Abstract
Cryptosporidium spp. are protozoan parasites that belong to subphylum apicomplexa and cause diarrhea in humans and animals worldwide. Data on the prevalence of Cryptosporidium spp. and its subtypes among calves in the Republic of Korea (KOR) are sparse. Hence, our study aimed to investigate the prevalence and association between the age of calf and the identified Cryptosporidium spp. and to determine the genotypes/subtypes of Cryptosporidium spp. in pre-weaned calves with diarrhea in the KOR. A total of 460 diarrheic fecal samples were collected from calves aged 1−60 days and screened for Cryptosporidium spp. by the 18S rRNA gene. Species identification was determined using the sequencing analysis of the 18S rRNA gene, and C. parvum-positive samples were subtyped via the sequence analysis of the 60-kDa glycoprotein (gp60) gene. Sequence analysis based on the 18S rRNA gene revealed the presence of three Cryptosporidium spp., namely, C. parvum (n = 72), C. ryanae (n = 12), and C. bovis (n = 2). Co-infection by these species was not observed. The infection rate was the highest in calves aged 11−20 days (26.1%, 95% CI 17.1−35.1), whereas the lowest rate was observed in calves aged 21−30 days (7.7%, 95% CI 0.0−16.1). The prevalence of C. parvum was detected exclusively in calves aged ≤20 days, and the highest infection rate of C. ryanae was seen in calves ≥31 days of age. The occurrence of C. parvum (χ2 = 25.300, P = 0.000) and C. ryanae (χ2 = 18.020, P = 0.001) was significantly associated with the age of the calves. Eleven different subtypes of the IIa family that belonging to C. parvum were recognized via the sequence analyses of the gp60 gene. Except for two (IIaA18G3R1 and IIaA15G2R1) subtypes, nine subtypes were first identified in calves with diarrhea in the KOR. IIaA18G3R1 was the most frequently detected subtype (72.2% of calves), followed by IIaA17G3R1 (5.6%), IIaA15G2R1 (4.2%), IIaA19G4R1 (4.2%), IIaA16G4R1 (2.8%), IIaA17G4R1 (2.8%), IIaA19G3R (2.8%), IIaA14G1R1 (1.4%), IIaA14G3R1 (1.4%), IIaA15G1R1 (1.4%), and IIaA19G1R1 (1.4%) These results suggest that the prevalence of Cryptosporidium spp. is significantly associated with calf age. Furthermore, the findings demonstrate the high genetic diversity of C. parvum and the widespread occurrence of zoonotic C. parvum in pre-weaned calves. Hence, calves are a potential source of zoonotic transmission with considerable public health implications.
Introduction
Cryptosporidium spp. are protozoan parasites that cause mild-to-severe diarrhea in humans and a wide range of animals [1]. Infections with these parasites occur via the fecal−oral route either by direct contact with infected animals or by the ingestion of infective oocysts from contaminated water or food [2–5]. To date, 40 Cryptosporidium spp. have been described [6], and among them, four species, namely, C. andersoni, C. bovis, C. parvum, and C. ryanae, have been identified in cattle. The distribution of these species is known to vary according to age [4, 7]. In particular, C. parvum is one of the most important pathogens causing diarrhea in neonatal calves worldwide and leads to severe economic losses owing to poor growth, decreased productivity, and even death [8]. Moreover, C. parvum is the major pathogenic species that affects humans [9, 10]. Unlike C. parvum, C. bovis, and C. ryanae usually infect post-weaned calves and yearlings without causing illness, and C. andersoni is mainly found in adult cattle [11–13]. The pathogenicity of C. bovis, and C. ryanae in post-weaned calves has not been established [9]. The oocysts of C. parvum, C. bovis, and C. ryanae are similar in size and shape. While C. ryanae is smaller than the others and requires molecular methods for its determination [14, 15], C. andersoni is larger in size and infects the abomasum [16].
According to the subtyping of C. parvum based on sequence analysis of the 60-kDa glycoprotein (gp60) gene, Ⅱa and Ⅱd subtypes have been detected in both humans and calves and can cause zoonotic cryptosporidiosis [17]. The Ⅱa subtype is mostly identified in calves, and IIaA15G2R1 is the predominant subtype [7] globally, including the Republic of Korea (KOR) [18]. The IId subtype is usually found in lambs and goat kids [4, 19] and has been described in calves in some countries such as Sweden, Turkey, Egypt, and China [20–23]. To date, most investigations of cryptosporidiosis in calves caused by C. parvum have focused on the IIa subtype in most countries. However, there are a few studies on C. parvum subtypes in calves in the KOR [18, 24].
Cryptosporidium parvum infects the intestinal mucosa and accounts for over 90% of Cryptosporidium infections in neonatal calves [23]. In contrast, in pre-weaned calves, the prevalence of C. bovis and C. ryanae and their effects on causing diarrhea remain unclear. Several studies have reported that C. bovis and C. ryanae are present in pre-weaned calves [23, 25, 26] and that C. ryanae infections are particularly associated with moderate diarrhea in pre-weaned calves [23]. However, little is known about the association between C. bovis and diarrhea. In addition, a previous study has indicated the high prevalence of C. bovis and C. ryanae in hemorrhagic diarrhea in the KOR [24]. Nevertheless, the pathogenicity of these organisms is still unclear.
So far, for the identification of Cryptosporidium spp., a nested polymerase chain reaction (PCR) technique based on the SSU rRNA gene has been the most widely used method [27]. However, in the present study, a conventional PCR method using species-specific primers was used [24]. Although the amplification had a short fragment compared with a previous method, this PCR technique enabled the differentiation between C. bovis and C. ryanae. Therefore, this study aimed to investigate the prevalence of Cryptosporidium spp. using species-specific primers in pre-weaned calves with diarrhea and to evaluate the association between the age of calf and the identified Cryptosporidium spp. Furthermore, we intended to determine the genotype of Cryptosporidium spp. and subtyping of C. parvum in calves in the KOR and to assess the significance of calves as a source of human infections.
Materials and methods
Ethics statement
All animal procedures were conducted according to ethical guidelines for the use of animal samples, and were approved by the Jeonbuk National University (Institutional Animal Care and Use Committee Decision No. CBNU 2020–052). All procedures and possible consequences were explained to the managers of the surveyed farm, and written consent was obtained.
Sample collection
Between August 2019 and August 2020, fresh fecal samples were collected directly from the rectum of 460 diarrheic pre-weaned calves (up to 60 days of age) by an experienced veterinarian using sterile plastic gloves in 11 different farms located in the KOR. The samples were placed in labeled sterile plastic tubes and transported to the Animal Immunology Laboratory of Kyungpook National University in a cooler with ice packs. Upon arrival, sampling date, age, animal identification number, and fecal consistency (pasty, loose, watery, or hemorrhagic) were recorded for each animal. The collected feces were mostly pasty or loose. Prior to DNA extraction, all feces were stored at 4°C for no more than 2 days without the additional treatment of preservation. The fecal samples were divided according to age as follows; 1−10 days (n = 271), 11−20 days (n = 92), 21−30 days (n = 39), and ≥31 days (n = 58). No microscopic examination was performed for the detection of oocysts.
DNA extraction, molecular analysis, and sequencing
DNA was extracted from 200 mg of each fecal sample using the QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. In brief, samples were suspended in lysis buffer, followed by boiling at 70°C for 5 min. Next, the inhibitors provided in the kit were added to the solution to remove substances that can degrade DNA and inhibit downstream enzymatic reactions. Supernatants were subsequently transferred into a tube containing proteinase K and then heated at 70°C for 10 min. A final volume of 200 μL of each DNA sample was then stored at −20°C until PCR amplification. The identification of Cryptosporidium spp. was first tested using the 18S rRNA gene [28]. Samples that yielded positive results for Cryptosporidium spp. via the sequence analysis were further screened to detect the four species using species-specific primers [24]. Positive samples for C. parvum were retested using the 60-kDa glycoprotein (gp60) gene to determine its subtype [4], whereas positive samples for C. bovis/C. ryanae were differentiated by sequence analysis. The subtypes of gp60 were named based on the repeated number of TCA (A), TCG (G), and ACATCA (R), as described previously [29]. All positive PCR products were purified using the AccuPower PCR Purification Kit (Bioneer, Daejeon, KOR) and employed for direct sequencing (Macrogen, Daejeon, KOR). The nucleotide sequences obtained in this study were analyzed using BioEdit (version 7.2.5) and compared with the reference sequences using the Basic Local Alignment Search Tool available at the National Center for Biotechnology Information database. As the sequences of C. bovis and C. ryanae are highly similar, all amplified samples were differentiated by comparing the sequences between the two species. To determine the subtype of C. parvum as well as the genotypes of C. bovis and C. ryanae, nucleotide sequences were aligned using ClustalX and then analyzed via direct comparison with reference sequences from GenBank. In this study, only samples showing a good sequencing result were considered positive for each Cryptosporidium spp. All nucleotide sequences generated in this study were deposited in the GenBank database with appropriate accession numbers (18S rRNA: MZ736386−MZ736399; gp60: MZ736314−MZ736385).
Statistical analysis
Statistical analysis was performed using SPSS Statistics 26 software package for Windows (SPSS Inc, Chicago, IL, USA). Chi-square test was used to determine the association between the prevalence of each species and age. Moreover, multinomial logistic regression analysis was used to determine any associations between the subtypes of C. parvum and age. A p-value of less than 0.05 was considered statistically significant.
Results
Prevalence of Cryptosporidium spp.
Among the 460 diarrheic fecal samples examined, 86 (18.7%) were positive for Cryptosporidium spp. on PCR analysis and sequencing based on the 18S rRNA gene. Three Cryptosporidium spp. were identified in pre-weaned Korean native calves (Table 1). No C. andersoni was detected in this study. Of these, C. parvum (15.7%, 72/460) was the most detected, followed by C. ryanae (2.6%, 12/460) and C. bovis (0.4%, 2/460). Co-infection of these species was not observed. The prevalence of the three Cryptosporidium spp. was compared according to the age groups. As shown in Table 1, the infection rate of Cryptosporidium spp. was highest in calves aged 11−20 days (26.1%, 95% CI 17.1−35.1), whereas the lowest infection rate was observed in calves aged 21−30 days (7.7%, 95% CI 0.0−16.1). All three Cryptosporidium spp. were detected only in calves aged 1−10 days (Table 1). The association between Cryptosporidium spp. and age-distribution was investigated. Interestingly, the identified Cryptosporidium spp. varied according to the age of the calves. C. parvum infection was detected exclusively in calves ≤20 days of age (Table 2). The prevalence peaked at the age of 11−20 days and decreased rapidly thereafter (Table 2). C. parvum infection was significantly associated with the age of the calves (χ2 = 25.300, P = 0.000). Unlike C. parvum, C. ryanae was found in all age groups, and the highest infection rate was observed at ≥31 days of age (Table 2). C. ryanae infection also had a significant age-related distribution (χ2 = 18.020, P = 0.001). In contrast, C. bovis was detected only in two calves aged 10 days and 35 days, and there was no statistical significance in the age-related distribution (P = 0.590).
Table 1. Prevalence and distribution of Cryptosporidium species according to age group in pre-weaned calves.
| Age (days) | Sample size | No. of positive (%) | 95% CI | Cryptosporidium species (No.) | ||
|---|---|---|---|---|---|---|
| C. parvum | C. ryanae | C. bovis | ||||
| 1−10 | 271 | 53 (19.6%) | 14.8–24.3 | 49 | 3 | 1 |
| 11−20 | 92 | 24 (26.1%) | 17.1–35.1 | 23 | 1 | 0 |
| 21−30 | 39 | 3 (7.7%) | 0.0–16.1 | 0 | 3 | 0 |
| 31−60 | 58 | 6 (10.3%) | 2.5–18.2 | 0 | 5 | 1 |
| Total | 460 | 86 (18.7%) | 15.1−22.3 | 72 | 12 | 2 |
Table 2. Distribution of Cryptosporidium species in pre-weaned Korean native calves according to age group.
| Age (days) | Frequency of C. parvum positivity (%) | χ2 (P-value) | Frequency of C. ryanae positivity (%) | χ2 (P-value) | Frequency of C. bovis positivity (%) | χ2 (P-value) |
|---|---|---|---|---|---|---|
| 1−10 | 49/271 (18.1%) | 25.300 (0.000) | 3/271 (1.1%) | 16.020 (0.001) | 1/271 (0.4%) | 2.824 (0.419) |
| 11−20 | 23/92 (25.0%) | 1/92 (1.1%) | 0 | |||
| 21−30 | 0 | 3/39 (7.7%) | 0 | |||
| 31−60 (Ref.) | 0 | 5/58 (8.6%) | 1/58 (1.7%) |
Distribution of Cryptosporidium spp. and C. parvum subtypes
All 72 C. parvum-positive samples were successfully amplified and subtyped by sequence analysis of the gp60 gene. A total of 11 different subtypes belonging to the family IIa were identified (Table 3). Subtype family IId was not detected. The distinction of each subtype within the IIa was in the number of trinucleotide region of TCA and TGA repeats (i.e., had one copy of sequence ACATCA immediately after the trinucleotide repeats). As shown in Table 3, in pre-weaned Korean native calves, the most frequently detected subtype was IIaA18G3R1 (72.2%), followed by IIaA17G3R1 (5.6%), and then IIaA15G2R1 (4.2%) and IIaA19G4R1 (4.2%). Other subtypes, namely, IIaA14G1R1 (1.4%), IIaA14G3R1 (1.4%), IIaA15G1R1 (1.4%), IIaA16G4R1 (2.8%), IIaA17G4R1 (2.8%), IIaA19G1R1 (1.4%), and IIaA19G3R1 (2.8%) were also identified. Except for the IIaA18G3R1, no statistical correlation was found between calf age and a specific subtype (Table 3). IIaA19G4R1 was observed only in calves aged 1−10 days, whereas IIaA17G3R1 was found exclusively in calves aged 11−20 days. Several more subtypes were found in calves aged 1−10 days (Table 3). The most predominant subtype, IIaA18G3R1, was seen in all ages.
Table 3. Distribution of Cryptosporidium parvum subtype according to age group.
| gp60 subtypes | Age groups (days) | No. of positive calves | P-value | |
|---|---|---|---|---|
| 1−10 | 11−20 | |||
| IIaA18G3R1 | 36 | 16 | 52 (72.2%) | 0.000 |
| IIaA17G3R1 | 1 | 3 | 4 (5.6%) | 0.753 |
| IIaA15G2R1 | 3 | 0 | 3 (4.2%) | 0.785 |
| IIaA19G4R1 | 3 | 0 | 3 (4.2%) | 0.785 |
| IIaA16G4R1 | 1 | 1 | 2 (2.8%) | 0.823 |
| IIaA17G4R1 | 1 | 1 | 2 (2.8%) | 0.823 |
| IIaA19G3R1 | 0 | 2 | 2 (2.8%) | 0.677 |
| IIaA14G1R1 | 1 | 0 | 1 (1.4%) | 0.874 |
| IIaA14G3R1 | 1 | 0 | 1 (1.4%) | 0.874 |
| IIaA15G1R1 | 1 | 0 | 1 (1.4%) | 0.874 |
| IIaA19G1R1 | 1 | 0 | 1 (1.4%) | 0.874 |
| Total | 46 | 26 | 72 | |
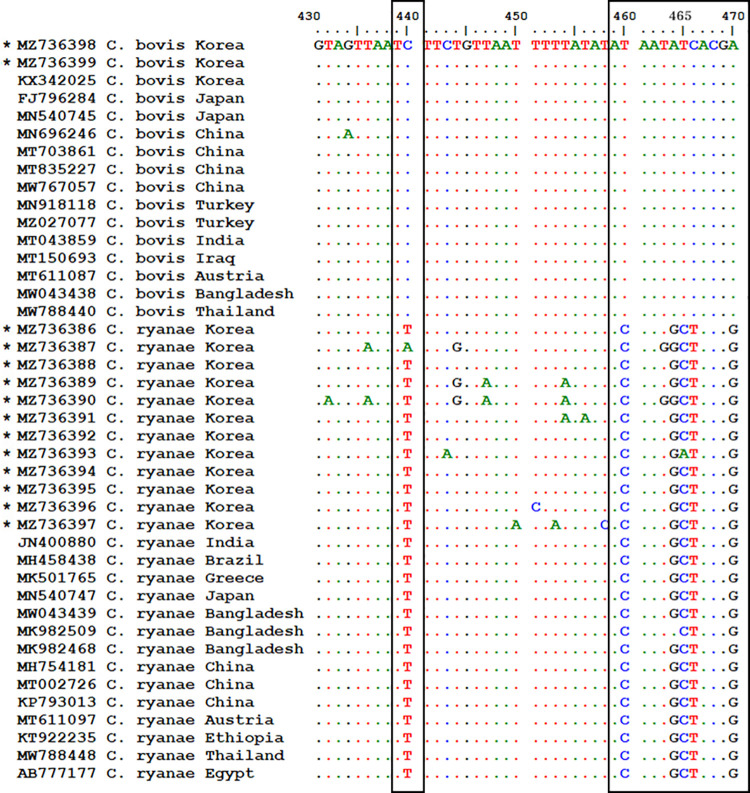
Based on the 18S rRNA gene, 14 (12 C. ryanae and 2 C. bovis) sequences were obtained and compared with the published literature. Twelve sequences of C. ryanae showed 95.1%−100% similarity with each other. The C. ryanae sequences shared 95.7%−100% identity with those found in Austria, China, India, Thailand, and Japan. Two sequences of C. bovis shared 94.1% similarity. These sequences demonstrated 95.5%−96.2% identity with those identified previously in the KOR and had 91.9%−96.2% homology with those from Austria, USA, Japan, and China. Interestingly, differences in nucleotides between C. ryanae and C. bovis were observed. As shown in Fig 1, the nucleotides in the six positions, i.e., 440, 460, 464−466, and 470, were different between the two species.
Fig 1. Sequence comparisons between C. bovis and C. ryanae for the partial18S rRNA gene from Korean sequences obtained in this study and reference strains.
Six nucleotide differences at 440, 460, 464−466, and 470 are shown. An asterisk indicates sequences obtained in this study.
Discussion
Cryptosporidium, along with rotavirus, has been well recognized as the main pathogen causing diarrhea in neonatal calves worldwide [30]. Our findings established the prevalence of Cryptosporidium spp. in pre-weaned diarrheic calves according to age, and the presence of various zoonotic subtypes of C. parvum in the KOR were identified. In the present study, the overall prevalence of Cryptosporidium spp. was found to be 18.7%, which is higher than that reported previously in the KOR [18, 24, 31]. These variations could be explained by the age of the animals, time of sample collection, and the differences in geographical location. However, the percentage of Cryptosporidium spp.-positive samples found in our study was lower than that reported in other countries such as Germany (88.9%), Japan (83.8%), China (38.4%), Italy (38.8%), Colombia (26.6%), Argentina (22.5%), and Estonia (22.6%) [25, 32–37].
In this study, the presence of three Cryptosporidium spp. in pre-weaned Korean native calves was ascertained: C. bovis, C. parvum, and C. ryanae. Of them, C. parvum was the most predominant species in the KOR. This finding agrees with the results observed in several other countries [7, 25, 33, 36, 38, 39]. Most studies have proven that C. parvum mainly infects calves up to 1 month of age [33, 40–43]. The results of the present study demonstrated that C. parvum was detected only in calves aged ≤20 days, and the infection rate was the highest in calves aged 11−20 days. This observation is consistent with a previous study performed by our group [18]. According to our findings, C. parvum was detected in calves aged ≤20 days. It is considered that calves in this age group are susceptible to C. parvum infection owing to their immature immune system [44]. In addition, it is well known that young calves can become infected with C. parvum and begin shedding the oocysts soon after birth [45–47]. This could be associated with cow-to-calf transmission. Several studies have reported that the possible source of infection in calves is transmission at birth from their mothers [48, 49]. However, at present, we do not have exact information on whether these calves were immediately removed from their mothers after birth, but the possibility of contamination via exposure to mother’s feces or the surroundings should be considered. Moreover, C. parvum is known to cause watery diarrhea [23, 30]. In this study, the number of animals with watery feces was small; hence, the association with diarrhea was not evaluated. Although we were not able to compare the occurrence of C. parvum with the diarrhea status, C. parvum was found to be the causative agent of diarrhea in young calves. Our results suggest that C. parvum infection is attributed to the significant age-related distribution (P = 0.000). Consequently, C. parvum was strongly associated with diarrhea in calves aged ≤20 days.
Cryptosporidium ryanae was the second most frequently detected species in pre-weaned Korean native calves. In general, C. ryanae is often found in post-weaned calves [15]. The results revealed that C. ryanae was detected in all age groups and that its occurrence increased with age. In particular, the infection rate of C. ryanae showed a low prevalence in calves aged <20 days, whereas it was rather high in calves aged ≥31 days (Table 2). The prevalence of C. ryanae found in this study was similar to that of a previous study performed in the KOR [24]. Our observation confirmed that C. ryanae has an age-associated distribution, similar to C. parvum. A recent study has reported that C. ryanae was common in pre-weaned as well as post-weaned calves and that the infection was associated with the occurrence of moderate diarrhea in pre-weaned calves [23]. In contrast, other studies have shown that C. ryanae was not associated with diarrhea [26, 39, 50]. So far, the pathogenicity of C. ryanae is controversial. A previous study conducted in the KOR demonstrated that although it is not a single infection, the prevalence of C. ryanae was significantly high in hemorrhagic diarrhea [24]. We could not arrive at a conclusion regarding the correlation with diarrhea since the number of C. ryanae-positive samples from diarrheic calves was small. Hence, C. ryanae infection may cause diarrhea in calves ≥21 days of age and should be considered as a causative agent of diarrhea in this age group. Further studies are necessary to clarify the pathogenicity of C. ryanae in pre-weaned calves.
We found that the prevalence of C. bovis was the lowest in pre-weaned Korean native calves. This observation is contradictory to the results reported by several studies in which C. bovis was the dominant species in pre-weaned calves [20, 47, 51–53]. In this study, C. bovis was detected only in two calves aged 10 and 35 days. Several studies have stated that C. bovis is common in 2−3-week-old calves [42, 50]. However, our result signified that C. bovis was not detected in this age (Table 1). Cai et al. mentioned that C. bovis usually appears after weaning and that the infection can last weeks or months and contribute to the small increase in Cryptosporidium infection rates soon after weaning [26]. This observation may also explain the low prevalence of C. bovis in the present study. To date, information on the prevalence and clinical signs of C. bovis infection in both pre-weaned and post-weaned calves is very limited in the KOR. C. bovis could have probably been considered to be less important than C. parvum and therefore overlooked as an etiological agent of diarrhea in calves. Moreover, the results revealed that infection by C. bovis, unlike the two other species, was not age-related. Most importantly, the involvement of C. bovis in diarrhea remains unclear. Unlike C. ryanae, many studies have suggested that C. bovis was associated with diarrhea [23, 26, 39, 54]. However, infection by C. bovis/C. ryanae may lead to clinical signs owing to the presence of C. parvum [33]. Therefore, the prevalence and pathogenicity of C. bovis in pre-weaned and post-weaned calves must be investigated through large-scale epidemiological surveys.
C. parvum IIa family is common in humans as well as calves and is considered potentially zoonotic. To date, three C. parvum subtypes have been detected in calves in the KOR [18, 24], whereas one subtype (IIaA16G3R1) was not found in this study. In addition to the two subtypes (IIaA15G2R1 and IIaA18G3R1) described above, nine other subtypes (IIaA14G1R1, IIaA14G3R1, IIaA15G1R1, IIaA16G4R1, IIaA17G3R1, IIaA17G4R1, IIaA19G1R1, IIaA19G3R1, and IIaA19G4R1) that have not previously been detected in the KOR were identified for the first time, showing the presence of high genetic diversity. Among them, IIaA18G3R1 was most commonly found in pre-weaned Korean native calves with diarrhea. This result is inconsistent with that of a previous study in which IIaA15G2R1 was shown as the predominant subtype [18]. This difference could be attributed to the fact that in the previous study, both normal and diarrheic feces were used and that IIaA15G2R1 was detected regardless of diarrhea [18]. Other variations are due to the differences in the season of sampling, regions, the number of samples, and herd management. IIaA15G2R1 has been known as the most prevalent C. parvum subtype infecting humans and cattle in many countries [7, 34, 55–59] and has also been detected in calves without diarrhea [18, 33, 60]. There seems to be no relationship between the subtype and diarrhea. In the present study, IIaA15G2R1 was detected only in three calves with diarrhea and was the third frequent subtype along with IIaA19G4R1.
Here, IIaA18G3R1 was the dominant subtype that accounted for 72.2% of C. parvum-infected pre-weaned Korean native calves and was the frequent cause of human cryptosporidiosis, besides being reported in calves and foals [61–66]. The second common subtype in the KOR, IIaA17G3R1, has been found in calves and humans in several countries [67–71]. IIaA19G4R1 was the third frequent subtype identified in the pre-weaned Korean native calves and was also detected in small ruminants and fish as well as humans and calves [61, 70, 72–74]. Interestingly, all sequences belonging to the IIaA19G4R1 subtype were identical to those reported from other countries previously. These subtypes are considered to be the most common ones in calves in the KOR.
The other seven subtypes were also identified in pre-weaned Korean native calves with diarrhea, but their prevalence was relatively low. Subtypes IIaA14G1R1, IIaA14G3R1, and IIaA15G1R1 were each detected in one calf. IIaA14G1R1 was identified in calves, goat kids, and humans [7, 12, 17, 19, 25, 34, 57, 58]. IIaA14G3R1 was found in humans, calf, lambs, and fresh molluscan shellfish [19, 25, 75, 76]. IIaA15G1R1 has been reported in humans [29, 57, 58, 77, 78] as well as in cattle and goat kids [22, 79–81]. Subtypes IIaA16G4R1 and IIaA17G4R1 were each found in two calves in the current study. Unlike the other subtypes, IIaA16G4R1 has so far been noted only in neonatal calf with diarrhea [82], which is consistent with our findings. Subtype IIaA16G4R1 has not yet been detected in humans; however, the possibility that this may represent a significant health risk cannot be excluded. The IIaA17G4R1 subtype has been identified in humans, cattle, and goats [32, 34, 65, 76, 82, 83] and has also been detected in diarrheic calves [32]. Finally, subtypes IIaA19G1R1 and IIaA19G3R1 have each been identified in one calf. IIaA19G1R1 has been reported in humans, cattle, and sheep [36, 58, 69, 84–86]. IIaA19G3R1 has been identified in humans, cattle, and deer [66, 87–90]. To the best of our knowledge, this is the first study to report the presence of various subtypes in pre-weaned calves in the KOR.
To detect C. bovis and C. ryanae, 18S rRNA and heat-shock protein 70 genes are generally used [15]. According to sequence analysis of the 18S rRNA gene, C. bovis and C. ryanae showed ≥99% identity, and it is not always possible to differentiate between them by PCR [91, 92]. However, in this study, we used only the 18S rRNA gene. Even without phylogenetic analysis, the difference between the two species could be confirmed via sequence analysis. At the six nucleotide positions of 440, 460, 464−466, and 470, C. bovis had C, T, A, T, C, and A, whereas C. ryanae had T, C, G, C, T, and G, respectively. These positions are representative markers that distinguish C. ryanae from C. bovis. Our results suggest that these two species can be discerned using the 18S rRNA gene.
Conclusion
Our results confirm the presence of three Cryptosporidium spp. in pre-weaned calves with diarrhea: C. bovis, C. parvum, and C. ryanae. C. parvum was found to be the dominant species in young calves in the KOR. The occurrence of C. ryanae and C. parvum, but not C. bovis, in pre-weaned Korean native calves was significantly related to age; the prevalence of C. parvum decreased with age, whereas that of C. ryanae increased with age. The most frequently detected subtype in calves with diarrhea was IIaA18G3R1, which was responsible for zoonotic transmission. This is the first report to identify nine potentially zoonotic subtypes belonging to the family IIa, which have not previously been reported in cattle in the KOR. This study establishes the high genetic diversity of C. parvum in diarrheic calves and the widespread distribution of zoonotic C. parvum in the KOR. Therefore, the results emphasize that young calves may be a potential source of infection and may serve as an important zoonotic reservoir for human cryptosporidiosis [47, 49].
Data Availability
All relevant data are within the paper.
Funding Statement
Kyoung-Seong Choi: This research was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, and Forestry (IPET) (Grant No. 321016-01-1-HD020). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Fayer R. Cryptosporidium: A water-borne zoonotic parasite. Vet Parasitol. 2004; 126:37–56. doi: 10.1016/j.vetpar.2004.09.004 [DOI] [PubMed] [Google Scholar]
- 2.Xiao L, Ryan UM. Cryptosporidiosis: An update in molecular epidemiology. Curr Opin Infect Dis. 2004; 17:483–90. doi: 10.1097/00001432-200410000-00014 [DOI] [PubMed] [Google Scholar]
- 3.McLauchlin J, Amar C, Pedraza-Diaz S, Nichols GL. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: Results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J Clin Microbiol. 2000; 38:3984–90. doi: 10.1128/JCM.38.11.3984-3990.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao L. Molecular epidemiology of cryptosporidiosis: An update. Exp Parasitol. 2010; 124:80–9. doi: 10.1016/j.exppara.2009.03.018 [DOI] [PubMed] [Google Scholar]
- 5.Wells B, Shaw H, Hotchkiss E, Gilray J, Ayton R, Green J, et al. Prevalence, species identification and genotyping Cryptosporidium from livestock and deer in a catchment in the Cairngorms with a history of a contaminated public water supply. Parasit Vectors. 2015; 8:66. doi: 10.1186/s13071-015-0684-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Y, Ryan UM, Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018; 34:997–1011, doi: 10.1016/j.pt.2018.07.009 [DOI] [PubMed] [Google Scholar]
- 7.Lichtmannsperger K, Harl J, Freudenthaler K, Hinney B, Wittek T, Joachim A. Cryptosporidium parvum, Cryptosporidium ryanae, and Cryptosporidium bovis in samples from calves in Austria. Parasitol Res. 2020; 119:4291–5. doi: 10.1007/s00436-020-06928-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dessi G, Tamponi C, Varcasia A, Sanna G, Pipia AP, Carta S, et al. Cryptosporidium infections in sheep farms from Italy. Parasitol Res. 2020; 119:4211–8. doi: 10.1007/s00436-020-06947-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomson S, Hamilton CA, Hope JC, Katzer F, Mabbott NA, Morrison LJ, et al. Bovine cryptosporidiosis: Impact, host-parasite interaction and control strategies. Vet Res. 2017; 48:42. doi: 10.1186/s13567-017-0447-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caffarena RD, Meireles MV, Carrasco-Letelier L, Picasso-Risso C, Santana BN, Riet-Correa F, et al. Dairy calves in Uruguay are reservoirs of zoonotic subtypes of Cryptosporidium parvum and pose a potential risk of surface water contamination. Front Vet Sci. 2020; 7:562. doi: 10.3389/fvets.2020.00562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aberg M, Emanuelson U, Troell K, Bjorkman C. A single-cohort study of Cryptosporidium bovis and Cryptosporidium ryanae in dairy cattle from birth to calving. Vet Parasitol Reg Stud Reports. 2020; 20:100400. doi: 10.1016/j.vprsr.2020.100400 [DOI] [PubMed] [Google Scholar]
- 12.Yildirim A, Adanir R, Inci A, Yukari BA, Duzlu O, Onder Z, et al. Prevalence and genotyping of bovine Cryptosporidium species in the Mediterranean and Central Anatolia Region of Turkey. Comp Immunol Microbiol Infect Dis. 2020; 69:101425. doi: 10.1016/j.cimid.2020.101425 [DOI] [PubMed] [Google Scholar]
- 13.Liang N, Wu Y, Sun M, Chang Y, Lin X, Yu L, et al. Molecular epidemiology of Cryptosporidium spp. in dairy cattle in Guangdong Province, South China. Parasitology. 2019; 146:28–32. doi: 10.1017/S0031182018001129 [DOI] [PubMed] [Google Scholar]
- 14.Fayer R, Santin M, Xiao L. Cryptosporidium bovis n. sp. (Apicomplexa: Cryptosporidiidae) in cattle (Bos taurus). J Parasitol. 2005; 91:624–9. doi: 10.1645/GE-3435 [DOI] [PubMed] [Google Scholar]
- 15.Fayer R, Santin M, Trout JM. Cryptosporidium ryanae n. sp. (Apicomplexa: Cryptosporidiidae) in cattle (Bos taurus). Vet Parasitol. 2008; 156:191–8. doi: 10.1016/j.vetpar.2008.05.024 [DOI] [PubMed] [Google Scholar]
- 16.Peng MM, Wilson ML, Holland RE, Meshnick SR, Lal AA, Xiao L. Genetic diversity of Cryptosporidium spp. in cattle in Michigan: Implications for understanding the transmission dynamics. Parasitol Res. 2003; 90:175–80. doi: 10.1007/s00436-003-0834-5 [DOI] [PubMed] [Google Scholar]
- 17.Khan A, Shaik JS, Grigg ME. Genomics and molecular epidemiology of Cryptosporidium species. Acta Trop. 2018; 184:1–14. doi: 10.1016/j.actatropica.2017.10.023 [DOI] [PubMed] [Google Scholar]
- 18.Lee YJ, Ryu JH, Shin SU, Choi KS. Prevalence and molecular characterization of Cryptosporidium and Giardia in pre-weaned native calves in the Republic of Korea. Parasitol Res. 2019; 118:3509–17. doi: 10.1007/s00436-019-06482-9 [DOI] [PubMed] [Google Scholar]
- 19.Kabir MHB, Ceylan O, Ceylan C, Shehata AA, Bando H, Essa MI, et al. Molecular detection of genotypes and subtypes of Cryptosporidium infection in diarrheic calves, lambs, and goat kids from Turkey. Parasitol Int. 2020; 79:102163. doi: 10.1016/j.parint.2020.102163 [DOI] [PubMed] [Google Scholar]
- 20.Silverlas C, Naslund K, Bjorkman C, Mattsson JG. Molecular characterisation of Cryptosporidium isolates from Swedish dairy cattle in relation to age, diarrhoea and region. Vet Parasitol. 2010; 169:289–95. doi: 10.1016/j.vetpar.2010.01.003 [DOI] [PubMed] [Google Scholar]
- 21.Muhid A, Robertson I, Ng J, Ryan U. Prevalence of and management factors contributing to Cryptosporidium sp. infection in pre-weaned and post-weaned calves in Johor, Malaysia. Exp Parasitol. 2011; 127:534–8. doi: 10.1016/j.exppara.2010.10.015 [DOI] [PubMed] [Google Scholar]
- 22.Taylan-Ozkan A, Yasa-Duru S, Usluca S, Lysen C, Ye J, Roellig DM, et al. Cryptosporidium species and Cryptosporidium parvum subtypes in dairy calves and goat kids reared under traditional farming systems in Turkey. Exp Parasitol. 2016; 170:16–20. doi: 10.1016/j.exppara.2016.06.014 [DOI] [PubMed] [Google Scholar]
- 23.Li N, Wang R, Cai M, Jiang W, Feng Y, Xiao L. Outbreak of cryptosporidiosis due to Cryptosporidium parvum subtype IIdA19G1 in neonatal calves on a dairy farm in China. Int J Parasitol. 2019; 49:569–77. doi: 10.1016/j.ijpara.2019.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee SH, VanBik D, Kim HY, Lee YR, Kim JW, Chae M, et al. Multilocus typing of Cryptosporidium spp. in young calves with diarrhea in Korea. Vet Parasitol. 2016; 229:81–9. doi: 10.1016/j.vetpar.2016.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabir MHB, Itoh M, Shehata AA, Bando H, Fukuda Y, Murakoshi F, et al. Distribution of Cryptosporidium species isolated from diarrhoeic calves in Japan. Parasitol Int. 2020; 78:102153. doi: 10.1016/j.parint.2020.102153 [DOI] [PubMed] [Google Scholar]
- 26.Cai M, Guo Y, Pan B, Li N, Wang X, Tang C, et al. Longitudinal monitoring of Cryptosporidium species in pre-weaned dairy calves on five farms in Shanghai, China. Vet Parasitol. 2017; 241:14–9. doi: 10.1016/j.vetpar.2017.05.005 [DOI] [PubMed] [Google Scholar]
- 27.Xiao L, Morgan UM, Limor J, Escalante A, Arrowood M, Shulaw W, et al. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl Environ Microbiol. 1999; 65:3386–91. doi: 10.1128/AEM.65.8.3386-3391.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheun HI, Choi TK, Chung GT, Cho SH, Lee YH, Kimata I, et al. Genotypic characterization of Cryptosporidium oocysts isolated from healthy people in three different counties of Korea. J Vet Med Sci. 2007; 69:1099–101. doi: 10.1292/jvms.69.1099 [DOI] [PubMed] [Google Scholar]
- 29.Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, et al. Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol. 2005; 43:2805–9. doi: 10.1128/JCM.43.6.2805-2809.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meganck V, Hoflack G, Piepers S, Opsomer G. Evaluation of a protocol to reduce the incidence of neonatal calf diarrhoea on dairy herds. Prev Vet Med. 2015; 118:64–70. doi: 10.1016/j.prevetmed.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SH, Kim HY, Choi EW, Kim D. Causative agents and epidemiology of diarrhea in Korean native calves. J Vet Sci. 2019; 20:e64. doi: 10.4142/jvs.2019.20.e64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Avendano C, Ramo A, Vergara-Castiblanco C, Sanchez-Acedo C, Quilez J. Genetic uniqueness of Cryptosporidium parvum from dairy calves in Colombia. Parasitol Res. 2018; 117:1317–23. doi: 10.1007/s00436-018-5818-6 [DOI] [PubMed] [Google Scholar]
- 33.Diaz P, Varcasia A, Pipia AP, Tamponi C, Sanna G, Prieto A, et al. Molecular characterisation and risk factor analysis of Cryptosporidium spp. in calves from Italy. Parasitol Res. 2018; 117:3081–90. doi: 10.1007/s00436-018-6000-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holzhausen I, Lendner M, Gohring F, Steinhofel I, Daugschies A. Distribution of Cryptosporidium parvum gp60 subtypes in calf herds of Saxony, Germany. Parasitol Res. 2019; 118:1549–58. doi: 10.1007/s00436-019-06266-1 [DOI] [PubMed] [Google Scholar]
- 35.Lombardelli JA, Tomazic ML, Schnittger L, Tiranti KI. Prevalence of Cryptosporidium parvum in dairy calves and gp60 subtyping of diarrheic calves in Central Argentina. Parasitol Res. 2019; 118:2079–86. doi: 10.1007/s00436-019-06366-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santoro A, Dorbek-Kolin E, Jeremejeva J, Tummeleht L, Orro T, Jokelainen P, et al. Molecular epidemiology of Cryptosporidium spp. in calves in Estonia: High prevalence of Cryptosporidium parvum shedding and 10 subtypes identified. Parasitology. 2019; 146:261–7. doi: 10.1017/S0031182018001348 [DOI] [PubMed] [Google Scholar]
- 37.Wu Y, Zhang K, Zhang Y, Jing B, Chen Y, Xu C, et al. Genetic diversity of Cryptosporidium parvum in neonatal dairy calves in Xinjiang, China. Pathogens. 2020; 9:692. doi: 10.3390/pathogens9090692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaupke A, Rzezutka A. Emergence of novel subtypes of Cryptosporidium parvum in calves in Poland. Parasitol Res. 2015; 114:4709–16. doi: 10.1007/s00436-015-4719-1 [DOI] [PubMed] [Google Scholar]
- 39.Qi M, Zhang K, Huang M, Wang S, Xu C, Wang T, et al. Longitudinal detection of Cryptosporidium spp. in 1-10-week-old dairy calves on a farm in Xinjiang, China. Parasitol Res. 2020; 119:3839–44. doi: 10.1007/s00436-020-06904-z [DOI] [PubMed] [Google Scholar]
- 40.Santin M, Trout JM, Xiao L, Zhou L, Greiner E, Fayer R. Prevalence and age-related variation of Cryptosporidium species and genotypes in dairy calves. Vet Parasitol. 2004; 122:103–17. doi: 10.1016/j.vetpar.2004.03.020 [DOI] [PubMed] [Google Scholar]
- 41.Santin M. Clinical and subclinical infections with Cryptosporidium in animals. N Z Vet J. 2013; 61:1–10. doi: 10.1080/00480169.2012.731681 [DOI] [PubMed] [Google Scholar]
- 42.Feng Y, Xiao L. Molecular epidemiology of cryptosporidiosis in China. Front Microbiol. 2017; 8:1701. doi: 10.3389/fmicb.2017.01701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tao W, Li Y, Yang H, Song M, Lu Y, Li W. Widespread occurrence of zoonotic Cryptosporidium species and subtypes in dairy cattle from Northeast China: Public health concerns. J Parasitol. 2018; 104:10–7. doi: 10.1645/17-140 [DOI] [PubMed] [Google Scholar]
- 44.Fayer R, Santin M, Trout JM. Prevalence of Cryptosporidium species and genotypes in mature dairy cattle on farms in eastern United States compared with younger cattle from the same locations. Vet Parasitol. 2007; 145:260–6. doi: 10.1016/j.vetpar.2006.12.009 [DOI] [PubMed] [Google Scholar]
- 45.Santin M, Trout JM, Fayer R. A longitudinal study of cryptosporidiosis in dairy cattle from birth to 2 years of age. Vet Parasitol. 2008; 155:15–23. doi: 10.1016/j.vetpar.2008.04.018 [DOI] [PubMed] [Google Scholar]
- 46.Silverlas C, Blanco-Penedo I. Cryptosporidium spp. in calves and cows from organic and conventional dairy herds. Epidemiol Infect. 2013; 141:529–39. doi: 10.1017/S0950268812000830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rieux A, Paraud C, Pors I, Chartier C. Molecular characterization of Cryptosporidium isolates from pre-weaned calves in western France in relation to age. Vet Parasitol. 2013; 197:7–12. doi: 10.1016/j.vetpar.2013.05.001 [DOI] [PubMed] [Google Scholar]
- 48.Faubert GM, Litvinsky Y. Natural transmission of Cryptosporidium parvum between dams and calves on a dairy farm. J Parasitol. 2000; 86:495–500. doi: 10.1645/0022-3395(2000)086[0495:NTOCPB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 49.Thomson S, Innes EA, Jonsson NN, Katzer F. Shedding of Cryptosporidium in calves and dams: Evidence of re-infection and shedding of different gp60 subtypes. Parasitology. 2019; 146:1404–13. doi: 10.1017/S0031182019000829 [DOI] [PubMed] [Google Scholar]
- 50.Wang R, Zhao G, Gong Y, Zhang L. Advances and perspectives on the epidemiology of bovine Cryptosporidium in China in the past 30 years. Front Microbiol. 2017; 8:1823. doi: 10.3389/fmicb.2017.01823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bjorkman C, Lindstrom L, Oweson C, Ahola H, Troell K, Axen C. Cryptosporidium infections in suckler herd beef calves. Parasitology. 2015; 142:1108–14. doi: 10.1017/S0031182015000426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng Y, Ortega Y, He G, Das P, Xu M, Zhang X, et al. Wide geographic distribution of Cryptosporidium bovis and the deer-like genotype in bovines. Vet Parasitol. 2007; 144:1–9. doi: 10.1016/j.vetpar.2006.10.001 [DOI] [PubMed] [Google Scholar]
- 53.Seppa-Lassila L, Orro T, Lassen B, Lasonen R, Autio T, Pelkonen S, et al. Intestinal pathogens, diarrhoea and acute phase proteins in naturally infected dairy calves. Comp Immunol Microbiol Infect Dis. 2015; 41:10–6. doi: 10.1016/j.cimid.2015.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu Z, Li N, Guo Y, Feng Y, Xiao L. Comparative genomic analysis of three intestinal species reveals reductions in secreted pathogenesis determinants in bovine-specific and non-pathogenic Cryptosporidium species. Microb Genom. 2020; 6:e000379. doi: 10.1099/mgen.0.000379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng Y, Torres E, Li N, Wang L, Bowman D, Xiao L. Population genetic characterisation of dominant Cryptosporidium parvum subtype IIaA15G2R1. Int J Parasitol. 2013; 43:1141–7. doi: 10.1016/j.ijpara.2013.09.002 [DOI] [PubMed] [Google Scholar]
- 56.Ichikawa-Seki M, Aita J, Masatani T, Suzuki M, Nitta Y, Tamayose G, et al. Molecular characterization of Cryptosporidium parvum from two different Japanese prefectures, Okinawa and Hokkaido. Parasitol Int. 2015; 64:161–6. doi: 10.1016/j.parint.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramo A, Quilez J, Vergara-Castiblanco C, Monteagudo L, Del Cacho E, Clavel A. Multilocus typing and population structure of Cryptosporidium from children in Zaragoza, Spain. Infect Genet Evol. 2015; 31:190–7. doi: 10.1016/j.meegid.2015.01.023 [DOI] [PubMed] [Google Scholar]
- 58.Soba B, Logar J. Genetic classification of Cryptosporidium isolates from humans and calves in Slovenia. Parasitology. 2008; 135:1263–70. doi: 10.1017/S0031182008004800 [DOI] [PubMed] [Google Scholar]
- 59.Valenzuela O, Gonzalez-Diaz M, Garibay-Escobar A, Burgara-Estrella A, Cano M, Durazo M, et al. Molecular characterization of Cryptosporidium spp. in children from Mexico. PLoS One. 2014; 9:e96128. doi: 10.1371/journal.pone.0096128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trotz-Williams LA, Martin DS, Gatei W, Cama V, Peregrine AS, Martin SW, et al. Genotype and subtype analyses of Cryptosporidium isolates from dairy calves and humans in Ontario. Parasitol Res. 2006; 99:346–52. doi: 10.1007/s00436-006-0157-4 [DOI] [PubMed] [Google Scholar]
- 61.Al Mawly J, Grinberg A, Velathanthiri N, French N. Cross sectional study of prevalence, genetic diversity and zoonotic potential of Cryptosporidium parvum cycling in New Zealand dairy farms. Parasit Vectors. 2015; 8:240. doi: 10.1186/s13071-015-0855-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grinberg A, Learmonth J, Kwan E, Pomroy W, Lopez Villalobos N, Gibson I, et al. Genetic diversity and zoonotic potential of Cryptosporidium parvum causing foal diarrhea. J Clin Microbiol. 2008; 46:2396–8. doi: 10.1128/JCM.00936-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Inacio SV, Widmer G, de Brito RL, Zucatto AS, de Aquino MC, Oliveira BC, et al. First description of Cryptosporidium hominis gp60 genotype IkA20G1 and Cryptosporidium parvum gp60 genotypes IIaA18G3R1 and IIaA15G2R1 in foals in Brazil. Vet Parasitol. 2017; 233:48–51. doi: 10.1016/j.vetpar.2016.11.021 [DOI] [PubMed] [Google Scholar]
- 64.Ng JS, Pingault N, Gibbs R, Koehler A, Ryan U. Molecular characterisation of Cryptosporidium outbreaks in Western and South Australia. Exp Parasitol. 2010; 125:325–8. doi: 10.1016/j.exppara.2010.02.012 [DOI] [PubMed] [Google Scholar]
- 65.Waldron LS, Power ML. Fluorescence analysis detects gp60 subtype diversity in Cryptosporidium infections. Infect Genet Evol. 2011; 11:1388–95. doi: 10.1016/j.meegid.2011.05.008 [DOI] [PubMed] [Google Scholar]
- 66.Zintl A, Proctor AF, Read C, Dewaal T, Shanaghy N, Fanning S, et al. The prevalence of Cryptosporidium species and subtypes in human faecal samples in Ireland. Epidemiol Infect. 2009; 137:270–7. doi: 10.1017/S0950268808000769 [DOI] [PubMed] [Google Scholar]
- 67.Del Chierico F, Onori M, Di Bella S, Bordi E, Petrosillo N, Menichella D, et al. Cases of cryptosporidiosis co-infections in AIDS patients: A correlation between clinical presentation and gp60 subgenotype lineages from aged formalin-fixed stool samples. Ann Trop Med Parasitol. 2011; 105:339–49. doi: 10.1179/1364859411Y.0000000025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Glaberman S, Moore JE, Lowery CJ, Chalmers RM, Sulaiman I, Elwin K, et al. Three drinking-water-associated cryptosporidiosis outbreaks, Northern Ireland. Emerg Infect Dis. 2002; 8:631–3. doi: 10.3201/eid0806.010368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mammeri M, Chevillot A, Chenafi I, Thomas M, Julien C, Vallee I, et al. Molecular characterization of Cryptosporidium isolates from diarrheal dairy calves in France. Vet Parasitol Reg Stud Reports. 2019; 18:100323. doi: 10.1016/j.vprsr.2019.100323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thompson HP, Dooley JS, Kenny J, McCoy M, Lowery CJ, Moore JE, et al. Genotypes and subtypes of Cryptosporidium spp. in neonatal calves in Northern Ireland. Parasitol Res. 2007; 100:619–24. doi: 10.1007/s00436-006-0305-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Waldron LS, Ferrari BC, Power ML. Glycoprotein 60 diversity in C. hominis and C. parvum causing human cryptosporidiosis in NSW, Australia. Exp Parasitol. 2009; 122:124–7. doi: 10.1016/j.exppara.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 72.Koinari M, Karl S, Ng-Hublin J, Lymbery AJ, Ryan UM. Identification of novel and zoonotic Cryptosporidium species in fish from Papua New Guinea. Vet Parasitol. 2013; 198:1–9. doi: 10.1016/j.vetpar.2013.08.031 [DOI] [PubMed] [Google Scholar]
- 73.Koinari M, Lymbery AJ, Ryan UM. Cryptosporidium species in sheep and goats from Papua New Guinea. Exp Parasitol. 2014; 141:134–7. doi: 10.1016/j.exppara.2014.03.021 [DOI] [PubMed] [Google Scholar]
- 74.Shrestha RD, Grinberg A, Dukkipati VS, Pleydell EJ, Prattley DJ, French NP. Infections with multiple Cryptosporidium species and new genetic variants in young dairy calves on a farm located within a drinking water catchment area in New Zealand. Vet Parasitol. 2014; 202:287–91. doi: 10.1016/j.vetpar.2014.03.034 [DOI] [PubMed] [Google Scholar]
- 75.Giangaspero A, Papini R, Marangi M, Koehler AV, Gasser RB. Cryptosporidium parvum genotype IIa and Giardia duodenalis assemblage A in Mytilus galloprovincialis on sale at local food markets. Int J Food Microbiol. 2014; 171:62–7. doi: 10.1016/j.ijfoodmicro.2013.11.022 [DOI] [PubMed] [Google Scholar]
- 76.Waldron LS, Dimeski B, Beggs PJ, Ferrari BC, Power ML. Molecular epidemiology, spatiotemporal analysis, and ecology of sporadic human cryptosporidiosis in Australia. Appl Environ Microbiol. 2011; 77:7757–65. doi: 10.1128/AEM.00615-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deshpande AP, Jones BL, Connelly L, Pollock KG, Brownlie S, Alexander CL. Molecular characterization of Cryptosporidium parvum isolates from human cryptosporidiosis cases in Scotland. Parasitology. 2015; 142:318–25. doi: 10.1017/S0031182014001346 [DOI] [PubMed] [Google Scholar]
- 78.Osman M, Benamrouz S, Guyot K, El Safadi D, Mallat H, Dabboussi F, et al. Molecular epidemiology of Cryptosporidium spp. in North Lebanon. J Infect Dev Ctries. 2018; 12:34S. doi: 10.3855/jidc.10014 [DOI] [PubMed] [Google Scholar]
- 79.Mahfouz ME, Mira N, Amer S. Prevalence and genotyping of Cryptosporidium spp. in farm animals in Egypt. J Vet Med Sci. 2014; 76:1569–75. doi: 10.1292/jvms.14-0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mi R, Wang X, Huang Y, Zhou P, Liu Y, Chen Y, et al. Prevalence and molecular characterization of Cryptosporidium in goats across four provincial level areas in China. PLoS One. 2014; 9:e111164. doi: 10.1371/journal.pone.0111164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Naguib D, El-Gohary AH, Mohamed AA, Roellig DM, Arafat N, Xiao L. Age patterns of Cryptosporidium species and Giardia duodenalis in dairy calves in Egypt. Parasitol Int. 2018; 67:736–41. doi: 10.1016/j.parint.2018.07.012 [DOI] [PubMed] [Google Scholar]
- 82.Mercado R, Pena S, Ozaki LS, Fredes F, Godoy J. Multiple Cryptosporidium parvum subtypes detected in a unique isolate of a Chilean neonatal calf with diarrhea. Parasitol Res. 2015; 114:1985–8. doi: 10.1007/s00436-015-4364-8 [DOI] [PubMed] [Google Scholar]
- 83.Al-Habsi K, Yang R, Williams A, Miller D, Ryan U, Jacobson C. Zoonotic Cryptosporidium and Giardia shedding by captured rangeland goats. Vet Parasitol Reg Stud Reports. 2017; 7:32–5. doi: 10.1016/j.vprsr.2016.11.006 [DOI] [PubMed] [Google Scholar]
- 84.Chalmers RM, Robinson G, Elwin K, Elson R. Analysis of the Cryptosporidium spp. and gp60 subtypes linked to human outbreaks of cryptosporidiosis in England and Wales, 2009 to 2017. Parasit Vectors. 2019; 12:95. doi: 10.1186/s13071-019-3354-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Connelly L, Craig BH, Jones B, Alexander CL. Genetic diversity of Cryptosporidium spp. within a remote population of Soay Sheep on St. Kilda Islands, Scotland. Appl Environ Microbiol. 2013; 79:2240–6. doi: 10.1128/AEM.02823-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Del Coco VF, Cordoba MA, Bilbao G, de Almeida Castro AP, Basualdo JA, Fayer R, et al. Cryptosporidium parvum gp60 subtypes in dairy cattle from Buenos Aires, Argentina. Res Vet Sci. 2014; 96:311–4. doi: 10.1016/j.rvsc.2013.12.010 [DOI] [PubMed] [Google Scholar]
- 87.Abeywardena H, Jex AR, Nolan MJ, Haydon SR, Stevens MA, McAnulty RW, et al. Genetic characterisation of Cryptosporidium and Giardia from dairy calves: Discovery of species/genotypes consistent with those found in humans. Infect Genet Evol. 2012; 12:1984–93. doi: 10.1016/j.meegid.2012.08.004 [DOI] [PubMed] [Google Scholar]
- 88.Nolan MJ, Jex AR, Koehler AV, Haydon SR, Stevens MA, Gasser RB. Molecular-based investigation of Cryptosporidium and Giardia from animals in water catchments in Southeastern Australia. Water Res. 2013; 47:1726–40. doi: 10.1016/j.watres.2012.12.027 [DOI] [PubMed] [Google Scholar]
- 89.O’Brien E, McInnes L, Ryan U. Cryptosporidium gp60 genotypes from humans and domesticated animals in Australia, North America and Europe. Exp Parasitol. 2008; 118:118–21. doi: 10.1016/j.exppara.2007.05.012 [DOI] [PubMed] [Google Scholar]
- 90.Quilez J, Torres E, Chalmers RM, Robinson G, Del Cacho E, Sanchez-Acedo C. Cryptosporidium species and subtype analysis from dairy calves in Spain. Parasitology. 2008; 135:1613–20. doi: 10.1017/S0031182008005088 [DOI] [PubMed] [Google Scholar]
- 91.Mirhashemi ME, Zintl A, Grant T, Lucy F, Mulcahy G, De Waal T. Molecular epidemiology of Cryptosporidium species in livestock in Ireland. Vet Parasitol. 2016; 216:18–22. doi: 10.1016/j.vetpar.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Santin M, Zarlenga DS. A multiplex polymerase chain reaction assay to simultaneously distinguish Cryptosporidium species of veterinary and public health concern in cattle. Vet Parasitol. 2009; 166:32–7. doi: 10.1016/j.vetpar.2009.07.039 [DOI] [PubMed] [Google Scholar]