Abstract
The molecular oscillator that keeps circadian time is generated by a negative feedback loop. Nuclear entry of circadian regulatory proteins that inhibit transcription from E-box-containing promoters appears to be a critical component of this loop in both Drosophila and mammals. The Drosophila double-time gene product, a casein kinase I ɛ (CKIɛ) homolog, has been reported to interact with dPER and regulate circadian cycle length. We find that mammalian CKIɛ binds to and phosphorylates the murine circadian regulator mPER1. Unlike both dPER and mPER2, mPER1 expressed alone in HEK 293 cells is predominantly a nuclear protein. Two distinct mechanisms appear to retard mPER1 nuclear entry. First, coexpression of mPER2 leads to mPER1-mPER2 heterodimer formation and cytoplasmic colocalization. Second, coexpression of CKIɛ leads to masking of the mPER1 nuclear localization signal and phosphorylation-dependent cytoplasmic retention of both proteins. CKIɛ may regulate mammalian circadian rhythm by controlling the rate at which mPER1 enters the nucleus.
The circadian rhythm is an intrinsic 24-h cycle that, in species from Neurospora to humans, is generated by an intracellular oscillating negative feedback loop that controls the periodic transcription of both regulatory and output genes. The molecular mechanism generating the circadian rhythm has been the object of intense study (reviewed in references 12, 42, and 57). Genetic investigations in the fruit fly Drosophila melanogaster, augmented by studies of circadian rhythm mutants in mammals, have led to a rapidly evolving understanding of the workings of the metazoan central clock. In Drosophila, a heterodimeric transcription factor composed of CLOCK and CYCLE binds to E-box-containing promoters and drives expression of the negative regulators PERIOD (dPER) and TIMELESS (dTIM). dPER and dTIM accumulate in the cytoplasm until they heterodimerize. Heterodimerization serves to mask their cytoplasmic localization domains, allowing the complex to enter the nucleus (46). Nuclear dPER-dTIM heterodimers repress the activity of the CLOCK/CYCLE transcription factor, thus causing a decrease in dPER and dTIM expression (10).
Although the mammalian circadian clock is composed of proteins homologous to those found in Drosophila, the mechanisms for regulating circadian rhythm in mammals appear to be more complex and in many aspects quite different from those in Drosophila. The increased complexity in the mammalian system is due in part to the expansion of the per gene family. Three mammalian period genes have been cloned; all are rhythmically expressed in the anatomic location of the central clock, the suprachiasmatic nucleus (SCN), as well as in diverse peripheral tissues (including heart, liver, and muscle) and cultured fibroblasts, with peak levels of transcripts detected during the circadian day in the mouse (1, 2, 47, 55, 62). The three mper genes differ in their transcriptional regulation. Several reports suggest that mper1 is expressed 4 to 8 h before mper2 and mper3 (1, 25, 53).
Regulated nuclear entry of the PER proteins is a common element in many but not all (50) of the observed circadian regulators (12, 42, 57). In the mouse, periodic nuclear accumulation of mPER1 protein has been demonstrated in the mouse SCN, peaking 4 to 6 h after mper1 mRNA expression (19). How and if mPER nuclear entry is regulated is less clear. In the murine system, heterodimerization of mPER proteins with mTIM has been controversial, being found by some but not all observers (48, 54, 61). However, each of the mPER proteins can homodimerize with itself and heterodimerize with other mPER proteins. Forced expression of mPER proteins alone can partially repress CLOCK/BMAL1-activated transcription (BMAL1 is the mammalian homolog of CYCLE) in the absence of coexpressed mTIM (25, 48). Recently, Kume et al. (31) reported that coexpression of cryptochrome proteins mCRY1 and mCRY2 facilitated the nuclear entry of mPER proteins and fully repressed transcription from CLOCK/BMAL1-driven promoters. Thus, PER nuclear entry seems to be periodic and regulated in mammals as well as in Drosophila.
Phosphorylation of the components of the circadian clock has been postulated to regulate the duration of the cycle. Treatment of the dinoflagellate Gonyaulax polyedra with either serine/threonine phosphatase or kinase inhibitors alters its circadian rhythm (6, 7). The frequency gene product, a negative regulator of the Neurospora circadian clock, is rhythmically phosphorylated (12), and its phosphorylation regulates both its stability and period duration (34). dCLOCK, dPER, and dTIM are phosphoproteins, and the level of dPER and dTIM phosphorylation increases steadily from the time of their synthesis until their degradation at dawn (13, 32). The first genetic evidence that a specific protein kinase regulates the Drosophila circadian rhythm was the discovery of the novel gene double-time (dbt), encoding a protein serine/threonine kinase (27, 41). dbt is coexpressed with per and tim in the fly brain lateral neurons that regulate circadian rhythm. Different missense alleles of dbt cause marked lengthening or shortening of the circadian period, while homozygosity for the null allele causes pupal lethality (27).
Examination of flies with mutations in the dbt gene led Kloss and coworkers (27) to conclude that the DBT kinase phosphorylated and regulated dPER. Drosophila larvae homozygous for the dbt-null allele manifest several distinct phenotypes. First, they accumulate high levels of dPER but not dTIM, suggesting a role for phosphorylation in the degradation of dPER. Second, the dPER that accumulates is hypophosphorylated, indicating a major role for DBT in the phosphorylation of dPER. In a final indication that DBT directly regulates dPER, DBT binds to an amino-terminal fragment of dPER (27).
Drosophila DBT is most similar in sequence (86% identical) in its kinase domain to the kinase domains of mammalian casein kinase I ɛ and δ (CKIɛ and CKIδ). The CKI gene family encodes a number of widely expressed kinases that localize to membranes, cytoplasm, and nucleus; and various members of the CKI family have been identified in plants, fungi, and mammals (14, 18, 44, 45). CKIɛ and CKIδ belong to a branch of the family that includes the yeast kinases HRR25 and Hhp1 and Hhp2, implicated in the response to DNA damage (11, 14, 21). Mammalian CKIδ and CKIɛ have closely related 123-amino-acid carboxy-terminal domains that can autoregulate kinase activity in a phosphorylation-dependent manner (5, 16, 17, 44). However, the carboxy-terminal domains of DBT and CKIɛ are unrelated.
Accumulating evidence suggests CKI family members can regulate the intracellular localization of specific substrates. For example, mammalian CKIα (71% identical to DBT over the kinase domain) binds to, phosphorylates, and inhibits the nuclear import of the transcription factor NF-AT4 (60). In Drosophila, a CKIα homolog shuttles from the cytoplasm into the larval nuclei in response to gamma irradiation (49). Finally, one of the few identified substrates of HRR25 is the yeast transcriptional regulator SWI6, a protein whose cytoplasmic retention is dependent on phosphorylation (20, 52).
Given the differences between the Drosophila and mammalian PER and TIM proteins and the higher level of complexity in the regulation and interactions of the mPER proteins, the interaction between CKI and the mammalian mPER1 protein was investigated. We find that specific and closely related isoforms of CKI bind to and phosphorylate mPER1 both in vitro and in vivo. Unexpectedly, overexpressed mPER1 was found to accumulate in the nuclei of transfected HEK 293 cells. Two distinct mechanisms appear to be capable of regulating mPER1 nuclear entry. First, coexpressed mPER2 prevents mPER1 nuclear accumulation. Second, CKIɛ or CKI∂ coexpression blocks mPER1 nuclear accumulation in a kinase-dependent manner, by masking its nuclear localization signal (NLS). These results suggest that a critical function of both mPER2 and CKI in circadian rhythm is to control the nuclear entry of mPER1.
MATERIALS AND METHODS
Plasmids and protein purification.
Cloning, expression, and purification of CKIɛ (GenBank accession no. L37043), CKIɛΔC320, and CKIɛ(K38A) have been described elsewhere (5, 14, 16). For in vitro kinase assays, CKIɛ was purified as described previously (5, 14). To generate Myc-tagged mPER1, cDNA encoding mPER1 (GenBank accession no. AB002108; a generous gift from M. Tei) was excised with EcoRI and SalI from pHSG396 and cloned into the EcoRI and XhoI sites in pCS2+MT. The resultant plasmid utilizes a cytomegalovirus promoter and encodes a polypeptide of 1,378 amino acids, with 86 amino-terminal amino acids from the six copies of the c-Myc epitope, and a calculated molecular mass of 146,492 Da. The Myc-tagged amino-terminal fragment of mPER1 was generated by excising the EcoRV-to-XbaI fragment of pCS2+MT-mPER1 and religating the blunted ends. Other amino-terminal fragments of mPER1, the NLS mutant constructs, and ST6A were made in pCS2+MT-mPER1 (above) by changing the indicated codons to either stop (truncations) or alanine with the QuikChange kit. The carboxyl-terminal fragments were PCR cloned into pCS2+MT between EcoRI and SalI. mPER2 (the generous gift from H. Okamura) was amplified by PCR with addition of NcoI and SalI sites and cloned into NcoI-XhoI sites in pCS2+MT. To make FLAG-mPER2, mPER2 was amplified using primers with engineered BglII and SalI sites and cloned into pFLAG-CMV2 cut with BglII and SalI. The double yellow fluorescent protein (YFP) fusions with mPER1 fragments were created by amplifying the indicated regions of mPER1 with addition of EcoRI and SalI sites. The PCR products were cloned into p2X-YFP (a generous gift from M. Morgan) carboxy terminal to the second YFP between EcoRI and SalI. All constructs were verified by sequence analysis.
Cell line maintenance, transfections, and immunofluorescence.
HEK 293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum in 5% CO2. Cells were transfected when 70 to 80% confluent in six-well dishes with 2 to 4 μg of total plasmid DNA, using Lipofectamine PLUS reagent (Life Technologies, Inc.) according to the manufacturer's directions. For immunofluorescence, cells were trypsinized and replated on glass coverslips in 35-mm-diameter dishes 24 h after transfection. After an additional 24 h, the cells were washed twice with phosphate-buffered saline (PBS) and then fixed with 4% paraformaldehyde for 30 min. After two washes with PBS, cells were permeabilized with 0.3% Triton X-100 in PBS for 10 min, washed twice with 0.1% Tween 20 in PBS, and then blocked in 1% bovine serum albumin, 10% normal goat serum, 0.1% thimerosal, and 0.1% Tween 20 in PBS (blocking solution) for 30 min at room temperature.
Anti-hemagglutinin epitope (anti-HA; 12CA5) and anti-Myc (9E10) monoclonal antibodies (MAbs) were directly conjugated to Alexa 594 (red) and Alexa 488 (green) dyes, respectively, as instructed by the manufacturer (Molecular Probes). For Fig. 3C, 9E10 and anti-FLAG (M2) MAbs were conjugated to Alexa 350 (blue) and Alexa 488, respectively. Primary MAbs were diluted into blocking solution and incubated with the cells 2 h to overnight with gentle rocking. The cells were then washed three times with 0.1% Tween 20 in PBS, and the nuclei were counterstained with either Hoechst 33258 (1 μg/ml) or 10 nM ToPro3 (Molecular Probes). The goat anti-rabbit secondary antibody coupled to Alexa 594 (Molecular Probes) was applied after the 0.1% Tween 20 washes for 2 h to overnight, followed by nuclear staining with Hoechst as above. Images were captured using the software package mFISH (Vysis, Inc.) at an initial magnification of ×60, using an Olympus BX50 fluorescence microscope equipped with a cooled charge-coupled device camera (Photometrics Ltd.) and appropriate filters sets (Chroma Technology Corp.). Images were assembled using Photoshop 3.04 (Adobe) and Canvas 6.0 (Deneba Software).
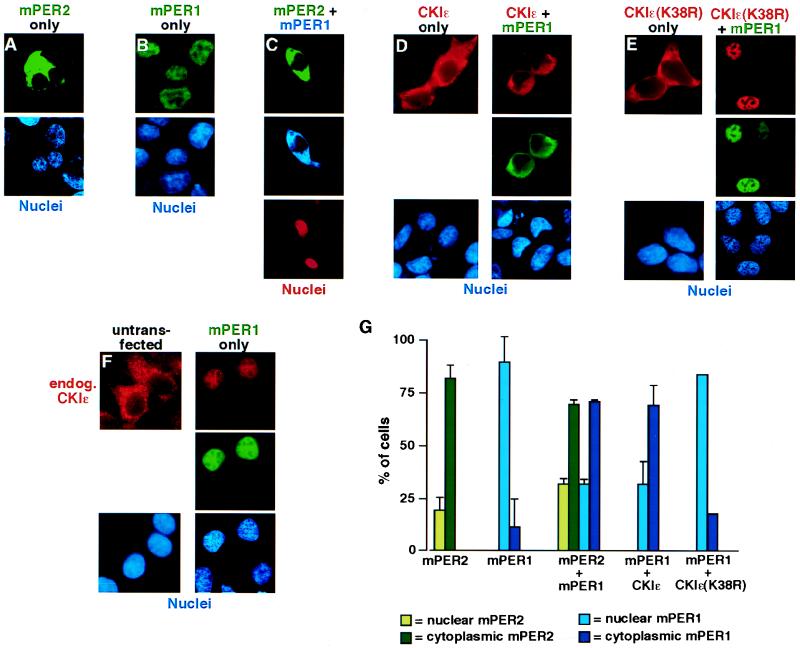
FIG. 3.
mPER2 and CKIɛ regulate mPER1 subcellular localization. (A to F) Representative micrographs illustrating subcellular localization of mPER2, mPER1, and CKIɛ. HEK 293 cells were transiently transfected with constructs encoding FLAG-mPER2 (A). Myc-mPER1 (B), FLAG-mPER2 and Myc-mPER1 (C), HA-CKIɛ without (D, left) or with (D, right) Myc-mPER1, HA-CKIɛ(K38R) without (E, left) or with (E, right) Myc-mPER1, and Myc-mPER1 (F). Forty-eight hours after transfection, the cells were fixed and epitope-tagged proteins were visualized by staining with Alexa 488 (green)-conjugated anti-FLAG (M2) (A and C), Alexa 350 (blue)-conjugated anti-Myc (9E10) (C), Alexa 488 (green)-conjugated MAb 9E10 (B, D, E, and F), Alexa 594 (red)-conjugated anti-HA MAb 12CA5 (D and E), and anti-CKIɛ MAb followed by an Alexa 594-conjugated goat anti-mouse secondary (F). Nuclei were visualized with Hoechst stain (A, B, D, E, and F) or ToPro3 stain (red; C). (G) Quantitation of the experiments illustrated above. Each bar is the result of at least two independent experiments (±standard deviation) in which 40 to 100 cells were counted. All immunofluorescence experiments were done at least twice, but where error bars are omitted experiments were quantitated only once.
Immunoprecipitations and kinase assays.
Coupled in vitro transcription-translation reactions (TnT; Promega) were carried out according to the manufacturer's instructions. Immunoprecipitations from TnT reactions were performed after preclearing with protein A-agarose beads (Gibco-BRL). Precleared reactions were incubated with 1.2 to 2.4 μg of anti-Myc MAb 9E10 (Santa Cruz Biotechnology, Inc.) and 30 to 60 μl of protein A-agarose beads. Following incubation and low-speed centrifugation, the beads were washed five times in incubation buffer (100 mM KCl, 25 mM HEPES [pH 7.5], 12.5 mM MgCl2, 100 μM EDTA, 20% glycerol, 0.1% NP-40, 1 mM dithiothreitol [DTT], 1 μg each of leupeptin and pepstatin per ml, 1 mM benzamidine, 0.5 mM phenymethylsulfonyl fluoride). Kinase reactions were performed with CKIɛΔC320 rather than with full-length kinase to avoid the kinase autoinhibition otherwise seen in vitro (5, 16). Kinase reactions were performed in HMB buffer (30 mM HEPES [pH 7.5], 7 mM MgCl2, 100 μg of BSA per ml, 25 μM ATP, 1 mM DTT) and stopped by washing the beads three times with HM buffer (30 mM HEPES [pH 7.5], 7 mM MgCl2).
For the coimmunoprecipitation experiments (Fig. 1C and 6A), 40-μl aliquots of each specific TnT reaction mixture were mixed and incubated for 30 min at 30°C; 120 μl of incubation buffer (see above) was then added to the combined TnT reaction mixtures, and Myc-mPER1 was immunoprecipitated with 600 ng of anti-Myc MAb 9E10 and 15 μl of protein A-agarose beads. The beads were then washed five times with incubation buffer, and the bound proteins were eluted with sodium dodecyl sulfate (SDS) sample buffer.
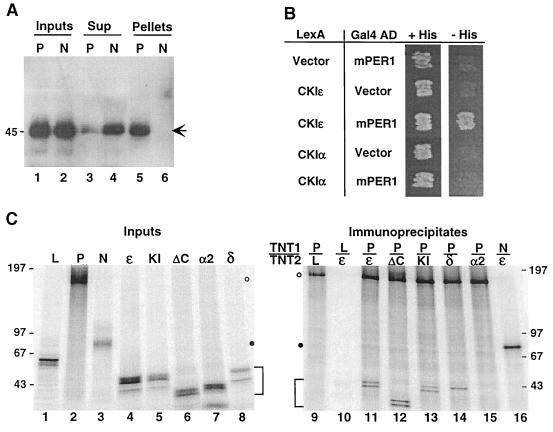
FIG. 1.
CKIɛ binds to mPER1 in vivo and in vitro. (A) Coimmunoprecipitation of mPER1 and endogenous CKIɛ. HEK 293 cells were transiently transfected with plasmids expressing either full-length Myc-mPER1 (P) or the amino-terminal fragment Myc-mPER1(1-485) (N). The PER proteins were immunoprecipitated from cell lysates with anti-Myc MAb 9E10; 20 μg of cell lysate protein (Inputs; lanes 1 and 2), the equivalent of 20 μg of the cell lysate supernatant following clarification and immunoprecipitation (Sup; lanes 3 and 4), and the immunoprecipitate pellet from 50 μg of cell lysate (Pellets; lanes 5 and 6) were analyzed by SDS-PAGE, followed by immunoblotting with anti-CKIɛ antibody UT31 (14). The arrow indicates the position of endogenous CKIɛ. (B) Specificity of the CKIɛ-mPER1 interaction assessed by two-hybrid assay. Yeast cotransformed with plasmids expressing the indicated proteins fused to either LexA or the Gal4 activation domain (AD) were grown on synthetic medium containing histidine (+His) or on medium containing 5 mM 3-aminotriazole and lacking histidine (−His) as previously described (37). Interaction between the indicated proteins was assessed by growth on −His plates. (C) Specificity of the CKIɛ-mPER1 interaction assessed by coimmunoprecipitation in vitro. In vitro-synthesized [35S]methionine-labeled proteins (Inputs; lanes 1 to 8) luciferase (L), Myc-mPER1 (P), truncated Myc-mPER1(1-485) (N), CKIɛ (ɛ), kinase-inactive CKIɛ(K38R) (KI), truncated CKIɛ(ΔC320) (ΔC), CKIα2 (α2), or CKIδ (δ) were mixed together (TNT1/TNT2) as indicated above lanes 9 to 16. Following a 30-min incubation, the protein mixtures were subjected to immunoprecipitation with anti-Myc MAb 9E10 and analyzed by SDS-PAGE (5 to 15% gel) (lanes 9 to 16). One-tenth of each of the in vitro synthesis reactions used for immunoprecipitation was loaded on the input gel (left). Data were collected and analyzed using a Molecular Dynamics PhosphorImager. Open and closed circles mark the positions of full-length and truncated Myc-mPER1, respectively; brackets mark positions of the various CKI proteins. Here and in subsequent figures, positions of the various protein molecular weight markers are indicated to the side of the gel, with the size of each marker expressed in kilodaltons.
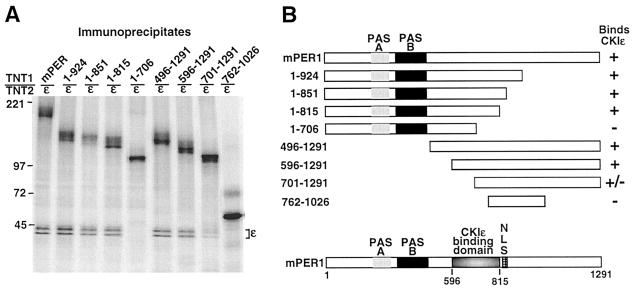
FIG. 6.
(A) Identification of CKIɛ binding site on mPER1. In vitro-synthesized [35S]methionine-labeled Myc-mPER1 (mPER1) and various amino-terminal (1-924, 1-851, 1-815, 1-706) and carboxyl-terminal (496-1291, 596-1291, 701-1291) fragments of mPER1 were mixed with in vitro-synthesized [35S]methionine-labeled full-length CKIɛ. After incubation, mPER1 was immunoprecipitated with the anti-Myc MAb 9E10 and analyzed for the presence of coimmunoprecipitating CKIɛ by SDS-PAGE and PhosphorImager analysis. The data presented are a subset of all the truncations tested; all results were repeated at least three times. (B) Diagrammatic representation of the constructs utilized and the results of the coimmunoprecipitation experiments.
For the mPER immunoprecipitations from transfected cells, HEK 293 cells were lysed in 10 mM HEPES (pH 7.5)–0.1% Triton X-100–150 mM NaCl–2 mM EDTA–2 mM EGTA, and then 200 μg of soluble protein was mixed with 600 ng of anti-Myc MAb 9E10. Immune complexes were removed from the lysate with 20 μl of protein A-agarose. The beads were then washed five times with cell lysis buffer (see above), and bound proteins were eluted with SDS sample buffer.
mPER1 gel shift assays.
Five microliters of a TnT reaction mixture containing [35S]methionine (Amersham Pharmacia Biotech)-labeled proteins was incubated with 50 ng of CKIɛΔC320 for the indicated length of time at 30°C in a final volume of 20 μl in a buffer containing 25 mM Tris-HCl (pH 7.5), 15% glycerol, 20 mM NaF, 0.5 mM Na3VO4, 2 mM DTT, and 150 μM ATP.
[35S]methionine pulse-chase.
Twenty-four hours after transfection, HEK 293 cells were incubated in methionine-free DMEM in the presence of Trans35S-label (400 μCi/ml; ICN Biomedicals) for 2 h. Following the pulse, fresh DMEM containing unlabeled methionine was added, and the cells were incubated for the indicated lengths of time. After the chase period, cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS), and then 120 μg of soluble protein was mixed with 400 ng of anti-Myc MAb 9E10. Immune complexes were removed from the lysate with 20 μl of protein A-agarose. The beads were then washed five times with radioimmunoprecipitation assay buffer, and bound proteins eluted with SDS sample buffer.
RESULTS
In vivo and in vitro interaction of CKIɛ and mammalian PER proteins.
To test whether endogenous mammalian CKIɛ interacted with mammalian PER proteins, Myc-mPER1 and an amino-terminal 485-amino-acid fragment of mPER1 [up to and including the PAS domains; Myc-mPER1(1-485)] were expressed in HEK 293 cells. After lysis, extracts were incubated with anti-Myc MAb and protein A-Sepharose beads, and both the immunoprecipitates and the supernatants were probed for the presence of CKIɛ. There was substantial coimmunoprecipitation of endogenous CKIɛ when full-length mPER1 was immunoprecipitated (Fig. 1A, lane 5), with depletion of CKIɛ from the supernatant (lane 3). We have found no other putative CKIɛ substrates that coimmunoprecipitate endogenous CKIɛ nearly as well (Z.-H. Gao and D. M. Virshup, unpublished data). When the amino-terminal region of mPER1 was expressed and then immunoprecipitated, there was no detectable coimmunoprecipitation of endogenous CKIɛ (lane 6). In control experiments, there was quantitative precipitation of both the Myc-mPER1 and the amino-terminal Myc-mPER1 (data not shown). Thus, mPER1 is capable of binding tightly to endogenous CKIɛ, and the amino terminus of mPER1 is not sufficient for this interaction. This is in contrast to the results seen with Drosophila PER and DBT, where a GST–amino-terminal region of dPER fusion was sufficient to bind to DBT (27).
The specificity of the interaction between mPER1 and CKIɛ was then assessed in a yeast two-hybrid assay. CKIɛ differs from CKIα2 both in the kinase domain, where the proteins are 77% identical, and in their carboxy-terminal regulatory domains, which differ in size (39 versus 123 amino acids) and sequence (no significant identity) (14). LexA-CKIɛ and LexA-CKIα2 fusions were tested for interaction with mPER1-Gal4 activation domain and control constructs. CKIɛ but not CKIα2 interacted specifically with mPER1 (Fig. 1B).
To determine if the carboxy-terminal regulatory domain of CKIɛ was required for the mPER1-CKI interaction, in vitro-synthesized [35S]methionine-labeled full-length mPER1 or an amino-terminal mPER1 fragment was incubated with various forms of CKI (Fig. 1C, Inputs). The mPER1 proteins were then immunoprecipitated, and their ability to coimmunoprecipitate CKI was determined. Full-length mPER1 coimmunoprecipitated with full-length CKIɛ (Fig. 1C, lane 11), and that interaction was not disturbed by removal of the CKIɛ carboxy-terminal regulatory domain (lane 12) or loss of kinase activity (lane 13). mPER1 also coimmunoprecipitated with the closely related CKIδ (lane 14) (97% identical to CKIɛ over the kinase domain) but again did not interact with the more distantly related CKIα2 (lane 15). CKIɛ again did not interact with the amino-terminal domain of mPER1 (lane 16). We conclude that it is the kinase domain of CKIɛ and CKIδ (each 86% identical to the DBT kinase domain) that interacts with mPER1.
The mPER1 protein is phosphorylated by CKIɛ.
To determine whether mPER1 was indeed a substrate for CKIɛ, three separate analyses were undertaken. First, kinase assays were performed with immunoprecipitated in vitro-synthesized mPER1 (full length or amino-terminal domain) as the substrate. Full-length mPER1 was readily phosphorylated by recombinant CKIɛΔ320 (lacking the carboxyl-terminal autoinhibitory domain [5, 44]) (Fig. 2A, lane 4). The amino-terminal fragment of mPER1, although expressed at much higher levels (compare lanes 7 and 8), was substantially less phosphorylated by CKIɛΔ320 (compare lanes 2 and 4). The phosphorylation of mPER1 was partially reversed by incubation of the labeled protein with protein phosphatase 2A (PP2A) (mPER1 radioactivity in lane 5 is 59% of that in lane 4). The PP2A-induced decrease in signal was due to dephosphorylation rather than proteolysis of mPER1, as the effect of PP2A was blocked by the inclusion of okadaic acid (lane 6). Chymotryptic phosphopeptide maps of in vitro-phosphorylated mPER1 showed several phosphopeptide spots, suggesting that CKIɛ phosphorylates mPER1 on multiple sites (E. Vielhaber, unpublished data).
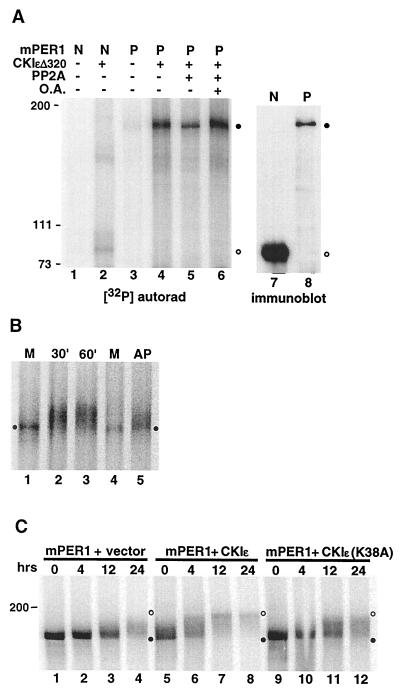
FIG. 2.
Phosphorylation of mPER1 by CKIɛ in vitro and in vivo. (A) CKIɛ phosphorylates immunoprecipitated mPER1. In vitro-synthesized unlabeled mPER1 (P) and an amino-terminal fragment (amino acids 1 to 485; N) were each immunoprecipitated from a single in vitro transcription-translation reaction (200 and 100 μl, respectively) with anti-Myc MAb 9E10, and the immunoprecipitation reactions were divided equally between the various experiments. The immunoprecipitated proteins were then incubated with (lanes 2 and 4 to 6) or without (lanes 1 and 3) 100 ng recombinant CKIɛΔ320 and 25 μM [γ-32P]ATP for 30 min at 37°C in a 20-μl volume. Following the kinase reaction, the samples in lanes 5 and 6 were washed and then incubated with 100 ng of catalytic subunit of PP2A in the absence (lane 5) or presence (lane 6) of 500 nM okadaic acid (O.A.). Phosphorylation was analyzed by SDS-PAGE followed by PhosphorImager analysis. Closed circles indicate the mobility of full-length mPER1; open circles indicate the mobility of the amino-terminal mPER1 fragment. The relative amounts of full-length mPER1 (P; lane 8) and the amino-terminal fragment (N; lane 7) were determined in parallel by immunoblotting with the anti-Myc MAb 9E10. (B) Mobility shift of phosphorylated mPER1 in vitro as assessed by kinase assay. In vitro-synthesized [35S]methionine-labeled mPER1 (from ∼1 mg of reticulocyte lysate) was incubated with 50 ng of CKIɛΔ320 (lanes 2, 3, and 5) for the indicated times, or without added kinase for 60 min (M; lanes 1 and 4). After the kinase reaction, the sample in lane 5 was incubated with 40 U of calf intestinal alkaline phosphatase (New England Biolabs) for an additional 30 min. The samples were then analyzed by SDS-PAGE and phosphorimaging. The closed circles indicate the mobility of unphosphorylated mPER1. (C) Mobility shift of mPER1 in vivo as assessed by pulse-chase experiment. HEK 293 cells expressing Myc-tagged mPER1 only (lanes 1 to 4), mPER1 and full-length active CKIɛ (lanes 5 to 8), or mPER1 and kinase-inactive CKIɛ (lanes 9 to 12) were pulse-labeled with [35S]methionine. At the indicated time points following addition of medium containing unlabeled methionine, cells were lysed and Myc-mPER1 was immunoprecipitated with anti-Myc MAb 9E10. Samples were all run on a single SDS-polyacrylamide gel, and the entire gel was visualized with a PhosphorImager at a constant intensity setting. Closed and open circles indicate the positions of unphosphorylated and phosphorylated mPER1, respectively.
Many proteins have an altered electrophoretic mobility after phosphorylation. To determine if this was true of mPER1, the protein was synthesized in vitro in the presence of [35S]methionine, and then purified CKIɛΔ320 (final concentration, approximately 30 nM) was added to the reticulocyte lysate. Control experiments demonstrated that reticulocyte lysates have low endogenous CKI activity, as assessed by their ability to phosphorylate a CKI substrate peptide (15). mPER1 electrophoretic mobility was then assessed by SDS-polyacrylamide gel electrophoresis (PAGE) on an 8% gel. Incubation of mPER1 in the reticulocyte lysate alone did not cause a significant shift in [35S]mPER1 migration (Fig. 2B, lane 1); however, when CKIɛΔ320 was added, there was a marked decrease in mPER1 mobility (lanes 2 and 3). The change in mobility was due to phosphorylation, since it was partially reversed by the addition of alkaline phosphatase to the lysate (lane 5). The failure of both PP2A and alkaline phosphatase to fully reverse the CKIɛ-dependent phosphorylation of mPER1 suggests that there are multiple phosphorylation sites on mPER1 and that only a subset are substrates for either phosphatase. The specificity of the kinase-substrate interaction is suggested by the fact that ∼30 nM CKIɛ was able to phosphorylate [35S]methionine-mPER1 in the presence of 1 mg of total reticulocyte lysate. The stoichiometry of mPER1 phosphorylation was not measured directly; however, virtually all of the mPER1 protein had a decreased electrophoretic mobility after addition of CKIɛ, which suggests that each mPER1 molecule had at least one molecule of phosphate added. This result is consistent with the findings in Drosophila, as dPER also has a significant shift in its electrophoretic mobility in SDS-PAGE upon phosphorylation. That shift disappears in dbt-null larvae, suggesting that phosphorylation of dPER by DBT leads to its retarded electrophoretic mobility (13, 41).
The above experiments demonstrate that CKIɛ can phosphorylate mPER1 in vitro. To determine if CKIɛ expression also caused a change in mPER1 phosphorylation in vivo, HEK 293 cells were transfected with vectors expressing Myc-tagged mPER1, along with either active or inactive full-length CKIɛ or an empty control vector (Fig. 2C). Twenty-four hours after transfection, the cells were pulse-labeled with [35S]methionine and mPER1 was immunoprecipitated from lysates prepared at various time points as indicated. Coexpression of active CKIɛ (Fig. 2C, lanes 5 to 8) led to both a more rapid decrease in the electrophoretic mobility of mPER1 and a greater distance of the mobility shift of mPER1 compared to cells expressing mPER1 alone (lanes 1 to 4) or coexpressing a kinase-inactive form of CKIɛ (lanes 9 to 12). The fact that overexpression of inactive CKIɛ did not abolish the time-dependent mobility shift of mPER1 suggests that cellular kinases other than CKI also phosphorylate mPER1. Preliminary phosphopeptide mapping in fact shows that CKIɛ phosphorylates less than one-third of the phosphopeptides present in overexpressed mPER1 (data not shown). The half-life of mPER1 coexpressed with CKIɛ in HEK 293 cells was somewhat shortened, from 12 to 9 h (mean of three experiments [data not shown]). This is in contrast to Drosophila, where it has been proposed that phosphorylation of dPER by DBT leads to accelerated degradation of the protein (27).
Regulation of mPER1 localization.
In Drosophila, the dPER protein when expressed alone is retained in the cytoplasm, and heterodimerization with the dTIM protein is required for the complex to enter the nucleus (8, 46). This inherent delay in dPER and dTIM nuclear entry and subsequent feedback inhibition of their own transcription is postulated to provide the lag required for stable circadian oscillations (10, 57). However, the mammalian per genes when expressed individually partially inhibit CLOCK/BMAL1-dependent transcription in the absence of mTIM (25). It was therefore of interest to determine where in the cell individually expressed mPER proteins were located. When mPER2 was expressed in HEK 293 cells, the protein was predominantly localized in the cytoplasm (Fig. 3A and G), similar to the behavior of dPER. However, when mPER1 was expressed, it was localized predominantly in the nuclei of transfected cells (Fig. 3B, F, and G). The unregulated and rapid nuclear entry of mPER1 would be predicted to lead to immediate rather than delayed negative feedback on circadian oscillations, and so we speculated that additional mechanisms might exist to retard mPER1 nuclear entry. mPER1 has been reported to heterodimerize with mPER2 (61), and we confirmed in coimmunoprecipitation experiments that mPER1 could both homodimerize and heterodimerize with mPER2, while no mPER1-hTIM interactions were detected (data not shown). When mPER1 was coexpressed with mPER2, both proteins localized to the cytoplasm (Fig. 3C and G). Thus, one mechanism to regulate mPER1 nuclear entry is heterodimerization with mPER2. However, since mper2 (and mper3) mRNAs and presumably proteins have in some studies been found to be expressed several hours after mPER1 in the SCN (1, 25, 53), this interaction might not prevent the first mPER1 molecules synthesized from entering the nucleus.
One emerging function of CKI family members is the regulation of nuclear entry of substrate proteins. To determine whether CKIɛ regulates the nuclear entry of mPER1, the proteins were coexpressed. Coexpression of mPER1 with active full-length CKIɛ led to accumulation of mPER1 in the cytoplasm and colocalization with overexpressed CKIɛ, which is normally localized to the cytoplasm when overexpressed alone (Fig. 3D and G). Identical results were seen with coexpression of CKIδ (data not shown). Coexpression of hTIM had no effect on mPER1 localization, both without and with coexpressed CKIɛ (data not shown). CKIɛ-mediated cytoplasmic retention of mPER1 was due to phosphorylation rather than just binding, since coexpression of mPER1 and kinase-inactive CKIɛ(K38R) failed to retain mPER1 in the cytoplasm (Fig. 3E). Furthermore, a stable interaction between CKIɛ and mPER1 is strongly suggested by relocalization of kinase-inactive CKIɛ(K38R) from the cytoplasm in the absence of mPER1 to the nucleus in the presence of mPER1 (Fig. 3E). Interestingly, endogenous CKIɛ also relocalized to the nucleus when mPER1 was overexpressed alone (Fig. 3F). The finding that overexpressed mPER1 relocalizes endogenous CKIɛ to the nucleus, while co-overexpression of mPER1 and CKIɛ leads to cytoplasmic localization of both proteins, suggests the ratio of the two proteins is important in determining subcellular localization (see discussion). Extending the results seen in the in vitro binding studies (Fig. 1C, lane 12), CKIɛ kinase lacking the carboxy-terminal regulatory domain was also able to cause cytoplasmic retention of mPER1 (data not shown). CKIɛ overexpression did not block the nuclear import of all substrate proteins, since we found that it had no effect on the nuclear localization of coexpressed p53 (28) (data not shown). The cytoplasmic retention of mPER1 caused by CKIɛ was not blocked by the addition of the nuclear export inhibitor leptomycin B (30) (data not shown), consistent with the hypothesis that phosphorylation of mPER1 leads to decreased nuclear import rather than increased nuclear export.
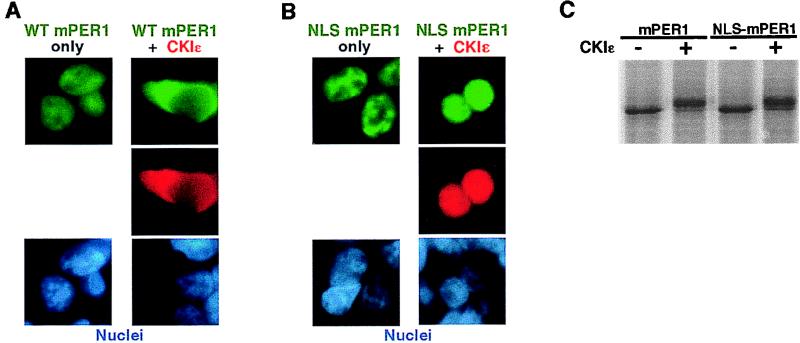
We then examined how coexpression of CKIɛ led to cytoplasmic accumulation of mPER1. Phosphorylation could prevent the nuclear import of mPER1 by masking an NLS. Such a mechanism has been described, for example, in the block to nuclear import of phosphorylated Pho4 in yeast (26). Alternatively, phosphorylation could strengthen binding of mPER1 to a cytoplasmic anchoring protein, as occurs with the Xenopus transcription factor Xnf7 (33, 51). In the case of Xnf7, addition of an exogenous NLS is unable to overcome the cytoplasmic binding, and NLS-Xnf7 remains in the cytoplasm. To discriminate between these possibilities, mPER1 with an simian virus 40 (SV40) large-T-antigen NLS added to the amino terminus (designated NLS-mPER1) was expressed in HEK 293 cells in the absence or presence of added CKIɛ (Fig. 4). As was observed for overexpressed wild-type mPER1 alone (Fig. 4A, left), NLS-mPER1 expressed alone was also nuclear (Fig. 4B, left). When CKIɛ was coexpressed with NLS-mPER1, both proteins accumulated in the nucleus (Fig. 4B, right), in contrast to the CKIɛ-mediated cytoplasmic accumulation of wild-type mPER1 (Fig. 4A, right). The ability of the added NLS to overcome the CKIɛ-induced cytoplasmic localization did not appear to be due to interference with phosphorylation, since both Myc-mPER1 and NLS-Myc-mPER1 had identical shifts in electrophoretic mobility upon incubation with CKIɛ (Fig. 4C). These results suggest that phosphorylation normally serves to mask an intrinsic NLS in mPER1, rather than promoting binding of mPER1 to a cytoplasmic anchoring protein. Additionally, the fact that coexpressed CKIɛ relocates to the nucleus with NLS-mPER1 strongly implies the two proteins are tightly associated in vivo.
FIG. 4.
A heterologous NLS overrides the CKIɛ-dependent cytoplasmic localization of mPER1. (A and B) HEK 293 cells were transiently cotransfected with constructs encoding either 4HA-CKIɛ or empty vector and Myc-mPER1 (A) or NLS-mPER1 (B). Forty-eight hours posttransfection, the epitope-tagged proteins were visualized with Alexa 488 (green)-conjugated anti-Myc MAb 9E10 and Alexa 594 (red)-conjugated anti-HA MAb 12CA5, and the nuclei were visualized with Hoechst staining. WT, wild type. (C) NLS-mPER1 is still a substrate for CKIɛ. In vitro-translated [35S]methionine NLS-mPER1 was incubated without or with added CKI. The addition of the amino-terminal NLS did not interfere with the kinase-dependent electrophoretic mobility shift.
Identification of an NLS in mPER1.
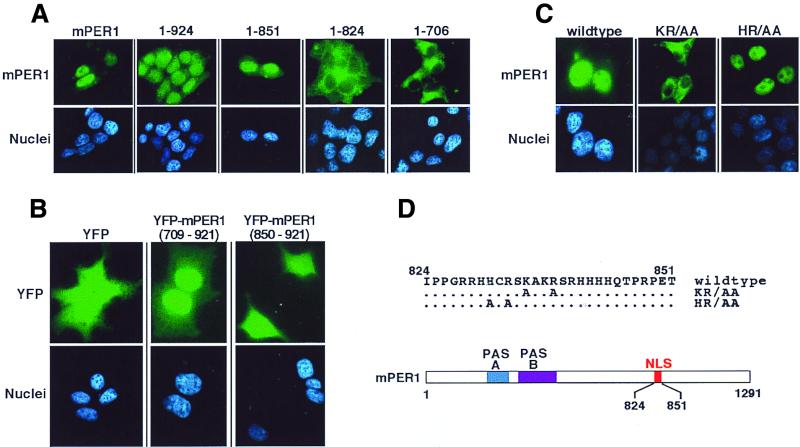
The nuclear accumulation of overexpressed mPER1 suggests it either contains an NLS or binds to a protein that contains an NLS. Visual inspection and computer-aided motif prediction programs found no obvious NLS in the mPER1 sequence. To functionally map the domain required for mPER1 nuclear accumulation, we therefore constructed a series of vectors expressing carboxy-terminal truncated forms of mPER1 and examined their intracellular localization in HEK 293 cells (Fig. 5A). A region of mPER1 from amino acids 824 to 851 appeared important for nuclear localization, since mPER1 containing only amino acids 1 to 851 was nuclear, while deletion of an additional 27 amino acids [mPER1(1-824)] resulted in a cytoplasmic protein. Suggesting this region was sufficient to direct nuclear import, a YFP-mPER1(709-921) construct localized predominantly to nuclei, while YFP alone and YFP-mPER1(850-921) constructs were diffusely distributed throughout the cell (Fig. 5B). Inspection of the sequence between residues 824 and 851 identified a highly basic region between amino acids 828 and 838 that might function as an NLS (Fig. 5D). Mutation of two basic residues K835 and R838 in this region to alanine abolished full-length mPER1 nuclear accumulation (Fig. 5C). Notably, mutation of two adjacent residues H831 and R833 to alanine had no effect on the nuclear import of mPER1. Interestingly, the homologous regions of mPER2 and mPER3 have predicted NLS function (mPER2 has a bipartite-type NLS, while mPER3 has an SV40 large-T-antigen-type NLS), although we found overexpressed mPER2 predominantly in the cytoplasm in HEK 293 cells.
FIG. 5.
Identification of the mPER1 NLS. (A) Progressive truncation of mPER1 reveals a potential NLS. HEK 293 cells were transiently transfected with constructs encoding either full-length Myc-mPER1(mPER1) or one of several carboxyl-terminal truncations of Myc-mPER1 (Fig. 6B). Forty-eight hours posttransfection, the proteins were visualized with Alexa 488 (green)-conjugated anti-Myc MAb 9E10 and the nuclei were visualized with Hoechst staining. Representative micrographs are shown. Quantitation of the nuclear localization of mPER1 and constructs encoding mPER1(1-924) and mPER1(1-851) is shown in Fig. 7. The cytoplasmic localization of mPER1(1-824) and mPER(1-706) was seen in >95% of transfected cells. (B) mPER1 amino acids 709 to 921 are sufficient to direct nuclear localization. HEK 293 cells were transiently transfected with constructs encoding double YFP, either alone or fused to mPER1(709-921) or mPER1(850-921). Double YFP alone and YFP-mPER1(850-921) were diffusely distributed in >95% of transfected cells, while YFP-mPER1(709-921) concentrated in the nuclei of >95% of transfected cells. (C and D) Point mutations identify essential residues in mPER1 NLS. HEK 293 cells were transiently transfected with constructs encoding wild-type or double point mutant K835A/R838A (KR/AA) or H831A/R833A (HR/AA) Myc-mPER1. After transfection, the epitope-tagged mPER1 was visualized as for panel A. A related NLS mutant, K837E/R838D-mPER1, was also cytoplasmic (data not shown). (D) Cartoon demonstrating location of NLS in mPER1 and mutations introduced to create KR/AA and HR/AA Myc-mPER1.
Identification of the CKIɛ binding site and an NLS-masking domain.
Having identified a functional NLS in mPER1, we speculated that CKIɛ might cause cytoplasmic retention of mPER1 by binding to and phosphorylating domains adjacent to the NLS. To determine the specific region of mPER1 required for CKIɛ binding, various fragments of mPER1 were synthesized in in vitro transcription-translation reactions, and the ability of the fragments to bind to CKIɛ was assessed by coimmunoprecipitation assays. As Fig. 6 illustrates, mPER1 amino acids 596 to 815 are required for CKIɛ binding. This region of mPER1 has areas of similarity to mPER2 and mPER3 but has no detectable similarity to other proteins reported to interact with CKI, including dPER, NF-T4, and the PDZ domain of dishevelled, nor to other sequences in GenBank (40, 60).
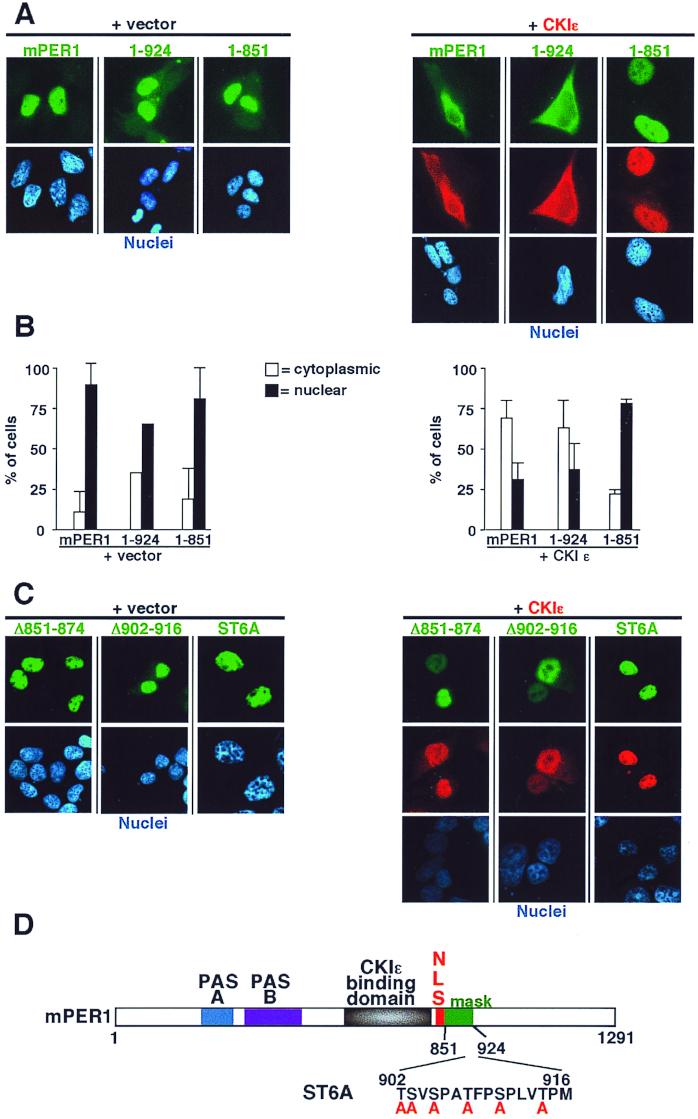
Having established that CKIɛ bound to mPER1 immediately upstream of the NLS, we examined the sequences required for the kinase-dependent cytoplasmic localization of mPER1 (Fig. 7). A construct expressing mPER1 amino acids 1 to 924 localized to the nucleus (Fig. 7A, left; Fig. 7B). Coexpression of CKIɛ led to the cytoplasmic localization of mPER1(1-924), similar to the pattern seen with full-length mPER1 (Fig. 7A, right; Fig. 7B). A slightly shorter construct, mPER1(1-851), alone also localized to the nucleus (Fig. 7A, left; Fig. 7B); and although mPER1(1-851) is competent to bind to CKIɛ (Fig. 6), addition of CKIɛ did not cause mPER1(1-851) to relocate to the cytoplasm (Fig. 7A, right; Fig. 7B). The truncated mPER1(1-851) protein appears to fold properly, since it was able to be transported into the nucleus, and it was able to bind to CKIɛ and relocalize it to the nucleus. These results indicate that mPER1 requires a domain between amino acids 851 and 924 for the CKIɛ-dependent masking of the NLS.
FIG. 7.
CKIɛ-mediated cytoplasmic retention of mPER1 requires a masking domain (amino acids 851-924). (A) Full-length (mPER1) and amino-terminal fragments (1-924 and 1-851) of Myc-mPER1 were expressed in HEK 293 cells without (left) or with (right) HA-CKIɛ. Forty-eight hours posttransfection, the localization of mPER1 and CKIɛ was assessed as described above. The mPER1(1-851) construct contains an NLS and can bind to CKIɛ but failed to relocalize to the cytoplasm. Full-length and mPER1(1-924) were retained in the cytoplasm by coexpression of CKIɛ. Representative micrographs are shown. (B) Quantitation of the experiments shown in panel A. (C) Internal deletions and mutation of potential phosphorylation sites disrupts the function of the masking domain. HEK 293 cells were transfected as above with Myc-mPER1 containing deletion of residues 851 to 874 (Δ851-874) or 902 to 916 (Δ902-916) or with simultaneous mutations of six serine and threonine residues between amino acids 902 to 916 region (ST6A). None of the mutations altered nuclear localization, while all abrogated the ability of CKIɛ to relocalize Myc-mPER1 to the cytoplasm. (D) Cartoon of mPER1 with identification of masking domain and mutant ST6A.
We noted that amino acids 902 to 916 of mPER1 bear similarity to the masking domain of NF-AT4. In NF-AT4, this region is required for the CKIα-mediated cytoplasmic retention of NF-AT4 (60). An mPER1 protein with a deletion of this region was localized to the nucleus and appeared to direct coexpressed CKIɛ to the nucleus as well (Fig. 7C and D). When all six serine and threonine residues in between amino acids 902 and 916 were mutated to alanine (mutant ST6A), once again the mutant full-length protein localized to the nucleus in the absence or presence of active CKIɛ (Fig. 7C and D). These results indicate this domain is required for the phosphorylation-mediated masking of the mPER1 NLS. Interestingly, a deletion of amino acids 851 to 874 of mPER1 also led to a constitutively nuclear mPER1 (Fig. 7C), although mutations of specific serine and threonine residues in the region did not alter the effect of CKIɛ (data not shown). The data suggest that alterations in phosphorylation, spacing, or conformation of this domain may inhibit its ability to mask the mPER1 NLS.
DISCUSSION
Protein phosphorylation has long been suspected of playing an important role in the regulation of circadian rhythm. A number of central clock proteins in insects and fungi are phosphoproteins, and kinase and phosphatase inhibitors alter circadian period in dinoflagellates. It was not until the genetic identification of the Drosophila dbt gene that a specific protein kinase was shown to be a regulator of the central clock. In the present study, CKIɛ, the mammalian homolog of the Drosophila kinase encoded by the dbt gene, was found to bind to and stimulate the phosphorylation of the murine mPER1 protein in vitro and in vivo. The closely related CKI∂ appears to similarly interact with mPER1. An unexpected finding was that mPER1 expressed in HEK 293 cells was predominantly nuclear, while mPER2 was cytoplasmic. Coexpression of mPER1 with mPER2 or with active (but not inactive) CKIɛ led to accumulation of mPER1 in the cytoplasm rather than the nucleus. The CKIɛ-dependent cytoplasmic localization required a domain adjacent to the NLS in mPER1, implying that phosphorylation led to a conformational change that masked the mPER1 NLS. These results suggest that both mPER2 and CKIɛ can regulate mPER1 nuclear entry. The mechanism by which mPER2 keeps mPER1 in the cytoplasm appears to be distinct, and a study of the mPER1-mPER2 interaction is ongoing. Both mechanisms may allow for a delay in the negative regulation of circadian transcriptional activators such as CLOCK and BMAL1.
Stable biologic oscillations can be generated by negative feedback loops with a fixed delay between the generation and the execution of the negative signal. In Drosophila, stable oscillations of circadian rhythm-regulated proteins appear to be determined by the delay between when the dPER and dTIM proteins are synthesized and when they actually enter the nucleus to repress their own transcription. Several groups have demonstrated that temporally regulated nuclear entry of dPER is correlated with the circadian clock in both the brain and eye of Drosophila, and mutations that delay or abolish the nuclear entry of dPER lengthen or abolish circadian cycle (8, 36, 56). Similarly, in mammals temporally regulated nuclear accumulation of mPER1 has been observed in the nuclei of SCN cells (19). Although there is as of yet no direct evidence that the regulated nuclear entry of mPER1 is important for the proper timing of the mammalian circadian clock, there is a clear correlation between the nuclear accumulation of mPER1 protein and the decline in mper1 mRNA levels (19). These observations are consistent with the negative feedback model that predicts that regulated nuclear entry of mPER proteins is important for repressing circadian transcription, thus setting up stable oscillations.
The unhindered accumulation of overexpressed mPER1 in the nucleus of HEK 293 cells was an unexpected result, and it initially was difficult to fit into the current model of a delayed negative feedback loop given that dPER expressed alone is a cytoplasmic protein (12). Unhindered nuclear entry of mPER1 might cause immediate negative feedback on CLOCK/BMAL1 activity, leading to steady rather than oscillatory transcription from CLOCK/BMAL1-driven promoters (25). Our results suggest that at least two mechanisms may regulate the rate of mPER1 nuclear entry. First, newly synthesized mPER1 binds to CKIɛ (and presumably CKI∂) and is retained in the cytoplasm by a kinase-dependent masking of the NLS. If CKIɛ remains tightly associated with mPER1 following masking of the NLS region, the amount of unbound CKIɛ will steadily decrease as the synthesis of mPER1 continues. When mPER1 is present in excess of CKIɛ, there is no free CKIɛ available to phosphorylate the excess mPER1 molecules, and they could start to accumulate in the nucleus. However, at this point the amount of mPER2 may be sufficient to bind free mPER1, so that the mPER1-mPER2 heterodimers could also remain in the cytoplasm.
What mechanism finally allows nuclear entry of mPER protein complexes, leading to inhibition of CLOCK/BMAL1 activity? In Drosophila, heterodimerization of dPER with dTIM allows nuclear import and subsequent inhibition of CLOCK/CYCLE transcription. However, we and others found no effect of mammalian TIM on mPER1 and mPER2 localization (reference 31 and data not shown). In mammals, mCRY1 and mCRY2 have recently been shown to relocalize mPER1 and mPER2 proteins to the nucleus and efficiently repress transcription from E-box-containing promoters, although the mechanism by which mCRY proteins mediate this relocalization is not yet known (31). mCRY proteins may supply an NLS, although our data raise the possibility that mCRY proteins could also allow unmasking of the mPER1 NLS by inhibition of CKIɛ or recruitment of a specific phosphatase such as PP5 (58).
One limitation of the present study is the reliance on overexpression of proteins. However, several recent reports also support the suggestion that regulated nuclear entry of mPER1 is important in the control of circadian rhythm, as has been proposed for dPER in Drosophila. Endogenous mPER1 protein has been shown to accumulate in the nuclei of mouse SCN cells several hours after the peak of mper1 mRNA (19). The mammalian CRY proteins, essential for circadian rhythm, appear to regulate the rate at which mPER1 and mPER2 transit from cytoplasm to nucleus (31). Supporting an in vivo interaction of endogenous mPER1 and mPER2, mice with mutant mper2 have a shortened circadian period and significantly reduced mper1 oscillations in the SCN (59). Thus, the results with overexpressed proteins are consistent with findings for endogenous proteins, overexpression of mCRY and mPER proteins, and mutation of mPER2 in vivo. It will be important in future studies to examine the circadian phenotype of mice with deletions of the CKIɛ and CKI∂ genes. However, since CKI∂ and CKIɛ both interact with mPER1 in vitro, there may be functional redundancy that may complicate analysis of individual knockout mice. Mice with mutations in mPER1 that alter CKI binding or phosphorylation sites may therefore be more informative as to the role of CKI in the mammalian circadian rhythm.
We found that when mPER1 is overexpressed, it accumulates in the nucleus, and that endogenous CKIɛ is also concentrated in the nucleus of transfected cells. Why is endogenous CKIɛ not able to maintain a subset of mPER1 in the cytoplasm? One potential explanation is that since mPER1 can form homodimers, CKIɛ-dependent phosphorylation and masking of both NLS sequences in the dimer may be required for cytoplasmic retention. Thus, when the number of mPER1 molecules in the cytoplasm exceeds the amount of available CKIɛ, dimeric mPER1 may enter the nucleus, dragging a single bound CKIɛ along. This implies that CKIɛ may phosphorylate mPER1 only in cis and not in trans, leaving one unmasked NLS. Alternatively, overexpressed CKIɛ might alter the import kinetics of mPER1 by interfering with nuclear import pathways. If this were the case, CKIɛ expression might block the nuclear import of other proteins. However, CKIɛ expression did not block the import of the NLS-mPER1 fusion protein, nor did it block the nuclear accumulation of p53. To more rigorously exclude this model, it will be important in future studies to identify a specific mPER1 nuclear import pathway.
Kume and coworkers recently examined the localization of overexpressed mPER proteins in COS-7 and NIH 3T3 cells (31). Unlike seen in our HEK 293 cells, they found mPER1 and mPER2 localized to both nucleus and cytoplasm. Since mPER2 and mCRY1 can alter the localization of mPER1, we speculate that these differences may be due to differences in levels of endogenous circadian proteins, as well as differences in overexpression levels. We have previously shown that levels of CKIɛ expression can vary widely in different cell lines (14).
Multiple assays demonstrate that CKIɛ binds to mPER1. Interestingly, CKIɛ binds to mPER1 in a region that has no obvious sequence similarity to the suggested DBT-binding region of dPER (27). PER proteins contain a protein-protein interaction domain termed the PAS domain (23). CKIɛ bound to the central region of the mPER1 protein, carboxy terminal to the PAS domain and adjacent to the NLS. Kloss et al. reported that DBT bound to an amino-terminal region of dPER, in the same region as the dPER NLS but amino terminal to the PAS domains (27). CKI family members have recently been reported to bind to other proteins including NF-AT4, dishevelled, and the yeast transcriptional regulator Swi6 (20, 40, 60). No apparent regions of sequence homology exist between these proteins in the kinase-binding domains. It remains possible that there are structural similarities in the substrate-binding sites; in fact, we found that CKIɛ also binds to the dPER protein although the binding site has not yet been mapped. The fact that CKI binding of dPER and mPER1 has been preserved while the location and sequence of the binding site may have been shuffled suggests there is strong selective pressure to maintain the interaction of the two proteins. We note that CKIɛ, CKI∂, and the kinase domain fragment of CKIɛ bound to mPER1 whereas CKIα2 did not, consistent with the mPER1 interaction taking place via the kinase domain and not via the carboxy-terminal regulatory domain. However, this does not exclude a role for the kinase regulatory domain in the regulation of circadian rhythm in vivo, as previous studies have shown that this domain can regulate kinase activity in vitro and in vivo (5, 16, 17, 44).
Phosphorylation mediated nucleocytoplasmic trafficking.
It has become increasingly clear that proteins whose functions are tightly regulated by phosphorylation are often maintained in close proximity to their regulatory kinases and phosphatases, either by colocalization (e.g., binding to common anchoring proteins) or by direct association (22, 38). Protein phosphorylation controls the nuclear import of a number of proteins, including Cdc25 and Pho4 in yeast and SV40 large T antigen, Dorsal, Cdc25C, Xnf7, NF-κB, NF-AT4, and FKHRL1 in vertebrates (3, 4, 9, 24, 29, 33, 35, 39, 43, 60). In many of these cases tight association between the kinase and the substrate has been shown. The mechanisms by which phosphorylation regulates nuclear trafficking appear to be diverse. Phosphorylation may alter binding of an importin (in the cases of SV40 large T antigen, Pho4, and Dorsal) or a 14-3-3 protein with a nuclear export signal (CDC25 or FKHRL1), promote binding to cytoplasmic anchoring structures (Xnf7), stimulate degradation of a cytoplasmic anchoring protein (NF-κB), or cause a conformational change that masks an NLS (NF-AT4). Our results suggest that CKIɛ prevents mPER1 nuclear entry by the last mechanism, utilizing a region carboxy terminal of the NLS to mask the NLS in a phosphorylation-dependent manner. In many of the cases discussed above a cellular phosphatase is able to reverse the effects of phosphorylation, thus regulating the subcellular localization of the substrate. Genetic or biochemical investigations in the future may identify such a regulator in the circadian system.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank M. Tei, H. Okamura, M. Young, J. Takahashi, and M. Morgan for generously providing plasmids, Rebecca Shepard and Aurelia Meloni-Ehrig for assistance with immunofluorescence, Bob Schackman for oligonucleotide synthesis, and L. Ptacek, D. Ayer, B. Graves, E. Raetz, and K. Ullman for constructive criticism of the manuscript.
This work was supported by grant R01 CA71074 from the NIH to D.M.V. Oligonucleotide synthesis was supported by Cancer Center Support grant 3P30 CA42014.
ADDENDUM IN PROOF
Supporting an essential role for casein kinase Iɛ in mammalian circadian rhythm, Lowrey et al. have reported that the tau hamster locus encodes casein kinase Iɛ (P. L. Lowrey et al., Science 288:483–491, 2000). Furthermore, Keesler et al. have reported in interaction of casein kinase Iɛ with human mPER1 (Keesler et al., NeuroReport 2:1–5, 2000).
REFERENCES
- 1.Albrecht U, Sun Z S, Eichele G, Lee C C. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 2.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 3.Briggs L J, Stein D, Goltz J, Corrigan V C, Efthymiadis A, Hubner S, Jans D A. The cAMP-dependent protein kinase site (Ser312) enhances dorsal nuclear import through facilitating nuclear localization sequence/importin interaction. J Biol Chem. 1998;273:22745–22752. doi: 10.1074/jbc.273.35.22745. [DOI] [PubMed] [Google Scholar]
- 4.Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 5.Cegielska A, Gietzen K F, Rivers A, Virshup D M. Autoinhibition of casein kinase I ɛ (CKIɛ) is relieved by protein phosphatases and limited proteolysis. J Biol Chem. 1998;273:1357. doi: 10.1074/jbc.273.3.1357. [DOI] [PubMed] [Google Scholar]
- 6.Comolli J, Taylor W, Rehman J, Hastings J W. Inhibitors of serine/threonine phosphoprotein phosphatases alter circadian properties in Gonyaulax polyedra. Plant Physiol. 1996;111:285–291. doi: 10.1104/pp.111.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comolli J C, Hastings J W. Novel effects on the Gonyaulax circadian system produced by the protein kinase inhibitor staurosporine. J Biol Rhythms. 1999;14:11–19. doi: 10.1177/074873099129000399. [DOI] [PubMed] [Google Scholar]
- 8.Curtin K D, Huang Z J, Rosbash M. Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron. 1995;14:365–372. doi: 10.1016/0896-6273(95)90292-9. [DOI] [PubMed] [Google Scholar]
- 9.Dalal S N, Schweitzer C M, Gan J, DeCaprio J A. Cytoplasmic localization of human cdc25C during interphase requires an intact 14-3-3 binding site. Mol Cell Biol. 1999;19:4465–4479. doi: 10.1128/mcb.19.6.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darlington T K, Wager-Smith K, Ceriani M F, Staknis D, Gekakis N, Steeves T D L, Weitz C J, Takahashi J S, Kay S A. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 11.Dhillon N, Hoekstra M F. Characterization of two protein kinases from Schizosaccharomyces pombe involved in the regulation of DNA repair. EMBO J. 1994;13:2777–2788. doi: 10.1002/j.1460-2075.1994.tb06571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunlap J C. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 13.Edery I, Zwiebel L J, Dembinska M E, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci USA. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fish K, Cegielska A, Getman M, Landes G, Virshup D M. Isolation and characterization of human casein kinase I epsilon, a novel member of the casein kinase I gene family. J Biol Chem. 1995;270:14875–14883. doi: 10.1074/jbc.270.25.14875. [DOI] [PubMed] [Google Scholar]
- 15.Gao Z-H, Metherall J, Virshup D M. Identification of casein kinase I substrates by in vitro expression cloning screening. Biochem Biophys Res Commun. 2000;268:562–566. doi: 10.1006/bbrc.2000.2168. [DOI] [PubMed] [Google Scholar]
- 16.Gietzen K F, Virshup D M. Identification of inhibitory autophosphorylation sites on casein kinase I ɛ. J Biol Chem. 1999;274:32063–32070. doi: 10.1074/jbc.274.45.32063. [DOI] [PubMed] [Google Scholar]
- 17.Graves P R, Roach P J. Role of COOH-terminal phosphorylation in the regulation of casein kinase I delta. J Biol Chem. 1995;270:21689–21694. doi: 10.1074/jbc.270.37.21689. [DOI] [PubMed] [Google Scholar]
- 18.Gross S D, Anderson R A. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signalling. 1998;10:699–711. doi: 10.1016/s0898-6568(98)00042-4. [DOI] [PubMed] [Google Scholar]
- 19.Hastings M H, Field M D, Maywood E S, Weaver D R, Reppert S M. Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: new insights into a core clock mechanism. J Neurosci. 1999;19:RC11. doi: 10.1523/JNEUROSCI.19-12-j0001.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho U, Mason S, Kobayashi R, Hoekstra M, Andrews B. Role of the casein kinase I isoform, Hrr25, and the cell cycle-regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:581–586. doi: 10.1073/pnas.94.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoekstra M F, Liskay R M, Ou A C, DeMaggio A J, Burbee D G, Heffron F. HRR25, a putative protein kinase from budding yeast: association with repair of damaged DNA. Science. 1991;253:1031–1034. doi: 10.1126/science.1887218. [DOI] [PubMed] [Google Scholar]
- 22.Holland P M, Cooper J A. Docking sites for kinases. Curr Biol. 1999;9:R329–R331. doi: 10.1016/s0960-9822(99)80205-x. [DOI] [PubMed] [Google Scholar]
- 23.Huang Z J, Edery I, Rosbash M. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature. 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 24.Hübner S, Xiao C Y, Jans D A. The protein kinase CK2 site (Ser111/112) enhances recognition of the simian virus 40 large T-antigen nuclear localization sequence by importin. J Biol Chem. 1997;272:17191–17195. doi: 10.1074/jbc.272.27.17191. [DOI] [PubMed] [Google Scholar]
- 25.Jin X, Shearman L P, Weaver D R, Zylka M J, Vries G J D, Reppert S M. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 26.Kaffman A, Rank N M, O'Shea E K. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 1998;12:2673–2683. doi: 10.1101/gad.12.17.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kloss B, Price J L, Saez L, Blau J, Rothenfluh A, Wesley C S, Young M W. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase I epsilon. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- 28.Knippschild U, Milne D M, Campbell L E, DeMaggio A J, Christenson E, Hoekstra M F, Meek D W. p53 is phosphorylated in vitro and in vivo by the delta and epsilon isoforms of casein kinase 1 and enhances the level of casein kinase 1 delta in response to topoisomerase-directed drugs. Oncogene. 1997;15:1727–1736. doi: 10.1038/sj.onc.1201541. [DOI] [PubMed] [Google Scholar]
- 29.Komeili A, O'Shea E K. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 30.Kudo N, Wolff B, Sekimoto T, Schreiner E P, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 31.Kume K, Zylka M J, Sriram S, Shearman L P, Weaver D R, Jin X, Maywood E S, Hastings M H, Reppert S M. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 32.Lee C, Bae K, Edery I. The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron. 1998;21:857–867. doi: 10.1016/s0896-6273(00)80601-7. [DOI] [PubMed] [Google Scholar]
- 33.Li X, Shou W, Kloc M, Reddy B A, Etkin L D. Cytoplasmic retention of Xenopus nuclear factor 7 before the mid blastula transition uses a unique anchoring mechanism involving a retention domain and several phosphorylation sites. J Cell Biol. 1994;124:7–17. doi: 10.1083/jcb.124.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Loros J, Dunlap J C. Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc Natl Acad Sci USA. 2000;97:234–239. doi: 10.1073/pnas.97.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez-Girona A, Furnari B, Mondesert O, Russell P. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature. 1999;397:172–175. doi: 10.1038/16488. [DOI] [PubMed] [Google Scholar]
- 36.Matsumoto A, Tomioka K, Chiba Y, Tanimura T. timrit lengthens circadian period in a temperature-dependent manner through suppression of PERIOD protein cycling and nuclear localization. Mol Cell Biol. 1999;19:4343–4354. doi: 10.1128/mcb.19.6.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCright B, Virshup D M. Identification of a new family of protein phosphatase 2A regulatory subunits. J Biol Chem. 1995;270:26123–26128. doi: 10.1074/jbc.270.44.26123. [DOI] [PubMed] [Google Scholar]
- 38.Pawson T, Scott J D. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 39.Peng C Y, Graves P R, Thoma R S, Wu Z, Shaw A S, Piwnica-Worms H. Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science. 1997;277:1501–1505. doi: 10.1126/science.277.5331.1501. [DOI] [PubMed] [Google Scholar]
- 40.Peters J M, McKay R M, McKay J P, Graff J M. Casein kinase I transduces Wnt signals. Nature. 1999;401:345–350. doi: 10.1038/43830. [DOI] [PubMed] [Google Scholar]
- 41.Price J L, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young M W. Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell. 1998;94:83–95. doi: 10.1016/s0092-8674(00)81224-6. [DOI] [PubMed] [Google Scholar]
- 42.Reppert S M. A clockwork explosion! Neuron. 1998;21:1–4. doi: 10.1016/s0896-6273(00)80234-2. [DOI] [PubMed] [Google Scholar]
- 43.Rihs H-P, Peters R. Nuclear transport kinetics depend on phosphorylation-site-containing sequences flanking the karyophilic signal of the Simian virus 40 T antigen. EMBO J. 1989;8:1479–1484. doi: 10.1002/j.1460-2075.1989.tb03531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rivers A, Gietzen K F, Vielhaber E, Virshup D M. Regulation of casein kinase 1 ɛ and δ by an in vivo futile phosphorylation cycle. J Biol Chem. 1998;273:15980–15984. doi: 10.1074/jbc.273.26.15980. [DOI] [PubMed] [Google Scholar]
- 45.Rowles J, Slaughter C, Moomaw C, Hsu J, Cobb M H. Purification of casein kinase I and isolation of cDNAs encoding multiple casein kinase I-like enzymes. Proc Natl Acad Sci USA. 1991;88:9548–9552. doi: 10.1073/pnas.88.21.9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saez L, Young M W. Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 47.Sakamoto K, Nagase T, Fukui H, Horikawa K, Okada T, Tanaka H, Sato K, Miyake Y, Ohara O, Kako K, Ishida N. Multitissue circadian expression of rat period homolog (rPer2) mRNA is governed by the mammalian circadian clock, the suprachiasmatic nucleus in the brain. J Biol Chem. 1998;273:27039–27042. doi: 10.1074/jbc.273.42.27039. [DOI] [PubMed] [Google Scholar]
- 48.Sangoram A M, Saez L, Antoch M P, Gekakis N, Staknis D, Whiteley A, Fruechte E M, Vitaterna M H, Shimomura K, King D P, Young M W, Weitz C J, Takahashi J S. Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron. 1998;21:1101–1113. doi: 10.1016/s0896-6273(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 49.Santos J, Logarinho E, Tapia C, Allende C, Allende J, Sunkel C. The casein kinase 1 alpha gene of Drosophila melanogaster is developmentally regulated and the kinase activity of the protein induced by DNA damage. J Cell Sci. 1996;109:1847–1856. doi: 10.1242/jcs.109.7.1847. [DOI] [PubMed] [Google Scholar]
- 50.Sauman I, Reppert S M. Circadian clock neurons in the silkmoth Antheraea pernyi: novel mechanisms of Period protein regulation. Neuron. 1996;17:889–900. doi: 10.1016/s0896-6273(00)80220-2. [DOI] [PubMed] [Google Scholar]
- 51.Shou W, Li X, Wu C, Cao T, Kuang J, Che S, Etkin L D. Finely tuned regulation of cytoplasmic retention of Xenopus nuclear factor 7 by phosphorylation of individual threonine residues. Mol Cell Biol. 1996;16:990–997. doi: 10.1128/mcb.16.3.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sidorova J M, Mikesell G E, Breeden L L. Cell cycle-regulated phosphorylation of Swi6 controls its nuclear localization. Mol Biol Cell. 1995;6:1641–1658. doi: 10.1091/mbc.6.12.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takumi T, Matsubara C, Shigeyoshi Y, Taguchi K, Yagita K, Maebayashi Y, Sakakida Y, Okumura K, Takashima N, Okamura H. A new mammalian period gene predominantly expressed in the suprachiasmatic nucleus. Genes Cells. 1998;3:167–176. doi: 10.1046/j.1365-2443.1998.00178.x. [DOI] [PubMed] [Google Scholar]
- 54.Takumi T, Nagamine Y, Miyake S, Matsubara C, Taguchi K, Takekida S, Sakakida Y, Nishikawa K, Kishimoto T, Niwa S I, Okumura K, Okamura H. A mammalian ortholog of Drosophila timeless, highly expressed in SCN and retina, forms a complex with mPER1. Genes Cells. 1999;4:67–75. doi: 10.1046/j.1365-2443.1999.00238.x. [DOI] [PubMed] [Google Scholar]
- 55.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 56.Vosshall L B, Price J L, Sehgal A, Saez L, Young M W. Block in nuclear localization of period protein by a second clock mutation, timeless. Science. 1994;263:1606–1609. doi: 10.1126/science.8128247. [DOI] [PubMed] [Google Scholar]
- 57.Young M W. The molecular control of circadian behavioral rhythms and their entrainment in Drosophila. Annu Rev Biochem. 1998;67:135–152. doi: 10.1146/annurev.biochem.67.1.135. [DOI] [PubMed] [Google Scholar]
- 58.Zhao S, Sancar A. Human blue-light photoreceptor hCRY2 specifically interacts with protein serine/threonine phosphatase 5 and modulates its activity. Photochem Photobiol. 1997;66:727–731. doi: 10.1111/j.1751-1097.1997.tb03214.x. [DOI] [PubMed] [Google Scholar]
- 59.Zheng B, Larkin D W, Albrecht U, Sun Z S, Sage M, Eichele G, Lee C C, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- 60.Zhu J, Shibasaki F, Price R, Guillemot J C, Yano T, Dotsch V, Wagner G, Ferrara P, McKeon F. Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell. 1998;93:851–861. doi: 10.1016/s0092-8674(00)81445-2. [DOI] [PubMed] [Google Scholar]
- 61.Zylka M J, Shearman L P, Levine J D, Jin X, Weaver D R, Reppert S M. Molecular analysis of mammalian Timeless. Neuron. 1998;21:1115–1122. doi: 10.1016/s0896-6273(00)80628-5. [DOI] [PubMed] [Google Scholar]
- 62.Zylka M J, Shearman L P, Weaver D R, Reppert S M. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]