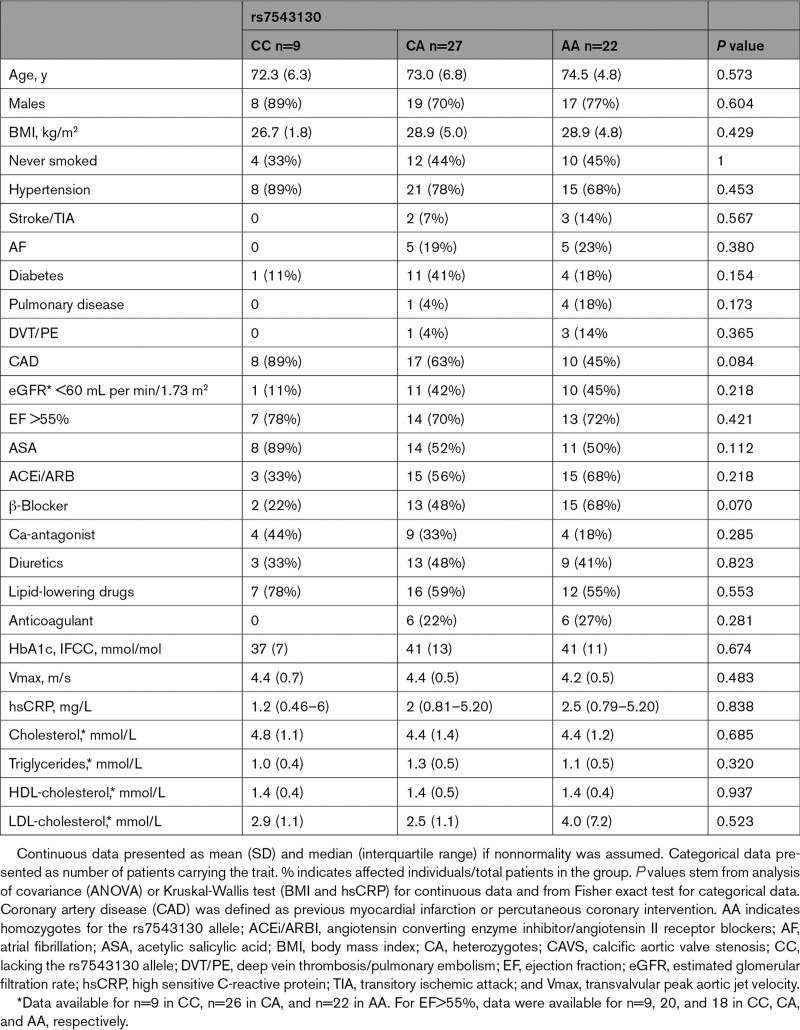
A cohort of 58 patients with severe tricuspid CAVS undergoing surgical valve replacement was examined for the presence of the PALMD SNP rs7543130 (Table 2), which strongly correlates with the disease.6 PALMD mRNA levels were compared among patient genotypes: CC, lacking the rs7543130 allele; CA, heterozygotes; and AA, homozygotes for the rs7543130 allele. PALMD mRNA expression was also compared among healthy, intermediate, and calcified parts of the valve. Figure 5E depicts the gradual decrease in PALMD levels from CC to AA genotypes, and from the healthy to the calcified regions between and within the genotypes. A 4-fold difference in PALMD expression was observed, from the highest in the CC healthy tissue to the lowest levels in the AA calcified part of the cusp (Figure 5E, note logarithmic scale).
Table 2.
Characteristics of Patients in CAVS Cohort
DEGs identified in the healthy and calcified parts of stenotic valves of AA versus CC patients were compared with those identified in siPALMD versus siControl cells in static or flow-exposed conditions (Data Set S1). Expressions of interleukin 33 (IL33), transforming growth factor-β receptor 1 (TGFBR1), TGFBR3, and LCP1/L-Plastin (lymphocyte cytosolic protein 1) were all affected in the CAVS transcriptome (Figure S4A), with effects progressively correlating with the calcification process and with the presence of the SNP. IL33 has been implicated in CAVS, acting on VICs.30 TGF-β1 activation has been associated with the calcification process.31 TGFBR3 is a negative regulator of TGF-β function,32 which may explain why it was inversely regulated compared with TGFBR1, an established transducer of TGF-β signals. LCP1 bundles actin in a Ca2+ -dependent manner and is implicated in negative regulation of integrin-dependent adhesion.33,34 LCP1 was consistently upregulated in PALMD-deficient HUVECs (Figure S2B) as well as in calcified human valve tissues (Figure S4A). Overall, the overlap between specific DEGs and their corresponding changes in expression in the siPALMD cultured cells and DEGs identified in the patient valve samples was modest (Data Set S1), likely reflecting the gradual accumulation of compensatory changes during the human disease process compared with the acute effects in a tissue culture model. Moreover, the patient valve samples used for the microarrays included multiple cell types in addition to endothelial cells. Of note, there was a gradual enrichment of DEGs (CC versus AA) present in actin-related GO terms corresponding to disease progression from healthy to calcified within the valve (Figure S4B), mirroring the upregulation of actin-regulating processes detected in the siPALMD HUVECs.
Changes in expression of other genes predisposing to CAVS, such as IL6, ALPL, and NAV1, were not detected.35 It is important that the levels of endothelial markers such as CDH5, KDR, and PECAM1 did not decrease from CC to AA genotypes (Figure S4C), confirming stable endothelial presence in the damaged tissues. Data Set S2 lists all GO:BP terms affected by PALMD deficiency in siPALMD HUVECs and the rs7543130 SNP in healthy and calcified tissue.
Strikingly, scanning electron microscopy analysis of the different regions in CAVS valves showed a progressive increase in EC nuclear misalignment relative to the flow direction (Figure 5F and 5G; Figure S5). These results support a correlation among PALMD expression, nuclear alignment, and CAVS. Further analyses were performed using primary hVECs identified as negative (CC) or homozygote (AA) for the rs7543130 SNP. There was significant accumulation of perinuclear RANGAP1 in the hVEC-AA cells carrying the SNP rs7543130 compared with the hVEC-CC (Figure 5H and 5I). Patient hVEC-CC and -AA were moreover analyzed by immunostaining for p53 and p21. In support of a role for PALMD in nucleocytoplasmic transport also in a clinically relevant condition, the hVEC-AA isolate, carrying the PALMD SNP rs7543130, showed preferential nuclear localization of both p53 and p21 (Figure 5J–5M).
Discussion
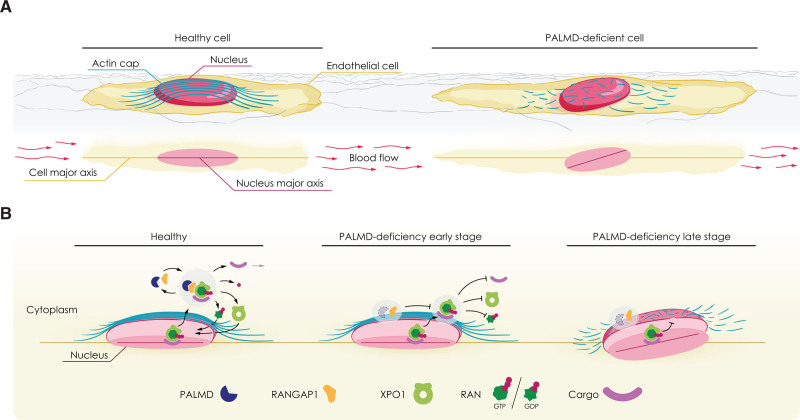
Here we provide evidence that endothelial PALMD regulates nucleocytoplasmic shuttling of XPO1 cargo, as indicated by (1) complex formation between PALMD and RANGAP1, (2) the loss of cytoplasmic RANGAP1 and XPO1, and specific reduction in levels of sumoylated RANGAP1 in siPALMD ECs, and (3) the nuclear arrest of p53 and p21 in the absence of PALMD. The delivery and release of XPO1 cargo from the nucleus to the cytoplasm is dependent on RANGAP1-induced GTPase activity of RAN.36 The role for PALMD in nucleocytoplasmic shuttling presented here is in agreement with analysis of the XPO1 proteome in HeLa cells, which identifies PALMD in association with XPO1 in a RAN-GTP–dependent manner.37 As XPO1 delivers a wide range of cargo involved in diverse biological processes such as autophagy, cytoskeletal organization, ribosome maturation, translation, and mRNA degradation, disturbed XPO1 function would have far-reaching consequences for transcriptional and translational activities.37 Thus, the altered gene regulation seen in PALMD-silenced ECs and in patient valve samples is likely to result from interference with RANGAP1-dependent release of XPO1 cargo. Bioinformatics analyses showed that cytoskeletal arrangements, likely involving not only actin, are among the top biological processes affected by loss of PALMD, in turn leading to impaired nuclear resilience to mechanical stress. In particular, PALMD was required for nuclear actin cap formation in response to mechanical stress. The actin cap is essential for cellular mechanosensation and mechanotransduction, and lack of cap formation correlates with gene regulatory changes.11,38 In accordance, nuclear alignment was disturbed in the absence of PALMD in EC cultures, the mouse aorta, and patient samples. See the schematic outline in Figure 6 for a summary of the consequences of PALMD deficiency.
Figure 6.
PALMD regulates nucleocytoplasmic shuttling via RANGAP1; short- and long-term consequences of PALMD deficiency. Schematics outlining effects of PALMD deficiency. A, Flow-exposed healthy cell (left) with actin cap protecting the nucleus and alignment of cell body and nuclear major axes. In PALMD-deficiency (right), the actin cap fails to form, and the nucleus becomes misaligned. B, In the healthy nucleus, the XPO1-cargo complex binds RAN-GTP for export to the cytoplasm, where PALMD/RANGAP1 catalyzes RAN’s GTPase activity (left). XPO1 releases its cargo followed by recycling of XPO1 and RAN-GDP back to the nucleus. In early-stage PALMD deficiency (middle), RANGAP1 assumes a perinuclear localization, RAN remains GTP-bound, and XPO1 retains its cargo. In late-stage PALMD deficiency (right), disturbed nucleocytoplasmic shuttling results in loss of actin cap, reduced nuclear resilience, and nuclear misalignment.
The gene regulatory changes established in PALMD deficiency were not dependent on flow per se. Thus, PALMD-dependent gene regulation was established also in static cultures (Table 1). Furthermore, siPALMD cells were restricted in their ability to enter and pass microchannel constrictions, which exert mechanical stress to the passing cell in a manner dependent on the size of the constriction.26 In conjunction with passing the constriction, a large fraction of PALMD-deficient cells died. Mechanical stress induced by passage through narrow constrictions may lead to nuclear membrane rupture and cell death.39 Nuclear plasticity is disturbed in pathologies with mutations in genes that regulate anchoring of the nucleus, such as laminopathies.40 The expression level of different lamins, however, was not affected by PALMD silencing (Data Set S1). Moreover, PALMD-deficient cells did not display changes in nuclear stiffness, which is a typical feature in laminopathies.41
XPO1 cargoes p53 and p21 are cell cycle regulators and tumor suppressors, which, when mutated or lost, can promote growth of tumor cells.42 The application of XPO1 inhibitors such as KPT-185 in cancer therapy is ongoing.14 Of note, in conjunction with DNA damage, p53 phosphorylated on Ser45 can induce expression of PALMD,5 providing a potential rescue mechanism to restore PALMD levels in disease. It was also suggested that PALMD may be translocated to the nucleus to regulate apoptosis,5 which was not further explored here because there was no apparent loss of cell numbers in PALMD-silenced ECs (Figure S1G), albeit under relatively short periods of analysis.
The present study identifies diminished levels of PALMD expression in ECs in aortic valves derived from patients with CAVS, which is the most common valvular heart disease in the elderly.43 CAVS is genetically linked to PALMD polymorphism,35 which here is extended to the correlation of genotype and disease stage with valvular PALMD expression. Different regions in the same CAVS valve are differently affected, with parts appearing healthy whereas others are heavily calcified. However, this separation probably reflects differences in disease progression rather than leaving certain cusps entirely unaffected; apparently healthy regions may be affected on a single-cell level. It is likely that the pathologies that become established as a consequence of loss of PALMD expression are gradual and that clear morphological changes including calcification represent insults that have moved over a “tipping point.” In agreement, when analyzing DEG accumulation in actin-related GO:BPs, a gradual increase was noted in aortic valve pathology advancing from healthy to calcified tissue (Figure S4B). Of note, the differential mechanical stresses exerted by the blood flow on blood vessels and valves may represent a predilection of the establishment of CAVS as a consequence of PALMD SNPs, despite resulting in reduced PALMD levels in all ECs. Whether oscillatory and laminar blood flow differently promote establishment of PALMD-dependent pathology remains to be investigated.
Gene regulation was complex in the human samples, likely reflecting the time span to establish the changes and the cell type heterogeneity including interstitial, smooth muscle cells, and inflammatory cells. In particular, the accompanying inflammatory process in the human condition may also contribute to the fibrotic calcifications and changes in metabolism engaging several cell types in the stenotic valve.44
The PALMD-associated SNP 6702619 was recently shown to promote valve fibrogenesis by modifying DNA shape leading to suppressed NFAT (nuclear factor of activated T cells) C2–medated transcriptional regulation of PALMD.45 Moreover, PALMD was shown to bind to actin, controlling cell shape in valve interstitial cells.45 In keeping with the present finding that PALMD expression in hVECs is 10-fold higher than in human VICs, an early endothelial dysfunction may constitute a primary event in the development of CAVS, subsequently involving other cell types such as VICs, which have been assigned a critical role in the calcification process.46
In conclusion, the data presented here show that PALMD is highly expressed in valvular and vascular ECs, where it is critical in nucleocytoplasmic transport. We demonstrate striking similarities in the effects of altered Palmd/PALMD expression between PALMD-silenced ECs, mouse aorta ECs after genetic deletion of Palmd, and hVECs isolated from samples from patients with CAVS with genetic PALMD variations, all showing disrupted nuclear alignment. The presence of misaligned nuclei correlated with reduced ability to form a protective perinuclear actin cap. These findings suggest a scenario in which the arrest in nucleocytoplasmic transport leading to nuclear confinement of XPO1 cargo exemplified by p21 and p53 predisposes to loss of perinuclear cap formation, which promotes nuclear misalignment and thereby accelerated changes in gene regulation. These findings support endothelial dysfunction as an important contributor to the development and progression of CAVS.
Article Information
Acknowledgments
The authors gratefully acknowledge Christoph Limbach (Uppsala University) and Pavel Juda (Max Planck Institute) for immune-electron microscopy; Pernilla Martinsson (Uppsala University) for biochemical analyses, and Drs Ying Sun and Liqun He (Uppsala University) for advice on bioinformatics. Concept and illustration of the artwork in Figure 6 was done by Philipp Jordan (www.JordanGraphics.eu). The authors further acknowledge expert services supplied by the BioVis imaging core facility (Uppsala), the SciLifeLab Clinical Proteomics Mass Spectrometry facility/the Chemical Proteomics and Proteogenomics facility, the Array and Analysis Facility/the SNP&SEQ Technology facility, and the Swedish National Genomics Infrastructure. For conceptualization, M.W.K. initially suggested studying Palmd function in ECs, prompted by immuno-electron microscopy observations. L.C.-W. and M.S.-J. conceptualized the presented study. The authors made the following contributions to the study: methodology, M.S.-J.; software, R.O.S., O.P., and S.N.; validation, M.S.-J., Y.J., S.-C.P., Y.D., M.H., and G.B.; formal analysis, M.S.-J., R.O.S., O.P., S.-C.P., G.B., F.A., and Q.L.; investigation, M.S.-J., R.O.S., O.P., S.-C.P., Y.J., Y.D., P.A., M.H., G.B., F.A., Q.L., C.S.D., D.F., and J.K.; resources, P.A., G.D., A.F.-C., M.F., M.W.K., M.B., and L.C.-W.; data curation, M.S.-J., R.O.S., O.P., and S.N.; writing original draft, L.C.-W.; writing and editing, all authors; visualization, M.S.-J., R.O.S., O.P., S.-C.P., Y.J., S.N., G.B., F.A., and Q.L.; supervision, L.C.-W., M.B., T.V.P., M.F., P.A., and M.W.K.; and funding acquisition, M.W.K., M.B., and L.C.-W.
Sources of Funding
This study was supported by the Swedish Research Council (2020-01349), the Knut and Alice Wallenberg Foundation (KAW 2015.0030 and KAW 2015.0275), and a Fondation Leducq Transatlantic Network of Excellence Grant in Neurovascular Disease (17 CVD 03) to L.C.-W. M.W.K. was supported by the Swedish Research Council (2003–3398) and Deutsche Forschungsgemeinschaft (Ki 324/14-1+2). Studies on human aortic valves were supported by the Swedish Research Council (grant No. 2019-01486 to M.B.), the Swedish Heart and Lung Foundation (grant No. 20180571 to M.B.), and a donation to A.C.-F. from Mr Fredrik Lundberg. S.-C.P. was supported by the Professor Nanna Svartz Foundation.
Disclosures
None.
Supplemental Materials
Supplemental Methods
Figures S1–S5
References 47–49
Data Sets S1–S3
Supplementary Material
Nonstandard Abbreviations and Acronyms
- CAVS
- calcific aortic valve stenosis
- CRM1
- chromosomal region maintenance 1
- COS
- cosine
- CoSMC
- human coronary vascular smooth muscle cell
- DEG
- differentially expressed gene
- EC
- endothelial cell
- GO
- gene ontology
- GTPase
- guanine triphosphate hydrolase
- hVEC
- human valve EC
- hVIC
- human valve interstitial cell
- FoV
- fields of view
- HUVEC
- human umbilical vein EC
- MA
- major axis
- MAL
- major axis length
- OE
- overexpression
- PALM
- paralemmin
- PALMD
- palmdelphin
- PDMS
- polydimethylsiloxane
- RANGAP1
- RAN GTPase activating protein1
- RNAseq
- RNA sequence
- SNP
- single nucleotide polymorphism
- TEM
- transmission electron microscopy
- XPO1
- Exportin1
M.W. Kilimann and M. Bäck contributed equally.
Supplemental Material is available with this article at https://www.ahajournals.org/doi/suppl/10.1161/CIRCULATIONAHA.121.054182.
For Sources of Funding and Disclosures, see page 1644.
Contributor Information
Miguel Sáinz-Jaspeado, Email: miguel.sainz@igp.uu.se.
Ross O. Smith, Email: ross.smith@igp.uu.se.
Oscar Plunde, Email: oscar.persson@ki.se.
Sven-Christian Pawelzik, Email: Sven-Christian.Pawelzik@ki.se.
Yi Jin, Email: yi.jin@igp.uu.se.
Sofia Nordling, Email: sofia.nordling@igp.uu.se.
Yindi Ding, Email: Yindi.ding@igp.uu.se.
Pontus Aspenström, Email: pontus.aspenstrom@igp.uu.se.
Marie Hedlund, Email: marie.hedlund@igp.uu.se.
Giulia Bastianello, Email: giulia.bastianello@ifom.eu.
Flora Ascione, Email: flora.ascione@ifom.eu.
Qingsen Li, Email: qingsen.li@ifom.eu.
Cansaran Saygili Demir, Email: cansaran.saygilidemir@unil.ch.
Dinesh Fernando, Email: dinesh.fernando@slu.se.
Geoffrey Daniel, Email: Geoffrey.Daniel@slu.se.
Anders Franco-Cereceda, Email: Anders.Franco-Cereceda@ki.se.
Jeffrey Kroon, Email: j.kroon@amc.uva.nl.
Marco Foiani, Email: marco.foiani@ifom.eu.
Tatiana V. Petrova, Email: tatiana.petrova@unil.ch.
Manfred W. Kilimann, Email: kilimann@em.mpg.de.
Magnus Bäck, Email: Magnus.Back@ki.se.
References
- 1.Hultqvist G, Ocampo Daza D, Larhammar D, Kilimann MW. Evolution of the vertebrate paralemmin gene family: ancient origin of gene duplicates suggests distinct functions. PLoS One. 2012; 7:e41850. doi: 10.1371/journal.pone.0041850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu B, Petrasch-Parwez E, Laue MM, Kilimann MW. Molecular characterization and immunohistochemical localization of palmdelphin, a cytosolic isoform of the paralemmin protein family implicated in membrane dynamics. Eur J Cell Biol. 2005; 84:853–866. doi: 10.1016/j.ejcb.2005.07.002 [DOI] [PubMed] [Google Scholar]
- 3.Kalebic N, Gilardi C, Stepien B, Wilsch-Bräuninger M, Long KR, Namba T, Florio M, Langen B, Lombardot B, Shevchenko A, et al. Neocortical expansion due to increased proliferation of basal progenitors is linked to changes in their morphology. Cell Stem Cell. 2019; 24:535–550.e9. doi: 10.1016/j.stem.2019.02.017 [DOI] [PubMed] [Google Scholar]
- 4.Nie Y, Chen H, Guo C, Yuan Z, Zhou X, Zhang Y, Zhang X, Mo D, Chen Y. Palmdelphin promotes myoblast differentiation and muscle regeneration. Sci Rep. 2017; 7:41608. doi: 10.1038/srep41608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dashzeveg N, Taira N, Lu ZG, Kimura J, Yoshida K. Palmdelphin, a novel target of p53 with Ser46 phosphorylation, controls cell death in response to DNA damage. Cell Death Dis. 2014; 5:e1221. doi: 10.1038/cddis.2014.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helgadottir A, Thorleifsson G, Gretarsdottir S, Stefansson OA, Tragante V, Thorolfsdottir RB, Jonsdottir I, Bjornsson T, Steinthorsdottir V, Verweij N, et al. Genome-wide analysis yields new loci associating with aortic valve stenosis. Nat Commun. 2018; 9:987. doi: 10.1038/s41467-018-03252-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z, Gaudreault N, Arsenault BJ, Mathieu P, Bossé Y, Thériault S. Phenome-wide analyses establish a specific association between aortic valve PALMD expression and calcific aortic valve stenosis. Commun Biol. 2020; 3:477. doi: 10.1038/s42003-020-01210-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thériault S, Gaudreault N, Lamontagne M, Rosa M, Boulanger MC, Messika-Zeitoun D, Clavel MA, Capoulade R, Dagenais F, Pibarot P, et al. A transcriptome-wide association study identifies PALMD as a susceptibility gene for calcific aortic valve stenosis. Nat Commun. 2018; 9:988. doi: 10.1038/s41467-018-03260-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang Y, Wu D, Birukov KG. Mechanosensing and mechanoregulation of endothelial cell functions. Compr Physiol. 2019; 9:873–904. doi: 10.1002/cphy.c180020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mammoto A, Mammoto T, Ingber DE. Mechanosensitive mechanisms in transcriptional regulation. J Cell Sci. 2012; 125(pt 13):3061–3073. doi: 10.1242/jcs.093005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim DH, Chambliss AB, Wirtz D. The multi-faceted role of the actin cap in cellular mechanosensation and mechanotransduction. Soft Matter. 2013; 9:5516–5523. doi: 10.1039/C3SM50798J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xylourgidis N, Fornerod M. Acting out of character: regulatory roles of nuclear pore complex proteins. Dev Cell. 2009; 17:617–625. doi: 10.1016/j.devcel.2009.10.015 [DOI] [PubMed] [Google Scholar]
- 13.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019; 47(D1):D442–D450. doi: 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Camacho SC, Silvers TR, Razak AR, Gabrail NY, Gerecitano JF, Kalir E, Pereira E, Evans BR, Ramus SJ, et al. Inhibition of the nuclear export receptor XPO1 as a therapeutic target for platinum-resistant ovarian cancer. Clin Cancer Res. 2017; 23:1552–1563. doi: 10.1158/1078-0432.CCR-16-1333 [DOI] [PubMed] [Google Scholar]
- 15.Gould RA, Butcher JT. Isolation of valvular endothelial cells. J Vis Exp. 2010; 46:e2158. doi: 10.3791/2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carracedo M, Persson O, Saliba-Gustafsson P, Artiach G, Ehrenborg E, Eriksson P, Franco-Cereceda A, Bäck M. Upregulated autophagy in calcific aortic valve stenosis confers protection of valvular interstitial cells. Int J Mol Sci. 2019; 20:E1486. doi: 10.3390/ijms20061486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kidiyoor GR, Li Q, Bastianello G, Bruhn C, Giovannetti I, Mohamood A, Beznoussenko GV, Mironov A, Raab M, Piel M, et al. ATR is essential for preservation of cell mechanics and nuclear integrity during interstitial migration. Nat Commun. 2020; 11:4828. doi: 10.1038/s41467-020-18580-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagy E, Andersson DC, Caidahl K, Eriksson MJ, Eriksson P, Franco-Cereceda A, Hansson GK, Bäck M. Upregulation of the 5-lipoxygenase pathway in human aortic valves correlates with severity of stenosis and leads to leukotriene-induced effects on valvular myofibroblasts. Circulation. 2011; 123:1316–1325. doi: 10.1161/CIRCULATIONAHA.110.966846 [DOI] [PubMed] [Google Scholar]
- 19.Schaum N, Karkanias J, Neff NF, May AP, Quake SR, Wyss-Coray T, Darmanis S, Batson J, Botvinnik O, Chen MB, et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018; 562:367–372. doi: 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spiering D, Hodgson L. Dynamics of the Rho-family small GTPases in actin regulation and motility. Cell Adh Migr. 2011; 5:170–180. doi: 10.4161/cam.5.2.14403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci U S A. 2009; 106:19017–19022. doi: 10.1073/pnas.0908686106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bischoff FR, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc Natl Acad Sci U S A. 1994; 91:2587–2591. doi: 10.1073/pnas.91.7.2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matunis MJ, Wu J, Blobel G. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol. 1998; 140:499–509. doi: 10.1083/jcb.140.3.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turner JG, Sullivan DM. CRM1-mediated nuclear export of proteins and drug resistance in cancer. Curr Med Chem. 2008; 15:2648–2655. doi: 10.2174/092986708786242859 [DOI] [PubMed] [Google Scholar]
- 25.Alam S, Lovett DB, Dickinson RB, Roux KJ, Lele TP. Nuclear forces and cell mechanosensing. Prog Mol Biol Transl Sci. 2014; 126:205–215. doi: 10.1016/B978-0-12-394624-9.00008-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thiam HR, Vargas P, Carpi N, Crespo CL, Raab M, Terriac E, King MC, Jacobelli J, Alberts AS, Stradal T, et al. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat Commun. 2016; 7:10997. doi: 10.1038/ncomms10997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beaudouin J, Gerlich D, Daigle N, Eils R, Ellenberg J. Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell. 2002; 108:83–96. doi: 10.1016/s0092-8674(01)00627-4 [DOI] [PubMed] [Google Scholar]
- 28.Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002; 108:97–107. doi: 10.1016/s0092-8674(01)00628-6 [DOI] [PubMed] [Google Scholar]
- 29.Lindman BR, Clavel MA, Mathieu P, Iung B, Lancellotti P, Otto CM, Pibarot P. Calcific aortic stenosis. Nat Rev Dis Primers. 2016; 2:16006. doi: 10.1038/nrdp.2016.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He YB, Guo JH, Wang C, Zhu D, Lu LM. IL-33 promotes the progression of nonrheumatic aortic valve stenosis via inducing differential phenotypic transition in valvular interstitial cells. J Cardiol. 2020; 75:124–133. doi: 10.1016/j.jjcc.2019.06.011 [DOI] [PubMed] [Google Scholar]
- 31.Jian B, Narula N, Li QY, Mohler ER, III, Levy RJ. Progression of aortic valve stenosis: TGF-beta1 is present in calcified aortic valve cusps and promotes aortic valve interstitial cell calcification via apoptosis. Ann Thorac Surg. 2003; 75:457–465. doi: 10.1016/s0003-4975(02)04312-6 [DOI] [PubMed] [Google Scholar]
- 32.Chu W, Li X, Li C, Wan L, Shi H, Song X, Liu X, Chen X, Zhang C, Shan H, et al. TGFBR3, a potential negative regulator of TGF-β signaling, protects cardiac fibroblasts from hypoxia-induced apoptosis. J Cell Physiol. 2011; 226:2586–2594. doi: 10.1002/jcp.22604 [DOI] [PubMed] [Google Scholar]
- 33.Namba Y, Ito M, Zu Y, Shigesada K, Maruyama K. Human T cell L-plastin bundles actin filaments in a calcium-dependent manner. J Biochem. 1992; 112:503–507. doi: 10.1093/oxfordjournals.jbchem.a123929 [DOI] [PubMed] [Google Scholar]
- 34.Tseng HY, Samarelli AV, Kammerer P, Scholze S, Ziegler T, Immler R, Zent R, Sperandio M, Sanders CR, Fässler R, et al. LCP1 preferentially binds clasped αMβ2 integrin and attenuates leukocyte adhesion under flow. J Cell Sci. 2018; 131:jcs218214. doi: 10.1242/jcs.218214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thériault S, Dina C, Messika-Zeitoun D, Le Scouarnec S, Capoulade R, Gaudreault N, Rigade S, Li Z, Simonet F, Lamontagne M, et al. ; D.E.S.I.R. Study Group. Genetic association analyses highlight IL6, ALPL, and NAV1 as 3 new susceptibility genes underlying calcific aortic valve stenosis. Circ Genom Precis Med. 2019; 12:e002617. doi: 10.1161/CIRCGEN.119.002617 [DOI] [PubMed] [Google Scholar]
- 36.Boudhraa Z, Carmona E, Provencher D, Mes-Masson AM. Ran GTPase: a key player in tumor progression and metastasis. Front Cell Dev Biol. 2020; 8:345. doi: 10.3389/fcell.2020.00345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirli K, Karaca S, Dehne HJ, Samwer M, Pan KT, Lenz C, Urlaub H, Görlich D. A deep proteomics perspective on CRM1-mediated nuclear export and nucleocytoplasmic partitioning. Elife. 2015; 4:e11466. doi: 10.7554/eLife.11466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tojkander S, Gateva G, Lappalainen P. Actin stress fibers–assembly, dynamics and biological roles. J Cell Sci. 2012; 125(pt 8):1855–1864. doi: 10.1242/jcs.098087 [DOI] [PubMed] [Google Scholar]
- 39.Maciejowski J, Hatch EM. Nuclear membrane rupture and its consequences. Annu Rev Cell Dev Biol. 2020; 36:85–114. doi: 10.1146/annurev-cellbio-020520-120627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hah J, Kim DH. Deciphering nuclear mechanobiology in laminopathy. Cells. 2019; 8:E231. doi: 10.3390/cells8030231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen B, Co C, Ho CC. Cell shape dependent regulation of nuclear morphology. Biomaterials. 2015; 67:129–136. doi: 10.1016/j.biomaterials.2015.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol. 2010; 2:a000935. doi: 10.1101/cshperspect.a000935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nkomo VT, Gardin JM, Skelton TN, Gottdiener JS, Scott CG, Enriquez-Sarano M. Burden of valvular heart diseases: a population-based study. Lancet. 2006; 368:1005–1011. doi: 10.1016/S0140-6736(06)69208-8 [DOI] [PubMed] [Google Scholar]
- 44.Cho KI, Sakuma I, Sohn IS, Jo SH, Koh KK. Inflammatory and metabolic mechanisms underlying the calcific aortic valve disease. Atherosclerosis. 2018; 277:60–65. doi: 10.1016/j.atherosclerosis.2018.08.029 [DOI] [PubMed] [Google Scholar]
- 45.Chignon A, Rosa M, Boulanger MC, Argaud D, Devillers R, Bon-Baret V, Mkannez G, Li Z, Rufiange A, Gaudreault N, et al. Enhancer-associated aortic valve stenosis risk locus 1p21.2 alters NFATC2 binding site and promotes fibrogenesis. iScience. 2021; 24:102241. doi: 10.1016/j.isci.2021.102241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donato M, Ferri N, Lupo MG, Faggin E, Rattazzi M. Current evidence and future perspectives on pharmacological treatment of calcific aortic valve stenosis. Int J Mol Sci. 2020; 21:E8263. doi: 10.3390/ijms21218263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li QS, Lee GY, Ong CN, Lim CT. AFM indentation study of breast cancer cells. Biochem Biophys Res Commun. 2008; 374:609–613. doi: 10.1016/j.bbrc.2008.07.078 [DOI] [PubMed] [Google Scholar]
- 48.Liebermeister W, Noor E, Flamholz A, Davidi D, Bernhardt J, Milo R. Visual account of protein investment in cellular functions. Proc Natl Acad Sci U S A. 2014; 111:8488–8493. doi: 10.1073/pnas.1314810111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005; 102:15545–15550. doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings reported in this study are available from the corresponding author on reasonable request.
The mass spectrometry analysis data set has been deposited to the ProteomeXchange Consortium via the PRIDE13 partner repository with the dataset identifier PXD024887.
The RNA sequence metadata on human ECs have been deposited with the Swedish Science for Life Laboratories and can be accessed through https://doi.org/10.17044/scilifelab.14270159.
Requests for the human CAVS transcript array data set should be directed to Magnus Bäck (magnus.back@ki.se).
Requests for Palmd gene-modified mice and Palmd antimouse antibodies should be directed to Manfred W. Kilimann (kilimann@em.mpg.de).