Abstract
Bromodomain protein 4 (BRD4) is a transcriptional and epigenetic regulator that is a therapeutic target in many cancers and inflammatory diseases. BRD4 plays important roles in transcription as an active kinase, which phosphorylates the carboxy-terminal domain (CTD) of RNA polymerase II (Pol II), the proto-oncogene c-MYC, and transcription factors TAF7 and CDK9. BRD4 is also a passive scaffold that recruits transcription factors. Despite these well-established functions, there has been little characterization of BRD4’s biophysical properties or its kinase activity. We report here that the 156 kD mouse BRD4 exists in an extended dimeric conformation with a sedimentation coefficient of ∼6.7 S and a high frictional ratio. Deletion of the conserved B motif (aa 503–548) disrupts BRD4’s dimerization. BRD4 kinase activity maps to amino acids 351 to 598, which span bromodomain-2, the B motif, and the BID domain (BD2-B-BID) and contributes to the in vivo phosphorylation of its substrates. As further assessed by analytical ultracentrifugation, BRD4 directly binds purified Pol II CTD. Importantly, the conserved A motif of BRD4 is essential for phosphorylation of Pol II CTD, but not for phosphorylation of TAF7, mapping its binding site to the A motif. Peptides of the viral MLV integrase (MLVIN) protein and cellular histone lysine methyltransferase, NSD3, which have been shown by NMR to bind to the extra-terminal (ET) domain, also are phosphorylated by BRD4. Thus, BRD4 has multiple distinct substrate-binding sites and a common kinase domain. These results provide new insights into the structure and kinase function of BRD4.
Keywords: BRD4, kinase, extended dimer, CTD
Abbreviations: A and B, conserved BET motifs; AUC, analytical ultracentrifugation; BD1 and BD2, bromodomains 1 and 2; BET, bromodomain and extra terminal; BID, basic residue-rich interaction domain; BRD4, bromodomain protein 4; CK2, casein kinase 2; CTD, RNA polymerase II carboxy terminal domain; CTM, C-terminal motif; DDM, n-dodecyl β-D-maltoside; ET, extra terminal domain; Km, Michaelis–Menten constant; MLVIN, murine leukemia virus integrase; NPS and CPS, N and C-terminal phosphorylation sites respectively; PTEFb, positive transcription elongation factor b; SEC, size-exclusion chromatography; SEED, Ser/Glu/Asp-rich region; TCEP, Tris(2-carboxyethyl) phosphine hydrochloride; Vmax, maximal rate
BRD4 is a bromodomain and extra terminal (BET) family member that has pleiotropic functions throughout cell cycle. It interacts with chromatin modifying factors and transcription factors, among them the positive transcription elongation factor b (PTEFb), and recruits them to enhancers and promoters (1). BRD4 is a mitotic bookmark that remains associated with chromosomes throughout mitosis (2). It also functions in DNA repair and contributes to the regulation of alternative splicing (3). Importantly, BRD4 is a kinase that regulates transcription, both directly and indirectly. It directly regulates transcription by phosphorylating the Ser2 of the RNA Pol II C-terminal domain (CTD) and facilitates the transition of RNA Pol II from initiation to elongation (4). It also phosphorylates both the TFIID component, TAF7, and the transcription elongation factor, PTEFb/CDK9 (5). It indirectly regulates transcription through its phosphorylation of the transcription factor, c-MYC, leading to c-MYC degradation (6). Thus, BRD4 is an active participant in transcription.
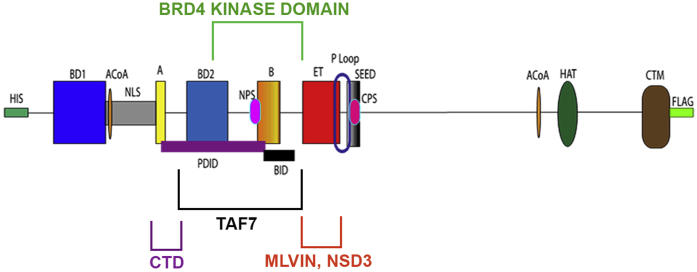
Although the functions of BRD4, particularly as they relate to cancer, have been extensively studied, less is known about its biophysical properties in general and its kinase activity, specifically. Within the molecule, the N-terminal half of BRD4 is the better characterized (Fig. 1A). The kinase activity of BRD4, an atypical kinase, maps to the N-terminal half of the molecule. However, the position of the active site within the N-terminus has not been determined. In addition to the kinase domain, the N-terminal segment also encompasses two bromodomains, whose structures have both been resolved, which bind to acetylated lysines on histones and other proteins (7). The ET domain, whose structure has been determined, is one of the defining domains of the BET family within this region (8). Additional N-terminal elements A and B motifs, BID and SEED domains are largely uncharacterized, both structurally and functionally. The B motif has been reported to be important for chromatin binding; the BID domain is the target of phosphorylation by the kinase, casein kinase 2 (CK2) (9, 10). Little is known about the structure of the C-terminal half of BRD4. It is predicted to be intrinsically disordered since it is responsible for forming phase-separated condensates, both in vitro and in vivo (11). Despite the separate characterizations of the two halves of BRD4, the biophysical properties of the full-length intact molecule are currently unknown.
Figure 1.
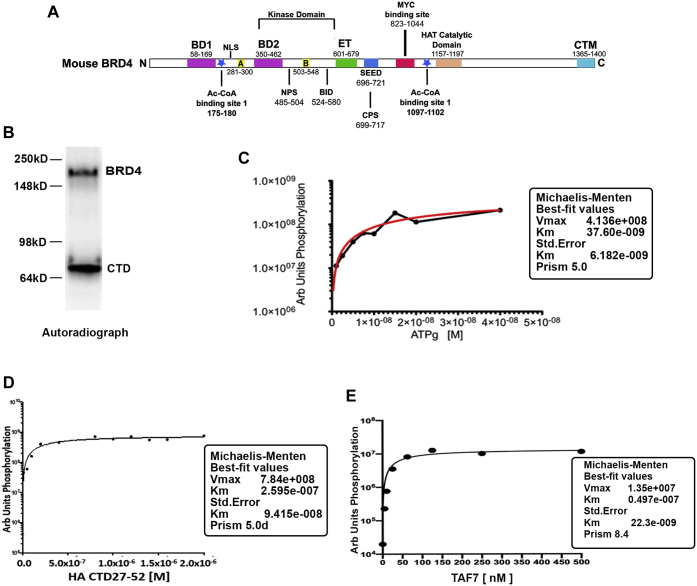
BRD4 kinase follows classical enzyme kinetics.A, map of mouse BRD4 showing the locations of various domains, the numbers indicate the amino acid residues encompassing the domain. B, in vitro kinase assay using 10uCi radiolabeled ATP showing the auto and trans kinase activities of 10 nM BRD4; 25 nM Pol II CTD27–52 was used as a substrate. Orange line represents the standard error of the best-fit parameters, as determined by Prism. C, Michaelis–Menten plot representing the kinetics of BRD4 autophosphorylation in the presence of increasing concentrations of radiolabeled ATP. D, Michaelis–Menten plot representing the kinetics of 10 nM BRD4 phosphorylation of increasing concentrations of substrate HA-CTD27–52. E, Michaelis–Menten plot representing the kinetics of 10 nM BRD4 phosphorylation of increasing concentration of TAF7 C-terminal fragment. Arbitrary quantification units on Y-axis were plotted against increasing concentrations of TAF7 on X-axis. All Km and Vmax values were calculated as described in Experimental procedures. A and B, conserved motifs; BD1 and BD2, bromodomains 1 and 2; BID, basic residue-rich interaction domain; CTM, C-terminal motif; ET, extra terminal domain; Km, Michaelis–Menten constant; NPS and CPS, N and C-terminal phosphorylation sites respectively; SEED, Ser/Glu/Asp-rich region; Vmax, maximal rate.
The present studies were undertaken to begin to map and characterize the kinase domain within BRD4 and to characterize its biophysical properties. We report that BRD4 exists in solution as an extended dimer whose structure is maintained by the conserved B motif. We locate the BRD4 kinase activity within the BRD4 region spanning the BD2-B-BID domains. The BRD4 kinase activity behaves with classical Michaelis–Menten kinetics both to autophosphorylate and to trans-phosphorylate its CTD and TAF7 substrates. In vivo phosphorylation of the CTD at Ser2 by BRD4 depends on the kinase activity. Interestingly, BRD4 has distinct binding sites for its different substrates. Thus, BRD4 directly binds its Pol II CTD substrate and phosphorylation of the CTD depends on the A motif, consistent with that being the binding site. In contrast, TAF7 phosphorylation does not depend on the A motif, indicating it has distinct binding requirements. Furthermore, we report that BRD4 phosphorylates both the viral murine leukemia virus integrase (MLVIN) and the cellular histone lysine methyltransferase, NSD3, which bind to the ET domain. Taken together, these studies provide important new insights into BRD4 structure and function.
Results
BRD4 kinase activity follows Michaelis–Menten kinetics
Functional domains of BRD4 have been well characterized. The amino terminal half of the mouse BRD4, a 1400 aa molecule, contains two bromodomains that bind acetylated histone lysines, anchoring BRD4 to chromatin, as well as other acetylated proteins (Fig. 1A). The carboxy-terminal half of BRD4 contains the HAT domain, the MYC-binding site, and the major binding site for PTEFb, as well as other factors (Fig. 1A) (12). Structural analysis has only been achieved for the two bromodomains and ET domain (7, 8). Computational modeling of BRD4’s secondary structure predicts that the amino-terminal half of the molecule is highly ordered, whereas the carboxy terminal half is extensively intrinsically disordered.
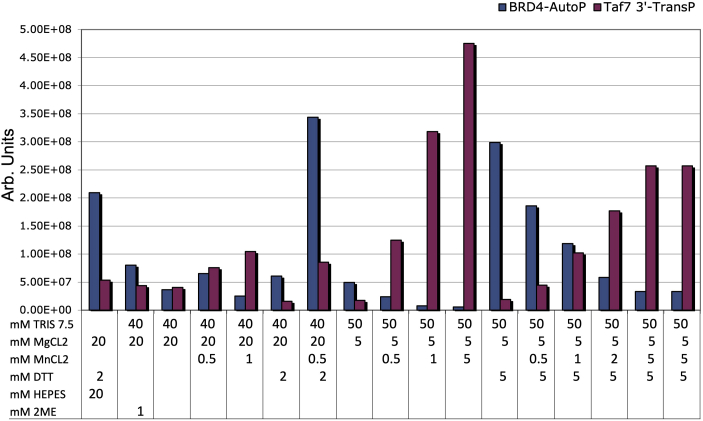
We have previously reported that BRD4 is a kinase that phosphorylates RNA polymerase II (Pol II) and localized the kinase domain to the amino terminal segment of BRD4 (4). In contrast to most typical kinases, BRD4 is an atypical kinase that does not have a classical active site (4). We have shown previously that BRD4 phosphorylates the carboxy-terminal domain of RNA polymerase II (CTD), which consists of 52 repeats of the heptad YSPTSPS. The CTD plays a critical role in regulating transcription initiation and elongation through the differential phosphorylation of its serine, threonine, and tyrosine residues by multiple kinases, including BRD4 (4, 13, 14). BRD4 phosphorylates the CTD of Pol II at Ser2 and preferentially phosphorylates heptad repeats 26 to 33, out of the 52 total (15). Although the kinase activity of BRD4 has been well documented, its enzyme kinetics have not been previously determined. To extend the characterization of BRD4 kinase activity, we determined the Km for two of its substrates, Pol II CTD and TAF7. Since BRD4 has both auto- and trans-phosphorylation activities (Fig. 1B), we monitored the efficiency of both activities under different assay conditions. Trans-phosphorylation kinetics were determined using Pol II CTD and TAF7 as substrates (5). Interestingly, while optimal auto-phosphorylation of BRD4 was observed in low or absent MnCl2 buffers, optimal trans-phosphorylation of the TAF7 substrate occurred in buffers with 5 mM MnCl2 (Fig. S1). Intermediate concentrations supported both activities. These findings suggest metal ion induced conformational changes that favor the binding of its trans substrates.
Subsequent experiments to determine whether the BRD4 kinase activity followed Michaelis–Menten kinetics were done in conditions to optimize the relevant activity. We first focused on autophosphorylation to determine the kinetics of hydrolysis of ATP. In the absence of any exogenous substrate, BRD4 hydrolyzed ATP efficiently with a Km of 37.6 nM ± 6.8 nM, as assessed by the extent of autophosphorylation (Fig. 1C). We observed a hyperbolic relationship between ATP concentration and velocity of the reaction indicating that BRD4 follows classical enzyme kinetics for its autophosphorylation.
To assess the kinetics of trans-phosphorylation, we examined two of BRD4’s substrates, the CTD and TAF7. BRD4 phosphorylation of the Pol II CTD was assessed using an HA-tagged fragment spanning repeats 27 to 52, which contains the preferred phosphorylation site. Phosphorylation occurred with a Km of 259 ± 94 nM. (Fig. 1D). This Km suggests an interaction of low-to-intermediate affinity, but may also reflect the fact that only a fragment of the CTD was used. BRD4 phosphorylation of the C-terminal fragment of TAF7, which spans its phosphorylation sites, occurred with a significantly lower Km of 49.7 nM indicative of a higher affinity of interaction (Fig. 1E). Both followed classical Michaelis–Menten kinetics. The above results establish that, despite the fact that BRD4 is an atypical kinase, BRD4 kinase activity follows classical enzyme kinetics.
BRD4 kinase activity is contained within the BD2-B-BID region
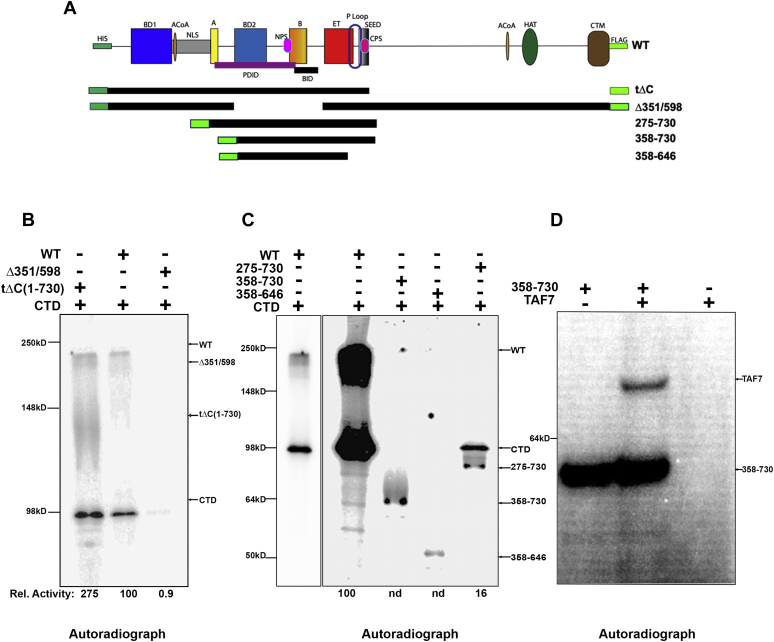
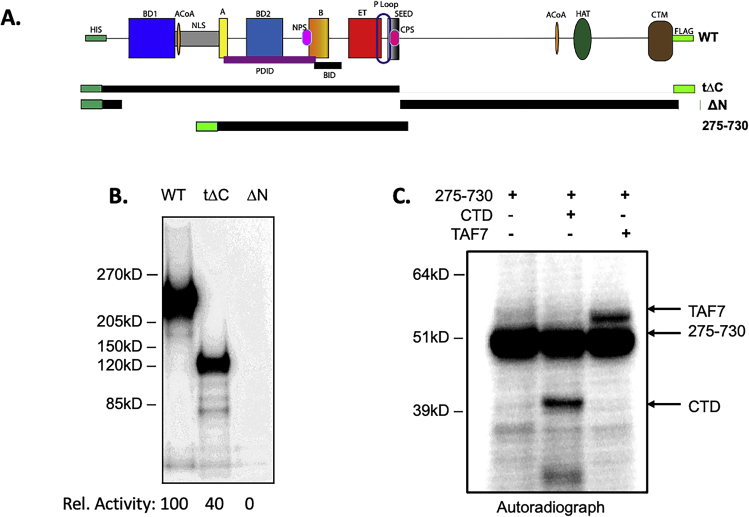
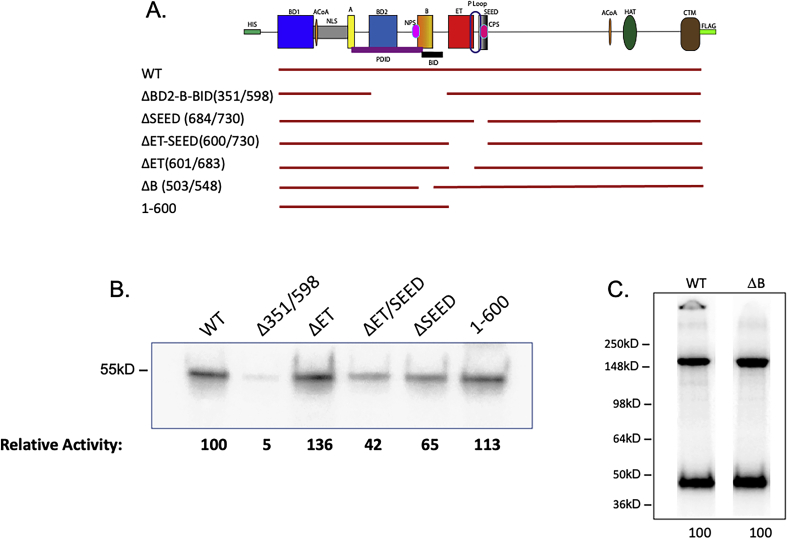
To map a minimal segment required for activity, we generated a series of truncation and deletion mutants (Figs. 2A, S2 and S3). As we previously reported, the N-terminal half of BRD4 (1–722)—which encompasses the two bromodomains as well as the A and B motifs, BID domain, ET and SEED domains—retains kinase activity, whereas the C-terminal half (730–1400, ΔN) has no kinase activity (4). Here we have extended these studies to further define the kinase domain. Consistent with our previous findings, WT and tΔC (1–730) efficiently phosphorylated full-length GST-CTD (Fig. 2B). Indeed, the tΔC fragment was more active than wild type, suggesting that BRD4 autoregulates its kinase activity, perhaps through conformational changes. A further truncation to generate a 1 to 600 aa segment retained full activity, mapping the kinase activity entirely within this region (Fig. S3B). (Deletion of the SEED domain (696–721 aa) from full-length BRD4 appeared to modestly reduce activity (Figs. S3 and S4). The reason for this is not known.) To further map the kinase activity within the 1 to 600 aa segment, we generated a BRD4 mutant deleted of the segment 351 to 598 (Δ351–598), which spans bromodomain 2 and the B-BID domains (BD2-B-BID) (Fig. 2A). Deletion of this region (Δ351–598) abrogated BRD4’s auto and trans phosphorylation activities by 97.5% ± 1.5% (Fig. 2B) indicating that the BD2-B-BID domain is necessary for BRD4’s kinase activity. However, within the BD2-B-BID segment, the B motif is not necessary for kinase activity (Fig. S3C).
Figure 2.
BRD4 kinase domain is contained within the BD2-B-BID regions.A, schematic map of WT, deletion mutants, and fragments of BRD4 (see Fig. 1 for definitions of BRD4 domains). B, in vitro kinase assay using radiolabeled ATP showing both auto and trans kinase activities of WT, tΔC, and a Δ351 to 598 recombinant BRD4 proteins, all at 50 nM, using 100 nM GST-CTD1–52 as substrate. Kinase activity of deletion mutant proteins on CTD is shown relative to WT BRD4. C, in vitro kinase assay using radiolabeled ATP showing both the auto and trans kinase activities of WT, A-to-SEED fragment of BRD4 (275–730), BD2-to-SEED (358–730), BD2-to-part of ET (358–646), all at 50 nM, using 100 nM GST-CTD1–52 as substrate. Kinase activity of deletion mutant proteins on CTD is shown relative to full-length WT BRD4. N.d., none detected. D, in vitro kinase assay using radiolabeled ATP showing both the auto and trans kinase activities of 200 nM BD2-SEED (358–730), using 100 nM GST-TAF7 as substrate.
The above experiments indirectly map the kinase domain. To directly demonstrate that the 351 to 598 aa segment is sufficient for kinase activity, peptide fragments of BRD4 spanning the segments 275 to 730, 358 to 730, and 358 to 646 aa (all-encompassing the BD2-B-BID region) were generated and tested for their ability to auto-phosphorylate and to trans-phosphorylate the CTD or TAF7 (Fig. 2C). All three fragments were able to auto-phosphorylate, although their activities were significantly reduced relative to wild type (Figs. 2C and S4). These results fine map both the kinase domain and sites of phosphorylation within this segment. The autophosphorylation by all three fragments is consistent with previous reports of phosphorylation of the BID domain (10).
The patterns of trans-phosphorylation were markedly different when the CTD or TAF7 was used as substrate. Among the fragments, only the largest fragment 275 to 730 aa could trans-phosphorylate the CTD (Figs. 2C and S4). Surprisingly, both 275 to 730 and 358 to 730 aa fragments could efficiently phosphorylate TAF7 (Figs. 2D, S2 and S4).
Taken together, these results indicate that 351 to 598 aa segment, which spans the BD2, B, and BID domains, is both necessary and sufficient for kinase activity. The inability of the 358 to 730 fragment to phosphorylate the CTD, but not TAF7, suggests that the CTD-binding site maps to the 275 to 358 aa region. In contrast, the TAF7-binding site maps to the region 358 to 730 aa. The finding of different binding sites of the CTD and TAF7 on BRD4 is consistent with their observed differences in enzyme kinetics, observed above.
BRD4 phosphorylates substrates that bind to its ET region
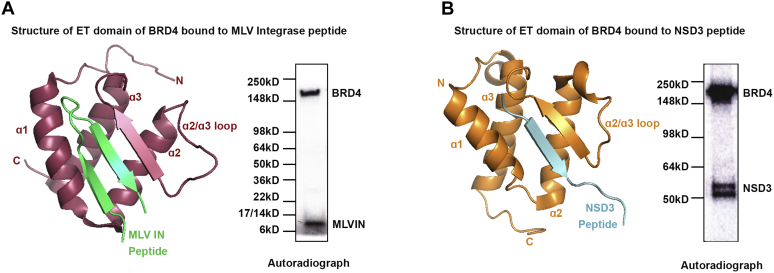
The finding that different BRD4 substrates bind to different regions of the molecule is consistent with earlier characterizations of BRD4 as a scaffold and extends our previous report that another substrate, MYC, binds to the C-terminal domain of BRD4 (6). The ET domain of BRD4 is highly conserved among BET proteins and known to interact with the MLV integrase (MLV IN) and NSD3, a histone lysine methyltransferase. NMR studies of the BRD4 ET domain documented that BRD4 binds peptides derived from both MLV integrase and NSD3 (16, 17). These findings led us to ask whether these peptides could also be substrates for BRD4 phosphorylation. Indeed, BRD4 efficiently phosphorylated both peptides (Figs. 3 and S4).
Figure 3.
BRD4 phosphorylates substrates that bind to its ET domain.A, left: ribbon diagram of ET domain of BRD4 (pink) bound to MLV integrase peptide (residues 389–405), (green) modified from PDB:2N3K. Right: in vitro kinase assay showing 400 nM MLV IN peptide phosphorylation by 70 nM full-length BRD4. B, left: ribbon diagram of ET domain of BRD4 (orange) bound to NSD3 peptide (residues 152–163) modified from PDB:2NCZ (cyan). Right: in vitro kinase assay showing 750 nM NSD3 peptide phosphorylation by 70 nM full-length BRD4.
Thus, BRD4 is capable of phosphorylating MLV IN and NSD3 peptides that bind to the ET domain, CTD and TAF7 that bind to the N-terminal region, as well as MYC, which binds to C-terminal domain, demonstrating that BRD4 kinase activity is independent of the substrate-binding site.
Taken together, these results demonstrate that the BRD4 kinase activity maps to a region between 351 and 598 aa and is capable of both autophosphorylation and trans-phosphorylation of a variety of substrates binding to various sites on the molecule.
BRD4 forms a complex with Pol II CTD
Although BRD4 phosphorylates the Pol II CTD, the Km of Pol II phosphorylation by BRD4 that was determined above suggested a very weak interaction. Therefore, we undertook to determine whether a direct interaction between BRD4 and the CTD could be observed. Relatively little is known about the biophysical properties of BRD4, although it is predicted that the C-terminal half is intrinsically disordered (Fig. 4A). We first analyzed BRD4s pattern of elution in size-exclusion chromatography (SEC). With a molecular weight of 156 kD, BRD4 would be expected to elute from a Sepharose 6 column with a retention factor of approximately 0.89. Surprisingly, we repeatedly observed purified recombinant BRD4 eluting as a broad peak much earlier than expected with a retention factor of 0.28 (Fig. 4B). This corresponds to a molecular weight of ∼900 kD, as determined by molecular weight standard markers. Similarly, aberrant migration of BRD4 was observed on native-PAGE gels, where there was a major BRD4 band at ∼900 kD; a minor band was also observed at ∼480 kD (Fig. 4C). These findings were particularly surprising in light of our observation that BRD4 migrates as a single band with a molecular weight of 156 kD in denaturing gels, as would be expected of a molecule of 1400 aa (Fig. 4D).
Figure 4.
BRD4 is an extended dimer in solution.A, plot showing intrinsically disordered regions in BRD4 as predicted by PONDR (Predictor of Natural Disordered Regions); VL-XT scores are plotted on the Y axis, and amino acid residue numbers are plotted on the X axis. The arrowheads indicate the locations of functional domains of BRD4. B, size exclusion profile of mouse BRD4 on Superose 6 increase column at 4 °C. Inset shows the standard calibration curve run on same condition. Arrowhead indicates the approximate predicted elution volume of monomeric BRD4. C, 3 to 12% gradient blue native page showing the migration of mouse BRD4. Arrowhead indicates the expected monomeric BRD4 migration. D, 8% SDS-PAGE showing the migration of mouse BRD4. E, sedimentation boundaries of 3 μM mouse BRD4 sedimenting at 50,000 rpm, as observed by absorbance at 280 nm (for clarity symbols show only every third data point of every second scan) and c(s) sedimentation coefficient distribution model (lines). The color temperature from purple to red indicates temporal evolution. F, normalized sedimentation coefficient distributions c(s) distribution plots of mouse BRD4 at different concentrations, 3 μM (purple), 1 μM (blue), and 0.3 μM (cyan) calculated on the basis of absorbance scans at 280 nm as shown in (F).
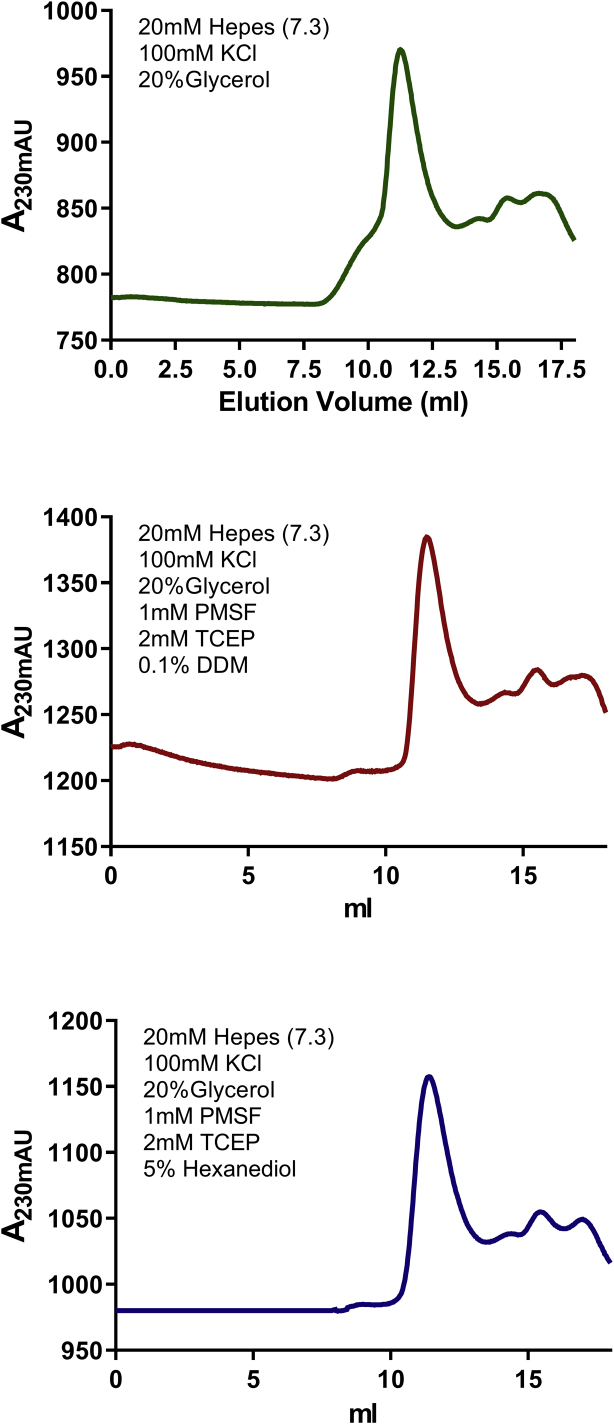
Therefore, these findings suggested three possibilities: 1) monomeric BRD4 exists in an aberrant conformation, 2) BRD4 forms oligomers in solution or 3) BRD4 undergoes phase separation under our experimental conditions. To address the third possibility, BRD4 was treated with the combination of TCEP and 5% 6-hexanediol, which disaggregates phase-separated condensates, and eluted from an SEC column equilibrated with TCEP and 5% 6-hexanediol. That treatment did not affect the elution pattern of BRD4 in SEC, indicating that BRD4 had not phase-separated. To address the second possibility of oligomerization, BRD4 was treated with reducing agents or nonionic detergents, which might have been able to disaggregate the oligomer. The presence of 2 mM Tris(2-carboxyethyl) phosphine hydrochloride (TCEP) together with 0.1% n-Dodecyl β-D-maltoside (DDM) was unable to affect the elution pattern of BRD4 in SEC (Fig. S5). These results suggested that BRD4 exists in an aberrant conformation.
Since BRD4 is predicted to be an intrinsically disordered protein, it is likely to exhibit asymmetry and a larger Stokes radius than expected for a folded protein, which would also be consistent with its aberrant behaviors in SEC and native gels. To further characterize the conformation of BRD4 in solution, we performed sedimentation velocity experiments using analytical ultracentrifugation (AUC), which allowed us to examine the in-solution properties of BRD4 without the effect of external factors (e.g., gel pore size). An example of the evolution of sedimentation boundaries at 280 nm of BRD4 is shown in Figure 4E. Analysis of sedimentation velocity experiments at three different concentrations from 0.3 to 3 μM resulted in a sedimentation coefficient distribution with a concentration-independent major peak at s20,w of 6.7 S and a best-fit frictional ratio of 1.87 (±0.27) (Fig. 4, E and F). This extended conformation would retard the elution of BRD4 in SEC and its migration in native gels. Theoretically, a smooth compact spherical monomeric or dimeric protein would sediment with s20,w of 8.5 S or 13.5 S, respectively, which jointly with the experimentally observed s-value would imply frictional ratios of 1.27 or 2.0, respectively. Therefore, considering the experimentally measured frictional ratio for BRD4 of 1.87 and the anomalous migration in SEC, our observations are most consistent with the conclusion that BRD4 is a dimer in solution. Based on the lack of apparent dissociation at 0.3 μM, we would expect the KD for dimerization to be in the low nM range.
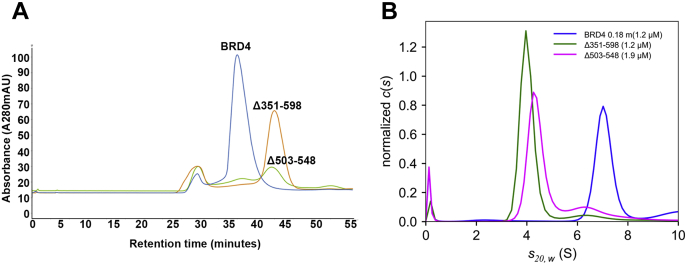
To further examine the solution structure of BRD4, we examined both the SEC elution profiles and sedimentation in AUC of two deletion mutants of BRD4, spanning the region reported to mediate dimerization of BRD2. As shown in Figure 5A, mutants deleted of segments spanning the BD2-B-BID (351–598 aa) domain or the B motif (503–548 aa) alone eluted in SEC with retention factors of 0.51 and 0.49, respectively, compared with the BRD4 retention factor of 0.28. This is a much larger increase than would be expected for BRD4 deletions of only 28 kDa and 5.4 kDa, respectively, suggesting a large shift in stokes radius. Indeed, the sedimentation in AUC of the two deletion constructs corresponded to that expected of monomers based on their molecular weights (Fig. 5B). Taken together, these studies lead to the conclusion that BRD4 in solution exists as an extended dimer. Importantly, these studies further demonstrate that dimerization is anchored through interactions mediated by the B motif of the molecule.
Figure 5.
Deletion of the BRD4 B motif disrupts dimerization.A, size exclusion profile on Superose 6 increase column at 4 °C of full-length mouse BRD4 and BRD4 deletion mutants spanning the B motif alone (Δ503–548) or the BD2-BID-B domain (Δ351–598). B, normalized sedimentation coefficient c(s) distribution plots of full-length mouse BRD4 and BRD4 deletion mutants spanning the B motif alone (Δ503–548) or the BD2-BID-B domain (Δ351–598), at the indicated concentrations.
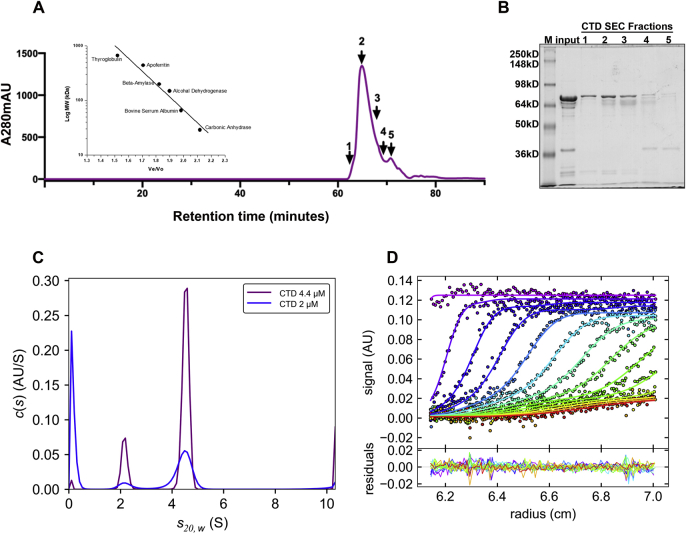
Having established the biophysical properties of BRD4 alone, we next determined the stoichiometry and affinity of interaction between BRD4 and purified CTD1–52 in solution by AUC. We first determined the behavior of the CTD alone in all three physical analyses. In SDS-PAGE, the 45.9 kDa CTD migrated with an apparent molecular weight of 65 kDa. It eluted in the SEC with a retention factor of 0.98, equivalent to a molecular weight of 76.6 kDa (Fig. 6, A and B). Similarly, in AUC we measured a sedimentation coefficient sw,20 of 4.52 S and a frictional coefficient of ≈1.3, indicating a moderately compact conformation (Fig. S6, C and D). Taken together, these data suggest that the CTD is a partially extended monomer.
Figure 6.
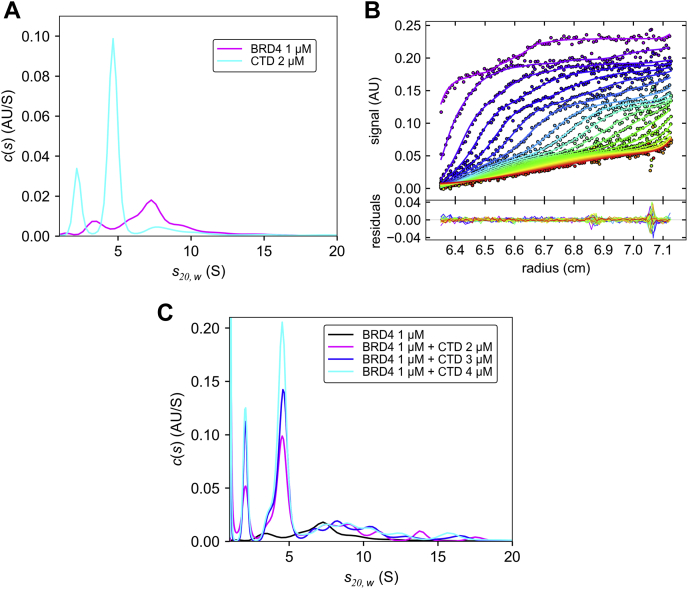
BRD4 forms a complex with CTD in a concentration-dependent manner.A, the normalized c(s) distribution plots of BRD4 alone (magenta) and CTD alone (cyan). The slower sedimenting peak of CTD represents a minor degradation component. B, absorbance scans at 280 nm of a mixture of 1 μM BRD4 and 2 μM CTD sedimenting at 50,000 rpm. Every third data point is shown, with solid lines representing the c(s) model with the associated sedimentation coefficient distribution in panel C. C, comparison of normalized c(s) distribution plots of BRD4 alone (black), BRD4 with CTD in 1:2 M ratio (magenta), 1:3 M ratio (blue), and 1:4 M ratio (cyan).
The sedimentation coefficient distribution plots for CTD alone and BRD4 alone indicate that their sedimentations are well separated and thus well suited for studying their interaction using AUC (Fig. 6A). Sedimentation velocity data of BRD4 and CTD together at an initial ratio of 1:2 are shown in Figure 6B. Four boundaries may be visually discerned, including the two slow boundaries from CTD (at signals below approximately 0.13 OD), a faster boundary comprising BRD4 species and mixed oligomers between 6 and 20 S (with a midpoint at approximately 0.15 OD), and a very fast boundary from very large particles (visible in the early, purple scans only). The latter is similar to large species in BRD4 alone, but in the mixture comprises a larger signal fraction. The sedimentation coefficient distribution (Fig. 6C) clearly shows a range of BRD4/CTD complexes in the mixture. Their precise oligomeric state cannot be determined due to their polydispersity and the characteristic oscillations in the calculated distributions caused by limited signal/noise ratio. Nonetheless, a reliable quantitative measure can be extracted in the form of the weighted-average sedimentation coefficient of BRD4-containing species over the range from 6 S to 20 S. This results in sw of 8.5 S for BRD4 alone increasing to 9.8 S in the 1:2 M ratio mixture. While it is difficult to visually discern single reliable features, the measured sw of BRD4-containing species (in the range of 6 S to 20 S) increases to 10.0 S and 10.4 S as the molar ratios increase to 1:3 and 1:4, respectively (Fig. 6C). This pattern of sedimentation is consistent with a reversible binding reaction driven by concentration and following mass action law. The extended sedimentation coefficient range of BRD4/CTD mixtures indicates the presence of a series of higher mixed oligomers.
Thus, these results clearly document a direct interaction between BRD4 and CTD, albeit of low affinity and driven by concentration. They also raise the possibility that the higher BRD4/CTD ratios lead to condensate formation, consistent with earlier reports showing condensate formation for both BRD4 and CTD individually (11, 18).
BRD4 kinase activity regulates phosphorylation of the Pol II CTD in vivo
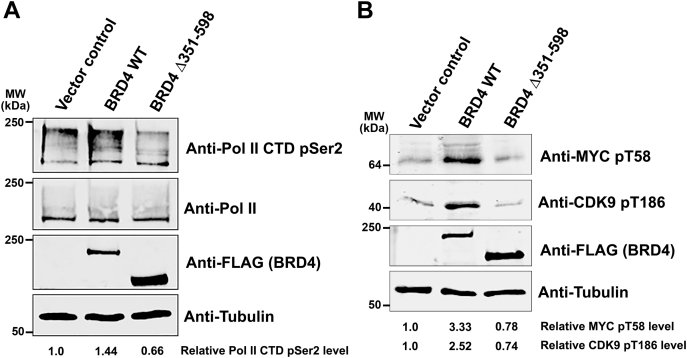
The above studies all focused on in vitro aspects of BRD4 biophysical properties and kinase activity, leading to the question of the functional consequences in vivo of the BRD4 kinase. We predicted that cellular overexpression of BRD4 wild type, but not of the kinase deficient mutant, would result in increased phosphorylation of Ser2 of the Pol II CTD, MYC, and CDK9, above the endogenous levels. To test this prediction, HCT-116 cells were transfected with either wild-type BRD4, the kinase-deficient mutant Δ351 to 598, or control vector, and the effect on in vivo levels of phosphorylation of endogenous Pol II CTD Ser2, MYC, and CDK9 was determined. As predicted, overexpression of WT BRD4 led to increased levels of phosphorylation of all three endogenous substrates (Fig. 7). In sharp contrast, introduction of the Δ351 to 598 BRD4 mutant into cells did not increase endogenous levels of phosphorylation of the BRD4 substrates, confirming the requirement for the 351 to 598 aa region for in vivo BRD4 kinase activity. Therefore, the kinase activity of BRD4 is required to mediate its phosphorylation of those substrates in vivo.
Figure 7.
Deletion of BRD4 kinase domain (351–598) leads to a loss of Pol II CTD phosphorylation in vivo. HCT116 cells were transfected with 3 μg of BRD4 WT, Δ351 to 598, or vector plasmids. Cells were harvested after 48 h, and level of phosphorylation measured by immunoblotting. A, immunoblot with antibodies for specific for P-Ser2-CTD, Pol II, FLAG and tubulin. The level of Ser2-CTD phosphorylation was calculated relative to control after normalizing for tubulin. The experiment shown is representative of two independent experiments. B, immunoblot with antibodies specific for pT58 MYC and pT186 CDK9, FLAG, and tubulin. The level of phosphorylation was calculated relative to control after normalizing for tubulin. The experiment shown is representative of two independent experiments.
Discussion
BRD4 contributes to the regulation of transcription, both as a scaffold that recruits various transcriptional cofactors including the transcription elongation factor PTEFb and as an active kinase that mediates transcriptional pause release and elongation. At the promoter, BRD4 phosphorylates the Pol II CTD on Ser2, a modification that is associated with pause release and transcriptional elongation. Although other CTD Ser2 kinases participate in regulating transcriptional elongation in mature cells, BRD4 is the only CTD Ser2 kinase in stem cells and embryonic cells (4). BRD4 kinase also contributes to transcriptional elongation through its phosphorylation of the transcription factor TAF7 (5). Importantly, BRD4 not only drives Myc gene transcription, it also phosphorylates MYC protein leading to its degradation (6). Thus, understanding the mechanisms by which BRD4 accomplishes these functions is critical. In this study, BRD4 kinase activity maps to a region spanning the BD2-B-BID segment of the molecule (Fig. 8). Further, we have demonstrated that substrate-binding sites occur across the length of the BRD4 molecule, including the ET domain. We have also established that BRD4 in solution exists in an extended dimer that depends on the presence of the B motif and that it directly interacts with its Pol II CTD substrate.
Figure 8.
BRD4 kinase domain and its flanking regions are required for the phosphorylation of its substrates. Schematic summarizing the locations of the BRD4 kinase domain and substrate-binding sites. The BRD4 kinase domain location (BD2-B-BID) is indicated by the green bracket. The predicted CTD-binding location (Motif A- 5′ PDID) is indicated by the purple bracket. The TAF7-binding region (5′PDID-BID) is indicated by the gray bracket, and the MLV IN, NSD3-binding domain (ET) is indicated by orange bracket.
Although there have been studies on the in vitro conformation of the isolated bromodomains of BRD4, our studies are the first to document that full-length BRD4 is an extended dimer in solution with high frictional ratios that retard migration, as assessed by AUC, gel filtration, and gel electrophoresis. These findings are consistent with previous studies, which reported dimerization of BRD4 in vivo, both by BiFc technology and coimmunoprecipitation experiments with differentially labeled molecules (19). Our finding that dimerization of full-length BRD4 is dependent on the B motif is also consistent with an earlier report that the B motif of BRD2, a related BET protein, contributes to dimerization (9). Of note, deletion of the B motif does not affect kinase activity in vitro but abrogates it in vivo (6), suggesting dimerization contributes to BRD4’s in vivo function. The finding that BRD4 is in an extended conformation, although the N-terminus of BRD4 is highly organized containing two bromodomains, suggests a disordered C-terminus with the flexibility and capture radius capable of mediating BRD4’s diverse interactions and functions.
Among BRD4’s diverse functions is its ability to phosphorylate a variety of substrates. In the present study, we have demonstrated and delineated the direct interaction with one of its substrates, the Pol II CTD. The extent of association of BRD4 with CTD is concentration-dependent, a phenomenon often observed for phase separating proteins (18). As detected by both AUC and the kinetics experiments, the affinity of the interaction in solution is weak. This may reflect the fact that these assay conditions do not promote cocondensation, as observed in transcription preinitiation complexes in vivo. Our mapping studies of BRD4 kinase activity are consistent with the CTD binding to a segment of BRD4 between amino acids 275 and 358. Interestingly, this region encompasses the A motif, which has been highly conserved during evolution, although nothing is known about its function. We speculate that the A motif contributes to BRD4’s regulation of transcription through an interaction with the Pol II CTD, leading to its phosphorylation.
In contrast to the CTD, TAF7 phosphorylation by BRD4 does not depend on the A motif. Thus, a fragment spanning the segment of BRD4 between aa 358 and 730 efficiently phosphorylates TAF7 but not the CTD. Although TAF7’s precise binding site on BRD4 has not been mapped, it is unlikely to bind to the ET domain, where MLV IN and NSD3 bind, since deletion of the ET domain does not affect TAF7 phosphorylation. The more efficient phosphorylation of the TAF7 fragment, relative to the CTD, may reflect its different binding site and higher affinity of interaction. Future experiments will address this issue.
The combination of deletion constructs of BRD4 and isolated fragments together map the kinase activity to a discrete region of BRD4 between 358 and 598 aa, which must contain the active site and ATP-binding site, as follows: The 1 to 600 aa segment is fully active. A BRD4 construct deleted of aa 351 to 598 has no kinase activity, whereas a fragment extending from aa 358 to 646 is capable of autophosphorylation. The finding that the kinase activity maps to a large segment of the molecule is consistent with the fact that BRD4 is an atypical kinase. Interestingly, the kinase domain includes bromodomain 2 (BD2), which is a binding site for acetylated histone lysines. That the kinase domain overlaps BD2 predicts that the binding of BRD4 to chromatin would repress its kinase activity. Indeed, as we have reported, chromatin-associated BRD4 is unable to phosphorylate MYC protein (6). Furthermore, deletion of the kinase domain leads to a loss of both Pol II CTD and MYC phosphorylation in vivo, documenting its functional importance.
In summary, the present studies have localized the kinase domain of BRD4 within its BD2-B-BID domain. The biophysical characterization of BRD4 suggests that the flexibility of BRD4 structure enables it to interact with and phosphorylate multiple substrates, which bind to different motifs within the molecule. These studies of the biophysical and biochemical features of BRD4 will inform future approaches to understanding its function in vivo and to the development of novel inhibitors.
Experimental procedures
Constructs
Flag-tagged BRD4 deletion constructs ΔN, tΔC, Δ351/598, ΔET, ΔET/SEED, ΔSEED, ΔB, and 1 to 600 were made utilizing oligos flanking desired locations on WT BRD4 in insect vector pVL1393. Constructs were sequenced and then packaged into SF9 insect cells to make viral stocks (Mirus flashBAC ULTRA system). The flag-tagged BRD4 protein fragments 275/730, 358/730, 358/646 were subcloned into pET 11d vector from PCR reactions on WT BRD4. The pCDNA3.1 WT BRD4 plasmid was constructed as described previously (6); the pCDNA3.1 Δ351/598 construct was made using flanking oligos at desired locations on WT BRD4. All constructs were entirely sequenced. GST-CTD1–52, GST-CTD27–52, GST-TAF7, FLAG-TAF7, FLAG-TAF7 3′ fragment, TAF7 with no tag were as previously described (20).
Protein purification
FLAG BRD4 was purified using cell extract prepared from Sf9 cells and with the anti-DYKDDDDK G1 Affinity Resin (Genscript) (4). Flag peptide was eliminated on a microcon column (Millipore), and proteins were recovered in 20 mM HEPES, pH 7.9, 150 mM KCl, and 20% vol/vol glycerol followed by preparative AKTA FPLC (GE Healthcare) using Superose 6 Increase, 10/300 GL column to remove the traces of contaminants. The quality and purity of FLAG-BRD4 were assessed on an SDS-PAGE and confirmed by mass spectrometric analysis (MS/MS; NCI Laboratory of Proteomics). FLAG ΔB (Δ503–548) and FLAG Δ351 to 598 were purified similarly as above. GST-CTD was purified as described previously (21). Briefly, the E. coli cells (Rosetta-Gami B(DE3) pLysS, EMD Millipore) expressing GST-CTD1–52 protein were induced using 0.5 mM isopropyl_β-D-1-thiogalactopyranoside (IPTG) overnight at 25 °C and purified using GST Sepharose beads. The GST tag was removed by thrombin cleavage for the AUC experiments in Figures 2 and S2. The proteins were concentrated on a microcon column (Millipore) and recovered in 20 mM HEPES, pH 7.9, 150 mM KCl, and 10% vol/vol glycerol. GST-CTD27–52, full-length GST-TAF7, and GST-TAF7204–349 were purified similarly except that the GST tags were not removed for kinase assays.
Flag-tagged BRD4 fragments (275/730, 358/730, 358/646) were grown in E. coli (Rosetta-Gami B(DE3) pLysS, EMD Millipore) as described above but purified with the anti-DYKDDDDK G1 Affinity Resin. The purity of these protein fragments was verified as described above. Flag-tagged TAF7 and TAF73′ were grown and purified similarly. 6X-His tagged MLVIN peptide (residues 289–408) was expressed and purified as previously described with a final buffer consisting of 500 mM NaCl, 7.5 mM CHAPS, 50 mM HEPES, pH 7.4, 2 mM DTT (22, 23). GST-tagged NSD3 peptide (residues 100–263) was expressed and purified with a final buffer consisting of 40 mM Tris pH 7, 300 mM NaCl, 5 mM DTT.
Size-exclusion chromatography
The oligomerization and molecular weight determinations of the recombinant BRD4, B (Δ503–548) and FLAG Δ351 to 598 and CTD were carried out by SEC experiments on a Superose 6 Increase, 10/300 GL column (manufacture's exclusion limit 1000 kD) on AKTA FPLC (GE Healthcare). The column was pre-equilibrated and run with 20 mM HEPES pH 7.8, 150 mM KCl, 5% glycerol at 4 °C with a flow rate of 0.3 ml/min, with detection at 280 nm and 230 nm. The column was calibrated with standard molecular weight markers. Retention factors were calculated according to Hong et al. (24).
Native-PAGE analysis
In total, 100 ng of SEC purified BRD4 was loaded onto a 3 to 12% Bis-Tris Native-PAGE (Thermo Fisher Scientific) and the blue native-PAGE was run following manufacturer’s protocol followed by silver staining (Pierce Silver Stain Kit) to visualize the bands. NativeMark Unstained Protein Standard marker (Thermo Fisher Scientific) was loaded in parallel lanes to compare the sizes.
Sedimentation velocity analytical ultracentrifugation experiments
Sedimentation velocity experiments were performed with an Optima XL-I Analytical Ultracentrifuge (Beckman Coulter) with an AnTi-50 eight-hole rotor. Cells with double sector centerpieces were used to analyze BRD4 alone at a concentration ranging from 0.3 μM to 3 μM in 20 mM HEPES pH 7.8, 150 mM KCl and 5% glycerol. In total, 360 μl samples were loaded into the sample channels of double-channel 12 mm centerpieces, and 360 μl of buffer in the reference channels. After temperature equilibration at nominal 20 °C, the rotor was accelerated to 50,000 rpm, and radial absorbance scans were collected using absorbance optics at 280 and 230 nm in continuous scan mode, in 2 min intervals with a redial step size of 0.003 cm. Absorbance scans were analyzed with the sedimentation coefficient distribution c(s) in SEDFIT (25). Similarly, ΔB (Δ503–548) and Δ351 to 598 were analyzed with the indicated concentrations. For analysis of the BRD4/CTD mixtures, due to low signal/noise ratios of the acquired data, the meniscus was constrained to the graphically determined value, and the frictional coefficient was fixed to the best-fit value of 1.87 from BRD4 alone. Maximum entropy regularization with p = 0.9 was applied. Scaling factors for transformation of experimental s-values to the reference values in water at 20 °C were calculated with the software SEDNTERP 3.0.3, kindly provided by Dr John Philo, after AUC calibration (26).
Kinase assays
In vitro kinase assays were performed as described previously (4). Briefly, 20 μl reactions were carried out in the kinase assay buffer (50 mM Tris, pH 8.0, 5 mM DTT, 5 mM MnCl2, and 5 mM MgCl2, 5% glycerol with 10 μCi γ32P ATP (6000 Ci/mM)). The kinase reactions were incubated for 1 h at 30 °C. The products were resolved by SDS PAGE in Tris/SDS/Glycine Buffer, or Bis/Tris PAGE in MES buffer; the extent of phosphorylation was quantitated by a GE Typhoon. For kinetic measurements, the quantitated values were determined using Prism software [GraphPad] to calculate Vmax and Km values.
Transfection assays
HCT116 cells were grown in DMEM media supplemented with 10% fetal bovine serum at 37 °C, 7.5% CO2. Transient transfections of the wild-type BRD4-FLAG and Δ351 to 598 BRD4-FLAG mutant constructs or empty vector only (3 μg each) were done in HCT116 cells using the Lipofectamine 3000 reagent (Invitrogen). Transfected cells were harvested 18 h posttransfection and whole cell extracts (WCEs) made with tissue extraction reagent I (Invitrogen).
Western blotting
WCEs were analyzed through immunoblotting. For immunoblotting, WCEs were resolved on SDS–polyacrylamide gels and transferred onto nitrocellulose for immunoblots. The primary antibodies used were anti-Pol II phosphoSer2: mAb 3E10 (Millipore), anti-RNA Pol II: mAb 8WG16 (Santa Cruz), anti-FLAG: DYKDDDDK-tag antibody (Genscript), anti-β-Tubulin (Abcam), anti-CDK9 phospho-T186 (Cell Signaling), and anti-MYC phospho-T58 (Abcam). All immunoblot analyses were performed using the Odyssey infrared scanner and secondary antibodies from Li-Cor.
Data availability
All data are contained within the manuscript and the supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors gratefully acknowledge Drs Ananda Roy, Paul Roche, and Sheetal Uppal for their critical reading of the manuscript. We also acknowledge the CCR genomics core for routine sequencing of plasmid constructs. We also thank Petria Thompson and Sabina Hlavaty for preliminary experiments and the members of the Singer lab for helpful discussions throughout these studies.
Author contributions
A. K. S., J. D. W., B. N. D., and D. S. S. conceptualization; A. K. S., J. D. W., and D. S. S. formal analysis; A. K. S., J. D. W., B. N. D., and P. S. investigation; R. C. L. resources; A. K. S., J. D. W., and D. S. S. writing–original draft.
Funding and additional information
R. C. L. was supported by KL2 TR002734. J. D. W., A. K. S., B. N. D., and D. S. S. were supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Craig Cameron
Supporting information
Figure S1.
BRD4 kinase activity is affected by buffer conditions. In vitro kinase assays of 10 nM BRD4 under different buffer conditions show differential optima for auto and trans kinase activities using 75 nM TAF7 3′ fragment as a substrate.
Figure S2.
Kinase activity is contained within BRD4 subregions spanning BD2-B-BID domains. A, schematic map of WT, deletion mutants, and fragments of BRD4. B, in vitro kinase assay using radiolabeled ATP showing autophosphorylation of WT, tΔC, and ΔN recombinant BRD4 proteins, all at 50 nM. Kinase activity relative to WT is shown. C, in vitro kinase assays of 200 nM BRD4 275 to 730 aa fragment using radiolabeled ATP and 100 nM GST-CTD27-52 and 100 nM GST-TAF7 as substrates.
Figure S3.
BRD4 kinase activity maps with the segment 351 to 598 aa, but does not require the B motif. A, schematic map of WT, deletion mutants, and fragments of BRD4. B, in vitro kinase assay using radiolabeled ATP showing kinase activities of WT, Δ351 to 598, ΔET (601–683), ΔET/SEED (600–730), ΔSEED (684–730), and 1 to 600 recombinant BRD4 fragment, all at 50 nM, using 60 nM TAF7 as substrate. Kinase activity relative to WT is shown. C, in vitro kinase assay using radiolabeled ATP showing kinase activities of 25 nM WT and ΔB (503–548), using 100 nM TAF7 as substrate. Kinase activity of ΔB is shown relative to WT.
Figure S4.
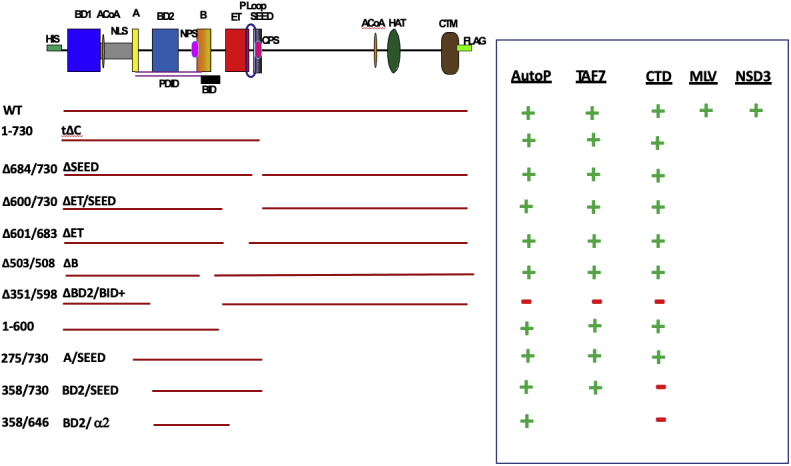
BRD4 kinase activity is contained within the BD2-B-BID domains. The summary of the auto and trans kinase activities for the recombinant BRD4 proteins is shown. +, indicates phosphorylation, −, indicates no phosphorylation.
Figure S5.
BRD4 Elution Profile is Unaffected by Treatment with Nonionic Detergents or Hexanediol. Top: Size exclusion profile of mouse BRD4 on SuperoseTM 6 increase column at 4 °C in standard buffer conditions. Middle: The SEC profile of mouse BRD4 on SuperoseTM 6 increase column at 4 °C in standard buffer conditions with 1 mM PMSF, 2 mM TCEP, and 0.1% DDM. Bottom: The SEC profile of mouse BRD4 on SuperoseTM 6 increase column at 4 °C in standard buffer conditions with 1 mM PMSF, 2 mM TCEP, and 5% hexanediol.
Figure S6.
Pol II CTD is a moderately compact monomer. A, size exclusion profile of CTD1-52 on SuperoseTM 6 increase column at 4 °C. The inset shows the standard calibration curve run on same condition. Fraction 2 was used for further AUC experiments. B, 10% SDS-PAGE of the input and SEC fractions of CTD. Full-length CTD migrates at ∼65 kD. A minor degradation product of ∼22 kD was also observed at the bottom of the gel. C, the normalized c(s) distribution plots using two different concentrations of CTD1-52 are shown 4.4 μM (purple) and 2 μM (blue). Data were acquired using A280 absorbance. Additionally, a slower sedimenting population at 2 S is observed (Figs. S2, C and D), which reflects a smaller molecular weight protein species resolved in SDS-PAGE from SEC fractions, as shown in Fig. S2B. Absorbance scans of 4.4 μM CTD sedimenting at 50,000 rpm as observed by absorbance at 280 nm (symbols, for clarity showing only every third data point of every second scan) and c(s) sedimentation coefficient distribution model (lines). The color temperature from purple to red indicates temporal evolution.
References
- 1.Devaiah B.N., Gegonne A., Singer D.S. Bromodomain 4: A cellular Swiss army knife. J. Leukoc. Biol. 2016;100:679–686. doi: 10.1189/jlb.2RI0616-250R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dey A., Ellenberg J., Farina A., Coleman A.E., Maruyama T., Sciortino S., Lippincott-Schwartz J., Ozato K. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol. 2000;20:6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uppal S., Gegonne A., Chen Q., Thompson P.S., Cheng D., Mu J., Meerzaman D., Misra H.S., Singer D.S. The bromodomain protein 4 contributes to the regulation of alternative splicing. Cell Rep. 2019;29:2450–2460.e5. doi: 10.1016/j.celrep.2019.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devaiah B.N., Lewis B.A., Cherman N., Hewitt M.C., Albrecht B.K., Robey P.G., Ozato K., Sims R.J., Singer D.S. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc. Natl. Acad. Sci. U. S. A. 2012;109:6927–6932. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devaiah B.N., Singer D.S. Cross-talk among RNA polymerase II kinases modulates C-terminal domain phosphorylation. J. Biol. Chem. 2012;287:38755–38766. doi: 10.1074/jbc.M112.412015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devaiah B.N., Mu J., Akman B., Uppal S., Weissman J.D., Cheng D., Baranello L., Nie Z., Levens D., Singer D.S. MYC protein stability is negatively regulated by BRD4. Proc. Natl. Acad. Sci. U. S. A. 2020;117:13457–13467. doi: 10.1073/pnas.1919507117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filippakopoulos P., Picaud S., Mangos M., Keates T., Lambert J.-P., Barsyte-Lovejoy D., Felletar I., Volkmer R., Müller S., Pawson T. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin Y.J., Umehara T., Inoue M., Saito K., Kigawa T., Jang M.K., Ozato K., Yokoyama S., Padmanabhan B., Güntert P. Solution structure of the extraterminal domain of the bromodomain-containing protein BRD4. Protein Sci. 2008;17:2174–2179. doi: 10.1110/ps.037580.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Gutierrez P., Mundi M., Garcia-Dominguez M. Association of bromodomain BET proteins with chromatin requires dimerization through the conserved motif B. J. Cell Sci. 2012;125:3671–3680. doi: 10.1242/jcs.105841. [DOI] [PubMed] [Google Scholar]
- 10.Wu S.-Y., Lee A.-Y., Lai H.-T., Zhang H., Chiang C.-M. Phospho switch triggers Brd4 chromatin binding and activator recruitment for gene-specific targeting. Mol. Cell. 2013;49:843–857. doi: 10.1016/j.molcel.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabari B.R., Dall’Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., Abraham B.J., Hannett N.M., Zamudio A.V., Manteiga J.C., Li C.H., Guo Y.E., Day D.S., Schuijers J., Vasile E., et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361 doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devaiah B.N., Case-Borden C., Gegonne A., Hsu C.H., Chen Q., Meerzaman D., Dey A., Ozato K., Singer D.S. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat. Struct. Mol. Biol. 2016;23:540. doi: 10.1038/nsmb.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eick D., Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 2013;113:8456–8490. doi: 10.1021/cr400071f. [DOI] [PubMed] [Google Scholar]
- 14.Nemec C.M., Singh A.K., Ali A., Tseng S.C., Syal K., Ringelberg K.J., Ho Y.-H., Hintermair C., Ahmad M.F., Kar R.K. Noncanonical CTD kinases regulate RNA polymerase II in a gene-class-specific manner. Nat. Chem. Biol. 2019;15:123–131. doi: 10.1038/s41589-018-0194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baranello L., Wojtowicz D., Cui K., Devaiah B.N., Chung H.-J., Chan-Salis K.Y., Guha R., Wilson K., Zhang X., Zhang H. RNA polymerase II regulates topoisomerase 1 activity to favor efficient transcription. Cell. 2016;165:357–371. doi: 10.1016/j.cell.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crowe B.L., Larue R.C., Yuan C., Hess S., Kvaratskhelia M., Foster M.P. Structure of the Brd4 ET domain bound to a C-terminal motif from γ-retroviral integrases reveals a conserved mechanism of interaction. Proc. Natl. Acad. Sci. U. S. A. 2016;113:2086–2091. doi: 10.1073/pnas.1516813113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q., Zeng L., Shen C., Ju Y., Konuma T., Zhao C., Vakoc C.R., Zhou M.-M. Structural mechanism of transcriptional regulator NSD3 recognition by the ET domain of BRD4. Structure. 2016;24:1201–1208. doi: 10.1016/j.str.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boehning M., Dugast-Darzacq C., Rankovic M., Hansen A.S., Yu T., Marie-Nelly H., McSwiggen D.T., Kokic G., Dailey G.M., Cramer P. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 2018;25:833. doi: 10.1038/s41594-018-0112-y. [DOI] [PubMed] [Google Scholar]
- 19.Wang R., Li Q., Helfer C.M., Jiao J., You J. Bromodomain protein Brd4 associated with acetylated chromatin is important for maintenance of higher-order chromatin structure. J. Biol. Chem. 2012;287:10738–10752. doi: 10.1074/jbc.M111.323493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gegonne A., Weissman J.D., Zhou M., Brady J.N., Singer D.S. TAF7: A possible transcription initiation check-point regulator. Proc. Natl. Acad. Sci. U. S. A. 2006;103:602–607. doi: 10.1073/pnas.0510031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh A.K., Rastogi S., Shukla H., Asalam M., Rath S.K., Akhtar M.S. Cdc15 phosphorylates the C-terminal domain of RNA polymerase II for transcription during mitosis. J. Biol. Chem. 2017;292:5507–5518. doi: 10.1074/jbc.M116.761056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larue R.C., Plumb M.R., Crowe B.L., Shkriabai N., Sharma A., DiFiore J., Malani N., Aiyer S.S., Roth M.J., Bushman F.D. Bimodal high-affinity association of Brd4 with murine leukemia virus integrase and mononucleosomes. Nucleic Acids Res. 2014;42:4868–4881. doi: 10.1093/nar/gku135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma A., Larue R.C., Plumb M.R., Malani N., Male F., Slaughter A., Kessl J.J., Shkriabai N., Coward E., Aiyer S.S. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc. Natl. Acad. Sci. U. S. A. 2013;110:12036–12041. doi: 10.1073/pnas.1307157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong P., Koza S., Bouvier E.S. A review size-exclusion chromatography for the analysis of protein biotherapeutics and their aggregates. J. Liq. Chromatogr. Relat. Technol. 2012;35:2923–2950. doi: 10.1080/10826076.2012.743724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophysical J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao H., Nguyen A., To S.C., Schuck P. Calibrating analytical ultracentrifuges. Eur. Biophys. J. 2021;50:353–362. doi: 10.1007/s00249-020-01485-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the manuscript and the supporting information.