Abstract
Chemotherapy is one of the most effective cancer treatments. Starting from the discovery of new molecular entities, it usually takes about 10 years and 2 billion U.S. dollars to bring an effective anti-cancer drug from the benchtop to patients. Due to the physiological differences between animal models and humans, more than 90% of drug candidates failed in phase I clinical trials. Thus, a more efficient drug screening system to identify feasible compounds and pre-exclude less promising drug candidates is strongly desired. For their capability to accurately construct in vitro tumor models derived from human cells to reproduce pathological and physiological processes, microfluidic tumor chips are reliable platforms for preclinical drug screening, personalized medicine, and fundamental oncology research. This review summarizes the recent progress of the microfluidic tumor chip and highlights tumor vascularization strategies. In addition, promising imaging modalities for enhancing data acquisition and machine learning-based image analysis methods to accurately quantify the dynamics of tumor spheroids are introduced. It is believed that the microfluidic tumor chip will serve as a high-throughput, biomimetic, and multi-sensor integrated system for efficient preclinical drug evaluation in the future.
INTRODUCTION
Today, cancer has become one of the most atrocious threats to human health. It is estimated that by 2021, there will be nearly 2 × 106 new cancer cases and more than 600 000 cancer deaths in the United States.1 Besides radiotherapy and surgical resection, chemotherapy is the most common and reliable anti-cancer treatment.2 Although cancer pharmacological treatment has made great progress in the past decade, it is still challenging to effectively discover and develop new anti-cancer drugs.3 In general, it takes over 10 years to bring an effective drug from research to clinic.4 In the drug development process, drug candidate screening is one of the most important but time-consuming stages. Herein, an appropriate preclinical tumor model can effectively eliminate unqualified drug candidates and improve screening efficiency. Traditionally, animal models have played a vital role in oncology research and drug testing. However, these methods are limited by the long experimental period, excessive number of animals consumed, high maintenance cost of the facility, and social ethics. In addition, the tumor characteristics from clinical patients and animal models vary greatly. Therefore, there is still a huge disparity between the efficacy of drug candidates in animal models and the final results of clinical patients, which often leads to misjudgments in drug test, increases in R&D costs, and extensions of the development cycle.5
Since the new century, cellular-level drug screening technology has gradually matured. Tumor models derived from human cells are used to quantitatively characterize cell viability, morphogenesis, proliferation, and gene expression under the action of different drug candidates.6 For a long time, the two-dimensional (2D) monolayer cancer model has been a mainstream in vitro oncology model for early preclinical drug screening and testing. However, 2D adherent growth is not the actual way of cell growth in the human body. Lack of cell-to-cell and cell-to-extracellular matrix (ECM) interactions lead to altered cell behaviors, such as gene expression, differentiation, and cell metabolism.7 Significantly, the physiological correlation between the in vitro model used for drug evaluation and the real tumor from patients is very critical. The more realistic the in vitro model, the more reliable the preclinical drug screening.8 Currently, most of the anti-cancer drug candidates screened by 2D models failed to pass clinical trials in the later stages.9
Nowadays, three-dimensional (3D) cellular models for drug testing have been widely studied and are attracting more and more attention.10 Compared to the conventional 2D monolayer cell culture model, 3D cell culture allows cells to grow in all directions and recapitulate more in vivo behaviors and functions, such as self-organization, proliferation, and protein expression.11 Among them, 3D multicellular tumor spheroid is one of the most used models for preclinical anti-cancer drug screening (Fig. 1). Based on microfluidic technology, tumor spheroids can be manufactured through standardized on-chip production, thereby reducing the individual differences between models, and improving reproducibility and consistency. In addition, the tumor microenvironment (TME) can also be restored in the on-chip microphysiological system to accurately reproduce the tumor–microenvironment interaction.12 Eventually, the microfluidic tumor chip technology is believed to play an important role in advancing fundamental cancer biology research, high-throughput preclinical drug screening, and precise personalized medicine.13
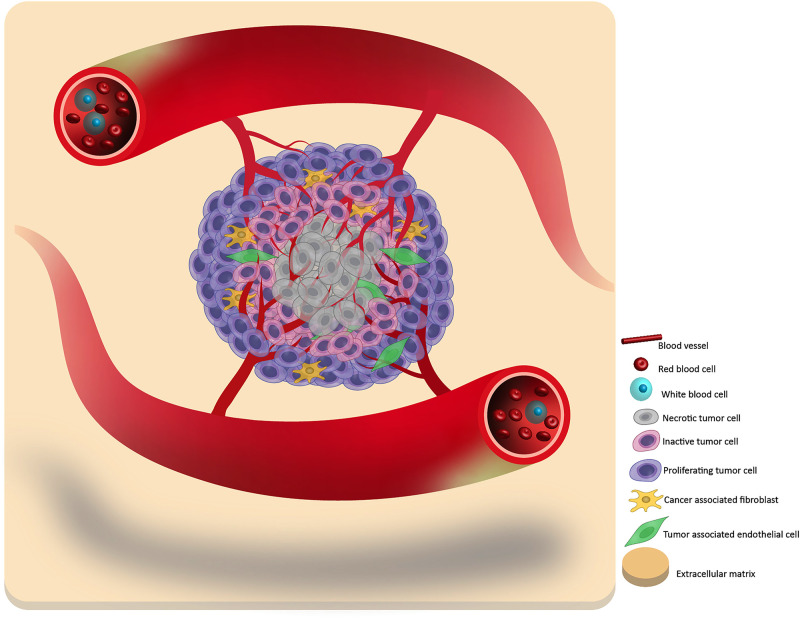
FIG. 1.
A schematic of the primary tumor structure and composition. In general, a tumor can be considered to contain three layers: the necrotic core, the inactive quiescent middle layer, and the proliferative outer layer. By hijacking the surrounding blood vessels, the tumor cells can obtain oxygen and nutrients to promote their growth. In addition, some tumors can also incorporate the surrounding tissue into a part of itself, such as cancer-associated fibroblasts and tumor-associated endothelial cells, which increases the tumor heterogeneity and complexity.
In view of the vigorous development of the field of microfluidic tumor chips, many review articles have been published.14 Some of them introduced the biological background of tumor spheroid in detail,15,16 some were devoted to the advanced fabrication technology,17,18 and some were focused on constructing the complex tumor microenvironment.19,20 This review will serve as a bridge to link the preceding accomplishment and the following advancement. Besides summarizing the latest representative achievements of microfluidic tumor chips, we will make a comprehensive review of microfluidic tumor chips in the order from homotypic tumor spheroid fabrication to vascularization-based tumor microenvironment construction. Meanwhile, as the source technology of microfluidic tumor chips, organ on a chip technology will also be briefly introduced. Finally, we will present some classic cases of novel imaging methods for data acquisition and machine learning-based image analysis to accurately quantify tumor spheroid dynamics. We hope that this review will help researchers get the basic engineering concepts for constructing microfluidic tumor models and encourage new ideas in the future.
CONVENTIONAL TUMOR CELL SPHEROID MODEL CONSTRUCTION TECHNIQUES
Cell self-assembly has always been the natural law guiding the formation of all living organisms, whether in vitro or in vivo.21 When cancer cells cannot adhere to the substrate in the in vitro environment, they will spontaneously aggregate and secrete extracellular matrix to wrap themselves into multicellular tumor spheroids.22 In early studies, the manufacture of tumor spheroids was often out of touch with the subsequent drug screening process. For instance, agitation-based approach was one of the early methods for generating tumor spheroids.23 However, most of these methods lack guarantees on the size and viability of mass-produced tumor spheroids. When the tumor spheroids were collected for further experiments, these by-effects would cause a certain degree of interference to the final experimental results.
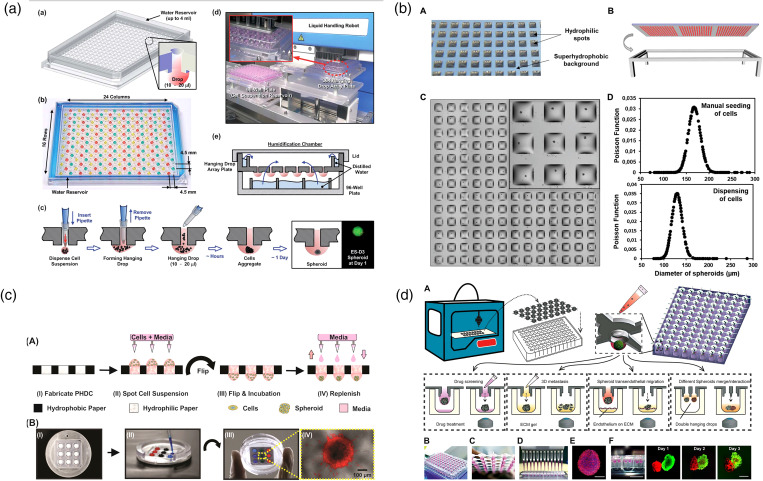
In the past ten years, microfluidic tumor chip technologies have emerged rapidly. The basic idea of microfluidic tumor spheroids manufacturing is to build micro-reactors to create non-attachable space, leading to cell self-assembly.24–31 Initially, the hanging drop method is one of the most classic and well-established tumor spheroid fabrication methods.32,33 The balance between surface tension and gravity keeps the droplets converging on the surface of the substrate without falling. Here, the droplets function as the micro-reactors and the liquid–air interface functions as the non-attachable substrate to prevent cell adhesion. The suspended cells will aggregate into spheroids at the bottom of the droplet under gravity, and the spheroid size will be dependent on the droplet dimension and initial cell concentration. Tung et al. reported a hanging drop based 3D tumor spheroid culture platform in 2011 [Fig. 2(a)].34 Developed tumor spheroids were able to form in one day, and diverse preclinical drug screening could be executed in each hanging droplet respectively.
FIG. 2.
Conventional “hanging drop method” for producing tumor spheroids for drug screening. (a) A 384-well format hanging drop culture plate that allows both tumor spheroid formation and subsequent drug testing.34 Reprinted with permission from Tung et al., Analyst 136, 473 (2011). Copyright 2011 Royal Society of Chemistry. (b) A droplet-microarray platform based on hydrophilic–superhydrophobic patterning for the formation of single tumor spheroids array.35 Reprinted with permission from Popova et al., Small 15, e1901299 (2019). Copyright 2019 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (c) A paper-based hanging drop chip for large scale tumor spheroids formation at low cost.36 Reprinted with permission from Michael et al., ACS Appl. Mater. Interfaces 10, 33839 (2018). Copyright 2018 American Chemical Society. (d) A 3D-printed hanging drop dripper for automated tumor spheroids generation, metastatic migration study, and drug screening.37 Reprinted with permission from Zhao et al., Sci. Rep. 9, 19717 (2019). Copyright 2019 Springer Nature.
Nowadays, to break through the limitation of tedious manual operation, the hanging drop technology continues to develop in the direction of automation and high-throughput. Recently, Popova et al. reported a “droplet-microarray platform” for one-stop tumor spheroid formation and drug screening [Fig. 2(b)].35 They used hydrophilic–superhydrophobic patterning to create a hanging droplet array on hydroxyethyl-methacrylate (HEMA)–ethylene dimethacrylate (EDMA) slides. Due to the great difference in water affinity between the hydrophilic and superhydrophobic parts of the surface, an array of separated homogeneous droplets could spontaneously form on the device surface. Also, Michael et al. reported a paper-based hanging drop chip for tumor spheroid fabrication [Fig. 2(c)].36 Zhao et al. announced a 3D-printed hanging drop dripper product. The printed artifact is simple and practical and can be directly mounted on the conventional 96/384-well plate [Fig. 2(d)].37 Compared to the traditional hanging drop methods, these innovative works simplify device fabrication, prove the possibility of further reducing costs, and pave the way for potential commercialization in the future. However, the hanging drop technology is usually limited by its inherent flaws. Due to the fragile structure of the liquid droplets, it was still difficult to change the culture medium for long-term tumor spheroid culture, which leads to fast nutrient depletion and waste accumulation. Moreover, all the spheroids were cultured in a purely static environment, while some mechanical stimulations, such as interstitial flow, have been demonstrated to better support tumor growth.38 Although not perfect, the hanging drop approaches are still considered the pioneering work to link the tumor spheroids generation and the following drug testing.
MICROFLUIDIC TUMOR-ON-A-CHIP
Microfluidic fabrication of homotypic tumor spheroid-on-a-chip
Since the new century, the maturity of microelectromechanical systems (MEMS) technology greatly boosts the development of the microfluidic tumor chip.39,40 Photolithography, vapor deposition, and other micro-processing techniques have enabled the preparation of some complex microstructures, which are necessary for precise manipulation of microscale chemical reagents and perfusion. Moreover, more and more electronic biochemical sensors are also coupled into the microfluidic tumor chip system.41 Consequently, real-time in situ monitoring of tumor response allows drug screening with much higher temporal and spatial resolution. It can be said that now the microfluidic tumor chip technology is right in the golden ages.
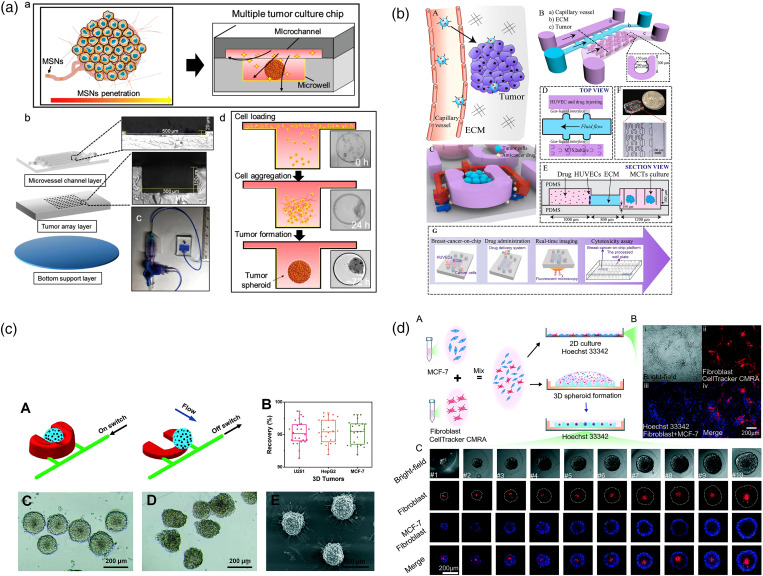
The most common application of a microfluidic tumor chip is serving as a one-stop platform, seamlessly connecting tumor spheroid model construction and preclinical drug screening for oncology research.45–50 In such systems, single tumor spheroids can be generated and maintained at fixed locations for drug screening and follow-up characterization. The basic microfluidic tumor chips can be classified into the following two categories: microwell42,51–60 [Fig. 3(a)] and U-shaped microanchor44,61–66 [Fig. 3(b)]. These two designs are similar in function but different in shape. In general, these chips are composed of the main fluid channel and the structure array.67 After being loaded into the main fluid channel, suspended tumor cells are captured by each microstructure. The trapped cells have no place to attach but to self-assemble into the tumor spheroids. After tumor spheroid formation, the function of the main channel will turn to medium exchange and drug administration [Fig. 3(a)].42 Since the established tumor spheroids are firmly stuck by the microstructures, dynamic fluid flow can continue to be applied in the main channel without harassing the spheroids or carrying them away. This perfusion function facilitates the long-term tumor spheroids culture, which is usually limited in the hanging drop methods. Some studies also reported that appropriate mechanical perturbation of the flow can promote tumor growth since it mimics the interstitial flow in in vivo tumor tissue to a certain extent.68
FIG. 3.
Classic “microfluidic tumor chip” designs. (a) Microwell-based multiple tumor spheroids culture chip (MTC-chip) cultured tumor under various flow conditions.42 Reprinted with permission from Zhuang et al., Adv. Sci. 6, 1901462 (2019). Copyright 2019 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (b) A U-shaped array based 3D breast-cancer-on-a-chip for the evaluation of nanoparticle-based drug delivery systems.43 Reprinted with permission from Chen et al., Anal. Chim. Acta 1036, 97 (2018). Copyright 2018 Elsevier B.V. (c) Pneumatically actuated U-shaped arrays for one-step cell localization, spheroids self-assembly, and real-time analysis.44 Reprinted with permission from Liu et al., Lab Chip 15, 1195 (2015). Copyright 2015 The Royal Society of Chemistry. (d) Liquid dome-assisted formation of gradient-sized spheroids on an agarose chip.29 Reprinted from Fang et al., Lab Chip 19, 4093 (2019). Copyright 2019 The Royal Society of Chemistry.
With the advancement of fabrication technology, the structure of the microfluidic tumor chips becomes more and more dynamic and flexible. In these specific cases, the conventional sealed microfluidic tumor chips described above would be inconvenient for retrieving the tumor spheroids for downstream analysis. To solve this problem, Liu et al. integrated a flexible pneumatic microanchor array design with the conventional U-shaped tumor chip [Fig. 3(c)].44,66 When the pneumatic switch was turned on, the inflatable anchor would stand upright to capture the suspended tumor cells. After nanomedicine treatment, the tumor spheroids from the chip could be selectively carried out by turning off the pneumatic microstructures. The recovery feature builds up a practical bridge between tumor-on-a-chip modeling and off-chip pathological characterization. In addition to pursuing a complex microfluidic tumor chip design, some groups, on the contrary, further simplify the chip manufacturing process and make it easier to operate. For example, Fang et al. reported an agar microfluidic tumor chip that can produce a tumor spheroid with only 250 μl cell suspension [Fig. 3(d)].29 Benefiting from a 3D-printed mold, an agar chip can be easily generated through the molding method, which is more advantageous for routine laboratories and easy to popularize.
High-throughput fabrication of tumor spheroid via the microfluidic droplet technique
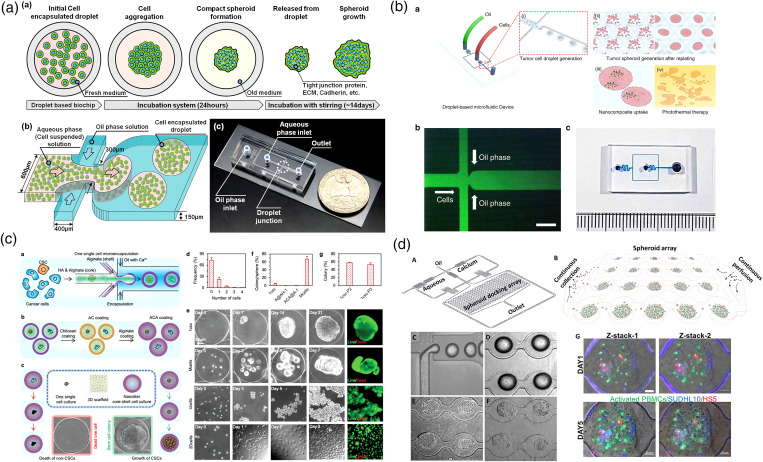
Besides solid microstructures, liquid microstructures can also be engaged in tumor spheroid modeling. Microfluidic droplet-based techniques stand out for their high-throughput and uniformity.70,73–76 In the channel of the microfluidic chip, two continuous fluids that are immiscible with each other will generate a stable and ordered non-continuous flow at the interface, which is the so-called microdroplets. By adjusting the chip design and the flow rate of the two-phase liquid, the size, shape, and frequency of the microdroplets can be precisely regulated.77 In particular, the water-in-oil (W/O) emulsion is the most used system for tumor spheroid preparation. A suspended cell-laden medium is wrapped into microdroplets and immersed in a continuous oil phase. These microdroplets can function as the bioreactors leading to the tumor spheroid formation in days. In some way, microdroplet-based tumor spheroid construction methods are like a “fully automatic version” of the conventional “hanging drop” method. The difference lies in that the cell-laden droplets are immersed in the culture medium instead of sticking to the plate. Kwak et al. reported a uniform-sized 3D tumor spheroid fabrication system in 2018 [Fig. 4(a)].69 Cell-loaded microdroplets could be generated at a rate of 1000 droplets/min. After 24-hour incubation, mature tumor spheroids were released from the oil capsule and recollected for downstream anti-cancer drug testing. Therefore, the microdroplet is considered one of the most efficient techniques to fabricate tumor spheroids. However, the oil capsule shells limit the exchange of the nutrient medium. The mature tumor spheroids need to be timely released from the microdroplets after one day; otherwise, the nutrients in the droplets will be exhausted. Without the microfluidic chamber's restriction, the tumor spheroids that were mass-produced in a short time can be recollected, transferred, and applied to a variety of tests, such as photothermal therapy (PTT) [Fig. 4(b)].70
FIG. 4.
Microdroplet technique for high-throughput tumor spheroid generation. (a) A droplet-based microfluidic system that generates cancer cells containing microdroplets at high yield (1000 droplets/min).69 Reprinted with permission from Kwak et al., J. Controlled Release 275, 201 (2018). Copyright 2018 Elsevier B.V. (b) Size-tunable brain tumor spheroids generated by microfluidic droplets are applied for photothermal therapy (PTT) testing.70 Reprinted with permission from Lee et al., Microsyst. Nanoeng. 6, 52 (2020). Copyright 2020 Springer Nature. (c) A droplet-based microfluidic device for constructing tumor spheroids from single cancer cells.71 Reprinted with permission from Wang et al., Adv. Sci. 7, 2000259 (2020). Copyright 2020 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (d) An integrated droplet-based microfluidic platform for dynamic analysis of cellular interaction, proliferation, and therapeutic efficacy via spatiotemporal profiling on chip.72 Reprinted with permission from Sabhachandani et al., J. Controlled Release 295, 21 (2019). Copyright 2018 Elsevier B.V.
Moreover, if the water phase is replaced with the pre-gel solution, the cell-laden microdroplets can be cured into 3D cell-laden hydrogel beads later [Fig. 4(c)].71 It has been reported that the cured hydrogel can serve as a scaffold, which facilitates the tumor spheroid formation.72,78,79 After being washed out from the oil phase, the tumor structures can still be maintained stably in hydrogel beads until spheroid maturation. Sabhachandani et al. improved the microfluidic droplet technique with an “anchor array” design [Fig. 4(d)].72 In this system, cells are loaded in alginate beads. Once the alginate gets solidified, the oil phase can be washed away with a medium without disrupting the cell spheroids. The tumor spheroids in alginate beads can be trapped on the chip for long-term culture until spheroid maturation for dynamic drug screening.80 This technique inherits the advantages of rapid fabrication of the microdroplet system and the conventional microwell-based tumor chip, while being more flexible, practical, and straightforward for drug testing.
Device-less manufacturing of tumor spheroids
Recently, more innovative materials and interdisciplinary elements are introduced into the microfluidic “tumor chip” design. Based on the concept of microfluidics, some device-free and device-less technologies have been developed accordingly for tumor spheroid fabrication.81 For example, Samara et al. tried to substitute traditional polydimethylsiloxane (PDMS) material with paper to fabricate a “tumor chip” for drug screening, which is claimed to be a more cost-efficient experimental procedure.82 Also, Shi et al. reported a device-free way to generate tumor spheroids for drug screening.83 By rolling up detached cell sheets, a large quantity of tumor spheroids can be formed with controllable size and high uniformity. As for device-less approaches, magnetic levitation is one of the latest emerging trends for scaffold-free tumor spheroid model construction.84–88 Activated by an external magnetic field, cells labeled with magnetic nanoparticle are gathered into cell clusters. Ho et al. can even directly magnetically pattern HeLa tumor spheroids into the desired topography.85 Similarly, acoustics is another popular element that frequently shows up in tumor spheroid-related studies.89–92 In acoustic scenarios, cells are compressed and reallocated together by the mechanical sound waves propagating in the medium. Rather than passively waiting cells to migrate together, these proactive manipulations can assemble cells in a rapid way, which greatly shortens the bio-fabrication time. These device-less methods rely more on external physics systems rather than sophisticated microfluidic chamber designs. However, these interdisciplinary technologies still need to be optimized. For example, magnetic bead-labeled on cells are usually hard to be removed later. It is difficult to guarantee that these remaining beads will not have any effect on the subsequent growth of the cells. In general, there is still a long way to go to integrate these new elements into the desired “all-in-one” tumor chip system (tumor formation, drug screening, and post-analysis).93,94
MODELING TUMOR–VASCULATURE INTERACTIONS IN MICROFLUIDIC PHYSIOLOGICAL SYSTEMS
Importance of incorporating a functional vascular system into tumor models for drug screening
Although tumors start as a small group of cells through a series of acquired mutations and epigenetic changes, the final tumor is a complex tissue composed of numerous heterogeneous tumor cells and tumor-related cell groups.95 Compared with regular 2D monolayer models, the 3D homotypic tumor spheroids can indeed restore some characteristics of solid tumors, such as their spatial constructure, physiological response, and part of gene expression.96,97 However, they cannot truthfully represent the tumor heterogeneity and the intricate the interaction between the tumor and the local TME,12,98–100 which are all sources of chemotherapy resistance.
In addition to its own complex cell composition, the in vivo tumor can also recruit neighboring normal tissue cells and transform them into parts of itself, such as cancer-associated fibroblasts101 and tumor-associated endothelial cells.102 Furthermore, the in vivo tumor can even directly incorporate an existing blood vessel to realize its own vascularization, seizing oxygen and nutrients needed for growth and eliminate waste of metabolism. The tumor vasculature can not only deliver nutrients inward but also help convey circulating tumor cells (CTCs) outward for cancer metastasis,103,104 which accounts for 90% of cancer death.105 As for cancer treatment, both chemotherapy and immunotherapy rely on the vasculature to transport medicine cargoes to tumors. Unlike the situation in normal tissues, the vasculature distribution inside of the tumor is random and chaotic, which makes it difficult to deliver drugs to the whole tumor and reach effective concentration.106 Moreover, the tumor vasculature may flush away the drug cargoes before they can diffuse into the surrounding tumor tissue, which greatly weakens the medication effect. None of these above-mentioned phenomena are reflected in homotypic tumor models. Therefore, integrating proper vasculature network into the in vitro tumor model should facilitate the following preclinical pharmaceutical investigation. In this pursuit, the focus of microfluidic tumor chip research has gradually transitioned from high-throughput homotypic tumor spheroids array-on-a-chip to TME reconstruction. Researchers hope to have a deeper understanding of tumorigenesis and improve drug screening efficiency by studying the process of tumor formation in vitro.107
Well-established tumor–vessel microfluidic models
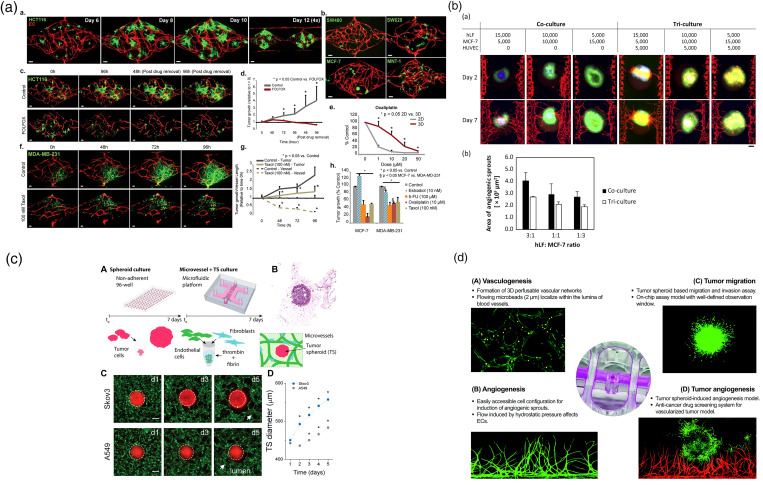
For constructing more realistic in vitro tumor models, researchers started with co-culture/tri-culture fibroblasts, endothelial cells together with tumor cells to fabricate heterotypic tumor spheroids.108–112 Although versatile cell–cell interaction is established, different types of cells were just rearranged and stratified into different layers rather than forming perfusable tumor vasculature inside.113,114 To date, the most effective in vitro tumor vascularization strategy is to restructure TME and induce interaction between the tumor spheroid and the nearby existing blood vessel.115–119 The tumor spheroids can both secrete growth factors to remotely manipulate blood vessels and physically interact with contacted blood vessels, which will be assimilated into tumor-associated vasculature.120
According to the scales of the vascular tissue, the microfluidic vessel–tumor models can be classified into three broad categories: capillary vessel–tumor model,115,116,121 single-lumen vessel–tumor model,118,122 and endothelium–tumor model.123 In these three models, the relative sizes of tumor and blood vessel tissues are different, thus simulating the interaction between tumor and blood vessels at different cancer stages.
Microvessel network interacting with tumor spheroids is a widely reported method to generate a vascularized tumor spheroid model [Fig. 5(a)].115,124,127–130 The developed tumor spheroids can secrete a series of growth factors, such as vascular endothelial growth factor (VEGF), to attract endothelial cells, self-assemble, and invade into the tumor spheroids. The diameter of the formed vessels is usually around 10 μm, and it is considered an in vitro reappearing of the tumor-activated angiogenesis process. After invasion, these capillary vessels will overspread and converge together in the core area of the spheroids, resulting in a perfusable vascularized tumor. Nashimoto et al. reported a “vascularized cancer-on-a-chip” work in 2020 [Fig. 5(b)].125 In their study, they started with tri-culturing the tumor cells with fibroblasts and endothelial cells. Most of fibroblasts and endothelial cells were dispersed in the outer layer of the tumor core, and no vascular tube was found to form inside. Then, they planted the heterotypic tumor spheroids in the center of their microfluidic culture chip. As the spheroids grew, the fibroblasts interacted with the tumor cells and secreted angiogenic factors. Under the stimulation of these growth factors, dispersed endothelial cells (ECs) at two sides self-assembled into microvessels and then grew toward the tumor spheroids. After invading into the tumor spheroids, the vessels growing from both sides were naturally connected, which significantly increased the proliferative activity of tumor cells and reduced cell death in the spheroids. Furthermore, due to the transport function of vessels and additional drug resistance from stroma cells, these vascularized tumors showed significantly different responses to anti-cancer drugs compared to the homotypic tumor spheroid. Contrary to the results collected under static conditions, drug administrated under perfusion conditions did not show a dose-dependent effect of anti-cancer drugs on tumor activity. The potential reason may be the blood flow flushed away the drug before it could be taken up by tumor tissue via either active transport or passive diffusion. Also, Haase et al. found that the tumor spheroids can regulate vessel density and barrier function, thereby affecting drug transport [Fig. 5(c)].119 Overall, these studies show that tumor vascularization significantly affects drug screening outcomes.
FIG. 5.
Capillary vessel–tumor interaction. (a) An in vitro vascularized microtumor (VMT) platform to test anti-angiogenesis drugs for cancer treatment.124 Reprinted with permission from Sobrino et al., Sci. Rep. 6, 31589 (2016). Copyright 2016 Springer Nature. (b) A tumor-on-a-chip platform that enables the evaluation of tumor activities with intraluminal flow in an engineered tumor vascular network. The fibroblasts in the tumor spheroid induced angiogenic sprouts, which constructed a perfusable vascular network in a tumor spheroid.125 Reprinted with permission from Nashimoto et al., Biomaterials 229, 119547 (2020). Copyright 2019 Elsevier Ltd. (c) A 3D vascularized tumor-on-a-chip model is used to examine drug delivery in a relevant TME within a large bed of perfusable vasculature.119 Reprinted with permission from Haase et al., Adv. Funct. Mater. 30, 2002444 (2020). Copyright 2020 WILEY-VCH Verlag GmbH & Co. KGaA Weinheim. (d) A standardized microfluidic culture platform for investigating tumor angiogenesis. The platform is made of polystyrene (PS) in a standardized 96-well plate and has the potential to be translated into market oriented preclinical applications.126 Reprinted with permission from Ko et al., Lab Chip 19, 2822 (2019). Copyright 2019 The Royal Society of Chemistry.
Notably, the current in vitro vascularized tumor models are most used for fundamental study since their throughput is much lower than homotypic “tumor chip” platforms. A limited number of vascularized tumors can be produced each time, and reproducibility of the method still needs to be improved. However, it is worth mentioning that there are already some tentative efforts on increasing the production of vascularized tumors for commercial applications [Fig. 5(d)].126 In the future, a standard automated microfluidic chip platform is expected to be invented for vascularized tumor spheroid array generation and high-throughput parallel screening of multiple drugs.131,132
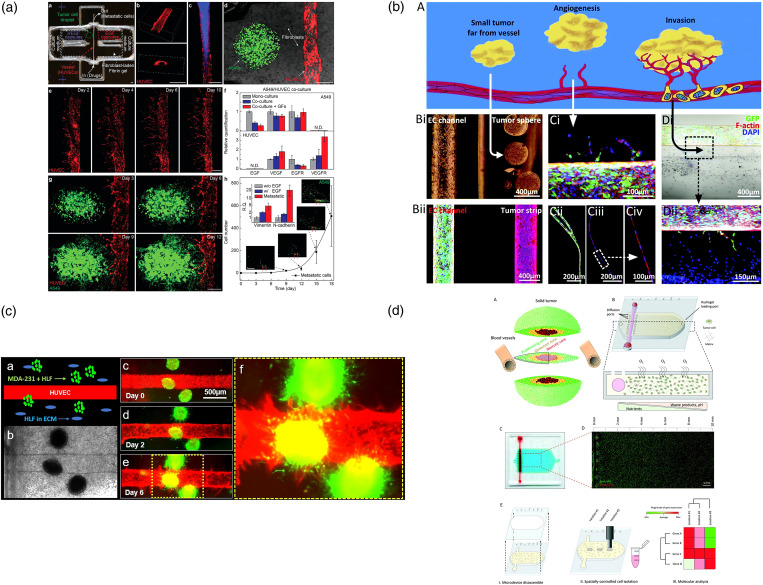
Another popular tumor–vessel interaction model is to co-culture tumor spheroids with a straight single-lumen blood vessel [Fig. 6(a)].134–136 In general, the diameter of the single-lumen vessel tubes is around several hundred micrometers, which are small arteriole-scale blood vessels. Different from the EC self-assembled vessel network, this single-lumen vessel is constructed by covering ECs in a pre-formed hollow channel, which is built in the hydrogel with mold embedding methods. After the mold (usually needles) is removed, endothelial cell suspension will be loaded inside. Once all the endothelial cells attach and cover the whole inner surface of the channel, the endothelialization process will end. Then, tumor spheroids/organoids will be seeded around the completed vascular channel [Fig. 6(b)].122 Based on the tumor types, matrix properties, and spatial distribution of tumors and blood vessels, different forms of tumor–vessel interactions can be recapitulated with such an approach. Kwak and Lee reported that tumor spheroids can induce capillary vessels sprouting from the single-lumen vascular channel [Fig. 6(c)].133 Additionally, Ayuso et al. constructed a vessel incorporated tumor slicing model to study cell behavior under metabolic starvation gradients [Fig. 6(d)].134 Significantly, a group from Johns Hopkins University found that breast tumors can directly fuse with normal vasculature into mosaic vessels.137 The researchers cocultured breast tumor organoids with a straight vasculature (around 200 μm in diameter) in a matrix hydrogel. As the tumor grew, the blood vessels were constantly squeezed until they merged. Part of the tumor even became the blood vessel wall. The observed “vascular mosaicism” phenomenon has also been demonstrated in clinical gastric cancer, glioblastoma, melanoma, and other cancer cases.138 Once tumor cells become part of blood vessels, the so-called “mosaic vessel” can provide a “highway” for circulating tumor cells (CTCs) and CTC clusters to be peeled off from the primary tumors and escape into the bloodstream. Compared with the other vascularized tumor models, this type of vessel–tumor model is simple to construct and is of great significance for understanding the cancer development.
FIG. 6.
Straight macro vessel–tumor interaction. (a) A bioprinted tumor–vessel co-culture model provides a platform to explore the molecular mechanisms of tumor progression and screen anti-cancer drugs.118 Reprinted with permission from Meng et al., Adv. Mater. 31, e1806899 (2019). Copyright 2019 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim. (b) A 3D bioprinted bulk breast tumor tissue with a functional vascular network is built for physiology study and therapy evaluation.122 Reprinted with permission from Nie et al., Mater. Horiz. 7, 82 (2020). Copyright 2020 The Royal Society of Chemistry. (c) A microfluidic model of the solid tumor–vascular interface is applied to study molecular crosstalk in tumorigenesis and cancer metastasis.133 Reprinted with permission from Kwak and Lee, Sci. Rep. 6, 20142 (2016). Copyright 2016 Springer Nature. (d) A microfluidic tumor slice model with a lumen on the flank of the chamber to perfuse media, mimicking the vasculature.134 Reprinted with permission from Ayuso et al., Lab Chip 19, 3461 (2019). Copyright 2019 The Royal Society of Chemistry.
The third type is monolayer endothelium–tumor co-culture model, which is usually exploited to model early-stage primary tumor. This model is usually composed of upper and lower two-tier structures.131,139,140,143–148 The upper and lower layers are usually separated by a porous membrane.142 Endothelium and epithelium are seeded on the upper and lower sides of the membrane149 [Fig. 7(a)]. Tumor aggregates are planted above the epithelium to mimic the initial phase of cancer.150 From an engineering point of view, it can be considered a microfluidic version of “trans-well assays.”151 At this early stage, the primary tumor is relatively small, and the pathological changes mainly occur on tumor–epithelium/endothelium interfaces. The cells cultured on surfaces of the membrane deposit their own basement membrane matrix crossing the membrane pores to help the upper and lower layers of cells to form direct interaction. The design of these devices is quite flexible. For example, hollow side chambers can be set on both sides of the tumor model. By applying circulatory ventilation in these airways, the entire device is stretched rhythmically, thereby simulating mechanical stimuli related to organs, such as blood flow and related physiological shear stress [Fig. 7(b)].140 Choi et al. described a biomimetic microengineering strategy to reconstitute 3D human breast tumors located in the corresponding TME [Fig. 7(c)]. In this design, a normal mammary epithelium inlaid with the tumor clusters was reproduced on the top surface of the membrane, and a mammary fibroblast-laden stromal layer was formed on the back side. Fresh culture medium was perfused in the upper and lower channels to provide nutrition and mechanical stimulation in the bloodstream. This engineered tumor model represented the first critical step toward recapitulating pathophysiological complexity of breast cancer and may serve as a tool to systematically examine the tumor initiation, progression, and metastasis in local TME. In addition to the above-mentioned study, this membrane based double-layer module can be further applied in a multi-organ system or a body-on-a-chip (BOC) system. Xu et al. used this design to mimic the lung tissue and integrated it with three downstream “distant organs” [Fig. 7(d)].142 In this way, cancer cell metastasis to the different organs, such as brain, bone, and liver, can be studied in an integrated system. Although these three types of microfluidic tumor/vascular models are different in spatial structure and cell composition, they simulate the interaction between tumors and blood vessels at different stages of cancer, facilitating tumor research at different stages and different purposes.
FIG. 7.
Endothelium monolayer–tumor interaction. (a) A biomimetic glioma perivascular niche model on-a-chip. Glioma cancer cells and endothelial cells are planted on the upper and lower sides of a PC membrane, respectively.139 Reprinted with permission from Lin et al., Anal. Chem. 90, 10326 (2018). Copyright 2018 American Chemical Society. (b) A microfluidic human orthotopic lung cancer-on-a-chip model. The endothelium monolayer covers the four walls of the lower channel to mimic the vessel tube. Epithelium and cancer cell aggregates are located on the top surface of the membrane.140 Reprinted with permission from Hassell et al., Cell Rep. 21, 508 (2017). Copyright 2017 Elsevier B.V. (c) A basement membrane based microfluidic model to mimic early-stage cancerous tissue. The breast cancer spheroids are planted on the top of epithelium tissue monolayer.141 Reprinted with permission from Choi et al., Lab Chip 15, 3350 (2015). Copyright 2015 The Royal Society of Chemistry. (d) A bilayer device to study the cancer metastasis. Cancer cells flow through the channel within the artificial blood and get trapped on the epithelium monolayer.142 Reprinted with permission from Xu et al., ACS Appl. Mater. Interfaces 8, 25840 (2016). Copyright 2016 American Chemical Society.
MICROFLUIDIC ORGANOID-ON-A-CHIP
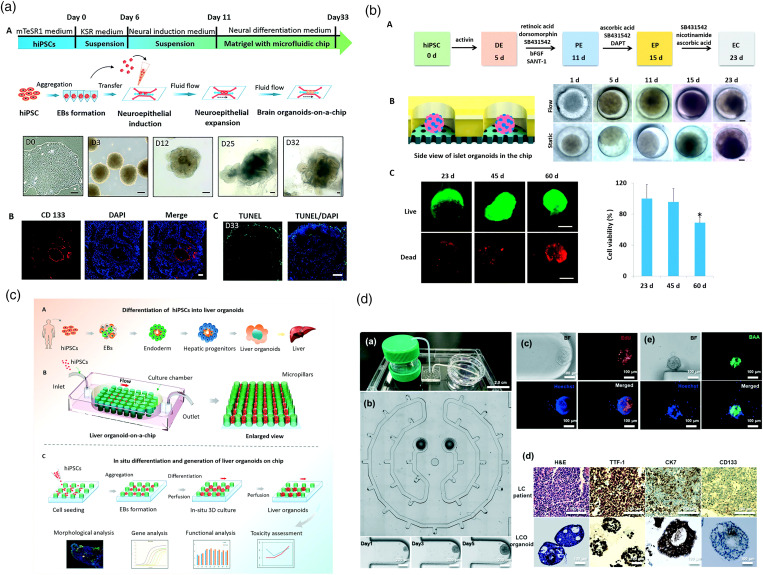
From the historical development of technology, a microfluidic tumor chip is a special branch of organ chip technology in the field of oncology.13 Organoids are miniaturized in vitro organ models that are mainly developed from human stem cells to mimic the critical characteristics and physiological functions of in vivo organs with high fidelity.8 Compared with time-consuming and elaborate animal models, organoids allow human-derived materials to be quickly cultivated for both in vitro developmental biology and pathology research.156 In order to produce organoids in a more uniform and high-throughput way, the cultivation process gradually shifted from “in-dish” to “on-a-chip,” which is the so-called “organoid-on-a-chip.”157 Here, the microfluidic chips can help strictly regulate the local microenvironment, thereby improving the reproducibility while reducing individual difference, such as morphology and gene expression. Nowadays, organoid-on-a-chip technology has attracted widespread attention. Various organ models have been reported, including, but not limited to, brain organoids152,158,159 [Fig. 8(a)], islet organoids153,160,161 [Fig. 8(b)], liver organoids154,162,163 [Fig. 8(c)], and so on. In particular, if the organoids are derivative of patients' biopsy or surgically removed diseased tissue, rather than stem cells, they will be referred to as patient-derived organoids.164 The patient-derived organoid can be used as disease model for individual patients, contributing to personal therapy and drug screening.165 For example, Jung et al. reported a lung cancer organoid-on-a-chip model in 2019155 [Fig. 8(d)]. Besides recapitulating primary small-cell lung cancer-specific characteristics, the patient-derived organoid was produced in a size-controllable manner, benefiting consecutive standard drug testing. In the next stage research, researchers will be confronted with some identical challenges, organ microenvironment reconstruction and tissue vascularization. Although tumor vascularization has been demonstrated, the fragility and complexity of organ organoids make it more difficult for them to be vascularized.
FIG. 8.
Microfluidic organ/cancer organoid-on-a-chip. (a) A brain organoid-on-a-chip system is established to model neurodevelopmental disorders under nicotine exposure.152 Reprinted with permission from Wang et al., Lab Chip 18, 851 (2018). Copyright 2018 The Royal Society of Chemistry. (b) An islet organoid-on-a-chip system enables controllable maturation of islet organoids from induced pluripotent stem cells and real-time imaging and in situ tracking of islet organoid growth.153 Reprinted with permission from Tao et al., Lab Chip 19, 948 (2019). Copyright 2019 The Royal Society of Chemistry. (c) A 3D perfusable liver organoid-on-a-chip system displays improved cell viability and high-fidelity reduction of liver functions.154 Reprinted with permission from Wang et al., Lab Chip 18, 3606 (2018). Copyright 2018 The Royal Society of Chemistry. (d) A microphysiological system reproducibly yields patient-derived small-cell lung cancer organoids in a size-controllable manner and efficiently predicts anti-cancer drug efficacy.155 Reprinted with permission from Jung et al., Lab Chip 19, 2854 (2019). Copyright 2019 The Royal Society of Chemistry.
PROMISING MICROSCOPIC CHARACTERIZATION AND IMAGE ANALYSIS OF TUMOR SPHEROIDS
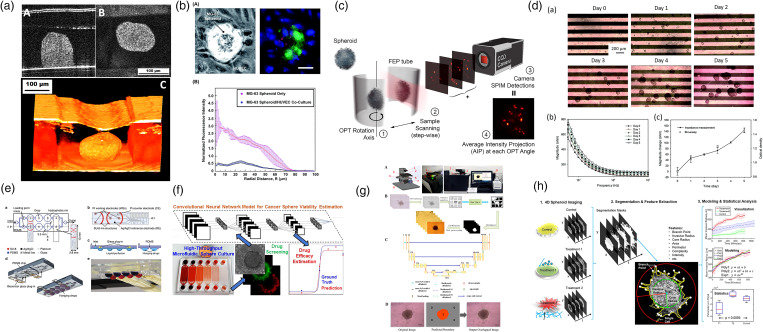
With the progress of microfluidic technology and cancer biology, significant improvements have been made in tumor-on-a-chip model. Subsequently, the characterization methods that can collect abundant information from these tumor models have become one of the research concerns. In addition to conventional bright-field and fluorescence-assisted morphological characterization, more and more promising imaging modalities have been used to investigate microfluidic tumor models to obtain both physiological and pathological information. For example, optical coherence tomography (OCT)144,173 has been used as a non-invasive approach to characterize the 3D structures deep inside tumor spheroids [Fig. 9(a)]. Based on the increased intrinsic optical attenuation, necrotic areas in these tumor spheroids were successfully detected, indicating a promising alternative to label-free viability testing in tumors. Moreover, multi-photon laser scanning microscopy (MPLSM) [Fig. 9(b)]166 has been used to study oxygen tension variation within live tumor spheroids.174 Phase-retrieved tomography [Fig. 9(c)]167 has been applied to improve the resolution of mesoscopic imaging on dense tumor spheroids. Along with the introduction of novel imaging approaches, various biosensors have also been integrated into microfluidic tumor chips for measuring and monitoring tumor behaviors and related microenvironments.175 For example, Lei et al. incorporated interdigitated electrodes for real-time and label-free impedimetric analysis of the formation and chemosensitivity of tumor spheroids [Fig. 9(d)]. Misun et al. presented enzyme-based multi-analyte biosensors to enable continuous monitoring of lactate and glucose levels in cancer tissues [Fig. 9(e)]. These novel characterization technologies not only provide new perspectives for understanding tumors but also bring much unprecedented information that inspires future innovative therapies.
FIG. 9.
Novel microtechnology-based characterization techniques integrated with tumor chips. (a) Optical coherence tomography (OCT) is employed to characterize a microfluidic bilayer vasculature/tumor co-culture model.144 Reprinted with permission from Shi et al., Biomicrofluidics 13, 044108 (2019). Copyright 2019 AIP Publishing LLC. (b) Multi-photon laser scanning microscopy (MPLSM) is used to study oxygen tension within cocultured MG-63 spheroids and human umbilical vein endothelial cells (HUVECs) monolayers.166 Reprinted with permission from Sarkar et al., RSC Adv. 8, 30320 (2018). Copyright 2018 The Royal Society of Chemistry. (c) Phase-retrieved tomography (PRT) is applied to radically improve mesoscopic scanning imaging of tumor spheroids.167 Reprinted with permission from Ancora et al., Sci. Rep. 7, 11854 (2017). Copyright 2017 Springer Nature. (d) Impedance measurements are used to monitor and record the number and size of tumor spheroids in a label-free manner.168 Reprinted with permission from Lei et al., RSC Adv. 7, 13939 (2017). Copyright 2017 The Royal Society of Chemistry. (e) Microfluidic hanging drop platform with multi-analyte biosensor for in situ monitoring of 3D cancer microtissue metabolism.169 Reprinted with permission from Misun et al., Microsyst. Nanoeng. 2, 16022 (2016). Copyright 2016 Springer Nature. (f) A convolutional neural network was trained with images of LIVE/DEAD stained tumor spheroids and used to estimate spheroid viability based on its bright-field image.170 Reprinted with permission from Zhang et al., Anal. Chem. 91, 14093 (2019). Copyright 2019 American Chemical Society. (g) A new tumor spheroid boundary detection algorithm based on convolutional neural network.171 Reprinted with permission from Chen et al., Biomaterials 272, 120770 (2021). Copyright 2021 Elsevier Ltd. (h) Open source software for end-to-end time analysis of tumor imaging, used to automatically segment spheroid images; extract features that describe their shape, growth, and invasiveness; and perform mathematical modeling and statistical analysis on these features.172 Reprinted with permission from Hou et al., Sci. Rep. 8, 7248 (2018). Copyright Springer Nature.
In addition to exploring characterization methods, how to analyze collected data in more detail has also received widespread attention. In recent years, the use of machine learning-based image processing technology on drug efficacy evaluation has become one of the research hotspots in the field of microfluidic tumor chips.176–178 For example, Zhang et al. invented a microfluidic tumor-on-a-chip model, in which more than 1900 tumor spheroids were constructed for drug testing simultaneously [Fig. 9(f)].170 The LIVE/DEAD viability staining images of the tumor spheroids were automatically collected to train a convolutional neural network (CNN), which could be used to estimate tumor spheroid viability based on its bright-field image later. Here, the microfluidic tumor chip facilitates the tumor model construction and large reliable data acquisition, and the advanced algorithms collect and process data intelligently to draw valid conclusions. It is a classic case of combining artificial intelligence (AI) and microfluidics for anti-cancer drug screening. Moreover, artificial intelligence also has unique advantages in analyzing bright-field photos of tumor spheroids. In addition to immunofluorescence staining, measuring tumor size and morphological changes is one of the most used tumor research methods. However, manual tumor imaging and hand-sketched tumor spheroid boundaries are time-consuming and subjective, and slight morphological changes are easy to be ignored. There is an urgent need for an accurate, reliable, automated, and high-throughput 3D tumor imaging analysis system. Chen et al. developed an automated spheroid monitoring and AI-based recognition technique (“SMART”) for analyzing tumor spheroid behavior [Fig. 9(g)].171 Auto-recognition, auto-focusing, and an improved CNN algorithm are integrated together on this platform for automated detection of tumor spheroid boundaries. How to transform abstract morphological information into precise quantitative data has always been an unmet need of cancer researchers in the development of anti-cancer drugs. Ultimately, machine learning-based image processing is expected to help bridge the gap between physiological feature extraction and mathematical data analysis [Fig. 9(h)]. Such systems can make accurate and automatic quantitative assessments of the physiology and pathology of tumors and are of great significance for advancing emerging cell-based anti-cancer medicine.
PERSPECTIVES AND CONCLUSION
For further development, microfluidic tumor chip technology is expected to help develop cutting-edge cancer immunotherapies and achieve commercially available high-throughput preclinical drug screening. So far, only a handful of cancer immunotherapies have been approved on market, and most of them can only work on blood cancer. There are many difficulties in cell therapy for solid tumors, such as the complex heterogeneity of solid tumors, the difficulty in maintaining the viability of immune cells for a long time, and the tumor microenvironment that can inhibit immunity.179 Recently, the immune system has been imported in some tumor-on-a-chip models for oncology immunotherapy study.180–183 Researchers hope to figure out how to induce immune cells to break through the peripheral extracellular matrix and TME to destroy solid tumors,184 whereas most of these immune system-tumor-on-a-chip models are still in the basic exploratory stage and the research focus remains on the direct interaction between T cells and tumors. The immunosuppressive TME and other pivotal elements, such as functional blood and lymphatic vessels, need to be incorporated into the model. Based on previous experience in building in vitro models for drug screening, microfluidic tumor chips can be a reliable platform to accelerate an immuno-oncology study. As for the translation of “tumor-on-a-chip” to market, the commercialization has just been unfolding. Many start-up companies and their products have sprung up, while it may take some time before pharmaceutical industries actually accept these products for preliminary drug testing.185
Overall, the microfluidic tumor chip is a revolutionary technology for anti-cancer drug development. Benefiting from this interdisciplinary platform, different research topics can be integrated together, such as biomechanics, immunology, and molecular biology, allowing researchers to understand cancer from new perspectives. In the foreseeable future, microfluidic tumor chip technology is promising to upgrade preclinical tumor models and shorten the translational gap between early drug discovery and clinical trials in drug developments.
ACKNOWLEDGMENTS
This work was supported by the National Institute of Health (Grant No. R01HL131750), the National Science Foundation (Grant No. CBET 2039310), and the Pennsylvania Department of Health Commonwealth Universal Research Enhancement Program (CURE).
AUTHOR DECLARATIONS
Conflicts of Interest
The authors have no conflicts to disclose.
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1.Siegel R. L., Miller K. D., Fuchs H. E., and Jemal A., CA Cancer J. Clin. 71, 7 (2021). 10.3322/caac.21654 [DOI] [PubMed] [Google Scholar]
- 2.Conklin K. A., Integr. Cancer Ther. 3, 294 (2004). 10.1177/1534735404270335 [DOI] [PubMed] [Google Scholar]
- 3.Falzone L., Salomone S., and Libra M., Front. Pharmacol. 9, 1300 (2018). 10.3389/fphar.2018.01300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prasad V. and Mailankody S., JAMA Intern. Med. 177, 1569 (2017). 10.1001/jamainternmed.2017.3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tammela T. and Sage J., Annu. Rev. Cancer Biol. 4, 99 (2020). 10.1146/annurev-cancerbio-030419-033413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitaeva K. V., Rutland C. S., Rizvanov A. A., and Solovyeva V. V., Front. Bioeng. Biotechnol. 8, 322 (2020). 10.3389/fbioe.2020.00322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapałczyńska M., Kolenda T., Przybyła W., Zajączkowska M., Teresiak A., Filas V., Ibbs M., Bliźniak R., Łuczewski Ł, and Lamperska K., Arch. Med. Sci. 14(4), 910–919 (2016). 10.5114/aoms.2016.63743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skardal A., Shupe T., and Atala A., Drug Discov. Today 21, 1399 (2016). 10.1016/j.drudis.2016.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Powley I. R., Patel M., Miles G., Pringle H., Howells L., Thomas A., Kettleborough C., Bryans J., Hammonds T., MacFarlane M., and Pritchard C., Br. J. Cancer 122, 735 (2020). 10.1038/s41416-019-0672-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langhans S. A., Front. Pharmacol. 9, 6 (2018). 10.3389/fphar.2018.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan D. and Weinberg R. A., Cell 144, 646 (2011). 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 12.Whiteside T. L., Oncogene 27, 5904 (2008). 10.1038/onc.2008.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sontheimer-Phelps A., Hassell B. A., and Ingber D. E., Nat. Rev. Cancer 19, 65 (2019). 10.1038/s41568-018-0104-6 [DOI] [PubMed] [Google Scholar]
- 14.Katt M. E., Placone A. L., Wong A. D., Xu Z. S., and Searson P. C., Front. Bioeng. Biotechnol. 4, 12 (2016). 10.3389/fbioe.2016.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nath S. and Devi G. R., Pharmacol. Ther. 163, 94 (2016). 10.1016/j.pharmthera.2016.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nunes A. S., Barros A. S., Costa E. C., Moreira A. F., and Correia I. J., Biotechnol. Bioeng. 116, 206 (2019). 10.1002/bit.26845 [DOI] [PubMed] [Google Scholar]
- 17.Shao C., Chi J., Zhang H., Fan Q., Zhao Y., and Ye F., Adv. Mater. Technol. 5, 2000183 (2020). 10.1002/admt.202000183 [DOI] [Google Scholar]
- 18.Cui P. and Wang S., J. Pharm. Anal. 9, 238 (2019). 10.1016/j.jpha.2018.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oh H. J., Kim J., Kim H., Choi N., and Chung S., Adv. Healthcare Mater. 10, 2002122 (2021). 10.1002/adhm.202002122 [DOI] [PubMed] [Google Scholar]
- 20.Shang M., Soon R. H., Lim C. T., Khoo B. L., and Han J., Lab Chip 19, 369 (2019). 10.1039/C8LC00970H [DOI] [PubMed] [Google Scholar]
- 21.Jakab K., Norotte C., Marga F., Murphy K., Vunjak-Novakovic G., and Forgacs G., Biofabrication 2, 022001 (2010). 10.1088/1758-5082/2/2/022001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller-Klieser W., J. Cancer Res. Clin. Oncol. 113, 101 (1987). 10.1007/BF00391431 [DOI] [PubMed] [Google Scholar]
- 23.Santo V. E., Estrada M. F., Rebelo S. P., Abreu S., Silva I., Pinto C., Veloso S. C., Serra A. T., Boghaert E., Alves P. M., and Brito C., J. Biotechnol. 221, 118 (2016). 10.1016/j.jbiotec.2016.01.031 [DOI] [PubMed] [Google Scholar]
- 24.Mosaad E. O., Chambers K. F., Futrega K., Clements J. A., and Doran M. R., Sci. Rep. 8, 253 (2018). 10.1038/s41598-017-18050-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vadivelu R. K., Ooi C. H., Yao R.-Q., Tello Velasquez J., Pastrana E., Diaz-Nido J., Lim F., Ekberg J. A. K., Nguyen N.-T., and St John J. A., Sci. Rep. 5, 15083 (2015). 10.1038/srep15083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim W., Hoang H.-H., You D., Han J., Lee J. E., Kim S., and Park S., Analyst 143, 5841 (2018). 10.1039/C8AN01752B [DOI] [PubMed] [Google Scholar]
- 27.Kingsley D. M., Roberge C. L., Rudkouskaya A., Faulkner D. E., Barroso M., Intes X., and Corr D. T., Acta Biomater. 95, 357 (2019). 10.1016/j.actbio.2019.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park S. M., Lee S. J., Lim J., Kim B. C., Han S. J., and Kim D. S., ACS Appl. Mater. Interfaces 10, 37878 (2018). 10.1021/acsami.8b15821 [DOI] [PubMed] [Google Scholar]
- 29.Fang G., Lu H., Law A., Gallego-Ortega D., Jin D., and Lin G., Lab Chip 19, 4093 (2019). 10.1039/C9LC00872A [DOI] [PubMed] [Google Scholar]
- 30.Lee J. M., Park D. Y., Yang L., Kim E.-J., Ahrberg C. D., Lee K.-B., and Chung B. G., Sci. Rep. 8, 17145 (2018). 10.1038/s41598-018-35216-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao S.-P., Ma Y., Lou Q., Zhu H., Yang B., and Fang Q., Anal. Chem. 89, 10153 (2017). 10.1021/acs.analchem.7b02267 [DOI] [PubMed] [Google Scholar]
- 32.Foty R., J. Vis. Exp. e2720 (2011). 10.3791/2720(2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ganguli A., Mostafa A., Saavedra C., Kim Y., Le P., Faramarzi V., Feathers R. W., Berger J., Ramos-Cruz K. P., Adeniba O., Diaz G. J. P., Drnevich J., Wright C. L., Hernandez A. G., Lin W., Smith A. M., Kosari F., Vasmatzis G., Anastasiadis P. Z., and Bashir R., Sci. Adv. (17), eabc1323 (2021). 10.1126/sciadv.abc1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tung Y.-C., Hsiao A. Y., Allen S. G., Torisawa Y.-S., Ho M., and Takayama S., Analyst 136, 473 (2011). 10.1039/C0AN00609B [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popova A. A., Tronser T., Demir K., Haitz P., Kuodyte K., Starkuviene V., Wajda P., and Levkin P. A., Small 15, 1901299 (2019). 10.1002/smll.201901299 [DOI] [PubMed] [Google Scholar]
- 36.Michael I. J., Kumar S., Oh J. M., Kim D., Kim J., and Cho Y.-K., ACS Appl. Mater. Interfaces 10, 33839 (2018). 10.1021/acsami.8b08778 [DOI] [PubMed] [Google Scholar]
- 37.Zhao L., Xiu J., Liu Y., Zhang T., Pan W., Zheng X., and Zhang X., Sci. Rep. 9, 19717 (2019). 10.1038/s41598-019-56241-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munson J. M. and Shieh A. C., Cancer Manage. Res. 6, 317 (2014). 10.2147/CMAR.S65444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bashir R., Adv. Drug Delivery Rev. 56, 1565 (2004). 10.1016/j.addr.2004.03.002 [DOI] [PubMed] [Google Scholar]
- 40.Liu X., Fang J., Huang S., Wu X., Xie X., Wang J., Liu F., Zhang M., Peng Z., and Hu N., Microsyst. Nanoeng. 7, 50 (2021). 10.1038/s41378-021-00277-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luka G., Ahmadi A., Najjaran H., Alocilja E., DeRosa M., Wolthers K., Malki A., Aziz H., Althani A., and Hoorfar M., Sensors 15, 30011 (2015). 10.3390/s151229783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhuang J., Zhang J., Wu M., and Zhang Y., Adv. Sci. 6, 1901462 (2019). 10.1002/advs.201901462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y., Gao D., Wang Y., Lin S., and Jiang Y., Anal. Chim. Acta 1036, 97 (2018). 10.1016/j.aca.2018.06.038 [DOI] [PubMed] [Google Scholar]
- 44.Liu W., Wang J.-C., and Wang J., Lab Chip 15, 1195 (2015). 10.1039/C4LC01242A [DOI] [PubMed] [Google Scholar]
- 45.Fan Y., Nguyen D. T., Akay Y., Xu F., and Akay M., Sci. Rep. 6, 25062 (2016). 10.1038/srep25062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee K., Kim C., Young Yang J., Lee H., Ahn B., Xu L., Yoon Kang J., and Oh K. W., Biomicrofluidics 6, 014114 (2012). 10.1063/1.3687409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marimuthu M., Rousset N., St-Georges-Robillard A., Lateef M. A., Ferland M., Mes-Masson A.-M., and Gervais T., Lab Chip 18, 304 (2018). 10.1039/C7LC00970D [DOI] [PubMed] [Google Scholar]
- 48.Xu Y., Xie F., Qiu T., Xie L., Xing W., and Cheng J., Biomicrofluidics 6, 016504 (2012). 10.1063/1.3687399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anada T., Masuda T., Honda Y., Fukuda J., Arai F., Fukuda T., and Suzuki O., Sens. Actuators B 147, 376 (2010). 10.1016/j.snb.2010.01.065 [DOI] [Google Scholar]
- 50.Liu W., Sun M., Lu B., Yan M., Han K., and Wang J., Sens. Actuators B 292, 111 (2019). 10.1016/j.snb.2019.04.121 [DOI] [Google Scholar]
- 51.Li H., Garner T., Diaz F., and Wong P. K., Small 15, 1901910 (2019). 10.1002/smll.201901910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mulholland T., McAllister M., Patek S., Flint D., Underwood M., Sim A., Edwards J., and Zagnoni M., Sci. Rep. 8, 14672 (2018). 10.1038/s41598-018-33055-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patra B., Peng C.-C., Liao W.-H., Lee C.-H., and Tung Y.-C., Sci. Rep. 6, 21061 (2016). 10.1038/srep21061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao L., Shi M., Liu Y., Zheng X., Xiu J., Liu Y., Tian L., Wang H., Zhang M., and Zhang X., Anal. Chem. 91, 4307 (2019). 10.1021/acs.analchem.9b00376 [DOI] [PubMed] [Google Scholar]
- 55.Zhao L., Mok S., and Moraes C., Biofabrication 11, 045013 (2019). 10.1088/1758-5090/ab30b4 [DOI] [PubMed] [Google Scholar]
- 56.Kwapiszewska K., Michalczuk A., Rybka M., Kwapiszewski R., and Brzózka Z., Lab Chip 14, 2096 (2014). 10.1039/C4LC00291A [DOI] [PubMed] [Google Scholar]
- 57.Ziółkowska K., Stelmachowska A., Kwapiszewski R., Chudy M., Dybko A., and Brzózka Z., Biosens. Bioelectron. 40, 68 (2013). 10.1016/j.bios.2012.06.017 [DOI] [PubMed] [Google Scholar]
- 58.Zhao L., Liu Y., Liu Y., Zhang M., and Zhang X., Anal. Chem. 92, 7638 (2020). 10.1021/acs.analchem.0c00408 [DOI] [PubMed] [Google Scholar]
- 59.Dadgar N., Gonzalez-Suarez A. M., Fattahi P., Hou X., Weroha J. S., Gaspar-Maia A., Stybayeva G., and Revzin A., Microsyst. Nanoeng. 6, 93 (2020). 10.1038/s41378-020-00201-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khot M. I., Levenstein M. A., de Boer G. N., Armstrong G., Maisey T., Svavarsdottir H. S., Andrew H., Perry S. L., Kapur N., and Jayne D. G., Sci. Rep. 10, 15915 (2020). 10.1038/s41598-020-72952-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pang L., Ding J., Ge Y., Fan J., and Fan S.-K., Anal. Chem. 91, 8318 (2019). 10.1021/acs.analchem.9b01084 [DOI] [PubMed] [Google Scholar]
- 62.Yu L., Chen M. C. W., and Cheung K. C., Lab Chip 10, 2424 (2010). 10.1039/c004590j [DOI] [PubMed] [Google Scholar]
- 63.Sabhachandani P., Motwani V., Cohen N., Sarkar S., Torchilin V., and Konry T., Lab Chip 16, 497 (2016). 10.1039/C5LC01139F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu W., Xu J., Li T., Zhao L., Ma C., Shen S., and Wang J., Anal. Chem. 87, 9752 (2015). 10.1021/acs.analchem.5b01915 [DOI] [PubMed] [Google Scholar]
- 65.Fu C.-Y., Tseng S.-Y., Yang S.-M., Hsu L., Liu C.-H., and Chang H.-Y., Biofabrication 6, 015009 (2014). 10.1088/1758-5082/6/1/015009 [DOI] [PubMed] [Google Scholar]
- 66.Liu W., Liu D., Hu R., Huang Z., Sun M., and Han K., Analyst 145, 6447 (2020). 10.1039/D0AN01229G [DOI] [PubMed] [Google Scholar]
- 67.Chen Y., Gao D., Liu H., Lin S., and Jiang Y., Anal. Chim. Acta 898, 85 (2015). 10.1016/j.aca.2015.10.006 [DOI] [PubMed] [Google Scholar]
- 68.Huang Y. L., Ma Y., Wu C., Shiau C., Segall J. E., and Wu M., Sci. Rep. 10, 9648 (2020). 10.1038/s41598-020-66528-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kwak B., Lee Y., Lee J., Lee S., and Lim J., J. Controlled Release 275, 201 (2018). 10.1016/j.jconrel.2018.02.029 [DOI] [PubMed] [Google Scholar]
- 70.Lee J. M., Choi J. W., Ahrberg C. D., Choi H. W., Ha J. H., Mun S. G., Mo S. J., and Chung B. G., Microsyst. Nanoeng. 6, 52 (2020). 10.1038/s41378-020-0167-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang H., Agarwal P., Jiang B., Stewart S., Liu X., Liang Y., Hancioglu B., Webb A., Fisher J. P., Liu Z., Lu X., Tkaczuk K. H. R., and He X., Adv. Sci. 7, 2000259 (2020). 10.1002/advs.202000259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sabhachandani P., Sarkar S., Mckenney S., Ravi D., Evens A. M., and Konry T., J. Controlled Release 295, 21 (2019). 10.1016/j.jconrel.2018.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoon S., Kim J. A., Lee S. H., Kim M., and Park T. H., Lab Chip 13, 1522 (2013). 10.1039/c3lc41322e [DOI] [PubMed] [Google Scholar]
- 74.Agarwal P., Wang H., Sun M., Xu J., Zhao S., Liu Z., Gooch K. J., Zhao Y., Lu X., and He X., ACS Nano 11, 6691 (2017). 10.1021/acsnano.7b00824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cui X., Liu Y., Hartanto Y., Bi J., Dai S., and Zhang H., Colloids Interface Sci. Commun. 14, 4 (2016). 10.1016/j.colcom.2016.09.001 [DOI] [Google Scholar]
- 76.Wang Y. and Wang J., Analyst 139, 2449 (2014). 10.1039/C4AN00015C [DOI] [PubMed] [Google Scholar]
- 77.Huebner A., Sharma S., Srisa-Art M., Hollfelder F., Edel J. B., and Demello A. J., Lab Chip 8, 1244 (2008). 10.1039/b806405a [DOI] [PubMed] [Google Scholar]
- 78.Wu Z., Gong Z., Ao Z., Xu J., Cai H., Muhsen M., Heaps S., Bondesson M., Guo S., and Guo F., ACS Appl. Bio Mater. 3, 6273–6283 (2020). 10.1021/acsabm.0c00768 [DOI] [PubMed] [Google Scholar]
- 79.Sun Q., Tan S. H., Chen Q., Ran R., Hui Y., Chen D., and Zhao C.-X., ACS Biomater. Sci. Eng. 4, 4425 (2018). 10.1021/acsbiomaterials.8b00904 [DOI] [PubMed] [Google Scholar]
- 80.Sart S., Tomasi R. F.-X., Amselem G., and Baroud C. N., Nat. Commun. 8(1), 469 (2017). 10.1038/s41467-017-00475-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li S., Yang K., Chen X., Zhu X., Zhou H., Li P., Chen Y., Jiang Y., Li T., Qin X., Yang H., Wu C., Ji B., You F., and Liu Y., Biofabrication 13, 045013 (2021). 10.1088/1758-5090/ac1ea8 [DOI] [PubMed] [Google Scholar]
- 82.Samara B., Deliorman M., Sukumar P., and Qasaimeh M. A., Lab Chip 21, 844-854 (2021). 10.1039/D0LC01300E [DOI] [PubMed] [Google Scholar]
- 83.Shi W., Kwon J., Huang Y., Tan J., Uhl C. G., He R., Zhou C., and Liu Y., Sci. Rep. 8(1), 6837 (2018). 10.1038/s41598-018-25203-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tseng H., Gage J. A., Shen T., Haisler W. L., Neeley S. K., Shiao S., Chen J., Desai P. K., Liao A., Hebel C., Raphael R. M., Becker J. L., and Souza G. R., Sci. Rep. 5, 13987 (2015). 10.1038/srep13987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ho V. H. B., Müller K. H., Barcza A., Chen R., and Slater N. K. H., Biomaterials 31, 3095 (2010). 10.1016/j.biomaterials.2009.12.047 [DOI] [PubMed] [Google Scholar]
- 86.Tocchio A., Durmus N. G., Sridhar K., Mani V., Coskun B., El Assal R., and Demirci U., Adv. Mater. 30, 1705034 (2018). 10.1002/adma.201705034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Perez J. E., Nagle I., and Wilhelm C., Biofabrication 13, 015018 (2020). 10.1088/1758-5090/abc670 [DOI] [PubMed] [Google Scholar]
- 88.Souza G. R., Molina J. R., Raphael R. M., Ozawa M. G., Stark D. J., Levin C. S., Bronk L. F., Ananta J. S., Mandelin J., Georgescu M.-M., Bankson J. A., Gelovani J. G., Killian T. C., Arap W., and Pasqualini R., Nat. Nanotechnol. 5, 291 (2010). 10.1038/nnano.2010.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hu X., Zhao S., Luo Z., Zuo Y., Wang F., Zhu J., Chen L., Yang D., Zheng Y., Zheng Y., Cheng Y., Zhou F., and Yang Y., Lab Chip 20, 2228 (2020). 10.1039/D0LC00255K [DOI] [PubMed] [Google Scholar]
- 90.Chen B., Wu Y., Ao Z., Cai H., Nunez A., Liu Y., Foley J., Nephew K., Lu X., and Guo F., Lab Chip 19, 1755 (2019). 10.1039/C9LC00135B [DOI] [PubMed] [Google Scholar]
- 91.Olofsson K., Carannante V., Ohlin M., Frisk T., Kushiro K., Takai M., Lundqvist A., Önfelt B., and Wiklund M., Lab Chip 18, 2466-2476 (2018). 10.1039/C8LC00537K [DOI] [PubMed] [Google Scholar]
- 92.Chen K., Wu M., Guo F., Li P., Chan C. Y., Mao Z., Li S., Ren L., Zhang R., and Huang T. J., Lab Chip 16, 2636 (2016). 10.1039/C6LC00444J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lim W. and Park S., Molecules 23, 3355 (2018). 10.3390/molecules23123355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Petreus T., Cadogan E., Hughes G., Smith A., Pilla Reddy V., Lau A., O’Connor M. J., Critchlow S., Ashford M., and Oplustil O’Connor L., Commun. Biol. 4, 1001 (2021). 10.1038/s42003-021-02526-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meacham C. E. and Morrison S. J., Nature 501, 328 (2013). 10.1038/nature12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim H., Phung Y., and Ho M., PLoS ONE 7, e39556 (2012). 10.1371/journal.pone.0039556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Luca A. C., Mersch S., Deenen R., Schmidt S., Messner I., Schäfer K.-L., Baldus S. E., Huckenbeck W., Piekorz R. P., Knoefel W. T., Krieg A., and Stoecklein N. H., PLoS ONE 8, e59689 (2013). 10.1371/journal.pone.0059689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yi H.-G., Jeong Y. H., Kim Y., Choi Y.-J., Moon H. E., Park S. H., Kang K. S., Bae M., Jang J., Youn H., Paek S. H., and Cho D.-W., Nat. Biomed. Eng. 3, 509 (2019). 10.1038/s41551-019-0363-x [DOI] [PubMed] [Google Scholar]
- 99.Chen K., Jiang E., Wei X., Xia Y., Wu Z., Gong Z., Shang Z., and Guo S., Lab Chip 21, 1604-1612 (2021). 10.1039/D1LC00003A [DOI] [PubMed] [Google Scholar]
- 100.Tang M., Tiwari S. K., Agrawal K., Tan M., Dang J., Tam T., Tian J., Wan X., Schimelman J., You S., Xia Q., Rana T. M., and Chen S., Small 17, 2006050 (2021). 10.1002/smll.202006050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xing F., Front. Biosci. 15, 166 (2010). 10.2741/3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hida K., Hida Y., Amin D. N., Flint A. F., Panigrahy D., Morton C. C., and Klagsbrun M., Cancer Res. 64, 8249 (2004). 10.1158/0008-5472.CAN-04-1567 [DOI] [PubMed] [Google Scholar]
- 103.Sleeman J. P. and Thiele W., Int. J. Cancer 125, 2747 (2009). 10.1002/ijc.24702 [DOI] [PubMed] [Google Scholar]
- 104.Wang S., Zhou Y., Qin X., Nair S., Huang X., and Liu Y., Sci. Rep. 10, 12226 (2020). 10.1038/s41598-020-69056-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Seyfried T. N. and Huysentruyt L. C., Crit. Rev. Oncog. 18, 43 (2013). 10.1615/CritRevOncog.v18.i1-2.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Siemann D. W., Cancer Treat. Rev. 37, 63 (2011). 10.1016/j.ctrv.2010.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim H. N., Habbit N. L., Su C., Choi N., Ahn E. H., Lipke E. A., and Kim D., Adv. Funct. Mater. 29, 1807553 (2019). 10.1002/adfm.201807553 [DOI] [Google Scholar]
- 108.Herter S., Morra L., Schlenker R., Sulcova J., Fahrni L., Waldhauer I., Lehmann S., Reisländer T., Agarkova I., Kelm J. M., Klein C., Umana P., and Bacac M., Cancer Immunol. Immunother. 66, 129 (2017). 10.1007/s00262-016-1927-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Antunes J., Gaspar V. M., Ferreira L., Monteiro M., Henrique R., Jerónimo C., and Mano J. F., Acta Biomater. 94, 392 (2019). 10.1016/j.actbio.2019.06.012 [DOI] [PubMed] [Google Scholar]
- 110.Yakavets I., Jenard S., Francois A., Maklygina Y., Loschenov V., Lassalle H.-P., Dolivet G., and Bezdetnaya L., J. Clin. Med. 8, 1686 (2019). 10.3390/jcm8101686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Carey S. P., Starchenko A., McGregor A. L., and Reinhart-King C. A., Clin. Exp. Metastasis 30, 615 (2013). 10.1007/s10585-013-9565-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.De Moor L., Merovci I., Baetens S., Verstraeten J., Kowalska P., Krysko D. V., De Vos W. H., and Declercq H., Biofabrication 10, 035009 (2018). 10.1088/1758-5090/aac7e6 [DOI] [PubMed] [Google Scholar]
- 113.Lazzari G., Nicolas V., Matsusaki M., Akashi M., Couvreur P., and Mura S., Acta Biomater. 78, 296 (2018). 10.1016/j.actbio.2018.08.008 [DOI] [PubMed] [Google Scholar]
- 114.Wu Z., Chen B., Wu Y., Xia Y., Chen H., Gong Z., Hu H., Ding Z., and Guo S., Lab Chip 21, 3498-3508 (2021). 10.1039/D1LC00496D [DOI] [PubMed] [Google Scholar]
- 115.Ehsan S. M., Welch-Reardon K. M., Waterman M. L., Hughes C. C. W., and George S. C., Integr. Biol. 6, 603 (2014). 10.1039/c3ib40170g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hachey S. J., Movsesyan S., Nguyen Q. H., Burton-Sojo G., Tankazyan A., Wu J., Hoang T., Zhao D., Wang S., Hatch M. M., Celaya E., Gomez S., Chen G. T., Davis R. T., Nee K., Pervolarakis N., Lawson D. A., Kessenbrock K., Lee A. P., Lowengrub J., Waterman M. L., and Hughes C. C. W., Lab Chip 21, 1333-1351 (2021). 10.1039/D0LC01216E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Singh M. S., Goldsmith M., Thakur K., Chatterjee S., Landesman-Milo D., Levy T., Kunz-Schughart L. A., Barenholz Y., and Peer D., Nanoscale 12, 1894 (2020). 10.1039/C9NR09572A [DOI] [PubMed] [Google Scholar]
- 118.Meng F., Meyer C. M., Joung D., Vallera D. A., McAlpine M. C., and Panoskaltsis-Mortari A., Adv. Mater. 31, 1806899 (2019). 10.1002/adma.201806899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Haase K., Offeddu G. S., Gillrie M. R., and Kamm R. D., Adv. Funct. Mater. 30, 2002444 (2020). 10.1002/adfm.202002444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pluda J. M., Semin. Oncol. 24, 203 (1997). [PubMed] [Google Scholar]
- 121.Sano E., Mori C., Nashimoto Y., Yokokawa R., Kotera H., and Torisawa Y.-S., Biomicrofluidics 12, 042204 (2018). 10.1063/1.5027183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nie J., Gao Q., Xie C., Lv S., Qiu J., Liu Y., Guo M., Guo R., Fu J., and He Y., Mater. Horiz. 7, 82 (2020). 10.1039/C9MH01283D [DOI] [Google Scholar]
- 123.Wang H.-F., Ran R., Liu Y., Hui Y., Zeng B., Chen D., Weitz D. A., and Zhao C.-X., ACS Nano 12, 11600 (2018). 10.1021/acsnano.8b06846 [DOI] [PubMed] [Google Scholar]
- 124.Sobrino A., Phan D. T. T., Datta R., Wang X., Hachey S. J., Romero-López M., Gratton E., Lee A. P., George S. C., and Hughes C. C. W., Sci. Rep. 6, 31589 (2016). 10.1038/srep31589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nashimoto Y., Okada R., Hanada S., Arima Y., Nishiyama K., Miura T., and Yokokawa R., Biomaterials 229, 119547 (2020). 10.1016/j.biomaterials.2019.119547 [DOI] [PubMed] [Google Scholar]
- 126.Ko J., Ahn J., Kim S., Lee Y., Lee J., Park D., and Jeon N. L., Lab Chip 19, 2822 (2019). 10.1039/C9LC00140A [DOI] [PubMed] [Google Scholar]
- 127.Paek J., Park S. E., Lu Q., Park K.-T., Cho M., Oh J. M., Kwon K. W., Yi Y.-S., Song J. W., Edelstein H. I., Ishibashi J., Yang W., Myerson J. W., Kiseleva R. Y., Aprelev P., Hood E. D., Stambolian D., Seale P., Muzykantov V. R., and Huh D., ACS Nano 13, 7627 (2019). 10.1021/acsnano.9b00686 [DOI] [PubMed] [Google Scholar]
- 128.Oh S., Ryu H., Tahk D., Ko J., Chung Y., Lee H. K., Lee T. R., and Jeon N. L., Lab Chip 17, 3405 (2017). 10.1039/C7LC00646B [DOI] [PubMed] [Google Scholar]
- 129.Dey M., Ayan B., Yurieva M., Unutmaz D., and Ozbolat I. T., Adv Biol. 5, 2100090 (2021). 10.1002/adbi.202100090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li C., Li S., Du K., Li P., Qiu B., and Ding W., ACS Appl. Mater. Interfaces 13, 19768 (2021). 10.1021/acsami.1c03740 [DOI] [PubMed] [Google Scholar]
- 131.Lee S. W., Hong S., Jung B., Jeong S. Y., Byeon J. H., Jeong G. S., Choi J., and Hwang C., Biotechnol. Bioeng. 116, 3041 (2019). 10.1002/bit.27114 [DOI] [PubMed] [Google Scholar]
- 132.Järvinen P., Bonabi A., Jokinen V., and Sikanen T., Adv. Funct. Mater. 30, 2000479 (2020). 10.1002/adfm.202000479 [DOI] [Google Scholar]
- 133.Kwak T. J. and Lee E., Sci. Rep. 10, 20142 (2020). 10.1038/s41598-020-77180-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ayuso J. M., Virumbrales-Munoz M., McMinn P. H., Rehman S., Gomez I., Karim M. R., Trusttchel R., Wisinski K. B., Beebe D. J., and Skala M. C., Lab Chip 19, 3461 (2019). 10.1039/C9LC00270G [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kerr S. C., Morgan M. M., Gillette A. A., Livingston M. K., Lugo-Cintron K. M., Favreau P. F., Florek L., Johnson B. P., Lang J. M., Skala M. C., and Beebe D. J., Integr. Biol. 12, 250 (2020). 10.1093/intbio/zyaa020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Humayun M., Ayuso J. M., Brenneke R. A., Virumbrales-Muñoz M., Lugo-Cintrón K., Kerr S., Ponik S. M., and Beebe D. J., Biomaterials 270, 120640 (2021). 10.1016/j.biomaterials.2020.120640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Silvestri V. L., Henriet E., Linville R. M., Wong A. D., Searson P. C., and Ewald A. J., Cancer Res. 80, 4288 (2020). 10.1158/0008-5472.CAN-19-1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chang Y. S., di Tomaso E., McDonald D. M., Jones R., Jain R. K., and Munn L. L., Proc. Natl. Acad. Sci. U.S.A. 97, 14608 (2000). 10.1073/pnas.97.26.14608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lin C., Lin L., Mao S., Yang L., Yi L., Lin X., Wang J., Lin Z.-X., and Lin J.-M., Anal. Chem. 90, 10326 (2018). 10.1021/acs.analchem.8b02133 [DOI] [PubMed] [Google Scholar]
- 140.Hassell B. A., Goyal G., Lee E., Sontheimer-Phelps A., Levy O., Chen C. S., and Ingber D. E., Cell Rep. 21, 508 (2017). 10.1016/j.celrep.2017.09.043 [DOI] [PubMed] [Google Scholar]
- 141.Choi Y., Hyun E., Seo J., Blundell C., Kim H. C., Lee E., Lee S. H., Moon A., Moon W. K., and Huh D., Lab Chip 15, 3350 (2015). 10.1039/C5LC00514K [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xu Z., Li E., Guo Z., Yu R., Hao H., Xu Y., Sun Z., Li X., Lyu J., and Wang Q., ACS Appl. Mater. Interfaces 8, 25840 (2016). 10.1021/acsami.6b08746 [DOI] [PubMed] [Google Scholar]
- 143.Tang Y., Soroush F., Sheffield J. B., Wang B., Prabhakarpandian B., and Kiani M. F., Sci. Rep. 7, 9359 (2017). 10.1038/s41598-017-09815-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shi W., Reid L., Huang Y., Uhl C. G., He R., Zhou C., and Liu Y., Biomicrofluidics 13, 044108 (2019). 10.1063/1.5108681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Song J. W., Cavnar S. P., Walker A. C., Luker K. E., Gupta M., Tung Y.-C., Luker G. D., and Takayama S., PLoS ONE 4, e5756 (2009). 10.1371/journal.pone.0005756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Marturano-Kruik A., Nava M. M., Yeager K., Chramiec A., Hao L., Robinson S., Guo E., Raimondi M. T., and Vunjak-Novakovic G., Proc. Natl. Acad. Sci. U.S.A. 115, 1256 (2018). 10.1073/pnas.1714282115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jing B., Luo Y., Lin B., Li J., Wang Z. A., and Du Y., RSC Adv. 9, 17137 (2019). 10.1039/C9RA02069A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Uhl C. G. and Liu Y., Lab Chip 19, 1458 (2019). 10.1039/C8LC01250D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Thomas A., Daniel Ou-Yang H., Lowe-Krentz L., Muzykantov V. R., and Liu Y., Biomicrofluidics 10, 014101 (2016). 10.1063/1.4936672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Mondrinos M. J., Yi Y.-S., Wu N.-K., Ding X., and Huh D., Lab Chip 17, 3146 (2017). 10.1039/C7LC00317J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gioeli D., Snow C. J., Simmers M. B., Hoang S. A., Figler R. A., Allende J. A., Roller D. G., Parsons J. T., Wulfkuhle J. D., Petricoin E. F., Bauer T. W., and Wamhoff B. R., Lab Chip 19, 1193 (2019). 10.1039/C8LC00755A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang Y., Wang L., Zhu Y., and Qin J., Lab Chip 18, 851 (2018). 10.1039/C7LC01084B [DOI] [PubMed] [Google Scholar]
- 153.Tao T., Wang Y., Chen W., Li Z., Su W., Guo Y., Deng P., and Qin J., Lab Chip 19, 948 (2019). 10.1039/C8LC01298A [DOI] [PubMed] [Google Scholar]
- 154.Wang Y., Wang H., Deng P., Chen W., Guo Y., Tao T., and Qin J., Lab Chip 18, 3606 (2018). 10.1039/C8LC00869H [DOI] [PubMed] [Google Scholar]
- 155.Jung D. J., Shin T. H., Kim M., Sung C. O., Jang S. J., and Jeong G. S., Lab Chip 19, 2854 (2019). 10.1039/C9LC00496C [DOI] [PubMed] [Google Scholar]
- 156.Lancaster M. A. and Knoblich J. A., Science 345, 1247125 (2014). 10.1126/science.1247125 [DOI] [PubMed] [Google Scholar]
- 157.Park S. E., Georgescu A., and Huh D., Science 364, 960 (2019). 10.1126/science.aaw7894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Karzbrun E., Kshirsagar A., Cohen S. R., Hanna J. H., and Reiner O., Nat. Phys. 14, 515 (2018). 10.1038/s41567-018-0046-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ao Z., Cai H., Havert D. J., Wu Z., Gong Z., Beggs J. M., Mackie K., and Guo F., Anal. Chem. 92, 4630 (2020). 10.1021/acs.analchem.0c00205 [DOI] [PubMed] [Google Scholar]
- 160.Patel S. N., Ishahak M., Chaimov D., Velraj A., LaShoto D., Hagan D. W., Buchwald P., Phelps E. A., Agarwal A., and Stabler C. L., Sci. Adv. 7, eaba5515 (2021). 10.1126/sciadv.aba5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sokolowska P., Zukowski K., Janikiewicz J., Jastrzebska E., Dobrzyn A., and Brzozka Z., Biosens. Bioelectron. 183, 113215 (2021). 10.1016/j.bios.2021.113215 [DOI] [PubMed] [Google Scholar]
- 162.Yu F., Deng R., Hao Tong W., Huan L., Chan Way N., IslamBadhan A., Iliescu C., and Yu H., Sci. Rep. 7, 14528 (2017). 10.1038/s41598-017-13848-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Takebe T., Sekine K., Enomura M., Koike H., Kimura M., Ogaeri T., Zhang R.-R., Ueno Y., Zheng Y.-W., Koike N., Aoyama S., Adachi Y., and Taniguchi H., Nature 499, 481 (2013). 10.1038/nature12271 [DOI] [PubMed] [Google Scholar]
- 164.Driehuis E., Kretzschmar K., and Clevers H., Nat. Protoc. 15, 3380 (2020). 10.1038/s41596-020-0379-4 [DOI] [PubMed] [Google Scholar]
- 165.Carvalho M. R., Barata D., Teixeira L. M., Giselbrecht S., Reis R. L., Oliveira J. M., Truckenmüller R., and Habibovic P., Sci. Adv. 5, 1317 (2019). 10.1126/sciadv.aaw1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sarkar S., Peng C.-C., Kuo C. W., Chueh D.-Y., Wu H.-M., Liu Y.-H., Chen P., and Tung Y.-C., RSC Adv. 8, 30320 (2018). 10.1039/C8RA05505J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ancora D., Di Battista D., Giasafaki G., Psycharakis S. E., Liapis E., Ripoll J., and Zacharakis G., Sci. Rep. 7, 11854 (2017). 10.1038/s41598-017-12193-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lei K. F., Lin B.-Y., and Tsang N.-M., RSC Adv. 7, 13939 (2017). 10.1039/C7RA00209B [DOI] [Google Scholar]
- 169.Misun P. M., Rothe J., Schmid Y. R. F., Hierlemann A., and Frey O., Microsyst. Nanoeng. 2, 16022 (2016). 10.1038/micronano.2016.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhang Z., Chen L., Wang Y., Zhang T., Chen Y.-C., and Yoon E., Anal. Chem. 91, 14093 (2019). 10.1021/acs.analchem.9b03896 [DOI] [PubMed] [Google Scholar]
- 171.Chen Z., Ma N., Sun X., Li Q., Zeng Y., Chen F., Sun S., Xu J., Zhang J., Ye H., Ge J., Zhang Z., Cui X., Leong K., Chen Y., and Gu Z., Biomaterials 272, 120770 (2021). 10.1016/j.biomaterials.2021.120770 [DOI] [PubMed] [Google Scholar]
- 172.Hou Y., Konen J., Brat D. J., Marcus A. I., and Cooper L. A. D., Sci. Rep. 8, 7248 (2018). 10.1038/s41598-018-25337-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Huang Y., Wang S., Guo Q., Kessel S., Rubinoff I., Chan L. L.-Y., Li P., Liu Y., Qiu J., and Zhou C., Cancer Res. 77, 6011 (2017). 10.1158/0008-5472.CAN-17-0821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wu H., Yang Y., Bagnaninchi P. O., and Jia J., Analyst 143, 4189-4198 (2018). 10.1039/C8AN00729B [DOI] [PubMed] [Google Scholar]
- 175.Patra B., Sharma M., Hale W., and Utz M., Sci. Rep. 11, 53 (2021). 10.1038/s41598-020-79693-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mittler F., Obeïd P., Rulina A. V., Haguet V., Gidrol X., and Balakirev M. Y., Front. Oncol. 7, 293 (2017). 10.3389/fonc.2017.00293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Benning L., Peintner A., Finkenzeller G., and Peintner L., Sci. Rep. 10, 11071 (2020). 10.1038/s41598-020-67960-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Grexa I., Diosdi A., Harmati M., Kriston A., Moshkov N., Buzas K., Pietiäinen V., Koos K., and Horvath P., Sci. Rep. 11, 14813 (2021). 10.1038/s41598-021-94217-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Xin Yu J., Hubbard-Lucey V. M., and Tang J., Nat. Rev. Drug Discovery 18, 821 (2019). 10.1038/d41573-019-00090-z [DOI] [PubMed] [Google Scholar]
- 180.Lee S. W. L., Adriani G., Ceccarello E., Pavesi A., Tan A. T., Bertoletti A., Kamm R. D., and Wong S. C., Front. Immunol. 9, 416 (2018). 10.3389/fimmu.2018.00416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cui X., Ma C., Vasudevaraja V., Serrano J., Tong J., Peng Y., Delorenzo M., Shen G., Frenster J., Morales R.-T. T., Qian W., Tsirigos A., Chi A. S., Jain R., Kurz S. C., Sulman E. P., Placantonakis D. G., Snuderl M., and Chen W., eLife 9, e52253 (2020). 10.7554/eLife.52253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Aung A., Kumar V., Theprungsirikul J., Davey S. K., and Varghese S., Cancer Res. 80, 263 (2020). 10.1158/0008-5472.CAN-19-0342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jenkins R. W., Aref A. R., Lizotte P. H., Ivanova E., Stinson S., Zhou C. W., Bowden M., Deng J., Liu H., Miao D., He M. X., Walker W., Zhang G., Tian T., Cheng C., Wei Z., Palakurthi S., Bittinger M., Vitzthum H., Kim J. W., Merlino A., Quinn M., Venkataramani C., Kaplan J. A., Portell A., Gokhale P. C., Phillips B., Smart A., Rotem A., Jones R. E., Keogh L., Anguiano M., Stapleton L., Jia Z., Barzily-Rokni M., Cañadas I., Thai T. C., Hammond M. R., Vlahos R., Wang E. S., Zhang H., Li S., Hanna G. J., Huang W., Hoang M. P., Piris A., Eliane J.-P., Stemmer-Rachamimov A. O., Cameron L., Su M.-J., Shah P., Izar B., Thakuria M., LeBoeuf N. R., Rabinowits G., Gunda V., Parangi S., Cleary J. M., Miller B. C., Kitajima S., Thummalapalli R., Miao B., Barbie T. U., Sivathanu V., Wong J., Richards W. G., Bueno R., Yoon C. H., Miret J., Herlyn M., Garraway L. A., Van Allen E. M., Freeman G. J., Kirschmeier P. T., Lorch J. H., Ott P. A., Hodi F. S., Flaherty K. T., Kamm R. D., Boland G. M., Wong K.-K., Dornan D., Paweletz C. P., and Barbie D. A., Cancer Discovery 8, 196 (2018). 10.1158/2159-8290.CD-17-0833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Driehuis E., Kolders S., Spelier S., Lõhmussaar K., Willems S. M., Devriese L. A., de Bree R., de Ruiter E. J., Korving J., Begthel H., van Es J. H., Geurts V., He G.-W., van Jaarsveld R. H., Oka R., Muraro M. J., Vivié J., Zandvliet M. M. J. M., Hendrickx A. P. A., Iakobachvili N., Sridevi P., Kranenburg O., van Boxtel R., Kops G. J. P. L., Tuveson D. A., Peters P. J., van Oudenaarden A., and Clevers H., Cancer Discovery 9, 852 (2019). 10.1158/2159-8290.CD-18-1522 [DOI] [PubMed] [Google Scholar]
- 185.Ma C., Peng Y., Li H., and Chen W., Trends Pharmacol. Sci. 42, 119 (2021). 10.1016/j.tips.2020.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.