Abstract
Established genetic risk factors for Alzheimer’s disease (AD) account for only a portion of AD heritability. The aim of this study was to identify novel associations between genetic variants and AD-specific brain atrophy. We conducted genome-wide association studies for brain magnetic resonance imaging measures of hippocampal volume and entorhinal cortical thickness in 2643 Koreans meeting the clinical criteria for AD (n = 209), mild cognitive impairment (n = 1449) or normal cognition (n = 985). A missense variant, rs77359862 (R274W), in the SHANK-associated RH Domain Interactor (SHARPIN) gene was associated with entorhinal cortical thickness (p = 5.0 × 10−9) and hippocampal volume (p = 5.1 × 10−12). It revealed an increased risk of developing AD in the mediation analyses. This variant was also associated with amyloid-β accumulation (p = 0.03) and measures of memory (p = 1.0 × 10−4) and executive function (p = 0.04). We also found significant association of other SHARPIN variants with hippocampal volume in the Alzheimer’s Disease Neuroimaging Initiative (rs3417062, p = 4.1 × 10−6) and AddNeuroMed (rs138412600, p = 5.9 × 10−5) cohorts. Further, molecular dynamics simulations and co-immunoprecipitation indicated that the variant significantly reduced the binding of linear ubiquitination assembly complex proteins, SHPARIN and HOIL-1 Interacting Protein (HOIP), altering the downstream NF-κB signaling pathway. These findings suggest that SHARPIN plays an important role in the pathogenesis of AD.
Subject terms: Biomarkers, Genetics
Introduction
Alzheimer’s disease (AD), the most common type of dementia, is a progressive disorder that causes cognitive dysfunction and memory loss. Major risk factors for AD include age, family history, and lifestyle [1]. AD has a strong genetic component with an estimated 58–79% of its liability explained by genetic factors [2]. The APOE ɛ4 allele accounts for a large portion of the heritability [3–6]. Genome-wide association studies (GWAS) have identified more than 30 independent loci [7, 8], yet, more than half of the phenotypic variance still remains unexplained [5]. Detection of additional AD risk loci can be enhanced through studies of diverse populations [9–11], demonstrated by GWAS focused on African Americans [12], Hispanics [13, 14], Japanese [15, 16], Chinese [17], and transethnic approaches [18]. Studies of diverse populations can leverage allele frequency differences that often result in variable strengths in association signals with particular variants and associations that are population-specific. Furthermore, effects of disease susceptibility loci (DSL) may be modulated by environmental risk factors that differ in exposure across populations.
Some of the genetic architecture of AD is likely too difficult to uncover by GWAS with samples as large as 100,000 subjects because of the mechanistic complexity underlying the disease [7]. Dissecting AD into biologically simpler disease-related outcomes (i.e., endophenotypes) has identified many significant genetic associations with measures of cognitive performance, structural brain atrophy (quantified by magnetic resonance imaging (sMRI)), and AD proteins in CSF or brain tissue [19–24]. Here, we report findings from a GWAS of several AD-related sMRI traits in a Korean sample including individuals with AD, mild cognitive impairment (MCI), and cognitively normal (CN) functioning. We found a genome-wide significant association of a missense variant in SHARPIN with measures of hippocampal atrophy and cortical thickness. Subsequent analysis revealed that this variant is also significantly associated with amyloid-beta (Aβ) levels in the brain, measured by positron emission tomography (PET) scan and confirmed by assessments of cognitive function and memory. Furthermore, we demonstrated experimentally that SHARPIN functionally affects NF-kB signaling in the nervous system, suggesting plays a role in the pathophysiology of AD.
Materials and methods
Genome-wide association study participants
The study sample included 5570 subjects who have enrolled in the Gwangju Alzheimer’s & Related Dementia (GARD) cohort registry at Chosun University in Gwangju, Korea. At baseline, there were 2030 CN, 2184 MCI, and 1356 AD subjects. The clinical definition of each group is provided in supplementary Text 1. A subset of 629 CN and 247 MCI subjects had at least one follow-up exam between 2010 and 2020 (mean follow-up interval = 28.8 months). After follow-up, 53 CN and 21 MCI subjects were re-classified as MCI and AD, respectively, resulting in a final sample of 1927 CN, 2216 MCI, and 1377 AD subjects.
The study protocol was approved by the Institutional Review Board of Chosun University Hospital, Korea (CHOSUN 2013–12–018–070). All volunteers or authorized guardians for cognitively impaired individuals gave written informed consent before participation.
Association analysis methods
Genotype data were preprocessed by the standard downstream quality control and imputation (Supplementary Text 2 and Supplementary Figs. S1 and S2). MRI traits (Supplementary Text 3 and Supplementary Table S1) were transformed by inverse normal transformation and GWAS were conducted for each trait using PLINK [25], ONETOOL [26], and linear regression models including imputed single nucleotide polymorphisms (SNP) genotype and covariates for age, sex, APOE genotype, a term for the log-transformed measure of intracranial volume (ICV), and the first three principal components (PCs) to adjust for population stratification. PC analysis was performed accounting for a genetic relationship matrix using EIGENSOFT [27]. APOE genotype was coded as a class variable with ε3/ε3 set as the reference and five dummy variables for ε2/ε2, ε2/ε3, ε2/ε4, ε3/ε4, and ε4/ε4. A total of 3930,740 SNPs with minor allele frequency (>0.01) were tested. The genome-wide significance (GWS) threshold was set as p < 5.0 × 10−8. We used LocusZoom [28] to generate regional plots and R software v.3.6 (R Development Core Team, Vienna, Austria) to create QQ, Manhattan, and regional plots. Follow-up analyses were performed in the ADNI [29] (n = 1566) and AddNeuroMed datasets [30] (n = 288) to replicate or extend GWS findings using similar models like those employed in the GWAS.
Gene-based association analyses with rare variants
Gene-based analyses for hippocampal volume (HV) and entorhinal thickness were performed using the MAGMA software tool [31] and models that included the same covariates as those described in tests of individual variants. These analyses included 9,784,321 functional SNPs with MAF < 0.01 that were annotated using the SNP2GENE function implemented in the FUMA program [32]. The significance threshold was set at p < 2.6 × 10−6 to correct for 19,231 gene-based tests.
Mediation analyses
The SHARPIN SNP showing GWS association with MRI traits was further evaluated in a sample CN and AD subjects (n = 985 and n = 209, respectively) to determine whether its influence on AD risk was mediated through a particular MRI trait. Mediation models were evaluated using linear regression with AD as the outcome, SNP as the predictor, and the MRI trait variables as mediators. Models also included sex, age, three PCs, and log-transformed ICV (logICV) as covariates. Mediation analyses were conducted using the PROCESS macro [33] implemented in SPSS by selecting four and 10,000 bias-corrected bootstrap samples.
Statistical methods for testing the association between PET imaging measures of Aβ accumulation and cognitive performance
Accumulation of Aβ in the brain was measured via PET in 1377 subjects (162 AD, 587 MCI, and 628 CN subjects), using a dedicated Discovery ST PET-CT scanner (General Electric Medical Systems, Milwaukee, WI, USA). PET images were obtained from subjects 90–100 min after IV injection of a mean dose of 303 MBq 20% florbetaben, a fluorin-18-labeled stilbene derivative with the trade name of NeuraCeq [34]. Preprocessing of Aβ-PET images was performed using previously described data [35]. Visual assessment of transaxial PET images was performed by a trained reader (B. Kim), using a gray scale. Each brain region (frontal cortex, lateral temporal cortex, parietal cortex, and posterior cingulate cortex/precuneus) was visually assessed and scored according to the brain amyloid plaque load (BAPL) scoring system for each PET scan. BAPL scores of 1 were classified as Aβ-negative PET scans, while BAPL scores of 2 and 3 were classified as Aβ-positive PET scans. Among 1377 subjects, 418 had a positive BAPL and 959 had a negative BAPL (Supplemental Table S2). We employed a logistic regression model to evaluate the association of the GWS SHARPIN missense variant with the derived binary BAPL variable adjusted for age and sex.
Association of this SNP with five domains of cognitive performance (attention, frontal/executive function, language, memory, and visuospatial skills) assessed by the Seoul Neuropsychological Screening Battery (SNSB) [36] was tested using linear regression models including age and sex as covariates. The SNSB cognitive data were available for all 2643 subjects used for our discovery GWAS.
Results
Multiple genes are associated with HV and entorhinal thickness in Koreans
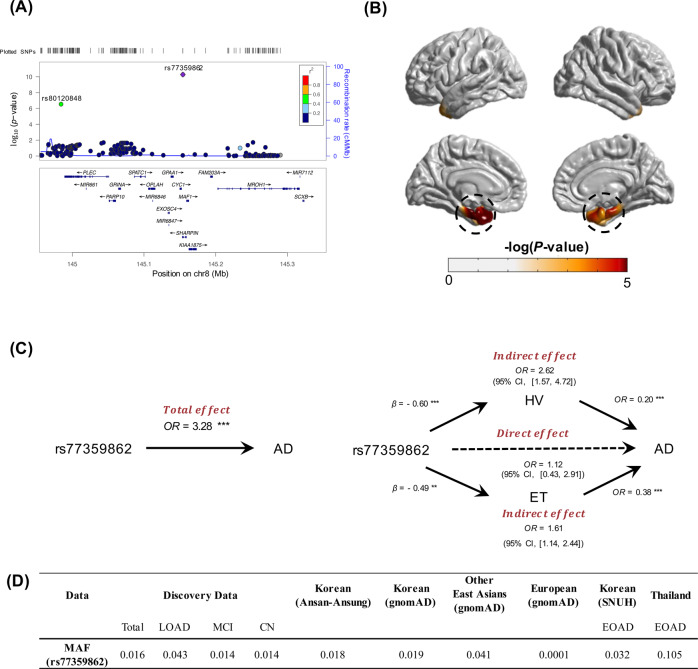
GWAS conducted for the five sMRI traits revealed GWS (p < 5.0 × 10−8) and suggestive (p < 1.0 × 10−6) associations for HV, entorhinal cortical thickness (ET), superior frontal cortical thickness, middle temporal cortical thickness and inferior parietal cortical thickness with SNPs in multiple regions (Supplemental Figs. S3 and S4). We found that the APOE genotype was significantly associated with the measures for the entorhinal (p = 5.1 × 10−11), superior frontal (p = 3.9 × 10−8), middle temporal (p = 5.8 × 10−7) and inferior parietal (p = 4.2 × 10−8) cortex regions, and the HV (p = 6.3 × 10−20) (Supplemental Table S3). These results are explained almost entirely by the dose-dependent effects of the ε4 allele on AD risk compared to the ε3ε3 reference genotype. The ε2 allele was protective but this effect was not significant, potentially due to the low frequency of this allele in Koreans. GWS associations were also observed for a missense variant (rs77359862) in SHARPIN with decreased ET (β = 0.59, p = 5.0 × 10−9) and HV (β = 0.62, p = 5.1 × 10−12) even after accounting for the APOE genotype (Table 1). Rs80120848, located ~5 kb apart from PLEC, was also associated with HV at the GWS level (p = 2.3 × 10−8, β = 0.53). Rs80120848 is 189 kb apart from and moderately correlated with rs77359862 (r = 0.68). To determine whether these are independent association signals, we tested a conditional model including both variants, and found that rs80120848 was not associated with HV (p = 0.32), whereas the association with rs77359862 was still significant (p = 4.2 × 10−4) indicating that these effects may not be independent association signals and are most likely driven by the SHARPIN locus (Fig. 1A). Suggestive associations were observed for ET with two SNPs (rs7160806, p = 7.1 × 10−7; rs1956822, p = 5.8 × 10−7) located in NOVA-AS1 that encodes a long intergenic non-protein-coding RNA 2588 and for HV with rs150912768 (p = 6.9 × 10−7) located in LOC1053722, a gene of unknown function that has an overlapping but reversely transcribed start site with SMAD4. Genome-wide analyses that were not adjusted for the APOE genotype did not reveal any additional GWS or suggestive associations outside of the APOE region (Supplementary Table S4). Our GWAS results for previously identified SNPs from other cohorts are provided as supplementary information (Supplementary Text 4, Supplementary Table S5).
Table 1.
Significant GWAS results (p < 1.0 × 10−6) for entorhinal cortical thickness and hippocampal volume adjusted for APOE genotype.
| Trait | Chr | Position | SNP | MA | MAF | IQS | β | SE | p-value | Locus |
|---|---|---|---|---|---|---|---|---|---|---|
| Entorhinal thickness | 8 | 145154282 | rs77359862 | A | 0.01 | G | −0.59 | 0.10 | 5.0 × 10−9 | SHARPIN |
| 14 | 27221601 | rs7160806 | G | 0.39 | 0.992 | −0.13 | 0.02 | 7.1 × 10−7 | NOVA1-AS1 | |
| 14 | 27219914 | rs1956822 | G | 0.39 | 0.995 | −0.13 | 0.02 | 5.8 × 10−7 | NOVA1-AS1 | |
| Hippocampal volume | 8 | 145154282 | rs77359862 | A | 0.01 | G | −0.62 | 0.09 | 5.1 × 10−12 | SHARPIN |
| 8 | 144984345 | rs80120848 | A | 0.01 | G | −0.53 | 0.10 | 2.3 × 10−8 | EPPK1/PLEC | |
| 18 | 48554594 | rs150912768 | T | 0.01 | 0.953 | −0.45 | 0.09 | 6.9 × 10−7 | SMAD4/ELAC1 |
Chr chromosome, MA minor allele, MAF minor allele frequency, IQS imputation quality score, G genotyped SNP, SE standard error
Fig. 1. Association of SHARPIN missense variant rs77359862 with AD and related traits.
A Regional plot for the association of SHARPIN with hippocampal volume (HV). B Illustration of cortical atrophy in left and right hemispheres. A general linear model was applied to detect point-wise differences in whole-brains of 84 carriers and 2559 non-carriers of the rs77359862 missense variant. Rs77359862 carriers (bottom) show greater atrophy in the entorhinal cortex and hippocampus (highlighted in dotted black circles) compared to non-carriers. C Mediation analysis shows that the effect of rs77359862 on HV and entorhinal cortical thickness (ET) mediates the association of rs77359862 and AD risk. The total effect represents the impact of rs77359862 on the odds of AD without considering indirect effects of the variant through the hippocampus and entorhinal region, whereas the direct effect is calculated controlling for its effect on HV and ET which are both associated with AD. Each indirect effect was obtained by adjusting for the other mediator; β regression coefficient. OR odds ratio, 95% CI 95% confidence interval of 10,000 bootstraps; **p-value < 0.01, ***p-value < 0.001. D Frequency of the rs77359862 missense variant in late-onset AD cases, early-onset AD (EOAD) cases, mild cognitive impairment (MCI) cases, and cognitively normal (CN) persons in clinical samples from Korea and Thailand and in several reference populations. Other East Asians exclude Korean and Japanese. Detailed information for EOAD data is provided in supplementary Text 6. SNUH Seoul National University Hospital.
Gene-based analyses of 18,950 protein-coding genes including rare variants with minor allele frequencies (MAF) < 0.01 after adjusting for the APOE genotype revealed several gene-wide significant (2.6 × 10−6) associations with HV and ET (Supplementary Fig. S5a) and little evidence for genomic inflation (Supplementary Fig. S5b) including COX7A2L (234 SNPs, p = 1.9×10−6) with ET and genes with HV: GUCA1A (64 SNPs, p = 7.7 × 10−7), VIT (289 SNPs, p = 7.8 × 10−9), and METTL6 (163 SNPs, p = 2.0 × 10−6). The association of HV with GABRR2 almost reached the gene-wide significance threshold (108 SNPs, p = 3.2 × 10−6).
SHARPIN missense variant rs77359862 indirectly affects AD risk through its impact on AD-specific brain damages
To evaluate the effect of rs77359862 on cortical atrophy, cortical thickness measures derived from multiple points spanning the entire cortex were separately regressed on rs77359862 with a generalized linear model (GLM). The 84 carriers of the rs77359862 missense variant including 28 CN subjects, 40 MCI, and 16 AD patients displayed significantly greater atrophy in the entorhinal cortex and hippocampus, than the 2559 non-carriers, whereas the differences between carriers and non-carriers were not significantly different in any other cortical regions (Fig. 1B) (see Supplementary Text 5 for Methods). Next, we conducted mediation analysis to estimate the indirect effect of rs77359862 on AD risk through its impact on HV and ET. As shown in Fig. 1C, rs77359862 is significantly associated with AD (total effect, OR = 3.28, p = 1.2 × 10−4), but after controlling for its association with HV and ET, the strength of this relationship was attenuated (OR = 1.12, p = 0.82), suggesting that the mechanism underlying the effect of rs77359862 on AD risk is mediated by its direct contribution to neurodegeneration particularly in the hippocampal and entorhinal cortex regions. Routes through the hippocampus and entorhinal cortex account for 67% and 33%, respectively of the indirect effect of rs77359862 on AD risk. Consistent with the finding of an attenuated effect of rs77359862 on AD risk after adjusting for the indirect routes, the rs77359862 minor allele increases AD risk 2.62-fold (CI: [1.57, 4.72]) via the hippocampus and 1.61-fold (CI: [1.14, 2.44]) via the entorhinal cortex.
SHARPIN missense variant rs77359862 is associated with AD-related clinical measures and biomarkers
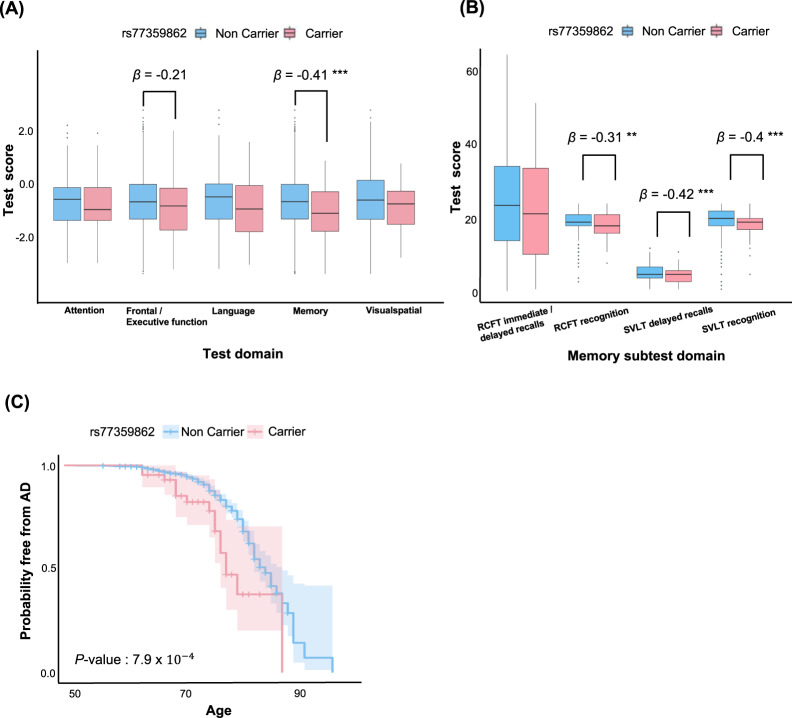
We evaluated the association of the rs77359862 missense variant and multiple measures of cognitive function using linear regression models that included covariates for age and sex. Significant associations were observed in measures of memory (β = −0.41, p = 1.0 × 10−4) and, to a lesser degree, in frontal/executive function (β = −0.21, p = 0.04), but not in attention (β = −0.09, p = 0.38), language (β = −0.18, p = 0.09) or visuospatial ability (β = −0.10, p = 0.33, Fig. 2A). Four subtests in memory measurement include Rey-complex Figure Test (RCFT) immediate/delayed recalls (β = −0.31, p = 0.16), RCFT recognition (β = −0.31, p = 0.006), Seoul Verbal Learning Test (SVLT) delayed recalls (β = −0.42, p = 1.6 × 10−4), and SVLT recognition (β = −0.40, p = 4.1 × 10−4) (Fig. 2B). Next, we investigated the effect of rs77359862 on age of AD symptom onset using the Kaplan–Meier approach to estimate a survival curve. This analysis showed that AD onset among individuals with the rs77359862 mutant variant was on average 1.5 years earlier than among those with the G allele (log-rank test p = 7.9 × 10−4, Fig. 2C).
Fig. 2. Association of rs77359862 with brain imaging, cognitive test measures and AD onset.
A Boxplots of Neuropsychological test (Seoul Neuropsychological Screening Battery (SNSB)) scores for attention, executive function, language, memory and visuospatial ability among rs77359862 carriers and non-carriers. Rs77359862 carriers had significantly lower scores for executive function and memory. **p-value < 0.01, ***p-value < 0.001. B Boxplots of subtests of memory domain in SNSB. Four subtests of memory function were assessed including visual memory (Rey-Complex Figure Test (RCFT) immediate/delayed recalls and recognition) and verbal memory (Seoul Verbal Learning Test (SVLT) delayed recalls and recognition). Rs77359862 carriers had significantly lower scores for all subtests, except RCFT immediate/delayed recalls. **p-value < 0.01, ***p-value < 0.001. C The rs77359862 missense variant significantly lowers age of AD onset. The effect of rs77359862 genotype on onset age was evaluated using Kaplan–Meier analysis and the survival curves for carriers and non-carriers of the missense variants were compared using the log-rank test.
In a subset of 876 subjects who were classified as CN or MCI at baseline and followed longitudinally (on average for 28.8 months), we also examined the impact of rs77359862 on the progression across clinical stages leading to AD. Within this group, 53 CN (10.1% of the CN) and 21 MCI participants (6.9% of the MCI) converted to MCI and AD, respectively (8.4% of the total) (Supplemental Table S6). The frequency of the mutant allele among the converters (6/74 = 8.1%) was higher than among the non-converters (26/802 = 3.2%). Analysis of the effect of rs77359862 genotype on the likelihood of conversion using a proportional hazards model adjusting for APOE genotype showed that participants with the mutant allele were 2.66 times as likely to progress to the next stage of cognitive decline (p = 0.023).
Analysis of the association of rs77359862 and Aβ accumulation in the brain using a logistic regression model with covariates for age and sex demonstrated that carriers of the rs77359862 missense variant had greater Aβ accumulation than non-carriers (p = 0.02, OR = 1.84).
Association of SHARPIN missense variant rs77359862 with HV in other cohorts
In our study, the frequency of the rs77359862 missense variant was consistently over 1% in CN Koreans in our study (1.4%) and in the Ansan-Ansung cohort (1.8%) [37], as well as in other East Asians (other than Korean and Japanese), included in the gnomAD database (4.1%) [38] (Fig. 1D). We observed a higher frequency of the variant among Koreans with late-onset AD (LOAD) in our study (4.3%) and in 77 early-onset AD (EOAD) patients who were diagnosed at the Seoul National University Bundang Hospital (3.2%). This variant was also more prevalent in a sample of Thailand EOAD patients (10.5%) [39]. Detailed information of EOAD patients was provided in supplementary Text 6. In contrast, the rs77359862 missense variant was virtually absent in non-Finnish persons of European ancestry (MAF = 0.0001) [38]. Due to rarity, we were unable to evaluate the rs77359862/AD association in populations with European ancestry. Therefore, we hypothesized that other variants in SHARPIN may be associated with MRI traits and AD in non-Asians. We conducted gene-based analyses by testing the association of HV with SHARPIN including 20 kb beyond the gene boundaries using imputed GWAS data from the ADNI [29] and AddNeuroMed cohorts [30]. Gene-based analyses revealed significant associations with SHARPIN in both the ADNI (86 SNPs, p = 0.002) and AddNeuroMed (93 SNPs, p = 0.04) datasets. Other suggested significant variants with HV from Table 1 were not significant in the ADNI dataset (EPPK1: 35 SNPs, p = 0.05; PLEC: 169 SNPs, p = 0.58; SMAD4: 130 SNPs, p = 0.52; ELAC1: 24 SNPs, p = 0.24).
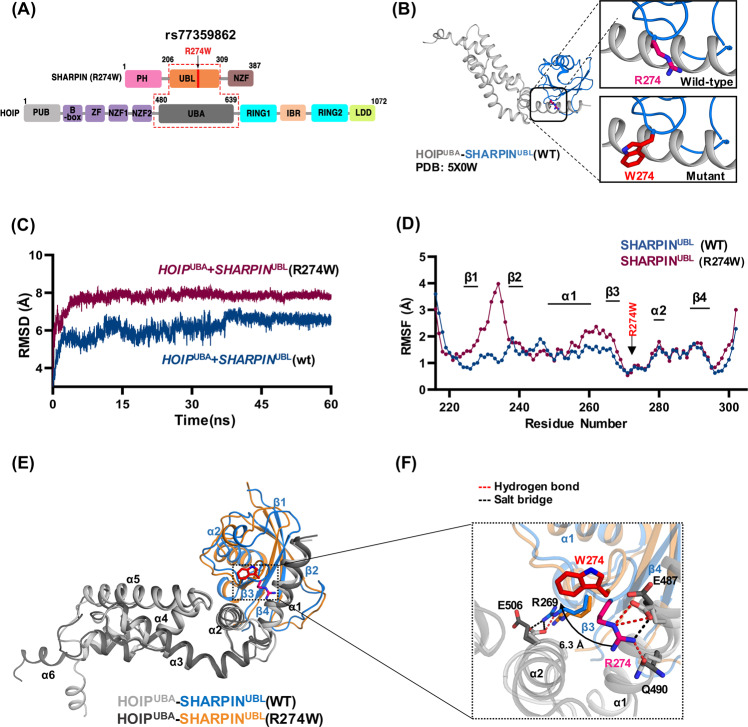
Rs77359862 missense variant alters the stability of SHARPIN complex structure
The rs77359862 variant is located in the domain mediating the binding of SHARPIN to its ligand HOIL-1-interacting protein (HOIP), which encodes the RING-between-RING (RBR) domain type ε3 ligase. The binding sites of these two proteins are the HOIP N-terminal UBA domain (HOIPUBA) and the UBL domain of SHARPIN (SHARPINUBL) [40] (Fig. 3A). The variant of rs77359862 located in SHARPINUBL leads to the substitution of polar R274 to hydrophobic tryptophan (NP_112236.3:p.R274W) (Fig. 3B). This switch in the chemical properties of the amino acids at the interface seems to affect the stability of the bound HOIPUBA-SHARPINUBL complex. To understand the effect of this variant, we performed a molecular dynamic (MD) simulation for the wild-type (WT) complex (PDB:5X0W) and an in silico SHARPIN mutated (R274W) complex. As shown in Fig. 3C, over time, the global root mean square deviation (RMSD) value of the mutant HOIPUBA-SHARPINUBL(R274W) was ~2 Å higher than the WT complex over time mainly due to the fluctuation in structural elements, such as the loops between β1-β2 and α1-β3 of the mutant SHARPINUBL (R274W) complex (Fig. 3D). Accordingly, amino acids in those regions were highly variable compared to those in the other parts of the protein as deduced by the root mean square fluctuation (RMSF) plot.
Fig. 3. Molecular dynamics simulation modeling of the effect of the SHARPIN R274W mutation on the HOIPUBA-SHARPINUBL complex.
A Domain map of SHARPIN and HOIP proteins. Protein binding occurs at the HOIPUBA and SHARPINUBL domains. B Crystal structure of the HOIPUBA-SHARPINUBL (PDB: 5X0W) binding complex indicating the location of the SHARPIN R274 residue in the wild type (WT) and manually mutated W274. C The root mean square deviation (RMSD) plot indicates the overall global deviation of the protein complex during 60 ns in WT and mutant. D The root mean square fluctuation (RMSF) plot shows the fluctuations of each residue of the SHARPIN (R274W) in complex during the last 20 ns of the equilibrated run. The structural elements corresponding to those residues were indicated above the plot. E Structural alignment of the structures obtained by averaging the atomic coordinates of the last 20 ns of the simulated run between WT complex and the mutant complex. The color code is maintained throughout the figure as per the legend between HOIPUBA-SHARPINUBL WT and R274W mutant protein complexes which are represented as cartoons and the residues in stick representation. The structural elements were marked starting from N-terminal ends of the protein. F The mutation site region is zoomed in from (E) (indicated by rectangular box) and the break in salt bridge (black dashed line) and hydrogen bonds (red dashed line) just with respect to the mutation is shown. The deviation of the residue from the WT to the mutant is marked by the arrow and the distance mentioned in Å.
Interaction on the interface between HOIPUBA WT and SHARPINUBL WT is strengthened by residues that contribute to hydrogen bonds and salt bridges [40]. Upon 60 ns MD simulation, we compared structural changes by using the averaged atomic coordinates for the stabilized last 20 ns (Fig. 3E, F). The comparative analysis revealed that the WT complex interface was built with ten hydrogen bonds and seven salt bridges, while the number of hydrogen bonds and salt bridges in the mutant complex HOIPUBA-SHARPINUBL (R274W) had decreased to 4 and 0, respectively (Supplemental Table S7), indicating that the mutant in SHARPIN induced a weaker interaction between the HOIPUBA and SHARPINUBL (R274W) (Fig. 3F and supplemental Fig. S6) (detailed information is provided in supplementary Text 7 and supplemental Figs. S7–8).
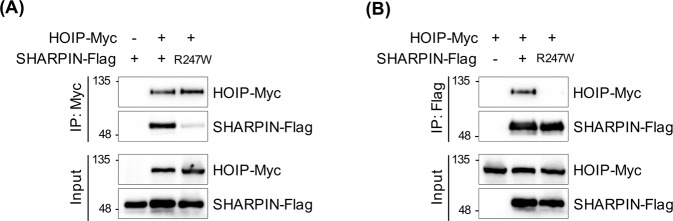
These observations were further supported by co-immunoprecipitation (co-IP) experiments (the detailed method is provided in supplementary Text 8). To determine whether the SHARPIN mutant R274W affected interactions with HOIP, flag-tagged SHARPIN WT and R274W mutant were co-immunoprecipitated with Myc-tagged HOIP WT (Fig. 4A) and Myc-tagged HOIP WT was co-immunoprecipitated with flag-tagged SHARPIN WT and R274W (Fig. 4B). The binding between SHARPINUBL (R274W) and HOIPUBA was significantly reduced compared with that of SHARPIN WT.
Fig. 4. Effect of mutant SHARPIN (R274W) on binding with HOIP.
293T cells transiently co-transfected with Flag-tagged WT and mutant SHARPIN and Myc-tagged HOIP were immunoprecipitated using anti-Myc antibody (A) and anti-Flag antibody (B). The binding affinity between SHARPIN variants and HOIP was confirmed through western blot analysis using anti-Myc and anti-Flag antibody, respectively.
In summary, our studies revealed that the SHARPIN mutant R274W might render the interaction between HOIPUBA and SHARPINUBL unstable, and thus destabilize the downstream SHARPIN-mediated pathway.
Discussion
Previous GWAS have identified many GWS loci for AD risk with GWS, but they have not been consistently replicated [7, 8, 15, 18, 41, 42]. The case/control study design has multiple limitations, including the power to detect associations with variants conferring small effects and biological complexity underlying, even well-defined, disease phenotypes. Genetic studies of AD are further complicated by mis-diagnosis, phenotypic heterogeneity (e.g., co-existing cerebrovascular disease and other brain pathologies, variable deficits in memory, language, and executive function), and misclassification of “controls” who may develop AD later in life. Studies of AD-related endophenotypes offer several advantages to gene discovery because statistical power is greater for quantitative traits compared to dichotomous outcomes, are not subject to misclassification, and are likely to have a less complex genetic architecture. Although our study is much smaller than previous GWAS of brain MRI traits conducted in population-based cohorts [43], our sample includes relatively larger AD and MCI cases, compared to CN cases, and significant associations with structural brain changes are more likely related to AD than normal aging.
Kang et al. performed GWASs for cases/controls with AD (n = 2291) in a Korean sample [44], but novel AD-related loci were not discovered. However, we identified a GWS association of a missense variant (rs77359862) in SHARPIN with decreasing ET and HV in a Korean population. This variant is infrequent in Koreans (MAF = 0.018), and virtually absent in populations outside of East Asia. Hence, we were unable to replicate this finding using non-Asian datasets. However, we found significant associations of HV with other rare and infrequent functional SHARPIN variants evaluated using a gene-based test in two large European ancestries (ADNI and AddNeuroMed) cohorts. Furthermore, Soheili-Nezhad et al. found a GWS association between another SHARPIN non-synonymous variant (rs34173062), located 4,325 base pairs away from rs77359862 and very rare in Koreans (MAF = 0.00035), and a composite MRI measure of limbic degeneration in the ADNI cohort (p = 2.1 × 10−10) [45]. The same variant was significantly associated with the thickness of the left entorhinal cortex (p = 0.002), right entorhinal cortex (8.6 × 10−4), and a history of AD in both parents (p = 2.3 × 10−6) in a UK Biobank (UKB) cohort dataset (n = 8428) [45]. Another recent large GWAS (n = 409,435) that included UKB data identified a GWS association between AD and yet another SHARPIN missense variant (rs34674752, p = 1.0 × 10−9) [46]. These results suggest that SHARPIN contributes to the development of AD in Koreans and Caucasians of European ancestry.
Mediation analysis findings suggest that the SHARPIN rs77359862 variant increases the risk of AD more than threefold, primarily through its effects on the entorhinal cortex and hippocampus. Our PET findings indicate that the same variant also affects the accumulation of Aβ, the main component of the amyloid plaques observed in PET images, which is a cardinal pathological feature of AD and is functionally related to frontal and memory regions [47]. According to Jung et al. [48], decreased executive function and memory, but not language or visuospatial impairment, were associated with a higher risk of cognitive decline, which is consistent with our observation that rs77359862 was significantly associated with executive and memory, but not language, visuospatial or attentional abilities. Longitudinal follow-up in this study revealed that CN and MCI carriers of the rs77359862 missense variant were significantly more likely to progress to MCI and AD, respectively. Finally, previous studies have shown that variants in several genes (e.g., ABCA7, SORL1, TREM2) are associated with both early-onset and late-onset AD [39]. The EOAD-associated variants are rare (MAF«0.01) and the LOAD-associated variants are infrequent (MAF > 0.01) or common (MAF > 0.05) in the populations in which they occur. With the notable example of carriers of a rare and highly penetrant mutation of the presenilin 2 gene, who developed AD symptoms between 45 and 88 years old [49], where we observed one of a few documented instances of a variant (rs77359862) that is associated with both EOAD and LOAD.
SHARPIN is a component of the linear ubiquitination assembly complex (LUBAC) [50] and, together with HOIP, suppresses NF-κB signaling [51, 52]. To study the mutational effect (R274W) of SHARPINUBL on its complex formation with HOIPUBA, we performed a 60 ns MD simulation using the crystal structure [40] (PDB: 5X0W), for both WT and mutant variants (R274W). Our MD analysis strongly suggests that the mutant complex HOIPUBA-SHARPINUBL(R274W) destabilizes the complex at the interface, as preserving the minimal integrity of the complex structure, which was validated with co-IP experiments. Therefore, the physical interaction between HOIP and R274W mutant SHARPIN was significantly reduced compared to the interaction with SHARPIN WT, and this unstable complex did not affect the downstream NF-κB signaling pathway [53]. NF-κB function in primary microglia isolated from SHARPIN mutant mice, rather than using an overexpression system will be evaluated in our future work.
NF-κB signaling in the nervous system plays a crucial role in the pathophysiology of AD, including neuroinflammation, deficits in memory consolidation, Aβ clearance, and neuronal cell death [54]. A rare functional variant (rs572750141, NM_030974.3:p.Gly186Arg) of SHARPIN has previously been found in a Japanese population and is significantly associated with an increased risk of LOAD [16]. This mutant of SHARPIN showed reduced NF-κB activation in HEK293 cells. SHARPIN is enriched at synaptic sites in mature neurons where it colocalizes with SHANK1 [55]. It is well known that activated NF-κB can be transported from activated synapses to the soma, which is essential for long-term memory. The reduction of neuronal NF-κB activity by the SHARPIN variant can inhibit anti-apoptosis pathways and lead to apoptosis or necroptosis in neurons [54, 56]. A recent study reported that knocking down SHARPIN using siRNA, inhibits Aβ-induced phagocytosis in macrophages [57], supporting our result of a marked increase in the accumulation of amyloid plaques in subjects with the R274W mutant SHARPIN.
There is mounting evidence that rare variants (MAF < 0.01) have large effects on AD risk [58–60] and may account for much of the missing heritability of the disorder [5]. We identified significant associations with several novel genes (COX7A2L, GUCA1A, VIT, GABRR2, and METTL6) through gene-based tests of aggregated rare variants with predicted functional consequences. Mitochondrial dysfunction has been widely reported in AD [61–63]. It has been reported that AD patients have a deficit of cytochrome C oxidase (COX) in both peripheral and brain tissue [64]. mRNA levels of several Cox genes including Cox7a2 correlated significantly with the Aβ plaque burden in the hippocampus of an AD mouse model [65]. VIT is known to be involved in brain asymmetry [66]. Gamma-aminobutyric acid (GABA), the major inhibitory neurotransmitter in the brain, is widely distributed in neurons of the cortex and contributes to many cortical functions by binding to GABA receptors, ligand-gated chloride channels. There is a large body of evidence suggesting that expression of the GABA receptor subunit alpha 1 gene (GABRA1) receptor gene is altered in the hippocampus of AD cases [67]. A SNP in the gene encoding GABA receptor subunit rho-2 (GABRR2) was associated with the general cognitive ability [68]. METTL6 was identified as a marker of the proliferation of luminal breast cancers [69, 70], but the biological role of this RNA-modifying methyltransferase is not well understood.
Our study has several limitations. The sample had limited power to detect genome-wide significant associations for rare variants with small effects, and this concern was exacerbated by the smaller number of AD cases compared to MCI cases and control subjects. However, the sample was adequately powered to detect larger effects, evidenced by the GWS association with the SHARPIN missense variant. In addition, longitudinal brain MRI data were available for only a small portion of individuals, thus limiting our ability to perform genome-wide scans for atrophy in brain measures, over time. Furthermore, experiments in cell or mouse models will need to be conducted to demonstrate the effects of rs77359862 on AD-specific pathological damages such as Aβ accumulation or tangle formation.
Supplementary information
Supplementary Figure legends and Table tiles
Acknowledgements
This study was supported by Healthcare AI Convergence Research & Development Program through the National IT Industry Promotion Agency of Korea (NIPA) funded by the Ministry of Science and ICT (No.1711120216), the Korea Brain Research Institute basic research program funded by the Ministry of Science and ICT (21-BR-03–05), the Original Technology Research Program for Brain Science of the National Research Foundation (NRF) funded by the Korean government, MSIT (NRF-2014M3C7A1046041), the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (NRF-2016M3A9E9941946), Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2020R1F1A01072033), a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea (HI18C0041) and the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (NRF-2016M3A9E9941946). Also, this work was supported by the National Research Foundation of Korea Grant funded by the Korean Government (No.21B20151213037). Additional support for data analyses was provided by NIH grants U01 AG024904, R01 LM012535, P30 AG010133, R01 AG019771, 2R01-AG048927, P30-AG13846, U01-AG032984, RF1-AG057519, and U01-AG062602. Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. Data collection and sharing for this project were funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01AG024904) and DODADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. The AddNeuroMed study was part of InnoMed (Innovative Medicines in Europe), an integrated project funded by the European Union as part of the Sixth Framework programme priority (FP6-2004-LIFESCIHEALTH-5). Clinical leads for the AddNeuroMed programme were Simon Lovestone, Hilkka Soininen, Patrizia Mecocci, Iwona Kloszewska, Magda Tsolaki, and Bruno Vellas.
Author contributions
JYP performed all the statistical analyses and wrote the manuscript. DL conducted to GWAS and QC/imputation for genomic data with ARD. JJ, JG, G, SK, and KYG contributed to data collection and MRI preprocessing. JP, JJ, and KN contributed to replication in ADNI and UK Biobank. ID, SG, and SHL supervised MD simulation and analyses. SK, JHC, and SR were responsible for IP assays. SYK helped obtaining EOAD samples. GJS, GRJ, and LAF critically revised the manuscript drafts. SW and KHL designed and co-wrote all drafts of the manuscript. All authors critically revised all manuscripts drafts, read, and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wpcontent/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Contributor Information
Sungho Won, Email: won1@snu.ac.kr.
Kun Ho Lee, Email: leekho@chosun.ac.kr.
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-021-01680-5.
References
- 1.2018 Alzheimer’s disease facts and figures. Alzheimer’s Dementia. 2018;14:367–429.
- 2.Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–74. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 3.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 4.Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA. 1993;90:1977–81. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ridge PG, Mukherjee S, Crane PK, Kauwe JS, Alzheimer’s Disease Genetics C. Alzheimer’s disease: analyzing the missing heritability. PLoS ONE. 2013;8:e79771. doi: 10.1371/journal.pone.0079771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stocker H, Möllers T, Perna L, Brenner H. The genetic risk of Alzheimer’s disease beyond APOE ε4: systematic review of Alzheimer’s genetic risk scores. Transl Psychiatry. 2018;8:166. doi: 10.1038/s41398-018-0221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51:414–30. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrews SJ, Fulton-Howard B, Goate A. Interpretation of risk loci from genome-wide association studies of Alzheimer’s disease. Lancet Neurol. 2020;19:326–35. doi: 10.1016/S1474-4422(19)30435-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010;11:356–66. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaitlen N, Paşaniuc B, Gur T, Ziv E, Halperin E. Leveraging genetic variability across populations for the identification of causal variants. Am J Hum Genet. 2010;86:23–33. doi: 10.1016/j.ajhg.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Liu X, Sim X, Xu H, Khor CC, Ong RT, et al. A statistical method for region-based meta-analysis of genome-wide association studies in genetically diverse populations. Eur J Hum Genet. 2012;20:469–75. doi: 10.1038/ejhg.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reitz C, Jun G, Naj A, Rajbhandary R, Vardarajan BN, Wang LS, et al. Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E 4,and the risk of late-onset Alzheimer disease in African Americans. JAMA. 2013;309:1483–92. doi: 10.1001/jama.2013.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JH, Cheng R, Barral S, Reitz C, Medrano M, Lantigua R, et al. Identification of novel loci for Alzheimer disease and replication of CLU, PICALM, and BIN1 in Caribbean Hispanic individuals. Arch Neurol. 2011;68:320–8. doi: 10.1001/archneurol.2010.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vardarajan BN, Barral S, Jaworski J, Beecham GW, Blue E, Tosto G, et al. Whole genome sequencing of Caribbean Hispanic families with late-onset Alzheimer’s disease. Ann Clin Transl Neurol. 2018;5:406–17. doi: 10.1002/acn3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyashita A, Koike A, Jun G, Wang LS, Takahashi S, Matsubara E, et al. SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PLoS ONE. 2013;8:e58618. doi: 10.1371/journal.pone.0058618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asanomi Y, Shigemizu D, Miyashita A, Mitsumori R, Mori T, Hara N, et al. A rare functional variant of SHARPIN attenuates the inflammatory response and associates with increased risk of late-onset Alzheimer’s disease. Mol Med. 2019;25:20. doi: 10.1186/s10020-019-0090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou X, Chen Y, Mok KY, Zhao Q, Chen K, Chen Y, et al. Identification of genetic risk factors in the Chinese population implicates a role of immune system in Alzheimer’s disease pathogenesis. Proc Natl Acad Sci USA. 2018;115:1697–706. doi: 10.1073/pnas.1715554115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jun GR, Chung J, Mez J, Barber R, Beecham GW, Bennett DA, et al. Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimers Dement. 2017;13:727–38. doi: 10.1016/j.jalz.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braskie MN, Thompson PM. Understanding cognitive deficits in Alzheimer’s disease based on neuroimaging findings. Trends Cogn Sci. 2013;17:510–6. doi: 10.1016/j.tics.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherva R, Tripodis Y, Bennett DA, Chibnik LB, Crane PK, de Jager PL, et al. Genome-wide association study of the rate of cognitive decline in Alzheimer’s disease. Alzheimer’s Dement: J Alzheimer’s Assoc. 2014;10:45–52. doi: 10.1016/j.jalz.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elliott LT, Sharp K, Alfaro-Almagro F, Shi S, Miller KL, Douaud G, et al. Genome-wide association studies of brain imaging phenotypes in UK Biobank. Nature. 2018;562:210–6. doi: 10.1038/s41586-018-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blennow K. Cerebrospinal fluid protein biomarkers for Alzheimer’s disease. NeuroRx. 2004;1:213–25. doi: 10.1602/neurorx.1.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu J, Patassini S, Rustogi N, Riba-Garcia I, Hale BD, Phillips AM, et al. Regional protein expression in human Alzheimer’s brain correlates with disease severity. Commun Biol. 2019;2:43. doi: 10.1038/s42003-018-0254-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saykin AJ, Shen L, Yao X, Kim S, Nho K, Risacher SL, et al. Genetic studies of quantitative MCI and AD phenotypes in ADNI: progress, opportunities, and plans. Alzheimers Dement. 2015;11:792–814. doi: 10.1016/j.jalz.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song YE, Lee S, Park K, Elston RC, Yang HJ, Won S. ONETOOL for the analysis of family-based big data. Bioinformatics. 2018;34:2851–3. doi: 10.1093/bioinformatics/bty180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 28.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74:201–9. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovestone S, Francis P, Kloszewska I, Mecocci P, Simmons A, Soininen H, et al. AddNeuroMed–the European collaboration for the discovery of novel biomarkers for Alzheimer’s disease. Ann N. Y Acad Sci. 2009;1180:36–46. doi: 10.1111/j.1749-6632.2009.05064.x. [DOI] [PubMed] [Google Scholar]
- 31.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bolin JH, Hayes, Andrew F. (2013). Introduction to Mediation, Moderation, and Conditional Process Analysis: A Regression-Based Approach. New York, NY: The Guilford Press. Journal of Educational Measurement, 2014;51:335–7.
- 34.Becker GA, Ichise M, Barthel H, Luthardt J, Patt M, Seese A, et al. PET quantification of 18F-florbetaben binding to β-amyloid deposits in human brains. J Nucl Med. 2013;54:723–31. doi: 10.2967/jnumed.112.107185. [DOI] [PubMed] [Google Scholar]
- 35.Choi KY, Lee JJ, Gunasekaran TI, Kang S, Lee W, Jeong J, et al. APOE promoter polymorphism-219T/G is an effect modifier of the influence of APOE ε4 on Alzheimer’s disease risk in a multiracial sample. J Clin Med. 2019;8, 10.3390/jcm8081236. [DOI] [PMC free article] [PubMed]
- 36.Ahn H-J, Chin J, Park A, Lee BH, Suh MK, Seo SW, et al. Seoul Neuropsychological Screening Battery-dementia version (SNSB-D): a useful tool for assessing and monitoring cognitive impairments in dementia patients. J Korean Med Sci. 2010;25:1071–6. doi: 10.3346/jkms.2010.25.7.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moon S, Kim YJ, Han S, Hwang MY, Shin DM, Park MY, et al. The Korea Biobank Array: design and identification of coding variants associated with blood biochemical traits. Sci Rep. 2019;9:1382. doi: 10.1038/s41598-018-37832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karczewski KJ, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. BioRxiv. 2019;531210.
- 39.Giau V, Senanarong V, Bagyinszky E, An S, Kim S. Analysis of 50 neurodegenerative genes in clinically diagnosed early-onset Alzheimer’s disease. Int J Mol Sci. 2019;20:1514. doi: 10.3390/ijms20061514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu J, Wang Y, Gong Y, Fu T, Hu S, Zhou Z, et al. Structural insights into SHARPIN-mediated activation of HOIP for the linear ubiquitin chain assembly. Cell Rep. 2017;21:27–36. doi: 10.1016/j.celrep.2017.09.031. [DOI] [PubMed] [Google Scholar]
- 41.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marioni RE, Harris SE, Zhang Q, McRae AF, Hagenaars SP, Hill WD, et al. GWAS on family history of Alzheimer’s disease. Transl Psychiatry. 2018;8:99. doi: 10.1038/s41398-018-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melville SA, Buros J, Parrado AR, Vardarajan B, Logue MW, Shen L, et al. Multiple loci influencing hippocampal degeneration identified by genome scan. Ann Neurol. 2012;72:65–75. doi: 10.1002/ana.23644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang S, et al. East Asian-specific novel loci associated with late-onset Alzheimer’s disease: the GARD cohort Genome-wide study. medRxiv. 2020; 2020.07.02.20145557.
- 45.Soheili-Nezhad S, Jahanshad N, Guelfi S, Khosrowabadi R, Saykin AJ, Thompson PM, et al. Imaging genomics discovery of a new risk variant for Alzheimer’s disease in the postsynaptic SHARPIN gene. Hum Brain Mapp. 2020;41:3737–48. doi: 10.1002/hbm.25083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Rojas I, et al. Common variants in Alzheimer’s disease: novel association of six genetic variants with AD and risk stratification by polygenic risk scores. medRxiv. 2020;19012021.
- 47.Selkoe DJ. Alzheimer’s disease results from the cerebral accumulation and cytotoxicity of amyloid ß-protein. J Alzheimer’s Dis. 2001;3:75–80. doi: 10.3233/jad-2001-3111. [DOI] [PubMed] [Google Scholar]
- 48.Jung YH, Park S, Jang H, Cho SH, Kim SJ, Kim JP, et al. Frontal-executive dysfunction affects dementia conversion in patients with amnestic mild cognitive impairment. Sci Rep. 2020;10:772. doi: 10.1038/s41598-020-57525-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherrington R, Froelich S, Sorbi S, Campion D, Chi H, Rogaeva EA, et al. Alzheimer’s disease associated with mutations in presenilin 2 is rare and variably penetrant. Hum Mol Genet. 1996;5:985–8. doi: 10.1093/hmg/5.7.985. [DOI] [PubMed] [Google Scholar]
- 50.Lancour D, Naj A, Mayeux R, Haines JL, Pericak-Vance MA, Schellenberg GD, et al. One for all and all for One: Improving replication of genetic studies through network diffusion. PLoS Genet. 2018;14:e1007306. doi: 10.1371/journal.pgen.1007306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niu J, Shi Y, Iwai K, Wu ZH. LUBAC regulates NF-κB activation upon genotoxic stress by promoting linear ubiquitination of NEMO. Embo J. 2011;30:3741–53. doi: 10.1038/emboj.2011.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aksentijevich I, Zhou Q. NF-κB pathway in autoinflammatory diseases: dysregulation of protein modifications by ubiquitin defines a new category of autoinflammatory diseases. Front Immunol. 2017;8:399. doi: 10.3389/fimmu.2017.00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, et al. SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–6. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 54.Kaltschmidt B, Kaltschmidt C. NF-kappaB in the nervous system. Cold Spring Harb Perspect Biol. 2009;1:a001271. doi: 10.1101/cshperspect.a001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim S, Sala C, Yoon J, Park S, Kuroda S, Sheng M, et al. Sharpin, a novel postsynaptic density protein that directly interacts with the shank family of proteins. Mol Cell Neurosci. 2001;17:385–97. doi: 10.1006/mcne.2000.0940. [DOI] [PubMed] [Google Scholar]
- 56.Yuan J, Amin P, Ofengeim D. Necroptosis and RIPK1-mediated neuroinflammation in CNS diseases. Nat Rev Neurosci. 2019;20:19–33. doi: 10.1038/s41583-018-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krishnan D, Menon RN, Mathuranath PS, Gopala S. A novel role for SHARPIN in amyloid-β phagocytosis and inflammation by peripheral blood-derived macrophages in Alzheimer’s disease. Neurobiol Aging. 2020;93:131–41. doi: 10.1016/j.neurobiolaging.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 58.Bis JC, Jian X, Kunkle BW, Chen Y, Hamilton-Nelson KL, Bush WS, et al. Whole exome sequencing study identifies novel rare and common Alzheimer’s-Associated variants involved in immune response and transcriptional regulation. Molecular Psychiatry. 2020;25:1859–75. doi: 10.1038/s41380-018-0112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patel H, Dobson RJB, Newhouse SJ. A meta-analysis of Alzheimer’s disease brain transcriptomic data. J Alzheimers Dis. 2019;68:1635–56. doi: 10.3233/JAD-181085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature. 2019;570:332–7. doi: 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Devine MJ, Kittler JT. Mitochondria at the neuronal presynapse in health and disease. Nat Rev Neurosci. 2018;19:63–80. doi: 10.1038/nrn.2017.170. [DOI] [PubMed] [Google Scholar]
- 62.Du H, Guo L, Yan S, Sosunov AA, McKhann GM, Yan SS. Early deficits in synaptic mitochondria in an Alzheimer’s disease mouse model. Proc Natl Acad Sci USA. 2010;107:18670–5. doi: 10.1073/pnas.1006586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DuBoff B, Feany M, Götz J. Why size matters - balancing mitochondrial dynamics in Alzheimer’s disease. Trends Neurosci. 2013;36:325–35. doi: 10.1016/j.tins.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 64.Parker WD, Jr., Parks JK. Cytochrome c oxidase in Alzheimer’s disease brain: purification and characterization. Neurology. 1995;45:482–6. doi: 10.1212/wnl.45.3.482. [DOI] [PubMed] [Google Scholar]
- 65.Bi R, Zhang W, Zhang DF, Xu M, Fan Y, Hu QX, et al. Genetic association of the cytochrome c oxidase-related genes with Alzheimer’s disease in Han Chinese. Neuropsychopharmacology. 2018;43:2264–76. doi: 10.1038/s41386-018-0144-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tadayon SH, Vaziri-Pashkam M, Kahali P, Ansari Dezfouli M, Abbassian A. Common genetic variant in VIT Is associated with human brain asymmetry. Front Hum Neurosci. 2016;10:236. doi: 10.3389/fnhum.2016.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steiger JL, Russek SJ. GABAA receptors: building the bridge between subunit mRNAs, their promoters, and cognate transcription factors. Pharm Ther. 2004;101:259–81. doi: 10.1016/j.pharmthera.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 68.Ma Z, Niu B, Shi Z, Li J, Wang J, Zhang F, et al. Genetic polymorphism of GABRR2 modulates individuals’ general cognitive ability in healthy Chinese Han People. Cell Mol Neurobiol. 2017;37:93–100. doi: 10.1007/s10571-016-0347-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gatza ML, Silva GO, Parker JS, Fan C, Perou CM. An integrated genomics approach identifies drivers of proliferation in luminal-subtype human breast cancer. Nat Genet. 2014;46:1051–9. doi: 10.1038/ng.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boriack-Sjodin PA, Ribich S, Copeland RA. RNA-modifying proteins as anticancer drug targets. Nat Rev Drug Discov. 2018;17:435–53. doi: 10.1038/nrd.2018.71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure legends and Table tiles