Abstract
Heterodimeric transcription factors, including the basic region-leucine zipper (bZIP) protein ATF-2–c-jun, are well-characterized components of an enhanceosome that mediates virus induction of the human beta interferon (IFN-β) gene. Here we report that within the IFN-β enhanceosome the ATF-2–c-jun heterodimer binds in a specific orientation, which is required for assembly of a complex between ATF-2–c-jun and interferon regulatory factor 3 (IRF-3). We demonstrate that correct orientation of the ATF-2–c-jun binding site is required for virus induction of the IFN-β gene and for IRF-3-dependent activation of a composite ATF-2– c-jun–IRF site in the IFN-β promoter. We also show that in vitro the DNA-bound ATF-2–c-jun heterodimer adopts a fixed orientation upon the binding of IRF-3 at an adjacent site in the IFN-β enhancer and that the DNA-binding domain of IRF-3 is sufficient to mediate this effect. In addition, we show that the DNA-binding domain of ATF-2 is necessary and sufficient for selective protein-protein interactions with IRF-3. Strikingly, in vivo chromatin immunoprecipitation experiments with IFN-β reporter constructs reveal that recruitment of IRF-3 to the IFN-β promoter upon virus infection is dependent on the orientation of the ATF-2–c-jun heterodimer binding site. These observations demonstrate functional and physical cooperativity between the bZIP and IRF transcription factor families and illustrate the critical role of heterodimeric transcription factors in formation of the IFN-β enhanceosome.
The expression of specific sets of genes in response to different extracellular signals is required for regulation of the immune and inflammatory responses. Most extracellular signals activate a number of different transcription factors. Moreover, individual transcription factors can be activated in response to many different signals. Thus, a mechanism for integrating signals and transcription factors is required for normal cellular regulation. One mechanism for signal integration is the assembly of multicomponent transcriptional enhancer complexes known as enhanceosomes (4). The coordinate activation of specific sets of transcription factors, their assembly into a higher-order complex, and their interaction with coactivator proteins and components of the basal transcription machinery ensure that the appropriate genes are activated in response to a given signal. An excellent example of this is the activation of the beta interferon (IFN-β) gene in response to virus infection (35).
Type I IFNs, including IFN-β, are synthesized and secreted by mammalian cells in response to virus infection and thus mediate the establishment of an antiviral state (59). Induction of IFN-β gene transcription depends on a remarkably compact enhancer region, spanning a region between 105 and 55 bp upstream of the start site of mRNA transcription. Indeed, virtually every DNA base pair in this region is contacted by a transcription factor through the major or minor groove. Induction of IFN-β transcription does not require de novo protein synthesis; rather, it occurs through virus-mediated posttranslational modification of transcription factors, which bind to specific positive regulatory domains (PRDs) within the IFN-β promoter (Fig. 1A). A heterodimer of NF-κB p50 and p65 and a heterodimer of the basic region-leucine zipper (bZIP) proteins ATF-2 and c-jun bind to PRDII and PRDIV, respectively. PRDIII and PRDI are recognized by a protein complex containing IFN regulatory factor 3 (IRF-3) and IRF-7, but the stoichiometry of these proteins within the enhanceosome is not known (35). Two molecules of the high-mobility-group protein I(Y) [HMGI(Y)] also recognize the IFN-β enhancer, one each at PRDII and PRDIV, and bind cooperatively with NF-κB and ATF-2–c-jun (15, 17, 53, 65). A formaldehyde cross-linking and chromatin immunoprecipitation (ChIP) procedure has been used to detect transcription factors that bind the endogenous IFN-β promoter before and after virus infection. These studies have shown that NF-κB p50 is bound to the IFN-β promoter prior to virus induction, but the full complement of activators—ATF-2, c-jun, IRF-3, IRF-7, and NF-κB p50 and p65—associates with the promoter following virus infection (61).
FIG. 1.
Orientation of asymmetric ATF-2–c-jun binding sites in the context of the IFN-β enhancer is critical for virus induction. (A) Diagram of the binding sites for ATF-2–c-jun, IRF proteins, NF-κB, and HMGI(Y) in the IFN-β enhancer. (B) Nucleotide sequences and activities of −110 IFN-β enhancer reporters used in this study. Numbering indicates nucleotide positions relative to the start site of transcription in the wild-type promoter. The core 5′ and 3′ half-sites in each sequence are shaded. (C) Histogram showing the levels of CAT activities from uninduced and Sendai virus-induced HeLa cells transfected with the indicated reporters. Average results of multiple independent experiments (total number as noted) are shown for each reporter; error bars indicate the standard error of the mean. For each experiment, wild-type (WT) reporter was included and wild-type virus-induced activity was normalized to 100. The average CAT activities (percent conversion of unacetylated to acetylated chloramphenicol) for uninduced and virus-induced wild-type reporter (± standard error of the mean) were 1.10 ± 0.280 and 10.68 ± 3.68, respectively.
Recently, the CREB-binding protein (CBP) and p300 coactivator proteins were shown to be required for IFN-β expression (30, 36, 49, 61, 67). Following virus infection, CBP and p300 associate with IRF-3 (30, 31, 49, 61, 62, 67) and IRF-7 (61). Recruitment of CBP and p300, in turn, is required for the recruitment of components of the preinitiation complex to the promoter, including TFIIB, TFIIA, and TFIID (23, 24). CBP and p300 are histone acetyltransferases, which promote transcription via the formation of open chromatin structure (52); indeed, histones at the IFN-β promoter become hyperacetylated after virus infection (40). Thus, a specific set of transcription factors, coactivators, and the architectural protein HMG I(Y) assemble into a higher-order nucleoprotein complex, the enhanceosome, at the IFN-β enhancer and activate transcription of the IFN-β gene in response to virus infection (4, 35).
Assembly of the IFN-β enhanceosome depends on proper spatial alignment and orientation of the PRDs along the DNA helix, and formation of the complex is accompanied by a remodeling of DNA conformation (17, 55). Moreover, multiple protein-protein interactions have been identified among the components of the enhanceosome (15, 36, 53, 66), suggesting that the specific positioning of transcription factors within the complex is critical for enhanceosome assembly. The half-sites of PRDII and PRDIV are in fact asymmetric, and in the case of PRDII, the NF-κB p50 subunit recognizes the 5′ half-site and the p65 subunit recognizes the 3′ half-site (55, 57). The positioning of heterodimeric transcription factors thus presents a potential basis for correct assembly of the IFN-β enhanceosome.
Here, we report the critical role of ATF-2–c-jun heterodimer orientation in the assembly and function of the IFN-β enhanceosome. Using site-directed mutagenesis of IFN-β reporter constructs, we demonstrated that the orientation of cyclic AMP response element (CRE) consensus and nonconsensus half-sites within PRDIV is critical for virus induction and that asymmetric ATF-2–c-jun sites from other gene promoters can exhibit similar effects in the context of the IFN-β promoter. We also show that PRDIV and PRDIII-I function as a strongly virus-inducible composite element (PRD431), critically dependent on the orientation of PRDIV, when placed upstream from a heterologous promoter.
Using site-specific photo-cross-linking of recombinant proteins to the IFN-β promoter, we have examined the orientation of the ATF-2–c-jun heterodimer bound to PRDIV. Although ATF-2–c-jun binds to PRDIV with only modest intrinsic preference in orientation, this orientation is fixed in the presence of IRF-1 or IRF-3, such that c-jun and ATF-2 recognize the 5′ consensus and 3′ nonconsensus half-sites, respectively, which are in turn distal and proximal to the IRF site. Furthermore, we show that the DNA-binding domain of IRF-3 and the IRF binding site adjacent to PRDIV are required for fixing of ATF-2–c-jun heterodimer orientation by IRF-3. Using glutathione S-transferase (GST) association assays, we found that ATF-2 preferentially interacts with IRF-3, consistent with the positions predicted by the cross-linking studies. Strikingly, the 66-amino-acid bZIP DNA-binding domain of ATF-2 is necessary and sufficient for interaction with IRF-3. We thus demonstrate direct interactions between proteins of the IRF and bZIP families.
An interesting result from our in vitro studies is that IRF-3 can fix the orientation of ATF-2–c-jun at PRD431 only when PRDIV is in the wild-type orientation. IRF-1, on the other hand, which exhibits equal affinity for ATF-2 and c-jun in GST association assays, fixes ATF-2–c-jun in the opposite orientation when PRDIV is reversed, such that c-jun is proximal to the IRF site. Consistent with this finding, transfection of exogenous IRF-3 activates a PRD431 reporter with PRDIV in the wild-type but not the reverse orientation, while exogenous IRF-1 activates both reporters, indicating that formation of a functional IRF-3–ATF-2–c-jun complex at the IFN-β promoter in vivo depends on the orientation of PRDIV. We have directly examined the effect of PRDIV orientation on the binding of transcription factors at the IFN-β promoter in vivo using ChIP assays of cells cotransfected with wild-type and mutant IFN-β promoter constructs. Our modification of the ChIP technique permits direct comparison of the binding of endogenous factors to wild-type and mutant forms of gene promoters of interest. Remarkably, IRF-3 is recruited to the wild-type IFN-β promoter but not an IFN-β promoter with the ATF-2–c-jun binding site in reverse orientation. Thus, we have demonstrated that the orientation of a heterodimeric transcription factor within the context of an enhancer can be critical for recruitment of other transcription factors to a functional enhancer complex. Given the importance of heterodimeric transcription factors in the activation of numerous genes, other functional nucleoprotein complexes may prove to exhibit transcription factor orientation-dependent assembly.
MATERIALS AND METHODS
Plasmids.
Site-directed mutagenesis of the −110 IFN-β CAT (chloramphenicol acetyltransferase) reporter at PRDIV was performed by a two-step PCR method described previously (53). The XbaI-BspDI fragments of the PCR products were subcloned into the −110 IFN-β CAT reporter. The PRD431 and PRD4rev31 SEAP (secreted alkaline phosphatase) constructs were made by subcloning one to three copies of the oligonucleotides 5′-CTAGCTAAATGTAAATGACATAGGAAAACTGAAAGGGAGAAGTGAAAGTGTctag-3′ and 5′-CTAGCTAAATGTAAACTATGTCAGAAACTGAAAGGGAGAAGTGAAAGTGTctag-3′, respectively, into the XbaI site of E1b TATA CAT (29). The XhoI-EcoRI fragment was then subcloned into pSEAP2-Basic (Clontech). The PRD31 SEAP constructs were made by subcloning the XhoI-EcoRI fragment from the PRD31 CAT reporters (61) into pSEAP2-Basic. All sequences were verified either by dideoxy sequencing or with an ABI 310 Genetic Analyzer (Perkin-Elmer).
The expression vectors pcDNA1-ATF-2, pcDNA1-c-jun, and pcDNA1-IRF-1 have been described previously (15, 54). The IRF-3 E5 expression vector, which contains serine-to-glutamic acid changes at positions 396, 398, 402, and 405 and a threonine-to-glutamic acid change at position 404, was prepared from wild-type H6-hIRF-3 expression vector (61) by standard PCR mutagenesis (C. H. Lin, M. G. Wathelet, and T. Maniatis, unpublished data).
Cell culture and transfections.
HeLa and P19 cells (American Type Culture Collection, Manassas, Va.) were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml in a 37°C incubator at 5% CO2. Transfections in HeLa cells were performed in six-well plates using Lipofectamine (Life Technologies) at 25 μg/ml according to the manufacturer's protocol and induced with Sendai virus as described elsewhere (61). For each experiment, 1.5 μg of CAT or SEAP reporter plasmid was used along with 0.5 μg of β-galactosidase control reporter plasmid (pCMVβ; Clontech). P19 cells were seeded in six-well plates (3 × 105 cells per well) and grown to approximately 75% confluency. Transfections were performed using FuGene6 (Boehringer Mannheim) at 2 μl of reagent per ml of medium according to the manufacturer's protocol, and media and cells were assayed 48 h posttransfection. PRD4312 SEAP or PRD4rev312 SEAP reporters (200 ng) were cotransfected with pCMVβ (200 ng), 270 ng of pcDNA1, 30 ng of ATF-2–c-jun expression vector, and up to 300 ng of IRF expression vector as indicated in the relevant figure legend; the total amount of DNA was kept constant with pcDNA1 (Invitrogen). Extracts were assayed for β-galactosidase activity or CAT activity as described elsewhere (48). Colorimetric SEAP assays were performed essentially as described previously; DMEM lacking phenol red was used following transfection (9).
Recombinant proteins and protein-DNA photo-cross-linking.
Proteins were expressed as hexahistidine or GST fusions in Escherichia coli BL21(DE3) using the expression vectors pET15-p50, pET25-p65, pET-25-HMGI, pET25-IRF-1, pGEX-IRF-3, pET15-ATF-2195, and pET15-c-jun and purified as described elsewhere (50, 53).
Oligonucleotides of the PRDIV site with or without an intact IRF site at PRDIII (5′-AAT*GTAAATGACATAGGAAA*CTGA-3′ or 5′-T*GTAAATGACATAGTCTAA*C-3′), the PRDII site (see Fig. 4A), or the IFN-β promoter containing single phosphorothioate substitutions at the positions indicated by the asterisks on the sense (5′) or antisense (3′) strand were synthesized by Operon Technologies. p-Azidophenacyl (AzP) bromide (Sigma) was coupled to the phosphorothioate moieties as follows. Phosphorothioate-substituted single-stranded oligonucleotide (1 nmol) was incubated in 100 μl (final volume) of 20 mM NaHCO3 (pH 9.0)–45% dimethyl sulfoxide–5 mM AzP bromide (diluted from a 0.33 M stock in methanol) for 2 h at room temperature in the dark. The reaction mixture was filtered through 2 columns of Sephadex G-50 (Pharmacia Biotech) by centrifugation, ethanol precipitated, and resuspended in 15 μl of H2O. The DNA was then labeled with 2 μl of [γ-32P]ATP and 1 μl of polynucleotide kinase in 1× PNK buffer (New England BioLabs) for 1 h at 37°C. The labeled DNA was then purified on a G-50 column as before, 0.5 nmol of the complementary strand was added, and the oligonucleotides were annealed by heating to 85°C and cooling to room temperature.
FIG. 4.
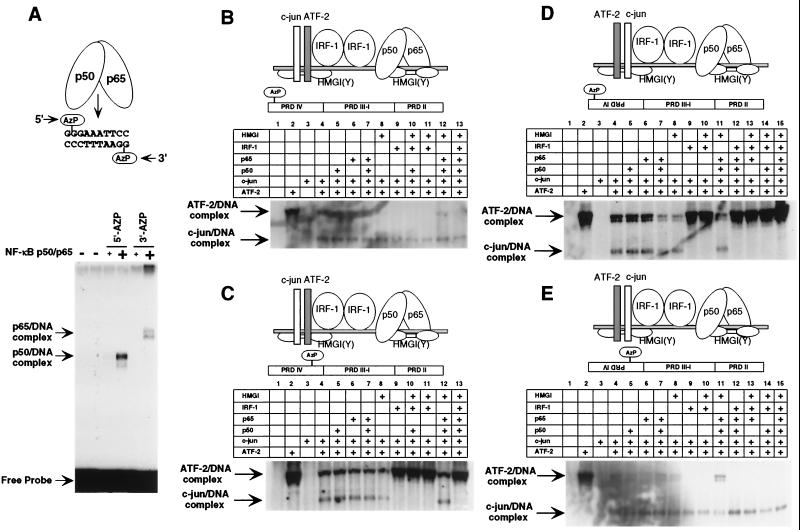
IRF-1 fixes the orientation of ATF-2–c-jun at PRDIV in vitro. (A) Photo-cross-linking of NF-κB p50-p65 to AzP-modified PRDII oligonucleotide as a control for cross-linking specificity. Recombinant p50-p65 heterodimer was incubated with radiolabeled PRDII probe bearing a single AzP group at the 5′ or 3′ half-site as indicated and resolved by SDS-PAGE. (B) Photo-cross-linking of ATF-2 (upper complex; top band in lane 8) and c-jun (lower complex) to the PRDIV site of the IFN-β enhancer in the presence or absence of other components of the enhanceosome. A radiolabeled oligonucleotide spanning the entire IFN-β promoter and bearing a single AzP group at the 5′ half-site of PRDIV was used as a probe, and protein-DNA complexes were resolved by SDS-PAGE. The proteins included in the binding reaction prior to photo-cross-linking are indicated above the lanes, and relative positions of the proteins and the AzP group within the IFN-β enhancer are indicated schematically. The same experiment was performed using an IFN-β enhancer with AzP at the 3′ half-site of PRDIV (C), using an IFN-β enhancer with the PRDIV reverse mutation and AzP at the 5′ half-site (D), and using an IFN-β enhancer with the PRDIV reverse mutation and AzP at the 3′ half-site (E).
Approximately 100 pmol of DNA was then used in binding reactions with the recombinant proteins. The binding reaction mixture contained 10 mM Tris HCl (pH 7.5), 50 mM NaCl, 1 mM EDTA, 5% glycerol, 0.1% NP-40, and 100 μg of bovine serum albumin (BSA)/ml. All proteins were titrated on the DNA probes to ensure minimally saturating conditions as determined by electrophoretic mobility shift assay (B. S. Parekh and T. Maniatis, unpublished data). After incubation of the proteins and probes for 20 min, reactions were exposed to UV radiation on ice for 5 min with an Ultra-Lum UVC-515 UV multilinker set at 310 nm; wavelengths below 300 nm were filtered out using a polystyrene culture dish. After the cross-linking reaction, loading buffer containing sodium dodecyl sulfate (SDS), formamide, and bromophenol blue was added, and reactions were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on 10% gels with molecular weight markers (Bio-Rad). Gels were dried and visualized by autoradiography.
Protein-protein interaction assays.
GST fusion expression vectors for ATF-2195 and c-jun (15) and the series of GST–ATF-2 constructs used in the ATF-2 deletion studies, GST–ATF-2(1-505), GST–ATF-2(1-349), GST–ATF-2(350-505), and GST–ATF-2(350-415) (32), have been described previously. GST and GST fusion proteins were expressed in E. coli BL21 cells, purified as recommended by the manufacturer (Pharmacia), and dialyzed against 1× phosphate-buffered saline–0.1% Triton X-100–10% glycerol. 35S-labeled full-length or truncated derivatives of hexahistidine-tagged IRF-3 protein were synthesized by in vitro transcription-translation of pcDNA constructs in rabbit reticulocyte lysate using a TnT kit (Promega). GST or the GST fusion proteins were immobilized to glutathione-agarose beads and incubated with in vitro-translated 35S-labeled proteins for 2 h at 4°C in binding buffer (50 mM KCl, 20 mM Tris [pH 8.0], 0.2% NP-40, 50 μg of ethidium bromide/ml, 5 mM dithiothreitol, 0.2% BSA), followed by two washes with binding buffer and two washes with binding buffer without BSA. Interactions were then analyzed by SDS-PAGE on 10% gels.
ChIP assays.
The ChIP assays were modified from the original protocol (38, 61) as follows. HeLa cells were seeded at 2.5 × 106 cells per 10-cm-diameter plate (eight plates per condition, uninduced and virus induced), grown to approximately 60% confluency, and transfected with 12 μg of −110 IFN-β SEAP and 12 μg of −110 IFN-β PRDIV reverse CAT per plate as described above. After induction with Sendai virus for 6 h, cells were fixed by adding formaldehyde to 1% (final concentration; diluted from a 10% stock in 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, and 50 mM HEPES [pH 8.0]) for 30 min at 37°C, followed by addition of glycine to 125 mM (final concentration) to quench the reaction. Fixed cells were washed twice in phosphate-buffered saline, harvested, and resuspended in 10 ml of ice-cold lysis buffer (0.25% Triton X-100, 0.5% NP-40, 10 mM EDTA, 0.5 mM EGTA, 10 mM Tris HCl [pH 8.0], 1 mM phenylmethylsulfonyl fluoride [PMSF]) for 10 min. Nuclei were pelleted by centrifugation and washed successively at room temperature in 0.2 mM NaCl–1 mM EDTA–0.5 mM EGTA–10 mM Tris HCl–1 mM PMSF and in 1 mM EDTA–0.5 mM EGTA–10 mM Tris HCl–1 mM PMSF. Samples were then sonicated 10 times with a 20-s constant burst using a Branson Sonifier 450. Supernatants were then purified by using a CsCl gradient and dialysis with 1 mM EDTA–0.5 mM EGTA–10 mM Tris HCl–5% glycerol. Approximately 50 μg of purified chromatin samples was immunoprecipitated with 1 μg of anti-ATF-2, anti-c-jun, anti-NF-κB p50, anti-NF-κB p65 (Santa Cruz Biotechnology), or anti-IRF-3 (SL14) (61). DNA isolated from immunoprecipitated material following reversal of formaldehyde cross-linking was amplified by PCR (cycles of 1 min each at 94, 60, and 72°C) using T7 and CAT reverse primer or primers flanking the pSEAP2 multiple cloning site (Clontech). PCR cycles were titrated to ensure that amplification was in the linear range. Formaldehyde cross-linking was reversed for 50 μg of purified chromatin as a control for amplification of the transfected DNA by PCR. PCR products were resolved on 1.5% agarose gels and visualized using a Stratagene Eagle Sight imaging system.
RESULTS
The orientation of an asymmetric ATF-2–c-jun site is required for virus induction of the IFN-β enhancer.
The central DNA sequence of PRDIV contains an asymmetric ATF/CREB site, which upon virus induction is bound by a heterodimer of ATF-2 and c-jun (14, 15, 61). The core ATF-2–c-jun binding site, 5′-ATGACATAGG-3′, consists of a half-site (underlined) identical to that of the consensus CRE, 5′-ATGACGTCAT-3′ (21), and a nonconsensus half-site. We previously speculated that c-jun and ATF-2 might interact specifically with the consensus and nonconsensus half-sites of PRDIV (17), because both proteins can bind the CRE as homodimers but only ATF-2 can bind PRDIV as a homodimer (13, 15). To test the functional importance of the relative positions of the CRE consensus and nonconsensus half-sites, we reversed the orientation of the core ATF-2–c-jun site in PRDIV in the context of the IFN-β enhancer fused to the CAT gene. All mutations were made within the central eight base pairs of the site, preserving the flanking sequences (Fig. 1B). Reversal of the site results in a sharp decrease of virus induction of the IFN-β reporter gene (Fig. 1C), even though it does not impair binding of recombinant ATF-2–c-jun heterodimer to the reporter (see Fig. 4 and 5; J. V. Falvo and T. Maniatis, unpublished data). It is important to note that this decrease is not the consequence of the T-to-C mutation near the HMGI(Y) binding site at the 5′ end of PRDIV in the reverse construct. An IFN-β enhancer reporter construct bearing a G-to-A mutation at position −91 exhibits wild-type inducibility by virus (Fig. 1C) (14), and reversal of the site abrogates inducibility while preserving the 5′ PRDIV sequence (Fig. 1C).
FIG. 5.
The DNA-binding domain of IRF-3 fixes the orientation of ATF-2–c-jun at PRDIV in vitro. (A) Photo-cross-linking of ATF-2 and c-jun to the PRDIV site of the IFN-β enhancer in the presence or absence of other components of the enhanceosome using an IFN-β enhancer probe with the AzP group at the 3′ half-site of PRDIV as in Fig. 3C, using a GST fusion protein containing the DNA-binding domain of IRF-3 (amino acids 1 to 133). The same experiment was performed using an IFN-β enhancer with the PRDIV reverse mutation and the AzP at the 5′ half-site (B). (C) Photo-cross-linking of ATF-2 and c-jun to a minimal PRDIV site in the absence (lanes 1 to 10) or presence (lanes 11 to 20) of an intact 3′ IRF binding site from PRDIII. Radiolabeled oligonucleotides with AzP at the 5′ half-site (lanes 1 to 5 and 11 to 15) or 3′ half-site (lanes 6 to 10 and 16 to 20) were used as probes.
Substitution of the ATF-2–c-jun site in PRDIV with a consensus CRE site in the context of the IFN-β enhancer results in a marked increase in both basal and virus-induced levels of activity (Fig. 1C). This is consistent with the observations that single base substitutions in PRDIV that result in a more CRE-like sequence result in higher levels of virus induction of IFN-β reporters (14) and that ATF-2–c-jun binds with higher affinity to the CRE than to PRDIV (13). Furthermore, replacement of the PRDIV ATF-2–c-jun site with an AP-1 site results in lower levels of virus inducibility (Fig. 1C), which is consistent with the lower affinity of ATF-2–c-jun for the AP-1 site compared to the CRE site (19).
The asymmetry of the ATF-2–c-jun site within PRDIV is a feature observed in functional ATF-2–c-jun binding sites in the promoter regions of a number of genes, including c-jun (58), the urokinase gene (10), and the E-selectin gene (46). In addition, the palindromic ATF-2–c-jun binding site in the tumor necrosis factor alpha (TNF-α) promoter (TNF-α CRE) varies from the consensus CRE at its central dinucleotide (37, 56). The ATF-2–c-jun binding sites from the c-jun (cjun2), urokinase, and E-selectin gene promoters, but not the TNF-α CRE, are all functional in the context of the IFN-β enhancer, resulting in wild-type levels of virus induction (Fig. 1C). Notably, the sites from the c-jun and urokinase genes are not functional when placed in the opposite orientation, while the E-selectin site remains functional in reverse orientation (Fig. 1C). None of the mutations impaired binding of ATF-2–c-jun to the promoter (Falvo and Maniatis, unpublished). When the sites tested in the context of the IFN-β enhancer are examined, a correlation emerges between the polarity of the ATF-2–c-jun site, including the central dinucleotide, and virus induction of the reporter. First, in the functional reporters a CRE core half-site (5′-TGA-3′) is present at the 5′ position. In the case of the ATF-2–c-jun sites in PRDIV, PRDIV G(−91)A, and cjun2, the CRE core half-site is in the 5′ position and a nonconsensus core half-site is in the 3′ position, and reversal of these sites results in a nonfunctional reporter (Fig. 1B). When both half-sites are CRE core half-sites, as is the case of the urokinase, E-selectin, and TNF-α ATF-2–c-jun sites, the central dinucleotide appears to be a critical determinant of function. For all of the sites examined, functional reporters contained purine-purine or pyrimidine-purine, but not pyrimidine-pyrimidine or purine-pyrimidine, as the central dinucleotide (Fig. 1B). Thus, the polarity of ATF-2–c-jun sites in the context of the IFN-β enhancer is a critical determinant of virus inducibility.
PRDIV and PRDIII-I function as a composite element in vivo.
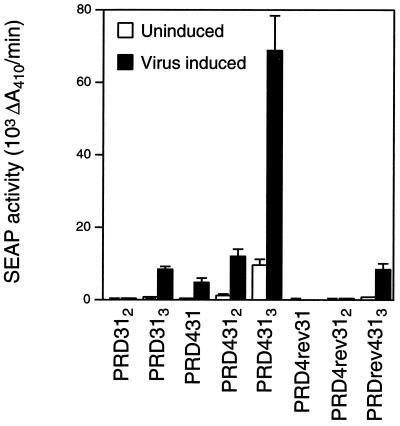
The PRDIV site and the neighboring PRDIII-I site are, in essence, a composite site for ATF-2–c-jun, HMGI(Y), and IRF-3 or IRF-7. Insertion of half a helical turn of DNA between PRDIV and PRDIII-I abolishes virus inducibility of the IFN-β enhancer, while insertion of a full helical turn of DNA preserves inducibility (55). To characterize the function of the PRDIV and PRDIII-I composite site (PRD431) in vivo, one to three copies of PRD431 containing PRDIV in the forward or reverse orientation were placed upstream of the E1b TATA box minimal promoter (29) driving expression of the SEAP gene. The activity of these constructs was compared to that of isogenic reporters driven by two or three copies of the PRDIII-I site (61). Multimers of PRD431 were strongly virus inducible, while multimers of PRD4rev31 were much less inducible (Fig. 2). Thus, both in the context of the IFN-β enhancer and as a reiterated element before a heterologous promoter, PRDIV and PRDIII-I function as a composite virus-inducible element, critically dependent on PRDIV orientation.
FIG. 2.
PRDIV and PRDIII-I function as a composite element in vivo. SEAP activity in media from uninduced and Sendai virus-induced HeLa cells transfected with the indicated reporters is shown. Histograms display the average results of three independent experiments, and error bars indicate the standard error of the mean. Activity of SEAP reporters containing one to three copies of PRD31, PRD431, or PRD4rev31 is indicated.
Selective interactions between ATF-2 and IRF-3 are mediated by the bZIP region of ATF-2.
Given the orientation-dependent functional cooperativity between the ATF-2–c-jun and IRF binding sites in PRDIV and PRDIII-I, we examined the possibility that ATF-2 and c-jun interact directly with IRF proteins using GST association assays. We tested two IRF proteins previously implicated in IFN-β regulation, IRF-1 and IRF-3 (35). IRF-1 activates the IFN-β promoter in a cooperative fashion with ATF-2–c-jun, NF-κB p50-p65, and HMGI in cotransfection experiments (55) and in vitro (24). The requirement of IRF-3 for virus induction of the IFN-β gene in vivo has recently been demonstrated by several laboratories (30, 49, 50, 61, 67).
Full-length IRF-1 interacted to a similar extent with ATF-2 and c-jun, while full-length IRF-3 did not interact with ATF-2 or c-jun (Fig. 3A). IRF-3 contains a C-terminal autoinhibitory domain; relief of this intramolecular interaction by phosphorylation or truncation exposes its DNA-binding and protein-protein interaction domains (31, 61). Indeed, a truncated form of IRF-3 lacking this C-terminal domain interacts with ATF-2 but, strikingly, not with c-jun. It is worth noting that when we compared the amount of retained IRF protein to input, the interaction between IRF-3 and ATF-2 was consistently stronger (10 to 11% of input) than the interaction between IRF-1 and ATF-2 or c-jun (3 to 5% of input [Fig. 3A]).
FIG. 3.
Protein-protein interactions between ATF-2–c-jun and IRF proteins. (A) Selective interaction between IRF-3 and ATF-2. In vitro-translated, 35S-labeled, hexahistidine-tagged IRF proteins were incubated with GST, GST–ATF-2 (murine ATF-2195 splice variant), or GST–c-jun immobilized on glutathione-agarose beads. Protein that remained bound following washing is shown with 20% of the total 35S-labeled fraction for comparison. (B) The bZIP region of ATF-2 is necessary and sufficient for interaction with IRF-3. GST association assays were performed as for panel A, with GST fusion proteins containing full-length human ATF-2 (amino acids 1 to 505; lane 3) or ATF-2 with the indicated deletions (lanes 4 to 6). These results are summarized schematically in panel C.
To further examine interactions between the activators bound at PRDIV and PRDIII-I, we mapped the region of ATF-2 that interacts with IRF-3 (Fig. 3B and C). As expected, the truncated IRF-3 protein interacted with full-length ATF-2 (Fig. 3B, lane 3). Strikingly, deletion of the ATF-2 bZIP region abrogated interactions between ATF-2 and IRF-3 (Fig. 3B, lane 4), while GST fusion proteins containing as few as 66 amino acids of the bZIP region retained the ability to interact with IRF-3 (Fig. 3B, lanes 5 and 6). Thus, the DNA-binding domain of ATF-2 mediates its interaction with IRF-3.
IRF-1 and binding site orientation fix the orientation of ATF-2–c-jun heterodimer bound to the IFN-β enhancer.
The experiments described above demonstrate specific ATF-2–c-jun-IRF interactions as well as the critical influence of ATF-2–c-jun binding site orientation on virus inducibility of the IFN-β enhancer. We next examined the effect of binding site orientation on the position of the ATF-2–c-jun heterodimer itself, using recombinant proteins. We used a protein-DNA photo-cross-linking method in which a reactive AzP group is introduced into oligonucleotides at specific positions in the phosphate backbone (27, 63). Recombinant proteins were incubated with modified, radioactively labeled DNA probes and exposed to UV light, and the cross-linked protein-DNA complexes were resolved by SDS-PAGE and visualized by autoradiography. The suitability of this technique was confirmed by incorporating AzP groups at the 5′ or 3′ half-site of a PRDII oligonucleotide and examining the photo-cross-linked product after incubation with recombinant NF-κB p50-p65 heterodimer. As expected from earlier photo-cross-linking experiments with 5-bromodeoxyuridine-substituted PRDII oligonucleotides (57), p50 bound specifically to the 5′ half-site and p65 bound specifically to the 3′ half-site (Fig. 4A). Oligonucleotide probes consisting of the entire IFN-β enhancer, with wild-type sequence or with a reversed ATF-2–c-jun site at PRDIV, were synthesized to contain phosphorothioate substitutions at positions flanking PRDIV at the 5′ or 3′ position. A set of recombinant proteins, including those sufficient for enhanceosome assembly and activation of the IFN-β gene in vitro (24), were prepared: NF-κB p50 homodimer, p50-p65 heterodimer, and p65 homodimer; IRF-1; ATF-2 homodimer, c-jun homodimer, and ATF-2–c-jun heterodimer; and HMGI. We confirmed that ATF-2 homodimer and ATF-2–c-jun heterodimer, but not c-jun homodimer, bind to PRDIV (Parekh and Maniatis, unpublished).
The distinct molecular weights of the cross-linked ATF-2-DNA and c-jun–DNA complexes provided a rapid and sensitive method of observing the effects of other IFN-β enhanceosome components upon ATF-2–c-jun heterodimer orientation (Fig. 4B to E). Surprisingly, ATF-2–c-jun bound to PRDIV with almost no specific orientation, although the CRE consensus half-site was slightly favored by c-jun, regardless of its orientation within the IFN-β promoter (Fig. 4B to E, lanes 4). We next examined the effects of other components of the enhanceosome upon the orientation of ATF-2–c-jun. HMGI, NF-κB p50, NF-κB p65, and NF-κB p50-p65 failed to influence the binding orientation of ATF-2–c-jun (Figs. 4B to E, lanes 5 to 8). IRF-1, however, was sufficient to fix the orientation of ATF-2–c-jun such that regardless of the orientation of PRDIV in the probe, c-jun was bound exclusively to the consensus half-site and ATF-2 was bound to the nonconsensus half-site (Fig. 4B to E, lanes 9). This is consistent with the ability of IRF-1 to interact with both ATF-2 and c-jun (Fig. 3A). In complementary experiments, the ATF-2–c-jun heterodimer bound in a fixed orientation in the presence of HMGI, IRF-1, and p50-p65, and removal of each component, with the exception of IRF-1, did not abolish this orientation (Figs. 4B and C, lanes 10 to 13; Fig. 4D and E, lanes 10 to 15). Thus, the combined effects of the orientation of the asymmetric PRDIV site and the presence of IRF-1 protein are critical for fixing the orientation of ATF-2–c-jun in the context of the IFN-β enhanceosome in vitro.
Interaction of the IRF-3 DNA-binding domain with PRDIII fixes the orientation of the ATF-2–c-jun heterodimer at PRDIV.
We next determined if IRF-3, like IRF-1, is necessary and sufficient to fix the orientation of ATF-2–c-jun bound to the IFN-β enhancer in vitro. A recombinant GST–IRF-3 fusion protein (50) was substituted for IRF-1 in the photo-cross-linking assays. Strikingly, IRF-3 is also necessary and sufficient to fix the orientation of ATF-2–c-jun on the wild-type IFN-β enhancer probe (Fig. 5A, lanes 4 to 15). In the wild-type orientation, the nonconsensus half-site of PRDIV is adjacent to the IRF site, so ATF-2 is proximal to IRF-3. When PRDIV is placed in reverse orientation in the context of the IFN-β enhancer, however, IRF-3 cannot fix the orientation of ATF-2–c-jun (Fig. 5B, lanes 4 to 14). This is consistent with the observations that IRF-3, unlike IRF-1, selectively interacts with ATF-2 (Fig. 3A) and that IRF-3 and ATF-2–c-jun bind cooperatively to the wild-type IFN-β enhancer but not to the IFN-β enhancer with the PRDIV reverse mutation (T. H. Kim and T. Maniatis, unpublished data).
Our experiments with recombinant IRF-3 utilized the DNA-binding domain of the protein (amino acids 1 to 133), which when expressed by stable transfection is recruited along with the endogenous components of the IFN-β enhanceosome during virus infection (40). Thus, we have established that the DNA-binding domain of IRF-3 mediates its ability to fix the orientation of ATF-2–c-jun at PRDIV. The cross-linking experiments also indicated that binding of IRF-3 to DNA was required to establish a fixed binding orientation of ATF-2–c-jun, as the presence of IRF-3 alone was not sufficient for fixing ATF-2–c-jun orientation. This is of note because some proteins can influence the DNA-binding properties of bZIP transcription factors by protein-protein interactions alone, such as hepatitis B virus X protein (34) and human T-cell lymphotropic virus type 1 Tax protein (60). To examine this in detail, we performed photo-cross-linking experiments with a minimal PRDIV oligonucleotide with point mutations in the 3′ IRF binding site (Fig. 5C, lanes 1 to 10) and with a PRDIV oligonucleotide containing an intact IRF binding site (Fig. 5C, lanes 11 to 20). Remarkably, while IRF-3 fails to fix ATF-2–c-jun heterodimer binding orientation to PRDIV in the absence of an IRF binding site (Fig. 5C, compare lanes 4 and 5 and lanes 9 and 10), the addition of an IRF binding site restores the ability of IRF-3 to fix ATF-2–c-jun binding orientation (Fig. 5C, compare lanes 14 and 15 and lanes 19 and 20). Thus, the ability of IRF-3 to determine the orientation of ATF-2–c-jun bound at PRDIV depends on the binding of IRF-3 to the adjacent IRF binding site at PRDIII.
IRF-1 and IRF-3 activate PRD431 with differential dependence on ATF-2–c-jun binding site orientation.
Our in vitro cross-linking experiments with IRF-1 and the IRF-3 DNA-binding domain indicate that while ATF-2–c-jun forms an oriented complex with IRF-1 on either the wild-type or PRDIV-reverse IFN-β promoter, this orientated complex only forms on the wild-type promoter with IRF-3. This is, in turn, consistent with the observations that ATF-2–c-jun and IRF-3 bind cooperatively to the wild-type but not PRDIV-reverse IFN-β promoter (Kim and Maniatis, unpublished), while binding of ATF-2–c-jun and IRF-1 to the IFN-β promoter is not cooperative (55). These in vitro distinctions in interactions between ATF-2–c-jun and the IRF proteins might be reflected in IRF-dependent transactivation of the PRD431 composite element; thus, we next examined the effects of ATF-2–c-jun binding site orientation on transactivation mediated by full-length IRF-1 and IRF-3 in vivo.
To uncouple IRF-1- and IRF-3-dependent transactivation from other virus-mediated activation pathways in vivo, we transfected full-length IRF-1 and IRF-3 into P19 embryonal carcinoma cells. These cells do not express IRF proteins (20) and have been used to study IFN-β promoter-dependent activation in response to exogenous activators (54, 55). IRF-3 contains a C-terminal autoinhibitory domain which is phosphorylated in response to virus infection (30, 31, 61), so in these experiments we expressed a constitutively nuclear form of IRF-3 containing phosphomimetic glutamic acid substitutions (Lin, Wathelet, and Maniatis, unpublished).
Transfection of increasing amounts of both IRF-1 (Fig. 6A) and IRF-3 (Fig. 6B) activates a minimal reporter consisting of two copies of PRD431. IRF-1 activates two copies of PRD4rev31 to a lesser extent than that achieved with two copies of PRD431; IRF-3, however, essentially fails to activate two copies of PRD4rev31 (compare Fig. 6A to Fig. 6B). Thus, IRF-3 activates the PRD431 composite element only when the ATF-2–c-jun binding site is in the correct orientation, while IRF-1 retains the ability to activate PRD431 if the ATF-2–c-jun binding site is in the reverse orientation, albeit to a lesser extent.
FIG. 6.
Activation of PRD431 and PRD4rev31 by IRF-1 and IRF-3. SEAP activity (absorbance at 410 nm after 16 h of incubation at 37°C) in media from murine P19 embryonal carcinoma cells cotransfected with PRD4312SEAP or PRD4rev312SEAP reporters is shown. Empty vector (pcDNA1, 300 ng) or increasing amounts (10, 30, 100, or 300 ng) of IRF-1 (A) or IRF-3 (B) were cotransfected with 30 ng of ATF-2–c-jun expression vector; the amount of DNA was kept constant with additional pcDNA1. Histograms display the average results of three independent experiments, and the error bars indicate the standard error of the mean.
A correctly oriented ATF-2–c-jun heterodimer binding site is required to recruit IRF-3 to the IFN-β promoter in vivo.
The ChIP assay has been used to detect the binding of endogenous and stably expressed exogenous transcription factors to the endogenous IFN-β promoter (40, 61). The assay can also be used to detect the binding of endogenous or exogenous factors to transiently transfected reporter plasmids (25, 33). To test the effect of reversal of the PRDIV ATF-2–c-jun core element in vivo, we cotransfected wild-type (−110 IFN-β SEAP) and PRDIV-reverse (−110 IFN-β PRDIVrev CAT) IFN-β reporters into HeLa cells and performed ChIP assays on chromatin samples from uninduced and virus-induced cells. Because the PCR products for the wild-type and mutant reporter are amplified from the same immunoprecipitated material, this permits direct comparison of transcription factor binding to each construct before and after virus infection in vivo; relative levels of amplified product provide a readout for binding. Control PCRs were performed with buffer (Fig. 7, lanes 2 and 3) and with genomic DNA from untransfected HeLa cells (Fig. 7, lanes 4 and 5) or from the transfected HeLa cells used in the immunoprecipitations (Fig. 7, lanes 6 to 9).
FIG. 7.
ATF-2–c-jun binding site orientation-dependent recruitment of IRF-3 to the IFN-β promoter in vivo. HeLa cells were transiently cotransfected with a SEAP reporter containing the wild-type (WT) IFN-β promoter (−110 IFN-β SEAP) and with a CAT reporter containing the PRDIV reverse mutant IFN-β promoter (−110 IFN-β PRDIVrev CAT) and were left uninduced or treated with Sendai virus. Following formaldehyde cross-linking and chromatin purification, DNA immunoprecipitated with the indicated antibodies was amplified by PCR using primers specific for −110 IFN-β SEAP or −110 IFN-β PRDIVrev CAT as indicated (lanes 10 to 29). PCR product amplification from immunoprecipitated material with specific primers correlates with protein binding to the indicated reporter. A 123 bp ladder (Life Technologies) is included as a marker (lane 1). Control PCRs with buffer or genomic DNA from untransfected and transfected HeLa cells are included as controls (lanes 2 to 9).
As shown in Fig. 7, ATF-2 (lane 11), c-jun (lane 15), IRF-3 (lane 19), and NF-κB p50 and p65 (lanes 23 and 27) associate with transfected wild-type IFN-β promoter following virus infection, as is the case with the endogenous gene (40, 61). Binding of NF-κB p50 is not apparent with the transfected promoter prior to virus infection (Fig. 7, lane 22), in contrast to the endogenous promoter (40, 61), possibly reflecting the influence of endogenous chromatin structure and/or the high levels of reporter DNA present following transient transfection. Strikingly, after virus infection every transcription factor except for IRF-3 (Fig. 7, lane 21) associates with the transfected IFN-β promoter containing a reversed PRDIV ATF-2–c-jun site. This demonstrates that the orientation of the ATF-2–c-jun binding site is critical for recruitment of IRF-3 to the IFN-β promoter upon virus infection.
DISCUSSION
Heterodimerization of transcription factors plays a critical role in the regulation of eukaryotic gene transcription, providing a wide range of combinations of factors for temporal and stimulus-specific patterns of gene expression. Indeed, homodimers and heterodimers may have opposite effects on gene expression, as illustrated by the positive and negative effects of the c-jun homodimer and the c-fos–c-jun heterodimer, respectively, on the function of a composite glucocorticoid response element (11). At the level of DNA recognition, binding site affinity or site specificity of a heterodimeric complex may differ from that of a homodimeric complex. In some cases, heterodimerization either permits DNA binding, as is the case with dimerization of myc with Max (2) and c-fos with c-jun (43), or prevents DNA binding, as is the case with Id proteins heterodimerizing with E-box proteins (1).
Heterodimerization can also modulate affinity for a single binding site: dimerization of c-jun with c-fos or with ATF-2 specifies binding to the AP-1 or the CRE site, respectively (19), and dimerization of c-jun with ATF-2 permits recognition of PRDIV by c-jun (15). Half-site spacing and asymmetry within a DNA-binding site can fix the binding orientation of its cognate transcription factor. This is elegantly illustrated by the binding of different heterodimeric combinations of nuclear hormone receptors to specific response elements, such that spacing between direct repeats determines the compositions of the bound heterodimer (45). DNA-directed binding orientation is also observed in the binding polarity of the NF-κB p50-p65 heterodimer, which is dictated by half-sites in the κB motif (5, 54, 57). The structural flexibility of sequences flanking the AP-1 site has been linked to the orientation of bound c-fos–c-jun heterodimer (28). Furthermore, heterodimer orientation can be directed by the binding of another protein. A well-characterized example of this is the complex formed between NF-AT and the c-fos–c-jun heterodimer at a composite NF-AT–AP-1 site in the interleukin-2 promoter, the distal antigen-receptor response element ARRE2 (44). Through extensive contacts between the DNA-binding domains of the two proteins, NF-AT recruits c-fos–c-jun to the ARRE2 site in a fixed orientation (6, 7, 12). Orientation of c-fos–c-jun within the NF-AT–AP-1 complex may be important for the establishment of enhanceosomes at the interleukin-2 promoter and promoters of other genes regulated by NF-AT–AP-1 elements, but the details of such higher-order complexes remain unclear (44, 47).
DNA-binding-site orientation has been associated with the transactivation potential of cognate heterodimeric transcription factors. For example, the orientation of reiterated response elements for a vitamin D3 receptor-retinoid X receptor heterodimer was shown to have a strong influence on ligand-dependent induction of transcription (51). Through the use of a yeast reporter system, transcriptional activation mediated by c-fos–c-jun bZIP domains fused to heterologous activation domains and engineered to bind in a fixed orientation to AP-1 sites was also shown to be dependent on binding site orientation (8). In the human immunodeficiency virus type 1 long terminal repeat, the orientation of a κB site adjacent to an Sp1 site is critical for induction of the long terminal repeat by phorbol-12-mystrate-13-acetate or TNF-α, consistent with preferential interactions between Sp1 and the NF-κB p65 subunit (41, 42). Similarly, in the IFN-β enhancer the orientation of the NF-κB p50-p65 binding site, PRDII, is critical for induction by virus (17).
Here, using in vitro and in vivo methods, we have shown that the orientation of a heterodimeric transcription factor determines assembly and function of the IFN-β enhanceosome. We have shown that the orientation of the ATF-2–c-jun binding site at PRDIV is critical for virus induction of the IFN-β gene and for the function of the ATF-2–c-jun–IRF composite element. Unlike the examples described above, however, the orientation of ATF-2–c-jun at PRDIV is not directed solely by binding site sequence or by the influence of a protein bound at an adjacent site. Instead, the combined effect of the polarity of the ATF-2–c-jun binding site at PRDIV and IRF protein bound at PRDIII establishes an ATF-2–c-jun–IRF nucleoprotein complex in which the ATF-2–c-jun heterodimer adopts a fixed orientation. When the ATF-2–c-jun–IRF complex contains IRF-1, which interacts with both ATF-2 and c-jun, the heterodimer can be fixed in either orientation depending on ATF-2–c-jun binding site polarity. On the other hand, if the complex contains IRF-3, which interacts selectively with ATF-2, the oriented complex forms only when the ATF-2–c-jun binding site is in the wild-type orientation.
This is consistent with our in vivo results with exogenous IRF-1 and IRF-3; IRF-1 can activate a PRD431 reporter when PRDIV is in either orientation, while IRF-3 can only activate the PRD431 reporter when PRDIV is in the wild-type orientation. This indicates that a functional complex between ATF-2–c-jun and IRF-3 fails to form at PRD431 when PRDIV is in the reverse orientation, potentially from inhibition of IRF-3 binding by the configuration of this composite site. This possibility is strongly supported by our ChIP assays with wild-type and mutant IFN-β reporters, which show that recruitment of IRF-3 to the IFN-β promoter in vivo depends on ATF-2–c-jun binding site orientation. Thus, reversal of ATF-2–c-jun binding site orientation in the context of the IFN-β enhancer abolishes recruitment of IRF-3 in vivo and results in reversed ATF-2–c-jun heterodimer orientation in the presence of IRF-1 in vitro (Fig. 8A). The relative positions of transcription factor activation domains are critical for recruitment of CBP and p300 proteins to the IFN-β promoter; thus, a specific activation domain surface appears to be required for correct interactions between transcription factors and coactivators in the context of the IFN-β enhanceosome (36). Reversal of the ATF-2–c-jun binding site at PRDIV strongly inhibits transcriptional activation mediated by the IFN-β promoter in vivo by virus infection, as we have shown, or by recombinant NF-κB, ATF-2–c-jun, and IRF-1 in vitro (24). A critical effect of reversal of the ATF-2–c-jun binding site thus appears to be alteration of the activation domain surface of the IFN-β enhanceosome, whether by a lack of IRF-3, which in turn fails to fix the orientation of ATF-2–c-jun, or by reversal of the orientation of ATF-2–c-jun in the presence of IRF-1. Reversal of ATF-2–c-jun orientation in the presence of IRF-1 may impair the ability of these factors to recruit coactivators even at the isolated PRD431 element, as we have shown that exogenous IRF-1 activates a PRD431 reporter more efficiently than a PRD4rev31 reporter.
FIG. 8.
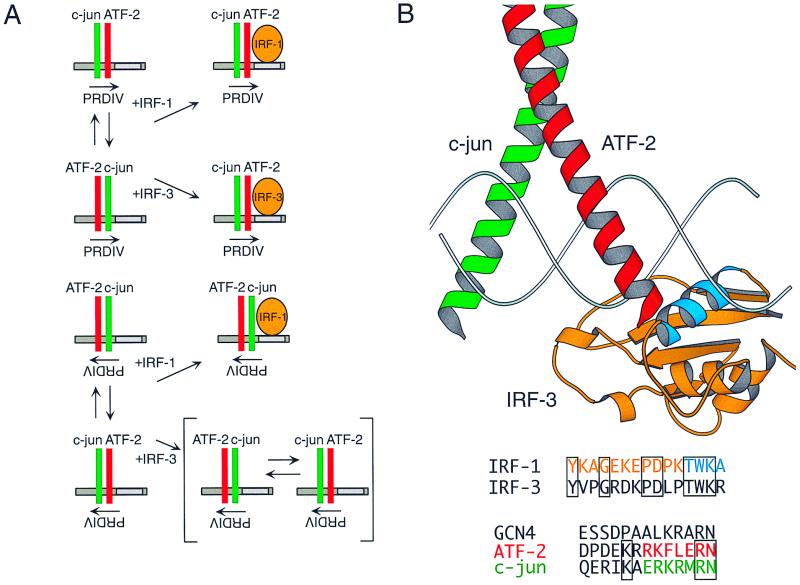
Model of ATF-2–c-jun–IRF complex formation at the IFN-β PRDIV and PRDIII elements. (A) Summary of the effect of IRF protein binding on ATF-2–c-jun heterodimer binding orientation. In the absence of IRF protein, ATF-2 (red) and c-jun (green) do not adopt a fixed orientation at PRDIV. In the presence of IRF-1, the position of the ATF-2–c-jun heterodimer becomes fixed such that ATF-2 is bound to the nonconsensus half-site and c-jun is bound to the nonconsensus half-site, regardless of whether PRDIV is in the wild-type or reverse orientation. In the presence of IRF-3, ATF-2–c-jun orientation is fixed only when PRDIV is in the forward orientation, and in vivo only the wild-type binding site orientation permits recruitment of IRF-3 to the IFN-β promoter. (B) Three-dimensional model of the ATF-2–c-jun–IRF-3 complex bound to the IFN-β promoter. The DNA-binding domains of IRF-1 bound to PRDI (16) and of the GCN4 homodimer bound to a CRE site (22) were used to model IRF-3 (orange) bound to PRDIII and ATF-2 (red) and c-jun (green) bound to PRDIV, respectively. The DNA recognition helix of IRF-3, which makes a close approach to ATF-2, is shown in cyan. The sequence from the amino acids in IRF-1 in this helix are shown in cyan, and those in the neighboring loop are shown in orange, along with the corresponding IRF-3 sequence. The N-terminal amino acid sequence of the GCN4 bZIP region and the corresponding amino acids from ATF-2 (starting with amino acid 350) and c-jun are also shown for comparison; green and red amino acids correspond to the positions found at the N terminus of the helix in the diagram. The figure was prepared using the program MOLSCRIPT (26).
Notably, we have shown that the DNA-binding domain of IRF-3 is sufficient to form an oriented ATF-2–c-jun–IRF complex in vitro. Consistent with this, the IRF-3 DNA-binding domain is recruited to the IFN-β promoter along with endogenous ATF-2–c-jun in the apparent absence of CBP and p300, based on lack of localized histone hyperacetylation (40). This further illustrates the importance of interactions among transcription factor DNA-binding domains in the assembly of the IFN-β enhanceosome. Recruitment of IRF-3 to the IFN-β promoter by ATF-2–c-jun and interactions between the DNA-binding domains of these factors draw an interesting parallel to recruitment of IRF-4 (also known as Pip, LSIRF, NF-EM5, or ICSAT) to B-cell-specific enhancers by the ETS domain protein PU.1. The binding of phosphorylated PU.1 permits the formation of a stable ternary complex with IRF-4 on DNA (3). Phosphorylation-independent interaction between the DNA-binding domains of the two proteins contributes to the stability of the ternary complex, and correct spacing between the IRF and ETS binding sites is critical for complex formation (3, 64). A critical interaction, however, occurs between the phosphorylated PEST domain of PU.1 and a putative alpha-helical region in IRF-4, which in the absence of PU.1 inhibits IRF-4 binding to DNA (3, 39).
Using protein-DNA co-crystal structures of related IRF and bZIP proteins, we have constructed a model of the ATF-2–c-jun–IRF-3 complex bound to the IFN-β enhancer (Fig. 8B). The DNA-binding domain of IRF-1 (16) was placed at the 5′-GAAA-3′ sequence in PRDIII. Given the strongly enhanced inducibility of the IFN-β enhancer when CRE was substituted for PRDIV (Fig. 1C), the structure of GCN4 homodimer bound to CRE (22) was placed at the sequence 5′-TGACATAG-3′. Based on our results, we have designated the bZIP subunits proximal and distal to IRF-3 as ATF-2 and c-jun, respectively. It is interesting that at the region of closest approach between the two proteins, the bZIP N terminus and the junction of the IRF DNA recognition helix and a neighboring loop, there are several differences in amino acid sequence between ATF-2 and c-jun and between IRF-1 and IRF-3. Differences may also exist in the overall structure of DNA-bound IRF-1 and IRF-3, as has been observed for IRF-1 and IRF-2 (18). Such structural differences may underlie the distinct features that we have observed in the interactions of IRF-1 and IRF-3 with ATF-2–c-jun. It is important to note that binding of IRF proteins at PRDIII does not simply fix an intrinsic preferential orientation of ATF-2–c-jun at PRDIV; fixing of intrinsic DNA binding orientation would result in the same distribution of orientations found in the absence of IRF proteins. Within the ATF-2–c-jun–IRF complex, interaction of IRF protein at PRDIII must further influence the properties of ATF-2–c-jun binding orientation at PRDIV, for example, through changes in protein structure induced by protein-protein contacts, alteration of DNA structure, or occlusion of DNA phosphate or base pair contacts. Structural analysis of the ATF-2–c-jun–IRF-3 complex bound to the PRDIV or PRDIII element is currently in progress and promises to reveal the details of these interactions at the atomic level. Given the complexity of many gene regulatory elements, the emerging role for enhanceosomes in transcription, and the array of potential heteromeric factors that function in eukaryotic gene regulation, the orientation of DNA-bound transcription factors as illustrated by the ATF-2–c-jun–IRF complex may have general importance in the establishment of functional higher-order nucleoprotein complexes.
ACKNOWLEDGMENTS
J.V.F. and B.S.P. contributed equally to this work.
We thank Paul Clemons for his work in the initial stages of the ATF-2–c-jun protein-DNA cross-linking experiments and Aseem Ansari for suggesting the AzP bromide photo-cross-linking technique. The IRF-3 E5 expression vector was prepared by C.H.L. and Marc Wathelet. We thank Michael Green and Paula Pitha for generously providing GST–ATF-2 and GST–IRF-3 expression vectors. We thank Tae Hoon Kim for critical reading of the manuscript and for sharing unpublished observations, and we thank Judith Grisham for editorial assistance. Special thanks go to Stephen C. Harrison, in whose laboratory E.F. is a postdoctoral fellow.
This work was supported by grant AI20642 from the NIH to T.M. J.V.F. acknowledges the support of a National Defense Science and Engineering Graduate Research Fellowship. E.F. is supported by the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation Fellowship, DRG 1547.
REFERENCES
- 1.Benezra R, Davis R L, Lassar A, Tapscott S, Thayer M, Lockshon D, Weintraub H. Id: a negative regulator of helix-loop-helix DNA binding proteins. Control of terminal myogenic differentiation. Ann N Y Acad Sci. 1990;599:1–11. doi: 10.1111/j.1749-6632.1990.tb42359.x. [DOI] [PubMed] [Google Scholar]
- 2.Blackwood E M, Eisenman R N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 3.Brass A L, Zhu A Q, Singh H. Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers. EMBO J. 1999;18:977–991. doi: 10.1093/emboj/18.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 5.Chen F E, Huang D-B, Chen Y-Q, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-κB bound to DNA. Nature. 1998;391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Glover J N M, Hogan P G, Rao A, Harrison S C. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Oakley M G, Glover J N M, Jain J, Dervan P B, Hogan P G, Rao A, Verdine G L. Only one of the two DNA-bound orientations of AP-1 found in solution cooperates with NFATp. Curr Biol. 1995;5:882–889. doi: 10.1016/s0960-9822(95)00178-3. [DOI] [PubMed] [Google Scholar]
- 8.Chytil M, Peterson B R, Erlanson D A, Verdine G L. The orientation of the AP-1 heterodimer on DNA strongly affects transcriptional potency. Proc Natl Acad Sci USA. 1998;95:14076–14081. doi: 10.1073/pnas.95.24.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen B R, Malim M H. Secreted placental alkaline phosphatase as a eukaryotic reporter gene. Methods Enzymol. 1992;216:362–368. doi: 10.1016/0076-6879(92)16033-g. [DOI] [PubMed] [Google Scholar]
- 10.De Cesare D, Vallone D, Caracciolo A, Sassone-Corsi P, Nerlov C, Verde P. Heterodimerization of c-Jun with ATF-2 and c-Fos is required for positive and negative regulation of the human urokinase enhancer. Oncogene. 1995;11:365–376. [PubMed] [Google Scholar]
- 11.Diamond M I, Miner J N, Yoshinaga S K, Yamamoto K R. Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science. 1990;249:1266–1272. doi: 10.1126/science.2119054. [DOI] [PubMed] [Google Scholar]
- 12.Diebold R J, Rajaram N, Leonard D A, Kerppola T K. Molecular basis of cooperative DNA bending and oriented heterodimer binding in the NFAT1-Fos-Jun-ARRE2 complex. Proc Natl Acad Sci USA. 1998;95:7915–7920. doi: 10.1073/pnas.95.14.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du W. Synergistic activation of the human interferon-β gene promoter. Ph.D. dissertation. Cambridge, Mass: Harvard University; 1993. [Google Scholar]
- 14.Du W, Maniatis T. An ATF/CREB binding site is required for virus induction of the human interferon β gene. Proc Natl Acad Sci USA. 1992;89:2150–2154. doi: 10.1073/pnas.89.6.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du W, Thanos D, Maniatis T. Mechanisms of transcriptional synergism between distinct virus-inducible enhancer elements. Cell. 1993;74:887–898. doi: 10.1016/0092-8674(93)90468-6. [DOI] [PubMed] [Google Scholar]
- 16.Escalante C R, Yie J, Thanos D, Aggarwal A K. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature. 1998;391:103–106. doi: 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- 17.Falvo J V, Thanos D, Maniatis T. Reversal of intrinsic DNA bends in the IFNβ gene enhancer by transcription factors and the architectural protein HMG I(Y) Cell. 1995;83:1101–1111. doi: 10.1016/0092-8674(95)90137-x. [DOI] [PubMed] [Google Scholar]
- 18.Fujii Y, Shimizu T, Kusumoto M, Kyogoku Y, Taniguchi T, Hakoshima T. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO J. 1999;18:5028–5041. doi: 10.1093/emboj/18.18.5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci USA. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harada H, Willison K, Sakakibara J, Miyamoto M, Fujita T, Taniguchi T. Absence of the type I IFN system in EC cells: transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell. 1990;63:303–312. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- 21.Karin M, Liu Z-G, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 22.Keller W, König P, Richmond T J. Crystal structure of a bZIP/DNA complex at 2.2 Å: determinants of DNA specific recognition. J Mol Biol. 1995;254:657–667. doi: 10.1006/jmbi.1995.0645. [DOI] [PubMed] [Google Scholar]
- 23.Kim T K, Kim T H, Maniatis T. Efficient recruitment of TFIIB and CBP-RNA polymerase II holoenzyme by an interferon-β enhanceosome in vitro. Proc Natl Acad Sci USA. 1998;95:12191–12196. doi: 10.1073/pnas.95.21.12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim T K, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-β enhanceosome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 25.Koipally J, Renold A, Kim J, Georgopoulos K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 1999;18:3090–3100. doi: 10.1093/emboj/18.11.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraulis P J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 27.Lagrange T, Kim T-K, Orphanides G, Ebright Y W, Ebright R H, Reinberg D. High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photocrosslinking: organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc Natl Acad Sci USA. 1996;93:10620–10625. doi: 10.1073/pnas.93.20.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leonard D A, Kerppola T K. DNA bending determines Fos-Jun heterodimer orientation. Nat Struct Biol. 1998;5:877–881. doi: 10.1038/2316. [DOI] [PubMed] [Google Scholar]
- 29.Lillie J W, Green M R. Transcription activation by the adenovirus E1a protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 30.Lin R, Heylbroeck C, Pitha P M, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin R, Mamane Y, Hiscott J. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol Cell Biol. 1999;19:2465–2474. doi: 10.1128/mcb.19.4.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu F, Green M R. Promoter targeting by adenovirus E1a through interaction with different cellular DNA-binding domains. Nature. 1994;368:520–525. doi: 10.1038/368520a0. [DOI] [PubMed] [Google Scholar]
- 33.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 34.Maguire H F, Hoeffler J P, Siddiqui A. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science. 1991;252:842–844. doi: 10.1126/science.1827531. [DOI] [PubMed] [Google Scholar]
- 35.Maniatis T, Falvo J V, Kim T H, Kim T K, Lin C H, Parekh B S, Wathelet M G. Structure and function of the interferon-β enhanceosome. Cold Spring Harbor Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 36.Merika M, Williams A J, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 37.Newell C L, Deisseroth A B, Lopez-Berestein G. Interaction of nuclear proteins with an AP-1/CRE-like promoter sequence in the human TNF-α gene. J Leukoc Biol. 1994;56:27–35. doi: 10.1002/jlb.56.1.27. [DOI] [PubMed] [Google Scholar]
- 38.Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 39.Ortiz M A, Light J, Maki R A, Assa-Munt N. Mutation analysis of the Pip interaction domain reveals critical residues for protein-protein interactions. Proc Natl Acad Sci USA. 1999;96:2740–2745. doi: 10.1073/pnas.96.6.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parekh B S, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-β promoter. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 41.Perkins N D, Agranoff A B, Pascal E, Nabel G J. An interaction between the DNA-binding domains of RelA(p65) and Sp1 mediates human immunodeficiency virus gene activation. Mol Cell Biol. 1994;14:6570–6583. doi: 10.1128/mcb.14.10.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perkins N D, Edwards N L, Duckett C S, Agranoff A B, Schmid R M, Nabel G J. A cooperative interaction between NF-κB and Sp1 is required for HIV-1 enhancer activation. EMBO J. 1993;12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ransone L J, Verma I M. Association of nuclear oncoproteins fos and jun. Curr Opin Cell Biol. 1989;1:536–540. doi: 10.1016/0955-0674(89)90017-3. [DOI] [PubMed] [Google Scholar]
- 44.Rao A, Luo C, Hogan P G. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 45.Rastinejad F, Perlmann T, Evans R M, Sigler P B. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 46.Read M A, Whitley M Z, Gupta S, Pierce J W, Best J, Davis R J, Collins T. Tumor necrosis factor α-induced E-selectin expression is activated by the nuclear factor-κB and c-JUN N-terminal kinase/p38 mitogen-activated protein kinase pathways. J Biol Chem. 1997;272:2753–2761. doi: 10.1074/jbc.272.5.2753. [DOI] [PubMed] [Google Scholar]
- 47.Rothenberg E V, Ward S B. A dynamic assembly of diverse transcription factors integrates activation and cell-type information for interleukin 2 gene regulation. Proc Natl Acad Sci USA. 1996;93:9358–9365. doi: 10.1073/pnas.93.18.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 49.Sato M, Tanaka N, Hata N, Oda E, Taniguchi T. Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-β gene. FEBS Lett. 1998;425:112–116. doi: 10.1016/s0014-5793(98)00210-5. [DOI] [PubMed] [Google Scholar]
- 50.Schafer S L, Lin R, Moore P A, Hiscott J, Pitha P M. Regulation of type I interferon gene expression by interferon regulatory factor-3. J Biol Chem. 1998;273:2714–2720. doi: 10.1074/jbc.273.5.2714. [DOI] [PubMed] [Google Scholar]
- 51.Schräder M, Nayeri S, Kahlen J-P, Müller K M, Carlberg C. Natural vitamin D3 response elements formed by inverted palindromes: polarity-directed ligand sensitivity of vitamin D3 receptor-retinoid X receptor heterodimer-mediated transactivation. Mol Cell Biol. 1995;15:1154–1161. doi: 10.1128/mcb.15.3.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 53.Thanos D, Maniatis T. The high mobility group protein HMG I(Y) is required for NF-κB-dependent virus induction of the human IFN-β gene. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 54.Thanos D, Maniatis T. Identification of the Rel family members required for virus induction of the human beta interferon gene. Mol Cell Biol. 1995;15:152–164. doi: 10.1128/mcb.15.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thanos D, Maniatis T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 56.Tsai E Y, Jain J, Pesavento P A, Rao A, Goldfeld A E. Tumor necrosis factor alpha gene regulation in activated T cells involves ATF-2/Jun and NFATp. Mol Cell Biol. 1996;16:459–467. doi: 10.1128/mcb.16.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Urban M B, Schreck R, Baeuerle P A. NF-κB contacts DNA by a heterodimer of the p50 and p65 subunit. EMBO J. 1991;10:1817–1825. doi: 10.1002/j.1460-2075.1991.tb07707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Dam H, Duyndam M, Rottier R, Bosch A, de Vries-Smits L, Herrlich P, Zantema A, Angel P, van der Eb A J. Heterodimer formation of c-Jun and ATF-2 is responsible for induction of c-jun by the 243 amino acid adenovirus E1A protein. EMBO J. 1993;12:479–487. doi: 10.1002/j.1460-2075.1993.tb05680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vilcek J, Sen G C. Interferons and other cytokines. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 375–399. [Google Scholar]
- 60.Wagner S, Green M R. HTLV-I Tax protein stimulation of DNA binding of bZIP proteins by enhancing dimerization. Science. 1993;262:395–399. doi: 10.1126/science.8211160. [DOI] [PubMed] [Google Scholar]
- 61.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 62.Weaver B K, Kumar K P, Reich N C. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang S-W, Nash H A. Specific photocrosslinking of DNA-protein complexes: identification of contacts between integration host factor and its target DNA. Proc Natl Acad Sci USA. 1994;91:12183–12187. doi: 10.1073/pnas.91.25.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yee A A, Yin P, Siderovski D P, Mak T W, Litchfield D W, Arrowsmith C H. Cooperative interaction between the DNA-binding domains of PU.1 and IRF4. J Mol Biol. 1998;279:1075–1083. doi: 10.1006/jmbi.1998.1838. [DOI] [PubMed] [Google Scholar]
- 65.Yie J, Liang S, Merika M, Thanos D. Intra- and intermolecular cooperative binding of high-mobility-group protein I(Y) to the beta-interferon promoter. Mol Cell Biol. 1997;17:3649–3662. doi: 10.1128/mcb.17.7.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yie J, Merika M, Munshi N, Chen G, Thanos D. The role of HMG I(Y) in the assembly and function of the IFN-β enhanceosome. EMBO J. 1999;18:3074–3089. doi: 10.1093/emboj/18.11.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]