Abstract
During entry, herpes simplex virus type 1 (HSV-1) releases its capsid and the tegument proteins into the cytosol of a host cell by fusing with the plasma membrane. The capsid is then transported to the nucleus, where it docks at the nuclear pore complexes (NPCs), and the viral genome is rapidly released into the nucleoplasm. In this study, capsid association with NPCs and uncoating of the viral DNA were reconstituted in vitro. Isolated capsids prepared from virus were incubated with cytosol and purified nuclei. They were found to bind to the nuclear pores. Binding could be inhibited by pretreating the nuclei with wheat germ agglutinin, anti-NPC antibodies, or antibodies against importin β. Furthermore, in the absence of cytosol, purified importin β was both sufficient and necessary to support efficient capsid binding to nuclei. Up to 60 to 70% of capsids interacting with rat liver nuclei in vitro released their DNA if cytosol and metabolic energy were supplied. Interaction of the capsid with the nuclear pore thus seemed to trigger the release of the viral genome, implying that components of the NPC play an active role in the nuclear events during HSV-1 entry into host cells.
Many of the viruses that enter the nucleus of their host cells to replicate have capsids that are too large to pass through the nuclear pore complexes (NPCs). Consequently, they have to either wait until cell division occurs or undergo a disassembly in the cytosol prior to nuclear import (50). In the case of Herpesviridae, Adenoviridae, and baculoviruses, electron microscopic (EM) analysis has shown that the incoming capsids are transported in intact form to the NPCs. Once bound to the cytosolic side of the NPC, these capsids apparently release their DNA through the pore into the nucleus. To learn more about the events that take place at the nuclear membrane, we have in this study analyzed the interactions between herpes simplex virus type 1 (HSV-1) capsids and the nuclear envelope in vivo and in vitro.
HSV-1 is an enveloped virus with an icosahedral capsid, 125 nm in diameter, that contains the viral DNA. The capsid shell contains six different proteins (44) with VP5 as the main structural component (32, 33, 44). In the virus particle, the capsid is covered by a layer of proteins called the tegument and by the viral envelope. These contain several additional proteins. The major tegument proteins are VP1-3 (also called VP1/2), VP11/12, VP13/14, VP16, VP18.8, and VP22 (17, 43).
HSV-1 penetrates into the cell directly through the plasma membrane. First, it interacts via its spike glycoproteins gB and/or gC with heparan sulfate chains on cell surface proteoglycans (reviewed in reference 41). The following fusion reaction requires the concerted action of three additional viral glycoproteins, gB, gH, and gL, and appears to be triggered by the binding of gD to its cognate receptors (38). The human gD receptors identified to date include a member of the tumor necrosis factor receptor family, designated HVEM or herpesvirus entry protein A (HveA) and officially named TNFRSF14, and two members of the immunoglobulin superfamily (19, 28). Another surface glycoprotein, HveB, has been shown to mediate entry of certain mutant strains that cannot use HVEM (48). Moreover, a human member of the immunoglobulin superfamily, designated HveC, was recently identified as a coreceptor in mucosal epithelia (8, 19).
After virus binding, the viral envelope fuses with the plasma membrane in a reaction mediated by the viral spike glycoproteins. The capsid and the tegument proteins are thus transferred into the cytosol. Next, the capsid is transported through the cytosol to the nucleus where it binds to NPCs (24, 37, 42). Transport occurs along microtubules, and there is evidence that it might be mediated by dynein, a minus-end-directed motor complex (40). The EM images of the infection process indicate that the DNA is rapidly and efficiently ejected from the NPC-bound capsid, leaving behind an empty capsid that is eventually released into the cytosol (1, 40, 46). Inside the nucleus, the incoming viral DNA localizes adjacent to the nuclear domain ND10 (23) and results in its disruption (3, 21, 22).
We have here analyzed the interaction of HSV-1 capsids with the NPC and characterized the DNA release step biochemically. To this end, we have measured the uncoating efficiency in living cells. In addition, we reconstituted in vitro the binding and uncoating events using isolated nuclei, cytosol, and purified capsids. This allowed us to determine some of the requirements for capsid docking and DNA expulsion.
MATERIALS AND METHODS
Cells, antibodies, and virus.
BHK-21 cells were grown in Glasgow's minimum essential medium with 5% fetal calf serum and 10% tryptose phosphate broth, and Vero cells were grown in minimum essential medium with 7.5% fetal calf serum and nonessential amino acids. The following rabbit polyclonal antibodies were used: anti-VP19c (NC-2) and anti-DNA-containing capsids (anti-HC; both provided by Roselyn Eisenberg and Gary Cohen, University of Pennsylvania, Philadelphia) anti-VP13/14 (R220; provided by D. Meredith, University of Leeds, Leeds, United Kingdom), anti-VP16 (SW1; obtained from Amy Sheaffer and Dan Tenney, Bristol-Myers Squibb, Wallingford, Conn.), anti-VP22 (AGV30; obtained from Gillian Elliott and Peter O'Hare, Marie Curie Research Institute, Oxted, United Kingdom), anti-VP5 RomV (Romulus, bleed V; this study), anti-importin β (provided by A. Radu and G. Blobel, Rockefeller University, New York, N.Y.), and anticalnexin (15). We also used mouse monoclonal antibodies (MAbs) 414 (BabCo Inc., Berkeley, Calif.), anti-p97 (3E9; Affinity Bioreagents, Inc.), RL1 and RL2 (both provided by F. Melchior and L. Gerace, Scripps Institute, San Diego, Calif.), and antitransportin (Transduction Laboratories, San Diego, Calif.). Fluorescently labeled secondary antibodies (tetramethyl rhodamine isothiocyanate- or fluorescein isothiocyanate-conjugated goat anti-rabbit or goat anti-mouse) were obtained from Jackson ImmunoResearch Laboratories (West Grove, Pa.).
To raise polyclonal rabbit anticapsid antibodies, capsids were purified from virions as described below and concentrated by centrifugation in a Beckman SW28 rotor at 24,000 rpm for 1 h at 4°C. The protein concentration was 1.1 mg/ml as determined by the bicinchoninic acid assay (Pierce). Two rabbits (Romulus and Remus) were immunized with 100 μg of the antigen emulsified in complete Freund's adjuvant for the initial intradermal injection. The subsequent booster injections were given subcutaneously at 14-day intervals using incomplete Freund's adjuvant emulsified with 50 to 100 μg of the antigen. Specificity of the obtained antisera was determined by enzyme-linked immunosorbent assay using purified capsids and by Western blotting using HSV-1-infected and noninfected cell lysates on nitrocellulose membranes and visualized by ECL detection (Amersham). The obtained antisera recognized capsids in enzyme-linked immunosorbent assay but reacted very little against whole virions. Moreover, both antisera recognized several capsid bands and some tegument bands in infected cells but nothing in noninfected cells by Western blotting (data not shown).
Viruses, capsids, and nuclei.
Virus was prepared essentially as previously described (40). Titers of 109 PFU/ml and protein concentrations of 0.5 to 1 mg/ml in the purified peak fraction were obtained. Preparation of radioactively [3H]thymidine- or [35S]cysteine-methionine-labeled virus was performed essentially as previously described (40). These preparations commonly had titers of 107 PFU/ml and contained about 0.1 mg of protein per ml.
Capsids were purified from virions collected from infected-cell medium by centrifugation and resuspended in MNT buffer (30 mM MES [morpholineethanesulfonic acid], 100 mM NaCl, 20 mM Tris, pH 7.4). All steps were carried out at 4°C. The virions were stripped of their envelopes and some of the tegument components by incubation in a lysis buffer (500 mM NaCl, 20 mM Tris [pH 7.4], 1% Triton X-100, 1 mM EDTA) for 30 min on ice in the presence of protease inhibitors (1 mM phenylmethylsulfonyl fluoride [PMSF] and 1× CLAP cocktail [chymotrypsin, leupeptin, aprotinin, and pepstatin; 10 μg/ml each]). After lysis, the sample was sonicated in a water bath (three times, 30 s each), layered onto a linear 20 to 45% sucrose gradient (in MNT supplemented with 400 mM NaCl, 1 mM EDTA, and 0.5 mM dithiothreitol [DTT]), and centrifuged in a Beckman SW50.1 rotor at 30,000 rpm for 25 min. Capsids were collected as a light scattering zone from the gradient and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis.
To release DNA and penton proteins by the procedure of Newcomb and Brown (31), the virions (a mixture of nonlabeled and [3H]thymidine-labeled virus pellets) were pretreated with 0.6 M guanidine-HCl (GuHCl) for 20 min at room temperature prior to capsid isolation in the sucrose gradient (see above). In a similar manner, after the 30-min lysis on ice, virions (a mixture of nonlabeled and [35S]cysteine-methionine-labeled virus pellets without protease inhibitors) were subjected to limited proteolysis with trypsin. Virus was treated with 10 μg of trypsin per ml for 5 min at 37°C. The reaction was stopped with a 10× molar excess of soybean trypsin inhibitor as well as with 1 mM PMSF and the 1× CLAP cocktail, and the resulting capsids were isolated in a sucrose gradient as described above. Both the GuHCl- and trypsin-treated capsids were analyzed by SDS-PAGE.
Nuclei from rat liver were purified essentially as described previously (2). Livers were homogenized in 0.25 M STKM buffer (0.25 M sucrose, 25 mM HEPES-KOH [pH 7.4], 25 mM KOAc, 5 mM MgCl2, 0.1 mM EDTA, 1 mM PMSF, 1× CLAP cocktail, and 1 mM DTT) in a Dounce homogenizer with a motor-driven Teflon pestle and filtered through cheesecloth. The homogenate was mixed with 2.3 M STKM buffer (2.3 M sucrose, 25 mM HEPES-KOH [pH 7.4], 25 mM KOAc, 5 mM MgCl2, 0.1 mM EDTA, 1 mM PMSF, 1× CLAP cocktail, and 1 mM DTT) to raise the sucrose concentration to approximately 1.6 M. Nuclei were then separated from the cytoplasmic components by underlaying the homogenate with 2.3 M STKM buffer followed by centrifugation in a Beckman SW28 rotor at 25,000 rpm for 40 min at 4°C. The top layer was discarded with a curved spatula, and the supernatant was decanted. The white nuclear pellets were harvested with a clean curved spatula and resuspended into 0.25 M STKM. The nuclei were washed once by centrifugation (1,000 × g, 10 min, 4°C) through 0.93 M STKM. The number of nuclei were determined using a hemocytometer, and the result was confirmed by measuring optical density at 260 nm in a spectrophotometer. One unit of optical density at 260 nm corresponds to 3 × 106 nuclei per ml. The nuclei were divided into 10 or 25 optical density aliquots, underlaid with 30% STKM, and centrifuged (1,000 × g, 10 min, 4°C). The supernatant was aspirated, and the nuclear pellets were snap frozen in liquid nitrogen and stored at −80°C. The intactness of nuclei was confirmed by light microscopy and EM and by their ability to exclude fluorescently tagged (tetramethyl rhodamine isothiocyanate) 70-nm dextran (data not shown).
Sucrose gradient analysis of entry intermediates from infected cells.
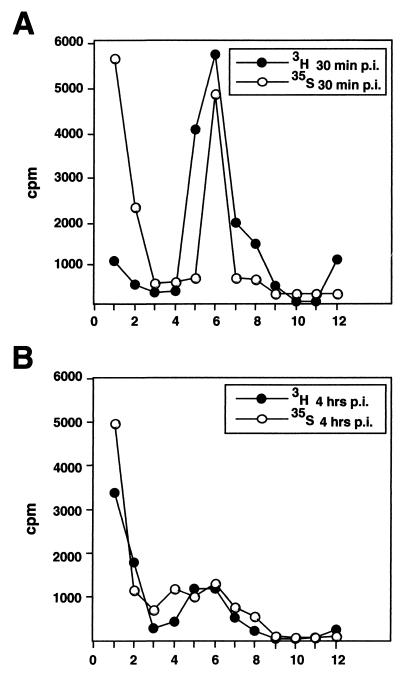
Virus internalization prior to isolation of entry intermediates was assayed essentially as previously described (40). Vero cells on 60-mm-diameter dishes grown to just confluency for 2 days were set on a metal plate on ice and washed three times with ice-cold RPMI medium-bovine serum albumin (BSA). The cells were inoculated with [3H]thymidine or [35S]cysteine-methionine-labeled virus at a multiplicity of infection (MOI) of 10 PFU/cell, and the virus was allowed to bind to the cells for 2 h on ice. The cells were washed to remove unbound virus and shifted to normal medium at 37°C and 5% CO2 for various lengths of time. Before harvesting at various time points (except the 0-min time point), the cells were briefly treated with 5 μM cytochalasin D for the last 10 min in complete medium at 37°C and 5% CO2, transferred back to ice, washed, and stored at 4°C until the last time point. The cells were washed with phosphate-buffered saline (PBS) and incubated with proteinase K (2 mg/ml in PBS; American Bioanalytical) on ice. After 1 h, the reaction was stopped by adding ice-cold stop solution (1.25 mM PMSF and 3% [wt/vol] BSA in PBS). The cells were collected in an excess of ice-cold wash buffer (PBS containing 0.2% BSA and 0.5 mM PMSF) and spun at 1,500 rpm for 10 min at 4°C. The pellet was resuspended into 100 μl of PBS, and 1/20 was counted in a liquid scintillation counter to measure the amount of internalized virus (cell-associated radioactivity). Control cells that were not warmed up after virus binding showed that the proteinase K treatment removed 90 to 95% of cell-bound virus as described earlier (40). The rest of the sample was then lysed in 10 mM Tris (pH 7.4)–150 mM NaCl–1% NP-40–1% Na-deoxycholate–1 mM PMSF–1× CLAP cocktail for 15 min on ice, and the nuclear debris was removed by centrifugation in a microcentrifuge at 5,000 rpm for 5 min (4°C). The pellet was counted in a liquid scintillation counter, and the supernatant was layered onto a linear 20 to 45% (wt/vol) sucrose gradient in 30 mM MES–500 mM NaCl–20 mM Tris (pH 7.4)–1 mM EDTA–0.5 mM DTT. The gradient was centrifuged in a Beckman TLS-55 rotor at 30,000 rpm for 25 min at 4°C, fractionated by hand into 12 fractions, and counted in a liquid scintillation counter. The calculation of sedimentation coefficients for capsids was carried out by the method of McEwen (25). In Fig. 2, sedimentation profiles from one experiment are shown. However, the analysis was repeated three times, and mean values (with standard deviations) of two independent experiments presented as percentages of total counts are shown in Table 1.
FIG. 2.
Uncoating of HSV-1 in Vero cells. Vero cells were infected with [3H]thymidine- or [35S]methionine-labeled virus at an MOI of 10 PFU/cell. At various time points, the extracellular viruses were removed by proteinase K and the cells were lysed. The postnuclear supernatants were loaded onto a linear sucrose gradient for ultracentrifugation analysis of the internalized capsids. The sedimentation profiles of internalized [3H]thymidine-labeled and [35S]methionine-labeled capsids at 30 min (A) and 4 h (B) p.i. are shown. Sedimentation profiles from one experiment are shown. However, the analysis was repeated three times, and mean values (with standard deviations) of two independent experiments with [3H]thymidine-labeled virus presented as percentages of total counts are shown in Table 1. The x axes indicate fractions.
TABLE 1.
Sedimentation profiles
| Fraction | Value (%) at time p.i.:
|
|||
|---|---|---|---|---|
| 30 min
|
4 h
|
|||
| Mean | SD | Mean | SD | |
| Top | 3.0 | 0.35 | 12.7 | 3.54 |
| Peak | 26.5 | 3.68 | 10.95 | 1.2 |
In vitro binding and uncoating assays.
Nuclei were thawed on ice into capsid binding buffer (CBB; 20 mM HEPES-KOH [pH 7.3], 80 mM K-acetate, 2 mM DTT, 1 mM EGTA, 2 mM Mg-acetate, 1 mM PMSF, and 1× CLAP cocktail) to a final concentration of about 5 × 107 nuclei/ml. A 20-μl aliquot of the suspension was used per assay. In addition, the standard assay contained rabbit reticulocyte lysate (Promega; final concentration about 2.5 mg/ml) or rat liver cytosol (final concentration, about 350 to 500 μg/ml), 10 mg of BSA per ml, viral capsids, and CBB. In some experiments, an ATP-regenerating system was included in the form of 5 mM creatine phosphate (Sigma), 20 U of creatine phosphokinase (Sigma), 1 mM ATP, and 0.2 mM GTP. Assays (50-μl volume) were performed in duplicate, and assay mixtures were incubated at 37°C for 40 min. For ATP depletion experiments, ATP, GTP, creatine phosphate, and creatine phosphokinase were omitted and the mixture was preincubated for 15 min at room temperature in the presence of 5 mM glucose and 16 U of hexokinase per ml before addition of capsids. For inhibition studies with antibodies or wheat germ agglutinin (WGA), the nuclei were preincubated in the complete binding mixture for 20 min on ice before addition of capsids. For incubation of capsids in the absence of cytosol, BSA was added instead of cytosol to the same protein concentration. In some experiments, importin β, importin α, or RanQ69L was diluted into CBB and preincubated with capsids for 15 min at room temperature prior to addition to the binding reaction. Preparation of the following soluble transport factors was carried out as previously described: C-terminally His-tagged Xenopus importin α (11), C-terminally His-tagged importin β (20), C-terminally His-tagged transportin (9), and N-terminally His-tagged RanQ69L (9).
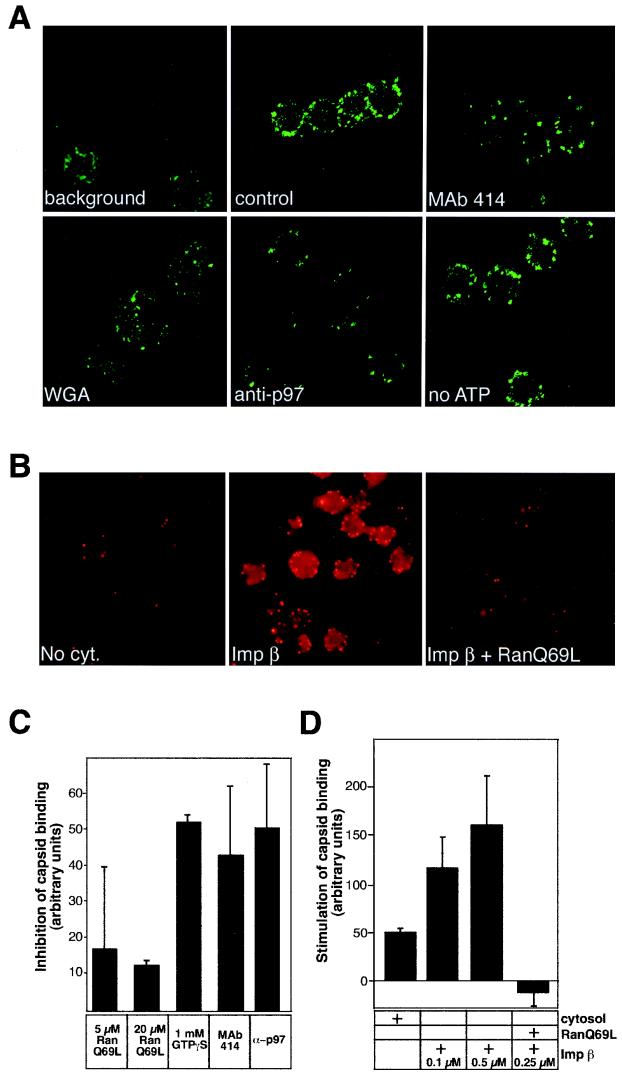
For immunofluorescence assays, the samples were diluted after binding into 1 ml of ice-cold wash buffer (CBB supplemented with 10 mg of BSA per ml and 10% [vol/vol] goat serum). The coverslips were pretreated with Cell-Tak (Collaborative Biomedical Products) to improve the attachment of nuclei. The nuclei were collected onto coverslips in a 24-well dish by centrifugation at 800 rpm (100 × g) for 5 min and washed twice with the ice-cold wash buffer. After washing, the nuclei were fixed with 4% (wt/vol) paraformaldehyde–0.1% glutaraldehyde for 20 min at room temperature followed by quenching of the remaining fixative using 2 mg of Na-borohydride per ml (three times for 2 min each). Immunolabeling was performed as described previously (40). The samples were examined with an Axiophot fluorescence microscope or a Zeiss Confocal LSM fluorescence microscope. Image processing was done using Adobe Photoshop. To quantify capsid binding to the nuclei, the capsids were detected by conventional immunofluorescence microscopy using antisera against purified capsids (RomV). Thereafter, the fluorescently labeled spots on the nuclear rim from 100 to 150 nuclei were counted to get an average of capsids bound per nucleus. This average amount was then compared to the average obtained in the control sample, which was capsids incubated with nuclei in either the presence (see Fig. 4C) or the absence (see Fig. 4D) of cytosol. The quantitation was performed with data from three to five independent experiments.
FIG. 4.
HSV-1 capsids bind to rat liver nuclei in vitro. (A) Confocal immunofluorescence microscopy of isolated HSV-1 capsids bound to purified rat liver nuclei in the presence of rat liver cytosol (control panel). The capsids were detected with RomV antibodies, which were generated against capsids isolated from virions. The background panel indicates a sample where capsids were omitted. Binding to nuclei pretreated with MAb 414, WGA, anti-importin β (anti-p97), or an ATP-depletion system (no ATP) is indicated in the other panels. (B) Conventional immunofluorescence microscopy of capsids binding to rat liver nuclei in the absence of cytosol (No cyt.), in the presence of 0.1 μM importin β (Imp β), or with 0.25 μM importin β and 10 μM RanQ69L (Imp β + RanQ69L). The capsids were detected as described for panel A. (C) Inhibition of capsid binding to nuclei pretreated with the indicated inhibitors in the presence of cytosol was quantitated as detailed in Materials and Methods. Inhibition of binding is shown relative to that of the control sample (without inhibitors) which represents the zero level in the graph. The mean values of at least triplicate samples with standard deviations are shown. (D) Stimulation of capsid binding to rat liver nuclei in vitro. Capsid binding to nuclei in the absence of cytosol (normalized to zero) or after addition of the components indicated on the right was quantitated as detailed in Materials and Methods. The mean values of at least triplicate samples with standard deviations are shown.
The uncoating assays were performed as described above except that, after the incubation of nuclei with capsids for 40 min at 37°C, the samples were transferred onto ice and the nuclei were lysed with 0.5% Triton X-100 for 15 min. Uncoating of capsids was measured by the conversion of viral DNA from a DNase-resistant to a DNase-sensitive form. After the lysis, 10 mM MgCl2 and 100 μg of DNase I (Boehringer Mannheim) per ml were added to the samples, which were always done in triplicate. For the DNase control, capsids in one sample were disrupted with 1% SDS by incubating the sample for 5 min at room temperature. SDS was then inactivated by addition of 10% Triton X-100. Samples were incubated with DNase for 60 min in a 37°C water bath. The reaction was stopped by the addition of ice-cold trichloroacetic acid (TCA) to a final concentration of 5%. After 30 min on ice, the precipitate was collected on Whatman GF/C filters and washed three times with ice-cold 5% TCA, twice with ice-cold 99% ethanol, and finally with acetone. The filters were air dried, and their radioactivity was determined by liquid scintillation counting.
EM.
Conventional Epon embedding (40) was used for thin-section EM of the capsid binding in vitro assay. After binding, the samples were washed once with the wash buffer (1,000 rpm for 8 min in a microcentrifuge), fixed with 1% glutaraldehyde in 200 mM cacodylate (pH 7.4) for 1 h at room temperature, treated with 1% OsO4–1.5% potassium ferrocyanide for 1 h and with 2% uranyl acetate in 50 mM maleate buffer (pH 5.2) for 1 h, and dehydrated using a graded ethanol series and propylenoxid. The nuclear pellets were embedded in Epon prior to cutting. The Epon sections were further contrasted using lead citrate and uranyl acetate (36).
RESULTS
HSV-1 uncoating in Vero cells.
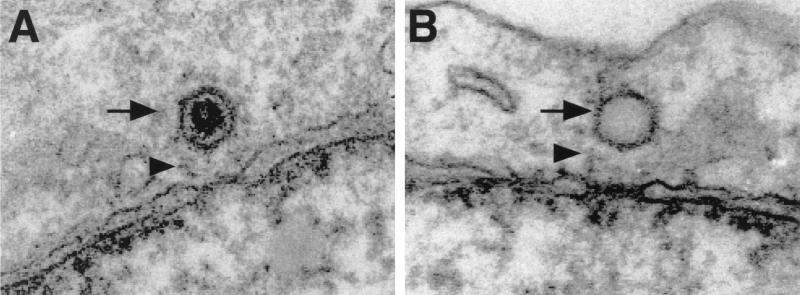
EM analysis shows that the capsids begin to accumulate on the nuclear membrane of Vero cells 1 h after penetration (40). They localize exclusively at the cytosolic aspect of the NPCs, at a distance of about 50 nm from the outer ring of the pore, and seem to be attached to the filaments emanating from the pores (Fig. 1). Interestingly, the capsids were always oriented with a penton toward the nuclear pore, as also previously noted for pseudorabies virus (12). Judging by the absence of the densely stained material seen in intact capsids, most of them lose their DNA rapidly after arriving at the nuclear membrane (Fig. 1B). Moreover, by using quantitative EM analysis, the appearance of empty capsids at the NPC has previously been shown to coincide with their arrival at the nucleus between 2 and 4 h postinfection (p.i.) (40). The dimensions and the shape of the empty capsids appear identical to those of the filled ones. Some of them remain attached to the nuclear pores, while others are released into the cytosol.
FIG. 1.
Incoming HSV-1 capsids bind to NPCs in vivo. Ultrathin Epon sections of Vero cells infected with HSV-1 at an MOI of 500 PFU/cell in the presence of cycloheximide are shown. At later times of infection, both DNA-containing, filled capsids (black arrow in panel A) and uncoated, empty capsids (black arrow in panel B) are located in close proximity to the NPC. Occasionally, capsids can be seen associated with fibers emanating from the pores (arrowheads in panels A and B).
To analyze biochemically the DNA release process and the appearance of empty capsids in Vero cells, [3H]thymidine- and [35S]methionine-cysteine-labeled virus was used. These virus were allowed to bind to the cells in the cold, whereafter entry was initiated by warming cells to 37°C. The cells were then treated briefly with cytochalasin D to dissociate microfilaments and thus improve capsid recovery after lysis and subjected to proteinase K digestion on ice to remove extracellular virus particles (16). The cells were lysed after 30 min, at which time the capsids were still en route to the nucleus, or after 4 h, when more than 60% of them had reached the nucleus (40). After lysis of the cells with Triton X-100, also the nuclear content (including the uncoated viral genome) was released to the supernatant. The insoluble debris was removed by a brief centrifugation, and the lysates were analyzed by sucrose velocity gradient centrifugation. The insoluble fraction contained mostly nuclear debris and thus also (filled) capsids still attached to nuclei. When this fraction was analyzed by scintillation counting, the 4-h sample contained two to three times more radioactivity than the 30-min one (data not shown). This is in agreement with more capsids being transported to the nuclei at 4 h than at 30 min p.i.
The sedimentation pattern in the 30-min sample showed, as expected, no evidence of capsid uncoating (Fig. 2A). The DNA-containing capsids were recovered in fractions 5 and 6 with an estimated sedimentation coefficient of about 600S. The [3H]thymidine radioactivity at the top of the gradient corresponding to free viral DNA amounted to less than 2 to 3% of the total. In contrast, the 4-h sample (Fig. 2B) showed that uncoating had taken place; the amount of [3H]thymidine in the two top fractions had clearly increased, while the amount of DNA sedimenting with the capsids had correspondingly decreased (Fig. 2B). A new DNA-free peak of [35S]methionine-labeled viral protein was present in fraction 4 (Fig. 2B). It was composed of empty capsids because (i) it sedimented in the same position (about 350S) as did empty capsids generated in vitro by treating detergent-lysed virions with GuHCl (data not shown and reference 31), and (ii) it contained the capsid proteins when analyzed by SDS-PAGE (data not shown). From the ratio of radioactivity in the empty capsid peaks to that in the full capsid peaks, we estimated that 50 to 60% of the capsids present in the lysate had released their DNA. These results confirmed that release of DNA is relatively efficient and that it results in the formation of apparently stable capsid structures devoid of DNA.
Characterization of purified capsids.
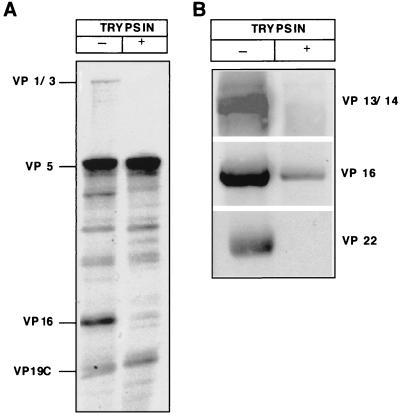
To analyze the processes occurring at the nuclear membrane in more detail, we developed in vitro assays for the binding of capsids to the nuclear envelope and for viral genome uncoating. For this, we used capsids isolated from mature viruses. These were expected to resemble more closely the authentic incoming capsids than capsids isolated from the nuclei of infected cells would have. To remove the envelope, virions were extracted with Triton X-100 in a high-salt-containing buffer and fractionated over a linear sucrose gradient. Figure 3A (lane 1) shows the overall protein pattern of such membrane-depleted capsid particles analyzed by SDS-PAGE. Figure 3B shows immunoblotting of the gels by antibodies against some of the structural components.
FIG. 3.
Characterization of HSV-1 capsids isolated from virions in vitro. (A) Coomassie blue staining of an SDS–10% PAGE analysis of the capsids. The capsids were isolated as a light-scattering zone in the sucrose gradient (left lane). Capsids were treated with 10 μg of trypsin per ml at 37°C for 5 min (right lane). The HSV-1 proteins identified by molecular weight are indicated on the left. (B) Western blot of the in vitro-isolated capsids (left lane) and capsids treated with 10 μg of trypsin per ml (right lane). The proteins identified with specific antibodies are indicated on the right.
The results revealed that, while the capsids prepared in this way were devoid of most glycoproteins, they were still associated with some of the tegument components (VP1-3, VP13/14, VP16, and VP22). EM images of the isolated capsids after negative staining showed additional external material particularly concentrated in the region of the pentons (see also Fig. 5, large arrows). Addition of DNase did not result in digestion of the 3H-labeled DNA, indicating that the capsids were intact and the DNA was fully protected (see below).
FIG. 5.
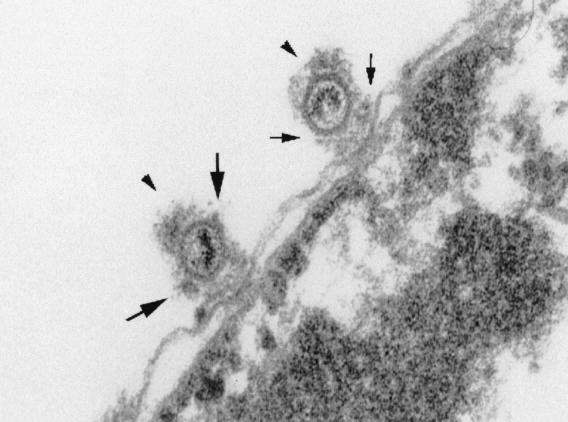
Preferred binding of HSV-1 capsids to NPCs. An electron micrograph of capsids (arrowheads) binding to rat liver nuclei in vitro is shown. Small arrows indicate the cytoplasmic fibrils emanating from the NPC, and large arrows show the additional electron-dense material at the capsid vertices.
When the isolated capsids were subjected to proteolysis with a low concentration of trypsin at 37°C, most of the tegument proteins were removed (Fig. 3A, lane 2). Coomassie blue staining and Western blotting revealed that VP1-3, VP13/14, and VP22 were completely removed as well as most of VP16. There was no detectable loss of VP5 or VP19c, the major capsid proteins (Fig. 3A). The capsid ultrastructure remained intact, and the viral DNA was inaccessible to DNase (data not shown). To further compare the trypsin-treated capsids to untreated capsids, we subjected them to immunoprecipitation by nonimmunized sera and the specific Romulus antisera. Both capsid preparations were prepared from [3H]thymidine-labeled virus, and they were both immunoprecipitated with the capsid-specific antibodies at least to the same extent as determined by liquid scintillation counting and Western blotting (data not shown).
Capsids bind to nuclei in vitro.
The isolated HSV-1 capsids were next incubated with rat liver nuclei in the presence or absence of cytosol. Binding was monitored by both conventional and confocal microscopy after indirect immunofluorescence staining using polyclonal antibodies against purified HSV-1 capsids. The bound capsids appeared as small intensely labeled spots giving a rim-like staining reminiscent of the pattern observed with antibodies against NPC proteins. Confocal immunofluorescence microscopy showed that, unlike the dim background staining inside the nucleus, the specific anticapsid staining was, indeed, located exclusively at the nuclear envelope and not inside (Fig. 4A, control panel).
Binding of capsids to nuclei in vitro was found to be independent of temperature (data not shown) and of energy in the form of ATP (Fig. 4A). Some capsid binding was observed even in the absence of cytosol, but it was clearly increased upon addition of exogenous cytosol (Fig. 4D). The somewhat surprising binding in the absence of cytosolic factors could in part be explained by residual cytosolic factors present in the nuclear preparation. In support of this, we found that the nuclei were brightly labeled by antibodies against importin β (anti-p97; data not shown).
Nuclear binding is mediated by importin β and inhibited by GTP-bound Ran.
That importin β was involved in capsid binding was suggested by the observation that pretreatment of nuclei with anti-importin β antibodies inhibited binding of capsids both in the presence (Fig. 4A, anti-p97) and in the absence (data not shown) of cytosol. To confirm the role of importin β in capsid binding, we performed in vitro binding in the presence of purified importin β (at 0.1 to 0.5 μM) when no other cytosolic components were added. Addition of importin β clearly stimulated the binding of capsids (Fig. 4B and D), indicating that importin β was sufficient and necessary for conferring capsid binding to the nuclei. Addition of importin α (at 0.25 μM) to the importin β-containing binding reaction had no effect on the binding (data not shown). When binding was assayed in the presence of excess nuclear localization signal (NLS) peptide coupled to BSA (NLS-BSA) and cytosol, only a slight decrease in the efficiency of binding was observed (data not shown). Taken together, these data suggest that capsid binding is presumably importin α independent. We also tested if another importin β-like factor, namely, transportin, could be involved in capsid binding. Addition of antitransportin antibodies or purified transportin (at 0.1 to 0.5 μM), either with or without cytosol, did not have any effect on capsid binding (data not shown), thus indicating that the binding is specific for importin β.
Nuclear import of most cargo is blocked by the addition of RanQ69L, a dominant-negative version of Ran unable to hydrolyze GTP (18). This Ran mutant is, therefore, predominantly in the GTP-bound state and causes premature dissociation of import complexes (10). To address the Ran dependence in the nuclear binding of capsids, we added RanQ69L to the in vitro binding reaction. In the presence of cytosol, addition of RanQ69L (at 5 to 10 μM) caused only a moderate decrease in the capsid binding (Fig. 4C). However, when RanQ69L was added to the importin β-containing binding reaction in the absence of cytosol, binding was almost completely blocked (Fig. 4B and D). Accordingly, in the presence of cytosol, binding was remarkably reduced by addition of 1 mM GTPγS, a nonhydrolyzable analog of GTP (Fig. 4C). Taken together, these data suggested that the importin β-mediated nuclear docking of HSV-1 capsids was dependent on the Ran GTPase cycle, too.
Preferred binding to NPCs.
Pretreatment of nuclei with MAb 414 against NPC proteins (Fig. 4A and C) efficiently reduced binding of capsids to nuclei. The same was observed with two other anti-nuclear pore protein antibodies, RL1 and RL2. MAb 414 interacts with the GFXFG repeats in the nucleoporins (5), and RL1 and RL2 interact with the nucleoporins that carry O-linked N-acetylglucosamine (39). These antibodies have been shown previously to block binding of karyophilic proteins to NPCs (14, 51). WGA, a lectin that associates with the glycoproteins in the NPC and inhibits nuclear import of many karyophilic proteins (7), was also found to decrease capsid binding (Fig. 4A, WGA). In contrast, incubation of the nuclei with control antibodies (anticalnexin; data not shown) had no effect on nuclear binding of capsids. The anticalnexin antibodies bind to the cytosolic tail of calnexin, a transmembrane protein present in the outer nuclear membrane (15).
The localization of the bound capsids on the nuclear envelope was also analyzed by EM. In ultrathin plastic sections, capsids were easily recognized (Fig. 5, arrowheads), and they were frequently detected in association with the NPCs. In some of the images, they seemed to be interacting with structures reminiscent of cytoplasmic fibrils emanating from the cytoplasmic face of NPCs (Fig. 5, small arrows). The viral DNA was still visible inside the capsids, and additional electron-dense material probably representing some of the tegument proteins or cytosolic components was also present on the capsid surface, especially concentrated at the capsid vertices (Fig. 5, large arrows). Taken together, the results indicated that the capsids bind to NPCs in vitro in a way similar to that observed in infected cells (1, 40).
Reduced binding with trypsin-treated capsids.
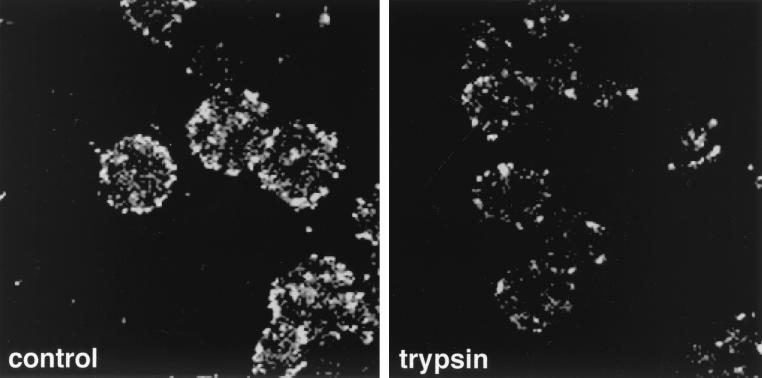
To address the role of individual HSV-1 proteins in the interaction of capsids with the NPC, we tested the ability of the trypsin-treated capsids to bind to nuclei. As shown in Fig. 6, their binding to nuclei was dramatically reduced (85% less) compared to that of untreated capsids. Therefore, tegument proteins VP1-3, VP13/14, VP16, and VP22, which were selectively affected by trypsinization (Fig. 3), emerged as the candidates for mediating capsid binding to the NPC.
FIG. 6.
Trypsin-treated capsids show reduced binding to nuclei in vitro. The figure shows confocal images of isolated, intact HSV-1 capsids (control) and trypsin-treated capsids (trypsin) bound to purified rat liver nuclei in the presence of rat liver cytosol.
In vitro uncoating of capsids.
To determine whether any of the capsids released their DNA upon binding to the NPC, we incubated [3H]thymidine-labeled capsids with nuclei under different conditions and analyzed thereafter the accessibility of the viral DNA to DNase. Prior to addition of DNase, the nuclei were lysed with 0.5% Triton X-100 to make the viral DNA in the nucleus also accessible to the added DNase. The validity of this assay was shown by the observation that the DNA in the purified capsid preparation was completely protected, while in the SDS-treated capsids it was fully digested to a TCA-soluble form (Fig. 7, columns 1 and 5).
FIG. 7.
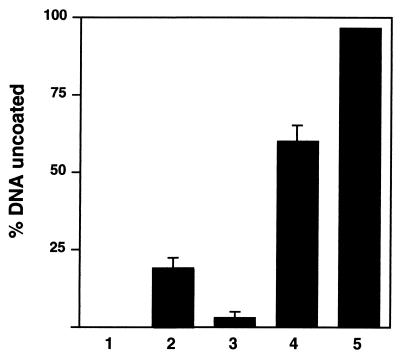
Uncoating of HSV-1 capsids in vitro. [3H]thymidine-labeled HSV-1 capsids were mixed either with BSA (column 1), BSA and rat liver nuclei (column 2), or BSA and rabbit reticulocyte lysate (column 3) or with BSA, reticulocyte lysate, and rat liver nuclei (column 4) in the presence of an ATP-regenerating system. The release of DNA was measured by assaying for DNase sensitivity using TCA precipitation. In untreated capsids (column 1), the DNA was completely protected, and in SDS-disrupted capsids (column 5), it was fully digested. The mean values of triplicate samples with standard deviations of at least three independent experiments are shown.
When capsids were incubated with cytosol in the presence of an ATP regeneration system but without nuclei, less than 5% of the DNA became DNase sensitive (Fig. 7, column 3). With nuclei alone (i.e., without cytosol), about 20% of the DNA became accessible (Fig. 7, column 2). However, when cytosol, the ATP-regenerating system, and nuclei were all present, about 60 to 70% of the capsid-associated DNA became sensitive to DNase (Fig. 7, column 4). This was similar to the level of uncoating observed for incoming capsids in living cells. The result indicated that the viral DNA can be released from the capsids in vitro and that the optimal release depends on the presence of nuclei, cytosol, and an energy source.
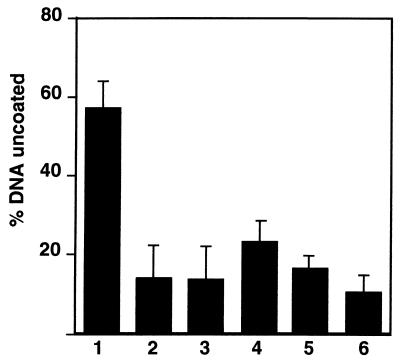
Preincubation of nuclei with antibodies against NPC components (MAb 414), against importin β, or with WGA decreased the extent of uncoating (Fig. 8). Since these antibodies also inhibited capsid binding to the nuclei, it was likely that specific binding of the capsids to the NPCs was required for the efficient release of viral DNA.
FIG. 8.
Inhibition of in vitro uncoating of HSV-1 capsids. [3H]thymidine-labeled HSV-1 capsids were mixed with BSA, reticulocyte lysate, and rat liver nuclei in the presence of an ATP-regenerating system (column 1). The release of DNA was measured by assaying for DNase sensitivity using TCA precipitation. The inhibitors were tested by pretreating the nuclei with MAb 414 (column 2), anti-importin β (column 3), WGA (100 μg/ml) (column 4), hexokinase and glucose for ATP depletion (column 5), and GTPγS (1 mM) (column 6). See Materials and Methods for the details. The mean values of triplicate samples with standard deviations of at least three independent experiments are shown.
To characterize the energy requirements of uncoating in more detail, we depleted the cytosol of energy with hexokinase and glucose prior to its addition to the uncoating mixture. Alternatively, we added GTPγS to the reaction mixture. Both of these treatments led to significant reduction in the efficiency of uncoating (Fig. 8). The results confirmed the need for metabolic energy but did not allow us to define which of the nucleotides was required. Nuclear import of karyophilic proteins is, however, known to require GTP hydrolysis by the small GTPase Ran/TC4 (27, 29), and GTP may constitute the sole energy for translocation (49).
In summary, we found that specific interaction of capsids with NPCs in vitro triggered the release of viral genome. The efficiency of release was enhanced by metabolic energy and factors present in the cytosol.
DISCUSSION
The uncoating process that renders the incoming genome competent for transcription and replication remains poorly understood for most animal viruses. Typically, it is, however, thought to involve a series of progressive changes in the incoming particle (13, 50). These depend on specific cues given by the host cell and on specific interactions between viral and cellular components. The dissociation steps are often coupled to intracellular transport and targeting events. In this way, the genome is activated only upon entry and only upon arrival in the correct cellular location. Our results in live cells and in vitro are fully consistent with such a stepwise process for HSV-1.
Disassembly begins at the plasma membrane where the envelope and many of the tegument proteins are lost (30, 40). Some of the tegument proteins, however, remain associated with the capsids, and these may play a role in capsid association with the dynein motor and later with the NPCs (40). Our analysis indicated that the tegument proteins that resisted detergent extraction included VP1-3, VP13/14, VP16, and VP22.
In Vero cells, the capsids start to arrive at the nuclear membrane about 1 h after penetration (40). They bind to the nuclear membrane and associate specifically with cytoplasmic surface components of the NPCs. Our in vitro studies indicated that capsid binding occurs specifically at the NPC and is independent of metabolic energy. When the tegument proteins were removed by mild trypsinization, capsids no longer bound efficiently to the NPCs in vitro, suggesting that one or more of them mediate NPC targeting or attachment. Using the in vitro binding assay, it was confirmed that capsid docking was mediated by importin β. Our preliminary results, however, suggest that docking is presumably independent of importin α. In this respect, HSV-1 capsid docking at the NPC is similar to that of cyclin B1 and human immunodeficiency virus type 1 Tat and Rev proteins that were recently reported to be imported by importin β in the absence of importin α (35, 45, 47). It further suggests that viruses replicating in the host cell nucleus, as HSV-1, may have adopted importin α-independent import pathways in order to achieve efficient nuclear targeting. By using importin β directly, they can avoid relying on importin α as a bridging factor upon docking at the NPC. Finally, our observation that the GTP-bound form of Ran (RanQ69L or addition of GTPγS to the cytosol) inhibits capsid binding to the NPC implies that the HSV-1 capsid docking is also Ran dependent.
Capsid association with the nucleus resulted in DNA release in living cells as well as in vitro when capsids were incubated with nuclei, cytosol, and an ATP-generating system. The efficiency was about 60% in both cases. In vitro uncoating was inhibited by WGA and by antibodies against importin β or nucleoporins, indicating that binding of the capsid to the NPC is necessary for uncoating. The inhibitory effect of GTPγS on uncoating suggested that HSV-1 uncoating is dependent on the integrity of the Ran GTPase cycle. During the import of karyophilic proteins, Ran GTPase binding to importin β causes it to dissociate from its cargo (34). It is an essential component in the import machinery.
It is noteworthy that a mutant of HSV-1, called tsB7, that is able to bind to NPC but fails to release the DNA at a nonpermissive temperature, has a mutated VP1-3 gene (1). As mentioned above, VP1-3 is one of the tightly bound, trypsin-sensitive tegument proteins. It is a large protein (270 kDa) that occurs in the virus in about 12 copies (26). Interestingly, the VP1-3 protein sequence contains four putative, although quite weak, bipartite NLSs, as well as several arginine-rich sequences (ProfileScan; the ExPASy proteomics server of the Swiss Institute of Bioinformatics), which also could be involved in the nuclear targeting as reported previously for certain human immunodeficiency virus type 1 proteins (35, 47). Moreover, VP1-3 is likely to constitute part of the tegument material observed around the pentons of isolated capsids (52). Since a modified penton probably provides a channel for DNA extrusion (31), it is tempting to speculate that one of the functions of VP1-3 is to interact with components of the NPC and trigger a change that allows DNA exit.
In the HSV-1-infected cell, progeny capsids can be seen by EM in the cytosol (4). Unlike incoming capsids, they do not appear to have affinity for the nuclei or for NPCs (4, 46). Passage of the progeny capsids from the nucleus through the cytosol to the Golgi complex is, in fact, suggested to be the main pathway of HSV-1 egress from the cell (reviewed in reference 6). In contrast, incoming capsids are efficiently targeted to the nuclear membrane (40). The changes that allow such altered capsid behavior might be triggered by events that occur during the final assembly and budding of the virus particle, during the extracellular phase, or as part of the entry program. Our lysis conditions mimic the changes necessary to make the capsids karyophilic. That these capsids isolated from virus particles were indeed karyophilic and uncoating competent in vitro suggested that the capsids in the virus already had undergone the necessary changes and therefore behaved like authentic incoming capsids. The in vitro assay developed here will hopefully allow more detailed molecular analysis of the complex events that take place at the nuclear pores including the targeted release of DNA through the NPCs.
ACKNOWLEDGMENTS
We thank Brian Burke, Frauke Melchior, Larry Gerace, Aurelian Radu, Junona Moroianu, Günter Blobel, David Meredith, Amy Sheaffer, Dan Tenney, Gillian Elliott, Peter O'Hare, Roselyn Eisenberg, and Gary Cohen for reagents. Erika Samoff and Ryan Price are acknowledged for their contributions during the development of in vitro assays. Michael Kann, Urs Greber, and Gary Whittaker as well as the other members of the Helenius lab are acknowledged for fruitful discussions, and Birgitta Tjäder is acknowledged for excellent technical assistance.
This study was supported by a National Institutes of Health grant to A.H. (AI 18599), an EMBO postdoctoral fellowship (ALTF 254-1993) to B.S., and grants from the Academy of Finland to P.M.O. Additional funding was obtained via an SA/DAAD collaboration grant to P.M.D. and B.S.
REFERENCES
- 1.Batterson W, Furlong D, Roizman B. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blobel G, Potter V R. Nuclei from rat liver: isolation method that combines purity with high yield. Science. 1966;154:1662–1665. doi: 10.1126/science.154.3757.1662. [DOI] [PubMed] [Google Scholar]
- 3.Burkham J, Coen D M, Weller S K. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J Virol. 1998;72:10100–10107. doi: 10.1128/jvi.72.12.10100-10107.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Church G A, Wilson D W. Study of herpes simplex virus maturation during a synchronous wave of assembly. J Virol. 1997;71:3603–3612. doi: 10.1128/jvi.71.5.3603-3612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis L I, Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- 6.Enquist L W, Husak P J, Banfield B W, Smith G A. Infection and spread of alphaherpesviruses in the nervous system. Adv Virus Res. 1998;51:237–347. doi: 10.1016/s0065-3527(08)60787-3. [DOI] [PubMed] [Google Scholar]
- 7.Finlay D R, Newmeyer D D, Price T M, Forbes D J. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J Cell Biol. 1987;104:189–200. doi: 10.1083/jcb.104.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geraghty R J, Krummenacher C, Cohen G H, Eisenberg R J, Spear P G. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 9.Gorlich D, Dabrowski M, Bischoff F R, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorlich D, Pante N, Kutay U, Aebi U, Bischoff F R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 11.Gorlich D, Prehn S, Laskey R A, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 12.Granzow H, Weiland F, Jons A, Klupp B G, Karger A, Mettenleiter T C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greber U F, Singh I, Helenius A. Mechanisms of virus uncoating. Trends Microbiol. 1994;2:52–56. doi: 10.1016/0966-842x(94)90126-0. [DOI] [PubMed] [Google Scholar]
- 14.Greber U F, Suomalainen M, Stidwill R P, Boucke K, Ebersold M W, Helenius A. The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 1997;16:5998–6007. doi: 10.1093/emboj/16.19.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammond C, Helenius A. Quality control in the secretory pathway: retention of a misfolded viral membrane glycoprotein involves cycling between the ER, intermediate compartment, and Golgi apparatus. J Cell Biol. 1994;126:41–52. doi: 10.1083/jcb.126.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helenius A, Kartenbeck J, Simons K, Fries E. On the entry of Semliki Forest virus into BHK-21 cells. J Cell Biol. 1980;84:404–420. doi: 10.1083/jcb.84.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honess R W, Roizman B. Proteins specified by herpes simplex virus. XI. Identification and relative molar rates of synthesis of structural and nonstructural herpes virus polypeptides in the infected cell. J Virol. 1973;12:1347–1365. doi: 10.1128/jvi.12.6.1347-1365.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klebe C, Prinz H, Wittinghofer A, Goody R S. The kinetic mechanism of Ran—nucleotide exchange catalyzed by RCC1. Biochemistry. 1995;34:12543–12552. doi: 10.1021/bi00039a008. [DOI] [PubMed] [Google Scholar]
- 19.Krummenacher C, Nicola A V, Whitbeck J C, Lou H, Hou W, Lambris J D, Geraghty R J, Spear P G, Cohen G H, Eisenberg R J. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J Virol. 1998;72:7064–7074. doi: 10.1128/jvi.72.9.7064-7074.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kutay U, Izaurralde E, Bischoff F R, Mattaj I W, Gorlich D. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukonis C J, Burkham J, Weller S K. Herpes simplex virus type 1 prereplicative sites are a heterogeneous population: only a subset are likely to be precursors to replication compartments. J Virol. 1997;71:4771–4781. doi: 10.1128/jvi.71.6.4771-4781.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maul G G, Guldner H H, Spivack J G. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 23.Maul G G, Ishov A M, Everett R D. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 24.McClain D S, Fuller A O. Cell-specific kinetics and efficiency of HSV-1 entry are determined by two distinct phases of attachment. Virology. 1994;198:690–702. doi: 10.1006/viro.1994.1081. [DOI] [PubMed] [Google Scholar]
- 25.McEwen C R. Tables for estimating sedimentation through linear concentration gradients of sucrose solution. Anal Biochem. 1967;20:114–149. doi: 10.1016/0003-2697(67)90271-0. [DOI] [PubMed] [Google Scholar]
- 26.McNabb D S, Courtney R J. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology. 1992;190:221–232. doi: 10.1016/0042-6822(92)91208-c. [DOI] [PubMed] [Google Scholar]
- 27.Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 29.Moore M S, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 30.Morrison E E, Stevenson A J, Wang Y F, Meredith D M. Differences in the intracellular localization and fate of herpes simplex virus tegument proteins early in the infection of Vero cells. J Gen Virol. 1998;79:2517–2528. doi: 10.1099/0022-1317-79-10-2517. [DOI] [PubMed] [Google Scholar]
- 31.Newcomb W W, Brown J C. Induced extrusion of DNA from the capsid of herpes simplex virus type 1. J Virol. 1994;68:433–440. doi: 10.1128/jvi.68.1.433-440.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newcomb W W, Brown J C. Structure of the herpes simplex virus capsid: effects of extraction with guanidine hydrochloride and partial reconstitution of extracted capsids. J Virol. 1991;65:613–620. doi: 10.1128/jvi.65.2.613-620.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newcomb W W, Trus B L, Booy F P, Steven A C, Wall J S, Brown J C. Structure of the HSv capsid—molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 34.Ohno M, Fornerod M, Mattaj I W. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- 35.Palmeri D, Malim M H. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reynolds E S. The use of lead citrate at high pH as an electron opaque stain in electron microscopy. J Histochem Cytochem. 1963;26:163–169. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Roizman B, Whitley R J, Lopez C, editors. The human herpesviruses. New York, N.Y: Raven Press, Ltd.; 1993. pp. 11–68. [Google Scholar]
- 38.Shukla D, Liu J, Blaiklock P, Shworak N W, Bai X, Esko J D, Cohen G H, Eisenberg R J, Rosenberg R D, Spear P G. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 39.Snow C M, Senior A, Gerace L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J Cell Biol. 1987;104:1143–1156. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sodeik B, Ebersold M W, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spear P. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 42.Spear P G. Membrane fusion induced by HSV. In: Bentz J, editor. Viral fusion mechanism. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 201–232. [Google Scholar]
- 43.Spear P G, Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpes virion. J Virol. 1972;9:143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steven A, Spear P. Herpesvirus capsid assembly and envelopment. New York, N.Y: Oxford University Press; 1997. [Google Scholar]
- 45.Takizawa C G, Weis K, Morgan D O. Ran-independent nuclear import of cyclin B1-Cdc2 by importin beta. Proc Natl Acad Sci USA. 1999;96:7938–7943. doi: 10.1073/pnas.96.14.7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tognon M, Furlong D, Conley A J, Roizman B. Molecular genetics of herpes simplex virus. V. Characterization of a mutant defective in ability to form plaques at low temperatures and in a viral function which prevents accumulation of coreless capsids at nuclear pores late in infection. J Virol. 1981;40:870–880. doi: 10.1128/jvi.40.3.870-880.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Truant R, Cullen B R. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol Cell Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warner M S, Geraghty R J, Martinez W M, Montgomery R I, Whitbeck J C, Xu R, Eisenberg R J, Cohen G H, Spear P G. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 49.Weis K, Dingwall C, Lamond A. Characterization of the nuclear protein import mechanism using Ran mutants with altered nucleotide binding specificities. EMBO J. 1996;15:7120–7128. [PMC free article] [PubMed] [Google Scholar]
- 50.Whittaker G R, Helenius A. Nuclear import and export of viruses and virus genomes. Virology. 1998;246:1–23. doi: 10.1006/viro.1998.9165. [DOI] [PubMed] [Google Scholar]
- 51.Yamada M, Kasamatsu H. Role of nuclear pore complex in simian virus 40 nuclear targeting. J Virol. 1993;67:119–130. doi: 10.1128/jvi.67.1.119-130.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Z H, Chen D H, Jakana J, Rixon F J, Chiu W. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J Virol. 1999;73:3210–3218. doi: 10.1128/jvi.73.4.3210-3218.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]