Deshpande et al. use a single-embryo explant assay, RNAi, and micromanipulation to show that antiparallel microtubule cross-linking by Fascetto (Feo) and the kinesin-4, Klp3A, is required for regular nuclear distribution in the Drosophila melanogaster syncytial embryo.
Abstract
The early insect embryo develops as a multinucleated cell distributing the genome uniformly to the cell cortex. Mechanistic insight for nuclear positioning beyond cytoskeletal requirements is missing. Contemporary hypotheses propose actomyosin-driven cytoplasmic movement transporting nuclei or repulsion of neighbor nuclei driven by microtubule motors. Here, we show that microtubule cross-linking by Feo and Klp3A is essential for nuclear distribution and internuclear distance maintenance in Drosophila. Germline knockdown causes irregular, less-dense nuclear delivery to the cell cortex and smaller distribution in ex vivo embryo explants. A minimal internuclear distance is maintained in explants from control embryos but not from Feo-inhibited embryos, following micromanipulation-assisted repositioning. A dimerization-deficient Feo abolishes nuclear separation in embryo explants, while the full-length protein rescues the genetic knockdown. We conclude that Feo and Klp3A cross-linking of antiparallel microtubule overlap generates a length-regulated mechanical link between neighboring microtubule asters. Enabled by a novel experimental approach, our study illuminates an essential process of embryonic multicellularity.
Introduction
The nucleus relocates within the cell boundary in response to cell function (Bone and Starr, 2016; Gundersen and Worman, 2013). Aberrant nuclear positioning has been linked to failure of fundamental processes such as early embryo development, cell differentiation, cell migration, polarity determination, and homeostasis (Almonacid et al., 2015, 2019; Levy and Holzbaur, 2008; Minc et al., 2011; Neelam et al., 2015; Starr and Han, 2002). In mononuclear cells, cytoskeletal elements mechanically connect the nucleus to the cell cortex and act as the reference system for positioning (von Dassow et al., 2009; Pécréaux et al., 2016). One exception are large eggs in which cytoskeletal links between the nucleus and the distant cell cortex are not achieved (Wühr et al., 2009). Conversely, a multinucleated cell (coenocyte) undergoing nuclear proliferation has to disseminate positional information to each additional nucleus and requires a mechanism that adjusts the distance between neighboring nuclei (Gibeaux et al., 2017; Manhart et al., 2018). The early embryo of Drosophila melanogaster is both large and multinucleated but exhibits a surprising positional regularity of hundreds of nuclei perturbed by cycles of metasynchronous nuclear divisions (Foe and Alberts, 1983). During the first seven cycles, the nuclei spread axially from the anterior to the posterior end of the syncytial embryo and occupy the entire volume of the embryo (Baker et al., 1993). During nuclear cycles 7–9, most nuclei migrate to the embryo cortex, where they undergo additional rounds of division as they are anchored and prepared for cellularization (Lecuit and Wieschaus, 2000). Adequate numbers of nuclei and their proper positioning at the cortex determine cell size (Callaini et al., 1992) and precision of developmental patterning (Petkova et al., 2019), and are a result of regular distribution of ancestor nuclei during the preceding developmental phase (Kao and Megraw, 2009; Megraw et al., 1999; Vaizel-Ohayon and Schejter, 1999). The mechanisms required for maintaining uniform internuclear distances are not understood.
Drug inhibition and mutagenesis suggest that actomyosin-mediated cortical contractions drive cytoplasmic streaming and transport the nuclei predominantly along the longer axis of the embryo (Callaini et al., 1992; von Dassow and Schubiger, 1994; Deneke et al., 2019; Hatanaka and Okada, 1991; Royou et al., 2004; Wheatley et al., 1995). However, large-scale transport of cytoplasm explains neither how a uniform distribution emerges nor how nuclei are kept separate. Conversely, astral microtubules from neighboring nuclei may interact and generate repulsive force by motor binding and sliding antiparallel overlaps (Baker et al., 1993), which is reminiscent of the spindle midzone model explaining spindle elongation during anaphase B (Fu et al., 2009; Khmelinskii et al., 2009; Scholey et al., 2016). The effector Klp61F, a homotetrameric, bipolar kinesin-5, binds two overlapping microtubules and, when microtubules are antiparallel, slides them outward, reducing microtubule overlap length (Cheerambathur et al., 2013; Reinemann et al., 2017; Tao et al., 2006). Fascetto (Feo) is the Drosophila homologue of the Ase1p/Prc1/MAP65 family of homodimeric nonmotor MAPs that preferentially binds antiparallel microtubule overlaps (Bieling et al., 2010; Subramanian et al., 2010; Vernì et al., 2004). It accumulates at the spindle midzone from anaphase to telophase upon cyclin B degradation and controls the binding affinity of molecular motors in the spindle midzone (Hu et al., 2012; Khmelinskii et al., 2009; Kwon and Scholey, 2004; Wang et al., 2015; Zhu et al., 2006). One of these motors is Klp3A, a kinesin-4 homologue and inhibitor of microtubule dynamics (Bieling et al., 2010; Bringmann et al., 2004; Kwon et al., 2004; Subramanian et al., 2013; Williams et al., 1997). Prc1 and kinesin-4 are sufficient to form a stable microtubule overlap in vitro (Bieling et al., 2010). Kinesin-5 can reduce the overlapping, antiparallel microtubules cross-linked by Prc1 in vitro (Subramanian et al., 2010), which was proposed to contribute to force balance in the spindle midzone during anaphase B (Scholey et al., 2016).
Here, we investigated whether these three proteins are required for nuclear separation, lending support to an aster–aster interaction model (Baker et al., 1993) which has been reconstituted in Xenopus laevis egg extract (Nguyen et al., 2014, 2018). We performed a combination of gene knockdown, micromanipulation, and perturbation by exogenous protein addition in embryo explants to enable previously unachieved time-lapse visualization of nuclear and cytoskeletal dynamics.
Results
Feo localization confirms antiparallel microtubule overlaps between asters of nonsister nuclei
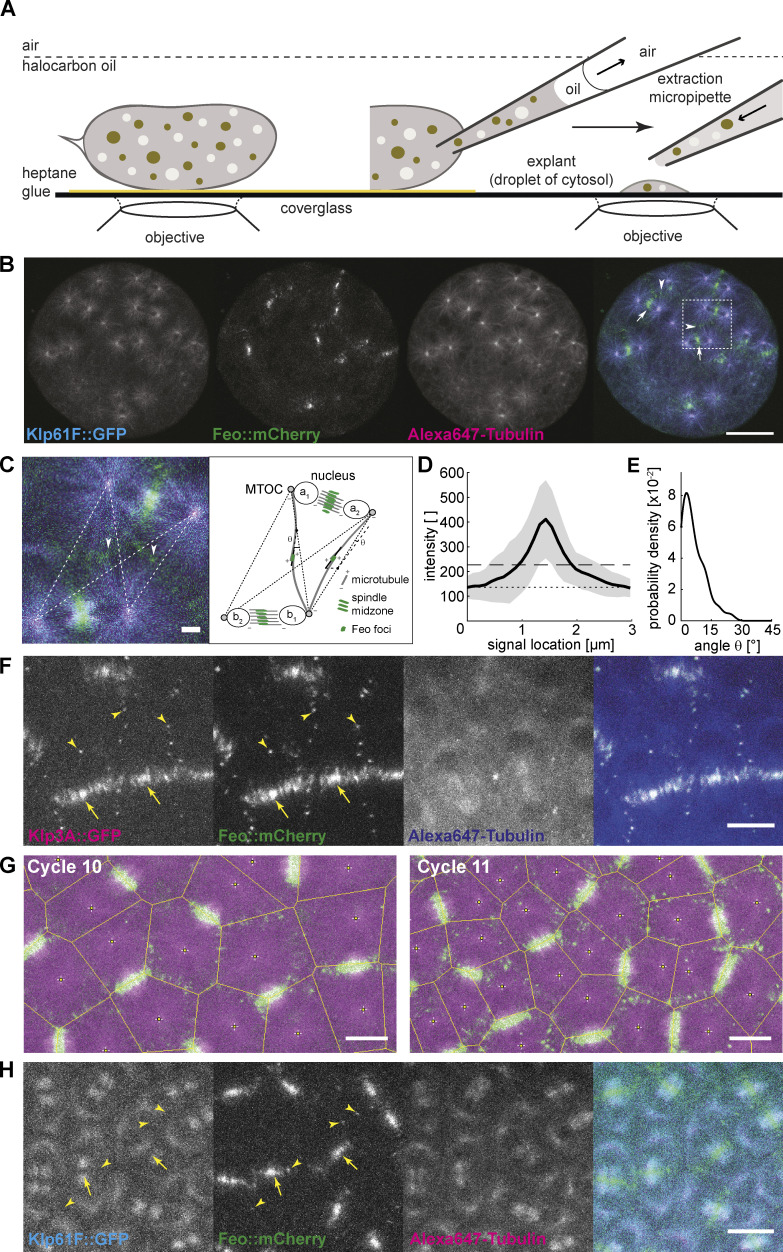
Molecular cross-linking between astral microtubules of neighboring nuclei during the preblastoderm embryo stage has largely been unexplored because of optical constraints in live imaging. Using an extraction method to generate embryo explants from individual preblastoderm embryos (de-Carvalho et al., 2018) expressing Klp61F::GFP and Feo::mCherry and injected with Alexa Fluor 647–labeled Tubulin (Fig. 1 A), we visualized the localization of Klp61F and Feo to study their binding to spindle microtubules (Fig. 1 B). Klp61F::GFP localized at the microtubule-organizing centers (MTOCs), the metaphase spindle, and the spindle midzone in anaphase, as described previously for the nuclear divisions at the blastoderm stage (Heck et al., 1993; Sharp et al., 1999; Tao et al., 2006; Cheerambathur et al., 2008, 2013; Video 1). Furthermore, during anaphase B and telophase, we observed Klp61F::GFP-decorated microtubules intercalating with those from the neighboring aster, raising the possibility of antiparallel alignment of these astral microtubules forming an overlap zone to which kinesin-5 binds. On the other hand, Feo::mCherry exhibited weak localization to the metaphase spindle but strong localization to the spindle midzone during anaphase B and telophase (Fig. 1 B, arrows), as previously described for blastoderm division cycles (Wang et al., 2015). Strikingly, Feo also localized as small foci to the region between the nuclei (Fig. 1, B and C, arrowheads), thus reporting the presence of antiparallel microtubule overlaps to which Feo binds with higher affinity than individual microtubules. In vitro, microtubule overlaps that are decorated by Feo homologues are length controlled through microtubule stabilization and sliding by kinesin-4 (Bieling et al., 2010). Thus, the signal of Feo along microtubule overlaps should have a consistent length for a given concentration or activity of Feo and Klp3A. Accordingly, we measured the length of Feo::mCherry signal foci during anaphase B (Fig. 1 D). Because individual microtubules were not resolved, we measured the orientation of the signal foci in the context of where microtubules were growing and radially emanating from the MTOCs at the spindle pole. In anaphase and telophase, the four nuclei emerging from any two neighboring spindles defined four MTOCs and, thus, four possible combinations of astral microtubule interaction (Fig. 1 C, right). We measured the angle between the long axes of the signal foci and the closest connecting line between two MTOCs (Fig. 1 E). deviated little from 0, supporting the notion that Feo reports microtubule overlaps along the shortest path between neighboring asters. Altogether, in explants from preblastoderm embryos, the relative position of nuclei and the length of astral microtubules led to the formation of short, antiparallel, Feo-decorated microtubule overlaps. Furthermore, as a consequence of Feo cross-linking astral microtubules, a mechanical connection was established that may control the distance between neighboring asters and their associated nuclei.
Figure 1.
Feo, Klp3A, and Klp61F localization confirm antiparallel microtubule overlaps between asters of nonsister nuclei. (A) Schematic showing a Drosophila syncytial embryo immobilized to the coverslip and covered with a thin layer of halocarbon oil ready for time-lapse microscopy. On the right, a preblastoderm embryo is punctured for extraction and deposition of cytosol on the coverslip using a micropipette, thereby generating a series of embryo explants. (B) Three-color snapshot from a time lapse (see also Video 1) of an explant generated from an embryo expressing Klp61F::GFP (cyan) and Feo::mCherry (green) and injected with Alexa Fluor 647–Tubulin (magenta). During the anaphase/telophase transition, Feo strongly localized to the spindle midzone (arrows) and to the intercalating microtubules from neighboring nuclei (arrowheads). Scale bar, 30 µm. (C) Zoom-in of the merged color channel image in B (dashed square). Feo localized as intense foci between neighboring spindles, where microtubules from nonsister nuclei meet (arrowheads show examples). The schematic on the right represents the configuration shown in the image, exemplifying the location of the two pairs of sister nuclei, a1–a2 and b1–b2, and two representative Feo foci. The dashed lines represent the shortest path of microtubule interactions between the MTOCs of nonsister nuclei. An intensity profile of the foci was generated by drawing a line (continuous) along the longest axis and centered to the foci. The angle θ relative to the dashed interaction line was determined. Scale bar, 2 µm. (D) The average intensity profile of Feo foci indicates foci length of 1.0 ± 0.35 µm. The gray area designates the SD, the dotted line marks the background level, and the dashed line marks two times SD above the background. Experimental repeats (N) = 7; n = 57. (E) The distribution of angles (θ) suggests that the antiparallel microtubule overlaps occur mostly along the connecting line between the neighboring nonsister nuclei. N = 7; n = 42. Cases where foci were symmetric and a long axis could not be determined were excluded from the analysis. (F) Three-color snapshot of a blastoderm embryo expressing Klp3A::GFP (magenta) and Feo::mCherry (green) and injected with Alexa Fluor 647–Tubulin (blue) showed that Klp3A colocalizes with Feo at the spindle midzone (arrows) and, more strikingly, as foci between neighboring nonsister nuclei (arrowheads). Scale bar, 10 µm. Refer to Video 3. (G) Two-color still images of a blastoderm embryo expressing RFP::β-tubulin (magenta) and Feo::GFP (green) during cycles 10 and 11, with Voronoi lines overlaid in yellow. The Voronoi segmentation was calculated with respect to the location of spindle poles and marks all locations with equidistant neighbors. Scale bar, 10 µm. (H) Three-color snapshot of a blastoderm embryo expressing Klp61F::GFP (cyan) and Feo::mCherry (green) and injected with Alexa Fluor 647–Tubulin (magenta) shows that Feo localized strongly between sister nuclei as part of the spindle midzone (arrows) and, unlike Klp61F, between neighboring nonsister nuclei as distinct foci (arrowheads). Scale bar, 10 µm. Refer to Video 2.
Video 1.
Maximum-intensity Z-projection from a 3D time-lapse video of an explant generated from an embryo expressing Klp61F::GFP (cyan) and Feo::mCherry (green) and injected with Alexa Fluor 647–Tubulin (magenta). Time in h:min:s; scale bar, 30 µm; acquired at 4 frames min−1; and displayed at 5 frames s−1. In support of Fig. 1.
During the last four syncytial nuclear cycles at the cortex, actin-based pseudo-compartments that drive membrane invagination—a physical barrier that is assembled and disassembled in every division cycle (Kotadia et al., 2010; Mavrakis et al., 2009a)—are thought to guarantee nuclear separation. Surprisingly, time-lapse confocal imaging of live embryos expressing Klp3A::GFP and Feo::mCherry (Fig. 1 F and Video 2) and injected with Alexa Fluor 647–Tubulin, revealed Feo colocalizing with Klp3A at the spindle midzone (arrows) and spot-like signals between neighboring spindles (arrowheads) in anaphase and telophase. On one hand, this observation suggests the combined activity of Feo and Klp3A, whereby Feo binding to microtubule overlaps recruits Klp3A to the overlap (Bieling et al., 2010; Subramanian et al., 2013). On the other hand, the signal foci indicate that antiparallel microtubule overlaps occur between neighboring nonsister nuclei across actin furrows and membrane invaginations. With consecutive division cycles and increasing nuclear packing, the foci maintain size but become more frequent (Fig. S1 A). Triangulation analysis revealed that foci appear at locations equidistant to neighboring spindle poles, in the center of the aster–aster overlap zone (Figs. 1 G and S1 B), and their size is comparable to that observed in explants (Fig. S1 C). In embryos expressing Klp61F::GFP and Feo::mCherry, the GFP signal is excluded at the spindle midzone, where Feo::mCherry localizes strongly. Although the Klp61F concentrates at the spindle poles, it does not show distinct localization at the aster–aster interaction zone (Fig. 1 H and Video 3). Altogether, these observations led us to question the current paradigm that actin pseudo-compartments prevent microtubule cross-linking between neighboring asters or nuclei. We hypothesize from this localization data that the microtubule-based mechanical connection plays a decisive role in nuclear positioning in preblastoderm- and early-blastoderm–stage embryos. Since microtubules are a prerequisite, we perturbed microtubule dynamics with low doses of the depolymerization drug nocodazole. We supplemented this drug after explant deposition, using a fine micropipette and buffer conditions described elsewhere (Telley et al., 2013). A concentration of 4 µM sustained spindle assembly and chromosome segregation but abolished daughter nuclei separation (Video 4). The microtubule perturbation caused local aggregation of dividing nuclei. We also observed chromosomes fusing and forming larger nuclei. Those nuclei that were more distant from the drug injection site still separated consistently, which we attribute to a diffusion gradient in drug concentration. Overall, this experiment demonstrates that nuclear separation depends on microtubules and supports a role of Feo and Klp3A in nuclear positioning.
Video 2.
Maximum-intensity Z-projection from a 3D time-lapse video of an embryo expressing Klp61F::GFP (cyan) and Feo::mCherry (green) and injected with Alexa Fluor 647–Tubulin (magenta). Time in h:min:s; scale bar, 10 µm; acquired at 4 frames min−1; and displayed at 5 frames s−1. In support of Fig. 1.
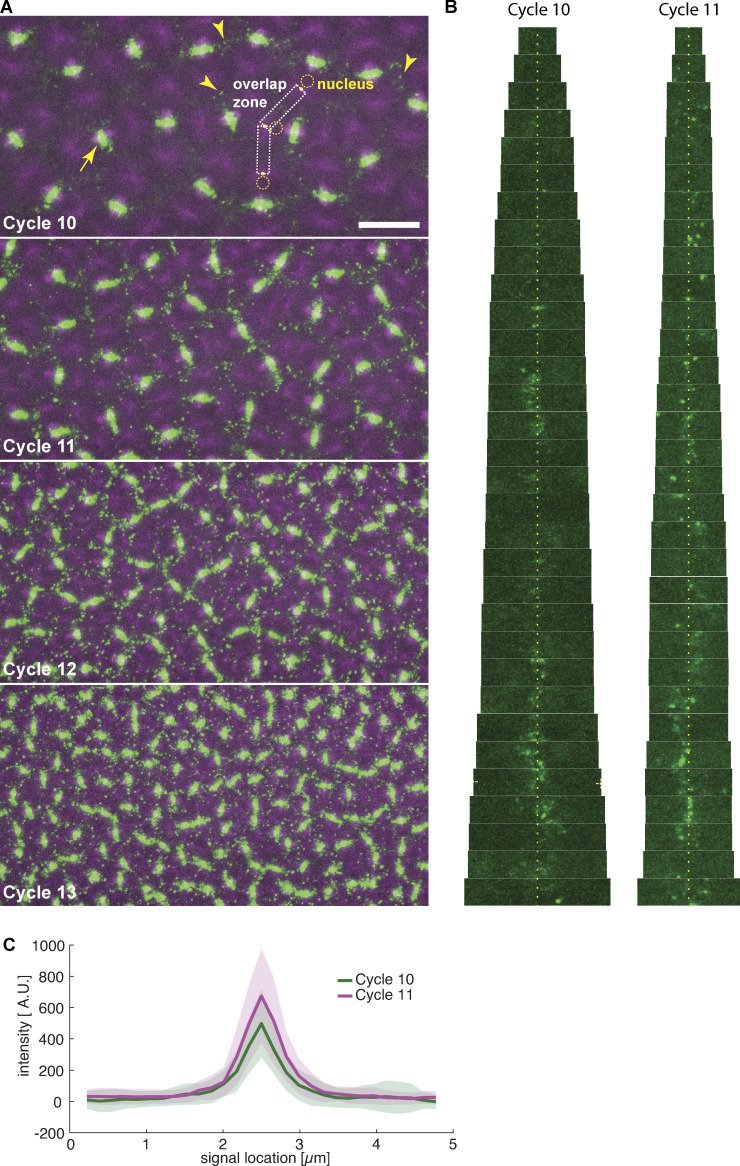
Figure S1.
Feo localization between neighboring nuclei in the developing embryo. Related to Fig. 1. (A) Two-color still images of a blastoderm embryo expressing RFP::β-tubulin (magenta) and Feo::GFP (green) during cycles 10, 11, 12, and 13, showing Feo localization between sister nuclei as part of the spindle midzone (arrow) and between neighboring nonsister nuclei as distinct foci (arrowheads). In cycle 10, very few Feo foci are visible; their appearance and density increase with every further division cycle. Qualitatively, their size does not change. For further quantification, we tracked nuclei (dashed circle; visible by the circular absence of tubulin signal) and the spindle pole (yellow dot; the peak signal in the tubulin channel next to the nuclei) and cropped the image into putative microtubule overlap zones (dashed rectangles). Scale bar, 20 µm. (B) Sequence of fluorescence images of Feo::GFP cropped from the putative overlap zones (see A), ordered by size, i.e., distance between neighboring spindle poles, for cycles 10 and 11 (n = 3 embryos). As expected, neighbor distance is overall smaller in cycle 11. Note that signals appear predominantly in the center of the overlap zones (yellow dotted line), representing overlaps equidistant from each neighbor spindle pole. (C) Quantification of foci size embryos of cycles 10 and 11 (where foci can still be reliably distinguished). The average size of foci is indistinguishable for these two cycles and matches the size of foci observed in explants (Fig. 1 D). Shaded areas represent SD (n = 39).
Video 3.
Maximum-intensity Z-projection from a 3D time-lapse video of an embryo expressing Klp3A::GFP (cyan) and Feo::mCherry (green) and injected with Alexa Fluor 647–Tubulin (blue). Time in h:min:s; scale bar, 10 µm; acquired at 4 frames min−1; and displayed at 5 frames s−1. In support of Fig. 1.
Video 4.
Single-plane time-lapse video of an explant generated from an embryo expressing Jupiter::GFP (green) and H2Av::RFP (magenta), after addition of Nocodazole on the left side of the explant using a fine micropipette, aiming for a final concentration of ∼4 µM. Time in min:s; scale bar, 50 µm; acquired at 2 frames min−1; and displayed at 5 frames s−1.
Knockdown of Feo, Klp3A, or Klp61F leads to defective nuclear delivery to the embryo cortex
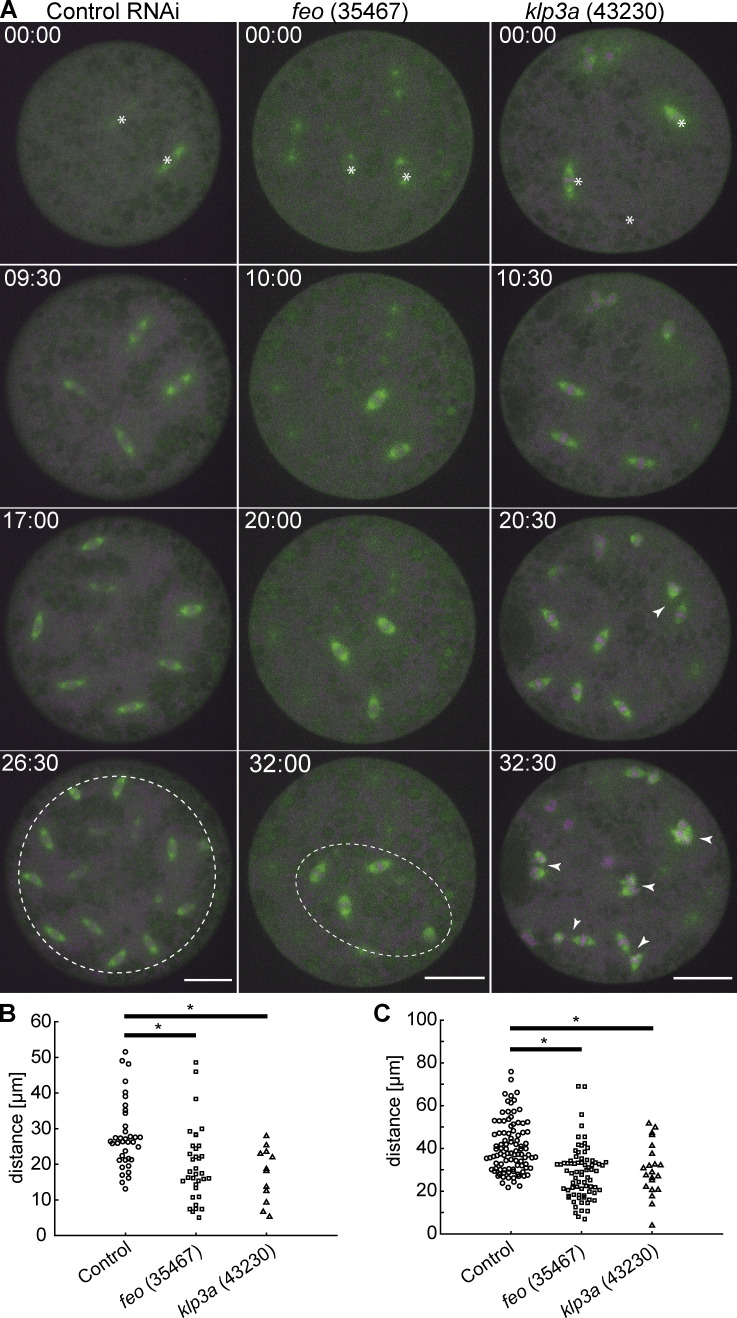
We wanted to understand the functional implication of the microtubule-binding proteins that localize between nonsister nuclei and if they are required for correct nuclear delivery to the cortex. To this end, we perturbed the protein levels of Feo or Klp3A using an RNAi approach and upstream activating sequence (UAS)–Gal4 expression in the germline (Staller et al., 2013). We also targeted Klp61F for phenotypic comparison. Using different available fly lines (TRiP), we expressed RNAi against these genes individually in the developing Drosophila oocyte (Fig. S2 A), while expressing Jupiter::GFP, a microtubule reporter (Morin et al., 2001), and H2Av::RFP, a chromatin reporter (Schuh et al., 2007). We exploited the expression kinetics of V32–Gal4 to drive the UASp–RNAi constructs with peak in late oogenesis to prevent undesirable defects during stem cell division. The efficiency of knockdown was measured at the RNA level using a quantitative PCR approach (Table 1 and Materials and methods). Fertilization in embryos inhibited for Feo, Klp3A, or Klp61F expression was similar to that of control embryos. However, we were unable to determine the exact cycle number when nuclei arrived at the cortex in knockdown embryos. Of note, the interval of division cycles occurring at the cortex and in the embryo explants remained unaltered when compared with controls. One RNAi construct against feo (TRiP 28926) did not show any phenotype. Under all other knockdown conditions, we observed nuclei arriving later on average: ∼45 min in knockdown condition versus ∼15 min in controls, following a 45-min egg laying period. In knockdown embryos, nuclei were irregularly distributed at the cortex, exhibiting clustering and sometimes missing entirely at the posterior end, in contrast to the regular distribution seen in the control RNAi embryo (Figs. 2 A and S2 B). The nuclear density is reduced after knockdown as compared with the control (Fig. 2 B) but exhibited considerable variability between embryos, which we attributed to variability in UAS-Gal4–mediated expression of RNAi between individual embryos. Reassuringly, in feo RNAi embryos, we observed a reduction (below detection) of fluorescence intensity of Klp3A::GFP at the midzone and between neighbor asters (Fig. S2 C and D). This confirms Klp3A being downstream of Feo binding to microtubule overlaps (Bieling et al., 2010).
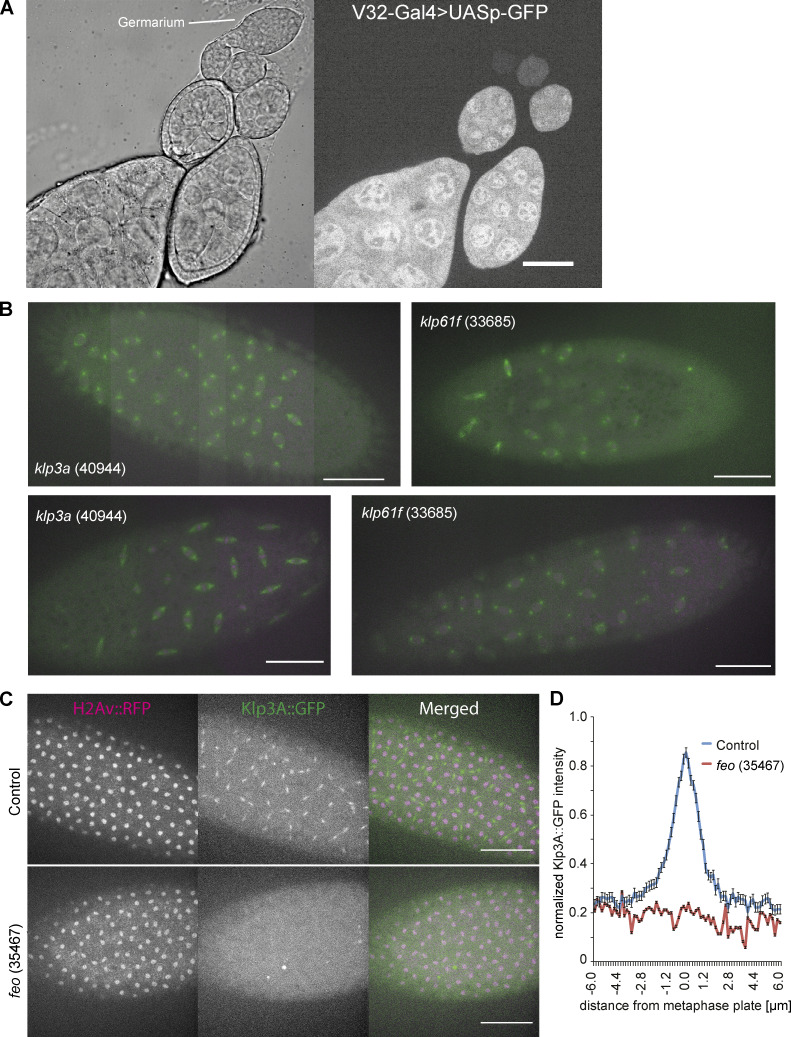
Figure S2.
Visualization of the GAL4 expression pattern during oogenesis driving the RNAi knockdown, images from embryos expressing two alternative RNAi constructs, and evidence for the delocalization of Klp3A at the spindle midzone when Feo is knocked down. (A) V32–Gal4 drives expression during late oogenesis. Related to Fig. 2. A construct expressing V32–Gal4 driving UASp–GFP expression specifically in the female germline indicates that the peak expression of GFP is achieved only at late stages of oogenesis. It illustrates the expression pattern of UASp constructs under the same Gal4 driver, including the various RNAi constructs described here with maximum effect in late oogenesis. Scale bar, 10 µm. (B) Knockdown of Klp3A (40944) or Klp61F (33685) by RNAi leads to defective nuclear delivery to the embryo cortex. Maximum-intensity projections from 3D time-lapse videos of embryos inhibited for Klp3A or Klp61F expression by RNAi while expressing Jupiter::GFP (green) marking microtubules and H2Av::RFP (magenta) marking chromatin. These two alternative RNAi-expressing fly lines exhibited similar irregularity in nuclear distribution during the first interphase occurring at the cortex. Scale bar, 50 µm. (C and D) Depletion of Feo abolishes recruitment of Klp3A to the spindle midzone. (C) Snapshots from a time lapse of embryos expressing H2Av::RFP (magenta, left) and Klp3A::GFP (green, middle) during anaphase B or telophase. Feo-knockdown embryos failed to recruit Klp3A at the spindle midzone when compared with the control embryos expressing no feo RNAi. (D) Quantification of Klp3A::GFP intensity measured at the spindle midzone along the spindle axis in control and Feo RNAi (35467) embryos. Scale bar, 50 µm.
Table 1. Quantitative PCR results of the RNAi-expressing fly lines examined in this study.
| Fly line | Fold-change | Average RNA depletion (%) | ||
|---|---|---|---|---|
| Replicate 1 | Replicate 2 | Replicate 3 | ||
| Feo (28926) | 0.748 | 0.219 | 0.379 | 55.1 |
| Feo (35467) | 0.002 | 0.002 | 0.002 | 99.8 |
| Klp3A (40944) | 0.484 | 0.514 | 0.459 | 51.4 |
| Klp3A (43230) | 0.835 | 0.967 | 0.922 | 9.2 |
| Klp61F (33685) | 0.003 | 0.002 | 0.003 | 99.7 |
| Klp61F (35804) | 0.253 | 0.349 | 0.299 | 70.0 |
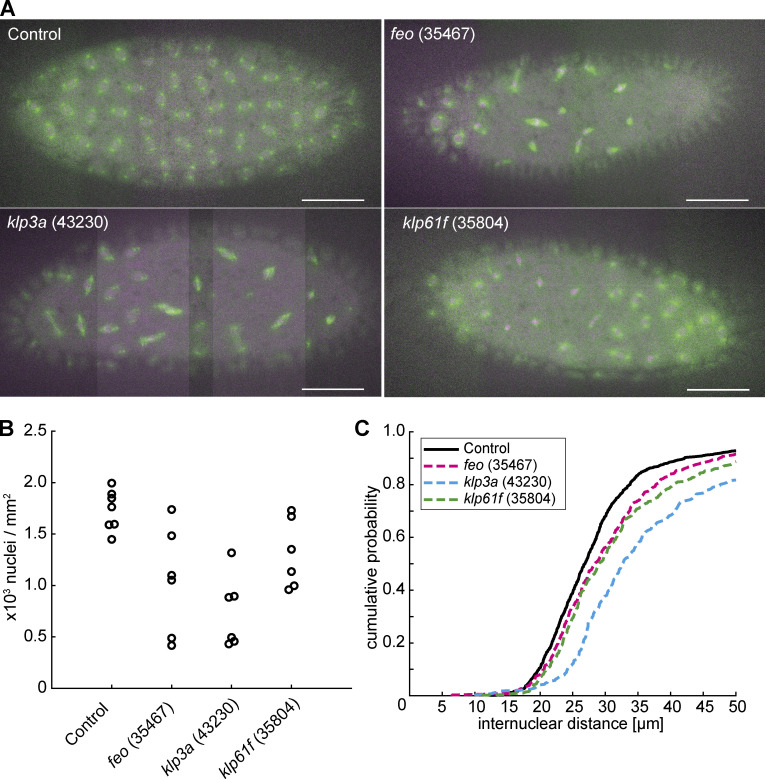
Figure 2.
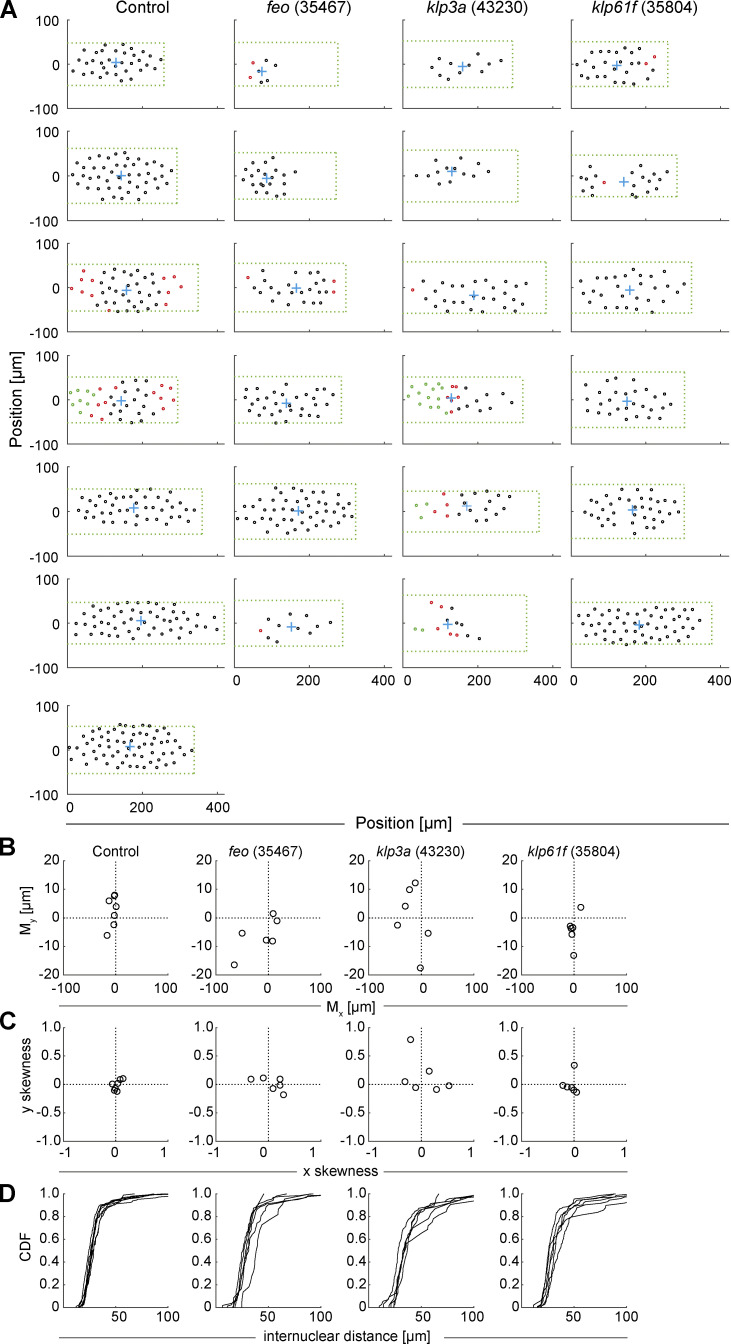
Knockdown of feo, klp3a, or klp61f by RNAi leads to defective nuclear delivery to the embryo cortex. (A) Maximum-intensity projections from 3D time-lapse videos of embryos expressing RNAi against mCherry, feo, klp3a, or klp61f and expressing Jupiter::GFP (green) marking microtubules and H2Av::RFP (magenta) marking chromatin. Knockdown embryos showed irregular nuclear distribution during the first interphase occurring at the cortex as compared with the regular nuclear distribution in control embryos (mCherry). Scale bar, 50 µm. (B) A quantification of the number of nuclei per square millimeter showed a higher degree of variation between the six embryos knocked down for either of the three genes, feo (35467), klp3a (43230), or klp61f (35804), as compared with control embryos. In all cases, the density decreased on average. Each data point represents one embryo. (C) The cumulative probability function of the internuclear distance between first-order neighbors in embryos inhibited for Feo, Klp3A, or Klp61F expression showed a flatter distribution and a median shifted to higher internuclear distance. Thus, the number of nuclei at the cortex was smaller with broader distribution, indicating greater irregularity and larger unoccupied areas with respect to the control. n = 7 (control), n = 6 (RNAi lines). Refer to Table 1, Fig. S2, and Video 5.
Our nuclear positioning analysis revealed embryos with larger areas lacking nuclei (Fig. S3 A) and with anatomically eccentric (Fig. S3 B) and asymmetric (Fig. S3 C) nuclear distribution. Overall, RNAi against feo resulted in larger distribution changes than RNAi against klp61f (Fig. S3) despite similar average internuclear distance (Fig. 2 C). The distribution of first-neighbor internuclear distance was broader and skewed toward longer distances for knockdown conditions, reflecting clusters and larger unoccupied areas, while RNAi against klp3a gave on average the strongest phenotype (Figs. 2 C and S3). The irregularity in nuclear position at the cortex increased as the cortical nuclear cycles progressed (Video 5). Occasionally, in feo knockdown embryos, we observed sister nuclei fusing after mitosis and nonsister nuclei moving toward each other, leading to fusion of the spindles and overcondensed chromatin. This likely occurs in preblastoderm cycles as well, leading to fewer nuclei arriving at the cortex and contributing to nonuniformity. Conversely, fusion was never seen in controls.
Figure S3.
Positions of nuclei in each of the analyzed embryos and distribution measurements highlight irregularity in the knockdown constructs. Related to Fig. 2. (A) The position (circle) of every nucleus arriving at the embryo cortex after the last preblastoderm division, relative to the axial and lateral borders of the embryo, for each condition: control (mCherry), Feo (35467), Klp3A (40320), and Klp61F (35804). The green dashed rectangle represents the area of the embryo bounded by the length and width of the visible embryo in the confocal stacks, with the anterior end at the coordinate origin. The blue cross represents the location of the 2D centroid determined from the position of all nuclei. The nuclei in interphase of the first division at the cortex are marked in black, the nuclei that have progressed to metaphase/anaphase are marked in magenta, and the nuclei in telophase/(next) interphase are marked in green. Note that the RNAi expression and penetration exhibit a variability, which we attribute as being responsible for the range of phenotypes. (B) Plot of the 2D centroid vector (Mx,My) of all cortical nuclei relative to the embryo center. The x axis designates the anterior-posterior axis, and the y axis is the dorsoventral axis of the embryo. Deviations from 0 mark an acentric delivery of nuclei to the cortex. (C) Skewness plot of the positional distribution of all nuclei along the anterior-posterior (x) and dorsoventral (y) axes. Feo RNAi and Klp3A RNAi embryos showed asymmetric nuclear distribution, while nuclei in Klp61F RNAi embryos were distributed symmetrically. (D) Cumulative distribution plot of the first-order neighbor distance between nuclei. All RNAi lines showed higher variability in internuclear distance as compared with the control. CDF, cumulative distribution function.
Video 5.
Maximum-intensity Z-projection from four 3D time-lapse videos of embryos expressing Jupiter::GFP (green) and H2Av::RFP (magenta) and expressing RNAi against mCherry (control), feo (35467), klp3a (43230), and klp61f (30804). Time in min:s; scale bar, 50 µm; acquired at 2 frames min−1; and displayed at 10 frames s−1. In support of Fig. 2.
In summary, the activity of all three microtubule-associated proteins is required in the preblastoderm embryo for correct delivery of nuclei to the embryo cortex. However, kinesin-5 is required for spindle assembly (Heck et al., 1993; Sawin et al., 1992), and thus the phenotype can emerge due to assembly defects rather than postmitotic nuclear separation. Because depletion of Klp61F led to a milder phenotype despite high knockdown efficiency, and because of the distinct localization of Feo and Klp3A in the aster–aster interaction zone (Fig. 1, F and G), we followed up on the role of the latter two genes in maintaining internuclear distance.
Developmental reset ex vivo reveals failure in nuclear distribution upon RNAi knockdown
Our analysis of nuclear distribution during knockdown in the embryo suggests that Feo and Klp3A are involved in nuclear delivery to the cortex. However, our knockdown approach in vivo has two drawbacks that can lead to misinterpretation: (1) the three proteins play a role in spindle midzone function, and their depletion may affect chromosome segregation in anaphase; and (2) the RNAi expression occurs chronically during late oogenesis. Thus, the irregular distribution of nuclei during cortical migration can be due to early sister chromatid separation errors, leading to missing nuclei in the embryo center and exponentially fewer in subsequent division cycles. Alternatively, inefficient nuclear separation following fertilization can lead to spindle fusion and mitotic errors. To circumvent the inability to detect accumulated effects, we performed time-lapse imaging of nuclear division cycles in embryo explants from preblastoderm embryos that were inhibited for either Feo or Klp3A protein expression. Because these explants contained only a few dividing nuclei, we followed their distribution, or the failure thereof, while mimicking the very beginning of preblastoderm embryo development. We tracked individual nuclei undergoing division cycles and registered the distribution and any fusion events between sister and nonsister nuclei (Fig. 3 A and Video 6). Importantly, the time-lapse observation of nuclear divisions allowed us to determine if nuclear distribution changes arise from reduced separation or form mitotic failures and arrest, which has different consequences for the delivery of nuclei to the embryo cortex. In explants from control embryos, nuclei divide and distribute regularly in the entire explant (Fig. 3 A, left, white dashed circle) until a saturated nuclear distribution is reached and occasional mitotic failures in the subsequent cycle are observed. The nuclear density at saturation is comparable to nuclear cycle 10 in the intact embryo (1,800–2,000 nuclei/mm2; Foe and Alberts, 1983), corresponding to an internuclear distance of ∼25 µm (hexagonal approximation). Strikingly, the nuclei from feo- and klp3a-knockdown embryos also divide consecutively. The average distance between sister nuclei and between nonsister nuclei was lower in the test RNAi as compared with the control (Fig. 3, B and C). However, the nuclear position after mitotic separation was maintained in the feo RNAi, while knockdown of klp3a led to frequent spindle fusion at a comparable nuclear density and accumulation of mitotic failure. Interestingly, spindle length decreased upon depletion of feo (Fig. 3 A, middle), but we did not observe a significant decrease in spindle length upon klp3a depletion as reported earlier, most likely due to inefficient knockdown as compared with deletion (Williams et al., 1995). In summary, the inhibition of Feo expression leads to reduced nuclear separation between sister nuclei and incomplete occupation of nuclei within the explant, a hallmark of the unoccupied spaces in the blastoderm embryo (Fig. 2). However, while a knockdown of feo sustains mitotic divisions, klp3a knockdown produces a spindle fusion phenotype. It is conceivable that the reduction of Klp3A protein causes microtubule overlap overgrowth, and despite cross-linking by Feo and other MAPs, these long overlaps allow for a net attractive force to build up between spindles (Gatlin et al., 2009). However, we cannot exclude that the partial knockdown of klp3a already causes mitotic defects related to spindle assembly and chromosome alignment. Because of these confounding effects, we decided to focus on Feo exclusively.
Figure 3.
Knockdown of feo, klp3a, or klp61f by RNAi leads to defective nuclear distribution in preblastoderm embryo explants. (A) Maximum-intensity projections from time-lapse videos of embryo explants under control conditions (mCherry) and RNAi against feo, klp3a, or klp61f while expressing Jupiter::GFP (green) marking microtubules and H2Av::RFP (magenta) marking chromatin. Each panel shows metaphase of consecutive division. White stars in the first frame mark the position of dividing nuclei (sometimes out of focus). The control explants underwent normal nuclear divisions and distributed the daughter nuclei within the entire explant volume (dashed circle). Explants from feo RNAi embryos underwent mitotic nuclear divisions, but daughter nuclei separated less efficiently, leading to a partial occupation of the cytoplasm (dashed ellipse). Explants from klp3a RNAi embryos underwent mitotic nuclear divisions with slightly less efficient distribution than in controls and with higher prevalence for spindle fusion (arrowheads). Scale bar, 30 µm; time in min:s. Refer to Video 6. (B) Separation distance between daughter nuclei after mitotic nuclear division under control conditions and under knockdown for feo (35467) and klp3a (43230) in embryo explants. Separation distance was significantly reduced in both knockdown conditions (control: experimental repeats [N] = 4, n = 38; feo [35467]: N = 2, n = 36; klp3a [43230]: N = 3, n = 23; *, P < 0.01, Wilcoxon signed-rank test). (C) Separation distance between first-neighbor nonsibling nuclei measured between mitotic divisions under control conditions and under knockdown for feo (35467) and klp3a (43230) in embryo explants. The separation distance was significantly shorter in both knockdown conditions (control: N = 3, n = 98; feo [35467]: N = 3, n = 77; klp3a [43230]: N = 3, n = 50; *, P < 0.05, Wilcoxon signed-rank test), although the effect was stronger under feo RNAi conditions.
Video 6.
Maximum-intensity Z-projection from four 3D time-lapse videos of explants expressing Jupiter::GFP (green) and H2Av::RFP (magenta) and expressing RNAi against mCherry (control), feo (35467), and klp3a (43230). Time in min:s; scale bar, 30 µm; acquired at 2 frames min−1; and displayed at 10 frames s−1. In support of Fig. 3.
Displacement of nuclei is rescued in control but not in feo RNAi embryo explants
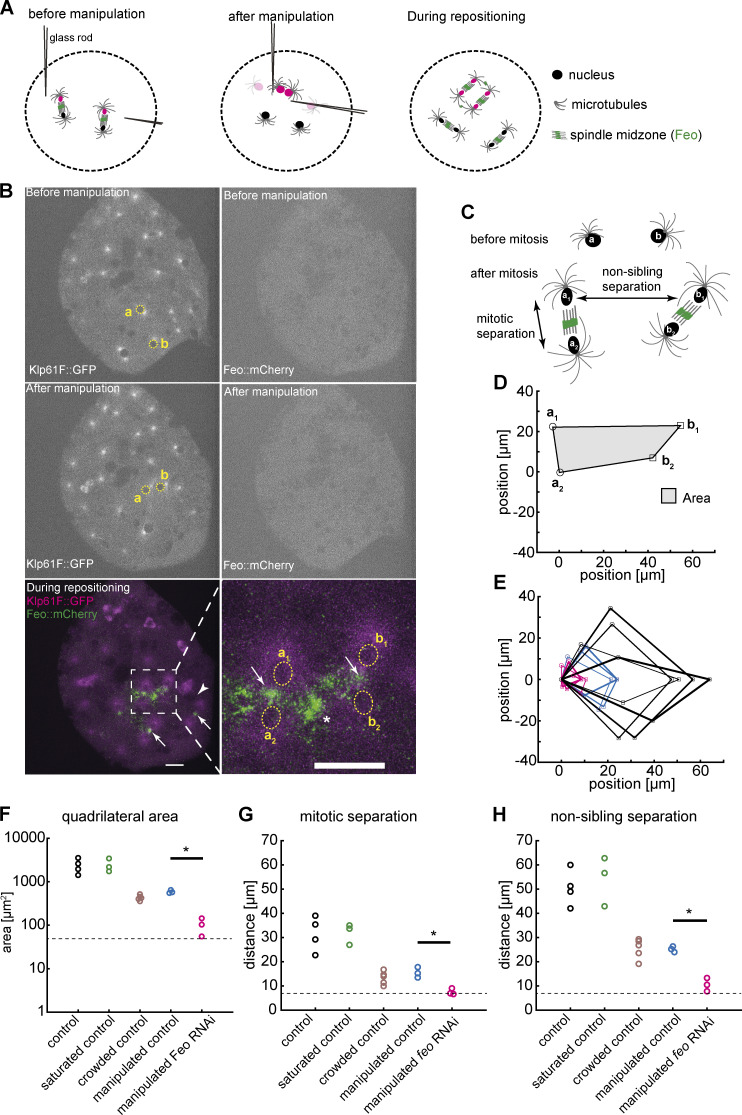
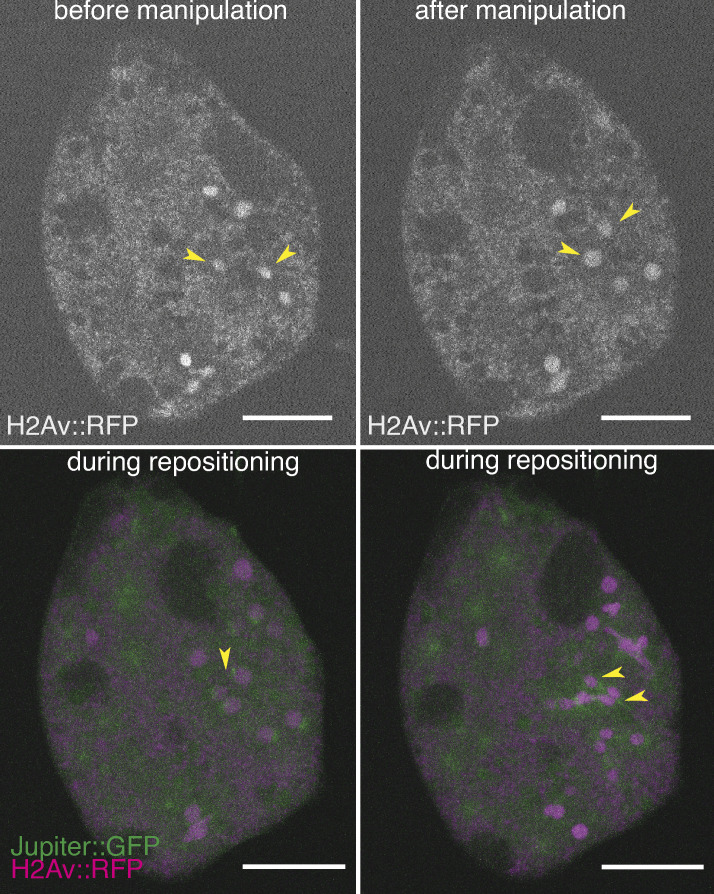
To test the model of an astral microtubule cross-linker–based separation mechanism for nonsister nuclei, we took advantage of the amenability of embryo explants for mechanical manipulation and designed an acute perturbation approach. We asked how Feo relocalizes when the distance between two interphase nuclei is manually reduced. Finally, we asked whether, under a feo-knockdown condition, nuclei could adjust their position when brought in close proximity before division. To address these questions, we performed contact micromanipulation and changed the positions of two nonsister nuclei that were exiting mitosis (Fig. 4 A, magenta; and Video 7), during the nonmitotic phase, which is amenable to manipulation (Telley et al., 2012). Following manipulation of two nuclei (Fig. 4 B, yellow), nuclei were typically in interphase, when Feo is not expected to localize due to cycle regulation. Nonetheless, we registered strong localization of Feo::mCherry in anaphase and telophase of the next cycle at midzones of all spindles, and strongly in the region between the manipulated nuclei (Fig. 4 B, bottom, asterisk), while asters from distant, nonmanipulated nuclei did not recruit the microtubule cross-linker detectably (Fig. 4 B, arrowhead). Next, we quantified nuclear separation of two neighboring nuclei dividing into four daughter nuclei by determining the four final positions (Fig. 4 C), arranging these positions as a quadrilateral; aligning, annotating, and overlaying them in a common coordinate system (Fig. 4, D and E); and calculating area (Fig. 4 F) and lateral distances (Fig. 4, G and H). We performed these measurements under the control RNAi condition for nuclei in a large unoccupied cytoplasmic space, in a saturated space where several nuclei have spread through the entire explant, and in a crowded explant representing one more division cycle after saturation; this differentiated comparison takes into account nuclear density dynamics in cycling explants, as described in the previous section (Fig. 3). We found that the area of nuclear separation after manipulation is lower than in the nonmanipulated and saturated space but indifferent from the crowded control (Fig. 4 F). The manipulated nuclei divided and separated their daughter nuclei at ∼15 µm, while the distance between nonsiblings was maintained at ∼25 µm (Fig. 4, G and H; Telley et al., 2012), phenocopying the minimal separation seen in crowded explants, for which distance maintenance is challenging. Interestingly, these separation distances are similar to what was reported for the blastoderm embryo (Kanesaki et al., 2011). Finally, we performed the manipulation of nuclear position in feo-knockdown explants expressing Jupiter::GFP and H2Av::RFP. In these experiments, after manipulation, the daughter nuclei moved toward each other rather than apart, forming nuclear aggregates (Fig. S4) and phenocopying the treatment with nocodazole as presented earlier (Video 4). The separation of siblings was approximately the nuclear diameter (∼7 µm; Fig. 4 G, dashed line) and the separation of nonsiblings was ∼10 µm (Fig. 4 H). We conclude that acute repositioning of nuclei is detected by the separation machinery, as reported by Feo, and is counteracted to prevent spindle fusion or aggregation of nuclei. In other words, Feo is required to prevent nuclear collisions.
Figure 4.
Explants fail to maintain nuclear separation distance following acute physical manipulation under feo knockdown. (A) Scheme showing the manipulation of internuclear distance in embryo explants. After a mitotic division and nuclear separation, two nonsister nuclei (magenta) were brought close to each other during anaphase B–telophase by means of two glass rods. Subsequently, nuclei divided again, and daughter nuclei separated at defined distances. (B) Fluorescence images illustrating physical manipulation of nuclear position in an explant made from an embryo expressing Klp61F::GFP (green) marking microtubules positively and nuclei negatively due to exclusion (dark disks), together with Feo::mCherry (magenta). The top row shows the GFP and mCherry signal before manipulation, and the second row shows after manipulation. Physical manipulation decreased the distance selectively between two nuclei (labeled a and b and marked with yellow circles). In the subsequent mitosis and during repositioning of the daughter nuclei (bottom row), Feo localized at spindle midzones (arrows) and between the daughters of the manipulated nucleus (star in zoomed view), indicating that microtubule overlaps had formed. In contrast, Feo localization is not detectable between manipulated nuclei before or immediately after manipulation or between nuclei that have not been moved and are further apart (bottom, arrowhead). Scale bars, 15 µm. Refer to Video 7. (C) Schematic of the mitotic separation distance and nonsister separation distance. Nuclei a and b were brought close to each other and, following a division, give rise to daughters a1, a2, b1, and b2, respectively. (D) Schematic of the quadrilateral area defined by the four nuclei a1, a2, b1, and b2 after mitosis. (E) Overlay of quadrilaterals aligned for coordinate a2 and rotated so that the vector b1–a2 matches the x axis. Control RNAi experiments without manipulation and with ample space in the explant are in black (n = 4); experiments involving manipulation under control RNAi conditions are shown in blue (n = 3); and manipulations experiments under knockdown of Feo are shown in magenta (n = 3). The quadrilateral area for nuclei manipulated in feo RNA–depleted explants was smaller when compared with control. (F) Quadrilateral area for five different experimental conditions. The color code of E applies; additional control conditions without manipulation in explants almost saturated with nuclei (n = 3) and in explants crowded with nuclei (n = 5) are shown in green and brown, respectively. The dashed line designates the lower boundary where the four nuclei touch each other. The quadrilateral area for nuclei manipulated in feo RNA–depleted explants was smaller when compared with control; *, P < 0.05, Wilcoxon signed-rank test. (G) The average mitotic separation distance between the dividing nuclei (|a1 − a2|; |b1 − b2|) was reduced in the manipulated feo RNAi condition (*, P < 0.05, Wilcoxon signed-rank test), and was close to the lower limit of separation (nuclear diameter) where the nuclei are touching each other. In contrast, sister nuclei were separated in all the control conditions. The color code is the same as in F. (H) The average nonsister separation between the dividing nuclei (|a1 − b1|; |a2 − b2|) was reduced in the manipulated Feo RNAi condition (*, P < 0.05, Wilcoxon signed-rank test), and was close to the lower limit of separation where the nuclei are touching each other. In the control, the distance between the nonsister nuclei was ∼25 µm. The color code is the same as in F.
Video 7.
Maximum-intensity Z-projection from a 3D time-lapse video of an embryo expressing Klp61F::GFP (green) and Feo::mCherry (magenta). Note that during manipulation there is additional signal from bright-field illumination, which helped visualize the glass cantilever. Time in min:s; scale bar, 30 µm; acquired at 1 frame s−1 in the first part and 2 frames min−1 in the second part; and displayed at 10 frames s−1. In support of Fig. 4.
Nuclear separation in the syncytium requires astral microtubule cross-linking by Feo
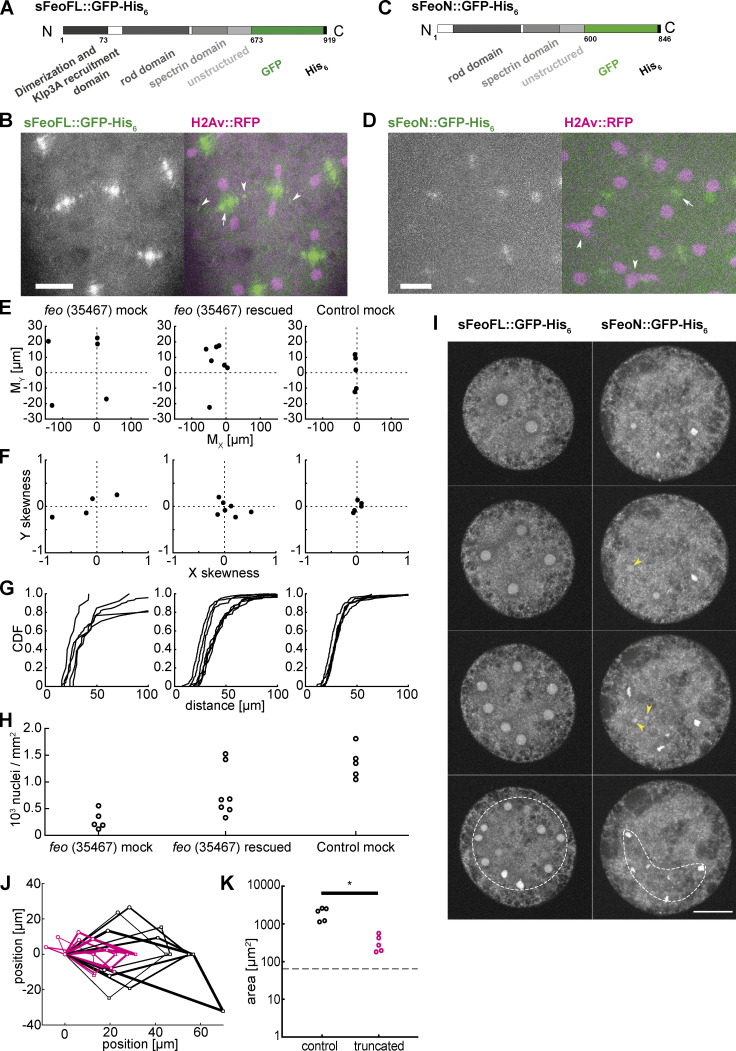
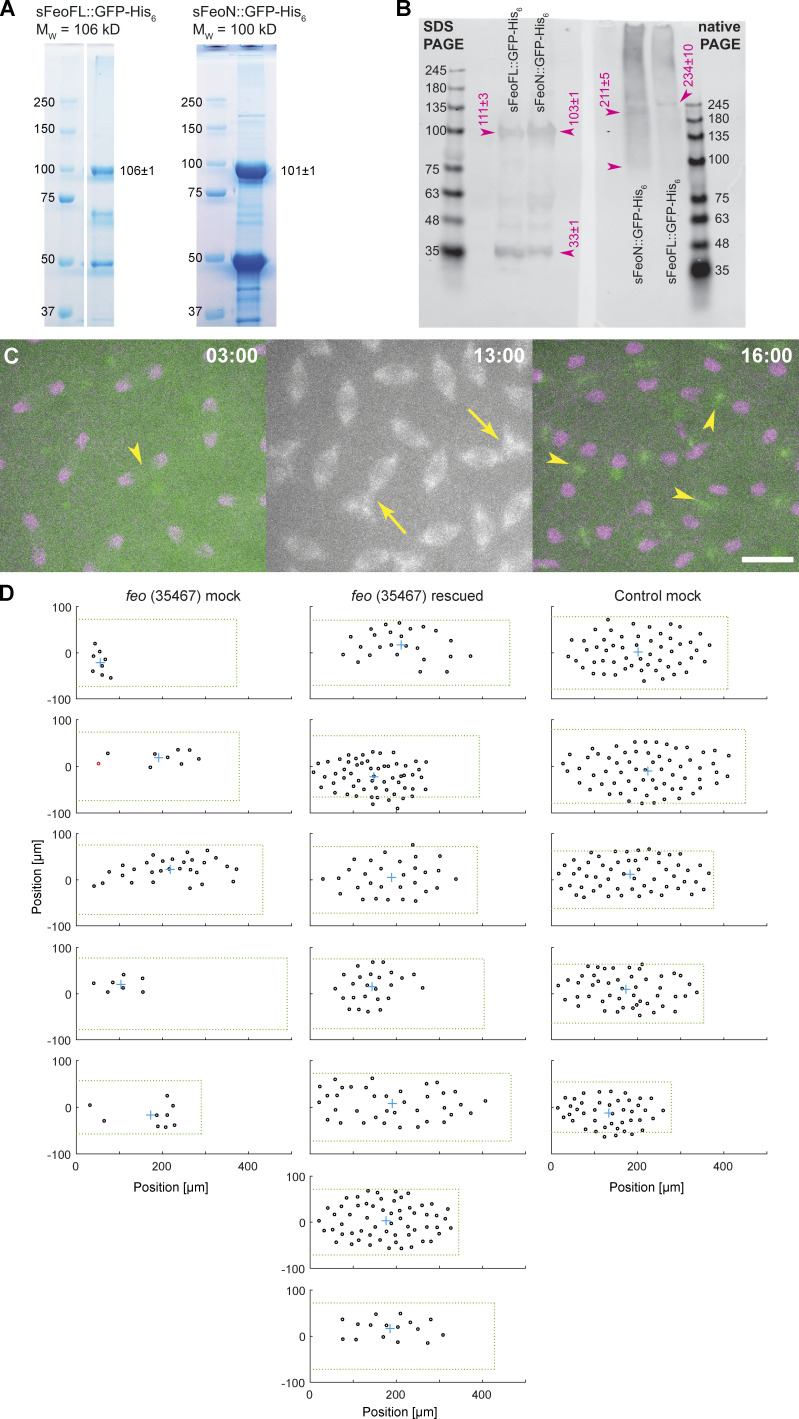
Feo is a dimer and, in vitro, has high affinity for binding two antiparallel microtubules (Bieling et al., 2010; Subramanian et al., 2013, 2010). In this function, Feo can generate a repulsive mechanical link—an apparent stiffness—that prevents concentric movement and eventual contact of neighboring nuclei. This model predicts a lower repulsion stiffness in the presence of a monomeric construct of Feo, which binds to the same microtubule lattice binding site as the full-length dimer but does not cross-link the antiparallel microtubules. We expect that this molecular competition leads to weaker cross-linking and can be measured as shorter internuclear distance, irregular separation, or frequent nuclear contacts. Thus, we designed two synthetic protein expression constructs, one containing the full feo coding sequence (sFeoFL) and the other lacking the N-terminal, putative dimerization domain (sFeoN; Subramanian et al., 2013), and fused them to a C-terminal GFP and a His6 tag sequence (Fig. 5, A and C). By design, the truncated construct should have unaltered microtubule-binding affinity, while dimerization and thus cross-linking are abolished. Proteins were expressed in Escherichia coli, affinity-purified, and dialyzed into embryo explant–compatible buffer (Telley et al., 2013; Fig. S5 A). Full-length Feo::GFP forms dimers, as was reported for vertebrate homologues (Bieling et al., 2010; Subramanian et al., 2013, 2010), while the truncated protein is dimerization deficient and forms monomers or weak dimers, as shown in a native-PAGE (Fig. S5 B). When the full-length protein was injected at nanomolar concentration into embryos, GFP signal localized at the central spindle (Fig. 5 B, arrow). As in transgenic embryos (Fig. 1, F and G, arrowheads), we also detected small foci of green fluorescence between neighboring nuclei (Fig. 5 B, arrowheads), suggesting that the purified protein and the transgenic construct localize identically and under cell cycle control, with fluorescence disappearing in interphase and reappearing during telophase of the following cycle (Video 8). Though less strong, the truncated protein construct also localized at the spindle midzone, however not in the cycle following injection but in the subsequent division cycle (Fig. 5 D, Fig. S5 C, and Video 9). The lower intensity suggests that the truncated construct competes weakly with endogenous Feo. Injection of full-length Feo::GFP maintained regular nuclear delivery to the embryo cortex, while injection of truncated FeoN::GFP caused unnatural spindle contacts and nuclear separation defects (Fig. S5 C, middle; and Video 8). Furthermore, when the full-length protein was injected into feo RNAi embryos, the defective nuclear distribution was rescued to a large extent (Fig. S5 D). Nuclei arrived at the embryo cortex more symmetrically between anterior and posterior ends (Fig. 5 E), in a less skewed distribution (Fig. 5 F), and with more uniform internuclear distance (Fig. 5 G) as compared with mock-injected feo RNAi embryos. Owing to the variability of injection and knockdown efficiency, we fully recovered nuclear density to a normal level in two embryos and significantly increased nuclear density in the remaining five embryos (Fig. 5 H). Notably, the injected protein pool was stable for ≥90 min, throughout several division cycles. In summary, we show that a GFP-tagged full-length protein construct localizes correctly and rescues the gene knockdown in the germline. We conclude that it is functionally identical to the endogenous protein that is maternally deposited in the egg and stable during syncytial development.
Figure 5.
Purified Feo protein rescues nuclear separation in feo RNAi embryos, and an N-terminally truncated Feo abolishes nuclear separation. (A) Scheme of the synthesized full-length Feo protein fusion construct containing a C-terminal GFP. The domains were determined based on sequence similarity from reported domains of the human construct. The N-terminal end induces dimerization and binds Klp3A, and the spectrin domain binds to microtubule lattice. (B) Fluorescence image of the GFP-tagged full-length Feo protein in a (control) blastoderm embryo in telophase after protein injection. The GFP signal alone (left) is shown merged with H2Av::RFP in magenta (right). sFeoFL::GFP-His6 localized correctly, under cell cycle control, at the spindle midzone (arrow) and between daughter nuclei (arrowhead) as observed in the transgenic overexpression fly line shown in Fig. 1. Scale bar, 10 µm. Refer to Video 8. (C) Scheme of a truncated Feo construct lacking the first 73 amino acids of the putative dimerization and Klp3A recruiting domain, fused to a C-terminal GFP. (D) Fluorescence image of the GFP-tagged truncated Feo protein in a (control) blastoderm embryo after protein injection. The GFP signal alone (left) is shown merged with H2Av::RFP in magenta (right). A faint GFP signal localized at the spindle midzone (arrow) in the second division after injection (Fig. S5 C). Nuclear separation defects manifest as neighboring nuclei touching or fusing after division (arrowheads). Scale bar, 10 µm. Refer to Video 9. (E) Plot of the 2D centroid vector (MX,MY) of all cortical nuclei relative to the embryo center for Feo RNAi embryos either mock injected (left; n = 5) or injected with sFeoFL::GFP-His6 protein (middle; n = 7), compared with mock injected control (mCherry) RNAi embryos (n = 5). The x axis designates the anterior-posterior axis, and the y axis is the dorsoventral axis of the embryo. Deviations from zero mark acentric delivery of nuclei to the cortex. Along the anterior-posterior axis, the injection of Feo full-length protein in Feo RNAi embryos partially rescued centering (middle); mock-injected Feo RNAi embryos had anatomically eccentric nuclei (left); and mock-injected control (mCherry) RNAi embryos exhibited strong centering. (F) Skewness plot of the positional distribution of all nuclei along the anterior-posterior (x) and dorsoventral (y) axes for the same conditions as in E. The asymmetric distribution in mock-injected Feo RNAi embryos (left) is partially rescued by Feo protein injection (middle), while mock-injected control embryos show little asymmetry. (G) Cumulative distribution plot of the first-order neighbor distance between nuclei, for the same conditions as in E and F. The irregular internuclear distances in mock-injected Feo RNAi embryos (left) are rescued to a considerable extent after full-length protein injection (middle), while mock-injected control (mCherry) RNAi embryos exhibit uniform internuclear distances (right). CDF, cumulative distribution function. (H) The low nuclear density arriving at the cortex in mock-injected Feo RNAi embryos is partially rescued when full-length Feo protein is injected in preblastoderm Feo RNAi embryos. (I) Addition of full-length GFP-tagged Feo protein to embryo explants supports normal nuclear division and regular distribution within the explant space (left, white circle), while addition of truncated Feo protein reduces nuclear separation (arrowheads), causing occasional spindle fusion, and abolishes nuclear distribution (dashed envelope). Scale bar, 30 µm. (J) Overlay of aligned quadrilaterals describing the nuclear separation after division in explants, as described in Fig. 4. Explants were generated from wild-type embryos and had ample space for the first few divisions. Experiments involving addition of full-length GFP-tagged Feo protein to the explant are in black (n = 5); experiments involving addition of N-terminally truncated, GFP-tagged Feo protein are shown in magenta (n = 5). (K) The truncated Feo protein significantly reduced nuclear separation, as measured by the area of quadrilaterals shown in J, as compared with the full-length protein construct (black).
Figure S4.
In explants inhibited for Feo protein expression, nuclear separation fails after manipulation. Related to Fig. 4. Still images during physical manipulation of two non-sister nuclei (arrowheads in top panels) in an explant made from an embryo expressing RNAi against feo and expressing Jupiter::GFP (green) marking microtubules and H2Av::RFP (magenta) marking chromatin. After manipulation (bottom panels), the nuclei failed to elicit an efficient repositioning response as observed in the control (Fig. 4). Instead, sister and nonsister nuclei failed to separate sufficiently, and nuclei came into contact or formed clusters (arrowheads in bottom panels). Scale bar, 30 µm.
Video 8.
Maximum-intensity Z-projection from a 3D time-lapse video of an embryo expressing H2Av::RFP (magenta) and injected with sFeoFL::GFP protein (green). Time in h:min:s; scale bar, 10 µm; acquired at 2 frames min−1; and displayed at 5 frames s−1. In support of Fig. 5.
Video 9.
Maximum-intensity Z-projection from a 3D time-lapse video of an embryo expressing H2Av::RFP (magenta) and injected with sFeoN::GFP (green) and Alexa Fluor 647–Tubulin (gray). The merge shows the GFP and RFP only. Time in h:min:s; scale bar, 30 µm; acquired at 2 frames m−1; and displayed at 7 frames s−1. In support of Fig. 5.
Finally, having designed and purified the truncated and full-length proteins with identical procedures, we asked how nuclear separation changes upon excess of the dimerization-deficient Feo protein, added at 100–200 nM final concentration to wild-type embryo explants containing one or two nuclei. As a control condition, we injected the full-length protein at the same final concentration into embryo explants, and despite this perturbation, the explant supported normal nuclear separation and distribution (Fig. 5 I, left). Conversely, adding the truncated protein construct worsened nuclear separation considerably after chromosomes segregated. Here, in contrast to the control condition, nuclei did not occupy the entire explant space after consecutive divisions. The short internuclear distance led to unnatural chromosome aggregation and fusion and eventually to mitotic failure. Nuclear separation of two neighboring nonsister nuclei, as measured by the quadrilateral area defined by their position, was significantly smaller than in control divisions in the presence of full-length Feo protein (Fig. 5, J and K). We conclude that microtubule cross-linking by Feo generates a repulsive mechanical link between microtubule asters. Thus, it lies at the heart of nuclear separation maintenance during the multinucleated one-cell stage of Drosophila embryo development.
Discussion
A cornerstone of embryonic development is the formation of a polarized epithelium. Plants and many invertebrates achieve this developmental stage with a unicellular embryo undergoing nuclear proliferation followed by cellularization, a specialized form of cytokinesis (Hehenberger et al., 2012; Lecuit and Wieschaus, 2000). Recently, the molecular building blocks and morphogenetic characteristics of cellularization have also been identified as part of the life cycle of a nonanimal eukaryote (Dudin et al., 2019). The offspring of Sphaeroforma arctica arises from nuclear proliferation, compartmentalization, and plasma membrane invagination, generating a proto-epithelium from which newborn cells detach. These observations support the hypothesis that epithelia evolutionarily predate animals (Dickinson et al., 2012). We propose that correct compartmentalization and generation of uninuclear offspring necessitates robust nuclear separation. If warranted true, then a separation mechanism must have coevolved with the origin of epithelia and was essential for the emergence of multicellularity.
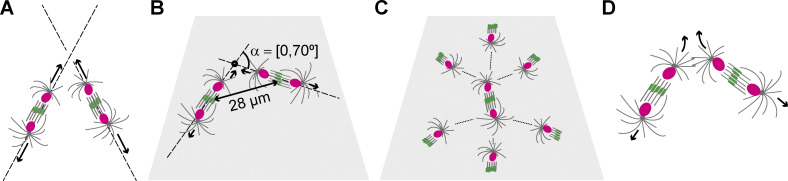
Nuclear proliferation in a coenocyte poses a new challenge: how does the cell safeguard the separation and prevent contact of nuclei while their number increases? Two solutions seem plausible. On one hand, the cell may control the division axes and separate daughter nuclei along linear paths that do not cross. On the other hand, the cell may constrain internuclear distance independently of separation trajectories. Consider two nuclei that are about to divide and separate their progeny along the spindle axis (Fig. 6 A). In a 3D space, none of the daughter nuclei may collide unless the spindle axes are both coplanar and nonparallel. Typically, nuclei migrate only 10–15 µm away from the original spindle center before dividing again (Telley et al., 2012). This geometric constraint reduces configurations that produce colliding trajectories in a 2D topology to ∼40% of all possible spindle axis orientations, so that axes intersect at an angle between 0° (collinear) and 70° (Fig. 6 B). Adding complexity, spindles in a network with optimal packing face a number of neighbors (6 in 2D and 12 in 3D; Fig. 6 C). Thus, a synchronously dividing spindle network will inevitably produce colliding trajectories of daughter nuclei. It is therefore necessary that, instead of controlling division axes, the cell controls nuclear proximity independently of the relative orientations they divide (Fig. 6 B). This enables the syncytial embryo to divide hundreds of nuclei synchronously and distribute them to any unoccupied position. Here, we demonstrate a molecular mechanism that responds to short internuclear distances in the syncytium with microtubule-dependent repulsion. Each nucleus is associated with a radial array of microtubules nucleated by the centrosome, which duplicates and forms the two spindle poles in the next division. First, however, this microtubule aster guides nuclear migration and grows large enough to encounter microtubules from neighboring asters that migrate as well. This encounter leads to interdigitation of the microtubule plus ends (antiparallel overlaps) and forms binding sites for cross-linking proteins. Our data shows that Feo, the Prc1 homologue in Drosophila and antiparallel microtubule cross-linker, plays a central role in defining a minimal internuclear distance in the syncytial Drosophila preblastoderm embryo.
Figure 6.
Feo and Klp3A prevent collision trajectories of dividing nuclei in space and on 2D topologies. (A) Two neighboring spindles with division axes that are oblique. Nuclei separate along the spindle axis, which do not have an intersecting point and do not cause nuclear collision. (B) Two neighboring spindles with coplanar spindle axes. If these axes are not parallel, they will always form an intersection point. However, because of the short nuclear migration from the previous spindle center (∼14 µm), the nuclear diameter (∼5 µm), and the average interspindle distance (∼28 µm), two nonsibling nuclei will collide only if the relative angle α between spindle axes is ≤70°. (C) In a 2D topology of spindles with optimal packing, each spindle has six neighbors. In this configuration, and considering the geometric constraints shown in B, any orientation of spindle axis for the spindle in the center will lead to collisions with nonsibling nuclei. (D) Model of aster-mediated repulsion between neighboring nuclei on a colliding trajectory after mitosis. Astral microtubule cross-linking by Feo and Klp3A generates a repulsive mechanical element that deviates the direction of separating nuclei from the spindle axis.
Vertebrate Prc1 is a microtubule-binding protein with high turnover kinetics and ≥28 times higher affinity for antiparallel microtubule overlaps than for single microtubules (Bieling et al., 2010). This biochemical property, together with fluorescent labeling, renders Prc1 homologues reliable reporters for microtubule aster overlaps in live-cell imaging assays (Nguyen et al., 2014). Prc1 cross-linking antiparallel microtubules generates a high-affinity binding site for the motor protein kinesin-4 (Kif4/Xklp1/Klp3A) at the overlap (Bieling et al., 2010; Kurasawa et al., 2004). In Xenopus eggs and early embryos, where asters are unusually large, Prc1 and Kif4A define the radial organization and dynamics of microtubules and prevent invasion of neighboring asters by antiparallel microtubule cross-linking (Nguyen et al., 2018). The same protein module is responsible for recruiting cytokinesis signaling complexes and formation of the cleavage furrow (Nguyen et al., 2014). In vitro, in addition to maintaining a stable overlap length, coactivity of Prc1 and Xklp1 causes buckling of overlapping microtubules, which are immobilized at their minus ends (Bieling et al., 2010). In a sliding assay of taxol-stabilized microtubules in which microtubules in solution and glass-immobilized microtubules form pairs cross-linked by Prc1, the antiparallel pairs of microtubules are slid apart by Kif4 (Wijeratne and Subramanian, 2018). This is reminiscent of plus end–directed sliding of kinesin-5 (Eg5; Kapitein et al., 2005) and explains the requirement of Prc1 orthologues for spindle elongation in several species (Khmelinskii et al., 2009; Schuyler et al., 2003; Vukušić et al., 2021; Wang et al., 2015; Zhu et al., 2006). Indeed, plus end–overlapping microtubules have an apparent mechanical stiffness that is governed by molecular friction and motor activity (Forth et al., 2014; Wijeratne and Subramanian, 2018). An assembly of tens of such microtubule pairs generates sufficient mechanical resistance against compressive forces in the nanonewton range, enough to keep two spherical organelles of 5–8-µm diameter attached to the microtubule minus ends well separated (Lele et al., 2018). Thus, modular upscaling of a single pair into overlapping radial arrays illustrates how the stabilization and sliding mechanism of a Feo- and Klp3A-decorated antiparallel microtubule pair produces repulsion between two syncytial nuclei.
Feo::GFP or Feo::mCherry expressed in the transgenic line, or supplemented as purified protein, exhibited focal fluorescence signals in the blastoderm embryo and in the explant from preblastoderm embryos. Here, we showed that the length of these signal foci is surprisingly short and uniform. According to in vitro data, and neglecting any regulation other than affinity and stabilization activity for the underlying microtubule overlap to maintain such a short length, the concentration of kinesin-4 in the cytoplasm must be at least one magnitude in excess of Feo (Bieling et al., 2010). Moreover, knockdown of feo by RNAi abolished the signal of Klp3A::GFP below detection, thus considerably reducing the bound fraction of Klp3A at the central spindle. In the embryo, while confirming their already established localization at the spindle midzone (D’Avino et al., 2007; Kwon et al., 2004; Page and Hawley, 2005; Wang et al., 2015; Williams et al., 1995), we recorded Klp3A::GFP signal colocalizing with Feo::mCherry in areas between neighboring spindle asters. However, we could not clearly assess the localization of Klp3A in explants from preblastoderm embryos because of the low signal intensity. A single copy–tagged Klp3A construct expressed with the endogenous promoter failed to provide sufficient signal, and we decided to work with overexpression constructs (Sarov et al., 2016). This indicates that the microtubule overlap–bound fraction of endogenous Klp3A is comparatively small despite the molar excess in the cytosol as derived from overlap length. Together, these observations point at a protein interaction network localized at antiparallel microtubule overlaps that is sensitive to small changes of Feo. As Feo binds microtubule overlaps independently (Bieling et al., 2010), the phenotypes in intact embryos and in explants can arise due to disproportionate Klp3A perturbation downstream of Feo.
In summary, our live-cell microscopy data from blastoderm embryos and preblastoderm embryo explants support a “central spindle model” built from individual pairs of microtubules cross-linked and length-regulated by Feo and Klp3A (Bieling et al., 2010), and potentially through stabilization/sliding activities of Klp3A (Vukušić et al., 2021). More importantly, we show how overlapping microtubules in the aster–aster interaction zone (Nguyen et al., 2014) form midzone-analogous cytoskeletal assemblies that persist throughout blastoderm development. This is particularly intriguing given that, at the embryo cortex from cycle 10 onward, actin-based pseudo-furrows are thought to form precellular compartments that prevent nuclear contact (Karr and Alberts, 1986; Kellogg et al., 1988; Lecuit, 2004; Mavrakis et al., 2009b). In the early blastoderm cycles, however, this compartmentalization may not yet be efficient enough to safeguard nuclear separation, and astral microtubule cross-linking persists as the dominant mechanism. This interpretation is further supported by an earlier observation in mutants of the maternal-effect gene sponge, embryos of which do not form actin caps and pseudo-furrows in blastoderm stage but depict a homogeneous nuclear distribution in cycles 10 and 11 (Postner et al., 1992).
Feo is essential for central spindle assembly and cytokinesis in somatic cells, containing two Cdk phosphorylation sites (Vernì et al., 2004). Feo, like Prc1 in human cells and Ase1p in fission yeast, is under cell cycle control and undergoes phosphorylation-dependent localization, from low-intensity decoration of metaphase spindle microtubules to a strong localization at the central spindle in anaphase and telophase (Khmelinskii et al., 2009; Polak et al., 2017; Subramanian et al., 2013; Wang et al., 2015; Zhu et al., 2006). In the present work, we showed that the focal localization of Feo and Klp3A between neighboring nuclei is in synchrony with central spindle localization. It is in this phase of the division cycle that expanding spindles and separating nuclei cause a large spatial perturbation to the positional distribution (Kanesaki et al., 2011; Lv et al., 2020). Thus, a dual role for Feo under cell cycle control emerges: while it targets the central spindle at anaphase onset, forming the spindle midbody, it also binds to astral microtubule overlaps in a phase during which collision prevention is most needed.
In Drosophila embryos, spindle elongation at anaphase B is powered by the sliding activity of Klp61F (Brust-Mascher et al., 2009). Following the mechanism proposed by Baker et al. (1993), and because Klp61F is a candidate cross-linker and slider of overlapping astral microtubules, we performed RNAi knockdown in the germline. Inhibition of Klp61F expression led to lower density and nonuniform delivery of nuclei to the embryo cortex, confirming its essential role during preblastoderm development. However, owing to the established role of Klp61F in spindle assembly, the RNAi phenotype can emerge because of multiple chromosome segregation failures that were undetectable in the preblastoderm embryo. Here, the embryo explant assay overcomes an experimental limitation and enables time-lapse image acquisition of uni- or binuclear explants undergoing multiple divisions. Consequently, we could confirm that klp61f knockdown led to more frequent division failures rather than shorter nuclear separation. Still, Klp61F and Feo can functionally cooperate in cross-linking astral microtubules because both proteins recognize and bind to microtubule pairs, although with different preference for microtubule orientation (Bieling et al., 2010; Kapitein et al., 2005; Kellogg et al., 2016; Subramanian et al., 2010). In human cells, Prc1-dependent Kif4A motor activity and the microtubule sliding by Eg5 are redundant for spindle elongation during anaphase (Vukušić et al., 2021). Interestingly, in Drosophila, while Feo modulates binding and localization of Klp61F at the spindle midzone in anaphase, Klp61F cannot functionally rescue the absence of Feo (Wang et al., 2015). Presumably, Ase1p/Prc1/Feo binding to microtubule overlaps creates a protein binding hub for motors and regulators (Bieling et al., 2010; D’Avino et al., 2007; Hu et al., 2012; Khmelinskii et al., 2009; Sasabe and Machida, 2006; Subramanian et al., 2013). This property has not been demonstrated for kinesin-5 orthologues. Together, the collection of evidence from us and others suggests that Klp61F is not at the core of astral microtubule-driven nuclear separation.
Lastly, the reader may wonder how astral microtubule overlap cross-linking by Feo and Klp3A defines the internuclear distance metric, leading to a distribution of syncytial nuclei with high regularity. In an earlier study, Telley et al. (2012) showed that microtubule aster size varies throughout the nuclear division cycle, reaching a maximum of 11 ± 3 µm in telophase. Herein, the aster size represents the length distribution of microtubules which, for dynamic microtubules with nongrowing minus ends, is well approximated with an exponential distribution (Howard, 2001). We assume that two microtubules from neighboring asters grow at least to average length, overlap with their plus ends, and are collinear. If the stabilized overlap length is ∼1 µm, then the total length from centrosome to centrosome is on average 21 ± 4 µm. Considering that a centrosome is ∼1 µm in size, and that a nucleus in late telophase is 5 ± 1 µm in diameter, the total distance between the centers of neighboring nuclei is 28 ± 4 µm. This estimate is in good agreement with the internuclear distance distribution measured from center to center of each nucleus (Fig. 3 C), the minimal nonsibling internuclear distance in extract (Fig. 4 H), and earlier reported separation distances of daughter nuclei (Telley et al., 2012). Thus, the short antiparallel overlap length of microtubules from neighboring asters and the microtubule length distribution are sufficient to explain the geometry of nuclear distribution in the Drosophila syncytial embryo.
Materials and methods
Drosophila
Rearing of flies for general maintenance was done as previously described (Stocker and Gallant, 2008). The following fly lines were used to make recombinants or trans-heterozygotes: Jupiter::GFP (Bloomington Drosophila Stock Center [BDSC] 6836), Jupiter::mCherry (generated by and obtained from Nick Lowe in D. St. Johnston’s laboratory, The Gurdon Institute, Cambridge, UK), pUbq>Spd2::GFP (gift from M. Bettencourt-Dias, Instituto Gulbenkian de Ciência), H2Av::RFP (BDSC 23650), Feo::GFP (BDSC 59274), Feo::mCherry (BDSC 59277), Klp61F::GFP (BDSC 35509), Klp3A::GFP (Vienna Drosophila Resource Center 318352), RNAi targeting feo (BDSC 28926 and 35467), RNAi targeting klp3a (BDSC 40944 and 43230), RNAi targeting klp61f (BDSC 33685 and 35804), RNAi targeting mcherry (BDSC 35785), and UASp–GFP (BDSC 35786).
RNAi experiments
Knockdown experiments were performed using the TRiPGermline fly lines for RNAi in germline cells (Perkins et al., 2015). The UAS-hairpin against a gene of interest was expressed using V32–Gal4 (gift from M. Bettencourt-Dias) at 25°C. The expression profile of V32–Gal4 in the oocyte was assessed by dissecting ovaries of flies expressing UASp–GFP at 25°C and comparing GFP expression at different developmental stages with fluorescence microscopy.
Sample preparation and extraction
Embryos were collected from apple juice agar plates mounted on a fly cage. They were dechorionated in 7% sodium hypochlorite solution, aligned, and immobilized on a clean coverslip using adhesive dissolved in heptane and covered with halocarbon oil (Voltalef 10S). Extraction of cytoplasm from individual embryos and generation of explants were performed on a custom-made microscope as previously described (de-Carvalho et al., 2018; Telley et al., 2013).
Quantitative PCR
To measure the transcript levels of feo, klp3a, and klp61f, total RNA was extracted following standard procedures (PureLink RNA Mini Kit; Ambion) from embryos collected after 40 min of egg laying. A cDNA library was made from Oligo(dT)12–18 as described in the manufacturer's protocol (Transcriptor First Strand cDNA Synthesis Kit; Roche). Quantitative PCR was performed using Quantifast SYBR Green PCR Kit (204052) and QuantiTect Primers for feo (QT00919758) in feo RNAi (35467 and 28926), klp3a (QT00497154) in klp3a RNAi (40944 and 43230), and klp61f (QT00955822) in klp61f RNAi (35804 and 33685). actin (QT00498883) was used as a housekeeping gene control. Results are presented in Table 1.
Purification of sFeoFL::GFP and sFeoN::GFP
The full coding sequence of the feo gene fused to a C-terminal GFP tag, synthesized, and codon optimized by NZYTech, and is referred to herein as sFeoFL::GFP. The DNA was cloned into the vector pET-21a containing a C-terminal His6-tag using restriction enzymes NheI/XhoI and transformed into E. coli Rosetta cells. The coding sequence of the feo gene without the initial 73 N-terminal residues fused to a C-terminal GFP tag, referred here as truncated sFeoN::GFP construct, was amplified from the synthesized sFeoFL::GFP construct and recloned into the pET-21a vector using restriction enzymes XhoI/BamHI. Both proteins were produced by induction with 0.5 mM IPTG at 25°C. After 4 h of incubation, the cells were harvested and resuspended in lysis buffer (100 mM K-Hepes, pH 7.4, 500 mM NaCl, 10% glycerol, 0.1% Triton X-100, and 3 M urea, supplemented with protease inhibitors [Roche] and 100 U of DNase type I [NZYTech]). The cells were lysed using a digital sonifier (SLPe; Branson) at 70% amplitude with six pulses of 30 s on/30 s off and clarified by centrifugation at 30,000 g for 45 min at 4°C. For purification of the truncated construct, the supernatant was loaded onto a 5-ml HiTrap Chelating HP (GE Healthcare), charged with 0.1 mM NiCl2, equilibrated with wash buffer (100 mM K-Hepes, pH 7.4, 500 mM NaCl, 10% glycerol, 40 mM imidazole, and 1 mM 2-mercaptoethanol), extensively washed with this wash buffer, and eluted with elution buffer (100 mM K-Hepes, pH 7.4, 500 mM NaCl, 10% glycerol, 500 mM imidazole, and 1 mM 2-mercaptoethanol) throughout a gradient of 6 column volumes. For purification of the full-length construct, the supernatant was loaded onto a 1-ml HiTrap TALON crude (GE Healthcare), charged with 50 mM CoCl2, equilibrated with wash buffer (100 mM K-Hepes, pH 7.2, 500 mM NaCl, 10% glycerol, 5 mM imidazole, 1 mM 2-mercaptoethanol), extensively washed with this wash buffer, and eluted with elution buffer (100 mM K-Hepes, pH 7.2, 500 mM NaCl, 10% glycerol, 150 mM imidazole, and 1 mM 2-mercaptoethanol), throughout a gradient of 20 column volumes. Fractions containing the protein of interest were pooled, and the buffer was exchanged into embryo explant–compatible buffer (100 mM K-Hepes, pH 7.8, 1 mM MgCl2, and 100 mM KCl) using a PD-10 desalting column (GE Healthcare) and concentrated using a 50K MWCO Amicon Ultracentrifugal filter (Merck). The purifications were performed using the ÄKTApurifier protein purification system (Cytiva), and the chromatographic profile of both proteins was followed by measuring the absorbance at 280, 254, and 488 nm in the UV-900 monitor. The size exclusion method resulted in Feo constructs strongly associated with an unknown contaminant at ∼50 kD. The concentration of each construct was estimated as ∼50% of the total measured protein concentration based on band analysis of SDS-PAGE. Total protein concentrations were measured with a NanoDrop2000 UV-Vis spectrophotometer (Thermo Fisher Scientific).
SDS-PAGE, native-PAGE, and blotting with quantification
Polyacrylamide gels for electrophoresis and Coomassie staining were made with 15% SDS. Molecular mass was estimated by linear regression of log10(molecular mass in daltons) as a function of the migration distances (centimeters) of the ladder (161-0375; Bio-Rad). To determine the dimerization of sFeoFL::GFP-His6 and sFeoN::GFP-His6, a Western blot was generated using a PVDF membrane (Bio-Rad) from a 4–12% gradient SDS and a native polyacrylamide gel side by side using a mouse anti-GFP antibody (11814460001; Roche) at 1:500 dilution. The molecular mass of the protein samples was estimated using a linear regression of log10(molecular mass in daltons) as a function of the migration distances (centimeters) of the ladder (MB090; NZYTech); for the SDS gel, a regression was performed on the entire ladder; for the native gel, only the higher molecular mass ladder (100–245 kD) was considered. See Fig. S5.
Figure S5.
N-terminally truncated Feo::GFP weakly dimerizes and causes nuclear separation defects, and full-length Feo::GFP protein partially rescues nuclear delivery to the cortex of feo RNAi embryos. Related to Fig. 5. (A) Coomassie-stained 15% SDS-PAGE of heterologous expressed and purified full-length sFeo::GFP-His6 (left) and an N-terminally truncated sFeo::GFP-His6 construct missing the putative dimerization and Klp3A-binding domain (right). The expected molecular mass is at the top, and the measured mass is on the right of the bands (details in Materials and methods). The lower bands are contaminants that were not separated by gel filtration and are of bacterial origin as determined by mass spectrometry. (B) Western blot of a 4–12% gradient SDS-PAGE (left) next to a native-PAGE (right) with sFeoFL::GFP-His6 and sFeoN::GFP-His6, using mouse anti-GFP antibody. Both protein constructs migrate according to expected molecular mass of a monomer in the SDS-PAGE gel. In the native-PAGE gel, the full-length Feo migrates as expected from a dimer. The slight deviation from the expected mass of a dimer (212 kD) can be explained by the rod-like structure of the protein, as was shown for the human homologue Prc1 (Subramanian et al., 2010). Conversely, the truncated construct migrates like a weak dimer, causing a wide distribution ranging between 100 and 200 kD. (C) Time-lapse fluorescence images of a (control) blastoderm embryo expressing H2Av::RFP (magenta) after injection of sFeoN::GFP-His6 protein (green) and Alexa Fluor 647–Tubulin (gray). During the cycle just after injection (left), the localization of sFeoN::GFP-His6 at the spindle midzone was not detectable (arrowhead). However, the effect of protein addition manifests in nuclear separation defects, and in the subsequent division (middle), spindles are observed in unnatural proximity, leading to spindle fusion (arrow). In telophase of the same cycle (right), sFeoN::GFP-His6 is detected at the spindle midzone (arrowheads). This delayed localization after injection is not observed for sFeoFL::GFP-His6 (Fig. 5 B). Time is in min:s. Scale bar, 10 µm. (D) The position (circle) of every nucleus arriving at the embryo cortex after the last preblastoderm division, relative to the axial and lateral borders of the embryo, for each condition: feo (35467) mock-injected (buffer), feo (35467) rescued by protein injection, and control (mcherry) mock-injected. The green dashed rectangle represents the area of the embryo bounded by the length and width of the visible embryo in the confocal stacks, with the anterior end at the coordinate origin. The cyan cross represents the location of the 2D centroid defined from the position of all nuclei. The nuclei in interphase of the first division at the cortex are marked in black, and nuclei that have progressed to metaphase/anaphase are marked in magenta.
Addition of exogenous purified proteins
Purified porcine Tubulin (T240; Cytoskeleton) was labeled with Alexa Fluor 647 (Invitrogen, Thermo Fisher Scientific) following a published protocol (Hyman, 1991) and injected into embryos or explants at 0.3–0.8 mg/ml. Freshly purified sFeoFL::GFP-His6 and sFeoN::GFP-His6 were injected at 2 mg/ml in EC buffer in embryos or explants. This concentration was a result from a series of titrations over a magnitude of different concentrations assessing phenotype or rescue. For embryos, the injected volume assumed a spherical shape with diameter D ≈ 0.018 mm, resulting an injection volume of 3.05 × 10−6 mm3. The average length and width of the embryo are 0.5 and 0.2 mm, respectively (Markow et al., 2009). Assuming an ellipsoid geometry for the embryo, its volume is ∼10−2 mm3. Thus, the final concentration of injected protein after equilibration in the entire embryo was 5–6 nM. For explants, both protein constructs were added to explant cytoplasm at 1:200 (vol/vol), resulting in a final concentration in the cytoplasm of 100–200 nM. Importantly, such an excess of full-length Feo protein preserved nuclear divisions and distribution.
Treatment of embryo explants with nocodazole
Nocodazole (M-1404, Sigma Aldrich) was dissolved in DMSO and diluted to 200 µM in electrical conductivity (EC) buffer at pH 7.8, with a concentration of DMSO in EC buffer of 2% (vol/vol). This buffer mixture was added to explants at ∼1:50 (vol/vol), estimated by measuring the diameters of the buffer droplet in the explant and the explant itself, calculating their area, and scaling according to area change (Telley et al., 2013). The final concentration of nocodazole in the explant was ∼4 µM. Control experiments were conducted by adding the buffer mixture without the drug to explants at similar ratios.
Image acquisition, processing, and analysis
Transmission light microscopy images were obtained with a 10× 0.25-NA objective and the polarizer and analyzer of the microscope were in crossed configuration. Time-lapse confocal fluorescence Z stacks were acquired on a Yokogawa CSU-W1 spinning disk confocal scanner with 488-, 561-, and 640-nm laser lines. Images of whole embryos were acquired with a 40× 1.3-NA oil immersion objective. Images of embryo explants were acquired with a 60× 1.2-NA or a 40× 1.15-NA water immersion objective. Images were recorded with an Andor iXon3 888 EMCCD 1024 × 1024 camera with 13-µm pixel2 size and 2× magnification in front of the camera.
Image processing (i.e., making Z-projections, image cropping, image down-sampling, and video generation) was performed in Fiji (Schindelin et al., 2012). Whole-embryo images for knockdown experiments were obtained by pairwise stitching using a plugin in Fiji.
The fluorescence signal of Feo::GFP in explants was analyzed with the line profile tool in Fiji. First, images of dividing nuclei during anaphase or telophase were filtered with a Gaussian kernel Spot-like signals located between nonsibling nuclei were identified and, where spots were noncircular, a line was drawn along the longer axis. The angle of the line relative to the image coordinate system was recorded, and an intensity profile was generated. Profiles were aligned relative to the position of highest intensity and averaged. For each image, an intensity profile from a location void of microtubule signal was generated to obtain the background and the SD of Feo::GFP intensity. Finally, the size of the spot was determined by calculating the width of the curve where the intensity was higher than two times SD of the background. The angle of every profile line was transformed relative to the closer of the two axes that connect the centrosome of one nucleus with the centrosome of the two neighboring sister nuclei (Fig. 1 C), termed angle A probability density plot from all measured angles was generated in Matlab. Voronoi segmentation was performed by determining the position of the spindle poles manually in Fiji and providing these coordinates to the voronoi function implemented in Matlab.
The nuclear density in whole-embryo images was obtained by measuring the area of the visible part of the embryo after manually tracing the border and dividing the number of nuclei by this area. The localization of nuclei in whole embryos was performed manually in Fiji. The precision of localization was 0.25 µm (intraoperator variability). Localization coordinates were imported into Matlab and transformed with respect to the coordinate system of the embryo, as defined by the anterior pole as coordinate origin and the anterior-posterior axis as x axis. The first-order internuclear distances were obtained from the triangulation connectivity list (delaunay function), while excluding any edge connections, and by calculating the distance between the remaining connections. The cumulative distribution function of internuclear distances from individual embryos was obtained with the ecdf function in Matlab. An average cumulative distribution function from several embryos was generated after pooling all distances together. Next, the deviation of the centroid of nuclear positions from the anatomical center of the embryo was obtained using the formula
whereby an estimate for the anatomic center of the embryo, with respect to the embryo coordinate system, is given by half the pole-to-pole distance on the x axis and 0 on the y axis. The third-order moment of the distribution of nuclear coordinates was calculated with the skewness function in Matlab, providing a measure for left-right asymmetry.
The measurement of internuclear distances in embryo explants was performed manually in Fiji. The precision of distance measurement was ±0.12 µm as determined by repeated measurement (intraoperator variability). The intensity profile plots of Klp3A::GFP in the Feo RNAi background were obtained using the line profile tool in Fiji, by drawing a line between daughter nuclei in the red (H2Av::RFP) channel and generating an intensity profile plot in the green channel, aligning these profiles according to the peak intensity, and averaging profiles from different locations and embryos.
Plots of aligned quadrilaterals were generated with Matlab by coordinate transformation. The area was calculated using the Gauss trapezoidal formula for general polygons,
while for quadrilaterals. For each quadrilateral, representing two sets of dividing nuclei, the average of the two involved mitotic separation distances and the average of the two involved nonsibling separations were calculated and plotted. All graphs were made with Matlab.
Statistical analysis
Unless otherwise mentioned, we plotted the distribution of raw data or cumulative probabilities. Sample size (n) and the number of experimental repeats (N) are reported in the figure legends. A Wilcoxon rank-sum test was performed with Matlab starting with a significance level of
Online supplemental material
Fig. S1 provides a detailed analysis of the localization of Feo between neighbor nuclei along subsequent division cycles. Fig. S2 provides a visualization of the GAL4 expression pattern during oogenesis driving the RNAi knockdown, images from embryos expressing two alternative RNAi constructs, and evidence for the delocalization of Klp3A at the spindle midzone when Feo is knocked down. Fig. S3 provides the full dataset of nuclear positions in embryos for the control and test RNAi, together with various distribution parameters. Fig. S4 shows images from an explant before and after micromanipulation, generated from an embryo under Feo knockdown conditions. Fig. S5 provides SDS-PAGE and native-PAGE gels of the full-length and truncated Feo protein construct, time-lapse images from an embryo after injection of the truncated construct FeoN::GFP, and the full dataset of nuclear positions in embryos where endogenous Feo is depleted and the phenotype is partially rescued by injection of full-length Feo::GFP. Video 1 shows fluorescence of Feo, Klp61F, and microtubules in a cycling embryo explant. Video 2 shows fluorescence of Feo, Klp61F, and microtubules in a cycling embryo. Video 3 shows fluorescence of Klp3A, Feo, and microtubules in a cycling embryo. Video 4 shows fluorescence of microtubules and chromatin in a cycling embryo explant after nocodazole treatment. Video 5 shows fluorescence of microtubules and chromatin in cycling embryos under RNAi knockdown. Video 6 shows fluorescence of microtubules and chromatin in cycling embryo explants under RNAi knockdown. Video 7 shows a micromanipulation experiment and subsequent time lapse of fluorescence of Feo. Video 8 shows fluorescence of purified sFeoFL::GFP protein injected in an embryo. Video 9 shows fluorescence of purified sFeoN::GFP protein injected in an embryo.
Supplementary Material
Acknowledgments
We thank members of the Telley laboratory for fruitful discussions, Jonathon Scholey (University of California, Davis, Davis, CA) for constructive comments on the manuscript and for fly stocks, and Thomas Surrey for discussions throughout the project. We thank the staff of the Fly Facility, the Advanced Imaging Facility, and the Technical Support Service at the Instituto Gulbenkian de Ciência.
Transgenic fly stocks were obtained from the Vienna Drosophila Resource Center and Bloomington Drosophila Stock Center (National Institutes of Health P40OD018537). We acknowledge financial support provided by Fundação Calouste Gulbenkian, European Commission Seventh Framework Programme Marie Curie Career Integration Grant to I.A. Telley (PCIG13-GA-2013-618743), Human Frontier Science Program Young Investigators’ Grant to I.A. Telley (RGY0083/2016), and a doctoral fellowship from Fundação para a Ciência e a Tecnologia (SFRH/BD/52174/2013) to O. Deshpande, and a project grant (PTDC/BIA-BQM-31843/2017) supporting D. Vieira. We acknowledge Programa Operacional Regional de Lisboa (LISBOA-01-0145-FEDER-007654) supporting Instituto Gulbenkian de Ciência’s core operation, Consortium for Genetically Tractable Organisms (CONGENTO; LISBOA-01-0145-FEDER-022170) supporting the Fly Facility, and Portuguese Platform of BioImaging (PPBI-POCI-01-0145-FEDER-022122) supporting the Advanced Imaging Facility, all co-financed by Fundação para a Ciência e a Tecnologia (Portugal) and Lisboa2020, under the PORTUGAL2020 agreement (European Regional Development Fund).
The authors declare no competing financial interests.
Author contributions: O. Deshpande, J. de-Carvalho, and I.A. Telley conceived and designed the project. O. Deshpande and I.A. Telley designed experiments, and O. Deshpande performed them with occasional support from J. de-Carvalho. D.V. Vieira and O. Deshpande designed, purified, and characterized the protein constructs. I.A. Telley designed and assembled the instrument for explant generation and manipulation. I.A. Telley designed the image analysis procedures. O. Deshpande, J. de-Carvalho, and I.A. Telley analyzed the image data. O. Deshpande, J. de-Carvalho, D.V. Vieira, and I.A. Telley prepared the figures. O. Deshpande and I.A. Telley wrote the draft manuscript, which was edited by all authors.
References
- Almonacid, M., Al Jord A., El-Hayek S., Othmani A., Coulpier F., Lemoine S., Miyamoto K., Grosse R., Klein C., Piolot T., et al. 2019. Active Fluctuations of the Nuclear Envelope Shape the Transcriptional Dynamics in Oocytes. Dev. Cell. 51:145–157.e10. 10.1016/j.devcel.2019.09.010 [DOI] [PubMed] [Google Scholar]
- Almonacid, M., Ahmed W.W., Bussonnier M., Mailly P., Betz T., Voituriez R., Gov N.S., and Verlhac M.-H.. 2015. Active diffusion positions the nucleus in mouse oocytes. Nat. Cell Biol. 17:470–479. 10.1038/ncb3131 [DOI] [PubMed] [Google Scholar]
- Baker, J., Theurkauf W.E., and Schubiger G.. 1993. Dynamic changes in microtubule configuration correlate with nuclear migration in the preblastoderm Drosophila embryo. J. Cell Biol. 122:113–121. 10.1083/jcb.122.1.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling, P., Telley I.A., and Surrey T.. 2010. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 142:420–432. 10.1016/j.cell.2010.06.033 [DOI] [PubMed] [Google Scholar]
- Bone, C.R., and Starr D.A.. 2016. Nuclear migration events throughout development. J. Cell Sci. 129:1951–1961. 10.1242/jcs.179788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann, H., Skiniotis G., Spilker A., Kandels-Lewis S., Vernos I., and Surrey T.. 2004. A kinesin-like motor inhibits microtubule dynamic instability. Science. 303:1519–1522. 10.1126/science.1094838 [DOI] [PubMed] [Google Scholar]
- Brust-Mascher, I., Sommi P., Cheerambathur D.K., and Scholey J.M.. 2009. Kinesin-5-dependent poleward flux and spindle length control in Drosophila embryo mitosis. Mol. Biol. Cell. 20:1749–1762. 10.1091/mbc.e08-10-1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaini, G., Dallai R., and Riparbelli M.G.. 1992. Cytochalasin induces spindle fusion in the syncytial blastoderm of the early Drosophila embryo. Biol. Cell. 74:249–254. 10.1016/0248-4900(92)90035-Y [DOI] [PubMed] [Google Scholar]
- Cheerambathur, D.K., Brust-Mascher I., Civelekoglu-Scholey G., and Scholey J.M.. 2008. Dynamic partitioning of mitotic kinesin-5 cross-linkers between microtubule-bound and freely diffusing states. J. Cell Biol. 182:429–436. 10.1083/jcb.200804100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheerambathur, D.K., Gassmann R., Cook B., Oegema K., and Desai A.. 2013. Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science. 342:1239–1242. 10.1126/science.1246232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino, P.P., Archambault V., Przewloka M.R., Zhang W., Lilley K.S., Laue E., and Glover D.M.. 2007. Recruitment of Polo kinase to the spindle midzone during cytokinesis requires the Feo/Klp3A complex. PLoS One. 2:e572. 10.1371/journal.pone.0000572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de-Carvalho, J., Deshpande O., Nabais C., and Telley I.A.. 2018. A cell-free system of Drosophila egg explants supporting native mitotic cycles. Methods Cell Biol. 144:233–257. 10.1016/bs.mcb.2018.03.011 [DOI] [PubMed] [Google Scholar]
- Deneke, V.E., Puliafito A., Krueger D., Narla A.V., De Simone A., Primo L., Vergassola M., De Renzis S., and Di Talia S.. 2019. Self-Organized Nuclear Positioning Synchronizes the Cell Cycle in Drosophila Embryos. Cell. 177:925–941.e17. 10.1016/j.cell.2019.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, D.J., Nelson W.J., and Weis W.I.. 2012. An epithelial tissue in Dictyostelium challenges the traditional origin of metazoan multicellularity. BioEssays. 34:833–840. 10.1002/bies.201100187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudin, O., Ondracka A., Grau-Bové X., Haraldsen A.A., Toyoda A., Suga H., Bråte J., and Ruiz-Trillo I.. 2019. A unicellular relative of animals generates a layer of polarized cells by actomyosin-dependent cellularization. eLife. 8:e49801. 10.7554/eLife.49801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe, V.E., and Alberts B.M.. 1983. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 61:31–70. 10.1242/jcs.61.1.31 [DOI] [PubMed] [Google Scholar]
- Forth, S., Hsia K.-C., Shimamoto Y., and Kapoor T.M.. 2014. Asymmetric friction of nonmotor MAPs can lead to their directional motion in active microtubule networks. Cell. 157:420–432. 10.1016/j.cell.2014.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, C., Ward J.J., Loiodice I., Velve-Casquillas G., Nédélec F.J., and Tran P.T.. 2009. Phospho-regulated interaction between kinesin-6 Klp9p and microtubule bundler Ase1p promotes spindle elongation. Dev. Cell. 17:257–267. 10.1016/j.devcel.2009.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatlin, J.C., Matov A., Groen A.C., Needleman D.J., Maresca T.J., Danuser G., Mitchison T.J., and Salmon E.D.. 2009. Spindle fusion requires dynein-mediated sliding of oppositely oriented microtubules. Curr. Biol. 19:287–296. 10.1016/j.cub.2009.01.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaux, R., Politi A.Z., Philippsen P., and Nédélec F.. 2017. Mechanism of nuclear movements in a multinucleated cell. Mol. Biol. Cell. 28:645–660. 10.1091/mbc.e16-11-0806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen, G.G., and Worman H.J.. 2013. Nuclear positioning. Cell. 152:1376–1389. 10.1016/j.cell.2013.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka, K., and Okada M.. 1991. Retarded nuclear migration in Drosophila embryos with aberrant F-actin reorganization caused by maternal mutations and by cytochalasin treatment. Development. 111:909–920. 10.1242/dev.111.4.909 [DOI] [PubMed] [Google Scholar]
- Heck, M.M., Pereira A., Pesavento P., Yannoni Y., Spradling A.C., and Goldstein L.S.. 1993. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J. Cell Biol. 123:665–679. 10.1083/jcb.123.3.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehenberger, E., Kradolfer D., and Köhler C.. 2012. Endosperm cellularization defines an important developmental transition for embryo development. Development. 139:2031–2039. 10.1242/dev.077057 [DOI] [PubMed] [Google Scholar]
- Howard, J. 2001. Mechanics of Motor Proteins and the Cytoskeleton. Sinauer Associates, Sunderland, MA; 384 pp. [Google Scholar]
- Hu, C.-K., Ozlü N., Coughlin M., Steen J.J., and Mitchison T.J.. 2012. Plk1 negatively regulates PRC1 to prevent premature midzone formation before cytokinesis. Mol. Biol. Cell. 23:2702–2711. 10.1091/mbc.e12-01-0058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman, A.A. 1991. Preparation of marked microtubules for the assay of the polarity of microtubule-based motors by fluorescence. J. Cell Sci. Suppl. 14(Suppl 14):125–127. 10.1242/jcs.1991.Supplement_14.25 [DOI] [PubMed] [Google Scholar]
- Kanesaki, T., Edwards C.M., Schwarz U.S., and Grosshans J.. 2011. Dynamic ordering of nuclei in syncytial embryos: a quantitative analysis of the role of cytoskeletal networks. Integr. Biol. 3:1112–1119. 10.1039/c1ib00059d [DOI] [PubMed] [Google Scholar]
- Kao, L.-R., and Megraw T.L.. 2009. Centrocortin cooperates with centrosomin to organize Drosophila embryonic cleavage furrows. Curr. Biol. 19:937–942. 10.1016/j.cub.2009.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein, L.C., Peterman E.J.G., Kwok B.H., Kim J.H., Kapoor T.M., and Schmidt C.F.. 2005. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 435:114–118. 10.1038/nature03503 [DOI] [PubMed] [Google Scholar]
- Karr, T.L., and Alberts B.M.. 1986. Organization of the cytoskeleton in early Drosophila embryos. J. Cell Biol. 102:1494–1509. 10.1083/jcb.102.4.1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, D.R., Mitchison T.J., and Alberts B.M.. 1988. Behaviour of microtubules and actin filaments in living Drosophila embryos. Development. 103:675–686. 10.1242/dev.103.4.675 [DOI] [PubMed] [Google Scholar]
- Kellogg, E.H., Howes S., Ti S.-C., Ramírez-Aportela E., Kapoor T.M., Chacón P., and Nogales E.. 2016. Near-atomic cryo-EM structure of PRC1 bound to the microtubule. Proc. Natl. Acad. Sci. USA. 113:9430–9439. 10.1073/pnas.1609903113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii, A., Roostalu J., Roque H., Antony C., and Schiebel E.. 2009. Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev. Cell. 17:244–256. 10.1016/j.devcel.2009.06.011 [DOI] [PubMed] [Google Scholar]
- Kotadia, S., Crest J., Tram U., Riggs B., and Sullivan W.. 2010. Blastoderm Formation and Cellularisation in Drosophila melanogaster. American Cancer Society, Chichester, UK, 10.1002/9780470015902.a0001071.pub2 [DOI] [Google Scholar]
- Kurasawa, Y., Earnshaw W.C., Mochizuki Y., Dohmae N., and Todokoro K.. 2004. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 23:3237–3248. 10.1038/sj.emboj.7600347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, M., and Scholey J.M.. 2004. Spindle mechanics and dynamics during mitosis in Drosophila. Trends Cell Biol. 14:194–205. 10.1016/j.tcb.2004.03.003 [DOI] [PubMed] [Google Scholar]
- Kwon, M., Morales-Mulia S., Brust-Mascher I., Rogers G.C., Sharp D.J., and Scholey J.M.. 2004. The chromokinesin, KLP3A, dives mitotic spindle pole separation during prometaphase and anaphase and facilitates chromatid motility. Mol. Biol. Cell. 15:219–233. 10.1091/mbc.e03-07-0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit, T. 2004. Junctions and vesicular trafficking during Drosophila cellularization. J. Cell Sci. 117:3427–3433. 10.1242/jcs.01312 [DOI] [PubMed] [Google Scholar]
- Lecuit, T., and Wieschaus E.. 2000. Polarized insertion of new membrane from a cytoplasmic reservoir during cleavage of the Drosophila embryo. J. Cell Biol. 150:849–860. 10.1083/jcb.150.4.849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lele, T.P., Dickinson R.B., and Gundersen G.G.. 2018. Mechanical principles of nuclear shaping and positioning. J. Cell Biol. 217:3330–3342. 10.1083/jcb.201804052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, J.R., and Holzbaur E.L.F.. 2008. Dynein drives nuclear rotation during forward progression of motile fibroblasts. J. Cell Sci. 121:3187–3195. 10.1242/jcs.033878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv, Z., Rosenbaum J., Mohr S., Zhang X., Kong D., Preiß H., Kruss S., Alim K., Aspelmeier T., and Großhans J.. 2020. The Emergent Yo-yo Movement of Nuclei Driven by Cytoskeletal Remodeling in Pseudo-synchronous Mitotic Cycles. Curr. Biol. 30:2564–2573.e5. 10.1016/j.cub.2020.04.078 [DOI] [PubMed] [Google Scholar]
- Manhart, A., Windner S., Baylies M., and Mogilner A.. 2018. Mechanical positioning of multiple nuclei in muscle cells. PLOS Comput. Biol. 14:e1006208. 10.1371/journal.pcbi.1006208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow, T.A., Beall S., and Matzkin L.M.. 2009. Egg size, embryonic development time and ovoviviparity in Drosophila species. J. Evol. Biol. 22:430–434. 10.1111/j.1420-9101.2008.01649.x [DOI] [PubMed] [Google Scholar]
- Mavrakis, M., Rikhy R., and Lippincott-Schwartz J.. 2009a. Plasma membrane polarity and compartmentalization are established before cellularization in the fly embryo. Dev. Cell. 16:93–104. 10.1016/j.devcel.2008.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis, M., Rikhy R., and Lippincott-Schwartz J.. 2009b. Cells within a cell: Insights into cellular architecture and polarization from the organization of the early fly embryo. Commun. Integr. Biol. 2:313–314. 10.4161/cib.2.4.8240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw, T.L., Li K., Kao L.R., and Kaufman T.C.. 1999. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 126:2829–2839. 10.1242/dev.126.13.2829 [DOI] [PubMed] [Google Scholar]
- Minc, N., Burgess D., and Chang F.. 2011. Influence of cell geometry on division-plane positioning. Cell. 144:414–426. 10.1016/j.cell.2011.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin, X., Daneman R., Zavortink M., and Chia W.. 2001. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA. 98:15050–15055. 10.1073/pnas.261408198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelam, S., Chancellor T.J., Li Y., Nickerson J.A., Roux K.J., Dickinson R.B., and Lele T.P.. 2015. Direct force probe reveals the mechanics of nuclear homeostasis in the mammalian cell. Proc. Natl. Acad. Sci. USA. 112:5720–5725. 10.1073/pnas.1502111112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, P.A., Groen A.C., Loose M., Ishihara K., Wühr M., Field C.M., and Mitchison T.J.. 2014. Spatial organization of cytokinesis signaling reconstituted in a cell-free system. Science. 346:244–247. 10.1126/science.1256773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, P.A., Field C.M., and Mitchison T.J.. 2018. Prc1E and Kif4A control microtubule organization within and between large Xenopus egg asters. Mol. Biol. Cell. 29:304–316. 10.1091/mbc.E17-09-0540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, S.L., and Hawley R.S.. 2005. The Drosophila meiotic mutant mei-352 is an allele of klp3A and reveals a role for a kinesin-like protein in crossover distribution. Genetics. 170:1797–1807. 10.1534/genetics.105.041194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pécréaux, J., Redemann S., Alayan Z., Mercat B., Pastezeur S., Garzon-Coral C., Hyman A.A., and Howard J.. 2016. The Mitotic Spindle in the One-Cell C. elegans Embryo Is Positioned with High Precision and Stability. Biophys. J. 111:1773–1784. 10.1016/j.bpj.2016.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, L.A., Holderbaum L., Tao R., Hu Y., Sopko R., McCall K., Yang-Zhou D., Flockhart I., Binari R., Shim H.-S., et al. 2015. The Transgenic RNAi Project at Harvard Medical School: Resources and Validation. Genetics. 201:843–852. 10.1534/genetics.115.180208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova, M.D., Tkačik G., Bialek W., Wieschaus E.F., and Gregor T.. 2019. Optimal Decoding of Cellular Identities in a Genetic Network. Cell. 176:844–855.e15. 10.1016/j.cell.2019.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak, B., Risteski P., Lesjak S., and Tolić I.M.. 2017. PRC1-labeled microtubule bundles and kinetochore pairs show one-to-one association in metaphase. EMBO Rep. 18:217–230. 10.15252/embr.201642650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postner, M.A., Miller K.G., and Wieschaus E.F.. 1992. Maternal effect mutations of the sponge locus affect actin cytoskeletal rearrangements in Drosophila melanogaster embryos. J. Cell Biol. 119:1205–1218. 10.1083/jcb.119.5.1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinemann, D.N., Sturgill E.G., Das D.K., Degen M.S., Vörös Z., Hwang W., Ohi R., and Lang M.J.. 2017. Collective Force Regulation in Anti-parallel Microtubule Gliding by Dimeric Kif15 Kinesin Motors. Curr. Biol. 27:2810–2820.e6. 10.1016/j.cub.2017.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royou, A., Field C., Sisson J.C., Sullivan W., and Karess R.. 2004. Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos. Mol. Biol. Cell. 15:838–850. 10.1091/mbc.e03-06-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov, M., Barz C., Jambor H., Hein M.Y., Schmied C., Suchold D., Stender B., Janosch S., K J V.V., Krishnan R.T., et al. 2016. A genome-wide resource for the analysis of protein localisation in Drosophila. eLife. 5:e12068. 10.7554/eLife.12068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe, M., and Machida Y.. 2006. MAP65: a bridge linking a MAP kinase to microtubule turnover. Curr. Opin. Plant Biol. 9:563–570. 10.1016/j.pbi.2006.09.010 [DOI] [PubMed] [Google Scholar]
- Sawin, K.E., LeGuellec K., Philippe M., and Mitchison T.J.. 1992. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature. 359:540–543. 10.1038/359540a0 [DOI] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey, J.M., Civelekoglu-Scholey G., and Brust-Mascher I.. 2016. Anaphase B. Biology (Basel). 5:51. 10.3390/biology5040051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh, M., Lehner C.F., and Heidmann S.. 2007. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr. Biol. 17:237–243. 10.1016/j.cub.2006.11.051 [DOI] [PubMed] [Google Scholar]
- Schuyler, S.C., Liu J.Y., and Pellman D.. 2003. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J. Cell Biol. 160:517–528. 10.1083/jcb.200210021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D.J., Yu K.R., Sisson J.C., Sullivan W., and Scholey J.M.. 1999. Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat. Cell Biol. 1:51–54. 10.1038/9025 [DOI] [PubMed] [Google Scholar]
- Staller, M.V., Yan D., Randklev S., Bragdon M.D., Wunderlich Z.B., Tao R., Perkins L.A., Depace A.H., and Perrimon N.. 2013. Depleting gene activities in early Drosophila embryos with the “maternal-Gal4-shRNA” system. Genetics. 193:51–61. 10.1534/genetics.112.144915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr, D.A., and Han M.. 2002. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 298:406–409. 10.1126/science.1075119 [DOI] [PubMed] [Google Scholar]
- Stocker, H., and Gallant P.. 2008. Getting started : an overview on raising and handling Drosophila. Methods Mol. Biol. 420:27–44. 10.1007/978-1-59745-583-1_2 [DOI] [PubMed] [Google Scholar]
- Subramanian, R., Wilson-Kubalek E.M., Arthur C.P., Bick M.J., Campbell E.A., Darst S.A., Milligan R.A., and Kapoor T.M.. 2010. Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell. 142:433–443. 10.1016/j.cell.2010.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, R., Ti S.-C., Tan L., Darst S.A., and Kapoor T.M.. 2013. Marking and measuring single microtubules by PRC1 and kinesin-4. Cell. 154:377–390. 10.1016/j.cell.2013.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, L., Mogilner A., Civelekoglu-Scholey G., Wollman R., Evans J., Stahlberg H., and Scholey J.M.. 2006. A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr. Biol. 16:2293–2302. 10.1016/j.cub.2006.09.064 [DOI] [PubMed] [Google Scholar]
- Telley, I.A., Gáspár I., Ephrussi A., and Surrey T.. 2012. Aster migration determines the length scale of nuclear separation in the Drosophila syncytial embryo. J. Cell Biol. 197:887–895. 10.1083/jcb.201204019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telley, I.A., Gáspár I., Ephrussi A., and Surrey T.. 2013. A single Drosophila embryo extract for the study of mitosis ex vivo. Nat. Protoc. 8:310–324. 10.1038/nprot.2013.003 [DOI] [PubMed] [Google Scholar]
- Vaizel-Ohayon, D., and Schejter E.D.. 1999. Mutations in centrosomin reveal requirements for centrosomal function during early Drosophila embryogenesis. Curr. Biol. 9:889–898. 10.1016/S0960-9822(99)80393-5 [DOI] [PubMed] [Google Scholar]
- Vernì, F., Somma M.P., Gunsalus K.C., Bonaccorsi S., Belloni G., Goldberg M.L., and Gatti M.. 2004. Feo, the Drosophila homolog of PRC1, is required for central-spindle formation and cytokinesis. Curr. Biol. 14:1569–1575. 10.1016/j.cub.2004.08.054 [DOI] [PubMed] [Google Scholar]
- von Dassow, G., and Schubiger G.. 1994. How an actin network might cause fountain streaming and nuclear migration in the syncytial Drosophila embryo. J. Cell Biol. 127:1637–1653. 10.1083/jcb.127.6.1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow, G., Verbrugghe K.J.C., Miller A.L., Sider J.R., and Bement W.M.. 2009. Action at a distance during cytokinesis. J. Cell Biol. 187:831–845. 10.1083/jcb.200907090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukušić, K., Ponjavić I., Buđa R., Risteski P., and Tolić I.M.. 2021. Microtubule-sliding modules based on kinesins EG5 and PRC1-dependent KIF4A drive human spindle elongation. Dev. Cell. 56:1253–1267.e10. 10.1016/j.devcel.2021.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Brust-Mascher I., and Scholey J.M.. 2015. The microtubule cross-linker Feo controls the midzone stability, motor composition, and elongation of the anaphase B spindle in Drosophila embryos. Mol. Biol. Cell. 26:1452–1462. 10.1091/mbc.E14-12-1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley, S., Kulkarni S., and Karess R.. 1995. Drosophila nonmuscle myosin II is required for rapid cytoplasmic transport during oogenesis and for axial nuclear migration in early embryos. Development. 121:1937–1946. 10.1242/dev.121.6.1937 [DOI] [PubMed] [Google Scholar]
- Wijeratne, S., and Subramanian R.. 2018. Geometry of antiparallel microtubule bundles regulates relative sliding and stalling by PRC1 and Kif4A. eLife. 7:e32595. 10.7554/eLife.32595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B.C., Riedy M.F., Williams E.V., Gatti M., and Goldberg M.L.. 1995. The Drosophila kinesin-like protein KLP3A is a midbody component required for central spindle assembly and initiation of cytokinesis. J. Cell Biol. 129:709–723. 10.1083/jcb.129.3.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B.C., Dernburg A.F., Puro J., Nokkala S., and Goldberg M.L.. 1997. The Drosophila kinesin-like protein KLP3A is required for proper behavior of male and female pronuclei at fertilization. Development. 124:2365–2376. 10.1242/dev.124.12.2365 [DOI] [PubMed] [Google Scholar]
- Wühr, M., Dumont S., Groen A.C., Needleman D.J., and Mitchison T.J.. 2009. How does a millimeter-sized cell find its center? Cell Cycle. 8:1115–1121. 10.4161/cc.8.8.8150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, C., Lau E., Schwarzenbacher R., Bossy-Wetzel E., and Jiang W.. 2006. Spatiotemporal control of spindle midzone formation by PRC1 in human cells. Proc. Natl. Acad. Sci. USA. 103:6196–6201. 10.1073/pnas.0506926103 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.