Abstract
Purpose:
The survival of women with brain metastases (BM) from breast cancer remains very poor, with over 80% dying within a year of their diagnosis. Here, we define the function of IL13Rα2 in outgrowth of breast cancer brain metastases (BCBM) in vitro and in vivo, and postulate IL13Rα2 as a suitable therapeutic target for BM.
Experimental Design:
We performed IHC staining of IL13Rα2 in BCBM to define its prognostic value. Using inducible shRNAs in TNBC and HER2+ breast–brain metastatic models, we assessed IL13Rα2 function in vitro and in vivo. We performed RNAseq and functional studies to define the molecular mechanisms underlying IL13Rα2 function in BCBM.
Results:
High IL13Rα2 expression in BCBM predicted worse survival after BM diagnoses. IL13Rα2 was essential for cancer-cell survival, promoting proliferation while repressing invasion. IL13Rα2 KD resulted in FAK downregulation, repression of cell cycle and proliferation mediators, and upregulation of Ephrin B1 signaling. Ephrin-B1 (i) promoted invasion of BC cells in vitro, (ii) marked micrometastasis and invasive fronts in BCBM, and (iii) predicted shorter disease-free survival and BM-free survival (BMFS) in breast primary tumors known to metastasize to the brain. In experimental metastases models, which bypass early tumor invasion, downregulation of IL13Rα2 before or after tumor seeding and brain intravasation decreased BMs, suggesting that IL13Rα2 and the promotion of a proliferative phenotype is critical to BM progression.
Conclusions:
Non-genomic phenotypic adaptations at metastatic sites are critical to BM progression and patients' prognosis. This study opens the road to use IL13Rα2 targeting as a therapeutic strategy for BM.
Translational Relevance.
The adaptation of disseminated cancer cells to the brain microenvironment involves the transition between invasive phenotype (required for dissemination) to proliferative phenotype (required for outgrowth). Yet, the mediators of this change at metastatic sites remain poorly understood. Here, we provide evidence that upregulation of IL13Rα2 cells in brain metastases (BM) promotes proliferation and dampens invasion, and knocking down IL13Rα2 in early and late BM decreases brain metastatic outgrowth. Phase I clinical trials for IL13Rα2-targeted chimeric antigen receptors (NCT02208362) have shown initial promise against recurrent multifocal leptomeningeal glioblastoma. Thus, this study provides proof of principle that emerging therapies targeting IL13Rα2 have the potential to benefit patients with breast cancer BM. Finally, levels of IL13Rα2 protein expression in the brain—but not mRNA expression levels at primary tumors—predicted worse survival following the diagnosis of BMs. Thus, interrogating non-genomic phenotypic adaptations at metastatic sites has the potential to provide novel therapeutic targets for BMs.
Introduction
Breast cancer brain metastases (BCBM) develop in 15% to 50% of patients with metastatic breast cancer depending on breast cancer subtype. The current treatment options for BM (surgery, radiation, chemo or targeted therapies) have limited success and may worsen neurological function (1). The survival of women with BM from breast cancer remains very poor, and more than 80% will die within a year of their diagnosis (2, 3). Defining the mechanisms underlying metastatic colonization and rapid progression of BM remains an unmet and critical need to identify new therapeutic strategies for these patients.
Despite the substantial progress in defining general mechanisms by which cancer cells in solid tumors acquire the hallmark capability to metastasize (4), the extent to which phenotypic plasticity at metastatic sites contributes to metastatic progression remains poorly understood. Brain metastatic colonization is a relatively rare event in which circulating tumor cells are arrested at brain capillaries, extravasate into the brain parenchyma, survive antitumorigenic effects of brain immunesurveillance cells (microglia and astrocytes), and colonize the brain by growing around existing vessels and adapting to the unique brain microenvironment forming the tumor niche (5–10). This metastatic colonization and adaptation of metastatic cells requires a critically important step, a change between highly invasive phenotypes (for extravasation) to proliferative phenotype (for outgrowth; ref. 11). Many aspects of the phenotypic change leading to tumor progression in the brain metastatic niche remain unknown.
IL13Rα2, a high-affinity receptor for IL-13, is upregulated in several tumors, and studies showed that IL13Rα2 overexpression facilitates the progression and metastasis of various cancer types, including glioblastoma (12–14). In breast cancer, IL13RA2 mRNA was first identified as upregulated during adaptation to lung and brain metastatic cell lines, and knockdown of IL13Rα2 decreased the metastatic colonization of breast cancer cells into lungs (5, 15). However, the function of IL13Rα2 during brain metastatic colonization and its value as therapeutic target in late brain metastasis remained unexplored. Initially, IL13Rα2 was thought to function mainly as a decoy receptor due to its lack of an intracellular kinase domain (16). However, recent studies show more complex functions for IL13Rα2 in brain tumor progression (17). Thus, here we investigated the clinical value and function of IL13Rα2 protein expression in BM and demonstrate that IL13Rα2 promotes proliferation of disseminated cancer cells and plays a critical role in brain metastatic progression.
Materials and Methods
Human-derived BM
De-identified human BM samples (N = 96) were obtained from archival paraffin-embedded tissue from consenting breast cancer patient-donors, under approved IRB protocols at the University of Colorado.
Cell lines
Triple-negative brain trophic cells 231BR (cells that colonize the brain at high frequency, derived from MDA-MB-231 cells, a kind gift from Dr. Patricia Steeg, Center for Cancer Research, National Cancer Institute) and HER2+ brain trophic JmT1BR3 (derived from JIMT1 cell line) were cultured in DMEM high glucose supplemented with 10% of FBS and penicillin–streptomycin (P/S). ER+HER2+ BT474 cells (parental BT474m1 and brain-trophic derivative BT474m1BR, a kind gift of Dr. Dihua Yu, MD Anderson Cancer Center, Houston, Texas) cells were cultured in DMEM/F12 supplemented with 10% of FBS and P/S. Cell lines were free of Mycoplasma (MycoAlertTM PLUS-Lonza) and identity of human cells were validated within 6 months of receipt by STR analysis (University of Colorado Tissue Culture Core).
shRNAs and sgRNAs
shRNAs targeting human IL13RA2, EPHB1, and EFNB1 (Supplementary Table S1) and non-targeting controls in the pLKO1 vector (RRID:Addgene_27368) were purchased from the Sigma MISSION(R) TRC library through the Functional Genomics Facility at the University of Colorado Cancer Center. Lentiviral doxycycline-inducible systems to overexpress IL13RA2 with a hemagglutinin tag (HA) or to express IL13RA2-targeting shRNAs, were built using pTripz Sox4.965 vector (RRID:Addgene_101120). Briefly, the 21-base shRNAs targeting human IL13RA2 from two MISSION shRNA Library vectors (shIL13RA2–1 and shIL13RA2–2, Supplementary Table S1) were used to design sense and antisense 110-base oligonucleotides flanked by cohesive ends with XhoI and EcoRI restriction sites, according to the protocol described by Chang and colleagues (18). These sequences were cloned in pTripz Sox4.965 vector (Supplementary Table S2) and verified by sequencing. For IL13Rα2 overexpression, full sequence human IL13RA2 cDNA was amplified from JmT1BR3 cells, flanked at the 3′-end with the sequence encoding an HA-tag, and cloned in the pTripz Sox4.965 vector. Lentiviral particles were produced in HEK 293T cells (RRID: CVCL_1926). Breast cancer cells were transduced for 48 hours and selected using puromycin or cell sorting. When indicated, clonal populations were isolated using the serial dilution method (19).
In a subset of experiments, IL13RA2 was knocked out using a CRISPR-Cas9 approach (sgIL13RA2). For this, a guide-sequence (sg) targeting IL13RA2 Exon 3 (Supplementary Table S2) was cloned in the SpCas9 plasmid px459 v2.0 (RRID:Addgene_62988) and transfected in 231BR cells. Clonal populations expressing sgEV or sgIL13RA2 were selected by puromycin resistance and validated by qRT-PCR and WB.
IHC and immunofluorescence
For IHC, heat immobilized deparaffinized primary tumors or BM sections were incubated with 10 mmol/L Citrate buffer (pH 6.0) at 125°C and 25 psi for 10 minutes. After antigen retrieval, endogenous peroxidase was blocked with 3% H2O2 in PBS and tissue was blocked with 10% normal horse serum in TBST. Slides were incubated overnight with antibodies for IL13Rα2 or ephrin B1 (Supplementary Table S3), and AP-anti–Goat IgG and horseradish peroxidase (HRP)-anti–Mouse IgG (ImmPRESS) followed by AP or HRP substrates were used as appropriate. For immunofluorescence studies, fresh-frozen OCT-embedded brain tissues were sectioned at 10-μm thickness, fixed with methanol–acetone and blocked with 10% normal donkey serum in TBST, and incubated with antibodies against p-FAK, ephrin B, and fluorescently labeled secondary antibodies as indicated (Supplementary Table S3).
QRT-PCR
Total RNA from cultured cell lines was isolated using TRizol according to manufacturer instructions. cDNA was synthesized using the Verso cDNA Synthesis Kit (Thermo Fisher Scientific, Inc.). Primers used for amplification of IL13RA2, EFNB1, and EPHB1 are described in Supplementary Table S2. Human RPLO gene expressions or mouse β-actin were used as normalization controls. Relative gene expression was calculated using the 2–ΔΔCt method.
Western blot
Total protein was collected using RIPA buffer (cOmplete Protease Inhibitor Cocktail, PhosSTOP Phosphatase inhibitor, 100 mmol/L Na3VO4 and 1 mol/L NaF). Extracts were sonicated (5 pulses, 1 second each pulse, using 20% amplitude) and centrifuged at 7,000 rpm, at 4°C for 10 minutes. Supernatant was collected and total protein was quantified using the DC Protein Assay (Bio-Rad). Protein extracts were denatured in 1X Laemmli Buffer at 95°C for 5 minutes. Proteins were separated in 10% SDS-PAGE and transferred overnight at 25 V to 0.45-μm PVDF membranes. Antibodies used for western blot are listed in Supplementary Table S3. Immunoreactive bands were acquired using a Li-COR Odissey CLx and analyzed using Image Studio Software V5.2.
Proliferation assays
Cells were plated (1,000–3,000 cells/well) on 96-well plates and treated with vehicle or 1 μg/mL doxycycline as indicated. Treatments were performed in 6-replicates in at least two independent experiments. Proliferation was measured using IncuCyte live Imaging (Essen Bioscience) using the percentage of confluence in the phase-channel or total red object area (μm²/Image) in red-fluorescent channel as readouts. When indicated, data were normalized to time 0 and reported as fold-changes.
BrdUrd incorporation and cell cycle analysis
Cells were plated in 4 replicates (250K cells/plate) and incubated 36 hours in regular media, and experiments repeated at least twice. Cells were then incubated with 15 μmol/L BrdUrd for 1–2 hours (231BR and BT474, respectively), and BrdUrd incorporation measured using flow cytometry. Briefly, BrdUrd was removed, cells washed with PBS and fixed with 70% ethanol, DNA was denatured with HCl 2 N/0.5% Triton X-100 and incorporated BrdUrd was detected using rat anti-BrdUrd followed by anti–rat-DyLight 488 antibodies (Supplementary Table S3). The percentage of BrdUrd-positive cells and cell-cycle analysis was performed using Gallios Flow Cytometer with Kaluza Analysis Software (Beckman Coulter, Inc.). BrdUrd incorporation was also measured using IF-BrdUrd labeling on slides from mice treated with BrdUrd 2 hours before euthanasia. The percentage of BrdUrd+ cells per BM was tabulated from individual metastases in sections from 4 different mice per group.
Matrigel-filled scratch-wound invasion assays
Cells were plated (35,000 cells/well) in 5% Charcoal Stripped FBS in regular media, on 96-well Essen ImageLock plates. Adherent cells were serum-starved for 6 hours and a scratch wound was made using a 96-pin WoundMaker (Essen Bioscience), and wounds were filled with Matrigel Growth Factor Reduced (GFR) Basement Membrane Matrix (Corning). Wound images were taken every 4 hours for 24 hours and the relative wound confluency was calculated at each time point using an IncuCyte S3 System (Essen BioScience). When indicated, cells were pretreated with doxycycline (1 μg/mL) for 48 hours before scratch wound and during assay. Treatments were performed in 6-replicates, and each experiment repeated at least twice.
Transwell invasion assay
Serum starved 231BR cells (150,000 cells) were seeded in the upper chamber of a Boyden Transwell chamber (Neuro Probe) in 500 μL of serum-free media, separated from the chemoattractant (10% FBS supplemented media) by 8-μm pore size PCTE-PVP-free membrane (Neuro Probe) coated with growth-factor reduced (GFR) Matrigel (2.5 mg/mL). After 24 hours, cells invading through the lower chamber were collected and quantified using a hemocytometer. Each treatment was done by triplicate in at least two independent experiments. When indicated, cells were pretreated 48 hours with doxycycline (1 μg/mL).
Invasion in organotypic brain slices
The invasive ability of cells in the brain microenvironment was assessed using organotypic brain slices as we have previously described (20). Other studies have shown that cancer cells invade in all directions into the organotypic brain slices (21). Briefly, 300-μm coronal brain slices from adult mice were obtained using a VT1000 S vibrating blade microtome (LEICA). Brain slices were plated on 8-μm pore 12-well/Transwells with 700 μL of media (MEM/HEPES 50%, horse serum 20%, NaHCO3 2 mmol/L, glucose 6.5 mg/mL, glutamine 2 mmol/L and P/S). 231BR and BT474M1 GFP+ cancer cells were cultured as mammospheres for 10 days in ultra-low attachment plates, resuspended in serum-free media and plated on top of brain slices (5–20 mammospheres/slice). GFP+ spheres were imaged using a fluorescent MVX10 stereo microscope (OLYMPUS) at 0 and 24 hours after plating. Images were exported to ImageJ software (RRID:SCR_003070) and invasion of GFP+ cells (green mask) away from the edge of the mammosphere were quantified using Scholl analysis (20, 22).
Experimental BM
Animal studies were approved by University of Colorado Institutional Animal Care and Use Committee. Inducible EV or shIL13RA1 231BR-GFP-luciferase cells (175,000 cells/mouse) were injected in the left ventricle of 8–12-week-old female NSG mice, and mice administered 1 mg/mL doxycycline in 10% sucrose/drinking water: (i) from two days before cell injection, or (ii) 7 days after intracardiac injection. Investigators blinded to the experimental groups performed cell injections and metastasis quantification. Inducible EV or OE-IL13RA2-BT474M1-GFP-luciferase cells (175,000 cells mouse) were injected in 4–16 week NSG mice supplemented with E2-release pellets (1 mg), and mice administered doxycycline from two days before cell injection. Sample size was calculated at 80% power, two-sided tests and α = 0.05. Metastatic burden was quantified at day 0, 7, 14, and 19 following intracardiac injection, using IVIS Spectrum in vivo imaging system. To assess the proliferative state of BMs, mice were injected intraperitoneally with 10 mg/mL BrdUrd 2 hours before euthanasia. Histological quantification of metastases was performed as previously described (23, 24). Briefly, six hematoxylin and eosin (H&E)–stained serial sections (10-μm thick), one every 300 μmol/L in a sagittal plane through the right hemisphere of the brain were analyzed at ×4 magnification using an ocular grid. Every micrometastases (≤300 μmol/L) and metastatic cluster (defined as 4 or more micrometastases together with a >300 μm along the longest axis) in each section was tabulated.
Global RNA sequencing and gene expression analysis
Total RNA was collected using TRizol and purified with the RNeasy MinElute Cleanup Kit (Qiagen). Messenger RNA (mRNA) was enriched by oligo dT Selection and cDNA was created by reverse transcription using random primers. cDNA was sequenced using paired-end libraries in the BGISEQ platform to get around 70 million of cleaned 100-base reads per sample. Reads were mapped against the human genome assembly (NCBI assembly accession GCF_000001405.39 and assembly name GRCh38.p13) using hisat2 v2.1.0 (25), and genes differentially expressed between groups (EV vs. shIL13RA2) were determined from the triplicates with Cuffdiff (26) and confirmed with DESeq2 (27). Differentially expressed genes were defined as those genes with logarithmic fold change (logFC) with absolute value greater than 1.5 and P value less than 0.05. Normalized expression levels to each sample were calculated using Cuffnorm v2.2.1 (28) and reported as fragments per kilobase of transcript per million mapped fragments (FPKM). GEO# GSE165898. Ingenuity Pathway Analysis (IPA) and GSEA (29) were used to determine canonical pathways significantly altered by downregulation of IL13RA2.
Digital imaging
For IF analysis images were collected using a Nikon Eclipse Ti-S inverted microscope or an Olympus MVX fluorescent scope. Minor linear adjustments to brightness and contrast were performed identically and in parallel. IHC slides were scanned using Aperio Scanscope T3 and analyzed using Aperio analysis tool.
Statistical analysis
Statistics were done using GraphPad Prism 9.0.0 software. Two-tailed t test, one way ANOVA, repeated measures mixed analysis or repeated measures ANOVA followed by multiple comparisons post hoc tests were used as appropriate. When samples did not comply any normality assumption, non-parametric test (Two-tailed Mann–Whitney or Kruskal–Wallis test) were used. For animal studies, data were normalized to time 0 (fold change) and log-transformed to fit normality assumptions. P < 0.05 was considered significant and test assumptions were checked for all analysis. Statistical analysis is reported with each figure legend. Adjusted P values are shown in all graphs.
Results
High IL13Rα2 expression in BCBMs is a predictor of decreased survival
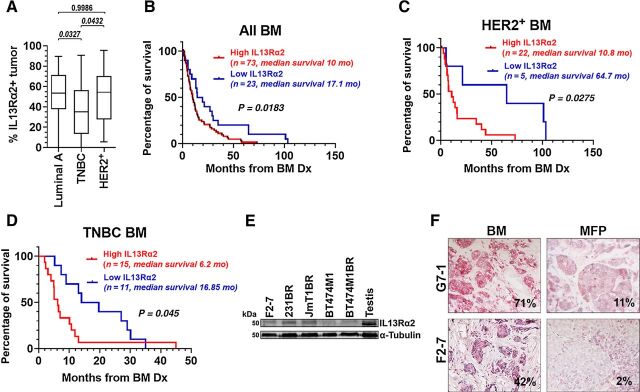
To assess the extent of IL13Rα2 protein expression in late BCBM, we performed IL13Rα2 IHC in a cohort of 96 BCBM and quantified its tumoral expression using Aperio digital pathology. IL13Rα2-positive staining was found in BCBM from Luminal A (ER+/PR±/HER2−, n = 32), HER2+ (n = 27) or TNBC (triple-negative breast cancer, n = 26), subtypes (Fig. 1A). Luminal A and HER2+ BM showed higher IL13Rα2 expression than TNBC BMs (P = 0.033 and P = 0.043, respectively). High IL13Rα2 expression (>25% strong positive tumor) predicted for a significantly shorter survival following BM diagnosis in all BCBM (n = 96, including 11 cases of unknown BC subtype) compared with those with low IL13Rα2 expression (<25% strong positive tumor; median survival 10 vs. 17.1 months, respectively, P = 0.0183, Fig. 1B). High IL13Rα2 expression also predicted poor survival within HER2+ BM (P = 0.027, Fig. 1C) and TNBC BM (P = 0.045, Fig. 1D) but not in luminal BM (where only 2/32 cases with known time-to-death had low IL13Rα2 expression, not shown). Consistent with previously reported expression of IL13Rα2 in brain trophic breast cancer cells derived from TN MDA231, IL13Rα2 expression was found on human cancer cells derived from BCBM (F2–7), and cells that had been selected for their ability to form BM at high frequencies (231BR, JmT1BR, BT474M1BR, Fig. 1E). Interestingly, expression of IL13Rα2 in two BCBM (F2–7, G7–1) was lower when tumors grew as patient-derived xenografts (PDX) in the mammary fat pad of NSG mice (Fig. 1F), suggesting plasticity in the expression of IL13Rα2 in the brain. Together, these data suggest that IL13Rα2 increased expression at late stages of BM progression.
Figure 1.
IL13Rα2 expression is high in BM, and increased levels predict worse survival after brain metastasis diagnoses. A, IL13Rα2 IHC staining was scored using Aperio Digital Imaging, and tumoral areas with strong intensity scores were considered positive. Graph shows percentage of positive tumor area for a cohort of BM samples from Luminal A (n = 32), HER2+ (n = 27), and TNBC (n = 26) breast cancer subtypes. Data were analyzed using ANOVA. Adjusted P value is shown. B, All BMs (n = 96) were classified as high (≥25% + tumor, blue) versus low IL13Rα2 (<25% + tumor, red), and percentage of survival following BM diagnoses was plotted. C, HER2+ BMs (n = 27) were classified as high or low IL13Rα2 as in B. D, TNBC BMs (N = 26) were classified as high or low IL13Rα2 expression as in B. Kaplan–Meier curves (B–D) were analyzed using the log-rank test. E, WB shows IL13Rα2 expression in breast cancer cells. F, The percentage of tumoral area expressing IL13Rα2 in clinical BM (F2–7, G7–1) as compared with the same tumors growing as PDXs in the mammary fat pad (MFP) of NSG mice.
IL13Rα2 promotes proliferation of brain-trophic breast cancer cells
Prior studies have shown opposite roles for IL13RA2 in breast cancer tumor progression. IL13RA2 knockdown decreased the ability of lung-trophic MDA231 cells to colonize the lungs without altering primary tumor formation (15, 30). By contrast, independent studies showed that overexpression of IL13RA2-inhibited tumorigenicity of human breast cancer MDA231 cells (31). To assess the role of IL13RA2 in the context of brain metastatic colonization and progression, we used CRISPR/cas9 to knockout IL13RA2 expression in 231BR cells (Supplementary Fig. S1A). However, stable knockout of IL13RA2 in multiple cell lines (4T1BR5, 231BR, JMT1Br3) resulted in cell death over passaging or selected clones would emerge with regained expression, suggesting critical roles for IL13RA2 on cell survival. We reasoned that this survival disadvantage of IL13RA2 KD cells and emergence of resistant clones could account for the previously reported discrepancies in IL13Rα2 function in breast cancer.
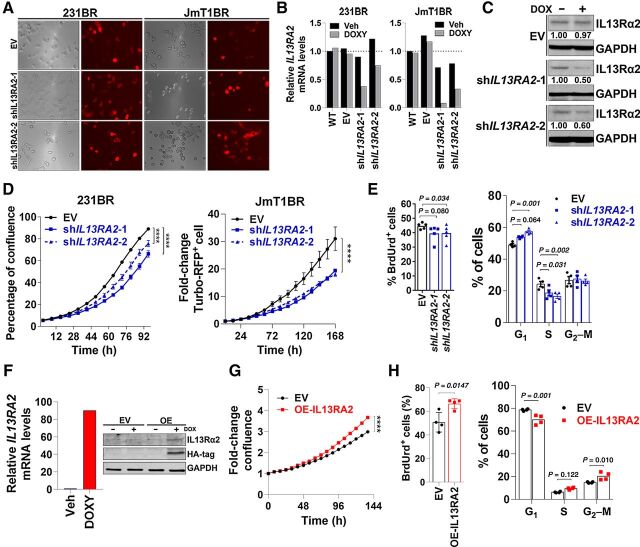
To address this question, shRNAs targeting IL13RA2 (shIL13RA2#1 and #2) were cloned downstream of a doxycycline-inducible promoter and linked via an IRES to Turbo-RFP reporter, which allows for transient and time-specific modulation of IL13Rα2 levels. Cells with high IL13Rα2 endogenous expression (231BR, JmT1BR3, Fig. 1E) were transduced with shEV or shIL13RA2, then induced with doxycycline and selected using cell sorting. Doxycycline at the concentration used in these experiments did not affect cell proliferation in parental cells (Supplementary Fig. S1B). 231BR and JmT1BR3 transduced with inducible EV, shIL13RA2–1 or shIL13RA2 showed strong Turbo-RFP expression as early as 48 hours following induction with doxycycline (Fig. 2A). IL13RA2 mRNA and protein levels decreased by 50% in 231BR cells and 80% in JmTBR3 cells expressing shIL13RA2 within 72 hours following doxycycline induction (Fig. 2B and C). To assess whether IL13Rα2 influenced survival and proliferation of brain trophic cell lines, 231BR or JmT1BR3 cells expressing EV, shIL13RA2–1 or shIL13RA2–2 were treated with doxycycline and cell confluence was measured over time using live cell imaging (Fig. 2D). In clonal populations of 231BR cells, downregulation of IL13RA2 decreased cell confluence from 89.1% ± 2.1% in EV to 66.2% ± 6.4% in shIL13RA2#1 and 75.7% ± 6.3% in shIL13RA2#2 cells (P < 0.0001 at 96h). Similarly, in non-clonal JmT1BR3 cells downregulation of IL13RA2 significantly reduced cell confluence of Turbo-RFP–expressing cells (Fig. 2D). Downregulation of IL13RA2 decreased the percentage of BrdUrd+ cells as compared with EV (39.6 ± 5.6 in shIL13RA2–1 vs. 44.7 ± 2.1 BrdUrd+ cells in EV, respectively, Fig. 2E). shIL13RA2 231BR cells also showed a significant decrease in the percentage of cells in S-phase and an increase in cells arrested in G1-phase of the cell cycle (Fig. 2E). Together, these data suggest that the survival disadvantage conferred by IL13RA2 downregulation results in part from G1-arrest and decreased proliferation.
Figure 2.
IL13Rα2 promotes proliferation of breast cancer cells. A, 231BR and JmT1BR3 cells were transfected with a lentiviral vector expressing the empty vector (EV) or shRNAs targeting IL13RA2 (shIL13RA2-1 or -2) upstream of Turbo-RFP reporter under doxycycline control. Image shows brightfield and RFP expression after 72 hours treatment with 1 μg/mL doxycycline. B, 231BR and JmT1BR3 cells expressing EV or shIL13RA2 were treated for 48 hours with vehicle or 1 μg/mL doxycycline. Graph shows IL13RA2 mRNA levels normalized to GAPDH and relative to WT (veh). C, Cells were cultured as in (B) for 72 hours, and IL13Rα2 protein expression was assessed by Western blot. GAPDH was used as loading control. Numbers indicate IL13Rα2 fold change relative to vehicle-treated cells. D, 231BR and JmT1BR3 cells expressing EV or shIL13RA2 were treated with doxycycline (1 μg/mL), and percentage of confluence (for clonal 231BR cells) or Turbo-RFP expression (for pooled JmT1BR3 cells) was measured over time using Incucyte live imaging (n = 5/6 treatment). Data analyzed with repeated measures ANOVA followed by multiple comparison post hoc corrections. ****, P < 0.001 at the last time point. E, 231BR cells expressing EV or shIL13RA2 were induced with doxycycline for 72 hours then treated with BrdUrd by 1 hour. BrdUrd incorporation (%, left) and cell-cycle analysis by PI (right) were measured by flow cytometry (n = 4). Adjusted P values are shown. F, Human IL13Rα2 with a hemagglutinin (HA) tag was overexpressed (OE) in a doxycycline inducible system in BT474M1 cells. IL13RA2 mRNA levels normalized to GAPDH and relative to the vehicle-treated cells (left). WB shows anti-HA and anti-IL13Rα2 (right). Induction with 0.5 μg/mL of doxycycline was allowed by 48 and 96 hours for qRT-PCR and WB, respectively. G, BT474M1 EV and OE-IL13RA2 cells were treated with 0.5 μg/mL of doxycycline and percentage of confluence measured over time as in D. Fold change in confluence relative to day 0 ± S.E.M. H, DNA replication and cell cycle were determined by BrdUrd (2 hours pulse) and propidium iodide (PI) incorporation in BT474M1 cells as in (E). ****, P < 0.001.
To determine whether IL13RA2 upregulation was sufficient to promote proliferation, an inducible IL13RA2 overexpression vector was transduced in HER2+ breast cancer cells BT474m1 (non-brain trophic parental cells that lack endogenous IL13Rα2, Fig. 1C). Inducible-overexpression (OE) of hemagglutinin-tagged IL13RA2 (Fig. 2F) significantly increased cell confluence (a 22.6% increase compared with EV by 144 hours P < 0.0001; Fig. 2G). Furthermore, IL13RA2–OE BT474 cells had an increased percentage of BrdUrd+ cells following a 2-hour BrdUrd, a significant decrease in G1 arrested and an increase in G2–M cells (Fig. 2H). Taken together, these studies suggest that high levels of IL13Rα2 promote proliferation of breast cancer cells.
IL13RA2 represses invasion of brain-trophic breast cancer cells
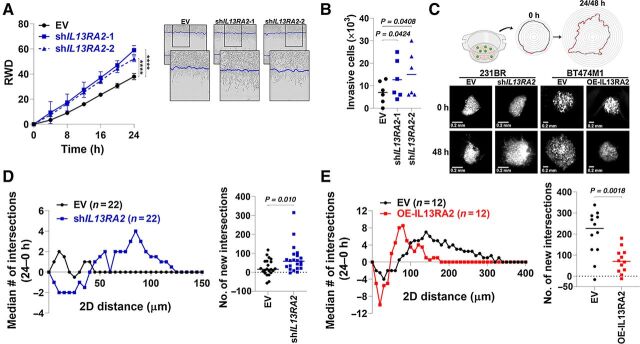
Development of BM depends on the ability of disseminated cancer cells to extravasate and colonize the brain parenchyma. To determine whether IL13Rα2 would modulate invasion of brain trophic cells, the invasive ability of EV or shIL13RA2 was assessed using a Matrigel-filled scratch-wound assay. Surprisingly, shIL13RA2 cells were more invasive as compared with shEV-231BR cells [37.9 ± 2.5 relative wound confluence (RWC) in EV vs. 59.0 ± 3.6 RWC in shIL13RA2 by 24 hours, P < 0.0001; Fig. 3A]. Likewise, downregulation of IL13Rα2 increased the number of cells capable to invade though an 8-μm pore in Matrigel-coated Boyden Chamber assays (Fig. 3B). To assess whether IL13Rα2-modulation of invasion also occurred in the brain extracellular matrix, 231BR cells expressing EV or shIL13RA2 were grown as spheres for 7 days, shRNAs induced with doxycycline for 48 hours and then spheres were plated on top of organotypic brain slices as we previously described (20). Invasion of cancer cells away from the sphere and into the brain slice were measured 24 hours later and quantified as a function of the formation of new intersections away from the initial sphere edge (Fig. 3C). Downregulation of IL13Rα2 resulted in spheres with an increased number of invasive fronts farther away from the edge (Fig. 3D, left) and significantly more intersections after 24 hours (median # of intersections 24.4 in EV vs. 69.4 in shIL13RA2, P = 0.01, Fig. 3D, right). Conversely, upregulation of IL13Rα2 in BT474m1 cells decreased their invasive ability in organotypic brain slices (Fig. 3E), suggesting a role for IL13Rα2 in repressing the invasive ability of cancer cells in vitro. Together, these data suggest a novel and more complex role of IL13Rα2 in modulating phenotypic plasticity of breast cancer cells, with dual roles promoting proliferation while repressing invasion of breast cancer cells.
Figure 3.
High levels of IL13Rα2 repress invasion of BC cells. A, 231BR cells expressing EV and shIL13RA2 were plated in a confluent monolayer and serum-starved overnight, and a modified Matrigel-filled scratch-wound was used to assess invasion. Left, Graph shows relative wound density (RWD) over time (left). Data analyzed with repeated measures ANOVA followed by multiple comparison post hoc corrections. ****, P < 0.001 at the last time point. Right, Representative images show invasive front. Blue line marks initial wound-edge. B, 231BR serum-starved cells were assessed for their ability to invade through a Matrigel-coated filter (8-μm pore size) in Boyden chambers. Graph shows number of cells in the lower reservoir after 24 hours. C, Invasion assay in organotypic brain slices. 231BR and BT474M1 eGFP+ spheres expressing EV, shIL13RA2 (for 231BR cells) or OE-IL13RA2 (for BT474M1 cells) were seeded on top of organotypic brain slices. Edges of spheres were monitored over time by fluorescence microcopy and new intersections to concentric circles determined. Representative images for the same sphere/treatment at 0 and 48 hours are shown. D, 231BR EV or shIL13RA2 cells were assayed as in C and new intersections quantified after 24 hours. Left, Median number of new intersections/sphere in 5-μm increments from initial sphere edge 24 hours. Right, Total number of new intersections per sphere after 24 hours. E, Invasion of BT474M1 EV or OE-IL13RA2 cells was analyzed as in D.
Downregulation of IL13Rα2 decreases FAK signaling and promotes Ephrin B1 pathway upregulation in cancer cells
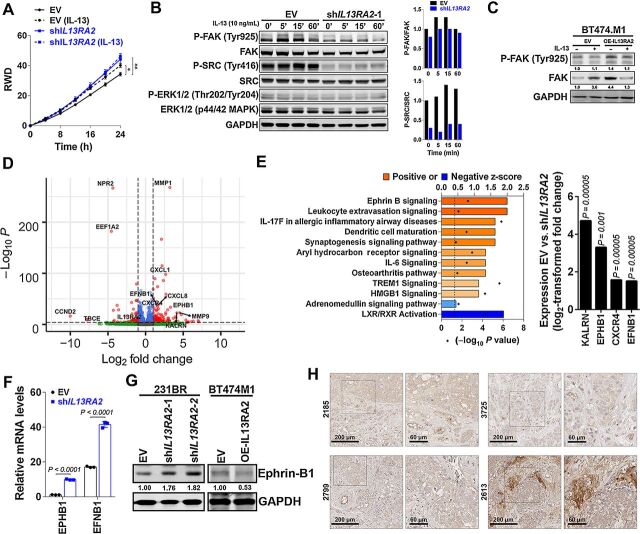
Prior studies in colon cancer have shown that stimulation of IL13Rα2 with its cognate ligand IL-13 signals through recruitment and activation of Focal Adhesion Kinase (FAK), SRC and Pi3K/AKT signaling (32). To assess whether similar signaling pathways mediated IL13Rα2 function in breast cancer cells, 231BR and BT474M1 cells expressing shIL13RA2 or OE-IL13RA2 were induced with doxycycline and then treated with IL-13. Stimulation of 231BR EV cells with human IL-13 promoted invasion of cancer cells (Fig. 4A) and led to activation of FAK and SRC (but not AKT or ERK activation; Fig. 4B). However, downregulation of IL13Rα2 alone promoted invasion to levels similar to those induced by IL-13 (Fig. 4A), and resulted in a decrease in FAK and SRC expression and activation even in the absence of ligand (Fig. 4B). Conversely, overexpression of IL13Rα2 in BT474.M1 cells resulted in increased FAK recruitment and activation, even in the absence of IL-13 (Fig. 4C). Given that these cells do not express known IL13Rα2 ligands (IL-13 or putative ligand Chi3L1, as measured by qRT-PCR and RNAseq, not shown), these data suggest that upregulation of IL13Rα2 promotes ligand-independent recruitment and activation of FAK/SRC in breast cancer cells.
Figure 4.
Downregulation of IL13RA2 decreases FAK/SRC but upregulates Ephrin B signaling. A, 231BR cells expressing EV or shIL13RA2 were induced with doxycycline for 48 hours, and treated with vehicle or 10 ng/mL human recombinant IL-13 (hrIL-13) in an invasion assay. Graph shows relative wound density (RWD) over time (n = 6–10 wells per group), in a modified Matrigel-filled scratch-wound assay. Data analyzed with repeated measures ANOVA followed by multiple comparison post hoc corrections. Adjusted P value at last time point *, P < 0.05; **, P < 0.01. B, WB show signaling pathways in 231BR EV or shIL13RA2 cells induced with doxycycline for 72 hours, serum starved overnight, and then treated with 10 ng/mL hrIL-13 for the indicated times. Plot shows quantification of pFAK/FAK and P-SRC/SRC from two independent experiments. C, WB shows P-FAK and FAK in BT474.M1 cells expressing EV or OE-IL13RA2, induced with doxycycline for 72 hours and treated with vehicle or 10 ng/mL hrIL-13 for 5 minutes. GAPDH is loading control. D, Global RNA sequencing was performed in doxycycline-induced 231BR EV versus 231BR shIL13RA2 cells 48 hours after doxycycline induction (n = 3/group). Volcano plot shows differentially expressed genes in EV versus shIL13RA2 cells. Red dots are genes with log2 FC>1.5 and P < 0.05. E, Ingenuity Pathway Analysis (IPA). The P value (calculated with Fischer's exact test) and Z-score were determined to establish any probable association between our set of genes and a specific pathway. Left, Ephrin B signaling is the only pathway modulated in shIL13RA2 cells with a significant P value and ≥2 positive Z-score. Right, The log2-transformed fold change in expression of upregulated genes in the Ephrin B1 signaling pathway (KALRN, EPHB1, CXCR4, and EFNB1). F, Relative mRNA expression of EphB1 receptor (EPHB1) and ephrin B1-ligand (EFNB1) in EV versus shIL13RA2 231BR cells (n = 3). G, WB shows ephrin B1 expression in 231BR expressing EV or shIL13RA2, and BT474 cells expressing EV or OE-IL13RA2. GAPDH was used as loading control. Numbers show ephrin B1 levels normalized to GAPDH and relative to EV control. H, IHC staining of ephrin B1 expression in BCBMs. Representative images of the tumor invasive front in late BM are shown in zoom-in.
To further investigate mechanisms downstream of IL13Rα2 that promote proliferation while decreasing invasion, 231BR EV versus 231BR shIL13RA2 cells were induced with 1 μg/mL doxycycline for 96 hours, and global RNA sequencing was performed (n = 3/group). Consistent with the opposite roles of IL13Rα2 in promoting proliferation while repressing invasion, top genes differentially downregulated in shIL13RA2 included pro-proliferative EEF1A2 (Log2FC −4.52, P = 0.00005) and cell cycle and proliferation mediator CCND2 (Cyclin D2, Log2FC −7.61, P = 0.046). Furthermore, top upregulated genes included well-known invasion mediators MMP1 (Log2FC 3.2, P = 0.00005) and MMP9 (Log2FC 4.5, P = 0.0035; Fig. 4D; Supplementary Table S4A). IPA showed that functions associated with cell migration were upregulated, whereas functions associated with proliferation were repressed in shIL13RA2 cells compared with shEV control (Supplementary Fig. S2A). Among pathways differentially regulated among EV and shIL13RA2 cells, ephrin B signaling was the top significantly upregulated pathway in shIL13RA2 cells with a >2 positive Z-score (P = 0.0026; Fig. 4E). Ephrin B signaling is a key axonal guidance molecule during brain development and can play proinvasive and/or pro-proliferative roles in cancer depending on context. Thus, we sought to further determine the extent to which ephrin B1 upregulation contributes to the promotion of invasion and repression of proliferation in BCBM. Genes differentially expressed within the Ephrin B signaling pathway included Ephrin Type B-receptor (EPHB1 and EphB1) and its membrane-bound ligand ephrin B1 (EFNB1; Fig. 4E), qRT-PCR confirmed both EPHB1 and EFNB1 were significantly upregulated in shIL13RA2 cells (Fig. 4F), with EFNB1 being more abundantly expressed than EPHB1. Furthermore, western blot analysis confirmed that downregulation of IL13Rα2 promotes ephrin B1 expression in 231BR cells whereas overexpression of IL13Rα2 downregulates ephrin B1 in BT474 cells (Fig. 4G). Although expression of IL13Rα2 and ephrin B1 proteins are not mutually exclusive in cancer cells or tumors (Fig. 4G), IHC staining of a subset of clinical BM with high levels of IL13Rα2, shows various degrees of ephrin B1 expression, with smaller cell clusters and invasive fronts showing highest ephrin B1 expression (Fig. 4H). Ephrin B1 was also expressed in endothelial cells in the brain niche. Thus, these data suggest that high ephrin B1 marks a subpopulation of highly invasive tumor cells, whereas high IL13Rα2+ preferentially marks highly proliferative cancer cells in BMs.
Ephrin B1 promotes invasion of BCBM cells
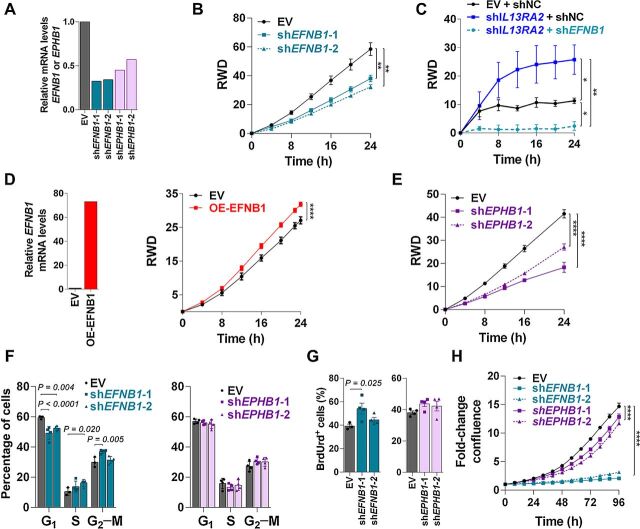
To assess whether ephrin B1 ligand (EFNB1) or EphB1 receptor (EPHB1) contributes to increased invasion in vitro, two shRNAs were used to downregulate ephrin B1 (shEFNB1–1, 2) or EphB1 receptor (shEPHB1–2; Fig. 5A) in 231BR cells. Partial downregulation of EFNB1 resulted in a significant decrease in the invasive ability of 231BR cells as measured in a Matrigel-filled scratch-wound assay (Fig. 5B). To assess whether EFNB1 upregulation was linked to the promotion of invasiveness in the context of IL13Rα2 loss, shIL13RA2 were transduced with shEFNB1 to prevent EFNB1 upregulation. Blockage of EFNB1 in shIL13RA2 cells strongly decreased the invasive ability of cancer cell in vitro (Fig. 5C). Conversely, forced expression of EFNB1 in EV cells (which mimics the EFNB1 upregulation in shIL13RA2 cells) significantly increased invasion as compared with shEV cells (Fig. 5D). Despite its lower relative expression, downregulation of EPHB1 also diminished invasion as compared with EV-expressing cells (Fig. 5E), suggesting that both ephrin B1 ligand and its receptor EphbB1 modulate the invasive ability of cancer cells.
Figure 5.
Downregulation of EFNB1 impairs invasion and promotes DNA synthesis. A, 231BR cells expressing EV or shRNAs targeting EFNB1 (shEFNB1–1/2) or EPHB1 (shEPHB1–1/2) were cultured for 48 hours. EFNB1 and EPHB1 mRNA levels were detected by RT-qPCR, normalized to GAPDH, and reported as relative to EV. B, 231BR cells expressing EV and shEFNB1–1/2 were plated in a confluent monolayer and serum starved overnight, and a modified Matrigel-filled scratch wound was used to assess invasion. Graph shows relative wound density (RWD) over time (n = 6–10 wells per group). Data analyzed with repeated measures ANOVA followed by multiple comparison post hoc corrections. **, P < 0.01; ****, P < 0.001 at the last time point. C, 231BR cells expressing EV or shIL13RA2 were transduced with shNC or shEFNB1 as indicated, and induced with doxycycline for 72 hours before plating. Invasion was measured as in B. *, P < 0.05; **, P < 0.01. D, 231BR cells were transfected with a lentiviral empty vector (EV) or overexpressing EFNB1 gene (OE-EFNB1). Left, Graph shows EFNB1 mRNA levels normalized to GAPDH and relative to EV cells. Right, Graphs show invasion of 231-EV and OE-EFNB1 analyzed as in B. ****, P < 0.001 at the last time point. E, 231BR cells expressing EV or shEPHB1–1/2 were plated and analyzed for invasion as in B. F, 231BR cells expressing EV and shEFNB1–1/2 or shEPHB1–1/2 were plated for 48 hours, treated with BrdUrd, 1 hour stained with PI for cell-cycle analysis. Graph shows cell-cycle analysis by flow cytometry (n = 4). Adjusted P values are shown. G, BrdUrd incorporation measured in cells from F. H, 231BR cells expressing EV and shEFNB1–1/2 or shEPHB1–1/2 were plated, and percentage of confluence was measured over time using Incucyte live imaging (n = 5 wells per treatment). Data analyzed with repeated measures ANOVA followed by multiple comparison post hoc corrections. ****, P < 0.001 at the last time point.
Loss of ephrins has a cell type and context-dependent effect on cancer cell proliferation. We therefore assessed whether EFNB1 or EPHB1 plays a role in the proliferative ability of breast cancer cells and can explain the phenotypic changes induced by KD of IL13Rα2. Downregulation of EFNB1 but not EPHB1 significantly increased the percentage of cells in S and G2–M, increased the percentage of G1-arrested cells (Fig. 5F) and increased BrdUrd incorporation (Fig. 5G), suggesting a role for EFNB1 in repressing the ability of cells to undergo DNA synthesis and replication. However, an increased percentage of cells incorporating DNA and entering the cell cycle did not increase overall cell growth, as long-term analysis showed a significantly decreased confluence of shENFB1 and shEPHB1 cells upon 96 hours in vitro (Fig. 5H). Together, these data suggest that EFNB1 expression in breast cancer cells promotes their invasion and survival.
Downregulation of IL13Rα2 decreases BM outgrowth after cancer cell dissemination
Given the observed dual role of IL13Rα2 in promoting proliferation but repressing invasion, we reasoned that upregulation of IL13Rα2 and its promotion of proliferation would be critical for brain metastatic outgrowth and progression and explain the clinical correlation between high IL13Rα2 and worse survival following BM diagnoses. Conversely, downregulation of IL13Rα2 (and/or increases in ephrin B1) would be necessary for cells to invade and be most relevant to early stages of tumor dissemination and early extravasation in the brain. Analysis of a public dataset of primary breast cancer with known development of BM (33) shows that IL13RA2 mRNA levels in the primary tumor do not predict different outcomes in disease-free survival (DFS) or brain metastases-free survival (BMFS; Supplementary Fig. S3A). However, high levels of EFNB1 mRNA in the primary tumor predicted a worse DFS (P = 0.0135) and BMFS (P = 0.043; Supplementary Fig. S3B). Moreover, patients with primary tumors expressing both high EFNB1/low IL13RA2 (predicted to be more invasive) had a worse DFS (P = 0.0067) and BMFS (P = 0.034) as compared with patients that had both low EFNB1/High IL13RA2 (predicted to be less invasive; Supplementary Fig. S3C). Thus, these data suggest that a proinvasive phenotype at the primary site may predict dissemination and seeding to metastatic sites, but the ability to proliferate at distant sites is critical for disseminated cancer cells to outgrowth as metastases.
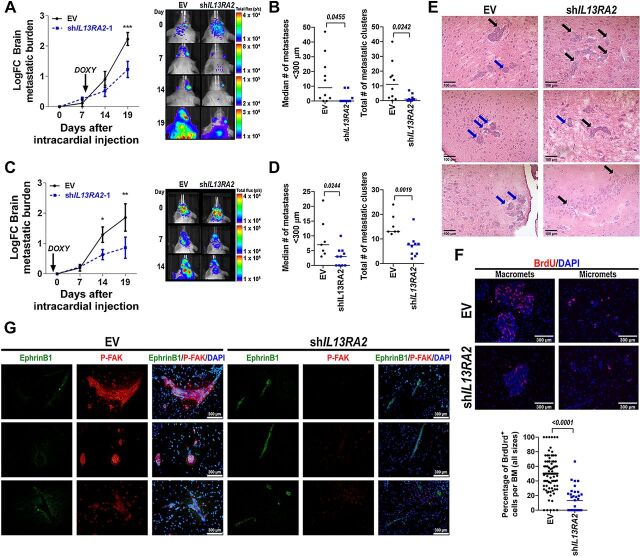
Because analysis of IL13Rα2 expression in late BMs showed that increased IL13Rα2 predicted worse survival after BMs diagnoses (Fig. 1), we next tested the effect of IL13Rα2 downregulation in the outgrowth of disseminated cancer cells. First, doxycycline-inducible shEV or shIL13RA2 231BR cells expressing luciferase reporter were injected intracardially in female NSG mice (N = 10/group), and cancer cells were allowed to invade and colonize the brain parenchyma for 7 days. At this time point in this experimental metastases model, cancer cells have extravasated the brain parenchyma and formed micrometastasis (6). Mice were then treated with doxycycline for additional 2 weeks to assess how downregulation of IL13Rα2 would influence late brain metastatic outgrowth. Consistent with its proliferative function in late BM, downregulation of IL13Rα2 resulted in a significant delay in the outgrowth of brain metastases and reduction of BM burden by 19 days following injection, as measured by IVIS (Fig. 6A). Histological quantification of BM at euthanasia showed that downregulation of IL13Rα2 decreased the median number of micrometastases per mouse brain (<300 μm, average 14.3 ± 16.30 in EV vs. 2.5 ± 4.07 in shIL13RA2, P = 0.045) as well as the total number of metastatic clusters per brain (average 12.8 ± 12.9 in EV vs. 2.0 ± 2.6 in shIL13RA2, P = 0.024, Fig. 6B).
Figure 6.
Downregulation of IL13Rα2 reduces brain metastatic progression. A, Female NSG mice were injected intracardially with 175.000 231BR-EV (n = 10) or shIL13RA2 cells (N = 9) expressing luciferase, and cells were allowed to seed and colonize for 7 days before induction with doxycycline. Brain metastatic burden was measured via in vivo imaging immediately after cell injection and the indicated times. Head total flux for each animal was normalized to brain signal at time 0 (Fold Change FC). Left, Graph shows Log-transformed FC ± SEM over time for EV versus shIL13RA2-injected mice. Arrow indicates start point for doxycycline treatment. Normally distributed Log-transformed FC values were analyzed using Repeated Measures Mixed effects. Right, Representative image of brain metastatic burden in mice injected with shEV and shIL13RA2. ***, P = 0.0007 at the indicated time point. B, Histologic quantification of BMs from mice in A. Left, Each dot represents the median number of micrometastases (<300 μm) per mouse, and the line designates the group median. Right, Each dot represents the total number of metastatic clusters per mouse. Data were analyzed using the Mann–Whitney test. C, Female NSG mice were injected as in A (n = 10/group), but cells and mice had been pretreated with doxycycline for 2 days before cell injection. Left, Graph shows Log-transformed FC ± SEM over time for EV versus shIL13RA2 injected mice. Normally distributed Log-transformed FC values were analyzed using Repeated Measures Mixed effects. *, P = 0.045; **, P = 0.0052 at the indicated time points. Right, Representative image of brain metastatic burden in mice injected with shEV and shIL13RA2 in this experiment. D, Histologic quantification of BMs for mice in C surviving at day 19 (n = 7 for EV, N = 10 for shIL13RA2). E, Representative images of BM in EV versus shIL13RA2 from A to B. Blue arrows denote BMs with a “less invasive” growth pattern, black arrows BMs with invasive fronts. F, Mice from A were injected with BrdUrd 2 hours before euthanasia, and BrdUrd incorporation in BMs was quantified by IF. Top, Representative image of BrdUrd staining in macro- and micromets from EV versus shIL13RA2 mice. Bottom, Percentage of BrdUrd+ cells quantified in individual metastases from four mice with histologically detectable BMs per group. G, Double-IF staining of p-FAK (red) and ephrin B1 (green) in BMs from EV versus shIL13RA2 mice. Blue is DAPI.
To assess whether earlier downregulation of IL13Rα2 (and the increase in pro-invasive ephrin B1) would promote—rather than block—brain metastatic colonization, the experiment was repeated as described, but both cancer cells and mice were pre-treated with doxycycline to ensure downregulation of IL13Rα2 from early stages of brain metastatic colonization (Fig. 6C). No changes in brain metastatic burden were observed at day 7 following intracardiac injection, but shIL13RA2 cells showed a significant decrease in brain metastatic IVIS signal by 14 and 19 days after intracardiac injection. Histological quantification of BMs at euthanasia also showed a significant decrease in the median number of micrometastases (9.0 ± 6.7 in EV vs. 3 ± 3.2 in shIL13RA2, P = 0.02) and the total number of metastatic clusters (15.5 ± 4.3 in EV vs. 7.1 ± 4.5 in shIL13RA2, P = 0.0019) in shIL13RA2 compared with EV cells at euthanasia (Fig. 6D). To assess whether overexpression of IL13Rα2 in poorly metastatic BT474.M1 cells would be sufficient to promote brain metastatic colonization, doxycycline–induced EV or OE-IL13RA2 cells were injected intracardically in doxycycline-treated NSG mice (n = 10/group) and metastatic outgrowth was measured via IVIS. Albeit an earlier disadvantage for OE-IL13RA2 BT474M1 cells to seed in the brain (possibly due to a decreased invasive ability), there were no differences in brain metastatic progression in EV versus OE-IL13RA2 (Supplementary Fig. S4). Thus, downregulation of IL13Rα2 before or after seeding in the brain decreased brain metastatic colonization and progression of BC cells in vivo, but overexpression of IL13Rα2 in a model with poor invasive potential was not sufficient to promote brain metastatic colonization.
We next evaluated the extent to which downregulation of IL13Rα2 in BMs in vivo, reflected the phenotypes and signaling pathways identified in vitro. Although there are not well-established markers for exclusively labeling invasive breast cancer cells (particularly from basal-like TNBC), BMs from shIL13RA2 cells were generally smaller and showed various invasive fronts (Fig. 6E, black arrows) compared with BMs from shEV cells, which tended to appear as rounded more delimited metastases (Fig. 6E, blue arrows). To quantify the proliferative status of EV versus shIL13RA2 BMs, mice in which IL13Rα2 was induced 7 days after intracardiac injection (from Fig. 6A) were injected with 10 mg/mL BrdUrd 2 hours before euthanasia and the percentage of brain metastatic cancer cells incorporating BrdUrd was quantified by immuofluorescence. Consistent with the decrease in proliferation in shIL13RA2 observed in vitro, BM from shIL13RA2 cells showed lower percentage of cancer cells incorporating BrdUrd per metastasis (Fig. 6F). Furthermore, BMs from EV cells (with high levels of IL13Rα2) showed high expression of p-FAK and low expression of ephrin B1, whereas BM from shIL13RA2 cells showed increased expression of ephrin B1 and lacked expression of p-FAK (Fig. 6G). Collectively, these results support the notion that IL13Rα2 promotes proliferation of brain metastatic cells through a mechanism involving activation of FAK signaling and modulation of ephrin B1.
Discussion
An increased invasive ability is better known to impact the ability of cancer cells to move away from primary tumors and seed at metastatic sites (34), but a complete or partial restoration of a more proliferative phenotype has been proposed as critical for the growth of disseminated cancer cells at metastatic sites, particularly BM (35). Here we show that (i) high levels of IL13Rα2 promote proliferation and repress invasion; (ii) high expression of IL13Rα2 protein in BM predicted worse survival after BM diagnosis, and iii), downregulation of IL13Rα2 decreased brain metastatic burden in vivo. These findings support the notion that the ability of cancer cells to acquire (at least temporarily) a less invasive, more epithelial and proliferative phenotype is critical for brain metastatic outgrowth. Consistent with prior reports showing upregulation of IL13RA2 in breast cancer cells selected for brain tropism (5), we found high expression of IL13Rα2 in brain metastasis but not in matched primary tumors growing in the mouse mammary fat pad. IL13RA2 mRNA levels in primary tumors do not predict for worse clinical prognosis, further supporting the upregulation of IL13Rα2 as a late event occurring at the site of metastasis, which cannot be predicted by interrogating the primary tumor.
Although our studies did not aim to address the dynamics of IL13Rα2 expression at primary versus metastatic sites, our results suggest a dynamic nature of IL13Rα2 expression similar to other molecular markers such as NDRG1 (36). Using multiphoton laser scanning microscopy, NDRG1 was shown to be critical for the arrest of slow-cycling cells to the brain capillary and the initiation of metastasis, yet loss of NDRG1 promoted the growth of primary tumors in at least one model (36). Similarly, prior studies showed controversial roles for IL13Rα2 as suppressor of tumorigenesis in models of primary breast cancers (31) and a tumor and metastasis promoter in a model of breast to lung metastasis (15). Our results provide an explanation for such discrepancies as sustained inhibition of IL13Rα2 (such as used in these prior studies; refs. 15, 30, 31) resulted in cell death or emergence of resistant phenotypes, and the observed dual role of IL13Rα2 in invasion versus proliferation can differentially influence primary tumor versus metastasis. Nonetheless, our results support a prometastatic function for high levels of IL13Rα2 in late-stage BM and in highly aggressive TNBC and Her2+ BC subtypes.
Mechanistically, our findings point to integrin and cell–cell communication as likely triggers for the upregulation of IL13Rα2 and the promotion of proliferation during metastatic progression. Similar to previous findings in colorectal cancers (4, 32), upregulation of IL13Rα2 was accompanied by increased levels of total and activated FAK, and numerous studies have demonstrated a regulatory role for FAK/SRC in cell-cycle progression driven through transcriptional activation of cyclin D1 (37). FAK/integrin signaling is well known to influence epigenetic plasticity. Interestingly, IPA analysis of EV versus shIL13RA2 cells showed KDM3A (a known activator of gene expression through removal of repressive H3K9 histone methyl 1 and 2 marks), as the top transcriptional regulator being activated in cells losing IL13Rα2 (2.06 Activation Z-score, P = 7.08 × 10–12, Supplementary Table S4B). KDM3A has been shown to promote anoikis (38), migration and invasion of breast cancer cells (39), as well as increase of KDM3A in breast cancer cells has been shown to result from loss of integrin/FAK signaling (38). Thus, it is likely that IL13Rα2 promotes proliferation trough downstream KDM3A activation.
Our in vitro studies demonstrate a ligand-independent function of IL13Rα2 in BM [as we did not observe expression of its high affinity ligand IL-13 (40, 41) or the putative ligand CHI3L1 (42) in human breast cancer cells]. However, we observed increases in FAK/SRC signaling in IL13Rα2-expressing cells in response to IL-13, suggesting that the function of IL13Rα2 is likely to be modulated by heterogeneous expression of IL-13 in cancer cells or local availability of IL-13 in the tumor microenvironment (43). It is unclear to what extent murine IL-13 binds to human IL13Rα2; therefore, our xenograft model may not fully inform how IL-13 or other IL13RA2 ligands contribute to the prometastatic function of IL13Rα2 in the brain. Moreover, IL-13 induced FAK activation in BT474 cells lacking IL13Rα2, indicating that not all IL-13–induced FAK recruitment is dependent on IL13Rα2. Because breast cancer cells have been reported to express the other cognate ligands for IL-13 (IL13RA1 and IL4R; ref. 44), these are likely to contribute to the overall signaling elicited by IL-13 in BC cells. Further studies are warranted to assess how IL-13/IL13Rα2 dependent signaling may affect the proliferative or invasive function of IL13Rα2.
Knockdown of IL13Rα2 in brain trophic cells increased expression of multiple members of the bidirectional ephrin B signaling pathway, including EphB1 (EPHB1, a receptor tyrosine kinase that mediates forward signaling) and ephrin B1 (EFNB1, an ephrinB1 transmembrane ligand, which mediates reverse signaling). Ephrin B1 was been identified as one of 5 potential biomarkers for brain metastatic cells in vitro (45), but their function was not fully elucidated. Ephrin-mediated signaling plays key roles in migration, invasion, and proliferation depending on the cell types and cellular contexts (46, 47). Eph-forward signaling induces cytoskeleton changes via the modulation of integrin expression and function, and the modulation of focal adhesion signaling (46, 48), whereas the activation of ephrin reverse signaling promotes cell polarization and mesenchymal–epithelial transition during development through inactivation of cell division control protein 42 (CDC42; ref. 49). SRC homology 2 (SH2)-adaptor proteins are thought to link Ephs to cytoskeletal and focal adhesion proteins and to facilitate actin cytoskeletal changes by modulating the activity of RHO family proteins (50). Here, we observed that ephrin B1 expressed in small metastatic clusters or invasive fronts of clinical BM, and downregulation of ephrin B1 significantly impaired the invasive ability of breast cancer cells, consistent with a proinvasive role for ephrin B1 in breast cancer cells. Moreover, high ephrin B1 mRNA levels at primary tumors predicted worse DFS and BMFS, supporting the hypothesis that invasive phenotypes are critical for metastatic dissemination. The fact that upregulation of IL13Rα2 in BT474M1 cells (with concomitant downregulation in endogenous ephrin B1 levels) was not sufficient to promote brain colonization in this model, further supports the notion that the ability to invade and extravasate is required for metastatic colonization. Yet, the proliferative ability of cancer cells at the metastatic sites remains a critical driver for late metastatic outgrowth, as downregulation of IL13Rα2 (albeit increasing the invasive ability of cancer cells) was sufficient to decrease brain metastatic progression in vivo.
Finally, our findings have important therapeutic implications. Multiple targeted therapies are being developed against IL13Rα2, including bacterial toxins conjugated to IL-13 (51), nanoparticles (52), oncolytic virus (53), as well as immunotherapies using monoclonal antibodies (54), IL13Rα2-pulsed dendritic cells (55), and IL13Rα2-targeted chimeric antigen receptors (56, 57). Phase I clinical trials for IL13Rα2-targeted chimeric antigen receptors (NCT02208362) showed that delivery of IL13Rα2-targeted CAR-T cells into the cerebrospinal fluid was well tolerated and proved effective against recurrent multifocal leptomeningeal glioblastoma, according to a case report (58). Given our findings that IL13Rα2 is expressed at various levels in all BCBMs and other studies have shown that IL13Rα2 promotes lung metastasis, emerging therapies targeting IL13Rα2 have the potential to benefit patients with brain and lung metastasis.
Authors' Disclosures
M.J. Contreras-Zárate reports grants from U.S Department of Defense during the conduct of the study. S.D. Karam reports grants from Roche and AstraZeneca outside the submitted work. P. Kabos reports grants from NIH during the conduct of the study as well as grants from Eli Lilly, Pfizer, Sanofi, Genentech, AstraZeneca, and Radius Health outside the submitted work. D.M. Cittelly reports grants from Department of Defense Breast Cancer Research Program and NIH/NCI during the conduct of the study. No disclosures were reported by the other authors.
Supplementary Material
Supplementary Table 4.
Supplementary Table 1. List of shRNAs Supplementary Table 2. List of primers and sgRNAs Supplementary Table 3: List of antibodies
Supplementary Figure 1. A. 231BR cells were transfected with an empty vector (gEV) or using sgRNAs targeting IL13Rα2 (gIL13Ra2). Cells were selected for clonal populations and IL13Rα2 protein expression assessed by western blot. α-tubulin was used as loading control. B. 231BR and JmT1BR3 cells transfected with a DOXY inducible system were assessed to determine any possible side proliferation effect after DOXY induction. Cells harboring the EV were treated with DOXY at 1 µg/mL or 2 µg/mL and % of confluence was measured over time using Incucyte live imaging. (n=4 treatment). There were no significant changes in proliferation.
Supplementary Figure 2. Global RNA sequencing was performed in doxycycline-induced 231BR EV vs 231BR shIL13RA2 cells 48 h after DOXY-induction (n=3/group). Ingenuity® Pathway Analysis (IPA®). The p-value (calculated with the Fischer's exact) and Z-score were determined to stablish any probable association between our set of genes and a function. Migration and replication biofunctions are predicted as being activated and repressed, respectively, when IL13Rα2 is downregulated with a high absolute Z-score ({greater than or equal to}2) and a significant p-value (<0.05).
Supplementary Figure 3. Predictive value of IL13RA2 or EFNB1 mRNA levels in primary tumors known to metastasize to brain. Dataset contains 21 matched breast cancer primary and brain metastasis samples, reported in (https://pubmed.ncbi.nlm.nih.gov/29961873/). A. Kaplanmeier plots for IL13RA2 mRNA expression in primary tumors. B. Kaplan-Meier plots to EFNB1 mRNA in primary tumors. C. Kaplan-meier plots for primary tumors with low-IL13RA2/highEFNB1 compared to samples with high-IL13RA2/low-EFNB1. For all plots, average mRNA expression was determined for each target and expression ranked as low (below mean) or high (above mean). Survival curves comparisons were made using Log-rank (Mantel-Cox) test. p<0.05 was considered significant (italic bold). OS: overall survival; DFS: disease free survival; BMFS: brain metastasis free survival; SPBM: survival post brain metastasis.
Supplementary Figure 4. Overexpression of IL13Rα2 does not increase metastatic colonization in poorly invasive BT474M1 BC cells. Poorly metastatic ER+HER2+ BT474M1 cells expressing EV or OE-IL13RA2 and a luciferase reporter, were induced with doxycycline for 48 h prior to injecting in dox-treated NSG mice implanted with 1 mg E2 pellets (N=12 per group). Brain metastatic colonization was measured via IVIS at the indicated times. Head total flux for each animal was normalized to brain signal at time 0 (Fold Change FC). Graph shows Log-transformed FC {plus minus}SEM over time for EV vs OE-IL13RA2 injected mice. Normally distributed Log-transformed FC values were analyzed using Repeated Measures Mixed effects.*P=0.02 at 7 days. Ns at other time points
Acknowledgments
This work was supported by DoD-BCRP W81XWH-15–1-0352, R37 CA227984, Cancer League of Colorado, and the Metavivor Research Foundation (to D.M. Cittelly). P. Kabos is supported by NIH R01-CA20544. M.J. Contreras-Zárate is supported by DoD BCRP W81XWH-19–1-0033. S.D. Karam is supported by the NIDCR (R01 DE028529–01, R01 DE028282–01). We thank the University of Colorado Cancer Center Animal Imaging Shared Resources, Tissue Culture Core, Cytometry and Cell Sorting Shared Resource, Functional Genomics Facility and Biostatistics and Bioinformatics Share Resource supported by NCI P30CA046934 and CTSA UL1TR001082 Center grants. We thank Dr. A. Van Bokhoven at the Biorepository Core Facility, and personnel at the UC Brain Tumor Biorepository for providing de-identified human tissues. We thank Dr. D. Yu for providing breast cancer cells BT474m1 and BT474m1Br1, and Dr. P. Steeg for providing 231BR and JmT1BR3 cell lines.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Contributions
R.A. Márquez-Ortiz: Conceptualization, data curation, formal analysis, validation, writing–original draft, writing–review and editing. M.J. Contreras-Zárate: Conceptualization, formal analysis, validation, investigation, writing–review and editing. V. Tesic: Investigation. K.L.F. Alvarez-Eraso: Validation, investigation. G. Kwak: Investigation. Z. Littrell: Investigation. J.C. Costello: Data curation, formal analysis, writing–review and editing. V. Sreekanth: Data curation, formal analysis, writing–review and editing. D.R. Ormond: Resources, funding acquisition, writing–review and editing. S.D. Karam: Conceptualization, investigation, writing–review and editing. P. Kabos: Conceptualization, funding acquisition, writing–review and editing. D.M. Cittelly: Conceptualization, resources, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Lee SS, Ahn J-H, Kim MK, Sym SJ, Gong G, Ahn SD, et al. Brain metastases in breast cancer: prognostic factors and management. Breast Cancer Res Treat 2008;111:523–30. [DOI] [PubMed] [Google Scholar]
- 2. Palmieri D, Smith QR, Lockman PR, Bronder J, Gril B, Chambers AF, et al. Brain metastases of breast cancer. Breast Dis 2006;26:139–47. [DOI] [PubMed] [Google Scholar]
- 3. Morris PG, Murphy CG, Mallam D, Accordino M, Patil S, Howard J, et al. Limited overall survival in patients with brain metastases from triple-negative breast cancer. Breast J 2012;18:345–50. [DOI] [PubMed] [Google Scholar]
- 4. Barderas R, Bartolomé RA, Fernandez-Aceñero MJ, Torres S, Casal JI. High expression of IL-13 receptor α2 in colorectal cancer is associated with invasion, liver metastasis, and poor prognosis. Cancer Res 2012;72:2780–90. [DOI] [PubMed] [Google Scholar]
- 5. Bos PD, Zhang XH-F, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature 2009;459:1005–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lorger M, Felding-Habermann B. Capturing changes in the brain microenvironment during initial steps of breast cancer brain metastasis. Am J Pathol 2010;176:2958–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Priego N, Zhu L, Monteiro C, Mulders M, Wasilewski D, Bindeman W, et al. STAT3 labels a subpopulation of reactive astrocytes required for brain metastasis. Nat Med 2018;24:1024–35. [DOI] [PubMed] [Google Scholar]
- 8. Valiente M, Obenauf AC, Jin X, Chen Q, Zhang XH-F, Lee DJ, et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell 2014;156:1002–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Neman J, Termini J, Wilczynski S, Vaidehi N, Choy C, Kowolik CM, et al. Human breast cancer metastases to the brain display GABAergic properties in the neural niche. Proc Natl Acad Sci U S A 2014;111:984–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kienast Y, von Baumgarten L, Fuhrmann M, Klinkert WEF, Goldbrunner R, Herms J, et al. Real-time imaging reveals the single steps of brain metastasis formation. Nat Med 2010;16:116–22. [DOI] [PubMed] [Google Scholar]
- 11. Christofori G. New signals from the invasive front. Nature 2006;441:444–50. [DOI] [PubMed] [Google Scholar]
- 12. Debinski W, Gibo DM. Molecular expression analysis of restrictive receptor for interleukin 13, a brain tumor-associated cancer/testis antigen. Mol Med 2000;6:440–9. [PMC free article] [PubMed] [Google Scholar]
- 13. Joshi BH, Plautz GE, Puri RK. Interleukin-13 receptor alpha chain: a novel tumor-associated transmembrane protein in primary explants of human malignant gliomas. Cancer Res 2000;60:1168–72. [PubMed] [Google Scholar]
- 14. Wykosky J, Gibo DM, Stanton C, Debinski W. Interleukin-13 receptor alpha 2, EphA2, and Fos-related antigen 1 as molecular denominators of high-grade astrocytomas and specific targets for combinatorial therapy. Clin Cancer Res 2008;14:199–208. [DOI] [PubMed] [Google Scholar]
- 15. Papageorgis P, Ozturk S, Lambert AW, Neophytou CM, Tzatsos A, Wong CK, et al. Targeting IL13Ralpha2 activates STAT6-TP63 pathway to suppress breast cancer lung metastasis. Breast Cancer Res 2015;17:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karmele EP, Pasricha TS, Ramalingam TR, Thompson RW, Gieseck RL, Knilans KJ, et al. Anti-IL-13Ralpha2 therapy promotes recovery in a murine model of inflammatory bowel disease. Mucosal Immunol 2019;12:1174–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Newman JP, Wang GY, Arima K, Guan SP, Waters MR, Cavenee WK, et al. Interleukin-13 receptor alpha 2 cooperates with EGFRvIII signaling to promote glioblastoma multiforme. Nat Commun 2017;8:1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang K, Marran K, Valentine A, Hannon GJ. Creating an miR30-based shRNA vector. Cold Spring Harb Protoc 2013;2013:631–5. [DOI] [PubMed] [Google Scholar]
- 19. Longo PA, Kavran JM, Kim MS, Leahy DJ. Single-cell cloning of a stable mammalian cell line. Methods Enzymol 2014;536:165–72. [DOI] [PubMed] [Google Scholar]
- 20. Contreras-Zárate MJ, Day NL, Ormond DR, Borges VF, Tobet S, Gril B, et al. Estradiol induces BDNF/TrkB signaling in triple-negative breast cancer to promote brain metastases. Oncogene 2019;38:4685–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eisemann T, Costa B, Strelau J, Mittelbronn M, Angel P, Peterziel H. An advanced glioma cell invasion assay based on organotypic brain slice cultures. BMC Cancer 2018;18:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ferreira TA, Blackman AV, Oyrer J, Jayabal S, Chung AJ, Watt AJ, et al. Neuronal morphometry directly from bitmap images. Nat Methods 2014;11:982–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sartorius CA, Hanna CT, Gril B, Cruz H, Serkova NJ, Huber KM, et al. Estrogen promotes the brain metastatic colonization of triple-negative breast cancer cells via an astrocyte-mediated paracrine mechanism. Oncogene 2016;35:2881–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gril B, Palmieri D, Bronder JL, Herring JM, Vega-Valle E, Feigenbaum L, et al. Effect of lapatinib on the outgrowth of metastatic breast cancer cells to the brain. JNCI J Natl Cancer Inst 2008;100:1092–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 2013;31:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28:511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kalli M, Mpekris F, Wong CK, Panagi M, Ozturk S, Thiagalingam S, et al. Activin a signaling regulates IL13Ralpha2 expression to promote breast cancer metastasis. Front Oncol 2019;9:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kawakami K, Kawakami M, Snoy PJ, Husain SR, Puri RK. In vivo overexpression of IL-13 receptor alpha2 chain inhibits tumorigenicity of human breast and pancreatic tumors in immunodeficient mice. J Exp Med 2001;194:1743–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bartolomé RA, García-Palmero I, Torres S, López-Lucendo M, Balyasnikova IV, Casal JI. IL13 receptor alpha2 signaling requires a scaffold protein, FAM120A, to activate the FAK and PI3K pathways in colon cancer metastasis. Cancer Res 2015;75:2434–44. [DOI] [PubMed] [Google Scholar]
- 33. Varešlija D, Priedigkeit N, Fagan A, Purcell S, Cosgrove N, O'Halloran PJ, et al. Transcriptome characterization of matched primary breast and brain metastatic tumors to detect novel actionable targets. J Natl Cancer Inst 2019;111:388–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lourenco AR, Ban Y, Crowley MJ, Lee SB, Ramchandani D, Du W, et al. Differential contributions of pre- and post-EMT tumor cells in breast cancer metastasis. Cancer Res 2020;80:163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chao Y, Wu Q, Acquafondata M, Dhir R, Wells A. Partial mesenchymal to epithelial reverting transition in breast and prostate cancer metastases. Cancer Microenviron 2012;5:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Berghoff AS, Liao Y, Karreman MA, Ilhan-Mutlu A, Gunkel K, Sprick MR, et al. Identification and characterization of cancer cells that initiate metastases to the brain and other organs. Mol Cancer Res 2021;19:688–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhao JH, Reiske H, Guan JL. Regulation of the cell cycle by focal adhesion kinase. J Cell Biol 1998;143:1997–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pedanou VE, Gobeil S, Tabariès S, Simone TM, Zhu LJ, Siegel PM, et al. The histone H3K9 demethylase KDM3A promotes anoikis by transcriptionally activating pro-apoptotic genes BNIP3 and BNIP3L. Elife 2016;5:e16844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun L, Yuan Y, Chen J, Ma C, Xu Y. Brahma related gene 1 (BRG1) regulates breast cancer cell migration and invasion by activating MUC1 transcription. Biochem Biophys Res Commun 2019;511:536–43. [DOI] [PubMed] [Google Scholar]
- 40. Fichtner-Feigl S, Strober W, Kawakami K, Puri RK, Kitani A. IL-13 signaling through the IL-13alpha2 receptor is involved in induction of TGF-beta1 production and fibrosis. Nat Med 2006;12:99–106. [DOI] [PubMed] [Google Scholar]
- 41. Lupardus PJ, Birnbaum ME, Garcia KC. Molecular basis for shared cytokine recognition revealed in the structure of an unusually high affinity complex between IL-13 and IL-13Ralpha2. Structure 2010;18:332–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. He CH, Lee CG, Dela Cruz CS, Lee C-M, Zhou Y, Ahangari F, et al. Chitinase 3-like 1 regulates cellular and tissue responses via IL-13 receptor alpha2. Cell Rep 2013;4:830–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wood N, Whitters MJ, Jacobson BA, Witek J, Sypek JP, Kasaian M, et al. Enhanced interleukin (IL)-13 responses in mice lacking IL-13 receptor alpha 2. J Exp Med 2003;197:703–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park MH, Kwon HJ, Kim J-R, Lee B, Lee SJ, Bae YK. Elevated interleukin-13 receptor alpha 1 expression in tumor cells is associated with poor prognosis in patients with invasive breast cancer. Ann Surg Oncol 2017;24:3780–7. [DOI] [PubMed] [Google Scholar]
- 45. Dun MD, Chalkley RJ, Faulkner S, Keene S, Avery-Kiejda KA, Scott RJ, et al. Proteotranscriptomic profiling of 231-BR breast cancer cells: identification of potential biomarkers and therapeutic targets for brain metastasis. Mol Cell Proteomics 2015;14:2316–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kandouz M. The Eph/Ephrin family in cancer metastasis: communication at the service of invasion. Cancer Metastasis Rev 2012;31:353–73. [DOI] [PubMed] [Google Scholar]
- 47. Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer 2010;10:165–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miao H, Burnett E, Kinch M, Simon E, Wang B. Activation of EphA2 kinase suppresses integrin function and causes focal-adhesion-kinase dephosphorylation. Nat Cell Biol 2000;2:62–69. [DOI] [PubMed] [Google Scholar]
- 49. Barquilla A, Pasquale EB. Eph receptors and ephrins: therapeutic opportunities. Annu Rev Pharmacol Toxicol 2015;55:465–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kania A, Klein R. Mechanisms of ephrin-Eph signalling in development, physiology and disease. Nat Rev Mol Cell Biol 2016;17:240–56. [DOI] [PubMed] [Google Scholar]
- 51. Debinski W, Obiri NI, Powers SK, Pastan I, Puri RK. Human glioma cells overexpress receptors for interleukin 13 and are extremely sensitive to a novel chimeric protein composed of interleukin 13 and pseudomonas exotoxin. Clin Cancer Res 1995;1:1253–8. [PubMed] [Google Scholar]
- 52. Madhankumar AB, Slagle-Webb B, Mintz A, Sheehan JM, Connor JR. Interleukin-13 receptor-targeted nanovesicles are a potential therapy for glioblastoma multiforme. Mol Cancer Ther 2006;5:3162–9. [DOI] [PubMed] [Google Scholar]
- 53. Candolfi M, Xiong W, Yagiz K, Liu C, Muhammad AKMG, Puntel M, et al. Gene therapy-mediated delivery of targeted cytotoxins for glioma therapeutics. Proc Natl Acad Sci U S A 2010;107:20021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Balyasnikova IV, Wainwright DA, Solomaha E, Lee G, Han Y, Thaci B, et al. Characterization and immunotherapeutic implications for a novel antibody targeting interleukin (IL)-13 receptor α2. J Biol Chem 2012;287:30215–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Okada H, Kalinski P, Ueda R, Hoji A, Kohanbash G, Donegan TE, et al. Induction of CD8+ T-cell responses against novel glioma-associated antigen peptides and clinical activity by vaccinations with {alpha}-type 1 polarized dendritic cells and polyinosinic-polycytidylic acid stabilized by lysine and carboxymethylcellulose in patients with recurrent malignant glioma. J Clin Oncol 2011;29:330–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sengupta S, Thaci B, Crawford AC, Sampath P. Interleukin-13 receptor alpha 2-targeted glioblastoma immunotherapy. Biomed Res Int 2014;2014:952128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kahlon KS, Brown C, Cooper LJN, Raubitschek A, Forman SJ, Jensen MC. Specific recognition and killing of glioblastoma multiforme by interleukin 13-zetakine redirected cytolytic T cells. Cancer Res 2004;64:9160–6. [DOI] [PubMed] [Google Scholar]
- 58. Brown CE, Alizadeh D, Starr R, Weng L, Wagner JR, Naranjo A, et al. Regression of glioblastoma after chimeric antigen receptor T-cell therapy. N Engl J Med 2016;375:2561–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 4.
Supplementary Table 1. List of shRNAs Supplementary Table 2. List of primers and sgRNAs Supplementary Table 3: List of antibodies
Supplementary Figure 1. A. 231BR cells were transfected with an empty vector (gEV) or using sgRNAs targeting IL13Rα2 (gIL13Ra2). Cells were selected for clonal populations and IL13Rα2 protein expression assessed by western blot. α-tubulin was used as loading control. B. 231BR and JmT1BR3 cells transfected with a DOXY inducible system were assessed to determine any possible side proliferation effect after DOXY induction. Cells harboring the EV were treated with DOXY at 1 µg/mL or 2 µg/mL and % of confluence was measured over time using Incucyte live imaging. (n=4 treatment). There were no significant changes in proliferation.
Supplementary Figure 2. Global RNA sequencing was performed in doxycycline-induced 231BR EV vs 231BR shIL13RA2 cells 48 h after DOXY-induction (n=3/group). Ingenuity® Pathway Analysis (IPA®). The p-value (calculated with the Fischer's exact) and Z-score were determined to stablish any probable association between our set of genes and a function. Migration and replication biofunctions are predicted as being activated and repressed, respectively, when IL13Rα2 is downregulated with a high absolute Z-score ({greater than or equal to}2) and a significant p-value (<0.05).
Supplementary Figure 3. Predictive value of IL13RA2 or EFNB1 mRNA levels in primary tumors known to metastasize to brain. Dataset contains 21 matched breast cancer primary and brain metastasis samples, reported in (https://pubmed.ncbi.nlm.nih.gov/29961873/). A. Kaplanmeier plots for IL13RA2 mRNA expression in primary tumors. B. Kaplan-Meier plots to EFNB1 mRNA in primary tumors. C. Kaplan-meier plots for primary tumors with low-IL13RA2/highEFNB1 compared to samples with high-IL13RA2/low-EFNB1. For all plots, average mRNA expression was determined for each target and expression ranked as low (below mean) or high (above mean). Survival curves comparisons were made using Log-rank (Mantel-Cox) test. p<0.05 was considered significant (italic bold). OS: overall survival; DFS: disease free survival; BMFS: brain metastasis free survival; SPBM: survival post brain metastasis.
Supplementary Figure 4. Overexpression of IL13Rα2 does not increase metastatic colonization in poorly invasive BT474M1 BC cells. Poorly metastatic ER+HER2+ BT474M1 cells expressing EV or OE-IL13RA2 and a luciferase reporter, were induced with doxycycline for 48 h prior to injecting in dox-treated NSG mice implanted with 1 mg E2 pellets (N=12 per group). Brain metastatic colonization was measured via IVIS at the indicated times. Head total flux for each animal was normalized to brain signal at time 0 (Fold Change FC). Graph shows Log-transformed FC {plus minus}SEM over time for EV vs OE-IL13RA2 injected mice. Normally distributed Log-transformed FC values were analyzed using Repeated Measures Mixed effects.*P=0.02 at 7 days. Ns at other time points