Abstract
Klebsiella pneumoniae complex (KPC) accounts for approximately one-third of all Gram-negative infections. Moreover, it is highly resistant and can taxonomically be distributed into KpI, KpII, and KpIII phylogroups. This study aimed to investigate the distribution of phylogenetic groups and the relationship between them and antibiotic resistance patterns. For this purpose, we collected KPC isolates from Tabriz, Iran, between 2018 and 2020. Antimicrobial susceptibility testing was performed by disk diffusion agar, and phylogenetic groups were then examined using gyrA restriction fragment length polymorphism (RFLP) and parC PCR methods. A total of 100 KPC isolates were obtained from the clinical specimens (urine, respiratory secretion, blood, wounds, and trachea). The enrolled patients included 47 men and 53 women aged from 1 to 91 years old. The highest sensitivity was found related to fosfomycin as 85%, followed by amikacin as 66%. The three phylogenetically groups by the RFLP-PCR method were found in KPC, 96% (96 isolates) as KpI, 3% (3 isolates) as KpII, and 1% (1isolate) as KpIII. The highest antibiotic resistance was observed in KpI. It was shown that a valid identification of three phylogenetic groups of KPC can be done by combining both gyrA PCR-RFLP and parC PCR. Of note, the KpI group was also observed as the dominant phylogenetic group with the highest resistance to antibiotics.
1. Introduction
Edwin Klebs first described Klebsiella pneumoniae complex (KPC) organisms in 1875, while studying the airways of patients who died due to pneumonia [1]. Additionally, Carl Friedlander formally described the species in 1882 [1, 2]. KPC is a Gram-negative bacterium from the Enterobacteriaceae family that can consequently cause various types of infections in human beings and is also found in both animals and plants [2]. KPC has a large adjunct genome of plasmids and chromosomal gene loci. In addition, KPC strains can be divided into multidrug-resistant, hypervirulent, and opportunistic groups based on the accessory genome [3]. Unfortunately, no approved potential therapy currently exists for some hypervirulent strains of this organism. Therefore, KPC can be classified as ESKAPE organisms [4], which “is a group consisting of the six most important microorganisms resistant to antimicrobials worldwide (Enterococcus faecium, Staphylococcus aureus, KPC, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.)” [5]. Of note, the important virulence factors of KPC were indicated to be resistant to serum, lipopolysaccharide, siderophores, pili, capsular antigens, and adhesions [6]. This bacterium is responsible for approximately one-third of all Gram-negative infections such as surgical wound infections, cystitis, urinary tract infections, pneumonia, endocarditis, and sepsis [7]. Moreover, it is often found in inflammation of necrotic lungs, pyogenic liver abscesses, internal endophthalmitis, meningitis, necrotizing fasciitis, and splenic abscesses [8]. KPC limits the treatment options for several groups of antibiotics by acquiring antibiotic resistance and innate resistance genes [9]. Currently, KPC strains by producing various types of β-lactamases (ESBLs) and carbapenemases can be found almost in all around the world [9]. KPC is classically and taxonomically divided into the KpI, KpII, and KpIII phylogroups [9, 10]. Recently, it has been suggested that these phylogroups, which are collectively known as the KPC, can be classified as separate species as follows: K. pneumoniae (KpI), K. quasipneumoniae (KpII), and K. variicola (KpIII) [11]. Up to now, several methods such as restriction fragment length polymorphism (RFLP-PCR), phylogenetic analysis of the ori region [12], phylogenetic analysis by sequencing the dnaJ gene [13], gyrB gene sequence, and the parC gene [14] have been introduced in order to determine the phylogenetic type of Enterobacteriaceae. Sequence analyses of gyrA and parC genes have shown that these genes are the reliable phylogenetic markers because they are not prone to repeated horizontal translocation across clusters [14]. Therefore, this study aimed to investigate the distribution of phylogenetic groups in KPC obtained from Tabriz city as well as determining the relationship between antibiotic resistance patterns and phylogenetic groups using the RFLP-PCR method.
2. Materials and Methods
2.1. Bacterial Isolation and Identification
At this stage, a total of 100 KPC isolates were obtained from various clinical specimens (including urine, respiratory secretion, blood, wounds, and trachea) at Imam Reza Hospital, Tabriz, Iran, between 2018 and 2020. Thereafter, the organisms were identified by the microscopic feature and differential tests [15]. Finally, these were stored in the tryptic soy broth medium containing glycerol and preserved at −70°C [16]. Notably, informed consent was obtained from all the adult participants and their parents or from legal guardians of minors participating in this study.
2.2. Antibiotic Susceptibilities
At this stage, the susceptibility testing was performed using the modified Kirby–Bauer method in Mueller-Hinton Agar Merck Co. (Darmstadt, Germany) for classes of antibiotics (Mast, Merseyside, UK), including fluoroquinolones (ciprofloxacin (CIP, 5 μg)), aminoglycoside (amikacin (AK, 30 μg) and gentamicin (GEN, 10 μg)), beta-lactams (ampicillin (AMP, 10 μg), imipenem (IPM, 10 μg), ertapenem (ERT, 10 μg), ceftazidime (CAZ, 30 μg), cefepime (FEP, 30 μg), ceftriaxone (CRO, 30 μg), piperacillin (PIP, 100 μg), and piperacillin/tazobactam (PTZ, 100/12.5 μg)), antimetabolites (co-trimoxazole (SX, 30 μg)), and fosfomycin (FOS, 200/50 μg), which were interpreted in terms of the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [17]. Moreover, in the current study, K. pneumoniae ATCC 12022 was used as the quality control strain.
2.3. DNA Preparation and Identification by the RFLP-PCR of gyrA Gene
The DNA extraction process was performed using the standard cell lysis. The isolates were then confirmed by a 441-bp PCR fragment of the gyrA gene (5`′-CGCGTACTATACGCCATGAACGTA-3′) and gyrA-C (5′-ACCGTTGATCACTTCGGTCAGG-3′) [14]. Thereafter, the RFLP-PCR was performed using restriction enzymes TaqI and HaeIII [18]. Thereafter, in order to confirm the identification of the KpII group, PCR was performed using the parC primer (parC2-1 (5′-GGCGCAACCCTTCTCCTAT-3′) and parC2-3 (5′-GAGCAGGATGTTTGGCAGG-3′)), which is specific to the KpII genotype, a 232 bp PCR product of which is only sound in the KpII isolates [19]. Finally, the RFLP products underwent electrophoresis in 2.5% agarose gel, followed by staining with 0.5 μg/mL safe stain and visualizing under ultraviolet light.
2.4. Statistical Analysis
The results were analyzed using the descriptive statistics in SPSS software for Windows (Version 23 SPSS Inc., Chicago, IL, USA). P values ≤0.05 were considered as statistically significant.
3. Results
In this study, based on the statistical analyses, a total of 100 KPC were collected from 47 (47%) male and 53 (53%) female cases. The mean age of the included patients (47 men and 53 women) was 25 ± 41 years old. The sources of the collected samples were urine (58%), blood (16%), wounds (9%), trachea (9%), and respiratory secretion (8%). Notably, the obtained samples were isolated from different wards, including internal (27%), ICU (22%), infectious (18%), surgery (13%), pediatric (11%), and emergency (9%) wards.
Table 1 provides the susceptibility testing of the KPC isolates to different antibiotics. The phylogenetic typing of the clinical KPC isolates was successfully done using the RFLP-PCR method. In the present study, 96 isolates were identified as KpI, 3 isolates as KpII, and only 1 isolate as KpIII (Figure 1).
Table 1.
Proportions of K. pneumoniae susceptibility testing in the three phylogenetic groups.
| Sensitive | Resistant | Intermediate | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Antibiotics | KpI | KpII | KpIII | KpI | KpII | KpIII | KpI | KpII | KpIII |
| Ampicillin | 0 | 0 | 0 | 96 | 3 | 1 | 0 | 0 | 0 |
| Piperacillin | 5 | 0 | 1 | 82 | 1 | 0 | 19 | 2 | 0 |
| Piperacillin/tazoba | 34 | 3 | 1 | 43 | 0 | 0 | 9 | 0 | 0 |
| Imipenem | 46 | 3 | 1 | 30 | 0 | 0 | 20 | 0 | 0 |
| Ertapenem | 42 | 2 | 1 | 33 | 1 | 0 | 19 | 2 | 0 |
| Cefepime | 19 | 1 | 1 | 62 | 1 | 0 | 15 | 1 | 0 |
| Ciprofloxacin | 26 | 2 | 1 | 56 | 1 | 0 | 14 | 0 | 0 |
| Co-trimoxazole | 12 | 1 | 0 | 75 | 2 | 0 | 9 | 0 | 1 |
| Fosfomycin | 83 | 2 | 0 | 6 | 1 | 1 | 7 | 0 | 0 |
| Gentamicin | 22 | 2 | 1 | 62 | 1 | 0 | 12 | 0 | 0 |
| Amikacin | 63 | 2 | 1 | 21 | 1 | 0 | 12 | 0 | 0 |
| Ceftazidime | 10 | 1 | 1 | 83 | 1 | 0 | 3 | 1 | 0 |
| Ceftriaxone | 8 | 2 | 1 | 80 | 1 | 0 | 8 | 0 | 0 |
Figure 1.
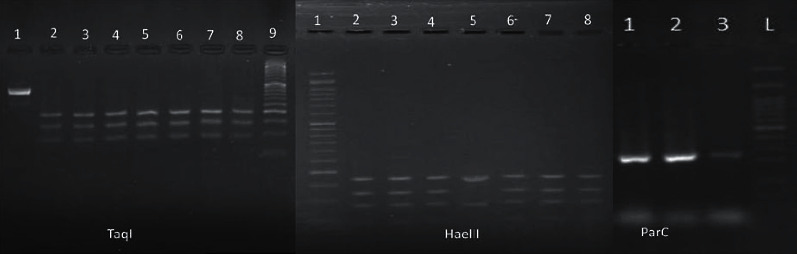
Taql, electrophoresis of the GyrA gene after adding the TaqI enzyme. Lane 1, electrophoresis of the GyrA gene prior to adding the enzyme with 441 bp bandwidth; lanes 2, 3, and 4, KpI group; lane 5, KpIII group; lanes 6, 7, and 8, KpII group; lane 9, 50 bp DNA ladder. HaeIII, electrophoresis of the GyrA gene after adding the HaeIII enzyme. Lane 1, 50 bp DNA ladder; lanes 2, 3, and 4, KpI group; lane 5, KpIII group; lanes 6, 7, and 8, KpII group. parC, parC gene electrophoresis to confirm the KpII group with 232 bp bandwidth. Lanes 1, 2, and 3, parC positive isolates with 232 bp bandwidth; lane 4, 100 bp DNA ladder. In KpI strains, the Taql enzyme created four bonds (including 197 bp, 142 bp, 93 bp, and 9 bp) and the HaeIII enzyme created four bonds (including 175 bp, 129 bp, 92 bp, and 45 bp). In KpII strains, the Taql enzyme created three bonds (including 197 bp, 151 bp, and 93 bp) and the HaeIII enzyme created four bonds (including 175 bp, 129 bp, 92 bp, and 45 bp). In KpIII strains, the Taql enzyme created four bonds (including 197 bp, 142 bp, 93 bp, and 9 bp) and the HaeIII enzyme created three bonds (including 175 bp, 174 bp, and 92 bp).
The number of bacteria (five strains of KpI, three strains of KpII, and one strain of KpIII) was confirmed by sequence analysis of both gyrA and parC genes. The highest percentage of antibiotic resistance was observed in the KpI group, followed by the KpII and KpIII groups (P value ≤0.05), respectively (Table 1). There was a significant statistical difference in the KPC groups in terms of resistance to antibiotics and wards (P values ≤0.05) (Tables 1 and 2).
Table 2.
The association among gender, wards, and sources in the phylogenetic groups.
| KPI | KPII | KPIII | P value | |
|---|---|---|---|---|
| Gender | ||||
| Male | 46 | 1 | 0 | 0.50 |
| Female | 50 | 2 | 1 | |
|
| ||||
| Sample | ||||
| Urine | 56 | 2 | 0 | 0.35 |
| Blood | 15 | 0 | 1 | |
| Wound | 9 | 0 | 0 | |
| Trachea | 9 | 0 | 0 | |
| Sputum | 7 | 1 | 0 | |
|
| ||||
| Wards | ||||
| Internal | 27 | 0 | 0 | 0.03 |
| ICU | 21 | 1 | 0 | |
| Infectious | 18 | 0 | 0 | |
| Surgery | 13 | 0 | 0 | |
| Emergency | 9 | 0 | 0 | |
| Pediatrics | 8 | 2 | 1 | |
4. Discussion
KPC is responsible for both nosocomial and community infections, and the resistance to antibiotics among KPC is very important in this regard. Correspondingly, KpI, KpII, and KpIII were found in 96 isolates, three isolates, and one isolate, respectively. The phylogenetic types have also been found to be associated with drug resistance.
Brisse et al. in their study have reported the frequency of group I as 82.1%, KpII as 6.9%, and KpIII as 11%. Additionally, resistance to all antibiotics was more common in KpI compared to the other types, and a significant relationship was also found between antibiotic resistances and both KpI and KpII [19]. Silva de Melo et al. in their study have analyzed phylogenetic groups KPC isolated from nosocomial infections, community-based infections, and natural microbiota. As a result, they found that KpI had the most dominant phylogeny, and the highest percentage of antibiotic resistance was observed in the KpI, KpII, and KpIII groups, respectively [10]. In a previous study performed in Iran by Pajand et al., the clonal relationship between blaNDM/OXA-48-producing strains and the phylogenetic type of KPC was examined, and as a result, 80.3% of the isolates belonged to KpI, 16.4% to KpII, and 2.5% to the KpIII. Furthermore, most of the isolates producing NDM-1 (81%) belonged to KpI and the rest of them belonged to KpII and KpIII [20]. In a study conducted by Brisse and van Duijkeren [18], the distributions of three phylogenetic groups of KPC were examined in clinical isolates obtained from animals. All multidrug-resistant isolates belonged either to the KpI or to KpII phylogenetic group, and a significant relationship was found between antibiotic resistances and KpI and KpII [18]. In 2010, Younes et al. have reported the prevalence of blaCTX-M-15 transmissible isolates of K. oxytoca in hospitals and communities in Scotland. Interestingly, the phylogenetic type of all 219 KPC was assigned to KpI [21]. Moreover, Atmani et al. in Algiers have studied the virulence factors' characteristics and genetic background of ESBL-producing KPC isolated from wastewater. Besides, by performing gyrA PCR-RFLP phylogenetic analysis, it was shown that almost all strains of wastewater, similar to our clinical strains, belong to the phylogenetic group KpI [22]. Correspondingly, this result also highlights the pathogenicity of wastewater strains, which was shown to have a significant relationship with KpI [22]. Although these results were not obtained from isolates from human beings, to the best of our knowledge, KPC is an enteric pathogen that can transfer from food, animals, and water to humans. In a study conducted by Pons et al. in Mozambique from 2004 to 2009, characterization of KPC-producing ESBLs causing bacteremia and urinary tract infection was investigated. Accordingly, they have shown that all isolates of KPC belong to the same phylogenetic group (KpI) by gyrA gene analysis, and a significant relationship was also found between the high prevalence of KpI isolates and antibiotic resistance [23]. Due to the health status of the host, geographic climatic conditions, dietary factors, the history of the use of antibiotics, and host genetic factors, the rate of the phylogenetic groups is different among various populations [24].
In silicon restriction analysis of gyrA sequences obtained previously, it was shown that gyrA RFLP-PCR assigns isolates reliably to one of the three KPC groups [14]. Brisse et al. have also reported that all their 100 studied isolates had a distinct gyrA RFLP-PCR profile, which then matched with the reference strains and both the gyrA and parC sequencing [19]. Nowadays, we can investigate type KPC using some molecular assays such as RFLP and direct sequencing. Besides, it should be noted that the use of the RFLP-PCR method is more cost-effective than sequencing [14].
In the current study, in terms of the modified Kirby–Bauer method, the highest resistance rates were found to be related to ceftazidime (88%) and piperacillin (84%), respectively. As well, the most effective antibiotic against KPC was fosfomycin (85%), followed by amikacin (66%) and imipenem (50%), respectively. Moreover, Jazayeri et al. in their study have examined the patterns of antibiotic resistance KPC in Semnan, Iran. Accordingly, blaSHV and blaTEM were observed in 46.8% and 33.5%, and the highest resistance belonged to ceftazidime and aztreonam, and the lowest one belonged to imipenem and cefepime [25]. In the current study, the highest rate of resistance was found to be related to ceftazidime followed by that of piperacillin. As well, the lowest resistance to antibiotics was discovered to belong to fosfomycin followed by amikacin. In 2019, Shahi et al. studied the patterns of antibiotic resistance of KPC strains isolated from different clinical specimens in the teaching hospitals of Tabriz. In this regard, of 104 strains of KPC, the highest antibiotic resistance was observed to be related to co-trimoxazole, and the lowest resistance belonged to colistin [26]. Fonseca et al. in their study have mentioned that KPC has three separate species, namely, K. pneumoniae (KpI), K. quasipneumoniae (KpII), and K. variicola (KpIII). The presence of genes such as blaKPC in K. variicola and K. quasipneumoniae strains as well as NDM-1 metallo-β-lactamase in K. variicola is more worrying because these genes confer resistance to many different β-lactam antibiotics. Of particular concern is the evidence of homologous recombination among these three species of Klebsiella [27]. From 2018 to 2020, various studies have been conducted on antibiotic susceptibility testing, showing a growing trend of antibiotic resistance [28–30].
Since strains of KPC cause various infections, it has different virulence factors, pathogenicity, and resistance to antibiotics. In addition, infections caused by KPC are mostly associated with high mortality, long-term hospitalization, and high costs [7], so phylogenotyping is recommended in this regard. Accordingly, to ensure proper treatment, having understanding on the phylogenetic groups, how to use the drugs correctly, and how it can help to prevent drug resistance and reduce treatment costs, phylogenetic typing of KPC may be essential.
The KPC has three separate species, including K. pneumoniae (KpI), K. quasipneumoniae (KpII), and K. variicola (KpIII). However, currently, there is no specific biochemical test that can detect these three different species in the usual clinical microbiology laboratories [31]. Therefore, the usual identification of Klebsiella species is still difficult, as a result of which it is now known as K. pneumoniae, and we need a strong marker to detect these species [31]. Considering that these species have distinct pathogenic and epidemiological characteristics, controlling Klebsiella infections could be improved by applying an effective method such as RFLP.
Despite our promising results, this study also had limitations such as the relatively small sample size, limitations in statistical analyses, the lack of the resistance genes study, the relationship among these resistance genes and three different genotypes, and confocal direct sequencing of all strains to further explore the types.
5. Conclusion
In conclusion, reliable identification of the three KPC phylogenetic groups can be performed by combining gyrA PCR-RFLP and parC PCR. As well, KpI was found as the most common phylogenetic type. Different distributions of these genotypes indicate their distinct pathogenicity and epidemiological characteristics, which may play a role in the control of KPC infections. In the present study, we found that the rate of KPC resistance to antimicrobial agents is high, and the highest percentage of antibiotic resistance belonged to KpI, KpII, and KpIII, respectively.
Acknowledgments
The authors thank the financial support by the Infectious and Tropical Diseases Research Center (IR, 63437), Tabriz University of Medical Sciences, Tabriz, Iran, and all those who helped them in this study.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Ristuccia P. A., Cunha B. A. Klebsiella. Infection Control . 1984;5(7):343–347. doi: 10.1017/s0195941700060549. [DOI] [PubMed] [Google Scholar]
- 2.de Oliveira Júnior N. G., Franco O. L. Promising strategies for future treatment of Klebsiella pneumoniae biofilms. Future Microbiology . 2020;15(1):63–79. doi: 10.2217/fmb-2019-0180. [DOI] [PubMed] [Google Scholar]
- 3.Martin R. M., Bachman M. A. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Frontiers in Cellular and Infection Microbiology . 2018;8(JAN):4–15. doi: 10.3389/fcimb.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shankar C., Veeraraghavan B., Nabarro L. E. B., Ravi R., Ragupathi N. K. D., Rupali P. Whole genome analysis of hypervirulent Klebsiella pneumoniae isolates from community and hospital acquired bloodstream infection. BMC Microbiology . 2018;18(1):6–9. doi: 10.1186/s12866-017-1148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernardini A., Cuesta T., Tomás A., Bengoechea J. A., Martínez J. L., Sánchez M. B. The intrinsic resistome of Klebsiella pneumoniae. International Journal of Antimicrobial Agents . 2019;53(1):29–33. doi: 10.1016/j.ijantimicag.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Ullmann U. Klebsiella spp . as nosocomial pathogens: epidemiology , taxonomy , typing methods , and pathogenicity factors. Clinical Microbiology Reviews . 1998;11(4):589–603. doi: 10.1128/cmr.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Effah C. Y., Sun T., Liu S., Wu Y. Klebsiella pneumoniae: an increasing threat to public health. Annals of Clinical Microbiology and Antimicrobials . 2020;19(1):1–9. doi: 10.1186/s12941-019-0343-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun B., Singhal S., Winslow D. L. Suppurative lymphadenitis caused by hypermucoid-variant Klebsiella in a Polynesian woman: a case report. Diagnostic Microbiology and Infectious Disease . 2020;98(4):p. 115166. doi: 10.1016/j.diagmicrobio.2020.115166. [DOI] [PubMed] [Google Scholar]
- 9.Doorduijn D. J., Rooijakkers S. H. M., van Schaik W., Bardoel B. W. Complement resistance mechanisms of Klebsiella pneumoniae. Immunobiology . 2016;221(10):1102–1109. doi: 10.1016/j.imbio.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 10.de Melo M. E. S., Cabral A. B., MacIel M. A. V., da Silveira V. M., de Souza Lopes A. C. Phylogenetic groups among Klebsiella pneumoniae isolates from Brazil: relationship with antimicrobial resistance and origin. Current Microbiology . 2011;62(5):1596–1601. doi: 10.1007/s00284-011-9903-7. [DOI] [PubMed] [Google Scholar]
- 11.Harada K., Shimizu T., Mukai Y., et al. Phenotypic and molecular characterization of antimicrobial resistance in Klebsiella spp. isolates from companion animals in Japan: clonal dissemination of multidrug-resistant extended-spectrum β-lactamase-producing Klebsiella pneumoniae. Frontiers in Microbiology . 2016;7(JUN):1021–1112. doi: 10.3389/fmicb.2016.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roggenkamp A. Phylogenetic analysis of enteric species of the family Enterobacteriaceae using the oriC-locus. Systematic and Applied Microbiology . 2007;30(3):180–188. doi: 10.1016/j.syapm.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Hong Nhung P., Ohkusu K., Mishima N., et al. Phylogeny and species identification of the family Enterobacteriaceae based on dnaJ sequences. Diagnostic Microbiology and Infectious Disease . 2007;58(2):153–161. doi: 10.1016/j.diagmicrobio.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 14.Brisse S., Verhoef J. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. International Journal of Systematic and Evolutionary Microbiology . 2001;51(3):915–924. doi: 10.1099/00207713-51-3-915. [DOI] [PubMed] [Google Scholar]
- 15.Hansen D. S., Aucken H. M., Abiola T., Podschun R. Recommended test panel for differentiation of Klebsiella species on the basis of a trilateral interlaboratory evaluation of 18 biochemical tests. Journal of Clinical Microbiology . 2004;42(8):3665–3669. doi: 10.1128/jcm.42.8.3665-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rohman A., Ijong F., Suwetja I. K. Viability of Edwardsiella tarda and Esherichia coli preserved with glycerol-tryptone soy broth (TSB) kept at freezing temperature. Aquatic Science & Management . 2013;1(2):p. 154. doi: 10.35800/jasm.1.2.2013.7278. [DOI] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. Performance standards for Antimicrobial Susceptibility Testing . 20th. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2020. [Google Scholar]
- 18.Brisse S., van Duijkeren E. Identification and antimicrobial susceptibility of 100 Klebsiella animal clinical isolates. Veterinary Microbiology . 2005;105(3–4):307–312. doi: 10.1016/j.vetmic.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Brisse S., van Himbergen T., Kusters K., Verhoef J. Research note: development of a rapid method for Klebsiella pneumoniae phylogenetic groups an analysis of 420 clinical isolates. Clinical Microbiology and Infection . 2004;10(10):942–945. doi: 10.1111/j.1469-0691.2004.00973.x. [DOI] [PubMed] [Google Scholar]
- 20.Pajand O., Darabi N., Arab M., et al. The emergence of the hypervirulent Klebsiella pneumoniae (hvKp) strains among circulating clonal complex 147 (CC147) harbouring bla NDM/OXA-48 carbapenemases in a tertiary care center of Iran. Annals of Clinical Microbiology and Antimicrobials . 2020;19(1):1–9. doi: 10.1186/s12941-020-00349-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Younes A., Hamouda A., Amyes S. G. B. First report of a novel extended-spectrum beta-lactamase KOXY-2 producing Klebsiella oxytoca that hydrolyses cefotaxime and ceftazidime. Journal of Chemotherapy . 2011;23(3):127–130. doi: 10.1179/joc.2011.23.3.127. [DOI] [PubMed] [Google Scholar]
- 22.Atmani S. M., Messai Y., Alouache S., et al. Virulence characteristics and genetic background of ESBL-producing Klebsiella pneumoniae isolates from wastewater. Fresenius Environmental Bulletin . 2015;24(1):103–112. [Google Scholar]
- 23.Pons M. J., Vubil D., Guiral E., et al. Characterisation of extended-spectrum β-lactamases among Klebsiella pneumoniae isolates causing bacteraemia and urinary tract infection in Mozambique. Journal of Global Antimicrobial Resistance . 2015;3(1):19–25. doi: 10.1016/j.jgar.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Derakhshandeh A., Firouzi R., Moatamedifar M., Motamedi A., Bahadori M., Naziri Z. Phylogenetic analysis of Escherichia coli strains isolated from human samples. Molecular Biology Research Communications . 2013;2(4):143–149. [Google Scholar]
- 25.Jazayeri Moghadas A., Kalantari F., Sarfi M., Shahhoseini S., Mirkalantari S. Evaluation of virulence factors and antibiotic resistance patterns in clinical urine isolates of Klebsiella pneumoniae in Semnan, Iran. Jundishapur Journal of Microbiology . 2018;11(7):7–12. doi: 10.5812/jjm.63637. [DOI] [Google Scholar]
- 26.Shahi A., Hasani A., Ahangarzadeh Rezaee M., Asghari Jafarabadi M., Hasani M., Kafil H. S. Klebsiella pneumoniae carbapenemase production among K. pneumoniae isolates and its concern on antibiotic susceptibility. Microbiology Research . 2019;10(1) doi: 10.4081/mr.2019.7587. [DOI] [Google Scholar]
- 27.Long S. W., Ojeda Saavedra M., Cantu C., Davis J. J., Brettin T., Olsen R. J. Whole-genome sequencing of human clinical Klebsiella pneumoniae isolates reveals misidentification and misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. mSphere . 2017;2(4):1–15. doi: 10.1128/mSphereDirect.00290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jafari Z., Harati A. A., Haeili M., et al. Molecular epidemiology and drug resistance pattern of carbapenem-resistant Klebsiella pneumoniae isolates from Iran. Microbial Drug Resistance . 2019;25(3):336–343. doi: 10.1089/mdr.2017.0404. [DOI] [PubMed] [Google Scholar]
- 29.Ranjbar R., Kelishadrokhi A. F., Chehelgerdi M. Molecular characterization, serotypes and phenotypic and genotypic evaluation of antibiotic resistance of the Klebsiella pneumoniae strains isolated from different types of hospital-acquired infections. Infection and Drug Resistance . 2019;12:603–611. doi: 10.2147/idr.s199639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moghadam M. T., Shariati A., Mirkalantari S., Karmostaji A. The complex genetic region conferring transferable antibiotic resistance in multidrug-resistant and extremely drug-resistant Klebsiella pneumoniae clinical isolates. New Microbes and New Infections . 2020;36 doi: 10.1016/j.nmni.2020.100693.100693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fonseca E. L., Ramos N. D. V., Andrade B. G. N., Morais L. L. C. S., Marin M. F. A., Vicente A. C. P. A one-step multiplex PCR to identify Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae in the clinical routine. Diagnostic Microbiology and Infectious Disease . 2017;87(4):315–317. doi: 10.1016/j.diagmicrobio.2017.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


