ABSTRACT
Microorganisms typically maintain cellular homeostasis despite facing large fluctuations in their surroundings. Microbes that reside on human mucosal surfaces may experience significant variations in nutrient and ion availability as well as pH. Whether the mechanisms employed by these microbial cells to sustain homeostasis directly impact on the interplay with the host’s mucosae remains unclear. Here, we report that the previously uncharacterized transcription regulator ZCF8 in the human-associated yeast Candida albicans maintains vacuole homeostasis when the fungus faces fluctuations in nitrogen. Genome-wide identification of genes directly regulated by Zcf8p followed by fluorescence microscopy to define their subcellular localization uncovered the fungal vacuole as a top target of Zcf8p regulation. Deletion and overexpression of ZCF8 resulted in alterations in vacuolar morphology and luminal pH and rendered the fungus resistant or susceptible to nigericin and brefeldin A, two drugs that impair vacuole and associated functions. Furthermore, we establish that the regulator modulates C. albicans attachment to epithelial cells in a manner that depends on the status of the fungal vacuole. Our findings, therefore, suggest that fungal vacuole physiology regulation is intrinsically linked to, and shapes to a significant extent, the physical interactions that Candida cells establish with mammalian mucosal surfaces.
KEYWORDS: Candida albicans, host-pathogen interactions, transcriptional regulation, vacuole
INTRODUCTION
Candida albicans is a prominent fungal species residing in humans. The majority of healthy adults carry this fungus as part of their normal gut microbiota (1, 2) although C. albicans can also thrive in other sites of the human body, including the oral cavity. Overgrowth of the fungus on the mouth’s mucosal surface causes a disease termed oropharyngeal candidiasis (OPC). As with other fungi, a debilitated immune system is the main risk factor associated with Candida infections. Indeed, OPC is one of the most prevalent manifestations in AIDS patients (3). In the last few years, the first unbiased and systematic searches for C. albicans genes involved in gut colonization and/or OPC have been reported (4–7). While a handful of the identified open reading frames (ORFs) could be linked to specific roles and cellular processes (based on previous studies in Candida or relying on the annotation of orthologs in the model yeast Saccharomyces cerevisiae), most of the “hits” derived from these genetic screenings remain uncharacterized. Investigating how this subset of uncharacterized Candida ORFs contribute to gut colonization and/or OPC is likely to reveal novel aspects of the biology of the fungus in the mammalian host.
The C. albicans ZCF8 gene encodes a putative zinc cluster transcription regulator with a narrow phylogenetic distribution (orthologous genes can be found only within the Candida CTG clade). Through genetic screenings, our laboratory found that the zcf8 null mutant strain displays two striking opposing phenotypes in mice: (i) impaired gut colonization after oral gavage (4) and (ii) increased fungal load in tongues in an OPC model (5). The molecular basis of these phenotypes, however, remained unexplored. The only other report on ZCF8 in the literature demonstrated a role for this gene in promoting C. albicans adherence to silicone surfaces (8). In large-scale phenotypic screenings conducted under a variety of laboratory growth conditions, the C. albicans zcf8 deletion mutant exhibited wild-type levels of proliferation across all conditions evaluated (9).
The fungal vacuole is a dynamic organelle with a range of functions, including the storage of amino acids, phosphate and calcium, glycoprotein turnover and hydrolysis, ion homeostasis, pH and osmotic regulation (reviewed in references 10 to 13). The vacuole lies at the center of fungal cell physiology, and as such, it is a highly responsive organelle that adjusts itself to both external and internal stimuli. Studies of S. cerevisiae indicate that the shape and number of vacuoles found in a cell often reflect a response to the surrounding environment. In proliferating cells, for example, high metabolic activity favors multiple medium-sized vacuoles, whereas nutrient limitation induces fusion events that result in one single organelle taking up most of the cell volume. A large size vacuole fits the needs of the cell to break down cellular material delivered to this organelle via autophagy and/or the multivesicular body pathway (11). Vacuole fission and fusion depend, at least in part, on the acidity inside the organelle which is typically maintained around pH 6.25 by the vacuolar ATPase. The activity of this proton pump generates the H+ ions that give the vacuole its characteristic acidic pH, which is critical for degradation processes, storage of metabolites, and energizing other vacuolar transporters (11, 14).
In this report, we investigate the role(s) that the ZCF8 gene play(s) in the biology of C. albicans. Employing genome-wide approaches to systematically identify ZCF8-regulated genes, we establish that Zcf8p is a regulator of vacuolar function. In support of this notion, we found that deletion or overexpression of the regulator resulted in alterations of various vacuolar properties. We also demonstrate that ZCF8 impinges upon the ability of the fungus to attach to mucosal surfaces and that the regulator’s effect on fungal adhesion depends, at least in part, on its regulation of the vacuole. Our results, therefore, establish a novel link between fungal attachment to mammalian host surfaces and the regulation of the fungal vacuole.
RESULTS
ChIP-Seq reveals that the transcription regulator Zcf8p binds upstream of genes encoding vacuolar and intracellular vesicle trafficking components.
The gene ZCF8 encodes a putative zinc cluster transcription regulator. While genetic screenings have linked mutations in this gene to altered patterns of C. albicans colonization and infection in mouse models of gastrointestinal colonization (4) and oropharyngeal candidiasis (5), respectively, as well as to reduced adherence to silicone-covered surfaces (8), how Zcf8p effectuates these phenotypes remains unknown. Phylogenetic reconstruction based on the conserved DNA binding domain of the protein family in Hemiascomycetes (a group that includes the model yeast Saccharomyces cerevisiae as well as the entire Candida clade) suggests that this gene arose by gene duplication (or horizontal gene transfer from an unidentified source) relatively recently in evolutionary timescales because true orthologs could be identified only in the Candida or CTG clade (see Fig. S1 in the supplemental material).
Closest homologs of ZCF8 in the Candida clade. (A) Cladogram depicting the phylogenetic relationships among extant species of the Candida and Saccharomyces clades. Colored arrows represent ZCF8 and its closest homologs according to the Candida Genome Database and BLAST searches conducted with either full-length or conserved DNA binding domain protein sequences. Gene orthology assignments (same colors) follow independent reconstructions generated by the Candida Gene Order Browser. OAF1 and HAP1 (striped lines) are retrieved as “best hits” when queried with ZCF34 and ZCF5 sequences, respectively, in S. cerevisiae (however, neither OAF1 nor HAP1 are considered true orthologs of the indicated Candida genes). (B) Protein sequence alignment of putative DNA binding domains of ZCF8 and its closest homologs. Alignment was performed by MUSCLE. Residues highlighted in red are the six cysteines characteristic of the zinc cluster domain. (C) Maximum likelihood phylogeny of ZCF8 and its closest homologs. Alignments were generated using MAFFT (v.7.475). IQ-TREE 2 (v.2.1.2) was used to predict the best-fitting model (VT+F+R4), and FigTree (v1.4.4) (http://tree.bio.ed.ac.uk/software/figtree/) was used to generate the tree. Download FIG S1, TIF file, 0.5 MB (501.3KB, tif) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
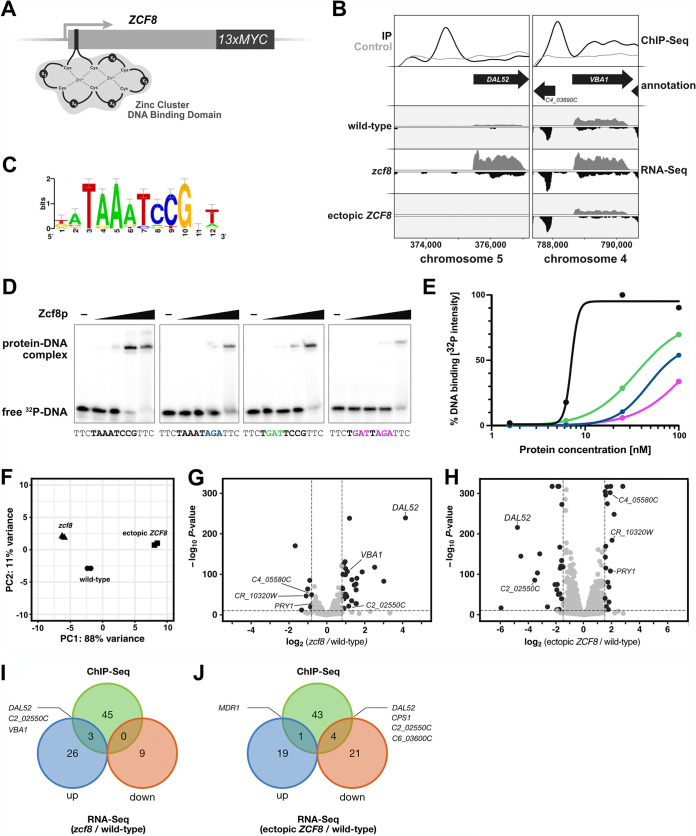
To identify genes regulated by Zcf8p in C. albicans, we carried out chromatin immunoprecipitation followed by deep (Illumina) sequencing (ChIP-Seq). The native ZCF8 locus was genetically modified to encode a C-terminal 13×myc epitope-tagged Zcf8 protein (Fig. 1A). The expression of the tagged gene remained under the control of the native promoter. We established that the activity of the tagged protein was only slightly diminished compared to the activity of the untagged version (Fig. S2A). Zcf8p constitutively localized in the Candida nucleus (Fig. S2B). Using stringent criteria to define segments enriched in the immunoprecipitated sample (see Materials and Methods), we identified 42 “peaks” or DNA regions bound by Zcf8p genome-wide (Fig. 1B; see also Table S1 in the supplemental material). The DNA sequences (500 nucleotides [nt] centered on the highest point of each peak) of the top scoring ∼1/4 of the bound regions were used to derive overrepresented DNA binding motifs de novo. The sequence 5′-TAAATCCG-3′ (Fig. 1C) was salient among the derived motifs because 5′-CCG-3′ is a well-established element of the sequences bound by other zinc cluster transcription regulators (15). We established that the derived sequence is indeed a bona fide binding site for Zcf8p because the purified recombinant protein bound a DNA fragment carrying this sequence in electrophoretic mobility shift assays (Fig. 1D). Furthermore, the introduction of point mutations in the 5′-AAA-3′ or 5′-CCG-3′ portion of the sequence impaired binding (Fig. 1D), although to different degrees (Fig. 1E).
FIG 1.
Identification of C. albicans genes controlled by the transcription regulator Zcf8p. (A) Schematic of the putative zinc cluster DNA binding domain found toward the 5′ end of the coding region of the C. albicans ZCF8 gene. A sequence encoding the 13× MYC tag was introduced in frame at the 3′ end of the native ZCF8 locus to facilitate immunoprecipitation. (B) ChIP-Seq and RNA-Seq tracks illustrating two genes directly regulated by Zcf8p. The top panel shows the IP (black) and control (gray) tracks resulting from ChIP-Seq. The bottom panels show the RNA-Seq tracks derived from three strains: wild-type strain, zcf8 deletion mutant, and a strain overexpressing ZCF8. (C) DNA motif overrepresented in the top scoring regions occupied by Zcf8p according to the ChIP-Seq experiment. (D) Gel shift assays showing binding of the Zcf8 protein to its predicted binding site. 32P-labeled DNA fragments (0.4 nM) containing either the wild-type or mutant binding site were incubated with increasing concentrations (0, 1.56, 6.25, 25, and 100 nM) of the purified DNA binding domain (amino acids 57 to 158) of the Zcf8 protein for 30 min at room temperature in standard electrophoretic mobility shift assay (EMSA) buffer and resolved in 6% polyacrylamide gels run with 0.5× Tris-glycine-EDTA buffer (TGE; 10× TGE contains 30.3 g Tris HCl, 142 g glycine, 37.2 g EDTA per 1 L of solution). The DNA fragment evaluated (30 nt in size) is located upstream of the DAL52 gene. Point mutations introduced at key positions are indicated in colors. (E) Quantification of the gel shifts shown in panel D. Colors correspond to DNA fragments with the wild-type sequence (black) or carrying point mutations introduced in different parts of the binding site (green, blue, and pink). Notice the log scale of the x axis. (F) Principal-component analysis (PCA) based on results of RNA-Seq experiment. Total RNA was prepared from the wild-type strain, zcf8 deletion mutant, and a strain overexpressing ZCF8. All three strains were grown in defined medium lacking amino acids and ammonium sulfate as described in Materials and Methods. (G and H) Transcripts up- or downregulated in the zcf8 deletion mutant (G) and in the strain overexpressing ZCF8 (H). Shown are volcano plots where each dot represents one transcript. Significantly up- or downregulated transcripts are shown in black. (I and J) Venn diagrams showing the overlap between ChIP-Seq and the indicated RNA-Seq data sets.
Activity of tagged ZCF8 allele, subcellular localization of Zcf8p, and transcriptional autoregulation. (A) Quantitative RT-PCR analysis comparing the activity of the tagged and untagged ZCF8 alleles. Shown is the level of repression of DAL52, the top target of regulation of Zcf8p. The DAL52 transcript levels measured in the homozygous deletion mutant (Δzcf8/Δzcf8) were used to estimate the percent repression achieved in each of the three indicated strains. The experimentally validated TAF10 transcript (M.-A. Teste, M. Duquenne, J. M. François, J.-L. Parrou, BMC Mol Biol 10:99, 2009, https://doi.org/10.1186/1471-2199-10-99) served as reference control to normalize the quantitative reverse transcription-PCR (qRT-PCR) data. Shown are the means ± SD (n = 3). The Δzcf8/ZCF8-MYC strain was used in the chromatin immunoprecipitation experiment. (B) Subcellular localization of GFP-tagged Zcf8p. Arrowheads indicate nuclei. Staining with FM4-64 delineates the vacuoles (marked with “v”). (C) Schematic of the reporter strains. In strain I (left), one ZCF8 allele was replaced by YFP. In strain II, the second ZCF8 allele was deleted. In both strains, the expression of the reporter is driven by the native ZCF8 promoter. (D) YFP fluorescence emitted by the reporter strains described in panel C. Cells were grown overnight in YPD and diluted in PBS. Fluorescence was measured using a flow cytometer. Ten thousand cells were evaluated per strain per experiment. Plotted are the means ± SD of three independent experiments. The Student’s t test was used in the statistical analysis. Download FIG S2, TIF file, 0.9 MB (939.2KB, tif) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of ChIP-Seq peaks and associated genes. This file includes a list of all Zcf8p ChIP peaks. The C. albicans chromosome and genome coordinates, fold enrichment as well as each peak’s nearest annotated ORF are indicated. Download Table S1, XLSX file, 0.02 MB (18.2KB, xlsx) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene Ontology (GO) analyses of the ∼50 ORFs associated with Zcf8 ChIP “peaks” in their upstream intergenic region revealed generic terms such as “sequence-specific DNA binding” (P = 0.00055), “biofilm formation” (P = 0.00033), “cell aggregation” (P = 0.00033), “transmembrane transport” (P = 0.00575), and “regulation of filamentous growth” (P = 0.0074) overrepresented in the data set. About half of the ∼50 ORFs remain unannotated in the Candida Genome Database. We noticed that about a quarter of the unambiguously annotated Zcf8p-regulated ORFs had roles connected to either vacuole or intracellular vesicle trafficking: for example, the vacuolar basic amino acid transporter VBA1 (16); the conserved oligomeric Golgi complex COG2 (17, 18), an essential component of a cytosolic tethering complex that functions in protein trafficking to mediate fusion of transport vesicles to Golgi compartments; the unconventional SNARE in the endoplasmic reticulum USE1 (19, 20), which is involved in retrograde traffic from the Golgi to the endoplasmic reticulum (ER); the putative oxidoreductase C1_10840C (21) which is involved in late endosome to Golgi transport; and C4_03050C which has homology to S. cerevisiae genes described as vacuole-related genes. These findings raised the possibility that Zcf8p may regulate some aspect of vacuole biology or intracellular trafficking in C. albicans.
Zcf8p’s top targets of regulation localize to the fungal vacuole.
To further define the transcripts regulated by Zcf8p, we conducted a transcriptome (RNA-Seq) experiment in which we included three C. albicans strains: the zcf8 deletion mutant, a strain overexpressing ZCF8 (i.e., in this strain ZCF8 expression is driven by the strong promoter TDH3) and the wild-type reference strain (Fig. 1F). This experiment identified 39 and 45 transcripts with significant changes in expression when comparing the wild type versus the deletion mutant (negative log10 P value > 10 and log2 fold change > |0.8|) or the wild type versus the ZCF8 overexpression strain (negative log10 P value > 10 and log2 fold change > |1.5|), respectively (Fig. 1G and H and Table S2). As highlighted in Fig. 1G and H, a subset of transcripts downregulated in the zcf8 deletion mutant were upregulated in the ZCF8 overexpression strain and vice versa (e.g., C4_05580C, CR_10320W, and DAL52), as expected. The inclusion of the strain overexpressing ZCF8, nonetheless, expanded the list of putative ZCF8-regulated transcripts. GO searches with the entire list of differently expressed ORFs retrieved “amino acid biosynthesis” (P = 0.00193) and “amino acid metabolism” (P = 0.00927) as terms overrepresented in the data set.
List of differentially expressed transcripts in zcf8 deletion and ZCF8 overexpression strains. This file includes a list of the C. albicans genes whose expression was up- or downregulated in the zcf8 deletion mutant or in a strain overexpressing ZCF8. Download Table S2, XLSX file, 0.02 MB (19.8KB, xlsx) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
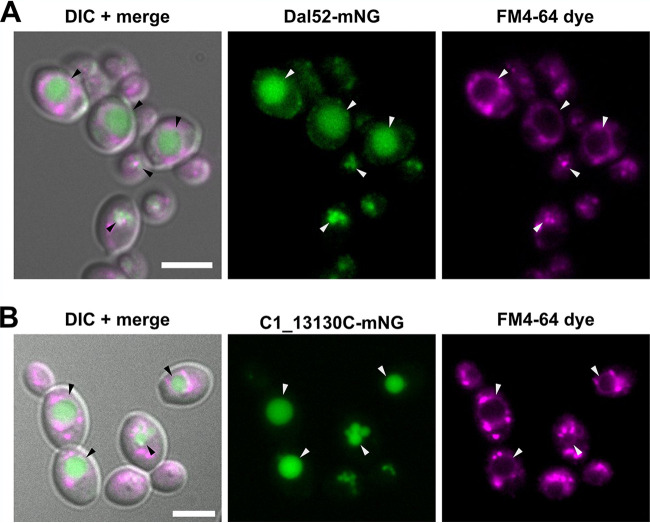
Two of the top targets of Zcf8p regulation (which we define here as the genes associated with a Zcf8p ChIP peak in their 5′ intergenic region and displaying the largest change in RNA-Seq reads when comparing a deletion mutant versus the wild-type reference strain) were DAL52, which is annotated as “putative allantoate permease” and VBA1, which encodes a vacuolar basic amino acid transporter (Fig. 1B, I, and J). To establish the subcellular localization of the product encoded by DAL52, we fused this gene in its native chromosomal location to a Candida-optimized version of the fluorescent reporter mNeonGreen (22). Strikingly, Dal52p displayed intravacuolar localization (the vacuole is outlined with the lipophilic dye FM4-64) (Fig. 2A). A similar intravacuolar distribution pattern was observed for C1_13130C (Fig. 2B), another gene that we identified as a target of Zcf8p regulation in the ChIP-Seq experiment and remains annotated as “putative histidine permease.” These findings further hinted at a connection between Zcf8p and the fungal vacuole.
FIG 2.
Putative allantoate and amino acid permeases regulated by Zcf8p are targeted to the fungal vacuole. (A and B) Subcellular localization of two gene products transcriptionally regulated by Zcf8p. The sequence encoding the C. albicans codon-optimized reporter mNeonGreen (mNG) was introduced in frame at the 3′ end of the DAL52 (A) and C1_13130C (B) genes. The FM4-64 dye stains the fungal vacuole (indicated by arrowheads) and intracellular vesicles resulting from endocytosis. DIC, differential interference contrast. Bars, 5 μm.
Zcf8p functions primarily as a transcriptional repressor.
Our RNA-Seq data indicated that the genes under direct control of Zcf8p, i.e., the genes that in addition to showing a change in transcript levels also displayed a ChIP “peak” in their 5′ intergenic region, were for the most part subject to transcriptional repression by this protein (Fig. 1B). A common feature found in transcriptional regulatory circuits is that the regulator controls not only other target genes but also its own expression by binding to its promoter (15). Our ChIP data indicated that the Zcf8 protein also bound to the intergenic region upstream of the ZCF8 ORF (Table S1), an arrangement that is suggestive of autoregulation. To test this model and determine whether Zcf8p exerted positive or negative regulation on itself, we constructed reporter strains in which one copy of the ZCF8 ORF was replaced by yellow fluorescent protein (YFP) (Fig. S2C). The second ZCF8 copy either remained intact or was deleted (Fig. S2C). The YFP levels (quantified by flow cytometry) were significantly lower in the heterozygous strain harboring an intact ZCF8 copy compared to the isogenic zcf8Δ strain, which is consistent with Zcf8p repressing its own expression (Fig. S2D). While it cannot be ruled out that ZCF8 may also act as an activator on a yet-to-be-determined set of genes, our results suggest that the Zcf8 protein acts primarily as a transcriptional repressor; as such, its autoregulation generates a negative-feedback loop.
zcf8 mutation and ZCF8 overexpression result in altered vacuole properties.
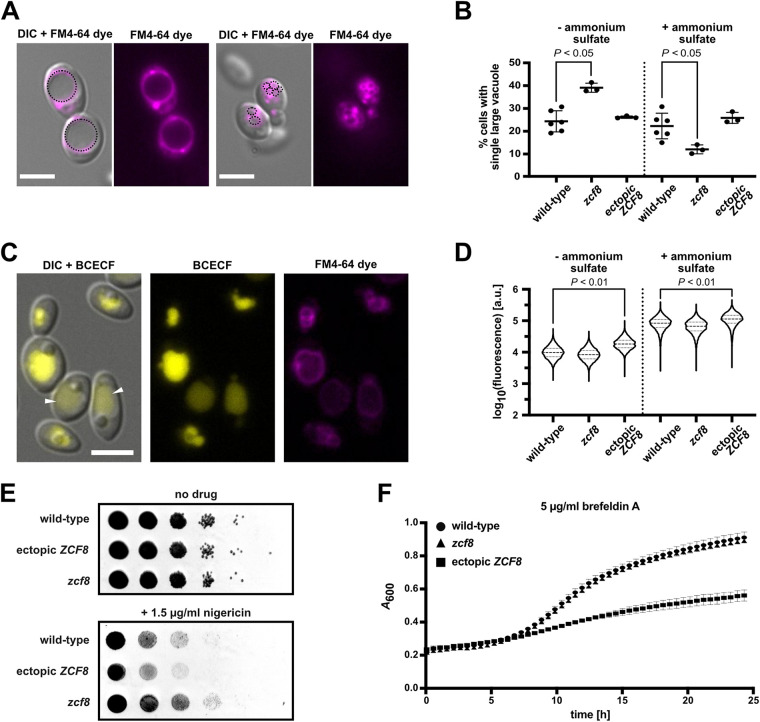
The fungal vacuole is a dynamic organelle. The presence of a single, large vacuole in yeast cells is thought to reflect nutrient limitation or other stress as these conditions induce vacuolar fusion events (11). Yet yeast cells can contain up to five vacuoles per cell (23) and the number can vary depending on extracellular and intracellular conditions (11). Because the presence of either a single large vacuole or multiple smaller compartments is a defining feature of the organelle, we first sought to evaluate this trait in Zcf8-deficient C. albicans cells and in a strain overproducing the regulator. We evaluated C. albicans cells grown in defined medium lacking any nitrogen source or supplemented with ammonium sulfate. The reasons for the choice of nitrogen as a variable in the growth conditions were twofold. First, our genome-wide ChIP and RNA-Seq data pointed to (intra)cellular transporters of amino acids or other nitrogen-rich molecules (e.g., allantoate) as top targets of Zcf8p’s regulation (Fig. 1 and 2). Second, the importance of mobilizing amino acids in and out of the vacuole is particularly evident when cells face nitrogen-poor conditions (24, 25). To visualize the vacuoles, we stained C. albicans cells with the lipophilic dye FM4-64 (Fig. 3A). As shown in Fig. 3B, the fraction of cells displaying a single large vacuole differed significantly in the zcf8 mutant compared to the wild-type reference strain: in the presence of ammonium sulfate, the zcf8 mutant had fewer single-vacuole cells, whereas under nitrogen starvation the fraction of single-vacuole cells increased almost threefold. These results suggest that ZCF8 is required for C. albicans to withstand large variations in vacuolar fusion and fission events due to changes in nitrogen availability.
FIG 3.
Zcf8p regulates vacuolar properties in C. albicans. (A and B) Vacuolar morphology in C. albicans cells is dependent on ZCF8. (A) The number of vacuoles within the fungal cell changes in response to conditions in the surroundings and can range from a single, large organelle to multiple, smaller compartments. (B) The fraction of cells (percentage) displaying a single large vacuole is plotted. The wild-type strain, zcf8 deletion mutant, and a strain overexpressing ZCF8 were evaluated. Cells were grown in the presence (+) and absence (−) of ammonium sulfate and stained using FM4-64 to visualize the vacuole(s). Each dot represents the mean of one experiment in which >500 cells were scored. The experiment was repeated at least three times. The Mann-Whitney U test was used in the statistical analysis. Bars, 5 μm. (C and D) Vacuolar pH increases in C. albicans cells overexpressing ZCF8. Cells were stained with the vacuolar pH indicator BCECF which emits higher fluorescence at higher pH. The microscopy images shown in panel C illustrate the variation in vacuolar fluorescence intensity (a proxy for variation in vacuolar pH) within a cell population. Arrowheads point to vacuoles with lower fluorescent signal (i.e., lower pH). (D) Quantification by flow cytometry of the fluorescence emitted by BCECF in single cells grown in media containing or lacking ammonium sulfate. Fluorescence is shown in arbitrary units. The median values and quartiles are indicated with dashed and dotted lines, respectively. Ten thousand cells were evaluated per strain in each individual experiment. Data shown are derived from at least four independent experiments. The Mann-Whitney U test was used in the statistical analysis. Bar, 5 μm. (E) Spot assay showing the increased resistance of the zcf8 mutant and increased susceptibility of the strain overexpressing ZCF8 to nigericin. Serially diluted cell suspensions were spotted on defined agar medium containing ammonium sulfate as the nitrogen source. Pictures were taken after 24 h of incubation. (F) Growth assay showing the susceptibility of the strain overexpressing ZCF8 to brefeldin A. Cells were grown in defined liquid medium containing ammonium sulfate as the nitrogen source.
Fungal vacuoles are acidic compartments. The acid pH is not only a defining feature of these organelles but can also be regulated and is important for vacuolar functions such as transport of ion and metabolites, fusion and fission events, and protein sorting (26). Thus, we sought to establish whether the vacuoles of Zcf8-deficient C. albicans cells or of a strain overproducing the regulator display alterations in pH. As described in the previous paragraph, we evaluated C. albicans cells grown in defined medium lacking any nitrogen source or supplemented with ammonium sulfate. The pH-sensitive fluorescent indicator BCECF [2′,7′‐bis‐(2‐carboxyethyl)‐5‐(and ‐6)‐carboxyfluorescein] was used to indirectly assess the pH within the vacuolar lumen (Fig. 3C). The fluorescence intensity emitted by this compound upon excitation is proportional to the pH in the respective compartment, within the pH range ∼6 to ∼9 (higher fluorescence intensity, more alkaline pH) (27, 28). We used flow cytometry to quantify the fluorescence in each population of cells, and the observed distributions are shown in Fig. 3D. This analysis revealed that ZCF8 overexpression resulted in cells containing vacuoles of more alkaline pH, particularly when the cells were grown in medium lacking ammonium sulfate. The zcf8 deletion mutant, on the other hand, displayed a trend toward vacuoles of more acidic pH. The pH or acidity of the vacuole, therefore, changes in response to alterations in ZCF8 expression.
We found that neither autophagy nor endocytosis, two cellular processes associated with the vacuole, are altered to a significant extent in the zcf8 mutant strain. Green fluorescent protein (GFP)-Atg8, a well-established marker of autophagy, was employed to assay bulk autophagy by monitoring vacuolar delivery of GFP-Atg8 by Western blotting analysis (Fig. S3) (29). The uptake of the fluorescent dye FM4-64 was used to visualize membrane internalization and endocytic delivery to the vacuole (Fig. S4) (30).
ZCF8 does not influence autophagy. (A) Western blotting of the autophagy marker GFP-Atg8p in wild-type background and in the zcf8 deletion mutant using an anti-GFP antibody. These two strains, together with the untagged control, were grown in defined liquid medium without amino acids and ammonium sulfate for 3 or 6 h. The accumulation of free GFP (due to Atg8p degradation) is a proxy for autophagy. Blotting with an anti-tubulin 1 antibody shows equal loading. (B) Vacuolar delivery of mNeonGreen-Atg8p by bulk autophagy visualized by fluorescence microscopy. Autophagosome is marked with white arrowhead. Bar, 5 μm. (C) Quantification of microscopy images shown in panel B. Plotted is the fraction of cells with a visible autophagosome as a proxy for autophagic activity. Every dot represents the percentage of cells per microscope field (only fields with >20 cells were scored). Twenty frames were scored in each experiment. Plotted are the results from five independent experiments. Download FIG S3, TIF file, 0.9 MB (915.6KB, tif) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Evaluating the role of ZCF8 in endocytosis. (A) Internalization of endocytic vacuoles in C. albicans. Cells grown in defined liquid medium without amino acids and ammonium sulfate were incubated with the lipophilic dye FM4-64 for 10 or 20 min, washed with ice-cold PBS, and kept on ice until evaluated by fluorescence microscopy. Representative images are shown. Bars, 5 μm. (B) Quantification of images shown in panel A. Plotted is the mean fluorescence (± SD) emitted by the internalized FM4-64 dye. The fluorescence emitted by each cell was measured using FIJI. At least 400 cells (derived from >20 microscope fields) were measured per strain per time point per experiment. Data collected from four independent experiments are shown. Download FIG S4, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Consistent with the observed alterations in vacuolar properties, we established that deletion of ZCF8 rendered C. albicans resistant to nigericin, a K+/H+ ionophore that targets, among other organelles, the fungal vacuole (31), whereas overexpression resulted in hypersensitivity to the drug (Fig. 3E). The ionophore introduces ion “channels” into membranes; as a consequence, it breaks down the pH gradient between cytoplasm and vacuole disrupting the organelle’s function. The ZCF8 overexpression strain also displayed increased sensitivity to brefeldin A, a drug that targets the Golgi and impairs vesicle formation and transport (Fig. 3F). Collectively, the findings described in this section demonstrate that key properties of the fungal vacuole such as number and pH are controlled by Zcf8p in C. albicans.
C. albicans upregulates amino acid transporters in response to nigericin.
Because the ZCF8 gene is connected to C. albicans susceptibility/resistance to nigericin, we were interested in establishing more broadly the response of the fungus to this drug. To do so, we conducted a RNA-Seq analysis comparing nigericin-treated versus untreated C. albicans cultures. To minimize secondary effects caused by drug treatment, we evaluated cells after 30 min of incubation with nigericin. To our knowledge, there is no published data set on the response of any yeast to this drug at the transcriptome level.
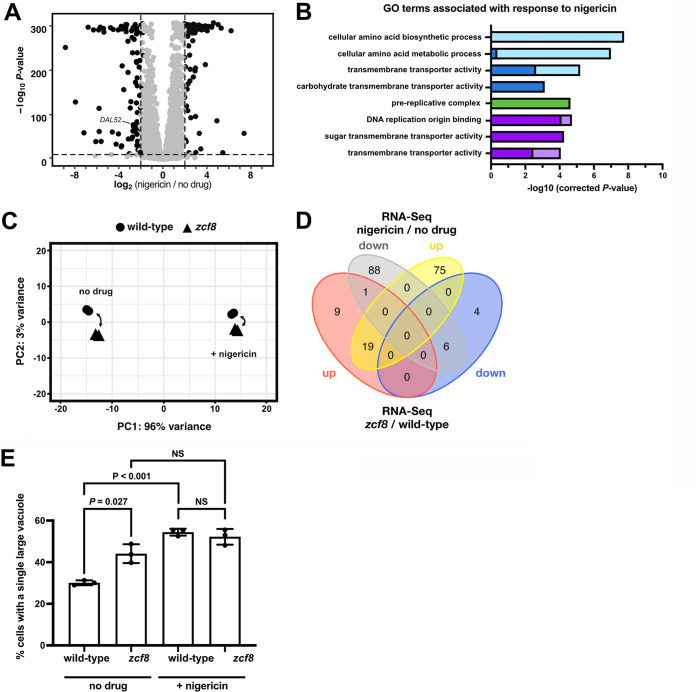
Our RNA-Seq experiment identified 188 protein-coding transcripts with altered expression in response to the drug (using the following cutoffs: −log10 P > 10 and expression changes >2-fold; Fig. 4A and Table S3). GO term searches of processes, functions, and components identified “cellular amino acid biosynthetic process” and “cellular amino acid metabolic process” as the top categories enriched in the set of nigericin-induced genes, whereas “prereplicative complex” was the top term overrepresented in the set of drug-repressed genes (Fig. 4B). An overall comparison of the responses to the drug at the transcriptome level between the wild-type reference strain and zcf8 mutant can be visualized in the principal-component analysis (PCA) shown in Fig. 4C. While the drug itself explains a substantial portion (96%) of the variance in this experiment, the wild type and zcf8 mutant are placed closer to each other in the presence of nigericin (compared to the samples derived from cultures without the drug). The distance that separates two samples in the PCA plot are proportional to their similarity/differences; therefore, this analysis indicates that, at a global level, the transcriptional changes effected by ZCF8 are diminished in the presence of the drug. Furthermore, the Venn diagram showing the overlap between nigericin- and ZCF8-regulated genes (Fig. 4D) indicates that more than half of the upregulated transcripts in the ZCF8 deletion mutant (19 of 29 transcripts) were also upregulated in response to the drug.
FIG 4.
C. albicans upregulates amino acid transporters in response to nigericin. (A) Transcripts up- or downregulated in response to nigericin treatment. Wild-type cells were grown in medium without amino acids and ammonium sulfate and treated for 30 min with the drug (at a final concentration of 15 μg/ml). Shown is a volcano plot where each dot represents one transcript. Significantly up- or downregulated transcripts are shown in black. (B) GO terms enriched in the set of transcripts that changed their expression level in response to nigericin. Light and dark colors indicate enrichment in the set of drug-induced or drug-repressed genes, respectively. Blue, green, and purple indicate “process”, “function,” or “component” categories. (C) Principal-component analysis based on results of RNA-Seq experiment conducted with the wild-type strain and zcf8 deletion mutant in the presence and absence of nigericin. (D) Venn diagram showing the overlap between the set of nigericin-regulated genes and up- or downregulated transcripts following ZCF8 deletion. (E) Altered vacuole morphology in response to nigericin. Cells cultured under nitrogen starvation conditions were treated with the drug (15 μg/ml) for 30 min and washed twice with PBS before staining with FM4-64. Plotted is the percentage of cells displaying a single large vacuole. Bars represent the means ± standard deviations (SD) (error bars) of three independent experiments in which >150 cells were scored per strain per condition. The Student’s t test was used in the statistical analysis. NS, not significant.
List of differentially expressed transcripts in response to nigericin. This file includes a list of the C. albicans genes whose expression was significantly altered in response to nigericin treatment in the wild-type reference strain and the zcf8 deletion mutant. Download Table S3, XLSX file, 0.03 MB (30.8KB, xlsx) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Consistent with the findings in the transcriptome data, we established that nigericin altered the fraction of Candida cells displaying a large single vacuole or multiple smaller organelles (Fig. 4E). Of importance to this report, the drug had no significant effect in the zcf8 mutant, further supporting a connection between the vacuole and regulator.
Zcf8p modulates Candida interactions with host epithelial cells in a vacuole-dependent manner.
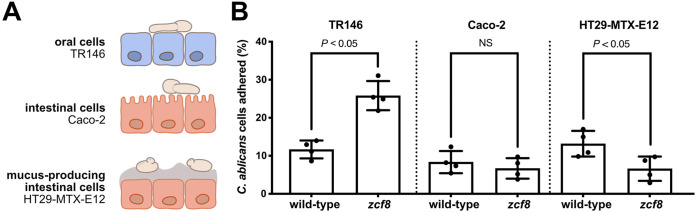
In previous studies, we had established that the C. albicans zcf8 null mutant strain displays two striking opposing phenotypes in mice: (i) impaired gut colonization after oral gavage (4) and (ii) increased fungal load in tongues in the oropharyngeal candidiasis model (5). Because Finkel et al. (8) had reported that the zcf8 mutant shows reduced fungal adherence to silicone, we hypothesized that the mouse-associated phenotypes may also be due to alterations in the attachment of the deletion mutant strain to mucosal surfaces. To test this idea, we probed the attachment of the fungus to intestinal (Caco-2 cells and the mucus-producing HT29-MTX-E12 cell line) and oral epithelial (TR146) cells (Fig. 5A). The inclusion of a mucus-secreting intestinal cell line follows the observation that, in the murine colon, C. albicans cells do not directly contact epithelial cells but rather remain adjacent to, or immersed within, the intestinal mucous layer (4, 32). We found that, compared to the wild-type reference strain, the isogenic zcf8 mutant adhered significantly less to the mucus-producing intestinal cells (Fig. 5B). Strikingly, the same mutant displayed increased adherence to the oral epithelial cells (Fig. 5B). These findings are consistent with, and provide a basis to, the opposing phenotypes reported in mice (impaired gut colonization on the one hand and higher fungal load in oral infection on the other hand).
FIG 5.
Zcf8p modulates Candida adherence to host epithelial cells. (A) Diagram illustrating the three epithelial cell systems employed to probe interactions with C. albicans cells. (B) Adhesion of C. albicans to intestinal and oral epithelial cells. Host cells were coincubated with C. albicans wild‐type or zcf8 deletion mutant for 1 h. After extensive washing, cells were fixed and stained. Adhered cells were visualized in a fluorescence microscope and quantified in 50 fields for each replicate. Each black circle represents the percentage of adhered cells in one of four independent experiments. Bars represent the means ± SD. The Mann-Whitney U test was used in the statistical analysis.
The results described so far in this report link Zcf8p to two traits: on the one hand, altered attachment to mucosal surfaces (Fig. 5) and, on the other hand, regulation of the fungal vacuole (Fig. 3). A logical question, therefore, is whether the two traits are linked. To probe whether impairing vacuolar function has any consequence on fungal adherence, we treated Candida cells with the drug nigericin and, after washing away the drug, brought Candida and oral epithelial cells together to quantify fungal adhesion. As shown in Fig. 6A and B, the enhanced adherence caused by the zcf8 mutation was abolished in the presence of the drug. Furthermore, we found that the vacuole number differed between wild-type and mutant strains upon contact with host epithelial cells (Fig. 6C). There is, therefore, good correlation between Zcf8’s effects on fungal adhesion and vacuolar function.
FIG 6.
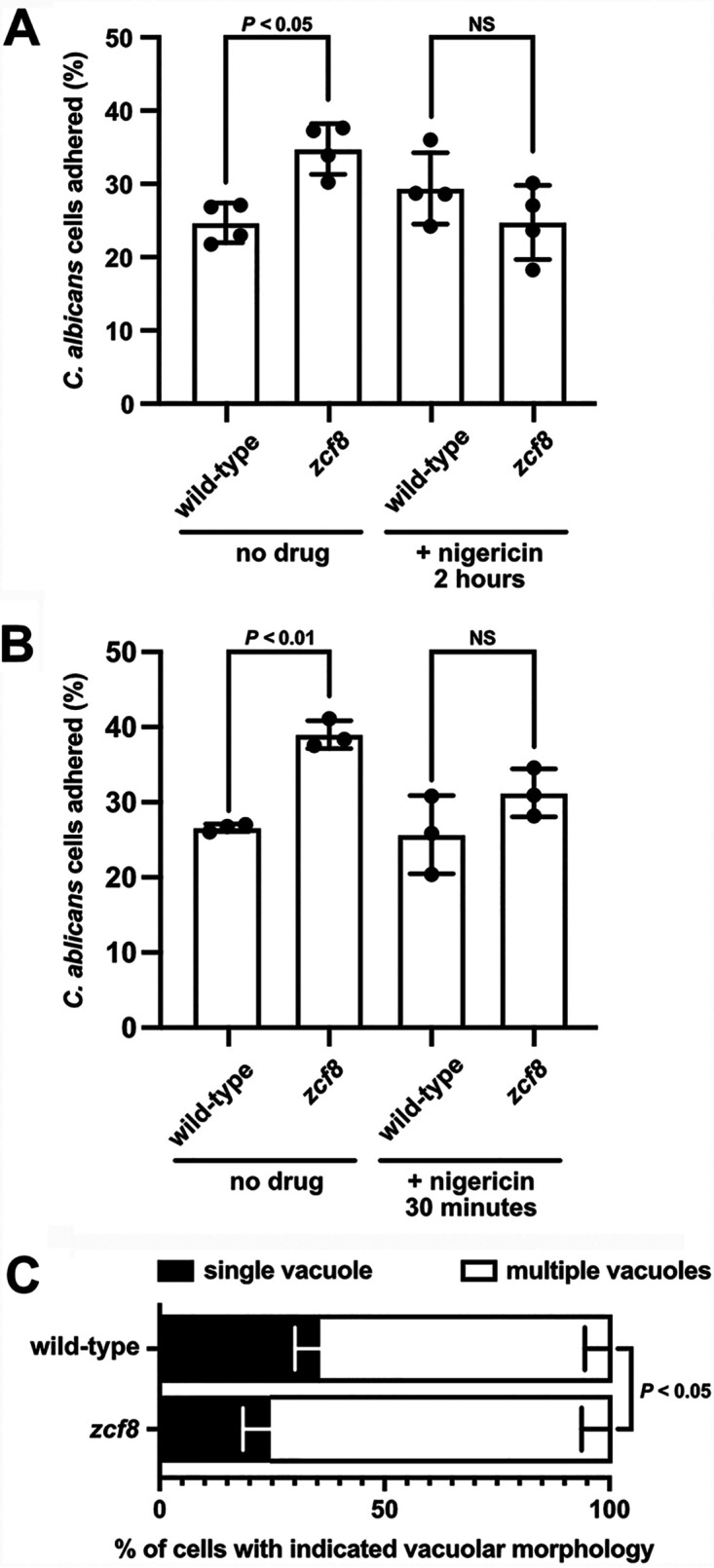
Increased adherence of zcf8 mutant cells to oral epithelial cells requires intact vacuole function. (A and B) C. albicans adhesion to oral epithelial cells. Candida cultures (wild‐type strain or zcf8 deletion mutant) were treated with the drug nigericin (at a final concentration of 15 μg/ml) for 2 h (A) or 30 min (B) (or left untreated in the control experiment). The drug was eliminated by washing twice the fungal cells with PBS before adding Candida to the oral epithelial cells. Adherence was monitored as described in the legend to Fig. 5B. Each black circle represents the percentage of adhered cells in one of four independent experiments. Bars represent the means ± SD. The Mann-Whitney U test was used in the statistical analysis. (C) C. albicans vacuolar morphology upon attachment to host epithelial cells. Candida cells (wild‐type strain or zcf8 deletion mutant) were added to oral epithelial cells and incubated for 1 h. Nonadhered fungal cells were washed away. Vacuoles in the adhered Candida population were visualized by FM4-64 staining. Plotted is the percentage of cells displaying the indicated vacuolar morphology. Bars represent the means ± SD of five independent experiments. At least 30 cells were evaluated per strain per replicate. The Mann-Whitney U test was used in the statistical analysis.
ZCF8 is a negative regulator of Candida filamentation in host tissues.
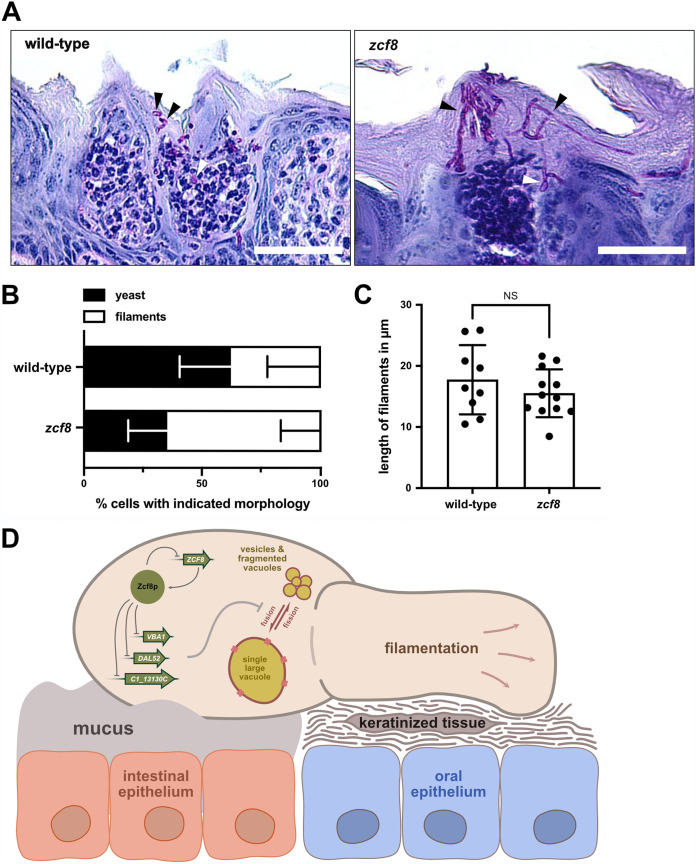
Mutations in genes that encode major structural components of the vacuole often lead to defects in C. albicans filamentation (12, 33). Thus, we sought to investigate whether the zcf8 mutant cells displayed any alteration in morphology during colonization of the murine host. We evaluated mouse tongue sections prepared from animals infected with either wild-type Candida or the isogenic zcf8 mutant strain (Fig. 7A). Quantification of the oval-shaped “yeast” cells as well as filaments in the tissues showed an increased proportion of the latter morphology in the tongues infected with the zcf8 mutant (Fig. 7A and B). The average length of the filaments did not vary between both strains (Fig. 7C). The results of this experiment indicate that ZCF8 is a negative regulator of C. albicans filamentation in host tissues.
FIG 7.
ZCF8 is a negative regulator of Candida filamentation in host tissue. (A) Shown are representative images of sites along the tongue where C. albicans established infection. PAS‐stained sagittal sections were prepared from tongues 72 h after infection with either the wild‐type reference strain (left) or the zcf8 deletion mutant. Bars, 100 μm. (B) Percentage of fungal cells found in the murine tongue in the oval-shaped “yeast” form or the filamentous form. Cells were considered filamentous when the length of the fungal cell exceeded by at least 2 times its diameter. At least 35 fungal cells were evaluated per strain per mouse (n = 4 animals). Bars represent the means ± SD. The Mann-Whitney U test was used in the statistical analysis. (C) Length of C. albicans filamenting cells in infected tongues. Clearly distinguishable single filamenting cells in PAS-stained sections were randomly chosen and their length measured. Each dot represents one cell. One hundred fifty cells were evaluated per strain. Bars represent means ± SD. The Mann-Whitney U test was used in the statistical analysis. (D) Model depicting how Zcf8p-regulated products targeted to the vacuole impinge upon the morphology of the organelle (i.e., presence of a single large vacuole or smaller, fragmented compartments) and subsequently on Candida attachment to mucosal surfaces. The yeast form of the fungus attaches better to intestinal mucus (4), whereas the filamentous form adheres more efficiently to oral cells (5).
DISCUSSION
In this report, we have investigated the molecular function of a previously uncharacterized fungal gene which is involved in C. albicans colonization of the murine oral cavity and intestinal tract. We establish that this gene, ZCF8, encodes a bona fide transcription regulator that governs multiple vacuole-associated traits in this fungus and modulates Candida interactions with mammalian epithelial cells. Several lines of evidence support this notion. (i) Zcf8p’s top targets of transcriptional regulation encode proteins that localize to the vacuole (Fig. 1 and 2). (ii) The number of vacuoles per fungal cell and the pH of the organelle depended on ZCF8 (Fig. 3A to D). (iii) Deletion or overexpression of ZCF8 altered the resistance/sensibility of the fungus to nigericin and brefeldin A, two drugs that disrupt vacuolar function and vesicle formation/transport, respectively (Fig. 3E and F). (iv) ZCF8 modulated the adherence of Candida cells to epithelial cells (Fig. 5). (v) Vacuole fragmentation (in vitro and in vivo) is correlated with the regulator’s effect on fungal adhesion (Fig. 6). On the basis of these findings, we posit that Candida employs Zcf8p to maintain vacuole homeostasis and that such activity impinges upon the physical contacts that fungal cells establish with mucosal surfaces (Fig. 7D).
The overlap between the Zcf8p ChIP and transcriptome data sets is rather low (Fig. 1I and J). Several reasons, some of them concerning the methodology, could explain this. For example, the laboratory culture conditions may not have been ideal to promote binding of the Zcf8 protein to some target promoters, particularly those with relatively low occupancy. Also, the thresholds that we used to call peaks was rather high; such strategy minimizes false-positive results (our main goal) but some true signal is lost as well. Beyond these technical considerations, it is also plausible that to promote or repress transcription on certain promoters, Zcf8p must act together with other unidentified transcription factors. In the absence of conditions that promote the activity of these additional factors, no expression changes would be observed. It is important to point out, however, that the main finding of our study, i.e., the discovery of Zcf8 as a regulator of the fungal vacuole, relies for the most part on the characterization of target genes for which we observed both Zcf8p binding and transcript changes (Fig. 1B).
A major function of the fungal vacuole is the storage and recycling of amino acids (12). The importance of mobilizing amino acids in and out of the vacuole is particularly evident when cells face nitrogen-poor environments (24, 25). Under these conditions, cells use amino acids produced by autophagic degradation of proteins in the vacuole. As illustrated in Fig. 7D, we have identified multiple transporters of amino acids or nitrogen-rich molecules (e.g., allantoate) as targets of Zcf8p regulation: for example, VBA1, a major vacuolar transporter of amino acids, and DAL52, a putative allantoate transporter that localizes to the vacuole (Fig. 2). Whether these transporters operate by moving their substrates into, or out of, the vacuole (or are bidirectional) remains to be established. Our results, however, imply that the misregulation of these transporters (and/or related products) can lead to significant vacuolar fusion and fission events in response to extracellular nitrogen availability (Fig. 3B). In agreement with this idea, it has been shown that deletion of VBA4, a nonessential transporter localized to the S. cerevisiae vacuole, can result in significant changes to the number of vacuoles within the yeast cell (34). ZCF8, therefore, appears to play a key role in limiting these events and, thus, preventing large fluctuations in the number of vacuoles per cell (i.e., maintaining vacuole homeostasis) (Fig. 7D). Further studies are needed, however, to establish whether ZCF8 governs nitrogen and/or amino acid metabolism in addition to the described vacuole-associated phenotypes.
How does the regulation of products targeted to the vacuole impinge upon attachment to mucosal surfaces? We propose that the misregulation of vacuole-associated products leads to alterations in the morphology and cell surface of the fungus, which ultimately dictate the attachment of the fungal cell to host surfaces (Fig. 7D). In support of this idea, it has been demonstrated that mutations in genes that encode major structural components of the vacuole often lead to defects in C. albicans filamentation (12, 33). Furthermore, a well-established response to nitrogen starvation in C. albicans is filamentation (35–37). We found a larger proportion of filamenting cells in host tissues infected with the zcf8 mutant strain (Fig. 7A and B), indicating that ZCF8 modulates this trait in vivo. Our ChIP-Seq data revealed that Zcf8p bound upstream of several well-established regulators of yeast-to-filament transition in C. albicans (e.g., BRG1 and RFG1). Thus, it is plausible that ZCF8’s effects on morphology are in part mediated by altering the expression of those regulators (although we found no evidence of changes in transcript levels in any of those ORFs in our RNA-Seq experiments). As a negative regulator of filamentation, ZCF8 is expected to have contrasting effects on C. albicans attachment to intestinal surfaces versus oral mucosa. That is because filaments bind more efficiently to keratinocytes (5), whereas the round yeast form attaches better to mucus (4) (Fig. 7D). Our results indeed support this prediction.
Mutations affecting vacuole function, biogenesis, and inheritance often lead to defects in fungal virulence (12, 38). The reports that have established a connection between the vacuole and Candida virulence, however, have relied almost exclusively on mutations that eliminate core structural components of the organelle such as the vacuolar proton-translocating ATPase (V-ATPase). The V-ATPase is a multisubunit complex that generates a proton gradient which is the driving force for the transport of metabolites into the vacuole and for the maintenance of the low pH in the vacuolar lumen and other endocytic vesicles (39). Loss of C. albicans V-ATPase activity alkalinizes the vacuolar lumen and has pleiotropic effects, including pH-dependent growth, calcium sensitivity, and cold sensitivity (14). Depletion of one of the V-ATPase subunits, VMA3, leads to alkalinization of the lumen, abnormal vacuole morphology, suppressed filamentation, and attenuated macrophage killing (14). Null mutations in VMA7, a different subunit, also resulted in defective vacuole acidification, reduced growth at alkaline pH and sensitivity toward the presence of metal ions (40). The vma7 null mutant strain was also avirulent in a mouse model of systemic candidiasis (40). The results described in this report establish that several properties of the Candida vacuole are under regulation of the Zcf8 protein and that mutations in this regulator (rather than in structural components of the vacuole) can also result in alterations of the fungus-host interplay.
While multiple protein kinases are known to regulate vacuolar functions, only a few transcription regulator proteins have been shown thus far to impinge upon the functioning of this organelle. One of these proteins is the transcriptional repressor Mot3p in S. cerevisiae (41). Mot3 represses transcription of genes needed for ergosterol biosynthesis. Ergosterol is required for endocytosis and homotypic vacuole fusion, providing a link between Mot3 and these processes. Indeed, mot3 mutant cells display a number of related phenotypes, including abnormal vacuolar morphology and poor growth on alkaline media (41). The MOT3 ortholog in C. albicans is termed CAS5 (C4_01190W) and has been reported to regulate cell wall stability and cell cycle dynamics in this species (42). The S. cerevisiae Forkhead box proteins Fkh1p and Fkh2p may represent another example, as they have been shown to bind upstream of the VMA6 and VMA8 genes, which encode subunits of the vacuolar ATPase (43). C. albicans harbors a single Forkhead box protein, Fkh2p, which does not appear to regulate transcription of any VMA gene. The Candida Fkh2p rather promotes the expression of hypha-specific and pathogenesis-related genes (44, 45). Another regulatory protein that has been linked to the fungal vacuole is Ume6p (46). In S. cerevisiae, Ume6p negatively regulates autophagy (47, 48), a process tightly linked to the function of the fungal vacuole. In C. albicans, Ume6p promotes hyphal formation, and its ectopic expression has been shown to stimulate the generation of vacuolated subapical compartments in the filamenting cells (49). Yet it remains to be established whether Ume6p directly controls the expression of any vacuolar component in Candida. While it may seem counterintuitive that an apparently important function for fungi in general (i.e., vacuole homeostasis) be under the control of different transcription regulators across species, the “rewiring” of transcriptional networks, at least in ascomycete yeasts, seems to be rather common (50). Indeed, it has been documented that key biological processes such as yeast mating, meiosis, and biofilm formation have undergone extensive modifications in their regulation over evolutionary timescales (51–53).
Finally, our finding that ZCF8 promotes Candida attachment to intestinal mucus but weakens binding to keratinocytes may explain the rather contradictory phenotypes that had been reported for this gene in mice. On the one hand, the zcf8 mutant had been shown to be impaired in gut colonization. The reduced attachment of the mutant strain to intestinal mucus could account for this phenotype. On the other hand, the same mutation caused higher fungal loads in tongues in a murine OPC model, which correlates well with the enhanced binding of the zcf8 mutant to oral cells. Our results, therefore, illustrate how divergent outcomes in the fungus-host interplay can be traced back to the function of a single regulatory gene.
MATERIALS AND METHODS
Strains and growth conditions.
The C. albicans strains used in this study are derivatives of clinical isolate SC5314 (54) and are listed in Table S4 in the supplemental material. Oligonucleotides and plasmids are listed in Tables S5 and S6, respectively. Gene deletions were carried out as described previously (55, 56). YFP-, GFP-, mNeonGreen- and MYC-tagging strategies for C. albicans have been described (6, 57). The TDH3 promoter-driven overexpression strains were generated using the plasmids and procedures described before (58). C. albicans strains were routinely grown at 30°C in yeast-peptone-dextrose (YPD) medium. When defined medium was employed, cells from overnight cultures were collected by centrifugation, washed twice with phosphate-buffered saline (PBS), diluted 1:10 in nitrogen starvation medium (BD Difco YNB without amino acids and ammonium sulfate, supplemented with 2% glucose) or medium containing ammonium sulfate as the only nitrogen source (DB Difco YNB without amino acids, supplemented with 2% glucose), and incubated for 6 h at 30°C with shaking at 220 rpm. Nigericin (catalog no. SML1779; Merck, Germany) and brefeldin A (catalog no. B6542; Merck, Germany) were used at a final concentration of 1.5 μg/ml or 5 μg/ml, respectively (unless otherwise indicated).
List of Candida albicans strains used in this study. Download Table S4, PDF file, 0.2 MB (194.1KB, pdf) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of oligonucleotides used in this study. Download Table S5, PDF file, 0.1 MB (121.7KB, pdf) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of plasmids used in this study. Download Table S6, PDF file, 0.1 MB (111.5KB, pdf) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Chromatin immunoprecipitation followed by deep sequencing (ChIP-Seq).
The ZCF8-MYC-tagged strain along with an untagged control were grown in YPD broth overnight at 30°C. The chromatin immunoprecipitation procedure was carried out as described previously (6, 57). Processing of raw data was conducted following the procedures outlined in other report (59).
Peak calling and DNA motif analysis.
The R package “bPeaks” (v.1.2) (60) was employed to detect significant peaks using an untagged immunoprecipitation (IP) as control. Based on the selected parameters (T1: 2, T2: 1, T3: 1.8, T4: 0.9), the program spotted 148 regions as potential peaks. We visualized all these peaks in MochiView (v.1.46) (61) and refined the list based on the following criteria. (i) Only peaks located in intergenic regions were maintained. (ii) Peaks adjacent to highly expressed genes were removed because these places tend to bind to almost all DNA binding proteins nonspecifically (6, 62–64). (iii) Only peaks that were consistent in at least two of four replicates were kept. This curation resulted in 42 peaks representing 51 genes (some peaks lie in divergently transcribed intergenic regions). To derive de novo putative DNA binding motifs, 500-nt sequences centered on the midpoint of the top ∼1/4 of scoring peaks were fed into the Regulatory Sequence Analysis Tools (RSAT) for fungal organisms (http://rsat.sb-roscoff.fr). The utility ran on default parameters with the background model set for C. albicans.
Recombinant protein purification and electrophoretic mobility gel shift assay.
The putative DNA binding domain of Zcf8p (amino acids 57 to 158) was amplified by PCR using oligonucleotides JCP2014 and JCP2026 and C. albicans genomic DNA (gDNA) as the template. The PCR fragment was subsequently digested and ligated into the SmaI/XhoI sites of plasmid pLIC-H3 (65). This plasmid is designed to overproduce 6×His-tagged proteins in Escherichia coli. The E. coli strain BL21 was used as the host of the expression plasmid. Cells were grown to an optical density at 600 nm (OD600) of ∼0.8, and expression was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After ∼3-h induction, cells were pelleted and stored at −80°C. Cells were lysed by sonication. The His-tagged protein was affinity purified from the lysate using nickel-nitrilotriacetic acid (Ni-NTA) agarose beads (Qiagen, Netherlands). Amicon Ultra-15 centrifugal filters (Merck) were used to exchange buffer and concentrate the protein. Protein concentration was estimated in Rothi-blue (Carl Roth, Germany)-stained gels using known amounts of bovine serum albumin as standards. Gel shift assays were carried out as described in reference 66.
Transcriptome analyses (RNA-Seq).
Cells harvested from overnight cultures in YPD were washed three times with PBS and diluted 1:10 in 3 ml yeast nitrogen base (YNB) without amino acids and ammonium sulfate. After 5.5 h of incubation at 30°C in a shaker, nigericin (at a final concentration of 15 μg/ml) or an equal volume of dimethyl sulfoxide (DMSO)/ethanol solution (mock) were added and incubated for an additional 30 min. Cells were collected by centrifugation. RNA was prepared using the RiboPure kit for yeast (Thermo Fisher Scientific, Waltham, USA). Illumina library preparation and strand-specific sequencing were carried out by Novogene (Cambridge, UK) following standard operating procedures. We obtained 41 to 48 million reads per sample (paired-end, 150-nt reads). Read processing, quality control, mapping, and differential gene expression analyses followed standard computational procedures described by our laboratory (59, 67). Briefly, Illumina adaptor sequences were removed using the Trimmomatic tool (v.0.39) (68). Trimmed reads were aligned to the C. albicans reference genome assembly 21 (www.candidagenome.org) using STAR (v.2.7.3a) (69). Count files were generated using HTSeq (v.0.11.2) (70). Raw read counts were loaded into R and analyzed with the DESeq2 package (v.1.28.1) (71), and volcano plots were created using the R package EnhancedVolcano (v.1.6.0) (https://github.com/kevinblighe/EnhancedVolcano). Two biological replicates were included in each RNA-Seq analysis.
Microscopy analyses.
Live-cell imaging of fungal cells at high resolution was performed using a wide field epifluorescence microscopy (Leica, DMI 6000 B). A 63× oil plan fluor objective (numerical aperture, 1.4) was employed. Excitation at 520 nm was used to visualize the subcellular localization of the mNeonGreen-tagged proteins. Excitation at 440 nm was used to visualize BCECF (18 μM) in the vacuolar lumen, and excitation at 550 nm was employed to detect vesicle/vacuolar membranes with FM4-64 (10 μM). Images were processed using FIJI (v.2.1.0).
Drug sensitivity assay.
Cells grown overnight in YPD liquid medium were washed with PBS, and serially diluted cell suspensions (initial cell concentration, 5 × 108 cells/ml) were spotted onto YNB agar containing ammonium sulfate and nigericin at a final concentration of 1.5 μg/ml (2 μM).
Flow cytometry analysis.
Cells were stained with 18 μM BCECF for 30 min at 30°C as established in reference 28 and analyzed using the BD Accuri C6 flow cytometer and the appurtenant software (v.1.0.264.21). The R package “flowCore” (v.1.11.20) was used to extract event-level information from the flow cytometry standard (FCS) files, and subsequent data manipulation and visualization were done using GraphPad (v.9.1.0).
Epithelial cell culture.
The human oral buccal cell line TR146 (Sigma-Aldrich, St. Louis, MO, USA) was cultured in Dulbecco modified Eagle medium (DMEM)/F12 medium, supplemented with 10% fetal bovine serum (FBS). The human Caucasian colon adenocarcinoma cell line Caco-2 (kindly provided by Cynthia Sharma, Institute for Molecular Infection Biology [IMIB], Würzburg, Germany) was cultivated in DMEM supplemented with 10% FBS. HT29-MTX-E12 (Sigma-Aldrich, St. Louis, MO, USA) were seeded on transwell membranes (Corning Transwell 0.4-μm-pore-size polycarbonate membrane; 12-mm inserts purchased from Sigma-Aldrich, Germany) and cultured in DMEM supplemented with 10% FBS, 5 mM glutamine, and 5 mM nonessential amino acids (Thermo Fisher Scientific) and allowed to grow for 21 days with regular medium exchange twice per week. All cell lines were cultivated in a humidified incubator at 37°C and 5% CO2.
Adherence assays.
Attachment of C. albicans to epithelial cells was measured either through fluorescence microscopy as described earlier for TR146 cells (5), which was also used for the Caco-2 cell line, or plated in the case of the HT29-MTX-E12 cell line. Briefly, cells of all three epithelial cell lines were challenged with C. albicans cells at a multiplicity of infection (MOI) of 1. After 1 h, cells were washed three times with PBS to remove nonadherent C. albicans cells. TR146 and Caco-2 cells were fixed with 4% paraformaldehyde and incubated overnight with a primary anti-Candida antibody (Invitrogen, Carlsbad, CA, USA). On the next day, cells were washed and stained with a secondary Alexa Fluor 594-conjugated goat anti-rabbit antibody (Life Technologies, Carlsbad, USA). Imaging was done using a Leica DMI 6000B microscope (Leica, Wetzlar, Germany). The number of adhered fungal cells was acquired in 50 frames per strain. Candida adherence to mucus covering the HT29-MTX-E12 transwells was measured by scraping all cells from wells (after extensive washing to remove nonadhered cells). Cells were pelleted, plated on YPD plates, and incubated for 48 h at 30°C. Adherence was determined in four independent experiments for each cell line.
C. albicans cells treated with nigericin were also assayed for attachment to the oral epithelial cells TR146. Briefly, fungal cells were incubated with nigericin or a mock solution (DMSO/ethanol [EtOH]) for 2 h at 30°C, washed twice, and added to TR146 cells. After the addition of Candida, cells were incubated for 2 h at 37°C with 5% CO2. Afterwards, cells were washed with PBS (to remove nonadhered cells) and fixed, stained, and quantified as described above.
OPC infection model.
The animal experiments were approved by the local government of Lower Franconia (Regierung von Unterfranken), Germany (protocol number 55.2-2531.01-50/14). Murine oral infections were conducted as described previously (5). Briefly, the C. albicans inoculum for infection was prepared using a 1-ml overnight culture in YPD, washed twice with sterile PBS, and serially diluted to a final concentration of 5 × 106 cells/ml. A sterile cotton ball (2.5 mg) was soaked for 1 h in 100 μl of this suspension. Six- to 9-week-old female C57BL/6J mice (Charles River, Sulzfeld, Germany) were anaesthetized with a mixture of ketamine (100 mg/kg of body weight; Zoetis, NJ, USA) and xylazine (6 mg/kg; CP-Pharma, Burgdorf, Germany) in sterile PBS which was injected intraperitoneally. A cotton ball saturated with C. albicans cells was placed sublingually for 90 min. Animal weight was monitored on a daily basis. Three days after inoculation, mice were euthanized, and tongues were removed, split sagittally, and fixed for histology.
Histology and PAS staining.
Mouse tongues were removed and split sagittally. Fixation of one half was done overnight in 10% buffered formalin (10% of a 37% formaldehyde solution, 0.65% Na2HPO4, and 0.4% NaH2PO4) and then washed several times with PBS. The tissue was embedded in paraffin on a paraffin embedding machine using a standard program consisting of washing steps in increasing ethanol concentrations, xylol baths, and 3 h in melted paraffin at 62°C. The resulting blocks were stored at room temperature. For microscopy, 4-μm sections were prepared and stained following standard periodic acid-Schiff (PAS) staining protocols. Fungal cells clearly distinguishable in PAS-stained sections were scored as filamenting if their length was >2 times their diameter (5).
Gene Ontology.
The Gene Ontology term finder tool of the Candida Genome Database (www.candidagenome.org) was used to identify overrepresented terms in the data sets.
Data availability.
The ChIP-Seq and RNA-Seq data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database (accession numbers GSE181652, GSE181653, and GSE181655).
ACKNOWLEDGMENTS
We thank Phillip Brandt and Slavena Vylkova from the Friedrich Schiller University Jena for technical support and helpful discussions; Elena Lindemann-Perez from the McGovern Medical School for conducting the quantitative reverse transcription-PCR (qRT-PCR) analysis; and Barbara Conrad, Christoph Schoen, and Oliver Kurzai from the Julius Maximilians University Würzburg for their support and generosity while carrying out article revisions.
This work was supported by funding from the Deutsche Forschungsgemeinschaft (DFG) (project TRR 124 FungiNet, number 210879364, subproject C01, to J.C.P.) and start-up funds from the University of Texas McGovern Medical School (to J.C.P.).
J.C.P. conceived the study and directed the project. P.R.-W. and J.M. conducted experiments. P.R.-W. analyzed ChIP-Seq and RNA-Seq data sets. P.R.-W. and J.C.P. wrote the paper.
Footnotes
Citation Reuter-Weissenberger P, Meir J, Pérez JC. 2021. A fungal transcription regulator of vacuolar function modulates Candida albicans interactions with host epithelial cells. mBio 12:e03020-21. https://doi.org/10.1128/mBio.03020-21.
Contributor Information
J. Christian Pérez, Email: jose.c.perez@uth.tmc.edu.
Reeta Prusty Rao, Worcester Polytechnic Institute.
Judith Berman, Tel Aviv University.
REFERENCES
- 1.Fiers WD, Gao IH, Iliev ID. 2019. Gut mycobiota under scrutiny: fungal symbionts or environmental transients? Curr Opin Microbiol 50:79–86. doi: 10.1016/j.mib.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pérez JC. 2021. Fungi of the human gut microbiota: roles and significance. Int J Med Microbiol 311:151490. doi: 10.1016/j.ijmm.2021.151490. [DOI] [PubMed] [Google Scholar]
- 3.Fidel PL, Noverr MC. 2011. Mucosal immunity to Candida albicans, p 137–157. In Calderone RA, Clancy CJ (ed), Candida and candidiasis, 2nd ed. ASM Press, Washington, DC. [Google Scholar]
- 4.Böhm L, Torsin S, Tint SH, Eckstein MT, Ludwig T, Pérez JC. 2017. The yeast form of the fungus Candida albicans promotes persistence in the gut of gnotobiotic mice. PLoS Pathog 13:e1006699. doi: 10.1371/journal.ppat.1006699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meir J, Hartmann E, Eckstein MT, Guiducci E, Kirchner F, Rosenwald A, LeibundGut-Landmann S, Pérez JC. 2018. Identification of Candida albicans regulatory genes governing mucosal infection. Cell Microbiol 20:e12841. doi: 10.1111/cmi.12841. [DOI] [PubMed] [Google Scholar]
- 6.Pérez JC, Kumamoto CA, Johnson AD. 2013. Candida albicans commensalism and pathogenicity are intertwined traits directed by a tightly knit transcriptional regulatory circuit. PLoS Biol 11:e1001510. doi: 10.1371/journal.pbio.1001510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witchley JN, Penumetcha P, Abon NV, Woolford CA, Mitchell AP, Noble SM. 2019. Candida albicans morphogenesis programs control the balance between gut commensalism and invasive infection. Cell Host Microbe 25:432–443. doi: 10.1016/j.chom.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finkel JS, Xu W, Huang D, Hill EM, Desai JV, Woolford CA, Nett JE, Taff H, Norice CT, Andes DR, Lanni F, Mitchell AP. 2012. Portrait of Candida albicans adherence regulators. PLoS Pathog 8:e1002525. doi: 10.1371/journal.ppat.1002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Homann OR, Dea J, Noble SM, Johnson AD. 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet 5:e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klionsky DJ, Herman PK, Emr SD. 1990. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev 54:266–292. doi: 10.1128/mr.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li SC, Kane PM. 2009. The yeast lysosome-like vacuole: endpoint and crossroads. Biochim Biophys Acta 1793:650–663. doi: 10.1016/j.bbamcr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Veses V, Richards A, Gow NA. 2008. Vacuoles and fungal biology. Curr Opin Microbiol 11:503–510. doi: 10.1016/j.mib.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 13.Weber RWS. 2002. Vacuoles and the fungal lifestyle. Mycologist 16:10–20. doi: 10.1017/S0269915X02006110. [DOI] [Google Scholar]
- 14.Rane HS, Bernardo SM, Raines SM, Binder JL, Parra KJ, Lee SA. 2013. Candida albicans VMA3 is necessary for V-ATPase assembly and function and contributes to secretion and filamentation. Eukaryot Cell 12:1369–1382. doi: 10.1128/EC.00118-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacPherson S, Larochelle M, Turcotte B. 2006. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev 70:583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimazu M, Sekito T, Akiyama K, Ohsumi Y, Kakinuma Y. 2005. A family of basic amino acid transporters of the vacuolar membrane from Saccharomyces cerevisiae. J Biol Chem 280:4851–4857. doi: 10.1074/jbc.M412617200. [DOI] [PubMed] [Google Scholar]
- 17.Loh E, Hong W. 2004. The binary interacting network of the conserved oligomeric Golgi tethering complex. J Biol Chem 279:24640–24648. doi: 10.1074/jbc.M400662200. [DOI] [PubMed] [Google Scholar]
- 18.Ram RJ, Li B, Kaiser CA. 2002. Identification of Sec36p, Sec37p, and Sec38p: components of yeast complex that contains Sec34p and Sec35p. Mol Biol Cell 13:1484–1500. doi: 10.1091/mbc.01-10-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burri L, Varlamov O, Doege CA, Hofmann K, Beilharz T, Rothman JE, Söllner TH, Lithgow T. 2003. A SNARE required for retrograde transport to the endoplasmic reticulum. Proc Natl Acad Sci USA 100:9873–9877. doi: 10.1073/pnas.1734000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dilcher M, Veith B, Chidambaram S, Hartmann E, Schmitt HD, Von Mollard GF. 2003. Use1p is a yeast SNARE protein required for retrograde traffic to the ER. EMBO J 22:3664–3674. doi: 10.1093/emboj/cdg339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanneganti V, Kama R, Gerst JE. 2011. Btn3 is a negative regulator of Btn2-mediated endosomal protein trafficking and prion curing in yeast. Mol Biol Cell 22:1648–1663. doi: 10.1091/mbc.E10-11-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frazer C, Hernday AD, Bennett RJ. 2019. Monitoring phenotypic switching in Candida albicans and the use of next-gen fluorescence reporters. Curr Protoc Microbiol 53:e76. doi: 10.1002/cpmc.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warren G, Wickner W. 1996. Organelle inheritance. Cell 84:395–400. doi: 10.1016/s0092-8674(00)81284-2. [DOI] [PubMed] [Google Scholar]
- 24.Kawano-Kawada M, Kakinuma Y, Sekito T. 2018. Transport of amino acids across the vacuolar membrane of yeast: its mechanism and physiological role. Biol Pharm Bull 41:1496–1501. doi: 10.1248/bpb.b18-00165. [DOI] [PubMed] [Google Scholar]
- 25.Mukaiyama H, Nakase M, Nakamura T, Kakinuma Y, Takegawa K. 2010. Autophagy in the fission yeast Schizosaccharomyces pombe. FEBS Lett 584:1327–1334. doi: 10.1016/j.febslet.2009.12.037. [DOI] [PubMed] [Google Scholar]
- 26.Richards A, Gow NAR, Veses V. 2012. Identification of vacuole defects in fungi. J Microbiol Methods 91:155–163. doi: 10.1016/j.mimet.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 27.Boens N, Qin W, Basarić N, Orte A, Talavera EM, Alvarez-Pez JM. 2006. Photophysics of the fluorescent pH indicator BCECF. J Phys Chem A 110:9334–9343. doi: 10.1021/jp0615712. [DOI] [PubMed] [Google Scholar]
- 28.Johnson RM, Allen C, Melman SD, Waller A, Young SM, Sklar LA, Parra KJ. 2010. Identification of inhibitors of vacuolar proton-translocating ATPase pumps in yeast by high-throughput screening flow cytometry. Anal Biochem 398:203–211. doi: 10.1016/j.ab.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torggler R, Papinski D, Kraft C. 2017. Assays to monitor autophagy in Saccharomyces cerevisiae. Cells 6:23. doi: 10.3390/cells6030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vida TA, Emr SD. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jakubkova M, Dzugasova V, Truban D, Abelovska L, Bhatia-Kissova I, Valachovic M, Klobucnikova V, Zeiselova L, Griac P, Nosek J, Tomaska L. 2016. Identification of yeast mutants exhibiting altered sensitivity to valinomycin and nigericin demonstrate pleiotropic effects of ionophores on cellular processes. PLoS One 11:e0164175. doi: 10.1371/journal.pone.0164175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eckstein MT, Moreno-Velásquez SD, Pérez JC. 2020. Gut bacteria shape intestinal microhabitats occupied by the fungus Candida albicans. Curr Biol 30:4799–4807. doi: 10.1016/j.cub.2020.09.027. [DOI] [PubMed] [Google Scholar]
- 33.Theiss S, Kretschmar M, Nichterlein T, Hof H, Agabian N, Hacker J, Köhler GA. 2002. Functional analysis of a vacuolar ABC transporter in wild-type Candida albicans reveals its involvement in virulence. Mol Microbiol 43:571–584. doi: 10.1046/j.1365-2958.2002.02769.x. [DOI] [PubMed] [Google Scholar]
- 34.Kawano-Kawada M, Pongcharoen P, Kawahara R, Yasuda M, Yamasaki T, Akiyama K, Sekito T, Kakinuma Y. 2016. Vba4p, a vacuolar membrane protein, is involved in the drug resistance and vacuolar morphology of Saccharomyces cerevisiae. Biosci Biotechnol Biochem 80:279–287. doi: 10.1080/09168451.2015.1083401. [DOI] [PubMed] [Google Scholar]
- 35.Biswas S, Roy M, Datta A. 2003. N-acetylglucosamine-inducible CaGAP1 encodes a general amino acid permease which co-ordinates external nitrogen source response and morphogenesis in Candida albicans. Microbiology (Reading) 149:2597–2608. doi: 10.1099/mic.0.26215-0. [DOI] [PubMed] [Google Scholar]
- 36.Biswas K, Morschhäuser J. 2005. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol Microbiol 56:649–669. doi: 10.1111/j.1365-2958.2005.04576.x. [DOI] [PubMed] [Google Scholar]
- 37.Holmes AR, Shepherd MG. 1988. Nutritional factors determine germ tube formation in Candida albicans. J Med Vet Mycol 26:127–131. doi: 10.1080/02681218880000181. [DOI] [PubMed] [Google Scholar]
- 38.Olsen I. 2014. Attenuation of Candida albicans virulence with focus on disruption of its vacuole functions. J Oral Microbiol 6:23898. doi: 10.3402/jom.v6.23898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rollenhagen C, Mamtani S, Ma D, Dixit R, Eszterhas S, Lee SA. 2020. The role of secretory pathways in Candida albicans pathogenesis. J Fungi 6:26. doi: 10.3390/jof6010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poltermann S, Nguyen M, Günther J, Wendland J, Härtl A, Künkel W, Zipfel PF, Eck R. 2005. The putative vacuolar ATPase subunit Vma7p of Candida albicans is involved in vacuole acidification, hyphal development and virulence. Microbiology (Reading) 151:1645–1655. doi: 10.1099/mic.0.27505-0. [DOI] [PubMed] [Google Scholar]
- 41.Hongay C, Jia N, Bard M, Winston F. 2002. Mot3 is a transcriptional repressor of ergosterol biosynthetic genes and is required for normal vacuolar function in Saccharomyces cerevisiae. EMBO J 21:4114–4124. doi: 10.1093/emboj/cdf415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie JL, Qin L, Miao Z, Grys BT, Diaz JDLC, Ting K, Krieger JR, Tong J, Tan K, Leach MD, Ketela T, Moran MF, Krysan DJ, Boone C, Andrews BJ, Selmecki A, Ho Wong K, Robbins N, Cowen LE. 2017. The Candida albicans transcription factor Cas5 couples stress responses, drug resistance and cell cycle regulation. Nat Commun 8:499. doi: 10.1038/s41467-017-00547-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghavidel A, Baxi K, Prusinkiewicz M, Swan C, Belak ZR, Eskiw CH, Carvalho CE, Harkness TA. 2018. Rapid nuclear exclusion of HCM1 in aging Saccharomyces cerevisiae leads to vacuolar alkalization and replicative senescence. G3 (Bethesda) 8:1579–1592. doi: 10.1534/g3.118.200161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greig JA, Sudbery IM, Richardson JP, Naglik JR, Wang Y, Sudbery PE. 2015. Cell cycle-independent phospho-regulation of Fkh2 during hyphal growth regulates Candida albicans pathogenesis. PLoS Pathog 11:e1004630. doi: 10.1371/journal.ppat.1004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bensen ES, Filler SG, Berman J. 2002. A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot Cell 1:787–798. doi: 10.1128/EC.1.5.787-798.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmer GE. 2011. Vacuolar trafficking and Candida albicans pathogenesis. Commun Integr Biol 4:240–242. doi: 10.4161/cib.4.2.14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delorme-Axford E, Klionsky DJ. 2018. Transcriptional and post-transcriptional regulation of autophagy in the yeast Saccharomyces cerevisiae. J Biol Chem 293:5396–5403. doi: 10.1074/jbc.R117.804641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartholomew CR, Suzuki T, Du Z, Backues SK, Jin M, Lynch-Day MA, Umekawa M, Kamath A, Zhao M, Xie Z, Inoki K, Klionsky DJ. 2012. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci USA 109:11206–11210. doi: 10.1073/pnas.1200313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmer GE. 2010. Endosomal and AP-3-dependent vacuolar trafficking routes make additive contributions to Candida albicans hyphal growth and pathogenesis. Eukaryot Cell 9:1755–1765. doi: 10.1128/EC.00029-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson AD. 2017. The rewiring of transcription circuits in evolution. Curr Opin Genet Dev 47:121–127. doi: 10.1016/j.gde.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nocedal I, Johnson AD. 2015. How transcription networks evolve and produce biological novelty. Cold Spring Harbor Symp Quant Biol 80:265–274. doi: 10.1101/sqb.2015.80.027557. [DOI] [PubMed] [Google Scholar]
- 52.Nocedal I, Mancera E, Johnson AD. 2017. Gene regulatory network plasticity predates a switch in function of a conserved transcription regulator. Elife 6:e23250. doi: 10.7554/eLife.23250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mancera E, Nocedal I, Hammel S, Gulati M, Mitchell KF, Andes DR, Nobile CJ, Butler G, Johnson AD. 2021. Evolution of the complex transcription network controlling biofilm formation in Candida species. Elife 10:e64682. doi: 10.7554/eLife.64682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gillum AM, Tsay EYH, Kirsch DR. 1984. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 55.Noble SM, Johnson AD. 2005. Strains and strategies for large-scale gene deletion studies of the diploid human fungal pathogen Candida albicans. Eukaryot Cell 4:298–309. doi: 10.1128/EC.4.2.298-309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reuss O, Vik A, Kolter R, Morschhäuser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 57.Hernday AD, Noble SM, Mitrovich QM, Johnson AD. 2010. Genetics and molecular biology in Candida albicans. Methods Enzymol 470:737–758. doi: 10.1016/S0076-6879(10)70031-8. [DOI] [PubMed] [Google Scholar]
- 58.Nobile CJ, Solis N, Myers CL, Fay AJ, Deneault JS, Nantel A, Mitchell AP, Filler SG. 2008. Candida albicans transcription factor Rim101 mediates pathogenic interactions through cell wall functions. Cell Microbiol 10:2180–2196. doi: 10.1111/j.1462-5822.2008.01198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.del Olmo Toledo V, Puccinelli R, Fordyce PM, Pérez JC. 2018. Diversification of DNA binding specificities enabled SREBP transcription regulators to expand the repertoire of cellular functions that they govern in fungi. PLoS Genet 14:e1007884. doi: 10.1371/journal.pgen.1007884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Merhej J, Frigo A, Le Crom S, Camadro JM, Devaux F, Lelandais G. 2014. bPeaks: a bioinformatics tool to detect transcription factor binding sites from ChIPseq data in yeasts and other organisms with small genomes. Yeast 31:375–391. doi: 10.1002/yea.3031. [DOI] [PubMed] [Google Scholar]
- 61.Homann OR, Johnson AD. 2010. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol 8:49. doi: 10.1186/1741-7007-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD. 2007. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol 5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tuch BB, Galgoczy DJ, Hernday AD, Li H, Johnson AD. 2008. The evolution of combinatorial gene regulation in fungi. PLoS Biol 6:e38. doi: 10.1371/journal.pbio.0060038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cain CW, Lohse MB, Homann OR, Sil A, Johnson AD. 2012. A conserved transcriptional regulator governs fungal morphology in widely diverged species. Genetics 190:511–521. doi: 10.1534/genetics.111.134080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lohse MB, Zordan RE, Cain CW, Johnson AD. 2010. Distinct class of DNA-binding domains is exemplified by a master regulator of phenotypic switching in Candida albicans. Proc Natl Acad Sci USA 107:14105–14110. doi: 10.1073/pnas.1005911107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moreno-Velásquez SD, Tint SH, del Olmo Toledo V, Torsin S, De S, Pérez JC. 2020. The regulatory proteins Rtg1/3 govern sphingolipid homeostasis in the human-associated yeast Candida albicans. Cell Rep 30:620–629. doi: 10.1016/j.celrep.2019.12.022. [DOI] [PubMed] [Google Scholar]
- 68.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Closest homologs of ZCF8 in the Candida clade. (A) Cladogram depicting the phylogenetic relationships among extant species of the Candida and Saccharomyces clades. Colored arrows represent ZCF8 and its closest homologs according to the Candida Genome Database and BLAST searches conducted with either full-length or conserved DNA binding domain protein sequences. Gene orthology assignments (same colors) follow independent reconstructions generated by the Candida Gene Order Browser. OAF1 and HAP1 (striped lines) are retrieved as “best hits” when queried with ZCF34 and ZCF5 sequences, respectively, in S. cerevisiae (however, neither OAF1 nor HAP1 are considered true orthologs of the indicated Candida genes). (B) Protein sequence alignment of putative DNA binding domains of ZCF8 and its closest homologs. Alignment was performed by MUSCLE. Residues highlighted in red are the six cysteines characteristic of the zinc cluster domain. (C) Maximum likelihood phylogeny of ZCF8 and its closest homologs. Alignments were generated using MAFFT (v.7.475). IQ-TREE 2 (v.2.1.2) was used to predict the best-fitting model (VT+F+R4), and FigTree (v1.4.4) (http://tree.bio.ed.ac.uk/software/figtree/) was used to generate the tree. Download FIG S1, TIF file, 0.5 MB (501.3KB, tif) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Activity of tagged ZCF8 allele, subcellular localization of Zcf8p, and transcriptional autoregulation. (A) Quantitative RT-PCR analysis comparing the activity of the tagged and untagged ZCF8 alleles. Shown is the level of repression of DAL52, the top target of regulation of Zcf8p. The DAL52 transcript levels measured in the homozygous deletion mutant (Δzcf8/Δzcf8) were used to estimate the percent repression achieved in each of the three indicated strains. The experimentally validated TAF10 transcript (M.-A. Teste, M. Duquenne, J. M. François, J.-L. Parrou, BMC Mol Biol 10:99, 2009, https://doi.org/10.1186/1471-2199-10-99) served as reference control to normalize the quantitative reverse transcription-PCR (qRT-PCR) data. Shown are the means ± SD (n = 3). The Δzcf8/ZCF8-MYC strain was used in the chromatin immunoprecipitation experiment. (B) Subcellular localization of GFP-tagged Zcf8p. Arrowheads indicate nuclei. Staining with FM4-64 delineates the vacuoles (marked with “v”). (C) Schematic of the reporter strains. In strain I (left), one ZCF8 allele was replaced by YFP. In strain II, the second ZCF8 allele was deleted. In both strains, the expression of the reporter is driven by the native ZCF8 promoter. (D) YFP fluorescence emitted by the reporter strains described in panel C. Cells were grown overnight in YPD and diluted in PBS. Fluorescence was measured using a flow cytometer. Ten thousand cells were evaluated per strain per experiment. Plotted are the means ± SD of three independent experiments. The Student’s t test was used in the statistical analysis. Download FIG S2, TIF file, 0.9 MB (939.2KB, tif) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of ChIP-Seq peaks and associated genes. This file includes a list of all Zcf8p ChIP peaks. The C. albicans chromosome and genome coordinates, fold enrichment as well as each peak’s nearest annotated ORF are indicated. Download Table S1, XLSX file, 0.02 MB (18.2KB, xlsx) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of differentially expressed transcripts in zcf8 deletion and ZCF8 overexpression strains. This file includes a list of the C. albicans genes whose expression was up- or downregulated in the zcf8 deletion mutant or in a strain overexpressing ZCF8. Download Table S2, XLSX file, 0.02 MB (19.8KB, xlsx) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ZCF8 does not influence autophagy. (A) Western blotting of the autophagy marker GFP-Atg8p in wild-type background and in the zcf8 deletion mutant using an anti-GFP antibody. These two strains, together with the untagged control, were grown in defined liquid medium without amino acids and ammonium sulfate for 3 or 6 h. The accumulation of free GFP (due to Atg8p degradation) is a proxy for autophagy. Blotting with an anti-tubulin 1 antibody shows equal loading. (B) Vacuolar delivery of mNeonGreen-Atg8p by bulk autophagy visualized by fluorescence microscopy. Autophagosome is marked with white arrowhead. Bar, 5 μm. (C) Quantification of microscopy images shown in panel B. Plotted is the fraction of cells with a visible autophagosome as a proxy for autophagic activity. Every dot represents the percentage of cells per microscope field (only fields with >20 cells were scored). Twenty frames were scored in each experiment. Plotted are the results from five independent experiments. Download FIG S3, TIF file, 0.9 MB (915.6KB, tif) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Evaluating the role of ZCF8 in endocytosis. (A) Internalization of endocytic vacuoles in C. albicans. Cells grown in defined liquid medium without amino acids and ammonium sulfate were incubated with the lipophilic dye FM4-64 for 10 or 20 min, washed with ice-cold PBS, and kept on ice until evaluated by fluorescence microscopy. Representative images are shown. Bars, 5 μm. (B) Quantification of images shown in panel A. Plotted is the mean fluorescence (± SD) emitted by the internalized FM4-64 dye. The fluorescence emitted by each cell was measured using FIJI. At least 400 cells (derived from >20 microscope fields) were measured per strain per time point per experiment. Data collected from four independent experiments are shown. Download FIG S4, TIF file, 1.9 MB (1.9MB, tif) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of differentially expressed transcripts in response to nigericin. This file includes a list of the C. albicans genes whose expression was significantly altered in response to nigericin treatment in the wild-type reference strain and the zcf8 deletion mutant. Download Table S3, XLSX file, 0.03 MB (30.8KB, xlsx) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of Candida albicans strains used in this study. Download Table S4, PDF file, 0.2 MB (194.1KB, pdf) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of oligonucleotides used in this study. Download Table S5, PDF file, 0.1 MB (121.7KB, pdf) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of plasmids used in this study. Download Table S6, PDF file, 0.1 MB (111.5KB, pdf) .
Copyright © 2021 Reuter-Weissenberger et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The ChIP-Seq and RNA-Seq data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database (accession numbers GSE181652, GSE181653, and GSE181655).