Abstract
Bacteriocins are antimicrobial peptides produced by bacteria to inhibit competitors in their natural environments. Some of these peptides have emerged as commercial food preservatives and, due to the rapid increase in antibiotic resistant bacteria, are also discussed as interesting alternatives to antibiotics for therapeutic purposes. Currently, commercial bacteriocins are produced exclusively with natural producer organisms on complex substrates and are sold as semi-purified preparations or crude fermentates. To allow clinical application, efficacy of production and purity of the product need to be improved. This can be achieved by shifting production to recombinant microorganisms.
Here, we identify Corynebacterium glutamicum as a suitable production host for the bacteriocin pediocin PA-1. C. glutamicum CR099 shows resistance to high concentrations of pediocin PA-1 and the bacteriocin was not inactivated when spiked into growing cultures of this bacterium. Recombinant C. glutamicum expressing a synthetic pedACDCgl operon releases a compound that has potent antimicrobial activity against Listeria monocytogenes and Listeria innocua and matches size and mass:charge ratio of commercial pediocin PA-1. Fermentations in shake flasks and bioreactors suggest that low levels of dissolved oxygen are favorable for production of pediocin. Under these conditions, however, reduced activity of the TCA cycle resulted in decreased availability of the important pediocin precursor l-asparagine suggesting options for further improvement. Overall, we demonstrate that C. glutamicum is a suitable host for recombinant production of bacteriocins of the pediocin family.
Keywords: Corynebacterium glutamicum, Bacteriocin, Pediocin, Recombinant production, Rational design, Antimicrobial peptide, Oxygen limitation, Listeria sp
Highlights
-
•
C. glutamicum CR099 is resistant to high concentrations of pediocin PA-1.
-
•
Recombinant C. glutamicum CR099/pEKEx-pedACDCg produces of a compound with antimicrobial activity against Listeria sp.
-
•
The purified compound matches size and mass:charge ratio of commercial pediocin PA-1.
-
•
Low oxygen levels are favorable for production of active pediocin by C. glutamicum in batch fermentations.
1. Introduction
Bacteriocins are small, ribosomally synthesized antimicrobial peptides (AMPs) naturally produced by a wide range of bacteria that are able to suppress growth and/or directly kill target organisms in a specific manner (Chikindas et al., 2018; Cotter et al., 2013). Their biological role is to provide a competitive advantage for the producing organisms in an ecological niche (Chikindas et al., 2018). Due to their activity against pathogens and food-spoiling organisms, bacteriocins are used as preservatives in food and beverages and in animal and pet feed (Chikindas et al., 2018; Mills et al., 2011; Silva et al., 2018).
The widespread over- and misuse of antibiotics in livestock farming contributes massively to spread of antimicrobial resistances of a wide range of infectious agents, which is recognized as one of the most urgent public health threats (World Health Organization, 2017). In recent years, a number of novel bacteriocins have been identified in bacteria from various sources (Angelopoulou et al., 2020; Azevedo et al., 2015; Egan et al., 2018; Walsh et al., 2015). Some of them have shown promising activity against clinically relevant pathogens. For example, enterocin K1 is active against different strains of Enterococcus faecium including multidrug resistant clinical isolates (Ovchinnikov et al., 2017). Similarly, garvicins Q and KS display activity against Gram-positive pathogens of the genera Staphylococcus, Bacillus, Listeria, and Enterococcus (Ovchinnikov et al., 2016; Tosukhowong et al., 2012; Tymoszewska et al., 2017). Recently, garvicin KS has been developed into a formulation for topical application that was successfully used to treat skin infections with methicillin-resistant S. aureus (Ovchinnikov et al., 2020). Thus, replacement of antibiotics with naturally occurring bacteriocins may contribute to solve problems with antimicrobial resistant infections.
A considerable number of bacteriocins belongs to the pediocin family of class IIa bacteriocins. Pediocins are produced by lactic acid bacteria (LAB) of the species Pediococcus sp. and Lactobacillus sp. and the genes involved in production and immunity in the natural producers have been identified (Ennahar et al., 1996; Marugg et al., 1992; Motlagh et al., 1994; Venema et al., 1995). Class IIa bacteriocins kill target bacteria by formation of pores in the cytoplasmic membrane (Chikindas et al., 1993; Ríos Colombo et al., 2018). The receptors that facilitate binding of these peptides to their targets are sugar transporters of the mannose phosphotransferase system (PTSMan) family (Diep et al., 2007; Kjos et al, 2009, 2010; Tymoszewska et al, 2017, 2018).
The major bottlenecks that hamper new bacteriocins from entering the market and prevent their application in clinical settings are easy and cost-effective production and purification at industrially relevant titers (Abbasiliasi et al., 2017; Johnson et al., 2017; Mesa-Pereira et al., 2018). The media currently used for industrial scale production of bacteriocins using natural producer strains are complex and often supplemented with milk or whey (Abbasiliasi et al., 2017; de Arauz et al., 2009), which requires expensive downstream processing and/or results in low product purity. Another barrier for AMP production is the high energy demand for product formation in combination with the low energy yield of the fermentative metabolism of LAB (Kördikanlıoğlu et al., 2015). These drawbacks may be overcome by transferring production to industrial platform organisms.
An interesting candidate for biotechnological production of bacteriocins is C. glutamicum, one of the most extensively used platform organisms for biotechnological production of more than 70 compounds including high value active ingredients and therapeutic proteins (Becker et al., 2018; Eggeling and Bott, 2015; Wolf et al., 2021; Yim et al, 2014, 2016). This platform organism bears the advantage that well-defined, low-cost, minimal media as well as appropriate industry-scale production processes are available. Products obtained with C. glutamicum have GRAS (generally regarded as safe) status and a long tradition in industry with well-established, large-scale cultivation processes. C. glutamicum has no significant extracellular proteolytic activity avoiding degradation of secreted protein or peptide products (Becker and Wittmann, 2017; Lee and Kim, 2018). Moreover, the organism has been engineered for efficient growth and production on different renewable feedstocks and industrial waste streams including raw starches, pentoses from hemicellulosic wastes, mannitol from seaweed hydrolysates and amino sugars derived from chitin (Hoffmann et al., 2021; Matano et al., 2014; Meiswinkel et al., 2013; Seibold et al., 2006).
In the present study, we establish C. glutamicum as a suitable host for recombinant production of pediocin PA-1, an industrially relevant bacteriocin.
2. Materials and methods
2.1. Strains and growth conditions
C. glutamicum strains were generally grown in 2xTY medium with 2% glucose (Glc) at 30 °C with aeration in either glass tubes, baffled or non-baffled Erlenmeyer flasks as indicated in individual experiments. Listeria spp. strains were grown in BHI medium at 37 °C with aeration overnight (o/N; i.e. approx. 16 h). P. acidilactici 347 was cultivated o/N statically in MRS medium (Gibco) at 30 °C. Where indicated, media were supplemented with IPTG (0.2 mM) and/or antibiotics as appropriate (kanamycin: 50 μg mL−1; chloramphenicol: 30 μg mL−1 for E. coli and Listeria spp. or 12.5 μg mL−1 for C. glutamicum). All bacterial strains and plasmids used in the current study are listed in Supplementary Table S1.
2.2. Molecular biology procedures
Cloning procedures were conducted using standard reagents according to the manufacturer's recommendation. Sequences of primers and codon-optimized genes pedACg, pedCCg, pedDCg used in this study are listed in Supplementary Table S2 and were obtained from a commercial service provider (Eurofins Genomics).
The genes for pediocin PA-1 biosynthesis were amplified via PCR using Q5 high fidelity polymerase (New England Biolabs) and primers creating overlapping sequences and restriction sites. Gibson assembly (Gibson et al., 2009) was used to fuse all fragments into pEKEx2, which was linearized by restriction endonucleases SacI and SalI, resulting in pEKEx2-pedACDCg and pEKEx2-pedCDACg. The plasmid pEKEx2-pedAACDCg was generated by adding an additional copy of pedACg via Gibson assembly into NheI-linearized pEKEx2-pedACDCg. Further constructs were obtained by restriction of pEKEx2-pedACDCg and re-ligation using T4 DNA ligase and blunting enzyme (both Thermo Scientific). For pEKEx2-pedACg, KpnI and NheI were used to remove pedCCg and pedDCg from pEKEx2-pedACDCg. For pEKEx2-pedACCg, NcoI and KpnI were used, removing pedDCg. For pEKEx2-pedADCg, NheI and NcoI were used, removing pedCCg. For pEKEx2-pedCDCg, SalI and NheI were used, removing pedACg. All plasmids were introduced into competent E. coli DH5α and analyzed via sequencing prior to further use. For construction of pXMJ19-pedACDCg, the empty vector pXMJ19 was linearized with SacI and SalI and subsequently ligated with pedACDCg obtained by restriction of pEKEx2-pedACDCg with the same enzymes. Transformation of Listeria sp. strains with plasmids pNZ44 and pIMK2 was performed according to a previously published protocol (Monk et al., 2008).
2.3. Determination of bacteriocin activity
Bacteriocin activity was determined using a standard microtiter plate assay (Holo et al., 1991) with slight modifications. The indicator strains L. monocytogenes EGDe:pIMK2, L. monocytogenes EGDe/pNZ44 and L. innocua LMG2785:pIMK2 were grown o/N at 37 °C in BHI and diluted 1:25 in fresh BHI prior to the assay. Two-fold dilution series of samples (100 μl) were mixed with the indicator strain (100 μl) in sterile 96-well plates. This corresponds to a starting optical density at 600 nm (OD600) of 0.05 ± 0.01. The plates were incubated at 37 °C for 5–6 h and growth was monitored by measuring OD600 in an Infinite M200 plate reader (Tecan). Bacteriocin activity in supernatants and different fractions of the purification process was determined in a semi-quantitative manner as described previously (Holo et al., 1991) and calculated as bacteriocin units per ml (BU mL−1). According to this method, BUs are defined as the reciprocal of the highest dilution showing at least 50% inhibition of the indicator strain.
Resistance of C. glutamicum CR0999 to pediocin, nisin and bacitracin was determined using a similar assay. Bacteria of an o/N culture in 2xTY were diluted 1:25 in fresh medium and 100 μl of this suspension were mixed with 100 μl of samples of 2-fold serial dilutions of AMP standard solutions in microtiter plates. This corresponds to a starting OD600 of 0.1 ± 0.01 and the indicated AMP concentrations. OD600 was again measured after incubation at 30 °C with shaking for 6 h.
2.4. Purification and identification of pediocin
Purification of pediocin PA-1 was carried out as described previously (Ovchinnikov et al., 2016). For cation exchange (CIEX) chromatography, culture supernatants from 1 L cultivations were precipitated with ammonium sulfate (50% saturation at 4 °C) and the precipitate was collected via centrifugation (45 min, 10000 g, 4 °C). After resuspension of the precipitate in HPLC grade H2O (1/10 of the initial volume), the pH was adjusted to 3.9 with 2 M HCl. Another centrifugation step (20 min, 10000 g, 4 °C) was carried out to remove insoluble precipitate. The resulting supernatant was then applied to a HiPrep SP FF 16/10 column (GE Healthcare Life Sciences) equilibrated with 20 mM sodium phosphate buffer at pH 3.9. Wash out of unbound sample was carried out with 5 column volumes of 20 mM sodium phosphate buffer at pH 6.9. The bacteriocin was eluted in a single step using 5 column volumes of 20 mM sodium phosphate buffer at pH 6.9 with 2 M NaCl. The eluate was fractionated to 5 mL aliquots and fractions containing bacteriocin activity were stored at −20 °C. For size exclusion (SIEX) analysis, active fractions were applied to a Superdex peptide 10/300 GL SIEX column (GE Healthcare Life Sciences) with an isocratic elution at a flow rate of 0.5 mL min−1 (50 mM Tris-HCl pH 7.5, 200 mM NaCl, and 5% (w/v) glycerol).
For identification, the eluate from CIEX chromatography was applied to a Resource reverse-phase chromatography (RPC) column (1 ml) (GE Healthcare Biosciences) connected to an ÅKTA purifier system (Amersham Pharmacia Biotech). A linear gradient of 2-propanol (Merck) with 0.1% (vol/vol) trifluoroacetic acid (buffer B) at a flow rate of 1.0 mL min−1 was used for elution. Pediocin was eluted with 35% of buffer B. RPC fractions with antimicrobial activity were further analyzed by mass spectroscopy (MS). For MS analysis, the molecular mass of the purified bacteriocin was determined by an Ultraflex MALDI-TOF/TOF (Bruker Daltonics, Bremen, Germany) instrument operated in reflection mode with delayed extraction. Positively charged ions with a mass:charge ratio (m/z) between 700 and 6000 were analyzed using an acceleration voltage of 25 kV. The sample spectra were calibrated externally with a calibration standard covering the m/z range from 700 to 3100 (Bruker Daltonics, Bremen, Germany).
In some cases, CIEX fractions were analyzed by MS directly without RPC. In these cases, the samples were desalted using a C18 ZipTip® (ZTC18S, Millipore, Billerica, MA) before being applied to Ultraflex MALDI-TOF/TOF.
2.5. SDS-PAGE and silver staining
SDS-PAGE was performed using Tris-Tricine 10–20% gels (Thermo Scientific) according to the manufacturer's protocol. Bacteriocin samples were mixed with the provided sample buffer and heated to 80 °C for 2 min before they were applied to the gels. After electrophoresis, gels were fixed by incubation for 1 h in 50% (v/v) methanol, 12% (v/v) glacial acetic acid and 0.0185% (v/v) formaldehyde. Fixed gels were washed twice for 10 min in 50% (v/v) ethanol, rinsed with 0.02% (w/v) Na2S2O3 x 5 H2O solution for 1 min and again washed three times in deionized H2O (dH2O) for 30 s. After incubation for 25 min in 0.2% (w/v) AgNO3 solution with 0.028% (v/v) formaldehyde, the gels were washed twice in dH2O and then incubated in developing solution containing 6% (w/v) Na2CO3, 0.0004% (w/v) Na2S2O3 x 5 H2O and 0.028% (v/v) formaldehyde until clear bands were visible in the gel. The reaction was stopped with dH2O and 15 min incubation with 50 mM EDTA solution.
2.6. Extracellular amino acid quantification
Amino acid concentrations in culture supernatant were analyzed by HPLC (Agilent 1200 series, Agilent Technologies, Waldbronn, Germany) after pre-column derivatization (Krömer et al., 2005), using separation on a reversed phase column (Gemini 5 μm C18 110 Å, 150 × 4.6 mm, Phenomenex, Aschaffenburg, Germany) and a gradient of eluent A (20 mM NaH2PO4 x H2O, 0.5 g L−1 sodium azide, pH 7.8) and B (45% acetonitrile, 45% methanol, 10% dH2O) at a flow rate of 1 ml min−1 and 40 °C and the following profile: 0–44.5 min: 100% eluent A, 44.5–45 min: 100-55% eluent A, 45–47 min: 55-39% eluent A, 47–48 min: 39-18% eluent A, 48–48.5 min: 18-0% eluent A, 48.5–50.5 min: 0% eluent A, 50.5–51 min: 0–100% eluent A, 51.0–53 min: 100% eluent A. The amino acids were quantified using fluorescence detection (excitation/emission at 206/305 nm for l-proline and 340/450 nm for other amino acids) and α-aminobutyrate as internal standard.
2.7. Intracellular amino acid quantification
Sampling for intracellular amino acids was performed via fast filtration and subsequent extraction in boiling water as previously validated and applied for such analysis in C. glutamicum (Bolten et al., 2007; Kind et al., 2011; Wittmann et al., 2004). Briefly, cells were collected by fast filtration, including a washing step with 2.5% NaCl solution (Wittmann et al., 2002). Afterwards, free intracellular amino acids were extracted from the cells by incubation in boiling water (15 min, 100 °C) and quantified as described above.
2.8. Metabolic activity profiling using 13C tracer experiments
The relative fractions of proteinogenic amino acids, taken up by C. glutamicum from the complex medium ingredients (tryptone, yeast extract) or synthetized de novo from Glc, respectively, were estimated by a13C tracer study with subsequent gas chromatography-mass spectrometry (GC/MS) analysis (Schwechheimer et al., 2018). In short, cells were grown in 2xTY medium with 2% [13C6] Glc (99% enrichment, Omicron Biochemicals, South Bend, IN, USA). Cells, sampled during the cultivation, were analyzed for the 13C labelling pattern of amino acids from hydrolysed cell protein using GC/MS (GC 7890A, 5975C quadrupole detector, Agilent, Darmstadt, Germany) as described recently (Hoffmann et al., 2018). The measured labelling patterns were corrected for the non-labelled inoculum and for natural isotopes (Yang et al., 2009) and then used to derive the summed fractional labelling for each amino acid (Schwechheimer et al., 2018). The latter value directly reflected the relative amount that was synthetized de novo from Glc and the remaining fraction could be attributed to uptake form the medium, respectively.
2.9. Cultivation in bioreactors
Fed-batch cultivation was performed with C. glutamicum CR099/pEKEx-pedACDCg in a Labfors 5 (Infors AG, Switzerland) stirred tank bioreactor with an initial working volume of 1.6 L. Precultures were grown o/N in 2xTY medium supplemented with 50 μg mL−1 kanamycin at 30 °C with aeration in non-baffled Erlenmeyer flasks at 130 rpm. The preculture was harvested by centrifugation (10 min, 3240 g, room temperature), to inoculate the main batch culture to an initial OD600 of 1. For bioreactor fermentations, a complex medium based on 2xTY and CGXII medium (Keilhauer et al., 1993) with 2% Glc was used. This bioreactor medium contained tryptone (16 g L−1) and yeast extract (10 g L−1) as in normal 2xTY, the salts and vitamins of CGXII except urea and MOPS, and 50 μg mL−1 kanamycin. The composition was formulated to mimic nutritional conditions of the shake flask experiments as close as possible while at the same time preventing limitations in trace elements etc. at high cell densities. Additionally, MOPS was omitted from the medium as the bioreactor was equipped with a pH control. Reactor temperature was set at 30 °C and pH was controlled at 7 by addition of 6 M ammonium hydroxide and 1 M phosphoric acid. Dissolved oxygen was controlled by a cascaded adaption of stirrer speed (400–1200 rpm) and aeration rate (0.3–2 L min−1 compressed air). During the uninduced growth phase dissolved oxygen was kept above 20%. At an OD600 of 40, production was induced with IPTG (0.2 mM) and the dissolved oxygen level was reduced to 5%. During the production phase 1 L feed solution was supplied at a linear feed rate of 50 mL h−1. The feed solution contained 152 g L−1 Glc, 122 g L−1 tryptone, 76 g L−1 yeast extract and 50 μg m L−1 kanamycin. The substrate ratios correspond to the medium used for small scale production (2xTY + 2% Glc; Section 2.1). Biomass dry cell weight was determined gravimetrically by centrifuging 1.8 mL culture broth (10 min, 11,880 g, 4 °C), washing the pellet with phosphate buffered saline and drying the washed pellet at 105 °C for 72 h. Culture supernatants were analyzed for bacteriocin activity (Section 2.3), l-glutamate, Glc, acetate and lactate concentrations (Cedex Bio HT analyzer, Roche Diagnostics GmbH, Germany).
3. Results
3.1. C. glutamicum is a suitable host for recombinant production of pediocin PA-1
Pediocin PA-1 is a bacteriocin with potent antimicrobial activity against a range of Gram-positive microorganisms including important human pathogens such as Listeria monocytogenes (Ríos Colombo et al., 2018). As a first step towards recombinant production of pediocin PA-1, we established a growth-dependent assay using L. monocytogenes EGDe:pIMK2 or L. innocua LMG2785:pIMK2 as indicator strains, and commercially available, purified pediocin (Supplementary Fig. S3A). Using serial, 2-fold dilutions of a pediocin standard, 39 ng mL−1 of pediocin was determined as the minimal concentration required to completely inhibit growth of the indicator strains under the conditions of the assay. This concentration was used in further experiments as a calibrator to estimate product concentrations in supernatants of producer strains.
To validate the assay, anti-microbial activity in supernatants of P. acidilactici 347, a natural producer of pediocin, was determined following growth in MRS medium to early stationary growth phase (Supplementary Fig. S3B). Complete inhibition of the indicator strain was achieved with a 1:128 dilution. Thus, supernatants of P. acidilactici 347 contained at least 5 μg mL−1 of active pediocin, based on the estimation with the pediocin standard (Supplementary Fig. S3A). According to the method to determine bacteriocin activity described in the methods section, this corresponded to 10,240 BU mL−1. Additional control experiments using a spot-on-lawn assay and L. monocytogenes EGDe or its isogenic mutant EGY2 (carrying a 84 bp deletion in the mptD gene encoding the IID subunit of the PTSMan) showed that the antimicrobial activity in supernatants of P. acidilactici 347 is dependent on a functional PTSMan (Supplementary Fig. S3C). This confirms that pediocin PA-1 is the only compound produced by P. acidilactici 347 with antimicrobial activity against L. monocytogenes.
The receptors for bacteriocins of the pediocin family on target organisms are the IIC and IID subunits of the PTSMan family (Kjos et al., 2009; Tymoszewska et al, 2017, 2018). In silico analysis of the genome of C. glutamicum CR099 and other strains of the species indicated that C. glutamicum does not encode a PTSMan suggesting resistance against pediocin. To corroborate this assumption, we determined the resistance of C. glutamicum CR099 against different AMPs using an endpoint measurement of growth in 96-well microtiter plates (Fig. 1A).
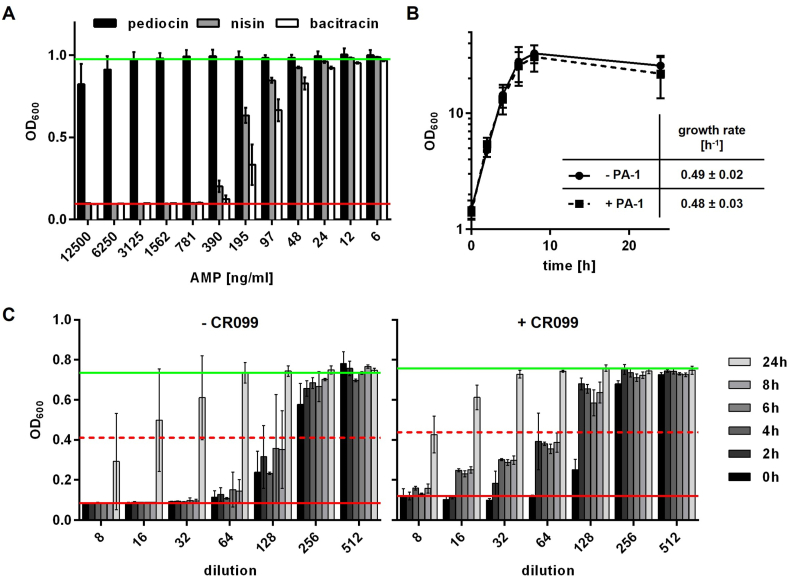
Fig. 1.
(A) Growth (OD600) of C. glutamicum CR099 in 96-well microtiter plates in the presence of pediocin (black bars), nisin (grey bars), or bacitracin (white bars) at the indicated concentrations. Values are mean ± standard deviation of n = 4–7 independent experiments. (B) Growth of C. glutamicum CR099 in standard batch culture in 2xTY medium with 2% Glc in the presence (+PA-1) or absence (- PA-1) of 2.0 μg mL−1 pediocin. Values are mean ± standard deviation of n = 4 independent experiments. Growth rates were calculated during exponential growth phase. (C) Growth inhibition of L. monocytogenes EGDe:pIMK2 by supernatants of C. glutamicum CR099 in 2xTY medium with 2% Glc plus 2 μg mL−1 pure pediocin (+CR099). Supernatants were collected during the experiment shown in (B) at the indicated time-points. As a control, pediocin was incubated under the same conditions in sterile medium (- CR099). Values are OD600 of the indicator strain L. monocytogenes EGDe at the indicated dilutions of the samples and are mean ± standard deviation of n = 3 independent experiments. In all graphs, the red and green lines indicate OD600 of the positive (i.e. complete inhibition of growth) or negative (i.e. in the absence of an AMP) control, respectively. The broken red lines represent growth inhibition of 50%, i.e. the threshold to calculate bacteriocin units.
Complete inhibition of growth was observed with bacitracin and nisin at concentrations of 390–781 ng mL−1. By contrast, C. glutamicum CR099 was able to grow in the presence of at least 12.5 μg mL−1 pediocin PA-1. Further experiments conducted in larger volumes under standard conditions of cultivation in baffled Erlenmeyer flask confirmed that 2 μg mL−1 of pediocin did not affect final optical density and growth rate (Fig. 1B). This indicates that production of pediocin by C. glutamicum CR099 at significant titers may be possible without adverse effects on growth.
Additionally, activity of pediocin was determined in the culture supernatant at different timepoints during the cultivation (Fig. 1C). Calculation of bacteriocin units (Supplementary Table S4) revealed that, after an initial reduction from 2560 to 1280 BU mL−1, activity remained stable for several hours in growing C. glutamicum CR099 cultures. After 24 h, activities dropped to 160 BU mL−1 in the presence of C. glutamicum CR099 and the same reduction was observed in control incubation in 2xTY without bacteria. In summary, C. glutamicum CR099 was able to grow in the presence of high concentrations of pediocin PA-1 and apparently did not degrade the bacteriocin in significant amounts during active growth. Thus C. glutamicum was considered a suitable host for recombinant pediocin PA-1 production.
3.2. Establishment of pediocin production in C. glutamicum CR099
To establish pediocin production in C. glutamicum CR099, the sequence of the biosynthetic operon of P. acidilactici PAC1.0 was retrieved from the NCBI GenBank database (accession no.: M83924.1; Fig. 2A). The operon comprises four genes located on a plasmid (pSRQ11): the structural gene pedA for the prepeptide of the bacteriocin, pedB for an immunity protein, and pedC and pedD encoding proteins required for processing, cleavage and export of the mature bacteriocin (Marugg et al., 1992).
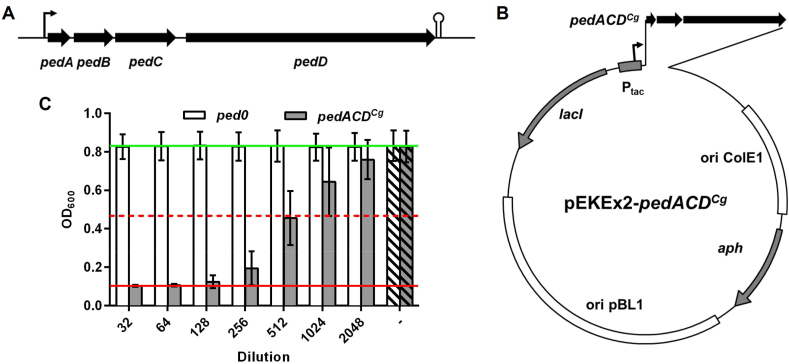
Fig. 2.
(A) Genetic organization of the operon of P. acidilactici PAC1.0 for biosynthesis of pediocin PA-1. (B) Plasmid map of pEKEx2-pedACDCg introduced into C. glutamicum CR099. (C) Growth inhibition of L. monocytogenes EGDe:pIMK2 by supernatants of recombinant C. glutamicum strains. C. glutamicum CR099/pEKEx2-pedACDCg (pedACDCg) or the empty vector control strain C. glutamicum CR099/pEKEx2 (ped0) were grown in 5 ml 2xTY medium in glass tubes and 2 h after inoculation Glc (2% w/v) and IPTG (0.2 mM) were added to induce production of pediocin. Activity was measured in 2-fold serial dilutions of supernatants harvested after o/N growth, i.e. early stationary growth phase, using L. monocytogenes EGDe:pIMK2 as indicator. Values are OD600 of the indicator strain and are mean ± standard deviation of n = 9 independent experiments. Hatched bars indicate growth of the indicator strain without addition of diluted supernatant or pediocin, i.e. maximum growth. The red and green lines indicate OD600 of the positive (i.e. complete inhibition of growth) or negative (i.e. in the absence of an AMP), respectively. The broken red line represents growth inhibition of 50%, i.e. the threshold to calculate bacteriocin units.
In P. acidilactici PAC1.0, all genes are transcribed from a single promoter upstream of pedA (Venema et al., 1995). The immunity protein confers resistance by a mechanism that depends on the PTSMan receptor (Diep et al., 2007), which is absent in C. glutamicum CR099. Thus, pedB was considered dispensable. A synthetic pedACDCgl operon for recombinant production of pediocin PA-1 was designed based on the protein sequences available on UniProt database (accession no.: P29430, P37249, and P36497). Gene sequences were codon-optimized for C. glutamicum, each equipped with a ribosome binding site, obtained as synthesized gene fragments, and cloned under the IPTG-inducible Ptac promoter into pEKEx2 by Gibson Assembly yielding pEKEx2-pedACDCg (Fig. 2B). This construct was successfully introduced into C. glutamicum CR099.
Supernatants of cultures of C. glutamicum CR099/pEKEx-pedACDCg grown to early stationary growth phase in 2xTY medium with 2% Glc and 0.2 mM IPTG in glass tubes contained up to 10,240 BU mL−1 of antimicrobial activity against L. monocytogenes. This corresponds to a concentration of approx. 5 μg mL−1 of pure pediocin (Fig. 2C), i.e. levels comparable to those of the natural producer P. acidilactici 347 (Supplementary Fig. S3B). By contrast, supernatants of the empty vector control strain C. glutamicum CR099/pEKEx-ped0 did not inhibit growth of L. monocytogenes EGDe:pIMK2 at all. Almost identical results were obtained with a second strain which contained the same operon and promoter in the pXMJ19 backbone (Supplementary Fig. S5).
To identify the compound responsible for inhibition of growth of the sensor strain, experiments in larger culture volumes were conducted. Surprisingly, only low activities were obtained when C. glutamicum CR099/pEKEx-pedACDCg was grown in baffled Erlenmeyer flasks for efficient oxygenation of the medium with a maximum of 640 BU mL−1 after 10 h of cultivation (Fig. 3, Supplementary Table S6).
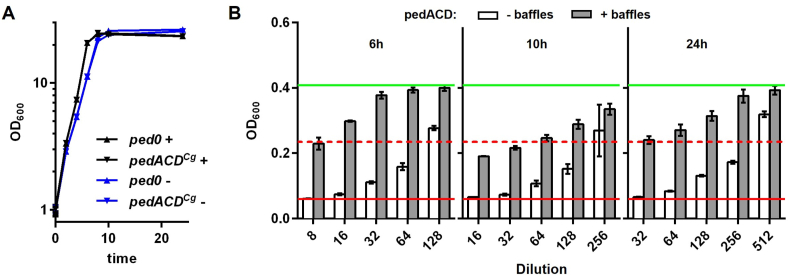
Fig. 3.
(A) Growth of C. glutamicum CR099/pEKEx-ped0 (ped0) and C. glutamicum CR099/pEKEx-pedACDCg (pedACDCg) in 2xTY medium in baffled (+) and non-baffled (−) Erlenmeyer flasks. 2h after inoculation Glc (2% w/v) and IPTG (0.2 mM) were added to induce production of pediocin. (B) Growth inhibition of L. innocua LMG2785:pIMK2 by different dilutions of supernatants of C. glutamicum CR099/pEKEx-pedACDCg collected at the indicated timepoints during the growth experiment shown in (A). All values are mean ± standard deviation of n = 3 independent experiments. The red and green lines indicate OD600 of the positive (i.e. complete inhibition of growth) or negative (i.e. in the absence of an AMP), respectively. The broken red line represents growth inhibition of 50%, i.e. the threshold to calculate bacteriocin units.
However, growth inhibitory activity of C. glutamicum CR099/pEKEx-pedACDCg supernatants was dramatically increased when cultivated under the same conditions in non-baffled Erlenmeyer flasks. Under these conditions, a maximum of 5120 BU mL−1 (equivalent to approx. 2.5 μg mL−1 of pure pediocin) were observed at the end of the cultivation (Fig. 3B and Supplementary Table S6).
3.3. Identification of pediocin in supernatants of recombinant C. glutamicum
For identification of pediocin, proteins in the supernatant of C. glutamicum CR099/pEKEx-pedACDCg grown o/N in non-baffled Erlenmeyer flasks in 2xTY medium with 2% Glc and 0.2 mM IPTG were precipitated with 50% ammonium sulfate and then separated via CIEX chromatography. Similar to supernatants of P. acidilactici 347, a single peak was observed at 280 nm at the onset of elution (Supplementary Fig. S7). Peak fractions F8–F10 were collected and further analyzed. All peak fractions strongly inhibited growth of L. monocytogenes EGDe:pIMK2 with a maximum of 204,800 BU mL−1, which is equivalent to at least 25 μg mL−1 active pediocin (Fig. 4A and Supplementary Table S8). In all fractions, activity was completely abolished by proteinase K treatment indicating that the active compound was a proteinaceous substance.
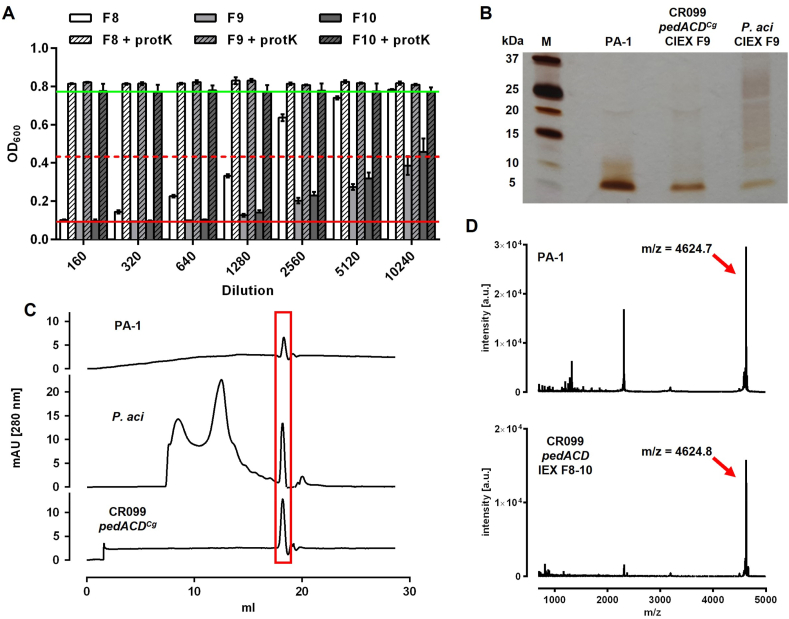
Fig. 4.
(A) Growth inhibitory activity in different dilutions of fractions F8-10 of CIEX chromatography of precipitated supernatant of C. glutamicum CR099/pEKEx-pedACDCg grow in 2xTY medium with 2% Glc in non-baffled Erlenmeyer flasks o/N. Samples were analyzed untreated or following incubation with proteinase K (+protK). All values are mean ± standard deviation of n = 3 independent experiments. The red and green lines indicate OD600 at baseline (i.e. complete inhibition of growth) or in the absence of AMPs, respectively. The broken red line represents growth inhibition of 50%, i.e. the threshold to calculate bacteriocin units. (B) SDS-PAGE of CIEX peak fractions F9 (IEX F9) of supernatants of C. glutamicum CR099/pEKEx-pedACDCg (CR099 pedACDCg) or P. acidilactici 347 (P. aci). Pure pediocin (PA-1) was analyzed on the same gel as control (M: molecular weight marker). (C) SIEX chromatography of pooled CIEX fractions F8–F10 from supernatants of P. acidilactici 347 (P. aci, middle panel) or C. glutamicum CR099/pEKEx-pedACDCg (CR099 pedACDCg; lower panel) or pure pediocin (PA-1) as a control. The red box indicates a signal in all samples with a size corresponding to pure pediocin. (D) Mass spectrometry of active fractions after RPC of pediocin PA-1 (upper panel) and pooled active CIEX fractions from supernatants C. glutamicum CR099/pEKEx-pedACDCg (lower panel) before applying to RPC. The red and black boxes highlight peaks observed in both samples with almost identical mass/charge ratio (m/z) that match the calculated m/z of pediocin PA-1.
Additionally, SDS-PAGE revealed a single protein band in CIEX peak fractions of both P. acidilactici 347 and C. glutamicum CR099/pEKEx2-pedACDCg at around 5 kDa, which corresponds to the size of pediocin PA-1 (Fig. 4B). SIEX analysis of CIEX preparations from P. acidilactici 347 and C. glutamicum CR099/pEKEx2-pedACDCg supernatants revealed a single peak at an elution volume of 18.3 mL, which is identical to the elution volume of pure, commercially available pediocin (Fig. 4C).
Interestingly, both SDS-PAGE and SIEX chromatography suggested that the preparations of C. glutamicum CR099/pEKEx2-pedACDCg contained pediocin as main product in high purity as indicated by a single band or peak. By contrast, samples prepared from P. acidilactici 347 supernatants contained several other peaks or signals indicative of further proteins/peptides possibly secreted by P. acidilactici 347 or present in MRS medium used for cultivation. To further confirm identity of the active compound, CIEX peak fractions were further analyzed by reverse phase chromatography coupled to mass spectrometry (Fig. 4D). This identified a peptide in the supernatants of C. glutamicum CR099/pEKEx2-pedACDCg with a m/z of 4624.7, which is almost identical to that of commercial pediocin PA-1 (m/z = 4624.8) and the theoretical m/z of pediocin PA-1 (m/z = 4624.3 (Johnsen et al., 2000);).
3.4. Gene complement and order for efficient production of pediocin by C. glutamicum
To assess the minimal operon for pediocin production by C. glutamicum further synthetic constructs with ped genes in different combinations (Supplementary Fig. S9) were cloned into pEKEx2 and corresponding plasmids were introduced into C. glutamicum CR099.
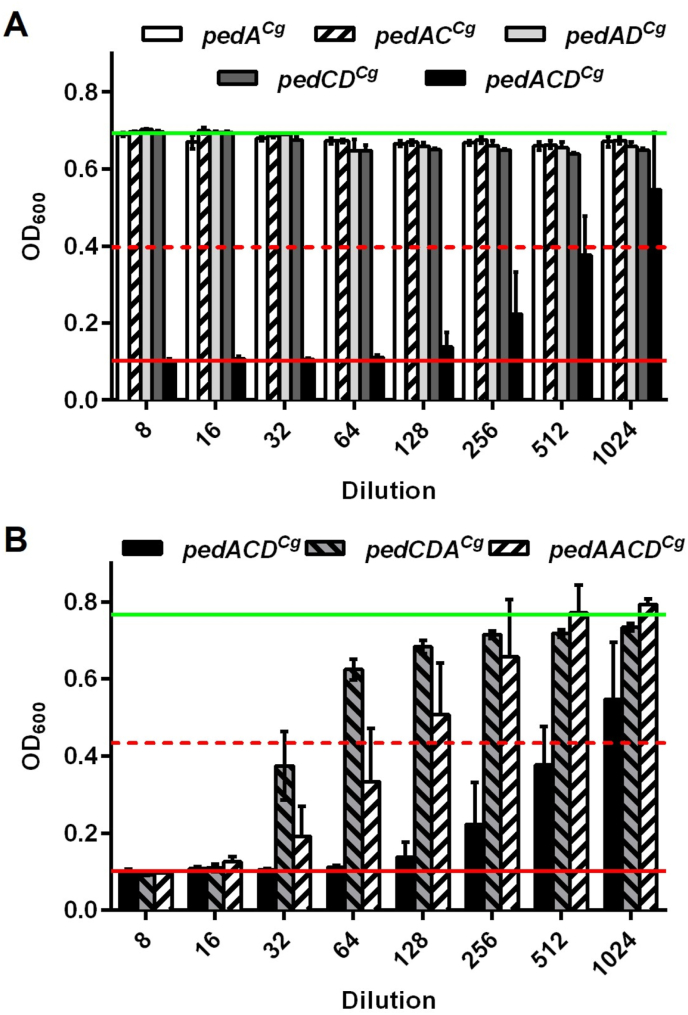
Measurement of activity in supernatants of strains with different combinations of the ped genes (pedACg, pedACCg, pedADCg, pedCDCg, pedACDCg) using L. monocytogenes EGDe as sensor strain revealed that strains that lack any of the three genes did not contain pediocin activity in their supernatants (Fig. 5A, Supplementary Table S10).
Fig. 5.
Growth inhibition of L. monocytogenes EGDe by different dilutions of supernatants of C. glutamicum CR099 strains harboring pEKEx2-derived constructs with ped genes in different combinations (A) or in different order and copy number (B). Bacteria were grown in 5 mL 2xTY medium in glass tubes and 2h after inoculation Glc (2% w/v) and IPTG (0.2 mM) were added. All values are mean OD600 ± standard deviation of n = 3 independent experiments. The red and green lines indicate the OD600 at baseline (i.e. complete inhibition of growth) or in the absence of AMPs, respectively. The broken red lines represent growth inhibition of 50%, i.e. the threshold to calculate bacteriocin units.
Similarly, altering the gene order by moving pedACg to the end of the operon (pedCDACg) or adding an additional copy of pedACg (pedAACDCg) resulted in reduced activity in the supernatants of the respective strains (Fig. 5B, Supplementary Table S10).
3.5. Metabolic physiology of C. glutamicum during pediocin production
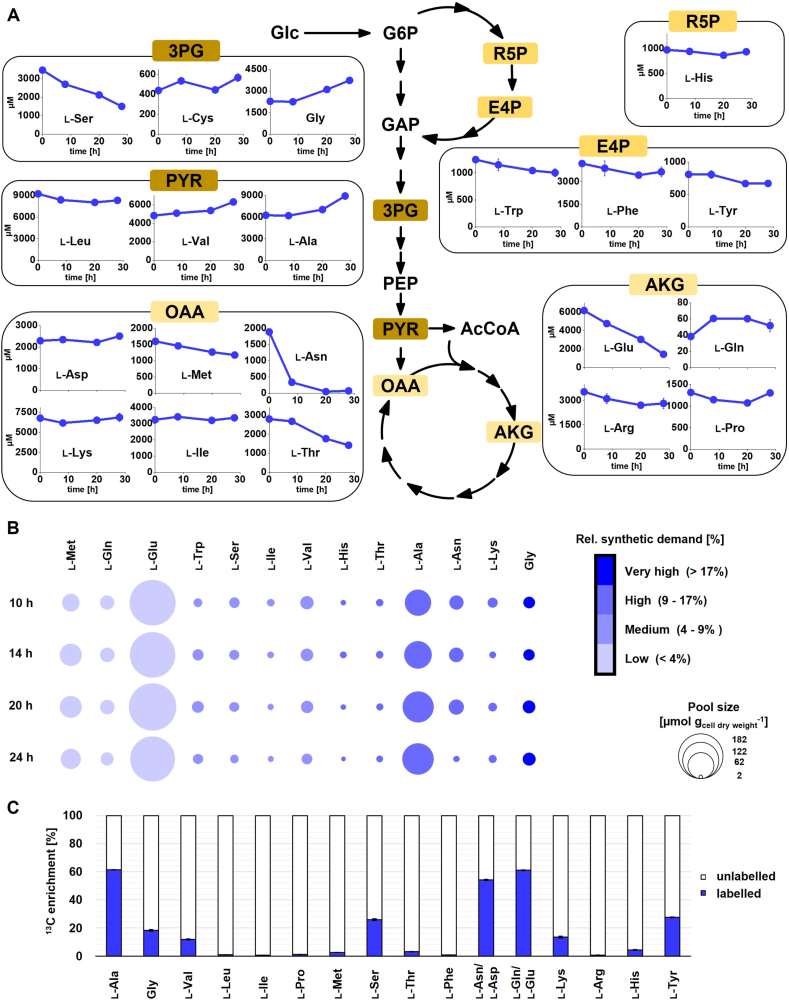
Next, the production process was studied in more detail to inspect the impact of plasmid-based pediocin production and the imposed growth conditions, i. e. complex 2xTY medium with 2% Glc under oxygen limitation, on the physiology of C. glutamicum CR099/pXMJ19-pedACDCg. First, extracellular amino acids were quantified at different time points of the pediocin production process. All 20 proteinogenic amino acids were detected in the medium and revealed a complex profile (Fig. 6A). TCA cycle derived amino acids, including l-glutamate, l-aspartate, l-asparagine, l-threonine, l-methionine, and l-arginine decreased over time, partly to complete depletion. Others, such as glycine, l-alanine and l-valine accumulated, whereas l-lysine, l-proline, and l-isoleucine remained rather unaffected.
Fig. 6.
Metabolic physiology of pediocin producing C. glutamicum CR099/pXMJ19-pedACDCg cultivated in 2xTY medium with 2% Glc. (A) Extracellular concentrations (in μM) of proteinogenic amino acids. For comparison, the corresponding data for the empty vector control strain CR099/pXMJ19 are shown in Supplementary Figure S11 (B) Intracellular concentrations of amino acids that serve as pediocin building blocks. Values represent absolute levels of the major pediocin precursors in μmol gcell dry weight−1 at different time points of the production process and are mean ± standard deviation of n = 3 independent experiments. The pool sizes of intracellular amino acids are visualized by circle areas corresponding to concentration, and the relative requirement of each amino acid for pediocin synthesis is indicated by color. Values are mean of n = 3 independent experiments. Corresponding data for the empty vector control strain CR099/pXMJ19 are shown in Supplementary Figure S12 (C) Metabolic activity profiling regarding the supply of amino acids for cell protein. The data display the relative 13C enrichment in the amino acids from a13C tracer study with 2xTY and 2% [13C6] glucose at the end of the growth phase after 15 h. The 13C enrichment reflects the relative contribution of de novo synthesis from 99% 13C enriched glucose and uptake from non-13C enriched complex 2xTY ingredients, respectively, to formation of an amino acid. Due to decomposition processes during hydrolysis of cellular protein prior to 13C labelling analysis, data for l-cysteine and l-tryptophan are not provided, while l-asparagine/l-aspartate and l-glutamine/l-glutamate are shown as lumped pool (Wittmann et al., 2007). Values are mean ± standard deviation of n = 3 independent experiments. Corresponding 13C enrichment data for all sampled time points (6 h, 11 h, 15 h, 25 h) are shown in Supplementary Fig. S13.
Free amino acids present in the cytoplasm of pediocin-producing C. glutamicum CR099/pXMJ19-pedACDCg and non-producing C. glutamicum CRO099/pXMJ19, strongly differed in concentration and changed over time (Fig. 6B, Supplementary Fig. S12). The concentration of the most abundant amino acid l-glutamate was almost hundred-fold higher than that of low-level compounds such as l-aspartate, l-isoleucine, and l-phenylalanine. Interestingly, the level of intracellular l-asparagine, one of the most abundant building blocks of pediocin, dramatically decreased towards the end of the process, when pediocin production had stopped. This was an unexpected limitation considering the rich medium used. Subsequent 13C tracer studies using [13C6]-Glc and naturally labelled 2xTY as substrate revealed that most amino acids were efficiently taken up from the medium, as indicated by their low 13C enrichment in cellular protein (Fig. 6C). However, TCA-cycle derived l-asparagine, l-aspartate, l-glutamine, l-glutamate, and glycolytic amino acids (l-alanine, glycine, l-serine) revealed substantial 13C enrichment and were thus largely synthetized de novo from Glc. This biosynthetic mode was rather constant throughout the cultivation (Supplementary Fig. S13). The strain C. glutamicum CR099/pXMJ19 (carrying the empty plasmid) revealed similar levels of extra- and intracellular amino acids as the producer (Supplementary Figs. S11 and S12). This suggests that strain physiology and partial amino acid depletion was mainly influenced by the imposed growth conditions but not by plasmid burden. A notable exception to this was a five-fold lower level of the pediocin precursor l-asparagine in the producer at later stages of the process.
3.6. Production of pediocin in bioreactors
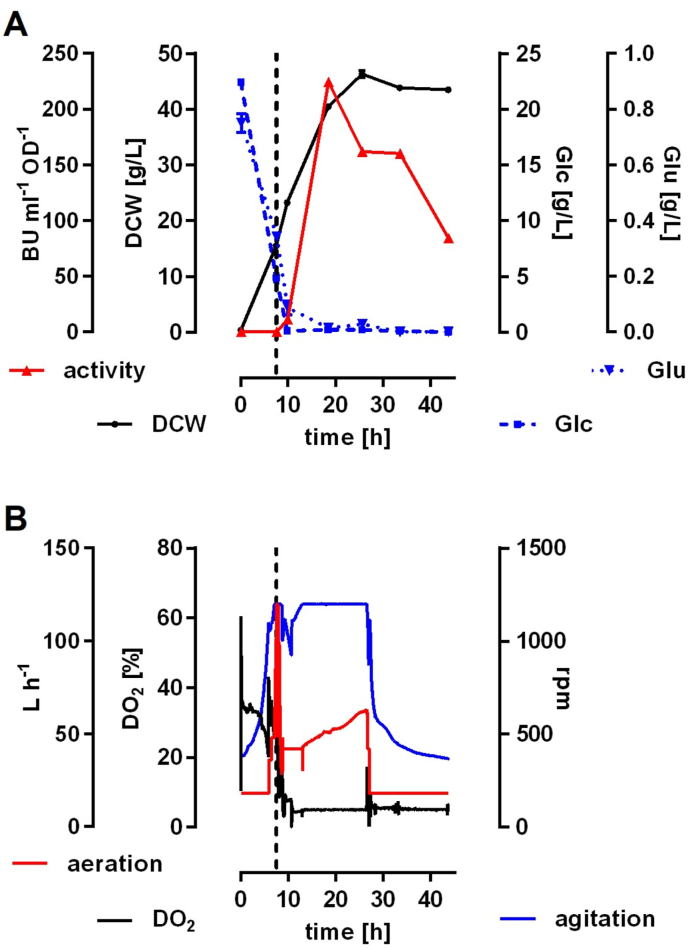
To demonstrate feasibility of recombinant pediocin production on a larger scale, further experiments were performed with C. glutamicum CR099/pEKEx-pedACDCg in bioreactors under fed-batch conditions (Fig. 7). Media composition and culture conditions (see section 2.9) were similar to those of the shake flask experiments. During the growth phase (0–7.5 h, i.e. in the absence of the inducer IPTG) provided nutrients were consumed and biomass increased with a growth rate of about 0.5 h−1 (Fig. 7A), which is in good correspondence with the results of shake flask experiments (Fig. 1, Fig. 3). When initial Glc approached depletion after 7.5 h, constant feed addition was started (50 mL h−1 and 1 L of feed) and production was induced with 0.2 mM IPTG. During the induction phase dissolved oxygen was reduced to 5% to prevent exhaustive oxidation of pediocin as indicated by baffled shake flask experiments (Fig. 3). Dissolved oxygen was tightly controlled by a split range control including stirrer speed and subsequent aeration rate adaption (Fig. 7B). During induction, fed glucose and l-glutamate were fully consumed and biomass was formed with a decreased growth rate (0.1 h−1 in average) before growth ceased at a biomass concentration of 46 g L−1 and after complete depletion of nutrients (Fig. 7A).
Fig. 7.
(A) Substrate, biomass and pediocin activity and (B) critical process parameters during fermentation of C. glutamicum CR099/pEKEx-pedACDCg grown in a bioreactor in bioreactor medium containing 2% Glc, 16 g L−1 tryptone and 10 g L−1 yeast extract under controlled fed-batch conditions. (A): offline measurements of biomass (DCW), biomass-specific bacteriocin activity (activity; BU mL−1 OD−1), glucose (Glc [g L−1]) and l-glutamate (Glu [g L−1]). (B): online measurements of dissolved oxygen (DO2 [%]), agitation (rpm) and aeration (L h−1). The timepoint of induction (t = 7.5 h) is indicated by the vertical broken black line.
No significant amounts of byproducts such as lactate or acetate were detected (data not shown). Measurements of pediocin activity indicate active pediocin already 2.5 h after induction reaching high levels of 20,480 BU mL−1 between t = 18.5 h and 33.5 h of the experiment (Supplementary Fig. S14 and Table S15). This activity corresponds to approx. 10 μg mL−1 of pure pediocin. A slight decrease in activity (10,240 BU mL−1) was observed at the end of the experiment (t = 43.5 h). Specific activity reached a maximum of 224.4 BU mL−1 OD−1 at t = 18.5 h, i.e. 11 h after induction. This is in good agreement with the specific activities observed at the end (t = 24 h) of the experiments in non-baffled Erlenmeyer flasks (200.9 BU mL−1 OD−1) and indicates a successful transfer of pediocin production to bioreactor conditions.
4. Discussion
Bacteriocins are an attractive, clean-label approach for preservation of food, beverages and animal feed (Chikindas et al., 2018; Mills et al., 2011; Silva et al., 2018). Moreover, they are increasingly considered for clinical applications (Kranjec et al., 2020; Ovchinnikov et al., 2020). Besides regulatory aspects, one of the main obstacles for market entry and clinical applications of novel bacteriocins are issues with economically viable production and purification to pharmaceutical-grade products. One possibility to overcome these obstacles is the transfer of bacteriocin production to industrial workhorse organisms.
Here, we implemented heterologous production of a commercially relevant bacteriocin into the biotechnological workhorse organism C. glutamicum. We demonstrated that C. glutamicum CR099 was able to grow in the presence of at least 12.5 μg mL−1 of pediocin (Fig. 1), i.e. levels of resistance comparable to a L. monocytogenes mutant lacking a PTSMan (Ramnath et al., 2000). In line, the genome of C. glutamicum CR099 does not harbor genes for a PTSMan. When spiked into growing cultures of C. glutamicum CR099 or sterile media, a considerable loss in activity was observed (Fig. 1C). As C. glutamicum does not show significant extracellular protease activity (Becker and Wittmann, 2017; Lee and Kim, 2018) this may be due to oxidative inactivation of pediocin as reported previously (Bédard et al., 2018; Johnsen et al., 2000). Additionally, adsorption to the glass surface of flasks via the hydrophobic C-terminus (Fimland et al., 1996) or electrostatic interactions of the cationic peptide with the negatively charged surface of bacteria may play a role.
To achieve recombinant production of pediocin, we designed a synthetic pedACDCg operon for pediocin biosynthesis with gene sequences optimized for codon usage of C. glutamicum. Supernatants of C. glutamicum CR099/pEKEx-pedACDCg contained a compound with antimicrobial activity against Listeria sp., which was identified as indeed pediocin (Fig. 4). To provide a more robust quantification of pediocin in the bioactivity assay, we established a calibration using a standard solution of a commercial pediocin preparation (Supplementary Fig. S3). Interestingly, pediocin purified from C. glutamicum supernatants contained considerably lower amounts of impurities than similar preparations of P. acidilactici supernatants. As C. glutamicum is already used for production of amino acids infusion solutions for human use (Bückle-Vallant et al., 2014; Krause et al., 2010), this suggests that pediocin (and possibly other bacteriocins) may be produced by C. glutamicum in sufficient purity for clinical applications.
In line with the proposed mechanisms of resistance mediated by PedB (Diep et al., 2007) and the lack of a PTSMan gene in the genome of C. glutamicum, pedB was not required for production of pediocin (Fig. 2, Fig. 5). The pedCCg and pedDCg genes were both required for production of detectable levels of pediocin by C. glutamicum strains (Fig. 5). By contrast, pedC was not required for production and release of pediocin by E. coli (Mesa-Pereira et al., 2017). PedC was recently shown to be involved in correct formation of disulfide bonds in pediocin (Oppegård et al., 2015) and pediocin analogues with incorrect disulfide bonds show reduced activity (Bédard et al., 2018). A possible explanation for the activity observed by Mesa-Pereira and colleagues could thus be spontaneous formation of different conformational variants of pediocin including the active form in the absence of PedC. Of note, a scrambled version of the operon (pedCDACg) and a construct with a second copy of pedACg (pedAACDCg) showed reduced activity compared to pedACDCg. This indicates that the stoichiometry of the structural peptide and biosynthesis proteins may be important for maximum production and points towards a bottleneck in the export apparatus rather than in expression of the peptide itself. However, this needs further investigation e.g. by transcriptional analysis and analysis of intracellular accumulation of the pediocin prepeptide.
Maximum levels of active pediocin achieved in lab-scale fermentations with C. glutamicum CR099/pEKEx2-pedACDCg and CR099/pXMJ-pedACDCg were at least 5 μg mL−1 according to the established calibration and are thus comparable to or higher than levels obtained with the natural producer P. acidilactici 347 reported here (Supplementary Fig. S3) or previously (Horn et al., 1999). Notably, lower titers were observed in baffled shake flasks again suggesting that excessive aeration of the culture broth may lead to oxidative inactivation of the product (Fig. 3). In further experiments in controlled bioreactors in larger volumes (Fig. 7), titers could be improved to at least 20,480 BU mL−1 or 10 μg mL−1 of active pediocin. Conditions in controlled bioreactors were set to match those in non-baffled shake flasks, i.e. low levels of dissolved oxygen (5%) during the production phase. The increase in absolute activity compared to experiments in non-baffled shake flask may be explained by higher biomass in the bioreactor compared to experiments in non-baffled shake-flasks. Specific activities were comparable in non-baffled shake flasks and the bioreactor (200.9 vs. 224.4 BU mL−1 OD−1) indicating successful upscaling from shake flask to lab-scale bioreactor conditions. As C. glutamicum can be cultivated at extremely high cell densities even under low oxygen conditions (Becker et al, 2011, 2013; Okino et al., 2005), this suggests that pediocin titers may be further increased simply by increasing biomass e.g. by providing additional substrate. Tuning of oxygen availability during growth and production phase provides opportunities for further improvements of the production process.
Limited availability of precursors is a potential bottleneck for product formation (Korneli et al., 2012; Kuhl et al., 2020). The producer strain revealed complex dynamics of extracellular and intracellular amino acids over time. Regarding extracellular amino acids, we observed with strong consumption of different TCA cycle-derived amino acids, obviously to fuel this cyclic pathway (Fig. 6A and B). In addition, glycolytic breakdown of Glc played an important role to replenish TCA cycle-related amino acids (Fig. 6C). Altogether, these reactions obviously compensated for the reduced TCA cycle operation and the reduced ATP formation under the imposed oxygen limiting conditions (Becker and Wittmann, 2017; Michel et al., 2015; Wittmann et al., 2007).
Overall, control strain and producer showed similar profiles of extra- and intracellular amino acids indicating that the environmental conditions had a generally higher impact than plasmid burden and product synthesis. However, one pronounced difference was observed between the two strains: the intracellular level of l-asparagine dramatically decreased in the medium and inside producing cells (Fig. 6A and B) and was more than five-fold lower in the producer than in the control strain after 24 h (Supplementary Fig. S12). As l-asparagine is one of the major building blocks of pediocin, this reduction is likely caused by the extra demand for pediocin synthesis. Thus, low availability of this amino acid towards the end of the process might be a bottleneck for product formation. This limitation was rather surprising considering the high levels of tryptone and yeast extract contained in the medium. In this regard, oxygen limitation and product formation unfavorably coincided in causing this limitation. The underling metabolic details deserve more attention in the future towards improved production. At present, we know only little about bacteriocin production and oxygen-limited growth of C. glutamicum.
In conclusion, we established recombinant production of pediocin using C. glutamicum at titers above 10 μg mL−1. Moreover, we provide clues towards optimization of fermentation process to increase yield. The developed approach may be adopted for production of other class IIa bacteriocins targeting the PTSMan and, upon integration of appropriate resistance mechanisms, may also be extended to other bacteriocin families. This adds another commercially interesting group of compounds that can be produced using C. glutamicum.
CRediT authorship contribution statement
O. Goldbeck: Conceptualization, Methodology, Validation, Investigation, Writing - original draft, Writing - review & editing. D.N. Desef: Investigation, Writing - review & editing. K.V. Ovchinnikov: Methodology, Investigation, Writing - review & editing. F. Perez-Garcia: Investigation, Writing - review & editing. J. Christmann: Methodology, Investigation, Writing - review & editing. P. Cao: Methodology, Investigation, Writing - review & editing. J. Becker: Methodology, Investigation, Writing - review & editing. M. Kohlstedt: Investigation, Writing - review & editing. P. Sinner: Methodology, Investigation, Writing - review & editing. P. Crauwels: Methodology, Writing - review & editing. J. Kager: Methodology, Investigation, Writing - review & editing. B.J. Eikmanns: Writing - review & editing. D. Weixler: Investigation, Writing - review & editing. G.M. Seibold: Conceptualization, Funding acquisition, Methodology, Writing - review & editing. C. Herwig: Conceptualization, Funding acquisition, Methodology. C. Wittmann: Conceptualization, Funding acquisition, Methodology, Validation, Formal analysis, Writing - review & editing. N.S. Bar: Conceptualization, Funding acquisition, Methodology, Validation, Formal analysis, Writing - review & editing. D.B. Diep: Funding acquisition, Methodology, Validation, Formal analysis, Writing - review & editing. C.U. Riedel: Conceptualization, Funding acquisition, Methodology, Validation, Formal analysis, Writing - original draft, Writing - review & editing.
Declaration of competing interest
OG, DW, GMS, CW, NSB, DBD, and CUR are co-inventors on a patent application related to this research.
Acknowledgements
This project has received funding from the Bio Based Industries Joint Undertaking under the European Union's Horizon 2020 research and innovation program under grant agreement No 790507. Judith Becker acknowledges funding by the Hans&Ruth-Giessen-Stiftung.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ymben.2021.09.002.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Abbasiliasi S., Tan J.S.S., Tengku Ibrahim T.A.A., Bashokouh F., Ramakrishnan N.R.R., Mustafa S., Ariff A.B.B. Fermentation factors influencing the production of bacteriocins by lactic acid bacteria: a review. RSC Adv. 2017;7:29395–29420. doi: 10.1039/C6RA24579J. [DOI] [Google Scholar]
- Angelopoulou A., Warda A.K., O'Connor P.M., Stockdale S.R., Shkoporov A.N., Field D., Draper L.A., Stanton C., Hill C., Ross R.P. Diverse bacteriocins produced by strains from the human milk microbiota. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo A.C., Bento C.B.P., Ruiz J.C., Queiroz M.V., Mantovani H.C. Distribution and genetic diversity of bacteriocin gene clusters in rumen microbial genomes. Appl. Environ. Microbiol. 2015;81:7290–7304. doi: 10.1128/AEM.01223-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J., Rohles C.M., Wittmann C. Metabolically engineered Corynebacterium glutamicum for bio-based production of chemicals, fuels, materials, and healthcare products. Metab. Eng. 2018 doi: 10.1016/j.ymben.2018.07.008. [DOI] [PubMed] [Google Scholar]
- Becker J., Schäfer R., Kohlstedt M., Harder B.J., Borchert N.S., Stöveken N., Bremer E., Wittmann C. Systems metabolic engineering of Corynebacterium glutamicum for production of the chemical chaperone ectoine. Microb. Cell Factories. 2013;12 doi: 10.1186/1475-2859-12-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J., Wittmann C. Industrial Microorganisms. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim, Germany: 2017. Industrial microorganisms: Corynebacterium glutamicum; pp. 183–220. [DOI] [Google Scholar]
- Becker J., Zelder O., Häfner S., Schröder H., Wittmann C. From zero to hero-Design-based systems metabolic engineering of Corynebacterium glutamicum for l-lysine production. Metab. Eng. 2011;13:159–168. doi: 10.1016/j.ymben.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Bédard F., Hammami R., Zirah S., Rebuffat S., Fliss I., Biron E. Synthesis, antimicrobial activity and conformational analysis of the class IIa bacteriocin pediocin PA-1 and analogs thereof. Sci. Rep. 2018;8:9029. doi: 10.1038/s41598-018-27225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolten C.J., Kiefer P., Letisse F., Portais J.C., Wittmann C. Sampling for metabolome analysis of microorganisms. Anal. Chem. 2007;79:3843–3849. doi: 10.1021/ac0623888. [DOI] [PubMed] [Google Scholar]
- Bückle-Vallant V., Krause F.S., Messerschmidt S., Eikmanns B.J. Metabolic engineering of Corynebacterium glutamicum for 2-ketoisocaproate production. Appl. Microbiol. Biotechnol. 2014;98:297–311. doi: 10.1007/s00253-013-5310-2. [DOI] [PubMed] [Google Scholar]
- Chikindas M.L., García-Garcerá M.J., Driessen A.J.M., Ledeboer A.M., Nissen-Meyer J., Nes I.F., Abee T., Konings W.N., Venema G. Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl. Environ. Microbiol. 1993;59:3577–3584. doi: 10.1128/AEM.59.11.3577-3584.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikindas M.L., Weeks R., Drider D., Chistyakov V.A., Dicks L.M. Functions and emerging applications of bacteriocins. Curr. Opin. Biotechnol. 2018;49:23–28. doi: 10.1016/j.copbio.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter P.D., Ross R.P., Hill C. Bacteriocins — a viable alternative to antibiotics? Nat. Rev. Microbiol. 2013;11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- de Arauz L.J., Jozala A.F., Mazzola P.G., Vessoni Penna T.C. Nisin biotechnological production and application: a review. Trends Food Sci. Technol. 2009;20:146–154. doi: 10.1016/j.tifs.2009.01.056. [DOI] [Google Scholar]
- Diep D.B., Skaugen M., Salehian Z., Holo H., Nes I.F. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2384–2389. doi: 10.1073/pnas.0608775104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan K., Field D., Ross R.P., Cotter P.D., Hill C. In silico prediction and exploration of potential bacteriocin gene clusters within the bacterial genus geobacillus. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggeling L., Bott M. A giant market and a powerful metabolism: l-lysine provided by Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2015;99:3387–3394. doi: 10.1007/s00253-015-6508-2. [DOI] [PubMed] [Google Scholar]
- Ennahar S., Aoude-Werner D., Sorokine O., Van Dorsselaer A., Bringel F., Hubert J.C., Hasselmann C. Production of pediocin AcH by Lactobacillus plantarum WHE 92 isolated from cheese. Appl. Environ. Microbiol. 1996;62:4381–4387. doi: 10.1128/aem.62.12.4381-4387.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimland G., Blingsmo O.R., Sletten K., Jung G., Nes I.F., Nissen-Meyer J. New biologically active hybrid bacteriocins constructed by combining regions from various pediocin-like bacteriocins: the C-terminal region is important for determining specificity. Appl. Environ. Microbiol. 1996;62:3313–3318. doi: 10.1128/aem.62.9.3313-3318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A., Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Hoffmann S.L., Jungmann L., Schiefelbein S., Peyriga L., Cahoreau E., Portais J.C., Becker J., Wittmann C. Lysine production from the sugar alcohol mannitol: design of the cell factory Corynebacterium glutamicum SEA-3 through integrated analysis and engineering of metabolic pathway fluxes. Metab. Eng. 2018;47:475–487. doi: 10.1016/j.ymben.2018.04.019. [DOI] [PubMed] [Google Scholar]
- Hoffmann S.L., Kohlstedt M., Jungmann L., Hutter M., Poblete-Castro I., Becker J., Wittmann C. Cascaded valorization of brown seaweed to produce l-lysine and value-added products using Corynebacterium glutamicum streamlined by systems metabolic engineering. Metab. Eng. 2021 doi: 10.1016/J.YMBEN.2021.07.010. [DOI] [PubMed] [Google Scholar]
- Holo H., Nilssen O., Nes I.F. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J. Bacteriol. 1991;173:3879–3887. doi: 10.1128/jb.173.12.3879-3887.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn N., Martínez M.I., Martinez J.M., Hernández P.E., Gasson M.J., Rodríguez J.M., Dodd H.M. Enhanced production of pediocin PA-1 and coproduction of nisin and pediocin PA-1 by Lactococcus lactis. Appl. Environ. Microbiol. 1999;65:4443–4450. doi: 10.1128/aem.65.10.4443-4450.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen L., Fimland G., Eijsink V., Nissen-Meyer J. Engineering increased stability in the antimicrobial peptide pediocin PA-1. Appl. Environ. Microbiol. 2000;66:4798–4802. doi: 10.1128/AEM.66.11.4798-4802.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.M., Jung D.Y.-G., Jin D.Y.-Y., Jayabalan D.R., Yang D.S.H., Suh J.W. Bacteriocins as food preservatives: challenges and emerging horizons. Crit. Rev. Food Sci. Nutr. 2017:1–25. doi: 10.1080/10408398.2017.1340870. [DOI] [PubMed] [Google Scholar]
- Keilhauer C., Eggeling L., Sahm H. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 1993;175:5595–5603. doi: 10.1128/jb.175.17.5595-5603.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind S., Kreye S., Wittmann C. Metabolic engineering of cellular transport for overproduction of the platform chemical 1,5-diaminopentane in Corynebacterium glutamicum. Metab. Eng. 2011;13:617–627. doi: 10.1016/j.ymben.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Kjos M., Nes I.F., Diep D.B. Class II one-peptide bacteriocins target a phylogenetically defined subgroup of mannose phosphotransferase systems on sensitive cells. Microbiology. 2009;155:2949–2961. doi: 10.1099/mic.0.030015-0. [DOI] [PubMed] [Google Scholar]
- Kjos M., Salehian Z., Nes I.F., Diep D.B. An extracellular loop of the mannose phosphotransferase system component IIC is responsible for specific targeting by class IIa bacteriocins. J. Bacteriol. 2010;192:5906–5913. doi: 10.1128/JB.00777-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kördikanlıoğlu B., Şimşek, Ö., Saris P.E.J. Nisin production of Lactococcus lactis N8 with hemin-stimulated cell respiration in fed-batch fermentation system. Biotechnol. Prog. 2015;31:678–685. doi: 10.1002/btpr.2075. [DOI] [PubMed] [Google Scholar]
- Korneli C., Bolten C.J., Godard T., Franco-Lara E., Wittmann C. Debottlenecking recombinant protein production in Bacillus megaterium under large-scale conditions-targeted precursor feeding designed from metabolomics. Biotechnol. Bioeng. 2012;109:1538–1550. doi: 10.1002/bit.24434. [DOI] [PubMed] [Google Scholar]
- Kranjec C., Ovchinnikov K.V., Grønseth T., Ebineshan K., Srikantam A., Diep D.B. A bacteriocin-based antimicrobial formulation to effectively disrupt the cell viability of methicillin-resistant Staphylococcus aureus (MRSA) biofilms. NPJ biofilms microbiomes. 2020;6:58. doi: 10.1038/s41522-020-00166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause F.S., Blombach B., Eikmanns B.J. Metabolic engineering of Corynebacterium glutamicum for 2-Ketoisovalerate production. Appl. Environ. Microbiol. 2010;76:8053–8061. doi: 10.1128/AEM.01710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krömer J.O., Fritz M., Heinzle E., Wittmann C. In vivo quantification of intracellular amino acids and intermediates of the methionine pathway in Corynebacterium glutamicum. Anal. Biochem. 2005;340:171–173. doi: 10.1016/j.ab.2005.01.027. [DOI] [PubMed] [Google Scholar]
- Kuhl M., Gläser L., Rebets Y., Rückert C., Sarkar N., Hartsch T., Kalinowski J., Luzhetskyy A., Wittmann C. Microparticles globally reprogram Streptomyces albus toward accelerated morphogenesis, streamlined carbon core metabolism, and enhanced production of the antituberculosis polyketide pamamycin. Biotechnol. Bioeng. 2020 doi: 10.1002/bit.27537. [DOI] [PubMed] [Google Scholar]
- Lee M.J., Kim P. Recombinant protein expression system in Corynebacterium glutamicum and its application. Front. Microbiol. 2018 doi: 10.3389/fmicb.2018.02523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marugg J.D., Gonzalez C.F., Kunka B.S., Ledeboer A.M., Pucci M.J., Toonen M.Y., Walker S.A., Zoetmulder L.C.M., Vandenbergh P.A. Cloning, expression, and nucleotide sequence of genes involved in production of pediocin PA-1, and bacteriocin from Pediococcus acidilactici PAC1.0. Appl. Environ. Microbiol. 1992;58:2360–2367. doi: 10.1128/AEM.58.8.2360-2367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matano C., Uhde A., Youn J.-W., Maeda T., Clermont L., Marin K., Krämer R., Wendisch V.F., Seibold G.M. Engineering of Corynebacterium glutamicum for growth and l-lysine and lycopene production from N-acetyl-glucosamine. Appl. Microbiol. Biotechnol. 2014;98:5633–5643. doi: 10.1007/s00253-014-5676-9. [DOI] [PubMed] [Google Scholar]
- Meiswinkel T.M., Gopinath V., Lindner S.N., Nampoothiri K.M., Wendisch V.F. Accelerated pentose utilization by Corynebacterium glutamicum for accelerated production of lysine, glutamate, ornithine and putrescine. Microb. Biotechnol. 2013;6:131–140. doi: 10.1111/1751-7915.12001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa-Pereira B., O'Connor P.M., Rea M.C., Cotter P.D., Hill C., Ross R.P. Controlled functional expression of the bacteriocins pediocin PA-1 and bactofencin A in Escherichia coli. Sci. Rep. 2017;7:3069. doi: 10.1038/s41598-017-02868-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa-Pereira B., Rea M.C., Cotter P.D., Hill C., Ross R.P. Heterologous expression of biopreservative bacteriocins with a view to low cost production. Front. Microbiol. 2018 doi: 10.3389/fmicb.2018.01654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel A., Koch-Koerfges A., Krumbach K., Brocker M., Bott M. Anaerobic growth of Corynebacterium glutamicum via mixed-acid fermentation. Appl. Environ. Microbiol. 2015;81:7496–7508. doi: 10.1128/AEM.02413-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills S., Stanton C., Hill C., Ross R.P. New developments and applications of bacteriocins and peptides in foods. Annu. Rev. Food Sci. Technol. 2011;2:299–329. doi: 10.1146/annurev-food-022510-133721. [DOI] [PubMed] [Google Scholar]
- Monk I.R., Gahan C.G.M., Hill C. Tools for functional postgenomic analysis of Listeria monocytogenes. Appl. Environ. Microbiol. 2008;74:3921–3934. doi: 10.1128/AEM.00314-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motlagh A., Bukhtiyarova M., Ray B. Complete nucleotide sequence of pSMB 74, a plasmid encoding the production of pediocin AcH in Pediococcus acidilactici. Lett. Appl. Microbiol. 1994;18:305–312. doi: 10.1111/j.1472-765X.1994.tb00876.x. [DOI] [PubMed] [Google Scholar]
- Okino S., Inui M., Yukawa H. Production of organic acids by Corynebacterium glutamicum under oxygen deprivation. Appl. Microbiol. Biotechnol. 2005;68:475–480. doi: 10.1007/s00253-005-1900-y. [DOI] [PubMed] [Google Scholar]
- Oppegård C., Fimland G., Anonsen J.H., Nissen-Meyer J. The pediocin PA-1 accessory protein ensures correct disulfide bond formation in the antimicrobial peptide pediocin PA-1. Biochemistry. 2015;54:2967–2974. doi: 10.1021/acs.biochem.5b00164. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov K.V., Chi H., Mehmeti I., Holo H., Nes I.F., Diep D.B. Novel group of leaderless multipeptide bacteriocins from gram-positive bacteria. Appl. Environ. Microbiol. 2016;82:5216–5224. doi: 10.1128/AEM.01094-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov K.V., Kranjec C., Thorstensen T., Carlsen H., Diep D.B. Successful development of bacteriocins into therapeutic formulation for treatment of MRSA-skin infection in a murine model. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/aac.00829-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov K.V., Kristiansen P.E., Straume D., Jensen M.S., Aleksandrzak-Piekarczyk T., Nes I.F., Diep D.B. The leaderless bacteriocin enterocin K1 is highly potent against Enterococcus faecium: a study on structure, target spectrum and receptor. Front. Microbiol. 2017;8:774. doi: 10.3389/fmicb.2017.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramnath M., Beukes M., Tamura K., Hastings J.W. Absence of a putative mannose-specific phosphotransferase system enzyme IIAB component in a leucocin a-resistant strain of Listeria monocytogenes, as shown by two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Appl. Environ. Microbiol. 2000;66:3098–3101. doi: 10.1128/AEM.66.7.3098-3101.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ríos Colombo N.S., Chalón M.C., Navarro S.A., Bellomio A. Pediocin-like bacteriocins: new perspectives on mechanism of action and immunity. Curr. Genet. 2018;64:345–351. doi: 10.1007/s00294-017-0757-9. [DOI] [PubMed] [Google Scholar]
- Schwechheimer S.K., Becker J., Peyriga L., Portais J.C., Sauer D., Müller R., Hoff B., Haefner S., Schröder H., Zelder O., Wittmann C. Improved riboflavin production with Ashbya gossypii from vegetable oil based on 13C metabolic network analysis with combined labeling analysis by GC/MS, LC/MS, 1D, and 2D NMR. Metab. Eng. 2018;47:357–373. doi: 10.1016/j.ymben.2018.04.005. [DOI] [PubMed] [Google Scholar]
- Seibold G., Auchter M., Berens S., Kalinowski J., Eikmanns B.J. Utilization of soluble starch by a recombinant Corynebacterium glutamicum strain: growth and lysine production. J. Biotechnol. 2006;124:381–391. doi: 10.1016/j.jbiotec.2005.12.027. [DOI] [PubMed] [Google Scholar]
- Silva C.C.G., Silva S.P.M., Ribeiro S.C. Application of bacteriocins and protective cultures in dairy food preservation. Front. Microbiol. 2018;9:594. doi: 10.3389/fmicb.2018.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosukhowong A., Zendo T., Visessanguan W., Roytrakul S., Pumpuang L., Jaresitthikunchai J., Sonomoto K. Garvieacin Q, a novel class II bacteriocin from lactococcus garvieae BCC 43578. Appl. Environ. Microbiol. 2012;78:1619–1623. doi: 10.1128/AEM.06891-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymoszewska A., Diep D.B., Aleksandrzak-Piekarczyk T. The extracellular loop of Man-PTS subunit IID is responsible for the sensitivity of Lactococcus garvieae to garvicins A, B and C. Sci. Rep. 2018;8:15790. doi: 10.1038/s41598-018-34087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymoszewska A., Diep D.B., Wirtek P., Aleksandrzak-Piekarczyk T. The non-lantibiotic bacteriocin garvicin Q targets man-PTS in a broad spectrum of sensitive bacterial genera. Sci. Rep. 2017;7:8359. doi: 10.1038/s41598-017-09102-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema K., Kok J., Marugg J.D., Toonen M.Y., Ledeboer A.M., Venema G., Chikindas M.L. Functional analysis of the pediocin operon of Pediococcus acidilactici PAC1.0: PedB is the immunity protein and PedD is the precursor processing enzyme. Mol. Microbiol. 1995;17:515–522. doi: 10.1111/j.1365-2958.1995.mmi_17030515.x. [DOI] [PubMed] [Google Scholar]
- Walsh C.J., Guinane C.M., Hill C., Ross R.P., O'Toole P.W., Cotter P.D. In silico identification of bacteriocin gene clusters in the gastrointestinal tract, based on the Human Microbiome Project's reference genome database. BMC Microbiol. 2015;15:183. doi: 10.1186/s12866-015-0515-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann C., Hans M., Heinzle E. In vivo analysis of intracellular amino acid labelings by GC/MS. Anal. Biochem. 2002;307:379–382. doi: 10.1016/S0003-2697(02)00030-1. [DOI] [PubMed] [Google Scholar]
- Wittmann C., Krömer J.O., Kiefer P., Binz T., Heinzle E. Impact of the cold shock phenomenon on quantification of intracellular metabolites in bacteria. Anal. Biochem. 2004;327:135–139. doi: 10.1016/j.ab.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Wittmann C., Weber J., Betiku E., Krömer J., Böhm D., Rinas U. Response of fluxome and metabolome to temperature-induced recombinant protein synthesis in Escherichia coli. J. Biotechnol. 2007;132:375–384. doi: 10.1016/j.jbiotec.2007.07.495. [DOI] [PubMed] [Google Scholar]
- Wolf S., Becker J., Tsuge Y., Kawaguchi H., Kondo A., Marienhagen J., Bott M., Wendisch V.F., Wittmann C. Advances in metabolic engineering of Corynebacterium glutamicum to produce high-value active ingredients for food, feed, human health, and well-being. Essays Biochem. 2021 doi: 10.1042/ebc20200134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2017. WHO | Global Action Plan on Antimicrobial Resistance. [Google Scholar]
- Yang T.H., Bolten C.J., Coppi M.V., Sun J., Heinzle E. Numerical bias estimation for mass spectrometric mass isotopomer analysis. Anal. Biochem. 2009;388:192–203. doi: 10.1016/j.ab.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Yim S.S., An S.J., Choi J.W., Ryu A.J., Jeong K.J. High-level secretory production of recombinant single-chain variable fragment (scFv) in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2014;98:273–284. doi: 10.1007/s00253-013-5315-x. [DOI] [PubMed] [Google Scholar]
- Yim S.S., Choi J.W., Lee R.J., Lee Y.J., Lee S.H., Kim S.Y., Jeong K.J. Development of a new platform for secretory production of recombinant proteins in Corynebacterium glutamicum. Biotechnol. Bioeng. 2016;113:163–172. doi: 10.1002/bit.25692. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.