Abstract
Objective(s):
Duchene muscular dystrophy (DMD) is a progressive neuromuscular disease caused by mutations in the DMD gene, resulting in the absence of dystrophin expression leading to membrane fragility and myofibril necrosis in the muscle cells. Because of progressive weakness in the skeletal and cardiac muscles, premature death is inevitable. There is no curative treatment available for DMD. In recent years, advances in genetic engineering tools have made it possible to manipulate gene sequences and accurately modify disease-causing mutations. CRISPR/Cas9 technology is a promising tool for gene editing because of its ability to induce double-strand breaks in the DNA.
Materials and Methods:
In this study for the exon-skipping approach, we designed a new pair of guide RNAs (gRNA) to induce large deletion of exons 48 to 53 in the DMD gene in the human skeletal muscle cell line (HSkMC), in order to correct the frame of the gene.
Results:
Data showed successful editing of DMD gene by deletion of exons 48 to 53 and correction of the reading frame in edited cells. Despite a large deletion in the edited DMD gene, the data of real-time PCR, immune florescent staining demonstrated successful expression of truncated dystrophin in edited cells.
Conclusion:
This study demonstrated that the removal of exons 48-53 by the CRISPR / Cas9 system did not alter the expression of the DMD gene due to the preservation of the reading frame of the gene.
Key Words: CRISPR/Cas9, DMD, Dystrophin, Gene editing, HSkMC
Introduction
Duchene muscular dystrophy (DMD) is one of the most common lethal neuromuscular disorders that approximately affects 1 in 3500 to 5000 male births. The inheritance pattern of DMD is X-linked recessive (1, 2). DMD is mainly caused by a frame-shift mutation in the 2.2 megabase pair DMD gene, which prevents expression of the dystrophin protein (3). Dystrophin is a member of the dystrophin-glycoprotein complex (DGC) and has an essential role in the stability of sarcolemma by connecting the actin cytoskeleton to the extracellular matrix (4). Disruption of DGC in the lack of dystrophin causes damage to muscle fibers resulting in progressive muscle degeneration leading to heart and respiratory failure and premature death (5, 6). Although all 79 exons of the DMD gene can bear mutation, exons 45 to 55 are mutational hotspot regions, which contain more than 60% of deleterious mutations (7). Different types of mutations have been described in DMD, the most common of these are large deletions and duplications, and small point mutations like frameshift, missense, and nonsense mutations consist of the rest of the mutations (8-10).
In contrast to DMD, Becker muscular dystrophy (BMD) is caused by in-frame mutations in dystrophin encoding gene- DMD - resulting in the expression of functional truncated dystrophin; therefore, BMD patients show the moderate form of disease with late-onset muscular dystrophy (11). At present, there is no effective treatment for DMD and common approaches are typically palliative and supportive (12). Recently, some proposed treatments for DMD have focused on modulating the disease and inducing symptoms similar to BMD. Exon skipping strategy by antisense oligonucleotide and delivery of truncated dystrophin such as mini- and micro-dystrophin are some of these approaches (13-16).
On the other hand, emerging gene-editing technologies such as ZFN (Zinc finger nuclease), TALEN (transcription activator-like effectors), and CRISPR (clustered regularly interspaced short palindromic repeats) present a promising approach toward editing and treating genetic disorders like DMD (17-20). CRISPR-Cas9 is a bacterial adaptive immune system that has been optimized as a powerful gene-editing technology (21, 22). This tool consists of a short guide RNA (sgRNA) which guides Cas9 nuclease to a specific genome sequence near a protospacer adjacent motif (PAM) and induces double-strand breaks (DSB) in DNA (23-25). Consequently, DSBs trigger endogenous DNA repair pathways: NHEJ (non-homologous end joining) or HDR (homology-directed repair) (26-28). We aimed to utilize CRISPR/Cas9 to efficiently edit the dystrophin gene and compare the expression of the edited dystrophin vs no edited dystrophin in the human skeletal muscle cell line. In this study we unlike others deleted 6 exons of dystrophin gene exons from 48 to exon 53, this part of the DMD gene is the most prevalent site for deletion. Using these sgRNAs enables us to edit a broad range of deletions. The cell line that we used in this study was the human skeletal muscle cell line that is myoblast cell and suitable for dystrophin gene expression and dystrophin gene editing. To the best of our knowledge, this is the first study using this cell line for dystrophin gene editing.
Materials and Methods
gRNA design and cloning
According to the DMD gene sequence (NG_012232.1 RefSeqGene, NCBI), gRNAs were designed by https://zlab.bio/guide-design-resources and validated using a Cas-OFFinder (www.rgenome.net/cas-offinder/)(Figure 1). We chose a pair of gRNAs, intDel 47 and intDel 53 to conduct further experiments in the human skeletal muscle cell line (HSkMC). The gRNAs were cloned into pSpCas9 (BB)-2A-GFP (PX458) (Cat No.48138.addgene) using the BbsI enzyme (Thermo Scientific). Selected gRNA sequences were presented in Table 1. The plasmid constructs were transformed into the DH5a Escherichia coli and extracted using the Miniprep kit (Qiagen. USA).
Figure 1.
Schematic construct map and primer site according to our design for edition of dystrophin
Table 1.
Selected gRNA for dystrophin gene edition
| Sequence | Guides |
|---|---|
| F : 5’-CACCGAACTGCAAAGGAAGCGCGTA- 3’ R: 3’-CTTGACGTTTCCTTCGCGCATCAAA- 5’ |
Int del 47 |
| F: 5’ –CACCGCCGCACATGGTGGTGCGGAC- 3’ R: 3’-CGGCGTGTACCACCACGCCTGCAAA -5’ |
Int del 53 |
Cell culture
Human embryonic kidney-293 cells (ATCC: CRL-1573) were cultured in F12 DMEM (Dulbecco’s Modified Eagle’s Medium) media that was supplemented with 10% FBS (fetal bovine serum) (Gibco) and 1% pen-strep (penicillin-streptomycin) (Thermo Fisher Scientific). After 2 subcultures, the cells were transferred to the 6 well plates and seeded with Opti-mem media (Reduced Serum Media Thermo Fisher Scientific); after 24 hr, the cloned vector was transfected to the cells (with 60% confluency) by lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. After 8 hr of transfection, Opti-mem media were discarded and complete DMEM media containing 10% FBS and 1% pen-strep were added to the cells. After 48 to 72 hr, the cells were observed by fluorescence microscopy.
The human skeletal muscle cell line (HSkMC) (NCBI code: C598) was purchased from the Pasteur Institute of Iran. HSCkM cells were cultured in DMEM high glucose supplemented with 10% FBS, 1% penicillin/streptomycin, and 5 µg/ml basic fibroblast growth factor (bFGF). Cells were incubated at 37 °C, 5% CO2. One day before transfection, the cells were seeded into the 6-well plates and cultured with F12 media, supplemented by 5% FBS. Due to the low transfection efficiency of lipofectamine in the human skeletal muscle cells, we used another transfection agent, TRANSFECTimin (Dara Zistfan Eram, Iran ). HSkMC were transfected with 10 μg of the plasmid constructs by transfection agent TRANSFECTimin. To compare the expression of edited (truncated) dystrophin with the wild type, we transfected the HSkMC cells by 3 different strategies, that were named according to the following code: eHSkMC (edited HSkMC transfected by intDel47 and intDel53), bHSkMC (HSkMC transfected by empty backbone PX458) and wHSkMC (nontransfected or wild type).
Fluorescence-activated cell sorting and single-cell isolation
Two days after transfection, the cells were trypsinized and collected for Fluorescence-Activated Cell Sorting and single-cell isolation using FACS AryaIII instrument (BD Bioscience). GFP-positive single cells were collected and expanded for the following molecular analysis.
Genomic DNA extraction and PCR-base assay
To detect the induced deletion in edited cells, we used primers for the flanking region of the deleted area, “check F” and “check R” (Table 2). Genomic DNA was extracted (DNA extraction Kit QIAGEN, USA) and PCR was performed by the following program: 95 °C for 2 min, 30 cycles (95 °C for 30 sec, 58 °C for 30 sec, and 72 °C for 40 sec), 72 °C for 5 min; followed by holding at 4 °C.
Table 2.
Primer set for detection of edition in dystrophin edited cells by CRISPR/Cas9
| Product size | Sequence | Name |
|---|---|---|
| 500bp | F:5-GACTCACAAACTATAGCTCACA-3’ R :5’-TCAAGGTAGAGAATAGAGG-3’ |
Check primer |
mRNA analysis and real-time PCR
The total RNA was extracted from HSkMC cells using the RNeasy Plus Mini Kit (QIAGEN, USA) according to the manufacturer’s instructions. Reverse transcription of 1 µg total RNA was carried out by cDNA synthesis (Takara, Japan). cDNA was synthesized in the following conditions according to the instructions: 15 min at 37 °C and 5 sec at 85 °C. Two sets of primers were designed; one pair contained forward primer 15-F (complementary to exon 15) and reverse primer 16-R (reverse complementary to exon 16) as control of dystrophin expression in both edited and non-edited cells, and the other pairs consisted of 47-F (complementary to exon 47) and 48-R (reverse complementary to exon 48) (Table 3). Amplification was done in an Applied Biosystem 7500 Real-Time PCR system.
Table 3.
Real-time PCR primer for confirmation of correct edition
| Size | Sequence | Name |
|---|---|---|
| 141bp | F: 5’- AGATTCACACAACTGGCTT-3’ R: 5’- TTCTGGGTCACTGACTTATTC-3’ |
Primer 15-16 |
| 131bp | F: 5’-AGGACCCGTGCTTGTAAGTG-3’ R: 5’-AAGCTGCCCAAGGTCTTTTA-3’ |
Primer 47-48 |
Immune florescent staining
To analyze dystrophin expression in the edited cells, the cells were coated on glass slides and fixed with 4% PFA (paraformaldehyde)(Sigma) for 5 min at room temperature. After permeabilization using 0.2% Triton X-100, the samples were washed twice with cold TPBS and incubated with a blocking solution containing 1% BSA in PBS for 1 hr at room temperature. The cells were stained using a primary antibody (ab15277; Abcam) overnight at 4 °C and then washed by cold TPBS and incubated with a secondary antibody (IgG-FITC-SantaCruz) for 1 hr at room temperature. Nucleic acids were labeled with DAPI.
Results
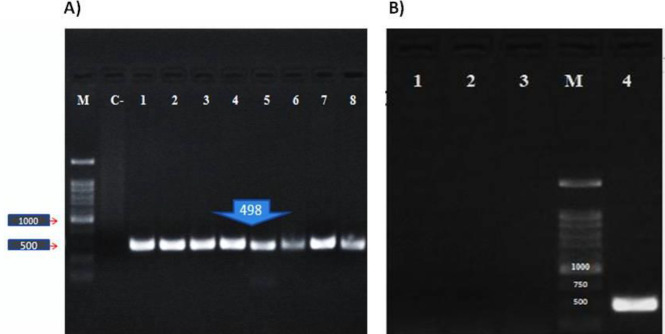
According to our study design, we chose exons 48-53 located in the hotspot mutational region of the DMD gene. The edited cells showed deletion in exons 48 to 53 in the DMD gene and resulted in the attachment of exon 47 to exon 54 which produced a shortened in-frame dystrophin gene. Transfection efficiency was 70%. 72 hr after transfection, the total genomic DNA was isolated. We directly analyzed genomic DNA in the transfected and nontransfected cells. To do this, specific PCR was performed by a forward primer located in intron 47 (check F) and reverse primer (check R) located in intron 53. These primers resulted in amplification of a 498bp fragment in the edited cells; no amplification was seen in control cells (Figure 2). Direct sequencing of the 498bp amplicon indicated the deletion of exon 48 to 53. By performing agarose gel electrophoresis, a 500bp fragment was seen in eHSkMC, while we did not have any product in bHSkMC and wHSkMC (Figure 3). This result indicated correct deletion of exons 48 to 53 in eHSkMC in comparison with bHSkMC and wHSkMC. To study dystrophin expression in mRNA level, total RNA was extracted from eHSkMC, bHSkMC, and wHSkMC. After quantitative PCR, amplification was detected in edited (eHSkMC) and control (wHSkMC) cells by 15-16 F&R primers. According to the design of 47-48 primers (48-R reverse complementary to exon 48), if edition occurs in the cells, the reverse primer cannot anneal to the exon 48; therefore, amplification was not seen. As expected, amplification was detected only in wHSkMC, which indicated deletion of exon 48 to 53 in eHSkMC. After agarose gel electrophoresis, the amplicon was extracted and cloned in PMinT plasmid (data not shown) and sent for sequencing. mRNA analysis results demonstrated dystrophin expression in the edited and non-edited cells (Figure 3). Base on bioinformatics analysis (www.edystrophin.genouest.org), deletion of these exons results in the production of a truncated and partially functional protein with a size of 390 kDa which is 37 kDa shorter than wild dystrophin. Deletion of exons 48-55 was modeled using I-Tasser (Iterative Threading Assembly Refinement) from protein modeling databank. (Table 4, Figure 4). Immune fluorescent staining of dystrophin (Figure 5) in eHSkMC in comparison with wHSkMC showed that truncated dystrophin could be expressed in the edited cell.
Figure 2.
Detection of the edited cells by PCR. A) Amplification of 498bp in edited cells. M (DNA Marker 1kb, Cinnagen), C-(negative control or non-edited HEK-293 cells), 1-8 (edited HEK-293 cells). B) Negative control (1), w- HSkMC or no edited cells (2), b-HSkMC, cells infected with PX458 empty or backbone (3), Amplification of 498bp fragment in e-HSkMC or edited cells (4), and DNA marker 1kb (Cinnagen. Iran) (M)
Figure 3.
Dystrophin expression in the edited cells (e-HSkMC) and non-edited cells (w-HSkMC). GAPDH was used as an internal control gene. A) Plot demonstrates the total RNA expression in edited (e-HSkMC) and non-edited cells (w-HSkMC) checked by primer designed for exons 15 and 16. B) Plot demonstrated deletion of exons 48-53 in the edited cell, so amplification was not seen by primer for exons 47 and 48 in edited cells (P-value >0.05)
Table 4.
Comparison of length and size of wild dystrophin with edited dystrophin
| Full length dystrophin | Deletedexon 48-56* dystrophin |
|
|---|---|---|
| Protein size | 3685 aa | 3365 aa |
| Protein weigh | 427kDa | 390 kDa |
Figure 4.
According to analysis of the DMD gene in www.edystrophin.genouest.org, deletion of exons 48-53 resulted in removal of a part of R18 to R 22 that is located in the rod domain. Rod domain of dystrophin is composed of 24 repeats (R1 to R24) homologue to spectrin repeats and 4 hinges. Deletion of exons 48-55 modeling from repeat 17 to repeat 23 from I-Tasser ( score: -0.94). Blue: R17, Violet: R18, Cyan: R22, Green: R23
Figure 5.
Immune fluorescent staining using dystrophin Ab and DAPI as counter-stain in edited cells (e-HSkMC) and non-edited cells (w-HSkMC). A) e-HSkMC. B) w-HSkMC. Both eHSkMC and wHSkMC showed a positive result in immune fluorescent staining using dystrophin Ab. These results demonstrate that dystrophin could express in the edited cells
Discussion
The dystrophin gene, which mainly expresses in the skeletal muscle cells, is a large gene in the human genome that spans about 2.2 Mbp, located on the X chromosome in the Xp21 region (29). Rod shaped dystrophin protein has four domains: N-terminal or actin-binding domain, central rod domain, cysteine-rich domain, and C-terminal domain. Two domains, N-terminal and C-terminal, have a crucial role in dystrophin function in connecting actin proteins to the cytoskeleton, thus stabilizing the sarcolemma (30-32). Mutation in the DMD gene causes progressive degeneration of the skeletal and cardiac muscles that leads to premature death due to cardiopulmonary complications (33-35). There is no definite treatment for DMD and current approaches including administration of corticosteroids and physical therapy are mostly supportive and alleviate the symptoms of the patients, but do not target the underlying cause of diseases (36, 37). Although the large size of the DMD gene and abundant affected tissues are two main obstacles in DMD gene therapy, gene therapy of DMD was more considered in the past decades (38). Two main approaches for gene therapy in DMD are delivery of engineered mini dystrophin (lacking non-essential domains) and up-regulation of utrophin that is one of the dystrophin isoforms (39, 40). Another strategy is the application of Antisense Oligo Nucleotides (AONs) which induce exon skipping that restores the reading frame of dystrophin transcript (41-43). AONs have shown promising results, but due to the transient nature of this treatment, patients should treat constantly during their lifetime (33). The use of RNA-DNA chimeric oligonucleotide (RDOs) or oligodeoxynucleotides (ODNs) is another strategy in DMD treatment (34, 35). Advancements in molecular technologies have paved the way for developing definite treatments for genetic diseases. Meganucleases, ZFNs, and TALENs were used for DMD gene editing in vivo and in vitro. They have achieved promising results but complicated the manufacturing procedures, and lower sequence target ability is the challenge yet to be addressed (44, 45). Recently, RNA-guided endonucleases, such as CRISPR/Cas9, which have several unique features in comparison with other gene-editing tools, have been considered more applicable for correcting genetic disorders (39, 40, 44).
Compared to other methods, the CRISPR cas9 construction technique is more straightforward, and gRNA could mediate cleavage in any region of the genome adjacent to a PAM sequence, providing targeted gene editing (46,47). Deletions, point mutations, and duplications occur frequently in the DMD gene. Considering the efficiency rate of CRISPR cas9, using a pair of gRNAs for editing of mutated exons is more preferred (48-50).
In one study Tabebordbar et al., deleted exon 23 in mouse muscle and stem cells of mdx mice by CRISPR/Cas9. Their results indicated restoration of dystrophin in the edited cells (35). In another study Young et al., targeted exons 45 to 55 in DMD patient derived hiPSCs by CRISPR/Cas9. In this study, a 750 kb fragment was deleted in the DMD gene in order to produce an in-frame mutation by which dystrophin expression was restored in the edited cells (37). Min et al. deleted exon 44 in cardiomyocytes differentiated from hiPSCs cells by CRISPR/Cas9 and restored dystrophin expression in the edited cells (36). In the present study, we targeted exons 48 to 53 that are located in the hotspot mutational region of the DMD gene, using the CRISPR/Cas9 system, and analyzed the expression of the truncated dystrophin in comparison with the wild type dystrophin in the human skeletal muscle cells. Our designed gRNAs could successfully delete exons 48-53 and acted as universal gRNA to target approximately 60% of mutations in the DMD gene. Moreover, we directly used human skeletal muscles cells rather than mouse muscle cells or hi-PSCs. Functional validation of truncated dystrophin would require in vivo studies. Further in vivo studies on the efficacy of the CRISPR cas9 gene-editing tool are needed in order to use this method as a promising treatment for DMD in the near future.
Conclusion
Our result indicates that successful deletion of exon 48 to 53, leads to expression of truncated dystrophin in the edited cells. A pair of designed gRNAs in this study was selected using molecular assays to be used as universal gRNAs for editing any type of mutation that could occur in this particular hotspot region. Finally, this study demonstrated that the removal of exons 48-53 by the CRISPR / Cas9 system did not alter the expression of the Dmd gene due to the preservation of the reading frame of the gene.
Acknowledgment
The results presented in this paper were part of the PhD thesis of Ms. Mahintaj Dara and supported by a grant (No. 95-01-74-13795) from Shiraz University of Medical Science, Shiraz, Iran. The authors would like to thank Ms. Mehboodi for the western blot assay and the RCC center of Shiraz University of Medical Sciences for editing this manuscript.
Authors’ Contributions
MD, MD: Study conception and design; MD: Data analyzing and draft manuscript preparation; MD, MD: Critical revision of the paper; MD: Supervision of the research; MD, VR, MM, MR, MN and MD: Final approval of the version to be published.
Conflicts of Interest
The authors have no conflicts of interest to disclose.
References
- 1.Falzarano M, Scotton C, Passarelli C, Ferlini A. Duchenne muscular dystrophy: From diagnosis to therapy. J Molecules. 2015;20:18168–18184. doi: 10.3390/molecules201018168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryder S, Leadley R, Armstrong N, Westwood M, De Kock S, Butt T, et al. The burden, epidemiology, costs and treatment for Duchenne muscular dystrophy: An evidence review. Orphanet J Rare Dis. 2017;12:1–21. doi: 10.1186/s13023-017-0631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aartsma-Rus A, Ginjaar IB, Bushby K. The importance of genetic diagnosis for Duchenne muscular dystrophy. World J Med Genet. 2016;53:145–151. doi: 10.1136/jmedgenet-2015-103387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walter MC, Reilich P. Recent developments in Duchenne muscular dystrophy: facts and numbers. J Cachexia Sarcopenia Muscle. 2017;8:681–685. doi: 10.1002/jcsm.12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garry DJ. Dystrophin-deficient cardiomyopathy. JACC CardioOncol. 2016;67:2533–2546. doi: 10.1016/j.jacc.2016.02.081. [DOI] [PubMed] [Google Scholar]
- 6.Birnkrant DJ, Bushby K, Bann CM, Alman BA, Apkon SD, Blackwell A, et al. Diagnosis and management of Duchenne muscular dystrophy, part 2: respiratory, cardiac, bone health, and orthopaedic management. Lancet Neurol. 2018;17:347–361. doi: 10.1016/S1474-4422(18)30025-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelson CE, Gersbach CA. Genome editing for duchenne muscular dystrophy. Mus Gen Thera: Springer. 2019;2: 383–403. [Google Scholar]
- 8.Suthar R, Sankhyan N. Duchenne muscular dystrophy: A practice update. Indian J Pediatr. 2018;85:276–281. doi: 10.1007/s12098-017-2397-y. [DOI] [PubMed] [Google Scholar]
- 9.White S, Kalf M, Liu Q, Villerius M, Engelsma D, Kriek M, et al. Comprehensive detection of genomic duplications and deletions in the DMD gene, by use of multiplex amplifiable probe hybridization. Am J Hum Genet. 2003;71:365–371. doi: 10.1086/341942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hegde MR, Chin EL, Mulle JG, Okou DT, Warren ST, Zwick ME. Microarray-based mutation detection in the dystrophin gene. Hum Mutat. 2008;29:1091–1099. doi: 10.1002/humu.20831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrews JG, Wahl RA. Duchenne and Becker muscular dystrophy in adolescents: current perspectives. Adolesc Health Med Ther. 2018;9:53–63. doi: 10.2147/AHMT.S125739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crone M, Mah JK. Current and emerging therapies for Duchenne muscular dystrophy. Curr Treat Options Neurol. 2018;20:1–17. doi: 10.1007/s11940-018-0513-6. [DOI] [PubMed] [Google Scholar]
- 13.Shimizu-Motohashi Y, Komaki H, Motohashi N, Takeda Si, Yokota T, Aoki Y. Restoring dystrophin expression in Duchenne muscular dystrophy: current status of therapeutic approaches. J Pers Med. 2019;9:1–14. doi: 10.3390/jpm9010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salmaninejad A, Valilou SF, Bayat H, Ebadi N, Daraei A, Yousefi M, et al. Duchenne muscular dystrophy: an updated review of common available therapies. Int J Neurosci. 2018;128:854–864. doi: 10.1080/00207454.2018.1430694. [DOI] [PubMed] [Google Scholar]
- 15.Guiraud S, Chen H, Burns DT, Davies KE. Advances in genetic therapeutic strategies for Duchenne muscular dystrophy. Ex Physiol. 2015;100:1458–1467. doi: 10.1113/EP085308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarmin S, Kymalainen H, Popplewell L, Dickson G. New developments in the use of gene therapy to treat Duchenne muscular dystrophy. Expert Opin Biol Ther. 2014;14:209–230. doi: 10.1517/14712598.2014.866087. [DOI] [PubMed] [Google Scholar]
- 17.Mendell JR, Rodino-Klapac LR. Duchenne muscular dystrophy: CRISPR/Cas9 treatment. Cell res. 2016;26:513–514. doi: 10.1038/cr.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aartsma-Rus A, Fokkema I, Verschuuren J, Ginjaar I, Van Deutekom J, van Ommen GJ, et al. Theoretic applicability of antisense-mediated exon skipping for duchenne muscular dystrophy mutations. Hum Muta. 2009;30:293–299. doi: 10.1002/humu.20918. [DOI] [PubMed] [Google Scholar]
- 19.Xu L, Park KH, Zhao L, Xu J, El Refaey M, Gao Y, et al. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol Thera. 2016;24:564–569. doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gee P, Xu H, Hotta A. Cellular reprogramming, genome editing, and alternative CRISPR Cas9 technologies for precise gene therapy of Duchenne muscular dystrophy. Stem Cells Int. 2017;9:1–12. doi: 10.1155/2017/8765154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrangou R. Cas9 targeting and the CRISPR revolution. Sci. 2014;344 :707–708. doi: 10.1126/science.1252964. [DOI] [PubMed] [Google Scholar]
- 24.Jiang F, Doudna JA. CRISPR–Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 25.Thurtle-Schmidt DM, Lo TW. Molecular biology at the cutting edge: A review on CRISPR/Cas9 gene editing for undergraduates. Biochem Mol Biol Educ. 2018;46:195–205. doi: 10.1002/bmb.21108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farboud B, Severson AF, Meyer BJ. Strategies for efficient genome editing using CRISPR-Cas9. J Genet. 2019;211:431–457. doi: 10.1534/genetics.118.301775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindahl T, Barnes D. Repair of endogenous DNA damage. Cold Spring Harbor Laboratory Press; 2000. [DOI] [PubMed] [Google Scholar]
- 28.Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9:1–13. doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frontera WR, Ochala J. Skeletal muscle: A brief review of structure and function. Calcif Tissue Int. 2015;96:183–195. doi: 10.1007/s00223-014-9915-y. [DOI] [PubMed] [Google Scholar]
- 30.Hotta A. Genome editing gene therapy for duchenne muscular dystrophy. J Neuromuscul Dis. 2015;2:343–345. doi: 10.3233/JND-150116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rando TA. Non-viral gene therapy for Duchenne muscular dystrophy: Progress and challenges. Biochim Biophys Acta Mol Basis Dis. 2007;1772:263–271. doi: 10.1016/j.bbadis.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 32.Bertoni C, Morris GE, Rando TA. Strand bias in oligonucleotide-mediated dystrophin gene editing. Hum Mol Genet. 2004;14:221–233. doi: 10.1093/hmg/ddi020. [DOI] [PubMed] [Google Scholar]
- 33.Ousterout DG, Kabadi AM, Thakore PI, Perez-Pinera P, Brown MT, Majoros WH, et al. Correction of dystrophin expression in cells from Duchenne muscular dystrophy patients through genomic excision of exon 51 by zinc finger nucleases. Mol Thera. 2015;23:523–532. doi: 10.1038/mt.2014.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ousterout DG, Perez-Pinera P, Thakore PI, Kabadi AM, Brown MT, Qin X, et al. Reading frame correction by targeted genome editing restores dystrophin expression in cells from Duchenne muscular dystrophy patients. Mol Thera. 2013;21:1718–1726. doi: 10.1038/mt.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tabebordbar M, Zhu K, Cheng JK, Chew WL, Widrick JJ, Yan WX, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Sci. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Min Y-L, Li H, Rodriguez-Caycedo C, Mireault AA, Huang J, Shelton JM, et al. CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci adv. 2019;5-3:324–336. doi: 10.1126/sciadv.aav4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young CS, Hicks MR, Ermolova NV, Nakano H, Jan M, Younesi S, et al. A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. J Stem Cell. 2016;18:533–540. doi: 10.1016/j.stem.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fortunato F, Rossi R, Falzarano MS, Ferlini A. Innovative Therapeutic Approaches for Duchenne Muscular Dystrophy. J Clin Med. 2021;10:820–841. doi: 10.3390/jcm10040820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Babačić H, Mehta A, Merkel O, Schoser B. CRISPR-cas gene-editing as plausible treatment of neuromuscular and nucleotide-repeat-expansion diseases: A systematic review. PloS One. 2019;14:e0212198. doi: 10.1371/journal.pone.0212198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chemello F, Bassel-Duby R, Olson EN. Correction of muscular dystrophies by CRISPR gene editing. J Clin Invest. 2020;130:2766–2776. doi: 10.1172/JCI136873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dara M, Razban V, Talebzadeh M, Moradi S, Dianatpour M. Using CRISPR/Cas9 system to knock out exon 48 in DMD gene. Avicenna J Med Biotechnol. 2021;13:54–57. doi: 10.18502/ajmb.v13i2.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salmaninejad A, Jafari Abarghan Y, Bozorg Qomi S, Bayat H, Yousefi M, Azhdari S, et al. Common therapeutic advances for Duchenne muscular dystrophy (DMD) Int J Neurosci . 2020:1–20. doi: 10.1080/00207454.2020.1740218. [DOI] [PubMed] [Google Scholar]
- 43.Dokholyan NV. Experimentally-driven protein structure modeling. J Proteomics. 2020;220:103777–103800. doi: 10.1016/j.jprot.2020.103777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hillary VE, Ceasar SA, Ignacimuthu S. Genome engineering in insects: focus on the CRISPR/Cas9 system. In: Genome engineering via CRISPR-Cas9 system: Academic Press. 2020;1:219–249. [Google Scholar]
- 45.Knott GJ, Doudna JA. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361:866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koeks Z, Bladen CL, Salgado D, Van Zwet E, Pogoryelova O, McMacken G, et al. Clinical outcomes in duchenne muscular dystrophy: a study of 5345 patients from the treat-nmd dmd global database. J Neuromuscul Dis. 2017;4:293–306. doi: 10.3233/JND-170280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manghwar H, Li B, Ding X, Hussain A, Lindsey K, Zhang X, et al. CRISPR/Cas systems in genome editing: methodologies and tools for sgRNA design, off-target evaluation, and strategies to mitigate off-target effects. Adv Sci. 2020;7:1902312–19002328. doi: 10.1002/advs.201902312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reinig AM, Mirzaei S, Berlau DJ. Advances in the treatment of duchenne muscular dystrophy: New and emerging pharmacotherapies. Pharmacotherapy: J Huma Pharmacol Drug Ther. 2017;37:492–499. doi: 10.1002/phar.1909. [DOI] [PubMed] [Google Scholar]
- 49.Mata López S, Balog-Alvarez C, Vitha S, Bettis AK, Canessa EH, Kornegay JN, et al. Challenges associated with homologous directed repair using CRISPR-Cas9 and TALEN to edit the DMD genetic mutation in canine Duchenne muscular dystrophy. PloS One. 2020;15:e0228072. doi: 10.1371/journal.pone.0228072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyamoto M, Tochinai R, Sekizawa S-i, Shiga T, Uchida K, Tsuru Y, et al. Cardiac lesions in duchenne muscular dystrophy model rats with out-of-frame Dmd gene mutation mediated by CRISPR/Cas9 system. J Toxicol Pathol. 2020;33:227–236. doi: 10.1293/tox.2020-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]