Abstract
Bulbophyllum is the largest genus in Orchidaceae with a pantropical distribution. Due to highly significant diversifications, it is considered to be one of the most taxonomically and phylogenetically complex taxa. The diversification pattern and evolutionary adaptation of chloroplast genomes are poorly understood in this species-rich genus, and suitable molecular markers are necessary for species determination and phylogenetic analysis. A natural Asian section Macrocaulia was selected to estimate the interspecific divergence of chloroplast genomes in this study. Here, we sequenced the complete chloroplast genome of four Bulbophyllum species, including three species from section Macrocaulia. The four chloroplast genomes had a typical quadripartite structure with a genome size ranged from 156,182 to 158,524 bp. The chloroplast genomes included 113 unique genes encoding 79 proteins, 30 tRNAs and 4 rRNAs. Comparison of the four chloroplast genomes showed that the three species from section Macrocaulia had similar structure and gene contents, and shared a number of indels, which mainly contribute to its monophyly. In addition, interspecific divergence level was also great. Several exclusive indels and polymorphism SSR loci might be used for taxonomical identification and determining interspecific polymorphisms. A total of 20 intergenic regions and three coding genes of the most variable hotspot regions were proposed as candidate effective molecular markers for future phylogenetic relationships at different taxonomical levels and species divergence in Bulbophyllum. All of chloroplast genes in four Bulbophyllum species were under purifying selection, while 13 sites within six genes exhibited site-specific selection. A whole chloroplast genome phylogenetic analysis based on Maximum Likelihood, Bayesian and Parsimony methods all supported the monophyly of section Macrocaulia and the genus of Bulbophyllum. Our findings provide valuable molecular markers to use in accurately identifying species, clarifying taxonomy, and resolving the phylogeny and evolution of the genus Bulbophyllum. The molecular markers developed in this study will also contribute to further research of conservation of Bulbophyllum species.
Keywords: Chloroplast genome, Sequence divergence, DNA markers, SSR, Bulbophyllum, Orchidaceae
Highlights
-
•
The first study on comparative chloroplast genomes analysis in four Chinese Bulbophyllum species.
-
•
The chloroplast genomes of Bulbophyllum section Macrocaulia showed both conservative and divergence.
-
•
A number of exclusive SSRs were identified for taxonomical identification and interspecific polymorphisms determination.
-
•
A number of hotspot regions were identified for phylogenetic and evolutionary studies.
1. Introduction
Bulbophyllum Thouars is probably the largest pantropical genus of Orchidaceae, with ca. 2200 species (Pridgeon et al., 2014). The Paleotropical region is the richest in species of this genus, with ca. 1700 species occurring in Asian (sub) tropical area, followed by Africa and then the neotropics (Pridgeon et al., 2014). It is well known that Bulbophyllum represents one of the most significant diversifications genera in the orchid family (Gamisch and Comes, 2019). Bulbophyllum species are notoriously difficult to classify taxonomically, due to highly morphological variation (Seidenfaden, 1979). Many independent and closely related genera established then combined. Numerous sectional classifications remained controversial (Pridgeon et al., 2014).
Several studies have been reported in recent years examining the phylogeny of Bulbophyllum using molecular data (Chase et al., 2015; Hosseini et al., 2012, 2016; Gravendeel et al., 2014; Fischer et al., 2007; Smidt et al., 2011). The finding of these works well supported the monophyly of continental groups, e.g., Asian, African and Neotropical clade, but contributed little to elucidating the relationships at infrageneric, interspecific level. Very recently, Hu et al. (2020) combined DNA and floral characters to reconstruct the phylogenetic relationship in Asian Cirrhopetalum alliance, and further indicated that the majority of traditional sectional classifications were shown to be highly artificial. The elucidation of the phylogenic relationships within Bulbophyllum still requires more detailed study.
The chloroplast (cp) genomes are considered to be useful and informative genetic resources for studies on evolutionary relationships in the plant kingdom at various taxonomic levels (Asaf et al., 2017; Wu et al., 2020), because of their uniparentally inherited and relatively conserved genome structure and higher evolutionary rate (Shaw et al., 2014; Wicke et al., 2011). Especially in Orchidaceae, the size and gene content of cp genome vary greatly. The size of the cp genome within orchids is ranging from 10 to 170 kb (Dong et al., 2018; Lin et al., 2015; Kim et al., 2015). The most reduced chloroplast genomes have been found in several mycoheterotrophic genera, e.g., Epipogium, Gastrodia, and Aphyllorchis (Feng et al., 2016; Schelkunov et al., 2015; Yuan et al., 2018), which have lost many photosynthetic genes (Graham et al., 2017). There are more than 100 orchid cp genomes, including Dendrobium 40 spp., Holcoglossum 11 spp., Corallorhiza 10 spp., Cymbidium 9 spp., Neottia 7 spp., have been reported and available in the recent years (Barrett and Davis, 2012; Kim et al., 2020a; Li et al., 2019; Logacheva et al., 2011; Niu et al., 2017; Shao et al., 2020; Yang et al., 2013; Zhu et al., 2018). However, the cp genomes are still little known in Bulbophyllum. Only nine cp genomes of Bulbophyllum species (eight neotropical and one Asian) were reported until now (Kim et al., 2020a; Zavala-Páez et al., 2020). Comparative cp genome studies at different taxonomical level are needed to clarify the diversification pattern in this species-rich group, and then to develop suitable molecular markers to study speciation, evolutionary adaptation and phylogeny. For taxonomists, uncertainty in taxonomic status of the Bulbophyllum species is quite often, which also requires suitable molecular markers for taxonomic revision (Hosseini et al., 2016; Zavala-Páez et al., 2020).
The Asian section Macrocaulia (Bl.) Aver. is one of acceptable natural sections in Bulbophyllun supported both by morphological characters and molecular evidence, and it is a medium section with more or less 68 species, mainly distributed in Southeastern Asia (Averyanov and Komarov, 1994; Vermeulen, 2014). There are six species recorded in (sub) tropical regions of China, which are the most northern parts for section Macrocaulia (Chen and Vermeulen, 2009; Jin et al., 2019; Huang et al., 2020). The section Macrocaulia is characterized by mini-miniature sized, epiphytes with creeping, close set pseudobulbs carrying a single, apical, usually persistent leaf that blooms on a single flower that have morphological diverse labellum and sepals. Based on the nrITS, the section Macrocaulia is monophyletic and regarded as the basal group sister to the rest of Asian Bulbophyllum clade (Gravendeel et al., 2014).
In this study, the complete chloroplast genomes of three species from section Macrocaulia were sequenced, assembled and characterized, then to investigate chloroplast genome diversification pattern and examine species divergence and evolutionary adaptation. The main objectives of the present study were to (1) understand the interspecific variation pattern in the cp genomes of closely related species in the genus of Bulbophyllum; (2) test the monophyly of section Macrocaulia using chloroplast genome; (3) identify genes underlying positive selection as potentially genes for adaptive evolution, radiation and speciation; (4) examine hotspot regions as candidate sequences for species delimitation and phylogenetic analysis in the genus Bulbophyllum.
2. Materials and methods
2.1. Plant material
Three species, Bulbophyllum lingii M.Z. Huang, G.S. Yang & J.M. Yin, B. pantenurum Seidenf., and B. menghaiense Z.H. Tsi, are closely related species from section Macrocaulia. B. lingii is newly found (Huang et al., 2020) and B. pantenurum is a new record (unpublished) in China recently. Bulbophyllum gedangense Y. Luo, J. P. Deng & Jian W. Li, which is also newly found in China recently, might belonging to section Desmosanthes (Bl.) J. J. Smith (Luo et al., 2020). Fresh leaves of four Bulbophyllum species were collected from the living collections at the greenhouse of Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences and Yunnan Fengchunfang Biotechnology Co. Ltd. The voucher specimens were deposited in the herbarium of Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences (HITBC) (Table 1). In addition, we downloaded the available complete cp genomes of nine other Bulbophyllum species from GenBank (Table 1) to analyze phylogenetic relationships.
Table 1.
Sampling and accession information for 15 Bulbophyllum and Dendrobium species in this study.
| Taxa | Section | Locality | Voucher | Genbank accession | Reference |
|---|---|---|---|---|---|
| Bulbophyllum menghaiense Z.H. Tsi | Macrocaulia | Yunnan, China | XY Wang & ZF Xu 202,003 | MW161050 | This study |
| Bulbophyllum lingii M. Z. Huang, G. S. Yang & J.M. Yin | Macrocaulia | Xizang, China | Y. Luo et al., 2247 | MW161051 | This study |
| Bulbophyllum pentanurum Seidenf. | Macrocaulia | Xizang, China | Y. Luo et al., 2252 | MW161052 | This study |
| Bulbophyllum gedangense Y. Luo, J. P. Deng & Jian W. Li | uncertain | Xizang, China | Y. Luo et al., 1239 | MW161053 | This study |
| Bulbophyllum inconspicuum Maxim. | uncertain | Japan | MN221389 | Kim et al., 2020a, Kim et al., 2020b | |
| Bulbophyllum weddellii (Lindl.) Rchb.f | Didactyle | Brazil | MN604059 | Zavala-Páez et al., 2020 | |
| Bulbophyllum exaltatum Lindl. | Didactyle | Brazil | MN604054 | Zavala-Páez et al., 2020 | |
| Bulbophyllum steyermarkii Foldats | Furvescens | Brazil | MN604058 | Zavala-Páez et al., 2020 | |
| Bulbophyllum epiphytum Barb. Robr. | Micranthae | Brazil | MN737573 | Zavala-Páez et al., 2020 | |
| Bulbophyllum mentosum Barb. Robr. | Micranthae | Brazil | MN604056 | Zavala-Páez et al., 2020 | |
| Bulbophyllum regnellii Rchb. f | Napelli | Brazil | MN604057 | Zavala-Páez et al., 2020 | |
| Bulbophyllum granulosum Barb. Rodr | Napelli | Brazil | MN604055 | Zavala-Páez et al., 2020 | |
| Bulbophyllum plumosum Barb. Rodr. | Xiphizusa | Brazil | MN580547 | Zavala-Páez et al., 2020 | |
| Dendrobium wattii (Hooker) Reichenbach | Yunnan, China | MN913340 | Wang et al. (2020) | ||
| Dendrobium wangliangii G.W.Hu, C. L. Long & X. H. Jin | Yunnan, China | MT798588 | Shao et al. (2020) |
2.2. DNA extraction and sequencing
Total genomic DNA from fresh leaves was extracted by using TiangenDNA kit (TIANGEN, China). An Illumina paired-end DNA library was constructed using the IlluminaTruSeq Library Preparation Kit (San Diego, CA, USA) following the manufacturer's instructions. The library was sequenced by the Illumina Hiseq 2500 sequencing platform (Illumina, CA, USA) at Personal Biotechnology Co., Ltd (Shanghai, China).
2.3. Chloroplast genome assembly and annotation
The total 3 G data of sequencing reads were trimmed to high quality reads (Table 2), and then GetOrganelle pipeline were used for de novo chloroplast genome assembly (Jin et al., 2020). CPGAVAS2 (Shi et al., 2019) and GeSeq (Tillich et al., 2017) were used to annotate the assembled chloroplast genome using default parameters to predict protein coding, rRNA, and tRNA genes. The annotation results were checked plus manual corrections and codon positions were adjusted by comparing them with the more similar six Bulbophyllum reference species present in the database using Geneious prime. The genome map of the species was illustrated with the help of OGDRAW (Greiner et al., 2019), and the annotated chloroplast genome sequences were submitted to NCBI (Table 1). GC content was calculated using Geneious prime.
Table 2.
The basic characteristics of the chloroplast genomes of four Bulbophyllum species.
| B. pentaneurum | B. lingii | B. menghaiense | B. gedangense | |
|---|---|---|---|---|
| Raw reads number | 24,698,734 | 24,785,884 | 24,012,502 | 22,330,566 |
| Raw data size (bp) | 3,704,810,100 | 3.717,882,600 | 3,601,875,300 | 3,349,584,900 |
| HQ Reads number | 24,645,614 | 24,625,976 | 23,945,784 | 22,284,110 |
| HQ Data size (bp) | 3,690,233,928 | 3,682,524,097 | 3,584,614,507 | 3,336,468,056 |
| genome size (bp) | 156,182 | 156,689 | 156,550 | 158,524 |
| LSC length (bp) | 84,240 | 84,607 | 84,663 | 86,200 |
| SSC length (bp) | 18,266 | 18,244 | 18,105 | 18,632 |
| IR length (bp) | 26,838 | 26,919 | 26,891 | 26,846 |
| Number of genes | 113 | 113 | 113 | 113 |
| Protein-coding genes | 79 | 79 | 79 | 79 |
| tRNA genes | 30 | 30 | 30 | 30 |
| rRNA genes | 4 | 4 | 4 | 4 |
| GC content (%) | 36.8 | 36.8 | 36.7 | 36.8 |
| GC content in LSC (%) | 34.4 | 34.4 | 34.2 | 34.5 |
| GC content in SSC (%) | 29.4 | 29.3 | 29.3 | 29.3 |
| GC content in IR (%) | 43.0 | 43.1 | 43.0 | 43.0 |
| GC content in Protein-coding (%) | 37.4 | 37.4 | 37.3 | 37.6 |
| GC content in tRNA (%) | 52.8 | 52.8 | 52.7 | 53 |
| GC content in rRNA IR (%) | 55.2 | 55.1 | 55.2 | 55.2 |
2.4. Simple repeat sequence analysis
We identified simple sequence repeats (SSRs) of Bulbophyllum with MISA (Beier et al., 2017) by setting the minimum number of repeats to 10, 5, 4, 3, 3, and 3 for mono-, di-, tri-, tetra-, penta-, and hexa-nucleotides, respectively.
2.5. Comparative genome analysis
The complete chloroplast genomes of four Bulbophyllum species were compared using the program mVISTA (Frazer et al., 2004). We also compared the borders of large single copy (LSC), small single copy (SSC), and inverted repeat (IR) regions in the genomes of the four species. The nucleotide diversity value (Pi) of the cp genomes of all four Bulbophyllum species was calculated using DnaSP v5.10 (Librado and Rozas, 2009). The number of mutations and indels events was calculated by DnaSP v5.0 (Rozas et al., 2004), and their rate was calculated per 100 bp. Due to indels having higher levels of homoplasy than mutations, indels were considered as events instead of sites in the alignment (Ingvarsson et al., 2003).
2.6. Selective pressure analysis
In order to detect the protein coding genes under selection in four Bulbophyllum species, the synonymous (dS), non-synonymous (dN) nucleotide substitution rates and the dN/dS ratio were calculated using the model in DnaSP v.5.0 (Librado and Rozas, 2009). To explore the selection patterns and identify positive selection on the protein coding genes, six models, model M0, M3, M1a, M2a, M7 and M8, implemented in the PAMLX Codeml program (Xu and Yang, 2013). The codon frequencies were determined by the F3 × 4 model. 79 common genes were examined. The Bayes empirical Bayes (BEB) method was used to statistically identify sites under positive selection with posterior probabilities≥0.95. Pairwise distances and branch lengths were computed using the maximum likelihood (ML) criterion under a codon model. The likelihood ratio test (LRT) was used to estimate the quality of each model (Yang and Nielsen, 2002; Yang et al., 2005).
2.7. Phylogenetic analysis
The complete chloroplast genomes of the four newly sequenced Bulbophyllum species, together with one Asian (from Japan) and eight neotropical Bulbophyllum species, and two species from Dendrobium (Table 1), which download from GenBank, were selected to reconstruct phylogenetic tree. Phylogenetic analyses were performed for two datasets: complete plastid genome sequences and the 68 protein coding sequences. The chloroplast DNA sequences were aligned with MAFFT v.7 (auto strategy) (Katoh and Standley, 2013). Maximum parsimony (MP), Maximum Likelihood (ML) and Bayesian (BI) analyses were run for each data set. Parsimony analyses were performed with MegaX (Kumar et al., 2008). Heuristic searches were conducted with tree bisection-reconnection (TBR) branch swapping, and random stepwise addition with 1000 replications. ML trees with 1000 BS were produced with IQ-tree (Trifinopoulos et al., 2016). Bootstrap analyses were used to evaluate the support for individual clades with 1000 replicates. BI tree used MrBayes 3.2.4 (Ronquist et al., 2012) in PhyloSuite (Zhang et al., 2020). The Markov chain Monte Carlo (MCMC) algorithm was run with two independent chains with a random starting tree and default priors for 2 × 106 generations, with trees sampled every 100 generations. The first 25% of all generations were excluded, and a consensus phylogenetic tree was obtained based on majority rule from the remaining trees.
3. Results
3.1. Characteristics of Bulbophyllum chloroplast genome
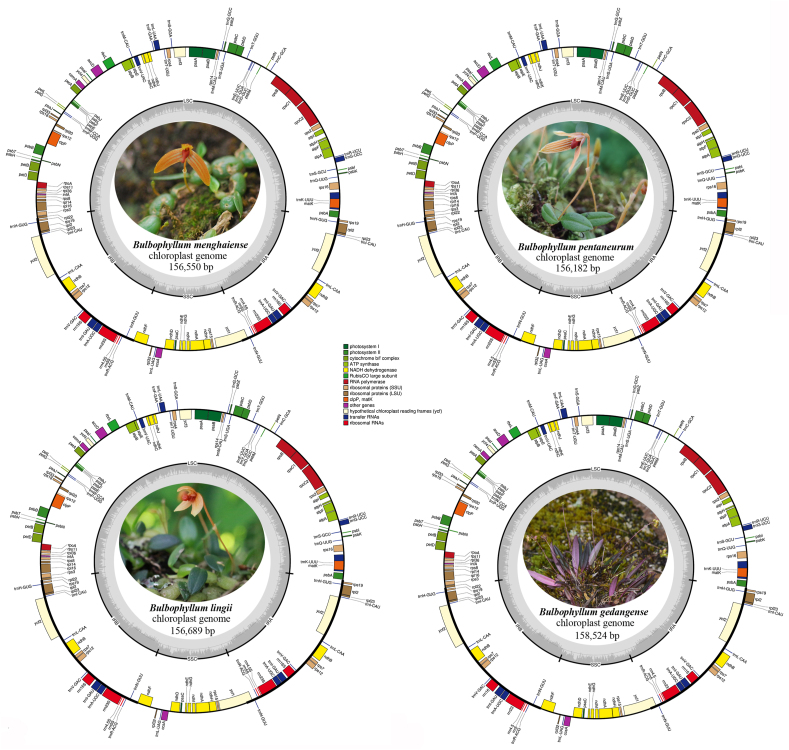
The cp genomes of three species of section Macrocaulia ranged in size from 156,182 bp (Bulbophyllum pentaneurum) to 156,689 bp (B. lingii), and that of B. gedangense is 158,524 bp (Fig. 1, Table 2). The genome length differences between the largest and smallest genomes were the 2342 bp. All of the cp genomes had a typical quadripartite structure, similar to those of most land plants, consisting two inverted repeats (IRs, 26,838 to 26,919 bp) separated by large (LSC, 84,240 to 86,200 bp) and small (SSC, 18,105 to 18,632 bp) single copy regions, respectively (Fig. 1, Table 2). GC content was similar (36.6–36.8%; Table 2). In addition, the gene composition and order in the cp genomes of four Bulbophyllum species are nearly identical (Fig. 1, Table 2). Four each cp genome, 113 unique genes were annotated, including 79 protein-coding genes, 30 tRNAs, and 4 rRNAs (Table 2). Most of genes occur as a single copy in LSC or SSC, while 20 genes are duplicated in the IRs, including eight protein coding genes, eight tRNAs and four rRNAs (Table 3). Eight protein coding genes and six tRNA genes included one intron, while three genes (ycf3, clpP, rps12) included two introns (Table 3).
Fig. 1.
Chloroplast genome map for four Bulbophyllum species. Genes located outside the outer rim are transcribed in a counterclockwise direction, whereas genes inside the outer rim are transcribed in a clockwise direction. The colored bars indicate known different functional groups. The dashed gray area in the inner circle shows the percentage GC contents of the corresponding genes. LSC, SSC, and IR denote large single copy, small single copy, and inverted repeat, respectively.
Table 3.
Gene contents in the chloroplast genomes of four Bulbophyllum species.
| Group of genes | Gene names |
|---|---|
| 1 Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| 2 Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
| 3 Cytochrome b/f complex | petA, petBa, petDa, petG, petL, petN |
| 4 ATP synthase | atpA, atpB, atpE, atpFa, atpH, atpI |
| 5 NADH dehydrogenase | ndhAa, ndhBa(× 2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| 6 RubisCO large subunit | rbcL |
| 7 RNA polymerase | rpoA, rpoB, rpoC1a, rpoC2 |
| 8 Ribosomal proteins (SSU) | rps2, rps3, rps4, rps7 (× 2), rps8, rps11, rps12b(× 2), rps14, rps15, rps16a, rps18, rps19 (× 2) |
| 9 Ribosomal proteins (LSU) | rpl2a (× 2), rpl14, rpl16a, rpl20, rpl22, rpl23 (× 2), rpl32, rpl33, rpl36 |
| 10 Other genes | clpPb, matK, accD, ccsA, infA, cemA |
| 11 Proteins of unknown function | ycf1, ycf2 (× 2), ycf3b, ycf4, ycf15 (× 2) |
| 12 Ribosomal RNAs | rrn4.5 (× 2), rrn5S (× 2), rrn16S (× 2), rrn23S (× 2) |
| 13 Transfer RNAs | trnA-UGCa(× 2), trnC-GCA, trnD-GUC,trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCCa, trnH-GUG (× 2), trnI-CAU( × 2), trnI-GAUa(× 2), trnK-UUUa, trnL-CAA (× 2), trnL-UAAa, trnL-UAG, trnM-CAU, trnN-GUU(× 2), trnP-UGG, trnQ-UUG, trnR-ACG (× 2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC (× 2), trnV-UACa, trnW-CCA, trnY-GUA, trnfM-CAU |
Gene containing one intron.
Gene containing two introns, (×2) Gene has two copies.
3.2. Characters of simple sequence repeats
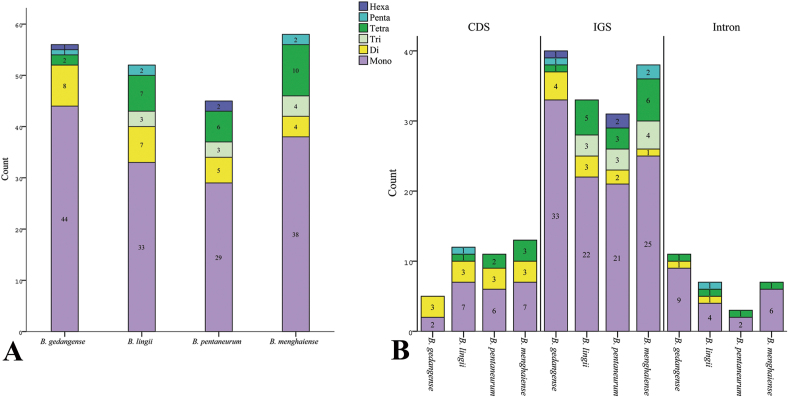
A total of 45 (Bulbophyllum pentanurum) to 58 (B. menghaiense) SSRs were detected in the cp genome of four Bulbophyllum species, and six categories of SSRs (mono-, di-, tri-, tetra-, penta-, hexa-nucleotide repeats) were found with lengths ranged from 10 to 26 bp (Fig. 2, Table 4). In the four Bulbophyllum species, the most abundant SSRs were mono-nucleotide repeats, accounting for 63.64%–78.57%, and number varies from 28 to 44, followed by 4 (6.9%) to 8 (14.29%) di-nucleotide repeats, 0 to 4 (6.9%) tri-nucleotide repeats, 2 (3.57%) to 10 (17.24%) tetra-nucleotides repeats, 0 to 2 (3.45%) penta-nucleotide repeats, and 0 to 2 (3.45%) hexa-nucleotide repeats (Fig. 2A, Table 4). Interestingly, all mono-nucleotide SSRs belonged to A or T type and the majority of di-, tri-, tetra-, penta-, and hexa-nucleotide SSRs were especially rich in A or T (Table S1). Of the SSR regions, 31 to 40 SSRs were identified in intergenic spaces, while 5 to 13 SSRs were located in the coding DNA sequence and 3 to 11 SSRs in the coding sequence introns (Fig. 2B).
Fig. 2.
Simple sequence repeats (SSRs) in chloroplast genomes of four Bulbophyllum species. (A) Frequency distribution of different types of SSRs in the cp genomes. (B) Frequency of SSRs identified in the coding regions (CDS), intergenic spacers (IGS) and introns of the cp genomes.
Table 4.
Types and amounts of simple sequence repeats (SSRs) in the chloroplast genomes of four Bulbophyllum species.
| SSR Type |
B. menghaiense |
B. pentaneurum |
B. lingii |
B. gedangense |
||||
|---|---|---|---|---|---|---|---|---|
| Number | Ration (%) | Number | Ration (%) | Number | Ration (%) | Number | Ration (%) | |
| mono- | 38 | 65.52 | 29 | 64.44 | 33 | 63.46 | 44 | 78.57 |
| di- | 4 | 6.90 | 5 | 11.11 | 7 | 13.46 | 8 | 14.29 |
| tri- | 4 | 6.90 | 3 | 6.67 | 3 | 5.77 | 0 | 0.00 |
| tetra- | 10 | 17.24 | 6 | 13.33 | 7 | 13.46 | 2 | 3.57 |
| penta- | 2 | 3.45 | 0 | 0.00 | 2 | 3.85 | 1 | 1.79 |
| hexa- | 0 | 0.00 | 2 | 4.44 | 0 | 0.00 | 1 | 1.79 |
| Total | 58 | 100.00 | 45 | 100.00 | 52 | 100.00 | 56 | 100.00 |
According to the criteria of SSRs, i.e., presence of the same repeat units, being located in the homologous regions, and having different number of the repeat units, 35 SSRs were identified as polymorphic SSRs between Bulbophyllum species (Table 5). Most of SSR loci were specific for each species. B. menghaiense had 10 exclusive SSRs, e.g., (TTA)4 in trnS-trnG and atpB-rbcL regions; B. lingii had six exclusive SSRs, e.g., (AT)5 in trnL-UAA and (TTATA)3 in clpP genes; B. gedangense had six exclusive SSRs e.g., (AT)6 in trnT-trnL and (TA)13 in trnE-trnT regions; B. pentanurum had five exclusive SSRs, e.g., (TA)5 in psbM-trnD and (CTT)4 in rpl14-rpl16 regions (Table 5).
Table 5.
The polymorphic simple sequence repeats (SSRs) in the chloroplast genomes of four Bulbophyllum species. Number indicated SSR repeats.
| SSR Type | B. menghaiense/B. pentaneurum/B. lingii/B. gedangense | Location | Regions |
|---|---|---|---|
| AT | 5/6/5/0 | atpH-atpI | LSC |
| AT | 0/0/5/0 | trnL-UAA | LSC |
| AT | 0/0/0/6 | trnT-UGU-trnL-UAA | LSC |
| AT | 0/0/5/8 | psaJ-rpl33 | LSC |
| AT | 0/0/0/5 | psbB-psbT | LSC |
| TA | 0/5/0/0 | psbM-trnD-GUC | LSC |
| TA | 0/0/0/13 | trnE-UUC-trnT-GGU | LSC |
| TA | 0/0/5/0 | clpP-psbB | LSC |
| CT | 0/0/0/5 | trnL-UAA | LSC |
| CTT | 0/4/0/0 | rpl14-rpl16 | LSC |
| TTA | 4/0/0/0 | trnS-GCU-trnG-UUC | LSC |
| TTA | 3/3/3/0 | atpF-atpH | LSC |
| TTA | 4/0/0/0 | atpB-rbcL | LSC |
| TTC | 4/0/0/0 | trnT-UGU-trnL-UAA | LSC |
| TTG | 0/4/4/0 | trnT-GGU-psbD | LSC |
| AAGT | 3/0/0/0 | rpoB-trnC-GCA | LSC |
| AATA | 0/3/0/0 | atpF | LSC |
| ATAA | 3/3/3/0 | ndhG-ndhI | SSC |
| ATGA | 3/0/0/0 | trnN-GUU-trnR-ACG | IR |
| ATTA | 3/0/0/0 | trnL-UAA | LSC |
| GGGA | 0/3/0/0 | rrn16S-trnI-GAU | IR |
| GGGA | 3/0/3/0 | trnI-GAU-rrn16S | IR |
| TCCC | 0/0/3/3 | rrn16S-trnI-GAU | IR |
| TCCC | 0/3/0/0 | trnI-GAU-rrn16S | IR |
| TTCA | 3/0/0/0 | trnR-ACG-trnN-GUU | IR |
| TTCT | 0/0/3/0 | rpl16 | LSC |
| TTGA | 3/0/0/0 | ndhE | SSC |
| TTTA | 0/0/3/0 | rpl33-rps18 | LSC |
| AATAA | 0/0/0/3 | rpl16-rps3 | LSC |
| ATCAA | 3/0/0/0 | trnS-GGA-rps4 | LSC |
| TACTG | 3/0/0/0 | matK-rps16 | LSC |
| TCTTA | 0/0/3/0 | rpoC1 | LSC |
| TTATA | 0/0/3/0 | clpP | LSC |
| TATCTA | 0/0/0/4 | trnP-UGG-psaJ | LSC |
| TCTGTC | 0/3/0/0 | rps7-trnV-GAC | IR |
3.3. Comparative genomic analysis
3.3.1. Sequence divergence
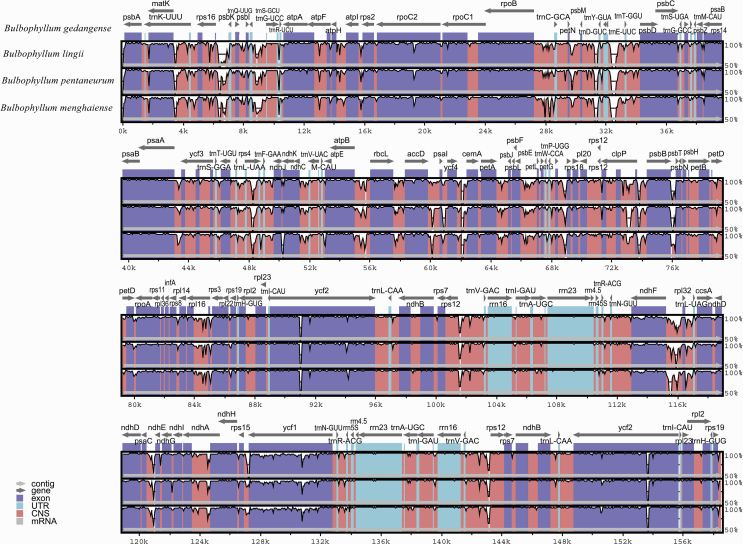
The divergence of sequences in the cp genomes of four Bulbophyllum species was plotted by mVISTA program using the annotated B. gedangense sequence as a reference (Fig. 3). A whole genome alignment revealed an overall high sequence similarity among the four Bulbophyllum species, and also showed lower sequence divergence between three species from section Macrocaulia. Furthermore, LSC and SSC regions were more divergent compared with IR regions. The regions with highest level of divergence were rps16-trnQ, rrn16, ndhF-rpl32, psbI-trnS, psaA-ycf3, trnS-rps4, ndhF-rpl32, and rps15-ycf1 (Fig. 3).
Fig. 3.
Alignment of the cp genome sequences of four Bulbophyllum species, with B. gedangense as a reference. The X-axis corresponds to coordinates within the cp genome. The Y-axis shows the percentage identity in the range 50%–100%.
3.3.2. Mutations and indels
The number of mutations and indels events were compared among these four Bulbophyllum cp genomes. There were 4600 mutations and 943 indels calculated. The intergenic regions had the most 2320 mutations and 710 indels respectively. The intergenic regions had the highest mutation rate, 4.65 mutations per 100 bp, following by introns and the coding regions with 3.32 and 2.11, respectively (Table 6). We further analyzed the details of indels (indel size>10 bp) of the four Bulbophyllum species based on multiple whole cp genome alignment. 189 positions showed DNA indels in at least one of the cp genomes. The three species from section Macrocaulia shared 67 deletions, mostly located in intergenic regions, which showed low similarity to B. gedangense (Table S2). These regions are related to large deletions, which become obvious in a multiple alignment of related whole cp genome sequences. The largest deletion, which is in trnE-trnT linker, is of a size of about 298 bp (at position 32,368 bp of the B. gedangense sequence). The additional large deletions were in different intergenic regions, e.g., trnV-rps12 (237 bp), rps16-trnQ (155 bp), and ycf4-cemA intergenic regions (189 bp) (Table S2). Several shared deletions were in introns, e.g., the intron of atpF, the intron of trnL-UAA. Only a very few deletions are in the coding regions, e.g. ycf2 gene (115 bp) (Table S2).
Table 6.
Mutations, indel events, and mutation rates for the chloroplast genomes of four Bulbophyllum species. CDS: protein-coding genes; IGS: intergenic spacers.
| Region | Mutations | Indels | Total length | Mutations/100 bp | Indels/100 bp |
|---|---|---|---|---|---|
| CDS | 1662 | 53 | 78,608 | 2.114288622 | 0.06742 |
| IGS | 2320 | 710 | 49,918 | 4.6476221 | 1.42233 |
| intron | 618 | 180 | 18,638 | 3.315806417 | 0.96577 |
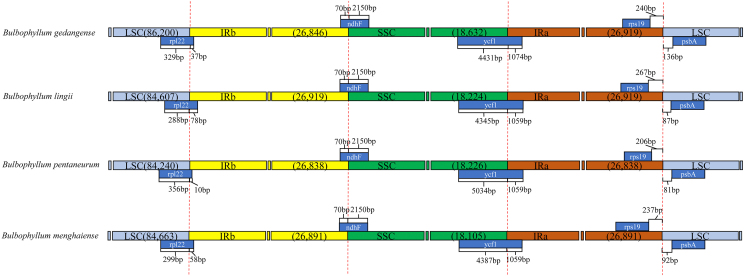
3.3.3. Border contraction and extension
We analyzed the IR/single copy (SC) region border positions and their adjacent genes in the four Bulbophyllum cp genomes (Fig. 4). In all cp genomes of the four Bulbophyllum species, the genes rpl22, ndhF, ycf1, rps19, and psbA were located at the junction of the LSC/IRb, IRb/SSC, SSC/IRa, and IRa/LSC borders (Fig. 4). The expansion and contraction of the border regions of cp genomes were similar in the four Bulbophyllum species. There were 10–78 bp extension of rpl22 gene into IRb region. Moreover, the IRb/SSC borders were well conserved among the four cp genomes, of which the IRb region expanded into the gene ndhF with 70 bp. The ycf1 gene crossed the SSC/IRa junction expanding 1059 bp to IRb regions in three species of section Macrocaulia, while 1074 bp in B. gedangense. There were 206–267 bp between rps19 and the LSC/IRa border, meanwhile, the psbA generated a distance of 81–136 bp to another LSC/IRa junction.
Fig. 4.
Comparison of the borders between neighboring genes and junctions of LSC, SSC, and IR regions in the chloroplast genomes in four Bulbophyllum species. Boxes above or below the main line indicate genes adjacent to borders.
3.3.4. Nucleotide diversity
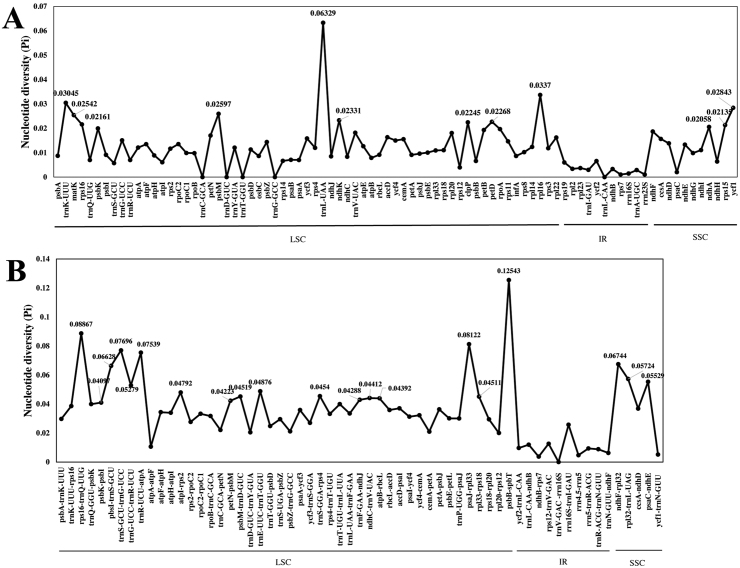
The nucleotide diversity value (Pi) of the coding regions and intergenic regions were calculated using DnaSP for understanding the sequence divergence among these four Bulbophyllum species. The coding regions were more conserved with the Pi value ranging from 0 to 0.06329, compared to the intergenic spacer with the Pi value ranging from 0 to 0.12543 (Fig. 5). Most of highly variable sites were located in the LSC regions, followed SSC regions. The sequences in IR regions exhibited lower nucleotide diversity value. Among the coding regions, the highest nucleotide diversity (Pi = 0.06329) was in trnL-UAA gene. A total of 12 genes were detected as highly variable coding regions, e.g., trnL-UAA, rpl16, trnK-UUU (Pi > 0.03), and ycf1, psbM, matK, ndhK, petD, clpP, rps16, rps15, ndhA (Pi > 0.02) (Fig. 5A). Among the non-coding regions, the highest nucleotide diversity (Pi = 0.12543) was in the psbB-psbT region. A total of 20 intergenic regions, psbB-psbT, rps16-trnQ, psaJ-rpl33, trnS-trnG, trnR-atpA, ndhF-rpl32, psbI-trnS, rpl32-trnL, psaC-ndhE, trnG-trnR, trnE-trnT, atpI-rps2, trnS-rps4, psbM-trnD, rpl33-rps18, ndhC-trnV, atpB-rbcL, trnF-ndhJ, petN-psbM, and psbK-psbI were recognized as hotspot regions with nucleotide diversity >0.04 (Fig. 5B).
Fig. 5.
Comparative analysis of nucleotide diversity (Pi) values among the cp genome sequences of four Bulbophyllum species. (A) Nucleotide diversity (Pi) values of coding genes in the LSC, SSC, and IR regions; (B) Nucleotide diversity (Pi) values of intergenic regions in the LSC, SSC, and IR regions.
3.4. Selection pressure analysis of evolution
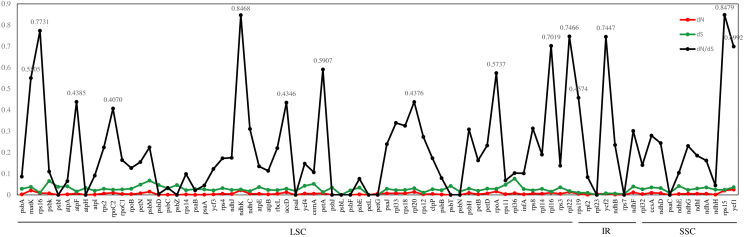
Synonymous and non-synonymous nucleotide substitution patterns are very important markers for gene evolution studies. A ratio of dN/dS < 1 indicates purifying selection, dN/dS > 1 denotes probable positive selection, and dN/dS values close to one indicates neutral evolution. We estimated the selective pressure among four Bulbophyllum species. 79 protein-coding genes were used to calculate the ratio of synonymous and non-synonymous substitution, and thereby to determine whether the selection pressure has acted on individual sequences. Thus, we found all 79 genes of the dN/dS average ratio were lower than 1.0 might be under purifying selection, and no gene under positive selection in four Bulbophyllum species (Fig. 6). We found that ten genes (matK, rps15, rps16, ndhK, petA, rpoA, rpl16, rpl22, yfc1, and ycf2) of the dN/dS average ratio above 0.5, indicating relaxed selection (Fig. 6). Fourteen most conserved genes (psaC, psaI, psbF, psbI, psbJ, psbL, psbN, psbT, psbZ, petL, petG, atpH, rpl23 and rps7) with the dN/dS average values of 0.0, suggesting very strong purifying selective pressure. The remaining genes showed the dN/dS average value were ranged from 0.0 to 0.5, suggesting strong purifying selective pressure. In addition, according to the models analysis, we found a total of 6 genes 13 sites exhibiting site-specific selection (Table 7). Of these genes, the ndhK gene harbored 6 sites under positive selection, with three in ycf2, and the other four genes (ndhF, rbcL, rps4, and rps16) each had only one selection site (Table 7).
Fig. 6.
Non-synonymous (dN), synonymous (dS) and dN/dS average ratios of four Bulbophyllum species.
Table 7.
Positive selection sites identified in the chloroplast genomes of four Bulbophyllum species.
| Gene | Model | Ln L | LRT P-value | Positive sites |
|---|---|---|---|---|
| ndhF | M0 | −3367.871835 | M0 vs. M3: 4.99 × 10−3 M1a vs. M2a: 1.57 × 10−1 M7 vs. M8: 8.57 × 10−2 |
588 E 0.976∗ |
| M3 | −3360.438925 | |||
| M1a | −3362.318789 | |||
| M2a | −3360.405802 | |||
| M7 | −3363.243214 | |||
| M8 | −3360.786314 | |||
| ndhK | M0 | −1047.915524 | M0 vs. M3: 2.00 × 10−8 M1a vs. M2a: 1.16 × 10−6 M7 vs. M8: 9.87 × 10−7 |
214 P 0.961∗, 216 G 0.954∗, 217 Q 0.971∗, 218 Y 0.993∗∗, 219 L 0.996∗∗, 220 P 0.995∗∗ |
| M3 | −1027.108483 | |||
| M1a | −1040.774810 | |||
| M2a | −1027.108483 | |||
| M7 | −1041.140233 | |||
| M8 | −1027.311588 | |||
| ycf2 | M0 | −9561.901080 | M0 vs. M3: 1.87 × 10−5 M1a vs. M2a: 2.12 × 10−5 M7 vs. M8: 1.15 × 10−5 |
563 G 0.973∗, 564 C 0.981∗, 774 L 0.951∗, 919 I 0.951∗ |
| M3 | −9548.340210 | |||
| M1a | −9559.101790 | |||
| M2a | −9548.340212 | |||
| M7 | −9559.710001 | |||
| M8 | −9548.339220 | |||
| rbcL | M0 | −2128.95659 | M0 vs. M3: 4.01 × 10−5 M1a vs. M2a: 5.33 × 10−4 M7 vs. M8: 7.95 × 10−5 |
96 A 0.952∗, 98 V 0.964∗, 101 E 0.975∗ |
| M3 | −2125.041535 | |||
| M1a | −2150.834797 | |||
| M2a | −2143.297217 | |||
| M7 | −2152.737025 | |||
| M8 | −2143.297216 | |||
| rps4 | M0 | −917.908875 | M0 vs. M3: 3.01 × 10−3 M1a vs. M2a: 3.33 × 10−3 M7 vs. M8: 1.04 × 10−3 |
183 L 0.998∗∗ |
| M3 | −909.907038 | |||
| M1a | −915.613890 | |||
| M2a | −909.909196 | |||
| M7 | −916.783516 | |||
| M8 | −909.913519 | |||
| rps16 | M0 | −366.198755 | M0 vs. M3: 4.00 × 10−9 M1a vs. M2a: 0.00 M7 vs. M8: 0.00 |
21 K 0.986∗ |
| M3 | −343.729536 | |||
| M1a | −368.201071 | |||
| M2a | −343.729536 | |||
| M7 | −370.023492 | |||
| M8 | −343.729564 | |||
∗P < 0.05, ∗∗P < 0.001.
3.5. Phylogenetic analysis
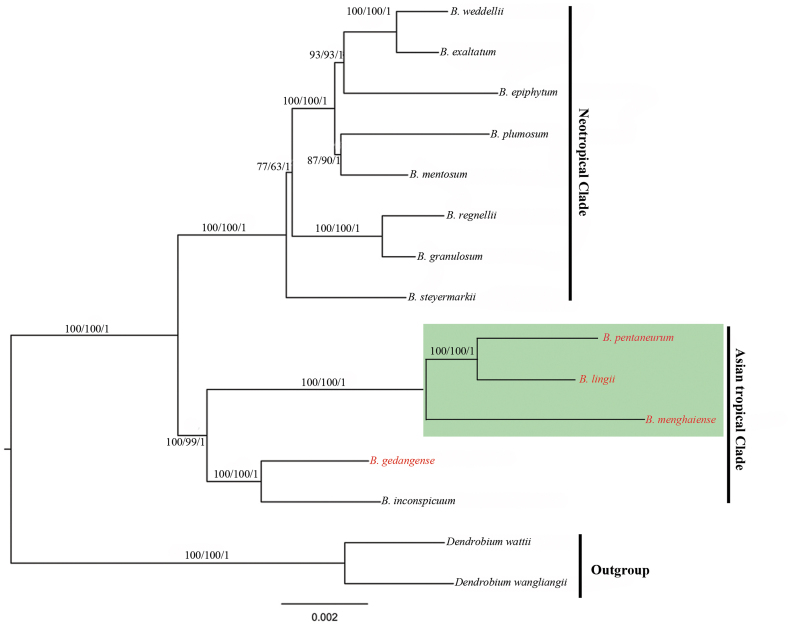
The protein coding gene sequences and all the complete chloroplast genome sequences of 15 Malaxideae members were used to reconstruct the phylogenetic relationships, including the 13 Bulbophyllum cpDNA sequences, and two Dendrobium species (Table 1, Fig. 7). In our analysis, the topologies obtained based on the maximum likelihood (ML), the parsimony (MP) and Bayesian (BI) analyses were largely consistent with the different two datasets, and the bootstrap values and posterior probabilities were very high for each lineage (Fig. 7). These results suggested that there was no conflict among the entire genome data set and protein coding gene sequences. All the sampled species of Bulbophyllum were clustered into one clade with 100% bootstrap value or the Bayesian posterior probability. Within the Bulbophyllum clade, the Asian tropical and neotropical Bulbophyllum form a monophyletic clade, respectively. Within the Asian tropical clade, B. pentaneurum was sister to B. lingii with B. menghaiense as their closest relative, which strongly supported that section Macrocauliais was monophyletic; In another clade, B. gedangense and B. inconspicuum (native to Japan) grouped together (Fig. 7).
Fig. 7.
Phylogenetic tree conducted using Maximum Likelihood (ML), Maximum Parsimonious (MP) and Bayesian Inference (BI) methods based on 68 protein-coding chloroplast genes from different species. The numbers above branches represent bootstrap percentage (BP) of ML/BP of MP/posterior probability (PP) of BI.
4. Discussion
4.1. Conservation and divergence of chloroplast genome in the section Macrocaulia
Based on our comprehensive comparative genomics analysis, the chloroplast genomes of section Macrocaulia showed both conservative and divergent. In terms of genome size, all of four cp genomes fell within the size of Orchidaceae species (Kim et al., 2015; Lin et al., 2015). The three species of the section Macrocaulia cp genomes were similar and smaller than that of B. gedangense. In addition, although loss of ndh genes has been previously in some orchids (Feng et al., 2016; Niu et al., 2017; Yang et al., 2013; Zavala-Páez et al., 2020), all the cp genomes of the section Macrocaulia had no evidence of ndh genes loss. The total length of angiosperm cp genomes is influenced by contractions and expansions of the IRs (Green, 2011; Weng et al., 2017). However, the decrease of cp genomes size in three species of section Macrocaulia might have been due to the large deletions. Several large deletions, more than 100 bp, were detected in the cp genomes of these miniature Bulbophyllum species, mostly in intergenic regions (Table S2). Compared with B. gedangense, 84 indels were shared in the three species of section Macrocaulia. The similarity between cp DNA sequences indicated the cp genome are highly conserved in the section Macrocaulia, and contributed to their monophyletic relationships. However, several exclusive large indels were also identified for the three species of section Macrocaulia respectively, e.g., an 87 bp-indel in rpoB-trnC intergenic region for B. menghaiense, a 65 bp-indel in the intron of rps16 gene for B. lingii, and a 43 bp indels in petN-psbM intergenic region for B. pentaneurum (Table S2). The differences between cp DNA sequences indicated interspecific differentiation in the section Macrocaulia, and might contributed to the species divergence and adaptive evolution. Large indels easily identified in whole cp genome alignments might be used for species-specific molecular markers as recently shown for a Populus tremula-specific marker that has been developed based on a 96 bp-indel (Kersten et al., 2016). The indels identified among the four Bulbophyllum cp genomes in this study might also serve as species-specific molecular marker for species delimitation and identification.
4.2. Simple sequence repeats diversity
Simple sequence repeats (SSRs) are the most frequent in genome sequences with high mutation rates, and play important roles in plants (Qin et al., 2015). The most abundant SSR type was the mononucleotide repeat, and the majority of SSRs in four Bulbophyllum species was composed of polyT and polyA repeats, corresponding with the angiosperm cp genome composition (Kim et al., 2015; Qin et al., 2015). In our study, the number and types of SSRs identified from the cp genomes were variable in four Bulbophyllum species (Fig. 2). Of di- to hexa-nucleotide SSRs among four Bulbophyllum species, most of SSRs were specific to each species, and only five SSRs shared in the section Macrocaulia. These findings indicated that SSRs exhibiting highly variable even in closely related species, might have few phylogenetic significance. Most of the previous studies revealed that the richness of SSR types is various in different species, which may contribute to the genetic variations differently among species (Kuang et al., 2011). SSRs are also effective molecular markers for interspecific and intraspecific polymorphisms and extensively used for species identification, speciation, population genetic diversity and phytogeography studies (Powell et al., 1995). In four Bulbophyllum species, SSR loci in the cp genome exhibited high level of interspecific polymorphisms. Several exclusive SSR loci presented in each species, e.g., B. menghaiense with 10 exclusive SSRs, B. lingii with six, and B. pentanurum with five. Thus, these specific SSRs are useful molecular markers for species identification and detecting genetic diversity.
4.3. Hotspot regions
We found the nucleotide sequence diversity was higher in the non-coding regions than the coding regions, which is generally consistent with most previous studies of the chloroplast genomes of angiosperms (Clegg et al., 1994). A number of cpDNA, e.g., matK, rbcL, psbA-trnH, trnS-trnG, trnL-trnF, trnF-ndhJ, trnE-trnD are previously used within Bulbophyllum to reconstruct phylogenetic relationships, but have not been shown useful for detecting infragenic or interspecific relationships (Fischer et al., 2007; Hosseini et al., 2012, 2016; Smidt et al., 2011). Smidt et al. (2011), for example, used two non-coding regions of cpDNA, psbA-trnH and trnS-trnG to examine phylogenetic relationships among Neotropical sections of Bulbophyllum, but did not support the monophyly of the neotropical group, offering very little phylogenetic value. Fischer et al. (2007), who studied Madagascan Bulbophyllum using four plastid markers, did not resolved some terminal nodes. In recent years, numerous orchid plastid markers based on the comparative cp genome have been proposed for Orchidaceae phylogeny at different taxonomical level (Dong et al., 2018; Li et al., 2019; Niu et al., 2017; Yang et al., 2013; Zavala-Páez et al., 2020; Zhu et al., 2018).
Based on comprehensive comparative analysis, a total of 20 sequence divergence hotspot regions located in intergenic regions will be helpful to analyze phylogenetic relationships especially between closely related taxa, or where recent divergence, rapid radiation in Bulbophyllum (Daniell et al., 2016; Tonti-Filippini et al., 2017). Compared with the previous study of cp genomes of Neotropical Bulbophyllum (Zavala-Páez et al., 2020), where 10 potential markers proposed for barcoding and phylogenetic studies in Bulbophyllum, there are four regions identical (psbB-psbT, trnS-trnG, trnR-atpA, rpl32-trnL) with our work. In addition, three protein coding genes (trnL-UAA, rpl16, trnK-UUU) also showed remarkably higher Pi values (>0.03). These genes might be undergoing rapid nucleotide substitution at the species level, and might be applied for molecular evolution and adaptive evolution analysis.
4.4. Adaptive evolution
The dN/dS ratios have been widely used to infer the evolutionary dynamics and identify adaptive signatures among species (Yang and Bielawski, 2000). The dN/dS value in all of 79 protein coding genes in four Bulbophyllum species were lower than 1. The dN/dS values are higher in protein-coding genes as a result of more frequent non-synonymous nucleotide substitutions than the synonymous substitutions, and the adaptation evolution may have contributed to the elevated dN/dS ratios (Makałowski and Boguski, 1998). Thus, the elevated dN/dS ratios indicated some unknown selective forces on Bulbophyllum species and results in species divergence (Hurst, 2002), which may increase our understanding of the evolutionary speciation and environment adaptation in Bulbophyllum species. We found that ten genes under relaxed selection (Fig. 6) and six genes under site-specific selection (Table 7). Of these genes, petA encoding cytochrome f, ndhF and ndhK encoding subunits of NADH dehydrogenase, rbcL encoding the large subunit of ribulose-bisphosphate carboxylase, all functioned in photosynthesis (Green, 2011; Table 3). A previous study showed that rbcL is often under positive selection in land plants, and evolved under strong positive selection after the C3–C4 photosynthetic transition in grasses (Kapralov and Filatov, 2007; Piot et al., 2017). The genes rpoA, rps15, rps16, rpl16, rpl22 all functioned in self-replication. The other genes are matK acting as a maturase, ycf1 and ycf2 with unknown function (Daniell et al., 2016; Green, 2011). These genes play important role in improving energy conversion efficiency, therefore they might have some evolutionary and adaptive significance for Bulbophyllum species. Different from our results, selection analysis in eight Neotropical Bulbophyllum species showed that four genes (ycf1, ycf2, accD, and rps3) were under site-specific selection (Zavala-Páez et al., 2020). The differences of site-specific selection pattern between Asian and neotropical Bulbophyllum indicated that they might each had various adaptive strategies under unpredictable environmental conditions. Our findings from selective pressure analysis could lead to a better understanding of the evolution of Bulbophyllum species.
4.5. Phylogenetic relationship
Complete chloroplast DNA sequences were valuable for analyzing phylogenetic relationships (Daniell et al., 2016; Tonti-Filippini et al., 2017). Kim et al. (2020a), for example, utilized the chloroplast genomes of 124 species of orchids to evaluate the phylogenomics and relationship within the Orchidaceae family. Moreover, the whole cp genome phylogeny that was generated at different taxonomical level in several groups of Orchidaceae, such as subtribe Aeridinae (Kim et al., 2020b), subtribe Calypsoinae (Li et al., 2020), and Holcoglossum (Li et al., 2019). Considering that most species-level diversity of Orchidaceae, a whole cp genome phylogeny is highly desired for Bulbophyllum. We examined phylogenetic relationships among 13 Bulbophyllum species from two continental groups in this study. Both the complete cp genome sequences and 65 protein coding genes datasets produced the same topology with similar support. In addition, all the ML, BI and MP phylogenetic results strongly supported that B. pentaneurum is sister to B. lingii with B. menghaiense as their closest relative, and all 13 Bulbophyllum species were closely clustered into one lineage, which strongly support that section Macrocaulia and the genus of Bulbophyllum are monophyletic, respectively (Fig. 7). The monophyly of section Macrocaulia was also supported by Gravendeel et al. (2014) using nuclear ITS sequence. However, our phylogenetic analysis did not show that section Macrocaulia was the basal clade in Asian Bulbophyllum group as their work, might due to sampling limited. Furthermore, the phylogenetic relationships among the Neotropical clade were also in accordance with recent phylogeny research (Zavala-Páez et al., 2020). In the future, more complete cpDNA sequences of member individuals of Bulbophyllum are needed to exploit the power of the whole cp genome phylogenies, especially for differentiation at lower taxonomic levels.
Author contributions
YL and LL designed the study and revised the manuscript. HQT, LT drafted the manuscript and prepared tables and figures. HQT, LT and SCS performed comparative genome analysis. YLP performed phylogenetic analysis. All authors discussed the results and commented on the manuscript.
Declaration of competing interest
The authors declared no conflict of interest.
Acknowledgements
We are grateful to Wenbin Yu, Yu Song, from Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Fei Zhao from Kunming Institute of Botany, Chinese Academy of Sciences, for their help in genomic data analysis. We also thank Lin Li, from South China Botanical Garden, Chinese Academy of Sciences, Xiaoyun Wang and Zhifeng Xu from the Orchid Conservation Center of Yunnan Fengchunfang Biotechnology Co., Ltd, for kindly providing photos and sample materials. This work was supported by the National Natural Science Foundation of China (No. 31870183, No.U1702235) and Southeast Asia Biodiversity Research Institute, Chinese Academy of Sciences (Y4ZK111B01).
Footnotes
Peer review under responsibility of Editorial Office of Plant Diversity.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.pld.2021.01.003.
Contributor Information
Lu Li, Email: lilu@swfu.edu.cn.
Yan Luo, Email: luoyan@xtbg.org.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Asaf S., Khan A.L., Aaqil Khan M., et al. Comparative analysis of complete plastid genomes from wild soybean (Glycine soja) and nine other Glycine species. PLoS One. 2017;12 doi: 10.1371/journal.pone.0182281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averyanov L.V., Komarov V.L. Botanical Institute, Russian Academy of Sciences; St. Petersburg, Russia: 1994. Identification Guide to Vietnamese Orchids (Orchidaceae Juss.) p. 432. [Google Scholar]
- Barrett C.F., Davis J.I. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am. J. Bot. 2012;99:1513–1523. doi: 10.3732/ajb.1200256. [DOI] [PubMed] [Google Scholar]
- Beier S., Thiel T., Münch T., et al. MISA-web, a web server for microsatellite prediction. Bioinformatics. 2017;33:2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase M.W., Cameron K.M., Freudenstein J.V., et al. An updated classification of Orchidaceae. Bot. J. Linn. Soc. 2015;177:151–174. [Google Scholar]
- Chen X.Q., Vermeulen J.J. In: Flora of China. Wu Z.Y., Raven P.H., Hong D.Y., editors. vol. 25. Science Press and St. Louis, Missouri Botanical Garden Press; Beijing, China: 2009. Bulbophyllum thouars; pp. 404–440. [Google Scholar]
- Clegg M.T., Gaut B.S., Learn G.H., et al. Rates and patterns of chloroplast DNA evolution. Proc. Natl. Acad. Sci. U.S.A. 1994;91:6795–6801. doi: 10.1073/pnas.91.15.6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H., Lin C.S., Yu M., et al. Chloroplast genomes, diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W.L., Wang R.N., Zhang N.Y., et al. Molecular evolution of chloroplast genomes of orchid species, insights into phylogenetic relationship and adaptive evolution. Int. J. Mol. Sci. 2018;19:716. doi: 10.3390/ijms19030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer G.A., Gravendeel B., Sieder A., et al. Evolution in resupination of Malagasy species of Bulbophyllum (Orchidaceae) Mol. Phylogenet. Evol. 2007;45:358–376. doi: 10.1016/j.ympev.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Feng Y.L., Wicke S., Li J.W., et al. Lineage-specific reductions of plastid genomes in an orchid tribe with partially and fully mycoheterotrophic species. Genome Biol. Evol. 2016;8:2164–2175. doi: 10.1093/gbe/evw144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A., Pachter L., Poliakov A., et al. VISTA, computational tools for comparative genomics. Nucleic Acids Res. 2004;1:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamisch A., Comes H.P. Clade-age-dependent diversification under high species turnover shapes species richness disparities among tropical rainforest lineages of Bulbophyllum (Orchidaceae) BMC Evol. Biol. 2019;19:93. doi: 10.1186/s12862-019-1416-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham S.W., Lam V.K.Y., Merckx V.S.F.T. Plastomes on the edge, the evolutionary breakdown of mycoheterotroph plastid genomes. New Phytol. 2017;214:48–55. doi: 10.1111/nph.14398. [DOI] [PubMed] [Google Scholar]
- Gravendeel B., Fischer G.A., Vermeulen J.J. In: Genera Orchidacearum. Pridgeon A.M., Cribb P.J., Chase M.W., Rasmussen F.N., editors. vol. 6. Oxford University Press; Oxford, England: 2014. Bulbophyllum thouars. [Google Scholar]
- Green B.R. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011;66:34–44. doi: 10.1111/j.1365-313X.2011.04541.x. [DOI] [PubMed] [Google Scholar]
- Greiner S., Lehwark P., Bock R. Organellar Genome DRAW (OGDRAW) version 1.3.1, expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47:W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini S., Dadkhah K., Go R. Molecular systematics of genus Bulbophyllum (Orchidaceae) in Peninsular Malaysia based on combined nuclear and plastid DNA sequences. Biochem. Systemat. Ecol. 2016;65:40–48. [Google Scholar]
- Hosseini S., Go R., Dadkhah K., et al. Studies on maturase K sequences and systematic classification of Bulbophyllum in Peninsular Malaysia. Pakistan J. Bot. 2012;44:2047–2054. [Google Scholar]
- Hu A.Q., Gale S.W., Liu Z.J., et al. Molecular phylogenetics and floral evolution of the Cirrhopetalum alliance (Bulbophyllum, Orchidaceae): evolutionary transitions and phylogenetic signal variation. Mol. Phylogenet. Evol. 2020;143:106689. doi: 10.1016/j.ympev.2019.106689. [DOI] [PubMed] [Google Scholar]
- Huang M.Z., Yang G.S., Lan S.R., et al. Bulbophyllum lingii, a new species (Malaxideae, epidendroideae, Orchidaceae) from Hainan, China. Phytotaxa. 2020;452:185–188. [Google Scholar]
- Hurst L.D. The Ka/Ks ratio, diagnosing the form of sequence evolution. Trends Genet. 2002;18:486–487. doi: 10.1016/s0168-9525(02)02722-1. [DOI] [PubMed] [Google Scholar]
- Ingvarsson P.K., Ribstein S., Taylor D.R. Molecular evolution of insertions and deletion in the chloroplast genome of Silene. Mol. Biol. Evol. 2003;20:1737–1740. doi: 10.1093/molbev/msg163. [DOI] [PubMed] [Google Scholar]
- Jin J.J., Yu W.B., Yang J.B., et al. GetOrganelle, a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21:241. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X.H., Li J.W., Ye D.P. Henan science and Technology Press; Zhengzhou, China: 2019. Atlas of Native Orchids in China; pp. 585–707. [Google Scholar]
- Kapralov M.V., Filatov D.A. Wide spread positive selection in the photosynthetic Rubisco enzyme. BMC Evol. Biol. 2007;7:73. doi: 10.1186/1471-2148-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7, improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten B., Rampant P.F., Mader M., et al. Genome sequences of Populus tremula chloroplast and mitochondrion, implications for holistic poplar breeding. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.S., Kim H.T., Kim J.H. The largest plastid genome of monocots, a novel genome type containing AT residue repeats in the slipper orchid Cypripedium japonicum. Plant Mol. Biol. Rep. 2015;33:1210–1220. [Google Scholar]
- Kim Y.K., Jo S., Cheon S.H., et al. Plastome evolution and phylogeny of Orchidaceae, with 24 new sequences. Front. Plant Sci. 2020;11:22. doi: 10.3389/fpls.2020.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.K., Jo S., Cheon S.H., et al. Plastome evolution and phylogeny of subtribe Aeridinae (Vandeae, Orchidaceae) Mol. Phylogenet. Evol. 2020;144:106721. doi: 10.1016/j.ympev.2019.106721. [DOI] [PubMed] [Google Scholar]
- Kuang D.Y., Wu H., Wang Y.L., et al. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae), implication for DNA barcoding and population genetics. Genome. 2011;54:663–673. doi: 10.1139/g11-026. [DOI] [PubMed] [Google Scholar]
- Kumar S., Nei M., Dudley J., et al. MEGA, a biologist centric software for evolutionary analysis of DNA and protein sequences. Briefings Bioinf. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P.J.R., Rozas Julio. DnaSP v5, a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Li Z.H., Ma X., Wang D.Y., et al. Evolution of plastid genomes of Holcoglossum (Orchidaceae) with recent radiation. BMC Evol. Biol. 2019;19:63. doi: 10.1186/s12862-019-1384-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.H., Jiang Y., Ma X., et al. Plastid genome evolution in the subtribe Calypsoinae (epidendroideae, Orchidaceae) Genome Biol. Evol. 2020;12:867–870. doi: 10.1093/gbe/evaa091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.S., Chen J.J.W., Huang Y.T., et al. The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci. Rep. 2015;5:1–10. doi: 10.1038/srep09040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logacheva M.D., Schelkunov M.I., Penin A.A. Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidusavis. Genome Biol. Evol. 2011;3:1296–1303. doi: 10.1093/gbe/evr102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Deng J.P., Peng Y.L., et al. Bulbophyllum gedangense (Orchidaceae, Epidendroideae, Malaxideae), a new species from Tibet, China. Phytotaxa. 2020;453:145–150. [Google Scholar]
- Makałowski W., Boguski M.S. Evolutionary parameters of the transcribed mammalian genome, an analysis of 2820 orthologous rodent and human sequences. Proc. Natl. Acad. Sci. U.S.A. 1998;95:9407–9412. doi: 10.1073/pnas.95.16.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Z., Xue Q., Zhu S., et al. The complete plastome sequences of four orchid species, insights into the evolution of the Orchidaceae and the utility of plastomic mutational hotspots. Front. Plant Sci. 2017;8:715. doi: 10.3389/fpls.2017.00715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piot A., Hackel J., Christin P.A., et al. One-third of the plastid genes evolved under positive selection in PACMAD grasses. Planta. 2017;247:255–266. doi: 10.1007/s00425-017-2781-x. [DOI] [PubMed] [Google Scholar]
- Powell W., Morgante M., McDevitt R., et al. Polymorphic simple sequence repeat regions in chloroplast genomes, applications to the population genetics of pines. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7759–7763. doi: 10.1073/pnas.92.17.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridgeon A.M., Cribb P.J., Chase M.W., et al. vol. 6. Oxford University Press; Oxford, England: 2014. Genera Orchidacearum. [Google Scholar]
- Qin Z., Wang Y.P., Wang Q.M., et al. Evolution analysis of simple sequence repeats in plant genome. PLoS One. 2015;10 doi: 10.1371/journal.pone.0144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., et al. MrBayes 3.2, Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J., Sánchez-DelBarrio J.C., Messeguer X., et al. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2004;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Schelkunov M.I., Shtratnikova V.Y., Nuraliev, et al. Exploring the limits for reduction of plastid genomes, a case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biol. Evol. 2015;7:1179–1191. doi: 10.1093/gbe/evv019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidenfaden G. Orchid genera in Thailand VIII, Bulbophyllum Thou. Dan. Bot. Ark. 1979;33:1–228. [Google Scholar]
- Shao S.C., Tang L., Luo Y. The complete chloroplast genome sequence of Dendrobium wangliangii (Orchidaceae) Mitochondrial DNA B. 2020;5:3513–3515. doi: 10.1080/23802359.2020.1827063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smidt E.C., Borba E.L., Gravendeel B. Molecular phylogeny of the Neotropical sections of Bulbophyllum (Orchidaceae) using nuclear and plastid spacers. Taxon. 2011;60:1050–1064. [Google Scholar]
- Shaw J., Shafer H.L., Leonard O.R., et al. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms, the tortoise and the hare IV. Am. J. Bot. 2014;101:1987–2004. doi: 10.3732/ajb.1400398. [DOI] [PubMed] [Google Scholar]
- Shi L., Chen H., Jiang M., et al. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019;47:W65–W73. doi: 10.1093/nar/gkz345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M., Lehwark P., Pellizzer T., et al. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonti-Filippini J., Nevill P.G., Dixon K., et al. What can we do with 1000 plastid genomes? Plant J. 2017;90:808–818. doi: 10.1111/tpj.13491. [DOI] [PubMed] [Google Scholar]
- Trifinopoulos J., Nguyen L.T., Haeseler A., et al. W-IQ-TREE, a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016;44(W1):W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen J.J. In: Genera Orchidacearum. Pridgeon A.M., Cribb P.J., Chase M.W., Rasmussen F.N., editors. vol. 6. Oxford University Press; Oxford,England: 2014. Asian sections of Bulbophyllum Thou; pp. 19–40. [Google Scholar]
- Wang Y.P., Ai J., Luo Y., et al. The complete chloroplast genome of Dendrobium wattii (Orchidaceae) Mitochondrial DNA B. 2020;5(2):1325–1326. [Google Scholar]
- Weng M.L., Ruhlman T.A., Jansen R.K. Expansion of inverted repeat does not decrease substitution rates in Pelargonium plastid genomes. New Phytol. 2017;214:842–851. doi: 10.1111/nph.14375. [DOI] [PubMed] [Google Scholar]
- Wicke S., Schneeweiss G.M., Müller K.F., et al. The evolution of the plastid chromosome in land plants, gene content, gene order, gene function. Plant Mol. Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Ma P.F., Li H.T., et al. Comparative plastomic analysis and insights into the phylogeny of Salvia (Lamiaceae) Plant Divers. 2021;43:15–26. doi: 10.1016/j.pld.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Yang Z. PAMLX, a graphical user interface for PAML. Mol. Biol. Evol. 2013;30:2723–2724. doi: 10.1093/molbev/mst179. [DOI] [PubMed] [Google Scholar]
- Yang J.B., Tang M., Li H., et al. Complete chloroplast genome of the genus Cymbidium, lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol. 2013;13:84. doi: 10.1186/1471-2148-13-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Bielawski J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.H., Nielsen R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002;19:908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- Yang Z.H., Wong W.S., Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Yuan Y., Jin X., Liu J., et al. The Gastrodia elata genome provides insights into plant adaptation to heterotrophy. Nat. Commun. 2018;9:1–11. doi: 10.1038/s41467-018-03423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala-Páez M., Vieira L.N., Baura V.A., et al. Comparative plastid genomics of neotropical Bulbophyllum (Orchidaceae; epidendroideae) Front. Plant Sci. 2020;11:799. doi: 10.3389/fpls.2020.00799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.F., Gao I., Jakovlić H., et al. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020;20:348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- Zhu S., Niu Z., Xue Q., et al. Accurate authentication of Dendrobium officinale and its closely related species by comparative analysis of complete plastomes. Acta Pharm. Sin. B. 2018;8:969–980. doi: 10.1016/j.apsb.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.