Abstract
During investigations into freshwater fungi from the Great Mekong Subregion, four Distoseptispora taxa were collected from China and Thailand. Based on morphological characteristics, and phylogenetic analyses of combined LSU, ITS, SSU, TEF1-α, and RPB2 sequence data, two new species Distoseptisporabangkokensis and D.lancangjiangensis are introduced, and two known species D.clematidis and D.thysanolaenae were first reported in freshwater habitat. Illustrations and descriptions of these taxa are provided, along with comparisons with extant taxa in the genus.
Keywords: 2 new taxa, Distoseptisporales, freshwater fungi, morphology, phylogeny, taxonomy
Introduction
Distoseptisporaceae was introduced by Su et al. (2016) based on morphological and phylogenetic analyses, with Distoseptispora as type genus. Distoseptisporaceae is placed in Distoseptisporales, which was introduced by Luo et al. (2019), and currently comprises two families, Aquapteridosporaceae and Distoseptisporaceae (Luo et al. 2019; Wijayawardene et al. 2020; Hyde et al. 2021). Species of both families are commonly reported from freshwater habitats (Yang et al. 2015, 2018; Su et al. 2016; Li et al. 2021; Hyde et al. 2016a, 2019, 2020; Luo et al. 2018, 2019; Song et al. 2020; Dong et al. 2021).
Distoseptispora as a single genus in Distoseptisporaceae was introduced by Su et al. (2016) with D.fluminicola as the type species. The genus is characterized by monoblastic, cylindrical, conidiogenous cells, with percurrent proliferation, acrogenous, solitary, brown or yellowish/reddish brown, olivaceous, distoseptate or euseptate, cylindrical, obclavate, rostrate conidia, truncate base, with rounded apices, basal cell with a cross wall and basal scar. This genus is not known for its sexual morph (Su et al. 2016; Yang et al. 2018; Hyde et al. 2019, 2020; Luo et al. 2019; Sun et al. 2020). Currently, 32 species are accepted in the genus of which 13 from terrestrial habitats and 19 were reported from freshwater environments (Su et al. 2016; Hyde et al. 2016a, 2019, 2020; Xia et al. 2017; Yang et al. 2018; Luo et al. 2018, 2019; Monkai et al. 2020; Song et al. 2020; Sun et al. 2020; Li et al. 2021; Index Fungorum 2021http://www.indexfungorum.org).
During our ongoing study of freshwater fungi along the north-south gradient in the Asian/Australian region (Hyde et al. 2016b), we collected four species in the genus. Two new species, Distoseptisporabangkokensis and D.lancangjiangensis, are introduced in this study, D.clematidis and D.thysanolaenae are newly recorded from freshwater habitats for the first time in China. Morphological descriptions and illustrations of the species and an updated multi-gene phylogenetic tree are provided to reveal their taxonomic position among the species in the Distoseptisporales, and also provided the comparison of morphological characteristics, habitats and hosts information of species newly added to Distoseptispora after Monkai et al. (2020) (Table 2).
Table 2.
Comparison of morphological characteristic, habitats and hosts’ information of species added to Distoseptispora after Monkai et al. (2020) (for other species see Monkai et al. 2020).
| Species | Conidiophore (μm) | Conidia (μm) | Conidia septation | Conidia characteristic | Habitat | Host | Reference |
|---|---|---|---|---|---|---|---|
| Distoseptisporabangkokensis | 37–55 × 3–4 | 400–568 × 13–16 | Multi-distoseptate | Elongate, obclavate, rostrate, dark olivaceous to dark brown | Freshwater | Unidentified submerged wood | This study |
| D.lancangjiangensis | 30–41 × 5–6 | 83–220 × 12–14 | 16–41-distoseptate | Obclavate, cylindrical, elongated, straight or curved, brown to greenish-brown | Freshwater | Unidentified submerged wood | This study |
| D.euseptata | 19–28 × 4–5 | 37–54 × 8–9 | 4–7-euseptate | Obpyriform to obclavate, straight or curved, olivaceous | Freshwater | Unidentified submerged wood | Li et al. 2021 |
| D.fasciculata | 12–16 × 5–6 | 46–200 × 10–16.5 | 10–40-distoseptate | Subcylindrical to obclavate, mostly curved, olivaceous when young, dark brown when mature | Freshwater | Unidentified submerged wood | Dong et al. 2021 |
| D.longispora | 17–37 × 6–10 | 189–297 × 16–23 | 31–56-distoseptate | Obclavate, elongated, straight or slightly curved, to yellowish brown | Freshwater | Unidentified submerged wood | Song et al. 2020 |
| D.saprophytica | 50–140 × 3.2–4.2 | 14.5–30 × 4.5–7.5 | 2–6-distoseptate | Subcylindrical to obclavate, straight or curved, olivaceous to brown | Freshwater | Unidentified submerged wood | Dong et al. 2021 |
| D.songkhlaensis | 70–90 × 4–5.5 | 44–125 × 9–14.5 | 9–16-distoseptate | Obclavate, straight or curved, olivaceous to brown | Freshwater | Unidentified submerged wood | Dong et al. 2021 |
| D.yunnanensis | 131–175 × 6–7 | 58–108 × 8–10 | 6–10-euseptate | Obclavate, rostrate, straight or slightly curved, mid olivaceous to brown | Freshwater | Unidentified submerged wood | Li et al. 2021 |
Materials and methods
Isolation and morphology
Specimens of submerged decaying wood were collected from Dulongjiang, Nanpanjiang, Lancangjiang and Chao Phraya River in China and Thailand respectively. Multiple samples will be collected at each collection site at different times, allowing more strains to be obtained for each species. Methods of morphological observation and isolation follow Luo et al. (2018) and Senanayake et al. (2020). IFW (Tarosoft(R) Image Frame Work) was used for measurement of photomicrograph, and Adobe Photoshop CS5 software was used to process images for making photo-plates (Adobe Systems Inc., USA). Single spore isolation was performed according to the following steps: The conidia suspension from specimens, absorbed with a sterilized pipette, was placed on potato dextrose agar (PDA) and incubated at room temperature overnight. Germinated conidia were transferred to new PDA/MEA (Beijing land bridge technology CO., LTD., China) plates and incubated in an incubator at room temperature (25 °C). Specimens were deposited in the Kunming Institute of Botany, Academia Sinica herbarium (KUN-HKAS), and Mae Fah Luang University herbarium (MFLU). Cultures were deposited in the Dali University Culture Collection (DLUCC), China General Microbiological Culture Collection Center (CGMCC), and Mae Fah Luang University Culture Collection (MFLUCC). Facesoffungi number was obtained as described in Jayasiri et al. (2015) and Index Fungorum number was also registered (http://www.indexfungorum.org/Names/Names.asp). In this study, multiple samples were collected for each sample site and related environment, but unfortunately, there were still no more strains for the two new species in the paper.
DNA extraction, PCR amplification, and sequencing
DNA extraction, PCR amplification, sequencing and phylogenetic analysis follow Dissayanake et al. (2020) with the following modifications. Fungal mycelia (200–500 mg) were scraped from grown on PDA/MEA plates using sterile scalpel, transferred to microcentrifuge tube with sterilized needles, and then grind with liquid nitrogen or quartz sand to break the cells. DNA was extracted using the TreliefTM Plant Genomic DNA Kit (TSP101) according to the manufacturer’s instructions.
Five gene regions, LSU, ITS, SSU, TEF1-α, and RPB2 were amplified using LR0R/LR5, ITS5/ITS4, NS1/NS4, 983F/EF1-2218R, and RPB2-5F/RPB2-7cR (Vilgalys and Hester 1990; White et al. 1990; Liu et al. 1999) primer pairs respectively. Primer sequences are available at the WASABI database at the AFTOL website (aftol.org). The PCR mixture contained 12.5 μL of 2 × Power Taq PCR Master Mix (a premix and ready to use solution, including 0.1 Units/μL Taq DNA Polymerase, 500μm dNTP Mixture each (dATP, dCTP, dGTP, dTTP), 20 mm Tris-HCl pH 8.3, 100 Mm KCl, 3 mM MgCl2, stabilizer and enhancer), 1 μL of each primer including forwarding primer and reverse primer (10 μm), 1 μL template DNA extract and 9.5 μL deionized water (Luo et al. 2018). The PCR cycling conditions of LSU, ITS, SSU and TEF1-α were as follows: 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 30s, annealing at 55 °C for 50s, elongation at 72 °C for 1 min, and a final extension at 72 °C for 10 min. The PCR thermal cycle of RPB2 has a total of 40 cycles, and the conditions are as follows: initially denature at 95 °C for 5 min, and then enter 40 cycles: denaturation at 95 °C for 1 min, annealing at 52 °C for 2 min, extension at 72 °C for 90s, and finally at 72 °C for 10 min. PCR products were then purified using minicolumns, purification resin, and buffer according to the manufacturer’s protocols (Amersham product code: 27–9602–01). The sequences were carried out at Beijing Tsingke Biotechnology Co., Ltd. (Beijing, P.R. China).
Phylogenetic analysis
Preliminary identification of genes obtained from fresh strains by GenBank database. The LSU, ITS, SSU, TEF1-α, and RPB2 used for phylogenetic analysis are selected based on the preliminary identification results and the related publications (Yang et al. 2018; Monkai et al. 2020). The sequences were aligned using MAFFT online service: Multiple alignment program for amino acid or nucleotide sequences MAFFT version 7 (Katoh and Standley 2013: http://mafft.cbrc.jp/alignment/server/index.html), and edited manually in BioEdit v. 7.0 (Hall 1999). The sequence dataset was combined using SquenceMatrix v.1.7.8 (Vaidya et al. 2011). The alignment formats were change to PHYLIP and NEXUS formats by ALigment Transformation EnviRonment (ALTER) website (http://sing.ei.uvigo.es/ALTER/).
Maximum likelihood (ML) analysis was carried out using the RAxML-HPC2 on XSEDE (8.2.12) (Stamatakis 2006; Stamatakis et al. 2008) of CIPRES Science Gateway website (Miller et al. 2010: http://www.phylo.org/portal2) and the estimated proportion of invariant sites is (GTRGAMMA+I) model.
Bayesian analyses were performed in MrBayes 3.2.6 (Ronquist et al. 2012) and the best-fit model (LSU, ITS, SSU, TEF1-α, and RPB2 are all GTR+I+G) of sequences evolution was estimated via MrModeltest 2.2 (Guindon and Gascuel 2003; Nylander 2004; Darriba et al. 2012). The Markov Chain Monte Carlo (MCMC) sampling approach was used to calculate posterior probabilities (PP) (Rannala and Yang 1996). Bayesian analyses of six simultaneous Markov chains were run for 10000000 generations with trees sampled every 1000 generations.
Phylogenetic trees were visualized using FigTree v1.4.0 (Rambaut 2012: http://tree.bio.ed.ac.uk/software/figtree/), editing and typesetting using Adobe Illustrator (AI) (Adobe Systems Inc., the United States). The new sequences were submitted in GenBank and the strain information used in this paper is provided in Table 1. The alignments and phylogenetic trees were deposited in TreeBASE (http://www.treebase.org/, accession number: 28758).
Table 1.
Strains used for phylogenetic analysis and their corresponding GenBank numbers. The type strain are in bold font.
*1 Ex-type strain of Distoseptisporasubmersa.
Results
Phylogenetic analysis
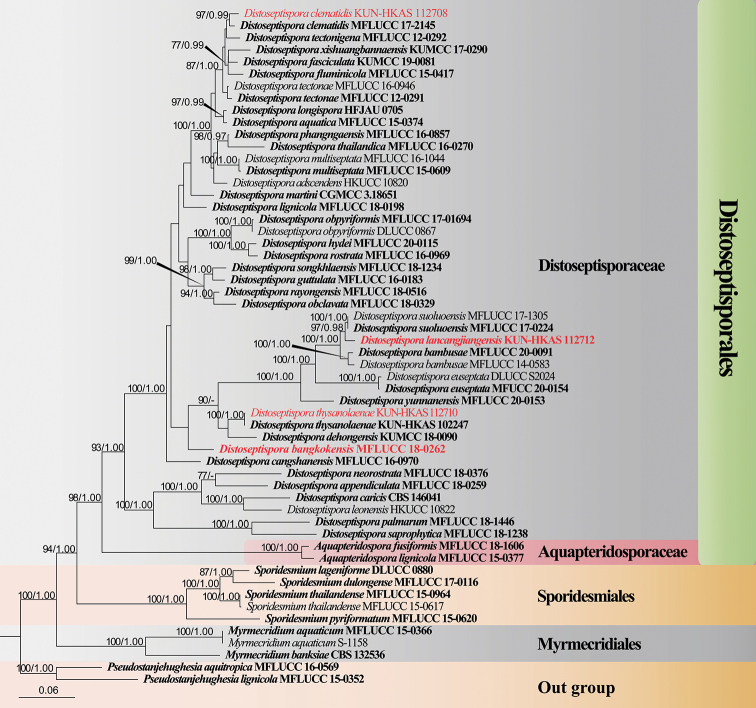
The dataset composed of LSU (1–744 bp), ITS (745–1310 bp), TEF1-α (1311–2161 bp), RPB2 (2162–3178 bp), and SSU (3179–4199 bp) gene, comprising a total of 4199 characters (including gaps), including 56 taxa with Pseudostanjehughesiaaquitropica (MFLUCC 16-0569) and P.lignicola (MFLUCC 15-0352) as the outgroup taxa (Figure 1). The ML and BI phylogenetic analyses produced similar topology. The combined dataset analysis of RAxML generates a best-scoring tree (Figure 1), with the final ML optimization likelihood value of -30393.557997. The aligned matrix had 1624 distinct alignment patterns, with 36.44% completely undetermined characters or gaps. The base frequency and rate are as follows: A = 0.243915, C = 0.259360, G = 0.279029, T = 0.217696; rate AC = 1.166355, AG = 2.813539, AT = 1.110401, CG = 0.796371, CT = 5.621229, GT = 1.000000; gamma distribution shape: α = 0.221933. Bootstrap support values with a maximum likelihood (ML) greater than 70%, and Bayesian posterior probabilities (PP) greater than 0.97 are given above the nodes.
Figure 1.
Maximum likelihood (ML) tree is based on combined of LSU, ITS, SSU, TEF1-α, and RPB2 sequence data. Bootstrap support values with an ML greater than 70% and Bayesian posterior probabilities (PP) greater than 0.97 given above the nodes, shown as “ML/PP”. The tree is rooted with Pseudostanjehughesiaaquitropica (MFLUCC 16-0569) and P.lignicola (MFLUCC 15-0352). New species are indicated in red and type strains are in bold.
The phylogenetic tree shows that the new species Distoseptisporabangkokensis (MFLUCC 18-0262) was placed as a sister taxon to D.bambusae (MFLUCC 14-0583 and MFLUCC 20-0091), D.dehongensis (KUMCC 18-0090), D.euseptata (MFUCC 20-0154 and DLUCC S2024), D.lancangjiangensis (KUN-HKAS 112712), D.suoluoensis (MFLUCC 17-0224 and MFLUCC 17-1305), D.thysanolaenae (KUN-HKAS 102247 and KUN-HKAS 112710), and D.yunnanensis (MFLUCC 20-0153) with low bootstrap support with low bootstrap support (Figure 1), whereas D.lancangjiangensis clustered with D.suoluoensis with 97%ML/0.98PP support. Distoseptisporathysanolaenae (KUN-HKAS 112710) and D.clematidis (KUN-HKAS 112708) clustered with the ex-type strain of D.thysanolaenae (KUN-HKAS 102247) and D.clematidis (MFLUCC 17-2145), respectively, with 100%ML/1.00PP and 97%ML/0.99PP bootstrap support.
Taxonomy
Distoseptispora bangkokensis
H.W. Shen, D.F. Bao, K.D. Hyde & Z.L. Luo sp. nov.
E08487C9-F52C-559D-B571-BFE767D78DD4
Index Fungorum Number No: IF558556
Facesoffungi Number No: FoF09993
Figure 2.
Distoseptisporabangkokensis (MFLU 21-0110, holotype) A colonies on the substratum B conidiophores C conidiophores with conidia D conidiogenous cell E-G conidia H germinating conidium Scale bars: 20 μm (B, C, H); 10 μm (D); 50 μm (E-G).
Etymology.
Referring to the collecting location, Bangkok, Thailand.
Holotype.
MFLU 21-0110
Description.
Saprobic on submerged wood in freshwater stream. Sexual morph: Undetermined. Asexual morph: Colonies effuse, glistening, hairy, brown to dark brown. Mycelium partly superficial in the substratum, composed of hyaline to pale brown, septate, branched hyphae. Conidiophores 37–55 × 3–4 μm (x¯ = 46 × 3 μm, n = 15) macronematous, mononematous, solitary or in a small group of 2–4, cylindrical, straight or slightly flexuous, 3–8-septate, dark brown, paler at the apical part, rounded at the apex. Conidiogenous cells 6–8 × 3–4 μm (x¯ = 7 × 3 μm, n = 15), integrated, terminal, monoblastic, cylindrical, brown. Conidia 400–568 × 13–16 μm (x¯ = 484 × 15 μm, n = 20), 6–7 μm at the narrowest apical region, acrogenous, solitary, elongate, obclavate, rostrate, multi-distoseptate, tapering towards the apex, truncate at the base, rounded at apex, dark olivaceous to dark brown, straight or slightly curved, guttulate, thick-walled, smooth, conidia percurrent proliferation which forms another conidium at the apex.
Culture characteristics. Conidia cultivated on PDA within 12h and germ tubes produced at the ends. Colonies on PDA, reaching 6 cm in 1 month at room temperature (25 °C). Mycelium loose, flocculent, smooth edge, brown to dark brown, dark brown on the reverse.
Material examined.
Thailand, Bangkok Province, Khwaeng Phra Khanong Nuea, 13°42'41"N; 100°36'03"E, on submerged decaying wood, 1 October 2017, Zonglong Luo, S–3083 (MFLU 21-0110, holotype), ex-type living culture (MFLUCC 18-0262).
Notes.
Distoseptisporabangkokensis is comparable to D.cangshanensis and D.multiseptata in having elongate, obclavate, or rostrate conidia (Su et al. 2016; Hyde et al. 2016a; Yang et al. 2018). However, D.bangkokensis has shorter and narrower conidiophores than those of D.cangshanensis (37–55 × 3–4 μm vs. 44–68 × 4–8 μm), but has longer conidia (400–568 μm vs. 58–166 μm); D.multiseptata (MFLU 17-0856) is similar to D.bangkokensis in conidial morphology, with conidia mostly 300–600 μm long (up to 700 μm) and significantly longer than those of the holotype (up to 380 μm long). However, Yang et al. (2018) did not give a detailed description of D.multiseptata (MFLU 17-0856). Phylogenetic analyses showed that D.bangkokensis clustered with D.bambusae, D.dehongensis, D.euseptata, D.lancangjiangensis, D.suoluoensis, D.thysanolaenae, and D.yunnanensis with low bootstrap support (26%ML/0.53PP, Figure 1). Distoseptisporabangkokensis is distoseptate conidia, and it is easily distinguished from D.bambusae, D.euseptata, D.lancangjiangensis, D.suoluoensis, and D.yunnanensis, which are euseptate. Distoseptisporabangkokensis is resemble to D.dehongensis and D.thysanolaenae in having obclavate, distoseptae conidia, but are distinguished by conidia characteristics, D.bangkokensis has elongate, obclavate, rostrate, multi-distoseptat, and longer conidia than D.dehongensis (400–568 × 13–16 μm vs. 17–30 × 7.5–10 μm) and D.thysanolaenae (400–568 × 13–16 μm vs. 30–70 × 5–8 μm), respectively.
Distoseptispora lancangjiangensis
H.W. Shen, H.Y. Su, K.D. Hyde & Z.L. Luo sp. nov.
BB47E43B-AC2E-525D-A336-5ED94C89D927
Index Fungorum Number No: IF558555
Facesoffungi Number No: FoF09994
Figure 3.
Distoseptisporalancangjiangensis (KUN-HKAS 112712, holotype) A colonies on the substratum B conidiophore and conidium C-E conidiophores F, G conidiogenous cells H conidiogenous cell with conidium I-Q conidia R germinating conidium S, T culture on PDA. Scale bars: 50 μm (B-E); 20 μm (F-R).
Etymology.
Referring to the collecting location, Lancangjiang River in China.
Holotype.
KUN-HKAS 112712
Description.
Saprobic on submerged wood in freshwater River. Sexual morph: Undetermined. Asexual morph: Colonies effuse, hairy, glistening, brown to dark. Mycelium partly immersed in the substratum, composed of hyaline to pale brown, septate, branched hyphae. Conidiophores 144–204 × 5–6 μm (x¯ = 175 × 6 μm, n = 20) macronematous, mononematous, solitary, inflate at the base, cylindrical, straight or slightly flexuous, 6–11-septate, dark brown, hyaline and rounded at apex. Conidiogenous cells 12–24 × 4–5 μm (x¯ = 18 × 5 μm, n = 20) integrated, terminal, monoblastic, cylindrical, brown. Conidia 64–84 × 9–10 μm (x¯ = 74 × 10 μm, n = 20), acrogenous, solitary, narrowly obclavate or obspathulate, tracted at base, tapering towards apex, 3–10-euseptate, brown to dark brown, thin-walled, becoming paler or hyaline towards apex, guttulate, with a darkened scar at base, smooth-walled.
Culture characteristics.
Conidia cultivated on PDA within 12h and germ tubes produced at the apex. Colonies on PDA, reaching 4.5 cm in 1 month at room temperature (25 °C). Mycelium loose, flocculent, smooth edges, convex middle, pale brown to dark brown on the surface of PDA. Smooth, black on the reverse.
Material examined.
China, Yunnan Province, Dali City, Lancangjiang River, 22°36'36"N; 100°37'59"E, on submerged decaying wood, 20 July 2017, Qishan Zhou and Qingxiong Ruan S–1864 (KUN-HKAS 112712, holotype; MFLU 21-0111, isotype), ex-type living culture (DLUCC 1864 = CGMCC 3.20265).
Notes.
Phylogenetic analysis showed that Distoseptisporalancangjiangensis clustered as a sister taxon to D.suoluoensis with 97%ML/0.98PP support. Distoseptisporalancangjiangensis is similar to D.suoluoensis in having long conidiophores, monoblastic conidiogenous cells, and obclavate to rostrate, euseptate conidia. However, D.suoluoensis has yellowish-brown or dark olivaceous, verrucose conidia, while in D.lancangjiangensis conidia are brown to dark brown and smooth-walled. Moreover, D.lancangjiangensis has smaller conidia than those of D.suoluoensis (64–84 × 9–10 μm vs. 80–125 × 8–13 μm) (Yang et al. 2018). Distoseptisporalancangjiangensis and D.bambusae have similar conidial shapes, but D.lancangjiangensis having longer conidia (64–84 × 9–10 μm vs. 45–74 × 5.5–10 μm) and longer conidiophores (144–204 × 5–6 μm vs. 40–96 × 4–5.5 μm). Furthermore, D.bambusae has polyblastic or monoblastic conidiogenous cells and olivaceous or brown conidia, while D.lancangjiangensis only has monoblastic conidiogenous cells and brown to dark brown conidia (Sun et al. 2020).
Distoseptispora clematidis
Phukhams., M.V. de Bult & K.D. Hyde, in Phukhamsakda et al., Fungal Diversity 102: 168 (2020)
AC7F9A70-BD17-5567-BC09-C62CA24F0F50
Index Fungorum Number No: IF557301
Facesofungi Number No: FoF07261
Figure 4.
Distoseptisporaclematidis (KUN-HKAS 112708) A colonies on the substratum B-C conidiophores with conidia D conidiogenous cells E-H conidia I germinating conidium J culture on PDA Scale bars: 30 μm (B, C, E-I); 20 μm (D).
Description.
Saprobic on submerged wood in freshwater River. Sexual morph: Undetermined. Asexual morph: Colonies on the substratum superficial, effuse, scattered, hairy, dark brown. Mycelium partly immersed in substrate, composed of branched, smooth, septate, brown to dark brown hyphae. Conidiophores 30–41 × 5–6 μm (x¯ = 36 × 6 μm, n = 15), macronematous, mononematous, single or in a small group, straight or slightly flexuous, unbranched, septate, erect, 2–4-septate, cylindrical, smooth, dark brown to brown. Conidiogenous cells 7–9 × 5–6 μm (x¯ = 8 × 5 μm, n = 15), monoblastic, integrated, determinate, terminal, cylindrical, pale brown to brown. Conidia 83–220 × 12–14 μm (x¯ = 151 × 13 μm, n = 20), acrogenous, solitary, obclavate, cylindrical, elongated, straight or curved, truncate at base, rounded at apex, 16–41-distoseptate, slightly constricted at some septa, smooth, brown to greenish-brown, thick-walled.
Culture characters.
Conidia cultivated on PDA within 12h and germ tubes produced at the ends. Colonies on PDA, attaining 4 cm after 1 month at room temperature (25 °C), gray at first, later becoming dark gray, loose, flocculent, smooth edge, dark brown on the reverse.
Material examined.
China, Yunnan Province, Kunming City, Yiliang County, Nanpanjiang River, 24°38'28"N; 103°09'38"E, on submerged decaying wood, 12 June 2018; Hongwei Shen and Xiu He, S–1797 (KUN-HKAS 112708), living culture (DLUCC 1797).
Notes.
Our new isolate clustered with the ex-type strain of Distoseptisporaclematidis (MFLU 17-1501) (Phukhamsakda et al. 2020) with 97%ML/0.99PP bootstrap support (Figure 1). Distoseptisporaclematidis (MFLU 17-1501) was collected on dead culms of Thysanolaenamaxima (Roxb. ex Hornem.) Honda in Yunnan Province, China. Based on morphological analysis, the size and shape of the conidia and conidiophores of our new isolate are similar to D.clematidis. Therefore, we identified our new isolate as D.clematidis and it is a new record from freshwater habitats in China.
Distoseptispora thysanolaenae
Goonas., Dayarathne, Phookamsak & K.D.Hyde, in Phookamsak et al., Fungal Diversity 95: 126 (2019)
68BD6629-9E92-5AC8-BE36-175569F80A7A
Index Fungorum Number No: IF555408
Facesoffungi Number No: FoF05011
Figure 5.
Distoseptisporathysanolaenae (KUN-HKAS 112710) A colonies on the substratum B-D conidiophores with conidia E, F conidiogenous cells G-N conidia O germinating conidium P, Q culture on PDA Scale bars: 30 μm (B-D); 10 μm (E, F); 20 μm (G-O).
Description.
Saprobic on submerged wood in freshwater River. Sexual morph: Undetermined. Asexual morph: Colonies on the substratum superficial, effuse, scattered, hairy, dark brown. Mycelium partly immersed, composed of branched, septate, smooth, brown to dark brown hyphae. Conidiophores 41–59 × 4–5 μm (x¯ = 50 × 5 μm, n = 20) macronematous, mononematous, unbranched, single, erect, straight or slightly curved, smooth, 3–6-septate, pale brown to brown. Conidiogenous cells monoblastic, integrated, determinate, terminal, dark brown, cylindrical. Conidia 46–87 × 9–12 μm (x¯ = 67 × 10 μm, n = 25) acrogenous, solitary, dry, smooth, obclavate, elongated, straight or slightly curved, truncate at base, tapering towards apex, 6–19-septate, dark grayish-brown to light yellow-green, thick-walled.
Culture characteristics.
Conidia cultivated on PDA within 12 h and germ tubes produced at the apex. Colonies on PDA, reaching 6 cm after 6 weeks at room temperature (25 ℃). Mycelium loose, flocculent, neat edges, convex in middle, pale brown to dark brown. Black, smooth on the back.
Material examined.
China, Yunnan Province, Lushui City, Nujiang River, 26°23'12"N; 98°53'94"E, on submerged decaying wood, 3 May 2016, Zonglong Luo and Songming Tang, S-876 (KUN-HKAS 112710), living culture (DLUCC 876 = KUNCC 21-10710)
Notes.
Our new collection is identical to Distoseptisporathysanolaenae in characters of the conidiophores, conidiogenous cell, and conidia (Phookamsak et al. 2019). Furthermore, our new isolate phylogenetically clusters with the ex-type strain of D.thysanolaenae (KUN-HKAS 102247) with 100%ML/1.00PP support (Figure 1). Distoseptisporathysanolaenae was collected from terrestrial habitats in China, while, our new isolate was collected from freshwater habitat in China. Therefore, we identified our new collection as D.thysanolaenae, and it is new to freshwater habitats in China.
Discussion
Distoseptispora has been reported from both freshwater and terrestrial habitats. Of these, species have been collected from freshwater environments (Su et al. 2016; Hyde et al. 2016a, 2019, 2020; Luo et al. 2018; Xia et al. 2017, 2019; Yang et al. 2018; Tibpromma et al. 2018; Crous et al. 2019; Phookamsak et al. 2019; Monkai et al. 2020; Phukhamsakda et al. 2020; Song et al. 2020; Sun et al. 2020; Li et al. 2021). To date, 18 species of Distoseptispora have been reported from Thailand, 14 species from China. In this study, we collected four distoseptispora-like taxa from rivers and streams in China and Thailand. Phylogenetic analysis showed that all four species were well-placed in Distoseptispora (Figure 1). Two new species and records are introduced based on morphological and phylogenetic analysis.
Species of Distoseptispora are highly diverse in morphology, especially the conidial shape. Conidia of most species are obclavate to cylindrical or rostrate (e.g. D.aquatica, D.tectonae, and D.suoluoensis), but a few are ellipsoid to subglobose (e.g. D.martinii), lanceolate (e.g. D.guttulata and D.multiseptata), and some species have conidia with a sheath at the apex (e.g. D.appendiculata) (Hyde et al. 2016a; Su et al. 2016; Xia et al. 2017; Yang et al. 2018; Luo et al. 2018, 2019). Some species also differ in the conidiogenous cells (D.palmarum, D.dehongensis, and D.bambusae are monoblastic or polyblastic, while the others are monoblastic) and conidial septate (D.bambusae, D.euseptatensis, D.guttulata, D.lignicola, D.rayongensis, D.suoluoensis, and D.yunnanensis are euseptate, while other species are distoseptate) (Yang et al. 2018; Hyde et al. 2019; Luo et al. 2019; Sun et al. 2020; Dong et al. 2021; Li et al. 2021).
Based on the key morphological characteristics, viz. conidiophores, conidiogenous cells, and conidia, Subramanian (1992) redisposed seven genera, viz., Sporidesmium, Polydesmus, Sporidesmiella, Stanjehughesia, Repetophragma, Penzigomyces, and Ellisembia to accommodate several Sporidesmium-like taxa. Based on multi-gene phylogenetic analysis and morphology, Su et al. (2016) introduced a new Sporidesmium-like genus Distoseptispora. Some Sporidesmium-like taxa were introduced in different lineages and synonymized Ellisembia under Sporidesmium. Although Distoseptispora was only introduced from submerged wood in freshwater habitat in 2016 (Su et al. 2016), the genus has previously been reported from both freshwater and terrestrial habitats as species in other genera. For example, Cai et al. (2002), Ho et al. (2001, 2002) and Luo et al. (2004) reported Distoseptispora as other species (Ellisembia, Sporidesmiella, and Sporidesmium) from submerged wood in freshwater habitats, and Kodsueb et al. (2016), Mena-Portales et al. (2016) and Zhou et al (2001) reported from terrestrial habitats. However, none of these records had molecular data and it is impossible to consider the placement of these isolates. In these species distoseptispora/sporidesmium-like genera, it is therefore better to describe taxa based on molecular data.
Based on phylogenetic analysis, Xia et al. (2017) transferred Acrodictysmartinii to Distoseptispora as Distoseptisporamartinii. The species is characterized by solitary erect, unbranched conidiophores, monoblastic conidiogenous cells with percurrent extensions and subhyaline to pale brown and solitary, transversal ellipsoid, oblate or subglobose, muriform conidia, separated by septa, sometimes with pores in the septa and pale brown to brown. However, the current understanding of Distoseptisporaceae, D.martinii is significantly different from other Distoseptispora taxa; thus, needs to be verified in the future (Luo et al. 2018; Sun et al. 2020).
Supplementary Material
Acknowledgment
We would like to thank the National Natural Science Foundation of China (Project ID: 32060005 and 31970021), and Fungal Diversity Conservation and Utilization Innovation Team of Dali University (ZKLX2019213) for financial support. Kevin D Hyde thanks the Thailand Research Fund for the grant (RDG6130001MS), Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion. Hongwei Shen thanks Saranyaphat Boonmee for her help in sample collection and herbarium deposit. Qishan Zhou, Qingxiong Ruan, Songming Tang, and Xiu He are thanked for their help on sample collection. We are grateful to Yanmei Zhang, Longli Li, and Wenli Li for their help on DNA extraction and PCR amplification.
Citation
Shen H-W, Bao D-F, Hyde KD, Su H-Y, Bhat DJ, Luo Z-L (2021) Two novel species and two new records of Distoseptispora from freshwater habitats in China and Thailand. MycoKeys 84: 79–101. https://doi.org/10.3897/mycokeys.84.71905
References
- Cai L, Tsui CKM, Zhang KQ, Hyde KD. (2002) Aquatic fungi from Lake Fuxian, Yunnan, China. Fungal Diversity 9: 57–70. [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Summerell BA, Shivas RG, Burgess TI, Decock CA, Dreyer LL, Granke LL, Guest DI, Hardy GEStJ, Hausbeck MK, Hüberli D, Jung T, Koukol O, Lennox CL, Liew ECY, Lombard L, McTaggart AR, Pryke JS, Roets F, Saude C, Shuttleworth LA, Stukely MJC, Vánky K, Webster BJ, Windstam ST, Groenewald JZ. (2012) Fungal Planet description sheets: 107–127. Persoonia 28: 138–182. 10.3767/003158512X652633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772–772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Hyde KD, Jeewon R, Doilom M, Yu XD, Wang GN, Liu NG, Hu DM, Nalumpang S. (2021) Towards a natural classification of Annulatascaceae-like taxa Ⅱ: introducing five new genera and eighteen new species from freshwater. Mycosphere 12: 1–88. 10.5943/mycosphere/12/1/1 [DOI] [Google Scholar]
- Guindon S, Gascuel O. (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52: 696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. http://wwwmbio-ncsuedu/bioedit/bioedithtml [Google Scholar]
- Ho WH, Hyde KD, Hodgkiss IJ. (2001) Fungal communities on submerged wood from streams in Brunei, Hong Kong, and Malaysia. Mycological 105: 1492–1501. 10.1017/S095375620100507X [DOI] [Google Scholar]
- Ho WH, Yanna, Hyde KD, Hodgkiss IJ. (2002) Seasonality and sequential occurrence of fungi on wood submerged in Tai Po Kau Forest Stream, Hong Kong. Fungal Diversity 10: 21–43. [Google Scholar]
- Hyde KD, Bao DF, Hongsanan S, Chethana KWT, Yang J, Suwanarach N. (2021) Evolution of freshwater Diaporthomycetidae (Sordariomycetes) provides evidence for five new orders and six new families. Fungal Diversity 107: 71–105. 10.1007/s13225-021-00469-7 [DOI] [Google Scholar]
- Hyde KD, Fryar S, Tian Q, Bahkali AH, Xu JC. (2016b) Lignicolous freshwater fungi along a north-south latitudinal gradient in the Asian/Australian region; can we predict the impact of global warming on biodiversity and function? Fungal Ecology 19: 190–200. 10.1016/j.funeco.2015.07.002 [DOI]
- Hyde KD, Hongsanan S, Jeewon R, Bhat DJ, McKenzie EHC, Jones EBG, Phookamsak R, Ariyawansa HA, Boonmee S, Zhao Q, Abdel-Aziz FA, Abdel-Wahab MA, Banmai S, Chomnunti P, Cui BK, Daranagama DA, Das K, Dayarathne MC, de Silva NI, Dissanayake AJ, Doilom M, Ekanayaka AH, Gibertoni TB, Góes-Neto A, Huang SK, Jayasiri SC, Jayawardena RS, Konta S, Lee HB, Li WJ, Lin CG, Liu JK, Lu YZ, Luo ZL, Manawasinghe IS, Manimohan P, Mapook A, Niskanen T, Norphanphoun C, Papizadeh M, Perera RH, Phukhamsakda C, Richter C, de A. Santiago ALCM, Drechsler-Santos ER, Senanayake IC, Tanaka K, Tennakoon TMDS, Thambugala KM, Tian Q, Tibpromma S, Thongbai B, Vizzini A, Wanasinghe DN, Wijayawardene NN, Wu HX, Yang J, Zeng XY, Zhang H, Zhang JF, Bulgakov TS, Camporesi E, Bahkali AH, Amoozegar MA, Araujo-Neta LS, Ammirati JF, Baghela A, Bhatt RP, Bojantchev D, Buyck B, da Silva GA, de Lima CLF, de Oliveira RJV, de Souza CAF, Dai YC, Dima B, Duong TT, Ercole E, Mafalda-Freire F, Ghosh A, Hashimoto A, Kamolhan S, Kang JC, Karunarathna SC, Kirk PM, Kytövuori I, Lantieri A, Liimatainen K, Liu ZY, Liu XZ, Lücking R, Medardi G, Mortimer PE, Nguyen TTT, Promputtha I, Raj KNA, Reck MA, Lumyong S, Shahzadeh-Fazeli SA, Stadler M, Soudi MR, Su HY, Takahashi T, Tangthirasunun N, Uniyal P, Wang Y, Wen TC, Xu JC, Zhang ZK, Zhao YC, Zhou JL, Zhu L. (2016a) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. 10.1007/s13225-016-0373-x [DOI] [Google Scholar]
- Hyde KD, Norphanphoun C, Maharachchikumbura SSN, Bhat DJ, Jones EBG1, Bundhun D, Chen YJ, Bao DF, Boonmee S, Calabon MS, Chaiwan N, Chethana KWT, Dai DQ, Dayarathne MC, Devadatha B, Dissanayake AJ, Dissanayake LS, Doilom M, Dong W, Fan XL, Goonasekara ID, Hongsanan S, Huang SK, Jayawardena RS, Jeewon R, Karunarathna A, Konta S, Kumar V, Lin CG, Liu JK, Liu NG, Luangsa-ard J, Lumyong S, Luo ZL, Marasinghe DS, McKenzie EHC, Niego AGT, Niranjan M, Perera RH, Phukhamsakda C, Rathnayaka AR, Samarakoon MC, Samarakoon SMBC, Sarma VV, Senanayake IC, Shang QJ, Stadler M, Tibpromma S, Wanasinghe DN, Wei DP, Wijayawardene NN, Xiao YP, Yang J, Zeng XY, Zhang SN, Xiang MM. (2020) Refined families of Sordariomycetes. Mycosphere 11: 305–1059. 10.5943/mycosphere/11/1/7 [DOI] [Google Scholar]
- Hyde KD, Tennakoon DS, Jeewon R, Bhat DJ, Maharachchikumbura SSN, Rossi W, Leonardi M, Lee HB, Mun HY, Houbraken J, Nguyen TTT, Jeon SJ, Frisvad JC, Wanasinghe DN, Lücking R, Aptroot A, Cáceres MES, Karunarathna SC, Hongsanan S, Phookamsak R, de Silva NI, Thambugala KM, Jayawardena RS, Senanayake IC, Boonmee S, Chen J, Luo ZL, Phukhamsakda C, Pereira OL, Abreu VP, Rosado AWC, Bart B, Randrianjohany E, Hofstetter V, Gibertoni TB, Soares AMdS, Plautz HL, Sotão HMP, Xavier WKS, Bezerra JDP, de Oliveira TGL, de Souza-Motta CM, Magalhães OMC, Bundhun D, Harishchandra D, Manawasinghe IS, Dong W, Zhang SN, Bao DF, Samarakoon MC, Pem D, Karunarathna A, Lin CG, Yang J, Perera RH, Kumar V, Huang SK, Dayarathne MC, Ekanayaka AH, Jayasiri SC, Xiao Y, Konta S, Niskanen T, Liimatainen K, Dai YC, Ji XH, Tian XM, Mešić A, Singh SK, Phutthacharoen K, Cai L, Sorvongxay T, Thiyagaraja V, Norphanphoun C, Chaiwan N, Lu YZ, Jiang HB, Zhang JF, Abeywickrama PD, Aluthmuhandiram JVS, Brahmanage RS, Zeng M, Chethana T, Wei D, Réblová M, Fournier J, Nekvindová J, do Nascimento Barbosa R, dos Santos JEF, de Oliveira NT, Li GJ, Ertz D, Shang QJ, Phillips AJL, Kuo CH, Camporesi E, Bulgakov TS, Lumyong S, Jones EBG, Chomnunti P, Gentekaki E, Bungartz F, Zeng XY, Fryar S, Tkalčec Z, Liang J, Li G, Wen TC, Singh PN, Gafforov Y, Promputtha I, Yasanthika E, Goonasekara ID, Zhao RL, Zhao Q, Kirk PM, Liu JK, Yan J, Mortimer PE, Xu J, Doilom M. (2019) Fungal diversity notes 1036–1150: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 96: 1–242. 10.1007/s13225-019-00429-2 [DOI] [Google Scholar]
- Index Fungorum (2021) Index Fungorum. www.indexfungorum.org [accessed 4 September 2021]
- Jayasiri SC, Hyde KD, Abd-Elsalam KA, Abdel-Wahab MA, Ariyawansa HA, Bhat J, Buyck B, Dai YC, Ertz D, Hidayat I, Jeewon R, Jones EBG, Karunarathna SC, Kirk P, Lei C, Liu JK, Maharachchikumbura SSN, McKenzie E, Ghobad-Nejhad M, Nilsson H, Pang KL, Phookamsak R, Rollins AW, Romero AI, Stephenson S, Suetrong S, Tsui CKM, Vizzini A, Wen TC, De Silva NI, Promputtha I, Kang JC. (2015) The Facesoffungi database: fungal names linked with morphology, molecular and human attributes. Fungal Diversity 74: 3–18. 10.1007/s13225-015-0351-8 [DOI] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodsueb R, Lumyong S, McKenzie EHC, Bahkali AH, Hyde KD. (2016) Relationships between terrestrial and freshwater lignicolous fungi. Fungal Ecology 19: 155–168. 10.1016/j.funeco.2015.09.005 [DOI] [Google Scholar]
- Li WL, Dissanayake AJ, Luo ZL, Su HY, Liu JK. (2021) Additions to Distoseptispora (Distoseptisporaceae) associated with submerged decaying wood in China. Phytotaxa 520: 75–86. 10.11646/PHYTOTAXA.520.1.5 [DOI] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution 16: 1799–1808. 10.1093/oxfordjournals.molbev.a026092 [DOI] [PubMed] [Google Scholar]
- Luo J, Yin JF, Cai L, Zhang KQ, Hyde KD. (2004) Freshwater fungi in Lake Dianchi, a heavily polluted lake in Yunnan, China. Fungal diversity 16: 93–112. [Google Scholar]
- Luo ZL, Hyde KD, Liu JK, Bhat DJ, Bao DF, Li WL, Su HY. (2018) Lignicolous freshwater fungi from China II: Novel Distoseptispora (Distoseptisporaceae) species from northwestern Yunnan Province and a suggested unified method for studying lignicolous freshwater fungi. Mycosphere 9: 444–461. 10.5943/mycosphere/9/3/2 [DOI] [Google Scholar]
- Luo ZL, Hyde KD, Liu JK, Maharachchikumbura SSN, Jeewon R, Bao DF, Bhat DJ, Lin CG, Li WL, Yang J, Liu NG, Lu YZ, Jayawardena RS, Li JF, Su HY. (2019) Freshwater Sordariomycetes. Fungal Diversity 99: 451–660. 10.1007/s13225-019-00438-1 [DOI] [Google Scholar]
- Mckenzie EHC. (1995) Dematiaceous Hyphomycetes on Pandanaceae. 5. Sporidesmium sensu lato. Mycotaxon 56: 9–29. [Google Scholar]
- Mena-Portales J, Hernandez-Restrepo M, Guarro J, Minters DW, Gené J. (2016) New species of Penzigomyces, Sporidesmium and Stanjehughesia from plant debris in Spain. Nova Hedwigia 103: 359–371. 10.1127/nova_hedwigia/2016/0355 [DOI] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE 2010), 14: 1–8. 10.1109/GCE.2010.5676129 [DOI]
- Monkai J, Boonmee S, Ren GC, Wei DP, Phookamsak R, Mortimer PE. (2020) Distoseptisporahydei sp. nov. (Distoseptisporaceae), a novel lignicolous fungus on decaying bamboo in Thailand. Phytotaxa 459: 93–107. 10.11646/phytotaxa.459.2.1 [DOI] [Google Scholar]
- Nylander J. (2004) MrModeltest v2. Program distributed by the author: Evolutionary Biology Centre. Uppsala University, Sweden: http://www.ebc.uu.se/systzoo/staff/nylander.html
- Phookamsak R, Hyde KD, Jeewon R, Bhat DJ, Jones EBG, Maharachchikumbura SSN, Raspé O, Karunarathna SC, Wanasinghe DN, Hongsanan S, Doilom M, Tennakoon DS, Machado AR, Firmino AL, Ghosh A, Karunarathna A, Mešić A, Dutta AK, Thongbai B, Devadatha B, Norphanphoun C, Senwanna C, Wei D, Pem D, Ackah FK, Wang GN, Jiang HB, Madrid H, Lee HB, Goonasekara ID, Manawasinghe IS, Kušan Cano J, Gené J, Li J, Das K, Acharya K, Raj KNA, Latha KPD, Chethana KWT, He MQ, Dueñas M, Jadan M, Martín MP, Samarakoon MC, Dayarathne MC, Raza M, Park MS, Telleria MT, Chaiwan N, Matočec N, de Silva NI, Pereira OL, Singh PN, Manimohan P, Uniyal P, Shang QJ, Bhatt RP, Perera RH, Alvarenga RLM, Nogal-Prata S, Singh SK, Vadthanarat S, Oh SY, Huang SK, Rana S, Konta S, Paloi S, Jayasiri SC, Jeon SJ, Mehmood T, Gibertoni TB, Nguyen TTT, Singh U, Thiyagaraja V, Sarma VV, Dong W, Yu XD, Lu YZ, Lim YW, Chen Y, Tkalčec Z, Zhang ZF, Luo ZL, Daranagama DA, Thambugala KM, Tibpromma S, Camporesi E, Bulgakov T, Dissanayake AJ, Senanayake IC, Dai DQ, Tang LZ, Khan S, Zhang H, Promputtha I, Cai L, Chomnunti P, Zhao RL, Lumyong S, Boonmee S, Wen TC, Mortimer PE, Xu J. (2019) Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 95: 1–273. 10.1007/s13225-019-00421-w [DOI] [Google Scholar]
- Phukhamsakda C, McKenzie E, Phillips AJL, Jones EBG, Bhat DJ, Marc S, Bhunjun CS, Wanasinghe DN, Thongbai B, Camporesi E, Ertz D, Jayawardena RS, Perera RH, Ekanayake AH, Tibpromma S, Doilom M, Xu J, Hyde KD. (2020) Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Diversity 102: 1–203. 10.1007/s13225-020-00448-4 [DOI] [Google Scholar]
- Rambaut A. (2012) FigTree v1. 4. http://tree.bio.ed.ac.uk/[accessed January 14, 2019].
- Rannala B, Yang Z. (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311. 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systems Biology 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake IC, Rathnayaka AR, Marasinghe DS, Calabon MS, Gentekaki E, Wanasinghe DN, Lee HB, Hurdeal VG, Pem D, Dissanayake LS, Wijesinghe SN, Bundhun D, Nguyen TT, Goonasekara ID, Abeywickrama PD, Bhunjun CS, Chomnunti P, Boonmee S, Jayawardena RS., Wijayawardene NN, Doilom M, Jeewon R, Bhat JD, Zhang HX, Xie N. (2020) Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11: 2678–2754. https://doi 10.5943/mycosphere/11/1/20 [DOI] [Google Scholar]
- Shenoy BD, Jeewon R, Wu WP, Bhat DJ, Hyde KD. (2006) Ribosomal and RPB2 DNA sequence analyses suggest that Sporidesmium and morphologically similar genera are polyphyletic. Mycological 110: 916–928. 10.1016/j.mycres.2006.06.004 [DOI] [PubMed] [Google Scholar]
- Shoemaker RA, White GP. (1985) Lasiosphaeriacaesariata with Sporidesmiumhormiscioides and L.triseptata with S.adscendens. Sydowia 38: 278–283 [Google Scholar]
- Song HY, El Sheikha AF, Zhai ZJ, Zhou JP, Chen MH, Huo GH, Huang XG, Hu DM. (2020) Distoseptisporalongispora sp. nov. from freshwater habitats in China. Mycotaxon 135: 513–523. 10.5248/135.513 [DOI] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. (2008) A rapid bootstrap algorithm for the RAxML web servers. Systems Biology 57: 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- Su HY, Hyde KD, Maharachchikumbura SSN, Ariyawansa HA, Luo Z, Promputtha I, Tian Q, Lin C, Shang Q, Zhao Y, Chai H, Liu X, Bahkali AH, Bhat JD, McKenzie EHC, Zhou D. (2016) The families Distoseptisporaceae fam. nov., Kirschsteiniotheliaceae, Sporormiaceae and Torulaceae, with new species from freshwater in Yunnan Province, China. Fungal Diversity 80: 375–409. 10.1007/s13225-016-0362-0 [DOI] [Google Scholar]
- Subramanian CV. (1992) A reassessment of Sporidesmium (Hyphomycetes) and some related taxa. Proceedings of the Indian National Science Academy Part B Biological sciences 58: 179–190. [Google Scholar]
- Sun YR, Goonasekara ID, Thambugala KM, Jayawardena RS, Wang Y, Hyde KD. (2020) Distoseptisporabambusae sp. nov. (Distoseptisporaceae) on bamboo from China and Thailand. Biodiversity Data Journal 8: e53678. 10.3897/BDJ.8.e53678.figure4 [DOI] [PMC free article] [PubMed]
- Tibpromma S, Hyde KD, McKenzie EHC, Bhat DJ, Phillips AJL, Wanasinghe DN, Samarakoon MC, Jayawardena RS, Dissanayake AJ, Tennakoon DS, Doilom M, Phookamsak R, Tang AMC, Xu J, Mortimer PE, Promputtha I, Maharachchikumbura SSN, Khan S, Karunarathna SC. (2018) Fungal diversity notes 840–928: micro-fungi associated with Pandanaceae. Fungal Diversity 93: 1–160. 10.1007/s13225-018-0408-6 [DOI] [Google Scholar]
- Vaidya G, Lohman DJ, Meier R. (2011) SequenceMatrix: concatenation software for the fast assembly of multi‐gene datasets with character set and codon information. Cladistics 27: 171–180. 10.1111/j.1096-0031.2010.00329.x [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. 10.1128/jb.172.8.4238-4246.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns TD, Lee SB, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications: 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Wijayawardene NN, Hyde KD, Al-Ani LKT, Tedersoo L, Haelewaters D, Rajeshkumar KC, Zhao RL, Aptroot A, Leontyev DV, Saxena RK, Tokarev YS, Dai DQ, Letcher PM, Stephenson SL, Ertz D, Lumbsch HT, Kukwa M, Issi IV, Madrid H, Phillips AJL, Selbmann L, Pfliegler WP, Horváth E, Bensch K, Kirk P, Kolaříková Z, Raja HA, Radek R, Papp V, Dima B, Ma J, Malosso E, Takamatsu S, Rambold G, Gannibal PB, Triebel D, Gautam AK, Avasthi S, Suetrong S, Timdal E, Fryar SC, Delgado G, Réblová M, Doilom M, Dolatabadi S, Pawłowska J, Humber RA, Kodsueb R, Sánchez-Castrov I, Goto BT, Silva DKA, de Souza FA, Oehl F, da Silva GA, Silva IR, Błaszkowski J, Jobim K, Maia LC, Barbosa FR, Fiuza PO, Divakar PK, Shenoy BD, Castañeda-Ruiz RF, Somrithipol S, Karunarathna SC, Tibpromma S, Mortimer PE, Wanasinghe DN, Phookamsak R, Xu J, Wang Y, Fenghua T, Alvarado P, Li DW, Kušan I, Matočec N, Maharachchikumbura SSN, Papizadeh M, Heredia G, Wartchow F, Bakhshi M, Boehm E, Youssef N, Hustad VP, Lawrey JD, Santiago ALCM, Bezerra JDP, Souza-Motta CM, Firmino AL, Tian Q, Houbraken J, Hongsanan S, Tanaka K, Dissanayake AJ, Monteiro JS, Grossart HP, Suija A, Weerakoon G, Etayo J, Tsurykau A, Kuhnert E, Vázquez V, Mungai P, Damm U, Li QR, Zhang H, Boonmee S, Lu YZ, Becerra AG, Kendrick B, Brearley FQ, Motiejūnaitė J, Sharma B, Khare R, Gaikwad S, Wijesundara DSA, Tang LZ, He MQ, Flakus A, Rodriguez-Flakus P, Zhurbenko MP, McKenzie EHC, Stadler M, Bhat DJ, Liu JK, Raza M, Jeewon R, Nassonova ES, Prieto M, Jayalal RGU, Yurkov A, Schnittler M, Shchepin ON, Novozhilov YK, Liu Pu, Cavender JC, Kang Y, Mohammad S, Zhang LF, Xu RF, Li YM, Dayarathne MC, Ekanayaka AH, Wen TC, Deng CY, Lateef AA, Pereira OL, Navathe S, Hawksworth DL, Fan XL, Dissanayake LS, Erdoğdu M. (2020) Outline of fungi and fungus-like taxa. Mycosphere 11: 1060–1456. 10.5943/mycosphere/11/1/8 [DOI] [Google Scholar]
- Xia JW, Ma YR, Li Z, Zhang XG. (2017) Acrodictys-like wood decay fungi from southern China, with two new families Acrodictyaceae and Junewangiaceae. Scientific Reports 7: 78–88. 10.1038/s41598-017-08318-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Maharachchikumbura SSN, Hyde KD, Bhat DJ, McKenzie EH, Bahkali AH, Gareth Jones EB, Liu ZY. (2015) Aquapteridosporalignicola gen. et sp. nov., a new hyphomycetous taxon (Sordariomycetes) from wood submerged in a freshwater stream. Cryptogamie Mycologie 36: 469–478. 10.7872/crym/v36.iss4.2015.469 [DOI] [Google Scholar]
- Yang J, Maharachchikumbura SSN, Liu JK, Hyde KD, Gareth Jones EB, Al-Sadi AM, Liu ZY. (2018) Pseudostanjehughesiaaquitropica gen. et sp. nov. and Sporidesmiumsensu lato species from freshwater habitats. Mycological Progress 17: 591–616. 10.1007/s11557-017-1339-4 [DOI] [Google Scholar]
- Zhang H, Dong W, Hyde KD, Maharachchikumbura SS, Hongsanan S, Bhat DJ, Al-Sadi AM, Zhang D. (2017) Towards a natural classification of Annulatascaceae-like taxa: introducing Atractosporales ord. nov. and six new families. Fungal Diversity 85: 75–110. 10.1007/s13225-017-0387-z [DOI] [Google Scholar]
- Zhaxybayeva O, Gogarten JP. (2002) Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC Genomics 3: 1–15. 10.1186/1471-2164-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou DQ, Hyde KD, Wu XL. (2001) New records of Ellisembia, Penzigomyces, Sporidesmium and Repetophragma species on bamboo from China. Acta Botanica Yunnanica. 1: 45–w51.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.