ABSTRACT
Acinetobacter baumannii is a nosocomial pathogen that exhibits substantial genomic plasticity. Here, the identification of two variants of A. baumannii ATCC 17978 that differ based on the presence of a 44-kb accessory locus, named AbaAL44 (A. baumannii accessory locus 44 kb), is described. Analyses of existing deposited data suggest that both variants are found in published studies of A. baumannii ATCC 17978 and that American Type Culture Collection (ATCC)-derived laboratory stocks comprise a mix of these two variants. Yet, each variant exhibits distinct interactions with the host in vitro and in vivo. Infection with the variant that harbors AbaAL44 (A. baumannii 17978 UN) results in decreased bacterial burdens and increased neutrophilic lung inflammation in a mouse model of pneumonia, and affects the production of interleukin 1 beta (IL-1β) and IL-10 by infected macrophages. AbaAL44 harbors putative pathogenesis genes, including those predicted to encode a type I pilus cluster, a catalase, and a cardiolipin synthase. The accessory catalase increases A. baumannii resistance to oxidative stress and neutrophil-mediated killing in vitro. The accessory cardiolipin synthase plays a dichotomous role by promoting bacterial uptake and increasing IL-1β production by macrophages, but also by enhancing bacterial resistance to cell envelope stress. Collectively, these findings highlight the phenotypic consequences of the genomic dynamism of A. baumannii through the evolution of two variants of a common type strain with distinct infection-related attributes.
KEYWORDS: Acinetobacter, host-pathogen interactions, pathogenesis, variable phenotypes
INTRODUCTION
Acinetobacter baumannii is a Gram-negative, opportunistic pathogen that is a common cause of infections such as pneumonia, wound infections, and sepsis (1–3). Infections with A. baumannii are often severe, and mortality rates associated with A. baumannii pneumonia are as high as 60% (4, 5). Further complicating infections with A. baumannii is the emergence of multidrug resistant (MDR) isolates. Isolates resistant to aminoglycosides (6–8), carbapenems (9–12), and the last-resort antibiotic colistin (13–17) have emerged as the causative agents of human disease over the last few decades. Panresistant strains of A. baumannii that are resistant to all clinically available antimicrobials are also encountered at an increased frequency (18, 19). Because of this, the Centers for Disease Control and Prevention have indicated carbapenem-resistant A. baumannii as an urgent threat (20). Studies of antibiotic-resistant A. baumannii primarily rely on clinical isolates (21, 22), whereas studies of bacterial pathogenesis and infection biology often rely on type strains. Type strains are descendants of the original isolates that exhibit all of the relevant phenotypic and genotypic properties cited in the original published taxonomic circumscriptions (23). Strains 17978, 19606, and AB5075 are examples of type strains used to study A. baumannii pathogenesis. A. baumannii 17978 was isolated in 1951 from an infant with meningitis and is susceptible to most antibiotics (24), A. baumannii 19606 was isolated in 1948 from the urine of a patient with a urinary tract infection (25), and A. baumannii AB5075 was isolated in 2008 from the tibia of a patient with osteomyelitis (26). These type strains have been used to identify A. baumannii genes required for persistence in the lung and to study A. baumannii virulence factors (26–33).
Strains of A. baumannii exhibit genomic plasticity through the incorporation and loss of genetic material (2, 34), underscored by genomic analyses of A. baumannii that have revealed an unusually large number of “singleton” genes that are unique to a given strain of A. baumannii and do not occur in other strains (35). The majority of exogenous genetic material incorporated by A. baumannii constitutes antibiotic resistance genes (36–38), selected for in part by the use of antimicrobials in health care settings (39, 40). This suggests that gene acquisition contributes to the emergence of MDR isolates of A. baumannii. Gene loss also contributes to differential patterns of antimicrobial resistance among A. baumannii, as a partial deletion of Tn1548 in A. baumannii ABUH315100 resulted in the deletion of the armA amikacin resistance gene (21). A. baumannii genomic plasticity also facilitates changes that affect virulence and interactions with its environment, including competing bacteria and health care settings (36). For instance, disruption of the gtr6 gene by spontaneous transposon (Tn) insertion eliminates a branch point in the capsular carbohydrate structure of the clinical isolate HUMC1 and renders it hypervirulent in a mouse model of bacteremia (41). Furthermore, several MDR strains of A. baumannii have acquired the plasmid pAB3 and related plasmids, which harbor multiple antibiotic resistance genes and encode repressors of the A. baumannii type 6 secretion system (T6SS), which can be used to directly kill competing bacteria (42, 43). Other clinical isolates of A. baumannii have lost genes encoding the T6SS (38, 44). This suggests that subpopulations of A. baumannii that have lost T6SS genes or harbor pAB3 have a competitive advantage in environments where antibiotics are present, such as health care facilities. In contrast, subpopulations of A. baumannii that harbor T6SS genes and lack pAB3 are able to actively attack competing bacteria.
Here, the identification of two variants of A. baumannii ATCC 17978 is described. These variants have been used interchangeably in pathogenesis studies but differ by the presence of an accessory locus and have unique attributes when interacting with host factors both in vitro and in vivo.
RESULTS
Two common variants of A. baumannii ATCC 17978 differ by the presence of an accessory genetic locus and by multiple single-nucleotide polymorphisms.
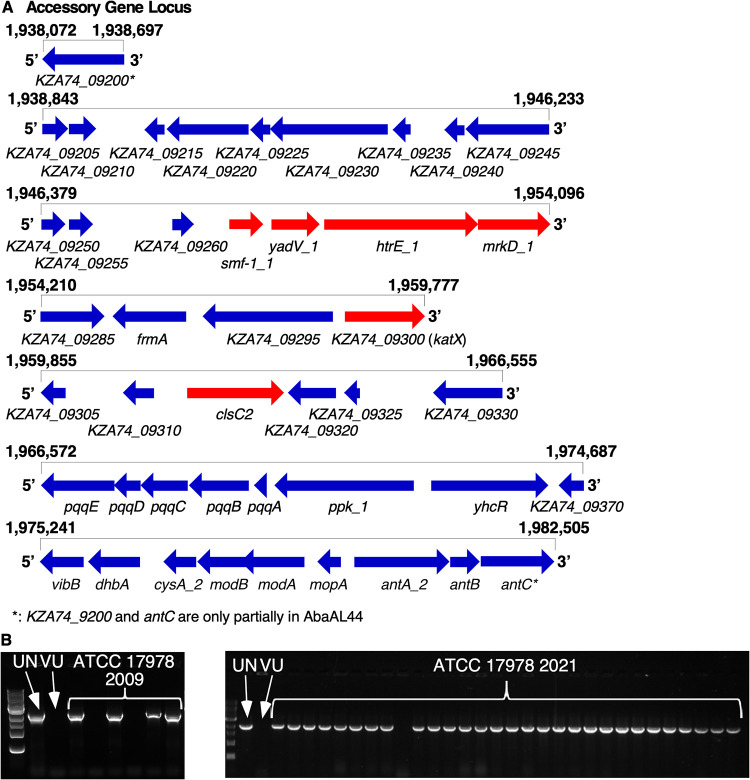
During investigations into the genetic determinants of A. baumannii pathogenesis using a Tn5 transposon mutant library of A. baumannii 17978, conserved phenotypes were observed among several transposon (Tn) mutants containing insertions into unrelated genes. In a macrophage model of A. baumannii infection, murine bone marrow-derived macrophages (BMDMs) produced differential quantities of the proinflammatory cytokine IL-1β upon infection with multiple Tn mutants than when infected with wild-type (WT) A. baumannii 17978 (see Fig. S1A in the supplemental material). In addition, phenotypic profiling of these mutants using scanning electron microscopy (SEM) revealed the presence of pili on the surface of multiple Tn mutants but not on the WT strain (Fig. S1B and C). As the Tn5 library had been constructed at a different institution, the finding of conserved phenotypes among multiple Tn insertion mutants raised concern that the parental A. baumannii 17978 variant used for library construction differed from the A. baumannii 17978 variant used as a comparator for the BMDM infection and SEM experiments. To test this hypothesis, the genome sequences of the parental variant used for library construction (A. baumannii 17978 UN) and of the variant used as the comparator in the BMDM infection and SEM assays (A. baumannii 17978 VU) were determined by PacBio sequencing (45). Structural comparison of these genomes revealed several differences, including differences in the size of the genomes, as well as different locations of an IS701-like element within each genome (Table 1). Notably, the genome of A. baumannii 17978 UN contained a 44-kb locus, encompassing multiple accessory genes, that was not present in the genome of A. baumannii 17978 VU (Table 1, Fig. 1A, and Table S2). This locus was named AbaAL44 (Acinetobacter baumannii accessory locus 44 kb). The accessory genes present within AbaAL44 include putative biosynthesis genes, transcriptional regulators, and genes associated with replication, as well as several putative bacterial pathogenesis genes. AbaAL44 includes smf-1, yadV, htrE, and mrkD, which together are predicted to encode a type I pilus or fimbriae; a gene predicted to encode a catalase (KZA74_09300); and a gene predicted to encode a cardiolipin synthase (clsC2) (Fig. 1A and Table S2). Additional genetic differences between A. baumannii 17978 UN and 17978 VU variants including single-nucleotide polymorphisms (SNPs), are shown in Table S3.
TABLE 1.
Summary data for two variants of A. baumannii 17978a
| Characteristic or descriptor | Variant of A. baumannii 17978 |
|
|---|---|---|
| VU | UN | |
| Accession no. for NCBI reference genome(s), chromosome (source or reference) | NZ_CP018664 (104); CP012004 (42) | CP079931 (this study) |
| UIC lab stock composition (no. of colonies/total no. of colonies)b | 1/36 | 35/36 |
| VUMC lab stock composition (no. of colonies/total no. of colonies)c | 2/6 | 4/6 |
| No. of bases | 4,066,914 | 4,100,908 |
| No. of genes | 3,910 | 3,938 |
| Avg nucleotide identity (%) | 99.99 | 99.99 |
| AbaAL44 | Absent | Present |
| IS701-like ISAba11 family transposase upstream of pitA_2 | Absent | Present |
| ISAba18 downstream of ACX60_00585 | Present | Absent |
Single-nucleotide polymorphisms are shown in Table S3 in the supplemental material.
ATCC 2021.
ATCC 2009.
FIG 1.
PacBio sequencing reveals structural differences between the genomes of Acinetobacter baumanii 17978 VU and 17978 UN variants. The genome sequences of A. baumanii 17978 VU and 17978 UN variants were determined by PacBio and Nanopore sequencing and subsequently compared to the genome of the reference strain, A. baumannii ATCC 17978-mff (RefSeq accession number NZ_CP012004.1). (A) Visual representation of AbaAL44, encompassing multiple accessory genes present in the genome of A. baumannii 17978 UN, based on analysis and inspection of a new A. baumannii 17978 UN reference genome (NCBI accession number CP079931). The relative position (in terms of base pair number) within the assembled genome of A. baumannii 17978 UN is indicated above each row, and gene names are indicated below each gene. Locus tags refer to CP079931. Arrows indicate the relative transcriptional direction of each gene. Genes indicated in red represent putative pathogenesis genes. (B) Images of agarose gel electrophoresis indicating the differential presence of the accessory clsC2 gene within individual colonies of A. baumannii 17978 laboratory stocks based on PCR detection of clsC2. The VUMC laboratory stock (left) was obtained from ATCC in 2009, and the UIC laboratory stock (right) was obtained from ATCC in 2021. Presence of accessory clsC2 indicates that an isolate belongs to A. baumannii 17978 UN variant.
The identification of two variants of A. baumannii ATCC 17978 within a laboratory raised the possibility that these two variants are present in other research laboratories. AbaAL44 is present in the A. baumannii 17978 genome published by the American Type Culture Collection (ATCC) in 2019 (https://genomes.atcc.org/genomes/e1d18ea4273549a0), but absent in two other A. baumannii 17978 reference genomes available in the NCBI database (accession numbers NZ_CP018664 and CP012004; see Table 1), indicating that both A. baumannii 17978 VU and 17978 UN variants are represented among the published A. baumannii genomes. Frozen stocks of A. baumannii ATCC 17978 obtained from ATCC in 2009 and a different stock obtained in 2021 were streaked for individual colonies, which were screened using PCR for the presence of the accessory cardiolipin synthase gene (clsC2) included in AbaAL44. The accessory gene clsC2 was present in 4 out of 6 colonies screened from the 2009 laboratory stock and in 35 out of 36 colonies screened from the 2021 laboratory stock, demonstrating that these culture collection stocks included a mixed population of A. baumannii 17978 variants VU and UN (Table 1 and Fig. 1B). To provide a high-quality reference genome, we performed Nanopore long-read sequencing on A. baumannii 17978 UN, isolated from an ATCC stock in 2021 (NCBI accession numbers CP079931, CP079932, CP079933, and CP079934; see Table 1). Illumina sequencing of A. baumannii 17978 UN isolates from 2009 and 2021 laboratory stocks found no predicted mutations compared to this new 17978 UN reference genome. These data indicate that the A. baumannii 17978 UN and 17978 VU variants differ from one another at a genetic level and that at least two ATCC-derived laboratory stocks of A. baumannii 17978 include these variants.
A. baumannii 17978 VU and 17978 UN variants exhibit differential pathogenicity in a mouse model of pneumonia.
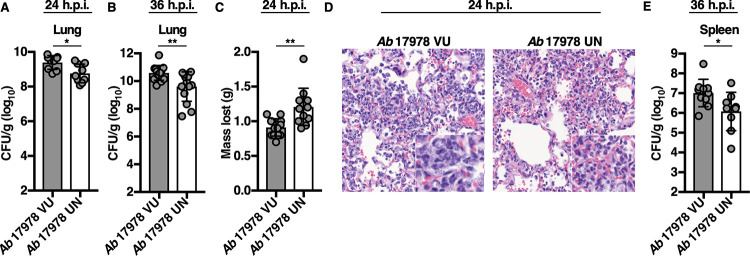
A. baumannii 17978 has been used extensively in studies of A. baumannii pathogenesis (30–32). To determine whether A. baumannii 17978 VU and 17978 UN variants exhibit differential fitness in vivo, both variants were assessed in a murine model of pneumonia, and bacterial burdens in the lungs and spleens of infected mice were compared at 24 and 36 h postinfection (h.p.i.). Mice infected with A. baumannii 17978 UN had a statistically significant decrease in lung bacterial burdens at both 24 and 36 h.p.i. compared to those of mice infected with A. baumannii 17978 VU (Fig. 2A and B). At 24 h.p.i., mice infected with A. baumannii 17978 UN had lost significantly more weight (Fig. 2C), suggesting an increase in morbidity relative to that of mice infected with A. baumannii 17978 VU. Histological examination of lungs from mice infected with A. baumannii 17978 UN revealed fewer visible bacteria within the alveolar spaces, and mice infected with A. baumannii 17978 UN exhibited reduced extrapulmonary dissemination to the spleen relative to that in mice infected with A. baumannii 17978 VU (Fig. 2D and E). Combined, these data suggest that A. baumannii 17978 UN exhibits decreased fitness in a murine model of pneumonia compared to that of A. baumannii 17978 VU.
FIG 2.
A. baumannii 17978 VU and 17978 UN variants exhibit differential virulence in a mouse model of pneumonia. Mice were challenged intranasally with 3 × 108 CFU of mid-log-phase A. baumannii 17978 VU or 17978 UN variants. At 24 (A and C) or 36 (B and E) h.p.i., mice were euthanized, and organs were harvested. (A and B) Bacterial burdens in the lungs of mice at 24 and 36 h.p.i., respectively. (C) Mean number of grams of total body weight lost by infected mice at 24 h.p.i. (D) Representative hematoxylin and eosin-stained lung sections of mice infected with A. baumannii 17978 VU (left) or A. baumannii 17978 UN (right) at 24 h.p.i. (E) Bacterial burdens in the spleens of mice at 36 h.p.i. (A, B, C, and E) Circles represent individual animals, columns depict the mean, and error bars show standard deviation of the mean. Means were compared using an unpaired Welch’s t test. CFU/g, CFU per gram of organ homogenate. *, P < 0.05; **, P < 0.01. H&E, hematoxylin and eosin.
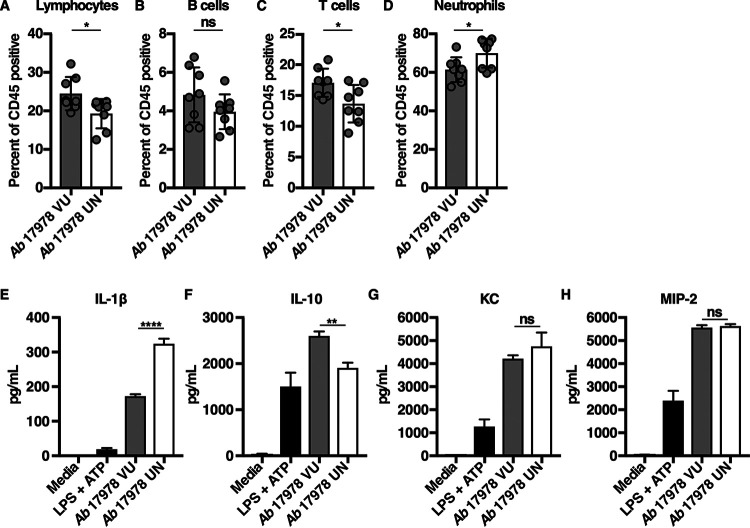
Mice infected with A. baumannii 17978 UN lost more weight than mice infected with A. baumannii 17978 VU (Fig. 2C), which could indicate inappetence or increased metabolic demand due to increased inflammation. Neutrophilic inflammation is a characteristic response to A. baumannii infection and is essential for host resistance to infection (46–48). To determine if A. baumannii 17978 VU and 17978 UN variants induce differential neutrophil recruitment, mice were challenged with either variant, and the relative abundances of immune cell populations in both the lungs and blood were determined. Relative to mice infected with A. baumannii 17978 VU, mice infected with A. baumannii 17978 UN exhibited a decrease in the abundance of T cells, B cells, and overall lymphocytes, and a statistically significant increase in the relative abundance of neutrophils in the lungs (Fig. 3A to D). However, no significant differences in the number or percentage of neutrophils or lymphocytes were detected in the blood of mice infected with A. baumannii 17978 VU relative to those in mice infected with A. baumannii 17978 UN (Fig. S2). These data demonstrate that A. baumannii 17978 UN promotes a shift toward neutrophilic inflammation upon pneumonic infection of mice relative to that in A. baumannii 17978 VU.
FIG 3.
Mice infected with A. baumannii 17978 UN exhibit increased neutrophilic inflammation in comparison to that of mice infected with A. baumannii 17978 VU. (A to D) Mice were challenged intranasally with 3 × 108 CFU of mid-log-phase A. baumannii 17978 VU or 17978 UN. At 24 h.p.i., mice were euthanized, lungs were harvested, and immune cell recruitment to the lungs was quantified using flow cytometry analysis. (E to H) Murine bone marrow-derived macrophages (BMDMs) were infected with mid-log-phase cultures of A. baumannii 17978 VU or 17978 UN at a multiplicity of infection (MOI) of 10 and incubated at 37°C. At 18 h.p.i., supernatants of infected BMDMs were collected, and the concentrations of IL-1β (E), interleukin 10 (IL-10) (F), KC (G), or MIP-2 (H) in the supernatants of infected BMDMs were determined by enzyme-limited immunosorbent assay (ELISA). (A to D) Circles represent individual animals, columns depict the mean, and error bars show standard deviation of the mean. Means were compared using an unpaired Welch’s t test. (E to H) N = 3 or 4 biological replicates per group per experiment. Experiments were repeated for a total of at least two times, with graphs depicting representative data. (E to H) Columns depict the mean, and error bars show standard deviation of the mean. Means were compared to the mean of the A. baumannii 17978 VU column using a one-way analysis of variance (ANOVA) adjusted for multiple comparisons. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, not significant.
Neutrophil recruitment to the site of infection is dependent upon the production of cytokines and chemokines. Interleukin 1 beta (IL-1β) is a potent proinflammatory cytokine that promotes neutrophil recruitment and stimulates neutrophil survival (49–52), whereas IL-10 is an anti-inflammatory cytokine that impedes neutrophil recruitment to the site of infection (53, 54). To test the hypothesis that infection with A. baumannii 17978 UN potentiates neutrophilic lung inflammation by inducing differential cytokine or chemokine production by host cells, murine bone marrow-derived macrophages (BMDMs) were infected with A. baumannii 17978 VU or 17978 UN variants, and cytokines and chemokines were quantified from cell culture supernatants. BMDMs infected with A. baumannii 17978 UN produced significantly more IL-1β and less IL-10 compared to that produced by BMDMs infected with A. baumannii 17978 VU (Fig. 3E and F). In contrast, the production of the murine neutrophil chemokines KC and MIP-2 did not differ between BMDMs infected with A. baumannii 17978 UN and 17978 VU variants (Fig. 3G and H). No differences in serum or lung IL-1β levels were detected at 24 h.p.i. between mice infected with A. baumannii 17978 VU and 17978 UN variants (Fig. S3). These data demonstrate that host responses to two common variants of A. baumannii 17978 differ in terms of induced cytokine production by macrophages, as well as in the degree of induced neutrophilic lung inflammation, bacterial persistence in the lung, and dissemination to the spleen in a mouse model of pneumonia.
The AbaAL44 katX gene promotes bacterial resistance to hydrogen peroxide stress and neutrophil-mediated killing.
To interrogate the role of putative pathogenesis genes contained within AbaAL44 in the differential pathogenicity of A. baumannii 17978 UN, the candidate accessory pilus assembly locus comprising smf-1, yadV, htrE, and mrkD; the accessory catalase gene (KZA74_09300; katX); or the accessory cardiolipin synthase gene (clsC2) was replaced with a kanamycin resistance cassette to create A. baumannii 17978 UN Δsmf-1_1-mrkD_1::kan (A. baumannii 17978 UN Δpilus), 17978 UN ΔKZA74_09300::kan (17978 UN ΔkatX), and 17978 UN ΔclsC2::kan (17978 UN ΔclsC2) strains, respectively (55). Type I pili are bacterial surface appendages involved in bacterial uptake by macrophages, biofilm formation, attachment to host cells, motility, and agglutination of red blood cells (56–59). However, the A. baumannii 17978 UN Δpilus strain did not differ from A. baumannii 17978 UN in terms of bacterial uptake by macrophage-like RAW 264.7 cells (Fig. S4A), biofilm formation (Fig. S4B), bacterial attachment to A549 epithelial cells (Fig. S4C), bacterial surface-associated motility (Fig. S4D), hemagglutination of human erythrocytes (Fig. S4E), or differential production of cytokines by infected BMDMs (Fig. S4F and G). These data suggest that the type I pilus locus does not contribute to the interactions with host cells under these in vitro conditions.
Catalases are enzymes that catalyze the decomposition of hydrogen peroxide (H2O2) to water and oxygen and thereby protect bacteria from reactive oxygen species (ROS)-mediated oxidative stress and killing by innate immune cells (60–62). To test the hypothesis that the AbaAL44 accessory catalase present in A. baumannii 17978 UN contributes to bacterial resistance to oxidative stress, the relative susceptibilities of A. baumannii 17978 VU, 17978 UN, 17978 UN ΔkatX/empty vector (EV), and 17978 UN ΔkatX in which the AbaAL44 katX gene was expressed in trans (A. baumannii 17978 UN ΔkatX/pkatX) to hydrogen peroxide stress in vitro was determined. Thirty minutes after incubation of 1 × 107 CFU/ml of bacteria in the presence of 1 mM hydrogen peroxide, the density of viable A. baumannii 17978 UN bacteria remained virtually unchanged at approximately 7 log10 CFU/ml. In contrast, the densities of viable A. baumannii 17978 VU and 17978 UN ΔkatX/EV bacteria were significantly reduced to approximately 5.5 log10 CFU/ml. Expression of the AbaAL44 katX in trans restored hydrogen peroxide stress resistance in the A. baumannii 17978 UN ΔkatX/pkatX strain to levels approximating those in the parental strain (Fig. 4A). These data indicate that the katX present in AbaAL44 enhances bacterial resistance to oxidative stress in vitro. Neutrophils are innate immune effector cells that phagocytose and subsequently kill engulfed pathogens using an array of antimicrobial molecules, including ROS produced by the enzyme NADP (NADPH) oxidase (63, 64). Based on this, we hypothesized that the katX present in A. baumannii 17978 UN promotes bacterial resistance to neutrophil-mediated killing. To test this, differentiated neutrophil-like HL-60 cells were incubated with A. baumannii 17978 UN, 17978 VU, 17978 UN ΔkatX/EV, or 17978 UN ΔkatX/pkatX strains and bacterial resistance to neutrophil-mediated extracellular and intracellular killing was determined by measuring relative bacterial survival postincubation. After incubation in the presence of differentiated neutrophil-like HL-60 cells, relative survival of A. baumannii 17978 UN was significantly greater than that of both the 17978 VU and 17978 UN ΔkatX/EV strains (Fig. 4B). Similar results were obtained with murine bone marrow-derived neutrophils (Fig. S5). In trans expression of AbaAL44 katX restored survival of the A. baumannii 17978 UN ΔkatX/EV strain to the levels of the parental strain (Fig. 4B). Deletion of the AbaAL44 katX gene negatively affected the growth of A. baumannii 17978 UN in vitro in both rich (lysogeny broth [LB]) and minimal (Tris minimal succinate [TMS]) media, which was restored by expressing AbaAL44 katX in trans (Fig. 5). Taken together, these data suggest that the katX present in AbaAL44 contributes to A. baumannii 17978 UN growth in vitro, as well as to bacterial resistance to oxidative stress and neutrophil-mediated killing.
FIG 4.

The AbaAL44 katX gene promotes bacterial resistance to hydrogen peroxide stress and neutrophil-mediated killing. (A) Mid-log-phase A. baumannii 17978 VU, 17978 UN, 17978 UN ΔkatX/EV, or 17978 UN ΔkatX/pkatX variants were incubated in LB medium supplemented with 1 mM hydrogen peroxide at a starting inoculum of 1 × 107 CFU/ml. Following a 30-min incubation, remaining viable bacteria were enumerated. (B) Differentiated neutrophil-like HL-60 cells were incubated with mid-log-phase A. baumannii 17978 VU, 17978 UN, 17978 UN ΔkatX/EV, or 17978 UN ΔkatX/pkatX variants at an MOI of 2. At 90 min postincubation, HL-60 cells were lysed, and neutrophil-mediated killing of bacteria was assessed by determining the number of viable bacteria in each well. Data are represented as bacterial survival relative to that for A. baumannii 17978 VU. In both panels, N = 3 or 4 biological replicates per group per experiment. Experiments were repeated for a total of at least two times, with graphs depicting average data. Columns depict the mean, and error bars show standard deviation of the mean. All means were compared to all other means using a one-way ANOVA adjusted for multiple comparisons. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
FIG 5.
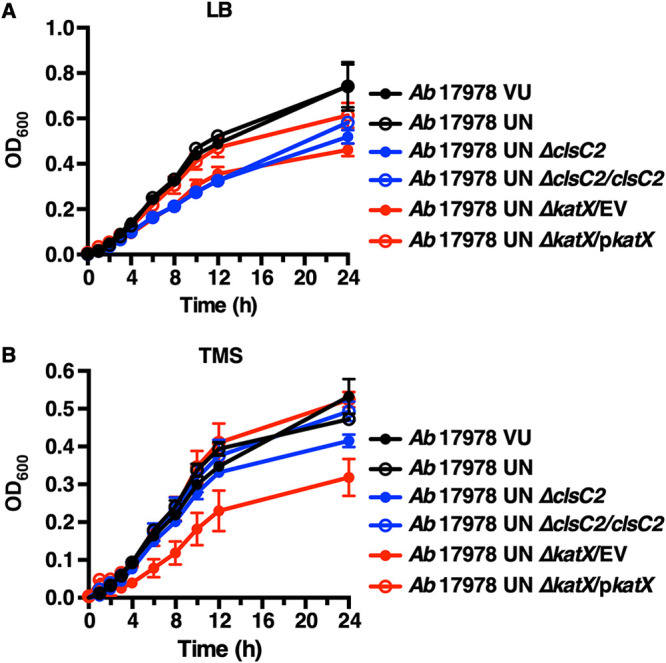
AbaAL44 clsC2 and katX mutants generated in the A. baumannii 17978 UN background exhibit growth defects in vitro. (A and B) Stationary-phase cultures of the indicated strains of A. baumannii were normalized to a similar optical density at 600 nm (OD600) and inoculated into fresh rich medium (LB) or minimal medium (Tris minimal succinate [TMS]). Growth was assayed by measuring OD600 over time. Symbols depict the mean, and error bars show standard deviation of the mean. N = 3 or 4 biological replicates per group per experiment. Graphs depict representative data from two independent experiments. For 8-, 10-, and 12-h time points, the means of A. baumannii 17978 UN ΔclsC2, 17978 UN ΔclsC2/clsC2, 17978 UN ΔkatX/EV, 17978 UN ΔkatX/pkatX, and 17978 UN variants were compared using a one-way ANOVA adjusted for multiple comparisons. (A) Differences between A. baumannii 17978 UN and 17978 UN ΔclsC2, 17978 UN ΔclsC2/clsC2, or 17978 UN ΔkatX/EV variants: P < 0.0001 for each comparison for each time point. Difference between A. baumannii 17978 UN and 17978 UN ΔkatX/pkatX variants: P < 0.05 for the10-h time point and P > 0.05 for 8- and 12-h time points. (B) Differences between A. baumannii 17978 UN and 17978 UN ΔclsC2, 17978 UN ΔclsC2/clsC2, or 17978 UN ΔkatX/pkatX variants: P > 0.05 for each comparison for each time point. Difference between A. baumannii 17978 UN and 17978 UN ΔkatX/EV variants: P < 0.0001, P < 0.0001, or P < 0.001 for 8-, 10-, and 12-h time points, respectively.
AbaAL44 clsC2 contributes to bacterial resistance to cell envelope stress and affects bacterial interactions with host immune cells.
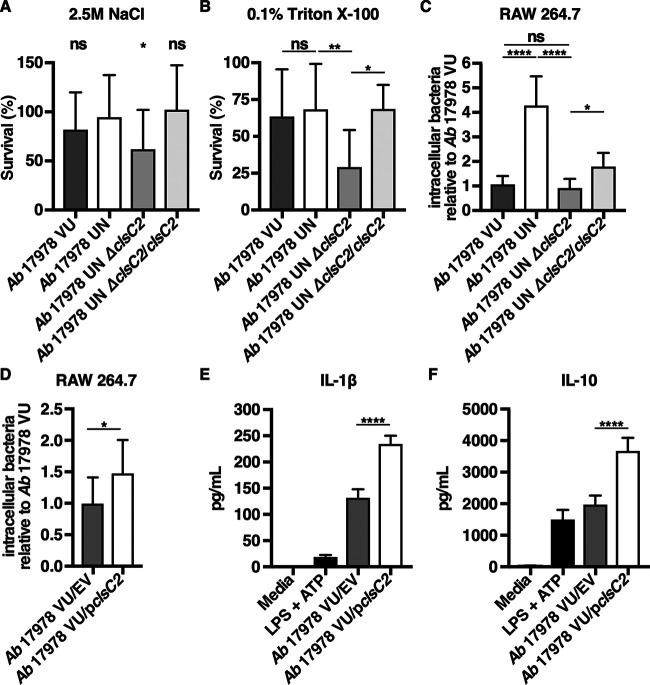
Bacterial cardiolipin synthases are enzymes that utilize phosphatidylglycerol (PG) and/or phosphatidylethanolamine (PE) as substrates to catalyze the formation of cardiolipin (65, 66). Cardiolipin is one of several anionic phospholipids that comprise the Gram-negative cell envelope, and, in Escherichia coli, it has been demonstrated to localize to the cellular poles (67). Because cardiolipin promotes bacterial resistance to envelope stressors such as high salinity (68, 69), it was hypothesized that A. baumannii 17978 UN is more resistant to conditions of high salinity than the A. baumannii 17978 UN ΔclsC2 strain. To test this, the susceptibility to conditions of high salinity of A. baumannii 17978 VU, 17978 UN, 17978 UN ΔclsC2, and 17978 UN ΔclsC2 in which clsC2 has been reintroduced chromosomally under its native promoter by mini-Tn7 integration (A. baumannii 17978 UN ΔclsC2/clsC2) was determined by comparing bacterial survival after incubation in LB supplemented with 2.5 M NaCl. After 2 h of incubation in high salinity, bacterial survival of the A. baumannii 17978 UN ΔclsC2 strain was significantly reduced by approximately 30% compared to the survival of 17978 UN. After incubation in high salinity, survival of the A. baumannii 17978 UN ΔclsC2/clsC2 strain did not differ significantly from that of 17978 UN (Fig. 6A). Perturbations in bacterial membrane phospholipid homeostasis involving cardiolipin have been demonstrated to increase bacterial susceptibility to molecular detergents such as surfactants, which are found in high concentrations in the lungs and may be relevant to bacterial pneumonia pathogenesis (70). To determine if the AbaAL44 clsC2 present in A. baumannii 17978 UN promotes bacterial resistance to detergents, the susceptibility of A. baumannii 17978 VU, 17978 UN, 17978 UN ΔclsC2, and 17978 UN ΔclsC2/clsC2 variants to the detergent Triton X-100 was assessed. After 6 h of incubation in 0.1% Triton X-100, bacterial survival of A. baumannii 17978 UN ΔclsC2 was significantly less than that of 17978 UN. Survival of A. baumannii 17978 UN ΔclsC2/clsC2 was significantly greater than that of 17978 UN ΔclsC2, and similar to that of 17978 UN (Fig. 6B). Taken together, these data indicate that the accessory cardiolipin synthase gene present in A. baumannii 17978 UN contributes to this strain’s resistance to conditions of high salinity and detergent stress.
FIG 6.
The AbaAL44 clsC2 gene contributes to bacterial resistance to cell envelope stress and affects bacterial interactions with host immune cells. (A) Mid-log-phase A. baumannii 17978 VU, 17978 UN, 17978 UN ΔclsC2, or 17978 UN ΔclsC2/clsC2 variants were inoculated into LB medium supplemented with 2.5 M NaCl at a starting inoculum of 1 × 107 CFU/ml, and incubated at 37°C for 2 h. After 2 h of incubation, bacterial viability was determined for each strain, and bacterial survival was calculated as the percentage of viable bacteria postincubation relative to the number of viable bacteria preincubation. (B) Mid-log-phase A. baumannii 17978 VU, 17978 UN, 17978 UN ΔclsC2, or 17978 UN ΔclsC2/clsC2 variants were inoculated into phosphate-buffered saline (PBS) supplemented with 0.1% Triton X-100 at a starting inoculum of 1 × 1010 CFU/ml and incubated at 37°C for 6 h. After incubation, bacterial viability was determined for each strain, and survival was calculated as described above. (C and D) Macrophage-like RAW 264.7 cells were infected with mid-log-phase A. baumannii 17978 VU, 17978 UN, 17978 UN ΔclsC2, 17978 UN ΔclsC2/clsC2, 17978 VU/EV, or 17978 VU/pclsC2 at an MOI of 15, and extracellular bacteria were killed with gentamicin at 30 min postinfection. Thirty minutes after the addition of gentamicin, RAW cells were washed and lysed, and intracellular bacterial burdens were determined. (E and F) Murine bone marrow-derived macrophages were infected with mid-log-phase cultures of A. baumannii 17978 VU/EV or A. baumannii 17978 VU/pclsC2 at an MOI of 10 and incubated at 37°C. At 18 h.p.i., supernatants of infected BMDMs were collected, and the concentrations of IL-1β (E) or IL-10 (F) in the supernatants of infected BMDMs were determined by ELISA. (A to D) N = 3 or 4 biological replicates per group per experiment. Graphs depict average results from at least three independent experiments. (E and F) N = 4 biological replicates per group per experiment. Graphs depict representative data. In all panels, columns depict the mean, and error bars show standard deviation of the mean. Means were compared to the mean of A. baumannii 17978 UN (A) or to all other means (B to F) using Welch’s t test (D) or one-way ANOVA adjusted for multiple comparisons (A, B, C, E and F). *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, not significant.
Previous work has demonstrated that cardiolipin promotes uptake of bacteria by professional phagocytes such as macrophages through the scavenger receptor CD36 (71). Therefore, it was hypothesized that the previously observed increase in intracellular bacterial burden in macrophage-like RAW cells infected with A. baumannii 17978 UN is mediated by the AbaAL44 clsC2 (Fig. S4A). To test this, the relative intracellular bacterial burdens of RAW cells infected with A. baumannii 17978 VU, 17978 UN, 17978 UN ΔclsC2, or 17978 UN ΔclsC2/clsC2 variants were determined. The intracellular bacterial burden of RAW cells infected with the A. baumannii 17978 UN ΔclsC2 variant was significantly reduced by approximately 75% relative to the intracellular bacterial burden of RAW cells infected with A. baumannii 17978 UN and was similar to the intracellular bacterial burden of RAW cells infected with 17978 VU. There was a small but statistically significant increase in the intracellular bacterial burden of RAW cells infected with the A. baumannii 17978 UN ΔclsC2/clsC2 variant relative to that of RAW cells infected with the 17978 UN ΔclsC2 variant (Fig. 6C). Expression of clsC2 in trans in A. baumannii 17978 VU (A. baumannii 17978 VU/pclsC2) also significantly increased the intracellular bacterial burden of infected RAW cells compared to that of RAW cells infected with A. baumannii 17978 VU/EV (Fig. 6D). These data suggest that the AbaAL44 clsC2 gene contributes to A. baumannii 17978 UN phagocytosis by macrophages.
Chromosomal reintroduction of clsC2 in the A. baumannii 17978 UN ΔclsC2 variant did not fully restore phagocytosis by infected RAW cells to the levels observed with A. baumannii 17978 UN (Fig. 6C). The deletion of clsC2 negatively affected the growth of A. baumannii 17978 UN in vitro in rich medium (LB), but not in minimal medium (TMS) (Fig. 5). The growth defect of A. baumannii 17978 UN ΔclsC2 in rich medium was also not restored by chromosomal reintegration of clsC2 under its native promoter (Fig. 5A). Furthermore, the supernatants of RAW cells infected with the A. baumannii 17978 UN ΔclsC2 or 17978 UN ΔclsC2/clsC2 variants contained small, punctate colonies (Fig. S6B). These colonies were also present in lysates of RAW cells infected with A. baumannii 17978 UN ΔclsC2 or 17978 UN ΔclsC2/clsC2 variants that were collected prior to treatment with gentamicin, but were absent from the starting inoculums (Fig. S6A and C). Small, punctate colonies were not observed in any samples collected from RAW cells infected with the A. baumannii 17978 UN parental strain (Fig. S6). These findings suggest a reduction in the fitness of the A. baumannii 17978 UN ΔclsC2 strain that is not restored by integration of clsC2 elsewhere on the chromosome and are indicative of another genetic difference between the parental A. baumannii 17978 UN and the ΔclsC2 derivative or polar effects of the inserted kanamycin cassette. As expression of clsC2 in A. baumannii 17978 VU resulted in an increase in macrophage phagocytosis, the differential growth phenotype between the 17978 UN and 17978 UN ΔclsC2 variants was not pursued further.
Mitochondrial cardiolipin activates the NLRP3 inflammasome, resulting in the production of IL-1β, and saturated cardiolipins increase IL-1β production through Toll-like receptor 4 (TLR4) signaling (72, 73). This raised the hypothesis that the increase in IL-1β production observed in macrophages infected with A. baumannii 17978 UN is due to the presence of AbaAL44 clsC2. Given the reduced fitness of the ΔclsC2 mutant in the A. baumannii 17978 UN background, this hypothesis was tested by expressing the AbaAL44 clsC2 gene in trans in 17978 VU, which naturally lacks AbaAL44 and thus clsC2. BMDMs were infected with A. baumannii 17978 VU/EV or 17978/pclsC2, and the concentrations of proinflammatory IL-1β and anti-inflammatory IL-10 in the supernatants of infected BMDMs were determined at 18 h.p.i. Relative to infection with A. baumannii 17978 VU/EV, infection with 17978 VU/pclsC2 significantly increased the production of IL-1β by infected BMDMs (Fig. 6E). These findings indicate that the presence of AbaAL44 clsC2 increases proinflammatory IL-1β production by macrophages infected with A. baumannii 17978 UN. Infection with A. baumannii 17978 UN decreases the production of IL-10 by infected macrophages relative to that after infection with 17978 VU (Fig. 3F). In contrast, expression of clsC2 in A. baumannii 17978 VU significantly increased IL-10 production by infected macrophages (Fig. 6F). This finding suggests that the decrease in IL-10 production observed in macrophages infected with A. baumannii 17978 UN cannot be explained by the presence of clsC2 alone. Together, these data indicate that AbaAL44 influences the interactions between A. baumannii and the host.
DISCUSSION
The process by which A. baumannii acquires or loses resistance to antimicrobials from gain or loss of genetic material is well characterized (21, 35). In contrast, less is known about how such genomic plasticity affects A. baumannii pathogenicity. Here, the discovery that two variants of A. baumannii ATCC 17978 differ based on the presence of the AbaAL44 accessory locus encompassing several pathogenesis genes is described. Review of the 5,301 publicly available A. baumannii genome assemblies in the NCBI database revealed that genetic elements similar to AbaAL44 are present in at least 100 strains of A. baumannii. Of the 8 A. baumannii ATCC 17978 genome sequences published in NCBI, AbaAL44 is not present in any of them, but AbaAL44 is present in the A. baumannii 17978 genome sequence published by the ATCC in 2019 (https://genomes.atcc.org/genomes/e1d18ea4273549a0). However, if laboratories assemble sequenced genomes of their A. baumannii 17978 laboratory stocks to a reference genome that does not include AbaAL44 (e.g., NCBI accession number NZ_CP018664), the presence of AbaAL44 in these sequenced genomes would not be detected. When combined with the data presented here, these findings suggest that the differential presence of AbaAL44 is a common structural genetic variant among A. baumannii strains and that laboratories studying A. baumannii ATCC 17978 may be working with either variant without awareness or may be working with cultures containing a mix of the two variants. The differential presence of AbaAL44 among two variants of a type strain is most likely due to loss of AbaAL44 by A. baumannii 17978 VU during the course of laboratory propagation. Adaptive loss of genetic material has been previously described for Acinetobacter spp. (74). Furthermore, genetic differences among lineages of a given laboratory strain have previously been demonstrated for other species. Continuous maintenance and propagation of Pseudomonas aeruginosa strain PAO1 in laboratories throughout the world has led to substantial genotypic differences among sublines of this strain (75), and two lineages of Clostridioides difficile R20291 differ based on a small number of single-nucleotide genomic changes, which lead to distinct phenotypic differences (76).
A. baumannii 17978 VU and 17978 UN variants exhibit differential fitness in a mouse model of pneumonia, which is likely due in part to the differential presence of AbaAL44. Putative pathogenesis genes present in AbaAL44, including the katX and clsC2 genes, increase A. baumannii 17978 UN resistance to oxidative stress, neutrophil-mediated killing, and surfactant stress. A. baumannii encounters hydrogen peroxide, the substrate of catalase enzymes, and surfactant inside the mouse lung (77, 78), and neutrophils are an essential component of the host immune response to infection with this pathogen (46–48). As such, the presence of accessory katX and clsC2 genes may confer some in vivo advantage to A. baumannii 17978 UN. A. baumannii 17978 UN also potentiates neutrophilic lung inflammation in vivo and increases the production of proinflammatory IL-1β by infected macrophages. This increase in inflammation and neutrophil recruitment to the site of infection may overwhelm any increased bacterial resistance to neutrophil-mediated killing in A. baumannii 17978 UN, resulting in net decreased bacterial survival inside the mouse lung. The effects of other genes contained in AbaAL44 or polymorphisms elsewhere on the chromosome on A. baumannii fitness have not been interrogated but may contribute to the net differences in pathogenesis between these variants.
As IL-1β promotes the recruitment of neutrophils to the site of infection (49), the increased production of proinflammatory IL-1β by macrophages infected with A. baumannii 17978 UN is likely to play a role in the shift toward neutrophilic inflammation observed in infected mice. Although differences in IL-1β concentration at 24 h.p.i. between mice infected with A. baumannii 17978 VU and mice infected with A. baumannii 17978 UN were not observed, such differences may occur earlier in the course of infection, culminating in the differential abundance of neutrophils observed at 24 h.p.i. Mitochondrial cardiolipin is a known activator of the NLRP3 inflammasome resulting in the production of IL-1β, and saturated cardiolipins increase IL-1β production in a TLR4-dependent manner (72, 73). In trans expression of clsC2 in A. baumannii 17978 VU increased IL-1β production by infected macrophages, suggesting that the presence of the AbaAL44 clsC2 gene in A. baumannii 17978 UN may increase cell envelope cardiolipin content, resulting in NLRP3 inflammasome activation and enhanced IL-1β production by macrophages.
Infection with A. baumannii 17978 UN resulted in decreased IL-10 production by infected macrophages relative to that produced during infection with A. baumannii 17978 VU. Mitochondrial cardiolipin inhibits IL-10 production by recruiting a repressor complex to the IL-10 promoter. The reduction in anti-inflammatory IL-10 promotes persistent inflammation in experimental bacterial pneumonia (79, 80). It is conceivable that bacterial cardiolipin similarly inhibits IL-10 production and that modulation of bacterial cardiolipin content through the AbaAL44 clsC2 gene is responsible for modulating macrophage IL-10 production. However, macrophage infection with a strain of A. baumannii 17978 VU in which clsC2 was expressed in trans did not recapitulate this finding. Paradoxically, macrophage infection with A. baumannii 17978 VU/pclsC2 increased IL-10 production. This suggests that the presence of clsC2 alone cannot explain the relative decrease in IL-10 production observed in macrophages infected with A. baumannii 17978 UN. Regulation of IL-10 production by immune cells is complex, and multiple factors affect its expression (81). Bacterial products such as cell wall components and lipopolysaccharide (LPS) increase macrophage IL-10 production through TLR2- and TLR4-mediated signaling, respectively (81, 82). In contrast, interferon gamma (IFN-γ) inhibits IL-10 production (81). IFN-γ production by macrophages is induced by IL-18, which, like IL-1β, is a product of NLRP3 inflammasome activation, and IL-1β production is increased in macrophages infected with A. baumannii 17978 UN (83–85). Therefore, any combination of these factors may account for the differences in IL-10 production between macrophages infected with A. baumannii 17978 UN and macrophages infected with 17978 VU/pclsC2. Other genes contained in AbaAL44 or other genetic differences between the two A. baumannii 17978 variants may also account for the differential cytokine production by macrophages infected with 17978 UN.
This study demonstrates that the AbaAL44 katX and clsC2 genes are advantageous to A. baumannii 17978 UN survival in vitro. Specifically, the AbaAL44 katX gene present in A. baumannii 17978 UN increases bacterial resistance to oxidative stress. In addition to the AbaAL44 katX gene described in this study, two catalase genes—katE and katG—have previously been identified in A. baumannii (86). Increased transcription of katG as a consequence of upstream insertion of ISAba1 has been demonstrated to increase A. baumannii resistance to oxidative stress (87). Taken together, these findings highlight the importance of catalases in A. baumannii pathogenesis and provide an example of how increasing bacterial catalase gene abundance can contribute to A. baumannii pathogenicity.
The findings presented here implicate a dichotomous role for the AbaAL44 clsC2 gene in A. baumannii 17978 UN. While this gene promotes the uptake of A. baumannii 17978 UN by phagocytes, its presence also increases 17978 UN resistance to envelope stressors, such as surfactants. These findings are consistent with previous reports demonstrating that membranes with bacterial cardiolipin on their surfaces promote macrophage phagocytosis through the scavenger receptor CD36 (71) and that bacterial cardiolipin promotes resistance to osmotic and surfactant stress (69, 70). Cardiolipin has also been demonstrated to increase A. baumannii resistance to cationic antimicrobial peptides, another type of envelope stressor, suggesting that increasing cell envelope cardiolipin content is a relevant survival strategy for this pathogen (88). In addition to the accessory clsC2 described in this study, both A. baumannii 17978 VU and 17978 UN variants contain two additional cardiolipin synthase genes. To our knowledge, however, the role of these genes in A. baumannii biology and pathogenesis remains to be fully elucidated. It has previously been reported that A. baumannii cell membranes contain a variety of unique, coexisting cardiolipin species (89). In E. coli, different cardiolipin synthase genes are active during distinct stages of bacterial growth (66). It is possible that the three cardiolipin synthase genes present in A. baumannii may be expressed during different growth phases and/or may be responsible for synthesizing distinct species of cardiolipin. This notion is supported by recent findings demonstrating that individual enzymes of the same family are implicated in the production of either cardiolipin or monolysocardiolipin, an intermediate species in the cardiolipin remodeling pathway, in A. baumannii (88, 90). The in vitro growth defect and reduced uptake by RAW cells observed in the A. baumannii 17978 UN ΔclsC2 variant could not be restored to the levels of the parental strain by chromosomally reintroducing clsC2 under its native promoter. This may be due to unidentified genetic differences between the ΔclsC2 mutant and the parental A. baumannii 17978 UN strain, or due to polar effects of the ΔclsC2::kan deletion-insertion on nearby genes. However, expression of AbaAL44 clsC2 in the Ab 17978 VU strain increased phagocytosis and IL-1β production by macrophages indicating that clsC2 impacts A. baumannii interactions with the host.
Together, these findings indicate that two genotypically and phenotypically distinct variants of the common type strain, A. baumannii ATCC 17978, exist and are indiscriminately used in published studies of A. baumannii (91). As such, careful genotyping and phenotyping of A. baumannii type strains by individual laboratories may be warranted. Furthermore, this study demonstrates how A. baumannii genomic plasticity can affect pathogenicity and interactions with the host.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. A. baumannii 17978 UN and 17978 VU variants were received from ATCC in 2009 and in March 2021. Strains were maintained as frozen stocks in lysogeny broth (LB) supplemented with 30% (vol/vol) glycerol at −80°C. Unless noted otherwise, bacteria were grown on LB agar (LBA) at 37°C, and single, isolated colonies were resuspended in LB and incubated overnight at 37°C with constant agitation. Overnight cultures were diluted 1:100 in fresh LB and incubated for 3.5 h at 37°C with constant agitation until the exponential phase. Exponential-phase cultures were washed twice with cold phosphate-buffered saline (PBS) and diluted to 1 × 1010 CFU/ml in PBS. Bacterial cultures were diluted further as appropriate for each experiment.
Genome sequencing and analyses.
The genomes of A. baumannii strains 17978 VU and 17978 UN variants were sequenced using PacBio sequencing by Genewiz (South Plainfield, NJ) (45). Pangenomes were assembled and annotated as follows. The raw sequence reads of the strains of A. baumannii were assembled using Flye version 2.4.2 with the genome size set to 3.86 Mb, choosing the option for plasmid assembly and three polishing iterations. An Amazon Web Services (AWS) EC2 m5.2xlarge instance was used for all of the assembly runs. Assembly statistics were compared and analyzed using Quast version 5.0.2 against the reference A. baumannii ATCC 17978-mff genome from NCBI (RefSeq accession number NZ_CP012004.1). The assembled genomes were annotated using Prokka version 1.13.5. Pangenomes were created from these annotated genomes using Roary version 3.12.0 with the options “-e -n -v.” Average nucleotide identity (ANI) of the genomes of A. baumannii 17978 VU and 17978 UN variants was determined using an online ANI calculator (http://enve-omics.ce.gatech.edu/ani/). The assembled genomes of A. baumannii 17978 VU and 17978 UN variants are available in the NCBI database (accession numbers CP079213 for A. baumannii 17978 VU and CP079212 for A. baumannii 17978 UN). Additionally, A. baumannii 17978 UN received from ATCC in 2021 was sequenced using Nanopore long-read sequencing technology by the Microbial Genome Sequencing Center (MiGS; Pittsburgh, PA). This reference genome sequence was deposited in NCBI and annotated using the Prokaryotic Genome Annotation Pipeline (PGAP) (accession numbers as follows: chromosome, CP079931; pAB3, CP079932; pAB1, CP079933; pAB2, CP079934).
To screen for other differences between the genomes of A. baumannii 17978 VU and 17978 UN variants, such as SNPs, the genomes of both variants were sequenced as follows. DNA was isolated from A. baumannii 17978 UN and 17978 VU variants using a Wizard genomic DNA purification kit (Promega) or a DNeasy blood and tissue kit (Qiagen). DNA concentration was estimated with NanoDrop (Thermo). Library preparation by Nextera Flex and sequencing by Illumina NovaSeq 2 × 150-bp reads were performed at University of Illinois at Chicago Sequencing Core (UICSQC) or 2 × 100 bp reads at MiGS. The resulting reads were mapped to the ATCC A. baumannii 17978 VU genome (NCBI accession numbers NZ_CP012004, NZ_CP012005, CP000522.1, and CP000523.1) or to the A. baumannii 17978 UN genome (accession numbers CP079931, CP079932, CP079933, and CP079934) using breseq version 0.35.5 with default settings (92, 93).
To screen for the presence of AbaAL44 in Vanderbilt University Medical Center (VUMC) and University of Illinois Chicago (UIC) laboratory A. baumannii 17978 stocks, individual colonies were screened for the presence of the accessory clsC2 gene by standard PCR using primers specific to this gene (forward primer, TCTTTCTGGCTGGTTGCTTACTCAGC; reverse primer, CCGCAGCTTTCTGATTGAGACAGGC). PCR products were visualized on an agarose gel. The presence of a PCR product indicated the presence of clsC2, signifying that that particular colony belonged to the A. baumannii 17978 UN variant. The absence of a PCR product indicated the absence of clsC2, signifying that that particular colony belonged to the A. baumannii 17978 VU variant.
Construction of A. baumannii 17978 UN Δpilus, 17978 UN ΔkatX, and 17978 UN ΔclsC2 variants and complemented strains.
A. baumannii 17978 UN Δpilus, 17978 UN ΔkatX, and 17978 UN ΔclsC2 variants were constructed by recombineering using the protocol by Tucker et al. (55). Briefly, primers were designed that include approximately 125 bp of sequence upstream of the gene/locus to be deleted fused to the kanamycin resistance (kanr) cassette forward primer, CCGGAATTGCCAGCTGGG, and 125 bp of sequence downstream of the gene/locus to be deleted fused to the kanr cassette reverse primer, TTCAGAAGAACTCGTCAAG (Integrated DNA Technologies). These primer pairs were used to amplify the kanr cassette from pCR2.1 plasmid DNA (Invitrogen). After purification, approximately 10 μg of each amplification product was electroporated into electrocompetent A. baumannii 17978 UN cells containing the recombinase plasmid pAT02. Transformants were recovered at 37°C with shaking in LB containing 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h to induce recombinase expression prior to plating onto LB agar containing kanamycin. Kanamycin-resistant colonies were purified by serial passage and confirmed to have the appropriate deletion in the following two ways: first, by the difference in size of PCR amplification products using primers that amplify the recombineered locus by binding the chromosome outside the flanking sequences included in the recombineering primers, and second, by PCR amplification using a primer that binds within the kanr cassette and a primer that binds the chromosome outside of the flanking sequence included in the recombineering primer.
For the A. baumannii 17978 UN ΔkatX variant, the genetic defect was restored by expressing the accessory katX gene in trans as follows. katX was PCR amplified using a forward primer annealing to the region upstream of katX (GTAATTGGATGAGGTGATGTATTAG) and a reverse primer annealing to the region downstream of katX (GGCCATCCTCATCAAATACTG). Forward and reverse primers contained a 20-nucleotide sequence that overlapped the BamHI- or SalI-digested plasmid, respectively. Subsequently, the amplicon was ligated into the BamHI and SalI restriction sites of the E. coli-A. baumannii shuttle vector pWH1266 under its native promoter using NEBuilder HiFi DNA assembly (New England Biolabs, Ipswich, MA) to create pWH1266(katX) (77). Appropriate plasmid constructs were electroporated into the A. baumannii 17978 UN ΔkatX variant as described above.
For the A. baumannii 17978 UN ΔclsC2 variant the genetic defect was restored by reintegrating the accessory clsC2 gene under its native promoter into a different part of the bacterial chromosome using methods described elsewhere (94). Briefly, clsC2 was cloned into the pKNOCK-mTn7 plasmid to make pKNOCK(clsC2) as described above, using a forward primer annealing to the region upstream of clsC2 (CGCATCTTATAACGACAAAGAGAAC) and a reverse primer annealing to the region downstream of clsC2 (GGGCGATAAAGCCACAGATAC). Next, the E. coli λpir116 strain was transformed with pKNOCK(clsC2), and clsC2 was integrated into the A. baumannii 17978 UN ΔclsC2 variant by setting up a four-way mating between E. coli λpir116/pKNOCK(clsC2), E. coli HB101/pRK2013, E. coli 100D/pTNS2, and the A. baumannii 17978 UN ΔclsC2 strain to make the A. baumannii 17978 UN ΔclsC2/clsC2 strain. Successful integration of clsC2 into the correct spot on the A. baumannii 17978 UN ΔclsC2 chromosome was verified in the following two ways: first, by PCR amplification using a forward primer that binds the A. baumannii chromosome near the target insertion site (GTCGTTTTCGCTGATGAAAATAG) and a reverse primer that binds within the Tn7 element (CACAGCATAACTGGACTGATTTC), and second, by PCR amplification of the clsC2 gene inserted into the Tn7 element.
To express clsC2 in trans in A. baumannii 17978 VU, clsC2 was cloned into pWH1266 as described above, using a forward primer annealing to the region upstream of clsC2 (AACCTTCTTAGATTATAACAAAATCATACAGTTATTG) fused to a 20-nucleotide sequence overlapping with the BamHI-digested plasmid and a reverse primer annealing to the region downstream of clsC2 (GCTCTCGAGAAAAAAGCAGAAG) fused to a 20-nucleotide sequence overlapping with the SalI-digested plasmid to generate pWH1266(clsC2). This plasmid construct (or the empty vector) was electroporated into A. baumannii 17978 VU as described above.
Murine infection models.
All animal experiments were approved by the Vanderbilt University Medical Center (VUMC) Institutional Care and Use Committee and conformed to policies and guidelines established by VUMC, the Animal Welfare Act, the National Institutes of Health, and the American Veterinary Medical Association. Wild-type, female, 8-week-old C57BL/6 mice were purchased from Jackson Laboratories. The murine model of A. baumannii pneumonia was performed as previously described (95). Briefly, A. baumannii was back-diluted 1:100 from overnight culture and grown in LB for 3.5 h at 37°C with constant agitation, washed twice with cold PBS, and resuspended in PBS at an appropriate cell density for infection. Mice were infected intranasally with 3 × 108 CFU of A. baumannii in 30 μl PBS. At 24 or 36 h postinfection (h.p.i.), mice were euthanized and organs were harvested. Lungs and spleens were homogenized, serially diluted in PBS, and plated onto LBA for bacterial enumeration. For histological analyses, lungs were inflated with 1 ml of 10% formalin, fixed, embedded, and stained as described previously (96). Mice were randomized to treatment groups using the GraphPad QuickCalcs online randomization software (https://www.graphpad.com/quickcalcs/randomize1.cfm).
Histology.
Paraffin-embedded mouse tissue sections were stained with hematoxylin and eosin by the Vanderbilt University Medical Center Translational Pathology Shared Resource. Specimens were examined and imaged by K. L. Boyd, who was blind to the treatment conditions. Images are representative of at least 2 independent experiments.
Bacterial growth assays.
Overnight cultures of single, isolated colonies were generated as described above. To prepare for growth assays, overnight cultures were standardized to a similar optical density at 600 nm (OD600) and subsequently diluted 1:100 in 200 μl of rich medium (LB) or minimal medium (Tris minimal succinate [TMS]) in 96-well flat-bottomed tissue culture plates (Costar; Corning). Plates were incubated for 24 h at 37°C with shaking and growth was assayed by measuring OD600 at the indicated times using a Synergy 2 multimode reader (BioTek).
Scanning electron microscopy analyses.
Bacterial cells were analyzed by scanning electron microscopy as previously described, with some modifications (97). Briefly, bacteria were cultured in LB at 37°C overnight. The following day, samples were fixed with 2.0% paraformaldehyde (Electron Microscopy Sciences) and 2.5% glutaraldehyde (Electron Microscopy Sciences) in 0.05 M sodium cacodylate (Electron Microscopy Sciences) buffer for 24 h. After primary fixation, samples were secondarily fixed in fresh 2.0% paraformaldehyde and 2.5% glutaraldehyde in 0.05 M sodium cacodylate buffer on poly-l-lysine-coated glass coverslips (Thermo Fisher) for 4 h. Samples were washed three times with 0.05 M sodium cacodylate buffer before sequential dehydration with increasing concentrations of absolute ethanol. After ethanol dehydration, samples were dried at the critical point using a critical point dryer machine (Tousimis), mounted onto aluminum SEM sample stubs (Electron Microscopy Sciences), and sputter coated with 5 nm of gold-palladium. Afterward, samples were painted with a thin strip of colloidal silver (Electron Microscopy Sciences) at the edge to facilitate charge dissipation. Bacteria were imaged with an FEI Quanta 250 field-emission gun scanning electron microscope. Micrographs shown are representative of at least four biological replicates. Pili were quantified as the fraction of cells expressing pili (no. of cells with pili/total no. of cells in each image) and the number of pili per piliated cell. At least 12 representative images of each strain were scored in a blind manner for each of four independent experiments.
Hemagglutination assay.
Hemagglutination experiments were performed based on a protocol by Greene et al. (98), with modifications. A. baumanii 17978 VU, 17978 UN, and 17978 UN Δpilus strains were grown overnight on LBA or in LB, as indicated, for 48 h at 37°C without agitation. E. coli UTI 89 (positive control) and E. coli UTI89 ΔfimAH (negative control) strains were grown in LB for 24 h at 37°C without agitation, diluted 1:1,000 in LB, and incubated for another 24 h at 37°C without agitation. Bacteria were normalized to an OD600 of 1.0, after which 1 ml of each culture was pelleted and resuspended in 100 μl of PBS. Aliquots (25 μl) of bacteria were diluted serially in wells of a 96-well U-bottomed plate containing 25 μl of PBS, after which an equal volume of a 1% suspension of washed human erythrocytes (Innovative Research, Novi, MI) was added to each well. Wells were incubated overnight at 4°C without agitation, after which the hemagglutination titer (i.e., the maximum dilution at which bacteria are able to agglutinate erythrocytes) was recorded for each strain.
Biofilm formation.
Stationary-phase cultures of A. baumanii 17978 VU, 17978 UN, and 17978 UN Δpilus strains were diluted 1:10 in LB in wells of a 96-well plate and incubated statically at 37°C for 24 to 48 h. After incubation, half of the wells were used for the determination of biomass by measuring OD600, and the other half was used for the measurement of biofilm formation. Biofilm formation was determined as follows. Supernatants were aspirated from each well, cells were permeabilized with 100% ethanol (EtOH), and biofilms were stained with crystal violet solution (0.41% in 12% EtOH) for 20 min. After staining, wells were washed vigorously with PBS, and biofilms were solubilized with 33% acetic acid, after which OD580 was measured for each well. Biofilm formation was recorded as the ratio of biofilm to biomass (i.e., OD580/OD600) (99).
A549 epithelial cell adhesion assay.
Immortalized human A549 epithelial cells were maintained as frozen stocks in liquid nitrogen. Prior to infection experiments, A549 cells were maintained in Dulbecco’s modified Eagle medium (DMEM; Gibco, Thermo Fisher Scientific) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin-streptomycin (Pen-Strep). A549 cells were grown until ∼75% confluence, after which they were split 1:3 in fresh medium. To prepare for infection experiments, A549 cells were seeded onto wells of 12-well plates and grown until confluence. Next, medium was aspirated from each well, and A549 cells were washed twice with prewarmed PBS. A549 cells were then infected with mid-log-phase bacteria suspended in DMEM without antibiotics at a multiplicity of infection (MOI) of 100, after which wells were incubated at 37°C for 2 h. At 2 h.p.i., medium was aspirated from each well, and nonadherent bacteria were removed by washing wells three times with PBS. A549 cells were then lysed by adding 1% Triton X-100 to each well and incubating plates at room temperature (RT) for 10 min. After lysis, 800 μl of ice-cold PBS was added to each well, and the contents of each well were pipetted up and down several times to suspend bacteria. The number of cell-associated bacteria was then determined via serial dilution in PBS and plating onto LBA. Data are represented as the percentage of cell-associated bacteria relative to that for A. baumanii 17978 VU.
Bacterial surface-associated motility assay.
Stationary-phase cultures of A. baumanii 17978 VU, 17978 UN, and 17978 UN Δpilus strains were washed once in LB, resuspended in LB, and standardized to a similar OD600 using LB as a diluent. Next, 1 μl of standardized bacterial cultures was inoculated on the surface of motility agar plates at the center. Bacterial inoculums were allowed to dry, after which plates were wrapped in parafilm and incubated overnight, upside down, single file at 37°C. After incubation, the maximum motility radius was measured.
Hydrogen peroxide susceptibility assay.
A. baumanii 17978 VU, 17978 UN, 17978 UN ΔkatX/EV, and 17978 UN ΔkatX/pkatX strains were grown to the mid-exponential phase as previously described, then inoculated into LB supplemented with 1 mM H2O2 at a starting inoculum of 1 × 107 CFU/ml. Bacteria were then incubated at 37°C for 30 min with constant agitation, after which the remaining number of viable bacteria was determined via serial dilution in PBS and plating onto LBA.
Total neutrophil-mediated killing (extracellular and intracellular).
Neutrophil-like HL-60 cells were maintained in RPMI medium (25 mM HEPES buffer) supplemented with 10% heat-inactivated FBS and 1% Pen-Strep. Prior to infection experiments, HL-60 cells were differentiated in media supplemented with 1.3% dimethyl sulfoxide (DMSO) for 6 days. On the day of infection, differentiated HL-60 cells were seeded at a density of 1.4 × 105 to 1.8 × 105 cells/well in wells of a 96-well plate, suspended in RPMI supplemented with 0.5% heat-inactivated FBS, and incubated at 37°C for 30 min. Bacteria for the infection were prepared as follows. A. baumanii 17978 VU, 17978 UN, 17978 UN ΔkatX/EV, and 17978 UN ΔkatX/pkatX strains were grown to the mid-log phase and washed and diluted in PBS as previously described. Prior to infection of neutrophils, bacteria were opsonized by resuspension in 20% normal human serum in RPMI and subsequent incubation at 37°C for 30 min with constant shaking at 50 rpm. Immediately following opsonization, bacteria were resuspended in RPMI supplemented with 0.5% heat-inactivated FBS and diluted further to the appropriate cell density for infection. Next, bacteria were added to each well at an MOI of 2, and the infection was synchronized by centrifuging the 96-well plate at 200 × g for 9 min at 4°C. Inoculums were verified by serially diluting in PBS and plating on LBA. The plate was then incubated for 90 min at 37°C, after which infected HL-60 cells were lysed by adding saponin to a final concentration of 0.1% to each well and incubating on ice for 15 min. Next, lysates were serially diluted in PBS and plated onto LBA for bacterial enumeration.
Murine neutrophils were isolated as follows. Murine hind limbs were dissected and bone marrow was isolated as previously described (100). Briefly, bone marrow was isolated from the long bones of C57BL/6 mouse hind limbs via centrifugation, washed, red blood cells were lysed, and the bone marrow suspension was passed through a cell strainer prior to counting. Neutrophils were isolated using a mouse neutrophil isolation kit (Miltenyi Biotec) according to the manufacturer’s protocol. Infection of murine neutrophils was performed as described above with modifications. Purified neutrophils were seeded at a density of 1.6 × 105 to 3.2 × 105 cells/well in wells of a 96-well plate pretreated with 100 μl of 20% normal human serum, after which they were allowed to attach to well bottoms by incubating plates at room temperature for 30 min. A. baumanii 17978 VU, 17978 UN, and 17978 UN ΔkatX strains were opsonized by resuspension in 50% C57BL/6 mouse complement serum (Innovative Research, Novi, MI) in RPMI medium supplemented with 10 mM HEPES buffer (RPMI/H). Immediately following opsonization, bacteria were washed twice with RPMI/H and diluted in RPMI/H to the appropriate cell density for infection. Bacteria were added to each well at an MOI of 1 and, following synchronization of the infection, the plate was incubated for 2 h at 37°C as described above. Neutrophils were lysed and viable bacteria were enumerated as described above.
Bacterial uptake by macrophages.
Immortalized macrophage-like RAW 264.7 cells were maintained as frozen stocks in liquid nitrogen. Prior to infection experiments, RAW cells were maintained in DMEM supplemented with 10% heat-inactivated FBS and 1% Pen-Strep. RAW cells were grown until ∼75% confluence, after which they were split 1:10 to 1:20 in fresh medium. To prepare for infection experiments, RAW cells were seeded onto wells of 12-well plates at a seeding density of 5 × 105 cells/well and activated with 100 ng/ml of LPS, after which they were incubated overnight at 37°C. On the day of infection, mid-log-phase cultures of A. baumannii were prepared as previously described, after which bacteria were resuspended in DMEM plus 10% FBS devoid of antibiotics at the appropriate bacterial density to reach a target MOI of 15. Medium was then aspirated from all wells, RAW cells were washed once with prewarmed PBS, and 1 ml of appropriate bacteria suspended in medium was added to each well. Inoculums were verified by serially diluting in PBS and plating on LBA. Infected RAW cells were then incubated at 37°C for 30 min, after which medium was aspirated and wells were washed twice with prewarmed PBS. Next, extracellular bacteria were killed by adding 1 ml of medium supplemented with 200 μg/ml gentamicin to each well and incubating at 37°C. Thirty minutes after the addition of gentamicin, medium was aspirated, cells were washed once with PBS, and cells were lysed by adding 200 μl of 0.01% triton to each well. Each well was scraped with a sterile cell scraper, after which the lysate was serially diluted in PBS and plated onto LBA for the enumeration of intracellular bacterial burden.
Bacterial cell envelope stress assays.
Bacterial susceptibility to osmotic stress was assayed as follows. A. baumannii 17978 VU, 17978 UN, 17978 UN ΔclsC2, and 17978 UN ΔclsC2/clsC2 strains were grown to the mid-exponential phase as previously described and inoculated into LB supplemented with 2.5 M NaCl at a starting inoculum of 1 × 107 CFU/ml. Bacteria were then incubated at 37°C for 2 h with constant agitation, after which bacterial viability was determined via serial dilution in PBS and plating onto LBA. Bacterial survival was calculated as the percentage of viable bacteria postincubation relative to the number of viable bacteria preincubation. Bacterial susceptibility to detergent stress was assayed as follows. A. baumannii 17978 VU, 17978 UN, A17978 UN ΔclsC2, and 17978 UN ΔclsC2/clsC2 strains were prepared as described above, and resuspended in PBS supplemented with 0.1% Triton X-100 at a starting inoculum of 1 × 1010 CFU/ml. Bacteria were then incubated at 37°C for 6 h with constant agitation, after which bacterial viability and survival were determined and calculated as described above.
Detection of cytokines produced by infected macrophages.
BMDMs were isolated and maintained as previously described (101). Briefly, bone marrow was isolated from the long bones of C57BL/6 mouse hind limbs via centrifugation and washed, red blood cells were lysed, and the bone marrow suspension was passed through a cell strainer prior to counting. The bone marrow cell suspension was plated at a density of 3 × 106 cells/ml in BMDM medium (RPMI medium supplemented with 10% FBS and 20% L929 cell conditioned medium [LCCM]) and incubated at 37°C with 5% CO2. Every 48 h, medium was aspirated and fresh BMDM medium was added to each plate. On day six, BMDMs were collected and seeded onto 12-well plates at a density of 1 × 106 to 2.5 × 106 cells/well in BMDM medium and incubated overnight at 37°C. On day 7, mid-log-phase cultures of A. baumannii were prepared as previously described and diluted to the appropriate bacterial density in RPMI medium supplemented with 10% FBS for a target MOI of 10. Medium was then aspirated from all wells, and BMDMs were washed once with prewarmed PBS, after which bacteria suspended in 1 ml of medium were added. As a negative control, BMDMs were infected with medium alone, and as a positive control, BMDMs were treated with medium supplemented with 100 ng/ml of LPS or 100 ng/ml of LPS plus 2 mM ATP. Plates were then incubated at 37°C and the supernatant was collected at 18 h.p.i. Cytokines and chemokines (IL-1β, IL-10, KC, and MIP-2) in BMDM supernatants were quantified using an enzyme-limited immunosorbent assay (ELISA) kit (R&D Systems) according to the manufacturer’s protocol for each.
Complete blood counts.
Complete blood counts with five-part differential were performed on EDTA-treated whole mouse blood by the Vanderbilt University Medical Center Translational Pathology Shared Resource.
Immune cell recruitment.
Flow cytometric analyses were performed with total erythrocyte-free lung cells isolated at 24 h.p.i. from individual mice infected with A. baumannii 17978 VU or 17978 UN variants, as indicated. Lungs were minced, digested with collagenase and DNase for 30 min, and passed through a 70-μm cell strainer prior to erythrocyte lysis. Cells were stained with a myeloid panel that included antibodies against CD45 (clone 104, fluorescein isothiocyanate [FITC]; eBioscience), CD103 (clone 2E7, PerCP-Cyanine5.5 [Cy5.5]; BioLegend), CD64 (clone X54-5/7.1, phycoerythrin [PE]; BioLegend), CD11c (clone HL-3, PE-Cy7; BD Pharmingen), Siglec F (clone E50-2440, Horizon PE-CF594; BD Pharmingen), CD11b (clone M1/70, eFluor-450; eBioscience), MHCII (clone M5/114.15.2, BV605; BD Pharmingen), CD24 (clone M1/69, allophycocyanin [APC]; eBioscience), Ly6C (clone AL-21, APC-Cy7; BD Pharmingen), and Ly6G (clone 1A8, Alexa Fluor 700; BD Pharmingen). Analyses were carried out on the 5-laser LSR Fortessa instrument (BD Biosciences) at the Vanderbilt Flow Cytometry Shared Resource, and analyses were performed using FlowJo software (Treestar, Inc.). Myeloid populations were gated according to the strategy of Misharin and colleagues (102, 103).
Quantification and statistical analysis.
Statistical analyses were performed using Prism version 6 (GraphPad Software, Inc.). Mean comparisons were performed using unpaired Welch’s t test or one-way analysis of variance (ANOVA) adjusted for multiple comparisons, as indicated. P values of less than 0.05 were considered statistically significant. Statistical details of the experiments can be found in the figure legends.
Data availability.
The assembled genomes of A. baumannii 17978 VU and 17978 UN variants that were sequenced using PacBio sequencing technology are available in the NCBI database under accession numbers CP079213.1 (VU variant) and CP079212.1 (UN variant). The newly deposited reference genome of the A. baumannii 17978 UN variant that was sequenced using Nanopore long-read sequencing technology is available in the NCBI database under accession numbers CP079931 (chromosome), CP079932 (pAB3), CP079933 (pAB1), and CP079934 (pAB2).
ACKNOWLEDGMENTS
We thank the Vanderbilt University Medical Center (VUMC) Division of Animal Care, the VUMC Translational Pathology Shared Resource, the VUMC flow cytometry shared resource, the VUMC Molecular Cell Biology Resource Core, and the Vanderbilt Institute for Infection, Immunology, and Inflammation (VI4). Additional support for this work was provided by Padmini Komalavilas, Ph.D.
This work was funded by Cystic Fibrosis Foundation award NOTO17Q0 and National Institutes of Health award 1R01HL152210-01 to M.J.N., by National Institutes of Health Award HD090061 to J.A.G., and by National Institutes of Health award R01AI101171 to E.P.S.
Footnotes
Supplemental material is available online only.
Contributor Information
Michael J. Noto, Email: michael.james.noto@emory.edu.
Andreas J. Bäumler, University of California, Davis
REFERENCES
- 1.Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5:939–951. 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 2.Antunes LC, Visca P, Towner KJ. 2014. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis 71:292–301. 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 3.Fournier PE, Richet H. 2006. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin Infect Dis 42:692–699. 10.1086/500202. [DOI] [PubMed] [Google Scholar]
- 4.Inchai J, Pothirat C, Bumroongkit C, Limsukon A, Khositsakulchai W, Liwsrisakun C. 2015. Prognostic factors associated with mortality of drug-resistant Acinetobacter baumannii ventilator-associated pneumonia. J Intensive Care 3:9. 10.1186/s40560-015-0077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanafani ZA, Zahreddine N, Tayyar R, Sfeir J, Araj GF, Matar GM, Kanj SS. 2018. Multi-drug resistant Acinetobacter species: a seven-year experience from a tertiary care center in Lebanon. Antimicrob Resist Infect Control 7:9. 10.1186/s13756-017-0297-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson SE, Sherman EX, Weiss DS, Rather PN. 2018. Aminoglycoside heteroresistance in Acinetobacter baumannii AB5075. mSphere 3:e00271-18. 10.1128/mSphere.00271-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGann P, Courvalin P, Snesrud E, Clifford RJ, Yoon EJ, Onmus-Leone F, Ong AC, Kwak YI, Grillot-Courvalin C, Lesho E, Waterman PE. 2014. Amplification of aminoglycoside resistance gene aphA1 in Acinetobacter baumannii results in tobramycin therapy failure. mBio 5:e00915. 10.1128/mBio.00915-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tada T, Miyoshi-Akiyama T, Shimada K, Shimojima M, Kirikae T. 2014. Dissemination of 16S rRNA methylase ArmA-producing Acinetobacter baumannii and emergence of OXA-72 carbapenemase coproducers in Japan. Antimicrob Agents Chemother 58:2916–2920. 10.1128/AAC.01212-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu LY, Apisarnthanarak A, Khan E, Suwantarat N, Ghafur A, Tambyah PA. 2017. Carbapenem-resistant Acinetobacter baumannii and Enterobacteriaceae in South and Southeast Asia. Clin Microbiol Rev 30:1–22. 10.1128/CMR.00042-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans BA, Hamouda A, Amyes SG. 2013. The rise of carbapenem-resistant Acinetobacter baumannii. Curr Pharm Des 19:223–238. 10.2174/138161213804070285. [DOI] [PubMed] [Google Scholar]
- 11.Pogue JM, Mann T, Barber KE, Kaye KS. 2013. Carbapenem-resistant Acinetobacter baumannii: epidemiology, surveillance and management. Expert Rev Anti Infect Ther 11:383–393. 10.1586/eri.13.14. [DOI] [PubMed] [Google Scholar]
- 12.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, Group W, WHO Pathogens Priority List Working Group. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 13.Park YK, Jung SI, Park KH, Cheong HS, Peck KR, Song JH, Ko KS. 2009. Independent emergence of colistin-resistant Acinetobacter spp. isolates from Korea. Diagn Microbiol Infect Dis 64:43–51. 10.1016/j.diagmicrobio.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Dickstein Y, Lellouche J, Ben Dalak Amar M, Schwartz D, Nutman A, Daitch V, Yahav D, Leibovici L, Skiada A, Antoniadou A, Daikos GL, Andini R, Zampino R, Durante-Mangoni E, Mouton JW, Friberg LE, Dishon Benattar Y, Bitterman R, Neuberger A, Carmeli Y, Paul M, Group AS, AIDA Study Group. 2019. Treatment outcomes of colistin- and carbapenem-resistant Acinetobacter baumannii infections: an exploratory subgroup analysis of a randomized clinical trial. Clin Infect Dis 69:769–776. 10.1093/cid/ciy988. [DOI] [PubMed] [Google Scholar]
- 15.Trebosc V, Gartenmann S, Tötzl M, Lucchini V, Schellhorn B, Pieren M, Lociuro S, Gitzinger M, Tigges M, Bumann D, Kemmer C. 2019. Dissecting colistin resistance mechanisms in extensively drug-resistant Acinetobacter baumannii clinical isolates. mBio 10:e01083-19. 10.1128/mBio.01083-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL. 2006. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis 6:589–601. 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 17.Xie R, Zhang XD, Zhao Q, Peng B, Zheng J. 2018. Analysis of global prevalence of antibiotic resistance in Acinetobacter baumannii infections disclosed a faster increase in OECD countries. Emerg Microbes Infect 7:31. 10.1038/s41426-018-0038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsueh PR, Teng LJ, Chen CY, Chen WH, Yu CJ, Ho SW, Luh KT. 2002. Pandrug-resistant Acinetobacter baumannii causing nosocomial infections in a university hospital, Taiwan. Emerg Infect Dis 8:827–832. 10.3201/eid0805.020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nowak J, Zander E, Stefanik D, Higgins PG, Roca I, Vila J, McConnell MJ, Cisneros JM, Seifert H, Mwg WP4, MagicBullet Working Group WP4. 2017. High incidence of pandrug-resistant Acinetobacter baumannii isolates collected from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J Antimicrob Chemother 72:3277–3282. 10.1093/jac/dkx322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CDC. 2019. Antibiotic resistance threats in the United States, 2019. CDC, Atlanta, GA. [Google Scholar]
- 21.Wright MS, Iovleva A, Jacobs MR, Bonomo RA, Adams MD. 2016. Genome dynamics of multidrug-resistant Acinetobacter baumannii during infection and treatment. Genome Med 8:26. 10.1186/s13073-016-0279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boone RL, Whitehead B, Avery TM, Lu J, Francis JD, Guevara MA, Moore RE, Chambers SA, Doster RS, Manning SD, Townsend SD, Dent L, Marshall D, Gaddy JA, Damo SM. 2021. Analysis of virulence phenotypes and antibiotic resistance in clinical strains of Acinetobacter baumannii isolated in Nashville, Tennessee. BMC Microbiol 21:21. 10.1186/s12866-020-02082-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kyrpides NC, Hugenholtz P, Eisen JA, Woyke T, Göker M, Parker CT, Amann R, Beck BJ, Chain PS, Chun J, Colwell RR, Danchin A, Dawyndt P, Dedeurwaerdere T, DeLong EF, Detter JC, De Vos P, Donohue TJ, Dong XZ, Ehrlich DS, Fraser C, Gibbs R, Gilbert J, Gilna P, Glöckner FO, Jansson JK, Keasling JD, Knight R, Labeda D, Lapidus A, Lee JS, Li WJ, Ma J, Markowitz V, Moore ER, Morrison M, Meyer F, Nelson KE, Ohkuma M, Ouzounis CA, Pace N, Parkhill J, Qin N, Rossello-Mora R, Sikorski J, Smith D, Sogin M, Stevens R, Stingl U, Suzuki K, et al. 2014. Genomic encyclopedia of bacteria and archaea: sequencing a myriad of type strains. PLoS Biol 12:e1001920. 10.1371/journal.pbio.1001920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antunes LC, Imperi F, Carattoli A, Visca P. 2011. Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS One 6:e22674. 10.1371/journal.pone.0022674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hugh R, Reese R. 1968. A comparison of 120 strains of Bacterium anitratum Schaub and Hauber with the type strain of this species. Int J Syst Evol Microbiol 18:1–3. [Google Scholar]
- 26.Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, Gancz HY, Corey BW, Moon JK, Si Y, Owen MT, Hallock JD, Kwak YI, Summers A, Li CZ, Rasko DA, Penwell WF, Honnold CL, Wise MC, Waterman PE, Lesho EP, Stewart RL, Actis LA, Palys TJ, Craft DW, Zurawski DV. 2014. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio 5:e01076-14–e01014. 10.1128/mBio.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang N, Ozer EA, Mandel MJ, Hauser AR. 2014. Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. mBio 5:e01163-14–e01114. 10.1128/mBio.01163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato Y, Unno Y, Kawakami S, Ubagai T, Ono Y. 2017. Virulence characteristics of Acinetobacter baumannii clinical isolates vary with the expression levels of omps. J Med Microbiol 66:203–212. 10.1099/jmm.0.000394. [DOI] [PubMed] [Google Scholar]
- 29.Ramirez MS, Penwell WF, Traglia GM, Zimbler DL, Gaddy JA, Nikolaidis N, Arivett BA, Adams MD, Bonomo RA, Actis LA, Tolmasky ME. 2019. Identification of potential virulence factors in the model strain Acinetobacter baumannii A118. Front Microbiol 10:1599. 10.3389/fmicb.2019.01599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hesse LE, Lonergan ZR, Beavers WN, Skaar EP. 2019. The Acinetobacter baumannii Znu system overcomes host-imposed nutrient zinc limitation. Infect Immun 87:e00746-19. 10.1128/IAI.00746-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lonergan ZR, Nairn BL, Wang J, Hsu YP, Hesse LE, Beavers WN, Chazin WJ, Trinidad JC, VanNieuwenhze MS, Giedroc DP, Skaar EP. 2019. An Acinetobacter baumannii, zinc-regulated peptidase maintains cell wall integrity during immune-mediated nutrient sequestration. Cell Rep 26:2009–2018.e6. 10.1016/j.celrep.2019.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood CR, Ohneck EJ, Edelmann RE, Actis LA. 2018. A light-regulated type I pilus contributes to Acinetobacter baumannii biofilm, motility, and virulence functions. Infect Immun 86:e00442-18. 10.1128/IAI.00442-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Williams CL, Neu HM, Alamneh YA, Reddinger RM, Jacobs AC, Singh S, Abu-Taleb R, Michel SLJ, Zurawski DV, Merrell DS. 2020. Characterization of Acinetobacter baumannii copper resistance reveals a role in virulence. Front Microbiol 11:16. 10.3389/fmicb.2020.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobrindt U, Hacker J. 2001. Whole genome plasticity in pathogenic bacteria. Curr Opin Microbiol 4:550–557. 10.1016/s1369-5274(00)00250-2. [DOI] [PubMed] [Google Scholar]
- 35.Imperi F, Antunes LC, Blom J, Villa L, Iacono M, Visca P, Carattoli A. 2011. The genomics of Acinetobacter baumannii: insights into genome plasticity, antimicrobial resistance and pathogenicity. IUBMB Life 63:1068–1074. 10.1002/iub.531. [DOI] [PubMed] [Google Scholar]
- 36.Traglia G, Chiem K, Quinn B, Fernandez JS, Montaña S, Almuzara M, Mussi MA, Tolmasky ME, Iriarte A, Centrón D, Ramírez MS. 2018. Genome sequence analysis of an extensively drug-resistant Acinetobacter baumannii indigo-pigmented strain depicts evidence of increase genome plasticity. Sci Rep 8:16961. 10.1038/s41598-018-35377-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu F, Zhu Y, Yi Y, Lu N, Zhu B, Hu Y. 2014. Comparative genomic analysis of Acinetobacter baumannii clinical isolates reveals extensive genomic variation and diverse antibiotic resistance determinants. BMC Genomics 15:1163. 10.1186/1471-2164-15-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright MS, Haft DH, Harkins DM, Perez F, Hujer KM, Bajaksouzian S, Benard MF, Jacobs MR, Bonomo RA, Adams MD. 2014. New insights into dissemination and variation of the health care-associated pathogen Acinetobacter baumannii from genomic analysis. mBio 5:e00963-13–e00913. 10.1128/mBio.00963-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams MD, Wright MS, Karichu JK, Venepally P, Fouts DE, Chan AP, Richter SS, Jacobs MR, Bonomo RA. 2019. Rapid replacement of Acinetobacter baumannii strains accompanied by changes in lipooligosaccharide loci and resistance gene repertoire. mBio 10:e00356-19. 10.1128/mBio.00356-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaidane N, Naas T, Mansour W, Radhia BB, Jerbi S, Boujaafar N, Bouallegue O, Bonnin RA. 2018. Genomic analysis of in vivo acquired resistance to colistin and rifampicin in Acinetobacter baumannii. Int J Antimicrob Agents 51:266–269. 10.1016/j.ijantimicag.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 41.Talyansky Y, Nielsen TB, Yan J, Carlino-Macdonald U, Di Venanzio G, Chakravorty S, Ulhaq A, Feldman MF, Russo TA, Vinogradov E, Luna B, Wright MS, Adams MD, Spellberg B. 2021. Capsule carbohydrate structure determines virulence in Acinetobacter baumannii. PLoS Pathog 17:e1009291. 10.1371/journal.ppat.1009291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber BS, Ly PM, Irwin JN, Pukatzki S, Feldman MF. 2015. A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc Natl Acad Sci USA 112:9442–9447. 10.1073/pnas.1502966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weber BS, Miyata ST, Iwashkiw JA, Mortensen BL, Skaar EP, Pukatzki S, Feldman MF. 2013. Genomic and functional analysis of the type VI secretion system in Acinetobacter. PLoS One 8:e55142. 10.1371/journal.pone.0055142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J, Lee JY, Lee H, Choi JY, Kim DH, Wi YM, Peck KR, Ko KS. 2017. Microbiological features and clinical impact of the type VI secretion system (T6SS) in Acinetobacter baumannii isolates causing bacteremia. Virulence 8:1378–1389. 10.1080/21505594.2017.1323164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jensen T, Tellgren-Roth C, Redl S, Maury J, Jacobsen SAB, Pedersen LE, Nielsen AT. 2019. Genome-wide systematic identification of methyltransferase recognition and modification patterns. Nat Commun 10:3311. 10.1038/s41467-019-11179-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Breslow JM, Meissler JJ, Hartzell RR, Spence PB, Truant A, Gaughan J, Eisenstein TK. 2011. Innate immune responses to systemic Acinetobacter baumannii infection in mice: neutrophils, but not interleukin-17, mediate host resistance. Infect Immun 79:3317–3327. 10.1128/IAI.00069-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Faassen H, KuoLee R, Harris G, Zhao X, Conlan JW, Chen W. 2007. Neutrophils play an important role in host resistance to respiratory infection with Acinetobacter baumannii in mice. Infect Immun 75:5597–5608. 10.1128/IAI.00762-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu H, KuoLee R, Harris G, Chen W. 2009. High susceptibility to respiratory Acinetobacter baumannii infection in A/J mice is associated with a delay in early pulmonary recruitment of neutrophils. Microbes Infect 11:946–955. 10.1016/j.micinf.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 49.Mahmutovic Persson I, Menzel M, Ramu S, Cerps S, Akbarshahi H, Uller L. 2018. IL-1β mediates lung neutrophilia and IL-33 expression in a mouse model of viral-induced asthma exacerbation. Respir Res 19:16. 10.1186/s12931-018-0725-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller LS, Pietras EM, Uricchio LH, Hirano K, Rao S, Lin H, O’Connell RM, Iwakura Y, Cheung AL, Cheng G, Modlin RL. 2007. Inflammasome-mediated production of IL-1β is required for neutrophil recruitment against Staphylococcus aureus in vivo. J Immunol 179:6933–6942. 10.4049/jimmunol.179.10.6933. [DOI] [PubMed] [Google Scholar]
- 51.Biondo C, Mancuso G, Midiri A, Signorino G, Domina M, Lanza Cariccio V, Mohammadi N, Venza M, Venza I, Teti G, Beninati C. 2014. The interleukin-1β/CXCL1/2/neutrophil axis mediates host protection against group B streptococcal infection. Infect Immun 82:4508–4517. 10.1128/IAI.02104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mantovani A, Dinarello CA, Molgora M, Garlanda C. 2019. Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity 50:778–795. 10.1016/j.immuni.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andrade EB, Alves J, Madureira P, Oliveira L, Ribeiro A, Cordeiro-da-Silva A, Correia-Neves M, Trieu-Cuot P, Ferreira P. 2013. TLR2-induced IL-10 production impairs neutrophil recruitment to infected tissues during neonatal bacterial sepsis. J Immunol 191:4759–4768. 10.4049/jimmunol.1301752. [DOI] [PubMed] [Google Scholar]
- 54.Sun L, Guo RF, Newstead MW, Standiford TJ, Macariola DR, Shanley TP. 2009. Effect of IL-10 on neutrophil recruitment and survival after Pseudomonas aeruginosa challenge. Am J Respir Cell Mol Biol 41:76–84. 10.1165/rcmb.2008-0202OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tucker AT, Nowicki EM, Boll JM, Knauf GA, Burdis NC, Trent MS, Davies BW. 2014. Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. mBio 5:e01313-14–e01314. 10.1128/mBio.01313-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Avalos Vizcarra I, Hosseini V, Kollmannsberger P, Meier S, Weber SS, Arnoldini M, Ackermann M, Vogel V. 2016. How type 1 fimbriae help Escherichia coli to evade extracellular antibiotics. Sci Rep 6:18109. 10.1038/srep18109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lo AW, Van de Water K, Gane PJ, Chan AW, Steadman D, Stevens K, Selwood DL, Waksman G, Remaut H. 2014. Suppression of type 1 pilus assembly in uropathogenic Escherichia coli by chemical inhibition of subunit polymerization. J Antimicrob Chemother 69:1017–1026. 10.1093/jac/dkt467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. 2000. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J 19:2803–2812. 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sarkar S, Vagenas D, Schembri MA, Totsika M. 2016. Biofilm formation by multidrug resistant Escherichia coli ST131 is dependent on type 1 fimbriae and assay conditions. Pathog Dis 74: ftw013. 10.1093/femspd/ftw013. [DOI] [PubMed] [Google Scholar]
- 60.Imlay JA. 2013. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11:443–454. 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wan B, Zhang Q, Ni J, Li S, Wen D, Li J, Xiao H, He P, Ou HY, Tao J, Teng Q, Lu J, Wu W, Yao YF. 2017. Type VI secretion system contributes to enterohemorrhagic Escherichia coli virulence by secreting catalase against host reactive oxygen species (ROS). PLoS Pathog 13:e1006246. 10.1371/journal.ppat.1006246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goulart CL, Barbosa LC, Bisch PM, von Krüger WMA. 2016. Catalases and PhoB/PhoR system independently contribute to oxidative stress resistance in Vibrio cholerae O1. Microbiology (Reading) 162:1955–1962. 10.1099/mic.0.000364. [DOI] [PubMed] [Google Scholar]
- 63.El-Benna J, Hurtado-Nedelec M, Marzaioli V, Marie JC, Gougerot-Pocidalo MA, Dang PM. 2016. Priming of the neutrophil respiratory burst: role in host defense and inflammation. Immunol Rev 273:180–193. 10.1111/imr.12447. [DOI] [PubMed] [Google Scholar]
- 64.Nguyen GT, Green ER, Mecsas J. 2017. Neutrophils to the ROScue: mechanisms of NADPH oxidase activation and bacterial resistance. Front Cell Infect Microbiol 7:373. 10.3389/fcimb.2017.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tropp BE. 1997. Cardiolipin synthase from Escherichia coli. Biochim Biophys Acta 1348:192–200. 10.1016/s0005-2760(97)00100-8. [DOI] [PubMed] [Google Scholar]
- 66.Tan BK, Bogdanov M, Zhao J, Dowhan W, Raetz CR, Guan Z. 2012. Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc Natl Acad Sci USA 109:16504–16509. 10.1073/pnas.1212797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Renner LD, Weibel DB. 2011. Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc Natl Acad Sci USA 108:6264–6269. 10.1073/pnas.1015757108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rowlett VW, Mallampalli VKPS, Karlstaedt A, Dowhan W, Taegtmeyer H, Margolin W, Vitrac H. 2017. Impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J Bacteriol 199 :e00849-16. 10.1128/JB.00849-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.López CS, Alice AF, Heras H, Rivas EA, Sánchez-Rivas C. 2006. Role of anionic phospholipids in the adaptation of Bacillus subtilis to high salinity. Microbiology (Reading) 152:605–616. 10.1099/mic.0.28345-0. [DOI] [PubMed] [Google Scholar]
- 70.López GA, Heredia RM, Boeris PS, Lucchesi GI. 2016. Content of cardiolipin of the membrane and sensitivity to cationic surfactants in Pseudomonas putida. J Appl Microbiol 121:1004–1014. 10.1111/jam.13238. [DOI] [PubMed] [Google Scholar]
- 71.Balasubramanian K, Maeda A, Lee JS, Mohammadyani D, Dar HH, Jiang JF, St Croix CM, Watkins S, Tyurin VA, Tyurina YY, Klöditz K, Polimova A, Kapralova VI, Xiong Z, Ray P, Klein-Seetharaman J, Mallampalli RK, Bayir H, Fadeel B, Kagan VE. 2015. Dichotomous roles for externalized cardiolipin in extracellular signaling: promotion of phagocytosis and attenuation of innate immunity. Sci Signal 8:ra95. 10.1126/scisignal.aaa6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iyer SS, He Q, Janczy JR, Elliott EI, Zhong Z, Olivier AK, Sadler JJ, Knepper-Adrian V, Han R, Qiao L, Eisenbarth SC, Nauseef WM, Cassel SL, Sutterwala FS. 2013. Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity 39:311–323. 10.1016/j.immuni.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pizzuto M, Lonez C, Baroja-Mazo A, Martínez-Banaclocha H, Tourlomousis P, Gangloff M, Pelegrin P, Ruysschaert JM, Gay NJ, Bryant CE. 2019. Saturation of acyl chains converts cardiolipin from an antagonist to an activator of Toll-like receptor-4. Cell Mol Life Sci 76:3667–3678. 10.1007/s00018-019-03113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sahl JW, Gillece JD, Schupp JM, Waddell VG, Driebe EM, Engelthaler DM, Keim P. 2013. Evolution of a pathogen: a comparative genomics analysis identifies a genetic pathway to pathogenesis in Acinetobacter. PLoS One 8:e54287. 10.1371/journal.pone.0054287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klockgether J, Munder A, Neugebauer J, Davenport CF, Stanke F, Larbig KD, Heeb S, Schöck U, Pohl TM, Wiehlmann L, Tümmler B. 2010. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J Bacteriol 192:1113–1121. 10.1128/JB.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Monteford J, Bilverstone TW, Ingle P, Philip S, Kuehne SA, Minton NP. 2021. What's a SNP between friends: the lineage of Clostridioides difficile R20291 can effect research outcomes. Anaerobe 71:102422. 10.1016/j.anaerobe.2021.102422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Juttukonda LJ, Green ER, Lonergan ZR, Heffern MC, Chang CJ, Skaar EP. 2019. OxyR regulates the transcriptional response to hydrogen peroxide. Infect Immun 87:e00413-18. 10.1128/IAI.00413-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Palmer LD, Minor KE, Mettlach JA, Rivera ES, Boyd KL, Caprioli RM, Spraggins JM, Dalebroux ZD, Skaar EP. 2020. Modulating isoprenoid biosynthesis increases lipooligosaccharides and restores Acinetobacter baumannii resistance to host and antibiotic stress. Cell Rep 32:108129. 10.1016/j.celrep.2020.108129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chakraborty K, Raundhal M, Chen BB, Morse C, Tyurina YY, Khare A, Oriss TB, Huff R, Lee JS, St Croix CM, Watkins S, Mallampalli RK, Kagan VE, Ray A, Ray P. 2017. The mito-DAMP cardiolipin blocks IL-10 production causing persistent inflammation during bacterial pneumonia. Nat Commun 8:13944. 10.1038/ncomms13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Garg M, Johri S, Sagar S, Mundhada A, Agrawal A, Ray P, Chakraborty K. 2021. Cardiolipin-mediated PPARγ S112 phosphorylation impairs IL-10 production and inflammation resolution during bacterial pneumonia. Cell Rep 34:108736. 10.1016/j.celrep.2021.108736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saraiva M, O'Garra A. 2010. The regulation of IL-10 production by immune cells. Nat Rev Immunol 10:170–181. 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 82.Moreira LO, El Kasmi KC, Smith AM, Finkelstein D, Fillon S, Kim YG, Núñez G, Tuomanen E, Murray PJ. 2008. The TLR2-MyD88-NOD2-RIPK2 signalling axis regulates a balanced pro-inflammatory and IL-10-mediated anti-inflammatory cytokine response to Gram-positive cell walls. Cell Microbiol 10:2067–2077. 10.1111/j.1462-5822.2008.01189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Castro F, Cardoso AP, Gonçalves RM, Serre K, Oliveira MJ. 2018. Interferon-gamma at the crossroads of tumor immune surveillance or evasion. Front Immunol 9:847. 10.3389/fimmu.2018.00847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Darwich L, Coma G, Peña R, Bellido R, Blanco EJ, Este JA, Borras FE, Clotet B, Ruiz L, Rosell A, Andreo F, Parkhouse RM, Bofill M. 2009. Secretion of interferon-γ by human macrophages demonstrated at the single-cell level after costimulation with interleukin (IL)-12 plus IL-18. Immunology 126:386–393. 10.1111/j.1365-2567.2008.02905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Robinson CM, O’Dee D, Hamilton T, Nau GJ. 2010. Cytokines involved in interferon-γ production by human macrophages. J Innate Immun 2:56–65. 10.1159/000247156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun D, Crowell SA, Harding CM, De Silva PM, Harrison A, Fernando DM, Mason KM, Santana E, Loewen PC, Kumar A, Liu Y. 2016. KatG and KatE confer Acinetobacter resistance to hydrogen peroxide but sensitize bacteria to killing by phagocytic respiratory burst. Life Sci 148:31–40. 10.1016/j.lfs.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wright MS, Mountain S, Beeri K, Adams MD. 2017. Assessment of insertion sequence mobilization as an adaptive response to oxidative stress in Acinetobacter baumannii using IS-seq. J Bacteriol 199:e00833-16. 10.1128/JB.00833-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pfefferle K, Lopalco P, Breisch J, Siemund A, Corcelli A, Averhoff B. 2020. In vivo synthesis of monolysocardiolipin and cardiolipin by Acinetobacter baumannii phospholipase D and effect on cationic antimicrobial peptide resistance. Environ Microbiol 22:5300–5308. 10.1111/1462-2920.15231. [DOI] [PubMed] [Google Scholar]
- 89.Lopalco P, Stahl J, Annese C, Averhoff B, Corcelli A. 2017. Identification of unique cardiolipin and monolysocardiolipin species in Acinetobacter baumannii. Sci Rep 7:2972. 10.1038/s41598-017-03214-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boyd KJ, Alder NN, May ER. 2018. Molecular dynamics analysis of cardiolipin and monolysocardiolipin on bilayer properties. Biophys J 114:2116–2127. 10.1016/j.bpj.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Powers MJ, Simpson BW, Trent MS. 2020. The Mla pathway in Acinetobacter baumannii has no demonstrable role in anterograde lipid transport. Elife 9:e56571. 10.7554/eLife.56571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barrick JE, Colburn G, Deatherage DE, Traverse CC, Strand MD, Borges JJ, Knoester DB, Reba A, Meyer AG. 2014. Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genomics 15:1039. 10.1186/1471-2164-15-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kumar A, Dalton C, Cortez-Cordova J, Schweizer HP. 2010. Mini-Tn7 vectors as genetic tools for single copy gene cloning in Acinetobacter baumannii. J Microbiol Methods 82:296–300. 10.1016/j.mimet.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 95.Hood MI, Mortensen BL, Moore JL, Zhang Y, Kehl-Fie TE, Sugitani N, Chazin WJ, Caprioli RM, Skaar EP. 2012. Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLoS Pathog 8:e1003068. 10.1371/journal.ppat.1003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jacobs AC, Hood I, Boyd KL, Olson PD, Morrison JM, Carson S, Sayood K, Iwen PC, Skaar EP, Dunman PM. 2010. Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect Immun 78:1952–1962. 10.1128/IAI.00889-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gaddy JA, Tomaras AP, Actis LA. 2009. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun 77:3150–3160. 10.1128/IAI.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Greene SE, Hibbing ME, Janetka J, Chen SL, Hultgren SJ. 2015. Human urine decreases function and expression of type 1 pili in uropathogenic Escherichia coli. mBio 6:e00820. 10.1128/mBio.00820-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Álvarez-Fraga L, Pérez A, Rumbo-Feal S, Merino M, Vallejo JA, Ohneck EJ, Edelmann RE, Beceiro A, Vázquez-Ucha JC, Valle J, Actis LA, Bou G, Poza M. 2016. Analysis of the role of the LH92_11085 gene of a biofilm hyper-producing Acinetobacter baumannii strain on biofilm formation and attachment to eukaryotic cells. Virulence 7:443–455. 10.1080/21505594.2016.1145335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Amend SR, Valkenburg KC, Pienta KJ. 2016. Murine hind limb long bone dissection and bone marrow isolation. J Vis Exp (110):53936. 10.3791/53936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang X, Goncalves R, Mosser DM. 2008. The isolation and characterization of murine macrophages. Curr Protoc Immunol Chapter 14:Unit 14.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hood-Pishchany MI, Pham L, Wijers CD, Burns WJ, Boyd KL, Palmer LD, Skaar EP, Noto MJ. 2020. Broad-spectrum suppression of bacterial pneumonia by aminoglycoside-propagated Acinetobacter baumannii. PLoS Pathog 16:e1008374. 10.1371/journal.ppat.1008374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GR, Perlman H. 2013. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am J Respir Cell Mol Biol 49:503–510. 10.1165/rcmb.2013-0086MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, Gerstein M, Snyder M. 2007. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev 21:601–614. 10.1101/gad.1510307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download iai.00454-21-s0001.pdf, PDF file, 0.2 MB (165.2KB, pdf)
Supplemental material. Download iai.00454-21-s0002.pdf, PDF file, 2.5 MB (2.5MB, pdf)
Data Availability Statement
The assembled genomes of A. baumannii 17978 VU and 17978 UN variants that were sequenced using PacBio sequencing technology are available in the NCBI database under accession numbers CP079213.1 (VU variant) and CP079212.1 (UN variant). The newly deposited reference genome of the A. baumannii 17978 UN variant that was sequenced using Nanopore long-read sequencing technology is available in the NCBI database under accession numbers CP079931 (chromosome), CP079932 (pAB3), CP079933 (pAB1), and CP079934 (pAB2).