Abstract
We describe the cloning and characterization of a new family of nuclear receptor coregulators (NRCs) which modulate the function of nuclear hormone receptors in a ligand-dependent manner. NRCs are expressed as alternatively spliced isoforms which may exhibit different intrinsic activities and receptor specificities. The NRCs are organized into several modular structures and contain a single functional LXXLL motif which associates with members of the steroid hormone and thyroid hormone/retinoid receptor subfamilies with high affinity. Human NRC (hNRC) harbors a potent N-terminal activation domain (AD1), which is as active as the herpesvirus VP16 activation domain, and a second activation domain (AD2) which overlaps with the receptor-interacting LXXLL region. The C-terminal region of hNRC appears to function as an inhibitory domain which influences the overall transcriptional activity of the protein. Our results suggest that NRC binds to liganded receptors as a dimer and this association leads to a structural change in NRC resulting in activation. hNRC binds CREB-binding protein (CBP) with high affinity in vivo, suggesting that hNRC may be an important functional component of a CBP complex involved in mediating the transcriptional effects of nuclear hormone receptors.
Nuclear hormone receptors comprise a superfamily of ligand-dependent transcription factors involved in controlling diverse cellular processes, including growth, differentiation, development, and homeostasis (37). The nuclear hormone receptor superfamily includes type I receptors which mediate the effects of glucocorticoids (glucocorticoid receptor [GR]), estrogens (estrogen receptor [ER]), mineralocorticoids (mineralocorticoid receptor [MR]), progestins (progestin receptor [PR]), and androgens (androgen receptor [AR]) and type II receptors for thyroid hormone (thyroid hormone receptors [TRs]), all-trans-retinoic acid (retinoic acid receptors [RARs]), 9-cis-retinoic acid (9-cis-RA) (the RARs and RXRs), vitamin D (vitamin D receptor [VDR]), and the PPARs. These receptors share a similar modular structure consisting of an N-terminal A/B domain, a DNA-binding C domain, and a D, E, and F ligand-binding domain (LBD) (4, 37). The DNA-binding C domain is highly conserved among members of type I and type II nuclear hormone receptors. Although the LBDs of nuclear hormone receptors (∼300 amino acids) are diverse in sequence, accounting for ligand specificity, they exhibit certain similarities in their overall structure (4, 13). Thus, the LBDs of all nuclear receptors are organized into 12 helical regions which play an important role in determining the conformation of the LBD in the presence and absence of ligand (59) and in mediating heterodimerization of type II receptors with the RXRs (2, 13).
Ligand-dependent conformational changes in the LBD are thought to recruit coactivators or coregulators to the DNA-bound receptor, which leads to transcriptional activation (37). The activation function mediated through the LBD has been referred to as activation function 2 (AF2) (38, 54). The majority of the coactivators, identified in a yeast two-hybrid screen, fall into two main groups: the p160/SRC family (SRC-1 [NCoA-1] [28, 41, 55], TIF-2 [also known as GRIP-1 or NCoA-2] [23, 24, 55, 58], and AIB1 [also known as p/CIP, ACTR, RAC3, and TRAM-1] [1, 8, 34, 52, 55]) and the CREB-binding protein (CBP)/p300 family (5, 20, 28). Coactivators which fall outside these groups include PGC-1 (44), ARA70 (62), p/CAF (3, 60), and NRIF3, which exhibits specificity for only the TRs and the RXRs (33). Using a biochemical approach, another class of factors have been identified to interact with nuclear hormone receptors in the presence of ligand. The VDR-interacting proteins (DRIPs) (46, 47) were identified in nuclear extracts interacting with ligand-bound VDR in vitro, while the TR-associated proteins (TRAPs) were identified in HeLa cells as factors associating with TRα in the presence of ligand (12, 26). The DRIPs and TRAPs are multiprotein complexes which appear to be similar, if not identical, in composition and, interestingly, are devoid of the coactivators described above. The TRAP and DRIP components were also reported to be a part of the SMCC complex, which consists of human homologues of yeast mediator or RNA polymerase II holoenzyme factors (26). The DRIP and TRAP protein complexes bind to ligand-bound receptors through the DRIP205 and TRAP220 protein components of the complex (DRIP205 and TRAP220 appear to be identical proteins) (46).
In addition to activation through the LBD, certain receptors contain an independent activation function in the variable N-terminal A/B domain referred to as AF1 (38, 54). Although the A/B domain of TRα does not contain an independent activation function, it participates in transcriptional activation by the full-length receptor through its interaction with TFIIB (18). The independent AF1 in the A/B domains of the receptors such as ER, PR, and AR appears to also interact with known coactivators such as SRC-1 and GRIP-1 (35, 40, 56). In addition, SRA, which is an RNA, has been reported to selectively coactivate the AF1 function of steroid receptors through association with the N terminus and SRC-1 (30). Although certain nuclear receptor A/B domains appear to contain an independent activation function, the association of the A/B domain with the LBD in the context of full-length receptors results in a mutually dependent function of AF1 and AF2.
The association of regulatory coactivators with the LBDs of nuclear hormone receptors occurs through their LXXLL motifs (9, 21, 36). The LXXLL motif binds to a hydrophobic cleft in the receptor, which is formed by several regions, including helix 12 of the LBD, as a result of a ligand-dependent conformational change (9, 11, 39). The affinity and relative specificity of interaction of coactivators with ligand-bound receptor are also influenced by the amino acid sequences flanking the LXXLL motif (9, 36). Interestingly, the TR-RXR-interacting motif in NRIF3 is a variant (LXXIL) of the LXXLL motif, indicating that isoleucine can substitute for leucine in coactivator-receptor interactions (33). Members of the p160/SRC family of coactivators contain several LXXLL modules, which may serve to enhance the affinity of binding of the coactivator to receptor dimers (9, 36). In addition, the different LXXLL modules within a p160 coactivator (e.g. SRC-1 or GRIP-1) appear to exhibit different receptor specificities (9, 36). CBP and p300 have been shown to bind many transcription factors and are thought to function as transcriptional integrators for multiple transcriptional factors, including NF-κB (42), p53 (16), the STATs (63), and nuclear hormone receptors (5, 20, 28), as well as members of the p160/SRC family of coactivators (55, 57). Thus, CBP and p300 appear to facilitate the integration of signals from a variety of factors, including nuclear transcription factors and cell surface receptors.
In this study we describe the cloning and characterization of a new family of nuclear receptor coregulators (NRCs) from rat and human cells which modulate the function of nuclear hormone receptors in a ligand-dependent manner. NRCs are expressed as alternatively spliced isoforms which may exhibit different intrinsic activities and receptor specificities. The NRCs are organized into several modular structures that appear to play an important role in their function as coactivators and coregulators for nuclear hormone receptors. NRCs contain a single functional LXXLL motif which associates with type I and type II nuclear hormone receptors with high affinity. Human NRC (hNRC) harbors a potent N-terminal activation domain (AD1), which is as active as the herpesvirus VP16 activation domain, and a second activation domain (AD2) which overlaps with the receptor-interacting LXXLL region. The C-terminal region of hNRC appears to function as a modulatory domain which influences the overall transcriptional activity of the protein. Our results indicate that NRC may bind to liganded receptors as a dimer and that this association mediates a conformational change in NRC leading to activation, possibly by exposing an activation domain(s). hNRC binds CBP with high affinity in vivo, suggesting that hNRC may be an important functional component of a CBP complex involved in mediating the transcriptional effects of nuclear hormone receptors.
MATERIALS AND METHODS
Construction of a yeast two-hybrid cDNA library from GH4C1 cells.
Poly(A)+ RNA isolated from GH4C1 cells was used for the synthesis of cDNA by using a Stratagene cDNA synthesis system. Prior to ligation, cDNA was size fractionated using Sephacryl S200. Fractions above 0.4 kb were pooled, precipitated with ethanol, and ligated with EcoRI-XhoI-digested pJG4-5, which conditionally expresses the cDNA as a fusion with the B42 activation domain in yeast (17). The pJG4-5 cDNA was transformed into Sure bacteria (Stratagene) by electroporation. This cDNA library contains about 107 independent transformants, and the average insert size was estimated to be 1.5 kb.
Yeast two-hybrid screen.
cTRα was used as a bait by cloning the full-length cTRα into the EcoRI site of pEGΔPL, a yeast LexA expression vector. pEGΔPL was derived from the parent vector, pEG202 (17), by inserting a new polylinker, NcoI-SalI-BamHI-KpnI-EcoRI-BglII-SacI-XhoI, which also inactivated the original EcoRI site in pEG202. The yeast strain EGY48 (17), harboring the LacZ reporter plasmid pSH18-34 (17) and pEGΔPL-cTRα constitutively expressing LexA-cTRα (33), was used to transform the pJG4-5 cDNA library. Transformants were directly plated on X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) SD-galactose-raffinose plates which contained 1 μM thyroid hormone (T3) but lacked Trp, Ura, His, and Leu. Putative positive clones were further purified and plated on SD-dextrose plates lacking Trp, Ura, and His to suppress expression of the cDNAs. Ligand-dependent interaction was verified upon replica plating each clone on X-Gal SD-galactose-raffinose plates, lacking Trp, Ura, and His, with and without T3. Putative positive clones were also plated on X-Gal SD-dextrose plates, lacking Trp, Ura, and His, with and without T3. Yeast cells were considered to harbor interesting clones if they exhibited a positive LacZ (X-Gal) response with T3 on galactose-raffinose plates and no LacZ response on dextrose plates. The cDNAs from positive clones were sequenced and subjected to restriction digestion and size determination.
Plasmids and transfections into mammalian cells.
Expression plasmids for nuclear receptors and various reporters have been described earlier (33). All plasmids described below were generated by either PCR or restriction enzyme digestion and verified by sequencing and expression studies. The hNRC cDNA insert (KIAA0181) was provided by the Kazusa Research Institute (Chiba, Japan) as a pBluescript SKII plasmid. hNRC was cloned into the HindIII-NotI site of a mammalian expression plasmid, pExpress (pEX) (14). All Gal4 DNA-binding domain (DBD) fusion constructs were generated using pSG424 (48). These include Gal4-hNRC (full length), Gal4-hNRC(1-783), Gal4-hNRC(1-1076), Gal4-hNRC(771-1986), and Gal4-hNRC(1352-1986). Rat NRC.1 (rNRC.1) was cloned into pSG424 (Gal4-rNRC.1) as an EcoRI fragment from clone 15 in pJG4-5. Gal4-rNRC.1ΔC was created by digesting Gal4-rNRC.1 with XbaI. The Gal4-rNRC.1 LXXLL-1 mutant was generated by two-step PCR (22) where LVNLL was changed to AVNAA. Glutathione S-transferase (GST)–rNRC.1 was constructed by cloning the EcoRI insert from clone 15 in pJG4-5 into pGEX4T-1. GST-hNRC and GST-hNRC(1-852) were constructed by cloning the appropriate fragments into pEBG, a GST mammalian expression vector (43, 53). Transfections were performed with appropriate controls using calcium phosphate coprecipitation. Various ligands, such as T3 for TR, 9-cis-RA for RAR and RXR, 1,25-dihydroxy-vitamin D3 for VDR, and dexamethasone (Dex) for GR, were used at 0.5 μM. TTNPB, which is specific for RAR, and LG100153, which is specific for RXR, were used at 200 nM unless otherwise indicated. Typically 50 ng of the Gal4 reporter plasmid pBL-G5-CAT2 and 2 μg of Gal4 fusion plasmids were used for transfections. Unless otherwise mentioned, all other chloramphenicol acetyltransferase (CAT) reporters were used at 0.5 μg/sample and pEX-hNRC was used at 2 μg/sample. All samples were in duplicate or triplicate, and each experiment described has been repeated two or three times.
Yeast plasmids and β-galactosidase assays.
All yeast plasmids were sequenced after cloning. pJG4-5ΔPL was derived from pJG4-5 (17) by inserting the same polylinker described above for pEGΔPL into the EcoRI-XhoI sites of pJG4-5. These vectors differ only in the polylinker cloning sequence. hNRC was also cloned into pEGΔPL and pJG4-5ΔPL after release of the full insert from pEX-hNRC. An AF2 mutant of hRXRα (amino acids 1 to 545) was constructed in pJGΔPL by PCR, while an AF2 mutant of the cTRα LBD (pJGΔPL/cTRα120-398) was constructed by restriction digestion. In certain experiments (see Fig. 8) we cloned the hRXRα LBD into pJG4-6. This vector is similar to pJG4-5 but lacks the B42 activation domain. It includes the same hemagglutinin (HA) epitope, and we introduced the same simian virus 40 viral nuclear localization signal as found in pJG4-5. To study the function of hNRC in yeast, various deletion constructs of hNRC were generated using pJG4-5ΔPL and pEGΔPL vectors. The pJG4-5ΔPL (B42) hNRC constructs were produced from pEX-hNRC by digestion and purification of appropriate fragments followed by cloning into pJG4-5ΔPL. These constructs express the following hNRC amino acids fused to B42: 1 to 783, 771 to 1986, and 1352 to 1986. We also constructed B42 and LexA fusions of hNRC(771-1076) which is analogous to the residues found in rNRC.1. LexA-rNRC.1 was generated by releasing the fragment from pJG4-5-rNRC.1 and cloning it into pEGΔPL. The LXXLL-1 rNRC.1 mutant in pJG4-5ΔPL was produced by releasing an insert from the Gal-rNRC.1 mutant and cloning into pJG4-5ΔPL. LexA-hGR-LBD, LexA-hRXRα-LBD, LexA-hTRβ-LBD (31, 51), LexA-hRARα-LBD, and LexA-cTRα (full length) (33) have been described previously. LexA-hVDR-LBD was generously provided by David Moore (Baylor). LexA-hERα (full length) was from Michael Garabedian (New York University). All β-galactosidase assays were performed at least two times in duplicate or triplicate. Various ligands, such as T3 for the TRs, 9-cis-RA for RXR and RAR, 1,25-dihydroxy-vitamin D3 for VDR, and estradiol (E2) were used at 1 μM, while deoxycorticosterone for GR was used at 10 μM. Yeast colonies were first grown in SD-dextrose medium lacking Ura, His, and Trp. The yeast cells at log phase were then washed, diluted to the appropriate density, and incubated in SD-galactose-raffinose medium lacking Trp, Ura, and His to induce cDNA expression, followed by quantitation of β-galactosidase as described previously (33). β-Galactosidase units are expressed as (optical density at 420 nm × 1,000)/(minutes of incubation × optical density at 600 nm of yeast suspension).
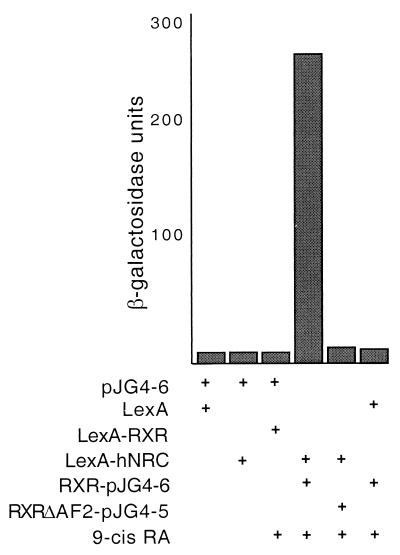
FIG. 8.
Receptor binding induces a conformational change in hNRC. The hRXRα LBD was cloned into pJG4-6. This vector is similar to pJG4-5 but lacks the B42 activation domain. It includes the same HA epitope and same simian virus 40 viral nuclear localization signal as found in pJG4-5, which we introduced. RXR-pJG4-6 expressing the hRXRα LBD was cotransformed into yeast along with LexA-hNRC as indicated. pJG4-6 alone or with pEG202 (LexA), LexA-hRXR-LBD, or LexA-hNRC was transformed into yeast as controls. RXRΔAF2-pJG4-5, contains the hRXRα LBD AF2 mutation expressed as a fusion protein with the B42 activation domain. RXRΔAF2-pJG4-5 was cotransformed with LexA or LexA-hNRC as shown. Yeast cells were incubated with or without 9-cis-RA, and the activity of β-galactosidase was determined.
In vitro and in vivo binding studies using GST fusions of NRC.
GST-rNRC.1 was expressed in Escherichia coli SG117 by induction with IPTG (isopropyl-β-d-thiogalactopyranoside) and then purified and immobilized to glutathione-agarose as described previously (18, 19). hGR, hVDR, hRARα, and hRXRα were labeled with [35S]methionine in vitro in the presence and absence of their cognate ligands by using rabbit reticulocyte lysate. cTRα was labeled in the absence of T3, which was added later in the in vitro binding assays. Typically 200 to 400 ng of GST protein bound to glutathione-agarose was used per assay. 35S-labeled nuclear receptors were mixed with GST or GST-rNRC.1. The samples were first incubated at 25°C for 15 min with cognate ligand in binding buffer (20 mM Tris-HCl [pH 7.7, 25°C], 4 mM MgCl2, 100 mM NaCl, 1 mM dithiothreitol, 0.25% bovine serum albumin, 0.5 mM phenylmethylsulfonyl fluoride, 20 μg of pepstatin and leupeptin per ml, and 0.25% NP-40). This was followed by a 20-min incubation at 4°C. The samples were then washed with same incubation buffer, and the bound 35S-labeled receptors were analyzed by sodium dodecyl sulfate [SDS] gel electrophoresis followed by autoradiography. Similar experiments were carried out with GST-rNRC.1 and 35S-labeled rNRC.1 to provide evidence for homodimerization of rNRC.1. Mammalian expression plasmids (pEBG) (43) for GST and GST fusions of hNRC were transiently transfected in 293T cells. Proteins bound to the expressed GST proteins were purified by lysing cells in buffer containing 50 mM Tris-HCl (pH 7.4), 250 mM KCl, 1 mM dithiothreitol, 0.25% NP-40, and protease inhibitors. Extracts were prepared by incubation of lysates on ice followed by freeze-thawing and centrifugation at high speed. Supernatants were incubated at 4°C with glutathione-agarose beads. The bound proteins were washed in lysis buffer and processed for SDS gel electrophoresis and Western blotting. Antibodies used for Western blotting include those against SRC-1 (from Bert O'Malley [Baylor] and Miles Brown [Dana-Farber]) and p/CAF (from Pat Nakatani [National Institutes of Health]). Antibodies against CBP (no. 06-294), p300 (no. 05-257), the Gal4 DBD (no. 06-262), and the LexA DBD (no. 06-719) were from Upstate Biotechnology.
Nucleotide sequence accession number.
The extended sequence of hNRC has been deposited in GenBank under accession number AF245115. The sequence of rNRC.1 has also been deposited in GenBank under accession number AF228043.
RESULTS
Cloning of NRC, a novel coactivator for nuclear hormone receptors.
GH4C1, a rat somatotrophic pituitary cell line, was used to construct a cDNA library for use in a yeast two-hybrid screen to identify novel coregulators for nuclear hormone receptors. We chose GH4C1 cells to construct a yeast two-hybrid cDNA library for several reasons. First, these cells contain virtually all nuclear hormone receptors at levels (∼10,000 per cell) similar to that found in somatotrophs and other hormone responsive tissues in vivo (15, 45, 49). Second, regulation of the endogenous growth hormone gene by thyroid hormone (T3) and glucocorticoids, and of the prolactin gene by estrogens, mimics the response of these genes by these hormones in pituitary cells in vivo (25, 49, 61). Last, although GH4C1 cells express only low levels of each nuclear hormone receptor, both endogenous and transiently transfected genes regulated by these receptors are highly responsive to their cognate ligands (45, 49, 61). This suggests that GH4C1 cells contain high levels of known coactivators and/or novel factors that play an important role in receptor function.
The GH4C1 cDNA library was constructed in pJG4-5, a yeast vector which conditionally expresses the cDNAs as a fusion with the B42 activation domain (17). Full-length cTRα was cloned into pEG202, which constitutively expresses the receptor as a LexA fusion in yeast (33). An EGY48 yeast strain expressing LexA-cTRα and harboring the LacZ reporter plasmid pSH18-34 was used for transformation of the pJG4-5 cDNA library (17, 33). Transformants were directly screened for ligand-dependent TR-interacting clones on X-Gal plates containing T3. Thirty-five pJG4-5 recombinant plasmids expressing strong T3-dependent interactors were isolated and subjected to secondary screening, followed by sequencing. Of these 35 clones, 13 were identified as rSRC-1, some of which appeared to represent splice variants. Three clones were rRXRβ, and 17 clones contained novel independent overlapping sequences representing three different cDNAs. A database search indicated that the sequences of two ∼1-kb clones (designated clone 14 and clone 15), which interacted with ligand-bound LexA-cTRα more strongly than the rSRC-1 clones, showed a striking homology with a 6,504-bp human cDNA of unknown function which was previously deposited as KIAA0181 (accession no. D80003) by the Kazusa Research Institute.
The KIAA0181 cDNA contains an apparent ATG initiation codon 60 bp from the 5′ end and an open reading frame which encodes a protein with a predicted molecular mass of 210 kDa. Upon closer inspection of the human cDNA clone, we identified several potentially important domains, which include two prominent glutamine-rich (Q-rich) regions (one of which is also rich in prolines [Q-P]), two putative nuclear hormone receptor interaction LXXLL motifs, and a C-terminal serine-threonine-leucine (S-T-L)-rich region (Fig. 1). Studies with rat clone 15 and the human cDNA clone in yeast and mammalian cells indicated that both cDNAs express proteins which exhibit features typical of a nuclear hormone receptor coregulators. A comparison of the amino acid sequences of the rat and human clones indicates that the rat clone is likely partial and missing N-terminal sequences. Based on our in vitro and in vivo findings, we designated the human cDNA (KIAA0181) hNRC and the ∼1-kb clone 15 rNRC.1.
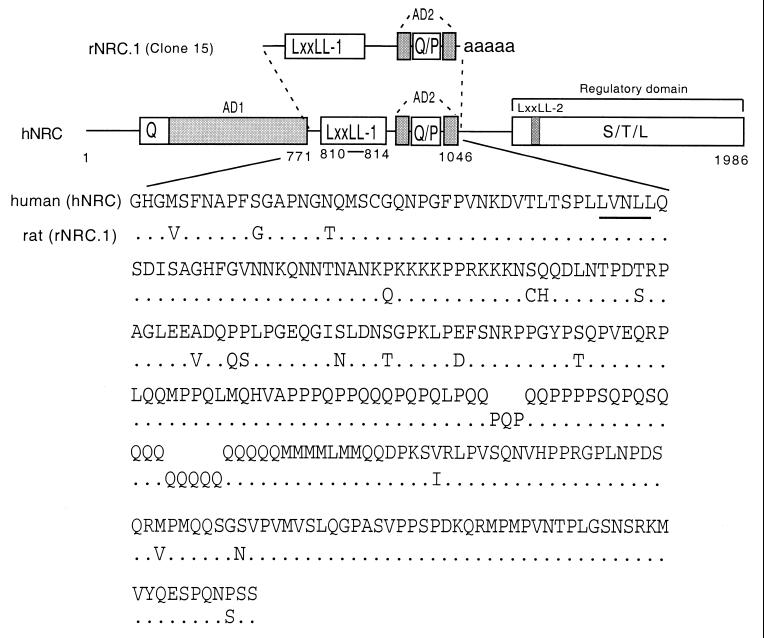
FIG. 1.
Structures and comparison of the hNRC and rNRC.1 isoforms. The upper section shows a schematic representation of the functional domains identified in hNRC and rNRC.1, a rat homologue, isolated from the GH4C1 pJG4-5 cDNA library. rNRC.1 is homologous to amino acids 771 to 1046 of hNRC. AD1 and AD2 are two activation domain regions, Q and P represent a glutamine- and proline-rich stretch of amino acids. LXXLL-1 is a nuclear receptor interaction motif. S-T-L is a serine-, threonine-, and leucine-rich sequence in the C-terminal part of hNRC which may be part of regulatory domain. LXXLL-2 is a second LXXLL motif which plays a less important role in nuclear receptor interactions. The lower section shows the amino acid sequence of rNRC.1 and its alignment with the corresponding region in hNRC. LXXLL-1 is underlined. The sequence of KIAA0181 is in GenBank under accession number D80003. We have extended the N terminus of hNRC and have deposited the sequence containing 58 additional amino acids in GenBank (accession number AF245115). The sequence of rNRC.1 has also been deposited in GenBank (accession number AF228043).
In vitro transcription-translation of the hNRC cDNA identified a protein of the expected size (data not shown). The deduced amino acid sequence of rNRC.1 revealed over 85% amino acid identity with a similar region of hNRC (Fig. 1). Interestingly, hNRC exhibited regions of about 30% similarity scattered around the 550-amino-acid proline- and glutamine-rich regions at the C termini of p300 and CBP. Analysis of the amino acid sequence of hNRC revealed two LXXLL motifs, which we refer to as LXXLL-1 and LXXLL-2, beginning at amino acids 810 and 1414 (Fig. 1). The two LXXLL motifs are separated by 604 amino acids, and the motifs and residues surrounding them are not identical to any of the LXXLL modules identified in the various members of the p160/SRC family of coactivators or CBP/p300.
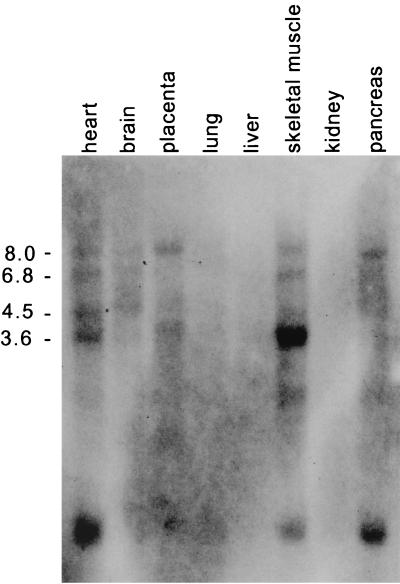
Interestingly, rNRC.1 contains only a single LXXLL motif (the LXXLL-1 module). Remarkably, rNRC.1 and hNRC are identical in sequence in the putative receptor interaction region spanning the LXXLL-1 motif (Fig. 1). The LXXLL-1 region of rNRC.1 is followed by a Q-P-rich region, a termination codon, and a polyadenylated tail, indicating that rNRC.1 represents an alternatively spliced C-terminal variant of the rat homologue of full-length hNRC. The possibility of alternatively spliced forms of hNRC is further suggested by the results of Northern blotting of RNAs derived from various human tissues probed with 32P-labeled hNRC. This study identified a prominent mRNA of 8 kb in most of the tissues examined (Fig. 2). mRNA species of 6.8, 4.5, and 3.6 kb with various abundances were also detected, and the 3.6-kb transcript was the major RNA species detected in skeletal muscle. In addition to the tissues shown in Fig. 2, a Northern blot shown on the Kazusa Research Institute's website (www.kazusa.or.jp) indicates that spleen, thymus, testis, ovary, and peripheral blood leukocytes also express hNRC mRNAs of various sizes in addition to a prominent 8-kb mRNA.
FIG. 2.
Northern blot of hNRC mRNAs in different tissues. hNRC mRNAs were detected using an MTN blot (Stratagene) containing RNAs from the various tissues indicated. The various-sized hNRC-related mRNAs were detected by probing the blot with 32P-labeled hNRC cDNA. Numbers on the left are kilobases.
Ligand-dependent interaction of the NRCs with various nuclear hormone receptors requires the LXXLL-1 motif.
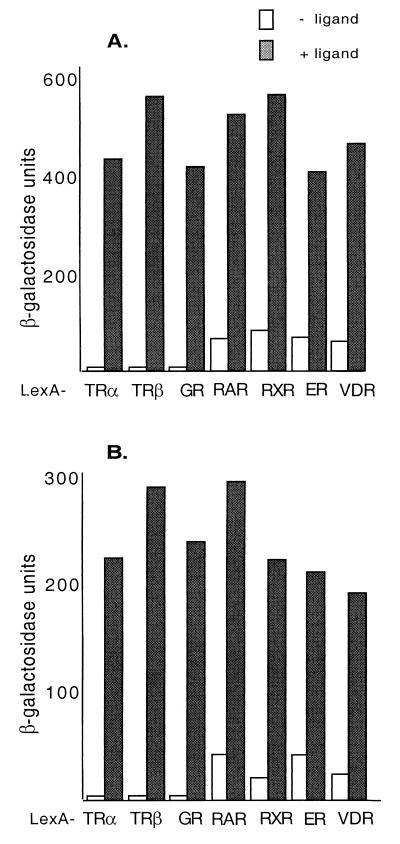
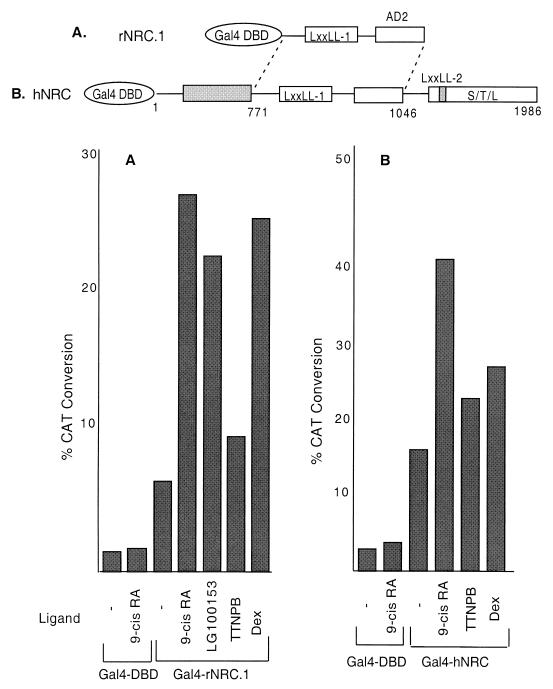
Although rNRC.1 was originally isolated using LexA-cTRα as a bait, we examined the ligand-dependent interaction of rNRC.1 in yeast with LexA chimeras containing various full-length nuclear hormone receptors or their LBDs (Fig. 3A). rNRC.1 interacted with all of the nuclear hormone receptors tested, and this interaction was greatly enhanced by their cognate ligands. LexA-cTRα, LexA-hTRβ-LBD, and LexA-hGR-LBD displayed strictly ligand-dependent interactions. We found that the LBDs of hRARα, hRXRα, and hVDR and full-length hERα exhibited some weak interactions with rNRC.1 in yeast in the absence of ligand. Such ligand-independent interactions in yeast have also been reported for GRIP-1 and TIF-2 and were speculated to be due to spontaneous conversion of receptors into an active form in yeast (10, 57). These results indicate that rNRC.1 interacts with various nuclear hormone receptors, and thus, the NRCs could potentially serve as coactivators with broad receptor specificity as reported for the p160/SRC coactivator family and CBP/p300 (37). The interaction of hNRC, cloned in pJG4-5, with LexA-nuclear hormone receptors in yeast was examined as described above for rNRC.1 (Fig. 3B). Although hNRC is not efficiently expressed in yeast as a fusion with the B42 activation domain, presumably because of its size, ligand-dependent interactions of 10- to 100-fold were found with the receptors tested. The interaction was strictly ligand dependent with the TRs and hGR, while, as with rNRC.1, a low level of interaction was noted for hRARα, hRXRα, hERα, and hVDR in the absence of ligands.
FIG. 3.
Ligand-dependent interaction of rNRC.1 and hNRC with nuclear hormone receptors in yeast. (A) rNRC.1 was expressed as a B42 fusion protein and tested against LexA fusions of cTRα, hTRβ-LBD, hGR-LBD, hRARα-LBD, hRXRα-LBD, hERα, and hVDR-LBD. Both ligand-dependent and ligand-independent interactions were quantified by measuring the activity of β-galactosidase in yeast extracts. Details are given in Materials and Methods, along with the ligands used. (B) Same as panel A except that hNRC was expressed as a B42 fusion and examined for interaction with the various LexA-receptor chimeras.
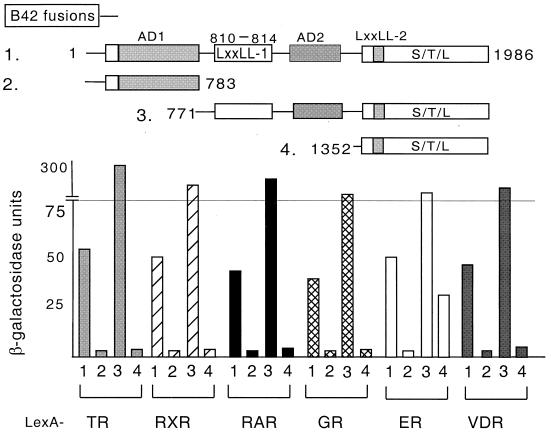
The receptor interaction domain of hNRC was next mapped by deletion analysis. hNRC and rNRC.1 contain a single LXXLL motif in common (LXXLL-1) (Fig. 1) and exhibit similar ligand-dependent receptor interactions (Fig. 3). This suggests that of the two LXXLL motifs in hNRC, LXXLL-1 plays a more direct and important role in mediating an interaction of hNRC with the various receptors. Various deletion mutants of hNRC were constructed in pJG4-5 and expressed as fusions with the B42 activation domain. As shown in Fig. 4, the N-terminal region of hNRC (amino acids 1 to 783) lacking the two LXXLL domains failed to interact with any of the nuclear hormone receptors. The clone expressing amino acids 771 to 1986, containing LXXLL-1 and LXXLL-2, interacted with high affinity with all of the receptors in a ligand-dependent manner. The contribution of each LXXLL motif and the specificity of interaction with each receptor were addressed by studying the C-terminal region of the protein containing only LXXLL-2 (amino acids 1352 to 1986) (Fig. 4). Interestingly, no interaction was detected with any of the nuclear hormone receptors except LexA-hERα (Fig. 4), the interaction of which was about 20% of that of the hNRC clones containing both LXXLL motifs (Fig. 4). These results strongly suggest that, with the exception of hERα, hNRC interacts with all of the nuclear receptors tested through only the single N-terminal LXXLL-1 motif.
FIG. 4.
Mapping the region of hNRC necessary for interaction with various receptors. As depicted, various deletion constructs of hNRC were generated as B42 fusions for two-hybrid interaction assays with the LexA-receptor chimeras indicated in Fig. 3.
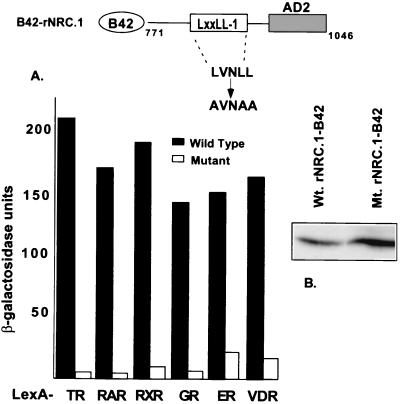
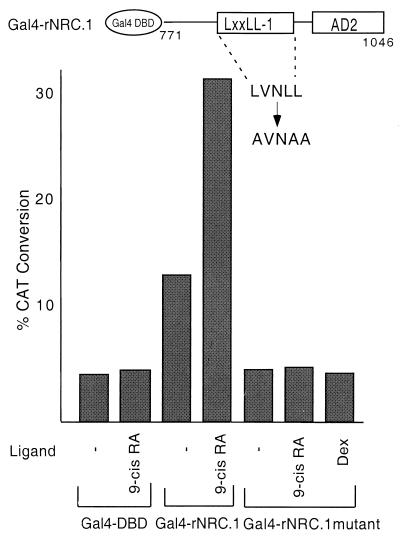
The critical role of leucines in the core LXXLL motif for interaction with the hydrophobic cleft formed in ligand-bound nuclear receptors has been established by both mutagenesis and crystallographic studies (9, 11, 36, 39). Therefore, mutagenesis studies were performed to provide direct evidence for a role for LXXLL-1 in the interaction of the NRCs with ligand-bound receptors. The LVNLL motif of LXXLL-1 in rNRC.1 was changed to AVNAA, and the interaction of B42 fusions of rNRC.1 and the mutant rNRC.1 was determined with LexA fusions of various nuclear receptors. As shown in the Fig. 5A, conversion of LXXLL-1 from LVNLL to AVNAA markedly reduced ligand-dependent interactions of rNRC.1 with all of the nuclear hormone receptors used in the study. Western blotting verified that the mutant rNRC.1 was induced by galactose and was expressed at levels equal to or greater than that of rNRC.1 (Fig. 5B). Thus, the experiments illustrated in Fig. 4 and 5 establish an essential and dominant role of the LXXLL-1 motif in mediating ligand-dependent interactions of nuclear hormone receptors with the NRCs.
FIG. 5.
Ligand-dependent interaction of rNRC.1 with nuclear receptors requires LXXLL-1. (A) LexA fusions of various nuclear hormone receptors as indicated were tested in yeast two-hybrid assays against wild-type and mutant rNRC.1 expressed as B42 fusion proteins. The amino acids indicated for rNRC.1 are those which are homologous to amino acids 771 to 1046 of hNRC. LXXLL-1 (LVNLL) was changed to AVNAA. The interaction was quantified by determining the activity of β-galactosidase with and without cognate ligands as described in Materials and Methods. (B) Western blotting using anti-HA antibody indicates the expression of wild-type (Wt.) and mutant (Mt.) proteins as shown.
NRCs form homodimers in vivo and in vitro.
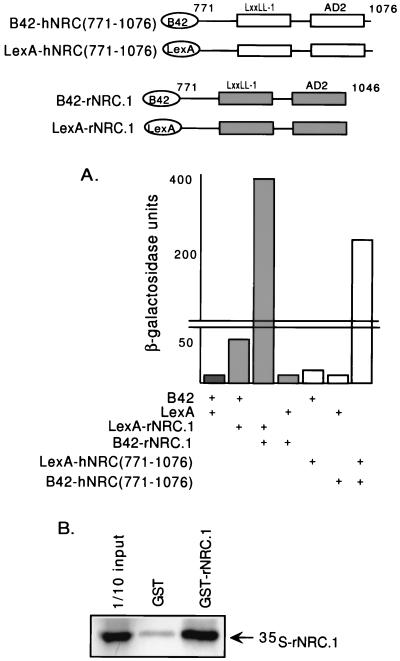
Studies with members of the p160/SRC family of coactivators have identified multiple LXXLL motifs which have been proposed to be important in the binding of a single coactivator molecule to receptor homo- and/or heterodimers (36). Since rNRC.1 contains only one LXXLL motif but interacts with receptors with high affinity, we considered the possibility that a dimer of rNRC.1 contributing two LXXLL motifs is necessary for a high-affinity receptor interaction. To explore whether rNRC.1 forms dimers in vivo, we expressed LexA and B42 fusions of rNRC.1 and, similarly, the analogous region of hNRC (amino acids 771 to 1076) (Fig. 6A). This study indicates that LexA-rNRC.1 and B42-rNRC.1 and the comparable regions of hNRC form a strong interaction in the cell, and thus, the NRCs may exist as a homodimer (or higher-order oligomers) in vivo. Binding studies with GST-rNRC.1 in vitro (Fig. 6B) also provide evidence for an rNRC.1-rNRC.1 interaction. To our knowledge, this is a first report demonstrating that a coactivator forms dimers in vivo and in vitro. Although further work will be needed to precisely map the dimerization domain, the results shown in Fig. 6 strongly suggest that NRC dimers may play an important role in high-affinity receptor dimer interactions.
FIG. 6.
NRCs form homodimers in vivo and in vitro. (A) LexA-rNRC.1 was transformed into yeast along with pJG4-5, which expresses rNRC.1 as a B42 fusion protein (B42-rNRC.1). pJG4-5 (B42) and pEG202 (LexA), pJG4-5 (B42) and LexA-rNRC.1, and B42-rNRC.1 and pEG202 (LexA) were also transformed into yeast and served as controls. Similar studies were carried out with hNRC(771-1076) cloned into LexA and B42 expression vectors. (B) GST-rNRC.1 or GST alone was incubated with 35S-rNRC.1 labeled with l-[35S]methionine in rabbit reticulocyte lysates, and in vitro binding was carried out as described in Materials and Methods. The samples were then electrophoresed in SDS gels, and the amount of 35S-rNRC.1 which bound to GST-rNRC.1 was visualized by fluorography. One-tenth of the input amount of 35S-labeled rNRC.1 used in the incubation with GST-rNRC.1 was electrophoresed in the same gel.
Ligand-dependent interactions of NRCs with nuclear hormone receptors in vitro and in vivo require an intact AF2 receptor domain.
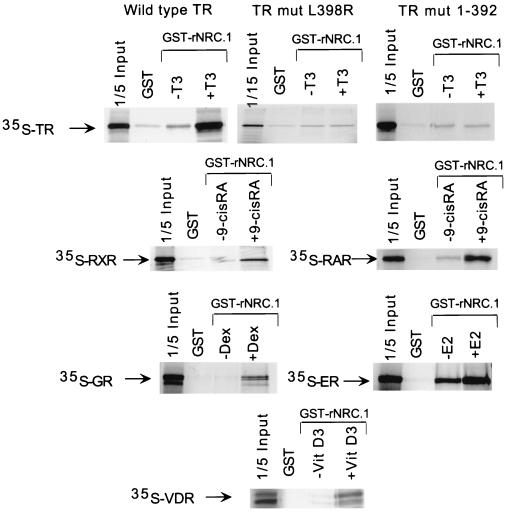
Ligand-dependent interactions and the role of the helix 12 AF2 region were studied by binding of nuclear hormone receptors to NRC in vitro. GST fusions of hNRC and rNRC.1 were expressed in bacteria. However, only GST-rNRC.1 was expressed at levels sufficient for in vitro binding studies. GST or GST-rNRC.1 bound to glutathione-agarose beads was incubated with various 35S-labeled receptors with or without cognate ligands, and the bound receptors were electrophoresed and visualized by autoradiography. As shown in Fig. 7, binding of the various receptors to GST-rNRC.1 was markedly enhanced by their cognate ligands. AF2 mutants of the 408-amino-acid cTRα, L398R and 1-392 (50), failed to bind GST-rNRC.1 in the presence of T3, indicating a requirement for an intact AF2 function for ligand-dependent binding of receptor to rNRC.1. Similarly, very little binding of hRXRα, hGR, or hRARα was observed without cognate hormones. Interestingly, some binding of hERα was observed with GST-rNRC.1 in the absence of ligand, although this association was greatly enhanced in the presence of an appropriate ligand. This ligand-independent interaction of hERα with rNRC.1 in vitro may be biologically relevant, since, as described below, ligand-independent enhancement of hERα activity with NRC was also found in transfection experiments with mammalian cells.
FIG. 7.
Ligand-dependent binding of nuclear receptors with rNRC.1 in vitro. All receptors were labeled with l-[35S]methionine by in vitro transcription-translation using reticulocyte lysates. Bacterially expressed and purified GST-rNRC.1 bound to glutathione-agarose beads was incubated with 35S-labeled receptors with or without the indicated cognate ligands. The samples were then electrophoresed in SDS gels, and the amount of 35S-receptor which bound to GST-rNRC.1 was visualized by fluorography. One-fifth of the amount of 35S-labeled receptor used in the incubation with GST-rNRC.1 was electrophoresed in the same gel. E2, estradiol; VitD3, 1,25-dihydroxy-vitamin D3.
The importance of an intact AF2 region of the nuclear hormone receptors was also assessed in vivo in yeast using AF2 mutants of hRXRα and cTRα lacking helix 12. After confirmation of expression of the mutant proteins, the deletion mutants were expressed in pJG4-5 as B42 fusions and examined for their interactions with LexA-rNRC.1 and LexA-hNRC. The AF2 mutants of hRXRα and cTRα were unable to interact with hNRC and rNRC.1 in a ligand-dependent manner (data not shown).
Receptor binding mediates a conformational change in hNRC.
Although LexA-rNRC.1 exhibits moderate intrinsic activity in yeast (Fig. 6), we found that the LexA fusion of the entire hNRC sequence exhibits no intrinsic basal activity (Fig. 8). This raises the interesting possibility that the full-length coactivator undergoes a conformational change upon receptor binding leading to activation. To provide evidence for this possibility, we cloned the hRXRα LBD in pJG4-6, a modified pJG4-5 plasmid lacking the heterologous B42 activation domain. As shown in Fig. 8, LexA-hRXRα-LBD, LexA-hNRC, or hRXRα-LBD-pJG4-6 expressed alone is not active in yeast with or without 9-cis-RA. Although the hRXRα LBD is inactive in yeast in the presence of ligand, coexpression of hRXRα-pJG4-6 and LexA-hNRC led to a 100-fold increase in activation in the presence of 9-cis-RA (Fig. 8). This suggests that receptor binding induces a conformational change in hNRC that results in exposure of its activation domain(s) (see Fig. 12), leading to enhanced activity in yeast.
FIG. 12.
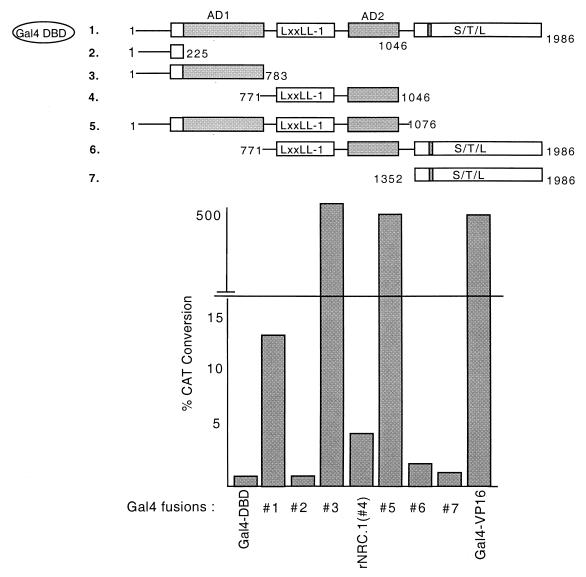
hNRC harbors a strong activation domain (AD1) and an inhibitory S-T-L region at the C terminus. The indicated Gal4 fusions of hNRC and rNRC.1 were transfected in HeLa cells with pBL-G5-CAT2. The amino acids indicated for the rNRC.1s are those which are homologous to amino acids 771 to 1046 of hNRC. The transfections were analyzed in duplicate, and the experiment was repeated at least three times with similar results. Gal4-VP16 was used as a control for comparison.
NRC enhances the transactivation function of endogenous nuclear hormone receptors in mammalian cells.
The preceding in vitro binding studies and the experiments in yeast provide strong support for the NRCs as coregulatory factors for various receptors. To provide further evidence for a role of the NRCs with receptors in mammalian cells, we first expressed hNRC and rNRC.1 as fusion proteins with the yeast Gal4 DBD. This would allow us to assess whether hNRC or rNRC.1 exhibited an independent activation function in mammalian cells and might also allow for studies of the interaction of hNRC or rNRC.1 with nuclear hormone receptors in vivo. As shown in Fig. 9A, compared with the Gal4 DBD, expression of Gal4-rNRC.1 in HeLa cells resulted in a threefold increase in the activity of a Gal4-CAT reporter alone, suggesting that rNRC.1 contains an intrinsic activation domain. However, the activity found with Gal4-rNRC.1 was enhanced about fivefold further when the cells were incubated with 9-cis-RA, suggesting a ligand-dependent association of hRXR and/or hRAR with Gal4-rNRC.1 in vivo. To study the individual contributions of hRAR and hRXR, we used ligands specific for these receptors. LG100153, an RXR-specific ligand, resulted in a further 3.5-fold stimulation, while the RAR-specific ligand, TTNPB, resulted in a small 1.25-fold stimulation. Gal4-rNRC.1 also interacted with endogenous hGR as determined by the fivefold increase in activity in cells incubated with Dex. Estradiol incubation resulted in a similar fivefold increase in activity compared with the stimulation found with Gal4-rNRC.1 alone (data not shown). Figure 9B shows the extent of stimulation and the ligand-dependent enhancement by endogenous receptors of a Gal4 chimera with hNRC. As with Gal4-rNRC.1, expression of Gal4-hNRC resulted in a fivefold increase in the activity of the Gal4-CAT reporter when compared with the Gal4 DBD alone. Incubation with 9-cis-RA resulted in a further 2.5-fold stimulation, while the RAR-specific ligand TTNPB and Dex resulted in only a 1.5-fold increase. These results indicate that hNRC and rNRC.1 contain an intrinsic activation domain(s) and associate with and enhance the activity of endogenous nuclear receptors in HeLa cells. The binding of NRCs with receptors through LXXLL-1 would be expected to preclude the association of other coactivators directly with the hydrophobic groove formed by ligand-bound receptor. Thus, we interpret the enhanced stimulation of Gal4-NRC by ligand-bound receptor to indicate that the receptor mediates a conformational change in NRC leading to further activation. This conclusion, based on mammalian cell studies, is consistent with the studies in yeast as shown in Fig. 8.
FIG. 9.
Ligand-dependent interaction of endogenous nuclear receptors with rNRC.1 and hNRC in mammalian cells. (A) HeLa cells were co-transfected with a Gal4-responsive CAT reporter plasmid (pBL-G5-CAT2) and pSG424 vectors which express the Gal4 DBD or Gal4-rNRC.1 with and without the various ligands as indicated. The amino acids indicated for rNRC.1 are those which are homologous to amino acids 771 to 1046 of hNRC. Each sample was analyzed in duplicate, and the experiment was repeated at least three times (see Materials and Methods for details about plasmids and the concentrations of the various ligands). (B) Same as panel A except that a Gal4-hNRC fusion was used instead of Gal4-rNRC.1.
Mutations of LXXLL-1 abolish transcriptional enhancement of nuclear hormone receptors by rNRC.1.
Interaction studies in yeast (Fig. 4 and 5) document the involvement of LXXLL-1 in the ligand-dependent interaction of hNRC and rNRC.1 with nuclear hormone receptors. The importance of LXXLL-1 of the NRCs in the ligand-dependent interaction of nuclear receptors in mammalian cells was studied using Gal4-rNRC.1 and the corresponding Gal4-rNRC.1 mutant in which LXXLL-1 was changed from LVNLL to AVNAA (Fig. 10). Both were expressed at similar levels based on Western blotting using antibody against Gal4. As expected, Gal4-rNRC.1 showed a ligand-dependent interaction with nuclear receptors. However, the Gal-rNRC.1 mutant showed no evidence for ligand-dependent enhancement by endogenous hRAR, hRXR, or hGR. Interestingly, ligand-independent intrinsic activity was not detected with the mutant. This suggests that the intrinsic basal activity of rNRC.1 may result from a factor which binds to the LXXLL-1 motif and that ligand-bound receptor displaces the factor and binds to the same site. We considered the possibility that the factor might be hER, since it is extremely difficult to remove all traces of estrogenic activity from the medium. However, this possibility seems unlikely, since the basal activity of Gal4-rNRC.1 was not reduced by the pure antiestrogen ICI 182780 (data not shown).
FIG. 10.
LXXLL-1 influences both the ligand-dependent and intrinsic basal activities of rNRC.1. The Gal4 DBD, Gal4-rNRC.1, and the LXXLL-1 Gal4-rNRC.1 mutant were cotransfected with pBL-G5-CAT2 with or without various ligands as indicated. The amino acids indicated for rNRC.1 are those which are homologous to amino acids 771 to 1046 of hNRC. Each sample was analyzed in duplicate, and the experiment was repeated at least two times with similar results.
hNRC contains a C-terminal inhibitory region, a potent N-terminal activation domain (AD1), and a central activation domain (AD2) that influence the transcriptional activity of nuclear hormone receptors bound to the LXXLL-1 region.
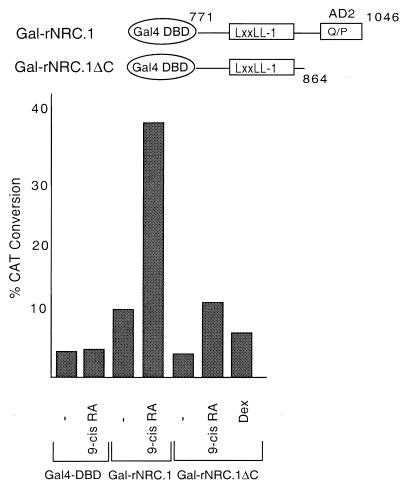
hNRC and rNRC.1 are transcriptionally active in mammalian cells and enhance the ligand-dependent transcriptional activation of nuclear hormone receptors, possibly through a common activation domain. We considered the Q-P-rich region common to hNRC and rNRC.1, designated AD2 in Fig. 1, as such an activation domain. To explore this possibility, this C-terminal AD2 region in rNRC.1 was deleted, the deletion mutant was expressed as a Gal4 chimera (Gal4-rNRC.1ΔC), and its activity was compared with that of Gal4-rNRC.1 (Fig. 11). The intrinsic activity of Gal4-rNRC.1ΔC was reduced to the level seen with the Gal4 DBD alone, indicating that C-terminal 184-amino-acid Q-P-rich region that we refer to as AD2 plays an important role in the intrinsic activation function of Gal4-rNRC.1. In addition to a reduction in basal expression, ligand-dependent enhancement of nuclear receptors by 9-cis-RA is also reduced, suggesting that AD2 plays a role in enhancing the activation function of liganded receptor which binds to the nearby LXXLL-1 region.
FIG. 11.
AD2 influences both the ligand-dependent and intrinsic basal activities of rNRC.1. pBL-G5-CAT2 was cotransfected in HeLa cells with the Gal4 DBD and the two Gal4 fusions of rNRC.1 as depicted. The amino acids indicated for the rNRC.1s are those which are homologous to amino acids 771 to 864 and 771 to 1046 of hNRC. The ligands used are as indicated.
Although the AD2 domain is also found in hNRC, Gal4-hNRC exhibits more intrinsic activity in mammalian cells than Gal4-rNRC.1 (Fig. 9). Therefore, we constructed Gal4 chimeras of other regions of hNRC to determine whether additional regions of hNRC might contribute to its higher intrinsic activity (Fig. 12). Gal4-hNRC(771-1986), which is analogous to Gal4-rNRC.1 but also contains the C-terminal serine-threonine-leucine-rich region, exhibited ligand-dependent stimulation but lower intrinsic (basal) expression than Gal4-rNRC.1. Gal4-hNRC(1352-1986), which contains LXXLL-2 but lacks AD2, exhibited no intrinsic activity. Gal4-hNRC(1-225), containing an N-terminal Q-rich region, was also without activity. In contrast, Gal4-hNRC(1-783) and Gal4-hNRC(1-1076) were as active as Gal4-VP16 and at least 50- to 100-fold greater in activity than Gal4-hNRC or Gal4-rNRC.1 (Fig. 9 and 12). We refer to the activation function contributed by hNRC(1-783) as AD1 (Fig. 1). The finding that deletion of the C-terminal serine-threonine-leucine-rich domain of hNRC can lead to a 50- to 100-fold increase in transcriptional activity [e.g., Gal4-hNRC(1-1076)], suggests that this C-terminal domain acts to repress or modulate the overall transcriptional activity of hNRC.
hNRC acts as a coactivator for various nuclear receptors by enhancing transactivation through cognate HREs.
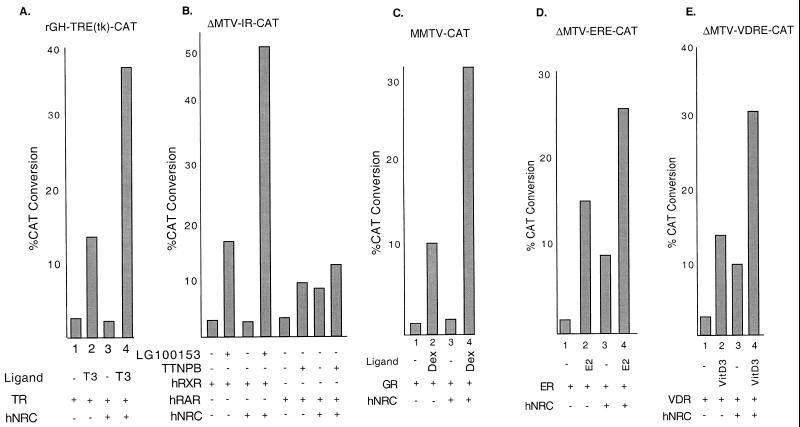
Our results indicate that hNRC harbors at least two activation domains, AD2 in the center of the protein and a potent activation domain, AD1, at the N terminus. These findings, along with the LXXLL-1 ligand-dependent receptor region, strongly support the notion that hNRC functions as a transcriptional coactivator for nuclear hormone receptors. To provide further evidence for this possibility, the effect of hNRC on ligand-dependent activation was studied with transfected wild-type nuclear receptors and various CAT reporter genes regulated by their hormone response elements. Figure 13 shows that expression of hNRC enhances the ligand-dependent activity of almost all of the nuclear receptors examined. Interestingly, in case of hRARα, hERα, and hVDR, expression of hNRC also resulted in a ligand-independent stimulation of 2.5- to 4-fold. This was not an effect of hNRC on the basal activity of the CAT reporter gene and was found only when these receptors were expressed. The mechanism(s) accounting for this ligand-independent activity of hRARα, hERα, and hVDR is unclear. hRARα and hVDR are thought to bind to response elements as heterodimers with the RXRs in the absence of their ligands. Thus, ligand-independent activation of these receptors by hNRC may result from the stabilization of a transcriptionally active conformation of these receptors by hNRC which can occur in the absence of ligand. For the other receptors, such as cTRα and hRXRα, which also bind their response elements without ligand, the adoption of an active conformation and the binding of hNRC is strictly ligand dependent. In contrast, GR does not bind its response element in the absence of ligand, and thus, the effect of hNRC on GR would be expected to be strictly ligand dependent. The ligand-independent effect of hNRC on hERα is surprising, since hERα is not thought to bind its response elements in the absence of ligand. This effect of hNRC on hERα does not appear to be due to residual estrogens in the medium, since a pure antiestrogen (ICI 182780) did not reduce this activity. Thus, the effect may reflect a ligand-independent interaction of hNRC with hERα in the cell. This conclusion is supported by the studies of Fig. 7 indicating that hERα can efficiently bind to GST-rNRC.1 in the absence of ligand. Although, the mechanism(s) of ligand-independent activation of hERα by hNRC is unclear, it may provide insights into the observed hormone-independent effects of ER in cells which have been ascribed to the AF1 region of the receptor.
FIG. 13.
hNRC functions as a coactivator for wild-type nuclear receptors on native hormone response elements. (A) HeLa cells were transfected with an rGH-TRE-tk-CAT reporter and expression vectors for cTRα and hNRC as indicated. T3 was at 1 μM. (B) Same as panel except that the CAT reporter was ΔMTV-IR-CAT and the receptors expressed were hRXRα and hRARα. The RXR-specific ligand LG100153 and the RAR-specific ligand TTNPB were each used at 200 nM. (C, D, and E) Same as panel except that the corresponding CAT reporters depicted were cotransfected with vectors expressing hGR, hERα, and hVDR and the cells were incubated with and without their cognate ligands at 200 nM. All samples were analyzed in duplicate, and the CAT activity shown is an average of three experiments.
hNRC associates with CBP in vivo.
Since hNRC exhibits properties characteristic for a coactivator of nuclear hormone receptors, we sought to determine whether it might exist as part of a complex with other factors thought to be important in nuclear receptor function. To explore this, we expressed full-length hNRC and an N-terminal region (amino acids 1 to 852) containing AD1 and the LXXLL-1 region as fusion proteins with GST, using the mammalian GST expression vector pEBG (43, 53). Full-length GST-hNRC, GST-hNRC(1-852), and a GST control were expressed in 293T cells. Initial Western blotting studies with antibody against GST indicated that GST-hNRC specifically localized to the cell nucleus. To ensure that GST and GST-hNRC(1-852) were also localized to the cell nucleus, they were expressed as a GST fusion which incorporates a nuclear localization signal just C terminal of GST. Thirty-six hours after transfection, the cells were lysed in buffer containing 0.25% NP-40 and 250 mM KCl, and after centrifugation, the cell extracts were incubated with glutathione-agarose beads and then washed twice in the same buffer. The proteins remaining bound to glutathione-agarose were electrophoresed, transferred to nitrocellulose, and analyzed for CBP, p300, SRC-1, and p/CAF by Western blotting.
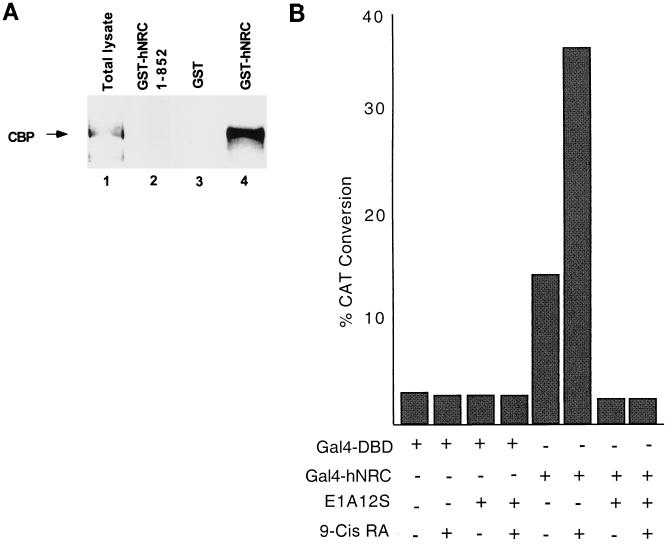
CBP was identified bound to GST-hNRC but not to GST-hNRC(1-852) or GST alone (Fig. 14A). Western blotting with antibody against GST indicated that GST-hNRC(1-852) was expressed at much higher levels than full-length GST-hNRC. This suggests that CBP selectively interacts with the region of hNRC which is C terminal of amino acid 852 or that this C-terminal region is required for appropriate folding of hNRC to allow for an hNRC-CBP interaction in vivo. The interaction of CBP with hNRC appears to be of high affinity, since the amount of CBP was only slightly reduced when the glutathione-agarose beads were washed with buffer containing 500 mM KCl (data not shown). Although the antibodies against SRC-1 and p/CAF detected these proteins in extracts, neither was detected to interact with full-length GST-hNRC (data not shown). We detected a very low level of association of SRC-1 (compared with the amount in the extract) with the more highly expressed GST-hNRC(1-852), suggesting an indirect or nonspecific interaction. To provide additional support for a functional interaction with CBP in cells, we examined the effect of the adenovirus 12S E1A protein in transcriptional activation by Gal4-hNRC (Fig. 14B). Expression of E1A, which binds to and inhibits transcriptional activation by CBP/p300 (6, 60), completely blocked both basal and ligand-dependent transcriptional activation by Gal4-hNRC.
FIG. 14.
hNRC associates with CBP in vivo, and EIA abrogates both the intrinsic activity and ligand-dependent activation function of hNRC. (A) pEBG vectors expressing GST, a GST fusion of hNRC, or GST-hNRC(1-852) were transfected into 293T cells. Extracts were prepared as described in Materials and Methods and incubated with glutathione-agarose. After washing, the proteins bound to the glutathione-agarose beads were resolved by SDS gel electrophoresis and analyzed for CBP by Western blotting using a polyclonal anti-CBP antibody. Lane 1, a sample of the total lysate prior to incubation with glutathione-agarose beads; lane 4, amount of CBP retained by GST-hNRC; lane 3, GST alone, which does not bind CBP; lane 2, hNRC(1-852), which also does not bind CBP. (B) pBL-G5-CAT2 was cotransfected with expression vectors for the Gal4 DBD or for Gal4-hNRC with or without a vector expressing adenovirus 12S E1A. The cells were incubated with or without 500 nM 9-cis-RA. All samples were analyzed in duplicate, and the experiment was repeated two times with similar results.
DISCUSSION
The NRCs appear to be a new class of nuclear receptor coregulators that are unrelated to the p160/SRC family or the DRIPs and TRAPs. Interestingly, a best-fit alignment of amino acid sequences indicates that hNRC shares about 30% dispersed similarity over a 550-amino-acid region with CBP and p300, suggesting that hNRC is more closely related to CBP/p300 than to other factors which regulate nuclear receptor function. Our studies with hNRC indicate an interesting structure (Fig. 1) consisting of (i) an N-terminal activation domain (AD1) which appears to be as potent as the activation domain of VP16, (ii) a receptor-interacting domain containing an LXXLL motif (LXXLL-1) which plays an essential role in ligand-dependent interaction with all nuclear receptors examined, (iii) a second Q-P-rich activation domain (AD2), (iv) a dimerization region, and (v) a C-terminal serine-threonine-leucine-rich region (containing a second LXXLL motif [LXXLL-2]) which appears to inhibit and/or modulate the activity of the AD1 and AD2 regions.
The LXXLL-1 region contains a proline in the −2 position (PLLVNLL) and resembles the class II group of LXXLL motifs identified in peptide libraries to interact with hERα (7). The class II LXXLL module was also shown to interact with TR, RXR, RAR, GR, ERβ, and VDR (7). The class II sequence is also found in DRIP205 and TRAP220, which were identified with VDR and TR, respectively. Interestingly, unlike the p160/SRC family of coactivators, which contain multiple LXXLL modules which interact with different nuclear receptors, rNRC.1 appears to contain only one LXXLL motif which interacts with nuclear receptors. Our findings shown in Fig. 6 support the notion that NRCs exist as a dimer and thus could bind to a receptor dimer through two LXXLL-1 motifs, thereby stabilizing the receptor-NRC interaction. A model depicting this is shown in Fig. 15. Furthermore, our studies using mutants of LXXLL-1 (LVNLL to AVNAA) in yeast (Fig. 5) and mammalian (Fig. 10) cells document the critical role of LXXLL-1 in ligand-dependent interactions of rNRC.1 with all of the nuclear receptors studied. This conclusion is further supported by studies using deletion mutants of hNRC in yeast (Fig. 4). Moreover, this study also indicates that the region containing AD1 (amino acids 1 to 783) does not interact with any of the nuclear hormone receptors or any of the cofactors studied. Figure 4 also shows that the C-terminal region of hNRC, which includes LXXLL-2, interacts only with hERα, suggesting that this domain may function as a specific interacting region for ER. Interestingly, SRC-1a, an isoform of SRC1, also contains an ER-specific interaction motif in the C terminus (27).
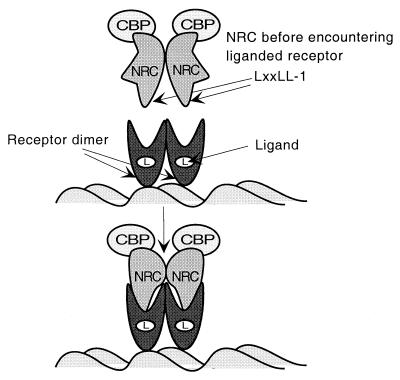
FIG. 15.
Model for transcriptional activation of nuclear hormone receptors by NRC. The LXXLL-1 motifs of NRC dimers interact with the hydrophobic grooves of ligand-bound receptor homo- or heterodimers. This results in a conformational change in NRC which may expose an activation domain(s), which leads to transcriptional activation. The finding that NRC interacts with CBP in the apparent absence of ligand (Fig. 14A) suggests that a preformed complex of NRC and CBP binds receptors. This model does not exclude the possibility that other factors or metabolic processes influence the association of hNRC with CBP.
One interesting finding is that LXXLL-1 of rNRC.1 appears to influence both ligand-dependent activity and the intrinsic basal activity mediated by the AD2 region (Fig. 10). The Q-P-rich region in AD2 is about 100 amino acids C terminal of LXXLL-1. However, we have not mapped the precise boundaries of AD2. Therefore, LXXLL-1 may be contiguous with AD2 and/or influence the conformation of the protein to allow for intrinsic AD2 activity. This raises the interesting possibility that the association of ligand-bound receptors with LXXLL-1 alters the conformation of rNRC.1, allowing for enhanced AD2 activity or the recruitment of other coactivators to NRC in the complex. This conclusion is supported by the findings shown in Fig. 8, indicating that the binding of liganded hRXRα-LBD to hNRC in vivo results in the strong activation of transcription by the coregulator. In addition, the results obtained from Fig. 9 indicate that hNRC and rNRC.1 contain an intrinsic activation domain(s) and associate with and enhance the activity of endogenous nuclear receptors in HeLa cells. The binding of NRCs with receptors through LXXLL-1 would be expected to preclude the association of other coactivators directly with the hydrophobic groove formed by ligand-bound receptor. We interpret these results with yeast and mammalian cells to indicate that receptor binding to NRC results in a conformational change leading to activation, possibly by exposing an activation domain(s) of NRC.
As coregulators, the NRCs might act through changes in chromatin structure, through a direct interaction with the basal transcription apparatus, or through an association with other coactivators or transcription factors. Western blotting indicated that GST-hNRC interacted with CBP with high affinity in mammalian cells. Although GST-hNRC could be barely detected with antibody against GST, it bound substantial amounts of CBP even at high salt concentrations. That this interaction is relevant is supported by the E1A inhibition studies (Fig. 14B). Similar Western blotting studies using antibodies against SRC-1 and p/CAF did not identify any apparent interaction with hNRC. The finding that CBP interacts with GST-hNRC but not GST-hNRC(1-852) suggests that CBP interacts with the region C terminal of residue 852 or that the C-terminal region allows proper folding of hNRC for interaction with CBP. The interaction of GST-hNRC with p300 was much weaker than that with CBP (data not shown), suggesting that hNRC may preferentially interact with CBP. This specificity for the CBP versus p300 may provide an explanation for differences in the biologic effects of these two closely related proteins (29). Based on the affinity of interaction of CBP with hNRC, we suggest that hNRC may be an important component of a functional CBP complex which mediates the transcriptional effects of nuclear hormone receptors and possibly other known or yet-to-be defined factors as well. Although CBP forms a complex with hNRC in vivo, we have observed only a weak synergistic effect resulting from cotransfecting expression vectors for CBP and hNRC in cells. Although this may appear to be inconsistent with our in vivo binding studies, a large synergistic response might not occur if hNRC binds CBP with high affinity and if the levels of endogenous CBP are already greater than the levels of expressed hNRC.
During the preparation of this paper, Lee et al. (32) described the cloning of a Xenopus homologue of the KIAA0181 cDNA clone and a related human cDNA (ASC-2) which is 58 amino acids longer at its N terminus than hNRC and may represent a longer version of hNRC or an alternative spliced product. We have extended the N terminus of KIAA0181 and also identified these additional sequences. hNRC/ASC-2 is homologous to a sequence (AIB-3) which was recently deposited in GenBank and reported to be amplified in many human cancers. Since there appears to be more than one isoform of hNRC proteins (see also Fig. 2), we suggest that hNRC/ASC-2 be referred to as hNRC-1. In AIB-3, the first 30 amino acids of the KIAA0181 hNRC clone is replaced with a 26-amino-acid novel sequence, the functional relevance of which is unknown. In the present study using mammalian cells and yeast, we have defined the modular structures of hNRC necessary for activation and for interaction with nuclear hormone receptors. Our results differ substantially with regard to the interaction regions for nuclear hormone receptors and for CBP from those reported for ASC-2 (32). Thus, based on qualitative X-Gal plate assays, ASC-2 amino acids 586 to 860 (equivalent to hNRC amino acids 509 to 783) were reported to contain an essential interacting domain for ligand-bound receptor. This region contains no LXXLL motifs or any LXXLL-like motifs containing other hydrophobic amino acids that might substitute for leucine. In contrast, using quantitative LacZ assays, we have consistently found no evidence to support an interaction of amino acids in this region with any nuclear hormone receptors (Fig. 4). In yeast, the N-terminal half of ASC-2 was reported to interact with different regions of CBP (amino acids 1 to 446, 452 to 721, 722 to 1166, 1161 to 1752, and 1867 to 2441). In contrast, our results indicate that in mammalian cells, CBP interacts with hNRC with high affinity but does not interact with the more highly expressed N-terminal part of hNRC (amino acids 1 to 852). The question of whether these different results reflect alterations in the interaction surfaces of full-length CBP and hNRCs in mammalian cells compared with fragments of CBP for regions of ASC-2 in yeast is important and requires further study. A model which summarizes our results (Fig. 15) depicts that the LXXLL-1 motifs of NRC dimers interact with the hydrophobic grooves of ligand-bound receptor homo- or heterodimers. This results in a conformational change in NRC which may expose an activation domain(s) which leads to transcriptional activation. The finding that NRC interacts with CBP in the apparent absence of ligand (Fig. 14A) suggests that a preformed complex of NRC and CBP binds receptors. The model does not distinguish whether this conformational change in NRC mediates a change in the structure in CBP and/or whether it recruits other factors to the receptor complex.
Northern blots of various human tissues suggest that multiple forms of hNRC are expressed. A comparison of the C-terminal regions of rNRC.1 and hNRC indicates that rNRC.1 represents an alternatively spliced variant that lacks the LXXLL-2 region and the C-terminal serine-threonine-leucine-rich domain. Variation in splicing of the NRC gene might result in NRC molecules that exhibit different activities or receptor specificities. Thus, a full-length version of rNRC.1 (or its human homologue) lacking the C-terminal inhibitory domain but containing AD2, the LXXLL-1 region, and the very strong AD1 activity might result in an extremely potent nuclear receptor coactivator. Furthermore, an alternatively spliced NRC which retains the activation domains and the C-terminal LXXLL-2 region, but which lacks the LXXLL-1 region, might be expected to function as an ER-specific coregulator.
It is interesting to also consider that the structure of the NRCs may allow for coupling cell surface signaling systems with nuclear hormone receptor function. Thus, the C-terminal serine-threonine-leucine-rich domain in hNRC, which appears to repress the activation function of AD1 and possibly AD2, contains 9 predicted protein kinase A phosphorylation sites in two clusters and 14 predicted Casein kinase II sites in four clusters. If such changes act to modulate or alter the inhibitory effect of the C terminus of hNRC on the activity of AD1 and/or AD2, this would lead to a more potent coregulator of ligand-dependent nuclear hormone receptor in vivo. In addition, although there are no apparent leucine zipper-like structures, the high leucine, isoleucine, and valine content of the C-terminal region may facilitate certain hydrophobically driven protein interactions which might modulate hNRC function. Our findings with rNRC.1 and hNRC provide strong evidence that this new family of coactivators-coregulators play an important role in nuclear receptor function. They also provide the basis for the future studies to understand the receptor-transduced changes in coactivators, the integrative role of the different hNRC regions on its activity, and the role of the different NRC isoforms in the function of nuclear receptors and other transcription factors in gene regulation.
ACKNOWLEDGMENTS
We thank Bert O'Malley for polyclonal antibody against SRC-1, Myles Brown for monoclonal antibody against SRC-1, Pat Nakatani for antibody against p/CAF, Michael Garabedian for Gal4-hERα, and David Moore for LexA-hTRβ-LBD, LexA-hGR-LBD, LexA-hRXRα-LBD, LexA-hRXRα-LBD, and LexA-hVDR-LBD. We thank Takahiro Nagase of the Kazusa Research Institute for sending us the KIAA0181 cDNA. We thank Richard Heyman of Ligand, Inc., for providing 9-cis-RA, TTNPB, and LG100153. We also thank Shahana Mahajan and Bruce Raaka for help with some of the studies.
This research was supported by NIH grant DK16636 (to H.H.S.). H.H.S. is a member of the New York University Medical Center Cancer Center (grant CA16087). Sequence analysis and database searches were through the New York University Medical Center Research Computing Resource which received support from the National Science Foundation (grant DIR-8908095).
REFERENCES
- 1.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 2.Au-Fliegner M, Helmer E, Casanova J, Raaka B M, Samuels H H. The conserved ninth C-terminal heptad in thyroid hormone and retinoic acid receptors mediates diverse responses by affecting heterodimer but not homodimer formation. Mol Cell Biol. 1993;13:5725–5737. doi: 10.1128/mcb.13.9.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanco J C G, Minucci S, Lu J, Yang X J, Walker K K, Chen H, Evans R M, Nakatani V, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carson-Jurica M A, Schrader W T, O'Malley B W. Steroid receptor family: structure and functions. Endocrine Rev. 1990;11:201–218. doi: 10.1210/edrv-11-2-201. [DOI] [PubMed] [Google Scholar]
- 5.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 6.Chakravarti D, Ogryzko V, Kao H Y, Nash A, Chen H, Nakatani Y, Evans R M. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- 7.Chang C Y, Norris J D, Gron H, Paige L A, Hamilton P T, Kenan D J, Fowlkes D, McDonnell D P. Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors α and β. Mol Cell Biol. 1999;19:8226–8239. doi: 10.1128/mcb.19.12.8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 9.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding X F, Anderson C M, Ma H, Hong H, Uht R M, Kushner P J, Stallcup M R. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 11.Feng W, Ribeiro R C, Wagner R L, Nguyen H, Apriletti J W, Fletterick R J, Baxter J D, Kushner P J, West B L. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 12.Fondell J D, Ge H, Roeder R G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forman B M, Samuels H H. Interactions among a subfamily of nuclear hormone receptors: the regulatory zipper model. Mol Endocrinol. 1990;4:1293–1301. doi: 10.1210/mend-4-9-1293. [DOI] [PubMed] [Google Scholar]
- 14.Forman B M, Samuels H H. pEXPRESS: a family of expression vectors containing a single transcription unit active in prokaryotes, eukaryotes and in vitro. Gene. 1991;105:9–15. doi: 10.1016/0378-1119(91)90507-8. [DOI] [PubMed] [Google Scholar]
- 15.Forman B M, Yang C-R, Stanley F, Casanova J, Samuels H H. c-erbA protooncogenes mediate thyroid hormone-dependent and -independent regulation of the rat growth hormone and prolactin genes. Mol Endocrinol. 1988;2:902–911. doi: 10.1210/mend-2-10-902. [DOI] [PubMed] [Google Scholar]
- 16.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 17.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 18.Hadzic E, Desai-Yajnik V, Helmer E, Guo S, Wu S, Koudinova N, Casanova J, Raaka B M, Samuels H H. A 10-amino-acid sequence in the N-terminal A/B domain of thyroid hormone receptor α is essential for transcriptional activation and interaction with the general transcription factor TFIIB. Mol Cell Biol. 1995;15:4507–4517. doi: 10.1128/mcb.15.8.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadzic E, Habeos I, Raaka B M, Samuels H H. A novel multifunctional motif in the N-terminal A/B domain of T3Rα modulates DNA-binding and receptor dimerization. J Biol Chem. 1998;273:10270–10278. doi: 10.1074/jbc.273.17.10270. [DOI] [PubMed] [Google Scholar]
- 20.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 22.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 23.Hong H, Kohli K, Garabedian M, Stallcup M R. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingraham H A, Chen R, Mangalam J, Elsholtz P, Flynn S E, Lin C, Simmons D, Swanson L, Rosenfeld M G. A tissue-specific transcription factor containing a homeodomain specifies pituitary phenotype. Cell. 1988;55:519–529. doi: 10.1016/0092-8674(88)90038-4. [DOI] [PubMed] [Google Scholar]
- 26.Ito M, Yuan C X, Malik S, Gu W, Fondell J D, Yamamura S, Fu Z Y, Zhang X, Qin J, Roeder R G. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 27.Kalkhoven E, Valentine J E, Heery D M, Parker M G. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 29.Kawasaki H, Eckner R, Yao T P, Taira K, Chiu R, Livingston D M, Yokoyama K K. Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature. 1998;393:284–289. doi: 10.1038/30538. [DOI] [PubMed] [Google Scholar]
- 30.Lanz R B, McKenna N J, Onate S A, Albrecht U, Wong J, Tsai S Y, Tsai M J, O'Malley B W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 31.Lee J W, Choi H-S, Gyuris J, Brent R, Moore D D. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 32.Lee S K, Anzick S L, Choi J E, Bubendorf L, Guan X Y, Jung Y K, Kallioniemi O P, Kononen L, Trent J M, Azorsa D, Jhun B H, Cheong J H, Lee Y C, Meltzer P S, Lee J W. A nuclear factor, ASC-2, is a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J Biol Chem. 1999;274:34283–34293. doi: 10.1074/jbc.274.48.34283. [DOI] [PubMed] [Google Scholar]
- 33.Li D, Desai-Yajnik V, Lo E, Schapira M, Abagyan R, Samuels H H. NRIF3 is a novel coactivator mediating functional specificity of nuclear hormone receptors. Mol Cell Biol. 1999;19:7191–7202. doi: 10.1128/mcb.19.10.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Gomes P J, Chen J D. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma H, Hong H, Huang S M, Irvine R A, Webb P, Kushner P J, Coetzee G A, Stallcup M R. Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol Cell Biol. 1999;19:6164–6173. doi: 10.1128/mcb.19.9.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenna N J, Lanz R B, O'Malley B W. Nuclear receptor coregulators: cellular and molecular biology. Endocrine Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 38.Nagpal S, Friant S, Nakshatri H, Chambon P. RARs and RXRs: evidence for two autonomous transactivation functions (AF-1 and AF-2) and heterodimerization in vivo. EMBO J. 1993;12:2349–2360. doi: 10.1002/j.1460-2075.1993.tb05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 40.Onate S A, Boonyaratanakornkit V, Spencer T E, Tsai S V, Tsai M J, Edwards D P, O'Malley B W. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J Biol Chem. 1998;15:12101–12108. doi: 10.1074/jbc.273.20.12101. [DOI] [PubMed] [Google Scholar]
- 41.Onate S A, Tsai S Y, Tsai M-J, O'Malley B W. Sequence and characterization of a coactivator of the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 42.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Regulation of NF-kappaB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 43.Powers C A, Mathur M, Raaka B M, Ron D, Samuels H H. TLS (Translocated-in-Liposarcoma) is a high-affinity interactor for steroid, thyroid hormone, and retinoid receptors. Mol Endocrinol. 1998;12:4–18. doi: 10.1210/mend.12.1.0043. [DOI] [PubMed] [Google Scholar]
- 44.Puigserver P, Wu Z, Park C W, Graves R, Wright M, Spiegelman B M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 45.Qi J-S, Desai-Yajnik V, Greene M E, Raaka B M, Samuels H H. The ligand binding domains of the thyroid hormone/retinoid receptor gene subfamily function in vivo to mediate heterodimerization, gene silencing, and transactivation. Mol Cell Biol. 1995;15:1817–1825. doi: 10.1128/mcb.15.3.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Naar A M, Erdjument-Bromage H, Tempst P, Freedman L P. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 47.Rachez C, Suldan Z, Ward J, Chang C P, Burakov D, Erdjument-Bromage H, Tempst P, Freedman L P. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadowski I, Ptashne M. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samuels H H, Stanley F, Shapiro L E. Control of growth hormone synthesis in cultured GH1 cells by 3,5,3′-triiodo-l-thyronine and glucocorticoid agonists and antagonists: studies on the independent and synergistic regulation of the growth hormone response. Biochemistry. 1979;18:715–721. doi: 10.1021/bi00571a025. [DOI] [PubMed] [Google Scholar]
- 50.Selmi-Ruby S, Casanova J, Malhotra S, Roussett B, Raaka B M, Samuels H H. Role of the conserved C-terminal region of thyroid hormone receptor-α in ligand-dependent transcriptional activation. Mol Cell Endocrinol. 1998;138:105–114. doi: 10.1016/s0303-7207(98)00016-1. [DOI] [PubMed] [Google Scholar]
- 51.Seol W, Choi H-S, Moore D D. Isolation of proteins that interact specifically with the retinoid X receptor: two novel orphan receptors. Mol Endocrinol. 1995;9:72–85. doi: 10.1210/mend.9.1.7760852. [DOI] [PubMed] [Google Scholar]
- 52.Takeshita A, Cardona G R, Koibuchi N, Suen C S, Chin W W. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka M, Gupta R, Mayer B J. Differential inhibition of signaling pathways by dominant-negative SH2/SH3 adapter proteins. Mol Cell Biol. 1995;15:6829–6837. doi: 10.1128/mcb.15.12.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 55.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 56.Tremblay A, Tremblay G B, Labrie F, Giguere V. Ligand-independent recruitment of SRC-1 to estrogen receptor-β through phosphorylation of activation function AF-1. Mol Cell. 1999;4:513–519. doi: 10.1016/s1097-2765(00)80479-7. [DOI] [PubMed] [Google Scholar]
- 57.Voegel J J, Heine M J, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voegel J J, Heine M J S, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 59.Wurtz J M, Bourguet W, Renaud J P, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 60.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 61.Ye Z-S, Forman B M, Aranda A, Pascual A, Park H Y, Casanova J, Samuels H H. Rat growth hormone gene expression: both cell-specific and thyroid hormone response elements are required for thyroid hormone regulation. J Biol Chem. 1988;263:7821–7829. [PubMed] [Google Scholar]
- 62.Yeh S, Chang C. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath C M, Darnell J E., Jr Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]